Abstract
DNA topoisomerases are the targets of important anticancer and antibacterial drugs. Camptothecins and novel noncamptothecins in clinical development (indenoisoquinolines and ARC-111) target eukaryotic type IB topoisomerases (Top1), whereas human type IIA topoisomerases (Top2α and Top2β) are the targets of the widely used anticancer agents etoposide, anthracyclines (doxorubicin, daunorubicin), and mitoxantrone. Bacterial type II topoisomerases (gyrase and Topo IV) are the targets of quinolones and aminocoumarin antibiotics. This review focuses on the molecular and biochemical characteristics of topoisomerases and their inhibitors. We also discuss the common mechanism of action of topoisomerase poisons by interfacial inhibition and trapping of topoisomerase cleavage complexes.
Topological Constraints of DNA
DNA strand separation for transcription and replication, the flawless segregation of two identical copies of entire genomes in two daughter cells following replication, and the formidable genomic compaction in cells exemplify the critical actions of DNA topoisomerases (Wang, 2009b).
DNA compaction is such that the entire genome of a single human cell (3 × 109 base pairs corresponding to approximately 1.8 m) needs to be squeezed into a nucleus with an average diameter of 6 μm. In other words, the circumference of an average mammalian cell nucleus is almost one million times smaller than the length of the genome that needs to be packed into it. Even the smaller circular Escherichia coli genome (4.7 × 106 base pairs) needs to be compacted within a bacterial cell whose average circumference is 3000 times smaller. To maintain DNA compacted, topoisomerases are required to avoid superhelical tension and knots.
DNA strand separation is obligatory to transcribe and replicate genomes by copying each base by RNA and DNA polymerases. Because of the DNA double-helical structure and lack of free rotation in cells, strand separation generates DNA supercoiling in the flanking regions where the two strands are separated by the polymerase-helicase complexes. As a result, positive supercoiling is generated in front of the replication or transcription sites and negative supercoiling behind (Liu and Wang, 1987). Without the action of DNA topoisomerases, positive supercoiling rapidly stalls replication and transcription and negative supercoiling favors the formation of abnormal DNA structures including D loops (invasion of a DNA duplex by a complementary single-stranded DNA segment), R loops (persistent annealing of RNA with its DNA template behind RNA polymerase), guanosine quartets, and Z-DNA, all of which interfere with normal DNA metabolism.
Replicating the whole genome needs to be accomplished within one cell cycle (i.e., by multiple replication forks with an average velocity of 2 kilobases per minute per fork in a human cell). Because of DNA’s double-helical structure, replication generates catenated progenies that have to be unlinked by topoisomerases prior to cytokinesis. In addition, in circular bacterial or viral genomes, replication forms obligatory catenanes (this can easily be realized by cutting a Mobius strip along its longitudinal axis).
Classification and Nomenclature
The human genome encodes six topoisomerases and E. coli four (Figure 1A). Topoisomerases are classified as type I and type II (Forterre et al., 2007; Schoeffler and Berger, 2008; Wang, 2009b). Type I enzymes cleave one DNA strand at a time and type II both strands to perform their catalytic functions (Figures 1C–1H). All topoisomerases cleave the DNA phosphodiester backbone by nucleophilic attack from a catalytic tyrosine residue which becomes linked to the phosphate end (P-Y) of the DNA break (Figures 1C–1H). Those reactions are highly reversible and leave the DNA sequence unchanged following topoisomerization.
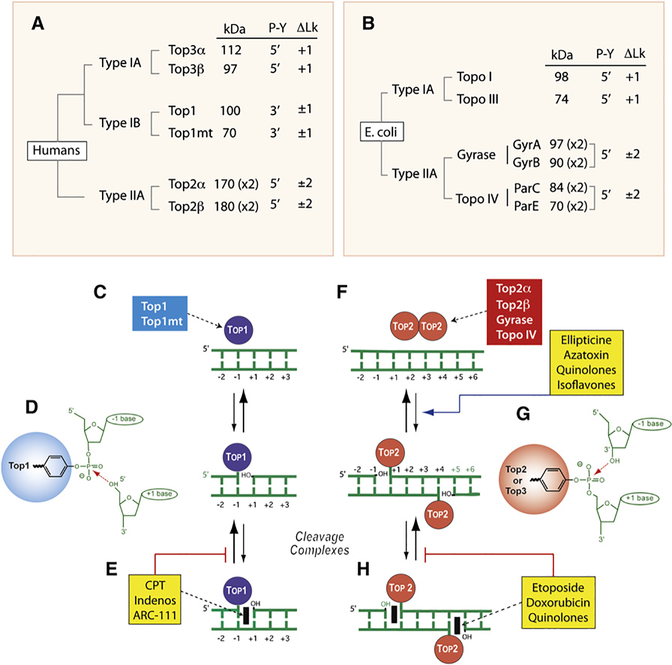
Figure 1. Overview of DNA Topoisomerases.
(A) Classification of human DNA topoisomerases. kDa: molecular masses calculated from polypeptide composition. (x2): dimer. Type IB are the only enzymes that form cleavage complexes (cc) with 3′-phosphotyrosyl (3′-P-Y) intermediates. ΔLk: linking number change produced by each catalytic cycle. Type I enzymes change Lk in steps of one and type II in steps of two as they cleave one and both strands of DNA, respectively. Type IA enzymes are the only enzymes that relax only negative but not positive supercoiling (ΔLk = +1).
(B) Classification of E. coli DNA topoisomerases.
(C) Noncovalent binding of type IB enzymes.
(D) Scheme of the 3′-phosphotyrosine covalent bond in the Top1cc. The arrow indicates the reversible (religation) reaction, which is favored under normal conditions.
(E) Trapping of the cleavage complex by camptothecin (CPT) and the Top1 inhibitors (see Figure 3).
(F) Noncovalent binding of type IIA enzyme homodimers (A2B2 and C2E2 in the case of gyrase and Topo IV, respectively).
(G) Scheme of the 5′-phosphotyrosine covalent bond in the Top2cc. The arrow indicates the reversible (religation) reaction, which is favored under normal conditions. Note the four base pair stagger with 5′-overhang on opposite strands characteristic of all type IIA enzymes.
(H) Trapping of the cleavage complex by etoposide, doxorubicin, or quinolones (see Figure 3). Note that some Top2 poisons increase the steady-state levels of Top2cc by increasing cleavage (ellipticines, azatoxin, quinolones, and isoflavones).
Type I topoisomerases are further subdivided into type IA and IB. E. coli type IA topoisomerase (Figure 1B), the first topoisomerase discovered, was initially named “ω protein” because it was purified by ultracentrifugation that uses ω as a parameter of angular velocity (Wang, 1971). Eukaryotic Top1 (Figure 1A) (Champoux and Dulbecco, 1972) is classified as type IB because of two key differences with E. coli Topo I. First, it relaxes both negative and positive supercoils, whereas E. coli Topo I only relaxes negative supercoils. Second, it cleaves DNA by forming a tyrosyl-DNA covalent catalytic intermediate at the 3′ end of the break (3′-P-Y; Figure 1D), whereas E. coli Topo I forms a 5′-P-Y intermediate. Eukaryotic type IA, named Top3, was identified through genomic homology search (Hanai et al., 1996). Vertebrates encode two Top3 enzymes (Top3α and β). Both form 5′-P-Y covalent intermediates and only relax negatively supercoiled DNA (only increasing the DNA linking number; Lk) (Figure 1A). The last discovered human topoisomerase is Top1mt, a type IB enzyme encoded in the nuclear genome but which operates on mitochondrial DNA (Zhang et al., 2001). In vertebrates, only Top1 and Top1mt form covalent linkages to the 3′ end of the breaks (Figure 1A). All the other topoisomerases form 5′-phosphotyrosyl covalent bonds. In that respect, Top1 and Top1mt (type IB eukaryotic topoisomerases) belong to the family of tyrosine recombinases which also includes XerCD of E. coli, Cre of the P1 phage, Flip (FLP) of yeast Saccharomyces cerevisiae, and λ integrase of lambda phage. However, no recombinase function has been assigned to Top1 in cells.
Bacteria tend to have a simpler organization than vertebrates, with only four topoisomerases in E. coli (Figure 1B). However, bacteria have two type IA enzymes (Topo I and Topo III) and two type IIA enzymes (gyrase and Topo IV).
Functional Size of Topoisomerases
Topoisomerases are large proteins (Figures 1A and 1B). The functional sizes of the bacterial and eukaryotic enzyme complexes range from 70 kDa (for Top1mt and bacterial Topo I) to 370–400 kDa (for the type II enzymes that function as multimers) (Figures 1F–1H). In comparison, the size of a nucleosome is ∼120 kDa without histone H1 and 180 kDa with H1 (i.e., more than 2-fold smaller).
Note regarding the following sections: we will not discuss further the type IA topoisomerases (bacterial Topo I and human Top3) and type IIB topoisomerases because they are not yet clinical therapeutic targets. Further information regarding type IA and IIB enzymes and the targeting of bacterial type IA enzymes can be found in several detailed reviews (Schoeffler and Berger, 2008; Tse-Dinh, 2009; Wang, 2002, 2009a).
