Abstract
Genome sequencing and bioinformatics tools have facilitated the identification and expression of an increasing number of cryptic biosynthetic gene clusters (BGCs). However, functional analysis of all components of a metabolic pathway to precisely determine biocatalytic properties remains time-consuming and labor intensive. One way to speed this process involves microscale cell-free protein synthesis (CFPS) for direct gene to biochemical function analysis, which has rarely been applied to study multicomponent enzymatic systems in specialized metabolism. We sought to establish an in vitro transcription/translation (TT)-assay to assess assembly of cyanobacterial-derived hapalindole-type natural products (cNPs) because of their diverse bioactivity profiles and complex structural diversity. Using a CFPS system including a plasmid bearing famD2 prenyltransferase from Fischerella ambigua UTEX 1903, we showed production of the central prenylated intermediate (3GC) in the presence of exogenous geranyl-pyrophosphate (GPP) and cis-indole isonitrile. Further addition of a plasmid bearing the famC1 Stig cyclase resulted in synthesis of both FamD2 and FamC1 enzymes, which was confirmed by proteomics analysis, and catalyzed assembly of 12-epi-hapalindole U. Further combinations of Stig cyclases (FamC1–C4) produced hapalindole U and hapalindole H, while FisC identified from Fischerella sp. SAG46.79 generated 12-epi-fischerindole U. The CFPS system was further employed to screen six unnatural halogenated cis-indole isonitrile substrates using FamC1 and FisC, and the reactions were scaled-up using chemoenzymatic synthesis and identified as 5- and 6-fluoro-12-epihapalindole U, and 5- and 6-fluoro-12-epi-fischerindole U, respectively. This approach represents an effective, high throughput strategy to determine the functional role of biosynthetic enzymes from diverse natural product BGCs.
Keywords: biosynthetic gene cluster (BGC), cell-free protein synthesis (CFPS), hapalindole, fischerindole, in vitro TT-assay
Graphical Abstract
Natural products (NPs) continue to be invaluable molecules for developing front-line drugs against infectious diseases, parasites, and cancers.1,2 Current advances in bioinformatics tools for mining genomic and metagenomic data have facilitated our ability to rapidly decode biosynthetic pathways to identify and exploit novel structural diversity. Heterologous expression and refactoring of NP biosynthetic gene clusters (BGCs), is also unlocking cryptic pathways and expanding our ability to discover novel molecules of pharmaceutical interest.1,3–7 It is also evident that evolutionary processes give rise to subtle DNA sequence variations8 especially in the complex multicomponent enzymatic systems including bacterial polyketide synthases (PKSs), nonribosomal polyketide synthases (NRPSs),9 the ribosomally synthesized and post-translationally modified peptides (RiPPs),10,11 and cyanobacterial indole alkaloid systems,12–17 leading to diverse product profiles. A remaining challenge is to rapidly predict from genome sequencing data the likely change in catalytic function and metabolite profile alterations mediated by closely related biosynthetic enzymes. In addition, defining the functional consequences continues to require rigorous, time-consuming biochemical experiments.
It is often impractical to express and purify all components of a biosynthetic pathway and corresponding families of related proteins to decipher biochemical and functional properties. Thus, cell-free protein synthesis (CFPS) has emerged18,19 with direct synthetic biology applications including production of metalloenzymes,20–22 biosynthesis of mevalonate23 and limonine,24 assembly of bioplastics,25 NP biosynthesis from NRPS,26 characterizing and prototyping genetic networks,27,28 and most recently for the study of modular construction of protein glycosylation pathways.29 In the current study, we decided to explore Stig cyclases using microscale CFPS as (1) an alternative approach for screening hapalindole-related biosynthetic enzyme function, and (2) as a means to rapidly assess production of new hapalindoles using synthetic unnatural substrates.
Cyanobacterial indole alkaloid BGCs represent a multi-component enzyme system that biosynthesizes hapalindole-type metabolites as we and others have shown. These molecules display a diverse array of stereochemical configurations at C10, C11, C12, and C15 chiral centers. This region and stereochemical result of Stig cyclases and additional late-stage enzymes are responsible for the production of more than 80 hapalindoles, fischerindoles, ambiguines, and welwitindolinones that exhibit antibacterial, antimycotic, insecticidal, and cytotoxic activities.30–33 Although each hapalindole-type metabolite subgroup has a distinct ring system, they are derived from a common intermediate 3 comprising a geranyl monoterpene unit 1 and a cis-indole isonitrile core 2 (Figure 1). Our recent studies have established the detailed steps of assembly and reaction mechanisms involved in hapalindole core formation (Figure 1).12,14–16,34–37 Hapalindole biosyn-thesis proceeds by combining cis-indole isonitrile 2 with geranyl-pyrophosphate (GPP) 1 to form cis-indole isonitrile GPP (3GC) intermediate 3. This unstable intermediate is cyclized by a specific Stig cyclase to produce the hapalindole or fischerindole core molecules.14,34–40 (Figure 1). Following formation of the initial tetracycle (and in some cases the hapalindole tricycle) the alkaloid ring contains four chiral centers, and may undergo late-stage tailoring reactions (e.g., halogenation, oxidation, prenylation) leading to formation of the ambiguines and welwitindolinones.15,41,42 Although 3GC 3 is a common intermediate, strain specific Stig cyclases catalyze formation of site- and stereoselective hapalindoles and fischerindoles. For instance, the biosynthetic gene cluster (fam) of Fischerella ambigua UTEX 1903 produces 12-epihapalindole U (C10S, 11R, 12S, and 15S) 4, hapalindole U (C10S, 11R, 12R, and 15S) 5, and hapalindole H (C10R, 11R, 12R, and 15R) 6 by the combination of four cyclase genes (famC1–C4)34 (Figure 1). However, when reacted with 3, the single cyclase gene (fisC) identified in the Fischerella sp. SAG46.79 BGC generated 12-epi-fischerindole U (C10S, 11R, 12S, and 15S) 7 and 12-epi-hapalindole C (C10S, 11R, 12S, and 15S) 8 as major and minor products, respectively34 (Figure 1).
Figure 1.
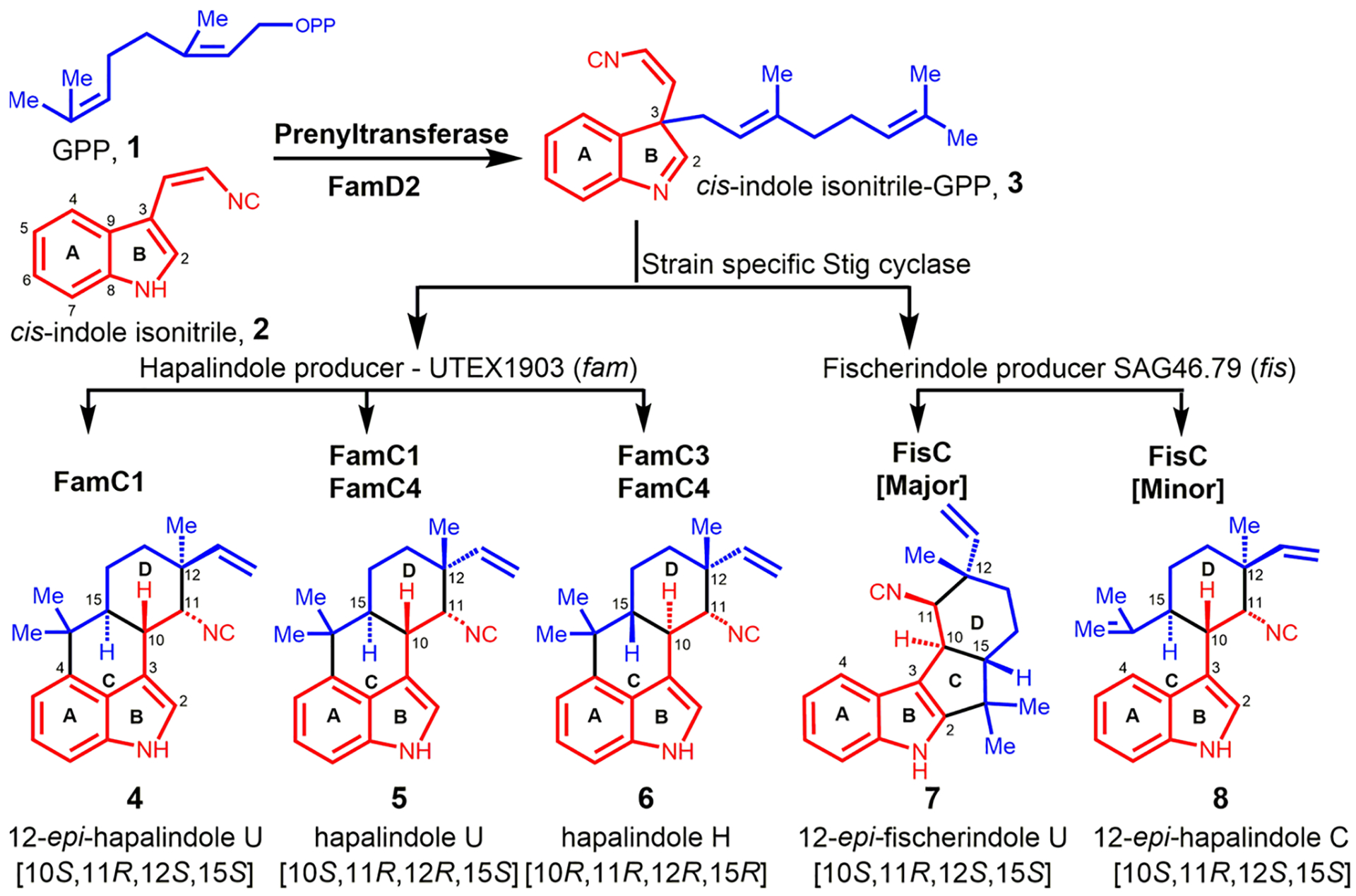
Biogenesis of cyanobacterial-derived hapalindole and fischerindole metabolites.
