ABSTRACT
Enteric illnesses remain the second largest source of communicable diseases worldwide, and wild birds are suspected sources for human infection. This has led to efforts to reduce pathogen spillover through deterrence of wildlife and removal of wildlife habitat, particularly within farming systems, which can compromise conservation efforts and the ecosystem services wild birds provide. Further, Salmonella spp. are a significant cause of avian mortality, leading to additional conservation concerns. Despite numerous studies of enteric bacteria in wild birds and policies to discourage birds from food systems, we lack a comprehensive understanding of wild bird involvement in transmission of enteric bacteria to humans. Here, we propose a framework for understanding spillover of enteric pathogens from wild birds to humans, which includes pathogen acquisition, reservoir competence and bacterial shedding, contact with people and food, and pathogen survival in the environment. We place the literature into this framework to identify important knowledge gaps. Second, we conduct a meta‐analysis of prevalence data for three human enteric pathogens, Campylobacter spp., E. coli, and Salmonella spp., in 431 North American breeding bird species. Our literature review revealed that only 3% of studies addressed the complete system of pathogen transmission. In our meta‐analysis, we found a Campylobacter spp. prevalence of 27% across wild birds, while prevalence estimates of pathogenic E. coli (20%) and Salmonella spp. (6.4%) were lower. There was significant bias in which bird species have been tested, with most studies focusing on a small number of taxa that are common near people (e.g. European starlings Sturnus vulgaris and rock pigeons Columba livia) or commonly in contact with human waste (e.g. gulls). No pathogen prevalence data were available for 65% of North American breeding bird species, including many commonly in contact with humans (e.g. black‐billed magpie Pica hudsonia and great blue heron Ardea herodias), and our metadata suggest that some under‐studied species, taxonomic groups, and guilds may represent equivalent or greater risk to human infection than heavily studied species. We conclude that current data do not provide sufficient information to determine the likelihood of enteric pathogen spillover from wild birds to humans and thus preclude management solutions. The primary focus in the literature on pathogen prevalence likely overestimates the probability of enteric pathogen spillover from wild birds to humans because a pathogen must survive long enough at an infectious dose and be a strain that is able to colonize humans to cause infection. We propose that future research should focus on the large number of under‐studied species commonly in contact with people and food production and demonstrate shedding of bacterial strains pathogenic to humans into the environment where people may contact them. Finally, studies assessing the duration and intensity of bacterial shedding and survival of bacteria in the environment in bird faeces will help provide crucial missing information necessary to calculate spillover probability. Addressing these essential knowledge gaps will support policy to reduce enteric pathogen spillover to humans and enhance bird conservation efforts that are currently undermined by unsupported fears of pathogen spillover from wild birds.
Keywords: agroecology, Campylobacter spp., E. coli, enteric illness, food safety, Salmonella spp., wild birds
I. INTRODUCTION
Enteric pathogens cause millions of illnesses and hundreds of thousands of deaths worldwide each year (Scallan et al., 2011; Batz, Hoffmann & Morris, 2012; Havelaar et al., 2015). The majority of these cases involve three enteric bacteria – Salmonella spp., Escherichia coli, and Campylobacter spp. – that originate in human, livestock, or wildlife waste (Havelaar et al., 2015). Among wildlife, birds have been the focus of many studies concerning enteric pathogens for several reasons. First, Salmonella spp. can cause mass die‐offs of songbirds among other taxa, causing large conservation concerns (Fichtel, 1978; Daoust et al., 2000; Tizard, 2004; Hall & Saito, 2008). Hall & Saito (2008) estimated that Salmonella spp. were involved in 21.5% of passerine and 5.4% of total bird mortality events in the United States from 1985 to 2004. Second, wild birds are highly mobile and can carry pathogens across large distances, especially during migration, which produces a risk of spreading pathogens beyond local outbreaks (Hussong et al., 1979; Altizer, Bartel & Han, 2011; Gardner et al., 2011; Callaway, Edrington & Nisbet, 2014). For example, sandhill cranes (Grus canadensis), which are commonly infected with Campylobacter spp. during migration, increase enteric bacteria levels in water bodies where they forage (Pacha et al., 1988; Lu et al., 2013; Vogel et al., 2013). Indeed, the only foodborne illness outbreak traced back to a wild bird source occurred from migrating sandhill cranes stopping over at a pea farm in Alaska (Gardner et al., 2011). Third, wild birds are extremely abundant across human‐inhabited landscapes (e.g. urban landscapes, agroecosystems), potentially leading to high contact rates with people and food.
Wild birds are thought to transmit enteric pathogens to humans via several routes. First, people may contact wild birds directly through hunting and consuming the contaminated meat (Navarro‐Gonzalez et al., 2016) or via intentional interactions with birds (e.g. feeding geese at a park). For example, many studies test carcasses of hunted waterfowl or gamebirds and often find high enteric pathogen prevalence (number positive/number tested) (e.g. Luechtefeld et al., 1980; Nebola, Borilova & Steinhauserova, 2007), which may be consumed and cause infection if improperly prepared (Navarro‐Gonzalez et al., 2016). Perhaps more commonly, people may contact surfaces contaminated with faeces from wild birds in locations where birds aggregate (e.g. parks, playgrounds, and beaches) (Strachan et al., 2013; Abdollahpour et al., 2015; Cody et al., 2015). Canada geese (Branta canadensis) can reach high densities at urban parks or beaches where people, particularly young children, may contact faeces either directly with hands or indirectly with clothing or toys (Feare et al., 1999). Children, in particular, are more likely to then place hands in mouths and ingest enteric bacteria (Feare et al., 1999; Strachan et al., 2013). Wild birds may also contaminate drinking, irrigation, or recreational water. An investigation of an E. coli O157:H7 outbreak at Battle Ground Lake in Washington State, USA found identical pulsed‐field gel electrophoresis/restriction fragment length polymorphism patterns between isolates from wild duck faeces, water samples, and case patients, suggesting the ducks may have introduced the bacteria to the recreational water (Samadpour et al., 2002). Finally, wild birds can infect livestock or defecate on crops, leading to foodborne illness (Carlson et al., 2011; Gardner et al., 2011).
To date, most literature has focused on a small number of bird species in a narrow range of habitat settings [e.g. European starlings (Sturnus vulgaris) and rock pigeons (Columba livia) in cities or intensified livestock operations (Carlson et al., 2011, 2015; Haesendonck et al., 2016; Marenzoni et al., 2016)]. Conversely, few data are available for the majority of wild bird species, including many of those commonly found in contact with people or agriculture [e.g. American robins (Turdus migratorius)]. Further, most studies provide data limited to enteric pathogen prevalence (proportion of individuals infected) and not transmission per se (movement of the pathogen). Indeed, a systematic review of 442 modelling studies covering 85 zoonotic pathogens conducted by Lloyd‐Smith et al. (2009) found that disease ecology literature often fails to account for multi‐host ecology of pathogens, with only six studies examined including a mechanistic model of zoonotic spillover. This constrains our ability conclusively to identify sources of pathogens and weakens assessment of risks that wildlife, including birds, pose to human health. For example, mallard ducks (Anas platyrhynchos) often have high prevalence of Campylobacter spp. [e.g. 9.2–52.2% in Colles et al., 2011 and 34% in Luechtefeld et al., 1980]. Yet, experimental infection data suggest Campylobacter spp. are highly host‐adapted, and mallards are poor reservoir hosts for non‐mallard strains (Atterby et al., 2018). In fact, Colles et al. (2011) found only 1 of 109 Campylobacter isolates from wild mallard ducks were a sequence type associated with human disease, suggesting a low risk of transmission despite high prevalence. Further, the few studies that attempt to trace human cases to their origin suggest that although prevalence may be high, crossover is rare (Strachan et al., 2013; Cody et al., 2015; Seguino et al., 2018). For example, Seguino et al. (2018) found that wild bird isolates accounted for only 0.23% of human C. jejuni and C. coli infections. Thus, reliance on prevalence data alone may be overestimating the risk of enteric pathogen spillover between wild birds and humans.
Although many reviews at least briefly discuss enteric pathogen transmission between wildlife, livestock, and/or humans (e.g. Hancock et al., 1998; Haag‐Wackernagel & Moch, 2004; Hubálek, 2004; Tizard, 2004; Wassenaar, 2011; Clark, 2014; Navarro‐Gonzalez et al., 2016), they lack a robust framework from which to develop future risk models (e.g. frameworks provided by Plowright et al., 2017; Cross et al., 2019; Washburne et al., 2019). Further, the lack of systematic or meta‐analytic approaches used in prior syntheses could lead to erroneous conclusions about pathogen prevalence and transmission if the narrow subset of species and habitats considered are subject to selection bias and are not representative of broader trends (Haddaway & Watson, 2016). Therefore, to establish a better understanding of the relationship between wild birds and human enteric illness, we developed a framework for understanding transmission, which has largely been ignored in the literature (Lloyd‐Smith et al., 2009), and summarize what is currently known throughout. Our conceptual framework builds upon those provided by Lloyd‐Smith et al. (2009) and Plowright et al. (2017) and captures the complex processes involved in regulating the spillover of enteric pathogens from wild birds to humans, including pathogen exposure, reservoir competence, contact with people or food, bacterial survival in the environment, and transmission to human hosts. Second, we conducted a comprehensive meta‐analysis of enteric pathogen prevalence in 431 North American breeding birds that focused on three enteric pathogens with large human health burdens known to occur frequently in wild birds: Campylobacter spp., E. coli, and Salmonella spp. We conducted this meta‐analysis using prevalence data since it is the most commonly reported proxy for transmission risk. Throughout our conceptual framework and meta‐analysis, we assess ideas about biological and ecological variables that may affect risk that are commonly found throughout the literature (e.g. juvenile birds will have higher prevalence because of adaptive immunity). We synthesize our results to identify the most important future research avenues needed to quantify the risk wild birds pose to human health.
II. LITERATURE SEARCH AND ANALYSIS
(1). Literature search
We began by acquiring papers concerning aspects of transmission of enteric pathogens from wild birds to livestock and/or humans, most of which focused on prevalence data. We searched the literature for studies reporting data on Campylobacter spp., E. coli, and Salmonella spp. in North American breeding birds (see online Supporting information, Table S1 and Data S1). First, we gathered a list of North American breeding birds from the USGS Breeding Bird Survey (Sauer et al., 2017) and supplemented this list with any species observed on West Coast farms by Smith et al. (2019), which yielded a list of 431 species (Table S2 and Data S2). We assigned each bird species to a taxonomic order and family using the Birds of North America online database taxonomy as of November 2018 (Table S2; Data S2; Rodewald, 2015). We then assigned each species to a diet guild and foraging strata using De Graaf, Tilghman & Anderson (1985), Rodewald (2015), and Wilman et al. (2014). We next searched the ISI Web of Knowledge for studies reporting the presence of Campylobacter spp., E. coli, and/or Salmonella spp. or other aspects of transmission for each North American breeding bird species and saved all review papers acquired through the search. Our search terms included ‘“Salmonel*” OR “E* coli” OR “Campylobacter” AND “[bird common name]” OR “[bird scientific name]”’. We searched for additional papers in references in all review papers acquired through the search and selected primary publications reporting estimates for under‐studied species. Due to a lack of data for most species included in our meta‐analysis (see Section IV.2), we gathered estimates from studies conducted outside of North America if they included estimates for North American breeding birds (Figs S1 & S2). We had six criteria for inclusion in prevalence analyses in our meta‐analysis section. The paper must (1) report if one or more of the 431 North American breeding birds was/were tested for Campylobacter spp., E. coli, and/or Salmonella spp., (2) present primary data that were not duplicated from other studies included in the meta‐analysis, (3) report the bird species tested (e.g. Larus spp. was not sufficient but Larus argentatus was), (4) report on natural infections (i.e. no experimental infection data), (5) report data from free‐ranging wild birds (we did not include estimates from farm, long‐term rehabilitation centre, or laboratory animals for prevalence estimates), and (6) be in English, Spanish, or French or have all data extractable from an English language abstract. We gathered data on generic E. coli when available but considered it an unsuitable proxy for pathogenic E. coli and marked it ‘Reject (7)’ in our study log (Data S1). Data were further considered unsuitable for generating pathogen prevalence estimates but suitable for reporting presence/absence of bacteria if they: (8) did not report the number of individuals tested or positive (including only reporting number of isolates) or (9) only reported data on birds collected after death to avoid overestimating prevalence in birds that died from enteric pathogens (excluding hunted birds which we assumed to be a random sample of wild bird populations), or were brought to a rehabilitation centre within 24 h of testing to avoid overestimating prevalence due to infections acquired after capture or underestimating prevalence due to treatment. A total of 211 papers fitted our full nine criteria for inclusion (Figs S1 and S2; Table S1; Data S1).
We gathered binary data from each study on 30 variables we classified as related to exposure (N = 6 variables), reservoir competence (N = 14 variables), contact with humans or food (N = 4 variables), or bacterial survival and transmission (N = 6 variables); bacterial species included; prevalence; substance tested; condition (live, sick, etc.) at testing; bacterial identification method(s); habitat setting(s) of study; and geographical location (Table S1; Data S1). For each study and species reported, we gathered data on number of individuals that tested positive, total number of individuals tested, and whether anti‐bacterial resistance was reported (Tables S2 and S3; Data S2 and S3). If Campylobacter spp. or Salmonella spp. serovar (bacterial groups with unique cell surface antigen variants) were reported, we recorded how many individuals had each species or serovar. E. coli were separated into pathogenic and generic forms.
(2). Statistical analysis
(a). Meta‐regressions
We estimated pathogen prevalence in two ways. First, we estimated prevalence for each bacterium in each bird species by summing the total number of individuals with positive samples divided by the total number of individuals tested across studies (Data S2). We then estimated overall prevalence by summing number of positive individuals/number of individuals tested for each pathogen. Second, we estimated pathogen prevalence using random effects models in the rma.mv function in the metafor package in R (Viechtbauer, 2010; R Core Team, 2018) for individual bird species. We included study as a random effect and estimated pathogen prevalence across bird species by including study and species nested within family as random effects. We only estimated species prevalence using random effects models when data came from two or more studies and we estimated that sufficient observations were available based on the Thrusfield (2007) formula. To estimate if sufficient observations were available with the Thrusfield (2007) formula, we assumed an infinite study population, used the expected overall pathogen prevalence calculations described above, used a confidence interval of 95%, and used 5% desired absolute precision. In the main text we present prevalence estimates derived from models including study as a random effect, while also providing estimates calculated by summing across studies in the supporting information.
We tested for differences in pathogen prevalence by sex and age using log risk ratios in the escalc function in the metafor package. We tested for differences in prevalence by age for Campylobacter spp., pathogenic E. coli, and Salmonella spp., but limited analyses on sex to Salmonella spp. due to data availability (Tables S4 and S5). Next, we compared the prevalence of Salmonella spp. for three species with the most estimates across studies (European starling, house sparrow (Passer domesticus), and rock pigeon, all introduced to North America and, therefore, not protected) by type of sample tested for bacteria [cloacal swab, faeces, blood, and dissected internal organs (‘necropsy’)] using mixed‐effects models in the rma.mv function in the metafor package including study as a random effect. We conducted pairwise comparisons using Tukey HSD tests. We hypothesized that studies that tested internal organs would find higher prevalence of pathogens because a bird would not have to be shedding bacteria in order to obtain a positive result; this, in turn, may erroneously suggest that commonly necropsied birds have higher prevalence than protected natives that generally cannot be necropsied. Finally, we conducted comparisons of pathogen prevalence by order, diet guild, and foraging strata using mixed‐effects models in the rma.mv function in the metafor package in R, followed by Tukey HSD tests for pairwise differences. Study and species nested within family were used as random effects to account for multiple observations from some studies and taxonomic relatedness, respectively. We suggest caution in interpreting P values from our analyses for two reasons. First, the data available are largely biased to a small number of commonly studied species (see Section IV.2) that may not be representative of most wild birds. Second, some pathogen–bird combinations have very few observations compared to others. Thus, lack of statistical differences in pathogen prevalence between some bird species or groups might often reflect low power, based on small sample sizes, rather than no differences in underlying biology. Conversely, data on Salmonella spp. prevalence were comparatively common in the literature, giving us greater power to detect differences.
(b). Representativeness of the literature
We evaluated representation of the wild bird species studied across the literature using a two‐part comparison. First, we compared the taxonomic orders and species studied for each pathogen to the percentage of species each taxon represented in the North American Breeding Bird Survey list of reported birds (Sauer et al., 2017). Second, we searched the eBird database for reported sightings of each species as a measure of relative abundance (Sullivan et al., 2009). Then, using estimated prevalence and the minimal sample size needed to determine prevalence with 5% precision, we classified species into those with no pathogen observations, those with 1+ observations but insufficient numbers to determine prevalence for any pathogens, and those with enough data to determine prevalence for one, two, or three of the pathogens. We then calculated the percentage of species falling into each category. Finally, we summed the total sightings in eBird of species in each category to calculate the relative abundances of individuals in each group. For the second comparison, we accessed a farm bird database from Smith et al. (2019) that surveyed birds on 52 small‐scale, diversified organic farms (23 that integrated livestock and 29 crop‐only) and two cattle feedlots. Organic produce farming is often thought to be a hotspot of enteric pathogen transmission, and increasingly, farmers are encouraged to remove wildlife habitat from their farms (Beretti & Stuart, 2008). Adhering to these Good Agricultural Practice recommendations can be extremely cost prohibitive to small‐scale growers (Bovay & Sumner, 2018). Therefore, understanding risk of enteric pathogen spillover within this system is particularly important. We compared our meta‐data to the percentage of species in each taxon represented within this farm population and the average on‐farm densities each taxon represented. We repeated our analyses described above for percent of North American breeding bird species and relative abundances using eBird with the farm bird data and classified farm bird species into those with no pathogen observations, those with 1+ observations but insufficient numbers to determine prevalence for any pathogens, and those with enough data to determine prevalence for one, two, or three of the pathogens. We compared the proportion of observations of each species for each of the four comparisons (North American breeding birds, eBird abundances, farm bird species, and farm bird abundances) with their proportions in the data collected for the meta‐analysis using Chi‐square goodness‐of‐fit tests.
III. CONCEPTUAL FRAMEWORK FOR UNDERSTANDING SPILLOVER OF ENTERIC PATHOGENS FROM WILD BIRDS TO HUMANS
Lloyd‐Smith et al. (2009) developed a framework for understanding zoonotic spillover that included prevalence of infection in animal reservoirs, the rate of human contact with reservoirs, and the probability that humans become infected when contact occurs. Plowright et al. (2017) expanded upon these ideas, noting that a hierarchical series of barriers must align for spillover to occur. The framework of Plowright et al. (2017) included pathogen pressure (determined by reservoir distribution, pathogen prevalence, and pathogen release), human and vector behaviour leading to route and dose of exposure, and attributes of recipient hosts which affect the probability and severity of infection (genetics, physiological, and immunological factors). Here, we expand these frameworks to create a wild bird–enteric pathogen‐specific framework for understanding the factors that influence the likelihood of enteric pathogens contacting and infecting humans (Fig. 1A). To accomplish this, we consider factors that influence wild bird exposure to enteric pathogens, reservoir competence, contact with humans and food, and probability of pathogens surviving in the environment, colonizing, and causing disease in a human host. Briefly, for wild birds to become transporters or reservoirs of a pathogen, they must first be exposed to bacteria in the environment. Exposure may vary based on ecological traits, including habitat associations and foraging traits. At the same time, an individual's susceptibility to being colonized by a pathogen will vary based on reservoir competence, which can differ based on a number of physiological factors. Lastly, for enteric pathogens to be transmitted from wild birds to humans, humans must either come into direct contact with the pathogens (e.g. from wild bird meat or faeces through direct hand‐to‐mouth contact) or indirectly from crops or water contaminated with faeces. If directly consumed, the bacteria must be a strain that can effectively colonize and cause disease in a human host and be ingested at an infectious dose. If indirectly consumed, the bacteria must also survive in the environment and through food preparation for long enough to remain at an infectious dose. Many factors influence each stage of the pathogen life cycle and the subsequent likelihood of wild birds transmitting enteric pathogens that may cause disease in humans. We describe these processes in more detail below. We suggest that research approaches that integrate all four stages will be most informative, and studies that only consider prevalence are likely to overestimate risk of enteric pathogen spillover from wild birds to humans.
Figure 1.
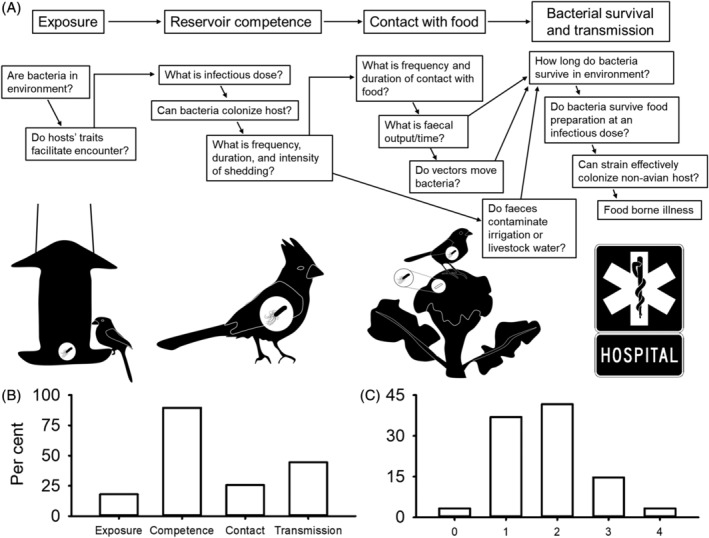
(A) Conceptual diagram outlining steps from bacterial acquisition to human infection. Icons below flow chart show a bird being exposed to E. coli at a bird feeder, E. coli replicating within a bird host, a bird defecating E. coli on a broccoli plant, and a hospital sign to indicate human enteric illness. (B) Percentage of studies included in meta‐analysis that reported data pertaining to exposure, reservoir competence, contact, and bacterial survival and transmission. (C) Percentage of studies that reported data on 0, 1, 2, 3, or 4 of the aspects in the conceptual diagram (exposure, reservoir competence, contact, and bacterial survival and transmission).
(1). Exposure
Wild birds must come into contact with bacteria in the environment to be colonized by enteric pathogens. If pathogen prevalence varies by landscape context, habitat associations may make species that tend to inhabit pathogen‐sparse landscapes less likely to encounter pathogens than species that inhabit pathogen‐rich landscapes (Taff et al., 2016). Within a species, individuals that inhabit pathogen‐rich versus pathogen‐poor landscapes may experience different exposure levels and have variable pathogen prevalence and shedding intensity (Barron et al., 2015; Taff et al., 2016). Several landscape contexts are thought to be hotspots for transmission. A large body of literature has demonstrated transmission of enteric pathogens from refuse sites to gull and corvid species, including Salmonella serovars known to infect humans (Butterfield et al., 1983; Ito, Totake & Ogawa, 1988; Tizard, 2004). Wild birds can also acquire and transmit bacteria in water bodies (Levesque et al., 2000; Fogarty et al., 2003; Lu et al., 2013). Species that interact with livestock are further thought to have high prevalence of enteric pathogens (Skov et al., 2008; Carlson et al., 2011; Callaway et al., 2014; Hald et al., 2016). Birds foraging close to livestock have been demonstrated to have higher prevalence of Campylobacter spp. than those foraging further away (Hald et al., 2016). Proximity to urban habitats (Hernandez et al., 2016) and the use of bird feeders (Fichtel, 1978; Daoust et al., 2000; Tizard, 2004) are also thought to increase prevalence. Hernandez et al. (2016) found Salmonella spp. prevalence was highest in white ibis (Eudocimus albus) in urban landscapes and decreased in natural landscapes. Conversely, neither Rouffaer et al. (2016), Brobey, Kucknoor & Armacost (2017), nor Hamer, Lehrer & Magle (2012) found variation in Salmonella spp. prevalence across an urbanization gradient. However, these studies primarily tested songbirds which have low Salmonella spp. prevalence (~4.8%; see below) compared to the Pelecaniformes (egrets, ibis, pelicans; ~17%), limiting ability to make landscape comparisons. It is generally thought that species such as finches and sparrows that gather at high densities at feeders are prone to mass mortality following contamination of a feeding station with Salmonella spp., especially in harsh weather conditions when birds are forced to aggregate for food (Fichtel, 1978; Daoust et al., 2000; Tizard, 2004). A review conducted by Brearley et al. (2013) found fairly inconsistent results in the impacts of human‐modified land usage on pathogen prevalence and suggested one reason may be the need to consider habitat fragmentation in addition to its loss (Allan, Keesing & Ostfeld, 2003; Brearley et al., 2013), which would be a novel approach in assessing how land usage influences enteric pathogen prevalence in wild birds.
