Abstract
Membrane fusion mediates a number of fundamental biological processes such as intracellular membrane trafficking, fertilization, and viral infection. Biological membranes are composed of lipids and proteins; while lipids generally play a structural role, proteins mediate specific functions in the membrane. Likewise, although proteins are key players in the fusion of biological membranes, there is emerging evidence supporting a functional role of lipids in various membrane fusion events. Intracellular membrane fusion is mediated by two protein families: SNAREs and membrane-bound GTPases. SNARE proteins are involved in membrane fusion between transport vesicles and their target compartments, as well as in homotypic fusion between organelles of the same type. Membrane-bound GTPases mediate mitochondrial fusion and homotypic endoplasmic reticulum fusion. Certain membrane lipids, known as regulatory lipids, regulate these membrane fusion events by directly affecting the function of membrane-bound GTPases, instead of simply changing the biophysical and biochemical properties of lipid bilayers. In this review, we provide a summary of the current understanding of how regulatory lipids affect GTPase-mediated intracellular membrane fusion by focusing on the functions of regulatory lipids that directly affect fusogenic GTPases.
Keywords: regulatory lipid, membrane fusion, GTPase, astlastin, mitofusin, endoplasmic reticulum, mitochondria
Introduction
Membrane fusion is a vital step of a variety of fundamental processes in the cell and can be defined as a merger of two membrane-enclosed compartments into a single compartment. Membrane fusion is catalyzed by either a single protein or a series of proteins. Two types of fusogenic proteins are involved in most intracellular fusion events: SNAREs catalyze most of the membrane fusion events that occur during intracellular vesicle trafficking, while membrane-bound GTPases mediate the homotypic fusion of organelles such as the endoplasmic reticulum (ER) and mitochondria. These GTPases belong to a dynamin-like GTPase superfamily with conserved domain compositions and structures (Yan et al., 2015). The members of this family are mechanochemical GTPases that participate in the fusion and fission of membranes (Praefcke and McMahon, 2004). Here, we focus on dynamin-like fusogenic GTPases, including mitofusins (MFNs) and atlastins (ATLs), which share common features but act in different parts of the cell.
While proteins generally act as catalysts during membrane fusion, lipids have been long known to play a structural role. However, there is emerging evidence that lipids can also regulate membrane fusion events directly. These lipids, such as diacylglycerol, phosphatidic acid, phosphoinositides, and sterols, play more functional roles than structural roles during membrane fusion and thus are termed “regulatory lipids” (Fratti et al., 2004). The structures and physical properties of these regulatory lipids often differ from those of structural phospholipids; specifically, structural phospholipids take the form of cylinders with a typical phosphate head group and two acyl chains, while regulatory lipids display differential head group sizes and numbers of acyl chains and charges, resulting in different overall shapes of the lipids. In addition, regulatory lipids often contribute to the formation of microdomains on membranes, thereby affecting their physiochemical properties (Munro, 2003). These microdomains play an important role in membrane fusion by serving as fusion sites at which lipid rearrangement and bilayer mergers occur (Lang et al., 2008). Regulatory lipid-containing microdomains are believed to control membrane fusion mainly by changing the fluidity and curvature of the membrane, making it more prone to fusion (Zhukovsky et al., 2019). However, recent studies revealed that regulatory lipids also control membrane fusion by physically interacting with fusogenic proteins and thereby affecting their functions. There is indeed evidence for the direct involvement of regulatory lipids in GTPase-induced ER fusion and mitochondrial fusion through protein–lipid interactions. In this review, we describe current knowledge of the mechanisms by which certain regulatory lipids affect GTPase-induced intracellular membrane fusion.
Mitofusin Is Involved in Mitochondrial Outer-Membrane Fusion
Mitochondria play a vital role in cellular homeostasis and survival by functioning as the key player in cellular ATP production, apoptosis regulation, and cell aging. Mitochondria normally exist as elongated tubules in the cytoplasm, undergoing constant fusion and fission (Bereiter-Hahn and Voth, 1994; Sesaki and Jensen, 1999; Shaw and Nunnari, 2002). Maintenance of the normal mitochondrial morphology is critical for their function, and mitochondrial dysfunction is associated with neurodegenerative disorders such as Parkinson’s and Huntington’s diseases (Chen and Chan, 2009). Because mitochondria are enclosed by outer- and inner-membranes with distinct roles, the mechanism by which fusion and fission of these two membranes are coordinated is a long-standing question. Fusion of the mitochondrial outer-membrane is controlled by the dynamin-like GTPases MFN1 and MFN2 in mammals and Fzo1p in yeast (Hermann et al., 1998; Rapaport et al., 1998; Ishihara et al., 2004; Koshiba et al., 2004), whereas OPA1/Mgm1p controls fusion of the inner-membrane (Alexander et al., 2000; Delettre et al., 2000; Olichon et al., 2003; Wong et al., 2003). Although fusion of the outer- and inner-membranes are mechanistically distinct events (Meeusen et al., 2004), they are tightly inter-regulated (Cipolat et al., 2004). In yeast, Fzo1p and Mgm1p cooperate to coordinate outer-membrane fusion and inner-membrane fusion (Sesaki et al., 2003; Sesaki and Jensen, 2004; Coonrod et al., 2007), and these two events are thought to be synchronized by Ugo1p (Hermann et al., 1998; Wong et al., 2003; Sesaki and Jensen, 2004). However, the exact mechanism involved in this process is still largely unknown, and a mammalian orthologue of Ugo1p is yet to be identified.
