Abstract
Cancer cells encounter numerous stresses that pose a threat to their survival. Tumor microenviroment stresses that perturb protein homeostasis can produce endoplasmic reticulum (ER) stress, which can be counterbalanced by triggering the unfolded protein response (UPR) which is considered the canonical ER stress response. The UPR is characterized by three major proteins that lead to specific changes in transcriptional and translational programs in stressed cells. Activation of the UPR can induce apoptosis, but also can induce cytoprotective programs such as autophagy. There is increasing appreciation for the role that UPR-induced autophagy plays in supporting tumorigenesis and cancer therapy resistance. More recently several new pathways that connect cell stresses, components of the UPR and autophagy have been reported, which together can be viewed as non-canonical ER stress responses. Here we review recent findings on the molecular mechanisms by which canonical and non-canonical ER stress responses can activate cytoprotective autophagy and contribute to tumor growth and therapy resistance. Autophagy has been identified as a druggable pathway, however the components of autophagy (ATG genes) have proven difficult to drug. It may be the case that targeting the UPR or non-canonical ER stress programs can more effectively block cytoprotective autophagy to enhance cancer therapy. A deeper understanding of these pathways could provide new therapeutic targets in cancer.
Keywords: Canonical endoplasmic reticulum stress, non-canonical endoplasmic reticulum stress, unfolded protein response, autophagy, cancer
Introduction
The endoplasmic reticulum (ER) is often the largest organelle in eukaryotic cells, organized into netlike labyrinth of membrane-enclosed tubules and flattened cisternae (sacs) that stretch out from the nuclear membrane throughout the cytosol (1, 2). Approximately 11% of the 25,000 secretory proteins and 20% of single-pass or multi-pass transmembrane proteins are driven into the ER lumen by N-terminal signal sequence present on them (3, 4). Once inside the ER lumen, nascent proteins are properly folded, attaining their three-dimensional structure, while also undergoing a wide array of post-translational modifications with the assistance of ER-resident chaperones, oxidoreductases and glycosylating enzymes, (5, 6). When the ER Ca2+ levels change, or misfolded or mutant proteins accumulate, ER stress commences (7). ER stress triggers an evolutionarily conserved cascade known as unfolded protein response (UPR) to counteract the deleterious consequence of ER stress and restore ER homeostasis, or if not possible to activate apoptosis (8). In this review we are defining the “canonical ER stress response” as the UPR pathway regulated by ER chaperone protein GRP78/BiP and three traditional UPR sensors proteins R (PKR)-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1α (IREα), and activating transcription factor 6 (ATF6). What has become clear is that other pathways intersect with components of the UPR to drive cytoprotection in the face of stress. We categorize these pathways reviewed extensively below as “non-canonical ER stress response”.
The discovery of lysosomes by Christian de Duve was recognized with the Nobel Prize in Physiology and Medicine in 1974 (9). More than twenty years later in 2016 the Nobel Prize was awarded to Yoshinori Ohsumi for the discovery of the genes that control autophagy, a lysosome dependent catabolic program that cells employ to degrade and recycle protein and organelles (10–12). Autophagy is an evolutionary conserved orchestrated program that includes initiation and membrane nucleation, phagophore formation and expansion, fusion with the lysosomes and degradation (13). Autophagy has gained a spotlight for its physiological importance in human health and diseases. Three defined types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) all converge at a common node of the lysosomal degradation pathway where all cytosolic components are targeted for degradation (14). Macroautophagy is the best studied form of autophagy and hereafter in this review the term autophagy will refer only to macroautophagy. Delivery of cargo proteins through double membrane-bound vesicles, referred to as autophagosomes is the characteristic feature of macro-autophagy (15). During microautophagy, the cytosolic components are directly engulfed by the invagination of the lysosomal membrane (16). In contrast, to macro- and microautophagy which can be both selective or non-selective, CMA involves the recognition of a sequence-motif in the target proteins by cytosolic chaperon proteins such us shock-cognate (Hsc-70) which bind and escort them across the lysosomal membrane. There the complex is recognized by the lysosomal membrane receptor lysosomal-associated membrane protein 2A (LAMP-2A) and after multimerization the target protein is unfolded, translocated in the lysosome and subsequently degraded (17). Autophagy is thought to be primarily associated with the UPR by removing unfolded proteins to favor the survival of stressed cells. However, organelle-specific autophagy mechanisms such as reticulophagy (ER), nucleophagy (nucleus), peroxyphagy (peroxisomes), mitophagy (mitochondria), xenophagy (bacteria) and others are also being investigated for their pathophysiological roles in various diseases (18–21).
Conceptually, the UPR and autophagy can be considered two different programs for cellular homeostasis that either work independently in the specified cellular locations, or work in co-ordination to protect the cellular physiology against a diverse array of stress. Here we review the mechanistic underpinnings of how canonical- and non-canonical ER stress responses regulate autophagy in cancer cells.
The canonical ER stress response: the UPR
The ER is a multifunctional organelle, and various subregions within the ER support distinct pathways involved in cellular homeostasis and survival. Rough ER is associated with protein synthesis and degradation, trafficking of secreted and membrane proteins, and harbors oxidoreductases. Mitochondria-associated ER membrane (MAM), the nuclear envelope, peroxisomal components, Russell bodies, and lipid droplets are examples of specialized ER compartments (22). Smooth ER is responsible for lipid synthesis and metabolism, and calcium storage. In addition, smooth ER is decorated with specialized domains, including plasma membrane-associated ER and regions associated with MAM formation, autophagosomes, and lipid droplets (23, 24). Accumulation of folding-incompetent or misfolded proteins above a critical threshold sets in motion a remedial signal transduction pathway known as the UPR, which is relayed through three ER transmembrane sensors: IRE1α, PERK, and ATF6. A key ER chaperone HSPA5/GRP78/Bip is typically bound to each of these UPR sensors, preventing downstream signaling. When unfolded protein stress reaches a critical threshold enough GRP78 dissociates from the sensors to allow downstream signaling activation that regulates transcription and translation in manner that restores ER homeostasis. The common theme across the UPR effectors is that global mRNA translation is severely limited while transcription and translation of very specific cell fate effectors is upregulated (25). The UPR-dependent cell fate (i.e. cell death or cell survival) across the diverse array of cellular organization depends on the nature of the stimulus and duration of stress. Details of the mechanistic actions of these sensors are discussed below (Figure 1).
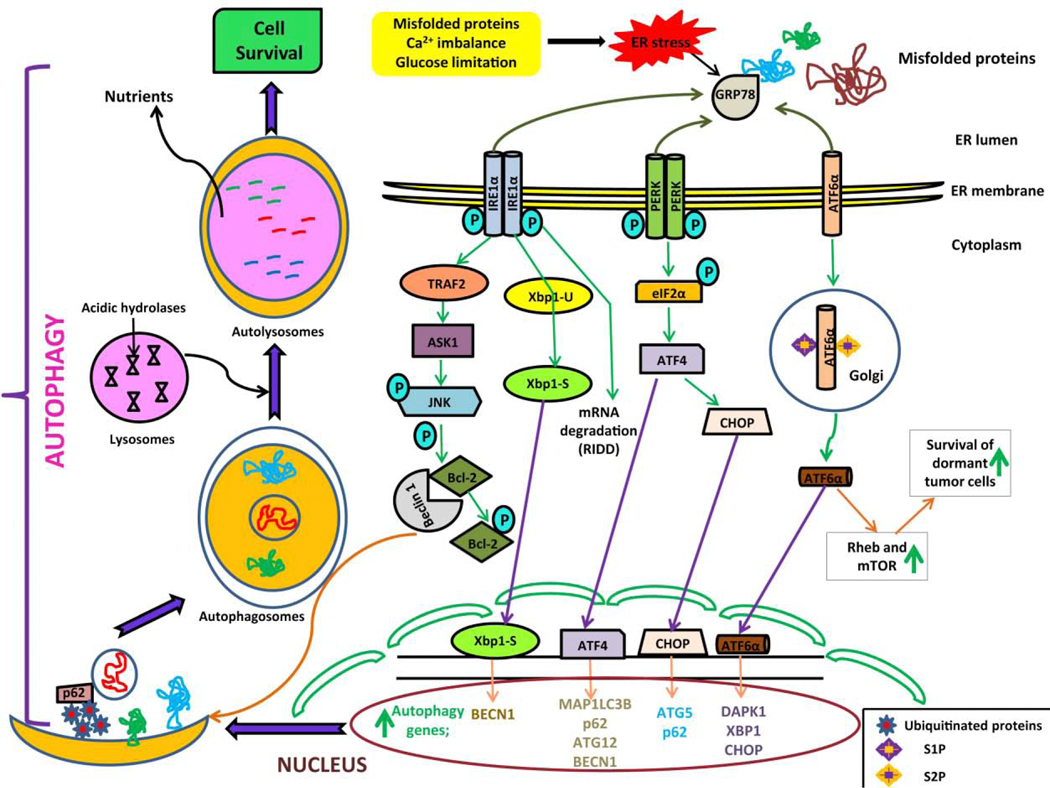
Figure 1: The unfolded protein response (UPR) signaling mechanisms and autophagy.
Perturbances in endoplasmic reticulum (ER) environment due to misfolded proteins, Ca2+ imbalance, or glucose limitation causes ER stress. Following ER stress, the UPR is executed by three major proteins and their downstream effectors: inositol-requiring enzyme 1α (IRE1α), protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), and the activating transcription factor 6 α (ATF6α). The IRE1α arm of UPR produces spliced X box-binding protein 1 (XBP1-S) enhances the expression of Beclin-1 by binding with BECN1 gene. IRE1-dependent decay (RIDD) activation degrades UPR target mRNAs. Activated IRE1α forms a complex with tumor necrosis factorα (TNFα) receptor-associated factor 2 (TRAF2) and apoptosis signaling-regulating kinase (ASK1), which in turn activates c-Jun N-terminal kinase (JNK). The IRE1α-JNK arm mediates phosphorylation of B-cell lymphoma 2 (Bcl-2), which causes Beclin-1 dissociation and induction of autophagy. Autophagy is induced via expression of p62, autophagy regulated (ATG) 5, ATG12, and microtubule associated protein 1 light chain 3 beta (MAP1LC3B) genes by the activating transcription factor 4 (ATF4) transcription factor and CCAAT/enhancer binding protein homologous protein (CHOP) which is mediated by PERK arm of the UPR. The ATF6α arm of UPR is believed to induce autophagy by regulating the transcriptional levels of death-associated kinase 1 (DAPK1), CHOP and XBP1 genes. In addition, ATF6α upregulates Ras homology enriched in brain (Rheb) and mammalian target of rapamycin (mTOR) to promote survival of dormant tumor cells.
IRE1-XBP1 signaling
The most conserved and oldest branch of the UPR is governed by a type-1 single pass transmembrane protein IRE1, which consist of a serine/threonine kinase domain and an endoribonuclease (RNase) domain. The two IRE1 homologs encoded by mammalian genome: IRE1α (26, 27) and IRE1β which are expressed ubiquitously and in limited tissues respectively (28, 29). Activation of IRE1 has been proposed to occur in two ways. The competition model indicates that release of GRP78 from IRE1 ER luminal domain allows GRP78 to bind to unfolded proteins in the ER lumen while promoting dimerization of IRE1α. The second model suggests that the direct binding of unfolded proteins to core luminal domain of IRE1 induces its dimerization (30, 31). Between the two, the first model is more widely accepted and once dimerization or oligomerization occurs, the IRE1α kinase domains become juxtaposed and trans-autophosphorylation ensues. Phosphorylation of IRE1α is required for its RNase activation. The RNase activity initiates the cleavage of mRNA encoding X box-binding protein 1 (XBP1) to the spliced form XBP1-S, which can function as an active transcription factor. XBP1-S translocates to the nucleus and initiates the expression of a diverse set of genes by binding to the UPR element (UPRE) and to the ER stress-response elements I and II (ERSE-I and ERSE-II) in the promoter region of multiple genes (32). These target genes orchestrate ER protein folding, secretion, ERAD, lipid biosynthesis and ER expansion to alleviate ER stress. In addition, IRE1-dependent decay (RIDD) activation degrades UPR target mRNAs, including Grp78 and other mRNAs that negatively regulate apoptosis, cell proliferation and differentiation (33–35).The IRE1α-XBP1 pathway critically regulates development and lineage fates of several cell types. Development of dendritic cells, zymogen processing and maturation, homeostasis of Paneth cells for innate immunity and host defense and several other physiological processes are all regulated by XBP1s (36, 37).
