Abstract
The transcription factor GLI1 (GLI family zinc finger 1) plays a key role in the development and progression of multiple malignancies. To date, regulation of transcriptional activity at target gene promoters is the only molecular event known to underlie the oncogenic function of GLI1. Here, we provide evidence that GLI1 controls chromatin accessibility at distal regulatory regions by modulating the recruitment of SMARCA2 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 2) to these elements. We demonstrate that SMARCA2 endogenously interacts with GLI1 and enhances its transcriptional activity. Mapping experiments indicated that the C-terminal transcriptional activation domain of GLI1 and SMARCA2's central domains, including its ATPase motif, are required for this interaction. Interestingly, similar to SMARCA2, GLI1 overexpression increased chromatin accessibility, as indicated by results of the micrococcal nuclease assay. Further, results of assays for transposase-accessible chromatin with sequencing (ATAC-seq) after GLI1 knockdown supported these findings, revealing that GLI1 regulates chromatin accessibility at several regions distal to gene promoters. Integrated RNA-seq and ATAC-seq data analyses identified a subset of differentially expressed genes located in cis to these regulated chromatin sites. Finally, using the GLI1-regulated gene HHIP (Hedgehog-interacting protein) as a model, we demonstrate that GLI1 and SMARCA2 co-occupy a distal chromatin peak and that SMARCA2 recruitment to this HHIP putative enhancer requires intact GLI1. These findings provide insights into how GLI1 controls gene expression in cancer cells and may inform approaches targeting this oncogenic transcription factor to manage malignancies.
Keywords: GLI family zinc finger 1 (GLI1), SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 2 (SMARCA2), enhancer, gene transcription, cancer cells, chromatin, epigenetic regulation, transcriptomics, ATAC-seq, chromatin, cancer, transcription enhancer, chromatin remodeling
The glioma-associated oncogene GLI1 was first identified in malignant glioma as a gene duplication (1). Subsequently it was found to be elevated in many tumor types including prostate (2), breast (3), and pancreas (4) and correlated to poor prognosis (5, 6). It has been demonstrated that GLI1 is a driver of oncogenesis by Fiaschi et al. (7), who demonstrated that GLI1 overexpression promotes tumorigenesis in murine mammary tissue. Mills et al. (8) showed the pivotal nature of GLI1 in tumorigenesis using the established pancreatic cancer mouse model driven by constitutively active KRAS. In this model, knockout of GLI1 disrupts tumor formation. Although there are established roles for GLI1 in several cancers, the mechanisms by which GLI1 operates to regulate gene transcription are not well-understood. We therefore approached the role of GLI1 in cancer by studying the gene transcriptional regulation mechanisms by which GLI1 controls gene expression.
The central role of transcription factors in converting upstream signaling to downstream gene expression depends on their binding to specific DNA sequences and recruiting co-factors that modify the chromatin landscape and the activity of RNA polymerase. The zinc finger domain of GLI1 recognizes a GC-rich nine-nucleotide sequence motif. The binding of GLI1 to that motif and the effect on gene regulation has been extensively characterized for the promoters of many target genes activated via Hedgehog (HH) (9), transforming growth factor β (10), and SULF2 (11) signaling.
Here, using a combination of a Gal4–UAS reporter system (12), mapping experiments, and immunoprecipitation studies, we identified the ATPase SMARCA2, a component of the BAF (SWI-SNF) chromatin remodeling complex (13), as a co-regulator of GLI1-mediated transcription. We found that ectopic expression of GLI1 increases chromatin accessibility using a MNase assay. Further, ATAC-seq assays supported the MNase data and indicated genome-wide changes in chromatin accessibility regulated by GLI1. It also showed that GLI1 is involved in regulation of chromatin opening predominately at sites distal to promoters. Similarly, SMARCA2 modifies chromatin access at sites distal to the promoters. We further evaluated the link between GLI1-mediated gene expression and these distal chromatin sites by integrating RNA-seq with the ATAC-seq data. We identified 32 genes differentially regulated by GLI1 with cis chromatin sites regulated by both GLI1 and SMARCA2. At the GLI1-regulated HHIP gene, we confirmed the presence of both GLI1 and SMARCA2 at the putative enhancer and a dependence upon GLI1 for SMARCA2 occupation at this site. Together, these results point to a transcription regulation mechanism in which GLI1 and SMARCA2 cooperatively alter chromatin accessibility at sites located distally to target genes.
Results
GLI1-mediated transcription is regulated through multiple regions in its C-terminal transactivation domain (TAD)
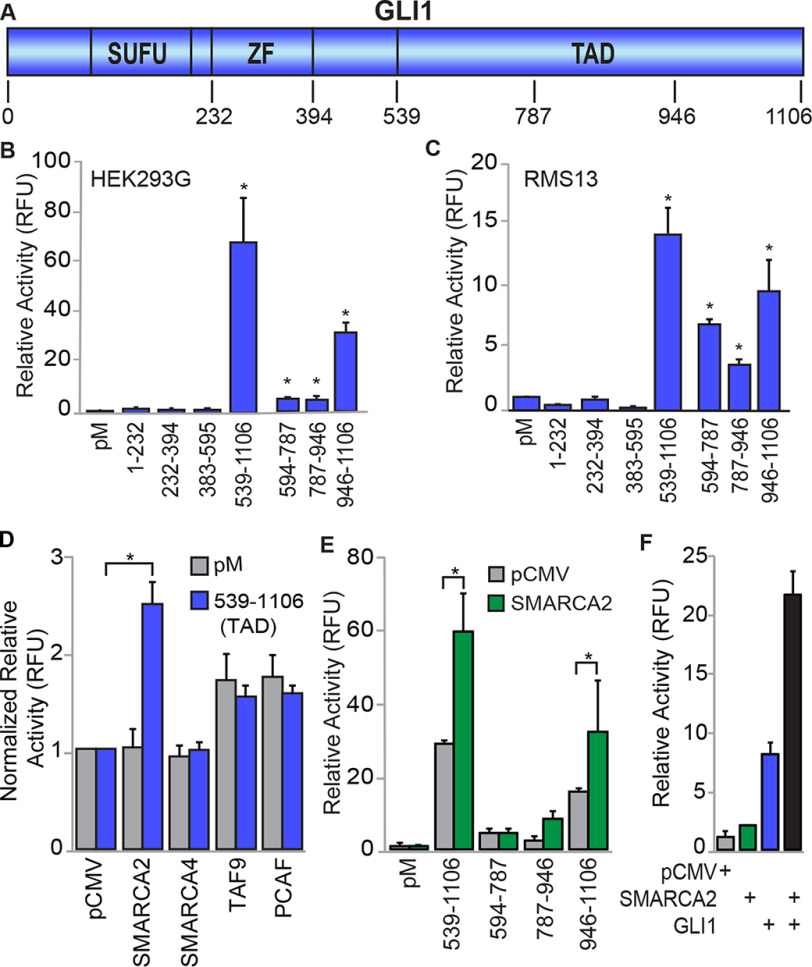
We initially sought to identify which GLI1 domains have transcriptional activity using GLI1–Gal4–DBD chimeras (Fig. 1A) in HEK293G cells stably transfected with a Gal4-responsive UAS–luciferase reporter. Using this system, the C-terminal TAD region (539–1106 aa) of GLI1 was shown to possess very strong transcriptional activity, 68-fold higher than control expressing only the Gal4–DBD (Fig. 1B). In contrast, the regions N-terminal to the TAD, which encode the SUFU (Suppressor of Fused) binding, DNA binding, and nuclear localization motifs, had transcriptional activity similar to the control, indicating little to no activator function in these domains. Further mapping shows that smaller fragments within the 539–1106 aa TAD retain activity at reduced levels. In particular, the chimera including GLI1 aa 946–1106 retains ∼50% activity compared with the chimera with the full 539–1106 aa TAD, (Fig. 1B). Reporter assays in RMS13 and PANC-1 cells transiently transfected to express both the UAS reporter and GLI1–peptide/Gal4–DBD chimeric proteins similarly demonstrated that transcriptional activity of GLI1 is found in its TAD and within TAD subdomains (Fig. 1C and Fig. S1A).
Figure 1.
GLI1-mediated transcription is modulated by SMARCA2 through C-terminal domains of GLI1. A, a representation of GLI1 and its domains and fragments tested for transcriptional regulation. ZF, zinc finger. The numbers correspond to protein amino acid positions for expression constructs. B and C, multiple C-terminal GLI1 domains mediate reporter transcriptional expression. Chimeric Gal4–GLI1 plasmid constructs were transfected into HEK293G cells (n = 6) containing the stable integration of the Gal4 luciferase reporter (B) or co-transfected into RMS13 cells (n = 3) along with the Gal4 luciferase reporter plasmid construct (C). The transcriptional activity was measured as the luminescence signal for each sample normalized to the total protein measured by the Bradford protein assay. D, impact of co-factor overexpression on activity of the full GLI1–TAD in PANC-1 cells (n = 3). Interacting proteins were transiently co-expressed in cells with either the pM control or GLI1–TAD. The luminescence signal for each sample was normalized to the total protein. The transcriptional activities are normalized to the pCMV controls. E, SMARCA2 acts on multiple GLI1 domains. In HEK293G cells, SMARCA2 was expressed with the GLI1–TAD or with different domains within the GLI1–TAD. The expression is normalized to the pM control group (n = 3). F, The transcriptional activity of full-length GLI1 is enhanced by SMARCA2. Transiently transfected plasmids for full-length SMARCA2, GLI1, or both were evaluated for their enhanced gene expression with 8× GLI1 reporter in PANC-1 cells (n = 3). The results are expressed as means ± S.E. *, statistical significance, p < 0.05.
