Abstract
Genomewide molecular profiling has revealed new subtypes of lymphoma that originate from lymphocytes that differ in developmental stage and that use distinct oncogenic programs, yet are indistinguishable under the microscope. In this review, we discuss recent progress in the molecular genetics of aggressive lymphomas and focus on the most common form of this disease, diffuse large-B-cell lymphoma, which accounts for 30 to 40% of newly diagnosed lymphomas.
B-CELL DEVELOPMENT AND LYMPHOMAGENESIS
Non-Hodgkin’s B-cell lymphomas co-opt the regulatory biologic features of normal B cells for their own malignant purpose. This means that the function of such neoplasms depends considerably on the differentiation state of the B cells from which they originate (Fig. 1). During B-cell development in the bone marrow, recombination of V, D, and J gene segments assembles immunoglobulin heavy-chain (IgH) and light-chain (IgL) genes. In this process, two enzymes encoded by recombinase-activating genes (RAG1 and RAG2) cause breaks in double-stranded DNA, but DNA repair processes (nonhomologous end-joining) resolve them. However, such breaks can contribute to chromosomal translocations in lymphoma.1
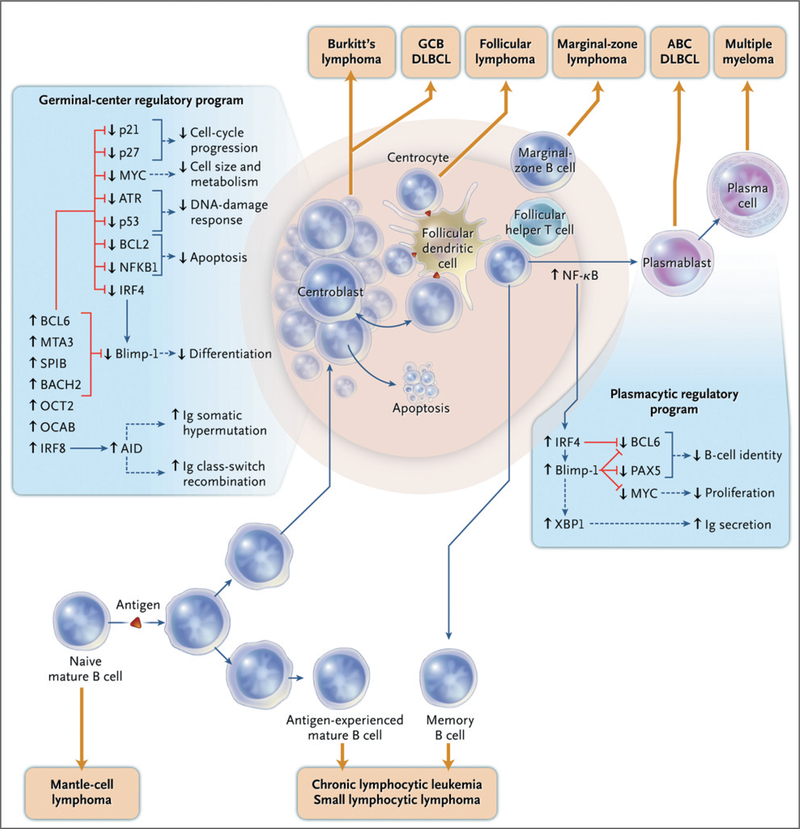
Figure 1 (facing page). B-Cell Differentiation and Lymphomagenesis.
Malignant lymphomas can arise at multiple stages of normal B-cell development. After the stimulation of a mature naive B cell with a T-cell-dependent antigen, the germinal-center reaction is initiated. The germinal-center B cell represents a discrete, quasi-stable differentiation stage that is characterized by a unique regulatory network and the action of activation-induced cytidine deaminase (AID), which induces both immunoglobulin (Ig) somatic hypermutation and heavy-chain class switching. Several transcription factors are required to establish and maintain the identity and function of the germinal-center B cell, including BCL6, MTA3, SPIB, BACH2, OCT2, OCAB, and IRF8. Red lines indicate that a regulatory factor inhibits the indicated gene or cellular function, and blue lines indicate positive regulation. In concert, these factors block plasmacytic differentiation by repressing Blimp-1. They also promote cell-cycle progression without cell growth while blocking the DNA damage response evoked by AID-dependent mutations and DNA breaks. Within the germinal center, the rapidly proliferating centroblasts are prone to cell death. Periodically, centroblasts travel to a subcompartment of the germinal center that is rich in follicular dendritic cells and follicular helper T cells, where they become centrocytes. Centrocytes may be rescued from cell death as a result of stimulation by antigen on follicular dendritic cells and CD40 ligand on T cells and may then revert to the centroblast state and resume proliferation. IRF4 initiates plasmacytic differentiation by establishing a characteristic regulatory network, which extinguishes the mature B-cell program while promoting terminal differentiation and immunoglobulin secretion. The putative origins of various non-Hodgkin’s lymphomas — including the germinal-center B-cell—like (GCB) and activated B-cell—like (ABC) subtypes of diffuse large-B-cell lymphoma (DLBCL) — are indicated. Lymphomas that are derived from germinal-center B cells have recurrent genetic abnormalities that circumvent the normal genetic program in order to block plasmacytic differentiation, promote cell growth, and evade apoptosis. NF-κB denotes nuclear factor-κB.
The germinal center is the probable source of many types of lymphoma.2 The germinal-center reaction begins when antigen, in conjunction with signals from T cells, activates mature B cells. Antigen-specific T cells and follicular dendritic cells bearing antigen surround germinal-center B cells. Centroblasts (rapidly dividing B cells with a noncleaved nucleus) in the dark zone of the germinal center expand at a remarkable rate. These cells periodically enter the light zone of the germinal center, where they morph into centrocytes (nondividing B cells with a cleaved nucleus). These centrocytes strip antigen from follicular dendritic cells and process it for presentation to nearby T cells.3 Centrocytes can revert to centroblasts and reinitiate proliferation, or they can differentiate into memory B cells or plasma cells.
During the germinal-center reaction, two distinct modifications of B-cell DNA alter the B-cell receptor: somatic hypermutation and class-switch recombination, both of which require activation-induced cytidine deaminase (AID).4 Class-switch recombination changes the immunoglobulin heavy-chain class from IgM to IgG, IgA, or IgE, whereas somatic hypermutation entails immunoglobulin-variable-region mutations, which create a population of B cells with increased (or decreased) affinity for a particular antigen. These genetic modifications are essential for a normal immune response, but they are also a source of DNA damage that can become pathologic in lymphomas.
