Abstract
The unique isocyclic E ring of chlorophylls contributes to their role as light-absorbing pigments in photosynthesis. The formation of the E ring is catalyzed by the Mg-protoporphyrin IX monomethyl ester cyclase, and the O2-dependent cyclase in prokaryotes consists of a diiron protein AcsF, augmented in cyanobacteria by an auxiliary subunit Ycf54. Here, we establish the composition of plant and algal cyclases, by demonstrating the in vivo heterologous activity of O2-dependent cyclases from the green alga Chlamydomonas reinhardtii and the model plant Arabidopsis thaliana in the anoxygenic photosynthetic bacterium Rubrivivax gelatinosus and in the non-photosynthetic bacterium Escherichia coli. In each case, an AcsF homolog is the core catalytic subunit, but there is an absolute requirement for an algal/plant counterpart of Ycf54, so the necessity for an auxiliary subunit is ubiquitous among oxygenic phototrophs. A C-terminal ∼40 aa extension, which is present specifically in green algal and plant Ycf54 proteins, may play an important role in the normal function of the protein as a cyclase subunit.
Keywords: chlorophyll, cyclase, photosynthesis, tetrapyrroles
Introduction
All chlorophototrophic organisms rely on the unique chemical properties of (bacterio)chlorophyll [(B)Chl] molecules for light harvesting and photochemical reactions, so the elucidation of the (B)Chl biosynthesis pathways is of great importance. In the common (B)Chl biosynthetic pathway, the least well-characterized step is the formation of the isocyclic E ring, via the oxidation and cyclization of the C13 methyl propionate group of Mg-protoporphyrin IX monomethyl ester (MgPME), producing 3,8-divinyl protochlorophyllide a (DV PChlide a). This step is catalyzed by two mechanistically unrelated cyclases: an O2-sensitive, radical SAM enzyme containing [4Fe–4S] and cobalamin cofactors [1] is encoded by the bchE gene in most anoxygenic phototrophic bacteria and some cyanobacteria [2], whereas an O2-dependent diiron monooxygenase is found in many purple bacteria, as well as cyanobacteria, algae and plants [3]. The catalytic subunit of the O2-dependent cyclase was first identified from the purple betaproteobacterium Rubrivivax (Rvi.) gelatinosus and named AcsF (aerobic cyclization system Fe-containing subunit) [4]. Following the identification of two auxiliary subunits, Ycf54 [5,6] and BciE, the O2-dependent cyclase has been delineated into three distinct classes: the Ycf54-dependent enzyme from oxygenic phototrophs, the AcsF-only enzyme in most anoxygenic phototrophs (e.g. in Rvi. gelatinosus) and the BciE-dependent alphaproteobacterial enzyme [3] (Figure 1B). The activity of the O2-dependent cyclase has been demonstrated in Escherichia coli using heterologously expressed enzymes from Rvi. gelatinosus and the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) [7], thus completing the identification of the O2-dependent cyclase components in prokaryotic phototrophs.
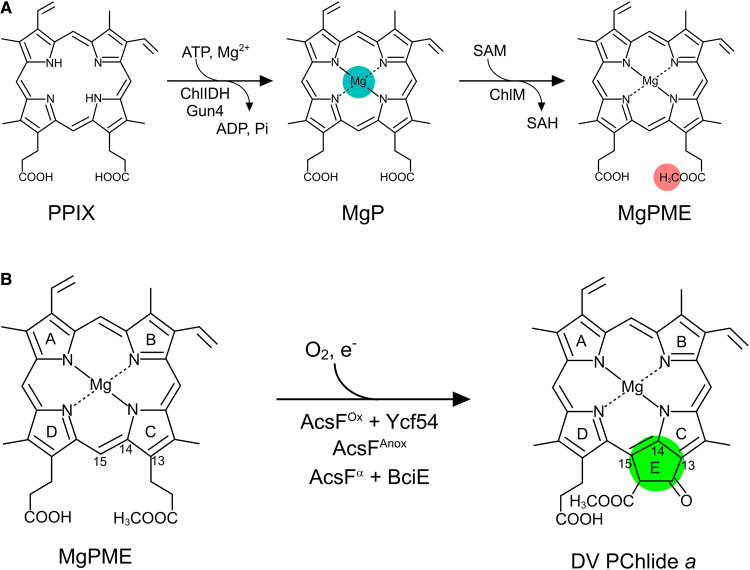
Figure 1. The early (B)Chl biosynthetic steps from PPIX to DV PChlide a.
(A) Formation of MgPME from PPIX catalyzed by the magnesium chelatase (ChlIDH, Gun4) and the MgP methyltransferase (ChlM). ATP, adenosine triphosphate; ADP, adenosine diphosphate; Pi, inorganic phosphate; SAM, S-adenosine-l-methionine; SAH, S-adenosyl-l-homocysteine. (B) Formation of the isocyclic E ring (highlighted) of (B)Chl is catalyzed by three classes of O2-dependent cyclase: AcsFOx denotes AcsF from oxygenic phototrophs, which requires Ycf54; AcsFAnox denotes AcsF from anoxygenic phototrophs excluding Alphaproteobacteria; and AcsFα denotes alphaproteobacterial AcsF, which requires BciE. e− represents the electron donor to the diiron center of AcsF. International Union of Pure and Applied Chemistry numbering of the relevant macrocycle carbons is indicated.
Oxygenic phototrophs, in particular higher plants, rely solely on the O2-dependent cyclase for Chl biosynthesis. Early research work was focused on biochemical characterization of the enzyme using fractions from cucumber developing chloroplasts, resolving the enzyme into soluble and membrane components [8,9]; a similar subcellular distribution was found subsequently for the Synechocystis enzyme [10]. The model plant Arabidopsis thaliana (hereafter Arabidopsis) contains a single AcsF protein, CHL27 [11], a homolog of AcsF from Rvi. gelatinosus [4], whereas the green alga Chlamydomonas reinhardtii (hereafter Chlamydomonas) has two AcsF isoforms, CRD1 and CTH1. Genes encoding these isoforms are expressed reciprocally; CRD1 has a basal level of expression that increases under copper- or O2-deplete conditions, whereas CTH1 is produced under copper- and O2-replete conditions [12,13]. Similarly, Synechocystis also has two AcsF homologs, CycI and CycII; CycI is constitutively active under both aerobic and micro-oxic conditions, whilst CycII is additionally required under micro-oxic conditions [14,15]. AcsF proteins have been shown to be membrane-bound in various organisms [5,11,16,17]. In the search for the ‘missing’ soluble component, Ycf54/LCAA was identified as an auxiliary cyclase subunit from Synechocystis by in vivo pulldown experiments using FLAG-tagged CycI and CycII as bait [5], and from tobacco by screening antisense plants with Chl deficiency [6]. The key role of Ycf54 proteins in cyclase function is exemplified by its ubiquity in oxygenic phototrophs and the severe phenotypes exhibited by knockout mutants including impaired AcsF protein levels, accumulation of MgPME, and lowered amounts of DV PChlide a and Chl [5,6,18–21].
