Florfenicol belongs to a class of phenicol antimicrobials widely used as feed additives and for the treatment of respiratory infections. In recent years, increasing resistance to florfenicol has been reported in Campylobacter spp., the leading foodborne enteric pathogens causing diarrheal diseases worldwide. Here, we reported the identification of fexA, a novel mobile florfenicol resistance gene in Campylobacter. Of the 100 Campylobacter jejuni strains isolated from poultry in Zhejiang, China, 9 were shown to be fexA positive, and their whole-genome sequences were further determined by integration of Illumina short-read and MinION long-read sequencing.
KEYWORDS: Campylobacter, fexA, multidrug resistance, food safety
ABSTRACT
Florfenicol belongs to a class of phenicol antimicrobials widely used as feed additives and for the treatment of respiratory infections. In recent years, increasing resistance to florfenicol has been reported in Campylobacter spp., the leading foodborne enteric pathogens causing diarrheal diseases worldwide. Here, we reported the identification of fexA, a novel mobile florfenicol resistance gene in Campylobacter. Of the 100 Campylobacter jejuni strains isolated from poultry in Zhejiang, China, 9 were shown to be fexA positive, and their whole-genome sequences were further determined by integration of Illumina short-read and MinION long-read sequencing. The fexA gene was found in the plasmid of one strain and chromosomes of eight strains, and its location was verified by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting. Based on comparative analysis, the fexA gene was located within a region with the tet(L)-fexA-catA-tet(O) gene arrangement, demonstrated to be successfully transferable among C. jejuni strains. Functional cloning indicated that acquisition of the single fexA gene significantly increased resistance to florfenicol, whereas its inactivation resulted in increased susceptibility to florfenicol in Campylobacter. Taken together, these results indicated that the emerging fexA resistance is horizontally transferable, which might greatly facilitate the adaptation of Campylobacter in food production environments where florfenicols are frequently used.
INTRODUCTION
Florfenicol is a fluorinated thiamphenicol derivative belonging to the broad-spectrum antimicrobial agents exclusively approved for use in veterinary medicine (1). It has been licensed in China for the control of respiratory tract diseases and enteric infections in food-producing animals since 1999 (2). However, the excessive use of florfenicol as an antimicrobial chemotherapeutic agent has resulted in bacterial species acquiring resistance to this agent. Several phenicol resistance-specific genes have been reported for florfenicol-resistant bacteria of animal and human origin, including the fexA, fexB, pexA, and floR phenicol-specific exporter genes, optrA, a ribosomal protection protein gene, the gene encoding RE-CmeABC, a functionally enhanced multidrug efflux pump variant (3–5), and cfr, the multidrug resistance gene encoding a 23S rRNA methyltransferase that confers resistance to phenicols, as well as 4 other structurally unrelated antimicrobial agents (lincosamides, oxazolidinones, pleuromutilins, and streptogramin A) (6).
Campylobacter is the leading cause of bacterial foodborne illnesses worldwide. According to data from the World Health Organization, the estimated incidence of gastroenteritis due to infections by Campylobacter spp. in high-income countries is between 4.4 and 9.3 per 1,000 people (7). Contaminated undercooked poultry meat is the main source of infection for human campylobacteriosis, with ruminant Campylobacter also being a significant contributor to foodborne illnesses. Numerous reports have indicated that Campylobacter has become increasingly resistant to antimicrobial agents used in animals and clinical settings (8–10). As a foodborne pathogen transmitted via foodborne routes, Campylobacter is constantly exposed to antimicrobial agents used for food production. Hence, in dealing with antimicrobial selection, Campylobacter has evolved various mechanisms of resistance to such antimicrobials. Some of the mechanisms have been shown to confer resistance to a specific class of antimicrobials, whereas others might confer multidrug resistance (11). In addition to the previously characterized intrinsic mechanisms in mediating antibiotic resistance, several new antibiotic resistance mechanisms have emerged in Campylobacter in recent years. These mechanisms include the Erm(B) rRNA methylase, mediating macrolide resistance (12–14), RE-CmeABC, a functionally enhanced multidrug efflux pump variant (5), a novel fosXCC gene conferring fosfomycin resistance (15), and the Cfr(C) rRNA methyltransferase, mediating multidrug resistance (16, 17). It should be noted that the presence of RE-cmeABC not only confers resistance to florfenicol but also contributes to the increased resistance to clinically important antimicrobial agents, such as ciprofloxacin and erythromycin (5). In addition, multiple characterized resistance mechanisms could form multidrug resistance genomic islands (MDRGIs), which could confer a fitness advantage under various antimicrobial selections in Campylobacter (10, 12, 17, 18).