A suite of transcription factors shapes the phenotype of the germinal-center B cell (Fig. 1). These cells selectively express B-cell lymphoma 6 (BCL6), a repressor of transcription. Genetic translocations in diffuse large-B-cell lymphoma and other lymphomas5 deregulate BCL6, and disruption of the gene abolishes the germinal-center response in mice.6–8 BCL6 protein represses many genes involved in the differentiation of plasma cells, cell-cycle progression, responses to DNA damage, and cell death. Notably, one of these genes is Blimp-1,9,10 a master regulator of plasma-cell differentiation that extinguishes the gene-expression program of mature B cells.11,12 As germinal-center B cells begin to differentiate into plasma cells, they upregulate interferon regulatory factor 4 (IRF4), a transcription factor required for the conversion to plasma cells. IRF4 in turn increases the expression of Blimp-1,13–15 and this increase represses BCL6, thus tipping differentiation in favor of plasma cells.11
Germinal-center B cells give rise to many types of lymphoma, including diffuse large-B-cell lymphoma, follicular lymphoma, and Burkitt’s lymphoma. These types of lymphoma carry the differentiation program of the normal B cell from which they arise,16–18 but oncogenic abnormalities in the lymphoma often subvert the normal program. For example, normal centroblasts lack the antiapoptotic activities of B-cell lymphoma 2 (BCL2) and the nuclear factor-κB (NF-κB) pathway and hence are poised to die.17,19 Malignant centroblasts, however, avoid cell death by acquiring activating translocations of BCL2 or by constitutively activating NF-κB. Likewise, BCL6 suppresses the MYC oncogene in most normal centroblasts,17,18,20 but malignant centroblasts evade this control by translocating or amplifying MYC, thereby allowing expression of a potent regulator of cell metabolism and growth.
ALTERATIONS OF B-CELL DNA IN LYMPHOMAS
RAG RECOMBINASE
The normal mechanisms of V(D)J recombination, somatic hypermutation, and class-switch recombination can alter the genome of lymphomas. RAG-mediated chromosomal breaks in pro-B cells can persist over many cell divisions, thereby creating the potential for a translocation in which immunoglobulin loci fuse to DNA breaks in other genes.21 The activation of RAG recombinase in mature B cells during immunoglobulin-receptor editing can also promote translocations.22 The t(14;18) chromosome translocation, which occurs in most follicular lymphomas and some diffuse large-B-cell lymphomas, places BCL2 under the control of IgH locus enhancers. This translocation requires RAG recombinase, which cleaves DNA in the IgH locus and in an unusual DNA structure in BCL2.23 In mantle-cell lymphoma, RAG recombinase allows a t(11;14) translocation that joins the CCND1 gene to the IgH locus, causing overexpression of cyclin D1 and hence deregulation of the cell cycle. The translocation breakpoints in t(14;18) and t(11;14) suggest that AID conspires with RAG recombinase to create double-stranded DNA breaks in BCL2 and CCND1.24
AID
The mutator enzyme AID plays several roles in lymphomagenesis. In mouse models, the development of diffuse large-B-cell lymphoma requires AID,25 and transgenic overexpression of AID causes B-cell lymphomas.26 AID can introduce mutations into nonimmunoglobulin genes of germinal-center B cells in mice at frequencies that are many orders of magnitude above the background rate of mutation.27–29 Diffuse large-B-cell lymphomas accumulate AID-dependent mutations in many genes, including MYC and PIM1 oncogenes.30 These mutations may accrue because of a deficiency in DNA mismatch repair or selection for cells with advantageous mutations.29 Regions that incur AID-mediated mutations coincide with chromosomal translocation breakpoints, suggesting that AID introduces double-stranded breaks that promote translocations.30
Class-switch recombination, which is mediated by AID, introduces DNA double-stranded breaks in IgH switch regions that can cause MYC-IgH translocations and breaks within the MYC locus.31–36 The activated B-cell-like (ABC) subtype of diffuse large-B-cell lymphoma not only has exceedingly high levels of AID but also undergoes aberrant class-switch recombination in which the IgH switch regions sustain deletions, insertions, and mutations without participating in a physiological class-switch event.32 Perhaps, as a consequence, this lymphoma subtype accumulates chromosomal translocations involving the IgH switch regions.32
PATHOGENESIS OF DIFFUSE LARGE-B-CELL LYMPHOMA
Diffuse large-B-cell lymphoma, the most common form of lymphoma, accounts for 30 to 40% of newly diagnosed lymphomas. Combination chemotherapy plus rituximab can cure approximately 50% of cases. Gene-expression profiling can divide this type of lymphoma into three histologically indistinguishable molecular subtypes: the ABC subtype, the germinal-center B-cell-like (GCB) subtype, and primary mediastinal B-cell Lymphoma (PMBL).16,37–40 These subtypes differ in the expression of thousands of genes and apparently arise from B cells that are at separate stages of differentiation. In addition, the process of malignant transformation differs for each subtype, as evidenced by distinctive genetic abnormalities, and the three subtypes differ in clinical presentation, in cure rates after chemotherapy, and in responsiveness to targeted therapies. For these reasons, we view each of the subtypes as a distinct neoplasm.
