Abstract
Whole genome sequencing (WGS) holds great potential as a diagnostic test. However, the majority of patients currently undergoing WGS lack a molecular diagnosis, largely due to the vast number of undiscovered disease genes and our inability to assess the pathogenicity of most genomic variants. The CAGI SickKids challenges attempted to address this knowledge gap by assessing state-of-the-art methods for clinical phenotype prediction from genomes. CAGI4 and CAGI5 participants were provided with WGS data and clinical descriptions of 25 and 24 undiagnosed patients from the SickKids Genome Clinic Project, respectively. Predictors were asked to identify primary and secondary causal variants. Additionally, for CAGI5, groups had to match each genome to one of three disorder categories (neurologic, ophthalmologic, connective), and separately to each patient. The performance of matching genomes to categories was no better than random but two groups performed significantly better than chance in matching genomes to patients. Two of the ten variants proposed by two groups in CAGI4 were deemed to be diagnostic, and several proposed pathogenic variants in CAGI5 are good candidates for phenotype expansion. We discuss implications for improving in silico assessment of genomic variants and identifying new disease genes.
Keywords: CAGI, SickKids, phenotype prediction, variant interpretation, whole genome sequencing data, pediatric rare disease
INTRODUCTION
Next generation sequencing (NGS) is a disruptive technology that provides more comprehensive tests and several fold higher diagnostic yields than conventional methods for diagnosing genetic disorders. Looking to the future, deploying NGS-based whole genome sequencing (WGS) as a first tier diagnostic test has the potential to revolutionize the diagnosis of genetic disorders, given that the diagnostic yield of WGS for children suspected of a Mendelian disorder currently averages over 40% and continues to increase with time (Clark et al., 2018; Scocchia et al., 2019). Nonetheless, the majority of patients who now undergo WGS after first-line genomic testing failed to yield an answer remain without a molecular diagnosis. This gap between the current performance of WGS and its ultimate potential as a diagnostic test is primarily due to the large number of undiscovered disease genes and our inability to assess the pathogenicity of most genomic variants.
WGS identifies ~3.8 million variants in the average individual (Shen et al., 2013) with over 650 million variants already cataloged in a small proportion of the world’s population (dbSNP Build 151, April 2018 release). Correctly assigning one or a few of these variants as the cause of disease in an individual suspected of a genetic disorder is a herculean task, in large part because the majority of rare and low frequency variants are of unknown clinical significance and non-coding variants are rarely classifiable (Giral, Landmesser, & Kratzer, 2018; Gloss & Dinger, 2018; Zhu, Tazearslan, & Suh, 2017). Beyond the sheer numbers of variants, the complexity of this endeavor is compounded by locus heterogeneity, in which pathogenic variants in multiple genes can yield overlapping phenotypes. Furthermore, many genetic disorders are likely oligogenic or polygenic in nature (Jordan & Do, 2018; Kousi & Katsanis, 2015).
Bioinformatics-guided analysis of clinical WGS data is essential to overcome these challenges. While there have been significant improvements in the detection and classification of single nucleotide variants, copy number variants, and other structural variation from WGS data, there is a critical need to substantially improve the accuracy and efficiency of computer algorithms designed to predict a patient’s phenotype from their genotype and distinguish a phenotype’s causal variant(s) from millions of others. Overcoming the current limitations of WGS variant interpretation will not only improve clinical diagnosis, it also will advance our understanding of the etiology of genetic disorders and facilitate the development of better therapeutics, which will ultimately lessen the burden of genetic disease.
The Hospital for Sick Children’s (SickKids) Genome Clinic Project was designed to pilot the diagnostic and predictive use of whole genome sequencing (WGS) in children (Bowdin, Hayeems, Monfared, Cohn, & Meyn, 2016). The Project’s first cohort involved testing the performance of WGS vs. diagnostic chromosomal microarray in 100 children referred to clinical geneticists for suspected genetic disease. The second cohort compared WGS against targeted panel sequencing in 103 children seen in pediatric specialty clinics. The initial diagnostic rates for WGS were 38% for the microarray cohort and 43% for the targeted panel cohort.
Genome Clinic patients that remained undiagnosed after clinical assessment of their WGS data form useful cohorts for trialing novel approaches to molecular diagnosis and gene discovery. In that regard, The Genome Clinic Project collaborated with the Critical Assessment of Genome Interpretation (CAGI) to create open bioinformatics challenges for CAGI4 and CAGI5. The challenge cohorts consisted of patients for whom previous diagnostic assessment of their WGS data had yielded no causal variants. The bioinformatics teams who participated were provided with demographic information and clinical descriptions for each patient, as well as assembled WGS data with variant calling for each genome.
The SickKids CAGI4 challenge involved 25 undiagnosed patients from the Genome Clinic microarray cohort (Stavropoulos et al., 2016). Bioinformatics teams were provided with linked WGS data and HPO-based clinical descriptions for each patient. The primary challenge task was to identify the causal genomic variant(s) responsible for the patient’s phenotype.
The SickKids CAGI5 challenge involved 24 undiagnosed patients from the Genome Clinic panel test cohort who were being evaluated for one of three disease categories: ophthalmologic disorders, neurologic disorders, or connective tissue disorders (Lionel et al., 2018). Importantly, unlike CAGI4, the WGS data were not linked to specific patients. Teams had three primary tasks: a) match each genome to one of the three broad disease categories (ophthalmologic, neurologic, or connective tissue disease); b) match each genome to a specific patient’s clinical phenotype; and c) propose one or more causal variants that would explain the selected patient’s disease phenotype. In addition, teams were encouraged to submit variants they considered secondary findings (pathogenic disease-causing variants not related to the patient’s current phenotype). Variants from genomes assigned by the teams to the correct patient as potentially causative of the phenotype were assessed and classified according to the American College of Medical Genetics (ACMG) guidelines (Richards et al., 2015). The results of the CAGI4 and CAGI5 challenges are presented here.
METHODS
Patient data
For CAGI5, the SickKids Genome Clinic project provided de-identified clinical phenotypic information and whole genome sequencing data for 24 cases that were selected from the SickKids Genome Clinic panel sequencing cohort. The 24 patients consisted of 13 girls and 11 boys, ranging from 3 to 18 years in age. Sequencing and data analysis were performed as described in Lionel et al., 2018. These 24 cases remained unsolved after initial screening by the project’s clinical molecular geneticists for plausible coding, splicing, non-coding, and structural variants. The challenge cohort consisted of 6 patients with ophthalmologic disorders, 7 with neurologic disorders, and 11 with connective tissue disorders (Supporting Information). Predictors were provided with the phenotypic descriptions as shared with the diagnostic laboratory.
In CAGI4, the SickKids challenge involved 25 children with a wide range of suspected genetic disorders who were referred for clinical genome sequencing, but remained unsolved after initial screening. Phenotypic data were provided by the referring physicians and entered into Phenotips, a Human Phenotype Ontology-based database (Girdea et al., 2013). Detailed information and description of these cases is provided in Stavropoulos et al., 2016 and Pal et al., 2017.
To model the clinical testing environment, phenotypic information was limited to that routinely obtained from clinicians prior to molecular testing, rather than from an iterative, genotype-driven assessment of the patient. The diversity of phenotypes in the dataset represents the range of clinical presentations typically seen in children referred for diagnostic evaluations in subspecialty clinics at SickKids. All patients in the CAGI cohorts were consented for sharing of de-identified genomic and phenotypic data with external research projects. The original Genome Clinic project and data sharing with CAGI were approved by the Research Ethics Board at The Hospital for Sick Children (REB Protocol #1000037726).
CAGI5 SickKids challenge format
The CAGI5 SickKids challenge was divided into several tasks. First, teams had to match genomes to a broad phenotype category (ophthalmologic, neurologic, or connective tissue disorder). Second, genomes had to be matched to individual patients based on their clinical phenotype descriptions. In addition, teams could report primary variant(s) underlying each prediction (i.e., diagnostic variants) and secondary variants predicted to confer high risk of other disorders not present in the clinical phenotypic description.
Groups were required to provide a probability (0–1; 0 = no match, 1 = match) that a genome sequence matched a broad phenotype class as well as a probability that it belonged to a specific patient. Each predicted probability of a match included a standard deviation indicating confidence in the prediction. Organizers provided a template file, which had to be used for submission. Up to six distinct submissions were allowed from each group.
Bioinformatics groups:
The CAGI5 SickKids challenge went live on the genomeinterpretation.org web site in December 2017 and submissions closed in April 2018. Seven bioinformatics groups provided a single submission, while one team (Group 6) provided two separate submissions. The CAGI4 SickKids challenge went live in December 2015 and submissions closed in February 2016. Four groups participated in this challenge.
Assessment for CAGI5 challenge
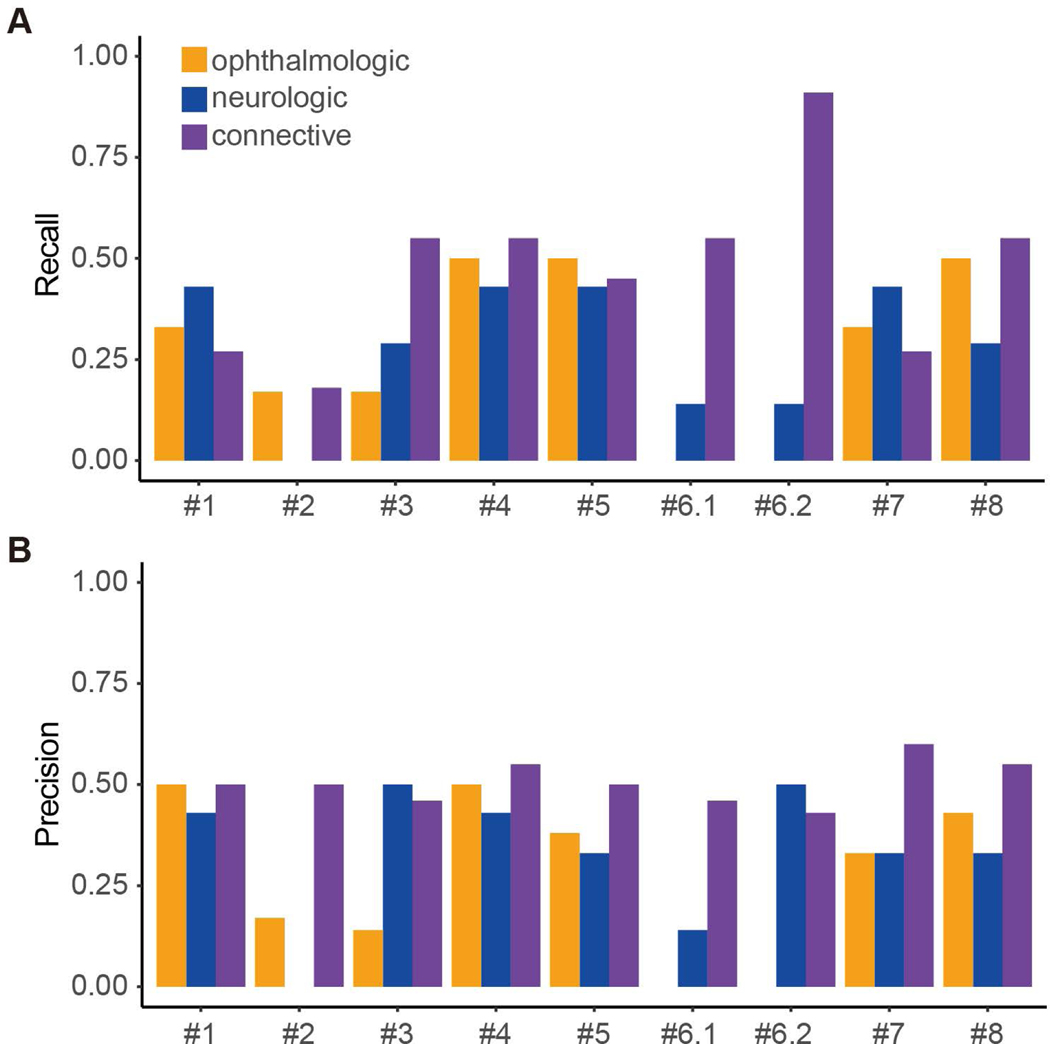
Predicted broad phenotype categories and specific phenotype-genotype matches in each submission were assessed against the SickKids answer key. Firstly, assessors calculated the number of correct predictions for broad phenotype categories. Only the highest probability predictions were included in the assessment. If probabilities for two categories were equal and one of them was correct, it was scored as correct (giving full credit to ties). The number of matches with no credit to ties are also shown. For probability-based assessment, probabilities were normalized in each submission for each genome to sum to 1.0. Mean probability assigned by the submissions to the correct disease category provides an assessment of assigned probabilities not dependent on whether the highest probability predictions were correct. For each disease category, recall and precision were defined as TP/(TP+FN) and TP/(TP+FP), respectively. Of note, SID#1, 2, and 7 did not provide predictions for all genomes (8, 8, and 6 genomes not predicted, respectively) (Supp. Table S1).
