Abstract
Lecanosticta acicola causes the disease known as brown spot needle blight (BSNB), on Pinus species. The pathogen is thought to have a Central American centre of origin. This was based on the morphological variation between isolates believed to represent L. acicola from native Pinus spp. Two species of Lecanosticta, L. brevispora and L. guatemalensis, have recently been described from Mexico and Guatemala respectively based on morphology and sequence-derived phylogenetic inference. However, the putative native pathogen, L. acicola, was not found in those areas. In this study, the species diversity of a large collection of Lecanosticta isolates from Central America was considered. Phylogenetic analyses of the BT1, ITS, MS204, RPB2 and TEF1 gene regions revealed six species of Lecanosticta, four of which represented undescribed taxa. These are described here as Lecanosticta jani sp. nov. from Guatemala and Nicaragua, L. pharomachri sp. nov. from Guatemala and Honduras, L. tecunumanii sp. nov. from Guatemala and L. variabilis sp. nov. from Guatemala, Honduras, and Mexico. New host and country records were also found for the previously described L. brevispora and L. guatemalensis. Lecanosticta acicola was not found in any of the samples from Central America, and we hypothesize that it could be a northern hemisphere taxon. The high species diversity of Lecanosticta found in Mesoamerica suggests that this is a centre of diversity for the genus.
Electronic supplementary material
The online version of this article (10.1186/s43008-019-0004-8) contains supplementary material, which is available to authorized users.
Keywords: Brown spot needle blight, Lecanosticta, Mesoamerica, Pinus pathogens, phylogeny
INTRODUCTION
Brown spot needle blight (BSNB) or Lecanosticta needle blight is an important needle disease on Pinus species. The disease is characterised by brown spots on necrotic yellow lesions at the points of infection and die-back of the needles from the apex, which often leads to premature defoliation (Ivory 1987). BSNB is caused by the fungal pathogen, Lecanosticta acicola (Siggers 1944). The fungus is a well-known pathogen in the USA and has also been recorded in Central America, Colombia, Europe as well as Asian countries including China, Japan and Korea. Lecanosticta acicola is regarded as an A2 quarantine pathogen in Europe and Colombia where it is present as well as an A1 quarantine pathogen in the rest of South America (COSAVE), Africa (IASPC) and the Eurasian Economic Union countries where it has yet to be recorded (https://gd.eppo.int/taxon/SCIRAC/categorization). Despite its quarantine status, L. acicola has been discovered in various new locations and on new hosts in Europe during the past decade (Jankovsky et al. 2009; Markovskaja et al. 2011; Anonymous 2012; Hintsteiner et al. 2012; Adamson et al. 2015; Janoušek et al. 2016; Ortíz de Urbina et al. 2017; Mullett et al. 2018; Cleary et al. 2019; Sadiković et al. 2019).
Siggers (1944) and Evans (1984) summarised the taxonomic and nomenclatural history of Lecanosticta acicola, which was complicated by the former system which allowed asexual and sexual morphs of the same species of fungi to be given separate scientific names (Kais 1971; Evans 1984). From 1972 to 2012, the name Mycosphaerella dearnessii was widely used for the causal agent of BSNB. It was, however, recently recognised that Mycosphaerella is polyphyletic and should be strictly used for fungi in Ramularia (Crous et al. 2007; Crous 2009). Following the One Fungus One Name (1F1N) convention (Hawksworth et al. 2011), the nomenclatural rules were changed in July 2011, and included in subsequent editions of the International Code of Nomenclature for algae, fungi, and plants (ICN) (Turland et al. 2018). Lecanosticta was taken up as the appropriate name, with L. acicola as type species of the genus (Crous et al. 2009a; Quaedvlieg et al. 2012).
Five species of Lecanosticta have been described: Lecanosticta acicola, L. brevispora, L. guatemalensis (Quaedvlieg et al. 2012), L. gloeospora (Evans 1984), and L. longispora (Marmolejo 2000). Lecanosticta acicola remains the best-known species and records suggest that it has a wide distribution in North and South America, Europe, and Asia (https://gd.eppo.int/taxon/SCIRAC/distribution). The remaining four species are known only from Mesoamerica (Evans 1984; Marmolejo 2000; Quaedvlieg et al. 2012). Lecanosticta gloeospora was described, based only on morphology, from disease symptoms on Pinus pseudostrobus from Iturbide, Nuevo León, Mexico (Evans 1984). It was subsequently reported on P. pseudostrobus collected in 1990 in Mexico (Marmolejo 2000). Lecanosticta longispora was originally described from Pinus culminicola in Nuevo León, Mexico, based on morphology (Marmolejo 2000). Quaedvlieg et al. (2012) redescribed and epitipified L. longispora based on DNA sequence and morphological data. Quaedvlieg et al. (2012) delineated Mycosphaerella species of quarantine significance in Europe, including isolates believed to be L. acicola from Central America. Those isolates were distinct taxa and were named L. brevispora and L. guatemalensis from Pinus sp. in Mexico and from P. oocarpa in Guatemala.
Names assigned to Lecanosticta species prior to 2012 were based only on morphological characteristics. Cryptic diversity in Lecanosticta is illustrated by L. guatemalensis (IMI281598), which was initially identified as L. acicola (Evans 1984; Quaedvlieg et al. 2012). Identifications made utilising only morphological characteristics should clearly be re-evaluated using DNA sequence data and phylogenetic inference.
Central America is believed to be the centre of origin of L. acicola. This hypothesis was first proposed by Evans (1984), when the fungus was isolated from native trees in pristine forests. In a recent phylogenetic study, high levels of diversity were found in the Translation Elongation 1-α gene region (TEF1) of isolates from Mexico and Guatemala (Janoušek et al. 2016). Furthermore, Central American isolates did not group in the same clade as isolates from Asia, Europe, and North America. Likewise, Janoušek et al. (2016) reported poor amplification of microsatellite regions that had been developed for L. acicola suggesting that the isolates could represent cryptic species. The present study emerged from an opportunity to collect pine needles infected with Lecanosticta spp. in Guatemala, Honduras and Nicaragua from 2010 to 2012. Specimens were identified based on DNA sequence comparisons and an attempt was made to confirm whether L. acicola occurs in Central America.
MATERIALS AND METHODS
Collections used in the study
Specimens prepared from ex-type cultures and other representatives of all known Lecanosticta species and closely related species (Quaedvlieg et al. 2012) were obtained from the culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands (CBS), and from the UK National Fungus Collection maintained by CABI Bioscience (Egham, UK: IMI). Living cultures or DNA of six isolates from Central America examined by Evans (1984), and believed to represent L. acicola, were also acquired from IMI (Table 1). Furthermore, isolates of Dothistroma septosporum, D. pini, Phaeophleospora eugenia, P. gregaria, and Amycosphaerella africana that represent genera in Mycosphaerellaceae closely related to Lecanosticta (Quaedvlieg et al. 2012) were included for comparative purposes. These cultures were obtained from CBS and the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI) in Pretoria, South Africa (Table 1).
Table 1.
Details of isolates used in this study
| Species | CMW numbera | Other collection numberb | Sampling site (Country, Region, Location) | Host | Collection date | Collector | GenBank accession numbersd | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | TEF1 | BT1 | MS204 | RPB2 | |||||||
| Amycosphaerella africana | 45395 | CBS 110843 | South Africa, Western Cape Province, Pampoenvlei | Eucalyptus cladocalyx | Nov 1994 | Crous PW | KF901702 | JX901653 | MK015047 | MK015515 | MK015290 |
| A. africana | 45396 | CBS 680.95 | South Africa, Western Cape Province, Stellenbosch mountain | E. viminalis | Oct 1994 | Crous PW | AY626981 | KF903117 | MK015048 | MK015516 | MK015291 |
| Dothistroma pini | 10930 | CBS 116485 | USA, Michigan, Montcalm County, Crystal Lake | Pinus nigra | 2001 | Adams G, Barnes I | AY808301 | AY808266 | AY808196 | NA | MK015292 |
| D. pini | 10951 | CBS 116487 | USA, Michigan, Montcalm County, Stanton | P. nigra | 2001 | Adams G, Barnes I | AY808302 | AY808267 | AY808197 | NA | MK015293 |
| D. septosporum | 44656 | CBS 140339 | Russia, St. Petersburg, Park Sosnovka | P. sylvestris | Nov 2013 | Drenkhan R, Musolin D, Adamson K | KU948400 | MK015397 | MK015049 | MK015517 | MK015294 |
| D. septosporum | 44657 | CBS 141531 | Russia, St. Petersburg, Park Sosnovka | P. sylvestris | Nov 2013 | Drenkhan R, Musolin D, Adamson K | KU948401 | MK015398 | MK015050 | MK015518 | MK015295 |
| Lecanosticta acicola | 9985 | CBS 871.95 | France | P. radiata | Apr 1995 | Morelet M | GU214663 | MK015399 | MK015051 | MK015519 | MK015296 |
| L. acicola | 45426 | CBS133790 | Lithuania | P. mugo | 2009 | Markovskaja S, Kacergius A, Treigiene A | HM367708 | JX901645 | MK015052 | MK015520 | MK015297 |
| L. acicola | 45427 | CBS 133791 | USA, New Hampshire, Blackwater | P. strobus | Jun 2011 | Ostrofsky B | KC012999 | KC013002 | MK015053 | MK015521 | MK015298 |
| L. acicola | 45428 | CBS 322.33 | USA | P. palustris | Feb 1933 | Siggers PV | MK015156 | MK015400 | MK015054 | MK015522 | MK015299 |
| L. acicola | 50541 | Lithuania, Curonian Spit, Juodkrante | P. mugo | Sep 2014 | Markovskaja S | MK015157 | MK015401 | MK015055 | MK015523 | MK015300 | |
| L. acicola | 50542 | Lithuania, Curonian Spit, Juodkrante | P. mugo | Sep 2014 | Markovskaja S | MK015158 | MK015402 | MK015056 | MK015524 | MK015301 | |
| L. brevispora | - e | 1A.N5S2 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015159 | MK015403 | – | – | – |
| L. brevispora | - e | 1C.N1S3 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015160 | MK015404 | MK015057 | NA | NA |
| L. brevispora | - e | 1C.N5S4 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015161 | MK015405 | MK015058 | MK015525 | MK015302 |
| L. brevispora | - e | 1C.N6S2 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015162 | MK015406 | – | – | – |
| L. brevispora | - e | 1D.N1S3 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015163 | MK015407 | MK015059 | NA | NA |
| L. brevispora | - e | IB31.4a | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015164 | MK015408 | MK015060 | MK015526 | MK015303 |
| L. brevispora | 36894 | Guatemala, Finca La Soledad (near Jalapa), Mataquescuintla | P. pseudostrobus | Oct 2010 | Barnes I | MK015165 | MK015409 | MK015061 | NA | MK015304 | |
| L. brevispora | 37123 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015166 | MK015410 | NA | NA | MK015305 | |
| L. brevispora | 42646 | Honduras | P. oocarpa | – | – | MK015167 | MK015411 | MK015062 | MK015527 | MK015306 | |
| L. brevispora | 42647 | Guatemala, Lugar, La Soledad, Jalapa | P. oocarpa | Oct 2010 | Barnes I | MK015168 | MK015412 | MK015063 | MK015528 | MK015307 | |
| L. brevispora | 45424 | CBS 133601 | Mexico | Pinus sp. | Oct 2009 | Yanes-Morales M | JX901763 | JX901649 | MK015064 | MK015529 | MK015308 |
| L. brevispora | 46499 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015169 | MK015413 | – | – | – | |
| L. brevispora | 46500 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015170 | MK015414 | – | – | – | |
| L. brevispora | 46501 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015171 | MK015415 | MK015065 | NA | NA | |
| L. brevispora | 46502 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015172 | MK015416 | – | – | – | |
| L. brevispora | 46503 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015173 | MK015417 | MK015066 | MK015530 | MK015309 | |
| L. brevispora | 46504 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015174 | MK015418 | MK015067 | MK015531 | MK015310 | |
| L. brevispora | 46505 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015175 | MK015419 | NA | NA | MK015311 | |
| L. brevispora | 46506 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015176 | MK015420 | – | – | – | |
| L. brevispora | 46507 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015177 | MK015421 | – | – | – | |
| L. brevispora | 46508 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015178 | MK015422 | – | – | – | |
| L. brevispora | 46509 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015179 | MK015423 | – | – | – | |
| L. brevispora | 46510 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015180 | MK015424 | NA | NA | MK015312 | |
| L. brevispora | 46511 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015181 | MK015425 | – | – | – | |
| L. brevispora | 46512 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015182 | MK015426 | – | – | – | |
| L. brevispora | 46807 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015183 | MK015427 | MK015068 | MK015532 | MK015313 | |
| L. brevispora | 49291 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015184 | MK015428 | MK015069 | NA | MK015314 | |
| L. brevispora | 49292 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015185 | MK015429 | MK015070 | MK015533 | MK015315 | |
| L. brevispora | 49293 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015186 | NA | MK015071 | MK015534 | MK015316 | |
| L. brevispora | 49294 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015187 | NA | MK015072 | MK015535 | MK015317 | |
| L. brevispora | 49295 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015188 | NA | MK015073 | MK015536 | MK015318 | |
| L. brevispora | 49296 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015189 | MK015430 | MK015074 | MK015537 | MK015319 | |
| L. brevispora | 49297 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015190 | MK015431 | MK015075 | MK015538 | MK015320 | |
| L. brevispora | 49298 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015191 | MK015432 | MK015076 | MK015539 | MK015321 | |
| L. brevispora | 50523 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015192 | MK015433 | – | – | – | |
| L. brevispora | 50526 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015193 | MK015434 | MK015077 | NA | NA | |
| L. brevispora | 50527 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015194 | MK015435 | NA | NA | MK015322 | |
| L. brevispora | 50528 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015195 | MK015436 | MK015078 | NA | NA | |
| L. brevispora | 50529 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015196 | MK015437 | – | – | – | |
| L. brevispora | 50530 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015197 | MK015438 | MK015079 | MK015540 | MK015323 | |
| L. brevispora | 50531 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015198 | MK015439 | MK015080 | MK015541 | MK015324 | |
| L. brevispora | 50532 | Guatemala, Chimaltenango, Tecpán, Finca La Esperanza | P. pseudostrobus | Jun 2011 | Barnes I | MK015199 | MK015440 | – | – | – | |
