Significance
The Hippo pathway plays a crucial role in maintaining tissue homeostasis. Generally, activated Hippo pathway effectors, YAP/TAZ, induce the transcription of their negative regulators, NF2 and LATS2, and this negative feedback loop maintains homeostasis of the Hippo pathway. However, YAP and TAZ are consistently hyperactivated in various cancer cells, enhancing tumor growth. Our study found that LATS2, a direct-inhibiting kinase of YAP/TAZ and a core component of the negative feedback loop in the Hippo pathway, is modified with O-GlcNAc. LATS2 O-GlcNAcylation inhibited its activity by interrupting the interaction with the MOB1 adaptor protein, thereby activating YAP and TAZ to promote cell proliferation. Thus, our study suggests that LATS2 O-GlcNAcylation is deeply involved in Hippo pathway dysregulation in cancer cells.
Keywords: Hippo pathway, LATS2, MOB1, O-GlcNAcylation, cancer
Abstract
The Hippo pathway controls organ size and tissue homeostasis by regulating cell proliferation and apoptosis. The LATS-mediated negative feedback loop prevents excessive activation of the effectors YAP/TAZ, maintaining homeostasis of the Hippo pathway. YAP and TAZ are hyperactivated in various cancer cells which lead to tumor growth. Aberrantly increased O-GlcNAcylation has recently emerged as a cause of hyperactivation of YAP in cancer cells. However, the mechanism, which induces hyperactivation of TAZ and blocks LATS-mediated negative feedback, remains to be elucidated in cancer cells. This study found that in breast cancer cells, abnormally increased O-GlcNAcylation hyperactivates YAP/TAZ and inhibits LATS2, a direct negative regulator of YAP/TAZ. LATS2 is one of the newly identified O-GlcNAcylated components in the MST-LATS kinase cascade. Here, we found that O-GlcNAcylation at LATS2 Thr436 interrupted its interaction with the MOB1 adaptor protein, which connects MST to LATS2, leading to activation of YAP/TAZ by suppressing LATS2 kinase activity. LATS2 is a core component in the LATS-mediated negative feedback loop. Thus, this study suggests that LATS2 O-GlcNAcylation is deeply involved in tumor growth by playing a critical role in dysregulation of the Hippo pathway in cancer cells.
The balance between cellular proliferation and apoptosis is crucial for the development and maintenance of tissue homeostasis in multicellular organisms. Cancer can result when this balance is disrupted. The Hippo pathway is a signal transduction pathway that controls cell proliferation, cell self-renewal, and apoptosis (1, 2). The Hippo pathway is mediated by phosphorylation, as the pivotal elements of the mammalian Hippo pathway consist of serine/threonine kinases, such as mammalian Ste20 like kinase1/2 (MST1/2) and large tumor suppressor 1/2 (LATS1/2), in addition to the effectors Yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) (3–6). Cell–cell adhesion, cell–matrix adhesion, and contractile tension are physical cues for turning on the Hippo pathway (7, 8). In addition, various hormonal cues, growth factors, and stressors, such as hypoxia and energy depletion, also regulate the Hippo pathway (9). Some of those stimuli activate MST1/2 by inducing phosphorylation at MST1/2 Thr183/Thr180 (10–12). Mps one binder 1 (MOB1) binds activated MST1/2 and this interaction induces a conformational change in MOB1 to enable it to bind to LATS1/2 (13, 14). LATS1/2 are recruited to MST-MOB1 in the membrane by neurofibromatosis 2 (NF2) (15). After formation of the MST-MOB1-LATS complex, MST1/2 phosphorylate MOB1 and the LATS1/2 hydrophobic motif (Thr1079 for LAST1 and Thr1041 for LATS2) (14). The phosphorylated MOB1-LATS1/2 complex is detached from MST1/2 and then LATS1/2 are fully activated after autophosphorylation of its activation loop (Ser909 for LAST1 and Ser872 for LATS2) (14, 16). As a result, activated LATS1/2 phosphorylates serine on the YAP/TAZ HXRXXS motifs (5, 7, 17–19). Phosphorylation at YAP Ser127 and TAZ Ser89 causes their cytoplasmic sequestration by inducing an interaction with 14-3-3 (5, 7, 17). In addition, phosphorylation at YAP Ser381 and TAZ Ser311 leads to their proteasomal degradation (18, 19). Consequently, YAP/TAZ cannot act as a transcriptional coactivator (5), so the transcription levels of essential genes for cell proliferation and antiapoptosis decrease. The Hippo pathway is dysregulated in cancer cells, which contributes to tumor growth, development, and metastasis (6). It has been assumed that the Hippo pathway might be suppressed by a nonmutational mechanism because mutations in the core components of the Hippo pathway are rare in cancer patients (6, 20–23).
Many studies have found that O-GlcNAcylation and O-GlcNAc transferase (OGT) levels are abnormally high in cancer cells (24, 25). O-GlcNAcylation is a posttranslational modification that is mediated by OGT (26–28), and O-GlcNAc is removed from proteins by O-GlcNAcase (OGA) (29). O-GlcNAcylation affects various cellular processes, such as cell growth, mobility, metabolism, and development, by regulating protein stability, localization, activity, and protein–protein interactions (30–36). In particular, it is well known that O-GlcNAcylation regulates phosphorylation-mediated signaling pathways through interplay with phosphorylation (37, 38). Therefore, it was hypothesized that the phosphorylation-mediated Hippo pathway might be masked by an abnormal increase of O-GlcNAcylation in cancer cells.
It was reported that O-GlcNAcylation enhances YAP activity (32, 39, 40). This finding establishes a correlation between abnormally high levels of O-GlcNAcylation and dysregulation of the Hippo pathway in cancer cells. Activated YAP induces transcription of NF2 and LATS2, which are involved in inactivating YAP/TAZ (41, 42). However, the mechanism that blocks this intrinsic negative feedback in the Hippo pathway remains to be elucidated in cancer cells. This study discloses the molecular inhibitory mechanism of the Hippo pathway by LATS2 O-GlcNAcylation. This finding suggests the possibility that LATS2 O-GlcNAcylation blocks the negative feedback loop in the Hippo pathway and contributes to the excessive proliferative potential of cancer cells by sustaining hyperactivation of YAP/TAZ.
Results
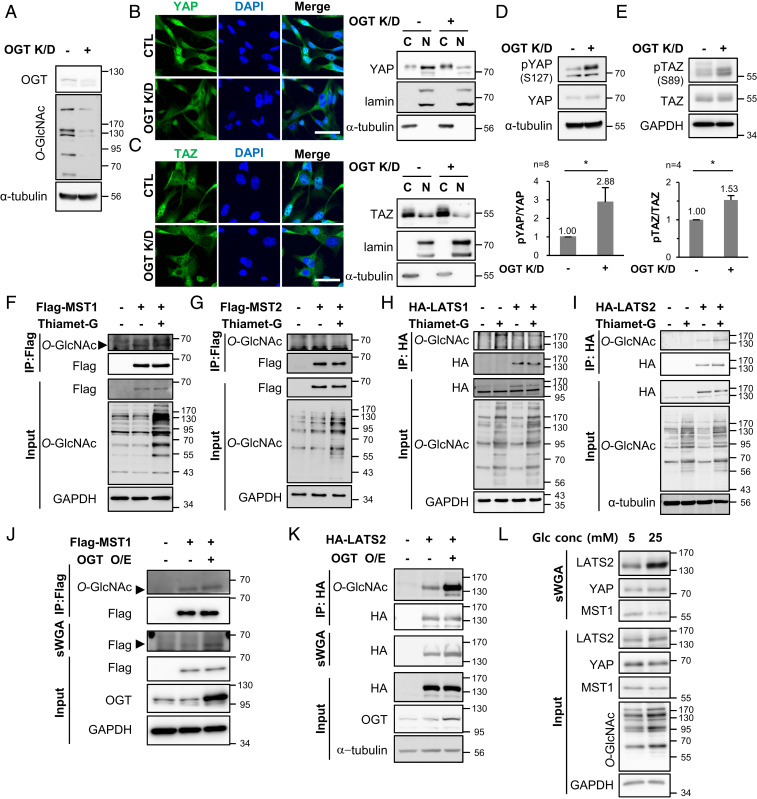
MST1 and LATS2 Are Modified with O-GlcNAc.