During our routine surveillance, we noticed that several Campylobacter isolates showed extremely high levels of resistance to florfenicol. In this study, we aimed to identify and characterize novel florfenicol resistance mechanisms in Campylobacter. A natural transformation assay indicated that florfenicol resistance could be transferred among Campylobacter organisms. Whole-genome sequencing of transformants revealed the presence of the previously uncharacterized fexA florfenicol resistance gene in Campylobacter isolated from poultry. We further analyzed the function and genetic environments of fexA.
RESULTS
Identification of fexA associated with resistance to florfenicol.
Between March 2018 and January 2019, a total of 100 Campylobacter jejuni strains were isolated from 3 poultry farms, 2 slaughterhouses, and 6 supermarkets in Zhejiang Province, China. During our surveillance study regarding Campylobacter from poultry, we noticed that 7 of the C. jejuni isolates exhibited a florfenicol MIC value of ≥64 mg/liter, which was higher than what was previously reported for Campylobacter. To search for the mechanisms mediating high resistance to florfenicol, we performed PCR analysis of the previously characterized florfenicol resistance genes in Campylobacter, namely, cfr(C) and RE-cmeABC. However, these 2 genes were absent in some of our florfenicol-resistant isolates, indicating the presence of a potentially novel mechanism conferring resistance to florfenicol. In addition, natural transformation assays revealed that florfenicol resistance in C. jejuni ZS005 was transferable to C. jejuni NCTC 11168. Specifically, the MIC value for florfenicol was shown to be increased 32-fold in the ZS005NT11168 transformant compared with that of the parent, C. jejuni NCTC 11168 (Table 1). This finding indicated that the performed transformation resulted in the transfer of florfenicol resistance mechanisms from C. jejuni ZS005 to ZS005NT11168. To examine this possibility, we performed whole-genome sequence analysis of ZS005NT11168. Comparative analysis of the draft genome of ZS005NT11168 with the complete genome of NCTC 11168 (accession no. NC_002163) revealed the presence of a 9,982-bp segment containing a 1,428-bp gene with 99.51% (7 single nucleotide polymorphisms [SNPs]) similarity to the fexA gene in Staphylococcus lentus (19), which could encode a phenicol-specific efflux pump conferring resistance to florfenicol (see Fig. S1 in the supplemental material). Beyond this replacement, no other insertions/deletions or plausible SNPs were observed in ZS005NT11168. Further PCR amplification screening indicated that all the 7 C. jejuni strains mentioned above are positive for fexA. In addition, the presence of fexA was identified in another 2 C. jejuni strains with a florfenicol MIC value of 32 mg/liter. These results indicated that the natural transformation led to the allelic exchange of fexA-containing multidrug resistance genomic islands (MDRGIs) between the donor and recipient strains, suggesting that the acquisition of the fexA gene from ZS005 might contribute to the elevated resistance to florfenicol in ZS005NT11168.
TABLE 1.
Key bacterial strains used in this study and MIC values of florfenicol for various C. jejuni strains as determined by the broth dilution method
| Bacterial strain | Description or relevant genotype | MIC (mg/liter)a |
|---|---|---|
| NCTC 11168 | Wild-type C. jejuni lacking fexA | 1 |
| ZS005NT11168 | NCTC 11168 derivative transformants | 32 (↑32) |
| 11168+fexA | NCTC 11168 derivative; rrs::fexA | 32 (↑32) |
| ZS005 | C. jejuni isolate containing fexA | 64 |
| ZS005 ΔfexA | ZS005 derivative; ΔfexA::Kanr | 0.5 (↓128) |
| ZS006 | C. jejuni isolate containing fexA | 64 |
| ZS006 ΔfexA | ZS005 derivative; ΔfexA::Kanr | 0.5 (↓128) |
Numbers in parentheses indicate fold changes over the wild-type control, either increased (↑) or decreased (↓).
