Environmental fatty acids can be harvested to supplement endogenous fatty acid synthesis to produce membranes and circumvent fatty acid biosynthesis inhibitors. However, how the inability to use these fatty acids impacts lipids is unclear. Our results reveal lipid composition changes in response to fatty acid addition and when S. aureus is unable to activate fatty acids through FakA. We identify concentration-dependent utilization of oleic acid that, when combined with previous work, provides evidence that fatty acids can serve as a signal to S. aureus. Furthermore, using mouse skin homogenates as a surrogate for in vivo conditions, we showed that S. aureus can incorporate host fatty acids. This study highlights how exogenous fatty acids impact bacterial membrane composition and function.
KEYWORDS: MRSA, cell membranes, fatty acids, phospholipids
ABSTRACT
Staphylococcus aureus can utilize exogenous fatty acids for phospholipid synthesis. The fatty acid kinase FakA is essential for this utilization by phosphorylating exogenous fatty acids for incorporation into lipids. How FakA impacts the lipid membrane composition is unknown. In this study, we used mass spectrometry to determine the membrane lipid composition and properties of S. aureus in the absence of fakA. We found the fakA mutant to have increased abundance of lipids containing longer acyl chains. Since S. aureus does not synthesize unsaturated fatty acids, we utilized oleic acid (18:1) to track exogenous fatty acid incorporation into lipids. We observed a concentration-dependent incorporation of exogenous fatty acids into the membrane that required FakA. We also tested how FakA and exogenous fatty acids impact membrane-related physiology and identified changes in membrane potential, cellular respiration, and membrane fluidity. To mimic the host environment, we characterized the lipid composition of wild-type and fakA mutant bacteria grown in mouse skin homogenate. We show that wild-type S. aureus can incorporate exogenous unsaturated fatty acids from host tissue, highlighting the importance of FakA in the presence of host skin tissue. In conclusion, FakA is important for maintaining the composition and properties of the phospholipid membrane in the presence of exogenous fatty acids, impacting overall cell physiology.
IMPORTANCE Environmental fatty acids can be harvested to supplement endogenous fatty acid synthesis to produce membranes and circumvent fatty acid biosynthesis inhibitors. However, how the inability to use these fatty acids impacts lipids is unclear. Our results reveal lipid composition changes in response to fatty acid addition and when S. aureus is unable to activate fatty acids through FakA. We identify concentration-dependent utilization of oleic acid that, when combined with previous work, provides evidence that fatty acids can serve as a signal to S. aureus. Furthermore, using mouse skin homogenates as a surrogate for in vivo conditions, we showed that S. aureus can incorporate host fatty acids. This study highlights how exogenous fatty acids impact bacterial membrane composition and function.
INTRODUCTION
Despite decades of intense research, Staphylococcus aureus remains a tremendous cause of infection and morbidity in the human population (1). Approximately 30% of the population are asymptomatic carriers of S. aureus (2); however, this bacterium can cause infection in numerous anatomical sites, including skin and soft tissues, bones, lungs, and the heart, as well as foreign implants, such as catheters and prosthetic joints (3). While S. aureus infection was originally characterized as a typically hospital-acquired infection, the incidence of infections in the community has increased concern and awareness of this pathogen, as community-associated strains have become dominant in the United States (4, 5). Thus, a thorough understanding of how S. aureus can establish infection, fend off the immune system, and maintain infection is needed to combat this pathogen.
Phospholipids lie at the interface of the host-pathogen interaction. Membrane-associated products, such as lipopolysaccharides, lipoteichoic acids, and lipoproteins, are sensed by the germ line-encoded pattern recognition receptors that induce the activity of numerous host immune cells (6, 7). In addition to these membrane products, phospholipids themselves can play a role in evading the immune system. For example, lysyl-phosphatidylglycerol (LPG) has been shown to be important for evading neutrophils and antimicrobial peptides (8, 9). The composition of the phospholipid membranes of bacteria can also dictate if antimicrobial treatment during infection is successful. Resistance to daptomycin, a lipopeptide antimicrobial, can result from the mutation of cardiolipin (CL) synthase (cls2) and increased abundance of cardiolipin (10). Microbial lipids can also serve as antigens for the immune system (11), further emphasizing the role that lipids play during the infection process. More recently, the identification of bacterial extracellular vesicles has also become a topic of interest; these vesicles could contribute to host-pathogen interactions (12). These data clearly suggest that the composition of the phospholipid membrane is a vital component of the host-pathogen interface.
Synthesis of lipids is preceded by the production of fatty acids. S. aureus endogenously synthesizes fatty acids via the fatty acid synthesis type II system (FASII) (13). Due to the differences between fatty acid synthesis enzymes of bacteria and humans, FASII has been the subject of antimicrobial targeting (14–17). Bacteria, including S. aureus, can supplement endogenous fatty acid synthesis by utilizing exogenous fatty acids (exoFAs) (18). These exoFAs are predicted to passively diffuse into the phospholipid membrane. In S. aureus, exoFAs are removed from the membrane by fatty acid-binding proteins FakB1 and FakB2 (18). Once removed from the membrane, fatty acid kinase, FakA, then phosphorylates the carboxyl head group of the fatty acid, creating an acyl phosphate (18) that can then be used for lipid synthesis. S. aureus synthesizes predominantly three classes of phospholipids: phosphatidylglycerol (PG), LPG, and CL (8, 19). One interesting caveat to fatty acid and lipid synthesis in S. aureus is the inability of this bacterium to synthesize unsaturated fatty acids (20). Instead, S. aureus utilizes branched-chain fatty acids (BCFAs), derived from the branched-chain amino acids isoleucine, leucine, and valine, to help modulate the membrane in response to environmental stimuli (21, 22). A large portion of the BCFAs produced by S. aureus include odd-numbered iso and anteiso BCFAs, with an acyl chain length of 15 being the most abundant (22, 23).
FakA was first identified as a regulator of virulence due to the decrease in α-hemolysin activity, increased protease activity, and increased dermonecrosis in a murine model of infection (24). Originally named virulence factor regulator B (VfrB) due to this altered virulence, FakA was eventually identified to be a fatty acid kinase (18). Subsequently, the altered virulence factor profile of a fakA mutant was identified to be due, in part, to altered activity of the SaeRS two-component system (25, 26). The current model for the FakA-dependent alteration of SaeRS signaling is due to the accumulation of fatty acids within the cell (26). A mechanism for how these accumulated fatty acids within the cell decrease SaeRS signaling is still undetermined. The absence of FakA affects global metabolism (27) and increases the resistance of S. aureus to toxic fatty acids (28, 29).
How the inability to use exoFAs affects the overall membrane lipid composition has not been evaluated. In the current study, we aimed to determine the changes in membrane lipid composition in the absence of FakA. We tested how these overall membrane lipid changes affect the properties of the membrane itself. Lastly, we provide evidence that S. aureus grown in the presence of host tissue can use unsaturated fatty acids to supplement phospholipid synthesis, extending our observations from standard laboratory media to include fatty acids found in murine skin. These results increase the importance that the fatty acid utilization system has in the presence of host-derived tissues.
RESULTS
Cellular and extracellular fatty acid profiles of a fakA mutant.