GCB lymphomas express hundreds of genes that define germinal-center B cells.16,37,41 The malignant clone continues to undergo somatic hypermutation,42 and the cells have often switched IgH classes.32 By contrast, ABC lymphomas have the plasma-cell expression program, including the transcription factor XBP1, the master regulator of immunoglobulin secretion.38,43 Constitutive activation of the NF-κB pathway causes ABC lymphomas to express the transcription factor IRF4, and this may push them toward differentiation into plasma cells.44,45 However, ABC lymphomas acquire genetic lesions that interfere with Blimp-1, thereby blocking full differentiation into plasma cells.9,46–50
A block in differentiation seems to be an important early step in the pathogenesis of the ABC subtype, but the nature of the precursor cell is unclear. These lymphomas contain high amounts of AID, and their IgH genes have been heavily mutated.32,38,42,51 Nevertheless, most ABC lymphomas have not undergone class-switch recombination, and they express IgM, unlike most normal germinal-center B cells and other GCB lymphomas.32,38 They could originate from IgM-positive post-germinal-center memory cells52 or from a pre-germinal-center B cell that expresses AID, a characteristic of certain extrafollicular B cells.53
PMBL, the third subtype, typically presents as a mediastinal mass in a young woman (median age, 30 to 35 years). The tumor mass often contains a thymic remnant, suggesting that it originates from a rare thymic B cell. Although the ABC and GCB subtypes can seed the gastrointestinal tract and bone marrow, PMBL spreads by direct extension to adjacent thoracic structures.39 Clinical features alone cannot reliably distinguish PMBL from other subtypes, but gene-expression profiling can readily distinguish them.39,40 The gene-expression signature of PMBL contains a molecular link with Hodgkin’s lymphoma, which may also arise from a thymic B cell.39,40 However, these two lymphomas differ because PMBL typically expresses genes of mature B cells, whereas Hodgkin’s Lymphoma does not.39
GENETIC ABERRATIONS
ONCOGENIC PATHWAYS IN LYMPHOMA SUBTYPES
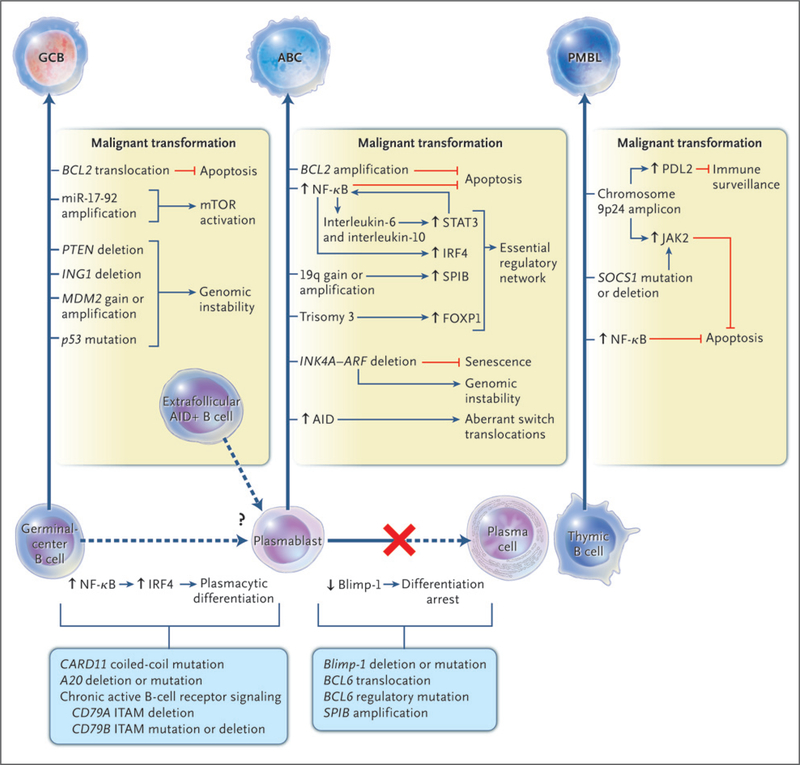
Some oncogenic abnormalities occur in more than one subtype of diffuse large-B-cell lymphoma,50,54 but many oncogenic pathways are predominantly or exclusively used by only one subtype (Fig. 2). Genetic lesions that are specific to GCB lymphoma are the t(14;18) translocation,37,50 deletion of the tumor suppressor PTEN, amplification of the microRNA cluster miR-17–92 (which down-regulates PTEN55), and p53 mutations.56
Figure 2. Oncogenic Pathways for Three Subtypes of Diffuse Large-B-Cell Lymphoma.
On the basis of gene-expression profiling, diffuse large-B-cell lymphoma can be divided into three molecular subtypes: the germinal-center B-cell–like (GCB) subtype, the activated B-cell–like (ABC) subtype, and primary mediastinal B-cell lymphoma (PMBL). These subtypes originate from various stages of B-cell differentiation and acquire distinct oncogenic abnormalities. The abnormalities that are listed are preferentially or exclusively observed in the indicated subtypes. AID denotes activation-induced cytidine deaminase, ITAM immunoreceptor tyrosine-based activation motifs, mTOR mammalian target of rapamycin, and NF-κB nuclear factor-κB. Blue lines indicate activation, and red lines indicate inhibition.
Numerous genetic abnormalities in the ABC subtype are rare or absent in the other subtypes. Most ABC lymphomas overexpress BCL2 and many amplify the BCL2 locus.50 Many delete the INK4A-ARF locus, which encodes p16, an inhibitor of senescence, and p14ARF, an inhibitor of p53 activation.50,57 Loss of these tumor suppressors blocks the action of chemotherapy, and this could contribute to the poor prognosis that is associated with the ABC subtype.50
A genetic hallmark of PMBL is amplification of a region on chromosome 9p24, which occurs in almost half the patients with this subtype and also in those with Hodgkin’s lymphoma.39,50,58 This region encodes JAK2, a tyrosine kinase that phosphorylates and activates the transcription factor STAT6.59 Moreover, SOCS1, a suppressor of JAK signaling, is regularly deleted in PMBL and Hodgkin’s lymphoma.60–62 The amplified region in 9p24 encompasses many megabases of DNA, suggesting involvement of other genes. For example, the amplification of PDL1 (also called CD274) and PDL2 (also called CD273), which encode inhibitors of T-cell responses, could allow the malignant clone to survive in the thymus.39