However, this signaling pathway can also activate cell death. Activated IRE1α forms a complex with tumor necrosis factorα (TNFα) receptor-associated factor 2 (TRAF2) and apoptosis signaling-regulating kinase (ASK1). Activated ASK1 in turn, activates a number of transcription factors including NF-κB, c-Jun N-terminal kinase (JNK) and transcription factor AP1, which together promote pro-inflammatory and pro-apoptotic responses (38–41). These apoptotic signals converge on both the mitochondria, by suppressing Bcl-2 and activating BIM, as well as on the cytoplasm to activate downstream effector caspases (caspase-9, caspase-3) (42, 43). IRE1α activated JNK phosphorylates insulin receptor substrate 1 and 2 to induce insulin resistance and apoptosis (44). Hyperactivation of IRE1α rapidly upregulates pro-oxidant protein TXNIP (thioredoxin-interacting protein), that drives inflammasome-caspase-1 cell death pathway with concomitant attenuation of levels of miRNAs by IRE1α’s RNase (45, 46). IRE1α and PERK oligomerization can activate stress-activated kinase p38 MAPK which upregulates the expression of UPR genes serving as a feed forward ER Stress response (47). Thus, IRE1α acts as a molecular switch that regulates both adaptive and suicide genes response during prolonged activation of ER stress.
PERK-eIF2α-ATF4-CHOP signaling
Another key effector of the canonical UPR is PERK. The structural and mechanistic activation of PERK is similar to that of IRE1α, involving GRP78 competition between PERK and unfolded proteins driving activation. Activated PERK attenuates translational initiation by phosphorylating Ser51 of the α subunit of eukaryotic translation initiation factor 2 (eIF2α). Phosphorylaton of eIF2α serves two purposes. First, it transiently inhibits global protein synthesis in order to minimize the protein-folding load in the ER, conserve adenosine-5’-triphosphate (ATP) and amino acids (48). Phosphorylation of eIF2α increases its affinity for the eIF2β guanine nucleotide exchange factor, and this binding contributes to global mRNA translation suppression (49, 50). The second effect of phosphorylation of eIF2α is the preferential translation of select mRNAs containing short upstream opening reading frames (uORFs) in their 5’ untranslated region (UTR). One of the proteins whose translation is upregulated is the activating transcription factor 4 (ATF4) that activates the expression of genes that aim to relieve ER stress.
The critical role of PERK-p-eIF2α axis in organismal physiology is most well understood in pancreatic β cells where this axis controls insulin secretion and cellular metabolism (51). Mutation of the Ser51 eIF2α phosphorylation site is associated with hypoglycemia and defective gluconeogenesis in the liver of transgenic mice (51–53). To prevent total collapse of protein translation, growth arrest and DNA damage-inducible protein 34 (GADD34) and protein phosphatase 1 (PP1) direct the dephosphorylation of eIF2α, fine tuning mRNA translation to restore adaptive functions for ER homeostasis (54–56). This includes the enhanced translation of cellular apoptotic inhibitors (X-linked inhibitor of apoptosis proteins (XIAP), cellular inhibitor of apoptosis proteins 1 (cIAP1) and 2 (cIAP2)) and ATF4 (57, 58). ATF4 contributes to multiple facets of cellular homeostasis including amino acid biosynthesis and antioxidant programs. Importantly, it is clear that ATF4 may be the critical link between ER stress response pathways and the expression of autophagy genes as it directly binds the promoters of, and upregulates expression of multiple autophagy genes (MAP1LC3B, ATG12, and BECN1) (59, 60). ATF4 also binds to the promoter region and activates transcription of is a 29 kDa bZIP transcription factor known as CCAAT/enhancer binding protein homologous protein (CHOP) (61). A well-known mediator of ER stress-mediated cell death, CHOP activates a plethora of pro-apoptotic factors and accentuates oxidative stress (by ER reductase gene ERO1α), when protein folding is compromised in multiple cell types (62). Thus, much like IRE1a signaling, PERK signalling can activate cell survival and cell death pathways depending on the severity and resolution of ER stress.
ATF6 signaling
ATF6 is a type-II ER transmembrane protein consisting of a CREB/ATF bZIP transcription factor domain at the amino terminus. The initial step in the activation of ATF6 during ER stress is similar to that of IRE1 and PERK as the dissociation of constitutively bound GRP78 from ATF6 results in activation of ATF6. However unlike PERK and IRE1α, ATF6 activation requires regulated intramembrane proteolysis (RIP), producing a cleaved protein domain that is no longer membrane bound (63). Upon activation, ATF6 is translocated from the ER to the Golgi apparatus for sequential cleavage at the transmembrane site by site-1 protease (S1P) and site-2 protease (S2P) to yield ATF6 p50, an N-terminal cytosolic fragment required for activation of gene expression in the nucleus (64–66). Like IRE1, ATF6 exists in mammals in two isoforms: ATF6α and ATF6β. ATF6α is required for the regulation of protein homeostasis, during ER stress, yet deletion of ATF6β is not embryonic lethal and ATF6β−/− mice exhibit no phenotype suggesting that redundancy in the ER stress program can compensate for this loss (31, 67, 68). As homodimer, ATF6α transactivates and constitutively controls the levels of the ER chaperone GRP94 and co-chaperone p58IPK in dopaminergic neurons (69). When ATF6α forms a heterodimer with XBP1s, this complex directs the expression of multiple components of the ERAD machinery (EDEM, HERP and HRD1) (70–72). In addition, ATF6α also promotes the expression of ER quality control genes by recruiting the CREB regulated transcription coactivator 2 (CRTC2) to ER stress response elements in gene promoters (73). In contrast, deletion of ATF6β do not produce a significant phenotype and there has not been repertoire of consistent ATF6β target genes reported (31, 67). However, the overlapping functions of ATF6α and ATF6β are essential for transcriptional upregulation of GRP78, CHOP and XBP1 (68). Mounting evidence suggests that ATF6α drives the up-regulation of Ras homology enriched in brain (Rheb) and mTOR in an Akt-independent manner and increases the sensitivity of anti-tumor activity of rapamycin during tumor dormancy (74–78). Apart from ATF6, other bZIP transcription factors such as Luman/CREB3, OASIS/CREB3L1, and CREBH/CREB3L3 are also regulated by RIP during ER stress (79, 80).
The UPR controls apoptosis and autophagy in response to cell stress in cancer.
Within the nutrient and oxygen-limited tumor microenvironment, oxidative stress is often a major driver of the cancer phenotype (81). Glucose limitation affects protein glycosylation which leads to ER-Ca2+ imbalance and increased GRP78 levels. Tumor cells adapt to low glucose environment via UPR and progressing to a high rate of aerobic glycolysis known as Warburg effect (82–85). The UPR regulates oxidative stress by reducing global translation, while simultaneously increasing the expression of specific proteins. In breast cancer cells, ERO1α enhances the oxidative protein folding capacity by elevating the biosynthesis of the antioxidant glutathione (86, 87). Moreover, PERK is responsible for resistance to oxidative stress in radiotherapy by preventing the oxidative damage to DNA and promoting adaptation to preconditioned ER stress in breast cancer (88–90). Furthermore, one of the major antioxidant responses downstream of the UPR is upregulation of the transcription factor NRF2, which lies in a dormant state by binding with Kelch-like ECH-associated protein 1 (Keap1) (91, 92). In addition, the UPR confers cytoprotection against cellular insults via inhibition of inflammatory and apoptotic signaling pathways (93). Collectively, these three pathways converge in a coordinated way as a homeostatic feedback loop to alleviate ER stress when the stress is acute or mild. To succeed this all three pathways aim to block global translation, expand the ER and selectively activate the expression of genes that contribute to relieving the stress and increasing the folding capacity, such as chaperon proteins. If successful in reducing the amount of misfolded proteins, the UPR is attenuated and the cell survives. However, if the stress is severe and prolonged and homeostasis cannot be reestablished, then the UPR will be sustained and transform into the terminal UPR that promotes cell death (8, 71). Although the proapoptotic character of the UPR is not yet fully understood molecularly, hyper-activation of PERK and prolonged inhibition of translation can lead the cells to death. Additionally, PERK hyperactivation can upregulate CHOP transcription factor which inhibits expression of the gene that encodes for the antiapoptotic BCL-2 (94, 95).
Mounting evidence suggests that activation of the UPR can induce cytoprotective autophagy, which promotes cell survival by recycling damaged organelles and proteins. In hypoxic tumor xenografts, expression of autophagic factor LC3 has been reported to enhance autophagy survival mechanism (89). During hypoxia-induced ER stress, PERK, promotes autophagosome formation by transcriptional upregulation of microtubule associated protein 1 light chain 3 beta (MAP1LC3) and autophagy regulated (ATG) 5 through ATF4 and CHOP respectively (89, 96, 97). Moreover, eIF2α/ATF4/CHOP axis regulates the transcriptional levels of p62 and promotes autophagy induction through binding with the amino acid response element (AARE) sequence of p62 promoter (59). PERK-mediated cytoprotective autophagy in MYC driven lymphomagenesis is driven by cell cycle deregulation and enhanced protein biosynthesis (98, 99). The IRE1α-XBP1-S axis increases the conversion of LC3 I to LC3 II in endothelial cells to induce autophagy (100). In addition, XBP1-S directly binds to the −537 and −755 region of BECN1 gene promoter which enhances the expression of Beclin-1 and autophagy induction (96, 100). ATF6α also regulates induction of autophagy by formation of a transcriptional heterodimer complex with C/EBP-B and binds with CRE/ATF elements of the death-associated kinase 1 (DAPK1) gene promoter (101). Activated DAPK1 promotes phosphorylation of Beclin-1 and drives the formation of autophagosomes which results in activation of autophagy (102). ATF6α-induced autophagy is mediated by upregulation of CHOP and XBP1 genes (103). Therefore, mechanistic role of both UPR and autophagy must be taken into account during cancer treatment. While both processes can lead to cell death if persistently activated, overwhelming evidence suggests that activation of these pathways in the tumor microenvironment typically supports tumor growth and survival.
Non-Canonical ER Stress Responses
The canonical ER stress response is described above and directed by the three ER stress sensors and downstream effects, collectively referred to as the UPR. It has become clear recently that many other pathways converge on the UPR, or specific components of the UPR, that may have an effect on cell fate independent of the classical UPR. Here we review the Integrated stress response (ISR), ER translocation and ERK reactivation, endoplasmic-reticulum-associated protein degradation (ERAD), ERphagy, and other pathways that are just beginning to be dissected. Our focus is on how these non-canonical ER stress response programs converge on autophagy to drive cytoprotection. Autophagy has been identified as a druggable pathway. However, the components of autophagy regulated (ATG) genes have proven difficult to drug. It may be the case that targeting the UPR or non-canonical ER stress programs can more effectively block cytoprotective autophagy to enhance cancer therapy.
The Integrated Stress Response (ISR)
The integrated stress response (ISR) serves as a supplemental route to drive the activation of the UPR by the three sensors which have been discussed above. The ISR is an essential adaptive mechanism with respect to extrinsic (hypoxia, bioenergetics demands etc.) and intrinsic factors (ER stress or oncogenic stress) in human malignancies (104, 105). Apart from PERK-mediated eIF2α phosphorylation, double-stranded RNA-activated protein kinase (PKR), general control non-derepressible-2 (GCN2) and the heme-regulated inhibitor kinase (HRI) also phosphorylate Ser51 of eIF2α during viral infection, amino-acid starvation and heme-deprivation in erythroid cells respectively (Figure 2). One hallmark of the ISR is the activation of GCN2 by uncharged tRNAs. Among the intrinsic factors, c-MYC oncogene (MYC)-driven cancers are transformed by the enhanced protein synthesis along with an increase in size and folding efficiency of the ER to sustain survival of malignant cells during tumor development. In MYC-driven cancers, MYC activation enhances the protein synthesis causing ER stress. In response, PERK-mediated eIF2α phosphorylation limits the rate of protein translation and activates pro-survival autophagy to circumvent proteotoxicity during tumor growth (99). Simultaneously, MYC activates the GCN2 arm of ISR resulting in further phosphorylation of eIF2α to conserve amino acid metabolism and ATF4 induction to prevent nutrient depletion. A recent study by Tameire and colleagues (106) unraveled a novel pathway of ISR-induced signaling in MYC-dependent cancer progression. This study showed that both PERK and GCN2 kinases of ISR are necessary for optimal ATF4 expression by MYC activity in various cancer cell lines. Expression of MYC was associated with the accumulation of uncharged tRNAs which activate GCN2. Activated GCN2, in turn, drives the activation of transcription factor ATF4 which coordinates a gene expression program that recalibrates protein translation through mTOR effectors p70S6K and 4EBP1 (Figure 2). Alleviation of MYC-induced ER stress is driven by enhanced phosphorylated levels of p70S6K and EIF4EBP1 (4E-BP1), a downstream substrate of mammalian target of rapamycin complex 1 (mTORC1) which hyperphosphorylate 4E-BP1 by its dissociation from eIF4E. Targeting the ISR can be achieved by drugging the translational machinery (107) or by targeting autophagy (59, 108), but recently more specific inhibitors of ISR have entered clinical trials. ONC201 (also known as TIC10) a small molecule inhibitor that produces tumor growth impairment by activating an eIF2α-ATF4-dependent apoptosis pathway (107). In addition, a new generation of GCN2 inhibitors can sensitize cancer cells with low basal-level expression of asparagine synthetase (ASNS) to the L-asparaginase (ASNase), an anti-leukemic agent by inducing MAPKs pathway and apoptosis (109). Recently, a small molecule ISR inhibitor (ISRIB) has been identified which facilitates the assembly of active eIF2B and rescues translation in the presence of intracellular p-eIF2α levels. The pharmacological activation of eIF2B to inhibit the ISR by ISRIB could be a promising approach for cancer treatment (110).
