Abstract
Age-related macular degeneration (AMD) is the leading cause of blindness among the elderly. Currently, there are no treatments for dry AMD, which is characterized by the death of retinal pigment epithelium (RPE) and photoreceptors. Reports from human donors with AMD suggest that RPE mitochondrial defects are a key event in AMD pathology. Thus, the most effective strategy for treating dry AMD is to identify compounds that enhance mitochondrial function and subsequently, preserve the RPE. In this study, primary cultures of RPE from human donors with (n = 20) or without (n = 8) AMD were used to evaluate compounds that are designed to protect mitochondria from oxidative damage (N-acetyl-l-cysteine; NAC), remove damaged mitochondria (Rapamycin), increase mitochondrial biogenesis (Pyrroloquinoline quinone; PQQ), and improve oxidative phosphorylation (Nicotinamide mononucleotide, NMN). Mitochondrial function measured after drug treatments showed an AMD-dependent response; only RPE from donors with AMD showed improvements. All four drugs caused a significant increase in maximal respiration (p < 0.05) compared to untreated controls. Treatment with Rapamycin, PQQ, or NMN significantly increased ATP production (p < 0.05). Only Rapamycin increased basal respiration (p < 0.05). Notably, robust responses were observed in only about 50% of AMD donors, with attenuated responses observed in the remaining AMD donors. Further, within the responders, individual donors exhibited a distinct reaction to each drug. Our results suggest drugs targeting pathways involved in maintaining healthy mitochondria can improve mitochondrial function in a select population of RPE from AMD donors. The unique response of individual donors to specific drugs supports the need for personalized medicine when treating AMD.
Keywords: Age-related macular degeneration, Retinal pigment epithelium, Mitochondrial function, Rapamycin, Pyrroloquinoline quinone, Nicotinamide mononucleotide
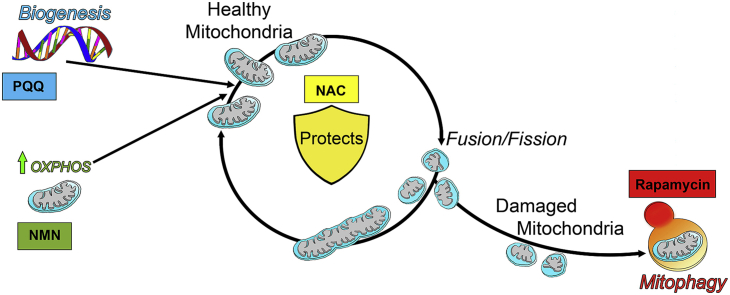
Graphical abstract
Highlights
-
•
Human primary RPE cultures were used to test the efficacy of drugs on mitochondrial function.
-
•
Drugs targeting mitochondrial homeostasis pathways improved mitochondrial function in AMD RPE.
-
•
The donor-specific response to drugs suggests personalized medicine is needed to treat AMD.
Abbreviations
- AMD
Age-related macular degeneration
- ANOVA
analysis of variance
- BCA
Bicinchoninic acid
- BR
basal respiration
- CMST
Cell Mito Stress Test
- COX IV
cytochrome c oxidase subunit IV
- LC
liquid chromatography
- LC3
microtubule-associated proteins 1A/1B light chain 3B
- MGS
Minnesota Grading System
- MS
mass spectrometry
- MR
maximal respiration
- Mt
mitochondrial
- NAC
N-acetyl-l-cysteine
- NAD
Nicotinamide adenine dinucleotide
- NMN
Nicotinamide mononucleotide
- NMNAT
Nicotinamide/nicotinate-nucleotide adenylyltransferase
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
- PGC-1α
Peroxisome Proliferator-Activated Receptor-gamma Coactivator 1α
- PQQ
Pyrroloquinoline quinone
- RPE
Retinal Pigment Epithelium
- SRC
spare respiratory capacity
- VDAC
voltage-dependent anion channel
1. Introduction
Age-related macular degeneration (AMD), the leading cause of vision loss in adults, globally affects approximately 20–30% of individuals over 75 years old in the developed world [17]. This disease is characterized by deterioration of the macula, a small area at the retina's center. The result is central vision loss, making it difficult to read, recognize faces, or perform tasks that require visualizing fine detail. Current treatment options for AMD are available only for a small percentage of the patient population with “wet” AMD (~10%), which manifests as the abnormal growth of blood vessels into the retina from the choroid, a fenestrated blood vessel network that provides oxygen and nutrients to the outer retina [47]. A key feature of the more prevalent “dry” AMD is the loss of retinal pigment epithelial (RPE) cells and photoreceptors. The RPE is a pigmented monolayer that is located between the retina and choroid. RPE perform tasks critical for maintaining retinal health, including transport of nutrients from the choroid, degradation of shed photoreceptor outer segments, and secretion of factors that are crucial for the structural integrity of the retina [52]. Treatment for dry AMD is limited to a nutritional supplement determined by two separate clinical trials, coined the Age-Related Eye Disease Study (AREDS), to reduce disease progression in a limited number of patients. The most efficacious formulations were composed of high doses of vitamin C and E plus the minerals zinc and copper [2,3]. While a portion of patients showed reduced odds of developing advanced AMD, the treatment effect was modest and for most participants, and vision loss continued to progress [7].
A comprehensive meta-analysis of 39 population-based studies reported the global estimate of people with AMD in 2020 is 196 million and will increase to 288 million by 2040 [62]. These statistics highlight the impeding epidemic of vision loss due to AMD. The significant negative impact on families and the global health care industry creates an urgent need to develop treatment strategies for patients with dry AMD. To be effective, new strategies must target the disease mechanism, which likely involves mitochondrial (Mt) dysfunction in the RPE [14,15,27,33,44,45]. The mitochondria are organelles that produce energy (ATP), regulate cell death, control redox signaling, and buffer calcium. Since RPE rely almost exclusively on mitochondria as an energy source [25], disruption of these essential processes could cause RPE cell death [16]. In support of this idea, two recent studies found that RPE cells cultured from human donors with AMD had reduced oxidative phosphorylation (OXPHOS) compared with RPE cells from non-diseased donors [15,21]. This altered RPE metabolism could create a bioenergetic crisis in the retina, driving AMD pathology [16].
Identifying compounds that enhance Mt function and preserve the RPE may provide the most effective treatment strategy for dry AMD. For this proof of concept study, we selected four drugs that specifically target key processes in maintaining optimal Mt function. N-acetyl-l-cysteine (NAC) has anti-oxidant properties that protect cells from oxidative damage by acting both as a free radical scavenger and as a precursor for glutathione, a tripeptide that helps restore cellular redox balance [64]. NAC is currently used to treat acetaminophen overdose and is available as both a prescription and over-the-counter supplement [10,26,60]. The macrolide antibiotic Rapamycin is used to prevent organ transplant rejection [28]. This drug inhibits mTOR and activates autophagy, a degradative process that eliminates defective organelles, including mitochondria. The drug Pyrroloquinoline quinone (PQQ) is classified as an essential micronutrient [65]. Small amounts of PQQ have been detected in fruits and vegetables; larger amounts are found in fermented products such as cocoa powder [50]. PQQ is a redox cofactor that induces Mt biogenesis [48]. Nicotinamide mononucleotide (NMN) is a key intermediate of nicotinamide adenine dinucleotide (NAD+), which transports electrons to facilitate Mt respiration and ATP generation [9,39]. Similar to PQQ, NMN is also present in various types of food [40], and both compounds are available as an over-the-counter supplement.
In this study, age-matched primary cultures of RPE from human donors with or without AMD were used to evaluate how NAC, Rapamycin, PQQ, and NMN affect Mt function. We have previously shown that RPE from donors with AMD had reduced Mt function, thereby providing a useful model system for testing the efficacy of drugs [15]. Our results show a drug-dependent effect on Mt function as well as a donor-specific response to the treatments. Of note, these positive results were mainly limited to a select portion of RPE from AMD donors.
2. Materials and methods
2.1. Human eye procurement and grading for AMD
Human eyes were procured from Lions Gift of Sight (formerly known as Minnesota Lions Eye Bank, Saint Paul, MN) with the written consent of the donor or the donor's family for use in medical research in accordance with the Declaration of Helsinki. Lions Gift of Sight is licensed by the Eye Bank Association of America (accreditation #0015204) and accredited by the FDA (FDA Established Identifier 3000718528). Donor tissue is considered pathological specimens and is therefore exempt from the process of Institutional Review Board approval.
Evaluation of the presence or absence of AMD was determined by a Board Certified Ophthalmologist (Sandra R. Montezuma) from stereoscopic fundus photographs of the RPE macula using the criteria (RPE pigment changes and the presence, size and location of drusen) established by the Minnesota Grading System [46]. Demographics (age, gender, cause of death, and time from death to cell harvesting) for the donors used to generate RPE primary cultures or for RPE proteomic analysis was provided by Eye Bank records and are summarized in Table 1 and Supplemental Table 1, respectively.
Table 1.
Donor characteristics and clinical information a.
| Disease State b | Samplec (n) | Sex Male (n) |
Sex Female (n) |
Age (Mean ± SD) | Timed (Mean ± SD) | Cause of Death e (n) |
|---|---|---|---|---|---|---|
| No AMD | 8 | 5 | 3 | 65 ± 9.7 | 19 ± 4.1 | AE (1), Cancer (3), ESLD (1), ICH (1), Respiratory Failure (1), Sepsis (1) |
| AMD (MGS2) (MGS3) |
20 16 4 |
12 11 1 |
8 5 3 |
70 ± 10.1 67 ± 9.5 80 ± 5.9 |
19 ± 4.0 18 ± 4.2 20 ± 3.3 |
AAA (1), ABI (1), Cancer (4), COPD (1), CVA (1), Heart failure (2), ICH (1), MI (2), MS (1), Pneumonia (2), Sepsis (4) |
AAA = abdominal aortic aneurysm; ABI = anoxic brain injury; AE = Anoxic Encephalopathy; COPD = chronic obstructive pulmonary disease; CVA = cerebrovascular accident (stroke); ESLD = End-Stage Liver Disease; ICH = Intracerebral hemorrhage; MI = myocardial infarction; MS = Multiple Sclerosis.
Information supplied by Lions Gift of Sight, St. Paul, MN (formerly known as Minnesota Lions Eye Bank).