CONSTITUTIVE NF-κB SIGNALING
The ABC subtype has gene-expression characteristics of normal B cells that were activated by cross-linking the B-cell receptor.16 Subsequent studies directly implicated B-cell-receptor signaling in the pathogenesis of this lymphoma. In resting normal B cells, the inhibitory protein IκBα sequesters NF-κB transcription factors in the cytoplasm.63 The stimulation of the cells through certain surface receptors leads to phosphorylation of IκBα, causing its degradation in proteasomes. Cytoplasmic NF-κB factors can then move to the nucleus and activate their target genes. In normal B cells, cross-linking the B-cell receptor transiently engages the NF-κB pathway, but in the ABC subtype, the NF-κB pathway is constitutively active (Fig. 2 and 3A).44 Interference with NF-κB signaling kills ABC but not GCB cells, which shows that the ABC subtype is dependent on the constitutive activity of this pathway.44,45
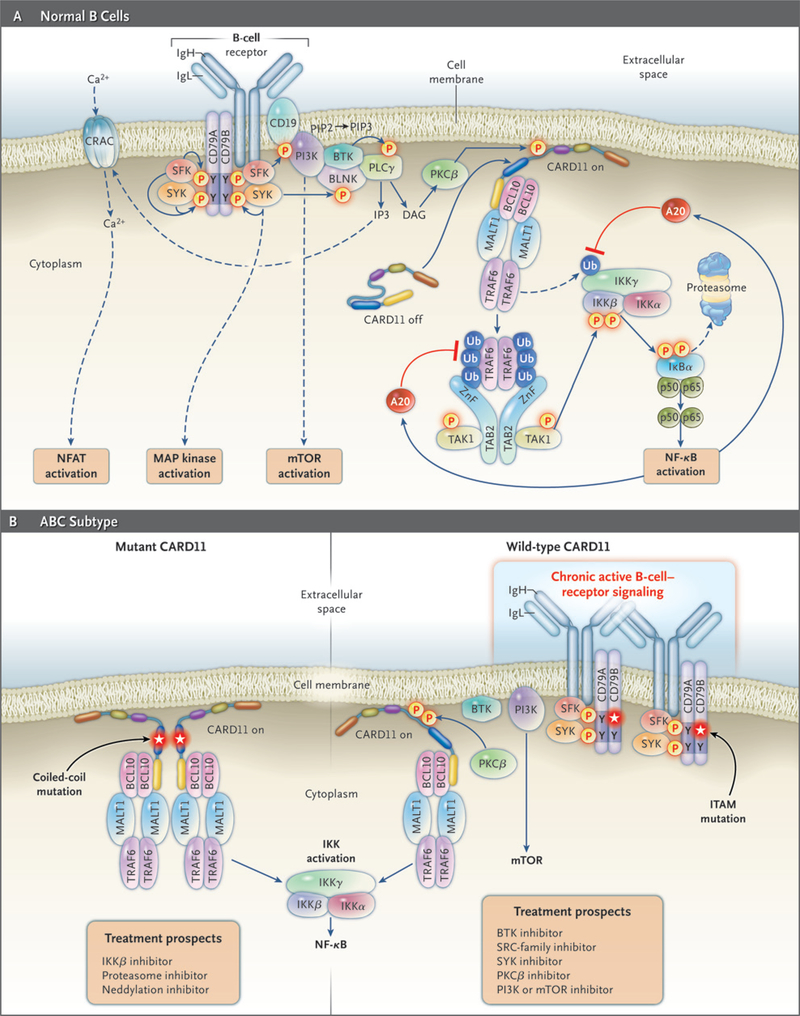
Figure 3 (facing page). B-Cell–Receptor and Nuclear Factor-κB (NF-κB) Signaling Pathways in Normal and Malignant Lymphocytes.
Panel A shows B-cell-receptor and NF-κB signaling in normal B cells. The engagement of the B-cell receptor by antigen evokes a signaling cascade that culminates in the activation of the NF-κB, mTOR (mammalian target of rapamycin), ERK MAP kinase, and nuclear factor of activated T cells (NFAT) pathways. Signaling is initiated when a SRC-family kinase (SFK) phosphorylates tyrosines in immunoreceptor tyrosine-based activation motifs (ITAMs) of the B-cell–receptor subunits CD79A and CD79B. The tyrosine kinase SYK is recruited to the ITAMs through its tandem SH2 domains and becomes enzymatically active. SYK phosphorylates many downstream targets, including the adapter BLNK, and initiates ERK MAP kinase signaling. In parallel, phosphatidylinositol 3-kinase (PI3K) is activated by recruitment to B-cell coreceptor CD19, generating membrane lipid phosphatidylinositol 3,4,5-triphosphate (PIP3) and activating the mTOR pathway. BTK is recruited to the membrane by binding to PIP3 and forms a complex with BLNK and phospholipase Gγ (PLCγ). PLCγ generates the second messenger IP3, which initiates an influx of calcium ions through the calcium-release-activated calcium (CRAC) channel and activation of the NFAT pathway, as well as diacylglycerol (DAG), which activates protein kinase Cβ (PKCβ). The signaling scaffold protein CARD11 is present in a latent state in the cytoplasm of resting B cells but translocates to the plasma membrane after phosphorylation by PKCβ. There, CARD11 nucleates a multiprotein complex, including MALT1, BCL10, and TRAF6. In the process, the ubiquitin ligase TRAF6 is activated, leading to ubiquitination of the IκB kinase (IKK) γ subunit, a necessary step toward IKK activation. TRAF6 becomes autoubiquitinated, leading to its association with a complex consisting of TAB2 and the kinase TAK1. TAK1 phosphorylates the IKKβ subunit in its activation loop, thereby activating IKK to phosphorylate IκBα and initiate NF-κB signaling. A20, a target gene of NF-κB, inhibits NF-κB signaling through the deubiquitination of IKKγ and TRAF6. Panel B shows pathways to the activation of NF-κB in the activated B-cell-like (ABC) subtype of diffuse large-B-cell lymphoma. Roughly 10% of ABC lymphomas have activating mutations of CARD11 in its coiled-coil domain (left subpanel). These mutations cause CARD11 to form large cytoplasmic aggregates that recruit other signaling components, including IKK, leading to constitutive activation of the NF-κB pathway. Other ABC lymphomas have wild-type CARD11 and instead have a chronic active form of B-cell-receptor signaling (right subpanel). In these cases, the B-cell receptors form immobile clusters in the plasma membrane and trigger multiple downstream pathways, including NF-κB. In 21% of ABC lymphomas, chronic active B-cell-receptor signaling is associated with somatic mutations in the ITAM signaling motifs of CD79B or CD79A, the majority of which alter the first tyrosine of the CD79B ITAM. Treatment prospects for patients with the ABC subtype will depend on the mechanism by which NF-κB is activated, as indicated. Blue lines indicate activation, and red lines indicate repression.
