Summary
Coronavirus disease 2019 (COVID-19) is a pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 is defined by respiratory symptoms, but cardiac complications including viral myocarditis are also prevalent. Although ischemic and inflammatory responses caused by COVID-19 can detrimentally affect cardiac function, the direct impact of SARS-CoV-2 infection on human cardiomyocytes is not well understood. Here, we utilize human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) as a model to examine the mechanisms of cardiomyocyte-specific infection by SARS-CoV-2. Microscopy and RNA sequencing demonstrate that SARS-CoV-2 can enter hiPSC-CMs via ACE2. Viral replication and cytopathic effect induce hiPSC-CM apoptosis and cessation of beating after 72 h of infection. SARS-CoV-2 infection activates innate immune response and antiviral clearance gene pathways, while inhibiting metabolic pathways and suppressing ACE2 expression. These studies show that SARS-CoV-2 can infect hiPSC-CMs in vitro, establishing a model for elucidating infection mechanisms and potentially a cardiac-specific antiviral drug screening platform.
Keywords: COVID-19, SARS-CoV-2, coronavirus, induced pluripotent stem cells, viral myocarditis, cardiomyocytes, cardiovascular biology, heart, stem cell, cardiology
Graphical Abstract
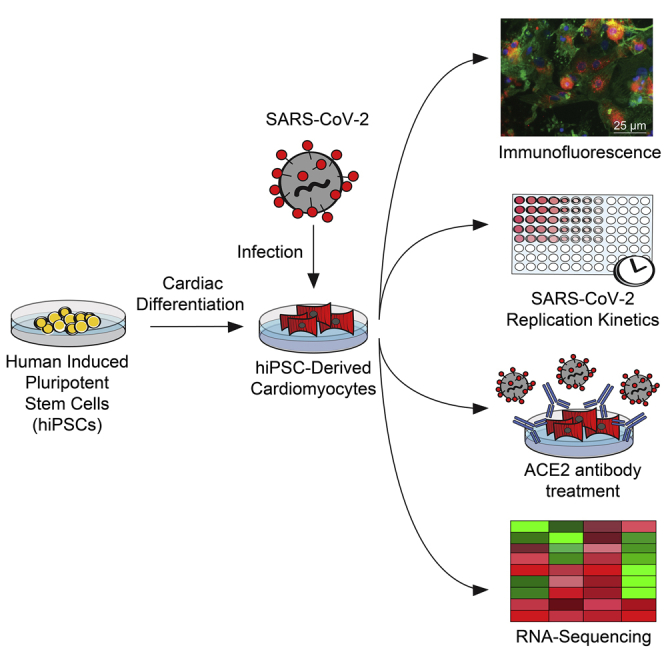
Highlights
Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection
ACE2 antibody blunts SARS-CoV-2 infection in cardiomyocytes
Infected human iPSC-derived cardiomyocytes activate viral clearance pathways
Sharma et al. demonstrate that human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are susceptible to SARS-CoV-2 infection. This establishes a platform for understanding the mechanisms of cardiac-specific infection by SARS-CoV-2 in vitro and could potentially be employed to develop antiviral compounds.
Introduction
Coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been declared a worldwide pandemic, causing the hospitalization and deaths of hundreds of thousands of people worldwide.1 SARS-CoV-2, a single-stranded enveloped RNA virus, is known to use the ACE2 receptor to enter host lung tissue, followed by rapid viral replication.2 Clinical presentation demonstrates predominantly pulmonary symptoms, including cough, shortness of breath, pneumonia, and acute respiratory distress syndrome.3 However, there is mounting evidence that SARS-CoV-2 infection may cause cardiac complications including elevated cardiac stress biomarkers, arrhythmias, and heart failure.4 A recent study demonstrated significantly elevated troponin levels among some COVID-19 patients, indicating cardiac injury, and notably, cardiac injury was associated with increased risk of mortality.5 The etiology of cardiac injury in COVID-19, however, remains unclear. Cardiac injury may be ischemia mediated, and the profound inflammatory and hemodynamic impacts seen in COVID-19 have been hypothesized to cause atherosclerotic plaque rupture or oxygen supply demand mismatch resulting in ischemia.3 Alternatively, cardiac tissue expresses the ACE2 receptor, further suggesting the feasibility of direct SARS-CoV-2 internalization in cardiomyocytes (CMs).6 ACE2 is also upregulated specifically in CMs during dilated and hypertrophic cardiomyopathy.7 Importantly, the related SARS-CoV virus has been noted to localize within the myocardium,8 and recent studies9 have shown 79% sequence conservation in the viral genomes of SARS-CoV and SARS-CoV-2. Structurally, both viruses use the ACE2 receptor to enter cells and bind with similar affinities to ACE2.10
Given these similarities to SARS-CoV, it is plausible that SARS-CoV-2 could also use ACE2 to enter adult CMs. Preliminary COVID-19 clinical case reports have raised suspicion for cardiac injury mediated by direct myocardial SARS-CoV-2 infection and resulting fulminant myocarditis.11, 12, 13 A recent multi-organ autopsy study of COVID-19 patients detected SARS-CoV-2 viral RNA in the heart via PCR.14 However, the gold standard for confirming SARS-CoV-2 localization in CMs is endomyocardial biopsy (EMB), an invasive procedure, and additional safety precautions also must be taken to conduct EMB in the setting of COVID-19. Thus, there are currently limited clinical data demonstrating localization of SARS-CoV-2 to adult human CMs. In order to gain further insights into the cardiac pathophysiology of COVID-19, it will be critical to determine whether SARS-CoV-2 can directly infect isolated human CMs. Elucidating the pathogenic mechanism of cardiac injury in COVID-19 in vitro could ultimately guide therapeutic strategies. Antiviral agents could potentially mitigate cardiac complications if the underlying mechanism of cardiac injury is direct myocardial viral infection.