Despite this recent progress with the O2-dependent cyclase from purple bacteria and cyanobacteria, there has been no activity-based assignment of algal or plant cyclase components. We have shown previously that Rvi. gelatinosus and E. coli are suitable hosts for heterologous expression of cyclase components and demonstration of cyclase activity [3,7], and in this study we extend this approach to the O2-dependent cyclase subunits from Synechocystis, Chlamydomonas and Arabidopsis, representative of cyanobacteria, green algae and higher plants, respectively. The detection of in vivo cyclase activity from recombinant Arabidopsis and Chlamydomonas enzymes shows that the eukaryotic cyclase consists of AcsF and Ycf54 subunits and Ycf54 is, therefore, an essential component of cyclases in oxygenic phototrophs.
Materials and methods
Synthesized genes
Gene fragments encoding Arabidopsis CHL27 (AT3G56940.1), YCF54 (AT5G58250.1), Chlamydomonas CRD1(CHLREDRAFT_183476), CTH1 (CHLREDRAFT_205856), and CGL78 (CHLREDRAFT_162021) proteins, lacking the N-terminal chloroplast transit peptides predicted by the ChloroP 1.1 Server [22], were synthesized (Integrated DNA Technologies) with codons optimized for expression in E. coli. It is worth noting that, based on the sequence alignments of Ycf54 proteins and experimental tests, Arabidopsis YCF54 was assumed to contain a 72 aa chloroplast transit peptide instead of the predicted 80 aa. The nucleotide sequences of synthesized genes are listed in Supplementary Table S1.
Plasmids and bacterial strains
The pBB[gene] plasmids were made by cloning the indicated gene fragment into the BglII/NotI or BglII/XhoI sites of pBBRBB-Ppuf843–1200 [23]. The Synechocystis cycI and cycI-ycf54 genes were amplified using the pK18[cycI-ycf54] plasmid [3] as a template. Overlap extension PCR was used to generate a CHL27-YCF54 gene fragment with a ribosome binding site (TATAGGAGCTTGGATT) placed between the two genes. To apply the link and lock method [24], genes were first cloned individually into the NdeI/SpeI sites of a modified pET3a vector (containing an added SpeI site immediately upstream of the BamHI site). Then the genes were cut from the pET3a plasmids and adjoined consecutively in the described order using the procedures described previously [7]. Primers used in this study are listed in Supplementary Table S2. Plasmids were sequenced by Eurofins Genomics and are listed in Supplementary Table S3. E. coli strains were grown in LB medium and, if required, antibiotics were supplemented at 30 µg ml−1 for kanamycin and 100 µg ml−1 for ampicillin. Rvi. gelatinosus strains were grown in polypeptone-yeast extract-sodium succinate (PYS) medium [25] at 30°C and, when required, antibiotics were added at 40 µg ml−1 for rifampicin and 50 µg ml−1 for kanamycin. A spontaneous rifampicin-resistant mutant, ΔbchEΔacsF RifR, was isolated from the Rvi. gelatinosus ΔbchEΔacsF [3] mutant and served as a platform strain to test foreign cyclases from Synechocystis and Arabidopsis. Cyclase genes were cloned into the expression vector pBBRBB-Ppuf843–1200 to get the pBB[gene] plasmids, which were conjugated into the ΔbchEΔacsF RifR strain via the E. coli S17-1 strain. E. coli S17-1 cells containing the plasmid were grown in LB medium with 30 µg ml−1 kanamycin at 37°C for 24 h and 30 µl of the resulting culture were mixed with Rvi. gelatinosus cells harvested from 30 ml culture and resuspended in 100 µl of LB medium. The mating mixture was placed onto a well-dried LB agar medium and incubated at 30°C overnight before streaking out onto PYS agar medium with 50 µg ml−1 kanamycin and 40 µg ml−1 rifampicin to select for transconjugants. Bacterial strains used in this study are listed in Supplementary Table S3.
Phenotypic analysis of Rvi. gelatinosus strains
Rvi. gelatinosus strains were grown in 10 ml PYS medium, supplemented with 50 µg ml−1 kanamycin if the strain contains a pBBRBB-puf843–1200-based plasmid, in 50 ml Falcon tubes at 30°C with shaking at 175 rpm for 2 days. Harvested cells were either used for recording whole-cell absorption spectra or subjected to pigment extraction. Absorption spectra were recorded on a Cary 60 UV–Vis spectrophotometer with cells resuspended in 60% (w/v) sucrose, and normalized by the absorbance at 950 nm, followed by subtraction of the ΔbchEΔacsF RifR spectrum to correct for light scattering. Pigment extraction and subsequent HPLC analysis were performed as described previously [26].
In vivo cyclase assay in E. coli
E. coli C43(DE3) [27] was transformed with the pET3a-based plasmids and selected on LB agar supplemented with 100 µg ml−1 ampicillin. A single colony was inoculated to 10 ml LB medium with 100 µg ml−1 ampicillin and grown overnight at 37°C with shaking at 220 rpm. The next day, 50 µl of the resulting culture were used to inoculate 10 ml of LB medium with 100 µg ml−1 ampicillin and grown at 37°C for 3 h. Then isopropyl-β-D-thiogalactopyranoside (IPTG) was added at 0.5 mM to induce gene expression. δ-Aminolevulinic acid (ALA) and Mg2+ (equimolar mixture of MgCl2 and MgSO4) were also added at 10 mM. The culture was incubated at 30°C with shaking at 175 rpm for 24 h before pigment extraction. Pigment extraction and high-performance liquid chromatography (HPLC) analysis were conducted as described previously [7] with slight modifications. The column was a Phenomenex Luna C18(2) reversed-phase column (particle size, 5 µm; pore size, 100 Å; 250 × 4.6 mm). The wash with 100% solvent B was shortened from 15 to 5 min. Elution was additionally monitored by fluorescence at 640 nm excited at 440 nm.
SDS–PAGE and immunodetection
E. coli strains containing the pET3a-based plasmids were incubated and induced as for the in vivo cyclase assay. After 5 h induction cells were harvested and resuspended in 50 mM Tris–HCl, pH 8.0, 5 mM EDTA, supplemented with Proteinase Inhibitor Cocktail (Sigma–Aldrich), and lysed by sonication on ice (3 × 20 s with 20 s resting between cycles). The cell lysate was clarified by centrifugation at 3000×g at 4°C for 5 min to remove unbroken cells and cell debris, followed by SDS–PAGE analysis using NuPAGE 12% Bis-Tris Protein Gels (Thermo Fisher Scientific). Protein bands were transferred to a polyvinylidene difluoride (PVDF) membrane, which was probed with specific primary antibodies raised against Arabidopsis CHL27 (Agrisera), YCF54 (Agrisera) and Synechocystis Ycf54 [5], and then with a secondary anti-rabbit antibody conjugated with horseradish peroxidase (Sigma–Aldrich). Chemiluminescent signal was developed using the WESTAR SUN enhanced chemiluminescence substrate (Cyanagen) and detected by an Amersham Imager 600 (GE Healthcare).
