Abstract
Infectious bronchitis virus (IBV), a gamma-coronavirus, causes infectious bronchitis (IB), a major respiratory disease of chicken. Its high mutation rate in conjunction with recombination of the RNA genome constantly creates IBV variants that are difficult to control by currently available vaccines. In this study, we addressed the question whether small-scale holdings might harbor IBV variants that serve as a reservoir for newly emerging variants. Egyptian IBV isolate EGY/NR725/2016 (NR725/16) from a small-scale broiler farm was assigned to genotype I, clade 23 (S1:GI-23), based on partial S1 gene sequences and corroborated by full genome sequencing. Analysis of the S1 gene established three subclades for historical IBV strains (S1:GI-23.1, S1:GI-23.2.1 and S1:GI-23.2.2) and confirmed NR725/16 as being part of a separate fourth subclade (S1:GI-23.3). Samples from the years 2018 and 2019 revealed that the new subclade prevails in Egypt, carrying fixed mutations within the hypervariable regions (HVR) 1–3 of the S1 protein that affect two neutralization sensitive epitopes at sites 294F, 297S and 306Y (48.2) and 329R (62.1). In addition, recombination was recognized in isolate NR 725/16, with intra-subtype mixing for the entire genes 3ab and E and inter-subtype mixing for the entire gene 6b with a close match to QX like viruses of genotype GI-19. Further analysis of gene 3ab detected the homologous gene pool to NR725/16 in samples from 2013 (3ab:C) and closely related 3ab genotypes in IBV Egyptian isolates from 2016, 2018 and 2019. These data prove a flourishing exchange between poultry holdings with a common gene pool. The continued circulation of viruses harboring genes S1:GI-23.3 and 3ab:C indicates an evolutionary advantage of this combination possibly by combining antigenic escape with modulated pathogenicity to facilitate IBV spread in the vaccinated poultry population in Egypt.
Keywords: Infectious bronchitis, Coronavirus, Chicken, Recombination
1. Introduction
Avian infectious bronchitis (IB) is an economically important infectious disease caused by infectious bronchitis virus (IBV), with clinical manifestations in the upper respiratory tract, oviduct and kidneys. Infection is highly contagious with morbidity reaching 100%, but mortality can vary between 0% and 80% and depends on the age and immune status of the birds and the circulating virus strain (Jackwood and de Wit, 2013). Clinical manifestation of IBV infection in Egypt is mainly observed in broilers and dominated by respiratory distress like gasping, coughing, sneezing or tracheal rales. Alterations in the respiratory tract are dominated by accumulation of excessive amounts of mucus in the trachea and thickened, turbid air sacs. Coinfections with bacterial or viral pathogens may lead to complicated chronic respiratory disease (CCRD) with morbidity and mortality rates of up to 50%. In particular, nephropathogenic IBV strains induce acute to subacute nephritis with polyuria evident by watery droppings. Pathological findings are dominated by congested, swollen kidneys with paleness in some cases and deposition of urates (Abdel-Moneim et al., 2012, Abd El Rahman et al., 2015, Sultan et al., 2015, Zanaty et al., 2016a).
IBV is an enveloped virus of about 120 nm in diameter and classified as gammacoronavirus, subgenus Igacovirus within the family of Coronaviridae (King et al., 2018). The genome consists of a single-stranded positive-sense RNA of approximately 27.6 kb. Indispensable for RNA replication and transcription is gene 1, with two open reading frames, the products of which provide RNA dependent RNA polymerase functions (Cavanagh, 2007, Masters and Perlman, 2013). Intrinsic to coronaviruses is the limited proof reading capacity of this viral polymerase which results in a high mutation rate with an estimated average rate of synonymous mutations of approximately 1.2 × 10–3 substitutions/site/year (Hanada et al., 2004, Holmes, 2009). Besides, RNA recombination is recognized as an important factor for viral evolution of IBV and coronaviruses in general (Thor et al., 2011, Jackwood et al., 2012). Even though the specific mechanisms are yet unknown, generation of a set of subgenomic plus-strand RNAs during replication is considered to facilitate homologous recombination among closely related genes from different lineages by template switching (Masters and Perlman, 2013). Beside gene 1, IBV comprises four genes coding for nonstructural proteins (3, 4, 5 and 6b) and four genes encoding structural proteins, i.e. the spike protein (S), integral membrane glycoprotein (M), membrane associated envelope protein (E) and nucleoprotein (N) (Cavanagh, 2007).
In terms of protective immune response against IBV, the S protein is of uppermost importance as it facilitates attachment and entry into the host cell: Cleaved into two subunits, the amino-terminal S1 is responsible for attachment of the virus to cells, whilst carboxy-terminal S2, anchored in the membrane, is responsible for membrane fusion. Accordingly, S1 has been reported to be the most important protein in terms of antigenic differentiation (Kusters et al., 1989, Thor et al., 2011, Jackwood et al., 2012), harboring at least five neutralization sites (S1-A to S1-E) (Kant et al., 1992). By means of cross-neutralization using polyclonal sera, serotypes can be distinguished with the Massachusetts and Connecticut serotypes as the best known standard strains (Cook, 1984, Toro et al., 1987). However, due to the intrinsic high mutation rate of IBV in conjunction with selection pressure generated by the immune response, the S1 subunit of S protein is highly variable and numerous antigenic variants are circulating. (Mockett et al., 1984, Minskaia et al., 2006, De Wit et al., 2011, Jackwood et al., 2012, Jackwood and Lee, 2017). In consequence, serotyping of IBV is difficult to standardize and has been replaced by genotyping, with particular focus on the S1 gene (Jackwood and de Wit, 2013). Recognition of three clusters of amino acid (aa) substitutions within the S1 gene, designated as hypervariable regions (HVR) 1–3 spanning in between amino acid residues 38–67, 91–141 and 274–387, respectively, draws attention of analysis to these sites (Niesters et al., 1986, Cavanagh et al., 1988, Kant et al., 1992, Moore et al., 1997). In an attempt to harmonize IBV classification, sequence information of the complete S1 gene fragment was used in order to distinguish six IBV genotypes (I–VI) that comprise altogether 32 lineages. Genotype S1:GI-1 includes the Massachusetts serotype vaccine strains like Mass/Mass41/41, Mass/H120/55 or Ma 5, whereas vaccine strain D274 belongs to genotype S1:GI-12. The attenuated vaccine strains CR88 and 4/91, also known as 793B type, belong to genotype S1:GI-13. So-called QX IBV strains segregate into genotype S1:GI-19, while genotype S1:GI-23 is represented by Israeli variant 2 and Egyptian variant 1 and 2 respectively (Valastro et al., 2016).
The latter, genotype S1:GI-23, became most important for North Africa and the Middle East. First reported in Egypt in 2001 and subsequently called Egyptian variant 1 (EGY-var1) (Abdel-Moneim et al., 2002), this genotype became endemic in Egypt (El-Mahdy et al., 2010), and subsequently a new cluster within genotype S1:GI-23 emerged, that was labeled Egyptian variant 2 (EGY-var2) (Abdel-Moneim et al., 2012). Beside the dominant genotype S1:GI-23 strains, other IBV strains like Mass and 793B were still circulating (Selim et al., 2013, Sultan et al., 2015, Rohaim et al., 2019a). In line with the co-circulation of different IBV genotypes, recombination events were detected within genotype S1:GI-23 strains (Zanaty et al., 2016b) as well aswith S1:GI-19, S1:GI-13 or S1:GI-1 strains (Abozeid et al., 2017). In addition, intra-genotypic recombinant S1:GI-23 IBV strains were identified not only in poultry but were recovered also from wild bird species, like house sparrow (Passer domesticus), quail (Coturnix coturnix), teal (Anas crecca) and cattle egret (Bubulcus ibis) (Rohaim et al., 2019b), indicating a flourishing exchange of IBV within and between poultry and wild birds.