Expression of individual chimeras and UAS–promoter binding for the above experiments was evaluated in HEK293G cells by Western blotting (WB) and ChIP–PCR assays, respectively. An antibody against Gal4 revealed the expression of chimeras aa 1–232, 232–394, 383–595, 594–787, and 787–946, whereas chimeras 539–1106 and 946–1106 were only detected using an antibody against the C terminus of GLI1 (Fig. S1B). All chimeras were detected, with 232–394 and 787–946 having the lowest signals, weakly above background. Using ChIP–PCR assays to detect binding to the UAS promoter, all chimeras were found to be enriched 10–40-fold over background using the Gal4 or GLI1 antibodies, except the aa 594–787 construct, which exhibited a much higher level of enrichment (Fig. S1C). Thus, all constructs appear to be expressed and bind to the UAS promoter. However, no conclusion could be reached regarding their relative expression because of the apparent variable recognition of the different chimeras by the Gal4 antibody in the WB and ChIP assays. Therefore, we cannot rule out the possibility that differential expression of the chimeras affects the results of the reporter assays. Nonetheless, our results indicate that all transcriptional activating function resides in different subdomains within the 539–1106-aa C terminus of GLI1.
SMARCA2 interacts with GLI1 and promotes its transcriptional activity
To define mediators of GLI1 activity through these newly identified TAD domains, we used the aforementioned Gal4–UAS reporter system to screen a panel of potential transcriptional co-regulators. We co-transfected the GLI1–TAD with the chromatin remodeling ATPases SMARCA2 and SMARCA4 (14), the histone acetyl transferase P300/CBP-associated factor (PCAF) (10, 15, 16) and the transcription initiation complex subunit TAF9 (17). These co-factors have been shown to interplay with GLI1 and regulate GLI1 target expression (10, 14, 15, 18). Expression control for co-regulators was determined by WB (Fig. S1D). Interestingly SMARCA2 exhibited a 2.5-fold increase in GLI1 transcription activity above the GLI1–TAD alone (Fig. 1D). In contrast, the related ATPase SMARCA4 did not increase the transcriptional activity of the GLI1–TAD. PCAF and TAF9 increased transcriptional activity, but this increase was less than 50% above the GLI1–TAD threshold (Fig. S1E). SMARCA2 co-expressed with Gal4-chimera–containing fragments of the GLI1–TAD has significantly increased luciferase activity of the 946–1106-amino acid region of GLI1 (Fig. 1E), indicating a specific interaction of SMARCA2 with the region of GLI1 having the highest transcriptional activity. Finally, we investigated the functional interaction between full-length GLI1 and SMARCA2 with a luciferase reporter containing an 8× GLI-binding site (GLI-BS) in the promoter (19). Following transient transfection in PANC-1 cells, SMARCA2 had little effect on transcriptional activity however, GLI1 alone stimulated activity ∼8-fold (Fig. 1F). When SMARCA2 and GLI1 were expressed together, luciferase expression was increased dramatically to over 2-fold compared with GLI1 alone, indicating that SMARCA2 enhances full-length GLI1 transcriptional activity at a GLI1-BS–containing promoter (Fig. 1F). Thus, this screening identifies SMARCA2 as a co-regulator of GLI1 transcriptional activity.
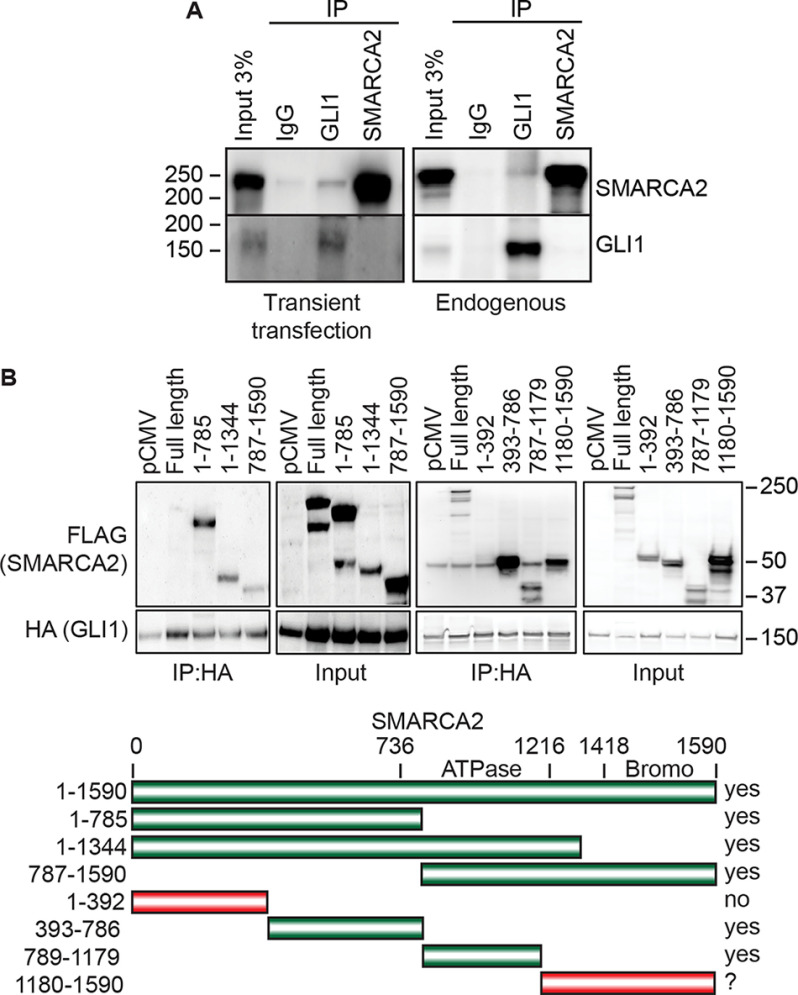
We next determined whether GLI1 and SMARCA2 have a physical interaction using co-immunoprecipitation (co-IP) assays. In RMS13 cells, the IP of GLI1 resulted in the co-IP of SMARCA2, for both co-expressed transfected proteins (Fig. 2A, left panel) and endogenous GLI1 and SMARCA2 (Fig. 2A, right panel). Further mapping of the GLI1–SMARCA2 interaction was performed by co-expressing full-length GLI1–HA with fragments of SMARCA2 tagged with FLAG in RMS13 cells. GLI1 was found to co-IP multiple regions of SMARCA2 including fragments 393–786 and 786–1179, with a possible weaker interaction with the bromodomain fragment (amino acids 1180–1590) of SMARCA2 (Fig. 2B). Interestingly, there is a stronger co-IP of the SMARCA2 fragments compared with the full-length proteins; this may be due to improved exposure of GLI1-binding regions or increased stability of the SMARCA2 fragments that facilitate the interaction. The lower diagram in Fig. 2B summarizes the domains of SMARCA2 that were tested. Together, these data indicate SMARCA2 is co-regulator of GLI1 activity at target genes, and multiple regions of SMARCA2 are important for SMARCA2–GLI1 interaction.
Figure 2.
GLI1 and SMARCA2 interact with each other through the central domains of SMARCA2 including the ATPase domain. A, GLI1 and SMARCA2 co-immunoprecipitation was shown in both ectopic and endogenous conditions in RMS13 cells. A representative WB is shown (n = 3). B, co-immunoprecipitation of GLI1 with domains of SMARCA2 shows the interaction through central domains of SMARCA2 including the ATPase domain and the bromodomain at the C terminus. A representative WB is shown, and the bar diagram summarizes the domains that are co-IPed (n = 3).