The breast cancer cells MCF7, MDA-MB-468, and MDA-MB-231 expressed higher levels of YAP/TAZ and had higher cellular O-GlcNAcylation levels than those in normal MCF10A cells (SI Appendix, Fig. S1). YAP/TAZ activity is also known to increase in these breast cancer cells (43). This is a common feature in various cancer cells (20, 44–46). YAP O-GlcNAcylation enhances its activity, and activated YAP increases the cellular O-GlcNAcylation level by promoting OGT transcription, creating a self-perpetuating cycle (32, 39). However, the correlation between O-GlcNAcylation and TAZ has been unknown. Activated YAP/TAZ induce transcription of their negative regulators, NF2 and LATS2 (41, 42). Therefore, YAP and TAZ reciprocally inhibit each other’s activation through this intrinsic negative feedback loop (41). Hyperactivated YAP by O-GlcNAcylation may repress TAZ activation, so it would be unexpected that TAZ would be hyperactivated in cancer cells with abnormally high levels of O-GlcNAcylation. Therefore, it was hypothesized that abnormally increased cellular O-GlcNAcylation might cause hyperactivation of TAZ in cancer cells. This hypothesis was tested using MDA-MB-231 cells which have abnormally high levels of cellular O-GlcNAcylation and in which YAP/TAZ are hyperactivated. RNA interference was employed to decrease the expression of OGT, which effectively decreased the cellular O-GlcNAcylation level (Fig. 1A). The decrease in O-GlcNAcylation induced cytoplasmic sequestration of TAZ as well as YAP (Fig. 1 B and C). The phosphorylation at YAP Ser127 and TAZ Ser89 by LATS1/2 leads to cytoplasmic sequestration of YAP/TAZ (5, 7, 17). Therefore, phosphorylation levels at the specific sites of both proteins were observed under an OGT knockdown condition. OGT knockdown increased phosphorylation at YAP Ser127 by 2.88-fold (Fig. 1D) and TAZ Ser89 by 1.53-fold (Fig. 1E). These results indicate that abnormally increased O-GlcNAcylation in MDA-MB-231 cells induced activation of both effectors, YAP and TAZ.
Fig. 1.
MST1 and LATS2, negative regulators of YAP/TAZ, are O-GlcNAcylated. (A–E) MDA-MB-231 cells were transfected with OGT siRNA or control (CTL) siRNA for 48 h. (A) The cell lysates were immunoblotted with the relevant antibodies. (B and C) Localization of endogenous YAP (B) and TAZ (C) was detected by immunofluorescence. (Scale bar, 50 μm.) Nuclear and cytoplasmic fractions were analyzed by immunoblotting with relevant antibodies. Lamin A/C and α-tubulin were used as nuclear and cytoplasmic markers, respectively. (D and E) The phosphorylation levels at YAP Ser127 (D) and at TAZ Ser89 (E) were determined by Western blotting. Each phosphorylation level was normalized to the total YAP or TAZ level. Data are presented as mean ± SEM from at least three independent experiments. Statistical significance was determined by the two-tailed Student’s t test (*P < 0.05). (F–I) HEK293 cells were transfected with Flag-MST1 (F), Flag-MST2 (G), HA-LATS1 (H), and HA-LATS2 (I) independently and then treated with 10 μM Thiamet-G for 24 h. The immunoprecipitated exogenous proteins were immunoblotted with anti-O-GlcNAc antibody. (J and K) HEK293 cells were cotransfected Flag-MST1 (J) or HA-LATS2 (K) with OGT. Immunoprecipitated Flag-MST1 and HA-LATS2 were immunoblotted with anti-O-GlcNAc antibody, and the sWGA precipitates were immunoblotted with indicated antibodies. (L) MDA-MB-231 cells were incubated in media containing 5 or 25 mM glucose for 3 d. The sWGA precipitates were immunoblotted with the relevant antibodies. Abbreviations: O/E (overexpression), K/D (knockdown), C (cytosol), N (nucleus).
O-GlcNAcylation of TAZ was not detected (32), so this study was conducted to identify O-GlcNAc-modified proteins in the MST-LATS kinase cascade upstream of YAP/TAZ. For this, MST1/2 and LATS1/2 were independently transfected into HEK293 cells, and immunoprecipitated MST1/2 and LATS1/2 were probed with anti-O-GlcNAc antibody. Immunoprecipitated MST1 and LATS2 were O-GlcNAcylated, and O-GlcNAcylation of these proteins increased in response to treatment with the OGA inhibitor Thiamet-G (Fig. 1 F and I). However, O-GlcNAc was not detected on MST2 and LATS1 (Fig. 1 G and H). In addition, it was confirmed that O-GlcNAcylation on MST1 and LATS2 increased under an OGT overexpressing condition by the terminal GlcNAc-specific lectin (sWGA) precipitation and immunoprecipitation (Fig. 1 J and K).
Glucose is the main source for O-GlcNAcylation, and extracellular glucose status controls activation of the Hippo pathway (47). Therefore, we confirmed whether the O-GlcNAcylation levels of endogenous MST1 and LATS2 in MDA-MD-231 cells change depending on extracellular glucose status. As a result, the amount of sWGA-precipitated LATS2 dramatically increased under the 25-mM glucose condition compared to the 5-mM glucose condition (Fig. 1L). These results indicate that MST1 and LATS2 are O-GlcNAc modified components of the Hippo pathway and that LATS2 O-GlcNAcylation is strongly affected by extracellular glucose status.
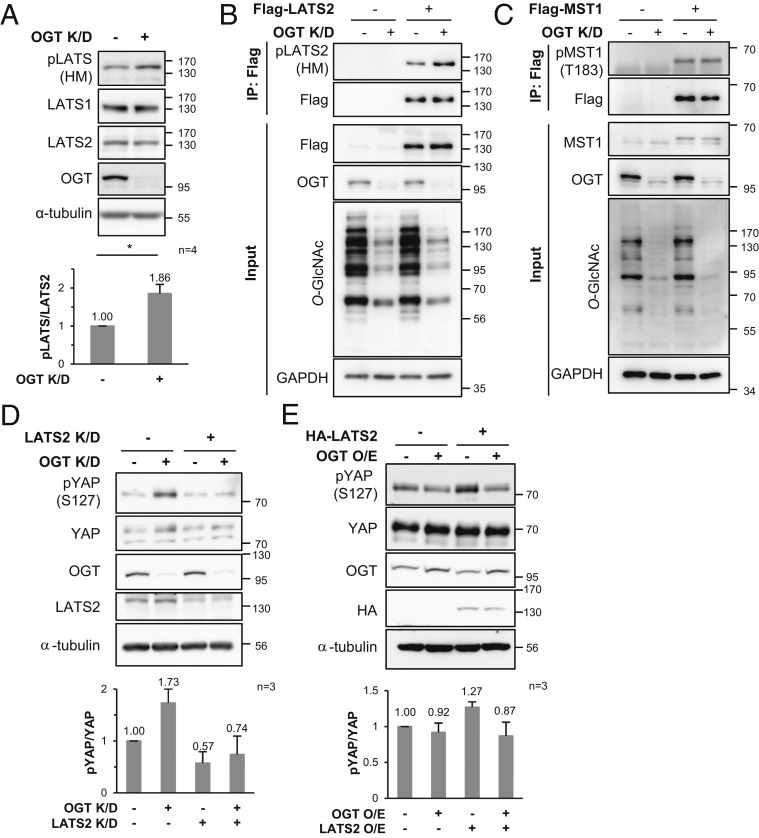
LATS2 Is a Key Regulator Controlling the Hippo Pathway Depending on the Cellular O-GlcNAcylation Level.
LATS1/2 phosphorylates YAP and TAZ at Ser127 and Ser89, respectively, and therefore acts as a direct regulator of YAP/TAZ (5, 7, 17). Thus, it was checked whether the phosphorylation level of the endogenous LATS1/2 hydrophobic motif, which is necessary for LATS1/2 activation, changed depending on the cellular O-GlcNAcylation level. OGT knockdown increased the amount of protein probed with the anti-p-hydrophobic motif-LATS antibody (Fig. 2A). The phosphorylation level of the LATS1 hydrophobic motif is not affected by OGT knockdown (32, 39), so it was inferred that phosphorylation of the LATS2 hydrophobic motif would increase under an OGT knockdown condition. To confirm this inference, exogenous LATS2 was immunoprecipitated and probed with anti-p-hydrophobic motif-LATS antibody. Phosphorylation at the hydrophobic motif of exogenous LATS2 was increased by OGT knockdown (Fig. 2B).
