Abstract
Glioblastoma is the most common primary malignant brain tumor. Although current standard therapy extends median survival to ~15 months, most patients do not have sustained response to treatment. While O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status is accepted as a prognostic and promising predictive biomarker in glioblastoma, its value in informing treatment decisions for glioblastoma patients is still debatable. Discrepancies between MGMT promoter methylation status and treatment response in some patients may stem from inconsistencies between MGMT methylation and expression levels in glioblastoma. Here, we discuss MGMT as a biomarker and elucidate the discordance between MGMT methylation, expression, and patient outcome, which currently challenges the implementation of this biomarker in clinical practice.
Keywords: Glioblastoma, MGMT, biomarker, neuro-oncology, precision medicine
Background of glioblastoma and MGMT
Glioblastoma, a grade IV astrocytoma (see Glossary), is the most common primary malignant brain tumor in adults [1, 2]. The current standard treatment for newly diagnosed patients includes maximum safe surgical resection followed by radiation therapy (RT) with concomitant and adjuvant temozolomide (TMZ) [2]. The addition of TMZ to RT for newly diagnosed glioblastoma has resulted in a significant survival benefit in the general glioblastoma patient population; yet, only a quarter of patients survive 2 years after initial diagnosis, suggesting a high variability of patient response to the standard therapy [3]. The identification of molecular biomarkers to successfully predict patient response to therapy is a crucial goal in neuro-oncology research.
MGMT promoter methylation status has emerged as one of the leading determinants of prognosis and potential predictor of response to TMZ [4, 5]. However, this biomarker has not yet been implemented in routine treatment decision-making, and the best method to determine MGMT status in patients remains under debate [6]. Current methods of evaluating MGMT status are summarized in Table 1. Specifically, there is strong evidence of discordance between methylation status and protein expression level, with variable reports of correlation with patient outcome [4, 7–15]. Here, we outline the challenge of implementing MGMT methylation status as a clinical biomarker for glioblastoma patients. We specifically emphasize the inconsistencies between MGMT promoter methylation status, MGMT protein/gene expression, and patient outcome. Further, we propose the need to evaluate additional parameters in combination with MGMT methylation status in order to likely improve prediction of patient outcome. The function of the MGMT protein and the role of MGMT promoter methylation are described in Box 1.
Table 1.
Current methods for evaluating MGMT status
| Parameter evaluated | Method | Description | Reference |
|---|---|---|---|
| Promoter methylation | Non-quantitative methylation-specific polymerase chain reaction (MSP) | DNA is treated with bisulfite, which converts unmethylated cytosine to uracil without modifying 5-methylcytosine. Using primers that are specific to methylated or unmethylated sequences, PCR is performed and analyzed by gel electrophoresis to provide qualitative results | [4, 6] |
| Quantitative methylation-specific PCR (qMSP) | qMSP is similar to the non-quantitative MSP assay but provides quantitative results after normalization to an unmethylated gene | [6, 40] | |
| Pyrosequencing | A method of DNA sequencing, also based on bisulfite conversion and PCR, offering a quantitative determination of methylation of each individual CpG site sequenced | [13] | |
| Methylation-sensitive multiplex ligation-dependent probe amplification (MS-MPLA) | Uses methylation-sensitive restriction enzymes to give semi-quantitative results for methylation status | [4, 13] | |
| Infinium Methylation EPIC BeadChip Array | Genome-wide methylation profiling of 850,000 CpG sites, including the MGMT genomic region | [6] | |
| mRNA expression | Quantitative real-time polymerase chain reaction (qRT-PCR) | Measurement of MGMT transcript expression | [13, 43] |
| Protein expression | Immunohistochemistry | Tumor cells with nuclear staining are counted as MGMT-positive, and the percentage of positive cells is assessed to determine MGMT status | [13, 50] |
| Protein activity | Enzymatic assays | Sample is incubated with 3H-labeled O6 methyl-guanine, and the transfer of 3H-labeled methyl groups to the MGMT protein is measured by isolation of the 3H-labeled MGMT molecules | [4, 40] |
Box 1. MGMT function and the role of promoter methylation.
Temozolomide (TMZ) is an alkylating agent that damages DNA by adding methyl groups to the N7 and O6 positions of guanine and the N3 position of adenine [44, 53]. Though the O6-methylguanine (O6-MeG) adduct is the least frequent lesion, it is the primary mechanism of temozolomide cytotoxicity [43, 44, 54]. This methylation results in inaccurate pairing of the methylated guanine with newly incorporated thymine during replication. Futile cycling of the mismatch repair (MMR) pathway, which removes the thymine but leaves the methylated guanine, results in DNA double-stranded breaks, irreparable genomic damage, and activation of cell death [21, 23, 36]. As illustrated in Figure IA, MGMT prevents this from happening by removing and transferring the methyl group from O6-MeG to an internal cysteine residue in an irreversible suicidal reaction. This activity effectively reverses the alkylation-induced DNA damage, thus blunting the cytotoxic effects of TMZ. This is one of the mechanisms that explains why patients whose cancer cells express MGMT do not typically respond to TMZ (Figure IB) [53, 55].
The MGMT promoter region contains a high frequency of repetitive CpG sequences [4]. Hypermethylation at CpG sites within this region typically results in epigenetic silencing of MGMT transcripts. The resulting lack of MGMT-mediated DNA repair promotes sensitivity to temozolomide when MMR function is intact (Figure IB) [12, 44, 55]. An unmethylated promoter often results in high MGMT protein expression, which allows the repair of O6-MeG and promotes resistance to TMZ (Figure IB). The landmark EORTC-NCIC clinical trial established the association between MGMT promoter methylation and increased survival benefit in newly diagnosed glioblastoma patients treated with radiation plus concomitant and adjuvant TMZ. Although the addition of TMZ to radiotherapy brought significant survival benefit in patients with MGMT methylated glioblastoma, a modest benefit was also noted in patients with an unmethylated promoter [3, 55]. Subsequent studies have confirmed the link between MGMT promoter methylation and patient outcome, and thus, MGMT promoter methylation has emerged as the primary marker for lack of MGMT function to determine prognosis and potential response to chemotherapy [6, 56]. However, the correlation between MGMT promoter methylation status and mRNA or protein expression level is not absolute [12, 44]. Indeed, in cancer cell lines, only ~50% of MGMT-negative cells show promoter methylation [44]. Hence, there is ongoing debate regarding the best method of classifying MGMT status in order to determine accurate prognosis and prediction of treatment response. Moreover, even in cells lacking MGMT function, MMR needs to be active for the cells to respond to TMZ (Figure IB) [44].