GLI1 and SMARCA2 regulate chromatin accessibility at distal regions to gene transcription start sites
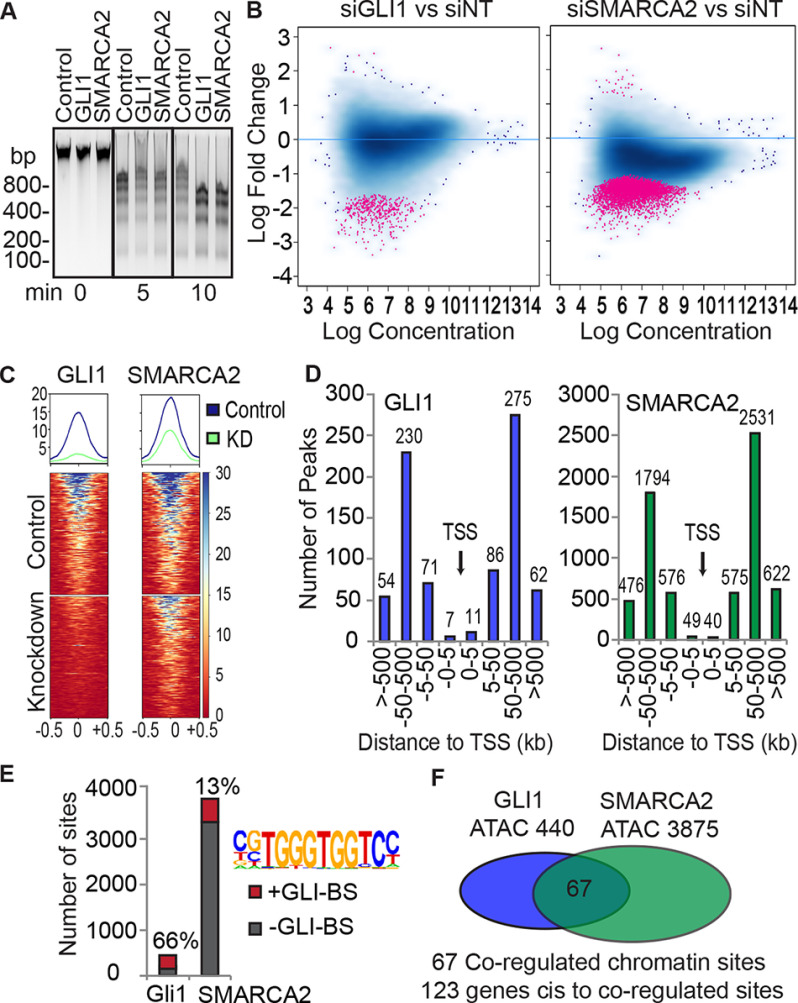
With the identification of SMARCA2 as a co-regulator of GLI1, we tested whether GLI1, like SMARCA2, can modulate chromatin accessibility using a MNase digestion assay. In triplicate assays, a time course showed, interestingly, that GLI1 overexpression caused a reduction in the overall nucleosome fragment sizes similar to the SMARCA2 overexpression (Fig. 3A). Expression of the transfected constructs was confirmed by WB (Fig. S2A). These results show that increased GLI1 expression enhances chromatin accessibility and led us to inquire what regions GLI1 and SMARCA2 co-regulate via alteration of chromatin density.
Figure 3.
ATAC-seq shows that GLI1 and SMARCA2 regulate chromatin density at positions distal to gene transcription start sites and enriched with GLI-BS. A, GLI1 and SMARCA2 increase chromatin accessibility. GLI1 and SMARCA2 were ectopically expressed for 48 h in HEK293G cells followed by MNase digestion over a time course. The control was the empty plasmid vectors used to express GLI1 and SMARCA2. A representative agarose blot with ethidium bromide detection is shown for 0-, 5-, and 10-min MNase digestion (n = 3). Molecular mass markers are indicated in kDa. B–F, changes in chromatin accessibility was evaluated through ATAC-seq in RMS13 cells. GLI1 and SMARCA2 were knocked down using targeted siRNAs and prepared for ATAC-seq. Chromatin accessibility changes were determined relative to the nontargeting siRNA control samples. Triplicate experiments were sequenced. B, the significantly changed peaks (log fold change) identified with the DiffBind package (FDR = 0.1) are shown in red on the MA plot (440 peaks for GLI1 and 3875 for SMARCA2). Chromatin accessibility is predominately reduced as shown by most points falling in the lower portion of the graph. C, the 440 significantly changed ATAC-seq peaks in the GLI1 data set are sorted by signal average using DeepTools computeMatrix (reference point – center) and visualized with plotHeatMap. The same 440 peaks and sorting order was applied to the SMARCA2 data set and visualized with plotHeatMap. The reduction of chromatin accessibility for the GLI1 knockdown also occurs in response to SMARCA2 knockdown. D, the distribution of the differentially regulated peaks by GLI1 and SMARCA2 is determined relative to gene TSSs using the GREAT package with basal parameters. E, the enrichment of GLI1 consensus sequences at the differentially accessible sites are determined by HOMER. Matrices from GLI2 and GLI3 ChIP experiments available in the HOMER package were used to identify the GLI-BS. F, the common differentially accessible sites (n = 67) between GLI1 and SMARCA2 were identified using bedtools intersect with minimum overlap set at 10%.
We assessed the genome-wide chromatin accessibility site alterations utilizing ATAC-seq. In triplicate experiments in RMS13 cells, GLI1 and SMARCA2 were knocked down with siRNAs. The efficacy of knockdown was confirmed for the ATAC-seq assays with the average mRNA reduction being greater than 85% for both siRNAs and corresponding strong protein reduction (Fig. S2B). The number of significantly changed peaks was statistically determined among three replicates comparing the siGLI1 or siSMARCA2 to their matched controls (siNTs) using the DiffBind program with a 0.1 FDR. There were 440 and 3875 significantly changed ATAC-seq peaks (156,745 and 158,446 total peaks) for GLI1 and SMARCA2 siRNA treatment, respectively. Only a few peaks, 5 of 440 and 28 of 3875, showed an increase in chromatin accessibility, with the majority showing a decrease in accessibility upon GLI1 and SMARCA2 knockdown. This is revealed in Fig. 3B, in which significantly changed peaks (in red) are predominately in the lower portion of the graph, and only a few red points occur in the upper portions of the graphs (indicating increased chromatin accessibility). These results, along with the MNase data, provide evidence that GLI1 and SMARCA2 predominantly enhance chromatin accessibility.
We next characterized the chromatin sites regulated by GLI1 and SMARCA2. First, we determined whether GLI1 sites are also regulated by SMARCA2. The average peak score for the 440 GLI1 ATAC-seq peaks was sorted and plotted on a heat map using Deeptools (20) (Fig. 3C, left panel). The same peaks and sort order were then used to show that SMARCA2 knockdown also reduces the chromatin accessibility at the same 440 sites (Fig. 3C, right panel). We then mapped the differentially regulated ATAC-seq peak locations relative to gene transcription start sites (TSSs) using GREAT (21) with basal parameters. Although regulatory regions can be very distal to their respective genes, most characterized GLI1-BSs have been in promoter regions (9, 22). Unexpectedly, we found most of the identified GLI1- and SMARCA2-regulated ATAC peaks to be outside the 5-kb promoter regions of the nearest cis-gene, in either direction (Fig. 3D). To compare the distribution of regulated peaks to all peaks, a comprehensive list was generated from the MACS2 peak lists by combining lists and merging overlapping peaks to generate a single, nonredundant list for each group, GLI1 and SMARACA2, which includes peaks in both the control and knockdown samples from all replicates. Mapping all the ATAC-seq peaks (Fig. S2C) showed a similar pattern, but the percentage of peaks in the 0–5 kb regions is higher at 7.5 and 7.3% compared with 2.3 and 1.3% for the differentially regulated peaks, GLI1 and SMARCA2, respectively. There is also an enrichment of GLI-BS in 66 and 13% of these respective GLI1 and SMARCA2 regulated ATAC-seq peaks (Fig. 3E). Next, we examined the overlap between GLI1- and SMARCA2-regulated ATAC-seq peaks and identified 67 sites (Fig. 3F) with at least 10% overlap (average overlap, 515 bp; range, 197–940 bp). We explored whether there were additional chromatin regions in which GLI1 and SMARCA2 work in very close proximity. However, expansion of all the peak domains to include 500 additional bases in each direction (1000 bp added) did not add additional shared peak regions. Of the 67 co-regulated sites, 38 (56.7%) contain GLI-BS. Using GREAT with basal setting, 123 genes were identified to be cis-located (5′ or 3′) to these 67 overlapping peaks. Together, these data identify the 123 genes associated with sites in which GLI1 and SMARCA2 together regulate chromatin accessibility and identify these chromatin sites to be mostly outside the promoters, possibly enhancers.
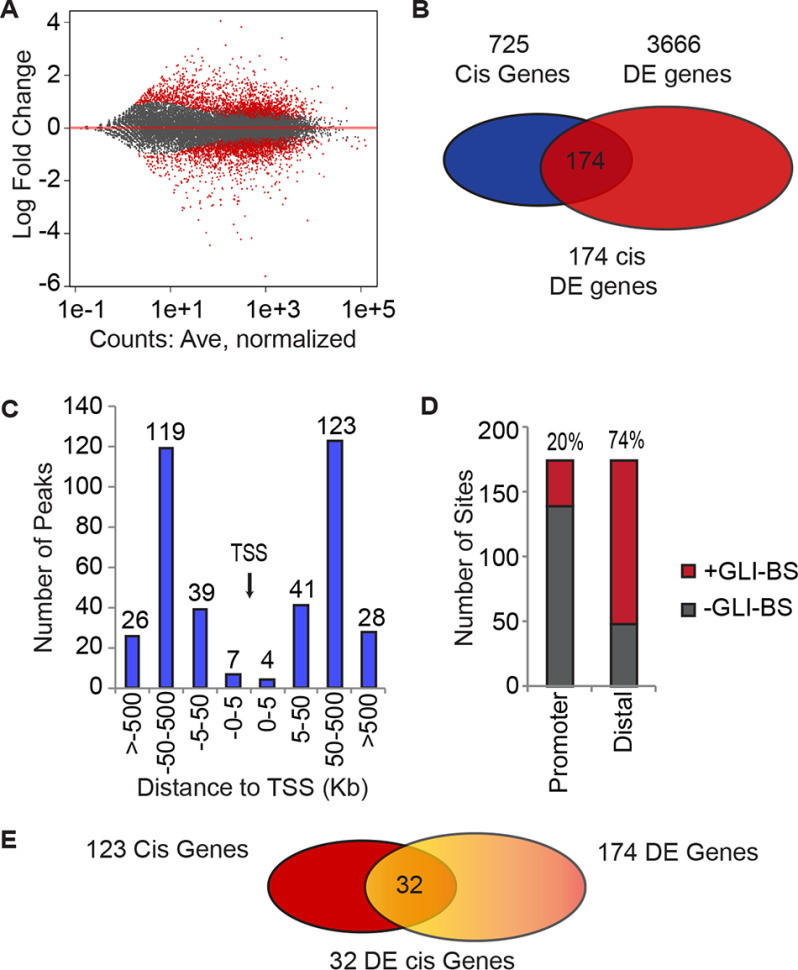
We next investigated the association between GLI1 regulation of gene expression and chromatin accessibility modulated by GLI1 and SMARCA2 depletion. We focused on GLI1 as the key regulator of gene expression because SMARCA2 alone showed little gene expression change in our reporter assays (Fig. 1F). GLI1 was depleted using siRNA in RMS13 cells in triplicate. Genes differentially regulated (DE) by GLI1 siRNA compared with control siRNA were determined using DESeq2 with a 1.5-fold cutoff and a p value at less than or equal to 0.05. GLI1 mRNA and protein reduction was confirmed by RT-PCR and WB (Fig. S2B). There were 3666 significantly DE genes as shown in red in Fig. 4A; 49% of the DE genes were down-regulated, including the known GLI1 target genes PTCH1, PTCH2, and HHIP (9, 23). To focus on chromatin accessibility, we limited our search to genes near the 440 identified GLI1-regulated ATAC-seq peaks. As diagramed in Fig. 4B, 725 genes located cis to the 440 peaks were identified using GREAT. This list of 725 genes was intersected with the list of 3666 DE genes from RNA-seq to identify 174 DE genes in cis to the GLI1-regulated peaks.