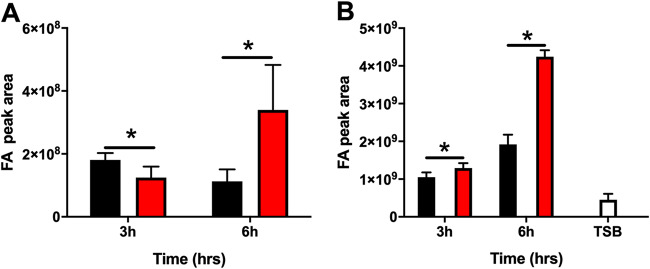
FakA is necessary for S. aureus to incorporate exoFAs into acyl chains of membrane lipids. Thus, it would be expected that the absence of FakA would alter the abundance of FAs in the cell. Indeed, a previous study determined that a fakA mutant accumulates fatty acids (26). Our studies are typically performed in tryptic soy broth (TSB), and therefore, we sought to recapitulate this accumulation of fatty acids at different phases of growth in the fakA mutant under our growth conditions and in our S. aureus strain of choice, AH1263, a derivative of the USA300 strain LAC. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS), we quantified the available free fatty acids in TSB and found that 16:0 and 18:0 constitute the most abundant fatty acids (see Fig. S1 in the supplemental material). Next, we determined how FakA affects fatty acid pools. To this end, the free fatty acid profiles of cellular and extracellular fatty acids for the wild type (WT) and fakA mutant was determined. This was tested at two time points representing the early (3 h) and late (6 h) exponential phases of growth, times that we have previously characterized the metabolic changes in the fakA mutant (27). In agreement with previous research, we observed a significant increase in cellular free fatty acids in the fakA mutant compared to that in the wild type (Fig. 1A) at 6 h. However, the cellular fatty acid levels were slightly, but significantly, decreased at 3 h. The identity of the individual fatty acids was also determined, and we observed differences in several fatty acid species between the wild type and fakA mutant after 3 h of growth (Fig. S2A) and 6 h of growth (Fig. S2B). Specifically, we found a significant (P < 0.05) increase in the proportion of 15:0, 17:0, 19:0, and 20:0 in the fakA mutant during the late exponential (6 h) growth phase. We reasoned that an accumulation of fatty acids in the cell could lead to release of fatty acids into the supernatant. We observed an increased abundance of supernatant fatty acids in both strains over time, which was enhanced in the fakA mutant (Fig. 1B). These data demonstrate that the fakA mutant accumulates fewer fatty acids during the early exponential phase (3 h) of growth but accumulates more fatty acids during the late exponential phase (6 h) of growth than the wild type. They also show that the fakA mutant possesses altered fatty acid pools compared to those of the wild-type strain.
FIG 1.
Cellular (A) and supernatant (B) free fatty acids (FA) of the wild type (black) and fakA mutant (red) grown for 3 and 6 h in TSB plus 14 mM glucose. The empty bar represents free fatty acids from sterile TSB plus 14 mM glucose. Data represent the means (n = 4) with standard deviations. *, P < 0.05 by Student’s t test.
Lipid profile for the wild type and fakA mutant in TSB.
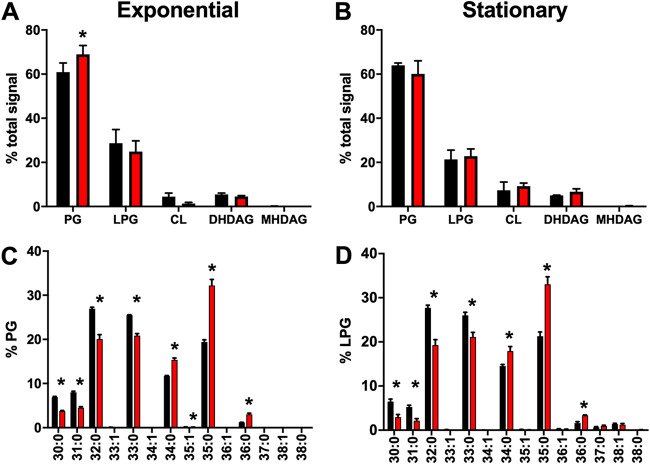
An in-depth analysis of how FakA affects the composition of the lipid membrane of S. aureus has not been undertaken. To do this, we performed a comprehensive analysis of the membrane lipids of fakA mutant compared to those of the wild type when grown in TSB. Lipids were extracted using a modified Bligh and Dryer liquid-liquid lipid extraction protocol (30). Since lipid ratios are known to vary by growth phase (31, 32), we examined cells during exponential phase (5 h) and stationary phase (24 h). As expected, PG and LPG were the primary lipids in both strains (Fig. 2A and B). We observed little difference between the major lipid species between the two strains, with only a modest significant increase in PG and a corresponding decrease in CL in the fakA mutant during the exponential phase of growth (Fig. 2A). Since LPG and CL are synthesized from PG, the lack of major differences was not unexpected, as FakA is important for insertion of exogenous fatty acids as phospholipid acyl side chains and not, as far as we know, the activity of the enzymes responsible for lipid species generation. Considering this, we determined the total carbon and saturations of the lipid acyl side chains for PG and LPG in the wild type and fakA mutant. During growth in TSB, lipids containing an unsaturated acyl chain were near absent at either phase of growth in both strains (Fig. 2C and D). We did identify significant changes in the lipid profiles between the two strains, with the fakA mutant tending to contain a higher abundance of longer acyl sides chains, predominantly 35:0, than the parent strain.
FIG 2.
Lipid analysis for the wild type (black) and fakA mutant (red). (A and B) Percent total signal for each lipid class was determined at the exponential (A) and stationary (B) phases of growth for phosphatidylglycerol (PG), lysyl-PG (LPG), cardiolipin (CL), dihex-diacylglycerol (DHDAG), and monohex-diacylglycerol (MHDAG). (C and D) Individual PG (C) and LPG (D) species from cells in exponential phase were determined and are presented as percentages of the total PG or LPG signal. Data represent the means (n = 4) with standard deviations. *, P < 0.05 by Student’s t test.
Since the data in Fig. 2C and D represent the total carbon length of both acyl side chains, we performed product ion analysis on pooled samples to determine the most abundant acyl side chain pairings that comprise each lipid (Table S1). This analysis revealed that both the wild type and the fakA mutant use 15:0 as one acyl side chain, as expected. The second acyl moiety was the same for each lipid species between the two strains, i.e., 35:0 possessed 15:0 and 20:0 fatty acids. Thus, despite using the same fatty acids for membrane generation, the mutant possesses a higher percentage of lipids containing longer acyl chains.
Lipid profiles of the wild type and fakA mutant in the presence of exogenous fatty acid.
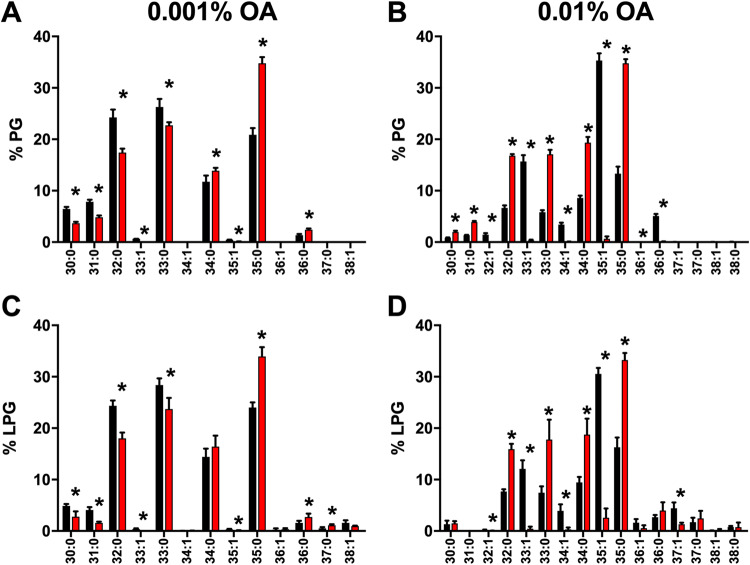
S. aureus is unable to endogenously synthesize unsaturated fatty acids but can incorporate them from the environment through FakA. This allows us to use exogenously added fatty acids to track exogenous fatty acid utilization and observe where exoFAs are incorporated. Using this approach, we determined the lipid composition of both the wild type and a fakA mutant when grown in the presence of oleic acid (18:1), which was chosen due its relatively low toxicity (Fig. S3) (28). Previously, we demonstrated that the addition of 0.001% (∼31 μM) oleic acid elicits a transcriptional response in S. aureus (25); therefore, we determined the lipid profiles in TSB supplemented with 0.001% oleic acid. We observed little, if any, incorporation of oleic acid into PG and LPG when grown at this concentration (Fig. 3A and C). Indeed, the lipid profiles of the wild type and fakA mutant grown with added 0.001% oleic acid were indistinguishable from growth in TSB alone (Fig. 2). We used fatty acid methyl ester (FAME) analysis followed by gas chromatography to examine the total fatty acid content of the cells, and in agreement with the above-described lipid profiles, we found little oleic acid (18:1) associated with the cells grown in TSB plus 0.001% oleic acid (Table S2). Thus, while S. aureus responds transcriptionally to 0.001% additional oleic acid, this concentration was too low to affect incorporation.