Discordance between promoter methylation and expression
A negative correlation between MGMT promoter methylation and expression has been demonstrated in numerous studies; however, there is consistent evidence of discordance. For example, multiple glioblastoma cell lines have an unmethylated MGMT promoter, but exhibit low mRNA expression (Figure 1A). The subgroup of patients exhibiting inconsistency are of particular interest. A systematic review and meta-analysis of 52 studies determining the correlation between immunohistochemistry (IHC) and methylation-specific PCR (MSP) for MGMT in human tumors concluded that the results found by IHC were not in close concordance with those found by MSP [12]. Though non-brain tumor studies were included in the analysis, greater disagreement between the two tests was found in brain tumors [12]. Similarly, no correlation was found between MGMT protein expression and methylation by MSP (p=0.903) when 76 glioblastoma samples were tested. 52.4% of unmethylated tumors showed low MGMT expression and 41.2% of methylated tumors showed high MGMT expression [16]. When comparing IHC and pyrosequencing in another study, the concordance rate was only 30.8% (N=350) [17].
Figure 1.
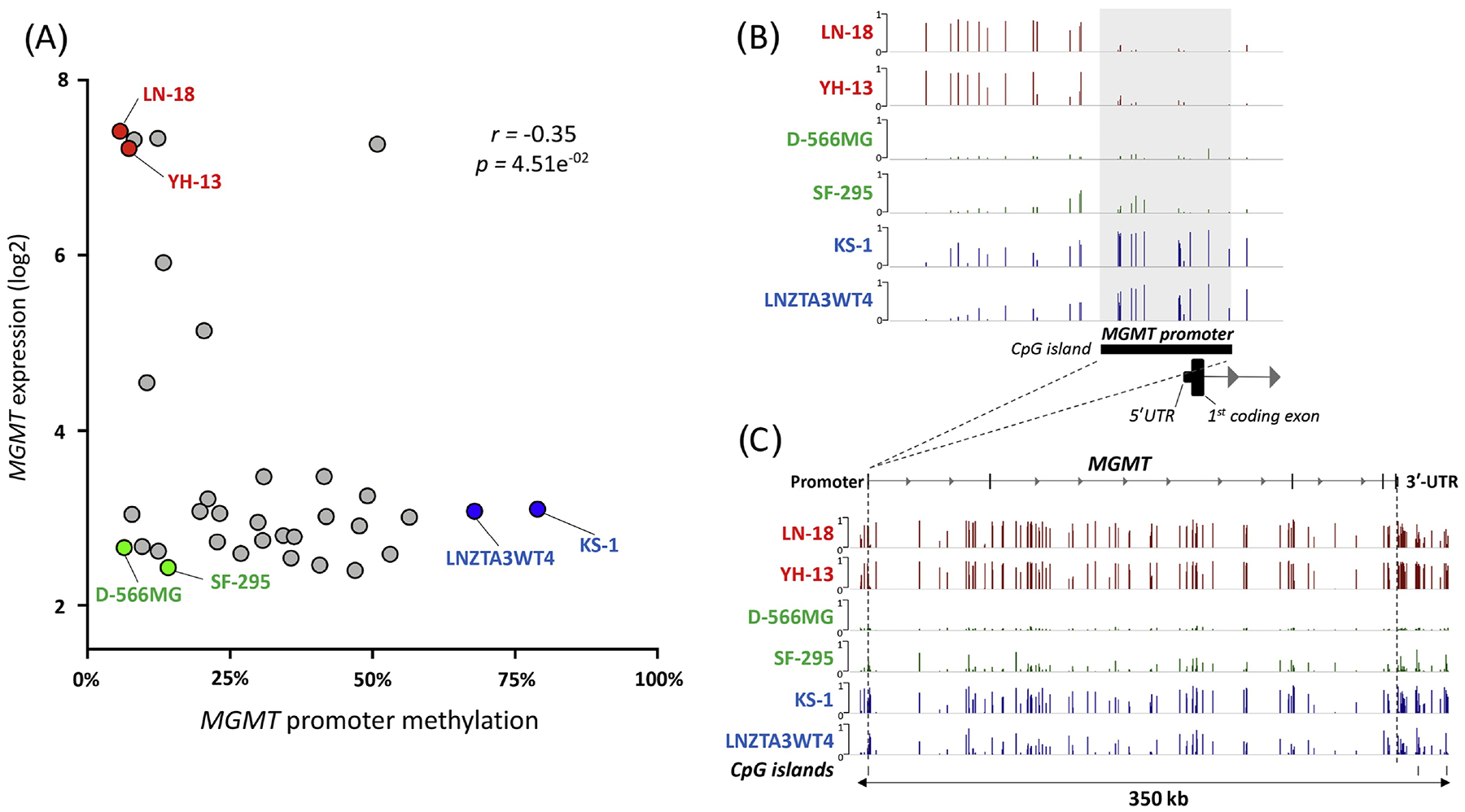
Association of MGMT mRNA expression with DNA methylation. A) Plot of MGMT gene expression levels and promoter methylation levels in glioblastoma cell lines using Genomics of Drug Sensitivity in Cancer Project (GDSC) data from CellMinerCDB (http://discover.nci.nih.gov/cellminercdb) [52]. Six cell lines were selected based on expression and promoter methylation for further visualization: 1) high expression and low promoter methylation (red), 2) low expression and low promoter methylation (green) and 3) low expression and high promoter methylation (blue). B) Promoter and C) gene body probe level methylation (beta values) of MGMT gene for the six selected cell lines.
