Significance
DNA repair is essential to life and to avoidance of genome instability and cancer. Xeroderma pigmentosum group G (XPG) protein acts in multiple DNA repair pathways, both as an active enzyme and as a scaffold for coordinating with other repair proteins. We present here the structure of the catalytic domain responsible for its DNA binding and nuclease activity. Our analysis provides structure-based hypotheses for how XPG recognizes its bubble DNA substrate and predictions of the structural impacts of XPG disease mutations associated with two phenotypically distinct diseases: xeroderma pigmentosum (XP, skin cancer prone) or Cockayne syndrome (XP/CS, severe progressive developmental defects).
Keywords: endonuclease, ERCC5, electron microscopy, crystallography, crystal structure
Abstract
Xeroderma pigmentosum group G (XPG) protein is both a functional partner in multiple DNA damage responses (DDR) and a pathway coordinator and structure-specific endonuclease in nucleotide excision repair (NER). Different mutations in the XPG gene ERCC5 lead to either of two distinct human diseases: Cancer-prone xeroderma pigmentosum (XP-G) or the fatal neurodevelopmental disorder Cockayne syndrome (XP-G/CS). To address the enigmatic structural mechanism for these differing disease phenotypes and for XPG’s role in multiple DDRs, here we determined the crystal structure of human XPG catalytic domain (XPGcat), revealing XPG-specific features for its activities and regulation. Furthermore, XPG DNA binding elements conserved with FEN1 superfamily members enable insights on DNA interactions. Notably, all but one of the known pathogenic point mutations map to XPGcat, and both XP-G and XP-G/CS mutations destabilize XPG and reduce its cellular protein levels. Mapping the distinct mutation classes provides structure-based predictions for disease phenotypes: Residues mutated in XP-G are positioned to reduce local stability and NER activity, whereas residues mutated in XP-G/CS have implied long-range structural defects that would likely disrupt stability of the whole protein, and thus interfere with its functional interactions. Combined data from crystallography, biochemistry, small angle X-ray scattering, and electron microscopy unveil an XPG homodimer that binds, unstacks, and sculpts duplex DNA at internal unpaired regions (bubbles) into strongly bent structures, and suggest how XPG complexes may bind both NER bubble junctions and replication forks. Collective results support XPG scaffolding and DNA sculpting functions in multiple DDR processes to maintain genome stability.
Xeroderma pigmentosum group G (XPG) protein acts in multiple DNA damage response (DDR) pathways, and mutations in its ERCC5 (excision repair cross-complementing rodent repair deficiency, complementation group 5) gene are associated with two distinct diseases. Functionally, it has a structure-specific endonuclease activity (1) and interacts with multiple DNA processing proteins in nucleotide excision repair (NER), transcription-coupled repair (TCR), base excision repair (BER), and homologous recombination (HR). In NER, XPG enzymatic activity cleaves the damaged DNA strand 3′ to the lesion. NER is initiated by lesion recognition requiring XPC, opening of the DNA around the lesion by the helicase activities of the TFIIH repair/transcription complex, and binding the resulting single-stranded (ss)DNA by replication protein A (RPA). XPG is recruited to the NER complex on DNA containing an unpaired bubble (bubble DNA) through its direct interactions with TFIIH, the XPD helicase activity of which is strongly stimulated by the interaction (2). Requiring the physical presence of XPG, the XPF-ERCC1 heterodimer is recruited and incises the double-stranded (ds)DNA 5′ to the lesion (3–6). XPG incises the dsDNA on the 3′ side of the lesion at a position 1 nt from the bubble junction (3–6). Defects in XPG’s NER nucleolytic and scaffolding roles are linked to the hereditary and highly skin cancer-prone disease xeroderma pigmentosum (XP) (7).
Different mutations in ERCC5 cause a particularly severe form of the fatal developmental disorder Cockayne syndrome (CS), which (in contrast to XP) features pronounced neurodegeneration and very early death, but no cancer predisposition (7, 8). The hallmark of CS is loss of the preferential repair of DNA damage in transcribed strands by TCR. While the mechanistic connection of XPG loss to CS is not entirely clear, it is evident that it extends beyond the enzymatic function of XPG in NER. The requirement for XPG in TCR likely involves its demonstrated interactions with RNA polymerase II (RNAPII), TFIIH, and CS group B (CSB) proteins, and biochemical evidence suggests that XPG may participate in allowing lesion removal in the presence of stalled RNAPII (3, 4, 9).
XPG also plays important roles in other DDR pathways through multiple protein interactions distinct from its NER functions. XPG depletion causes DNA double-strand breaks, chromosomal abnormalities, cell-cycle delays, defective HR repair (HRR), sensitivity to poly(ADP-ribose) polymerase inhibition, inability to overcome replication fork stalling, and replication stress (10). These phenotypes reflect its function in HRR mediated by its direct interactions with BRCA1, BRCA2, PALB2, and the RAD51 recombinase, which likely promote loading of RAD51 onto RPA-coated ssDNA. In BER, XPG stimulates glycosylase activity of NTH1 in vitro (11–13). XPG interacts with and stimulates Werner syndrome helicase (WRN) in vitro, and the two proteins colocalize during replication (14). XPG’s incision activity is implicated in cleavage of R-loops, including those generated by loss of WRN in the premature aging disorder Werner syndrome (15–17). While the multiple roles of XPG in DDR depend on its many protein–protein interactions, some require its enzymatic activity but others apparently do not. It is less clear how its specific binding to ss/dsDNA junctions might be involved.
The type of mutations that lead to cancer-prone XP (XP-G) and the fatal neurodevelopmental disorder CS (XP-G/CS) are distinct, although both are rare autosomal recessive (18, 19). All of the ERCC5 changes causing XP-G are missense point mutations, whereas the originally described XP-G/CS mutations were predicted to be severely truncating, leading to the model that loss of XPG protein causes XP-G/CS, whereas XP-G results from an incision-defective mutant protein causing loss of global NER (20). This idea received strong support from mouse models, since mice with knockin catalytically inactivating point mutations in ERCC5 are UV-sensitive but otherwise normal (21, 22), whereas knockout mice display CS-like progressive progeroid features, extensive neurodegeneration, and early death (8, 23). More recently, however, several new XP-G/CS patients have been described with missense mutations in ERCC5 (24, 25), suggesting that the situation is complicated with respect to causal changes in XPG protein for disease outcome (i.e., XP vs. XP/CS). It is also presently unclear to what extent the requirement for participation of XPG in DDR processes other than NER or TCR, in particular BER and HRR, is relevant to development of CS when XPG function is lost.
XPG binding and incision of Y and bubble DNA structures is well studied (1, 4, 26–28), yet the structural mechanisms underlying XPG activities including DNA binding in DDR are not known. XPG is a founding member of the primordial FEN superfamily, also known as the 5′ nuclease or Rad2 superfamily, that includes human FEN1, EXO1, and GEN1 (29, 30), and yeast ortholog Rad2 (31–34). FEN1, involved in resolution of 5′ flaps in lagging-strand replication, is one of the most well-studied in the family and can serve as an example to broadly understand 5′ nuclease mechanisms. In FEN1, incision requires at least a four-step process of: 1) Initial binding to dsDNA regions of the 5′ flap; 2) specific substrate validation of target features (dsDNA, single-strand 5′ flap, and a 1-nt 3′ flap) with concomitant conformation shifts in both the protein and DNA conformations; 3) shifting the scissile phosphate into the catalytic site; and finally 4) two-metal–based phosphodiester bond cleavage (29, 35–39). Thus, only after substrate testing by coordinated DNA and protein conformational changes (35) is FEN1 licensed to cut DNA, making it distinct from other nuclease classes where substrate validation is at the binding step. Furthermore, for XPG some substrates are bound but not incised (4, 28). As incision appears not to be required for all XPG functions, could some DDR pathways involve its structured DNA binding capability?
To address questions regarding XPG structural mechanisms in the DDR, we solved the crystal structure of the human XPG catalytic domain (XPGcat). We furthermore combined biochemical, small angle X-ray scattering (SAXS), and electron microscopy (EM) analyses of full-length protein with experimental data on disease-causing mutations both in XPG in patient cells and in bacterially expressed XPGcat. We show that XPG structure shares key metal-binding and basic residues with FEN1 and is likely to work through an analogous disorder-to-order transition requiring the properly folded XPGcat structure for an analogous multistep process to that associated with specificity in FEN1 and EXO1. We furthermore found that human XPG unexpectedly formed a stable dimer in solution and on DNA that appears to be required for catalytic activity and for protein stability. Rotary shadowing EM data showed that XPG bound specifically to bubble DNA and replication forks and strongly bent bubble DNA. We examine XP-G and XP-G/CS missense mutations and characterize the impact of these mutations on expression and endonuclease activity, prompting a reexamination of how these mutations result in the different disease phenotypes. The collective findings provide fundamental mechanistic implications for the molecular roles of XPG in multiple DDR pathways.
