Abstract
Lgr5+ intestinal stem cells (ISCs) exhibit self‐renewal and differentiation features under homeostatic conditions, but the mechanisms controlling Lgr5 + ISC self‐renewal remain elusive. Here, we show that the chromatin remodeler SRCAP is highly expressed in mouse intestinal epithelium and ISCs. Srcap deletion impairs both self‐renewal of ISCs and intestinal epithelial regeneration. Mechanistically, SRCAP recruits the transcriptional regulator REST to the Prdm16 promoter and induces expression of this transcription factor. By activating PPARδ expression, Prdm16 in turn initiates PPARδ signaling, which sustains ISC stemness. Rest or Prdm16 deficiency abrogates the self‐renewal capacity of ISCs as well as intestinal epithelial regeneration. Collectively, these data show that the SRCAP‐REST‐Prdm16‐PPARδ axis is required for self‐renewal maintenance of Lgr5 + ISCs.
Keywords: intestinal stem cell, PPARδ, REST, self‐renewal, SRCAP
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Regenerative Medicine
SRCAP recruitment of transcriptional coregulator REST to the Prdm16 promoter induces its expression, in turn activating PPARδ expression and signaling to sustain ISC self‐renewal.
Introduction
Intestine epithelium of adult mammals maintains strong and long‐term self‐renewal capacity for rapid turnover of tissues driven by intestinal stem cells (ISCs) (Barker, 2014). The ISC population resides in specialized niches at the bottom of the crypts and harbors both self‐renewal and differentiation features, giving rise to Paneth cells and proliferating TA cells in crypts and differentiating epithelial cells, including enterocytes, goblet, enteroendocrine, and tuft cells in villi (Sato et al, 2011; Clevers, 2013; Barker, 2014). A Wnt target Lgr5 was initially identified as the first marker of ISCs. Lgr5+ ISCs were also called crypt base columnar cells (CBCs), whose self‐renewal and multipotent capacities were confirmed by lineage tracing (Barker et al, 2007; Huch et al, 2013). Many signaling pathways including Wnt, Notch, BMP, PPAR, and Yap signaling are implicated in the regulation of self‐renewal and differentiation of Lgr5+ ISCs (Clevers et al, 2014; Gregorieff et al, 2015; Beyaz et al, 2016). ISCs are responsible for intestinal homeostasis, tissue regeneration, and intestinal tumorigenesis (Zhou et al, 2013b; Shimokawa et al, 2017). Although several signaling pathways and some niche factors have been defined for maintaining ISC stemness, the epigenetic regulation of ISC self‐renewal still remains largely unknown.
Chromatin remodeling represents a critical layer of epigenetic regulation of eukaryotic gene transcription (Hartley & Madhani, 2009; Zhang et al, 2011). Chromatin remodeling complexes are divided into four major subfamilies, including SWI/SNF, ISWI, CHD, and INO80, according to SWI2/SNF2‐related catalytic ATPase subunits (Tosi et al, 2013; Eustermann et al, 2018). The chromatin remodeling complexes participate in modulating chromatin accessibility to genetic reading, gene transcription, and functions in a wide range of biological processes (Bajpai et al, 2010; Pan et al, 2018; Wei et al, 2018). The SRCAP complex belongs to the INO80 subfamily that can exchange histone H2A to its variant H2A.Z in nucleosomes for activation of gene transcription (Watanabe et al, 2013). It is composed of eleven protein subunits in mammalian cells (Cuadrado et al, 2010). The SRCAP protein (known as SWR1 in yeast) confers ATPase activity (Mizuguchi et al, 2004), which is a core subunit of SRCAP remodeling complex. SRCAP mutations are implicated in human Floating–Harbor syndrome (Hood et al, 2012). We recently showed that SRCAP participates in the regulation of self‐renewal maintenance of embryonic stem cells (ESCs) and lymphoid lineage commitment (Ye et al, 2017, 2018b). However, how SRCAP modulates self‐renewal of ISCs remains poorly understood.
Peroxisome proliferator‐activated receptors (PPARs) belong to a ligand‐activated nuclear hormone receptor protein superfamily, comprising three members of ɑ, γ, and δ. PPAR binds to liver/retinoid X receptor (LXR/RXR) as its heterodimeric partners and functions as transcription factors (TFs) to regulate gene expression, which plays important roles in differentiation, development, lipid metabolism, and tumorigenesis (Lee et al, 2015; Prost et al, 2015). In gut, PPARδ, a predominant member of PPAR family, is expressed in ISCs and progenitor cells. High‐fat diet can promote stemness and tumorigenicity of intestinal progenitors in a PPARδ‐dependent manner (Beyaz et al, 2016). Here, we show that SRCAP is required for the self‐renewal maintenance of ISCs. SRCAP recruits REST onto Prdm16 promoter to induce its transcription, which initiates PPARδ signaling activation to sustain ISC stemness.
Results
SRCAP is highly expressed in intestinal epithelium and ISCs
We previously demonstrated that SRCAP regulates the self‐renewal of ESCs (Ye et al, 2018b). We also showed that the SRCAP chromatin remodeling complex modulates lymphoid lineage commitment (Ye et al, 2017). We then sought to explore the physiological role of SRCAP in the regulation of self‐renewal maintenance of ISCs. We observed that Srcap mRNA was highly expressed in liver, spleen, bone marrow, and gut through real‐time PCR and Northern blot (Figs 1A and EV1A). We next established SRCAP‐C‐HA tag mice by androgenetic haploid technology (Fig EV1B and C). We further validated that SRCAP was really mainly distributed in liver, spleen, and intestine (Fig 1B). We noticed that SRCAP was localized in the nucleus of intestinal epithelial cells through whole‐mount intestine immunostaining and clearmap 3D imaging (Fig 1C). Furthermore, SRCAP was also expressed in Lgr5+ ISCs (Fig 1D). In addition, SRCAP was also detectable in intestinal crypt‐villus organoids formed by in vitro culture (Fig 1E). Collectively, SRCAP is distributed in intestinal epithelium and ISCs, predominantly expressed in crypt progenitors including Lgr5 + ISCs.
Figure 1. SRCAP is highly expressed in intestinal epithelium and ISCs.
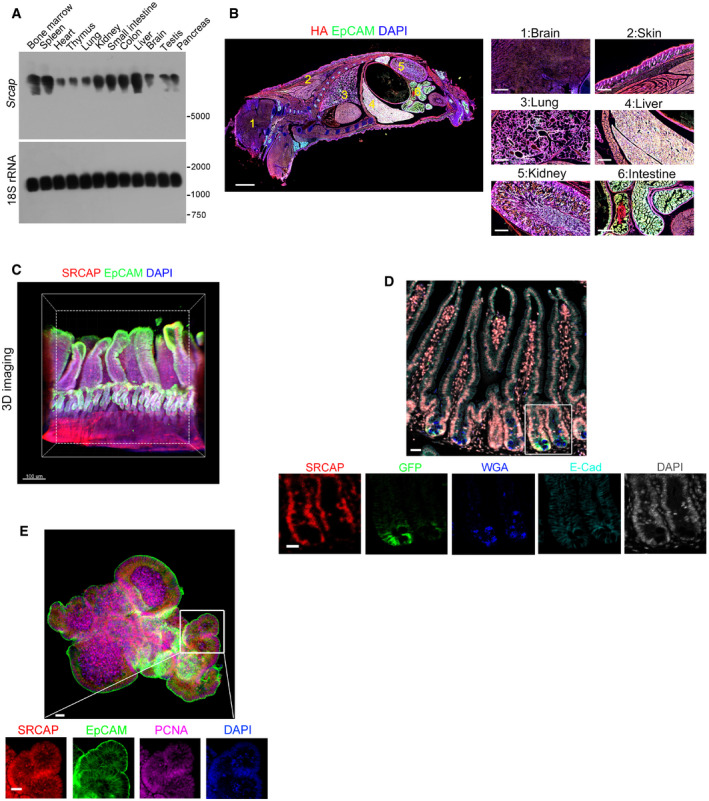
- Srcap expression in murine different tissues was examined by Northern blot. A 277 nt probe of Srcap (81–358 nt) was labeled for Northern blot analysis. 18S RNA was used as a loading control.
- Two‐week‐old SRCAP‐C‐HA tag mice were sacrificed for longitudinal sections followed by immunofluorescence staining. A global look of the section was shown. Scale bar: 500 μm. Green: EpCAM, red: HA tag, nuclei were counterstained by DAPI.
- Murine intestine tissues were fixed and stained according to iDISCO staining protocol. SRCAP was highly expressed in vast majority of crypts in mouse intestine. Scale bar: 50 μm. Green: EpCAM, red: SRCAP, nuclei were counterstained by DAPI.
- Duodenum region of intestinal tissues was obtained from Lgr5 GFP‐CreERT2 (Lgr5 GFP) mice for immunofluorescence staining by indicated antibodies with Opal™ 7‐color fIHC kit. Nuclei were counterstained with DAPI. Scale bar: 50 μm.
- ISCs from Lgr5 GFP mice were collected for organoid formation. Organoids were fixed for immunofluorescence staining with indicated antibodies. Scale bar: 50 μm.
Figure EV1. Generation strategies of Srcap KO mice.
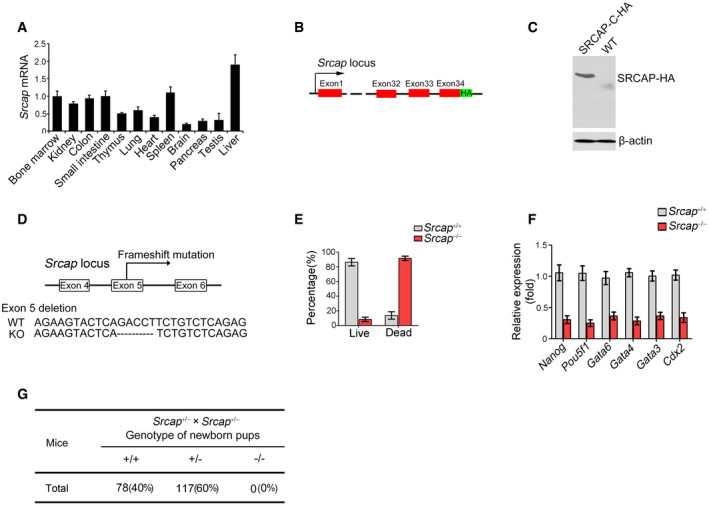
- Srcap expression in murine different tissues was examined by real‐time PCR. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates.
- Schematic diagram of SRCAP‐C‐HA tag construction for reporter mice.
- Expression of HA‐tagged SRCAP protein was confirmed by immunoblotting. β‐actin was used as a loading control.
- Schematic diagram of Srcap KO construction by CRISPR/Cas9 technology.
