Abstract
A fundamental question is how memory is stored for several weeks and even longer. A long-lasting increase in gene transcription has been suggested to mediate such long-term memory storage. Here, we used contextual fear conditioning in mice to search for lasting transcription that may contribute to long-term memory storage. Our study focussed on hippocampal area CA1, which has been suggested to have a role for at least one week in contextual fear memory. Using an unbiased microarray analysis followed by confirmatory quantitative real-time PCR, we identified an upregulation of two transcription factors, Fosl2 and Nfil3, which lasted for seven days after conditioning. To our knowledge these are the longest transcriptional changes ever detected in the hippocampus after contextual fear conditioning. Thus, our findings suggest novel transcriptional candidates for long-term memory storage.
Keywords: Hippocampus, Contextual Fear Conditioning, Transcription, Fosl2, Nfil3
Introduction
A fundamental question in brain research is how memory is stored for a lifetime. Particular emphasis has been given to the idea that memory storage could be mediated by persistent kinase activity at synapses overcoming molecular turnover (Bear et al., 2018; Crick, 1984; Lisman, 1985; 2017; Sacktor & Fenton, 2018; Smolen, Baxter, & Byrne, 2019). However, it is unknown whether such feedforward kinase signaling is sufficient for memory storage. In addition to synaptic signaling, it has been suggested that epigenetic mechanisms in the nucleus contribute to memory storage (Crick, 1984; Smolen, Baxter, & Byrne, 2019; Zovkic, Guzman-Karlsson, & Sweatt, 2013). In this case, involved epigenetic mechanisms would need to cause lasting changes in gene transcription. However, the vast majority of transcriptional studies associated with learning and memory have identified only transient transcriptional events, induced shortly after training and dissipating within 24 hours (Crick, 1984; Zovkic, Guzman-Karlsson, & Sweatt, 2013; Peixoto, & Abel, 2013; Alberini, & Kandel, 2015). Such transient transcription is essential for cellular memory consolidation, which is a prerequisite for long-term memory, but it cannot maintain long-term memory due to its short duration and molecular turnover. Nonetheless, training-induced transcription that lasts longer than the cellular consolidation window has been identified. For example, after contextual fear conditioning, a memory task in which an animal associates a novel environment with an aversive stimulus, transcription of the memory suppressor gene calcineurin is downregulated for at least 30 days in prefrontal cortex (Miller et al., 2010). At this time point also various changes in DNA methylation have been detected (Miller et al., 2010; Halder et al., 2016). Although long-term storage of contextual fear memory involves prefrontal cortex (Frankland, & Bontempi, 2005), it has emerged that the hippocampus may also involved, although this debated (Doron, & Ghoshes, 2018; Lux, Atucha, Kitsukawa, & Sauvage, 2016; Wiltgen, & Tanaka, 2013; Yonelinas, Ranganath, Ekstrom, & Wiltgen, 2019).
Here, we searched for hippocampal transcription lasting at least 7 days after contextual fear conditioning, using an unbiased microarray analysis. The 7-day time point was chosen as there is clear consensus that the hippocampus is required for contextual fear memory (Frankland & Bontempi, 2005). We focussed on hippocampal area CA1, as this region is essential for contextual fear memory (Goshen et al., 2011; Lux, Atucha, Kitsukawa, & Sauvage, 2016; Matsuo, Reijmers, & Mayford, 2008; Rampon, Tang, Goodhouse, Shimizu, Kyin, & Tsien, 2000). We detected and validated an upregulation of two transcription factors, fos-related antigen 2 (Fosl2/Fra-2) and nuclear factor interleukin-3 regulated (Nfil3/E4BP4), persisting for at least 7 days.
Materials and Methods
Subjects:
Experiments were conducted with 3–6 month-old male C57BL/6J mice (Charles River, UK), which were group-housed with food ad libitum with a 12:12 light-dark cycle. The experiments were performed in accordance with the UK Animals Scientific Procedures Act 1986.