| L. brevispora | 51050 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015200 | MK015441 | NA | MK015542 | MK015325 | |
| L. gloeospora c | 42645 | IMI 283812 | Mexico, Nuevo León, Iturbide-Galeana | P. pseudostrobus | May 1983 | Evans HC | KU948431 | MK015442 | MK015081 | MK015543 | MK015326 |
| L. guatemalensis | - e | IB30/2d | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015201 | MK015443 | – | – | – |
| L. guatemalensis | - e | IB32/1a | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015202 | MK015444 | – | – | – |
| L. guatemalensis | - e | IB32/2e | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015203 | MK015445 | MK015082 | NA | NA |
| L. guatemalensis | - e | IB35/2e | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015204 | MK015446 | MK015083 | NA | NA |
| L. guatemalensis | - e | IB35/2j | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015205 | MK015447 | – | – | – |
| L. guatemalensis | - e | IB35/9a | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015206 | MK015448 | MK015084 | NA | NA |
| L. guatemalensis c | IMI 275573 | Honduras, Yoro | P. oocarpa | Oct 1980 | Evans HC | MK015207 | MK015449 | NA | NA | NA | |
| L. guatemalensis c | IMI 281563 | Honduras | P. caribaea | May 1982 | Evans HC | MK015208 | NA | NA | NA | NA | |
| L. guatemalensis c | IMI 281596 | Nicaragua | P. tecunumanii | Nov 1981 | Evans HC | MK015209 | MK015450 | NA | NA | NA | |
| L. guatemalensis | - e | N3/1c | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015210 | MK015451 | MK015085 | MK015544 | MK015327 |
| L. guatemalensis | 36811 | Guatemala, Jalapa, Finca Forestal Soledad | P. maximinoi | Oct 2010 | Barnes I | MK015211 | MK015452 | MK015086 | NA | MK015328 | |
| L. guatemalensis | 36812 | Guatemala, Coban, San Juan Chamelco | P. maximinoi | Oct 2010 | Barnes I | MK015212 | MK015453 | MK015087 | MK015545 | MK015329 | |
| L. guatemalensis | 37121 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015213 | MK015454 | – | – | – | |
| L. guatemalensis | 37122 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015214 | MK015455 | MK015088 | MK015546 | MK015330 | |
| L. guatemalensis | 37124 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015215 | MK015456 | – | – | – | |
| L. guatemalensis | 37126 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015216 | MK015457 | MK015089 | MK015547 | MK015331 | |
| L. guatemalensis | 37127 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015217 | MK015458 | – | – | – | |
| L. guatemalensis c | 42206 | IMI 281598 | Guatemala | P. oocarpa | 1983 | Evans HC | JX901764 | JX901650 | MK015090 | MK015548 | MK015332 |
| L. guatemalensis | 43890 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015218 | MK015459 | – | – | – | |
| L. guatemalensis | 43891 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015219 | MK015460 | MK015091 | NA | NA | |
| L. guatemalensis | 43892 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015220 | MK015461 | MK015092 | NA | NA | |
| L. guatemalensis | 43893 | Guatemala, Chiquimula, San José la Arada | P. oocarpa | Oct 2010 | Barnes I | MK015221 | MK015462 | MK015093 | NA | NA | |
| L. guatemalensis | 43894 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015222 | MK015463 | MK015094 | NA | NA | |
| L. guatemalensis | 43895 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015223 | MK015464 | MK015095 | MK015549 | MK015333 | |
| L. guatemalensis | 45386 | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015224 | MK015465 | – | – | – | |
| L. guatemalensis | 45387 | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015225 | MK015466 | MK015096 | MK015550 | MK015334 | |
| L. guatemalensis | 45391 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015226 | MK015467 | MK015097 | NA | NA | |
| L. guatemalensis | 45392 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2011 | Barnes I | MK015227 | MK015468 | MK015098 | MK015551 | MK015335 | |
| L. guatemalensis | 45393 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015228 | MK015469 | – | – | – | |
| L. guatemalensis | 45394 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015229 | NA | – | – | – | |
| L. guatemalensis | 46811 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015230 | MK015470 | MK015099 | MK015552 | MK015336 | |
| L. guatemalensis | 46817 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015231 | MK015471 | MK015100 | MK015553 | MK015337 | |
| L. guatemalensis | 46819 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015232 | NA | – | – | – | |
| L. guatemalensis | 47108 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015233 | MK015472 | – | – | – | |
| L. guatemalensis | 49400 | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015234 | MK015473 | MK015101 | MK015554 | MK015338 | |
| L. guatemalensis | 49402 | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015235 | MK015474 | MK015102 | MK015555 | MK015339 | |
| L. guatemalensis | 51052 | Guatemala, Chiquimula, San José la Arada | P. oocarpa | Oct 2010 | Barnes I | MK015236 | MK015475 | MK015103 | MK015556 | MK015340 | |
| L. guatemalensis | 51142 | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015237 | MK015476 | MK015104 | MK015557 | MK015341 | |
| L. jani | - e | 267.44.N1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015238 | MK015477 | MK015105 | MK015558 | MK015342 |
| L. jani | - e | 267.47.N1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015239 | MK015478 | MK015106 | MK015559 | MK015343 |
| L. jani | - e | 267.47.N2 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015240 | MK015479 | MK015107 | MK015560 | MK015344 |
| L. jani | - e | 267.51.N2S1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015241 | MK015480 | NA | NA | MK015345 |
| L. jani | - e | 267.52.N1S1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015242 | MK015481 | MK015108 | MK015561 | MK015346 |
| L. jani | - e | 267.52.N2S1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015243 | MK015482 | MK015109 | MK015562 | MK015347 |
| L. jani | - e | IB30/2b | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015244 | MK015483 | MK015110 | MK015563 | NA |
| L. jani | - e | IB35/3c | Guatemala, Chiquimula | P. oocarpa | Oct 2010 | Barnes I | MK015245 | MK015484 | MK015111 | MK015564 | MK015348 |
| L. jani | - e | IB13/2f | Guatemala | P. maximinoi | Oct 2010 | Barnes I | MK015246 | MK015485 | MK015112 | MK015565 | MK015349 |
| L. jani | - e | N3/2c | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015247 | NA | MK015113 | MK015566 | MK015350 |
| L. jani | 36808 | Guatemala, Jalapa, Finca Forestal Soledad | P. maximinoi | Oct 2010 | Barnes I | MK015248 | NA | MK015114 | MK015567 | MK015351 | |
| L. jani | 36810 | Guatemala, Jalapa, Finca Forestal Soledad | P. maximinoi | Oct 2010 | Barnes I | MK015249 | NA | MK015115 | MK015568 | MK015352 | |
| L. jani | 37128 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015250 | MK015486 | MK015116 | MK015569 | MK015353 | |
| L. jani | 38950 | CBS 144446; PREM 62186 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015251 | MK015487 | MK015117 | MK015570 | MK015354 |
| L. jani | 38958 | CBS 144456; PREM 62185 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015252 | MK015488 | MK015118 | MK015571 | MK015355 |
| L. jani | 38959 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015253 | NA | NA | NA | NA | |
| L. jani | 38968 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015254 | NA | MK015119 | NA | NA | |
| L. jani | 45388 | Guatemala | P. maximinoi | Oct 2010 | Barnes I | MK015255 | NA | MK015120 | MK015573 | MK015356 | |
| L. jani | 45389 | Guatemala | P. maximinoi | Oct 2010 | Barnes I | MK015256 | MK015489 | MK015121 | MK015574 | MK015357 | |
| L. jani | 47109 | Guatemala | P. maximinoi | Oct 2010 | Barnes I | MK015257 | MK015490 | MK015122 | MK015575 | MK015358 | |
| L. jani | 48830 | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015258 | NA | MK015123 | MK015576 | MK015359 | |
| L. jani | 48831 | CBS 144447; PREM 62187 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015259 | MK015491 | MK015124 | MK015577 | MK015360 |
| L. jani | 49401 | Guatemala | P. maximinoi | Oct 2010 | Barnes I | MK015260 | MK015492 | NA | MK015578 | MK015361 | |
| L. jani | 51051 | Guatemala | P. maximinoi | Oct 2010 | Barnes I | MK015261 | MK015493 | MK015125 | MK015579 | MK015362 | |
| L. jani | 51058 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015262 | MK015494 | MK015126 | MK015580 | MK015363 | |
| L. jani | 51059 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. tecunumanii | Sep 2012 | Barnes I | MK015263 | MK015495 | MK015127 | MK015581 | MK015364 | |
| L. jani | 51143 | Nicaragua, Matagalpa | P. oocarpa | Jun 2011 | Barnes I | MK015264 | NA | MK015128 | MK015582 | MK015365 | |
| L. longispora | 45429 | CBS 133602 | Mexico | Pinus sp. | Oct 2009 | Yanes-Morales M | JX901766 | JX901651 | MK015129 | MK015583 | MK015366 |
| L. longispora | 45430 | CPC 17941 | Mexico | Pinus sp. | Oct 2009 | Yanes-Morales M | JX901765 | JX901652 | MK015130 | MK015584 | MK015367 |
| L. pharomachri | - e | 267.8A.N2S1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015265 | MK015496 | NA | NA | MK015368 |
| L. pharomachri | - e | 267.12.N1S2 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015266 | NA | NA | NA | MK015369 |
| L. pharomachri | - e | 267.30.MD.N1 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015267 | NA | NA | NA | MK015370 |
| L. pharomachri | - e | 267.30.MD.N2 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015268 | MK015497 | MK015131 | NA | MK015371 |
| L. pharomachri | - e | 267.30.N4 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015269 | MK015498 | MK015132 | MK015585 | MK015372 |
| L. pharomachri | 37132 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015270 | MK015499 | MK015133 | MK015586 | MK015373 | |
| L. pharomachri | 37133 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015271 | MK015500 | MK015134 | MK015587 | MK015374 | |
| L. pharomachri | 37134 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015272 | MK015501 | MK015135 | MK015588 | MK015375 | |
| L. pharomachri | 37136 | CBS 144448; PREM 62188 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015273 | MK015502 | MK015136 | MK015589 | MK015376 |
| L. pharomachri | 38947 |
CBS 144695; PREM 62189 |
Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015274 | MK015503 | MK015137 | MK015590 | MK015377 |
| L. pharomachri | 38974 | CBS 144449; PREM 62190 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015275 | MK015504 | MK015138 | MK015591 | MK015378 |
| L. pharomachri | 38975 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015276 | NA | NA | NA | NA | |
| L. pharomachri | 38976 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015277 | MK015505 | MK015139 | NA | MK015379 | |
| L. pharomachri | 46810 | Honduras | P. oocarpa | – | – | MK015278 | MK015506 | MK015140 | MK015592 | MK015380 | |
| L. pharomachri | 46813 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015279 | MK015507 | MK015141 | MK015593 | MK015381 | |
| L. pharomachri | 51053 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015280 | NA | MK015142 | NA | MK015382 | |
| L. pharomachri | 51054 | Guatemala, Jalapa, Finca La Soledad, Mataquescuintla | P. oocarpa | Sep 2012 | Barnes I | MK015281 | MK015508 | NA | NA | MK015383 | |
| L. tecunumanii | 46805 | CBS 144450; PREM 62191 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015282 | MK015509 | MK015143 | MK015594 | MK015384 |
| L. tecunumanii | 46812 | CBS 144452; PREM 62193 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015283 | MK015510 | MK015144 | MK015595 | MK015385 |
| L. tecunumanii | 49403 | CBS 144451; PREM 62192 | Guatemala, Baja Verapaz, San Jerónimo, Salamá | P. tecunumanii | Oct 2010 | Barnes I | MK015284 | MK015511 | MK015145 | MK015596 | MK015386 |
| L. variabilis | 36809 | CBS 144455; PREM 62195 | Guatemala, Jalapa, Finca Forestal Soledad | P. maximinoi | Oct 2010 | Barnes I | MK015285 | MK015512 | MK015146 | MK015597 | MK015387 |
| L. variabilis | 37125 | CBS 144454; PREM 62194 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015286 | KJ938446 | MK015147 | MK015598 | MK015388 |
| L. variabilis | 37129 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015287 | KJ938445 | MK015148 | MK015599 | MK015389 | |
| L. variabilis c | 42205 | IMI 281561; CBS 144453; PREM 62196 | Honduras, Santa Barbara, Lago Yojoa | P. caribaea | Oct 1980 | Evans HC | MK015288 | MK015513 | MK015149 | MK015600 | MK015390 |
| L. variabilis | 45390 | Guatemala, Alta Verapaz, Santa Cruz Verapaz, near Tactíc | P. oocarpa | Oct 2010 | Barnes I | MK015289 | MK015514 | MK015150 | MK015601 | MK015391 | |
| L. variabilis | 45425 | CBS 133789 | Mexico | Pinus sp. | Nov 2009 | Yanez-Morales M | JX901762 | JX901648 | MK015151 | MK015602 | MK015392 |
| Phaeophleospora eugeniae | 45432 | CPC15159 | Brazil, Vicosa, Paraiso | Eugenia uniflora | Mar 2008 | Alfenas AC | FJ493189 | JX901667 | MK015152 | MK015603 | NA |
| P. eugeniae | 45433 | CPC 15143 | Brazil, Vicosa, Paraiso | E. uniflora | Mar 2008 | Alfenas AC | FJ493188 | JX901666 | MK015153 | MK015604 | NA |
| P. gregaria | 45434 | CBS 111166 | South Africa, Western Cape Province, de Hoop Nature Reserve | Eucalyptus cladocalyx | Sep 1995 | Wood A | JX901773 | JX901664 | MK015154 | MK015605 | MK015393 |
| P. gregaria | 45435 | CBS 114662 | South Africa, Western Cape Province, Devon Valley, Stellenbosch | Eucalyptus sp. | Jun 1995 | Crous PW | DQ302953 | JX901654 | MK015155 | MK015606 | MK015394 |
aCMW Culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa;
bCBS Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands, CPC Personal collection of Pedro Crous housed at CBS, IMI The UK National Fungus Collection, CABI Bioscience, Egham, UK, PREM The dried herbarium collection of the South African National Collection of Fungi, Mycology Unit, Biosystematics Division, Plant Protection Institute, Agricultural Research Council, Pretoria, South Africa
cCultures were collected by HC Evans in Central America
d‘-‘= was not amplified; ‘NA‘= amplification unsuccessful;
e = no viable culture available that could be submitted to CMW
Ex-type isolate of each species is indicated in bold
Pine needles, showing symptoms of brown spots or bands, were collected from Pinus species native to Central America from 2010 to 2012 in Guatemala, as well as from Honduras and Nicaragua in 2011 (Table 1). Conidiomata formed on the needles were aseptically excised, rolled onto 2% Dothistroma Sporulating Media (DSM: 5 g yeast extract (Biolab, Merck, Modderfontein, South Africa), 20 g malt extract (Biolab) and 15 g agar (BD Difco™, Sparks, MD) per litre of distilled water) with 100 mg/L streptomycin (Sigma-Aldrich, St Louis, MO) in order to release conidia from the conidiomata as described by Barnes et al. (2004). The isolated conidiomata were incubated for one to two days at 23 °C. The plates were examined using a dissection microscope and single germinating conidia were selected and replated onto 2% DSM. The single conidial isolates were grown for 4–6 wk. on a natural day light cycle, at 23 °C.