The first factor identified as a regulator of mitochondrial morphology was fuzzy onions (fzo) in Drosophila (Hales and Fuller, 1997). The mammalian homologues of fzo, MFN1 and MFN2, are similar in structure to each other, but these proteins seem to play separate roles in mitochondrial fusion (Santel and Fuller, 2001). Overexpression of MFN2 suppresses MFN1-induced mitochondrial tubulation (Eura et al., 2003). MFNs consist of a large N-terminal GTPase domain followed by two heptad repeat (HR) domains. Although it is generally accepted that the HR domains are separated by two transmembrane domains, thus both face the cytoplasm (Rojo et al., 2002; Li et al., 2019), a different topology of MFNs was also suggested (Mattie et al., 2018). In a working model for MFN1-induced fusion, MFN1 proteins in the fusing membranes form a homodimer via their GTPase domains upon GTP hydrolysis (Cao et al., 2017; Yan et al., 2018). This homodimerization induces a drastic conformational change of MFN1, resulting in close apposition and the subsequent merger of the membranes (Yan et al., 2018). The HR domains of MFNs (HR1 and HR2) are structurally similar to the SNARE domain of SNARE proteins, well-characterized fusogens involved in intracellular vesicle fusion (Bonifacino and Glick, 2004). Structural studies revealed that the HR domains of MFNs which consist of repeats of seven amino acids, form amphiphilic helices that potentially interact with each other by building coiled-coil structures, similar to the formation of trans-SNARE complexes between apposed membranes (Koshiba et al., 2004; Daste et al., 2018). Notably, HR1 and HR2 play distinct roles as follows: the HR2 domain forms an antiparallel dimer with another HR2 domain on the opposing membrane, which mediates docking between the two membranes (Koshiba et al., 2004), whereas the amphiphilic property of the HR1 domain enables it to bind to the surface of the membrane and perturb its structure, thereby facilitating membrane fusion (Daste et al., 2018). Although this working model by which MFN1 mediates mitochondrial membrane fusion has been widely accepted, the exact mechanism by which the HR domains facilitate fusion remains largely unclear.
Phosphatidic Acid and Mitofusin-Mediated Fusion
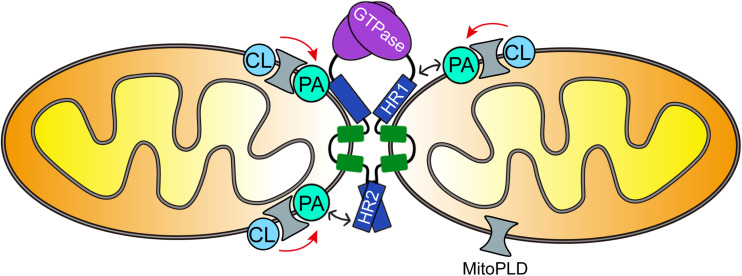
Phosphatidic acid (PA) constitutes approximately 5% of the mitochondrial membrane. PA has a relatively small head group and thus becomes a cone-shaped lipid that spontaneously induces negative membrane curvature when present in lipid bilayers (Kooijman et al., 2005). There are two ways through which PA is incorporated into the mitochondrial membrane: first, the majority of PA molecules are transferred from the ER to the mitochondrial outer-membrane, presumably through ER-mitochondrial contact sites, such as ERMES in yeasts (Murley and Nunnari, 2016; Petrungaro and Kornmann, 2019); second, a smaller number of PA molecules are generated in the mitochondrial membrane directly through enzymatic conversion of cardiolipin (CL) by mitochondrial phospholipase D (MitoPLD) (Choi et al., 2006). PA influences both fusion and fission of the mitochondrial outer-membrane, although its exact roles in these processes remain poorly characterized (Choi et al., 2006; Adachi et al., 2016). One plausible role of PA in membrane fusion is the introduction of negative curvature into the membrane, making its shape more favorable for fusion (Frohman, 2015). MitoPLD also seems to be important for mitochondrial outer-membrane fusion as follows: overexpression of MitoPLD aggregates mitochondria, indicating fusion of these structures, and RNAi-mediated knockdown of MitoPLD dramatically decreases mitochondrial fusion (Choi et al., 2006).
Although there is no direct evidence that PA physically interacts with MFN1 to mediate membrane fusion, overexpression of phospholipase A1, which converts PA to lysophosphatidic acid, triggers mitochondrial fragmentation, while its suppression induces elongation of mitochondria (Baba et al., 2014), suggesting that mitochondrial fusion and fission depend on the level of PA in the mitochondrial outer-membrane. Notably, PA interacts directly with the N-terminal amphipathic helix of the SNARE Spo20p, a yeast homologue of mammalian SNAP25, recruiting it to the site of fusion (Nakanishi et al., 2004; Horchani et al., 2014). Since the HR domains of MFN also contain 2 conserved amphipathic helices and bind to the lipid bilayer, it is possible that they also associate with PA directly to facilitate mitochondrial outer-membrane fusion (Figure 1; Cohen and Tareste, 2018). A direct interaction between PA and Ugo1p, a protein involved in the coordination of mitochondrial inner- and outer-membrane fusion, has been reported in yeast, and PA is required for the biosynthesis of Ugo1p (Vogtle et al., 2015). Thus, it can be speculated that PA promotes the generation of Ugo1p, thereby enriching Ugo1p at the fusion site where the yeast MFN Fzo1p is also recruited. Taken together, these studies suggest that PA can regulate MFN-induced mitochondrial outer-membrane fusion, although the exact mode of action remains yet to be clarified.
FIGURE 1.
Schematic model of mitochondrial outer-membrane fusion. Mitochondrial phospholipase D (MitoPLD, gray) converts cardiolipin (CL) to phosphatidic acid (PA) on the mitochondrial outer-membrane, thereby increasing the concentration of PA at the site of fusion. PA then interacts with the HR domains of mitofusins (blue) and recruits them to the fusion site to facilitate membrane fusion.
OPA1 Is Involved in Mitochondrial Inner-Membrane Fusion
OPA1 is a major regulator of mitochondrial inner-membrane fusion, and its genetic mutation is the main cause of optic atrophy (Alexander et al., 2000; Delettre et al., 2000). Deletion or mutation of the genes encoding OPA1 and its yeast orthologue Mgm1p results in abnormal mitochondrial morphology (Olichon et al., 2003; Wong et al., 2003). OPA1/Mgm1p belongs to the dynamin-like GTPase family and includes a GTPase domain in the middle section, a transmembrane domain at the N-terminus, and a membrane-binding domain, called a paddle domain, at the C-terminus (Faelber et al., 2019). Although encoded by a single gene, OPA1/Mgm1p exists in the following two forms: the long isoform L-OPA1/Mgm1p and the short isoform S-OPA1/Mgm1p. Short isoforms are produced by proteolytic cleavage (MacVicar and Langer, 2016) and lack the transmembrane domain, thereby existing as soluble proteins in the intermembrane space of mitochondria. Although both the short and long forms participate in inner-membrane fusion (Meeusen et al., 2006; DeVay et al., 2009; Zick et al., 2009), they seem to play distinct roles. The short form readily hydrolyzes GTP to initiate membrane tethering, and its drastic conformational change triggers membrane fusion (Zick et al., 2009; Faelber et al., 2019). By contrast, although the long form lacks GTPase activity, it associates with and activates the GTPase activity of the short form. Furthermore, the transmembrane domain of the long form is required for its precise targeting to the mitochondrial inner-membrane (DeVay et al., 2009). However, a recent study revealed that the long form of OPA1 is sufficient to drive liposome fusion in a GTP-dependent manner (Ban et al., 2017), indicating that it also plays a direct role in fusion. Thus, although both forms of OPA1/Mgm1 are required for mitochondrial inner-membrane fusion (DeVay et al., 2009; Ban et al., 2017; Ge et al., 2020), it is unclear how they cooperate to mediate this process.