FIG 3.
Lipid profile for the wild type (black) and fakA mutant (red) of PG (A and B) and LPG (C and D) grown in TSB plus 0.001% oleic acid (A and C) or TSB plus 0.01% oleic acid (B and D). Data represent the means (n = 4) with standard deviations. *, P < 0.05 by Student’s t test.
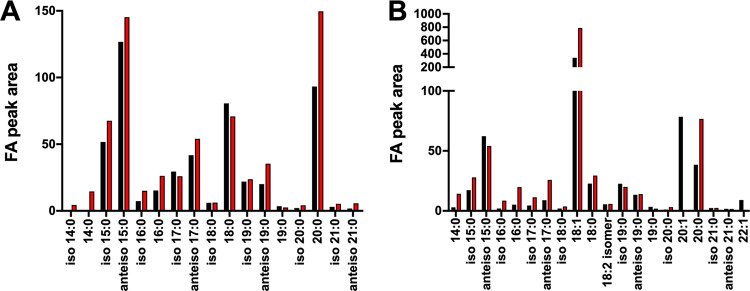
Next, we increased the oleic acid concentration to 0.01% (∼314 μM). At this concentration, the wild-type strain readily used the oleic acid and was observed as the appearance of a single unsaturated fatty acid chain primarily in 32:1, 33:1, 34:1, and 35:1 lipids (Fig. 3B and D). We had predicted that oleic acid (18:1) would be found primarily paired with 15:0, the most common fatty acid lipid side chain in S. aureus. While we did readily detect 33:1, the most abundant lipid was 35:1, which composed >30% of total lipid species. As expected, there was no unsaturated fatty acid in the lipids isolated from the fakA mutant. To confirm that the absence of oleic acid in fakA mutant lipids is not due to oleic acid not associating with the cell, we performed FAME analysis followed by gas chromatography on the same extracts as used for lipid analysis. Our FAME analysis revealed that in TSB, the wild type and fakA mutant have similar total fatty acid profiles, with increased 20:0 in the fakA mutant (Fig. 4A). When grown with the additional 0.01% oleic acid, this fatty acid was readily detectable from both strains (Fig. 4B and Table S2). FAME identifies all fatty acids, both free and lipid associated. Since oleic acid was identified by FAME analysis but was not found as a component of the fakA lipids, we can conclude that oleic acid associates with the cells but cannot be incorporated into the fakA mutant. Thus, the lack of oleic acid in mutant membranes is due to a lack of incorporation and not because oleic cannot interact with or enter the cell.
FIG 4.
Fatty acid methyl esterase analysis of lipid extracts from the wild type (black) and fakA mutant (red) grown in TSB (A) or TSB plus 0.01% oleic acid (B). Data are from pooled replicates (n = 4) from each strain and growth environment.
Our lipid profiling indicated that wild-type cells can pair oleic acid with more than one fatty acid to produce a lipid. Moreover, we had expected 33:1 to be the most prevalent lipid species by pairing oleic acid with 15:0. We again used product ion analysis to determine the fatty acid pairing in each lipid (Table S1). It was expected that the fatty acid paired with oleic acid would be a saturated fatty acid and that this fatty acid would be equivalent to the total lipid carbon length minus the oleic acid. This prediction was true for 32:1 (14:0 and 18:1) and 33:1 (15:0 and 18:1). Considering that 18:1 was the added fatty acid, we anticipated 35:1 to be comprised of 17:0 and 18:1 fatty acids. This was not the case, as the major 35:1 fatty acid pair consisted of 15:0 and 20:1. The included 20:1 was also the fatty acid associated in the lower-abundance lipid 34:1.
While S. aureus membranes comprise both straight-chain fatty acids (SCFAs) and branched-chain fatty acids (BCFAs), the organism relies on BCFAs to modulate the rigidity of its lipid membrane (23). BCFAs in S. aureus are comprised of odd-numbered iso and anteiso fatty acids, and the proportions of BCFAs and SCFAs are highly dependent on the growth environment (21). We analyzed our FAME data to determine if the absence of fakA or the addition of oleic acid impacts BCFA or SCFA preference (Table S2). In TSB alone, the wild type and the fakA mutant have similar ratios of anteiso and iso fatty acids (∼1.5) and BCFAs and SCFAs (1.65 for the wild type and 1.48 for the fakA mutant). Not surprising considering our other analysis, when the wild type and fakA mutant were grown in TSB plus 0.001% oleic acid, the fatty acid profiles of both were nearly identical to those for growth in TSB. In contrast, growth in TSB plus 0.01% oleic acid resulted in accumulation of oleic acid in both wild-type and fakA mutant bacteria. When comparing the BCFA/SCFA ratio under this condition, we observed a dramatic shift, most likely due to the added SCFA oleic acid. For example, wild-type cells grown in TSB contain ∼38% SCFA, while growth in 0.01% oleic acid led to the cells altering the fatty acid pools to ∼78% SCFA. A similar shift (40% to 84%) was also seen in the fakA mutant. As noted in previous studies (33), the wild-type S. aureus unsaturated fatty acid pool was comprised of both 18:1 and 20:1. These data, along with our lipid and product ion data, indicate that wild-type S. aureus elongates oleic acid (18:1) to 20:1. This was not observed in the fakA mutant, as the majority of oleic acid remained oleic acid. In conclusion, the fatty acid composition of the membrane is dramatically altered in the presence of incorporation-level oleic acid seen in the wild type as the incorporation of oleic acid into PG, LPG, and CL.
Membrane function is affected by the inability to utilize exoFAs.
The membrane lies at the interface of cellular processes and the external environment. We have now shown that fakA influences membrane composition. We previously reported that the fakA mutant produces decreased pigment (28), which is associated with membrane fluidity (34–36). Also, our previous metabolomics study found that the fakA mutant has a more reduced cellular environment (27). The electron transport chain is embedded in the phospholipid membrane, suggesting that fatty acid accumulation and the composition of the membrane may affect respiration. Finally, membrane potential is indicative of metabolic state and membrane function. Therefore, we wanted to determine the role FakA and exoFAs play in membrane fluidity, permeability, membrane potential, and respiratory chain activity.
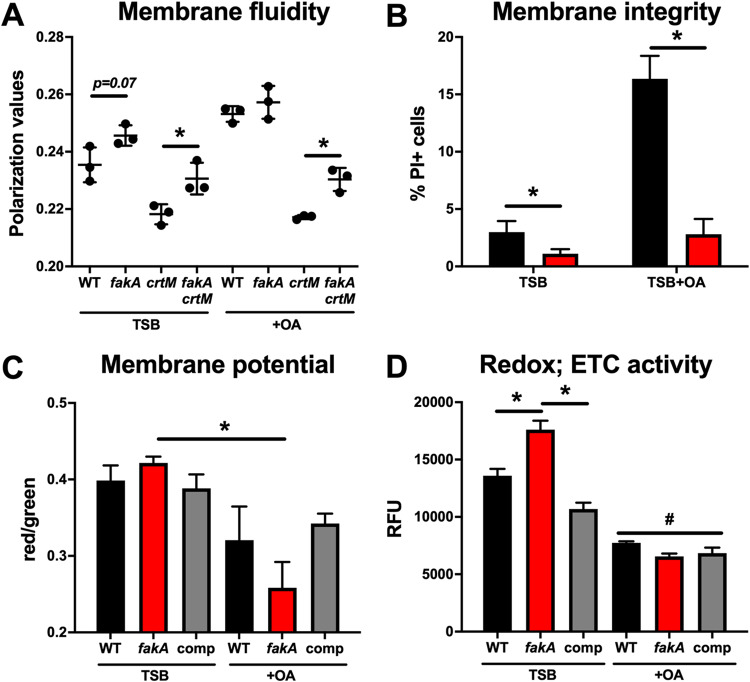
First, membrane fluidity was determined using the fluorescent probe 1,6-diphenyl-1,3,5-hexatriene (DPH) (23, 34) in the wild type and fakA mutant grown to exponential phase (5 h) in TSB with and without incorporation-level oleic acid (0.01%). While the value did not reach significance (P = 0.07), the fakA mutant had decreased membrane fluidity compared to that of the wild type (Fig. 5A). The Crt-derived pigment staphyloxanthin is known to affect membrane fluidity (37), and fakA mutants produce less pigment (28). To remove this complication, we tested a crtM mutant with and without fakA. We observed that a crtM mutant has more membrane fluidity than its isogenic parents, both in the case of the wild type and in the case of the fakA mutant. Interestingly, a fakA crtM double mutant has significantly decreased membrane fluidity compared to that of the crtM mutant. The addition of 0.01% oleic acid decreased membrane fluidity for both the wild type and fakA mutant. This was not due to incorporation as part of phospholipids, since the wild type and fakA mutants were equivalent. For reasons that are not clear, adding oleic acid to crtM mutants had no effect on membrane fluidity (compare the growth of the crtM mutant with and without oleic acid), and, again, we observed lower membrane fluidity in the fakA crtM mutant than in the crtM mutant. Thus, we determined that the absence of FakA affects membrane fluidity and that this is not due to the altered pigment levels in the mutant.