Type IB Topoisomerases and Their Targeting by Anticancer Drugs
Eukaryotic Nuclear Top1
All eukaryotes encode at least one type IB topoisomerase gene (TOP1). TOP1 is essential in Drosophila (Lee et al., 1993) and vertebrates but not in yeast (Goto and Wang, 1985; Uemura and Yanagida, 1984). Mouse embryos lacking TOP1 die early before the fertilized egg gets to the eight-cell stage (Morham et al., 1996). Only one murine cell line (p388/CPT45) selected for resistance to camptothecin (CPT) grows with almost undetectable levels of Top1 (Eng et al., 1990; Tuduri et al., 2009), and the two human cell lines recently generated by stable Top1 knockdown express 10%–20% of normal Top1 levels (Miao et al., 2007). Those cells accumulate spontaneous genomic alterations and replication defects (Miao et al., 2007; Tuduri et al., 2009). They also exhibit altered gene expression (Miao et al., 2007), reflecting the critical importance of Top1 nicking-closing activity via formation of Top1-DNA cleavage complexes (Top1cc) (Figure 2C) for DNA relaxation during replication and transcription. Human Top1 also has transcriptional activities independent of DNA nicking-closing activity: promoter regulation (Merino et al., 1993) and phosphorylation/activation of splicing factors (Rossi et al., 1998; Soret et al., 2003).
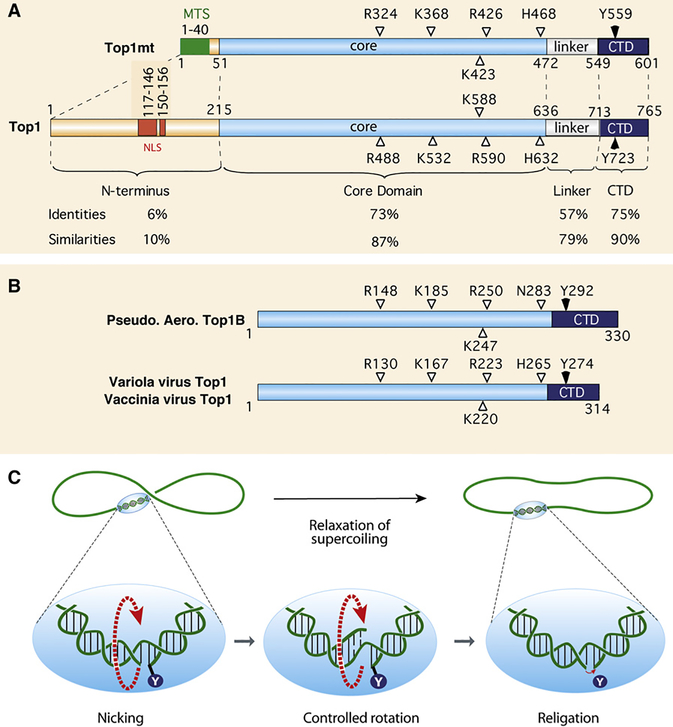
Figure 2. Overview of Type IB Topoisomerases and Relaxation by Nicking-Closing Activity.
(A) Schematic representation of the two human type IB enzymes. Top1mt has a much shorter N-terminal segment consisting primarily of a mitochondrial targeting sequence (MTS) and lacking the nuclear localization sequences (NLS) of Top1 (nuclear). The catalytic homologous residues are indicated with their position.
(B) Schematic representation of the poxvirus (variola and vaccinia) Top1s and comparison with the recently discovered bacterial Top1B (Pseudomonas aeroginosa).
(C) Top1-mediated DNA relaxation by controlled rotation. By contrast to type IA or II enzymes, this reaction does not require an energy cofactor or divalent metal. Top1 tends to bind DNA crossovers (supercoils) and nicks DNA by transesterification (see Figure 1D). The enzyme then allows the DNA to swivel by controlled rotation (Koster et al., 2005; Stivers et al., 1997). Upon DNA realignment by base pairing and stacking across the nick, the DNA 5′-hydroxyl end (OH in lower panels) removes the tyrosyl linkage by reverse transesterification (see Figure 1D).
Vertebrate Mitochondrial Top1
TOP1mt is present in the nuclear genome of vertebrates (Zhang et al., 2004, 2007) (Figure 2A). Similarities between TOP1mt and TOP1 (Figure 2A) suggest they probably originated from the duplication of an ancestral gene still present in chordates (Zhang et al., 2004). Top1mt binds to the region flanking the end of the replication D loop at the putative attachment site of nucleoids to the mitochondrial inner membrane (Zhang and Pommier, 2008). In spite of its conservation in all vertebrates, TOP1mt is dispensable in mice (Zhang et al., 2007), indicating complementation by another topoisomerase (Low et al., 2003; Zhang et al., 2007). Top1mt is sensitive to CPT (Zhang et al., 2001; Zhang and Pommier, 2008) but is unlikely to be targeted by the drug in cells because the mitochondria matrix is alkaline (Zhang et al., 2001), which inactivates CPT (see Figure 3 and section below).
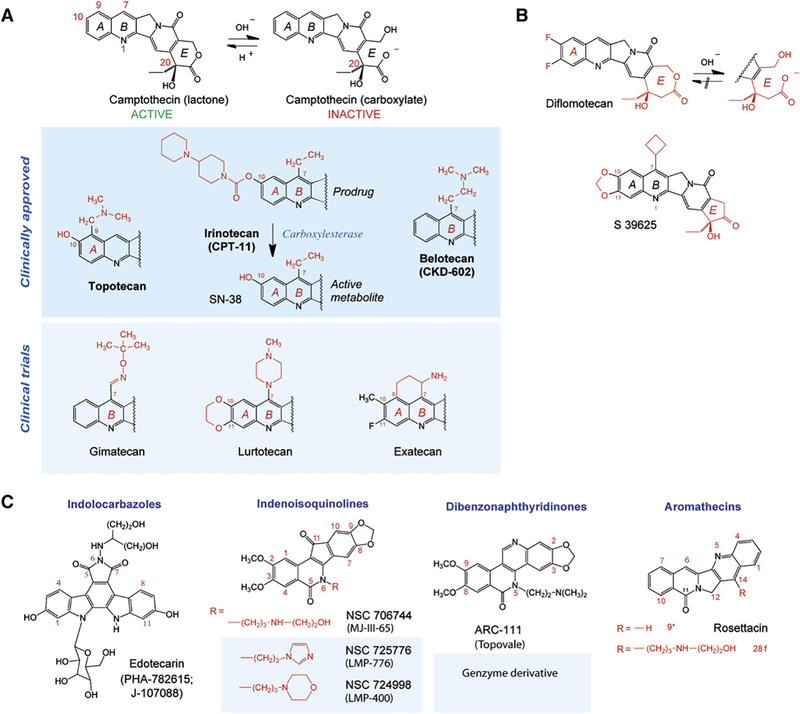
Figure 3. Top1 Inhibitors.
(A) Camptothecin and its clinical derivatives. The facile and reversible opening of the α-hydroxylactone E ring of camptothecin is shown at the top. Topotecan and irinotecan are the two FDA-approved camptothecins. Irinotecan is a prodrug; its active metabolite is SN-38. Belotecan is approved in South Korea.
(B) Synthetic E-ring-modified camptothecin derivatives. Whereas diflomotecan (a homocamptothecin) can still be converted irreversibly to a carboxylate, the α-keto derivative S39625 is chemically stable while still being a potent Top1 inhibitor.
(C) Noncamptothecins. The indenoisoquinolines and one dibenzonaphthyridinone are beginning clinical trials.
Viral and Bacterial Type IB Enzymes
Poxviruses (vaccinia and variola) encode one topoisomerase, which is the smallest topoisomerase (314 residues; ∼37 kDa) (Figure 2B). Vaccinia and variola Top1s are similar except for three amino acid residues. They contain the conserved catalytic residues R, K, R, H found in the core domain of eukaryotic Top1 and Top1mt, but lack the linker region (Figure 2A). The poxvirus Top1s are required because the host Top1 is restricted to the nucleus and poxviruses replicate in the cytoplasm. Poxvirus Top1s cleave DNA with greater sequence selectivity (at CCTTT recognition sequences; Shuman, 1998) than eukaryotic Top1s and are immune to camptothecins. Vaccinia Top1 acts as a very efficient recombinase and is used as a cloning tool (TOPO-TA Cloning; Invitrogen) (Shuman, 1989). Poxvirus Top1s are potential weapons in the context of bioterrorism.
Type IB genes have been found in some bacteria including Pseudomonas aeroginosa and the radiation-resistant Deinococcus radiodurans. They are structurally related to the poxvirus Top1s, suggesting they arose from a common ancestor strand transferase with tyrosine recombinases (Krogh and Shuman, 2002). Whether the bacterial (and viral) type IB topoisomerases act as recombinases and/or DNA-relaxing enzymes remain to be determined. No effective inhibitor exists, although they may represent novel antibacterial targets.
Anticancer Top1 Inhibitors
CPT was first identified from the Chinese tree Camptotheca acuminata (Wall et al., 1966; Wall and Wani, 1995). It poisons Top1cc by reversibly inhibiting its religation (Hsiang et al., 1985) (Figure 1E). Four lines of evidence demonstrate the selective poisoning of Top1 by CPT (reviewed in Pommier, 2006, 2009): (1) only the natural CPT isomer is active against Top1 (Hsiang et al., 1989; Jaxel et al., 1989); (2) genetically modified yeast deleted for TOP1 (Top1Δ) is immune to CPT (Eng et al., 1988); (3) cells selected for CPT resistance harbor precise and causal point mutations in the TOP1 gene (Pommier et al., 1999); and (4) CPT-producing plants bear a point mutation in Top1 (N722S) (Sirikantaramas et al., 2008) that also renders the enzyme immune to CPT (Fujimori et al., 1995).
Three water-soluble CPT derivatives are approved for clinical use: topotecan, irinotecan (worldwide), and belotecan (in South Korea) (Figure 3A). In spite of their established anticancer activity, camptothecins have a major limitation. They are inactivated within minutes at physiological pH by lactone E ring opening (Figure 3A). Derivatives (gimatecan, belotecan, lurtotecan, and exatecan) (Figure 3A) have been developed to improve solubility and clinical tolerability and allow oral administration. However, they retain the chemical instability of camptothecins.