Primary human CMs are difficult to obtain and maintain for research use. Improved methods to convert human induced pluripotent stem cells (hiPSCs) to multiple somatic lineages have enabled in vitro mass production of patient-specific cells, including hiPSC-derived CMs (hiPSC-CMs).15 The hiPSC-CMs produce relevant proteins found in adult human CMs, can spontaneously contract, can be made in weeks using defined differentiation protocols, and can be genetically customized using genome editing.16 HiPSC-CMs express ACE2, which increases in expression over 90 days of differentiation.17 The hiPSC-CMs are also responsive to inotropic drugs such as norepinephrine, and beating rates can be controlled via electrical stimulation.18 Because hiPSC-CMs can be purified and replated for downstream applications, research groups in academia and industry now utilize these cells for cardiovascular disease modeling and high-throughput drug screening assays. HiPSC-CMs can recapitulate cellular phenotypes for cardiovascular diseases including various forms of cardiomyopathy19,20 and drug-induced cardiotoxicity.21,22 Notably, hiPSC-CMs have also shown promise as an in vitro model for studying the mechanisms of direct CM viral infection in the context of viral myocarditis. A previous study demonstrated that coxsackievirus B3 (CVB3), one of the major causative agents for viral myocarditis, can rapidly infect and proliferate within hiPSC-CMs.23 CVB3, like SARS-CoV-2, is a positive-sense, single-stranded RNA virus, although unlike SARS-CoV-2, it does not have a viral envelope. The hiPSC-CMs produce the coxsackie and adenovirus receptor protein, which is needed for infection by CVB3. A detrimental virus-induced cytopathic effect was observed in hiPSC-CMs within hours of CVB3 infection, manifesting in cell death and contractility irregularities. Importantly, this study also established hiPSC-CMs as a cardiac-specific antiviral drug screening platform and demonstrated that drugs such as interferon beta and ribavirin can stymie CVB3 proliferation in vitro.23 Interferon beta was able to transcriptionally activate viral clearance gene networks in CVB3-infected hiPSC-CMs.
Here, the aforementioned foundational viral myocarditis study is extended to assess the effect of SARS-CoV-2 on hiPSC-CMs. We show that hiPSC-CMs are susceptible to SARS-CoV-2 infection, resulting in functional alterations, transcriptional changes, and cytopathic effects. These cellular phenotypes occurred in the absence of systemic inflammatory and hemodynamic impacts, establishing an in vitro cardiac platform to study SARS-CoV-2 infection.
Results
hiPSCs Can Differentiate into CMs
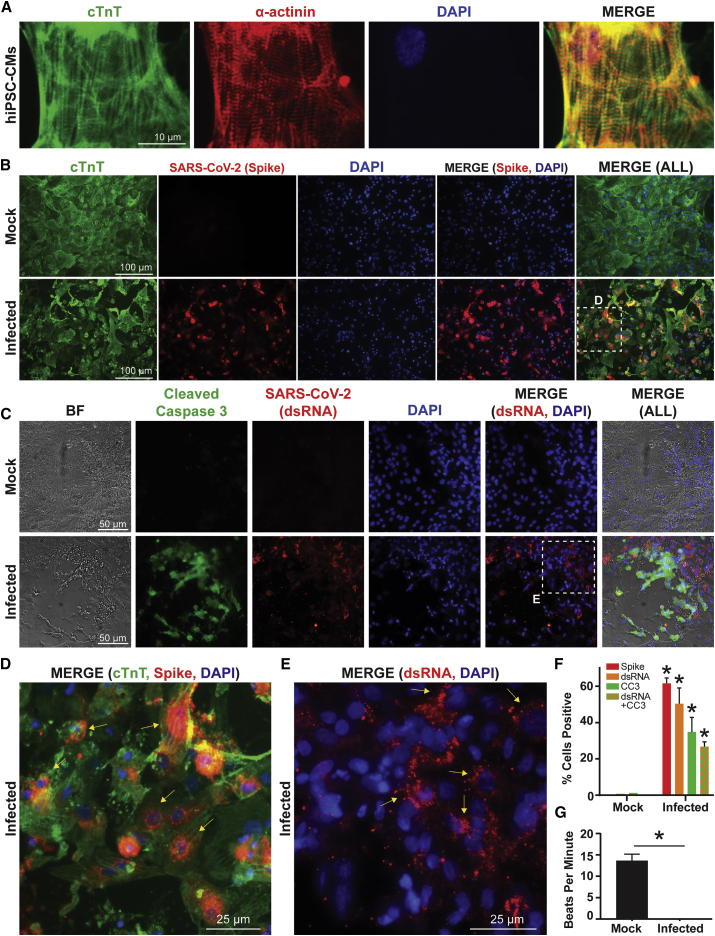
An hiPSC control line (02iCTR) was generated by the Cedars-Sinai Medical Center iPSC Core from peripheral blood mononuclear cells and shown to be fully pluripotent.24 The hiPSCs were differentiated into CMs using an established monolayer differentiation protocol utilizing small molecule modulators of Wnt signaling.25 Differentiated hiPSC-CMs were metabolically purified by depriving cells of glucose, as previously demonstrated.25 Purified hiPSC-CMs expressed standard cardiac sarcomeric markers cardiac troponin T (cTnT) and α-actinin (Figure 1A).
Figure 1.
SARS-CoV-2 Internalizes and Replicates within hiPSC-CMs In Vitro, Eliciting Cytopathic Effect and Contractility Alterations
(A) Human iPSC-CMs exhibit standard sarcomeric markers including cardiac troponin T (cTnT) and α-actinin with DAPI as nuclear counterstain.
(B) Immunofluorescence for cTnT and SARS-CoV-2 “spike” protein demonstrates that hiPSC-CMs can be infected by SARS-CoV-2. SARS-CoV-2 spike protein is not present in mock infected cultures.
(C) HiPSC-CMs after SARS-CoV-2 infection, but not mock infection, exhibit signs of cellular apoptosis, indicated by morphological changes seen in brightfield (BF) and cleaved caspase-3 (CC3) production. A second SARS-CoV-2 antibody marks a viral-specific double-stranded intermediate RNA (dsRNA).
(D) Magnified inset from (B) shows a merged immunofluorescence image for SARS-CoV-2 spike protein, cTnT, and DAPI. Arrows indicate perinuclear accumulation of viral particles and suggest active viral protein translation at perinuclear ribosomes.
(E) Magnified inset from (C) shows a merged immunofluorescence image for SARS-CoV-2 dsRNA and DAPI. Arrows indicate dsRNA stain.
(F) Quantification of immunofluorescence indicates percentage of total DAPI-positive cells that are positive for spike protein, viral dsRNA, CC3, and dsRNA+CC3 in hiPSC-CMs infected with SARS-CoV-2, compared to mock infection. n = 5–7 images (technical replicates) quantified for each stain for mock and infected conditions. ∗p < 0.05.
(G) Quantification of beats per minute in wells containing hiPSC-CMs with mock infection versus wells containing hiPSC-CMs infected with SARS-CoV-2. n = 6 videos (technical replicates) recorded for each condition. See Video S1 for representative video clips. ∗p < 0.05.