DNA extractions and sequencing
Fungal tissue was scraped from the surface of the cultures on 2% DSM with a sterile scalpel blade and lyophilized. The freeze-dried mycelium was homogenized using a Retsch MM301 mixer mill (Haan, Germany) and approximately 20 ng of the crushed mycelium was used as starting material for DNA extractions. DNA was extracted using a Zymo Research ZR Fungal/Bacterial DNA MiniPrep™ kit (Irvine, CA) and eluted into a final volume of 50 μl. The quality and quantity of the extracted DNA was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Fischer Scientific, Waltham, MA). DNA concentrations were diluted to 20 ng/μl working stock for polymerase chain reaction (PCR) amplifications and stored at − 20 °C until further use.
The nuclear rDNA region encompassing the internal transcribed spacers (ITS) 1 and 2, along with the 5.8S rDNA region was amplified using primers ITS1 and ITS4 (White et al. 1990) and a portion of the translation elongation factor 1-α gene (TEF1) using primers EF1-728F (Carbone and Kohn 1999) and EF2 (O’Donnell et al. 1998) for all the isolates. The Beta-tubulin-2 gene region (BT2) was amplified using the primer pair T1 (O’Donnell and Cigelnik 1997) and β-Sandy-R (Stukenbrock et al. 2012) or the primers Bt2A and Bt2B (Glass and Donaldson 1995). The Beta-tubulin-1 gene region (BT1) was amplified using primers Bt1A and Bt1B (Glass and Donaldson 1995), the RNA polymerase II second largest subunit (RPB2) gene region using primers RPB2-5f2 (Sung et al. 2007) and RPB2-7cR (Liu et al. 1999) and the guanine nucleotide-binding protein subunit beta (MS204) using primers MS204F.cerato and MS204R.cerato (Fourie et al. 2015).
PCR reactions for each of the six regions contained 20 ng DNA, 2.5 μl 10x PCR reaction buffer, 2.5 mM MgCl2, 400 nM of each primer, 200 μM of each dNTP and 1 U Faststart Taq DNA Polymerase (Roche Diagnostics, Indianapolis, IN). Reaction volumes were adjusted to 25 μl with sterile SABAX water (Adcock Ingram, Midrand, South Africa). PCR reactions were carried out on an Applied Biosystems® Veriti® 96 well Thermal cycler (Thermo Fisher Scientific, Waltham, MA). The cycling conditions for all six gene regions included an initial denaturation step at 95 °C for 4 min, 10 cycles consisting of 94 °C for 20 s (denaturation), a 45 s annealing step according to the primer pair annealing temperature (Table 2) and an elongation step of 45 s at 72 °C. This was followed by a further 25 cycles of 94 °C for 20 s, 45 s with a 5 s extension step per cycle at the indicated annealing temperature, a 72 °C extension for 45 s and a final step of 72 °C for 10 min. The annealing temperature was set at 56 °C for ITS, 52 °C for TEF1, 50 °C for BT1, 52 °C for BT2, 55 °C for MS204 and 56 °C for RPB2. To visualise amplified products, 5 μl PCR products were stained with 1 μl GelRed™ nucleic acid gel stain (Biotium, Fremont, CA) and separated on 2% SeaKem® LE agarose gel (Lonza, Rockland, ME) for 20 min at 100 V and viewed under a UV light using the GelDoc™ EZ Imager (BioRad, Hercules, CA). PCR products were cleaned with a 6.65% G-50 Sephadex solution (Sigma-Aldrich, St Louis, MO) following the manufacturer’s instructions using Centri-sep spin columns (Princeton Separations, Freehold, NJ).
Table 2.
Primers used for PCR amplification and sequencing in this study
| Locus | Primer name | Direction | Primer sequence 5′ to 3’ | Annealing temperature used (°C) | Amplification success | Reference |
|---|---|---|---|---|---|---|
| BT1 | Bt1a | Forward | TTC CCC CGT CTC CAC TTC TTC ATG | 50 | 87.4% | Glass and Donaldson 1995 |
| Bt1b | Reverse | GAC GAG ATC GTT CAT GTT GAA CTC | 50 | Glass and Donaldson 1995 | ||
| BT2a | T1 | Forward | AAC ATG CGT GAG ATT GTA AGT | 52 | – | O’Donnell and Cigelnik 1997 |
| β-Sandy-R | Reverse | GCR CGN GGV ACR TAC TTG TT | 52 | Stukenbrock et al. 2012 | ||
| Bt2a | Forward | GGT AAC CAA ATC GGT GCT GCT TTC | 52 | – | Glass and Donaldson 1995 | |
| Bt2b | Reverse | ACC CTC AGT GTA GTG ACC CTT GGC | 52 | Glass and Donaldson 1995 | ||
| TEF1 | EF1-728F | Forward | CAT CGA GAA GTT CGA GAA GG | 52 | 88.2% | Carbone and Kohn 1999 |
| EF-2 | Reverse | GGA RGT ACC AGT SAT CAT GTT | 52 | O’Donnell et al. 1998 | ||
| ITS | ITS1 | Forward | GAA GTA AAA GTC GTA ACA AGG | 56 | 100% | White et al. 1990 |
| ITS4 | Reverse | TCC TCC GCT TAT TGA TAT GC | 56 | White et al. 1990 | ||
| MS204 | MS204F.cerato | Forward | AAG GGC ACC CTC GAG GGC CAC | 55 | 71.7% | Fourie et al. 2015 |
| MS204R.cerato | Reverse | GAT GGT RAC GGT GTT GAT GTA | 55 | Fourie et al. 2015 | ||
| RPB2 | RPB2-5f2 | Forward | GGG GWG AYC AGA AGA AGG C | 56 | 82.7% | Sung et al. 2007 |
| fRPB2-7cR | Reverse | CCC ATR GCT TGY TTR CCC AT | 56 | Liu et al. 1999 |
aBT2 amplification success using all primer combinations was very low and abandoned
The concentrations of the cleaned PCR products were determined using a NanoDrop ND-1000 spectrophotometer and 60–100 ng of DNA and products were sequenced in both the forward and reverse direction using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) on an ABI PRISM 3500xl capillary auto sequencer (Thermo Fisher Scientific).
Forward and reverse sequences were aligned and consensus sequences generated in CLC Main workbench version 8.0 (CLC Bio, https://www.qiagenbioinformatics.com/products/clc-main-workbench/). All consensus sequences generated in this study were deposited in GenBank that is hosted by the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genbank/) (Table 1).
Data analyses
Five datasets (BT1, ITS, MS204, RPB2 and TEF1) were generated and analysed individually. A partition homogeneity test (PHT) was performed with the software package PAUP* 4.0b10 (Swofford 2003) to test congruence between the five gene regions and a sixth dataset, where sequences were available for all five gene regions, was compiled and analysed. The BT1, ITS, MS204 and RPB2 datasets included all of the sequences generated in this study and additional sequences available from GenBank (Table 1). The TEF1 dataset included all of the sequence data generated in this study as well as additional sequences representing 14 different TEF1 haplotypes of L. acicola (including possible cryptic species) (Janoušek et al. 2016) that were downloaded from GenBank (Table 3). Sequences for all datasets were aligned with the online version of MAFFT Version 7 (Katoh and Standley 2013; http://mafft.cbrc.jp/alignment/server/) using default settings. Aligned data were imported into MEGA 7.0.14 (Kumar et al. 2016) and manually checked and adjusted.
Table 3.