Two approaches have been taken to overcome the E ring instability of camptothecins. Addition of a methylene group in the E ring, as in the homocamptothecins (Figure 3B), limits E ring opening (Lavergne et al., 1998) but, once this happens, they become irreversibly converted to an inactive carboxylate (Urasaki et al., 2000). In the second approach, conversion of the E ring to a five-membered ring completely stabilizes the drug. These α-keto derivatives (Hautefaye et al., 2003) exemplified by S39625 (Figure 3C) (Takagi et al., 2007) are highly potent synthetic compounds against Top1 and in cancer cells.
The first noncamptothecin Top1 inhibitor (Figure 3C) reaching clinical trials was edotecarin. However, like other indolocarbazoles, it affects other cellular targets besides Top1, including DNA itself. Two families of noncamptothecin Top1 inhibitors are beginning clinical development (Pommier and Cushman, 2009; Teicher, 2008): the indenoisoquinolines and the dibenzonaphthyridinones (Figure 3C). Both are synthetic and chemically stable, thereby overcoming the E-ring-opening inactivation of camptothecins. They also overcome the drug efflux-associated resistance of camptothecins and produce more stable cleavage complexes at different genomic locations compared to camptothecins (Pommier and Cushman, 2009; Teicher, 2008). The aromathecins (Figure 3C) are at an earlier stage of development (Cinelli et al., 2009).
Determinants of the Anticancer Activity of Top1 Inhibitors: Importance of Repair and Checkpoint Defects
In spite of the fact that Top1 is the sole target of camptothecins, the molecular determinants of their anticancer activity are complex. Top1 is required for both normal and cancer cells, and intrinsic defects in DNA repair and checkpoints, which are the landmarks of cancer cells, are likely the Achilles’ heels of cancer cells treated with anti-Top1 drugs (Pommier, 2006; Pommier et al., 2006a).
Drug-stabilized Top1cc are converted into DNA damage by two main processes: DNA replication and transcription. Collision of DNA or RNA polymerase complexes into stalled Top1cc Disrupts the architecture of the Top1cc. The double-strand end generated by the stalled polymerase cannot reverse the tyrosine-phosphodiester bond (see reverse transesterification reaction in Figure 1D) and thereby generates an irreversible Top1-DNA crosslink associated with a double-strand end. The repair of such lesions is only partially understood. At least two steps need to take place: removal of the Top1 from the 3′-DNA end and relegation of the free 5′ end.
The best-characterized pathway for the removal of Top1-DNA adducts implicates tyrosyl-DNA phosphodiesterase 1 (Tdp1) (Yang et al., 1996) (reviewed in Dexheimer et al., 2008). Additional cellular processes are required in association with Tdp1. Top1 needs to be proteolyzed by the 26S proteasome (Desai et al., 2001; Lin et al., 2009) before Tdp1 can efficiently process the tyrosyl-DNA adduct (Debethune et al., 2002; Interthal et al., 2005; Pouliot et al., 1999). Polynucleotide kinase phosphatase (PNK) has to remove the 3′-phosphate left by Tdp1 to allow the actions of DNA polymerases and ligases (Pommier et al., 2006a).
Studies in yeast have also demonstrated alternative endonucleolytic repair pathways for removing Top1-DNA complexes. Rad1XPF/Rad10ERCC1, Mus81/Mms4Emi1, Mre11/Xrs2Nbs1, and Slx4/Slx1 (reviewed in Pommier et al., 2006a) (human orthologs in superscripts) can catalyze the endonucleolytic cleavage of DNA immediately upstream from the Top1-DNA adduct.
A third possibility for the reversal of the trapped Top1cc associated with replication fork collisions is by backtracking the DNA polymerase complex together with the newly replicated DNA. The Bloom (BLM) helicase has been implicated in such a process by allowing the reannealing and religation of the cleavage complex (Pommier et al., 2006a). Consequently, BLM together with Top3α can resolve the associated double-Holliday junction (Wu and Hickson, 2006).
One approach to augment the selectivity of Top1 poisons toward cancer cells is to combine them with Tdp1 inhibitors. The rationale is derived from yeast experiments showing that knocking out Tdp1 has minimal impact on the toxicity of CPT unless the cell is also deficient for Rad9, a key checkpoint gene in yeast encoding a BRCT-containing protein (Pouliot et al., 2001). Because cancer cells are commonly defective for BRCA1, one of the human orthologs of Rad9, Tdp1 inhibitors are expected to selectively enhance the activity of Top1 inhibitors in cancer cells, which are checkpoint defective, whereas having minimal impact on normal cells (Dexheimer et al., 2008).
Another approach to increase the selectivity of Top1 inhibitors toward cancer cells is to combine them with checkpoint kinase inhibitors. The proof of principle for this combination was initially established with UCN-01 (7-hydroxystaurosporine), which shows a remarkable synergy with camptothecin in p53-deficient cells (Shao et al., 1997). The ongoing development of specific Chk1 and Chk2 inhibitors should provide promising combinations with Top1 inhibitors (Lapenna and Giordano, 2009; Pommier et al., 2006b).
Several mechanisms are commonly associated with cellular resistance to camptothecins: drug efflux pump overexpression (Brangi et al., 1999), reduction of Top1 protein levels (Eng et al., 1990; Fujimori et al., 1995), and Top1 mutations (Pommier et al., 1999). Multifactorial resistance is likely in human cancers, underlining the importance of a multiparametric approach for the rational use of Top1 inhibitors.
Eukaryotic Top2 Enzymes and Their Targeting by Anticancer Drugs
Eukaryotic Top2 Enzymes
Both Top1 and Top2 can remove DNA supercoiling. In yeast, inactivation of Top1 is compensated by the other topoisomerases (Goto and Wang, 1985; Uemura et al., 1987). However, Top2-deficient yeast strains die at mitosis because Top2 is essential for chromosome condensation and segregation (Goto and Wang, 1985). This is because only Top2 can separate interlinked duplex DNA circles (catenanes) (see next section). In all cells, decatenation is essential at the end of replication to enable the segregation of newly replicated chromosomes.
Mammals (but not yeast or Drosophila) have two isoenzymes, Top2α and β (Figures 1A and 4A). Top2α is tightly linked to cell proliferation, whereas Top2β is also expressed in nondividing cells. Top2α increases 2- to 3-fold during G2/M and is orders of magnitude higher in rapidly proliferating than in quiescent cells. Top2α relaxes positively supercoiled DNA more efficiently than negatively supercoiled DNA. In contrast, Top2β acts on both similarly (McClendon et al., 2005). Knocking out Top2β is compatible with embryonic development, but Top2β−/− mice die at birth from neurological defects probably related to the critical role of Top2β in transcription (Ju et al., 2006; Perillo et al., 2008).
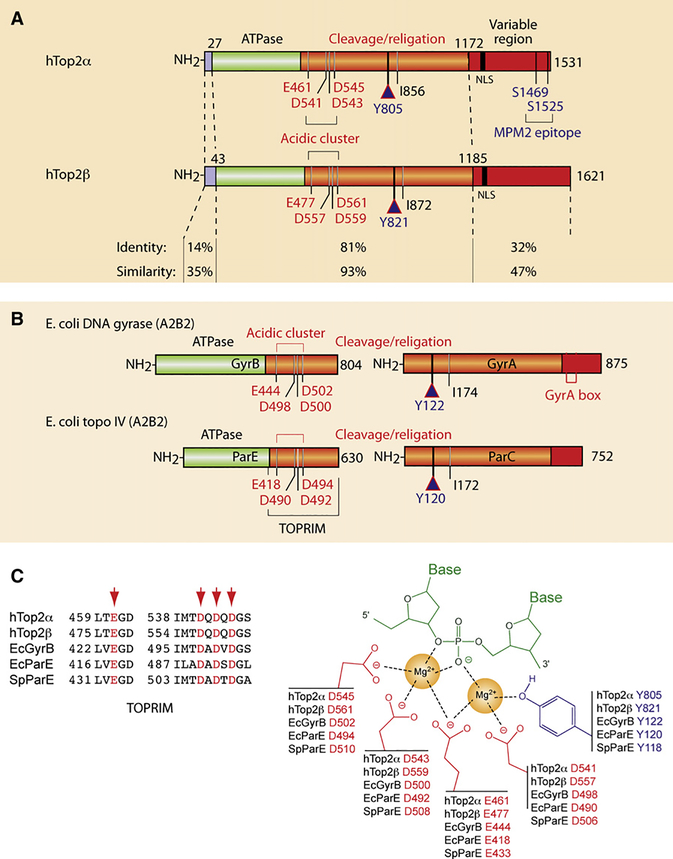
Figure 4. Structural Overview of the Type IIA Topoisomerases Targeted by Anticancer and Antibacterial Drugs.
(A) Schematic representation of the human Top2 enzymes Top2α and β. Note the high similarity of their ATPase and cleavage/religation domains, whereas their N- and C-terminal regions are more divergent. Conserved catalytic residues are indicated including the catalytic tyrosines Y805 and Y821 in Top2α and β, respectively.
(B) Schematic representation of the bacterial Top2 enzymes gyrase and Topo IV. Gyrase is a heterotetramer encoded by two genes: GyrA and GyrB (A2B2; see Figure 1B). Topo IV is also a heterotetramer encoded by two genes: ParC and ParE (C2E2; see Figure 1B). Conserved catalytic residues are indicated including the catalytic tyrosines Y122 andY120 ingyrase and Topo IV, respectively.
(C) Conserved interaction of type IIA enzymes with DNA by their TOPRIM motifs (alignment at left). The right scheme represents the two-metal-binding model between the TOPRIM motif and DNA (modified from Deweese et al., 2008).