Foraging traits may alter enteric pathogen prevalence in wild birds by altering rates of exposure to faecal contamination (Waldenström et al., 2002; Skov et al., 2008; Hald et al., 2016). Waldenström et al. (2002) tested Campylobacter spp. prevalence in 1794 migrating birds and found shoreline‐foraging birds, opportunistic feeders, and non‐granivorous ground‐foragers had the highest prevalence. A study conducted in Danish livestock farms that tested 1607 individuals found that birds whose diets consisted primarily of animals or mixed animals and vegetables, those foraging on the ground, and those foraging near livestock stables were more likely to carry Campylobacter spp. than aerial foragers and other guilds (Hald et al., 2016). Similarly, Sensale et al. (2006) found higher Campylobacter spp. prevalence in ground foragers and arboreal/herbaceous insectivores and no Campylobacter spp. in granivores or aerial insectivores. Interestingly, Broman et al. (2004) found C. jejuni strains exhibited high similarities within foraging guilds, suggesting shared sources of transmission. Our literature review yielded no studies that compared E. coli or Salmonella spp. prevalence by foraging guild. We examine the relationship between diet guild and foraging strata using our meta‐data (see Section IV.4).
Daily and seasonal movements may also impact exposure rates and ability to disseminate and maintain pathogens at new locations. For example, European starlings are often cited as a risk for food safety (Carlson et al., 2011, 2015) and have large daily movement patterns from roost to feedlot sites. Further, starlings occasionally will forage between multiple feedlots, disseminating pathogens to new herds (Lejeune et al., 2007; Gaukler et al., 2012; Bray, Larsen & Mott, 2018). Birds often stop and forage along the route, including in agricultural fields, where they could contaminate produce with pathogens acquired from the feedlots. Raptors and crows represent other groups with large daily movement patterns (Rodewald, 2015) that may act as disseminators of pathogens into new environments. Studies on species that have large daily movement abilities are of interest and could help elucidate mechanisms that contribute to the introduction and maintenance of pathogens at important contamination points.
Migratory species are generally thought to have high exposure to pathogens during migration, but others have hypothesized that migration could cause individuals to leave pathogen‐rich areas and reduce exposure levels (Altizer et al., 2011). Some evidence exists that migration increases enteric pathogen prevalence, particularly for migratory water birds, likely due to large aggregations of birds from across wide areas with high faecal outputs (Hussong et al., 1979; Waldenström et al., 2002; Hubálek, 2004; Skov et al., 2008; Lu et al., 2013). In addition to exposing birds to a variety of pathogens, migratory behaviours can impact prevalence through changes in host physiology, stress, and immune function and can contribute to dissemination of pathogens across large distances (Hubálek, 2004; Altizer et al., 2011; Callaway et al., 2014). Migratory waterfowl and other water birds have long been known to cause seasonal peaks in enteric pathogens when they aggregate in water bodies during migration and at overwintering sites (e.g. Hussong et al., 1979; Lu et al., 2013). Similarly, Taff et al. (2016) found Campylobacter spp. prevalence was highest in American crows (Corvus brachyrhynchos) during winter when migratory individuals return and crows form large communal roosts. Migratory distance can also impact pathogen prevalence: short‐distance migrants have been found to have higher prevalence than long‐distance migrants for both Campylobacter spp. and Salmonella spp. (Waldenström et al., 2002; Sensale et al., 2006; Skov et al., 2008). Skov et al. (2008) further compared migratory to resident species and found the lowest Salmonella spp. prevalence in resident birds. However, results are certainly not ubiquitous as Hald et al. (2016) found no correlation between migratory status and Campylobacter spp. prevalence.
(2). Reservoir competence
While habitat usage and species traits may alter exposure levels, once an individual is exposed to a pathogen, susceptibility to infection will vary based on reservoir competence. For instance, individuals could function simply as temporary transporters with a low probability of transmitting bacteria or become infected and shed bacteria for prolonged periods, increasing the risk of transmission to new hosts. Here, we define reservoir competence as the probability that an infected host will transmit an infection to a new host (Barron et al., 2015). Reservoir competence is influenced by factors such as exposure, host immune response, shedding intensity, and shedding duration. Sex, age, body size, body condition, microbiome, coinfection (simultaneous infection of a host by multiple pathogen species), pace of life, variation in innate immunity, daily and seasonal movement, and host density, among other factors, could all impact colonization by bacteria, duration of infection, and intensity of shedding (Mills, Lombardo & Thorpe, 1999; Waldenström et al., 2002; Benskin et al., 2009; Colles et al., 2011; Ostfeld et al., 2014; Owen et al., 2014; Taff et al., 2016; Grond et al., 2018). Although microbiome could affect enteric pathogen prevalence in wild birds (Peachey, Jenkins & Cantacessi, 2017; Grond et al., 2018), we found no studies that tested this idea. Microbiome is influenced by physiology, diet, environment, and phase of the annual cycle (Grond et al., 2018), suggesting that species traits may influence pathogen prevalence if a diverse microbial community were to alter pathogen establishment. Coinfection has been demonstrated to decrease (Johnson & Hoverman, 2012; Johnson et al., 2013) or increase infection success (Wang et al., 2018), although results are inconsistent (Peachey et al., 2017; Wang et al., 2018) and could depend on factors such as host immune response (Peachey et al., 2017) or order of infection (Johnson & Hoverman, 2012; Atterby et al., 2018). Twenty‐two papers gathered through our literature review reported coinfection data, although their observational nature makes it difficult to make inferences (Table S6). Experiments examining the impacts of microbiome diversity or coinfection on enteric pathogen colonization, shedding intensity, or shedding duration would be novel contributions to the field.
Sex could influence susceptibility to infection if differential parental investment affects condition, immune investment, or alters habitat usage and subsequent exposure (Monaghan et al., 1985; Martin, Weil & Nelson, 2008; Girard, Goldberg & Hamer, 2011). We tested the impact of sex on Salmonella spp. prevalence in 711 female (11.6% prevalence summed across studies) and 985 male (7.9% prevalence summed across studies) individuals from six studies and found females had 1.45 times (95% CI: 1.08, 1.94) higher Salmonella spp. prevalence [estimated average log relative risk () = 0.37 ± 0.15 (SE), Z = 2.46, P = 0.014; Fig. S3; Table S4]. Three studies reported Campylobacter spp. prevalence estimates by sex, and all found prevalence did not differ by sex but did not report sample sizes needed to conduct analyses across studies. Similarly, studies reporting generic E. coli estimates by sex did not report sample sizes needed to conduct analyses but stated results were not significant. One study reported sample sizes of male and female birds tested for pathogenic E. coli and found that prevalence was higher in female (14.3%, N = 7) versus male (7.1%, N = 14) birds, but no statistical analysis was conducted.
Age may impact bacterial colonization, intensity, and shedding through differences in acquired immunity (Levesque et al., 2000; Colles et al., 2009, 2011; Taff et al., 2016). We tested the impact of age on Campylobacter spp. prevalence from 10 estimates including 1138 juvenile (19.0% prevalence summed across studies) and 1273 adult (35.5% prevalence summed across studies) individuals from seven studies and found juveniles had 1.04 times higher prevalence (95% CI: 0.60, 1.79), but the difference was not significant [ = 0.036 ± 0.28 (SE), Z = 0.13, P = 0.90; Fig. S4; Table S5]. We tested the impact of age on pathogenic E. coli prevalence from two estimates from two studies including 171 juvenile (18.1% prevalence summed across studies) and 493 adult (8.3% prevalence summed across studies) individuals and found adults had 1.57 times higher prevalence (95% CI: 0.37, 6.68), but the difference was not significant = 0.45 ± 0.74 (SE), Z = 0.61, P = 0.54; Fig. S5; Table S5]. Two studies reported differences in generic E. coli by age but did not report the number of individuals sampled (Table S5). Finally, we tested the impact of age on Salmonella spp. prevalence from 13 estimates from 12 studies including 1104 juvenile (13.9% prevalence) and 2946 adult (5.0% prevalence) individuals and found juveniles had 1.91 times higher prevalence [95% CI: 1.17, 3.12; = 0.65 ± 0.25 (SE), Z = 2.57, P = 0.010; Fig. S6; Table S5]. Our finding that juvenile birds have higher prevalence of Salmonella spp. supports the hypothesis that acquired immunity may decrease prevalence in wild birds (Levesque et al., 2000; Colles et al., 2009, 2011; Taff et al., 2016). Salmonella Typhimurium is known to cause mass mortalities in wild birds (Tizard, 2004; Connolly et al., 2006; Hall & Saito, 2008), but Campylobacter spp. are generally thought to be a natural commensal (Wassenaar, 2011; Griekspoor et al., 2013), potentially explaining our finding that age does not appear to influence Campylobacter spp. prevalence but it does appear to influence Salmonella spp. prevalence.
Poor body condition may increase susceptibility to infection, though current evidence is mixed (Espinosa‐Arguelles et al., 2010; Fukui et al., 2014; Sánchez et al., 2018). We found 36 estimates from 15 studies that attempted to relate condition metrics to enteric pathogen prevalence (Table S7). Twenty‐one estimates were reported for Campylobacter spp. prevalence. Seven of these estimates were reported as significant: four reported a negative effect (combined condition index, mass, and wing cord) while three reported a positive effect (body score, tarsus length, and mass). The remaining 14 were non‐significant (combined condition indices, skeletal size, metatarsus length, tarsus asymmetry and length, left wing length, mass, fat score, packed cell volume, wing ectoparasites). One study reported estimates for pathogenic E. coli (STEC) and found mass was negatively correlated with prevalence in juvenile/subadult rock pigeons but was not correlated with prevalence in adults. One study reported generic E. coli prevalence in relation to European starling mass and found no effect. Five studies related Salmonella spp. prevalence to mass, three of which concerned Salmonellosis cases. One of the two studies conducted on free‐living populations found a significant negative relationship while the other found no effect. All three studies concerning Salmonellosis cases found negative relationships between infection and mass, although only one conducted a statistical analysis. Standardization of methods across the literature would facilitate comparison, and experimental work may help disentangle cause and effect (Waldenström et al., 2010; Taff & Townsend, 2017; Sánchez et al., 2018). A recent meta‐analysis by Sánchez et al. (2018) found that the method used to evaluate body condition was one of the strongest predictors of positive, negative, or null condition–infection relationships, suggesting the importance of choosing appropriate condition metrics. Sánchez et al. (2018) recommend utilizing multiple condition metrics appropriate for the host–parasite biology being studied and pairing these with experimental methods when possible. For example, it is likely commensal Campylobacter spp. and avian pathogenic serovars of Salmonella Typhimurium have vastly differing impacts on host condition, but more work is needed to demonstrate this.
Innate immunity can vary among individuals both within and among species. Differences in innate immunity leading to differences in bacteria‐killing ability and bacterial colonization within a species have been observed in tree swallows (Tachycineta bicolor) (Mills et al., 1999; Schmitt & Bélisle, 2017). A cross‐fostering experiment of tree swallows conducted by Morrison, Ardia & Clotfelter (2009) suggested heritable immunity was more important in bacteria‐killing ability against E. coli than body condition or brood size of the foster nest. Girard et al. (2011) found variation in bacteria‐killing ability against E. coli between American robins, house sparrows, and gray catbirds (Dumetella carolinensis), suggesting that species could vary in susceptibility to infection due to innate differences in ability to defend against infection, leading to differences in risk of causing enteric pathogen outbreaks. Further, physiological differences among species such as full versus rudimentary caeca may affect susceptibility to pathogen colonization (Albuquerque et al., 2013). The ecoimmunological ‘pace of life’ hypothesis predicts that bird species with early maturity, rapid breeding, short longevity, etc., face a trade‐off between resources devoted to these life‐history characteristics and those allotted to anti‐pathogen defences (Ostfeld et al., 2014). Additionally, immunological trade‐offs occur throughout the annual cycle during energetically costly events including breeding, migration, and moult periods (Martin, 2005; Martin et al., 2008; Altizer et al., 2011). Thus, reservoir competence is likely to vary both within and among species due to differences in innate immunity, season, life‐history events, and anatomical factors.
Infectious dose (the minimum number of microorganisms sufficient to establish an infection) may also vary among individuals and species. However, few studies have experimentally infected birds to determine what constitutes an infectious dose (Table S8). Rock pigeons orally inoculated with 9.5 × 107 colony forming units/ml (CFU/ml) of Salmonella Enteritidis began to shed Salmonella spp. 3 days post inoculation, but lower doses were not tested, leaving uncertainty around the infectious dose. Rock pigeons shed between 1.5 × 104 and 2 × 109 CFU/ml up to 14 days post inoculation during a 35‐day trial (Albuquerque et al., 2013). In a 10‐day trial, house sparrows orally inoculated with 102 CFU of a songbird outbreak strain of Salmonella Typhimurium shed on days 1 and 5, birds given 103 CFU shed on days 1–2 and 6–10, birds given 105 CFU shed most days (2/6 died on days 8 and 10), and birds given 108 CFU shed every day until their death (6/6 birds died on days 3–8) (Connolly et al., 2006). In another study, herring gulls (Larus argentatus) already infected with Salmonella spp. were captured and maintained in captivity for 3 weeks and shed 170 most probable numbers/gram (MPN/g) for up to 4 days (Girdwood et al., 1985). Infectious dose is important because bird species and individuals within species that are susceptible to lower infectious doses are more likely to become infected, but few data are available to characterize these differences or evaluate their impacts on disease risk in the field.
Further, many strains appear to be host adapted which can alter infectious dose and duration/intensity of shedding (Atterby et al., 2018). Mallards inoculated with 5 × 104 CFU of C. jejuni of mallard, chicken, or song thrush (Turdus philomelos) origin per ml of water showed interesting and differing responses to the strains. Birds exposed to the mallard strain excreted around 104–106 CFU/ml throughout the 18‐day experiment. Birds exposed to the chicken strain excreted an average peak level of 104 CFU/ml, and at the end of the experiment, only two of six birds continued to shed the bacteria. Mallards exposed to the song thrush strain shed 103–104 CFU/ml for the first few days after exposure then shedding declined rapidly (Atterby et al., 2018). In another experiment, European robins (Erithacus rubecula) were exposed to either a song thrush C. jejuni strain or a C. jejuni strain of human origin and monitored for 25 days. Robins inoculated with the song thrush strain shed bacteria for 6.8 days on average, whereas those given the human isolate were not colonized, and bacteria were only detected in three of eight birds in the human isolate treatment group for up to 3 days post inoculation (Waldenström et al., 2010). This suggests that songbirds may not be competent hosts of human‐adapted strains. European starlings inoculated with doses ranging from 1 × 100.6 to 5 × 106 CFU of E. coli O157:H7 had an ID50 (number of microbes necessary to infect a host in 50% of the exposed population) of log10 4.5 CFU for one strain and log10 5.5 CFU for another strain. As dose increased, duration of shedding increased; shedding intensity ranged from around 101 to 106 CFU/g. High exposure levels resulted in shedding up to the final day of the 14‐day trial (Kauffman & Lejeune, 2011). Altogether, this body of studies suggest infectious dose, shedding intensity, and shedding duration may vary by bacterial strain.
Few data exist on naturally occurring bacterial shedding intensity, and current data suggest wide variation in shedding intensity by bacterial species and avian taxa. For example, naturally occurring wild bird faeces have been found to have concentrations of Campylobacter spp. ranging from 340 cell equivalents per gram (CE/g) [California gull (Larus californicus)] to 1 × 108 CFU/g [ring‐necked pheasants (Phasianus colchicus)], averaging between 4.8 × 103 CFU/g (Canada goose) to 6.7 × 106 CE/g (California gull; Table S8). Naturally occurring wild bird faeces have been found to have concentrations of generic E. coli ranging from 1.9 × 102 CFU/g to 2.5 × 109 CFU/g (herring gull), averaging between 2.8 × 104 MPN/g (sandhill crane) and 4.9 × 108 CFU/g (Larus spp.). Naturally occurring wild bird faeces have been found to have concentrations of Salmonella spp. averaging from 22 MPN/g to 2.4 × 109 CFU/g (herring gull). We found no reports of naturally occurring pathogenic E. coli concentrations. We only found reports of naturally occurring enteric bacteria concentrations for herring gull, California gull, sandhill crane, Canada goose, mallard, ring‐necked pheasant, cattle egret (Bubulcus ibis), and unspecified Larus spp., with no reports for Passeriformes (songbirds, flycatchers). Colles et al. (2009) reported carriage among recaptured European starlings and found 18.2% were shedding Campylobacter spp. on each capture occasion (1 and 588 days apart), 40.1% were negative on each occasion (1 to 364 days apart), and 41.7% changed status (1–392 days). The majority (83.8%) of C. jejuni isolates from starlings shedding on more than one occasion were of a different genotype between surveys, suggesting rapid turnover and re‐colonization.
The virulence of bacteria to bird species and individuals within a species is variable but can affect the duration and intensity of shedding and subsequent likelihood of infecting new hosts. For example, oral inoculations of Salmonella pullorum [a bacterium common in Galliformes (pheasants/quail)] at 1 × 104 CFU/ml was sufficient to cause mortality of northern bobwhite quail (Colinus virginianus) whereas mallards given up to 1 × 1010 CFU/ml showed no signs of disease, although bacteria were isolated from mallard tissues, indicating successful colonization (Buchholz & Fairbrother, 1992). Species that are asymptomatic carriers of bacteria may transmit bacteria for a longer duration over a wider area than species that are killed rapidly by the pathogen (Tizard, 2004). Salmonella Typhimurium tends to cause mass mortalities among finches, sparrows, and cowbirds (Faddoul, Fellows & Baird, 1966; Daoust et al., 2000; Hall & Saito, 2008), whereas other bird species such as raptors and pigeons tend to survive infection, although mortality can occur (Tizard, 2004; Albuquerque et al., 2013). Thus, species or individuals that survive infection may pose a greater risk than individuals killed rapidly. Wild birds are generally considered asymptomatic carriers of Campylobacter spp. [but see Waldenström et al., 2010 and Taff & Townsend, 2017], and although avian pathogenic E. coli (APEC) can cause avian disease, it is generally associated with environmental and predisposing factors (Dho‐Moulin & Fairbrother, 1999); thus, the severity of infection with Campylobacter spp. and E. coli is unlikely to compare to avian Salmonellosis. Wild birds may be more competent hosts of Campylobacter spp. if they remain largely unaffected, while species highly vulnerable to die‐offs due to Salmonella Typhimurium may be poor hosts due to rapid death and less time for dissemination.
(3). Contact
For enteric pathogens to spill over from wild birds to humans, humans must either come into direct contact with the pathogens (i.e. direct hand to mouth contact) or consume food items or water contaminated with faeces. Direct contact with faeces may be the greatest source of enteric pathogen spillover from wild birds to humans (Strachan et al., 2013; Cody et al., 2015). Direct faecal contact often occurs when children in playgrounds, parks, or beaches touch bird faeces and then place their hands in their mouths (Strachan et al., 2013; Abdollahpour et al., 2015; Cody et al., 2015). Handling and consuming undercooked game meat is another direct source of enteric pathogens (Navarro‐Gonzalez et al., 2016). Indirect sources include contaminated produce (Gardner et al., 2011), infected livestock (Carlson et al., 2011; Hald et al., 2016), contaminated water (Lu et al., 2013; Strawn et al., 2013; Clark, 2014; Marine et al., 2015), and domesticated household cats infected as a result of consuming birds (Fichtel, 1978; Tizard, 2004), among other sources (e.g. Penfold, Amery & Morley Peet, 1979; Neal & Slack, 1997; Ejidokun et al., 2006).
Avian faecal contact with produce or other crops can occur in production fields, food wash and packing structures, storage facilities, or during preparation (Penfold et al., 1979; Gardner et al., 2011). Wild birds can directly contaminate produce by defecating while in fields (resting, foraging, etc.) or while flying over. Few data are available comparing faecal outputs per unit time during different activities for wild bird species, but activities that generate greater rates of faecal output could increase the risk of crop contamination (Feare et al., 1999). Canada geese are known to have higher rates of faecal output while feeding compared to loafing (Feare et al., 1999), suggesting that birds actively foraging in fields for insects or crops may have higher defecation rates that could increase contamination risk. Birds actively foraging in patches likely have higher faecal output rates than birds in flight (Guillemette, 1994), and more sustained contact times could lead to greater accumulation of faeces in production areas on or near crops (Feare et al., 1999). Further, gut retention time can vary by diet items, which could increase or decrease faecal output rates for particular diet guilds (Levey & Karasov, 1994).
Intraspecific differences in habitat associations could alter bird contact with crops that differ in susceptibilities to facilitating food‐borne illness (Brandl, 2006; Berger et al., 2010). For example, arboreal bird species likely spend more time foraging in tree fruit than row crops, whereas ground‐ and understorey foraging species may be more likely to forage in crops such as lettuces, brassicas, and cucurbits. Understorey foragers that forage on leaves or stems may be more likely to deposit droppings on edible parts of plants whereas ground foragers may primarily contaminate soil. Grassland species may be more likely to inhabit livestock pastures, and species that nest on structures may have greater contact with livestock near livestock shelters, increasing likelihood of transmission to livestock. Species nesting within food wash/packing structures may also contaminate produce before it leaves the farm. Species with low pathogen prevalence but high contact rates with sensitive areas of food production may pose greater risk than species with high prevalence but low contact rates, and testing this idea would be an interesting avenue for future research.
In addition to direct faecal contact with crops and livestock, birds may cause indirect contamination. For example, European starlings have been shown to mechanically vector Salmonella enterica within feedlots when cattle excrement adheres to feathers and feet (Carlson et al., 2015), although data are limited on the ability of other species to mechanically vector enteric pathogens. Flies could also mechanically vector pathogens from wild bird faeces to sensitive crop areas or to chickens that consume the contaminated arthropods (Skov et al., 2008). Further, wild birds such as geese and ducks could contaminate produce by defecating in irrigation water (Lu et al., 2013; Strawn et al., 2013; Clark, 2014; Marine et al., 2015).
The increase in enteric pathogen prevalence during migration (see Section III.1) is a concern because farmland habitat is recognized as important stopover habitat for a variety of birds, which may increase their direct and indirect contact rates with produce concurrent with a spike in prevalence (Hussong et al., 1979; Dänhardt et al., 2010; Lu et al., 2013; Callaway et al., 2014; Taff et al., 2016). Indeed, the only food‐borne illness outbreak directly linked to wild birds occurred during sandhill crane migration when cranes were using a produce farm as a stopover site (Gardner et al., 2011). Although Marine et al. (2015) found autumn had the highest prevalence of Salmonella spp. on leafy greens, irrigation water, compost, field soil, and pond sediment samples, neither Strawn et al. (2013) nor Benjamin et al. (2013) found higher prevalence of Salmonella spp. or E. coli in on‐farm water or soil samples in autumn. More research could help elucidate whether increased prevalence of pathogens in wild birds and increased crop contact rates during migration translate to higher on‐farm pathogen contamination.
The seasonality of bird contact with people or food production versus the timing of pathogen acquisition by birds could mediate spillover probability if pathogen shedding is short in duration (Table S9). For example, finches, blackbirds, and sparrows are often infected with Salmonella spp. when they aggregate at feeders during cold weather (Fichtel, 1978; Daoust et al., 2000; Tizard, 2004). However, people are less likely to come into direct contact with bird faeces during winter (Strachan et al., 2013; Cody et al., 2015), and crop production is generally reduced or absent. Nevertheless, if birds infected during the winter can shed pathogens during warmer months when humans have greater direct contact with wild bird faeces and when birds forage in agricultural production areas, birds could remain a year‐round risk. In addition to seasonal behavioural changes such as using bird feeders more often, many resident bird species alter habitat usage throughout the annual cycle, which may alter exposure rates (Zuckerberg et al., 2016) and cause fluctuations in risk levels. For example, European starlings have the highest abundances in feedlots in winter (Fischl & Caccamise, 1985) where the birds can acquire enteric pathogens and spread bacteria between herds (Carlson et al., 2011; Kauffman & Lejeune, 2011), but the likelihood of harbouring the pathogens over the winter and shedding pathogens on produce in the crop growing season is unknown. Glunder, Neumann & Braune (1992) observed Campylobacter spp. shedding in 27 herring gull chicks in captivity for 58 weeks and failed to detect Campylobacter spp. after 4 weeks. Albuquerque et al. (2013) inoculated rock pigeons with Salmonella Enteritidis and observed shedding up to day 14 of the 35 day trial, but we know of no other studies examining how long birds can shed enteric pathogens, leaving uncertainty if species infected during the winter are likely to shed pathogens during the growing season.