Figure 4.
GLI1-regulated genes have distal putative enhancer regions with GLI-binding site enrichment. A, MA plot showing an approximately equal distribution of up and down-regulated genes following GLI1 knockdown as determined by RNA-seq analysis using DESeq2 at 1.5-fold change (n = 3). B, the differentially expressed genes located cis to GLI1-responsive ATAC peaks were identified. Using GREAT, 725 genes were identified cis to the GLI1 ATAC-seq peaks and then intersected with the 3666 DE GLI1 genes to identify 174 genes. C, the positions of the GLI1-responsive ATAC peaks were mapped relative to the transcription start site for the 174 genes using the GREAT package. D, the frequency of GLI-BS in the promoters and the distal sites of the 174 gene was determined using the HOMER package to identify the GLI-BS by their matrices. E, the differentially expressed genes (32) with distal GLI1- and SMARCA2-responsive ATAC peaks (overlapping) were identified by comparison of the gene lists, 123 genes cis to overlapping GLI1- and SMARCA2-responsive ATAC peaks to the 174 DE genes near a GLI1-responsive ATAC peak.
We further characterized the ATAC-seq peaks associated with these 174 genes and found that 97% (376 of 387) of these peaks are located outside the 5-kb promoter regions of the associated genes (Fig. 4C), and 74% (126/174) are enriched with GLI-BS (Fig. 4D). In contrast, we found the presence of GLI-BS in the promoter regions (2000 bp upstream and 100 bp downstream) of these 174 genes to be low at only 20% (35/174). In summary, we identified 174 GLI1-regulated genes that also possess GLI1-regulated chromatin sites enriched with GLI-BS. Finally, we found the intersection between the 174 GLI1-centric genes and the 123 genes identified near GLI1 and SMARCA2 co-regulated ATAC-seq peaks. This identifies 32 GLI1-regulated genes (Fig. 4E and Table 1) potentially controlled through enhancers in which GLI1 and SMARCA2 modulate the chromatin accessibility.
Table 1.
Differentially expressed gene cis to chromatin sites regulated by GLI1 and SMARCA2
| Gene | Description | GLI1 RNA Log2FC | GLI1 peak fold change | SMARCA2 peak fold change |
|---|---|---|---|---|
| MMP11 | Matrix metallopeptidase 11 | −2.72 | −2.02 | −1.5 |
| MRAP2 | Melanocortin 2 receptor accessory protein 2 | −1.91 | −1.79 | −1.77 |
| A4GALT | α1,4-Galactosyl-transferase (P blood group) | −1.80 | −2.38 | −1.55 |
| HHIP | Hedgehog interacting protein | −1.72 | −3 | −1.6 |
| CHCHD10 | Coiled-coil–helix–coiled-coil–helix domain containing 10 | −1.10 | −2.02 | −1.5 |
| GORASP2 | Golgi reassembly stacking protein 2 | −1.07 | −3.01 | −1.79 |
| DCC | DCC netrin 1 receptor | −0.84 | −2.28 | −1.73 |
| ZCCHC8 | Zinc finger CCHC-type containing 8 | −0.74 | −2.01 | −1.45 |
| FSTL4 | Follistatin like 4 | −0.73 | −1.77 | −1.48 |
| FRMD4A | FERM domain containing 4A | −0.72 | −1.81 | −1.66 |
| NFIA | Nuclear factor I A | −0.67 | −1.99 | −1.57 |
| LRCH1 | Leucine-rich repeats and calponin homology domain containing 1 | 0.58 | −1.88 | −1.21 |
| ACVR2A | Activin A receptor type 2A | 0.61 | −2.22 | −1.49 |
| PDCD6IP | Programmed cell death 6 interacting protein | 0.62 | −2.36 | −1.5 |
| FBXO11 | F-box protein 11 | 0.68 | −1.85 | −1.44 |
| SPRY2 | Sprouty RTK signaling antagonist 2 | 0.73 | −1.99 | −1.7 |
| PRKG1 | Protein kinase cGMP-dependent 1 | 0.76 | −1.82 | −1.88 |
| MEF2C | Myocyte enhancer factor 2C | 0.80 | −1.97 | −1.68 |
| CDC42EP3 | CDC42 effector protein 3 | 0.84 | −1.88 | −1.58 |
| DARS | Aspartyl-tRNA synthetase | 0.84 | −2.24 | −1.6 |
| FOXN2 | Forkhead box N2 | 0.89 | −1.85 | −1.38 |
| NOG | Noggin | 1.07 | −1.87 | −1.44 |
| CXCR4 | CXC motif chemokine receptor 4 | 1.21 | −2.24 | −1.78 |
| ANXA3 | Annexin A3 | 1.29 | −2.05 | −1.79 |
| ELAVL2 | ELAV like RNA binding protein 2 | 1.46 | −2 | −1.33 |
| OTUD1 | OTU deubiquitinase 1 | 1.46 | −1.78 | −1.54 |
| GALNT10 | Polypeptide N-acetyl-galactosaminyltransferase 10 | 1.51 | −2.2 | −1.94 |
| DCX | Doublecortin | 1.57 | −1.67 | −1.77 |
| KLHL31 | Kelch like family member 31 | 1.62 | −2.11 | −1.74 |
| RND3 | Rho family GTPase 3 | 1.68 | −1.63 | −1.56 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif 5 | 1.69 | −2.38 | −1.78 |
| SEMA3E | Semaphorin 3E | 1.82 | −2.1 | −1.58 |
GLI1 and SMARCA2 are enriched at distal chromatin sites for HHIP, a GLI1-regulated gene
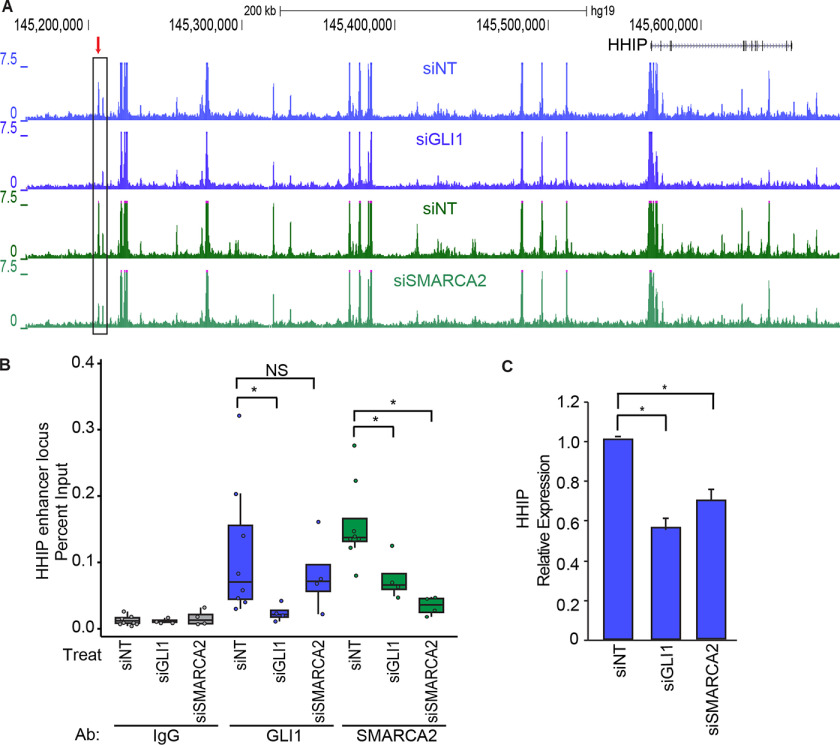
With the identification of putative regulatory regions where GLI1 and SMARCA2 both regulate accessibility, we looked for the presence of GLI1 and SMARCA2 at these sites to confirm their role. Fig. 5A show the chromatin accessibility averaged for three independent ATAC-seq experiments in RMS13 cells and displayed as custom tracks in the UCSC genome browser for the HHIP locus (chromosome 4). Each knockdown has its own matched siNT from the same manufacturer as the specific siRNA. Using the three replicates for each condition, the DiffBind package identified the GLI1- and SMARCA2-regulated ATAC-seq peak (marked with an arrow) 360 kb upstream of the HHIP TSS (Fig. 5A). This peak contains a GLI-BS and corresponds with ENCODE H3K4Me1 enrichment but not H3K27ac (Fig. S3), suggesting an inactive enhancer state. This peak region is also identified in ENCODE ChromHMM (24) to have a candidate enhancer chromatin state. With the identification of a putative regulatory region in which GLI1 and SMARCA2 both regulate accessibility, we looked for the presence of GLI1 and SMARCA2 at this HHIP putative distal regulatory region. Using ChIP–PCR in RMS13 cells (n = 4), we demonstrate GLI1 and SMARCA2 occupancy of this ATAC-seq–identified chromatin site with robust median percentage input of 0.093 and 0.137 (7.4 × and 11.2 × median fold enrichment above the median IgG controls, respectively) (Fig. 5B). The enrichment of GLI1 and SMARCA2 at this site was clearly susceptible to targeted siRNA knockdowns resulting in a respective median percentage input close to the median levels of the nonspecific IgG control. Interestingly, the presence of SMARCA2 at the HHIP putative enhancer depends on GLI1 as shown by the 2.2-fold reduction in SMARCA2 presence in the GLI1 knockdown cells. In contrast, GLI1 occupancy was not dependent on SMARCA2 (Fig. 5B). RT-PCR demonstrated that HHIP gene expression was reduced 1.8- and 1.5-fold in response to GLI1 and SMARCA2 siRNA knockdown, respectively (Fig. 5C). These observations show that GLI1 and SMARCA2 localize to this distal chromatin site upstream of the HHIP gene and that GLI1 affects the recruitment of SMARCA2 and provide evidence that GLI1 and SMARCA2 work together to regulate this putative HHIP enhancer.