In this respect, small-scale holdings with limited biosecurity measures might be an important reservoir where new IBV variants emerge. To address this question, we were interested in characterizing IBV strains circulating in small-scale farms in Egypt, identified in a former study in which IBV was detected in 3 out of 16 farms from sector 3, i.e. commercial poultry production systems with low to minimal biosecurity (FAO, 2004) farms (Moharam et al, 2019). From the same study, five IBV positive samples were derived from sector 2 farms, i.e. commercial poultry production system with moderate to high biosecurity. Poultry in all farms suffered from respiratory distress with mortality rates of 10 to 70%.
2. Material and methods
2.1. Samples
IBV positive tracheal samples from a previous study were derived from chicken flocks with respiratory distress from sector 3 (n = 3) and sector 2 (n = 5) and were described in detail elsewhere (Moharam et al., 2019). These samples were collected during the year 2016. For comparison, 15 historical Egyptian IBV isolates from year 2012/2013 IBV (Sultan et al., 2015) were included as well as recent IBV positive samples form large-scale production enterprises: three samples from 2018 (Hassan et al., 2019) and three samples from 2019. Details of all used IBV isolates and samples are given in Table 1 ..
Table 1.
IBV positive samples
| NR | Gov | Type | sample | Date | Flock size 1 | Grouping by specific gene /segments | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3ab | 3a | 3b | HVR1-2 | HVR3 3 | S1 2 | S1 | ||||||
| NR 715/16 | Monufia | Broiler | Trachea | 3\2016 | 20000 | C | B/C | C | nd | GI-23.2 | EGY-var2 | |
| NR 718/16 | Giza | Broiler | Trachea | 5\2016 | 25000 | C | B/C | C | nd | GI-23.2 | EGY-var2 | |
| NR 719/16 | Giza | Broiler | Trachea | 5\2016 | 15000 | C | B/C | C | nd | GI-23.2 | EGY-var2 | |
| NR 725/16* | Monufia | Broiler | Trachea | 4\2016 | 300 | B | B/C | B | GI-23.3 | GI-23.3 | outgroup | GI-23.3 |
| NR 730/16 | Behiera | Layer | Trachea | 7\2016 | 1000 | C | B/C | C | nd | GI-23.2 | EGY-var2 | |
| SCU-1/2013 | Monufia | Broiler | AAF | 2\2013 | 20000 | B | B/C | B | nd | Vac | ||
| SCU-2/2013 | Cairo | Broiler | AAF | 10\2012 | 10000 | B | B/C | B | nd | GI-23.2 | EGY-var2 | |
| SCU-3/2013 | Giza | Broiler | AAF | 5\2012 | 10000 | B | B/C | B | nd | GI-23.2 | EGY-var2 | |
| SCU-4/2013 | Cairo | Broiler | AAF | 7\2012 | 14000 | A | A | A | nd | GI-23.2 | EGY-var2 | |
| SCU-6/2013 | Behera | Broiler | AAF | 11\2012 | 60000 | A | A | A | nd | GI-23.2 | EGY-var2 | |
| SCU-7/2013 | Behera | Broiler | AAF | 11\2012 | 70000 | A | A | A | nd | GI-23.2 | EGY-var2 | |
| SCU-11/2013 | Cairo | Broiler | AAF | 7\2012 | 10000 | A | A | A | nd | GI-23.2 | EGY-var2 | |
| SCU-12/2013 | Gharbia | Broiler | AAF | 5\2012 | 12000 | B | B/C | B | nd | GI-23.2 | EGY-var2 | |
| SCU-13/2013 | Gharbia | Broiler | AAF | 5\2012 | 50000 | B | B/C | B | nd | GI-23.2 | EGY-var2 | |
| SCU-14/2013 | Giza | Broiler | AAF | 11\2012 | 20000 | A | A | A | nd | GI-23.2 | EGY-var2 | |
| USC-2/2013 | Gharbia | Broiler | AAF | 5\2012 | 15000 | B | B/C | B | nd | GI-23.2 | EGY-var2 | |
| USC-3/2013 | Giza | Broiler | AAF | 2\2012 | 20000 | Vac | Vac | B | nd | GI-23.2 | EGY-var2 | |
| USC-4/2013 | Monufia | Broiler | AAF | 11\2012 | 15000 | A | A | A | nd | GI-23.2 | EGY-var2 | |
| USC-5/2013* | Monufia | Broiler | AAF | 11\2012 | 80000 | A | A | A | GI-23.1 | GI-23.1 | EGY-var2 | GI-23.2.2 |
| USC-6/2013 | Monufia | Broiler | AAF | 11\2012 | 80000 | Vac | Vac | B | nd | Vac | ||
| AR 545/18 | Qualiobia | Broiler | Trachea | 05/2018 | 5000 | C | B/C | C | outgroup | GI-23.3 | nd | |
| AR 563/18 | Giza | Broiler | Trachea | 04/2018 | 9000 | C | noSeq | C | outgroup | noSeq | nd | |
| AR 593/18 | Fayoum | Broiler | Trachea | 05/2018 | 7000 | C | B/C | C | outgroup | GI-23.3 | nd | |
| AR 90/19 | Beni-Suef | Broiler | Trachea | 03/2019 | 5000 | C | B/C | C | outgroup | noSeq | nd | |
| AR 93/19 | Beni-Suef | Broiler | Trachea | 04/2019 | 3500 | C | B/C | C | outgroup | noSeq | nd | |
| AR 97/19 | Beni-Suef | Broiler | Trachea | 05/2019 | 8000 | C | B/C | C | noSeq | noSeq | nd | |
| Pol/G052/2016 | B | B/C | B | GI-23.1 | GI-23.1 | EGY-var1 | GI-23.1 | |||||
| ck/ISR/1494/2006 | (B/C)) | B/C | A | GI-23.1 | GI-23.1 | EGY-var1 | GI-23.1 | |||||
2.2. Screening of samples for IBV, NDV and AIV by qRT- PCR
Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), and RNA extracts were screened by real time RT-qPCR for different respiratory pathogens: Newcastle disease virus (NDV), lentogenic and velogenic (Wise et al., 2004, Moharam et al., 2019); AIV H5N1 (Naguib et al., 2017b), H9N2 (Monne et al., 2008), and IBV (Naguib et al., 2017a). All reactions were performed using the QIAGEN One-Step RT-qPCR kit (Qiagen, Hilden, Germany), and the reactions were carried out in a BioRad CFX96 Real-Time cycler (BioRad, Hercules, CA, USA). Results with a Ct value ≤ 38 were considered positive.
2.3. Virus propagation
Isolation attempts and IBV propagation were carried out in specific pathogen free (SPF) embryonated chicken eggs (ECE) by the allantoic route, following routine procedures (EU/92/66/EEC, 1992) as described earlier (Moharam et al., 2019). Up to three serial passages were conducted for each specimen.
2.4. Purification of IBV from co-infected samples
Initially, isolate (USC-5/2013) was recognized to be co-infected with NDV that could be eliminated by pretreatment with NDV-specific hyper-immune serum. Shortly: Virus isolate USC-5/2013 was diluted 1:1000 in MEM cell culture medium and subsequently mixed with an equal volume of 1:4 pre-diluted NDV specific antiserum (HI-titer: 512) in MEM. After incubation for one hour at 37 °C, the mixture was inoculated into SPF eggs as described above. Three egg passages were performed with antibody pretreatment, followed by two egg passages without. All passages were tested for NDV by HA and for both IBV and NDV by qRT-PCR.