In our literature review, we found 58 estimates from 45 studies on seasonal variation in prevalence (Table S9). It is generally thought that outbreaks of Salmonellosis in wild birds occur in winter when birds aggregate at feeders during harsh weather (Daoust et al., 2000; Hall & Saito, 2008). Eleven of twelve studies investigating mortality from suspected Salmonellosis outbreaks reported the highest incidence in the winter months. Twelve studies conducted statistical analyses testing for seasonal variation in Salmonella spp. prevalence in populations of live birds: seven found no difference, three found higher prevalence in summer, one found higher prevalence in autumn, and one found higher prevalence in winter. Seven studies conducted statistical analyses testing for seasonal variation in Campylobacter spp.: one found no difference, three found higher prevalence in summer, one found higher prevalence in winter, one found higher prevalence in winter and spring, and one found higher prevalence in spring. The seven studies testing for seasonal variation in pathogenic E. coli had the most consistent trends: five studies found the lowest prevalence in winter, three found the highest prevalence in summer, two found the highest prevalence in autumn, one reported the highest prevalence in a combined summer/autumn period, and one found no significant difference.
(4). Bacterial survival and transmission
In order for pathogens in bird droppings to spill over into humans, the pathogens must survive in faeces, water, or food until faecal–oral contact occurs. If contact occurs with food, the bacteria must successfully colonize livestock or survive in crop fields on produce, survive through washing, shipment, food processing at plants, preparation, and finally enter and establish within a human host. Data on pathogen survival in wild bird faeces are limited. The most information about pathogen survival in wild bird faeces has come from studies on Canada geese. A 28‐day trial conducted in parks in London, England that inoculated Canada goose droppings with 104–105 CFU/g of Salmonella Newport found that the bacteria were able to survive through the full 28‐day trial, despite heavy rain (Fontaine et al., 1980). A 77‐day trial in New Zealand that inoculated Canada goose faeces with 108 CFU/g C. jejuni, placed faeces in pasture, and measured survival in both summer and winter found C. jejuni fell below the limit of detection by day 2 in summer. In winter, C. jejuni were reduced to below 1% of the original concentration by day 4 and were last detected on day 9. Naturally occurring generic E. coli averaging around 3.5 × 106 CFU/g in summer and 4.9 × 104 CFU/g in winter were also monitored in the faeces. In summer, concentrations doubled on day 2 then steadily decreased until day 42 when concentrations were 1% of original levels and dropped to <0.005% on the final day (day 77) of the trial. In winter, E. coli concentrations declined 10‐fold by day 2, fell below the detection limit on day 14, and remained below detection until days 56 and 63 when E. coli were again detected (Moriarty et al., 2012). Bacterial survival in Canada goose faeces may be greater than survival in songbird faeces, which tend to be smaller with greater relative surface area, leaving faeces subject to greater desiccation, but data are lacking to support this assumption. E. coli O157:H7 have remained viable in laboratory‐kept pooled and homogenized European starling faeces stored in the dark at 22°C for up to 76 days (Kauffman & Lejeune, 2011), but outdoor conditions could vastly alter survival. We know of no other published estimates on Campylobacter spp., E. coli, or Salmonella spp. in songbird faeces. Research examining bacterial survival in a diverse range of wild bird faeces is a high priority. Shorter versus longer survival times in faeces of various species of wild birds could cause vastly differing likelihoods of viable bacteria reaching a human consumer at an infectious dose.
Once deposited, bacteria could leave faeces and enter soil or water where survival may differ based on both biotic and abiotic factors. Soil or water with diverse microorganisms could suppress pathogens through predation or competition (Flint, 1987; Jiang, Morgan & Doyle, 2002; Liang et al., 2011; Jacobsen & Bech, 2012). However, the relationship is not always straightforward: biofilms could also protect pathogens from ultraviolet exposure and increase bacterial survival (Brandl et al., 2005). Salinity, pH, nutrient sources, soil type, temperature, and moisture can all also influence bacterial survival (Van Donsel, Geldreich & Clarke, 1967; Reddy, Khalell & Overcash, 1981; Ogden et al., 2001; Natvig et al., 2002; Ma et al., 2011, 2012). Thus, the relationships between biotic and abiotic factors and bacterial survival are complex and still not well understood.
Salmonella spp. generally have the highest survival in soil, followed by E. coli, then Campylobacter spp. (Guan & Holley, 2003; Winfield & Groisman, 2003). Salmonella Typhimurium, the most commonly reported serovar in wild birds, can survive up to 231 days in naturally occurring soil (Islam et al., 2004a), and Salmonella Newport can survive up to 405 days in manure‐amended autoclaved soil if microbial competition has been eliminated (You et al., 2006). E. coli can survive in soil for up to 217 days (Islam et al., 2004c). Further, E. coli have been found to survive in sheep manure piles for up to 21 months (Kudva, Blanch & Hovde, 1998). Campylobacter spp. can survive in manure‐amended soils for up to 120 days (Hutchison et al., 2004). Thus, all three bacteria can have long survival times in soils where they could remain viable until encountered.
Most studies examining bacterial survival in water are limited by short experimental durations (<2 weeks), but some papers have examined survival over periods of several months. E. coli can survive up to 260 days in autoclaved and filtered water (Flint, 1987). Campylobacter spp. can survive up to 4 months (~122 days) in stream water (Rollins & Colwell, 1986). Salmonella Typhimurium can survive up to 14 weeks (~98 days) in water heavily contaminated with sheep faeces (Tannock & Smith, 1972). Survival of all bacteria has been found to be highest at the lowest temperatures, with decreasing survival as temperatures increase (Rollins & Colwell, 1986; Flint, 1987; Mezrioui, Baleux & Troussellier, 1995). Studies of longer durations examining both biotic and abiotic mediators would help improve our understanding of the long‐term viability of enteric pathogens in the environment prior to encounter by animal hosts.
Many factors can influence pathogen survival on crop surfaces including the physicochemical nature of plant surfaces, biofilm formation, microbe–microbe interactions, and plant–microbe interactions (Brandl, 2006; Berger et al., 2010). Salmonella Typhimurium can survive on lettuce for up to 63 days, on parsley for up to 231 days, on radishes for up to 84 days, and on carrots for up to 203 days (Islam et al., 2004a, b ). E. coli can survive on lettuce for up to 77 days, on parsley for up to 177 days, on onions for up to 74 days, and on carrots for up to 168 days (Islam et al., 2004c, 2005). Campylobacter spp. can survive on spinach up to 9 days, on lettuce for up to 1 day, and in radish roots up to 23 days (Brandl et al., 2004). Crop–bacteria combinations that have greater survival durations are more likely to reach human consumers, although data are still limited to a few crop–bacteria examples. Further, if the outer skin or shells are removed prior to consumption, pathogens may be less likely to infect a human consumer.
Once contaminated produce or livestock leaves the farm environment, the risk of causing foodborne illness could differ depending on transport, slaughter practices, food preparation, and proper cooking (Cesare et al., 2003; Wassenaar, 2011; Batz et al., 2012). Batz et al. (2012) found Campylobacter–poultry had the highest annual disease burden of pathogen–food combinations examined, with S. enterica–poultry in fourth place, S. enterica–complex foods in seventh, S. enterica–produce in eighth, and S. enterica–eggs in tenth. E. coli O157:H7–beef was the highest E. coli–food combination in 21st place. Salmonella spp. can have an infectious dose as low as 1.7 × 101 CFU in humans (Blaser & Newman, 1982). Campylobacter spp. have an infectious dose as low as 8 × 10 2 CFU in humans (Black et al., 1988). E. coli also have an extremely low infectious dose with 4–45 bacteria being sufficient to cause enteric illness in humans (Tilden et al., 1996). As summarized above for birds, the infectious dose for humans could vary among strains and individual people due to variation in genetic factors, age, immunological status, physiological state, and health (Blaser & Newman, 1982; Feare et al., 1999; Plowright et al., 2017). Further, different types of foods can differ in risk not only due to differences in hospitableness to bacteria and preparation but also due to differences in fat and protein content, which can affect bacterial survival inside a human stomach (Fontaine et al., 1980). While there may be wide variation in the likelihoods of food–pathogen combinations resulting in illness (Fontaine et al., 1980; Batz et al., 2012), the infectious doses could be extremely low for some individuals (Blaser & Newman, 1982; Black et al., 1988; Tilden et al., 1996).
A growing body of research examining genetic relatedness between bacteria of wild birds, livestock, and human origin suggests that crossover is rare and most strains are host adapted (Table S10) (Waldenström et al., 2002; Colles et al., 2009; Kauffman & Lejeune, 2011; Weis et al., 2016; Atterby et al., 2018). Current literature suggests isolates from wild birds often exhibit sub‐types with higher levels of similarity to isolates from birds of the same species or guild than to isolates from other groups of wild birds, livestock, or humans (Broman et al., 2004; Griekspoor et al., 2013). Both laboratory and field studies have demonstrated that wild birds can harbour strains that infect livestock and humans (Skov et al., 2008; Kauffman & Lejeune, 2011). However, host adaptation of bacterial strains may cause wild birds to have low reservoir competence for isolates from livestock and humans, and vice versa (Waldenström et al., 2010; Atterby et al., 2018). Thus, spillover of enteric pathogens from birds may be less likely than suggested by high prevalence values for a given bacterial genus or species (Fig. 2). There is generally a low prevalence of enteric bacteria in livestock or humans that have genetic similarity to bird isolates, suggesting that spillovers are rare [Table S10; e.g. no similar strains in starling isolates in Colles et al., 2009, 0.9% of mallard isolates in Colles et al., 2011, and 2.2% of migrating bird isolates in Broman et al., 2004]. For example, although mallards have a high prevalence of Campylobacter spp. (~26%; see Section IV.1a), if only 0.9% isolates are known to cause human disease (Colles et al., 2011), then only about 2.3 in 1000 mallards would carry a human disease isolate, much lower than the genus‐level estimate suggests (Fig. 2A). Moreover, the rare mallards carrying isolates that can cause human disease must also contact humans or food for spillover to occur. Using the Smith et al. (2019) farm bird database, we estimated the number of farm survey points (100 m radius point count survey) with one or more mallards observed over the two‐year study. Mallards were only observed in or flying over 18/217 (8.3%) survey points during one or more survey occasion. Thus, only about 2 in 10000 survey points were likely to harbour a mallard with a disease isolate (Fig. 2B). Further, extrapolating Campylobacter spp. survival in Canada goose faeces to mallards from Moriarty et al. (2012), bacteria are unlikely to survive for more than 2 days in summer (Fig. 2C). Thus, what appears to be a high likelihood of spillover based on prevalence data alone decreases when multiple transmission parameters are considered together (Figs 1 and 2D).
Figure 2.
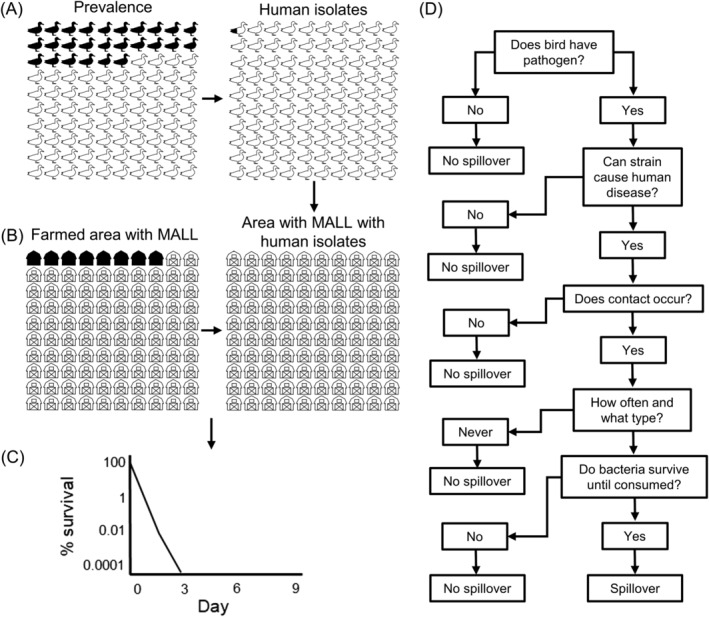
Conceptual diagram of likelihood of Campylobacter spp. spillover from mallards (MALL) to humans. (A) Campylobacter spp. prevalence in mallards estimated from meta‐analysis (26%) on left and prevalence of Campylobacter isolates matched with human disease cases estimated from Colles et al. (2011) (26% prevalence of which 0.9% of isolates are known to cause human disease) on right. (B) Estimated prevalence of mallards in farmland from Smith et al. (2019) farm bird database (8.3% of points) on left and area likely to have mallards with human isolates on left (about 2 in 10000). (C) Estimated survival time of Campylobacter spp. in mallard faeces modified from Canada goose faeces study in Moriarty et al. (2012). (D) Flow chart to determine whether spillover will occur.
Another approach that has been used to estimate spillover is to analyse the number of livestock isolates or human disease isolates of wild bird origin. Pennycott, Park & Mather (2006) found Salmonella spp. isolates from livestock of wild duck/goose origin accounted for 3% of isolates, while all other wild bird isolates accounted for <1%. Similarly, strains isolated from human disease cases tend to show high similarity to livestock strains not of wild bird origin and low similarity to wild bird isolates (Griekspoor et al., 2013). Seguino et al. (2018) found that wild bird isolates accounted for 0.23% of human C. jejuni and C. coli infections while livestock sources accounted for 16%. Cody et al. (2015) found that wild bird C. jejuni isolates accounted for between 2.1 and 3.5% of cases in the UK annually, with the largest proportion occurring in the summer months when humans frequent parks and beaches. Similarly, Strachan et al. (2013) found the highest proportion of Campylobacter spp. human clinical isolates of wild bird origin in the summer from young children (9% of cases were wild bird isolates). However, studies that have quantified relatedness using more than one technique often find contradictory results, leading to uncertainty in robustness of conclusions. For example, Sanad et al. (2013) used both pulse‐field gel electrophoresis and multi‐locus sequence typing to compare European starling and cattle isolates and obtained highly variable phylogenies using the two techniques. This points to the need to determine which methods yield the most robust results for future comparisons. Research quantifying the risk of wild birds transmitting enteric pathogens should compare wild bird isolates to human or livestock disease isolates robustly, given that shared strains are likely a minority of cases (Broman et al., 2004; Colles et al., 2011; Seguino et al., 2018).
(5). Integrating from exposure to transmission
We collected data on 30 binary variables from each study gathered through our literature review that met inclusion criteria 1–6, which we classified as related to exposure (N = 6 variables), reservoir competence (N = 14 variables), contact (N = 4 variables), or bacterial survival and transmission (N = 6 variables) (Data S1 and S2; Tables S1 and S2). We then quantified the number of studies that presented data on one or more aspects within each category and the number of categories each study covered. Thirty‐eight (18.0%) studies reported data on one or more aspects of exposure, 189 (89.6%) studies reported data on one or more aspects of reservoir competence, 54 (25.6%) studies reported data on one or more aspects of contact, and 94 (44.5%) studies reported data on one or more aspects of bacterial survival and transmission (Fig. 1B). Seven studies (3.3%) did not report data beyond simple prevalence estimates, 78 (37.0%) reported data on one of the four categories, 88 (41.7%) reported data on two of the four categories, 31 (14.7%) reported data on three of the four categories, and 7 (3.3%) reported data on all four categories (Fig. 1C). Studies which integrate all stages of our framework described above will be most effective at generating useful data to develop risk models and to develop policy to reduce pathogen spillover from wild birds into humans. Indeed, this appears to be a problem across the disease literature: a systematic review of 442 modelling studies covering 85 zoonotic pathogens conducted by Lloyd‐Smith et al. (2009) found the disease ecology literature often fails to account for the full ecology of pathogens, with only six of the 442 studies examined including a mechanistic model of zoonotic spillover.
Models integrating data across the entire process are no trivial undertaking (Plowright et al., 2017; Childs et al., 2019; Cross et al., 2019; Washburne et al., 2019). Cross et al. (2019) present case studies to highlight challenges and potential solutions to estimating spatiotemporal variation in spillover risk. For example, data sets on multiple host species collected in similar locations, seasons, and at similar resolutions along with data sets collected at all levels of the spillover process are rare. Cross et al. (2019) describe mechanistic approaches where researchers use data on host density, pathogen prevalence, transmission, and shedding to make predictions about spillover events. Conversely, phenomenological models use spillover events to estimate risk covariates that are correlated with host and pathogen distributions which are useful when data on host density and pathogen shedding are lacking. Washburne et al. (2019) present percolation models as a tool to model spillover whereby pathways to spillover are represented as directed graphs as pathogens move from reservoirs to people. Pathogens shed by reservoirs progress through the stages of transmission where pathogens diminish at each stage along the pathway due to failure to persist in the environment, failure to contact humans, failure to infect humans given contact, and failure to be detected by researchers. Each stage is represented as a series of probabilistic models. However, these models may fail if there are environmental feedbacks between environmental factors and wildlife reservoirs, which may be the case in enteric pathogen systems. Finally, Childs et al. (2019) present an environmental risk model to examine spillover probability of yellow fever virus from non‐human primates to humans that may be successfully modified to model enteric pathogen spillover. However, the models presented by Childs et al. (2019) are fairly data intensive.
IV. META‐ANALYSIS OF ENTERIC PATHOGEN PREVALENCE IN WILD BIRDS
Next, we conducted a meta‐analysis on enteric pathogen prevalence in North American breeding bird species with several primary objectives. We sought to (i) generate more robust prevalence estimates than available in individual studies and compile estimates into an easily accessible database for wild bird community enteric pathogen transmission models (Data S2), (ii) identify which species and guilds should be the focus of future research, while pointing to under‐studied groups, and (iii) use our meta‐data to test for differences in enteric pathogen prevalence by taxa and foraging guilds. We use prevalence as a proxy for transmission due to the much greater availability of prevalence data than other types of data, but we caution the reader in extrapolations from prevalence to risk of spillover (Fig. 2).
(1). Prevalence estimates
(a). Campylobacter spp.
14.8% (64/431) of North American breeding birds had Campylobacter spp. prevalence data (1+ observations) meeting our inclusion criteria 1–9 (Data S2). The species with the most observations meeting our inclusion criteria 1–9 were rock pigeon [N = 3659 from 15 studies, range 6–1800 individuals tested, 0.1–70% reported prevalence, estimated prevalence 16 ± 5.3% (SE)], European starling [N = 2094 from 12 studies, range 1–957 individuals tested, 0–75% reported prevalence, estimated prevalence 28 ± 6.0% (SE)], mallard [N = 1941 from 11 studies, range 5–716 individuals tested, 0–79% reported prevalence, estimated prevalence 26 ± 7.0% (SE)], Canada goose [N = 1322 from 8 studies, range 44–357 individuals tested, 0–52% reported prevalence, estimated prevalence 16 ± 6.7% (SE)], and ring‐necked pheasant [N = 932 from 8 studies, range 1–287 individuals tested, 0–37% reported prevalence, estimated prevalence 18 ± 5.6% (SE)]. Estimated Campylobacter spp. prevalence across all birds (N = 13606 individuals of 64 species) and studies (N = 56) was 27 ± 3.5% (SE). To determine prevalence with 5% precision with 27% average prevalence, we estimate that a study would have to test at least 303 individuals, but only 1.6% (7/431) of species examined met this threshold. The estimated prevalence for these seven species was 24 ± 4.4% (SE), with American crow, European starling, and mallard having the highest prevalence [52 ± 12% (SE), 28 ± 6.0% (SE), 26 ± 7.0% (SE), respectively; Fig. S7]. Antibiotic‐resistant Campylobacter spp. were reported from five species (European starling, mallard, Canada goose, ring‐necked pheasant, and house sparrow), all of which had >600 observations. 76% of reported campylobacters were identified as C. jejuni, 7.1% were identified as C. coli, 1.9% were identified as C. lari, and 0.08% were identified as C. canadensis. C. peloridis was reported once in herring gulls (Larus argentatus).
(b). E. coli
5.3% (23/431) of North American breeding birds had pathogenic E. coli prevalence data (1+ observations) meeting our inclusion criteria 1–9. The species with the most observations of data meeting inclusion criteria 1–9 on pathogenic E. coli were rock pigeon [N = 4954 from 17 studies, range 14–1800 individuals tested, 0–71% reported prevalence, estimated prevalence 8.5 ± 1.5% (SE)], European starling [N = 1081 from 5 studies, range 7–434 individuals tested, 0–14% prevalence, estimated prevalence 2.1 ± 0.6% (SE)], Canada goose [N = 1076 from 8 studies, range 1–600 individuals tested, 0–93% prevalence, estimated prevalence 33 ± 11% (SE)], house sparrow [N = 556 from 4 studies, range 40–237 individuals tested, 0–27% prevalence, estimated prevalence 8.4 ± 5.9% (SE)], and brown‐headed cowbird (Molothrus ater, N = 309 from 1 study, 3.6% prevalence across the 309 individuals tested). Estimated pathogenic E. coli prevalence across birds (N = 9185) and studies (N = 36) was 20 ± 6.3% (SE). To determine prevalence of pathogenic E. coli with 5% precision and 20% average prevalence for a given species or community, we estimate that a study would have to test at least 246 individuals; yet, only 1.6% (7/431) of species examined met this threshold. We estimated the prevalence for these seven was 14 ± 6.1% (SE). Of those species, mallard, Canada goose, and Eurasian tree sparrow (Passer montanus) had the highest prevalence [41 ± 18% (SE), 33 ± 11% (SE), 12.9 ± 5.6% (SE), respectively; Fig. S7]. Including other studies that did not report data from which prevalence could be estimated, including necropsy studies, 7.4% (32/431) of species were tested for pathogenic E. coli. 8.1% (35/431) of North American breeding birds had any data allowing calculation of prevalence of generic E. coli, which we estimated to have an overall prevalence of 54 ± 5.9% (SE). Antibiotic resistance of any E. coli was reported in 20/431 (4.6%) species.
(c). Salmonella spp.
Salmonella spp. were the most studied bacteria with 33% (141/431) of North American breeding birds having prevalence data (1+ observations) meeting our inclusion criteria 1–9. The species with the most observations of data meeting inclusion criteria 1–9 were herring gull [N = 12470 from 10 studies, range 1–5324 individuals tested, 0–22% prevalence, estimated prevalence 8.2 ± 2.2% (SE)], house sparrow [N = 5581 from 19 studies, range 2–1124 individuals tested, 0–21% prevalence, estimated prevalence 2.5 ± 0.7% (SE)], rock pigeon [N = 5458 from 30 studies, range 4–1800 individuals tested, 0–100% prevalence, estimated prevalence 4.0 ± 0.9% (SE)], wild turkey [Meleagris gallopavo, N = 2401 from 4 studies, range 70–1164 individuals, 0–22.5% prevalence, estimated prevalence 5.5 ± 4.7% (SE)], and European starling [N = 2288 from 20 studies, range 4–1800 individuals tested, 0–100% prevalence, estimated prevalence 2.7 ± 1.0% (SE)]. Estimated Salmonella spp. prevalence across all birds (N = 40295) and studies (N = 102) was 6.4 ± 0.9% (SE). To determine prevalence with 5% precision with 6.4% average prevalence, a study would have to test at least 93 individuals, but only 7.4% (32/431) of species examined met this minimum threshold. Of these 32 species, we estimated Salmonella spp. prevalence was 6.3 ± 1.0% (SE). Of these species, Franklin's gull (Leucophaeus pipixcan), white‐winged dove (Zenaida asiatica), and cattle egret had the highest prevalence [41 ± 34% (SE), 26 ± 3.3% (SE), 22 ± 15% (SE), respectively; Fig. S7]. Including necropsy studies or other studies that did not report data from which prevalence could be estimated, 36% (157/431) were tested for Salmonella spp. Antibiotic‐resistant bacteria were reported from 17 bird species. Studies reported 105 serovars, the most common of which was Typhimurium (738/2298 individual birds from studies that reported serovar), followed by Virchow (410/2298), and Bredeney (87/2298).