FIG 5.
(A) Membrane fluidity in the wild type, fakA mutant, crtM mutant, and fakA crtM mutant grown in the presence (+OA) or absence (TSB) of 0.01% oleic acid. (B) Membrane integrity measured using propidium iodide (PI) as an indicator for the wild type (black) and fakA mutant (red). (C) Membrane potential was analyzed using DiOC(3)2 staining of the wild type, fakA mutant, and complemented fakA mutant (comp). (D) Respiratory activity of the wild type, fakA mutant, and complemented fakA mutant as measured using CTC as an indicator. Data represent the means (n = 3) with SD. *, P < 0.05 by Student’s t test. “#” indicates that all samples treated with OA differed significantly from the corresponding strain in TSB alone. Data are representative of those from at least three independent experiments.
We next assessed whether the absence of FakA altered membrane integrity using propidium iodide (PI) as an indicator, since this stain cannot diffuse into cells unless membranes are compromised. Cells were grown in TSB and membrane integrity was determined in the presence and absence of 0.01% oleic acid. When grown in TSB alone until mid-exponential phase, the wild type and fakA mutant had approximately 3% and 1.1% PI-positive cells, respectively (Fig. 5B). However, when cells were grown in the presence of 0.01% oleic acid, 16.4% of wild-type cells were PI positive, compared to 2.8% for the fakA mutant. Thus, when grown in TSB, the fakA mutant has less permeable membranes and the permeability is less affected by the presence of oleic acid than for the wild-type strain.
Membrane potential is important for driving ATP production and transporting ions and metabolites into and out of the cell. We determined the membrane potential (ΔΨ) of wild-type and fakA mutant S. aureus using the fluorescent reporter 3,3′-diethyloxacarbocyanine iodide [DiOC2(3)] (38, 39). The wild type and fakA had similar membrane potentials when grown in TSB alone (Fig. 5C). When grown in the presence of 0.01% oleic acid, both strains had decreased membrane potentials compared to that when grown in TSB alone, but this was enhanced in the fakA mutant, with a 40% reduction compared to a 20% reduced potential in the wild type. Thus, under these conditions, oleic acid decreases membrane potential and this may be enhanced in a fakA mutant.
Electron transport chain activity was measured using the fluorescent reporter 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) (39). This reporter is reduced by respiratory dehydrogenases and is thus indicative of cellular respiration. In TSB alone, the fakA mutant had significantly more respiratory activity than that of the wild type (Fig. 5D). However, both the wild type and fakA mutant display decreased respiratory activity in the presence of incorporation-level oleic acid, which correlates with reduced growth (Fig. S3). Overall, these studies demonstrate that the fakA mutant displays phenotypes that are associated with membrane changes.
Lipid profile is altered in the presence of mouse skin homogenate.
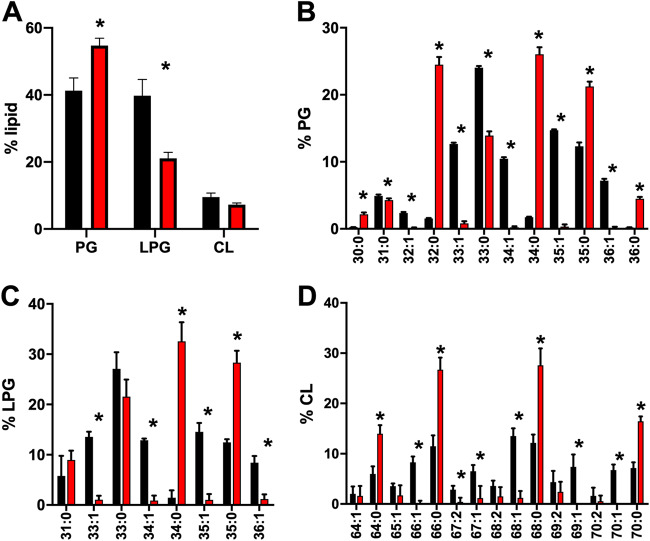
The most common infection site of S. aureus is the skin, which contains high levels of various fatty acids and lipids (40). These molecules have antimicrobial properties and limit the growth of certain bacteria. It is unknown if S. aureus can utilize fatty acids from mouse skin, though it harvests fatty acids from other host molecules, such as low-density lipoprotein (41). To gain insight into this, we determined (i) if S. aureus alters its lipids in response to mouse skin and (ii) if S. aureus incorporates host fatty acids. To this end, we grew wild-type and fakA mutant S. aureus in a combination of 75% mouse skin homogenate and 25% TSB. This ratio was chosen based on relatively high bacterial growth (optical density at 600 nm [OD600] of 3.5 to 4.5) to allow for quality extraction of bacterial lipids. After 24 h of growth in 75% mouse skin homogenate, cells were isolated and lipids were extracted and analyzed via LC-MS/MS. To ensure that we did not collect mouse skin lipids without bacterial cells, one sample of 75% mouse skin homogenate and 25% TSB without bacterial cells was treated identically to the bacterial cells we extracted. This sample without bacteria was also analyzed via LC-MS/MS, and we found only trace amounts of lipids (Fig. S4), demonstrating that our data represent lipids isolated from S. aureus and not contaminants from the mouse skin. Under this growth condition, we observed equal levels of PG and LPG in the wild-type strain. Unlike in TSB, we saw a pronounced difference in the fakA mutant compared to the wild type. In this case, the fakA mutant resembled membranes of cells grown in TSB alone, with PG being the most abundant, followed by LPG, and CL being the least abundant lipid. The most abundant saturated PG species were 33:0 and 35:0 in the wild type, while the fakA mutant had a relatively even distribution of 32:0, 34:0, and 35:0. Using the natural unsaturated fatty acids found in mouse skin as a marker of host fatty acid utilization, we observed considerable incorporation of unsaturated fatty acids as a component of the wild-type cell lipids (Fig. 6B to D). The unsaturated PG species 33:1, 34:1, 35:1, and 36:1 were all found in S. aureus grown in mouse skin homogenate. Similar trends were observed for LPG and CL species. We did not identify lipids containing more than one fatty acid unsaturation. As expected, we did not find unsaturated fatty acids in the lipids of the fakA mutant. In conclusion, we showed that S. aureus can utilize unsaturated fatty acids found in mouse skin homogenate for lipid synthesis.
FIG 6.
Lipid profile of the wild type (black) and fakA mutant (red) grown in 75% mouse skin homogenate. (A) Total percent lipid signal of the wild type and fakA mutant by class of lipid. (B to D) Identification of PG (B), LPG (C), and CL (D) represented by the percentage of the total from each class. Data represents the means (n = 4) with standard deviations from a representative experiment. *, P < 0.05 by Student’s t test.