2.5. Sanger sequencing
For initial genotyping of IBV viruses, sequencing for IBV HVR 1, 2, and 3 of the S1 gene was done using primer sets and reaction conditions as described by (Adzhar et al., 1997, Naguib et al., 2017a) by direct amplification of the RNA extracted from the sample homogenate. Primer sensitivity was tested using RNA stock from IBV isolate NR725/16 with an initial Ct value of 13 that could be diluted × 10–5. For sequencing of gene 3ab gene a specific primer set was generated to amplify gene 3ab from diagnostic samples consisting of two forward primers (IBV/3ab/FW1: CAA TAC AGA CCT AAA AAG TCT G, IBV/3ab/FW2: TAT TAA GTG GCC TTG GTA TGT) and one reverse primer (IBV/3ab/RV: CCA CTA CCC ATG TRT ACC A). The SuperScript®III One-Step RT-PCR kit (Invitrogen, Carlsbad, CA, USA) was used for amplification with the following cycling conditions: RT step 45 °C for 10 min, followed by an initial denaturation step at 95 °C for 10 min. PCR amplification was done for 35 cycles each with 95 °C:15 s, 55 °C:15 s, and 68 °C:30 s with a final extension step of 68 °C for 5 min. Agarose gel size separated PCR products were excised and purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The product was analyzed by nucleotide sequencing with the BigDye Terminator Cycle Sequencing Kit 1.1 (PE Applied Biosystems, Weiterstadt, Germany).
2.6. Next generation sequencing
Aamnio allantoic fluid (AAF) from the third virus passage (150 µl) was added to 750 µl TRIzol® (Invitrogen, Carlsbad, CA, USA) for RNA extraction and further genome analysis by Ilumina platform next generation sequencing (NGS). All the sequencing and data analysis procedures were done as described by (Wylezich et al., 2018).
2.7. Phylogenetic analysis of IBV
From the obtained partial and full genome, S1 gene sequences were aligned using ClustalW and analyzed in MEGA 7 (Kumar et al., 2016) with sequences representing all IBV genotypes and all sequences related to genotype 23 as extracted from the NCBI Genbank database. Phylogenetic analysis, pairwise distance analysis as well as between group distance analysis were performed by maximum likelihood analysis (1,000 bootstrap replicates) and the Tamura-Nei model (Tamura and Nei, 1993) as the most appropriate model. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology by superior log likelihood values. A discrete Gamma distribution was used to model evolutionary rate differences among sites (4 categories (+G, parameter = 1.1333). The rate variation model allowed for some sites to be invariable ([+I], 22.6138% sites).
Similarly, the relationship of other NR725/16 genes was compared to USC-5/2013 and other related viral sequences derived from GenBank. Therefore, sequences were aligned using ClustalW and pairwise distances were calculated for each gene. The distances were then used to generate a heatmap using the function “pheatmap” as implemented in R (version 3.6.1) package “pheatmap” using R Studio (version 1.2.5019).
For further differentiation within clade 23 of genotype I, we applied the criteria used for genotyping of NDV (Dimitrov et al., 2019): nucleotide distance between the sub-clades had to be above 5% and the bootstrap support value for the nodes had to be 70% or above after at least 1000 bootstrap replicates. Throughout the figures virus names include the genotype according to S1 gene, followed by accession number, species the virus was detected from, with abbreviation for chicken (ck), country code according ISO 3166-1 alpha-3, laboratory identification number and year of publication.
2.8. Recombination analysis
Bootscan analysis included in the Recombination Detection software package RDP4 (Martin et al., 2015) was used to identify recombination events implicating NR725/16 against three viruses (USC-5/2013, POL/G052/16 and CH/LJS/110439). The selection of these viruses was suggested by phylogenic analysis and analysis of pairwise distances for different genes. Bootscan analysis was performed on the basis of pairwise distances, modelled with a window size of 200 nt, step size of 50 nt, and 100 bootstrap replicates as parameters.
3. Results
3.1. Recent Egyptian IBV viruses from sector 2 and 3 segregated into GI-23
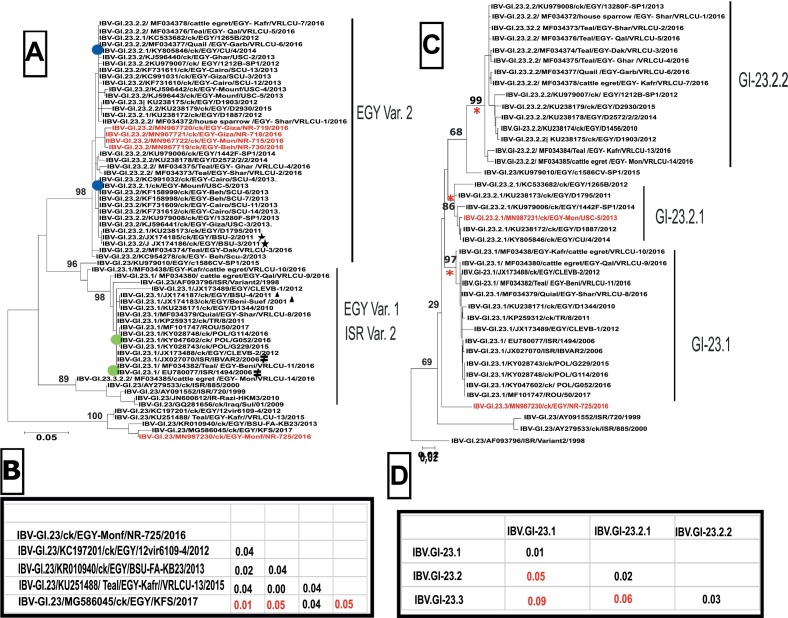
From eight IBV RT-qPCR positive samples, only one IBV isolate could be recovered (NR725/16) and a full genome sequence was generated. However, for four additional samples partial sequence information for the S1 gene (nt 750–1069) including the HVR 3 region could be obtained (Table 1). Phylogenetic analysis indicated that all five viruses belong to genotype I clade 23 (GI-23) (Fig. S1A). Within genotype GI-23 one virus from a small scale poultry farm (NR730/16) grouped with viruses from industrial sector 2 poultry farms (NR715/16, NR718/16 and NR719/16) within the EGY-Var2 group (Fig. 1 A). The IBV isolate NR725/16 was in an outgroup of genotype GI-23. This separate phylogenetic grouping is also reflected by the level of nucleotide diversity: While diversity between the four EGY-var2 viruses was only 0.3 to 0.6%, diversity between the EGY-var2 viruses and the sector 3 virus NR725/16 was 20 to 21%. Within the branch of NR725/16, four other viruses from Egypt detected in 2012, 2013, 2015 and 2017 are grouped, that have a diversity within the branch of 1 to 5% (Fig. 1A). Genotyping was corroborated based on sequence information of the entire S1 gene. Following the suggested nomenclature by Valastro and colleagues (Valastro et al., 2016) the isolate as well as the historical reference strain USC-5/2013 (Sultan et al., 2015) included in the NGS analysis, was verified as genotype I clade 23 (S1:GI-23) (Fig. S1B). However, by applying criteria for clade differentiation of NDV (Dimitrov et al., 2019), clade 23 could be divided into three subclades (Fig. 1C). Viruses formerly combined as EGY-var2 are now separated into two subclades: the historical IBV strain USC-5/2013 was part of a branch (n = 6) forming sub-clade 2.1 (S1:GI-23.2.1). Subclade 2.2 (S1:GI-23.2.2, n = 15) harbors other viruses from Egypt from 2010 until 2016. A third subclade, S1:GI-23.1 (n = 14) combines EGY-Var1 and IS-Var2 viruses from Israel, Egypt, Turkey, Romania and Poland from the years 2010–2017. Group distances between subclades were 5%, 6% and 9% respectively (Fig. 1D). According to this analysis, isolate NR725/16 was not within any of the three subclades and has to be considered an out-group, as only one sequence was available (Fig. 1B). At the level of the S1, NR725/16 had the highest nucleotide similarity (94 %) with both CU/4/2014 (GI-23.2.1) and POL/G052/16 (GI-23.1).