Importantly, even within studies that demonstrate a significant correlation between promoter methylation status and gene/protein expression, there are often several inconsistencies. For example, in a set of 53 glioblastoma and 10 anaplastic astrocytoma, a strong correlation between mRNA expression level and promoter methylation status was found (p<0.0001); however, 6 patients with a methylated promoter expressed high mRNA levels and 6 patients with an unmethylated promoter expressed low mRNA levels [18]. Remarkably, these discordant findings only occurred in glioblastoma patients. Moreover, when extending the analysis to a validation set using TCGA-GBM data, inconsistent findings were observed in 46 out of 209 samples (22%), similar to the study population (19%) [18]. In a larger cohort, IHC was negatively correlated with promoter methylation by MSP (p<0.0001); yet 37% of 215 patients with an unmethylated promoter had low MGMT expression [10]. Multiple other studies report differential protein expression regardless of MGMT methylation status [7, 9, 16].
Potential explanations for inconsistent correlation between methylation status, expression level, and/or clinical outcome
Although promoter methylation is a major mechanism of gene silencing, additional factors may affect the correlation between MGMT methylation, expression, and patient outcome. In addition to the explanations below, alternative mechanisms, such as post-transcriptional modulation of MGMT by microRNAs or the association of MGMT methylation with IDH mutation or the glioma CpG island methylator phenotype, may explain these inconsistent correlations [19–22].
Limitations of IHC analysis
Though IHC is a common and inexpensive assay used in many cancer types, including glioblastoma, the value of using IHC to determine MGMT status has been controversial [13]. The limitations of the assay could partially explain the findings of poor correlation between protein expression by IHC and methylation or clinical outcome. One challenge is contamination of non-neoplastic, MGMT-expressing cells in the tissue sample, such as microglia, tumor-infiltrating lymphocytes, and vascular endothelium, which could lead to false positives in scoring the tumor sample [23]. Another major challenge includes variation in interobserver agreement of IHC determinations [7, 12]. Other limitations could result from intratumoral variation in protein expression and the standard characterization of MGMT expression by positive cell count, which overlooks the level of protein within each cell [21]. Though the limitations of IHC are evident, this method is widely used for glioma diagnosis based on immunoreactivity of other molecular biomarkers (e.g., IDH1 R132H, ATRX). Development and standardization of the use of an antibody clone with maximum specificity and reliability and exclusion of non-neoplastic cells could aid in improving the measurement of MGMT protein, the reproducibility of the assay, and the correlation of IHC results with methylation status and patient outcome. Alternatively, transitioning to the use of quantitative fluorescent IHC may give a better determination of MGMT level compared to traditional IHC [14].
Lack of standardized cutoff values in diagnostic assays
Perhaps one of the most considerable limitations is the lack of standard cutoff values for determining MGMT status for methylation and expression assays. The most common cutoff values for methylation by pyrosequencing are 8 – 10% [24, 25]. However, binary cutoffs neglect patients with an intermediate level of methylation. A pooled analysis of quantitative MSP results from four clinical trials evaluated both an unsupervised technical cutoff and a clinically optimized cutoff supervised by overall survival [26]. Importantly, the 9.5% of patients who fell within the “gray zone” of intermediate methylation between the optimal and technical cutoffs had a significant survival benefit compared to those patients who were “truly” unmethylated [26]. This demonstrates the importance of standardizing a reliable clinically relevant cutoff for diagnostic assays rather than a strict technical cutoff, considering that patients with low or intermediate levels of methylation may still receive some benefit from temozolomide. Moreover, not all studies use the same cutoff values. Some studies found that the correlation of MGMT methylation status even depends on different cutoff values within their own study [27, 28]. Cutoffs for protein expression also vary, with high and low expression stratified at the median or 10%, 25%, 30%, or 50% cutoff levels [7, 9, 10, 14, 16, 29, 30]. This lack of standardization complicates the interpretation of correlation with patient outcome and the clinical usability of these assays [6, 31].
Correlation between MGMT methylation, expression, and/or clinical outcome is CpG location-dependent
The MGMT promoter and gene include many CpG dinucleotides; however, methylation of specific CpG sites have been shown to be more relevant for gene silencing than others. The −228, −186, +125, and +137 CpG positions (relative to the transcription start site) were previously identified as most relevant for expression and this was confirmed as >80% concordant in a separate study, which also identified additional critical CpG sites at +95, +113, and +135 [32, 33]. Interestingly, this second study further found that the region commonly investigated by MSP did not have the best correlation with gene expression, observing 28% discordant results, which could partially explain poor correlation between expression and methylation status by MSP in some patients [32]. Two genomic regions, differentially methylated region 1 (DMR1) and 2 (DMR2), have been identified as being most strongly concordant with expression and patient outcome [34, 35]. A BeadChip-based MGMT-STP27 MGMT classification model identified these two regions as highly significant for gene silencing and predicting outcome in chemo-radiotherapy-treated patients [34]. However, further research should attempt to establish a more targeted region for methylation probing; though the region investigated by MSP is located in the DMR2 region, methylation of particular CpG sites within DMR2 could explain the better correlation between clinical outcome and methylation within this region, compared to methylation of the specific CpG sites interrogated using MSP [35].
Gene body methylation
Methylation also occurs in the MGMT gene body, and methylation of exonic regions may result in increased MGMT expression in some patients, which could partially explain why MGMT transcript levels may differ from what is expected by the promoter methylation status [36]. The effect of gene body cytosine modifications was analyzed in 91 glioblastoma [37]. In patients with an unmethylated promoter, gene body hypomethylation resulted in decreased MGMT expression to a similar degree of those with a methylated promoter [37]. Moreover, gene body hypermethylation in these patients was correlated with increased MGMT expression. However, this phenomenon was not observed in patients with a methylated promoter. Assessing cytosine modification levels in both the promoter region and gene body may improve prediction of MGMT expression and response to TMZ. For example, hypomethylation of the gene body in D-566MG and SF-295 cells may possibly explain the decreased MGMT expression in these cells with an unmethylated promoter. It would be interesting to know whether the high MGMT expression in LN-18 and YH-13 cells occurs primarily as a result of an unmethylated promoter or hypermethylation of the gene body (Figure 1A–C).
