Abstract
Background
The molecular mechanisms underlying chronic kidney disease (CKD) transition to end-stage renal disease (ESRD) and CKD acceleration of cardiovascular and other tissue inflammations remain poorly determined.
Methods
We conducted a comprehensive data analyses on 7 microarray datasets in peripheral blood mononuclear cells (PBMCs) from patients with CKD and ESRD from NCBI-GEO databases, where we examined the expressions of 2641 secretome genes (SG).
Results
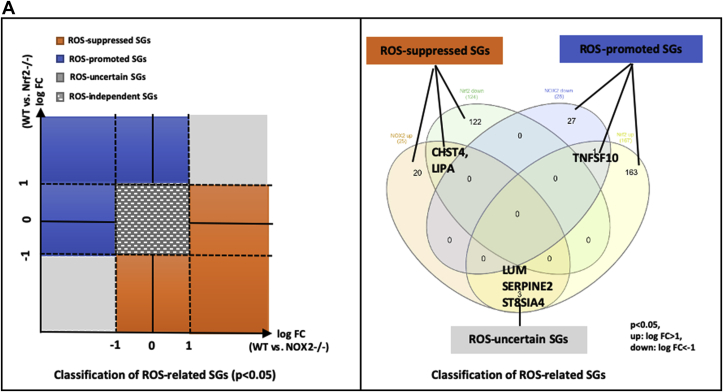
1) 86.7% middle class (molecular weight >500 Daltons) uremic toxins (UTs) were encoded by SGs; 2) Upregulation of SGs in PBMCs in patients with ESRD (121 SGs) were significantly higher than that of CKD (44 SGs); 3) Transcriptomic analyses of PBMC secretome had advantages to identify more comprehensive secretome than conventional secretomic analyses; 4) ESRD-induced SGs had strong proinflammatory pathways; 5) Proinflammatory cytokines-based UTs such as IL-1β and IL-18 promoted ESRD modulation of SGs; 6) ESRD-upregulated co-stimulation receptors CD48 and CD58 increased secretomic upregulation in the PBMCs, which were magnified enormously in tissues; 7) M1-, and M2-macrophage polarization signals contributed to ESRD- and CKD-upregulated SGs; 8) ESRD- and CKD-upregulated SGs contained senescence-promoting regulators by upregulating proinflammatory IGFBP7 and downregulating anti-inflammatory TGF-β1 and telomere stabilizer SERPINE1/PAI-1; 9) ROS pathways played bigger roles in mediating ESRD-upregulated SGs (11.6%) than that in CKD-upregulated SGs (6.8%), and half of ESRD-upregulated SGs were ROS-independent.
Conclusions
Our analysis suggests novel secretomic upregulation in PBMCs of patients with CKD and ESRD, act synergistically with uremic toxins, to promote inflammation and potential disease progression. Our findings have provided novel insights on PBMC secretome upregulation to promote disease progression and may lead to the identification of new therapeutic targets for novel regimens for CKD, ESRD and their accelerated cardiovascular disease, other inflammations and cancers. (Total words: 279).
Keywords: Chronic kidney disease, (CKD), End-stage renal disease, (ESRD), PBMC secretome, Reactive oxygen species, (ROS), Trained immunity
1. Introduction
The incidence of chronic kidney disease (CKD) is increasing worldwide [1]. A major cause of mortality in patients with CKD has been found to be atherosclerosis-related cardiovascular disease (CVD) [2]. Our and others’ recent reports showed that CVD stressors and risk factors such as hyperlipidemia [3,4], hyperglycemia [5], hyperhomocysteinemia [6,7], and chronic kidney disease [[10], [8], [9]], promote atherosclerosis and vascular inflammation via several mechanisms. These mechanisms include endothelial cell (EC) activation [3,[11], [12], [13], [14]] and injury [15]; caspase-1/inflammasome activation [8,10], mitochondrial reactive oxygen species (ROS) [4]; Ly6Chigh mouse monocyte and CD40+ human monocyte differentiation [7,[16], [17], [18]]; decreased/transdifferentiated regulatory T cells [[19], [20], [21], [22]] (Treg); impaired vascular repairability of bone marrow-derived progenitor cells [23,24]; downregulated histone modification enzymes [25] and increased expressions of trained immunity pathway enzymes [26].
CKD is classified into five stages [27] based on glomerular filtration rate (GFR, mL/min. per 1.73 m2); ≥90 mL/min (stage 1), 60–89 mL/min (stage 2), 30–59 mL/min (stage 3), 15–29 mL/min (stage 4) and <15 mL/min (stage 5). At stage 5, the patient develops end-stage renal disease (ESRD) and requires life-long renal replacement therapy (RRT). Clinical evaluations for kidney function include creatinine level, blood urea nitrogen (BUN) assessment and cystatin C level (MedlinePlus, NIH https://medlineplus.gov/kidneytests.html). CVD risk increases significantly according to the stages of CKD, ranging from 1.5-fold in stage 2, to between 20 and 1000-folds with ESRD [28]. Indeed, CVD accounts for 50% of deaths in patients receiving dialysis [29], demonstrating that CKD accelerates atherosclerotic pathology [28], which along with its complications such as myocardial infarction, stroke and peripheral artery disease, are the leading cause of morbidity and mortality in this country, and account for 75% of all CKD deaths from CVD [30]. The molecular and cellular mechanisms underlying CKD-accelerated atherosclerotic pathology, remain unknown.
It has been suggested that CKD-derived uremic toxins (UTs) [31], in combination with other risk factors, cause oxidative stress including mitochondrial ROS [4], low-grade inflammation with increased circulating cytokines and endothelial dysfunction [28,32]. Recently, in novel UT metabolomics/gene databases, we analyzed the expression changes of UT receptors and UT synthases in CKD and CVD. We made the following observations: 1) UTs represent only 1/80th of the human serum small-molecule metabolome; 2) Increased in CKD and CVD, some UTs induce or suppress the expression of inflammatory molecules; 3) The expression of UT genes are significantly modulated in CKD patients, and coronary artery disease (CAD) patients; and 4) The expressions of UT genes are upregulated by pathogen/danger associated molecular pattern receptors (PAMPs/DAMPs)/inflammasome-caspase-1 as we reported [8] and tumor necrosis factor-α (TNF-α) pathways but are inhibited in CD4+Foxp3+ regulatory T cells (Treg). These results demonstrate that UTs are selectively increased, and serve as DAMPs and homeostasis-associated molecular patterns (HAMPs) that modulate inflammation [33]; and that some UT genes are upregulated in CKD and CAD rather than by purely passive accumulation [10]. One well-characterized UT example is carbamylated low-density lipoprotein (cLDL) [34]. Protein carbamylation has been found in atherosclerotic plaque; and serum level of cLDL is increased significantly in patients with ESRD, which has been shown to have all of the major biological effects relevant to atherosclerosis, including EC injury and dysfunction [35] by binding to oxidized low-density lipoprotein (oxLDL) receptor (LOX-1) [36], increased expression of cell adhesion molecules, monocyte adhesion, and vascular smooth muscle cell (VSMC) proliferation [34]. However, an important question remains whether additional secretory proteins participate in the pathogenesis and inflammatory acceleration of CKD and ESRD.
The secretome, defined as a portion of total proteins secreted by cells to the extracellular space, secures a proper micro-environmental niche, thus maintaining tissue homeostasis [37,38]. Secreted molecules are key mediators in cell-cell interactions and influence the cross-talk with the surrounding tissues in addition to their endocrine functions in long-distance as previously demonstrated by hormones, growth factors, cytokines, adipokines, myokines, cardiokines [39], and chemokines [40]. There is strong evidence supporting that crucial cellular functions such as proliferation, differentiation, communication and migration are strictly regulated from the cell secretome [41]. The major difference between our current study and previous reports on the roles of cytokines and chemokines in CKD pathology is that secretome analyses provide a panoramic view on all the secreted genes in the human genome modulated in CKD and ESRD, as opposed to focusing on only one or a few cytokines/chemokines. Recent reports showed that aberrant endothelial secretome in kidney diseases contribute to fibroblast reprogramming [40]. More importantly, peripheral blood mononuclear cells (PBMCs) are first tier of sensors to uremic toxins and other proinflammatory molecules in serum during kidney dysfunction [42,43]. Gene expression profile, metabolite profile, monocyte counts of PBMCs are identified to provide an access to evaluate and predict the settings of CVD and CKD [7,[44], [45], [46]]. The PBMC morphology, Treg/Th17 disequilibrium and activation of TLRs on membrane of PBMCs promote vascular calcification and endothelial dysfunction, which are closely related to cardiovascular risk in CKD patients [[47], [48], [49]]. Meanwhile, glomerular inflammation is correlated with IL-6 and IL-1β secretion in the peripheral blood [50]. However, an important question remains whether CKD and ESRD upregulate the secretome gene expressions in innate immune cells such as PBMCs, by which chronic systemic and tissue inflammations get accelerated.
In order to broaden our understanding of CKD and ESRD-accelerated inflammation, we hypothesized that CKD and ESRD induce differential secretomic gene (SG) expression patterns in PBMCs [51], by which CKD and ESRD accelerate inflammation. We conducted a comprehensive data analyses on a microarray dataset (GEO ID:GSE15072) containing genomic screening of PBMCs from patients with CKD and ESRD from the NIH-NCBI-GEO databases (https://www.ncbi.nlm.nih.gov/gds/), where we examined expressions of 2641 secretome genes (SG). We made the following findings: 1) 86.7% middle class UTs were encoded by SGs; 2) Upregulations of SGs in PBMCs in patients with ESRD (121 SGs) were significantly higher than that of CKD (44 SGs); 3) ESRD-induced SGs had strong proinflammatory pathways; 4) Proinflammatory cytokines-based UTs such as IL-1β and IL-18 promote ESRD modulation of SGs; 5) ESRD-upregulated co-stimulation receptors CD48 and CD58 increase secretomic upregulation in the PBMCs, which are magnified enormously in tissues; 6) M1-, and M2-macrophage polarization signals contribute to ESRD- and CKD-upregulated SGs; 7) ESRD- and CKD-upregulated SGs contain senescence-promoting regulators by upregulating proinflammatory IGFBP7 and downregulating anti-inflammatory TGF-β1 and telomere stabilizer SERPINE1/PAI-1; and 8) ROS pathways play bigger roles in mediating ESRD-upregulated SGs (11.6%) than that in CKD-upregulated SGs (6.8%), and half of ESRD-upregulated SGs are ROS-independent. Novel PBMC-secretome acts synergistically with uremic toxins, to promote inflammation and potential disease progression. Our findings provided novel insights on secretomic upregulation in PBMCs of patients with CKD and ESRD and identification of new therapeutic targets on CKD, various inflammations and cancers.
2. Materials and methods
2.1. Expression profile of secretomic genes (SGs) and innate immunomic genes (IIGs) in PBMC from patients with CKD and with ESRD
Microarray datasets were collected from National Institutes of Health (NIH)-National Center for Biotechnology Information (NCBI)-Gene Expression Omnibus (GEO) databases (https://www.ncbi.nlm.nih.gov/gds/) and analyzed with an online software GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). The numbers of 7 GEO datasets were listed in Table 2A. The detailed information of these GEO datasets was shown in Table 2A and other tables.
Table 2a.
Seven microarray datasets collected from the NIH-NCBI-GeoData Sets database and were analyzed in this study (https://www.ncbi.nlm.nih.gov/gds/). Nine canonical housekeeping genes (ACTB, GAPDH, PGK1, PPIA, B2M, YWHAZ, SDHA, HMBS, TBP) were used to verify the quality of all the datasets with their expression variation listed in the supplemental “Supplement table of Housekeeping Genes”.
| No. | GEO ID | Species | cell type/tissue | Disease Comparison/Treatmenta | PMID |
|---|---|---|---|---|---|
| 1 | GSE15072 | Human | Peripheral blood mononuclear cells | ESRD vs. CKD vs. health | 24189015/19698090 |
| 2 | GSE103500 | human | human blood leukocytes | WT vs. IL-1b-treated | N/A |
| 3 | GSE103500 | human | human blood leukocytes | WT vs. IL-18-treated | N/A |
| 4 | GSE15215 | Human | human plasmacytoid dendritic cells | CD2+ DCs vs. CD2− DCs | 19454677 |
| 5 | GSE85346 | Human | Macrophage | M1 vs. M0, M2a vs. M0, M2b vs. M0 | 27990286 |
| 6 | GSE100671 | Human | leukemic cells | WT vs. NOX2−/− PLB-985 cells | 29967760 |
| 7 | GSE7810 | mouse | type II cells isolated from lungs | WT vs. Nrf2−/− mice Type II cell | 17895394 |
WT: wild type; IL: Interleukin; DC: dendritic cells; M1: M1 Macrophages; M0: M0 Macrophages; NOX2: NADPH Oxidase 2; Nrf2: nuclear factor, erythroid 2 like 2.
2.2. Statistical analysis of microarray data
As we reported [26,52], we applied a statistical method similar to that meta-analysis and analyzed the expressions of 9 house-keeping genes including ACTB, GAPDH, PGK1, PPIA, B2M, YWHAZ, SDHA, HMBS, TBP (Supplement Table of Housekeeping Genes) in all GEO datasets regardless of species that were chosen for this study. The house-keeping gene list was extracted from the list provided by Eisenberg and Levanon [53]. Briefly, the mean log fold change (LogFC) of house-keeping genes between treatment and control groups vary from −1.27 to 1.28. As this variation was very narrow, we concluded that the datasets (Table 2A) are of high quality. The target genes with expression changes more than 2-folds in CKD and ESRD were defined as the upregulated genes, while genes with their expression decreased more than 2-fold in CKD and ESRD were defined as downregulated genes |logFC|>1).
2.3. Ingenuity Pathway Analysis
We utilized Ingenuity Pathway Analysis (IPA, Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) to characterize clinical relevance and molecular and cellular functions related to the identified genes in our microarray analysis. Differentially expressed genes were identified and uploaded into IPA for analysis. The core and pathways analysis was used to identify molecular and cellular pathways, as we have previously reported [52,54].
3. Results
3.1. 86.7% middle class (molecular weight >500 Daltons) uremic toxins (UTs) were encoded by secretomic genes (SGs)
We recently reported that UTs, classified in three major groups including 1) small solutes, 2) protein-bound uremic toxins, 3) middle molecules, are significantly upregulated and modulated in patients with chronic kidney disease (CKD) [10,[55], [56], [57]]. To improve our understanding of how many uremic toxins are encoded by secretomic genes, we hypothesized that SGs encode the majority of middle class uremic toxins (UTs). To test this hypothesis, we collected a comprehensive list of human SGs containing 2641 secreted protein genes as predicted by majority decision‐based method for secreted proteins (MDSEC) used for protein classification within the Protein Atlas (https://www.proteinatlas.org/search/protein_class:Secreted+proteins+predicted+by+MDSEC, accessed December 4, 2019) [51]. As others reported, the expression levels of mRNAs are strongly correlated to that of proteins when comparing samples of the same cell type/tissue [58], justifying for us to estimate SG changes in the diseases using peripheral blood mononuclear cells (PBMC) transcriptomic changes for the SG changes. A list of 30 middle class UTs were collected from the European Uremic Toxins (EUTox) Database (www.uremic-toxins.org, assessed on December 4, 2019) [[59], [60], [61]]. As shown in Table 1A, 26 out of 30 middle molecular class UTs (86.7%) were well-characterized cytokines and encoded by SGs. In addition, as shown in Table 1B, the Ingenuity Pathway Analysis (IPA) showed that at least five out of top 10 pathways of middle class UTs were closely related with cytokine-associated signaling functions including apelin (adipokine) liver signaling [62], role of hypercytokinemia and hyperchemokinemia, role of cytokines in mediating communication between immune cells, differential regulation of cytokine production in macrophages and T helper cells by IL-17A and IL-17F, and IL-10 signaling. If we associate the rest of five UTs top pathways with innate immune responses, and tissue inflammation including communication between innate and adaptive immune cells, glucocorticoid receptor signaling, graft-versus-host disease signaling [63], triggering receptor expressed on myeloid cells-1 (TREM1) signaling [64], and cardiac hypertrophy signaling [65], then we can classify all the top 10 pathways of UTs play significant roles in promoting inflammations related to ESRD. Therefore, these results suggest that secretomic changes in the PBMCs contributed to the generation of middle class UTs and therefore may play significant roles in the progression of CKD and end-stage renal disease (ESRD); and that secretomic changes detected in transcriptomic approaches reflected secretomic changes in protein levels, demonstrated by UTs, detected on protein levels, as examples, at least partially.
Table 1a.
Middle class uremic toxins (UTs) classified into secretomic proteins and non-secretomic toxins from the EUTox Work Group (http://www.uremic-toxins.org/). Middle class UTs defined as molecular weight >500 Daltons. All 2641 Secretomic Genes (SGs) and UTs are listed in Supplement Table 1.
| name | related gene symbol | |
|---|---|---|
| Secretomic UTs (86.67%) | Adiponectin | ADIPOQ |
| Adrenomedullin | ADM | |
| Atrial Natriuretic Peptide (ANP) | NPPA | |
| Basic fibroblast growth factor (BFGF) | FGF2 | |
| Calcitonin gene-related peptide (CGRP) | CALCA | |
| Cholecystokinin | CCK | |
| Clara cell protein (CC16) | SCGB1A1 | |
| Complement Factor D | CFD | |
| Cystatin C | CST3 | |
| Endothelin | EDN1, EDN2, EDN3 | |
| Guanylin | GUCA2A,GUCA2B, GUCA2C | |
| Interleukin-18 | IL18 | |
| Interleukin-1β | IL1B | |
| Interleukin-6 | IL6 | |
| Methionine-Enkephalin | PENK | |
| Motiline | MLN | |
| Neuropeptide Y | NPY | |
| Parathyroid hormone | PTH | |
| Resistin | RETN | |
| Substance P | TAC1 | |
| Tumor Necrosis Factor Alpha (TNF) | TNF | |
| Uroguanylin | GUCA2A, GUCA2B | |
| Vasoactive intestinal peptide (VIP) | VIP | |
| Vasopressin (ADH) | AVP | |
| β-2-Microglobulin | B2M | |
| β-Endorphin |
POMC |
|
| Degranulation Inhibiting Protein I | ||
| non-secretomic UTs (13.33%) | Delta-sleep Inducing Peptide | TSC22D3 |
| Hyaluronic acid (Hyaluronan) | HAS1,HAS2 | |
| λ-Ig Light Chain | IGLC1, IGLC2, IGLC3, IGLC7 | |
Table 1b.
Top 10 pathways of middle class UTs classified by Ingenuity Pathway Analysis (IPA) are closely related with cytokine-associated signaling functions. Top 10 pathways of all of 2641 SGs are shown in Supplement Fig. 1 with the full list of pathways of middle class UTs and SGs are shown in Supplement Table 2.
| Ingenuity Canonical Pathways | -log (p-value) | Ratio∗ |
|---|---|---|
| Apelin Liver Signaling Pathway | 7.13 | 0.154 |
| Communication between Innate and Adaptive Immune Cells | 6.41 | 0.0521 |
| Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza | 6.23 | 0.093 |
| Glucocorticoid Receptor Signaling | 6.12 | 0.0208 |
| Graft-versus-Host Disease Signaling | 6.03 | 0.0833 |
| Role of Cytokines in Mediating Communication between Immune Cells | 5.82 | 0.0741 |
| Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F | 5.55 | 0.167 |
| IL-10 Signaling | 5.39 | 0.058 |
| TREM1 Signaling | 5.25 | 0.0533 |
| Cardiac Hypertrophy Signaling (Enhanced) | 5.06 | 0.0144 |
The number of SGs classed/total genes in this pathway.
3.2. Upregulations of SGs in PBMCs in patients with ESRD (121 SGs) were significantly higher than that of CKD (44 SGs); and among ESRD specifically modulated 975 SGs, ESRD upregulated 116 SGs (11.9%) but downregulated 859 SGs (88.1%), respectively
To improve our understanding of how many secreted molecules are generated in the prototypic innate immune cell types, PBMCs, in patients with CKD and ESRD, we hypothesized that the expressions of secretome in PBMCs from patients with CKD and ESRD are modulated in comparing to that of healthy controls. As shown in Table 2B, we identified significant secretomic mRNA expression changes in PBMCs, from patients with CKD and patients with ESRD (see the references for the information regarding the classification of CKD and ESRD, patients and controls) [[66], [67], [68]]. Total 44 out of 2641 SGs (1.67%) were upregulated in PBMCs from patients with CKD; in comparison, 121 out of total 2641 SGs (4.58%) were upregulated in the PBMCs from patients with ESRD. In addition, 55 out of 2641 SGs (2.08%) were downregulated in the PBMCs from patients with CKD; and 928 out of 2641 SGs (35.14%) were downregulated in the PBMCs from patients with ESRD. These results suggest that 1) panoramic view of secretomic changes in PBMCs can be generated by analyzing microarray data from patients with CKD and ESRD; 2) secretomic changes in PBMCs may contribute significantly to generation of UTs in patients with chronic kidney disease (CKD) and end-stage renal disease (ESRD) (also see Fig. 3A); and 3) UTs-based secretome in PBMCs in patients with CKD and ESRD may play significant roles in promoting CKD- and ESRD-accelerated systemic and tissue inflammations (also see Table 4D).
Table 2b.
The numbers of SGs upregulated in the PBMCs from patients with ESRD (121 SGs, 4.58%) were significantly higher than that in CKD (44 SGs, 1.67%) while more dramatic changes of the numbers of downregulated SGs (928 SGs, 35.14%) were observed in the PBMCs from patients with ESRD than that in CKD (55 SGs, 2.08%). (Gene list for all these up- and downregulated SGs in CKD and ESRD are listed in Supplement Table 3.)
| CKD | ERSD | ||
|---|---|---|---|
| up-regulated | number | 44 | 121 |
| Cutoff: P < 0.05, log FC > 1 |
percentage |
1.67% (44/2641) |
4.58% (121/2641) |
| down-regulated | number | 55 | 928 |
| Cutoff: P < 0.05, log FC < −1 | percentage | 2.08% (55/2641) | 35.14% (928/2641) |
Fig. 3a.