Results
XPGcat Crystal Structure Reveals 5′ Nuclease Fold with XPG-Specific Features.
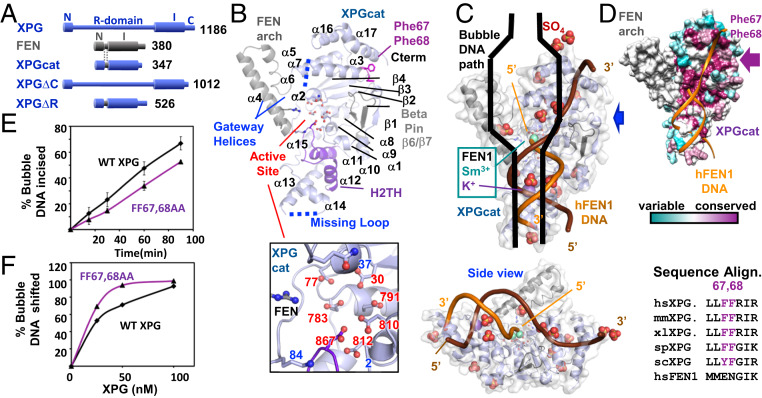
To reveal the structural basis for XPG’s DNA incision and binding activities, we created a construct “XPGcat” with the XPG catalytic domain for crystallographic analysis. Between the superfamily-conserved catalytic domain composed of two regions, N and I, XPG has an additional ∼680-amino acid insert (called the “R-domain” or spacer region) as well as an extended C terminus following the I region (Fig. 1A) (4, 40). The R-domain and the C terminus are predicted to contain coiled coils and large regions of disorder that would likely interfere with crystallization (41). For XPGcat, we replaced the XPG R-domain with the corresponding 40-amino acid helical gateway/cap or arch region from an archaeal FEN (4, 42) and truncated the C terminus (Fig. 1A). To show that XPGcat is active for incision, we compared incision activity of full-length XPG or XPGcat on Y DNA (DNA forming a Y-like structure with one dsDNA and two ssDNA regions) or 15-nt bubble DNA, where 15 nt refers to a central, unpaired region between two dsDNA arms (SI Appendix, Fig. S1 A–C). As previously observed, full-length XPG could incise both, with greater activity for bubble DNA (4). XPGcat had robust incision activity for Y DNA but no detectable activity on 15-nt bubble DNA, consistent with the inserted FEN cap limiting active site access to only those substrates having a free 5′ terminus (43).
Fig. 1.
Conserved features in XPGcat structure. (A) Domain schematic of XPG, FEN, and XPG deletion mutants. In XPGcat and XPG∆R, XPG N and I catalytic regions have replaced the R-domain with the corresponding region in FEN. (B) XPGcat cartoon denoting FEN superfamily elements. Zoom-in view shows XPGcat active site carboxylates and gateway basic residues and Gln37. (C) Orthogonal views of overlay of FEN1:DNA (PDB ID code 3Q8K) onto XPGcat surface representation and SO4’s. Only DNA, K+, and catalytic metals from the FEN1:DNA complex are shown. Blue arrow gives side view perspective. Black cartoon of bubble DNA suggests that only half a bubble DNA would fit on the DNA binding face of XPGcat. (D) XPGcat surface representation colored by conservation. Sequence alignment shows that Phe67 and Phe68 are highly conserved residues in XPG family, but not in FEN1. (E) Incision activity of 15-nt bubble DNA (HP15T/C) by FF67,68AA was decreased relative to WT XPG. Single turnover assays done in triplicate with 33.3-nM flag-tagged protein and 3.3-nM DNA. Error bars show SD. Representative gel in SI Appendix, Fig. S1F. (F) FF67,68AA had higher affinity for 15-nt bubble DNA (5 nM) compared to WT XPG in EMSA. Representative gel in SI Appendix, Fig. S1G.
We crystallized XPGcat, solved the structure, and refined the model to a resolution of 2.1 Å with an R and Rfree of 0.22 and 0.25, respectively (Fig. 1B and SI Appendix, Table S1). The overall structure is a mixed α/β single domain containing a central seven-stranded twisted β-sheet core with α-helices on either side. This fold resembles members of the FEN superfamily and the catalytic domain of its yeast ortholog Rad2 (RMSD 1.0 Å) (33), with which it shares 30% identity (29). A highly conserved α15/random coil (residues 937 to 955) on the back side (not fully visible in Fig. 1B) connects one side of the protein to the other. The XPGcat structure contains the two C-terminal helices (α16 and α17) found in Rad2 and FEN1, but not in EXO1 nor GEN1 (31, 33, 34, 36). These helices form the interface for DNA recognition on the nonincised strand near the junction. For FEN1, this is a dsDNA arm with a 1-nt 3′ flap. For XPG, this would be the ssDNA region of its bubble DNA substrate. In EXO1, this region is not a required feature of its substrate.
Structurally conserved in all FEN superfamily members, seven carboxylates in the β-sheet core and basic residues in the gateway helices (α2 and α4) in XPG are similarly positioned in the active site (29). As observed for FEN1 and EXO1, mutation of one of the conserved basic residues (K84A) in XPGcat drastically reduced incision activity on Y DNA compared to WT XPGcat, with only a relatively minor effect on DNA binding (SI Appendix, Fig. S1 D and E). Although we cannot assess part of α4 and α5 derived from the FEN protein, residue conservation and the similarity of the XPG α2 support a gateway-based mechanism for selection of the ssDNA on the strand that will be incised. Suggesting a similar disorder-to-order regulation of the active site, α2 is partially disordered as found for DNA-free FEN1 (44). XPGcat also includes the helix-2turn-helix (H2TH) that helps form the K+ binding site and the β-pin implicated in DNA binding in FEN (36). No K+ was bound, not unexpectedly given the absence of DNA substrate.
All FEN superfamily members interact with ss/dsDNA junction substrates, primarily bind the dsDNA portions, and position the 5′ ssDNA region near to the active site (29). As FEN1 is similar to XPG in overall structure and a FEN1 crystal structure exists with DNA on either side of the scissile phosphate, we modeled our XPGcat domain with FEN1 DNA. The alternative of modeling with DNA in the ortholog Rad2 structure would not be ideal, as Rad2 crystal structures with DNA contained only one dsDNA arm (33). Like other superfamily members, our structure indicates that XPG could bind dsDNA at two positions, the active site and near the H2TH/K+ site (Fig. 1 B and C).
A key difference between XPGcat and other FEN superfamily members comes from XPG-specific features of its bubble DNA substrate. XPGcat does not contain the 3′ flap binding pocket or the acid block observed in the FEN1 complex, not unexpectedly, as these structural elements provide specificity for FEN-specific substrates. In this region, the XPG bubble DNA substrate would instead have ssDNA (Fig. 1C). Two Phe residues, highly conserved in XPG but not in other 5′ nucleases (Figs. 1 B and D and 2), stick out from the surface in this region, in a manner atypical for this hydrophobic side chain. These Phe residues are suitably positioned to intercalate with the bases of ssDNA. Mutation of these residues to alanine (FF67,68AA) reduced full-length XPG activity slightly in single turnover experiments (Fig. 1E and SI Appendix, Fig. S1F). Surprisingly, we observed that affinity for bubble DNA increased (Fig. 1F and SI Appendix, Fig. S1G). Since there could be a number of explanations, such as interaction with other regions of XPG, we tested the FF67,68AA mutation in XPGcat. Although DNA binding also increased in the FF67,68AA XPGcat mutant (SI Appendix, Fig. S1E), incision was significantly increased in the mutant (SI Appendix, Fig. S1D), opposite to what we observed in full-length XPG. One possible explanation is that the double phenylalanine motif distorts the bubble DNA, as part of a FEN1-like substrate validation (SI Appendix, Fig. S1H). Elimination of these aromatic side chains reduces distortion and enhances DNA binding. However, substrate validation can no longer occur properly, so the gateway helices remain disordered and incision activity in full-length XPG is reduced. In the XPGcat construct, however, substrate validation has been bypassed by replacing the disordered gateway helices in XPG with the ordered thermophilic FEN helices.
Fig. 2.
Sequence alignment of XPGcat, Rad2, FEN1, EXO1, and GEN1.
Human XPG Forms a Dimer.