TMZ-induced upregulation of MGMT
Another potential explanation is the observation that MGMT expression and/or activity may be induced in response to TMZ [6, 38]. It has been reported that recurrent glioblastoma showed a significant increase in mean MGMT activity after chemotherapy with no significant increase after radiation alone [39]. Similarly, MGMT protein level, MGMT mRNA expression, and MGMT activity increased after TMZ treatment in unmethylated patient-derived glioblastoma xenografts, and this upregulation was associated with TMZ resistance [38]. Also likely is treatment selection of high-MGMT subclones, which would promote resistance to TMZ [40, 41]. Although it can change upon recurrence in some patients, MGMT promoter methylation is relatively stable during disease progression [42]. Upregulation of MGMT activity was observed in recurrent glioblastoma compared to pre-treatment, both with and without changes in promoter methylation status [30]. This may explain the discordance between methylation or protein status at initial diagnosis and clinical outcome.
Mismatch repair deficiency in recurrent tumors
As mentioned in Box 1, Figure IB, MMR activity is required for TMZ cytotoxicity. Tumors deficient in MMR proteins, including MLH1, MSH2, MSH6, and PMS2, are unable to recognize the mispairing of O6-MeG with thymine, and thus evade the cytotoxic effects of TMZ [43, 44]. Accordingly, a combined measure of low MGMT activity and functional MMR was demonstrated to best predict sensitivity to TMZ in patient-derived xenograft models of glioblastoma [45]. Though MMR deficiency is less common in newly diagnosed glioblastoma, recurrent tumors are often associated with reduced levels of MMR proteins, as observed in paired primary and recurrent glioblastoma [46–48]. This could partially explain observed resistance in recurrent tumors independent of MGMT methylation status.
Figure I.
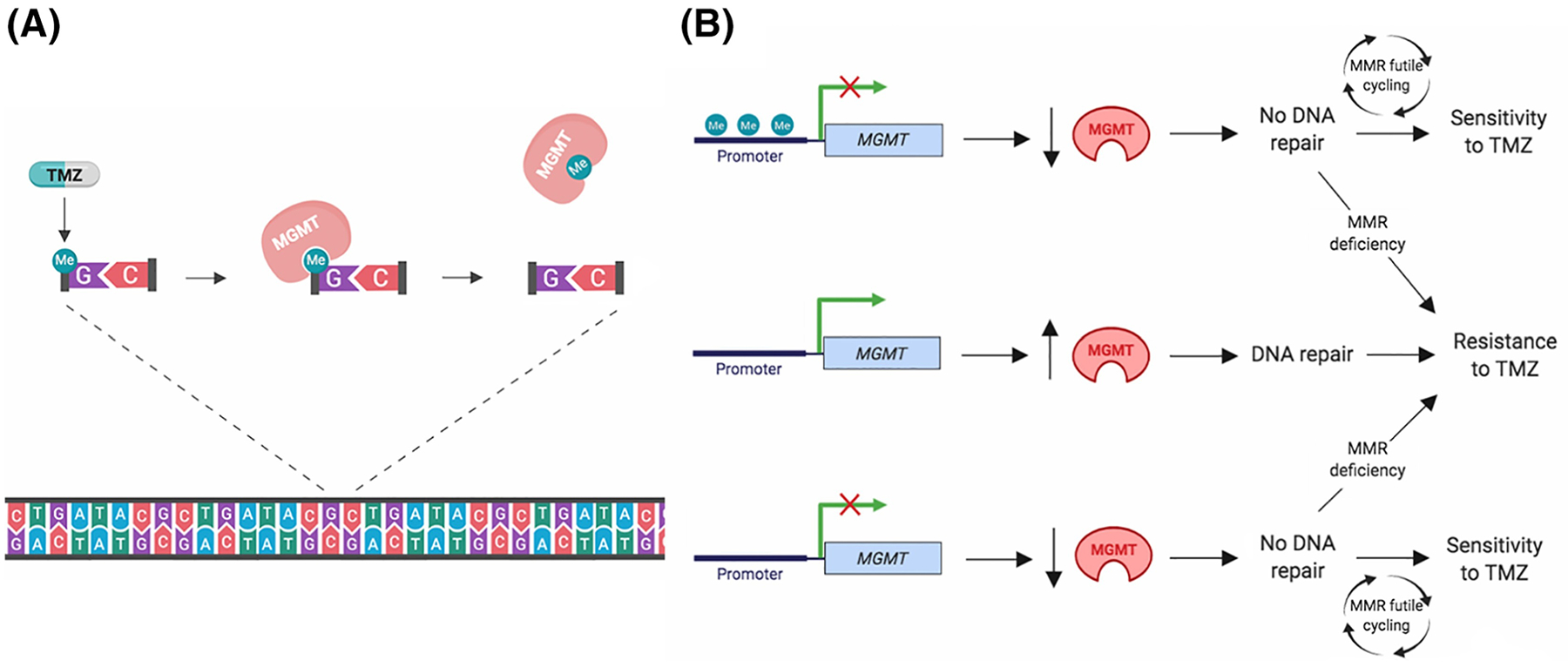
Function of MGMT as determinant of response to TMZ A) Mechanism of MGMT-mediated repair of TMZ-induced DNA damage. Methylation of the O6 position of guanine by TMZ is removed by MGMT, and prevents cell killing by MMR. B) Proposed role of MGMT promoter methylation and expression as determinant of response to TMZ. When the MGMT promoter is methylated (top), silencing of transcription results in low MGMT protein expression. This promotes sensitivity to temozolomide in MMR-proficient cells due to lack of MGMT-mediated DNA damage repair. MMR-deficient cells do not respond to TMZ due to evasion of MMR-dependent DNA double-stranded breaks. When the MGMT promoter is unmethylated (middle), transcription of the MGMT gene results in high expression of MGMT protein, which is able to remove the alkylation adducts, promoting resistance to temozolomide. In some glioblastomas (bottom), MGMT is not expressed in spite of lack of promoter methylation. This promotes sensitivity to TMZ in MMR-proficient cells and resistance in MMR-deficient cells. Created with BioRender.com
MGMT expression as a clinical biomarker?