(d). Prevalence by taxonomic order
We summarized pathogen prevalence by taxonomic order for the three pathogens (Fig. S8; Tables S11–S16), primarily to provide a basis for comparison of bird groups with high prevalence and to point to under‐studied avenues for future research. The Gruiformes (cranes, rails) had the highest Campylobacter spp. prevalence [N = 217; estimated prevalence = 76.6 ± 10.6% (SE); Tables S11 and S12]. Other orders did not differ significantly in Campylobacter spp. prevalence (Tukey HSD, P > 0.05; Table S12). There were few individuals tested for pathogenic E. coli, limiting comparisons (Tables S13 and S14). Galliformes had the highest prevalence of pathogenic E. coli [N = 70; estimated prevalence = 55.2 ± 16.6% (SE); Tables S13 and S14], making this group a high priority for continued research. The prevalence of pathogenic E. coli was significantly higher (Tukey HSD, P < 0.05; Table S14) in the Galliformes compared to the Charadriiformes [gulls; N = 93; estimated prevalence = 0 ± 20.9% (SE)], Pelecaniformes [N = 66; estimated prevalence = 10.3 ± 13.4% (SE)], and Anseriformes [ducks, geese; N = 1563; estimated prevalence = 22.1 ± 10.3% (SE)], but other pairwise comparisons were not significant. The Pelecaniformes had the highest estimated Salmonella spp. prevalence [Tables S15 and S16; N = 566, 16.8 ± 3.7% (SE)], which was significantly higher (Tukey HSD, P < 0.05; Table S16) than Salmonella spp. prevalence in the Passeriformes [N = 13997; estimated prevalence = 4.8 ± 1.0% (SE)], Anseriformes [N = 3136; estimated prevalence = 4.7 ± 1.2% (SE)], Columbiformes [doves, pigeons; N = 5724; estimated prevalence = 5.2 ± 1.3% (SE)], Charadriiformes [N = 13395; estimated prevalence = 8.4 ± 1.5% (SE)], Strigiformes [owls; N = 151; estimated prevalence = 7.4 ± 3.1% (SE)], and Galliformes [N = 2688; estimated prevalence = 8.0 ± 2.6% (SE)]. Salmonella spp. prevalence was higher in the Charadriiformes than the Columbiformes, Passeriformes, and Anseriformes.
(2). Taxonomic bias in research
To assess taxonomic bias in study species selection and representativeness of the current literature of birds that contact humans and food production, we used a two‐part comparison. First, we compared observations by taxon to both the percentage of North American breeding birds each order comprises (species richness comparison) and the percentage of eBird observations by order (abundance comparison). Second, we compared observations by taxon to a database on farm bird abundances from Smith et al. (2019) to assess representativeness of currently available literature with respect to wild birds present within a vulnerable farming system as a case study of one of several important transmission points of enteric pathogens. The analyses revealed similar trends, so we only present results from the first comparison below and refer the reader to Tables S17–S19 and Figs S9 and S10 for farm bird summaries.
Compared to the percentage of species of North American breeding birds each taxon represents, there was significant bias in the number of individuals tested by taxonomic order for Campylobacter spp. (χ2 17 = 70004, P < 0.0001), pathogenic E. coli (χ2 17 = 154,655, P < 0.0001), generic E. coli (χ2 17 = 39333, P < 0.0001), and Salmonella spp. (χ2 17 = 90432, P < 0.0001) (Fig. S11; Table S20). Compared to the percentage of eBird sightings reported for North American breeding birds within each taxon, there was significant bias in the number of individuals tested by taxonomic order for Campylobacter spp. (χ2 17 = 36553, P < 0.0001), pathogenic E. coli (χ2 17 = 57753, P < 0.0001), generic E. coli (χ2 17 = 21314, P < 0.0001), and Salmonella spp. (χ2 17 = 98876, P < 0.0001) (Fig. 3; Table S21). Anseriformes, Charadriiformes, and Columbiformes tended to be overrepresented, while Passeriformes were under‐represented in all categories. No data meeting inclusion criteria 1–6 or 8–9 existed for one or more of the bacteria for Accipitriformes (hawks, eagles, new world vultures), Caprimulgiformes (nighthawks, swifts, hummingbirds), Falconiformes (falcons), Gruiformes, Pelecaniformes, Piciformes (woodpeckers), Strigiformes, or Suliformes (cormorants). No data meeting inclusion criteria 1–6 or 8–9 existed for any bacteria for Ciconiiformes (storks), Coraciiformes (kingfishers), Cuculiformes (cuckoos), Gaviiformes (loons), or Podicipediformes (grebes). Passeriformes were the most diverse and abundant taxon within all data sets examined, suggesting that further research on this group should be a high priority. Accipitriformes and Piciformes were also common species in both data sets, but few studies have tested them for pathogens, again suggesting high priority for testing these taxa.
Figure 3.
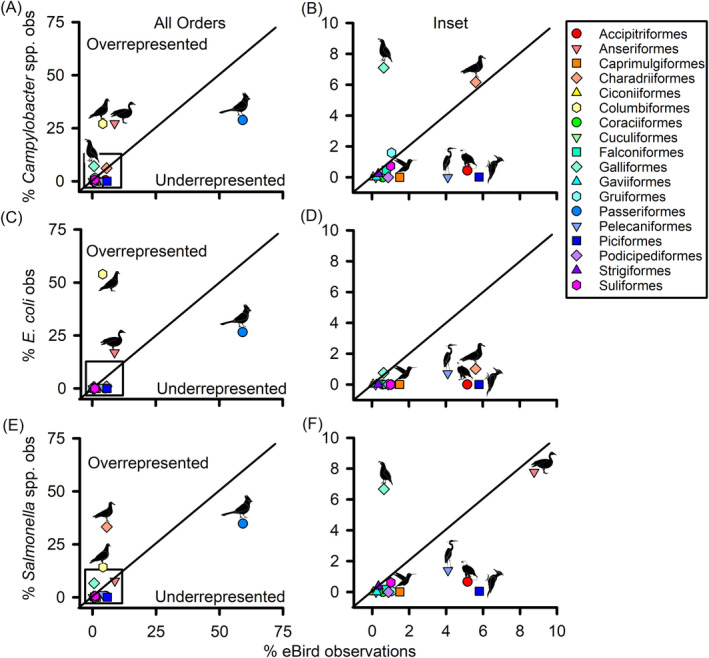
Scatterplot showing the percentage of pathogen observations (obs) belonging to each taxonomic order versus the % of eBird observations (eBird.org) each taxon comprises (Sullivan et al., 2009). (A), (C) and (E) show all orders. (B), (D) and (F) show orders that comprise less than 10% of pathogen observations and less than 10% of eBird observations (boxed regions in A, C and E, respectively).
Next, we examined what percentage of species and what proportion of sightings represented individuals with enough data to estimate prevalence with 5% precision for no, one, two, or three pathogens (Fig. 4; Tables S19 and S22). Of all North American breeding bird species, 5/431 (1.2%) had sufficient data to calculate prevalence for three pathogens, 4/431 (0.9%) had sufficient data to calculate prevalence for two pathogens, 23/431 (5.3%) had sufficient data to calculate prevalence for one pathogen, 119/431 (28%) had some data but insufficient observations to determine prevalence, and 280/431 (65%) had no data (Fig. 4A). Our second analysis considered the percentage of birds in North America that had enough data to estimate prevalence with 5% precision for no, one, two, or three pathogens. Thus, we weighted each species by the number of observations in eBird and divided each by the total number of individuals reported across species in eBird. 7.6% of all sightings were comprised of species that had sufficient data to calculate prevalence for three pathogens, 3.1% of sightings were comprised of species with enough data to calculate prevalence for two pathogens, 12% of sightings were comprised of species with enough data to calculate prevalence for one pathogen, 51% of sightings were comprised of species that had some data but insufficient observations to determine prevalence, and 26% of sightings were comprised of species that had no data (Fig. 4B). These trends point to large inadequacies of the current literature for the majority of species and individuals that may contact humans. Future work should focus on species that are currently under‐studied to understand whole‐community risk better (Fig. 5). Although prior work has focused on key globally abundant species, large portions of highly abundant species have few to no data (e.g. American robin), which hinders the ability of robust risk modelling efforts to control enteric pathogen transmission to humans.
Figure 4.
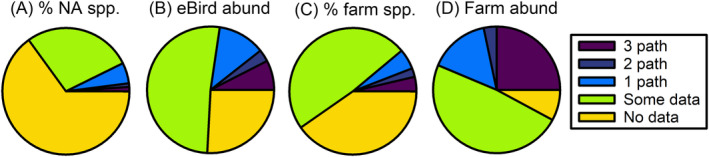
Pie charts showing (A, C) the proportion of species or (B, D) relative abundances for which enough observations exist to estimate pathogen prevalence for Campylobacter spp., pathogenic E. coli and Salmonella spp. (purple), two of the three pathogens (dark blue), one of these three pathogens (blue), species with some data but insufficient numbers to determine prevalence (green), and no observations (yellow). (A) North American (NA) breeding bird species found in the North American Breeding Bird Survey (Sauer et al., 2017), (B) eBird (ebird.org) relative abundances (Sullivan et al., 2009), (C) West Coast farm bird species observed by Smith et al. (2019), and (D) farm bird species relative abundances from the database in Smith et al. (2019). Path = pathogens.
Figure 5.
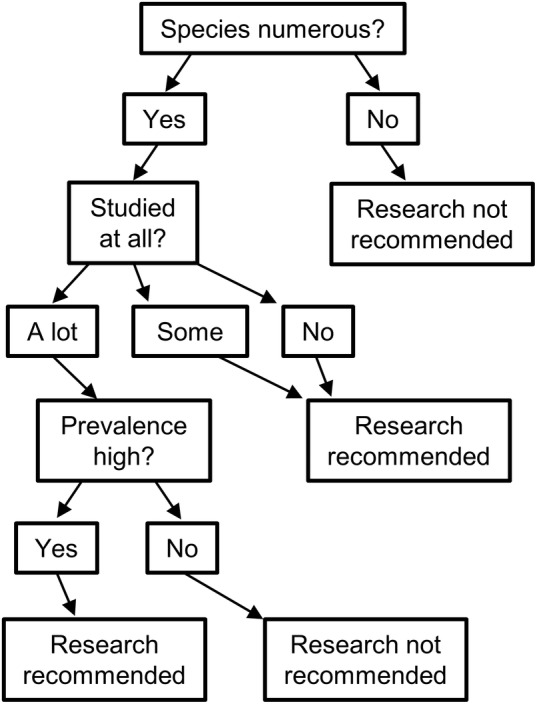
Flow chart suggesting directions for future research.
(3). Prevalence by testing method
Next, we compared the prevalence of Salmonella spp. for three of the most tested species (European starling, house sparrow, and rock pigeon) by substance tested for bacteria [cloacal swabs, faeces, blood, and dissected internal organs (‘necropsy’)]. We hypothesized that studies that tested internal organs would find higher prevalence of pathogens because a bird would not have to be shedding bacteria in order to obtain a positive result (Girdwood et al., 1985). If this were the case, species that are commonly tested by internal organ cultures after culling (invasive birds, hunted waterfowl, gulls, hunted upland game birds such as pheasants) may appear to have higher prevalence than species that require special permits to kill (native songbirds and other protected migratory birds that are not hunted for recreation). We did not make comparisons by substance tested for gamebirds or native species due to limited data.
Studies that tested house sparrow internal organs (model estimate 4.0 ± 0.6%) had higher prevalence than those that tested cloacal swabs (model estimate 0.7 ± 0.2%) or faeces (0.2 ± 0.2%) (Z = 5.33, P < 0.0001 and Z = 6.06, P < 0.0001, respectively) (Fig. 6; Table S23). Cloacal swabs had higher prevalence than faeces (Z = −2.1, P = 0.035). However, prevalence did not differ by substance tested for European starlings or rock pigeons (P > 0.05; Fig. 6; Tables S24 and S25). In line with our findings for house sparrows, Girdwood et al. (1985) found that 15.6% (N = 746) of gulls tested were positive for Salmonella spp. when the entire gut was tested whereas only 9.6% of the same gulls were positive when cloacal swabs were tested. Therefore, we suggest some caution when comparing prevalence between species that are often killed and dissected (geese, invasive species, gulls) compared to protected native species (finches, thrushes, etc.), but it does not appear that necropsies always yield higher prevalence estimates. We suggest that future work uses consistent methodology to facilitate comparisons among species.
Figure 6.
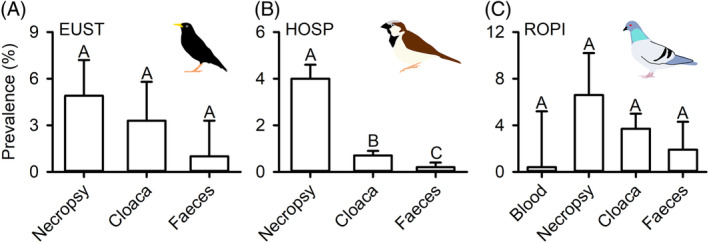
Comparison of estimated Salmonella spp. prevalence (+SE) by substance tested for the three species with data from the most individual studies. (A) European starling, (B) house sparrow, and (C) rock pigeon. Different letters indicate significant differences using pairwise Tukey HSD tests.
(4). Prevalence by ecological guild
Next, we used our meta‐data to assess the idea that foraging traits may alter enteric pathogen prevalence in wild birds by altering exposure rates to faecal contamination (Waldenström et al., 2002; Skov et al., 2008; Hald et al., 2016). Across 85 Tukey HSD pairwise comparisons of diet guild and foraging strata for the three enteric pathogens (Fig. 7; Fig. S12; Tables S26–S37), only two were significant: omnivores had higher Salmonella spp. prevalence [N = 20577; 6.9 ± 1.0% (SE)] than granivores [N = 13252; 5.4 ± 1.0% (SE); Z = 2.33; P = 0.020], and omnivores had higher Salmonella spp. prevalence than herbivores [N = 2642; 4.9 ± 1.3% (SE); Z = 1.99; P = 0.047]. We note the limitations in our data due to heavy representation of a few species that represent only a fraction of common urban and farm inhabitants and note the low number of observations for most guilds. Thus, we suggest extreme caution in extrapolating any ecological trends from our analyses. Our results may also be biased due to confounding factors such as species traits (e.g. habitat association), sampling biases resulting from lack of data for most species, or both. For example, as summarized in our literature review above, primary studies have generally found lower enteric pathogen prevalence in granivorous species and higher enteric pathogen prevalence in ground‐foraging species which may have lower and higher enteric pathogen exposure, respectively (Waldenström et al., 2002; Sensale et al., 2006; Skov et al., 2008; Hald et al., 2016). By contrast, we found only two differences across many comparisons. An alternative explanation is that few species forage exclusively within a single diet guild or strata and most exhibit some level of diet plasticity (Wilman et al., 2014). Thus, grouping species into discrete classifications may be of little use in quantifying an association between diet or foraging strata and enteric pathogen prevalence.
Figure 7.
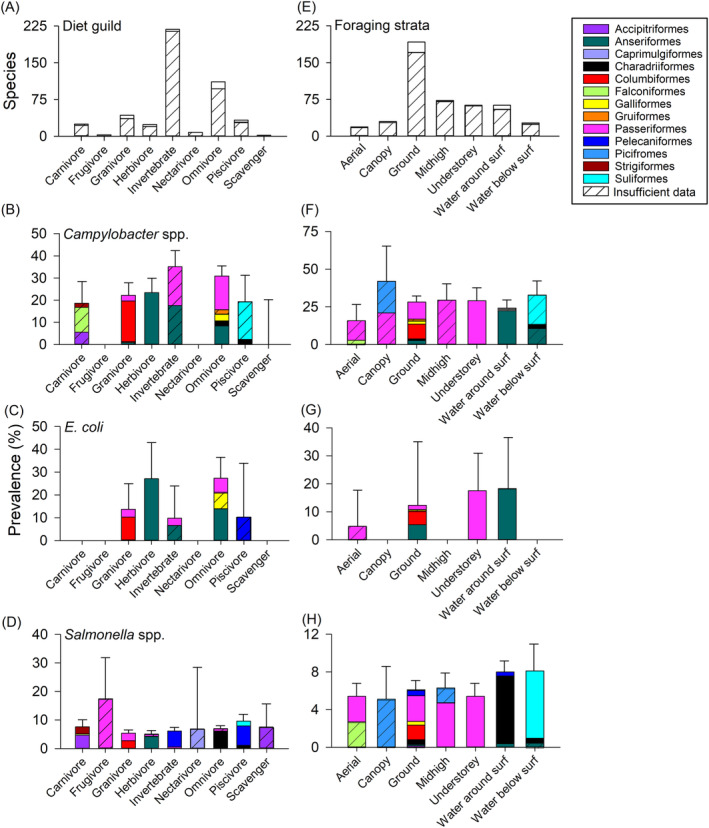
Estimated prevalence (+SE) of enteric pathogens by (B–D) diet guild and (F–H) foraging strata, and number of species within each diet guild (A) and foraging strata (E). In A and E, pattern indicates insufficient observations to determine prevalence for any of the three pathogens while white indicates species for which enough observations were available to determine prevalence for one or more pathogens. (B, F) Campylobacter spp., (C, G) pathogenic E. coli, and (D, H) Salmonella spp. for each diet guild or each foraging strata. Colour indicates proportion of positive individuals for each estimate from each taxonomic order. Pattern indicates insufficient observations to determine prevalence within an order. Spaces with no error bars indicate no observations in the literature.
V. FUTURE RESEARCH
In Section III, we developed a conceptual framework for developing risk models, expanding upon those proposed by Lloyd‐Smith et al. (2009) and Plowright et al. (2017) (Fig. 1), and suggested avenues for future research. Based on a literature review, we identified critical gaps in knowledge throughout the pathogen life cycle and recommend that future studies use an approach that integrates across all aspects of pathogen transmission, from environmental exposure to successful colonization of human hosts (Fig. 1) (e.g. Hernandez et al., 2016). In Section IV, we used a meta‐analytic approach to summarize pathogen prevalence by species and orders, and we assessed what species, orders, and ecological guilds should be priorities for future research. Here, we synthesize what we see as the highest priority areas for future research.
We recommend that research efforts quantify prevalence in common but understudied species as a top priority (Fig. 5). We found few to no estimates of prevalence for Passeriformes, Accipitriformes, Caprimulgiformes, Falconiformes, Gruiformes, Piciformes, and Strigiformes, despite being common in agricultural and anthropogenic settings and representing 74% of all North American breeding birds. By contrast, we found disproportionately high amounts of data focused on Anseriformes, Charadriiformes, and Columbiformes (Fig. 3; Figs S9–S11; Tables S17, S18, S20 and S21). A large proportion of our meta‐data included studies of a few highly abundant, globally distributed species, including rock pigeons, European starlings, Canada geese, mallards, herring gulls, and house sparrows (54.5% of studies were conducted outside of North America). We found few estimates on many common North American breeding birds (Fig. 4): 65% of North American breeding bird species had no pathogen data, and only 1.2% of species had enough data to determine prevalence for all three pathogens. Although data on a few key globally distributed species have the largest reach, some of the most abundant species in North America had few to no data, although necropsy studies indicate many of these species may be important reservoirs (e.g. American robin). This highlights a critical need for studies to focus on common, yet underrepresented species that could have disproportionately high influence on pathogen transmission within human‐dominated landscapes (Fig. 5).
Although we cannot eliminate the possibility that our taxonomic and foraging guild analyses simply reflect a bias in the research, we suggest they may, to some degree, reflect unique pathogen–host–environment relationships that create some highly competent systems, driven by the unique lifestyle and survivability of each enteric pathogen (Jones et al., 2013). Campylobacter spp. occurred at high prevalence (27%) throughout the groups tested compared to pathogenic E. coli (20%) and Salmonella spp. (6.4%) (Figs S7–S18; Tables S11, S13, S15). Campylobacter spp. are generally thought to be a natural commensal of wild birds with a more recent crossover into humans and livestock (Wassenaar, 2011; Griekspoor et al., 2013). As a commensal, there would be little immune investment to kill the bacteria, allowing them to remain at high prevalence. This might suggest that prevalence is dictated more by survival in the non‐host environment, which is thought to be low in comparison to Salmonella spp. and E. coli (see Section III.4). Because Campylobacter spp. generally have low survival on plant surfaces (Brandl et al., 2004) but high prevalence in bird hosts, human disease from Campylobacter spp. may be more likely from consumption of poultry, game, or direct contact with faeces. Indeed, Campylobacter infection in humans most commonly arises from consumption of poultry meat rather than from produce, although exceptions do occur (Batz et al., 2012). However, most cases of human Campylobacteriosis of avian origin are thought to occur from direct faecal contact (Strachan et al., 2013; Cody et al., 2015).
In contrast to Campylobacter spp., Salmonella spp. can cause mass mortalities in wild birds, with the highest documented mortalities in songbirds caused by Salmonella Typhimurium (Tizard, 2004; Connolly et al., 2006; Hall & Saito, 2008). It is, therefore, hypothesized that Salmonella spp. should exist at low prevalence in wild birds (Brittingham et al., 1988; Tizard, 2004; Hall & Saito, 2008), which our data supported (6.4% prevalence). Salmonella spp. are also thought to have high survival in the environment compared to the other pathogens (Winfield & Groisman, 2003). Salmonella spp. also have a large host range, including mammals, reptiles, birds, and amphibians (Navarro‐Gonzalez et al., 2016). It has been suggested that raptors may have high exposure to Salmonella spp. through consumption of contaminated rodents or infected birds (Tizard, 2004), which our data suggest may be the case (7.5% in Accipitriformes, 9.5% in Falconiformes, and 7.4% in Strigiformes; Table S15) and point to avenues of future research on this agriculturally important group (Shave et al., 2018). Thus, Salmonella spp. likely exist at lower prevalence in wild birds than Campylobacter spp. and E. coli due to negative impacts on host health (Tizard, 2004; Hall & Saito, 2008), but high survival in the environment and possibly lower impacts on large birds such as the Pelecaniformes (17% prevalence) and Charadriiformes (8.4% prevalence) may lead to higher prevalence in large, water‐associated birds (Fig. S8; Table S15) (Winfield & Groisman, 2003; Tizard, 2004).
We found the least data on pathogenic E. coli of the three pathogens examined. The Galliformes (55% prevalence;) appear to be especially competent hosts (Figs S7 and S8; Tables S13 and S14), although it is unclear what role other species play due to low sample sizes. Pheasants may have had high prevalence due to captive rearing and release for hunting (Nebola et al., 2007). Despite the associations between several bird taxa and particular pathogens described above, we find it likely that our results are a combination of research bias and competent pathogen–host–environment systems. Future research efforts will need to continue to determine what constitutes a highly competent pathogen–host–environment system by integrating all components of the host–pathogen cycle, from exposure to reservoir competence, contact, bacterial environmental survival, and colonization of a human host (Fig. 1).
We end with several conclusions from our review of the literature. First, the data are too limited and biased currently to make any data‐driven recommendations for managing wild birds to reduce enteric pathogen spillover to people or livestock. Beyond collecting data on understudied species, we suggest a few key pieces of data will be most crucial to advancing policy to reduce enteric pathogen transmission between humans and wild birds. First, robust studies demonstrating that wild birds and humans/livestock share the same strains are needed (Table S10). Experiments that inoculate wild birds with bacterial strains of human, livestock, and wild bird origin will provide the strongest evidence (e.g. Atterby et al., 2018). Current evidence suggests wild birds are often poor reservoir hosts of human strains (Waldenström et al., 2010; Atterby et al., 2018). Second, experiments determining the long‐term shedding potential of enteric pathogens by wild birds are crucial. Understanding long‐term shedding potential will help to determine if species commonly infected in winter such as birds at feeders (Tizard, 2004) can shed enteric bacteria in summer when children commonly contact faeces outdoors and most crops are grown. Third, studies must quantify contact rates, direct and indirect, in developing risk assessments. Finally, determining the shedding intensity and subsequent survival of bacteria in wild bird faeces, particularly for songbirds, is a key piece of missing information from the literature. Bacteria in bird faeces must survive long enough at an infectious dose until consumed by a human to potentially infect a human host, so shedding intensity and subsequent survival data are crucial for risk models. Without these four key pieces of information, studies could over‐estimate risk if only considering the most basic estimate of prevalence. It is likely that the current focus on pathogen prevalence has over‐inflated the perceived risk of wild birds to human health.