Fig. 1.
Phylogenetic analysis of IBV viruses within genotype GI-23 based on partial (a) and full (b) S1-gene sequences. The phyologenetic tree was generated using Maximum likelihood analysis for the partial S1 gene (290 nucleotides) including partial HVR3. Marked are reference strain for IS variant 2 viruses ( ), Egyptian variant 1 (
), Egyptian variant 1 ( ) and Egyptian variant 2 (
) and Egyptian variant 2 ( ). Prototype viruses used throughout the study include 2 EGY-var2 - (
). Prototype viruses used throughout the study include 2 EGY-var2 - ( ) and 2 EGY-var1-strains (
) and 2 EGY-var1-strains ( ). Virus from the small broiler flock (NR 725/16) and four additional viruses from the year 2016 are marked in red(A). In addition, the nucleotide distances between NR725/16 and related sequences in the gene bank are given (B). Phylogenetic analysis based on full S1 gene (1610 Nucleotide) resulted in establishing three sub-clades (GI-23.1, −23.2.1 and −23.2.2) (C). Viruses with full genome sequences in the study are highlighted in red. Distances between the proposed sub-clades are given below (D). Names include the genotype according to S1 gene, followed by accession number, species the virus was detected from with abbreviation for chicken (ck), country code according ISO 3166-1 alpha-3, laboratory identification number and year of publication.
). Virus from the small broiler flock (NR 725/16) and four additional viruses from the year 2016 are marked in red(A). In addition, the nucleotide distances between NR725/16 and related sequences in the gene bank are given (B). Phylogenetic analysis based on full S1 gene (1610 Nucleotide) resulted in establishing three sub-clades (GI-23.1, −23.2.1 and −23.2.2) (C). Viruses with full genome sequences in the study are highlighted in red. Distances between the proposed sub-clades are given below (D). Names include the genotype according to S1 gene, followed by accession number, species the virus was detected from with abbreviation for chicken (ck), country code according ISO 3166-1 alpha-3, laboratory identification number and year of publication.
3.2. Gene flow from genotypes GI-23.1 and GI-19 contributed to the emergence of NR725/16
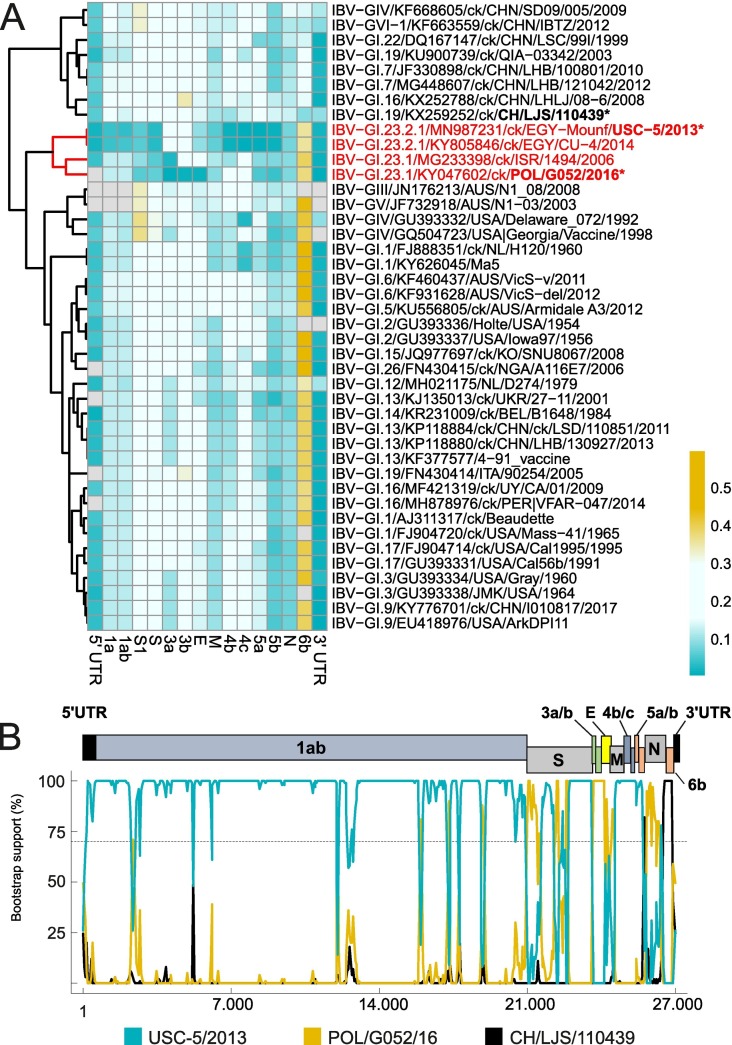
To verify the relationship of IBV isolate NR725/16 from a small-scale poultry farm, other genome regions were analyzed: At the level of the whole genome, NR725/16 had 95% and 96% similarity with EGY/CU/4/14 and USC-5/2013 respectively. Both IBV strains, EGY/CU/4/14 as well as USC-5/2013were characterized as genotype S1:GI-23.2.1, based on the S1 phylogeny. Likewise, genes 1ab, S2, 4bc and 5ab of NR725/16 are most similar to USC-5/2013 (Fig. 2 ), with similarities of 96%, 97%, 99% and 99%, respectively (Table S1). In contrast, gene 3ab and E of NR725/16 show the highest similarity (99%) with GI-23.1 virus POL/G052/16. Interestingly, another GI-23.1 prototype virus from Israel (ISR/1494/2006) has a closely related gene 3a (98%) but a higher degree of difference in genes 3b (81%) and E (82%). Striking differences of NR725/16 to GI-23 viruses were observed in gene 6b, with a homology of less than 65%. The closest match to NR725/16 was a virus from GI-19 (CH/LJS/110439 QX/2011) with a similarity of 91 %. However, gene 6b was not conserved within genotype GI-19, as a second virus from this subclade (ck/ITA/90254/2005) showed a homology of only 61%. The relationship was confirmed by phylogenetic analysis (Fig. S2), where NR725/16 clusters in gene 6b analysis together with QX CH/LJS/110439/2011, that is grouped to viruses from China obtained between 1999 to 2012 and belonging to diverse genotypes (GI-1, GI-16, GI-22 and GIV). The Italian GI-19 virus clusters together with GI-13 viruses including vaccine strain 4/91 (KF377577). Analogously to the heat map (Fig. 2A), NR 725/16 clusters with EGY/CU/4/14 and USC-5/2013 in analyses of genes 1, 4 and 5. In all three analyses, GI-23.1 viruses (POL/G052/16 and ISR/1494/2006) are in separate branches to each other and to the NR725/16 group. Inversely, analysis of genes 3 and E, NR725/16 clusters with the GI-23.1 virus described in Poland (POL/G052/16) but apart from the GI-23-2.1 viruses EGY/CU/4/14 and USC-5/2013. According to analyses for the E gene, GI-23.1 reference strain ISR/1494/2006 formed a separate group with viruses from Nigeria, Italy and China, identified as S genotype GI-26, GI-19 and GI-16, respectively. These data strongly suggested multiple recombination events for generating the NR725/16 strain, a hypothesis supported by bootscan analysis. Applying RDP recombination analysis package suggests recombination events within six genes (1a, 1b, S, N, 3a, 3b and 6b), indicating that genotypes GI-23.1 and GI-19 donated genes to circulating GI-23.2 strains (Fig. 2B) to give rise to isolate NR725/16.