Figure 5.
The HHIP gene locus is GLI1 and SMARCA2 responsive. A, in RMS13 cells, chromatin accessibility by ATAC-seq at the HHIP locus on the UCSC genome browser. An average of three replicates are shown for each track for the GLI1 (siGLI1) and SMARCA2 (siSMARCA2) knockdowns and their respective matched nontargeting controls. Highlighted with a red arrow is the chromatin peak that decreases in accessibility in response to GLI1 and SMARCA2 knockdown as determined statistically using the DiffBind package. The two siNTs are independent nontargeting controls matched by manufacturer to the siRNAs for GLI1 and SMARCA2. B, GLI1 and SMARCA2 presence at the HHIP putative enhancer position was determined by ChIP–PCR after siGLI1, siSMARCA2, or siNT treatment (n = 4). HHIP regulatory region detection in GLI1, SMARCA2, or IgG immunoprecipitates is expressed as a percentage of input using boxes encompassing the first and third quartiles, with the median indicated by the horizontal line, and individual data points are overlaid. The statistically significant difference of the means was calculated with the Wilcoxon rank sum test. GLI1 and SMARCA2 presence at this site is responsive to their respective knockdowns, and SMARCA2 presence at this site is partially dependent on GLI1. NS, not significant. C, the down-regulation of HHIP mRNA expression in response to GLI1 and SMARCA2 knockdown was confirmed by RT-PCR using the HPRT and TBP genes for normalization. The results are expressed as means ± S.E. (n = 3). *, statistical significance using Student's t test, p < 0.05.
Discussion
GLI1 is a regulator of gene transcription implicated in promoting tumor formation and progression. Although multiple signaling mechanisms tightly control its activity in normal cells, it can be difficult to know which pathway to target in cancer and whether an alternative pathway will be used in developing drug resistance. Rather than controlling GLI1 at the level of upstream signaling, we propose that targeting GLI1's ability to regulate transcription may provide a novel therapeutic approach for cancer. To do this, we need to understand how GLI1 interacts with its transcriptional co-activators. Therefore, we sought to identify active GLI1 domains and transcriptional co-factors that interact with these regions of GLI1. In this study, we identified multiple active domains within the GLI1–TAD and demonstrated the transcriptional regulation of the GLI1–TAD by the co-regulator SMARCA2. In addition, we identified the novel role of GLI1 as a regulator of chromatin accessibility at sites outside promoter regions.
Although the transcriptional activity of GLI1 had been previously determined to be a large portion of the protein at the C-terminal region, the important subdomains and co-factors for regulating its target gene expression are less well-defined. It is consistent in normal and cancer cell lines that the transcriptional activity of GLI1 is restricted to its TAD (539–1106 amino acids), within which we identified multiple active subdomains. A general pattern indicates higher transcriptional activity toward the more C-terminal end. This may be due in part to the acidic domain of GLI1 at residues 1038–1055. Acidic domains are important for protein–protein interactions, particularly in complexes.
We have identified the ATPase SMARCA2 as a co-activator of GLI1 activity that acts through specific domains in the GLI1–TAD. Although SMARCA4, PCAF, and TAF9 have been shown to interact with GLI1 in other publications (10, 14, 15, 17), our study provides novel insight into the roles of transcriptional co-factors in the regulation of GLI1 activity. Although SMARCA2 and SMARCA4 are very similar proteins with central roles in the BAF chromatin remodeling complexes, they differ in the genes they regulate (25) and the cells they are expressed in (26). We found that SMARCA2 but not SMARCA4 is able to stimulate the activity of the GLI1–TAD. In our study, TAF9 acted to generally increase transcription rather than specifically for the GLI1–TAD. In contrast, Yoon et al. (17, 18) identified the TAF9 component of the TFIID complex as interacting with the 1020–1091 region of GLI1 and activating GLI1 transcription activity. In addition, Bosco-Clement (27) reported that a full-length GLI1 plus TAF9 had enhanced transcription. However, in our study, we found TAF9 to behave as a general transcription factor but did not specifically enhance the transactivity of the GLI1–TAD in the context of the Gal4/UAS reporter assay. The discrepancy between studies might be explained by differences in GLI1 chimeric constructs, reporter assay systems, or cell types. In other studies, PCAF was shown to regulate—in a transforming growth factor β–dependent manner—the BCL2 promoter through a GLI1–SMAD–PCAF (10) axis. We found that PCAF expression did not enhance the transcriptional activity of the GLI1–TAD in reporter assays, suggesting that PCAF may require other regions of GLI1 for co-activation impact.
It was surprising to find that GLI1, like SMARCA2, increased the accessibility of chromatin in normal cells as shown in MNase digestion assays. Equally interestingly was that the regulated chromatin sites determined by ATAC-seq predominately occurred beyond regions considered as promoters. This finding suggests control of potential enhancer sites as a mechanism for gene regulation by GLI1, with cooperative regulatory activity by SMARCA2. We use the term “enhancer” in this paper in the general sense to describe regulatory regions outside the promoters. Follow-up studies (e.g. ChIP-seq) will be needed to fully define these sites as enhancers looking at the enrichment of H3K4me1 and H3K27ac marks in these regions and CRISPR-Cas9 experiments coupled with chromosomal capture assays to determine the functionality of these distal regulatory elements. The possibility of direct control is supported with confirmation of robust enrichment of GLI-BS in these putative enhancers and a decrease in chromatin accessibility at overlapping loci between GLI1 and SMARCA2 knockdown. We identified GLI1-mediated DE genes with cis-located GLI1- and SMARCA2-regulated ATAC-seq chromatin sites. Although many transcription-controlling sites fall relatively close or consecutive to their regulated gene, many regulatory sites can be very distal, separated by several genes and even on different chromosomes. Therefore, it is probable that some identified altered chromatin sites regulate genes that we could not identify by proximity. There may be broader gene regulation events controlled through enhancer on enhancer effects, secondary gene regulation following primary perturbation of TFs or co-regulators, or through enhancer RNA promotion of transcription or complex stabilization. Secondary regulation events may explain the high number (3666 genes) of DE genes up- or down-regulated by GLI1 knockdown. To identify regulated genes located further than our cis-limited search, a chromatin conformation study would be beneficial. We identified an ATAC-seq site regulated by both GLI1 and SMARCA2 in a region distal to the known GLI1 target gene, HHIP. We confirmed by RT-PCR the siGLI1-dependent HHIP down-regulation and by ChIP–PCR the occupancy of GLI1 and SMARCA2 at the HHIP putative enhancer, and, interestingly, a dependence upon GLI1 for SMARCA2 occupation at the HHIP enhancer. This site is not currently listed in the GeneCards (28) GeneHancer (29) registry. We also demonstrated that knockdown of either GLI1 or SMARCA2 decreased the expression of HHIP. To further characterize the role of GLI1 and SMARCA2 at additional ATAC-seq peaks, ChIP-seq for GLI1 and SMARCA2 would provide additional evidence for their interaction in chromatin accessibility. Taken together, our data show that GLI1 activity can be modulated by interaction with SMARCA2 and that the two proteins can cooperatively control chromatin accessibility at the HHIP putative enhancer and possibly other distal regulatory regions. The identification of this new interaction between GLI1 and SMARCA2 expands the molecular function of GLI1 as playing a role in distal regulatory elements and chromatin remodeling. Our study furthers the understanding of both GLI1- and SMARCA2- regulated mechanisms in cancer models and has potential relevance in normal cells where both proteins are expressed. Understanding GLI1/SMARCA2 interaction in both normal and disease states will increase our knowledge of the mechanisms underlying disease pathobiology and may provide potential targets to be exploited for cancer therapy.