VI. CONCLUSIONS
(1) Wild birds are often thought to pose a risk to human health through transmission of enteric pathogens, yet few data are available to assess this idea. Here, we developed a comprehensive framework for understanding spillover of enteric pathogens from wild birds to humans and identified where important knowledge gaps remain.
(2) Only 3% of studies included in our meta‐analysis provided data from all phases in the pathogen transmission process included in our conceptual framework. Most studies provided data limited to pathogen prevalence, but even prevalence data were limited to a small number of common species (e.g. rock pigeons, European starlings, mallards, Canada geese, house sparrows, herring gulls). No pathogen prevalence data were available for 65% of North American breeding bird species, including many commonly in contact with humans and agricultural production (e.g. many Passeriformes and raptors).
(3) We found an overall Campylobacter spp. prevalence of 27%, pathogenic E. coli prevalence of 20%, and Salmonella spp. prevalence of 6.4%. These estimates were derived from data from only 14.8% of North American breeding bird species for Campylobacter spp., 5.3% of bird species for pathogenic E. coli, and 33% of bird species for Salmonella spp. Given that most bird species in North America are understudied or entirely untested as reservoirs of enteric bacteria, it remains unknown how important a role most birds play in the risk of enteric pathogen transmission to humans.
(4) The primary focus in the literature on pathogen prevalence data likely overestimates the probability of enteric pathogen spillover from wild birds to humans because a pathogen from bird faeces must survive at an infectious dose until consumed by humans and be a strain able to cause disease in humans. For example, although we estimated a Campylobacter spp. prevalence of 26% in mallards (apparently high risk), Colles et al. (2011) estimated that only 0.9% of mallard Campylobacter spp. isolates cause human disease, suggesting the prevalence of human pathogenic Campylobacter spp. strains is about 2.3 in 1000 mallards, much lower than the Campylobacter genus prevalence alone suggests.
(5) We conclude that the current research is not sufficient for estimating the risk of enteric pathogen spillover from wild birds to humans. Future research should focus on the large number of under‐studied species commonly in contact with people and food production, identify if bacteria are human‐pathogenic strains, and model the entire transmission process. Fully assessing the probability of enteric pathogen spillover from wild birds to humans is complex and data intensive. Yet, the only way to develop data‐driven and effective management strategies for reducing pathogen transmission is to understand the entire transmission process fully. Only upon doing this can effective policy be implemented to reduce spillover.
Supporting information
S1. Supplementary tables and figures.
Data S1. List of studies and their methods included in meta‐analysis.
Data S2. Meta‐data on enteric pathogen prevalence in North American breeding bird species.
Data S3. Meta‐data of enteric pathogen prevalence for each study included in meta‐regressions.
VII. ACKNOWLEDGEMENTS
We thank J. Brunner, K. Cornell, and C. Latimer for helpful comments on earlier versions of this review.
REFERENCES
References marked with asterisk have been cited within the supporting information
- Abdollahpour, N. , Zendehbad, B. , Alipour, A. & Khayatzadeh, J. (2015). Wild–bird feces as a source of Campylobacter jejuni infection in children's playgrounds in Iran. Food Control 50, 378–381. [Google Scholar]
- * Adesiyun, A. A. , Seepersadsingh, N. , Inder, L. & Caesar, K. (1998). Some bacterial enteropathogens in wildlife and racing pigeons from Trinidad. Journal of Wildlife Diseases 34, 73–80. [DOI] [PubMed] [Google Scholar]
- * Aguirre, A. A. , Quan, T. J. , Cook, R. S. & Mclean, R. G. (1992). Cloacal flora isolated from wild black‐bellied whistling ducks (Dendrocygna autumnalis) in Laguna La Nacha, Mexico. Avian Diseases 36, 459–462. [PubMed] [Google Scholar]
- Albuquerque, A. , Cardoso, W. , Teixeira, R. , Lopes, E. , Sales, R. , Horn, R. , Rocha‐E‐Silva, R. , Bezerra, W. & Gomes‐Filho, V. (2013). Dissemination of Salmonella Enterititids by experimentally–infected pigeons. Brazilian Journal of Poultry Science 15, 211–215. [Google Scholar]
- * Alcalá, L. , Alonso, C. A. , Simón, C. , González‐Esteban, C. , Orós, J. , Rezusta, A. , Ortega, C. & Torres, C. (2016). Wild birds, frequent carriers of extended‐spectrum β‐lactamase (ESBL) producing Escherichia coli of CTX‐M and SHV‐12 types. Microbial Ecology 72, 861–869. [DOI] [PubMed] [Google Scholar]
- * Al‐Harbi, A. H. (2003). Faecal coliforms in pond water, sediments and hybrid tilapia Oreochromis niloticus × Oreochromis aureus in Saudi Arabia. Aquaculture Research 34, 517–524. [Google Scholar]
- Allan, B. F. , Keesing, F. & Ostfeld, R. S. (2003). Effect of forest fragmentation on Lyme disease risk. Conservation Biology 17, 267–272. [Google Scholar]
- * Alley, M. R. , Connolly, J. H. , Fenwick, S. G. , Mackereth, G. F. , Leyland, M. J. , Rogers, L. E. , Haycock, M. , Nicol, C. & Reed, C. E. M. (2002). An epidemic of Salmonellosis caused by Salmonella Typhimurium DT160 in wild birds and humans in New Zealand. New Zealand Veterinary Journal 50, 170–176. [DOI] [PubMed] [Google Scholar]
- * Alroy, K. & Ellis, J. C. (2011). Pilot study of antimicrobial‐resistant Escherichia coli in herring gulls (Larus argentatus) and waste‐water in the Northeastern United States. Journal of Zoo and Wildlife Medicine 42, 160–163. [DOI] [PubMed] [Google Scholar]
- Altizer, S. , Bartel, R. & Han, B. A. (2011). Animal migration and infectious disease risk. Science 331, 296–303. [DOI] [PubMed] [Google Scholar]
- * Antilles, N. , Sanglas, A. & Cerdà‐Cuéllar, M. (2015). Free‐living waterfowl as a source of zoonotic bacteria in a dense wild bird population area in Northeastern Spain. Transboundary and Emerging Diseases 62, 516–521. [DOI] [PubMed] [Google Scholar]
- Atterby, C. , Mourkas, E. , Méric, G. , Pascoe, B. , Wang, H. , Waldenström, J. , Sheppard, S. K. , Olsen, B. & Järhult, J. D. (2018). The potential of isolation source to predict colonization in avian hosts: a case study in Campylobacter jejuni strains from three bird species. Frontiers in Microbiology 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Badouei, M. A. , Salehi, T. Z. , Koochakzadeh, A. , Kalantari, A. & Tabatabaei, S. (2014). Molecular characterization, genetic diversity and antibacterial susceptibility of Escherichia coli encoding Shiga toxin 2f in domestic pigeons. Letters in Applied Microbiology 59, 370–376. [DOI] [PubMed] [Google Scholar]
- * Báez, J. , Hernández‐García, M. , Guamparito, C. , Díaz, S. , Olave, A. , Guerrero, K. , Cantón, R. , Baquero, F. , Gahona, J. , Valenzuela, N. , del Campo, R. & Silva, J. (2014). Molecular characterization and genetic diversity of ESBL‐producing Escherichia coli colonizing the migratory Franklin's gulls (Leucophaeus pipixcan) in Antofagasta, north of Chile. Microbial Drug Resistance 21, 111–116. [DOI] [PubMed] [Google Scholar]
- Barron, D. G. , Gervasi, S. S. , Pruitt, J. N. & Martin, L. B. (2015). Behavioral competence: how host behaviors can interact to influence parasite transmission risk. Current Opinion in Behavioral Sciences 6, 35–40. [Google Scholar]
- Batz, M. B. , Hoffmann, S. & Morris, J. G. (2012). Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. Journal of Food Protection 75, 1278–1291. [DOI] [PubMed] [Google Scholar]
- Benjamin, L. , Atwill, E. R. , Jay‐Russell, M. , Cooley, M. , Carychao, D. , Gorski, L. & Mandrell, R. E. (2013). Occurrence of generic Escherichia coli, E. coli O157 and Salmonella spp. in water and sediment from leafy green produce farms and streams on the Central California coast. International Journal of Food Microbiology 165, 65–76. [DOI] [PubMed] [Google Scholar]
- Benskin, C. M. H. , Wilson, K. , Jones, K. & Hartley, I. R. (2009). Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biological Reviews 84, 349–373. [DOI] [PubMed] [Google Scholar]
- Beretti, M. & Stuart, D. (2008). Food safety and environmental quality impose conflicting demands on Central Coast growers. California Agriculture 62, 68–73. [Google Scholar]
- Berger, C. N. , Sodha, S. V. , Shaw, R. K. , Griffin, P. M. , Pink, D. , Hand, P. & Frankel, G. (2010). Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environmental Microbiology 12, 2385–2397. [DOI] [PubMed] [Google Scholar]
- Black, R. E. , Levine, M. M. , Clements, M. L. , Hughes, T. P. & Blaser, M. J. (1988). Experimental Campylobacter jejuni infection in humans. The Journal of Infectious Diseases 157, 472–479. [DOI] [PubMed] [Google Scholar]
- Blaser, M. J. & Newman, L. S. (1982). A review of human Salmonellosis: I. Infective dose. Reviews of Infectious Diseases 4, 1096–1106. [DOI] [PubMed] [Google Scholar]
- * Bonnedahl, J. , Stedt, J. , Waldenström, J. , Svensson, L. , Drobni, M. & Olsen, B. (2015). Comparison of extended‐spectrum β‐lactamase (ESBL) CTX‐M genotypes in Franklin gulls from Canada and Chile. PLoS One 10, e0141315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Borges, C. A. , Cardozo, M. V. , Beraldo, L. G. , Oliveira, E. S. , Maluta, R. P. , Barboza, K. B. , Werther, K. & Ávila, F. A. (2017). Wild birds and urban pigeons as reservoirs for diarrheagenic Escherichia coli with zoonotic potential. Journal of Microbiology 55, 344–348. [DOI] [PubMed] [Google Scholar]
- * Borilova, G. , Nebola, M. & Steinhauserova, I. (2007). Occurrence and antibiotic resistance of Campylobacter spp. isolated from pheasants (Phasianus colchicus spp. torquatus). Archiv für Lebensmittelhygiene 58, 183–187. [Google Scholar]
- Bovay, J. & Sumner, D. A. (2018). Economic effects of the U.S. Food Safety Modernization Act. Applied Economic Perspectives and Policy 40, 402–420. [Google Scholar]
- Brandl, M. T. (2006). Fitness of human enteric pathogens on plants and implications for food safety. Annual Review of Phytopathology 44, 367–392. [DOI] [PubMed] [Google Scholar]
- Brandl, M. T. , Haxo, A. F. , Bates, A. H. & Mandrell, R. E. (2004). Comparison of survival of Campylobacter jejuni in the phyllosphere with that in the rhizosphere of spinach and radish plants. Applied and Environmental Microbiology 70, 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl, M. T. , Rosenthal, B. M. , Haxo, A. F. & Berk, S. G. (2005). Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Applied and Environmental Microbiology 71, 1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, O. E. , Larsen, K. H. & Mott, D. F. (2018). Winter movements and activities of radio‐equipped starlings. The Journal of Wildlife Management 39, 795–801. [Google Scholar]
- Brearley, G. , Rhodes, J. , Bradley, A. , Baxter, G. , Seabrook, L. , Lunney, D. , Liu, Y. & Mcalpine, C. (2013). Wildlife disease prevalence in human‐modified landscapes. Biological Reviews 88, 427–442. [DOI] [PubMed] [Google Scholar]
- Brittingham, M. C. , Temple, S. A. , Duncan, R. M. , Temple, A. & Duncan, M. (1988). A survey of the prevalence of selected bacteria in wild birds. Journal of Wildlife Diseases 24, 299–307. [DOI] [PubMed] [Google Scholar]
- Brobey, B. , Kucknoor, A. & Armacost, J. (2017). Prevalence of Trichomonas, Salmonella, and Listeria in wild birds from Southeast Texas. Avian Diseases 61, 347–352. [DOI] [PubMed] [Google Scholar]
- Broman, T. , Waldenstrom, J. , Dahlgren, D. , Carlsson, I. , Eliasson, I. & Olsen, B. (2004). Diversities and similarities in PFGE profiles of Campylobacter jejuni isolated from migrating birds and humans. Journal of Applied Microbiology 96, 834–843. [DOI] [PubMed] [Google Scholar]
- Buchholz, A. P. S. & Fairbrother, A. (1992). Pathogenicity of Salmonella pullorum in northern bobwhite quail and mallard ducks. Avian Diseases 36, 304–312. [PubMed] [Google Scholar]
- Butterfield, J. , Coulson, J. C. , Kearsey, S. V. , Monaghan, P. , Mccoy, J. H. & Spain, G. E. (1983). The herring gull Larus argentatus as a carrier of Salmonella . The Journal of Hygiene 91, 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Caballero, M. , Rivera, I. , Jara, L. M. , Ulloa‐Stanojlovic, F. M. & Shiva, C. (2015). Isolation and molecular identification of potentially pathogenic Escherichia coli and Campylobacter jejuni in feral pigeons from an urban area in the city of Lima, Peru. Revista do Instituto de Medicina Tropical de São Paulo 57, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, T. R. , Edrington, T. S. & Nisbet, D. J. (2014). Isolation of Escherichia coli O157:H7 and Salmonella from migratory brown‐headed cowbirds (Molothrus ater), common grackles (Quiscalus quiscula), and cattle egrets (Bubulcus ibis). Foodborne Pathogens and Disease 11, 791–794. [DOI] [PubMed] [Google Scholar]
- Carlson, J. C. , Franklin, A. B. , Hyatt, D. R. , Pettit, S. E. & Linz, G. M. (2011). The role of starlings in the spread of Salmonella within concentrated animal feeding operations. Journal of Applied Ecology 48, 479–486. [Google Scholar]
- Carlson, J. C. , Hyatt, D. R. , Ellis, J. W. , Pipkin, D. R. , Mangan, A. M. , Russell, M. , Bolte, D. S. , Engeman, R. M. , Deliberto, T. J. & Linz, G. M. (2015). Mechanisms of antimicrobial resistant Salmonella enterica transmission associated with starling – livestock interactions. Veterinary Microbiology 179, 60–68. [DOI] [PubMed] [Google Scholar]
- * Carroll, D. , Wang, J. , Fanning, S. & Mcmahon, B. J. (2015). Antimicrobial resistance in wildlife: implications for public health. Zoonoses and Public Health 62, 534–542. [DOI] [PubMed] [Google Scholar]
- * Cass, J. S. & Williams, J. E. (1947). Salmonella pullorum recovered from a wild pheasant in Minnesota. Journal of the American Veterinary Medical Association 111, 282. [PubMed] [Google Scholar]
- Cesare, A. D. E. , Sheldon, B. W. , Smith, K. S. & Jaykus, L. (2003). Survival and persistence of Campylobacter and Salmonella species under various organic loads on food contact surfaces. Journal of Food Protection 66, 1587–1594. [DOI] [PubMed] [Google Scholar]
- * Chandran, A. & Mazumder, A. (2014). Occurrence of diarrheagenic virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various avian hosts in British Columbia, Canada. Applied and Environmental Microbiology 80, 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Charlton, K. G. (2000). Antibodies to selected disease agents in translocated wild turkeys in California. Journal of Wildlife Diseases 36, 161–164. [DOI] [PubMed] [Google Scholar]
- Childs, M. L. , Nova, N. , Colvin, J. & Mordecai, E. A. (2019). Mosquito and primate ecology predict human risk of yellow fever virus spillover in Brazil. Philosophical Transactions of the Royal Society B: Biological Sciences 374, 20180335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Čížek, A. , Literák, I. , Hejlíček, K. , Treml, F. & Smola, J. (1994). Salmonella contamination of the environment and its incidence in wild birds. Journal of Veterinary Medicine. Series B 41, 320–327. [DOI] [PubMed] [Google Scholar]
- Clark, L. (2014). Disease risks posed by wild birds associated with agricultural landscapes In The Produce Contamination Problem: Causes and Solutions (ed. Matthews, G.M. Sapers, & C.P. Gerba K. R.), pp. 139–166. Oxford, England: Academic Press. [Google Scholar]
- * Clavijo, A. , Robinson, Y. & López, J. (2001). Isolation of Newcastle disease virus and Salmonella typhimurium from the brain of double‐crested cormorants (Phalacrocorax auritus). Avian Diseases 45, 245–250. [PubMed] [Google Scholar]
- * Clegg, F. G. & Hunt, A. E. (1975). Salmonella infection in mute swans (Cygnus olor). The Veterinary Record 97, 373. [DOI] [PubMed] [Google Scholar]
- Cody, A. J. , Mccarthy, N. D. , Bray, J. E. , Wimalarathna, H. M. L. , Colles, F. M. , Van Rensburg, M. J. J. , Dingle, K. E. , Waldenström, J. & Maiden, M. C. J. (2015). Wild bird‐associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environmental Microbiology Reports 7, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles, F. M. , Ali, J. S. , Sheppard, S. K. , Mccarthy, N. D. & Maiden, M. C. J. (2011). Campylobacter populations in wild and domesticated mallard ducks (Anas platyrhynchos). Environmental Microbiology Reports 3, 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles, F. M. , Mccarthy, N. D. , Hoew, J. C. , Devereux, C. L. , Gosler, A. G. & Maiden, M. C. J. (2009). Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European starling). Environmental Microbiology 11, 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, J. H. , Dutton, G. J. , Rogers, L. E. , Connolly, J. H. , Dutton, G. J. & Rogers, L. E. (2006). Infectivity and persistence of an outbreak strain of Salmonella enterica serotype Typhimurium DT160 for house sparrows (Passer domesticus) in New Zealand. New Zealand Veterinary Journal 54, 329–332. [DOI] [PubMed] [Google Scholar]
- * Coulson, J. C. , Butterfield, J. & Thomas, C. (1983). The herring gull Larus argentatus as a likely transmitting agent of Salmonella montevideo to sheep and cattle. The Journal of Hygiene 91, 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Craven, S. E. , Stern, N. J. , Line, E. , Bailey, J. S. , Cox, N. A. & Fedorka‐Cray, P. (2000). Determination of the incidence of Salmonella spp., Campylobacter jejuni, and Clostridium perfringens in wild birds near broiler chicken houses by sampling intestinal droppings. Avian Diseases 44, 715–720. [PubMed] [Google Scholar]
- Cross, P. C. , Prosser, D. J. , Ramey, A. M. , Hanks, E. M. & Pepin, K. M. (2019). Confronting models with data: the challenges of estimating disease spillover. Philosophical Transactions of the Royal Society B: Biological Sciences 374, 20180435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Dakman, A. , Yapicier, Ö. Ş. , Yaşarer, A. & Güleç, M. (2017). First isolation of Salmonella Hessarek from Sturnus vulgaris in Turkey: a case report. Kafkas Universitesi Veteriner Fakultesi Dergisi 23, 343–346. [Google Scholar]
- Dänhardt, J. , Green, M. , Lindström, Å. , Rundlöf, M. & Smith, H. G. (2010). Farmland as stopover habitat for migrating birds – effects of organic farming and landscape structure. Oikos 119, 1114–1125. [Google Scholar]
- Daoust, P. , Busby, D. G. , Ferns, L. , Goltz, J. , Mcburney, S. , Poppe, C. & Whitney, H. (2000). Salmonellosis in songbirds in the Canadian Atlantic provinces during winter–summer 1997–98. Canadian Veterinary Journal 41, 54–59. [PMC free article] [PubMed] [Google Scholar]
- De Graaf, R. M. , Tilghman, N. G. & Anderson, S. H. (1985). Foraging guilds of North American birds. Environmental Management 9, 493–536. [Google Scholar]
- * De Oliveira, M. C. V. , Camargo, B. Q. , Cunha, M. P. V. , Saidenberg, A. B. , Teixeira, R. H. F. , Matajira, C. E. C. , Moreno, L. Z. , Gomes, V. T. M. , Christ, A. P. G. , Barbosa, M. R. F. , Sato, M. I. Z. , Moreno, A. M. & Knöbl, T. (2018). Free‐ranging synanthropic birds (Ardea alba and Columba livia domestica) as carriers of Salmonella spp. and diarrheagenic Escherichia coli in the vicinity of an urban zoo. Vector‐Borne and Zoonotic Diseases 18, 65–69. [DOI] [PubMed] [Google Scholar]
- * De Sousa, E. , Junior, A. B. , Pinto, A. A. , Machado, R. Z. , Carrasco, A. O. T. , Marciano, J. A. & Werther, K. (2010). Prevalence of Salmonella spp. antibodies to Toxoplasma gondii, and Newcastle disease virus in feral pigeons (Columba livia) in the city of Jaboticabal, Brazil. Journal of Zoo and Wildlife Medicine 41, 603–607. [DOI] [PubMed] [Google Scholar]
- * Dell'Omo, G. , Morabito, S. , Quondam, R. , Agrimi, U. , Ciuchini, F. , Macrì, A. & Caprioli, A. (1998). Feral pigeons as a source of verocytotoxin‐producing Escherichia coli . The Veterinary Record 142, 309–310. [DOI] [PubMed] [Google Scholar]
- Dho‐Moulin, M. & Fairbrother, J. M. (1999). Avian pathogenic Escherichia coli (APEC). Veterinary Research 30, 299–316. [PubMed] [Google Scholar]
- * Dobbin, G. , Hariharan, H. , Daoust, P. Y. , Hariharan, S. , Heaney, S. , Coles, M. , Prices, L. & Muckle, C. A. (2005). Bacterial flora of free‐living double‐crested cormorant (Phalacrocorax auritus) chicks on Prince Edward Island, Canada, with reference to enteric bacteria and antibiotic resistance. Comparative Immunology, Microbiology & Infectious Diseases 28, 71–82. [DOI] [PubMed] [Google Scholar]
- * Dobeic, M. (2003). Regulation of population size of street pigeons in Ljubljana, Slovenia. Acta Veterinaria Beograd 53, 171–182. [Google Scholar]
- * Doumandji, A. , Setbel, S. , Saidi, N. , Doumandji, S. , Voisin, J. F. & Voisin, C. (2010). Flore microbienne dans les déjections et dans le tube digestif du héron garde‐bœufs Bubulcus ibis (Ardeidae, Aves). Revue d'Écologie 65, 377–383. [Google Scholar]
- * Dovč, A. , Zorman‐Rojs, O. , Vergles Rataj, A. , Bole‐Hribovšek, V. , Krapež, U. & Dobeic, M. (2004). Health status of free‐living pigeons (Columba livia domestica) in the city of Ljubljana. Acta Veterinaria Hungarica 52, 219–226. [DOI] [PubMed] [Google Scholar]
- * Duncan, R. M. , Stroud, R. K. & Locke, L. N. (1983). Salmonella enteritidis isolated from an eared grebe (Podiceps nigricollis). Journal of Wildlife Diseases 19, 63–64. [DOI] [PubMed] [Google Scholar]
- Ejidokun, O. O. , Walsh, A. , Barnett, J. , Hope, Y. , Ellis, S. , Sharp, M. W. , Paiba, G. A. , Logan, M. , Willshaw, G. A. & Cheasty, T. (2006). Human Vero cytotoxigenic Escherichia coli (VTEC) O157 infection linked to birds. Epidemiology & Infection 134, 421–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Erickson, M. C. (2016). Overview: foodborne pathogens in wildlife populations In Food Safety Risks from Wildlife (eds Jay‐Russell M. and Doyle M. P.), pp. 1–30. Cham, Switzerland: Springer International Publishing Switzerland. [Google Scholar]
- Espinosa‐Arguelles, A. , de la Cruz‐Hernandez, N. I. , Infante‐Rodriguez, F. , Flores‐Maldonada, J. , Zertuche‐Rodriguez, J. & Flores‐Gutierrez, G. (2010). Seroprevalence of antibodies against Salmonella enterica subsp. enterica serovar Gallinarum‐Pullorum in wild doves (Zenaida asiatica and Zenaida macroura) from the Northeast of Mexico. Preventive Veterinary Medicine 93, 77–79. [DOI] [PubMed] [Google Scholar]
- * Euden, P. R. (1990). Salmonella isolates from wild animals in Cornwall. British Veterinary Journal 146, 228–232. [DOI] [PubMed] [Google Scholar]
- * Ewers, C. , Guenther, S. , Wieler, L. H. & Schierack, P. (2009). Mallard ducks – a waterfowl species with high risk of distributing Escherichia coli pathogenic for humans. Environmental Microbiology Reports 1, 510–517. [DOI] [PubMed] [Google Scholar]
- Faddoul, G. , Fellows, G. & Baird, J. (1966). A survey on the incidence of salmonellae in wild birds. Avian Diseases 10, 89–94. [Google Scholar]
- * Fallacara, D. M. , Monahan, C. M. , Morishita, T. Y. & Wack, R. F. (2001). Fecal shedding and antimicrobial susceptibility of selected bacterial pathogens and a survey of intestinal parasites in free‐living waterfowl. Avian Diseases 45, 128–135. [PubMed] [Google Scholar]