Fig. 2.
Heatmap and Bootscan analysis. Heatmap showing the relationship of IBV isolate NR725/16 on the basis of pairwise distances for each gene in relation to other viruses representing different subclades. The highest related genes have a deep blue color that converts to light blue, light yellow and deep yellow with increasing distance (A). The Bootscan analysis showing different recombination events within NR725/16 genome compared to viruses suggested by phylogenetic analysis of different genes, with arrangement of the IBV genome given (B).
3.3. Mobility of gene 3ab in Egyptian IBV strains
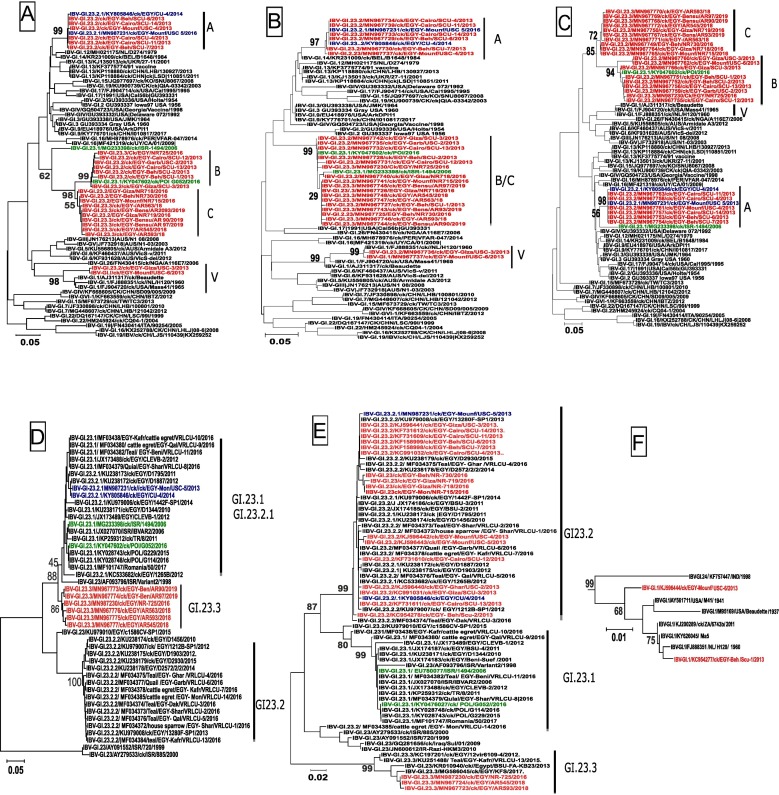
The similarity in genes 3 and E of NR725/16 to a virus identified in Poland (POL/G052/2016) raised the question whether the IBV strain from a small-scale farm reflects the introduction of a new gene pool to Egypt, or the gene pool was present already before 2016 and may have spread to other farms/sectors in Egypt. By specifically designed PCR protocols for gene 3ab we were able to sequence 14 historical GI-23 IBV strains from the years 2012 and 2013 (Table 1). It became apparent that seven strains cluster together with prototype S1:GI-23.2.1 strain EGY/USC-5/2013 (Fig. 3 A, gene 3ab:A) (Table 1). However, 6 historical IBV strains group together with isolate NR 725/2016 and POL/G052/2016, clearly demonstrating that this particular 3ab genotype (Fig. 3A, 3ab:B) already was present in IBV strains in Egypt before 2016. In addition, by analyzing new samples from large-scale broiler farms in Egypt, from the years 2018 and 2019 (n = 6) (Table 1), we recognized a closely related 3ab genotype to NR725/16 (Fig. 3A, 3ab:C), that was already present in IBV-positive samples from large-scale poultry farms, taken in 2016.
Fig. 3.
Evolution of gene 3ab and S-gene in Egyptian IBV strains. IBV-positive samples from 2012 until 2019 were sequenced specifically for 3ab gene and S1 genes by applying specific RT-PCRs. Phylogenetic analysis was done for combined gene 3ab (A) and separately for 3a (B) and 3b (C). In addition, two fragments of the S1 gene are given with a fragment spanning part of the HVR1 and 2 (D). The second fragment encloses partial HVR 3 region and was analyzed in the context of subclade GI-23 (E) and for two viruses in the context of vaccine strains of subclade GI-1 (F). The Egyptian subclade GI-23.2.1 prototype sequences are colored in blue, whereas subclade GI-23.1 prototype viruses are colored in green. Historical and recent IBV samples sequenced in this study are depicted in red.
Separate analysis of gene 3a and 3b indicated an independent evolution of both genes (Fig. 3B/3C). When analyzing gene 3a separately, historical as well as recent Egyptian IBV strains form one group (Fig. 3B, 3a:B/C). In contrast, in the analysis of gene 3b (Fig. 34C), all 6 strains from 2013 (SCU-1, −2, −3, −12, −13/2013 and USC-2/2013) cluster together with NR725/16 and POL/G052/2016 (3b:B) and apart from recent IBV strains from sector 2 (Fig. 3C, 3b:C). The independence of gene 3a and 3b is even more evident for prototype S1:GI-23.1 strain ISR/1494/2006: While for gene 3a, ISR/1494/2006 is within the group of newly recognized genotype 3a:B/C (Fig. 3B), for gene 3b it clusters with historical Egyptian viruses in 3b:A (Fig. 3C) like EGY/USC-5/2013. A pronounced differentiation between genes 3a and 3b is also evident for historical strains EGY/ USC-3/2013 and EGY/ USC-6/2013. Both strains cluster for gene 3a with vaccine type viruses of genotype S1:GI-1 and are closely related to strain NL/H120/1960 (nucleotide pairwise similarity 99%) (Table 1). In contrast to gene 3b, both strains cluster together within the same group as NR725/16 (3b:B). These data support the notion that recombination of gene 3a and 3b can take place as a separate event and coexist in strains with different S gene configuration.
3.4. New IBV strains represent combination 3ab: C and new S-subclade GI-23.3
To address the question whether the recent IBV strains had kept the historical S gene (S1:GI-23.2) or switched to the outgroup constellation found in sector 3 isolate NR725/16, we attempted to sequence the IBV positive samples from 2018 and 2019. We succeeded in obtaining sequence information on S1 hypervariable regions 1 and 2 (HVR1-2) for five IBV positive samples (Fig. 3D) and for HVR3 of two samples (Fig. 3E). In all instances the recent viruses cluster together with “outgroup” strain NR725/16 forming a forth subclade GI-23.3. The fact that all obtained IBV-positive samples harvested the new subclade GI-23.3, indicates that the new S gene might be beneficial to spread within a vaccinated poultry population. This notion is supported further when analyzing the amino acid composition of the obtained partial HVR 1–3 fragments (Table S3). All together prototype strain NR725/16 displayed 99 mutations within the stretch of 224 amino acids determined to known sequences of genotype GI-23 and vaccine type viruses. Within GI-23.3 “outgroup”, these mutations were stable and differences were observed only at five positions. In contrast, compared to vaccine strains GI.12/D274, GI.13/4/91, GI.1/Ma5 and GI.1/H120, 50, 59, 74 and 75 mutations were detected, respectively. Compared to the other subclades of genotype GI-23, subclade GI-23.3 viruses accumulated additional mutations that included defined epitopes within the S protein: Unique changes were present at site 48.2 (294F, 297S and 306Y) and 62.1 (329R).
