ABSTRACT
Tendons and ligaments are crucial components of the musculoskeletal system, yet the pathways specifying these fates remain poorly defined. Through a screen of known bioactive chemicals in zebrafish, we identified a new pathway regulating tendon cell induction. We established that statin, through inhibition of the mevalonate pathway, causes an expansion of the tendon progenitor population. Co-expression and live imaging studies indicate that the expansion does not involve an increase in cell proliferation, but rather results from re-specification of cells from the neural crest-derived sox9a+/sox10+ skeletal lineage. The effect on tendon cell expansion is specific to the geranylgeranylation branch of the mevalonate pathway and is mediated by inhibition of Rac activity. This work establishes a novel role for the mevalonate pathway and Rac activity in regulating specification of the tendon lineage.
KEY WORDS: Tendon, Scleraxis, Chemical screen, Statin, Zebrafish
Summary: A screen-based and chemical genetics approach in zebrafish reveals that statin, an inhibitor of the mevalonate pathway, and Rac GTPases regulate the poorly understood process of early tendon development.
INTRODUCTION
Tendons and ligaments are key components of the musculoskeletal system, transmitting and stabilizing high tensile forces. Exposed to such forces, adult tendons are often injured during overuse, trauma or age-related decline (Jozsa and Kannus, 1997). Knowledge of tendon fate determination has important implications for developing regenerative approaches to treating tendon injuries. Although the basic helix-loop-helix transcription factor scleraxis (Scx) provides a marker of early tendon and ligament progenitors (Schweitzer et al., 2001), the pathways leading to tendon cell fate induction remain largely unknown.
Scx marks tendon and ligament cells in all anatomical locations (Brent et al., 2003; Grenier et al., 2009; Schweitzer et al., 2001), and is required for the development of the limb force-transmitting tendons (Murchison et al., 2007). Scx also regulates the expression of extracellular matrix proteins – including Col1a1 – through its proximal promoter (Léjard et al., 2007). The identification of Scx as a marker of early tendon progenitors opened the door to studies of factors involved in tendon specification. TGFβ signals are sufficient to induce Scx expression in mesenchymal tissues (Havis et al., 2014; Pryce et al., 2009). However, mouse genetic loss-of-function studies indicate a role for this pathway in tendon cell maintenance, rather than induction (Oka et al., 2008; Pryce et al., 2009; Tan et al., 2020). An important role for FGF signaling was identified in the specification of axial tendon progenitors in the chick (Brent et al., 2003; Brent and Tabin, 2004), yet the tendon-promoting effect of FGF signaling is not conserved in mouse, as highlighted in studies showing that early FGF/ERK signaling represses Scx expression in mouse (Havis et al., 2014, 2016).
Although the crucial pathways responsible for tendon cell specification remain unknown, studies examining neighboring tissues provide insights into the interactions required for their specification. In the craniofacial, axial and limb regions, tendon and ligament cells form in close association with skeletal cells and are thought to arise from common progenitors (Soeda et al., 2010). The cranial skeletal and tendon progenitors are derived from the cranial neural crest (CNC) (Chen and Galloway, 2014; Couly et al., 1993; Kontges and Lumsden, 1996; Le Douarin and Kalcheim, 1999; Le Lievre, 1978; Lumsden et al., 1991; Noden, 1978; Schilling and Kimmel, 1994); in the limb, they are derived from the lateral plate mesoderm (Hurle et al., 1989, 1990; Kieny and Chevallier, 1979; Pearse et al., 2007; Ros et al., 1995; Saunders, 1948; Wortham, 1948); and in axial regions, tendon progenitors, which form the syndetome, are derived from the early sclerotome, a somitic compartment (Brand-Saberi and Christ, 2000; Brent et al., 2003; Monsoro-Burq and Le Douarin, 2000). Sox9 is expressed in chondrogenic cells, and common Sox9/Scx co-expression domains are found at tendon-bone attachment sites (Blitz et al., 2013; Sugimoto et al., 2013). Furthermore, loss of transcription factors important for cartilage differentiation results in ectopic expression of tendon markers within the axial rib cartilage structures (Brent et al., 2005). Although this effect was not observed in cranial or limb regions, mammalian in vitro studies suggest that tendon and cartilage are mutually exclusive lineages of common progenitor populations (Havis et al., 2014).
To discover new pathways that govern tendon cell induction, we turned to the zebrafish, a system that is highly amenable to high-throughput small molecule screening (Mathias et al., 2012). Previous zebrafish chemical screens have expanded our understanding of developmental processes and identified compounds of potential therapeutic benefit (Goessling and North, 2014; North et al., 2007), highlighting the conservation between zebrafish and humans. Zebrafish tendons share many similarities with those in mammals, including gene expression, morphology, collagen ultrastructural arrangement and developmental regulation (Chen and Galloway, 2014; Subramanian and Schilling, 2014). We therefore reasoned that chemical screens of biologically annotated bioactives in zebrafish could reveal new pathways regulating tendon development.