- Feare, C. J. , Saunders, M. F. , Blasco, R. & Bishop, J. D. (1999). Canada goose droppings as a potential source of pathogenic bacteria. The Journal of the Royal Society for the Promotion of Health 119, 146–155. [DOI] [PubMed] [Google Scholar]
- * Ferreira, V. L. , Dias, R. A. & Raso, T. F. (2016). Screening of feral pigeons (Columba livia) for pathogens of veterinary and medical importance. Brazilian Journal of Poultry Science 18, 701–704. [Google Scholar]
- Fichtel, C. C. (1978). A Salmonella outbreak in wild songbirds. North American Bird Bander 3, 146–148. [Google Scholar]
- Fischl, J. & Caccamise, D. F. (1985). Influence of habitat and season on foraging flock composition in the European starling (Sturnus vulgaris). Oecologia 67, 532–539. [DOI] [PubMed] [Google Scholar]
- Flint, K. (1987). The long‐term survival of Escherichia coli in river water. Journal of Applied Bacteriology 63, 261–270. [DOI] [PubMed] [Google Scholar]
- Fogarty, L. R. , Haack, S. K. , Wolcott, M. J. & Whitman, R. L. (2003). Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. Journal of Applied Microbiology 94, 865–878. [DOI] [PubMed] [Google Scholar]
- Fontaine, R. E. , Cohen, M. L. , Martin, W. T. & Vernon, T. M. (1980). Epidemic Salmonellosis from cheddar cheese: surveillance and prevention. American Journal of Epidemiology 111, 247–253. [DOI] [PubMed] [Google Scholar]
- Fukui, D. , Takahashi, K. , Kubo, M. , Une, Y. & Kato, Y. (2014). Mass mortality of Eurasian tree sparrows (Passer montanus) from Salmonella Typhimurium DT40 in Japan. Journal of Wildlife Diseases 50, 484–495. [DOI] [PubMed] [Google Scholar]
- * Fukuyama, M. , Furuhata, K. , Oonaka, K. , Sakata, S. , Hara, M. , Kakuno, Y. , Itoh, T. , Kai, A. , Obata, H. & Watanabe, T. (2003). Isolation and serotypes of Vero toxin‐producing Escherichia coli (VTEC) from pigeons and crows. The Journal of the Japanese Association for Infectious Diseases 77, 5–9. [DOI] [PubMed] [Google Scholar]
- * Gabriele‐Rivet, V. , Fairbrother, J. H. , Tremblay, D. , Harel, J. , Côté, N. & Arsenault, J. (2015). Prevalence and risk factors for Campylobacter spp., Salmonella spp., Coxiella burnetii, and Newcastle disease virus in feral pigeons (Columba livia) in public areas of Montreal, Canada. The Canadian Journal of Veterinary Research 80, 81–85. [PMC free article] [PubMed] [Google Scholar]
- * Galbraith, J. A. , Stanley, M. C. , Jones, D. N. & Beggs, J. R. (2017). Experimental feeding regime influences urban bird disease dynamics. Journal of Avian Biology 48, 700–713. [Google Scholar]
- Gardner, T. J. , Fitzgerald, C. , Xavier, C. , Klein, R. , Pruckler, J. , Stroika, S. & Mclaughlin, J. B. (2011). Outbreak of Campylobacteriosis associated with consumption of raw peas. Clinical Infectious Diseases 53, 26–32. [DOI] [PubMed] [Google Scholar]
- * Gargiulo, A. , Russo, T. P. , Schettini, R. , Mallardo, K. , Calabria, M. , Menna, L. F. , Raia, P. , Pagnini, U. , Caputo, V. , Fioretti, A. & Dipineto, L. (2014). Occurrence of enteropathogenic bacteria in urban pigeons (Columba livia) in Italy. Vector‐Borne and Zoonotic Diseases 14, 251–255. [DOI] [PubMed] [Google Scholar]
- * Gargiulo, A. , Sensale, M. , Marzocco, L. , Fioretti, A. , Menna, L. F. & Dipineto, L. (2011). Campylobacter jejuni, Campylobacter coli, and cytolethal distending toxin (CDT) genes in common teals (Anas crecca). Veterinary Microbiology 150, 401–404. [DOI] [PubMed] [Google Scholar]
- Gaukler, S. , Homan, H. , George, M. & Bleier, W. (2012). Using radio‐telemetry to assess the risk European starlings pose in pathogen transmission among feedlots. Human‐Wildlife Interactions 6, 30–37. [Google Scholar]
- * Gaukler, S. M. , Linz, G. M. , Sherwood, J. S. , Dyer, N. W. , Bleier, W. J. , Wannemuehler, Y. M. , Nolan, L. K. & Logue, C. M. (2009). Escherichia coli, Salmonella, and Mycobacterium avium subsp. paratuberculosis in wild European starlings at a Kansas cattle feedlot. Avian Diseases 53, 544–551. [DOI] [PubMed] [Google Scholar]
- * Ghanbarpour, R. & Daneshdoost, S. (2012). Identification of Shiga toxin and intimin coding genes in Escherichia coli isolates from pigeons (Columba livia) in relation to phylotypes and antibiotic resistance patterns. Tropical Animal Health and Production 44, 307–312. [DOI] [PubMed] [Google Scholar]
- * Gibbs, P. S. , Kasa, R. , Newbrey, J. L. , Petermann, S. R. , Wooley, R. E. , Vinson, H. M. & Reed, W. (2007). Identification, antimicrobial resistance profiles, and virulence of members from the family Enterobacteriaceae from the feces of yellow‐headed blackbirds (Xanthocephalus xanthocephalus) in North Dakota. Avian Diseases 51, 649–655. [DOI] [PubMed] [Google Scholar]
- Girard, J. , Goldberg, T. L. & Hamer, G. L. (2011). Field investigation of innate immunity in passerine birds in suburban Chicago, Illinois, USA. Journal of Wildlife Diseases 47, 603–611. [DOI] [PubMed] [Google Scholar]
- Girdwood, R. , Fricker, C. , Munro, D. , Shedden, C. & Monaghan, O. (1985). The incidence and significance of Salmonella carriage by gulls (Larus spp.) in Scotland. The Journal of Hygiene 95, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glunder, G. , Neumann, I. & Braune, S. (1992). Occurrence of Campylobacter spp. in young gulls, duration of Campylobacter infection and reinfection by contact. Journal of Veterinary Medicine. Series B 39, 119–122. [DOI] [PubMed] [Google Scholar]
- * González‐Acuña, D. , Silva, F. , Moreno, L. , Cerda, F. , Donoso, S. , Cabello, J. & López, J. (2007). Detección de algunos agentes zoonóticos en la paloma doméstica (Columba livia) en la ciudad de Chillán, Chile. Revista Chilena de Infectología 24, 199–203. [DOI] [PubMed] [Google Scholar]
- * Gorski, L. , Parker, C. T. , Liang, A. , Cooley, M. B. , Jay‐Russell, M. T. , Gordus, A. G. , Atwill, E. R. & Mandrell, R. E. (2011). Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Applied and Environmental Microbiology 77, 2734–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor, P. , Colles, F. M. , Mccarthy, N. D. , Hansbro, P. M. , Olsen, N. , Hasselquist, D. & Maiden, M. C. J. (2013). Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Molecular Ecology 22, 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Grigar, M. K. , Cummings, K. J. & Rankin, S. C. (2017). Prevalence of Salmonella among waterfowl along the Texas Gulf coast. Zoonoses and Public Health 64, 689–692. [DOI] [PubMed] [Google Scholar]
- * Grigar, M. K. , Cummings, K. J. , Rodriguez‐Rivera, L. D. , Rankin, S. C. , Johns, K. , Hamer, G. L. & Hamer, S. A. (2016). Salmonella surveillance among great‐tailed grackles (Quiscalus mexicanus) and other urban bird species in Eastern Texas. Vector‐Borne and Zoonotic Diseases 16, 752–757. [DOI] [PubMed] [Google Scholar]
- Grond, K. , Sandercock, B. K. , Jumpponen, A. & Zeglin, L. H. (2018). The avian gut microbiota: community, physiology and function in wild birds. Journal of Avian Biology 49, e01788. [Google Scholar]
- Guan, T. Y. & Holley, R. A. (2003). Pathogen survival in swine manure environments and transmission of human enteric illness — a review. Journal of Environmental Quality 32, 383–392. [DOI] [PubMed] [Google Scholar]
- * Guenther, S. , Grobbel, M. , Lübke‐Becker, A. , Goedecke, A. , Friedrich, N. D. , Wieler, L. H. & Ewers, C. (2010). Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Veterinary Microbiology 144, 219–225. [DOI] [PubMed] [Google Scholar]
- Guillemette, M. (1994). Digestive‐rate constraint in wintering common eiders (Somateria mollissima): implications for flying capabilities. The Auk 111, 900–909. [Google Scholar]
- * Gülhan, T. , Boynukara, B. , Durmuş, A. , Kiziroğlu, I. & Sancak, Y. C. (2012). Properties of Enterococcus faecalis, Enterococcus faecium and Escherichia coli strains isolated from wild ducks and gulls. Fresenius Environmental Bulletin 21, 1961–1966. [Google Scholar]
- Haag‐Wackernagel, D. & Moch, H. (2004). Health hazards posed by feral pigeons. Journal of Infection 48, 307–313. [DOI] [PubMed] [Google Scholar]
- Haddaway, N. R. & Watson, M. J. (2016). On the benefits of systematic reviews for wildlife parasitology. International Journal for Parasitology: Parasites and Wildlife 5, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Haemig, P. D. , Hernandez, J. , Waldenström, J. , Bonnedahl, J. & Olsen, B. (2008). Barn swallows (Hirundo rustica) test negative for Salmonella . Vector‐Borne and Zoonotic Diseases 8, 451–454. [DOI] [PubMed] [Google Scholar]
- Haesendonck, R. , Rasschaert, G. , Martel, A. , Verbrugghe, E. , Heyndrickx, M. , Haesebrouck, F. & Pasmans, F. (2016). Feral pigeons: a reservoir of zoonotic Salmonella Enteritidis strains? Veterinary Microbiology 195, 101–103. [DOI] [PubMed] [Google Scholar]
- Hald, B. , Skov, M. N. , Nielsen, E. M. , Rahbek, C. , Madsen, J. J. , Wainø, M. , Chriél, M. , Nordentoft, S. , Baggesen, D. L. & Madsen, M. (2016). Campylobacter jejuni and Campylobacter coli in wild birds on Danish livestock farms. Acta Veterinaria Scandinavica 58, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. J. & Saito, E. K. (2008). Avian wildlife mortality events due to Salmonellosis in the United States, 1985–2004. Journal of Wildlife Diseases 44, 585–593. [DOI] [PubMed] [Google Scholar]
- Hamer, S. A. , Lehrer, E. & Magle, S. B. (2012). Wild birds as sentinels for multiple zoonotic pathogens along an urban to rural gradient in Greater Chicago, Illinois. Zoonoses and Public Health 59, 355–364. [DOI] [PubMed] [Google Scholar]
- Hancock, D. D. , Besser, T. E. , Rice, D. H. , Ebel, E. D. , Herriott, D. E. & Carpenter, L. V. (1998). Multiple sources of Escherichia coli 0157 in feedlots and dairy farms in the Northwestern USA. Preventive Veterinary Medicine 35, 11–19. [DOI] [PubMed] [Google Scholar]
- * Hansen, D. L. , Ishii, S. , Sadowsky, M. J. & Hicks, R. E. (2009). Escherichia coli populations in Great Lakes waterfowl exhibit spatial stability and temporal shifting. Applied and Environmental Microbiology 75, 1546–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelaar, A. H. , Kirk, M. D. , Torgerson, P. R. , Gibb, H. J. , Hald, T. , Lake, R. J. & Praet, N. (2015). World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Medicine 12, e1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Hernandez, J. , Johansson, A. , Stedt, J. , Bengtsson, S. , Porczak, A. , Granholm, S. , González‐Acuña, D. , Olsen, B. , Bonnedahl, J. & Drobni, M. (2013). Characterization and comparison of extended‐spectrum β‐lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin's gulls (Leucophaeus pipixcan) and humans in chile. PLoS One 8, e76150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Hernandez, S. M. , Keel, K. , Sanchez, S. , Trees, E. , Gerner‐Smidt, P. , Adams, J. K. , Cheng, Y. , Ray, A. , Martin, G. , Presotto, A. , Ruder, M. G. , Brown, J. , Blehert, D. S. , Cottrell, W. & Maurer, J. J. (2012). Epidemiology of a Salmonella enterica subsp. enterica serovar Typhimurium strain associated with a songbird outbreak. Applied and Environmental Microbiology 78, 7290–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, S. M. , Welch, C. N. , Peters, V. E. , Lipp, E. K. , Curry, S. , Yabsley, M. J. , Sanchez, S. , Presotto, A. , Gerner‐Smidt, P. , Hise, K. B. , Hammond, E. , Kistler, W. M. , Madden, M. , Conway, A. L. , Kwan, T. , et al. (2016). Urbanized white ibises (Eudocimus albus) as carriers of Salmonella enterica of significance to public health and wildlife. PLoS One 11, e0164402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Horigan, V. , Davies, R. H. , Kelly, L. A. , Mead, G. C. , Irvine, R. M. & Simons, R. R. L. (2014). A qualitative risk assessment of the microbiological risks to consumers from the production and consumption of uneviscerated and eviscerated small game birds in the UK. Food Control 45, 127–137. [Google Scholar]
- * Houston, C. S. , Saunders, J. R. & Crawford, R. D. (1997). Aerobic bacterial flora of addled raptor eggs in Saskatchewan. Journal of Wildlife Diseases 33, 328–331. [DOI] [PubMed] [Google Scholar]
- * Howerth, E. W. (1985). Salmonellosis in a wild turkey. Journal of Wildlife Diseases 21, 433–434. [DOI] [PubMed] [Google Scholar]
- * Hsu, T. T. D. , Mitsch, W. J. , Martin, J. F. & Lee, J. (2017). Towards sustainable protection of public health: the role of an urban wetland as a frontline safeguard of pathogen and antibiotic resistance spread. Ecological Engineering 108, 547–555. [Google Scholar]
- * Hsu, T. T. D. , Rea, C. L. , Yu, Z. & Lee, J. (2016). Prevalence and diversity of Shiga toxin genes in Canada geese and water in western Lake Erie region. Journal of Great Lakes Research 42, 476–481. [Google Scholar]
- Hubálek, Z. (2004). An annotated checklist of pathogenic microorganisms associated with migratory birds. Journal of Wildlife Diseases 40, 639–659. [DOI] [PubMed] [Google Scholar]
- * Hubálek, Z. , Juřicová, Z. & Halouzka, J. (1995). A survey of free‐living birds as hosts and ‘lessors' of microbial pathogens. Folia Zoologica 44, 1–11. [Google Scholar]
- * Hudson, C. R. , Quist, C. , Lee, M. D. , Keyes, K. , Dodson, S. V. , Morales, C. , Sanchez, S. , White, D. G. & Maurer, J. J. (2000). Genetic relatedness of Salmonella isolates from nondomestic birds in Southeastern United States. Journal of Clinical Microbiology 38, 1860–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Hughes, L. A. , Bennett, M. , Coffey, P. , Elliott, J. , Jones, T. R. , Jones, R. C. , Lahuerta‐Marin, A. , Mcniffe, K. , Norman, D. , Williams, N. J. & Chantrey, J. (2009). Risk factors for the occurrence of Escherichia coli virulence genes eae, stx1, and stx2 in wild bird populations. Epidemiology & Infection 137, 1574–1582. [DOI] [PubMed] [Google Scholar]
- * Hughes, L. A. , Shopland, S. , Wigley, P. , Bradon, H. , Leatherbarrow, A. H. , Williams, N. J. , Bennett, M. , De Pinna, E. , Lawson, B. , Cunningham, A. A. & Chantrey, J. (2008). Characterisation of Salmonella enterica serotype Typhimurium isolates from wild birds in northern England from 2005–2006. BMC Veterinary Research 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Hughes, L. A. , Wigley, P. , Bennett, M. , Chantrey, J. & Williams, N. (2010). Multi‐locus sequence typing of Salmonella enterica serovar Typhimurium isolates from wild birds in northern England suggests host‐adapted strain. Letters in Applied Microbiology 51, 477–479. [DOI] [PubMed] [Google Scholar]
- * Hurvell, B. , Borg, K. , Gunnarsson, A. & Jevring, J. (1975). Studies on Salmonella Typhi‐murium infections in passerine birds in Sweden. International Union of Game Biologists 11, 493–497. [Google Scholar]
- Hussong, D. , Damare, J. , Limpert, R. , Sladen, W. , Weiner, R. & Colwell, R. (1979). Microbial impact of Canada geese (Branta canadensis) and whistling swans (Cygnus columbianus columbianus) on aquatic ecosystems. Applied and Environmental Microbiology 37, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, M. L. , Walters, L. D. , Moore, A. , Crookes, K. M. & Avery, S. M. (2004). Effect of length of time before incorporation on survival of pathogenic bacteria present in livestock wastes applied to agricultural soil. Applied and Environmental Microbiology 70, 5111–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Ingram, D. R. , Miller, D. L. , Baldwin, C. A. , Turco, J. & Lockhart, J. M. (2015). Serologic survey of wild turkeys (Meleagris gallapavo) and evidence of exposure to avian encephalomyelitis virus in Georgia and Florida, USA. Journal of Wildlife Diseases 51, 374–379. [DOI] [PubMed] [Google Scholar]
- Islam, M. , Doyle, M. P. , Phatak, S. C. , Millner, P. & Jiang, X. (2004c). Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. Journal of Food Protection 67, 1365–1370. [DOI] [PubMed] [Google Scholar]
- Islam, M. , Doyle, M. P. , Phatak, S. C. , Millner, P. & Jiang, X. (2005). Survival of Escherichia coli O157:H7 in soil and on carrots and onions grown in fields treated with contaminated manure composts or irrigation water. Food Microbiology 22, 63–70. [Google Scholar]
- Islam, M. , Morgan, J. , Doyle, M. P. , Phatak, S. C. & Millner, P. (2004b). Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Applied and Environmental Microbiology 70, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. , Morgan, J. , Doyle, M. P. , Phatak, S. C. , Millner, P. & Jiang, X. (2004a). Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathogens and Disease 1, 27–35. [DOI] [PubMed] [Google Scholar]
- Ito, K. , Totake, Y. V. & Ogawa, M. (1988). Occurrence of Campylobacter jejuni in free‐living wild birds from Japan. Journal of Wildlife Diseases 24, 467–470. [DOI] [PubMed] [Google Scholar]
- Jacobsen, C. S. & Bech, T. B. (2012). Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Research International 45, 557–566. [Google Scholar]
- * Janecko, N. , Čížek, A. , Halová, D. , Karpíšková, R. , Myšková, P. & Literák, I. (2014). Prevalence, characterization and antibiotic resistance of Salmonella isolates in large corvid species of Europe and North America between 2010 and 2013. Zoonoses and Public Health 62, 292–300. [DOI] [PubMed] [Google Scholar]
- * Jay‐Russell, M. T. , Madigan, J. E. , Bengson, Y. , Madigan, S. , Hake, A. F. , Foley, J. E. & Byrne, B. A. (2013). Salmonella Oranienburg isolated from horses, wild turkeys and an edible home garden fertilized with raw horse manure. Zoonoses and Public Health 61, 64–71. [DOI] [PubMed] [Google Scholar]
- Jiang, X. , Morgan, J. & Doyle, M. P. (2002). Fate of Escherichia coli O157:H7 in manure‐amended soil. Applied and Environmental Microbiology 68, 2605–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Jijón, S. , Wetzel, A. & Lejeune, J. (2007). Salmonella enterica isolated from wildlife at two Ohio rehabilitation centers. Journal of Zoo and Wildlife Medicine 38, 409–413. [DOI] [PubMed] [Google Scholar]
- Johnson, P. T. J. & Hoverman, J. T. (2012). Parasite diversity and coinfection determine pathogen infection success and host fitness. Proceedings of the National Academy of Sciences of the United States of America 109, 9006–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, P. T. J. , Preston, D. L. , Hoverman, J. T. & Lafonte, B. E. (2013). Host and parasite diversity jointly control disease risk in complex communities. Proceedings of the National Academy of Sciences of the United States of America 110, 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Jones, B. , Grace, D. , Kock, R. , Alonso, S. , Rushton, J. , Said, M. , Mckeever, D. , Mutua, F. , Young, J. , Mcdermott, J. & Pfeiffer, D. (2013). Zoonosis emergence linked to agricultural intensification and environmental change. Proceedings of the National Academy of Sciences of the United States of America 110, 8399–8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Jones, M. S. , Tadepalli, S. , Bridges, D. F. , Wu, V. C. H. & Drummond, F. (2015). Suppression of Escherichia coli O157:H7 by dung beetles (Coleoptera: Scarabaeidae) using the lowbush blueberry agroecosystem as a model system. PLoS One 10, e0120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Kalisińska, E. , Lisowski, P. , Czernomysy‐Furowicz, D. & Kavetska, K. (2008). Serratospiculiasis, mycosis, and haemosiderosis in wild peregrine falcon from Poland. A case report. Bulletin of the Veterinary Institute in Pulawy 52, 75–79. [Google Scholar]
- * Kapperud, G. & Rosef, O. (1983). Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Applied and Environmental Microbiology 45, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, M. D. & Lejeune, J. (2011). European starlings (Sturnus vulgaris) challenged with Escherichia coli O157 can carry and transmit the human pathogen to cattle. Letters in Applied Microbiology 53, 596–601. [DOI] [PubMed] [Google Scholar]
- * Keller, J. I. & Shriver, W. G. (2014). Prevalence of three Campylobacter species, C. jeuni, C. coli, and C. lari using multilocus sequence typing in wild birds of the Mid‐Atlantic Region, USA. Journal of Wildlife Diseases 50, 31–41. [DOI] [PubMed] [Google Scholar]
- * Kirk, J. H. , Holmberg, C. A. & Jeffrey, J. S. (2002). Prevalence of Salmonella spp in selected birds captured on California dairies. Journal of the American Veterinary Medical Association 220, 359–362. [DOI] [PubMed] [Google Scholar]
- * Kirkpatrick, C. E. & Colvin, B. A. (1986). Salmonella spp. in nestling common barn–owls (Tyto alba) from Southwestern New Jersey. Journal of Wildlife Diseases 22, 340–343. [DOI] [PubMed] [Google Scholar]
- * Kirkpatrick, C. E. & Trexler‐Myren, V. P. (1986). A survey of free‐living falconiform birds for Salmonella . Journal of the American Veterinary Medical Association 189, 997–998. [PubMed] [Google Scholar]
- * Kobayashi, H. , Kanazaki, M. , Hata, E. & Kubo, M. (2009). Escherichia coli from wild birds in the immediate environment of Tokyo Bay. Applied and Environmental Microbiology 75, 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Krawiec, M. , Woźniak‐Biel, A. , Bednarski, M. & Wieliczko, A. (2017). Antimicrobial susceptibility and genotypic characteristic of Campylobacter spp. isolates from free‐living birds in Poland. Vector‐Borne and Zoonotic Diseases 17, 755–763. [DOI] [PubMed] [Google Scholar]
- * Kuczkowski, M. , Krawiec, M. , Voslamber, B. , Książczyk, M. , Płoskońska‐Bugla, G. & Wieliczko, A. (2016). Virulence genes and the antimicrobial susceptibility of Escherichia coli, isolated from wild waterbirds, in the Netherlands and Poland. Vector‐Borne and Zoonotic Diseases 16, 528–236. [DOI] [PubMed] [Google Scholar]
- Kudva, I. T. , Blanch, K. & Hovde, C. J. (1998). Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Applied and Environmental Microbiology 64, 3166–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Kullas, H. , Coles, M. , Rhyan, J. & Clark, L. (2002). Prevalence of Escherichia coli serogroups and human virulence factors in faeces of urban Canada geese (Branta canadensis). International Journal of Environmental Health Research 12, 153–162. [DOI] [PubMed] [Google Scholar]
- * Laberski, N. , Hull, A. C. , Fish, A. M. , Beckmen, K. & Morishita, T. Y. (2003). A survey of the choanal and cloacal aerobic bacterial flora in free‐living and captive red‐tailed hawks (Buteo jamaicensis) and Cooper's hawks (Accipiter cooperii). Journal of Avian Medicine and Surgery 17, 131–135. [Google Scholar]
- * Lawson, B. , Howard, T. , Kirkwood, J. K. , Macgregor, S. K. , Perkins, M. , Robinson, R. A. , Ward, L. R. & Cunningham, A. A. (2010). Epidemiology of Salmonellosis in garden birds in England and Wales, 1993 to 2003. EcoHealth 7, 294–306. [DOI] [PubMed] [Google Scholar]
- * Lawton, S. J. , Weis, A. M. , Byrne, B. A. , Fritz, H. , Taff, C. C. , Townsend, A. K. , Weimer, B. C. , Mete, A. , Wheeler, S. & Boyce, W. M. (2018). Comparative analysis of Campylobacter isolates from wild birds and chickens using MALDI‐TOF MS, biochemical testing, and DNA sequencing. Journal of Veterinary Diagnostic Investigation 30, 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, J. , Homan, J. , Linz, G. & Pearl, D. L. (2007). Role of the European starling in the transmission of E. coli O157 on dairy farms. In Proceedings of the 23rd Vertebrate Pest Conference, pp. 31–34.
