Abstract
The level of microRNA‐9‐5p (miRNA‐9‐5p) in brain tissues is significantly changed after traumatic brain injury (TBI). However, the effect of miRNA‐9‐5p for brain function in TBI has not been elucidated. In this study, a controlled cortical impact model was used to induce TBI in Sprague–Dawley rats, and an oxygen glucose deprivation model was used to mimic the pathological state in vitro. Brain microvascular endothelial cells (BMECs) and astrocytes were extracted from immature Sprague–Dawley rats and cocultured to reconstruct blood–brain barrier (BBB) in vitro. The results show that the level of miRNA‐9‐5p was significantly increased in brain tissues after TBI, and up‐regulation of miRNA9‐5p contributed to the recovery of neurological function. Up‐regulation of miRNA‐9‐5p with miRNA agomir may significantly alleviate apoptosis, neuroinflammation, and BBB damage in rats after TBI. Moreover, a dual luciferase reporter assay confirmed that miRNA‐9‐5p is a post‐transcriptional modulator of Ptch‐1. In in vitro experiments, the results confirmed that up‐regulation of miRNA‐9‐5p with miRNA mimic alleviates cellular apoptosis, inflammatory response, and BBB damage mainly by inhibiting Ptch‐1. In addition, we found that the activation of Hedgehog pathway was accompanied by inhibition of NF‐κB/MMP‐9 pathway in the BMECs treated with miRNA‐9‐5p mimic. Taken together, these results indicate that up‐regulation of miRNA‐9‐5p alleviates BBB damage and neuroinflammatory responses by activating the Hedgehog pathway and inhibiting NF‐κB/MMP‐9 pathway, which promotes the recovery of neurological function after TBI.
Keywords: apoptosis, blood–brain barrier, microRNA‐9‐5p, neuroinflammation, Ptch‐1, traumatic brain injury
Hedgehog pathway and NF‐κB proteins play important roles in cellular apoptosis and inflammatory responses. Using bioinformatics techniques, we find that Ptch‐1, which is an inhibitor of Hedgehog pathway, and NF‐κB are potential targets for microRNA‐9‐5p. We propose the following cascade for microRNA‐9‐5p that alleviates the damage of blood–brain barrier and neuroinflammation after traumatic brain injury: over‐expression of microRNA‐9‐5p activates Hedgehog pathway and blocks NF‐κB/MMP‐9 pathway by inhibiting Ptch‐1 and NF‐κB, which inhibit cellular apoptosis and inflammatory responses of brain microvascular endothelial cells. We think these findings may provide a new target for the treatment of traumatic brain injury.
Abbreviations
- AKT
protein kinase B
- BBB
blood–brain barrier
- BMECs
brain microvascular endothelial cells
- BSA
bovine serum albumin
- CCI
control cortex injury
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- ELISA
enzyme‐linked immunosorbent assay
- GAPDH
glycerinaldehyde‐3‐phosphate‐dehydrogenase
- Gli‐1
glioma‐associated oncogene homolog 1
- GSK3β
glycogen synthase kinase‐3 β
- IL‐1β
interleukin‐1 beta
- IL‐6
Interleukin‐6
- miRNA
microRNA
- MMP‐9
matrix metalloproteinase 9
- mNSS
modified neurological severity score
- Mut
mutant type
- NF‐κB
transcription factors of the nuclear factor kappa B
- OGD
oxygen–glucose deprivation
- p‐AKT
phosphor‐Protein kinase B
- p‐GSK3β
phosphor‐Glycogen synthase kinase‐3β
- Ptch‐1
Patched 1
- qRT‐PCR
quantitative real‐time polymerase chain reaction
- RRID
research resource identifier
- SEM
standard error of mean
- TBI
traumatic brain injury
- TEER
transendothelial electrical resistance
- TNF‐α
tumor necrosis factor alpha
- UTR
untranslated region
- WT
wild‐type
- ZO‐1
zona occludens 1
1. INTRODUCTION
Traumatic brain injury (TBI) refers to craniocerebral injury caused by blunt, penetrating, accelerating, or decelerating force, which manifests as memory loss, unconsciousness, or even death (Ng & Lee, 2019). Worldwide, nearly 300 per 100,000 people suffer a TBI each year. According to the pathological process, TBI can be divided into two stages: the primary injury which is caused by external force, and the secondary injury which is caused by a series of pathophysiological changes in neurochemistry, metabolism, cells, and molecules (Amorini et al., 2016; Di Pietro et al., 2013). The secondary brain injury mainly includes excessive inflammatory response and blood–brain barrier (BBB) breakdown (Liu et al., 2019). Following TBI, microglia were activated and released a large number of inflammatory factors including IL‐1, IL‐6, and TNF‐α, which can destroy the blood–brain barrier and induce cellular apoptosis (Chen, Zhu, Hang, & Wang, 2019). The BBB is mainly composed of brain microvascular endothelial cells (BMECs), astrocytes, and basal membrane, which is the basic structural barrier to maintain normal brain function (Xu, Nirwane, & Yao, 2019). Brain edema caused by BBB damage is one of the main reasons for poor prognosis after TBI (Jha, Kochanek, & Simard, 2019). However, the specific mechanism of neuroinflammation and BBB damage in TBI have not been fully elucidated.
Recently, more and more studies report that non‐transcription factors play an important role in TBI (Chandran, Mehta, & Vemuganti, 2017). MicroRNAs (miRNAs) are short (19–28 nucleotides), endogenous, non‐coding RNAs which act at the post‐transcriptional level to regulate protein synthesis (Churov, Summerhill, Grechko, Orekhova, & Orekhov, 2019). Numerous studies have confirmed that the level of miRNAs is changed after TBI, and various miRNAs are involved in cellular apoptosis, BBB damage, and neuroinflammatory response. For example, the level of miRNA‐21‐5p is increased after TBI, and up‐regulation of miRNA‐21‐5p improves neurological function by alleviating BBB damage and neuroinflammatory response (Ge et al., 2016). Phosphatase and tensin homolog (PTEN), a well‐known regulator of the AKT/mTOR pathway, was found to be a direct target of miRNA‐23a‐3p. A recent study found that the level of miRNA‐23a‐3p was decreased after TBI and up‐regulation of miRNA‐23a‐3p inhibited cortical neuron apoptosis and improved the neural function (Li et al., 2020). MiRNA‐9 is highly expressed in the central nervous system and plays an important role in the differentiation of neural stem cells (Hu et al., 2014; Topol et al., 2016). Some studies have reported that the level of miRNA‐9 was significantly changed after brain injury (Lei, Li, Chen, Yang, & Zhang, 2009). Up‐regulation of miRNA‐9 inhibited neural apoptosis in rats after stroke (Chen et al., 2017; Nampoothiri & Rajanikant, 2019) and promoted synaptic remodeling in Alzheimer’s disease (Zhao, Pogue, & Lukiw ,2015). However, the effect of miRNA‐9 on TBI remains unclear.
In this study, we established the controlled cortical impact (CCI) model in rats and cultured BMECs and astrocytes in vitro to investigate the effect of miRNA‐9‐5p on cellular apoptosis, BBB damage, and inflammatory response after TBI.
2. MATERIALS AND METHODS
2.1. Animals
This study was not pre‐registered. All the animal procedures were approved by the Commission of Chongqing Medical University for the Ethics of Experiments on Animals and were in accordance with international standards. Animal care and killing was conducted according to the methods approved by the Chongqing Medical University Animal Experimentation Committee (Permit Number: 20141011). The adult male (aged approximately 16 weeks, weighing 250–300 g) and neonatal Sprague–Dawley rats (RRID: RGD_1566457) were purchased from the Experimental Animal Center of Chongqing Medical University (Chongqing, China). The adult rats were raised in temperature‐controlled room (22 ± 2°C) with the 12 hr light/12 hr dark cycle and were free to get food and water. Animals were identified by earmark numbers. Microsoft Excel 365 (Microsoft Corporation) was used to generate a table of random numbers that were assigned to each rat for simple random grouping. For example, 32 rats in experiment 2 were numbered 1 to 32, and 32 random numbers between 0 and 1 were generated using the RAND () command in EXCEL. Random numbers were sorted from small to large and assigned to groups of eight in turn. The rats in other experiments were randomly grouped using the above method.
2.2. Experimental design
This study was mainly divided into the following six experiments (Figure 1). A total of 205 adult and 10 juvenile Sprague‐Dawley rats were involved in this study. A total of 27 rats which died within 24 hr post‐CCI were excluded. Detailed information about animal distribution and usage is listed in Table 1. The alpha level was set to 0.05.
Figure 1.
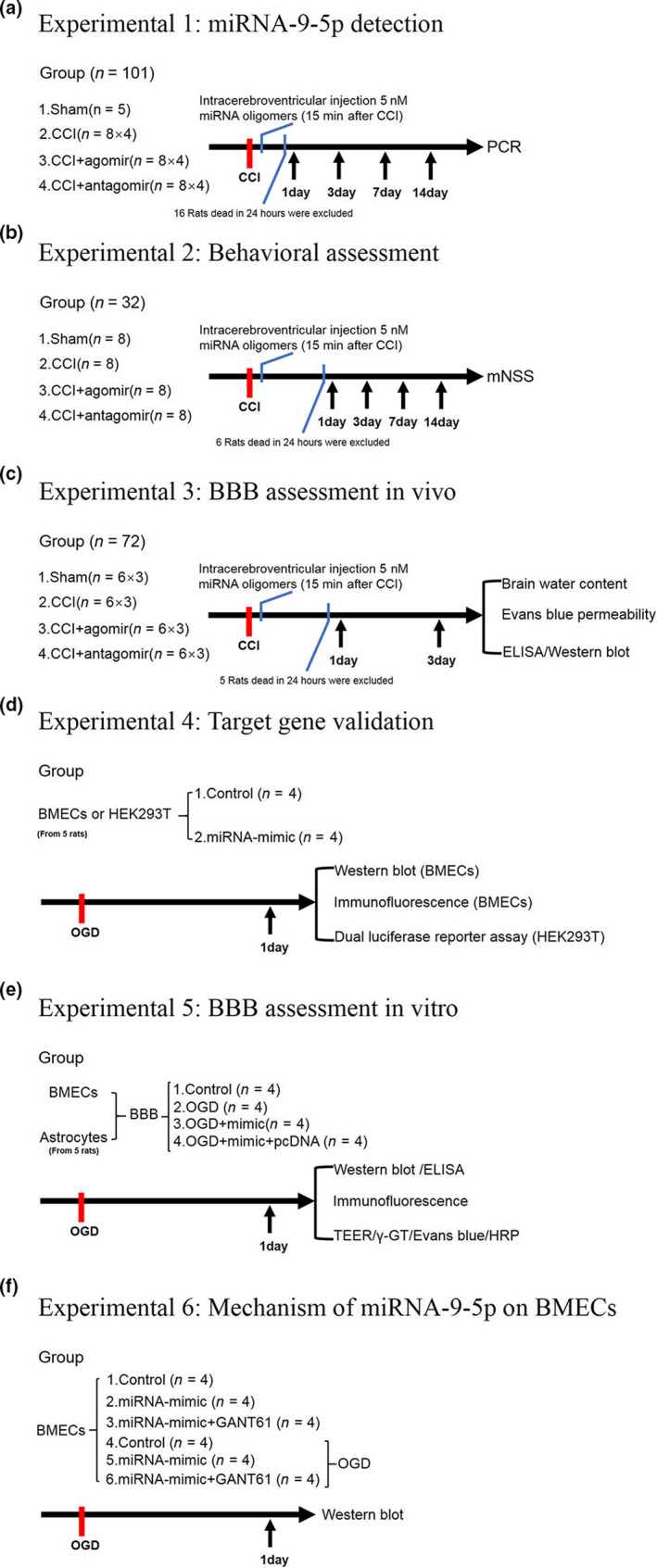
Schematic of the experimental design. (a) miRNA‐9‐5p detection in rats. (b) Behavioral assessment. (c) BBB assessment in vivo. (d) Target gene validation. (e) BBB assessment in vitro. (f) mechanism of miRNA‐9‐5p on BMECs. BBB: blood–brain barrier; BMECs: Brain microvascular endothelial cells; CCI: control cortex injury; ELISA: enzyme‐linked immunosorbent assay; mNSS: modified neurological severity score; OGD: oxygen glucose deprivation; PCR: polymerase chain reaction. (n: number of rats in each group or number of independent cell culture preparations in each group;)
Table 1.