Results
Complementation of a Rvi. gelatinosus ΔbchEΔacsF mutant with Synechocystis and Arabidopsis cyclase genes
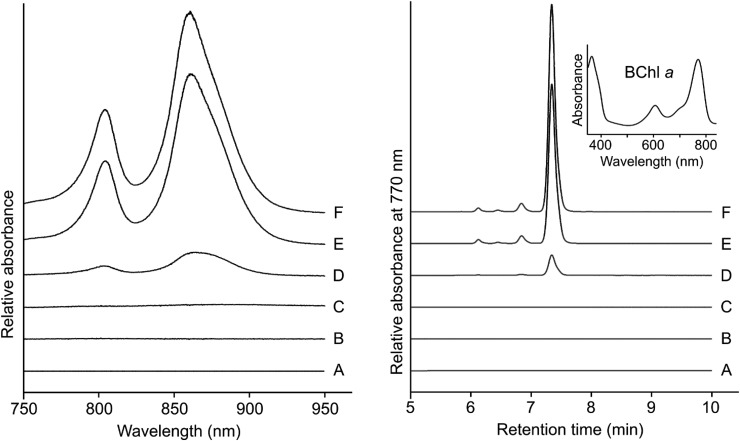
The catalytic subunit of O2-dependent cyclase, AcsF, containing a signature diiron binding motif (E-xn-E-x-x-H-xn-E-xn-E-x-x-H), is classified as a membrane-bound carboxylate diiron protein [28] (Figure 2). We used a ΔbchEΔacsF RifR mutant of the facultative phototroph Rvi. gelatinosus, which lacks both the O2-sensitive and -dependent cyclase enzymes [3], as a background strain for assaying plasmid-borne Synechocystis cycI and ycf54 genes, or genes encoding the Arabidopsis CHL27 and YCF54 proteins. Whole-cell absorption spectra of transconjugant strains showed that neither CycI nor CHL27 was functional but when partnered with Ycf54 or YCF54, respectively, each was able to replace the missing native cyclase and restore the assembly of photosynthetic complexes to variable levels (Figure 3). It is striking that the activity of the Synechocystis CycI-Ycf54 pair matched that of the native AcsF protein (Figure 3). We further analyzed the accumulation of BChl a in these strains by HPLC and the detected level is in good agreement with the abundance of photosynthetic complexes revealed by the corresponding whole-cell absorption spectra (Figure 3).
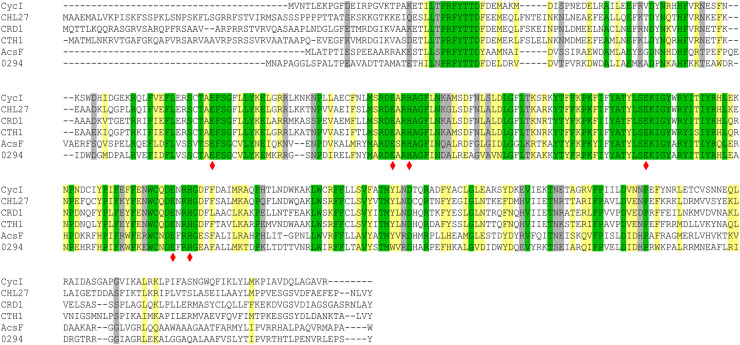
Figure 2. Amino acid sequence alignments of AcsF proteins.
Sequences are those from Synechocystis sp. PCC 6803 (CycI, BAA16583), Arabidopsis thaliana (CHL27, NP_191253), Chlamydomonas reinhardtii (CRD1, XP_001692557; CTH1, XP_001691047), Rubrivivax gelatinosus IL144 (AcsF, BAL96694) and Rhodobacter sphaeroides 2.4.1 (0294, abbreviated for RSP_0294, YP_353369). Conserved, highly similar and similar residues are highlighted in green, yellow and gray, respectively. The putative diiron binding ligands are marked with red diamonds.
Figure 3. Heterologous activity of Synechocystis and Arabidopsis cyclases in Rvi. gelatinosus.
Cyclase genes were tested in the Rvi. gelatinosus ΔbchEΔacsF RifR mutant by expression from a plasmid vector: (A) no plasmid, (B) pBB[cycI], (C) pBB[CHL27], (D) pBB[CHL27-YCF54], (E) pBB[acsF] and (F) pBB[cycI-ycf54]. Left, whole-cell absorbance spectra of the described strains, normalized and corrected for light scattering. Right, HPLC elution profiles of pigment extracts from the described strains standardized by cell number. The major peak in the traces was identified to be BChl a by the retention time and absorbance spectrum (inset).