The value of MGMT promoter methylation status has been widely accepted in the neuro-oncology field, though this biomarker is a surrogate of MGMT activity. Because multiple studies have identified discordance between methylation and expression, recent investigations (Table 2, Key Table) have started to analyze the correlation between gene and/or protein expression directly with patient survival and response to chemotherapy to determine the value of MGMT expression as a biomarker. Low MGMT protein or gene expression has been found to be significantly associated with improved patient survival or treatment response independently of MGMT promoter methylation, and further, has also been found to be an independent prognostic marker in glioblastoma patients by multivariate analysis [10, 11, 18, 25, 29, 41, 49]. For example, when promoter methylation status was analyzed by MSP and biSEQ and protein expression level by IHC, both markers were correlated with both overall survival (OS) and progression-free survival (PFS) [10]. Conversely, in a different study, promoter methylation by MSP, SQ-MSP, and pyrosequencing correlated with outcome, but mRNA or protein expression did not [50].
Table 2, Key Table.
Summary of studies that evaluate the prognostic or predictive value of MGMT protein or gene expressionin addition to promoter methylation status
| Study | Study goal relevant to this Opinion | Methods | Sample size | Relevant results | Conclusion | Reference |
|---|---|---|---|---|---|---|
| Lalezari et al. | To evaluate prognostic value of MGMT protein expression and optimize determination of MGMT status | Methylation: MSP Methylation: BiSEQ (cutoff: median number of methylated sites) Protein expression: IHC (cutoff: median) |
N=418 glioblastoma (IHC: 355, MSP: 02, BiSEQ: 312) | Multivariate analysis: Methylation by MSP is prognostic for OSa (p<0.0001c) and PFSb (p<0.0001) Methylation by BiSEQ is prognostic for OS (p<0.0001) and PFS (p=0.0006) Protein by IHC is prognostic for OS (p<0.0001) and PFS (p=0.001) Combination of methylation and low protein showed improved OS (p=0.0087) and PFS (p=0.0087) compared to other stratified groups |
Combined IHC and methylation analysis yields best assessment of MGMT status | [10] |
| Kreth et al. | To investigate prognostic and/or predictive value of MGMT mRNA expression | Methylation: MSP mRNA expression: qRT-PCR (cutoff: median) | N=63 (53 glioblastoma, 10 anaplastic astrocytoma) | Multivariate analysis: Methylation is prognostic for OS (p=0.0002) and PFS (p=0.0001) Low mRNA expression is prognostic for OS (p=0.0001) and PFS (p=0.0001) Notably, methylated tumors with high mRNA expression showed shorter OS (p<0.001) and PFS (p<0.001) than methylated tumors with low mRNA exp |
Methylation status alone is not sufficient for determining clinical outcome | [18] |
| Pandith et al. | To investigate the prognostic value of MGMT methylation and MGMT protein expression | Methylation: MSP Protein expression: IHC (cutoff: 10%) | N=63 (32 glioblastoma, 14 astrocytoma, 14 oligodendrogliom a, 3 other) | Multivariate analysis: MGMT promoter methylation was an independent prognostic factor (p<0.05) MGMT protein expression was an independent prognostic factor (p=0.002) Significant association between protein and patient outcome was found in groups treated with TMZ (p=0.040) and without TMZ (p=0.006) Significant association between promoter methylation and patient outcome was found in group treated with TMZ (p=0.029) but not without TMZ (p=0.706) |
Both methylation and protein expression should be evaluated | [29] |
| Uno et al. | To determine which method of evaluating MGMT status provides best prognostic and/or predictive value | Methylation: MSP Methylation: pyrosequencing (cutoff: 10%) mRNA expression: qRT-PCR (cutoff: median) Protein expression: IHC (cutoff: 10%) |
N=51 glioblastoma | Multivariate analysis: Only methylation by MSP (p=0.023) or pyrosequencing (p=0.005) was an independent prognostic factor | Promoter methylation status is a more reliable biomarker compared to mRNA or protein expression levels | [9] |
| Shah et al. | To correlate methylation and protein expression with clinical outcome | Methylation: MS- MLPA (3 region classification, cutoff: 0.1) Protein: IHC (cutoff: 15%) | N=70 glioblastoma (IHC: 31) | Multivariate analysis: Low protein expression was an independent predictive factor for PFS (p<0.0001) Methylation using 3R classification was an independent predictive factor for PFS (p<0.0001) |
Refinement of the best method to determine MGMT status is warranted | [11] |
| Cao et al. | To analyze the prognostic value of promoter methylation and protein expression | Methylation: MSP Protein expression: IHC (cutoff: 5%) |
N=83 glioblastoma (IHC: 80, MSP: 76, 73 with both IHC and MSP) | Univariate analysis: Methylation correlated with increased survival (p=0.014) Low protein expression was not prognostic (p=0.197) Notably, combination of methylation and low protein expression yielded longer survival compared to other subgroups (p=0.005) Multivariate analysis: best outcome in methylated- immunonegative compared to unmethylated- immunonegative (p=0.006) |
Combined evaluation of both Methylation status and negative protein expression may have more prognostic value |
[8] |
| Nagane et al. | To analyze the prognostic value of MGMT protein expression | Protein expression: Western blotting (cutoff: median) | N=30 glioblastoma (Western: 19, clinical outcome: 17) | Multivariate analysis: Low MGMT protein expression was an independent favorable prognostic factor for OS (p=0.040), but not PFS (p=0.060) | Low protein expression is a favorable prognostic factor for overall survival benefit in patients treated with TMZ | [41] |
| Sonoda et al. | To evaluate the prognostic value of MGMT promoter methylation and protein expression | Methylation: MSP Protein expression: IHC (cutoff: 20%) |
N=73 glioblastoma (MSP: 62) | Methylation (p=0.011) and negative expression (p=0.049) were independently associated with increased PFS, but not OS | Methylation and low expression are predictive markers for increased progression- free survival | [57] |
| Dahlrot et al. | To investigate the prognostic value of protein expression and combined evaluation of both protein and methylation status | Methylation: pyrosequencing (cutoff: 10%) Protein expression: double immunofluorescenc e assay (cutoff: median) |