- Percentages of live embryos and dead embryos were counted as means ± SD. 200 embryos were isolated and analyzed for each group.
- Expression levels of indicated genes were examined in Srcap KO embryos by real‐time PCR. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates.
- Srcap‐deficient pups were genotyped after heterozygotes crossing.
Source data are available online for this figure.
Srcap deficiency impairs the self‐renewal of ISCs and intestinal epithelial regeneration
We next generated Srcap knockout (KO) mice through a CRISPR/Cas9 approach (Fig EV1D). Srcap deficiency impaired blastocyst development at E3.5 and caused early embryonic lethality (Fig EV1E–G). We then generated Srcap flox/flox mice through insertion of loxP sequences flanking at the exon5 of Srcap gene locus (Fig EV2A). We established Srcap flox/flox;Lgr5 GFP‐CreERT2 mice through crossing Srcap flox/flox mice with Lgr5 GFP‐CreERT2 mice (Fig EV2A). With administration of tamoxifen (TAM), Srcap was completely deleted in Lgr5+ ISCs (Fig EV2B and C; hereafter, TAM‐treated Srcap flox/flox;Lgr5 GFP‐CreERT2 mice were referred as Srcap KO, whereas TAM‐treated Srcap fl/fl mice were called as Srcap WT). We found that Srcap conditional deficiency in ISCs caused body weight loss and more than half of these mice died 1 month after tamoxifen treatment (Fig EV2D and E). We found that Srcap KO mice displayed shorter crypts and villi in length compared with Srcap WT mice (Figs 2A and EV2F and G). Srcap flox/flox;Lgr5 GFP‐CreERT2 mice showed reduced numbers of Lgr5‐GFP+ ISCs. Reduced ISCs in Srcap KO mice were also confirmed by Olfm4 staining (Figs 2B and EV2H). In addition, numbers of proliferating TA cells were also decreased in Srcap KO mice by PCNA staining (Fig 2B). In addition, we found that Srcap depletion remarkably reduced organoid formation (Fig EV2I and J).
Figure EV2. Generation strategies of Srcap flox mice.
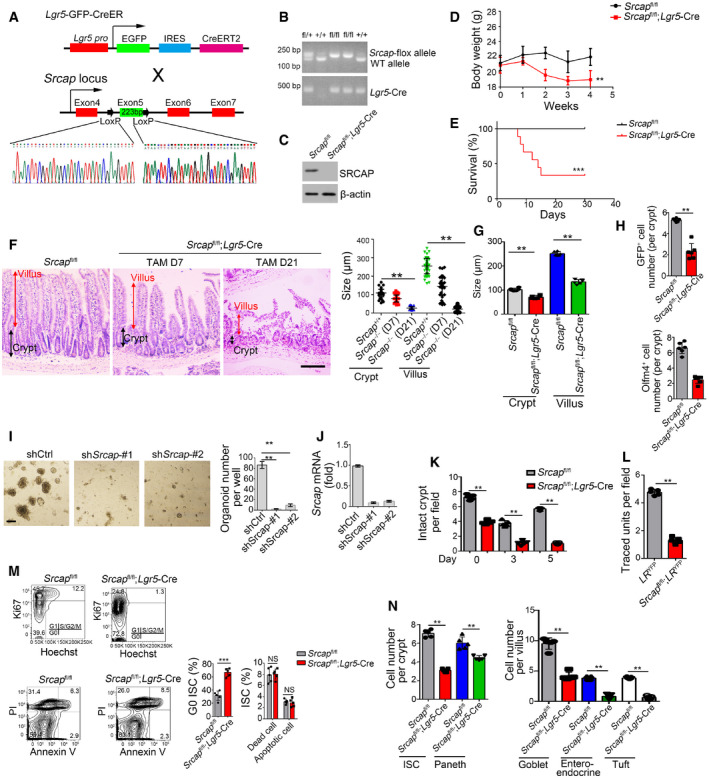
- Schematic diagram of Srcap flox construction by CRISPR/Cas9 technology.
- Srcap‐deficient pups were genotyped after crossing.
- SRCAP protein was examined in Srcap‐deleted ISCs by immunoblotting.
- Body weight changes of Srcap fl/fl and Srcap fl/fl; Lgr5 GFP‐CreERT2 mice after TAM treatment were shown as means ± SD. n = 10. **P < 0.01 by two‐tailed Student's t‐test.
- Kaplan–Meier survival curves of Srcap fl/fl and Srcap fl/fl; Lgr5 GFP‐CreERT2 mice after TAM treatment. n = 10. ***P < 0.001 by log‐rank test.
- Representative images of duodenum from 8‐week‐old Srcap fl/fl and Srcap fl/fl; Lgr5 GFP‐CreERT2 mice after TAM treatment for indicated time (n = 5 per group). Scale bar, 100 μm. Length of 100 villi and 100 crypts was measured and shown in right panel as means ± SD. **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests were shown as in Fig 2A. Length of villi and crypts (means per mouse) was measured and shown as means ± SD n = 6. **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests were shown as in Fig 2B. ISC numbers per crypt (means per mouse) were shown as means ± SD n = 6. **P < 0.01 by two‐tailed Student's t‐test.
- 1 × 104 ISCs were collected and infected with shSrcap lentivirus followed by organoid formation. Typical images of organoid formation were shown in left panel. Scale bar, 200 μm. Organoid numbers per well were counted as means ± SD in right panel. **P < 0.01 by two‐tailed Student's t‐test. n = 6 for each group.
- Srcap was examined in Srcap‐depleted cells by real‐time PCR. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD.
- Biological variations and statistical tests were shown as in Fig 2D. Numbers of intact crypts (means per mouse) were shown as means ± SD n = 5. **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests were shown as in Fig 2E. Numbers of traced crypt‐villus units (means per mouse) were shown as means ± SD n = 5. **P < 0.01 by two‐tailed Student's t‐test.
- Cell cycle and apoptosis of Srcap +/+ and Srcap –/– ISCs were examined by Hoechst/Ki67 and PI/Annexin V FACS and shown in left panel. n = 6 mice per group. Percentages of G0 stage ISCs and apoptotic ISCs were counted as means ± SD (right panel). **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests showed means per mouse as in Fig 2G. Numbers of each kind of epithelial cells from crypts or villi (means per mouse) were shown in right panel as means ± SD n = 5. **P < 0.01 by two‐tailed Student's t‐test.
Source data are available online for this figure.
Figure 2. Srcap deficiency impairs the self‐renewal of ISCs and intestinal epithelial regeneration.
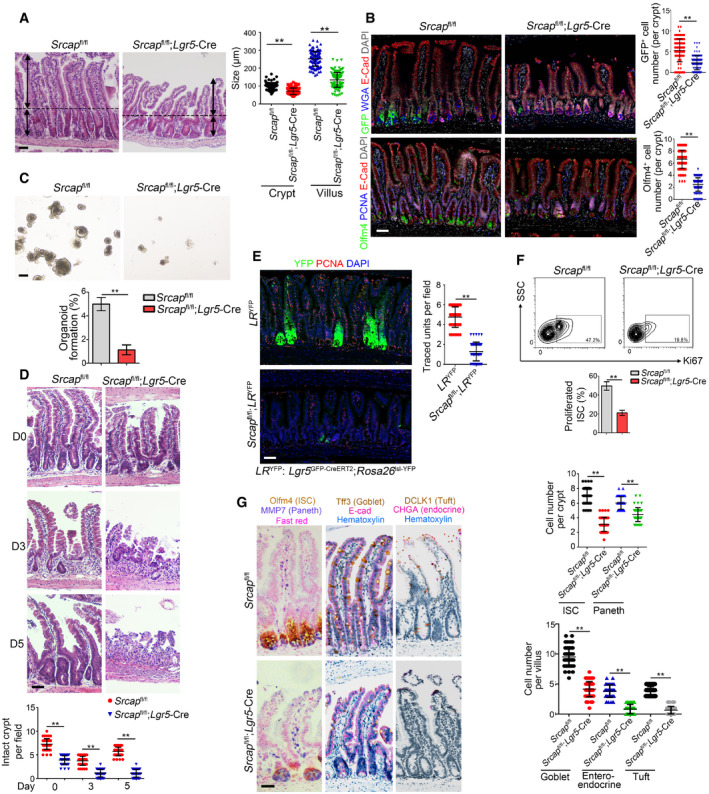
- Representative images of intestines from 8‐week‐old Srcap +/+ and Srcap –/– mice (n = 6 per group). Length of 120 villi and 120 crypts (20 villi and 20 crypts per mouse) was measured and shown in right panel as means ± SD. ** P < 0.01 by two‐tailed Student's t‐test. Scale bar, 100 μm. Dotted lines indicate the boundary between crypts and villi. Arrows indicate the length of crypts or villi.
- Intestinal tissues from 8‐week‐old Srcap +/+ and Srcap –/– mice (n = 6 per group) were collected for immunofluorescence staining with indicated antibodies. Typical images were shown in left panel and statistical ISC numbers from 120 crypts (20 crypts per mouse) were shown in right panel as means ± SD. ** P < 0.01 by two‐tailed Student's t‐test. Scale bar, 50 μm.
- 1 × 104 ISCs from indicated mice were collected for organoid formation. Scale bar, 200 μm. n = 3 for each group. Organoid formation ratios were shown as means ± SD. **P < 0.01 by two‐tailed Student's t‐test.
- Srcap +/+ and Srcap –/– mice were treated with 10 Gy's radiation and sacrificed at different time points. Intestinal tissues were collected for H&E staining. Numbers of intact crypts were shown in lower panel as means ± SD. ** P < 0.01 by two‐tailed Student's t‐test. 50 fields (10 fields per mouse) were taken for each group (n = 5). Scale bars, 100 μm.
- Lgr5 GFP‐CreERT2;Rosa26 lsl‐YFP (LR YFP) were crossed with Srcap flox/flox mice, followed by administration of tamoxifen (TAM) for lineage tracing analysis. Mice were sacrificed 7 days after TAM induction, and typical jejunum sections were stained. Numbers of traced crypt‐villus units were shown in right panel as means ± SD. **P < 0.01 by two‐tailed Student's t‐test. 50 fields (10 fields per mouse) were taken for each group (n = 5). Scale bars, 50 μm.
- Proliferation of Srcap +/+ and Srcap –/– ISCs was examined by Ki67 staining followed by flow cytometry analysis. Percentages of proliferated cells were counted as means ± SD (lower panel). **P < 0.01 by two‐tailed Student's t‐test. n = 5 per group.
- Multiple‐color staining was performed in Srcap +/+ and Srcap –/– intestine sections. AP‐red, DAB, and AP‐blue were used for immunohistochemistry coloration. Numbers of each kind of epithelial cells from 50 crypts or 50 villi (10 crypts and 10 villi per mouse) were shown in right panel as means ± SD. **P < 0.01 by two‐tailed Student's t‐test. Scale bars, 50 μm.