DISCUSSION
The exogenous fatty acid utilization system of S. aureus has recently emerged as a new metabolic pathway and is made up of at least the fatty acid kinase FakA and the fatty acid-binding partner proteins FakB1 and FakB2 (18, 20). Despite the characterization of this system, how inactivation of this pathway (via deletion of fakA) affects the composition of the phospholipid membrane has remained unknown. S. aureus is routinely cultivated in TSB, which is abundant in carbohydrates, protein, and fatty acids, primarily palmitic acid (16:0) and stearic acid (18:0). Not surprisingly, based on a previous report, we identified an accumulation of fatty acids in a fakA mutant under our growth conditions. However, our studies revealed that this changes over the course of growth and showed that not only do the fakA mutant cells accumulate fatty acids, but also fatty acids are released into the media, where they also increase in abundance. It is unknown whether this accumulation of fatty acids in the extracellular growth environment is an intentional process (i.e., release of fatty acids from the cell), due to cell death and turnover, or whether they are actively released by an efflux system that has been proposed previously (29). Since only free fatty acids were identified in this experiment, the fatty acids found in the supernatant were likely not part of extracellular membrane vesicles produced by S. aureus. The consequence of altered fatty acid metabolism in the fakA mutant was not a change in the ratio of primary lipid species but a reorganization of the fatty acid phospholipid side chains. Moreover, we demonstrate that the addition of excess 18:1 fatty acid led to its incorporation into wild-type cells but also a change in the fatty acid pools in both the wild type and fakA mutant.
S. aureus is known to produce primarily three phospholipids (8, 42–44). Phosphatidylglycerol (PG) is the primary phospholipid and is the precursor for the generation of additional phospholipids. Lysyl-PG (LPG) is produced by MprF, and while S. aureus has two cardiolipin (CL) synthases, cardiolipin is the least abundant species. As expected, our results demonstrated that both wild-type and fakA mutant membranes contained primarily PG and LPG. The amount of CL can vary by strain and condition, and we detected only 4.5% of total phospholipids as cardiolipin in our USA300 S. aureus strain at either the exponential or stationary phase of growth, showing that LAC derivatives grown under our conditions produce little CL.
S. aureus generally prefers endogenously produced 15:0 in the sn2 position of the phospholipids and inserts either a second endogenous fatty acid or exoFA in the sn1 position. Our product ion analysis of the wild-type and fakA mutant phospholipids most often showed a 15:0 fatty acid. However, the fakA mutant paired this 15:0 more frequently with longer-chain fatty acids (Fig. 2) than the parent strain. Phospholipids of lower total carbon abundance (i.e., 30:0, 31:0, 32:0, and 33:0) were more abundant in the wild type than in the fakA mutant. These phospholipids all contained 15:0 in one sn position, with the other acyl chain being 15:0, 16:0, 17:0, and 18:0 in phospholipids 30:0, 31:0, 32:0, and 33:0, respectively. The most abundant phospholipids observed in the wild type were 32:0 and 33:0. In contrast, the fakA mutant tended to have larger fatty acids as part of phospholipids, shifting to 35:0 in both PG and LPG due to the insertion of 20:0 alongside 15:0. This was also reflected in the free fatty acid pools in the fakA mutant (see Fig. S2 in the supplemental material), showing that this mutant possesses and uses a greater abundance of longer fatty acids, such as 19:0 and 20:0. However, the presence of exogenous oleic acid altered the normal relationship of use of 15:0 alongside a varying-length second fatty acid. In this case, the wild type generated lipids with 18:1 oleic acid in combination with not only 15:0 but also commonly 14:0. One observation made from this study, and in agreement with those of Parsons et al. (33) in strain RN4220, is the abundance of fatty acid 20:1 in wild-type S. aureus. Since oleic acid (18:1) was the exogenous fatty acid added to the media, this demonstrates that S. aureus can elongate exoFAs. This was a frequent event, since 35:1 composed of 15:0 and 20:1 was the most abundant lipid species identified in the wild type when grown in the presence of oleic acid (Fig. 3 and Table S1). Our FAME analysis confirmed that the lack of incorporation of 18:1 into the fakA mutant is not due to the inability to associate with the cells and is a result of the inability of the fakA mutant to phosphorylate the fatty acid, as expected. The presence of 20:1 has been observed previously, and while the exact mechanism behind exoFA elongation has not been elucidated, it has been proposed to occur through FabF, part of the fatty acid biosynthesis pathway (45). However, our observation that there was no 20:1 fatty acid in the fakA mutant demonstrates that elongation of exoFAs requires FakA and that only phosphorylated fatty acids can be elongated by S. aureus.
In previous studies, we found that oleic acid can inhibit the Sae two-component system when added exogenously to the medium at 0.001% (25). This was later confirmed by another group (26). Considering this transcriptional response, we anticipated that this concentration would yield membranes containing oleic acid. However, oleic acid-containing phospholipids were not detected when S. aureus was grown in the presence of 0.001% (∼31 μM) oleic acid. When the concentration of oleic acid was increased 10-fold to 0.01% (∼314 μM), we observed incorporation of oleic acid into lipids. Thus, there is an apparent concentration-dependent incorporation of exogenous unsaturated fatty acids by S. aureus. Moreover, these data revealed that S. aureus can sense and respond transcriptionally to levels of fatty acids that are lower than those used for membrane synthesis, further supporting fatty acids as potent signaling molecules in S. aureus.
The membrane is a dynamic structure that dictates multiple functions of the cell. We identified functional changes between wild-type and fakA mutant S. aureus. In TSB alone, the fakA mutant has more rigid membranes regardless of pigment production. Since pigment impacts membrane fluidity and the fakA mutant produces less pigment, this could confound membrane fluidity experiments with this strain. Our data confirm that pigment does impact membrane fluidity (Fig. 5), but they also demonstrate that there is a FakA-dependent component. This could possibly be due to increases in the acyl chain length of the phospholipids (Fig. 2C), as longer acyl chains result in a more rigid membrane. Additionally, a slight decrease in BCFA/SCFA ratios in the fakA mutant (Table S2) could increase rigidity. To our surprise, addition of 0.01% oleic acid decreased membrane fluidity of both the wild type and fakA mutant. While the unsaturated fatty acid linoleic acid (18:2) has been shown to increase membrane fluidity (46), our data clearly demonstrate that growth in the presence of oleic acid does not result in similar membrane fluidity. There may be two explanations for this result. First, this may be due to the difference in structure of the fatty acids. The presence of two unsaturations in linoleic acid may be more disruptive to fluidity than oleic acid. Second, the addition of 0.01% oleic acid dramatically decreases the BCFA/SCFA ratio in both the wild type and fakA mutant. Wild-type cells contained 38% and greater than 75% SCFAs when grown in TSB without and with 0.01% oleic acid, respectively. SCFAs, like oleic acid, are known to affect membrane fluidity, and our data are consistent with the increased abundance of SCFAs in the presence of oleic acid decreasing membrane fluidity. It appears that oleic acid impacts membrane fluidity without the requirement of being incorporated, and this is influenced by the presence of the pigment staphyloxanthin. Notably, previous work suggests that pigment levels are not influenced by fatty acid incorporation (47). Dissecting this relationship between fatty acid addition, fatty acid utilization, and S. aureus pigmentation will be the focus of future studies.