In this study, we sought to expand the application of CFPS for cyanobacterial NPs representing a multicomplex enzyme system (Figure 2). Two cyanobacterial derived indole alkaloid BGCs were selected, including Fischerella ambigua UTEX 190334 (hapalindole producer) and Fischerella sp. SAG 46.7914 (fischerindole producer). As a first step, we demonstrated that CFPS is able to express plasmid-borne FamD2 to synthesize 3GC intermediate 3 using native substrates 1 and 2. Intermediate 3 is subsequently cyclized in a three-step reaction by the combination of strain specific Stig cyclases FamC1-C4 and FisC to afford the corresponding fused ring system either at the 3,4- (hapalindole) or 2,3-position (fischerindole) of the indole ring, respectively (Figure 1). These data were supported by proteomics identification of the CFPS derived prenyltransferase (FamD2) and cyclase (FamC/FisC) enzymes. The in vitro transcription-translation assay (TT-assay) was further employed to screen a series of unnatural halogenated cis-indole isonitrile derivatives to generate new hapalindole and fischerindole metabolites using a second plasmid bearing FamC1 or FisC, respectively. The new indole alkaloid molecules were confirmed by MS and NMR following chemoenzymatic scale-up (Figure 2). This approach represents the first example of in vitro TT applied to a multicomponent pathway to generate a fully elaborated NP. With the rapidly growing natural product BGC database, we envision multiple applications for rapid functional analysis of biosynthetic enzymes, and substrate profiling to create and assess natural product structural diversity.
Figure 2.
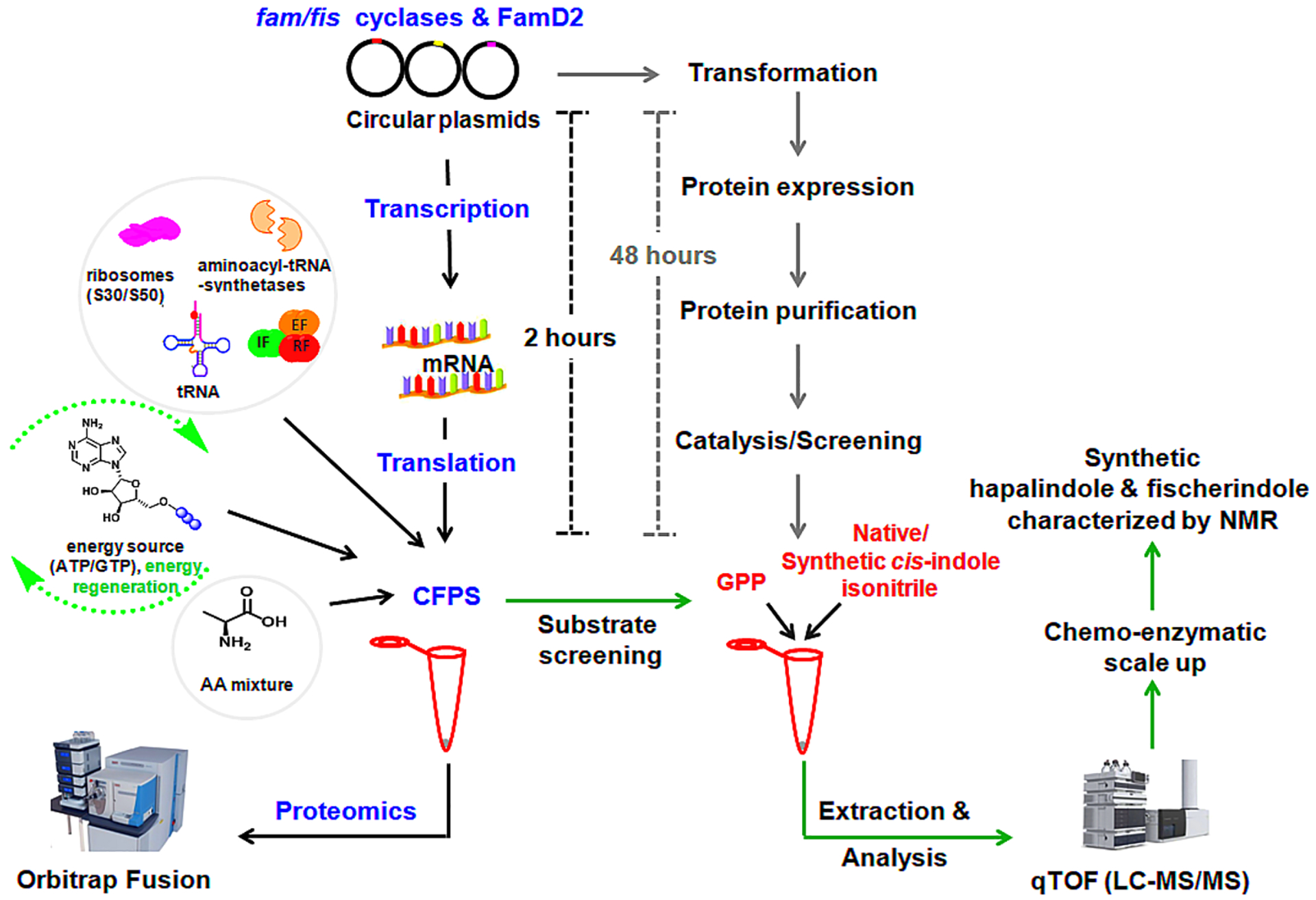
Overview of cell-free protein synthesis (CFPS) using exogenous plasmids for the synthesis of prenyltransferase and Stig cyclases, identification of expressed protein by proteomics analysis, and detection and analysis of secondary metabolites. The time-line of CFPS is compared with the traditional protein expression, purification, and catalysis.
RESULT AND DISCUSSION
Development of an In Vitro TT-Assay for the Biosynthesis of Hapalindole and Fischerindole Metabolites.
To establish an in vitro transcription–translation platform for hapalindole biosynthesis in Fischerella ambigua UTEX 1903, individual plasmids containing genes famD2, famC1, famC2, famC3, and famC4 were cloned into the pET28a or pET28H8T systems as described.14,34 Geranylated cis-indole isonitrile 3 is the key intermediate for subsequent formation of the tetracyclic core ring system of the hapalindoles.34 Thus, our in vitro TT-assay was performed using a plasmid bearing the aromatic prenyltransferase FamD2 in the presence of 1 and 2, with transcription and translation mediated through the T7 promoter (PureExpress system). Proteomics analysis of the in vitro TT-assay showed 25 unique FamD2 peptides covering 72.49% of the total amino acids sequence (Supporting Information Table S1 and Excel sheet 1), providing strong evidence for efficient in vitro expression of FamD2. LC–MS analysis of the extracted reaction products ([M + H]+ = 305) possessed identical retention time (tR) and MS-pattern compared to authentic 3 (Figure 3B, Figure S1B). Although enzymatic prenylation of 2 with GPP showed the production of geranylated C2 and C3 isocyano-indoles under pH dependent conditions favoring 3 at basic pH;34,36 LC–MS analysis of the TT-assay reaction showed no detectable formation of the C2 form as the reaction was performed in the presence of Tris-buffer at pH 7.8.
Figure 3.
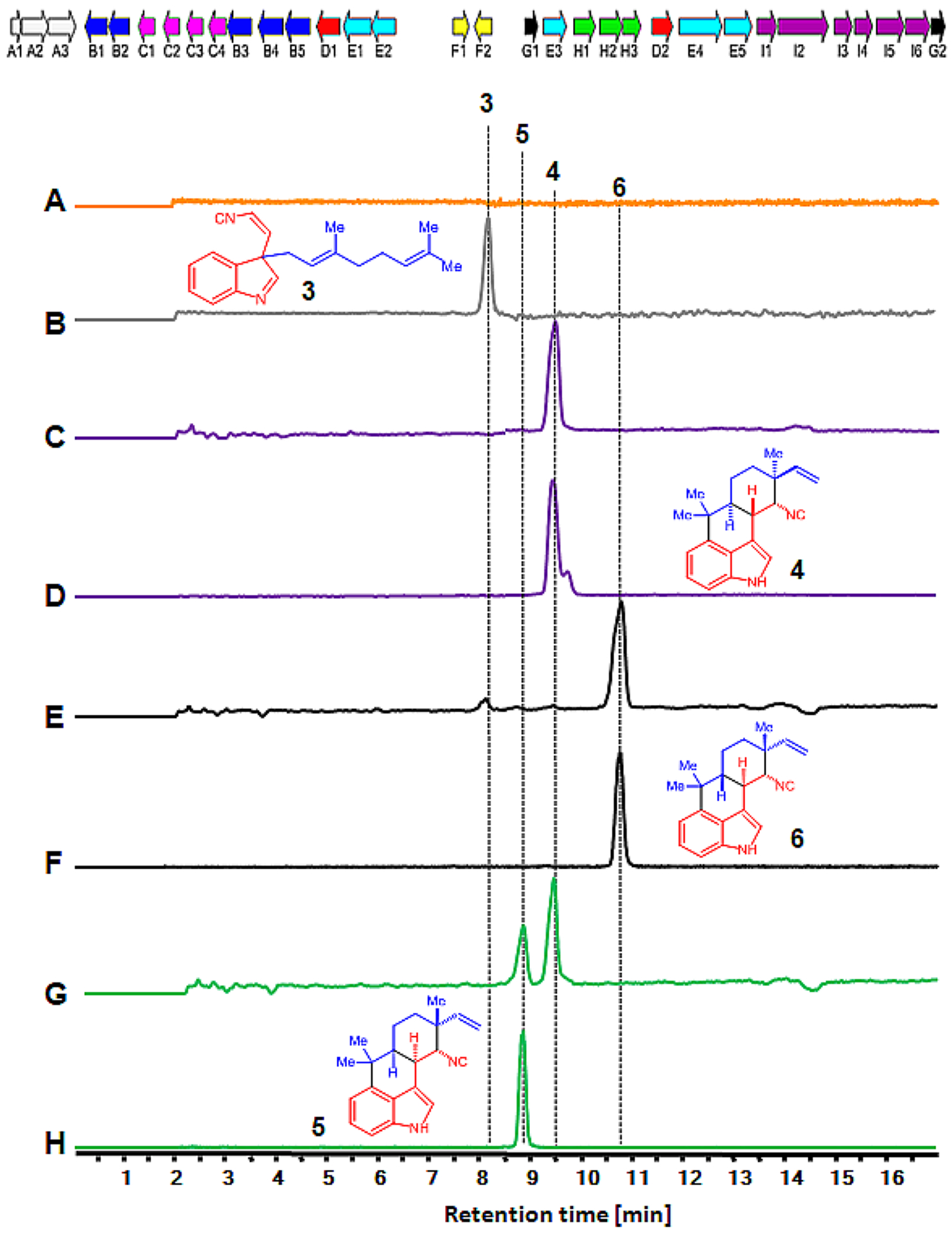
In vitro TT-assay and production of hapalindoles. Top: Biosynthetic gene cluster of Fischerella ambigua UTEX 1903 (fam). The genes for transporters (white, A1–3), oxygenases (blue, B1–6), cyclases (pink, C1–5), aromatic prenyltransferases (red, D1–2), GPP synthases (cyan, E1–5), regulators (yellow, F1–2), cis-indole isonitrile synthase (green, H1–3) and other core enzymes (black, G1–2) are shown. Bottom: Extracted ion chromatograms (EICs) are shown in all panels. Detection of in vitro synthesized hapalindole products showing EIC (m/z of 305) with no plasmid (A), FamD2 alone (B) for cis-indole isonitrile 3, FamD2 plus FamC1 (C) for 12-epi-hapalindole U 4, FamD2 plus FamC2 and FamC3 (E) for hapalindole H 6, and FamD2 plus FamC1 and FamC4 (G) for hapalindole U 5, and 12-epi-hapalindole U 4 production. The traces D, F, and H are the authentic standards of 12-epi-hapalindole U 4, hapalindole H 6, and hapalindole U 5, respectively.