The many downstream targets of NF-κB collectively prevent apoptosis and thereby block the action of many forms of chemotherapy.64 The constitutive activation of NF-kB pathways may thus contribute to the poor response of the ABC subtype to chemotherapy.16,37,65 NF-κB signaling in the ABC subtype also induces the cytokines interleukin-6 and interleukin-10, which act through JAK kinases and the STAT3 transcription factor as autocrine signals to the cell.66,67 Blockade of JAK signaling acts synergistically with the inhibition of NF-κB in killing ABC cells.66
An “Achilles heel” screen has been developed in the hope of finding new targets for therapeutic development by identifying genes that are required for the proliferation or survival of cancer cells.68 This screen revealed that the ABC subtype relies on three molecules to activate NF-κB: CARD11, BCL10, and MALT1 (CBM complex). When normal lymphocytes encounter antigen, B-cell-receptor signaling causes phosphorylation of CARD11, which allows it to assemble with BCL10 and MALT1 into a complex that activates IκB kinase (IKK), the enzyme that phosphoryates IκBα69 (Fig. 3A). This CBM complex forms transiently in normal B cells after antigen engagement but assembles constitutively in the ABC subtype.
Various genetic abnormalities activate CARD11 in the ABC subtype. In approximately 10% of patients, CARD11 is a bona fide oncogene that acquires mutations that activate NF-κB and prolong cell survival.70 Mutant CARD11 isoforms create large protein aggregates in the cytoplasm, the probable sites of constitutive NF-κB activation70 (Fig. 3B).
CHRONIC ACTIVE B-CELL-RECEPTOR SIGNALING
Many ABC lymphomas have wild-type CARD11 yet rely on CARD11 to activate NF-κB.68,70 In these lymphomas, there is a chronic active form of B-cell-receptor signaling that engages the CBM pathway.71 The B-cell receptor consists of the antigen-binding IgH and IgL chains and two signaling subunits, CD79A and CD79B (Fig. 3A). Engagement of the receptor by antigen triggers a cascade of kinases that activate multiple downstream pathways. The survival of the ABC subtype with wild-type CARD11 depends on the B-cell receptor and downstream kinases.71 Like antigen-stimulated normal B cells, these ABC cells display prominent clusters of B-cell receptors on their surface.71 About 20% of ABC lymphomas have mutations in CD79A or CD79B, which are rare or absent in GCB and other Lymphoma subtypes.71 The mutant CD79 proteins increase expression of the B-cell receptor and reduce activation of LYN,71 a negative regulator of B-cell-receptor signaling.72,73
It appears from this evidence that chronic active B-cell-receptor signaling is a critical step in the pathogenesis of the ABC subtype. Conceivably, ABC lymphomas could originate from a B cell that acquires a mutation in CD79A or CD79B, thereby fostering clonal expansion and increasing the probability of additional oncogenic events.
Inactivation of A20, a negative regulator of NF-κB signaling, increases the activity of this pathway in the ABC subtype.74–76 A20 aberrations do not occur commonly in the GCB subtype, but they are present in other lymphomas with NF-κB activity.74,75,77–79 The expression of A20 requires NF-κB signaling, suggesting that tumors with A20 inactivation rely on additional mechanisms to activate NF-κB signaling, such as chronic active B-cell-receptor signaling or a CARD11 mutation.
MOLECULAR SIGNATURES OF THERAPEUTIC RESPONSE
The subtypes of diffuse large-B-cell lymphoma are associated with distinctly different overall rates of survival after anthracycline-based chemotherapy: overall survival is favorable in patients with the GCB subtype and PMBL and inferior in those with the ABC subtype.37,39,80–82 The addition of rituximab to standard chemotherapy combining cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) has improved survival in patients with the ABC subtype,83 but this subtype remains less curable than the GCB subtype65 (Fig. 4B).
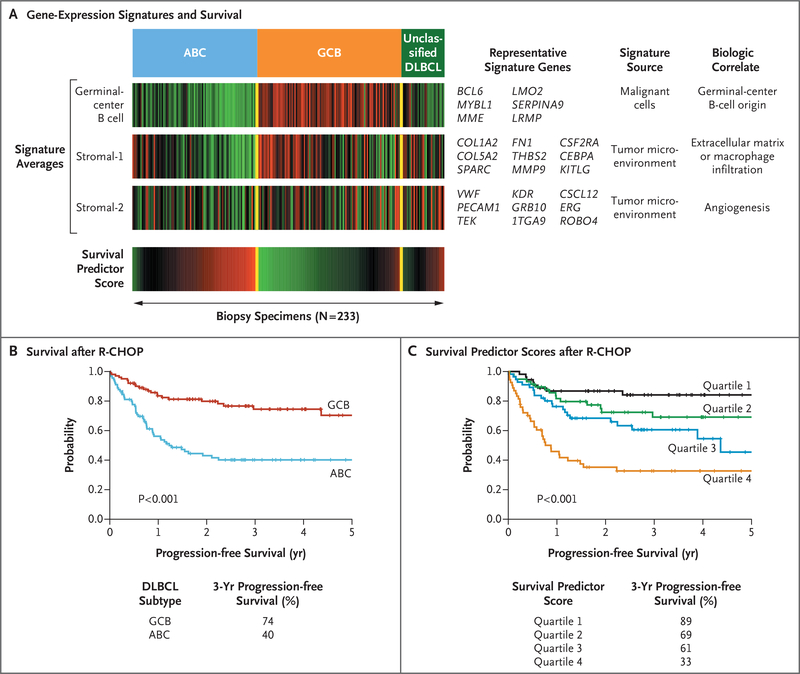
Figure 4. Prediction of Survival According to Gene Expression in Diffuse Large-B-Cell Lymphoma.