Functional confirmation of fexA.
To determine the role of fexA in florfenicol resistance in C. jejuni, insertional mutagenesis was used to construct isogenic mutations in ZS005 and ZS006. The generated fexA mutants were compared with the parent strains for susceptibility to florfenicol compounds using the broth dilution method. According to MIC results obtained from the broth dilution method, inactivation of fexA in these isolates resulted in 64- and 128-fold reductions in their MICs for florfenicol, respectively (Table 1).
To further confirm the function of fexA, its encoding sequence along with the promoter region was inserted into C. jejuni NCTC 11168 between the 16S and 23S rRNA genes. The sequences of promoter regions from eight chromosomes are consistent (Fig. S2). According to the MIC results, acquisition of the single fexA gene in NCTC 11168 resulted in a 32-fold increase in its MIC for florfenicol (Table 1) but had no effects on the MICs for other tested antimicrobial agents (data not shown), indicating that fexA specifically contributes to the resistance to florfenicol. Collectively, these results clearly demonstrated the specific role of fexA in the florfenicol resistance phenotype in Campylobacter.
Genetic environments of fexA.
To identify the associated genetic environments of fexA, the whole genomes of 9 fexA-containing C. jejuni isolates were sequenced. At least 100× coverage of raw reads from Illumina sequencing were obtained for each isolate. Draft genomes were de novo assembled using the CLC Genomics Workbench (version 8.5). The number of contigs ranged from 51 to 232, while the N50 of contigs ranged from 43 to 521 kb for the isolates assembled by the CLC Genomics Workbench. Due to the short reads generated by Illumina sequencing and the high number of insertion elements, the assembled fexA-carrying contigs for the 9 isolates were relatively short, ranging from 2.3 to 3.8 kb.
In order to obtain the genetic environments of fexA, these 9 isolates were resequenced by MinION long-read sequencing to generate complete chromosomes and plasmids. Draft genomes of these isolates were 1,612,562 to ∼1,828,288 bp in length, with a GC content of 30.15 to ∼30.64%.
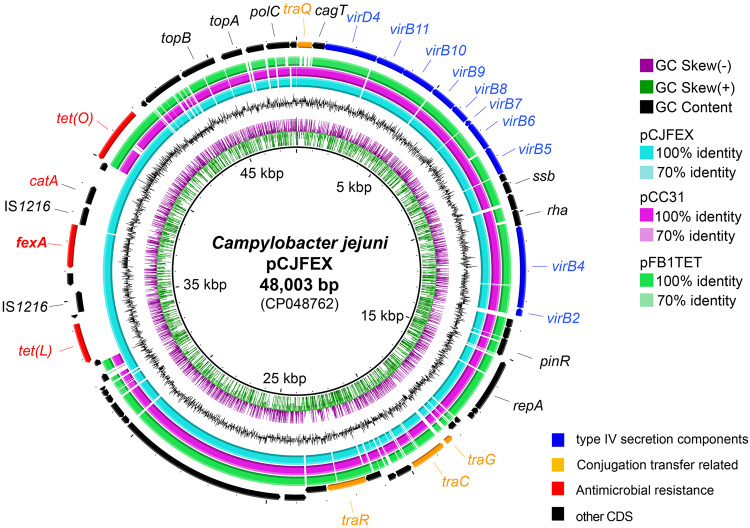
Comparative analysis of complete genomes revealed that the ZS004 C. jejuni strain had a 48,003-bp fexA-carrying pCJFEX plasmid (Fig. 1). Note that pCJFEX contains 58 predicted open reading frames (ORF) with a 29.98% GC content. In addition, this plasmid from the ZS004 strain can be aligned very well to pCC31 (GenBank accession no. AY394560) and pFB1TET (GenBank accession no. CP011017) (99% in both identity and coverage) recovered from Campylobacter coli, with a conserved backbone associated with the plasmid transfer by conjugation.
FIG 1.
Alignment of the fexA-bearing pCJFEX plasmid from the ZS004 C. jejuni strain against homologous plasmids. pCJFEX, labeled in cyan, was aligned to the pCC31 and pFB1TET plasmids using BLAST Ring Image Generator (BRIG) software. The tet(L)-fexA-catA-tet(O) gene arrangement in pCJFEX represents an insertion sequence compared with the other two plasmids.
