Abstract
Five new members of the salinilactone family, salinilactones D–H, are reported. These bicyclic lactones are produced by Salinispora bacteria and display extended or shortened alkyl side chains relative to the recently reported salinilactones A–C. They were identified by GC/MS, gas chromatographic retention index, and comparison with synthetic samples. We further investigated the occurrence of salinilactones across six newly proposed Salinispora species to gain insight into how compound production varies among taxa. The growth‐inhibiting effect of this compound family on multiple biological systems including non‐Salinispora actinomycetes was analyzed. Additionally, we found strong evidence for significant cytotoxicity of the title compounds.
Keywords: A-factor, biosynthesis, GC/MS, microbial volatiles, toxic compounds
Finding further function: Five new salinilactones have been identified by GC/MS and synthesis, thereby extending the family of salinilactones to eight compounds. Inhibition assays with actinomycetes, tracking of salinilactone production in Salinispora bacteria, and brine shrimp assays provided insight into the occurrence and biological purpose of salinilactones.
Introduction
Marine Salinispora bacteria are known for the production of a wide variety of natural products,1 often with promising biological activities, for example, salinipostins2 or salinosporamide A.3 Besides these metabolites, they also produce a wide variety of volatile compounds such as alcohols, ketones, esters, amides, ureas, imides, and sulfinamides.4 Recently, we discovered a new group of bicyclic lactones in the headspace of Salinispora cultures using a combination of GC/MS, coupled gas chromatography/direct deposition infrared spectroscopy (GC/DD‐FTIR), spectra calculations and synthesis.5 These compounds, called salinilactones A–C (Figure 1 A–C), are structurally related to the A‐factor family of γ‐butyrolactone (GBL) autoregulators6, 7 and also might have a signaling function. They display a unique bicyclo[3.1.0]lactone ring system not known from other natural products and are volatile in contrast to known GBL autoregulators.
Figure 1.
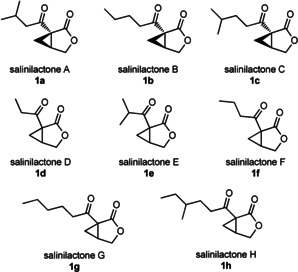
Structures of salinilactones A–H, 1 a–h.
Salinilactone biosynthesis has been linked to the spt gene cluster, with a knock‐out mutant of the afsA homologue spt9 in Salinispora arenicola CNS‐205 failing to produce both salinilactones and the related salinipostins.5, 8
Employing GC/MS analysis and synthesis, we were able to identify five additional salinilactones produced by Salinispora bacteria. We report here on the identification, synthesis and biological evaluation of these compounds.
Results and Discussion
Eleven Salinispora strains representing nine different species were analyzed for the presence of salinilactone derivatives in their headspace using closed‐loop‐stripping‐analysis (CLSA)9 and GC/MS. Five compounds with mass spectra similar to those of salinilactones A–C were detected and tentatively termed salinilactones D–H (Figure 1 D–H). Their structures were proposed based on their molecular mass, gas chromatographic retention indices I (Table 1) and mass spectra (Figure S1 in the Supporting Information).
Table 1.
Known salinilactones with gas chromatographic retention index I and molecular mass.
|
Compound |
I: nat./syn. |
Alkyl chain (R)[a] |
M r [Da] |
|---|---|---|---|
|
salinilactone A (1 a) |
1413/1410 |
2‐methylpropyl |
182 |
|
salinilactone B (1 b) |
1472/1468 |
n‐butyl |
182 |
|
salinilactone C (1 c) |
1528/1527 |
3‐methylbutyl |
196 |
|
salinilactone D (1 d) |
1287/1285 |
ethyl |
154 |
|
salinilactone E (1 e) |
1315/1315 |
isopropyl |
168 |
|
salinilactone F (1 f) |
1369/1368 |
n‐propyl |
168 |
|
salinilactone G (1 g) |
1566/1566 |
n‐pentyl |
196 |
|
salinilactone H (1 h) |
1634/1635 |
3‐methylpentyl |
210 |
|
1 i |
/1629 |
4‐methylpentyl |
210 |
[a] Referring to the general structure shown in Figure 2 A.