We have established that the fakA mutant has altered metabolism (27). Since the electron transport chain is embedded in the phospholipid membrane, we sought to determine whether the changes in membrane composition in the fakA mutant and the presence of 0.01% oleic acid altered cellular respiration. The fakA mutant had more respiratory activity than the wild type in TSB alone (Fig. 5D). This agrees with our metabolic study showing that the fakA mutant had an increase in NAD+/NADH and NADP+/NADPH levels at the same phase of growth. Growth in TSB plus 0.01% oleic acid resulted in decreased respiration in both the wild type and the fakA mutant. While the increased respiration of the fakA mutant in TSB did not affect membrane potential, growth in TSB plus 0.01% oleic acid depolarized both the wild type and fakA mutant; however, the fakA mutant was more greatly affected (Fig. 5C). Previous studies indicated that palmitoleic acid (16:1), and not oleic acid (18:1), depolarized membranes (45). It should be noted that we used approximately three times the concentration of oleic acid as the study mentioned, and this can likely account for the differences between experiments. Additionally, the study mentioned above was published prior to the identification of FakA, and therefore, incorporation versus free fatty acid exposure could not be tested. Thus, our data indicate that oleic acid will decrease respiration and depolarize membranes at 0.01% (314 μM) regardless of being incorporated into phospholipids. Why the fakA mutant has more respiratory activity in TSB is unknown; however, this observation occurred at a point where we identified enhanced growth in the fakA mutant during the switch between glucose and acetate/amino acid utilization (27). Thus, it may be not surprising to see increased respiratory activity during periods of enhanced growth. Why FakA influences metabolism has not been completely elucidated. This could be due to changes in the membrane that may influence nutrient diffusion or transport. Another possibility is that it is the result of the accumulation of fatty acids, which could have different activities or impact levels based on saturation. A recent study indicated that inactivation of type II NADH dehydrogenases (Ndh-2s), particularly NdhC, results in fatty acid accumulation and regulation of the SaeRS two-component system (48). Ndh-2s do not pump ions across the membrane, thus playing an indirect role in membrane potential. In Escherichia coli, Ndh-2s are associated with aerobic respiration, while Ndh-1s are associated with anaerobic respiration (49). Since the fakA mutant possesses an aeration-dependent growth enhancement and has increased respiratory activity without a significant increase in membrane potential, we predict that Ndh-2s play a pivotal role in altering metabolism when fatty acids are accumulated within the cell or in the presence of exogenous fatty acids. Studies testing how different types of exogenous fatty acids affect respiration through Ndh-2s, as well as other members of the respiratory chain, will help us understand the mechanism by which fatty acids affect bacterial physiology. It is clear that the inability of S. aureus to use fatty acids and/or exposure to exogenous fatty acids impacts physiology. This has been seen by us and others as changes in the glucose, acetate, amino acid metabolism, and energy or redox homeostasis that apparently involve at some level NADH dehydrogenases and regulatory proteins such as CcpA, as well as the transcriptional response in arginine deiminase and urease (27, 28, 45, 48, 50, 51). It is also possible that S. aureus identifies unsaturated fatty acids as a stress, as we previously saw that enhanced resistance to unsaturated fatty acids in a fakA mutant requires the alternative sigma factor σB (28). Future studies will be needed to determine the specific mechanism by which fatty acid addition modulates metabolism in S. aureus and whether this is a general fatty acid response or is restricted to certain fatty acid types.
We wanted to expand our observations to more closely mimic what the bacteria may encounter during infection. Since S. aureus commonly causes infections in the skin, we wanted to determine if S. aureus can actively scavenge unsaturated fatty acids for phospholipid synthesis from host tissue. To do this, we homogenized mouse skin and grew wild-type and fakA mutant S. aureus in 75% mouse skin homogenate supplemented with 25% TSB. While S. aureus can grow relatively well in 100% mouse skin homogenate, we added 25% TSB to ensure high bacterial growth. We confirmed that S. aureus can scavenge exogenous fatty acids when grown in mouse skin homogenate. We did observe lipidome changes in wild-type S. aureus grown in mouse skin compared to the organism grown in TSB plus 0.01% oleic acid. For example, growth in mouse skin resulted in wild-type S. aureus having relatively similar levels of PG 33:0, PG 33:1, PG 35:0, and PG 35:1. This differs from growth in TSB plus 0.01% oleic acid, in which PG 35:1 clearly became the most abundant phospholipid. While other studies have found that S. aureus can utilize fatty acids from a host source, specifically low-density lipoprotein (LDL) and pig liver homogenate (41, 52), none have used homogenized skin as a medium. Given the propensity of S. aureus to commonly infect the skin, our data provide a framework for future studies utilizing skin as a source of fatty acids and for how bacterial phospholipid membranes adapt to skin tissue. How S. aureus scavenges fatty acids from mouse skin and whether they are derived from free fatty acids, lipids, or LDL is one component of ongoing studies.
The exoFA utilization pathway of FakB1, FakB2, and FakA is newly described (18, 24). Despite this pathway being necessary for the incorporation of exoFAs into the membrane, a detailed analysis of how this system alters membrane composition has not been performed. Not surprisingly, our results show no change in the PG/LPG/CL ratio by the absence of FakA, but there was a reorganization of the fatty acid side chains, e.g., change in fatty acid length preference, of S. aureus phospholipids in the absence of FakA. These studies provide key details on how membrane composition changes in response to exoFAs regardless of whether they are used for phospholipid synthesis. Furthermore, this work expands our knowledge on how the activity of FakA influences S. aureus physiology.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. aureus was grown in tryptic soy broth (TSB) or tryptic soy agar supplemented with chloramphenicol (10 μg ml−1) or erythromycin (5 μg ml−1) when necessary. S. aureus cultures were then inoculated at a 1:10 medium-to-flask volume ratio at an initial optical density at 600 nm of 0.1 in TSB and grown at 37°C with shaking at 250 rpm. TSB was supplemented with oleic acid (Alfa Aesar) by adding the desired percentage (volume/volume) of oleic acid to media and vortexed vigorously before being aliquoted into flasks.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| AH1263 | USA300 CA-MRSA strain LAC without LAC-p03 | 58 |
| JLB2 | AH1263 ΔfakA | 24 |
| JLB112 | AH1263 crtM::ΝΣ | 28 |
| JLB129 | JLB2 crtM::ΝΣ | 28 |
| pJB165 | fakA complement plasmid | 24 |
“ΝΣ” indicates mutations originating from the Nebraska Transposon Mutant Library.
Fatty acid analysis.
Cell pellets and supernatants were flash frozen in liquid nitrogen and stored at –80°C until processed. The cell pellets were washed three times with 1 ml of 40 mM ammonium formate in water. The washed pellets were resuspended in 50 μl of 5 mM ammonium acetate and homogenized three times for 30 s with a BeadBeater, with 15-min rests on ice in between. Protein concentrations of the homogenates were measured, and each sample was normalized to 500 μg ml−1 and aliquoted at 25 μl for extraction. The supernatants were not normalized and aliquoted at 125 μl for extraction. All samples were extracted by protein precipitation with 1 ml of 80%:20% methanol-water for 20 min at 4°C. The samples were centrifuged at 20,000 × g for 10 min at 4°C. One milliliter of supernatant was collected and dried down under nitrogen gas at 30°C. The samples were reconstituted at 25 μl of 80:20 acetonitrile–5 mM ammonium acetate in water. Each sample was spiked with 1 μl of fatty acid-stable isotope standards.
Analysis of the free fatty acids was performed on a Thermo Q-Exactive Orbitrap mass spectrometer with a Dionex ultrahigh-performance liquid chromatograph (UHPLC) and autosampler. All samples were analyzed in negative heated electrospray ionization in data-dependent MS/MS with a mass resolution of 70,000 in full MS and 17,500 at m/z 200 in data-dependent MS/MS. Separation was achieved on an Acquity ultraperformance liquid chromatography (UPLC) high-strength silica T3 2.1- by 150-mm, 1.8-μm column with 1 mM ammonium acetate as mobile phase A and 0.1% acetic acid in acetonitrile as mobile phase B. The flow rate was 500 μl min−1, with a column temperature of 30°C. The injection volume was 4 μl. Peak areas were integrated for each analyte and internal standards using Xcalibur.
ESI-MS/MS lipid profiling.
An automated electrospray ionization-tandem mass spectrometry (ESI-MS/MS) approach was used, and data acquisition and analysis and acyl group identification were carried out as described previously (53), with modifications. Each sample was dissolved in 1 ml of chloroform and an aliquot of 2 to 39 μl of extract was used.