In vivo activity of the recombinant Synechocystis and Arabidopsis cyclases in E. coli
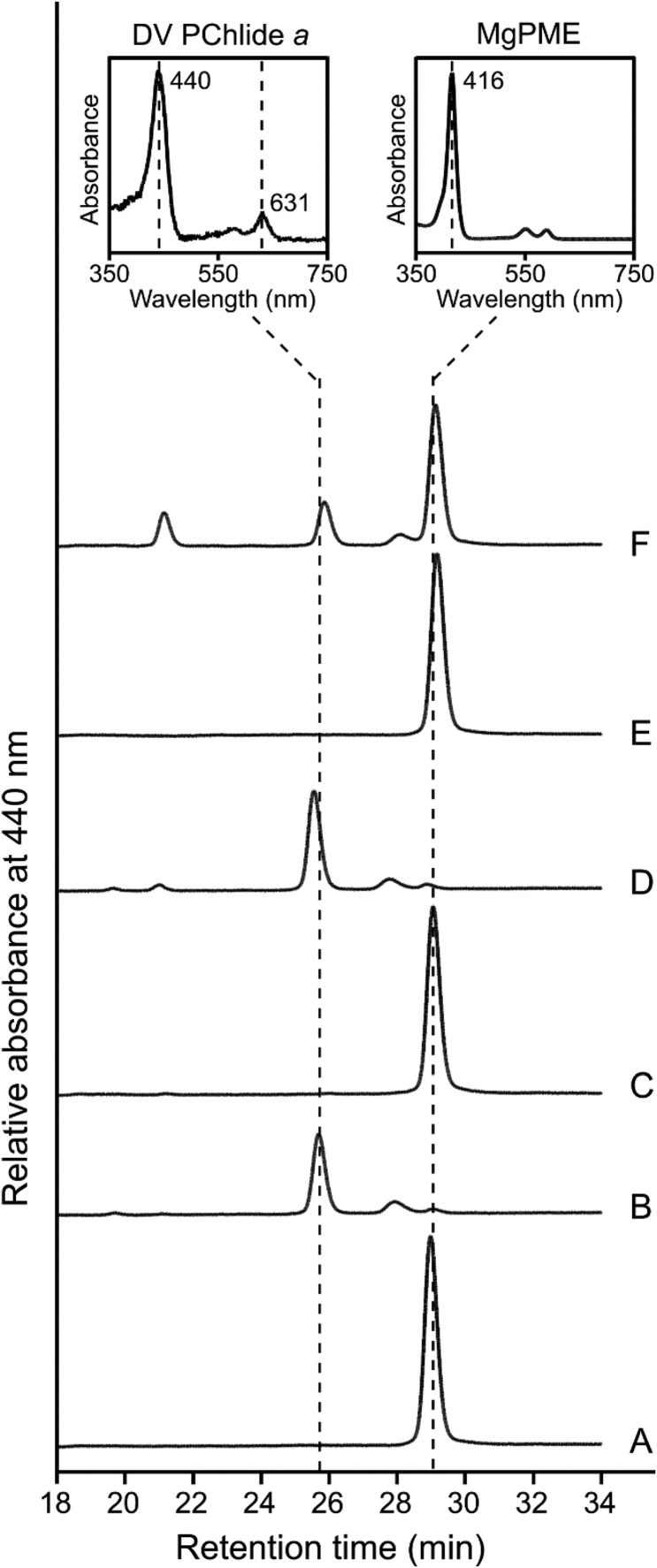
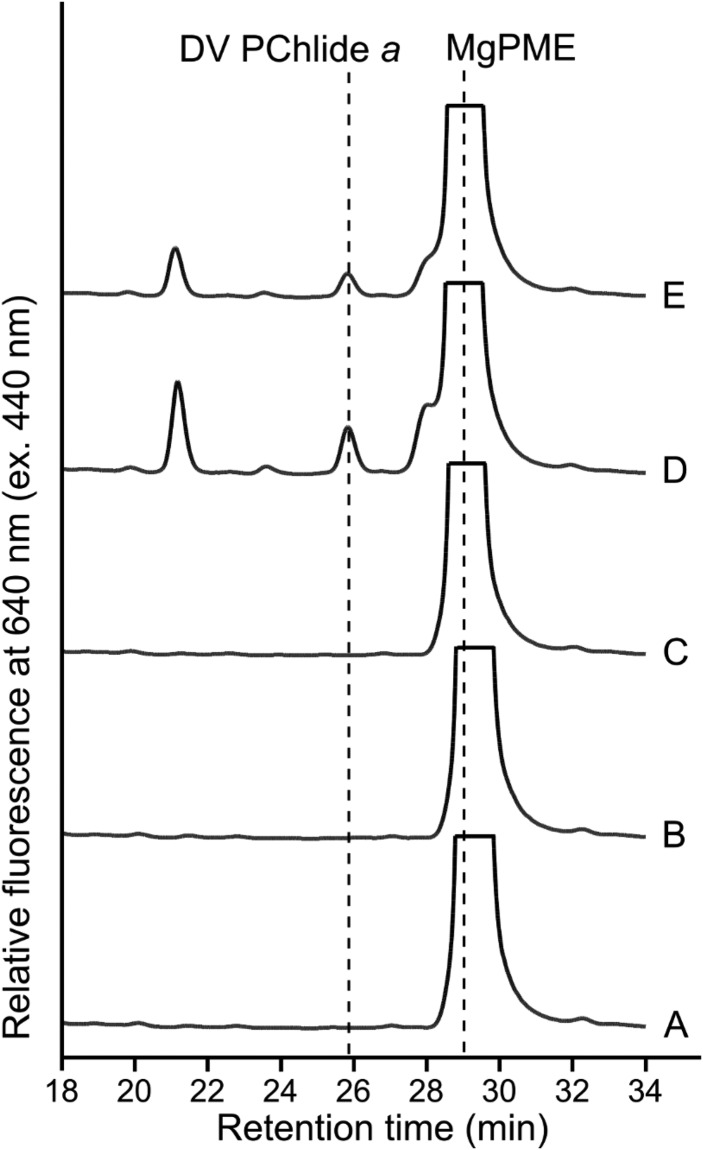
The Synechocystis CycI-Ycf54 and Arabidopsis CHL27-YCF54 pairs were clearly active in the purple phototrophic bacterium Rvi. gelatinosus. Next, we adopted a previously reported strategy [7] to demonstrate their activity in the non-photosynthetic bacterium E. coli. Cyclase encoding genes were cloned into the pET3a-based plasmid IM [7], which contains genes encoding the first two enzymes of the Chl pathway that convert native, endogenous protoporphyrin IX (PPIX) to Mg-protoporphyrin IX (MgP), then to the cyclase substrate, MgPME (Figure 1A). These constructs were transformed into the E. coli C43(DE3) strain, followed by in vivo cyclase assays. Pigments from the harvested E. coli cells were extracted and analyzed by HPLC with elution profiles monitored by absorbance, and also fluorescence for a higher sensitivity for detecting Chl biosynthesis intermediates. In the absence of a functional cyclase, only MgPME (Soret band at 416 nm; emission maximum at 593 nm) was detected, which eluted at ∼29 min (Figure 4). The E. coli strains expressing Rvi. gelatinosus AcsF (plasmid IA), Synechocystis CycI-Ycf54 (IM-cycI-ycf54) and Arabidopsis CHL27-YCF54 (IM-CHL27-YCF54), accumulated DV PChlide a (Soret/Qy bands at 440 and 631 nm, respectively; emission maximum at 639 nm) as represented by a peak at ∼26 min, which is accompanied by a decreased level of MgPME (Figure 4). Arabidopsis CHL27-YCF54 was less active than the Rvi. gelatinosus and Synechocystis enzymes, leaving a considerable amount of MgPME substrate not converted to product (Figure 4).
Figure 4. HPLC analysis of pigments extracted from E. coli strains expressing Synechocystis and Arabidopsis cyclases.
E. coli strains contained (A) IM (chlI-chlD-chlH-gun4-chlM), (B) IA (chlI-chlD-chlH-gun4-chlM-acsF), (C) IM-cycI, (D) IM-cycI-ycf54, (E) IM-CHL27 and (F) IM-CHL27-YCF54 plasmids. Protein expression was induced with IPTG, and ALA and Mg2+ were supplemented to boost the production of cyclase substrate, MgPME. Pigments were extracted from the same number of cells except for the strains containing IA and IM-cycI-ycf54 plasmids, for which only 1/10th of cells were used. Elution of cyclase substrate and product was monitored by absorbance at 440 nm. Retention times and spectra (inset) of peaks are used to identify pigment species.