We next isolated ISCs from Srcap fl/fl and Srcap fl/fl;Lgr5‐Cre mice, followed by organoid formation assay. We observed that Srcap KO ISCs showed impaired capacity of organoid formation (Fig 2C). Lgr5+ ISCs play a pivotal role in radiation‐induced intestinal regeneration. We noticed that Srcap KO mice displayed impaired capacity of intestinal epithelial regeneration with irradiation injury compared with Srcap WT mice (Figs 2D and EV2K). The self‐renewal and multipotent capacity of Lgr5+ ISCs from Srcap fl/fl;Lgr5‐Cre and Srcap fl/fl mice can be confirmed by lineage tracing assay (Barker et al, 2007). We crossed Srcap fl/fl;Lgr5 GFP‐CreERT2 mice with Rosa26 lsl‐YFP mice to generate Srcap +/+;Lgr5 GFP‐CreERT2;Rosa26 lsl‐YFP (termed as LR YFP) and Srcap fl/fl;Lgr5 GFP‐CreERT2;Rosa26 lsl‐YFP (termed as Srcap fl/fl;LR YFP) mice. Of note, Srcap fl/fl;LR YFP mice showed impaired intestinal epithelial regeneration after TAM treatment (Figs 2E and EV2L). In addition, Srcap KO ISCs displayed decreased proliferation compared with Srcap WT ISCs (Fig 2F). In addition, we found that Srcap knockout mainly suppressed cell cycling, but not affect their cell death (Fig EV2M), suggesting that the reduction of cell numbers in Srcap‐deficient mice was due to impairment of self‐renewal maintenance of ISCs. Besides their self‐renewal capacity, ISCs can differentiate into five major epithelial cell types in the intestinal epithelium, including enterocytes, goblet, enteroendocrine, Paneth, and tuft cells (Barker, 2014). We observed that Srcap fl/fl;Lgr5‐Cre mice showed reduced numbers of intestinal epithelial cells by multiple‐color staining (Figs 2G and EV2N). These results suggest that SRCAP is required for the maintenance of Lgr5+ ISC self‐renewal and intestinal epithelial regeneration.
SRCAP associates with REST in ISCs
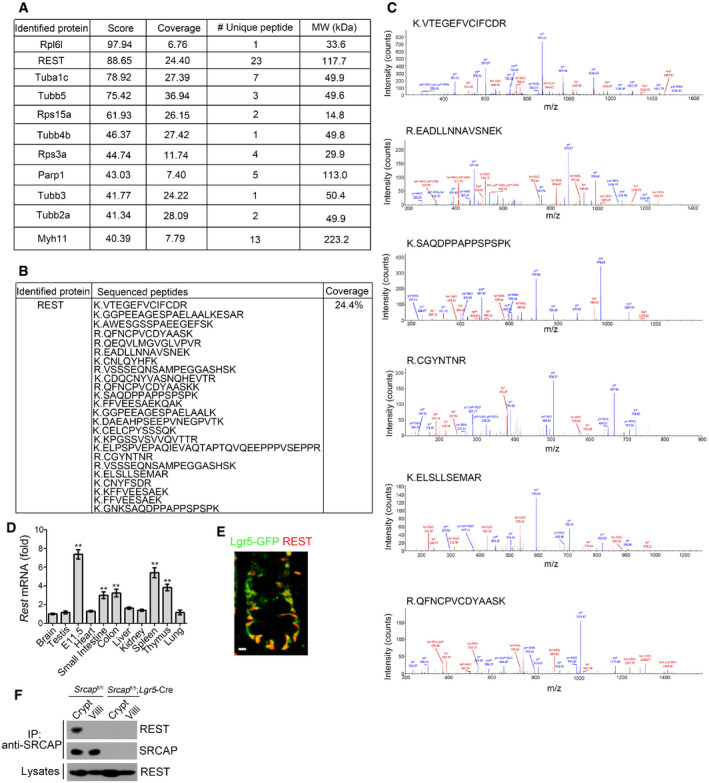
To further identify potential SRCAP‐associated protein candidates, we conducted a co‐immunoprecipitation (co‐IP) assay through mouse intestinal crypt lysates by anti‐SRCAP antibody. Of interest, RE‐1 silencing transcription factor (REST) was identified to bind SRCAP by mass spectrometry (Figs 3A and EV3A–C). REST and CoREST are components of a larger complex, consisting minimally of HDAC/CoREST/LSD1/REST (HCLR) repressor complex (Zhou et al, 2013a). It has been reported that REST is involved in the regulation of neurogenesis and insulin secretion via mediating transcriptional repression or activation in a cell type‐ and gene‐specific manner (Kuwabara et al, 2004; Thiel et al, 2015). Of note, Rest was highly expressed in mouse embryos, spleen, thymus, small intestine, and colon by real‐time PCR analysis (Fig EV3D). REST protein was predominantly distributed in the Lgr5‐GFP+ ISCs in the bottom of intestinal crypts compared with villus enterocytes (Fig EV3E). Moreover, REST was co‐localized with SRCAP in the nuclei of Lgr5‐GFP+ ISCs at the base of mouse intestinal crypt (Fig 3B). Through domain mapping, the C‐terminal domain of REST was sufficient and essential for SRCAP binding (Fig 3C–E). Of note, REST associated with SRCAP complex component proteins such as Yeats4 and YL1 in mouse intestinal crypt lysates, rather than its repressor complex partners CoREST, HDAC1, and LSD1 (Fig 3F). Furthermore, increased doses of SRCAP could competitively bind REST against CoREST (Fig 3G), suggesting the SRCAP protein inhibited the association of REST with CoREST. Interestingly, we observed that SRCAP really interacted with REST in crypt lysates, but not in villi lysates (Fig EV3F), suggesting that the interaction of SRCAP with REST exists in the crypt cells and is involved in the regulation of determining stem cell fitness. These data indicate that SRCAP associates with REST protein to form Yeats4‐contained active SRCAP complex rather than the HCLR repressor complex in ISCs.
Figure 3. SRCAP associates with REST .
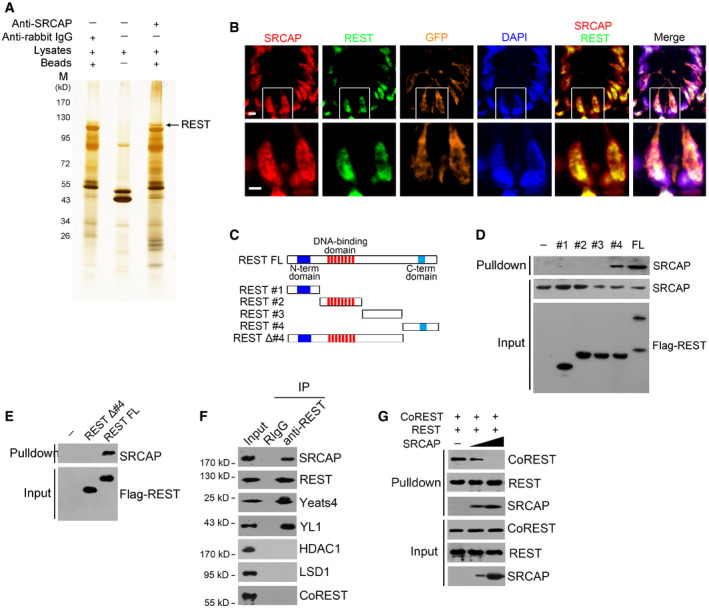
- Nuclear extracts of murine intestinal crypts were immunoprecipitated with anti‐SRCAP antibody or rabbit IgG followed by ADASDS–PAGE, sliver staining, and mass spectrometry.
- SRCAP and REST were stained in murine intestinal crypts. Scale bar, 10 μm.
- A schematic diagram of full length and truncated fragments of REST protein was shown.
- Flag‐tagged REST fragments were expressed in 293T and purified by immunoprecipitation with anti‐Flag antibody. REST proteins were incubated with crypt lysates for pulldown followed by immunoblotting.
- C‐terminal domain of REST was sufficient and essential for SRCAP association by pulldown assay.
- Intestinal crypt lysates were incubated with anti‐REST antibody, followed by immunoblotting with indicated antibodies.
- Intestinal crypt lysates were incubated with anti‐REST antibody and protein A/G beads in 4°C overnight. Recombinant SRCAP protein was added with different concentrations, followed by immunoblotting.
Figure EV3. SRCAP associates with REST.
-
A–CIdentification of Rest as a SRCAP binding protein. Candidate interactors of SRCAP were identified by mass spectrometry LTQ Orbitrap XL.
-
DRest expression in murine different tissues was examined by real‐time PCR. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
-
ERest expression was visualized in indicated intestinal tissues by immunofluorescence staining. Green: Lgr5‐GFP; red: Rest. Scale bar, 10 μm.
-
FIntestinal crypt and villi lysates were incubated with anti‐SRCAP antibody and protein A/G beads in 4°C overnight followed by immunoblotting.
Source data are available online for this figure.
REST recruits SRCAP onto Prdm16 promoter for its transcription
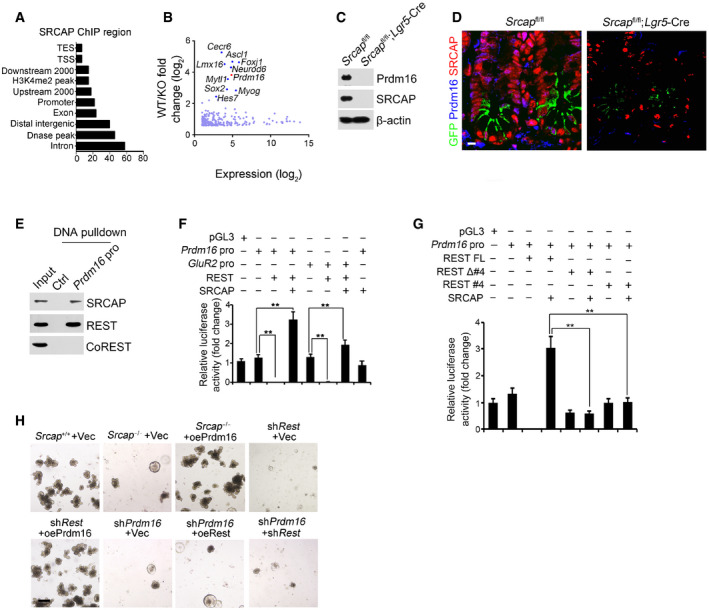
The major role of SRCAP complex can exchange the histone H2A to its variant H2A.Z in the nucleosomes for gene accessibility regulation (Mizuguchi et al, 2004). However, genome‐wide target screening of SRCAP has not been characterized yet. We next performed chromatin immunoprecipitation sequencing (ChIP‐seq) against SRCAP to identify SRCAP binding DNA regions in ISCs. Through analysis of our ChIP‐seq data, SRCAP was prone to interact with transcription‐coupled regions (Fig 4A). We also performed transcriptome RNA‐seq analysis between Srcap −/− and WT ISCs. 4,967 genes (3,399 upregulated and 1,568 downregulated, fold change > 1.5, FDR < 0.05) were differentially expressed in Srcap KO ISCs versus WT ISCs (Fig 4B). Of note, Prdm16 was one of dramatically downregulated transcription factors (TFs) in Srcap KO ISCs (Fig 4B). In addition, we compared TFs among SRCAP target genes in ISCs, ISC signature genes (GSE92332) (Haber et al, 2017), and REST target genes (Lunyak et al, 2002). Intriguingly, Prdm16 is the only TF fitted all of these features (Fig EV4A). It has been reported that Prdm16 is involved in the stemness regulation of hematopoietic stem cells, neural stem cells, and adipose tissue stem cells (Yin et al, 2013; Shimada et al, 2017; Corrigan et al, 2018). In intestinal epithelium, Prdm16 can maintain homeostasis by controlling fatty acid oxidation (Stine et al, 2019). Of note, SRCAP was enriched in Prdm16 promoter in ISCs (Fig EV4B). Through DNase I‐accessible chromatin assay, Srcap deficiency in ISCs caused reduced chromatin accessibility on Prdm16 locus (Fig EV4C). Consistently, we found that Srcap KO abolished Prdm16 expression in ISCs (Figs 4C and D, and EV4D).