Purified hiPSC-CMs Can Be Directly Infected by SARS-CoV-2
Purified hiPSC-CMs were replated into 96-well plates at 100,000 cells per well and allowed to regain contractility (Video S1) before being subjected to SARS-CoV-2 infection. The SARS-CoV-2 was obtained from the Biodefense and Emerging Infections (BEI) Resources of the National Institute of Allergy and Infectious Diseases (NIAID) and titered on Vero-E6 cells (see STAR Methods). A viral dose response and time course were conducted on hiPSC-CMs, and a multiplicity of infection (MOI) of 0.1 was chosen for all experiments unless otherwise specified (Figure S1). The hiPSC-CMs were infected with SARS-CoV-2 for 72 h in all experiments unless otherwise specified, with a mock treatment without virus serving as a control condition. Plaque formation assays conducted from supernatant harvested after hiPSC-CM infection confirmed active SARS-CoV-2 infection (Figure S1D). Cells from both infected and mock conditions were stained for cardiac marker cTnT and SARS-CoV-2 viral capsid “spike” protein (Figure 1B). The infected hiPSC-CMs stained positively for spike protein, suggesting that SARS-CoV-2 can establish active infection in hiPSC-CMs. As demonstrated previously,2 pre-treatment of infected hiPSC-CMs with an ACE2 antibody significantly diminished viral protein expression and plaque forming unit production, suggesting that ACE2 is critical for SARS-CoV-2 internalization in hiPSC-CMs (Figure S1).
Thirty-second video clips (in triplicate) were taken of mock and infected hiPSC-CMs at 72 h after viral infection. Cells were infected at MOI 0.1.
SARS-CoV-2 Infection of hiPSC-CMs Causes Apoptosis and Cessation of Beating
To determine if SARS-CoV-2 induced a cytopathic effect on hiPSC-CMs, mock and infected hiPSC-CMs were stained for the apoptosis marker cleaved caspase-3, as well as for the double-stranded RNA (dsRNA) intermediate unique to positive sense RNA virus infection (Figure 1C). The dsRNA and spike protein stains represent two independent assays for visualizing SARS-CoV-2 viral uptake and genome replication in hiPSC-CMs. A proportion of infected cells were positive for dsRNA and also stained positive for cleaved caspase-3, indicating that hiPSC-CMs were undergoing virus-induced apoptosis. Pre-treatment of infected hiPSC-CMs with an ACE2 antibody significantly reduced cleaved caspase-3 expression and blunted apoptosis (Figure S1). Notably, the SARS-CoV-2 spike protein localized at perinuclear regions in hiPSC-CMs (Figure 1D), consistent with prior results with coronavirus infection in non-CMs26 and coxsackievirus infection on hiPSC-CMs.23 The dsRNA intermediates exhibit cytoplasmic localization (Figure 1E). Quantification of stained cells demonstrated the percentage of total cells that were positive for dsRNA and spike protein, as well as for cleaved caspase-3 (Figure 1F). Simple beat rate contractility analysis was also conducted, whereby 30 s videos were taken of wells containing mock and infected hiPSC-CMs (Video S1). Functionally, infected hiPSC-CMs ceased beating after 72 h of SARS-CoV-2 infection, whereas mock wells continued to contract (Figure 1G). Taken together, these results indicate that hiPSC-CMs are susceptible to ACE2-mediated SARS-CoV-2 infection and downstream detrimental cytopathic effects, the SARS-CoV-2 may be able to replicate in distinct perinuclear locations within hiPSC-CMs by co-opting cellular organelles for viral protein translation, and the SARS-CoV-2 infection significantly reduces functional contractility in hiPSC-CMs.
Transcriptional Analysis of SARS-CoV-2-Infected hiPSC-CMs
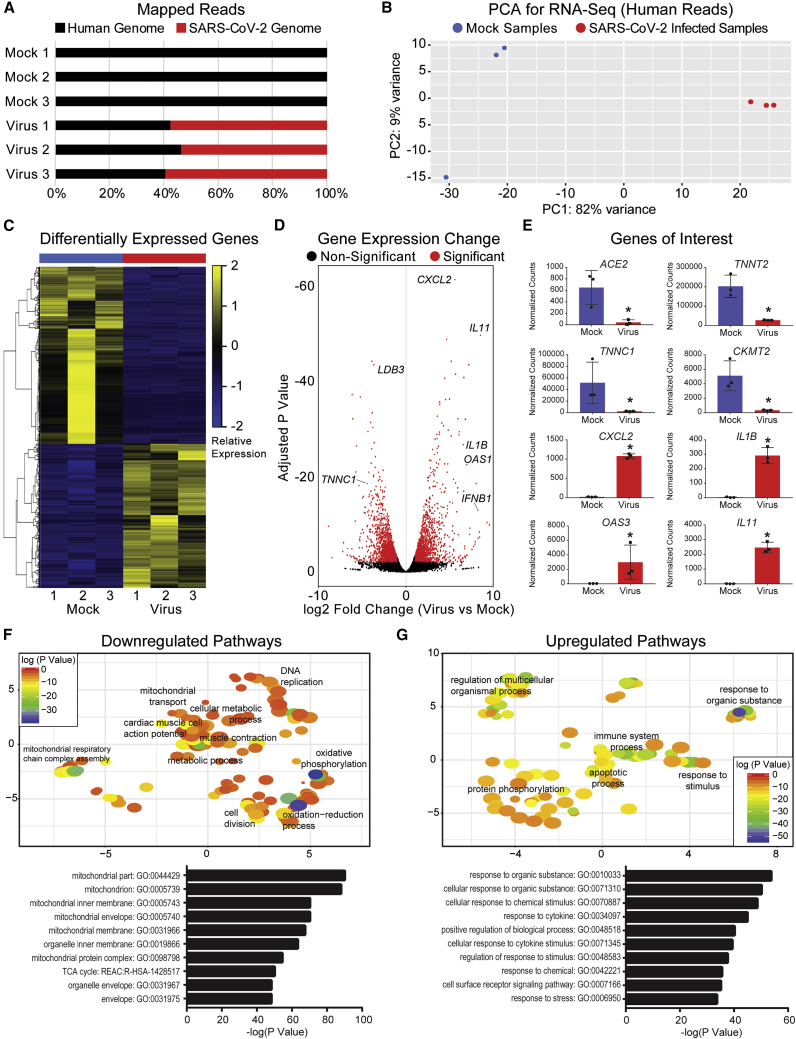
The hiPSC-CMs infected with 0.1 MOI SARS-CoV-2 for 72 h were also harvested for transcriptomic analysis via RNA sequencing. Upon mapping genomic reads in infected hiPSC-CMs, greater that 50% of all mapped reads were from the SARS-CoV-2 genome, further indicating active SARS-CoV-2 viral genome replication within infected hiPSC-CMs (Figure 2A). Principal component analysis and heatmaps of differentially expressed genes demonstrated clustering of samples by condition (mock or infected) and transcriptomic profile (Figures 2B and 2C). SARS-CoV-2 infection induced significant gene expression changes within hiPSC-CMs (Figure 2D). The most significant expression change by adjusted p value was for CXCL2, which encodes an immune cytokine known to be transcriptionally upregulated during SARS-CoV infection.27 Similarly, immunomodulators such as interleukins were significantly upregulated in infected samples, along with antiviral response pathways genes such as OAS3. Expression of CM markers TNNT2 and TNNC1 were significantly reduced in infected samples, along with mitochondrial genes related to oxidative phosphorylation, such as CKMT2, encoding for creatine kinase. Notably, ACE2 was significantly downregulated in SARS-CoV-2-infected hiPSC-CMs, consistent with prior reports examining SARS-CoV infection in myocardium.8 Gene pathway analysis confirmed downregulation of transcriptional pathways related to mitochondrial function, oxidative phosphorylation, and cardiac function, whereas upregulated pathways included responses to organic substance, immune system processes, and apoptosis (Figures 2F and 2G). Taken together, these results indicate that SARS-CoV-2 infection induces significant transcriptional changes within hiPSC-CMs in gene pathways related to cellular metabolism and immune response.