Panel A shows three gene-expression signatures that are associated with overall survival in patients with diffuse large-B-cell lymphoma (DLBCL) who have been treated with chemotherapy combining cyclophosphamide, doxorubicin, vincristine, and prednisone with rituximab (R-CHOP). Shown are the average expression levels of the genes in each signature in 233 biopsy specimens; each vertical line represents a single specimen. The germinal-center B-cell signature is prognostically favorable and mirrors the distinction between the two most prevalent subtypes, activated B-cell–like (ABC) and germinal-center B-cell–like (GCB) subtypes, as indicated. The stromal-1 and stromal-2 signatures reflect the character of the nonmalignant cells infiltrating the biopsy specimens. The stromal-1 signature is associated with favorable survival, and the stromal-2 signature is associated with inferior survival. These three signatures are combined in a mathematical model that provides a survival predictor score (bottom bar), which can be used to assess risk for each patient treated with R-CHOP. Gene-expression values and the survival predictor scores are shown over a range that varies by a factor of 16, according to a red-green color scale. For gene expression, red indicates high expression, and green indicates low expression. For the survival predictor score, red indicates unfavorable survival, and green indicates favorable survival. Shown are representative signature genes that are upregulated in expression in association with the indicated signature. Panel B shows survival rates for patients with a molecular diagnosis of the GCB or ABC subtype after R-CHOP therapy. Panel C shows a model for prediction of survival according to gene expression. Patients were ranked according to their survival predictor scores (as shown in Panel A) and divided into quartiles. In this Kaplan-Meier analysis, the gene-expression-based model defines differences in 3-year progression-free survival among patients treated with R-CHOP.
Biologic attributes that influence overall survival have been identified by gene-expression signatures. In one such approach, the signatures are integrated into a statistical model in which the risk for each patient is estimated by a survival predictor score65 (Fig. 4A and 4C). The expression of a signature for a germinal-center B cell in the GCB subtype is associated with favorable survival and mirrors the distinction between the GCB and ABC subtypes.65 Two other signatures reveal differences in nonmalignant cells in the microenvironment of tumors.65 A favorable stromal-1 signature reflects extracellular matrix deposition and infiltration of the tumors with macrophages. The fibrosis may be caused by connective-tissue growth factor, a cytokine that stimulates fibrotic responses of mesenchymal cells.65 The infiltrating macrophages may transmit trophic signals to the tumor, suppress antitumor immune responses, or both.84 Conversely, a stromal-2 signature is associated with inferior survival and identifies tumors with a high density of blood vessels.65 The angiogenesis of these tumors may be due to low expression of the angiogenesis inhibitor thrombospondin 2 and high expression of the chemokine stromal-cell-derived factor 1 (SDF-1), which can recruit endothelial precursors to the tumor.65
THERAPEUTIC OPPORTUNITIES
NF-κB PATHWAY
Therapies targeting the antiapoptotic NF-κB pathway seem especially attractive, given the evidence implicating this pathway in the ABC subtype, PMBL, marginal-zone lymphoma, and Hodgkin’s lymphoma.74,75,77–79 These tumors engage the classic NF-κB pathway, in which the IKK β subunit (IKKβ) phosphorylates IκBα, triggering its ubiquitination by ubiquitin ligase SCFβ-TrCP (Skp1-Cul1-F-box ubiquitin ligase with a beta-transducin repeat-containing protein subunit) and then degradation in the proteasome. Selective IKKβ inhibitors hold promise in the treatment of lymphoma45 but have yet to enter clinical trials. Inhibitors of heat shock protein 90 (HSP90)85 can also block IKK, since HSP90 is a component of the IKK macromolecular complex.86 Another strategy to inhibit NF-κB is to prevent the degradation of IκBα by the blockade of ubiquitin ligase SCFβ-TrCP by means of a small-molecule antagonist of the NEDD8 activating enzyme.87 Recently, MALT1 has emerged as a therapeutic target in this pathway since it has protease activity that is required for NF-κB signaling and the survival of ABC lymphoma cells.88,89
An indirect way to inhibit NF-κB is to block the degradation of IkBα in proteasomes with the use of bortezomib. In a phase 2 trial of bortezomib plus chemotherapy in patients with relapsed or refractory diffuse large-B-cell lymphoma,90 the response rate was higher in patients with the ABC subtype than in those with the GCB subtype (85% vs. 13%), and patients with the ABC subtype had superior overall survival. These results are notable, given the consistently poor response of ABC lymphoma to chemotherapy alone.16,37,65,80,81 The inhibition of NF-κB may have sensitized the tumor to the cytotoxic action of chemotherapy,64 although bortezomib affects many additional pathways that may be used differently by ABC and GCB subtypes.
B-CELL–RECEPTOR SIGNALING
Chronic active B-cell-receptor signaling in patients with the ABC subtype can be interrupted by inhibiting the SRC-family kinases BTK, SYK, and PKCβ, or the phosphatidylinositol 3-kinase (PI3K)-mTOR pathway (Fig. 3A and 3B).71 A selective BTK inhibitor kills ABC cells with chronic active B-cell-receptor signaling, as does the multikinase inhibitor dasatinib, which inhibits BTK and SRC-family kinases.71 A SYK inhibitor, R406, can produce remissions in patients with diffuse large-B-cell Lymphoma that last for several months.91,92 However, R406 has activity in vitro against diffuse large-B-cell lymphomas that are not SYK-dependent, suggesting that the drug’s in vivo activity may in some cases stem from the inhibition of other kinases.71 A small-molecule inhibitor of PKCβ produced complete and partial remissions in a small fraction of patients, but whether these tumors were ABC lymphomas was not determined.93 Since both PI3K and NF-κB signaling supply survival signals to ABC cells,71,94 combination therapies that block both pathways may prove synergistic in patients with chronic active B-cell-receptor signaling.
BCL2
Preclinical data from lymphoma cell lines and primary tumor samples indicate high efficacy of BCL2 inhibitor ABT-737 against lymphoma.95 The sensitivity to ABT-737 treatment in vitro can be predicted by the nature and relative abundance of the antiapoptotic BCL2 family members and the proapoptotic BH3-only proteins.96,97
BCL6
Many cells lines for diffuse large-B-cell lymphoma and Burkitt’s Lymphoma undergo growth arrest and apoptosis after inhibition of BCL6.9,98 BCL6 dependency occurs in ABC and GCB cell lines, with or without a BCL6 translocation, and thus probably reflects the dependency of normal germinal-center B cells on BCL6.6–8 The inhibition of BCL6 by a cell-permeable peptide that disrupts its interaction with corepressor proteins kills xenografts but is not overtly toxic to mice, suggesting that therapeutic targeting of BCL6 holds promise.98
TUMOR MICROENVIRONMENT
An unexplored opportunity in Lymphoma therapy is modulation of the tumor microenvironment. A subgroup of patients with high expression of the stromal-2 signature and increased blood-vessel density in tumors might respond to therapy against vascular endothelial growth factor.65 A component of the stromal-2 signature, SDF-1, is angiogenic, suggesting that agents that block its receptor (CXCR4) should be evaluated in patients with diffuse large-B-cell lymphoma.99 The myeloid-lineage cells that infiltrate some of these lymphomas may provide trophic stimuli to the malignant cells,65 raising the possibility that monoclonal antibodies against these cells might be efficacious. The fibrosis in some tumors could promote cytokine or chemokine signaling,65 and a monoclonal antibody against the profibrotic cytokine CTGF might block this effect.100
SUMMARY
Given the emergence of many new therapeutic agents that affect essential regulatory pathways in lymphomas, the challenge is to identify rational combinations that kill lymphoma cells synergistically. This effort requires both preclinical evaluation and the development of clinical-trial strategies that include molecular profiling. Gene-expression profiling could be used to determine the lymphoma subtype and to estimate the risk associated with standard therapy. Cancer-gene resequencing could be used to identify tumors that are likely to be dependent on a particular signaling pathway and to determine which step in the pathway to inhibit. Finally, the activity of the targeted pathways before and during therapy could be used to assess the efficacy of the treatment. We expect that this “divide-and-conquer” approach will provide increasingly effective and nontoxic therapies for patients with lymphoma.
