Abstract
N-acetyl-galactosamine (GalNAc) conjugation enhances liver specificity for therapeutic oligonucleotides. Here we report on a novel design with improved activity and stability compared with a triantennary design. We applied a versatile monovalent serinol-GalNAc conjugation strategy. First, 1–4 serial serinol-linked GalNAc units were conjugated to terminal positions of small interfering RNA (siRNA) molecules. In primary hepatocytes, 5′ antisense GalNAc conjugates were inactive, whereas 3′ antisense and 3′ or 5′ sense conjugates displayed low activity for single GalNAc units, while 2–4 serial GalNAc conjugates were all equally potent. In mice, 5′ sense conjugates with 2–4 serial GalNAc units were all as potent as a triantennary GalNAc control (1 mg/kg). Second, increased spacing between two serial 5′ sense-conjugated GalNAc units did not affect in vitro activity. Finally, two single GalNAc units were positioned at opposite ends of the sense strand. A single dose (0.3 mg/kg) of this novel conjugate in mice showed a 3-fold reduction of serum target protein level at day 7 and 4-fold lower serum level at day 27, relative to an equimolar dose of a triantennary GalNAc conjugate of the same siRNA. Improved tritosome stability (by liquid chromatography-mass spectrometry [LC-MS] analysis) can at least partially explain the increased activity and duration of action for the novel GalNAc conjugate.
Keywords: siRNA-conjugate, GalNAc-siRNA, serinol-GalNAc, asialoglycoprotein, GalNAc-cluster
Graphical Abstract
We systematically tested serinol-GalNAc siRNA conjugates with valences of 1–4 at the terminal positions. We demonstrate equal RNAi activity for two, three, and four GalNAc moieties. Interestingly, one serinol-GalNAc at either end of the sense strand increases siRNA stability in vitro and improves activity and duration of action in vivo.
Introduction
N-acetyl-galactosamine (GalNAc) conjugation for hepatocyte-targeted delivery of oligonucleotide therapeutics has become a success story with the promise of precision treatment of severe diseases with hepatic origin, as illustrated by the increasing number of GalNAc-conjugated RNAi and antisense oligonucleotide (ASO) drug candidates under evaluation in clinical trials,1 as well as by the recent FDA approval of GIVLAARI (givosiran) from Alnylam.
The asialoglycoprotein receptor (ASGP-R) has an important role in the homeostasis of glycosylated serum proteins.2 It belongs to the c-type family of lectins and recognizes a broad spectrum of carbohydrates, such as D-fucose, D-galactose, D-lactose, and GalNAc, with increasing affinity.3 The differences in affinity result in well-balanced levels of serum glycoproteins that are cleared at different rates. GalNAc conjugates bind with high affinity to ASGP-R and take advantage of the first step in clathrin-dependent endocytic uptake.2,4 Upon acidification in early endosomes, the ligand is released3 and further transported to the lysosomal compartment, while the receptor recycles back to the plasma membrane.
ASGP-R is a type II membrane protein that forms hetero-oligomers of two highly homologous subunits, ASGP-R H1 and ASGP-R H2. Reported ASGP-R oligomerization stoichiometries differ between species and methods applied, ranging from two to six subunits. However, early studies noted that when carbohydrate ligand clusters were tested for ASGP-R binding, the number of carbohydrate units had a positive correlation with receptor binding; for example, trivalent GalNAc clusters had a 16-fold higher affinity than divalent clusters,5,6 whereas only limited improvement was noticed by tetravalent galactose compared with trivalent galactose.5 Hence first attempts to deliver oligonucleotides by GalNAc conjugation were conducted with trimeric, or triantennary, GalNAc clusters.7 Recently, different linker modalities addressing valency and steric configuration of the GalNAc clusters have been reported.8,9 In addition, several teams have also focused on broader structure-function relationships while facilitating synthesis of GalNAc-conjugated oligonucleotides.10, 11, 12, 13, 14, 15
In the spirit of earlier work aiming at developing potent siRNA therapeutics with reduced complexity of synthesis, we investigated in vitro activity and stability of serial serinol-linked GalNAc conjugates (1–4 GalNAc units), sequentially testing the effect of conjugation to the four terminal positions of 21-mer blunt-ended siRNA targeting transcripts of the murine transthyretin (Ttr) gene. We concluded that in vitro activity was very similar for serinol-linked serial GalNAc conjugates with two, three, or four GalNAc units in the 3′ antisense, 3′ sense, or 5′ sense positions. We also observed that the distance between the two serial GalNAc units was not crucial for potent in vitro activity. Finally, we saw that two single serinol-linked GalNAc units conjugated to the respective terminal positions of the sense strand resulted in improved in vivo activity and duration of action compared with a triantennary GalNAc conjugate version of the same siRNA.
Results
Optimal Valency and Location of Serinol-Linked GalNAc Units
Most experiments were performed with GalNAc units conjugated to a blunt-end 21-mer siRNA sequence targeting Ttr derived from Nair et al.8 Sequences, conjugation designs, and purities are listed in Table S1. High-pressure liquid chromatography (HPLC) traces of selected siRNA conjugates are provided in Table S2. Exemplary conversions and yields for GalNAc conjugates are given in Table S3 for siRNAs 7–14, 24, and 25. In all cases, conversion was complete, and high purity of the product was achieved. Serially attached serinol-GalNAc structures, as well as triantennary conjugates, are illustrated in Figure S7.
Primary mouse hepatocytes were incubated for 24 h with 0.001- to 10-nM concentrations of the respective design variant. In all experiments, target transcript reduction was compared with a triantennary 5′-sense GalNAc conjugate of the same sequence (“positive control”), a triantennary 5′-sense GalNAc-conjugated anti-Luciferase siRNA (“non-targeting control” [NTC]), and untreated cells incubated in cell culture media alone (“UT”).
