A total of 2,283 Salmonella isolates were recovered from 18,334 samples, including samples from patients with diarrhea, food of animal origin, and pets, across 5 provinces of China. The highest prevalence of Salmonella spp. was detected in chicken meats (39.3%, 486/1,237). Fifteen serogroups and 66 serovars were identified, with Salmonella enterica serovars Typhimurium and Enteritidis being the most dominant. Most (85.5%, 1,952/2,283) isolates exhibited resistance to ≥1 antimicrobial, and 56.
KEYWORDS: CTX-M, MLST, PMQR, QRDRs, Salmonella spp., antimicrobial resistance
ABSTRACT
A total of 2,283 Salmonella isolates were recovered from 18,334 samples, including samples from patients with diarrhea, food of animal origin, and pets, across 5 provinces of China. The highest prevalence of Salmonella spp. was detected in chicken meats (39.3%, 486/1,237). Fifteen serogroups and 66 serovars were identified, with Salmonella enterica serovars Typhimurium and Enteritidis being the most dominant. Most (85.5%, 1,952/2,283) isolates exhibited resistance to ≥1 antimicrobial, and 56.4% were multidrug resistant (MDR). A total of 222 isolates harbored extended-spectrum β-lactamases (ESBLs), and 200 of these were of the CTX-M type and were mostly detected in isolates from chicken meat and turtle fecal samples. Overall, eight blaCTX-M genes were identified, with blaCTX-M-65, blaCTX-M-123, blaCTX-M-14, blaCTX-M-79, and blaCTX-M-130 being the most prevalent. In total, 166 of the 222 ESBL-producing isolates had amino acid substitutions in GyrA (S83Y, S83F, D87G, D87N, and D87Y) and ParC (S80I), while the plasmid-mediated quinolone resistance (PMQR)-encoding genes oqxA, oqxB, qepA, qnrB, and qnrS were detected in almost all isolates. Of the 15 sequence types (STs) identified in the 222 ESBLs, ST17, ST11, ST34, and ST26 ranked among the top 5 in number of isolates. Our study revealed considerable serovar diversity and a high prevalence of the co-occurrence of MDR determinants, including CTX-M-type ESBLs, quinolone resistance-determining region (QRDR) mutations, and PMQR genes. This is the first report of CTX-M-130 Salmonella spp. from patients with diarrhea and QRDR mutations from turtle fecal samples. Our study emphasizes the importance of actions, both in health care settings and in the veterinary medicine sector, to control the dissemination of MDR, especially the CTX-M-type ESBL-harboring Salmonella isolates.
INTRODUCTION
Salmonella infections have been proven to be a major cause of global morbidity and mortality in both humans and animals (1, 2). Worldwide, 93.8 million salmonellosis cases occur annually, with 155,000 resulting in death (3). In China, more than 20% of all foodborne illnesses are estimated to be caused by Salmonella spp. (4). In 2013, unpublished data from the China CDC surveillance system showed that the rate of human carriage of Salmonella spp. was 549 per 100,000 people. This is more than 30 times higher than the number of human Salmonella infections in Europe in 2017 (19.7 per 100,000) and the United States in 2018 (18.3 per 100,000) (5, 6). Moreover, the indiscriminate use of wide-spectrum antibiotics creates an additional threat, represented by the appearance and dissemination of multiantibiotic resistance profiles in the pathogen population. Multidrug-resistant (MDR) Salmonella spp. potentially arising due to selective pressure from sustained antimicrobial exposure are more likely to be the causative agents of invasive disease (7). Of concern is the increased incidence of infections caused by extended-spectrum β-lactamase (ESBL)-producing organisms, including Salmonella spp., because they are resistant not only to most of the β-lactam antimicrobials but also to other antimicrobial classes, leaving few treatment options and the potential for clinical outcomes worse than those of infections caused by non-ESBL-producing strains (8, 9).
During the last decade, the most frequently encountered (particularly in areas of Europe and Asia) ESBL genes were those encoding the CTX-M enzyme family, primarily carried by transferable plasmids and transposons (10). The emergence of CTX-M-type ESBL-producing Salmonella spp. has been reported in clinical cases, animals, and food samples worldwide, including China (11–13). Worryingly, the plasmids and transposons carrying the gene for CTX-M can also contain genes for resistance to other antimicrobials, like fluoroquinolones (14). This brings big challenges to clinical treatment, as extended-spectrum cephalosporins (ESCs) and fluoroquinolones are the drugs of choice for the treatment of invasive Salmonella infections (15). The mechanism of quinolone resistance has been elucidated to be plasmid-mediated quinolone resistance (PMQR) and chromosomal mutations in the quinolone resistance-determining regions (QRDRs) (16, 17). PMQR can be classified into three different resistance mechanisms, AAC(6′)-Ib-cr acetylating ciprofloxacin and norfloxacin, Qnr proteins mediating target protection, and OqxAB and QepA mediating drug efflux (17), while mutations in QRDR genes, encoding DNA gyrase or topoisomerase IV, are also frequently found in quinolone-resistant Salmonella isolates (16). The co-occurrence of ESBLs (especially the CTX-M gene on plasmids and transposons) and PMQR in Salmonella spp. is a cause for concern because plasmids can spread with relative ease between different reservoirs, and such spread may be extremely difficult to control.
In this study, we therefore investigated ESBL-producing Salmonella isolates collected from patients with diarrhea, food of animal origin, and pets across 5 provinces of China. We dissected the serovar diversity, the prevalence of antimicrobial resistance (AMR), the multilocus sequence types (MLSTs), and the co-occurrence of MDR determinants, including CTX-M-type ESBLs and quinolone resistance. Furthermore, we investigated the relatedness of blaCTX-M genes, amino acid substitutions in QRDRs, and determinants of PMQR across serovars and sources and provide evidence of the existence of Salmonella spp. harboring the blaCTX-M-130 gene and QRDR mutations in previously undescribed infection reservoirs, such as humans, food, and pets.
RESULTS
Salmonella isolates from patients with diarrhea, food of animal origin, and pet samples.
A total of 2,283 Salmonella isolates were recovered from 18,334 samples (12.5%) (Table 1) collected from 5 provinces in China. Of these 2,283 isolates, 1,572 (10.8%) of 14,579 were isolated from patients with diarrhea, 660 (19.4%, 660/3,405) were isolated from food of animal origin, and 51 (14.6%, 51/350) were isolated from pet fecal samples. Overall, the prevalence among pets was lower than that among food of animal origin (P < 0.05) but higher than that among patients with diarrhea (P < 0.05). Besides, the prevalence among patients ≤5 years old with diarrhea was significantly higher than that among other patients with diarrhea (P < 0.05). Finally, the results showed that the prevalence among chicken meat samples was significantly higher than that among the other types of samples (P < 0.05).
TABLE 1.
Prevalence of Salmonella isolates recovered from patients with diarrhea, food of animal origin, and pets in China
| Source | No. of samples tested | No. of positive samples | % prevalence |
|---|---|---|---|
| Patients with diarrhea ages (yr): | |||
| ≤5 | 5,515 | 717 | 13.0 |
| 5–59 | 6,654 | 646 | 9.7 |
| ≥60 | 2,410 | 209 | 8.7 |
| Total | 14,579 | 1,572 | 10.8 |
| Food of animal origin | |||
| Chicken meat | 1,237 | 486 | 39.3 |
| Pork | 1,354 | 122 | 9.0 |
| Aquatic products | |||
| Freshwater fish | 349 | 22 | 6.3 |
| Saltwater fish | 309 | 17 | 5.5 |
| Shrimp | 156 | 13 | 8.3 |
| Subtotal | 814 | 52 | 6.4 |
| Total | 3,405 | 660 | 19.4 |
| Pets | |||
| Turtle | 290 | 42 | 14.5 |
| Pigeon | 60 | 9 | 15.0 |
| Total | 350 | 51 | 14.6 |
| Overall | 18,334 | 2,283 | 12.5 |
Salmonella serovars and their distribution.