Minnesota Grading System (MGS) was used to evaluate the stage of AMD in eye bank eyes (Olsen and Feng, 2004). No AMD = MGS1; AMD = MGS2 and MGS3.
Sample number indicates the total donors with or without AMD used in the current study.
Provided is the time from death to RPE harvest in hours.
The number of donors for each cause of death is indicated in parentheses.
2.2. RPE cell culturing
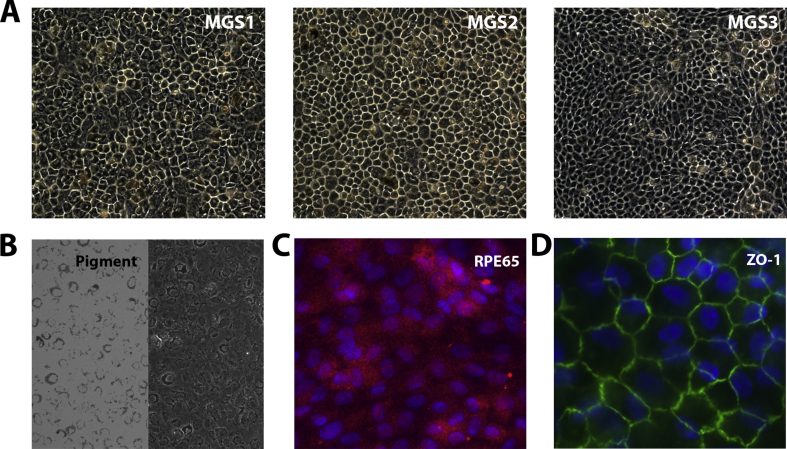
RPE cells, harvested from the peripheral region after removing a 5 mm punch of cells centered over the macula, were cultured using the conditions previously described [15]. Cells in passage 2 or 3 were used for analysis. Cell number and condition are indicated under each experimental protocol. Donor RPE were selected for various assays based on their availability. These cells were extensively characterized in our previous study [15] and in Supplemental Fig. 1.
2.3. Immunofluorescence/microscopy
Cells were grown on either chamber slides (Nunc) or transwell filters (Costar). Paraformaldehyde-fixed cells were blocked for 1 h in 10% donkey serum and then incubated in primary antibody, RPE65 (provided by Dr. T. Michael Redman – NEI) or ZO-1 (Life Technologies) overnight. The reaction was visualized using an appropriate secondary antibody. Cells were cover slipped with mounting medium containing DAPI (Vector Laboratories) and imaged with a fluorescence microscope (Leica DM4000B). For phase contrast images, confluent RPE cell cultures grown in Primaria 6-well plates were imaged with an Axiovert 200 M inverted microscope (Zeiss).
2.4. Optimization of drug treatment
RPE from several donors (n = 9 total) were used for preliminary assays to optimize drug treatment conditions. Drug concentrations and experimental timing were selected from a series of experiments comparing untreated to treated cells to determine the optimal dose from a range of concentrations (2.5–100 nM for Rapamycin; 10 nM to 1 μM for PQQ; 0.1–2 mM for NMN) and a range of times (2–48hr). The optimal concentration and time for each drug was determined from results of changes in protein or NAD+/NADH content and Mt function.
2.5. Western blotting
RPE cells (3 × 105) were grown to confluence in Primaria 6-well plates (Corning). Cells were treated with Rapamycin or PQQ and lysates were extracted at specific times. Protein concentrations were determined with the Bicinchoninic acid (BCA) assay using albumin as the standard. Western blots were performed as previously described [37]. Proteins were resolved on SDS-PAGE gels and transferred to PVDF membranes (Millipore) and incubated overnight with primary antibodies, LC3 (Novus Biologicals), VDAC (Cell Signal), COX IV (Cell Signal). β-Actin (Santa Cruz Biotechnology) was used as a loading control. Appropriate secondary antibodies conjugated to horseradish peroxidase were used to visualize immune reactions using chemiluminescence with SuperSignal West Dura Extended Duration substrate (Thermo Fisher). Immune reactions were imaged using a ChemiDoc XRS (Bio-Rad) and quantified using Quantity One software (Bio-Rad).
Autophagy in Rapamycin-treated cells was evaluated by dividing LC3-II densitometry values by LC3-I values, then normalizing the data to densitometry values of β-Actin. Biogenesis in PQQ-treated cells was evaluated by dividing VDAC or COX IV densitometry values by β-Actin (loading control) values.
2.6. Measurement of NAD+ and NADH
RPE cells were seeded (5 × 103 cells/well) in all white 96-well plates (Corning) and grown for 24hr. Cells were treated with varying concentrations (100 μM to 2 mM) of NMN for 24hr, and at various time points (2–32hr). NAD content was determined using NAD/NADH-Glo Assay kit (Promega) following the manufacturer's protocol. The concentration of NAD+ and NADH in the samples was calculated from standard curves. Luminescence was measured on a microplate reader (Biotek, Cytation One).
2.7. RNA isolation and RT-PCR
Donor eyes were enucleated 4 h or less from death. One eye was placed directly into RNAprotect cell reagent (Qiagen) and stored at −20 °C until RNA isolation. RNA quality was evaluated post isolation using an Agilent Bioanalyzer 2100 (Biomedical Genomics Center, U of MN). Fundus imaging was performed on the alternative eye within 6 h post-mortem to evaluate the presence and severity of AMD.
cDNA was synthesized from total RNA prepared from RPE tissue or RPE cultured cells as previously described [56]. The expression of NMNAT genes was determined using quantitative reverse transcription PCR using a BioRad iQ5 multicolor real time PCR detection system. Triplicate wells of 25 μL reactions contained 1 ng cDNA, 0.2 μM Forward and Reserve primers, and 13.5 μL BioRad iQ SYBR Green Supermix. The following primers were used: NMNAT1, forward 5′-TCCCATCACCAACATGCACC-3′ and reverse 5′-TGATGACCCGGTGATAGGCAG-3’; NMNAT2, forward 5′–TGGAGCGTTTCACCTTTGTA-3′ and reverse 5′-CAATCTCTTCATACCGCATC-3’; NMNAT3, forward 5′-AGTTTGCTGTGATGCCTC-3′ and reverse 5′-AGTTTGCTGTGATGATGCCTC-3’; HPRT1(housekeeping gene), forward 5′- TGCAGACTTTGCTTTCCTTGGTCAGG-3′ and reverse 5′-CCAACACTTCGTGGGGTCCTTTTCA-3’. The housekeeping gene HPRT1 was used to calculate ΔCt for each gene of interest. To determine fold change relative to No AMD, ΔΔCt of each AMD donor was calculated by subtracting the mean ΔCt of No AMD cells. A modified Livak method was used to calculate relative expression using efficiency for each primer.
PCR products were verified by the appearance of a single band migrating at the correct size on a 2% agarose gel using a 50bp DNA ladder as reference. PCR products were visualized under UV light and imaged using a ChemiDoc XRS (Bio-Rad).
2.8. Sample preparation and NanoLiquid chromatography (LC)-Mass spectrometry (MS/MS) analysis
Demographics of donors used in the RPE protein analysis performed using Mass Spectrometry is found in Supplemental Table 1. To isolate an organelle-enriched fraction, RPE cell pellets were suspended in isolation buffer (70 mM sucrose, 200 mM mannitol, 1 mM EGTA, 10 mM HEPES pH 7.4) and subjected to two freeze/thaw cycles prior to homogenization. Samples were centrifuged at 800×g for 8 min. The supernatant containing the organelles was centrifuged at 12,000×g for 10 min. The pellet containing the organelle-enriched fraction was resuspended in ice-cold Tris buffered saline and frozen at −80 °C.
All samples were extracted with an ice-cold lysis buffer (50 mM Tris-formic acid, 150 mM NaCl, 0.5% sodium deoxycholate, 2% SDS, 2% NP-40, pH 8.0) as described [58]. The same amount of protein (60 μg) was processed with an acetone precipitation/on-pellet-digestion approach. The peptide mixture of 4 μg was separated using a nano-LC system (Eksigent, Dublin, CA) and analyzed using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). A nano-LC column (75 μm ID x 75 cm, packed with 3-μm particles) with a 160-min gradient was used for separation. The MS data was acquired using the data-dependent product ion mode. Survey scans (m/z range 400–1500) were performed at a resolution of 120,000 with an automatic gain control target of 5 × 105. Tandem MS (MS2) was analyzed using isolation at 1.2 Th with the quadrupole for high energy collision dissociation fragmentation. An MS2 automatic gain control target was set to 5 × 104 and the maximum injection time was 50 ms. Dynamic exclusion was enabled with the settings: repeat count 1; repeat duration 50 s; exclusion duration 60 s. The MS instrument was run in top speed mode with a cycle time of 3 s.
For database search and validation, raw files were searched against the Swiss-Prot protein database (released January 2015) using Proteome Discoverer 1.4. Two missed cleavages were allowed for tryptic peptides. Mass tolerances for precursor ions and fragment ions were 20 ppm and 0.02 Da, respectively. Carbamidomethylation of cysteines was set as a fixed modification. Variable modifications of methionine oxidation and protein N-terminal acetylation were allowed. Scaffold 4.5 software was used for peptide and protein validation. The threshold of 0.1% peptide FDR, 1% protein FDR, and 2 distinct peptides was used to filter the peptide and protein identification. Gene Ontology (GO) software was used to obtain the cellular localization of identified proteins.
