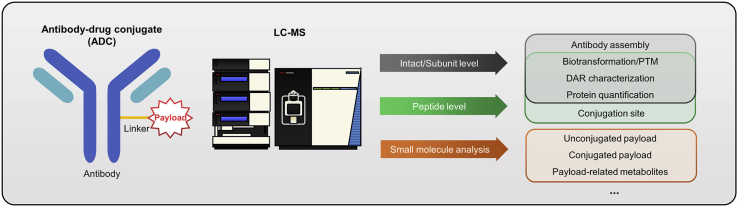
Abstract
The past few years have witnessed enormous progresses in the development of antibody-drug conjugates (ADCs). Consequently, comprehensive analysis of ADCs in biological systems is critical in supporting discovery, development and evaluation of these agents. Liquid chromatography-mass spectrometry (LC-MS) has emerged as a promising and versatile tool for ADC analysis across a wide range of scenarios, owing to its multiplexing ability, rapid method development, as well as the capability of analyzing a variety of targets ranging from small-molecule payloads to the intact protein with a high, molecular resolution. However, despite this tremendous potential, challenges persist due to the high complexity in both the ADC molecules and the related biological systems. This review summarizes the up-to-date LC-MS-based strategies in ADC analysis and discusses the challenges and opportunities in this rapidly-evolving field.
Keywords: Antibody-drug conjugate (ADC), Liquid chromatography-mass spectrometry (LCMS), Drug-to-antibody ratio (DAR)
Graphical abstract
Highlights
-
•
ADC is an important class of anti-cancer agents but optimal analytical approaches remain elusive despite years of efforts.
-
•
LC-MS shows promising capability for protein analysis at peptide, subunit and intact levels, as well as payload analysis.
-
•
LC-MS is a powerful tool for ADC DAR measurement because of its molecular-level resolution and the matrix-independency.
-
•
A score of novel LC-MS-based techniques have emerged in ADC analysis, both quantitatively and qualitatively.
1. Introduction
Antibody-drug conjugates (ADCs) constitute one of the most promising types of targeted cancer therapeutics. Typically, an ADC molecule includes an antibody targeting a tumor cell surface antigen, coupled with a number of potent cytotoxic payloads, via covalent conjugation (i.e., a linker). To date, eight ADCs that have been approved by U.S. Food and Drug Administration (FDA) include gemtuzumab ozogamicin (Mylotarg, Pfizer, Inc.), brentuximab vedotin (Adcetris, Seattle Genetics, Inc.), ado-trastuzumab emtansine (Kadcyla, Genentech, Inc.), inotuzumab ozogamicin (Besponsa, Pfizer, Inc.), polatuzumab vedotin-piiq (Polivy, Genentech, Inc.), enfortumab vedotin-ejfv (Padcev, Seattle Genetics, Inc.), trastuzumab deruxtecan (Enhertu, Daiichi Sankyo, Inc.) and sacituzumab govitecan (Trodelvy, Immunomedics, Inc.) and more than 100 in active clinical trials [1].
Though antibodies in ADCs rarely have antitumor activities by themselves, their specificity to target antigens often makes them useful delivery vehicles of payloads targeting tumor cells [2]. The toxic payloads used in most approved and clinical-stage ADCs are microtubule disruptors (e.g., maytansinoid and dolastatin analogs) or DNA-damaging agents (e.g., duocarmycins, pyrrolobenzodiazepines, and calicheamicins). The conjugating linkers in ADCs are generally classified into cleavable and non-cleavable ones, which behave quite differently in a biological system, and produce distinct in vivo forms of released toxin. Specifically, cleavable linkers (e.g., valine-citrulline dipeptide, hydrazine, and disulfide bridge) could be sensitive to cancer cell-specific intracellular properties, such as expression of certain protease, pH, and glutathione, and thus carrying the potential to achieve selective release of payloads [3]. By comparison, non-cleavable linkers contain no specific release mechanism and rely on intracellular proteolytic degradation following ADC internalization [3]. From an analytical perspective, while ADCs with cleavable linkers release free payload in a biological system, those with non-cleavable linkers usually release active payload-linker-amino acid moieties that are produced after the complete degradation of ADCs [3]; consequently, different analytes should be targeted.
Due to the high complexity of ADCs, analysis of these compounds is uniquely challenging, especially in a biological system. For instance, both the small-molecule payloads and the protein portion need to be analyzed. In addition, drug-to-antibody ratio (DAR) is an important parameter describing the number of payloads conjugated to the antibody since the DAR species could dynamically change in vivo, which brings additional challenges for bioanalysis. Compared to other traditional methods, liquid chromatography–mass spectrometry (LC-MS) has the unique capability in ADC analysis, since it can broadly analyze small molecules, intact proteins, digested proteins, as well as specific domain of proteins with the molecule-level resolution. Therefore, LC-MS represents a highly versatile and valuable tool and has played an indispensable role in ADC characterization, both quantitatively and qualitatively.
In the initial development stage, especially for linker and payload discovery, LC-MS is highly valuable in providing structure-activity relationship information [4]. LC-MS has also been widely employed in characterization of the physicochemical properties of ADCs, which have profound effects on the safety and efficacy profile [5]. For in vivo analysis, typically quantification in plasma, including enzyme-linked immunosorbent assay (ELISA), LC-MS, or a combination of the two methods are often employed. In general, ELISA is more commonly used in quantification of ADCs in plasma but is often matrix- and species-dependent. Moreover, the method development for ELISA is often time consuming and costly, which is impractical in the early phase of ADCs development. Another challenge for ELISA is that it cannot differentiate DAR species of ADCs. By comparison, LC-MS is often matrix- and species-independent, and method development is much faster, therefore providing a promising alternative to ELISA for ADC quantification [6,7]. Additionally, owing to the molecular-resolution of LC-MS, it is capable of determining DAR in biological samples directly and therefore is very helpful in characterizing DAR species [4].
Moreover, the therapeutic window is narrow for most of the current ADCs due to off-target toxicity [8]. It is critical to perform a comprehensive in vivo analysis of ADCs to understand the effects of various species produced by ADC (e.g., via biotransformation) on toxicity [9]. One paradigm is that measurement of tissue distribution of ADC-derived species at the off-target sites would be highly valuable for identifying perpetrators of toxic side effects. Radiolabeling approach has been used in previous studies to investigate ADC tissue distribution, which suffered from low accuracy and specificity owing to issues associated with radiolabeling [10,11]. By comparison, LC-MS-based method could be a promising solution to quantification of ADCs in tissue because of its high specificity and matrix independency [12].
With the ever-increasing interests in ADCs, a deeper understanding of the molecular characteristics and in vivo behaviors, as well as the connection between the two is urgently needed and appears to be a good fit for LC-MS. That being said, further technical improvements are essential for overcoming current hurdles and warranting reliable and practical investigation.
2. Analysis of the large-molecule portion
With the capability of protein analysis at peptide, subunit and intact levels, LC-MS can provide comprehensive protein characterization, both qualitatively and quantitatively. In this section, we review LC-MS analysis on whole ADC and protein levels, including conjugation site analysis, biotransformation characterization, localization of post-translational modification (PTM), and antibody structural integrity confirmation.
2.1. Qualitative characterization
Extensive characterization of ADC products is often required by FDA, such as DAR analysis, drug-load-distribution (DLD) assessment, conjugation sites characterization, and PTM evaluation. LC-MS-based techniques are one of the most important tools for these assays, and particularly useful for characterization of the protein component of ADCs. Depending on the purpose of analysis, LC-MS can analyze proteins at bottom-up (i.e., peptide mapping), middle-down (i.e., subunit analysis) or top-down (i.e., intact protein) levels. These different levels of protein analysis are also often performed in parallel to provide orthogonal and more comprehensive information [13]. Reversed-phase liquid chromatography (RPLC) is the most commonly used for separation because of high efficiency and robustness, and high-resolution MS such as Orbitrap, Quadrupole-Time-of-Flight (Q-TOF), and Fourier Transform Ion Cyclotron Resonance (FTICR) are routinely used for detection because their high resolving power benefits identification of biotransformation/PTM as well as differentiation of DAR species [[14], [15], [16]]. Typically, a 17,500 resolution could be sufficient for DAR characterization, and it has been reported that a resolution limited around 35,000 could be beneficial to DAR characterization when using Orbitrap [17]. Here we review the use of LC-MS with peptide mapping as well as the intact/subunit approaches for in-depth ADC characterization.
2.1.1. Peptide mapping for analysis of conjugation site and PTMs
Due to the high heterogeneity of ADCs, the analysis of antibody vehicle is often challenging. Moreover, PTMs such as glycosylation, phosphorylation, deamidation and methylation further compound this problem. Peptide mapping or bottom-up LC-MS represents a powerful tool for characterization of the sequence and PTMs of proteins with high specificity and reproducibility, and thereby has been extensively used in quality control [18]. The protein molecules are regularly prepared with or without protein purification depending on the complexity of sample, followed by denaturation, reduction, alkylation and enzyme digestion to produce relatively short, completely proteolyzed peptides. The digest is then analyzed on RPLC-High-Resolution-MS and a data processing tool (usually including a searching engine) to obtain detailed information on protein primary sequence and modifications.
With the ability to elucidate site-specific information, peptide mapping is the most frequently used tool for conjugation site identification and characterization. One study has utilized peptide mapping analysis on a UPLC-MS for confirmation of T-DM1 primary sequence and evaluation of site of conjugation/occupancy [19]. The work identified 82 conjugated lysine sites accounting for nearly 90% of available lysine residues and achieved much improved site coverage compared with several previous studies which had identified 38–44 conjugation sites [[19], [20], [21]]. A group employed peptide mapping with UPLC-Q-TOF to determine the stability of 26 conjugation sites of T-DM1 and compared the degradation rates at each site to provide guidance for quality control [22]. Another group developed a new procedure with enhanced depth-of-analysis, including improved reduction, immune-globulin degrading enzyme of Streptococcus pyogenes (IdeS) digestion protocol and the use of UPLC-TOF to identify payload positional isomers at cysteine residues of brentuximab vedotin [23].
However, one concern is that the conjugation of the payload often results in a significant increase in hydrophobicity of the conjugated peptides derived from ADCs and thus the LC separation of such conjugated peptides may be difficult [24,25]. A number of studies attempting to address these challenges have been reported. For example, capillary zone electrophoresis (CZE)/MS peptide mapping has been used as an orthogonal tool to help the separation of large hydrophobic peptides, hydrophilic di-/tri-peptides and glycopeptides from ADCs [26]. Another work adopted micro-pillar array columns in LC-nano-electrospray ionization (ESI)-Q-TOF for ado-trastuzumab emtansine (T-DM1) characterization, which offers highly efficient separations even for hydrophobic segments, and achieved a high sequence coverage of peptide mapping [27].