Cardiolipin and OPA1-Mediated Fusion
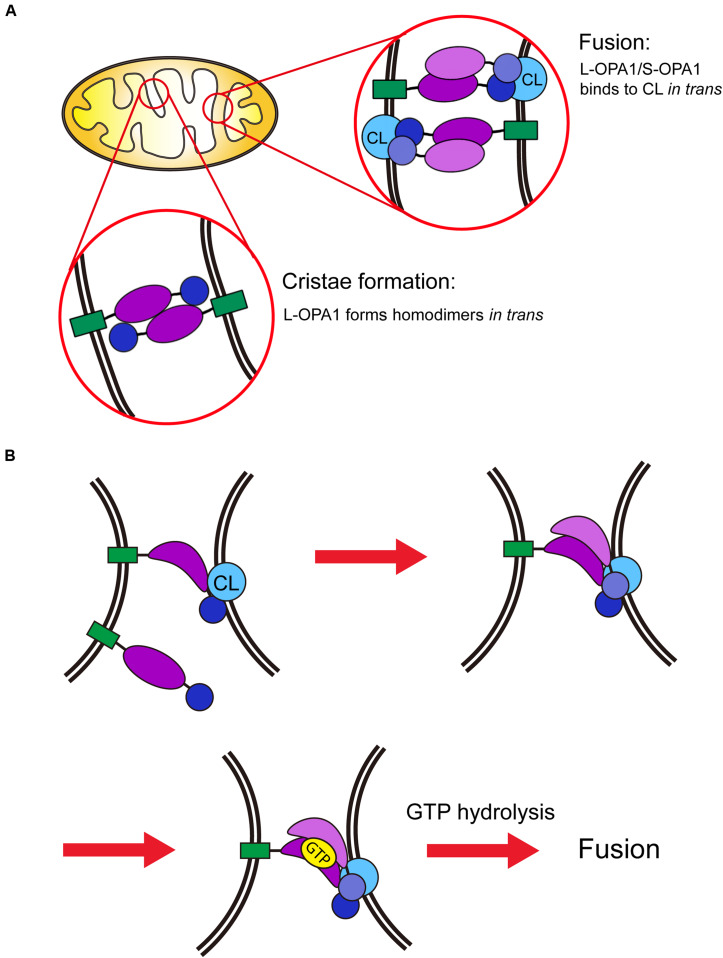
Cardiolipin is an important lipid that comprises approximately 25% of the inner-membrane and approximately 4% of the outer-membrane phospholipids (Ardail et al., 1990; Horvath and Daum, 2013). Unlike other phospholipids, CL has a unique chemical structure; it contains two phosphate head groups and four acyl chains, forming a symmetric structure. A number of reports have emphasized the importance of CL in mitochondrial inner-membrane fusion. For example, the inactivation of enzymes involved in CL synthesis generally causes morphological defects of mitochondria (Matsumura et al., 2018). In addition, CL regulates the mitochondrial morphology directly by facilitating the assembly of the dynamin-like GTPase OPA1/Mgm1p (DeVay et al., 2009; Rujiviphat et al., 2009; Joshi et al., 2012; Ban et al., 2017). Moreover, CL stimulates the GTPase activity of S-Mgm1p in a concentration-dependent manner, as evidenced by the finding that GTP hydrolysis by S-Mgm1p was higher in liposomes containing 20% CL than in liposomes containing 6% CL (DeVay et al., 2009). Similarly, enhanced GTP hydrolysis and S-OPA1 oligomerization were observed in the presence of CL (Ban et al., 2010). Compared with the short form of OPA1/Mgm1p, little is known about the long form, mainly because L-OPA1 is difficult to purify for biochemical studies. However, in a recent study, recombinant L-OPA1 was successfully purified from silk worm, and its function was assessed in vitro. Strikingly, this study reported that L-OPA1 was sufficient to drive fusion of liposomes containing 25% CL in a GTP-dependent manner. This fusion requires heterotypic interactions between L-OPA1 and CL in trans; specifically, L-OPA1 in a liposome binds to CL in another liposome (Ban et al., 2017; Ge et al., 2020). This result may explain why fusion was observed between mitochondria from OPA1-depleted cells and those from wild-type cells (Ban et al., 2017, 2018). Thus, CL may serve as a binding site for S/L-OPA1 heterodimers, thereby enabling these proteins to tether membranes and induce the subsequent fusion (Figure 2A).
FIGURE 2.
Schematic model of mitochondrial inner-membrane fusion. (A) GTP hydrolysis by OPA1/Mgm1p and the subsequent binding of OPA1/Mgm1p to CL are required for fusion of the mitochondrial inner-membrane. For cristae formation, the long form of OPA1/Mgm1p (L-OPA1/Mgm1p) forms a homodimer in trans. This process occurs independently of GTP hydrolysis. (B) L-OPA1/Mgm1p binds directly to CL in trans. This interaction induces the conformational change of L-OPA1/Mgm1p, allowing the short form of OPA1/Mgm1p (S-OPA1/Mgm1p) to associate with L-OPA1/Mgm1p. In turn, this interaction induces a conformational change of S-OPA1/Mgm1p to facilitate fusion.
L-OPA1 induces fusion only when it interacts with CL on the opposite membrane in trans. Therefore, it has been suggested that the CL-binding region of L-OPA1 is required for its recruitment to CL-enriched microdomains to facilitate fusion (Ban et al., 2017; Ge et al., 2020). Furthermore, a recent structural study of S-Mgm1p revealed that the CL-binding site lies on the GTPase domain, and its positively charged residues on the surface participate in electrostatic interactions between negatively charged lipids (Yan et al., 2020). Therefore, it is possible that the interaction of L-Mgm1p/OPA1 with CL induces a conformational change in the protein to facilitate the formation of S/L-OPA1/Mgm1p heterodimers. It is also possible that this interaction enhances the GTPase activity of Mgm1p/OPA, which then supports efficient fusion (Figure 2B).