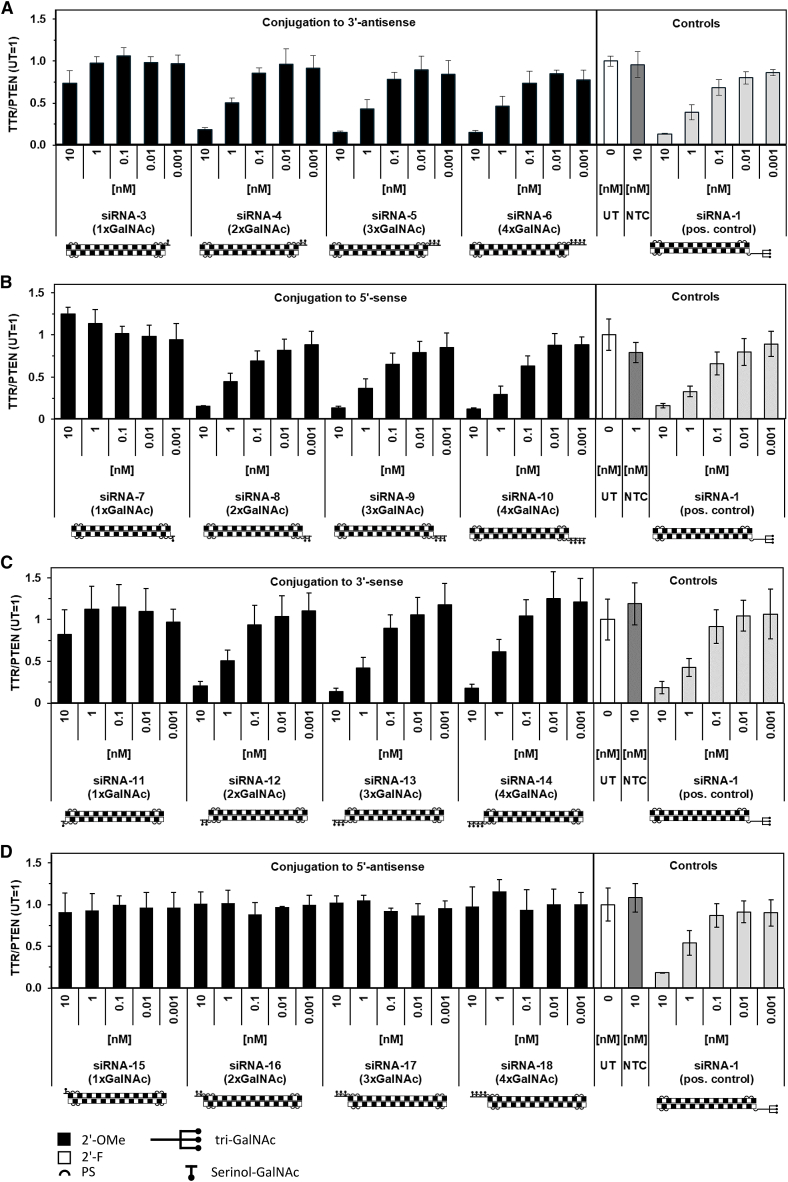
A single GalNAc unit conjugated siRNA had no, or minimal, effect on target transcript levels, regardless of where the GalNAc was positioned (Figure 1). All conjugates, regardless of valency, lacked in vitro activity if the GalNAc units were conjugated to the 5′ antisense end of the siRNA (Figure 1D). However, for all other possible end positions, i.e., for 3′ antisense, and 3′ or 5′ sense conjugation, there was little or no difference in in vitro activity between conjugation of two, three, and four serial GalNAc units. Note that for each series of serial conjugates, in vitro activity in primary murine hepatocytes was in the same range as for the positive, GalNAc triantennary, control (Figures 1A–1C).
Figure 1.
In Vitro Dose Response to GalNAc-Conjugated siRNA with Different Valency and Positioning of Serial Conjugation
(A–D) Primary mouse hepatocyte expression of Ttr after 24-h treatment with 1–4 serinol-linked GalNAc units in the (A) 3′ antisense, (B) 5′ sense, (C) 3′ sense, and (D) 5′ antisense end of the siRNA. Positive (pos.) control is a 5′ triantennary GalNAc conjugate of the same siRNA (siRNA-1), and negative control is a 5′ triantennary GalNAc conjugate of a non-targeting siRNA (NTC; siRNA-2). Data represent mean ± SD normalized to untreated cells (UT) in one experiment, representative of three biological replicates.
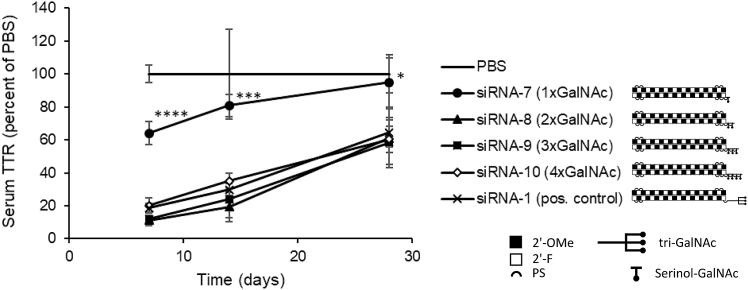
Next, mice were injected subcutaneously (s.c.) with the triantennary positive control siRNA-1 (1 mg/kg) or equimolar amounts of serial GalNAc compounds with 1–4 serinol-linked GalNAc units conjugated to the 5′ sense position of the same siRNA. Serum TTR was tracked over time after injection (Figure 2). Just as in the in vitro experiment, conjugation to a single GalNAc unit resulted in significantly reduced activity compared with the multivalent conjugates, whereas 2–4 serial GalNAc units conjugation resulted in the same range and time course of reduction of the serum biomarker. Importantly, there was no significant increase in activity when the number of serial GalNAc units increased from two to three or four, and no significant difference between triantennary GalNAc and any of the two, three, or four serial GalNAc conjugates at any of the analyzed time points.
Figure 2.