In a small-molecule whole-embryo screen, we identified statins as regulators of early tendon specification. Statins are a class of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) inhibitors that are prescribed as cholesterol-lowering agents (Istvan and Deisenhofer, 2001). In addition to preventing cardiovascular disease, statins and HMGCR pathway inhibition regulate cell proliferation in tumors (Jones et al., 1994) and cell migration during heart and germ cell development (D'Amico et al., 2007; Santos and Lehmann, 2004; Thorpe et al., 2004; Yi et al., 2006). Using chemical and genetic approaches, we show that Hmgcr inhibition expands the number of craniofacial and pectoral fin tendon progenitors in the zebrafish embryo. The expansion is not a consequence of increased proliferation of a pre-existing scleraxisa-positive (scxa+) cell population, but rather is caused by recruitment of additional neural crest sox9a+/sox10+ skeletal progenitors towards tendon fates. We further demonstrate through loss-of-function studies that the interaction is mediated by inhibition of the geranylgeranylation branch of the mevalonate pathway, and specifically Rac GTPases. Taken together, we establish that the mevalonate pathway via Rac activity is a crucial regulator of tendon fate specification.
RESULTS
Statin promotes tendon cell expansion in zebrafish craniofacial and pectoral fin regions
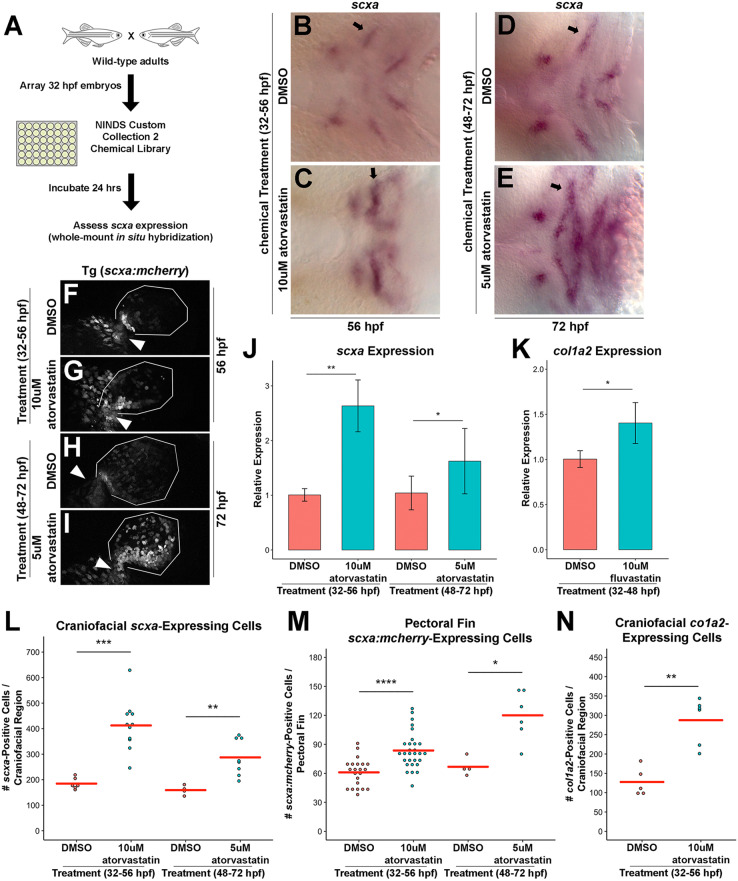
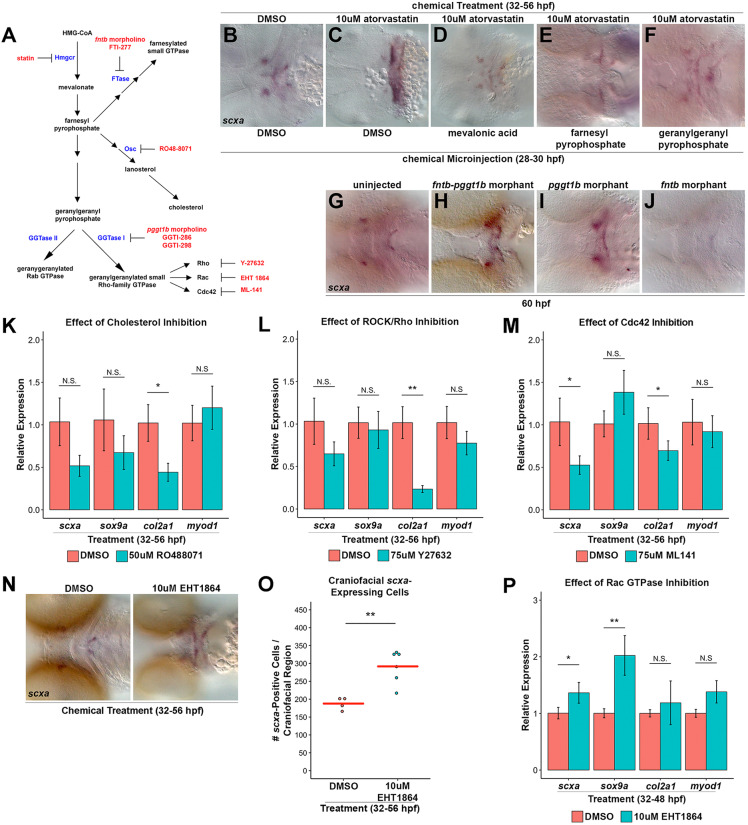
To identify new pathways regulating tendon development in zebrafish, we conducted a chemical screen using a known bioactive compound library: NINDS Custom Collection 2. Wild-type embryos were incubated with individual chemicals from 32 to 56 h post-fertilization (hpf) and examined at 56 hpf by whole-mount in situ hybridization for altered craniofacial expression of scxa (a scx paralogue in zebrafish) (Fig. 1A). These stages were chosen to identify pathways affecting tendon cell induction, as scxa is first detected in the zebrafish pharyngeal arches by 40 hpf (Chen and Galloway, 2014) (Fig. S1). Our screen identified lovastatin and simvastatin (Fig. S2A-D) as agents that increase craniofacial scxa expression. Additional statins, atorvastatin and fluvastatin, similarly expand craniofacial scxa expression (Fig. S2E-L). All statin compounds used in this study inhibit HMGCR and have some differences in their metabolism and pharmacokinetic properties (Klotz, 2003). Atorvastatin caused the most robust phenotype in the cranial and fin regions (Fig. 1B,C,F,G), and most analysis was performed with this compound. Atorvastatin treatment from 48 to 72 hpf resulted in a similar scxa expansion at 72 hpf when assessed by whole-mount in situ hybridization (Fig. 1D,E,H,I). Analysis of fluvastatin, which has the shortest half-life among statins (Igel et al., 2001), demonstrated the 32-48 hpf treatment window was sufficient to expand craniofacial scxa expression (Fig. S2M-R). The statin-mediated upregulation of scxa was further confirmed by