Identification and quantification of PTMs are imperative owing to its potential of altering protein functions such as deactivating proteins and leading to a decreased efficacy. LC-MS-based peptide mapping is the gold standard to evaluate PTMs of therapeutic antibodies owing to the ability of accurate site localization. Though no study has yet used peptide mapping for evaluation of PTMs in ADCs, many studies on therapeutic antibodies have been reported [[28], [29], [30]]. While the technique is proved to be very useful and widely practiced, some issues remain. Proper protocols in the database searching process are important for minimizing the false positive of the PTM annotation [29]. Additionally, multiple-step sample preparation may introduce artificial modifications [13]. The optimization of sample preparation protocols, especially the digestion step, has been conducted in attempt to address this problem [18,31,32]. Analysis on intact/subunit levels was also performed for evaluation of PTMs as well as other forms of biotransformation. Though these approaches may not be able to provide the exact location of PTMs, the reduced sample preparation step could decrease artificial modifications and improve throughput. Moreover, such methods can preserve information of the functional relationship of multiple PTMs and identify different proteoforms [33,34].
2.1.2. Intact/subunit analysis
Characterization of ADC at intact/subunit levels is an important component for LC-MS-based ADC analysis. Table 1 summarizes the analytical conditions of intact/subunit level LC-MS-based ADC analysis [17,24,25,[35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48]].
Table 1.
Recent applications of intact/subunit LC-MS to ADC analysis.
| ADC | Subject | Condition | Intact/Subunit | LC | Column | Mobile phase | MS | Ref. |
|---|---|---|---|---|---|---|---|---|
| Trastuzumab-vc-MMAE | In vitro DAR characterization | Denature | Subunit | RPLC | C4 (100 mm × 2.1 mm i.d., 3.5 μm, 450 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [17] |
| C4 (150 mm × 1 mm i.d., 5 μm, 300 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Orbitrap | ||||||
| Site-specific ADC | Conjugation site and glycosylation site identification | Denature | Subunit | RPLC | PS/DVB (100 mm × 1 mm i.d., 4 μm, 1500 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Orbitrap | [24] |
| Lys-conjugated ADC, site-specific ADC | In vitro DAR characterization | Denature | Intact | Organic SEC | Ethylene bridged hybrid-based particle, diol bonding (150 mm × 4.6 mm i.d., 1.7 μm,200 Å) | 30% ACN, 70% water with 0.05% TFA | Q-TOF | [25] |
| Cys-conjugated ADC | Native | Intact | SEC | 20 mM–40 mM ammonium acetate in water | ||||
| Cys-conjugated ADC, site-specific ADC | In vitro DAR characterization | Denature | Subunit | RPLC | C4 (100 mm × 2.1 mm i.d., 1.7 μm, 300 Å); Diphenyl (100 mm × 2.1 mm, 1.8 μm, 300 Å) |
Consisting of TFA, water, isopropanol, ACN | ||
| Site-specific ADC | Biotransformation characterization | Denature | Subunit | RPLC | C4 (50 mm × 2.1 mm i.d., 1.8 μm, 300 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [35] |
| Site-specific ADC | In vivo DAR characterization | Denature | Subunit | RPLC | PS/DVB (150 mm × 2.1 mm i.d., 8 μm, 1000 Å) | A: 0.1% TFA in water B: 0.08% TFA in ACN |
Q-TOF | [36] |
| Native | Intact | SEC | Ethylene bridged hybrid-based particle, diol bonding (150 mm × 2.1 mm i.d., 1.7 μm, 200 Å) | A: 0.1% FA/0.025%TFA in water B: 0.1% FA/0.025%TFA in ACN |
Q-TOF or Orbitrap | |||
| Site-specific ADC | In vivo DAR characterization and catabolite characterization | Denature | Subunit | RPLC | PS/DVB (50 mm × 500 μm i.d.) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [37] |
| Trastuzumab emtansine | Biotransformation characterization | Denature | Intact or subunit | RPLC | PS/DVB (5 cm × 500 μm i.d.), PS/DVB (25 cm × 200 μm i.d.) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Orbitrap | [38] |
| Cysteine-conjugated ADC, site-specific ADC, lysine-conjugated ADC | In vivo DAR characterization | Denature | Intact or subunit | RPLC | PS/DVB (50 mm × 0.3 mm i.d., 5 μm, 4000 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [39] |
| THIOMAB-vc-MMAE | In vivo DAR characterization | Denature | Intact | RPLC | PS/DVB (50 mm × 0.3 mm i.d., 5 μm, 4000 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [40] |
| Brentuximab Vedotin | In vitro DAR characterization, positional isomer characterization | Native | Intact | HIC × SEC | HIC: PA (100 mm × 4.6 mm i.d., 5 μm, 1000 Å); SEC: Silica-based particle, hydrophilic bonding (50 mm × 4.6 mm, 2.7 μm, 300 Å) |
HIC: 2.5 M of ammonium acetate and 0.1 M phosphate buffer (pH 7.0); 0.1 M phosphate buffer (pH 7.0); SEC: 100 mM ammonium acetate |
IM × MS | [41] |
| Trastuzamab Entansine, cysteine-conjugated ADC | In vitro DAR characterization | Native | Intact | SEC | Ethylene bridged hybrid-based particle, diol bonding (150 mm × 2.1 mm i.d., 1.7 μm, 200 Å) | 50 mM ammonium acetate in water | Q-TOF | [42] |
| Trastuzumab-DSEA- fluorophore | In vitro DAR characterization | Denature | Subunit | RPLC | PS/DVB (150 mm × 2.1 mm i.d., 8 μm, 1000 Å) | A: 0.05% TFA in water; B: 0.05% TFA in ACN |
Q-TOF | [43] |
| Trastuzumab-mc-MMAF, Trastuzumab-vc-MMAE | In vitro DAR characterization | Denature | Subunit | RPLC | Phenyl (5 mm × 2.1 mm i.d., 20 μm, 1000 Å) | Water; ACN; 1% FA | Q-TOF | [44] |
| Native | Intact | SEC | Ethylene bridged hybrid-based particle, diol bonding (150 mm × 4.6 mm i.d., 1.7 μm, 200 Å) | 10 mM ammonium acetate in water (pH 6.9) | ||||
| Lysine-conjugated ADCs, dual-payload ADC, site-specific ADC | In vitro DAR characterization | Denature | Intact | RPLC | PS/DVB (100 mm × 3.0 mm i.d., 4 μm, 1500 Å) | 20% ACN, 80% water with 0.1% FA | TOF | [45] |
| Cantuzumab-SPDB-DM4 | In vitro DAR characterization | Denature | Intact | SEC | Diol-bonded silica (30 cm × 4.6 μm i.d., 4 mm, 250 Å) | 50% aqueous ACN containing 0.02% TFA and 1% FA | TOF | [46] |
| Site-specific ADC | In vivo DAR characterization | Denature | Intact | RPLC | C4 (100 mm × 0.3 mm i.d., 1.7 μm, 300 Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [47] |
| Trastuzumab Emtansine | Quantification in biological samples | Denature | Intact | RPLC | C4 (50 mm × 2.1 mm i.d., 1.7 μm, 300Å) | A: 0.1% FA in water; B: 0.1% FA in ACN |
Q-TOF | [48] |
PS/DVB: Polystyrene Divinylbenzene, FA: formic acid, ACN: acetonitrile, TFA: trifluoroacetic acid, PA: Polyamide.
2.1.2.1. Biotransformation characterization
ADC in vivo biotransformations such as payload deconjugation, protein mass adduct/loss and payload metabolism often occur due to the nature of the in vivo environment, and the instability and complexity of the conjugation sites or linkers [35,49]. Such biotransformations could potentially decrease efficacy and increase off-target toxicity. Even though some new engineering technologies (e.g., cystine engineering, non-natural amino acid engineering, enzyme-mediated conjugation, peptide tags engineering, new heterobifunctional reagent) have been utilized in a site-specific manner to produce more homogenous and stable ADCs, in vivo biotransformations could very well persist [35,36]. Therefore, the analytical method to characterize ADC biotransformations is particularly important in helping guide the engineering efforts of ADCs as well as elucidate the paths of ADC metabolism/catabolism [16].
In this regard, intact and subunit LC-MS is the method of choice due to its ability to provide high-level sequence and structure information in a high-throughput manner [50]. While intact analysis measures the intact ADCs (e.g., ≥ 150 kDa) either under native or denaturing conditions, subunit analysis measures fragments (20–50 kDa) produced by reduction or enzyme digestion such as papain, IdeS and carboxypeptidase B digestion [51]. In plasma, affinity capture using specific capture agents is often employed for in vivo biotransformation analysis [35]. Previously, several subunit LC-MS studies surveyed biotransformations of Fab-conjugated ADCs using affinity capturing, but the developed technical procedures are often not applicable to Fc-conjugated ADCs [37]. More recently, an improved affinity capture LC-MS assay has been developed for analysis of biotransformation and conjugation sites in a variety of antibody types [35], which, according to the authors, could identify small-size modifications such as hydrolysis with enhanced sensitivity and resolution. The authors have applied this method to evaluate in vivo biotransformation of HC-Fc conjugated ADCs and discovered catabolites from deacetylation of conjugated tubulysin. Although LC-MS analysis of intact proteins or large protein fragments usually carries significantly lower sensitivity and limited site-specific information, this problem could be partially alleviated with the recent development with high-resolution MS. For example, one group has studied the catabolites of T-DM1 at intact level and found Orbitrap showed superior results compared with Q-TOF MS [38]. They have identified three types of biotransformation involving cysteine and glutathione adduct formation, loss of maytansinol and hydrolysis at the linker-drug sites.
It is often found that combined multi-level analysis is beneficial since it provides complementary information. For example, a study has shown that middle-level analysis combined with intact and native MS provided broader insights into the conjugation heterogeneities and DAR analysis of ADCs [36]. In the same way, intact and subunit LC-MSs are also frequently used for DAR and DLD analysis, which is to be discussed in the “DAR characterization” section.
2.1.2.2. Characterization of antibody assembly
One recent development in the field is the ADC with a bi-specific antibody. Generally, poor internalization rate has been a prominent issue for ADCs; the bi-specific ADCs emerged to enhance the internalization and trafficking to the lysosome by either targeting a fast internalizing receptor and tumor cell surface specific target using the two arms respectively, or targeting two different epitopes of the same antigen. Examples of bi-specific ADCs currently on clinical trials are MEDI4276 and ZW49 [52,53]. Given that ADCs containing bi-specific antibody represent a future trend, and that evaluating structural integrity of bi-specific antibody is important in ensuring safety and efficacy, here we review the utilization of LC-MS in assessment of the structural heterogeneity of bi-specific antibodies. Although all three components of ADCs (i.e., the antibody, payload and linker) are critical for structural integrity, this section will mainly discuss bi-specific antibody assembly, and payload conjugation will be discussed in detail in a later section [54,55]. Despite emerging strategies to promote correct chain pairing, undesirable antibody assembly persists, which must be characterized [56]. Intact-level LC-MS analysis is a promising technique in this regard, for instance, characterization of the assembly of heavy and light chains [57]. The correctly assembled antibodies can be separated from byproducts such as hole-hole dimer, hole half-antibody and hole half-antibody fragments by different chromatographic techniques including KappaSelect affinity chromatography, LambdaFabSelect affinity chromatography, hydrophobic interaction chromatography (HIC) and RPLC [58].