Time Course for Serum TTR in Mice following Single s.c. Dose of siRNA-GalNAc Conjugates of Different Valency
C57BL/6 mice were injected s.c. with 1 mg/kg siRNA-1 (pos. control), a 5′ sense triantennary GalNAc conjugate targeting Ttr, or equimolar amounts of the same siRNA sequence conjugated to 1–4 serinol-linked GalNAc units in the terminal 5′ sense position of the molecule. Data represent serum TTR levels normalized to mean values for PBS control animals at respective time points, mean ± SD, n = 4 animals/group. Asterisks represent significant differences between siRNA-7 and siRNA-1 (pos. control): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Varying Distance between Two Serinol-Linked GalNAc Units
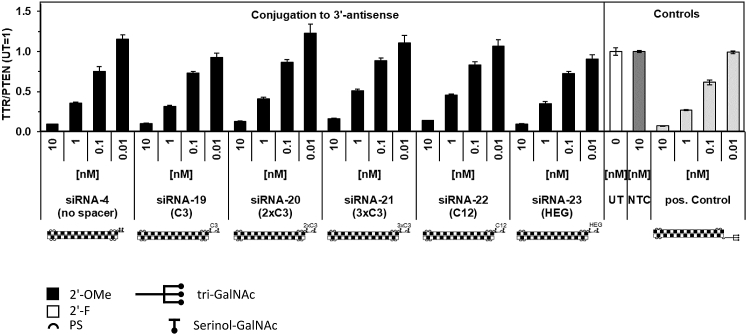
After demonstrating that conjugation of siRNA to two serial GalNAc units was sufficient for potent hepatocyte target transcript reduction in vitro and in vivo, we tested the effect of increased spacing between two single GalNAc units located at the 3′-antisense position. Surprisingly, the introduction of one, two, and three C3-linker units, or a C12- or hexaethylene glycol (HEG)-linker, had no apparent impact on GalNAc-mediated siRNA in vitro activity after 24-h incubation with primary mouse hepatocytes (Figure 3; further linker details are listed in Table S1).
Figure 3.
In Vitro Dose Response to Dimeric GalNAc Conjugates with Different Spacer Length
Ttr transcript levels in primary mouse hepatocytes 24 h posttreatment with siRNA conjugated to two serinol-linked GalNAc units at the 3′ antisense terminus, with varying spacers between the GalNAc units. Data represent mean ± SD in one experiment, representative of three biological replicates.
Single Serinol-Linked GalNAc Units at Respective Ends of the Sense Strand
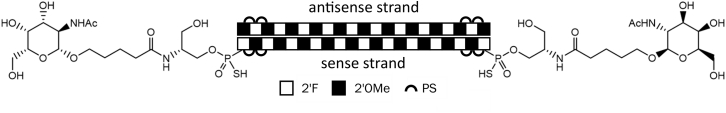
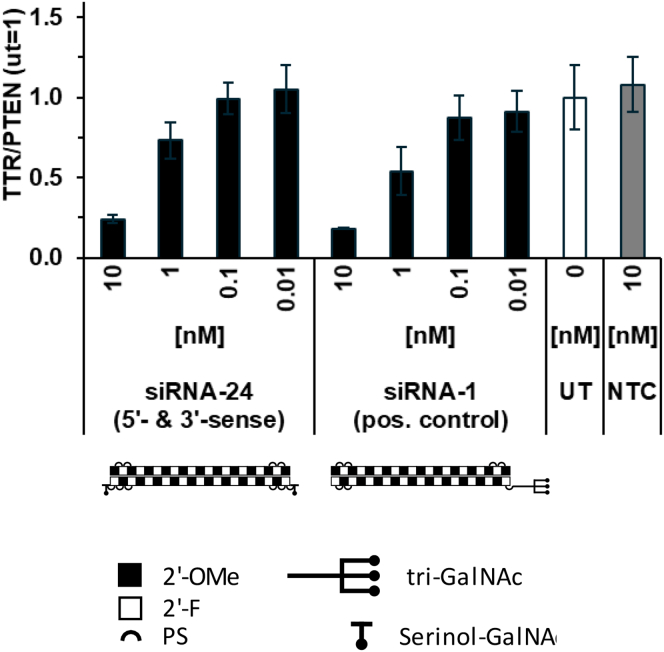
The spacers/linkers reported in Figure 3 can be assumed to be flexible in aqueous solution, and so the hydrocarbon chain length does not necessarily translate to the actual distance between the two GalNAc units in these different constructs. To test the limits for spacing between two single GalNAc units in relation to ASGP-R-mediated, GalNAc-conjugated siRNA uptake and activity, we therefore proceeded to separate two single GalNAc units by a solid and inflexible spacer, i.e., the annealed siRNA, by serinol-linked conjugation to both ends of the sense strand (siRNA-24; Figure 4).
Figure 4.
Structure of Serinol GalNAc Conjugate with Ligands at Both Sense Strand Termini
Illustration of serinol-GalNAc-linker structure and position of siRNA-24.
This molecular construct should separate the GalNAc units from each other by approximately 5–6 nm, as estimated by the structural characteristics of double-stranded RNA in the A-form (length of the dsRNA 19-mer and 21-mer calculated as 5.3 and 5.9 nm, respectively16). In vitro activity of this novel construct was tested in primary mouse hepatocytes. For the tested concentration range, reduction of target transcript levels by siRNA-24 was close to the effect seen with the positive control (Figure 5).
Figure 5.
In Vitro Dose Response to siRNA with Single GalNAc at Both Sense Strand Termini
Primary mouse hepatocyte expression of Ttr after 24-h treatment with a single serinol-linked GalNAc unit in each end of the sense strand. Pos. control is a 5′ triantennary GalNAc conjugate of the same siRNA, and negative control is a 5′ triantennary GalNAc conjugate of a non-targeting siRNA. Data represent mean ± SD in one experiment, representative of three biological replicates.
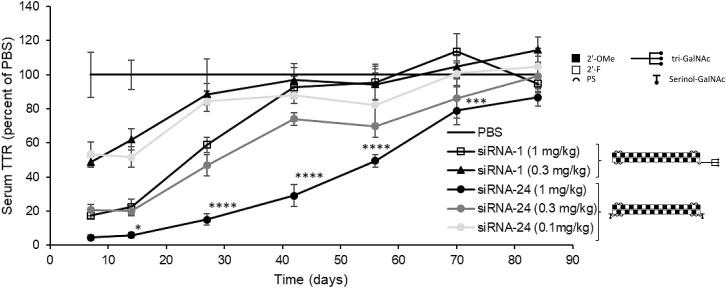
Translatability of this in vitro finding to in vivo activity was tested in mice. Animals were injected s.c. with equimolar amounts of siRNA-24 and the control compound. Serum TTR (as a biomarker of liver activity) was tracked for 84 days. Surprisingly, siRNA-24 demonstrated both improved activity and duration of action compared with equimolar amounts of the triantennary positive control (Figure 6).
Figure 6.