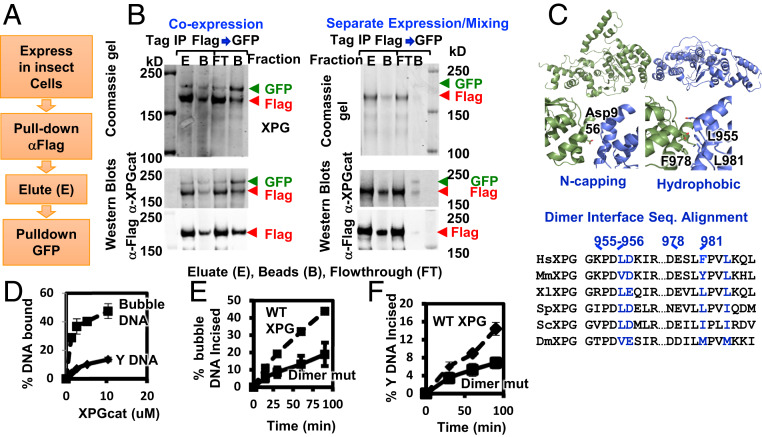
Earlier immunoprecipitation experiments suggested that XPG may be a multimer, although that conclusion was contradicted by biophysical measurements (45). We revisited this issue by examining full-length XPG recombinantly expressed in insect cells. We designed XPG with two different C-terminal tags, FLAG and GFP, so that the differentially tagged proteins could be distinguished by their mobility in SDS/PAGE. When the two tagged XPG proteins were coexpressed, anti-FLAG resin pulled down both XPG-FLAG and XPG-GFP, suggesting a complex had formed (Fig. 3A). The greater quantity of XPG-FLAG would be consistent with the stochastic presence of both homomeric XPG-FLAG multimers and heteromeric XPG multimers with both tags. Consistent with this interpretation, protein eluted from the FLAG beads and reisolated on GFP binder resin contained equivalent amounts of XPG-FLAG and XPG-GFP proteins (Fig. 3 A and B). These results establish XPG as a multimer. Significantly, when the differentially tagged XPGs were expressed separately in two different insect cell cultures and then mixed before lysis, we did not observe XPG-GFP bound with anti-FLAG resin nor XPG-FLAG bound to GFP binder beads (Fig. 3B), indicating that XPG multimers are relatively stable and do not readily exchange under these conditions.
Fig. 3.
XPG forms a dimer in solution. (A) Schematic of sequential coimmunoprecipitation strategy of XPG-FLAG and XPG-GFP. (B) Stable multimerization tested by coimmunoprecipitation of differently tagged XPG either coexpressed or separately expressed in insect cells. Coomassie and Westerns of FLAG peptide eluate, protein remaining on the α-FLAG beads, GFP binder flow-through (FT), and protein bound to GFP binder beads. (C) Cartoon of XPGcat dimer in crystal lattice. Zoom-in views show interface. XPG sequence alignment shows interface residues are conserved. (D) Quantitation of DNA binding analysis by EMSA shows XPGcat had higher affinity for 15 nt bubble DNA (HP15T/C) than for Y DNA. Representative gel shown in SI Appendix, Fig. S4B. (E and F) Dimer mutant 4A had reduced incision activity on 15-nt bubble DNA (HP15T/C) and Y DNA compared to WT XPG incision activity. Single turnover assays done in triplicate with 33.3-nM flag-tagged protein and 3.3 nM DNA. Error bars show SD. Representative gels in SI Appendix, Fig. S4 C and D.
We measured the stoichiometry of purified XPG in solution by SAXS, from which the mass of a protein can be calculated within ∼10% error (46). Prior to SAXS data collection, we purified XPG through size-exclusion chromatography (SEC). We observed two peaks of XPG (SI Appendix, Fig. S2 A and B), which had different mobility in native PAGE (SI Appendix, Fig. S2C). We collected SAXS data on the second, major peak, the only one for which we had enough protein concentration to get a reliable SAXS signal (SI Appendix, Figs. S2 and S3 and Table S3). The molecular mass calculated from the SAXS data was 265 kDa, consistent with a dimer (theoretical mass 266 kDa). The Porod exponent, a measure of the protein volume and thereby protein flexibility, was 2.9 (47). Globular, well-folded proteins have a Porod exponent of 4, flexibly linked globular regions 3, and unstructured proteins 2. Thus, a Porod exponent of 2.9 signifies a partially structured protein, in agreement with disorder prediction analyses. Indicating that XPG alone is always in a dimer or higher-order multimer in solution, we found no peak in SEC consistent with a monomer. To map the dimerization interface, we tested XPG truncation constructs (Fig. 1A and SI Appendix, Figs. S2 and S3 and Table S3). Elimination of the C terminus (XPG∆C) did not alter the dimerization state. Elimination of the R-domain (XPG∆R) gave mixed populations of monomer and dimer. Addition of 10-nt bubble DNA to XPG∆R increased the ratio of the dimer (SI Appendix, Fig. S3B). The DNA-induced dimerization for this construct was corroborated by multiangle light scattering (SI Appendix, Fig. S3C). The Porod exponents for XPG∆C and XPG∆R increased to 3.2, indicating a reduction in flexibility. The XPGcat construct without the R- and C-terminal domains had greatly reduced flexibility with a Porod exponent of 3.9. The experimental SAXS curve generally matched that predicted from the crystal structure (SI Appendix, Fig. S2E). Based on the residual differences between experimental and model SAXS curves, the best match came with an ensemble of monomer and dimer models, indicating that XPGcat is mostly monomeric with a small population of dimer. Together, these mass measurement results suggest that multiple regions in the R-domain, XPGcat, and the C terminus participate together in dimer stabilization.
As the crystal lattice can reveal protein–protein interfaces, we examined the XPGcat crystal lattice. A symmetric dimer interface (Fig. 3C) is formed by hydrophobic residues Leu955, Phe978, and Leu981 in α14 and α15 in one molecule with the same region from a crystallographically related molecule, and between the acidic side chain of Asp956 N-capping the positively charged N-terminal end of α14 (Figs. 2 and 3C). Automated assembly analysis independently identified the same dimer (48). These four residues are conserved in XPG but not in related superfamily member FEN1, although the helical structures themselves are conserved. FEN1 does not form a crystallographic contact at this position (36) and is a monomer in solution. In the yeast Rad2 ortholog of XPG, crystal contacts did not occur with this exact interface, but did form with nearby residues (33) (SI Appendix, Fig. S4A). The superfamily member GEN1 is a homodimer, but the C terminus to C terminus dimer interface in XPGcat is on a different side from the GEN1 dimer, where the gateway helices protecting the active site stack end-to-end and where helices α14 and α15 are packing with GEN’s C-terminal chromodomain (32).
Comparison of XPGcat structure to that of FEN1 bound to its DNA substrate indicated that XPGcat as a monomer could bind to only one dsDNA arm (i.e., a Y-structure) and not to both dsDNA arms of a bubble DNA. As a dimer (albeit at a low ratio relative to monomer), XPGcat might interact with both dsDNA arms of a full bubble DNA, even if it cannot incise (SI Appendix, Fig. S1 A–C). To test this possibility, we compared binding of XPGcat to Y DNA and to 15-nt bubble DNA. Although we showed earlier that XPGcat could not incise the bubble DNA (SI Appendix, Fig. S1 A–C), surprisingly it bound to bubble DNA up to fourfold better than to Y DNA (Fig. 3D and SI Appendix, Fig. S4B), albeit still more weakly than does full-length XPG (4). This seemingly antithetical DNA binding with no incision is consistent with FEN superfamily members physically separating their initial DNA binding from positioning for incision (35). An XPGcat dimer, which our SAXS mass measurements suggest exists at low frequency, would create two dsDNA-interacting surfaces to preferentially bind bubble DNA over Y DNA.
To test the functional relevance of this dimer interface, we made quadruple mutations (dimer mutant 4A: L955A/D956A/F978A/L981A) in full-length XPG. We observed significantly lower expression levels of the mutant protein when expressed in insect cells, suggesting protein destabilization. Although we were unable to purify enough mutant protein for SAXS or other biophysical analyses for protein stability, we were able to test for incision activity. Consistent with the dimerization interface being functionally relevant, the 4A dimer interface mutant showed an approximately twofold decrease in 15-nt bubble DNA incision activity (Fig. 3E and SI Appendix, Fig. S4C). We reasoned that incision of bubble DNA would be more affected than of Y DNA. Indeed, we observed a 40% smaller loss in incision activity with the Y DNA (Fig. 3F and SI Appendix, Fig. S4D). Nonetheless, the dimer interface was important for protein activity even with Y DNA. We observed a similar protein destabilization of the dimer interface mutant in the XP-G/CS patient cell line XPCS1RO that lacks detectable XPG protein. We transfected these cells with WT XPG expression construct, the 4A mutant, or a quadruple dimer interface mutant “A3E” with three glutamate substitutions, instead of alanine, at the hydrophobic residues. Western analysis done at 48 h after transfection revealed greatly diminished XPG protein amounts expressed in the human cells from the two dimer interface mutants compared to WT (SI Appendix, Fig. S4E). This destabilization is specific to the full-length protein, as we saw no apparent destabilization of the XPGcat dimer interface mutant, and the Porod exponent was 3.9 for both XPGcat and the XPGcat 4A mutant (SI Appendix, Figs. S2 and S3).