Top2 Catalytic Cycle
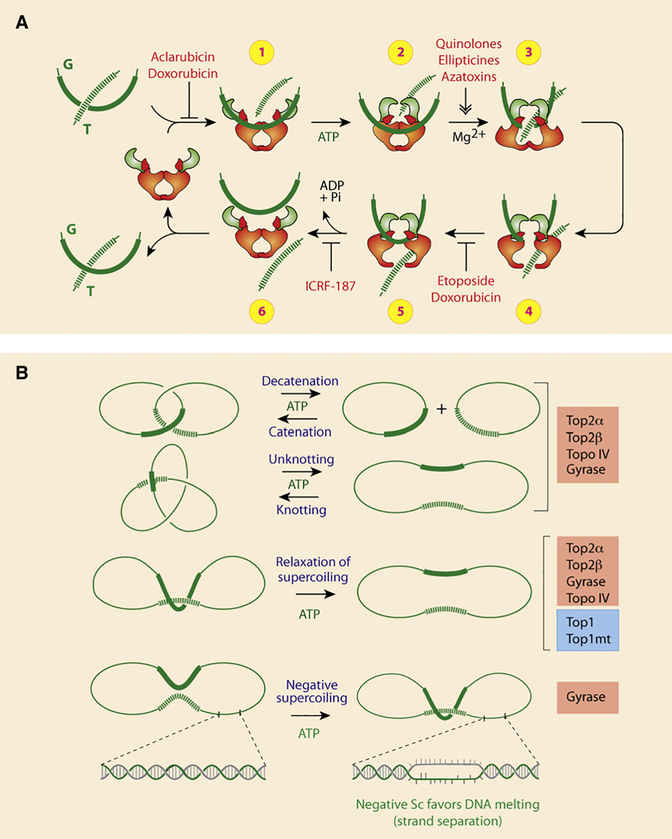
Top2α and β function as homodimers and require Mg2+ (Figures 4C and 5A) (for more details, see Deweese et al., 2009; Nitiss, 2009b; Schoeffler and Berger, 2008; Wang, 2009a). Like Top1, Top2 exhibits a relatively “relaxed” sequence selectivity, allowing its action at multiple sites of the genome. The enzyme dimer, however, preferentially binds DNA crossover regions (DNA supercoils, knots, catenanes) (Figure 5B). The two DNA segments are referred to as the G and T segments. The G (gate) segment is cleaved by the enzyme in order to pass the T (transported) segment through the enzyme-DNA complex (Figure 5A). Upon ATP binding, Top2 undergoes a conformational change from an open to a closed clamp form (Figure 5A, step 2). In the presence of Mg2+, the tyrosine from each Top2 monomer (tyrosine 804 for human Top2α or tyrosine 821 for Top2β; Figure 4A) attacks a DNA phosphodiester bond four bases apart on opposite strands of the G duplex and becomes covalently linked to the 5′ ends of the broken DNA (see Figure 1E). The T segment can then pass through the broken G segment (steps 3 and 4). Under normal conditions, the cleavage complex is short lived. After strand passage, the T segment is released from the clamp and the broken ends of the G segment are religated within the Top2 homodimer complex (steps 5 and 6). ATP hydrolysis converts the complex back to its open clamp form with release of the G segment (step 6). Thus, closing and opening of the Top2 clamp are coupled with ATP binding and hydrolysis.
Figure 5. Catalytic Cycle and Reactions Carried Out by Type IIA Topoisomerases.
(A) Schematic representation of the catalytic cycle for human Top2 enzymes (see text for details). Note that doxorubicin can block the catalytic cycle at two different steps. At low concentration (<1 μM), doxorubicin acts like etoposide (VP-16) by blocking DNA religation (between steps 4 and 5). At higher concentrations (>10 μM), doxorubicin acts like aclarubicin by interfering with Top2 binding to DNA (before step 1). ICRF-187 blocks ATP hydrolysis and inhibits reopening of the ATPase domain (between steps 5 and 6), thereby trapping topological complexes with DNA inside the enzyme.
(B) Reactions catalyzed by type IIA topoisomerase. Note that Top1 can only catalyze supercoiling relaxation. Also, in bacteria, Topo IV acts preferentially as the decatenation enzyme, whereas gyrase acts preferentially in removing positive supercoiling and selectively in generating negative supercoiling.
Through its ability to catalyze strand passage, Top2 can perform a variety of reactions. DNA relaxation is common to Top1, whereas catenation-decatenation, and knotting-unknotting, are Top2 specific (Figure 5B).
Anticancer Top2 Poisons and Top2 Inhibitors
Drugs (Figure 6A) poison Top2 cleavage complexes (Top2cc) by two mechanisms (Figures 1H and 5A). Etoposide (VP-16), teniposide (VM-26), and the DNA intercalators doxorubicin, daunorubicin, amsacrine (m-AMSA), and TAS-103 inhibit DNA religation, whereas the quinolone CP-115,953, the ellipticines, azatoxins, and the natural flavonoid genistein (Figure 6A) enhance the formation of Top2cc (Figure 5A) (Fortune and Osheroff, 2000).
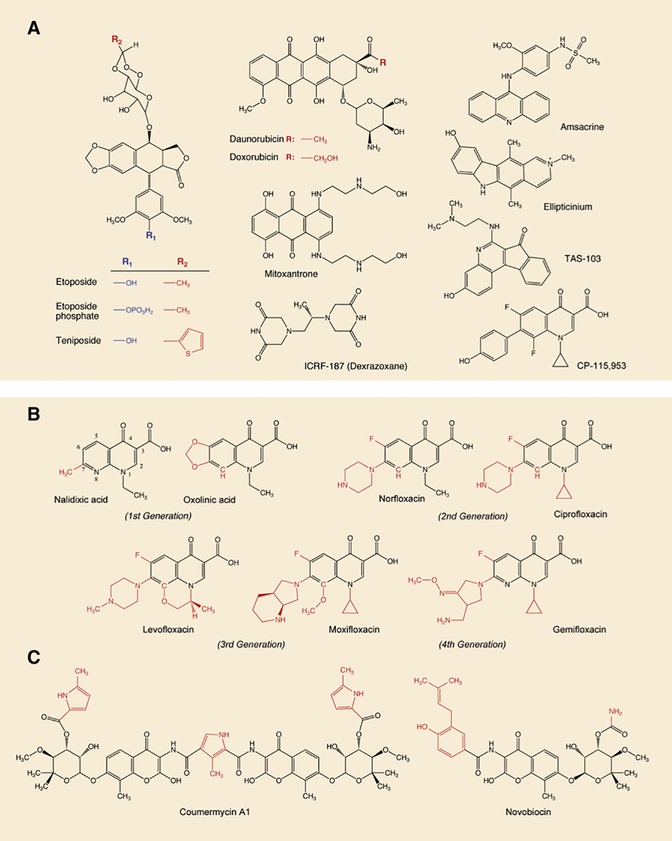
Figure 6. Inhibitors of Type IIA Topoisomerases.
(A) Inhibitors of eukaryotic Top2 enzymes. All drugs except ICRF-187 trap Top2 cleavage complexes and generate DNA breaks (see Figures 1F–1H). ICRF-187 is a catalytic inhibitor.
(B) Gyrase and Topo IV inhibitors from the quinolone family. All drugs trap cleavage complexes. Note that the quinolone CP-115,953 traps both eukaryotic and prokaryotic enzymes, which is not the case in the other quinolones shown in this panel. Also note that TAS-103 is a dual inhibitor of Top1 and Top2 (Wilson Byl et al., 1999).
(C) Gyrase catalytic inhibitors.
Inhibition of Top2 catalytic activity without trapping cleavage complexes is also observed for DNA intercalators at drug concentrations that alter DNA structure, thereby preventing Top2 from binding DNA (step 1 in Figure 5A) or from forming Top2cc (step 3) (Pommier et al., 1984; Tewey et al., 1984). This explains why doxorubicin poisons Top2cc at low drug concentrations but suppresses Top2cc at higher concentrations. Non-DNA binders, such as merbarone and bisdioxopiperazines (ICRF 159, 187 [dexrazoxane], and 193) (Figure 6A), also act by inhibiting ATP hydrolysis that produces “closed clamps” (steps 5 and 6 in Figure 5A) without trapping Top2cc.
Top2 poisons differ from each other. As mentioned above, some act by inhibiting the religation of Top2cc (VP-16, VM-26, anthracyclines, amsacrine), whereas others induce their formation (ellipticines, isoflavones, azatoxins, and the quinolone CP-115,953) (Fortune and Osheroff, 2000). The kinetics of formation and religation of cellular Top2cc vary from slow in the case of doxorubicin to very rapid in the case of VP-16, m-AMSA, and ellipticinium (Long et al., 1985; Zwelling et al., 1981). Drugs not only poison Top2-mediated DNA double-strand breaks (DSB) but also single-strand breaks (SSB). Ellipticines produce almost exclusively DSB, whereas VP-16 and amsacrine produce 7–20 SSB per DSB and anthracyclines an equal number of SSB and DSB (Long et al., 1985; Pommier et al., 1984; Zwelling et al., 1981). The genomic localization of the Top2cc varies among drug classes (Capranico et al., 1990; Capranico and Binaschi, 1998; Fortune and Osheroff, 2000; Pommier et al., 1991). Adenine at position −1 (see Figure 1F) tends to be preferred for doxorubicin; cytosine −1 for VP-16, VM-26, mitoxantrone, amonafide, and quinolones; thymine −1 for ellipticines and genistein; adenine +1 for amsacrine; and guanine +1 for saintopin. Finally, some inhibitors have additional targets besides Top2. The anthracyclines (doxorubicin, daunorubicin, and their derivatives) can affect a broad range of DNA processes by intercalation and undergo redox reactions that generate reactive oxygen species (ROS), which have been implicated in their dose-limiting cardiotoxicity.