In GC, an additional methylene group in an alkyl chain results in an increase of I of about 100 units. When a lower increase is observed, a methyl‐branched chain is present. This general pattern led to the proposal of the side chain length and branching of the newly discovered bicyclic lactones. The characteristic ions at m/z 140 and 122 in the mass spectra, already known from salinilactones A–C,5 arise from McLafferty rearrangement and subsequent loss of water and facilitated the identification of salinilactones F to H (Figure 2).
Figure 2.
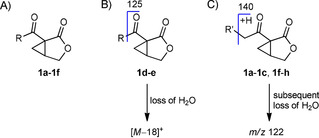
A) General structure of salinilactones 1 a–h; B) mass spectrometric fragmentation of short chain salinilactones; C) fragmentation of long chain salinilactones.
Salinilactone F showed an I=1369, 100 units lower than the value for salinilactone B with a butyl side chain, and was therefore proposed to be the n‐propyl analogue. Similarly, I=1566 indicated a n‐pentyl side chain in salinilactone G. Salinilactone H did not fit this pattern with I=1634. Therefore, a methyl branched side chain was proposed, either 3‐ or 4‐methylpentyl, because 2‐methylpentyl would likely show a different mass spectrum (Figure 3).
Figure 3.
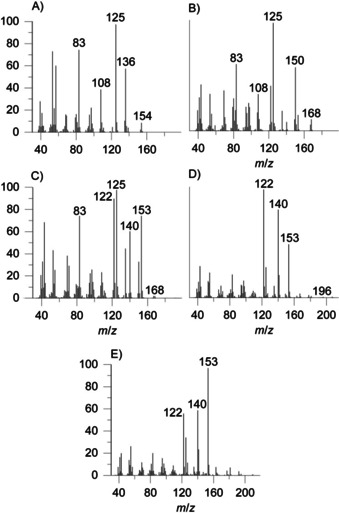
Mass spectra of synthetic salinilactones: A) 1 d (salinilactone D); B) 1 e (salinilactone E); C) 1 f (salinilactone F); D) 1 g (salinilactone G); E) 1 h (salinilactone H).
The mass spectra of the two salinilactones D and E do not display the ions m/z 140 and 122 because the alkyl chain is too short to undergo McLafferty rearrangement. Instead, an ion at m/z 125 is observed, resulting from ketone α‐cleavage (Figure 2). Also, both compounds display prominent fragments from water loss at m/z 136 and 150, respectively. Because salinilactone E has the same mass as salinilactone F, the only other possible side chain is isopropyl, fitting the shorter I=1315. Finally, salinilactone D with the lowest I=1287 likely had an ethyl side chain, evident by its M+ ion at m/z 154.
To support these structural proposals, compounds 1 d–i, were synthesized following the already established route using an intramolecular palladium‐catalyzed cyclization of enyne esters (Scheme 1).5, 10 The syntheses of the shorter side chain lactones started from the commercially available alkynes or alkynoic acids. The longer homologues 1 h and 1 i were synthesized from the corresponding alcohol or bromide from which the required alkynes 3 h and 3 i were produced by substitution with lithium acetylide. All these synthetic routes finally converged into the enyne ester synthesis. No asymmetric synthesis was undertaken, but so far all investigated salinilactones 1 a–c have a 1R,5S configuration.5
Scheme 1.
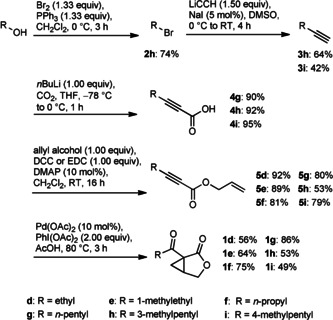
Synthesis of bicyclic lactones 1 d–i. The starting points of the synthetic routes differ depending on the commercial availability of precursors.