EM Analysis of DNA Binding.
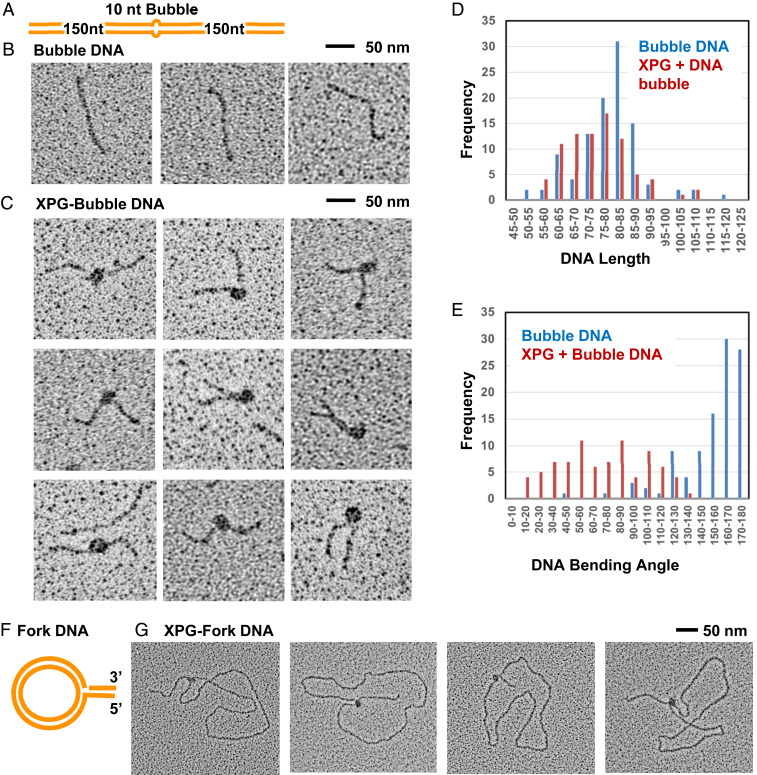
Previous EMSA revealed stable binding of XPG to bubble DNA but did not show where XPG was bound along the DNA nor whether binding might bend the DNA (4). To address these questions, we used direct EM with tungsten metal shadow casting to visualize XPG/DNA complexes (Fig. 4). XPG from the SEC dimer peak C3 (SI Appendix, Fig. S2 A and B) was incubated with a 300-bp dsDNA with a central 10-nt bubble (Fig. 4A). We purified XPG/DNA complexes by a second SEC. We adjusted the concentration of XPG such that only ∼50% of the DNA was bound by XPG as seen by EM, as higher ratios generated confounding multimolecule aggregates. We used mass ratios of 4 to 6 μg of XPG per microgram of the DNA substrate, equivalent to three to four XPG dimers per 300-bp DNA.
Fig. 4.
EM analysis shows direct interaction of XPG with bubble DNA and forks. EM analysis included tungsten rotary metal shadow casting following mounting of protein or DNA onto thin carbon foil substrates. (A) Schematic of the 10-nt bubble DNA EM substrate. (B) EM analysis showed bubble DNA alone was mostly straight. (C) XPG and bubble DNA with 5 mM Ca+2. A distinct protein particle of a size consistent with an XPG dimer was observed at the center of a large fraction of the DNA, but never at the end. Protein-bound DNA frequently had severe bends. (D) Histogram of measured XPG-free and XPG-bound DNA lengths. To determine if the DNA wrapped around XPG, the length of protein free DNA (blue), or DNA containing XPG bound as in C were measured. The two length distributions are similar arguing against DNA wrapping. (E) Histogram of XPG-free and XPG-bound DNA bending angles, determined from the EM images; “180” corresponds to a straight DNA, while a value of 0 corresponds to a DNA fully folded back on itself at the center. (F) Schematic of the single replication fork EM substrate. (G) The fork DNA was incubated with XPG with 5 mM Ca+2 and analyzed by EM. XPG protein particles, often oblong, were present only at the replication fork.
Examination of fields of molecules revealed three species. The predominant species 1 was 300-bp DNA bound by protein particles ∼10 to 12 nm in diameter, including the metal coating (n = 100 measurements) (Fig. 4C). This size, roughly 8 to 10 nm when corrected for metal coating, is most consistent with the particles being XPG dimers. Species 2 was DNA bound by discrete protein particles that appeared larger, and species 3 was linear protein-free 300-bp bubble DNA molecules (Fig. 4B). We interpret the smaller protein particles as representing dimers and the larger particles as higher-order XPG multimers. Of the several thousand XPG/DNA complexes visually examined, no XPG bound the DNA ends; rather, all XPG bound the DNA center, where the bubble was located.
XPG/DNA images were collected sequentially at the EM, and every DNA or XPG/DNA complex not in an aggregate was photographed. Images were then analyzed using Gatan digital Image software, and the length of DNA was determined for DNA alone or with XPG (n = 100 each). For measuring DNA within XPG/DNA complexes, the DNA path through the complex was taken to be straight from entry to exit. The lengths of DNA alone centered ∼80 to 85 nm, whereas that of the DNA in complexes centered at ∼75 to 80 nm, with skewing toward lower values (Fig. 4D). If the DNA had wrapped around the protein complex in a manner similar to that in nucleosomes, we would have expected a greater foreshortening of 10 to 20 nm. Therefore, we conclude that DNA does not wrap around the protein when bound by XPG.
XPG Bends Bubble DNA Substrate.
We collected images of ∼200 examples each of DNA alone or XPG-bound DNA sequentially as they were encountered at the EM. In these incubations, most protein particles were species 1 as contrasted to the larger species 2. Most of the protein-bound DNAs were observed to have a significant bend (Fig. 4C). We determined the DNA bend angle at the center of the XPG/DNA complexes using ImageJ software (Fig. 4E). A 180° angle corresponds to a straight molecule with no bend or kink at the center, while a 0° angle would correspond to a molecule in which the two arms are folded fully back on themselves. Unexpectedly for a bubble DNA, the bending angle for protein-free DNA fell predominantly within values of 160° to 180°, consistent with a slight variation of 10 to 20° from the straight. The bending angle distribution for complexes of XPG on the 300-bp bubble DNA substrate was determined from two experiments conducted twice, 1 mo apart. The angle distributions were very similar. In the first experiment, the mean bending angle was 97 ± 43°; in the second (Fig. 4E), it was 80 ± 45°. Examination of the two distributions, however, showed that the peaks centered at about 80° (DNA being bent by 100° from the straight) in both cases, and we judge this to be the best value.
Breaking Stacking in Discontinuous dsDNA.
The observation that protein-free bubble DNA was linear, whereas XPG-bound DNA was bent with a wide range of angles, implied that XPG could break stacking at ss/ds junctions in DNA. The mechanism by which XPG might do this has precedence in the FEN superfamily. In FEN1, EXO1, and GEN1, a hydrophobic wedge formed by the second helix α2 packs against the dsDNA at the ss/dsDNA junction (32, 34, 36). We reasoned that XPG could similarly pack at the ss/dsDNA junction of the bubble and break the stacking at this position. To test this idea, we took advantage of the increase in fluorescence when 2-aminopurine (2AP) in DNA is no longer stacked against another base. We designed the bubble DNA with 2AP in the dsDNA at one ss/dsDNA junction (SI Appendix, Fig. S5A). Buffer contained 1 mM EDTA, to prevent incision. As expected, 2AP fluorescence rose from 13,000 Units (U) within bubble DNA to 22,000 U in ssDNA (the 2AP-containing strand by itself). When we incubated the bubble DNA with full-length XPG at equimolar concentration, we observed a stronger fluorescence of 32,000 U (SI Appendix, Fig. S5B). As the active site is highly negatively charged and could repulse dsDNA, we added 10 mM CaCl2, a divalent cation that does not support incision (28). We did not observe a significant increase in fluorescence compared to XPG without CaCl2, consistent with the major break in stacking occurring at the initial binding. These results are consistent with XPG breaking the base stacking at the ss/dsDNA junction in bubble DNA, as observed in the EM analysis.
XPG Binding to Three-Way Junctions.
XPG plays a key role in maintenance of genome stability by HRR at stalled replication forks (10), raising the possibility that XPG itself may bind to forks, as implied by EMSA on three-way DNA junctions (28). To test this possibility, we used EM (Fig. 4F). The substrate was an ∼3-kb dsDNA circle with a displaced 400-bp arm that created a single replication fork (49). The 3′ leading-strand terminus was near the fork, and the displaced (lagging) strand had an ssDNA gap of 15 nt. We found that XPG bound with high specificity to the fork, never along the duplex circle or at the end of the displaced arm (Fig. 4G). The ability to bind to substrates with discontinuous dsDNA may underlie XPG-specific functions in multiple DDR pathways in addition to NER.
Structural Biochemistry of Pathogenic Mutation Sites.