Determinants of the Anticancer Activity of Top2 Poisons
Drug-induced Top2cc are not sufficient to fully account for the anticancer activity of Top2 poisons, as both normal and cancer cells express Top2. In contrast to camptothecin, DNA synthesis inhibition provides only partial protection against VP-16 (Holm et al., 1989). Interferences between the trapped Top2cc and transcription may play a prominent role, as the cytotoxicity of VP-16 is decreased by RNA synthesis inhibitors (D’Arpa et al., 1990). The dependence of Top1 and Top2 inhibitors on ongoing replication and transcription probably explains why simultaneous treatment with camptothecin and VP-16 is antagonistic (Bertrand et al., 1992; D’Arpa et al., 1990; Kaufmann, 1991). VP-16 may antagonize camptothecin by inhibiting replication, and camptothecin may suppress the effects of VP-16 by inhibiting transcription. The involvement of ROS (Pommier et al., 1983) remains unexplained, albeit reminiscent of bacterial killing by quinolones (Drlica et al., 2009).
The repair of Top2cc is poorly understood. As in the case of Top1, proteosomal degradation of the covalently linked Top2 has been proposed to enable access of DNA repair enzymes to the broken DNA (Zhang et al., 2006). Recently, a 5′-tyrosine phosphodiesterase (TTRAP/Tdp2) has been identified for the excision of Top2-DNA adducts (Cortes Ledesma et al., 2009). In addition, as in the case of Top1, endonucleolytic cleavage may involve the removal of the trapped Top2 by cleavage of the DNA near the point of attachment of Top2 to the 5′-DNA end. The 5′-flap endonuclease Rad27FEN1 and the end-processing nucleases Mre11 and CtIP have been proposed to act in this way (Malik and Nitiss, 2004; Nitiss, 2009b).
The cellular response to Top2 inhibitors is complex, and several mechanisms are commonly associated with cellular resistance to Top2 inhibitors: Pgp and/or MRP overexpression, reduction of Top2 protein levels, changes in subcellular localization, Top2 phosphorylation, and Top2 mutations (Nitiss, 2009a, 2009b).
Secondary Malignancies Induced by Top2 Poisons: Implication of Top2β
Etoposide induces treatment-related acute myelocytic leukemia (t-AML) and treatment-related myelodysplastic syndromes (t-MDS), which often progress to t-AML (Pedersen-Bjergaard et al., 2002). Those t-AML and t-MDS are characterized by balanced translocations involving chromosome bands 11q23 or 21q22 (Ratain and Rowley, 1992). The 11q23 pathway involves activation of the MLL gene following its translocation and has been directly related to the trapping of Top2cc in the MLL gene (Lovett et al., 2001). The exact molecular mechanisms by which the Top2cc induce the translocations remain to be determined (Azarova et al., 2007; Felix et al., 2006). Top2 subunit dissociation and exchanges have been invoked (Pommier et al., 1988). However, the translocation mechanisms are likely to be more complex. The carcinogenicity of natural isoflavones has been related to the mutagenicity of Top2cc (Deweese and Osheroff, 2009; Strick et al., 2000).
Recent studies in mice lacking Top2β suggest that etoposide-induced carcinogenic lesions mainly involve Top2β (Azarova et al., 2007). The development of Top2α-specific drugs is therefore a legitimate and novel avenue for novel inhibitors with reduced risk of secondary malignancies.
Bacterial Type II Topoisomerases: Gyrase and Topo IV Inhibitors Used as Antibacterials
Bacterial Type II Enzymes
DNA gyrase was identified in extracts from E. coli in 1976 by Gellert, Mizuuchi, Nash, and coworkers during studies on the integrative recombination of phage λ (Gellert et al., 1976a). It was named gyrase because the enzyme was able to negatively supercoil DNA in the presence of ATP. Gyrase’s enzymatic activity is essential for the regulation of DNA superhelicity, transcription initiation and elongation, and bacterial replication. Gyrase functions as a heterotetramer composed of two GyrA subunits (97 kDa, 875 aa) and two GyrB subunits (90 kDa, 804 aa) (see Figures 1B, 4B, and 4C).
The second bacterial type II enzyme is Topo IV. It was discovered in 1990 in a cytological screen for temperature-sensitive mutations conferring a partition phenotype (hence the name “par” for the two subunits) characterized by chromosomes that can be replicated but not partitioned, resulting in the accumulation of large nucleoids in the middle of filamentous cells. This phenotype established Topo IV as the primary decatenase in the cell (Kato et al., 1990). Like gyrase, Topo IV functions as a heterotetramer composed of two ParC (84 kDa, 752 aa) and two ParE (70 kDa, 630 aa) subunits (see Figures 1B, 4B, and 4C). However, unlike gyrase, Topo IV cannot generate negative supercoiling.
Both gyrase and Topo IV are capable of removing negative or positive supercoiling (Figure 5) (Wang, 2009a, 2009b). However, Topo IV preferentially binds and relaxes positive supercoils (Crisona et al., 2000; Stone et al., 2003). The two enzymes, although closely related with respect to amino acid sequence (approximately 40% sequence identity and a much higher level of homology; see Figure 4B), differ strikingly in their ability to catalyze inter- versus intramolecular strand passage reactions. Topo IV has higher specificity for intermolecular (decatenation) reactions, whereas gyrase favors intramolecular (supercoiling) reactions.
Antibacterial Topoisomerase Inhibitors
Bacterial type II topoisomerases are good targets for antibacterial chemotherapy for the following reasons: (1) they are essential in all bacteria for replication and cell division; (2) an accumulation of cleavage complexes has bactericidal (not just bacteriostatic) effect; (3) targeting bacterial type II topoisomerases is not poisonous for human enzymes (antibacterial agents show a level of specificity/selectivity for prokaryotic enzymes at least three orders of magnitude higher than for eukaryotic enzymes); and (4) due to the high degree of homology between gyrase and Topo IV, antibacterial topoisomerase inhibitors tend to target both enzymes.
Quinolones are synthetic drugs based on the 4-oxo-1,4-dihydroquinolone skeleton (Figure 6B). Nalidixic acid was discovered 14 years before gyrase (Gellert et al., 1976b) as an impurity in a batch of antimalarial chloroquine (Lesher et al., 1962). The antibacterial activity of the first generation of quinolones (nalidixic and oxolinic acids) as well as their simple synthetic route appeared immediately interesting. However, their antibacterial spectrum was limited to Gram-negative bacteria and their clinical use was restricted to urinary tract infections. The introduction of a fluorine atom at position 6 (Figure 6B) and of a bulky piperidine at position 7 broadened the antimicrobial spectrum to Pseudomonas species and some Gram-positive organisms (including Staphylococcus aureus). The third generation of quinolones presents substitutions at the 7 as well as at the 8 position, resulting in enhanced activity against Gram-positive bacteria (and in most cases higher specificity for Topo IV). Moxifloxacin and levofloxacin (Figure 6B) are active against S. aureus and S. pneumoniae, pathogens responsible for upper and lower respiratory tract infections, acute otitis, and meningitis (Keating and Scott, 2004). Moxifloxacin is also active against Mycobacterium tuberculosis, which lacks Topo IV (Mdluli and Ma, 2007). Recently, the fourth generation of fluoroquinolones has been developed, with enhanced potency and a broader spectrum including anaerobic coverage. Examples of this class include gemifloxacin (Figure 6B), approved by the FDA in 2003, and currently one of the most potent fluoroquinolones against community-acquired pneumonia and acute bronchitis. Trovafloxacin is also used against intra-abdominal and pelvic infections. The current new inhibitors under preclinical and clinical investigation are mainly molecules based on the quinolone structure; some examples are fluoroquinolones with bulkier 7 substitutions (Bradbury and Pucci, 2008) and the 8-cyano-fluoroquinolones, naphtiridones, and nonfluorinated quinolones (Black and Coleman, 2009). Promising nonquinolone molecules such as the quinolines still target GyrA and ParC and show broad activity spectrum as well as coverage against the current quinolone-resistant strain (Black and Coleman, 2009). Other agents such as the class of “quinolone hybrids” (Bradbury and Pucci, 2008) are under investigation, but the clinical relevance of all these new molecules has still to be proved.
As for eukaryotic anti-Top2 agents, antibacterial quinolones convert bacterial type II enzymes into potent cellular toxins, stabilizing the cleavage complexes in an open form with generation of chromosome breaks, which culminate in cell death (Drlica et al., 2009). Quinolones selectively poison the GyrA subunit of gyrase and the ParC subunit of Topo IV and interact with both GyrA and B and ParC and E (see next section on interfacial inhibition and Figure 7) (Drlica et al., 2009; Heddle et al., 2000; Laponogov et al., 2009). The target preference for gyrase versus Topo IV (as well as the appearance of mutations) seems to depend upon two factors: the species of organism and the structure of the drug. In Gram-negative bacteria, gyrase is the primary target of fluoroquinolones, whereas Topo IV is their preferential target in Gram-positive bacteria (Drlica et al., 2009; Ferrero et al., 1994; Pan and Fisher, 1998).