qPCR at both stages (Fig. 1J). Quantification of scxa+ cells using whole-mount in situ hybridization or Tg(scxa:mcherry) zebrafish (Fig. S3I-Q) verified that statin treatment increases the number of scxa+ cells in the craniofacial (Fig. 1L) and pectoral fin (Fig. 1M, Fig. S3B) regions compared with controls. At the stages examined, the Tg(scxa:mcherry) line was found to recapitulate endogenous scxa expression (Fig. S1). Although early (32-56 hpf) and later (48-72 hpf) treatments resulted in expanded scxa expression, it is possible that the cellular mechanism underlying expansion is not identical between the stages. Because the early treatment was better tolerated by embryos and resulted in less toxicity, we focused on dissecting the effects of statin treatment in the 32-56 hpf stages.
Fig. 1.
A zebrafish chemical screen identifies statins as regulators of craniofacial and pectoral fin tendon progenitors. (A) Design of the small-molecule screen. Wild-type embryos were incubated with individual compounds from 32 to 56 hpf and alterations in craniofacial scxa expression were assessed at 56 hpf. (B-E) Craniofacial scxa expression at 56 hpf, after incubation from 32 to 56 hpf (B,C) and at 72 hpf after incubation from 48 to 72 hpf (D,E). Atorvastatin expanded scxa expression in the pharyngeal arches (arrows) compared with controls. (F-I) Pectoral fin scxa:mcherry expression at 56 hpf (F,G) and 72 hpf (H,I) upon incubation from 32 to 56 hpf and 48 to 72 hpf, respectively. Atorvastatin expanded scxa:mcherry expression at the cleithrum base (arrowhead), extending distally to the actinotrichia. (J) qPCR quantification revealed that atorvastatin increased scxa expression at 56 hpf and 72 hpf upon incubation from 32 to 56 hpf and 48 to 72 hpf, respectively, compared with controls. n=4, whole embryo (32-56 hpf) and head region (48-72 hpf); Welch's two-tailed t-test. (K) qPCR quantification revealed that fluvastatin increased col1a2 expression at 72 hpf, after incubation from 32 to 48 hpf compared with controls. The combination of fluvastatin, which is characterized by a shorter half-life and a shortened exposure window (Fig. S2M-R), mitigated the toxic effects observed at 72 hpf with atorvastatin treatment. n=3, head region, Welch's two-tailed t-test. (L) Atorvastatin increased the quantity of craniofacial scxa+ cells at 56 hpf and 72 hpf after incubation from 32 to 56 hpf and 48 to 72 hpf, respectively, compared with controls. (M) Atorvastatin increased the quantity of pectoral fin scxa:mcherry+ cells at 56 hpf and 72 hpf upon incubation from 32 to 56 hpf and 48 to 72 hpf, respectively, compared with controls. (N) Atorvastatin increased the quantity of col1a2+ cells at 80 hpf, after incubation from 32 to 56 hpf compared with controls. (J,K) Data are mean±s.d. (L-N) Red bars indicate mean; individual points represent values for individual embryos; Mann–Whitney-Wilcoxon test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Ventral (B-E) and lateral (F-I) views, anterior towards the left.