Mass spectra as well as I values of the synthetic samples 1 d–g matched those of the naturally occurring salinilactones d–G (Figure S1), confirming their identity.
Compounds 1 h and 1 i were synthesized as candidate structures of salinilactone H. The position of the methyl branch had to be assigned by comparison of I of the two synthetic compounds with the naturally occurring lactone because the mass spectra of both compounds were similar and the natural compound spectra were of poor quality due to the miniscule amounts in the samples (Figures S2 and S3). Lactone 1 h thus proved to be identical with salinilactone H, exhibiting an anteisomethyl group.
We recently proposed the designation of six new Salinispora species in addition to the three that have already been described.11 Consequently, we tracked salinilactone production in members of all nine species as represented by 11 different strains. For this purpose, CLSA of liquid cultures and agar plates was performed (Table S2). Salinilactone production was widespread in the Salinispora genus. Nine out of 11 strains produced salinilactones, although in variable amounts. The two strains that did not produce any compounds both lacked the spt gene cluster (Figure 4). These strains are dispersed in the genus suggesting two independent loss events of the gene cluster. The amounts of natural 1 i were so small that it was only detected after prolongation of the extraction time from 24 to 48 h in the high producing strain S. arenicola CNS‐205. Hence it is not included in Table S2 or Figure 4.
Figure 4.

Species phylogeny in relation to salinilactone production and BGC organization. Salinispora species tree generated from 77 concatenated, single‐copy genes using autoMLST;16 with 1000 bootstraps and M. nigra DSM 43818 as an outgroup. Production of salinilactones A–G is indicated by black circles of various size corresponding to Table S2. When the salinipostin BGC is present, 20 kb up‐ and downstream are shown. Genes are colored based on COG function. Black asterisks indicate regulatory genes: transcriptional regulators include tetR (pink), arsR (purple), gntR (grey), fadR (maroon), luxR (light blue), ompR (light purple), and XRE (dark blue) families.
The bouquet of compounds is usually very similar among strains, although the amount detected varied (Figure 4). Salinilactones A, B, and F often were the most abundant compounds, with both strains of Salinispora pacifica and S. arenicola producing the highest yields and greatest diversity of salinilactones.
The gene cluster is highly conserved across all nine strains in which it was detected although observed in different genomic environments that include varied numbers and types of regulatory elements that could play a role in the amounts and types of salinilactones produced. Differences in production of agar and liquid cultures were usually small (Table S2).
We also analyzed liquid and agar plate CLSA‐extracts of a range of actinomycetes including Streptomyces griseus, Streptomyces lavendulae, and Amycolatopsis mediterranei, all of which are known to produce GBLs.7, 12
None of these strains produced salinilactones (Table 2). The genus Micromonospora is a close relative to the genus Salinispora.13 The absence of salinilactone production in Micromonospora nigra and Micromonospora echinospora suggests it is not a common feature outside of the genus Salinispora.
Table 2.
Actinomycetes tested for salinilactone production and inhibitory activity in agar diffusion assays. Minimum loading of salinilactone B needed to observe growth inhibition.
|
Actinomycete |
Salinilactone production |
Inhibition/loading |
|---|---|---|
|
S. arenicola CNH‐996A |
yes (≈500 μg L−1 1 b)5 |
yes/50 μg5 |
|
S. arenicola CNS‐205 |
yes |
no |
|
S. violaceoruber A3(2) |
no |
yes/50 μg5 |
|
S. griseus DSM40236 |
no |
no |
|
M. nigra DSM43818 |
no |
yes/30 μg |
|
M. echinospora DSM43816 |
no |
yes/100 μg |
|
S. lavendulae DSM40069 |
no |
no |
|
A. mediterranei DSM43304 |
no |
no |
|
A. teichomyceticus DSM43866 |
no |
yes/100 μg |
The structural relation of the salinilactones to the GBL autoregulators points to a specific function in the ecology of Salinispora. Therefore, we performed several assays to probe the biological activity of the salinilactones. Inhibition assays showed inhibitory activity of salinilactone B against Actinoplanes teichomyceticus DSM 43866 and M. echinospora DSM 43816 at higher concentrations matching our previous results5 for Salinispora arenicola CNH‐996A and Streptomyces violaceoruber A3(2). Surprisingly, M. nigra DSM 43818 was impacted quite heavily by salinilactone B, while the remaining other actinomycetes S. griseus, S. lavendulae and A. mediterranei were not affected (Figure S3, Table 2).