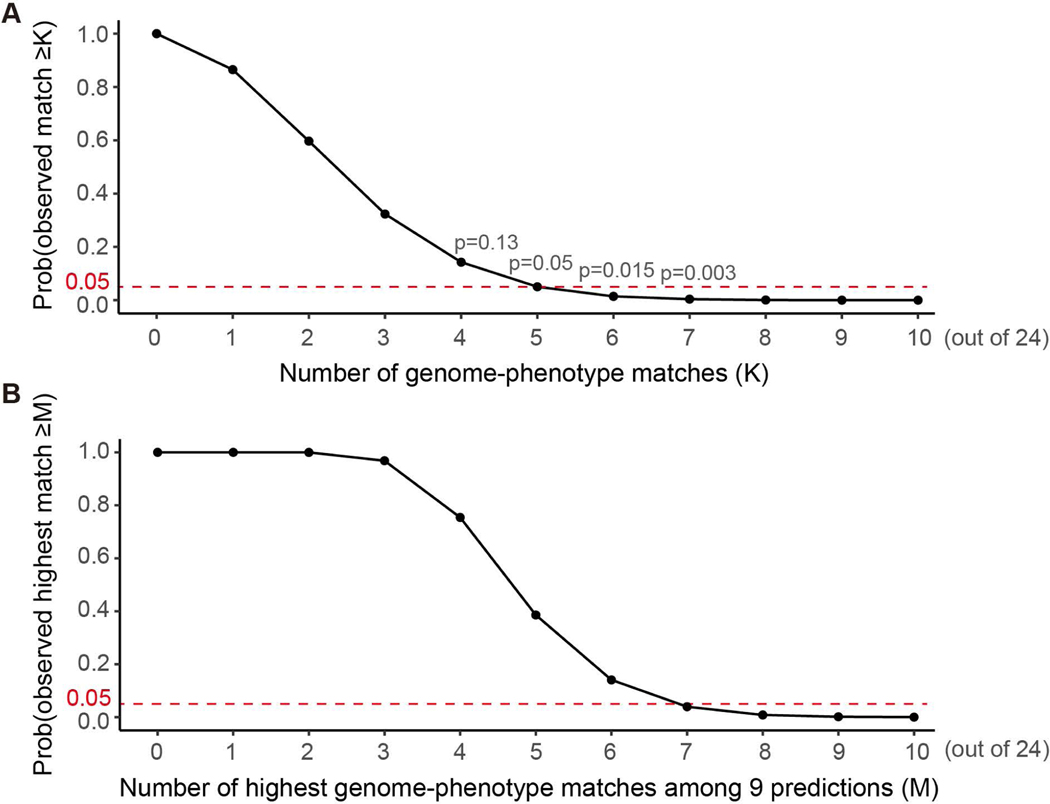
In order to assess the statistical significance of the submissions, random predictions were simulated 10,000 times (Figure 1A and C). Each time, disease categories were assigned for 6 ‘ophthalmologic’, 7 ‘neurologic’ and 11 ‘connective’ purely based on their composition, i.e., the probabilities we assign a genome ‘ophthalmologic’, ‘neurologic’ and ‘connective’ are 6/24, 7/24 and 11/24 respectively; then the numbers of both overall correct assignments and correct assignments for each category were calculated. Moreover, in order to evaluate all the submitted predictions as a community, we also simulated the random predictions from a community containing 9 independent predictions (to match the number of submissions in this challenge) (Figure 1B). Specifically, each time we simulated the highest match number of 9 random predictions (as described above) and this process was conducted 10,000 times.
Figure 1.
The expected number of genome-disease category matches inferred by in silico simulation based on composition of disease categories. (A) Distribution of overall number of matches. (B) Distribution of the highest match number observed in nine predictions (to simulate nine submissions of this challenge). (C) Distribution of the number of matches for each disease category.
The second part of the challenge was to match the phenotype information given for each patient to the correct genome. Probabilities were normalized in each submission for each genome to sum to 1.0. Only the highest probabilities were considered in the assessment. If probabilities for two phenotype descriptions were equal and one of them was correct, it was scored as correct. This only affected SID#3, which had 1 match instead of 2 if ties were not taken into account. Equal probability for ≥5 phenotype descriptions including the correct one was not considered as a match. SID#2, 5, and 7 did not provide predictions for 12, 12, and 3 genomes, respectively. Similar to the broad disease category matching, the random number of genome-phenotype matches was inferred by in silico simulation. The distribution was calculated from 10,000 simulation runs. Each time, we assumed that the 24 genomes were ordered as G=G1, G2,..,G24; a prediction can be treated as a random rearrangement of the above orders: P= Ga1, Ga2,..,Ga24, a1,a2,..,a24 ∈{1,2,…,24}; then the number of matched genomes was recorded. As sex information, which can be accurately inferred from genomes, was listed in the phenotypes (11 males and 13 females), we simulated the number of genome-phenotype matches considering sex. This was conducted by summing the match numbers of two independent simulations, which were similar to the process described above, while 11 and 13 genomes were included in each simulation respectively. In addition, to evaluate all the submissions as a community, we also simulated the highest number of matches from 9 independent predictions.
Phenotypic informational content scores for each patient were generated by PhenoTips from Monarch Initiative phenotypic profile analyses of the HPO terms contained in their supplied clinical description (Girdea et al., 2013). Correct genome-patient match scores were based on the number of highest probability matches for the correct genome with one match receiving one point.
Clinical assessment and classification of predicted variants
The proposed primary diagnostic and secondary variant(s) submitted by each group with correct genome patient matches were evaluated. Variants were classified as pathogenic, likely pathogenic, uncertain, likely benign, or benign according to ACMG diagnostic guidelines (Richards et al., 2015) by trained clinical genomic scientists. ACMG guidelines provide a framework for determining the level of evidence that a particular variant is a clinically actionable finding. The majority of information for variant classification was gleaned from VarSome (Kopanos et al., 2019). VarSome has links and information from the clinical variant curation database ClinVar (Landrum et al., 2018), the population database gnomAD (Karczewski et al., 2019), and references to relevant publications. The seventeen in silico predictions available in VarSome were also taken into consideration. The ACMG classification information in VarSome was not used to classify variants as they are generally not curated. Human Splicing Finder (Desmet et al., 2009) was used to determine the impact of variants on splicing.
Prediction methods
A detailed description of the methods used for the challenge accompanied each submission file. A brief summary of each CAGI5 prediction method is provided here, and detailed descriptions as well as CAGI4 methods are included in Supporting Information.
Group 1:
For phenotype matching, text mining for HPO terms (TPX software) was used, followed by manual QC. Gene prioritization was done by querying HANRD (Heterogeneous Association Network for Rare Diseases) and TPXRD (PubMed text mining) that give a set of ranked genes based on the input phenotype. Variant prioritization was achieved by using an in-house method (VPR). MAF (minor allele frequency), evolutionary conservation, in silico predictions, and ClinVar data were considered. Matching of genotypic to phenotypic case was done manually using the best possible intermediary disease.
Group 2:
Group 2 used eDGAR (Babbi et al., 2017) that collects known associations among genes and diseases, and PhenPath (Babbi et al., 2019), which groups diseases in terms of HPO terms and OMIM classifications and provides associations among phenotypes and genes, were used. For variant prioritization, SNPs&GO and UniProt were utilized. Sex of patients was also used to guide and validate the matching.
Group 3:
VCF files were analyzed using standard parameters, including variant quality, allele frequency, functional damage prediction and gene-phenotype associations, using a variety of tools and databases. Gender was considered in phenotype-genotype matches, but ethnic origin was not taken into account.
Group 4:
Group 4 used a phenotype-weighted subjective scoring of phenotypic profile (HPO and dbNSFP databases) together with gender information to guide matching. African ethnicity was also checked. The reasoning for choosing the strategy of phenotype-weighted scoring was to extract the pathogenic genetic information relevant to a particular profile out from each of the genomes. MAF, reported and predicted pathogenicity were considered.
Group 5:
Group 5 used an evolutionary action approach. In order to predict the disorder class for each individual, the predictors calculated the effect of the genetic variants on the fitness of each gene (evolutionary action). This fitness effect was used as the input of a diffusion process over a network of genes and diseases. The diffusion signal on each of the three disorder classes was used to calculate the probability of each genome to be linked to each disease. In order to match each individual’s genome to a clinical report, the predictors used again the diffusion process, and manual matching. Sex and ethnic origin information were also used.
Group 6:
Group 6 provided 2 separate submissions based on two different approaches. Clinical notes were searched against a gene-phenotype database (Monarch initiative for submission 6.1 and eRAM for submission 6.2), and the genes were sorted by the highest number of matching terms. Prediction of sex and ethnic origin was implemented. For variant prioritization, MAF filtering of protein altering variants was performed.