Figure 4. REST recruits SRCAP onto Prdm16 promoter for its transcription.
-
AAnalysis for genome‐wide SRCAP binding regions in ISCs from ChIP‐seq was shown. Coordinate information for sequence alignment was from UCSC (intron and exon), Ensemble GRCM38 (mm10, release‐90) (upstream 2,000 bp, downstream 2,000 bp, distal intergenic region, TSS (transcription start site), TES, and promoter), GSE57919 (Dnase‐seq peaks, liftover to mm10), and GSE34568 (H3K4me2, liftover to mm10). Distribution of SRCAP peaks in ChIP‐seq and its overlap with each kind of region was analyzed by bedtools.
-
BTranscriptome microarray analysis between Srcap −/− ISCs and WT ISCs. Top ten downregulated TFs in Srcap −/− ISCs were listed.
-
CPrdm16 expression was tested in Srcap +/+ and Srcap −/− ISCs by Western blotting.
-
DPrdm16 expression was visualized in indicated intestinal tissues by immunofluorescence staining. Green: GFP; red: SRCAP; blue: Prdm16. Scale bar, 10 μm.
-
EPrdm16 promoter (−875˜−854) region was used as probe to perform DNA‐pulldown assay with murine intestinal crypt lysates followed by immunoblotting.
-
F, GIndicated promoter activation was analyzed with luciferase assay. DKK1 was used as a control. Results of relative fold changes are shown as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
-
H1 × 104 ISCs from indicated mice were collected for organoid formation. For gene depletion or restoration, ISCs were infected with lentivirus. Scale bar, 200 μm.
Figure EV4. SRCAP deposits onto Prdm16 promoter for its transcription and activate downstream Ppard expression.
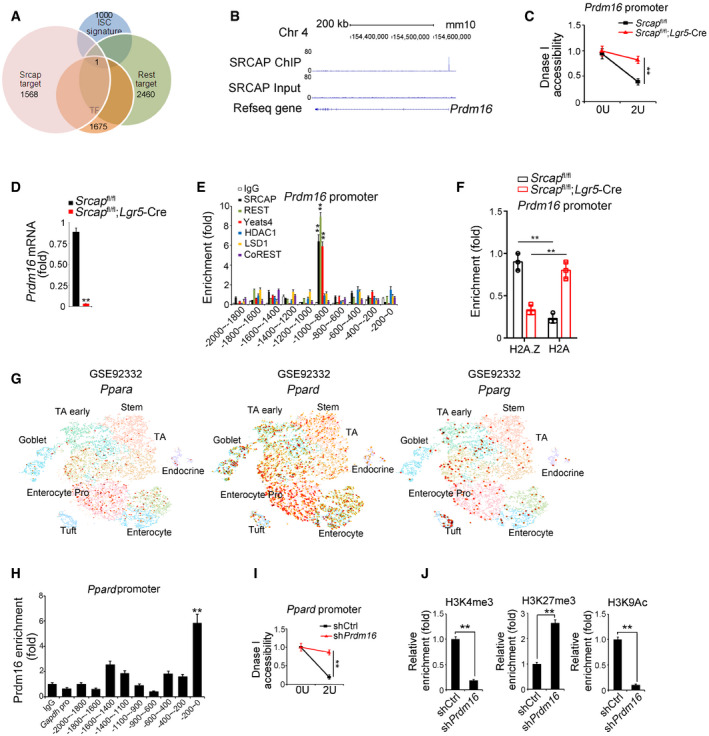
- Target genes of SRCAP and REST in mouse ISCs were analyzed by intersection analysis. Gene clusters of downregulation genes in Srcap KO ISCs, transcriptional factors, genes harbor REST binding site in promoter, and ISC signature genes identified by intestinal epithelium single‐cell RNA‐seq (GSE92332) were used for analysis. Prdm16 was the only one of SRCAP‐targeted TF that harbors REST binding site in its promoter, which was also identified to be one of ISC signature genes by intestinal epithelium single‐cell RNA‐seq (GSE92332).
- SRCAP deposited to Prdm16 promoter region near the TSS from ChIP‐seq data. Prdm16 promoter regions from ChIP‐seq data were analyzed by IGV software.
- Nuclei were extracted from ISCs followed by DNase I digestion. Total DNA was extracted and quantitated by qPCR with Prdm16 promoter‐specific primers. Primers were listed in Table EV1. Relative fold changes were counted as means ± SD. **P < 0.01 by two‐tailed Student's t‐test.
- Prdm16 expression was analyzed by real‐time qPCR in Srcap +/+ and Srcap −/− ISCs. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
- ISCs were lysed for ChIP assays. Indicated proteins enriched to Prdm16 promoter were examined by real‐time qPCR. Signals were normalized to input DNA. Relative fold changes were calculated as means ± SD. Data represent five independent replicates. ** p < 0.01 **P < 0.01 by two‐tailed Student's t‐test.
- Indicated ISCs were lysed for ChIP assays. H2A.1 and H2A enriched to Prdm16 promoter were examined by real‐time qPCR. Signals were normalized to input DNA. Relative fold changes were calculated as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
- Expression levels and patterns of PPAR components in murine intestinal epithelium were analyzed based on a single‐cell RNA‐seq dataset (GSE92332).
- Prdm16 was recruited near the TSS region of Ppard promoter in ISCs. ISCs were lysed for ChIP assay with anti‐Prdm16 antibody. Prdm16 protein enriched to Ppard promoter was examined by real‐time qPCR. Signals were normalized to input DNA. Primers were listed in Table EV1. IgG and Prdm16 proteins enriched on Gapdh promoter were used as negative controls. Relative fold changes were calculated as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
- Nuclei were extracted from ISCs followed by DNase I digestion. Total DNA was extracted and quantitated by qPCR with Ppard promoter‐specific primers. Primers were listed in Table EV1. Relative fold changes were counted as means ± SD. **P < 0.01.
- ISCs were lysed for ChIP assay. Indicated proteins enriched to Ppard promoter were examined by real‐time qPCR. Signals were normalized to input DNA. Enrichment fold changes were counted as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
Through ChIP‐qPCR assay, SRCAP protein bound to −1,000~−800 bp region of Prdm16 promoter (Fig EV4E). The binding region of Prdm16 promoter also enriched Yeats4, another SRCAP complex component (Fig EV4E). By contrast, the HCLR repressor complex partners such as CoREST, HDAC1, and LSD1 did not bind to this region of Prdm16 promoter in ISCs (Fig EV4E). The Prdm16 promoter contains a potential RE‐1 site at −875~−854 region. We used this fragment as a probe to conduct DNA‐pulldown assay through mouse intestinal crypt lysates. As expected, Prdm16 promoter associated with SRCAP and REST (Fig 4E). Moreover, H2A.Z was enriched on Prdm16 promoter in ISCs (Fig EV4F). Srcap depletion impaired enrichment of H2A.Z on Prdm16 promoter, while enhanced enrichment of H2A on this promoter, which suggested that SRCAP regulated histone H2A.Z/H2A exchange in nucleosomes for initiation of Prdm16 transcription in ISCs (Fig EV4F).
Through luciferase assay, SRCAP and REST could enhance Prdm16 transcription (Fig 4F). Additionally, REST‐induced Prdm16 transcription was dependent on its SRCAP binding and DNA binding domains (Fig 4G). To further explore the mechanism of SRCAP and REST in regulating Prdm16 expression, we depleted or overexpressed these genes in ISCs and followed by intestinal organoid formation assay. We observed that Srcap deletion remarkably inhibited ISC organoid formation (Fig 4H). Notably, restoration of Prdm16 in Srcap −/− ISCs was able to rescue ISC self‐renewal (Fig 4H). Moreover, Rest depletion dramatically impaired organoid formation, which could be rescued by Prdm16 overexpression (Fig 4H). Finally, Prdm16 knockdown also abrogated organoid formation, which could not be rescued by restoration or depletion of Rest (Fig 4H). These results suggest that Prdm16 is a downstream target gene of Rest and SRCAP. Collectively, SRCAP recruits REST protein onto the Prdm16 promoter to initiate its transcription.
Prdm16 initiates PPARδ expression to promote ISC stemness
Prdm16 takes part in the regulation of multiple signaling pathways such as PPAR and transforming growth factor‐β (TGFβ) (Ohno et al, 2012; Hasegawa et al, 2018). We noticed that PPARδ target genes were markedly downregulated in Srcap KO ISCs through gene set enrichment analysis (GSEA) (Fig 5A). PPARs are a group of nuclear receptor proteins that function as transcription factors. PPARs play important roles in the regulation of development, metabolism, and tumorigenesis (Dubois et al, 2017). It has been reported that PPARδ is implicated in high‐fat diet‐induced stemness enhancement of ISCs (Beyaz et al, 2016). Through analysis of single‐cell RNA‐seq dataset (GSE92332), PPARδ was the most highly expressed PPAR molecules in mouse intestinal epithelium (Fig EV4G). We observed that Srcap‐deficient ISCs drastically reduced expression levels of PPARδ target genes (Fig 5B). Of note, PPARδ was substantially decreased in Srcap KO crypts versus Srcap WT crypts (Fig 5C).
Figure 5. Prdm16 initiates PPARδ expression to promote ISC stemness.
