Abstract
Retinal ganglion cell (RGC) degeneration is the root cause for vision loss in glaucoma as well as in other forms of optic neuropathy. A variety of studies have implicated abnormal mitochondrial quality control (MQC) as contributing to RGC damage and degeneration in optic neuropathies. The ability to differentiate human pluripotent stem cells (hPSCs) into RGCs provides an opportunity to study RGC MQC in great detail. Degradation of damaged mitochondria is a critical step of MQC, and here we have used hPSC-derived RGCs (hRGCs) to analyze how altered mitochondrial degradation pathways in hRGCs affect their survival. Using pharmacological methods, we have investigated the role of the proteasomal and endo-lysosomal pathways in degrading damaged mitochondria in hRGCs and their precursor stem cells. We found that upon mitochondrial damage induced by the proton uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP), hRGCs more efficiently degraded mitochondria than did their precursor stem cells. We further identified that for degrading damaged mitochondria, stem cells predominantly use the ubiquitine-proteasome system (UPS) while hRGCs use the endo-lysosomal pathway. UPS inhibition causes apoptosis and cell death in stem cells, while hRGC viability is dependent on the endo-lysosomal pathway but not on the UPS pathway. These findings suggest that manipulation of the endo-lysosomal pathway could be therapeutically relevant for RGC protection in treating optic neuropathies associated with mitophagy defects. Endo-lysosome dependent cell survival is also conserved in other human neurons as we found that differentiated human cerebral cortical neurons also degenerated upon endo-lysosomal inhibition but not with proteasome inhibition.
Keywords: Stem cells, Human retinal ganglion cells (hRGCs), Glaucoma, Mitophagy, Autophagy, Ubiquitine-proteasome system (UPS)
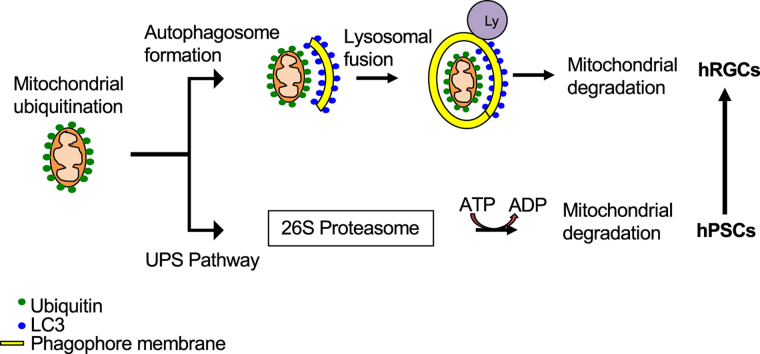
Graphical abstract
The above graphical abstract represents a summary of the data presented in this work. Our data shows human pluripotent stem cells (hPSCs) predominantly use the ubiquitin-proteasome system (UPS) for maintaining mitochondrial homeostasis. However, during the course of differentiation to human retinal ganglion cells (hRGCs), cellular preference for maintaining mitochondrial homeostasis shifts towards the endo-lysosomal pathway, which becomes essential for hRGC survival.
Highlights
-
•
Human retinal ganglion cells (hRGCs) degrade damaged mitochondria more efficiently than the origin stem cells.
-
•
Human stem cells rely on the ubiquitin proteasome system (UPS) for damaged mitochondrial clearance and survival.
-
•
hRGCs rely on the endo-lysosomal pathway for mitochondrial clearance and survival.
-
•
Unlike stem cells, proteasomal inhibition did not cause severe cell death for hRGCs.
-
•
Transition from the UPS to endo-lysosomal pathway during differentiation was also observed for cerebral cortical neurons.
1. Introduction
Optic neuropathies such as glaucoma, Leber's hereditary optic neuropathy (LHON), dominant optic atrophy (DOA) [1] and several other neurodegenerative diseases have been reported to be associated with abnormal mitochondrial quality control (MQC) [2,3]. Almost all LHON patients carry a mutation in one of the complex I subunits of the mitochondrial respiratory chain at 3460G > A (ND1), 11778G > A (ND4) or 14484T > C (ND6). Identification of such mutations is in fact used for definitive LHON diagnosis [4]. Nearly 75% of DOA patients are diagnosed with mutations in the OPA1 gene, which is a nuclear encoded inner mitochondrial membrane protein involve in mitochondrial fusion [5]. Genetic analysis has identified several gene mutations associated with the primary open-angle glaucoma (POAG) patients [6,7]. Additionally, a small subset POAG patients have been found to have mutations in the Optineurin (OPTN) gene [8], which is a key player in mitochondrial autophagy (mitophagy) [9]. In almost all of these optic neuropathies, irreversible damage of retinal ganglion cells (RGCs) leads to vision loss and can result in complete blindness [1].
MQC involves mitochondrial dynamics (fission/fusion), biogenesis and degradation. While each step of MQC is important for mitochondrial homeostasis, defects in mitochondrial degradation are particularly severe, as they result in an accumulation of damaged mitochondria and can ultimately lead to cell death through apoptosis [[10], [11], [12]]. Macroautophagy is a conserved catabolic process in which damaged proteins or organelles are degraded by forming a double membrane structure around them, allowing complex protein interactions known as autophagosomes. These autophagosomes then fuse with the lysosomes where the damaged materials are degraded [[13], [14], [15]]. Selective degradation of damaged mitochondria through the lysosome-mediated autophagic pathway is called mitophagy [16,17]. Apart from cell autonomous mitophagy, a recent report has also shown that RGCs in mice shed mitochondria at the optic nerve head (ONH), a process mediated by adjacent astrocytes referred to as transmitophagy [18].
Advanced imaging techniques have been very powerful to understand the details of retinal ganglion cell layer in living eye [19], however these techniques still lack the ability to identify mitochondrial abnormality in RGC layer. In vivo models as well as cultured cells have been instrumental in understanding molecular details of MQC pathways and the pathophysiology associated with abnormal MQC [20]. However, mitochondrial abnormalities have different consequences in different cells, and one powerful example of this is the propensity for certain mitochondrial mutations to specifically affect RGCs in mitochondrial optic neuropathies [4,5,8]. Also, recent single-cell transcriptomic studies further suggest that there are many basic differences between rodent and primate retinal cells [21]. Hence, an increased understanding of MQC in human RGCs could be therapeutically important for the mitochondrial optic neuropathies. Therefore, in order to promote the understanding and treatment of human optic neuropathies we feel it is important to study MQC in the context of human RGCs, and to do so we have been studying stem-cell derived human RGCs using in vitro models of mitochondrial stress. Furthermore, a stem cell-based approach will enable us to study the adaption of the MQC pathways during the course of RGC differentiation by comparing the process in stem cells versus in differentiated hRGCs.
Healthy mitochondrial homoeostasis in adult human stem cells is required to prevent stem cell aging and to maintain pluripotency [22]. The endo-lysosomal and proteasomal pathways are the two major cellular quality control pathways for clearing damaged organelles and proteins. However, it is unclear how hRGCs and their origin stem cells use either pathway for maintaining mitochondrial homeostasis. Studies in mice have shown that mitophagy is required for the self-renewal [23,24] and differentiation [25] of hematopoietic stem cells (HSCs) as well as for cancer stem cell maintenance [26,27] in humans. The ubiquitin proteasome system (UPS) is highly active in hPSCs and upon cellular differentiation, the proteasome remains active but at a reduced level [28,29]. It is still unclear if hPSCs use the UPS system for degrading damaged mitochondria.
Several studies in mice have shown that programmed mitophagy is required for RGC differentiation [30,31], and the E50K mutation in the autophagy adaptor protein optineurin (OPTN) has been shown to cause mitochondrial defects and RGC death [32]. Additionally, the OPTNE50K mutation is associated with some cases of so-called normal-tension glaucoma (NTG) [8]. It is well accepted that mitochondrial dynamics and quality control are central to mouse RGC viability [33]; however, the role of the lysosomal-autophagy and proteasomal pathways in degrading damaged mitochondria in hRGCs and its effect on hRGC survival are not yet understood.
In this study, we used small molecule-based hPSC differentiation and bead-based immunopurification to obtain highly pure, well-characterized hRGCs [34]. These hRGCs were used to investigate the role of the endo-lysosomal and the proteasomal pathways in clearing damaged mitochondria in comparison to their precursor stem cells. Our study indicates that hRGCs predominantly use the endo-lysosomal pathway for degrading damaged mitochondria to prevent apoptosis, whereas hPSCs primarily use the proteasomal pathway for mitochondrial clearance and cell survival.
2. Materials and methods
2.1. Reporter line generation
H9 and H7 (WiCell, Madison, https://www.wicell.org) human embryonic stem cells with the BRN3B–P2A-tdTomato-P2A-THY1.2 reporters were developed in our lab [34] and the H7 reporter line contains additional TRE:Cas9 cassette as described in Ref. [35] but without Cas9 induction for this study. Both the reporter lines are referred here as H9-ESCs and H7-ESCs and the differentiated RGCs are H9-RGCs and H7-RGCs respectively. iPSCs (EP1) were developed in our lab [36] and used here as EP1-iPSCs. EP1 with the BRN3B–P2A-tdTomato-P2A-THY1.2 reporter was made by CRISPR/Cas9-based gene editing using a gRNA plasmid with Cas9 and puromycin selection, and the donor plasmid with the reporter genes as used before [34]. In Brief, EP1-iPSCs were transfected with the DNA-In stem reagent (MTI-GlobalStem/ThermoFisher) followed by fresh media change after 24 h. After 40 h of transfection, cells were selected against puromycin (0.9 μg/ml) for 24 h and recovered for 4–5 days with fresh media without puromycin. To isolate positive clones, cells were re-plated at clonal density, and single colonies were genotyped by PCR as described in the previous study [34].