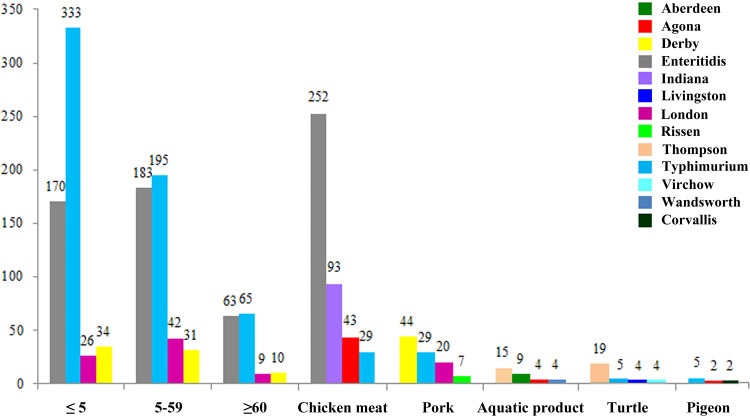
The 2,283 Salmonella isolates were serologically divided into 15 serogroups and 66 serovars (see Table S1 in the supplemental material), with serogroup B (n = 1,044, 45.7%) representing the most common serogroup identified, followed by serogroup D1 (n = 684, 30.0%), serogroup C1 (n = 227, 9.9%), serogroup E1 (n = 155, 6.8%), and serogroup C2-C3 (n = 109, 4.8%). These five serogroups comprised 52 (52/66, 78.8%) serovars and 2,219 isolates (2,219/2,283, 97.2%). The distribution of the serogroups and serovars derived from different sources is shown in Table S1. A total of 13 serogroups and 56 serovars were identified from patients with diarrhea. The distribution of serovars varied among different the sources (Table S1 and Fig. 1).
FIG 1.
Prevalence of the top 4 Salmonella serovars among the total isolates recovered from patients with diarrhea, food of animal origin and pet samples in China. The number of isolates is indicated on the y axis. Colors indicate different serovars. Each of the histograms represents the number of isolates of each serovar. The sample classes (patients with diarrhea, food of animal origin, and pet samples) are shown. The patient age distribution is indicated (ages, ≤5, 5 to 59, and ≥60 years).
Antimicrobial susceptibility testing of 2,283 Salmonella isolates.
Out of 2,283 Salmonella isolates, 331 (14.5%) were susceptible to all antimicrobial agents tested (pan-susceptible), while 1,952 (85.5%) exhibited resistance to at least one compound (Table 2). The top three dominant antimicrobial agents to which the isolates were resistant were ampicillin (AMP) (64.6%), nalidixic acid (NAL) (62.0%), and tetracycline (TET) (58.7%). Notably, three isolates (0.1%, 3/2,283) (two Salmonella enterica serovar Enteritidis isolates from patients ≤5 years old with diarrhea and one S. Indiana isolate from chicken meat) showed resistance to carbapenems (imipenem [IPM] and meropenem [MEM]) (Table 2 and Table S2). Additionally, the isolates cultured from food of animal origin showed significantly higher levels of resistance to NAL (75.9%), cefotaxime (CTX) (18.3%), and ceftazidime (CAZ) (14.1%) than those cultured from the other samples (P < 0.05). The isolates cultured from pet fecal samples showed significantly higher levels of resistance to TET (80.4%), ampicillin-sulbactam (SAM) (62.7%), chloramphenicol (CHL) (60.8%), and trimethoprim-sulfamethoxazole (SXT) (60.8%) than those cultured from the other samples (P < 0.05). The isolates cultured from samples from patients with diarrhea showed significantly lower levels of resistance to SAM (26.2%), SXT (23.6%), gentamicin (GEN) (16.1%), and ciprofloxacin (CIP) (10.0%) than those cultured from the other samples (P < 0.05).
TABLE 2.
Antimicrobial resistance of Salmonella isolates recovered from patients with diarrhea, food of animal origin, and pets in China
| Antimicrobial or no. of antimicrobial classes to which isolate is resistant | No. (%) of isolates resistant to the tested antimicrobial agents or the indicate no. of antimicrobial classesa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients with diarrhea, by age (yr) |
Food of animal origin |
Pets (n = 51) | Overall (n = 2,283) | |||||||
| ≤5 (n = 717) | 5–59 (n = 646) | ≥60 (n = 209) | Total (n = 1,572) | Aquatic products (n = 52) | Chicken meat (n = 486) | Pork (n = 122) | Total (n = 660) | |||
| Ampicillin | 509 (71.0) | 425 (65.8) | 135 (64.6) | 1,069 (68.0) | 22 (42.3) | 261 (53.7) | 83 (68.0) | 366 (55.5) | 40 (78.4) | 1,475 (64.6) |
| Cefotaxime | 84 (11.7) | 63 (9.8) | 29 (13.9) | 176 (11.2) | 2 (3.8) | 114 (23.5) | 5 (4.1) | 121 (18.3) | 3 (5.9) | 300 (13.1) |
| Ceftazidime | 51 (7.1) | 27 (4.2) | 15 (7.2) | 93 (5.9) | 1 (1.9) | 90 (18.5) | 2 (1.6) | 93 (14.1) | 2 (3.9) | 188 (8.2) |
| Ciprofloxacin | 83 (11.6) | 57 (8.8) | 17 (8.1) | 157 (10.0) | 8 (15.4) | 136 (28.0) | 38 (31.1) | 182 (27.6) | 14 (27.5) | 353 (15.5) |
| Nalidixic acid | 387 (54.0) | 374 (57.9) | 126 (60.3) | 887 (56.4) | 19 (36.5) | 399 (82.1) | 83 (68.0) | 501 (75.9) | 28 (54.9) | 1,416 (62.0) |
| Ampicillin-sulbactam | 212 (29.6) | 144 (22.3) | 56 (26.8) | 412 (26.2) | 15 (28.8) | 236 (48.6) | 46 (37.7) | 297 (45.0) | 32 (62.7) | 741 (32.5) |
| Gentamicin | 139 (19.4) | 89 (13.8) | 25 (12.0) | 253 (16.1) | 1 (1.9) | 108 (22.2) | 31 (25.4) | 140 (21.2) | 13 (25.5) | 406 (17.8) |
| Chloramphenicol | 281 (39.2) | 170 (26.3) | 54 (25.8) | 505 (32.1) | 9 (17.3) | 163 (33.5) | 59 (48.4) | 231 (35.0) | 31 (60.8) | 767 (33.6) |
| Trimethoprim-sulfamethoxazole | 204 (28.5) | 127 (19.7) | 40 (19.1) | 371 (23.6) | 25 (48.1) | 182 (37.4) | 64 (52.5) | 271 (41.1) | 31 (60.8) | 673 (29.5) |
| Tetracycline | 473 (66.0) | 410 (63.5) | 134 (64.1) | 1,017 (64.7) | 29 (55.8) | 148 (30.5) | 104 (85.2) | 281 (42.6) | 41 (80.4) | 1,339 (58.7) |
| Imipenem | 2 (0.3) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 3 (0.1) |
| Meropenem | 2 (0.3) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 3 (0.1) |
| Pan-susceptible | 100 (13.9) | 121 (18.7) | 34 (16.3) | 255 (16.2) | 18 (34.6) | 43 (8.8) | 8 (6.6) | 69 (10.5) | 7 (13.7) | 331 (14.5) |
| ≥1 class | 617 (86.1) | 525 (81.3) | 175 (83.7) | 1,317 (83.8) | 34 (65.4) | 443 (91.2) | 114 (93.4) | 591 (89.5) | 44 (86.3) | 1,952 (85.5) |
| ≥3 classes | 407 (56.8) | 357 (55.3) | 118 (56.5) | 882 (56.1) | 24 (46.2) | 252 (51.9) | 89 (73.0) | 365 (55.3) | 41 (80.4) | 1,288 (56.4) |
| ≥4 classes | 295 (41.1) | 198 (30.7) | 73 (34.9) | 566 (36.0) | 18 (34.6) | 197 (40.5) | 66 (54.1) | 281 (42.6) | 31 (60.8) | 878 (38.5) |
| ≥5 classes | 220 (30.7) | 135 (20.9) | 50 (23.9) | 405 (25.8) | 13 (25.0) | 178 (36.6) | 49 (40.2) | 240 (36.4) | 30 (58.8) | 675 (29.6) |
| ≥6 classes | 141 (19.7) | 90 (13.9) | 32 (15.3) | 263 (16.7) | 6 (11.5) | 146 (30) | 36 (29.5) | 188 (28.5) | 17 (33.3) | 468 (20.5) |
| ≥7 classes | 65 (9.1) | 43 (6.7) | 10 (4.8) | 118 (7.5) | 0 (0.0) | 110 (22.6) | 27 (22.1) | 137 (20.8) | 12 (23.5) | 267 (11.7) |
| ≥8 classes | 20 (2.8) | 13 (2.0) | 3 (1.4) | 36 (2.3) | 0 (0.0) | 29 (6.0) | 3 (2.5) | 32 (4.8) | 1 (2.0) | 69 (3.0) |
| ≥9 classes | 2 (0.3) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 3 (0.1) |
n, total number of isolates tested for susceptibility in different samples.