Group 7:
Group 7 utilized Ingenuity Variant Analysis (QIAGEN), which utilizes curated content from the literature as well as external databases. Genes known to be associated with patients’ phenotypes were selected. Phred score, MAF and ACMG classification (pathogenic and likely pathogenic) were taken into account.
Group 8:
To predict correspondence between phenotypes and genomes, group 8 used calculated scores for all genome-phenotype pairs and assigned the most likely connections using a bipartite matching algorithm. FunctionalFlow (Nabieva, Jim, Agarwal, Chazelle, & Singh, 2005) was used to predict risk genes with scores calibration based on the proportion of disease genes estimated by the AlphaMax algorithm (Jain, White, Trosset, & Radivojac, 2016; Jain, White, & Radivojac, 2016). MutPred2 (Pejaver et al., 2017) and MutPred-LOF (Pagel et al., 2017) were used to assign pathogenicity scores to variants. The final scores were assigned by combining gene scores and variant scores. Sex and ethnic origin information were also considered.
RESULTS
CAGI5 SickKids Challenge
The CAGI5 SickKids challenge was primarily designed to test how well bioinformatics algorithms are able (1) to match 24 genomes to three broad phenotype categories, and (2) to match each of the 24 genomes to a specific patient based on typical phenotype information. Groups could also identify diagnostic variants that underlie the predictions as well as secondary variants conferring high risk of other diseases. VCF files containing WGS data (SNVs and indels) from 24 patients and 24 unlinked phenotype descriptions were provided. Eight teams submitted predictions to this challenge (Table 1), with two distinct predictions from group 6.
Table 1.
A list of participating teams
| ID | Submission ID | PI |
|---|---|---|
| CAGI5 | ||
| Group 1 | SID#1 | Aditya Rao |
| Group 2 | SID#2 | Rita Casadio |
| Group 3 | SID#3 | Rehovot group |
| Group 4 | SID#4 | Lipika R. Pal/John Moult |
| Group 5 | SID#5 | Olivier Lichtarge |
| Group 6 | SID#6.1, SID#6.2 | Aashish Adhikari |
| Group 7 | SID#7 | Jennifer Poitras |
| Group 8 | SID#8 | Sean Mooney/Predrag Radivojac |
| CAGI4 | ||
| Group 9 | SID#9 | Chris Mungall |
| Group 10 | SID#10 | Julian Gough |
| Group 11 | SID#11 | Aditya Rao |
| Group 12 | SID#12 | Lipika R. Pal/John Moult |
Broad phenotype category matching
The first part of the CAGI5 SickKids challenge was to match 24 genomes to three broad phenotype categories: ophthalmologic (n=6), neurologic (n=7), or connective (n=11). Groups were allowed to give probabilities for one, two or all three categories for a given genome; however, it was noted in the challenge description that every genome sequence matches exactly one clinical phenotypic category. Table 2 shows which submissions provided the highest category probability to the correct genome. The highest probability was assigned to the correct genome only by 3.3 out of 9 submissions on average. Genome 81 had the correct category predicted in 8 out of 9 submission files, while genome 71 had the correct prediction in only one, SID#3 (Table 2). Intriguingly, connective tissue disorder was the correct category for both of these genomes.
Table 2.
Summary of the performance of all groups of matching the broad phenotype category to each genome and predicting which specific clinical description corresponds to which genome for CAGI5 challenge.
| Genome code | Patient code | Correct category | SID# with correct disease category match | SID# with correct genome-patient match |
|---|---|---|---|---|
| 7 | X | ophthalmologic | 7, 8 | 5, 7, 8 |
| 9 | W | ophthalmologic | 4, 5 | |
| 17 | H | ophthalmologic | 4, 5 | 4 |
| 18 | U | neurologic | 1, 3 | |
| 21 | G | neurologic | 5, 7 | 6.1 |
| 30 | R | neurologic | 1, 6.2 | |
| 39 | P | neurologic | 1, 4, 5, 6.1, 7 | |
| 42 | O | ophthalmologic | 3, 7 | 3 |
| 56 | N | connective | 3, 4, 5, 6.1, 6.2, 8 | 4, 8 |
| 57 | T | connective | 6.1, 6.2, 8 | |
| 67 | M | ophthalmologic | 1, 5, 8 | 1 |
| 68 | J | neurologic | 3, 8 | 3, 5, 8 |
| 71 | L | connective | 3 | 1, 5 |
| 76 | Q | connective | 3, 4, 5, 6.1, 6.2 | |
| 78 | V | connective | 1, 2, 3, 4, 6.2, 7, 8 | 6.2 |
| 79 | K | connective | 4, 5, 6.2 | |
| 81 | I | connective | 1, 2, 3, 5, 6.1, 6.2, 7, 8 | |
| 91 | E | neurologic | 4, 5, 7 | |
| 92 | S | connective | 1, 3, 4, 6.1, 6.2 | |
| 93 | F | connective | 4, 6.2, 7, 8 | 4, 8 |
| 95 | C | ophthalmologic | 1, 2, 4, 8 | 2, 4, 5 |
| 97 | D | connective | 5, 6.1, 6.2 | |
| 99 | B | neurologic | 4, 8 | 4, 8 |
| 102 | A | connective | 6.2, 8 | 8 |
The nine submissions reached an average accuracy of 37% when the category with the highest probability was considered (giving ties full credit). This accuracy weighted by the submitted probabilities was even lower (25%). SID#4 performed the best by assigning the correct category to 50% of the genomes (Tables 2, 3), whereas SID#5, 6.2 and 8 did not lag far behind by correctly predicting the broad phenotype category for 11 of the 24 (45.8%) genomes. SID#8 predicted the correct matches with a higher probability among the best-performing groups and overall had the highest mean probability (0.49) assigned to the correct disease category (Table 3). When giving no credit to ties, SID#4 and 8 both ranked first with 11 matches (Table 3) and the mean accuracy of all submissions was 33%. These aforementioned five submissions all considered gender and ethnic information to guide matching; however, the strategies used were rather different, from evolutionary action to phenotype-weighed subjective scoring of phenotypic profile (see Methods for details).
Table 3.
Summary of the performance of each group’s submission(s) in broad disease category matching for CAGI5 challenge
| Submission ID | Number of matches | Sum of probabilities for matches | Number of matches, no credit to ties | Mean probability assigned to the correct class |
|---|---|---|---|---|
| SID#1 | 8 | 7.0 | 7 | 0.44 |
| SID#2 | 3 | 3.0 | 3 | 0.25 |
| SID#3 | 9 | 4.7 | 9 | 0.34 |
| SID#4 | 12 | 7.6 | 11 | 0.42 |
| SID#5 | 11 | 5.2 | 9 | 0.36 |
| SID#6.1 | 7 | 2.7 | 7 | 0.35 |
| SID#6.2 | 11 | 7.8 | 9 | 0.40 |
| SID#7 | 8 | 6.0 | 6 | 0.39 |
| SID#8 | 11 | 9.9 | 11 | 0.49 |
In order to evaluate the statistical significance of the submissions, we simulated 10,000 random predictions based solely on the composition of disease categories (Figure 1A). A random prediction, on average, can correctly match 9 genomes (9/24=37.5%) to the corresponding category, and a submission would have to match at least 13 genomes to perform significantly better than random chance, as indicated by a p-value cutoff of 0.05. These results indicate that the submitted methods did not perform better than expected for random chance, as the average accuracy of the nine submissions was equal to the expected accuracy of a random prediction. There were equal numbers of submissions with accuracy higher or lower than 9, the median matches of random predictions, with none of the individual submissions performing significantly better than chance. Moreover, the simulation results showed that, the expected highest match number of nine random predictions was 12 (Figure 1B), the same as we observed here. Another strategy of a random prediction would be to give the highest prediction to the largest disease category (connective tissue), which would always result in an accuracy of 11 out of 24 (46%). Based on this, the average performance of the submissions was even lower than random chance.
Looking at the different disease categories, genomes belonging to the connective tissue category were the easiest to match with 47% of the genomes correctly assigned by all groups on average (Table 2). For ophthalmologic and neurologic disease categories, none of the groups performed significantly better than random. Only matching 4, 5, and >8 genomes for ophthalmologic, neurologic and connective tissue category respectively would achieve a p-value less than 0.05 (Figure 1C). SID#6.2 achieved the highest recall (0.91) in the connective tissue category by matching 10 out of 11 genomes correctly (Figure 2). This result is statistically significant compared to random prediction (Figure 1C); however, the submission had rather low precision (0.43) for the same category and failed to match any eye category genomes. Overall, most submissions achieved low true positive rates as well as low positive predictive values in this part of the challenge (Figure 2).
Figure 2.
(A) Recall and (B) precision values shown for each broad phenotype category by submissions.
Matching specific genomes to specific patients using phenotypic descriptions
The second part of the CAGI5 SickKids challenge was to use each patient’s phenotypic information to match the child to the correct genome. Groups provided as many probabilities as they wished, but only the highest probabilities were considered in the assessment (described in Methods). Table 4 shows which submissions provided the highest probability phenotype descriptions to the correct genome. On average only 1 out of 9 submissions made the correct genome to patient match (Table 2). Three genomes (7, 68, and 95) all had the most matches with the highest probability from three submissions, while 11 genomes (9, 18, 30, 39, 57, 76, 79, 81, 91, 92 and 97) were not matched by any group. Contrary to the broad phenotype matching, the ophthalmologic category was the easiest to predict correctly here: 83% of genomes were matched by at least one submission, compared to 43–45% for other categories.
Table 4.
Summary of the performance of each group’s submission(s) in the specific genome to patient matching
| Submission ID | Number of matches | Sum of probabilities for matches | Mean probability assigned to the correct class |
|---|---|---|---|
| SID#1 | 2 | 2.00 | 0.08 |
| SID#2 | 1 | 1.00 | 0.10 |
| SID#3 | 2 | 0.38 | 0.08 |
| SID#4 | 5 | 2.32 | 0.14 |
| SID#5 | 4 | 3.59 | 0.34 |
| SID#6.1 | 1 | 0.06 | 0.06 |
| SID#6.2 | 1 | 0.36 | 0.07 |
| SID#7 | 1 | 1.00 | 0.05 |
| SID#8 | 6 | 4.52 | 0.26 |
The nine submissions achieved a mean accuracy of 11% considering the highest probability predictions. If weighted by the submitted probabilities, the accuracy dropped to 7%. Groups 1, 3, 4, 6, and 8 each predicted the phenotype description correctly for one genome that no other submission predicted (Table 2). The matches that groups 2, 5, and 7 predicted correctly were also assigned correctly by at least one other submission. Most of the groups considered gender based on their method descriptions; however, only three submissions (SID#3, 4, and 8) made the accurate sex prediction for all 24 genomes (Supp. Table S2).