As a next step to test the cyclization of 3 using Stig cyclases (FamC1-C4), we tested equal concentrations of FamC1 and FamD2 plasmids for the in vitro TT-assay incubated in the presence of 1 and 2. Proteomics analysis of the reaction revealed 20 and 11 unique peptides covering 67.96% and 62.11% of the total protein sequences for FamD2 and FamC1, respectively (Table 1, Excel sheet 2). LC–MS analysis of the TT-assay demonstrated the formation of a metabolite with m/z of 305, and its tR and MS-pattern were identical to the standard 12-epi-hapalindole U 4 (Figure 3C, Figure S1C).
Table 1.
Proteomics Analysis of FamD2 and FamC1 Expression from in vitro TT-Assay Containing Exogenous Plasmids FamD2 and FamC1, and Substrates 1 and 2 for 12-epi-Hapalindole U 4 Biosynthesisa
| FamD2 | FamC1 | ||||
|---|---|---|---|---|---|
| Total peptide | 20 | Total peptide | 11 | ||
| Unique peptide | 20 | Unique peptide | 11 | ||
| Coverage (%) | 67.96 | Coverage (%) | 62.11 | ||
| Annotated Peptide Sequence | Sequence Positions | PSM FamD2 | Annotated Peptide Sequence | Sequence Position | PSM FamC1 |
| [R].IRTDIINVAK.[T] | FamD2 [7–16] | 10 | [K].RTTWTSNNGVGYVGPGTQFYNQLAPEGR.[N] | FamC1 [70–97] | 2 |
| [R].TDIINVAK.[T] | FamD2 [9–16] | 6 | [R].TTWTSNNGVGYVGPGTQFYNQLAPEGR.[N] | FamC1 [70–97] | 8 |
| [K].TFGAEYSEK.[V] | FamD2 [17–25] | 3 | [R].NIGYIYLSQNPGSGVAGFEQILDATLEPDTK.[Y] | FamC1 [98–128] | 35 |
| [K].VLDEVFQVFGEQFADNSFMIR.[T] | FamD2 [26–16] | 19 | [K].YTLTVDVGNLAGTFK.[G] | FamC1 [129–143] | 87 |
| [R].TSNKQPDK.[L] | FamD2 [47–54] | 1 | [K]. GLSFAGFPGYR. [V] | FamC1 [144–154] | 19 |
| [K].LGCYFR.[Y] | FamD2 [55–60] | 3 | [R].VELLAGDTVLAADHNNLFIK.[E] | FamC1 [155–174] | 306 |
| [R].YHEEDESQLGLAWDIAR.[K] | FamD2 [61–77] | 4 | [K].TSTVTYTSTAK. [D] | FamC1 [180–190] | 11 |
| [R].KSGLLSDQGRPVDQLIPEICETFPIMADGVDFDVK.[H] | FamD2 [78–112] | 6 | [K].DLHLGQK.[L] | FamC1 [191–197] | 20 |
| [K].SGLLSDQGRPVDQLIPEICETFPIMADGVDFD VK. [H] | FamD2 [79–112] | 8 | [R].LVNLLQDK.[F] | FamC1 [202–209] | 2 |
| [K].IWQSIK.[G] | FamD2 [118–123] | 3 | [K].FSGLDFDNVR.[L] | FamC1 [210–219] | 15 |
| [K].GWPVQDAFK.[L] | FamD2 [124–133] | 5 | [R].LVNLLQDKFSGLDFDNVR. [L] | FamC1 [202–219] | 2 |
| [K].LSLPASVTTHSDFLK.[N] | FamD2 [134–148] | 7 | |||
| [R].LCFYLPFLNR.[E] | FamD2 [224–233] | 2 | |||
| [R].EAVPQNLLNPLLK.[K] | FamD2 [234–246] | 1 | |||
| [R].EAVPQNLLNPLLKK.[Y] | FamD2 [234–247] | 2 | |||
| [K].KYINEAPALVDNPGFILGWSFGPQGGK.[G] | FamD2 [247–273] | 4 | |||
| [K].YINEAPALVDNPGFILGWSFGPQGGK.[G] | FamD2 [248–273] | 2 | |||
| [K].VDVDYHGR.[T] | FamD2[279–286] | 1 | |||
| [R].TVPLFMK.[V] | FamD2 [287–293] | 1 | |||
| [K].VHSQPLPK.[A] | FamD2 [294–301] | 2 | |||
| FamD2 | FamC1 | ||||
| MNDVNRIRTDIINVAKTFGAEYSEKVLDEVFQVFGEQFADNSFMIRTSNKQPDKLGCYFRYHEEDESQLGLAWDIARKSGLLSDQGRPVDQLIPEICETFPIMADGVDFDVKHGLAKIWQSIKGWPVQDAFKLSLPASVTTHSDFLKNHHLDALYAFGIDYHHSSVNLYFDTYHPKHHTSEYYKNLLQDLQFQPPSDELLELLTNNGEIALTFNFASPRIERLCFYLPFLNREAVPQNLLNPLLKKYINEAPALVDNPGFILGWSFGPQGGKGTYTKVDVDYHGRTVPLFMKVHSQPLPKAADFALAQ, 309 residues | MKRKLIVAVWLIFICLGINTPAHATSAVSIPINNAGFENPFMDWDDYTIDTPPGWTTYDPNNLVPEKRTTWTSNNGVGYVGPGTQFYNQLAPEGRNIGYIYLSQNPGSGVAGFEQILDATLEPDTKYTLTVDVGNLAGTFKGLSFAGFPGYRVELLAGDTVLAADHNNLFIKEGEFKTSTVTYTSTAKDLHLGQKLGIRLVNLLQDKFSGLDFDNVRLTTEPTET, 227 residues | ||||
The sequence highlighted in yellow represents the coverage of the synthesized protein using proteomic analysis.
We have shown that the chemoenzymatic reaction using FamD2 and FamC1 proteins also converts intermediate 3 into 4 through the three step process of (1) Cope rearrangement43 that sets the stereochemistry at positions C11 and C12, followed by (2) a 6-exo-trig cyclization introducing stereo-chemistry at positions C10 and C15 and (3) terminal electrophilic aromatic substitution (C-ring formation) to generate 12-epi-hapalindole U 4.35 Our current TT-assay using two plasmids (FamD2 and FamC1) enabled microscale assembly of 4 in the absence of cell-based protein production. We have demonstrated previously that the function of alternative homodimeric and heterodimeric Stig cyclases controls the relative stereochemistry of hapalindoles and fischerindoles during chemoenzymatic synthesis.14,34,35 Thus, we sought to ascertain the ability of in vitro TT-assay to process multiple Stig cyclase plasmids to synthesize heterodimeric, functional pools of these enzymes.
While performing CFPS of FamD2, FamC1, and FamC4 plasmids containing exogenous 1 and 2 in the presence of supplementary calcium (20 mM), the synthesized heterodimeric cyclases showed production of hapalindole U 5 and predominantly 12-epi-hapalindole U 4, which were confirmed by LC–MS and correlated with authentic standards (Figure 3G, Figure S1G). However, the enzymatic reaction with the purified FamC1 and FamC4 under the same conditions showed the reverse product profile with 5 as the predominate product. We reasoned that the stoichiometry of FamC1 is relatively high compared to the calcium-dependent FamC4 resulting in increased production of 4 relative to 5 in the TT-assay. The heterodimerization of FamC1 and FamC4 has been shown previously to produce hapalindole U 5.39 To study further the effect of alternative cyclases on stereocontrol of hapalindole molecules, we tested FamC2 and FamC3 combined with FamD2 (to form central intermediate 3 using exogenous 1 and 2) using supplementary calcium (20 mM). This combination resulted in formation of hapalindole H 6 (Figure 3E), which was confirmed by an authentic standard (Figure S1E) and corroborates our earlier findings using chemoenzymatic synthesis.14 In this multiplasmid format, we found that a lower amount of calcium ions resulted in failed hapalindole production (Figure S1D,F), which further confirms the dependence of Stig cyclase heterodimerization and catalysis on this metal cofactor.
Following conclusive demonstration of hapalindole-type core ring system production by CFPS, we sought to employ FisC cyclase from Fischerella sp. SAG 46.79 to generate the fischerindole core (Figure 1). Although the biosynthetic gene cluster of this strain contains only one cyclase (FisC) and a single prenyltransferase (FisD, a homologue of FamD2), it is known to produce 12-epi-fischerindole U 7, having the same core stereochemistry (C10S, 11R, 12S, 15S) of 12-epihapalindole U 4, bearing the indole C2/3 ring connection rather than C3/4.14,35 In this case, the in vitro TT-assay was designed using two plasmids encoding individually FamD2 and FisC, and supplying exogenous 1 and 2. Proteomics analysis of the in vitro TT-assay product showed 22 and 11 unique peptides covering 70.55% and 78.07% of FamD2 and FisC amino acids sequences, respectively (Table 2, Excel data 3), demonstrating in vitro production of the prenyltransferase and cyclase enzymes. The LC–MS analysis of the reaction showed production of 12-epi-fischerindole U 7, which matched an authentic standard (Figure 4, Figure S1G). This result also correlated with our previous results derived from chemoenzymatic synthesis of 12-epi-fischerindole U 7.14
Table 2.