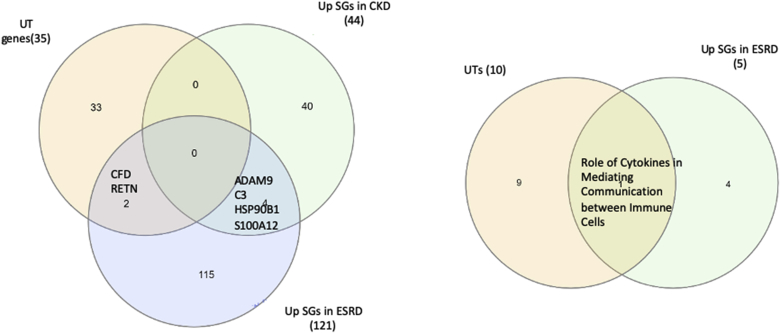
The Venn Diagram results on the three groups such as 35 UT genes (encode total 30 UTs), 44 CKD upregulated SGs and 121 ESRD-upregulated SGs showed that 1) UTs have no overlaps with CKD-upregulated SGs; 2) UTs have two toxins (CFD, and RETN) overlapped with ESRD-upregulated SGs; 3) ESRD-upregulated SGs have four SGs (ADAM9, C3, HSP90B1, and S100A12) overlapped with CKD-upregulated SGs. In addition, one signaling pathway “Role of Cytokines in Mediating Communication between Immune Cells” was shared by the top 10 pathways associated with UTs and the five active pathways upregulated by SGs in ESRD.
Table 4d.
Proinflammatory and profibrotic molecules, pathways and transcription factors played important roles in pathophysiological process of ESRD.
| Method | Table/Figure | Result |
|---|---|---|
| IPA | Fig. 2B | upregulated SGs in ESRD can active proinflammatory pathways such as IL8 signaling, neuroinflammation signaling pathway. |
| IPA | Fig. 2D | downregulated SGs in ESRD can active profibrotic pathways such as inhibition of matrix metalloporteases |
| TRANSFAC | Table 3C | upregulated SGs in ESRD may be modulated by transcription factors involved in inflammation and fibrosis (Egr-1, POU1F1) |
| ClueGo | Fig. 3B | UT-encoded genes and up-regulated SGs in ESRD are shared in some pro-inflammatory pathways。 |
| GEO2R | Table 4a, Table 4bA and 4B | Proinflammatory uremic cytokines IL1b and IL18 in UTs can amplify the upregulation of SGs in ESRD |
| IPA | Table4C | Cytokines can be modulated during CKD progression to induce imbalance of anti-inflammatory and proinflammatory function. |
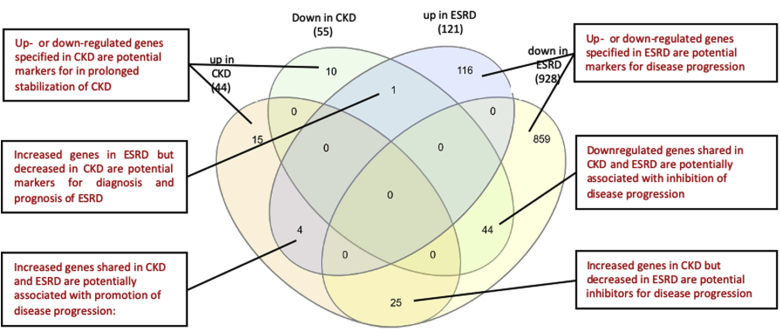
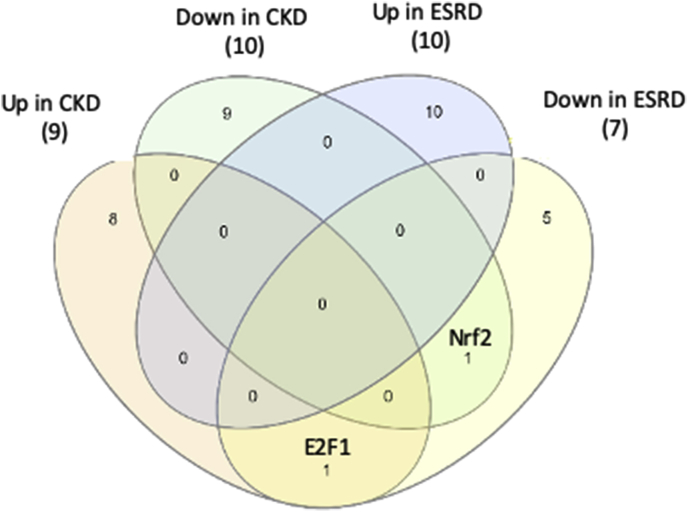
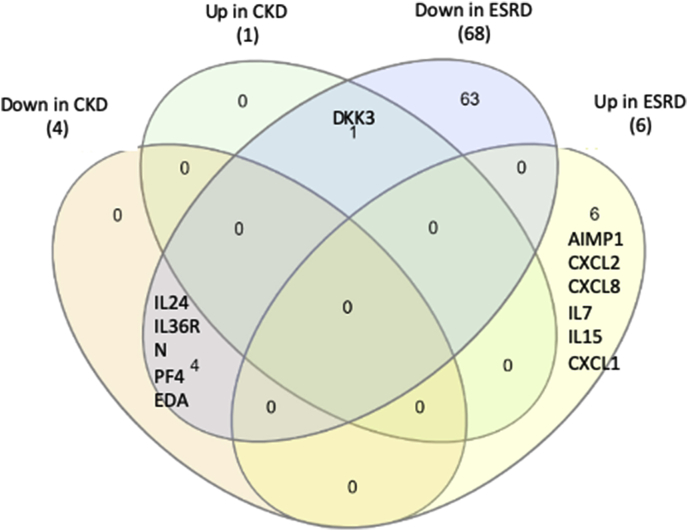
Secretomic upregulation and downregulation in the PBMCs from patients with CKD and ESRD can be categorized into four groups by Venn Diagram analysis, Fig. 1. The results showed that: 1) 25 SGs upregulated in CKD (25/44, 56.8%) shared with that downregulated in ESRD, and shared four SGs with that upregulated in ESRD. The 25 SGs upregulated in CKD but downregulated in ESRD had potential in inhibiting the progression of ESRD. The four SGs downregulated in CKD but upregulated in ESRD suggest: a) their regulatory pathways switched from downregulation in CKD to upregulation in ESRD; and b) their potential roles in promoting ESRD; 2) 44 out of 55 SGs (80%) downregulated in CKD shared with that downregulated in ESRD, suggesting their potential roles in suppressing the progression of CKD and ESRD. One SG, endoplasmic reticulum calcium-binding protein reticulocalbin 2 (RCN2) was increased in ESRD but decreased in CKD, suggesting its potential markers for diagnosis and prognosis of ESRD; 3) 116 out of 121 SGs (95.9%) upregulated in ESRD were ESRD-specific, suggesting their roles in promoting the progression of ESRD; and 4) 859 out of 928 SGs (92.6%) downregulated in ESRD were ESRD-specific, suggesting their potential roles in inhibiting the ESRD progression.
Fig. 1.
Venn diagram analysis of the secretomic upregulation and downregulation in the PBMCs from patients with CKD and ESRD. This secretomic regulation can be broken into six categories. (Gene lists for each category are listed in Supplement Table 4.)
Therefore, we have demonstrated for the first time that ESRD upregulates 116 SGs that may promote ESRD progression and downregulates 859 SGs that may inhibit ESRD progression in the PBMCs in patients with ESRD. If ESRD-specific upregulated 116 SGs plus ESRD-specific downregulated 859 SGs as 100% (975 SGs), this might suggest that ESRD upregulates specifically only 11.9% SGs to promote disease progression and downregulates 88.1% SGs for disease progression, indicating for the first time that as high as 88.1% SGs in the PBMCs may play homeostatic functions which could potentially contribute to the inhibition of ESRD progression.
3.3. ESRD-upregulated SGs had 2 folds higher percentages of the cytoplasm and nucleus subcellular groups than the controls; and had the higher percentages of five out of 13 SG functional groups including enzyme, kinase, peptide, transcription regulator, and transmembrane in comparison to the controls
We used IPA to map the subcellular locations for CKD- and ESRD-modulated SGs. As shown in Table 3A, CKD-upregulated SGs had higher percentages of extracellular space SGs (56.82%) and plasma membrane SGs (20.45%) than that of total SGs controls, 49% and 16.19% respectively; but CKD-downregulated SGs had decreased percentages of extracellular space SGs (41.82%) in comparison to that total SGs control (49%). ESRD-upregulated SGs had higher percentages of cytoplasm SGs (47.11%) and nucleus SGs (8.26%) than that of total SGs controls, 22.08% and 4.46%, respectively. In contrast, ESRD-downregulated SGs had decreased percentages of cytoplasm SGs (15.95%) and other group SGs (3.23%) in comparison to that total SGs control (cytoplasm, 22.08%), and other group SGs (8.27%), respectively.
Table 3a.
All of the percentages of five subcellular location groups of SGs and 14 functional groups of SGs were significantly changed in CKD and ESRD compared with that of total SG controls according to IPA results. Gene list for all these up- and downregulated SGs in CKD and ESRD as well as all of the SGs are listed in Supplement Table 3. (# - number of SGs, % - percentage in each location).
| classification | Total SGs (control) |
up in CKD* |
down in CKD* |
up in ESRD* |
down in ESRD* |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| group | # | % | # | % | # | % | # | % | # | % | |
| Location | cytoplasm | 574 | 22.08% | 8 | 18.18% | 12 | 21.82% | 57 | 47.11% | 148 | 15.95% |
| extracellular space | 1274 | 49.00% | 25 | 56.82% | 23 | 41.82% | 31 | 25.62% | 532 | 57.33% | |
| nucleus | 116 | 4.46% | 1 | 2.27% | 4 | 7.27% | 10 | 8.26% | 31 | 3.34% | |
| other | 215 | 8.27% | 1 | 2.27% | 4 | 7.27% | 1 | 0.83% | 30 | 3.23% | |
| plasma membrane |
421 |
16.19% |
9 |
20.45% |
12 |
21.82% |
22 |
18.18% |
187 |
20.15% |
|
| total |
2600 |
100.00% |
44 |
100.00% |
55 |
100.00% |
121 |
100.00% |
928 |
100% |
|
| Functional group | cytokine | 156 | 6.00% | 1 | 2.27% | 4 | 7.27% | 6 | 4.96% | 68 | 7.33% |
| enzyme | 464 | 17.85% | 5 | 11.36% | 9 | 16.36% | 32 | 26.45% | 151 | 16.27% | |
| G-protein coupled receptor | 18 | 0.69% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 11 | 1.19% | |
| growth factor | 120 | 4.62% | 2 | 4.55% | 3 | 5.45% | 6 | 4.96% | 69 | 7.44% | |
| ion channel | 10 | 0.38% | 1 | 2.27% | 1 | 1.82% | 0 | 0.00% | 8 | 0.86% | |
| kinase | 47 | 1.81% | 2 | 4.55% | 4 | 7.27% | 4 | 3.31% | 21 | 2.26% | |
| other | 1279 | 49.19% | 18 | 40.91% | 22 | 40.00% | 39 | 32.23% | 389 | 41.92% | |
| peptide | 232 | 8.92% | 8 | 18.18% | 2 | 3.64% | 13 | 10.74% | 108 | 11.64% | |
| phosphatase | 25 | 0.96% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 11 | 1.19% | |
| transcription regulator | 46 | 1.77% | 2 | 4.55% | 3 | 5.45% | 5 | 4.13% | 12 | 1.29% | |
| translation regulator | 2 | 0.08% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% | |
| transmembrane receptor | 115 | 4.42% | 4 | 9.09% | 4 | 7.27% | 11 | 9.09% | 45 | 4.85% | |
| transporter |
86 |
3.31% |
1 |
2.27% |
3 |
5.45% |
5 |
4.13% |
35 |
3.77% |
|
| total | 2600 | 100.00% | 44 | 100.00% | 55 | 100.00% | 121 | 100.00% | 928 | 100.00% | |
*P < 0.05.
We then used IPA to map the 13 function groups for CKD- and ESRD-modulated SGs (Table 3A). In CKD-upregulated SGs, the percentages of five out of 13 functional groups including ion channel, kinase, peptide, transcription regulator, and transmembrane were increased. In ESRD-upregulated SGs, the percentages of five out of 13 functional groups including enzyme, kinase, peptide, transcription regulator, and transmembrane were increased in comparison to that of total SGs. These results demonstrated that CKD upregulated SGs have upregulated functional groups of SGs similar to that of ESRD except for the functional groups of enzyme and ion channel. Of note, some PBMC secretome proteins identified with a transcriptomic approach localized in the subcellular locations other than the supernatants of cultured cells and plasma that conventional secretomic analyses sampled and examined. Therefore, our data have also demonstrated that transcriptomic analyses of PBMC secretome have advantages to identify more comprehensive secretome than conventional secretomic analyses [69].
3.4. Although CKD- and ESRD-upregulated SGs were highly diversified in signaling, ESRD-induced SGs had strong proinflammatory pathways
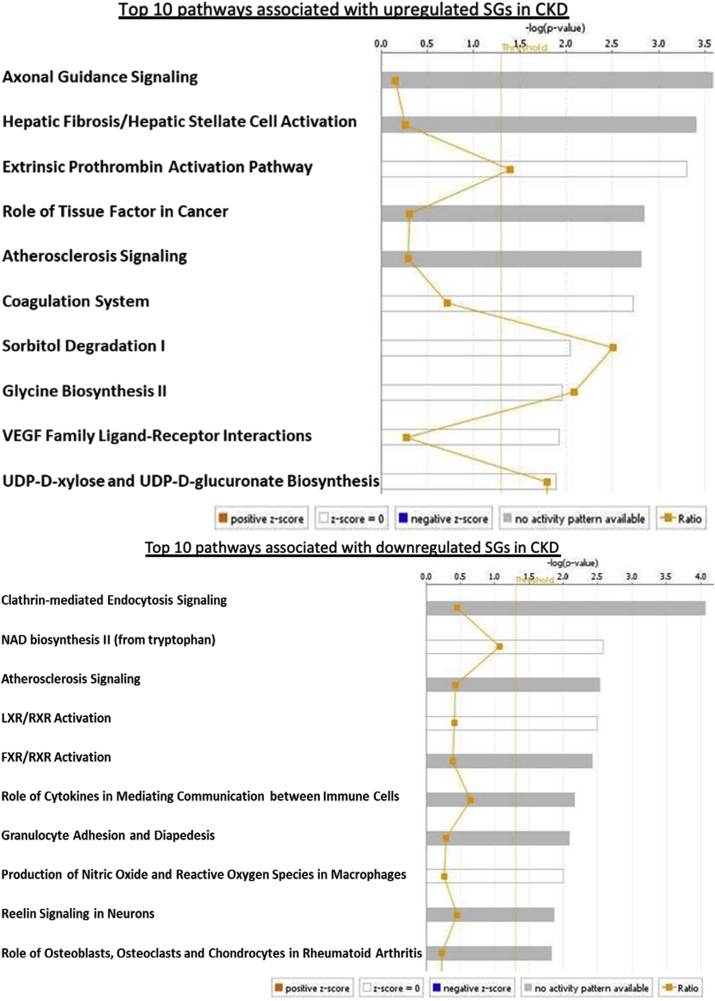
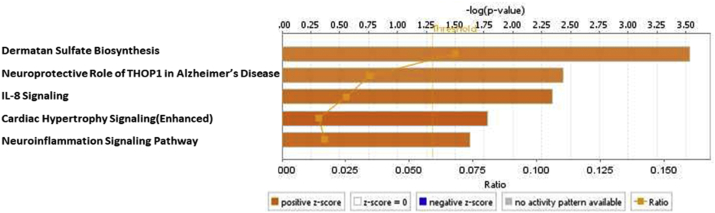
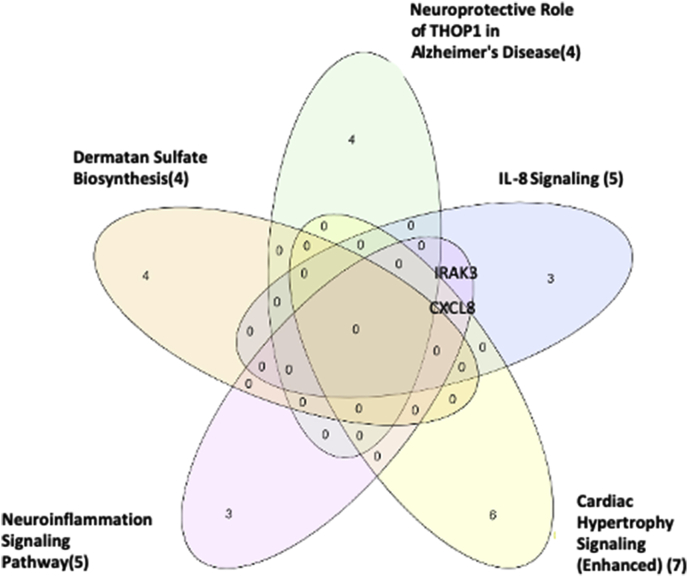
To characterize the signaling pathways that CKD- and ESRD-modulated SGs are involved in, we adapted IPA to map SGs pathways. As shown in Fig. 2A, IPA indicated that upregulated and downregulated SGs in CKD were highly diversified in signaling pathways and were not classified into any signaling pathways in a statistically significant manner. The results suggest that there are diversified and multi-regulatory factor-based signals involved in controlling SGs modulations in CKD. In Fig. 2B, the IPA results showed that 22 out of total 121 (18.2%) upregulated SGs in the PBMCs from patients with ESRD were classified in five active pathways according to IPA core analysis including dermatan sulfate biosynthesis, neuroprotective role of Thimet oligopeptidase (THOP1) [70] in Alzheimer’s disease, IL-8 signaling, cardiac hypertrophy and neuroinflammation signaling pathway. The rest of 99 SGs (81.8%) upregulated SGs in the PBMCs from patients with ESRD were in a diversified manner similar to that observed in CKD. Of note, the chondroitin sulfate/dermatan sulfate (CS/DS)-containing proteoglycans (CS/DSPGs) are extracellular matrix (ECM) molecules, which mediate the accumulation of lipoproteins in the sub-intimal spaces, a key event occurring during the pathobiology of atherosclerosis and the progression of vascular damage [71]. Further Venn Diagram analysis shown in Fig. 2C and, 18 SGs in the four out of five pathways indicated that the SGs upregulated in ESRD have strong proinflammatory roles in PBMCs, especially newly identified chondroitin sulfate/dermatan sulfate (CS/DS)-containing proteoglycans (CS/DSPGs).
Fig. 2a.
Top 10 pathways of upregulated and downregulated SGs in CKD from IPA. These pathways were highly diversified in signaling pathways and were not classified into any signaling pathways in a statistically significant manner (cutoff: P value < 0.05, |z-score|>2). (Lists of all pathways associated with these up- and downregulated SGs in CKD via IPA are listed in Supplement Table 5.)
Fig. 2b.
22 SGs out of total 121 (18.2%) upregulated SGs in the PBMCs from patients with ESRD were classified in five active pathways according to IPA core analysis (cutoff: |z-score|>2). The other 99 SGs (81.8%) were in a diversified manner similar to that in CKD. (Lists for all pathways associated with these up- and downregulated SGs in ESRD via IPA are listed in Supplement Table 5.)
Fig. 2c.
The Venn Diagram Analysis of the five signaling pathways specified in Fig. 2B. Interleukin 1 receptor-associated kinase 3 (IRAK3) is involved in two pathways (IL-8 signaling and Neuroinflammation Signaling Pathway). C-X-C Motif Chemokine Ligand 8 (CXCL8) is involved in three pathways (IL-8 Signaling, Cardiac Hypertrophy and Neuroinflammation Signaling Pathway). (Gene list for these five pathways associated with these up- and downregulated SGs in ESRD via IPA are listed in Supplement Table 6.)
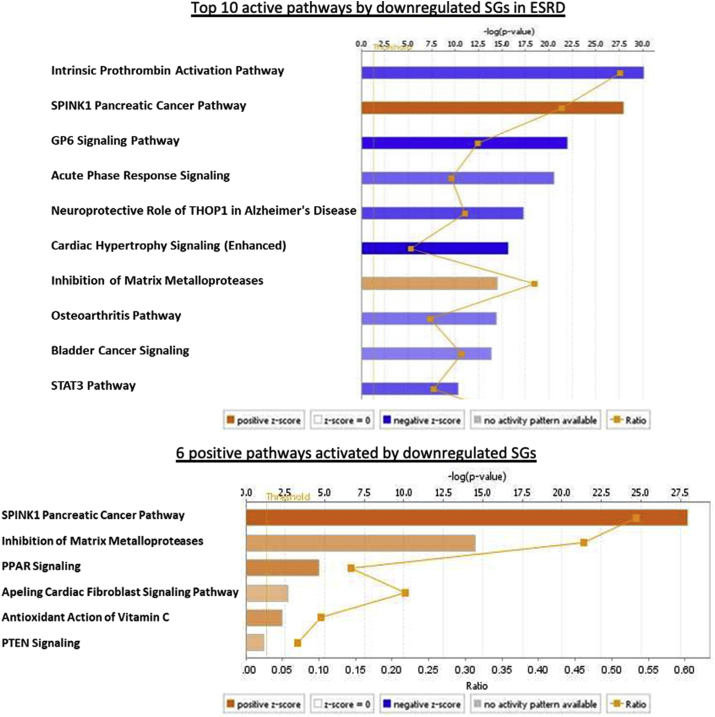
A total of 113 active pathways were identified in ESRD down-regulated SGs via IPA analysis. Six out of 113 pathways (5.31%) were positively activated by downregulated SGs in ESRD, including SPINK1 Pancreatic Cancer Pathway, Inhibition of Matrix Metalloproteases, PPAR Signaling, Apelin Cardiac Fibroblast Signaling Pathway, Antioxidant Action of Vitamin C, and PTEN Signaling. The rest of the pathways were downregulated, suggesting that a large number of SGs pathways in the PBMCs from patients with ESRD were downregulated for ESRD progression, which potentially drives physiological functions and homeostasis. Of note, the five active pathways induced by upregulated SGs in ESRD (Fig. 2B) were also included in 113 active pathways induced by downregulated SGs in ESRD according to Venn Diagram, which showed that some components in these ESRD-upregulated pathways can be fully functional in the absence of the other components in these pathways downregulated in ESRD.