Materials and methods
General cell culture
Standard incubation conditions of 37 °C and 5% CO2 were used in all experiments. Purchased medium was enriched with 10% fetal bovine serum. Dulbecco's modified Eagle's medium was used for HEK293G and PANC-1 cell lines. RPMI was used for RMS13 cells. The HEK293G cell line was kindly provided by Dr. Robert G. Roeder (Rockefeller University, New York, NY, USA). PANC-1 and RMS13 were obtained from the American Type Culture Collection (Manassas, VA, USA).
Plasmid preparation
Plasmids were constructed by subcloning DNA fragments obtained by PCR into pM (Clontech, Mountain View, CA, USA) for the Gal4–GLI1 constructs and p3×FLAG CMV14 (Sigma) for SMARCA2. The primers are listed in Table 2. GLI1–HA is in the PRK5 vector. SMARCA4, TAF9, and PCAF constructs are in 3× FLAG, PSG5 FLAG, and pCI FLAG vectors, respectively.
Table 2.
Primers for DNA subcloning Gal4–GLI1 constructs
| Amino acid region | Sense primer (5′ to 3′) | Antisense primer (5′ to 3′) |
|---|---|---|
| GLI1 | ||
| Full-length | CCCAAGCTTATGTTCAACTCGATGACCCCA | CTAGTCTAGAGGCACTAGAGTTGAGGAATTCTGT |
| 1–234 | CCCAAGCTTCATGTTCAACTCGATGACCCCA | CTAGTCTAGATTATTCATACACAGATTCAGGCTCA |
| 232–393 | CCCAAGCTTCGTGTATGAAACTGACTGCCGT | CTAGTCTAGATTACACATGGGCGTCAGGACCATGC |
| 383–595 | CCCAAGCTTCGTCAAGACAGTGCATGGTCCTGACGCC | CTATCTAGAAGCATATCTTGCCCGAAGCAGGTAGTG |
| 539–1106 | CCCAAGCTTCCTTGAACGCCGCAGCAGCAGC | CTATCTAGATTAGGCACTAGAGTTGAGGAATTC |
| 594–787 | CCCAAGCTTCGCTTCAGCCAGAGGGGGTGGTACTTCG | CTATCTAGAAGAGTGGGAAGGGAACTCACCCCATGT |
| 748–946 | CCCAAGCTTCCCAGGCTCTCTGCCTCTTGGG | CTATCTAGAGTTCACTGGAGCTTTAGCACGGCT |
| 946–1106 | CCCAAGCTTCAACACATATGGACCTGGCTTT | CTATCTAGATTAGGCACTAGAGTTGAGGAATTC |
| SMARCA2 | ||
| Full-length | CAGTTCGAATTCATGTCAACGCCCACAGACCCTGGTGCG | GCTCTAGACTCATCATCCGTCCCACTTCCTTCTGAC |
| 1–1344 | CGGAATTCATGTCAACGCCCACAGACCCTG | GCTCTAGACTTAAGCCGTACTTCCTCTTCCATTTCCTCC |
| 1–785 | CGGAATTCATGTCCACGCCCACAGACCC | GCTCTAGATAGAGTCGAAAGGGGAACAATGATGAGATAGGG |
| 786–1590 | GGAATTCATGTCTAACTGGACATATGAATTTGACAAATGGGCTCCT | GCTCTAGACTCATCATCCGTCCCACTTCCTTCTGAC |
| 1–394 | CGTAGAATTCATGTCCACGCCCACAGACCCTGGTG | CGTCTAGACAGCTGACGCTGGAAATTGAGTAACC |
| 393–786 | CGGAATTCATGAGACAGGAGGTGGTGGCCTGCATG | GCTCTAGATAGAGTCGAAAGGGGAACAATGATGAGATAGGG |
| 786–1179 | CGGAATTCATGTCTAACTGGACATATGAATTTGACAAATGGGCTCCT | GCTCTAGAGCTGTTCACGGTACAGAGCCTCAG |
| 1180–1590 | CGGAATTCATGGTGGAGGAAAAGATCCTCGCGG | GCTCTAGACTCATCATCCGTCCCACTTCCTTCTGAC |
Transfection conditions
Reverse transfections were used in all experiments except where noted. In general, OptiMEM was added to an empty cell culture plate followed by plasmids or siRNA and finally XtremeGene (Roche) or RNAiMax (Invitrogen), respectively. XtremeGene was used in a 2:1 ratio (μl XtremeGene: μg plasmid). RNAiMax was used at 1 μl/10 pmol siRNA. The components were mixed, distributed to cover the entire plate surface, and allowed to complex for 30 min The appropriate cells numbers were added to each plate with dilution in their appropriate media, mixed, and placed in an incubator. Standard transfections were prepared in a tube with addition, in order, of OptiMEM, plasmid, and XtremeGene. After mixing and complex formation for 30 min, the mixture was added dropwise to cells plated the previous day, with fresh medium, and placed in an incubator. Expression cultures were harvested after 48 h, whereas siRNA knockdowns were collected after 72 h.
To measure GLI1 domain transcriptional activity, HEK293G, RMS13, and PANC-1 cells were reverse-transfected with 3.3E-6, 4.4E-6, and 2.0E-6 μg of the Gal4–GLI1 plasmids per cell respectively. For RMS13 and PANC-1 cells, the Gal4-luciferase reporter was transfected at 2.1E-6 and 2.0E-6 μg plasmid/cell, respectively. For Gal4–GLI1 and SMARCA2 co-transcriptional activity, HEK293G cells were reverse-transfected with plasmid DNA at 5.4E-6 μg of each DNA (10.8E-6 μg total) per cell. For co-IP in RMS13 cells, 4.25E-6 μg plasmid per cell for GLI1 and SMARCA2 were prepared for a reverse transfection. The co-IP in HEK293G cells had 1.67E-6 μg of each plasmid per cell reversed transfected. Knockdown of GLI1 and SMARCA2 in RMS13 cells for RNA-seq and ATAC-seq analysis was set up as reverse transfection with 6.68E-6 μg plasmid per cell.
Luciferase assays
Transfected cells prepared for transcriptional activity analysis by luciferase detection were carefully washed twice with PBS, lysed in passive lysis buffer (Promega), frozen on dry ice, and stored at −20°C until analysis with Promega luciferase assay system per the kit instructions. All bioluminescent signals are normalized to total protein determined by the Bradford assay (Bio-Rad).
Isolation of protein and WB
The cells for protein analysis and WB were trypsinized, centrifuged, and washed twice with PBS containing protease inhibitors, and the cell pellet was frozen on dry ice and stored at −20°C until processing and analysis. The cells were thawed on ice and suspend in lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 20 mm MgCl2, 0.5% Nonidet P-40) and 1× cOmplete protease inhibitor (Roche) for 10 min with occasional gentle mixing. The nuclei were disrupted with five strokes through a fine-gauge tuberculin syringe. The solution was centrifuged for 10 min at 4 °C and 10,000 × g to pellet the debris. The supernatant was transferred to clean tube, total protein was quantitated by the BCA (Pierce) method, and the remaining solution was refrozen until PAGE and WB analysis. Loading was normalized to total protein. Mini-PROTEAN precast gels were purchased from Bio-Rad for PAGE protein separation. The separated proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore) using a semidry transfer (Pierce G2). The following antibodies were used for WB detection: GLI1 from Novus Biologicals NB600-600 or Cell Signaling 2643, SMARCA2 from Cell Signaling 11966, FLAG from Sigma 1804, HA from Sigma 11867423001, and PCAF from Abnova 12188. Dilutions were 1:1000. Secondary antibodies were diluted 1:10,000. Digital images of the chemiluminescence signal were captured using the Bio-Rad Chemidoc MP or Thermo MYECL.
MNase digestion
HEK293G cell were transfected with plasmid containing GLI1 or SMARCA2 and allowed to grow for 48 h. The cells were trypsinized then counted and aliquoted to 1.0E6 cells per reaction. The cells were washed and suspended in a MNase digestion buffer (20 mm Tris-HCl, pH 7.5, 15 mm NaCl, 60 mm KCl, 1 mm CaCl2) containing cOmplete (Roche) protease inhibitors. The samples were placed in a 37 °C heating block for 5 min before the addition of 5 μl of MNase diluted 1:150. After 10 min of incubation with rotation, the digestion was terminated with the addition of a stop buffer (100 mm Tris-HCl, pH 8.1, 20 mm EDTA, 200 mm NaCl, 2% Triton X-100, and 0.2% sodium deoxycholate) and placed on ice. The DNA was isolated using a QiaAMP DNA mini kit (Qiagen), separated on a 1.5% agarose gel, and visualized with ethidium bromide.