SID#8 ranked the first in this part of the challenge, having the highest number of matches to correct phenotype descriptions (6 genomes out of 24). As for the broad disease category predictions, this submission also assigned the highest mean probability (0.26) to the correct phenotype descriptions among the best-performing groups (Table 4). As noted previously, the groups did not perform better than chance for broad disease category matching, however two teams performed significantly better than chance for matching patients to genomes. 10,000 random predictions were again simulated purely based on the number of genomes (Figure 3) to assess the significance of the submitted predictions. Sex of the patients was also included in the simulation. A random prediction was found on average to correctly match 2 genomes (2/24=8.3%) to the corresponding patient, which is slightly lower than the average of all groups (2.6 genomes) (Figure 3A). If a submission matched at least 5 genomes, the performance would be significantly better than random chance (p-value ≤0.05). The assessment shows that SID#4 and 8 performed significantly better than expected for random chance by matching 5 and 6 genomes to correct phenotype descriptions, respectively. Additionally, the simulation results revealed that, the expected highest match number of nine random predictions was 4, which is lower than the predictors achieved, although not statistically significant (Figure 3B).
Figure 3.
The expected number of genome-phenotype matches inferred by 10,000 times in silico simulation. (A) Distribution of overall number of matches. (B) Distribution of the highest match number observed in nine predictions (to simulate nine submissions of this challenge). In the simulation, we assume that sex for all genomes can be predicted correctly.
Effect of phenotype informational content on genome-patient matching
Diagnostic laboratories routinely use phenotypic descriptions provided by clinicians to guide their assessment of variants in patients undergoing exome and genome sequencing. In this regard, there is a general assumption that the more detailed the clinical description, the more useful it is in aiding molecular diagnosis. To see if this applied to the CAGI5 SickKids assessments, we examined the relationship between the informational content of a patient’s phenotypic description and the number of times they were matched to the correct genome. We found that there was a modest correlation (R2=0.478, p>0.05) between the informational content of the clinical descriptions and number of correct matches of genome to ophthalmologic patients (Figure 4). In contrast, for both neurologic and connective tissue patients, there was almost no correlation between the informational content of the phenotypic descriptions and the number of correct genome-patient matches (R2=0.042 and R2=0.029 respectively). Although the numbers of patients were small, these results suggest that rich phenotypic descriptions may aid genome-patient matching for specific categories of disease.
Figure 4.
Correlation between the informational content of the patient’s clinical description and the number of correct genome-patient matches by all submissions.
Classification and evaluation of predicted diagnostic variants
The third part of the CAGI5 challenge was to submit variants predicted to be causative of the patient’s phenotype. The variants associated with the highest probability for the correct genome were evaluated and classified by trained clinical geneticists using the 2015 ACMG clinical interpretation guidelines (Richards et al., 2015). Consistent with these patients already having gone through a diagnostic laboratory assessment of their genomes (described in Methods) without finding any clearly causal variants, none of the variants proposed by the groups were deemed to meet clinical criteria for being returnable to the clinicians as the causal variants for the patients’ disorders. However, predicted variants classified as likely pathogenic and certain variants of unknown significance (VUS) could be included in a clinical report. These variants are discussed below with regards to the plausibility of their contributing to the patients’ phenotypes (Table 5). Predicted variants that were clinically classified as benign or likely benign are listed in Supp. Table S3.
Table 5.
Nominated diagnostic variants predicted with the highest probability for correct genome-patient matches
| Genome (patient) | Phenotype category | SID# | Genomic position (hg19) | Gene | Transcript | Nucleotide change | Protein change | Plausible gene for category? | Gene Phenotype Correlation | Classification |
|---|---|---|---|---|---|---|---|---|---|---|
| 7(X) | Ophthalmologic | 7 | 4:16024957:A:G | PROM1 | NM_001145849.1 | c.776T>C | p.Met259Arg | Yes | Poor | VUS |
| 67(M) | Ophthalmologic | 1 | 11:89017973:C:T | TYR | NM_000372.4 | c.1217C>T | p.Pro406Leu | Yes | Poor | Likely pathogenic |
| 68(J) | Neurological | 3 | 1:156105054:G:T | LMNA | NM_170707.3 | c.887G>T | p.Arg296Leu | Yes | Partial | VUS |
| 78(V) | Connective | 6.2 | 8:11615803:C:T | GATA4 | NM_002052.4 | c.1148C>T | p.Thr383Met | No | None | VUS |
| 93(F) | Connective | 4 | 22:41574743:A:G | EP300 | NM_001429.3 | c.7028A>G | p.Gln2343Arg | No | Poor | VUS |
| 99(B) | Neurological | 4 | 20:44044789:C:T | PIGT | NM_015937.5 | c.−8C>T | 5’UTR | Yes | Poor | VUS |
Importantly, while one or more groups identified potential variant(s) in 13 out of 24 genomes, none of the groups identified the same variant or gene as disease-causing in the same genome (Table 5 and Supp. Table S3.) In addition, while several candidate causal variants were associated with recessive disease, all proposed variants were heterozygous and no additional variants, excluding likely benign and benign variants, were nominated in the same genes. Of note, Group 5 did not participate in this part of the challenge and Group 2 did not predict a variant for its correct patient-genome match (genome 95/patient C). The performance of each group based on the laboratory geneticist evaluation (Methods) is described below.
Group 1:
Group 1 proposed the highest probability matches of patients and genomes for 71 (L) and 67(M). The variant predicted for genome 71 could not be evaluated clinically as it is an intergenic variant that is not associated with any known gene or disease state. Group 1 correctly classified genome 67 as having an ophthalmologic disorder and nominated a missense variant (c.776T>G, p.Pro406Leu) in the TYR gene as a potential causal variant. Pathogenic variants in this gene are known to be associated with a form of autosomal recessive oculocutaneous albinism (OCA type 1). The p.Pro406Leu variant has been reported in at least 5 homozygous and 7 compound heterozygous individuals with clinical features of OCA type 1 (ClinVar). In addition, while this variant has been identified in ~1% of the Finnish population (gnomAD, http://gnomAD.broadinstitute.org), it has been reported as pathogenic/likely pathogenic by multiple submissions in ClinVar (Variation ID: 3777). In vitro functional studies provide evidence that the p.Pro406Leu variant may impact protein function (Giebel et al., 1991; Spritz et al., 1997; Toyofuku, Wada, Spritz, & Hearing, 2001). Based on the above evidence, the laboratory geneticists classified this TYR variant as likely pathogenic.
However, while this variant is associated with an ophthalmologic phenotype, patient M’s phenotype of retinitis pigmentosa is not consistent with the current known phenotype spectrum of OCA type 1 or the biological function of gene product, tyrosinase. Additionally, as TYR-associated OCA is autosomal recessive, a single pathogenic TYR variant would not, by itself, be predicted to result in a disorder. Hence this variant would not be included in a clinical report as a causative variant but could be considered reportable as a carrier variant. Of note, this gene has a pseudogene complicating accurate variant calling and requiring orthogonal testing modalities.
Group 3:
Probability scores obtained by group 3’s method also yielded two genome patient matches: 68 (J), and 42 (O). The IFT140 gene proposed for patient O was a good fit for the patient’s phenotype of retinal dysfunction. However, the variant has a relatively high MAF (1.57% in the Finnish gnomAD cohort) yet is not a major cause of retinal disease in the Finns (Avela et al., 2018). It was classified therefore as benign.
The missense variant (c.887G>T, p.Arg296Leu) in LMNA identified for the neurologic patient J is very rare as it is not found in the gnomAD database nor described in the literature. It is predicted by multiple in silico algorithms to be damaging, but no functional evidence is available for this particular variant. Based on this, the variant was clinically classified as a VUS.
A structural analysis of the Arg296Leu variant, locates it in the coil 2 region of LMNA (residues 243–383). This substitution could potentially disrupt the intra-helical ion pair formation observed in intermediate filament coiled-coils, thereby leading to protein destabilization (Letai & Fuchs, 1995) and aggregation (Sylvius et al., 2008). Abnormal processing of LMNA can cause mitochondrial dysfunction (Bereziat et al., 2011), which was the working clinical hypothesis for patient J, and is thought to contribute to the variety of disease phenotypes observed in laminopathies (Sieprath et al., 2015). Additionally, lamin-A/C is required for osteoblastogenesis and bone formation in vivo, organ development and tissue differentiation (Zuela, Bar, & Gruenbaum, 2012), and has emerged as a regulator of the immune response (Gonzalez-Granado et al., 2014). These observations support additional clinical assessment of this variant, and encourage further investigations in a research setting.
Of note, the nominated gene is associated with at least 10, mostly dominant, genetic disorders affecting multiple organ systems and the patient has a complex phenotype that includes retinal/corneal dystrophies, extreme short stature with normal BMI, myopathy with abnormal mitochondria, chronic renal failure, cerebellar abnormalities, and type 1 diabetes. While the patient’s phenotype does not directly overlap with one of the known LMNA syndromes, the phenotypic heterogeneity of LMNA variants suggests further research investigation might be fruitful. E.g., the parents and the patient’s similarly affected identical twin could be assessed to determine if the variant is de novo in the affected twins. If true, then the twins could undergo careful reverse phenotyping in order to more fully assess the potential involvement of this LMNA variant in the patient’s clinical presentation and the possible delineation of a novel LMNA-related syndrome.
Group 4 (ref to Pal et al CAGI5 predictor paper):
The bioinformatics approach used by group 4 resulted in five correct genome to patient matches: 17 (H), 56(N), 93(F), 95(C), 99(B) and yielded candidate variants for each (Table 5, Supp. Table S3). Of note, ethnicity was stated for three of the five cases. In all five cases, the correct phenotypic category was also selected and in four of five instances the variant was in a gene that could possibly fit that category at the root level overlap of HPO terms. Two of the proposed variants were assessed as benign/likely benign and two were considered VUS.
Patient F was classified as having a connective tissue disorder and a novel missense variant (c.7028A>G, p.Gln2343Arg) in EP300 was identified as a potential disease causing variant. The amino acid glutamine at codon 2343 is conserved among species and the variant has not been previously reported in the general population (gnomAD). In silico analyses provide conflicting predictions of pathogenicity. While the rarity of the variant and its evolutionary conservation suggested possible pathogenicity, the variant was clinically classified as a VUS due to limited information and a lack of functional evidence. Additionally, pathogenic variants in EP300 are usually associated with Rubinstein-Taybi syndrome 1 (RSTS1, MIM# 180849). While several of the root level HPO terms of the Rubinstein-Taybi phenotype overlap with the patient’s described phenotype, the patient lacks the intellectual disability seen in RSTS1 as well as its distinctive dysmorphic features (e.g., broad thumbs/toes, arched eyebrows, down-slanting palpebral fissures, and a convex nasal ridge with low hanging columella). Neither does the patient’s phenotype fit the other known EP300-associated syndrome: Menke-Hennekam syndrome 2 (Menke et al., 2018). Hence, it is unlikely that this variant would be considered as clinically reportable based on the current knowledge.