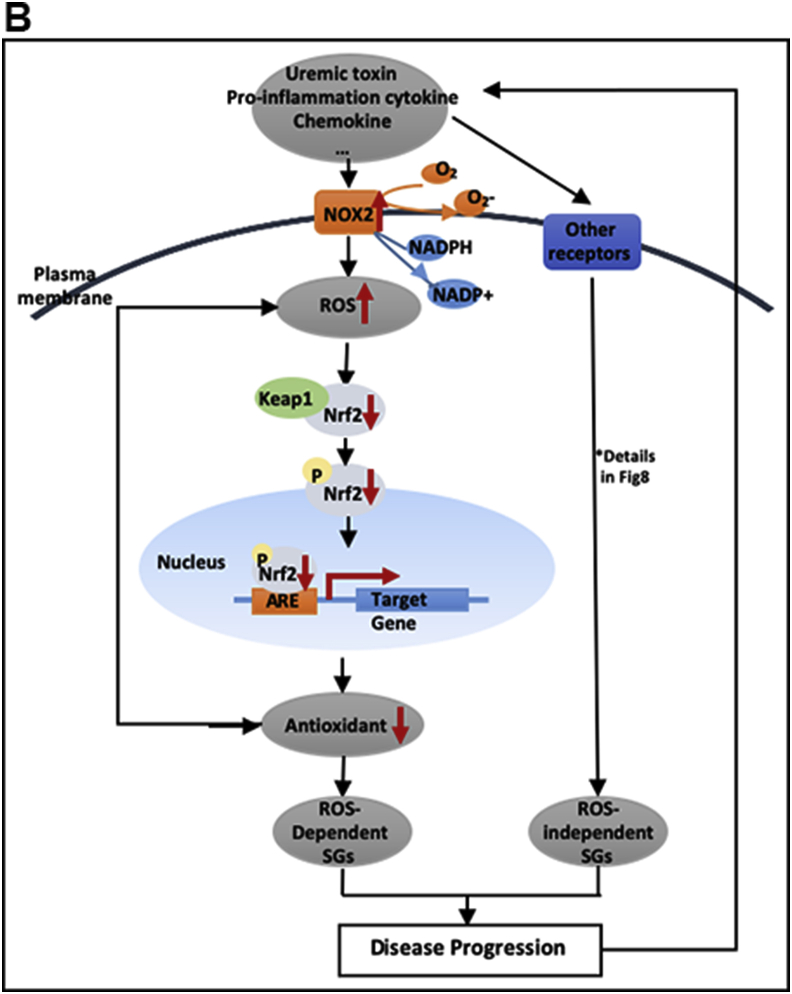
In addition, to identify the upstream regulating transcription factors (TFs) for CKD- and ESRD-modulated SGs, we used the GATHER database (https://gather.genome.duke.edu/) [72] to map the TF binding in the promoters of the modulated SGs. As shown in Table 3B and Fig. 2F, the top 10 TFs bound to the promoters of CKD-upregulated SGs were E2F1, FoxJ2, NFkBp65, E2F1, ROX, DP-1 heterodimer, E2A, LEF1TCF1, upstream stimulating factor and activator protein 1, which were different from that of ESRD-upregulated SGs. The top 10 TFs bound to the promoters of ESRD-upregulated SGs included Egr-1 (inflammation and fibrosis) [73], Egr-2 (maintenance of peripheral nerve myelin), CCAAT/enhancer-binding protein (CEBPgamma) (antioxidant regulator) [74], phosphate inorganic transporter 1 (PIT1) (obesity and insulin resistance) [75], Ikaros 3 (T helper cell 2 transcription factor) [76], hepatic nuclear factor 1 (HNF1) (transcription inducer for proinflammatory molecules such as C-reactive protein, IL-6, HNF1a and HNF4a in CKD) [77], early growth response gene 3 (IL-1β co-expressed inflammatory gene) [78], POU1F1 (inflammation/immunity and hormone regulator) [79], E4BP4 (obesity and insulin resistance) [80], and cell-division control protein 5. In addition, we noticed that nuclear respiratory factor 2 (different from the antioxidant transcription factor nuclear factor-erythroid-derived 2-like 2, Nrf2) [81] was downregulated in ESRD-downregulated SGs (Table 3B and Fig. 2F). In summary, the seven TFs out of ten TFs identified with the Gather database indicated that ESRD-upregulated SGs promote inflammation, obesity and insulin resistance and fibrosis.
Table 3b.
Transcription factor analysis using TRANSFAC database (http://genexplain.com/transfac/) through the access of the GATHER database (https://changlab.uth.tmc.edu/gather/) showed that top 10 transcription factors filtered by P value were involved in each of the four groups of SGs such as CKD upregulated SGs, CKD-downregulated SGs, ESRD-upregulated SGs and ESRD-downregulated SGs. Of note, five out of ten transcription factors that are involved in ESRD downregulated SGs are isoforms of E2F1.
| Annotation | -ln (p value)* | main function/effect | PMID | |
|---|---|---|---|---|
| up SGs in CKD | E2F1(V$E2F1_Q6_01) | 5.94 | control cell-cycle progression from G1 to S phase | 7969176 |
| fork head box J 2(V$FOXJ2_01) | 5.94 | suporession of migration and invasion | 25873280 | |
| NF-kappaB (p65) (V$NFKAPPAB65_01) | 5.13 | proinflammation to disease progression | 30135182 | |
| E2F1(V$E2F1_Q6) | 5.08 | control cell-cycle progression from G1 to S phase | 7969176 | |
| KROX (V$KROX_Q6) | 5.01 | tooth development | 12489153 | |
| DP-1 heterodimer (V$E2F1DP1_01: E2F-1) | 4.31 | cell cycle | 8405995 | |
| E2A (V$E2A_Q2) | 4.31 | pathogenesis of lymphocytic leukemia. | 26301816 | |
| LEF1(V$LEF1TCF1_Q4) | 3.89 | cell proliferation | 31623618 | |
| upstream stimulating factor (V$USF_Q6) | 3.84 | lipid metabolism and atherosclerosis | 19910639 | |
| activator protein 1(V$AP1_Q4) |
3.84 |
cell growth, differentiation, and apoptosis |
15564374 |
|
| down SGs in CKD | nuclear respiratory factor 2(V$NRF2_01) | 5.56 | oxidative stress | 27646262 |
| SREBP(V$SREBP_Q3) | 5.51 | glucose metabolism | 28920951 | |
| NFKB(V$NFKB_Q6_01) | 5.24 | proinflammatory response | 31101940 | |
| c-ETS-1 binding site (V$ETS1_B) | 4.9 | cell differentiation | 30566881 | |
| AP-1 binding site (V$AP1_C) | 4.84 | cell growth, differentiation, and apoptosis | 15564374 | |
| c-Ets-2 binding sites (V$ETS2_B) | 4.84 | osteogenesis | 11175361 | |
| ZTA (V$ZTA_Q2) | 4.78 | Epstein-Barr Virus Reactivation | 27708396 | |
| c-Rel (V$CREL_01) | 4.61 | tumorigenesis | 26757421 | |
| DEAF1(V$DEAF1_01) | 4.61 | intellectual disability | 24726472 | |
| TEL2(V$TEL2_Q6) |
4.49 |
hematopoiesis |
28693791 |
|
| up SGs in ESRD | Egr-1(V$EGR1_01) | 8.42 | inflammation and fibrosis | 21511034 |
| Egr-2(V$EGR2_01) | 7.4 | maintenance of peripheral nerve myelin | 15836632 | |
| CEBPGAMMA (V$CEBPGAMMA_Q6) | 6.61 | antioxidant regulator | 26667036 | |
| PIT1(V$PIT1_Q6) | 6 | obesity and insulin resistance | 27568561 | |
| Ikaros 3(V$IK3_01) | 5.94 | T helper cell 2 transcription factor | 21469117 | |
| Hepatic nuclear factor 1(V$HNF1_C) | 5.79 | transcription inducer for proinflammatory molecules such as C-reactive protein, IL-6, HNF1a and HNF4a in CKD | 29330688 | |
| early growth response gene 3 product (V$EGR3_01) | 5.18 | IL-1b co-expressed inflammatory gene | 31612215 | |
| POU1F1(V$POU1F1_Q6) | 5.13 | inflammation/immunity and hormone regulator | 27709372 | |
| E4BP4(V$E4BP4_01) | 4.96 | obesity and insulin resistance | 27050305 | |
| cell division control protein 5(V$CDC5_01) |
4.72 |
unguarded cellular proliferation |
18583928 |
|
| down SGs in ESRD | E2F1(V$E2F1_Q6) | 12.62 | control cell-cycle progression from G1 to S phase | 7969176 |
| nuclear respiratory factor 2(V$NRF2_01) | 12.62 | oxidative stress | 27646262 | |
| E2F1(V$E2F1_Q6_01) | 12.62 | control cell-cycle progression from G1 to S phase | 7969176 | |
| E2F1(V$E2F1_Q3_01) | 12.62 | control cell-cycle progression from G1 to S phase | 7969176 | |
| E2F(V$E2F_Q6) | 12.62 | control cell-cycle progression from G1 to S phase | 7969176 | |
| E2F(V$E2F_Q3_01) | 12.62 | control cell-cycle progression from G1 to S phase | 7969176 | |
| KROX (V$KROX_Q6) | 12.62 | tooth development | 12489153 | |
| CREB(V$CREBATF_Q6) | 12.62 | formation of long-lasting memories | 20223527 | |
| CREB(V$CREB_Q4_01) | 12.62 | formation of long-lasting memories | 20223527 | |
| NRF1(V$NRF1_Q6) | 12.62 | Cholesterol Homeostasis | 29149604 |
*P value were calculated based on the probability of seeing a Bayes factor of a particular magnitude in a query.
Fig. 2f.
The Venn Diagram of transcript factors analysis in Table 3B. E2F1 was shared by CKD-upregulated SGs and ESRD-downregulated SGs in ESRD. In addition, nuclear respiratory factor 2 (Nrf2), a key transcription factor in Redox Oxygen Species (ROS), was shared in two groups of SGs, CKD-downregulated SGs and ESRD-downregulated SGs, indicating these two transcription factors may serve as an important inhibitor of disease progression.
The Venn Diagram Analysis results (Fig. 3A) on the three secretory gene groups such as 35 UT genes (encoded for 30 UTs in Table 1A), 44 CKD upregulated SGs and 121 ESRD-upregulated SGs showed that: 1) UTs have no overlaps with CKD-upregulated SGs; 2) UTs have two toxins (CFD, and RETN) overlapped with ESRD-upregulated SGs; 3) ESRD-upregulated SGs have four SGs (ADAM Metallopeptidase Domain 9 (ADAM9), complement C3 (C3), Heat Shock Protein 90 Beta Family Member 1 (HSP90B1), and S100 Calcium Binding Protein A12 (S100A12)) overlapped with CKD-upregulated SGs. In addition, one signaling pathway “Role of Cytokines in Mediating Communication between Immune Cells” was shared by the top 10 pathways associated with UTs and the five active pathways upregulated by SGs in ESRD. These results suggest that the signaling pathway “Role of Cytokines in Mediating Communication between Immune Cells” may be significant in ESRD progression.
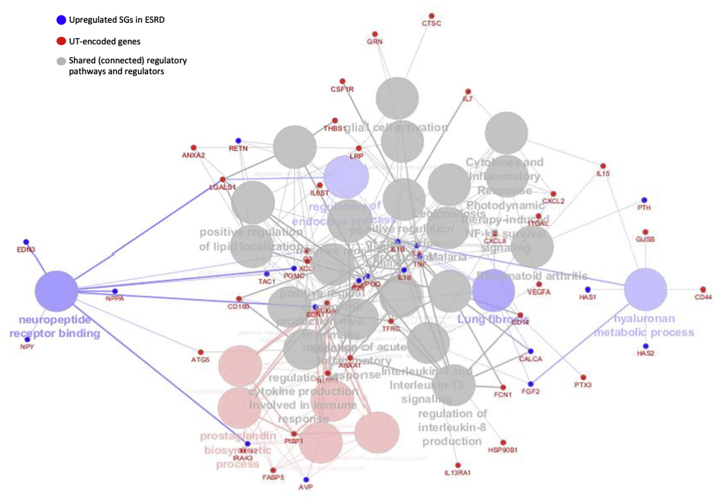
ClueGo (v2.5.4) from Cytoscape (v3.7.2) was also used to verify a close functional relationship between UT genes further and up-regulated SGs in ESRD (Fig. 3B) [[82], [83], [84]]. Of note, the small dots were the genes that connected UT gene group and upregulated SGs in ESRD. ClueGo identified four ESRD-upregulated SGs pathways (big balls in blue color), neuropeptide receptor finding, regulation of endocrine process, lung fibrosis and hyaluronan metabolic process. In addition, ClueGo identified five UT genes pathways, such as prostaglandin biosynthetic process (big balls in red color). Moreover, ClueGo found 20 shared (big balls in grey color, connected) regulatory pathways (Supplemental Table 8).
Fig. 3b.
The ClueGo v2.5.4 from Cytoscape v3.7.2 used as a secondary software to confirm a close functional relationship between UT-encoded genes and up-regulated SGs in ESRD. (Group-specific and connective function are listed in supplement Table 8.)
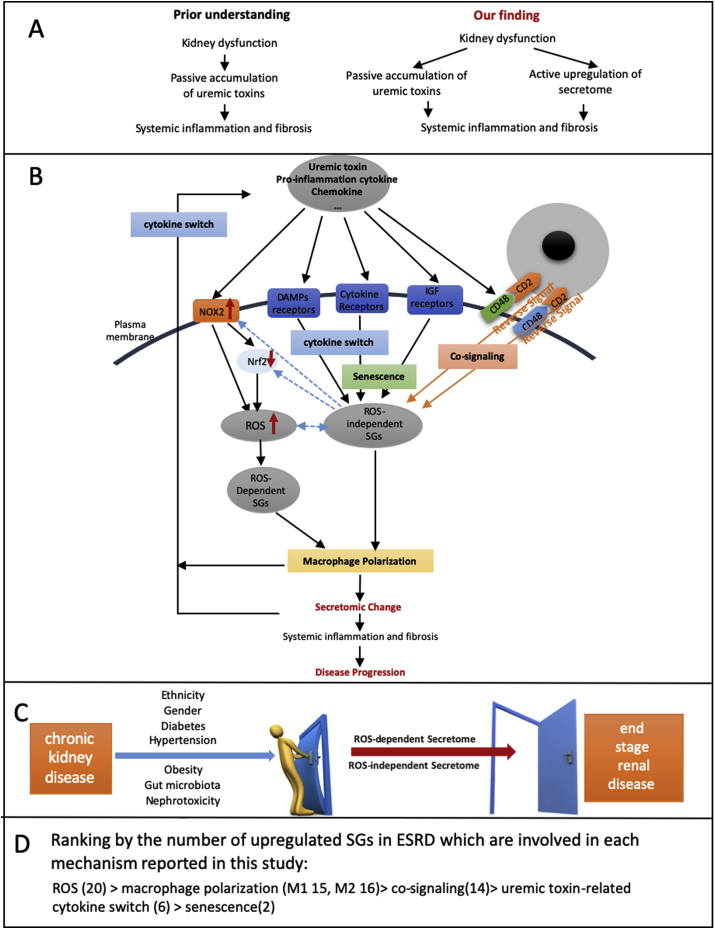
In summary (Table 4D), based on the four aspects above including 1) IPA analyses on proinflammatory signaling pathways of ESRD-upregulated SGs (Fig. 2B) and ) IPA analyses on activating profibrotic pathways in ESRD-downregulated SGs (Fig. 2D); 3) Gather/TRANSFAC-identified TFs involved in proinflammatory and profibrotic pathways (Table 3B); 4) ClueGo (Cytoscape)-identified ESRD-upregulated SGs shared with that of UTs (Fig. 3B), our results demonstrated that UT genes and ESRD-upregulated SGs share many signaling pathways, especially in some pro-inflammatory/profibrotic pathways. These results were correlated well with our recent report on our new model of inflammation-driven upregulation for uremic toxin generation rather than the traditional model of passive accumulation of metabolites fully due to kidney dysfunction to generate uremic toxins [10].
Table 4b.
As a novel mechanism, cytokine-based m.w. UTs can amplify secretomic changes in ESRD. The second example is that 4.13% upregulated SGs in ESRD were upregulated in IL-18-treated human blood leukocytes (GEO ID: GSE103500) and 11.31% were downregulated (the full gene list is attached in supplement Table 9).
| Uremic Toxin | upregulated ESRD SGs | Gene | P.Value | logFC |
|---|---|---|---|---|
| IL-18 | 5 (4.13%) |
CFD | 0.007418 | 1.755116 |
| IL6ST | 0.007849 | 3.737921 | ||
| IRAK3 | 0.005272 | 1.357006 | ||
| PDGFC | 0.01745 | 1.366625 | ||
| SIL1 |
0.024563 |
1.612347 |
||
| downregulated ESRD SGs | ADAM12 | 0.047778 | −3.15133 | |
| 105 (11.31%) | ALPPL2 | 0.036972 | −1.34452 | |
| AMBN | 0.048698 | −1.73612 | ||
| AMELX | 0.010632 | −2.25732 | ||
| ANTXR1 | 0.032292 | −2.91483 | ||
| ARHGAP6 | 0.017996 | −3.93008 |
Fig. 2d.
A total of 113 active pathways were identified in ESRD down-regulated SGs according to IPA (cutoff: |z-score|>2). The top 10 active pathways are shown (top). Only six active pathways (5.31%) were positively activated by downregulated SGs in ESRD (bottom), including SPINK1 Pancreatic Cancer Pathway, Inhibition of Matrix Metalloproteases, PPAR Signaling, Apelin Cardiac Fibroblast Signaling Pathway, Antioxidant Action of Vitamin C, and PTEN Signaling. The rest of the pathways were downregulated. (Gene list for these 113 pathways associated with these up- and downregulated SGs in ESRD via IPA are listed in Supplement Table 7.)
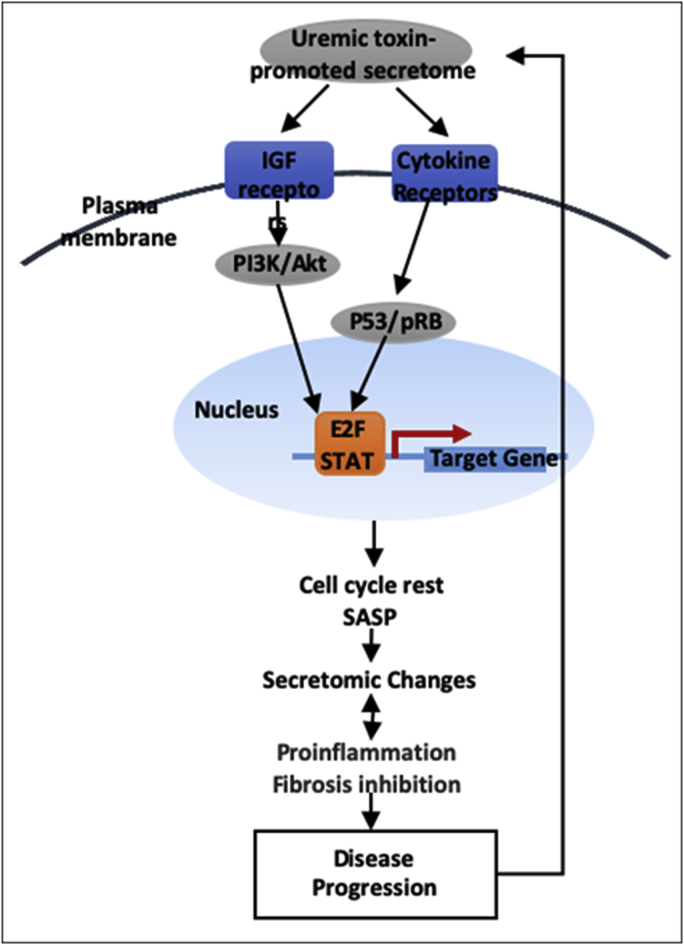
3.5. Proinflammatory cytokines-based middle class UTs such as interleukin-1β (IL-1β) and IL-18 promote ESRD modulation of SGs
To identify the molecular mechanisms underlying ESRD modulation of SGs, we selected proinflammatory cytokine-based UTs such as IL-1β and IL-18 stimuli to demonstrate proof of principle. As shown in Table 4A, IL-1β stimulation of PBMCs resulted in upregulation of five out of 121 ESRD-upregulated SGs such as C-X-C Motif Chemokine Ligand 2 (CXCL2), Interleukin 1 Receptor Associated Kinase 3 (IRAK3), Phospholipase A2 Group VII (PLA2G7), Sphingomyelin Phosphodiesterase Acid Like 3A (SMPDL3A) and Thrombospondin 1 (THBS1). Of note, the Geo dataset (microarray experiments) used as many as 17 innate immune stimuli, IL-1β stimulated data were chosen for our analysis since the rest of innate immune stimuli were not the reported UTs. In addition, IL-1β stimulation downregulated 29 out of 928 ESRD-downregulated SGs (Table 2B) (3.13%). Moreover, IL-18 stimulation upregulated five out of 121 ESRD-upregulated SGs (4.13%) such as complement factor D (CFD), Interleukin-6 Receptor Subunit Beta (IL6st), IRAK3, Platelet-Derived Growth Factor C (PDGFC), and SIL1 Nucleotide Exchange Factor (SIL1); and downregulated 29 out of 928 ESRD-downregulated SGs (Table 2B) (3.13%). Furthermore, IL-18 downregulated 105 out of 928 ESRD-downregulated SGs (11.31%). Our results have demonstrated for the first time that as a novel mechanism underlying the phenotype of ESRD-induced SG changes in the PBMCs from patients with ESRD, proinflammatory cytokines-based middle class UTs such as IL-1β and IL-18 promote ESRD modulation of SGs; and that the ESRD-downregulated SGs contribute to physiological functions. Their downregulation by proinflammatory cytokines IL-1β and IL-18 further strengthen this conclusion.