Proteomics Analysis of FamD2 and FisC Expression from in Vitro TT-Assay Containing Exogenous Plasmids FamD2 and FisC, and the Substrates 1 and 2 for 12-epi-Fischerindole U 8 Biosynthesisa
| FamD2 | FisC | ||||
|---|---|---|---|---|---|
| Total peptide | 22 | Total peptide | 11 | ||
| Unique peptide | 22 | Unique peptide | 11 | ||
| Coverage (%) | 70.55 | Coverage (%) | 78.07 | ||
| Annotated Peptide Sequence | Sequence Positions | PSM FamD2 | Annotated Peptide Sequence | Sequence Position | PSM FisC |
| [R].IRTDITNVAK.[T] | FamD2 [7–16] | 8 | [K].NAGFEEPSLTVEDYYTIDTPPGWITYDPNGLVPAK.[R] | FisC [36–70] | 8 |
| [R].TDIINVAK.[T] | FamD2 [9–16] | 6 | [K].NAGFEEPSLTVEDYYTIDTPPGWITYDPNGLVPAKR.[T] | FisC [36–71] | 1 |
| [K].TFGAEYSEK.[V] | FamD2 [17–25] | 10 | [R].ITSNNGVGYTGPNSAYYNHK.[A] | FisC [74–93] | 110 |
| [K].VLDEVFQVFGEQFADNSFMIR.[T] | FamD2 [26–46] | 5 | [R].ITSNNGVGYTGPNSAYYNHKAPEGR.[N] | FisC [74–98] | 6 |
| [R].TSNKQPDK.[L] | FamD2 [47–54] | 5 | [R].NVAYVYLAQEIGSGIAGLEQTLDAVLKPNTK.[Y] | FisC [99–129] | 1 |
| [R].TSNKQPDKLGCYFR.[Y] | FamD2 [47–60] | 1 | [K].YTLTVDIGNSGGSFQGFPLDGFPGYR.[V] | FisC [130–155] | 2 |
| [K].LGCYFR.[Y] | FamD2 [55–60] | 5 | [R].VELLAGDTVLAADQNNLYIK.[E] | FisC [156–175] | 107 |
| [R].YHEEDESQLGLAWDIAR.[K] | FamD2 [61–77] | 30 | [K].TTTVTFIATPESPYLGQHLGIR.[L] | FisC [181–202] | 32 |
| [R].KSGLLSDQGRPVDQLIPEICETFPIMADGVDFDYK.[H] | FamD2 [78–112] | 4 | [R].LINPLQGIC.[F] | FisC [203–210] | 27 |
| [K].SGLLSDQGRPVDQLIPEICETFPIMADGVDFDVK.[H] | FamD2 [79–112] | 2 | [R].LINPLQGKFSGVDFDNVR.[L] | FisC [203–220] | 1 |
| [K].IWQSIK.[G] | FamD2 [118–123] | 5 | [K].FSGVDFDNVR.[L] | FisC [211–220] | 20 |
| [K].GWPVQDAFKL.[L] | FamD2 [124–133] | 8 | |||
| [K].LSLPASVTTHSDFLK.[N] | FamD2 [134–148] | 8 | |||
| [K].HHTSEYYK.[N] | FamD2 [178–185] | 1 | |||
| [R].LCFYLPFLNR.[E] | FamD2 [224–233] | 1 | |||
| [R].EAVPQNLLNPLLK.[K] | FamD2 [234–246] | 15 | |||
| [R].EAVPQNLLNPLLKK.[Y] | FamD2 [234–247] | 4 | |||
| [K].KYINEAPALVDNPGFILGWSFGPQGGK.[G] | FamD2 [247–273] | 1 | |||
| [K].YINEAPALVDNPGFILGWSFGPQGGK.[G] | FamD2 [248–273] | 2 | |||
| [K].VDVDYHGR.[T] | FamD2 [279–286] | 1 | |||
| [R].TVPLFMK.[V] | FamD2 [287–293] | 10 | |||
| [K].VHSQPLPK.[A] | FamD2 [294–301] | 4 | |||
| FamD2 | FisC | ||||
| MNDVNRIRTDIINVAKTFGAEYSEKVLDEVFQVFGEQFADNSFMIRTSNKQPDKLGCYFRYHEEDESQLGLAWDIARKSGLLSDQGRPVDQLIPEICETFPIMADGVDFDVKHGLAKIWQSIKGWPVQDAFKLSLPASVTTHSDFLKNHHLDALYAFGIDYHHSSVNLYFDTYHPKHHTSEYYKNLLQDLQFQPPSDELLELLTNNGEIALTFNFASPRIERLCFYLPFLNREAVPQNLLNPLLKKYINEAPALVDNPGFILGWSFGPQGGKGTYTKVDVDYHGRTVPLFMKVHSQPLPKAADFALAQ, 309 residues | MKRNFIIAAIVLLVYICFGISISANAASAVSIPIKNAGFEEPSLTVEDYYTIDTPPGWITYDPNGLVPAKRTRITSNNGVGYTGPNSAYYNHKAPEGRNVAYVYLAQEIGSGIAGLEQTLDAVLKPNTKYTLTVDIGNSGGSFQGFPLDGFPGYRVELLAGDTVLAADQNNLYIKEKDFKTTTVTFIATPESPYLGQHLGERLINPLQGKFS GVDFDNVRTAEPAET, 228 residues | ||||
The sequence highlighted in yellow shows the coverage of the corresponding desired translation products. PSM represents the Total peptide to spectrum match.
Figure 4.
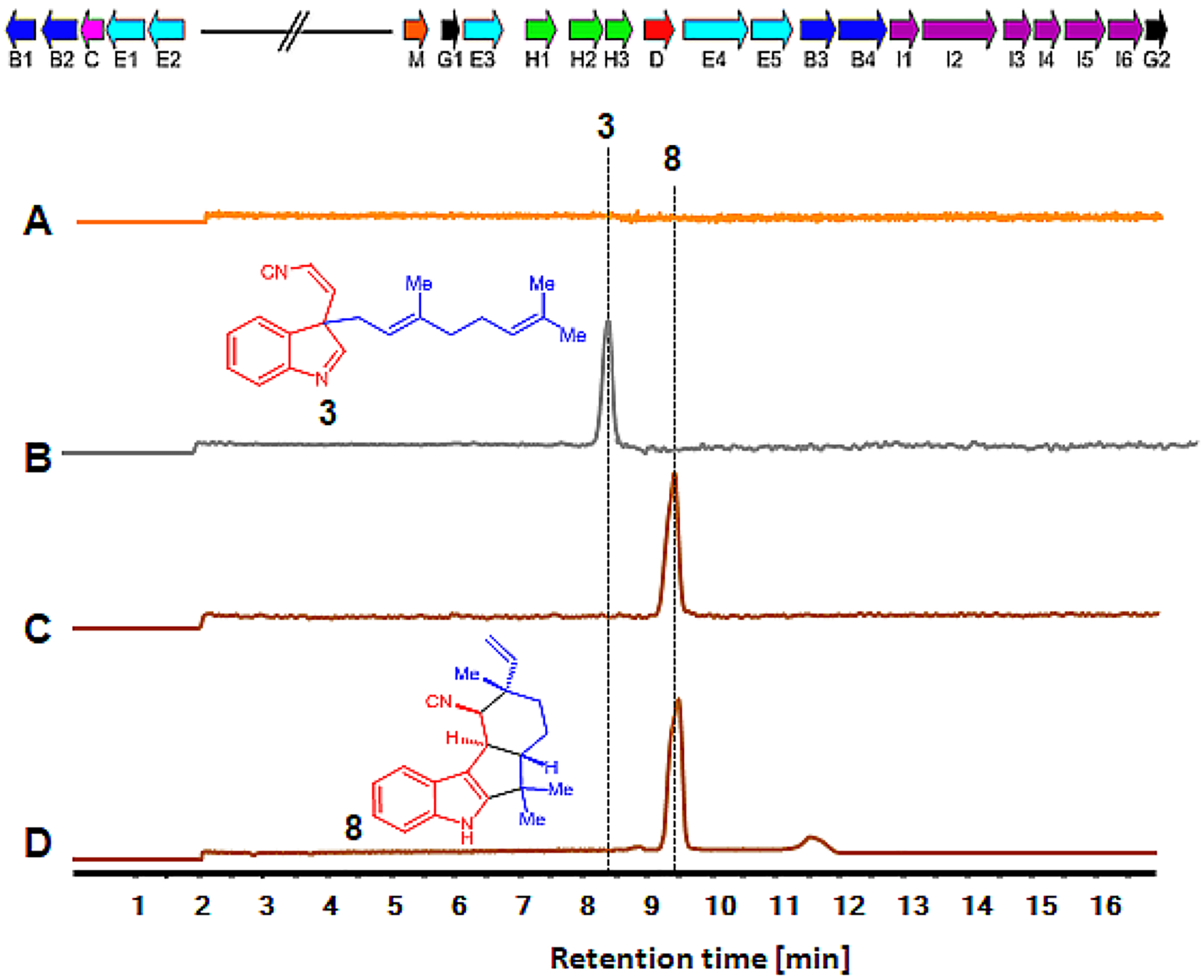
In vitro TT-assay and production of fischerindole. Top: Biosynthetic gene cluster of Fischerella sp. SAG 46.79 (fis). The genes for oxygenases (blue, B1–4), cyclase (pink, C), aromatic prenyltransferases (red, D), GPP synthases (cyan, E1–5), methyl transferase (red, M), tryptophan synthase (magenta, I1–6), isonitrile synthase (green, H1–3) and other core enzymes (black, G1–2) are shown. Bottom: Extracted ion chromatograms (EICs) are shown in all panels. Detection of in vitro synthesized fischerindole products showing EIC (m/z of 305) with no plasmid (A), FamD2 alone (B) for cis-indole isonitrile 3, and FamD2 plus FisC (C) for 12-epi-fischerindole U 8 production. The trace D is the authentic standards of 12-epi-fischerindole U 8.
Generation of Unnatural Hapalindoles.
Once we were able to employ CFPS to produce prenyltransferase and cyclase enzymes for assembly of C3-geranylated indole isonitrile 3 for hapalindole and fischerindole assembly, we sought to test this system as a tool for rapid screening of non-native substrates. Toward this end, we synthesized a series unnatural substrates including 5- and 6-halogenated cis-indole isonitrile derivatives, specifically 5-fluoro-9, 6-fluoro-10, 6-chloro-11, 5-bromo-12, 6-bromo-13, and 5-iodo-cis-indole isonitrile 14 (Figure 5). For these experiments, we employed the in vitro TT-assay to assess the unnatural cis-indole isonitriles using plasmids encoding FamC1 and FisC individually with the goal of generating the corresponding unnatural hapalindoles/fischerindoles linked at C3/4 or C2/3, respectively. The metabolite profile for each reaction was initially determined using mass spectrometry, and halogenated products were further characterized by chemoenzymatic scale-up methods for full NMR structure elucidation.
Figure 5.