Animal allocation and usage
| Experiments | Groups | PCR | mNSS | WB | BWC | EB |
Cell culture |
Death | Mortality |
|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Sham | 5 | – | – | – | – | – | 0 | 0 |
| CCI | 8 × 4 | – | – | – | – | – | 4 | 12.5% | |
| CCI + antagomir | 8 × 4 | – | – | – | – | – | 6 | 18.75% | |
| CCI + agomir | 8 × 4 | – | – | – | – | – | 6 | 18.75% | |
| Experiment 2 | Sham | – | 8 | – | – | – | – | 0 | 0 |
| CCI | – | 8 | – | – | – | – | 2 | 25% | |
| CCI + antagomir | – | 8 | – | – | – | – | 3 | 37.5% | |
| CCI + agomir | – | 8 | – | – | – | – | 1 | 12.5% | |
| Experiment 3 | Sham | – | – | 6 | 6 | 6 | – | 0 | 0 |
| CCI | – | – | 6 | 6 | 6 | – | 1 | 5.56% | |
| CCI + antagomir | – | – | 6 | 6 | 6 | – | 2 | 11.11% | |
| CCI + agomir | – | – | 6 | 6 | 6 | – | 2 | 11.11% | |
| Experiment 4–6 | BMEC/Astrocyte | – | – | – | – | – | 10 | – | – |
| Total number | Adult: 205 | 101 | 32 | 24 | 24 | 24 | – | 27 | 13.17% |
| immature:10 | – | – | – | – | – | 10 | – | – |
Detailed information about animal allocation and usage in this study. Although compared with the rats which suffered control cortex impact, the mortality number in each subexperiment and overall mortality number were higher in CCI + antagomir group, no significant difference was achieved.
Abbreviations: CCI, control cortex impact; WB, western blot; BWC, brain water content; EB, Evans Blue; BMEC, brain microvascular endothelial cell.
2.2.1. Experiment 1
First, 37 rats were randomized into two groups: sham (n = 5) and control cortex injury (CCI) (n = 32) which including CCI‐1 day (n = 8), CCI‐3 day (n = 8), CCI‐7 day (n = 8), and CCI‐14 hr (n = 8). The time course of miRNA‐9‐5p expression was determined by qRT‐PCR (Figure 1a).
Second, 64 rats were equally randomized by the method described above into two groups with agomir and antagomir: CCI + agomir‐1 day (n = 8) and CCI + antagomir‐1 day (n = 8), CCI + agomir‐3 day (n = 8) and CCI + antagomir‐3 day (n = 8), CCI + agomir‐7 day (n = 8) and CCI + antagomir‐7 day (n = 8), and CCI + agomir‐14 day (n = 8) and CCI + antagomir‐14 day (n = 8). The time course of miRNA‐9‐5p expression in both groups was also detected by qRT‐PCR.
At last, 16 rats were dead and excluded in this step. Five rats were randomly selected and included in each group for this experiment.
2.2.2. Experiment 2
The neurological function was evaluated by modified neurological severity score at different time point after brain injury. In total, 32 rats were separated randomly into four groups: sham (n = 8), CCI(n = 8), CCI + agomir (n = 8), and CCI + antagomir (n = 8) (Figure 1b).
At last, six rats were dead and excluded in this step. Five rats were randomly selected and included in each group for this experiment.
2.2.3. Experiment 3
First, 24 rats were separated randomly into four groups: sham (n = 6), CCI (n = 6), CCI + agomir (n = 6), and CCI + antagomir (n = 6). The changing of brain water content was evaluated by dry‐wet weight method in different groups at day 3 after injury (Figure 1c).
Second, 24 rats were separated randomly into four groups: sham (n = 6), CCI (n = 6), CCI + agomir (n = 6), and CCI + antagomir (n = 6). The evans blue permeability was evaluated by detecting evans blue content at day 3 in the injured cerebral hemisphere.
The integrity of BBB in vivo was evaluated by detecting brain water content and evans blue permeability in rats after brain injury.
Third, 24 rats were separated randomly into four groups: sham (n = 6), CCI (n = 6), CCI + agomir (n = 6), and CCI + antagomir (n = 6). The western blot and enzyme‐linked immunsorbent assay (ELISA) were performed at day 3 after CCI.
At last, 5 rats were dead and excluded in this step. Five rats were randomly selected and included in each group for this experiment.
2.2.4. Experiment 4
Brain microvascular endothelial cells (BMECs) were extracted from five 1‐week‐old rats and the cells were divided into two groups: control (n = 4) and mimic (n = 4), which was treated with vehicle and miRNA‐9‐5p mimic for 24 hr, respectively. The expression of Ptch‐1 and glioma‐associated oncogene homolog 1 (Gli‐1) was detecting by western blot and immunofluorescence in both groups (Figure 1d).
2.2.5. Experiment 5
Astrocytes were extracted from five 1‐day‐old rats. The blood–brain barrier (BBB) model in vitro, which was reconstructed by BMECs and astrocytes, was divided into four groups: control (n = 4), oxygen‐glucose deprivation (OGD), OGD + mimic (n = 4), and OGD + mimic+pcDNA (n = 4). The integrity of BBB in vitro was evaluated by calculating the number of tight junction structure and detecting transendothelial electrical resistance (TEER) values, γ‐GT activity, evans blue, and HRP permeability. The effect of miRNA‐9‐5p on apoptosis and inflammation in vitro was evaluated by western blot and ELISA in BMECs or cell medium (Figure 1e).
2.2.6. Experiment 6
The BMECs were separated into six groups: CON (control) (n = 4), MIC (mimic)(n = 4), MIC + GAN (GANT61) (n = 4), OGD (n = 4), OGD + MIC (n = 4), and OGD + MIC+GAN (n = 4). The mechanism of miRNA‐9‐5p on BMECs was detected by western blot (Figure 1f).
After the experiment, we analyzed the obtained gene expression data to confirm this assumption, evaluating whether the agreed upon sample size was sufficient to reach a power level above 0.95 and a significance level below 0.05. To estimate statistical power, we employed the post hoc (a posteriori) analysis of G*Power 3.1 to calculate Cohen's effect size f from the group size, the mean values for each group, and the average standard deviance (Faul, Erdfelder, Lang, & Buchne, 2007). We employed the F test: fixed‐effects one‐way ANOVA routine of G‐power software to calculate the Cohen's effect size f from the data about group size, number of groups, mean value for each group, and average standard deviation. Parameters employed were as follows: number of groups = 4, sample size = 5/group, significance level = 0.05, estimated effect size = Cohen's f. Using these data, we estimated an effect size of f = 4.38 and a power = 0.99 for miRNA‐9‐5p expression data and an effect size of f = 1.73 and a power = 0.99 for the mNSS data.
2.3. Controlled cortical impact model and intracerebroventricular injection
The controlled cortical impact (CCI) model was applied to stimulate moderate TBI (Cao et al., 2016) by using the controlled impactor device (TBI‐0310 TBI Model System; Precision Systems and Instrumentation, Chongqing, China) in rats. All rats were anesthetized by intraperitoneal injection 10% chloral hydrate (3ml/kg) (Sigma‐Aldrich, Cat# 47335‐U) and sufentanil (50 μg/kg) (Sigma‐Aldrich, Cat#S‐008‐1Ml) and maintained at 36–37°C. The chloral hydrate was mainly used to anesthesia for rats, which has no analgesic effect. So, the sufentanil which is a powerful analgesic was used to assist anesthesia. The impact parameters were set as velocity 3.5 m/s, depth 2.5 mm, and dwell time 200 ms. The thrombin (500 units/ml) (Solarbio, Beijing, China, Cat#T8021) was locally perfused into the wound to stop bleeding. After the operation, the bone window was closed with bone wax (Johnson & Johnson, USA, Cat#W810T) and the wound was sutured. The rats in the sham group underwent the same surgical procedure excluding controlled cortical impact. Brian tissues were collected from the impacted area with a diameter of 8 mm for subsequent testing.
MiRNA‐9‐5p agomir (or inhibitor) (RiboBio) was diluted to 5 nM (5 µl) according to the manufacturer's instructions. The Hamilton brain infusion syringe was stereotaxically injected into the contralateral ventricle through a bone hole (Ge et al., 2014) (coordinates: 1.5 mm caudal to the bregma; 1.1 mm lateral to the midline; 4.5 mm deep from the surface of skull) at 15min post‐CCI. The mixture of oligomers (5 µl) was continuously injected into the contralateral ventricle within 5 min and the needle was withdrawn within 3 min after infusion. The speed of insertion and withdrawn was controlled at 1.5 mm/min.
Some measures were taken to reduce suffering for rats post‐CCI. The ibuprofen (50 mg/kg) (Solarbio, Beijing, China, Cat# I8430) was taken orally for 3 days to alleviate pain and penicillin sodium (15 mg/kg) (Solarbio, Beijing, China, Cat#C8250) was injected intraperitoneally once a day for 7 days to avoid infection. All rats were returned to the cage with free access to food and water.