2.2. Human RGC differentiation and immunopurification
RGC reporter lines were plated on 1% (vol/vol) Matrigel-GFR (BD Biosciences) coated dishes and differentiated using small molecules as described in the previous study [34]. In brief, stem cells were plated on Matrigel coated plate at ~53K/cm2 density in mTeSR media (Stemcell Technologies) with 5 μM blebbistatin and incubated in the hypoxia incubator (37 °C, 10% CO2, 5% O2), this day referred as day minus one (d-1). After 24 h, on d0 cells were transferred into the normoxia incubator (37 °C, 5% CO2, 20% O2) for differentiation and mTeSR was replaced with the differentiation media (N2B27: 1:1 mixture of DMEM/F12 and Neurobasal, 1X GlutaMAX supplement, 1X antibiotic-antimycotic, 1% N2 supplement and 2% B27 supplement (all from ThermoFisher Scientific). On d1, the following small molecules were added with full exchange of N2B27 media: Forskolin (FSK; 25 μM, Cell Signaling Technology), Dorsomorphin (DSM; 1 μM, Stemcell Technologies), IDE2 (2.5 μM, R&D Systems) and Nicotinamide (NIC; 10 mM, Sigma-Aldrich). All small molecules were treated with full exchange of N2B27 media every other day until d6, between d6 to d10 cells were treated with FSK and NIC, d10-18 with FSK, d18-30 with FSK and DAPT (10 μM, Sigma-Aldrich), and from d30 onwards only with N2B27 media. Successful RGC differentiation was monitored by tdTomato expression. RGC purification was done during day 40–45 after dissociation with Accumax cell dissociation solution (Sigma-Aldrich) followed by purification using CD90.2 (THY1.2) MicroBeads and magnetic activated cell sorting system (MACS, Miltenyi Biotec) as described in the previous studies [34,37].
2.3. hPSC and RGC maintenance and drug treatments
hPSCs and RGCs were cultured and maintained on 1% (vol/vol) Matrigel-GFR (BD Biosciences) coated dishes in mTeSR and N2B27 media [34] respectively. Stem cells and RGCs were cultured in 37 °C hypoxia (10% CO2, 5% O2) and normoxia (5% CO2, 20% O2) incubators, respectively. The following drugs were used in this study: CCCP (Sigma), bafilomycin A1 (Baf, Sigma), hydroxychloroquine (HCQ, Fisher Scientific), bortezomib (Selleckchem), oligomycin (Millipore), antimycin (Sigma), oligomycin-antimycin (OA) drug combination used at 10 μM and 4 μM concentrations respectively, and MG132 (Sigma). All the drugs were dissolved in DMSO except hydroxychloroquine which is soluble in water and the stock solutions were prepared in a way such that each condition requires similar volume of drug solution. For hydroxychloroquine treatment similar volume of DMSO was added to the culture followed by drug addition. All the treatments were compared to the corresponding DMSO group.
2.4. Mitochondrial DNA quantification by qPCR
After purification, RGCs were plated on Matrigel-coated tissue culture plates and grown for three days prior to the indicated drug treatments. Cells were dissociated using Accumax for 15min and quenched with the N2B27 media followed by centrifugation at 300×g for 5min. DNA was isolated from the cell pellets using DNeasy Blood and Tissue kit (Qiagen) followed by simultaneous quantification of the mitochondrial and nuclear DNA content within the same sample using TaqMan chemistry (Thermo Fisher) with StepOnePlus Real-Time PCR system (Applied Biosystems). Human mitochondrial DNA was detected via measurement of the very stable region on the mitochondrial ND1 gene [38] using following primers [39];
Forward: 5′ CCTTCGCTGACGCCATAAA3′, reverse: 5′TGGTAGATGTGGCGGGTTTT3′, ND1-probe: 6FAM-5′TCTTCACCAAAGAGCC3′-MGBNFQ (6FAM and the MGBNFQ are the fluorescence reporter and quencher respectively). For an internal control, nuclear DNA content was measured using the human RNase P gene (TaqMan Copy Number Reference assay Catalog # 4403326).
For hPSCs (H9, H7-ESCs and EP1-iPSCs), 15,000 cells were plated on each well of a Matrigel-coated 96-well dish. After 24 h of recovery, cells were treated with the indicated drugs and dissociated with Accutase (Millipore-Sigma) and quenched with mTeSR media containing blebbistatin (Sigma), followed by centrifugation at 300×g for 5min to pellet the cells. Mitochondrial content for each sample was measured as explained above.
2.5. Mitochondrial quantification by flow cytometry
Flow cytometry-based measurements were done using a mitochondria specific dye, mito tracker deep red (MTDR, Molecular probes) using the cell sorter SH800 (Sony) on analyzer mode. 10,000–20,000 cells were analyzed at the FL-4 channel (far-red) to measure MTDR intensity. For 3 h CCCP treatments (Fig. 1G–I), cells were labelled before treatment, while for 24 h treatments, cells were labelled afterwards with media containing 10 nM MTDR for 15 min at 37 °C. Cells were dissociated and centrifuged as explained in the qPCR method followed by suspension in media without MTDR for flow analysis.
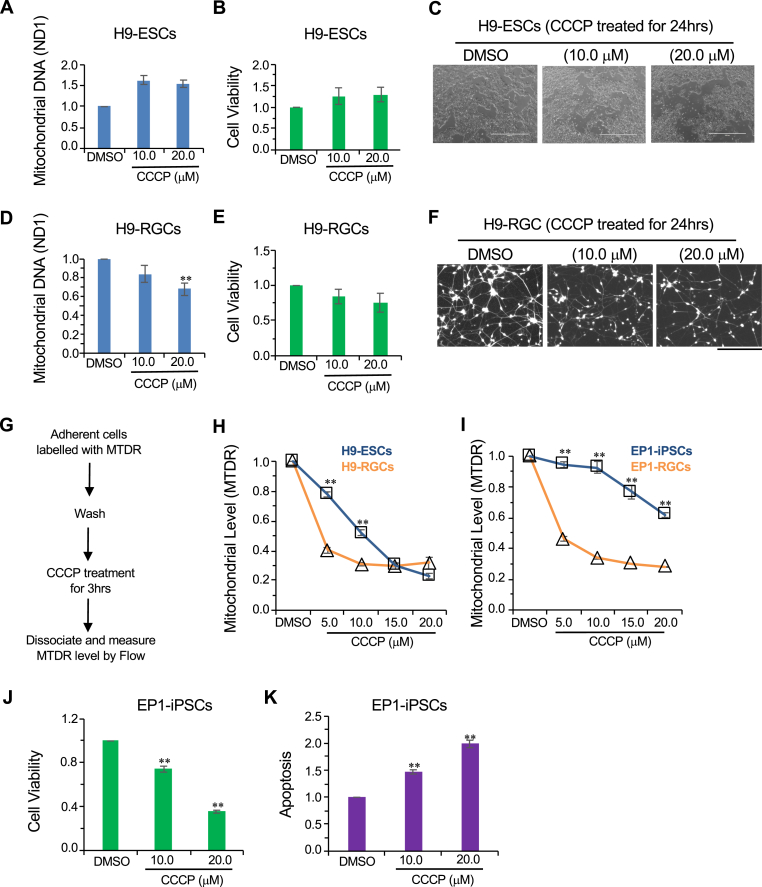
Fig. 1.
Mitochondrial degradation in stem cells and hRGCs upon CCCP treatment. (A, D) Mitochondrial content analyzed by qPCR for the mitochondrial gene ND1 and normalized with-respect-to (w.r.t) the nuclear gene RNase P. Shown are ΔΔct fold change relative to the DMSO control after 3 h treatment with the indicated CCCP doses for both h9-ESCs (A) and H9-RGCs (D). Data shown are from three independent biological replicates with three technical replicates for each biological replicate. (B, E) Cell viability measurements of H9-ESCs (B) and H9-RGCs (E) after 24 h treatment with the indicated doses of CCCP. Cell viability was measured using the fluorescence based ApoTox-Glo triplex assay kit and normalized w.r.t DMSO control. Data shown are from three independent biological replicates. (C, F) Brightfield images shown are H9-ESCs after 24 h treatment with CCCP (C), fluorescence images shown are in the red channel for tdTomato expressing H9-RGCs after 24 h CCCP treatments (F). (G–I) Mitochondrial level analyzed by flow cytometry using the mitochondria specific dye MTDR followed by CCCP treatments for 3 h. Chart (G) shows the experimental design while the graphs show loss of mitochondria labelled MTDR intensity normalized w.r.t DMSO control at different CCCP doses for H9-ESCs (H) and EP1-iPSCs (I) compared to the corresponding RGCs. Data presented are from 3 to 10 independent biological replicates and statistical analysis is done between stem cells and corresponding RGCs at the indicated treatments. (J, K) Cell viability (J) and apoptosis by luminescence-based caspase-3/7 activity (K) were measured for EP1-iPSCs using ApoTox-Glo triplex assay kit and normalized w.r.t DMSO control after 24 h treatment with the indicated drugs using three independent biological replicates. Scale bars, 1000 μm (C) and 200 μm (F). Error bars are SEM. **, p-value < 0.005. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.6. Cell viability and apoptosis measurements
Cell viability and apoptosis were measured using ApoTox-Glo Triplex assay kit (Promega) following manufacturer's guideline with the CLARIOstar microplate reader (BMG LABTECH). Cell viability was measured by the ratio of fluorescence intensity between 400 nm (viability) and 482 nm (cytotoxicity) channels, and apoptosis was measured by luminescence-based caspase-3/7 activity. hPSCs (10,000/well) and RGCs (15,000/well) were plated in each well of Matrigel-coated 96-well dishes in mTeSR containing blebbistatin and N2B27 media respectively. After one day (hPSCs) and three days (RGCs) of recovery, cells were treated with the indicated drugs for 24 h and analyzed for cell viability and apoptosis.