4. Discussion
Vaccination-directed control of IB is hampered by the high mutation rate of IBV as well as by homologous recombination of the viral RNA genome, leading to a broad variety of circulating IBV strains (Cavanagh, 2007, De Wit et al., 2011). As a gene reservoir, backyard holdings and wild birds were previously discussed (Sultan et al., 2015, Hassan et al., 2016, Pauly et al., 2019, Rohaim et al., 2019b). In this study, we investigated an IBV strain from a small-scale broiler farm of sector 3 (EGY/NR 725/2016) in order to elucidate the relation to IBV strains from large-scale poultry enterprises. Initial comparison to four samples from sector 2 broiler farms from the same study (Moharam et al., 2019) indicated that all five viruses belong to genotype G1-23, but that isolate NR725/16 from the small-scale farm, was different from previously described EGY-var1 and EGY-var2 genotypes (Fig. 1a). For further full genome analysis, we were able to obtain an IBV isolate from the sector 3 but not from the sector 2 samples. For comparison, we therefore chose a different historic IBV isolate from a previous study in 2012 (Sultan et al., 2015). The virus (EGY/USC-5/2012) was obtained from a large-scale broiler production farm in 2012 and was previously described to belong to sub-group EGY-var2.
4.1. Origin of new IBV variant
The obtained IBV isolate NR725/16 enabled full genome sequencing by NGS. Initial genotyping was based on the established S1 gene, clearly verifying that NR725/16 and the historical IBV USC-5/2013 belong to genotype S1:GI-23, the major IBV genotype circulating in Egypt in recent years (Abdel-Moneim et al., 2012, Sultan et al., 2015, Zanaty et al., 2016a, Abozeid et al., 2017). Our deeper phylogenetic analysis, however, revealed, that considerable variability exists within clade 23 of genotype I (GI). For distinguishing different subclades within genotype GI-23, we applied criteria of nucleotide diversity adapted for NDV genotyping (Dimitrov et al., 2019) and could identify three separate subclades. The historic isolate EGY/USC-5/2013 that before was part of the EGY-var2 group, was assigned to subclade GI-23.2.1. The second subclade GI23.2.2 separates group EGY-var2 and comprises IBV from poultry as well as wild birds from Egypt detected between 2010 and 2016. Viruses that before were assigned to EGY-var1/ISR-var2 group are now gathered within subclade GI-23.1, a subclade that contains not only viruses from Egypt or the Middle East but also from Turkey, Poland and Romania. In contrast, the IBV isolate from sector 3 (NR725/16) was not integrated in the three subclades and forms a separate branch in genotype GI-23. Furthermore, three viruses from Israel that are considered to be ancestor viruses for GI-23, form another separate branch. According to the criteria that at least sequences of three independent virus sequences should be available to form a separate subclade, also the three early Israeli viruses are outgroup viruses, not assigned to a specific subclade. However, our analysis of partial S1 sequences indicates that isolate NR725/16 is a prototype of currently circulating viruses. Distance values of isolate NR725/16 of 6% to GI-23.1 as well GI-23.2.1 clearly are above the threshold of 5% distinguishing a new subclade and suggest that the emerged IBV viruses will form a fourth subclade, GI-23.3. Taken together our phylogenetic analysis is a further proof of the ongoing evolution of the S gene of IBV strains in Egypt.
However, when analyzing further individual genes of the IBV genome, it became clear that NR725/2016 is not a direct progeny of subclade GI-23.2 or GI-23.1, but a recombinant: Intra-genotypic gene exchange is indicated for genes S1, 3ab and E, while gene 6b was acquired from inter-genotypic recombination. Previous studies reported recombination of Egyptian viruses on the level of S1 (Zanaty et al., 2016b, Rohaim et al., 2019b), but this is the first report of intra-genotypic recombination events for an Egyptian IB virus affecting the whole genome. This additional information revealed a puzzling high similarity of genes 3ab and E for NR725/16 to a virus detected in Poland in 2016 (Lisowska et al., 2017), indicating a possible link between European and Egyptian IBV viruses (Fig. 2). However, a direct introduction of the IBV strain detected in Poland in 2016 to Egypt, or vice versa, seems unlikely. This was confirmed by our analyses of historic Egyptian samples which carried the particular 3ab:B genotype already in 2013. Back then, this genome constellation existed within the context of the dominating Egyptian S1 genotype (Fig. 3).
A highly similar 3ab genotype (3ab:C, Fig. 3) was present also in sector 2 farms sampled in 2016. This 3ab:C genotype prevailed in IBV and was present in positive samples from 2018 and 2019. However, the IBV strains from 2016 carried the S1:GI-23.2 genotype and all recent viruses carry the S1:GI-23.3 genotype (Fig. 3). These data indicate that viruses within the newly designated subclade GI23.3 do not have a single ancestor but have emerged by successive recombination events at least since 2013 and were successful in spreading in the Egyptian poultry population until at least 2019.
4.2. New IBV genotype GI-23.3 prevailed in Egypt and accumulated specific mutations in S1 protein
The finding that recombinants with 3ab:C in conjunction with S1 GI-23.3 were successfully maintained in the vaccinated Egyptian poultry population for at least the last 3 years suggests a selective advantage of these viruses. The S1 protein likely represents one major factor facilitating entry of the virus into the host cell and being a key target for protective immune responses. Defined conformational epitopes for neutralizing antibodies help to connect sequence data to possible antigenic escape variants (Kant et al., 1992). For the emerging S1:GI-23.3 viruses we could demonstrate an accumulation of point mutations within HVR 1–3: All S1:GI-23.3 viruses share mutations to vaccine strains with an average of 28.8 % amino acids changed (99/224). Point mutations are also evident compared to genotype GI-23.1 and GI-23.2.1 with 13.6% and 12.9% of the analyzed amino acids mutated in NR725/16 respectively. It is striking that 23.2 % (23/99) of overall mutations are conserved in other Egyptian IBV strains, with two of these sites also changed in GI-23.1 viruses (aa: 255, 282). Five additional sites are different between NR725/16 and vaccine viruses as well as GI-23.2.2 but are identical in GI-23.1 viruses and the majority of GI-23.2.1 viruses (aa 88, 108, 127, 132 (epitope 52.1), 168). These findings further support the notion that S1:GI-23.3 viruses emerged from viruses of subclade GI-23.1 and/or GI-23.2.1. In addition, the analysis indicates that the new viruses have accumulated mutations that facilitate immune escape. This notion is corroborated by the finding that two additional mutated neutralizing epitopes (48.2 and 62.1) in NR725/16 are sustained in all investigated S1:GI-23.3 outgroup viruses. Further functional studies will have to test the hypothesis of an antigenic escape and investigate whether current vaccines are able to mediate protection.
4.3. Possible contribution of genes 3ab for evolutionary advantage
The contribution of protein 3ab to a possible evolutionary advantage for these new viruses remains unclear. It is known that the gamma-coronavirus-specific protein 3ab is non-structural and non-essential for virus replication in ovo (Hodgson et al., 2006). New evidence, however, indicates that protein 3a and 3b might, like protein 5b, play a role in viral pathogenesis in chickens. The first line of evidence is provided by in vitro observations demonstrating that protein 3a is involved in resistance of IBV to the cellular antiviral status induced by type I interferon (IFN) by modulating the IFN response at transcriptional and translational levels (Kint et al., 2015a, Kint et al., 2015b). Secondly, recombinant IBV 3ab deletion mutants in the background of virulent IBV strain H52 or a QX strain displayed a lower pathogenicity (van Beurden et al., 2018, Zhao et al., 2019). The latter study differentiated between contribution of genes 3a and 3b and concluded that protein 3b has a greater effect on pathogenicity than protein 3a. In this respect it is interesting to note that gene 3a is more uniform compared to 3b, which can be used to divide Egyptian strains into two subgroups (3b:B and 3b:C). Further studies will have to elucidate whether specific pathotypes can be associated with specific 3a or 3b genotype or with specific motifs in the proteins.