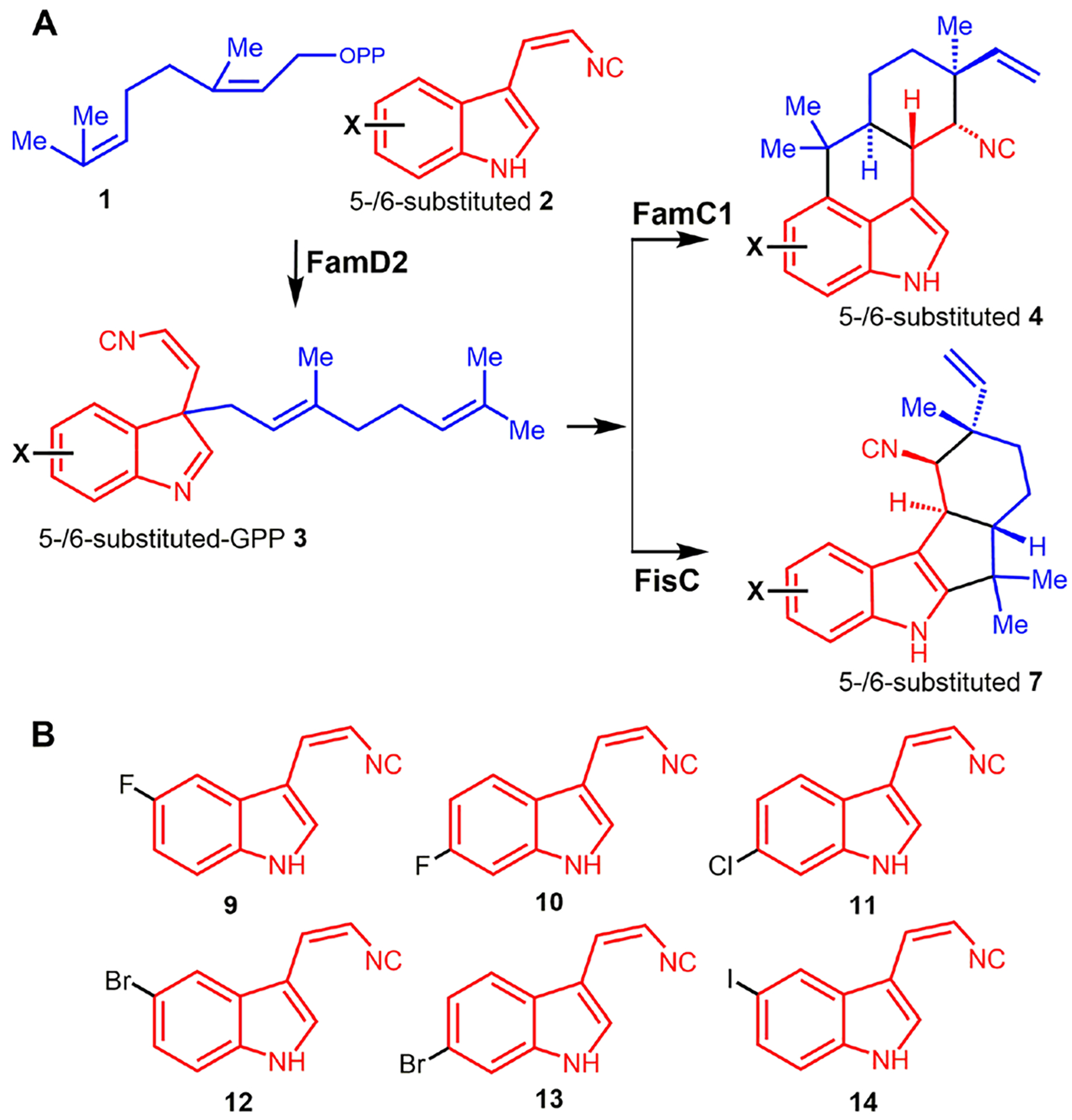
(A) Scheme showing the screening of unnatural 5- or 6- halogenated cis-indole isonitrile by CFPS for FamC1 or FisC plasmids in the presence of FamD2. (B) Structure of the halogenated cis-indole isonitrile -5-fluoro- 9, 6-fluoro- 10, 6-chloro- 11, 5-bromo- 12, 6- bromo- 13, and 5-iodo-cis-isonitrile 14 are shown.
LC–MS analysis of the in vitro TT-assay comprising plasmids encoding FamD2 and FamC1 in the presence of substrate 1 and the fluorinated cis-indole isonitrile 8 or 9 revealed a cyclized product (m/z of 323.18) (Figure 6A,B, Figure S2). The new metabolite was further validated by employing heterologously purified FamD2 and FamC1 enzymes for in vitro chemoenzymatic reactions of unnatural cis-indole isonitrile 9 or 10 containing 1, and the molecule was isolated with an identical tR and MS-pattern compared to CFPS-derived product. Similarly, the in vitro TT-assay performed with the exogenous FamD2 and FisC with the substrate 1 and 9 or 10, also showed the cyclized product (m/z of 323.19) identical with the molecule derived from standard chemoenzymatic synthesis (Figure 6C,D, Figure S3). However, screening of the other halogenated cis-indole isonitrile 11, 12, 13, and 14 for both FamC1 and FisC failed to show cyclized products. This suggested the inability of FamC1 and FisC to cyclize more sterically demanding halogenated (F < Cl < Br <I) cis-indole isonitrile substrates, despite forming the corresponding C3-geranylated cis-indole isonitrile derivatives. The structure–function study of aromatic prenyltransferase and its biocatalytic potential to synthesize hapalindole family related molecules has recently been reported.17 However, this study using CFPS represents the first reported effort to examine the substrate scope of Stig cyclases using unnatural cisindole isonitrile derivatives.
Figure 6.
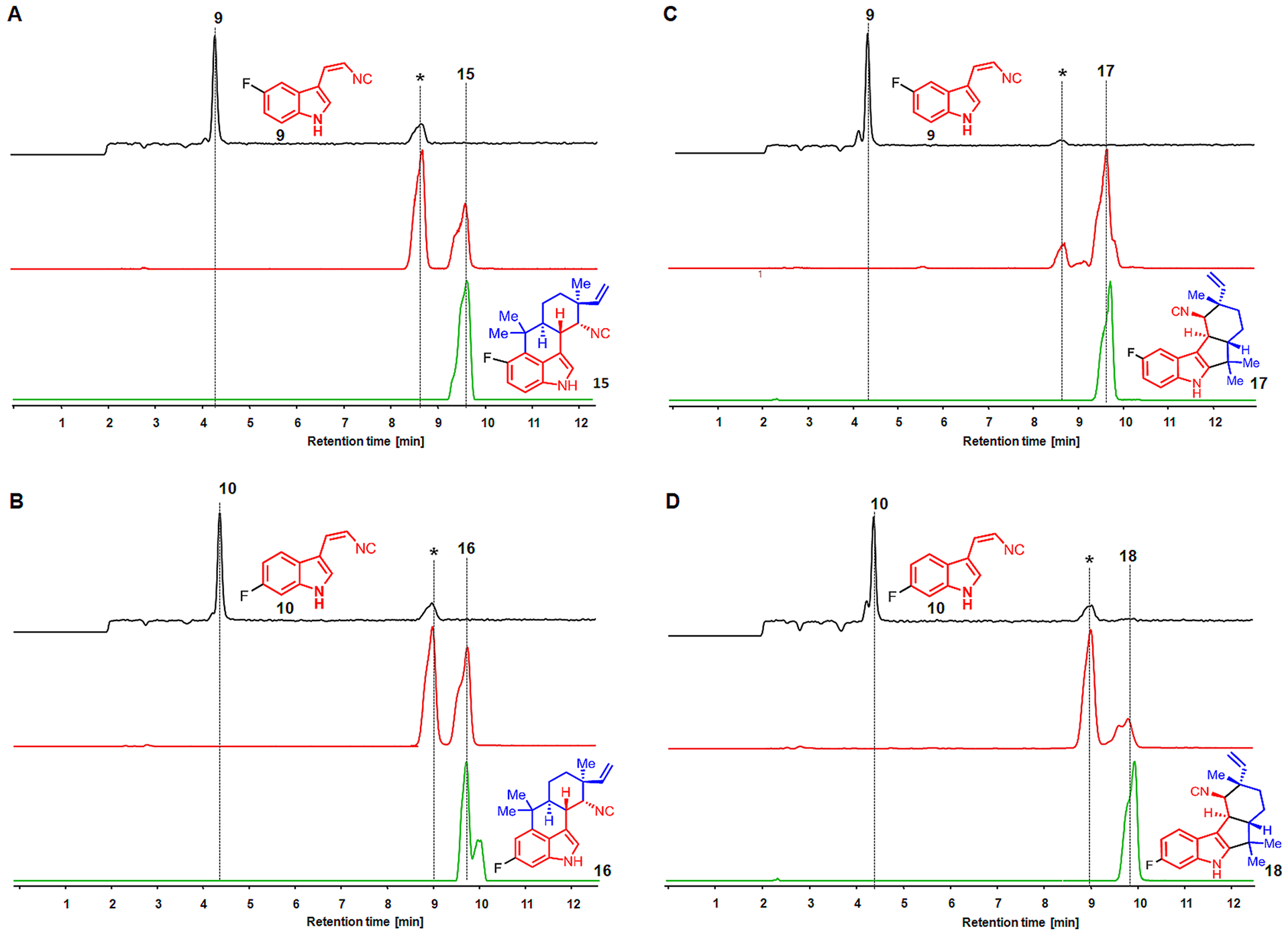
Screening of unnatural substrate 5-fluoro- 9 (panels A and C) and 6-fluoro-cis-indole isonitrile 10 (panels B and D) using CFPS. The substrate 9 and 10 are converted into 5-fluoro-12-epi-hapalindole U 15 and 6-fluoro-12-epi-hapalindole U 16 using FamD2 and FamC1 plasmids (panels A and B) in the presence of GPP. Where as the combination of exogenous plasmids FamD2 and FisC showed the production of 5-fluoro-12-epi-fischerindole U 17 and 6-fluoro-12-epi-fischerindole U 18 (panels C and D). Extracted ion chromatograms (EIC) for the substrate 9 or 10 (black line, m/z of 187.02) and TT-assay (brown line, m/z of 323.18) are shown. EICs of authentic product-standard 15, 16, 17, and 18 (green line, m/z of 323.18) obtained by corresponding chemoenzymatic reaction are shown. The asterisk (*) represents 5F-/6F-12-epi-hapalindole-GPP (panels A and B) and 5F-/6F-12-epi-fischerindole-GPP intermediates.