2.7. Image acquisition to show cell viability
Images were acquired after indicated treatments using an EVOS FL Imaging System (ThermoFisher Scientific).
2.8. Lysosome/acidic vesicles inhibition assay
20,000 H9-RGCs were plated and grown on Matrigel-coated glass-bottom dishes (MatTek) in N2B27 media for three days in the 37 °C normoxia incubator. After 24 h of treatment with the endo-lysosomal inhibitors Baf or HCQ, media was replaced with 100 μl of media containing pH sensitive pHrodo-green conjugated dextran solution (ThermoFisher Scientific) (100 μg/ml) for 20min followed by a fresh media exchange after a brief wash. Confocal (Zeiss LSM 710) live images were acquired using the live cell set up with plan-apochromat 40x/oil objective with 1.4 numerical aperture. pHrodo-Green and tdTomato-expressing RGCs were detected with the 488 nm and 560 nm laser lines respectively.
2.9. Human cortical neuron differentiation
Cortical neurons were differentiated from H1-ESCs as described in Xu et al. [40]. Experiments were performed on 100–120 days of post-differentiated cortical neurons.
2.10. Immunofluorescence and imaging
20,000 purified RGCs were plated on matrigel-coated glass-bottom dishes (MatTek) for three days followed by 24 h treatment with the indicated drugs. Cells were fixed with 4% paraformaldehyde in PBS for 15 min at 37 °C followed by 1 h of blocking at room temperature with blocking solution (PBS with 5% donkey serum and 0.2%Triton X-100). The following primary antibodies were used overnight at 4 °C in blocking solution: Rabbit-anti-ubiquitin (Cell Signaling Technology, 1:200 dilution), Rabbit-anti-RBPMS (GeneTex, 1:200 dilution) and Mouse-anti-TUBB3 (BioLegend, 1:200 dilution). Samples were washed for three times for 5min each with washing solution (PBS with 1% donkey serum and 0.05%Triton X-100) and incubated with the secondary antibodies (1:500 dilution) in blocking solution for 1 h at room temperature. Following the secondary antibody, samples were again washed three times, with DAPI added in the second wash.
Cultured human cortical neurons of 100–120 days post-differentiation were immunostained as above with primary antibodies against MAP2 (Mouse-anti-MAP2, Sigma, 1:200 dilution), VGLUT1 (Mouse-anti-VGLUT, SYSY, 1:2500 dilution) and VGAT (Rabbit-anti-VGAT, SYSY, 1:500 dilution). Confocal images were acquired using LSM 710 (Zeiss) as done for pHrodo-Green, but without the live set-up.
2.11. Statistical analysis
Statistical comparisons between two data sets were done with the Student's t-test. One-way ANOVA tests were performed for analysis containing three or more independent groups followed by post hoc t-test for individual groups with respect to the control DMSO group (Table-1).
Table 1.
One-way ANOVA test for group of data containing three or more data sets and post hoc test with respect to the control DMSO data set.
| Fig | One-way ANOVA (p value) | Post hoc test with respect to control (p value) |
|---|---|---|
| Fig. 1A | 1.3226E-05 | CCCP (10.0 μM): 1.9045E-05 CCCP (20.0 μM): 1.32723E-05 |
| Fig. 1B | 0.36823588 | CCCP (10.0 μM): 0.24992561 CCCP (20.0 μM): 0.15533524 |
| Fig. 1D | 0.00696819 | CCCP (10.0 μM): 0.102379 CCCP (20.0 μM): 0.00018268 |
| Fig. 1E | 0.27528422 | CCCP (10.0 μM): 0.20314162 CCCP (20.0 μM): 0.14190736 |
| Fig. 1J | 6.288E-07 | CCCP (10.0 μM): 0.00068674; CCCP (20.0 μM): 1.07961E-06 |
| Fig. 1K | 1.6562E-05 | CCCP (10.0 μM): 0.00027468 CCCP (20.0 μM): 0.00014416 |
| Fig. 2A | 0.00333067 | Baf (50.0 nM): 0.02347454 HCQ (30.0 μM): 0.19815269 CCCP (10.0 μM): 0.89069482 CCCP (10.0 μM) + Baf (50.0 nM): 0.15636769 OA: 0.00972516 OA + Baf (50.0 nM): 0.03139634 |
| Fig. 2B | 1.6741E-08 | Baf (50.0 nM): 0.00010464 HCQ (30.0 μM): 0.00105926 CCCP (10.0 μM): 0.24992561 CCCP (10.0 μM) + Baf (50.0 nM): 5.65264E-05; OA: 2.70799E-07 OA + Baf (50.0 nM): 1.94566E-08 |
| Fig. 2C | 2.4542E-06 | Baf (50.0 nM): 0.0060723 HCQ (30.0 μM): 0.06146984 CCCP (10.0 μM): 0.10332896 CCCP (10.0 μM) + Baf (50.0 nM): 0.0001987 OA: 0.00134962 OA + Baf (50.0 nM): 0.00029999 |
| Fig. 2H | 0.00193593 | Baf (50.0 nM): 0.26729767 HCQ (30.0 μM): 0.01818717 CCCP (10.0 μM): 0.07470509 CCCP (10.0 μM) + Baf (50.0 nM): 0.49075726; CCCP (10.0 μM) + HCQ (30.0 μM): 6.50831E-09 |
| Fig. 2I | 3.41207E-06 | Baf (50.0 nM): 0.0005702 HCQ (30.0 μM): 0.00035478 CCCP (10.0 μM): 0.20314162 CCCP (10.0 μM) + Baf (50.0 nM): 0.00033786; CCCP (10.0 μM) + HCQ (30.0 μM): 1.10269E-05 |
| Fig. 2J | 6.8186E-05 | Baf (50.0 nM): 0.00259663; HCQ (30.0 μM): 1.30404E-05; CCCP (10.0 μM): 0.00146037; CCCP (10.0 μM) + Baf (50.0 nM): 0.00278443; CCCP (10.0 μM) + HCQ (30.0 μM): 0.00392949 |
| Fig. 3A | 0.00080816 | Bort (50.0 nM): 0.21089911 Bort (500.0 nM): 0.00263605 Bort (1000.0 nM): 0.00060078 |
| Fig. 3B | 0.03867105 | Bort (50.0 nM): 0.25931389 Bort (500.0 nM): 0.02038087 Bort (1000.0 nM): 0.00686592 |
| Fig. 3C | 0.29285313 | Bort (50.0 nM): 0.25931389 CCCP (10.0 μM): 0.89069482 CCCP (10.0 μM) + Bort (50.0 nM): 0.00879123 |
| Fig. 3I | 5.9384E-08 | Bort (15.0 nM): 1.7877E-06; Bort (50.0 nM): 0.00015752; Bort (500.0 nM): 7.07749E-05; Bort (1000.0 nM): 4.18868E-05 |
| Fig. 3L | 9.6407E-05 | Bort (15.0 nM): 1.00596E-05 Bort (50.0 nM): 0.00030353 |
| Fig. 3 N | 0.00954914 | MG132 (10.0 μM): 3.14391E-05; MG132 (20.0 μM): 0.11225478 |
| Fig. 3O | 0.0737521 | MG132 (10.0 μM): 0.06210585 MG132 (20.0 μM): 0.04643869 |
| Fig. 4B | 0.2133412 | HCQ (30.0 μM): 0.43894442 HCQ (60.0 μM): 0.02783898 |
| Fig. 4C | 0.06621655 | HCQ (30.0 μM): 0.03771928 HCQ (60.0 μM): 0.00060646 |
| Fig. 4D | 0.36246771 | Bort (50.0 nM): 0.39282229 Bort (500.0 nM): 0.32810538 |
| Fig. 4E | 5.8274E-05 | Bort (50.0 nM): 0.00011123 Bort (500.0 nM): 0.19615009 |
| Fig. S5A | 3.6587E-05 | Bort (15.0 nM): 5.369E-05; Bort (50.0 nM): 0.00012986 Bort (500.0 nM): 0.00030233 Bort (1000.0 nM): 0.0032803 |
| Fig. S5B | 6.9674E-05 | Bort (15.0 nM): 0.00018375 Bort (50.0 nM): 0.00498865; Bort (500.0 nM): 4.76206E-06; Bort (1000.0 nM): 0.00020643 |
| Fig. S5C | 3.4096E-05 | Bort (15.0 nM): 0.00022683 Bort (50.0 nM): 0.00016114 Bort (500.0 nM): 0.00018035 Bort (1000.0 nM): 0.00413441 |
3. Results
3.1. RGCs are more efficient in degrading damaged mitochondria than their precursor stem cells
To investigate mitochondrial degradation in both differentiated human RGCs and in the stem cells from which they originated, we used a CRISPR/Cas9 mediated genetically engineered human embryonic stem cell (hESC-H9) reporter line with a P2A-tdTomato-P2A-Thy1.2 construct introduced into the endogenous RGC-specific POU4F2 (BRN3B) locus [34]. Small molecule-based differentiation followed by immunopurification of Thy1.2-expressing cells yields highly enriched RGCs, which are positive for RGC specific markers such as RBPMS [41] and the neuronal marker TUBB3 (Supplemental Fig. S1) and that have been well-characterized transcriptomically and electrophysiologically [34,42]. The reporter line will be referred as “H9-ESCs” and the corresponding RGCs as “H9-RGCs.” To study the effect of mitochondrial damage on RGCs and stem cells, we have used the mitochondrial uncoupler CCCP. Upon mitochondrial damage with CCCP for 3 h, H9-ESCs showed no reduction in their mitochondrial level (Fig. 1A), as measured by a qPCR assay that compares the level of mitochondrial gene ND1 DNA to that of nuclear gene RNase P DNA. This result was surprising because CCCP has been reported to induce mitophagy within 1 h of exposure [43], and hence we expected to see a decrease in mitochondrial content. We hypothesized that due to this apparent lack of mitochondrial clearance in the H9-ESC cells, which would presumably lead to a buildup of CCCP-induced damaged mitochondria, there would be an increase of cell death. However, even with 24 h treatment with CCCP there was no detectable cell death in H9-ESCs (Fig. 1B and C). Contrary to the situation with H9-ESCs, CCCP treatment for 3 h reduced the mitochondrial content of H9-RGCs (Fig. 1D). As with the H9-ESCs, CCCP treatment did not result in RGC cell death at 24 h (Fig. 1E and F). These results suggest that RGCs may be more efficient in degrading damaged mitochondria than their precursor stem cells.