2.4. Modified neurological severity score
The modified neurological severity score (mNSS), which includes motor, sensory, reflex, and balance tests, was used to evaluate the effect of miRNA oligomers on TBI as previously reported (Ge et al., 2014). The data of mNSS were examined and corrected from rats before suffering CCI and the time point of 1, 3, 7 and 14 days post‐CCI. All data were analyzed by a person who was blinded to animal group assignment to avoid any bias from observers.
2.5. Cell culture
In the cell extraction experiment, isoflurane was used to anesthetize the juvenile rats. According to a previous study, isoflurane can alleviate inflammatory response in many diseases (An, 2019; Lian, Fang, Zhou, Jiang, & Xie, 2019). Therefore, we thought it can promote the survival of primary cells after extraction.
Brain microvascular endothelial cells (BMECs) were extracted from five 1‐week‐old rats as we previously described (Tian et al., 2016). The rats were anesthetized by inhalation of 4% isoflurane for 2 min (Sigma‐Aldrich, Cat# 792632). The brain was collected and grey matter was separated into 1 mm3 pieces which were minced and digested with 0.1% collagenase type‐2 (Thermo Fisher Scientific, Cat# 17101015) and DNase I (Thermo Fisher Scientific, Cat# 18047019) (39 U/mL) at 37°C for 30 min and the mixtures were centrifuged with 50 g for 8 min. Then, the precipitates were collected, resuspended with 20% bovine serum albumin (Sigma‐Aldrich, Cat# SRE0096), and centrifuged at 200 g for 10 min again. The re‐precipitates were digested with collagenase/dispase (1 mg/ml) (Sigma‐Aldrich, Cat# 10269638001) and DNase I (39 U/mL) at 37°C for 30 min, and the mixtures were centrifuged at 50 g for 8 min again. All the cell clusters were maintained in Dulbecco's Modified Eagle's Medium Nutrient Mixture F‐12 (DMEM/F12) (Gibco, Cat# 11330057) with 15% Fetal Bovine Serum (FBS) (Gibco, Cat# 10099141). The cell culture plate was placed in the incubator at 37°C with 5% carbon dioxide and the medium was changed every 3 days.
Astrocytes were extracted from brain cortex from five 1‐day‐old rats as we previously described (Tian et al., 2016). The rats were also anesthetized with inhalation of 4% isoflurane for 2 min. The grey matter was minced and digested analogous to the processing of BMECs. After centrifugation, the precipitates were resuspended in DMEM/F12 medium with 10% FBS and 1% glutamine (2 mM) (Thermo Fisher Scientific, Cat# 21051024). When the rate of cell fusion reached 80%, the cells were shaken at 250 rpm for 18 hr to purify for astrocytes. The astrocytes were maintained in DMEM/F12 with 10% FBS and placed in the incubator at 37°C with 5% carbon dioxide, and the medium was changed every 3 days.
2.6. Reconstructed blood–brain barrier in vitro
The blood–brain barrier is mainly composed of astrocytes and BMECs (Greenberg & Jin, 2005). The reconstruction procession (Figure 6a) for BBB in vitro as we previously reported (Tian et al., 2016). Astrocytes (5 × 105 cells/cm2) were plated on the exterior of poly‐L‐lysine‐coated inserts (polyethylene terephthalate membrane, 1.0 μm, Millipore, Germany) and cultured at 37°C with 5% carbon dioxide for 5 hr. Then, the inserts were placed into matching wells and cultured for 24 hr. Next, BMECs (5.0 × 105 cells/cm2) were plated on the inner side of the insert membrane coated with Matrigel matrix (BD, Cat# 354234). BMECs and astrocytes were cocultured for 72 hr to establish the BBB in vitro.
Figure 6.
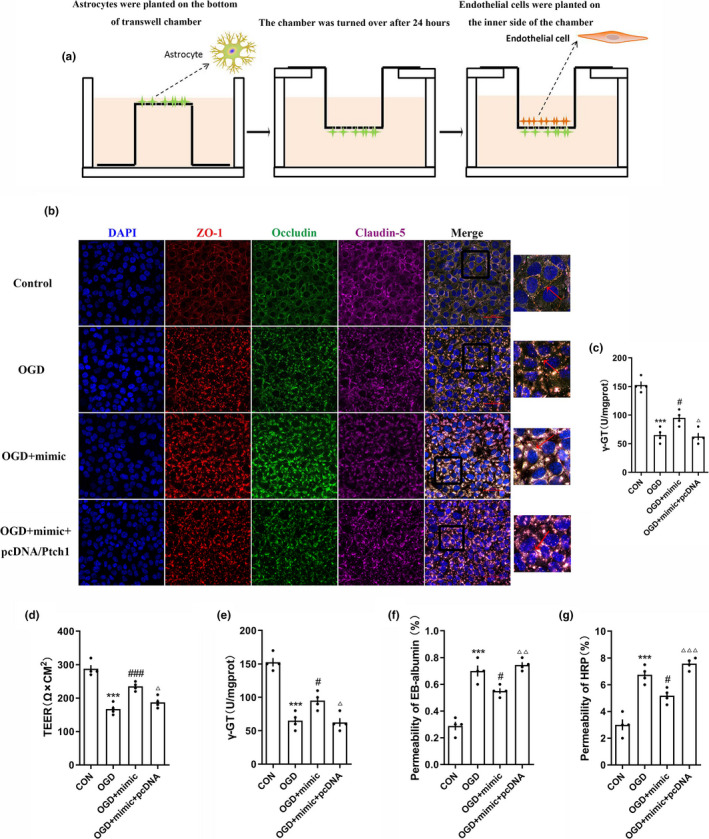
Over‐expression of miRNA‐9‐5p improved the stability of blood–brain barrier in vitro. (a) Reconstruction of the BBB model in vitro. (b) The immunofluorescence and (c) quantitative data of tight junction structure between BMECs (Bar = 50 μm). The quantitative data of miRNA‐9‐5p mimic on (d) TEER, (e) γ‐GT activity, (f) Evans blue, and (g) HRP permeability at 12 hr post‐injury induced by OGD (n = 4/group). ( ***p < .001 vs. the untreated control group; # p < .05, ### p < .001 vs. the OGD group.) n: number of independent cell culture preparations in each group; CON: untreated control group; BBB: blood–brain barrier; OGD: oxygen–glucose deprivation
2.7. Oxygen–glucose deprivation
The oxygen–glucose deprivation (OGD) model was established to stimulate the injured state in vivo (Tian et al., 2016; Zhong et al., 2017). We constructed the BBB in vitro and replaced the cell medium with glucose‐free medium (Gibco, Cat# 11966025) without serum. Then, the plate was transferred into the incubator and cultivated with 94% N2, 5% CO2, and 1% O2 at 37°C for 6 hr. The BMECs in the BBB model and the medium in the plate were collected for testing.
2.8. Mimic and inhibitor transfection in vitro
The concentration of miRNA‐9‐5p mimic (or inhibitor) (RiboBio) in the cell culture medium was 50 nM (or 100 nM). The specific method was according to the instruction: 2.5 μL stock solutions of the miRNA mimic or inhibitor were diluted and gently blended with 50 μL RiboFect™ CP buffer (RiboBio, Guangzhou, China) after the diluent was incubated at 25°C for 5 min. Five μL RiboFectTM CP reagent was added to the diluent, and the mixture was incubated at 25°C for 15 min. Then, the 7.5 μL mixture was blended with 442.5 μL cell culture medium, and the transfection of mimic or inhibitor was performed at 37°C for 24 hr.
2.9. Plasmid and siRNA transfection in vitro
We used plasmid and siRNA to regulate the expression of Ptch‐1 in BMECs. Transfection of plasmid (pcDNA3.1‐Ptch1) (GenePharma) or siRNA (siRNA‐Ptch1) (GenePharma) began when the fusion rate of BMECs reached 80% on 24‐well cell culture plate according to the manufacturer's instructions.
For the transfection of plasmids, 2 μg plasmid was dissolved in 200 μL (50 μL/well × 4well) DMEM/F12 without serum in 1.5 ml tube and 1.6 μl Lipofectamin 2000 was diluted with 200 μL (50 μL/well × 4well) DMEM/F12 without serum in another tube. Both the mixtures were incubated at 25 ℃ for 5 min and then were mixed gently. The final mixtures were also incubated at 25℃ for 20 min. pcDNA/lipofectamine mixtures of 100 μL and 900 μL medium without serum were added to each well. For siRNA‐Ptch1, 20 pmol (0.8 g) siRNA was dissolved in 50 μL DMEM/F12 serum‐free medium, and 1 μL lipofectamin 2000 was also diluted with 50 μL DMEM/F12 without serum. Both the mixtures were incubated at 25℃ for 5 min. Then, the diluted siRNA was mixed with RNAi‐Mate reagent and gently mixed. A total volume of 100 μL mixtures were incubated at 25℃ for 20 min to form the siRNA/lipofectamine compound. siRNA/lipofectamine compounds of 100 μL were added to 24‐well cell culture plate containing BMECs and medium. The BMECs were incubated in 5% CO2 in an incubator at 37°C for 24 hr and then collected for further study.
2.10. Drug treatment
In this study, 0.1 μM GANT61 (Selleck, Cat# S8075), which is an inhibitor of Gli (Lauth, Bergstrom, Shimokawa, & Toftgard, 2007), was used to inhibit the Hedgehog pathway. The drug was dissolved in cell culture medium with 0.1% dimethyl sulfoxide (Sigma‐Aldrich, Cat# D2650). The BMECs were randomly separated into three groups: (1) the untreated control group, which was incubated with cell medium; (2) the mimic group, which was incubated with 50 nM miRNA‐9‐5p mimic; and (3) the M + G group, which was incubated with 50 nM miRNA‐9‐5p mimic and 0.1 μM GANT61. All groups were incubated in both normal and OGD state for 24 hr before testing.
In addition, 200nM SC75741 (Selleck, Cat# S7273) was also used to inhibit the activation of NF‐κB in BMECs (Zhong et al., 2020). The BMECs were randomly separated into four groups: (1) the untreated control group; (2) the mimic group, which was incubated with 50 nM miRNA‐9‐5p mimic; (3) the inhibitor group, which was incubated with 100 nM miRNA‐9‐5p inhibitor; and (4) the inhibitor + SC group, which was incubated with 100 nM miRNA‐9‐5p inhibitor and 200 nM SC75741. All cells were incubated at 37°C with 5% carbon dioxide for 24 hr before testing.