Improved Dose Response of Serum TTR in Mice to siRNA with Single GalNAc at Both Sense Strand Termini
Serum TTR in mice at indicated time points following s.c. administration of 1, 0.3, or 0.1 mg/kg siRNA conjugates with single GalNAc units at both termini of the sense strand (siRNA-24) or a triantennary GalNAc-cluster at the 5′-sense (pos. control; siRNA-1). Data represent mean ± SD, n = 4/group, data normalized to PBS control for each time point. Asterisks represent significant differences between siRNA-24 and siRNA-1 at 1 mg/kg: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Because the single 5′, 3′-sense GalNAc conjugate (siRNA-24) showed improved in vivo activity, we proceeded to investigate whether activity could be further improved by adding two serial serinol-linked GalNAc units at both ends of the sense strand. There was no difference in in vitro activity or tritosome stability between “2×1 GalNAc” (siRNA-24) and “2×2 GalNAc” (siRNA-25, data not shown). Equimolar amounts of 2×1 GalNAc and 2×2 GalNAc compounds were injected s.c. in mice, and serum TTR was tracked for 42 days. Comparing in vivo performance of the two compounds, we observed no increase in activity by adding two additional GalNAc units (Figure S2).
Stability Analysis of GalNAc Conjugates
We hypothesized that the increased activity of siRNA-24 compared with the triantennary control could in part be explained by increased stability in the endosomal/lysosomal compartment. The rat liver tritosome fraction is enriched for lysosomal and endosomal proteins,17 and is a highly representative model for lysosomal enzymes used to study chemical modifications that stabilize siRNA molecules against nuclease degradation.18, 19, 20 Stability of conjugated siRNA was analyzed in vitro by incubation with acidified rat liver tritosome lysates at 37°C for up to 72 h, a method intended to mimic processing in endosomes and lysosomes. Already after 4-h incubation with tritosome lysate (pH 4), a band shift versus untreated material was apparent for all tested GalNAc conjugates (Figure S3; Table S4). In addition to this basal degradation, positioning of the serinol-linked GalNAc units appeared to influence RNA degradation when comparing across different constructs (Figure S3; Table S4). Surprisingly, 3′ antisense conjugation destabilized the molecule, whereas conjugation to all other termini resulted in stabilizing the siRNA in this experimental model of endosomal/lysosomal degradation (Figure S3, comparison with unconjugated control molecule). For each GalNAc unit positioning on the two strands, a similar degree of stabilization was demonstrated for both single and serial serinol-GalNAc conjugates (data not shown). siRNA-24 stands out when compared with the other tested serinol-linked GalNAc conjugates, as well as the triantennary positive control siRNA-1, by demonstrating an unusually high level of stability even at the 72-h time point (Figure S3).
To better understand this degradation process, the 5′, 3′ sense conjugate (siRNA-24) and the triantennary positive control (siRNA-1) were analyzed by HPLC-MS after a 24-h incubation in acidified tritosome lysates and RNA extraction. Stable metabolites, identified by mass changes, were compared with individual full-length single strands. UV signals of tritosome-treated samples show a loss of 1, 4, and 5 nucleotides from the 3′ end of the antisense strand (A annotations in Figures S4A and S4B, right). The sense strand (B annotations in Figures S4A and S4B, right) underwent complete processing within the incubation time. However, the mass delta of 406 and 609 Da indicates only the loss of two and three GalNAc units cleaved at the glycosidic bond, respectively, leaving the actual sequence intact. Both conjugates (siRNA-1 and siRNA-24) share the nature and distribution of stable metabolites. Thus, the conjugates differ only in the rate of their breakdown by lysosomal enzymes. In summary, there is a difference in stability for the two different constructs, and this difference may at least partially explain the increased activity and duration of action for the novel GalNAc conjugate.
Discussion
We have systematically investigated the impact of valency and position of serinol GalNAc-conjugated siRNA molecules for targeting and reduction of a hepatocyte transcript. Using GalNAc conjugation for siRNA delivery means piggybacking on a scavenger receptor-mediated mechanism evolved to maintain homeostasis of serum glycoprotein levels via recognition of exposed terminal carbohydrates with different affinities for the receptor, and thus a balanced clearance of the different proteins from blood. This mechanism has been investigated for decades as a means of liver-targeted delivery of a range of different cargos before the surge of GalNAc-conjugated ASOs and siRNAs intended for therapeutic use.21 After reports on optimal conformation of triantennary clusters reached the public domain,5,7,22 many teams working in the field have used such structures for experimental purposes.
The surprising find of strongly increased activity and duration of action for the 5′, 3′ sense conjugate (siRNA-24) described above originated with an intent to develop a simple and flexible synthesis path for GalNAc-conjugated siRNA, and to assure that the new constructs were at least as potent as the same siRNA conjugated to a triantennary GalNAc cluster. A variety of synthetic solutions have been developed over the last two decades to facilitate oligonucleotide conjugation to multivalent GalNAc clusters. The main synthetic approaches so far described for synthesis of multivalent GalNAc conjugates can be divided into three major categories: (1) the use of pre-assembled bi-, tri-, or tetravalent clusters, which are introduced as a non-nucleosidic building block, either as a loaded solid support or a phosphoramidite;8 (2) the use of branching amidite building blocks, followed by multiple coupling of a single GalNAc amidite;23 and (3) the serial use of nucleosidic11 and non-nucleosidic GalNAc amidites.24 These approaches can all be, and have been, adapted to post-synthetic conjugation procedures in order to allow for structure-function relationship studies with different cluster architectures.25, 26, 27
Reported conclusions on optimal GalNAc valency have varied with selected chemistry and modality, but as a rule a minimum of two GalNAc units are needed for achieving in vivo activity, and in most studies, hepatocyte uptake and activity increase with further increased valency. In a comparison of biantennary and triantennary structures, Nair et al.8 reported on improved uptake of fluorescently labeled siRNA molecules with triantennary GalNAc clusters over the biantennary. Interestingly, ASOs showing a 5-fold reduction of ED50 in mice have been reported for molecules with a single GalNAc conjugation as compared with an unconjugated ASO. At higher valences (bi- and tri-GalNAc), an additional 3-fold reduction in ED50, equal for both conjugates, was shown.9 Yamamoto et al.,13 using serial GalNAc monomer-conjugated ASO gapmers, demonstrated valency (1 < 3 < 5)-dependent targeting of apolipoprotein B in mice, both with respect to effect and to liver exposure. Recently, Sharma et al.14 reported a comparison of triantennary GalNAc clusters and 2–4 serial linked GalNAc monomers to siRNA, where the triantennary cluster conjugates were superior over any of the other tested conjugates in mice. In contrast with these publications, we observed that serial conjugates of 2–4 monomeric ligands demonstrated similar activity both in vitro and in vivo.