Figure 2.
SARS-CoV-2 Alters hiPSC-CM Transcriptomic Profiles after Infection
(A) Graph of mapped reads during RNA sequencing. In virus-infected hiPSC-CMs, more than 50% of genomic reads mapped to the SARS-CoV-2 genome, suggesting viral genome presence within infected hiPSC-CMs.
(B) Principal component analysis (PCA) for 3 mock hiPSC-CM samples and 3 SARS-CoV-2-infected hiPSC-CM samples illustrates transcriptional clustering by condition (mock versus infected). Samples represent biological replicates of mock and infected conditions, respectively.
(C) Heatmap of differentially expressed genes shows that samples cluster based on transcriptomic profile and condition (mock versus infected).
(D) Volcano plot of gene expression change in virus-infected versus mock samples. Significantly changed gene are defined with adjusted p < 0.01 and absolute value fold change >2. Genes of interest noted include cardiac markers LDB3 and TNNC1, cytokines CXCL2, IL11, IL1B, and IFNB1, and antiviral response gene OAS1.
(E) Expression of genes of interest in mock versus infected conditions. TNNT2 and TNNC1 represent cardiac markers, whereas CKMT2 represents mitochondrial enzymes. CXCL2, IL1B, IL11, and OAS3 represent innate immune response and viral clearance genes. ∗p < 0.05.
(F) Top: downregulated transcriptional pathways based on Gene Ontology (GO) analysis, visualized using REViGO. Significantly downregulated pathways include mitochondrial transport, oxidative phosphorylation, oxidation-reduction processes, and muscle contraction. Bottom: top 10 most significant GO terms associated with downregulated pathways are related to mitochondrial function.
(G) Top: upregulated transcriptional pathways based on GO analysis, visualized using REViGO. Significantly upregulated pathways include response to organic substance, immune system process, and apoptotic process. Bottom: top 10 most significant GO terms associated with upregulated pathways are related to response to organic stimulus.
Discussion
Results presented here establish that human iPSC-derived CMs are susceptible to ACE2-mediated direct infection by SARS-CoV-2 and that the virus may induce detrimental cytopathic effects in these cells. At an MOI of 0.1, hiPSC-CMs experienced cytopathic effect after 72 h of infection. The virus presumably enters the cell through the ACE2 receptor, expressed by the hiPSC-CMs in this study. An ACE2 receptor antibody was able to significantly blunt viral protein expression, replication, and cell death. However, in spite of the ACE2 antibody treatment, cells were still susceptible to infection, suggesting potential alternative mechanisms for SARS-CoV-2 internalization. Immunostaining of two SARS-CoV-2 components, the unique dsRNA intermediate and the “spike” capsid protein responsible for viral entry and virion particle assembly, demonstrates that this novel coronavirus can enter hiPSC-CMs to unleash its RNA cargo and hijack host hiPSC-CM translational machinery to produce new viral components. Notably, the “spike” protein stain localized to perinuclear regions, likely near the endoplasmic reticulum. Not only has this perinuclear viral component accumulation been reported in other in vitro coronavirus infection studies,26 but it parallels what was seen in hiPSC-CMs infected with CVB3.23 Presumably, this increased viral load within hiPSC-CMs activated downstream apoptotic events. This was indicated by alterations in cellular morphologies, increased production of cleaved caspase-3, and activation of cell death gene networks.
In parallel with these stains showing viral accumulation, there was a functional alteration in hiPSC-CM contractility where the cells ceased beating. The cause of this dramatic functional change in hiPSC-CM beating must be further investigated, but perhaps cytopathic effect and associated hiPSC-CM apoptosis significantly disrupted cell-cell gap junctions and therefore impaired proper excitation-contraction coupling within and between CMs. Indeed, significant alterations in calcium handling and contractility were similarly observed in CVB3-infected hiPSC-CMs.23 Additionally, SARS-CoV-2 induced widespread transcriptional changes in infected hiPSC-CMs. Paralleling findings from SARS-CoV infection studies, SARS-CoV-2 infection on CMs downregulated the expression of ACE28 and upregulated expression of CXCL2,27 along with other immunomodulatory cytokines. Reduction in cardiac-specific contractile genes such as TNNT2 likely reflects the impairment in contractility of infected hiPSC-CMs, as well as overall CM death. Infected hiPSC-CMs also experienced profound downregulation in gene networks associated with oxidative phosphorylation and mitochondrial function, consistent with the dramatic metabolic impacts that viral infections elicit.28
We predict that future studies will be able to use hiPSC-CMs as a cardiac-specific antiviral drug screening platform, in similar fashion to previous studies with CVB3 on hiPSC-CMs.23 A variety of antiviral approaches ranging from repurposed small molecules, such as nucleoside analogs or viral polymerase inhibitors, to novel antibodies and antisense oligonucleotides have been proposed for COVID-19 and are currently being tested in vitro and in clinical trials.29 HiPSC-CMs could serve as a cardiac-specific auxiliary cell type for in vitro pre-clinical efficacy studies for any drug aiming to stymie SARS-CoV-2 proliferation. In parallel, drug-induced arrhythmias and QT-interval prolongation can also be examined for antiviral compounds in pre-clinical development, given that some existing COVID-19 drug treatments exhibit these off-target cardiotoxicities.30 Due to the enormity of the current COVID-19 pandemic, and the increasing evidence for associated cardiac symptoms, innovative approaches using technologies such as the hiPSC-CMs presented here will be critical to further understand and counteract SARS-CoV-2 infection.