Table 4a.
As a novel mechanism, cytokine-based m.w. UTs can amplify the ESRD signals in inducing secretomic changes in the PBMCs from patients with ESRD. The first example is that 4.13% of upregulated SGs in ESRD were upregulated in Interleukine-1 beta (IL-1B)-treated human blood leukocytes (GEO ID: GSE103500) and 3.13% were downregulated.
| Uremic Toxin | Primary Change | Gene | P.Value | logFC |
|---|---|---|---|---|
| IL-1B | upregulated ESRD SGs | CXCL2 | 6.53E-05 | 4.959039 |
| GSE103500 | 5 (4.13%) |
IRAK3 | 0.002333 | 1.783057 |
| PLA2G7 | 0.033483 | 1.751108 | ||
| SMPDL3A | 0.038285 | 1.081 | ||
| THBS1 |
0.02777 |
1.307772 |
||
| downregulated ESRD SGs | ADAM12 | 0.003898 | −2.62067 | |
| 29 (3.13%) | B3GNT3 | 0.02258 | −3.46046 | |
| BRINP2 | 0.044496 | −5.65328 | ||
| CAMP | 0.024428 | −1.31643 | ||
| CLPS | 0.049925 | −4.62095 | ||
| EGFL7 | 0.042206 | −2.06849 | ||
| FGF6 | 0.015567 | −4.70715 | ||
| FN1 | 0.018602 | −2.11926 | ||
| GREM1 | 0.031547 | −1.73607 | ||
| INSL6 | 0.012816 | −3.0913 | ||
| IZUMO4 | 0.029209 | −5.18373 | ||
| KLK10 | 0.017674 | −2.11833 | ||
| KLK11 | 0.037963 | −2.50173 | ||
| LIPF | 0.028216 | −1.33852 | ||
| LRRC17 | 0.015359 | −7.35399 | ||
| MIA | 0.013393 | −1.3311 | ||
| MMP11 | 0.021633 | −1.8703 | ||
| MMP28 | 0.004459 | −3.13363 | ||
| MUC2 | 0.034662 | −2.09434 | ||
| NRG1 | 0.038783 | −1.98528 | ||
| OPRPN | 0.049001 | −3.26088 | ||
| PGC | 0.034397 | −1.69163 | ||
| POFUT1 | 0.011287 | −1.93524 | ||
| PON1 | 0.036631 | −4.57456 | ||
| PRL | 0.015419 | −4.23378 | ||
| PRLR | 0.044216 | −1.79944 | ||
| PVR | 0.034772 | −4.70854 | ||
| SERPINE1 | 0.012631 | −1.57299 | ||
| SPARCL1 | 0.023002 | −2.49064 |
In Table 3a, Table 4cC, IPA classified the parts of the SGs modulated in the PBMCs from patients with CKD and patients with ESRD as the cytokine group. The results in Table 4C showed that 4 out of 55 SGs (7.2%) downregulated in CKD (also shown in Table 2B) were cytokines such as IL-24, interleukin 36 receptor antagonist (IL36RN), platelet factor 4 (PF4), and ectodysplasin A (EDA); one Dickkopf WNT signaling pathway inhibitor 3 (DKK3, a tumor suppressor) out of 44 SGs (2.27%) upregulated in CKD was cytokine; 68 out of 928 SGs (7.3%) downregulated in ESRD were cytokines; and 6 out of 121 SGs (4.96%) upregulated in ESRD were cytokines including inflammation-modulating aminoacyl tRNA synthetase complex interacting multifunctional protein 1 (AIMP1), inflammatory C-X-C motif chemokine ligand 2 (CXCL2), inflammatory CXCL8, T cell and B cell promoting IL7, T cell and natural killer cell-activating IL-15 and inflammatory X–C motif chemokine ligand 1 (XCL1).
Table 4c.
The IPA classified the parts of the SGs modulated in the PBMCs from patients with CKD and patients with ESRD as the cytokine group. The results showed that 4 out of 55 SGs (7.2%) downregulated in CKD (also shown in Table 2B) were cytokines; one out of 44 SGs (2.27%) upregulated in CKD was cytokine; 68 out of 928 SGs (7.3%) downregulated in ESRD were cytokines; and 6 out of 121 SGs (4.96%) upregulated in ESRD were cytokines.
| Symbol | Entrez Gene Name | Expr p-value | Expr Log Ratio | Location | |
|---|---|---|---|---|---|
| down in CKD | IL24 | interleukin 24 | 0.0327 | −1.174 | Extracellular Space |
| [4] | IL36RN | interleukin 36 receptor antagonist | 0.0319 | −1.2 | Extracellular Space |
| PF4 | platelet factor 4 | 0.0149 | −1.11 | Extracellular Space | |
| EDA |
ectodysplasin A |
0.00949 |
−1.069 |
Plasma Membrane |
|
| up in CKD (1) |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
0.0094 |
1.827 |
Extracellular Space |
| down in ESRD | BMP8A | bone morphogenetic protein 8a | 0.000409 | −2.355 | Extracellular Space |
| [68] | CCL1 | C–C motif chemokine ligand 1 | 0.0184 | −1.28 | Extracellular Space |
| CCL8 | C–C motif chemokine ligand 8 | 0.0221 | −1.682 | Extracellular Space | |
| CCL13 | C–C motif chemokine ligand 13 | 5.57E-05 | −1.797 | Extracellular Space | |
| CCL17 | C–C motif chemokine ligand 17 | 0.0196 | −1.465 | Extracellular Space | |
| CCL18 | C–C motif chemokine ligand 18 | 0.00866 | −1.367 | Extracellular Space | |
| CCL19 | C–C motif chemokine ligand 19 | 0.0044 | −2.113 | Extracellular Space | |
| CCL21 | C–C motif chemokine ligand 21 | 0.00242 | −2.011 | Extracellular Space | |
| CCL22 | C–C motif chemokine ligand 22 | 3.47E-05 | −1.656 | Extracellular Space | |
| CCL23 | C–C motif chemokine ligand 23 | 0.00434 | −2.54 | Extracellular Space | |
| CCL24 | C–C motif chemokine ligand 24 | 0.00354 | −1.942 | Extracellular Space | |
| CCL25 | C–C motif chemokine ligand 25 | 0.0037 | −1.042 | Extracellular Space | |
| CRH | corticotropin releasing hormone | 0.00196 | −1.518 | Extracellular Space | |
| CSF1 | colony stimulating factor 1 | 7.44E-05 | −1.84 | Extracellular Space | |
| CSF2 | colony stimulating factor 2 | 0.00115 | −1.158 | Extracellular Space | |
| CX3CL1 | C-X3-C motif chemokine ligand 1 | 0.00352 | −1.231 | Extracellular Space | |
| CXCL5 | C-X-C motif chemokine ligand 5 | 0.0264 | −1.939 | Extracellular Space | |
| CXCL9 | C-X-C motif chemokine ligand 9 | 0.014 | −1.063 | Extracellular Space | |
| CXCL11 | C-X-C motif chemokine ligand 11 | 0.0439 | −1.101 | Extracellular Space | |
| CXCL12 | C-X-C motif chemokine ligand 12 | 0.000564 | −1.422 | Extracellular Space | |
| CXCL14 | C-X-C motif chemokine ligand 14 | 0.00645 | −1.535 | Extracellular Space | |
| DKK3 | dickkopf WNT signaling pathway inhibitor 3 | 0.00168 | −1.867 | Extracellular Space | |
| EDN1 | endothelin 1 | 0.0271 | −1.424 | Extracellular Space | |
| EPO | erythropoietin | 0.00154 | −2.113 | Extracellular Space | |
| FASLG | Fas ligand | 0.000484 | −1.074 | Extracellular Space | |
| IFNA5 | interferon alpha 5 | 0.0351 | −1.309 | Extracellular Space | |
| IFNA7 | interferon alpha 7 | 0.0259 | −1.167 | Extracellular Space | |
| IFNA16 | interferon alpha 16 | 0.0243 | −1.327 | Extracellular Space | |
| IFNB1 | interferon beta 1 | 0.0248 | −1.679 | Extracellular Space | |
| IFNW1 | interferon omega 1 | 0.000648 | −1.794 | Extracellular Space | |
| IL2 | interleukin 2 | 0.00237 | −1.245 | Extracellular Space | |
| IL3 | interleukin 3 | 0.00158 | −1.82 | Extracellular Space | |
| IL4 | interleukin 4 | 0.000549 | −2.658 | Extracellular Space | |
| IL5 | interleukin 5 | 0.00255 | −1.791 | Extracellular Space | |
| IL9 | interleukin 9 | 0.00207 | −1.852 | Extracellular Space | |
| IL11 | interleukin 11 | 0.00046 | −1.87 | Extracellular Space | |
| IL16 | interleukin 16 | 0.000374 | −1.476 | Extracellular Space | |
| IL19 | interleukin 19 | 0.00647 | −1.564 | Extracellular Space | |
| IL21 | interleukin 21 | 0.000431 | −2.258 | Extracellular Space | |
| IL22 | interleukin 22 | 0.0124 | −1.315 | Extracellular Space | |
| IL24 | interleukin 24 | 0.000213 | −2.527 | Extracellular Space | |
| IL25 | interleukin 25 | 0.00486 | −1.597 | Extracellular Space | |
| IL26 | interleukin 26 | 0.00285 | −1.789 | Extracellular Space | |
| IL37 | interleukin 37 | 0.031 | −1.296 | Extracellular Space | |
| IL17A | interleukin 17A | 0.0004 | −1.944 | Extracellular Space | |
| IL1A | interleukin 1 alpha | 1.52E-07 | −2.584 | Extracellular Space | |
| IL1RN | interleukin 1 receptor antagonist | 4.04E-05 | −1.628 | Extracellular Space | |
| IL36A | interleukin 36 alpha | 0.00185 | −1.492 | Extracellular Space | |
| IL36G | interleukin 36 gamma | 0.0143 | −1.615 | Extracellular Space | |
| IL36RN | interleukin 36 receptor antagonist | 0.0281 | −1.175 | Extracellular Space | |
| LIF | LIF interleukin 6 family cytokine | 0.0323 | −1.402 | Extracellular Space | |
| LTA | lymphotoxin alpha | 2.73E-05 | −2.497 | Extracellular Space | |
| OSM | oncostatin M | 0.00294 | −2.002 | Extracellular Space | |
| PF4 | platelet factor 4 | 0.0432 | −1.106 | Extracellular Space | |
| PRL | prolactin | 0.00443 | −1.693 | Extracellular Space | |
| PRLH | prolactin releasing hormone | 0.00346 | −1.191 | Extracellular Space | |
| SCG2 | secretogranin II | 0.00633 | −2.162 | Extracellular Space | |
| SCGB1A1 | secretoglobin family 1A member 1 | 0.0166 | −1.372 | Extracellular Space | |
| SLURP1 | secreted LY6/PLAUR domain containing 1 | 0.000616 | −1.435 | Extracellular Space | |
| SPP1 | secreted phosphoprotein 1 | 0.000139 | −2.907 | Extracellular Space | |
| THPO | thrombopoietin | 0.00506 | −1.895 | Extracellular Space | |
| TNFSF11 | TNF superfamily member 11 | 0.000298 | −1.936 | Extracellular Space | |
| TNFSF14 | TNF superfamily member 14 | 1.85E-05 | −3.778 | Extracellular Space | |
| WNT1 | Wnt family member 1 | 0.00424 | −1.889 | Extracellular Space | |
| WNT2 | Wnt family member 2 | 0.0082 | −1.681 | Extracellular Space | |
| WNT4 | Wnt family member 4 | 0.00799 | −1.371 | Extracellular Space | |
| WNT5A | Wnt family member 5A | 0.00932 | −2.049 | Extracellular Space | |
| EDA |
ectodysplasin A |
3.23E-06 |
−1.747 |
Plasma Membrane |
|
| up in ESRD | AIMP1 | aminoacyl tRNA synthetase complex interacting multifunctional protein 1 | 8.15E-09 | 1.927 | Extracellular Space |
| [6] | CXCL2 | C-X-C motif chemokine ligand 2 | 0.000396 | 3.051 | Extracellular Space |
| CXCL8 | C-X-C motif chemokine ligand 8 | 1.78E-06 | 4.136 | Extracellular Space | |
| IL7 | interleukin 7 | 0.0188 | 1.258 | Extracellular Space | |
| IL15 | interleukin 15 | 5.13E-06 | 1.645 | Extracellular Space | |
| XCL1 | X–C motif chemokine ligand 1 | 0.00893 | 1.131 | Extracellular Space |
In Fig. 3C, the Venn Diagram Analysis results showed that four cytokines downregulated in CKD were shared with that downregulated in ESRD. The expanded list of 68 cytokines downregulated in ESRD can be split into two groups. First are the chemokine subset of cytokines. Chemokines are involved in cell migration, activation and tissue injury and thus key mediators of inflammation, especially in cardiovascular disease [85]. According to previous studies, the majority of these proteins were involved in the homeostatic function of immune cells due to their ligand promiscuity [86]. The profile of chemokines downregulated in ERSD were mostly “homeostatic” compartments rather than “inflammatory” ones. Typically, CCL2, 3, 4, 5, 11, CXCL1, 2, 8, and 10 played proinflammatory roles in kidney disease; and these chemokines were not included in this list [[87], [88], [89]]. Secondly, pro-inflammatory cytokine drivers were “common” in kidney disease including IL6, IL8, IL10, IL17 and IL18 and were not included in our downregulated cytokine list [88]. These cytokines may be contributed by cell types other than PBMCs during kidney diseases. Interestingly, both proinflammatory and anti-inflammatory members (IL-1Ra and IL-36G) were all downregulated, indicating that a compensatory balance weighed by PBMCs was associated with disease progression [90]. Of note, IL10 family members, including IL-19, IL-22, IL-24, IL-26 and IL-28 were downregulated [90]. Based on the overall proinflammatory phenotype associated with secretomic changes during ESRD, these results suggested that in addition to the modulation by cytokines and chemokines, additional secretomic changes modulated by other mechanisms may play significant roles in disease progression.
Fig. 3c.
The Venn Diagram Analysis results showed that 1) all of the four cytokines (IL24, IL36RN, PF4, EDA) downregulated in CKD are overlapped that downregulated in ESRD; 2) the one cytokine upregulated in CKD is overlapped with that downregulated in ESRD; and 3) six cytokines upregulated in ESRD are not overlapped with the other three groups.
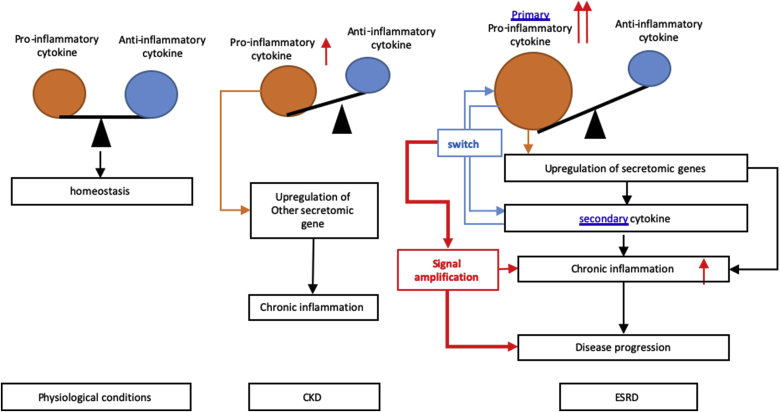
One cytokine upregulated in CKD, DKK3, was shared with that downregulated in ESRD. The six cytokines upregulated in ESRD were ESRD-specific. The results suggest that first, the numbers of SGs in the cytokine groups upregulated in ESRD are significantly higher than that of CKD; and second, highly focused six cytokines and chemokines upregulated in ESRD may play significant roles in promoting ESRD progression and systemic inflammations. To integrate all the findings on proinflammatory cytokines from UTs and from SGs upregulated in ESRD, we proposed a new mechanism in Fig. 3D. Proinflammatory cytokines (primary, upstream) play significant roles in combination with other uremic toxins and other mechanisms in upregulating SGs (secondary, downstream), promoting the pathogenesis of ESRD and inflammations. Of note, the classification of primary/upstream cytokines and secondary/downstream cytokines is conceptual to demonstrate the cytokine interaction as the proof of principle. We used the proinflammatory cytokines IL-1β and IL-18 from UTs as prototypic secretomic proteins to demonstrate the mutual promotion and modulation among the secretomic proteins as the role-switching of “primary” or “secondary” cytokines during ESRD. Future time course experiments will be needed to characterize chronological upregulation of cytokines in upregulated UTs and SGs in ESRD.
Fig. 3d.
Novel mechanism I. Proinflammatory cytokines (primary) play significant roles in combination with uremic toxins and other mechanisms in upregulating SGs (secondary), promoting the pathogenesis of ESRD and inflammations. We used the proinflammatory cytokines as prototypic secretomic proteins to demonstrate the mutual promotion and modulation among the secretomic proteins as the role-switching of “primary” and “secondary” cytokines during ESRD.
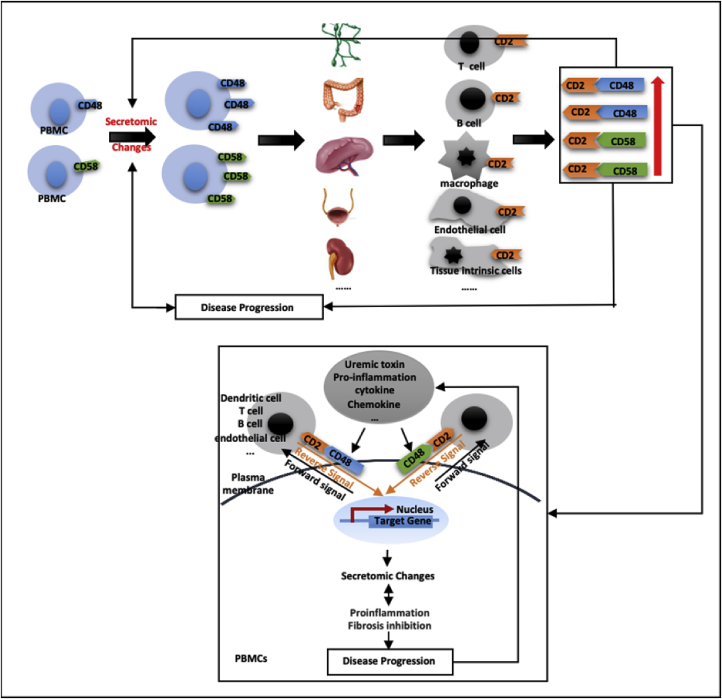
3.6. ESRD-upregulated co-stimulation receptors CD48 and CD58 increase secretomic upregulation in the PBMCs; may not be even limited in the PBMCs, CD48/CD58-CD2 signaling can be actually magnified enormously in tissues
We hypothesized that CKD- and ESRD-upregulated T cell activation co-stimulation receptors (CSRs) and co-inhibition receptors (immune checkpoint receptors, ICRs) [91], as prototypic cell membrane receptors in mediating cell-cell interactions, upregulate SGs in PBMCs (antigen-presenting cells, APC) via reverse signaling as we reported [22,92]. As shown in Table 5A, the expression of 14 CSRs, 4 dual-function receptors, and 10 ICRs were examined in the microarrays of the PBMCs from patients with ESRD and CKD (Table 2A) as we reported [92]. The results show that 1) ESRD upregulates CSRs CD48 and CD58 but downregulates seven out of 14 CSRs including Inducible T Cell Costimulator Ligand (ICOSLG), CD70, TNF Superfamily Member 14 (TNFSF14), CD40, TNFSF15, TNFSF18, and Signaling Lymphocytic Activation Molecule Family Member 1 (SLAMF1); 2) ESRD downregulated one out of four dual function receptors poliovirus receptor (PVR) (con-stimulation at naïve T cells but co-inhibitory at activated T cells); 3) ESRD downregulated four out of 10 immune checkpoint receptors (co-inhibition receptors) such as nectin cell adhesion molecule 3 (NECTIN3), programmed cell death 1 ligand 2 (PDCD1LG2), human endogenous retrovirus-H long terminal repeat-associating protein 2 (HHLA2) and butyrophilin like 2 (BTNL2); and 4) CKD upregulated one immune checkpoint receptor HHLA2 but downregulated one co-stimulation receptor TNFSF8.
Table 5a.