A similar mode of action in promoting yeast vacuole fusion was observed for the Phox homology domain of the SNARE Vam7p. This domain binds phosphatidylinositol 3-phosphate (PI(3)P) on the vacuolar membrane, resulting in the accumulation of Vam7p at PI(3)P-rich regions, where it forms trans-SNARE complexes with other SNARE proteins to promote vacuole fusion (Cheever et al., 2001). In addition, the interaction between PI(3)P and the Phox homology domain of Vam7p is thought to cause a conformational change in Vam7p, which may enhance its interaction with other fusion components (Cheever et al., 2001; Miner et al., 2016).
Atlastin Is Involved in ER Fusion
The ER, a large but single organelle that spreads throughout the cytoplasm, is the major site of lipid synthesis, protein folding, and protein quality control (Baumann and Walz, 2001; Ellgaard and Helenius, 2003). Although enclosed by a single, continuous lipid bilayer, the ER exists in the following two distinct forms: a sheet like structure surrounding the nucleus and a tubular network dispersed throughout the cytoplasm (Voeltz et al., 2002). The tubular ER is a dynamic structure that constantly undergoes elongation, retraction, and fusion (Lee and Chen, 1988). The tubular structure of the ER seems to be important for its function because it enables distinct membrane contact sites with various organelles (Phillips and Voeltz, 2015). Maintenance of the proper morphology of the ER is thought to be important for normal cell physiology, and its disruption is often associated with neurological disorders such as hereditary spastic paraplegia (Namekawa et al., 2006; Salinas et al., 2008; Park et al., 2010).
Although the mechanism by which the tubular ER network is formed and maintained remains poorly understood, Yop1/DP1 and a class of proteins called reticulons are thought to play a critical role in generating the high membrane curvature required to form ER tubules (Voeltz et al., 2006; Hu et al., 2008). In addition, ATLs, which belong to the family of dynamin-like GTPases, are also thought to mediate the fusion of ER tubules (Orso et al., 2009) by forming three-way junctions of the tubules and thus generating the mesh-like structure of the ER. Drosophila ATL alone or yeast ATL (Sey1p) with either reticulon or DP1 is sufficient to recapitulate formation of the tubular ER network structure in vitro when reconstituted into synthetic liposomes (Powers et al., 2017). Furthermore, proteoliposomes reconstituted with purified Drosophila ATL, Sey1p, or the plant ATL Root Hair Defective 3 are able to fuse with each other, confirming that these proteins can function as genuine fusogens (Orso et al., 2009; Anwar et al., 2012; Zhang et al., 2013). However, human ATL1 is unable to induce liposome fusion, suggesting that additional proteins are required for ER membrane fusion in human cells (Wu et al., 2015). The fusogenic activities of the other human ATLs (ATL2 and ATL3) have not yet been investigated.
Atlastin family proteins contain a large N-terminal GTPase domain followed by three helical bundles, two transmembrane domains, and a short α-helix at the C-terminal end (Bian et al., 2011; Yan et al., 2015). The current model for ATL-induced membrane fusion is that upon GTP hydrolysis, the GTPase domain of ATL forms a homodimer with that of another ATL molecule on the apposed membrane, and their helix bundles then undergo dramatic conformational changes that bring the membranes into close proximity, which eventually induces the fusion of ER tubules (Bian et al., 2011; Yan et al., 2015; O’Donnell et al., 2017; Winsor et al., 2017). Although it is widely accepted that ATLs are sufficient to drive liposome fusion and are therefore the major fusogens for ER membrane fusion (Orso et al., 2009; Anwar et al., 2012; Zhang et al., 2013), a recent study using purified yeast ER microsomes suggested that additional factors are required for efficient ER fusion in vivo, at least in yeast (Lee et al., 2015). In this study, ER-resident SNAREs were critical for ER microsome fusion in vitro and for normal ER morphology in vivo. This finding is consistent with the observation that human ATL1 alone is insufficient to induce liposome fusion.
Cholesterol and Atlastin-Mediated Fusion
Cholesterol has a small hydrophilic head group and a bulky steroid backbone, and is a vital component of biological membranes. Accumulating evidence supports the importance of cholesterol in various fusion events, such as exocytosis (Wasser et al., 2007; Linetti et al., 2010) and viral fusion (Klug et al., 2017; Lee et al., 2017). Cholesterol is thought to participate in membrane fusion mainly by altering the biophysical properties of the membrane, such as the fluidity, thickness, curvature, and stability of lipid bilayers (Yang et al., 2016). In addition, cholesterol may also regulate membrane fusion by interacting directly with fusogenic proteins. Consistent with this idea, cholesterol promotes clustering of SNARE proteins at the site of fusion (Murray and Tamm, 2011; Enrich et al., 2015). Furthermore, some SNARE proteins contain cholesterol-binding motifs, such as CRAC [Cholesterol Recognition/interaction Amino acid Consensus sequence, (L/V)-X1–5-Y-X1–5-(K/R)] and CARC [an inverted CRAC motif, (K/R)-X1–5-(Y/F)-X1–5-(L/V)], in or near their transmembrane regions (Enrich et al., 2015), suggesting that cholesterol affects the function of SNAREs to facilitate membrane fusion by binding to them directly.