Fig. 2.
Phosphorylation of LATS2 hydrophobic motif changes depending on cellular O-GlcNAcylation status, but phosphorylation at MST1 Thr183 does not. (A) MDA-MB-231 cells were transfected with OGT siRNA or CTL siRNA for 48 h, and the cell lysates were immunoblotted with the relevant antibodies. The amount of phosphorylation of the LATS hydrophobic motif was normalized to that of LATS2. Data are presented as mean ± SEM from four independent experiments. Statistical significance was determined by the two-tailed Student’s t test (*P < 0.05). (B) Endogenous OGT in Flag-LATS2 expressing MDA-MB-231 cells was knocked down by siRNA. Immunoprecipitated Flag-LATS2 was immunoblotted with anti-p-hydrophobic motif-LATS antibody. (C) MDA-MB-231 cells were transfected with Flag-MST1 and OGT siRNA for 48 h. Immunoprecipitated Flag-MST1 was immunoblotted with anti-p-T183-MST1 antibody. (D) MDA-MB-231 cells were transfected with OGT siRNA and LATS2 siRNA for 48 h. (E) MDA-MB-231 cells were transfected with HA-LATS2 and OGT for 48 h. (D and E) Phosphorylation at YAP Ser127 was detected with anti-p-S127-YAP antibody. The amount of phosphorylation at YAP Ser127 was normalized to that of YAP. Data are presented as mean ± SEM from three independent experiments. Abbreviations: O/E (overexpression), K/D (knockdown), HM (hydrophobic motif).
MST1, another newly discovered O-GlcNAcylated component, is a serine/threonine kinase that phosphorylates the LATS2 hydrophobic motif (14). If MST1 activity is regulated by its O-GlcNAcylation status, an increase in phosphorylation of the LATS2 hydrophobic motif could be induced by activated MST1 under the OGT knockdown condition. However, phosphorylation levels at MST1 Thr183, which are essential for MST1 activation, were unchanged despite a significant global O-GlcNAcylation decrease caused by OGT knockdown (Fig. 2C). These results indicate that the differences in LATS2 phosphorylation caused by changes in intracellular O-GlcNAcylation occurred regardless of MST1 O-GlcNAcylation.
Thus, it was hypothesized that LATS2 O-GlcNAcylation might inhibit phosphorylation of its hydrophobic motif. If this hypothesis is correct, it is also possible that OGT knockdown enhances phosphorylation at YAP Ser127 by activating LATS2. Thus, this study examined phosphorylation at YAP Ser127 in LATS2/OGT double-knockdown (dKO) cells. The increase in YAP phosphorylation levels caused by OGT knockdown was lower in LATS2 knockdown cells than in LATS2 nonknockdown cells (Fig. 2D). In addition, the reduction in phosphorylation at YAP Ser127 in response to OGT overexpression was enhanced by LATS2 overexpression (Fig. 2E). This result indicates that LATS2 is deeply involved in the OGT-induced phosphorylation of YAP. Taken together, there is a high probability that the O-GlcNAcylation of LATS2 inhibits LATS2 activity by decreasing the phosphorylation of its hydrophobic motif. Given that LATS2 is not only a direct negative regulator of YAP/TAZ but also a crucial component of the negative feedback loop in the Hippo pathway (41, 42), it is possible that LATS2 O-GlcNAcylation has a significant effect on dysregulation of the Hippo pathway in cancer cells. Therefore, this study focused on the O-GlcNAc modification of LATS2.
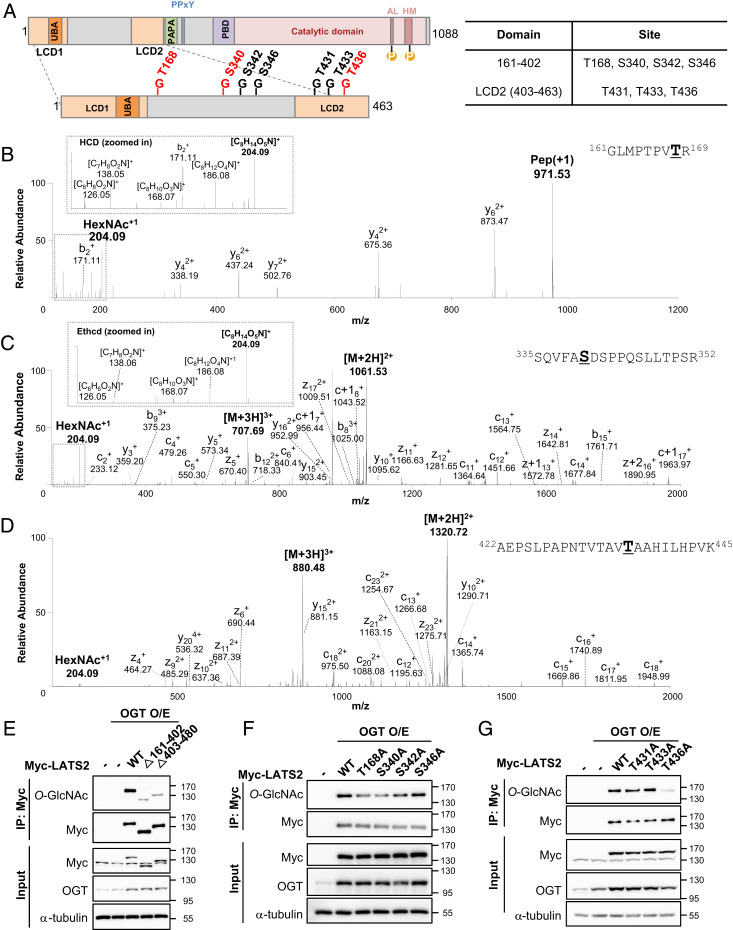
Thr168, Ser340, and Thr436 Are the Major LATS2 O-GlcNAcylation Sites.
Myc-LATS2 and pCMV-OGT were transiently overexpressed in HEK293 cells to identify the LATS2 O-GlcNAcylation sites. Immunoprecipitated Myc-LATS2 was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and in-gel digestion with trypsin and then analyzed by Orbitrap Fusion mass spectrometry in high collision dissociation (HCD) and electron-transfer/higher-energy collision dissociation (Ethcd) fragmentation modes. The identified putative LATS2 O-GlcNAcylation sites are summarized in Fig. 3A. Seven putative LATS2 O-GlcNAcylation sites were identified (Fig. 3 B–D and SI Appendix, Fig. S2). Four of these sites (Thr168, Ser340, Ser342, and Ser346) existed on the linker region and the remainder (Thr431, Thr433, and Thr436) occurred on LCD2. To verify that the linker region and LCD2 were actually O-GlcNAcylated, LATS2 deletion mutants (Δ161-402, deletion of the linker region between LCD1 and LCD2; Δ403-480, deletion of LCD2 and a PAPA repeat) were transfected with pCMV-OGT. Both deletion mutants exhibited a significant decrease in O-GlcNAcylation levels (Fig. 3E), indicating that O-GlcNAc is modified in the linker region and LCD2.
Fig. 3.
LATS2 O-GlcNAcylation sites were identified. (A) A schematic drawing of the LATS2 O-GlcNAcylation regions based on mass spectrometry data (Fig. 3 B–D and SI Appendix, Fig. S2). Abbreviations: LCD1 (LATS conserved domain 1), LCD2 (LATS conserved domain 2), PDB (protein binding domain), UBA (ubiquitin-associated domain), AL (activation loop), HM (hydrophobic motif), PAPA (sequence with repeats of proline-alanine residues), PPxY (proline-proline-any amino acid-tyrosine). Red letters indicate major LATS2 O-GlcNAcylation sites. (B–D) O-GlcNAcylation was detected at Thr168 (B), Ser340 (C), and Thr436 (D). (B) This peptide corresponds to residues 161 to 169, GLMPTPVTR. The O-GlcNAc oxonium ion (m/z, 204.09) and a series of its fragments (m/z, 186.08, 168.07, 138.05, and 126.05) were also assigned. (C) This peptide corresponds to residues 335 to 352, SQVFASDSPPQSLLTPSR. The O-GlcNAc oxonium ion (m/z, 204.09) and a series of its fragments (m/z, 186.08, 168.07, 138.06, 126.05) were also assigned. (D) This peptide corresponds to residues 422 to 445, AEPSLPAPNTVTAVTAAHILHPVK. The O-GlcNAc oxonium ion (m/z, 204.09) was also assigned. (B–D) O-GlcNAcylation detected residues are bolded and underlined. (E–G) HEK293 cells were cotransfected with each indicated Myc-LATS2 and OGT. (E) Each immunoprecipitated LATS2 wild-type and deletion mutants was immunoblotted with anti-O-GlcNAc antibody. Abbreviations: Δ161-402 (deletion of the linker region between LCD1 and LCD2), Δ403-480 (deletion of LCD2 and a PAPA repeat). (F and G) Each overexpressed Myc-LATS2 in HEK293 cells was immunoprecipitated with Myc antibody-conjugated A/G agarose beads. Western blotting was performed with anti-O-GlcNAc antibody.