As this is a novel variant, it is possible that this patient represents a new EP300-related syndrome. While the evidence for this hypothesis is currently lacking, a plausible molecular argument can be made. The p.Gln2343Arg residue change in histone acetyltransferase p300 (p300 HAT), the product of EP300, resides in the glutamine-rich (Q-rich) region of the C-terminal transactivation domain of the molecule. This region shares similarities with the Q-rich transcriptional activation domains found in a number of transcriptional activators (Kraus, Manning, & Kadonaga, 1999). Variable Q-rich repeats modulate transcription activity (Gemayel et al., 2015), so this variant may affect the transcriptional function of p300 HAT. Furthermore, recent research indicates strong support for a role of p300 HATin autophagy regulation in connective tissue (Kang, Sun, & Zhang, 2019; Leung et al., 2017; Sacitharan, Lwin, Gharios, & Edwards, 2018), so mutations in this gene could be implicated in connective tissue disorders. This variant is therefore another potential candidate for further research.
Group 4 also identified an intronic variant in PIGT c.−8C>T as potentially disease causing for patient B. Splicing algorithms (Human Splice Finder, http://www.umd.be/HSF) predict that this alteration may disrupt splicing. The variant’s MAF is low enough to be plausibly pathogenic (0.0015% in gnomAD) but no additional information could be found for this variant. Consequently, due to insufficient evidence, this variant was clinically classified as a VUS. Again, this is a VUS in a gene that fits the correct phenotype category. In this regard, germ line sequence variants in this gene are associated with an autosomal recessive multiple congenital anomalies-hypotonia-seizures syndrome (MIM# 615398). However, like the EP300 case above, the patient’s described phenotype lacks many of the HPO terms associated with the known disorder, making it an unlikely clinical diagnosis. In addition, the lack of a second pathogenic PIGT variant further diminishes the likelihood of being causative of this recessive disorder.
Group 6, SID#6.1 and 6.2:
Both of group 6’s submissions matched one genome to the correct patient. However, the predicted disease causing variant for SID#6.1 was clinically classified as benign due to a 2.57% MAF. SID#6.2 correctly matched genome 78(V) and proposed candidate variants in four genes (MYO1E, COL9A2, COL9A1, GATA4) for this patient, who had a clinical diagnosis of hypermobility type Ehlers-Danlos syndrome. One of the diseases associated with COL9A1 and COL9A2, Stickler syndrome, has a phenotype that includes both ophthalmologic and connective tissue components, while the other disease, epiphyseal dysplasia, could be considered a pure connective tissue disorder. The potential variants identified in these two genes were classified as likely benign. Given that the MYO1E and GATA4 are associated with focal glomerulosclerosis and cardiac malformations respectively, they would not be considered likely candidate genes for this patient’s Ehlers-Danlos syndrome. In addition, because of lack of information, the deep intronic variant identified in MYO1E was clinically classified as likely benign.
Group 7:
Group 7 proposed two candidate variants for a single ophthalmologic patient (7/X), whose phenotype included bilateral retinal hamartomas, nystagmus and severe myopia: a missense alteration in PROM1 (c.776T>G, p.Met259Arg) and a benign 5’UTR variant in GNAQ. Sequence variants in PROM1 have been associated with autosomal recessive retinitis pigmentosa (MIM# 612095) and autosomal dominant Stargardt disease (MIM# 603786). Of note, the amino acid methionine at codon 259 is not well conserved among species but this variant is novel, as it has not been previously observed in the general population (gnomAD) and in silico models predict the variant to be damaging. Based on this limited information and lack of functional evidence, the variant was clinically classified as a VUS. Importantly, because hamartomas have yet to be associated with PROM1 variants, PROM1 would not be considered a high priority candidate for assessment as a disease gene.
Group 8:
The computational algorithms used by group 8 to match genomes to patients yielded six matches: 7(X), 56(N), 68(J), 93(F), 99(B), 102(A). However, all of the variants predicted to be associated with the patient phenotypes were classified as benign or likely benign, again, primarily due to high allele frequencies in the general population. In four matches, the selected variants fell in genes that could be considered to match the phenotype category; 56(N) TNXB connective, 68(J) ATM neurologic, 99(B) SLC25A22 neurologic, and 102(A) COL5A2 connective. In addition, for all but 68(J), the proposed disease gene could plausibly explain the patient’s phenotype, particularly if one allowed for some phenotypic expansion.
Variants of interest not matched to the correct patient
During the clinical assessment of the nominated variants it became clear that matching genomes to patients based on the information in the phenotype descriptions was difficult at best. Therefore we examined the highest probability incorrectly matched variants and evaluated the ones identified in more than one submission for a given genome. Only variants classified as VUS or above were considered. This yielded a number of interesting variants discussed below (Table 6).
Table 6.
Highest probability, incorrectly matched variants identified by more than one submission.
| SID# (incorrectly attributed patients) | Correct genome (patient) - Category | Genomic position (hg19) | Gene | Disorders | Transcript | Nucleotide change | Protein change | gnomAD frequency | Classification |
|---|---|---|---|---|---|---|---|---|---|
| 2(97-O); 3(97-O); 4(97-W); 8(67-C) | 97(D) -Connective, 67(M) - Eye | 7:128040571:G:A | IMPDH1 | Retinitis pigmentosa 10, Leber congenital amaurosis 11 | NM_000883.4 | c.602C>T | p.Thr201Met | 0.002% (0.013% in AA) | VUS |
| 3(78-S); 4(78-Q); 6.1(78Q) | 78(V) - Connective | 7:94049587:C:T | COL1A2 | Ehlers-Danlos syndrome, Osteogenesis imperfecta | NM_000089.3 | c.2122C>T | p.Arg708Trp | 0.001% | VUS |
| 6.1(9-V); 6.2(9-V) | 9(W) - Eye | 1:2235335:C:T | SKI | Shprintzen-Goldberg syndrome | NM_003036.4 | c.1268C>T | p.Pro423Leu | 0.001% (0.002% EU) | VUS |
| 1 (95-H&M); 6.2(95-M) | 95(C) - Eye | 11:76905405:G:A | MYO7A | Usher syndrome, type 1B | NM_000260.4 | c.4159G>A | p.Asp1387Asn | 0.001% (0.007% AA) | VUS |
| 6.1(39-K); 6.2(39-R) | 39(P) - Neurologic | 15:49089695:G:A | CEP152 | Seckel syndrome 5, primary microcephaly 9 | NM_001194998.2 | c.343C>T | p.Arg115Ter | 0.0004% (0.003% Lat.) | Pathogenic |
| 6.1(79-F); 6.2(79-F) | 79(K) - Connective | 15:90174856:T:C | KIF7 | Acrocallosal syndrome, Joubert syndrome 12 | NM_198525.3 | c.2981A>G | p.Gln994Arg | 0.165% (0.306% Lat.) 2 homozygous | VUS |
| 3(21-V); 4(21-T) | 21(G) - Neurologic | X:153579297:T:C | FLNA | FLNA-related disorders | NM_001110556.2 | c.7136A>G | p.Tyr2379Cys | none | VUS |
Variants in this table may have been identified in additional submissions, but only highest probability variants per genome were considered (3 equal matches allowed).
A rare missense VUS (c.602C>T, p.Thr201Met) in IMPDH1 was identified by groups 2, 3, 4, and 8 in genome 97 and 67, although this variant was not present in the VCF file of genome 67. IMPDH1 is associated with autosomal dominant retinitis pigmentosa (MIM #180105) which is consistent with retinitis pigmentosa found in patient M. This variant was incorrectly attributed to other patients (O, W, and C) all of which had eye phenotypes that could be considered overlapping with retinitis pigmentosa. However, the p.Thr201Met is found in six heterozygous individuals in gnomAD. This number of supposed healthy adult individuals in gnomAD would exclude most variants from clinical consideration in the case of a highly penetrant autosomal dominant disease such as retinitis pigmentosa indicating that if by chance this variant is pathogenic, other factors must be at play.
For genome 78, a missense variant in the COL1A2 gene (c.2122C>T, p.Arg708Trp) was identified as a potential candidate in SID# 3, 4, and 6.1. The correct patient match for genome 78 is patient V, who has hypermobility type Ehlers-Danlos syndrome, and variants in COL1A2 are associated with autosomal dominant Ehlers-Danlos syndrome (MIM# 120160). Ehlers-Danlos syndrome was also the primary phenotype for patients S and Q to which this genome was incorrectly attributed. The amino acid arginine at codon 708 is highly conserved among species and most in silico algorithms predict the variant to be damaging. This particular variant has been observed in the general population at a frequency of 0.0012% (gnomAD) and was clinically classified as a variant of uncertain significance (VUS) in agreement with ClinVar. As a VUS, this variant might be included in a clinical report depending on how the population frequency and penetrance of disease are considered by the individual diagnostic laboratory, but would provide little impetus for clinical action. Hypermobility type Ehlers-Danlos syndrome is likely an under-reported phenotype and variable phenotypic presentation may be dependent on many factors, which means that three heterozygous individuals in gnomAD should definitely not exclude this variant as a possible cause of disease. However, it would be difficult to resolve the VUS status of the variant without additional evidence from functional, case/control, or segregation studies.
Group 6 selected a SKI missense variant (c.1268C>T, p.Pro423Leu) in both submissions for genome 9. This variant was clinically assessed to be a VUS, mainly due to limited information. Pathogenic alterations in SKI cause Shprintzen-Goldberg syndrome, a severe, congenital, mainly connective tissue disorder. Patient W, who is the correct match for genome 9, has an ophthalmologic phenotype rather than Shprintzen-Goldberg syndrome. While the penetrance of Shprintzen-Goldberg syndrome due to SKI alterations is not known, most SKI alterations are de novo (Greally, 1993), which is consistent with it being a severe dominant congenital disorder. Consequently, the presence of two heterozygous carriers of this variant in gnomAD casts serious doubts that this alteration could cause Shprintzen-Goldberg syndrome. This variant was incorrectly attributed to patient V, a child with primarily an Ehlers-Danlos syndrome phenotype, but with other features that could have been consistent with Shprintzen-Goldberg syndrome had this variant belonged to patient V.
Usher syndrome is an autosomal recessive disease characterized by retinitis pigmentosa and sensorineural hearing loss. Usher syndrome 1B is caused by pathogenic alterations in MYO7A, which account for over half of all Usher syndrome cases (Lentz & Keats, 1993). SID#1 and 6.2 selected a p.Asp1387Asn MYO7A alteration for genome 95, which belongs to patient C and was clinically classified as VUS due to lack of information. Interestingly, patient C has vision problems that could be consistent with Usher syndrome and also had hearing issues and speech delay, but the hearing issues have reportedly resolved. While patient C has some possible phenotype overlap with Usher syndrome the MYO7A variant would only account for one allele and a second candidate variant was not identified in C’s genome for this recessive disorder. Hence, this variant is unlikely to be causative. Of note, this variant was incorrectly attributed to two other retinitis pigmentosa patients, H and M, neither of whose phenotypic descriptions included hearing issues.