To test whether statin expands expression of additional tendon genes, as opposed to simply upregulating scxa expression, we examined expression of col1a2, which is expressed during tendon differentiation and is a major component of the tendon extracellular matrix (Brent et al., 2003; Chen and Galloway, 2014). We found that embryos incubated with atorvastatin also have expanded col1a2 expression (Fig. S2W-X). Likewise, statin causes >2-fold increase in the number of craniofacial col1a2+ tendon progenitors compared with controls (Fig. 1N) and increases col1a2 expression by qPCR (Fig. 1K). As forming bone is rich in type I collagen, we examined expression of runx2a, a marker of osteoblasts (Burns et al., 2002), following statin treatment. We found that runx2a expression was not qualitatively expanded at 56 hpf and was decreased at 72 hpf (Fig. S2S-V), suggesting that the increase in col1a2 expression is due to expanded tendon fates. Besides the head and fin, scxa+ tendon progenitors in zebrafish exist in the myosepta, connecting the muscles of the trunk. In contrast to the pectoral fin and craniofacial region, atorvastatin does not affect the quantity of scxa+ (Fig. S2Y) or scxa:mcherry+ (Fig. S2Y-C′) cells in the myosepta. Furthermore, atorvastatin treatment from 32-56 hpf reduced myoseptal col1a2 expression at 74 hpf (Fig. S2D′-E′). Although the differences in response to statin treatment in the different anatomical locations may be derived from tissue-specific differences in drug efficaciousness, we hypothesize the difference is more reflective of region-specific programs of tendon development (Brent et al., 2003; Chen and Galloway, 2014).
Statin-mediated expansion of tendon progenitors does not depend on increased proliferation
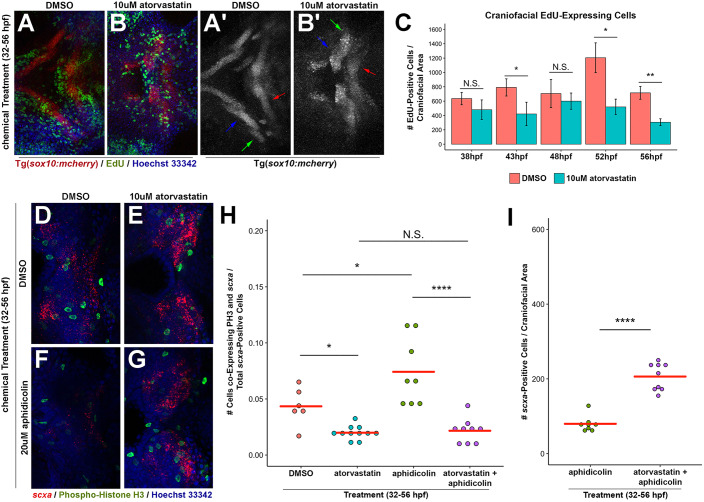
In principle, the increased quantity of tendon progenitors could arise from increased proliferation of scxa+ cells. To determine whether statin increases tendon cell proliferation, we performed EdU labeling (Salic and Mitchison, 2008) and phospho-histone H3 (PH3) staining. EdU pulse-labeling at 4 or 5 h intervals of chemically treated embryos transgenic for a neural crest and cranial cartilage reporter, sox10:mcherry (Fig. 2A-B′), indicated that atorvastatin does not increase the number of proliferative cells in anatomically defined regions compared with corresponding regions in controls (Fig. 2C). Rather, we observed a trend of decreased proliferation following atorvastatin treatment at 43 hpf, 52 hpf and 56 hpf, suggesting that the mevalonate pathway regulates proliferation in the craniofacial region. The mevalonate pathway negatively affects proliferation in other cell types (Corsini et al., 1993; Gong et al., 2019). Given that subtle changes in proliferation within the scxa+ domain may not be detected with this analysis, we performed two sets of experiments to address this concern. First, we quantified the number of PH3+ mitotic cells in atorvastatin-treated and control embryos. Consistent with our EdU findings, we determined that atorvastatin does not promote proliferation in the craniofacial or fin regions (Fig. S3A,C). More importantly, we did not detect any increase in the ratio of PH3+/scxa+ cells in either region (Fig. 2D,E,H and Fig. S3D,I,L-Q). Second, we hypothesized that if the statin-mediated expansion were acting through increased cell proliferation, an antimitotic agent would abrogate its affect. We analyzed the effect of aphidicolin, a DNA polymerase inhibitor (Spadari et al., 1984), on proliferation and craniofacial scxa expression with and without statin treatment. The concentration of aphidicolin used reduces the quantity of PH3+ cells (Fig. 2D,F and Fig. S3A) (Ladstein et al., 2010) as well as the total number of scxa+ cells in DMSO-treated embryos (Figs 1L, 2D,F,I and Fig. S3E,F). This indicates that proliferation contributes to the expansion of scxa+ cells during normal development in control DMSO-treated embryos. Strikingly, atorvastatin and aphidicolin co-treatment from 32 to 56 hpf caused >2-fold expansion in craniofacial scxa+ cells (Fig. 2E,G,I and Fig. S3G,H), compared with aphidicolin treatment alone, indicating that inhibiting proliferation has no effect on the ability of statin to expand tendon cell number.