GenBank numbers of Lecanosticta acicola TEF1 haplotypes included in the TEF1 phylogenetic analysis (Fig. 2) as well as additional locations represented by the haplotypes
| Species name assigned in this studya | GenBank Accession number | Country | State / Region | Location | Host | Date of collection | Collector / Supplier |
|---|---|---|---|---|---|---|---|
| Lecanosticta acicola | KJ938442 | Japan | Shimane | Matsue, Hamanogi | Pinus thunbergii | Feb 2010 | Suto Y |
| L. acicola | KJ938439 | Mexico | Nuevo León | Iturbide, Bosque Escuela | Pinus halepensis | May 2010 | Marmolejo JG |
| L. acicola | KJ938440 | Mexico | Nuevo León | Iturbide, Bosque Escuela | Pinus halepensis | May 2010 | Marmolejo JG |
| L. acicola | KJ938441 | Mexico | Nuevo León | Iturbide, Bosque Escuela | Pinus halepensis | May 2010 | Marmolejo JG |
| L. acicola | KJ938438 | USA | Maine | York, Lyman | Pinus strobus | Jun 2011 | Ostrofsky W |
| L. acicola | KJ938443 | USA | Mississippi | Harrison County | Pinus palustris | Oct 2012 | Bartlett B, Burdine C |
| L. acicola | KJ938444 | USA | Mississippi | Harrison County | Pinus palustris | Oct 2012 | Bartlett B, Burdine C |
| L. acicola | KJ938450 | USA | Mississippi | Harrison County | Pinus palustris | Oct 2012 | Bartlett B, Burdine C, Roberds J |
| L. acicola | KJ938451 | USA | Mississippi | Harrison County | Pinus palustris | Oct 2012 | Bartlett B, Burdine C |
| Lecanosticta variabilis | KJ938445 | Guatemala | Alta Verapaz | Santa Cruz Verapaz, near Tactíc | Pinus oocarpa | Oct 2010 | Barnes I |
| L. variabilis | KJ938446 | Guatemala | Alta Verapaz | Santa Cruz Verapaz, near Tactíc | Pinus oocarpa | Oct 2012 | Barnes I |
| L. variabilis | KJ938447 | Mexico | Nuevo León | Piñal de los Amoles, Querétaro | Pinus sp. | 2011 | Kunte L |
| L. variabilis | KJ938448 | Mexico | Nuevo León | Iturbide, Bosque Escuela | Pinus halepensis | May 2010 | Marmolejo JG |
| L. variabilis | KJ938449 | Mexico | Nuevo León | Galeana, Cerro del Potosí | Pinus arizonica var. stormiae | Apr 2010 | Marmolejo JG |
| Countries, regions, locations and hosts represented by the above isolatesb | |||||||
| the same as KJ938438 | Austria | Lower Austria | Hollenstein an der Ybbs | Pinus mugo | Oct 2004 | Kirisits T, Barnes I | |
| the same as KJ938438 | Austria | Lower Austria | Opponitz | Pinus mugo | 2010 | Hintsteiner M | |
| the same as KJ938438 | Austria | Lower Austria | Saimannslehen | Pinus sp. | 2010 | Hintsteiner M | |
| the same as KJ938438 | Austria | Lower Austria | Sankt Gallen | Pinus mugo | 2010 | Hintsteiner M | |
| the same as KJ938438 | Austria | Lower Austria | Steyer, Pestalozzistraße | Pinus mugo | 2010 | Hintsteiner M | |
| the same as KJ938438 | Austria | Lower Austria | Waidehofen an der Ybbs | Pinus mugo | Aug 2010 | Janoušek J | |
| the same as KJ938438 | Austria | Upper Austria | Gmunden | Pinus nigra | Jun 2012 | Kirisits T | |
| the same as KJ938438 | Canada | Québec | Demers-Centre | Pinus strobus | Jun 2011 | Harvey L | |
| the same as KJ938438 | Canada | Québec | Lake Aberdeen | Pinus strobus | Jun 2011 | Harvey L | |
| the same as KJ938438 | Canada | Québec | Lake Pinseault | Pinus strobus | Jun 2011 | Harvey L | |
| the same as KJ938438 | Canada | Québec | Montréal | Pinus mugo | Jun 2011 | Harvey R | |
| the same as KJ938438 | Canada | Québec | Waltham | Pinus strobus | Jun 2011 | Harvey L | |
| the same as KJ938442 | China | Fujie | Pinus elliottii | 1988 | Zheng-Yu H | ||
| the same as KJ938451 | Colombia | Refocosta L-75 | Villanueva, Casanare | Pinus caribaea | Mar 2011 | Rodas C, Barnes I | |
| the same as KJ938438 | Croatia | Zadar | Pinus halapensis | Sep 2009 | Diminic D | ||
| the same as KJ938438 | Czech Republic | Southern Bohemia | Borkovická Blata | Pinus uncinata subsp. uliginosa | Oct 2011 | Janoušek J | |
| the same as KJ938438 | Czech Republic | Southern Bohemia | Červená Blata | Pinus uncinata subsp. uliginosa | Aug 2009 | Dvořák M, Janoušek J | |
| the same as KJ938438 | Estonia | Harju maakond | Tallin | Pinus ponderosa | Jul 2008 | Cech T | |
| the same as KJ938451 | France | Pyrénées-Atlantiques | Pinus radiata | 2012 | Kersaudy E, Ioos R | ||
| the same as KJ938438 | Germany | Bavaria | Grassau | Pinus mugo | 2000 | Blaschke FR, Wulf | |
| the same as KJ938438 | Germany | Bavaria | Murnau | Pinus mugo | Feb 2010 | Nannig A | |
| the same as KJ938438 | Germany | Bavaria | Murnauer Filze | Pinus mugo | Nov 2011 | Nannig A | |
| the same as KJ938438 | Germany | Bavaria | Pfrűhlmoos | Pinus mugo | Nov 2011 | Nannig A | |
| the same as KJ938438 | Italy | Brecia | Gardone | Pinus mugo | Jun 2008 | Cech T | |
| the same as KJ938438 | Lithuania | Klaipėdský kraj | Curonian Spit, Juodkrante | Pinus mugo | 2010 | Markovskaja S | |
| the same as KJ938438 | Slovenia | Upper Carniola | Bled | Pinus mugo | Jul 2009 | Jurc D | |
| the same as KJ938442 | South Korea | Naju | Sanpo-myeon | Pinus thunbergii | 2010 | KACC, Seo ST | |
| the same as KJ938451 | Spain | Cantabria | San Sebastián de Garabandal | Pinus radiata | Oct 2012 | Jankovský L, Janoušek J | |
| the same as KJ938438 | Switzerland | Canton St Gallen | Walensee | Pinus mugo | Oct 1999 | Wulf | |
| the same as KJ938438 | USA | Maine | Androscoggin, Leeds | Pinus strobus | Jun 2011 | Ostrofsky W | |
| the same as KJ938438 | USA | Maine | Piscataquis, Sangerville | Pinus strobus | Jun 2011 | Weimer J | |
| the same as KJ938438 | USA | Maine | York, Lyman | Pinus strobus | Jun 2011 | Ostrofsky W | |
| the same as KJ938438 | USA | Michigan | Wexford County, Springville Township | Pinus sylvestris | 2011 | Odonnell J | |
| the same as KJ938444 | USA | Mississippi | Harrison County | Pinus palustris | Oct 2012 | Bartlett B, Burdine C, Roberds J | |
| the same as KJ938438 | USA | New Hampshire | Hillsboro, Fox State Park | Pinus strobus | Jun 2011 | Weimer J | |
| the same as KJ938438 | USA | New Hampshire | Merrimack, Black Water Reserve | Pinus strobus | Jun 2011 | Weimer J | |
| the same as KJ938438 | USA | New Hampshire | Merrimack, Hopkinton-Everett | Pinus strobus | Jun 2011 | Weimer J | |
| the same as KJ938438 | USA | Vermont | Washington, Waterbury | Pinus strobus | Jun 2011 | Lackey J | |
| the same as KJ938438 | USA | Vermont | Windsor, Bethel | Pinus strobus | Jul 2011 | Munck I | |
| the same as KJ938438 | USA | Wisconsin | Merrillan | Pinus sylvestris | Apr 2010 | Stanosz G | |
aLecanosticta variabilis was previously identified as L. acicola but is now defined as a new species
bInformation adapted from Janoušek et al. (2016), Table S1
Three separate analyses were performed for each of the six datasets: Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian inference (BI). The MP analysis were performed with the software package PAUP* 4.0b10 (Swofford 2003). Gaps were treated as a fifth character state. One thousand random stepwise addition heuristic searches were performed with tree-bisection-reconnection (TBR) as the branch-swapping algorithm. Uninformative characters were excluded and the consistency index (CI), homoplasy index (HI), rescaled consistency index (RC), retention index (RI) and tree length (TL) were determined for the resulting trees (Table 4). The confidence levels were estimated by performing 1000 bootstrap replicates.
Table 4.
PCR amplification size, phylogenetic data and the substitution models used in the phylogenetic analysis for each gene region and for the combined datasets
| ITS | TEF1 | BT1 | MS204 | RPB2 | Combined datasets | |
|---|---|---|---|---|---|---|
| Approximate amplicon size (bp) | 550 | 520 | 420 | 760 | 940 | – |
| Number of taxa analysed | 153 | 147 | 111 | 91 | 105 | 76 |
| Aligned characters (bp) | 734 | 586 | 440 | 785 | 929 | 3344 |
| Number of parsimony-uninformative characters | 621 | 143 | 357 | 519 | 538 | 2438 |
| Number of parsimony-informative characters | 114 | 423 | 82 | 266 | 371 | 1121 |
| Number of trees retained | 108 | 396 | 1 | 2448 | 420 | 100 |
| Consistency index | 0.865 | 0.499 | 0.739 | 0.791 | 0.738 | 0.607 |
| Homoplasy index | 0.135 | 0.501 | 0.261 | 0.209 | 0.262 | 0.393 |
| Rescaled consistency index | 0.850 | 0.459 | 0.703 | 0.748 | 0.696 | 0.555 |
| Retention index | 0.982 | 0.919 | 0.951 | 0.946 | 0.943 | 0.914 |
| Tree Length | 163 | 1675 | 138 | 546 | 722 | 2642 |
| Substitution model | TPM2uf + G | GTR + G | GTR + G | TVM + G | TrN + G | GTR + G |
In order to determine the ML and BI, the best fit substitution model for each of the data sets were determined using jModelTest 0.1.1 (Posada 2008). Maximum likelihood analysis was performed with the program PhyML 3.0 (Guindon et al. 2010). The confidence levels were estimated with 1000 bootstrap replicates.
MrBayes 3.1.2 (Ronquist et al. 2012) was used to determine the BI for each data set by applying the Markov Chain Monte Carlo (MCMC) method. For each dataset, four independent MCMC chains were randomly started and run for six million generations, applying the best substitution model determined by jModelTest 0.1.1. Trees were sampled every 100 generations. Burn-in values were determined using Tracer 1.6 (Rambaut et al. 2014) by comparing the log likelihoods. Trees sampled in the burn-in phase were discarded. The remaining trees were used to construct majority rule consensus trees and to determine posterior probabilities for the tree topology.
Morphological characterization
Cultures were grown on 2% Malt Extract Agar (MEA), Oatmeal Agar (OA) and Potato Dextrose Agar (PDA) (Crous et al. 2009b; Quaedvlieg et al. 2012) at 20 °C for 2 wk. in darkness in order to examine the morphology and colour of the cultures of each species. Cultures on MEA were used for microscopic measurements of the conidiophores, conidiogenous cells and conidia. Slides were mounted in SABAX water (Adcock Ingram, Midrand, South Africa) for microscopy and examined using a Zeiss Axioskop 2 Plus compound microscope (Zeiss, Oberkochen, Germany). Photographic images were captured with a Nikon DS-Ri2 camera with the NIS Element BR v4.3 software package (Nikon, Tokyo, Japan). Up to 50 measurements of each morphologically characteristic structure was taken for each ex-type isolate and ten measurements were made for each of the paratypes examined. The mean, standard deviation, minimum and maximum were calculated for each morphological structure and the measurements presented as (minimum–) (mean – standard deviation) – (mean + standard deviation) (−maximum) for the conidia and conidiogenous cells. For the conidiophores, the maximum observed length was indicated together with the width as (minimum–) (mean) (−maximum).
Temperature requirements for growth in culture was studied on representative isolates selected for each of the novel species. Four by four millimeter blocks of each culture were plated, in triplicate, onto the centres of 2% MEA plates per temperature (10, 15, 20, 25, and 30 °C) and incubated in darkness. The diameters of each colony were recorded weekly along perpendicular axes for 4 wk. The colour and shape of each colony was recorded after 2 wk. of growth at 20 °C. Culture colour was determined using Rayner’s colour chart (Rayner 1970).
Accession of cultures and types
Holotype specimens of the new species, which are dried cultures, are deposited in the National Mycological Herbarium in Pretoria (PREM). Cultures are deposited in the culture collection (CBS) of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands, and ex-type cultures, as well as all other isolates included in this study, are maintained in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI) in Pretoria, South Africa (Table 5).
Table 5.
Specimens for which the morphology was examined for the description of Lecanosticta jani, L. pharomachri, L. tecunumanii and L. variabilis
| Species | CMW numbera | Status of specimen | Herbarium specimenb | Ex-type isolatesc |
|---|---|---|---|---|
| Lecanosticta jani | CMW 38950d | Paratype | PREM 62186 | CBS 144446 |
| CMW 38958d | Holotype | PREM 62185 | CBS 144456 | |
| CMW 48831e | Paratype | PREM 62187 | CBS 144447 | |
| CMW 51058d | Additional material examined | |||
| CMW 51059d | Additional material examined | |||
| CMW 51143e | Additional material examined | |||
| CMW47109e | Additional material examined | |||
| Lecanosticta pharomachri | CMW 37136 | Holotype | PREM 62188 | CBS 144448 |
| CMW 38947 | Paratype | PREM 62189 | CBS 144695 | |
| CMW 38974 | Paratype | PREM 62190 | CBS 144449 | |
| CMW 38976 | Additional material examined | |||
| CMW 51053 | Additional material examined | |||
| CMW 51054 | Additional material examined | |||
| Lecanosticta tecunumanii | CMW 46805 | Holotype | PREM 62191 | CBS 144450 |
| CMW 46812 | Paratype | PREM 62193 | CBS 144452 | |
| CMW 49403 | Paratype | PREM 62192 | CBS 144451 | |
| Lecanosticta variabilis | CMW 42205 | Holotype | PREM 62196 | CBS 144453, IMI 281561 |
| CMW 37125 | Paratype | PREM 62194 | CBS 144454 | |
| CMW 36809 | Paratype | PREM 62195 | CBS 144455 | |
| CMW 45425 | Additional material examined | CBS H-21112 | CBS 133789 | |
| CMW 37129 | Additional material examined | |||
aCMW Culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa; bThe herbarium deposits are dried cultures that serve as holotype and paratype specimens. PREM = The dried herbarium collection of the South African National Collection of Fungi, Mycology Unit, Biosystematics Division, Plant protection Institute, Agricultural Research Council, Pretoria, South Africa; cThe ex-type cultures are living cultures linked to the holotype and paratype specimens. CBS = The culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; IMI = The UK National Fungus Collection maintained by CABI Bioscience, Egham, UK; d Lecanosticta jani cultures with the Type 2 morphology; e Lecanosticta jani cultures with the Type 1 morphology
RESULTS
Fungal collections
Twenty-six isolates or DNA samples were obtained from culture collections to include in the study. An additional 127 isolates of putative Lecanosticta species were obtained from symptomatic needles collected from 36 different trees in Guatemala, Nicaragua and Honduras (Table 1). In Guatemala, 22 isolates were obtained from Pinus oocarpa, P. maximinoi, and P. tecunumanii needles that were collected in the Alta Verapaz District, 16 isolates were obtained from P. oocarpa needles collected in Chiquimula, 35 isolates from P. pseudostrobus needles collected in the Chimaltenango District in the Tecpán Municipality, eight isolates from P. tecunumanii needles collected in the Baja Verapaz District, 29 isolates from P. tecunumanii and P. oocarpa needles collected in the Jalapa District, and seven isolates from P. maximinoi needles in Coban and other regions (Table 1). Two isolates were obtained from P. oocarpa needles collected in Honduras and eight isolates were made from P. oocarpa needles collected in Matagalpa, Nicaragua.