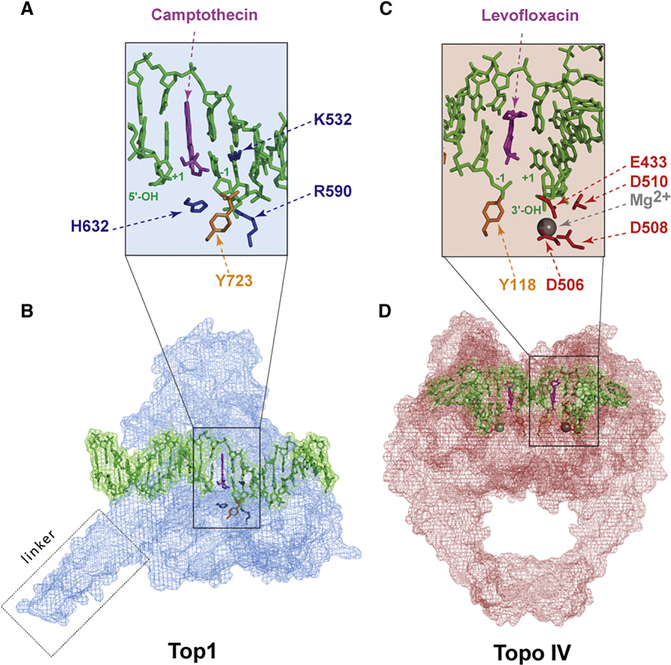
Figure 7. Interfacial Inhibition by Topoisomerase Inhibitors.
(A) Intercalation of camptothecin in the DNA break between the base pairs flanking the Top1 cleavage complex (positions −1 and +1; see Figures 1C–1E). The basic residues that catalyze the nucleophilic attack of DNA by the catalytic tyrosine (Y743 in gold) are shown in blue (see Figure 2A). The Top1 amino acid residues that make hydrogen-bond interactions with the camptothecin E ring are not shown (see Figure 4 in Marchand et al., 2006).
(B) Overview of the Top1-DNA cleavage complex.Top1 is in blue, DNA is in green, and camptothecin is in purple (Ioanoviciu et al., 2005; Marchand et al., 2006) (Protein Data Bank ID code 1T8I).
(C) Intercalation of levofloxacin in the DNA break between the base pairs that flank the Topo IV cleavage complex (−1 and +1; see Figure 1). The acidic residues that coordinate the Mg2+ (see Figures 4B and 4C) are shown in red and the catalytic tyrosine (Y118) in gold.
(D) Overview of the Topo IV-DNA cleavage complex. Topo IV is in red, the DNA is in green, and the levofloxacin molecules in both cleavage sites are in purple (Laponogov et al., 2009) (Protein Data Bank ID code 3K9F).
Resistance to fluoroquinolones is caused by stepwise mutations in Topo IV and gyrase, leading to alterations of their binding site. Such mutations occur primarily in an area called the quinolone resistance-determining region (positions 67–106 in E. coli GyrA). Mutations in this region greatly reduce the affinity of quinolones for the cleavage complex. Other mechanisms of resistance include altered drug uptake, increased drug efflux, and/or chemical drug inactivation before the quinolone reaches its intracellular target (Drlica et al., 2009).
Gyrase and to a lesser extent Topo IV can also be targeted in their ATP-binding sites (GyrB and ParE, respectively) (see Figure 4). Aminocoumarins such as coumermycin A1 and novobiocin (Figure 6C) are antibiotics isolated from Streptomyces strains (Heide et al., 2008). They were discovered in the 1950s, well before the quinolones. They bind to the ATP-binding pocket of GyrB and ParE and thereby block enzyme activity (Lewis et al., 1996). In spite of its potent antigyrase activity, novobiocin was discontinued as a clinical drug because of its low bioavailability and solubility and because of side effects. Aminocoumarins are also toxic toward eukaryotic cells, which could be related to their activity against the host Top2 enzymes or other ATP-binding enzymes. Efforts are still ongoing to obtain more selective and less toxic GyrB inhibitors (Charifson et al., 2008; Oblak et al., 2007).
Interfacial Inhibition Revealed by Topoisomerase Inhibitors
The hypothesis that topoisomerase inhibitors trap cleavage complexes by binding at the enzyme-DNA interface was first proposed to account for the results of DNA-sequencing experiments showing that drugs from different chemical families trap different sites depending on the bases flanking the cleavage sites (Capranico et al., 1990; Capranico and Binaschi, 1998; Fortune and Osheroff, 2000; Jaxel et al., 1991; Pommier et al., 1991). In the early 1990s, the drugs were proposed to act by stacking between the flanking base pairs (see schemes in Figures 1E and 1H). A second component of the drug selectivity was related to hydrogen bonds formed between the drug and the enzyme amino acids in the immediate vicinity of the cleaved DNA. The stacking interfacial model accounts for the fact that Top1 and Top2 inhibitors enhance the steady-state levels of cleavage complexes (see Figure 1) and for the rapid reversibility of the cleavage complexes upon drug removal.
Interfacial Complexes for Top1 Inhibitors
The interfacial stacking hypothesis was validated with the Top1 inhibitors after the resolution of cocrystals including topotecan (Staker et al., 2002), camptothecin (Ioanoviciu et al., 2005), indenoisoquinolines (Ioanoviciu et al., 2005), and an indolocarbazole derivative (Ioanoviciu et al., 2005; Marchand et al., 2006; Pommier and Marchand, 2005) (Figures 7A and 7B). The interfacial drug binding was also borne out from the mapping of camptothecin-resistance point mutations (Pommier et al., 1999), which turned out to affect those amino acid residues that form the hydrogen bonds between the drugs and Top1 (Chrencik et al., 2004). In the case of camptothecins, three H bonds are formed: the first between the camptothecin N-1 position (see Figure 3A) and arginine 364, the second between the camptothecin 20-hydroxyl and aspartate 533, and the third between the camptothecin 17-carbonyl and asparagine 722 (see Figure 4 in Marchand et al., 2006). Mutation of that single residue (i.e., N722S) renders Top1 immune to camptothecins, albeit without prohibiting the formation of cocrystals (Chrencik et al., 2004), which emphasizes the importance of each of the H bonds for stabilizing the drug in the cleavage complex.
Interfacial Complexes for Topoisomerase II Inhibitors
The interfacial inhibitor paradigm has recently been validated for the quinolones and Topo IV (Figures 7C–7D) (Laponogov et al., 2009). Consistent with the studies with the anticancer Top2 inhibitors (Capranico et al., 1990; Freudenreich and Kreuzer, 1994; Pommier et al., 1991; Pommier and Marchand, 2005) and quinolone resistance (Yoshida et al., 1991), and in agreement with the findings with Top1 inhibitors (see above), moxifloxacin or levofloxacin (Figures 7C and 7D) stacks between the base pairs flanking the cleavage site and binds at the interface of the enzyme and DNA. As in the case of Top1 inhibitors, this mechanism accounts for the inhibition of religation and the accumulation of cleavage complexes produced by the drugs.
Conclusions and Prospects
In spite of the remarkable elucidation of topoisomerase structures, enzymatic mechanisms, biological functions, and mechanisms of action of inhibitors as antibacterial and anticancer agents over the past 30 years, we only know a small number of molecular partners (cellular proteins) of topoisomerases. There are still no relevant inhibitors for viral and bacterial type I topoisomerases. The discovery of novel antibacterials not only against type II enzymes but also against type I bacterial topoisomerases is warranted. Such drugs could provide new therapies against human diseases, including bacterial infections associated with the emergence of highly resistant strains. Hopefully, the structural insight into the cleavage complexes formed by Topo IV with DNA and quinolones will open new doors for the rational development of more effective drugs. Novel noncamptothecin Top1 inhibitors are beginning clinical trials and the search for Top2 inhibitors specific for the α isoform should limit the occurrence of drug-induced secondary cancers. There are still no available inhibitors for eukaryotic Top3. Such drugs would be valuable as molecular tools and as potential novel anticancer agents. Finally, ongoing progress in molecular genetics and biology should enable clinicians to personalize treatments with antitopoisomerase drugs against cancers and infectious diseases.
ACKNOWLEDGMENTS
Our studies have been supported by the Center for Cancer Research, Intramural Program of the National Cancer Institute, National Institutes of Health. We wish to thank Kurt Kohn as outstanding mentor and colleague, and Elie Pommier for help with chemical structures.