Limitations of Study
The cellular immaturity of hiPSC-derivative cell types must be considered in this model. It is known that hiPSC-CMs are electrophysiologically, structurally, and genetically immature in comparison to their adult counterparts,31 although biomechanical efforts to mature these cells toward an adult phenotype have recently gained traction.32 However, even in these immature cells, the current study showed overt virus-induced cardiotoxicity and physiological effects at SARS-CoV-2 MOIs ranging from 0.001 to 0.1. Since expression of ACE2 increases as hiPSC-CMs mature in in vitro culture,17 one could hypothesize that after maturation, older hiPSC-CMs would be even more susceptible to SARS-CoV-2 infection. Significant additional data are needed to test these possibilities.
The results presented here are in an isolated in vitro system, devoid of any immune system cellular components that are thought to be critical to the many aspects of the COVID-19 viral response in vivo.33 The resulting myocarditis, by definition, is an inflammation of the myocardium caused by immune cell infiltration, which may or may not be the result of direct CM viral infection. However, as shown here, hiPSC-derived CMs are clearly susceptible to SARS-CoV-2 infection even in the absence of a cellular immune response. Still, current clinical data have not definitively established direct infection of the heart playing a role in the clinical syndrome. Although studies presenting EMBs from COVID-19 patients are rare at this point, biopsied heart tissue has shown the SARS-CoV-2 virus localizing in the heart of a COVID-19 patient, albeit not specifically in the CMs.13 A recent multi-organ autopsy study also showed SARS-CoV-2 viral RNA in the heart.14 Additional clinical data are needed, but this raises the question of whether the SARS-CoV-2 enters the heart via a viremic phase or through macrophages clearing the virus from the lungs. It remains to be seen whether direct CM infection may contribute to the variety of in vivo cardiac clinical sequelae presented by patients with COVID-19, such as arrhythmias, elevated cardiac troponin biomarkers, or heart failure. Nonetheless, we establish that hiPSC-CMs in vitro are susceptible to SARS-CoV-2 infection.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-actinin | Sigma-Aldrich | Cat# A7811, RRID: AB_476766 |
| cardiac troponin T | Abcam | Cat# ab45932, RRID: AB_956386 |
| SARS-CoV-2 spike (S) protein | BEI Resources | NR-616 Monoclonal Anti-SARS-CoV S Protein (Similar to 240C) SARS coronavirus |
| SARS-CoV-2 double stranded RNA | Absolute Antibody Inc | J2 clone |
| cleaved caspase-3 | Cell Signaling Technology | Cat# 9661, RRID: AB_2341188 |
| donkey anti-rabbit 488 | Thermo Fisher Scientific | at# R37118, RRID: AB_2556546 |
| goat anti-mouse 555 | Thermo Fisher Scientific | Cat# A32727, RRID: AB_2633276 |
| donkey anti-mouse 594 | Thermo Fisher Scientific | Cat# R37115, RRID: AB_2556543 |
| ACE2 | R&D Systems | Cat# AF933, RRID: AB_355722 |
| TIM4 | R&D Systems | Cat# AF2929, RRID: AB_2240431 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 | Biodefense and Emerging Infections (BEI) Resources of the National Institute of Allergy and Infectious Diseases (NIAID) | isolate USA-WA1/2020 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| mTeSR1 | STEMCELL Technologies | mTeSR1 |
| CHIR99021 | Cayman Chemical | 252917-06-9 |
| Wnt-C59 | Cayman Chemical | 1243243-89-1 |
| RPMI 1640 | Thermo Fisher Scientific | 11875093 |
| RPMI 1640 no glucose | Thermo Fisher Scientific | 11879020 |
| B27 supplement minus insulin | Thermo Fisher Scientific | A1895601 |
| B27 supplement with insulin | Thermo Fisher Scientific | 17504044 |
| DMEM/F-12, HEPES | Thermo Fisher Scientific | 11330057 |
| Matrigel | Corning | cat. # 354230 |
| Phosphate Buffered Saline | GIBCO | cat. # 10010023 |
| Fetal Bovine Serum | Millipore Sigma | F2442 |
| HEPES Buffer Solution | Millipore Sigma | 83264 |
| Penicillin/Streptomycin | Millipore Sigma | P4333 |
| 4% paraformaldehyde solution | Fisher Scientific | AAJ19943K2 |
| Bovine Serum Albumin | Millipore Sigma | A1933 |
| Normal Donkey Serum | Abcam | Ab7475 |
| Normal Goat Serum | Abcam | Ab7481 |
| Triton X-100 | Millipore Sigma | X100 |
| DAPI | Millipore Sigma | MBD0015 |
| Trizol | Thermo Fisher Scientific | 15596026 |
| L-Glutamine | Thermo Fisher Scientific | 25030081 |
| Critical Commercial Assays | ||
| RNeasy mini kit | QIAGEN | cat. # 74104 |
| SMART-Seq V4 Ultra Low RNA Input Kit | Takara Bio USA | R400752 |
| Nextera XT Library Preparation kit | Illumina | FC-131-1024 |
| Deposited Data | ||
| Raw genomic data | This paper | GSE150392 |
| Experimental Models: Cell Lines | ||
| 02iCTR hiPSC line | Cedars-Sinai Medical Center (Laperle et al., 2020) | 02iCTR |
| Vero-E6 | ATCC | CRL-1586 |
| Software and Algorithms | ||
| ImageJ (Fiji) | NIH | https://imagej.nih.gov/ij/docs/guide/146-2.html |
| GraphPad Prism (8.0) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Microsoft Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/p/excel/cfq7ttc0k7dx?activetab=pivot%3aoverviewtab |
| Adobe Photoshop(CC) | Adobe | https://www.adobe.com/products/photoshop.html |
| Adobe Illustrator (CC) | Adobe | https://www.adobe.com/products/illustrator.html |
| STAR (version 2.5.0) | Github | https://github.com/alexdobin/STAR/releases |
| SARS-CoV-2 viral genome MT246667.1 | NIH GenBank | https://www.ncbi.nlm.nih.gov/nuccore/MT246667.1 |
| R version 3.6.3 | R project | https://www.r-project.org/ |
| REViGO | Rudjer Boskovic Institute, Croatia | http://revigo.irb.hr/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Clive Svendsen (Clive.Svendsen@cshs.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The gene expression data from this study are found at Gene Expression Omnibus with accession number GSE150392.