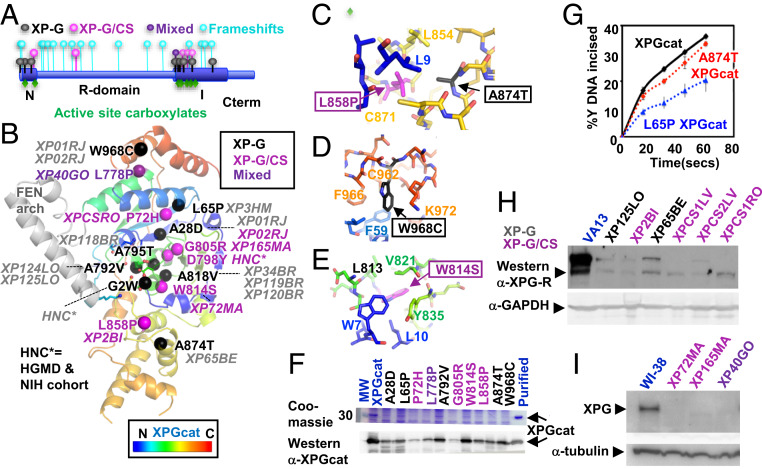
To examine XPG pathogenic mutations in their molecular context, we mapped all known missense mutations onto our structure (nine XP-G, five XP-G/CS, and one mixed/uncertain) (24, 25, 50–59). Although deletions and truncations that cause XP-G/CS map throughout the ERCC5 gene, 14 of 15 missense mutations in both XP-G and XP-G/CS map to the N and I catalytic domains (Fig. 5A). Within the catalytic domain, mutations do not cluster by disease in sequence or location (Fig. 5 A and B). Interestingly, none are of a known, conserved catalytic residue. For both XP-G and XP-G/CS mutations, the side chains are generally in well-packed areas with side chains pointing into the protein. All but two (residues 818 and 968) are located within or at the ends of helices (nine residues) or β-strands (three residues) (SI Appendix, Table S4). Similar to many XP and XP/CS mutations in XPD (60, 61), five mutations (of residues 2, 778, 792, 798, and 805) are at N- or C-terminal ends of helices or β-strands that control tertiary organization.
Fig. 5.
ERCC5 mutation analysis. (A) XPG schematic shows most missense XP-G and XP-G/CS pathogenic mutations map within XPGcat, with frameshifts throughout XPG. (B) Missense pathogenic mutations mapped onto XPGcat structure. Patient cell lines or source are in italics. (C) Zoom-in view of two neighboring residues that when mutated cause different diseases. XP-G mutation A874T would likely disrupt local packing of the central region, while the XP-G/CS mutation L858P would globally disrupt interaction between N- and C-terminal regions. (D) Zoom-in view showing surface exposed location of XP-G mutation W968C. Mutation is likely to disrupt local C-terminal packing. (E) Zoom-in view showing core location of XP-G/CS mutation W814S. Mutation would likely globally disrupt interaction of the N-terminal and central regions. (F) XPGcat expression level decreased with XP-G (black), XP-G/CS (plum), or mixed (purple) missense mutations relative to XPGcat (blue). Coomassie and Western analyses of crude supernatant after recombinant expression of different XPGcat mutants. Western analysis confirmed the decreased expression but showed residual levels existed. The full gel and blot are shown in SI Appendix, Fig. S6. (G) A874T and L65P XPGcat retained significant incision activity. Single turnover assays done in triplicate with 10 nM Y DNA and 25 nM protein. Error bars show SD (H and I) Western analyses of XP-G (black), XP-G/CS (plum), or mixed (purple) patient cell lines show significant reduction in XPG protein levels, compared to SV40-transformed WT control VA13 fibroblasts or hTERT-immortalized WT control WI-38 fibroblasts (blue), as appropriate. XPG detected with antibody R2; 97727 (4). The same membrane was reprobed with α-GAPDH antibody or α-tubulin for loading control. Shorter exposure of Left Upper is shown in SI Appendix, Fig. S6.
Analysis of our structure reveals that both XP-G and XP-G/CS mutations are positioned to perturb protein stability, but with distinct differences (SI Appendix, Table S4). First, XP-G mutations change residue interactions for sites adjacent in sequence that would cause local disruption. For example, an XP-G mutation might disrupt packing with another in the next helix. In contrast, XP-G/CS mutation sites lie at interfaces that connect sequence-distant regions likely to cause domain impact. Second, XP-G disease mutations substitute larger side chains that would be expected to disrupt structural elements in neighboring regions acting in DNA binding or incision. In contrast, XP-G/CS mutations are within secondary structures key for domain stability. Four of the five either mutate glycine or proline to another residue or vice versa, thereby causing dramatic changes in flexibility or rigidity. The fifth XP-G/CS mutation, W814S, would create a core destabilizing cavity. Two sets of XP-G and XP-G/CS mutations exemplify these features: 1) XP-G mutation A874T and XP-G/CS mutation L858P (Fig. 5C) and 2) XP-G mutation W968C and XP-G/CS mutation W814S (Fig. 5 D and E). Due to the β-branched Thr, XP-G mutation A874T is likely to break local packing in the H2TH motif acting in K+ and dsDNA binding. In contrast, the proline in the XP-G/CS mutation L858P is likely to redirect the key turns and globally disrupt the N- and C-terminal interface between residues 9 and 858. Analogously in the second example (Fig. 5 D and E), the XP-G mutation W968C is likely to disrupt a surface-exposed hydrophobic channel and local binding of the 3′ ssDNA region. In contrast, the XP-G/CS mutation W814S removes a buried Trp to form a hydrophilic cavity and is likely to disrupt the junction of N- and C-terminal regions (residues 7, 10, and 814). Thus, structure analysis predicts that XP-G mutations are locally disrupting and impact functional regions, whereas XP-G/CS mutations globally disrupt the XPG core domain, thereby impacting the full-length protein and consequently likely all of its interactions.
To test these observations, we made five XP-G, four XP-G/CS, and one “mixed” missense mutations individually in XPGcat to test catalytic activity and protein stability. The XP-G mutations were A28D, L65P, A792V, A874T, and W968C. The XP-G/CS mutations were P72H, G805R, W814S, and L858P. The mutation L778P was defined as mixed, since it exhibits as XP in homozygous patients, but cells from a phenotypically uncharacterized patient in which L778P is heterozygous with a frameshift mutation in ERCC5 have biochemical properties that are CS-like (SI Appendix, Table S4) (24, 50). All 10 pathogenic mutations reduced recombinant expression of XPGcat to different extents compared to the WT (Fig. 5F and SI Appendix, Fig. S6), consistent with all 10 destabilizing the structure (Fig. 5F and SI Appendix, Fig. S6). We were able to purify a small amount of protein for two XP-G mutants for biochemical analysis. The generally accepted model is that XP-G mutations will directly disrupt the incision activity and that it is this reduction in catalytic activity that leads to defects in NER (45). XP-G mutations in XPGcat did cause decreased incision activity on Y DNA compared to an equal protein amount as WT, but not as dramatic a decrease as expected (Fig. 5G) (45). It is quite possible that pathogenic mutations may have a bigger effect in full-length XPG than in XPGcat, due to the folded archaeal FEN arch in place of the unstructured R-domain (SI Appendix, Fig. S1H). However, if both XP-G and XP-G/CS mutations destabilize XPG protein in vivo, we should observe lowered expression of full-length XPG in cells from both patient categories. Reduced expression in XP-G/CS cells has been observed (20), but it has not previously been documented for XP-G.
Indeed, as we saw for XPGcat mutant proteins, we observed greatly reduced XPG expression in two XP-G patient cell lines (XP125LO, XP65BE) compared to a WT cell line VA13 (Fig. 5 H and I). This is especially surprising given that both patients were only mildly affected. Three XP-G/CS cell lines (XPCS1LV, XPCS2LV, and XPCS1RO) with severely truncating mutations in ERCC5 and three with missense mutations (XP2BI, XP72MA, and XP165MA) had more dramatic changes: All lacked detectable XPG protein, with the exception of a potential trace amount in XP165MA. XP40GO, a cell line with no associated patient information but biochemical analysis of which was consistent with XP-G/CS (24), also lacked detectable XPG. We did not have cells for analysis from sibling patients XP174-1 and -2, which carry the same L788P mutation as XP40GO but are homozygous for it and are XP in phenotype (SI Appendix, Table S4) (50). Nonetheless, our results suggest that both XP-G and XP-G/CS mutations disrupt XPG protein stability.
Discussion
Multiple specialized structure-specific nucleases act in DDRs and maintenance of genome integrity, and the key to their different activities and functions is in their DNA and protein binding, catalytic specificities, and regulation. Besides its important roles in NER, XPG has key functions in BER (11–13) and HRR (10), and its loss is implicated in accumulation of R-loops (15–17, 62). A molecular understanding of the diverse XPG functions will be greatly facilitated by the combined XPGcat crystal structure and solution measurements (SAXS, EM, and biochemical, mutational, and cell biology analyses) presented here that reveal DNA-specificity elements, dimer assembly, bubble DNA bending, and the location of pathogenic mutations.