To identify the major O-GlcNAcylation sites in LATS2, site-specific O-GlcNAcylation-deficient mutants were generated by substituting alanine for serine/threonine at each of the identified sites. These mutants and pCMV-OGT were overexpressed in HEK293 cells. A decrease in O-GlcNAcylation levels was observed in the T168A, S340A, and T436A mutants (Fig. 3 F and G). These results indicate that Thr168 and Ser340 are major O-GlcNAcylation sites in the linker region and that Thr436 is the major O-GlcNAcylation site in LCD2.
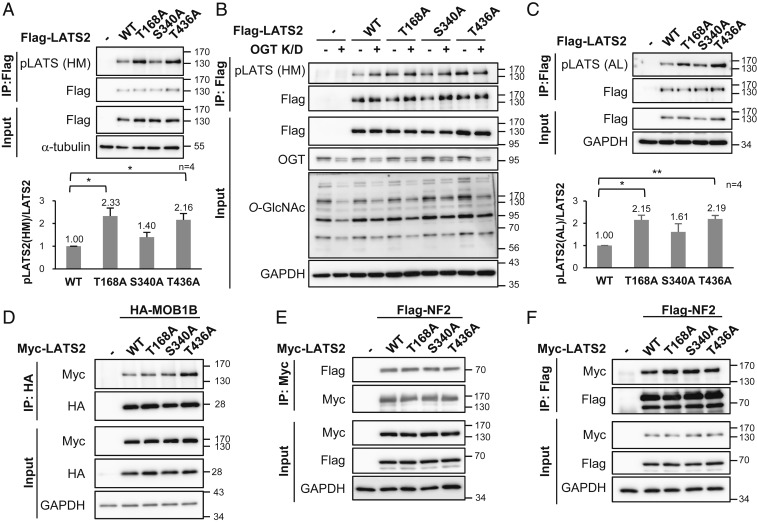
O-GlcNAcylation of LATS2 Prevents Its Own Phosphorylation by Disturbing the Interaction with MOB1.
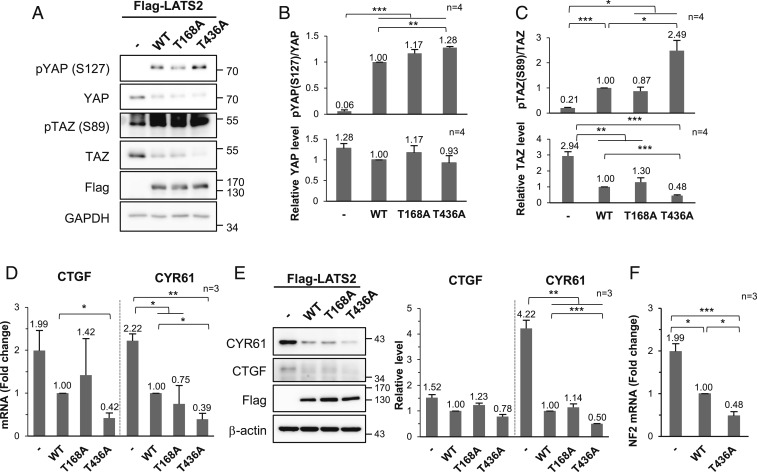
To verify whether LATS2 O-GlcNAcylation inhibits phosphorylation at Thr1041 of the LATS2 hydrophobic motif, wild-type LATS2 and each site-specific O-GlcNAcylation-deficient mutant (T168A, S340A, and T436A) were stably expressed in MDA-MB-231. Phosphorylation levels at the hydrophobic motif of the T168A mutant and the T436A mutant were more than twice as high as that of the wild type. The increase in phosphorylation level at the hydrophobic motif of S340A mutant was less than that of T168A and T436A and was not statistically significant (Fig. 4A). To clarify the effect of O-GlcNAcylation at each site, OGT was knocked down in each of the wild-type LATS2 and mutant LATS2-expressing cells and phosphorylation of the LATS2 hydrophobic motif was checked. Unlike the wild-type LATS2, phosphorylation at the hydrophobic motif of the LATS2 mutants did not significantly increase under the OGT knockdown condition (Fig. 4B). LATS2 requires additional phosphorylation at Ser872 located in the activation loop to be fully activated (14, 16). Phosphorylation of the activation loop showed similar results as phosphorylation of the hydrophobic motif (Fig. 4C). These results indicate that LATS2 O-GlcNAcylation, particularly at the Thr168 and Thr436 residues, inhibited its phosphorylation at Ser872 and Thr1041, which has a decisive effect on LATS2 activation.
Fig. 4.
O-GlcNAcylation of LATS2 inhibits its phosphorylation by blocking its interaction with MOB1B. (A) Flag-tagged wild-type (WT) LATS2 or LATS2 mutants was stably expressed in MDA-MB-231 cells, and immunoprecipitated Flag-LATS2 was detected with anti-p-hydrophobic motif-LATS antibody. Phosphorylation levels at Thr1041 in hydrophobic motif of Flag-LATS2 were normalized to the Flag-LATS2 level. (B) Endogenous OGT in the indicated Flag-LATS2-expressing MDA-MB-231 cells was knocked down by siRNA. Immunoprecipitated Flag-LATS2 was immunoblotted with anti-p-hydrophobic motif-LATS antibody. (C) Wild-type LATS2 or LATS2 mutants were stably expressed in MDA-MB-231 cells. Immunoprecipitated Flag-LATS2 was detected with anti-p-activation loop-LATS antibody. Phosphorylation levels at Ser872 in the activation loop of Flag-LATS2 were normalized to the Flag-LATS2 level. (D) HA-MOB1B was expressed with Myc-tagged wild-type LATS2 or LATS2 mutants in LATS1/2-dKO HEK293A cells, and 10 μM Thiamet-G was added for 24 h. HA-MOB1B was pulled down and Myc-LATS2 was detected by immunoblotting. (E and F) Indicated Myc-LATS2 and Flag-NF2 were cotransfected into LATS1/2-dKO HEK293A and 10 μM Thiamet-G was added for 24 h. Cell lysates were subjected to immunoprecipitation using anti-Myc antibody, followed by immunoblotting for Flag-NF2 (E), or vice versa (F). (A and C) Data are presented as mean ± SEM from four independent experiments. Statistical significance was determined by one-way ANOVA (*P < 0.05, **P < 0.01). Abbreviations: HM (hydrophobic motif), AL (activation loop).
Next, this study focused on the interaction between LATS2 and MOB1 to identify the molecular mechanism underlying how LATS2 O-GlcNAcylation prevents phosphorylation of its hydrophobic motif. MOB1 helps form the MST-MOB1-LATS2 complex by connecting MST to LATS2 (13, 14). This complex formation is an indispensable step to phosphorylate the LATS2 hydrophobic motif by MST. There are two kinds of human MOB1 proteins, MOB1A and MOB1B, and they share 95% amino acid sequence identity (48). Therefore, the effect of O-GlcNAcylation of LATS2 on the LATS2-MOB1B interaction was verified. LATS1/2-dKO HEK293A cells were chosen to rule out the influence of endogenous LATS1/2. They showed similar results as MDA-MB-231 in which O-GlcNAcylation of LATS2 inhibits its phosphorylation at Ser872 and Thr1041 (SI Appendix, Fig. S3). Wild-type LATS2 or mutants were cotransfected with HA-MOB1B into LATS1/2-dKO cells and treated with Thiamet-G for 24 h. Immunoprecipitated HA-MOB1B showed a stronger interaction with LATS2 mutants compared to wild-type LATS2, especially the T436A mutant (Fig. 4D). These results indicate that O-GlcNAcylation of LATS2, particularly O-GlcNAcylation at the Thr436 residue, blocked the LATS2-MOB1 interaction. Before LATS2 interacts with MOB1, NF2 recruits LATS2 to the MST-MOB1 complex in the cell membrane by directly interacting with LATS2 (15). Therefore, the effects of LATS2 O-GlcNAcylation on the NF2-LATS2 interaction were evaluated. There was no significant difference in the degree of NF2-wild-type LATS2 interaction and the NF2-LATS2 mutant interaction (Fig. 4 E and F). These results suggest that LATS2 O-GlcNAcylation directly interrupted the interaction with MOB1.