The Chlamydomonas AcsF isoform CRD1, but not CTH1, is functional when expressed in E. coli
Unlike the single AcsF found in Arabidopsis, the green alga Chlamydomonas contains two AcsF isoforms, CRD1 and CTH1 [13]. Genes encoding CRD1, CTH1 and the Chlamydomonas Ycf54 protein, CGL78, were synthesized and cloned into the IM plasmid [7] for in vivo cyclase assay in E. coli. HPLC analysis of pigments extracted from the assays reveals that CRD1 was functional only when CGL78 was co-expressed (Figure 5). In the current test system, no activity was detected for CTH1 even in the presence of CGL78 (Figure 5).
Figure 5. HPLC analysis of pigments extracted from E. coli strains expressing Chlamydomonas cyclases.
Analysis of E. coli strains containing (A) IM-CRD1, (B) IM-CRD1-CGL78 and (C) IM-CTH1-CGL78 plasmids. Pigments were extracted from the same number of cells. Elution of cyclase substrate and the product was monitored by absorbance at 440 nm and fluorescence at 640 nm excited at 440 nm.
Arabidopsis and Chlamydomonas Ycf54 proteins differ from their Synechocystis homolog
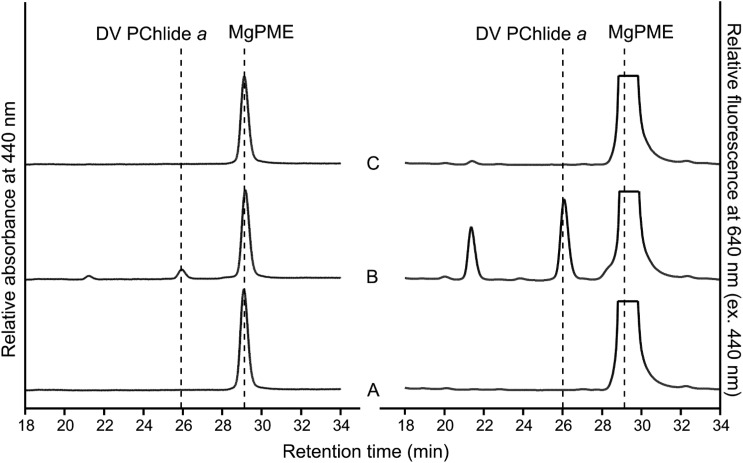
Our in vivo cyclase assays using enzymes from representative species of cyanobacteria, green algae and plants, reveal the universal requirement of Ycf54 for cyclase activity among oxygenic phototrophs. Sequence alignments of Ycf54 proteins clearly identify a conserved Ycf54 domain (designated PF10674/DUF2488 in the Pfam database; Figure 6A). The alignments also reveal C-terminal extensions present only in green algal and plant Ycf54 proteins (Figure 6A), containing nine conserved residues (F180, W186, A188, P189, Y190, Y193, W197, W198 and W201; numbering in Arabidopsis YCF54, NP_200633) among these species (Figure 6B). To investigate the potential role of the C-terminal extension, we truncated the Arabidopsis YCF54 gene to generate a YCF54* mutant, with the C-terminal 37 aa sequence removed. We also swapped Ycf54 proteins between Arabidopsis, Chlamydomonas and Synechocystis cyclases to check whether Ycf54 is functional with a foreign AcsF protein. CHL27-YCF54* behaved essentially the same as CHL27 alone in the in vivo cyclase assays, indicating the inactive nature of YCF54* (Figure 7). The Synechocystis Ycf54 protein was unable to replace the Arabidopsis or Chlamydomonas counterpart whereas Arabidopsis YCF54 and Chlamydomonas CGL78 supported some cyclase activity (detectable by fluorescence) when partnered with Synechocystis CycI (Figure 7). These results suggest green algal and plant Ycf54 proteins differ from the proteins found in other organisms, probably by their C-terminal extensions.
Figure 6. Amino acid sequence alignments of Ycf54 proteins.
Conserved, highly similar and similar residues are highlighted in green, yellow and gray, respectively. (A) Sequences are those from the primordial cyanobacterium Gloeobacter violaceus PCC 7421 (NP_923828); cyanobacteria, Prochlorococcus marinus MED4 (CAE19565), Synechococcus sp. PCC 7002 (ACA98109), Synechocystis sp. PCC 6803 (BAA16792), Acaryochloris marina MBIC11017 (ABW27358); red algae, Porphyra purpurea (NP_053814), Bangia fuscopurpurea (AKE98807); green algae, Bathycoccus prasinos (XP_007514179), Ostreococcus tauri (XP_022839105), Chlamydomonas reinhardtii (XP_001691121); the moss Physcomitrella patens (XP_001756877); higher plants, Cucumis sativus (XP_004139926), Arabidopsis thaliana (NP_200633), Nicotiana tobacum (XP_016480530), Hordeum vulgare L. cv. Bonus (BAJ91312), Oryza sativa L. ssp. japonica (XP_015628146). The conserved Ycf54 domain is indicated. The C-terminal 37 aa sequence deleted in the YCF54* mutant is underlined. (B) Sequence alignments showing the conservation of the C-terminal extensions present specifically in green algal and plant Ycf54 proteins. Additional green algal and plant Ycf54 sequences are included, which are from the green alga Coccomyxa subellipsoidea C-169 (XP_005642819); the liverwort Marchantia polymorpha (PTQ33664); higher plants, Brachypodium distachyon (XP_003557990), Zea mays (NP_001131876), Spinacia oleracea (XP_021852323), Vitis vinifera (XP_010650790), Erythranthe guttata (XP_012838088), Ipomoea nil (XP_019188624), Solanum lycopersicum (XP_004240451), Medicago truncatula (XP_013460499), Lotus japonicus (AFK37846). Full-length sequences were used for alignments but for clarity, only the C-terminal extension regions with the residue range indicated, are shown.
Figure 7. HPLC analysis of pigments extracted from E. coli strains expressing YCF54* and swapped ycf54 genes.
E. coli strains contained (A) IM-YCF54*, (B) IM-CHL27-ycf54, (C) IM-CRD1-ycf54, (D) IM-cycI-YCF54 and (E) IM-cycI-CGL78 plasmids. Pigments were extracted from the same number of cells. Elution of cyclase substrate and the product was monitored by fluorescence at 640 nm excited at 440 nm.