Most of 28 co-stimulation receptors and immune checkpoint (co-inhibition) receptors (expressed in the antigen presenting cell (APC) surface) are modulated in the PBMCs from patients with CKD and ESRD. The results showed that: 1) seven out of 14 co-stimulation receptors, one out of four dual receptors (functional as co-stimulation for naïve T cells and co-inhibition receptors for activated T cells), and four out of 10 immune checkpoint receptors were downregulated in ESRD, respectively; 2) two co-stimulation receptors CD48 and CD58 were upregulated in ESRD; 3) one immune checkpoint receptor HHLA2 was upregulated in CKD; and 4) one co-stimulation receptor TNFSF8 was downregulated in CKD.
| Effect | up in CKD |
down in CKD |
up in ESRD |
down in ESRD |
|
|---|---|---|---|---|---|
| Gene symbol | logFC | logFC | logFC | logFC | |
| co-stimulation | ICOSLG | −1.9552 | |||
| CD70 | −1.5467 | ||||
| TNFSF14 | −3.77804 | ||||
| CD40 | −2.93849 | ||||
| TNFSF9 | |||||
| TNFSF4 | |||||
| TNFSF15 | −1.11762 | ||||
| TNFSF18 | −1.25532 | ||||
| TNFSF8 | −1.36894 | ||||
| TIMD4 | |||||
| SLAMF1 | −1.51147 | ||||
| CD48 | 1.089174 | ||||
| SEMA4A | |||||
| CD58 |
1.68807 |
||||
| co-stimulation at naïve TC and co-inhibitory at activated TC | CD80 | ||||
| CD96 | |||||
| PVR | −1.21184 | ||||
| IL2RB |
|||||
| co-inhibition | LGALS9 | ||||
| NECTIN3 | −1.6273 | ||||
| TNFRSF14 | |||||
| PDCD1LG2 | −1.4391 | ||||
| CD274 | |||||
| CD276 | |||||
| VTCN1 | |||||
| VSIR | |||||
| HHLA2 | 1.191813 | −1.41072 | |||
| BTNL2 | −2.12092 |
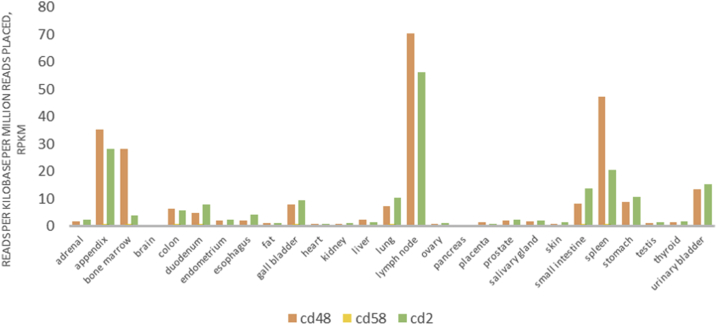
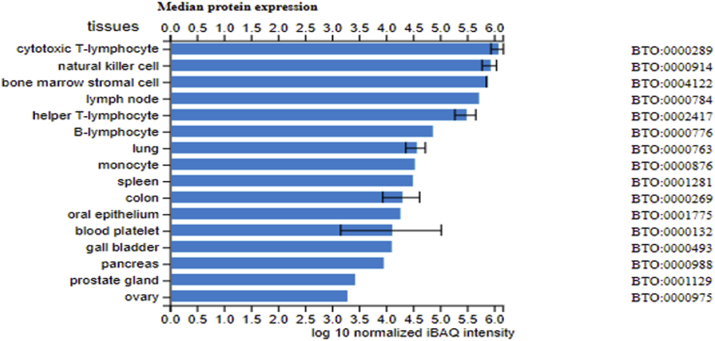
To determine whether ESRD upregulated co-stimulation receptors [92], CD48 and CD58, play any causative roles in regulating ESRD-modulated SG expressions, we tried to find available microarray or RNA-sequencing datasets associated with overexpression or deficiency of CD48 and CD58. As no such datasets are available at the time of this writing, we used the GEO datasets (GSE15215) related to CD2, a membrane protein acting as the ligand for both CD48 and CD58 on dendritic cells (DCs, CD2+ DCs versus CD2− DCs), to determine whether the forward signaling of CD48 and CD58 (from antigen-presenting cells toward T cells) can modulate the expression of SGs modulated in ESRD. These results showed that CD2 upregulates 14 out of 121 SGs (11.6%) upregulated in ESRD; and CD2 downregulates 25 out of 928 SGs (2.7%) downregulated in ESRD (see Table 5b). We found that CD48/CD58-CD2 signaling may amplify the SGs alteration in ESRD. These results suggest that CD48/CD58-CD2 signaling promotes SGs upregulation in ESRD. Of note, the justifications for this analysis are: 1) DCs can be the parts of PBMCs in patients with ESRD [93]; 2) CD2 protein is also expressed in monocytes, B lymphocytes, CD4+ T cells, CD8+ T cells NK cells, platelets, bone marrow stromal cells, which were the CD2 expression data collected from the GeneCards database (https://www.genecards.org/cgi-bin/carddisp.pl?gene=CD2#protein_expression); 3) new RNA-sequencing data from Human Protein Atlas Database (https://www.proteinatlas.org) indicated that CD48 and CD58 are expressed in every one of 27 tissue examined; and that their ligand (CD2) is also expressed in every one of 27 tissues examined, which are correlated with the CD2 protein expression data collected in the GeneCards database shown above (see Fig. 4a); and 4) as shown in Fig. 4bB, the protein expression data from the Proteomics Database (https://www.proteomicsdb.org/) showed that CD2 protein can be highly enriched in cytotoxic T-lymphocyte, natural killer cell, bone marrow stromal cell, helper T-lymphocyte, B lymphocyte and monocyte, which indicated the possibility of the signal amplification induced by co-stimulation of CD48/CD58-CD2 pathway. Of note, we reported previously that CD40+ proinflammatory monocytes accelerate inflammation in CKD [7]. Taken together, these data suggest that first, ESRD-upregulated CD48 and CD58 increase secretomic upregulation in the PBMCs, whose signals may not be even limited in the PBMCs from patients with ESRD examined in this study; second, CD48/CD58-CD2 pathway can be actually magnified enormously in tissue levels so that the CD48/CD58-CD2 pathway-activated PBMCs in blood circulation accelerate vascular and other inflammations; and third, reverse signaling from CD2+ T cells to CD48+/CD58+ PBMCs play significant roles in modulating PBMC secretomic changes in ESRD (see Fig. 4c).
Table 5b.
Since there are no datasets of CD48 and CD58 deficient/overexpressed microarray datasets available in the NIH-NCBI-GEO database, we use the GEO datasets (GSE15215) related to CD2, a membrane protein acting as CD48/CD58 ligand on dendritic cells (DCs), CD2+ DCs versus CD2− DCs to determine whether the forward signaling of CD48 and CD58 can modulate the expression of SGs modulated in ESRD. These results showed that CD2 upregulates 14 out of 121 SGs (11.6%) upregulated in ESRD; and CD2 downregulates 25 out of 928 SGs (2.7%) downregulated in ESRD.
| upregulated in ESRD (14/121, 11.6%) |
downregulated in ESRD (25/928, 2.7%) |
||||
|---|---|---|---|---|---|
| Gene | p value | log FC | Gene | p value | log FC |
| ANXA1 | 0.018331 | 2.18431 | ADM2 | 0.004031 | −1.64776 |
| ANXA2 | 0.006552 | 1.797703 | BMP1 | 0.027839 | −1.26577 |
| ASGR1 | 0.022441 | 2.235951 | C2CD2 | 0.028997 | −1.03495 |
| CTSH | 0.004221 | 1.910232 | COL4A6 | 0.007275 | −1.47307 |
| EREG | 0.020802 | 1.079933 | COL8A1 | 0.035575 | −1.64511 |
| HS2ST1 | 0.001885 | 2.614528 | CRISP1 | 0.003 | −2.27406 |
| IGFBP7 | 0.001049 | 2.731112 | CRLF2 | 0.003679 | −2.21456 |
| IRAK3 | 0.006719 | 2.871895 | CXCL5 | 0.00911 | −2.13001 |
| MTHFD2 | 0.01361 | 1.340201 | CYP2A13 | 0.010832 | −1.52688 |
| NLRP3 | 0.011059 | 2.662663 | EXOG | 0.0476 | −1.9656 |
| PIGK | 0.014052 | 1.876857 | GPC3 | 0.010239 | −1.53284 |
| RNASE4 | 0.007783 | 2.112123 | IGFBP2 | 0.044615 | −1.30936 |
| TPP1 | 0.033834 | 1.607275 | IL21 | 0.03692 | −2.36894 |
| XCL1 | 0.003066 | 1.997939 | KLKB1 | 0.023261 | −1.42336 |
| LIPF | 0.020219 | −1.48391 | |||
| MSMB | 0.003153 | −2.3955 | |||
| NAGLU | 0.018426 | −1.20719 | |||
| PHLPP1 | 0.000438 | −2.88351 | |||
| PLA2G2A | 0.001974 | −2.3566 | |||
| PSG1 | 0.003256 | −2.95988 | |||
| RAB26 | 0.017058 | −1.88871 | |||
| SOSTDC1 | 0.014098 | −1.25801 | |||
| SPINK1 | 0.002876 | −2.26369 | |||
| TTR | 0.026675 | −1.29499 | |||
| WNT2 | 0.008818 | −1.73443 | |||
Fig. 4a.
New RNA-seq (RNA-sequencing) data from Human Protein Atlas (https://www.proteinatlas.org) indicated that CD48 and CD58 are expressed in every one of 27 tissue examined; and that their ligand (CD2) is also expressed in every one of 27 tissues examined, which are correlated with the CD2 protein expression data collected in the GeneCards database shown above.
Fig. 4b.
CD2 protein can be highly enriched in cytotoxic T-lymphocyte, natural killer cell, bone marrow stromal cell, helper T-lymphocyte, B lymphocyte and monocyte according to Proteomics Database (https://www.proteomicsdb.org/).
Fig. 4c.
Novel mechanism. Co-stimulation receptors CD48 and CD58 can initiate signaling cascades via their interactions with their ligand CD2 to amplify the expression changes of SGs upregulated in the PBMCs in patients with ESRD.
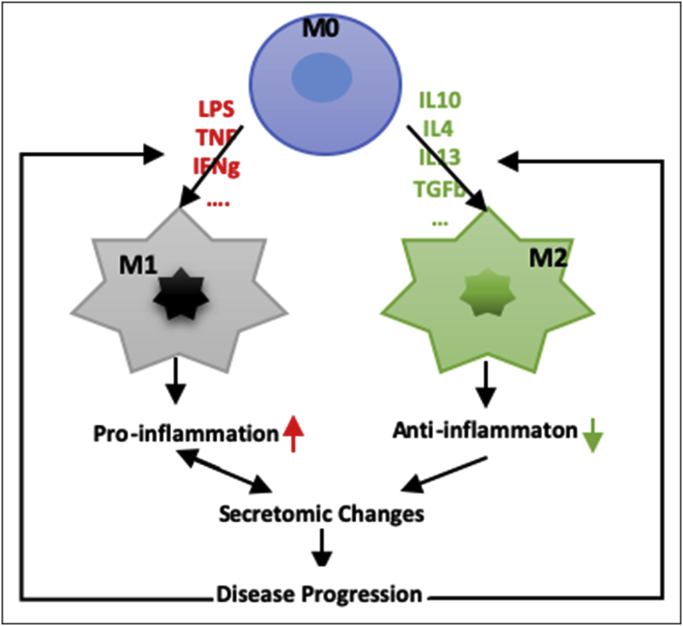
3.7. Classically activated macrophages (M1)-, and alternatively activated macrophages (M2)- macrophage polarization signals contribute to ESRD- and CKD-upregulated SGs
We recently identified 20 new disease group-specific and 12 new shared pathways in macrophages in eight groups of 34 diseases including 24 inflammatory organ diseases and 10 types of cancers [94,95]. It has also been reported that M1 proinflammatory macrophages contribute to infection clearance, inflammation and renal injury, and M2 anti-inflammatory macrophages can contribute to the resolution phase of the response to injury [96,97]. We hypothesized that M1 and M2 macrophage polarization signals contribute to CKD- and ESRD-upregulated PBMC SG expressions. As shown in Table 6, eight out of 44 (18.18%) CKD-upregulated SGs were found in M1 macrophage polarization dataset; six out of 44 (13.64%), including three out of 44 (6.8%, M2a), four out of 44 (9.1%, M2b), one out of 44 (2.3%, M2c), CKD-upregulated SGs, were found in M2a, M2b, and M2c macrophage subset polarization, respectively. In addition, 15 out of 121 (12.4%) ESRD-upregulated SGs were found in M1 macrophage polarization dataset; 16 out of 121 (13.2%), including one out of 121 (0.83%, M2a), 11 out of 121 (9.1%, M2b), and 9 out of 121 (7.4%) ESRD-upregulated SGs were found in M2a, M2b, and M2c macrophage polarization, respectively. When examining those modulated SGs with IPA, no significant pathways were found. As shown in Fig. 5, these results suggest that: 1) macrophage polarization pathways participate CKD-, and ESRD-upregulated secretomic changes in PBMCs in patients with CKD and ESRD; 2) M1 proinflammatory macrophage polarization signal may play more important roles in facilitating PBMC secretomic upregulations in CKD and ESRD than the signals mediating three M2 macrophage subset polarizations and 3) M1-, and M2-polarization signaling pathways involving in upregulating SGs are diversified.
Table 6.
A Eight out of 44 SGs (18.18%) upregulated in CKD were found in M1 macrophage polarization dataset; and six out of 44 SGs (13.64%) upregulated in CKD were found in M2 macrophage polarization. 15 out of 121 SGs (12.4%) upregulated in ESRD were found in M1 macrophage polarization dataset; 16 out of 121 SGs (13.2%) were found in M2 macrophage polarization. These results suggest that macrophage polarization pathways participate CKD-, and ESRD-upregulated secretomic changes in the PBMCs in patients with CKD and ESRD (PMID: 30827512).
| GEO ID |
GSE85346 |
M2 (6) |
||||||
|---|---|---|---|---|---|---|---|---|
| M1 (8) | ||||||||
| M1/M0 |
M2a/M0 |
M2b/M0 |
M2c/M0 |
|||||
| Gene Symbol | Log FC | Gene Symbol | Log FC | Gene Symbol | FC | Gene Symbol | Log FC | |
| CKD | ADAM23 | 3.347203 | APCS | 1.077464 | C3 | 2.819101 | S100A12 | 2.702538 |
| APCS | 2.623466 | CELA2B | 1.065595 | CELA2B | 1.216281 | |||
| CGREF1 | 3.072414 | PLA2G5 | 1.280399 | S100A12 | 4.50631 | |||
| LAMB1 | 3.023362 | TNFAIP6 | 5.233126 | |||||
| NRP2 | 1.740985 | |||||||
| PI3 | 3.458997 | |||||||
| S100A12 | 3.80053 | |||||||
| TNFAIP6 |
8.844792 |
|||||||
| M1 (15) |
M2 (16) |
|||||||
| ESRD | CD44 | 1.014886 | CTSC | 3.295263 | ASGR1 | 1.066338 | CTSC | 1.434133 |
| CD48 | 1.739752 | C3 | 2.819101 | DSE | 1.554709 | |||
| CXCL2 | 2.43399 | CTSC | 1.612899 | HEG1 | 1.066944 | |||
| DSE | 1.893179 | CXCL2 | 2.70138 | IL7 | 2.068031 | |||
| EREG | 3.423501 | IRAK3 | 1.930381 | MGAT4A | 1.202096 | |||
| IGFBP7 | 1.388798 | PTX3 | 1.792836 | S100A12 | 2.702538 | |||
| IL15 | 2.544436 | RETN | 4.306012 | S100A8 | 3.187036 | |||
| IRAK3 | 1.341773 | RNASE2 | 1.692809 | VCAN | 2.876119 | |||
| MGAT4A | 1.055701 | S100A12 | 4.50631 | XCL1 | 1.001179 | |||
| MTHFD2 | 1.605983 | S100A8 | 5.25634 | |||||
| PTX3 | 3.304702 | VCAN | 1.791611 | |||||
| S100A12 | 3.80053 | |||||||
| S100A8 | 3.323343 | |||||||
| TXN | 1.426231 | |||||||
| VEGFA | 2.795328 | |||||||
Fig. 5.
Novel mechanism. Macrophage polarization pathways participate CKD-, and ESRD-upregulated secretomic changes in the PBMCs in patients with CKD and ESRD; M1-, and M2-polarization signaling pathways involving in upregulating SGs are diversified.
3.8. ESRD- and CKD-upregulated SGs in PBMCs contain senescence-promoting regulators by upregulating proinflammatory growth factor IGFBP7 and downregulating anti-inflammatory cytokine TGF-β1 and leukocyte telomere length stabilizer SERPINE1/PAI-1
The senescence program is implicated in diverse biological processes, including embryogenesis, tissue regeneration and repair, tumorigenesis, aging and inflammation. Two main classes of senescent cells have been identified: acute and chronic senescent cells. Acute senescent cells are generated during coordinated, beneficial biological processes characterized by a defined senescence trigger, transient senescent-cell signaling functions, and eventual senescent-cell clearance. In contrast, chronic senescent cells arise more slowly from cumulative, diverse stresses and are inefficiently eliminated, leading to their accumulation and deleterious effects through a secretory phenotype [98]. Senescent cells secrete a variety of proteins collectively known as the senescence-associated secretory phenotype (SASP) [99]. Recent murine studies have shown that depletion of chronically senescent cells extends healthy lifespan and delays age-associated disease, implicating senescence and the senescence-associated secretory phenotype as drivers of organ (kidney) dysfunction [100]. Previous reports suggest that secretomic changes in CKD and ESRD modulate cellular senescence and disease-modulated aging process, renal fibrosis and cancers. In addition to senescence in the kidney, senescent vascular cells, both endothelial and smooth muscle cells, participate in atherosclerosis; senescent preadipocytes and adipocytes have been shown to lead to insulin resistance [101]. Thus, we hypothesize that CKD- and ESRD-related senescence signaling contributes to upregulated SGs in PBMCs. To examine this hypothesis, we compared 71 senescence regulators [102] with CKD-regulated SGs and ESRD-upregulated SGs, respectively. As shown in Table 7, first, no senescence regulators matched with CKD-upregulated SGs; second, two out of 71 senescence regulatory genes were decreased in CKD downregulated SGs [102], specifically ID1 (a member of ID family of helix-loop-helix transcriptional regulatory proteins, a kidney damage inhibitor and target of bone morphogenetic proteins [103]) and secreted protein acidic and rich in cysteine (SPARC). It has been reported that SPARC accelerates disease progression in experimental crescentic glomerulonephritis [104]; and SPARC leads to a progressive reduction in podocyte number, thus fueling the future development of glomerulosclerosis [105]. In addition, two out of 71 senescence regulatory genes including cellular repressor of E1A stimulated genes 1 (CREG1) and insulin-like growth factor-binding protein 7 (IGFBP7) were increased in ESRD-upregulated SGs. Of note, CREG1 haploinsufficiency confers increased susceptibility of adipose tissue to inflammation, leading to aggravated obesity and insulin resistance when challenged with a high fat diet [106]. IGFBP7 is one of the growth factors upregulated in patients with inflammatory breast cancer [107]. Moreover, five out of 71 senescence regulators such as serpin family E member 1, (SERPINE1, plasminogen activator inhibitor 1, PAI-1), SPARC, transforming growth factor β1 (TGFβ1), insulin-like growth factor 1 (IGF1), and insulin-like growth factor-binding protein 5 (IGFBP5) were downregulated in ESRD-downregulated SGs. Of note, TGF-β1 is a key reactive oxygen species (ROS) promoting cytokine in renal fibrosis [108]. TGF-β1 promotes the cell cycle G2/M arrest based senescence-associated secretory phenotype (SASP) rather than DNA-damage based G1/S arrest [109]. TGFβ1-Smads form an anti-proliferation pathway [110]. Anti-aging gene Klotho deficiency exacerbates early diabetic nephropathy via enhancing TGFβ1 signaling in kidneys, which is a strong inducer of cellular senescence in a mouse model of chronic kidney injury [111]. Insulin-like grow factor 1 (IGF1) were increased in secretomic genes upregulated in ESRD. Circulating IGF-1 forms a complex with two other proteins – the IGF binding protein (IGFBP) and the acid labile subunit (ALS). High level concentrations of circulating IGF-1 are related to a higher risk of prostate, colorectal and breast cancers [112]. The results suggest that secretomic genes modulated in PBMCs from patients with ESRD modulate senescence via the following mechanisms: first, increasing expressions of proinflammatory growth factor IGFBP7; second, promoting inflammation and inhibiting fibrosis by decreasing TGF-β1 [113]; third, on the other hand, as potential negative feedback mechanisms, inhibiting inflammation and kidney injury by upregulating anti-inflammatory CREG1 and downregulating proinflammatory SPARC, respectively; fourth, inhibiting higher risk of prostate, colorectal and breast cancers by decreasing IGF1 and IGFBP5; and fifth, promoting senescence by decreasing leukocyte telomere length (LTL) [114] via inhibiting SERPINE1/PAI-1 expression [115]. Therefore, our data suggest that modulating secretomic gene expressions in PBMCs may have beneficial therapeutic effects in the treatment of ESRD-related cancer and aging-related diseases [116] (see Fig. 6).
Table 7.
Among all SGs we investigated, no senescence regulators out of 71 were upregulated in CKD; two out of 71 senescence regulator genes such as Inhibitor Of DNA Binding 1 (ID1) and secreted protein acidic and rich in cysteine (SPARC) were downregulated in CKD; two out of 71 senescence regulator genes such as Cellular Repressor Of E1A Stimulated Genes 1 (CREG1) and Insulin Like Growth Factor Binding Protein 7 (IGFBP7) were upregulated in ESRD while five were downregulated in ESRD such as Serpin Family E Member 1 (SERPINE1), SPARC, transforming growth factor b1 (TGFB1), insulin-like growth factor (IGF1) and Insulin Like Growth Factor Binding Protein 5 (IGFBP5).
| Group | Gene Symbol | p value | log FC | Function |
|---|---|---|---|---|
| Upregulated in CKD |
N/A |
|||
| Downregulated in CKD | ID1 | 0.042591 | −1.10654 | p53/pRb signaling & cell cycle |
| SPARC |
0.028006 |
−1.00435 |
p53/pRb signaling & cell cycle |
|
| Upregulated in ESRD | CREG1 | 7.63E-05 | 1.463896 | p53/pRb signaling & cell cycle |
| IGFBP7 |
5.81E-05 |
2.059804 |
Interferon Signaling; insulin growth factor related |
|
| Downregulated in ESRD | SERPINE1 | 0.0234 | −1.17965 | Other Senescence Response Gene |
| SPARC | 0.00428 | −1.93975 | p53/pRb signaling & cell cycle | |
| TGFB1 | 3.62E-06 | −2.48915 | p53/pRb signaling & cell cycle; Cell Adhesion Molecules | |
| IGF1 | 0.000375 | −2.37204 | insulin growth factor related | |
| IGFBP5 | 0.000673 | −1.7563 | insulin growth factor related |
Fig. 6.
Novel mechanism. Uremic toxins-promoted secretome accelerated renal disease and inflammation by inducing cellular senescence and senescence-associated secretory phenotype (SASP) according to p53 signaling and insulin growth factor (IGF) related pathways (PI3K/Akt) in ESRD.