With the unexpected result of no change in mitochondrial content with CCCP and yet no cell death for H9-ESCs, we next asked whether ESCs might be clearing up damaged mitochondria while simultaneously synthesizing more mitochondria to keep up with their metabolic needs for rapid cell division, and this simultaneous new synthesis might be masking possible mitochondrial degradation. To test this possibility, we tracked mitochondrial levels upon CCCP treatment using the mitochondria-specific dye mitotracker deep red (MTDR) followed by flow cytometry [44] (Supplemental Fig. S2). MTDR covalently binds to the reduced thiols within the mitochondria matrix proteins and once bound, MTDR remains in the mitochondria independent of mitochondrial membrane potential [45,46]. CCCP lowers mitochondrial membrane potential which could affect initial MTDR binding; to avoid this potential artifact mitochondria were labelled with the MTDR dye prior to CCCP treatment. After which appropriate incubation was followed with flow cytometry-based analysis of mitochondrial content (Fig. 1G). With this experimental paradigm, mitochondria synthesized after CCCP treatment will not be labelled, and thus will not be detected, and hence they will not mask possible degradation of pre-existing damaged mitochondria. Interestingly, we observed reduced mitochondria levels with increasing doses of CCCP for H9-ESCs (Fig. 1H) and saw similar but more dramatic reduction in mitochondria in H9-RGCs at 5 and 10 μM CCCP (Fig. 1H). To make sure this was not a cell line-specific effect, we performed a parallel experiment with an iPSC-derived POU4F2 reporter line (EP1-iPSCs), again examining both undifferentiated stem cells and differentiated RGCs (EP1-RGCs). In agreement with the H9 results, the iPSCs showed mitochondrial degradation with increasing doses of CCCP and the corresponding RGCs degraded mitochondria more efficiently compared to the undifferentiated iPSCs (Fig. 1I). Of note, EP1-iPSCs showed relatively reduced mitochondrial clearance compared to the H9-ESCs (Fig. 1H and I) and upon CCCP damage, correspondingly showed reduced cell viability and increased apoptosis (Fig. 1J and K), further supporting the hypothesis that inefficient degradation of damaged mitochondria may lead to apoptotic cell death. It should be noted that in this experiment, based upon prior protocols, the stem cells were cultured and treated with CCCP in the hypoxic (10% CO2, 5% O2) condition while the RGCs were cultured under normoxic condition (5% CO2, 20% O2). Given this difference, we asked whether the observed difference in mitochondrial degradation between stem cells and corresponding RGCs might be due to the difference in culture conditions. To test this, both H9-ESCs and the corresponding RGCs were cultured and treated with CCCP under similar normoxic conditions followed by mitochondrial measurements. In agreement with our prior findings (Fig. 1H and I), we observed increased mitochondrial degradation in the H9-RGCs compared to their precursor stem cells (Supplemental Fig. S3), suggesting that hRGCs are more efficient in degrading damaged mitochondria irrespective of their culture conditions. Investigating the pathways involved in degrading damaged mitochondria in hRGCs could be therapeutically important as modulation of the pathways involved could potentially be used to enhance hRGC survival under specific conditions.
3.2. The endo-lysosomal pathway is required for hRGCs but not for hESCs to degrade damaged mitochondria
To better define possible cell type-specific mechanisms of mitochondrial quality control in hRGCs, we first tested the role of the endo-lysosomal pathway in degrading damaged mitochondria in hPSCs and hRGCs. Mitochondrial levels were measured after CCCP treatment both in the presence and absence of the endo-lysosomal inhibitors hydroxychloroquine (HCQ) [47] and Bafilomycin A1 (Baf) [48]. qPCR-based analysis showed that individual treatment with CCCP, HCQ, or Baf, as well as CCCP with Baf, did not affect mitochondrial level in H9-ESCs (Fig. 2A). Tracking MTDR labelled mitochondrial content upon CCCP treatment showed mitochondrial degradation in hPSCs (Fig. 1H and I). Our inability to detect an increase in mitochondria levels upon endo-lysosomal inhibition suggests the existence of an alternative pathway in H9-ESCs for degrading damaged mitochondria. We observed significant cell death and apoptosis when cells were treated with Baf but did not observe similar cell death with HCQ treatment (Fig. 2B and C). This could be due to the requirement of endo-lysosomal activity and the autophagy pathway for other cellular functions, such as non-mitochondrial protein and organelle homeostasis [49,50]. The differential effects of Baf and HCQ on H9-ESC survival could be due to the distinct modes of action of the two inhibitors [51].
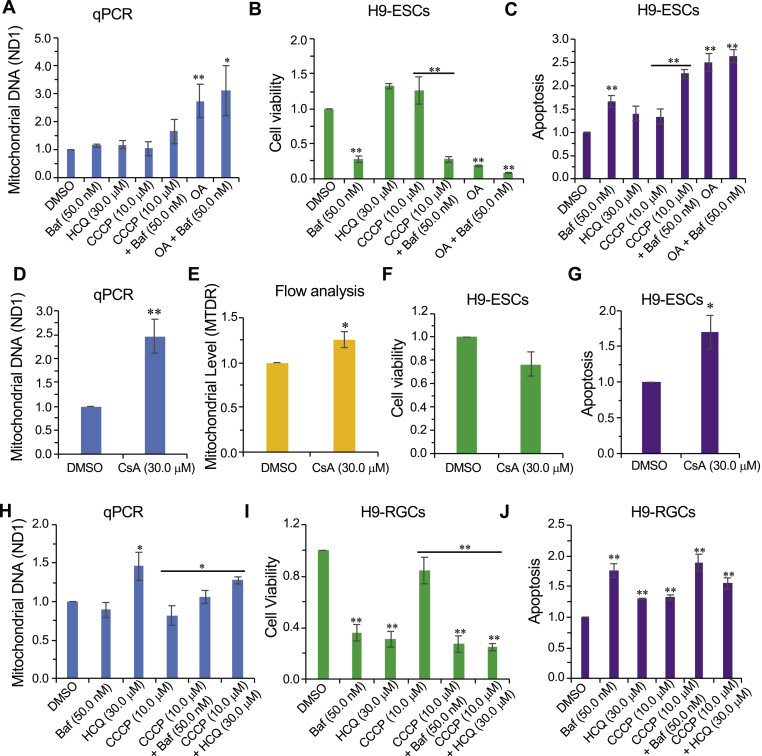
Fig. 2.
hRGCs, but not hESCs, predominantly use the endo-lysosomal pathway for degrading mitochondria. (A, D, H) qPCR-based analysis of the mitochondrial content for H9-ESCs (A, D) and H9-RGCs (H) after 24 h treatment with the indicated drugs, the quantification and analysis are done as in (Fig. 1). Data shown are from 3 to 5 independent biological replicates with three technical replicates for each biological replicate. (B, F, I) Shown are cell viability measurements after 24 h treatment with the indicated drugs for H9-ESCs (B, F) and H9-RGCs (I), and are done as in (Fig. 1). (C, G, J) Quantifications represent cellular apoptosis, measured by luminescence-based caspase-3/7 activity for H9-ESCs (C, G) and H9-RGCs (J). Data shown are from three independent biological replicates. (E) Flow cytometry-based analysis of the MTDR labelled mitochondria for H9-ESCs after 24 h treatment with the indicated drug, quantification shows normalized average intensity in the MTDR channel w.r.t DMSO from nine independent biological replicates. Error bars are SEM. **, p-value < 0.007; *, p-value < 0.05.
We next tested if inhibition of the mitochondrial electron transport chain (mETC) with oligomycin-antimycin (OA) would cause mitochondrial degradation in ESCs. Interestingly, we did not observe reduced mitochondrial levels with OA treatment; on the contrary, we observed increased mitochondrial levels when cells were treated alone or in combination with Baf (Fig. 2A). This finding is consistent with a prior report that inhibition of the mETC is associated with the inhibition of autophagy [52], which could account for the observed increase in mitochondrial content, and the increased cell death and apoptosis with OA treatment (Fig. 2B and C). Since oligomycin has been reported to increase inner mitochondria membrane potential (ΔΨm) [53,54], we further asked if increasing ΔΨm by inhibition of the mitochondrial permeability transition pore (mPTP) with cyclosporin A (CsA) [54] could also block mitochondrial degradation. In agreement with the OA result, we observed increased mitochondrial levels in H9-ESCs treated with CsA (Fig. 2D and E). Presumably because inhibition of damaged mitochondrial degradation can be toxic, CsA treatment also caused increased cell death and activation of apoptosis (Fig. 2F and G).