Hence, the inhibitory activities of salinilactone B varies considerably both among strains of the same genus and, in the case of S. arenicola, between strains of the same species.
To get some insight into the chemical ecology of the marine sediment affiliated genus Salinispora, a toxicity assay with brine shrimps was performed with the racemic synthetic products to evaluate the protective value of these compounds. Interestingly, salinilactones A–C and G exhibited significant activity in the assays, which are commonly used for evaluation of cytotoxicity of bioactive compounds (Table 3).14
Table 3.
LC50 values of salinilactones in standard brine shrimp (Artemia salina) cytotoxicity assays.
|
Compound |
LC50 [μm][a] |
Compound |
LC50 [μm][a] |
|---|---|---|---|
|
salinilactone A (1 a) |
117±51 |
salinilactone B (1 b) |
91±26 |
|
salinilactone C (1 c) |
85±24 |
salinilactone F (1 f) |
576±140 |
|
salinilactone G (1 g) |
87±33 |
berberine chloride14 |
25.2±2.9 |
[a] Values are the mean±SD, n=3.
The brine shrimp activity is similar to that of berberine chloride, a compound well‐known for its cytotoxic effects.15 This indicates a potential defensive function of the salinilactones against grazers. The lower activities of salinilactones A and F might be the result of the compounds volatility. As the activity of salinilactones B, C and G is very similar, the non‐polar side chain seems to have negligible effect relative to the bicyclic core. A reaction of the highly strained three membered ring with a nucleophile is conceivable as this kind of compound is known for reactions of this type at elevated temperatures employing alcohols or thiols.17
Conclusions
In summary, we identified five new members of the salinilactone family, salinilactones D–H, by GC/MS studies and synthesis of candidate structures. The production of salinilactones in a wide variety of Salinispora strains was confirmed. The absence of salinilactone production in strains lacking AfsA, the salinipostin biosynthetic gene cluster, supports our previously postulated biosynthetic pathway. Additionally, salinilactone B inhibits a range of actinomycetes including salinilactone and other GBL producers. Finally, salinilactones exhibit significant cytotoxicity in brine shrimp assays demonstrating the ability of salinilactones to affect higher organisms, even though the specific biological function of the salinilactones remains unclear.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank Jessica Grube for the cultivation of several actinomycetes, their extraction and inhibition testing. P.R.J. acknowledges funding from the US National Institutes of Health (NIH) under award R01 GM085770.
C. Schlawis, T. Harig, S. Ehlers, D. G. Guillen-Matus, K. E. Creamer, P. R. Jensen, S. Schulz, ChemBioChem 2020, 21, 1629.
References
- 1.
- 1a. Jensen P. R., Williams P. G., Oh D.-C., Zeigler L., Fenical W., Appl. Environ. Microbiol. 2007, 73, 1146; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b. Nett M., Ikeda H., Moore B. S., Nat. Prod. Rep. 2009, 26, 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schulze C. J., Navarro G., Ebert D., DeRisi J., Linington R. G., J. Org. Chem. 2015, 80, 1312. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Gulder T. A. M., Moore B. S., Angew. Chem. Int. Ed. 2010, 49, 9346; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 9534; [Google Scholar]
- 3b. Feling R. H., Buchanan G. O., Mincer T. J., Kauffman C. A., Jensen P. R., Fenical W., Angew. Chem. Int. Ed. 2003, 42, 355; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2003, 115, 369. [Google Scholar]
- 4.