Among the 2,283 Salmonella isolates, 1,288 (56.4%) were resistant to three or more classes of antimicrobials, and these were classified as MDR (Table 2). Notably, isolates recovered from pet fecal samples showed the highest percentages of resistance to ≥3 (80.4%), ≥4 (60.8%), ≥5 (58.8%), ≥6 (33.3%), and ≥7 (23.5%) classes of antimicrobials, which were substantially higher than those for isolates recovered from the other samples (P < 0.05). In total, we identified 159 antimicrobial resistance profiles and 138 MDR profiles. Among the MDR profiles, the most common one was NAL-AMP-TET (8.0%, n = 183) (Table S2). Besides, the three carbapenem-resistant isolates were found to be resistant to all other antimicrobials tested in this study.
The antimicrobial susceptibility phenotypes of the isolates among different Salmonella serovars are shown in Table S3. Most of the isolates found to be resistant to ≥1 classes of antimicrobials were those of Salmonella serovar Indiana (97.9%, 93/95), followed by Salmonella serovars Enteritidis (97.6%, 656/672), Derby (93.8%, 121/129), Rissen (93.5%, 43/46), Typhimurium (92.0%, 611/664), and Corvallis (91.7%, 22/24), while MDR profiles were mostly observed among isolates of Salmonella serovar Indiana (94.7%, 90/95). Serovars with less than 10 isolates were not considered.
Prevalence of ESBL-producing and blaCTX-M-positive Salmonella isolates.
The prevalence of ESBL-producing Salmonella isolates was 9.7% (222/2,283), and of these, 102 isolates were collected from patients with diarrhea, 100 were collected from food of animal origin, and 20 were collected from pet fecal samples (Table 3). No ESBL-producing isolates were detected from aquatic products or pigeon fecal samples. The prevalence of ESBL-producing isolates in chicken meat samples (20.0%, 97/486) was lower than that in turtle fecal samples (47.6%, 20/42) (P < 0.05) but higher than that in the other samples (P < 0.05). Additionally, ESBL-producing S. Indiana isolates were most often detected (96.8%, 92/95). Notably, of these 92 ESBL-producing S. Indiana isolates, 90 were recovered from chicken meat samples, 1 was recovered from a patient with diarrhea, and 1 was recovered from a pork sample.
TABLE 3.
Prevalence of ESBL-producing and blaCTX-M positive isolates among Salmonella isolates recovered from patients with diarrhea, food of animal origin, and pets in Chinaa
| Source | No. of isolates |
||
|---|---|---|---|
| Total | ESBL producing | blaCTX-M positive | |
| Patients with diarrhea ages (yr): | |||
| ≤5 | 717 | 46 (6.4) | 46 (6.4) |
| 5–59 | 646 | 38 (5.9) | 36 (5.6) |
| ≥60 | 209 | 18 (8.6) | 17 (8.1) |
| Total | 1,572 | 102 (6.5) | 99 (6.3) |
| Food of animal origin | |||
| Aquatic product | 52 | —b | — |
| Chicken meat | 486 | 97 (20.0) | 95 (19.5) |
| Pork | 122 | 3 (2.5) | 3 (2.5) |
| Total | 660 | 100 (15.2) | 98 (14.8) |
| Pets | |||
| Turtle | 42 | 20 (47.6) | 3 (7.1) |
| Pigeon | 9 | — | — |
| Total | 51 | 20 (39.2) | 3 (5.9) |
| Serovars | |||
| S. Agona | 75 | 1 (1.3) | 1 (1.3) |
| S. Derby | 129 | 6 (4.7) | 5 (3.9) |
| S. Enteritis | 672 | 49 (7.3) | 47 (7.0) |
| S. Give | 26 | 8 (30.8) | 8 (30.8) |
| S. Havana | 1 | 1 (100%) | — |
| S. Hvittingfoss | 1 | 1 (100%) | — |
| S. Indiana | 95 | 92 (96.8) | 91 (95.8) |
| S. Saintpaul | 18 | 3 (16.7) | 3 (16.7) |
| S. Stanley | 36 | 1 (2.8) | 1 (2.8) |
| S. Thompson | 70 | 21 (30.0) | 7 (10.0) |
| S. Typhimurium | 664 | 39 (5.9) | 37 (5.6) |
| Overall | 2,283 | 222 (9.7) | 200 (8.8) |
The data are for 2,283 isolates tested.
—, not detected.
Of these 222 ESBL-producing isolates, 200 contained blaCTX-M genes (8.8%, 200/2,283) (Table 3). Most of the blaCTX-M-positive isolates were recovered from chicken meat samples (19.5%, 95/486), from which such isolates were recovered at a significantly higher rate than from the other samples (P < 0.05). S. Indiana was also the serovar harboring most of the blaCTX-M genes (95.8%, 91/95), followed by S. Give (30.8%, 8/26), S. Saintpaul (16.7%, 3/18), and S. Thompson (10.0%, 7/70).
Distribution of blaCTX-M genes, gyrA and parC mutations, and PMQR among ESBL-producing Salmonella isolates.
Overall, 8 blaCTX-M alleles (blaCTX-M-14, blaCTX-M-24, blaCTX-M-27, blaCTX-M-65, blaCTX-M-79, blaCTX-M-90, blaCTX-M-123, and blaCTX-M-130) were identified among the 222 ESBL-producing Salmonella isolates (Table 4 and Fig. 2). Approximately 53% (117/222) of the ESBL-producing isolates carried one of the blaCTX-M genes, while 37% (83/222) were found to cocarry two blaCTX-M genes (Fig. 2). The most prevalent gene detected was blaCTX-M-65, with 86 out of the 222 ESBL-producing Salmonella isolates harboring blaCTX-M-65 (38.7%, 86/222), followed by blaCTX-M-123 (27.9%, 62/222), blaCTX-M-14 (20.7%, 46/222), blaCTX-M-79 (19.8%, 44/222), and blaCTX-M-130 (16.7%, 37/222). The distribution of blaCTX-M genes varied across sources and serovars (Table S4 and Fig. 2). Notably, the blaCTX-M-130 gene was mostly detected from patients with diarrhea (25/37) and chicken meat samples (11/37), with S. Typhimurium, Indiana, and Enteritidis being the dominant serovars. Furthermore, the three carbapenem-resistant isolates were also found to carry CTX-M-type ESBLs; two of these isolates were S. Enteritidis isolates from patients with diarrhea (with one carrying the blaCTX-M-79 and blaCTX-M-130 genes and one carrying the blaCTX-M-79 and blaCTX-M-14 genes), and one was an S. Indiana isolate from chicken meat carrying the blaCTX-M-65 gene.
TABLE 4.
Prevalence of blaCTX-M genes, gyrA and parC mutations, and PMQR genes in ESBL-producing Salmonella isolates recovered from patients with diarrhea, food of animal origin, and pets in Chinaa
| Gene | No. of isolates | % of isolates |
|---|---|---|
| blaCTX-M | ||
| blaCTX-M-14 | 46 | 20.7 |
| blaCTX-M-24 | 6 | 2.7 |
| blaCTX-M-27 | 1 | 0.5 |
| blaCTX-M-65 | 86 | 38.7 |
| blaCTX-M-79 | 44 | 19.8 |
| blaCTX-M-90 | 1 | 0.5 |
| blaCTX-M-123 | 62 | 27.9 |
| blaCTX-M-130 | 37 | 16.7 |
| gyrA mutations | ||
| S83Y | 23 | 10.4 |
| S83F | 95 | 42.8 |
| D87G | 9 | 4.1 |
| D87N | 94 | 42.3 |
| D87Y | 8 | 3.6 |
| S80R parC mutation | 90 | 40.5 |
| PMQR genes | ||
| qnrB | 116 | 52.3 |
| qnrS | 222 | 100.0 |
| qepA | 52 | 23.4 |
| aac(6')-Ib | 222 | 100.0 |
| oqxA | 218 | 98.2 |
| oqxB | 218 | 98.2 |
The data are for 222 isolates tested.