DNA extraction and sequencing
The ITS and TEF1 regions were sequenced for all 153 isolates obtained and the BT1, MS204 and RPB2 regions were sequenced for 127 representatives of all monophyletic groups identified in the generated ITS and TEF1 phylogenetic trees. The selected representatives included all of the closely related Mycosphaerellaceae isolates, all the isolates that did not group with known Lecanosticta species, and a selection of isolates that grouped with known Lecanosticta species (Table 1). PCR fragments of approximately 550 bp were generated for ITS, 520 bp for TEF1, 420 bp for BT1, 760 bp for MS204 and 940 bp for RPB2. The amplification success of the TEF1, BT1, MS204 and RPB2 gene regions varied for the isolates that were selected and the amplification success rate of TEF1 was 88.2%, BT1 was 87.4%, MS204 was 71.7 and 82.7% for the RPB2 region (Table 2). The BT2 region did not amplify well across species of Lecanosticta. The amplification success rate and subsequent sequencing of the BT2 region using the T1 and β-Sandy-R primer pair, as well as Bt2a and Bt2b was very poor and further analysis of the BT2 region was abandoned.
Phylogenetic analyses
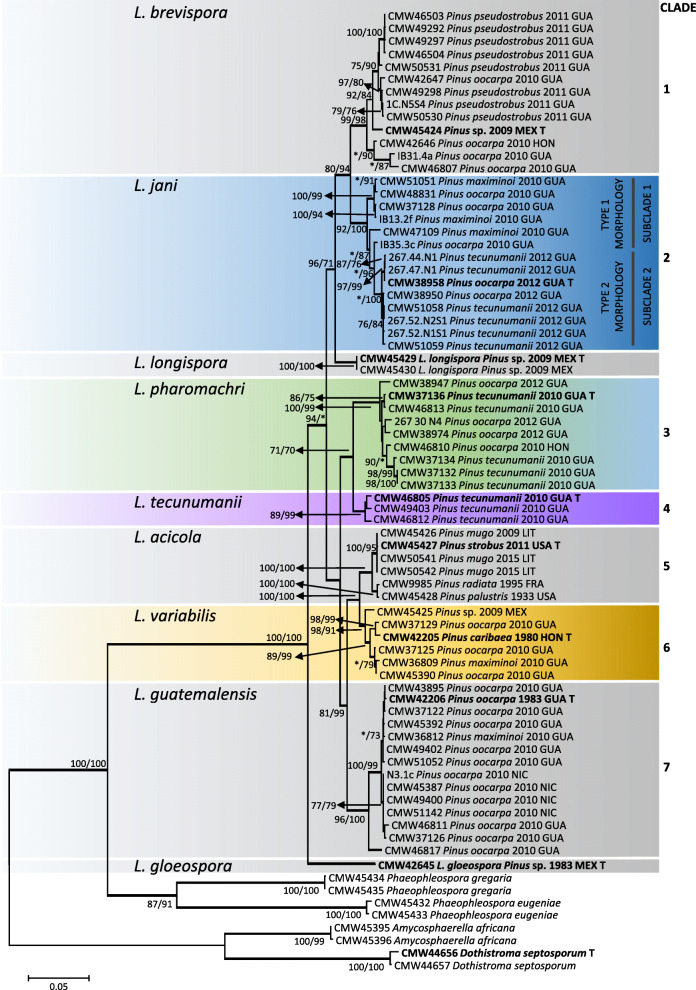
For the analyses, the datasets of the ITS region consisted of 153 taxa with 734 aligned nucleotides including gaps; the TEF1 dataset consisted of 147 taxa with 586 aligned nucleotides, the BT1 dataset consisted of 111 taxa with 440 aligned nucleotides; the MS204 dataset consisted of 91 taxa with 785 aligned nucleotides, and the RPB2 dataset consisted of 105 taxa with 929 aligned nucleotides, all including gaps. The PHT test yielded a P value = 0.01 and therefore the five datasets were considered incongruent. However, it was previously argued that a P value > 0.01 did not reduce phylogenetic accuracy (Cunningham 1997) and a combined phylogenetic tree representing the five gene regions ITS, TEF1, BT1, MS204 and RPB2 was constructed for presentation purposes (Fig. 1). The combined dataset consisted of 76 taxa with 3344 aligned nucleotides including gaps. Constant characters, parsimony-uninformative and informative characters, the consistency index (CI), homoplasy index (HI), rescaled consistency index (RC), retention index (RI) and tree length (TL) values for the maximum parsimony analyses are indicated in Table 4. For the parsimony analyses, 108 trees were retained for ITS, 396 for TEF1, 1 for BT1, 2448 for MS204 and 420 for RPB2. The best fit substitution models for ML and BI were selected by Akaike Information Criterion (AIC) and are indicated in Table 4. A 10% burn-in value was selected in the BI analysis for each of the data matrices for each of the analyses. Because the MP, ML and BI analysis all resulted in similar tree topologies, the ML trees were selected and chosen for presentation (Figs. 1 and 2, Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3 and Additional file 4: Figure S4).
Fig. 1.
Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the combined data of the ITS, TEF1, BT1, MS204 and RPB2 gene regions. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma septosporum was used as the outgroup taxa. The indicated clades are referred to in the text. All represented type species are indicated in bold and with a “T”
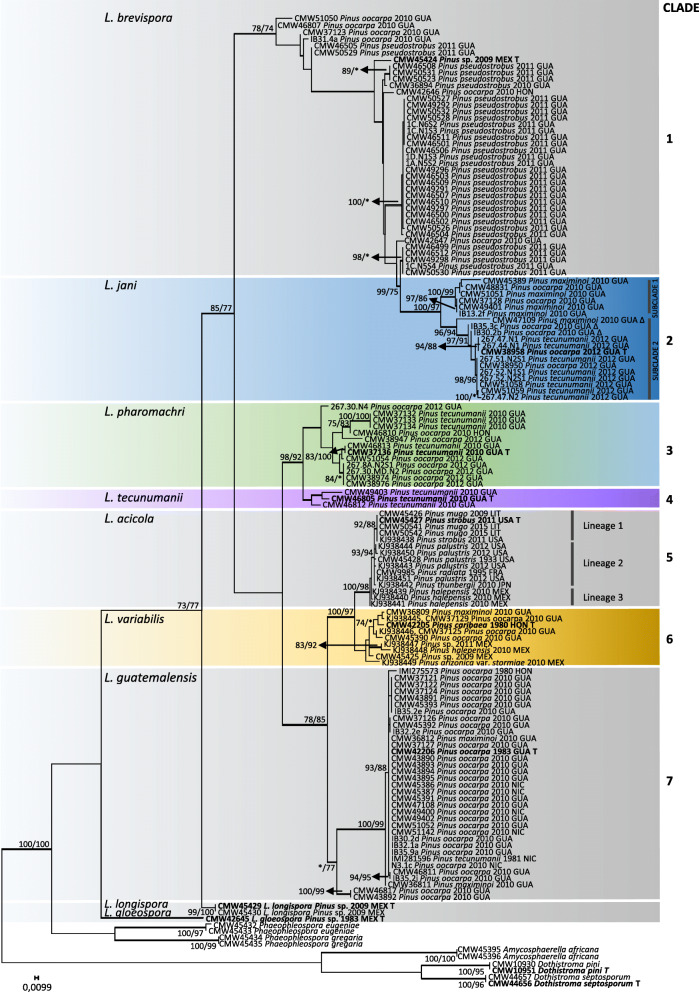
Fig. 2.
Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the TEF1 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate either exhibits Type 2 morphology and groups with Subclade 1, or, exhibits Type 1 morphology and groups with Subclade 2
Phylogenetic analyses of the combined dataset (Fig. 1), ITS (Additional file 1: Figure S1), TEF1 (Fig. 2) and MS204 (Additional file 3: Figure S3) consistently grouped the isolates sequenced in this study into seven distinct clades. The clades in Fig. 2 and Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3 and Additional file 4: Figure S4 are labelled according to the clades assigned in Fig. 1. In the case of RPB2 (Additional file 4: Figure S4) Clades 1–4, and 7 were also present but Clades 5 and 6 were not distinct from each other for this particular gene region. In the case of BT1 (Additional file 2: Figure S2), Clades 3, 5 and 6 could not be distinguished from each other. None of the isolates grouped with the types of L. gloeospora or L. longispora.
Forty-two of the isolates from Central America grouped in Clade 1 based on the ITS analysis (Additional file 1: Figure S1) and were identified as Lecanosticta brevispora. This was the most common species identified from the Central American collection and most isolates were from Chimaltenango on Pinus pseudostrobus. The pathogen was also isolated from P. oocarpa needles near Jalapa as well as near Tactíc in Guatemala and in Honduras. This clade was well supported for all five of the gene regions analysed.
Twenty-seven isolates grouped into Clade 2 in the ITS analyses (Additional file 1: Figure S1) and represent an undescribed species. Clade 2 resolved into two subclades in the five gene analyses. Subclade 1 was mostly isolated from Chiquimula and Alta Verapaz in Guatemala on P. oocarpa, P. maximinoi and P. tecunumanii as well as from P. oocarpa in Nicaragua. Isolates collected in Jalapa in Guatemala mostly grouped into Subclade 2. However, the topology of isolates CMW 47109 (Subclade 1 on Additional file 1: Figure S1, Additional file 3: Figure S3, Additional file 4: Figure S4; Subclade 2 on Fig. 2), CMW 51059 (Subclade 1 on Additional file 1: Figure S1, Additional file 3: Figure S3, Additional file 4: Figure S4), IB30.2b (Subclade 1 on Additional file 1: Figure S1, Additional file 3: Figure S3; Subclade 2 on Fig. 2) and IB30.2b (Subclade 1 on Additional file 1: Figure S1, Additional file 3: Figure S3, Additional file 4: Figure S4; Subclade 2 on Fig. 2) changed in the two subclades depending on the gene region analysed (Fig. 2, Additional file 1: Figure S1, Additional file 3: Figure S3, Additional file 4: Figure S4). Furthermore, the two subclades were not well supported for the BT1 gene region. Therefore, the two subclades are treated here as representing a single species.
Clade 3 also represented an undescribed Lecanosticta species. This clade included 11 isolates from P. oocarpa in Jalapa, Guatemala, one isolate from P. oocarpa in Honduras, as well as five isolates collected from Baja Verapaz in Guatemala on P. tecunumanii. This clade had high bootstrap support for TEF1, MS204 and RPB2 but was not well supported in the ITS and BT1 gene regions. Three isolates collected from different needles on a single P. tecunumanii tree in Baja Verapaz in Guatemala grouped together in Clade 4 and represent another undescribed species. With the exception of BT1, Clade 4 was statistically well supported in all the gene regions that were analysed.
Clade 5 accommodated sequences representing nine of the 14 known TEF1 haplotypes of L. acicola identified by Janoušek et al. (2016). These TEF1 haplotypes represent isolates collected from North America (Canada, USA, and Mexico), South America (Colombia), Europe (Spain, France, Switzerland, Slovenia, Lithuania, Italy, Germany, Estonia, Czech Republic, Croatia, and Austria) and Asia (South Korea, Japan, and China) (Table 3). This clade was clearly distinct from other clades in the ITS, TEF1, BT1 and MS204 phylogenetic analysis and statistically well supported in the ITS, TEF1, and MS204 analyses. Clade 5 included the ex-type of L. acicola and therefore is that species. None of the isolates from Central America obtained in the present study grouped with this clade in any of the gene regions analysed.
The remaining five assigned L. acicola TEF1 haplotypes considered by Janoušek et al. (2016), grouped together in Clade 6. This was together with an isolate obtained from P. caribaea in Honduras collected in 1983 (Evans 1984), four isolates obtained in the present study from Guatemala on P. oocarpa and P. maximinoi, and an isolate previously identified as L. acicola from Mexico on an unknown Pinus species (Quaedvlieg et al. 2012). In the present study, Clade 6 is treated as a novel taxon. The ITS, TEF1, BT1 and MS204 gene regions clearly distinguish Clades 5 and 6, however, RPB2 was not effective in resolving these two groups.
The second most abundant species collected in this study was Lecanosticta guatemalensis, represented by Clade 7 in the phylogenetic analyses. This clade was well supported in all five gene regions that were analysed. A total of 37 isolates from our collection grouped together with L. guatemalensis based in the ITS and TEF1 analyses. Lecanosticta guatemalensis was identified on P. maximinoi and P. oocarpa in various regions of Guatemala, as well as on P. oocarpa in Nicaragua. Isolates that had previously been collected in Nicaragua and Honduras and that were identified as L. acicola by Evans (1984) based on morphological characteristics also grouped with L. guatemalensis in the present study.
TAXONOMY
Using phylogenetic analyses, 51 of the Lecanosticta isolates obtained from Guatemala, Honduras and Nicaragua, one isolate obtained from CBS, and one isolate obtained from IMI, were found to include four undescribed species. These are described below as follows:
Lecanosticta jani van der Nest, M.J. Wingf. & I. Barnes, sp. nov.