Acknowledgments
Supported by the Intramural Research Program, the National Cancer Institute, and the Center for Cancer Research at the National Institutes of Health and a grant from the German Research Foundation (to Dr. Lenz).
Contributor Information
Georg Lenz, Metabolism Branch, National Cancer Institute, Bethesda, MD Charité–Humboldt University, Campus Virchow, Department of Hematology–Oncology, Berlin.
Louis M. Staudt, Metabolism Branch, National Cancer Institute, Bethesda, MD
REFERENCES
- 1.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D) J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol 2006;24:541–70. [DOI] [PubMed] [Google Scholar]
- 2.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity 2007;27:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista FD, Harwood NE. The who, how and where of antigen presentation to B cells. Nat Rev Immunol 2009;9:15–27. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000;102:553–63. [DOI] [PubMed] [Google Scholar]
- 5.Ye BH, Lista F, Lo Coco F, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 1993;262:747–50. [DOI] [PubMed] [Google Scholar]
- 6.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 1997;276:589–92. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda T, Yoshida T, Okada S, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med 1997;186:439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet 1997;16:161–70. [DOI] [PubMed] [Google Scholar]
- 9.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 2000;13:199–212. [DOI] [PubMed] [Google Scholar]
- 10.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med 2000;192:1841–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaffer AL, Lin KI, Kuo TC, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 2002; 17:51–62. [DOI] [PubMed] [Google Scholar]
- 12.Turner CA Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 1994;77:297–306. [DOI] [PubMed] [Google Scholar]
- 13.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 2006;25: 225–36. [DOI] [PubMed] [Google Scholar]
- 14.Klein U, Casola S, Cattoretti G, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 2006;7:773–82. [DOI] [PubMed] [Google Scholar]
- 15.Shaffer AL, Emre NC, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature 2008;454:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. [DOI] [PubMed] [Google Scholar]
- 17.Shaffer AL, Rosenwald A, Hurt EM, et al. Signatures of the immune response. Immunity 2001;15:375–85. [DOI] [PubMed] [Google Scholar]
- 18.Klein U, Tu Y, Stolovitzky GA, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A 2003;100:2639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basso K, Klein U, Niu H, et al. Tracking CD40 signaling during germinal center development. Blood 2004;104:4088–96. [DOI] [PubMed] [Google Scholar]
- 20.Ci W, Polo JM, Cerchietti L, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood 2009;113:5536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callén E, Jankovic M, Difilippantonio S, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell 2007;130:63–75. [DOI] [PubMed] [Google Scholar]
- 22.Wang JH, Gostissa M, Yan CT, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 2009;460:231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature 2004;428:88–93. [DOI] [PubMed] [Google Scholar]
- 24.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell 2008;135:1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasqualucci L, Bhagat G, Jankovic M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet 2008;40:108–12. [DOI] [PubMed] [Google Scholar]
- 26.Robbiani DF, Bunting S, Feldhahn N, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell 2009;36:631–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 1998; 280:1750–2. [DOI] [PubMed] [Google Scholar]
- 28.Pasqualucci L, Migliazza A, Fracchiolla N, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A 1998;95:11816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Duke JL, Richter DJ, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature 2008;451:841–6. [DOI] [PubMed] [Google Scholar]
- 30.Pasqualucci L, Neumeister P, Goos- sens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 2001;412:341–6. [DOI] [PubMed] [Google Scholar]
- 31.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci U S A 1996; 93:13931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz G, Nagel I, Siebert R, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med 2007;204:633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramiro AR, Jankovic M, Eisenreich T, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 2004; 118:431–8. [DOI] [PubMed] [Google Scholar]
- 34.Ramiro AR, Jankovic M, Callen E, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 2006;440:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franco S, Gostissa M, Zha S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell 2006;21:201–14. [DOI] [PubMed] [Google Scholar]
- 36.Robbiani DF, Bothmer A, Callen E, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 2008;135:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diflymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A 2008;105: 13520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A 2003; 100:9991–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenwald A, Wright G, Leroy K, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003;198:851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savage KJ, Monti S, Kutok JL, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 2003;102:3871–9. [DOI] [PubMed] [Google Scholar]
- 41.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med 2006;354:2431–42. [DOI] [PubMed] [Google Scholar]
- 42.Lossos IS, Alizadeh AA, Eisen MB, et al. Ongoing immunoglobulin somatic mutation in germinal center B cell-like but not in activated B cell-like diffuse large cell lymphomas. Proc Natl Acad Sci U S A 2000;97:10209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaffer AL, Shapiro-Shelef M, Iwako- shi NN, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 2004;21:81–93. [DOI] [PubMed] [Google Scholar]
- 44.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med 2001;194:1861–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam LT, Davis RE, Pierce J, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res 2005; 11:28–40. [PubMed] [Google Scholar]
- 46.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 2006;107:4090–100. [DOI] [PubMed] [Google Scholar]
- 47.Pasqualucci L, Compagno M, Houlds-worth J, et al. Inactivation of the PRDM1/ BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med 2006;203:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito M, Gao J, Basso K, et al. A signaling pathway mediating downregula- tion of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell 2007;12:280–92. [DOI] [PubMed] [Google Scholar]
- 49.Iqbal J, Greiner TC, Patel K, et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia 2007;21:2332–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell scription factor NF-kappaB. J Clin Invest 2001;107:241–6.11160144 [Google Scholar]
- 51.Pasqualucci L, Guglielmino R, Houlds- worth J, et al. Expression of the AID protein in normal and neoplastic B cells. Blood 2004;104:3318–25. [DOI] [PubMed] [Google Scholar]