Expression levels of recombinant cyclase subunits in E. coli
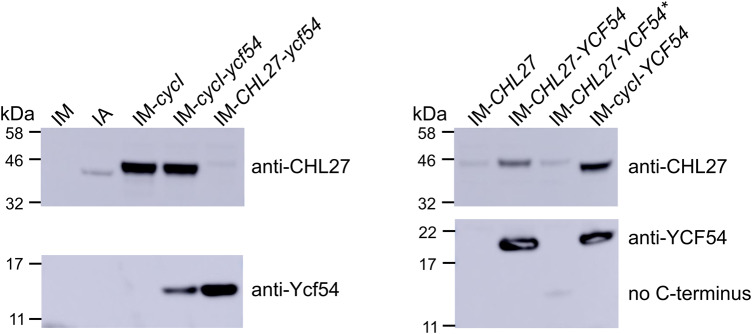
Our in vivo cyclase assays demonstrate the cyclase from oxygenic phototrophs is dependent on the Ycf54 protein for activity. To check whether Ycf54 affects the expression/stability of AcsF when expressed in E. coli, we detected the level of AcsF and Ycf54 proteins in clarified cell lysates using specific antibodies. The commercially available anti-CHL27 antibody also reacts with Synechocystis CycI but does not react with Chlamydomonas CRD1 or CTH1 so the immunodetection was limited to Arabidopsis and Synechocystis cyclases. In contrast with the indistinguishable CycI levels with and without Ycf54, the CHL27 level was clearly dependent on its native interacting partner YCF54 (Figure 8). The absence or C-terminal truncation of YCF54 significantly decreased the abundance of CHL27, which could not be rectified by the presence of Synechocystis Ycf54 (Figure 8). Additionally, the C-terminal truncation seems to destabilize YCF54, and/or affect the interaction between YCF54 and its antibody as YCF54* produced a much weaker signal than YCF54 (Figure 8).
Figure 8. Immunodetection of recombinant cyclase subunits expressed in E. coli.
E. coli strains containing the indicated plasmids were grown, induced as for the in vivo cyclase assay and cells were harvested after 5 h induction. Clarified cell lysates were analyzed by SDS–PAGE with loading standardized by OD600, followed by transfer to a PVDF membrane for immunodetection. Cyclase subunits were detected by using antibodies raised against Arabidopsis CHL27, YCF54 and Synechocystis Ycf54.
Discussion
The enigma of the subunit composition of the O2-dependent cyclase has been clarified by the discovery of three distinct classes of the enzyme among photosynthetic organisms [3]. Genetic complementation experiments conducted in the Rvi. gelatinosus cyclase mutant revealed the equivalence between the three classes of the enzyme, as indicated in Figure 1B. The next advance involved demonstration of heterologous cyclase activity in E. coli, with two of the three classes of cyclase, from Rvi. gelatinosus and Synechocystis [7], essentially completing the long endeavour to find the ‘missing’ cyclase components. It is widely accepted that cyanobacteria are evolutionarily related to the chloroplasts of algae and plants, endorsed by the conservation of photosynthetic components and processes between cyanobacteria and eukaryotic phototrophs. This conservation also exists in the O2-dependent cyclase as indicated by the shared subunits, AcsF and Ycf54, so the likelihood that eukaryotic cyclase would require extra subunits was low. In this study, we show that, indeed, no extra cyclase-specific subunits are required, and heterologously expressed Arabidopsis CHL27 and YCF54 function as an active cyclase in Rvi. gelatinosus (Figure 3) and E. coli (Figure 4). We did not detect any cyclase activity of Chlamydomonas CTH1 co-expressed with CGL78 in E. coli (Figure 5), possibly because CTH1 was not expressed or it was expressed but unstable in E. coli. Alternatively, the activity of CTH1 may depend on specific copper and/or oxygen levels that differ from those in the E. coli cellular context since it has been documented that CTH1 is produced under copper- and oxygen-replete conditions in Chlamydomonas [12,13]. Nevertheless, we were able to demonstrate the in vivo cyclase activity of Chlamydomonas CRD1 and CGL78 produced in E. coli (Figure 5). A reductase is likely required, but E. coli can provide this function for a variety of bacterial, algal and plant cyclases, and Rvi. gelatinosus can also supply reductase activity for a cyclase from a cyanobacterium [3] and a plant (Figure 3). Thus, this study has defined the cyclase-specific subunits of the eukaryotic cyclase.
At this point, it is necessary to mention the cyclase studies conducted with barley xantha-l and viridis-k mutants, which suggested that the enzyme in this organism contains at least one soluble and two membrane-bound components, of which one is encoded by the Xantha-l gene, an acsF orthologue, and the other could be defective in the viridis-k mutants [16]. The suggestion that barley Ycf54 is a membrane-associated cyclase component increased the potential number of components to at least four [29]. Analysis of barely mutants is a powerful tool, but it is likely to detect lesions in serially linked processes because Chl biosynthesis is co-ordinated with photosystem biogenesis through multiple regulation checkpoints such as the initiating magnesium chelatase and the light-dependent PChlide oxidoreductase steps, and potentially the O2-dependent cyclase step [30–32]. Thus, cyclase-deficient phenotypes are not necessarily due to lack of a cyclase subunit but can be a product of perturbed regulation or unstable cyclase components or even shortage of co-substrates/cofactors. Moreover, various factors can affect cyclase activity as indicated by the studies showing that the NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins [33,34], and the thylakoid plastoquinone pool, are linked with cyclase activity [33,35].
Analogous to other diiron enzymes, the diiron center of cyclase needs to be reduced from +3 to +2 during the catalytic cycle by a reductant or an electron donor, which is believed to be NADPH as suggested by in vitro cyclase assays conducted with chloroplast/plastid fractions from plants and Chlamydomonas [8,10,16] and with Synechocystis cell extracts [10]. However, both AcsF and Ycf54 lack an apparent NAD(P)H-binding motif, implying that the direct electron donor is yet to be determined. As a cellular electron currency, NADPH is utilized to reduce many redox-active components, and the biochemical fractions used for in vitro cyclase assays probably contained some of these redox-active components, one of which could be the direct electron donor to the diiron center of cyclase. Herbst et al. [20] recently proposed that the plastidal ferredoxin-NADPH reductase (FNR) could be involved in reducing the cyclase based on the interaction between FNR1 and YCF54, and the disturbed cyclase activity that resulted from FNR1 deficiency. In oxygenic phototrophs FNR utilizes the electrons from photosynthetically reduced ferredoxin (Fd) to produce NADPH, and when there is a shortage of reduced Fd, such as in the dark, the enzyme can also catalyze the reverse reaction. Reduced Fd serves as the electron donor to a few redox enzymes including some involved in (B)Chl biosynthesis: Fd-dependent 8-vinyl reductase [36], dark-operative PChlide oxidoreductase [37], chlorophyllide oxidoreductase [38] and Chl a oxygenase [39]. In particular, some diiron enzymes use reduced Fd as the reductant as in the case for plant stearoyl-acyl carrier protein Δ9 desaturase [40] and p-aminobenzoate N-oxygenase [41]. To identify the direct electron donor to cyclase, future work should test reduced Fd as well as other types of reductants using purified cyclase subunits rather than undefined complex cellular fractions. Considering the heterologous activity of cyclase in non-pigmented E. coli [7] (Figures 4 and 5), it is predictable that the as-yet unidentified reductant and its potentially existing oxidoreductase are generic and shared with other metabolic processes rather than being specific to the cyclase reaction.