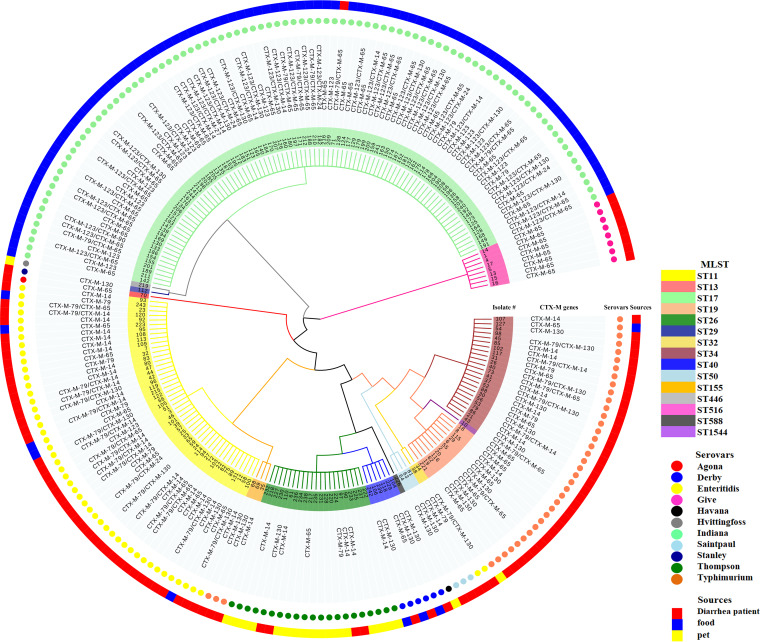
FIG 2.
Dendrogram of the whole ESBL-producing Salmonella cohort. Phylogenetic tree (minimum spanning tree) based on seven loci of 222 ESBL-producing Salmonella isolates recovered from patients with diarrhea, food of animal origin, and pet samples. The phylogenetic tree was developed with the MEGA5 program (www.megasoftware.net) and visualized by use of the Evolview program (www.evolgenius.info/evolview/). Sequence types (STs) are indicated by means of the colors marked in the branches and the backgrounds of the isolate names. The blaCTX-M genes (CTX-M genes), serovars, and sample types (sources) are indicated by text or color coded in the following rings.
As all of the 222 ESBL-producing isolates were resistant to quinolones (149 CIP-resistant isolates and 199 NAL-resistant isolates), we further investigated the isolates for the presence of QRDR mutations and PMQR genes (Table 4). Sequencing analysis resulted in the identification of six QRDR point mutations, five in gyrA (S83Y, S83F, D87G, D87N, and D87Y) and one in parC (S80I). The six QRDR mutations were found in 166 ESBL-producing isolates, including 97 isolates from chicken meat samples, 48 isolates from patients with diarrhea, 18 isolates from turtle fecal samples, and 3 isolates from pork samples. The oqxAB (n = 218), qepA (n = 52), qnrB (n = 116), and qnrS (n = 222) genes were found in the 222 ESBL-producing isolates. The aac(6′)-Ib gene was detected in all 222 isolates. Further analysis done by DNA sequencing and BLAST searches confirmed that this gene is the one conferring resistance to aminoglycosides, as no known aac(6′)-Ib variants that could lead to ciprofloxacin resistance were present.
MLST.
In total, 15 sequence types (STs), including ST11, ST13, ST17, ST19, ST26, ST29, ST32, ST34, ST40, ST50, ST155, ST446, ST516, ST588, and ST1544, were identified from the 222 ESBL-producing isolates (Fig. 2). ST17 (92/222) was the most prevalent sequence type among the ESBL-producing isolates. Notably, isolates cocarrying two of the blaCTX-M genes were identified to be Salmonella serovars Indiana, Enteritidis, and Typhimurium. Interestingly, 14 out of 21 S. Thompson isolates were found not to carry blaCTX-M genes. Besides, 8 ST516 Salmonella serovar Give isolates, 6 ST40 Salmonella serovar Derby isolates, 3 ST50 Salmonella serovar Saintpaul isolates, 1 ST13 Salmonella serovar Agona isolate, 1 ST29 Salmonella serovar Stanley isolate, 1 ST446 Salmonella serovar Hvittingfoss isolate, and 1 ST588 Salmonella serovar Havana isolate were also identified among the ESBL-producing isolates.
DISCUSSION
Globally, the burden of morbidity, mortality, and economic losses from human and animal enteric pathogenic bacteria, including Salmonella spp., is immense, despite the presence of antibiotic drugs (18). Worryingly, the emergence of MDR and ESBLs, especially in CTX-M-producing Salmonella spp., in humans, animals, pets, and foods is increasingly worldwide, including in China (1, 2, 4, 5, 8). In this study, we surveyed the prevalence, serovar distribution, MDR profiles, and occurrence of ESBL-producing Salmonella spp. in patients with diarrhea from 20 hospitals, samples of food of animal origin from a total of 20 supermarkets, and pet fecal samples from 5 veterinary clinics between 2014 and 2015 from 5 provinces (Beijing, Heilongjiang, Hubei, Jiangxi, and Shandong) in China. Furthermore, we investigated the characteristics of the CTX-M-type ESBL-producing Salmonella spp. at the genetic level.
Overall, 1,572 Salmonella isolates were recovered from 14,579 patients with diarrhea, showing a prevalence of 10.8% among isolates from humans with diarrhea, which concurs with previous findings obtained in Shanghai (8.2%) but which was higher than the prevalence in the provinces of Beijing (4.3% in children) and Guangzhou (4.5%) (19–21). Moreover, our results revealed that the prevalence of Salmonella spp. in children ≤5 years old (13.0%) was higher than that in adults (8.7% to 9.7%) (P < 0.05), which is consistent with the findings presented in the literature, showing that children are more susceptible to salmonellosis (22, 23). Therefore, our findings suggest that efforts to determine the risk factors causing such high rates of infection with Salmonella spp. should concentrate on children ≤5 years old in China.
Accordingly, more than 70% of foodborne disease outbreaks in China are attributed to Salmonella spp., and many diseases are linked to the consumption of food of animal origin, especially chicken and pork, which are considered the major reservoirs from which Salmonella spp. disseminate (24, 25). In our study, the prevalence (19.4%) of Salmonella spp. in food samples of animal origin concurs with previous findings obtained in China but was higher than that in Spain (8.9%) and Poland (5.5%) (26–29). The upper edge of the Salmonella prevalence range was observed in chicken meats (39.3%), similar to what was previously found in Henan (38%) but lower than what was found in Shaanxi (54%) and Guangdong (63.6%) (30–32). In contrast, we found a prevalence rate (9.0%) of Salmonella spp. in pork lower than that previously found in China (26, 30–32). The levels of contamination of aquatic products (prevalence rate, 6.4%) were lower than those previously found in China, Thailand, and Malaysia but higher than those found in Morocco (5, 26, 33–36). Our data show that food, especially chicken meat, is an important reservoir of Salmonella contamination and emphasize the importance of monitoring Salmonella infections in food-producing animals and the food chain supply.
Recently, more people have obtained pet animals, and consequently, the number of pet shops and pet clinics has increased in China. Notably, pet reptiles and birds have been proven to pose an important zoonotic potential, being important reservoirs for pathogens, including Salmonella spp., and with patients who are immunocompromised, young children, pregnant women, and older adults being at the greatest risk for transmission via direct and indirect contact (37, 38). However, pets are generally considered to be of little concern as a source of Salmonella spp. for humans (39). Our findings support the assertion that pets are important reservoirs of infections; specifically, we observed an overall prevalence of Salmonella spp. in pet fecal samples of 14.6% (pigeons, 15.0%; turtles, 14.5%), which were lower than that in Costa Rica (pigeons, 24.1%) and South Korea (turtles, 50%) but higher than that in a previous study in China (pigeons, 4.1%) (40–42). An estimated 11% of all Salmonella infections are attributed to animal exposure annually in the United States, with the highest rates of illness and death occurring among children (43). From 1990 to 2014, a total of 53 live poultry-associated salmonellosis (LPAS) outbreaks were reported, involving 2,630 illnesses, 387 hospitalizations, and 5 deaths. Since 2007, numerous outbreaks of human Salmonella infections linked to contact with animals and their environments have been investigated, including those involving contact with turtles and backyard poultry (44). Taken all together, these findings emphasize the importance of managing and studying animal-associated salmonellosis outbreaks, as they occur at the intersection of human and animal health.
Our data showed that Salmonella Typhimurium and Enteritidis were the most common serovars found among patients with diarrhea, which is consistent with the results obtained previously in China and other regions worldwide (3, 5, 19–21), while Salmonella serovars Enteritidis and Indiana, Derby and Typhimurium, and Thompson and Aberdeen were the most common serovars found in chicken meat, pork, and aquatic product, respectively, which is consistent with the findings in the literature (8, 31–33). In contrast, previous investigations covering the northern Chinese regions found Salmonella serovars Senftenberg, Meleagridis, Hadar, Derby, Corvallis, and Kentucky to be the most prevalent in chicken meat (26, 30). Such differences may result from variations in temperature both within and between seasons, local environmental conditions, and the sampling strategy.