DNA Sculpting and a Unified Basis for XPG DDR Activities.
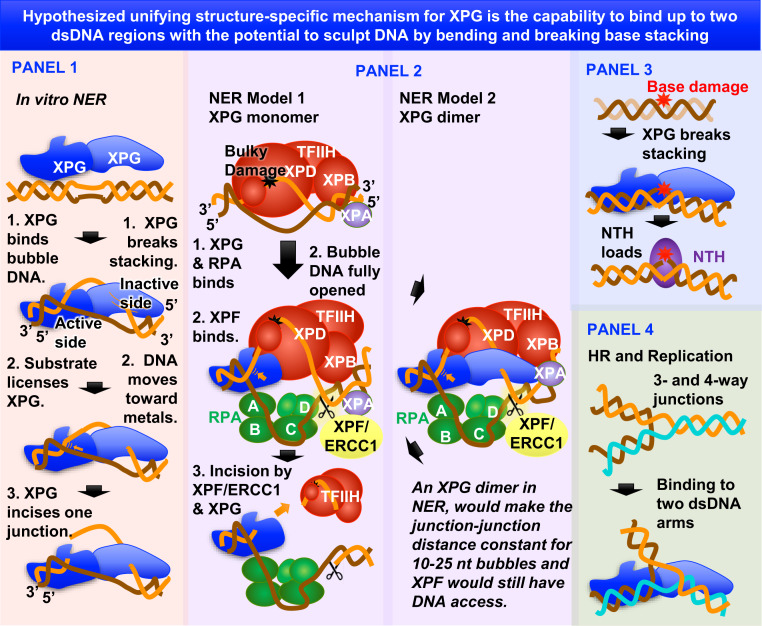
The reported findings and implications from XPG nuclease superfamily DNA complexes support sculpting of dsDNA having unpaired regions (discontinuous dsDNA) as a likely foundational basis for diverse XPG activities (Fig. 6). In NER (Fig. 6, panels 1 and 2), the DNA bubble is the discontinuous dsDNA that XPG binds, bends, validates as a substrate, and then incises, analogous to nucleases such as FEN1 and MRE11 that sculpt DNA for specific incision (36, 63). Notably, XPG’s substrate interaction is quite distinct from the XPC substrate recognition that initiates NER. In NER, XPD holds the lesion-containing ssDNA, while RPA binds the undamaged ssDNA. In BER (Fig. 6, panel 3), where XPG directly stimulates hNTH1 glycosylase activity, disrupted stacking by thymine glycol in the substrate (SI Appendix, Fig. S7) would be transiently recognized and sculpted by XPG to promote glycosylase loading. In this connection it is noteworthy that DNA bending by FEN superfamily members does not require stable binding (35). In HRR (Fig. 6, panel 4), discontinuous DNA could be three- or four-way junctions representing stalled replication forks, chicken foot structures from fork reversal, or Holliday junctions. In these situations, XPG binding may position dsDNA arms for downstream processing, as observed in the promotion of specific repair pathways by the alkylated DNA-binding protein Atl1 in fission yeast (64). Analogous to other FEN superfamily members, XPG incision activity can be disconnected from DNA binding, thus allowing DNA binding-only functions (4, 28).
Fig. 6.
Unifying models for XPG function, pathways, and cancer. Hypothesized unifying structure-specific mechanism based on XPG binding to discontinuous dsDNA and breaking base stacking.
XPG Dimer and DNA Recognition.
For the MRE11 nuclease dimer, a helix-bundle interface similar in size to that in the XPGcat crystal structure was assumed to be only a crystal contact, but it was later found functionally key for DNA binding and for distinguishing two-ended breaks versus one-ended stalled replication forks (65). Our combined results (from coimmunoprecipitation analysis, mutational analysis, EM, and SAXS) establish that XPG without TFIIH exists as a stable dimer, which appears suitable for its NER functions. Notably, it is not excluded that during NER, the second XPG dimer subunit could be exchanged with a TFIIH subunit in a heteromeric interaction, as seen for TFIIH p52 and p8 subunits (66). Distinguishing between these possibilities will require further structural information on the NER preincision complex. Nonetheless, possible participation of the previously unrecognized XPG dimer in NER has implications for bubble recognition that merit consideration.
First, rather than an XPG monomer binding only to the bubble junction 3′ to the lesion where it incises, an XPG dimer could bind both bubble junctions, explaining XPG preference for bubble DNA over Y DNA (4).
Second, XPG could bind the same ss/dsDNA junction as XPF, on opposite sides. FEN superfamily members bind the dsDNA region of their substrate at two distinct parts of the dsDNA helix, leaving the intervening DNA region arching out from the protein surface (29). As our structure implies that XPG binds DNA similarly, XPF-ERCC1 could bind this intervening region, which would contain the proper site for its incision (67) (Fig. 6). Thus, both XPG and XPF-ERCC1 could bind the same dsDNA arm, but at different places. Precedence within the superfamily for interacting on opposites sides of the same DNA comes from FEN1 and DNA ligase (68). Colocalization to the same junction might be a part of XPG licensing of XPF incision.
Third, only one ss/dsDNA junction of the bubble DNA could be incised by an XPG dimer. In our dimer model, one subunit positions the 5′ ssDNA near the active site, while for the other it is the 3′ ssDNA (Fig. 6). In the FEN1 superfamily, the spacing between the DNA binding sites (K+/H2TH and the active site) positions the substrate 5′ ssDNA properly for incision, but not 3′ ssDNA (36). Thus, only one junction of the bubble would be positioned for XPG catalysis.
Fourth, the XPG dimer could help define the distance and ssDNA path between the two bubble junctions, independent of bubble size from 10 to 30 nt. The distance between XPG dimer dsDNA regions is fixed and forces the ssDNA to expand outward in a U-shape during NER bubble expansion. This U-shape DNA conformation was observed in an RPA crystal (69) (Fig. 6). Although human RPA/ssDNA did not form this shape on its own in solution (70), an XPG dimer would be suitable to promote this conformation for RPA/DNA.
Implications from XP-G and XP-G/CS Mutations.
Based on observed ERCC5 frameshift mutations (20) and mouse models (8, 21, 22), XP-G/CS mutations are postulated to result in loss of XPG protein, which we have now confirmed. In contrast, XP-G mutations are all missense mutations and have been assumed primarily to cause defective nuclease activity with consequent NER loss, UV sensitivity, and cancer predisposition, but no presumed major destabilization of the protein (20). Indeed, a mouse model with incision-defective mutant protein exhibits XP characteristics (22). Surprisingly, our XPGcat biochemistry shows that many XP-G mutations also destabilize the catalytic domain, and a significant reduction in XPG protein was observed in two XP-G patient cell lines. Low XPG protein availability could in itself contribute to reduced NER activity in XP-G patient cells in addition to the effect of the mutation on catalysis per se. A related implication is that the residual levels of mutant protein in XP-G patients must still protect against CS. A possible partial explanation may relate to the known up-regulation of XPG in response to replication stress (10), particularly if it is also regulated by transcription stress. However, to what extent the effect of disease mutations on protein stability in XPGcat mirrors the effect of the same mutations on the endogenous human protein remains to be fully determined. In our construct, XP-G mutations generally appear to allow more stable XPGcat protein than do XP-G/CS mutations, but there is overlap between the range of expression levels. If this situation reflects the mutation effects on XPG in humans, it would not be possible to differentiate XP-G and XP-G/CS mutants simply by how much protein exists. Furthermore, given the genetic and environmental diversity in the human population, many other factors are likely involved in determining the disease outcome of particular mutations. It is also possible that heteroallelic effects may contribute to disease outcome. Indeed, the mutation L778P exhibits as XP in homozygous patients, but cells from a patient in which that missense mutation is heterozygous with a truncating frameshift mutation in Xpg have biochemical properties that are CS-like (SI Appendix, Table S4) (24, 50).
The structure of a protein is intimately linked to its stability, function, and interactions, and the effect of many human missense variants can be accurately interpreted from an analysis of experimental structures (71). Based on our XPGcat structure, we hypothesize that the residual proteins in missense mutants have different degrees of “intactness.” We predict that XP-G mutants will have limited local disruption of the catalytic domain, while XP-G/CS mutations, located at interfaces between distant sequence regions of the protein or in the β-sheet core, will disrupt much larger portions of the catalytic domain. These disease-related predictions are generally analogous to XP and XP/CS mutations in XPD, in that XP mutations appeared to locally target XPD functions while XP/CS mutations affected its flexibility/rigidity and community interfaces (60, 61).
Future Prospects.