Additionally, the use of high-resolution MS can differentiate the correctly assembled antibodies from its byproducts by mass shift, thus achieving unambiguous identification [59]. Schachner et al. [59] described an LC-MS method using Exactive Plus Extended Mass Range Orbitrap, which had identified low levels of mis-paired anti-interleukin (IL)-4/IL-13 bispecific immunoglobulin G (IgG). Due to the different masses of anti-IL-4 light chain and anti-IL-13 light chain, the mass analyzer readily distinguishes the correctly assembled form with the 2x IL-4 L form and 2x IL-13 L form. Additionally, Woods et al. [60] utilized an advanced ESI-Q-TOF MS and intact level analysis to evaluate the heterodimeric purity of a prototype asymmetric antibody containing two different heavy chains and two identical light chains. Moreover, Gomes et al. [61] employed Q-TOF MS to characterize heterogeneity of in-house produced antibody.
However, when the two different light chains show similar properties (e.g., molecular weight and/or polarity), discrimination of swapped dimeric products from the correctly assembled ones would be difficult using intact analysis. In such events, partial digestion and analysis of subunits become helpful. For example, Wang et al. [62] incubated a bispecific antibody product with the enzyme GingisKHAN, which specifically cuts between the K and T residues above the hinge region. In the digest, the two Fabs generated from the swapped light chains were clearly different from these from the correctly assembled antibody, which can be specifically analyzed by LC-MS.
2.2. DAR characterization
In most cases, payloads are conjugated to the ε-NH2 of surface-exposed lysine residues or the sulfhydryl group of interchain cysteine residues of the antibody, or specifically conjugated to engineered cysteine resides (THIOMAB) to form an ADC with well-defined DAR [3]. For the first two types of ADCs, since over 70 lysine residues and about 8 cysteine residues are available in an antibody molecule, linkages of cytotoxins result in a heterogeneous mixture of ADCs in the final product, with a wide distribution of DAR [[63], [64], [65]]. Because various DAR species could carry different efficacy and safety, the DAR value is a critical parameter for ADCs. In practice, weighted average DAR has become a key attribute to ADCs quality control, which is routinely measured in ADC products to validate homogeneity [57,66]. Furthermore, although ADCs are designed to remain stable until internalized, non-specific deconjugation occurs after ADCs administration, resulting in altered DAR in vivo [39,40]. Therefore, monitoring DAR changes after drug administration is important in determining ADC stability and assessing therapeutic effects. While DAR measurement is typically conducted at the protein level, average DAR, especially for in vivo systems, could also be alternatively calculated by separated quantification of conjugated payloads and total antibody [57,67,68]. In this section, we only focus on DAR measurement at intact protein/subunit level, and bottom-up strategy will be discussed in the “Conjugated payload in biological sample” section.
Traditional analytical techniques available for DAR characterization include ultraviolet/visible (UV/Vis) spectroscopy, absorbance spectroscopy coupled with chromatographic techniques including hydrophobic HIC, CE, capillary isoelectric focusing (cIEF), ion exchange chromatography (IEC), and RPLC [5,40,[69], [70], [71], [72], [73], [74], [75], [76], [77]]. More recently, LC-MS and other MS-based approaches, which provide far more accurate and detailed characterization of DAR, have been devised. These include intact/subunit LC-MS, matrix-assisted laser desorption/ionization (MALDI)-TOF-MS, CE-MS, ion mobility (IM)-MS et al. [17,57]. Selection of method should be based on the considerations of conjugation chemistry, characteristics of the linker and payload, and sample matrix. Given that several publications have discussed this topic in detail [2,4,57], here the focus is on the application and future trends of LC-MS-based technologies in DAR measurement.
LC-MS is a powerful tool for DAR measurement because of its molecular-level resolution and the compatibility with various matrices [78]. Moreover, compared with the widely-used HIC-UV/Vis, LC-MS consumes significantly less sample for DAR measurement, while delivers comparable or better analytical performance than other methods [17]. That being said, several important issues are worth noting. Firstly, the composition and pH of mobile phase should be adjusted for ADCs with acid-labile linkers such as hydrazone in order to maintain linker-drug integrity during analysis [3,25]. Secondly, ESI source parameters must be carefully optimized to minimize in-source dissociation of ADCs, and therefore maintain minimal analytical artifacts and sufficient sensitivity [4,25]. A general procedure of LC-MS-based DAR measurement at intact protein/subunit level involves deconvoluting the mass spectra to a series of “zero-charge” masses, and then obtaining DAR distribution or computing average DAR by integrating and weighting the spectral peak area or peak intensities [2]. Though widely practiced, one potential concern is that different states of payload conjugation or other modifications could change MS response, resulting in biased DAR [79]. Moreover, ionization of co-eluted DAR species might interfere with each other [2]. To address these concerns, one study suggested using an orthogonal approach to validate LC-MS-obtained DAR [17].
2.2.1. In vitro DAR characterization for product quality control
As mentioned above, the majority of ADCs are either Cys- or Lys-linked. Deglycosylation is commonly performed in sample preparation to reduce spectra complexity. For Cys-conjugated ADCs, conventional RPLC-MS-based DAR analysis using intact ADCs is not suitable, since the harsh, acidic mobile phase used in conventional RPLC-MS dissociates Cys-conjugated ADCs where the heavy and light chains may not be covalently bound [2]. Native MS is preferred for intact analysis of Cys-conjugated ADC because of its non-denaturing condition [[80], [81], [82]]. Size exclusion chromatography (SEC)-native MS using non-denaturing and MS-compatible mobile phase conditions enables direct DAR measurement of intact Cys-conjugated ADCs [25,42,83]. However, SEC could not resolve each DAR species, and the differentiation among various DAR species relies on high-resolution mass analyzers. Instead, HIC can resolve DAR species but involves the use of nonvolatile salts that are not compatible with ESI-MS. Although traditionally HIC is not preferred for MS analysis, it favors Cys-conjugated ADC analysis owing to its non-denaturing feature. More recently, efforts have been directed toward online coupling of HIC with native MS [41,84,85]. Nonetheless, despite potentials of native MS in DAR measurement of Cys-conjugated ADCs, native MS requires instruments to have extended mass range, and often demands strong expertise and laborious procedure, especially for complex samples [80,86]. Alternatively, reduction with dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP) is performed followed by RPLC-MS-based DAR measurement from the dissociated light and heavy chains, and RPLC is capable of separating chains with various payloads [2]. In addition, partial-digestion with IdeS followed by a reduction step generating various ∼25 kDa fragments, along with the non-conjugated Fc/2 fragment as internal reference, improves DAR measurement accuracy and provides supplementary structural information of ADCs such as C-terminal lysine truncation, pyroglutamylation, oxidation and degradation products [43,44].
Unlike Cys-linked ADCs, Lys-linked ADCs remain intact under the denaturing conditions of RPLC-MS, where the average DAR could be directly calculated from deconvoluted spectra [2,45]. A study comparing DAR obtained by LC-ESI-MS and a reference method (UV/Vis) for a Lys-conjugated ADC product (huC242-SPDB-DM4) suggested LC-MS produced an average DAR comparable to that by UV/Vis; the study also found it is important to use a full charge envelope for deconvolution of mass spectra [46]. Nonetheless, the interpretation of the mass spectra of Lys-conjugated ADCs is often challenging due to their high heterogeneity and high charge states under denaturing conditions (thus narrowly-spaced MS peaks). Therefore, C-terminal lysine removal to reduce charge-heterogeneity during sample preparation are recommended to reduce the spectral complexity [57]. Compared with RPLC-MS, SEC-native MS shifts the charge envelope to a higher mass window, which could improve mass spectrum quality for Lys-conjugated ADCs [42].
2.2.2. In vivo DAR characterization
The in vivo dynamic change of the average DAR reflects ADC stability after drug administration. Measurement of DAR in biological samples is highly challenging owing to problems associated with sensitivity and the complex biological matrices. Towards this end, a highly efficient and selective immunoaffinity enrichment of ADCs must be performed prior to intact LC-MS-based in vivo DAR measurement [37,39,40]. Though such strategy has been adopted in plenty of work of DAR measurement in plasma, it suffers from low sensitivity rooting from the intrinsic low MS response for intact analysis. Alternatively, reduced or limitedly-digested samples can be used to enhance sensitivity for in vivo DAR determination, as shown in a number of reports [47,[87], [88], [89]]. Furthermore, the intact analysis in vivo requires a highly specific and efficient capturing reagent, which may not be available in many cases. Finally, immunoaffinity enrichment does not work well in tissues, limiting the method only applicable to plasma analysis [90]. One new approach to determining in vivo DAR is to separately quantify conjugated payload and total antibody, currently applicable to ADCs containing cleavable linkers [67,68,[91], [92], [93], [94]]. Compared with intact DAR measurement, this method has much higher sensitivity favored by peptide-level protein quantification. Examples are described in the “conjugated payload in biological sample” part.
In order to further take the advantage of the superior ability of LC-MS in DAR measurement, a number of new techniques are under development at the moment. For example, to obtain improved separation, DAR measurement using 2-dimensional LC (2D-LC) such as HIC × RPLC and HIC × SEC was described [41,76,85]. Furthermore, IM technique that provides a new, orthogonal dimension for separation of ADC samples has attracted considerable interest [41,82]. IM-MS separates gas-phase ions based on their differential mobility against a buffer gas [95]. By providing a third dimension of separation, IM-MS could help to achieve unambiguous identification of DAR species and allow more accurate DAR measurement [41].