2.11. The BBB integrity assessment in vivo
BBB integrity in vivo was evaluated by measuring the change in brain water content and evans blue content in the injured cerebral hemisphere (Cao et al., 2016; Si et al., 2014). The brain water content was measured by the dry‐wet weight method. The rats were killed without perfusion at 3 days after TBI. The brain was divided into the left and right hemispheres, and the olfactory bulb and cerebellum were removed. The injured hemispheres were weighed and then placed in the oven at 100°C. After 48h, the dried brains were removed and weighed again. The brain water content was calculated as (wet weight ‐ dry weight)/wet weight × 100%.
The optical density (OD) of evans blue (OD/g) in brain tissue could also reflect the BBB integrity. 2% Evans blue solution (Sigma‐Aldrich, Cat# E2129) was injected via the tail vein with 50 µg/g. The rats were anesthetized and perfused from the left heart ventricle with PBS (Sigma‐Aldrich, Cat# P7059) at 3 hr after injection. The injured hemispheres were ground with PBS and 60% trichloroacetic acid (Sigma‐Aldrich, Cat# 8223420250). Then, the mixture was blended and centrifuged at 18,000 g, 4°C for 10 min. The supernatant absorbance was detected at 620 nm using microplate reader (Berthold, Cat#LB940).
2.12. The BBB integrity assessment in vitro
Transendothelial electrical resistance (TEER) values, γ‐GT activity, the permeability of HRP, and Evans blue were detected to evaluate the integrity of BBB in vitro as we previously reported (Tian et al., 2016). TEER values were obtained using the epithelial‐volt‐ohm resistance meter (Millipore, Cat# ERS‐2), and the resistance value (5Ω × cm2) of empty filter was subtracted from each measurement. For the assessment of γ‐GT activity, BMECs were scraped from the insert membrane, suspended in PBS, and centrifuged. The precipitates were dissociated with 1% Triton X‐100 and centrifuged again. The γ‐GT activity in the supernatant was determined with a γ‐GT kit (Jiancheng Company, Cat# C017‐2). For the HRP test, the upper insert was filled with fresh medium containing 50 μg/mL HRP (Sigma‐Aldrich, Cat# P8375). After incubation for 2 hr, O‐phenylenediamine was added to each group, and the reaction was terminated by adding 50 μL H2SO4. The absorbance was determined using microplate reader. For the Evans blue test, the culture medium in the upper insert was replaced with 200 μL DMEM‐F12 medium containing 165 μg/mL Evans Blue and 0.1% bovine serum albumin. After 20 min, the samples were removed from the plates and transferred to 96‐well plates, and the absorbance was also determined with microplate reader.
2.13. The prediction of target gene and dual luciferase reporter assay
The database of TargetScan (RRID:SCR_010845), PicTar (RRID:SCR_00334), and miRbase (RRID:SCR_003152) was used to predict the candidate target gene of miRNA‐9‐5p. The HEK293T cells (1 × 105) (ATCC Cat# CRL‐3216, RRID: CVCL_0063), a cell line which is not listed as a commonly misidentified cell line by the International Cell Line Authentication Committee, were cultured in 24‐well plates and a maximum of eight cell passages was used. Then, all the cells were transfected with Ptch1‐3′UTR wild‐type (wt) or Ptch1‐3′‐untranslated region (UTR) mutant (mt). The miRNA‐9‐5p mimic or miRNA‐9‐5p‐NC was transfected in HEK293 cells by Lipofectamine™ 2000 (GenePharma, Cat# 11668). The luciferase activity was measured with the Dual Luciferase Reporter Assay System (Promega) at 24 hr after transfection. The results were normalized to Renilla activity. All experimental steps were performed according to the manufacturer's instruction.
2.14. RNA isolation and real‐time quantitative PCR
Total RNA was extracted from the injured brain tissue by using the TRIzol reagent (Thermo Fisher Scientific Cat# 15596018) according to the manufacturer's instruction. Total RNA concentration was tested using the Nanodrop spectrophotometer (Nanodrop 2000, Thermo Fisher Scientific) and RNA integrity was assessed through gel electrophoresis. The total RNA was reverse transcribed to determine the expression of miRNA‐9‐5p, and the cDNA was mixed with specific Bulge‐Loop™ miRNA Primer (Ribobio) and miDETECT A Track miRNA qRT‐PCR Starter Kit (Ribobio). The level of U6 RNA was used as an endogenous control for normalization. Quantitative real‐time PCR was performed using the ABI PRISM 7500 Real‐Time PCR instrument (Applied Biosystems). All the steps were performed according to the manufacturer's instruction. The relative changes in miRNA were measured using the comparative threshold cycle (Ct) method and 2−ΔΔCt as previously described (Zhong et al., 2017).
2.15. Western blot analysis
There were five independent tissue samples and four independent cell culture preparations in each group, which were homogenized with RIPA lysis buffer (Beyotime, China) containing protease and phosphatase inhibitors. The debris was eliminated by centrifugation with 7200 g at 4°C for 15 min. The protein levels in the supernate were quantified by using the bicinchoninic acid (BCA) protein assay kit (Beyotime). Then, the supernate was mixed with 5X sodium dodecyl sulfate (SDS) loading buffer (Beyotime) and boiled at 100°C for 5 min. The Western blot test was performed as we previously described (He et al., 2018; Zhong et al., 2017). The primary antibodies included rabbit anti‐Ptch‐1 (1:1,000, Abcam Cat# ab53715, RRID:AB_882208), mouse anti‐Gli‐1 (1:1,000, Proteintech Cat# 66905‐1‐Ig), rabbit anti‐AKT (1:1,000, Cell Signaling Technology Cat# 4,691, RRID:AB_915783), rabbit anti‐Phospho‐AKT (1:1,000, Cell Signaling Technology Cat# 86758, RRID:AB_2800089), rabbit anti‐GSK3β (1:1,000, Cell Signaling Technology Cat# 12456, RRID:AB_2636978), rabbit anti‐Phospho‐GSK3β (1:1,000, Cell Signaling Technology Cat# 5558, RRID:AB_10013750), rabbit anti‐ZO‐1 (1:1,000, Proteintech Cat# 21773‐1‐AP), rabbit anti‐Occludin (1:1,000, Proteintech Cat# 27260‐1‐AP), mouse anti‐Claudin‐5 (1:1,000, Thermo Fisher Scientific Cat# 35‐2500, RRID:AB_2533200), rabbit anti‐NF‐κB p65 (1:1,000, Cell Signaling Technology Cat# 8,242, RRID:AB_10859369), rabbit anti‐MMP‐9 (1:1,000, Cell Signaling Technology Cat# 27,647, RRID:AB_2798945), rabbit anti‐Bcl‐2 (1:1,000, Proteintech Cat# 50599‐2‐Ig), rabbit anti‐Bax (1:1,000, Proteintech Cat# 50599‐2‐Ig), rabbit anti‐Cleaved Caspase‐3 (1:1,000, Proteintech Cat# 19677‐1‐AP), and rabbit anti‐GAPDH (1:1,000, Proteintech Cat# 10494‐1‐AP). Horseradish peroxidase‐conjugated goat anti‐rabbit (1:3,000, Proteintech Cat# SA00001‐15) or goat anti‐mouse IgG (1:3,000, Proteintech Cat# SA00001‐1) was used as the secondary antibody. The expression of glycerinaldehyde‐3‐phosphate‐dehydrogenase was used as the endogenous control. The immunoreactive bands were visualized by using the enhanced chemiluminescence (ECL) kit (Bio‐Rad, Shanghai, China), and the density of the band was quantified and analyzed using the Fusion‐FX7 system (Vilber Lourmat, France).
2.16. Immunofluorescence staining
The tight junction structure in vitro was tested by immunofluorescence (DeStefano, Xu, Williams, Yimam, & Searson, 2017; Katt, Linville, Mayo, Xu, & Searson, 2018). The primary antibody was goat anti‐ZO‐1 (1:500, Abcam Cat# ab190085), rabbit anti‐occludin (1:500, Thermo Fisher Scientific Cat# 33‐1500, RRID:AB_2533101), and mouse anti‐claudin‐5 (1:500, Thermo Fisher Scientific Cat# 35‐2500, RRID:AB_2533200). The secondary antibody was donkey anti‐goat Alexa Fluor® 555 (1:100, Thermo Fisher Scientific Cat# A32816, RRID:AB_2762839), donkey anti‐rabbit Alexa Fluor® 488 (1:100, Thermo Fisher Scientific Cat# A32790, RRID:AB_2762833), and donkey anti‐mouse Alexa Fluor® 647 (1:100, Thermo Fisher Scientific Cat# A32787, RRID:AB_2762830).
The expression of Ptch‐1 and Gli‐1 in BMECs was also detected by immunofluorescence. The primary antibody contained rabbit anti‐Ptch‐1 (1:1,000, Abcam Cat# ab53715, RRID: AB_882208) and mouse anti‐Gli‐1 (1:1,000, Proteintech Cat# 66905‐1‐Ig). Goat anti‐rabbit Alexa Fluor® 594 (1:100, Thermo Fisher Scientific Cat# A32740, RRID: AB_2762824) and goat anti‐mouse Alexa Fluor® 488 (1:100, Thermo Fisher Scientific Cat# A32723, RRID: AB_2633275) were used as the secondary antibody. The nucleus was stained with 4',6‐diamidino‐2‐phenylindole (DAPI, Thermo Fisher Scientific Cat# D3571, RRID: AB_2307445). All images were captured by fluorescence microscope (Eclipse Ti‐S, Nikon, Japan).
2.17. Enzyme‐linked immunosorbent assay
The contents of IL‐1β (RD, Cat# PRLB00), IL‐6 (RD, Cat# PR6000B), TNF‐α (RD, Cat# PRTA00), and MMP‐9 (BOSTER, Cat# EK1463) in different cell culture supernatants were tested by enzyme‐linked immunosorbent assay (ELISA) according to the manufacturer's instructions. All data were calculated as percentages relative to the untreated control group.
2.18. Statistical analysis
All statistical analyses were performed with SPSS software 19.0 (RRID:SCR_002865) and statistical figures were made using GraphPad Prism 8.00 software (RRID:SCR_002798). Values were expressed as mean ± standard error of mean (SEM). No outliers were excluded from the analysis. The Grubbs’ test was used to search for outliers among the analyses data. The Shapiro–Wilk test was applied to test for normality, and the data were normally distributed. Student's t‐test was used to compare two groups. Comparisons among multiple groups were calculated by one‐way or two‐way analysis of variance (ANOVA) using Tukey's HSD post hoc test and Holm‐Bonferroni correction. The differences were considered statistically significant at p < .05.