In each experiment where the in vitro TT-assay showed a new metabolite, we proceeded to chemoenzymatic scale-up for structure elucidation. Thus, reaction of purified FamD2 and FamC1 proteins in the presence of GPP 1 and unnatural cisindole isonitrile 9 and 10 provided new hapalindoles, 5-fluoro-15 and 6-fluoro-12-epi-hapalindole U 16, respectively (Figure S4 and Table S2). However, the scaled-up product of 1 with 9 and 10 for FisC was generated using homologous protein FimC5 from the fischerindole producing strain Fischerella muscicola UTEX 182914 because of its identical catalytic function14 and higher enzymatic activity. The corresponding novel identified products were indole C2/3 ring fused unnatural fischerindole, 5-fluoro- 17 and 6-fluoro-12-epi-fischerindole U 18 (Figure S5, Table S3). Stereochemistry of 15, 16, 17, and 18 were assigned by NMR association with authentic 4 and 8.
In this study, we sought to establish an in vitro TT-assay platform as a biosynthetic prototype for the synthesis and screening of natural and unnatural hapalindole-type molecule. By using detailed proteomics analysis, we have demonstrated that the assay comprising multiple plasmids showed production of a prenyltransferase and selected Stig cyclases from cyanobacterial indole alkaloid pathways. The assay was further applied to screen chemically synthesized halogenated cis-indole isonitrile substrates for assembly of unnatural hapalindole-type molecules. The TT-assay guided screening for metabolite production was scaled-up using chemoenzymatic synthesis, and the structural identities of the novel unnatural hapalindole-type molecules were determined. We believe this approach represents an effective, high throughput strategy to determine the functional role of homologous genes available in the large pool of diverse natural product BGCs.
METHODS
General.
All chemicals were purchased from Sigma-Aldrich, ACROS, and Combi-Blocks. Plasmid pET28a bought from Invitrogen was used for cloning and expression of full length FamD2, FamC3, and FamC4. Plasmid pET28H8T (pET28b variant generated in house containing an N-terminal 8xHis-tag followed by a TEV cleavage sequence upstream of the multiple cloning site) was used for cloning and expression of N-terminally truncated FamC1, full length FamC2, FimC5, and FisC. The protein expression host Escherichia coli strain BL21(DE3) and Ni-NTA agarose to purify His-tag proteins were also purchased from Invitrogen. The liquid chromatography–mass spectrometry (LC–MS) was performed on an Agilent G6545A quadrupole-time-of-flight (Q-TOF) or Agilent 6230 time-of-flight (TOF) using an XBridge Shield RP18 3.5 μm, 3.0 mm × 150 mm column from Waters. The chromatographic method for all substrates was from 60 to 100% acetonitrile water gradients over 27 min. Preparative-scale HPLC was performed on a Shimadzu 20-AT equipped with an LUNA C18 250 × 10 mm column, using a mobile phase gradient of 60–100% acetonitrile in water over 60 min. All NMR spectra were acquired on a Varian 400 and 600 MHz and Bruker 800 MHz spectrometers. Proton and carbon signals are reported in parts per million (δ) using residual solvent signals as an internal standard. Multiplicities are abbreviated as following: singlet (s), doublet (d), triplet (t), quartet (q), doublet–doublet (dd), triplet-doublet (td), doublet–doublet– doublet (ddd), triplet-doublet-doublet (tdd), and multiplet(m). Chemical abbreviations: ethyl acetate (EtOAc), tetrahydrofuran (THF), potassium bis(trimethylsilyl)amide (KHMDS), acetic acid (AcOH), sodium sulfate (Na2SO4), diethyl ether (Et2O), acetonitrile (CH3CN), magnesium chloride (MgCl2), calcium chloride (CaCl2), sodium chloride (NaCl).
In Vitro TT-Assay, Proteomics Analysis, and Substrate Conversion.
In Vitro TT-Assay.
The microscale in vitro synthesis of natural and unnatural hapalindole and fischerindole molecules was performed using a PURExpress In Vitro Protein Synthesis kit (New England Biolabs). This system is based on the PURE system originally developed by the Ueda group44 and later commercialized as PURESYSTEM by Biocomber (Tokyo, Japan).45The assay comprised synthetic substrates 1 and 2 or halogenated 5-/6-substituted 2 (9–14) and exogenous prenyltransferase (FamD2) and fam (FamC1-C4) or fis (FisC) Stig cyclase plasmids to imitate the hapalindole/fischerindole biosynthetic gene cluster.
The reaction consists of exogenous plasmid(s) constructed in pET28a or pET28H8T that allow transcription of a gene under the T7 promoter into mRNA and to be subsequently translated into a protein in the same reaction. Our typical TT-assay comprised 500 ng of freshly extracted plasmid (dissolved in nuclease free H2O, <2 μL added), 0.6 μL of murine RNase inhibitor (40 U/μL, New England Biolabs), commercially available solution A (12 μL), and solution B (8 μL) (New England Biolabs). Solution A is the premix of tRNAs, rNTPS, and amino acids mixture, while solution B comprises ribosomes and protein components such as T7 RNA polymerase, translation factors, aminoacyl-tRNA synthetases, and energy regeneration enzymes. In our assay, 1.2 μL of DTmix chaperone (provided by New England Biolabs) was also added. The noncommercial DTmix solution of 1 μL comprised 2.4 μM DnaK, 0.48 μM DnaJ, 1.4 μM GrpE, and 1.2 μM of Trigger Factor. The in vitro TT-assay was performed in a PCR tube at an end volume of 30 μL adjusted with 50 mM Tris HCl buffer, pH 7.8. The assay was incubated at 25 °C for 3 h with agitation at 150 rpm for the transcription and translation of exogenous plasmid(s). Depending on the target for biosynthetic assembly of natural or unnatural hapalindole and fischerindole compounds, the corresponding exogenous plasmid(s) were used (Table S1). The control TT-assay devoid of plasmid was employed as a negative control in which only the substrate 2 (or 5-/6-substituted 2) could be detected in the LC–MS, whereas the assay comprising FamD2 plasmid alone produces 3 (or 5-/6-substituted-GPP 3). The presence of FamD2 plus FamC1 plasmids produces 4, FamD2 plus FamC2 and FamC3 produce 6, and FamD2 plus FamC1 and FamC4 showed the production of 4 and 5. The combination of FamD2 and FisC plasmids results in production of 7. The use of unnatural 5-/6-fluorinated 2, FamD2 in combination with FamC1 or FisC showed the production of 5-/6-substituted 15, 16, 17, and 18. The sample was subjected to proteomics analysis and substrate conversion as described below. Assays were conducted with proteins known to be soluble from previous E. coli production. Proteins that may be insoluble in E. coli or other hosts were not assessed in the current study.
Proteomics Analysis by LC–Tandem MS.
Proteomics analysis of the in vitro TT-assay samples were performed in the Proteomics Resource Facility at the University of Michigan using an LC–MS based approach. Briefly, following reduction (10 mM DTT, 30 min) and alkylation (65 mM 2-chloroacetamide, 30 min), proteins were digested overnight with 500 ng of sequencing grade modified trypsin (Promega). The resulting peptides were resolved on a nanocapillary reverse-phase column (Acclaim PepMap C18, 2 μm, 50 cm, Thermo Fisher Scientific) using 0.1% formic acid/CH3CN gradient at 300 nL/min (2–25% CH3CN in 105 min; 25–40% CH3CN in 20 min followed by a 90% CH3CN wash for 10 min and a further 30 min re-equilibration with 2% CH3CN) and directly introduced into Q Exactive HF mass spectrometer (Thermo Fisher Scientific, San Jose CA).
MS1 scans were acquired at 60 K resolution. Data-dependent high-energy C-trap dissociation MS/MS spectra were acquired with top speed option (3 s) following each MS1 scan (relative CE ~28%). Proteins were identified by searching the data against E. coli database (UniProtKB, 4331 entries, downloaded on 11/07/2018) appended with FamD2, FamC1, and FisC protein sequences using Proteome Discoverer (v2.1, Thermo Fisher Scientific). Search parameters included MS1 mass tolerance of 10 ppm and fragment tolerance of 0.1 Da; two missed cleavages were allowed; carbamidimethylation of cysteine was considered fixed modification and oxidation of methionine, and deamidation of asparagine and glutamine were considered as potential modifications. FDR was determined using Percolator, and proteins/peptides with a FDR of ≤1% were retained.
Substrate Conversion and Analysis of Product.
Following a 3 h incubation period of in vitro TT-assay components with exogenous plasmid, the substrates GPP 1 and native/unnatural cis-indole isonitrile 2 were added, and the mixture was incubated further for 2 h, extracted, and analyzed as described below. Typically, for the TT-assay consisting of the exogenous plasmid FamD2 alone or the combination of FamD2 plus FamC1 or FamD2 plus FisC, 10 mM each of GPP 1 and cisindole isonitrile 2, MgCl2 (5 mM), CaCl2 (5 mM), and the energy mix (glucose-6-phosphate (5 μM), G-6-P-dehydrogenase (0.5 U), NADPH (1 mM), and glucose (1 mM) were used. For the assay consisting of three plasmids–FamD2 plus FamC1 and FamC4, or FamD2 plus FamC2 and FamC3–higher concentration of the CaCl2 (20 mM) was used.
To analyze the product, the reaction was quenched by adding equal volumes of EtOAc and extracted twice. The organic layers were combined, dried in a nitrogen stream, and reconstituted in 20 μL of CH3CN for LC–MS analysis. The analytical scale LC–MS run was performed on an Agilent G6545A quadrupole-time-of-flight (Q-TOF) or Agilent 6230 time-of-flight (TOF) mass spectrometer equipped with a dual AJS ESI source and an Agilent 1290 Infinity series diode array detector, auto sampler, and binary pump. An XBridge Shield RP18 3.5 μm, 3.0 mm × 150 mm from Waters was used for all separations. The chromatographic method for all substrate gradients was from 60% to 100% of CH3CN in water over 27 min. A volume of 1 μL injections was made for each sample.
Protein Expression and Purification.
Prenyltransferase and Stig Cyclase Expression and Purification.