To test whether H9-RGCs degrade damaged mitochondria via endo-lysosomes, we blocked endo-lysosomal activity with Baf and HCQ. HCQ (with and without CCCP), but not Baf, increased mitochondrial content (Fig. 2H), indicating that HCQ is a more potent inhibitor of mitophagy in H9-RGCs than in hESCs. As expected, presumably due to its inhibition of mitochondrial clearance, HCQ caused hRGC death and apoptosis (Fig. 2I and J). Although we did not observe increased levels of mitochondria with Baf treatment (Fig. 2H), we did observe increased H9-RGC apoptosis and cell death (Fig. 2I and J). As an indication that they were having their expected pharmacological activities, both Baf and HCQ increased the pH of the acidic endo-lysosomal vesicles as shown by a decrease in fluorescence dots of the pH-sensitive dye pHrodo-green dextran (Supplemental Fig. S4). The explanation of the differences between the effects of Baf and HCQ is unclear, but may reflect that the two drugs affect the endo-lysosomal compartments differently, which has been reported [51].
The above data suggest differentiated RGCs are different from their origin stem cells in terms of using endo-lysosomes for degrading damaged mitochondria. For stem cells, inhibition of endo-lysosomes was toxic but did not increase mitochondrial content. But in H9-RGCs inhibition of the endo-lysosomal pathway both inhibited mitochondrial degradation as well as reduced RGC survival, suggesting RGCs predominantly use the endo-lysosomal pathway for mitophagy and cellular homeostasis. The choice for using the endo-lysosomal pathway versus UPS to maintain healthy cellular homeostasis is critical for cell survival. With the apparent difference between hESCs and hRGCs in using the endo-lysosomal pathway and with the potential involvement of UPS in neurodegenerative diseases [55], led us to ask about the role of UPS for mitochondrial clearance and neuro-protection in hRGCs and their origin stem cells.
3.3. The UPS is required for mitochondrial degradation and cell survival for hPSCs but not for hRGCs
As an alternative to the endo-lysosomal pathway, the ubiquitin-proteasomal system (UPS) is the other major cellular quality control pathway for protein and organelle homeostasis [56]. We next investigated the role of UPS in mitochondrial clearance by using the drug bortezomib to inhibit the proteasome's 20S core particle [57]. Unexpectedly, we found that inhibiting proteasomes in H9-ESCs increased mitochondrial levels in a dose dependent manner (Fig. 3A and B). We next asked if proteasomal activity is required for the clearance of acutely damaged mitochondria in H9-ESCs. To test this, we induced mitochondrial damage by CCCP both in the presence and the absence of bortezomib. In agreement with our hypothesis, we observed an increase level of mitochondria when proteasomal clearance was blocked by bortezomib compared to CCCP alone (Fig. 3C), suggesting that hPSCs use the proteasomal pathway to degrade damaged mitochondria.
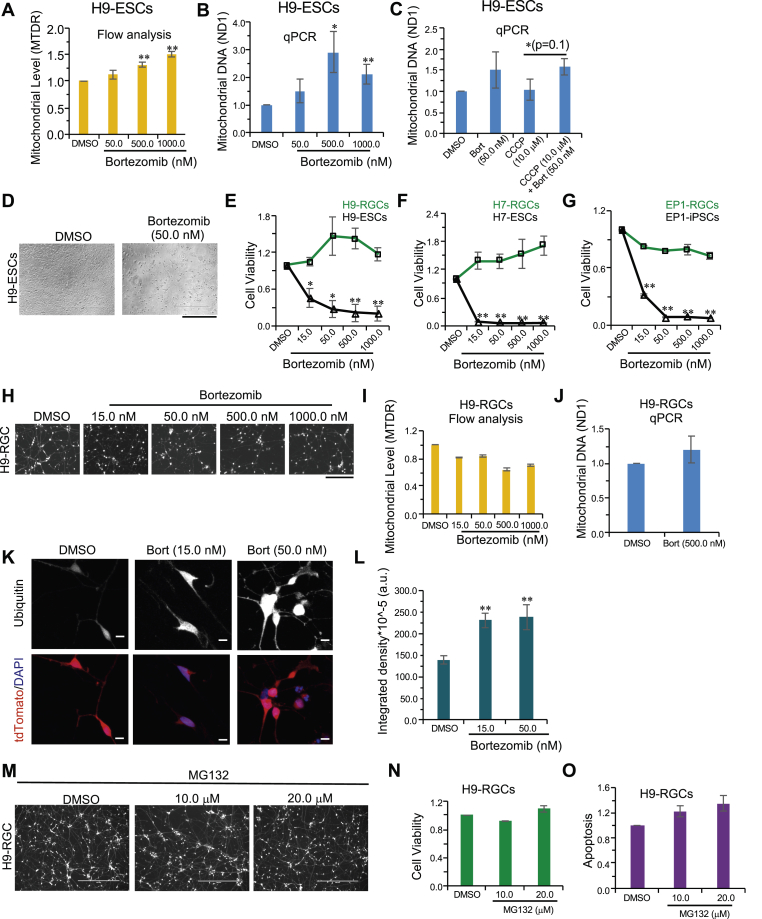
Fig. 3.
hESCs but not hRGCs predominantly use the proteasomal pathway for degrading mitochondria. (A, I) Flow cytometry-based analysis of the MTDR labelled mitochondria in H9-ESCs (A) and H9-RGCs (I) after 24 h treatment with the indicated doses of bortezomib from three independent biological replicates. (B, C, J) qPCR analysis of the mitochondrial content in H9-ESCs (B, C) and H9-RGCs (J) after 24 h treatment with the indicated drugs, as done in (Fig. 1). Data shown are from three independent biological replicates with three technical replicates for each biological replicate. (D) Brightfield images showing cell death in the H9-ESCs after 24 h treatment with the indicated bortezomib (Bort) dose. (E–G) Shown are cell viability measurements after 24 h treatment with bortezomib to the indicated stem cells and corresponding RGCs as done in (Fig. 1). Data presented are from 3 to 4 independent biological replicates and statistical analysis is done between stem cells and corresponding RGCs at the indicated treatments. (H, M) Fluorescence images shown are in the red channel for the tdTomato expressing H9-RGCs after 24 h bortezomib (Bort) (H) and MG132 (M) treatments with the indicated doses. (K, L) Images shown are the sum projections of the confocal z-stacks on immunofluorescence against ubiquitin in H9-RGCs after 24 h treatment with the indicated bortezomib doses (K), and the quantification shows the integrated fluorescence intensity from the sum-projections averaged over 14 to 32 individual cells for each condition (L). (N, O) Quantifications represent cell viability (N) and cellular apoptosis (O), measured by using the fluorescence based ApoTox-Glo triplex assay kit and luminescence-based caspase-3/7 activity for H9-RGCs at the indicated treatment. Data presented are from three independent biological replicates. Scale bars, 400 μm (D, M), 200 μm (H) and 10 μm (K). Error bars are SEM. **, p-value < 0.007; *, p-value < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As accumulation of damaged mitochondria could lead to cellular toxicity, we investigated whether inhibiting proteasomal activity could lead to cell death for stem cells, and then examined the effect of this same inhibition on the corresponding hRGCs. We observed severe cell death for H9-ESCs with bortezomib treatment (Fig. 3D and E); however, this effect was largely absent for the corresponding H9-RGCs (Fig. 3E, H). To test if the observed effect was cell type specific, we also inhibited proteasome function in two additional lines; EP1-iPSCs and H7-ESCs, as well as their corresponding RGCs. Much to our surprise we saw a similar high level of cell death for both the stem cell lines but not for the corresponding differentiated RGCs (Fig. 3F and G; there was a slight increase in cell death for EP1-RGCs but significantly less than that seen with the undifferentiated EP1-iPSCs). In agreement with the increase in cell death, proteasome inhibition also induced cellular apoptosis for all of these three stem cell lines (Supplemental Figs. S5A–C). These data suggest that proteasomal activity is critical for basal level mitochondrial clearance and survival of hPSCs, while hRGC survival is not dependent upon proteasome activity.
To investigate if proteasomal activity is required for mitochondrial degradation in hRGCs, we treated H9-RGCs with different doses of bortezomib for 24 h followed by mitochondrial level measurements. Interestingly, unlike hPSCs, inhibiting the UPS with bortezomib did not increase the mitochondrial level in H9-RGCs (Fig. 3I and J). With the observed differences in proteasomal regulation between hPSCs and hRGCs, we next asked if the UPS is still active in the hRGCs. Ubiquitinated proteins and organelles are degraded through the proteasome [58,59], hence inhibiting UPS activity should increase total ubiquitinated protein levels where the system is normally active. In support of our hypothesis, we found a bortezomib dose-dependent increase in the ubiquitinated protein level in H9-RGCs (Fig. 3K and L). We further tested the bortezomib results using another very potent proteasome inhibitor, MG132 [57,60]. In agreement with the bortezomib data, MG132 negligibly effected cell death or apoptosis in H9-RGCs (Fig. 3M − O).