Group 6 selected a loss of function alteration (c.343C>T, p.Arg115Ter) in CEP152 in both submissions for genome 39. As a loss of function alteration, the p.Arg115Ter alteration was clinically classified as pathogenic and loss of function alterations in CEP152 cause two autosomal recessive syndromes: primary microcephaly 9 and Seckel syndrome 5 (MIM# 613529). Patient P, the patient associated with genome 39, has a neurological phenotype and the known CEP152 related diseases encompass neurological phenotypes, so the gene is plausible for the broad neurologic category, but patient’s overall clinical picture does not overlap well with the two diseases associated with CEP152. Patient P’s primary phenotype, epileptic encephalopathy, is not a feature of either primary microcephaly 9 and Seckel syndrome 5. In addition, patient P does not have severe microcephaly, a constant feature of the known CEP152-associated disorders and a second disease-causing CEP152 allele was not identified. The CEP152 variant was incorrectly attributed to patients K and R. Patient K was considered to belong to the connective tissue category and has little if any phenotype correlation with CEP152 related diseases. Patient R belonged to the neurologic category with some limited phenotype correlation with CEP152 related diseases. However, neither patient K nor R have severe microcephaly.
For both submissions, group 6 identified a missense variant (c.2981A>G, p.Gln994Arg) in the KIF7 gene for genome 79. Genome 79 belongs to patient K, who has a clinical diagnosis of Ehlers-Danlos syndrome and possibly ADHD (attention-deficit/hyperactivity disorder). There are two 2 homozygous individuals in gnomAD and MAF’s for this variant range as high as 1/320, casting doubt on its ability to cause any disease. In addition, the disorders associated with KIF7 are not consistent with patient K’s phenotype. However, due to conflicting information, it was classified as a VUS (ClinVar variation ID: 194572). Of note, this variant was incorrectly attributed to patient F, most likely because F’s neurological findings (seizures and hearing impairment, motor delay) and Ehlers-Danlos syndrome characteristics partially overlap with those seen in KIF7-associated disorders.
Group 3 and 4 predicted a FLNA missense variant (c.7136A>G, p.Tyr2379Cys) for genome 21 but incorrectly attributed this genome to patient V and T respectively. The p.Tyr2379Cys alteration is listed in VarSome, but was not found in any other control or disease database that was searched. Patient G, the matching patient for genome 21, is a female with infantile epileptic encephalopathy and global developmental delay. As with LMNA, FLNA variants are associated with a broad range of diverse phenotypes (MIM# 300017). The FLNA phenotype with the best phenotype correlation with patient P is periventricular heterotopia 1, an X-linked dominant disorder, which is associated with refractory seizures (MIM# 300049). However, MRI abnormalities reported in patient G (delayed myelination and thin corpus callosum) are not consistent with reported brain abnormalities in periventricular heterotopia 1. In addition, the majority of FLNA variants associated with periventricular heterotopia are LOF variants, not missense variants. Patient V and T both were considered to belong to the connective tissue category due to a clinical suspicion of Ehlers-Danlos syndrome, but also had histories of developmental delays/learning disabilities. Because this combination of features is suggestive of case reports of FLNA variants causing an Ehlers-Danlos/periventricular heterotopia phenotype, it likely drove the misattribution of genome 21 to patients V and T.
Secondary variants
Three groups (1, 4 and 7) submitted secondary predictive variants that confer high risks for other diseases whose phenotypes were not reported in the clinical descriptions (see Supp. Table S4). 35 distinct variants were submitted, 8 of which were clinically classified as pathogenic, 4 as likely pathogenic, 16 VUS, 4 likely benign, and 3 benign alterations. Overall, the variants chosen as secondary findings were much more likely to be truly pathogenic than the predicted primary diagnostic variants. This task was also easier as it did not require any genotype-phenotype matching; however, secondary variants do require a higher burden of proof for inclusion in a clinical report. Only variants that provide strong evidence that a person will develop a disease are reported, thus VUS alterations are not reported clinically. Of the eight pathogenic variants, seven are associated with autosomal recessive disorders and would only be considered as clinically reportable as heterozygous carrier variants. A pathogenic variant in G6PD was identified but would not be reported by all clinical laboratories, as it is a low penetrance gene and it causes favism, a very mild disorder.
Only one alteration represented a clearly reportable clinical finding, a MSH2 splice alteration reported several times in ClinVar as a likely pathogenic variant, predicted by SID#1 and 4 in genome 91 and by SID#4 also in genome 81. Of note, this variant is absent from gnomAD and is not a common pathogenic MSH2 variant, yet it was present in 2/24 CAGI5 patients. Such a coincidence raises the possibility of sequencing artifacts requiring Sanger verification, particularly since this was a one base pair insertion. If verified by confirmatory clinical sequencing, this c.942+2delT alteration would confer a high risk of Lynch syndrome and its associated cancers (MIM# 12434).
CAGI4 SickKids challenge - solely variant prediction
The CAGI4 SickKids challenge took place in 2016 and focused on identifying diagnostic variants and predictive secondary variants in unsolved cases, without the need for genome to phenotype matching. This challenge involved 25 children with a wide range of suspected genetic disorders who were referred for clinical genome sequencing, but remained unsolved after initial analysis (Pal et al., 2017; Stavropoulos et al., 2016). Predictors were given patients’ WGS data and clinical phenotypic descriptions in the form of Phenotips annotations, based on HPO terms. Four teams participated in this challenge (Table 1); two of them took part also in the CAGI5 SickKids challenge described above (groups 11 and 12).
Altogether 191 potential diagnostic variants were proposed for 25 patients, including 60 variants in genes not associated with OMIM disease phenotypes. The majority were variants for dominant disorders and no structural variants were proposed. Of note, half of the variants proposed by SID#10 and over 20% of the diagnostic variants proposed by SID#12 had MAF >1%, rendering them unlikely to be causal variants. Although variants generally were chosen for their ability to explain the patient’s phenotype, for the majority, the phenotypic abnormalities associated with the variant only partially overlapped with those of the patient. In many of these cases, the degree of mismatch between predicted phenotype and patient’s phenotype was sufficient to make causality highly unlikely. E.g., a novel MECP2 missense variant (p.Glu55Lys) was proposed by SID#9 and SID#11 for patient 1041, who presented with microcephaly, hypotonia, and developmental delays (Table 7). While these terms are also seen in Rett syndrome (the condition associated with MECP2), they were present at birth in the child, while girls affected by Rett syndrome appear normal at birth and only become symptomatic with age.
Table 7.
Potential diagnostic variants predicted by multiple submissions in CAGI4.
| Patient ID | SID# | Genomic position (hg19) | Gene | Transcript | Nucleotide change | Protein change |
|---|---|---|---|---|---|---|
| 1011 | 12, SickKids | 17:48266777:AGGGCCAGG:A | COL1A1 | NM_000088.3 | c.2782_2789delCCTGGCCC | p.Pro928CysfsTer10 |
| 1014 | 9, 11 | 13:100623343:T:A | ZIC5 | NM_033132.4 | c.587A>T | p.Asp196Val |
| 1024 | 12, SickKids | 16:3778350:TG:AA | CREBBP | NM_004380.2 | c.6697_6698delCAinsTT | p.Gln2233Leu |
| 1025 | 12, SickKids | 5:36985083:AA:A | NIPBL | NM_133433.4 | c.1808dupA | p.Ser604ValfsTer2 |
| 1028 | 9, 11 | 7:155595841:C:A | SHH | NM_000193.4 | c.1142G>T | p.Arg381Leu |
| 1041 | 9, 11 | X:153297872:C:T | MECP2 | NM_004992.3 | c.163G>A | p.Glu55Lys |
| 1060 | 9, 12, SickKids | 9:140648627:C:T | EHMT1 | NM_024757.5 | c.1253C>T | p.Ser418Leu |
| 1064 | 11, 12 | 11:6654044:G:A | DCHS1 | NM_003737.4 | c.2699C>T | p.Thr900Met |
| 1076 | 9, 11 | 11:47470494:T:A | RAPSN | NM_005055.5 | c.23A>T | p.Gln8Leu |
| 1083 | 9, 11 | 11:118307298:G:C | KMT2A | NM_001197104.1 | c.71G>C | p.Arg24Pro |
| 1099 | 9, 12 | 5:88056849:T:TT | MEF2C | NM_001193347.1 | c.412dupA | p.Ile138AsnfsTer2 |
| 1105 | 9, 11 | 10:112361827:A:T | SMC3 | NM_005445.3 | c.2996A>T | p.Lys999Met |
| 1106 | 11, 12 | 20:47991474:A:G | KCNB1 | NM_004975.4 | c.623T>C | p.Leu208Pro |
For CAGI4, most variants were predicted by a single submission only. However, 10 variants were nominated for the same patients by more than one group (Table 7). SID#9 and 11 had the largest overlap (six variants), this is somewhat expected as they both used Exomizer for variant prioritization. In four cases, the predictor’s candidate variant also had been classified as a candidate variant by the SickKids bioinformatics pipeline but discarded upon initial manual review by the SickKids Genome Clinic diagnostic team. Nomination by the CAGI4 participants prompted reassessment of these variants by the SickKids Genome Clinic team, which performed validation and reverse phenotyping for several of the proposed causal variants. For patient 1024, the predicted variant was in a gene that did not fit the clinical phenotype and for patient 1025, the gene was a good phenotypic fit but the variant did not validate by Sanger sequencing (both were predicted by SID#12 and the SickKids bioinformatics pipeline). Prioritized variants for patients 1011 (by SID#12 and SickKids) and 1060 (by SID#9, 12, and SickKids) were located in genes that had partial overlap with the clinical phenotype and were successfully validated. In two instances (patients 1105, 1106) when two teams picked the same variant (Table 6), the patient’s referring clinical geneticist re-assessed the patient in light of the proposed disease gene and concluded that it was a good fit for the patient’s phenotype, meaning that the CAGI participants provided a clinical diagnosis for these two cases (Pal et al., 2017).
In addition to primary diagnostic variants, three groups also predicted secondary findings. No variants were proposed by more than one team. Variants submitted by SID#10 and 11 did not fulfill the 2015 ACMG criteria for pathogenic or likely pathogenic variants and would hence not be returned as medically actionable. Four of the variants predicted by SID#12 had also been picked by the SickKids diagnostic assessment team as potentially medically actionable (data not shown). Of note, three of them were discarded due to low read depth and two did not validate by Sanger sequencing. This left only one variant in FBN1 (c.G1027A, p.Gly343Arg) that has conflicting interpretations in ClinVar (Variation ID: 161244), meaning that ultimately none of the nominated predictive secondary variants were reportable clinical findings.
Discussion
One of the greatest barriers to fully realizing the potential of genomic medicine to transform clinical practice is variant interpretation. While current technologies allow us to identify the vast majority of variants in the human genome, we can only interpret the phenotypic and clinical significance of a few. This is due to lack of knowledge about the impact/pathogenicity of variation in most parts of the human genome as well as insufficient clinical descriptions or per contra heterogeneity/diversity of the phenotype under examination. This calls for a multidisciplinary approach that enlists computational biologists, clinical experts and research geneticists to tackle these challenges. The CAGI SickKids challenges were designed to begin to address this complex problem.