3.9. Reactive oxygen species (ROS) pathways play much bigger roles in ESRD-upregulated SGs (11.6%) than that in CKD-upregulated SGs (6.8%), and half of ESRD-upregulated SGs are ROS-independent
It has been well documented that reactive oxygen species (ROS) plays a key role in regulating pathophysiological signaling in endothelial cell activation [117], cardiovascular diseases [118] and chronic kidney disease/end-stage renal disease [119]. We also reported that mitochondrial ROS plays a significant role in mediating EC activation [4,120,121]. Overproduction of ROS by impaired mitochondria can lead to positive feedback to enhance the cellular damage and generate uremic toxins, especially those produced by oxidation or peroxidation, such as creatine, urea and Melatonin [122,123]. This process aggravated by the accumulation of uremic toxins was hallmarked by mitochondria dysfunction defined as increased proton leaks as we reported [4,120,121,124,125], impaired mitochondria dynamics, alteration of mitochondria morphology and remodeling, which lead to dysfunction of podocytes and endothelial cells in the kidney. Mitochondria is both a source and a target for uremic toxins. In addition, it has been reported that uremia is associated with a reduction in the numbers and functions of lymphoid cells, whereas numbers of myeloid cells in uremic patients are either normal or increased with increased production of inflammatory cytokines and ROS [119]. Moreover, to find the evidence that ROS pathway genes are modulated by CKD and ESRD, the 84 oxidative and anti-oxidative regulatory genes [126] were examined. As shown in Table 8A, ESRD upregulated an antioxidant enzyme peroxiredoxin 4 (PRDX4) and a potential neuron development regulator prion protein (PRNP), and downregulated eight oxidative/anti-oxidative genes including antioxidant glutathione peroxidase 3 (GPX3), anti-oxidant glutathione peroxidase 5 (GPX5), antimicrobial lactoperoxidase (LPO), microbicidal myeloperoxidase (MPO), anti-oxidant superoxide dismutase 3 (SOD3), anti-inflammatory cytokine IL19, proinflammatory cytokine IL-22, anti-inflammatory apolipoprotein E (APOE) in PBMCs, respectively. These results suggest that ESRD downregulates more antioxidant enzymes/proteins than upregulate them, whereby promoting ROX generation. In addition, CKD downregulated one anti-oxidative gene SOD3. These results suggest that ESRD and CKD modulate ROS regulatome (oxidative and anti-oxidative regulatory genes).
Table 8a.
Oxidative stress-related gene expressions (reactive oxygen species, ROS, regulatome) contributes to the progression of kidney dysfunction. We analyzed the gene expression of total 84 ROS regulatom84e (shown in supplement table 11) in CKD and ESRD. We found that 2 out of total 121 (1.65%) secretomic genes in ESRD while none of SGs in CKD are upregulated. Meanwhile, 8 out of 928 (0.8%) in ESRD and 1 out of 55 (1.82%) in CKD are downregulated.
| Group | Gene Symbol | p value | log FC |
|---|---|---|---|
| Upregulated in CKD |
N/A |
||
| Downregulated in CKD |
SOD3 |
0.031602 |
−1.04179 |
| Upregulated in ESRD | PRDX4 | 3.25E-05 | 1.308793 |
| PRNP |
8.80E-08 |
1.915437 |
|
| Downregulated in ESRD | GPX3 | 4.56E-04 | −1.13831 |
| GPX5 | 0.0101 | −1.48694 | |
| LPO | 0.00185 | −1.8362 | |
| MPO | 4.01E-03 | −1.85159 | |
| SOD3 | 0.0214 | −1.23203 | |
| IL19 | 0.00647 | −1.56396 | |
| IL22 | 0.0124 | −1.3153 | |
| APOE | 4.61E-06 | −1.78856 |
However, an important question remains whether ROS signaling and antioxidant signaling mediate CKD and ESRD-modulation of SGs. Thus, we examined a novel hypothesis that ROS signaling and anti-oxidant signaling mediate CKD-, and ESRD-, modulation of SGs. By mining the microarray datasets in the NIH-NCBI-GeoDataset database, we found several microarray datasets with the inhibition of nicotinamide adenine dinucleotide phosphate oxidase 2 (NOX2) [118] and the deficiency of antioxidant transcription factor nuclear factor erythroid 2–related factor 2 (Nrf2) [118]. Nrf2 was chosen since Nrf2 was considered as a novel therapeutic target for mitochondria dysfunction in chronic kidney disease by animal experimental researches [121,[127], [128], [129], [130], [131], [132]]. In Fig. 7A, we classified all the SGs into four groups: first, the SGs increased in NOX2 inhibited cells and decreased in Nrf2 deficient cells are ROS-suppressed genes; second, the SGs expressions decreased in NOX2 inhibited cells and increased in Nrf2 deficient cells are ROS-promoted genes; third, the SGs either promoted by ROS pathways in one group of microarray datasets or suppressed by ROS pathways in another groups of microarray datasets are ROS pathways-unsettled genes; and fourth, the SGs that were not significantly modulated in NOX2 inhibited and Nrf2 deficient are ROS-independent genes (Fig. 7A). Of note, only 1030 out of 2641 SGs were found in the NOX2-, Nrf2-deficient datasets and were focused in our analyses. As shown in Table 8B, 144 out of 1030 SGs (14.0%) were ROS-suppressed SGs; 191 out of 1030 SGs (18.5%) were ROS-promoted SGs; 3 out of 1030 SGs (0.29%) were ROS-unsettled SGs; and 692 out of 1030 SGs (67.2%) were ROS-independent. In addition, 17 out of CKD-upregulated 44 SGs (38.6%) were found to be modulated in ROS-related manners including 5 as ROS-suppressed SGs (29.41%), 3 as ROS-promoted SGs (17.65%), 0 as ROS-unsettled SGs, and 9 as ROS-independent SGs (52.94%); moreover, 26 out of CKD-downregulated 55 SGs (47.3%) were found to be modulated in ROS-related manners including 2 as ROS-suppressed SGs (7.69%), 8 as ROS-promoted SGs (30.77%), 0 as ROS-unsettled SGs, and 16 as ROS-independent SGs (61.54%); furthermore, 84 out of ESRD-upregulated 121 SGs (69.4%) were found to be modulated in ROS-related manners including 6 as ROS-suppressed SGs (7.14%), 14 as ROS-promoted SGs (16.67%), 0 as ROS-unsettled SGs, and 64 as ROS-independent SGs (76.19%); and finally, 341 out of ESRD-downregulated 928 SGs (36.7%) were found to be modulated in ROS related manners including 59 as ROS-suppressed SGs (17.30%), 74 as ROS-promoted SGs (21.70%), 2 as ROS-unsettled SGs (0.59%), and 206 as ROS-independent SGs (60.41%). These results suggest that first, ROS play much bigger roles in ESRD-upregulated SGs (69.4%) than that in CKD-upregulated SGs (38.6%); second, ROS-independent ESRD-upregulated SGs (76.19%) are much more than ROS-independent CKD-upregulated SGs (52.94%), indicating that ESRD uses more ROS-independent mechanisms than CKD in upregulating SGs in the PBMCs; and third, ROS-suppressed CKD-upregulated SGs (29.41%) are much higher than that of ESRD-upregulated SGs (7.14%) (see Fig. 7b).
Fig. 7a.
All of the 2641 SGs were testified in NOX2 and Nrf2 knockout GEO datasets (GSE7810, GSE100671) and total of 1030 SGs (39.0% of all SGs) were found in these two datasets (cutoff: P value < 0.05). We classified these 1030 SGs into 4 categories: 1) ROS-suppressed SGs, 2) ROS-promoted SGs, 3) ROS-uncertain SGs, and 4) ROS-independent SGs by analyzing expression changes in NOX2 and Nrf2 knockout GEO.
Table 8b.
Using the classification in Fig. 7A, four SG subsets were compared based on the regulation of these CKD and ESRD modulated SGs by reactive oxidative species (ROS) generated by NOX2 and suppressed by NRF2. The results showed that 1) ROS-suppressed SGs (29.41%) upregulated in CKD were much higher than that of total SGs control (7.14%); 2) ROS-promoted SGs (30.77%) downregulated in CKD were much higher than that in total SGs control (18.54%); 3) ROS-independent SGs (76.19%) upregulated in ESRD were much higher than that in total SGs control and that in CKD (67.18% and 52.94%), respectively. These results suggest that ROS play much bigger roles in ESRD-upregulated SGs than that in other groups of SGs; and ROS play much more significant roles in ESRD pathologies.
| Group | classification | ROS-suppressed SGs | ROS-promoted SGs | ROS-uncertain SGs | ROS-independent SGs | all ROS-related genes |
|---|---|---|---|---|---|---|
| ROS-related SGs | Number | 144 | 191 | 3 | 692 | 1030 |
| Percentage |
13.98% |
18.54% |
0.29% |
67.18% |
100.00% |
|
| up in CKD | Number | 5 | 3 | 0 | 9 | 17 |
| Percentage |
29.41% |
17.65% |
0.00% |
52.94% |
100.00% |
|
| down in CKD | Number | 2 | 8 | 0 | 16 | 26 |
| Percentage |
7.69% |
30.77% |
0.00% |
61.54% |
100.00% |
|
| up in ESRD | Number | 6 | 14 | 0 | 64 | 84 |
| Percentage |
7.14% |
16.67% |
0.00% |
76.19% |
100.00% |
|
| down in ERSD | Number | 59 | 74 | 2 | 206 | 341 |
| Percentage | 17.30% | 21.70% | 0.59% | 60.41% | 100.00% |
Fig. 7b.
Novel Mechanism: Reactive oxygen species (ROS)-related mechanisms, regulated by ROS generating enzyme NADPH oxidase 2 (NOX2) and antioxidant transcription factor Nrf2 pathways, modulated the secretomic changes during kidney dysfunction. The modulation was closely related to the balance of ROS and antioxidants and the imbalance contributes to the alteration of ROS-dependent SGs which could promote disease progression.
4. Discussion
Since UTs have been first identified via low throughput mass spectrometry, 130 UTs are documented in the European Uremic Solutes (EUTox) Database (http://www.uremic-toxins.org/DataBase.html). It remains unknown whether secreted proteins generated in innate immune cells in response to UTs stimulation can contribute to the pathogenesis of CKD and ESRD. We recently reported that uremic toxins (UTs) are selectively increased and serve as danger signal-associated molecular patterns (DAMPs) and homeostasis-associated molecular patterns (HAMPs) that modulate inflammation. These results also show that some UT genes are upregulated in CKD and CAD via caspase-1/inflammatory cytokine pathways, rather than by purely passive accumulation. Our findings raised the possibility that UTs-stimulated innate immune cells and other types of cells increase their secretome, which contributes to the CKD/ESRD progression. As reported, the secretory proteins are important for maintaining cell-cell communication and proliferation. Examples of secretory proteins include hormones, digestive enzymes, cytokines, chemokines, interferons (IFNs), colony-stimulating factors (CSFs), growth factors, and tumor necrosis factors (TNFs) [133]. However, secretomic studies in CKD and other metabolic diseases have been at a low pace due to the low throughput technologies [134]. Our previous reports demonstrated that innate immune cells, PBMCs containing such as Ly6Chigh (mice) [7,17,18,[135], [136], [137]] and CD40+ (human) [7] monocytes, contribute significantly to the pathogenesis of metabolic vascular diseases including CKD [7]. However, three important questions remained, first, whether UTs are the only soluble molecular drivers for the progression of CKD to ESRD; second, whether secretomic changes in innate immune cells, PBMCs, upregulated in CKD and ESRD contribute to the pathogenesis and progression of CKD and ESRD; and third, whether CKD and ESRD differentially modulate the secretomic changes via disease stage-specific pathways. To fill in these important knowledge gaps, in this study, we used cutting-edge molecular database mining approaches that we pioneered in 2004 [25,113,138,139] and analyzed PBMC secretomic (all the signal peptide sequence-containing secreted protein genes) changes in induced by CKD and ESRD. Our data analyses have made for the first time the following significant findings: 1) 86.7% middle class (molecular weight >500 Daltons) uremic toxins (UTs) were encoded by secretomic genes (SGs); 2) Upregulations of SGs in PBMCs in patients with ESRD (121 SGs) were significantly higher than that of CKD (44 SGs); and among ESRD specifically modulated 975 SGs, ESRD upregulated 116 SGs (11.9%) but downregulates 859 SGs (88.1%), respectively; 3) ESRD-upregulated SGs had 2 folds higher percentages of the cytoplasm and nucleus subcellular groups than the controls; and had the higher percentages of five out of 13 SG functional groups including enzyme, kinase, peptide, transcription regulator, and transmembrane in comparison to the controls. Transcriptomic analyses of PBMC secretome have advantages to identify more comprehensive secretome than conventional secretomic analyses; 4) Although CKD-, and ESRD-upregulated SGs were highly diversified in signaling, ESRD-induced SGs had strong proinflammatory pathways; 5) Proinflammatory cytokines-based middle class UTs such as interleukin-1β (IL-1β) and IL-18 promote ESRD modulation of SGs; 6) ESRD-upregulated co-stimulation receptors CD48 and CD58 increase secretomic upregulation in the PBMCs; may not be even limited in the PBMCs, CD48/CD58-CD2 signaling can be actually magnified enormously in tissues; 7) Classically activated macrophages (M1)-, and alternatively activated macrophages (M2)- macrophage polarization signals contribute to ESRD- and CKD-upregulated SGs; 8) ESRD- and CKD-upregulated SGs in PBMCs contain senescence-promoting regulators by upregulating proinflammatory growth factor IGFBP7 and downregulating anti-inflammatory cytokine TGF-β1 and leukocyte telomere length stabilizer SERPINE1/PAI-1; and 9) Reactive oxygen species (ROS) pathways play much bigger roles in ESRD-upregulated SGs (11.6%) than that in CKD-upregulated SGs (6.8%); and half of ESRD-upregulated SGs are ROS-independent.
Clinical and epidemiological studies have identified more than 10 risk factors in accelerating CKD progression and potential transition to ESRD as shown in Table 9A. However, the molecular pathways driving the pathogenesis of ESRD remained poorly characterized. Based on our findings, we propose a new working model (Fig. 8), under the stimulation of uremic toxins, more than 121 novel secreted proteins are significantly upregulated in innate immune cells, PBMCs, in patients with ESRD, which makes PBMCs the major cell types in upregulating secretomes. This is the first time for us to understand that in addition to uremic toxins identified, significant secretomic changes may play highly significant roles in driving ESRD pathogenesis. In addition, since some PBMC secretome proteins identified with transcriptomic approach are localized in the subcellular locations other than the supernatants of cultured cells and plasma that conventional secretomic analyses sampled and examined, therefore, our data have also demonstrated that transcriptomic analyses of PBMC secretome have advantages to identify more comprehensive secretome than conventional secretomic analyses [69]. To determine the mechanisms underlying the SGs, we identified several novel molecular mechanisms: first, UTs play significant roles in upregulating PBMC SGs in patients with CKD and ESRD; and ESRD-induced SGs have strong proinflammatory pathways. In addition, our IPA results indicate that SGs in PBMCs upregulated in ESRD have a novel proinflammatory signaling pathway overlapped with that of UTs, role of cytokines in mediating communication between immune cells (Fig. 3A). We also used the Cytoscape and found that UTs pathways and ESRD-upregulated pathways share 20 regulatory or regulators. These results have demonstrated for the first time that ESRD-upregulated PBMC SGs have synergistic effects with that of UTs, which contribute to the disease progression significantly; second, we found strong functional evidence that proinflammatory cytokines-based middle class UTs such as interleukin-1β (IL-1β) and IL-18 promote ESRD modulation of SGs, which also serve as a new working model for UTs and PBMC secretome interactions in patients with ESRD and CKD; third, as a novel membrane protein mechanism, ESRD-upregulated co-stimulation receptors CD48 and CD58 increase secretomic upregulation in the PBMCs; may not be even limited in the PBMCs, CD48/CD58-CD2 signaling can be actually magnified enormously in tissues; fourth, as another novel membrane Toll-like receptor-mediated signaling and interferon-g receptor signaling mechanisms, we found that classically activated macrophages (M1)-, and alternatively activated macrophages (M2)- macrophage polarization signals contribute to ESRD- and CKD-upregulated SGs; fifth, ESRD- and CKD-upregulated SGs in PBMCs contain senescence-promoting regulators by upregulating proinflammatory growth factor IGFBP7 and downregulating anti-inflammatory cytokine TGF-β1 and leukocyte telomere length stabilizer SERPINE1/PAI-1. Therefore, our data have demonstrated for the first time that controlling senescence-associated inflammation cell by targeting specific inflammatory mediators may have a beneficial therapeutic effect in treatment of ESRD-related cancers, aging and inflammatory diseases [116]; and sixth, Reactive oxygen species (ROS) pathways play much bigger roles in ESRD-upregulated SGs (11.6%) than that in CKD-upregulated SGs (6.8%); and half of ESRD-upregulated SGs are ROS-independent.
Table 9a.
Multiple risk factors accelerate CKD progression to ESRD, which have been reported by clinical studies and scientific literature. Our report provides novel data analyses evidences and mechanisms to support the multi-hit model for the development of CKD.
| clinical risk factor for CKD progression | scientific mechanism | PMID |
|---|---|---|
| ethnicity | APOL1 mutant in African | 27312436 |
| Epigenetically heritable changes |
25993323 |
|
| gender | direct effects of sex steroids on kidney | 29355169 |
| sex differences in NO metabolism and oxidative stress | 29355169 | |
| gender-differential impact of comorbidities and lifestyle risk factors |
29355169 |
|
| diabetes | Renal hemodynamic changes | 29486908 |
| ischemia and inflammation | 29486908 | |
| Overactive renin-angiotensin-aldosterone system |
29486908 |
|
| hypertension | Global RAS activation | 29144825 |
| renal inflammation |
29144825 |
|
| obesity | RAAS activation | 31015582 |
| renal compression | 31015582 | |
| Metabolic abnormalities |
31015582 |
|
| gut microbiota | production of uremic toxins | 30464044 |
| promoting translocation of bacterial component | 31243394 | |
| secreting metabolites favoring insulin resistance favoring insulin resistance and endothelial dysfunction |
31243394 |
|
| nephrotoxicity | inducing apoptosis, autophagy and necrosis | 29341864 |
| Oxidant-Induced Renal Injury | 15519281 | |
| mitochondria dysfunction in the proximal tubules |
29939355 |
|
| Hepatitis B and C infections | glomerular immune complex deposition | 28149647 |
| direct viral invasion of the renal parenchyma |
31155101 |
|
| dyslipidemia | Direct lipid-induced cellular injury | 22290079 |
| inflammatory cytokines |
29176657 |
|
| proteinuria | Increased intraglomerular hydraulic pressure | 22137726 |
| damage to glomerular filtration barrier |
22137726 |
|
| mineral bone disorder | VSMC dedifferentiation | 28119179 |
| neointimal atherosclerotic calcification | 28119179 |
Fig. 8.
A: Our new finding suggested a new model that not only passive accumulation of uremic toxins, but also other active upregulation of secretome during ESRD contributes to disease progression through proinflammatory and profibrotic pathways and molecules. B: This active accumulation modulated by both ROS-dependent and –independent pathways could promote systemic inflammation and fibrosis to accelerate disease progression. Uremic toxin-related cytokine switching, macrophage polarization and co-signaling by the interaction of CD48 and CD58 with CD2 were important pathways associated with ROS-independent SGs. Of note, there has been researches identified those pathways could modulate and interact with ROS-dependent pathway. C: The active accumulation of secretomic changes are the key mediators when combined with and modulated by other risk factors as multiple hits in the transition from CKD to ESRD. D: Ranking of all the mechanisms in our research by the numbers of SGs upregulated in each entry suggested that ROS serves as an important complementary role for prior knowledge of CKD progression, multiple-hit model of CKD progression.
Of note, the interactions of the pathways in our findings have been reported by other researches. These interactions may collectively lead to disease progression: First, cytokines exerted as key initiators, mediators and effectors for other pathway entries. Upregulated co-stimulating secretomic member CD48 can be induced under proinflammatory environment; secretion of IL2 was decreased in T cells isolated from CD48-deficient mice [140]. Second, co-signaling molecule CD48 and CD58 are widely reported to be closely related to cytokine synthesis: CD48−CD2 interaction can facilitate TCR signaling to promote production of pro-inflammatory cytokines such as IL2 [141]. Third, macrophages as a crucial component of innate immunity, can function as scavengers to clear the interstitial environment of extraneous cellular materials and also as antigen-presenting cells to stimulate adaptive immune response. Based on functionality, resident and infiltrating macrophages can produce a set of pro-inflammatory cytokines and other metabolites under the stimuli such as uremic toxins in ESRD and their phenotype can be reversely reprogrammed by the different subsets of cytokines and polarized into different subsets [[142], [143], [144]]; Fourth, senescence has been induced by signaling through a bevy of critical cytokines such as TNF-a, IFN-g as an important extrinsic pathway of senescence and those cytokines initiated an inflammatory network acted both cause and consequences during senescence [[145], [146], [147]]. Fifth, redox oxygen species has been widely reported to be a marker and an inducer to deteriorate diseases which could establish the inflammatory network [[148], [149], [150], [151], [152]]. This process could be carried out directly or indirectly. Direct pathways were according to the enhancing secretion of pro-inflammatory cytokines such as IFN‐γ, TNF‐α and IL‐1 by activating TCR and mTOR signaling while indirect pathways was activated by imbalance of M1-and M2-macrophages, SASP, and T cell signaling [[153], [154], [155], [156], [157], [158], [159], [160], [161], [162]]. In conclusion, these pathways cross talk directly or indirectly, and make ROS pathway as potential therapeutic targets to suppress disease progression.
One limitation of the current study is that due to the low throughput nature of verification techniques so that we could not verify every result we identified with the analyses of high throughput data (Table 10). We acknowledge that carefully designed in-vitro and in-vivo experimental models will be needed to verify the CKD-, and ESRD-upregulated PBMC secretomes further and underlying mechanisms we report here. Nevertheless, our findings provide novel insights on the roles of PBMC secretomes in the pathogenesis of ESRD and CKD, novel pathways underlying the multi-hit models as well as new targets for the future therapeutic interventions for CKD, ESRD and their related diseases, aging and cancers.
Table 10.