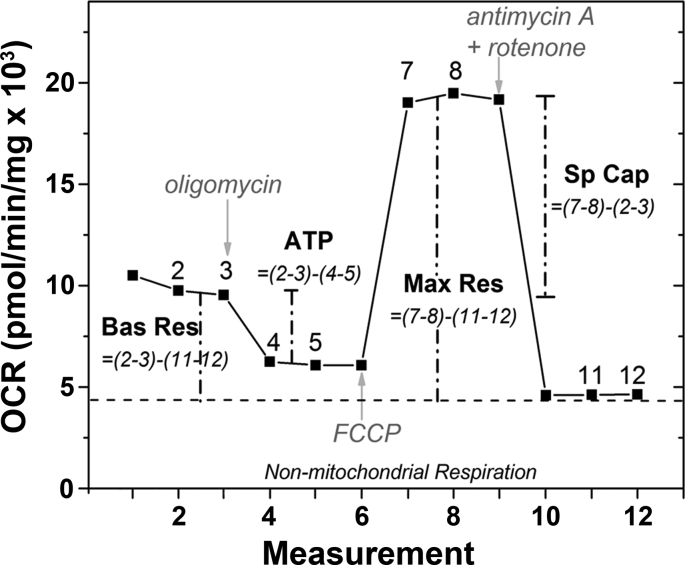
2.9. Measurement of mitochondrial function using the cell mito stress test (CMST)
Analysis of Mt function was performed on live cells using XFe96 Extracellular Flux Analyzer (Agilent Technologies) as previously described [15]. RPE cells (4 × 104 cells/well) were seeded and grown for 24hr, then treated with one of the following compounds: 500 μM NAC (N-acetyl-l-cysteine; Sigma-Aldrich), 100 nM Rapamycin (Sigma-Aldrich), or 100 nM PQQ (Pyrroloquinoline-quinone disodium salt; Sigma-Aldrich) for 48hr, or 500 μM NMN (Nicotinamide mononucleotide; Sigma Aldrich) for 24hr. The CMST assay protocol was performed as detailed by the manufacturer (Agilent Technologies) and our previous analysis [15]. A graphical description of the assay results and parameters measured are provided in Supplemental Fig. 2.
After the assay was complete, cells were incubated on ice and washed with PBS (2X). Cell lysates were made with RIPA buffer (50 μL/well). Protein concentration of each well was determined using a BCA assay kit (Thermo Fisher) with bovine serum albumin as the standard. The total amount (mg) of protein per well was used for normalization of the oxygen consumption rate (OCR) in the CMST assay.
2.10. Statistical analysis
All treatment data were normalized to the no treatment value for each donor (fold change relative to no treatment). Statistical analysis was performed on log transformed fold change values. One Way ANOVA with Dunnett's post hoc was used to determine optimal treatment (PQQ, Rapamycin, or NMN) dose and timing (Fig. 1). One-sample t-test or Wilcoxon t-test comparing all treatments to their untreated control were performed to determine effectiveness of drug treatments (Fig. 3). Unpaired t-tests of the ΔΔCt values were used to compare NMNAT gene expression levels between No AMD and AMD cells in Fig. 4B. Unpaired t-tests were used to compare drug responses from donors without AMD (No AMD) to donors with AMD (Fig. 5). Analyses were performed using the statistical software in GraphPad Prism 8. Data were reported as mean ± SEM for each group. p ≤ 0.05 was considered statistically significant.
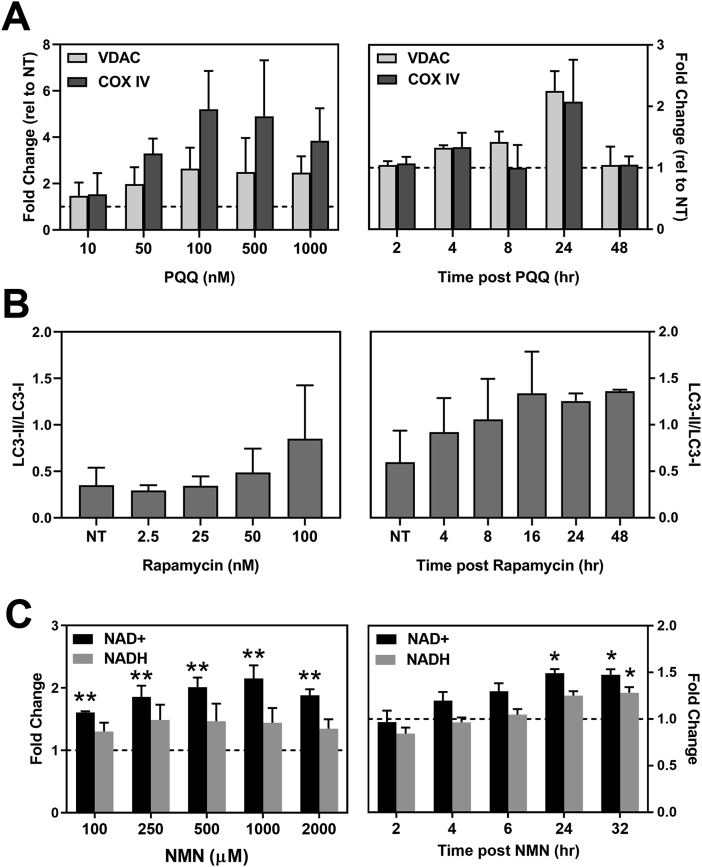
Fig. 1.
Characterization of drug treatment effect. (A,B) RPE cells were treated with each compound for the times or concentrations specified, lysates were collected, Western blots were run, and densitometry of protein bands was performed. (A) RPE (n = 2) were treated with PQQ for 24 h to determine the optimal drug concentration (left). The time-dependent response (n = 2) was determined following incubation with 100 nM PQQ (right). The amount of VDAC and COX IV content were estimated from densitometry data. Results are the fold change relative to untreated control (NT = 1) for each donor. No significant difference was observed comparing treatments to no treatment controls. VDAC, voltage-dependent anion channel; COX IV, cytochrome C oxidase 4. (B) RPE (n = 2) were treated with Rapamycin for 24 h to determine optimal drug concentration (left). The time-dependent response (n = 2) was determined following incubation with 100 nM Rapamycin (right). The ratio of delipidated (LC3-I) and lipidated (LC3-II) proteins were determined from densitometry data. β-Actin was used as the loading control. No significant difference was observed comparing treatments to no treatment controls. LC3, microtubule-associated proteins 1A/1B. (C) NAD/NADH content in RPE cells (n = 3) treated with NMN for 24 h was measured to determine the optimal drug concentration (left). The time-dependent response (n = 5) was determined following incubation with 500 μM NMN (right). Graphs show fold change relative to no treatment control (NT = 1) for each donor. *p < 0.05, **p < 0.01 were statistically different than untreated controls. Data in (A–C) are mean ± SEM. One Way ANOVA with Dunnett's post hoc was used to compare treatment to untreated control.
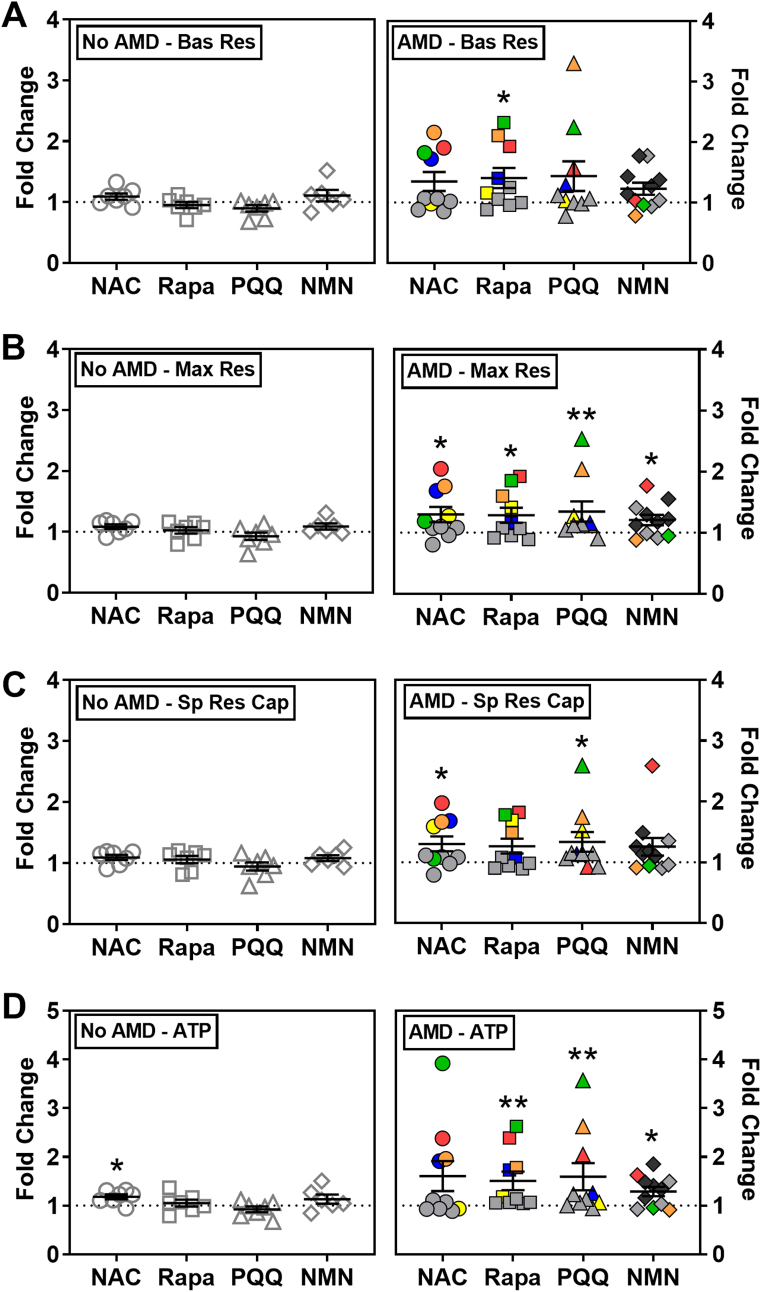
Fig. 3.