5. Conclusions
In general, recombination is considered to be advantageous in evolution, if there is a negative linkage between specific genes/alleles, an effect termed negative disequilibrium. In exchanging genes/alleles offspring might gain advantage through an immediate switch to more favorable combinations (Barton, 2010). For IBV circulation, mutations in the S1 protein could give rise to variants that are more efficiently transmitted in a vaccinated population. If the second allele is linked to a highly virulent pathotype, the combination would efficiently kill the birds and reduce the number of individuals in a population. In this respect, an allele that facilitates low pathogenicity in escape variants would be beneficial for sustaining virus circulation. Following this hypothesis, the current 3ab:C genotype protein would facilitate a weaker pathogenicity, possibly by a less efficient interference with IFN. From the evolutionary point of view, this could be an advantage, in particular in a finite population, which in general favors recombination (Felsenstein, 1974). Further studies are needed to evaluate a connection of different 3ab genotypes to pathogenicity and a possible application as diagnostic marker.
CRediT authorship contribution statement
Ibrahim Moharam: Investigation, Data curation, Formal analysis, Writing - original draft. Hesham Sultan: Supervision, Data curation. Kareem Hassan:. Mahmoud Ibrahim: Writing - original draft. Salama Shany: Investigation. Awad A. Shehata: Data curation. Mohammed Abo-ElKhair: Data curation. Florian Pfaff: Data curation. Dirk Höper:. Magdy EL Kady: Conceptualization, Data curation. Martin Beer: Funding acquisition, Writing - review & editing. Timm Harder: Conceptualization, Writing - review & editing. Hafez Hafez: Validation, Writing - review & editing. Christian Grund: Supervision, Validation, Visualization.
Acknowledgments
Acknowledgements
We thank Cornelia Illing for excellent technical assistance.
Funding
This work was supported by the German Ministry of Food and Agriculture through the budget of the FLI. This research did not receive any specific grant from funding agencies in the commercial or not-for-profit sectors. Ibrahim Moharam is supported by Ministry of Higher Education and Scientific Research, Egypt.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104433.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abd El Rahman S., Hoffmann M., Lueschow D., Eladl A.-F., Hafez H.M. Isolation and characterization of new variant strains of infectious bronchitis virus in northern Egypt. Adv. Anim. Veterin. Sci. 2015;3:362–371. doi: 10.14737/journal.aavs/2015/3.7.362.371. [DOI] [Google Scholar]
- Abdel-Moneim, A., Madbouly, H., Gelb, J. J., Ladman, B., 2002. Isolation and identification of Egypt/Beni-Suef/01 a novel genotype of infectious bronchitis virus. Vet.Med.J., Giza 50, 1065-1078.doi:https://www.researchgate.net/publication/257856074.
- Abdel-Moneim A.S., Afifi M.A., El-Kady M.F. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch. Virol. 2012;157:2453–2457. doi: 10.1007/s00705-012-1445-1. [DOI] [PubMed] [Google Scholar]
- Abozeid H.H., Paldurai A., Khattar S.K., Afifi M.A., El-Kady M.F., El-Deeb A.H., Samal S.K. Complete genome sequences of two avian infectious bronchitis viruses isolated in Egypt: evidence for genetic drift and genetic recombination in the circulating viruses. Infect. Genet. Evol. 2017;53:7–14. doi: 10.1016/j.meegid.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Adzhar A., Gough R.E., Haydon D., Shaw K., Britton P., Cavanagh D. Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian Pathol. 1997;26:625–640. doi: 10.1080/03079459708419239. [DOI] [PubMed] [Google Scholar]
- Barton N.H. Mutation and the evolution of recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1281–1294. doi: 10.1098/rstb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Mockett A.P. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988;11:141–150. doi: 10.1016/0168-1702(88)90039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.K. The classification of new serotypes of infectious bronchitis virus isolated from poultry flocks in Britain between 1981 and 1983. Avian Pathol. 1984;13:733–741. doi: 10.1080/03079458408418570. [DOI] [PubMed] [Google Scholar]
- De Wit S.J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov K.M., Abolnik C., Afonso C.L., Albina E., Bahl J., Berg M., Briand F.X., Brown I.H., Choi K.S., Chvala I., Diel D.G., Durr P.A., Ferreira H.L., Fusaro A., Gil P., Goujgoulova G.V., Grund C., Hicks J.T., Joannis T.M., Torchetti M.K., Kolosov S., Lambrecht B., Lewis N.S., Liu H., Liu H., McCullough S., Miller P.J., Monne I., Muller C.P., Munir M., Reischak D., Sabra M., Samal S.K., Servan de Almeida R., Shittu I., Snoeck C.J., Suarez D.L., Van Borm S., Wang Z., Wong F.Y.K. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 2019;74 doi: 10.1016/j.meegid.2019.103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mahdy S., El-Hady M.M., Soliman Y.A. Isolation and characterization of nephropathogenic strain of infectious bronchitis virus in EGYPT. J. Am. Sci. 2010;6:669–674. [Google Scholar]
- EU/92/66/EEC, 1992. Introducing community measures for the control of Newcastle disease. Official Journal of the European Communities. L260, 1–20.
- FAO, 2004. FAO recommendations on the prevention, control and eradication of highly pathogenic avian influenza (HPAI) in Asia, September 2004.
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Suzuki Y., Gojobori T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 2004;21:1074–1080. doi: 10.1093/molbev/msh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan K.E., El-Kady M.F., El-Sawah A.A.A., Luttermann C., Parvin R., Shany S., Beer M., Harder T. Respiratory disease due to mixed viral infections in poultry flocks in Egypt between 2017 and 2018: upsurge of highly pathogenic avian influenza virus subtype H5N8 since 2018. Transbound Emerg. Dis. 2019 doi: 10.1111/tbed.13281. [DOI] [PubMed] [Google Scholar]
- Hassan K.E., Shany S.A., Ali A., Dahshan A.H., El-Sawah A.A., El-Kady M.F. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult. Sci. 2016;95:1271–1280. doi: 10.3382/ps/pew068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. 1st ed. Oxford University Press Inc.; New York: 2009. The Evolution and Emergence of RNA Viruses. [Google Scholar]
- Jackwood, M., de Wit, J. J., 2013. Infectious bronchitis, in: J. R. G. S.E. Swayne, L.R. McDougald, L.K. Nolan, D.L. Suarez & V. Nair (Eds.), (Ed.), Diseases of poultry (13th ed). Ames, IA: Blackwell Publishing Professional, pp. 117-135.