There are several previous reports on the importance of GalNAc positioning for oligonucleotide conjugate activity. Triantennary GalNAc conjugation to the 5′ end of an ASO has been shown to increase both in vitro and in vivo potency compared with 3′ end conjugation of the same oligonucleotide.28 Optimal location has also been investigated for GalNAc conjugation to siRNAs, such as by Matsuda et al.,11 where three serial GalNAc units at the 3′ end of the sense strand conjugated via the 2′ or 3′-OH of the ribose, or the N-1 of pseudouridine base, showed minor differences in vitro and in vivo, whereas walking the three units, together or spaced apart, through the sequence all through to the 5′ end strongly reduced knockdown activity in vitro and in vivo. In line with these findings, we noted a difference in siRNA stability when incubating conjugates in the tritosomal fraction of rat liver lysates depending on the position of terminal conjugation (3′ or 5′ end of the sense strand). Not unexpectedly, we show that 5′-antisense conjugation results in loss of function, probably because of the residual linker that interferes with 5′-phosphorylation and RNA-induced silencing complex (RISC) loading. Based on this observation, we speculate that the new 5′ and 3′ sense conjugate will result in reduced risk for sense strand off-target activity, in addition to the used sense strand modification pattern that in and of itself drastically reduces the risk for sense strand RNAi activity.29
Schmidt et al.30 demonstrated that varying the phosphorothioate content in a 2'-O-methoxyethyl (MOE) gapmer conjugated to 1–3 GalNAc units affects binding to ASGP-R (in competition assays with GalNAc triclusters). It is interesting to note that some of the ASOs were duplexed with a complementary all-RNA strand. Conjugating two GalNAc units to the 3′ end or one GalNAc each to the 3′ and the 5′ ends of the ASO (i.e., making a double-stranded conjugate with a GalNAc conformation similar to siRNA-24 described above), resulted in higher ASGP-R binding than a single GalNAc alone. However, this ASO construct had reduced in vivo activity compared with a triantennary GalNAc cluster version of the same oligonucleotide. Here we note a clear difference to our novel 5′, 3′ sense GalNAc monomer siRNA conjugate, where in vitro activity was similar or reduced compared with the triantennary positive control, but both in vivo activity and duration of action were several-fold higher than for the control molecule (Figure 6). The ASGP-Rs on the hepatocyte are typically considered to form heterotrimers. It is tempting to speculate that for siRNA-24 the ligand positioning may allow for interactions between the two singular GalNAc units and two separate ASGP-R clusters at the hepatocyte plasma membrane.
Serial attachment of non-nucleoside building blocks gives greater flexibility, omits lengthy synthesis of cluster architectures, and harnesses the power of solid-phase oligonucleotide synthesis. With a minimum set of individual molecules, a medicinal and combinatoric approach can be used to assemble a broad range of siRNA conjugates. A preformed cluster architecture is currently part of the most mature clinical candidate GalNAc-conjugated siRNAs and ASOs. We propose that serial building block-based GalNAc conjugates can be beneficial over preformed triantennary building blocks when scaling up from laboratory-scale synthesis. The repeated use of smaller building blocks should allow for reduced costs and efforts in the synthesis of starting materials. Their ability to elongate a synthesis allows for the use of state-of-the-art universal solid supports.31 The presence of the 5′-DMT (4,4'-dimethoxytrityl) group may be exploited in both anion-exchange (AEX) and reverse-phase HPLC (RP-HPLC)-based methods to ease purification.
The serinol scaffold used in the studies described above has an additional benefit, because it can be easily produced in a stereochemically defined manner with high yield from a pool of chiral starting material (l-serine). We suggest that a process scale oligonucleotide synthesis could be performed with a preformed serinol-GalNAc phosphoramidite monomer of the same architecture. Hence the post-column conjugation protocol and second purification step can both be omitted. The yield of the individual conjugations varied and was largely dependent on the scale of the reaction and the selection of fraction pools. A yield optimization process was not performed.
Another potential benefit of siRNA conjugation to serinol-linked GalNAc units is the serinol itself. It has previously been described that serinol capping of terminal positions in 2-OMe RNA splice switching single-stranded oligonucleotides results in increased serum stability over 24 h,32 as well as increased anti-miR-activity for 3′/5′ serinyl capped 2′-OMe-RNA.33 Similar observations were made for siRNA with phosphodiester-linked serinol nucleic acids in the 3′ end of the antisense, as well as in the 5′ and 3′ ends of the sense strand, which can increase both in vitro stability against exonuclease attack and in vitro activity through preventing sense strand loading.29 Stabilization of terminal positions in modified siRNA, e.g., with phosphorothioates, has been demonstrated to improve stability against exonuclease attack, which, in turn, improves the in vivo dose response and duration of action for GalNAc conjugates.19 We have confirmed this stabilization against serum exonucleases by phosphodiester-linked serinol GalNAc (data not shown). However, possibly of higher importance is the observation that the phosphorothioate serinol GalNAc linker stabilizes the siRNA against degradation in tritosome lysates (Figure S3). Compared with other tested conjugates and the triantennary control molecule, the novel 5′, 3′ sense GalNAc monomer conjugate demonstrated a clear improvement both in activity and in duration of action in the in vivo experiments, but this was not reflected in the receptor-mediated 24-h in vitro activity for the same molecule (Figure 5). We hypothesized that the use of a serinol linker could affect siRNA stability after the receptor-mediated uptake in hepatocytes, i.e., along the endosomal-lysosomal pathway, and that this would manifest in stronger effects in a longer-term in vivo study than in a relatively short in vitro assay. The acidic tritosome lysate stability data (Figures S3 and S4) support this hypothesis, although they do not confirm the causality between increased endosomal/lysosomal stability and increased activity and duration of action for the novel GalNAc conjugate in vivo.