Fig. 2.
Statin-mediated expansion of craniofacial tendon progenitors is not due to increased proliferation of scxa-positive cells. (A,B) Tg(sox10:mcherry) embryos at 56 hpf were chemically treated from 32 to 56 hpf, labeled for EdU+ cells and counterstained with Hoechst. Arrows in A′,B′ mark Meckel's cartilage (blue), palatoquadrate cartilage (green) and ceratohyal cartilage (red). (C) Quantification of EdU+ cells in the craniofacial region upon incubation from 32 hpf. Atorvastatin does not increase the quantity of craniofacial EdU+ cells at timepoints examined, compared with controls. n=3, Welch's two-tailed t-test. Data are mean±s.d. (D-G) Craniofacial expression of scxa and phospho-histone H3 (PH3) at 56 hpf upon incubation with the indicated compounds from 32 to 56 hpf. Craniofacial scxa expression compared with controls (D) is reduced in aphidicolin-treated embryos (F), and expanded in embryos treated with atorvastatin alone (E) and in combination with aphidicolin (G). Data from these representative images are plotted in H,I and Fig. S3A. (H) Quantification of craniofacial cells co-expressing PH3 and scxa at 56 hpf after incubation from 32 to 56 hpf. (I) Quantification of scxa+ cells indicated that decreased proliferation does not hamper atorvastatin-mediated scxa expansion. (H,I) Red bars indicate mean; individual points represent values for individual embryos; Mann–Whitney-Wilcoxon test. N.S., no significance; *P<0.05; **P<0.01; ****P<0.0001. Representative craniofacial domain analyzed is shown in A-B′,D-G. Ventral (A-B′,D-G) views, anterior towards the left.
Statin increases the number of scxa-positive tendon cells derived from CNC skeletal progenitors
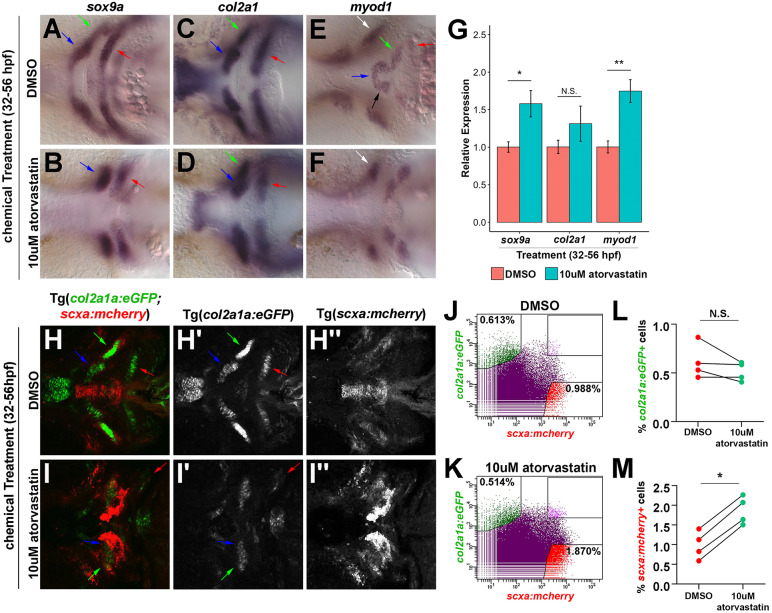
As increased proliferation cannot explain the statin-mediated tendon cell expansion, we explored the possibility that cells from adjacent tissues are recruited to tendon fates following statin treatment. In the craniofacial region, skeletal and tendon cells arise from the CNC and interact with the pharyngeal mesoderm-derived muscle to form the craniofacial musculoskeletal system (Chen and Galloway, 2014; Schilling and Kimmel, 1994). To determine whether statin alters musculoskeletal development, we examined muscle and cartilage tissues by whole-mount in situ hybridization. Atorvastatin treatment from 32 to 56 hpf altered the spatial expression patterns of sox9a (Fig. 3A,B), an early marker of skeletal progenitors and differentiating chondrocytes, col2a1 (Fig. 3C,D), a component of the cartilage matrix (Yan et al., 2002), and myod1 (Fig. 3E,F), a marker of myoblast progenitors (Lin et al., 2006). Specifically, the expression of col2a1 (Fig. 3C,D, arrows) and myod1 (Fig. 3E,F, white arrow) appear to mark groups of cells that are not properly organized in the midline compared with controls. In the pectoral fin, the expression of sox9a, col2a1, sox10:eGFP and myod1 (Fig. S4A-H) was present, but with an altered morphology following statin treatment. Quantification by qPCR showed increased expression of sox9a and myod1 and no significant effect on col2a1 expression (Fig. 3G). Together, these results suggest that statin alters the morphology of the forming cartilage and muscle but does not cause a loss in their respective gene expressions.