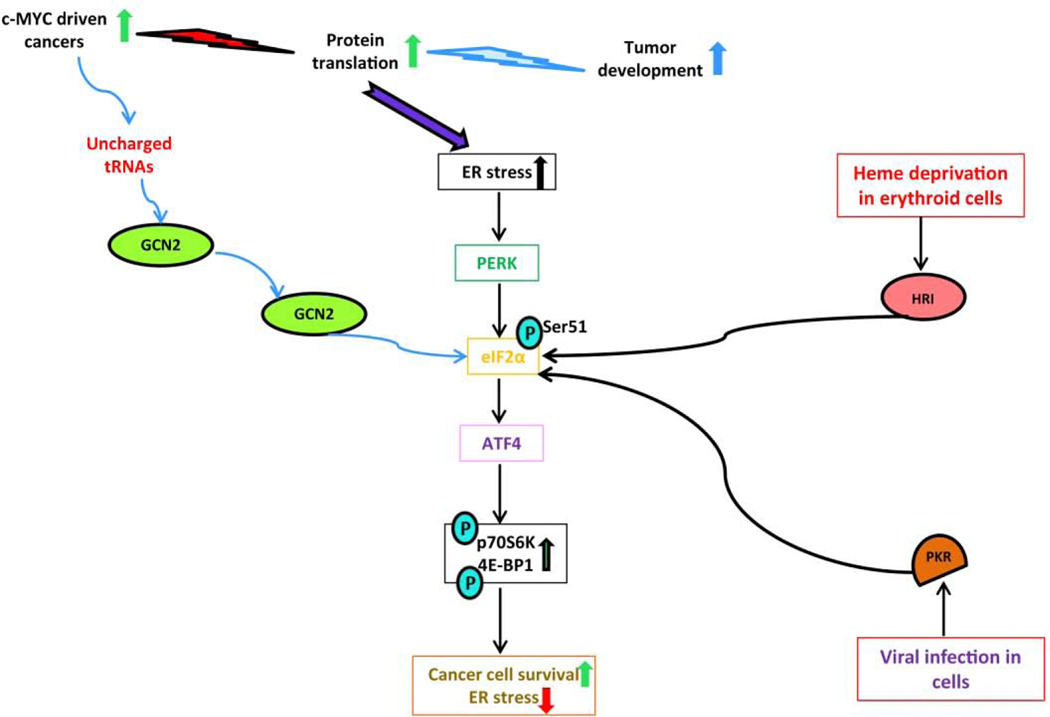
Figure 2: The Integrated stress response (ISR) in MYC-driven malignancies.
The ISR is activated by various intrinsic and extrinsic factors. Phosphorylation of eIF2α at Ser51 by double-stranded RNA-activated protein kinase (PKR), general control non-derepressible-2 (GCN2) and the heme-regulated inhibitor kinase (HRI) occurs during viral infection, amino-acid starvation and the heme-deprivation in erythroid cells respectively. During amino-acid starvation GCN2 is activated by uncharged tRNAs and phosphorylates eIF2α and triggers ATF4 induction in MYC driven cancers. Enhanced phosphorylation of p70S6K1 and 4E-BP1 abrogates ER stress and supports c-MYC driven cancer survival.
Therapeutic Stress drives ER Translocation of MAPK pathways during targeted therapy:
ER stress is not only activated by environmental stresses or by oncogenic stress, but also by therapeutic stress. Numerous cancer therapies activate ER stress but the mechanism by which this occurs is not always clear for drugs that are not well known to perturb protein folding. In BRAF mutant melanoma, BRAF inhibitors and combined BRAF and MEK inhibitors are the standard of care treatment for patients with metastatic or high risk BRAF mutant melanoma (111). These inhibitors are very effective initially with a 75% response rate in some cases, however, eventually most patients who respond recur. An intense effort has been underway to understand the mechanisms of resistance to BRAF and BRAF and MEK inhibition. Our group previously found that BRAF or BRAF and MEK inhibition activates cytoprotective autophagy by engaging the canonical UPR (112). Numerous groups have confirmed these findings in both BRAF mutant melanoma, but also in BRAF mutant colon cancer, and thyroid cancer. Since this publication numerous other gro ups have corroborated this finding (113–120). Meanwhile, other groups have found that numerous distinct resistance mechanisms impinge on ERK reactivation as a common mechanism of resistance in many patients (121–126). In cell culture immediately after BRAF and MEK inhibition, phosphorylation of ERK disappears, but within 4–12 hours in almost all cell lines tested, rephosphorylation of ERK emerges. Similar findings were found in biopsies of tumor that progressed on BRAF inhibitors (BRAFi) (127) where ERK phosphorylation was increased in resistant tumor specimens compared to either baseline samples or samples collected during initial tumor shrinkage. While drug-induced Raf hetero-dimerization has been proposed as a mechanism of ERK reactivation (128), the fact that this occurs in the presence of MEK inhibitor (MEKi) argued for another explanation for this phenomenon. Our groups had previously delineated the adaptive resistance mechanism of ERK reactivation and autophagy following BRAFi + MEKi treatment in BRAF mutant or BRAF/NRAS mutant melanoma tumors. ERK reactivation is critically dependent on ER translocation of the MAPK pathway (Figure 3). During BRAF and MEK inhibition a cytoplasmic pool of GRP78 binds to the scaffolding protein KSR2 which scaffolds NRAS, BRAF, MEK and ERK and shuttle this multiprotein complex to the ER membrane on RAB5+ early endosomes. MAPK components then translocate into the ER lumen via the ER translocase SEC61, as demonstrated by the trypsin protection assay and digitonin permeabilization assay in the literature (129). Sec/Sec61 dependent protein translocation can occur in two different ways. The first is co-translational translocation, which consists of arrested elongation of the polypeptide chain at the ribosome coupled with translocation through the Sec61 channel in the ER membrane. The second is post-translational translocation whereby the synthesis of the precursor is completed (or nearly completed) in the cytosol. Specific cytosolic chaperones prevent the precursor from attaining its native conformation and target the precursor to the translocon (130–132). The mechanism how a huge MAPK protein complex translocate through the Sec61 channel or engulfed into ER still needs to be studied extensively. We can speculate that MAPK precursors do not attain their native conformation or become unfolded as they are bound to KSR2 and hence are translocated in their non-native conformation. This sudden increase in ER luminal protein load provides a likely explanation as to why and how the UPR is activated following BRAF and MEK inhibition. After the ER translocation of MAPK components, ERK translocates from the ER back to the cytoplasm while NRAS, BRAF and MEK remain in the ER. It is still unclear about the fate of the remaining proteins inside the ER and what signal drives out the ERK from ER is a topic of future study. It may be that ERK is retrotranslocated from the ER to cytosol as previously described (133, 134). The other possible mechanism that ERK is associated with other ER luminal proteins Derlin-1 and Derlin-2 which may facilitate the subsequent movement of ERK from ER lumen to ER membrane by the cytosolic p97 ATPase (135, 136). Inhibition of the ERAD pathway does not impact MAPK components in the ER (unpublished data) as further experiments are needed to study this mechanism, hence we have not included in this review. Rephosphorylation of ERK2 by the cytoplasmic domain of PERK is critical for ERK2 reactivation following targeted therapy. Once rephosphorylated the cytoplasmic ERK species localizes to the nucleus (129). In the nucleus reactivated ERK phosphorylates ATF4, stabilizing this protein independently of eIF2alpha and the UPR. This non-canonical ER stress response driven by ATF4 phosphorylation results in the transcription of a number of autophagy genes and upregulation of autophagix flux. The critical nature of ATF4 phosphorylation in generating resistance to BRAF and MEK inhibitors was demonstrated by mutating the ERK phosphorylation site on ATF4, and reversing resistance to BRAFi + MEKi treatment in a highly treatment refractory patient derived xenograft. In this paper targeting any of the components of the ER translocation machinery (GRP78, KSR2, SEC61), PERK, ERK, or ATF4 abrogated therapy-induced autophagy and resistance to targeted therapy. Thus, targeting regulators of ER translocation would curb on the adaptive resistance mechanism in BRAF mutant cancer therapy.
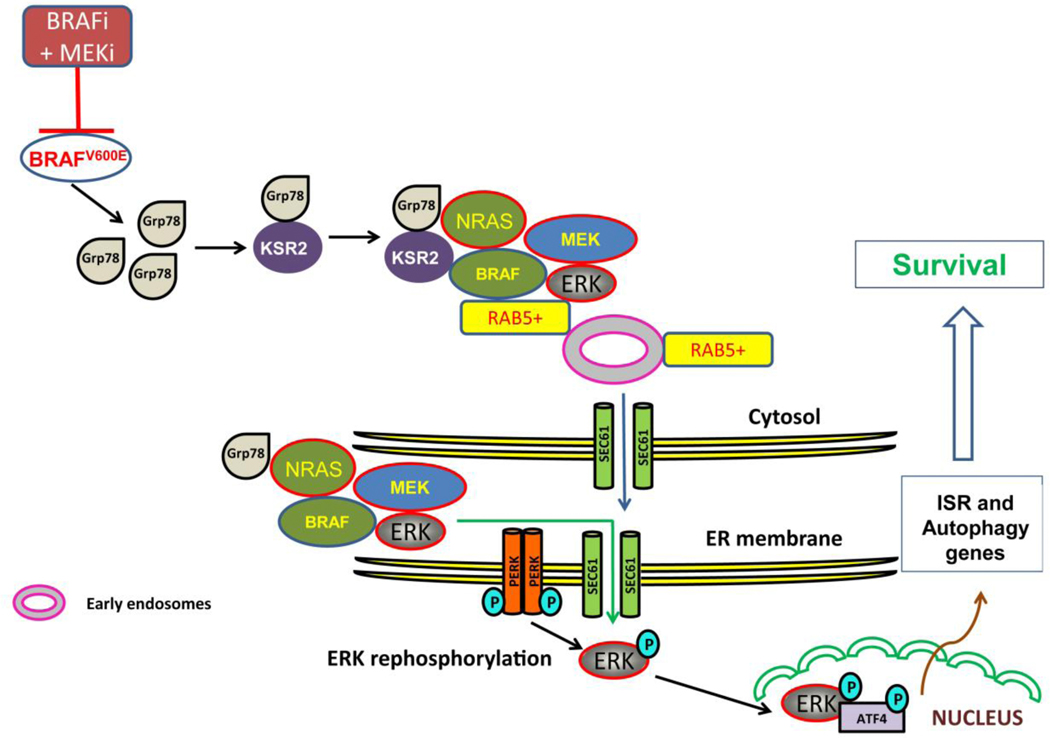
Figure 3: ER translocation of MAPK pathways and autophagy.
During targeted therapy with BRAF and MEK inhibition in BRAF mutant melanoma, a cytoplasmic pool of Grp78 binds to the scaffold protein KSR2 which recruits NRAS, BRAF, MEK and ERK. This multiprotein complex is shuttled to Rab5+ on early endosomes on ER membrane. MAPK components translocate to ER via the ER translocase SEC61. ERK translocates from ER back to the cytoplasm where ERK is phosphorylated by the cytoplasmic domain of PERK and ERK is reactivated. This reactivated ERK promotes phosphorylation of ATF4 independent of eIF2α and the UPR. ATF4 activates autophagy through transcriptional upregulation of multiple autophagy genes.
PERK-Nrf2 signaling can drive autophagy.
Besides proteotoxic stress, nutrient stress and therapeutic stress, another non-canonical ER stress response is driven by a unique relationship between PERK and the master antioxidant transcriptional factor NRF2 (Figure 4). PERK phosphorylates and stabilizes nuclear factor erythroid-derived 2-like 2 (NRF2), promoting cytoplasmic to nuclear translocation and expression of NRF2 target genes. NRF2 targets include antioxidant genes such as superoxide dismutase (Cu-Zn) 1 (SOD1) and catalase which increased the ratio of reduced to oxidized glutathione. NRF2 activation through PERK, also promotes drug resistance to cancer drugs through transcriptional upregulation of drug efflux pumps such as p-glycoprotein/multi-drug resistance (MDR) and confers enhanced oxidative stress buffering of both non-cancerous and cancerous de-differentiated cells (137). NRF2 activation has a complex effect on autophagy. The KEAP1 - NRF2 complex plays a major role in the oxidative stress defense system. Under normal conditions, KEAP1 binds with NRF2 and targets NRF2 for proteasomal degradation. Exposure to reactive oxygen species inactivates KEAP1 stabilizing NRF2 and promotes its nuclear translocation, where it binds with antioxidant response elements (AREs) and induces the transcription of numerous cytoprotective genes. The KEAP1-NRF2 pathway intersects with autophagy through competitive inhibition of the KEAP1-NRF2 interaction by p62/sequestosome 1 (SQSTM1). p62 binds to NRF2, and promotes the stabilization and activation of NRF2. NRF2, in turn, increases expression of autophagy cargo receptors p62 and NDP52. p62 serves as a selective autophagy substrate and a cargo receptor for autophagy as it binds both to ubiquitin and to LC3. Thus, a positive-feedback loop exists within the p62-KEAP1-NRF2 axis. At least one report demonstrates that NRF2 overexpression increases the formation of autophagosomes and autophagic flux (138). Activation of PERK can, therefore, drive autophagy through NRF2 stabilization (139–142).