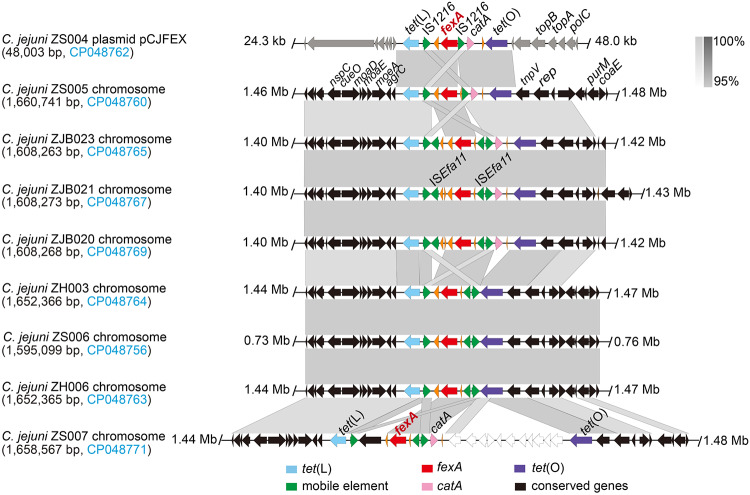
The fexA gene on the plasmid was demonstrated to be located within a 9,982-bp region, with a tet(L)-fexA-catA-tet(O) gene arrangement. The 9,982-bp region had 2 intact copies of IS1216 in the same orientation and contained 5 genes, including tet(L) and tet(O) for tetracycline resistance, fexA for florfenicol resistance, catA for phenicol resistance, and a gene encoding a hypothetical protein. The tet(L)-fexA-catA-tet(O) gene arrangement in pCJFEX represented an insertion sequence (IS) compared with the other 2 plasmids. Similar regions with ≥95% nucleotide sequence identity were also identified in the chromosomes of 8 other C. jejuni isolates (ZH003, ZH006, ZS005, ZS006, ZS007, ZJB020, ZJB021, and ZJB023) (Fig. 2). All these MDRGIs were shown to be located between the agrC and repA genes. Based on colinear alignment, these regions could be divided into at least 4 kinds of sequences according to the number of mobile elements inserted (Fig. 2). The presence of the IS element indicated the potential translocation of the fexA-carrying region between different plasmids and integration into the chromosome. Further, we found that the IS1216 around fexA and the IS1216 copies played the key role in integrating this fexA-carrying segment based on retrospective analysis (Fig. S3).
FIG 2.
Genetic environment of fexA in plasmid or genomes of C. jejuni isolates and comparison of the fexA-carrying regions. Arrows indicate the direction of transcription of the genes. Regions of >95% homology are marked by gray shading. Genes are differentiated by color. Brightly colored ORF represent the tet(L)-fexA-catA-tet(O) gene arrangement. These multidrug resistance genomic islands (MDRGIs) could be divided into 5 different types. ZJB023, ZJ021, and ZJ020 carry the same MDRGIs, whereas ZH003, ZS006, and ZH006 belong to a group containing the same MDRGIs.
Molecular typing and phylogenetic analysis of the fexA-carrying C. jejuni isolates.
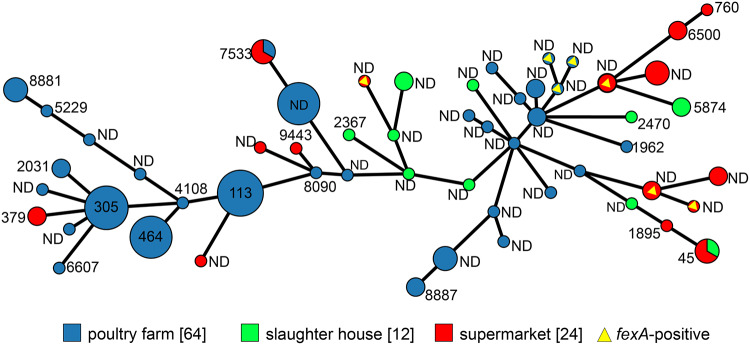
The combined data indicated the significant horizontal dissemination of the fexA gene through C. jejuni isolates of different origins (Fig. 3). Of 100 Campylobacter jejuni isolates, 52 were identified as known sequence types (STs), including 21 STs. Among them, ST113, ST305, and ST464 were shown to include a large number of isolates. In contrast, 48 strains did not match the known ST, including the 9 fexA-positive strains. These 9 strains were shown to belong to 7 different unknown STs, 3 on poultry farms and 4 in supermarkets, meaning that these 9 fexA-positive isolates were multisourced (Fig. 3).