2.3. Quantitative analysis in biological samples for pharmacokinetic investigation
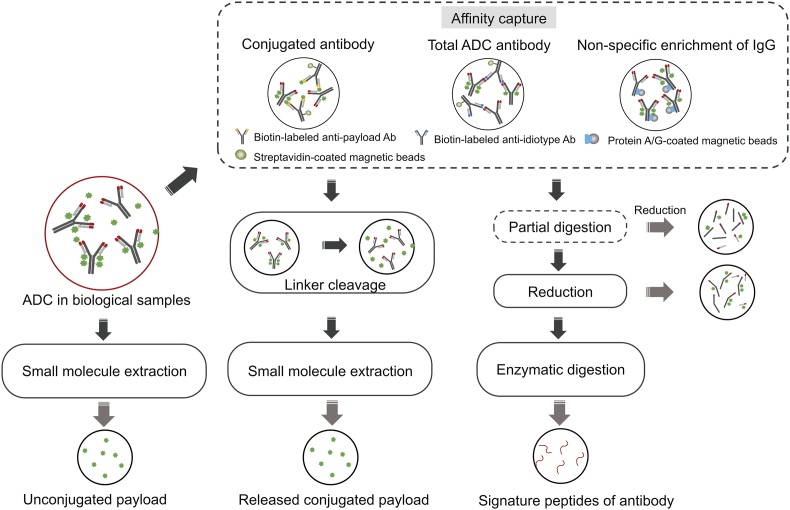
Pharmacokinetic investigation of ADC in vivo is complicated by nature. For instance, it is still under debate as to which ADC forms in plasma are the most representative of the exposure-response relationship, and consequently, efficacy and safety [96]. A position paper on ADCs bioanalysis has recommended that the following components should be measured at an early drug development stage for evaluation purpose: conjugated forms (conjugated antibody or antibody-conjugated drug), the total antibody, and unconjugated drug [66]. Correspondingly, sample preparation procedure is often quite complicated in order to achieve comprehensive ADC bioanalysis and fit-for-purpose protocols should be developed based on the desired target analytes as well as nature of the sample. The general sample preparation strategies for LC-MS-based ADC bioanalysis are summarized in Fig. 1. For LC-MS based quantification methods, peptide-level quantification of the antibody part is at the main stage, while intact ADC quantification is picking up albeit slowly. Here both the peptide- and intact-level quantification methods are reviewed. Analysis of unconjugated drugs and metabolites entities is discussed in a latter section.
Fig. 1.
Summarized schematic for the general sample preparation procedure for LC-MS-based ADC bioanalysis.
2.3.1. Protein quantification using peptide-level (bottom-up) approach
2.3.1.1. Total antibody
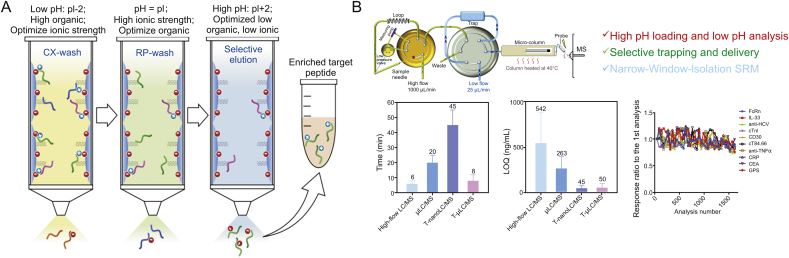
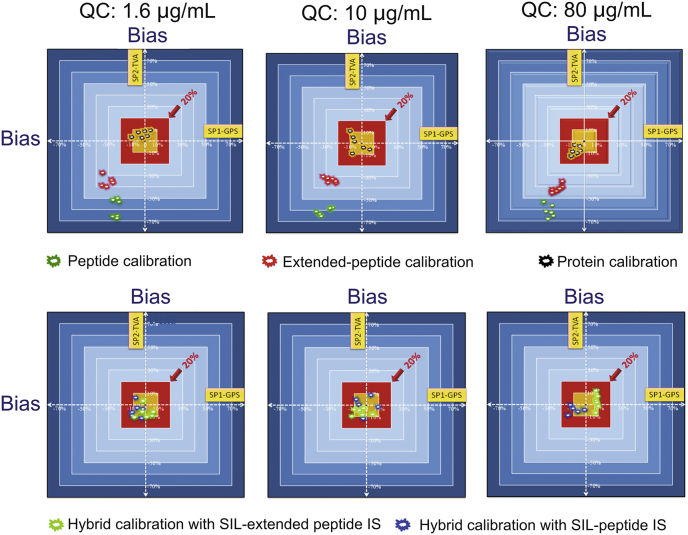
After drug administration, due to the change of DAR in vivo, the calibration curve prepared using the drug as the reference may not exactly represent the analytes in the study samples over the time course of PK measurement. Total antibody quantification targets all antibody forms regardless of the presence of conjugated drug, which is considered to be the most useful in characterization of antibody-related PK behavior of the ADCs [2,97]. Quantitative strategy for total antibody of ADCs is usually the same as these for general antibodies, where bottom-up LC-MS strategy is a widely used quantitative method [78,93,[97], [98], [99]]. Many factors such as LC conditions, sample preparation and charge states all affect the sensitivity for protein quantification [100]. To achieve desired sensitivity, affinity capture is typically incorporated. For instance, antibodies are isolated from the matrix by capture reagents such as anti-idiotype (anti-ID) antibody or protein A/G [94]. The isolated antibodies are denatured/reduced, alkylated and then proteolyzed. Based on several generally-accepted criteria, usually signature peptides (SP) from complementarity-determining region (CDR) will be selected and monitored by LC-MS [12,50,93,101]. One issue with the anti-ID antibody enrichment is that only the ADC species with at least one unbound variable region could be captured by anti-ID antibody. Therefore, binding to soluble target compromises anti-ID antibody capture efficiency [94]. Furthermore, protein-level enrichment only works well in plasma but not tissues [90]. Use of protein G/A also has further limitation because of their indiscriminately binding of endogenous IgG. One alternative is to develop a sensitive LC-MS strategy without protein-level enrichment. For instance, we described an immuno-enrichment-free procedure affording highly sensitive LC-MS-based quantification of antibodies directly from plasma or tissue homogenates, which includes the following technical advances: i) A high-throughput on-the-fly orthogonal array optimization (OAO) strategy [6,102,103], which utilizes a systematic experimental design to develop the optimal LC/SRM-MS conditions for multiple SP candidates in matrix, enabling experimental identification of the optimal SP in a high-throughput and accurate manner. Moreover, synthesis of potential SP candidates is not necessary for the SP selection in this method, which is time/cost effective. ii) A surfactant-aided precipitation/on-pellet-digestion (SOD) sample preparation procedure. The method enables high and reproducible peptide recovery regardless of the matrix (e.g., tissues or plasma), thus achieving accurate and sensitive antibody quantification with good robustness [104,105]. Surfactant treatment followed by precipitation achieves high and reproducible protein/peptide recovery from various matrices, because surfactant allows not only a high protein extraction efficiency but also extensive denaturation of proteins, rendering them more accessible to digestive enzymes. Moreover, it was found that surfactant greatly facilitates the removal of detrimental matrix components such as lipids and fatty acids [106]. iii) An antibody-free multiple-mechanism peptide-level enrichment via strategic regulation of pH, and ionic and solvent strengths (Fig. 2A). The retention of a target peptide on SPE cartridge via cation exchange (CX) and reversed phase (RP) mechanisms is relied on ionic and hydrophobic interactions, respectively; both are profoundly regulated by pH. Thus a highly specific method was developed to enrich SP by a series of selective wash and elution steps using buffers with strategically controlled pH, ionic strength and organic solvent composition. This method eliminates a majority of non-target peptides and matrix components, improving sensitivity and robustness significantly [107]. Moreover, unlike protein-level enrichment methods, this peptide-level strategy works well in tissues. iv) A trapping-micro-LC-MS method (Fig. 2B) [108]. The system enables selective trapping and delivery of the SP while specifically removing matrix peptides to a large extent. Meanwhile, the target peak is concentrated prior to the micro-flow LC/MS, coupled with narrow window isolation (NWI)-SRM, which further boosts the sensitivity. This method achieves high sensitivity comparable to nano-LC/MS while maintaining the comparable throughput to high-flow-LC/MS and excellent robustness. v) Hybrid calibration strategy with full-length protein calibrator and stable isotope labeled (SIL) peptide or extended peptide internal standard (IS) to enable highly accurate quantification of antibodies in a cost-effective manner (Fig. 3) [7]. It is worth noting that severe negative bias is almost inevitable when synthesized peptides or extended peptides are used as the calibrator [7].
Fig. 2.
Examples for non-immunoaffinity methods to improving sensitivity for LC-MS-based quantification of antibodies from plasma or tissue homogenates. The detailed procedures can be found in the corresponding publications. (A) General workflow of a selective antibody free, peptide-level CX-RP enrichment, to improve sensitivity for LC-MS quantification. Adapted with permission from Ref. [107]. Copyright (2020) American Chemical Society. (B) A trapping-micro-LC-MS workflow for quantitative analysis of mAb with high sensitivity, exceptional robustness and high throughput. Reprinted with permission from Ref. [108]. Copyright (2020) American Chemical Society.
Fig. 3.
Two-dimensional representations of the quantitative accuracy by peptide-, extended-peptide-, and protein-level calibration approaches and the “hybrid” calibration approaches, indicating the profound effects of calibration approaches on the accuracy for LC−MS targeted quantification of therapeutic protein. Reprinted with permission from Ref. [7]. Copyright (2020) American Chemical Society.
2.3.1.2. Conjugated antibody
Conjugated antibody refers to antibody forms with at least one conjugated payload. Though the correlation between levels of conjugated antibody and efficacy or toxicity of an ADC has not yet been fully established, considerable interests have been directed to measurement of the conjugated antibody, which is recognized as one of the important active species of ADCs. For conjugated antibody quantification, ligand-binding assay (LBA) using an anti-payload antibody is widely practiced, while a ‘hybrid’ assay combining immunoaffinity enrichment and LC-MS analysis also serves as an alternative [64,93,94]. In such hybrid assays format, the sample enriched by immunocapture with anti-payload antibody is digested and then the antibody is quantified with bottom-up LC-MS approach [94]. It should be noticed that peptides-linker-drug moieties might be generated after tryptic digestion of ADCs [2]. These species are often highly heterogenous in the digest, which should not be selected as the signature peptide for antibody quantification, especially for these ADCs with non-cleavable linkers. Another important issue worth noting is that during the early drug development stage, anti-payload reagents may not be available, which poses a challenge for both LBA and LC-MS-based hybrid assays format.
2.3.2. Protein quantification at intact level
As discussed earlier, advancement of high-resolution MS instruments greatly facilitated LC-MS based intact quantification [109]. Compared with bottom-up quantification, intact quantification, if properly carried out, can preserve the whole protein information instead of using an SP which could only partially represent the original target. For ADC quantification, LC-MS-based intact quantification is able to provide DAR information.
For intact quantification, major challenges remain such as the low sensitivity, requirement of highly specific and effective capturing reagents, as well as a high-resolution MS suitable for large, intact protein analysis (e.g., proper pressure and ion optic settings). Moreover, the spectra are usually hard to interpret in a quantitative manner, owing to the many charge states and isotopes forms, which further compounds the sensitivity problem [48]. The large molecule weight of antibody often results in low ionization efficacy and therefore low sensitivity for intact analysis. ADCs exhibit even lower sensitivity than that of a naked antibody because of the signal distribution into different DAR species. Therefore, the success heavily relies on the immunocapture process, which must provide a high recovery and effective removal of interference from biological matrix. For accurate quantification, internal standard (IS) is often indispensable [110]. However, not only the antibody IS is costly, other issues also exist. For example, one group has tried intact quantification on T-DM1, where the IS co-eluted with the target caused a much more complex spectra that was hard to deconvolute [48]. As a comparison, the authors further applied narrow-window XIC extraction and deconvolution method for quantification without IS and achieved acceptable quantitative performance within the concentration ranging from 5 to 100 μg/mL. Consequently, though intact-level quantification has been applied for antibodies, quantification of intact ADCs is still in its infancy, rooting from various technical challenges.