3. RESULTS
3.1. Alteration of miRNA‐9‐5p in the traumatic foci after TBI and up‐regulation of miRNA‐9‐5p improved the neurological outcome
We established the control cortex injury model in rats and detected the level of miRNA‐9‐5p in traumatic foci at different time points from 0 hr to 14 days post‐injury. The results showed that the level of miRNA‐9‐5p was continuously increased and reached the peak at days 14 after brain injury (Figure 2a). We also found that the level of miRNA‐9‐5p was significantly increased (or decreased) in the CCI + agomir (or CCI + antagomir) group at different time points compared with the CCI group after brain injury (Figure 2a).
Figure 2.
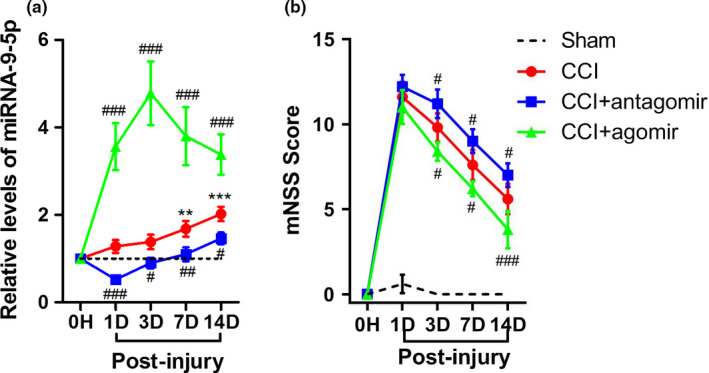
Altered miRNA‐9‐5p level in the injured brain tissues after TBI and intervention with miRNA‐9‐5p oligomers. (a) The temporal profile (from 1 day to 14 days post‐CCI) of miRNA‐9‐5p level in the traumatic foci was determined by qRT‐PCR. The quantitative data were analyzed using the 2−△△Ct method and the level of miRNA‐9‐5p in sham group was used as control. (n = 5/group) (b) The neurological outcome treated with miRNA‐9‐5p oligomers was evaluated by mNSS (n = 5/group). (**p < .01, ***p < .001 vs. the Sham group; # p < .05, ## p < .01, ### p < .001 vs. the CCI group.) n: number of rats in each group; CCI: control cortex impact
The neurological outcome of injured rats was evaluated using mNSS and the results showed that the mNSS score was decreased (or increased) in the CCI + agomir (or CCI + antagomir) group compared with the CCI group (Figure 2b). These data demonstrated that up‐regulation of miRNA‐9‐5p in brain is beneficial to improve neurological function after TBI.
3.2. miRNA‐9‐5p agomir inhibited the apoptosis and inflammation in rats after TBI
We detected the apoptosis‐related molecules and inflammatory cytokines in rats by western blotting and ELISA on day 3 post‐CCI to explore the effect of miRNA‐9‐5p on TBI. We found the expressions of apoptosis‐related molecules including Bcl‐2, Bax, and cleaved caspase 3 were altered by miRNA‐9‐5p agomir or antagomir (Figure 3a). Compared with the CCI group, the expression of Bcl‐2 was increased in the CCI + agomir group (Figure 3b) and decreased in the CCI + antagomir group, while reverse changes were observed on Bax (Figure 3c) and cleaved caspase‐3 (Figure 3d). We also observed that the levels of MMP‐9 (Figure 3e) and inflammatory cytokines including IL‐1β (Figure 3f), IL‐6 (Figure 3g), and TNF‐α (Figure 3h) were decreased in the CCI + agomir group or increased in the CCI + antagomir group compared with the CCI group.
Figure 3.
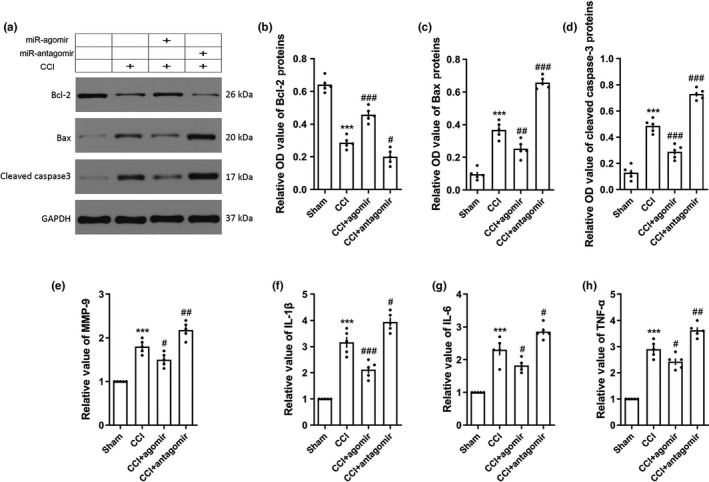
Over‐expression of miRNA‐9‐5p inhibited cellular apoptosis and inflammation after TBI. (a) The immunoblotting and quantitative data of apoptosis‐related molecules (b) Bcl‐2, (c) Bax, and (d) cleaved Caspase 3 acquired from the traumatic foci at 3‐day post‐CCI (n = 5/group). The level of (e) MMP‐9 and inflammatory cytokines including (f) IL‐1β, (g) IL‐6, and (h) TNF‐α was detected by ELISA at 3‐day post‐CCI (n = 5/group). (***p < .001 vs. the Sham group; # p < .05, ## p < .01, ### p < .001 vs. the CCI group.) n: number of rats in each group; CCI: control cortex impact
3.3. miRNA‐9‐5p agomir alleviated brain edema in rats after TBI
The effect of miRNA‐9‐5p on brain edema in rats was assessed by measuring the brain water content and Evans blue permeability. The brain water content (Figure 4a) and Evans blue permeability (Figure 4b) were significantly increased after TBI. However, compared with the CCI group, the brain water content (Figure 4a) and Evans blue permeability (Figure 4b) on day 3 post‐injury were decreased in the CCI + agomir group or increased in the CCI + antagomir group. The expressions of tight junction proteins (TJs) including ZO‐1, Occludin, and Claudin‐5 in the traumatic foci were quantified by western blotting. The results showed that the expression of ZO‐1, Occludin, and Claudin‐5 was decreased after brain injury (Figure 4c). We also found the expressions of TJs were increased (or decreased) in the CCI + agomir (or CCI + antagmir) group compared with the CCI group (Figure 4c).
Figure 4.
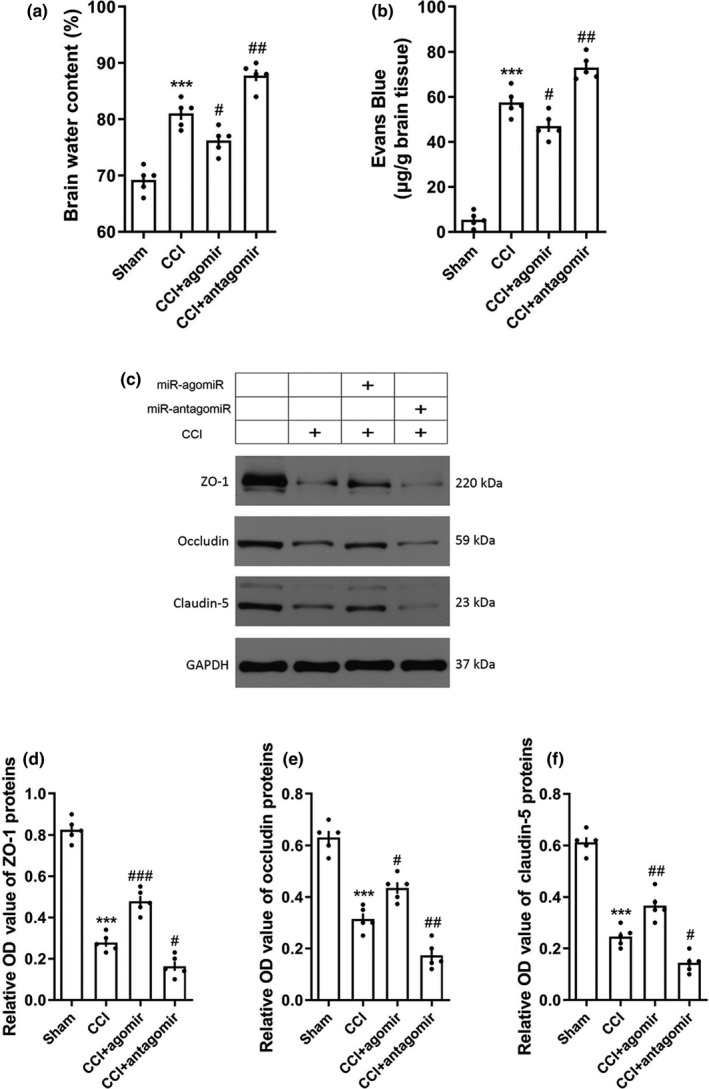
Over‐expression of miRNA‐9‐5p alleviated the destruction of blood–brain barrier after TBI. (a) The brain water content and (b) Evans blue permeability were detected to evaluate the brain edema at 3‐day post‐CCI (n = 5/group). (c) The immunoblotting and quantitative data of tight junction proteins (d) ZO‐1, (e) Occludin, and (f) Claudin‐5 acquired from traumatic foci at 3‐day post‐CCI (n = 5/group). (*** p < .001 vs. the Sham group; # p < .05, ## p < .01, ### p < .001 vs. the CCI group.) n: number of rats in each group; TBI: traumatic brain injury; CCI: control cortex impact
3.4. miRNA‐9‐5p targeted the 3’UTR of Ptch‐1 mRNA
We used bioinformatic database to explore the mechanism of miRNA‐9‐5p on TBI. We found that the Ptch‐1 was the potential target of miRNA‐9‐5p and the results of luciferase reporter assay showed that the miRNA‐9‐5p mimic decreased the luciferase activity in the WT group (Figure 5a), which certified that miRNA‐9‐5p targeted the 3’UTR of Ptch‐1 mRNA. Next, we also found miRNA‐9‐5p mimic decreased the Ptch‐1 and increased the gli‐1 in endothelial cells by immunoblotting (Figure 5b and c) and immunofluorescence (Figure 5d and e).
Figure 5.