REFERENCES
- Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, and Liu LF (2007). Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. USA 104, 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand R, O’Connor P, Kerrigan D, and Pommier Y (1992). Sequential administration of camptothecin and etoposide circumvents the antagonistic cytotoxicity of simultaneous drug administration in slowly growing human colon carcinoma HT-29 cells. Eur. J. Cancer 28A, 743–748. [DOI] [PubMed] [Google Scholar]
- Black MT, and Coleman K (2009). New inhibitors of bacterial topoisomerase GyrA/ParC subunits. Curr. Opin. Investig. Drugs 10, 804–810. [PubMed] [Google Scholar]
- Bradbury BJ, and Pucci MJ (2008). Recent advances in bacterial topoisomerase inhibitors. Curr. Opin. Pharmacol. 8, 574–581. [DOI] [PubMed] [Google Scholar]
- Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, Robey R, Pommier Y, Fojo T, and Bates SE (1999). Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 59, 5938–5946. [PubMed] [Google Scholar]
- Capranico G, and Binaschi M (1998). DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta 1400, 185–194. [DOI] [PubMed] [Google Scholar]
- Capranico G, Kohn KW, and Pommier Y (1990). Local sequence requirements for DNA cleavage by mammalian topoisomerase II in the presence of doxorubicin. Nucleic Acids Res. 18, 6611–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ, and Dulbecco R (1972). An activity from mammalian cells that untwists superhelical DNA—a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc. Natl. Acad. Sci. USA 69, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charifson PS, Grillot AL, Grossman TH, Parsons JD, Badia M, Bellon S, Deininger DD, Drumm JE, Gross CH, LeTiran A, et al. (2008). Novel dual-targeting benzimidazole urea inhibitors of DNA gyrase and topoisomerase IV possessing potent antibacterial activity: intelligent design and evolution through the judicious use of structure-guided design and structure-activity relationships. J. Med. Chem. 51, 5243–5263. [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, and Redinbo MR (2004). Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 339, 773–784. [DOI] [PubMed] [Google Scholar]
- Cinelli MA, Cordero B, Dexheimer TS, Pommier Y, and Cushman M (2009). Synthesis and biological evaluation of 14-(aminoalkyl-aminomethyl) aromathecins as topoisomerase I inhibitors: investigating the hypothesis of shared structure-activity relationships. Bioorg. Med. Chem. 17, 7145–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, and Caldecott KW (2009). A human 50-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678. [DOI] [PubMed] [Google Scholar]
- Crisona NJ, Strick TR, Bensimon D, Croquette V, and Cozzarelli NR (2000). Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14, 2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arpa P, Beardmore C, and Liu LF (1990). Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 50, 6919–6924. [PubMed] [Google Scholar]
- Debethune L, Kohlhagen G, Grandas A, and Pommier Y (2002). Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 30, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, and Liu LF (2001). Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 61, 5926–5932. [PubMed] [Google Scholar]
- Deweese JE, and Osheroff N (2009). The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese JE, Burgin AB, and Osheroff N (2008). Human topoisomerase IIa uses a two-metal-ion mechanism for DNA cleavage. Nucleic Acids Res. 36, 4883–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese JE, Osheroff MA, and Osheroff N (2009). DNA topology and topoisomerases: teaching a “knotty” subject. Biochem. Mol. Biol. Educ. 37, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer TS, Antony S, Marchand C, and Pommier Y (2008). Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer. Agents Med. Chem. 8, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, and Zhao X (2009). Quinolones: action and resistance updated. Curr. Top. Med. Chem. 9, 981–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng WK, Faucette L, Johnson RK, and Sternglanz R (1988). Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 34, 755–760. [PubMed] [Google Scholar]
- Eng WK, McCabe FL, Tan KB, Mattern MR, Hofmann GA, Woessner RD, Hertzberg RP, and Johnson RK (1990). Development of a stable camptothecin-resistant subline of P388 leukemia with reduced topoisomerase I content. Mol. Pharmacol. 38, 471–480. [PubMed] [Google Scholar]
- Felix CA, Kolaris CP, and Osheroff N (2006). Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst.) 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, and Blanche F (1994). Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13, 641–653. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, and Serre MC (2007). Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446. [DOI] [PubMed] [Google Scholar]
- Fortune JM, and Osheroff N (2000). Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 64, 221–253. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH, and Kreuzer KN (1994). Localization of an aminoacridine antitumor agent in a type II topoisomerase-DNA complex. Proc. Natl. Acad. Sci. USA 91, 11007–11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori A, Harker WG, Kohlhagen G, Hoki Y, and Pommier Y (1995). Mutation at the catalytic site of topoisomerase I in CEM/C2, a human leukemia cell resistant to camptothecin. Cancer Res. 55, 1339–1346. [PubMed] [Google Scholar]
- Gellert M, Mizuuchi K, O’Dea MH, and Nash HA (1976a). DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73, 3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, O’Dea MH, Itoh T, and Tomizawa J (1976b). Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 73, 4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, and Wang JC (1985). Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc. Natl. Acad. Sci. USA 82, 7178–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai R, Caron PR, and Wang JC (1996). Human TOP3: a single copy gene encoding DNA topoisomerase III. Proc. Natl. Acad. Sci. USA 93, 3653–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautefaye P, Cimetiere B, Pierre A, Leonce S, Hickman J, Laine W, Bailly C, and Lavielle G (2003). Synthesis and pharmacological evaluation of novel non-lactone analogues of camptothecin. Bioorg. Med. Chem. Lett. 13, 2731–2735. [DOI] [PubMed] [Google Scholar]
- Heddle JG, Barnard FM, Wentzell LM, and Maxwell A (2000). The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19, 1249–1264. [DOI] [PubMed] [Google Scholar]
- Heide L, Gust B, Anderle C, and Li SM (2008). Combinatorial biosynthesis, metabolic engineering and mutasynthesis for the generation of new aminocoumarin antibiotics. Curr. Top. Med. Chem. 8, 667–679. [DOI] [PubMed] [Google Scholar]
- Holm C, Covey JM, Kerrigan D, and Pommier Y (1989). Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 49, 6365–6368. [PubMed] [Google Scholar]
- Hsiang YH, Hertzberg R, Hecht S, and Liu LF (1985). Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260, 14873–14878. [PubMed] [Google Scholar]
- Hsiang Y-H, Liu LF, Wall ME, Wani MC, Nicholas AW, Manikumar G, Kirschenbaum S, Silber R, and Potmesil M (1989). DNA topoisomerase I-mediated DNA cleavage and cytotoxicity of camptothecin analogs. Cancer Res. 49, 4385–4389. [PubMed] [Google Scholar]
- Interthal H, Chen HJ, and Champoux JJ (2005). Human Tdp1 cleaves a broad spectrum of substrates including phosphoamide linkages. J. Biol. Chem. 280, 36518–36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioanoviciu A, Antony S, Pommier Y, Staker BL, Stewart L, and Cushman M (2005). Synthesis and mechanism of action studies of a series of norindenoisoquinoline topoisomerase I poisons reveal an inhibitor with a flipped orientation in the ternary DNA-enzyme-inhibitor complex as determined by X-ray crystallographic analysis. J. Med. Chem. 48, 4803–4814. [DOI] [PubMed] [Google Scholar]
- Jaxel C, Kohn KW, Wani MC, Wall ME, and Pommier Y (1989). Structure-activity study of the actions of camptothecin derivatives on mammalian topoisomerase I: evidence for a specific receptor site and a relation to antitumor activity. Cancer Res. 49, 1465–1469. [PubMed] [Google Scholar]
- Jaxel C, Capranico G, Kerrigan D, Kohn KW, and Pommier Y (1991). Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem. 266, 20418–20423. [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, and Rosenfeld MG (2006). A topoisomerase IIb-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802. [DOI] [PubMed] [Google Scholar]
- Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, and Suzuki H (1990). New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH (1991). Antagonism between camptothecin and topoisomerase II-directed chemotherapeutic agents in a human leukemia cell line. Cancer Res. 51, 1129–1136. [PubMed] [Google Scholar]
- Keating GM, and Scott LJ (2004). Moxifloxacin: a review of its use in the management of bacterial infections. Drugs 64, 2347–2377. [DOI] [PubMed] [Google Scholar]
- Koster DA, Croquette V, Dekker C, Shuman S, and Dekker NH (2005). Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674. [DOI] [PubMed] [Google Scholar]
- Krogh BO, and Shuman S (2002). A poxvirus-like type IB topoisomerase family in bacteria. Proc. Natl. Acad. Sci. USA 99, 1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapenna S, and Giordano A (2009). Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 8, 547–566. [DOI] [PubMed] [Google Scholar]
- Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, and Sanderson MR (2009). Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 16, 667–669. [DOI] [PubMed] [Google Scholar]
- Lavergne O, Lesueur-Ginot L, Pla Rodas F, Kasprzyk PG, Pommier J, Demarquay D, Prevost G, Ulibarri G, Rolland A, Schiano-Liberatore AM, et al. (1998). Homocamptothecins: synthesis and antitumor activity of novel E-ring-modified camptothecin analogues. J. Med. Chem. 41, 5410–5419. [DOI] [PubMed] [Google Scholar]
- Lee MP, Brown SD, and Hsieh T-S (1993). DNA topoisomerase I is essential in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 90, 6656–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesher GY, Froelich EJ, Gruett MD, Bailey JH, and Brundage RP (1962). 1,8-naphthyridine derivatives. A new class of chemotherapeutic agents. J. Med. Pharm. Chem. 91, 1063–1065. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Singh OM, Smith CV, Skarzynski T, Maxwell A, Wonacott AJ, and Wigley DB (1996). The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15, 1412–1420. [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Ban Y, Lyu YL, and Liu LF (2009). Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double-strand breaks at arrested replication forks. J. Biol. Chem. 284, 28084–28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, and Wang JC (1987). Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BH, Musial ST, and Brattain MG (1985). Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res. 45, 3106–3112. [PubMed] [Google Scholar]
- Lovett BD, Strumberg D, Blair IA, Pang S, Burden DA, Megonigal MD, Rappaport EF, Rebbeck TR, Osheroff N, Pommier Y, and Felix CA (2001). Etoposide metabolites enhance DNA topoisomerase II cleavage near leukemia-associated MLL translocation breakpoints. Biochemistry 40, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Low RL, Orton S, and Friedman DB (2003). A truncated form of DNA topoisomerase IIb associates with the mtDNA genome in mammalian mitochondria. Eur. J. Biochem. 270, 4173–4186. [DOI] [PubMed] [Google Scholar]