Our work further confirms that Ycf54 is an authentic cyclase subunit in oxygenic phototrophs. In their native hosts, Ycf54 is required for the accumulation of AcsF as demonstrated in Synechocystis [5,18,19], tobacco [6] and Arabidopsis [20] and its interaction with AcsF, mediated by the conserved R82 residue that forms part of a positively charged patch on Ycf54, is required for optimal PChlide formation as shown in Synechocystis [19]. The dependency of AcsF accumulation on Ycf54 was also found for the Arabidopsis enzyme when heterologously expressed in E. coli, but not for the Synechocystis cyclase as indicated by the unaffected level of CycI in the absence of Ycf54 (Figure 8). These results cannot rule out the possibility that Ycf54 plays a role in the assembly of the diiron center of CycI, which requires future characterization of purified CycI from a Ycf54-minus background. It is also possible that Ycf54 plays a role in substrate delivery/channelling or, alternatively, electron transfer during the catalytic cycle; in vitro biochemical assays using purified AcsF and Ycf54 proteins will be required to investigate a catalytic role for Ycf54. In addition, swapping Ycf54 proteins between different cyclase enzymes revealed the Synechocystis protein differs from its Arabidopsis and Chlamydomonas homologs as it was unable to stabilize CHL27 (Figure 8) and its co-expression with CHL27 or CRD1 did not result in a functional cyclase (Figure 7). This difference may be explained by the C-terminal extension, which is present only in green algal and plant Ycf54 proteins (Figure 6A), and which is highly conserved in eukaryotic phototrophs (Figure 6B). Despite its small size, ∼40 aa, this C-terminal extension is significant in the context of the ∼15 kDa Ycf54 protein and its removal abolishes cyclase activity (Figure 7), the basis for which appears to be the absence of any YCF54 protein, and also a significantly lowered level of CHL27 (Figure 8). We propose that the C-terminal extension plays a role in the normal function of green algal and plant YCF54 and CHL27 proteins. It has been reported that Arabidopsis and barley YCF54 proteins form oligomers [6,29] whereas the 1.3 Å structure of the Synechocystis protein indicates that it is monomeric [19]. The C-terminal extension may stimulate and/or maintain the oligomerisation of Ycf54 through hydrophobic interactions between its highly conserved residues, of which 7 out of 9 are aromatic (Figure 6B). Alternatively, the C-terminal extension may be involved in the interaction between green algal and plant Ycf54 and AcsF proteins. Our work also suggests that future studies involving the heterologous activity of CHL27, and possibly CRD1, in E. coli will benefit from co-expression of their cognate Ycf54 proteins.
Abbreviations
- MgP
Mg-protoporphyrin IX
- PPIX
protoporphyrin IX
- PVDF
polyvinylidene difluoride
- PYS
polypeptone-yeast extract-sodium succinate
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contributions
G.E.C. designed and performed research, analyzed data and wrote the paper. C.N.H. designed research and wrote the paper.
Funding
G.E.C. and C.N.H. gratefully acknowledge financial support from the Biotechnology and Biological Sciences Research Council [BB/M000265/1], and European Research Council Synergy Award 854126.
Supplementary Material
References
- 1.Gough S.P., Petersen B.O. and Duus J.Ø (2000) Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc. Natl. Acad. Sci. U.S.A. 97, 6908–6913 10.1073/pnas.97.12.6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanashi K., Minamizaki K. and Fujita Y. (2015) Identification of the chlE gene encoding oxygen-independent Mg-protoporphyrin IX monomethyl ester cyclase in cyanobacteria. Biochem. Biophys. Res. Commun. 463, 1328–1333 10.1016/j.bbrc.2015.06.124 [DOI] [PubMed] [Google Scholar]
- 3.Chen G.E., Canniffe D.P. and Hunter C.N. (2017) Three classes of oxygen-dependent cyclase involved in chlorophyll and bacteriochlorophyll biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 114, 6280–6285 10.1073/pnas.1701687114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinta V., Picaud M., Reiss-Husson F. and Astier C. (2002) Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J. Bacteriol. 184, 746–753 10.1128/JB.184.3.746-753.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingshead S., Kopečá J., Jackson P.J., Canniffe D.P., Davison P.A., Dickman M.J. et al. (2012) Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287, 27823–27833 10.1074/jbc.M112.352526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albus C.A., Salinas A., Czarnecki O., Kahlau S., Rothbart M., Thiele W. et al. (2012) LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 160, 1923–1939 10.1104/pp.112.206045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G.E., Canniffe D.P., Barnett S.F.H., Hollingshead S., Brindley A.A., Vasilev C. et al. (2018) Complete enzyme set for chlorophyll biosynthesis in Escherichia coli. Sci. Adv. 4, eaaq1407 10.1126/sciadv.aaq1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong Y.S. and Castelfranco P.A. (1984) Resolution and reconstitution of Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase, the enzyme system responsible for the formation of the chlorophyll isocyclic ring. Plant Physiol. 75, 658–661 10.1104/pp.75.3.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker C.J., Castelfranco P.A. and Whyte B.J. (1991) Synthesis of divinyl protochlorophyllide. Enzymological properties of the Mg-protoporphyrin IX monomethyl ester oxidative cyclase system. Biochem. J. 276, 691–697 10.1042/bj2760691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollivar D.W. and Beale S.I. (1996) The chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase (characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803). Plant Physiol. 112, 105–114 10.1104/pp.112.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tottey S., Block M.A., Allen M., Westergren T., Albrieux C., Scheller H.V. et al. (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. U.S.A. 100, 16119–16124 10.1073/pnas.2136793100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moseley J., Quinn J., Eriksson M. and Merchant S. (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151 10.1093/emboj/19.10.2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F. et al. (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 10.1105/tpc.010420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minamizaki K., Mizoguchi T., Goto T., Tamiaki H. and Fujita Y. (2008) Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 2684–2692 10.1074/jbc.M708954200 [DOI] [PubMed] [Google Scholar]