The expression and purification of FamD2, FamC1, and FimC5 were performed as described.14,34 Briefly, a single BL21(DE3) colony of FamD2/FamC1/FimC5 transformant was inoculated in LB medium containing 50 μg/mL kanamycin and grown overnight at 37 °C shaking at 200 rpm. The main culture (1 L) was prepared at the dilution of 1:100 in 2.8 L of Fernbach flask containing LB medium and the same concentration of antibiotic. The cells were grown (37 °C, 200 rpm) to an optical density (A600 nm) of 0.6. The culture flasks were chilled in ice for 30 min, induced with IPTG (0.2 mM), and incubated (16 °C, 200 rpm) further for 16 h. The cells were harvested (5000 rpm, 4 °C, 15 min), flash frozen in liquid nitrogen, and stored at −80 °C until purification.
For the protein purification, the cell pellets were resuspended in chilled lysis buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol), containing 0.5 mg/mL of lysozyme and 1 mL of 2 mg/mL DNase. The mixture was stirred for 30 min and sonicated on ice for 120 s total time using 10 s pulses followed by a 50 s pause. The cellular debris was removed by centrifugation (35 000g, 4 °C, 35 min). Imidazole (10 mM) was added in the clarified lysate and loaded onto Ni-NTA agarose column equilibrated with lysis buffer. The column was washed with five column volumes of wash buffer (10 mM HEPES, 300 mM NaCl, 0.2 mM TCEP, 10% glycerol, 20 mM imidazole) and the His-tagged protein was eluted with elution buffer (10 mM HEPES, 50 mM NaCl,0.2 mM TCEP, 10% glycerol, 300 mM imidazole). The fractions were pooled and dialyzed using a PD10-desalting column (GE Healthcare) using lysis buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol). The purified protein was analyzed by SDS-PAGE for purity, measured by Nanodrop for concentration, and flash-frozen in liquid nitrogen to store at −80 °C.
Scale-up Chemoenzymatic Reactions.
For the structure analysis of the enzymatic products, reactions were scaled-up to 10 mL containing FamD2 (5 μM), cyclase (20 μM), substrate (1 mM), GPP (1 mM), MgCl2 (5 mM), and CaCl2 (7.5 mM) in 50 mM Tris-HCl, pH 7.0 buffer. The reaction was incubated at 37 °C overnight, quenched and extracted with EtOAc, dried with Na2SO4 and concentrated. The extracted products were purified by HPLC. The purified compounds were concentrated, dissolved in C6D6, and analyzed using a Varian 600 MHz NMR and Bruker 800 MHz NMR.
Chemical Synthesis of cis-Indole Isonitrile Derivatives.
All derivatives were prepared using a method previously described.14 Briefly, in a 50 mL two-neck round-bottom flask purged with nitrogen at −78 °C (dry ice/acetone), diethyl (isocyanomethyl) phosphonate (0.20 mL, 1.248 mmol) was diluted with THF (5 mL). KHMDS (1 M THF, 2.60 mL, 2.60 mmol) was added dropwise, and the reaction was stirred at −78 °C for 30 min. To a separate 4 mL vial, indole-3-carboxaldehyde derivative (1.13 mmol) was dissolved in THF (5 mL), and the resulting solution was added dropwise to the KHMDS solution at −78 °C. The resulting mixture was stirred at 0 °C (cryocool) overnight. The resulting solution was quenched by the addition of AcOH (0.15 mL, 2.6 mmol) and concentrated. The resulting residue was diluted with EtOAc (20 mL), washed with 1 M aqueous potassium phosphate buffer (20 mL, pH 7), washed with brine, dried with Na2SO4, and concentrated to an oil. The residue was dissolved in Et2O and purified by flash chromatography (24%–100% pentane/ Et2O, SiO2) to afford the titled compound as reported below. Yields and spectral data are reported below.
(Z)-3-(2-Isocyanovinyl)-1H-indole (cis-Indole Isonitrile) (2).
Red solid, 21 mg, 11%. 1H NMR (600 MHz, acetone-d6): δ 5.37 (dd, J = 11.8, 1.1 Hz, 1H), 7.14−7.31 (m, 2H),7.50−7.60 (m, 1H), 7.67 (d, J = 11.7 Hz, 1H), 7.77−7.86 (m, 1H), 8.37 (s, 1H), 11.02 (s, 1H).
(Z)-5-Fluoro-3-(2-isocyanovinyl)-1H-indole (9).
Tan solid, 19 mg, 9%. 1H NMR (600 MHz, acetone-d6): δ 5.95 (d, J = 8.9 Hz, 1H), 6.94 (p, J = 4.8 Hz, 1H), 7.03 (td, J = 9.1, 2.5 Hz, 1H), 7.52 (td, J = 9.9, 3.5 Hz, 2H), 8.23 (s, 1H), 10.97 (s, 1H). 13C NMR (151 MHz, dmso): δ 168.74, 158.87, 157.32, 132.31, 128.84, 127.59, 127.52, 124.97, 113.67, 113.61, 111.19, 111.02, 109.61, 109.58, 103.90, 103.74.
(Z)-6-Fluoro-3-(2-isocyanovinyl)-1H-indole (10).
Tan solid, 14 mg, 7%. 1H NMR (600 MHz, acetone-d6): δ 5.97 (d, J = 8.9 Hz, 1H), 6.93–7.01 (m, 2H), 7.27 (dd, J = 9.7, 2.3 Hz, 1H), 7.77 (dd, J = 8.7, 5.2 Hz, 1H), 8.17 (s, 1H), 10.93 (s, 1H).
(Z)-6-Chloro-3-(2-isocyanovinyl)-1H-indole (11).
Yellow solid, 36 mg, 31%. 1H NMR (599 MHz, acetone-d6): δ 5.96 (d, J = 8.9 Hz, 1H), 6.94 (dt, J = 9.5, 4.9 Hz, 1H), 7.16 (dd, J = 8.5, 1.9 Hz, 1H), 7.57 (d, J = 1.8 Hz, 1H), 7.75 (d, J = 8.5 Hz, 1H), 8.17−8.20 (m, 1H), 11.02 (s, 1H). 13C NMR (151 MHz, acetone): δ 170.89, 137.02, 128.93, 128.64, 128.47, 126.68, 124.71, 121.72, 120.25, 112.65, 110.61.
(Z)-5-Bromo-3-(2-isocyanovinyl)-1H-indole (12).
Red solid, 51 mg, 37%. 1H NMR (400 MHz, acetone-d6): δ 5.96 (d, J = 8.9 Hz, 1H), 6.97 (dt, J = 9.2, 4.7 Hz, 1H), 7.33 (dd, J = 8.6, 1.9 Hz, 1H), 7.49 (d, J = 8.6 Hz, 1H), 7.95 (d, J = 1.8 Hz, 1H), 8.20 (d, J = 2.2 Hz, 1H), 11.08 (s, 1H). 13C NMR (101 MHz, acetone): δ 170.77, 135.27, 129.69, 128.97, 128.80, 126.21, 124.63, 121.61, 114.62, 114.27, 110.11
(Z)-6-Bromo-3-(2-isocyanovinyl)-1H-indole (13).
Red solid, 10 mg, 4%. 1H NMR (400 MHz, acetone-d6): δ 5.98 (d, J = 8.9 Hz, 1H), 6.85−7.00 (m, 1H), 7.29 (dd, J = 8.5, 1.8 Hz, 1H), 7.67−7.76 (m, 2H), 8.18 (d, J = 1.9 Hz, 1H), 10.99 (s, 1H). 13C NMR (151 MHz, acetone): δ 169.98, 136.50, 127.61, 126.03, 123.70, 123.38, 119.69, 115.57, 114.74, 109.70, 104.77.
(Z)-5-Iodo-3-(2-isocyanovinyl)-1H-indole (14).
Red solid, 10 mg, 6%. 1H NMR (400 MHz, acetone-d6): δ 5.98 (d, J = 8.9 Hz, 1H), 7.00 (d, J = 9.0 Hz, 1H), 7.40 (d, J = 8.5 Hz, 1H),7.51 (dd, J= 8.6, 1.7 Hz, 1H), 8.16 (d, J= 3.0 Hz, 2H), 11.03 (s, 1H). 13C NMR (151 MHz, acetone): δ169.93, 152.01, 134.82, 130.90, 129.54, 127.63, 127.06, 123.71, 114.15, 108.91, 83.50.
Chemical Synthesis of Geranyl Diphosphate, Triam-monium (1).
The synthesis was performed as described previously.46Briefly, in a 10 mL round-bottom flask purged with nitrogen, tris (tetrabutylammonium) hydrogen pyrophosphate (1.0 g, 1.04 mmol) was dissolved in CH3CN (1.0 mL). Geranyl chloride (0.09 mL, 0.475 mmol) was added, and the reaction mixture was stirred at room temperature for 2 h.
Dowex 50WX8 Resin Preparation.
Dowex 50WX8 resin (20 g, hydrogen form) was washed with half saturated aqueous ammonium chloride (5 × 50 mL) and water (5 × 50 mL) until the pH of the supernatant equaled 5. The slurry was rinsed twice with ion exchange buffer (2% isopropyl alcohol in 25 mM aqueous ammonium bicarbonate) and loaded into a flash column and equilibrated with ion exchange buffer.
Purification.
The reaction mixture was concentrated to afford an orange residue which was diluted with ion exchange buffer. The crude mixture was chromatographed with two column volumes of ion exchange buffer (75 mL). The fractions were combined and concentrated by rotary evaporation, flash frozen, and lyophilized for two days. The resulting white powder was diluted with 1 M ammonium bicarbonate (4 mL) and 50% isopropyl alcohol/CH3CN (10 mL), vortexed for 30 s, and centrifuged (2000 rpm, rt, 5 min). The organic layer was extracted and the residual 0.5 mL of yellow liquid was diluted with 50% isopropyl alcohol/CH3CN and the dilution/vortex/centrifugation process was repeated twice. The organic layers were combined and concentrated to afford a white solid (360 mg). The white solid was taken up in 50% isopropyl alcohol/ 25% CH3CN/25% 1 M aqueous ammonium bicarbonate and chromatographed with cellulose. The resulting fractions were combined and lyophilized affording the titled compound as a white powder (108 mg, 62.2%). 1H NMR (400 MHz, D2O/ND4OD): δ 1.92 (d, J = 1.3 Hz, 3H), 1.98 (s, 3H), 2.01 (d, J =1.3 Hz, 3H), 2.39 (d, J = 6.5 Hz, 2H), 2.41−2.49 (m, 2H),5.45–5.53 (m, 1H), 5.74 (dt, J = 6.1, 3.9 Hz, 1H). 13C NMR (151 MHz, D2O/ND4OD): δ 142.46, 133.67, 124.40, 120.46,62.61, 39.10, 25.92, 25.16, 17.26, 15.89. 31P NMR (162 MHz, D2O/ND4OD) δ −9.93 (d, J = 21.6 Hz), −6.08 (d, J = 21.6 Hz).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the National Institutes of Health (R35 GM118101), the National Science Foundation under the CCI Center for Selective C-H Functionalization (CHE-1700982), and the Hans W. Vahlteich Professorship (to D.H.S.) for financial support. The authors thank Dr. Venkatesha Basrur, at the Proteomics Resource Facility, Department of Pathology, University of Michigan for assistance with proteomic analysis. The authors thank Rajani Arora of the Life Sciences Institute, University of Michigan, for her work in preparing the table of contents graphic.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.0c00038.