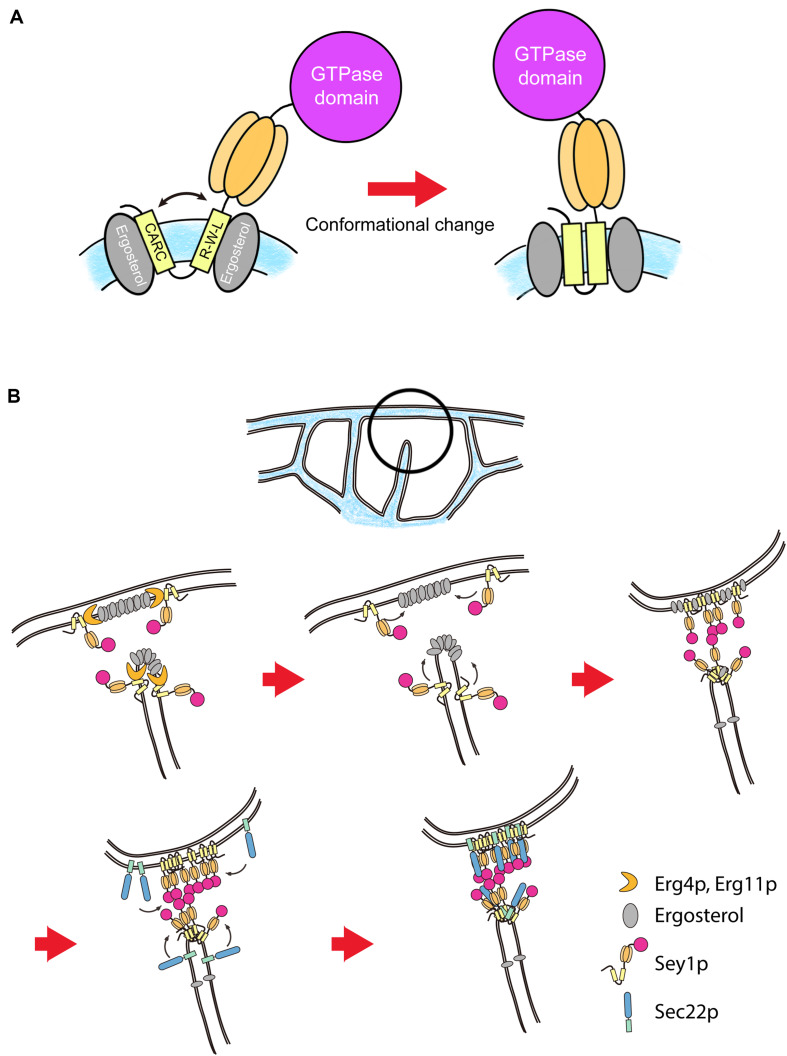
We recently revealed that ergosterol (yeast cholesterol) affects ER membrane fusion by interacting directly with Sey1p (Lee et al., 2019). The transmembrane domains of Sey1p contain two sterol-binding motifs, the R-W-L motif (a combination of basic [R], aromatic [W], and aliphatic [L/V] residues) and the CARC motif (Figure 3A). Furthermore, disruption of these sterol-binding motifs abolished the binding of sterols to Sey1p, severely reduced ER microsome fusion in vitro, and disrupted the normal ER morphology in vivo. Although the exact mechanism by which sterols stimulate Sey1p-medited ER fusion remains unclear, one possibility is that the interaction between the transmembrane domain of ATLs and cholesterol (or ergosterol in yeast) causes conformational changes of ATLs, making them more favorable for fusion. Consistent with this idea, mutant Sey1p lacking the sterol-binding motifs is unable to interact with Sec22p (Lee et al., 2019), an ER SNARE involved in Sey1p-dependent ER fusion (Lee et al., 2015), supporting the notion that the binding of cholesterol to Sey1p affects the overall conformation of the protein, resulting in modification of its fusogenic activity as well as of the profiles of its interacting proteins (Figure 3A). Notably, the transmembrane domain of the SNARE synaptobrevin-2 exists as two distinct forms, an open scissor form and a closed, parallel form, depending on the presence of cholesterol. This conformational transition modifies the fusogenic activity of the protein by changing the curvature of the surrounding membrane and possibly promotes complex formation with other SNAREs (Tong et al., 2009). Because ATLs contain two transmembrane domains, it is plausible that their conformations are affected by the presence of cholesterol similarly to that of the transmembrane domain of synatobrevin-2. Furthermore, because potential sterol-binding motifs are found in all human ATL proteins, regulation of ATL activity by direct binding of cholesterol is likely to be evolutionarily conserved.
FIGURE 3.
Schematic model of the role of ergosterol in Sey1p-mediated ER membrane fusion in yeast. (A) Sey1p interacts directly with ergosterol through its sterol-binding motifs, CARC and R-W-L. This interaction may promote transition of the transmembrane domains of Sey1p from an open, scissor-like configuration to a closed, parallel configuration. The conformational change may also increase the binding affinity of Sey1p for the ER SNARE Sec22p, recruiting more Sec22p proteins to the fusion site to enable efficient fusion. (B) Ergosterol is synthesized by a series of sterol biosynthetic enzymes, including Erg4p and Erg11p. Sey1p interacts with Erg4p/Erg11p and thus recruits them to the fusion site, increasing the local concentration of ergosterol. In turn, this process recruits more Sey1p and Sec22p proteins to the site of fusion, and this positive loop may greatly facilitate ER fusion.
We also found that Sey1p interacts physically with Erg4p and Erg11p, enzymes involved in the biosynthesis of ergosterol, which raises the possibility that Sey1p acts to increase the local concentration of ergosterol at the fusion site (Lee et al., 2019). In turn, this process not only stimulates the pre-existing Sey1p molecules for efficient fusion, but also recruits more Sey1p molecules and interacting proteins such as Sec22p to the site of fusion (Figure 3B). In support of this concept, ER subdomains containing Rab10, which reportedly mediates fusion between ER tubules in mammalian cells, are enriched in ER enzymes that regulate phospholipid synthesis, including phosphatidylinositol synthase and choline/ethanolamine phosphotransferase 1, which converts diacylglycerol precursors to phosphatidylethanolamine and phosphatidyl-choline (English and Voeltz, 2013).
In addition to the direct participation of cholesterol in ATL-mediated ER fusion, structural and biochemical studies of Drosophila ATL have suggested that a direct interaction of the C-terminal tail of ATL with lipid bilayers plays an important role in ER membrane fusion (Moss et al., 2011; Liu et al., 2012). In one of these studies, deletion of the short C-terminal tail of Drosophila ATL almost completely abolished the fusion of phosphatidylcholine:phosphatidyl-serine (PC:PS) proteoliposomes (Moss et al., 2011). The C-terminal tail of ATL is predicted to form an amphiphilic helix, which is very likely to be embedded into the lipid bilayer, thereby affecting the curvature and the stability of the membrane (Drin and Antonny, 2010). Indeed, the hydrophobic residues of the C-terminal tail of ATL interact directly with the hydrophobic side of the lipid bilayer (Liu et al., 2012). Similar observations were made for the plant ATL Root Hair Defective 3, which contains a conserved C-terminal tail that is required for ER targeting and efficient ER membrane fusion, implying that the C-terminal region is inserted into the lipid bilayer, as seen in Drosophila ATL-mediated fusion (Sun and Zheng, 2018). Although it is unclear how the C-terminal tail of ATL functions during ER membrane fusion, its insertion into the membrane may perturb the lipid bilayer, making it more prone to membrane fusion (Liu et al., 2012; Faust et al., 2015). However, it was reported that the necessity of the C-terminal tail of ATL for membrane fusion became less stringent when phosphatidylethanolamine (PE), a non-bilayer-prone lipid, was added to PC:PS proteoliposomes (Faust et al., 2015). This result suggests that although the C-terminal tail of ATL facilitates fusion, it is not essential for ER membrane fusion in vivo, as ER membranes contain significant amounts of non-bilayer-prone lipids such as phosphatidylethanolamine, cholesterol, and diacylglycerol (van Meer et al., 2008). In particular, Sey1p-mediated liposome fusion is highly susceptible to the omission of PE or ergosterol (Sugiura and Mima, 2016; Lee et al., 2019).