Contextual fear conditioning:
The mice were trained as described previously (Irvine, Vernon, & Giese, 2005; Mizuno, Dempster, Mill, & Giese, 2012). Briefly, a mouse was placed in a conditioning chamber (MedAssociates), after 148 s a mild footshock was provided (0.7 mA, 2 s). Subsequent footshocks were applied at 90 s intervals and this was repeated four times (five footshocks in total). Thirty seconds after the final footshock the mouse was returned to their home cage. The behavioral control group was tested for contextual fear memory 24 hours after conditioning, by exposing the mouse to the conditioning chamber and scoring freezing for 5 minutes.
RNA isolation:
The mice were killed at 7 days after contextual fear conditioning. Hippocampal area CA1 was micro-dissected and fresh-frozen on dry ice and stored at −80°C. Total RNA was isolated using the AllPrep DNA/RNA/Protein mini kit (Qiagen). Total RNA (2 μg) was reverse transcribed using superscript II reverse transcriptase (Invitrogen). The obtained cDNA was diluted 1:10 and stored at −20°C.
Microarray experiment:
The quality of RNA was checked using Agilent RNA 6000 Nano Kit on Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). cRNA samples were hybridized onto Illumina mouse array chip (mouse ref 6v3, Illumina, Inc.; CA, USA) and scanned (Genome Centre; Barts and The London). The resulting data was then extracted from GenomeStudio software (Illumina) and the lumi Bioconductor package (Du, Kibbe, & Lin; 2008) used to apply variance stabilizing transformation followed by robust spline normalization.
Quantitative real-time PCR (qPCR):
qPCR was performed on the same RNA used for the microarray analysis, in triplicate on 7900HT Fast Real-Time PCR System (Applied Biosystems; ABI, US) using SYBR Green (KAPAbiosystems; Roche, Switzerland) and analysed using SDA Software 2.3 (ABI) except Ankrd35. qPCR of Ankrd35 was performed using QuantStudio 7 (ABI). Reactions were performed in 384-wells PCR Plates (Thermo Scientific, Hampshire, UK) with optical adhesive covers (ABI). The cycle conditions were 95°C for 10 min, followed by 40 amplification cycles (95°C for 15s, 60°C for 1min). Analysis of relative gene expression were using delta deltaCt (ddCt) method. The specific primers used for qPCR are listed in Supplementary Table 1.
Statistical analysis:
Normalized microarray data were analysed using significance analysis of microarrays (SAM) (Tusher, Tibshirani, & Chu, 2001) with a false discovery rate (FDR)<0.01. qPCR data freezing scores were analysed by one-way ANOVA followed by Student Newman Keuls post hoc tests. Statistical tests were carried out on SigmaPlot v13.0.
Results
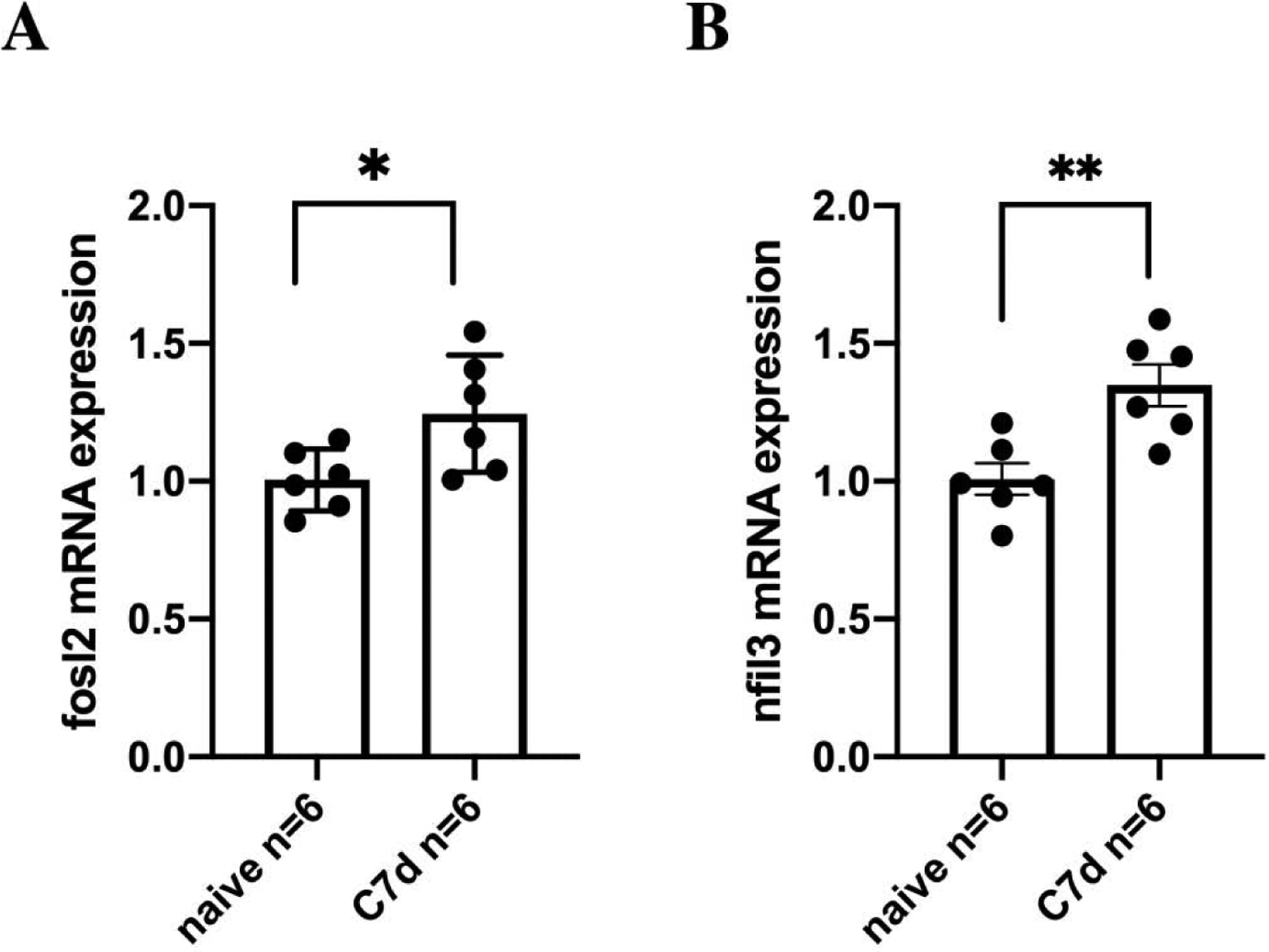
To identify long-lasting transcription in hippocampus that may contribute to memory storage, we trained adult C57BL/6J mice in a strong contextual fear conditioning paradigm (Irvine, Vernon, & Giese, 2005; Mizuno, Dempster, Mill, & Giese, 2012). This training generated contextual fear memory (Fig. 1). RNA from hippocampal area CA1 was isolated from naïve mice (N = 6) and mice killed 7 days after contextual fear conditioning (N = 6). These RNA samples were compared in a microarray analysis. This approach identified 6 candidate upregulations lasting for at least 7 days after contextual fear conditioning (Table 1). No downregulations were found. Six of these potential upregulations were studied for validation with qPCR. A long-lasting regulation of the transcription factors Fosl2 and Nfil3 was confirmed (Fig. 2), whereas expression changes for Andrd35, Pvalb, H2afj were not significant and Sdf2l1 could not be detected (Table 2). Fosl2 transcription was significantly upregulated after contextual conditioning (F1,10=6.12; p=0.033). Nfil3 transcription was also significantly upregulated after contextual conditioning (F1,10=12.82 p=0.005).