Full LCMS traces and data, NMR spectra and analysis, amino acid sequences (PDF)
Full proteomics analysis (XLSX)
The authors declare no competing financial interest.
Contributor Information
Yogan Khatri, Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States;.
Robert M. Hohlman, Life Sciences Institute and Department of Medicinal Chemistry, University of Michigan, Ann Arbor, Michigan 48109, United States;.
Johnny Mendoza, Department of Chemistry, University of Michigan, Ann Arbor, Michigan 48109, United States;.
Shasha Li, Life Sciences Institute and Department of Medicinal Chemistry, University of Michigan, Ann Arbor, Michigan 48109, United States.
Andrew N. Lowell, Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States;.
Haruichi Asahara, New England Biolabs, Inc., Ipswich, Massachusetts 01938, United States.
David H. Sherman, Life Sciences Institute, Department of Medicinal Chemistry, Department of Chemistry, and Department of Microbiology and Immunology, University of Michigan, Ann Arbor, Michigan 48109, United States;.
REFERENCES
- 1.Katz L, and Baltz RH (2016) Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol 43, 155–176. [DOI] [PubMed] [Google Scholar]
- 2.Patridge E, Gareiss P, Kinch MS, and Hoyer D (2016) An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discovery Today 21, 204–207. [DOI] [PubMed] [Google Scholar]
- 3.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, and Fischbach MA (2014) Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, and Metcalf WW (2014) A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol 10, 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de Los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, and Medema MH (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skinnider MA, Merwin NJ, Johnston CW, and Magarvey NA (2017) PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45, W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Dusterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJ, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kotter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nutzmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gomez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng YQ, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk WA, Dorrestein PC, Doyle S, Driessen AJ, Ehling-Schulz M, Entian KD, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Hofte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon HJ, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Mendez C, Metsa-Ketela M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Muller R, Neilan BA, Nett M, Nielsen J, O’Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet JL, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Sussmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, and Glockner FO (2015) Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol 11, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noda-Garcia L, Liebermeister W, and Tawfik DS (2018) Metabolite-enzyme coevolution: From single enzymes to metabolic pathways and networks. Annu. Rev. Biochem 87, 187–216. [DOI] [PubMed] [Google Scholar]
- 9.Fischbach MA, and Walsh CT (2006) Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev 106, 3468–3496. [DOI] [PubMed] [Google Scholar]
- 10.Hudson GA, and Mitchell DA (2018) RiPP antibiotics: biosynthesis and engineering potential. Curr. Opin. Microbiol 45, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, and Rebuffat S (2020) The manifold roles of microbial ribosomal peptide-based natural products in physiology and ecology. J. Biol. Chem 29, No. 006545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Micallef ML, Sharma D, Bunn BM, Gerwick L, Viswanathan R, and Moffitt MC (2014) Comparative analysis of hapalindole, ambiguine and welwitindolinone gene clusters and reconstitution of indole-isonitrile biosynthesis from cyanobacteria. BMC Microbiol. 14, 014–0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Q, and Liu X (2017) Molecular and genetic basis for early stage structural diversifications in hapalindole-type alkaloid biogenesis. Chem. Commun 53, 2826–2829. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Lowell AN, Newmister SA, Yu F, Williams RM, and Sherman DH (2017) Decoding cyclase-dependent assembly of hapalindole and fischerindole alkaloids. Nat. Chem. Biol 13, 467–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong CP, Awakawa T, Nakashima Y, Mori T, Zhu Q, Liu X, and Abe I (2018) Two distinct substrate binding modes for the normal and reverse prenylation of hapalindoles by the prenyltransferase AmbP3. Angew. Chem., Int. Ed 57, 560–563. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Chen CC, Yang Y, Liu W, Ko TP, Shang N, Hu X, Xie Y, Huang JW, Zhang Y, and Guo RT (2018) Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis. Biochem. Biophys. Res. Commun 495, 1782–1788. [DOI] [PubMed] [Google Scholar]
- 17.Awakawa T, Mori T, Nakashima Y, Zhai R, Wong CP, Hillwig ML, Liu X, and Abe I (2018) Molecular insight into the Mg(2+)-dependent allosteric control of indole prenylation by aromatic prenyltransferase AmbP1. Angew. Chem., Int. Ed 57, 6810–6813. [DOI] [PubMed] [Google Scholar]
- 18.Perez JG, Stark JC, and Jewett MC (2016) Cell-free synthetic biology: Engineering beyond the cell. Cold Spring Harbor Perspect. Biol 8, a023853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman AD, Karim AS, and Jewett MC (2020) Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet 28, 019–151. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Lawton TJ, Kostecki JS, Nisthal A, Fang J, Mayo SL, Rosenzweig AC, and Jewett MC (2016) Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J 11, 212–218. [DOI] [PubMed] [Google Scholar]
- 21.Boyer ME, Stapleton JA, Kuchenreuther JM, Wang CW, and Swartz JR (2008) Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol. Bioeng 99, 59–67. [DOI] [PubMed] [Google Scholar]
- 22.Kwon YC, Oh IS, Lee N, Lee KH, Yoon YJ, Lee EY, Kim BG, and Kim DM (2013) Integrating cell-free biosyntheses of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng 110, 1193–1200. [DOI] [PubMed] [Google Scholar]
- 23.Dudley QM, Anderson KC, and Jewett MC (2016) Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol 5, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudley QM, Nash CJ, and Jewett MC (2019) Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synth. Biol 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelwick R, Ricci L, Chee SM, Bell D, Webb AJ, and Freemont PS (2018) Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synth.Biol 3, 016 DOI: 10.1093/synbio/ysy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goering AW, Li J, McClure RA, Thomson RJ, Jewett MC, and Kelleher NL (2017) in vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol 6, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi MK, Chappell J, Hayes CA, Sun ZZ, Kim J, Singhal V, Spring KJ, Al-Khabouri S, Fall CP, Noireaux V, Murray RM, and Lucks JB (2015) Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth. Biol 4, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garamella J, Marshall R, Rustad M, and Noireaux V (2016) The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth. Biol 5, 344–355. [DOI] [PubMed] [Google Scholar]
- 29.Kightlinger W, Duncker KE, Ramesh A, Thames AH, Natarajan A, Stark JC, Yang A, Lin L, Mrksich M, DeLisa MP, and Jewett MC (2019) A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat. Commun 10, 019–12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acuna UM, Mo S, Zi J, Orjala J, and de Blanco EJC (2018) Hapalindole H induces apoptosis as an inhibitor of NF-kB and affects the intrinsic mitochondrial pathway in PC-3 androgen-insensitive prostate cancer cells. Anticancer Res. 38, 3299–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, Lantvit D, Hwang CH, Kroll DJ, Swanson SM, Franzblau SG, and Orjala J (2012) Indole alkaloids from two cultured cyanobacteria, Westiellopsis sp. and Fischerella muscicola. Bioorg. Med. Chem 20, 5290–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walton K, and Berry JP (2016) Indole alkaloids of the Stigonematales (cyanophyta): Chemical diversity, biosynthesis and biological activity. Mar. Drugs 14, No. 40073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo S, Krunic A, Chlipala G, and Orjala J (2009) Antimicrobial ambiguine isonitriles from the cyanobacterium Fischerella ambigua. J. Nat. Prod 72, 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Lowell AN, Yu F, Raveh A, Newmister SA, Bair N, Schaub JM, Williams RM, and Sherman DH (2015) Hapalindole/ambiguine biogenesis is mediated by a Cope rearrangement, C-C bond-forming cascade. J. Am. Chem. Soc 137, 15366–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newmister SA, Li S, Garcia-Borras M, Sanders JN, Yang S, Lowell AN, Yu F, Smith JL, Williams RM, Houk KN, and Sherman DH (2018) Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis. Nat. Chem. Biol 14, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoot CJ, Khatri Y, Hohlman RM, Sherman DH, and Pakrasi HB (2019) Engineered production of hapalindole alkaloids in the cyanobacterium Synechococcus sp. UTEX 2973. ACS Synth. Biol 8, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillwig ML, Zhu Q, and Liu X (2014) Biosynthesis of ambiguine indole alkaloids in cyanobacterium Fischerella ambigua. ACS Chem. Biol 9, 372–377. [DOI] [PubMed] [Google Scholar]
- 38.Brady SF, and Clardy J (2005) Systematic investigation of the Escherichia coli metabolome for the biosynthetic origin of an isocyanide carbon atom. Angew. Chem., Int. Ed 44, 7045–7048. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Q, and Liu X (2017) Discovery of a calcium-dependent enzymatic cascade for the selective assembly of hapalindole-type alkaloids: On the biosynthetic origin of hapalindole U. Angew. Chem., Int. Ed 56, 9062–9066. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Hillwig ML, Koharudin LM, and Gronenborn AM (2016) Unified biogenesis of ambiguine, fischerindole, hapalindole and welwitindolinone: identification of a monogeranylated indolenine as a cryptic common biosynthetic intermediate by an unusual magnesium-dependent aromatic prenyltransferase. Chem. Commun 52, 1737–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hillwig ML, Zhu Q, Ittiamornkul K, and Liu X (2016) Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: Implication for an evolvable enzyme family for late-stage halogenation of aliphatic carbons in small molecules. Angew. Chem., Int. Ed 55, 5780–5784. [DOI] [PubMed] [Google Scholar]
- 42.Hillwig ML, and Liu X (2014) A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol 10, 921–923. [DOI] [PubMed] [Google Scholar]
- 43.Cope AC, and Hardy EM (1940) The introduction of substituted vinyl groups. V. A rearrangement involving the migration of an allyl group in a three-carbon system. J. Am. Chem. Soc 62, 441–444. [Google Scholar]
- 44.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, and Ueda T (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol 19, 751–755. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu Y, and Ueda T (2010) PURE technology. Methods Mol. Biol 607, 11–21. [DOI] [PubMed] [Google Scholar]
- 46.Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, and Poulter CD (1986) Phosphorylation of isoprenoid alcohols. J. Org. Chem 51, 4768–4779. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