RNA isolation and quantitation
The cells for mRNA analysis were trypsinized, centrifuged, thoroughly mixed in TRIzol (Invitrogen) or RLT buffer (Qiagen), frozen on dry ice, and stored at −80 °C until processing and analysis. RNA isolation was done following the protocol in the Qiagen RNeasy kit including the DNase I treatment and quantitated using the Nanodrop UV-Vis spectrophotometer (ThermoScientific), and cDNA was prepared using the high-capacity cDNA transcriptions kit (Applied Biosystems). The linear range for RNA conversion to cDNA was determined. Specific genes were amplified using the PerfeCTa SYBR Green SuperMix-Quanta, Bio-Rad CFX384 RT-PCR instruments and validated primers specific for the genes of interest. TBP and HPRT1 were used as dual housekeeping genes where appropriate. Expression primers are listed in Table 3.
Table 3.
mRNA expression primers
| Gene | Forward | Reverse |
|---|---|---|
| GLI1 | TGCCTTGTACCCTCCTCCCGAA | GCGATCTGTGATGGATGAGATTCCC |
| HPRT | TGGAAAAGCAAAATACAAAGCCTAAGATGA | ATCCGCCCAAAGGGAACTGATAGTC |
| SMARCA2 | GGAGCAGGATGAACGTGAACAGTC | AAACAACCCAGTGCCTATATGACA |
| TBP | GGTTTGCTGCGGTAATCATGA | CTCCTGTGCACACCATTTTCC |
| HHIP | AGAACTGCAAAATGTGAGCCAG | TCTGATCAAGAATACCTGCCCTG |
RNA-seq and ATAC-seq
RMS13 cells were transfected with siRNAs targeting GLI1 (Qiagen FlexiTube SI03063641) or SMARCA2 (Invitrogen Stealth HSS110001) and the nontargeting controls from the same manufacture as the targeted siRNA (Qiagen 0001027281, Invitrogen 12935300). siRNAs transfections were prepared as described under “Transfection conditions.” Triplicate experiments were prepared independently.
The RNA was isolated following the protocol in the Qiagen RNeasy kit including the DNase I step and submitted to the Mayo Clinic Genomic Analysis Core. The unstranded mRNA library was prepared using the Illumina kit v2 and sequenced (paired-end) on the Illumina HiSeq 4000 to a depth of 30 million reads. The reads were aligned to hg19 using Bowtie2 analysis. Differential expression was done with DESeq2 (30) for RNA on Galaxy with the p value equal to 0.05 and the fold change cutoff at 1.5.
DNA for ATAC-seq was prepared from 20,000 cells following the OMNI-ATAC procedures from Corces et al. (31) and Buenrostro et al. (32) with modifications using the Nextera kit (Illumina). The cells were lysed for 3 min on ice and transposed for 30 min at 37 °C following cleanup. The DNA libraries were prepared with 5–9 cycles of PCR amplification with the NEB High Fidelity Master Mix (New England Biolabs). Clean up was done using the Zymo DNA clean and concentrator kit (Zymo Research) and followed with AMPure XP (Beckman Coulter) bead cleanup to remove primer dimers and under-digested chromatin. Sequencing was on an Illumina HiSeq 4000 to a depth of 50 million reads/sample. The reads were aligned to hg19 using Bowtie2 and peaks called with MACS2 with a 0.005 q value cutoff. Differential peak analysis was performed with DiffBind (33) at FDR of 0.1 within Galaxy (34). The packages from HOMER (35) were used to determine transcription factor enrichment and GLI-BS. The DeepTools2 (20) packages computeMatrix (reference point – center) and plotHeatmap were used to generate the sorted heat maps. The website GREAT (21) was used to identify cis genes and the peak locations relative to the TSS.
The additional bioinformatics packages used are SAMtools (36), MarkDuplicates (RRID:SCR_006525), awk (GitHub), FastQC (RRID:SCR_014583), Trim Galore (RRID:SCR_011847), MultiQC (37), GNU grep (RRID:SCR_012764), and Subread (38). The ATAC and RNA-seq data are available under GEO accession numbers GSE143681 and GSE143684, respectively.
ChIP analysis
For ChIP analysis, proteins and DNA were fixed with a final concentration of 10% formaldehyde in the cell medium for 10 min and quenched with glycine. The cells were gently washed twice with PBS containing 1× cOmplete (Roche) protease inhibitor and then removed by scraping the cells in cold PBS with 1× cOmplete and transferring to a clean tube. The cells were pelleted by centrifugation, suspended in ChIP lysis buffer (5 mm PIPES, 85 mm KCl, 0.5% Nonidet P-40, 20 mm Tris, pH 8.1, 1× cOmplete) for 20 min followed by centrifugation to pellet the nuclei. The nuclei were suspended in nuclear lysis buffer (150 mm Tris, pH 8.1, 10 mm EDTA, 1% SDS, 1 × cOmplete). Chromatin was sheared with sonication for 32 cycles (30-s on/off cycles) in a Diogenade Biorupter 300. After the shearing check, the samples were centrifuged to remove the debris, and the supernatant was transferred to a clean tube. The samples were then equally aliquoted with 2 E 6 cells per IP, diluted 1:10 with dilution buffer (0.01% SDS, 1% Triton, 1.2 mm EDTA, 16.7 mm Tris, pH 8.1, 167 mm NaCl, 1× cOmplete), and incubated with 2 μg of IgG control or a specific antibody overnight with slow rotation at 4 °C. Dynabeads Protein A/G (Thermo Fisher) were washed one time with dilution buffer, and 30 μl were added to each sample and allowed react for 4 h with slow rotation at 4 °C. The beads were gently washed three times with dilution buffer, and the DNA was eluted with 100 μl of elution Buffer (26.4 mm Tris-HCl, 0.01 n NaOH, 0.01% HCl, 200 mm NaCl, 1% SDS) plus 1 μl of proteinase K and incubation for 2 h at 62 °C and 95 °C for 10 min. DNA was purified using a DNA purification spin column kit (Qiagen). Extracted DNA was diluted 1:20 with water and PCR-amplified with validated ChIP primers and SYBR Green detection; Gal4 UAS domain forward, GCATGCGATATTTGCCGAC, and reverse, AGATGTAGCGACACTCCCAGTTG; and HHIP putative enhancer (Chr 4: 145206838–145207375) forward, TTGGAAGAGACTGATGTGGTAAA, and reverse, ATGTGCCATAGACAGACACC.
Immunoprecipitations
Cells were harvested by trypsinization, counted with a hemocytometer, pelleted by centrifugation, and washed twice with cold PBS containing 1× cOmplete (Roche) protease inhibitor. The cell pellet was frozen on dry ice then transferred to a −20 °C freezer until further preparation. The cell pellet was thawed and suspended in buffer (50 mm Tris, pH 7.5, 300 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, pH adjusted to 7.5 at 4 °C and 1× cOmplete) for 20 min with occasional mixing. The cell lysate was centrifuged for 10 min at 13,000 × g and 4 °C, and the supernatant was transferred to a clean tube. 50-μl aliquots were saved as input, the lysate protein was quantitated with the BCA method, and 2E6 cells were aliquoted for each IP/co-IP. The lysate was diluted 1:2 with dilution buffer (50 mm Tris, pH 7.5, 0.5% Nonidet P-40, pH adjusted to 7.5 at 4 °C with 1× cOmplete). 1 μg of antibody was bound to the protein overnight at 4 °C, and then 30 μl of Dynabeads added and incubated for 4 h at 4 °C. The beads were washed twice for 10 min with 200 μl of lysis buffer diluted 1:2 with the dilution buffer with cOmplete at 1 ×. 100 μl of this same diluted lysis buffer was used to suspend the beads and transfer everything to a clean tube. The diluted lysis buffer was completely removed, and 15 μl of PBS with 1 × cOmplete and 15 μl of PAGE loading buffer was added. The analysis continued with PAGE and WB.
Data Availability
All the described data are contained within this article. The ATAC and RNA-seq data have been deposited in the Gene Expression Omnibus under accession numbers GSE143681 and GSE143684.
Supplementary Material
Acknowledgments
We thank Terry Stephenson for secretarial assistance.
This article contains supporting information.
Author contributions—S. L. S., L. L. A., D. L. M., N. H.-A., and M. E. F.-Z. conceptualization; S. L. S., R. L. O. O., L. L. A., N. H.-A., A. G.-M., and M. E. F.-Z. data curation; S. L. S., R. L. O. O., L. L. A., D. L. M., N. H.-A., A. G.-M., and M. E. F.-Z. formal analysis; S. L. S., R. L. O. O., AMV, L. L. A., D. L. M., and N. H.-A. investigation; S. L. S., R. L. O. O., AMV, L. L. A., D. L. M., and N. H.-A. methodology; S. L. S., D. L. M., N. H.-A., and M. E. F.-Z. writing-original draft; S. L. S., R. L. O. O., AMV, L. L. A., D. L. M., N. H.-A., A. G.-M., and M. E. F.-Z. writing-review and editing; AMV, L. L. A., and M. E. F.-Z. resources; M. E. F.-Z. funding acquisition; M. E. F.-Z. visualization.