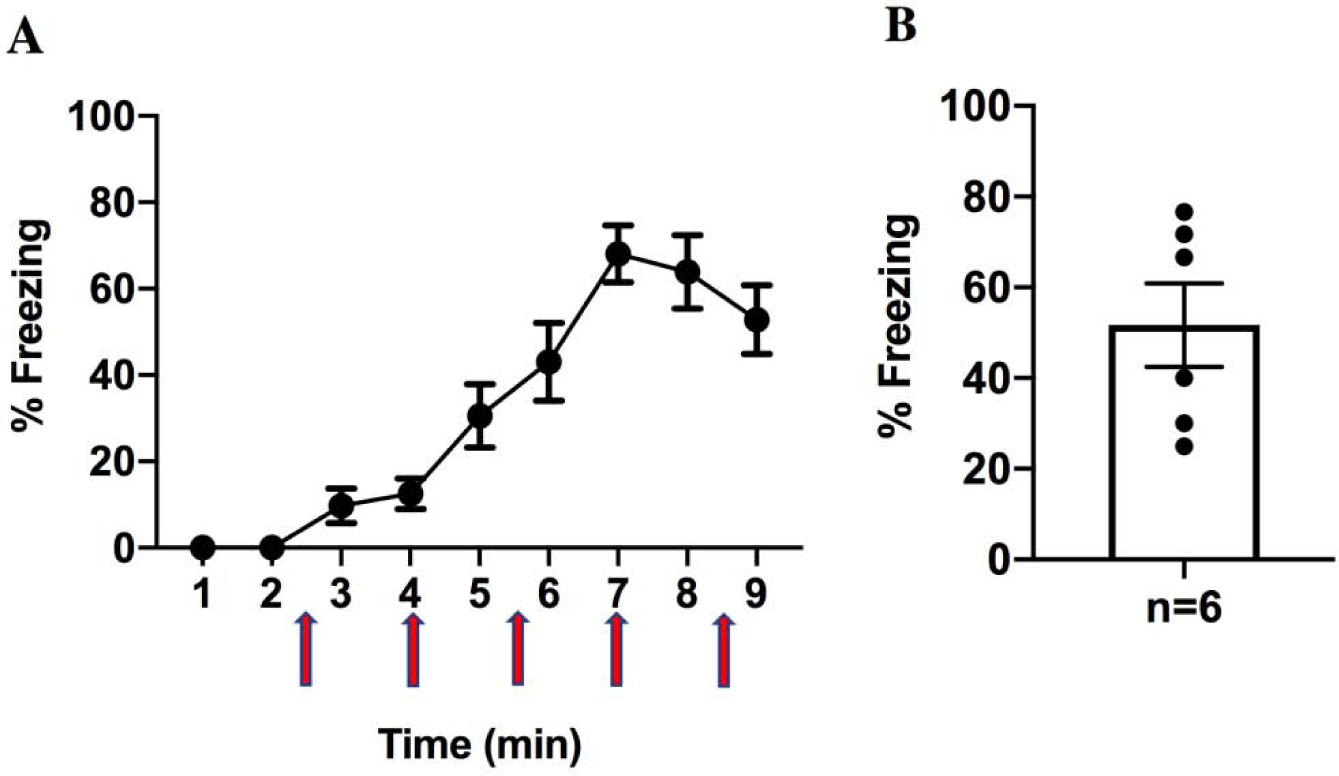
Figure 1. Contextual fear conditioning paradigm generated robust contextual fear memory.
(A) Mice were conditioned after placed in the conditioning chamber for 148s and a mild footshock was provided and followed 4 footshocks at 90s intervals. The mouse was returned to the home cage 30s after the final shock (footshocks are indicated in red arrows). (B) Mice underwent contextual fear conditioning and their 1-day memory was tested by scoring freezing to the trained context. N=6 Means ± sem are indicated.
Table 1.
Candidate genes upregulated 7d after tear conditioning in microarray analysis.