N=171 glioblastoma (pyrosequencing: 157, expression: 171) | Univariate analysis: Low MGMT expression in tumor resulted in increased overall survival compared to high expression overall (p=0.01) and within the subgroup receiving the Stupp regimendd(p=0.001), but this trend in multivariate analysis was not significant (p=0.11) Notably, in the patient group who received the Stupp regimen, combined methylation and low expression resulted in the best prognosis, whereas unmethylated promoter and high expression showed the poorest prognosis (p=0.0002) |
Both methylation status and protein expression status are important to evaluate | [25] |
| Melguizo et al. | To investigate the prognostic value of MGMT promoter methylation and protein expression | Methylation: MSP Protein expression: IHC (cutoff: 25%) |
N=78 glioblastoma (methylation and protein expression: 76) | Univariate analysis: Methylation was significantly associated with increased PFS (p=0.036) and OS (p=0.031). Protein expression was not significantly correlated with PFS (0.712) or OS (p=0.894) | Methylation status, but not protein expression, has significant prognostic value | [16] |
| Karayan- Tapon et al. | To identify the method of determining MGMT status that has best prognostic value | Methylation: MSP Methylation: SQ- MSP (cutoff: median) Methylation: pyrosequencing (cutoff: median) mRNA expression: qRT-PCR (cutoff: median) Protein expression: IHC (cutoff: median) |
N=81 glioblastoma | Univariate analysis: Methylation by MSP (p=0.005), SQ-MSP (p<10−4), and pyrosequencing (p<10−4) was significantly associated with increased OS Low mRNA expression was significantly associated with increased OS (p=0.028) Protein expression was not significantly associated with OS (p=0.595) Multivariate analysis: Only methylation at CpG4 by pyrosequencing was a significant factor for predicting overall survival (p<0.0001) |
Methylation status is the best approach | [50] |
| Preusser et al. | To assess the value of MGMT by IHC as a clinical biomarker | Methylation: MSP Protein: IHC (cutoff: 10%, 50%) |
N=164 glioblastoma (MSP: 122) | Univariate analysis: Methylation by MSP was significantly associated with increased survival (p=0.0001) Protein expression was not significantly associated with clinical outcome at any tested cutoff value |
Protein expression by IHC is a less clinically useful biomarker than promoter methylation | [7] |
| Trabelsi et al. | To determine the best method for assessing MGMT methylation status | Methylation: MS- MLPA (cutoff: 0.25) and HM-450K Protein expression: IHC (cutoff: 10%) |
N=55 glioblastoma | Univariate analysis: All gene methylation by MS- MLPA was significantly associated with increased overall survival (p=0.021) and relapse-free survival (p=0.02) in all glioblastoma and in TMZ- treated group (p=0.022 and p- 0.017, respectively) Promoter methylation by MS- MLPA was significantly associated with overall survival in TMZ-treated group (p=0.046), but not RFS No significant correlation was found between survival and expression by IHC, or methylation by HM-450K (small sample size for HM- 450K) |
MGMT methylation has predictive value | [58] |
| Hsu et al. | To compare the prognostic value of MGMT status by four different methods | Methylation: MSP, qMSP (cutoff: median), pyrosequencing (cutoff: 5%) Protein expression: IHC (cutoff: 10%) |
N=121 glioblastoma | Multivariate analysis: All methods had prognostic value for PFS and OS, respectively, including IHC- (p=0.003, p=0.047), MSP+ (p=0.002, p=0.001), qMSP+ (p=0.002, p=0.001), PSQ+ (p<0.001, p=0.001) Notably, patients with IHC- negative/methylation-positive tumors showed increased PFS and OS compared to those with IHC-positive/methylation- negative; further, the addition of pyrosequencing significantly improved prediction of prognosis in IHC-negative cases |
All methods were significantly associated with clinical outcome, and addition of methylation status evaluation may enhance the predictive power of IHC for some patients |
[49] |
OS = overall surviva
PFS = progression-free survival
Statistical significance indicated by p<0.05
Stupp regimen = protocol of glioblastoma treatment with radiation therapy and concomitant and adjuvant temozolomide
Stratifying patients into four subgroups based on combined analysis of methylation and expression (methylated + low expression, methylated + high expression, unmethylated + low expression, unmethylated + high expression) appears to give the most accurate prediction of patient outcome. Such studies concluded that in patients treated with RT and TMZ, those with both MGMT methylation and low protein expression had the longest survival [8, 10, 25, 49]. For example, in one study, no correlation was found between immunostaining and survival alone; however, after combining MSP with IHC analysis, the difference in patient outcome between each subgroup was significant with the greatest median survival in the methylated-immunonegative patient subgroup. Surprisingly, the unmethylated-immunonegative subgroup showed the poorest survival [8]. Combined analysis in a set of 121 glioblastoma patients revealed a better outcome in methylated, IHC-negative patients compared to unmethylated, IHC-positive patients [49]. Similarly, in a larger cohort, combined MGMT hypermethylation and low expression status was associated with both improved overall and progression-free survival compared to the other combinations [10]. In a series of 350 gliomas and gangliogliomas, including 154 glioblastoma, the sensitivity of IHC was 84.4%, and the specificity was only 45.7%; however, when combined with qMSP, the sensitivity and specificity of IHC for predicting MGMT status increased to 99.5% and 93.9%, respectively [17]. It would be important to further determine whether combined analysis also results in improved prediction of patient survival compared to IHC and qMSP alone in this study. Further research testing the correlation of combined methylation and expression status with outcome is warranted. Stratification of patients into subgroups incorporating both methylation and expression parameters may very likely enhance the prognostic and/or predictive value of MGMT methylation status (see Outstanding Questions).
Outstanding Questions.
Does the combined evaluation of MGMT methylation status and gene/protein expression better predict patient outcome compared to either parameter alone?
What other molecular characteristics beyond mismatch repair (MMR) contribute to patient response to temozolomide treatment which would enhance the predictive value of MGMT status?
How can we take the next step to implement these biomarkers in routine treatment decision-making for glioblastoma patients?