3. Analysis of payload
Payload related analytes include residual payloads and associated compounds in the drug product, as well as unconjugated payloads, conjugated payloads, and payload-related metabolites in biological samples after drug dosing [5,12,111]. Because of its unique advantages in specificity and sensitivity, LC-MS plays a pivotal role in payload analysis. For example, the high-sensitivity feature of LC-MS, which enables quantification of unconjugated payload that presents at a very low concentration in vivo making it highly valuable [112]. Also, LC coupled to high-resolution MS is commonly used in identification of payload-related metabolites.
3.1. Unconjugated payload
During ADC production, incomplete removal of unconjugated payload or payload-linker may pose a risk for toxicity due to the extreme potency of the payload toxin [112]. Therefore, measurement of residual unconjugated payload and related compounds is designated as a CQA for ADC products, which must be routinely monitored [113]. In an in vivo system, unconjugated payloads refer to the payload forms deconjugated in plasma or target tissues post-dosing [12]. Plasma level of unconjugated payload closely correlates with off-target toxicities [47]. Intra-tissue distribution of unconjugated payload is also of great importance in understanding efficacy and toxicity of ADCs [114]. In this regard, mass spectrometry imaging (MSI) has been proved to be valuable by visualizing spatial distribution of unconjugated payload [115].
The forms of unconjugated payload from cleavable and non-cleavable linkers are often different owing to the disparate release mechanisms. Specifically, deconjugation of an ADC with cleavable linkers releases the free cytotoxin, while an ADC with non-cleavable linker mainly produces more complicated formats such as amino acid-linker-payload moieties after near-complete degradation of the antibody [3]. It should be noted that catabolism of ADCs with non-cleavable linkers might produce free cytotoxins as well, which is also characterized in pharmacokinetics studies [116].
ADCs with cleavable linkers primarily release free cytotoxin; consequently, LC-MS-based analysis of unconjugated payload from these agents usually employs the same strategy as that for quantification of the cytotoxin. Due to the high hydrophobicity of typical cytotoxins, liquid-liquid extraction (LLE) and solid-phase extraction (SPE) are often carried out for extraction. Additionally, deconjugation of payload should be minimized during sample preparation to avoid positive bias. For example, to avoid deconjugation, the pH should be adjusted for acid-labile linkers such as hydrazone linker; adding protease inhibitors is preferred considering various proteases present in the sample may cleave enzyme-cleavable linkers, and sample extraction in an ice-water bath is recommend to minimize deconjugation [12,117]. A specific consideration for maytansinoid payloads is that the reactive thiol groups could undergo disulfide exchange with other thiol-containing molecules in the matrix, e.g., forming dimers after release [38,117,118]. Therefore, reduction and derivation of thiol are usually necessary before LC-MS analysis of this type of payloads.
For ADCs with non-cleavable linkers, several payload-containing forms could be produced, which should be quantified together. For example, in the PK and toxicokinetic (TK) studies of Kadcyla (ado-trastuzumab-mcc-emtansine), DM1, Lys-mcc-DM1 and mcc-DM1 are monitored [116,119]. LC-MS analysis of those species could be achieved in one run with satisfying resolution using a typical reversed-phase chromatography [118].
3.2. Conjugated payload in biological sample
The level of conjugated payload is generally considered as a valuable indicator related to ADC efficacy and toxicity [119]. As mentioned previously, LC-MS based quantitative analysis of conjugated ADCs is usually coupled with immunoaffinity pull-down [57,68,91,97]. For ADCs with cleavable linkers, conjugated payload is quantified after isolation of small molecules from proteins and then linker cleavage. For example, for the ADC utilizing an enzyme-sensitive dipeptide Val-Cit linker (e.g., brentuximab vedotin), the release of payload can be achieved by digestion with proteases such as cathepsin B and papain [67,68,93,94,[120], [121], [122]]. For the ADC with a disulfide bond linker (e.g., coltuximab ravtansin), reducing agent such as DTT and TCEP is employed for cleavage [123,124]. Conventionally, conjugated antibody and conjugated payload are measured by two independent assays using two aliquots of the same samples. Xu et al. [125] introduced an LC-MS approach enabling simultaneous measurement of total antibody and antibody-conjugated drug in plasma samples, followed by immunocapture enrichment. The strategy utilizes sequentially enzymatic digestion with cathepsin B and then Lys-C to release conjugated payload and then signature peptide of the antibody component. The assay platform was further applied in another ADC containing a polymer linker via an ester bond, which was cleaved by sodium hydroxide [99].
As mentioned previously, quantification of conjugated payloads of ADCs with cleavable linkers can be achieved after linker cleavage, which is an alternative method to intact LC-MS-based DAR measurement in biological samples [67,68,91,93,94]. The average DAR is calculated as the molar ratio of conjugated payload vs. total antibody, and the change of average DAR could indicate ADC deconjugation in vivo [94]. However, a highly specific immunoaffinity enrichment is commonly required to isolate free payload with conjugated ADCs, which not only is feasible in many projects, but also impedes conjugated payload quantification in tissue samples. An alternative and simpler method is using protein precipitation followed by on-pellet linker cleavage [91].
By comparison, given the inherent feature of non-cleavable linker, payloads conjugated by non-cleavable linkers are often indirectly determined by multiplying in vivo average DAR measure using intact LC-MS with total antibody concentration [57]. That being said, direct quantification of small-molecule forms of payloads conjugated with non-cleavable linkers has recently been explored, where the target analytes are payload-linker-amino-acids or payload-linker-peptides after extensive or site-specific digestion [93]. Such attempts are currently limited to ADCs with site-specific conjugation. Hyung et al. [126] developed an LC-MS method for quantification of conjugated payload of an engineered, cysteine-conjugated ADC with non-cleavable linkers in plasma sample. After a rough enrichment using protein A, the ADC was subjected to tryptic digestion, which produced a unique peptide-linker-payload moiety for quantification. In another case, an ADC that contained maytansinoid tubulin inhibitor DM1 conjugated to engineered cysteine residues through a tri-glycine-containing peptide linker (CX1) was investigated [127]. A tryptic peptide containing cystine-linker-payload was selected as the surrogate for quantification of the conjugated payload.
Apart from investigations on unconjugated and conjugated payloads, it appears that protein-payload adduct has attracted increasing attention, especially for cysteine-maleimide-based ADCs (e.g., brentuximab vedotin, T-DM1) which might undergo thiol-exchange reactions with matrix proteins [128]. The protein-payload adduct could originate from thiol-exchange reactions between matrix proteins and unconjugated payloads or conjugated payloads of maleimide-linker-containing ADCs. Characterization of these products provides important information on plasma stability of the ADC product [98].
4. Future perspective
The past decade has witnessed growing interest and accelerating development in ADCs. Although these agents have demonstrated improved clinical outcomes, the relationship between structural features of ADCs and clinical efficacy/toxicity is still poorly understood owing to the complicated nature of this therapeutic system. A comprehensive, integrated characterization of ADCs and the related pharmaceutical system is highly valuable in evaluating efficacy/safety of these agents as well as in directing both therapeutic and engineering efforts, which requires reliable analytical approaches to answering quantitative and qualitative questions from a wide range of aspects.
Among the techniques applicable for ADCs analysis, LC-MS emerges as a highly valuable and versatile tool. Over the past few years, a growing number of LC-MS strategies at protein-, subunit-, peptide-, and payload levels have been developed, which permitted a significantly improved understanding of the molecular characteristics and pharmacokinetic/pharmacodynamic of ADCs, and provided novel insights into the complicated albeit interesting therapeutic system. Additionally, these new analytical methods have discovered novel information that profoundly affects the efficacy and safety of ADCs, for example, the identification and quantification of albumin-adduct formation which accounts for a new mechanism for DAR loss in maleimide-containing ADCs [120,128,129].
Despite these tremendous technical advancements, challenges remain. To name a few: i) Analysis of ADCs in tissues is highly critical to understanding drug effects, but is still difficult; ii) lack of an optimal method to analyze conjugated payload for ADCs with non-cleavable linkers; iii) problems associated with analysis of various products of biotransformation and catabolism; and iv) suboptimal robustness and accuracy are often an intractable problem. Addressing these challenges would greatly accelerate drug discovery and development, and facilitate clinical efforts of ADCs. Consequently, we anticipate that in the near future, intense efforts will be directed toward the development of new LC-MS-based analytical strategies in order to meet these challenges. Conceivably, such efforts will also be markedly fueled by the ever-increasing requirements of defining new parameters of ADCs (e.g., average DAR in vivo, charge heterogeneity, and positional isomers) and the evolution of new ADC modalities (e.g., ADC for non-oncology indications, antibody-dual-drug conjugates, and biparatopic ADC) [4,12,94,130]. Finally, given the rapid advancement of LC-MS techniques, LC-MS will continue to improve as the most powerful tool for ADC analysis.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Wikipedia, Antibody-drug conjugate. 2020. https://en.wikipedia.org/wiki/Antibody-drug_conjugate 202011.