Funding and additional information—This work was supported by NCI, National Institutes of Health Grant CA136526 (to M. E. F.-Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MNase
- micrococcal nuclease
- ATAC-seq
- transposase-accessible chromatin with sequencing
- TAD
- transactivation domain
- DBD
- DNA-binding domain
- aa
- amino acid(s)
- WB
- Western blotting
- BS
- binding site
- IP
- immunoprecipitation
- FDR
- false discovery rate
- TSS
- transcription start site
- HA
- hemagglutinin
- PCAF
- P300/CBP-associated factor
- DE
- differential expression.
References
- 1. Kinzler K. W., Bigner S. H., Bigner D. D., Trent J. M., Law L., O'Brien S. J. O., Wong A. J., and Vogelstein B. (1987) Identification of an amplified, highly expressed gene in a human glioma. Science 236, 70–73 10.1126/science.3563490 [DOI] [PubMed] [Google Scholar]
- 2. Wu M., Ingram L., Tolosa E. J., Vera R. E., Li Q., Kim S., Ma Y., Spyropoulos D. D., Beharry Z., Huang J., Fernandez-Zapico M. E., and Cai H. (2016) Gli transcription factors mediate the oncogenic transformation of prostate basal cells induced by a Kras–androgen receptor axis. J. Biol. Chem. 291, 25749–25760 10.1074/jbc.M116.753129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu L., Kwon Y. J., Frolova N., Steg A. D., Yuan K., Johnson M. R., Grizzle W. E., Desmond R. A., and Frost A. R. (2010) Gli1 promotes cell survival and is predictive of a poor outcome in ERα-negative breast cancer. Breast Cancer Res. Treat. 123, 59–71 10.1007/s10549-009-0617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maréchal R., Bachet J. B., Calomme A., Demetter P., Delpero J. R., Svrcek M., Cros J., Bardier-Dupas A., Puleo F., Monges G., Hammel P., Louvet C., Paye F., Bachelier P., Le Treut Y. P., et al. (2015) Sonic hedgehog and Gli1 expression predict outcomein resected pancreatic adenocarcinoma. Clin. Cancer Res. 21, 1215–1224 10.1158/1078-0432.CCR-14-0667 [DOI] [PubMed] [Google Scholar]
- 5. Jiang H., Li F., He C., Wang X., Li Q., and Gao H. (2014) Expression of Gli1 and Wnt2B correlates with progression and clinical outcome of pancreatic cancer. Int. J. Clin. Exp. Pathol. 7, 4531–4538 [PMC free article] [PubMed] [Google Scholar]
- 6. Liao X., Siu M. K., Au C. W., Wong E. S., Chan H. Y., Ip P. P., Ngan H. Y., and Cheung A. N. (2009) Aberrant activation of hedgehog signaling pathway in ovarian cancers: effect on prognosis, cell invasion and differentiation. Carcinogenesis 30, 131–140 10.1093/carcin/bgn230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiaschi M., Rozell B., Bergström Å., and Toftgård R. (2009) Development of mammary tumors by conditional expression of GLI1. Cancer Res. 69, 4810–4817 10.1158/0008-5472.CAN-08-3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mills L. D., Zhang Y., Marler R. J., Herreros-Villanueva M., Zhang L., Almada L. L., Couch F., Wetmore C., Pasca Di Magliano M., and Fernandez-Zapico M. E. (2013) Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. J. Biol. Chem. 288, 11786–11794 10.1074/jbc.M112.438846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ågren M., Kogerman P., Kleman M. I., Wessling M., and Toftgård R. (2004) Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330, 101–114 10.1016/j.gene.2004.01.010 [DOI] [PubMed] [Google Scholar]
- 10. Nye M. D., Almada L. L., Fernandez-Barrena M. G., Marks D. L., Elsawa S. F., Vrabel A., Tolosa E. J., Ellenrieder V., and Fernandez-Zapico M. E. (2014) The transcription factor GLI1 interacts with SMAD proteins to modulate transforming growth factor β-induced gene expression in a p300/CREB-binding protein-associated factor (PCAF)-dependent manner. J. Biol. Chem. 289, 15495–15506 10.1074/jbc.M113.545194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carr R. M., Romecin Duran P. A., Tolosa E. J., Ma C., Oseini A. M., Moser C. D., Banini B. A., Huang J., Asumda F., Dhanasekaran R., Graham R. P., Toruner M. D., Safgren S. L., Almada L. L., Wang S., et al. (2020) The extracellular sulfatase SULF2 promotes liver tumorigenesis by stimulating assembly of a promoter looping GLI1–STAT3 transcriptional complex. J. Biol. Chem. 295, 2698–2712 10.1074/jbc.RA119.011146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brand A. H., and Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 13. Chiba H., Muramatsu M., Nomoto A., and Kato H. (1994) Two human homologues of saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22, 1815–1820 10.1093/nar/22.10.1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhan X., Shi X., Zhang Z., Chen Y., and Wu J. I. (2011) Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc. Natl. Acad. Sci. U.S.A. 108, 12758–12763 10.1073/pnas.1018510108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazzà D., Infante P., Colicchia V., Greco A., Alfonsi R., Siler M., Antonucci L., Po A., De Smaele E., Ferretti E., Capalbo C., Bellavia D., Canettieri G., Giannini G., Screpanti I., et al. (2013) PCAF ubiquitin ligase activity inhibits Hedgehog/Gli1 signaling in p53-dependent response to genotoxic stress. Cell Death Differ. 20, 1688–1697 10.1038/cdd.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malatesta M., Steinhauer C., Mohammad F., Pandey D. P., Squatrito M., and Helin K. (2013) Histone acetyltransferase PCAF is required for hedgehog-Gli–dependent transcription and cancer cell proliferation. Cancer Res. 73, 6323–6333 10.1158/0008-5472.CAN-12-4660 [DOI] [PubMed] [Google Scholar]
- 17. Yoon J. W., Lamm M., Iannaccone S., Higashiyama N., Leong K. F., Iannaccone P., and Walterhouse D. (2015) p53 modulates the activity of the GLI1 oncogene through interactions with the shared coactivator TAF9. DNA Repair (Amst.) 34, 9–17 10.1016/j.dnarep.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon J. W., Liu C. Z., Yang J. T., Swart R., Iannaccone P., and Walterhouse D. (1998) GLI activates transcription through a herpes simplex viral protein 16-like activation domain. J. Biol. Chem. 273, 3496–3501 10.1074/jbc.273.6.3496 [DOI] [PubMed] [Google Scholar]
- 19. Hyman J. M., Firestone A. J., Heine V. M., Zhao Y., Ocasio C. A., Han K., Sun M., Rack P. G., Sinha S., Wu J. J., Solow-Cordero D. E., Jiang J., Rowitch D. H., and Chen J. K. (2009) Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc. Natl. Acad. Sci. U.S.A. 106, 14132–14137 10.1073/pnas.0907134106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., and Manke T. (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., and Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., and Ishii S. (1999) Sonic hedgehog–induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274, 8143–8152 10.1074/jbc.274.12.8143 [DOI] [PubMed] [Google Scholar]
- 23. Diao Y., Rahman M. F., Vyatkin Y., Azatyan A., St Laurent G., Kapranov P., and Zaphiropoulos P. G. (2018) Identification of novel GLI1 target genes and regulatory circuits in human cancer cells. Mol. Oncol. 12, 1718–1734 10.1002/1878-0261.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., Epstein C. B., Zhang X., Wang L., Issner R., Coyne M., Ku M., Durham T., Kellis M., and Bernstein B. E. (2011) Systematic analysis of chromatin state dynamics in nine human cell types HHS public access. Nature 473, 43–49 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kadam S., and Emerson B. M. (2003) Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell. 11, 377–389 10.1016/S1097-2765(03)00034-0 [DOI] [PubMed] [Google Scholar]
- 26. Reisman D. N., Sciarrotta J., Bouldin T. W., Weissman B. E., and Funkhouser W. K. (2005) The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl. Immunohistochem. Mol. Morphol. 13, 66–74 10.1097/00129039-200503000-00011 [DOI] [PubMed] [Google Scholar]
- 27. Bosco-Clément G., Zhang F., Chen Z., Zhou H.-M., Li H., Mikami I., Hirata T., Yagui-Beltran A., Lui N., Do H. T., Cheng T., Tseng H.-H., Choi H., Fang L.-T., Kim I.-J., et al. (2014) Targeting Gli transcription activation by small molecule suppresses tumor growth. Oncogene 33, 2087–2097 10.1038/onc.2013.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T. I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., et al. (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1.30.1–1.30.33 10.1002/cpbi.5 [DOI] [PubMed] [Google Scholar]
- 29. Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M., Lancet D., and Cohen D. (2017) GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017, 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., et al. (2017) An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buenrostro J., Wu B., Chang H., and Greenleaf W. (2016) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stark R., and Brown G. (2016) DiffBind: differential binding analysis of ChIP-Seq peak data, Cambridge University, Cambridge, UK [Google Scholar]
- 34. Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Grüning B. A., Guerler A., Hillman-Jackson J., Hiltemann S., Jalili V., Rasche H., et al. (2018) The Galaxy platform for accessible, reproducible and colaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ewels P., Magnusson M., Lundin S., and Käller M. (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao Y., Smyth G. K., and Shi W. (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108–e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the described data are contained within this article. The ATAC and RNA-seq data have been deposited in the Gene Expression Omnibus under accession numbers GSE143681 and GSE143684.