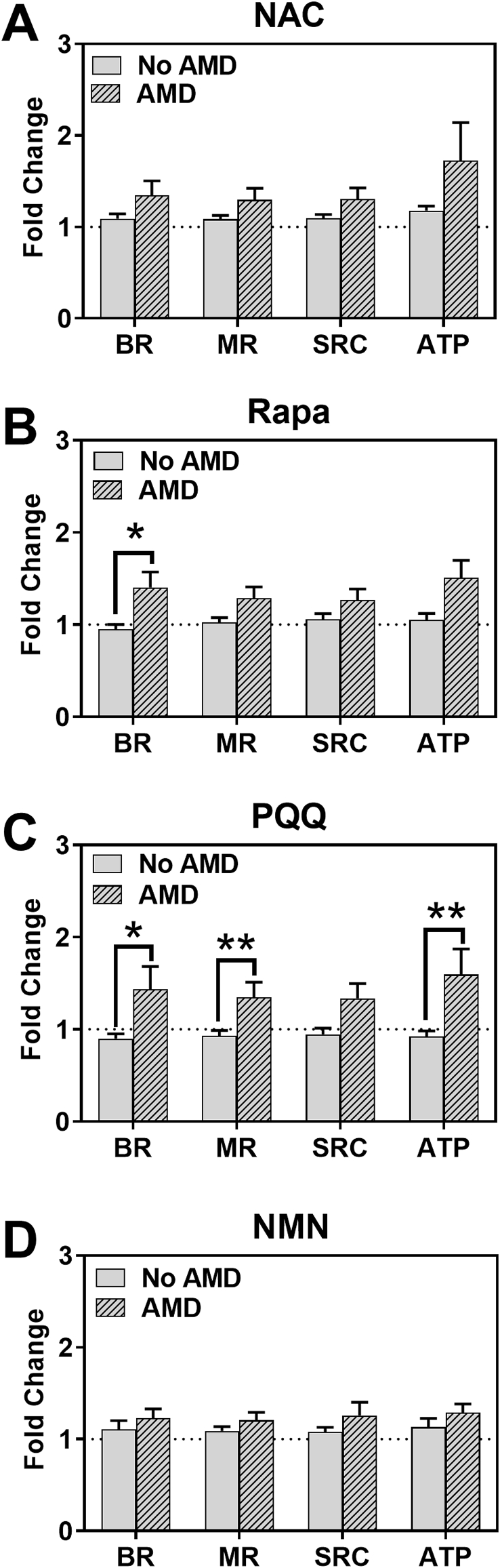
Effect of drugs on RPE mitochondrial function. RPE cells from donors without AMD (n = 7) or with AMD (n = 10; MGS2 n = 7, MGS3 n = 3) were treated with drugs for 24 or 48hr. Following treatment, parameters of mitochondrial function (A–D) were calculated from OCR measured using an XFe96 Extracellular Flux Analyzer. OCR data was normalized to mg protein per well. Graphs show fold change relative to each donor's no treatment control (dashed line). Colored data points indicate response from individual donors that exhibited improved function for at least one drug. Red = MGS3 donor. Blue, yellow, orange, and green = MGS2 donors. Bas Res = Basal Respiration; Max Res = Maximal Respiration; Sp Res Cap = Spare Respiratory Capacity; ATP = ATP production. The bar and whiskers represent the mean ± SEM (*p < 0.05, **p < 0.01 determined by one-sample t-test or Wilcoxon t-test).
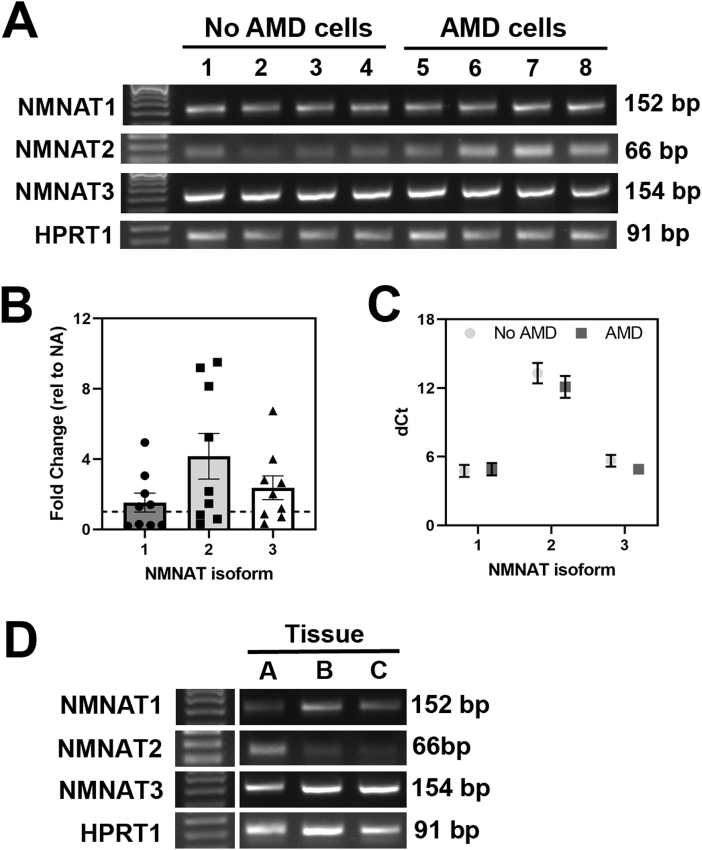
Fig. 4.
NMNAT gene expression in RPE cells and RPE tissue. (A) Gel image shows PCR products of NMNAT1, NMNAT2, and NMNAT3 in RPE cell cultures from donors without AMD (donors 1 to 4) or with AMD (donors 5 to 8). (B) mRNA expression of NMNAT family genes in No AMD (n = 9) and AMD (n = 9; MGS2 n = 6, MGS3 n = 3) cells was measured by quantitative RT-PCR. Results are fold change in expression relative to the average for No AMD (NA) samples (dotted line). Unpaired t-tests of ddCt values were used to compare basal expression of NMNAT genes in AMD cells to No AMD cells. There was no significant difference in expression comparing AMD to No AMD. (C) Expression of NMNAT family genes relative to housekeeping gene (dCt). Data are mean ± SEM in both B & C. (D) PCR products of NMNAT1, NMNAT2, NMNAT3 in RPE tissue harvested from three individual donors (A = MGS4, 87 year old female; B = MGS2, 87 year old male; C = MGS1; 56 year old male). In (A) & (D) 50bp ladder was used as a reference for PCR product size. HPRT1 was used as a housekeeping control.
Fig. 5.
Comparing drug response in RPE cells from donors with or without AMD. Drug response was compared in cells from AMD donors (n = 10; MGS2 n = 7, MGS3 n = 3) and No AMD donors (n = 7). The same data from Fig. 3 was plotted to investigate differences in average response to each drug according to disease state. Fold change relative to no treatment was plotted. Bars represent mean (±SEM) parameters of mitochondrial function (BR = Basal Respiration; MR = Maximal Respiration; SRC = Spare Respiratory Capacity; ATP = ATP Production) as measured by XFe96 Extracellular Flux Analyzer. *p < 0.05, **p < 0.01 determined by t-test with Welch's or Mann-Whitney corrections.
3. Results
A summary of clinical information and demographics for the donors used in the cell culture experiments is provided in Table 1. The average time of death to cell harvesting was 19 ± 4 h (mean ± SD) for both groups. The average age of donors with No AMD (65 ± 9.7 yr, n = 8) compared to AMD (70 ± 10.1 yr, n = 20) was not statistically different (p = 0.28). Our AMD group included MGS2 and MGS3 donors, which represent early and intermediate stages of the disease. As would be expected with disease progression, the average age of MGS3 (80 ± 5.9 yr) donors was significantly older than MGS2 (67 ± 9.5 yr) donors (p = 0.03).
Confluent primary RPE cell cultures from donors used in the experiments were pigmented, had a cobblestone appearance, expressed the RPE-specific marker, RPE65, and developed tight junctions as shown by ZO-1 staining (Supplemental Fig. 1). As seen in the photomicrographs, RPE from donors with or without AMD had similar shape and degree of pigmentation. These data are consistent with cell characterization in our previous study [15].
3.1. Characterization of compounds that target mitochondrial pathways
The optimal dose and timing for each compound was experimentally determined by comparing the drug response (increase in protein or NAD+/NADH content) in at least two donors. Initial experiments tested a range of drug concentrations (for PQQ, Rapamycin, NMN) to determine the optimal dose. Subsequent experiments narrowed down the optimal time of treatment.
The mechanism of action for PQQ involves the activation of transcription factors and coactivators that regulate genes involved in cell energy metabolism and Mt biogenesis [8,22,48]. To assess the effect of PQQ treatment on Mt biogenesis, we measured the content of voltage-dependent anion channel (VDAC) and cytochrome c oxidase subunit IV (COX IV) (Fig. 1A). These proteins are located on the outer and inner Mt membranes and are used as an estimate of Mt content. We found 100 nM PQQ elicited maximal upregulation of Mt proteins. In following the response over time, Mt protein content was increased starting at 4 h, with the most pronounced upregulation at 24 h. By 48 h, the Mt protein content returned to baseline.
To assess the effect of Rapamycin treatment on autophagy in RPE cells, we tracked the conversion of LC3-I to LC3-II, a well-described indicator of autophagic activity (Fig. 1B). During autophagy, LC3-I (cytosolic form) is processed and recruited to the autophagosomes, where LC3-II (the lipidated form of this protein) is generated [43]. We found the maximal response to treatment occurred with 100 nM rapamycin. At this dose, maximal upregulation of autophagy occurred from 16 to 48 h.
NMN is an intermediate of NAD + biosynthesis [63]. As a co-enzyme, NAD regulates ATP production through its NAD+/NADH redox state [67]. To assess the effect of NMN treatment on RPE cells, we measured intracellular content of NAD+ and NADH. At 500 and 1000 μM NMN, maximal NAD + content was observed, with a slight decrease in NAD + content at 2000 μM (Fig. 1C, left). Of note, NADH content remained slightly above baseline but did not change for all NMN doses. After treating RPE with 500 μM NMN, the largest increase in NAD + content was observed at 24 and 32 h (Fig. 1C, right).
In our previous investigation testing the effect of NAC treatment on RPE cells, we found that 500 μM NAC increased ATP content after 24 h of incubation [56]. We also found that pre-treatment with NAC protected RPE cells from oxidation-induced mitochondrial dysfunction [56]. Therefore, 500 μM NAC was adopted for the current study.
3.2. Assessing mitochondrial function to determine optimal treatment time
Having determined the dose for each compound that provided the optimal response in proteins or NAD + content, we next evaluated the time-dependent effect on mitochondrial function using an Extracellular Flux Analyzer. The Cell Mito Stress Test (CMST) assay was performed to measure oxygen consumption rate (OCR), which is a gauge of mitochondrial respiration. OCR measured after sequential injections of mitochondrial stressors (oligomycin, FCCP, and antimycin A/rotenone) was used to calculate basal respiration (BR), maximal respiration (MR), spare respiratory capacity (SRC), and ATP production (Supplemental Fig. 2). OCR was measured in RPE cells following 24 or 48 h of drug treatment. Fig. 2 (left) shows an example trace of RPE OCR from a single donor treated with NAC, Rapamycin, PQQ, or NMN. Fig. 2 (right) shows the average calculated values for Mt function associated with results from two donors.