Fig. 3.
Effect of statin on the craniofacial musculoskeleton. (A-F) Craniofacial expression of sox9a, col2a1 and myod1 at 56 hpf upon incubation from 32 to 56 hpf. At 56 hpf, atorvastatin altered the spatial expression patterns of sox9a (A,B), col2a1 (C,D) and myod1 (E,F) compared with controls. Arrows in A-D,H-I′ mark Meckel's cartilage (blue), palatoquadrate cartilage (green) and ceratohyal cartilage (red). Arrows in E,F mark intermandibularis anterior (blue), intermandibularis posterior (black), interhyal (green), hyohyal (red) and adductor mandibularis (white). (G) qPCR quantification revealed that atorvastatin increased sox9a and myod1 expression, but not col2a1 at 56 hpf after incubation from 32 to 56 hpf compared with controls. n=3, whole embryo, Welch's two-tailed t-test. Data are mean±s.d. (H,I) Craniofacial expression of col2a1a:eGFP;scxa:mcherry at 56 hpf upon incubation from 32 to 56 hpf. Corresponding views of eGFP (H′,I′) and mcherry (H″,I″). (J,K) Representative flow cytometry analysis of mcherry+ tendon and eGFP+ cartilage cells at 56 hpf in the head region of Tg(scxa:mcherry;col2a1a:eGFP) embryos after incubation from 32 to 56 hpf. Atorvastatin expanded mcherry+ tendon progenitors (0.988% in DMSO; 1.870% in atorvastatin). (L,M) Flow cytometry quantification of eGFP+ and mcherry+ cells at 56 hpf in the head region of Tg(scxa:mcherry;col2a1a:eGFP) embryos after incubation from 32 to 56 hpf. Atorvastatin increased mcherry+ cells but not eGFP+ cells (n=4; Mann–Whitney-Wilcoxon test). N.S., no significance; *P<0.05; **P<0.01. Ventral views (A-F,H-I″), anterior towards the left.
To further quantify the effects of statin on the numbers of chondrocyte and tendon cells, we performed flow cytometry on dissociated Tg(scxa:mcherry;col2a1a:eGFP) embryos treated with DMSO or statin (Dale and Topczewski, 2011). Consistent with our col2a1 whole-mount in situ hybridization results, the morphology of col2a1a:eGFP+ cranial chondrocytes appeared altered (Fig. 3H-I′) compared with controls. Using flow cytometry, we were able to identify singly positive col2a1a:eGFP+ or scxa:mcherry+ cell populations from dissociated transgenic embryos. Statin had no effect on the percentage of col2a1:eGFP+ cells, but significantly increased the percentage of scxa:mcherry+ cells (Fig. 3J-M). This approximate 2-fold expansion corresponds to the increase detected by qPCR and cell quantification analysis of stained embryos. Although a scxa:mcherry+/col2a1a:eGFP+ cell population was found in controls, this represented less than 0.1% of the total cells and changes to this population upon statin treatment were not statistically significant.
We next tested whether the increased craniofacial scxa+ cells of statin-treated embryos derive from the neural crest. We used expression of the Tg(sox10:kaede) (Dougherty et al., 2012) as a reporter to mark cells derived from the sox10:kaede+ lineage. Confocal images of craniofacial region of DMSO-treated (n=4 per timepoint) and atorvastatin-treated (n=8 per timepoint) embryos were analyzed for colocalized Kaede and scxa expression. By analyzing the optical sections from confocal images, we found that scxa+ cells co-expressed the Kaede protein in all Tg(sox10:kaede) embryos examined following DMSO and atorvastatin treatment from 32 to 56 hpf (Fig. S4I-L). This demonstrates that the statin-mediated expanded scxa+ domain in the craniofacial region is exclusively derived from the CNC. Given the significant increase in scxa+ cells and expression in the craniofacial region, we focused on the craniofacial tendons in subsequent analyses.