For analysis of PG, phosphatidic acid (PA), monogalactosyldiacylglycerol (monohexDAG), and digalactosyldiacylglycerol (dihexDAG), phospholipid and galactolipid internal standards, obtained and quantified as previously described (54), were added in the amounts described by Narayanan et al. (55), except for phosphatidylinositol (PI) (16:0/18:0) (0.28 nmol), PI (18:0/18:0) (0.11 nmol), digalactosyldiacylglycerol (DGDG) (18:0/16:0) (0.44 nmol), DGDG (18:0/18:0) (1.48 nmol), monogalactosyldiacylglycerol (MGDG) (18:0/16:0) (1.67 nmol), MGDG (18:0/18:0) (1.41 nmol), and cardiolipin (CL) (14:0/14:0/14:0/14:0) (0.015 nmol). The sample and internal standard mixture were combined with solvents such that the ratio of chloroform, methanol, and 300 mM ammonium acetate in water was 300/665/35 and the final volume was 1.4 ml. Analysis of PG, PA, monohexDAG, and dihexDAG was performed on unfractionated lipid extracts introduced by continuous infusion into the ESI source on a triple quadrupole MS/MS (API 4000; Applied Biosystems, Foster City, CA). Data were collected and analyzed as previously described for analysis of PG, PA, monohexDAG, and dihexDAG (55).
For CL and LPG analysis, phospholipid standards were added at one-fifth of the amounts indicated above. CL and LPG were analyzed on a triple quadrupole MS/MS (Xevo TQS; Waters Corp., Milford, MA). Samples were introduced to the Xevo TQS using a Waters 2777 Sample Manager autosampler. Lipid species were detected with the following scans: LPG [M-H]− in negative-ion mode with Precursor 145.0 and cardiolipin in negative ion mode with Precursor 153.0. The scan speed was 50 or 100 u per s. The Xevo TQS in negative-ion mode has the following parameters for CL and LPG: capillary, 2.8 kV; cone voltage, 40 V; source temperature, 150°C; desolvation temperature, 120°C; collision gas, 0.14 ml/min; and nebulizer gas flow, 7 bars. Collision energies on the Xevo TQS, with argon in the collision cell, were −75 V for CL and −35 V for LPG.
Data processing was similar to that described previously, including background subtraction, smoothing, integration of spectral peaks, isotopic deconvolution, correction for chemical or instrumental noise, and quantification (54, 56). Internal standards of the same class were used for PA, PG, and cardiolipin. LPG was quantified against PG (40:0) in negative Precursor mode, and monohexDAG and dihexDAG were quantified against PG (40:0) in positive neutral loss (NL) mode. Finally, the data were corrected for the fraction of the sample analyzed and normalized to signal per CFU to produce data in the units per nanomole per 1e10 CFU.
Product ion analyses were performed on pooled replicates of the treatments on a 4000 QTrap mass spectrometer in enhanced product ion mode. Aliquots of 11 to 14 μl were used. Collision energy was varied as needed for sufficient fragmentation, starting at −45 V. The curtain gas was 10 ml/min, collisionally activated dissociation (CAD) gas was the medium, and the electrospray capillary voltage was −4,500 V.
Membrane fluidity, integrity, potential, and electron transport chain activity.
Membrane fluidity for wild-type and fakA mutant strains was determined as previously described (57). Membrane integrity was determined using propidium iodide (PI) as an indicator. Wild-type and fakA mutant bacteria were grown in TSB with or without 0.01% (314 μM) oleic acid for 5 h. A total of 1.2 ml of culture was spun down and resuspended in 600 μl of phosphate-buffered saline (PBS). The cells were then diluted to an OD600 of 0.25 in 500 μl in PBS. As a control, one sample was diluted to an OD600 of 0.25 in 500 μl of 70% ethanol. One microliter of PI (50 mg/ml) was added to the cells and allowed to incubate at room temperature for 15 min. PI-positive cells were determined using flow cytometry.
Membrane potential was determined using the BacLight bacterial membrane potential kit (Molecular Probes, Invitrogen) (39). Wild-type and fakA mutant bacteria were grown for 5 h. Cultures were diluted to an OD600 of 0.1 in 1 ml of PBS. As a negative control, 10 μl of 500 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to one sample and vortexed. Ten microliters of 3 mM DiOC2(3) was then added to all samples, excluding a no-stain control, and vortexed. Samples were then allowed to incubate at room temperature for 30 min and analyzed via flow cytometry. The data are presented as the geometric mean ratios of red to green using Texas Red and fluorescein isothiocyanate (FITC) filters, respectively.
Electron transport chain activity (respiratory activity) was determined using the BacLight RedoxSensor CTC Vitality kit (Molecular Probes, Invitrogen) (39). Wild-type and fakA mutant bacteria were grown for 5 h. Cultures were diluted to an OD600 of 0.1 in 650 μl of PBS. One sample was diluted in 70% ethanol as a negative control. A total of 65 μl of 50 mM CTC was added to each sample, except for a no-stain control. The samples were then incubated in the dark at 37˚C for 30 min and analyzed using a Tecan Spark 10M plate reader (excitation wavelength, 485 ± 20 nm, and emission wavelength, 645 ± 40 nm).
Generation of mouse skin homogenates.
These studies were conducted under an approved protocol by the Institutional Animal Care and Use Committee (IACUC) of the University of Kansas Medical Center. We used mice of the C57BL/6 background. Following sacrifice, mice were shaved and treated with Nair to remove remaining hair. Next, skin was removed and homogenized in PBS using Lysing Matrix H with a FastPrep-24 5G using the manufacturer’s setting. Homogenates were pooled and sequentially passed through 40-μm and 0.45-μm filters.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by funding from National Institute of Allergy and Infectious Diseases (NIAID) award R01AI121073 to J.L.B. and a pilot project grant through award P20GM112117. Instrument acquisition and lipidomics method development were supported by the National Science Foundation (including support from the Major Research Instrumentation program; current award DBI-1726527), K-IDeA Networks of Biomedical Research Excellence (INBRE) of the National Institutes of Health (P20GM103418), USDA National Institute of Food and Agriculture (Hatch/Multi-State project 1013013), and Kansas State University. We are grateful to Tim Garrett at the Southeast Center for Metabolomics for the free fatty acid analysis done for this research (NIH grant number U24 DK097209).
We thank Miranda Ridder for the help in harvesting mouse skin for the mouse skin homogenate experiments. The lipid analyses described in this work were performed at the Kansas Lipidomics Research Center Analytical Laboratory and would not have been possible without Ruth Welti and Mary Roth.