It is interesting to note that after tritosome treatment at low pH, the 5′, 3′ sense conjugate samples contain only one major degradation product. We can only speculate at this time that the resulting molecule, which lacks only the GalNAc sugars at their glycosidic attachments site, either resides as a depot in the cell, increases endosomal escape to the cytosol, or increases RISC loading. The 5′, 3′ sense GalNAc conjugate molecular design has been used previously for two additional siRNA sequences targeting ALDH2 at different positions of the target mRNA and has demonstrated a high level of activity and duration of action for both; however, the degree of improvement over triantennary GalNAc cluster control molecules was seen to vary between the different siRNA sequences (Figures S5 and S6). Continued studies of uptake and intracellular processing of this interesting novel GalNAc conjugate will elucidate which part(s) of the processing on the way “from injection to action” is different from the triantennary GalNAc cluster constructs, and if there is a specific metabolite that can be used for further improvement of activity for new GalNAc-conjugated siRNAs for therapeutic or target validation purposes. Likewise, it is tempting to speculate that the 3′, 5′ sense GlaNAc conjugation in combination with advanced molecular designs20,34,35 might result in additional improvements of activity.
Materials and Methods
Cell Culture
Plateable, male mouse (CD-1) cryopreserved hepatocytes (MSCP10, various lots; Thermo Fisher Scientific) were thawed and cryo-preservation medium exchanged for Williams E medium (A1217601; Thermo Fisher Scientific) supplemented with Primary Hepatocyte Thawing and Plating Supplements (CM3000; Thermo Fisher Scientific). Cell density was adjusted to 250,000 cells per 1 mL. 100 μL per well of this cell suspension was seeded into collagen pre-coated 96-well plates. The test item was prediluted in the same medium (five times concentrated) for each concentration, and 25 μL of this prediluted siRNA or medium only were added to the cells. Cells were cultured at 37°C and 5% CO2. The supernatant was discarded 24 h after treatment, cells were washed in cold PBS, and 250 μL RNA-Lysis Buffer S (Stratec, Germany) was added. Following 15-min incubation at room temperature, plates were stored at −80°C until RNA isolation was performed according to the manufacturer’s protocol.
TaqMan Analysis
For Ttr and Pten MultiPlex TaqMan analysis, 10 μL isolated RNA for each treatment group was mixed with 10 μL PCR mastermix (UF-LP5X-RT0101; Eurogentec, Germany) containing 300 nM Ttr primer, 300 nM Pten primer, and 100 nM of each probe, as well as 0.5 U Euroscript II RT polymerase with 0.2 U RNase inhibitor. TaqMan analysis was performed in a 384-well plate with a 10-min RT step at 48°C, 3 min initial denaturation at 95°C, and 40 cycles of 95°C for 10 s and 60°C for 1 min. TaqMan primer and probes listed in Table S5 were purchased from BioTEZ and Eurogentech.
Tritosome Stability Assay
siRNA was incubated for 0, 4, 24, or 72 h in Sprague-Dawley Rat Liver Tritosomes (R0610.LT; Sekisui Xenotech, Kansas City, MO, USA). To mimic the acidified environment, we mixed tritosome lysates 10:1 with low pH buffer (1.5M acetic acid, 1.5M sodium acetate, pH 4.75). 30 μL of these acidified tritosomes was mixed with 10 μL siRNA (20 μM) and incubated for the indicated times at 37°C. Following incubation, RNA was isolated with the Clarity OTX Starter Kit-Cartridges (KSO-8494; Phenomenex, Germany) according to the manufacturer’s protocol for biological fluids. Lyophilized RNA was reconstituted in 30 μL H2O, mixed with 4× loading buffer, and 5 μL was loaded to a 20% Tris-borate-EDTA (TBE)-polyacrylamide gel electrophoresis (PAGE) for separation and qualitative semiquantitative analysis. PAGE was run at 120 V for 2 h, and RNA was visualized by ethidium-bromide staining, with digital images acquired with a Bio-Rad Gel Doc EZ Imager. Liquid chromatography-tandem mass spectrometry (LC-MS) analysis of the resulting metabolites was performed under desaturating conditions. For a detailed description, see Supplemental Information.
In Vivo Experiments
In vivo experiments with C57BL/6 mice were performed at Experimental Pharmacology & Oncology Berlin-Buch (EPO, Germany) in accordance with the United Kingdom Co-ordinating Committee on Cancer Research (UKCCCR) regulations for the Welfare of Animals36 and of the German Animal Protection Law and approved by the local responsible authorities.
Compounds were injected s.c. into the scapular region of mice. The administration volume was adjusted corresponding to the body weight (10 mL/kg). Blood was collected by orbital sinus bleeding, and serum was extracted by centrifugation using Microvette 500 Z-Gel (Sarstedt) and transferred into Eppendorf tubes for storage at −20°C.
Mouse TTR-ELISA
Serum TTR levels in mice were analyzed with a mouse Prealbumin ELISA kit (Alpco 41-PALMS-E01; ALPCO, Salem, NH, USA) according to the manufacturer’s protocol. Typically, serum was diluted 1:800 up to 1:8,000 in the recommended diluent.
Oligonucleotide Synthesis and GalNAc Conjugation
Assembly of the oligonucleotide chain and linker building blocks was performed applying phosphoramidite methodology. Necessary building blocks are depicted in Figure S1A. For the on-column conjugation of the triantennary GalNAc (positive control molecule), the final two synthesis cycles were performed using the necessary trivalent branching amidite (4a or 4b) followed by another round of the synthesis cycle using the C4GN amidite (5). Synthesis of serial GalNAc-conjugated oligonucleotides is also depicted in Figure S1A with the sense strand of siRNA-24 as an example. Synthesis was performed using DMT-(S)-serinol(TFA)-succinate-lcaa-CPG (1) and/or DMT-(S)-serinol(TFA)-CEP (2) in the respective synthesis cycles. Upon completion of the solid-phase synthesis, the crude product was cleaved from the solid support and then purified by AEX-HPLC to yield the pure precursor oligonucleotide (pre-siRNA-24B, Figure S1B, left). Conjugation of the GalNAc synthon was achieved by coupling GalNAc-NHS ester (3) to the serinol-amino function of the respective precursor. AEX analysis indicated full conversion after 30 min (Ac-siRNA-24B, Figure S1B, middle) by peak shift. LC-MS analysis confirmed successful conjugation multiplicity (data not shown). After precipitation of the crude product by addition of 10% volume of 2M NaCl, followed by 10× iPrOH, the crude product pellet was dissolved in 40% aqueous (aq.) MeNH2 and then further purified by AEX-HPLC to yield the final sense strand siRNA-24B with high purity (Figure S1B, right).
Detailed protocols are found in the Supplemental Information.