A novel research publication type with big-omics experimental database mining analyses leads to original new findings and generate anew hypotheses. A few aspects of comparisons were made within this study using big-omics experimental database mining approaches, the traditional literature reviews and the meta-analysis.
| category | Big-omics Database mining | Traditional literature review | meta analysis |
|---|---|---|---|
| Analysis of experimental Data (NIH-Geo-DataSets with microarray experimental data, etc.) | yes | no | yes |
| Original new Findings | yes | no | no |
| Association research (gene co-expression patterns at the same pathology or stimuli) | yes | no | yes |
| Causative research (upstream regulator gene deficient microarrays, …) | yes | no | no |
| Panoramic view at multiple mechanisms and pathways | yes | yes | yes |
| Improvement of our understanding | yes | yes | yes |
| Searchable Database requirements and tools | yes | no | yes |
| New publication types after –omics and high throughput experimental data generation | yes | no | yes |
| Different focuses from original papers | yes | no | no |
| Use of Ingenuity Pathway Analysis (IPA) to analyze experimental data | yes | no | no |
| Bioinformatic prediction | no | no | no |
| Future experimental verification | yes | yes | yes |
| Summary of previous reports | no | yes | yes |
| Example | PMID: 22438968 [our datamining paper focusing on IL-35 (highly cited by 173 papers) ] | PMID: 24060958 [a Nature Review paper of management of hypertriglyceridaemia] | PMID: 23083786 [a meta-analysis paper focusing on Effects of fibrates in kidney disease] |
| experimental papers verifying the findings originated from example paper | PMIDs: 26085094; 29371247 | N/A | PMID: 25419705 |
| Use of multiple NIH databases including PubMed database (https://www.ncbi.nlm.nih.gov/books/NBK143764/) yes | yes | no | no |
Authors’ contributions
RJZ carried out the data gathering, data analysis and prepared tables and figures. JS, YS, TY, LL, FS, WYY, YS, CJ, CDIV, HF, YL, KX, ML, JW, EC, DY, XJ, YL, RL, LW, ETC, HW aided with analysis of the data. XFY supervised the experimental design, data analysis, and manuscript writing. All authors read and approved the final manuscript.
Funding
This work was supported by the hospital fellowship to RJ Zhang.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by NIH RO1 HL131460.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101460.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Centers for Disease Control and Prevention . US Department of Health and Human Services, Centers for Disease Control and Prevention; Atlanta, GA: 2019. Chronic Kidney Disease in the United States, 2019.https://www.cdc.gov/kidneydisease/publications-resources/2019-national-facts.html [Google Scholar]
- 2.Reikes S.T. Trends in end-stage renal disease. Epidemiology, morbidity, and mortality. PGM (Postgrad. Med.) 2000;108(1):124–126. doi: 10.3810/pgm.2000.07.1152. 9-31, 35-6 passim. [DOI] [PubMed] [Google Scholar]
- 3.Yin Y., Li X., Sha X., Xi H., Li Y.F., Shao Y. Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arterioscler. Thromb. Vasc. Biol. 2015;35(4):804–816. doi: 10.1161/ATVBAHA.115.305282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Fang P., Li Y., Kuo Y.M., Andrews A.J., Nanayakkara G. Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arterioscler. Thromb. Vasc. Biol. 2016;36(6):1090–1100. doi: 10.1161/ATVBAHA.115.306964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang P., Zhang D., Cheng Z., Yan C., Jiang X., Kruger W.D. Hyperhomocysteinemia potentiates hyperglycemia-induced inflammatory monocyte differentiation and atherosclerosis. Diabetes. 2014;63(12):4275–4290. doi: 10.2337/db14-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi H., Zhang Y., Xu Y., Yang W.Y., Jiang X., Sha X. Caspase-1 inflammasome activation mediates homocysteine-induced pyrop-apoptosis in endothelial cells. Circ. Res. 2016;118(10):1525–1539. doi: 10.1161/CIRCRESAHA.116.308501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., Fang P., Yu D., Zhang L., Zhang D., Jiang X. Chronic kidney disease induces inflammatory CD40+ monocyte differentiation via homocysteine elevation and DNA hypomethylation. Circ. Res. 2016;119(11):1226–1241. doi: 10.1161/CIRCRESAHA.116.308750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer L.M., Monroy A.M., Lopez-Pastrana J., Nanayakkara G., Cueto R., Li Y.F. Caspase-1 plays a critical role in accelerating chronic kidney disease-promoted neointimal hyperplasia in the carotid artery. J Cardiovasc Transl Res. 2016;9(2):135–144. doi: 10.1007/s12265-016-9683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monroy M.A., Fang J., Li S., Ferrer L., Birkenbach M.P., Lee I.J. Chronic kidney disease alters vascular smooth muscle cell phenotype. Front Biosci (Landmark Ed) 2015;20:784–795. doi: 10.2741/4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y., Johnson C., Zhou J., Wang L., Li Y.F., Lu Y. Uremic toxins are conditional danger- or homeostasis-associated molecular patterns. Front Biosci (Landmark Ed) 2018;23:348–387. doi: 10.2741/4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sha X., Meng S., Li X., Xi H., Maddaloni M., Pascual D.W. Interleukin-35 inhibits endothelial cell activation by suppressing MAPK-AP-1 pathway. J. Biol. Chem. 2015;290(31):19307–19318. doi: 10.1074/jbc.M115.663286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao Y., Cheng Z., Li X., Chernaya V., Wang H., Yang X.F. Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction--a novel mechanism for maintaining vascular function. J. Hematol. Oncol. 2014;7:80. doi: 10.1186/s13045-014-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Wang L., Fang P., Sun Y., Jiang X., Wang H. Lysophospholipids induce innate immune transdifferentiation of endothelial cells, resulting in prolonged endothelial activation. J. Biol. Chem. 2018 doi: 10.1074/jbc.RA118.002752. jbc.RA118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li A., Sun Y., Drummer Ct, Lu Y., Yu D., Zhou Y. Increasing upstream chromatin long-range interactions may favor induction of circular RNAs in LysoPC-activated human aortic endothelial cells. Front. Physiol. 2019;10:433. doi: 10.3389/fphys.2019.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Pastrana J., Ferrer L.M., Li Y.F., Xiong X., Xi H., Cueto R. Inhibition of caspase-1 activation in endothelial cells improves angiogenesis: a novel therapeutic potential for ischemia. J. Biol. Chem. 2015;290(28):17485–17494. doi: 10.1074/jbc.M115.641191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C.E., Wei W., Liu Y.H., Peng J.H., Tian Q., Liu G.P. Hyperhomocysteinemia increases beta-amyloid by enhancing expression of gamma-secretase and phosphorylation of amyloid precursor protein in rat brain. Am. J. Pathol. 2009;174(4):1481–1491. doi: 10.2353/ajpath.2009.081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D., Fang P., Jiang X., Nelson J., Moore J.K., Kruger W.D. Severe hyperhomocysteinemia promotes bone marrow-derived and resident inflammatory monocyte differentiation and atherosclerosis in LDLr/CBS-deficient mice. Circ. Res. 2012;111(1):37–49. doi: 10.1161/CIRCRESAHA.112.269472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang P., Li X., Shan H., Saredy J.J., Cueto R., Xia J. Ly6C(+) inflammatory monocyte differentiation partially mediates hyperhomocysteinemia-induced vascular dysfunction in type 2 diabetic db/db mice. Arterioscler. Thromb. Vasc. Biol. 2019;39(10):2097–2119. doi: 10.1161/ATVBAHA.119.313138. ATVBAHA119313138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Z., Song J., Yan Y., Huang Y., Cowan A., Wang H. Higher expression of Bax in regulatory T cells increases vascular inflammation. Front. Biosci. 2008;13:7143–7155. doi: 10.2741/3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Z., Yan Y., Song J., Fang P., Yin Y., Yang Y. Expression of TCTP antisense in CD25(high) regulatory T cells aggravates cuff-injured vascular inflammation. Atherosclerosis. 2009;203(2):401–408. doi: 10.1016/j.atherosclerosis.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W.Y., Shao Y., Lopez-Pastrana J., Mai J., Wang H., Yang X.F. Pathological conditions re-shape physiological Tregs into pathological Tregs. Burns Trauma. 2015;3(1) doi: 10.1186/s41038-015-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K., Yang W.Y., Nanayakkara G.K., Shao Y., Yang F., Hu W. gaTa 3, hDac6, and Bcl 6 regulate FOXP3+ Treg Plasticity and Determine Treg conversion into either novel antigen-Presenting cell-like Treg or Th1-Treg. Front. Immunol. 2018;9:45. doi: 10.3389/fimmu.2018.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson J., Wu Y., Jiang X., Berretta R., Houser S., Choi E. Hyperhomocysteinemia suppresses bone marrow CD34+/VEGF receptor 2+ cells and inhibits progenitor cell mobilization and homing to injured vasculature-a role of beta1-integrin in progenitor cell migration and adhesion. Faseb. J. : official publication of the Federation of American Societies for Experimental Biology. 2015;29(7):3085–3099. doi: 10.1096/fj.14-267989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y.F., Huang X., Li X., Gong R., Yin Y., Nelson J. Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Front Biosci (Landmark Ed) 2016;21:178–191. doi: 10.2741/4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao Y., Chernaya V., Johnson C., Yang W.Y., Cueto R., Sha X. Metabolic diseases downregulate the majority of histone modification enzymes, making a few upregulated enzymes novel therapeutic Targets-"Sand out and gold stays. Journal of cardiovascular translational research. 2016;9(1):49–66. doi: 10.1007/s12265-015-9664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y., Sun Y., Drummer Ct, Nanayakkara G.K., Shao Y., Saaoud F. Increased acetylation of H3K14 in the genomic regions that encode trained immunity enzymes in lysophosphatidylcholine-activated human aortic endothelial cells - novel qualification markers for chronic disease risk factors and conditional DAMPs. Redox Biol. 2019;24:101221. doi: 10.1016/j.redox.2019.101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahn M.R., Robbins M.J., Kim M.C., Fuster V. Management of cardiovascular disease in patients with kidney disease. Nat. Rev. Cardiol. 2013;10(5):261–273. doi: 10.1038/nrcardio.2013.15. [DOI] [PubMed] [Google Scholar]
- 28.Schiffrin E.L., Lipman M.L., Mann J.F. Chronic kidney disease: effects on the cardiovascular system. Circulation. 2007;116(1):85–97. doi: 10.1161/CIRCULATIONAHA.106.678342. [DOI] [PubMed] [Google Scholar]
- 29.Moe S.M., Chen N.X. Pathophysiology of vascular calcification in chronic kidney disease. Circ. Res. 2004;95(6):560–567. doi: 10.1161/01.RES.0000141775.67189.98. [DOI] [PubMed] [Google Scholar]
- 30.Mestas J., Ley K. Monocyte-endothelial cell interactions in the development of atherosclerosis. Trends Cardiovasc. Med. 2008;18(6):228–232. doi: 10.1016/j.tcm.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moradi H., Sica D.A., Kalantar-Zadeh K. Cardiovascular burden associated with uremic toxins in patients with chronic kidney disease. Am. J. Nephrol. 2013;38(2):136–148. doi: 10.1159/000351758. [DOI] [PubMed] [Google Scholar]
- 32.Shao Y., Cheng Z., Li X., Chernaya V., Wang H., Yang X-f. Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction-a novel mechanism for maintaining vascular function. J. Hematol. Oncol. 2014;7(1):80. doi: 10.1186/s13045-014-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Li Y.F., Nanayakkara G., Shao Y., Liang B., Cole L. Lysophospholipid receptors, as novel conditional danger receptors and homeostatic receptors modulate inflammation-novel paradigm and therapeutic potential. J Cardiovasc Transl Res. 2016;9(4):343–359. doi: 10.1007/s12265-016-9700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apostolov E.O., Basnakian A.G., Ok E., Shah S.V. Carbamylated low-density lipoprotein: nontraditional risk factor for cardiovascular events in patients with chronic kidney disease. J. Ren. Nutr. : the official journal of the Council on Renal Nutrition of the National Kidney Foundation. 2012;22(1):134–138. doi: 10.1053/j.jrn.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Speer T., Owala F.O., Holy E.W., Zewinger S., Frenzel F.L., Stahli B.E. Carbamylated low-density lipoprotein induces endothelial dysfunction. Eur. Heart J. 2014;35(43):3021–3032. doi: 10.1093/eurheartj/ehu111. [DOI] [PubMed] [Google Scholar]
- 36.Mehta J.L., Basnakian A.G. Interaction of carbamylated LDL with LOX-1 in the induction of endothelial dysfunction and atherosclerosis. Eur. Heart J. 2014;35(43):2996–2997. doi: 10.1093/eurheartj/ehu122. [DOI] [PubMed] [Google Scholar]
- 37.Farhan H., Rabouille C. Signalling to and from the secretory pathway. J. Cell Sci. 2011;124(Pt 2):171–180. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- 38.Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27(3):230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Planavila A., Fernandez-Sola J., Villarroya F. Cardiokines as modulators of stress-induced cardiac disorders. Adv Protein Chem Struct Biol. 2017;108:227–256. doi: 10.1016/bs.apcsb.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Lipphardt M., Song J.W., Matsumoto K., Dadafarin S., Dihazi H., Muller G. The third path of tubulointerstitial fibrosis: aberrant endothelial secretome. Kidney Int. 2017;92(3):558–568. doi: 10.1016/j.kint.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makridakis M., Roubelakis M.G., Vlahou A. Stem cells: insights into the secretome. Biochim. Biophys. Acta. 2013;1834(11):2380–2384. doi: 10.1016/j.bbapap.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 42.Heine G.H., Ortiz A., Massy Z.A., Lindholm B., Wiecek A., Martinez-Castelao A. Monocyte subpopulations and cardiovascular risk in chronic kidney disease. Nat. Rev. Nephrol. 2012;8(6):362–369. doi: 10.1038/nrneph.2012.41. [DOI] [PubMed] [Google Scholar]
- 43.Naicker S.D., Cormican S., Griffin T.P., Maretto S., Martin W.P., Ferguson J.P. Chronic kidney disease severity is associated with selective expansion of a distinctive intermediate monocyte subpopulation. Front. Immunol. 2018;9:2845. doi: 10.3389/fimmu.2018.02845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ha C.Y., Kim J.Y., Paik J.K., Kim O.Y., Paik Y.H., Lee E.J. The association of specific metabolites of lipid metabolism with markers of oxidative stress, inflammation and arterial stiffness in men with newly diagnosed type 2 diabetes. Clin. Endocrinol. 2012;76(5):674–682. doi: 10.1111/j.1365-2265.2011.04244.x. [DOI] [PubMed] [Google Scholar]
- 45.Waehre T., Damas J.K., Pedersen T.M., Gullestad L., Yndestad A., Andreassen A.K. Clopidogrel increases expression of chemokines in peripheral blood mononuclear cells in patients with coronary artery disease: results of a double-blind placebo-controlled study. J. Thromb. Haemostasis. 2006;4(10):2140–2147. doi: 10.1111/j.1538-7836.2006.02131.x. [DOI] [PubMed] [Google Scholar]
- 46.Kim M., Kim M., Han J.Y., Lee S.H., Jee S.H., Lee J.H. The metabolites in peripheral blood mononuclear cells showed greater differences between patients with impaired fasting glucose or type 2 diabetes and healthy controls than those in plasma. Diabetes Vasc. Dis. Res. 2017;14(2):130–138. doi: 10.1177/1479164116678157. [DOI] [PubMed] [Google Scholar]
- 47.Chen D., Gan H., Huang X., Shen Q., Du X., Tang W. Effects of peripheral blood mononuclear cells morphology on vascular calcification in uremic patients on maintenance hemodialysis. Ther. Apher. Dial. 2012;16(2):173–180. doi: 10.1111/j.1744-9987.2011.01044.x. [DOI] [PubMed] [Google Scholar]
- 48.Lorenzen J.M., David S., Richter A., de Groot K., Kielstein J.T., Haller H. TLR-4+ peripheral blood monocytes and cardiovascular events in patients with chronic kidney disease--a prospective follow-up study. Nephrol. Dial. Transplant. 2011;26(4):1421–1424. doi: 10.1093/ndt/gfq758. [DOI] [PubMed] [Google Scholar]
- 49.Provenzano M., Coppolino G., Faga T., Garofalo C., Serra R., Andreucci M. Epidemiology of cardiovascular risk in chronic kidney disease patients: the real silent killer. Rev. Cardiovasc. Med. 2019;20(4):209–220. doi: 10.31083/j.rcm.2019.04.548. [DOI] [PubMed] [Google Scholar]
- 50.Batal I., De Serres S.A., Mfarrej B.G., Grafals M., Pinkus G.S., Kalra A. Glomerular inflammation correlates with endothelial injury and with IL-6 and IL-1beta secretion in the peripheral blood. Transplantation. 2014;97(10):1034–1042. doi: 10.1097/01.TP.0000441096.22471.36. [DOI] [PubMed] [Google Scholar]
- 51.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 52.Li Y.F., Nanayakkara G., Sun Y., Li X., Wang L., Cueto R. Analyses of caspase-1-regulated transcriptomes in various tissues lead to identification of novel IL-1beta-, IL-18- and sirtuin-1-independent pathways. J. Hematol. Oncol. 2017;10(1):40. doi: 10.1186/s13045-017-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 54.Wang L., Fu H., Nanayakkara G., Li Y., Shao Y., Johnson C. Novel extracellular and nuclear caspase-1 and inflammasomes propagate inflammation and regulate gene expression: a comprehensive database mining study. J. Hematol. Oncol. 2016;9(1):122. doi: 10.1186/s13045-016-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark W.R., Dehghani N.L., Narsimhan V., Ronco C. Uremic toxins and their relation to dialysis efficacy. Blood Purif. 2019;48(4):299–314. doi: 10.1159/000502331. [DOI] [PubMed] [Google Scholar]
- 56.Vanholder R., Boelaert J., Glorieux G., Eloot S. New methods and technologies for measuring uremic toxins and quantifying dialysis adequacy. Semin. Dial. 2015;28(2):114–124. doi: 10.1111/sdi.12331. [DOI] [PubMed] [Google Scholar]
- 57.Vanholder R.C., Eloot S., Glorieux G.L. Future avenues to decrease uremic toxin concentration. Am. J. Kidney Dis. 2016;67(4):664–676. doi: 10.1053/j.ajkd.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 58.Wang T., Cui Y., Jin J., Guo J., Wang G., Yin X. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res. 2013;41(9):4743–4754. doi: 10.1093/nar/gkt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanholder R., De Smet R., Glorieux G., Argiles A., Baurmeister U., Brunet P. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 60.Meert N., Schepers E., De Smet R., Argiles A., Cohen G., Deppisch R. Inconsistency of reported uremic toxin concentrations. Artif. Organs. 2007;31(8):600–611. doi: 10.1111/j.1525-1594.2007.00434.x. [DOI] [PubMed] [Google Scholar]
- 61.Duranton F., Cohen G., De Smet R., Rodriguez M., Jankowski J., Vanholder R. Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 2012;23(7):1258–1270. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feve B., Bastard C., Fellahi S., Bastard J.P., Capeau J. New adipokines. Ann. Endocrinol. 2016;77(1):49–56. doi: 10.1016/j.ando.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 63.Thiagarajan S., Neurath M.F., Hildner K. Resolution of acute intestinal graft-versus-host disease. Semin. Immunopathol. 2019;41(6):655–664. doi: 10.1007/s00281-019-00769-w. [DOI] [PubMed] [Google Scholar]
- 64.Pelham C.J., Agrawal D.K. Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expet Rev. Clin. Immunol. 2014;10(2):243–256. doi: 10.1586/1744666X.2014.866519. [DOI] [PubMed] [Google Scholar]
- 65.Schultheiss H.P., Fairweather D., Caforio A.L.P., Escher F., Hershberger R.E., Lipshultz S.E. Dilated cardiomyopathy. Nat Rev Dis Primers. 2019;5(1):32. doi: 10.1038/s41572-019-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Granata S., Zaza G., Simone S., Villani G., Latorre D., Pontrelli P. Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genom. 2009;10:388. doi: 10.1186/1471-2164-10-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaza G., Masola V., Granata S., Pontrelli P., Sallustio F., Gesualdo L. Dialysis-related transcriptomic profiling: the pivotal role of heparanase. Exp. Biol. Med. 2014;239(1):52–64. doi: 10.1177/1535370213506678. [DOI] [PubMed] [Google Scholar]
- 68.Zaza G., Granata S., Rascio F., Pontrelli P., Dell'Oglio M.P., Cox S.N. A specific immune transcriptomic profile discriminates chronic kidney disease patients in predialysis from hemodialyzed patients. BMC Med. Genom. 2013;6:17. doi: 10.1186/1755-8794-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anjo S.I., Manadas B. A translational view of cells' secretome analysis - from untargeted proteomics to potential circulating biomarkers. Biochimie. 2018;155:37–49. doi: 10.1016/j.biochi.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Santos N.B.D., Franco R.D., Camarini R., Munhoz C.D., Eichler R.A.S., Gewehr M.C.F. Thimet oligopeptidase (EC 3.4.24.15) key functions suggested by knockout mice phenotype characterization. Biomolecules. 2019;9(8) doi: 10.3390/biom9080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scuruchi M., Poti F., Rodriguez-Carrio J., Campo G.M., Mandraffino G. Biglycan and atherosclerosis: lessons from high cardiovascular risk conditions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019:158545. doi: 10.1016/j.bbalip.2019.158545. [DOI] [PubMed] [Google Scholar]