Antimicrobial resistance in foodborne pathogens, such as Salmonella spp., is a major concern for public health safety. Still more worrying is the fact that the incidence of Salmonella isolates with resistance to multiple drugs is rapidly increasing globally (8, 28, 34). In Europe, more than half of the Salmonella isolates (52.6%) collected from humans were found to be susceptible and only 28.6% of the isolates were found to be MDR (45). Conversely, 85.5% of the isolates in our investigation were resistant to at least one antimicrobial and 56.4% were MDR. A study of a total of 178 isolates related to human infections caused by invasive Salmonella spp. collected in five provinces of China between 2007 and 2016 revealed that 53.4% of the isolates were MDR (46). The high rates of MDR among Salmonella spp. could pose a significant challenge for the effective treatment of salmonellosis in China. Furthermore, our findings show that the prevalence of Salmonella isolates resistant to the conventional first-line antimicrobials (AMP, NAL, CHL, SXT, and TET) remains high (23.6% to 68.0%) in patients with diarrhea. In comparison, studies performed in patients with diarrhea in the United States showed lower rates of resistance (2.7% to 20%) of Salmonella isolates to AMP, CHL, and NAL (47). Although isolates showed low rates of resistance to some antimicrobials, like gentamicin (17.8%), in this study, they should not be used for clinical therapy, as they are not effective in either humans or animals. Overall, these circumstances render China particularly suitable to study the MDR Salmonella spp. that are found in the food chain.
Most of the isolates (>90%) identified as Salmonella serovars Indiana, Enteritidis, Derby, Rissen, Typhimurium, and Corvallis were resistant to at least one antimicrobial, which concurs with previous findings obtained in China (20, 27, 32, 33). The highest rates of MDR were observed in the above-mentioned serovars, as well as Salmonella Thompson and London. However, the highest percentage of MDR was observed among isolates identified as Salmonella serovars Kentucky, Typhimurium, and Infantis, while the Salmonella Enteritidis isolates were more susceptible in humans and animals in Europe (48). Of note, S. Indiana isolates, mainly recovered from chicken meats (93/95), were reported to be the serovar in China with the second highest percentage of MDR (49). More attention needs to be paid to MDR Salmonella isolates, especially S. Indiana isolates, in China among workers in the fields of veterinary medicine, workers who work with foods of animal origin, and workers in public health.
In our study, 102 (6.5%) of 1,572 Salmonella isolates recovered from patients with diarrhea were identified to be ESBL producers, and among those isolates, 99 were found to harbor blaCTX-M genes. The high prevalence of blaCTX-M genes among ESBL-producing isolates was consistent with previous findings obtained for children with diarrhea in China (8). Of interest, most ESBL-producing isolates were found in pet turtle fecal samples (47.6%), but only 3 of these isolates produced ESBLs of the CTX-M type. Few, if any, data on ESBL-producing Salmonella isolates in pet turtles are currently available. In 2019, 35 Salmonella isolates were recovered from 59 pet turtle samples, but none were identified to be ESBL producers (50). Similar to the findings of other studies, 20% of the Salmonella isolates from chicken meat samples were identified to be ESBL producers, and 97.9% of the ESBL-producing isolates carried blaCTX-M genes (9, 51, 52). In Asia, the daily intake of animal protein increased more than three times between 1960 and 2013 (53). To meet this demand, the scale of broiler farming increased very rapidly. With the high density of birds, the use of antimicrobials for disease prevention and treatment during animal growing in husbandry, especially in the chicken industry, is placing an ever greater selection pressure for the evolution of resistant strains of bacteria. The widespread misuse and overuse of antimicrobials may have led to the emergence of these MDR and ESBL-producing Salmonella strains in foods of animal origin.
Our findings highlight the presence of the blaCTX-M-65, blaCTX-M-79, and blaCTX-M-130 genes, in addition to the blaCTX-M-14 gene, which were the genes most commonly found in patients with diarrhea. Our results suggest that these CTX-M subtypes may have particular epidemic characteristics in different geographical regions (8, 54–56). In 2019, the blaCTX-M-130 gene was first found in Salmonella isolates recovered from food samples in China (57). However, to the best of our knowledge, this is the first study reporting the detection of the blaCTX-M-130 gene in Salmonella isolates recovered from patients with diarrhea in China. The copresence of the blaCTX-M-65 gene with the blaCTX-M-14, blaCTX-M-24, blaCTX-M-27, blaCTX-M-79, and blaCTX-M-90 genes in Salmonella isolates from chicken meat found in this study has been previously described (58, 59). The blaCTX-M-123 gene has recently been detected in Salmonella isolates recovered from patients with diarrhea and chicken meat but at levels lower than those of its ortholog found in 2013 in Escherichia coli isolates in China (19, 32, 60). Nevertheless, our findings provide evidence for the potential spread of the blaCTX-M-123 gene, which was found at a high prevalence among chicken meat samples in China. The blaCTX-M genes are known to be carried on transmissible plasmids, facilitating their transmission between different reservoirs, such as Salmonella spp. and other Enterobacterales (14). This has important implications for understanding the transmission dynamics and for evaluating control measures targeting blaCTX-M dissemination between animals and humans.
Finally, we also tested for the co-occurrence of quinolone and ESBL resistance traits. Six QRDR point mutations, five in GyrA (S83Y, S83F, D87G, D87N, and D87Y) and one in ParC (S80I), were found in 166 of the 222 ESBL-producing Salmonella isolates from different sources, as previously determined (12, 16, 30). Overall, 47.1% of the ESBL-producing isolates recovered from patients with diarrhea, all of which produced ESBLs of the CTX-M type, also had QRDR amino acid substitutions. The prevalence of QRDR amino acid substitutions among ESBL-producing isolates from patients with diarrhea was consistent with that in previous reports from Thailand but is in contrast to the findings gathered from patients with diarrhea in Senegal, in which the frequency was much lower (14, 55). Notably, all 97 ESBL-producing isolates recovered from chicken meat samples had QRDR amino acid substitutions, similar to the data from Henan Province in China (30). To date, several investigations have tried to identify QRDR mutations in Salmonella isolates recovered from turtles, without success (61). To our best knowledge, our results represent the first evidence of QRDR amino acid substitutions in ESBL-producing Salmonella isolates recovered from turtle fecal samples. Furthermore, all 222 ESBL-producing Salmonella isolates were found to carry at least three of the PMQR genes, including oqxA, oqxB, qepA, qnrB, and qnrS. Of note, it is quite common to have the oqxA and oqxB genes on plasmids carrying genes encoding MDR, along with other resistance genes, such as ESBL-encoding genes (62). The qepA gene was previously detected in Salmonella spp. from patients with diarrhea in China, but it was absent from isolates collected from patients with diarrhea in this study (46). The copresence of the qnrB and qnrS genes, the oqxA and oqxB genes, and the qepA gene in a single Salmonella isolate is seldom reported in Europe (63). Worryingly, our findings suggest that the incidence of PMQR genes in ESBL-producing Salmonella isolates is increasing in China.
MLST revealed a total of 15 STs among the 222 ESBL-producing Salmonella isolates. ST17 (92/222) was the most prevalent sequence type. All ST17 isolates were serotyped as S. Indiana, and 90 of these 92 isolates were detected in chicken meat from Shandong and Jiangxi Provinces, 1 was from pork, and 1 was from a patient with diarrhea. Our data are consistent with previous findings obtained in China, showing that the ST17 S. Indiana isolates with the highest percentage of MDR are mainly recovered from chicken and that chicken is considered the major reservoir of the ST17 S. Indiana clone in China (59, 64). Significantly, all isolates cocarrying two of the blaCTX-M genes were serotyped as S. Indiana, Enteritidis, and Typhimurium, while 14 out of 21 ST26 S. Thompson isolates were found to carry none of the blaCTX-M genes. The MLST results showed that the S. Indiana, Enteritidis, and Typhimurium isolates may pose a serious public health risk.
To the best of our knowledge, we are the first to report the detection of Salmonella spp. harboring the blaCTX-M-130 gene from patients with diarrhea and QRDR mutations from turtle fecal samples. Furthermore, antimicrobial resistance affects the development of the world economy and threatens public health. Considering the very high ESBL prevalence in China, we strongly suggest that the government initiate both clinical and veterinary testing for ESBLs when resistance to the first-line β-lactams is detected in Salmonella spp. in order to improve monitoring and support the selection of effective treatments. Based on the concept of One Health, our study emphasizes the importance of a holistic working approach for the animal, human, environment, and related sectors. Specifically, our results stress the pressing need for investigating antimicrobial usage (AMU) as well as antimicrobial resistance (AMR) across the entire food safety chain and the establishment of a national AMU and AMR surveillance network system. The results for AMU and AMR obtained will provide some knowledge for public communication and education. Besides, the rational and prudent use of antimicrobials should be propagandized in the community, health care settings, and animal farms to control the dissemination of MDR, especially that conferred by CTX-M-type ESBLs, in Salmonella spp. at the national level.