| Gene ID | Gene Name | Score(d) | Numerator(r) | Denominator(s + s0) | Fold change | q-value (%) | |
|---|---|---|---|---|---|---|---|
| H2afj | 770240 | 4.859465621 | 0.27775047 | 0.057156587 | 1.21230312 | < 0.01 | ILMN_1222767 |
| Fosl2 | 7400044 | 4.079573912 | 0.328429365 | 0.080505801 | 1.255645632 | < 0.01 | ILMN_1252481 |
| Ankrd 35 | 5220754 | 4.004673635 | 0.251250379 | 0.06273929 | 1.190238244 | < 0.01 | ILMN_3157692 |
| Pvalb | 5340064 | 3.721709584 | 0.301169958 | 0.080922477 | 1.232143219 | < 0.01 | ILMN_1218223 |
| Sdf2l1 | 3360010 | 3.713768409 | 0.292996474 | 0.078894654 | 1.225182338 | < 0.01 | ILMN_1221943 |
| Nfil3 | 3840521 | 3.709728092 | 0.395717585 | 0.10667024 | 1.315596965 | < 0.01 | ILMN_2595732 |
Figure 2. Regulation of Fosl2 and Nfil3 transcription in hippocampal area CA1 after contextual fear conditioning.
Results from a qPCR analysis of Fosl2 (A) and Nfil3 (B) in hippocampal CA1 of naïve mice (N=6) and 7 days (N=6) after contextual fear conditioning. Means ± sem are indicated. ** (p<0.01), * (p<0.05).
Table2.
Summary of confirmational qPCR 7d after fear conditioning
| Gene ID | p value naive vs 7dFC |
|---|---|
| Fosl2 | p<0.05 |
| Nfil3 | p<0.01 |
| Ankrd35 | p>0.05 |
| Pvalb | p>0.05 |
| H2afj | p>0.05 |
| Sdf2l1 | not detected |
Discussion
We have identified long-lasting upregulation of two transcription factors, Fosl2 and Nfil3, in hippocampal area CA1 after contextual fear conditioning. The upregulation lasted for at least 7 days after conditioning. Our finding is consistent with the idea that storage of contextual fear memory involves the hippocampus for at least one week (Doron, & Ghoshes, 2018; Goshen et al., 2011; Lux, Atucha, Kitsukawa, & Sauvage, 2016; Wiltgen, & Tanaka, 2013; Yonelinas, Ranganath, Ekstrom, & Wiltgen, 2019).
Our analysis did not include a context-only group, leaving it uncertain whether the observed long-lasting transcriptional changes are specific for a context-shock association. However, we think that this is rather unlikely when considering the role of the hippocampus in contextual fear conditioning. The hippocampus stores a contextual memory, whereas the amygdala, and not the hippocampus, is thought to store the context-shock association (Rudy, Huff & Mattus-Hamat, 2005; Keeley et al., 2006). Accordingly, transcriptional changes in the hippocampus that occur after conditioning, but not in a context only group, may not indicate transcription specific for the context-shock association, instead they may simply reflect that contextual memory is stronger after a shock presentation than without shock presentation.
Previous analyses of transcription after contextual fear conditioning have identified up- and downregulations that terminate within one day in the hippocampus. This is consistent with the widely held view that cellular consolidation is completed within one day. However, it has emerged that beyond this window still cellular consolidation processes occur (Bambah-Mukku, Travaglia, Chen, Pollobibi, & Alberini, 2014; Katche et a., 2010; Mizuno, Dempster, Mill, & Giese, 2012; Ryan et al., 2012). Therefore, in principle even 7-day transcriptional changes may be part of a late consolidation process. Alternatively, such late transcriptions may represent mechanisms of memory storage.
Fosl2 is a member of the c-Fos family of transcription factors (Nishina, Sato, Suzuki, Sato, & Iba, 1990). Phosphorylation of the transcription factor CREB and the CREB-regulated NR4A transcription factors, which are required for memory consolidation (Alberini, & Kandel, 2015), stimulate Fosl2 transcription (Hawk et al., 2012; Spessert, Rapp, Jastrow, Karabul, Blum, & Vollrath, 2000). In turn, Fosl2 transcription increases expression of the transcription factor C/EBPβ (Chang, Rewari, Centrella, & McCarthy, 2004), an effector of memory consolidation (Alberini, & Kandel, 2015; Bambah-Mukku, Travaglia, Chen, Pollobibi, & Alberini, 2014). Therefore, we suggest that Fosl2 upregulation may execute transcription needed for generation and maintenance of memory lasting for weeks.