Experimental Model and Subject Details
HiPSC derivation
The appropriate institutional review board (IRB) and stem cell research oversight committee (SCRO) were consulted at UCLA and Cedars-Sinai Medical Center. The 02iCTR hiPSC line (male, clinically normal, no known associated disease) was derived from human peripheral blood mononuclear cells and has been published previously.24 This cell line was obtained from the Cedars-Sinai Medical Center iPSC core facility and was derived using non-integrating episomal reprogramming under their IRB-SCRO protocol “Pro00032834: iPSC Core Repository and Stem Cell Program.” These hiPSCs were grown on Matrigel and maintained in mTeSR1 (STEMCELL Technologies). Cells were maintained at approximately passage 30-40 during differentiation, with daily replacement of mTeSR1 and manual passaging after reaching 80% confluency (approximately every 4 days).
SARS-CoV-2 isolate
SARS-CoV-2, isolate USA-WA1/2020, was obtained from the Biodefense and Emerging Infections (BEI) Resources of the National Institute of Allergy and Infectious Diseases (NIAID). Importantly, all studies involving SARS-CoV-2 infection of hiPSC-CMs were conducted within a Biosafety Level 3 facility at UCLA.
Vero-E6 cells
Vero-E6 cell line was obtained from the American Type Culture Collection (ATCC) cell line catalog, under catalog number VERO C1008 [Vero 76, clone E6, Vero E6] (ATCC CRL1586). These are immortalized adherent kidney epithelial cells from Cercopithecus aethiops, the African green monkey.
Method Details
HiPSC-CM differentiation
The hiPSC differentiation work in the Svendsen Lab is carried out under “Pro00021505: Svendsen Stem Cell Program,” authorized by the Cedars-Sinai Medical Center IRB. Initially, hiPSCs were grown on Matrigel and maintained in mTeSR1 (STEMCELL Technologies). The hiPSC-CMs were generated from hiPSCs using a small-molecule mediated differentiation approach that modulates Wnt signaling.25 Briefly, this approach uses the CHIR99021 GSK3β inhibitor from days 0-2 to initiate mesoderm specification, followed by Wnt-C59 Wnt inhibitor at days 3-5 to initiate cardiac specification. From days 0-7 of differentiation, cells are maintained in RPMI 1640 + B27 supplement without insulin. Cells began beating at approximately day 7 post-differentiation, at which point medium was changed to RPMI 1640 + B27 supplement with insulin. Cardiomyocytes were metabolically selected from other differentiated cells by conducting glucose deprivation in RPMI 1640 without glucose + B27 supplement with insulin as previously described.25 After selection, hiPSC-CMs were replated as a monolayer into 96-well plate format at 100,000 cells per well in hiPSC-CM culture medium, RPMI 1640 + B27 supplement with insulin.
SARS-CoV-2 expansion in Vero-E6 cells
SARS-CoV-2 was passaged once in Vero-E6 cells (ATCC) and viral stocks were aliquoted and stored at −80°C. Virus titer was measured in Vero-E6 cells by TCID50 assay. Vero-E6 cells were cultured in DMEM growth media containing 10% fetal bovine serum, 2 mM L-glutamine, penicillin (100 units/ml), streptomycin (100 units/ml), and 10 mM HEPES. Cells were incubated at 37°C with 5% CO2.
SARS-CoV-2 infection of hiPSC-CMs
For hiPSC-CM infection, viral inoculum (MOI of 0.1, or 1 plaque forming unit per 10 cells) was prepared using serum-free media, unless otherwise specified. Culture media from each well containing hiPSC-CMs was removed and replaced with 250 μL of prepared inoculum. For mock infection, serum-free media (250 μL/well) alone was added. The inoculated plates were incubated for 1 hour at 37°C with 5% CO2. The inoculum was spread by gently tilting the plate sideways every 15 minutes. At the end of incubation, the inoculum was replaced with fresh hiPSC-CM culture medium. Cells remained at 37°C with 5% CO2 for 72 hours before analysis.
Imaging and immunofluorescence
After 72 hours of SARS-CoV-2 infection (or mock), live cell images and videos were obtained by bright field microscope (Leica DMIL LED). For immunofluorescence, separate wells of hiPSC-CMs were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes. The fixed samples were then permeabilized and blocked for 1 hour in a “blocking solution” containing PBS with 2% bovine serum albumin, 5% donkey serum, 5% goat serum, and 0.3% Triton X-100. Primary antibodies were diluted in the blocking solution and added to samples overnight at 4°C. The following antibodies and dilutions were used: α-actinin (1:100, Sigma-Aldrich Cat# A7811, RRID: AB_476766); cardiac troponin T (cTnT, 1:100, Abcam Cat# ab45932, RRID: AB_956386); SARS-CoV-2 spike (S) protein (1:100, BEI Resources NR-616 Monoclonal Anti-SARS-CoV S Protein (Similar to 240C) SARS coronavirus); SARS-CoV-2 double stranded RNA (1:100, J2 clone; Absolute Antibody Inc.); cleaved caspase-3 (1:200, Cell Signaling Technology Cat# 9661, RRID: AB_2341188). Samples were then rinsed 5 times for 2 minutes each with PBS containing 0.3% Triton X-100, followed by incubation with fluorescent-conjugated secondary antibodies diluted 1:1000 in blocking buffer for 2 hours at room temperature. Antibodies were donkey anti-rabbit 488 (Thermo Fisher Scientific Cat# R37118, RRID: AB_2556546), goat anti-mouse 555 (Thermo Fisher Scientific Cat# A32727, RRID: AB_2633276), and donkey anti-mouse 594 (Thermo Fisher Scientific Cat# R37115, RRID: AB_2556543). Samples were then rinsed 5 times for 2 minutes each with PBS containing 0.3% Triton X-100, followed by DAPI diluted in PBS at 1:5000 for 10 minutes. Immunofluorescence images were quantified using ImageJ software. DAPI was used to count total cell numbers in order to obtain a percentage of cells positive for dsRNA, spike protein, or cleaved caspase-3.