Discussion
This review describes the role of regulatory lipids in GTPase-mediated intracellular membrane fusion, focusing on examples of how these lipids affect proteins involved in membrane fusion processes. Some regulatory lipids facilitate membrane fusion by serving as an anchoring site for partner proteins and thus concentrating them at the site of membrane fusion, while others may bind directly to fusion proteins and modulate their fusogenic activity. Although lipids and proteins are both key players of membrane fusion, we have only just started to understand how their interactions control membrane fusion, and much remains to be clarified. A number of fusogenic proteins have potential lipid-binding domains or motifs; however, further studies are required to determine whether they indeed bind to lipids and how their interactions affect membrane fusion. In a recent report (Lee et al., 2019), we demonstrated that the yeast ATL Sey1p contains two sterol-binding motifs near its transmembrane domains. Disruption of these motifs severely abrogates Sey1p-mediated ER fusion, suggesting that the binding of sterols affects the fusogenic function of Sey1p. We also found that all three human ATL proteins contain two potential sterol-binding motifs. It would be interesting to investigate whether human ATLs associate directly with cholesterols, and whether this interaction influences their fusogenic activity. A study by Joji Mima’s laboratory showed that Sey1p-mediated liposome fusion is stimulated by other regulatory lipids, such as phosphatidylinositol and PA (Sugiura and Mima, 2016). It would therefore also be interesting to investigate how these lipids regulate Sey1p-mediated fusion. Compared with current knowledge of the role of regulatory lipids in ATL-mediated ER fusion, much less is known about how regulatory lipids control GTPase-mediated mitochondrial fusion. Recent advances in research tools for lipid studies and microscopy will guarantee a deeper and more comprehensive understanding of how regulatory lipids dictate GTPase-mediated intracellular membrane fusion events.
Author Contributions
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a grant from the Cell Logistics Research Center (NRF-2016R1A5A1007318) and a grant (NRF-2019R1A2C2004874) from the National Research Foundation of Korea. This work was also supported by a grant from the GIST Research Institute (GRI) at the Gwangju Institute of Science and Technology.
References
- Adachi Y., Itoh K., Yamada T., Cerveny K. L., Suzuki T. L., Macdonald P., et al. (2016). Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell. 63 1034–1043. 10.1016/j.molcel.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Votruba M., Pesch U. E., Thiselton D. L., Mayer S., Moore A., et al. (2000). OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26 211–215. 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- Anwar K., Klemm R. W., Condon A., Severin K. N., Zhang M., Ghirlando R., et al. (2012). The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 197 209–217. 10.1083/jcb.201111115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardail D., Privat J. P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. (1990). Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265 18797–18802. [PubMed] [Google Scholar]
- Baba T., Kashiwagi Y., Arimitsu N., Kogure T., Edo A., Maruyama T., et al. (2014). Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J. Biol. Chem. 289 11497–11511. 10.1074/jbc.m113.531921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban T., Heymann J. A., Song Z., Hinshaw J. E., Chan D. C. (2010). OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19 2113–2122. 10.1093/hmg/ddq088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban T., Ishihara T., Kohno H., Saita S., Ichimura A., Maenaka K., et al. (2017). Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 19 856–863. 10.1038/ncb3560 [DOI] [PubMed] [Google Scholar]
- Ban T., Kohno H., Ishihara T., Ishihara N. (2018). Relationship between OPA1 and cardiolipin in mitochondrial inner-membrane fusion. Biochim. Biophys. Acta Bioenerg. 1859 951–957. 10.1016/j.bbabio.2018.05.016 [DOI] [PubMed] [Google Scholar]
- Baumann O., Walz B. (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205 149–214. 10.1016/s0074-7696(01)05004-5 [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J., Voth M. (1994). Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27 198–219. 10.1002/jemt.1070270303 [DOI] [PubMed] [Google Scholar]
- Bian X., Klemm R. W., Liu T. Y., Zhang M., Sun S., Sui X., et al. (2011). Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. U.S.A. 108 3976–3981. 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116 153–166. 10.1016/s0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Cao Y. L., Meng S., Chen Y., Feng J. X., Gu D. D., Yu B., et al. (2017). MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542 372–376. 10.1038/nature21077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever M. L., Sato T. K., De Beer T., Kutateladze T. G., Emr S. D., Overduin M. (2001). Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3 613–618. 10.1038/35083000 [DOI] [PubMed] [Google Scholar]
- Chen H., Chan D. C. (2009). Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet. 18 R169–R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., Frohman M. A. (2006). A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8 1255–1262. 10.1038/ncb1487 [DOI] [PubMed] [Google Scholar]
- Cipolat S., Martins De Brito O., Dal Zilio B., Scorrano L. (2004). OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 101 15927–15932. 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. M., Tareste D. (2018). Recent insights into the structure and function of Mitofusins in mitochondrial fusion. F1000Res 7:F1000 Faculty Rev-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonrod E. M., Karren M. A., Shaw J. M. (2007). Ugo1p is a multipass transmembrane protein with a single carrier domain required for mitochondrial fusion. Traffic 8 500–511. 10.1111/j.1600-0854.2007.00550.x [DOI] [PubMed] [Google Scholar]
- Daste F., Sauvanet C., Bavdek A., Baye J., Pierre F., Le Borgne R., et al. (2018). The heptad repeat domain 1 of Mitofusin has membrane destabilization function in mitochondrial fusion. EMBO Rep 19:e43637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J. M., Gigarel N., Lorenzo C., Belenguer P., et al. (2000). Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26 207–210. 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- DeVay R. M., Dominguez-Ramirez L., Lackner L. L., Hoppins S., Stahlberg H., Nunnari J. (2009). Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186 793–803. 10.1083/jcb.200906098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Antonny B. (2010). Amphipathic helices and membrane curvature. FEBS Lett. 584 1840–1847. 10.1016/j.febslet.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4 181–191. [DOI] [PubMed] [Google Scholar]