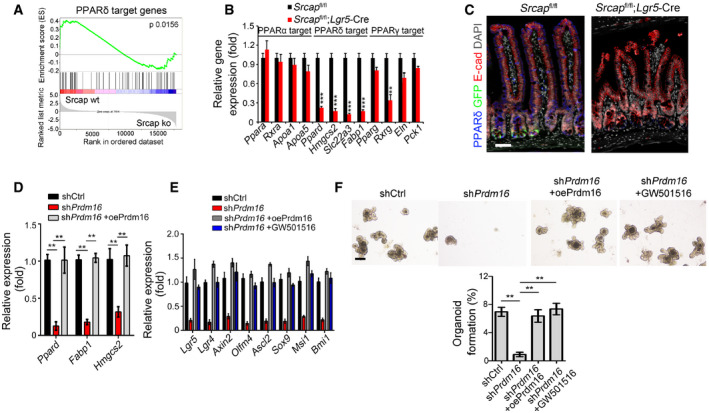
- WT and Srcap −/− ISCs were conducted for transcriptome microarray, followed by gene set enrichment analysis (GSEA) analysis for PPARδ target genes.
- Expression levels of PPARs and PPAR target genes were assessed in sorted ISCs by quantitative RT–PCR. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates. **P < 0.01 and ***P < 0.001 by two‐tailed Student's t‐test.
- PPARδ expression was visualized in intestinal tissues by immunofluorescence staining. Scale bar, 50 μm.
- ISCs were collected for organoid formation followed by quantitative RT–PCR. For Prdm16 overexpression, ISCs were infected with lentivirus carrying Prdm16 overexpressing plasmids. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates. **P < 0.01 by two‐tailed Student's t‐test.
- mRNA levels of ISC signature genes were examined by quantitative RT–PCR. Primers were listed in Table EV1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. Data represent five independent replicates.
- Representative images of organoid formation from indicated ISCs were shown (upper panel). Scale bar, 200 μm. Organoid formation ratios were shown as means ± SD. **P < 0.01 by two‐tailed Student's t‐test (lower panel). n = 6 for each group. GW501516 (10 μM) were added directly into organoid culture medium.
In addition, Prdm16 was enriched near the transcription start site (TSS) region (‐200~0 bp) of Ppard promoter in ISCs (Fig EV4H). We noticed that Prdm16 depletion in ISCs dramatically abolished chromatin accessibility of Ppard promoter (Fig EV4I). Moreover, Prdm16 depletion also impaired enrichment of H3K4 trimethylation (H3K4me3), and H3K9 acetylation (H3K9Ac) on Ppard promoter, while enhanced enrichment of H3K27 trimethylation (H3K27me3) on Ppard promoter (Fig EV4J). Consequently, Prdm16 depletion in ISCs dramatically suppressed expression of Ppard and its target genes (Fig 5D). In addition, Prdm16 depletion remarkably inhibited expression of ISC marker genes in Prdm16 silenced organoids (Fig 5E). Of note, restoration of Prdm16 could rescue the expression of ISC markers in Prdm16 silenced organoids (Fig 5F). In parallel, the PPAR agonist GW501615 could also restore the expression of ISC markers in Prdm16 silenced organoids (Fig 5F). Finally, both Prdm16 restoration and GW501615 could rescue the self‐renewal capacity of Prdm16‐depleted ISCs (Fig 5F). Taken together, Prdm16 initiates PPARδ expression to promote self‐renewal of ISCs.
Deletion of Rest or Prdm16 abrogates the stemness of ISCs
To further determine the physiological role of Rest in the self‐renewal regulation of ISCs, we generated Rest fl/fl;Lgr5 GFP‐CreERT2 mice through crossing Rest fl/fl mice with Lgr5 GFP‐CreERT2 mice. After TAM administration, Rest was completely deleted in Lgr5+ ISCs (Fig EV5A; hereafter, TAM‐treated Rest fl/fl mice are called as Rest WT, whereas TAM‐treated Rest fl/fl;Lgr5‐Cre mice are referred to as Rest KO). We observed that Rest deficiency in ISCs caused body weight loss and over half of these mice died 1 month after tamoxifen treatment (Fig EV5B and C). We observed that Rest KO mice displayed shorter crypts and villi in length compared with Rest WT mice, similar phenotype to Srcap KO mice (Figs 6A and EV5D). In addition, Rest KO ISCs showed impaired capacity of organoid formation (Fig 6B). Rest KO mice showed reduced number of Olfm4+ ISCs (Figs 6C and EV5E). We noticed that Rest KO mice showed reduced ISC numbers similar to Srcap KO mice (Fig 6D).
Figure EV5. Generation strategies of Rest KO, Prdm16 KO and Ppard KO mice. Generation strategies of Rest KO, Prdm16 KO and Ppard KO mice.
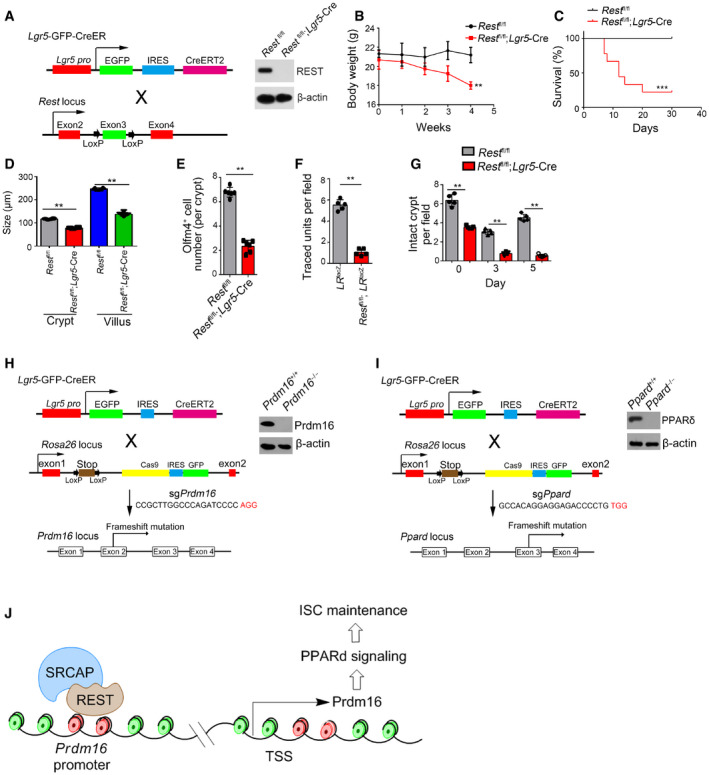
- Schematic diagram of Rest KO mice and validation by immunoblotting.
- Body weight changes of Rest fl/fl and Rest fl/fl; Lgr5 GFP‐CreERT2 mice after TAM treatment were shown as means ± SD n = 10. **P < 0.01 by two‐tailed Student's t‐test.
- Kaplan–Meier survival curves of Rest fl/fl and Rest fl/fl; Lgr5 GFP‐CreERT2 mice after TAM treatment. n = 10. ***P < 0.001 by log‐rank test.
- Biological variations and statistical tests were shown as in Fig 6A. Length of villi and crypts (means per mouse) was measured and shown as means ± SD n = 6. **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests were shown as in Fig 6C. OLFM4+ ISC numbers per crypt (means per mouse) were shown as means ± SD n = 6. **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests were shown as in Fig 6E. Numbers of traced crypt‐villus units (means per mouse) were shown as means ± SD n = 5. **P < 0.01 by two‐tailed Student's t‐test.
- Biological variations and statistical tests were shown as in Fig 6F. Numbers of intact crypts (means per mouse) were shown as means ± SD n = 5. **P < 0.01 by two‐tailed Student's t‐test.
- Schematic diagram of Prdm16 KO by CRISPR/Cas9 technology and validation by immunoblotting.
- Schematic diagram of Ppard KO by CRISPR/Cas9 technology and validation by immunoblotting.
- A work model represents the mechanism of SRCAP regulating ISC self‐renewal. SRCAP recruits REST protein onto the Prdm16 promoter to induce its transcription, which promotes PPAR‐δ signaling activation that maintains ISC self‐renewal.
Source data are available online for this figure.
Figure 6. Rest or Prdm16 deletion abrogates the stemness of ISCs.
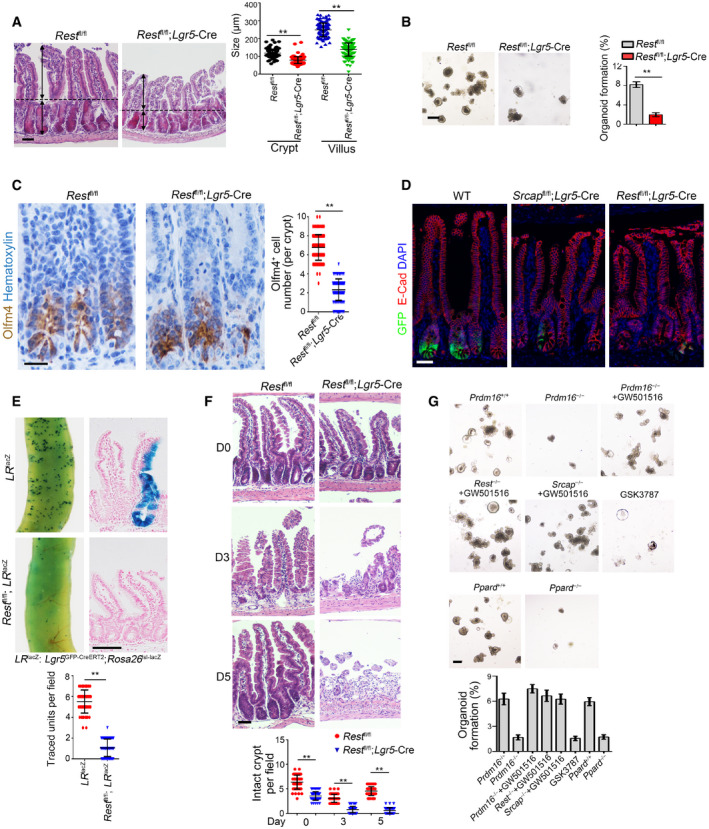
- Representative images of intestines from 8‐week‐old Rest +/+ and Rest –/– mice (n = 6 per group). Length of 120 villi and 120 crypts (20 villi and 20 crypts per mouse) was measured in right panel as means ± SD. **P < 0.01 by two‐tailed Student's t‐test. Scale bar, 100 μm. Dotted lines indicate the boundary between crypts and villi. Arrows indicate the length of crypts or villi.
- 1 × 104 ISCs from indicated mice were collected for organoid formation. Scale bar, 200 μm. n = 3 for each group. Organoid formation ratios were shown as means ± SD. **P < 0.01 by two‐tailed Student's t‐test.