- 15.Peter E., Salinas A., Wallner T., Jeske D., Dienst D., Wilde A. et al. (2009) Differential requirement of two homologous proteins encoded by sll1214 and sll1874 for the reaction of Mg protoporphyrin monomethylester oxidative cyclase under aerobic and micro-oxic growth conditions. Biochim. Biophys. Acta 1787, 1458–1467 10.1016/j.bbabio.2009.06.006 [DOI] [PubMed] [Google Scholar]
- 16.Rzeznicka K., Walker C.J., Westergren T., Kannangara C.G., von Wettstein D., Merchant S. et al. (2005) Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 5886–5891 10.1073/pnas.0501784102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen M.D., Kropat J. and Merchant S.S. (2008) Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem. Photobiol. 84, 1336–1342 10.1111/j.1751-1097.2008.00440.x [DOI] [PubMed] [Google Scholar]
- 18.Hollingshead S., Kopecna J., Armstrong D.R., Bucinska L., Jackson P.J., Chen G.E. et al. (2016) Synthesis of chlorophyll-binding proteins in a fully segregated Δycf54 strain of the cyanobacterium Synechocystis PCC 6803. Front. Plant Sci. 7, 292 10.3389/fpls.2016.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollingshead S., Bliss S., Baker P.J. and Neil Hunter C. (2017) Conserved residues in Ycf54 are required for protochlorophyllide formation in Synechocystis sp. PCC 6803. Biochem. J. 474, 667–681 10.1042/BCJ20161002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbst J., Girke A., Hajirezaei M.R., Hanke G. and Grimm B. (2018) Potential roles of YCF54 and ferredoxin-NADPH reductase for magnesium protoporphyrin monomethylester cyclase. Plant J. 94, 485–496 10.1111/tpj.13869 [DOI] [PubMed] [Google Scholar]
- 21.Yu N., Liu Q., Zhang Y., Zeng B., Chen Y., Cao Y. et al. (2019) CS3, a Ycf54 domain-containing protein, affects chlorophyll biosynthesis in rice (Oryza sativa L. Plant Sci. 283, 11–22 10.1016/j.plantsci.2019.01.022 [DOI] [PubMed] [Google Scholar]
- 22.Emanuelsson O., Nielsen H. and Von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 10.1110/ps.8.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tikh I.B., Held M. and Schmidt-Dannert C. (2014) BioBrick compatible vector system for protein expression in Rhodobacter sphaeroides. Appl. Microbiol. Biotechnol. 98, 3111–3119 10.1007/s00253-014-5527-8 [DOI] [PubMed] [Google Scholar]
- 24.McGoldrick H.M., Roessner C.A., Raux E., Lawrence A.D., McLean K.J., Munro A.W., et al. (2005) Identification and characterization of a novel vitamin B12 (cobalamin) biosynthetic enzyme (CobZ) from Rhodobacter capsulatus, containing flavin, heme, and Fe-S cofactors. J. Biol. Chem. 280, 1086–1094 10.1074/jbc.M411884200 [DOI] [PubMed] [Google Scholar]
- 25.Nagashima K.V., Shimada K. and Matsuura K. (1996) Shortcut of the photosynthetic electron transfer in a mutant lacking the reaction center-bound cytochrome subunit by gene disruption in a purple bacterium, Rubrivivax gelatinosus. FEBS Lett. 385, 209–213 10.1016/0014-5793(96)00382-1 [DOI] [PubMed] [Google Scholar]
- 26.Chen G.E., Canniffe D.P., Martin E.C. and Hunter C.N. (2016) Absence of the cbb3 terminal oxidase reveals an active oxygen-dependent cyclase involved in bacteriochlorophyll biosynthesis in Rhodobacter sphaeroides. J. Bacteriol. 198, 2056–2063 10.1128/JB.00121-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miroux B. and Walker J.E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 10.1006/jmbi.1996.0399 [DOI] [PubMed] [Google Scholar]
- 28.Berthold D.A. and Stenmark P. (2003) Membrane-bound diiron carboxylate proteins. Annu. Rev. Plant Biol. 54, 497–517 10.1146/annurev.arplant.54.031902.134915 [DOI] [PubMed] [Google Scholar]
- 29.Bollivar D., Braumann I., Berendt K., Gough S.P. and Hansson M. (2014) The Ycf54 protein is part of the membrane component of Mg-protoporphyrin IX monomethyl ester cyclase from barley (Hordeum vulgare L. FEBS J. 281, 2377–2386 10.1111/febs.12790 [DOI] [PubMed] [Google Scholar]
- 30.Reinbothe S. and Reinbothe C. (1996) The regulation of enzymes involved in chlorophyll biosynthesis. Eur. J. Biochem. 237, 323–343 10.1111/j.1432-1033.1996.00323.x [DOI] [PubMed] [Google Scholar]
- 31.Masuda T. and Fujita Y. (2008) Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 7, 1131–1149 10.1039/b807210h [DOI] [PubMed] [Google Scholar]
- 32.Stenbaek A. and Jensen P.E. (2010) Redox regulation of chlorophyll biosynthesis. Phytochemistry 71, 853–859 10.1016/j.phytochem.2010.03.022 [DOI] [PubMed] [Google Scholar]
- 33.Stenbaek A., Hansson A., Wulff R.P., Hansson M., Dietz K.J. and Jensen P.E. (2008) NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 582, 2773–2778 10.1016/j.febslet.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 34.Richter A.S., Peter E., Rothbart M., Schlicke H., Toivola J., Rintamäki E. et al. (2013) Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol. 162, 63–73 10.1104/pp.113.217141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steccanella V., Hansson M. and Jensen P.E. (2015) Linking chlorophyll biosynthesis to a dynamic plastoquinone pool. Plant Physiol. Biochem. 97, 207–216 10.1016/j.plaphy.2015.10.009 [DOI] [PubMed] [Google Scholar]
- 36.Saunders A.H., Golbeck J.H. and Bryant D.A. (2013) Characterization of BciB: a ferredoxin-dependent 8-vinyl-protochlorophyllide reductase from the green sulfur bacterium Chloroherpeton thalassium. Biochemistry 52, 8442–8451 10.1021/bi401172b [DOI] [PubMed] [Google Scholar]
- 37.Fujita Y. and Bauer C.E. (2000) Reconstitution of light-independent protochlorophyllide reductase from purified BchL and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J. Biol. Chem. 275, 23583–23588 10.1074/jbc.M002904200 [DOI] [PubMed] [Google Scholar]
- 38.Nomata J., Kitashima M., Inoue K. and Fujita Y. (2006) Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett. 580, 6151–6154 10.1016/j.febslet.2006.10.014 [DOI] [PubMed] [Google Scholar]
- 39.Tanaka A., Ito H., Tanaka R., Tanaka N.K., Yoshida K. and Okada K. (1998) Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. U.S.A. 95, 12719–12723 10.1073/pnas.95.21.12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox B.G., Lyle K.S. and Rogge C.E. (2004) Reactions of the diiron enzyme stearoyl-acyl carrier protein desaturase. Acc. Chem. Res. 37, 421–429 10.1021/ar030186h [DOI] [PubMed] [Google Scholar]
- 41.Choi Y.S., Zhang H., Brunzelle J.S., Nair S.K. and Zhao H. (2008) In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 6858–6863 10.1073/pnas.0712073105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.