- English A. R., Voeltz G. K. (2013). Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15 169–178. 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enrich C., Rentero C., Hierro A., Grewal T. (2015). Role of cholesterol in SNARE-mediated trafficking on intracellular membranes. J. Cell Sci. 128 1071–1081. 10.1242/jcs.164459 [DOI] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Yokota S., Mihara K. (2003). Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 134 333–344. 10.1093/jb/mvg150 [DOI] [PubMed] [Google Scholar]
- Faelber K., Dietrich L., Noel J. K., Wollweber F., Pfitzner A. K., Muhleip A., et al. (2019). Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 571 429–433. 10.1038/s41586-019-1372-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J. E., Desai T., Verma A., Ulengin I., Sun T. L., Moss T. J., et al. (2015). The Atlastin C-terminal tail is an amphipathic helix that perturbs the bilayer structure during endoplasmic reticulum homotypic fusion. J. Biol. Chem. 290 4772–4783. 10.1074/jbc.m114.601823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R. A., Jun Y., Merz A. J., Margolis N., Wickner W. (2004). Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 167 1087–1098. 10.1083/jcb.200409068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M. A. (2015). Role of mitochondrial lipids in guiding fission and fusion. J. Mol. Med. 93 263–269. 10.1007/s00109-014-1237-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Shi X., Boopathy S., Mcdonald J., Smith A. W., Chao L. H. (2020). Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. eLife 9:e50973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. G., Fuller M. T. (1997). Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90 121–129. 10.1016/s0092-8674(00)80319-0 [DOI] [PubMed] [Google Scholar]
- Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., et al. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143 359–373. 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horchani H., De Saint-Jean M., Barelli H., Antonny B. (2014). Interaction of the Spo20 membrane-sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PLoS One 9:e113484. 10.1371/journal.pone.0113484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. E., Daum G. (2013). Lipids of mitochondria. Prog. Lipid Res. 52 590–614. [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., et al. (2008). Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319 1247–1250. 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Eura Y., Mihara K. (2004). Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 117 6535–6546. 10.1242/jcs.01565 [DOI] [PubMed] [Google Scholar]
- Joshi A. S., Thompson M. N., Fei N., Huttemann M., Greenberg M. L. (2012). Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287 17589–17597. 10.1074/jbc.m111.330167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug Y. A., Rotem E., Schwarzer R., Shai Y. (2017). Mapping out the intricate relationship of the HIV envelope protein and the membrane environment. Biochim. Biophys. Acta Biomembr. 1859 550–560. 10.1016/j.bbamem.2016.10.012 [DOI] [PubMed] [Google Scholar]
- Kooijman E. E., Chupin V., Fuller N. L., Kozlov M. M., De Kruijff B., Burger K. N., et al. (2005). Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 44 2097–2102. 10.1021/bi0478502 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Detmer S. A., Kaiser J. T., Chen H., Mccaffery J. M., Chan D. C. (2004). Structural basis of mitochondrial tethering by mitofusin complexes. Science 305 858–862. 10.1126/science.1099793 [DOI] [PubMed] [Google Scholar]
- Lang T., Halemani N. D., Rammner B. (2008). Interplay between lipids and the proteinaceous membrane fusion machinery. Prog. Lipid Res. 47 461–469. 10.1016/j.plipres.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Lee C., Chen L. B. (1988). Dynamic behavior of endoplasmic reticulum in living cells. Cell 54 37–46. 10.1016/0092-8674(88)90177-8 [DOI] [PubMed] [Google Scholar]
- Lee J., Nyenhuis D. A., Nelson E. A., Cafiso D. S., White J. M., Tamm L. K. (2017). Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. U.S.A. 114 E7987–E7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Ko Y. J., Moon Y., Han M., Kim H. W., Lee S. H., et al. (2015). SNAREs support atlastin-mediated homotypic ER fusion in Saccharomyces cerevisiae. J. Cell Biol. 210 451–470. 10.1083/jcb.201501043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Moon Y., Lee S., Lee C., Jun Y. (2019). Ergosterol interacts with Sey1p to promote atlastin-mediated endoplasmic reticulum membrane fusion in Saccharomyces cerevisiae. FASEB J. 33 3590–3600. 10.1096/fj.201800779rr [DOI] [PubMed] [Google Scholar]
- Li Y. J., Cao Y. L., Feng J. X., Qi Y., Meng S., Yang J. F., et al. (2019). Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset. Nat. Commun. 10:4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linetti A., Fratangeli A., Taverna E., Valnegri P., Francolini M., Cappello V., et al. (2010). Cholesterol reduction impairs exocytosis of synaptic vesicles. J. Cell Sci. 123 595–605. 10.1242/jcs.060681 [DOI] [PubMed] [Google Scholar]
- Liu T. Y., Bian X., Sun S., Hu X., Klemm R. W., Prinz W. A., et al. (2012). Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. U.S.A. 109 E2146–E2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar T., Langer T. (2016). OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci. 129 2297–2306. 10.1242/jcs.159186 [DOI] [PubMed] [Google Scholar]
- Matsumura A., Higuchi J., Watanabe Y., Kato M., Aoki K., Akabane S., et al. (2018). Inactivation of cardiolipin synthase triggers changes in mitochondrial morphology. FEBS Lett. 592 209–218. 10.1002/1873-3468.12948 [DOI] [PubMed] [Google Scholar]
- Mattie S., Riemer J., Wideman J. G., Mcbride H. M. (2018). A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 217 507–515. 10.1083/jcb.201611194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., Devay R., Block J., Cassidy-Stone A., Wayson S., Mccaffery J. M., et al. (2006). Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127 383–395. 10.1016/j.cell.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Meeusen S., Mccaffery J. M., Nunnari J. (2004). Mitochondrial fusion intermediates revealed in vitro. Science 305 1747–1752. 10.1126/science.1100612 [DOI] [PubMed] [Google Scholar]
- Miner G. E., Starr M. L., Hurst L. R., Sparks R. P., Padolina M., Fratti R. A. (2016). The central polybasic region of the soluble SNARE (Soluble N-Ethylmaleimide-sensitive Factor Attachment Protein Receptor) Vam7 affects binding to Phosphatidylinositol 3-Phosphate by the PX (Phox Homology) Domain. J. Biol. Chem. 291 17651–17663. 10.1074/jbc.m116.725366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. J., Andreazza C., Verma A., Daga A., Mcnew J. A. (2011). Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc. Natl. Acad. Sci. U.S.A. 108 11133–11138. 10.1073/pnas.1105056108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. (2003). Lipid rafts: elusive or illusive? Cell 115 377–388. [DOI] [PubMed] [Google Scholar]