Nfil3 is transcriptional repressor, which is activated by CREB and competes with CREB and C/EBP isoforms for binding to target genes (MacGillavry et al., 2009; 2011). It has been suggested that this results in feedforward loops, which seems important for dynamic control of target gene expression. Therefore, we propose that Nfil3 upregulation may also execute transcription required for generation and maintenance of long-lasting memory.
Finally, there may be more long-lasting transcription changes after contextual fear conditioning, which our approach could not detect. For example, a recent study identified 7-day long upregulation of delta-isoform of calcium/calmodulin kinase II (CaMKIIδ) transcription in hippocampus after object location training (Zalcman et al., 2019). Our microarray analysis did not detect a significant CaMKIIδ upregulation 7 days after contextual fear conditioning. This discrepancy could be due to the fact that our analysis focussed on hippocampal area CA1, whereas the object location memory study considered the entire hippocampus. In future, sophisticated approaches, such single-cell RNA-sequencing, may succeed to identify more comprehensively long-lasting transcriptions after contextual fear conditioning.
In conclusion, we have identified for the first time transcription in hippocampal area CA1 that lasts for at least 7 days after contextual fear conditioning. This is an unexpected result, as most transcriptional studies of learning and memory assume that in the hippocampus training-induced transcriptional dissipate within one or two days. Our findings suggeststhat lasting alterations in transcription in hippocampus contribute to contextual long-term memory storage.
Supplementary Material
Highlights.
Unbiased microarray analysis of gene transcription in hippocampal area CA1 7 days after contextual fear conditioning
Fosl2 expression is upregulated for 7 days after conditioning.
Nfil3 expression is upregulated for 7 days after conditioning.
Acknowledgements
We thank Dr. Jyoti Chatterjee for helpful comments on an earlier version of this manuscript. This work was supported by NIH RO1 grant (2 RO1 MH087463-06).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
Authors declare no conflict of interest.
References
- Alberini CM, & Kandel ER (2015). The regulation of transcription in memory consolidation. Cold Spring Harbour Perspectives in Biology 7, a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambah-Mukku D, Travaglia A, Chen DY, Pollobibi G, & Alberini CM (2014). A positive autoregulatory BDNF feedback loop via C/EBPβ mediates hippocampal memory consolidation. Journal of Neuroscience 34, 12547–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Cooke S, Giese KP, Kaang BK, Kennedy MB, Kim JI, … Park P (2018) In memoriam: John Lisman - commentaries on CaMKII as memory molecule. Molecular Brain 11, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Rewari A, Centrella M, & McCarthy TL (2004). Fos-related antigen 2 controls protein kinase A-induced CCAATTenhancer-binding protein beta expression in osteoblasts. Journal of Biological Chemistry 279, 42438–42444. [DOI] [PubMed] [Google Scholar]
- Crick F (1984). Memory and molecular turnover. Nature 312, 101. [DOI] [PubMed] [Google Scholar]
- Doron I, & Ghoshen I (2018). Investigating the transition from recent to remote memory using advanced tools. Brain Research Bulletin 141, 35–43. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, & Lin SM (2008). lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548. [DOI] [PubMed] [Google Scholar]
- Frankland PW, & Bontempi B (2005). The organization of recent and remote memory. Nature Reviews in Neuroscience 6, 119–130. [DOI] [PubMed] [Google Scholar]
- Goshen I, Brodsky M, Prakash R, Wallace J., Gradinaru V, Ramakrishnan C, Deisseroth K (2011). Dynamics of retrieval strategies for remote memories. Cell 147, 678–689. [DOI] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, … Bonn S (2016). DNA methlyation changes in plasticity genes accompany the formation and maintenance of memory. Nature Neuroscience 19, 102–110. [DOI] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, … Abel T (2012). NR4A nuclear receptors support memroy enhancement by histone deacetylase inhibitors. Journal of Clinical Investigation 122, 3593–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine EE, Vernon J, Giese KP (2005). AlphaCaMKII autophosphorylation contributes to rapid learning but is not necessary for memory. Nature Neuroscience 8, 411–412. [DOI] [PubMed] [Google Scholar]