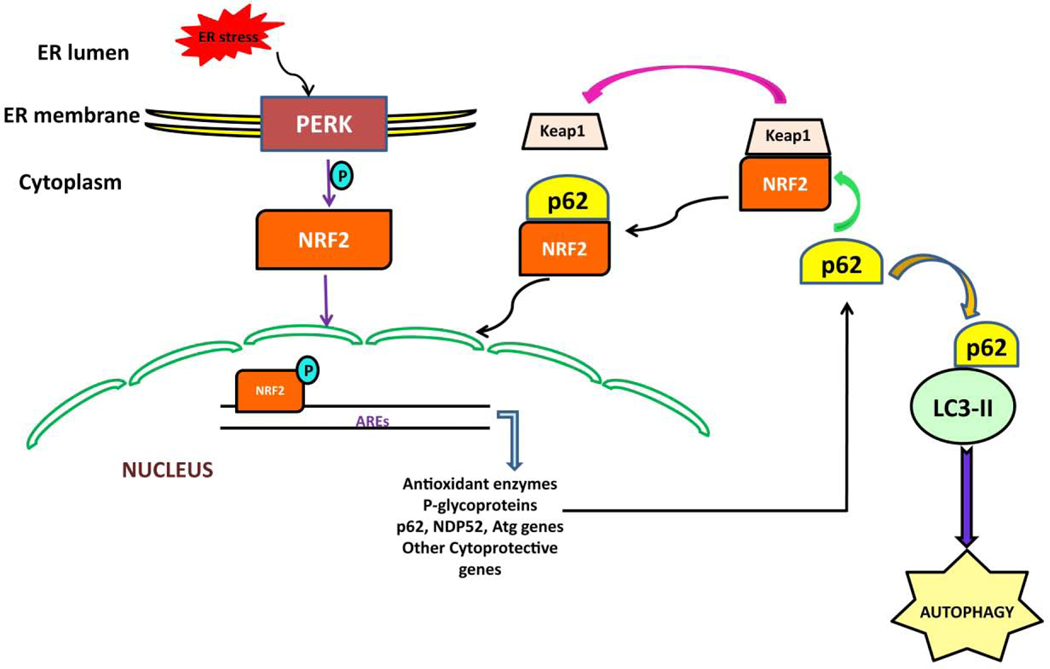
Figure 4: The PERK-NRF2 pathway and autophagy.
PERK triggers phosphorylation of NRF2 resulting in the translocation of NRF2 from the cytoplasm to the nucleus. Nuclear NRF2 binds with antioxidant response elements (AREs) and permits transcription of cytoprotective and autophagy related genes including p62. NRF2 forms a complex with KEAP1, which limits its access to the nucleus Autophagy cargo receptor p62 competitively binds NRF2 releasing it from KEAP1 inhibition promoting NRF2 stabilization and its translocation to nucleus. NRF2 increases expression of p62 which binds with LC3-II to promote autophagy.
PERK-p38-chaperone mediated autophagy (CMA)
When the UPR fails to expeditiously alleviate ER stress and restore ER homeostasis, persistent or late phase ER stress can lead to the activation of autophagy (143, 144). During the late phase of ER stress, p38MAPK/Hog1 phosphorylation is mediated by cytoplasmic regulator of osmolarity two-component system protein SSK1 (required during osmotic stress), canonical UPR components IRE1 and HAC1 (ortholog of XBP1 in yeast). Following its phosphorylation, high-osmolarity glycerol 1 (Hog1) translocates to the nucleus and governs the expression of late phase ER stress response genes such as HSP12, YMR103C and YPL088W. Eventually, Hog1 translocates back to the cytoplasm with elevated phosphorylation levels and stabilizes the essential autophagy protein LC3/Atg8 (145). More recently, the role of p38MAPK as a link between ER stress response and autophagy was further elucidated. This work found that ER stress induces phosphorylation of p38 MAPK and attenuates the levels of CMA substrate transcription factor monocyte enhancer 2D (MEF2D). Two key regulators of CMA, LAMP2A and Hsc70 were upregulated in lysosomal fractions during ER stress. Activation of lysosomal p38 MAPK by PERK causes phosphorylation at T211 and T213 of LAMP2A and leads to its oligomerization through recruitment of activated MKK4 to the lysosomes protecting the cell against the deleterious effects of ER stress. The PERK-p38 MAPK-CMA axis serves as a fundamental pathway for ER stress induced CMA (ERICA) where ER and lysosomes are associated with each other, and loss of CMA activity promotes ER stress-mediated cell death (146).
ERAD, ER-phagy and ERCQ-autophagy
ERAD, ER-phagy and ER-quality control-autophagy (ERQC-autophagy) are processes notably distinct from each other. ERAD is one of the best characterized cellular homeostasis pathways that monitors biosynthesis of membranous and secretory proteins in ER and regulated degradation of certain folded ER proteins (147) (Figure 5). ER-phagy is a selective process in which autophagic sequestration of ER fragments in autophagosomes occurs in eukaryotic cells (148, 149), whereas ERQC-autophagy is associated with transfer of conformer mutant of proteins from ER and their apparent clearance into autophagosomes (150, 151), RETREG1/FAM134B (149), RTN3 (152), and SEC62 (153) are known ER-phagy regulators. More recently, cell-cycle progression gene 1 (CCPG1) has been recognized as a vertebrate-specific protein with a cytosolic N-terminal region, a transmembrane domain within ER membrane and ER-luminal C-terminal region that traffics between autophagic vesicles and the ER membrane. Upon amino acid starvation or activation of ER stress, (154–156) CCGP1 localizes to the ER and binds both GABARAP/LC3 family members and focal adhesion kinase family interacting protein of 200 kD (FIP200), two different major autophagy proteins that drive ER-phagy and partition ER from the autophagy pathway (155). The GABARAP/LC3 family of autophagy proteins binds to the cytosolic domain of CCPG1 and via a conserved LC3-interacting region (LIR) motif protein in the cytosolic N-terminus of CCPG1, indicating CCPG1 is an autophagy cargo receptor for the ER. CCPG1 has direct interaction with FIP200 which is part of the UNC-51-like kinase 1 (ULk1) complex that readies membranes to become autophagosomes. Either ATG5 deletion or CCPG1 deletion prevented ER-phagy in HeLa cells. Thus, CCPG1 is an ER stress-mediated transcriptionally active gene which facilitates ER-phagy via interacting with ATG8 family members and FIP200.
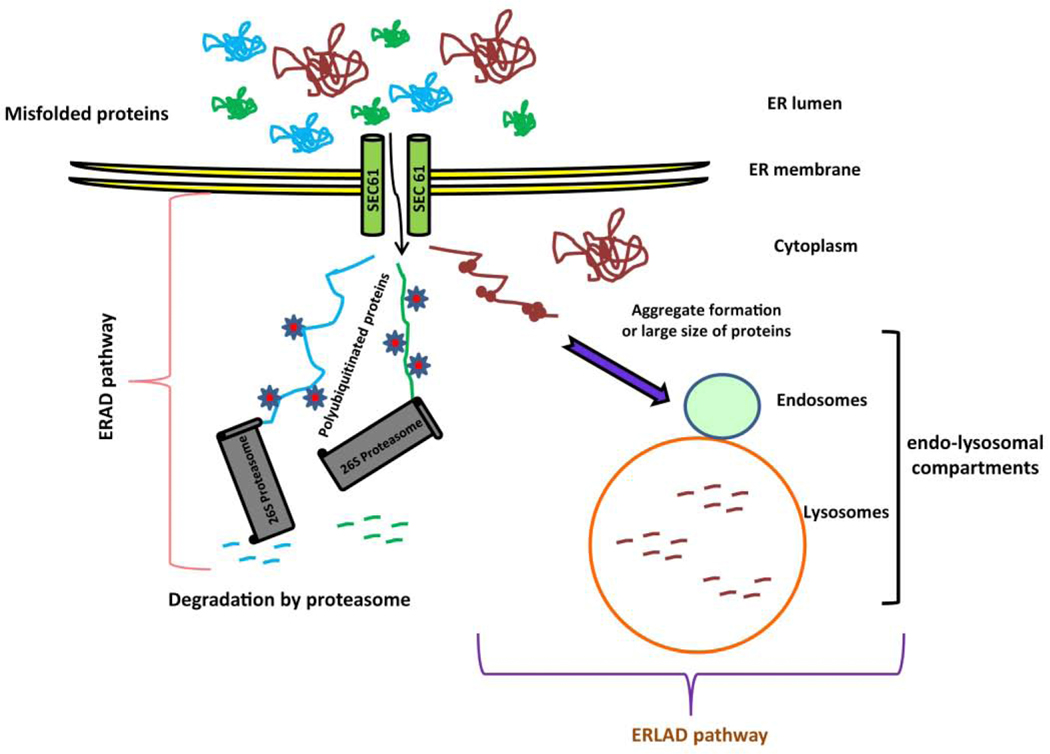
Figure 5: Endoplasmic-reticulum-associated protein degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathway.
Misfolded proteins are retrotranslocated from the ER to the cytoplasm via the ER translocase SEC61 and ubiquitinated. These ubiquitinated proteins are recognized and degraded by 26S proteasome which is known as ERAD pathway. In contrast, ERAD resistant proteins are transported from ER to endo-lysosomal compartments are degraded by autophagic and non-autophagic pathways which are known as ERLAD pathway.
The SEC61 translocon in ER membrane permits the unfolded proteins to cross the ER membrane for their folding inside the ER. The ERAD pathway removes misfolded proteins from the lumen or the membrane of the ER by retrotranslocation of the misfolded proteins back to the cytosol, where they are ubiquitinated. These ubiquitinated-proteins are recognized and degraded by the cytosolic 26S proteasome and maintain the ER protein quality control. Despite of the tightly regulated ERAD pathway, some misfolded proteins become resistance to the ERAD pathway due to their large size or aggregate formation. These proteasome resistant proteins are delivered from ER to endo-lysosomal compartments for degradation by autophagic and non-autophagic pathways, which are known as ER-to-lysosome-associated degradation (ERLAD) pathways (Figure 5). The involvement of ATG genes in disposal from the ER of proteasome-resistant proteins has been reported, yet their mechanism of signal transduction pathway remains to be explored. The clearance of misfolded proteins generated in the ER depends on their delivery via ERAD or ERLAD pathways to maintain ER homeostasis (151, 157–160).
Angiogenic factor with a G-patch domain and a Forkhead-associated domain (AGGF1) and CHOP
Initially identified as a susceptible gene in congenital vascular disorder Klippel-Trenaunay syndrome (161, 162), AGGF1 serves as an unfavorable prognostic factor and could play a pivotal role in tumor growth and metastasis (163–165). Elevated levels of APPG1 were significantly associated with the enhanced expression of CD34-labelled microvessel density and vascular endothelial growth factor (VEGF) in hepatocellular carcinoma (HCC). Additionally, expression levels of APPG1 were found to be an independent marker for the disease-free survival (DFS) of HCC patients (166).
Much of the work on ER stress and AGGF1 was performed in a cardiovascular model, but provides lessons that may be relevant in cancer. In heart failure, ER stress is activated in cardiac myocytes and leads to increased CHOP expression, which acts as a critical regulator of cardiac myocyte apoptosis (167). Recently it was reported that ER stress decreases levels of AGGF1 resulting in activation of ERK1/2, reduced levels of the transcriptional repressor ZEB1, with concomitant increased expression of miR-183–5p. miR-183–5p initiates post-transcriptional suppression of CHOP. Therefore ER Stress-regulated AGGF provides a built in negative feedback loop that favors ER stress response-associated autophagy by preventing CHOP mediated apoptosis. Apart from this mechanism, AGGF1 also governs other ER stress signaling markers, including sXBP1, GRP78, ERO1α, p-eIF2α, ATF4 and DR5, which were upregulated during ER stress (168).
Other examples of non-canonical ER stress response signaling
There are other pathways independent of the tradition UPR that can rescue the cancer cell from ER stress conditions. Mutant or misfolded proteins can be evacuated from the ER lumen by the Golgi reassembly stacking protein 1 (GRASP1)-mediated unconventional secretory pathway (169). The ER surveillance (ERSU) pathway is activated during mitosis to ensure inheritance of a functional ER to both daughter cells because the ER cannot be generated de novo. The ERSU pathway is regulated by MAPK Slt2 kinase, which works independently of UPR (170).