These results suggest a switch in the mitochondrial degradation pathways from the proteasome to the endo-lysosomal pathway during human RGC differentiation, making the lysosomal-autophagy pathway a potential therapeutic target for improving mitochondrial health and therefore hRGC survival in glaucoma and in other forms of optic neuropathy patients. While these findings could be important for improving hRGC health, we were additionally interested to see if these phenomena are specific to hRGCs, or if they are also true for other types of human neurons. To address this question, we differentiated human cortical neurons and tested the effect of endo-lysosomal and proteasomal inhibition on them.
3.4. Human cortical neurons are susceptible to endo-lysosomal inhibition but not proteasome inhibition
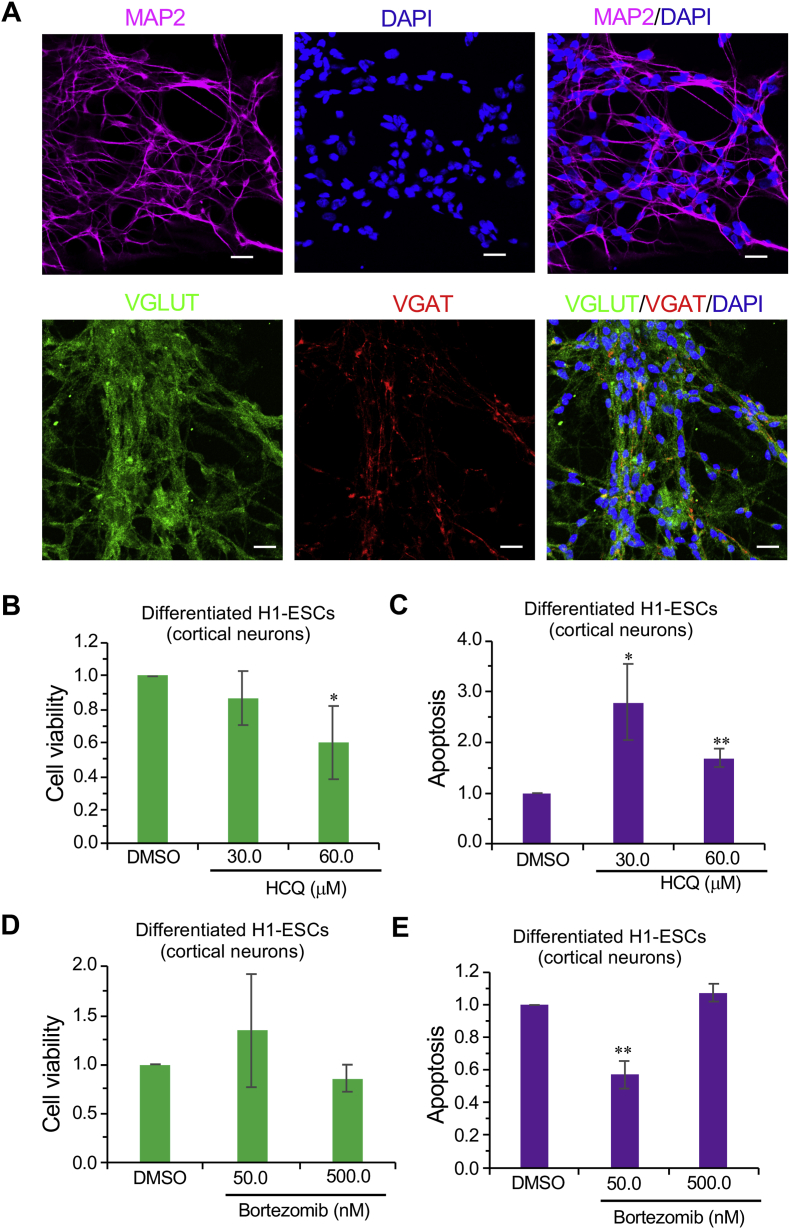
To study the role of proteasomal and endo-lysosomal pathways for cortical neuron survival, we differentiated cortical neurons from human stem cells (H1-ESCs) following published methods [61]. Cultured human cortical neurons were tested and shown to be positive for expression of the mature neuronal marker microtubule-associated protein 2 (MAP2), inhibitory marker vesicular GABA transporter (VGAT), and excitatory marker vesicular glutamate transporter (VGLUT) (Fig. 4A). To test the effect of endo-lysosomal inhibition, cells were treated with HCQ. Similar to hRGCs, we observed significant cell death and corresponding activation of apoptosis (Fig. 4B and C), suggesting the endo-lysosomal pathway is important for cellular homeostasis in cortical neurons.
Fig. 4.
Effect of UPS and endo-lysosomal pathway inhibition on human cortical neuron survival. (A) Shown are the confocal images of immunofluorescence against neuronal marker MAP2, excitatory marker VGLUT and the inhibitory marker VGAT in differentiated H1-ESCs. (B–E) After 24 h treatment with the indicated drugs, cell viability (B, D) and apoptosis (C, E) were measured using ApoTox-Glo triplex assay kit. Data presented are from 3 to 6 independent biological replicates. Scale bars are 20 μm. Error bars are SEM. **, p-value < 0.005; *, p-value < 0.05.
Next, we tested the effect of proteasomal inhibition by treating cortical neurons with the proteasome inhibitor bortezomib. Interestingly, like hRGCs, proteasome inhibition did not cause cell death for human cortical neurons (Fig. 4D and E).
Taking together, our data suggest that the endo-lysosomal pathway may be the predominant pathway for degrading damaged mitochondria and maintaining cellular homeostasis for not only human RGCs but also for cortical neurons suggesting such phenomena could be true for other differentiated human cell types.
4. Discussion
Our study identifies pathways important for maintaining healthy mitochondrial homeostasis in human RGCs and suggests a step forward for developing strategies to enhance RGC viability under disease conditions. The results shown here suggest three key points: first, human RGCs are more efficient in clearing up damaged mitochondria than their precursor stem cells; second, the proteasomal pathway is essential for stem cell survival but not for RGCs; and third, during RGC maturation from stem cells, the pathway for mitochondrial clearance and cellular homeostasis shifts from the proteasomal to the endo-lysosomal pathway.
While we observed that mitochondrial degradation in stem cells is dependent on UPS, a question still remains on how proteasomes degrade mitochondria? An elegant study by Chan et al. (2011) [62] suggests that this could happen by Parkin mediated activation of the UPS. Upon its translocation to mitochondria, Parkin activates the 26S proteasome, leading to the degradation of the mitochondrial outer membrane proteins. For hPSCs, a similar mechanism may lead to UPS mediated mitochondrial degradation as well. However, our data suggest UPS dependent mitochondrial degradation could depend on the mitochondrial inner membrane potential (ΔΨm). We have seen that when ESCs were treated with OA or CsA, which are known to increase ΔΨm [53,54], mitochondrial degradation was inhibited (Fig. 2A, D, E). However, when treated with the uncoupler CCCP, which abolishes ΔΨm [53], mitochondrial degradation was induced (Fig. 1H and I). This suggests ΔΨm could negatively regulate UPS mediated mitochondrial degradation. Further efforts will be required to understand the mechanism of how ΔΨm can regulate UPS mediated mitochondrial degradation. Even though we observed that inhibition of endo-lysosomes did not block mitophagy in ESCs, endo-lysosome involvement cannot be ruled out, as a previous report had shown inhibiting lysosomes increased mitochondrial content in hematopoietic stem cells (HSCs) [63].
Our study indicates that the endo-lysosomal pathway is the primary route for degrading damaged mitochondria in hRGCs. This is significant as it makes the endo-lysosomal pathway a potential therapeutic target to enhance mitochondrial homeostasis to increase hRGC survival. Consistent with our findings, genetic analyses have identified several mutations in MQC related genes in various human optic neuropathies. These mutations include: a mutation in the mitophagy adaptor protein Optineurin (OPTN) in primary open-angle glaucoma (POAG) patients [8]; mutations in the MQC pathway proteins Mitofusin1/2 (Mfn1/2) which have also been associated with normal-tension glaucoma (NTG) patients [64]; and mitochondrial DNA mutations affecting mitochondrial function have been shown to be causative for the other forms of optic neuropathy such as LHON and mutation in the mitochondrial fusion protein OPA1 for Dominant Optic Atrophy patients [65]. A cellular-level intervention to maintain healthy mitochondria and mitigate optic nerve disease progression will be aided by further increases in our understanding of the damaged mitochondrial clearance pathways in human RGCs. In support to this notion it has been found that improving mitochondrial health could serve as a retinal neuroprotection mechanism [66].
5. Conclusion
A switch from the ubiquitin proteasome system to the endo-lysosomal pathway for mitochondrial degradation and cell survival during RGC differentiation is potentially highly significant as it could reflect a general shift in the homeostasis of other proteins and organelles as well. While this study was done in vitro utilizing human stem cell derived RGC cultures, our findings may also reflect the behavior of RGCs and other retinal cells in vivo. The results presented here could have following implications; first, UPS-mediated protein degradation is an ATP-dependent process and requires energy [67], avoiding the UPS for protein and organelle homeostasis could represent an energy saving strategy for the highly energy-dependent RGCs [68]; second, the endo-lysosomal pathway being the primary mitochondria degradation pathway for RGCs implicates this pathway as a possible therapeutic target for RGC neuroprotection in mitochondria-based optic neuropathies. Additionally, modulating the endo-lysosome pathway under specific conditions could be effective for other neurodegenerative diseases, since in our study this pathway was found to be essential in stem-cell derived human cortical neurons, in addition to RGCs, suggesting the potential application of these findings to other neurons that may also use the endo-lysosomal pathway for their survival.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Drs. Jin-Chong Xu, Ted M. Dawson and Valina L. Dawson for providing differentiated human cortical neurons and Dr. Ping-Wu Zhang for the H7 reporter line. We also thank Drs. Sayantan Datta and James Tahara Handa for helping us to develop qPCR based mitochondrial measurements. We thank Michelle Surma for helping to edit and format the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101465.