XPG is important as a surprisingly multifunctional enzyme with the potential to help balance DDR at the crossroads with transcription and replication. From combined crystallography, biochemistry, SAXS, and EM data, a foundational feature for XPG biological function may be its ability to recognize and sculpt discontinuities in discontinuous dsDNA suitable to facilitate its repair with or without incision activity. Based upon disease-associated point mutations and XPG structural biochemistry, we posit that XPG protein stability, and consequently its specific functional interactions, depend upon the integrity of the XPGcat domain structure defined here. Furthermore, the newly defined dimeric structure of XPG provides insight into its multiple functions. From these collective findings, we propose that XPGcat-enforced discontinuous dsDNA binding, unstacking, and bending provide a testable unified mechanism (Fig. 6) whereby XPG, in combination with different protein partnerships, may determine repair function and cell fate in multiple key DDR transactions.
Materials and Methods
See SI Appendix for further materials and methods.
Antibodies.
Antibodies used in this study include α-GAPDH (Millipore), α-tubulin (Calbiochem), α-Flag (Sigma), α-XPG R2 # 97727 (4) and α-XPGcat.
Protein Constructs and Purification.
XPGcat was cloned with residues 79 to 785 replaced with residues 89 to 128 from Pyrococcus furiosus FEN-1 (42) and with a C-terminal truncation after residue 987 and recombinantly expressed in bacteria and purified by Q, SP, Heparin, and SEC. XPG∆R and untagged full-length XPG were purified as previously described (4). Full-length XPG with a C-terminal FLAG tag was expressed in High Five insect cells and purified by anti-FLAG M2 resin (Sigma), SP Sepharose, and Superose 6.
Incision Assays.
XPG is product-inhibited, and protein was assayed until single turnover questions with enzyme excess. The DNA substrates were formed as outlined in SI Appendix, Table S2. For untagged XPG, WT XPG-FLAG, or a mutant version of XPG-FLAG, labeled 15-nt (100 fmol: 3.3 nM) bubble substrates were incubated at 37 °C with XPG protein (33.3 nM) in a buffer (25 mM Tris, pH 6.8, 10% glycerol, 2.5 mM BME, 0.5 mg/mL BSA, 30 mM KCl, and 4 mM MgCl2) for 15, 30, 60, or 90 min. For XPGcat assay, 10-nM substrates were incubated with 25 nM XPGcat at 16 °C for 15, 30, 45, and 60 s in a buffer containing 25 mM Tris, pH 6.8, 10% glycerol, 2.5 mM BME, 0.5 mg/mL BSA, 0.3 mM EDTA, 30 mM KCl, and 2 mM MgCl2. Product DNA was separated from substrate DNA by denaturing PAGE. Assays were done in triplicate, on different days.
EMSA for DNA Binding of XPGcat or Mutant Constructs.
For EMSA, 5 nM of the 5′-32P-end-labeled bubble or Y-substrates were incubated with protein in a binding buffer containing 10 mM Hepes pH 7.5, 110 mM KCl, 1 mM EDTA, 1 mM DTT, 10 mM CaCl2, 100 ng of poly dI-dC, and 4% glycerol at room temperature for 15 min. For DNA binding of WT XPG or various mutants, all of the above binding conditions are the same, except 10 mM CaCl2 was replaced with 0.1% Nonidet P-40. Assays were done in triplicate on different days.
Crystallization and Structure Determination.
Data from three crystals of human XPGcat were merged for structure refinement. XPGcat (27 mg/mL) was incubated 1:1 in one of three mother liquors: 1) 40% AmSO4, 200 mM Imidizole/Malate Buffer pH 4.2,100 mM MgCl2; 2) 24% AmSO4, 200 mM Imidizole/Malate Buffer pH 4.2, 250 mM MgCl2, 0.5 mM SmSO4, 10 mM DTT; 3) 32% AmSO4, 10 mM DTT, 200 mM Imidizole/Malate Buffer pH 4.2, and 50 mM MgCl2. The crystal diffraction was highly anisotropic, which led to unusually high rejection of reflections (72).
Biochemical Analysis of Multimerization.
XPG-FLAG and XPG-GFP (full-length XPG with either a C-terminal FLAG or GFP tag) were expressed together or separately in High Five insect cells. When we expressed the differently tagged XPG separately, we mixed the cells postexpression. Cells were lysed by Dounce homogenizers. XPG-FLAG was purified from the lysate by anti-FLAG M2 resin (Sigma) and eluted by 3×-FLAG peptide. The eluant from the anti-FLAG resin was sequentially incubated with beads attached to GFP binder nanobodies (73). Beads and eluant were analyzed by PAGE and Western analysis.
SAXS Analysis of XPGcat, XPG∆R, and Full-Length XPG.
SAXS data on freshly purified full-length XPG-FLAG, XPG∆C-FLAG, XPG∆R, and XPGcat were collected at beamline 12.3.1 at the Advanced Light Source synchrotron on a MarCCD 165 detector (75) and included Porod exponent analysis for objective quantization of flexibility (76). SAXS data collection and data analysis details are provided in SI Appendix, Table S3.
Preparation of DNA Substrates for EM.
The substrate resembling a replication fork was created as previously described (49). The bubble DNA substrate consisted of a 300-bp DNA constructed to have a 10 base unpaired bubble at the exact center (Fig. 4A). The DNA was generated by annealing and ligating four oligo sequences (SI Appendix, Table S2).
Preparation of XPG/DNA Protein Complexes for EM Visualization.
XPG purified through SEC was incubated with 50-ng DNA in a buffer consisting of 10 mM Hepes pH 7.5, 110 mM KCl, 1 mM EDTA, and 4% glycerol. For most experiments these reactions contained the DNA at 2.5 μg/mL and XPG at 10 to 15 μg/mL equivalent to a ratio of three to four XPG dimers per 300-bp DNA. An equal volume of 1.2% glutaraldehyde was added for 5 min at room temperature followed by chromatography over a 2-mL column of 5% agarose beads (Agarose Bead Technology Inc Burgos, Spain) equilibrated in 10 mM Tris pH 7.5, 0.1 mM EDTA.
Preparation of Samples for EM Visualization.
Samples were prepared for EM analysis as previously described (74). Bending angles were measured on the micrographs using ImageJ software and lengths measured with Gatan Digital Image software.
2AP Experiments.
For 2AP experiments, 6 μM 20-nt bubble DNA with 2AP at the 3′-junction was incubated alone, with 6-μM untagged XPG protein, or protein and 10 mM CaCl2. The oligonucleotide containing 2AP was used for the ssDNA control. The experiment was done in triplicate
Cell Extract Preparation and Western Blotting Method.
Supernatants from immortalized WT VA13 or various XP-G or XP-G/CS lines were were analyzed by Western blot with rabbit anti-XPG 97727 antibody (R2) (4) and anti-GAPDH antibody (Millipore, MAB374) or anti-Actin (Calbiochem, Ab-1) to obtain loading controls. Westerns were done twice.
Accession Codes.
Coordinates and structure factors have been deposited in the Protein Data Bank under PDB ID code 6VBH (77).
Supplementary Material
Acknowledgments
We thank the researchers, patients, and families who have critically contributed to defining the genetic basis for xeroderma pigmentosum group G (XPG) pathophysiology and its window into XPG biology; Steffen Emmert for providing primary patient cells XP72MA, XP165MA, and XP40GO; and Samir Hamdan and members of the Structural Biology of DNA Repair program, including Patrick Sung and Orlando Schärer, for comments. Work on XPG was supported by National Cancer Institute P01 CA092584 (to S.E.T., P.K.C., and J.A.T.), R35 CA220430 (to J.A.T.); NIH R01GM110387 (to S.E.T.), NIH P01 AG017242 and R01 ES019935 (to P.K.C.), NIH R01GM31819 and R01 ES013773 (to J.D.G.), NIH T32 AG000266 (to A.G.R.); and King Abdullah University of Science and Technology CRG3 (to J.A.T.). J.A.T. is a Cancer Prevention and Research Institute of Texas Scholar in Cancer Research and acknowledges support by a Robert A. Welch Chemistry Chair. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Laboratory, supported by the Department of Energy (DOE), Office of Biological and Environmental Research, NIH, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences, and at Advanced Light Source SIBYLS Beamline 12.3.1, supported by NIH project ALS-ENABLE (P30 GM124169) and the Integrated Diffraction Analysis Technologies program of the US DOE Office of Biological and Environmental Research.
Footnotes
The authors declare no competing interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, http://www.pdb.org (PDB ID code 6VBH).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921311117/-/DCSupplemental.