Most of the studies on the ER stress response in cancer have focused on the cancer cell, but it is clear the host cells especially immune cells also undergo canonical and non-canonical ER stress responses. A comprehensive discussion of the ER stress response and autophagy in each cell type is beyond the scope of this review. However, a recent study shows how the ER stress response and a non-canonical aspect of it can regulate epithelial-to-immune cell interactions. Intestinal epithelial cells (IECs) are exposed to constant environmental challenges and function as a front-line defense in the mucosal barrier (171, 172). There is clear evidence that demonstrates that IECs are reliant on the UPR and autophagy for homeostasis, and to perform their secretory function in the gut (173, 174). For instance an intact UPR and autophagy are required for the survival of mucin-secreting goblet cells and Paneth cells in the small intestine, two cell types that together maintain the intestinal stem cell niche (175, 176). When elements of the UPR are disabled, unchecked ER stress in IEC leads to enteritis, or inflammatory bowel disease. Detection of IECs’ ER stress by intestinal immune cells could be hallmark of cancer surveillance in the intestine. Earlier reports suggested that specific deletion of XBP1 in the epithelium result in enhanced expression of homologue of MHC class I molecules known as natural-killer group 2 member D (NKG2D) ligand (NKG2DL). Activation of cytomegalovirus UL16-binding proteins (ULBPs) and murine ULBP-like transcript 1 (MULT1) in humans and mice respectively are driven by UPR component CHOP in the IECs (177). Enhanced expression of NKG2DL is related with concomitant increase in numbers of intraepithelial NKG2D-expressing group 1 innate lymphoid cells (ILC1), which promoted ER stress-induced inflammatory cytolysis in vitro and spontaneous small intestinal enteritis in vivo (178–180). Pharmacological inhibition of NKG2D and depletion of NK1.1-expressing cells in Rag1−/−Xbp1ΔIEC double-mutant mice dampens inflammation and showed the importance of NKG2D and ILCs as innate immune activators (177). In other prospective, a polyreactive IgA response induced during IEC-ER stress is barrier-protective immune response against enteric inflammation. This ER stress in IEC is peritoneal B1b cell-derived which causes T cell-independent and microbiota independent-expansion that culminates in the peritoneal cavity. Enhanced numbers of IgA-producing plasma cells is witnessed during defective autophagy is a “eustress” response for homeostatic function of epithelial ER stress (181).
Path to a clinical implementation for ER stress response inhibitors:
Canonical and non-canonical ER stress response pathways regulate cell fate through multiple mechanisms as described above. In most disease contexts, including cancer, inhibition of the ER stress response components is desirable. The one caveat is that that in certain contexts such as cancer therapy induced ER stress response the cell fate may be apoptosis and inhibition of the ER stress response would be detrimental. For the vast majority of therapeutic contexts in cancer patients with advanced therapy resistant cancer, the ER Stress response pathway and its intimate link to cytoprotective autophagy suggests it would be an attractive target for drug development. A growing number of tool compounds used to study canonical and non-canonical ER stress response in the laboratory have spawned some interest in modifying these or related compounds to take agents into clinical trials. Examples of these agents have been reviewed elsewhere (129, 182–184). Meanwhile increased expression of ER stress response signaling components are becoming evident across in clinical samples, further emphasizing the attractiveness of this pathway as a druggable pathway in cancer. Expression levels of GRP78 have been associated with poor prognosis (185) and resistance to chemotherapy (186, 187) in multiple cancers. In addition, expression of IRE1 (188–190), PERK (191) and other canonical and non-canonical ER stress markers have been associated with cancer progression (192–198). ER stress is associated with progression and chemoresistance of a variety of cancers through multiple cellular pathways, including autophagy, which makes ER-stress induced cytoprotective autophagy as molecular target for clinical therapeutics (199). DNA-damaging chemotherapeutic agent cisplatin (cis-diamminedichloroplatinum II), induces autophagy via activating IRE1α-JNK pathway in HeLa cells (200). Apatinib, a novel tyrosine kinase inhibitor (TKI) significantly increased the expression levels of UPR target marker GRP78, IRE1-α, and LC3-II proteins in colorectal cancer cells (CRC). The ER stress-autophagy in CRC is activated through IRE1α-JNK pathway, which is ameliorated by synergistic treatment of autophagy inhibitor chloroquine (CQ) and apatinib (201). Nelfinavir, an ER stress-inducing drug which is currently in clinical trials for cancer patients resulted in the expression of endogenous mTOR inhibitor sestrin-2 (SESN2) and autophagy in breast cancer cells. Ectopic expression of ATF4 caused transcriptional upregulation of SESN2 expression, resulted in mTOR inhibition and autophagy (202). In addition, nelfinavir induces ER morphological changes and stress response, along with an autophagic protective strategy (203). Sequential treatment with celecoxib, a cyclooxygenase-2 (COX-2)-specific inhibitor, and proteasome inhibitor ortezomib enhances the ER stress-mediated autophagy-associated cell death by inducing the intracellular Ca2+ release, leading to the generation of autophagosomes in colon cancer cells (204). Sensitivity of melanoma cells to dabrafenib and temozolomide (TMZ) is determined by ER-stress induced autophagy (205, 206). The mechanisms of ER stress-induced autophagy in chemotherapy resistance has been discussed in this manuscript is discussed in revised review manuscript. Different anticancer treatments, including those that stimulate ER stress, activate autophagy in tumor cells, which has been proposed to either enhance cancer cell death or act as a mechanism of resistance to chemotherapy (207–211).
Efforts to inhibit components of the ER stress response must be met with caution as this stress response pathway may be required to maintain homeostasis in non-cancerous tissues in cancer patients. Careful characterization of preclinical toxicity at pharmacologically active doses needs to be established before moving ER stress response inhibitors into the clinic.
Conclusions and Open Questions
With the increasing understanding of canonical and non-canonical ER stress responses and how they regulate autophagy and apoptosis, it is now possible for us to design new combinatorial approaches for the treatment of multiple cancers. Although, inhibitors (natural or chemical) or small molecules that target canonical ER stress-mediated UPR and autophagy are being considered for clinical development in cancer treatment, one most consider the important aspects of how these inhibitors would impact non-canonical ER stress response pathways. Taken together these stress response pathways are an attractive area for future drug development. Open questions remain about how therapeutically targeting the ER Stress response would impact tumor immunity? How can specific inhibitors be designed to promote cancer cell death and avoid toxicity to normal tissue? Is it more effective to target UPR components, non-canonical ER Stress response proteins, autophagy proteins or the lysosome? Is there an ideal cancer or cancer therapy combination to test the efficacy of ER Stress inhibitors? Much work remains to be done to answer these and other burning questions in the field.
Acknowledgements:
This work was supported by funding from 1R01CA198015 (RKA) and 2 P01 CA165997 (CK).
Footnotes
Conflict of interest statement: RKA is inventor on patents related to novel autophagy inhibitors and is founder of Pinpoint Therapeutics. He is on the scientific advisory board for Sprint Biosciences and Immunaccel, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.GM C The Cell: A Molecular Approach. 2nd edition. Sunderland (MA): Sinauer Associates; 2000. [Google Scholar]
- 2.Alberts B, Johnson A, Lewis J ea. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. [Google Scholar]
- 3.Kanapin A, Batalov S, Davis MJ, Gough J, Grimmond S, Kawaji H, Magrane M, Matsuda H, Schönbach C, Teasdale RD, Z; Y, Group; RG, Members. G. Mouse proteome analysis. Genome Res. 2003; 13(6B):1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AM B Protein secretion and the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2012; 4(8): a012872. doi: 10.1101/cshperspect.a012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oakes SA, FR P The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2015; 10:173–94. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu BP, JS W Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004; 164(3):341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Anken E, I. B Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005; 40(4):191–228. [DOI] [PubMed] [Google Scholar]
- 8.Shore GC, Papa FR, SA O Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011; 23(2):143–9. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B L, DJ K Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci U S A. 2017; 114(2):201–5. doi: 10.1073/pnas.1619876114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dikic I, Z E Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018; 19(6):349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 11.Yu L, Chen Y, SA T Autophagy pathway: Cellular and molecular mechanisms. . Autophagy. 2018; 14(2):207–15. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley JH, LN Y Mechanisms of Autophagy Initiation. Annu Rev Biochem. 2017; 86:225–44. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen M, Rubinsztein DC, DW W Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018; 19(9):579–93. doi: 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marinković M, Šprung M, Buljubašić M IN Autophagy Modulation in Cancer: Current Knowledge on Action and Therapy. Oxid Med Cell Longev. 2018; 2018::8023821. doi: 10.1155/2018/8023821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Ohsumi Y TY Autophagosome formation in mammalian cells. Cell Struct Funct. 2002; 27(6):421–9. [DOI] [PubMed] [Google Scholar]
- 16.Li WW, Li J, JK B Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012; 69(7):1125–36. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo AM EW Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014; 24(1):92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakatogawa H KM Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy. 2015; 11(12):2377–8. doi: 10.1080/15548627.2015.1106665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozpolat B, DM B. Targeting autophagy in cancer management - strategies and developments. Cancer Manag Res. 2015; 7:291–9. doi: 10.2147/CMAR.S34859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva Paz M, Cotán D, Garrido-Maraver J, Cordero MD, Oropesa-Ávila M, de La Mata M, Delgado Pavón A, de Lavera I, Alcocer-Gómez E, JA S-A Targeting autophagy and mitophagy for mitochondrial diseases treatment. Expert Opin Ther Targets. 2016; 20(4):487–500. doi: 10.1517/14728222.2016.1101068. [DOI] [PubMed] [Google Scholar]
- 21.Mao K, DJ K A battlefield between host and microbe, and a possible avenue for cancer treatment. Autophagy. 2017; 13(2):223–4. doi: 10.1080/15548627.2016.1267075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynes E TS Urban planning of the endoplasmic reticulum (ER): How diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta 2011; 1813(10):1893–905. doi: 10.1016/j.bbamcr.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.English AR, Zurek N, GK V. Peripheral ER structure and function. Curr Opin Cell Biol. 2009; 21(4):596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi T, Rizzuto R, Hajnoczky G, TP S. MAM: more than just a housekeeper. Trends Cell Biol. 2009; 19(2):81–8. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollien J, JS W Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006; 313(5783):104–7. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y FB IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013; 23(11):547–55. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, RJ K The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2006; 103(39):14343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura D, Tsuru A, Ikegami K, Imagawa Y, Fujimoto N KK Mammalian ER stress sensor IRE1β specifically down-regulates the synthesis of secretory pathway proteins. FEBS Lett. 2010; 585(1):133–8. doi: 10.1016/j.febslet.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Tsuru A, Fujimoto N, Takahashi S, Saito M, Nakamura D, Iwano M, Iwawaki T, Kadokura H, Ron D KK Negative feedback by IRE1β optimizes mucin production in goblet cells. Proc Natl Acad Sci U S A. 2013; 110(8):2864–9. doi: 10.1073/pnas.1212484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohno K How transmembrane proteins sense endoplasmic reticulum stress. Antioxid Redox Signal. 2007; 9(12):2295–303. [DOI] [PubMed] [Google Scholar]