Contributor Information
Arupratan Das, Email: arupdas@iu.edu.
Donald J. Zack, Email: dzack@jhmi.edu.
Funding sources
This work was supported by grants from the NIH, United States (P30 EY001765, K99 EY028223, and R01 EY026471), Research to Prevent Blindness, United States and generous gifts from the Guerrieri Family Foundation.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
hRGC differentiation from the H9-ESCs. (A) Images shown are representative of different time points for RGC differentiation; tdTomato positive cells at day 20, 26 and 43 indicates successful RGC differentiation. (B) Images shown are the H9-RGCs immunopurified against the surface antigen Thy1.2 and grown on matrigel coated tissue culture dishes. The high overlap between the brightfield and the tdTomato channel indicates highly pure RGC culture. (C) Shown are the confocal immunofluorescence images of H9-RGCs against RGC specific marker RBPMS and neuronal marker TUBB3. Scale bars, 400 μm (A, B) and 20 μm (C).
Flow cytometry-based analysis of the mitochondrial content. (A) Forward-backward scatter plot of the H9-ESCs labelled with the dead cell dye propidium iodide (PI). (B) Diagonally distributed green dots in ‘A’ are low in PI intensity representing live cells and red dots along the BSC axis in ‘A’ are high in PI intensity representing dead cells. This allowed empirical selection of the diagonally distributed live cell population for analysis. (C) Diagonally distributed live H9-ESCs were gated (red oval) for analysis. (D) Live H9-ESCs labelled with mitochondria dye MTDR (far-red) as shown in P-Q4 quadrant were analyzed for average MTDR intensity. (E) Diagonally distributed live H9-RGCs were gated (red oval) for analysis. (F) Live H9-RGCs positive for both tdTomato (red) and MTDR (far-red) distributed in the P-Q2 quadrant were analyzed for average MTDR intensity.
CCCP induced mitochondrial degradation in stem cells and corresponding RGCs at the normoxia culture condition. Stem cells and the corresponding RGCs were cultured and treated in the normoxia (5% CO2, 20% O2) condition with indicated CCCP doses. Graph shows loss of mitochondria labelled MTDR intensity normalized w.r.t DMSO control at different CCCP doses for H9-ESCs compared to the corresponding H9-RGCs. H9-RGC data also presented in Fig. 1H. Data shown are from 3 to 10 independent biological replicates and statistical analysis is done between stem cells and corresponding RGCs at the indicated treatments. Error bars are SEM. **p-value < 0.005; *-value < 0.05.
Bafilomycin A1 (Baf) and hydroxychloroquine (HCQ) increased pH in RGCs. Confocal images shown from live H9-RGCs after 24h treatment with the indicated drugs followed by 20 min incubation with the pH sensitive pHrodo-green conjugated dextran. Scale bar, 10 μm.
Activation of cellular apoptosis upon proteasomal inhibition in stem cells. Cellular apoptosis was measured after 24h treatment with bortezomib at the indicated doses for H9-ESCs (A), H7-ESCs (B) and EP1-iPSCs (C) by measuring luminescence-based caspase-3/7 activity. Data presented are from three independent biological replicates. Error bars are SEM. **p-value < 0.005; *-value < 0.05.
References
- 1.Carelli V., La Morgia C., Ross-Cisneros F.N., Sadun A.A. Optic neuropathies: the tip of the neurodegeneration iceberg. Hum. Mol. Genet. 2017;26:R139–R150. doi: 10.1093/hmg/ddx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Castro I.P., Martins L.M., Tufi R. Mitochondrial quality control and neurological disease: an emerging connection. Expet Rev. Mol. Med. 2010;12 doi: 10.1017/S1462399410001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arun S., Liu L., Donmez G. Mitochondrial biology and neurological diseases. Curr. Neuropharmacol. 2016;14:143–154. doi: 10.1353/crb.0.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu-Wai-Man P., Griffiths P.G., Hudson G., Chinnery P.F. Inherited mitochondrial optic neuropathies. J. Med. Genet. 2009;46:145–158. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenaers G., Hamel C., Delettre C., Amati-Bonneau P., Procaccio V., Bonneau D., Reynier P., Milea D. Dominant optic atrophy. Orphanet J. Rare Dis. 2012;7 doi: 10.1186/1750-1172-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Lin Y., Vithana E.N., Jia L., Zuo X., Wong T.Y., Chen L.J., Zhu X., Tam P.O.S., Gong B., Qian S., Li Z., Liu X., Mani B., Luo Q., Guzman C., Leung C.K.S., Li X., Cao W., Yang Q., Tham C.C.Y., Cheng Y., Zhang X., Wang N., Aung T., Khor C.C., Pang C.P., Sun X., Yang Z. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat. Genet. 2014;46:1115–1119. doi: 10.1038/ng.3078. [DOI] [PubMed] [Google Scholar]
- 7.Khawaja A.P., Cooke Bailey J.N., Wareham N.J., Scott R.A., Simcoe M., Igo R.P., Song Y.E., Wojciechowski R., Cheng C.Y., Khaw P.T., Pasquale L.R., Haines J.L., Foster P.J., Wiggs J.L., Hammond C.J., Hysi P.G. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 2018;50:778–782. doi: 10.1038/s41588-018-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rezaie T., Child A., Hitchings R., Brice G., Miller L., Coca-Prados M., Héon E., Krupin T., Ritch R., Kreutzer D., Crick R.P., Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 9.Wong Y.C., Holzbaur E.L.F. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tait S.W.G., Green D.R. Mitochondrial regulation of cell death. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a008706. a008706–a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser J.A., Biousse V., Newman N.J. The neuro-ophthalmology of mitochondrial disease. Surv. Ophthalmol. 2010;55:299–334. doi: 10.1016/j.survophthal.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tait S.W.G., Green D.R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 13.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–312. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Behrends C., Sowa M.E., Gygi S.P., Harper J.W. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiššová I., Deffieu M., Manon S., Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 17.Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress. Mitochondrial Dysfunc. Aging, Rejuvenat. Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 18.Davis C.O., Kim K.-Y., Bushong E.A., Mills E.A., Boassa D., Shih T., Kinebuchi M., Phan S., Zhou Y., Bihlmeyer N.A., Nguyen J.V., Jin Y., Ellisman M.H., Marsh-Armstrong N. Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi E.A., Granger C.E., Sharma R., Yang Q., Saito K., Schwarz C., Walters S., Nozato K., Zhang J., Kawakami T., Fischer W., Latchney L.R., Hunter J.J., Chung M.M., Williams D.R. Imaging individual neurons in the retinal ganglion cell layer of the living eye. Proc. Natl. Acad. Sci. U. S. A. 2017;114:586–591. doi: 10.1073/pnas.1613445114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodger C.E., McWilliams T.G., Ganley I.G. Mammalian mitophagy – from in vitro molecules to in vivo models. FEBS J. 2018;285:1185–1202. doi: 10.1111/febs.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Y.R., Shekhar K., Yan W., Herrmann D., Sappington A., Bryman G.S., van Zyl T., Do M.T.H., Regev A., Sanes J.R. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell. 2019;176:1222–1237. doi: 10.1016/j.cell.2019.01.004. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katajisto P., Döhla J., Chaffer C.L., Pentinmikko N., Marjanovic N., Iqbal S., Zoncu R., Chen W., Weinberg R.A., Sabatini D.M. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348(80-):340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warr M.R., Binnewies M., Flach J., Reynaud D., Garg T., Malhotra R., Debnath J., Passegué E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen M., Soilleux E.J., Djordjevic G., Tripp R., Lutteropp M., Sadighi-Akha E., Stranks A.J., Glanville J., Knight S., Jacobsen S.-E.W., Kranc K.R., Simon A.K. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Wu K., Xiao X., Liao J., Hu Q., Chen H., Liu J., An X. Autophagy as a regulatory component of erythropoiesis. Int. J. Mol. Sci. 2015;16:4083–4094. doi: 10.3390/ijms16024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K., Lee J., Kim J.Y., Wang L., Tian Y., Chan S.T., Cho C., Machida K., Chen D., Ou J.H.J. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol. Cell. 2017;68:281–292. doi: 10.1016/j.molcel.2017.09.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan C., Luo L., Guo C.Y., Goto S., Urata Y., Shao J.H., Li T.S. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Canc. Lett. 2017;388:34–42. doi: 10.1016/j.canlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Saez I., Koyuncu S., Gutierrez-Garcia R., Dieterich C., Vilchez D. Insights into the ubiquitin-proteasome system of human embryonic stem cells. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-22384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilchez D., Boyer L., Morantte I., Lutz M., Merkwirth C., Joyce D., Spencer B., Page L., Masliah E., Travis Berggren W., Gage F.H., Dillin A. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esteban‐Martínez L., Sierra‐Filardi E., McGreal R.S., Salazar‐Roa M., Mariño G., Seco E., Durand S., Enot D., Graña O., Malumbres M., Cvekl A., Cuervo A.M., Kroemer G., Boya P. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 2017;36:1688–1706. doi: 10.15252/embj.201695916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esteban-Martínez L., Boya P. BNIP3L/NIX-dependent mitophagy regulates cell differentiation via metabolic reprogramming. Autophagy. 2017:1–3. doi: 10.1080/15548627.2017.1332567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shim M.S., Takihara Y., Kim K.Y., Iwata T., Yue B.Y.J.T., Inatani M., Weinreb R.N., Perkins G.A., Ju W.K. Mitochondrial pathogenic mechanism and degradation in optineurin E50K mutation-mediated retinal ganglion cell degeneration. Sci. Rep. 2016;6 doi: 10.1038/srep33830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito Y.A., Di Polo A. Mitochondrial dynamics, transport, and quality control: a bottleneck for retinal ganglion cell viability in optic neuropathies. Mitochondrion. 