MycoBank MB 826875. (Fig. 3)
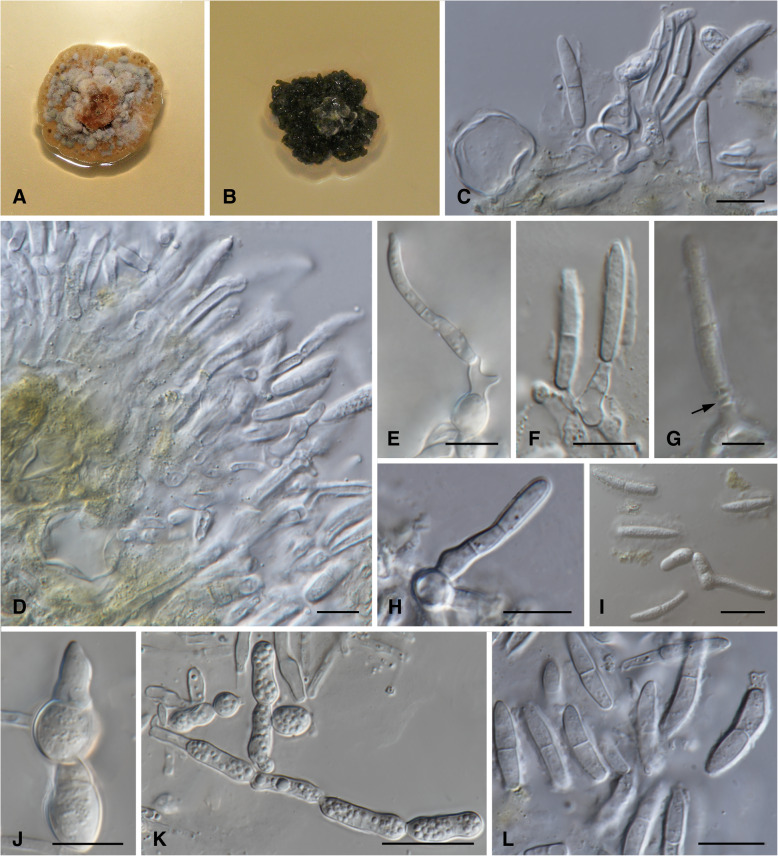
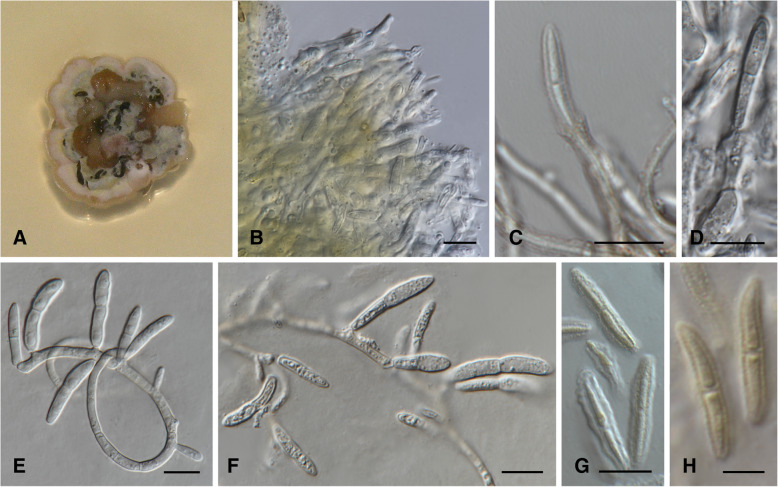
Fig. 3.
Lecanosticta jani (CMW38958; CMW38950; CMW48831; CMW47109; CMW51058; CMW51143) a-b Two wk. old colonies on MEA. A represents Type 1 colonies (CMW38950) and B represents Type 2 colonies (CMW48831). c-h Conidiogenous cells giving rise to conidia on MEA, with swollen globose basal cells of the conidiophores in E, F and H as well as annelations (see arrow) in G. i-k Swollen conidiogenous cells and conidia on MEA. Note endospore formation and germination in I. l Conidia on MEA. Bars: K = 50 μm; C-F and H-L = 10 μm; G = 5 μm
Etymology: The name is derived from Janus, the Roman god of gates and doorways having two faces or sides, and refers to the variable culture morphology ranging from light pink and fluffy to dark olive green and mucoid.
Diagnosis: Lecanosticta jani can be distinguished from the closely related L. brevispora by the distinct globose basal cells on the conidiophores that are mostly observed on MEA.
Type: Guatemala: Jalapa, Finca la Soledad, Mataquescuintla, on needles of Pinus oocarpa, 20 Sept 2012, I. Barnes (PREM 62185 – holotype; CMW 38958 = CBS 144456 – ex-type culture).
Description: Sexual morph unknown. Conidiomata isabelline to vinaceous brown on MEA. Conidiophores subcylindrical, often with a swollen globose basal cell, densely aggregated, honey to hyaline, smooth to verruculose, unbranched or branched at base, often encased in a yellow to light brown mucoid sheath, to 82 μm in length, 4.5–7.0 μm diam. Conidiogenous cells terminal, integrated, subcylindrical, honey to hyaline, smooth to verruculose, proliferating several times percurrently with visible annelations near apex, septate or aseptate, (8.5–)16.5(− 24.0) × (3.0–)4.5(− 6.5) μm. Conidia solitary, sub-cylindrical to narrowly fusoid-ellipsoidal, with subobtusely rounded apex, base truncate, brown, verruculose, frequently with mucoid sheath, two distinct sizes with conidial type one more abundant than conidial type two. Conidial type 1: 1–2-septate, base (1.5–)2.0–2.5(− 3.5) μm diam, (9.5–)14.5–21.5(− 30.0) × (2.0–)2.5–3.5(− 4.0) μm. Conidial type 2: 1–3-septate, base (1.5–)2.0–2.5(− 3.0) μm diam, (26.5–)30.5–37.0(− 38.0) × (2.0–)2.5–3.0(− 3.5) μm.
Culture characteristics: Colonies with two distinct morphologies. One type (Type 1), flat to somewhat erumpent, spreading with flat to fluffy aerial mycelium. A second type (Type 2) erumpent, mucoid and shiny, with irregular form and undulate to filiform edges. On MEA, the surface of Type 1 isolates pale to rosy vinaceous, reverse flesh to peach coloured. Type 2 isolates citrine to isabelline, reverse olivaceous to fuscuous black (Fig. 3). On PDA, Type 1 surface rosy vinaceous to peach in centre with dark brown edge, isabelline in reverse. Type 2, surface dark olivaceous with fuscious black centres and tufts of isabelline mycelium at edges, dark isabelline in reverse. On OA, Type 1 surface dirty white to pale vinaceous, fluffy mycelia to flat growth. Type 2 surface flat with smooth edge, fuscious black in centre at the point of inoculation with light apricot surrounding mycelium. Growth characteristics: optimal growth temperature for Type 1 isolates 25 °C, after 4 wk., colonies at 10, 15, 20, 25 and 30 °C reached maximum of 10.5, 22, 32, 32 and 10 mm respectively, with mean growth rate of 2.1, 5.1, 6.9, 7 and 1.8 mm / wk. respectively. Type 2 isolates optimal growth temperature 20 °C, after 4 wk., colonies at 10, 15, 20, 25 and 30 °C reached maximum of 12.5, 17, 29.5, 22 and 4.5 mm, with mean growth of 2.1, 3.3, 5.5, 5 and 1 mm / wk. respectively.
Notes: Lecanosticta jani resolved in a distinct clade (Clade 2, Figs. 1 and 2, Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3 and Additional file 4: Figure S4) based on all five gene regions considered. This clade divides into two subclades that were mostly represented by isolates obtained from Alta Verapaz and Chiquimula in Guatemala as well as in Nicaragua in subclade 1 and isolates obtained from Jalapa in Guatemala in subclade 2. Jalapa isolates all had the Type 2 morphology and the dark colour was associated with conidial production. Type 1 isolates produced few spores after 2 wk. The optimal growth temperature and growth rates were different for the two isolate types. However, the topology of some isolates changed between the two subclades depending on the gene region that is analysed and therefore the subclades are treated as one species. The morphological variation suggests that the two types could represent two ecotypes.
Additional material examined: Guatemala: Alta Verapaz, Santa Cruz Verapaz, near Tactíc, on needles of Pinus oocarpa, 21 Oct 2010, I. Barnes (culture CMW47109); loc. cit. I. Barnes (PREM 62187; CMW 48831 = CBS 144447 – culture); Jalapa, Finca la Soledad, Mataquescuintla, on needles of Pinus oocarpa, 20 Sept 2012, I. Barnes (PREM 62186, CMW 38950 = CBS 144446 – culture); Jalapa, Finca la Soledad, Mataquescuintla, on needles of Pinus tecunumanii, 20 Sept 2012, I. Barnes (cultures CMW 51058, CMW 51059). -Nicaragua: Matagalpa, on needles of Pinus oocarpa, 20 June 2011, I. Barnes (culture CMW 51143).
Lecanosticta pharomachri van der Nest, M.J. Wingf. & I. Barnes, sp. nov.
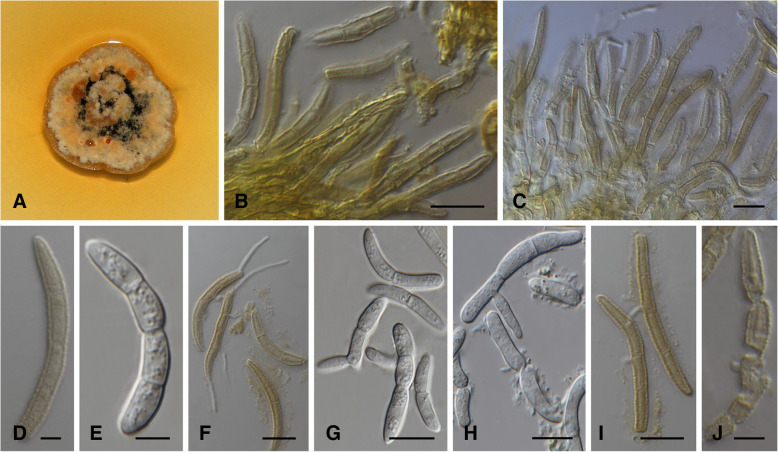
MycoBank MB 826876. (Fig. 4)
Fig. 4.
Lecanosticta pharomachri (CMW 37136; CMW38947). a, b Two wk. old colonies on MEA. c-e Conidiogenous cells giving rise to conidia on MEA. f, g Conjugation tube formation between conidia as well as conidia bearing smaller conidial cells. h-j Variation in conidia on MEA. Bars: D, F-H and J = 10 μm; C, E and I = 5 μm
Etymology: The epithet refers to the Resplendid Quetzal (Pharomachrus mocinno), which is the national bird of Guatemala and the spirit bird/companion of Tecún Umán; a Guatemalan legend.
Diagnosis: Lecanosticta pharomachri is distinguished from the other taxa in the genus by all five gene regions investigated but especially by sequences of TEF1, MS204 or RPB2. Conidia are also larger than those of L. guatemalensis and similar to L. acicola but differ from these species in that the conidia are frequently surrounded by a thick mucoid sheath and are mostly straight.
Type: Guatemala: Baja Verapaz, San Jerónimo, Salamá, on needles of Pinus tecunumanii, Nov 2010, I. Barnes (PREM 62188 – holotype; CMW 37136 = CBS 144448 – ex-type cultures).
Description: Sexual morph not observed. Conidiomata dark vinaceous brown on MEA. Conidiophores subcylindrical to cylindrical, densely aggregated, vinaceous brown to hyaline, smooth to verruculose, unbranched or branched at base, often encased in a light brown mucoid sheath, to 45 μm in length, 2.5–4.0 μm diam. Conidiogenous cells terminal, integrated, subcylindrical to cylindrical, luteus brown to hyaline, smooth to verruculose, surrounded by mucilage, holoblastic, proliferating several times percurrently with visible annelations near apex, septate or aseptate, (6.5–)9.5–13.5(− 16.0) × (1.5–)2.0–2.5(− 3.0) μm. Conidia released in a greenish olivaceous to honey mass, solitary, straight to slightly curved, cylindrical, with subobtusely rounded apex, base truncate, guttulate, hyaline to light brown, verruculose, frequently with thick mucoid sheath, 1–3-septate, base (1.5–)2.0–3.0(− 3.5) μm diam, (21.0)25.0–34.0(− 49.0) × (2.5–)3.0–4.0(− 5.0) μm. Germ tubes observed between conidia as well as conidial budding - secondary conidia sometimes produced from apical cell, 0–2-septate.
Culture characteristics: Colonies flat to erumpent, form irregular with undulate edge, spreading with fluffy aerial mycelium at centers. On MEA, surface apricot to cinnamon with isabelline and rosy buff mycelial mat at centers, reverse isabelline to dark brick in centre with cinnamon to apricot edges. On PDA, surfaces rosy to pale vinaceous with light isabelline to greenish white edges, reverse isabelline with cream edges. On OA, surface dirty white to isabelline to dark brown, fluffy mycelium to flat growth. Growth characteristics: optimal growth temperature 20 °C, after 4 wk., colonies at 10, 15, 20, 25, and 30 °C reaching a maximum of 9, 17, 18.5, 18.5 and 8.5 mm diam, with mean growth rates of 1.9, 3.6, 4.6, 4.4, and 1.9 mm / wk. respectively.
Notes: Some of the isolates, including the ex-type strain, produced a luteus exudate that diffused into MEA after 4–6 wk. Conjugation tubes were reported previously in L. acicola cultures as well as in needles (Siggers 1950; Crosby 1966). Conjugation tubes were also observed in this species (Fig. 4g) in the present study. Endospores as described by Crosby (1966) were also observed in some conidia.
Additional material examined: Guatemala: Jalapa, Finca la Soledad, Mataquescuintla, on needles of Pinus oocarpa, 20 Sept 2012, I. Barnes (cultures CMW 38976, CMW 51053 and CMW 51054); loc. cit., I. Barnes (PREM 62189; CMW 38947 = CBS 144695 – culture; PREM 62190, CMW 38974 = CBS 144449 – culture).
Lecanosticta tecunumanii van der Nest, M.J. Wingf. & I. Barnes, sp. nov.