- Levesque, B. , Brousseau, P. , Bernier, F. , Dewailly, E. & Joly, J. (2000). Study of the bacterial content of ring‐billed gull droppings in relation to recreational water quality. Water Research 34, 1089–1096. [Google Scholar]
- Levey, D. & Karasov, W. (1994). Gut passage of insects by European starlings and comparison with other species. The Auk 111, 478–481. [Google Scholar]
- * Levre, E. , Valentini, P. , Brunetti, M. & Sacchelli, F. (1989). Stationary and migratory avifauna as reservoirs of Salmonella, Yersinia and Campylobacter . Annali di Igiene Medicina Preventiva e di Comunità 1, 729–740. [PubMed] [Google Scholar]
- Liang, Z. , He, Z. , Powell, C. A. & Stoffella, P. J. (2011). Survival of Escherichia coli in soil with modified microbial community composition. Soil Biology & Biochemistry 43, 1591–1599. [Google Scholar]
- * Lillehaug, A. , Jonassen, C. M. , Bergsjø, B. , Hofshagen, M. , Tharaldsen, J. , Nesse, L. L. & Handeland, K. (2005). Screening of feral pigeon (Colomba livia), mallard (Anas platyrhynchos) and graylag goose (Anser anser) populations for Campylobacter spp., Salmonella spp., avian influenza virus and avian paramyxovirus. Acta Veterinaria Scandinavica 46, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Literak, I. , Dolejska, M. , Janoszowska, D. , Hrusakova, J. , Meissner, W. , Rzyska, H. , Bzoma, S. & Cizek, A. (2010). Antibiotic resistant Escherichia coli bacteria, including strains with genes encoding the extended‐spectrum beta‐lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Applied and Environmental Microbiology 76, 8126–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd‐Smith, J. O. , George, D. , Pepin, K. M. , Pitzer, V. E. , Pulliam, J. R. C. , Dobson, A. P. , Hudson, P. J. & Grenfell, B. T. (2009). Epidemic dynamics at the human‐animal interface. Science 326, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Locke, L. N. , Shillinger, R. B. & Jareed, T. (1973). Salmonellosis in passerine birds in Maryland and West Virginia. Journal of Wildlife Diseases 9, 144–145. [DOI] [PubMed] [Google Scholar]
- * Lombardo, M. P. , Thorpe, P. A. , Cichewicz, R. , Henshaw, M. , Millard, C. , Steen, C. & Zeller, T. K. (1996). Communities of cloacal bacteria in tree swallow families. The Condor 98, 167–172. [Google Scholar]
- * López‐Martín, J. , Junod, T. , Riquelme, F. , Contreras, C. & González‐Acuña, D. (2011). Detección de especies de Salmonella y Mycobacterium en gaviotas dominicanas (Larus dominicanus) y gaviotas de Franklin (Leucophaeus pipixcan) en la ciudad de Talcahuano, Chile. Revista Médica de Chile 139, 1496–1502. [PubMed] [Google Scholar]
- * Lu, J. , Ryu, H. , Santo Domingo, J. W. , Griffith, J. F. & Ashbolt, N. (2011). Molecular detection of Campylobacter spp. in California gull (Larus californicus) excreta. Applied and Environmental Microbiology 77, 5034–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Ryu, H. , Vogel, J. , Domingo, J. S. & Ashbolt, N. J. (2013). Molecular detection of Campylobacter and fecal indicators during the northern migration of sandhill cranes (Grus canadensis) at the Central Platte River. Applied and Environmental Microbiology 79, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld, N. A. W. , Blaser, M. J. , Reller, L. B. & Wang, W. L. (1980). Isolation of Campylobacter fetus subsp. jejuni from migratory waterfowl. Journal of Clinical Microbiology 12, 406–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Luque, I. , Escheita, A. , León, J. , Herrera‐León, S. , Tarradas, C. , González‐Sanz, R. , Huerta, B. & Astorga, R. J. (2009). Salmonella Indiana as a cause of abortion in ewes: genetic diversity and resistance patterns. Veterinary Microbiology 134, 396–399. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Ibekwe, A. M. , Crowley, D. E. & Yang, C. (2012). Persistence of Escherichia coli O157:H7 in major leafy green producing soils. Environmental Science and Technology 46, 12154–12161. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Ibekwe, A. M. , Yi, X. , Wang, H. , Yamazaki, A. , Crowley, D. E. & Yang, C. (2011). Persistence of Escherichia coli O157:H7 and its mutants in soils. PLoS One 6, e23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenzoni, M. L. , Morganti, G. , Moretta, I. , Crotti, S. , Agnetti, F. , Moretti, A. , Pitzurra, L. , Proietti, P. C. , Sechi, P. & Franciosini, M. P. (2016). Microbiological and parasitological survey of zoonotic agents in apparently healthy feral pigeons. Polish Journal of Veterinary Sciences 19, 309–315. [DOI] [PubMed] [Google Scholar]
- Marine, S. C. , Pagadal, S. , Wang, F. , Pahl, D. M. , Melendez, M. V. , Kline, W. L. , Oni, R. A. , Walsh, C. S. , Everts, K. L. , Buchanan, R. L. & Micallef, A. (2015). The growing season, but not the farming system, is a food safety risk determinant for leafy greens in the Mid‐Atlantic region of the United States. Applied and Environmental Microbiology 81, 2395–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, L. B. (2005). Trade‐offs between molt and immune activity in two populations of house sparrows (Passer domesticus). Canadian Journal of Zoology 83, 780–787. [Google Scholar]
- Martin, L. B. , Weil, Z. M. & Nelson, R. J. (2008). Seasonal changes in vertebrate immune activity: mediation by physiological trade‐offs. Philosophical Transactions of the Royal Society B: Biological Sciences 363, 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Medhanie, G. A. , Pearl, D. L. , Mcewen, S. A. , Guerin, M. T. , Jardine, C. M. , Schrock, J. & Lejeune, J. T. (2014). A longitudinal study of feed contamination by European starling excreta in Ohio dairy farms (2007–2008). Journal of Dairy Science 97, 5230–5239. [DOI] [PubMed] [Google Scholar]
- * Merkeviciene, L. , Ruzauskaite, N. , Klimiene, I. , Siugzdiniene, R. , Dailidaviciene, J. , Virgailis, M. , Mockeliunas, R. & Ruzauskas, M. (2017). Microbiome and antimicrobial resistance genes in microbiota of cloacal samples from European herring gulls (Larus argentatus). Journal of Veterinary Research 61, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezrioui, N. , Baleux, B. & Troussellier, M. (1995). A microcosm study of the survival of Escherichia coli and Salmonella Typhimurium in brackish water. Water Research 29, 459–465. [Google Scholar]
- * Middleton, J. H. & Ambrose, A. (2005). Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). Journal of Wildlife Diseases 41, 334–341. [DOI] [PubMed] [Google Scholar]
- * Mikaelian, I. , Daignault, D. , Duval, M. C. & Martineau, D. (1997). Salmonella infection in wild birds from Quebec. The Canadian Veterinary Journal 38, 385. [PMC free article] [PubMed] [Google Scholar]
- * Millán, J. , Aduriz, G. , Moreno, B. , Juste, R. A. & Barral, M. (2004). Salmonella isolates from wild birds and mammals in the Basque Country (Spain). Revue Scientifique et Technique‐Office International des Epizooties 23, 905–911. [DOI] [PubMed] [Google Scholar]
- Mills, T. K. , Lombardo, M. P. & Thorpe, P. A. (1999). Microbial colonization of the cloacae of nestling tree swallows. The Auk 116, 947–956. [Google Scholar]
- * Mitchell, T. R. & Ridgwell, T. (1970). The frequency of salmonellae in wild ducks. Journal of Medical Microbiology 4, 359–361. [DOI] [PubMed] [Google Scholar]
- * Mohan, V. , Stevenson, M. , Marshall, J. , Fearnhead, P. , Holland, B. R. , Hotter, G. & French, N. P. (2013). Campylobacter jejuni colonization and population structure in urban populations of ducks and starlings in New Zealand. MicrobiologyOpen 2, 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan, P. , Shedden, C. , Ensor, K. , Fricker, C. & Girdwood, R. (1985). Salmonella carriage by herring gulls in the Clyde Area of Scotland in relation to their feeding ecology. Journal of Applied Ecology 22, 669–679. [Google Scholar]
- * Morabito, S. , Dell'omo, G. , Agrimi, U. , Schmidt, H. , Karch, H. , Cheasty, T. & Caprioli, A. (2001). Detection and characterization of Shiga toxin‐producing Escherichia coli in feral pigeons. Veterinary Microbiology 82, 275–283. [DOI] [PubMed] [Google Scholar]
- * Moriarty, E. M. , Karki, N. , Mackenzie, M. , Sinton, L. W. , Wood, D. R. & Gilpin, B. J. (2011). Faecal indicators and pathogens in selected New Zealand waterfowl. New Zealand Journal of Marine and Freshwater Research 45, 679–688. [Google Scholar]
- Moriarty, E. M. , Weaver, L. , Sinton, L. W. & Gilpin, B. (2012). Survival of Escherichia coli, enterococci and Campylobacter jejuni in Canada goose faeces on pasture. Zoonoses and Public Health 59, 490–497. [DOI] [PubMed] [Google Scholar]
- * Morishita, T. Y. , Aye, P. P. , Ley, E. C. & Harr, B. S. (1999). Survey of pathogens and blood parasites in free‐living passerines. Avian Diseases 43, 549–552. [PubMed] [Google Scholar]
- Morrison, E. S. , Ardia, D. R. & Clotfelter, E. D. (2009). Cross‐fostering reveals sources of variation in innate immunity and hematocrit in nestling tree swallows Tachycineta bicolor . Journal of Avian Biology 40, 573–578. [Google Scholar]
- * Murphy, J. , Devane, M. L. , Robson, B. & Gilpin, B. J. (2005). Genotypic characterization of bacteria cultured from duck faeces. Journal of Applied Microbiology 99, 301–309. [DOI] [PubMed] [Google Scholar]
- Natvig, E. E. , Ingham, S. C. , Ingham, B. H. , Cooperband, L. R. & Roper, T. R. (2002). Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Applied and Environmental Microbiology 68, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro‐Gonzalez, N. , Ugarte‐Ruiz, M. , Domínguez, L. & Ruiz‐Fons, F. (2016). A European perspective on the transmission of foodborne pathogens at the wildlife–livestock–human interface In Food Safety Risks from Wildlife (eds Jay‐Russel M. and Doyle M. P.), pp. 59–88. Cham, Switzerland: Springer International Publishing Switzerland. [Google Scholar]
- Neal, K. R. & Slack, R. C. B. (1997). Diabetes mellitus, anti‐secretory drugs and other risk factors for Campylobacter gastro‐enteritis in adults: a case‐control. Epidemiology & Infection 119, 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebola, M. , Borilova, G. & Steinhauserova, I. (2007). Prevalence of Campylobacter subtypes in pheasants in the Czech Republic. Veterinarni Medicina 52, 496–501. [Google Scholar]
- * Nelson, M. , Jones, S. H. , Edwards, C. & Ellis, J. C. (2008). Characterization of Escherichia coli populations from gulls, landfill trash, and wastewater using ribotyping. Diseases of Aquatic Organisms 81, 53–63. [DOI] [PubMed] [Google Scholar]
- * Nielsen, E. M. , Skov, M. N. , Madsen, J. J. , Lodal, J. , Jespersen, J. B. & Baggesen, D. L. (2004). Verocytotoxin‐producing Escherichia coli in wild birds and rodents in close proximity to farms. Applied and Environmental Microbiology 70, 6944–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden, I. D. , Fenlon, D. R. , Vinten, A. J. A. & Lewis, D. (2001). The fate of Escherichia coli O157 in soil and its potential to contaminate drinking water. International Journal of Food Microbiology 66, 111–117. [DOI] [PubMed] [Google Scholar]
- * Osman, K. M. , Mehrez, M. , Erfan, A. M. & Nayerah, A. (2013). Salmonella enterica isolated from pigeon (Columba livia) in Egypt. Foodborne Pathogens and Disease 10, 481–483. [DOI] [PubMed] [Google Scholar]
- Ostfeld, R. S. , Levi, T. , Jolles, A. E. , Martin, L. B. , Hosseini, R. R. & Keesing, F. (2014). Life history and demographic drivers of reservoir competence for three tick‐borne zoonotic pathogens. PLoS One 9, e107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Oteo, J. , Mencía, A. , Bautista, V. , Pastor, N. , Lara, N. , González‐González, F. , García‐Peña, F. J. & Campos, J. (2018). Colonization with enterobacteriaceae‐producing ESBLs, AmpCs, and OXA‐48 in wild avian species, Spain 2015–2016. Microbial Drug Resistance 24, 932–938. [DOI] [PubMed] [Google Scholar]
- Owen, J. P. , Waite, J. L. , Holden, K. Z. & Clayton, D. H. (2014). Does antibody binding to diverse antigens predict future infection? Parasite Immunology 36, 573–584. [DOI] [PubMed] [Google Scholar]
- Pacha, R. E. , Clark, G. W. , Williams, E. A. & Carter, A. M. (1988). Migratory birds of central Washington as reservoirs of Campylobacter jejuni . Canadian Journal of Microbiology 34, 80–82. [DOI] [PubMed] [Google Scholar]
- * Padilla, L. R. , Santiago‐Alarcon, D. , Merkel, J. , Miller, R. E. & Parker, G. P. (2004). Survey for Haemoproteus spp., Trichomonas gallinae, Chlamydophila psittaci, and Salmonella spp. in Galapagos Islands Columbiformes. Journal of Zoo and Wildlife Medicine 35, 60–64. [DOI] [PubMed] [Google Scholar]
- * Palmgren, H. , Broman, T. , Waldenström, J. , Lindberg, P. , Aspán, A. & Olsen, B. (2004). Salmonella Amager, Campylobacter jejuni, and urease‐positive thermophilic Campylobacter found in free‐flying peregrine falcons (Falco peregrinus) in Sweden. Journal of Wildlife Diseases 40, 583–587. [DOI] [PubMed] [Google Scholar]
- * Palmgren, H. , Sellin, M. , Bergström, S. & Olsen, B. (1997). Enteropathogenic bacteria in migrating birds arriving in Sweden. Scandinavian Journal of Infectious Diseases 29, 565–568. [DOI] [PubMed] [Google Scholar]
- * Pao, S. , Hagens, B. E. , Kim, C. , Wildeus, S. , Ettinger, M. R. , Wilson, M. D. , Watts, B. D. , Whitley, N. C. , Porto‐Fett, A. C. S. , Schwarz, J. G. , Kaseloo, P. , Ren, S. , Long, W. , Li, H. & Luchansky, J. B. (2014). Prevalence and molecular analyses of Campylobacter jejuni and Salmonella spp. in co‐grazing small ruminants and wild‐living birds. Livestock Science 160, 163–171. [Google Scholar]
- Peachey, L. E. , Jenkins, T. P. & Cantacessi, C. (2017). This gut ain't big enough for both of us. Or is it? Helminth–microbiota interactions in veterinary species. Trends in Parasitology 33, 619–632. [DOI] [PubMed] [Google Scholar]
- * Pedersen, K. , Clark, L. , Andelt, W. & Salman, M. D. (2006). Prevalence of Shiga toxin‐producing Escherichia coli and Salmonella enterica in rock pigeons captured in Fort Collins, Colorado. Journal of Wildlife Diseases 42, 46–55. [DOI] [PubMed] [Google Scholar]
- * Pedersen, K. , Marks, D. R. , Arsnoe, D. M. , Afonso, C. L. , Bevins, S. N. , Miller, P. J. , Randall, A. R. & Deliberto, T. J. (2014). Avian paramyxovirus serotype I (Newcastle disease virus), avian influenza virus, and Salmonella spp. in mute swans (Cygnus olor) in the Great Lakes Region and Atlantic Coast of the United States. Avian Diseases 58, 129–136. [DOI] [PubMed] [Google Scholar]
- Penfold, J. B. , Amery, H. C. C. & Morley Peet, P. J. (1979). Gastroenteritis associated with wild birds in a hospital kitchen. British Medical Journal 2, 802. [PMC free article] [PubMed] [Google Scholar]
- Pennycott, T. W. , Park, A. & Mather, H. A. (2006). Isolation of different serovars of Salmonella enterica from wild birds in Great Britain between 1995 and 2003. The Veterinary Record 158, 817–820. [DOI] [PubMed] [Google Scholar]
- * Peterson, M. J. , Aguirre, R. , Ferro, P. J. , Jones, D. A. , Lawyer, T. A. , Peterson, M. N. & Silvy, N. J. (2002). Infectious disease survey of Rio Grande wild turkeys in the Edwards Plateau of Texas. Journal of Wildlife Diseases 38, 826–833. [DOI] [PubMed] [Google Scholar]
- * Phalen, D. N. , Drew, M. L. , Simpson, B. , Roset, K. , Dubose, K. & Mora, M. (2010). Salmonella enterica subsp. enterica in cattle egret (Bubulcus ibis) chicks from Central Texas: prevalence, serotypes, pathogenicity, and epizootic potential. Journal of Wildlife Diseases 46, 379–389. [DOI] [PubMed] [Google Scholar]
- * Pinowska, B. , Chyliński, G. & Gondek, B. (1976). Studies on the transmitting of salmonellae by house sparrows (Passer domesticus L.) in the region of Żuławy. Polish Ecological Studies 2, 113–121. [Google Scholar]
- * Pinowski, J. , Barkowska, M. , Kruszewicz, A. H. & Kruszewicz, A. G. (1994). The causes of the mortality of eggs and nestlings of Passer spp. Journal of Biosciences 19, 441–451. [Google Scholar]
- * Plant, C. W. (1978). Salmonellosis in wild birds feeding at sewage treatment works. Epidemiology & Infection 81, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright, R. K. , Parrish, C. R. , Mccallum, H. , Hudson, P. J. , Ko, A. I. , Graham, A. L. & Lloyd‐Smith, J. O. (2017). Pathways to zoonotic spillover. Nature Reviews Microbiology 15, 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Quessy, S. & Messier, S. (1992). Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring‐billed gulls (Larus delawarensis). Journal of Wildlife Diseases 28, 526–531. [DOI] [PubMed] [Google Scholar]
- * Quevedo, F. , Lord, R. D. , Dobosch, D. , Granier, I. & Michanie, S. C. (1973). Isolation of Salmonella from sparrows captured in horse corrals. The American Journal of Tropical Medicine and Hygiene 22, 672–674. [DOI] [PubMed] [Google Scholar]
- R Core Team (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- * Radimersky, T. , Frolkova, P. , Janoszowska, D. , Dolejska, M. , Svec, P. , Roubalova, E. , Cikova, P. , Cizek, A. & Literak, I. (2010). Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. Journal of Applied Microbiology 109, 1687–1695. [DOI] [PubMed] [Google Scholar]
- * Radwan, A. I. & Lampky, J. R. (1972). Enterobacteriaceae isolated from cowbirds (Molothrus ater) and other species of wild birds in Michigan. Avian Diseases 16, 343–350. [PubMed] [Google Scholar]
- * Ramey, A. M. , Hernandez, J. , Tyrlöv, V. , Uher‐Koch, B. D. , Schmutz, J. A. , Atterby, C. , Järhult, J. D. & Bonnedahl, J. (2018). Antibiotic‐resistant Escherichia coli in migratory birds inhabiting remote Alaska. EcoHealth 15, 72–81. [DOI] [PubMed] [Google Scholar]
- * Rausch, R. L. (1947). Pullorum disease in the coot. Journal of Wildlife Management 11, 189. [Google Scholar]
- * Reche, M. P. , Jiménez, P. A. , Alvarez, F. , García De Los Ríos, J. E. , Rojas, A. M. & De Pedro, P. (2013). Incidence of salmonellae in captive and wild free‐living raptorial birds in Central Spain. Journal of Veterinary Medicine. Series B 50, 42–44. [DOI] [PubMed] [Google Scholar]
- Reddy, K. R. , Khalell, R. & Overcash, M. R. (1981). Behavior and transport of microbial pathogens and indicator organisms in soils treated with organic wastes. Journal of Environmental Quality 10, 255–266. [Google Scholar]
- * Refsum, T. , Handeland, K. , Baggesen, D. L. , Holstad, G. & Kapperud, G. (2002). Salmonellae in avian wildlife in Norway from 1968 to 2000. Applied and Environmental Microbiology 68, 5595–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Refsum, T. , Holstad, G. , Kapperud, G. & Handeland, K. (2005). An investigation of Salmonella bacteria in waterfowls and migratory birds in Norway. Acta Veterinaria Scandinavica 46, 95–100. [PubMed] [Google Scholar]
- * Rivera, W. A. , Husic, J. S. , Gaylets, C. E. , Mosher, R. H. , Palestis, B. G. & Houlihan, A. J. (2012). Carriage of bacteria and protozoa in the intestinal tract of common tern chicks. Waterbirds 35, 490–494. [Google Scholar]
- * Robino, P. , Tomassone, L. , Tramuta, C. , Rodo, M. , Giammarino, M. , Vaschetti, G. & Nebbia, P. (2010). Prevalence of Campylobacter jejuni, Campylobacter coli and enteric Helicobacter in domestic and free living birds in North‐Western Italy. Schweizer Archiv fur Tierheilkunde 152, 425–431. [DOI] [PubMed] [Google Scholar]
- * Robinson, R. A. & Daniel, M. J. (1968). The significance of Salmonella isolations from wild birds and rats in New Zealand. New Zealand Veterinary Journal 16, 53–55. [DOI] [PubMed] [Google Scholar]
- * Rocha‐E‐Silva, R. C. , Cardoso, W. M. , Teixeira, R. S. C. , Albuquerque, Á. H. , Horn, R. V. , Lopes, E. S. , Gomes Filho, V. J. R. , Almeida, C. P. , Santos, I. C. L. , Machado, D. N. , Lima, S. V. G. & Carneiro, I. S. (2014). Isolation of Salmonella enterica subsp. enterica (O:4,5:i) and Salmonella enterica subsp. typhimurium from free‐living domestic pigeons (Columba livia). Arquivo Brasileiro de Medicina Veterinária e Zootecnia 66, 1435–1438. [Google Scholar]
- Rodewald, P. (2015). The Birds of North America. Cornell Laboratory of Ornithology, Ithaca. [Google Scholar]
- * Rodríguez, F. , Moreno, J. , Ortega, R. , Mathieu, C. , García, A. , Cerda‐Leal, F. & González‐Acuña, D. (2012). Evidence for kelp gulls (Larus dominicanus) and Franklin's gulls (Leucophaeus pipixcan) as carriers of Salmonella by real‐time polymerase chain reaction. Journal of Wildlife Diseases 48, 1105–1108. [DOI] [PubMed] [Google Scholar]
- Rollins, D. M. & Colwell, R. R. (1986). Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Applied and Environmental Microbiology 52, 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouffaer, L. O. , Lens, L. , Haesendonck, R. & Teyssier, A. (2016). House sparrows do not constitute a significant Salmonella Typhimurium reservoir across urban gradients in Flanders, Belgium. PLoS One 11, e0155366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Rutledge, M. E. , Siletzky, R. M. , Gu, W. , Degernes, L. A. , Moorman, C. E. , Deperno, C. S. & Kathariou, S. (2013). Characterization of Campylobacter from resident Canada geese in an urban environment. Journal of Wildlife Diseases 49, 1–9. [DOI] [PubMed] [Google Scholar]