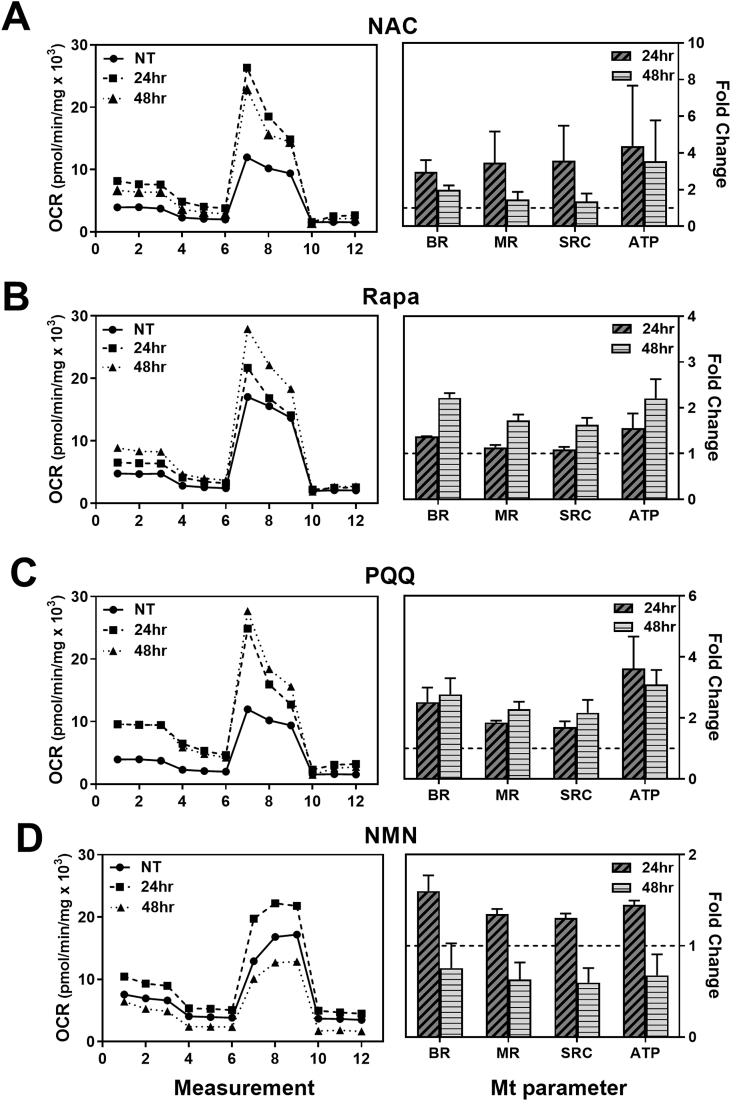
Fig. 2.
Measuring Mt function to determine the optimal treatment time. RPE cells were plated in a 96-well plate and incubated overnight. Cells were treated with compounds (NAC, Rapamycin, PQQ, or NMN) for 24 or 48 h. Mitochondrial function was evaluated using XFe96 Extracellular Flux Analyzer. An example trace of oxygen consumption rate (OCR) from a single donor treated with the specified drug is shown (left). Average calculated values (±StDev) of Mt function (compared to untreated controls) for two donors are shown (right). BR = basal respiration; MR = maximal respiration; SRC = spare respiratory capacity; ATP = ATP production.
In comparing the effect on Mt function, we found optimal treatment times varied between drugs (Fig. 2). RPE cells treated with NAC had increases in BR and ATP production at both 24 and 48hr; MR and SPR were highest at 24 h (Fig. 2A). In our previous study, we observed an overall improvement in Mt function with 24hr NAC treatment [56]. To extend these initial studies and test the longer-term effect of NAC treatment, 48hr was chosen as the experimental time point in the current study. RPE cells treated with Rapamycin showed increases in Mt function at 48hr (Fig. 2B). RPE cells treated with PQQ had increases in BR, MR, SRC, and ATP production at both 24 and 48 h (Fig. 2C). From these data, 48 h treatment (NAC, Rapamycin, or PQQ) was chosen for subsequent experiments. The improvement in Mt function following NMN treatment was higher at 24hr (Fig. 2D), so this time was selected as the treatment time for NMN.
3.3. Determining the optimal drug for improving mitochondrial function
To evaluate the efficacy of individual drugs on Mt function, OCR was measured following incubation with each drug at their optimal dose and time. We first examined individual donor response by comparing treated to untreated cells. In RPE from donors without AMD, treatments had no significant effect on Mt function since the response of individual donors clustered around 1 (no treatment control) (Fig. 3, left). One exception was the improvement in ATP production after NAC treatment (p = 0.01). In contrast, RPE from donors with AMD were more responsive (Fig. 3, right). Treatment with NAC significantly improved MR (p = 0.04) and SRC (p = 0.04). Rapamycin significantly increased BR (p = 0.03), MR (p = 0.05), and ATP production (p = 0.01). Treatment with PQQ significantly improved MR (p < 0.01), SRC (p = 0.02), and ATP production (p = 0.03). NMN treatment significantly increased MR (p = 0.04) and ATP production (p = 0.01).
An additional observation about the response of RPE from AMD donors is that approximately half of the individuals showed a 50%–350% improvement in Mt function after treatment. The other half of the AMD donors were much less responsive, exhibiting only ~5%–25% improvement in Mt function. It is important to note that the group of drug-responsive donors with AMD exhibited a very individualized response to the drugs (Fig. 3, right – individual donors are indicated by a single color.). As an example, the green (MGS2) and red-symbol (MGS3) donors had higher BR after exposure to NAC, Rapamycin, and PQQ, whereas robust improvement in BR was observed only after NAC treatment for the blue-symbol (MGS2) donor (Fig. 3A). Since the AMD group contains donors with early (MGS2) and intermediate (MGS3) AMD, we asked whether disease severity influenced the response to drugs. While our sample size is limited, the results suggest that the disease stage does not appear to influence the drug response; 1 out of 3 MGS3 donors responded to drugs and 4 out of 7 MGS2 donors responded. Rather, we conclude that the responders may be individuals with defects in specific processes and support the requirement for personalized treatment as the most efficacious approach for AMD.
3.4. Investigation of NMNAT in RPE
In our initial optimization experiments, we observed NMN treatment increased intracellular NAD+ in RPE cells. The enzymes involved in producing NAD+ from the precursor NMN are Nicotinamide/nicotinate-nucleotide adenylyltransferase (NMNAT) 1, 2, and 3. These isoforms are found in the nucleus, cytosol, and mitochondria, respectively. Under basal conditions, NMNAT1, NMNAT2, and NMNAT3 gene expression was observed in cultured RPE cells from both No AMD and AMD donors (Fig. 4A). Results in Fig. 3 show that NMN treatment caused a significant improvement in maximal respiration and ATP production in only AMD RPE compared to their untreated controls (Fig. 3). These results suggest differences in content of NMNAT could explain the functional improvement in AMD RPE in response to NMN supplementation. To test this idea, we compared NMNAT expression in RPE from donors with or without AMD by quantitative RT-PCR. We observed a 2- to 4-fold increased expression in AMD RPE, but this increase was not statistically different due to the high degree of variability between donors (Fig. 4B). We also compared the relative abundance of each NMNAT isoform by comparing their mean dCt values (lower dCt indicates higher expression). We observed expression of NMNAT1 and NMNAT3 was more abundant than NMNAT2 in RPE from both No AMD and AMD donors, indicating the highest levels of NMNAT are localized to the mitochondria and nucleus (Fig. 4C).
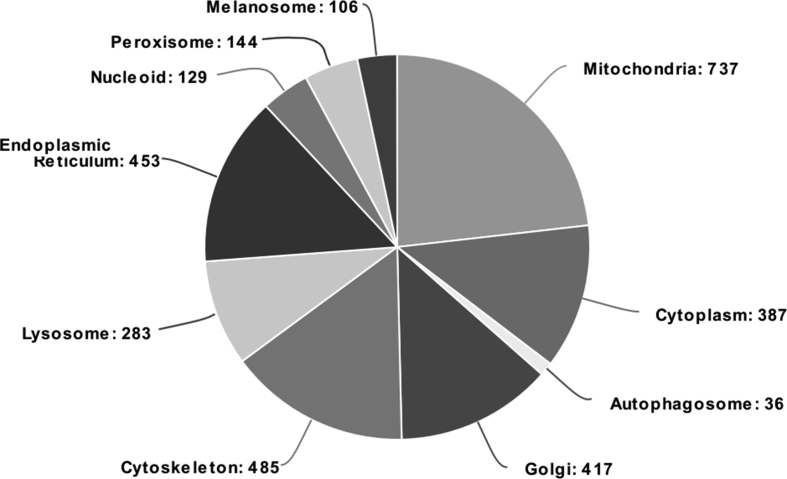
To test whether these enzymes are also present in RPE in vivo, we examined RPE tissue harvested from human donor eyes. Gene expression of all three isoforms of the NMNAT enzyme was also detected in RPE tissue harvested from human donor eyes using high-quality RNA (RNA integrity number = 8 ± 0.3, mean ± SEM) (Fig. 4D). A proteomic screen further confirmed the presence of NMNAT1 and NMNAT3 in organelle-enriched fractions from human RPE tissue (Table 2). These fractions contained proteins from the mitochondria, lysosomes, endoplasmic reticulum, nucleus, melanosomes, peroxisomes, and cytoplasm (Supplement Figure 3). In 5 out of 7 RPE samples, nuclear-localized NMNAT1 had 1 or 2 peptides covering ~10% of the amino acid sequence (Table 2). The mitochondrial NMNAT3 was found in all seven RPE tissue samples and had 4 to 7 peptides covering ~27% of the sequence (Table 2). The cytoplasmic NMNAT2 protein was not observed, likely because the organelle-enriched sample preparation contained few cytoplasmic proteins. This result is also consistent with the lower expression of NMNAT2 measured in RPE cultures (Fig. 4C).
Table 2.