CAGI4 and CAGI5 participants were provided with WGS data and phenotype descriptions for 25 and 24 patients respectively who had remained without a diagnosis after evaluation by the SickKids Genome Clinic project (Lionel et al., 2018; Stavropoulos et al., 2016). Participants were asked to predict primary and secondary causal variants. Additionally, for CAGI5, groups had to match each genome to one of three categories of disease (neurologic, ophthalmologic and connective), and separately to each patient.
A single category does not provide enough information to distinguish a genome
For the first task in CAGI5 SickKids challenge, matching genomes to one of the three clinical categories, groups performed no better than random prediction (Figure 1), assigning the correct phenotype category to an average of nine genomes. Only a third of all genomes were correctly matched by more than three submissions. Although half of the submissions had a higher accuracy than the nine matches expected of a random prediction, the results were not statistically significant. A comparison of the accuracy of predictions in each broad disease category showed that the connective tissue category was slightly easier to match, with one submission (SID#6.2) achieving a significantly better result compared to 10,000 random simulations by matching 10 out of 11 genomes correctly. However, this result would still rank lower than the random prediction where the highest probability would always be given to the connective tissue category (11 out of 24 genomes or 46%). This challenge was made all the more difficult by the presence of patients with complex phenotypes who had been assigned to a specific clinical category based on their most prominent clinical features, but had clinical features belonging to two or more categories. These results indicate that classifying patients into a single category may not provide sufficient information to distinguish a genome, or that there is insufficient knowledge about genomic variation to segregate genomes into broad categories.
Thorough understanding of the phenotypic descriptions leads to success in the specific genome to patient matching
From the outset, one of the greatest obstacles for the CAGI5 SickKids challenge was to match a genome to a patient. The only information that could be accurately assigned based on our current knowledge was gender and reported ancestry, with the latter available only for 19 of the 24 patients. Interestingly, not all groups utilized gender information to the full extent (Supp. Table S2). Despite the difficulty of matching a given genome to a specific clinical description and in contrast to the lack of success in broad category matching, two submissions (SID#4 and 8) performed significantly better than random chance. These two groups were able to correctly match five and six genomes respectively, whereas the expected highest match number of nine random predictions was four. A similar success range was achieved in another CAGI challenge (Cai et al., 2017), where the best prediction could correctly match up to 25% of genomes to self-reported phenotypes.
To understand whether the overall low accuracy of the predictor community was caused by lack of information in the clinical descriptions, we assessed the relationship between the number of correct genome-patient matches and the informational content of a patient’s phenotype description. Unexpectedly, except for ophthalmologic disorders, richer clinical descriptions did not correlate with higher correct prediction rates (Figure 4). The mismatch between patients and genomes in the presence of rich phenotypic information could be explained, at least in part, by the complexity of the clinical descriptions. Reflecting the reality of clinical diagnosis of genetic disorders, these often included terms belonging to more than one phenotypic category, potentially confounding prediction classification. Specifically, in 29% (7/24) of cases, clinical descriptions contain references to both connective and neurological defects (patients A, I, K, V), eye and connective (patient X) or eye and neurological (patient J) defects, or defects belonging to all three categories (patient D). In at least two cases (patients J and L), these descriptions reference conditions outside the three defined categories, including major, multi-system health concerns, abnormal organ morphology and physiology, abnormal immunity and metabolism.
Most of the predictors used HPO coding (or another similar gene-phenotype database) for gene prioritization in each clinical case. A recent paper has demonstrated that the use of specific HPO terms improves gene-ranking with the top 10% of HPO terms being sufficient to rank the causative gene (Tomar, Sethi, & Lai, 2019). Unlike other submissions, SID#4 weighed the clinical terms by scoring the most serious and definitive (to a presumed disease) term in the profile with the highest value (ref to Pal et al CAGI5 paper). SID#8 built eight gene sets related to the diseases of interest and classified each case as belonging to one of those categories, rather than using all the genes associated with any of the HPO terms derived from the clinical descriptions. Complex phenotypes (patients A, J and X) were among the correctly matched genomes by these best-performing submissions, suggesting that in this case, phenotypic diversity could present an advantage in genome matching if appropriately leveraged.
In summary, this part of the challenge suggests that exhaustive phenotyping can introduce noise in the selection process, and that focusing on specific phenotypic features of a disease most relevant to the patient under examination can be a more effective prediction strategy than collecting as much phenotypic information as possible. Therefore, in addition to considering gender and ethnicity, a thorough understanding of the phenotypic descriptions was likely a key success factor.
Considerations for improving variant prediction
The CAGI challenges were designed to test our ability to associate genotype and phenotype beyond our current limits, and to look at genomic and phenotypic data in a much more multifactorial and complex way than monogenic Mendelian approaches. In the past, genetics was, out of necessity, a phenotype-to-genotype driven field. With the advent of NGS, a genotype-to-phenotype approach became possible. For the CAGI SickKids challenges, groups mainly adopted a phenotype-to-genotype approach, where most groups started with gene prioritization by building gene lists associated with given phenotype descriptions and selected variants within those genes or gene ontologies. No group took a purely variant-to-phenotype approach where rare, predicted damaging variants were used to independently build a set of gene ontology and phenotype terms that were then matched to patients’ phenotype descriptions. In clinical practice, bioinformatic pipelines typically filter out common, benign, and non-coding variants, and the phenotypes previously associated with the genes in which the remaining variants are found are compared to the patient phenotype, assuming a single gene disease relationship. It would be interesting to see how combinations of various approaches might lead to an improvement in performance of computational methods and identification of multigenic possibilities in similar challenges in the future.
The variant prediction part of the CAGI5 challenge was also complicated by the fact that the genomes were unlinked from the respective clinical descriptions. In CAGI4 on the other hand, the specific genome-patient matches were known. The vast majority of variants in that challenge appeared to be chosen for the ability to partially explain phenotype. In two cases this approach led two groups to predict the same variant for the same genome which resulted in a diagnosis for these patients. Yet, for many proposed diagnostic variants, the phenotype associated with the variant had little enough overlap with the patient’s phenotype that the variant was considered implausible by the clinical assessors. This issue arose multiple times in both CAGI4 and CAGI5, suggesting that there may be systematic problems with how phenotypic information is used bioinformatically. One contributing cause is the tendency to match the patient’s phenotype and the phenotype associated with the variant at the root HPO term. E.g., retinal degeneration and cataracts match at the root term of eye disorder, but have fundamentally different etiologies and clinical significance. Another problem is matching one HPO term at a time, which can produce an apparent match between the candidate gene and the patient when they don’t share other major HPO terms. E.g., a patient with developmental delay is matched to a gene that is associated with developmental delay, but the patient lacks the severe microcephaly and malformations seen with the candidate gene.
Study limitations
Primary disease causing variants selected by groups were often excluded from further clinical analysis due to the population frequencies of the variant being too high to cause Mendelian disease, variants located in non-coding regions, or variants being synonymous. Population frequencies of variants involved in multifactorial genetic disease may be higher than for Mendelian disorders. Such high frequency, multifactorial variants may prove to be important for clinical phenotypes in the future, but evidence for such a role is currently lacking.
The phenotype spectrum caused by pathogenic variants in a particular gene often expands over time with new information and identification of additional patients. In addition, ~5% of children evaluated for rare genetic disorders have more than one causal genomic variant (Stavropoulos, et al., 2016, Smith et al., 2019). Furthermore, inheritance patterns for genes often change with improved knowledge and understanding. It is reasonable to suspect that some of the VUS identified in this study may, in the future, be reclassified as phenotype expansions (Masuda et al., 2015; Negri et al., 2015; Sellars, Sullivan, & Schaefer, 2016). Examples of such variants include the EP300 p.Gln2343Arg variant in patient F (genome 93) and the LMNA p.Arg296Leu variant in patient J (genome 68).
Finally, a major limitation of current genome sequence analysis methods, is the identification of significantly large number of variants of unknown clinical significance. This is in part due to limited functional information available for these variants. Integrating other “omics” data, such as transcriptomics and proteomics analyses, would enrich variant functional characterization and aid in the identification of causative variants.
Conclusion
Adopting WGS as a diagnostic tool requires addressing the current lack of understanding of the role of many genes and variants in disease. Our assessment of the CAGI SickKids challenges involving undiagnosed children, suggests that computational approaches are most successful in predicting genotype from phenotypic information when the associated clinical terms are weighted by relevance. This may be especially pertinent in the case of complex phenotypes. Reportable clinical findings were discovered in the linked genomes challenge (CAGI4), while several other variants were identified as good candidates for phenotypic expansion or further research in CAGI5. Introducing clinical methodologies, such as combining phenotype-to-gene with variant-to-phenotype information, and integrating different types of omics data could hold promise for future development of computational methods seeking to explore the genetic basis of disease.
Supplementary Material
Acknowledgements
We thank the CAGI organizers and data providers as well as the parents and patients without whom this project would have been impossible. The CAGI experiment coordination is supported by NIH U41 HG007346 and the CAGI conference by NIH R13 HG006650.
Funding information National Institutes of Health, Grant/Award Numbers: R01 GM079656, R01 GM120364, R01 GM104436, U41 HG007346, R01 LM009722, R13 HG006650, R01 GM066099, U19 HD077627, R01-AG061105; Eesti Teadusagentuur, Grant/Award Number: IUT34-12; The Hospital for Sick Children’s Centre for Genetic Medicine, Grant/Award Number: N/A
Footnotes
Conflict of interests:
Jesse M. Hunter is a paid employee of Ambry Genetics. Aditya Rao, Thomas Joseph, Uma Sundaram, Saipradeep VG, Sujatha Kotte and Naveen Sivadasan are employees of Tata Consultancy Services Ltd. All other authors declare no conflicts of interest.
Data availability statement
The data that support the findings of this study are available to registered users from the CAGI web site (https://genomeinterpretation.org/SickKids5_clinical_genomes). Restrictions apply to the availability of these data, which were used for this study under license from the Hospital for Sick Children. Data are available from the authors with the permission of the Hospital for Sick Children.