Statistical Analysis
Statistical analysis was carried out in GraphPad Prism with one-way ANOVA followed by Tukey’s multiple comparison tests. siRNA conjugate treatments were analyzed against the vehicle control of similar conditions, unless otherwise stated in the figure legends; p values were adjusted to account for multiple comparisons.
Author Contributions
A.W. designed and led in vitro and in vivo experiments. L.B. provided conjugation strategy and led syntheses of siRNA conjugates. L.W. and M.S. conducted in vitro experiments, as well as tissue samples analysis of in vivo experiments. M.W.L. conducted statistical analysis. A.W., M.W.L., and L.B. wrote the manuscript.
Conflicts of Interest
All authors are employed at Silence Therapeutics GmbH.
Acknowledgments
We thank Sybille Dames for designing TaqMan probes and primers; Gabi Anlauf and Jens Endruschat for siRNA conjugate synthesis; Pablo Lores Lareo for oligo characterization; and Steffen Schubert, Giles Campion, and Eliot Morrison for reviewing the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.05.026.
Supplemental Information
References
- 1.Benizri S., Gissot A., Martin A., Vialet B., Grinstaff M.W., Barthélémy P. Bioconjugated Oligonucleotides: Recent Developments and Therapeutic Applications. Bioconjug. Chem. 2019;30:366–383. doi: 10.1021/acs.bioconjchem.8b00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steirer L.M., Park E.I., Townsend R.R., Baenziger J.U. The asialoglycoprotein receptor regulates levels of plasma glycoproteins terminating with sialic acid α2,6-galactose. J. Biol. Chem. 2009;284:3777–3783. doi: 10.1074/jbc.M808689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onizuka T., Shimizu H., Moriwaki Y., Nakano T., Kanai S., Shimada I., Takahashi H. NMR study of ligand release from asialoglycoprotein receptor under solution conditions in early endosomes. FEBS J. 2012;279:2645–2656. doi: 10.1111/j.1742-4658.2012.08643.x. [DOI] [PubMed] [Google Scholar]
- 4.Ashwell G., Morell A.G. The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv. Enzymol. Relat. Areas Mol. Biol. 1974;41:99–128. doi: 10.1002/9780470122860.ch3. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y.C., Townsend R.R., Hardy M.R., Lönngren J., Arnarp J., Haraldsson M., Lönn H. Binding of synthetic oligosaccharides to the hepatic Gal/GalNAc lectin. Dependence on fine structural features. J. Biol. Chem. 1983;258:199–202. [PubMed] [Google Scholar]
- 6.Lee R.T., Lee Y.C. Preparation of Cluster Glycosides of N-Acetylgalactosamine That Have Subnanomolar Binding Constants Towards the Mammalian Hepatic Gal/GalNAc-specific Receptor. Glycoconj. J. 1987;4:317–328. [Google Scholar]
- 7.Biessen E.A.L., Vietsch H., Rump E.T., Fluiter K., Kuiper J., Bijsterbosch M.K., van Berkel T.J. Targeted delivery of oligodeoxynucleotides to parenchymal liver cells in vivo. Biochem. J. 1999;340:783–792. [PMC free article] [PubMed] [Google Scholar]
- 8.Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 9.Kinberger G.A., Prakash T.P., Yu J., Vasquez G., Low A., Chappell A., Schmidt K., Murray H.M., Gaus H., Swayze E.E., Seth P.P. Conjugation of mono and di-GalNAc sugars enhances the potency of antisense oligonucleotides via ASGR mediated delivery to hepatocytes. Bioorg. Med. Chem. Lett. 2016;26:3690–3693. doi: 10.1016/j.bmcl.2016.05.084. [DOI] [PubMed] [Google Scholar]
- 10.Manoharan M., Nair J.K., Kandasamy P., Matsuda S., Kelin A.V., Jayaraman M., Rajeev K.G. 2014. Oligonucleotide-ligand conjugates and process for their preparation related. International Patent WO 2015/006740 A2, filed July 11 and published January 15, 2015. [Google Scholar]
- 11.Matsuda S., Keiser K., Nair J.K., Charisse K., Manoharan R.M., Kretschmer P., Peng C.G., V Kel’in A., Kandasamy P., Willoughby J.L. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 2015;10:1181–1187. doi: 10.1021/cb501028c. [DOI] [PubMed] [Google Scholar]
- 12.Rajeev K.G., Nair J.K., Jayaraman M., Charisse K., Taneja N., O’Shea J., Willoughby J.L., Yucius K., Nguyen T., Shulga-Morskaya S. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem. 2015;16:903–908. doi: 10.1002/cbic.201500023. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto T., Sawamura M., Wada F., Harada-Shiba M., Obika S. Serial incorporation of a monovalent GalNAc phosphoramidite unit into hepatocyte-targeting antisense oligonucleotides. Bioorg. Med. Chem. 2016;24:26–32. doi: 10.1016/j.bmc.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 14.Sharma V.K., Osborn M.F., Hassler M.R., Echeverria D., Ly S., Ulashchik E.A., Martynenko-Makaev Y.V., Shmanai V.V., Zatsepin T.S., Khvorova A., Watts J.K. Novel Cluster and Monomer-Based GalNAc Structures Induce Effective Uptake of siRNAs in Vitro and in Vivo. Bioconjug. Chem. 2018;29:2478–2488. doi: 10.1021/acs.bioconjchem.8b00365. [DOI] [PubMed] [Google Scholar]
- 15.Prakash T.P., Yu J., Migawa M.T., Kinberger G.A., Wan W.B., Østergaard M.E., Carty R.L., Vasquez G., Low A., Chappell A. Comprehensive Structure-Activity Relationship of Triantennary N-Acetylgalactosamine Conjugated Antisense Oligonucleotides for Targeted Delivery to Hepatocytes. J. Med. Chem. 2016;59:2718–2733. doi: 10.1021/acs.jmedchem.5b01948. [DOI] [PubMed] [Google Scholar]
- 16.Arias-Gonzalez J.R. Single-molecule portrait of DNA and RNA double helices. Integr. Biol. 2014;6:904–925. doi: 10.1039/c4ib00163j. [DOI] [PubMed] [Google Scholar]