- Intestine tissues from 8‐week‐old Rest +/+ and Rest –/– mice (n = 6 per group) were collected for immunohistochemistry staining. Typical images were shown in left panel and statistical ISC numbers from 120 crypts (20 crypts per mouse) were shown in right panel as means ± SD. **P < 0.01 by two‐tailed Student's t‐test. Scale bar, 50 μm.
- Immunofluorescence staining of intestine tissues from Srcap fl/fl; Lgr5 GFP‐CreERT2 and Rest fl/fl; Lgr5 GFP‐CreERT2 mice. Scale bar, 50 μm.
- Lgr5 GFP‐CreERT2;Rosa26 lsl‐lacZ (LR lacZ) were crossed with Rest −/− mice, followed by TAM administration for lineage tracing for intestinal whole‐mount staining for β‐gal (upper panel). Mice were sacrificed after 1 week, and typical sections were shown (middle panel). Scale bar, 200 μm. Numbers of traced crypt‐villus units were calculated as means ± SD. **P < 0.01 by two‐tailed Student's t‐test in right panel. 50 fields (10 fields per mouse) were calculated for each group (n = 5) (lower panel).
- Rest +/+ and Rest –/– mice were treated with 10 Gy's radiation and sacrificed at the indicated time points. Intestinal tissues were collected for H&E staining. Scale bars, 100 μm. Numbers of intact crypts were shown in lower panel as means ± SD. **P < 0.01 by two‐tailed Student's t‐test. 50 fields (10 fields per mouse) were taken for each group (n = 5).
- Representative images of organoid formation from indicated ISCs were shown (upper panel). Scale bar, 200 μm. n = 6 for each group. Organoid number per well was counted as means ± SD (lower panel). Lentivirus‐carried sgPrdm16 and sgPpard were prepared in 293T cells and infected into ISCs derived from TAM‐treated LR Cas9 mice to obtain Prdm16 −/− and Ppard −/− ISCs. 1 × 104 ISCs from indicated mice were collected for organoid formation. Scale bar, 200 μm. n = 5 for each group.
We next generated Lgr5 GFP‐CreERT2;Rosa26 lsl‐LacZ (termed as LR LacZ) and Rest fl/fl; Lgr5 GFP‐CreERT2;Rosa26 lsl‐LacZ (termed as Rest fl/fl;LR LacZ) mice for lineage tracing assay through crossing Rest fl/fl; Lgr5 GFP‐CreERT2 mice with Rosa26 lsl‐LacZ mice. We observed that Rest fl/fl;LR LacZ mice showed reduced ISC numbers and impaired intestinal epithelial regeneration after TAM treatment (Figs 6E and EV5F). Post‐irradiation injury, Rest fl/fl; Lgr5 GFP‐CreERT2mice displayed impaired intestinal epithelial regeneration capacity compared with Rest fl/fl WT mice (Figs 6F and EV5G). Therefore, Rest deletion abolishes ISC self‐renewal capacity.
We then generated Rosa26 lsl‐Cas9;Lgr5 GFP‐CreERT2 (LR Cas9) mice through crossing Rosa26 lsl‐Cas9 with Lgr5 GFP‐CreERT2 mice and followed by organoid formation assay. We found that Prdm16 deletion abrogated organoid formation of ISCs (Figs 6G and EV5H). By contrast, the PPAR agonist GW501516 (Beyaz et al, 2016) could rescue the self‐renewal capacity impaired by Prdm16‐depleted ISCs (Fig 6G). However, the PPAR antagonist GSK3787 abrogated organoid formation of ISCs (Fig 6G). Finally, we observed that Ppard deletion also impaired organoid formation of ISCs (Figs 6G and EV5I). Collectively, deletion of Rest or Prdm16 abrogates the stemness of ISCs.
Discussion
Adult intestinal stem cells have self‐renewal and differentiation features under a homeostatic condition (Barker, 2014). The ability of ISCs to indefinitely self‐renew also in turn makes stem cells prime candidates for accumulating mutations that drives oncogenesis. In this study, we show that SRCAP is highly expressed in intestinal epithelium and ISCs. Srcap conditional knockout in ISCs impairs the self‐renewal of ISCs and intestinal epithelial regeneration. Mechanistically, SRCAP recruits REST onto the Prdm16 promoter to induce its transcription, which initiates PPARδ signaling activation. Rest or Prdm16 deficiency abrogates the self‐renewal of ISCs and intestinal epithelial regeneration (Fig EV5J).
Chromatin remodeling complexes (also known as remodelers) function in a wide range of biological processes including development, stemness maintenance, immune response, and tumorigenesis (Bajpai et al, 2010; Wei et al, 2018). Accumulating evidence shows that the chromatin remodeling complexes exert important roles in the regulation of gene expression. The SRCAP subunit confers its ATPase activity for the SRCAP complex, which exchanges histone H2A to its variant H2A.Z in nucleosomes for activation of gene transcription as an active remodeler (Watanabe et al, 2013). We recently showed that SRCAP protein participates in the regulation of self‐renewal maintenance of ESCs and lymphoid lineage commitment (Ye et al, 2017, 2018b). Another subunit of the SRCAP chromosome remodeling complex, ZNHIT1, has recently been reported to regulate intestinal homeostasis postnatal establishment by promoting H2A.Z interaction (Zhao et al, 2019). Here, we showed that Srcap knockout causes early embryonic lethality, suggesting SRCAP plays a pivotal role in development. Intriguingly, SRCAP is predominantly distributed in the bottom of crypts including Lgr5+ ISCs. SRCAP is required for the self‐renewal of ISCs and intestinal epithelial regeneration. Here, we reveal the in vivo physiological role of SRCAP protein in the regulation of ISC stemness maintenance.
REST binds to specific genomic regions containing RE1 consensus sites within target gene promoters for regulation of their expression. REST can recruit many epigenetic and transcriptional modulators, often with the assistance of scaffold protein CoREST (Ballas et al, 2005). The interaction of REST and CoREST can further assemble with histone deacetylase HDAC and demethylase LSD1 to form the HCLR repressor complex. The HCLR repressor complex is initially identified to repress neuronal genes in non‐neuronal cells (Lunyak et al, 2002). REST also contains a DNA binding domain that anchors proteins to DNA (Thiel et al, 2015). Of note, conformational change of REST can adjust its transcriptional activities, even converting REST from a transcriptional repressor to a transcriptional activating factor (Kuwabara et al, 2004). It has been reported that REST can regulate the balance of stemness versus differentiation and lineage specification of embryonic and neural stem cells (Singh et al, 2008; Mukherjee et al, 2016). These processes involve the direct repression of differentiation‐promoting genes, especially of the neural lineage. However, the physiological role of REST in intestinal stem cells has not been defined yet. Here, we show that REST binds to SRCAP to impede its association with CoREST, leading to assembly of the Yeats4‐contained active complex in ISCs. This active SRCAP complex recruits onto the Prdm16 promoter to trigger its transcription, which initiates PPARδ activation to maintain ISC self‐renewal. Thus REST is required for the self‐renewal maintenance of ISCs.
PPAR family members play critical roles in the regulation of development, differentiation, lipid metabolism, and tumorigenesis (Ohno et al, 2012; Lee et al, 2015). PPARδ has proposed to modulate the inflammatory response by NF‐κB‐mediated signaling (Zingarelli et al, 2010) and to play an inhibitory role in the development of atherosclerosis (Li et al, 2004). In gut, PPARδ is the predominant member of PPAR family and is expressed in ISCs and progenitor cells. High‐fat diet enhances stemness and tumorigenicity of intestinal progenitors in a PPARδ‐dependent manner, which induces Wnt/β‐catenin activation to sustain ISC self‐renewal maintenance (Beyaz et al, 2016). However, how PPARδ regulates ISC self‐renewal is still unknown. Here, we show that PPARδ is the most highly expressed PPAR molecules in murine intestinal epithelium and ISCs. Prdm16 initiates PPARδ expression as an upstream TF to promote self‐renewal of ISCs. In summary, our findings reveal that the SRCAP‐REST‐Prdm16‐PPARδ axis is required for the self‐renewal maintenance of Lgr5+ ISCs.
Materials and Methods
Antibodies and reagents
Antibodies against mouse Olfm4 (14369), DCLK1 (62257), MMP7 (3801), E‐cadherin (3195), H2A.Z (2718), H2A (12349), H3K9Ac (9649), H3K4me3 (9751), and H3K27me3 (9733) were purchased from Cell Signaling Technology (Danvers, USA). Antibody against mouse β‐actin (clone AC‐74, A2228), anti‐HA‐tag (clone HA‐7, H9658), and anti‐Flag‐tag (clone M2, F3165) were from Sigma‐Aldrich (St. Louis, USA). Antibodies against GFP (ab183735), Ki67 (ab15580), and PCNA (ab29) were purchased from Abcam (Cambridge, USA). Anti‐mouse or human SRCAP antibody (ESL104) was from Kerafast (Boston, USA). Anti‐YL1 antibody (orb315826) was from Biorbyt (Cambridge, UK). Antibodies against TFF3 (23277‐1‐AP), CHGA (23342‐1‐AP), PPARδ (10156‐2‐AP), CoREST (27686‐1‐AP), and LSD1 (20813‐1‐AP) were purchased from Proteintech (Chicago, USA). Anti‐mouse CD326 (EpCAM) (11‐5791‐80) were purchased from eBioscience (San Diego, USA). WGA Lectin (FITC) (GTX01502) and anti‐Yeats4 antibody (GTX16452) were from GeneTex (Irvine, USA). Anti‐Rest antibody (bs‐2590R) was purchased from Bioss. Anti‐YFP (orb334991) antibody was from Biorbyt (Cambridge, UK). Secondary antibodies conjugated with Alexa‐594, Alexa‐488, Alexa‐405, or Alexa‐649 were purchased from Molecular probes Inc (Eugene, USA). HRP‐conjugated secondary antibodies were from Sungene Biotech (Tianjin, China). OpalTM 7‐color fIHC kit (MEL797001KT) was from PerKinElmer (Waltham, USA). Polymer HRP and AP detection kits were from GBI labs (Bothell, USA). GW501516 (SML‐1491) was purchased from Sigma‐Aldrich (St. Louis, USA). GSK3787 (S8025) was purchased from Selleck (Houston, USA). DNase I was purchased from Roche Molecular Biochemicals (Basel, Switzerland). Protein A/G beads were from Santa Cruz Biotechnology (Santa Cruz, USA). Protein A agorose/Salmon sperm DNA beads for ChIP (16‐157) were from Millopore (Millerica, USA). SuperReal premix plus qPCR buffer was from TIANGEN Biotech (Beijing, China). N2 supplement and B27 supplement were from Invitrogen. DAPI, N‐acetylcysteine, Y27632, and hyaluronidase were from Sigma‐Aldrich. Wnt3a, RSPO1, Noggin, and EGF proteins were purchased from PeproTech (Suzhou, China). Matrigel (356234) was from Corning (Corning, USA). IntestiCult organoid growth medium Kit (06005) was from StemCell (Vancouver, Canada).