- 31.Cao SS, RJ K Unfolded protein response. Current Biol. 2012; 22(16):R622–R6. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Hou C, Cao Y, Cheng Q, Zhang L, Li H, Feng L YS XBP1 activation enhances MANF expression via binding to endoplasmic reticulum stress response elements within MANF promoter region in hepatitis B. Int J Biochem Cell Biol. 2018; 99:140–6. doi: 10.1016/j.biocel.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Moore K JH Ire1-mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol Biol Cell. 2015; 26(16):2873–84. doi: 10.1091/mbc.E15-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang CH, Chang S, Paton AW, Paton JC, Gabrilovich DI, Ploegh HL, Del Valle JR, CC H. Phosphorylation of IRE1 at S729 regulates RIDD in B cells and antibody production after immunization. J Cell Biol. 2018; 217(5):1739–55. doi: 10.1083/jcb.201709137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurel M, Chevet E, Tavernier J SG Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014; 39(5):245–54. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010; 140(6):900–17. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinon F, LH G Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr Opin Immunol. 2011; 23(1):35–40. doi: 10.1016/j.coi.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP DR Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000; 287(5453):664–6. [DOI] [PubMed] [Google Scholar]
- 39.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A HI ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002; 16(11):1345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabas I, D R Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011; 13(3):184–90. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu P, Han Z, Couvillon AD, Kaufman RJ, JH E Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006; 26(8):3071–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P AS ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007; 129(7):1337–49. [DOI] [PubMed] [Google Scholar]
- 43.Giam M, Huang DC PB BH3-only proteins and their roles in programmed cell death. Oncogene. 2008; 27 Suppl 1:: S128–36. [DOI] [PubMed] [Google Scholar]
- 44.Liang L, Chen J, Zhan L, Lu X, Sun X, Sui H, Zheng L, Xiang H, F. Z Endoplasmic reticulum stress impairs insulin receptor signaling in the brains of obese rats. PLoS One. 2015; 10(5):: e0126384. doi: 10.1371/journal.pone.0126384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, Papa FR, SA. O IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012; 338(6108):818–22. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, Heintz N, Greengard P, Hui S, Tang Q, Trusina A, Oakes SA, FR P IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012; 16(2):250–64. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koeberle A, Pergola C, Shindou H, Koeberle SC, Shimizu T, Laufer SA, O W Role of p38 mitogen-activated protein kinase in linking stearoyl-CoA desaturase-1 activity with endoplasmic reticulum homeostasis. FASEB J. 2015; 29(6):2439–49. doi: 10.1096/fj.14-268474. [DOI] [PubMed] [Google Scholar]
- 48.Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005; 16(1):3–12. [DOI] [PubMed] [Google Scholar]
- 49.Bogorad AM, Lin KY, A M Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res. 2017; 45(20):11962–79. doi: 10.1093/nar/gkx845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adomavicius T, Guaita M, Zhou Y, Jennings MD, Latif Z, Roseman AM, GD P. The structural basis of translational control by eIF2 phosphorylation. Nat Commun. 2019; 10(1):2136. doi: 10.1038/s41467-019-10167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Back SH, RJ. K Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012; 81:767–93. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, Jefferson LS, DR C The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002; 22(19):6681–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cnop M, Toivonen S, Igoillo-Esteve M, P S Endoplasmic reticulum stress and eIF2α phosphorylation: The Achilles heel of pancreatic β cells. Mol Metab. 2017; 6(9):1024–39. doi: 10.1016/j.molmet.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brush MH, Weiser DC, S S Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003; 23(4):1292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojas M, Vasconcelos G, TE D An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc Natl Acad Sci U S A. 2015; 112(27):E3466–75. doi: 10.1073/pnas.1501557112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, W P Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep. 2015; 11(12):1885–91. doi: 10.1016/j.celrep.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiramatsu N, Messah C, Han J, LaVail MM, Kaufman RJ, JH L Translational and posttranslational regulation of XIAP by eIF2α and ATF4 promotes ER stress–induced cell death during the unfolded protein response. Mol Biol Cell. 2014; 25(9):1411–20. doi: 10.1091/mbc.E13-11-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, JA D PERK-dependent regulation of IAP translation during ER stress. Oncogene. 2009; 28(6):910–20. doi: 10.1038/onc.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.B’chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, A B The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013; 41(16):7683–99. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luhr M, Torgersen ML, Szalai P, Hashim A, Brech A, Staerk J, N E The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress J Biol Chem. 2019; 294(20):8197–217. doi: 10.1074/jbc.RA118.002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, I M The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med. 2016; 16(6):533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu H, Tian M, Ding C, S Y The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front Immunol. 2019; 9:3083. doi: 10.3389/fimmu.2018.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hillary RF, U F A lifetime of stress: ATF6 in development and homeostasis. J Biomed Sci. 2018; 25(1):48. doi: 10.1186/s12929-018-0453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen J, Chen X, Hendershot L, R P Er stress regulation of atf6 localization by dissociation of bip/grp78 binding and unmasking of golgi localization signals. Dev Cell. 2002; 3(1):99–111. [DOI] [PubMed] [Google Scholar]
- 65.Teske BF, Wek SA, Bunpo P, Cundiff JK, McClintick JN, Anthony TG, RC W The eif2 kinase perk and the integrated stress response facilitate activation of atf6 during endoplasmic reticulum stress. Mol Biol Cell. 2011; 22(22):4390–405. doi: 10.1091/mbc.E11-06-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sykes EK, Mactier S, RI C Melanoma and the Unfolded Protein Response. Cancers (Basel). 2016; 8(3): pii:: E30. doi: 10.3390/cancers8030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kohl S, Zobor D, Chiang WC, Weisschuh N, Staller J, Gonzalez Menendez I, Chang S, Beck SC, Garcia Garrido M, Sothilingam V, Seeliger MW, Stanzial F, Benedicenti F, Inzana F, Héon E, Vincent A, Beis J, Strom TM, Rudolph G, Roosing S, Hollander AI, Cremers FP, Lopez I, Ren H, Moore AT, Webster AR, Michaelides M, Koenekoop RK, Zrenner E, Kaufman RJ, Tsang SH, Wissinger B, JH L Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet. 2015; 47(7):757–65. doi: 10.1038/ng.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Correll RN, Grimes KM, Prasad V, Lynch JM, Khalil H, JD M Overlapping and differential functions of ATF6α versus ATF6β in the mouse heart. Sci Rep. 2019; 9(1):2059. doi: 10.1038/s41598-019-39515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Egawa N, Yamamoto K, Inoue H, Hikawa R, Nishi K, Mori K, R T The endoplasmic reticulum stress sensor, ATF6α, protects against neurotoxin-induced dopaminergic neuronal death. . J Biol Chem. 2011; 286(10):7947–57. doi: 10.1074/jbc.M110.156430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto K, Suzuki N, Wada T, Okada T, Yoshida H, Kaufman RJ, K M Human HRD1 promoter carries a functional unfolded protein response element to which XBP1 but not ATF6 directly binds. J Biochem. 2008; 144(4):477–86. doi: 10.1093/jb/mvn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AF, S L Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol. 2013; 301:215–90. doi: 10.1016/B978-0-12-407704-1.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, K M Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007; 13(3):365–76. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Vera L, Fischer WH, M M The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009; 460(7254):534–7. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schewe DM, JA A-G ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A. 2008; 105(30):10519–24. doi: 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allen D, J S Stress Activates the TOR Pathway through Atf6. J Mol Signal. 2018; 13:1. doi: 10.5334/1750-2187-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu ZH, Shvartsman MB, Lee AY, Shao JM, Murray MM, Kladney RD, Fan D, Krajewski S, Chiang GG, Mills GB, JM A Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res. 2010; 70(8):3287–98. doi: 10.1158/0008-5472.CAN-09-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsu SK, Chiu CC, Dahms HU, Chou CK, Cheng CM, Chang WT, Cheng KC, Wang HD, IL L Unfolded Protein Response (UPR) in Survival, Dormancy, Immunosuppression, Metastasis, and Treatments of Cancer Cells. Int J Mol Sci. 2019; 20(10):pii: E2518. doi: 10.3390/ijms20102518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blackwood EA, Hofmann C, Santo Domingo M, Bilal AS, Sarakki A, Stauffer W, Arrieta A, Thuerauf DJ, Kolkhorst FW, Müller OJ, Jakobi T, Dieterich C, Katus HA, Doroudgar S, CC G ATF6 Regulates Cardiac Hypertrophy by Transcriptional Induction of the mTORC1 Activator, Rheb. Circ Res. 2019; 124(1):79–93. doi: 10.1161/CIRCRESAHA.118.313854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan CP, Kok KH, DY J CREB3 subfamily transcription factors are not created equal: Recent insights from global analyses and animal models. Cell Biosci. 2011; 1(1):6. doi: 10.1186/2045-3701-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fox RM, DJ A Transcriptional regulation of secretory capacity by bZip transcription factors. Front Biol (Beijing). 2015; 10(1):28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clarke HJ, Chambers JE, Liniker E, SJ M Endoplasmic Reticulum Stress in Malignancy. Cancer Cell. 2014; 25(5):563–73. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 82.Corazzari M, Gagliardi M, Fimia GM, M P Endoplasmic Reticulum Stress, Unfolded Protein Response, and Cancer Cell Fate. Front Oncol. 2017; 7:78. doi: 10.3389/fonc.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giampietri C, Petrungaro S, Conti S, Facchiano A, Filippini A, E Z Cancer Microenvironment and Endoplasmic Reticulum Stress Response. Mediators Inflamm. 2015; 2015:417281. doi: 10.1155/2015/417281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rufo N, Garg AD, P A The Unfolded Protein Response in Immunogenic Cell Death andCancerImmunotherapy.TrendsCancer.2017;3(9):643–58.doi: 10.1016/j.trecan.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 85.Ryder C, McColl K, Zhong F, CW D Acidosis promotes Bcl-2 family-mediated evasion of apoptosis: involvement of acid-sensing G protein-coupled receptor GPR65 signaling to Mek/Erk. J Biol Chem. 2012; 287(33):27863–75. doi: 10.1074/jbc.M112.384685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, D R An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003; 11(3):619–33. [DOI] [PubMed] [Google Scholar]
- 87.Kutomi G, Tamura Y, Tanaka T, Kajiwara T, Kukita K, Ohmura T, Shima H, Takamaru T, Satomi F, Suzuki Y, Torigoe T, Sato N, K H Human endoplasmic reticulum oxidoreductin 1-alpha is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 2013; 104(8):1091–6. doi: 10.1111/cas.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, JA D PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010; 29(27):3881–95. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, BG W The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010; 120(1):127–41. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rouschop KM, Dubois LJ, Keulers TG, van den Beucken T, Lambin P BJ, van der Kogel AJ, Koritzinsky M, BG W PERK/eIF2a signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS. Proc Natl Acad Sci U S A. 2013; 110(12):4622–7. doi: 10.1073/pnas.1210633110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, JA D Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003; 23(20):7198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cullinan SB, JA D PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004; 279(19):20108–17. [DOI] [PubMed] [Google Scholar]
- 93.Cubillos-Ruiz JR, Bettigole SE, LH G Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell. 2017; 168(4):692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCullough KD, Martindale JL, Klotz LO, Aw TY, NJ H Gadd153 Sensitizes Cells to Endoplasmic Reticulum Stress by Down-Regulating Bcl2 and Perturbing the Cellular Redox State. Mol Cell Biol. 2001; 21(4):1249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, D R CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004; 18(24):3066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan MM, Ni JD, Song D, Ding M, J H Interplay between unfolded protein response and autophagy promotes tumor drug resistance. Oncol Lett. 2015; 10(4):1959–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hosoi T, Nomura J, Tanaka K, Ozawa K, Nishi A, Y N Link between endoplasmic reticulum stress and autophagy in neurodegenerative diseases. Endoplasm Reticul Stress Dis. 2017; 4(1):37–45. doi: 10.1515/ersc-2017-0004. [DOI] [Google Scholar]
- 98.CV D Enigmatic MYC Conducts an Unfolding Systems Biology Symphony. Genes Cancer. 2010; 1(6):526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, Qiu B, Zhang H, Cerniglia G, Bi M, Li Y, Gao Y, Liu H, Li C, Maity A, Thomas-Tikhonenko A, Perl AE, Koong A, Fuchs SY, Diehl JA, Mills IG, Ruggero D, C K ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012; 122(12):4621–34. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Margariti A, Li H, Chen T, Martin D, Vizcay-Barrena G, Alam S, Karamariti E, Xiao Q, Zampetaki A, Zhang Z, Wang W, Jiang Z, Gao C, Ma B, Chen YG, Cockerill G, Hu Y, Xu Q, L Z XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem. 2013; 288(2):859–72. doi: 10.1074/jbc.M112.412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gade P, Ramachandran G, Maachani UB, Rizzo MA, Okada T, Prywes R, Cross AS, Mori K, DV K An IFN-γ-stimulated ATF6-C/EBP-β-signaling pathway critical for the expression of Death Associated Protein Kinase 1 and induction of autophagy. Proc Natl Acad Sci U S A. 2012; 109(26):10361–21. doi: 10.1073/pnas.1119273109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, A K DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009; 10(3):285–92. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mei Y, Thompson MD, Cohen RA, X T. Endoplasmic Reticulum Stress and Related Pathological Processes. J Pharmacol Biomed Anal. 2013; 1(2):1000107. [PMC free article] [PubMed] [Google Scholar]