- 2.Ducry L., editor. Antibody-drug Conjugates. Humana Press; New York: 2013. [Google Scholar]
- 3.Jain N., Smith S.W., Ghone S. Current ADC linker chemistry. Pharm. Res. (N. Y.) 2015;32:3526–3540. doi: 10.1007/s11095-015-1657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagh A., Song H., Zeng M. Challenges and new frontiers in analytical characterization of antibody-drug conjugates. mAbs. 2018;10:222–243. doi: 10.1080/19420862.2017.1412025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakankar A., Chen Y., Gokarn Y. Analytical methods for physicochemical characterization of antibody drug conjugates. mAbs. 2011;3:161–172. doi: 10.4161/mabs.3.2.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan X., Abuqayyas L., Dai L. High-throughput method development for sensitive, accurate, and reproducible quantification of therapeutic monoclonal antibodies in tissues using orthogonal array optimization and nano liquid chromatography/selected reaction monitoring mass spectrometry. Anal. Chem. 2012;84:4373–4382. doi: 10.1021/ac2034166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nouri-Nigjeh E., Zhang M., Ji T. Effects of calibration approaches on the accuracy for LC-MS targeted quantification of therapeutic protein. Anal. Chem. 2014;86:3575–3584. doi: 10.1021/ac5001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donaghy H. Effects of antibody, drug and linker on the preclinical and clinical toxicities of antibody-drug conjugates. mAbs. 2016;8:659–671. doi: 10.1080/19420862.2016.1156829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahalingaiah P.K., Ciurlionis R., Durbin K.R. Potential mechanisms of target-independent uptake and toxicity of antibody-drug conjugates. Pharmacol. Ther. 2019;200:110–125. doi: 10.1016/j.pharmthera.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Mandler R., Kobayashi H., Hinson E.R. Herceptin-geldanamycin immunoconjugates. Canc. Res. 2004;64:1460. doi: 10.1158/0008-5472.can-03-2485. [DOI] [PubMed] [Google Scholar]
- 11.Boswell C.A., Mundo E.E., Zhang C. Impact of drug conjugation on pharmacokinetics and tissue distribution of anti-STEAP1 antibody-drug conjugates in rats. Bioconjugate Chem. 2011;22:1994–2004. doi: 10.1021/bc200212a. [DOI] [PubMed] [Google Scholar]
- 12.Wei C., Su D., Wang J. LC–MS challenges in characterizing and quantifying monoclonal antibodies (mAb) and antibody-drug conjugates (ADC) in biological samples. Curr. Pharmacol. Rep. 2018;4:45–63. [Google Scholar]
- 13.Beck A., Wagner-Rousset E., Ayoub D. Characterization of therapeutic antibodies and related products. Anal. Chem. 2013;85:715–736. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- 14.Sandra K., Vandenheede I., Sandra P. Modern chromatographic and mass spectrometric techniques for protein biopharmaceutical characterization. J. Chromatogr. A. 2014;1335:81–103. doi: 10.1016/j.chroma.2013.11.057. [DOI] [PubMed] [Google Scholar]
- 15.Bergquist J., Palmblad M., Wetterhall M. Peptide mapping of proteins in human body fluids using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mass Spectrom. Rev. 2002;21:2–15. doi: 10.1002/mas.10016. [DOI] [PubMed] [Google Scholar]
- 16.Bobaly B., Fleury-Souverain S., Beck A. Current possibilities of liquid chromatography for the characterization of antibody-drug conjugates. J. Pharmaceut. Biomed. Anal. 2018;147:493–505. doi: 10.1016/j.jpba.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Kallsten M., Hartmann R., Artemenko K. Qualitative analysis of antibody-drug conjugates (ADCs): an experimental comparison of analytical techniques of cysteine-linked ADCs. Analyst. 2018;143:5487–5496. doi: 10.1039/c8an01178h. [DOI] [PubMed] [Google Scholar]
- 18.Mouchahoir T., Schiel J.E. Development of an LC-MS/MS peptide mapping protocol for the NISTmAb. Anal. Bioanal. Chem. 2018;410:2111–2126. doi: 10.1007/s00216-018-0848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Wang L., Shion H. In-depth structural characterization of Kadcyla(R) (ado-trastuzumab emtansine) and its biosimilar candidate. mAbs. 2016;8:1210–1223. doi: 10.1080/19420862.2016.1204502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arlotta K.J., Gandhi A.V., Chen H.N. In-depth comparison of lysine-based antibody-drug conjugates prepared on solid support versus in solution. Antibodies. 2018;7:6. doi: 10.3390/antib7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sang H., Lu G., Liu Y. Conjugation site analysis of antibody-drug-conjugates (ADCs) by signature ion fingerprinting and normalized area quantitation approach using nano-liquid chromatography coupled to high resolution mass spectrometry. Anal. Chim. Acta. 2017;955:67–78. doi: 10.1016/j.aca.2016.11.073. [DOI] [PubMed] [Google Scholar]
- 22.Wu G., Gao Y., Liu D. Study on the heterogeneity of T-DM1 and the analysis of the unconjugated linker structure under a stable conjugation process. ACS Omega. 2019;4:8834–8845. doi: 10.1021/acsomega.9b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janin-Bussat M.C., Dillenbourg M., Corvaia N. Characterization of antibody drug conjugate positional isomers at cysteine residues by peptide mapping LC-MS analysis. J. Chromatogr. B. 2015;981:9–13. doi: 10.1016/j.jchromb.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez-Alba O., Houel S., Hessmann S. A case study to identify the drug conjugation site of a site-specific antibody-drug-conjugate using middle-down mass spectrometry. J. Am. Soc. Mass Spectrom. 2019;30:2419–2429. doi: 10.1007/s13361-019-02296-2. [DOI] [PubMed] [Google Scholar]
- 25.Friese O.V., Smith J.N., Brown P.W. Practical approaches for overcoming challenges in heightened characterization of antibody-drug conjugates with new methodologies and ultrahigh-resolution mass spectrometry. mAbs. 2018;10:335–345. doi: 10.1080/19420862.2018.1433973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dada O.O., Zhao Y., Jaya N. High-resolution capillary zone electrophoresis with mass spectrometry peptide mapping of therapeutic proteins: peptide recovery and post-translational modification analysis in monoclonal antibodies and antibody-drug conjugates. Anal. Chem. 2017;89:11236–11242. doi: 10.1021/acs.analchem.7b03643. [DOI] [PubMed] [Google Scholar]
- 27.Sandra K., Vandenbussche J., Vandenheede I. Peptide mapping of monoclonal antibodies and antibody-drug conjugates using micro-pillar array columns combined with mass spectrometry. LC-GC Eur. 2018;31:155–166. [Google Scholar]
- 28.Wang Y., Li X., Liu Y.H. Simultaneous monitoring of oxidation, deamidation, isomerization, and glycosylation of monoclonal antibodies by liquid chromatography-mass spectrometry method with ultrafast tryptic digestion. mAbs. 2016;8:1477–1486. doi: 10.1080/19420862.2016.1226715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen M.R., Trelle M.B., Thingholm T.E. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques. 2006;40:790–798. doi: 10.2144/000112201. [DOI] [PubMed] [Google Scholar]
- 30.Jensen O.N. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Dick L.W., Jr., Mahon D., Qiu D. Peptide mapping of therapeutic monoclonal antibodies: improvements for increased speed and fewer artifacts. J. Chromatogr. B. 2009;877:230–236. doi: 10.1016/j.jchromb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Ren D., Pipes G.D., Liu D. An improved trypsin digestion method minimizes digestion-induced modifications on proteins. Anal. Biochem. 2009;392:12–21. doi: 10.1016/j.ab.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Donnelly D.P., Rawlins C.M., DeHart C.J. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat. Methods. 2019;16:587–594. doi: 10.1038/s41592-019-0457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilpatrick L.E., Kilpatrick E.L. Optimizing high-resolution mass spectrometry for the identification of low-abundance post-translational modifications of intact proteins. J. Proteome Res. 2017;16:3255–3265. doi: 10.1021/acs.jproteome.7b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotapati S., Passmore D., Yamazoe S. Universal affinity capture liquid chromatography-mass spectrometry assay for evaluation of biotransformation of site-specific antibody drug conjugates in preclinical studies. Anal. Chem. 2020;92:2065–2073. doi: 10.1021/acs.analchem.9b04572. [DOI] [PubMed] [Google Scholar]
- 36.Botzanowski T., Erb S., Hernandez-Alba O. Insights from native mass spectrometry approaches for top- and middle- level characterization of site-specific antibody-drug conjugates. mAbs. 2017;9:801–811. doi: 10.1080/19420862.2017.1316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su D., Ng C., Khosraviani M. Custom-designed affinity capture LC-MS F(ab’)2 assay for biotransformation assessment of site-specific antibody drug conjugates. Anal. Chem. 2016;88:11340–11346. doi: 10.1021/acs.analchem.6b03410. [DOI] [PubMed] [Google Scholar]
- 38.He J., Yu S.F., Yee S. Characterization of in vivo biotransformations for trastuzumab emtansine by high-resolution accurate-mass mass spectrometry. mAbs. 2018;10:960–967. doi: 10.1080/19420862.2018.1494487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu K., Liu L., Dere R. Characterization of the drug-to-antibody ratio distribution for antibody–drug conjugates in plasma/serum. Bioanalysis. 2013;5:1057–1071. doi: 10.4155/bio.13.66. [DOI] [PubMed] [Google Scholar]
- 40.Xu K., Liu L., Saad O.M. Characterization of intact antibody-drug conjugates from plasma/serum in vivo by affinity capture capillary liquid chromatography-mass spectrometry. Anal. Biochem. 2011;412:56–66. doi: 10.1016/j.ab.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Ehkirch A., D’Atri V., Rouviere F. An online four-dimensional HICxSEC-IMxMS methodology for proof-of-concept characterization of antibody drug conjugates. Anal. Chem. 2018;90:1578–1586. doi: 10.1021/acs.analchem.7b02110. [DOI] [PubMed] [Google Scholar]
- 42.Henry Shion Y.Q.Y., Chen Weibin. 2018. Analytical Scale Native SEC-MS for Antibody-Drug Conjugates (ADCs) Characterization, Waters Application Note. [Google Scholar]
- 43.Wagner-Rousset E., Janin-Bussat M.C., Colas O. Antibody-drug conjugate model fast characterization by LC-MS following IdeS proteolytic digestion. mAbs. 2014;6:173–184. doi: 10.4161/mabs.26773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Firth D., Bell L., Squires M. A rapid approach for characterization of thiol-conjugated antibody-drug conjugates and calculation of drug-antibody ratio by liquid chromatography mass spectrometry. Anal. Biochem. 2015;485:34–42. doi: 10.1016/j.ab.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y., Tang F., Yang Y. Real-time analysis on drug-antibody ratio of antibody-drug conjugates for synthesis, process optimization, and quality control. Sci. Rep. 2017;7:7763. doi: 10.1038/s41598-017-08151-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazar A.C., Wang L., Blattler W.A. Analysis of the composition of immunoconjugates using size-exclusion chromatography coupled to mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:1806–1814. doi: 10.1002/rcm.1987. [DOI] [PubMed] [Google Scholar]