Mass Spectrometry identification of proteins in RPEa.
| Protein | UniProt ID | Number of Peptidesb | Probabilityc | Exclusive Unique Peptidesd | Sequence Coveragee | |
|---|---|---|---|---|---|---|
| NMNAT1 | Q9HAN9 | Donor 1 Donor 2 Donor 3 Donor 4 Donor 5 Donor 6 Donor 7 |
2 2 0 2 1 1 0 |
100 100 100 45 98 |
2 2 2 1 1 |
14% 14% 11% 7% 5% |
| NMNAT3 | Q96T66 | Donor 1 Donor 2 Donor 3 Donor 4 Donor 5 Donor 6 Donor 7 |
7 7 5 4 5 7 6 |
100 100 100 100 100 100 100 |
6 6 5 4 4 6 5 |
37% 28% 26% 22% 19% 30% 29% |
Organelle-enriched fraction prepared from donor RPE (see methods).
The number of unique peptides (each with a different amino acid sequence) that are attributed to a single protein including those shared with other proteins.
Calculated probability that the protein identification is correct.
The number of peptides containing different amino acid sequences, regardless of any modification, that are attributed to a single protein/protein group.
Provides the percentage of amino acids in the whole protein sequence that was identified in the sample.
In summary, all three NMNAT isoforms are present in RPE from both tissue and cultured cells. Further, the similar expression between No AMD and AMD cell cultures suggests the disease-specific response to NMN supplementation is due to factors other than the availability of the NMNAT enzyme.
3.5. Comparing drug responses between RPE cells with or without AMD
To determine if there was a difference in response due to disease state of the donor, we compared the average response of RPE from AMD donors to No AMD donors for the four drugs. NAC treatment improved overall Mt function ~10% in No AMD donors compared to an approximate 30% increase in AMD RPE. However, this difference was not statistically significant (Fig. 5A). Rapamycin treatment significantly improved BR in RPE from AMD donors (30%) compared to RPE from No AMD donors (0%; p = 0.02; Fig. 5B). The most impressive differences in response were with PQQ. This drug significantly improved BR (44%; p = 0.03), MR (35%; p = 0.01) and ATP production (59%; p = 0.01) in RPE cells from donors with AMD compared to RPE from non-diseased donors (Fig. 5C). NMN treatment improved overall Mt function ~10% in No AMD RPE compared to ~20% in AMD RPE. However, this difference was not statistically significant (Fig. 5D).
4. Discussion
In this study, we used primary cultures of RPE from age-matched human donors with or without AMD to evaluate the efficacy of compounds that target key processes involved in maintaining optimal Mt function. NAC protects the mitochondria from oxidative damage, Rapamycin removes damaged mitochondria via mitophagy, PQQ stimulates production of new mitochondria, and NMN helps increase oxidative phosphorylation. Intervening in these four processes, which benefit Mt homeostasis, would help maintain the delicate metabolic ecosystem of the RPE [16]. Of note, we observed an AMD-dependent response to drug treatment. Only RPE from AMD donors responded to the drugs as detected by the increase in Mt function. In contrast, RPE from donors without AMD did not respond to the drugs since no changes in Mt function were detected with the exception of improved ATP production with NAC treatment. These results are consistent with the idea that Mt dysfunction in the diseased cells is ameliorated by the drug treatments targeted to Mt defects and that RPE from non-diseased donors did not share these defects. We also observed a highly variable response in RPE from AMD donors, with responsive donors exhibiting an individualized improvement in Mt function for specific drugs. These results suggest a more personalized approach is needed when developing treatments for AMD.
Although RPE cell cultures retain many of the prototypic characteristics of RPE in vivo, there are caveats that need to be mentioned with regards to the use of donor tissue and our model system. In using donor tissue as a source for our primary cultures, we have no control over the donor availability, including gender, disease severity, and age. While we strive for balance between age, gender, and disease stage, the distribution (i.e. early AMD and intermediate AMD donors) was not always equal in each assay and could potentially influence our results. Another consideration is how post-mortem time could influence mitochondrial function and drug response. In this investigation, the average time from death to cell harvest for both groups (No AMD and AMD) was 19 ± 4hr (±SD), so the time did not vary greatly between donors and any affect would be felt across all donor groups. Additionally, we use cells at passage 3, which means the cells are cultured for ~4 months, so initial differences in process times are likely diminished over time in culture. Our lab also follows strict “standard operating procedures” in handling and processing to minimize technical variability. However, even using identical procedures, RPE cultures exhibited heterogeneity in drug response, which could be due to general heterogeneity within the cultures and/or differences due to individual donor response. We favor donor intrinsic differences as the major explanation of our findings, which is consistent with a study comparing the phenotype and function of iPSC-RPE lines from 15 donors [42]. Results revealed the donor-to-donor variability exceeded the clone-related differences, suggesting donor-specific genetic or epigenetic differences were the major contributors to how cultures behave.
Another caveat of our model system is that RPE cells in culture do not fully replicate the retinal environment since they lack the overlying photoreceptors and underlying choroid. While this is a limitation of all cell culture systems, we acknowledge that the influence of other cell types could affect results. How the application of cell culture results translates to in vivo therapeutic interventions is also a challenge. A specific concern for AMD research is that our cell culture system uses only RPE cells from the periphery, not the macular region, which appears clinically to suffer the greatest consequences of the disease. However, recent advances in ocular imaging that permit observation of cells in the peripheral retina have reported retinal lesions (i.e. drusen, RPE hypopigmentation and degeneration) that are clinically similar to lesions observed in the macula of AMD patients [11]. Additionally, a study measuring mtDNA damage in 5 mm punches of RPE from the macula, inferior, and superior regions, found mtDNA damage was widespread and not limited to the macula [55]. Taken together, these results suggest the pathological effect of AMD involves the entire retina and so cells from the periphery are a valid source for primary RPE cultures. In support of this idea, we had previously reported primary RPE harvested from the periphery of AMD donors exhibited a significant reduction in Mt function [15]. Thus, while there are caveats, our model system replicates aspects of AMD that are essential for developing therapies to treat this disease.
Multiple studies examining RPE in human donors with AMD have reported defects in mitochondria, including a loss in Mt mass and intact internal structures [14] and decreased content of proteins in the electron transport chain [44,45]. These changes could disrupt Mt function within the RPE. Additional evidence from our lab [27,55] and others [33] have shown increased levels of mtDNA damage in donor RPE at stages of AMD preceding macular degeneration and vision loss. The mtDNA damage is likely due to increased production of reactive oxygen species, which may not only cause damage to mitochondrial structures, but could also alter redox signaling within the cell. Thus, protecting and improving Mt function in RPE is the most efficacious strategy for treating AMD and preserving vision.
NAC and other anti-oxidants play a protective role in oxygen radical-related stress injuries. NAC has many properties that make it a good candidate drug for AMD patients, including that it can be taken orally or applied topically to the cornea. Evidence that NAC can penetrate from the cornea to the posterior segment is provided by NACs protection of photoreceptors in rd10+/+ mice, a model of retinitis pigmentosa [31]. Direct application of NAC has also protected ARPE19 cells (an immortalized RPE cell line) and primary human adult RPE cells from tert-butyl hydroperoxide (t-BOOH)-induced oxidative stress [12,24,56]. In the current study, NAC treatment improved MR and SRC in RPE cells from AMD donors and significantly increased ATP production in RPE cells from donors without AMD (Fig. 3). These results contrast with our previous study where 24 h of NAC treatment significantly increased overall Mt function and protected from depletion of glutathione after an acute oxidative stress in RPE from donors with and without AMD [56]. The slightly less robust response observed at 48hr may indicate that NAC is depleted at this time point. The bioavailability and stability of NAC is limited because of its hydrophobicity and free sulfhydryl group, which can be easily oxidized. Previous analysis of NACs half-life after oral or intravenous administration was approximately 6 h [5]. This short half-life suggests more frequent applications of NAC may be required. Another approach would be to use NAC nanoparticles. In a study using LPS-activated microglia cells, the conjugation of NAC to nanoparticles increased its anti-oxidant and anti-inflammatory potential compared to free NAC [38]. Taken together, results suggest more frequent application or reformulation that stabilize the molecule are required to improve NAC efficacy and has important implications for NAC as a potential treatment for AMD.
Because of its ability to inhibit mTOR and induce autophagy, Rapamycin has been effective in reducing RPE dedifferentiation and preserved photoreceptor function in mice with an RPE-selective postnatal loss of OXPHOS [66]. Rapamycin was also effective in reducing injury in various models of neurodegenerative diseases [13,36] and in improved Mt function in a rat model of ischemic stroke [32]. Our rationale for testing rapamycin arose from the strong evidence that diminished autophagy flux may contribute to AMD pathology [21,41,59]. In the current study, we observed up-regulation of LC3-II and improved Mt function in RPE after rapamycin treatment in AMD RPE (Fig. 1, Fig. 2, Fig. 3, Fig. 5). Since rapamycin stimulates autophagy, it is possible that this improvement in Mt function is due to the elimination of dysfunctional mitochondria within the RPE through mitophagy. As a caveat, Rapamycin has been part of AMD clinical trials under the name “Sirolimus” [18,61]. Both studies were stopped early due to lack of efficacy, as determined by changes in geographic atrophy growth and visual acuity. Since dry AMD is a slow progressing disease, these stringent outcomes, which likely did not substantially change during the clinical trial, could have been responsible for the negative result. Additionally, our data show considerable variability in response for donors with AMD, suggesting that not every patient has a specific defect that is responsive to rapamycin treatment. The prevalence of non-responders in the clinical trials could contribute to the lack of statistically significant changes in outcomes. However, it may be worthwhile to take a second look at this drug or some of the newly developed “rapalogs” [6,30] to see if more favorable outcomes could be obtained.