- Katche C, Bekinschtein P, Slipczuk L, Goldin A, Izquierdo IA, Cammarota M, & Medina JH (2010). Delayed wave of c-Fos expression in the dorsal hippocampus involved specifically in persistence of long-term memroy storage. Proceedings of the National Academy of Sciences (USA) 107, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keelet MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S & Abel T (2006). Differential transcriptional response to nonassociative associative components of classical fear conditioning in the amygdala and the hippocampus. Learning & Memory 13, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman JE (1985). A mechanism for memroy storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proceedings of the National Academy of Sciences (USA) 82, 3055–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J (2017). Criteria for identifying the molecular basis of the engram (CaMKII, PKMzeta). Molecular Brain 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux V, Atucha E, Kitsukawa T, & Sauvage MM (2016). Imaging a memory trace over half a life-time in the medial temporal lobe reveals a time-limited role of CA3 neurons in retrieval. eLife 5, e11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry HD, Stam FJ, Sassen MM, Kegel L, Hendriks WT, Verhaagen J, … van Kesteren RE (2009). Nfil3 and cAMP response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. Journal of Neuroscience 29, 15542–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGallavry HD, Cornelis J, van der Kallen LR, Sassen MM, Verhaagen J, Smit AB, & van Kesteren RE (2011). Genome-wide expression and promoter binding analysis identifies Nfil3 as a repressor of C/EBP target genes in neuronal outgrowth. Molecular and Cellular Neuroscience 46, 460–468. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Reijmers L, Mayford M (2008). Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science 319, 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, … Sweatt JD (2010). Cortical DNA methylation maintains remote memory. Nature Neuroscience 13, 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, & Giese KP (2012). Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes, Brain & Behavior 11, 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina H, Sato H, Suzuki T, Sato M, & Iba H (1990). Isolation and characterization of fra-2, an additional member of the fos gene family. Proceedings of the National Academy of Sciences (USA) 87, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto L, & Abel T (2013). The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C, Tang YP, Goodhouse J, Shimizu E, Kyin M, & Tsien JZ (2000). Enrichment induces structural changes and recovery from nonspatial deficits in CA1 NMDAR-knockout mice. Nature Neuroscience 3, 238–244. [DOI] [PubMed] [Google Scholar]
- Rudy JW, Huff NC, & Matus-Amat P (2004). Understanding contextual fear conditioning: insights from a two-process model. Neuroscience and Biobehavioral Reviews 28, 675–685. [DOI] [PubMed] [Google Scholar]
- Ryan MM, Ryan B, Kyrke-Smith M, Logan B, Tate WP, Abraham WC, & Williams JM (2012). Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. Public Library of Science One 7, e40538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor TC, & Fenton AA (2018). What does LTP tell us about the roles of CaMKII and PKMζ in memory? Molecular Brain 11, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P, Baxter DA, & Byrne J (2019). How can memory last for days, years, or a lifetime? Proposed mechanisms for maintaining synaptic potentiation and memory. Learning & Memory 26, 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spessert R, Rapp M, Jastrow H, Karabul N, Blum F, & Vollrath L (2000). differential role of CREB phosphorylation in cAMP-inducible gene expression in the rat pineal. Brain Research 864, 270–280. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, & Chu G (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences (USA) 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltgen BJ, & Tanaka KZ (2013). Systems consolidation and the content of memory. Neurobiology of Learning and Memory 106, 365–371. [DOI] [PubMed] [Google Scholar]
- Yonelinas AP, Ranganath C, Ekstrom AD, & Wiltgen BJ (2019). A contextual binding theory of episodic memory: systems consolidation reconsidered. Nature Reviews in Neuroscience 20, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalcman G, Federman N, Fiszbein A, de la Fuente V, Ameneiro L, Schor I, & Romano A (2019). Sustained CaMKII delta gene expression is specifically required for long-lasting memories in mice. Molecular Neurobiology 56, 1437–1450. [DOI] [PubMed] [Google Scholar]
- Zovkic IB, Guzman-Karlsson MC, & Sweatt JD (2013). Epigenetic regulation of memory formation and maintenance. Learning & Memory 20, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