O-GlcNAcylation at LATS2 Thr436 Interrupts the Hippo Pathway by Inhibiting Its Own Activity.
O-GlcNAcylation at LATS2 Thr168 and Thr436 most effectively blocked phosphorylation of LATS2 crucial for its activation (Fig. 4 A and C). To confirm that O-GlcNAcylation of LATS2 at these sites reduced its activity, downstream targets of LATS2 were investigated in LATS1/2-dKO HEK293A cells, which were reconstituted with wild-type LATS2 or LATS2 mutants (T168A or T436A). As expected, the amount of TAZ decreased and phosphorylation at YAP Ser127 and TAZ Ser89 increased in LATS2-expressing cells (Fig. 5A). In particular, such changes were more noticeable in the LATS2 T436A mutant-expressing cells. The TAZ level was by 52% lower (Fig. 5 A and C) and phosphorylation levels at YAP Ser127 and TAZ Ser89 were higher in LATS2 T436A mutant-expressing cells than those in wild-type LATS2-expressing cells (Fig. 5 A–C). However, such changes were less noticeable in the LATS2 T168A mutant-expressing cells. LATS2 also phosphorylates YAP at Ser109. Therefore, the amount of phosphorylation at YAP Ser109 between the wild-type and the LATS2 mutant-expressing cells was additionally compared. Only LATS2 T436A mutant-expressing cells had greater levels of phosphorylation at YAP Ser109 than wild-type LATS2-expressing cells (SI Appendix, Fig. S4). These results support the conclusion that O-GlcNAcylation at LATS2 Thr436 inhibits its kinase activity.
Fig. 5.
O-GlcNAcylation at LATS2 Thr436 suppresses its kinase activity. (A–F) Wild-type (WT) LATS2 or LATS2 mutants were stably expressed in LATS1/2-dKO HEK293A cells. (A) Cell lysates were immunoblotted with the relevant antibodies. (B) The phosphorylation levels at YAP Ser127 were normalized to the YAP level, and the YAP expression level was normalized to the GAPDH level. (C) Phosphorylation levels at TAZ Ser89 were normalized to the TAZ level, and the TAZ expression level was normalized to the GAPDH expression level. (D) CTGF and CYR61 mRNA levels were determined by qRT-PCR. All values were normalized to the GAPDH mRNA level. (E) Lysates of the indicated Flag-LATS2 stably expressing LATS1/2-dKO HEK293A cells were immunoblotted with anti-CYR61 and anti-CTGF antibodies. CTGF and CYR61 expression levels were normalized to β-actin expression level. (F) NF2 mRNA level was determined by qRT-PCR. The NF2 mRNA level was normalized to GAPDH mRNA level. (B–F) Data are presented as mean ± SEM from at least three independent experiments. Statistical significance was determined by one-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.005).
Next, qRT-PCR was performed to measure the mRNA levels of connective tissue growth factor (CTGF) and cysteine-rich angiogenic inducer 61 (CYR61), which are YAP/TAZ transcriptional targets. The mRNA levels of both molecules were lower in LATS2 T436A mutant-expressing cells than those in wild-type LATS2-expressing cells (Fig. 5D). Similarly, the expression levels of CTGF and CYR61 were lower in cells expressing the T436A mutant compared to cells expressing the wild type (Fig. 5E). These results revealed the same tendency when intracellular O-GlcNAcylation in MDA-MB-231 cells was reduced by OGT knockdown condition (SI Appendix, Fig. S5 A and B). A strong positive correlation has been reported between CTGF/CYR61 and LATS2 mRNA levels in various cancer cells (41). LATS2 and NF2 are negative regulators and transcriptional targets of YAP/TAZ (41, 42). LATS2 and NF2 mRNA levels were reduced by OGT knockdown in MDA-MB-231 cells (SI Appendix, Fig. S5C), similar to the reductions in CTGF and CYR61 mRNA levels. In addition, NF2 transcription levels decreased significantly by expressing wild-type LATS2, and this decrease was enhanced more in LATS2 T436A mutant-expressing cells (Fig. 5F). These findings indicate that inactivation of LATS2 by O-GlcNAcylation at LATS2 Thr436 enhances YAP/TAZ activity. Taken together, it was concluded that O-GlcNAcylation at LATS2 Thr436 suppressed the Hippo pathway by inhibiting LATS2 activity.
LATS2 O-GlcNAcylation Promotes Tumor Growth by Enhancing Cell Proliferation.
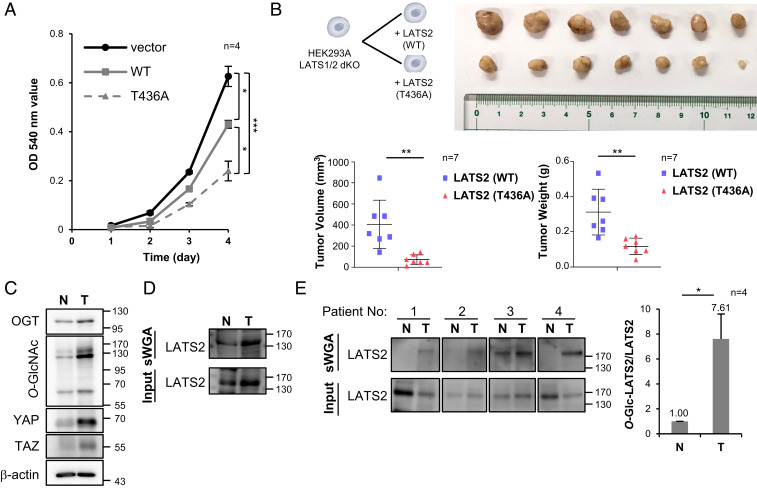
LATS2 suppresses cell proliferation by inhibiting transcription of YAP/TAZ target genes related to cell proliferation, such as CTGF and CYR61 (7, 49, 50). Therefore, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) cell assay was performed to confirm whether O-GlcNAcylation at LATS2 Thr436 promotes cell proliferation. The optical density (OD) 540-nm values obtained for wild-type LATS2 and T436A mutant-expressing LATS1/2-dKO HEK293 cells were 31% and 62% lower than those of LATS2-nonexpressing cells, respectively, 4 d after seeding (Fig. 6A). Additionally, the effect of LATS2 O-GlcNAcylation on tumor growth was investigated in vivo with male BALB/c nude mice. The size and weight of tumors formed from T436A mutant-expressing cells were 82% and 62% smaller than those of tumors formed from wild-type LATS2-expressing cells, respectively (Fig. 6B). These results indicate that O-GlcNAcylation at LATS2 Thr436 promotes tumor growth by enhancing cell proliferation.
Fig. 6.
LATS2 O-GlcNAcylation promotes tumor growth. (A) LATS1/2-dKO HEK293A cells, which were reconstituted with the wild-type (WT) LATS2 or T436A mutant, were seeded on 24-well plates at 2 × 104 cells/well. MTT assays were performed after growth for the indicated time. (B) BALB/c nude mice (n = 7/group) were injected with 1 × 107 of the indicated cells into the hypodermis. Tumor size and weight were measured 60 d after injection. (C and D) Mammary tissue samples were obtained from tumor-induced (T) and normal (N) C57BL/6NHsd mice. Tissue samples were examined by Western blot using the indicated antibodies (C). Tissue samples were precipitated by sWGA and then examined by Western blot analysis using an anti-LATS2 antibody (D). (E) Tumor tissue samples (T) and adjacent noncancerous tissue samples (N) obtained from breast cancer patients were precipitated by sWGA and examined by Western blot analysis using an anti-LATS2 antibody. (A, B, and E) Data are presented as mean ± SEM from at least three independent experiments. Statistical significance was determined by one-way ANOVA (A) or the two-tailed Student’s t test (B and E) (*P < 0.05, **P < 0.01, ***P < 0.005).