- 104.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, AM G The integrated stress response. EMBO Rep. 2016; 17(10):1374–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rabouw HH, Langereis MA, Anand AA, Visser LJ, de Groot RJ, Walter P, FJM vK. Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc Natl Acad Sci U S A. 2019; 116(6):2097–102. doi: 10.1073/pnas.1815767116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, Salas C, Chinga F, Monroy AM, Fu W, Wang P, Kossenkov A, Ye J, Amaravadi RK, Ignatova Z, Fuchs SY, Diehl JA, Ruggero D, C K ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumor progression. Nat Cell Biol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kline CL, Van den Heuvel AP, Allen JE, Prabhu VV, Dicker DT, WS E-D ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2α kinases. Sci Signal. 2016; 9(415): ra18. doi: 10.1126/scisignal.aac4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xia X, Lei L, Qin W, Wang L, Zhang G, J. H. GCN2 controls the cellular checkpoint: potential target for regulating inflammation. Cell Death Discov. 2018; 4:20. doi: 10.1038/s41420-017-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakamura A, Nambu T, Ebara S, Hasegawa Y, Toyoshima K, Tsuchiya Y, Tomita D, Fujimoto J, Kurasawa O, Takahara C, Ando A, Nishigaki R, Satomi Y, Hata A, T H Inhibition of GCN2 sensitizes ASNS-low cancer cells to asparaginase by disrupting the amino acid response. Proc Natl Acad Sci U S A. 2018; 115(33):E7776–E85. doi: 10.1073/pnas.1805523115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nguyen HG, Conn CS, Kye Y, Xue L, Forester CM, Cowan JE, Hsieh AC, Cunningham JT, Truillet C, Tameire F, Evans MJ, Evans CP, Yang JC, Hann B, Koumenis C, Walter P, Carroll PR, D R Development of a stress response therapy targeting aggressive prostatecancer. Sci Transl Med.2018;10(439):pii:eaar2036.doi: 10.1126/scitranslmed.aar2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.JJ L Comprehensive Clinical Trial Data Summation for BRAF-MEK Inhibition and Checkpoint Immunotherapy in Metastatic Melanoma. Oncologist. 2019: pii:theoncologist. 2018–0876. doi: 10.1634/theoncologist.2018-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G, Lazova R, Klump V, Pawelek JM, Xu X, Xu W, Schuchter LM, Davies MA, Herlyn M, Winkler J, Koumenis C, RK A Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014; 124(3):1406–17. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mulcahy Levy JM, Zahedi S, Griesinger AM, Morin A, Davies KD, Aisner DL, Kleinschmidt-DeMasters BK, Fitzwalter BE, Goodall ML, Thorburn J, Amani V, Donson AM, Birks DK, Mirsky DM, Hankinson TC, Handler MH, Green AL, Vibhakar R, Foreman NK, A T Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife. 2017; 6:pii : e19671. doi: 10.7554/eLife.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sueda T, Sakai D KK, Konno M, Nishida N, Koseki J, Colvin H, Takahashi H, Haraguchi N, Nishimura J, Hata T, Takemasa I, Mizushima T, Yamamoto H, Satoh T, Doki Y, Mori M, H I BRAFV600E inhibition stimulates AMP-activated protein kinase-mediated autophagy in colorectal cancer cells. Sci Rep. 2016; 6:18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin S, Dudek-Perić AM, Maes H, Garg AD, Gabrysiak M, Demirsoy S, Swinnen JV, P A Concurrent MEK and autophagy inhibition is required to restore cell death associated danger-signalling in Vemurafenib-resistant melanoma cells. Biochem Pharmacol. 2015; 93(3):290–304. doi: 10.1016/j.bcp.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 116.Li YY, Wu C, Chen SM, Shah SS, Wangpaichitr M, Feun LG, Kuo MT, Suarez M, Prince J, N S BRAF inhibitor resistance enhances vulnerability to arginine deprivation in melanoma. Oncotarget. 2016; 7(14):17665–80. doi: 10.18632/oncotarget.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xie X, Koh JY, Price S, White E, JM M Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015; 5(4):410–23. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goodall ML, Wang T, Martin KR, Kortus MG, Kauffman AL, Trent JM, Gately S, JP M. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy. 2014; 10(6):1120–36. doi: 10.4161/auto.28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mulcahy Levy JM, Foreman NK, A T Using BRAF (V600E) as a marker of autophagy dependence in pediatric brain tumors. Autophagy. 2014; 10(11):2077–8. doi: 10.4161/auto.36138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goulielmaki M, Koustas E, Moysidou E, Vlassi M, Sasazuki T, Shirasawa S, Zografos G, Oikonomou E, A P BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Autophagy. 2016; 7(8):9188–221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, N R BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006; 439(7074):358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Long GV, Fung C, Menzies AM, Pupo GM, Carlino MS, Hyman J, Shahheydari H, Tembe V, Thompson JF, Saw RP, Howle J, Hayward NK, Johansson P, Scolyer RA, Kefford RF, H R Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun. 2014; 5(5694). doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 123.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, Rosenberg M, Goetz EM, Sullivan RJ, Farlow DN, Friedrich DC, Anderka K, Perrin D, Johannessen CM, McKenna A, Cibulskis K, Kryukov G, Hodis E, Lawrence DP, Fisher S, Getz G, Gabriel SB, Carter SL, Flaherty KT, Wargo JA, LA G MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014; 4(1):61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Corcoran RB, Settleman J, JA E Potential therapeutic strategies to overcome acquired resistance to BRAF or MEK inhibitors in BRAF mutant cancers. Oncotarget. 2011; 2(4):336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, de Stanchina E, Abdel-Wahab O, Solit DB, Poulikakos PI, N R BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer cell. 2015; 28(3):370–83. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, Koya RC, Samatar AA, Khanlou N, Braun J, Ruchalski K, Seifert H, Larkin J, Dahlman KB, Johnson DB, Algazi A, Sosman JA, Ribas A, RS L Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 2015; 27(2):240–56. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Kim KB, Weber JS, Hersey P, Long GV, Lawrence D, Ott PA, Amaravadi RK, Lewis KD, Puzanov I, Lo RS, Koehler A, Kockx M, Spleiss O, Schell-Steven A, Gilbert HN, Cockey L, Bollag G, Lee RJ, Joe AK, Sosman JA, A R Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013; 31(14):1767–74. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 128.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Ribas A, Lo RS, Rosen N, DB S RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF (V600E). Nature. 2011; 480(7377):387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ojha R, Leli NM, Onorati A, Piao S, Verginadis II, Tameire F, Rebecca VW, Chude CI, Murugan S, Fennelly C, Noguera-Ortega E, Liu S, Xu X, Krepler C, Xiao M, Xu W, Wei Z, Frederick DT, Boland G, Mitchell TC, Karakousis GC, Schuchter LM, Flaherty KT, Zhang G, Herlyn M, Koumenis C, RK A ER Translocation of the MAPK Pathway Drives Therapy Resistance in BRAF-Mutant Melanoma. Cancer Discov. 2018; 9(3):396–415. doi: 10.1158/2159-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stephenson K Sec-dependent protein translocation across biological membranes: evolutionary conservation of an essential protein transport pathway (review). Mol Membr Biol. 2005; 22(1–2):17–28. [DOI] [PubMed] [Google Scholar]
- 131.Römisch K Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J Cell Sci. 1999; 112:4185–91. [DOI] [PubMed] [Google Scholar]
- 132.Araki K, K N Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol. 2012; 4(8):: a015438. doi: 10.1101/cshperspect.a015438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Afshar N, Black BE, BM P Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol Cell Biol. 2005; 25(20):8844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Petris G, Vecchi L, Bestagno M, OR B Efficient detection of proteins retrotranslocated from the ER to the cytosol by in vivo biotinylation. PLoS One. 2011; 6(8):: e23712. doi: 10.1371/journal.pone.0023712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ye Y, Shibata Y, Yun C, Ron D, TA R. A membrane protein complex mediates retrotranslocation from the ER lumen into the cytosol. Nature. 2004; 429(6994):841–7. [DOI] [PubMed] [Google Scholar]
- 136.Lilley BN, HL P Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005; 102(40):14296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Del Vecchio CA, Feng Y, Sokol ES, Tillman EJ, Sanduja S, Reinhardt F, PB G De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLos Biol. 2014; 12(9):: e1001945. doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang J, Liu Z, Hu T, Han L, Yu S, Yao Y, Ruan Z, Tian T, Huang T, Wang M, Jing L, Nan K, X L Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle. 2017; 16(11):1053–62. doi: 10.1080/15384101.2017.1312224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, M K Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013; 51(5):618–31. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 140.Zhou Y, Li Y, Ni HM, Ding WX, H Z Nrf2 but not autophagy inhibition is associated with the survival of wild-type epidermal growth factor receptor non-small cell lung cancer cells. Toxicol Appl Pharmacol. 2016; 310:140–9. doi: 10.1016/j.taap.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kageyama S, Saito T, Obata M, Koide RH, Ichimura Y, M K Negative Regulation of the Keap1-Nrf2 Pathway by a p62/Sqstm1 Splicing Variant. Mol Cell Biol. 2018; 38(7): pii: e00642–17. doi: 10.1128/MCB.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yoshinori K, Yoshinobu I, K M Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Current Opinion in Toxicology. 2016; 1:54–61. [Google Scholar]
- 143.Bernales S, McDonald KL, P W Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLos Biol. 2006; 4(12):: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yorimitsu T, Nair U, Yang Z, DJ K Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006; 281(40):30299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bicknell AA, Tourtellotte J, M N Late phase of the endoplasmic reticulum stress response pathway is regulated by Hog1 MAP kinase. J Biol Chem. 2010; 285(23):17545–55. doi: 10.1074/jbc.M109.084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li W, Zhu J, Dou J, She H, Tao K, Xu H, Yang Q, Z M Phosphorylation of LAMP2A by p38 MAPK couples ER stress to chaperone-mediated autophagy. Nat Commun. 2017; 8(1):: 1763. doi: 10.1038/s41467-017-01609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ruggiano A, Foresti O, P C Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014; 204(6):869–79. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lipatova Z, N S A Role for Macro-ER-Phagy in ER Quality Control. PLos Genet. 2015; 11(7):: e1005390. doi: 10.1371/journal.pgen.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, Katona I, Qualmann B, Weis J, Reggiori F, Kurth I, Hübner CA, I D Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015; 522(7556):354–8. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 150.Teckman JH, DH P Retention of mutant alpha (1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000; 279(5):961–74. [DOI] [PubMed] [Google Scholar]
- 151.Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, Janovick JA, Conn PM, DM C Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell. 2014; 54(1):166–79. doi: 10.1016/j.molcel.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grumati P, Morozzi G, Hölper S, Mari M, Harwardt MI, Yan R, Müller S, Reggiori F, Heilemann M, I D Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017; 6: pii: e25555. doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Soldà T, D’Antuono R, Raimondi A, Jung M, Melnyk A, Schorr S, Schreiber A, Simonelli L, Varani L, Wilson-Zbinden C, Zerbe O, Hofmann K, Peter M, Quadroni M, Zimmermann R, M M Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016; 18(11):1173–84. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 154.Kostenko EV, Olabisi OO, Sahay S, Rodriguez PL, IP W Ccpg1, a novel scaffold protein that regulates the activity of the Rho guanine nucleotide exchange factor Dbs. Mol Cell Biol. 2006; 26(23):8964–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, S W CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell. 2018; 44(2):217–32.e11. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smith MD, S W CCPG1, a cargo receptor required for reticulophagy and endoplasmic reticulum proteostasis. Autophagy. 2018; 14(6):1090–91. doi: 10.1080/15548627.2018.1441473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fregno I, M M Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit Rev Biochem Mol Biol. 2019; 14:1–11. doi: 10.1080/10409238.2019.1610351. [DOI] [PubMed] [Google Scholar]