Concluding remarks and future perspectives
Although MGMT status has been a biomarker in clinical trials for some time and has been implemented as a predictive marker for elderly patients, it has not yet been integrated for all patients in routine clinical practice for prognostic evaluation or treatment decision-making [51]. 30 – 60% of all glioblastoma patients have a methylated MGMT promoter; yet, those expressing MGMT due to an unmethylated promoter or alternate mechanism will likely respond poorly to standard alkylating therapy [4, 29]. Current practice does not withhold TMZ from unmethylated patients treated with standard protocols due to the uncertainty of the predictive value of MGMT methylation status and the lack of alternative treatment modalities, and this has precluded clinical application of research findings on correlation of MGMT status with patient outcome [31].
We suggest that MGMT methylation and MGMT protein expression should not be used interchangeably as single biomarkers. It is important to elucidate the molecular and genetic mechanisms regulating MGMT expression beyond promoter methylation and also establish standardized clinical cutoffs for assays evaluating MGMT status. It is likely that evaluation of both MGMT methylation and gene/protein expression is critical for most accurately predicting patient survival and treatment response, which should be considered in further research studies, such as clinical trial inclusion criteria and stratification between treatment arms, and routine clinical practice. Decisions based on both parameters will likely give a better indication of response for glioblastoma patients, especially for the patient subset with inconsistent MGMT methylation status and protein level. It also remains to be established whether the evaluation of other molecular markers in addition to MGMT status improves the prognostic and predictive value of this biomarker (see Outstanding Questions). Integrating additional molecular characteristics with MGMT methylation status, such as MGMT protein expression or MMR proficiency, to develop a combined biomarker status should be the next step in assisting the treatment decision-making of which patients should or should not receive temozolomide. It is critical to address these challenges in order to implement MGMT status as a reliable biomarker to identify all patients that are likely to benefit from TMZ, while avoiding unnecessary treatment toxicities in patients who are unlikely to respond; this will allow the use of personalized therapeutic strategies that are more likely to bring favorable outcomes.
Highlights.
MGMT promoter methylation status is a widely accepted biomarker in glioblastoma.
Inconsistencies between MGMT promoter methylation status and expression level have raised the question of the value of promoter methylation status in predicting patient response to temozolomide treatment in glioblastoma.
Combined evaluation of MGMT methylation and expression and/or MMR proficiency may provide better insight into a personalized treatment approach.
Understanding the molecular and genetic mechanisms regulating MGMT expression beyond promoter methylation is essential to enhance the utility of MGMT status as a biomarker in treatment decision-making for glioblastoma patients.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH.
Glossary
- Adjuvant therapy
Additional cancer treatment given after the primary treatment to decrease the risk of cancer recurrence
- Alkylating agent
A class of anti-cancer drugs that interferes with cell DNA to kill tumor cells
- Astrocytoma
A type of tumor originating from brain or spinal cord cells called astrocytes
- Glioma
A class of brain tumor that arises from cells called glia that surround and support nerve cells
- Methylation
An epigenetic mechanism where methyl groups are added to DNA molecules (in the context of DNA methylation), often modifying the gene expression and function; promoter methylation refers to the addition of methyl groups to DNA sequences within the promoter region of a gene
- Neuro-oncology
A specialty which involves the management and study of central nervous system cancers, including tumors of the brain and spinal cord
- TCGA-GBM
A dataset of glioblastoma as part of The Cancer Genome Atlas program that is built for cancer research
- Xenograft
The transplant of tissue or cells to an individual of another species. In this article, it refers to a model of human tumor grown in immunodeficient mice
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: All authors declare no conflicts of interest.
References
- 1.Ostrom QT et al. (2018) CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol 20 (suppl_4), iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY and Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359 (5), 492–507. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R et al. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352 (10), 987–96. [DOI] [PubMed] [Google Scholar]
- 4.Weller M et al. (2010) MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 6 (1), 39–51. [DOI] [PubMed] [Google Scholar]
- 5.Weller M et al. (2009) Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol 27 (34), 5743–50. [DOI] [PubMed] [Google Scholar]
- 6.Mansouri A et al. (2018) MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preusser M et al. (2008) Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol 18 (4), 520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao VT et al. (2009) The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery 65 (5), 866–75; discussion 875. [DOI] [PubMed] [Google Scholar]
- 9.Uno M et al. (2011) Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma. Clinics 66 (10), 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lalezari S et al. (2013) Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro Oncol 15 (3), 370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah N et al. (2011) Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One 6 (1), e16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brell M et al. (2011) O6-Methylguanine-DNA methyltransferase protein expression by immunohistochemistry in brain and non-brain systemic tumours: systematic review and meta-analysis of correlation with methylation-specific polymerase chain reaction. BMC Cancer 11, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason S and McDonald K (2012) MGMT testing for glioma in clinical laboratories: discordance with methylation analyses prevents the implementation of routine immunohistochemistry. J Cancer Res Clin Oncol 138 (11), 1789–97. [DOI] [PubMed] [Google Scholar]
- 14.Bell EH et al. (2017) Molecular-Based Recursive Partitioning Analysis Model for Glioblastoma in the Temozolomide Era: A Correlative Analysis Based on NRG Oncology RTOG 0525. JAMA Oncol 3 (6), 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dullea A and Marignol L (2016) MGMT testing allows for personalised therapy in the temozolomide era. Tumour Biol 37 (1), 87–96. [DOI] [PubMed] [Google Scholar]
- 16.Melguizo C et al. (2012) MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med 10, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L et al. (2017) Comparative assessment of three methods to analyze MGMT methylation status in a series of 350 gliomas and gangliogliomas. Pathol Res Pract 213 (12), 1489–1493. [DOI] [PubMed] [Google Scholar]