- * Sacristán, C. , Esperón, F. , Herrera‐León, S. , Iglesias, I. , Neves, E. , Nogal, V. , Muñoz, M. J. & De La Toree, A. (2014). Virulence genes, antibiotic resistance and integrons in Escherichia coli strains isolated from synanthropic birds from Spain. Avian Pathology 43, 172–175. [DOI] [PubMed] [Google Scholar]
- Samadpour, M. , Steingart, K. , Louderback, J. & Ellington, J. (2002). Laboratory investigation of an E. coli 0157:H7 outbreak associated with swimming in Battle Ground Lake, Vancouver, Washington. Journal of Environmental Health 64, 16–20. [PubMed] [Google Scholar]
- * Sambyal, D. S. & Sharma, V. K. (1972). Screening of free‐living animals and birds for Listeria, Brucella and Salmonella infections. British Veterinary Journal 128, 50–55. [DOI] [PubMed] [Google Scholar]
- Sanad, Y. M. , Closs, G. , Kumar, A. & Lejeune, J. T. (2013). Molecular epidemiology and public health relevance of Campylobacter isolated from dairy cattle and European starlings in Ohio, USA. Foodborne Pathogens and Disease 10, 229–236. [DOI] [PubMed] [Google Scholar]
- Sánchez, C. A. , Becker, D. J. , Teitelbaum, C. S. , Barriga, P. , Brown, L. M. , Majewska, A. A. , Hall, R. J. & Altizer, S. (2018). On the relationship between body condition and parasite infection in wildlife: a review and meta‐analysis. Ecology Letters 21, 1869–1884. [DOI] [PubMed] [Google Scholar]
- * Santaniello, A. , Gargiulo, A. , Borrelli, L. , Dipineto, L. , Cuomo, A. , Sensale, M. , Fontanella, M. , Calabria, M. , Musella, V. , Menna, L. F. & Fioretti, A. (2007). Survey of Shiga toxin‐producing Escherichia coli O157:H7 in urban pigeons (Columba livia) in the city of Napoli, Italy. Italian Journal of Animal Science 6, 313–316. [Google Scholar]
- Sauer, J. R. , Niven, D. , Hines, J. E. , Ziolkowski, D. , Pardieck, K. L. , Fallon, J. & Link, W. A. (2017). The North American Breeding Bird Survey, Results and Analysis 1966–2015. Version 2.07.2017. USGS Patuxent Wildlife Research Center, Laurel. [Google Scholar]
- * Sayah, R. S. , Kaneene, J. B. , Johnson, Y. & Miller, R. (2005). Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic‐ and wild‐animal fecal samples, human septage, and surface water. Applied and Environmental Microbiology 71, 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan, E. , Hoekstra, R. M. , Angulo, F. J. , Tauxe, R. V. , Widdowson, M. , Roy, S. L. , Jones, J. L. & Griffin, P. M. (2011). Foodborne illness acquired in the United States — major pathogens. Emerging Infectious Diseases 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, C. & Bélisle, M. (2017). Agricultural intensification is linked to constitutive innate immune function in a wild bird population. Physiological and Biochemical Zoology 90, 201–209. [DOI] [PubMed] [Google Scholar]
- * Schmitz, A. , Kronthaler, F. , Stein, K. , Rinder, M. & Korbel, R. (2017). Decline of game birds (Phasianus colchicus and Perdix perdix) in Bavaria: a survey of pathogenic bacteria, parasites, pesticide residues, and influence of set‐aside land and maize cultivation. Journal of Zoo and Wildlife Medicine 48, 18–30. [DOI] [PubMed] [Google Scholar]
- Seguino, A. , Chintoan‐Uta, C. , Smith, S. H. & Shaw, D. J. (2018). Public health significance of Campylobacter spp. colonisation of wild game pheasants (Phasianus colchicus) in Scotland. Food Microbiology 74, 163–171. [DOI] [PubMed] [Google Scholar]
- Sensale, M. , Cuomo, A. , Dipineto, L. , Santaniellos, A. , Calabria, M. , Menna, L. & Fioretti, A. (2006). Survey of Campylobacter jejuni and Campylobacter coli in different taxa and ecological guilds of migratory birds. Italian Journal of Animal Science 5, 291–294. [Google Scholar]
- * Sharma, V. D. , Sethi, M. S. & Singh, S. P. (1980). The occurrence of salmonellae in free‐flying‐avifauna: isolation and antibiogram. International Journal of Zoonoses 7, 54–57. [PubMed] [Google Scholar]
- Shave, M. E. , Shwiff, S. A. , Elser, J. L. & Lindell, C. A. (2018). Falcons using orchard nest boxes reduce fruit eating bird abundances and provide economic benefits for a fruit‐growing region. Journal of Applied Ecology 55, 2451–2460. [Google Scholar]
- * Silva, V. L. , Nicoli, J. R. , Nascimento, T. C. & Diniz, C. G. (2009). Diarrheagenic Escherichia coli strains recovered from urban pigeons (Columba livia) in Brazil and their antimicrobial susceptibility patterns. Current Microbiology 59, 302–308. [DOI] [PubMed] [Google Scholar]
- * Sinai, N. L. , Coates, P. S. , Andrle, K. M. , Jefferis, C. , Sentíes‐Cué, C. G. & Pitesky, M. E. (2017). A serosurvey of greater sage‐grouse (Centrocercus urophasianus) in Nevada, USA. Journal of Wildlife Diseases 53, 136–139. [DOI] [PubMed] [Google Scholar]
- * Singleton, D. R. & Harper, R. G. (1998). Bacteria in old house wren nests. Journal of Field Ornithology 69, 71–74. [Google Scholar]
- * Sippy, R. , Sandoval‐Green, M. J. , Sahin, O. , Plummer, P. , Fairbanks, W. S. , Zhang, Q. & Blanchong, J. A. (2012). Occurrence and molecular analysis of Campylobacter in wildlife on livestock farms. Veterinary Microbiology 157, 369–375. [DOI] [PubMed] [Google Scholar]
- Skov, M. N. , Madsen, J. J. , Rahbek, C. , Lodal, J. , Jespersen, J. B. , Jørgensen, J. C. & Dietz, H. H. (2008). Transmission of Salmonella between wildlife and meat‐production animals in Denmark. Journal of Applied Microbiology 105, 1558–1568. [DOI] [PubMed] [Google Scholar]
- Smith, O. , Kennedy, C. , Owen, J. , Northfield, T. , Latimer, C. & Snyder, W. (2019). Highly diversified crop‐livestock farming systems reshape wild bird communities. Ecological Applications 0, e02031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Smith, W. A. , Mazet, J. A. K. & Hirsh, D. C. (2002). Salmonella in California wildlife species: prevalence in rehabilitation centers and characterization of isolates. Journal of Zoo and Wildlife Medicine 33, 228–235. [DOI] [PubMed] [Google Scholar]
- * Smith, S. , Wang, J. , Fanning, S. & Mcmahon, B. J. (2014). Antimicrobial resistant bacteria in wild mammals and birds: a coincidence or cause for concern? Irish Veterinary Journal 67, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Snoeyenbos, G. H. , Morin, E. W. & Wetherbee, D. K. (1967). Naturally occurring Salmonella in "blackbirds" and gulls. Avian Diseases 11, 642–646. [PubMed] [Google Scholar]
- * Sovada, M. A. , Pietz, P. J. , Converse, K. A. , King, D. T. , Hofmeister, E. K. , Scherr, P. & Ip, H. S. (2008). Impact of West Nile virus and other mortality factors on American white pelicans at breeding colonies in the northern plains of North America. Biological Conservation 141, 1021–1031. [Google Scholar]
- * Stenkat, J. , Krautwald‐Junghanns, M. E. , Schmitz Ornés, A. , Eilers, A. & Schmidt, V. (2014). Aerobic cloacal and pharyngeal bacterial flora in six species of free‐living birds. Journal of Applied Microbiology 117, 1564–1571. [DOI] [PubMed] [Google Scholar]
- * Stenzel, T. , Tykałowski, B. , Mazur‐Lech, B. & Koncicki, A. (2008). Infections in wildlife birds — results of serological screening. Bulletin of the Veterinary Institute in Pulawy 52, 63–66. [Google Scholar]
- * Stoddard, R. A. , Delong, R. L. , Byrne, B. A. , Jang, S. & Gulland, F. M. D. (2008). Prevalence and characterization of Salmonella spp. among marine animals in the Channel Islands, California. Diseases of Aquatic Organisms 81, 5–11. [DOI] [PubMed] [Google Scholar]
- Strachan, N. J. C. , Rotariu, O. , Smith‐Palmer, A. , Cowden, J. , Sheppard, S. K. , O'brien, S. J. , Maiden, M. C. J. , Macrae, M. , Bessell, P. R. , Matthews, L. , Reid, S. W. J. , Innocent, G. T. , Ogden, I. D. & Forbes, K. J. (2013). Identifying the seasonal origins of human Campylobacteriosis. Epidemiology & Infection 141, 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn, L. K. , Fortes, E. D. , Bihn, E. A. , Nightingale, K. K. , Gröhn, Y. T. & Worobo, R. W. (2013). Landscape and meteorological factors affecting prevalence of three food‐borne pathogens in fruit and vegetable farms. Applied and Environmental Microbiology 79, 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B. L. , Wood, C. L. , Iliff, M. J. , Bonney, R. E. , Fink, D. & Kelling, S. (2009). eBird: a citizen‐based bird observation network in the biological sciences. Biological Conservation 142, 2282–2292. [Google Scholar]
- * Sulzner, K. , Kelly, T. , Smith, W. & Johnson, C. K. (2014). Enteric pathogens and antimicrobial resistance in turkey vultures (Cathartes aura) feeding at the wildlife–livestock interface. Journal of Zoo and Wildlife Medicine 45, 931–934. [DOI] [PubMed] [Google Scholar]
- * Swirski, A. L. , Pearl, D. L. , Williams, M. L. , Homan, H. J. , Linz, G. M. , Cernicchiaro, N. & Lejeune, J. T. (2014). Spatial epidemiology of Escherichia coli O157:H7 in dairy cattle in relation to night roosts of Sturnus vulgaris (European starling) in Ohio, USA (2007–2009). Zoonoses and Public Health 61, 427–435. [DOI] [PubMed] [Google Scholar]
- Taff, C. C. & Townsend, A. K. (2017). Campylobacter jejuni infection associated with relatively poor condition and low survival in a wild bird. Journal of Avian Biology 48, 1071–1076. [Google Scholar]
- Taff, C. C. , Weis, A. M. , Wheeler, S. , Hinton, M. G. , Weimer, B. C. , Barker, C. M. , Jones, M. , Logsdon, R. , Smith, W. A. , Boyce, W. M. & Townsend, A. K. (2016). Influence of host ecology and behavior on Campylobacter jejuni prevalence and environmental contamination risk in a synanthropic wild bird species. Applied and Environmental Microbiology 82, 4811–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Tanaka, C. , Miyazawa, T. , Watarai, M. & Ishiguro, N. (2005). Bacteriological survey of feces from feral pigeons in Japan. Journal of Veterinary Medical Science 67, 951–953. [DOI] [PubMed] [Google Scholar]
- Tannock, G. & Smith, J. (1972). Studies on the survival of Salmonella Typhimurium and Salmonella Bovismorbificans on soil and sheep faeces. Research in Veterinary Science 13, 150–153. [PubMed] [Google Scholar]
- Thrusfield, M. (2007). Surveys In Veterinary Epidemiology, 3rd Edition (ed. Thrusfield M.), pp. 228–242. Blackwell Science, Oxford. [Google Scholar]
- Tilden, J. , Young, W. , Mcnamara, A. , Custer, C. , Boesel, B. , Lambert‐Fair, M. , Majkowski, J. , Vugia, D. , Wemer, S. B. , Hollingsworth, J. & Morris, J. G. (1996). A new route of transmission for Escherichia coli: infection from dry fermented salami. Public Health Briefs 86, 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizard, I. (2004). Salmonellosis in wild birds. Seminars in Avian and Exotic Pet Medicine 13, 50–66. [Google Scholar]
- * Tizard, I. R. , Fish, N. A. & Harmeson, J. (1979). Free flying sparrows as carriers of Salmonellosis. The Canadian Veterinary Journal 20, 143–144. [PMC free article] [PubMed] [Google Scholar]
- * Ton, J. (2017). Determining the pathogenicity of bacteria present in wetland waters and crow feces in Bothell. O Mundo de Saúde, São Paulo 41, 378–384. [Google Scholar]
- * Toro, H. , Saucedo, C. , Borie, C. , Gough, R. E. & Alcaíno, H. (1999). Health status of free‐living pigeons in the city of Santiago. Avian Pathology 28, 619–623. [DOI] [PubMed] [Google Scholar]
- * Torres‐Mejía, A. M. , Blanco‐Peña, K. , Rodríguez, C. , Duarte, F. , Jiménez‐Soto, M. & Esperón, F. (2018). Zoonotic agents in feral pigeons (Columba livia) from Costa Rica: possible improvements to diminish contagion risks. Vector‐Borne and Zoonotic Diseases 18, 49–54. [DOI] [PubMed] [Google Scholar]
- * Tsubokura, M. , Matsumoto, A. , Otsuki, K. , Animas, S. B. & Sanekata, T. (1995). Drug resistance and conjugative R plasmids in Escherichia coli strains isolated from migratory waterfowl. Journal of Wildlife Diseases 31, 352–357. [DOI] [PubMed] [Google Scholar]
- * Une, Y. , Sanbe, A. , Suzuki, S. , Niwa, T. , Kawakami, K. , Kurosawa, R. , Izumiya, H. , Watanabe, H. & Kato, Y. (2008). Salmonella enterica serotype Typhimurium infection causing mortality in Eurasian tree sparrows (Passer montanus) in Hokkaido. Japanese Journal of Infectious Diseases 61, 166–167. [PubMed] [Google Scholar]
- Van Donsel, D. J. , Geldreich, E. E. & Clarke, N. A. (1967). Seasonal variations in survival of indicator bacteria in soil and their contribution to storm‐water pollution. Applied Microbiology 15, 1362–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Vasconcelos, R. H. , Bezerra, W. G. A. , Siqueira, R. A. S. , De Medeiros, P. H. Q. S. , Lucena, R. B. , Havt, A. , Da Silva, I. N. G. & Maciel, W. C. (2017). Natural coinfection of Pseudomonas aeruginosa and enteroaggregative Escherichia coli in a feral pigeon (Columba livia). Acta Scientiae Veterinariae 45, 1–4. [Google Scholar]
- * Vaughan‐Higgins, R. , Murphy, S. , Carter, I. , Pocknell, A. , Harris, E. & Sainsbury, A. W. (2013). Fatal epicarditis in a hen harrier (Circus cyaneus) a red‐listed bird of high conservation concern in Britain associated with Cyathostoma species and Escherichia coli infection. The Veterinary Record 173, 1–3. [DOI] [PubMed] [Google Scholar]
- * Vázquez, B. , Esperón, F. , Neves, E. , López, J. , Ballesteros, C. & Muñoz, M. J. (2010). Screening for several potential pathogens in feral pigeons (Columba livia) in Madrid. Acta Veterinaria Scandinavica 52, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Veatch, J. K. , Applegate, R. D. & Osborne, S. J. (1998). Serologic incidence of some diseases in Kansas wild turkeys. Avian Diseases 42, 393–396. [PubMed] [Google Scholar]
- Viechtbauer, W. (2010). Conducting meta‐analyses in R with the metafor package. Journal of Statistical Software 36, 1–48. [Google Scholar]
- * Vilela, S. M. O. , Júnior, J. W. P. , Silva, J. S. A. , De Pace, F. , Silveira, W. , Saukas, T. N. , Reis, E. M. F. & Mota, R. A. (2012). Research of Salmonella spp. and evaluation of pathogenicity, cytotoxicity of Escherichia coli isolates proceeding from sparrows (Passer domesticus). Pesquisa Veterinária Brasileira 32, 931–935. [Google Scholar]
- * Vlahović, K. , Matica, B. , Bata, I. , Pavlak, M. , Pavičić, Z. , Popović, M. , Nejedli, S. & Dovč, A. (2004). Campylobacter, Salmonella, and chlamydia in free‐living birds of Croatia. European Journal of Wildlife Research 50, 127–132. [Google Scholar]
- Vogel, J. R. , Griffin, D. W. , Ip, H. S. , Ashbolt, N. J. , Moser, M. T. , Lu, J. , Beitz, M. K. , Ryu, H. & Domingo, J. W. S. (2013). Impacts of migratory sandhill cranes (Grus canadensis) on microbial water quality in the Central Platte River, Nebraska. Water, Air, & Soil Pollution 224, 1576. [Google Scholar]
- * Vučemilo, M. , Vlahović, K. , Dovč, A. , Mužinić, J. , Pavlak, M. , Jerčić, J. & Župančić, Ž. (2003). Prevalence of Campylobacter jejuni, Salmonella Typhimurium, and avian Paramyxovirus type 1 (PMV‐1) in pigeons from different regions in Croatia. Zeitschrift für Jagdwissenschaft 49, 303–313. [Google Scholar]
- * Wahlström, H. , Tysén, E. , Engvall, E. O. , Brändström, B. , Eriksson, E. , Mörner, T. & Vågsholm, I. (2003). Survey of Campylobacter species, VTEC O157 and Salmonella species in Swedish wildlife. The Veterinary Record 153, 74–80. [DOI] [PubMed] [Google Scholar]
- Waldenström, J. , Axelsson‐Olsson, D. , Hasselquist, D. , Griekspoor, P. , Waldenstro, J. , Jansson, L. , Teneberg, S. , Svensson, L. & Ellstro, P. (2010). Campylobacter jejuni colonization in wild birds: results from an infection experiment. PLoS One 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström, J. , Broman, T. , Carlsson, I. , Hasselquist, D. , Achterberg, P. & Wagenaar, J. A. (2002). Campylobacter coli in different ecological guilds and taxa of migrating birds. Applied and Environmental Microbiology 68, 5911–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , He, Y. , Jin, X. , Zhou, Y. , Chen, X. , Zhao, J. , Zhang, H. & Chen, W. (2018). The effect of co‐infection of food‐borne pathogenic bacteria on the progression of Campylobacter jejuni infection in mice. Frontiers in Microbiology 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburne, A. D. , Crowley, D. E. , Becker, D. J. , Manlove, K. R. , Childs, M. L. & Plowright, R. K. (2019). Percolation models of pathogen spillover. Philosophical Transactions of the Royal Society B: Biological Sciences 374, 20180331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar, T. M. (2011). Following an imaginary Campylobacter population from farm to fork and beyond: a bacterial perspective. Letters in Applied Microbiology 53, 253–263. [DOI] [PubMed] [Google Scholar]
- * Weis, A. M. , Miller, W. A. , Byrne, B. A. , Chouicha, N. , Boyce, W. M. & Townsend, A. K. (2014). Prevalence and pathogenic potential of Campylobacter isolates from free‐living, human‐commensal American crows. Applied and Environmental Microbiology 80, 1639–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, A. M. , Storey, D. B. , Taff, C. C. , Townsend, A. K. , Huang, B. C. , Kong, N. T. & Clothier, K. A. (2016). Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Applied and Environmental Microbiology 82, 7165–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * White, F. H. & Forrester, D. J. (1979). Antimicrobial resistant Salmonella spp. isolated from double‐crested cormorants (Phalacrocorax auritus) and common loons (Gavia immer) in Florida. Journal of Wildlife Diseases 15, 235–237. [DOI] [PubMed] [Google Scholar]
- * Wieliczko, A. , Piasecki, T. , Dorrestein, G. M. , Adamski, A. & Mazurkiewicz, M. (2003). Evaluation of the health status of goshawk chicks (Accipiter gentilis) nesting in Wrocław vicinity. Bulletin of the Veterinary Institute in Pulawy 47, 247–257. [Google Scholar]
- * Williams, C. K. , Davidson, W. R. , Lutz, R. S. & Applegate, R. D. (2000). Health status of northern bobwhite quail (Colinus virginianus) in Eastern Kansas. Avian Diseases 44, 953–956. [PubMed] [Google Scholar]
- * Williams, B. M. , Richards, D. W. & Lewis, J. (1976). Salmonella infection in the herring gull (Lans argentatus). The Veterinary Record 98, 51. [DOI] [PubMed] [Google Scholar]
- * Williams, B. M. , Richards, D. W. , Stephens, D. P. & Griffiths, T. (1977). Transmission of S. livingstone to cattle by the herring gull (Larus argentatus). The Veterinary Record 100, 450–451. [DOI] [PubMed] [Google Scholar]
- Wilman, H. , Belmaker, J. , Jennifer, S. , de la Rosa, C. , Rivadeneira, M. M. & Jetz, W. (2014). EltonTraits 1.0: species‐level foraging attributes of the world's birds and mammals. Ecology 95, 2027. [Google Scholar]
- * Windsor, D. K. , Bloebaum, A. P. & Mathewson, J. J. (1981). Gram‐negative, aerobic, enteric pathogens among intestinal microflora of wild turkey vultures (Cathartes aura) in West Central Texas. Applied and Environmental Microbiology 42, 1123–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield, M. D. & Groisman, E. A. (2003). Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli . Applied and Environmental Microbiology 69, 3687–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Wobeser, G. A. & Finlayson, M. C. (1969). Salmonella Typhimurium infection in house sparrows. Archives of Environmental Health 19, 882–884. [DOI] [PubMed] [Google Scholar]
- * Wood, A. J. & Trust, T. J. (1972). Some qualitative and quantitative aspects of the intestinal microflora of the glaucous‐winged gull (Larus glaucescens). Canadian Journal of Microbiology 18, 1577–1583. [DOI] [PubMed] [Google Scholar]
- * Yogasundram, K. , Shane, S. M. & Harrington, K. S. (1989). Prevalence of Campylobacter jejuni in selected domestic and wild birds in Louisiana. Avian Diseases 33, 664–667. [PubMed] [Google Scholar]
- You, Y. , Rankin, S. C. , Aceto, H. W. , Benson, C. E. , Toth, J. D. & Dou, Z. (2006). Survival of Salmonella enterica serovar Newport in manure and manure‐amended soils. Applied and Environmental Microbiology 72, 5777–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerberg, B. , Fink, D. , La Sorte, F. A. , Hochachka, W. M. & Kelling, S. (2016). Novel seasonal land cover associations for eastern North American forest birds identified through dynamic species distribution modelling. Diversity and Distributions 22, 717–730. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Supplementary tables and figures.
Data S1. List of studies and their methods included in meta‐analysis.
Data S2. Meta‐data on enteric pathogen prevalence in North American breeding bird species.
Data S3. Meta‐data of enteric pathogen prevalence for each study included in meta‐regressions.