MycoBank MB 826877. (Fig. 5)
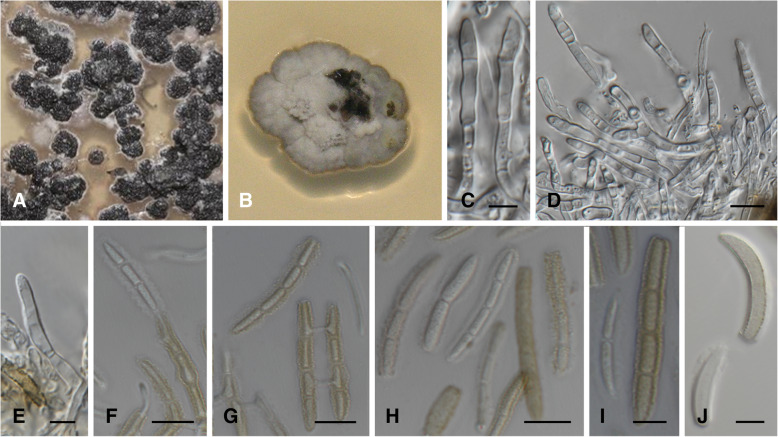
Fig. 5.
Lecanosticta tecunumanii (CMW46805; CMW46812). a Two wk. old colony on MEA. b-d Conidiogenous cells giving rise to conidia on MEA. e-f Micronematous conidiogenesis observed on MEA with conidia. g-h Uniseptate conidia with or without a mucoid sheath observed on MEA. Bars: B-G = 10 μm; H = 5 μm
Etymology: Name refers to the Guatemalan legend, Tecún Umán, and Pinus tecunumanii, the host plant from which the holotype was collected.
Diagnosis: Lecanosticta tecunumanii is distinguished from the other taxa by the ITS, TEF1, MS204 and RPB2 gene regions. Morphologically, it is distinct in having only 1-septate conidia after 2 wk. of incubation on MEA, but 2-septate and 3-septate conidia are occasionally observed in older cultures.
Type: Guatemala: Baja Verapaz, San Jerónimo, Salamá, on needles of Pinus tecunumanii, Oct 2011, I. Barnes (PREM 62191 – holotype; CMW 46805 = CBS 144450 – ex-type cultures).
Description: Sexual morph not observed. Conidiomata isabelline to visaceous brown on MEA. Conidiophores cylindrical, densely aggregated, hyaline to pale yellow-brown, smooth to slightly verruculose, unbranched or branched at base, to 120 μm in length, 2.0–5.0 μm diam. Conidiogenous cells terminal or indeterminate, integrated or discrete, cylindrical, hyaline to honey, smooth to verruculose, proliferating several times percurrently with visible annelations near apex or micronematous, septate or aseptate, (5.0–)7.0–14.5(− 15.5) × (1.5–)2.0–2.5(− 3.0) μm. Micronematous cells (6–)10.5–18.5(− 27.0) × (2.0–)2.0–2.5(− 3.0) μm. Conidia solitary, straight to slightly curved, subcylindrical to fusiform, with subobtusely rounded or sharply pointed apex, base truncate, guttulate, smooth to granulate, hyaline to cream buff to light brown, occasionally enclosed in mucoid sheath, 1-septate, base (1.5–)1.5–2.0(− 2.0) μm diam., (14.5–)16.0–21.0(− 24.0) × (2.0–)2.5–3.0(− 3.5) μm.
Culture characteristics: Colonies somewhat erumpent, spreading with flat to fluffy aerial mycelium. On MEA, surface olivaceous to isabelline with rosy buff mycelial tufts, reverse isabelline. On PDA, surface rosy vinaceous to peach in centre with a dark brown edge, isabelline in reverse. On OA, surface dirty white to pale vinaceous, fluffy mycelia to flat peach growth. Growth characteristics: optimal growth temperature 25 °C, after 4 wk., colonies at 10, 15, 20, 25, and 30 °C reached maximum of 9, 15.5, 24, 24, and 4.5 mm, with mean growth of 2.2, 3.8, 5.3, 5.7, and 1.1 mm / wk. respectively.
Notes: Micronematous conidiogenesis (Fig. 5E - F), observed more frequently than distinct conidiophores in culture.
Additional material examined: Guatemala: Baja Verapaz, San Jerónimo, Salamá, on needles of Pinus tecunumanii, Oct 2011, I. Barnes (PREM 62192, CMW 49403 = CBS 144451 – culture; PREM 62193, CMW 46812 = CBS 144452 – culture).
Lecanosticta variabilis van der Nest, M.J. Wingf. & I. Barnes, sp. nov.
MycoBank MB 826878. (Fig. 6)
Fig. 6.
Lecanosticta variabilis (CMW42205; CMW37125). a Colony on MEA with luteus exudate diffusing into medium. b-c Conidiogenous cells giving rise to conidia on MEA. d-h Various conidial shapes and sizes on MEA. f Germinating conidia on MEA. g-h Swollen conidial cells giving rise to smaller conidia. i Conjugation tube formation between two conidia. j Conidium disintegrating on MEA. Bars: B-C, F-I = 10 μm; E, J = 5 μm; D = 2,5 μm
Etymology: The epithet refers to the variable size and shape of the conidia.
Diagnosis: Lecanosticta variabilis is distinguished from the closely related species, L. acicola, by either ITS, TEF1 or MS204. Morphologically, it is distinguished from other species with the exception of L. acicola by the diffusion of sulphur-yellow to cinnamon metabolite into PDA and a luteus to sienna coloured metabolite produced on MEA within 2 wk. This species also has smaller conidia than those of L. acicola.
Type: Honduras: Santa Barbara, on needles of Pinus caribaea, 1980, H.C. Evans, (PREM 62196 – holotype; CMW 42205 = IMI 281561 = CBS 144453 – ex-type culture).
Description: Sexual state not observed. Conidiomata olivaceous to vinaceous brown on MEA. Conidiophores cylindrical, extending in densely aggregated palisade, hyaline to honey to pale vinaceous brown, smooth to verruculose, unbranched or branched at base, septate or aseptate, often encased in granular yellow to light brown mucoid sheath, length up to 60 μm, 2.0–5.0 μm diam. Conidiogenous cells terminal, integrated, subcylindrical to cylindrical, hyaline to light brown, smooth to verruculose, proliferating several times percurrently with visible annelations near apex, septate or aseptate, (4.5–)5.5–10.5(− 12.0) × (1.5–)2.0–3.5(− 5.0) μm. Conidia three different conidial types. All three types solitary, smooth to verruculose, subhyaline to honey to light brown, often enclosed in granular light luteus mucoid sheath. Type 1 straight to strongly curved, subcylindrical to cylindrical, subobtusely rounded apex, truncate, 1–4-septate, base (1.5–)2.0–2.5(− 3.0) μm diam. (22–)25.0–34.0(− 43.0) × (2.0–)2.5–3.0(− 3.5) μm. Type 2 slightly curved, cylindrical with both apex and base rounded, 0–2-septate, (14.5–)15.5–19.5(− 22.0) × (2.0–)2.5–3.0(− 3.5) μm. Type 3 buds from larger conidia (see notes) or from conidiogenous cells, hyaline, fusiform to cylindrical with subobtusely rounded apex and base, 0–1-septate, (10.0–)11.0–14.0(− 15.5) × (2.0–)2.0–2.5(− 3.0) μm.
Culture characteristics: Colonies flat to somewhat erumpent, spreading, with sparse aerial mycelium, surface folded, with smooth, lobate margins. On MEA, surface isabelline with patches of pale luteus to dark olivaceous green, reverse olivaceous to fuscous black. Mucoid yellow to peach to yellow-green exudate present. Luteus to sienna coloured metabolite diffusing into medium. On PDA, surface isabelline in centre, rosy buff in outer region, dark olivacous-brown on edges and isabelline in reverse. Sulphur yellow to cinnamon coloured metabolite diffuses into media. On OA, surface dirty white with diffuse umber outer region. Growth characteristics: optimal growth temperature 25 °C, after 4 wk., colonies at 10, 15, 20, 25 and 30 °C reached maximum of 11.5, 21, 31, 31.5 and 22.5 mm, with mean growth of 2.2, 4.5, 6.1, 6.9 and 3.6 mm / wk. respectively.
Notes: The cells in the conidia often swell and break off, forming endospores as described in L. acicola (Siggers 1950; Crosby 1966; Evans 1984). Secondary conidia were commonly produced in cultures of this species, similar to those previously described for L. acicola specimens examined directly from needles (Evans 1984).
Additional material examined: Guatemala: Alta Verapaz, Santa Cruz Verapaz, near Tactíc, on needles of Pinus oocarpa, 21 Oct 2010, I. Barnes (PREM 62194, CMW 37125 = CBS 144454 – culture); loc. cit., I. Barnes (culture CMW 37129); Jalapa, Finca Forestal Soledad, on needles of Pinus maximinoi, 21 Oct 2010, I. Barnes (PREM 62195, CMW 36809 = CBS 144455 – culture). –Mexico: on needles of a Pinus sp., 30 Nov 2009, M. de Jesús Yáñez-Morales (CBS H-21112; culture CMW45425 = CPC 17822 = CBS 133789);
DISCUSSION
Four novel species of Lecanostica from infected pine needles collected in Central America are reported and named as L. jani, L. pharomachri, L. tecunumanii, and L. variabilis. There are now nine species described in the genus and these can be distinguished based on a phylogenetic inference for multiple gene regions. The two previously described species, L. brevispora and L. guatemalensis, were also found in this study and they provide new host and country records. The well-known pine pathogen, L. acicola, was not found on any of the samples collected from five Pinus spp. in seven regions of Central America considered in this study. This suggests that the species is not native in that region.
Results of the present study support the view of Quaedvlieg et al. (2012) that a combination of the ITS and TEF1 should be used as barcoding loci to distinguish between species of Lecanosticta and other closely related species. Additionally, statistically well supported clades were obtained in this study using the MS204 gene region. However, genus-specific primers should ideally be designed to increase the amplification success rate for this gene region in Lecanosticta. Although the BT2 gene was also proposed as a possible barcoding region that could be used to distinguish between Lecanosticta species and other species of Mycosphaerellaceae (Quaedvlieg et al. 2012), it amplified poorly in the present study. The BT1 gene region distinguished most of the species, but not L. pharomachri and L. variabilis and provided low statistical support at all nodes.
The results of this study support the view of Evans (1984) that Lecanostica species are comprised of morphotypes or ecotypes. Based on phylogenetic analyses, we were able to define lineages for species also supported by morphological characteristics. The TEF1 sequences were highly variable but several well supported clades and subclades were observed within species (Fig. 2). These clades possibly represent additional new species but we lacked sufficient cultures and support to describe them. The clade with the most diversity in terms of unique TEF1 haplotypes, Clade 1, was L. brevispora (represented by 22.1% of TEF1 haplotypes in the genus) and this species was also represented by the largest number of isolates. High haplotype diversity was observed in the L. jani (16.1% of TEF1 haplotypes) and L. pharomachri (10.3% of TEF1 haplotypes) clades and different lineages were observed in the L. acicola (13.2% of TEF1 haplotypes), L. guatemalensis (17.6% of TEF1 haplotypes), and L. variabilis (13.2% of TEF1 haplotypes) clades. The other gene regions, especially MS204 and RPB2 were also highly variable in terms of distinguishing haplotypes. RPB2 is however, not recommended to distinguish between L. acicola and L. variabilis as these two species form paraphyletic groups in the tree for this gene region.
The paleo-geographic region that includes Mexico and extends into Central America is regarded as one of three centres of diversity of Pinus species (Farjon 1996). Pine needles that were sampled from Central America in this study were symptomatic but serious disease was not observed. This suggests that Lecanosticta species have co-speciated with their native pine hosts in this region. Of the nine known species, L. gloeospora and L. longispora have been identified only in Mexico and L. brevispora and L. variabilis have been identified in both Mexico and Central America. Lecanosticta guatemalensis, L. jani, L. pharomachri and L. tecunumanii are currently known only from Central America.
Lecanosticta acicola has been redefined in this study. All isolates from Central America that had previously been identified as L. acicola, based on morphological characteristics, are now treated as different species. This is based on newly available DNA sequence data and phylogenetic analyses emerging from this study as well as that of Quaedvlieg et al. (2012). L. acicola is, however, still considered as present in Mexico.
Based on TEF1 analyses, L. acicola resolves in three lineages. Janoušek et al. (2016) used microsatellites to show that a lineage of L. acicola from the northern USA was introduced into Central and Northern Europe, and a lineage from the southern USA was introduced into France, Spain, and Colombia. Similarly, Huang et al. (1995) reported that L. acicola was introduced into China from the southern part of the USA. Our analyses of the TEF1 sequences of isolates from the northern parts of the USA, Lithuania, and a representative sequence for Central and Northern Europe and Canada (KJ938438, Table 3), formed one distinct lineage with L. acicola (Fig. 2). All isolates from the southern parts of the USA, as well as representative sequences for Asia, France, Spain, and Colombia (Table 3), formed a second distinct lineage in the clade accommodating L. acicola (Fig. 2). The third lineage included only isolates from Mexico, which suggests that isolates in this lineage have remained in their area of origin and have not been introduced elsewhere. Because this Mexican lineage had strong bootstrap support separating it from the other two lineages, it could represent a further new species. Only TEF1 data are currently available for the Mexican collections (downloaded from GenBank) and other gene regions would need to be sequenced and analysed to determine whether this really represents a further novel taxon.