Cell culture and plasmid construction
Human 293T cells (ATCC, CRL‐3216) were cultured with DMEM supplemented with 10% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin. Human HCT‐116 cells (ATCC, CCL‐247) were cultured with RPMI‐1640 supplemented with 15% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin. Lentivirus infecting primary crypts or sorted Lgr5+ ISCs were produced in 293T cells by using the standard protocols. Transfection was performed using Lipofectin reagent (Invitrogen). Cell lines were obtained from ATCC and authenticated by PCR. Mycoplasma contamination had been tested by PCR and excluded. Experiments were approved by the Institutional Medical Research Ethics Committee at Institute of Biophysics, Chinese Academy of Sciences.
For shRNA knockdown and overexpression experiments, shRNAs were constructed into pSicoR plasmid. Full length of Rest or Prdm16 was constructed into pSIN plasmid. Lentivirus was produced in 293T cells using the standard protocols. Transfection was performed using Lipofectin (Invitrogen). Most efficient shRNA among 3 shRNA constructs was screened out for following experiments. At least four independent experiments were performed as biological replicates. shRNA sequences were listed in Table EV2. For pulldown assay, the mouse Rest cDNA (FL: 1–3,249 bp) and truncations (#1: 1–450 bp, #2: 451–1,290 bp, #3: 1,291–2,850 bp, #4: 2,851–3,249 bp, Δ#4: 1–2,850 bp) were cloned into p3Xflag‐CMV‐9 expression vector. Plasmids were transfected into 293T cells. After 48 h, cells were harvested and lysed. Flag‐tagged proteins were immunoprecipitated by anti‐Flag antibody followed by 3XFlag peptide elution. For CRISPR/Cas9‐mediated gene editing, sgRNA sequences were cloned into lentiCRISPRv2‐GFP plasmid, and gRNA sequences were listed in Table EV2. LentiCRISPRv2, pVSVg (8454), and psPAX2 (12260) were transfected into 293T cells to generate CRISPR/Cas9 lentivirus (Zhu et al, 2018). Rosa26 lsl‐Cas9 mice were crossed with Lgr5 GFP‐CreERT2 mice to generate Rosa26 lsl‐Cas9; Lgr5 GFP‐CreERT2 (LR Cas9) mice. TAM (10 μg/μl) was intraperitoneally injected into mice 3 days before ISC sorting. ISCs were infected with lentivirus followed by organoid culture.
Generation of knockout mice and mouse strains
Srcap flox/flox mice were generated by a CRISPR‐Cas9 approach (Zhu et al, 2014; Liu et al, 2017). Briefly, the DNA template corresponding to a single‐strand DNA fragment comprising LoxP cassette and homology arms derived from the exon5 of Srcap was purified and mixed with Cas9 mRNA and corresponding sgRNAs (Table EV2) as described (Yoshimi et al, 2016). The injected zygotes were transferred into the uterus of pseudopregnant ICR females. All mouse genotypes were identified by PCR screening and DNA sequencing. Rosa26 lsl‐Cas9 mice were purchased from the Jackson Laboratory. Lgr5 GFP‐CreERT2 mice were purchased from Model Animal Research Center of Nanjing University. Rosa26 YFP and Rest flox/flox mice were from Shanghai Biomodel Organism Science & Technology Development Co., Ltd. Rosa26 lacZ mice were obtained from Shanghai Bioray Laboratory. All the mice are C57BL/6 background and maintained under specific pathogen‐free conditions.
Srcap flox/flox mice were crossed with Lgr5 GFP‐CreERT2 mice to generate Srcap flox/flox ; Lgr5‐Cre+ mice. Srcap +/+ ; Lgr5‐Cre+ mice were used as littermate control. Rest flox/flox mice were crossed with Lgr5 GFP‐CreERT2 mice to generate Rest flox/flox ; Lgr5‐Cre+ mice. Rest +/+ ; Lgr5‐Cre+ mice were used as littermate control. For lineage tracing assay, Srcap flox/flox; Lgr5 GFP‐CreERT2 mice were crossed with Rosa26 lsl‐YFP mice and obtained Srcap +/+; Lgr5 GFP‐CreERT2; Rosa26 lsl‐YFP together with Srcap flox/flox; Lgr5 GFP‐CreERT2;Rosa26 lsl‐YFP mice. Rest flox/flox; Lgr5 GFP‐CreERT2 mice were crossed with Rosa26 lsl‐LacZ mice and obtained Rest +/+; Lgr5 GFP‐CreERT2; Rosa26 lsl‐LacZ together with Rest flox/flox; Lgr5 GFP‐CreERT2; Rosa26 lsl‐LacZ mice. 200 μl tamoxifen in sunflower oil (10 μg/μl) was intraperitoneally injected into Lgr5 CreERT2 mice at 60 days old to induce gene deletion. Phenotypes were observed 2 weeks after tamoxifen induction. Animal use and protocols were approved by the Institutional Animal Care and Use Committees at Institute of Biophysics, Chinese Academy of Sciences. We used littermates with the same age (8–12 weeks old) and gender for each group. We excluded the mice 5 g thinner than other littermates before any treatment or analysis. We did not use randomization in our animal studies. We were not blinded to the group in our animal studies.
Northern blot
Total RNA was extracted from different mouse tissues, intestinal organoids, or sorted ISCs with TRIzol. 10 μg RNA from each sample was subjected to formaldehyde denaturing agarose electrophoresis followed by transferring to positively charged NC film with 20 × SSC buffer (3.0 M NaCl and 0.3 M sodium citrate, pH 7.0). Membrane was UV cross‐linked and incubated with biotin‐labeled RNA probes generated by in vitro transcription. Biotin signals were detected with HRP‐conjugated streptavidin for the Chemiluminescent Nucleic Acid Detection Module according to the manufacturer's instruction (Han et al, 2014).
Histology and immunohistochemistry
For histology analysis, duodenum region of intestine tissues was fixed in 4% formaldehyde. Longitudinal paraffin sections were prepared for hematoxylin and eosin (H&E) staining according to standard procedures. Sections of intestine tissue were performed with immunohistochemical staining as the methods previously described (Zhu et al, 2018). For immunohistochemistry quantitation, positive cells in different fields were enumerated in 20 × lens.
Crypt and ISC isolation
Crypts were isolated from Lgr5 GFP mice as described with minor modifications (Miyoshi & Stappenbeck, 2013). Briefly, small intestines were flushed with PBS, opened longitudinally, and then washed with cold PBS by vigorous shaking. Villi were removed by scraping the mucosa with a glass slide, and then, tissues were minced to 1‐mm2 fragments. Tissues were then incubated in collagenase solution (DMEM/F12 medium, containing 0.1% type I collagenase (Invitrogen), 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 10 mM HEPES) and digested for about 30 min. Tissues were mixed vigorously, and the separation of crypts with microscope was checked until more than 50% crypts were separated. Crypts were filtered through 70‐μm cell strainer, centrifuged, and washed before seeding in Matrigel. For ISC isolation, the crypts were dissociated into individual cells with 1 × TrypLE express (Life Technologies) supplemented with 0.8 kU/ml DNase I (Roche) followed by sorting according to GFP intensity.
Intestinal organoid culture
Intestine organoid medium was used as previously described with minor modifications (Sato et al, 2009). Briefly, Dulbecco's modified Eagle's medium/F12 medium (Invitrogen) was supplemented with N2 (Invitrogen), B27 (Invitrogen), 1.25 mM N‐acetylcysteine (Sigma‐Aldrich), 1 mg/ml RSPO1 (peproTech), 20 ng/ml EGF (PeproTech), 20 μM Y27632 (Sigma‐Aldrich), and 100 ng/ml Noggin. Medium was replaced every 3 days, and pictures of organoids were taken 1 week later. Alternatively, primary crypts or sorted ISCs were seeded into Matrigel together with IntestiCult organoid growth medium Kit (Cat#: 06005, StemCell) according to the manufacturer's instruction.
RNA‐seq analysis
For transcriptome analysis, ISCs were pooled from Srcap −/− and Srcap +/+ mice by GFP sorting. RNAs from mouse ISCs were prepared for library construction and sequenced on BGISEQ500. Differentially expressed genes were identified as fold change cutoff > 1.5, FDR < 0.05. Data were from one biological replicate.
Immunofluorescence assay
The multiple‐color staining with Opal™ 7‐color fIHC kit was used according to the manufacturer's instruction. Briefly, paraffin sections of mouse were sequentially stained with primary antibodies and HRP‐conjugated secondary antibody, followed by Opal reagent staining, microwave treatment, and another round staining. Samples were observed using Vectra Automated Quantitative Pathology Imaging System (PerkinElmer) or LSM700 laser scanning confocal microscopy (Zeiss) (Zhu et al, 2018). Mouse whole‐mount body sections were prepared with PFA fixation, sucrose rehydration, and OCT‐embedded frozen sections. After sequentially staining with primary antibodies and fluorescence‐conjugated secondary antibodies, images were obtained with Axio Scan. Z1 slide scanner (Zeiss) (Zhu et al, 2018). Intestine tissue 3D images were obtained by iDISCO method. Briefly, mouse intestine tissues were dissected into little fragments and fixed by PFA. Tissues were pretreated with PBS/0.2% TritonX‐100/20% DMSO and PBS/0.1% Tween‐20/0.1% TritonX‐100/0.1% deoxycholate/0.1% NP40/20%DMSO buffer. After sequentially staining with primary antibodies and fluorescence‐conjugated secondary antibody, samples were cleared by incubating with tetrahydrofuran, dichloromethane, and dibenzyl ether. Images were obtained with Zeiss LSM700 laser scanning confocal microscopy (Renier et al, 2014).
Immunoprecipitation assay and mass spectrometry
Mouse intestinal crypts and villi were isolated as described. Cells were lysed with ice‐cold RIPA buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM EDTA, 2 mM PMSF, 20 mg/ml aprotinin, 20 mg/ml leupeptin, 10 mg/ml pepstatin A, 150 mM benzamidine, and 1% Nonidet P‐40) for 1 h. Lysates were incubated with antibodies followed by immunoprecipitation with protein A/G agarose beads. Immunoprecipitation components were separated with SDS–PAGE followed by silver staining. Differential bands enriched by SRCAP were analyzed by LTQ Orbitrap XL mass spectrometry (Ye et al, 2018a).