- Murley A., Nunnari J. (2016). The emerging network of mitochondria-organelle contacts. Mol. Cell. 61 648–653. 10.1016/j.molcel.2016.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D. H., Tamm L. K. (2011). Molecular mechanism of cholesterol- and polyphosphoinositide-mediated syntaxin clustering. Biochemistry 50 9014–9022. 10.1021/bi201307u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., De Los Santos P., Neiman A. M. (2004). Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15 1802–1815. 10.1091/mbc.e03-11-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa M., Ribai P., Nelson I., Forlani S., Fellmann F., Goizet C., et al. (2006). SPG3A is the most frequent cause of hereditary spastic paraplegia with onset before age 10 years. Neurology 66 112–114. 10.1212/01.wnl.0000191390.20564.8e [DOI] [PubMed] [Google Scholar]
- O’Donnell J. P., Cooley R. B., Kelly C. M., Miller K., Andersen O. S., Rusinova R., et al. (2017). Timing and reset mechanism of GTP Hydrolysis-Driven conformational changes of Atlastin. Structure 25 997–1010.e4. 10.1016/j.str.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., et al. (2003). Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278 7743–7746. 10.1074/jbc.c200677200 [DOI] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T. J., Faust J. E., et al. (2009). Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460 978–983. 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Park S. H., Zhu P. P., Parker R. L., Blackstone C. (2010). Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 120 1097–1110. 10.1172/jci40979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrungaro C., Kornmann B. (2019). Lipid exchange at ER-mitochondria contact sites: a puzzle falling into place with quite a few pieces missing. Curr. Opin. Cell Biol. 57 71–76. 10.1016/j.ceb.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Phillips M. J., Voeltz G. K. (2015). Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17:69. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers R. E., Wang S., Liu T. Y., Rapoport T. A. (2017). Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543 257–260. 10.1038/nature21387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G. J., McMahon H. T. (2004). The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5 133–147. 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. (1998). Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273 20150–20155. 10.1074/jbc.273.32.20150 [DOI] [PubMed] [Google Scholar]
- Rojo M., Legros F., Chateau D., Lombes A. (2002). Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115 1663–1674. [DOI] [PubMed] [Google Scholar]
- Rujiviphat J., Meglei G., Rubinstein J. L., Mcquibban G. A. (2009). Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284 28682–28686. 10.1074/jbc.m109.044933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas S., Proukakis C., Crosby A., Warner T. T. (2008). Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 7 1127–1138. 10.1016/s1474-4422(08)70258-8 [DOI] [PubMed] [Google Scholar]
- Santel A., Fuller M. T. (2001). Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114 867–874. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147 699–706. 10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. (2004). Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J. Biol. Chem. 279 28298–28303. 10.1074/jbc.m401363200 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S. M., Yaffe M. P., Jensen R. E. (2003). Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell 14 2342–2356. 10.1091/mbc.e02-12-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. M., Nunnari J. (2002). Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12 178–184. 10.1016/s0962-8924(01)02246-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura S., Mima J. (2016). Physiological lipid composition is vital for homotypic ER membrane fusion mediated by the dynamin-related GTPase Sey1p. Sci. Rep. 6:20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zheng H. (2018). Efficient ER Fusion Requires a Dimerization and a C-terminal tail mediated membrane anchoring of RHD3. Plant Physiol. 176 406–417. 10.1104/pp.17.01411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Borbat P. P., Freed J. H., Shin Y. K. (2009). A scissors mechanism for stimulation of SNARE-mediated lipid mixing by cholesterol. Proc. Natl. Acad. Sci. U.S.A. 106 5141–5146. 10.1073/pnas.0813138106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Voelker D. R., Feigenson G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9 112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124 573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Voeltz G. K., Rolls M. M., Rapoport T. A. (2002). Structural organization of the endoplasmic reticulum. EMBO Rep. 3 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtle F. N., Keller M., Taskin A. A., Horvath S. E., Guan X. L., Prinz C., et al. (2015). The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J. Cell Biol. 210 951–960. 10.1083/jcb.201506085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasser C. R., Ertunc M., Liu X., Kavalali E. T. (2007). Cholesterol-dependent balance between evoked and spontaneous synaptic vesicle recycling. J. Physiol. 579 413–429. 10.1113/jphysiol.2006.123133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor J., Hackney D. D., Lee T. H. (2017). The crossover conformational shift of the GTPase atlastin provides the energy driving ER fusion. J. Cell Biol. 216 1321–1335. 10.1083/jcb.201609071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. D., Wagner J. A., Scott S. V., Okreglak V., Holewinske T. J., Cassidy-Stone A., et al. (2003). The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160 303–311. 10.1083/jcb.200209015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Hu X., Bian X., Liu X., Hu J. (2015). Comparison of human and Drosophila atlastin GTPases. Protein Cell 6 139–146. 10.1007/s13238-014-0118-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Qi Y., Huang X., Yu C., Lan L., Guo X., et al. (2018). Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion. Nat. Struct. Mol. Biol. 25 233–243. 10.1038/s41594-018-0034-8 [DOI] [PubMed] [Google Scholar]
- Yan L., Qi Y., Ricketson D., Li L., Subramanian K., Zhao J., et al. (2020). Structural analysis of a trimeric assembly of the mitochondrial dynamin-like GTPase Mgm1. Proc. Natl. Acad. Sci. U.S.A. 117:201919116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Sun S., Wang W., Shi J., Hu X., Wang S., et al. (2015). Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion. J. Cell Biol. 210 961–972. 10.1083/jcb.201502078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. T., Kreutzberger A. J. B., Lee J., Kiessling V., Tamm L. K. (2016). The role of cholesterol in membrane fusion. Chem. Phys. Lipids 199 136–143. 10.1016/j.chemphyslip.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wu F., Shi J., Zhu Y., Zhu Z., Gong Q., et al. (2013). ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 163 713–720. 10.1104/pp.113.224501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovsky M. A., Filograna A., Luini A., Corda D., Valente C. (2019). Phosphatidic acid in membrane rearrangements. FEBS Lett. 593 2428–2451. 10.1002/1873-3468.13563 [DOI] [PubMed] [Google Scholar]
- Zick M., Duvezin-Caubet S., Schafer A., Vogel F., Neupert W., Reichert A. S. (2009). Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 583 2237–2243. 10.1016/j.febslet.2009.05.053 [DOI] [PubMed] [Google Scholar]