References
- Avela K, Sankila EM, Seitsonen S, Kuuluvainen L, Barton S, Gillies S, & Aittomaki K (2018). A founder mutation in CERKL is a major cause of retinal dystrophy in Finland. Acta Ophthalmol, 96(2), 183–191. doi: 10.1111/aos.13551 [DOI] [PubMed] [Google Scholar]
- Babbi G, Martelli PL, Profiti G, Bovo S, Savojardo C, & Casadio R (2017). eDGAR: a database of Disease-Gene Associations with annotated Relationships among genes. BMC Genomics, 18(Suppl 5), 554. doi: 10.1186/s12864-017-3911-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbi G, Martelli PL, Casadio R. (2019) PhenPath: a tool for characterizing biological functions underlying different phenotypes. BMC Genomics. (accepted, in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereziat V, Cervera P, Le Dour C, Verpont MC, Dumont S, Vantyghem MC, . . . Lipodystrophy Study, G. (2011). LMNA mutations induce a non-inflammatory fibrosis and a brown fat-like dystrophy of enlarged cervical adipose tissue. Am J Pathol, 179(5), 2443–2453. doi: 10.1016/j.ajpath.2011.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdin SC, Hayeems RZ, Monfared N, Cohn RD, & Meyn MS (2016). The SickKids Genome Clinic: developing and evaluating a pediatric model for individualized genomic medicine. Clin Genet, 89(1), 10–19. doi: 10.1111/cge.12579 [DOI] [PubMed] [Google Scholar]
- Cai B, Li B, Kiga N, Thusberg J, Bergquist T, Chen YC, . . . Mooney SD. (2017). Matching phenotypes to whole genomes: Lessons learned from four iterations of the personal genome project community challenges. Hum Mutat, 38(9), 1266–1276. doi: 10.1002/humu.23265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, & Kingsmore SF (2018). Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med, 3, 16. doi: 10.1038/s41525-018-0053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, & Beroud C (2009). Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res, 37(9), e67. doi: 10.1093/nar/gkp215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Chavali S, Pougach K, Legendre M, Zhu B, Boeynaems S, . . . Verstrepen KJ (2015). Variable Glutamine-Rich Repeats Modulate Transcription Factor Activity. Mol Cell, 59(4), 615–627. doi: 10.1016/j.molcel.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel LB, Tripathi RK, Strunk KM, Hanifin JM, Jackson CE, King RA, & Spritz RA (1991). Tyrosinase gene mutations associated with type IB (“yellow”) oculocutaneous albinism. Am J Hum Genet, 48(6), 1159–1167. [PMC free article] [PubMed] [Google Scholar]
- Giral H, Landmesser U, & Kratzer A (2018). Into the Wild: GWAS Exploration of Non-coding RNAs. Front Cardiovasc Med, 5, 181. doi: 10.3389/fcvm.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdea M, Dumitriu S, Fiume M, Bowdin S, Boycott KM, Chenier S, . . . Brudno M. (2013). PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat, 34(8), 1057–1065. doi: 10.1002/humu.22347 [DOI] [PubMed] [Google Scholar]
- Gloss BS, & Dinger ME (2018). Realizing the significance of noncoding functionality in clinical genomics. Exp Mol Med, 50(8), 97. doi: 10.1038/s12276-018-0087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Granado JM, Silvestre-Roig C, Rocha-Perugini V, Trigueros-Motos L, Cibrian D, Morlino G, . . . Andres V. (2014). Nuclear envelope lamin-A couples actin dynamics with immunological synapse architecture and T cell activation. Sci Signal, 7(322), ra37. doi: 10.1126/scisignal.2004872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally MT (1993). Shprintzen-Goldberg Syndrome. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, & Amemiya A (Eds.), GeneReviews((R)). Seattle (WA). [PubMed] [Google Scholar]
- Jain S, White M, Trosset MW, & Radivojac P (2016). Nonparametric semi-supervised learning of class proportions. arXiv preprint arXiv:160101944. Available from: http://arxiv.org/abs/1601.01944. [Google Scholar]
- Jain S, White M, & Radivojac P (2016). Estimating the class prior and posterior from noisy positives and unlabeled data. Advances in Neural Information Processing Systems, pp. 2693–2701. [Google Scholar]
- Jordan DM, & Do R (2018). Using Full Genomic Information to Predict Disease: Breaking Down the Barriers Between Complex and Mendelian Diseases. Annu Rev Genomics Hum Genet, 19, 289–301. doi: 10.1146/annurev-genom-083117-021136 [DOI] [PubMed] [Google Scholar]
- Kang X, Sun Y, & Zhang Z (2019). Identification of key transcription factors - gene regulatory network related with osteogenic differentiation of human mesenchymal stem cells based on transcription factor prognosis system. Exp Ther Med, 17(3), 2113–2122. doi: 10.3892/etm.2019.7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, . . . MacArthur DG. (2019). Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv, 531210. doi: 10.1101/531210 [DOI] [Google Scholar]
- Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, & Massouras A (2019). VarSome: the human genomic variant search engine. Bioinformatics, 35(11), 1978–1980. doi: 10.1093/bioinformatics/bty897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousi M, & Katsanis N (2015). Genetic modifiers and oligogenic inheritance. Cold Spring Harb Perspect Med, 5(6). doi: 10.1101/cshperspect.a017145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, & Kadonaga JT (1999). Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol, 19(12), 8123–8135. doi: 10.1128/mcb.19.12.8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, . . . Maglott DR (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res, 46(D1), D1062–D1067. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz J, & Keats BJB (1993). Usher Syndrome Type I. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, & Amemiya A (Eds.), GeneReviews((R)). Seattle (WA). [Google Scholar]
- Letai A, & Fuchs E (1995). The importance of intramolecular ion pairing in intermediate filaments. Proc Natl Acad Sci U S A, 92(1), 92–96. doi: 10.1073/pnas.92.1.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung VYL, Zhou L, Tam WK, Sun Y, Lv F, Zhou G, & Cheung KMC (2017). Bone morphogenetic protein-2 and −7 mediate the anabolic function of nucleus pulposus cells with discrete mechanisms. Connect Tissue Res, 58(6), 573–585. doi: 10.1080/03008207.2017.1282951 [DOI] [PubMed] [Google Scholar]
- Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, . . . Marshall CR. (2018). Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med, 20(4), 435–443. doi: 10.1038/gim.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Akiyama K, Arakawa M, Nishi E, Kitazawa N, Higuchi T, . . . Izumi K (2015). Exome Sequencing Identification of EP300 Mutation in a Proband with Coloboma and Imperforate Anus: Possible Expansion of the Phenotypic Spectrum of Rubinstein-Taybi Syndrome. Mol Syndromol, 6(2), 99–103. doi: 10.1159/000375542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke LA, study DDD, Gardeitchik T, Hammond P, Heimdal KR, Houge G, . . . Hennekam RC (2018). Further delineation of an entity caused by CREBBP and EP300 mutations but not resembling Rubinstein-Taybi syndrome. Am J Med Genet A, 176(4), 862–876. doi: 10.1002/ajmg.a.38626 [DOI] [PubMed] [Google Scholar]
- Nabieva E, Jim K, Agarwal A, Chazelle B, & Singh M (2005). Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics, 21 Suppl 1, i302–310. doi: 10.1093/bioinformatics/bti1054 [DOI] [PubMed] [Google Scholar]
- Negri G, Milani D, Colapietro P, Forzano F, Della Monica M, Rusconi D, . . . Gervasini C. (2015). Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin Genet, 87(2), 148–154. doi: 10.1111/cge.12348 [DOI] [PubMed] [Google Scholar]
- Pagel KA, Pejaver V, Lin GN, Nam HJ, Mort M, Cooper DN, . . . Radivojac P. (2017). When loss-of-function is loss of function: assessing mutational signatures and impact of loss-of-function genetic variants. Bioinformatics, 33(14), i389–i398. doi: 10.1093/bioinformatics/btx272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal LR, Kundu K, Yin Y, & Moult J (2017). CAGI4 SickKids clinical genomes challenge: A pipeline for identifying pathogenic variants. Hum Mutat, 38(9), 1169–1181. doi: 10.1002/humu.23257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejaver V, Urresti J, Lugo‐Martinez J, Pagel KA, Lin GN, Nam HJ, … Radivojac P (2017).MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv 134981; 10.1101/134981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, . . . Committee, A. L. Q. A. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacitharan PK, Lwin S, Gharios GB, & Edwards JR (2018). Spermidine restores dysregulated autophagy and polyamine synthesis in aged and osteoarthritic chondrocytes via EP300. Exp Mol Med, 50(9), 123. doi: 10.1038/s12276-018-0149-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocchia A, Wigby KM, Masser-Frye D, Del Campo M, Galarreta CI, Thorpe E, . . . Taft RJ. (2019). Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico. NPJ Genom Med, 4, 5. doi: 10.1038/s41525-018-0076-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellars EA, Sullivan BR, & Schaefer GB (2016). Whole exome sequencing reveals EP300 mutation in mildly affected female: expansion of the spectrum. Clin Case Rep, 4(7), 696–698. doi: 10.1002/ccr3.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li J, Zhang J, Xu C, Jiang Y, Wu Z, . . . Deng HW. (2013). Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians. PLoS One, 8(4), e59494. doi: 10.1371/journal.pone.0059494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieprath T, Corne TD, Nooteboom M, Grootaert C, Rajkovic A, Buysschaert B, . . . De Vos WH (2015). Sustained accumulation of prelamin A and depletion of lamin A/C both cause oxidative stress and mitochondrial dysfunction but induce different cell fates. Nucleus, 6(3), 236–246. doi: 10.1080/19491034.2015.1050568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Blanco K, Sajan SA, Hunter JM, Shinde DN, Wayburn B, . . . Radtke K. (2019). A retrospective review of multiple findings in diagnostic exome sequencing: half are distinct and half are overlapping diagnoses. Genet Med. doi: 10.1038/s41436-019-0477-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz RA, Oh J, Fukai K, Holmes SA, Ho L, Chitayat D, . . . Levin AV. (1997). Novel mutations of the tyrosinase (TYR) gene in type I oculocutaneous albinism (OCA1). Hum Mutat, 10(2), 171–174. doi: [DOI] [PubMed] [Google Scholar]
- Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, . . . Marshall CR. (2016). Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Pediatric Medicine. NPJ Genom Med, 1. doi: 10.1038/npjgenmed.2015.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvius N, Hathaway A, Boudreau E, Gupta P, Labib S, Bolongo PM, . . . Tesson F. (2008). Specific contribution of lamin A and lamin C in the development of laminopathies. Exp Cell Res, 314(13), 2362–2375. doi: 10.1016/j.yexcr.2008.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tazearslan C, & Suh Y (2017). Challenges and progress in interpretation of non-coding genetic variants associated with human disease. Exp Biol Med (Maywood), 242(13), 1325–1334. doi: 10.1177/1535370217713750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuela N, Bar DZ, & Gruenbaum Y (2012). Lamins in development, tissue maintenance and stress. EMBO Rep, 13(12), 1070–1078. doi: 10.1038/embor.2012.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S, Sethi R, & Lai PS (2019). Specific phenotype semantics facilitate gene prioritization in clinical exome sequencing. Eur J Hum Genet. doi: 10.1038/s41431-019-0412-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku K, Wada I, Spritz RA, & Hearing VJ (2001). The molecular basis of oculocutaneous albinism type 1 (OCA1): sorting failure and degradation of mutant tyrosinases results in a lack of pigmentation. Biochem J, 355(Pt 2), 259–269. doi: 10.1042/0264-6021:3550259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Database of Single Nucleotide Polymorphisms (dbSNP). Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine; (dbSNP Build ID: 151). Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- [dataset] CAGI; 2016/2018; SickKids challenge dataset; Dataset available to registered users from the CAGI web site (https://genomeinterpretation.org/SickKids5_clinical_genomes).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.