Chromatin immunoprecipitation (ChIP) and ChIP‐seq
Quantitative ChIP was performed according to a standard protocol (Upstate). Sheared chromatin (sonicated to 200–500 bp) from ISCs (5 × 105) fixed in 1% formaldehyde was incubated with 4 μg antibody overnight at 4°C followed by immunoprecipitation with salmon sperm DNA/protein agarose beads. After washing, elution, and cross‐link reversal, DNA from each ChIP sample and corresponding input sample was purified and analyzed using qPCR. Primers were listed in Table EV1. For ChIP‐seq DNA was prepared followed by library construction and sequenced on BGISEQ‐500. Data were from one biological replicate.
DNase accessibility assay
Nuclei were isolated from 5 × 105 ISCs using Nuclei Isolation Kit (Sigma‐Aldrich) according to the manufacturer's protocol. Nuclei were resuspended in 200 μl of DNase I digestion buffer (1 mM EDTA, 0.1 mM EGTA, 5% sucrose, 1 mM MgCl2, 0.5 mM CaCl2). Two equal aliquots of 100 μl nuclei were treated with the indicated units of DNase I (Sigma, USA) and incubated at 37°C for 5 min. Reactions were stopped by 2 × DNase I stop buffer (20 mM Tris ph 8.0, 4 mM EDTA, 2 mM EGTA). DNA was extracted and analyzed by qPCR. Primers were listed in Table EV1.
Luciferase assay
Mouse Prdm16 promoter region and GluR2 promoter region were cloned into pGL4 basic vector (Promega) for luciferase assay. Reporter plasmid, SRCAP, Rest FL, and Rest truncation plasmids were transfected into 293T cells (1 × 105) along with thymidine kinase (TK). 36 h later, cells were lysed and detected using substrates L and S according to the Promega Dual‐Luciferase system.
Statistical analysis
For statistical evaluation, an unpaired Student's t‐test was applied for calculating statistical probabilities in this study. Statistical calculation was performed using Excel 2010 or GraphPad Prism 6. P < 0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001); P > 0.05, non‐significant (NS). For survival analysis, the Kaplan–Meier survival analysis and log‐rank test were used.
Author contributions
BY designed and performed experiments, analyzed data, and wrote the paper; LY, GQ and BL performed experiments and analyzed data; XZ constructed plasmids and established mice tools. PZ, JM, WX, HL, TL, YW, SW, and ZW analyzed data; JJ and JL established HA‐SRCAP mice tool. YD, DF, SM, and JW performed some experiments; YT initiated the study, designed animal experiments, and analyzed data; ZF initiated the study, organized, designed, and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Acknowledgements
We thank Yihui Xu, Xiang Ding, Yan Teng, Xiaoyan Wang, Xudong Zhao, Xiang Shi, Jing Cheng, and Feng Yun for technical support. We thank J. Li (Cnkingbio Company Ltd, Beijing, China) for technical support. This work was supported by the National Natural Science Foundation of China (31930036, 81921003, 91640203, 31530093, 31670886, 31871494, 81672956, 31601189, 81772646, 31601189, 31870883), the Ministry of Science and Technology of China (2019YFA0508501), Beijing Natural Science Foundation (7181006, 5192018), and the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDB19030203).
EMBO Journal (2020) 39: e103786
Contributor Information
Buqing Ye, Email: bqye@moon.ibp.ac.cn.
Yong Tian, Email: ytian@ibp.ac.cn.
Zusen Fan, Email: fanz@moon.ibp.ac.cn.
Data availability
RNA‐seq data: Gene Expression Omnibus GSE143657 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143657).
ChIP‐seq data: Gene Expression Omnibus GSE144267 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144267).
References
- Bajpai R, Chen DA, Rada‐Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J (2010) CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463: 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G (2005) REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121: 645–657 [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ et al (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Barker N (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15: 19–33 [DOI] [PubMed] [Google Scholar]
- Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer‐Rowe KE, Xifaras ME, Akkad A, Arias E et al (2016) High‐fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2013) The intestinal crypt, a prototype stem cell compartment. Cell 154: 274–284 [DOI] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R (2014) Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346: 2 [DOI] [PubMed] [Google Scholar]
- Corrigan DJ, Luchsinger LL, Justino de Almeida M, Williams LJ, Strikoudis A, Snoeck HW (2018) PRDM16 isoforms differentially regulate normal and leukemic hematopoiesis and inflammatory gene signature. J Clin Invest 128: 3250–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Corrado N, Perdiguero E, Lafarga V, Munoz‐Canoves P, Nebreda AR (2010) Essential role of p18Hamlet/SRCAP‐mediated histone H2A.Z chromatin incorporation in muscle differentiation. EMBO J 29: 2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois V, Eeckhoute J, Lefebvre P, Staels B (2017) Distinct but complementary contributions of PPAR isotypes to energy homeostasis. J Clin Invest 127: 1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP (2018) Structural basis for ATP‐dependent chromatin remodelling by the INO80 complex. Nature 556: 386–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL (2015) Yap‐dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526: 715–718 [DOI] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y et al (2017) A single‐cell survey of the small intestinal epithelium. Nature 551: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ et al (2014) A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514: 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Ikeda K, Chen Y, Alba DL, Stifler D, Shinoda K, Hosono T, Maretich P, Yang Y, Ishigaki Y et al (2018) Repression of adipose tissue fibrosis through a PRDM16‐GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab 27: 180–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJ, Allanson J, Kim CA, Wieczorek D, Moilanen JS et al (2012) Mutations in SRCAP, encoding SNF2‐related CREBBP activator protein, cause Floating‐Harbor syndrome. Am J Hum Genet 90: 308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ et al (2013) In vitro expansion of single Lgr5 + liver stem cells induced by Wnt‐driven regeneration. Nature 494: 247–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH (2004) A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116: 779–793 [DOI] [PubMed] [Google Scholar]
- Lee HY, Gao X, Barrasa MI, Li H, Elmes RR, Peters LL, Lodish HF (2015) PPAR‐alpha and glucocorticoid receptor synergize to promote erythroid progenitor self‐renewal. Nature 522: 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AC, Binder CJ, Gutierrez A, Brown KK, Plotkin CR, Pattison JW, Valledor AF, Davis RA, Willson TM, Witztum JL et al (2004) Differential inhibition of macrophage foam‐cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J Clin Invest 114: 1564–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Ye B, Zhu X, Huang G, Yang L, Zhu P, Du Y, Wu J, Meng S, Tian Y et al (2017) IL‐7Ralpha glutamylation and activation of transcription factor Sall3 promote group 3 ILC development. Nat Commun 8: 017–00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G et al (2002) Corepressor‐dependent silencing of chromosomal regions encoding neuronal genes. Science 298: 1747–1752 [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Stappenbeck TS (2013) In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 8: 2471–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP‐driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Brulet R, Zhang L, Hsieh J (2016) REST regulation of gene networks in adult neural stem cells. Nat Commun 7: 13360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, Kajimura S (2012) PPARgamma agonists induce a white‐to‐brown fat conversion through stabilization of PRDM16 protein. Cell Metab 15: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P et al (2018) A major chromatin regulator determines resistance of tumor cells to T cell‐mediated killing. Science 359: 770–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost S, Relouzat F, Spentchian M, Ouzegdouh Y, Saliba J, Massonnet G, Beressi JP, Verhoeyen E, Raggueneau V, Maneglier B et al (2015) Erosion of the chronic myeloid leukaemia stem cell pool by PPARgamma agonists. Nature 525: 380–383 [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier‐Lavigne M (2014) iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159: 896–910 [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ et al (2009) Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada IS, Acar M, Burgess RJ, Zhao Z, Morrison SJ (2017) Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells. Genes Dev 31: 1134–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, Sato T (2017) Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 545: 187–192 [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker‐Thornburg J, Adams H, Majumder S (2008) REST maintains self‐renewal and pluripotency of embryonic stem cells. Nature 453: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine RR, Sakers AP, TeSlaa T, Kissig M, Stine ZE, Kwon CW, Cheng L, Lim HW, Kaestner KH, Rabinowitz JD et al (2019) PRDM16 maintains homeostasis of the intestinal epithelium by controlling region‐specific metabolism. Cell Stem Cell 25: 830–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, Ekici M, Rossler OG (2015) RE‐1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res 359: 99–109 [DOI] [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R et al (2013) Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154: 1207–1219 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Radman‐Livaja M, Rando OJ, Peterson CL (2013) A histone acetylation switch regulates H2A.Z deposition by the SWR‐C remodeling enzyme. Science 340: 195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yoshihara E, He N, Hah N, Fan W, Pinto AFM, Huddy T, Wang Y, Ross B, Estepa G et al (2018) Vitamin D switches BAF complexes to protect beta cells. Cell 173: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Liu B, Yang L, Huang G, Hao L, Xia P, Wang S, Du Y, Qin X, Zhu P et al (2017) Suppression of SRCAP chromatin remodelling complex and restriction of lymphoid lineage commitment by Pcid2. Nat Commun 8: 1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Liu B, Hao L, Zhu X, Yang L, Wang S, Xia P, Du Y, Meng S, Huang G et al (2018a) Klf4 glutamylation is required for cell reprogramming and early embryonic development in mice. Nat Commun 9: 1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W, Zhu P, Wang Y, Wang S, Xia P et al (2018b) LncKdm2b controls self‐renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J 37: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F et al (2013) MicroRNA‐133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 17: 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T (2016) ssODN‐mediated knock‐in with CRISPR‐Cas for large genomic regions in zygotes. Nat Commun 7: 10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF (2011) A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Chen Y, Jiang N, Yang L, Sun S, Zhang Y, Wen Z, Ray L, Liu H, Hou G et al (2019) Znhit1 controls intestinal stem cell maintenance by regulating H2A.Z incorporation. Nat Commun 10: 1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Du T, Roizman B (2013a) HSV carrying WT REST establishes latency but reactivates only if the synthesis of REST is suppressed. Proc Natl Acad Sci USA 110: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WJ, Geng ZH, Spence JR, Geng JG (2013b) Induction of intestinal stem cells by R‐spondin 1 and Slit2 augments chemoradioprotection. Nature 501: 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, Song G, Gao X, Yao L, Fan D et al (2014) An efficient genotyping method for genome‐modified animals and human cells generated with CRISPR/Cas9 system. Sci Rep 4: 6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, He L, Ye B, Wang S, Meng S et al (2018) LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol 20: 1134–1144 [DOI] [PubMed] [Google Scholar]
- Zingarelli B, Piraino G, Hake PW, O'Connor M, Denenberg A, Fan H, Cook JA (2010) Peroxisome proliferator‐activated receptor delta regulates inflammation via NF‐{kappa}B signaling in polymicrobial sepsis. Am J Pathol 177: 1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Data Availability Statement
RNA‐seq data: Gene Expression Omnibus GSE143657 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143657).
ChIP‐seq data: Gene Expression Omnibus GSE144267 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144267).