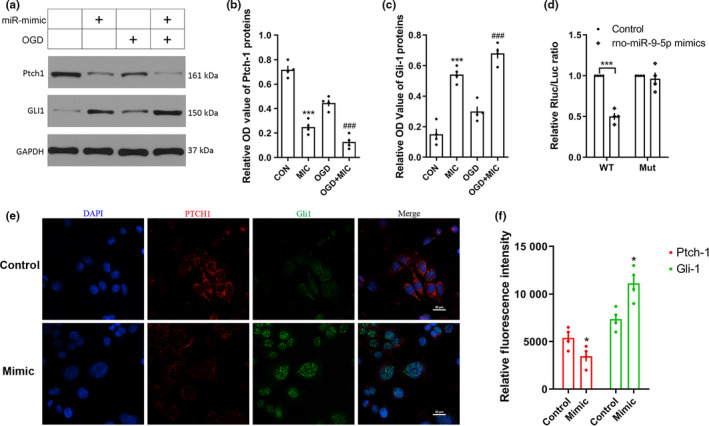
miRNA‐9‐5p targeted 3’UTR of ptch‐1and promoted the expression of Gli‐1. (a) The immunoblotting and quantitative data of (b) Ptch‐1 and (c) Gli‐1 acquired from BMECs. (d) The dual luciferase reporter assay. (e) The immunofluorescence and (f) quantitative data of Ptch‐1 and Gli‐1 in BMECs (Bar = 20 μm). (n = 4/group). (*p < .05, ***p < .001 vs. the untreated control group; ### p < .001 vs. the OGD group.) n: number of independent cell culture preparations in each group; CON: untreated control group; MIC: mimic; OGD: oxygen–glucose deprivation; WT: wild‐type; Mut: mutant type
3.5. miRNA‐9‐5p mimic improved BBB stability by inhibiting Ptch‐1 in vitro
To explore the protective role of miRNA‐9‐5p on BBB, the BMECs and astrocytes were cocultured to establish the BBB model in vitro (Figure 6a). We detected the tight junction structure between endothelial cells by immunofluorescence. The tight junction structure was colocated by ZO‐1, Occludin, and Claudin 5. We observed the completely and continuously intercellular connection structure in the untreated control group, while the structures were generally destroyed in the OGD group. Compared with the OGD group, the tight junction structures were partially retained in the OGD + mimic group (Figure 6b), we also found that the transcellular resistance (Figure 6c) and γ‐GT expression (Figure 6d) were decreased, and the Evans blue permeability (Figure 6e) and HRP permeability (Figure 6f) were increased in the OGD + mimic group. However, up‐regulated the Ptch‐1 by pcDNA reversed the effect of mimic, which indicated that miRNA‐9‐5p mimic improved BBB stability by inhibiting Ptch‐1.
3.6. miRNA‐9‐5p mimic inhibited cellular apoptosis and inflammation by inhibiting Ptch‐1 in vitro
To explore the protective role of miRNA‐9‐5p on cellular apoptosis and inflammation, we detected the apoptosis‐relatived protein including Bcl‐2, Bax in BMECs by western blotting (Figure 7a), and inflammatory cytokines including IL‐1β, IL‐6, and TNF‐α in cell medium by ELSAs. Compared with the OGD group, we found that the level of Bcl‐2 (Figure 7b) in BMECs was increased in the OGD + mimic group, whereas the Bax (Figure 7c) and cleaved caspase 3 (Figure 7d) were decreased. The results also showed that the levels of inflammatory cytokines including IL‐1β (Figure 7e), IL‐6 (Figure 7f), TNF‐α (Figure 7g), and MMP‐9 (Figure 7h) in cell medium were decreased in the OGD + mimic group compared with the OGD group. However, up‐regulated the Ptch‐1 by pcDNA reversed the effect of miRNA‐9‐5p mimic on cellular apoptosis and inflammation.
Figure 7.
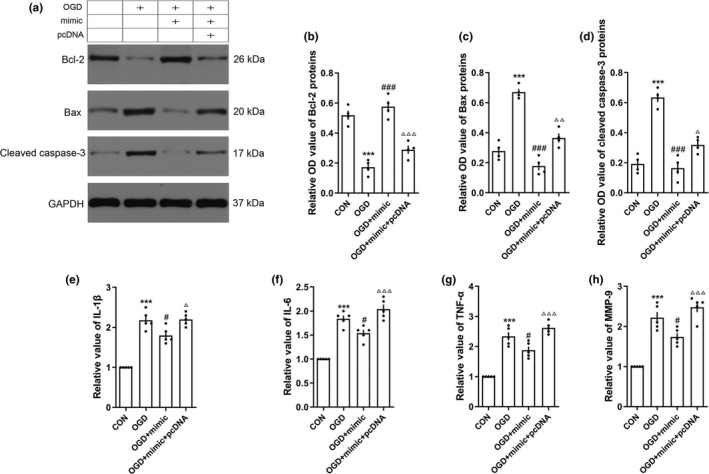
Over‐expression of miRNA‐9‐5p inhibited cellular apoptosis and inflammation in vitro. (a) The immunoblotting and quantitative data of (b) Bcl‐2, (c) Bax, and (d) cleaved Caspase‐3 acquired from BMECs. The level of (e) IL‐1β, (f) IL‐6, and (g) TNF‐α and (h) MMP‐9 was detected by ELISA in cell medium (n = 4/group). (***p < .001 vs. the untreated control group; # p < .05, ### p < .001 vs. the OGD group.) n: number of independent cell culture preparations in each group; CON: untreated control group; OGD: oxygen–glucose deprivation
3.7. miRNA‐9‐5p mimic promoted AKT/GSK3β pathway by activating Hedgehog pathway
As presented in Figure S1, we verified that the activation of Hedgehog pathway promoted the AKT/GSK3β pathway. To further reveal the protective mechanism of miRNA‐9‐5p on OGD‐induced cell damage, the expression changing in AKT/GSK3β pathway was detected in BMECs (Figure 8a). The results showed that the expression of Ptch‐1 (Figure 8b) in BMECs was decreased in both untreated and OGD condition, while the Gli‐1 (Figure 8c) was increased. We also found that the expression of p‐AKT (Figure 8d) and p‐GSK3β (Figure 8f) was significantly increased accompanied by the increasing of Gli‐1 treated with miRNA‐9‐5p mimic in both untreated and OGD condition. But the statistically significant difference on AKT (Figure 8e) and GSK3β (Figure 8g) levels was not observed. The expression of Bcl‐2 (Figure 8h) was also increased, while the Bax (Figure 8i) and cleaved caspase 3 (Figure 8j) were decreased treated with miRNA‐9‐5p mimic in both untreated and OGD condition. However, the effect of miRNA‐9‐5p mimic on promoting AKT/GSK3β pathway can be reversed by GANT61, which is an inhibitor of Hedgehog pathway. These data indicated that miRNA‐9‐5p mimic promoted the AKT/GSK3β pathway mainly by activating Hedgehog pathway.
Figure 8.
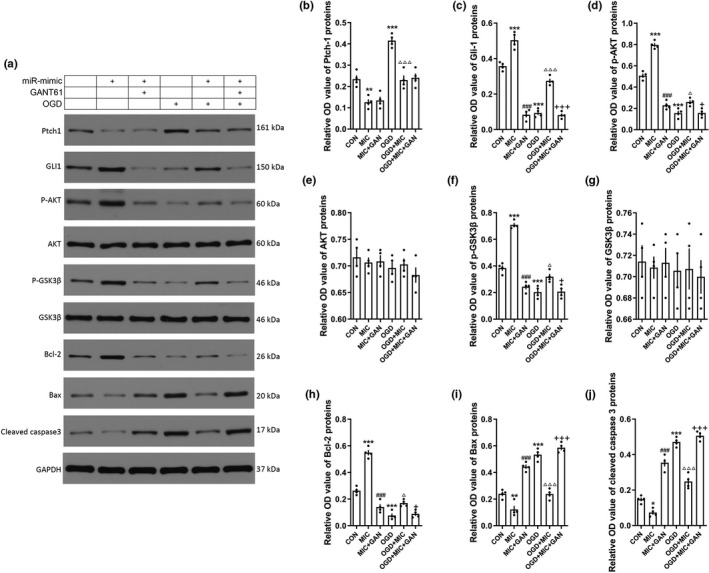
miRNA‐9‐5p mimic promoted AKT/GSK3β pathway by activating Hedgehog pathway. (a) The immunoblotting and quantitative data of (b) Ptch‐1, (c) Gli‐1, (d) p‐AKT, (e) AKT, (f) p‐GSK3β, (g) GSK3β, (h) Bcl‐2, (i) Bax, and (j) cleaved caspase 3 in BMECs treated with GANT61. (n = 4/group). (*p < .05, **p < .01, ***p < .001 vs. the untreated control group; ### p < .001 vs. the MIC group; △ p < .05,△△△ p < .001 vs. the OGD group; +p < .05,+++p < .001 vs. the OGD + MIC group.) n: number of independent cell culture preparations in each group; CON: untreated control group; OGD: oxygen–glucose deprivation; MIC: mimic; GAN: GANT61
3.8. miRNA‐9‐5p mimic regulated NF‐κB/MMP‐9 pathway
According to the results of Figure S1, we also found that activation of Hedgehog pathway promoted the NF‐κB/MMP‐9 pathway. However, the results of the miRNA‐9‐5p mimic on NF‐κB/MMP‐9 (Figure 9a) in BMECs were contradictory to the supplement which led to expression of NF‐κB (Figure 9b); and MMP‐9 (Figure 9c) was decreased when treated with the mimic in both untreated and OGD condition. These results indicate that miRNA‐9‐5p may directly inhibit the NF‐κB/MMP‐9 pathway. To further explore the effect of miRNA‐9‐5p on NF‐κB/MMP‐9 pathway, the miRNA‐9‐5p inhibitor was used to reduce the level of miRNA‐9‐5p and SC75741 was used to inhibit the expression of NF‐κB (Figure 9d). The results showed that the mimic inhibited the expression of NF‐κB (Figure 9e) and MMP‐9 (Figure 9f), whereas the inhibitor promoted it. The effect of miRNA‐9‐5p inhibitor on MMP‐9 can be reversed by SC75741. These data indicated that miRNA‐9‐5p mimic can directly regulate the NF‐κB/MMP‐9 pathway.
Figure 9.