To confirm whether LATS2 O-GlcNAcylation affects tumor size by inducing dysregulation of the Hippo pathway, tumor tissues were obtained from both tumor-induced mouse models and breast cancer patients. The tumor-induced mouse model was generated by expressing the polyomavirus middle T oncogene in C57BL/6NHsd mice. As commonly observed in various cancers, OGT and O-GlcNAcylation levels in mouse mammary tumor tissues were higher than that in normal mammary tissues, and YAP and TAZ expression was more dominant in tumor tissues (Fig. 6C). Additionally, the amount of LATS2 precipitated with sWGA was higher in tumor tissues than that in normal tissues (Fig. 6D). The same results were observed with a similar experiment using human clinical samples (Fig. 6E). These findings support our conclusion that LATS2 O-GlcNAcylation induces excessive cell proliferation contributing to tumor growth.
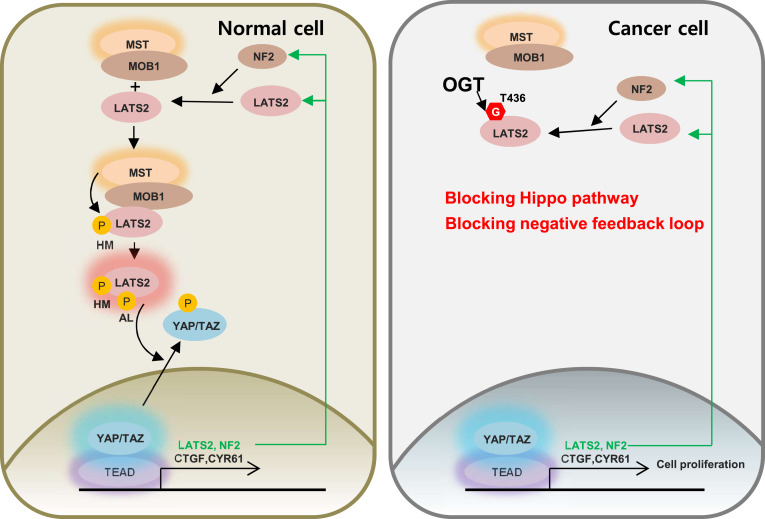
Overall, the conclusions of this study can be summarized as follows: LATS2 O-GlcNAcylation at Thr436 inhibits its phosphorylation, which is necessary for LATS2 activation, by blocking the interaction with MOB1. This blockage increases the quantity of activated YAP/TAZ. As a result, O-GlcNAcylation at LATS2 Thr436 enhances cell proliferation by inducing the transcription of YAP/TAZ target genes, which positively regulate cell proliferation. O-GlcNAcylation at LATS2 Thr436 increases NF2 and LATS2 transcription by inhibiting its activity. However, even if the newly synthesized NF2 recruits more LATS2 to the MST-MOB1 complex, activation of LATS2 might be impeded by O-GlcNAcylation at LATS2 Thr436 which blocks MOB1-LATS2 interactions. Similarly, even if LATS2 transcription rates increase, its effect on YAP/TAZ activity might be blocked by O-GlcNAcylation at LATS2 Thr436 in the same manner (Fig. 7). Thus, this study suggests that O-GlcNAcylation of LATS2 contributes to dysregulation of the Hippo pathway by inhibiting LATS2 activation.
Fig. 7.
Model for enhancing tumor growth due to the Hippo pathway being interrupted by LATS2 O-GlcNAcylation. The O-GlcNAcylation at LATS2 Thr436 by OGT interrupts the interaction with MOB1. Therefore, phosphorylation of the hydrophobic motif (HM) and activation loop (AL) essential for LATS2 activity decreases. Activated YAP and TAZ increase in response to LATS2 inactivation, which enhances transcription of CTGF, CYR61, NF2, and LATS2. However, even if the transcription rates of NF2 and LATS2 increase, the negative feedback loop would not effectively work by LATS2 O-GlcNAcylation in cancer cells. Consequently, O-GlcNAcylation at LATS2 Thr436 disrupts the Hippo pathway, possibly enhancing tumor growth.
Discussion
YAP and TAZ are highly expressed and more active in various cancer cells (20, 44–46). It was hypothesized that abnormally high O-GlcNAcylation levels in cancer cells might be the reason for dysregulation of the Hippo pathway. It has been recently found that YAP O-GlcNAcylation enhances its activity by blocking the interaction between YAP and LATS1 (32, 39). This study reports that O-GlcNAcylation is deeply involved in the Hippo pathway. Activated YAP/TAZ induces transcription of LATS2 and NF2, thereby increasing activation of LATS2 which inhibits YAP/TAZ activity (41). This process is a negative feedback loop in the Hippo pathway that helps maintain homeostasis in terms of intracellular YAP/TAZ levels and activities. Thus, it can be logically inferred that YAP O-GlcNAcylation decreases TAZ levels by stimulating LATS2 activity. However, the amount of TAZ remains high and LATS activity is inhibited in various cancer cells (43, 44, 51). Therefore, it was hypothesized that other components of the Hippo pathway are O-GlcNAcylated, leading to a network-level perturbation in cancer cells.
Our study revealed O-GlcNAc modified Hippo pathway components, MST1 and LATS2, and identified several LATS2 O-GlcNAcylation sites. Of those sites, O-GlcNAcylation at Thr436 in LCD2 showed the potential to hyperactivate YAP/TAZ and block an intrinsic negative feedback loop in the Hippo pathway by inhibiting LATS2 activation. Additionally, considering that O-GlcNAcylation competes with phosphorylation for the same YAP Ser109 residues (32), LATS2 O-GlcNAcylation is expected to have a synergistic effect on dysregulation of the Hippo pathway, as O-GlcNAcylation of LATS2 Thr436 increased the chance of O-GlcNAcylation at YAP Ser109 by inhibiting phosphorylation at YAP Ser109. Thus, findings about O-GlcNAcylation of LATS2 would be key to understanding how the Hippo pathway is dysregulated in cancer cells.
Hyperglycemia enhances breast tumor growth (52). O-GlcNAcylation levels of wild-type LATS2 dramatically increased under 25-mM glucose condition, whereas O-GlcNAcylation levels of the T436A mutant were not affected by the glucose concentration in the media (SI Appendix, Fig. S6). Thus, O-GlcNAcylation at LATS2 Thr436 may be involved in high-glucose-stimulated breast tumor growth.
Thr168 and Thr340 were identified as major O-GlcNAcylation sites in the linker region between LCD1 and LCD2 of LATS2. However, O-GlcNAcylation at these residues did not have a definite inhibitory effect on LATS2 activity. In particular, LATS2 T168A mutant kinase activity was not promoted (Fig. 5 A–C and SI Appendix, Fig. S4), even though phosphorylation levels of the hydrophobic motif and activation loop were significantly higher in the LATS2 T168A mutant than in the wild-type LATS2 (Fig. 4 A and C). LCD2 is essential for the function of LATS2 (53), whereas the exact function of the linker region is unknown. The LATS2-T168M mutation has been observed in a few cancer samples (54). In addition, the Ser172 residue adjacent to Thr168 is one of the sites that is phosphorylated by PKA (55). Phosphorylation by PKA does not affect phosphorylation at the hydrophobic motif and activation loop but enhances LATS2 activity (55). Therefore, we speculate that the kinase activity of the LATS2 T168A mutant did not increase, possibly due to the inhibition of phosphorylation at Ser172 or a conformational change in the LATS2 T168A mutant.
Studies about O-GlcNAcylation of the Hippo pathway have only focused on tumor growth. However, TAZ expression, which is strongly influenced by O-GlcNAcylation at LATS2 T436, is correlated with cancer invasiveness (44). This study also found that the transcription and expression levels of hyaluronan-mediated motility receptor (RHAMM), a target gene regulated by YAP/TAZ, and the phosphorylation levels of ERK essential for its activation decreased in response to OGT knockdown in MDA-MB-231 cells (SI Appendix, Fig. S7). RHAMM enhances cancer motility by activating ERK (56). These results suggest that O-GlcNAcylation might affect cancer metastasis by regulating the Hippo pathway. In addition, carcinogenesis can be induced by defects in the negative feedback loop of the Hippo pathway arising from LATS2 knockout (57), and the increase of intracellular O-GlcNAcylation level during malignant transformation is necessary for carcinogenesis (58, 59). Thus, the present results suggest that LATS2 O-GlcNAcylation might cause carcinogenesis by blocking the negative feedback loop of the Hippo pathway. Furthermore, the Hippo pathway is closely related to cellular development by regulating stem cell self-renewal (60), so O-GlcNAcylation would be expected to affect cellular development by regulating the Hippo pathway. Hence, we hope that this study will lead to new findings about various physiological functions of O-GlcNAcylation in the Hippo pathway.