FIG 3.
Minimum spanning tree of fexA-positive and fexA-negative C. jejuni isolates according to MLST and gene allele profile. The tree was created by BioNumerics (Applied Maths, Belgium). Each node within the tree represents a single ST. The size of the nodes is proportional to the number of isolates represented by said node. Selected nodes are labeled with corresponding ST, phylogenetic group, and number of represented isolates. ND, unknown ST.
DISCUSSION
In this study, we identified the fexA gene as an emerging mechanism in mediating resistance to florfenicol in C. jejuni. This finding was supported by multiple pieces of evidence. The presence of fexA was associated with elevated MIC values for florfenicol (Table 1), whereas its inactivation resulted in a 128-fold reduction in the MIC value. Concomitantly, cloning of fexA into a C. jejuni strain lacking fexA resulted in the recipient strain exhibiting a 32-fold increase in its MIC for florfenicol, whereas no effect was observed on the MIC values for other tested antimicrobial agents. These results indicated that fexA could function as a florfenicol-specific resistance mechanism. Importantly, this finding further enriched our knowledge of the various mechanisms of florfenicol resistance in Campylobacter.
Due to the use of florfenicol as well as other antimicrobial agents in poultry production, Campylobacter spp. in poultry must be able to deal with the toxicity and selective pressure deriving from these compounds. Accumulated studies have indicated that Campylobacter isolates of animal origin have shown increased resistance to various antimicrobial agents, with continuous acquisition of different mechanisms of resistance to such compounds (5, 11). In recent years, we have characterized different multidrug resistance genomic islands (MDRGIs), which confer resistance to macrolides, aminoglycosides, tetracyclines, fosfomycin, and so on (10, 12–15, 18). Those genomic islands showed different GC contents when compared with the whole genome of Campylobacter, suggesting that these contents originated from different species. In this study, the characterized fexA gene was shown to be associated with tet(L), tet(O), and catA, forming a novel multidrug resistance genomic island. Although a Basic Local Alignment Search Tool (BLAST) search against the GenBank database could not match the exact same MDRGI from the other species, similar MDRGIs containing fexA associated with IS1216 were found in Enterococcaceae (Fig. S3). The GC content for this MDRGIs was reported to be 36.4%, which was different from the GC content of the Campylobacter genome (∼30%), suggesting that Campylobacter might have obtained this MDRGI from other species. Interestingly, whole-genome analysis of the fexA-harboring isolates indicated that they were generally diverse, with fexA being either in the chromosome or on the plasmid, suggesting that fexA could spread by horizontal gene transfer. Accordingly, we demonstrated the transferability of this MDRGI in the laboratory. Moreover, searching the fexA gene in the GenBank database using BLAST revealed that one C. coli isolate from PulseNet in the United States contained the fexA gene; however, we could not obtain the genetic environment of the fexA gene, probably due to the short read produced by the second-generation sequencing method. During our preparation of the manuscript, the whole-genome sequence of another fexA-carrying C. coli strain, 16SHKX65C (GenBank accession no. CP038868), was released from China (Fig. S3). Given that Campylobacter is known to be naturally transformable, it is expected that the prevalence of fexA will continue to increase, which could confer a fitness advantage under selection from continued florfenicol usage.
MATERIALS AND METHODS
Campylobacter strains and antimicrobial susceptibility testing.
The Campylobacter isolates used in this study were isolated from cecal contents, carcasses, and feces of ducks and chickens in Zhejiang Province, China. All Campylobacter strains were grown on Mueller-Hinton (MH) agar (Sigma-Aldrich, St. Louis, MO) at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2).