- 158.Ju LG, Lin X, Yan D, Li QL, Wu M, LY L Characterization of WDR20: A new regulator of the ERAD machinery. Biochim Biophys Acta Mol Cell Res 2018; 1865(7):970–80. doi: 10.1016/j.bbamcr.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 159.Bernasconi R, M M ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr Opin Cell Biol. 2011; 23(2):176–83. doi: 10.1016/j.ceb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Calì T, Galli C, Olivari S, M M Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun. 2008; 371(3):405–10. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- 161.Tian XL, Kadaba R, You SA, Liu M, Timur AA, Yang L, Chen Q, Szafranski P, Rao S, Wu L, Housman DE, DiCorleto PE, Driscoll DJ, Borrow J, Q W Identification of an angiogenic factor that when mutated causes susceptibility to Klippel-Trenaunay syndrome. Nature. 2004; 427(6975):640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu Y, Li L, Seidelmann SB, Timur AA, Shen PH, Driscoll DJ, QK W Identification of association of common AGGF1 variants with susceptibility for Klippel-Trenaunay syndrome using the structure association program. Ann Hum Genet. 2008; 72(Pt 5):636–43. doi: 10.1111/j.1469-1809.2008.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen D, Li L, Tu X, Yin Z, Q W Functional characterization of Klippel-Trenaunay syndrome gene AGGF1 identifies a novel angiogenic signaling pathway for specification of vein differentiation and angiogenesis during embryogenesis. Hum Mol Genet. 2013; 22(5):963–76. doi: 10.1093/hmg/dds501. [DOI] [PubMed] [Google Scholar]
- 164.Yao HH, Wang BJ, Wu Y, Q H High Expression of Angiogenic Factor with G-Patch and FHA Domain1 (AGGF1) Predicts Poor Prognosis in Gastric Cancer. Med Sci Monit. 2017; 23:1286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang W, Li GY, Zhu JY, Huang DB, Zhou HC, Zhong W, CS J Overexpression of AGGF1 is correlated with angiogenesis and poor prognosis of hepatocellular carcinoma. Med Oncol. 2015; 32(4):131. doi: 10.1007/s12032-015-0574-2. [DOI] [PubMed] [Google Scholar]
- 166.Tu J, Ying X, Zhang D, Weng Q, Mao W, Chen L, Wu X, Tu C, Ji J, Y H High expression of angiogenic factor AGGF1 is an independent prognostic factor for hepatocellular carcinoma. Oncotarget. 2017; 8(67):111623–30. doi: 10.18632/oncotarget.22880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fu HY, Okada K, Liao Y, Tsukamoto O, Isomura T, Asai M, Sawada T, Okuda K, Asano Y, Sanada S, Asanuma H, Asakura M, Takashima S, Komuro I, Kitakaze M, T M Ablation of C/EBP homologous protein attenuates endoplasmic reticulum-mediated apoptosis and cardiac dysfunction induced by pressure overload. Circulation. 2010; 122(4):361–9. doi: 10.1161/CIRCULATIONAHA.109.917914. [DOI] [PubMed] [Google Scholar]
- 168.Yao Y, Lu Q, Hu Z, Yu Y, Chen Q, QK W A non-canonical pathway regulates ER stress signaling and blocks ER stress-induced apoptosis and heart failure. Nat Commun. 2017; 8(1):133. doi: 10.1038/s41467-017-00171-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gee HY, Noh SH, Tang BL, Kim KH, MG L Rescue of ΔF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011; 146(5):746–60. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 170.Babour A, Bicknell AA, Tourtellotte J, M N A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell. 2010; 142(2):256–69. doi: 10.1016/j.cell.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baker AM, Cereser B, Melton S, Fletcher AG, Rodriguez-Justo M, Tadrous PJ, Humphries A, Elia G, McDonald SA, Wright NA, Simons BD, Jansen M, TA G Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014; 8(4):940–7. doi: 10.1016/j.celrep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Peterson LW DA Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014; 14(3):141–53. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 173.Bartolome A, Guillen C, M B Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic β cell death. Autophagy. 2012; 8(12):1757–68. doi: 10.4161/auto.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Grootjans J, Kaser A, Kaufman RJ, RS B The unfolded protein response in immunity and inflammation. Nat Rev Immunol. 2016; 16(8):469–84. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.AJ O Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010; 26(6):547–53. doi: 10.1097/MOG.0b013e32833dccde. [DOI] [PubMed] [Google Scholar]
- 176.Hodin CM, Verdam FJ, Grootjans J, Rensen SS, Verheyen FK, Dejong CH, Buurman WA, Greve JW, K L Reduced Paneth cell antimicrobial protein levels correlate with activation of the unfolded protein response in the gut of obese individuals. J Pathol. 2011; 225(2):276–84. doi: 10.1002/path.2917. [DOI] [PubMed] [Google Scholar]
- 177.Hosomi S, Grootjans J, Tschurtschenthaler M, Krupka N, Matute JD, Flak MB, Martinez-Naves E, Gomez Del Moral M, Glickman JN, Ohira M, Lanier LL, Kaser A, R B Intestinal epithelial cell endoplasmic reticulum stress promotes MULT1 up-regulation and NKG2D-mediated inflammation. J Exp Med. 2017; 214(10):2985–97. doi: 10.1084/jem.20162041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, M C, Consortium IG Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015; 16(3):306–17. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Spits H, Bernink JH, L L NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol. 2016; 17(7):758–64. doi: 10.1038/ni.3482. [DOI] [PubMed] [Google Scholar]
- 180.Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, Munneke JM, Hazenberg MD, Villaudy J, Buskens CJ, Bemelman WA, Diefenbach A, Blom B, H S Interleukin-12 and −23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 2015; 43(1):146–60. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 181.Grootjans J, Krupka N, Hosomi S, Matute JD, Hanley T, Saveljeva S, Gensollen T, Heijmans J, Li H, Limenitakis JP, Ganal-Vonarburg SC, Suo S, Luoma AM, Shimodaira Y, Duan J, Shih DQ, Conner ME, Glickman JN, Fuhler GM, Palm NW, de Zoete MR, van der Woude CJ, Yuan GC, Wucherpfennig KW, Targan SR, Rosenstiel P, Flavell RA, McCoy KD, Macpherson AJ, Kaser A, RS B Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science. 2019; 363(6430):993–8. doi: 10.1126/science.aat7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ojha R, RK A Targeting the unfolded protein response in cancer. Pharmacol Res. 2017; 120:258–66. doi: 10.1016/j.phrs.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tang Z, Yuan X, Du R, Cheung SH, Zhang G, Wei J, Zhao Y, Feng Y, Peng H, Zhang Y, Du Y, Hu X, Gong W, Liu Y, Gao Y, Liu Y, Hao R, Li S, Wang S, Ji J, Zhang L, Li S, Sutton D, Wei M, Zhou C, Wang L, L L BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers. Mol Cancer Ther. 2015; 14(10):2187–97. doi: 10.1158/1535-7163. [DOI] [PubMed] [Google Scholar]
- 184.Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S, Middleton G, Muro K, Gordon MS, Tabernero J, Yaeger R, O’Dwyer PJ, Humblet Y, De Vos F, Jung AS, Brase JC, Jaeger S, Bettinger S, Mookerjee B, Rangwala F, E VC Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAF (V600E)-Mutant Colorectal Cancer. Cancer Discov. 2018; 8(4):428–43. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gifford JB RH GRP78 Influences Chemoresistance and Prognosis in Cancer. Curr Drug Targets. 2018; 19(6):701–8. doi: 10.2174/1389450118666170615100918. [DOI] [PubMed] [Google Scholar]
- 186.Wang Y WJ, Zhang XL, Wang XL, Yang L. Endoplasmic reticulum chaperone glucose-regulated protein 78 in gastric cancer: An emerging biomarker. Oncol Lett. 2018; 15(5):6087–93. doi: 10.3892/ol.2018.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Avril T, Vauléon E, E C Endoplasmic reticulum stress signaling and chemotherapy resistance in solid cancers. Oncogenesis. 2017; 6(8):: e373. . doi: 10.1038/oncsis.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yang Z, Zhang J, Jiang D, Khatri P, Solow-Cordero DE, Toesca DAS, Koumenis C, Denko NC, Giaccia AJ, Le QT, AC K A Human Genome-Wide RNAi Screen Reveals Diverse Modulators that Mediate IRE1α-XBP1 Activation. Mol Cancer Res. 2018; 16(5):745–53. doi: 10.1158/1541-7786.MCR-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sheng X, Nenseth HZ, Qu S, Kuzu OF, Frahnow T, Simon L, Greene S, Zeng Q, Fazli L, Rennie PS, Mills IG, Danielsen H, Theis F, Patterson JB, Jin Y, F S IRE1α-XBP1s pathway promotes prostate cancer by activating c-MYC signaling. Nat Commun. 2019; 10(1):323. doi: 10.1038/s41467-018-08152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Logue SE, McGrath EP, Cleary P, Greene S, Mnich K, Almanza A, Chevet E, Dwyer RM, Oommen A, Legembre P, Godey F, Madden EC, Leuzzi B, Obacz J, Zeng Q, Patterson JB, Jäger R, Gorman AM, A S Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat Commun. 2018; 9(1):3267. doi: 10.1038/s41467-018-05763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Alasiri G, Jiramongkol Y, Zona S, Fan LY, Mahmud Z, Gong G, Lee HJ, EW L Regulation of PERK expression by FOXO3: a vulnerability of drug-resistant cancer cells. Oncogene. 2019; 38(36):6382–98. doi: 10.1038/s41388-019-0890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cubillos-Ruiz JR BS, Glimcher LH. . Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell. 2017; 168(4):692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liu CY, Hsu CC, Huang TT, Lee CH, Chen JL, Yang SH, Jiang JK, Chen WS, Lee KD, HW T ER stress-related ATF6 upregulates CIP2A and contributes to poor prognosis of colon cancer. Mol Oncol. 2018; 12(10):1706–17. doi: 10.1002/1878-0261.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hanaoka M, Ishikawa T, Ishiguro M, Tokura M, Yamauchi S, Kikuchi A, Uetake H, Yasuno M, T K Expression of ATF6 as a marker of pre-cancerous atypical change in ulcerative colitis-associated colorectal cancer: a potential role in the management of dysplasia. J Gastroenterol. 2018; 53(5):631–41. doi: 10.1007/s00535-017-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cheng Y, Qiao Z, Dang C, Zhou B, Li S, Zhang W, Jiang J, Song Y, Zhang J, D D p38 predicts depression and poor outcome in esophageal cancer. Oncol Lett. 2017; 14(6):7241–49. doi: 10.3892/ol.2017.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang Y, Yang WK, Wen GM, Tang H, Wu CA, Wu YX, Jing ZL, Tang MS, Liu GL, Li DZ, Li YH, YJ D High expression of PRKDC promotes breast cancer cell growth via p38 MAPK signaling and is associated with poor survival. Mol Genet Genomic Med. 2019; 12(:e908). doi: 10.1002/mgg3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Adhikaree J, Franks HA, Televantos C, Vaghela P, Kaur AP, Walker D, Schmitz M, Jackson AM, PM P Impaired circulating myeloid CD1c+ dendritic cell function in human glioblastoma is restored by p38 inhibition - implications for the next generation of DC vaccines. Oncoimmunology. 2019; 8(7):: 1593803. doi: 10.1080/2162402X.2019.1593803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sahu V, Mohan A, S D p38 MAP kinases: plausible diagnostic and prognostic serum protein marker of non-small cell lung cancer. Exp Mol Pathol. 2019; 107:118–23. doi: 10.1016/j.yexmp.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 199.Thakur PC, Miller-Ocuin JL, Nguyen K, Matsuda R, Singhi AD, Zeh HJ, N B Inhibition of endoplasmic-reticulum-stress-mediated autophagy enhances the effectiveness of chemotherapeutics on pancreatic cancer. J Transl Med. 2018; 16(1):: 190. doi: 10.1186/s12967-018-1562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xu Y, Yu H, Qin H, Kang J, Yu C, Zhong J, Su J, Li H, L S Inhibition of autophagy enhances cisplatin cytotoxicity through endoplasmic reticulum stress in human cervical cancer cells. Cancer Lett. 2012; 314(2):232–43. doi: 10.1016/j.canlet.2011.09.034. [DOI] [PubMed] [Google Scholar]
- 201.Cheng X, Feng H, Wu H, Jin Z, Shen X, Kuang J, Huo Z, Chen X, Gao H, Ye F, Ji X, Jing X, Zhang Y, Zhang T, Qiu W, R Z Targeting autophagy enhances apatinib-induced apoptosis via endoplasmic reticulum stress for human colorectal cancer. Cancer Lett. 2019; 431(156):105–14. doi: 10.1016/j.canlet.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 202.Brüning A, Rahmeh M, K F Nelfinavir and bortezomib inhibit mTOR activity via ATF4-mediated sestrin-2 regulation. Mol Oncol. 2013; 7(6):1012–8. doi: 10.1016/j.molonc.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mahoney E, Maddocks K, Flynn J, Jones J, Cole SL, Zhang X, Byrd JC, AJ J Identification of endoplasmic reticulum stress-inducing agents by antagonizing autophagy: a new potential strategy for identification of anti-cancer therapeutics in B-cell malignancies. Leuk Lymphoma. 2013; 54(12):2685–92. doi: 10.3109/10428194.2013.781168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Park GB, Jin DH, D K Sequential treatment with celecoxib and bortezomib enhances the ER stress-mediated autophagy-associated cell death of colon cancer cells. Oncol Lett. 2018; 16(4):4526–36. doi: 10.3892/ol.2018.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ji C, Zhang Z, Chen L, Zhou K, Li D, Wang P, Huang S, Gong T, B C Endoplasmic reticulum stress-induced autophagy determines the susceptibility of melanoma cells to dabrafenib. Drug Des Devel Ther. 2016; 10:2491–8. doi: 10.2147/DDDT.S112740. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 206.Ryabaya O, Prokofieva A, Khochenkov D, Abramov I, Zasedatelev A, E S Inhibition of endoplasmic reticulum stress-induced autophagy sensitizes melanoma cells to temozolomide treatment. Oncol Rep. 2018; 40(1):385–94. doi: 10.3892/or.2018.6430. [DOI] [PubMed] [Google Scholar]
- 207.He Y, Su J, Lan B, Gao Y, J Z Targeting off-target effects: endoplasmic reticulum stress and autophagy as effective strategies to enhance temozolomide treatment. Onco Targets Ther. 2019; 12:1857–65. doi: 10.2147/OTT.S194770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, H P Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013; 4:: e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bahar E, Kim JY, H Y Chemotherapy Resistance Explained through Endoplasmic Reticulum Stress-Dependent Signaling. Cancers (Basel). 2019; 11(3): pii: E338. doi: 10.3390/cancers11030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Battista RA, Resnati M, Facchi C, Ruggieri E, Cremasco F, Paradiso F, Orfanelli U, Giordano L, Bussi M, Cenci S, E M Autophagy mediates epithelial cancer chemoresistance by reducing p62/SQSTM1 accumulation. PLoS One. 2018; 13(8):: e0201621. doi: 10.1371/journal.pone.0201621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Amaravadi RK, Kimmelman AC, J D Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019; 9(9):1167–81. doi: 10.1158/2159-8290.CD-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]