SARS-CoV-2 growth kinetics time course
The hiPSC-CMs were infected with SARS-CoV-2 at varying MOIs of 0.1, 0.01 and 0.001 in 96-well plate (Figure S1C). Mock infected cells received only the media used for preparing the SARS-CoV-2 inoculum. After 1-hour incubation at 37°C with 5% CO2, the inocula were replaced with fresh media followed by PBS (phosphate buffered saline) wash. At each time point (days 1, 2 and 3), cell culture supernatant samples from mock and SARS-CoV-2 infected wells were collected and stored at −80°C. Viral production by infected hiPSC-CMs at each time point was measured by quantifying TCID50 (Median Tissue Culture Infectious Dose). In brief, Vero-E6 cells were plated in 96-well plate at a density of 5 x103 cells/well. The next day, culture supernatants collected from hiPSC-CMs at various time points were subjected to 10-fold serial dilutions (10−1 to 10−6) and inoculated onto Vero-E6 cells. The cells were incubated at 37°C with 5% CO2. After 72 hours, each inoculated well was examined for presence or absence of viral cytopathic effect and percent infected dilutions immediately above and immediately below 50% were determined. TCID50 was calculated as described previously.34 TCID50 values of each viral MOIs at different time points were subjected to statistical analysis and plotted in a line graph.
Viral plaque formation assay
To quantify virus titer after 72 hour SARS-CoV-2 infection dose response on hiPSC-CMs (Figure S1), culture supernatants from each MOI condition were subjected to 10-fold serial dilutions and added to Vero-E6 cells in a 96-well plate. At 48 hours post infection on Vero-E6, the cells were fixed in methanol and subjected to immunostaining using dsRNA antibody. Immunostained viral plaques were counted in the highest viral dilution-containing wells. Plaque forming unit per milliliter (PFU/mL) of culture supernatant was calculated for each condition.
Blocking ACE2 with antibody
The hiPSC-CMs were pre-treated with anti-ACE2 antibody (R&D Systems Cat# AF933, RRID: AB_355722; 20 μg/well in 100 μL media) for 1 hour. Untreated hiPSC-CMs and non-specific antibody (anti-TIM4 antibody, R&D Systems Cat# AF2929, RRID: AB_2240431; 20 μg/well in 100 μL media) treated cells were included as controls. After 1 hour of antibody pre-treatment at 37°C, SARS-CoV-2 viral inoculum (MOI of 0.01) was added to each well. Subsequently at 1-hour post-infection (hpi), virus inoculum was replaced with fresh media. 72 hours later, the cells were fixed with 4% paraformaldehyde for immunofluorescence analysis.
RNA-Sequencing
After 72-hour viral treatment, cells designated for transcriptomic analysis were harvested in Trizol and total RNA extracted using a QIAGEN RNEasy Mini Kit. Total RNA was then used for library preparation and sequencing. The SMART-Seq V4 Ultra Low RNA Input Kit for Sequencing (Takara Bio USA, Inc., Mountain View, CA) was used for reverse transcription and generation of double stranded cDNA for library preparation using the Nextera XT Library Preparation kit (Illumina, San Diego, CA). Total RNA quality was analyzed via the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and RNA quantified using QubitTM (ThermoFisher Scientific, Waltham, MA) fluorometric quantitation. An input of 10 ng total RNA was used for oligo(dT)-primed reverse transcription, cDNA amplification, and cDNA cleanup. The cDNA was analyzed on the 4200 TapeStation (Agilent Technologies). Quantification of cDNA was performed using Qubit. cDNA normalized to 30 pg/mL was fragmented and sequencing primers added simultaneously. A limiting-cycle PCR added Index 1 (i7) adapters, Index 2 (i5) adapters, and sequences required for cluster formation on the sequencing flow cell. Indexed libraries were cleaned up, library size verification performed on the 4200 TapeStation, and library quantified via Qubit. Libraries were sequenced on a NextSeq 500 using with a 1x75 bp read length and coverage of ∼60M reads/cell.
Bioinformatics and data analysis
Raw reads obtained from RNA-Seq were aligned to the transcriptome using STAR (version 2.5.0)35 / RSEM (version 1.2.25)36 with default parameters, using a custom human GRCh38 transcriptome reference downloaded from https://www.gencodegenes.org, containing all protein coding and long non-coding RNA genes based on human GENCODE version 33 annotation. For SARS-CoV-2 viral genome alignment, GenBank: MT246667.1 reference sequence was used. Expression counts for each gene in all samples were normalized by a modified trimmed mean of the M-values normalization method and the unsupervised principal component analysis (PCA) was performed with DESeq2 Bioconductor package version 1.10.1 in R version 3.6.3. Each gene was fitted into a negative binomial generalized linear model, and the Wald test was applied to assess the differential expressions between two sample groups by DESeq2. Benjamini and Hochberg procedure was applied to adjust for multiple hypothesis testing, and differential expression gene candidates were selected with a false discovery rate less than 0.05. For visualization of coordinated gene expression in samples, a two-way hierarchical clustering with Pearson correlation distance matrix was performed with samples and differentially-expressed candidates using the Bioconductor g-plots package (version 3.0.3) in R. Gene ontology analysis and visualization conducted using REViGO online software.37
Beat rate analysis
Thirty-second video clips were taken of mock and infected hiPSC-CMs at 72 hours after viral infection. A beats-per-minute measurement was obtained by manually counting individual beats in a 30 s video and multiplying by 2 to extrapolate to 1 minute.
Quantification and statistical analysis
For statistical analyses, the Student’s t test was used for comparison between two datasets. Excel or Prism software was used for statistical analysis. Data are presented as mean ± standard deviation as indicated in figure legends. A p value of < 0.05 is considered statistically significant and is denoted by ∗. Images used for statistical analysis were randomly chosen within a well of interest. Other statistical details can be found in figure legends. No data were excluded.
Acknowledgments
We acknowledge that this work represents an area of study that is rapidly growing in relevance, and thus we apologize to any authors whose work we were not able to include here. We thank Dr. Soshana Svendsen for editing the manuscript. We thank the Cedars-Sinai Center for Bioinformatics and Functional Genomics for their assistance with RNA sequencing. Research from the Svendsen laboratory has been supported by the National Institutes of Health (5UG3NS105703) and the Cedars-Sinai Board of Governors Regenerative Medicine Institute. A.S. is supported by an institutional training grant (T32 HL116273). The gene expression data from this study are found at Gene Expression Omnibus with accession number GSE150392.
Author Contributions
A.S., V.A., and C.N.S. designed analyses, analyzed data, and drafted the manuscript. A.S. and G.G. conducted experiments and acquired data. Y.W. and J.T.P. assisted with genomics and bioinformatics analysis. K.M. assisted with the ACE2 antibody experiment. A.S., V.A., and C.N.S. analyzed data. All authors contributed to the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: June 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100052.
Contributor Information
Arun Sharma, Email: arun.sharma@cshs.org.
Vaithilingaraja Arumugaswami, Email: varumugaswami@mednet.ucla.edu.
Clive N. Svendsen, Email: clive.svendsen@cshs.org.