MATERIALS AND METHODS
Study setting, sample collection, and bacterial strains.
From January 2014 to December 2015, a Salmonella control program was conducted in China to monitor Salmonella infections across different sources and regions. A total of 14,579 fresh fecal samples were collected from patients with acute diarrhea from 20 days to 81 years of age (5,515 patients ≤5 years of age, 6,654 patients from 5 to 59 years of age, and 2,410 patients ≥60 years of age) at the enteric clinic setting of 20 hospitals in the Chinese provinces of Beijing, Heilongjiang, Hubei, Jiangxi, and Shandong. Clinical information for each patient was extracted from the archived medical records. In parallel, 3,405 food samples of animal origin, including 1,237 chicken meat, 1,354 pork, and 814 aquatic products, were also collected from 20 supermarket outlets, including 10 big department stores and 10 local agriculture markets, across the five aforementioned Chinese provinces. Alongside these samples, 350 fresh pet fecal samples, including 290 turtle and 60 pigeon fecal samples, were also collected from 5 veterinary clinics across the aforementioned Chinese provinces. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Research Ethics Committee of China National Center of Food Safety Risk Assessment, Beijing, China (approval no. 2014003).
Both human and pet fecal samples (the weight of each sample was ≥1 g) were placed in a sterile tube, then placed in a box maintained at a temperature lower than 4°C, and then immediately transported to the laboratory and subjected to microbiological analysis within 2 h. Fecal samples were cultured by streaking on xylose-lysine-desoxycholate agar (HopeBio, Qingdao, China) and CHROMagar Salmonella spp. (CHROMagar Microbiology, Paris, France), followed by incubation at 36°C ± 1°C for 18 h to 24 h. Three suspected Salmonella colonies were streaked onto Trypticase soy agar (HopeBio, Qingdao, China) and further incubated at 37°C for 18 h.
The animal food samples (the weight of each sample was ≥250 g) were collected at each sampling site, and all were stored inside tightly sealed aseptic bags surrounded by a biological ice bag and then placed in a box maintained at a temperature lower than 4°C. The samples were also immediately transported to the laboratory and subjected to microbiological analysis within 2 h. All samples were subjected to qualitative analysis for Salmonella spp. using an enrichment method described by the National Food Safety Standard of China—Food Microbiological Examination, Salmonella spp. (method GB 4789.4).
Finally, confirmation that the isolates recovered from the fecal samples and food of animal origin were Salmonella spp. was done through biochemical and molecular methods. Biochemical characterization was done using API 20E test identification test strips (bioMérieux, Marcy l’Etoile, France), while for molecular confirmation, we performed a PCR assay targeting the invA gene (65). For all the confirmed Salmonella isolates, serovars were determined by the slide agglutination test, using Salmonella antisera (Statens Serum Institute, Denmark), according to the Kauffmann-White scheme. All isolates confirmed to be Salmonella spp. were stored in brain heart infusion broth with 40% (vol/vol) glycerol (HopeBio, Qingdao, China) at −80°C. Each sample retained one isolate.
Antimicrobial susceptibility testing.
The antimicrobial susceptibility of all Salmonella isolates was determined using the broth dilution method and Biofosun Gram-negative bacterial panels (Shanghai Biofosun Biotech, China) according to CLSI guidelines (66). Susceptibility to the following antimicrobials was assessed: ampicillin (AMP; 1 to 32 mg/liter), ampicillin-sulbactam (SAM; 0.25/0.125 to 32/16 mg/liter), ceftazidime (CAZ; 0.25 to 32 mg/liter), cefotaxime (CTX; 0.25 to 32 mg/liter), imipenem (IPM; 0.125 to 16 mg/liter), meropenem (MEM; 0.125 to 16 mg/liter), trimethoprim-sulfamethoxazole (SXT; 0.125/2.38 to 16/304 mg/liter), gentamicin (GEN; 0.25 to 32 mg/liter), tetracycline (TET; 0.25 to 32 mg/liter), ciprofloxacin (CIP; 0.03 to 64 mg/liter), nalidixic acid (NAL; 0.25 to 128 mg/liter), and chloramphenicol (CHL; 0.25 to 32 mg/liter). Confirmation of the presence of a carbapenemase was done by the agar dilution method, for which the results were expressed as the MIC values for imipenem and meropenem, followed by the Etest (bioMérieux, Marcy l’Etoile, France).
Salmonella isolates expressing resistance to cephalosporins (CAZ or CTX) were further screened to detect their production of ESBLs, which was done by a combination disc diffusion test with cefotaxime and ceftazidime discs with and without clavulanic acid (HopeBio, Qingdao, China) according to CLSI guidelines (66). Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were applied as reference strains in antimicrobial susceptibility tests (AST). All identified isolates were preserved in brain heart infusion broth (HopeBio, Qingdao, China) containing 40% (vol/vol) glycerol at −80°C for subsequent study.
DNA purification.
The identified ESBL-producing Salmonella isolates were incubated for 18 h to 24 h at 37°C in Luria-Bertani broth (HopeBio, Qingdao, China). A commercial bacterial DNA extraction kit (OMEGA D3350; Guangzhou, China) was used to extract pure genomic DNA from the bacterial culture. A Qubit (version 3.0) fluorometer (Thermo Fisher Scientific, NH, USA) was used to detect the quality of the DNA. DNA samples were diluted to a concentration of 50 mg/liter with sterile deionized water for subsequent PCR assay.
PCR and DNA sequencing.
Genomic DNA extracted from the ESBL-producing Salmonella isolates was further screened for the blaCTX-M gene cluster by PCR (67). In addition, all ESBL-producing Salmonella isolates were screened via PCR amplification for the presence of QRDRs (gyrA, gyrB, parC, and parE) and PMQR determinants [qepA, aac(6′)-Ib, oqxAB, and qnrABCDS] (68–74). All PCR products were commercially sequenced (Thermo Fisher Scientific China, Shanghai, China) and subsequently analyzed with DNAStar software (DNAStar Inc., Madison, WI, USA), and then the resulting DNA sequences were compared with reference sequences from NCBI by BLAST analysis.
MLST.
MLST of all ESBL-producing Salmonella isolates was performed following the protocols described at the MLST website (https://enterobase.readthedocs.io/en/latest/mlst/mlst-legacy-info-senterica.html). Seven conserved housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) of Salmonella enterica were amplified and sequenced at Thermo Fisher Scientific (China) Co. Ltd. (Shanghai, China). The sequences were submitted to the Salmonella MLST database website (http://mlst.warwick.ac.uk/mlst/dbs/Senterica) to assign the sequence types (STs).
Statistical analysis.