Antimicrobial susceptibility testing was conducted using the standard broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (20). The C. jejuni ATCC 33560 strain was used for quality control. All experiments were repeated 3 times.
Natural transformation.
Natural transformation was conducted following the method previously described (21). Briefly, purified genomic DNA from the florfenicol-resistant C. jejuni or constructed suicide plasmids served as the donor DNA, and the naturally competent C. jejuni NCTC 11168 was used as the recipient strain. The genomic or plasmid DNA was spotted (10 μl) onto an overnight lawn of C. jejuni and incubated for 6 h at 37°C. After incubation, the lawn was harvested and plated on MH agar containing selective antimicrobial agents. Transformants were selected on MH agar plates containing florfenicol (4 μg/ml) or kanamycin (30 μg/ml). Transformation without donor DNA was used as a negative control. Transformants were confirmed using pulsed-field gel electrophoresis (PFGE) analysis, which was performed using SmaI as the restriction endonuclease following the protocol for Campylobacter (22). Consecutively, transformants were examined for mutations involved in florfenicol resistance.
Construction of the fexA mutant.
To identify its function, the fexA gene from wild-type C. jejuni was knocked out by insertional mutagenesis. Primers (Table 2) fexA-5′F and fexA-5′R were used to amplify a 920-bp fragment containing the 5′ part of fexA and its upstream gene, while primers fexA-3′F and fexA-3′R were used to amplify the 3′ region of the fexA gene with 209 bp immediately downstream of the gene. The aphA3 kanamycin resistance cassette was amplified from pMW10 using the Phusion high-fidelity DNA polymerase (New England BioLabs [NEB]). After purification, the PCR products of fexA-5′, aphA3, and fexA-3′ were ligated using the Gibson assembly, yielding a 3,175-bp fexA-5′-aphA3-fexA-3′ fragment. The purified PCR product was subsequently introduced into C. jejuni NCTC 11168 using natural transformation. Mutants were selected on MH agar containing 30 μg/ml of kanamycin. Insertion of the aphA3 cassette into the fexA gene was confirmed by PCR analysis.
TABLE 2.
Key primers used in this study
| Primer | Sequence | PCR product size (bp) |
|---|---|---|
| fexA-5′F | GTAATGGGAATTGATTTCATTAATGTCGA | 920 |
| fexA-5′R | CCACAATTATGATAGAATTTACTCCACCAAATATTGGACCAG | |
| aphA3-F | AAATTCTATCATAATTGTGGTTTCAAAATCGGCT | 1,215 |
| aphA3-R | ACCCTAAATATTGACCAACTAAAATGTCAAAAGTTGCCACC | |
| fexA-3′F | AGTTGGTCAATATTTAGGGTGGAATGC | 1,080 |
| fexA-3′R | TTCGCACCAATAAAACAGTGTACG | |
| fexA-F | AGGAGAACCTGCGGTTGGATCACCTCCTTTCTAGAGCAAAAATTTATGAATCTATTGCAT | 1,878 |
| fexA-R | TAATAGTTGTGAGACTTATTACTTTGTACTCTAGAGAAACGATCACCAATGTTTTCATTG | |
| fexA-IF | TTTTAATGATGGTACTCTCCCT | 529 |
| fexA-IR | GGTAACGCGTAGTAGGCACCAA |
Functional clone of fexA.
A wild-type copy of fexA was inserted into the chromosome between the 16S and 23S rRNAs of C. jejuni NCTC 11168 lacking the fexA gene, as previously described (23). The entire fexA gene, including its promoter region, was amplified from C. jejuni ZJB020 by PCR using primers fexA-F and fexA-R (Table 2). The pRRK plasmid containing an aphA3 cassette in the opposite orientation to the ribosomal genes was linearized by XbaI digestion. The homologous recombination method was used to fuse the fexA amplicon to pRRK to obtain the pRRK-fexA plasmid construct. The pRRK-fexA suicide plasmid construct was naturally transformed into the wild-type C. jejuni NCTC 11168 strain, resulting in the insertion of a copy of fexA in the chromosome. Transformants were selected on MH agar plates containing 30 μg/ml of kanamycin and confirmed by PCR using primers fexA-IF and fexA-IR. The obtained transformant strain was named 11168+fexA.
Genome sequencing and assembly.