Supplemental Information
References
- 1.Ramaiah A., Arumugaswami V. Insights into Cross-species Evolution of Novel Human Coronavirus 2019-nCoV and Defining Immune Determinants for Vaccine Development. bioRxiv. 2020 doi: 10.1101/2020.01.29.925867. [DOI] [Google Scholar]
- 2.Hoffmann, M., Kleine-Weber, H., Schroeder, S., Kruger, N., Herrler, T., and Erichsen, S., Schiergens, T.S., Herrler, G., Wu, N.-H., Nitsche, A., Müller, M.A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280. [DOI] [PMC free article] [PubMed]
- 3.Madjid M., Safavi-Naeini P., Solomon S.D., Vardeny O. Potential Effects of Coronaviruses on the Cardiovascular System: A Review. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1286. Published online: March 27, 2020. [DOI] [PubMed] [Google Scholar]
- 4.Fried J.A., Ramasubbu K., Bhatt R., Topkara V.K., Clerkin K.J., Horn E., Rabbani L., Brodie D., Jain S.S., Kirtane A.J. The Variety of Cardiovascular Presentations of COVID-19. Circulation. 2020;141:1930–1936. doi: 10.1161/CIRCULATIONAHA.120.047164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q. Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.0950. Published online: March 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116:1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker N.R., Chaffin M., Bedi K.C., Papangeli I., Akkad A.-D., Arduini A., Hayat S., Eraslan G., Muus C., Bhattacharyya R. Myocyte Specific Upregulation of ACE2 in Cardiovascular Disease: Implications for SARS-CoV-2 mediated myocarditis. medRxiv. 2020 doi: 10.1161/CIRCULATIONAHA.120.047911. doi: 2020.04.09.20059204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oudit G.Y., Kassiri Z., Jiang C., Liu P.P., Poutanen S.M., Penninger J.M., Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu H., Ma F., Wei X., Fang Y. Coronavirus fulminant myocarditis saved with glucocorticoid and human immunoglobulin. Eur. Heart J. 2020 doi: 10.1093/eurheartj/ehaa190. ehaa190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng J.H., Liu Y.X., Yuan J., Wang F.X., Wu W.B., Li J.X., Wang L.F., Gao H., Wang Y., Dong C.F. First case of COVID-19 complicated with fulminant myocarditis: a case report and insights. Infection. 2020 doi: 10.1007/s15010-020-01424-5. Published online April 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavazzi G., Pellegrini C., Maurelli M., Belliato M., Sciutti F., Bottazzi A., Sepe P.A., Resasco T., Camporotondo R., Bruno R. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur. J. Heart Fail. 2020;22:911–915. doi: 10.1002/ejhf.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2011400. Published online May 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A., Sances S., Workman M.J., Svendsen C.N. Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell. 2020;26:309–329. doi: 10.1016/j.stem.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma A., Toepfer C.N., Ward T., Wasson L., Agarwal R., Conner D.A. CRISPR/Cas9-Mediated Fluorescent Tagging of Endogenous Proteins in Human Pluripotent Stem Cells. Curr. Protoc. Hum. Genet. 2018;96:21.11.1–21.11.20. doi: 10.1002/cphg.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churko J.M., Garg P., Treutlein B., Venkatasubramanian M., Wu H., Lee J., Wessells Q.N., Chen S.Y., Chen W.Y., Chetal K. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat. Commun. 2018;9:4906. doi: 10.1038/s41467-018-07333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toepfer C.N., Sharma A., Cicconet M., Garfinkel A.C., Mücke M., Neyazi M., Willcox J.A.L., Agarwal R., Schmid M., Rao J. SarcTrack. Circ. Res. 2019;124:1172–1183. doi: 10.1161/CIRCRESAHA.118.314505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun N., Yazawa M., Liu J., Han L., Sanchez-Freire V., Abilez O.J., Navarrete E.G., Hu S., Wang L., Lee A. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A., Burridge P.W., McKeithan W.L., Serrano R., Shukla P., Sayed N., Churko J.M., Kitani T., Wu H., Holmström A. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 2017;9:eaaf2584. doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burridge P.W., Li Y.F., Matsa E., Wu H., Ong S.G., Sharma A., Holmström A., Chang A.C., Coronado M.J., Ebert A.D. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A., Marceau C., Hamaguchi R., Burridge P.W., Rajarajan K., Churko J.M., Wu H., Sallam K.I., Matsa E., Sturzu A.C. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ. Res. 2014;115:556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laperle A.H., Sances S., Yucer N., Dardov V.J., Garcia V.J., Ho R., Fulton A.N., Jones M.R., Roxas K.M., Avalos P. iPSC modeling of young-onset Parkinson’s disease reveals a molecular signature of disease and novel therapeutic candidates. Nat. Med. 2020;26:289–299. doi: 10.1038/s41591-019-0739-1. [DOI] [PubMed] [Google Scholar]
- 25.Sharma A., Li G., Rajarajan K., Hamaguchi R., Burridge P.W., Wu S.M. Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J. Vis. Exp. 2015 doi: 10.3791/52628. Published online: March 18, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J., de Haan C.A. Visualizing coronavirus RNA synthesis in time by using click chemistry. J. Virol. 2012;86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versteeg G.A., van de Nes P.S., Bredenbeek P.J., Spaan W.J. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;81:10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaker S.K., Ch’ng J., Christofk H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019;17:59. doi: 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 30.Roden D.M., Harrington R.A., Poppas A., Russo A.M. Considerations for Drug Interactions on QTc in Exploratory COVID-19 (Coronavirus Disease 2019) Treatment. Circulation. 2020;141:e906–e907. doi: 10.1161/CIRCULATIONAHA.120.047521. [DOI] [PubMed] [Google Scholar]
- 31.Bedada F.B., Wheelwright M., Metzger J.M. Maturation status of sarcomere structure and function in human iPSC-derived cardiac myocytes. Biochim. Biophys. Acta. 2016;1863(7 Pt B):1829–1838. doi: 10.1016/j.bbamcr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaBarre D.D., Lowy R.J. Improvements in methods for calculating virus titer estimates from TCID50 and plaque assays. J. Virol. Methods. 2001;96:107–126. doi: 10.1016/s0166-0934(01)00316-0. [DOI] [PubMed] [Google Scholar]
- 35.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Thirty-second video clips (in triplicate) were taken of mock and infected hiPSC-CMs at 72 h after viral infection. Cells were infected at MOI 0.1.
Data Availability Statement
The gene expression data from this study are found at Gene Expression Omnibus with accession number GSE150392.