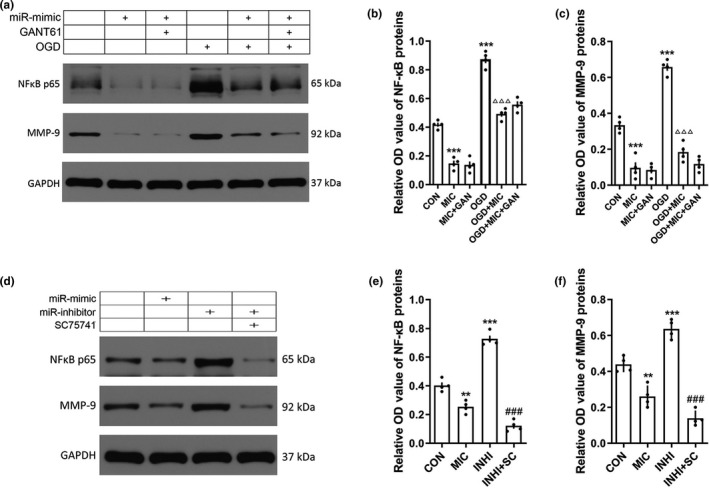
miRNA‐9‐5p mimic inhibited NF‐κB/MMP‐9 pathway. (a) The immunoblotting and quantitative data of (b) NF‐κB and (C)MMP‐9 in BMECs treated with GANT61. (**p < .01, ***p < .001 vs. the untreated control group; ### p < .001 vs. the MIC group; △△△ p < .001 vs. the OGD group.) (d)The immunoblotting and quantitative data of (e) NF‐κB and (f) MMP‐9 in BMECs treated with QNZ. (n = 4/group) (**p < .01, ***p < .001 vs. the untreated control group; ###p < .001 vs. the INHI group.) n: number of independent cell culture preparations in each group; CON: untreated control group; OGD: oxygen–glucose deprivation; MIC: mimic; GAN: GANT61; INHI: inhibitor
4. DISCUSSION
The BBB is a highly sealed barrier between endothelial cells that regulates the transportation of proteins, nutrients, and molecules from blood to brain, and metabolic waste products from the brain into circulation (Xu et al., 2019). Extensive BBB damage can lead to whole‐brain edema, leading to a sustained increase in intracranial pressure, which can lead to death (Zhang et al., 2019). The breakdown of the BBB is one of the main reasons for the poor prognosis of TBI. At present, the clinical assessment for BBB mainly depends on the clinical manifestation and visual examination through imaging techniques. Sometimes these subjective examination results do not reflect severity and prognosis of TBI in a timely and accurate way. Especially in patients with mild and moderate brain trauma, the imaging results are often inconsistent with the clinical manifestation.
In fact, studies of protein biomarkers as a tool for diagnosing, monitoring, and predicting the prognosis of TBI have increased markedly over the past decade (Papa et al., 2013). GFAP and UCH‐L1 have proven utility for discerning adult patients with TBI who may benefit from neuroimaging (Papa et al., 2016; Posti et al., 2016). However, the protein biomarkers from cerebral spinal fluid (CSF) into blood may be subject to degradation by endogenous proteases, which limit their utility to acute and subacute periods of TBI. In addition, many protein biomarkers are released following damage by myocytes and osteocytes, which could limit their specificity in patients with polytrauma (Hergenroeder, Redell, Moore, & Dash, 2008). Therefore, few biological measures can be used for personalized BBB assessment. The lack of sensitive, objective assessment indicator hinders the ability of physicians to provide accurate diagnoses and effective treatments for TBI patients.
The central nervous system contains the highest concentration and highest diversity of miRNAs (Petri, Malmevik, Fasching, Akerblom, & Jakobsson, 2014; Sengupta & Kar, 2018; Sun et al., 2014). Almost 70% miRNAs are expressed in brain, spinal cord, or peripheral nerves. Growing evidence has reported that miRNAs are essential mediators in TBI. MiRNA‐144 promoted cognitive impairments induced by β‐amyloid accumulation post‐TBI through suppressing of ADAM10 expression (Cheng et al., 2013). Sabirzhanov et al. also found that down‐regulation of miRNA‐711 protected neurons against TBI‐induced cell death (Sabirzhanov et al., 2016). Studies have found that miRNA can freely pass through the BBB in the exosomes and can be detected in the peripheral blood (Lu & Xu, 2016; Ramirez, Andrews, Paul, & Pachter, 2018; Valadi et al., 2007). Moreover, miRNA has the same physical and chemical properties in fresh and frozen serum and plasma, which provides a feasible basis for the study of miRNA as a blood biomarker. Therefore, it is important to clarify the role of each abnormally expressed miRNA after TBI.
In this study, we established a control cortex impact model in rats and explored the effect of miRNA‐9‐5p on TBI. The results showed that up‐regulation of miRNA‐9‐5p level attenuated the BBB damage and neuroinflammatory response for rats after TBI. The dual luciferase assay showed that Ptch‐1 was the target gene of miRNA‐9‐5p and up‐regulation of miRNA‐9‐5p activated the Hedgehog pathway by inhibiting the expression of Ptch‐1. To further verify the effect of miRNA‐9‐5p on BBB and neuroinflammation, we established a BBB model in vitro by coculturing rat BMECs and astrocytes. The OGD model was used to simulate the pathophysiological state of TBI in vitro. Previous studies have reported that the destruction of the BBB was significantly enhanced after TBI, which may suffer from increasing MMP activity and excessive inflammatory response (Wu et al., 2020). In this study, the results manifested that over‐expression of miRNA‐9‐5p protected the BBB in vitro by decreasing the damage of tight junction structure and reducing the permeability. We also found that miRNA‐9‐5p mimic can reduce the level of MMP‐9 and inflammatory cytokines including IL‐1β, IL‐6, and TNF‐α in cell medium. However, the effect of miRNA‐9‐5p on BBB can be reversed by increasing the expression of Ptch‐1. All these results suggested that miRNA‐9‐5p plays a protective role on BBB mainly by inhibiting Ptch‐1. The structural and functional integrity of endothelial cells is the basis for maintaining the integrity of the BBB. To explore the protective mechanism of miRNA‐9‐5p on BBB, we detected the relative changes in apoptosis and inflammatory pathways in BMECs. Here, we found that miRNA‐9‐5p promoted AKT/GSK3β pathway by activating Hedgehog pathway, which inhibited cellular apoptosis. According to results presented in our supplementary material, the activation of Hedgehog pathway may promote the expression of NF‐κB. However, although over‐expression of miRNA‐9‐5p activated the Hedgehog pathway, the NF‐κB has also been inhibited in BMECs. Previous studies have reported that NF‐κB is also a target gene for miRNA‐9 and miRNA‐9 reduces the expression of NF‐κB by directly acting on 3’UTR of mRNA (Chakraborty, Zawieja, Davis, & Muthuchamy, 2015). These results suggested that the inhibitory effect of miRNA‐9 is stronger than the activation effect of Hedgehog pathway and miRNA‐9 plays the protective role on BBB after TBI through various molecular pathways.
There are still some shortcomings in our research. This study mainly investigated the effect of miR‐9‐5p on the blood–brain barrier and neuroinflammation after TBI. However, the BBB structure was difficult to separate in vivo. The local brain tissue around the traumatic foci, which mixed other cellular components, was unable to accurately express these results. Secondly, we reconstructed the BBB in vitro by coculturing BMECs and astrocytes, but recent studies have shown that the intact function of blood–brain barrier also requires other cell, such as pericytes and microglia. However, the technique of recombination and culture various cells in vitro according to the structure of BBB in vivo is yet immature. Thus, the BBB model we reconstructed in vitro cannot completely mimic the BBB in vivo, which may lead to some limitations to these results. Thirdly, we used OGD model in vitro to stimulate the local state after TBI, which is generally used to simulate the state of ischemia and hypoxia. However, ischemia and hypoxia are only one part of the complex pathological process after TBI. Therefore, OGD might not be the best model in vitro for TBI. In a follow‐up study, we shall collect peripheral blood from patients with brain injury and establish more cell models to further explore the role of miRNA‐9‐5p on the diagnosis and treatment of TBI.
In conclusion, we demonstrated that miRNA‐9‐5p alleviated the destruction of BBB and neuroinflammatory responses by activating Hedgehog pathway and inhibiting NF‐κB/MMP pathway. This study extended our understanding of miRNA‐9‐5p and suggested that miRNA‐9‐5p can be a promising diagnosis and therapeutic target for TBI.
AUTHORSHIP
Jingchuan Wu and Xiaochuan Sun designed the experiments. Jingchuan Wu and Xiaocui Tian performed the experiments in vitro. Jingchuan Wu, Junchi He, Jianjun Zhong, Hongrong Zhang, and Hui Li performed the experiments in vivo. Jingchuan Wu, Yuetao Luo, and Bo Cen analyzed the data. Jingchuan Wu and Tao Jiang wrote and revised the manuscript.
Supporting information
ACKNOWLEDGMENTS AND CONFLICT OF INTEREST DISCLOSURE
We acknowledge the service provided by Chongqing Key Laboratory of Ophthalmology and Laboratory Research Central, the First Affiliated Hospital of Chongqing Medical University. We also acknowledge the assistance provided by Xiaoyun Dou (Institute of Life Sciences, Chongqing Medical University). All experiments were conducted in compliance with the ARRIVE guidelines. All authors confirm that they have no competing interests.
Wu J, He J, Tian X, et al. microRNA‐9‐5p alleviates blood–brain barrier damage and neuroinflammation after traumatic brain injury. J. Neurochem. 2020;153:710–726. 10.1111/jnc.14963
Funding information
This work was supported by grants from the National Natural Science Foundation of China (NO. 81571159), National Natural Science Foundation for Youth of China (No. 81801230), Health Commission of Wuhan Municipality scientific research Funding (No. WX19Q17), Health Commission of Hubei Province scientific research project (No. WJ2019H364), and Capital's Funds for Health Improvement and Research (CFH2018‐2‐2042).
Contributor Information
Tao Jiang, Email: zacharytaojiang@163.com.
Xiaochuan Sun, Email: sunxch1445@qq.com.