- 47.Grafmuller L., Wei C., Ramanathan R. Unconjugated payload quantification and DAR characterization of antibody–drug conjugates using high-resolution MS. Bioanalysis. 2016;8:1663–1678. doi: 10.4155/bio-2016-0120. [DOI] [PubMed] [Google Scholar]
- 48.Jin W., Burton L., Moore I. LC-HRMS quantitation of intact antibody drug conjugate trastuzumab emtansine from rat plasma. Bioanalysis. 2018;10:851–862. doi: 10.4155/bio-2018-0003. [DOI] [PubMed] [Google Scholar]
- 49.Kellie J.F., Karlinsey M.Z. Review of approaches and examples for monitoring biotransformation in protein and peptide therapeutics by MS. Bioanalysis. 2018;10:1877–1890. doi: 10.4155/bio-2018-0113. [DOI] [PubMed] [Google Scholar]
- 50.Kang L., Weng N., Jian W. LC-MS bioanalysis of intact proteins and peptides. Biomed. Chromatogr. 2019;34 doi: 10.1002/bmc.4633. [DOI] [PubMed] [Google Scholar]
- 51.Beck A., Terral G., Debaene F. Cutting-edge mass spectrometry methods for the multi-level structural characterization of antibody-drug conjugates. Expert Rev. Proteomics. 2016;13:157–183. doi: 10.1586/14789450.2016.1132167. [DOI] [PubMed] [Google Scholar]
- 52.Damelin M., editor. Innovations for Next-Generation Antibody-Drug Conjugates. Humana Press; New York: 2018. [Google Scholar]
- 53.Hamblett K.J., Barnscher S.D., Davies R.H. Abstract P6-17-13: ZW49, a HER2 targeted biparatopic antibody drug conjugate for the treatment of HER2 expressing cancers. Canc. Res. 2019;79 17-13. [Google Scholar]
- 54.Chen T., Chen Y., Stella C. Antibody-drug conjugate characterization by chromatographic and electrophoretic techniques. J. Chromatogr. B. 2016;1032:39–50. doi: 10.1016/j.jchromb.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 55.Sun M.M.C., Beam K.S., Cerveny C.G. Reduction− alkylation strategies for the modification of specific monoclonal antibody disulfides. Bioconjugate Chem. 2005;16:1282–1290. doi: 10.1021/bc050201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang J., Zhang X., Chen T. Removal of half antibody, hole-hole homodimer and aggregates during bispecific antibody purification using MMC ImpRes mixed-mode chromatography. Protein Expr. Purif. 2020;167:105529. doi: 10.1016/j.pep.2019.105529. [DOI] [PubMed] [Google Scholar]
- 57.Huang R.Y., Chen G. Characterization of antibody-drug conjugates by mass spectrometry: advances and future trends. Drug Discov. Today. 2016;21:850–855. doi: 10.1016/j.drudis.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Wang C., Vemulapalli B., Cao M. A systematic approach for analysis and characterization of mispairing in bispecific antibodies with asymmetric architecture. mAbs. 2018;10:1226–1235. doi: 10.1080/19420862.2018.1511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schachner L., Han G., Dillon M. Characterization of chain pairing variants of bispecific IgG expressed in a single host cell by high-resolution native and denaturing mass spectrometry. Anal. Chem. 2016;88:12122–12127. doi: 10.1021/acs.analchem.6b02866. [DOI] [PubMed] [Google Scholar]
- 60.Woods R.J., Xie M.H., Von Kreudenstein T.S. LC-MS characterization and purity assessment of a prototype bispecific antibody. mAbs. 2013;5:711–722. doi: 10.4161/mabs.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomes R.A., Almeida C., Correia C. Exploring the analytical power of the QTOF MS platform to assess monoclonal antibodies quality attributes. PloS One. 2019;14 doi: 10.1371/journal.pone.0219156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L., Amphlett G., Blattler W.A. Structural characterization of the maytansinoid-monoclonal antibody immunoconjugate, huN901-DM1, by mass spectrometry. Protein Sci. 2005;14 doi: 10.1110/ps.051478705. 2436-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panowski S., Bhakta S., Raab H. Site-specific antibody drug conjugates for cancer therapy. mAbs. 2014;6:34–45. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dere R., Yi J.-H., Lei C. PK assays for antibody–drug conjugates: case study with ado-trastuzumab emtansine. Bioanalysis. 2013;5:1025–1040. doi: 10.4155/bio.13.72. [DOI] [PubMed] [Google Scholar]
- 65.Tsuchikama K., An Z. Antibody-drug conjugates: recent advances in conjugation and linker chemistries. Protein Cell. 2018;9:33–46. doi: 10.1007/s13238-016-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorovits B., Alley S.C., Bilic S. Bioanalysis of antibody–drug conjugates: american association of pharmaceutical scientists antibody–drug conjugate working group position paper. Bioanalysis. 2013;5:997–1006. doi: 10.4155/bio.13.38. [DOI] [PubMed] [Google Scholar]
- 67.Sanderson R.J., Hering M.A., James S.F. In vivo drug-linker stability of an anti-CD30 dipeptide-linked auristatin immunoconjugate. Clin. Canc. Res. 2005;11:843–852. [PubMed] [Google Scholar]
- 68.Sanderson R.J., Nicholas N.D., Baker Lee C. Antibody-conjugated drug assay for protease-cleavable antibody–drug conjugates. Bioanalysis. 2016;8:55–63. doi: 10.4155/bio.15.230. [DOI] [PubMed] [Google Scholar]
- 69.Alley S.C., Anderson K.E. Analytical and bioanalytical technologies for characterizing antibody-drug conjugates. Curr. Opin. Chem. Biol. 2013;17:406–411. doi: 10.1016/j.cbpa.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 70.Khawli L.A., Goswami S., Hutchinson R. Charge variants in IgG1: isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs. 2010;2:613–624. doi: 10.4161/mabs.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fekete S., Beck A., Veuthey J.-L. Ion-exchange chromatography for the characterization of biopharmaceuticals. J. Pharmaceut. Biomed. Anal. 2015;113:43–55. doi: 10.1016/j.jpba.2015.02.037. [DOI] [PubMed] [Google Scholar]
- 72.Harrington S.J., Varro R., Li T.M. High-performance capillary electrophoresis as a fast inprocess control method for enzyme-labelled monoclonal antibody conjugates. J. Chromatogr. A. 1991;559:385–390. [Google Scholar]
- 73.Krull I.S., Liu X., Dai J. HPCE methods for the identification and quantitation of antibodies, their conjugates and complexes. J. Pharmaceut. Biomed. Anal. 1997;16:377–393. doi: 10.1016/s0731-7085(97)00071-x. [DOI] [PubMed] [Google Scholar]
- 74.Michels D.A., Salas-Solano O., Felten C. Imaged capillary isoelectric focusing for charge-variant analysis of biopharmaceuticals. BioProcess. Int. 2011;9:48–54. [Google Scholar]
- 75.Hamblett K.J., Senter P.D., Chace D.F. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin. Canc. Res. 2004;10:7063–7070. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 76.Birdsall R.E., Shion H., Kotch F.W. A rapid on-line method for mass spectrometric confirmation of a cysteine-conjugated antibody-drug-conjugate structure using multidimensional chromatography. mAbs. 2015;7:1036–1044. doi: 10.1080/19420862.2015.1083665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Redman E.A., Mellors J.S., Starkey J.A. Characterization of intact antibody drug conjugate variants using microfluidic capillary electrophoresis-mass spectrometry. Anal. Chem. 2016;88:2220–2226. doi: 10.1021/acs.analchem.5b03866. [DOI] [PubMed] [Google Scholar]
- 78.Qu M., An B., Shen S. Qualitative and quantitative characterization of protein biotherapeutics with liquid chromatography mass spectrometry. Mass Spectrom. Rev. 2017;36:734–754. doi: 10.1002/mas.21500. [DOI] [PubMed] [Google Scholar]
- 79.Krusemark C.J., Frey B.L., Belshaw P.J. Modifying the charge state distribution of proteins in electrospray ionization mass spectrometry by chemical derivatization. J. Am. Soc. Mass Spectrom. 2009;20:1617–1625. doi: 10.1016/j.jasms.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rogstad S., Faustino A., Ruth A. A retrospective evaluation of the use of mass spectrometry in FDA biologics license applications. J. Am. Soc. Mass Spectrom. 2016;28:786–794. doi: 10.1007/s13361-016-1531-9. [DOI] [PubMed] [Google Scholar]
- 81.Valliere-Douglass J.F., McFee W.A., Salas-Solano O. Native intact mass determination of antibodies conjugated with monomethyl Auristatin E and F at interchain cysteine residues. Anal. Chem. 2012;84:2843–2849. doi: 10.1021/ac203346c. [DOI] [PubMed] [Google Scholar]
- 82.Debaene F., Boeuf A., Wagner-Rousset E. Innovative native MS methodologies for antibody drug conjugate characterization: high resolution native MS and IM-MS for average DAR and DAR distribution assessment. Anal. Chem. 2014;86:10674–10683. doi: 10.1021/ac502593n. [DOI] [PubMed] [Google Scholar]
- 83.Shion H., Birdsall R., Kotch F.W. 2014. Development of Integrated Informatics Workflows for the Automated Assessment of Comparability for Antibody Drug Conjugates (ADCs) Using LC/UV and LC/UV/MS, Poster Session Presented at the 62nd Annual Conference on Mass Spectrometry and Allied Topics. Baltimore, MD. [Google Scholar]
- 84.Chen B., Peng Y., Valeja S.G. Online hydrophobic interaction chromatography–mass spectrometry for top-down proteomics. Anal. Chem. 2016;88:1885–1891. doi: 10.1021/acs.analchem.5b04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarrut M., Corgier A., Fekete S. Analysis of antibody-drug conjugates by comprehensive on-line two-dimensional hydrophobic interaction chromatography x reversed phase liquid chromatography hyphenated to high resolution mass spectrometry. I - optimization of separation conditions. J. Chromatogr. B. 2016;1032:103–111. doi: 10.1016/j.jchromb.2016.06.048. [DOI] [PubMed] [Google Scholar]
- 86.Leney A.C., Heck A.J. Native mass spectrometry: what is in the name? J. Am. Soc. Mass Spectrom. 2017;28:5–13. doi: 10.1007/s13361-016-1545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davis J.A., Kagan M., Read J. Immunoprecipitation middle-up LC–MS for in vivo drug-to-antibody ratio determination for antibody–drug conjugates. Bioanalysis. 2017;9:1535–1549. doi: 10.4155/bio-2017-0148. [DOI] [PubMed] [Google Scholar]
- 88.Kellie J.F., Thomson A.S., Chen S. Biotherapeutic antibody subunit LC-MS and peptide mapping LC-MS measurements to study possible biotransformation and critical quality attributes in vivo. J. Pharmaceut. Sci. 2019;108:1415–1422. doi: 10.1016/j.xphs.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 89.Rago B., Clark T., King Lindsay. Calculated conjugated payload from immunoassay and LC–MS intact protein analysis measurements of antibody-drug conjugate. Bioanalysis. 2016;8:2205–2217. doi: 10.4155/bio-2016-0160. [DOI] [PubMed] [Google Scholar]