- Malik M, and Nitiss JL (2004). DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot. Cell 3, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C, Antony S, Kohn KW, Cushman M, Ioanoviciu A, Staker BL, Burgin AB, Stewart L, and Pommier Y (2006). A novel norindenoisoquinoline structure reveals a common interfacial inhibitor paradigm for ternary trapping of topoisomerase I-DNA covalent complexes. Mol. Cancer Ther. 5, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon AK, Rodriguez AC, and Osheroff N (2005). Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 280, 39337–39345. [DOI] [PubMed] [Google Scholar]
- Mdluli K, and Ma Z (2007). Mycobacterium tuberculosis DNA gyrase as a target for drug discovery. Infect. Disord. Drug Targets 7, 159–168. [DOI] [PubMed] [Google Scholar]
- Merino A, Madden KR, Lane WS, Champoux JJ, and Reinberg D (1993). DNA topoisomerase I is involved in both repression and activation of transcription. Nature 365, 227–232. [DOI] [PubMed] [Google Scholar]
- Miao ZH, Player A, Shankavaram U, Wang YH, Zimonjic DB, Lorenzi PL, Liao ZY, Liu H, Shimura T, Zhang HL, et al. (2007). Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 67, 8752–8761. [DOI] [PubMed] [Google Scholar]
- Morham SG, Kluckman KD, Voulomanos N, and Smithies O (1996). Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 16, 6804–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL (2009a). DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL (2009b). Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblak M, Kotnik M, and Solmajer T (2007). Discovery and development of ATPase inhibitors of DNA gyrase as antibacterial agents. Curr. Med. Chem. 14, 2033–2047. [DOI] [PubMed] [Google Scholar]
- Pan XS, and Fisher LM (1998). DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42, 2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Andersen MK, Christiansen DH, and Nerlov C (2002). Genetic pathways in therapy-related myelodysplasia and acute myeloid leukemia. Blood 99, 1909–1912. [DOI] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, and Avvedimento EV (2008). DNA oxidation as triggered by H3K9me2 demethylation drives estrogeninduced gene expression. Science 319, 202–206. [DOI] [PubMed] [Google Scholar]
- Pommier Y (2006). Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802. [DOI] [PubMed] [Google Scholar]
- Pommier Y (2009). DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 109, 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, and Cushman M (2009). The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol. Cancer Ther. 8, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, and Marchand C (2005). Interfacial inhibitors of protein-nucleic acid interactions. Curr. Med. Chem. Anticancer Agents 5, 421–429. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Zwelling LA, Mattern MR, Erickson LC, Kerrigan D, Schwartz R, and Kohn KW (1983). Effects of dimethyl sulfoxide and thiourea upon intercalator-induced DNA single-strand breaks in mouse leukemia (L1210) cells. Cancer Res. 43, 5718–5724. [PubMed] [Google Scholar]
- Pommier Y, Schwartz RE, Kohn KW, and Zwelling LA (1984). Formation and rejoining of deoxyribonucleic acid double-strand breaks induced in isolated cell nuclei by antineoplastic intercalating agents. Biochemistry 23, 3194–3201. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Kerrigan D, Covey JM, Kao-Shan CS, and Whang-Peng J (1988). Sister chromatid exchanges, chromosomal aberrations, and cytotoxicity produced by antitumor topoisomerase II inhibitors in sensitive (DC3F) and resistant (DC3F/9-OHE) Chinese hamster cells. Cancer Res. 48, 512–516. [PubMed] [Google Scholar]
- Pommier Y, Capranico G, Orr A, and Kohn KW (1991). Local base sequence preferences for DNA cleavage by mammalian topoisomerase II in the presence of amsacrine or teniposide. Nucleic Acids Res. 19, 5973–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Pourquier P, Urasaki Y, Wu J, and Laco G (1999). Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resist. Updat. 2, 307–318. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, et al. (2006a). Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81, 179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Weinstein JN, Aladjem MI, and Kohn KW (2006b). Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin. Cancer Res. 12, 2657–2661. [DOI] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, and Nash HA (1999). Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–555. [DOI] [PubMed] [Google Scholar]
- Pouliot JJ, Robertson CA, and Nash HA (2001). Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 6, 677–687. [DOI] [PubMed] [Google Scholar]
- Ratain MJ, and Rowley JD (1992). Therapy-related acute myeloid leukemia secondary to inhibitors of topoisomerase II: from the bedside to the target genes. Ann. Oncol. 3, 107–111. [DOI] [PubMed] [Google Scholar]
- Rossi F, Labourier E, Gallouzi I. e., Derancourt J, Allemand E, Divita G, and Tazi J (1998). The C-terminal domain but not the tyrosine 723 of human DNA topoisomerase I active site contributes to kinase activity. Nucleic Acids Res. 26, 2963–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeffler AJ, and Berger JM (2008). DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101. [DOI] [PubMed] [Google Scholar]
- Shao R-G, Cao C-X, Shimizu T, O’Connor P, Kohn KW, and Pommier Y (1997). Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53. Cancer Res. 57, 4029–4035. [PubMed] [Google Scholar]
- Shuman S (1989). Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 86, 3489–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S (1998). Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim. Biophys. Acta 1400, 321–337. [DOI] [PubMed] [Google Scholar]
- Sirikantaramas S, Yamazaki M, and Saito K (2008). Mutations in topoisomerase I as a self-resistance mechanism coevolved with the production of the anticancer alkaloid camptothecin in plants. Proc. Natl. Acad. Sci. USA 105, 6782–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Gabut M, Dupon C, Kohlhagen G, Stevenin J, Pommier Y, and Tazi J (2003). Altered serine/arginine-rich protein phosphorylation and exonic enhancer-dependent splicing in mammalian cells lacking topoisomerase I. Cancer Res. 63, 8203–8211. [PubMed] [Google Scholar]
- Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB Jr., and Stewart L (2002). The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 99, 15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers JT, Harris TK, and Mildvan AS (1997). Vaccinia DNA topoisomerase I: evidence supporting free rotation mechanism for DNA supercoil relaxation. Biochemistry 36, 5212–5222. [DOI] [PubMed] [Google Scholar]
- Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, and Cozzarelli NR (2003). Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl. Acad. Sci. USA 100, 8654–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick R, Strissel PL, Borgers S, Smith SL, and Rowley JD (2000). Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc. Natl. Acad. Sci. USA 97, 4790–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Dexheimer TS, Redon C, Sordet O, Agama K, Lavielle G, Pierre A, Bates SE, and Pommier Y (2007). Novel E-ring camptothecin keto analogues (S38809 and S39625) are stable, potent, and selective topoisomerase I inhibitors without being substrates of drug efflux transporters. Mol. Cancer Ther. 6, 3229–3238. [DOI] [PubMed] [Google Scholar]
- Teicher BA (2008). Next generation topoisomerase I inhibitors: rationale and biomarker strategies. Biochem. Pharmacol. 75, 1262–1271. [DOI] [PubMed] [Google Scholar]
- Tewey KM, Chen GL, Nelson EM, and Liu LF (1984). Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J. Biol. Chem. 259, 9182–9187. [PubMed] [Google Scholar]
- Tse-Dinh YC (2009). Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 37, 731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, and Yanagida M (1984). Isolation of type I and II DNA topoisomer-ase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 3, 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Morino K, Uzawa S, Shiozaki K, and Yanagida M (1987). Cloning and sequencing of Schizosaccharomyces pombe DNA topoisomerase I gene, and effect of gene disruption. Nucleic Acids Res. 15, 9727–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki Y, Takebayashi Y, and Pommier Y (2000). Activity of a novel camptothecin analogue, homocamptothecin, in camptothecin-resistant cell lines with topoisomerase I alterations. Cancer Res. 60, 6577–6580. [PubMed] [Google Scholar]
- Wall ME, and Wani MC (1995). Camptothecin and taxol: discovery to clinic—Thirteenth Bruce F. Cain Memorial Award lecture. Cancer Res. 55, 753–760. [PubMed] [Google Scholar]
- Wall ME, Wani MC, Cooke CE, Palmer KH, McPhail AT, and Slim GA (1966). The isolation and structure of camptothecin, a novel alkaloidal leukemia and tumor inhibitor from Camptotheca acuminata. J. Am. Chem. Soc. 88, 3888–3890. [Google Scholar]
- Wang JC (1971). Interaction between DNA and an Escherichia coli protein u. J. Mol. Biol. 55, 523–533. [DOI] [PubMed] [Google Scholar]
- Wang JC (2002). Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440. [DOI] [PubMed] [Google Scholar]
- Wang JC (2009a). A journey in the world of DNA rings and beyond. Annu. Rev. Biochem. 78, 31–54. [DOI] [PubMed] [Google Scholar]
- Wang JC (2009b). Untangling the Double Helix (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Wilson Byl JA, Fortune JM, Burden DA, Nitiss JL, Utsugi T, Yamada Y, and Osheroff N (1999). DNA topoisomerases as targets for the anticancer drug TAS-103: primary cellular target and DNA cleavage enhancement. Biochemistry 38, 15573–15579. [DOI] [PubMed] [Google Scholar]
- Wu L, and Hickson ID (2006). DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40, 279–306. [DOI] [PubMed] [Google Scholar]
- Yang S-W, Burgin AB, Huizenga BN, Robertson CA, Yao KC, and Nash HA (1996). A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. USA 93, 11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Bogaki M, Nakamura M, Yamanaka LM, and Nakamura S (1991). Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35, 1647–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Pommier Y (2008). Mitochondrial topoisomerase I sites in the regulatory D-loop region of mitochondrial DNA. Biochemistry 47, 11196–11203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Barcelo JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, and Pommier Y (2001). Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. USA 98, 10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Meng LH, Zimonjic DB, Popescu NC, and Pommier Y (2004). Thirteen-exon-motif signature for vertebrate nuclear and mitochondrial type IB topoisomerases. Nucleic Acids Res. 32, 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Lyu YL, Lin CP, Zhou N, Azarova AM, Wood LM, and Liu LF (2006). A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 281, 35997–36003. [DOI] [PubMed] [Google Scholar]
- Zhang H, Meng LH, and Pommier Y (2007). Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie 89, 474–481. [DOI] [PubMed] [Google Scholar]
- Zwelling LA, Michaels S, Erickson LC, Ungerleider RS, Nichols M, and Kohn KW (1981). Protein-associated deoxyribonucleic acid strand breaks in L1210 cells treated with the deoxyribonucleic acid intercalating agents 4′-(9-acridinylamino) methanesulfon-m-anisidide and adriamycin. Biochemistry 20, 6553–6563. [DOI] [PubMed] [Google Scholar]