- 18.Kreth S et al. (2011) O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One 6 (2), e17156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molenaar RJ et al. (2014) The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol 16 (9), 1263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mur P et al. (2015) Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. J Neurooncol 122 (3), 441–50. [DOI] [PubMed] [Google Scholar]
- 21.Cabrini G et al. (2015) Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review). Int J Oncol 47 (2), 417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wick W et al. (2013) Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology 81 (17), 1515–22. [DOI] [PubMed] [Google Scholar]
- 23.Hegi ME et al. (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol 26 (25), 4189–99. [DOI] [PubMed] [Google Scholar]
- 24.Radke J et al. (2019) Predictive MGMT status in a homogeneous cohort of IDH wildtype glioblastoma patients. Acta Neuropathol Commun 7 (89). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahlrot RH et al. (2018) Prognostic value of O-6-methylguanine-DNA methyltransferase (MGMT) protein expression in glioblastoma excluding nontumour cells from the analysis. Neuropathol Appl Neurobiol 44 (2), 172–184. [DOI] [PubMed] [Google Scholar]
- 26.Hegi ME et al. (2019) MGMT Promoter Methylation Cutoff with Safety Margin for Selecting Glioblastoma Patients into Trials Omitting Temozolomide: A Pooled Analysis of Four Clinical Trials. Clin Cancer Res 25 (6), 1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brigliadori G et al. (2016) Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. J Neurooncol 128 (2), 333–9. [DOI] [PubMed] [Google Scholar]
- 28.Dunn J et al. (2009) Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer 101 (1), 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandith AA et al. (2018) Concordant association validates MGMT methylation and protein expression as favorable prognostic factors in glioma patients on alkylating chemotherapy (Temozolomide). Sci Rep 8 (1), 6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christmann M et al. (2010) MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int J Cancer 127 (9), 2106–18. [DOI] [PubMed] [Google Scholar]
- 31.Taylor JW and Schiff D (2015) Treatment considerations for MGMT-unmethylated glioblastoma. Curr Neurol Neurosci Rep 15 (1), 507. [DOI] [PubMed] [Google Scholar]
- 32.Everhard S et al. (2009) Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol 11 (4), 348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pieper RO et al. (1996) Methylation of CpG island transcription factor binding sites is unnecessary for aberrant silencing of the human MGMT gene. J Biol Chem 271 (23), 13916–24. [DOI] [PubMed] [Google Scholar]
- 34.Bady P et al. (2012) MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol 124 (4), 547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malley DS et al. (2011) A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol 121 (5), 651–61. [DOI] [PubMed] [Google Scholar]
- 36.Christmann M et al. (2011) O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim Biophys Acta 1816 (2), 179–90. [DOI] [PubMed] [Google Scholar]
- 37.Moen EL et al. (2014) The role of gene body cytosine modifications in MGMT expression and sensitivity to temozolomide. Mol Cancer Ther 13 (5), 1334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitange GJ et al. (2009) Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol 11 (3), 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiewrodt D (20008) MGMT in primary and recurrent human glioblastomas after radiation and chemotherapy and comparison with p53 status and clinical outcome. Int J Cancer 122, 1391–99. [DOI] [PubMed] [Google Scholar]
- 40.Silber JR et al. (2012) O(6)-methylguanine-DNA methyltransferase in glioma therapy: promise and problems. Biochim Biophys Acta 1826 (1), 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagane M et al. (2007) Prognostic significance of O6-methylguanine-DNA methyltransferase protein expression in patients with recurrent glioblastoma treated with temozolomide. Jpn J Clin Oncol 37 (12), 897–906. [DOI] [PubMed] [Google Scholar]
- 42.Brandes AA et al. (2017) Role of MGMT Methylation Status at Time of Diagnosis and Recurrence for Patients with Glioblastoma: Clinical Implications. Oncologist 22 (4), 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wick W et al. (2014) MGMT testing--the challenges for biomarker-based glioma treatment. Nat Rev Neurol 10 (7), 372–85. [DOI] [PubMed] [Google Scholar]
- 44.Thomas A et al. (2017) Temozolomide in the Era of Precision Medicine. Cancer Res 77 (4), 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagel ZD et al. (2017) DNA Repair Capacity in Multiple Pathways Predicts Chemoresistance in Glioblastoma Multiforme. Cancer Res 77 (1), 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felsberg J et al. (2011) Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer 129 (3), 659–70. [DOI] [PubMed] [Google Scholar]
- 47.McFaline-Figueroa JL et al. (2015) Minor Changes in Expression of the Mismatch Repair Protein MSH2 Exert a Major Impact on Glioblastoma Response to Temozolomide. Cancer Res 75 (15), 3127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Indraccolo S et al. (2019) Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clin Cancer Res 25 (6), 1828–1837. [DOI] [PubMed] [Google Scholar]
- 49.Hsu CY et al. (2017) Comparative Assessment of 4 Methods to Analyze MGMT Status in a Series of 121 Glioblastoma Patients. Appl Immunohistochem Mol Morphol 25 (7), 497–504. [DOI] [PubMed] [Google Scholar]
- 50.Karayan-Tapon L et al. (2010) Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol 97 (3), 311–22. [DOI] [PubMed] [Google Scholar]
- 51.Thon N et al. (2013) Personalized treatment strategies in glioblastoma: MGMT promoter methylation status. Onco Targets Ther 6, 1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajapakse VN et al. (2018) CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines. iScience 10, 247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erasimus H et al. (2016) DNA repair mechanisms and their clinical impact in glioblastoma. Mutat Res Rev Mutat Res 769, 19–35. [DOI] [PubMed] [Google Scholar]
- 54.Villano JL et al. (2009) Temozolomide in malignant gliomas: current use and future targets. Cancer Chemother Pharmacol 64 (4), 647–55. [DOI] [PubMed] [Google Scholar]
- 55.Hegi ME et al. (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352 (10), 997–1003. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y et al. (2013) MGMT promoter methylation and glioblastoma prognosis: a systematic review and meta-analysis. Arch Med Res 44 (4), 281–90. [DOI] [PubMed] [Google Scholar]
- 57.Sonoda Y et al. (2010) O(6)-Methylguanine DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression is correlated with progression-free survival in patients with glioblastoma. Int J Clin Oncol 15 (4), 352–8. [DOI] [PubMed] [Google Scholar]
- 58.Trabelsi S et al. (2016) MGMT methylation assessment in glioblastoma: MS-MLPA versus human methylation 450K beadchip array and immunohistochemistry. Clin Transl Oncol 18 (4), 391–7. [DOI] [PubMed] [Google Scholar]