- 4a. Groenhagen U., de Oliveira A. L. L., Fielding E., Moore B. S., Schulz S., ChemBioChem 2016, 17, 1978; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b. Harig T., Schlawis C., Ziesche L., Pohlner M., Engelen B., Schulz S., J. Nat. Prod. 2017, 80, 3289. [DOI] [PubMed] [Google Scholar]
- 5. Schlawis C., Kern S., Kudo Y., Grunenberg J., Moore B. S., Schulz S., Angew. Chem. Int. Ed. 2018, 57, 14921; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 15137. [Google Scholar]
- 6.
- 6a. Khokhlov A. S., Anisova L. N., Tovarova I. I., Kleiner E. M., Kovalenko I. V., Krasilnikova O. I., Kornitskaya E. Y., Pliner S. A., Z. Allg. Mikrobiol. 1973, 13, 647; [DOI] [PubMed] [Google Scholar]
- 6b. Horinouchi S., Beppu T., Mol. Microbiol. 1994, 12, 859. [DOI] [PubMed] [Google Scholar]
- 7. Willey J. M., Gaskell A. A., Chem. Rev. 2011, 111, 174. [DOI] [PubMed] [Google Scholar]
- 8. Amos G. C. A., Awakawa T., Tuttle R. N., Letzel A.-C., Kim M. C., Kudo Y., Fenical W., Moore B. S., Jensen P. R., Proc. Natl. Acad. Sci. USA 2017, 114, E11121–E11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Grob K., Zürcher F., J. Chromatogr. A 1976, 117, 285; [DOI] [PubMed] [Google Scholar]
- 9b. Citron C. A., Gleitzmann J., Laurenzano G., Pukall R., Dickschat J. S., ChemBioChem 2012, 13, 202; [DOI] [PubMed] [Google Scholar]
- 9c. Schulz S., Fuhlendorff J., Reichenbach H., Tetrahedron 2004, 60, 3863. [Google Scholar]
- 10. Welbes L. L., Lyons T. W., Cychosz K. A., Sanford M. S., J. Am. Chem. Soc. 2007, 129, 5836. [DOI] [PubMed] [Google Scholar]
- 11. Millán-Aguiñaga N., Chavarria K. L., Ugalde J. A., Letzel A.-C., Rouse G. W., Jensen P. R., Sci. Rep. 2017, 7, 3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Efremenkova O. V., Russ. J. Bioorg. Chem. 2016, 42, 457; [Google Scholar]
- 12b. Schulz S., Hötling S., Nat. Prod. Rep. 2015, 32, 1042; [DOI] [PubMed] [Google Scholar]
- 12c. Takano E., Nihira T., Hara Y., Jones J. J., Gershater C. J. L., Yamada Y., Bibb M., J. Biol. Chem. 2000, 275, 11010. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Jensen P. R., Mafnas C., Environ. Microbiol. 2006, 8, 1881; [DOI] [PubMed] [Google Scholar]
- 13b. Maldonado L. A., Fenical W., Jensen P. R., Kauffman C. A., Mincer T. J., Ward A. C., Bull A. T., Goodfellow M., Int. J. Syst. Evol. Microbiol. 2005, 55, 1759. [DOI] [PubMed] [Google Scholar]
- 14. Solis P. N., Wright C. W., Anderson M. M., Gupta M. P., Phillipson J. D., Planta Med. 1993, 59, 250. [DOI] [PubMed] [Google Scholar]
- 15. Jantová S., Cipák L., Cernáková M., Kostálová D., J. Pharm. Pharmacol. 2003, 55, 1143. [DOI] [PubMed] [Google Scholar]
- 16. Alanjary M., Steinke K., Ziemert N., Nucleic Acids Res. 2019, 47, W276–W282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.
- 17a. Skerry P. S., Swain N. A., Harrowven D. C., Smyth D., Bruton G., Brown R. C. D., Chem. Commun. 2004, 1772; [DOI] [PubMed] [Google Scholar]
- 17b. Swain N. A., Brown R. C. D., Bruton G., Chem. Commun. 2002, 2042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