Materials and Methods
Cell Culture and Reagents.
The nontumorigenic MCF10A breast epithelial cell line was cultured in Dulbecco's modified Eagle's medium/Ham's nutrient mixture F12 (DMEM/F12) (Thermo Fisher Scientific) supplemented with 5% horse serum, 10 µg/mL insulin, 20 ng/mL epidermal growth factor (EGF), 100 ng/mL cholera toxin, 500 ng/mL hydrocortisone, 100 U/mL penicillin, and 100 μg/mL streptomycin. The breast cancer cell lines MCF7, MDA-MB-468, and MDA-MB-231 were cultured in DMEM (Lonza) with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. The HEK293, HEK293FT, HEK293A, and LATS1/2-dKO HEK293A cell lines were cultured in DMEM with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. All cells used in this study were grown at 37 °C in 5% CO2. Thiamet-G was supplied by Injae Shin, Yonsei University, Seoul, Korea.
Transfection and Retrovirus Infection.
Cells were small interfering RNA (siRNA) transfected with Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer’s transfection protocol. The siRNA sequences used in this study are described in SI Appendix, Materials and Methods. Transfection was performed with polyethylenimine (Sigma-Aldrich) as described previously for transient overexpression of the target proteins (61). The mammalian expression plasmids for Myc-LATS1 and Myc-LATS2 were generated by inserting LATS1 or LATS2 cDNA into the pcDNA3.0 vector. The pcDNA3.0-Myc-LATS2 deletion mutants (Δ161-402 and Δ403-480) were kindly provided by Quan Chen, Nankai University, Tianjin, China. Various LATS2 mutants for the identified O-GlcNAcylation sites were generated using site-directed mutagenesis and the QuikChange site-directed mutagenesis kit (Agilent Technologies).
The wild-type and each O-GlcNAcylation-deficient LATS2 mutant DNAs were cloned into the Flag-tagged pMSCV vector. Two packaging plasmids (pCMV-VSV-G and pCMV-Gag-Pol) and retroviral constructs were cotransfected in HEK293FT cells with Lipofectamine 2000 (Invitrogen) for 48 h according to the manufacturer’s transfection protocol. MDA-MB-231 and LATS1/2-dKO HEK293A cells were infected in the presence of polybrene (4 μg/mL) using retroviral supernatants and the virally infected cells were selected with 1 to 2 μg/mL of puromycin (Sigma-Aldrich). The pMSCV-puro, pCMV-VSV-G, and pCMV-Gag-Pol vectors were kindly provided by Dae-Sik Lim, Korea Advanced Institute of Science and Technology, Daejeon, Korea.
Protein Extraction from Tissue Samples.
A total of four pairs of frozen deidentified human triple negative breast cancer and adjacent noncancerous tissues collected in 2015 were obtained from Yonsei University Cancer Center, Seoul, Korea. This study was approved by the Yonsei University Institutional Review Board (IRB No. 7001988–201910-BR-732-01). Mouse breast tumors were induced by expression of the polyomavirus middle T oncogene in C57BL/6NHsd mice, as described previously (62). Tissue samples from patients and mice were homogenized using a Dounce tissue homogenizer in NET buffer (150 mM NaCl, 50 mM Tris, pH 7.4, 1 mM EDTA, and 1% Nonidet P-40) supplemented with phosphatase inhibitor mixture (Roche) and protease inhibitor mixture (Roche). Homogenates were centrifuged at 14,000 rpm for 20 min at 4 °C, and the supernatants were collected to perform sWGA precipitation, and Western blotting.
Nuclear and Cytosol Fractionation.
Cells were incubated in ice-cold hypotonic buffer (5 mM Tris pH 7.5, 100 μM dithiothreitol (DTT), 50 μM ethylenediaminetetraacetic acid (EDTA), and 0.5% Nonidet P-40) for 15 min and centrifuged at 3,400 × g for 3 min at 4 °C. The supernatant was retained as the cytosolic fraction, the pellet was washed with PBS and centrifuged again at 3,400 × g for 3 min at 4 °C. To obtain the nuclear fraction, the pellet was incubated in RIPA buffer (150 mM NaCl, 50 mM Tris⋅HCl, pH 7.4, 2 mM EDTA, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, and 50 mM NaF) on ice for 10 to 15 min and centrifuged at 18,000 × g for 10 min at 4 °C. The supernatant of the lysed pellet was collected as the nuclear fraction.
Immunoprecipitation, sWGA Precipitation, and Western Blotting.
Cells were lysed in RIPA buffer supplemented with phosphatase inhibitor mixture and protease inhibitor mixture for immunoblotting. The cells for immunoprecipitation or sWGA precipitation were solubilized in lysis buffer (500 mM Tris, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, and 10 mM DTT) supplemented with phosphatase inhibitor mixture and protease inhibitor mixture or NET buffer supplemented with phosphatase inhibitor mixture and protease inhibitor mixture. Immunoprecipitation, sWGA precipitation, and Western blotting were performed as described previously (63). The antibodies used for Western blot and immunoprecipitation are described in SI Appendix, Materials and Methods. The results acquired by Western blotting were quantified using Multi Gauge ver. 3.1 software (Fuji Film Corp.).
Xenograft Mouse Experiment.
LATS1/2-dKO HEK293A cells which were reconstituted with wild-type LATS2 or T436A mutant were harvested using trypsin/EDTA solution and washed with PBS. Cells were suspended at 1 × 107 per 100 μL of PBS and injected into 5-wk-old male BALB/c nude mice (n = 7/group) purchased from DBL. The mice were housed in the Yonsei laboratory animal research center. Tumor size and weight of each group were measured 60 d after injection. Tumor volume was calculated as 0.5 × L (length) × W (width)2. This study was performed in accordance with the guidelines of the Yonsei University Institutional Animal Care Use Committee after review and approval (IACUC-A-202001-100102).
Immunofluoresence.
MDA-MB-231 cells were grown on 0.17 ± 0.01 mm thickness coverslips (Glaswarenfabik Karl Hecht GmbH and Co.). The cells were fixed with 3% formaldehyde (Fluka) for 5 min at 37 °C, then fixed again with 0.5% formaldehyde for 30 min at room temperature. The cells were washed with PBS and incubated in a quenching solution (50 mM NH4Cl in PBS) for 30 min at 4 °C. The cells were washed in PBS for 10 min and permeabilized with 0.2% Triton X-100 (Biotech) or 0.3% saponine (Sigma) in PBS containing 0.1% bovine serum albumin (BSA). Mouse anti-YAP1 antibody (1:50) or rabbit anti-TAZ antibody (1:50) was applied for 2 h to detect endogenous YAP or TAZ, followed by rinsing with PBS containing 0.1% BSA two times for 5 min. The cells were incubated with Alexa Fluor 488-conjugated secondary antibodies (1:500) (Invitrogen) for 1 h. After rinsing twice with PBS containing 0.1% BSA, the cells were mounted on a glass slide with Mowiol containing 4′, 6-diamidino-2-phenylindole. Images were collected on a Zeiss LSM 700 confocal microscope (Carl Zeiss) and analyzed with Zeiss ZEN software.
Cell Proliferation Assay.
LATS1/2-dKO HEK293A cells, which were reconstituted with wild-type LATS2 or T436A mutant, were seeded into 24-well plates at 2 × 104 cells per well. After 1, 2, 3, and 4 d, 600 μL of MTT solution (0.5 mg/mL MTT) was added to each well and the cells were incubated at 37 °C for 40 min. The MTT solution was removed, and 600 μL of DMSO was added to each well. Absorbance was measured at 540 nm using an Epoch microplate spectrophotometer (BioTek).
Quantitative RT-PCR Analysis.