REFERENCE
- Amorini, A. M. , Lazzarino, G. , di Pietro, V. , Signoretti, S. , Lazzarino, G. , Belli, A. , & Tavazzi, B. (2016). Metabolic, enzymatic and gene involvement in cerebral glucose dysmetabolism after traumatic brain injury. Biochimica Et Biophysica Acta, 1862, 679–687. [DOI] [PubMed] [Google Scholar]
- An, Y. , Zhao, L. , Wang, T. , Huang, J. , Xiao, W. , Wang, P. , … Chen, X. (2019). Preemptive oxycodone is superior to equal dose of sufentanil to reduce visceral pain and inflammatory markers after surgery: A randomized controlled trail. BMC Anesthesiology, 19, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, F. , Jiang, Y. , Wu, Y. , Zhong, J. , Liu, J. , Qin, X. , … Sun, X. (2016). Apolipoprotein E‐Mimetic COG1410 reduces Acute Vasogenic Edema following traumatic brain injury. Journal of Neurotrauma, 33, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S. , Zawieja, D. C. , Davis, M. J. , & Muthuchamy, M. (2015). MicroRNA signature of inflamed lymphatic endothelium and role of miR‐9 in lymphangiogenesis and inflammation. American Journal of Physiology. Cell Physiology, 309, C680–C692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran, R. , Mehta, S. L. , & Vemuganti, R. (2017). Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochemistry International, 111, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Wang, M. , Yang, H. , Mao, L. , He, Q. , Jin, H. , … Hu, B. (2017). LncRNA TUG1 sponges microRNA‐9 to promote neurons apoptosis by up‐regulated Bcl2l11 under ischemia. Biochemical and Biophysical Research Communications, 485, 167–173. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Zhu, J. , Hang, C. H. , & Wang, Y. H. (2019). The potassium SK channel activator NS309 protects against experimental traumatic brain injury through anti‐inflammatory and immunomodulatory mechanisms. Front Pharmacol, 10, 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C. , Li, W. , Zhang, Z. , Yoshimura, S. , Hao, Q. , Zhang, C. , & Wang, Z. (2013). MicroRNA‐144 is regulated by activator protein‐1 (AP‐1) and decreases expression of Alzheimer disease‐related a disintegrin and metalloprotease 10 (ADAM10). Journal of Biological Chemistry, 288, 13748–13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churov, A. , Summerhill, V. , Grechko, A. , Orekhova, V. , & Orekhov, A. (2019). MicroRNAs as potential biomarkers in atherosclerosis. International Journal of Molecular Sciences, 20, 5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destefano, J. G. , Xu, Z. S. , Williams, A. J. , Yimam, N. , & Searson, P. C. (2017). Effect of shear stress on iPSC‐derived human brain microvascular endothelial cells (dhBMECs). Fluids Barriers CNS, 14, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Pietro, V. , Amorini, A. M. , Tavazzi, B. , Hovda, D. A. , Signoretti, S. , Giza, C. C. , … Belli, A. (2013). Potentially neuroprotective gene modulation in an in vitro model of mild traumatic brain injury. Molecular and Cellular Biochemistry, 375, 185–198. [DOI] [PubMed] [Google Scholar]
- Faul, F. , Erdfelder, E. , Lang, A.‐G. , & Buchne, A. (2007). G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior Research Methods, 39, 175–191. [DOI] [PubMed] [Google Scholar]
- Ge, X. , Huang, S. , Gao, H. , Han, Z. , Chen, F. , Zhang, S. , … Zhang, J. (2016). miR‐21‐5p alleviates leakage of injured brain microvascular endothelial barrier in vitro through suppressing inflammation and apoptosis. Brain Research, 1650, 31–40. [DOI] [PubMed] [Google Scholar]
- Ge, X. T. , Lei, P. , Wang, H. C. , Zhang, A. L. , Han, Z. L. , Chen, X. , … Zhang, J. N. (2014). miR‐21 improves the neurological outcome after traumatic brain injury in rats. Scientific Reports, 4, 6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, D. A. , & Jin, K. (2005). From angiogenesis to neuropathology. Nature, 438, 954–959. [DOI] [PubMed] [Google Scholar]
- He, J. , Liu, H. , Zhong, J. , Guo, Z. , Wu, J. , Zhang, H. , … Cheng, C. (2018). Bexarotene protects against neurotoxicity partially through a PPARgamma‐dependent mechanism in mice following traumatic brain injury. Neurobiology of Diseases, 117, 114–124. [DOI] [PubMed] [Google Scholar]
- Hergenroeder, G. W. , Redell, J. B. , Moore, A. N. , & Dash, P. K. (2008). Biomarkers in the clinical diagnosis and management of traumatic brain injury. Molecular Diagnosis & Therapy, 12, 345–358. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Luo, M. , Ni, N. , Den, Y. , Xia, J. , Chen, J. , … Gu, P. (2014). Reciprocal actions of microRNA‐9 and TLX in the proliferation and differentiation of retinal progenitor cells. Stem Cells and Development, 23, 2771–2781. [DOI] [PubMed] [Google Scholar]
- Jha, R. M. , Kochanek, P. M. , & Simard, J. M. (2019). Pathophysiology and treatment of cerebral edema in traumatic brain injury. Neuropharmacology, 145, 230–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katt, M. E. , Linville, R. M. , Mayo, L. N. , Xu, Z. S. , & Searson, P. C. (2018). Functional brain‐specific microvessels from iPSC‐derived human brain microvascular endothelial cells: The role of matrix composition on monolayer formation. Fluids Barriers CNS, 15, 7 10.1186/s12987-018-0092-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth, M. , Bergstrom, A. , Shimokawa, T. , & Toftgard, R. (2007). Inhibition of GLI‐mediated transcription and tumor cell growth by small‐molecule antagonists. Proceedings of the National Academy of Sciences of the United States of America, 104, 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, P. , Li, Y. , Chen, X. , Yang, S. , & Zhang, J. (2009). Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Research, 1284, 191–201. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Xu, R. , Zhu, X. , Li, Y. , Wang, Y. , & Xu, W. (2020). MicroRNA‐23a‐3p improves traumatic brain injury through modulating the neurological apoptosis and inflammation response in mice. Cell Cycle, 19, 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, Y.-H. , Fang, J. , Zhou, H.-D. , Jiang, H.-F. , & Xie, K.-J. (2019). Sufentanil preconditioning protects against hepatic ischemia-reperfusion injury by suppressing inflammation. Medical Science Monitor : International Medical Journal of Experimental and Clinical Research, 25, 2265–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. , Jiang, Y. , Chung, J. Y. , Li, Y. , Yu, Z. , Kim, J. W. , … Wang, X. (2019). Annexin A2 deficiency exacerbates neuroinflammation and long‐term neurological deficits after traumatic brain injury in mice. International Journal of Molecular Sciences, 20, 6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D. , & Xu, A. D. (2016). Mini review: Circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front Genet, 7, 53 10.3389/fgene.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nampoothiri, S. S. , & Rajanikant, G. K. (2019). miR‐9 Upregulation Integrates Post‐ischemic Neuronal Survival and Regeneration In Vitro. Cellular and Molecular Neurobiology, 39, 223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, S. Y. , & Lee, A. Y. W. (2019). Traumatic brain injuries: Pathophysiology and potential therapeutic targets. Frontiers in Cellular Neuroscience, 13, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, L. , Brophy, G. M. , Welch, R. D. , Lewis, L. M. , Braga, C. F. , Tan, C. N. , … Hack, D. C. (2016). Time course and diagnostic accuracy of glial and neuronal blood biomarkers GFAP and UCH‐L1 in a large cohort of trauma patients with and without mild traumatic brain injury. JAMA Neurology, 73, 551–560. 10.1001/jamaneurol.2016.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, L. , Ramia, M. M. , Kelly, J. M. , Burks, S. S. , Pawlowicz, A. , & Berger, R. P. (2013). Systematic review of clinical research on biomarkers for pediatric traumatic brain injury. Journal of Neurotrauma, 30, 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri, R. , Malmevik, J. , Fasching, L. , Akerblom, M. , & Jakobsson, J. (2014). miRNAs in brain development. Experimental Cell Research, 321, 84–89. [DOI] [PubMed] [Google Scholar]
- Posti, J. P. , Takala, R. S. , Runtti, H. , Newcombe, V. F. , Outtrim, J. , Katila, A. J. , … Tenovuo, O. (2016). The levels of glial fibrillary acidic protein and ubiquitin C‐Terminal Hydrolase‐L1 during the first week after a traumatic brain injury: correlations with clinical and imaging findings. Neurosurgery, 79, 456–464. 10.1227/NEU.0000000000001226 [DOI] [PubMed] [Google Scholar]
- Ramirez, S. H. , Andrews, A. M. , Paul, D. , & Pachter, J. S. (2018). Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS, 15, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanov, B. , Stoica, B. A. , Zhao, Z. , Loane, D. J. , Wu, J. , Dorsey, S. G. , & Faden, A. I. (2016). miR‐711 upregulation induces neuronal cell death after traumatic brain injury. Cell Death and Differentiation, 23, 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, D. , & Kar, S. (2018). Alteration in MicroRNA expression governs the nature and timing of cellular fate commitment. ACS Chemical Neuroscience, 9, 725–737. [DOI] [PubMed] [Google Scholar]
- Si, D. , Li, J. , Liu, J. , Wang, X. , Wei, Z. , Tian, Q. , … Liu, G. (2014). Progesterone protects blood‐brain barrier function and improves neurological outcome following traumatic brain injury in rats. Experimental and Therapeutic Medicine, 8, 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T. Y. , Chen, X. R. , Liu, Z. L. , Zhao, L. L. , Jiang, Y. X. , Qu, G. Q. , … Liu, L. (2014). Expression profiling of microRNAs in hippocampus of rats following traumatic brain injury. Journal of Huazhong University of Science and Technology, 34, 548–553. [DOI] [PubMed] [Google Scholar]
- Tian, X. , Peng, J. , Zhong, J. , Yang, M. , Pang, J. , Lou, J. , … Dong, Z. (2016). beta‐Caryophyllene protects in vitro neurovascular unit against oxygen‐glucose deprivation and re‐oxygenation‐induced injury. Journal of Neurochemistry, 139, 757–768. [DOI] [PubMed] [Google Scholar]
- Topol, A. , Zhu, S. , Hartley, B. J. , English, J. , Hauberg, M. E. , Tran, N. , … Brennand, K. J. (2016). Dysregulation of miRNA‐9 in a subset of schizophrenia patient‐derived neural progenitor cells. Cell Reports, 15, 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekstrom, K. , Bossios, A. , Sjostrand, M. , Lee, J. J. , & Lotvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Wu, M.‐Y. , Gao, F. , Yang, X.‐M. , Qin, X. , Chen, G.‐Z. , Li, D. , … Chen, G. (2020). Matrix metalloproteinase‐9 regulates the blood brain barrier via the hedgehog pathway in a rat model of traumatic brain injury. Brain Research, 1727, 146553. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Nirwane, A. , & Yao, Y. (2019). Basement membrane and blood‐brain barrier. Stroke and Vascular Neurology, 4, 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Gu, Y. , Li, P. , Jiang, A. , Sheng, X. , Jin, X. , … Li, G. (2019). Matrix metalloproteases‐mediated cleavage on beta‐Dystroglycan may play a key role in the blood‐brain barrier after intracerebral hemorrhage in rats. Medical Science Monitor, 25, 794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Pogue, A. I. , & Lukiw, W. J. (2015). MicroRNA (miRNA) Signaling in the human CNS in sporadic Alzheimer's Disease (AD)‐novel and unique pathological features. International Journal of Molecular Sciences, 16, 30105–30116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, J. , Jiang, L. , Huang, Z. , Zhang, H. , Cheng, C. , Liu, H. , … Sun, X. (2017). The long non‐coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain, Behavior, and Immunity, 65, 183–194. [DOI] [PubMed] [Google Scholar]
- Zhong, J. H. , Xiang, X. , Wang, Y. Y. , Liu, X. , Qi, L. N. , Luo, C. P. , … Li, L. Q. (2020). The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. Journal of Cellular Physiology, 235, 1090–1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