Jeffrey L. Bose serves on the Scientific Advisory Board and is a consultant for Azitra, Inc. These activities did not financially support and are unrelated to this article.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 3.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr.. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klevens RM, Active Bacterial Core surveillance (ABCs) MRSA Investigators, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 5.Knox J, Uhlemann AC, Lowy FD. 2015. Staphylococcus aureus infections: transmission within households and the community. Trends Microbiol 23:437–444. doi: 10.1016/j.tim.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler CE, Ernst RK. 2017. Bacterial lipids: powerful modifiers of the innate immune response. F1000Res 6:1334. doi: 10.12688/f1000research.11388.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen MT, Gotz F. 2016. Lipoproteins of Gram-positive bacteria: key players in the immune response and virulence. Microbiol Mol Biol Rev 80:891–903. doi: 10.1128/MMBR.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KPM, van Strijp J. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor Mprf is based on modification of membrane lipids with l-lysine. J Exp Med 193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst CM, Peschel A. 2019. MprF-mediated daptomycin resistance. Int J Med Microbiol 309:359–363. doi: 10.1016/j.ijmm.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J-H, Bhuiyan MS, Shen H-H, Cameron DR, Rupasinghe TWT, Wu C-M, Le Brun AP, Kostoulias X, Domene C, Fulcher AJ, McConville MJ, Howden BP, Lieschke GJ, Peleg AY. 2019. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc Natl Acad Sci U S A 116:3722–3727. doi: 10.1073/pnas.1812066116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowds CM, Kornell S-C, Blumberg RS, Zeissig S. 2014. Lipid antigens in immunity. Biol Chem 395:61–81. doi: 10.1515/hsz-2013-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Defourny KAY, Smid EJ, Abee T. 2018. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9:1502. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White SW, Zheng J, Zhang Y-M, Rock CO. 2005. The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Ma S. 2013. Recent advances in inhibitors of bacterial fatty acid synthesis type II (FASII) system enzymes as potential antibacterial agents. ChemMedChem 8:1589–1608. doi: 10.1002/cmdc.201300209. [DOI] [PubMed] [Google Scholar]
- 15.Lu H, Tonge PJ. 2008. Inhibitors of FabI, an enzyme drug target in the bacterial fatty acid biosynthesis pathway. Acc Chem Res 41:11–20. doi: 10.1021/ar700156e. [DOI] [PubMed] [Google Scholar]
- 16.Omura S. 1976. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev 40:681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio Á, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 18.Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, Rock CO. 2014. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci U S A 111:10532–10537. doi: 10.1073/pnas.1408797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn S, Slavetinsky CJ, Peschel A. 2015. Synthesis and function of phospholipids in Staphylococcus aureus. Int J Med Microbiol 305:196–202. doi: 10.1016/j.ijmm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. 2014. Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol Microbiol 92:234–245. doi: 10.1111/mmi.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen S, Sirobhushanam S, Johnson SR, Song Y, Tefft R, Gatto C, Wilkinson BJ. 2016. Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PLoS One 11:e0165300. doi: 10.1371/journal.pone.0165300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser JC, Sen S, Sinha A, Wilkinson BJ, Heinrichs DE. 2016. The role of two branched-chain amino acid transporters in Staphylococcus aureus growth, membrane fatty acid composition and virulence. Mol Microbiol 102:850–864. doi: 10.1111/mmi.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh VK, Hattangady DS, Giotis ES, Singh AK, Chamberlain NR, Stuart MK, Wilkinson BJ. 2008. Insertional inactivation of branched-chain alpha-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl Environ Microbiol 74:5882–5890. doi: 10.1128/AEM.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose JL, Daly SM, Hall PR, Bayles KW. 2014. Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun 82:1813–1822. doi: 10.1128/IAI.01655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krute CN, Rice KC, Bose JL. 2017. VfrB is a key activator of the Staphylococcus aureus SaeRS two-component system. J Bacteriol 199:e00828-16. doi: 10.1128/jb.00828-16:JB.00828-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ericson ME, Subramanian C, Frank MW, Rock CO. 2017. Role of fatty acid kinase in cellular lipid homeostasis and SaeRS-dependent virulence factor expression in Staphylococcus aureus. mBio 8:e00988-17. doi: 10.1128/mBio.00988-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demars Z, Bose JL. 2018. Redirection of metabolism in response to fatty acid kinase in Staphylococcus aureus. J Bacteriol 200:e00345-18. doi: 10.1128/JB.00345-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krute CN, Ridder MJ, Seawell NA, Bose JL. 2019. Inactivation of the exogenous fatty acid utilization pathway leads to increased resistance to unsaturated fatty acids in Staphylococcus aureus. Microbiology 165:197–207. doi: 10.1099/mic.0.000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alnaseri H, Kuiack RC, Ferguson KA, Schneider JET, Heinrichs DE, McGavin MJ. 2018. DNA binding and sensor specificity of FarR, a novel TetR family regulator required for induction of the fatty acid efflux pump FarE in Staphylococcus aureus. J Bacteriol 201:e00602-18. doi: 10.1128/JB.00602-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 31.Koprivnjak T, Zhang D, Ernst CM, Peschel A, Nauseef WM, Weiss JP. 2011. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J Bacteriol 193:4134–4142. doi: 10.1128/JB.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst CM, Staubitz P, Mishra NN, Yang S-J, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. 2011. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci U S A 108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braungardt H, Singh VK. 2019. Impact of deficiencies in branched-chain fatty acids and staphyloxanthin in Staphylococcus aureus. BioMed Res Int 2019:2603435–2603438. doi: 10.1155/2019/2603435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subczynski WK, Wisniewska A. 2000. Physical properties of lipid bilayer membranes: relevance to membrane biological functions. Acta Biochim Pol 47:613–625. doi: 10.18388/abp.2000_3983. [DOI] [PubMed] [Google Scholar]
- 36.Gruszecki WI, Strzałka K. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim Biophys Acta 1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, Bayer AS. 2011. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother 55:526–531. doi: 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mogen AB, Carroll RK, James KL, Lima G, Silva D, Culver JA, Petucci C, Shaw LN, Rice KC. 2017. Staphylococcus aureus nitric oxide synthase (saNOS) modulates aerobic respiratory metabolism and cell physiology. Mol Microbiol 105:139–157. doi: 10.1111/mmi.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis AM, Matzdorf SS, Endres JL, Windham IH, Bayles KW, Rice KC. 2015. Examination of the Staphylococcus aureus nitric oxide reductase (saNOR) reveals its contribution to modulating intracellular NO levels and cellular respiration. Mol Microbiol 96:651–669. doi: 10.1111/mmi.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiezel-Tsugunova M, Kendall AC, Nicolaou A. 2018. Fatty acids and related lipid mediators in the regulation of cutaneous inflammation. Biochem Soc Trans 46:119–129. doi: 10.1042/BST20160469. [DOI] [PubMed] [Google Scholar]
- 41.Delekta PC, Shook JC, Lydic TA, Mulks MH, Hammer ND. 2018. Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of fatty acids. J Bacteriol 200:e00728-17. doi: 10.1128/JB.00728-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayami M, Okabe A, Kariyama R, Abe M, Kanemasa Y. 1979. Lipid composition of Staphylococcus aureus and its derived L-forms. Microbiol Immunol 23:435–442. doi: 10.1111/j.1348-0421.1979.tb00483.x. [DOI] [PubMed] [Google Scholar]
- 43.Mishra NN, Bayer AS. 2013. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:1082–1085. doi: 10.1128/AAC.02182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay K, Whitmire W, Xiong YQ, Molden J, Jones T, Peschel A, Staubitz P, Adler-Moore J, McNamara PJ, Proctor RA, Yeaman MR, Bayer AS. 2007. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153:1187–1197. doi: 10.1099/mic.0.2006/003111-0. [DOI] [PubMed] [Google Scholar]
- 45.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. 2012. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol 194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez MS, Tan IS, Yan D, Kang J, McCreary M, Modrusan Z, Austin CD, Xu M, Brown EJ. 2017. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc Natl Acad Sci U S A 114:11223–11228. doi: 10.1073/pnas.1700627114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiwari K, Gatto C, Wilkinson B. 2018. Interrelationships between fatty acid composition, staphyloxanthin content, fluidity, and carbon flow in the Staphylococcus aureus membrane. Molecules 23:1201. doi: 10.3390/molecules23051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schurig-Briccio LA, Parraga Solorzano PK, Lencina AM, Radin JN, Chen GY, Sauer JD, Kehl-Fie TE, Gennis RB. 2020. Role of respiratory NADH oxidation in the regulation of Staphylococcus aureus virulence. EMBO Rep 2020:e45832. doi: 10.15252/embr.201845832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 50.Neumann Y, Ohlsen K, Donat S, Engelmann S, Kusch H, Albrecht D, Cartron M, Hurd A, Foster SJ. 2015. The effect of skin fatty acids on Staphylococcus aureus. Arch Microbiol 197:245–267. doi: 10.1007/s00203-014-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenny JG, Ward D, Josefsson E, Jonsson I-M, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. doi: 10.1371/journal.pone.0004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morvan C, Halpern D, Kénanian G, Hays C, Anba-Mondoloni J, Brinster S, Kennedy S, Trieu-Cuot P, Poyart C, Lamberet G, Gloux K, Gruss A. 2016. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat Commun 7:12944. doi: 10.1038/ncomms12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. 2006. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67:1907–1924. doi: 10.1016/j.phytochem.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Welti R, Li M, Li W, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X. 2002. Profiling membrane lipids in plant stress response. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 55.Narayanan S, Tamura PJ, Roth MR, Prasad PV, Welti R. 2016. Wheat leaf lipids during heat stress: I. High day and night temperatures result in major lipid alterations. Plant Cell Environ 39:787–803. doi: 10.1111/pce.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A 94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh VK, Sirobhushanam S, Ring RP, Singh S, Gatto C, Wilkinson BJ. 2018. Roles of pyruvate dehydrogenase and branched-chain α-keto acid dehydrogenase in branched-chain membrane fatty acid levels and associated functions in Staphylococcus aureus. J Med Microbiol 67:570–578. doi: 10.1099/jmm.0.000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.