Genomic DNA of all fexA-positive isolates was extracted using the Wizard genomic DNA purification kit (Promega, Beijing, China), following the manufacturer’s instructions. Indexed Illumina sequencing libraries were prepared using the TruSeq DNA PCR-free sample preparation kit (Illumina Inc., San Diego, CA) following the standard protocol and sequenced on the Illumina HiSeq 2500 platform according to the manufacturer’s protocols, thus producing 150-bp paired-end reads. Draft genomes were assembled using SPAdes (24) and CLC Genomics Workbench (version 8.5; CLC Bio, Aarhus, Denmark) software.
To obtain the complete genome sequence, these 9 isolates were also selected for MinION long-read sequencing. Library preparation was performed using a rapid barcoding sequencing kit (SQK-RBK004) according to the standard protocol provided by the manufacturer (Oxford Nanopore). Guppy (version 3.2.4) was used for base calling and demultiplexing of MinION long-read sequencing raw data. Consecutively, we performed a hybrid strategy of de novo assembly by combining Illumina short-read data and MinION long-read data using Unicycler (version 0.4.4) in order to obtain high-quality complete genomes as previously described (25, 26).
Multilocus sequence typing.
Multilocus sequence typing (MLST) was performed by PCR and sequencing of 7 housekeeping genes (aspA, glnA, gltA, glyA, tkt, pgm, and uncA) (27), and analysis was performed based on the PubMLST web tool (http://pubmlst.org/campylobacter).
Analysis of fexA location.
The plasmid or chromosome location of the fexA gene in these 9 Campylobacter isolates was first determined by S1 nuclease pulsed-field gel electrophoresis (PFGE) and Southern blotting as previously described (9). To this end, S1 nuclease (TaKaRa, Dalian, China) was used to digest agarose gel plugs containing cells of fexA-carrying isolates. Southern blotting was performed to detect the location of the fexA gene. The probe was amplified using the specific primers (fexA-S1F, 5′-CTTATCTCCCTTCGTTGGC-3′, and fexA-S1R, 5′- TACTGCGGCGTTATTTGC-3′) for the fexA gene and then labeled using a DIG High Prime I DNA labeling and detection starter kit for hybridization. Signals from the bands were visualized using a nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) color detection kit (Roche Diagnostics, Mannheim, Germany) following the recommendations of the supplier.
The location of fexA was further confirmed through the assembly of whole-genome sequencing, by analyzing whether it was on a plasmid or chromosome.
Analysis of genetic context of fexA.
The fexA-carrying contigs were annotated using the RAST annotation server (28). Insertion sequences were identified by ISfinder (29). Gene prediction and annotation of the genomes were performed using the NCBI Prokaryotic Genome Annotation Pipeline. Acquired antimicrobial resistance genes were predicted using ResFinder 3.1 (https://cge.cbs.dtu.dk/services/ResFinder/). oriTfinder (http://202.120.12.134/oriTfinder/oriTfinder.html) was used to identify the origin of transfers in the genome. The BLAST Ring Image Generator (BRIG) and Easyfig software were used in the comparative analysis of plasmids.
Accession numbers.