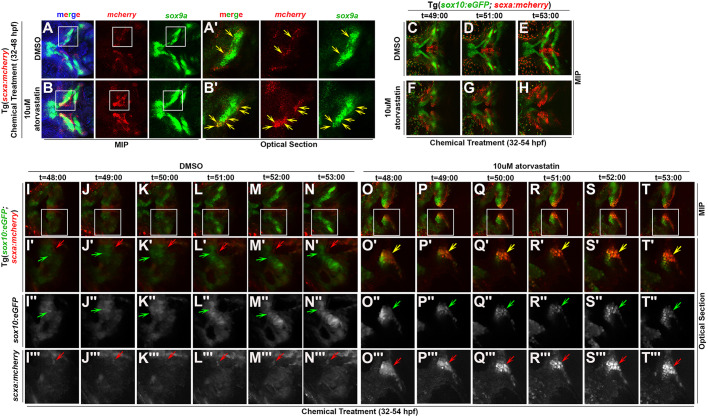
Previous studies using genetic lineage tracing in mice have shown that tendon and ligament cells are descendants of Sox9+ cells (Huang et al., 2019; Soeda et al., 2010). Zebrafish sox9a marks migrating CNC as well as later skeletal cell types (Yan et al., 2005). Therefore, we sought to explore the possibility that the statin phenotype results from more CNC sox9a+ progenitors being diverted to scxa+ tendon fates. As our results show statin increases sox9a transcripts (Fig. 3G), we wanted to determine whether the expanded scxa+ cells co-expressed sox9a. To examine co-expression of scxa and sox9a, we performed fluorescent whole-mount in situ hybridization for transcripts of sox9a and mcherry for identifying scxa:mcherry+ tendon progenitors. For most regions in the lower jaw, sox9a and mcherry occupied spatially distinct domains in DMSO-treated Tg(scxa:mcherry) embryos at 48 hpf. In these control embryos, limited co-expression of mcherry and sox9a in some tendon attachment regions, specifically near the forming palatoquadrate and maxilla were observed (Fig. 4A,A′, yellow arrows). In contrast, we observed significant and consistent co-expression of sox9a and mcherry in statin-treated Tg(scxa:mcherry) embryos at 48 hpf (Fig. 4B,B′, yellow arrows).
Fig. 4.
Statin expands the scxa-expressing cell population by promoting tenogenic specification of CNC. (A-B′) Expression of mcherry and sox9a in (A,A′) DMSO and (B,B′) atorvastatin-treated Tg(scxa:mcherry) embryos at 48 hpf. White boxes mark the craniofacial domain in maximum intensity projection (MIP) (A,B), magnified in corresponding optical sections (A′,B′). Yellow arrows mark mcherry+/sox9a+ cells. Atorvastatin expanded mcherry+/sox9a+ cells (B′, compare with A′). (C-T‴) Time-course of live imaging of tendon and cartilage development in (C-E,I-N‴) DMSO- and (F-H,O-T‴) atorvastatin-treated Tg(sox10:eGFP;scxa:mcherry) embryos (n=4 shown). Arrows mark sox10:eGFP+ cells of Meckel's cartilage (green), scxa:mcherry+ tendon progenitors (red) and scxa:mcherry+/sox10:eGFP+ cells (yellow). Atorvastatin expanded scxa:mcherry+/sox10:eGFP+ cells near the region where Meckel's cartilage forms (compare F-H and O-T‴ with C-E and I-N‴, respectively). White boxes mark the craniofacial domain in the MIPs (I-T) of high-resolution images, magnified in the corresponding MIPs (I′-T′) and optical sections: eGFP (I″-T″) and mcherry (I‴-T‴) expression. Ventral views, anterior towards the left.
sox9a and sox10:eGFP are expressed in CNC cells by 26 hpf, but their later expression becomes predominantly restricted to the forming cartilage elements by 48 hpf (Carney et al., 2006; Yan et al., 2005). Based on this, we wanted to determine whether statin causes sox10:eGFP+ CNC progenitors to adopt a tendon fate. To test this possibility, we performed live imaging of Tg(scxa:mcherry;sox10:eGFP) embryos treated with DMSO or statin from 32 to 53 hpf. Analysis of live images shows sox10:eGFP+ cells in and around Meckel's cartilage co-expressing scxa:mcherry in statin-treated embryos (Fig. 4F-H,O-T‴, arrows) whereas this was not observed in controls (Fig. 4C-E,I-N‴, arrows). Taken together, these results indicate that the statin-mediated increase in scxa+ cranial tendon cells results from the conversion of CNC sox9a+/sox10+ progenitors to scxa+ cells.
The effect of statin on patterning and known tendon-promoting signaling pathways
One potential mechanism by which multipotent CNC cells might be diverted to a different cell fate would be if the patterning of the craniofacial region were altered. To test this, we examined known markers of anterior-posterior and dorsal-ventral arch polarity. Despite statin-mediated scxa expansion (Fig. S5A,B), we did not identify any major patterning alterations in the expression of hoxa2b, hoxb2a, hand2, bapx1, shha, patched1 or gsc at the stages examined (Hunter and Prince, 2002; Miller et al., 2003), although the expression levels of some genes appeared reduced upon chemical treatment compared with controls (Fig. S5C-M,R). This suggested that dorsal-ventral and anterior-posterior arch polarity are not affected by statin. A second possibility was that statins could interact with and act through pathways known to maintain or promote tendon fate. We interrogated constituents of the BMP (bmp4), TGFβ (tgfbr2a and tgfbi), FGF (pea3) and Wnt (fzd7a and fzd7b) pathways (Brent and Tabin, 2004; Pryce et al., 2009; ten Berge et al., 2008; Yamamoto-Shiraishi and Kuroiwa, 2013), and observed no increase in expression of any markers in atorvastatin-treated embryos compared with controls (Fig. S5N-Q,S-X). Analysis of embryos transgenic for 6xTcf/LefBS-miniP:d2eGFP (Shimizu et al., 2012) and bre:egfp (Laux et al., 2011), which mark Wnt/β-catenin-mediated Tcf and BMP/Smad1/5 transcriptional activity, respectively, did not show expanded egfp expression in atorvastatin-treated embryos compared with controls (Figs S5Y-B′ and S6A-F). Although some differences in bre:egfp expression were observed near the mouth and posterior pharyngeal arch regions of statin-treated embryos, these alterations are consistent with a developmental delay in bre:egfp expression based on previous descriptions (Laux et al., 2011). To examine TGFβ signaling and its interaction with the statin-mediated scxa expansion, we treated embryos with an ALK5 inhibitor (SB-431542) (Inman et al., 2002), alone and in combination with atorvastatin. We found SB-431542 significantly decreased scxa expression and increased sox9a expression at 56 hpf compared with controls (Fig. S6G). In embryos co-treated with SB-431542 and atorvastatin, scxa expression is significantly increased compared with SB-431542 treatment alone to a level comparable to that of DMSO controls. The finding that statin rescues scxa expression in the presence of TGFβ inhibition suggests that the pathway targeted by statin acts independently of TGFβ signaling. Furthermore, we performed staining for phospho-Smad3 and observed differences in phospho-Smad3 staining surrounding the cartilage regions of statin-treated embryos compared with controls. Although this result indicates altered TGFβ signaling in statin-treated embryos, these phospho-Smad3+ regions did not co-express scxa:mcherry (Fig. S6H-S′). Together, these results suggest that statin may alter the activity of some signaling pathways in the craniofacial region, but it is unclear how these changes could directly cause the increase in tendon cells.