- Jackwood M.W., Hall D., Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Lee D.H. Different evolutionary trajectories of vaccine-controlled and non-controlled avian infectious bronchitis viruses in commercial poultry. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant A., Koch G., van Roozelaar D.J., Kusters J.G., Poelwijk F.A., van der Zeijst B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73(Pt 3):591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- King A.M.Q., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M.L., Rubino L., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018) Arch. Virol. 2018;163:2601–2631. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
- Kint J., Dickhout A., Kutter J., Maier H.J., Britton P., Koumans J., Pijlman G.P., Fros J.J., Wiegertjes G.F., Forlenza M. Infectious bronchitis coronavirus inhibits STAT1 signaling and requires accessory proteins for resistance to Type I interferon activity. J Virol. 2015;89:12047–12057. doi: 10.1128/JVI.01057-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint J., Fernandez-Gutierrez M., Maier H.J., Britton P., Langereis M.A., Koumans J., Wiegertjes G.F., Forlenza M. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J. Virol. 2015;89:1156–1167. doi: 10.1128/JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J.G., Niesters H.G., Lenstra J.A., Horzinek M.C., van der Zeijst B.A. Phylogeny of antigenic variants of avian coronavirus IBV. Virology. 1989;169:217–221. doi: 10.1016/0042-6822(89)90058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowska A., Sajewicz-Krukowska J., Fusaro A., Pikula A., Domanska-Blicharz K. First characterization of a middle-east GI-23 lineage (Var2-like) of infectious bronchitis virus in Europe. Virus Res. 2017;242:43–48. doi: 10.1016/j.virusres.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. In: Fields Virology, Sixth Edition. 2013. ed. Knipe D., Howley P., editors. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. Coronaviridae; pp. 825–858. [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3 '-> 5 ' exoribonuclease that is critically involved in coronavirus RNA synthesis. PNAS. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockett A.P., Cavanagh D., Brown T.D. Monoclonal antibodies to the S1 spike and membrane proteins of avian infectious bronchitis coronavirus strain Massachusetts M41. J. Gen. Virol. 1984;65(Pt 12):2281–2286. doi: 10.1099/0022-1317-65-12-2281. [DOI] [PubMed] [Google Scholar]
- Moharam I., Razik A.A.E., Sultan H., Ghezlan M., Meseko C., Franzke K., Harder T., Beer M., Grund C. Investigation of suspected Newcastle disease (ND) outbreaks in Egypt uncovers a high virus velogenic ND virus burden in small-scale holdings and the presence of multiple pathogens. Avian Pathol. 2019;48:406–415. doi: 10.1080/03079457.2019.1612852. [DOI] [PubMed] [Google Scholar]
- Monne I., Ormelli S., Salviato A., De Battisti C., Bettini F., Salomoni A., Drago A., Zecchin B., Capua I., Cattoli G. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J. Clin. Microbiol. 2008;46:1769–1773. doi: 10.1128/JCM.02204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.M., Jackwood M.W., Hilt D.A. Identification of amino acids involved in a serotype and neutralization specific epitope within the s1 subunit of avian infectious bronchitis virus. Arch. Virol. 1997;142:2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib M.M., El-Kady M.F., Luschow D., Hassan K.E., Arafa A.S., El-Zanaty A., Hassan M.K., Hafez H.M., Grund C., Harder T.C. New real time and conventional RT-PCRs for updated molecular diagnosis of infectious bronchitis virus infection (IBV) in chickens in Egypt associated with frequent co-infections with avian influenza and Newcastle Disease viruses. J. Virol. Methods. 2017;245:19–27. doi: 10.1016/j.jviromet.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib M.M., Graaf A., Fortin A., Luttermann C., Wernery U., Amarin N., Hussein H.A., Sultan H., Al Adhadh B., Hassan M.K., Beer M., Monne I., Harder T.C. Novel real-time PCR-based patho- and phylotyping of potentially zoonotic avian influenza A subtype H5 viruses at risk of incursion into Europe in 2017. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.1.30435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H.G., Lenstra J.A., Spaan W.J., Zijderveld A.J., Bleumink-Pluym N.M., Hong F., van Scharrenburg G.J., Horzinek M.C., van der Zeijst B.A. The peplomer protein sequence of the M41 strain of coronavirus IBV and its comparison with Beaudette strains. Virus Res. 1986;5:253–263. doi: 10.1016/0168-1702(86)90022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M., Snoeck C.J., Phoutana V., Keosengthong A., Sausy A., Khenkha L., Nouanthong P., Samountry B., Jutavijittum P., Vilivong K., Hubschen J.M., Black A.P., Pommasichan S., Muller C.P. Cross-species transmission of poultry pathogens in backyard farms: ducks as carriers of chicken viruses. Avian Pathol. 2019;48:503–511. doi: 10.1080/03079457.2019.1628919. [DOI] [PubMed] [Google Scholar]
- Rohaim M.A., El Naggar R.F., Hamoud M.M., Bazid A.I., Gamal A.M., Laban S.E., Abdel-Sabour M.A., Nasr S.A.E., Zaki M.M., Shabbir M.Z., Zahran O.K., Munir M. Emergence and genetic analysis of variant pathogenic 4/91 (serotype 793/B) infectious bronchitis virus in Egypt during 2019. Virus Genes. 2019;55:720–725. doi: 10.1007/s11262-019-01693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim M.A., El Naggar R.F., Helal A.M., Bayoumi M.M., El-Saied M.A., Ahmed K.A., Shabbir M.Z., Munir M. Genetic diversity and phylodynamics of avian coronaviruses in egyptian wild birds. Viruses. 2019;11 doi: 10.3390/v11010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim, K., Arafa, A. S., Hussein, H. A., El-Sanousi, A.-A., 2013. Molecular characterization of infectious bronchitis viruses isolated from broiler and layer chicken farms in Egypt during 2012. International Journal of Veterinary Science and Medicine.doi:http://dx.doi.org/10.1016/j.ijvsm.2013.10.002. [DOI] [PMC free article] [PubMed]
- Sultan, H., Abdel-Razik, A., Shehata, A., brahim, M., Talaat, S., Abo-Elkhair, M., Bazid, A., Moharam, I., Vahlenkamp, T., 2015. Characterization of Infectious Bronchitis Viruses Circulating in Egyptian chickens during 2012 and 2013. Journal of Veterinary Science & Medical Diagnosis 4:5.doi:http://dx.doi.org/2325-9590.1000180.
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Thor S.W., Hilt D.A., Kissinger J.C., Paterson A.H., Jackwood M.W. Recombination in avian gamma-coronavirus infectious bronchitis virus. Viruses. 2011;3:1777–1799. doi: 10.3390/v3091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro H., Schemera B., Kaleta E.F. Serological differentiation of avian infectious bronchitis field isolates using an enzyme immunoassay: presence of Dutch strains in West Germany. Avian Dis. 1987;31:187–192. [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beurden S.J., Berends A.J., Kramer-Kuhl A., Spekreijse D., Chenard G., Philipp H.C., Mundt E., Rottier P.J.M., Verheije M.H. Recombinant live attenuated avian coronavirus vaccines with deletions in the accessory genes 3ab and/or 5ab protect against infectious bronchitis in chickens. Vaccine. 2018;36:1085–1092. doi: 10.1016/j.vaccine.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M.G., Suarez D.L., Seal B.S., Pedersen J.C., Senne D.A., King D.J., Kapczynski D.R., Spackman E. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004;42:329–338. doi: 10.1128/jcm.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylezich C., Papa A., Beer M., Hoper D. A versatile sample processing workflow for metagenomic pathogen detection. Sci. Rep. 2018;8:13108. doi: 10.1038/s41598-018-31496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaty A., Arafa A.S., Hagag N., El-Kady M. Genotyping and pathotyping of diversified strains of infectious bronchitis viruses circulating in Egypt. World J. Virol. 2016;5:125–134. doi: 10.5501/wjv.v5.i3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaty A., Naguib M.M., El-Husseiny M.H., Mady W., Hagag N., Arafa A.S. The sequence of the full spike S1 glycoprotein of infectious bronchitis virus circulating in Egypt reveals evidence of intra-genotypic recombination. Arch. Virol. 2016;161:3583–3587. doi: 10.1007/s00705-016-3042-1. [DOI] [PubMed] [Google Scholar]
- Zhao X., Jiang Y., Cheng X., Yu Y., Gao M., Zhou S. Pathogenicity of a QX-like strain of infectious bronchitis virus and effects of accessory proteins 3a and 3b in chickens. Vet. Microbiol. 2019;239 doi: 10.1016/j.vetmic.2019.108464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.