- 52.Seifert M, Küppers R. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J Exp Med 2009;206:2659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cattoretti G, Büttner M, Shaknovich R, Kremmer E, Alobeid B, Niedobitek G. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood 2006;107:3967–75. [DOI] [PubMed] [Google Scholar]
- 54.Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009; 114:3533–7. [DOI] [PubMed] [Google Scholar]
- 55.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 2008;9:405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young KH, Leroy K, M0ller MB, et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood 2008;112:3088–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tagawa H, Suguro M, Tsuzuki S, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood 2005;106: 1770–7. [Erratum, Blood 2006;107:3052.] [DOI] [PubMed] [Google Scholar]
- 58.Joos S, Küpper M, Ohl S, et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000; 60:549–52. [PubMed] [Google Scholar]
- 59.Guiter C, Dusanter-Fourt I, Copie- Bergman C, et al. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood 2004;104:543–9. [DOI] [PubMed] [Google Scholar]
- 60.Weniger MA, Melzner I, Menz CK, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006;25:2679–84. [DOI] [PubMed] [Google Scholar]
- 61.Mestre C, Rubio-Moscardo F, Rosenwald A, et al. Homozygous deletion of SOCS1 in primary mediastinal B-cell lymphoma detected by CGH to BAC microarrays. Leukemia 2005;19:1082–4. [DOI] [PubMed] [Google Scholar]
- 62.Melzner I, Bucur AJ, Brüderlein S, et al. Biallelic mutation of SOCS-1 impairs JAK2 degradation and sustains phospho- JAK2 action in the MedB-1 mediastinal lymphoma line. Blood 2005;105:2535–42. [DOI] [PubMed] [Google Scholar]
- 63.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 2009;27:693–733. [DOI] [PubMed] [Google Scholar]
- 64.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the tranfuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–47.12075054 [Google Scholar]
- 65.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lam LT, Wright G, Davis RE, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008;111:3701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding BB, Yu JJ, Yu RY, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 2008;111:1515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ngo VN, Davis RE, Lamy L, et al. A loss- of-function RNA interference screen for molecular targets in cancer. Nature 2006; 441:106–10. [DOI] [PubMed] [Google Scholar]
- 69.Rawlings DJ, Sommer K, Moreno- García ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol 2006;6:799–812. [DOI] [PubMed] [Google Scholar]
- 70.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 2008;319:1676–9. [DOI] [PubMed] [Google Scholar]
- 71.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010;463:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol Immunol 2004;41:599–613. [DOI] [PubMed] [Google Scholar]
- 73.Gauld SB, Cambier JC. Src-family kinases in B-cell development and signaling. Oncogene 2004;23:8001–6. [DOI] [PubMed] [Google Scholar]
- 74.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009;459:717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood 2009; 114:2467–75. [DOI] [PubMed] [Google Scholar]
- 76.Wertz IE, O’Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004;430:694–9. [DOI] [PubMed] [Google Scholar]
- 77.Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009;459:712–6. [DOI] [PubMed] [Google Scholar]
- 78.Schmitz R, Hansmann ML, Bohle V, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 2009;206:981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Novak U, Rinaldi A, Kwee I, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood 2009;113:4918–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell zyme as a new approach to treat cancer. Nature 2009;458:732–6.19360080 [Google Scholar]
- 81.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med 2006;354:2419–30. [DOI] [PubMed] [Google Scholar]
- 82.Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 2004; 350:1828–37. [DOI] [PubMed] [Google Scholar]
- 83.Coiffier B, Lepage E, Briere J, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med 2002;346:235–42. [DOI] [PubMed] [Google Scholar]
- 84.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev 2008;22:559–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Workman P, Burrows F, Neckers L, Rosen N. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci 2007;1113:202–16. [DOI] [PubMed] [Google Scholar]
- 86.Chen G, Cao P, Goeddel DV. TNF- induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol Cell 2002;9:401–10. [DOI] [PubMed] [Google Scholar]
- 87.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating en-Metabolic catastrophe as a means to cancer cell death. J Cell Sci 2007;120:379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hailfinger S, Lenz G, Ngo V, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 2009; 106:19946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferch U, Kloo B, Gewies A, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med 2009;206:2313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortez- omib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 2009;113:6069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen L, Monti S, Juszczynski P, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood 2008;111:2230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Friedberg JW, Sharman J, Sweeten- ham J, et al. Inhibition of Syk with fosta- matinib disodium has significant clinical activity in non Hodgkin’s lymphoma and chronic lymphocytic leukemia. Blood 2009. November 17 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robertson MJ, Kahl BS, Vose JM, et al. Phase II study of enzastaurin, a protein kinase C beta inhibitor, in patients with relapsed or refractory diffuse large B-cell lymphoma. J Clin Oncol 2007;25:1741–6. [DOI] [PubMed] [Google Scholar]
- 94.Jin S, DiPaola RS, Mathew R, White E. lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005;105:1851–61. [DOI] [PubMed] [Google Scholar]
- 95.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005;435:677–81. [DOI] [PubMed] [Google Scholar]
- 96.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apop- totic blocks to predict response to ABT- 737 and conventional chemotherapeutic agents. Cancer Cell 2007;12:171–85. [DOI] [PubMed] [Google Scholar]
- 97.van Delft MF, Wei AH, Mason KD, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 2006;10:389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Polo JM, Dell’Oso T, Ranuncolo SM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med 2004;10:1329–35. [DOI] [PubMed] [Google Scholar]
- 99.Petit I, Jin D, Rafii S. The SDF-1-CX- CR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends Immunol 2007;28:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aikawa T, Gunn J, Spong SM, Klaus SJ, Korc M. Connective tissue growth factor-specific antibody attenuates tumor growth, metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol Cancer Ther 2006;5:1108–16. [DOI] [PubMed] [Google Scholar]