The most robust response in Mt function was observed after PQQ treatment. Bioactive compounds, like PQQ, have been shown to improve Mt respiration and stimulate Mt biogenesis by activating the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) signaling pathway [8,22,48]. PGC-1α is a transcriptional coactivator for an array of transcription factors that play roles in Mt biogenesis, protection from oxidative stress, fatty acid oxidation, and lipid metabolism [8,49,53]. Mt biogenesis involves PGC-1α activation of mtDNA transcription and replication by binding to nuclear respiratory factors (NRF-1 and NRF-2) and Mt transcription factor A (TFAM) [19]. In support of PQQs effect on Mt biogenesis, BALB/c mice fed a diet supplemented with PQQ exhibited increased Mt number and improved Mt function and respiration in liver [4,51]. PQQ can also protect cells from oxidative stress through PGC-1α activation of transcription factors that regulate anti-oxidant gene expression. These transcription factors include forkhead box O-3A (FOXO-3A), MAD box transcription enhancer factor (MEF), and nuclear factor (erythroid-derived 2)-like2 (Nrf2) [53]. Considering the highly oxidative environment of the diseased retina, upregulation of antioxidant genes elicited by PQQ could help protect the retina from further damage. Lastly, PQQ can improve fatty acid oxidation and lipid metabolism via PGC-1α binding to transcription factors, estrogen-related receptor (ERRα), and peroxisome proliferator-activated receptors (PPARα or PPARγ) [49]. While we did not investigate whether PQQ directly activates these transcription factors in our system, PQQs positive effect on lipid metabolism would be highly beneficial since phagocytosis and degradation of outer segments, comprised of 40–50% lipids [1,20], is one of the RPEs critical functions.
In the current study, PQQ was the only drug to improve MR, SRC, and ATP production when comparing untreated to treated cells (Fig. 3) and significantly improved BR, MR, and ATP production in RPE from AMD donors (Fig. 5). Since PQQ stimulates Mt biogenesis, the improvement in Mt function is likely due to increased Mt content. This idea is supported by the increases in Mt proteins, VDAC and COX IV, in initial experiments characterizing PQQs effect (Fig. 1) and reports by another group that found a dose-dependent increase in the level of mtDNA relative to nuclear DNA (mtDNA/nDNA) in PQQ-treated in NIH/3T3 fibroblasts [48]. Consistent with increased Mt content, Saihara et al. and our current study observed an enhanced level of ATP production. Considering PQQs positive effects not only on Mt respiration, but also in protecting from oxidative damage and improving lipid metabolism, this compound seems like a viable choice for further exploration of its potential use in combating the effects of AMD. PQQ treatment early in AMD could potentially stave off the detrimental effects of oxidation-induced stress, Mt dysfunction, and the ensuing “bioenergetic crisis” that occurs in advanced AMD [16].
The supplement NMN is a precursor for NAD+, a vital redox carrier that is central to metabolism and other cellular processes [54]. The decline in NAD levels with aging and neurodegenerative conditions has elicited intense investigations on ways to boost NAD content using a variety of supplements [23,29,35,39,40]. Although NAD + itself is difficult to administer directly to humans, its precursor molecule, NMN, is a promising natural compound that augments NAD levels in the body [57]. In vivo evidence showed NMN restores Mt function and NAD + content to normal levels in brain Mt pellets from a transgenic mouse model of Alzheimer's disease [35] and ameliorates retinal dysfunction in aged C57BL/6 N mice [40]. In vitro experiments reported treatment with NMN preserved NAD + levels in ARPE19 (an immortalized RPE cell line), primary mouse RPE cells, and 661W cone photoreceptor-like cells [23,34]. Consistent with results from previous studies, NMN treatment improved Mt function in primary RPE cultured from AMD donors (Fig. 3). NMN was taken up by the cells as shown by the increase in intracellular NAD + content (Fig. 1). Furthermore, NMN was taken up by the mitochondria, albeit through an unidentified carrier [54], as shown by the improvement in Mt function (Fig. 3). The presence of NMNAT3 in RPE cells (Fig. 4 and Table 2) suggests that this isozyme is responsible for the synthesis of NAD in the mitochondria. An increase in Mt NAD levels may have led to an increase in OXPHOS in RPE cells.
5. Conclusions
Mitochondria are highly active organelles. They are constantly undergoing biogenesis, fusion, fission, and mitophagy, processes that are important for maintaining a population of healthy mitochondria. Since alterations in these processes could be involved in AMD pathology, compounds that target processes involved in Mt homeostasis could be attractive therapies for the disease. In this proof-of-concept study, we used four drugs to target Mt homeostasis and tested whether they could improve Mt function in primary RPE cultures from donors with and without AMD. Of the four drugs, PQQ was the best at improving Mt function, especially in RPE cells from AMD donors. This may be due to PQQ's ability to activate PGC-1α, a transcriptional co-activator that plays a role in Mt biogenesis, protection from oxidative stress, and lipid metabolism. For these reasons, additional studies with PQQ or other bioactive molecules are warranted. One other significant finding was that, just as reported for patients in clinical trials, RPE cells from different donors exhibited significant variability in their response to the four drugs. Only RPE from a select group of donors with AMD responded by upregulating Mt function and among this group, considerable heterogeneity in drug response was observed. These results support the idea that an individualized approach to treating AMD is required. Developing ways to pre-screen patients to determine the optimal type of intervention is a critical linchpin in finding effective treatments for AMD.
Funding
This work was supported in part by the Foundation Fighting Blindness (grant number TA-NMT-0613-0620-UMN); National Institutes of Health/National Eye Institute (grant numbers R01EY026012, R01EY028554); Office of the NIH Director (T35OD011118 to JRP); the Elaine and Robert Larson Endowed Vision Research Chair (to DAF); Lindsay Family Foundation; an anonymous benefactor for Age-Related Macular Degeneration Research. None of the funding agencies had a role in study design, in the collection, analysis and interpretation of data, in writing the manuscript, or in the decision to submit the manuscript for publication.
Declaration of competing interest
All authors declare that there is no conflict of interest associated with this manuscript.
Acknowledgements
The authors acknowledge the contribution of the Lions Gift of Sight personnel for their assistance in procuring eyes, and to Kathy Goode and Sung Lee for photographing and processing eye tissue. Dr. T. Michael Redman (NEI) provided the antibody for RPE65. We thank Cody Fisher for his help in generating figures. The authors also thank the donors and their families for their essential contribution to this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101552.
Contributor Information
Mara C. Ebeling, Email: ebeli017@umn.edu.
Jorge R. Polanco, Email: polan070@umn.edu.
Jun Qu, Email: junqu@buffalo.edu.
Chengjian Tu, Email: chengjian.tu@thermofisher.com.
Sandra R. Montezuma, Email: smontezu@umn.edu.
Deborah A. Ferrington, Email: ferri013@umn.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Fig. 1.
Images of primary human RPE cell cultures derived from adult donors. (A) Phase microscopy images show cultured RPE cells exhibit cobblestone appearance. Pictures were taken with 10x objective. (B) Bright Field microscopy image (left) shows pigmentation in RPE cells. Matching phase contrast image is on right. (C) RPE cells, grown on a glass slide, were stained with anti-RPE65 (red) antibody and imaged with a fluorescence microscope. Picture was taken with 20x objective. (D) RPE cells, grown in a transwell, stain positive for the tight junction protein, ZO-1 (green). Picture was taken with 40x objective. Nuclei in (C) & (D) are stained with DAPI (blue).
Supplemental Fig. 2.
Determining mitochondrial function using an XFe Extracellular Flux Analyzer. Graph shows trace and methods for calculating mitochondrial function parameters. Arrows indicate time of injection for specific reagents. The formula for calculating each parameter is shown in italics. Numbers in parenthesis indicate the average of the two values used in the calculation. Oxygen consumption rate (OCR) following injections of oligomycin (2 μM final), FCCP (1 μM final), rotenone & antimycin A (1 μM final, each) perturbs mitochondria function. Calculation of basal respiration, ATP production, maximal respiration, and spare respiratory capacity are shown.
Supplemental Fig. 3.
Distribution of proteins in the RPE organelle-enriched fraction. Data shown represent compiled data of RPE from seven human donors. Proteins were identified by LC-MS and categorized by Gene Ontology (GO). Subcellular compartments can include proteins associated with multiple GO terms. Pie chart was created on meta-chart.com.