Statistical analysis was performed using SPSS (version 20.0; SPSS, Chicago, IL, USA) software. Differences between proportions were tested using the chi-square test. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Key R&D Program of China (grant 2016YFD0401102) and China Food Safety Talent Competency Development Initiative: CFSA 523 Program.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zheng B, Feng Y. 2019. MCR-1-producing Salmonella Typhimurium ST34 links animal foods to human community infections. EBioMedicine 42:10–11. doi: 10.1016/j.ebiom.2019.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reimschuessel R, Grabenstein M, Guag J, Nemser SM, Song K, Qiu J, Clothier KA, Byrne BA, Marks SL, Cadmus K, Pabilonia K, Sanchez S, Rajeev S, Ensley S, Frana TS, Jergens AE, Chappell KH, Thakur S, Byrum B, Cui J, Zhang Y, Erdman MM, Rankin SC, Daly R, Das S, Ruesch L, Lawhon SD, Zhang S, Baszler T, Diaz-Campos D, Hartmann F, Okwumabua O. 2017. Multilaboratory survey to evaluate Salmonella prevalence in diarrheic and nondiarrheic dogs and cats in the United States between 2012 and 2014. J Clin Microbiol 55:1350–1368. doi: 10.1128/JCM.02137-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian R, Im J, Lee JS, Jeon HJ, Mogeni OD, Kim JH, Rakotozandrindrainy R, Baker S, Marks F. 2019. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum Vaccin Immunother 15:1421–1426. doi: 10.1080/21645515.2018.1504717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu M, Cui S, Lin L, Xu B, Zhao J, Xia S, Deng W, Xie Z, Zhang J, Wang Z, Feng Z, Yang W, Ran L. 2013. Analysis of the aetiology of diarrhea in outpatients in 2007, Henan Province, China. Epidemiol Infect 141:540–548. doi: 10.1017/S0950268812000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). 2018. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 16:5500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Scallan E, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D'Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2019. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2015-2018. MMWR Morb Mortal Wkly Rep 68:369–373. doi: 10.15585/mmwr.mm6816a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okoro CK, Barquist L, Connor TR, Harris SR, Clare S, Stevens MP, Arends MJ, Hale C, Kane L, Pickard DJ, Hill J, Harcourt K, Parkhill J, Dougan G, Kingsley RA. 2015. Signatures of adaptation in human invasive Salmonella Typhimurium ST313 populations from sub-Saharan Africa. PLoS Negl Trop Dis 9:e0003611. doi: 10.1371/journal.pntd.0003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Z, Xu X, Yan M, Chang H, Li Y, Kan B, Zeng M. 2019. Salmonella Typhimurium and Salmonella Enteritidis infections in sporadic diarrhea in children: source tracing and resistance to third-generation cephalosporins and ciprofloxacin. Foodborne Pathog Dis 16:244–255. doi: 10.1089/fpd.2018.2557. [DOI] [PubMed] [Google Scholar]
- 9.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, Tolli R, D'Incau M, Staffolani M, Di Giannatale E, Hendriksen RS, Battisti A. 2015. Emergence of a clonal lineage of multidrug-resistant ESBL producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One 10:e0144802. doi: 10.1371/journal.pone.0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown AC, Chen JC, Watkins LKF, Campbell D, Folster JP, Tate H, Wasilenko J, Van Tubbergen C, Friedman CR. 2018. CTX-M-65 extended-spectrum β-lactamase-producing Salmonella enterica serotype Infantis. Emerg Infect Dis 24:2284–2291. doi: 10.3201/eid2412.180500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang CZ, Ding XM, Lin XL, Sun RY, Lu YW, Cai RM, Webber MA, Ding HZ, Jiang HX. 2019. The emergence of chromosomally located blaCTX-M-55 in Salmonella from foodborne animals in China. Front Microbiol 10:1268. doi: 10.3389/fmicb.2019.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moura Q, Fernandes MR, Silva KC, Monte DF, Esposito F, Dropa M, Noronha C, Moreno AM, Landgraf M, Negrão FJ, Lincopan N. 2018. Virulent nontyphoidal Salmonella producing CTX-M and CMY-2 β-lactamases from livestock, food and human infection, Brazil. Virulence 9:281–286. doi: 10.1080/21505594.2017.1279779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrois D, Breurec S, Seck A, Delauné A, Le Hello S, Pardos de la Gándara M, Sontag L, Perrier-Gros-Claude JD, Sire JM, Garin B, Weill FX. 2014. Prevalence and characterization of extended-spectrum beta-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin Microbiol Infect 20:O109–O116. doi: 10.1111/1469-0691.12339. [DOI] [PubMed] [Google Scholar]
- 15.Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D, Peixe L, Poirel L, Schuepbach-Regula G, Torneke K, Torren-Edo J, Torres C, Threlfall J. 2013. Public health risks of enterobacterial isolates producing extended spectrum beta-lactamases or AmpC beta-lactamases in food and food producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis 56:1030–1037. doi: 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 16.Hooper DC. 1999. Mechanisms of fluroquinolone resistance. Drug Resist Updat 2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Martinez L, Pascual A, Garcia I, Tran J, Jacoby GA. 2003. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother 51:1037–1039. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 18.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Jin H, Hu J, Yuan Z, Shi W, Ran L, Zhao S, Yang X, Meng J, Xu X. 2014. Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006–2010. Epidemiol Infect 142:826–832. doi: 10.1017/S0950268813001659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Z, Ke B, Deng X, Liang J, Ran L, Lu L, He D, Huang Q, Ke C, Li Z, Yu H, Klena JD, Wu S. 2015. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009–2012. BMC Infect Dis 15:53. doi: 10.1186/s12879-015-0784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu M, Lv B, Zhang X, Yan H, Huang Y, Qian H, Pang B, Jia L, Kan B, Wang Q. 2016. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhea in Beijing, China (2010-2014). Gut Pathog 8:31. doi: 10.1186/s13099-016-0116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng X, Ran L, Wu S, Ke B, He D, Yang X, Zhang Y, Ke C, Klena JD, Yan M, Feng Z, Kan B, Liu X, Mikoleit M, Varma JK. 2012. Laboratory-based surveillance of non-typhoidal Salmonella infections in Guangdong Province, China. Foodborne Pathog Dis 9:305–312. doi: 10.1089/fpd.2011.1008. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Xie X, Xu X, Wang X, Chang H, Wang C, Wang A, He Y, Yu H, Wang X, Zeng M. 2014. Nontyphoidal Salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis 11:200–206. doi: 10.1089/fpd.2013.1629. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Duan H, Zhang W, Li JW. 2007. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol Med Microbiol 51:8–13. doi: 10.1111/j.1574-695X.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 25.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. 2013. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg Infect Dis 19:1239–1244. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan H, Li L, Alam MJ, Shinoda S, Miyoshi S, Shi L. 2010. Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int J Food Microbiol 143:230–234. doi: 10.1016/j.ijfoodmicro.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 27.Zeng YB, Xiong LG, Tan MF, Li HQ, Yan H, Zhang L, Yin DF, Kang ZF, Wei QP, Luo LG. 2019. Prevalence and antimicrobial resistance of Salmonella in pork, chicken, and duck from retail markets of China. Foodborne Pathog Dis 16:339–345. doi: 10.1089/fpd.2018.2510. [DOI] [PubMed] [Google Scholar]
- 28.Doménech E, Jimenez-Belenguer A, Amoros JA, Ferrus MA, Escriche I. 2015. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in eastern Spain. Food Control 47:120–125. doi: 10.1016/j.foodcont.2014.06.043. [DOI] [Google Scholar]
- 29.Zadernowska A, Chajęcka-Wierzchowska W. 2017. Prevalence, biofilm formation and virulence markers of Salmonella sp. and Yersinia enterocolitica in food of animal origin in Poland. LWT Food Sci Technol 75:552–556. doi: 10.1016/j.lwt.2016.10.007. [DOI] [Google Scholar]
- 30.Bai L, Lan R, Zhang X, Cui S, Xu J, Guo Y, Li F, Zhang D. 2015. Prevalence of Salmonella isolates from chicken and pig slaughterhouses and emergence of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana in Henan, China. PLoS One 10:e0144532. doi: 10.1371/journal.pone.0144532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang B, Qu D, Zhang X, Shen J, Cui S, Shi Y, Xi M, Sheng M, Zhi S, Meng J. 2010. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int J Food Microbiol 141:63–72. doi: 10.1016/j.ijfoodmicro.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Fu Y, Xiong Z, Ma Y, Wei Y, Qu X, Zhang H, Zhang J, Liao M. 2018. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front Microbiol 9:2104. doi: 10.3389/fmicb.2018.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang XJ, Wu QP, Zhang JM, Huang JH, Chen L, Liu SR, Yu SB, Cai SZ. 2015. Prevalence, enumeration, and characterization of Salmonella isolated from aquatic food products from retail markets in China. Food Control 57:308–313. doi: 10.1016/j.foodcont.2015.03.046. [DOI] [Google Scholar]
- 34.Woodring J, Srijan A, Puripunyakom P, Oransathid W, Wongstitwilairoong B, Mason C. 2012. Prevalence and antimicrobial susceptibilities of Vibrio, Salmonella, and Aeromonas isolates from various uncooked seafoods in Thailand. J Food Prot 75:41–47. doi: 10.4315/0362-028X.JFP-11-211. [DOI] [PubMed] [Google Scholar]