2017;36:186–192. doi: 10.1016/j.mito.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Sluch V.M., Chamling X., Liu M.M., Berlinicke C.A., Cheng J., Mitchell K.L., Welsbie D.S., Zack D.J. Enhanced stem cell differentiation and immunopurification of genome engineered human retinal ganglion cells. Stem Cells Transl. Med. 2017;6:1972–1986. doi: 10.1002/sctm.17-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eldred K.C., Hadyniak S.E., Hussey K.A., Brenerman B., Zhang P.W., Chamling X., Sluch V.M., Welsbie D.S., Hattar S., Taylor J., Wahlin K., Zack D.J., Johnston R.J. Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science. 2018;80–:362. doi: 10.1126/science.aau6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhise N.S., Wahlin K.J., Zack D.J., Green J.J. Evaluating the potential of poly(beta-amino ester) nanoparticles for reprogramming human fibroblasts to become induced pluripotent stem cells. Int. J. Nanomed. 2013;8:4641–4658. doi: 10.2147/IJN.S53830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gill K.P., Hung S.S.C., Sharov A., Lo C.Y., Needham K., Lidgerwood G.E., Jackson S., Crombie D.E., Nayagam B.A., Cook A.L., Hewitt A.W., Pébay A., Wong R.C.B. Enriched retinal ganglion cells derived from human embryonic stem cells. Sci. Rep. 2016;6 doi: 10.1038/srep30552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abnet C.C., Huppi K., Carrera A., Armistead D., McKenney K., Hu N., Tang Z.-Z., Taylor P.R., Dawsey S.M. Control region mutations and the “common deletion” are frequent in the mitochondrial DNA of patients with esophageal squamous cell carcinoma. BMC Canc. 2004;4:30. doi: 10.1186/1471-2407-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta S., Chattopadhyay E., Ray J.G., Majumder M., Das Roy P., Roy B. D-loop somatic mutations and ~5 kb “common” deletion in mitochondrial DNA: important molecular markers to distinguish oral precancer and cancer. Tumor Biol. 2015;36:3025–3033. doi: 10.1007/s13277-014-2937-2. [DOI] [PubMed] [Google Scholar]
- 40.Xu J.C., Fan J., Wang X., Eacker S.M., Kam T.I., Chen L., Yin X., Zhu J., Chi Z., Jiang H., Chen R., Dawson T.M., Dawson V.L. Cultured networks of excitatory projection neurons and inhibitory interneurons for studying human cortical neurotoxicity. Sci. Transl. Med. 2016 doi: 10.1126/scitranslmed.aad0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez A.R., de Sevilla Müller L.P., Brecha N.C. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J. Comp. Neurol. 2014;522:1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sluch V.M., Davis C.O., Ranganathan V., Kerr J.M., Krick K., Martin R., Berlinicke C.A., Marsh-Armstrong N., Diamond J.S., Mao H.-Q., Zack D.J. Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci. Rep. 2015;5:16595. doi: 10.1038/srep16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauro-Lizcano M., Esteban-Martínez L., Seco E., Serrano-Puebla A., Garcia-Ledo L., Vieira H.L.A., Vieira H.L.A., Boya P. New method to assess mitophagy flux by flow cytometry. Autophagy. 2015;11:833–843. doi: 10.1080/15548627.2015.1034403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keij J.F., Bell-Prince C., Steinkamp J.A. Staining of mitochondrial membranes with 10-nonyl acridine orange MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry. 2000;39:203–210. doi: 10.1002/(SICI)1097-0320(20000301)39:3<203::AID-CYTO5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 46.Poot M., Zhang Y.Z., Krämer J.A., Wells K.S., Jones L.J., Hanzel D.K., Lugade A.G., Singer V.L., Haugland R.P. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 47.Ohkuma S., Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. U. S. A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 49.Vessoni A.T., Muotri A.R., Okamoto O.K. Autophagy in stem cell maintenance and differentiation. Stem Cell. Dev. 2012;21:513–520. doi: 10.1089/scd.2011.0526. [DOI] [PubMed] [Google Scholar]
- 50.Subramani S., Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X., Jin M., Cai Y., Xia H., Long K., Liu J., Yu Q., Yuan J. Mitochondrial electron transport chain complex III is required for antimycin A to inhibit autophagy. Chem. Biol. 2011;18:1474–1481. doi: 10.1016/j.chembiol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perry S.W., Norman J.P., Barbieri J., Brown E.B., Gelbard H.A. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 2011;50:98–115. doi: 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brustovetsky N., Dubinsky J.M. Limitations of cyclosporin A inhibition of the permeability transition in CNS mitochondria. J. Neurosci. 2000 doi: 10.1007/s10495-009-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dantuma N.P., Bott L.C. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front. Mol. Neurosci. 2014;7 doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dikic I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 57.Goldberg A.L. Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 2012;199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 59.Kleiger G., Mayor T. Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol. 2014;24:352–359. doi: 10.1016/j.tcb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 61.Xu J.C., Fan J., Wang X., Eacker S.M., Kam T.I., Chen L., Yin X., Zhu J., Chi Z., Jiang H., Chen R., Dawson T.M., Dawson V.L. Cultured networks of excitatory projection neurons and inhibitory interneurons for studying human cortical neurotoxicity. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aad0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L.J., Hess S., Chan D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Almeida M.J., Luchsinger L.L., Corrigan D.J., Williams L.J., Snoeck H.W. Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells. Cell Stem Cell. 2017;21:725–729. doi: 10.1016/j.stem.2017.11.002. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf C., Gramer E., Muller-Myhsok B., Pasutto F., Reinthal E., Wissinger B., Weisschuh N. Evaluation of nine candidate genes in patients with normal tension glaucoma: a case control study. BMC Med. Genet. 2009;10:91. doi: 10.4312/as.8.3-4.91-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu-Wai-Man P., Griffiths P.G., Chinnery P.F. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Prog. Retin. Eye Res. 2011;30:81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajala A., Gupta V.K., Anderson R.E., Rajala R.V.S. Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion. 2013;13:566–576. doi: 10.1016/j.mito.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gottesman S., Maurizi M.R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 1992;56:592–621. doi: 10.1128/mmbr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osborne N.N. Pathogenesis of ganglion “cell death” in glaucoma and neuroprotection: focus on ganglion cell axonal mitochondria. Prog. Brain Res. 2008;173:339–352. doi: 10.1016/S0079-6123(08)01124-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
hRGC differentiation from the H9-ESCs. (A) Images shown are representative of different time points for RGC differentiation; tdTomato positive cells at day 20, 26 and 43 indicates successful RGC differentiation. (B) Images shown are the H9-RGCs immunopurified against the surface antigen Thy1.2 and grown on matrigel coated tissue culture dishes. The high overlap between the brightfield and the tdTomato channel indicates highly pure RGC culture. (C) Shown are the confocal immunofluorescence images of H9-RGCs against RGC specific marker RBPMS and neuronal marker TUBB3. Scale bars, 400 μm (A, B) and 20 μm (C).
Flow cytometry-based analysis of the mitochondrial content. (A) Forward-backward scatter plot of the H9-ESCs labelled with the dead cell dye propidium iodide (PI). (B) Diagonally distributed green dots in ‘A’ are low in PI intensity representing live cells and red dots along the BSC axis in ‘A’ are high in PI intensity representing dead cells. This allowed empirical selection of the diagonally distributed live cell population for analysis. (C) Diagonally distributed live H9-ESCs were gated (red oval) for analysis. (D) Live H9-ESCs labelled with mitochondria dye MTDR (far-red) as shown in P-Q4 quadrant were analyzed for average MTDR intensity. (E) Diagonally distributed live H9-RGCs were gated (red oval) for analysis. (F) Live H9-RGCs positive for both tdTomato (red) and MTDR (far-red) distributed in the P-Q2 quadrant were analyzed for average MTDR intensity.
CCCP induced mitochondrial degradation in stem cells and corresponding RGCs at the normoxia culture condition. Stem cells and the corresponding RGCs were cultured and treated in the normoxia (5% CO2, 20% O2) condition with indicated CCCP doses. Graph shows loss of mitochondria labelled MTDR intensity normalized w.r.t DMSO control at different CCCP doses for H9-ESCs compared to the corresponding H9-RGCs. H9-RGC data also presented in Fig. 1H. Data shown are from 3 to 10 independent biological replicates and statistical analysis is done between stem cells and corresponding RGCs at the indicated treatments. Error bars are SEM. **p-value < 0.005; *-value < 0.05.
Bafilomycin A1 (Baf) and hydroxychloroquine (HCQ) increased pH in RGCs. Confocal images shown from live H9-RGCs after 24h treatment with the indicated drugs followed by 20 min incubation with the pH sensitive pHrodo-green conjugated dextran. Scale bar, 10 μm.
Activation of cellular apoptosis upon proteasomal inhibition in stem cells. Cellular apoptosis was measured after 24h treatment with bortezomib at the indicated doses for H9-ESCs (A), H7-ESCs (B) and EP1-iPSCs (C) by measuring luminescence-based caspase-3/7 activity. Data presented are from three independent biological replicates. Error bars are SEM. **p-value < 0.005; *-value < 0.05.