Complete genome sequences have been deposited in GenBank under BioProject number PRJNA605601 and GenBank accession numbers CP048756 to CP048774.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Key Research and Development Program of China (2017YFC1601501), the State Key Laboratory for Quality and Safety of Agro-products (2010DS700124-ZZ1703, ZZ1905), the Youth Program of the National Natural Science Foundation of China (31700007 and 31802240), the Key Research and Development Program of Zhejiang Province (2020C02031), and the National Natural Science Foundation of China (31761133004 and 31722057).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Q, Wang Y, Wang S, Wang Z, Du XD, Jiang H, Xia X, Shen Z, Ding S, Wu C, Zhou B, Wu Y, Shen J. 2016. Prevalence and abundance of florfenicol and linezolid resistance genes in soils adjacent to swine feedlots. Sci Rep 6:32192. doi: 10.1038/srep32192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li D, Wang Y, Schwarz S, Cai J, Fan R, Li J, Feßler AT, Zhang R, Wu C, Shen J. 2016. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J Antimicrob Chemother 71:1474–1478. doi: 10.1093/jac/dkw040. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Feßler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 5.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su CC, Wang B, Wang S, Wu C, Yu EW, Zhang Q, Shen J. 2016. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 7:e01543-16. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO. 2013. The global view of campylobacteriosis: report of an expert consultation, Utrecht, Netherlands, 9–11 July 2012. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 8.Wang Y, Dong Y, Deng F, Liu D, Yao H, Zhang Q, Shen J, Liu Z, Gao Y, Wu C, Shen Z. 2016. Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008–14. J Antimicrob Chemother 71:666–669. doi: 10.1093/jac/dkv382. [DOI] [PubMed] [Google Scholar]
- 9.Yao H, Liu D, Wang Y, Zhang Q, Shen Z. 2017. High prevalence and predominance of the aph(2″)-If gene conferring aminoglycoside resistance in Campylobacter. Antimicrob Agents Chemother 61:e00112-17. doi: 10.1128/AAC.00112-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D, Liu W, Lv Z, Xia J, Li X, Hao Y, Zhou Y, Yao H, Liu Z, Wang Y, Shen J, Ke Y, Shen Z. 2019. Emerging erm(B)-mediated macrolide resistance associated with novel multidrug resistance genomic islands in Campylobacter. Antimicrob Agents Chemother 63:e00153-19. doi: 10.1128/AAC.00153-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Z, Wang Y, Zhang Q, Shen J. 2018. Antimicrobial resistance in Campylobacter spp. Microbiol Spectr 6:ARBA-0013-2017. doi: 10.1128/microbiolspec.ARBA-0013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, Wu C, Wang S, Zhang J, Shen J. 2014. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother 69:964–968. doi: 10.1093/jac/dkt492. [DOI] [PubMed] [Google Scholar]
- 13.Deng F, Wang Y, Zhang Y, Shen Z. 2015. Characterization of the genetic environment of the ribosomal RNA methylase gene erm(B) in Campylobacter jejuni. J Antimicrob Chemother 70:613–615. doi: 10.1093/jac/dku418. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Zhang M, Deng F, Shen Z, Wu C, Zhang J, Zhang Q, Shen J. 2014. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother 58:5405–5412. doi: 10.1128/AAC.03039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Yao H, Deng F, Liu D, Zhang Y, Shen Z. 2015. Identification of a novel fosXCC gene conferring fosfomycin resistance in Campylobacter. J Antimicrob Chemother 70:1261–1263. doi: 10.1093/jac/dku488. [DOI] [PubMed] [Google Scholar]
- 16.Liu D, Li X, Liu W, Yao H, Liu Z, Feßler AT, He J, Zhou Y, Shen Z, Wu Z, Schwarz S, Zhang Q, Wang Y. 2019. Characterization of multiresistance gene cfr(C) variants in Campylobacter from China. J Antimicrob Chemother 74:2166–2170. doi: 10.1093/jac/dkz197. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Dai L, Sahin O, Wu Z, Liu M, Zhang Q. 2017. Emergence of a plasmid-borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter. J Antimicrob Chemother 72:1581–1588. doi: 10.1093/jac/dkx023. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, Deng F, Gao Y, Yao H, Shen Z, Wu C, Wang Y, Shen J. 2017. Dissemination of erm(B) and its associated multidrug-resistance genomic islands in Campylobacter from 2013 to 2015. Vet Microbiol 204:20–24. doi: 10.1016/j.vetmic.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 19.Kehrenberg C, Schwarz S. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob Agents Chemother 48:615–618. doi: 10.1128/aac.48.2.615-618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A3. CLSI, Wayne, PA. [Google Scholar]
- 21.Wang Y, Taylor DE. 1990. Natural transformation in Campylobacter species. J Bacteriol 172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol 39:1889–1894. doi: 10.1128/JCM.39.5.1889-1894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muraoka WT, Zhang Q. 2011. Phenotypic and genotypic evidence for l-fucose utilization by Campylobacter jejuni. J Bacteriol 193:1065–1075. doi: 10.1128/JB.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R, Xie M, Dong N, Lin D, Yang X, Wong MHY, Chan EW, Chen S. 2018. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7:1–9. doi: 10.1093/gigascience/gix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJ, Urwin R, Maiden MC. 2001. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.