- 90.Pu J., An B., Vazvaei F. Enrichment of protein therapeutics and biomarkers for LC–MS quantification. Bioanalysis. 2018;10:979–982. doi: 10.4155/bio-2018-0056. [DOI] [PubMed] [Google Scholar]
- 91.Zhu X., An B., Zhang M. 2019. Investigation of Ocular Tissue Disposition of Antibody-Drug Conjugates Using Novel LC-MS-based Strategies, Poster Session Presented at the 66th Annual Conference on Mass Spectrometry and Allied Topics. San Diego, CA, USA. [Google Scholar]
- 92.Myler H., Rangan V.S., Wang J. An integrated multiplatform bioanalytical strategy for antibody–drug conjugates: a novel case study. Bioanalysis. 2015;7:1569–1582. doi: 10.4155/bio.15.80. [DOI] [PubMed] [Google Scholar]
- 93.Liu A., Kozhich A., Passmore D. Quantitative bioanalysis of antibody-conjugated payload in monkey plasma using a hybrid immuno-capture LC–MS/MS approach: assay development, validation, and a case study. J. Chromatogr. B. 2015;1002:54–62. doi: 10.1016/j.jchromb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 94.Wang J., Gu H., Liu A. Antibody–drug conjugate bioanalysis using LB-LC–MS/MS hybrid assays: strategies, methodology and correlation to ligand-binding assays. Bioanalysis. 2016;8:1383–1401. doi: 10.4155/bio-2016-0017. [DOI] [PubMed] [Google Scholar]
- 95.Eiceman G.A., Karpas Z., Hill H.H., Jr. 3rd ed. CRC press; New York: 2013. Ion Mobility Spectrometry. [Google Scholar]
- 96.Kraynov E., Kamath A.V., Walles M. Current approaches for absorption, distribution, metabolism, and excretion characterization of antibody-drug conjugates: an industry white paper. Drug Metab. Dispos. 2016;44:617–623. doi: 10.1124/dmd.115.068049. [DOI] [PubMed] [Google Scholar]
- 97.Faria M., Peay M., Lam B. Multiplex LC-MS/MS assays for clinical bioanalysis of MEDI4276, an antibody-drug conjugate of tubulysin analogue attached via cleavable linker to a biparatopic humanized antibody against HER-2. Antibodies. 2019;8:11. doi: 10.3390/antib8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong L., Li C., Locuson C. A two-step immunocapture LC/MS/MS assay for plasma stability and payload migration assessment of cysteine-maleimide-based antibody drug conjugates. Anal. Chem. 2018;90:5989–5994. doi: 10.1021/acs.analchem.8b00694. [DOI] [PubMed] [Google Scholar]
- 99.Xu L., Zhang Z., Xu S. Simultaneous quantification of total antibody and antibody-conjugated drug for XMT-1522 in human plasma using immunocapture-liquid chromatography/mass spectrometry. J. Pharmaceut. Biomed. Anal. 2019;174:441–449. doi: 10.1016/j.jpba.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 100.Qu J., Straubinger R.M. Improved sensitivity for quantification of proteins using triply charged cleavable isotope-coded affinity tag peptides. Rapid Commun. Mass Spectrom. 2005;19:2857–2864. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- 101.Jenkins R., Duggan J.X., Aubry A.-F. Recommendations for validation of LC-MS/MS bioanalytical methods for protein biotherapeutics. AAPS J. 2015;17:1–16. doi: 10.1208/s12248-014-9685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cao J., Covarrubias V.M., Straubinger R.M. A rapid, reproducible, on-the-fly orthogonal array optimization method for targeted protein quantification by LC/MS and its application for accurate and sensitive quantification of carbonyl reductases in human liver. Anal. Chem. 2010;82:2680–2689. doi: 10.1021/ac902314m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duan X., Dai L., Chen S.-C. Nano-scale liquid chromatography/mass spectrometry and on-the-fly orthogonal array optimization for quantification of therapeutic monoclonal antibodies and the application in preclinical analysis. J. Chromatogr. A. 2012;1251:63–73. doi: 10.1016/j.chroma.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.An B., Zhang M., Johnson R.W. Surfactant-aided precipitation/on-pellet-digestion (SOD) procedure provides robust and rapid sample preparation for reproducible, accurate and sensitive LC/MS quantification of therapeutic protein in plasma and tissues. Anal. Chem. 2015;87:4023–4029. doi: 10.1021/acs.analchem.5b00350. [DOI] [PubMed] [Google Scholar]
- 105.Nouri-Nigjeh E., Zhang M., Ji T. Effects of calibration approaches on the accuracy for LC-MS targeted quantification of therapeutic protein. Anal. Chem. 2014;86:3575–3584. doi: 10.1021/ac5001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen S., An B., Wang X. Surfactant cocktail-aided extraction/precipitation/on-pellet digestion strategy enables efficient and reproducible sample preparation for large-scale quantitative proteomics. Anal. Chem. 2018;90:10350–10359. doi: 10.1021/acs.analchem.8b02172. [DOI] [PubMed] [Google Scholar]
- 107.An B., Zhang M., Pu J. High-throughput, sensitive LC-MS quantification of biotherapeutics and biomarkers using antibody-free, peptide-level, multiple-mechanism enrichment via strategic regulation of pH and ionic and solvent strengths. Anal. Chem. 2019;91:3475–3483. doi: 10.1021/acs.analchem.8b05046. [DOI] [PubMed] [Google Scholar]
- 108.Zhang M., An B., Qu Y. Sensitive, high-throughput, and robust trapping-micro-LC-MS strategy for the quantification of biomarkers and antibody biotherapeutics. Anal. Chem. 2018;90:1870–1880. doi: 10.1021/acs.analchem.7b03949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vasicek L.A., Zhu X., Spellman D.S. Direct quantitation of therapeutic antibodies for pharmacokinetic studies using immuno-purification and intact mass analysis. Bioanalysis. 2019;11:203–213. doi: 10.4155/bio-2018-0240. [DOI] [PubMed] [Google Scholar]
- 110.Jian W., Kang L., Burton L. A workflow for absolute quantitation of large therapeutic proteins in biological samples at intact level using LC-HRMS. Bioanalysis. 2016;8:1679–1691. doi: 10.4155/bio-2016-0096. [DOI] [PubMed] [Google Scholar]
- 111.de Mel N., Mulagapati S.H.R., Cao M. A method to directly analyze free-drug-related species in antibody-drug conjugates without sample preparation. J. Chromatogr. B. 2019;1116:51–59. doi: 10.1016/j.jchromb.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 112.Tumey L.N., editor. Antibody-Drug Conjugates: Methods and Protocols. Humana Press; New York: 2020. [Google Scholar]
- 113.Gong H.H., Ihle N., Jones M.T. Control strategy for small molecule impurities in antibody-drug conjugates. AAPS PharmSciTech. 2018;19:971–977. doi: 10.1208/s12249-017-0943-6. [DOI] [PubMed] [Google Scholar]
- 114.Lin K., Tibbitts J. Pharmacokinetic considerations for antibody drug conjugates. Pharm. Res. (N. Y.) 2012;29:2354–2366. doi: 10.1007/s11095-012-0800-y. [DOI] [PubMed] [Google Scholar]
- 115.Fujiwara Y., Furuta M., Manabe S. Imaging mass spectrometry for the precise design of antibody-drug conjugates. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep24954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen B.-Q., Bumbaca D., Saad O. Catabolic fate and pharmacokinetic characterization of trastuzumab emtansine (T-DM1): an emphasis on preclinical and clinical catabolism. Curr. Drug Metabol. 2012;13:901–910. doi: 10.2174/138920012802138598. [DOI] [PubMed] [Google Scholar]
- 117.Wei D., Sullivan M., Espinosa O. A sensitive LC–MS/MS method for the determination of free maytansinoid DM4 concentrations—method development, validation, and application to the nonclinical studies of antitumor agent DM4 conjugated hu-anti-Cripto MAb B3F6 (B3F6-DM4) in rats and monkeys. Int. J. Mass Spectrom. 2012;312:53–60. [Google Scholar]
- 118.Liu Y., Zhou F., Sang H. LC-MS/MS method for the simultaneous determination of Lys-MCC-DM1, MCC-DM1 and DM1 as potential intracellular catabolites of the antibody-drug conjugate trastuzumab emtansine (T-DM1) J. Pharmaceut. Biomed. Anal. 2017;137:170–177. doi: 10.1016/j.jpba.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 119.Kaur S., Xu K., Saad O.M. Bioanalytical assay strategies for the development of antibody–drug conjugate biotherapeutics. Bioanalysis. 2013;5:201–226. doi: 10.4155/bio.12.299. [DOI] [PubMed] [Google Scholar]
- 120.Rago B., Tumey L.N., Wei C. Quantitative conjugated payload measurement using enzymatic release of antibody–drug conjugate with cleavable linker. Bioconjugate Chem. 2017;28:620–626. doi: 10.1021/acs.bioconjchem.6b00695. [DOI] [PubMed] [Google Scholar]
- 121.Liu A. Immuno-capture LC-MS/MS hybrid assay methodology in ADC bioanalysis. Immunome Res. 2016;12:54. [Google Scholar]
- 122.Li Y., Gu C., Gruenhagen J. An enzymatic deconjugation method for the analysis of small molecule active drugs on antibody-drug conjugates. mAbs. 2016;8:698–705. doi: 10.1080/19420862.2016.1151590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen T., Su D., Gruenhagen J. Chemical de-conjugation for investigating the stability of small molecule drugs in antibody-drug conjugates. J. Pharmaceut. Biomed. Anal. 2016;117:304–310. doi: 10.1016/j.jpba.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 124.Hussain A., Gorovits B., Leal M. PK of immunoconjugate anticancer agent CMD-193 in rats: ligand-binding assay approach to determine in vivo immunoconjugate stability. Bioanalysis. 2013;6:21–32. doi: 10.4155/bio.13.278. [DOI] [PubMed] [Google Scholar]
- 125.Xu L., Packer L.E., Li C. A generic approach for simultaneous measurements of total antibody and cleavable antibody-conjugated drug by LC/MS/MS. Anal. Biochem. 2017;537:33–36. doi: 10.1016/j.ab.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 126.Hyung S.-J., Li D., Koppada N. Method development of a novel PK assay for antibody-conjugated drug measurement of ADCs using peptide-linker drug analyte. Anal. Bioanal. Chem. 2019;411:2587–2596. doi: 10.1007/s00216-019-01701-9. [DOI] [PubMed] [Google Scholar]
- 127.Shi C., Goldberg S., Lin T. Bioanalytical workflow for novel scaffold protein–drug conjugates: quantitation of total Centyrin protein, conjugated Centyrin and free payload for Centyrin–drug conjugate in plasma and tissue samples using liquid chromatography–tandem mass spectrometry. Bioanalysis. 2018;10:1651–1665. doi: 10.4155/bio-2018-0201. [DOI] [PubMed] [Google Scholar]
- 128.Wei C., Zhang G., Clark T. Where did the linker-payload go? A quantitative investigation on the destination of the released linker-payload from an antibody-drug conjugate with a maleimide linker in plasma. Anal. Chem. 2016;88:4979–4986. doi: 10.1021/acs.analchem.6b00976. [DOI] [PubMed] [Google Scholar]
- 129.Shen B.Q., Xu K., Liu L. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat. Biotechnol. 2012;30:184–189. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 130.Beck A., Goetsch L., Dumontet C. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]