References
- 1.Agbaga M.-P., Merriman D.K., Brush R.S. Differential composition of DHA and very-long-chain PUFAs in rod and cone photoreceptors. J. Lipid Res. 2018;59:1586–1596. doi: 10.1194/jlr.M082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch. Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Age-Related Eye Disease Study 2 Research Group Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: the Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. J. Am. Med. Assoc. 2013;309(19):2005–2015. doi: 10.1001/jama.2013.4997. [DOI] [PubMed] [Google Scholar]
- 4.Bauerly K.A., Storms D.H., Harris C.B. Pyrroloquinoline quinone nutritional status alters lysine metabolism and modulates mitochondrial content in the mouse and rat. Biochim. Biophys. Acta Gen. Subj. 2006;1760:1741–1748. doi: 10.1016/j.bbagen.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Bavarsad Shahripour R., Harrigan M.R., Alexandrov A.V. N-acetylcysteine (NAC) in neurological disorders: mechanisms of action and therapeutic opportunities. Brain Behav. 2014;4(2):108–122. doi: 10.1002/brb3.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagosklonny M. From rapalogs to anti-aging formula. Oncotarget. 2017;8:35492–35507. doi: 10.18632/oncotarget.18033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew E.Y., Clemons T.E., Agron E. Long-term effects of vitamins C, E, beta-carotene and zinc on age-related macular degeneration. Ophthalmology. 2013;120(8):1604–1611. doi: 10.1016/j.ophtha.2013.01.021. AREDS report no. 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowanadisai W., Bauerly K.A., Tchaparian E. Pyrroloquinoline quinone stimulates mitochondrial biogenesis through cAMP response element-binding protein phosphorylation and increased PGC-1α expression. J. Biol. Chem. 2010;285:142–152. doi: 10.1074/jbc.M109.030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davila, Liu L., Chellappa K. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. eLife. 2018;7 doi: 10.7554/eLife.33246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dilger R.N., Baker D.H. Oral N-acetyl-L-cysteine is a safe and effective precursor of cysteine. J. Anim. Sci. 2007;85:1712–1718. doi: 10.2527/jas.2006-835. [DOI] [PubMed] [Google Scholar]
- 11.Domalpally A., Clemons T.E., Danis R.P. Peripheral retina changes associated with age-related macular degeneration in the age-related eye disease study 2: age-related eye disease study 2 report number 12 by the age-related eye disease study 2 optos PEripheral RetinA (OPERA) study research group. Ophthalmol. Times. 2017;124(4):479–487. doi: 10.1016/j.ophtha.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Eichler W., Reiche A., Yafai Y., Lange J., Wiedemann P. Growth-related effects of oxidant-induced stress on cultured RPE and choroidal endothelial cells. Exp. Eye Res. 2008;87:342–348. doi: 10.1016/j.exer.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Erlich S., Alexandrovich A., Shohami E., Pinkas-Kramarski R. Rapamycin is a neuroprotective treatment for traumatic brain injury. Neurobiol. Dis. 2007;26:86–93. doi: 10.1016/j.nbd.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Feher J., Kovacs I., Artico M. Mitochondrial alterations of RPE in AMD. Neurobiol. Aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Ferrington D.A., Ebeling M.C., Kapphahn R.J. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol. 2017;13:255–265. doi: 10.1016/j.redox.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher C.R., Ferrington D.A. Perspective on AMD pathobiology: a bioenergetic crisis in the RPE. Invest. Ophthamol. Vis. Sci. 2018;59:AMD41–AMD47. doi: 10.1167/iovs.18-24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman D.S., O'Colmain B.J., Munoz B. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 18.Gensler G., Clemons T.E., Domalpally A. Treatment of geographic atrophy with intravitreal sirolimus: the Age-Related Eye Disease Study 2 ancillary study. Ophthalmol. Retina. 2018;2:441–450. doi: 10.1016/j.oret.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gleyzer N., Vercauteren K., Scarpulla R.C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg A.F., Moritz O.L., Williams D.S. Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 2016;55:52–81. doi: 10.1016/j.preteyeres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golestaneh N., Chu Y., Xiao Y.Y., Stoleru G.L., Theos A.C. Dysfunctional autophagy in RPE, a contributing factor in age-related macular degeneration. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris C.B., Chowanadisai W., Mishchuk D.O. Dietary pyrroloquinoline quinone (PQQ) alters indicators of inflammation and mitochondrial-related metabolism in human subjects. J. Nutr. Biochem. 2013;24:2076–2084. doi: 10.1016/j.nutbio.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Jadeja R.N., Powell F.L., Jones M.A. Loss of NAMPT in aging retinal pigment epithelium reduces NAD+ availability and promotes cellular senescence. Aging (Albany NY) 2018;10:1306–1323. doi: 10.18632/aging.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kagan D.B., Liu H., Hutnik C.M.L. Efficacy of various antioxidants in the protection of the retinal pigment epithelium from oxidative stress. Clin. Ophthamol. 2012;6:1471–1476. doi: 10.2147/OPTH.S35139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanow M.A., Giarmarco M.M., Jankowski C.S. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017;6 doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanter M.Z. Comparison of oral and i.v. acetylcysteine in the treatment of acetaminophen poisoning. Am. J. Health Syst. Pharm. 2006;63:1821–1827. doi: 10.2146/ajhp060050. [DOI] [PubMed] [Google Scholar]
- 27.Karunadharma P.P., Norgaard C.L., Olsen T.W., Ferrington D.A. Mitochondrial DNA damage as a potential mechanism for AMD. Invest. Ophthalmol. Vis. Sci. 2010;51:5470–5479. doi: 10.1167/iovs.10-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klawitter J., Nashan B., Christians U. Everolimus and Sirolimus in transplantation – related but different. Expet Opin. Drug Saf. 2015;14:1055–1070. doi: 10.1517/14740338.2015.1040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klimova N., Long A., Kristian T. Nicotinamide mononucleotide alters mitochondrial dynamics by SIRT3-dependent mechanism in male mice. J. Neurosci. Res. 2019;97:975–990. doi: 10.1002/jnr.24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamming D.W., Ye L., Sabatini D.M., Baur J.A. Rapalogs and mTOR inhibitors as anti-aging therapeutics. J. Clin. Invest. 2013;123:980–989. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S.Y., Usui S., Zafar A.-B. N-acetylcysteine promotes long-term survival of cones in a model of retinitis pigmentosa. J. Cell. Physiol. 2011;226:1843–1849. doi: 10.1002/jcp.22508. [DOI] [PubMed] [Google Scholar]
- 32.Li Q., Zhang T., Wang J. Rapamycin attenuates mitochondrial dysfunction via activation of mitophagy in experimental ischemic stroke. Biochem. Biophys. Res. Commun. 2014;444:182–188. doi: 10.1016/j.bbrc.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 33.Lin H., Xu H., Liang F.Q. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:3251–3259. doi: 10.1167/iovs.10-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J.B., Kubota S., Ban N. NAMPT-mediated NAD(+) biosynthesis is essential for vision in mice. Cell Rep. 2016;17:69–85. doi: 10.1016/j.celrep.2016.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long A.N., Owens K., Schlappal A.E. Effect of nicotinamide mononucleotide on brain mitochondrial respiratory deficits in an Alzheimer's disease-relevant murine model. BMC Neurol. 2015;15 doi: 10.1186/s12883-015-0272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malagelda C., Jin Z.H., Jackson-Lewis V., Przedborski S., Greene L.A. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson's disease. J. Neurosci. 2010;30:1166–1175. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maldonado M., Kapphahn R.J., Terluk M.R. Immunoproteasome deficiency modifies the alternative pathway of NFκB signaling. PloS One. 2013;8 doi: 10.1371/journal.pone.0056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markoutsa E., Xu P. Redox potential sensitive N-acetyl cysteine-prodrug nanoparticles inhibit the activation of microglia and improve neuronal survival. Mol. Pharm. 2017;14(5):1591–1600. doi: 10.1021/acs.molpharmaceut.6b01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massudi H., Grant R., Braidy N. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PloS One. 2012;7 doi: 10.1371/journal.pone.004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills K.F., Yoshida S., Stein L.R. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metabol. 2016;24:795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitter S.K., Rao H.V., Qi X. Autophagy in the retina: a potential role in age-related macular degeneration. Adv. Exp. Med. Biol. 2012;723:83–90. doi: 10.1007/978-1-4614-0631-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyagishima K.J., Wan Q., Corneo B. In pursuit of authenticity: induced pluripotent stem cell-derived retinal pigment epithelium for clinical applications. Stem Cells Transl. Med. 2016;5:1562–1574. doi: 10.5966/sctm.2016-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordgaard C.L., Berg K.M., Kapphahn R.J. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2006;47:815–822. doi: 10.1167/iovs.05-0976. [DOI] [PubMed] [Google Scholar]
- 45.Nordgaard C.L., Karunadharma P.P., Feng X., Olsen T.W., Ferrington D.A. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of AMD. Invest. Ophthalmol. Vis. Sci. 2008;49:2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsen T.W., Feng X. The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2004;45:4484–4490. doi: 10.1167/iovs.04-0342. [DOI] [PubMed] [Google Scholar]
- 47.Roth F., Bindewald A., Holz F.G. Key pathophysiologic pathways in age-related macular disease. Graefes Arch. Clin. Exp. Ophthalmol. 2004;242:710–716. doi: 10.1007/s00417-004-0976-x. [DOI] [PubMed] [Google Scholar]
- 48.Saihara K., Kamikubo R., Ikemoto K., Uchida K., Akagawa M. Pyrroloquinoline Quinone, a redox-active o-Quinone, stimulates mitochondrial biogenesis by activating the SIRT1/PGC-1α signaling pathway. Biochemistry. 2017;56:6615–6625. doi: 10.1021/acs.biochem.7b01185. [DOI] [PubMed] [Google Scholar]
- 49.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stites T.E., Mitchell A.E., Rucker R.B. Physiological importance of Quinoenzymes and the O-Quinone family of cofactors. J. Nutr. 2000;130:719–727. doi: 10.1093/jn/130.4.719. [DOI] [PubMed] [Google Scholar]
- 51.Stites T., Storms D., Bauerly K. Pyrroloquinoline quinone modulates mitochondrial quantity and function in mice. J. Nutr. 2006;136:390–396. doi: 10.1093/jn/136.2.390. [DOI] [PubMed] [Google Scholar]
- 52.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 53.St-Pierre J., Drori S., Uldry M. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 54.Stromland O., Niere M., Nikiforov A.A. Keeping the balance in NAD metabolism. Biochem. Soc. Trans. 2019;47:119–130. doi: 10.1042/BST20180417. [DOI] [PubMed] [Google Scholar]
- 55.Terluk M.R., Kapphahn R.J., Soukup L.M. Investigating mitochondria as a target for treating age-related macular degeneration. J. Neurosci. 2015;35(18):7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terluk M.R., Ebeling M.C., Fisher C.R. N-acetyl-L-cysteine protects human retinal pigment epithelial cells from oxidative damage: implications for age-related macular degeneration. Oxidative Med. Cell, Longev. 2019 doi: 10.1155/2019/5174957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsubota K. The first human clinical study for NMN has started in Japan. npj Aging Mech. Dis. 2016;2:16021. doi: 10.1038/npjamd.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tu C., Li J., Jiang X. Ion-current-based proteomic profiling of the retina in a rat model of Smith-Lemli-Opitz syndrome. Mol. Cell. Proteomics. 2013;12:3583–3598. doi: 10.1074/mcp.M113.027847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang A.L., Lukas T.J., Yuan M. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PloS One. 2009;4 doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whyte A.J., Kerhrl T., Brooks D.E., Katz K.D., Sokolowski D. Safety and effectiveness of acetadote for acetaminophen toxicity. J. Emerg. Med. 2010;39:607–611. doi: 10.1016/j.jemermed.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 61.Wong W.T., Dresner S., Forooghian F. Treatment of geographic atrophy with subconjunctival sirolimus: results of a phase I/II clinical trial. Invest. Ophthalmol. Vis. Sci. 2013;54:2941–2950. doi: 10.1167/iovs.13-11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong W.L., Su X., Li X. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 63.Yoshino J., Baur J.A., Imai S. NAD+ intermediates: the biology and therapeutic potential of NMN and NMR. Cell Metabol. 2018;27:513–528. doi: 10.1016/jcmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zafarullah M., Li W.Q., Sylvester J., Ahmad M. Molecular mechanisms of N-acetylcysteine actions. Cell. Mol. Life Sci. 2003;60:6–20. doi: 10.1007/s00018030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L., Liu J., Cheng C. The neuroprotective effect of pyrroloquinoline quinone on traumatic brain injury. J. Neurotrauma. 2008;29:851–864. doi: 10.1089/neu.2011.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao C., Yasumura D., Li X. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu X.H., Lu M., Lee B.Y. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2876–2881. doi: 10.1073/pnas.141792112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.