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). To synthesize cDNA, reverse transcription was carried out using ReverTra Ace qPCR RT Master Mix (Toyobo Co.) according to the manufacturer's instructions. Real-time PCR was performed with oligonucleotide primers using SYBR Premix Ex Taq (Takara) on an Applied Biosystems 7300 Sequence Detection System (Applied Biosystems). The following PCR program was used; initial denaturation at 95 °C for 2 min; the cDNA was amplified for 40 cycles with the following settings: denaturation at 95 °C for 5 s, annealing at 60 °C for 31 s, and elongation at 72 °C for 31 s. The DNA was dissociated at 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s, and 60 °C for 1 min. The mRNA levels of CTGF, CYR61, RHAMM, and NF2 were normalized to those of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The PCR primer sequences are described in SI Appendix, Materials and Methods.
Mapping of the LATS2 O-GlcNAcylation Sites Using Mass Spectrometry.
The eluted LATS2 sample was subjected to electrophoresis in a 4 to 12% Bis-Tris NuPAGE gel (NOVEX) and stained with Coomassie Brilliant Blue (Sigma). The band corresponding to LATS2 was excised from the gel and subjected to in-gel tryptic digestion according to a standard protocol (64). Trypsin/Lys-C Mix, mass spectometry grade (Promega) was used in this procedure. Mass spectra of extracted peptide samples were obtained using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) interfaced with a nanoAcquity UPLC (Waters) as described previously in published methods (65). The raw data were compared with a Uniprot human database (entry no. 173,324) using the SEQUEST HT search engine in Proteome Discoverer 2.2 (Thermo Fisher Scientific). Trypsin was selected as the proteolytic enzyme with a maximum allowance of up to two missed cleavages. The carbamidomethyl of cysteine was considered a fixed modification, and variable modification was set for oxidation of methionine and O-GlcNAcylation of serine and threonine with a maximum of three posttranslational modifications at one peptide. Weight of the b, y ions was set to 0.5 and weight of the c, z ions was set to 1, relatively. Peptides were identified with ≥95% confidence and filtered at a 1% false-discovery rate.
Statistical Analysis.
Data are expressed as mean ± SEM based on at least three independent experiments. The statistical analysis was performed with the two-tailed Student’s t test for two groups and by one-way analysis of variance for multiple groups. A P value <0.05 was considered significant.
Data Availability.
The authors confirm that all data supporting the findings of this study are available within the article and its supplementary information.
Supplementary Material
Acknowledgments
We thank Dr. Dae-Sik Lim and Dr. Wonyul Jang for experimental advice and for providing reagents. This study was supported by National Research Foundation of Korea Grants funded by the Korean Government (The Ministry of Science, ICT and Future Planning) (NRF-2016R1A5A1010764 and NRF-2015M3A9B6073840) to J.W.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.A.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913469117/-/DCSupplemental.
References
- 1.Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G., Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Lian I. et al., The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106–1118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey K. F., Pfleger C. M., Hariharan I. K., The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Huang J., Wu S., Barrera J., Matthews K., Pan D., The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Hao Y., Chun A., Cheung K., Rashidi B., Yang X., Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Harvey K. F., Zhang X., Thomas D. M., The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Zhao B. et al., Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halder G., Dupont S., Piccolo S., Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Meng Z., Moroishi T., Guan K. L., Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Praskova M., Khoklatchev A., Ortiz-Vega S., Avruch J., Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381, 453–462 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boggiano J. C., Vanderzalm P. J., Fehon R. G., Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21, 888–895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poon C. L., Lin J. I., Zhang X., Harvey K. F., The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 21, 896–906 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Praskova M., Xia F., Avruch J., MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni L., Zheng Y., Hara M., Pan D., Luo X., Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 29, 1416–1431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin F. et al., Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan E. H. et al., The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076–2086 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Lei Q. Y. et al., TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28, 2426–2436 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C. Y. et al., The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFbeta-TrCP E3 ligase. J. Biol. Chem. 285, 37159–37169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L., A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinhardt A. A. et al., Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 39, 1582–1589 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X. et al.; AOCS Study group , The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene 30, 2810–2822 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Park H. W., Guan K. L., Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol. Sci. 34, 581–589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y. et al., Comprehensive molecular characterization of the hippo signaling pathway in cancer. Cell Rep 25, 1304–1317.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akella N. M., Ciraku L., Reginato M. J., Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 17, 52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanover J. A., Chen W., Bond M. R., O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle. J. Bioenerg. Biomembr. 50, 155–173 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Haltiwanger R. S., Blomberg M. A., Hart G. W., Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 267, 9005–9013 (1992). [PubMed] [Google Scholar]
- 27.King D. T., Males A., Davies G. J., Vocadlo D. J., Molecular mechanisms regulating O-linked N-acetylglucosamine (O-GlcNAc)-processing enzymes. Curr. Opin. Chem. Biol. 53, 131–144 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Ong Q., Han W., Yang X., O-GlcNAc as an integrator of signaling pathways. Front. Endocrinol. (Lausanne) 9, 599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong D. L., Hart G. W., Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 (1994). [PubMed] [Google Scholar]
- 30.Yang W. H. et al., Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Olivier-Van Stichelen S. et al., O-GlcNAcylation stabilizes β-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 28, 3325–3338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng C. et al., Regulation of the hippo-YAP pathway by glucose sensor O-GlcNAcylation. Mol. Cell. 68, 591–604.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Lima V. V. et al., O-GlcNAc modification during pregnancy: Focus on placental environment. Front. Physiol. 9, 1263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X., Qian K., Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cork G. K., Thompson J., Slawson C., Real talk: The inter-play between the mTOR, AMPK, and hexosamine biosynthetic pathways in cell signaling. Front. Endocrinol. (Lausanne) 9, 522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abramowitz L. K., Hanover J. A., T cell development and the physiological role of O-GlcNAc. FEBS Lett. 592, 3943–3949 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Butkinaree C., Park K., Hart G. W., O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeidan Q., Hart G. W., The intersections between O-GlcNAcylation and phosphorylation: Implications for multiple signaling pathways. J. Cell Sci. 123, 13–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X. et al., The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat. Commun. 8, 15280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Lu Z., Shi Y., Sun F., AMOT is required for YAP function in high glucose induced liver malignancy. Biochem. Biophys. Res. Commun. 495, 1555–1561 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Moroishi T. et al., A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park G. S. et al., An evolutionarily conserved negative feedback mechanism in the Hippo pathway reflects functional difference between LATS1 and LATS2. Oncotarget 7, 24063–24075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamar J. M. et al., The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. U.S.A. 109, E2441–E2450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan S. W. et al., A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 68, 2592–2598 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Wang L. et al., Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One 8, e65539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W. et al., AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 17, 490–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hergovich A., MOB control: Reviewing a conserved family of kinase regulators. Cell. Signal. 23, 1433–1440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pu N. et al., Cell-intrinsic PD-1 promotes proliferation in pancreatic cancer by targeting CYR61/CTGF via the hippo pathway. Cancer Lett. 460, 42–53 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Zhao B. et al., TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strazisar M., Mlakar V., Glavac D., LATS2 tumour specific mutations and down-regulation of the gene in non-small cell carcinoma. Lung Cancer 64, 257–262 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Ryu T. Y., Park J., Scherer P. E., Hyperglycemia as a risk factor for cancer progression. Diabetes Metab. J. 38, 330–336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y. et al., Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene 22, 4398–4405 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Yu T., Bachman J., Lai Z. C., Mutation analysis of large tumor suppressor genes LATS1 and LATS2 supports a tumor suppressor role in human cancer. Protein Cell 6, 6–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim M. et al., cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 32, 1543–1555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z. et al., Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc. Natl. Acad. Sci. U.S.A. 111, E89–E98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He C. et al., YAP1-LATS2 feedback loop dictates senescent or malignant cell fate to maintain tissue homeostasis. EMBO Rep. 20, e44948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng Q. et al., O-linked GlcNAcylation elevated by HPV E6 mediates viral oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 113, 9333–9338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duan F. et al., O-GlcNAcylation of RACK1 promotes hepatocellular carcinogenesis. J. Hepatol. 68, 1191–1202 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Liu H., Jiang D., Chi F., Zhao B., The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein Cell 3, 291–304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boussif O. et al., A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guy C. T., Cardiff R. D., Muller W. J., Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang J. G. et al., O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J. Biol. Chem. 284, 34777–34784 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Seo H. G. et al., Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci. Rep. 6, 34614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all data supporting the findings of this study are available within the article and its supplementary information.