- 72.Chang J.T., Nevins J.R. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22(23):2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 73.Bhattacharyya S., Wu M., Fang F., Tourtellotte W., Feghali-Bostwick C., Varga J. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol. 2011;30(4):235–242. doi: 10.1016/j.matbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huggins C.J., Mayekar M.K., Martin N., Saylor K.L., Gonit M., Jailwala P. C/EBPgamma is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol. Cell Biol. 2015;36(5):693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forand A., Koumakis E., Rousseau A., Sassier Y., Journe C., Merlin J.F. Disruption of the phosphate transporter Pit1 in hepatocytes improves glucose metabolism and insulin signaling by modulating the USP7/IRS1 interaction. Cell Rep. 2016;17(7):1905. doi: 10.1016/j.celrep.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 76.Serre K., Mohr E., Benezech C., Bird R., Khan M., Caamano J.H. Selective effects of NF-kappaB1 deficiency in CD4(+) T cells on Th2 and TFh induction by alum-precipitated protein vaccines. Eur. J. Immunol. 2011;41(6):1573–1582. doi: 10.1002/eji.201041126. [DOI] [PubMed] [Google Scholar]
- 77.Sucajtys-Szulc E., Debska-Slizien A., Rutkowski B., Milczarek R., Pelikant-Malecka I., Sledzinski T. Hepatocyte nuclear factors as possible C-reactive protein transcriptional inducer in the liver and white adipose tissue of rats with experimental chronic renal failure. Mol. Cell. Biochem. 2018;446(1–2):11–23. doi: 10.1007/s11010-018-3268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bilchick K., Kothari H., Narayan A., Garmey J., Omar A., Capaldo B. Cardiac resynchronization therapy reduces expression of inflammation-promoting genes related to interleukin-1beta in heart failure. Cardiovasc. Res. 2019;pii: cvz232 doi: 10.1093/cvr/cvz232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan J., Zhang C., Chen Q., Zhou J., Franc J.L., Chen Q. Genomic analyses identify agents regulating somatotroph and lactotroph functions. Funct. Integr. Genom. 2016;16(6):693–704. doi: 10.1007/s10142-016-0518-8. [DOI] [PubMed] [Google Scholar]
- 80.Lee B.C., Kim M.S., Pae M., Yamamoto Y., Eberle D., Shimada T. Adipose natural killer cells regulate adipose tissue macrophages to promote insulin resistance in obesity. Cell Metabol. 2016;23(4):685–698. doi: 10.1016/j.cmet.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baldelli S., Aquilano K., Ciriolo M.R. Punctum on two different transcription factors regulated by PGC-1 alpha: nuclear factor erythroid-derived 2-like 2 and nuclear respiratory factor 2. Biochim. Biophys. Acta. 2013;1830(8):4137–4146. doi: 10.1016/j.bbagen.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 82.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lotia S., Montojo J., Dong Y., Bader G.D., Pico A.R. Cytoscape app store. Bioinformatics. 2013;29(10):1350–1351. doi: 10.1093/bioinformatics/btt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noels H., Weber C., Koenen R.R. Chemokines as therapeutic targets in cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 2019;39(4):583–592. doi: 10.1161/ATVBAHA.118.312037. [DOI] [PubMed] [Google Scholar]
- 86.Sozzani S., Del Prete A., Bonecchi R., Locati M. Chemokines as effector and target molecules in vascular biology. Cardiovasc. Res. 2015;107(3):364–372. doi: 10.1093/cvr/cvv150. [DOI] [PubMed] [Google Scholar]
- 87.Moreno J.A., Moreno S., Rubio-Navarro A., Gomez-Guerrero C., Ortiz A., Egido J. Role of chemokines in proteinuric kidney disorders. Expet Rev. Mol. Med. 2014;16:e3. doi: 10.1017/erm.2014.3. [DOI] [PubMed] [Google Scholar]
- 88.Lam C.W. 2. Inflammation, cytokines and chemokines in chronic kidney disease. EJIFCC. 2009;20(1):12–20. [PMC free article] [PubMed] [Google Scholar]
- 89.Chung A.C., Lan H.Y. Chemokines in renal injury. J. Am. Soc. Nephrol. 2011;22(5):802–809. doi: 10.1681/ASN.2010050510. [DOI] [PubMed] [Google Scholar]
- 90.Neirynck N., Vanholder R., Schepers E., Eloot S., Pletinck A., Glorieux G. An update on uremic toxins. Int. Urol. Nephrol. 2013;45(1):139–150. doi: 10.1007/s11255-012-0258-1. [DOI] [PubMed] [Google Scholar]
- 91.Simons K.H., de Jong A., Jukema J.W., de Vries M.R., Arens R., Quax P.H.A. T cell co-stimulation and co-inhibition in cardiovascular disease: a double-edged sword. Nat. Rev. Cardiol. 2019;16(6):325–343. doi: 10.1038/s41569-019-0164-7. [DOI] [PubMed] [Google Scholar]
- 92.Shen H., Wu N., Nanayakkara G., Fu H., Yang Q., Yang W.Y. Co-signaling receptors regulate T-cell plasticity and immune tolerance. Front Biosci (Landmark Ed) 2019;24:96–132. doi: 10.2741/4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kleiveland C.R. Peripheral blood mononuclear cells. In: Verhoeckx K., Cotter P., Lopez-Exposito I., Kleiveland C., Lea T., Mackie A., editors. The Impact of Food Bioactives on Health: in Vitro and Ex Vivo Models. CH); Cham: 2015. pp. 161–167. [PubMed] [Google Scholar]
- 94.Lai B., Wang J., Fagenson A., Sun Y., Saredy J., Lu Y. Twenty novel disease group-specific and 12 new shared macrophage pathways in eight groups of 34 diseases including 24 inflammatory organ diseases and 10 types of tumors. Front. Immunol. 2019;10(2612) doi: 10.3389/fimmu.2019.02612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan B.C.L., Lam C.W.K., Tam L.S., Wong C.K. IL33: roles in allergic inflammation and therapeutic perspectives. Front. Immunol. 2019;10:11. doi: 10.3389/fimmu.2019.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang P.M., Nikolic-Paterson D.J., Lan H.Y. Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 2019;15(3):144–158. doi: 10.1038/s41581-019-0110-2. [DOI] [PubMed] [Google Scholar]
- 97.Murray P.J. Macrophage polarization. Annu. Rev. Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 98.Sturmlechner I., Durik M., Sieben C.J., Baker D.J., van Deursen J.M. Cellular senescence in renal ageing and disease. Nat. Rev. Nephrol. 2017;13(2):77–89. doi: 10.1038/nrneph.2016.183. [DOI] [PubMed] [Google Scholar]
- 99.Ritschka B., Storer M., Mas A., Heinzmann F., Ortells M.C., Morton J.P. The senescence-associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes Dev. 2017;31(2):172–183. doi: 10.1101/gad.290635.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Docherty M.H., O'Sullivan E.D., Bonventre J.V., Ferenbach D.A. Cellular senescence in the kidney. J. Am. Soc. Nephrol. 2019;30(5):726–736. doi: 10.1681/ASN.2018121251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sikora E., Bielak-Zmijewska A., Mosieniak G. Cellular senescence in ageing, age-related disease and longevity. Curr. Vasc. Pharmacol. 2014;12(5):698–706. doi: 10.2174/1570161111666131219094045. [DOI] [PubMed] [Google Scholar]
- 102.Carnero A. Markers of cellular senescence. Methods Mol. Biol. 2013;965:63–81. doi: 10.1007/978-1-62703-239-1_4. [DOI] [PubMed] [Google Scholar]
- 103.Vigolo E., Marko L., Hinze C., Muller D.N., Schmidt-Ullrich R., Schmidt-Ott K.M. Canonical BMP signaling in tubular cells mediates recovery after acute kidney injury. Kidney Int. 2019;95(1):108–122. doi: 10.1016/j.kint.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 104.Sussman A.N., Sun T., Krofft R.M., Durvasula R.V. SPARC accelerates disease progression in experimental crescentic glomerulonephritis. Am. J. Pathol. 2009;174(5):1827–1836. doi: 10.2353/ajpath.2009.080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Durvasula R.V., Shankland S.J. Mechanical strain increases SPARC levels in podocytes: implications for glomerulosclerosis. Am. J. Physiol. Ren. Physiol. 2005;289(3):F577–F584. doi: 10.1152/ajprenal.00393.2004. [DOI] [PubMed] [Google Scholar]
- 106.Tian X., Yan C., Liu M., Zhang Q., Liu D., Liu Y. CREG1 heterozygous mice are susceptible to high fat diet-induced obesity and insulin resistance. PloS One. 2017;12(5) doi: 10.1371/journal.pone.0176873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bieche I., Lerebours F., Tozlu S., Espie M., Marty M., Lidereau R. Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature. Clin. Canc. Res. 2004;10(20):6789–6795. doi: 10.1158/1078-0432.CCR-04-0306. [DOI] [PubMed] [Google Scholar]
- 108.Richter K., Konzack A., Pihlajaniemi T., Heljasvaara R., Kietzmann T. Redox-fibrosis: impact of TGFbeta1 on ROS generators, mediators and functional consequences. Redox Biol. 2015;6:344–352. doi: 10.1016/j.redox.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valentijn F.A., Falke L.L., Nguyen T.Q., Goldschmeding R. Cellular senescence in the aging and diseased kidney. J Cell Commun Signal. 2018;12(1):69–82. doi: 10.1007/s12079-017-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsuura I., Denissova N.G., Wang G., He D., Long J., Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 111.Tasanarong A., Kongkham S., Khositseth S. Dual inhibiting senescence and epithelial-to-mesenchymal transition by erythropoietin preserve tubular epithelial cell regeneration and ameliorate renal fibrosis in unilateral ureteral obstruction. BioMed Res. Int. 2013;2013:308130. doi: 10.1155/2013/308130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tracz A.F., Szczylik C., Porta C., Czarnecka A.M. Insulin-like growth factor-1 signaling in renal cell carcinoma. BMC Canc. 2016;16:453. doi: 10.1186/s12885-016-2437-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li X., Mai J., Virtue A., Yin Y., Gong R., Sha X. IL-35 is a novel responsive anti-inflammatory cytokine--a new system of categorizing anti-inflammatory cytokines. PloS One. 2012;7(3) doi: 10.1371/journal.pone.0033628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Peng H., Yeh F., Lin J., Best L.G., Cole S.A., Lee E.T. Plasminogen activator inhibitor-1 is associated with leukocyte telomere length in American Indians: findings from the Strong Heart Family Study. J. Thromb. Haemostasis. 2017;15(6):1078–1085. doi: 10.1111/jth.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Freeberg M.A.T., Easa A., Lillis J.A., Benoit D.S.W., van Wijnen A.J., Awad H.A. Transcriptomic analysis of cellular pathways in healing flexor tendons of plasminogen activator inhibitor 1 (PAI-1/Serpine1) null mice. J. Orthop. Res. 2019;38(1):43–58. doi: 10.1002/jor.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lasry A., Ben-Neriah Y. Senescence-associated inflammatory responses: aging and cancer perspectives. Trends Immunol. 2015;36(4):217–228. doi: 10.1016/j.it.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 117.Pattillo C.B., Pardue S., Shen X., Fang K., Langston W., Jourd'heuil D. ICAM-1 cytoplasmic tail regulates endothelial glutathione synthesis through a NOX4/PI3-kinase-dependent pathway. Free Radic. Biol. Med. 2010;49(6):1119–1128. doi: 10.1016/j.freeradbiomed.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ray P.D., Huang B.W., Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24(5):981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Betjes M.G. Immune cell dysfunction and inflammation in end-stage renal disease. Nat. Rev. Nephrol. 2013;9(5):255–265. doi: 10.1038/nrneph.2013.44. [DOI] [PubMed] [Google Scholar]
- 120.Li X., Shao Y., Sha X., Fang P., Kuo Y.M., Andrews A.J. IL-35 (Interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of H3K14 (histone 3 lysine 14) Arterioscler. Thromb. Vasc. Biol. 2018;38(3):599–609. doi: 10.1161/ATVBAHA.117.310626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li X., Fang P., Sun Y., Shao Y., Yang W.Y., Jiang X. Anti-inflammatory cytokines IL-35 and IL-10 block atherogenic lysophosphatidylcholine-induced, mitochondrial ROS-mediated innate immune activation, but spare innate immune memory signature in endothelial cells. Redox Biol. 2019;28:101373. doi: 10.1016/j.redox.2019.101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Popkov V.A., Silachev D.N., Zalevsky A.O., Zorov D.B., Plotnikov E.Y. Mitochondria as a source and a target for uremic toxins. Int. J. Mol. Sci. 2019;20(12) doi: 10.3390/ijms20123094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Galvan D.L., Green N.H., Danesh F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92(5):1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li X., Fang P., Yang W.Y., Chan K., Lavallee M., Xu K. Mitochondrial ROS, uncoupled from ATP synthesis, determine endothelial activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells. Can. J. Physiol. Pharmacol. 2017;95(3):247–252. doi: 10.1139/cjpp-2016-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li X., Wang L., Fang P., Sun Y., Jiang X., Wang H. Lysophospholipids induce innate immune transdifferentiation of endothelial cells, resulting in prolonged endothelial activation. J. Biol. Chem. 2018;293(28):11033–11045. doi: 10.1074/jbc.RA118.002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gornicka A., Morris-Stiff G., Thapaliya S., Papouchado B.G., Berk M., Feldstein A.E. Transcriptional profile of genes involved in oxidative stress and antioxidant defense in a dietary murine model of steatohepatitis. Antioxidants Redox Signal. 2011;15(2):437–445. doi: 10.1089/ars.2010.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang W., Wang Y., Long J., Wang J., Haudek S.B., Overbeek P. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metabol. 2012;15(2):186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nishikawa M., Ishimori N., Takada S., Saito A., Kadoguchi T., Furihata T. AST-120 ameliorates lowered exercise capacity and mitochondrial biogenesis in the skeletal muscle from mice with chronic kidney disease via reducing oxidative stress. Nephrol. Dial. Transplant. 2015;30(6):934–942. doi: 10.1093/ndt/gfv103. [DOI] [PubMed] [Google Scholar]
- 129.Kirkman D.L., Muth B.J., Ramick M.G., Townsend R.R., Edwards D.G. Role of mitochondria-derived reactive oxygen species in microvascular dysfunction in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2018;314(3):F423–F429. doi: 10.1152/ajprenal.00321.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duni A., Liakopoulos V., Roumeliotis S., Peschos D., Dounousi E. Oxidative stress in the pathogenesis and evolution of chronic kidney disease: untangling ariadne's thread. Int. J. Mol. Sci. 2019;20(15) doi: 10.3390/ijms20153711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ruiz S., Pergola P.E., Zager R.A., Vaziri N.D. Targeting the transcription factor Nrf2 to ameliorate oxidative stress and inflammation in chronic kidney disease. Kidney Int. 2013;83(6):1029–1041. doi: 10.1038/ki.2012.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Granata S., Dalla Gassa A., Tomei P., Lupo A., Zaza G. Mitochondria: a new therapeutic target in chronic kidney disease. Nutr. Metab. 2015;12:49. doi: 10.1186/s12986-015-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mukherjee P., Mani S. Methodologies to decipher the cell secretome. Biochim. Biophys. Acta. 2013;1834(11):2226–2232. doi: 10.1016/j.bbapap.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abbasi-Malati Z., Roushandeh A.M., Kuwahara Y., Roudkenar M.H. Mesenchymal stem cells on horizon: a new arsenal of therapeutic agents. Stem Cell Rev Rep. 2018;14(4):484–499. doi: 10.1007/s12015-018-9817-x. [DOI] [PubMed] [Google Scholar]
- 135.Zhang D., Jiang X., Fang P., Yan Y., Song J., Gupta S. Hyperhomocysteinemia promotes inflammatory monocyte generation and accelerates atherosclerosis in transgenic cystathionine beta-synthase-deficient mice. Circulation. 2009;120(19):1893–1902. doi: 10.1161/CIRCULATIONAHA.109.866889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fang P., Zhang D., Cheng Z., Yan C., Jiang X., Kruger W.D. Hyperhomocysteinemia potentiates hyperglycemia-induced inflammatory monocyte differentiation and atherosclerosis. Diabetes. 2014:DB_140809. doi: 10.2337/db14-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meng S., Ciment S., Jan M., Tran T., Pham H., Cueto R. Homocysteine induces inflammatory transcriptional signaling in monocytes. Front Biosci (Landmark Ed). 2013;18:685–695. doi: 10.2741/4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ng B., Yang F., Huston D.P., Yan Y., Yang Y., Xiong Z. Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J. Allergy Clin. Immunol. 2004;114(6):1463–1470. doi: 10.1016/j.jaci.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yin Y., Yan Y., Jiang X., Mai J., Chen N.C., Wang H. Inflammasomes are differentially expressed in cardiovascular and other tissues. Int. J. Immunopathol. Pharmacol. 2009;22(2):311–322. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Elishmereni M., Levi-Schaffer F. CD48: a co-stimulatory receptor of immunity. Int. J. Biochem. Cell Biol. 2011;43(1):25–28. doi: 10.1016/j.biocel.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 141.McArdel S.L., Terhorst C., Sharpe A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016;164:10–20. doi: 10.1016/j.clim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cao Q., Harris D.C., Wang Y. Macrophages in kidney injury, inflammation, and fibrosis. Physiology. 2015;30(3):183–194. doi: 10.1152/physiol.00046.2014. [DOI] [PubMed] [Google Scholar]
- 143.Guiteras R., Flaquer M., Cruzado J.M. Macrophage in chronic kidney disease. Clin Kidney J. 2016;9(6):765–771. doi: 10.1093/ckj/sfw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Curi R., de Siqueira Mendes R., de Campos Crispin L.A., Norata G.D., Sampaio S.C., Newsholme P. A past and present overview of macrophage metabolism and functional outcomes. Clin Sci (Lond). 2017;131(12):1329–1342. doi: 10.1042/CS20170220. [DOI] [PubMed] [Google Scholar]
- 145.Li P., Gan Y., Xu Y., Song L., Wang L., Ouyang B. The inflammatory cytokine TNF-alpha promotes the premature senescence of rat nucleus pulposus cells via the PI3K/Akt signaling pathway. Sci. Rep. 2017;7:42938. doi: 10.1038/srep42938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wieder T., Brenner E., Braumuller H., Bischof O., Rocken M. Cytokine-induced senescence for cancer surveillance. Canc. Metastasis Rev. 2017;36(2):357–365. doi: 10.1007/s10555-017-9667-z. [DOI] [PubMed] [Google Scholar]
- 147.Freund A., Orjalo A.V., Desprez P.Y., Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med. 2010;16(5):238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lavieri R., Rubartelli A., Carta S. Redox stress unbalances the inflammatory cytokine network: role in autoinflammatory patients and healthy subjects. J. Leukoc. Biol. 2016;99(1):79–86. doi: 10.1189/jlb.3MR0415-159R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Voltan R., Secchiero P., Casciano F., Milani D., Zauli G., Tisato V. Redox signaling and oxidative stress: cross talk with TNF-related apoptosis inducing ligand activity. Int. J. Biochem. Cell Biol. 2016;81(Pt B):364–374. doi: 10.1016/j.biocel.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 150.Varga G., Gattorno M., Foell D., Rubartelli A. Redox distress and genetic defects conspire in systemic autoinflammatory diseases. Nat. Rev. Rheumatol. 2015;11(11):670–680. doi: 10.1038/nrrheum.2015.105. [DOI] [PubMed] [Google Scholar]
- 151.Sengupta M., Pal R., Nath A., Chakraborty B., Singh L.M., Das B. Anticancer efficacy of noble metal nanoparticles relies on reprogramming tumor-associated macrophages through redox pathways and pro-inflammatory cytokine cascades. Cell. Mol. Immunol. 2018;15(12):1088–1090. doi: 10.1038/s41423-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gorczynski R.M., Alexander C., Brandenburg K., Chen Z., Heini A., Neumann D. An altered REDOX environment, assisted by over-expression of fetal hemoglobins, protects from inflammatory colitis and reduces inflammatory cytokine expression. Int. Immunopharm. 2017;50:69–76. doi: 10.1016/j.intimp.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 153.Padgett L.E., Broniowska K.A., Hansen P.A., Corbett J.A., Tse H.M. The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann. N. Y. Acad. Sci. 2013;1281:16–35. doi: 10.1111/j.1749-6632.2012.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Diebold L., Chandel N.S. Mitochondrial ROS regulation of proliferating cells. Free Radic. Biol. Med. 2016;100:86–93. doi: 10.1016/j.freeradbiomed.2016.04.198. [DOI] [PubMed] [Google Scholar]
- 155.Belikov A.V., Schraven B., Simeoni L. T cells and reactive oxygen species. J. Biomed. Sci. 2015;22:85. doi: 10.1186/s12929-015-0194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tang H., Li J., Liu X., Wang G., Luo M., Deng H. Down-regulation of HSP60 suppresses the proliferation of glioblastoma cells via the ROS/AMPK/mTOR pathway. Sci. Rep. 2016;6:28388. doi: 10.1038/srep28388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim J.H., Chu S.C., Gramlich J.L., Pride Y.B., Babendreier E., Chauhan D. Activation of the PI3K/mTOR pathway by BCR-ABL contributes to increased production of reactive oxygen species. Blood. 2005;105(4):1717–1723. doi: 10.1182/blood-2004-03-0849. [DOI] [PubMed] [Google Scholar]
- 158.Malaquin N., Martinez A., Rodier F. Keeping the senescence secretome under control: molecular reins on the senescence-associated secretory phenotype. Exp. Gerontol. 2016;82:39–49. doi: 10.1016/j.exger.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 159.Zhu Y., Armstrong J.L., Tchkonia T., Kirkland J.L. Cellular senescence and the senescent secretory phenotype in age-related chronic diseases. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17(4):324–328. doi: 10.1097/MCO.0000000000000065. [DOI] [PubMed] [Google Scholar]
- 160.Shan M., Qin J., Jin F., Han X., Guan H., Li X. Autophagy suppresses isoprenaline-induced M2 macrophage polarization via the ROS/ERK and mTOR signaling pathway. Free Radic. Biol. Med. 2017;110:432–443. doi: 10.1016/j.freeradbiomed.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 161.Griffiths H.R., Gao D., Pararasa C. Redox regulation in metabolic programming and inflammation. Redox Biol. 2017;12:50–57. doi: 10.1016/j.redox.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tan H.Y., Wang N., Li S., Hong M., Wang X., Feng Y. The reactive oxygen species in macrophage polarization: reflecting its dual role in progression and treatment of human diseases. Oxid Med Cell Longev. 2016;2016:2795090. doi: 10.1155/2016/2795090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.