- 17.Bagshaw R.D., Mahuran D.J., Callahan J.W. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol. Cell. Proteomics. 2005;4:133–143. doi: 10.1074/mcp.M400128-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Zlatev I., Foster D.J., Liu J., Charisse K., Brigham B., Parmar R.G., Jadhav V., Maier M.A., Rajeev K.G., Egli M., Manoharan M. 5′-C-Malonyl RNA: Small Interfering RNAs Modified with 5′-Monophosphate Bioisostere Demonstrate Gene Silencing Activity. ACS Chem. Biol. 2016;11:953–960. doi: 10.1021/acschembio.5b00654. [DOI] [PubMed] [Google Scholar]
- 19.Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017;45:10969–10977. doi: 10.1093/nar/gkx818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parmar R., Willoughby J.L.S.S., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. 5′-(E)-Vinylphosphonate: A Stable Phosphate Mimic Can Improve the RNAi Activity of siRNA-GalNAc Conjugates. ChemBioChem. 2016;17:985–989. doi: 10.1002/cbic.201600130. [DOI] [PubMed] [Google Scholar]
- 21.Springer A.D., Dowdy S.F. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther. 2018;28:109–118. doi: 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X., Leroux J.C., Castagner B. Well-Defined Multivalent Ligands for Hepatocytes Targeting via Asialoglycoprotein Receptor. Bioconjug. Chem. 2017;28:283–295. doi: 10.1021/acs.bioconjchem.6b00651. [DOI] [PubMed] [Google Scholar]
- 23.Dubber M., Fréchet J.M.J. Solid-phase synthesis of multivalent glycoconjugates on a DNA synthesizer. Bioconjug. Chem. 2003;14:239–246. doi: 10.1021/bc0256244. [DOI] [PubMed] [Google Scholar]
- 24.Matulic-Adamic J., Serebryany V., Haeberli P., Mokler V.R., Beigelman L. Synthesis of N-acetyl-D-galactosamine and folic acid conjugated ribozymes. Bioconjug. Chem. 2002;13:1071–1078. doi: 10.1021/bc025525q. [DOI] [PubMed] [Google Scholar]
- 25.Spinelli N., Defrancq E., Morvan F. Glycoclusters on oligonucleotide and PNA scaffolds: synthesis and applications. Chem. Soc. Rev. 2013;42:4557–4573. doi: 10.1039/c2cs35406c. [DOI] [PubMed] [Google Scholar]
- 26.Prakash T.P., Brad Wan W., Low A., Yu J., Chappell A.E., Gaus H., Kinberger G.A., Østergaard M.E., Migawa M.T., Swayze E.E., Seth P.P. Solid-phase synthesis of 5′-triantennary N-acetylgalactosamine conjugated antisense oligonucleotides using phosphoramidite chemistry. Bioorg. Med. Chem. Lett. 2015;25:4127–4130. doi: 10.1016/j.bmcl.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Farzan V.M., Ulashchik E.A., Martynenko-Makaev Y.V., Kvach M.V., Aparin I.O., Brylev V.A., Prikazchikova T.A., Maklakova S.Y., Majouga A.G., Ustinov A.V. Automated Solid-Phase Click Synthesis of Oligonucleotide Conjugates: From Small Molecules to Diverse N-Acetylgalactosamine Clusters. Bioconjug. Chem. 2017;28:2599–2607. doi: 10.1021/acs.bioconjchem.7b00462. [DOI] [PubMed] [Google Scholar]
- 28.Østergaard M.E., Yu J., Kinberger G.A., Wan W.B., Migawa M.T., Vasquez G., Schmidt K., Gaus H.J., Murray H.M., Low A. Efficient Synthesis and Biological Evaluation of 5′-GalNAc Conjugated Antisense Oligonucleotides. Bioconjug. Chem. 2015;26:1451–1455. doi: 10.1021/acs.bioconjchem.5b00265. [DOI] [PubMed] [Google Scholar]
- 29.Kamiya Y., Takai J., Ito H., Murayama K., Kashida H., Asanuma H. Enhancement of stability and activity of siRNA by terminal substitution with serinol nucleic acid (SNA) ChemBioChem. 2014;15:2549–2555. doi: 10.1002/cbic.201402369. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt K., Prakash T.P., Donner A.J., Kinberger G.A., Gaus H.J., Low A., Østergaard M.E., Bell M., Swayze E.E., Seth P.P. Characterizing the effect of GalNAc and phosphorothioate backbone on binding of antisense oligonucleotides to the asialoglycoprotein receptor. Nucleic Acids Res. 2017;45:2294–2306. doi: 10.1093/nar/gkx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravikumar V.T., Kumar R.K., Olsen P., Moore M.N., Carty R.L., Andrade M., Gorman D., Zhu X., Cedillo I., Wang Z. UnyLinker: An Efficient and Scaleable Synthesis of Oligonucleotides Utilizing a Universal Linker Molecule: A Novel Approach To Enhance the Purity of Drugs. Org. Process Res. Dev. 2008;12:399–410. [Google Scholar]
- 32.Kotikam V., Arzumanov A.A., Gait M.J., Kumar V.A. Enhanced splice correction by 3′, 5′-serinol and 2′-(ω-O-methylserinol) guarded OMe-RNA/DNA mixmers in cells. Artif. DNA PNA XNA. 2013;4:77–83. doi: 10.4161/adna.27279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nahar S., Kotikam V., Kumar V.A., Maiti S. Inhibition of miR-21 by 3′/5′-Serinyl-Capped 2′-O-Methyl RNA Interspersed with 2′-O-(2-Amino-3-Methoxypropyl) Uridine Units. Nucleic Acid Ther. 2016;26:327–334. doi: 10.1089/nat.2015.0591. [DOI] [PubMed] [Google Scholar]
- 34.Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M. Advanced siRNA Designs Further Improve In Vivo Performance of GalNAc-siRNA Conjugates. Mol. Ther. 2018;26:708–717. doi: 10.1016/j.ymthe.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elkayam E., Parmar R., Brown C.R., Willoughby J.L., Theile C.S., Manoharan M., Joshua-Tor L. siRNA carrying an (E)-vinylphosphonate moiety at the 5' end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2017;45:3528–3536. doi: 10.1093/nar/gkw1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Workman P., Balmain A., McNally N.J., Mitchison N.A., Pierrepoint C.G., Raymond R., Rowlatt C., Stephens T.C., Wallace J. United Kingdom Co-ordinating Committee on Cancer Research (UKCCCR) Guidelines for the Welfare of Animals in Experimental Neoplasia (Second Edition) Br. J. Cancer. 1998;77:1–10. doi: 10.1038/bjc.1998.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.