- 35.Puah SM, Chua KH, Tan J. 2016. Prevalence of virulent resistant Salmonella enterica strains from sushi and sashimi samples in Malaysia. Trop Biomed 33:476–485. [PubMed] [Google Scholar]
- 36.Bouchrif B, Paglietti B, Murgia M, Piana A, Cohen N, Ennaji MM, Rubino S, Timinouni M. 2009. Prevalence and antibiotic-resistance of Salmonella isolated from food in Morocco. J Infect Dev Ctries 3:35–40. doi: 10.3855/jidc.103. [DOI] [PubMed] [Google Scholar]
- 37.Gambino-Shirley K, Stevenson L, Concepción-Acevedo J, Trees E, Wagner D, Whitlock L, Roberts J, Garrett N, Van Duyne S, McAllister G, Schick B, Schlater L, Peralta V, Reporter R, Li L, Waechter H, Gomez T, Fernández Ordenes J, Ulloa S, Ragimbeau C, Mossong J, Nichols M. 2018. Flea market finds and global exports: four multistate outbreaks of human Salmonella infections linked to small turtles, United States—2015. Zoonoses Public Health 65:560–568. doi: 10.1111/zph.12466. [DOI] [PubMed] [Google Scholar]
- 38.Söderlund R, Jernberg C, Trönnberg L, Pääjärvi A, Ågren E, Lahti E. 2019. Linked seasonal outbreaks of Salmonella Typhimurium among passerine birds, domestic cats and humans, Sweden, 2009 to 2016. Euro Surveill 24(34):pii=1900074. doi: 10.2807/1560-7917.ES.2019.24.34.1900074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoelzer K, Moreno Switt AI, Wiedmann M. 2011. Animal contact as a source of human non-typhoidal salmonellosis. Vet Res 42:34. doi: 10.1186/1297-9716-42-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres-Mejía AM, Blanco-Peña K, Rodríguez C, Duarte F, Jiménez-Soto M, Esperón F. 2018. Zoonotic agents in feral pigeons (Columba livia) from Costa Rica: possible improvements to diminish contagion risks. Vector Borne Zoonotic Dis 18:49–54. doi: 10.1089/vbz.2017.2131. [DOI] [PubMed] [Google Scholar]
- 41.Back DS, Shin GW, Wendt M, Heo GJ. 2016. Prevalence of Salmonella spp. in pet turtles and their environment. Lab Anim Res 32:166–170. doi: 10.5625/lar.2016.32.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong J, Zhang J, Xu M, Zhu C, Yu Y, Liu X, Kelly P, Xu B, Wang C. 2014. Prevalence and fimbrial genotype distribution of poultry Salmonella isolates in China (2006 to 2012). Appl Environ Microbiol 80:687–693. doi: 10.1128/AEM.03223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, Lathrop S, Tobin-D’Angelo M, Clogher P. 2012. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin Infect Dis 54:S472–S479. doi: 10.1093/cid/cis051. [DOI] [PubMed] [Google Scholar]
- 44.Basler C, Nguyen TA, Anderson TC, Hancock T, Behravesh CB. 2016. Outbreaks of human Salmonella infections associated with live poultry, United States, 1990–2014. Emerg Infect Dis 22:1705–1711. doi: 10.3201/eid2210.150765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). 2019. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J 17:5598. doi: 10.2903/j.efsa.2019.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan Z, Xu X, Gu Z, Meng J, Wufuer X, Wang M, Huang M, Chen J, Jing C, Xiong Z, Zeng M, Liao M, Zhang J. 2019. Molecular epidemiology and antimicrobial resistance of invasive non-typhoidal Salmonella in China, 2007–2016. Infect Drug Resist 12:2885–2897. doi: 10.2147/IDR.S210961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, Barzilay EJ, Emerging Infections Program NARMS Working Group . 2011. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother 55:1148–1154. doi: 10.1128/AAC.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). 2016. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2014. EFSA J 14:4380. doi: 10.2903/j.efsa.2016.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong J, Kelly P, Wang C. 2017. Prevalence and antimicrobial resistance of Salmonella enterica serovar Indiana in China (1984–2016). Zoonoses Public Health 64:239–251. doi: 10.1111/zph.12328. [DOI] [PubMed] [Google Scholar]
- 50.Hossain S, De Silva BCJ, Dahanayake PS, Shin GW, Heo GJ. 2019. Molecular characterization of virulence, antimicrobial resistance genes, and class one integron gene cassettes in Salmonella enterica subsp. enterica isolated from pet turtles in Seoul, Korea. J Exotic Pet Med 28:209–217. doi: 10.1053/j.jepm.2018.11.002. [DOI] [Google Scholar]
- 51.Hu YJ, He YY, Wang YR, Fanning S, Cui SH, Chen Q, Liu GH, Chen QX, Zhou G, Yang BW, Huang JL, Li FQ. 2017. Serovar diversity and antimicrobial resistance of non-typhoidal Salmonella enterica recovered from retail chicken carcasses for sale in different regions of China. Food Control 81:46–54. doi: 10.1016/j.foodcont.2017.05.031. [DOI] [Google Scholar]
- 52.Choi D, Chon JW, Kim HS, Kim DH, Lim JS, Yim JH, Seo KH. 2015. Incidence, antimicrobial resistance, and molecular characteristics of nontyphoidal Salmonella including extended-spectrum β-lactamase producers in retail chicken meat. J Food Prot 78:1932–1937. doi: 10.4315/0362-028X.JFP-15-145. [DOI] [PubMed] [Google Scholar]
- 53.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu F, Chen Q, Yu X, Li Q, Ding B, Yang L, Chen C, Qin Z, Parsons C, Zhang X, Huang J, Luo Y, Wang L, Pan J. 2011. High prevalence of extended-spectrum beta lactamases among Salmonella enterica Typhimurium isolates from pediatric patients with diarrhea in China. PLoS One 6:e16801. doi: 10.1371/journal.pone.0016801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luk-In S, Chatsuwan T, Pulsrikarn C, Bangtrakulnonth A, Rirerm U, Kulwichit W. 2018. High prevalence of ceftriaxone resistance among invasive Salmonella enterica serotype Choleraesuis isolates in Thailand: the emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. Choleraesuis isolates. Int J Med Microbiol 308:447–453. doi: 10.1016/j.ijmm.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 56.Doi Y, Iovleva A, Bonomo RA. 2017. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J Travel Med 24:S44–S51. doi: 10.1093/jtm/taw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen K, Chan EWC, Chen S. 2019. Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerg Microbes Infect 8:396–403. doi: 10.1080/22221751.2019.1585965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang WH, Lin XY, Xu L, Gu XX, Yang L, Li W, Ren SQ, Liu YH, Zeng ZL, Jiang HX. 2016. CTX-M-27 producing Salmonella enterica serotypes Typhimurium and Indiana are prevalent among food-producing animals in China. Front Microbiol 7:436. doi: 10.3389/fmicb.2016.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Zhang A, Yang Y, Lei C, Jiang W, Liu B, Shi H, Kong L, Cheng G, Zhang X, Yang X, Wang H. 2017. Emergence of Salmonella enterica serovar Indiana and California isolates with concurrent resistance to cefotaxime, amikacin and ciprofloxacin from chickens in China. Int J Food Microbiol 262:23–30. doi: 10.1016/j.ijfoodmicro.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 60.He D, Partridge SR, Shen J, Zeng Z, Liu L, Rao L, Lv L, Liu JH. 2013. CTX-M-123, a novel hybrid of the CTXM-1 and CTX-M-9 group β-lactamases recovered from Escherichia coli isolates in China. Antimicrob Agents Chemother 57:4068–4071. doi: 10.1128/AAC.00541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Silva BCJ, Hossain S, Wimalasena S, Pathirana H, Wendt M, Heo GJ. 2017. Quinolone susceptibility and genetic characterization of Salmonella enterica subsp. enterica isolated from pet turtles. Lab Anim Res 33:49–56. doi: 10.5625/lar.2017.33.2.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crofts TS, Gasparrini AJ, Dantas G. 2017. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol 15:422–434. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N, Bruneau M, Perrin-Guyomard A, Cerny T, De Frutos Escobar C, Guerra B, Schroeter A, Gutierrez M, Hopkins K, Myllyniemi AL, Sunde M, Wasyl D, Aarestrup FM. 2011. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother 66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 64.Cao Y, Shen Y, Cheng L, Zhang X, Wang C, Wang Y, Zhou X, Chao G, Wu Y. 2018. Combination of multilocus sequence typing and pulsed-field gel electrophoresis reveals an association of molecular clonality with the emergence of extensive-drug resistance (XDR) in Salmonella. Microbiol Res 207:170–176. doi: 10.1016/j.micres.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Malorny B, Hoorfar J, Bunge C, Helmuth R. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl Environ Microbiol 69:290–296. doi: 10.1128/aem.69.1.290-296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing, 25th ed M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 67.Xu L, Ensor V, Gossain S, Nye K, Hawkey P. 2005. Rapid and simple detection of blaCTX-M genes by multiplex PCR assay. J Med Microbiol 54:1183–1187. doi: 10.1099/jmm.0.46160-0. [DOI] [PubMed] [Google Scholar]
- 68.Ling JM, Chan EW, Lam AW, Cheng AF. 2003. Mutations in topoisomerase genes of fluoroquinolone-resistant Salmonella in Hong Kong. Antimicrob Agents Chemother 47:3567–3573. doi: 10.1128/AAC.47.11.3567-3573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamane K, Wachino J, Suzuki S, Arakawa Y. 2008. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob Agents Chemother 52:1564–1566. doi: 10.1128/AAC.01137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother 50:2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, Hooper DC. 2009. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother 53:639–645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cavaco LM, Hasman H, Xia S, Aarestrup FM. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother 53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.