References
- 1.O’Donovan A., Davies A. A., Moggs J. G., West S. C., Wood R. D., XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371, 432–435 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Kokic G. et al., Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 10, 2885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer N., Reagan M. S., Wu K. J., Canagarajah B., Friedberg E. C., Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry 35, 2157–2167 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Sarker A. H. et al., Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: Insights for transcription-coupled repair and Cockayne syndrome. Mol. Cell 20, 187–198 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Gary R., Ludwig D. L., Cornelius H. L., MacInnes M. A., Park M. S., The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem. 272, 24522–24529 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Staresincic L. et al., Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28, 1111–1120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marteijn J. A., Lans H., Vermeulen W., Hoeijmakers J. H., Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Barnhoorn S. et al., Cell-autonomous progeroid changes in conditional mouse models for repair endonuclease XPG deficiency. PLoS Genet. 10, e1004686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brueckner F., Cramer P., DNA photodamage recognition by RNA polymerase II. FEBS Lett. 581, 2757–2760 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Trego K. S. et al., Non-catalytic roles for XPG with BRCA1 and BRCA2 in homologous recombination and genome stability. Mol. Cell 61, 535–546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessho T., Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repairenzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 27, 979–983 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klungland A. et al., Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell 3, 33–42 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Oyama M. et al., Human NTH1 physically interacts with p53 and proliferating cell nuclear antigen. Biochem. Biophys. Res. Commun. 321, 183–191 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Trego K. S. et al., The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle 10, 1998–2007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marabitti V. et al., ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. 47, 3485–3502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sollier J. et al., Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasuhara T. et al., Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558–570.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Keijzer W. et al., A seventh complementation group in excision-deficient xeroderma pigmentosum. Mutat. Res. 62, 183–190 (1979). [DOI] [PubMed] [Google Scholar]
- 19.Moriwaki S. et al., DNA repair and ultraviolet mutagenesis in cells from a new patient with xeroderma pigmentosum group G and cockayne syndrome resemble xeroderma pigmentosum cells. J. Invest. Dermatol. 107, 647–653 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Nouspikel T., Lalle P., Leadon S. A., Cooper P. K., Clarkson S. G., A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: Implications for a second XPG function. Proc. Natl. Acad. Sci. U.S.A. 94, 3116–3121 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Shiomi N. et al., Identification of the XPG region that causes the onset of Cockayne syndrome by using Xpg mutant mice generated by the cDNA-mediated knock-in method. Mol. Cell. Biol. 24, 3712–3719 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian M., Jones D. A., Smith M., Shinkura R., Alt F. W., Deficiency in the nuclease activity of xeroderma pigmentosum G in mice leads to hypersensitivity to UV irradiation. Mol. Cell. Biol. 24, 2237–2242 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada Y. N. et al., Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol. Cell. Biol. 19, 2366–2372 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schäfer A. et al., Characterization of three XPG-defective patients identifies three missense mutations that impair repair and transcription. J. Invest. Dermatol. 133, 1841–1849 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Zafeiriou D. I. et al., Xeroderma pigmentosum group G with severe neurological involvement and features of Cockayne syndrome in infancy. Pediatr. Res. 49, 407–412 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Evans E., Fellows J., Coffer A., Wood R. D., Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 16, 625–638 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D., Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16, 6559–6573 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohl M., Thorel F., Clarkson S. G., Schärer O. D., Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 278, 19500–19508 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Grasby J. A., Finger L. D., Tsutakawa S. E., Atack J. M., Tainer J. A., Unpairing and gating: Sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem. Sci. 37, 74–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington J. J., Lieber M. R., Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: Implications for nucleotide excision repair. Genes Dev. 8, 1344–1355 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Lee S. H. et al., Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage. eLife 4, e12256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y. et al., Crystal structure of a eukaryotic GEN1 resolving enzyme bound to DNA. Cell Rep. 13, 2565–2575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miętus M. et al., Crystal structure of the catalytic core of Rad2: Insights into the mechanism of substrate binding. Nucleic Acids Res. 42, 10762–10775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orans J. et al., Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145, 212–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashid F. et al., Single-molecule FRET unveils induced-fit mechanism for substrate selectivity in flap endonuclease 1. eLife 6, e21884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutakawa S. E. et al., Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145, 198–211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsutakawa S. E. et al., Phosphate steering by flap endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat. Commun. 8, 15855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson M. J., Gotham V. J. B., Ciani B., Grasby J. A., A conserved loop-wedge motif moderates reaction site search and recognition by FEN1. Nucleic Acids Res. 46, 7858–7872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W., Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunand-Sauthier I. et al., The spacer region of XPG mediates recruitment to nucleotide excision repair complexes and determines substrate specificity. J. Biol. Chem. 280, 7030–7037 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Iakoucheva L. M., Brown C. J., Lawson J. D., Obradović Z., Dunker A. K., Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323, 573–584 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Hosfield D. J., Mol C. D., Shen B., Tainer J. A., Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: Coupling DNA and PCNA binding to FEN-1 activity. Cell 95, 135–146 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Patel N. et al., Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 40, 4507–4519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakurai S. et al., Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24, 683–693 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Constantinou A. et al., Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J. Biol. Chem. 274, 5637–5648 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Rambo R. P., Tainer J. A., Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambo R. P., Tainer J. A., Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Subramanian D., Griffith J. D., p53 Monitors replication fork regression by binding to “chickenfoot” intermediates. J. Biol. Chem. 280, 42568–42572 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Chikhaoui A. et al., Identification of a ERCC5 c.2333T>C (L778P) variant in two Tunisian siblings with mild xeroderma pigmentosum phenotype. Front. Genet. 10, 111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emmert S. et al., Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J. Invest. Dermatol. 118, 972–982 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Fassihi H. et al., Deep phenotyping of 89 xeroderma pigmentosum patients reveals unexpected heterogeneity dependent on the precise molecular defect. Proc. Natl. Acad. Sci. U.S.A. 113, E1236–E1245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lalle P., Nouspikel T., Constantinou A., Thorel F., Clarkson S. G., The founding members of xeroderma pigmentosum group G produce XPG protein with severely impaired endonuclease activity. J. Invest. Dermatol. 118, 344–351 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Moriwaki S. et al., Xeroderma pigmentosum complementation group G patient with a novel homozygous missense mutation and no neurological abnormalities. Exp. Dermatol. 21, 304–307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norris P. G., Hawk J. L., Avery J. A., Giannelli F., Xeroderma pigmentosum complementation group G—Report of two cases. Br. J. Dermatol. 116, 861–866 (1987). [DOI] [PubMed] [Google Scholar]
- 56.Nouspikel T., Clarkson S. G., Mutations that disable the DNA repair gene XPG in a xeroderma pigmentosum group G patient. Hum. Mol. Genet. 3, 963–967 (1994). [DOI] [PubMed] [Google Scholar]
- 57.Pugh J. et al., Use of big data to estimate prevalence of defective DNA repair variants in the US population. JAMA Dermatol. 155, 72–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soltys D. T. et al., Novel XPG (ERCC5) mutations affect DNA repair and cell survival after ultraviolet but not oxidative stress. Hum. Mutat. 34, 481–489 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Sun Z. et al., Genotype-phenotype correlation of xeroderma pigmentosum in a Chinese Han population. Br. J. Dermatol. 172, 1096–1102 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Fan L. et al., XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell 133, 789–800 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan C. et al., Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nat. Struct. Mol. Biol. 26, 397–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makharashvili N. et al., Sae2/CtIP prevents R-loop accumulation in eukaryotic cells. eLife 7, e42733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shibata A. et al., DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tubbs J. L. et al., Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459, 808–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams R. S. et al., Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135, 97–109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kainov D. E., Vitorino M., Cavarelli J., Poterszman A., Egly J. M., Structural basis for group A trichothiodystrophy. Nat. Struct. Mol. Biol. 15, 980–984 (2008). [DOI] [PubMed] [Google Scholar]
- 67.de Laat W. L., Appeldoorn E., Jaspers N. G., Hoeijmakers J. H., DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 273, 7835–7842 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Mayanagi K. et al., Direct visualization of DNA baton pass between replication factors bound to PCNA. Sci. Rep. 8, 16209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan J., Pavletich N. P., Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brosey C. A. et al., A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 41, 2313–2327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ittisoponpisan S. et al., Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? J. Mol. Biol. 431, 2197–2212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Read R. J., Detecting outliers in non-redundant diffraction data. Acta Crystallogr. D Biol. Crystallogr. 55, 1759–1764 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Meek K., Lees-Miller S. P., Modesti M., N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res. 40, 2964–2973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffith J. D., Christiansen G., Electron microscope visualization of chromatin and other DNA-protein complexes. Annu. Rev. Biophys. Bioeng. 7, 19–35 (1978). [DOI] [PubMed] [Google Scholar]
- 75.Classen S. et al., Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 46, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brosey C., Tainer J., Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr. Opin. Struct. Biol. 58, 197–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsutakawa S. E., Arvai A. S., Tainer J. A., Human XPG endonuclease catalytic domain RCSB Protein Data Bank. https://www.rcsb.org/structure/6VBH. Deposited 17 December 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.