Evans (1984) first speculated that Central America could be the centre of origin of Lecanosticta. The phylogenetic analyses conducted in the present study showed that there is a high diversity of species and lineages for this genus in Central America, which supports Evans’ hypothesis. This is the first study where all known species of Lecanosticta have been delineated based on DNA sequence data and phylogenetic analysis, and it has led to the recognition of additional new taxa from Central America and Mexico. Eight of the nine species of Lecanosticta have been reported only from this region, and our results consequently represent strong support for a Mesoamerican Lecanosticta centre of diversity and likely origin. Population genetic analyses for the most common of these species will serve to provide additional support for this hypothesis.
CONCLUSIONS
Phylogenetic inference based on DNA sequence data including new collections from Mexico and Central America revealed four novel species and reaffirmed the identity of the five previously described taxa. The most important of these species is the well-known pine pathogen L. acicola that was redefined as a North American taxon and for which at least three distinct lineages can be distinguished using the TEF1 gene region. New regions of occurrence and host range emerged for Lecanosticta spp. with eight of the nine species occurring in Mesoamerica. This suggests that Mesoamerica is the most likely centre of origin for Lecanosticta. Lecanosticta acicola was best known as the causal agent of the important brown spot needle blight of Pinus palustris in the southeastern USA but it has more recently spread within the USA and Europe where it has become an increasingly important pathogen of numerous Pinus spp. The other species of Lecanosticta, including those newly described, are of unknown importance but it seems likely that some of them could pose a threat to Pinus spp. if they were introduced into new environments in the future. The fact that various Mesoamerican Pinus spp. are increasingly being used for plantation development in the Southern Hemisphere implies that extreme caution should be applied not to introduce Lecanosticta spp. together with germplasm needed for future planting programmes.
Additional files
Figure S1. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the ITS region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate exhibits Type 2 morphology but it groups with Subclade 1 or exhibits Type 1 morphology but groups with Subclade 2. (PPTX 61 kb)
Figure S2. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the BT1 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. (PPTX 54 kb)
Figure S3. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the MS204 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma septosporum was used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate exhibits Type 2 morphology but it groups with Subclade 1 or exhibits Type 1 morphology but groups with Subclade 2. (PPTX 55 kb)
Figure S4. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the RPB2 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate exhibits Type 2 morphology but it groups with Subclade 1 or exhibits Type 1 morphology but groups with Subclade 2. (PPTX 61 kb)
Acknowledgements
We thank Jeff Garnas and Elmer Gutierrez from Camcore for their assistance in collecting pine needle samples. We also wish to thank Josef Janoušek and Yves du Toit for their assistance in isolating Lecanosticta spp. from the infected pine needles.
Funding
This project was financed by the National Research Foundation of South Africa (Thuthuka Grant no 80670, and Grant no 95875) as well as by members of the Tree Protection Cooperative Program (TPCP). AvdN was supported by a Scarce Skills Doctoral Scholarship (no 89086) provided by the National Research Foundation of South Africa. The NRF acknowledge that opinions, findings, conclusions and/or recommendations expressed in any publication generated by the NRF supported research are that of the author(s), and that the NRF accepts no liability whatsoever in this regard. The NRF had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated in this study are included in this published article and its supplementary files. The datasets analysed are available from the corresponding author on reasonable request.
Abbreviations
- 1F1N
One Fungus One Name
- AIC
Akaike Information Criterion
- BI
Bayesian inference
- BSNB
Brown spot needle blight
- BT1
Beta-tubulin-1 gene region
- BT2
Beta-tubulin-2 gene region
- CA
California
- CBS
The culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands
- CI
Consistency index
- CMW
The culture collection of the Forestry and Agricultural Biotechnology Institute
- COSAVE
El Comité de Sanidad Vegetal
- CPC
Personal collection of Pedro Crous housed at CBS
- DSM
Dothistroma Sporulating Media
- FABI
Forestry and Agricultural Biotechnology Institute
- HI
Homoplasy index
- IASPC
Inter-African Phytosanitary Council
- ICN
International Code of Nomenclature for algae, fungi, and plants
- IMI
The UK National Fungus Collection maintained by CABI Bioscience, Egham, UK
- ITS
Internal transcribed spacers
- MA
Massachusetts
- MB
MycoBank
- MCMC
Markov Chain Monte Carlo
- MD
Maryland
- ME
Maine
- MEA
Malt Extract Agar
- ML
Maximum likelihood
- MO
Missouri
- MP
Maximum parsimony
- MS204
The guanine nucleotide-binding protein subunit beta
- NCBI
National Centre for Biotechnology Information
- NJ
New Jersey
- OA
Oatmeal Agar
- PCR
Polymerase chain reaction
- PDA
Potato Dextrose Agar
- PHT
Partition homogeneity test
- PREM
The dried herbarium collection of the South African National Collection of Fungi
- RC
Rescaled consistency index
- RI
Retention index
- RPB2
RNA polymerase II second largest subunit
- TBR
Tree-bisection-reconnection
- TEF1
Translation elongation factor 1-α gene
- TL
Tree length
Authors’ contributions
Acquisition of sample material was performed by PO and IB. Fungal isolations were done by IB. Data collection and all analyses were performed by AvdN. Funding acquisition was done by IB and MJW. IB and MJW supervised the project. AvdN wrote the original draft, and review and editing was performed by AvdN, IB MJW and PO. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ariska van der Nest, Email: ariska.vandernest@fabi.up.ac.za.
Michael J. Wingfield, Email: mike.wingfield@fabi.up.ac.za
Paulo C. Ortiz, Email: portiz@inab.gob.gt
Irene Barnes, Phone: +27 12 420 5143, Email: irene.barnes@fabi.up.ac.za.
References
- Adamson K, Drenkhan R, Hanso M. Invasive brown spot needle blight caused by Lecanosticta acicola in Estonia. Scandinavian Journal of Forest Research. 2015;30:587–593. doi: 10.1080/02827581.2015.1041550. [DOI] [Google Scholar]
- Anonymous First report of Mycosphaerella dearnessii in Latvia. European and Mediterranean Plant Protection Organization Bulletin. 2012;8:5–6. [Google Scholar]
- Barnes I, Crous PW, Wingfield BD, Wingfield MJ. Multigene phylogenies reveal that red band needle blight of Pinus is caused by two distinct species of Dothistroma, D. septosporum and D. pini. Studies in Mycology. 2004;50:551–565. [Google Scholar]
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- Cleary Michelle, Laas Marili, Oskay Funda, Drenkhan Rein. First report of Lecanosticta acicola on non-native Pinus mugo in southern Sweden. Forest Pathology. 2019;49(3):e12507. doi: 10.1111/efp.12507. [DOI] [Google Scholar]
- Crosby ES. Endospores in Schirrhia acicola. Phytopathology. 1966;56:720. [Google Scholar]
- Crous PW. Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Diversity. 2009;38:1–24. [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. Mycosphaerella is polyphyletic. Studies in Mycology. 2007;58:1–32. doi: 10.3114/sim.2007.58.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter GC, et al. Unravelling Mycosphaerella: do you believe in genera? Persoonia. 2009;23:99–118. doi: 10.3767/003158509X479487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, Samson RA. Fungal biodiversity. CBS Laboratory manual series 1. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2009. [Google Scholar]
- Cunningham CW. Can three incongruence tests predict when data should be combined? Molecular Biology and Evolution. 1997;14:733–740. doi: 10.1093/oxfordjournals.molbev.a025813. [DOI] [PubMed] [Google Scholar]
- Evans HC. The genus Mycosphaerella and its anamorphs Cercoseptoria, Dothistroma and Lecanosticta on pines. Mycological Paper. 1984;153:1–102. [Google Scholar]
- Farjon A. Biodiversity of Pinus (Pinaceae) in Mexico: speciation and palaeo-endemism. Botanical Journal of the Linnean Society. 1996;121:365–384. [Google Scholar]
- Fourie A, Wingfield MJ, Wingfield BD, Barnes I. Molecular markers delimit cryptic species in Ceratocystis sensu stricto. Mycological Progress. 2015;14:1–18. doi: 10.1007/s11557-014-1020-0. [DOI] [Google Scholar]
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, et al. The Amsterdam declaration on fungal nomenclature. IMA Fungus : The Global Mycological Journal. 2011;2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintsteiner M, Cech TL, Halmschlager E, Stauffer C, Kirisits T. First report of Mycosphaerella dearnessii on Pinus nigra var. nigra in Austria. Forest Pathology. 2012;42:437–440. doi: 10.1111/j.1439-0329.2012.00794.x. [DOI] [Google Scholar]
- Huang Z-Y, Smalley EB, Guries RP. Differentiation of Mycosphaerella dearnessii by cultural characters and RAPD analysis. Phytopathology. 1995;85:522–527. doi: 10.1094/Phyto-85-522. [DOI] [Google Scholar]
- Ivory MH. In: Diseases and disorders of pines in the tropics: a field and laboratory manual. Institute ODAaOF, editor. Oxford: Oxford Forestry Institute; 1987. [Google Scholar]
- Jankovsky L, Palovcíková D, Tomsovsky M. Brown spot needle blight associated with Mycosphaerella dearnessii occurs on Pinus rotundata in the Czech Republic. Plant Pathology. 2009;58:398. doi: 10.1111/j.1365-3059.2008.01999.x. [DOI] [Google Scholar]
- Janoušek J, Wingfield MJ, Monsivais JG, Jankovsky L, Stauffer C, et al. Genetic analyses suggest separate introductions of the pine pathogen Lecanosticta acicola into Europe. Phytopathology. 2016;106:1413–1425. doi: 10.1094/PHYTO-10-15-0271-R. [DOI] [PubMed] [Google Scholar]
- Kais AG. Dispersal of Schirria acicola spores in southern Mississippi. Plant disease reporter. 1971;55:309–311. [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Markovskaja S, Kacergius A, Treigiene A. Occurrence of new alien pathogenic fungus Mycosphaerella dearnessii in Lithuania. Botanica Lithuanica. 2011;17:29–37. [Google Scholar]
- Marmolejo JG. The genus Lecanosticta from Nuevo Leon, Mexico. Mycotaxon. 2000;76:393–397. [Google Scholar]
- Mullett M. S., Adamson K., Bragança H., Bulgakov T. S., Georgieva M., Henriques J., Jürisoo L., Laas M., Drenkhan R. New country and regional records of the pine needle blight pathogens Lecanosticta acicola , Dothistroma septosporum and Dothistroma pini. Forest Pathology. 2018;48(5):e12440. doi: 10.1111/efp.12440. [DOI] [Google Scholar]
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz de Urbina E, Mesanza N, Aragonés A, Raposo R, Elvira-Recuenco M, et al. Emerging needle blight diseases in Atlantic Pinus ecosystems of Spain. Forests. 2017;8:1–18. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg W, Groenewald JZ, de Jesús Yáñez-Morales M, Crous PW. DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia. 2012;29:101–115. doi: 10.3767/003158512X661282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A, Suchard, MA, Xie, D, and Drummond, AJ. 2014. Tracer v1.6, Available from http://beast.community/tracer
- Rayner, RW. 1970. A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiković D, Piškur B, Barnes I, Hauptman T, Diminić D, Wingfield MJ, Jurc D (2019) Genetic diversity of the pine pathogen Lecanosticta acicola in Slovenia and Croatia. Plant Pathology. 10.1111/ppa.13017.
- Siggers PV. The brown spot needle blight of pine seedlings. United States department of agriculture Washington, DC Technical Bulletin. 1944;870:1–36. [Google Scholar]
- Siggers PV. Possible mechanism of variation in the imperfect stage of Scirrhia acicola. Phytopathology. 1950;40:726–728. [Google Scholar]
- Stukenbrock EH, Quaedvlieg W, Javan-Nichah M, Zala M, Crous PW, et al. Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola) Mycologia. 2012;104:1397–1407. doi: 10.3852/11-374. [DOI] [PubMed] [Google Scholar]
- Sung G-H, Sung J-M, Hywel-Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates.
- Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, et al. Regnum Vegetabile 159. Glashütten: Koeltz Botanical Books; 2018. International code of nomenclature for algae, fungi, and plants (Shenzhen code) adopted by the nineteenth international botanical congress Shenzhen, China, July 2017. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990;18:315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the ITS region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate exhibits Type 2 morphology but it groups with Subclade 1 or exhibits Type 1 morphology but groups with Subclade 2. (PPTX 61 kb)
Figure S2. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the BT1 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. (PPTX 54 kb)
Figure S3. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the MS204 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma septosporum was used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate exhibits Type 2 morphology but it groups with Subclade 1 or exhibits Type 1 morphology but groups with Subclade 2. (PPTX 55 kb)
Figure S4. Maximum likelihood tree representing the five known and four novel species of Lecanosticta generated from the RPB2 region. MP bootstrap support (> 70%) are indicated first, followed by ML bootstrap values (MP/ML, * = insignificant value). Bold branches indicate BI values > than 0.95. Dothistroma species were used as the outgroup taxa. All represented type species are indicated in bold and with a “T”. Clades indicated on the left correspond with the clades in Fig. 1. Within the L. jani clade a “∆” next to the isolate indicates that the isolate exhibits Type 2 morphology but it groups with Subclade 1 or exhibits Type 1 morphology but groups with Subclade 2. (PPTX 61 kb)
Data Availability Statement
All data generated in this study are included in this published article and its supplementary files. The datasets analysed are available from the corresponding author on reasonable request.