Inhibition of Hmgcr and mevalonate production expands the craniofacial tendon program
Taken together, our results suggest that statins expand the progenitor pool by altering CNC cell specification through a pathway previously unexplored in this context. To confirm the chemical inhibition results, we established that the expanded tendon cell phenotype results from statin inhibition of Hmgcr, the rate-limiting enzyme of the mevalonate pathway (Fig. 5A). Zebrafish have two HMGCR orthologs, hmgcra and hmgcrb. hmgcra is expressed in the larval liver and intestine; hmgcrb is maternally transcribed and ubiquitously expressed during early development (Thorpe et al., 2004). As hmgcrb is expressed during the spatial-temporal domain of interest, we focused on hmgcrb for further analysis. To test whether the statin-mediated expansion in craniofacial tendon progenitors is due to inhibition of Hmgcrb, we examined scxa and col1a2 expression in hmgcrb-deficient embryos by whole-mount in situ hybridization. In the hmgcr1bs617 mutants (D'Amico et al., 2007; Mapp et al., 2011), identified by their severe pericardial edema, scxa expression appears expanded at 60 hpf, and in a subset at 76 hpf, the pattern of col1a2 is expanded compared with controls (Fig. S7A-D, arrow). The remaining mutants had extreme morphological abnormalities and did not express col1a2. Morpholino-mediated knockdown of the maternal and zygotic hmgcrb in zebrafish resulted in severe morphological defects and death prior to the onset of craniofacial scxa expression. Consistent with the zebrafish phenotype, the Hmgcr knockout in mouse is embryonic lethal (Ohashi et al., 2003), suggestive of an essential role for HMGCR/Hmgcr in early mouse and zebrafish development. For this reason, we injected a sub-lethal concentration of morpholino directed against hmgcrb. We next exposed the same sensitized embryos to a dose of atorvastatin that alone does not cause a tendon phenotype from 32 to 56 hpf. In this manner, we were able to induce loss of Hmgcrb within a specified developmental time window and to circumvent its earlier embryonic requirement. We observed an expansion of scxa in such doubly treated embryos at 56 hpf compared with controls (Fig. S7E-H, arrow), suggesting an essential role for Hmgcrb inhibition in the statin-mediated expansion of craniofacial scxa. To obtain unequivocal evidence that inhibition of the mevalonate pathway is responsible for the phenotype observed, we rescued the expansion in craniofacial scxa expression to wild-type levels at 56 hpf with injection of mevalonic acid, the immediate metabolite targeted by statin (Fig. 5B-D). Together, these data establish the inhibition of Hmgcrb as the mechanism by which statin promotes expansion of craniofacial scxa+ tendon progenitors in the zebrafish.
Fig. 5.
Inhibition of geranylgeranylation branch causes statin-mediated tendon cell expansion. (A) Schematic of Hmgcr pathway with enzymes (blue), inhibitors (red) and morpholinos (red) indicated. (B,C) scxa expression in control DMSO-treated/DMSO-injected embryos at 56 hpf (B, 100%; n=30) is expanded upon atorvastatin treatment and DMSO injection (C, 100%; n=62). (D-F) scxa expression is rescued to wild-type levels in embryos incubated with atorvastatin from 32 to 56 hpf and injected at 28 hpf with mevalonic acid (D, 94% rescued; n=18), farnesyl pyrophosphate (E, 33% rescued; n=72) or geranylgeranyl pyrophosphate (F, 49% rescued; n=43). (G-J) At 60 hpf, craniofacial scxa expression, compared with controls (G, 100% wild type, n=41), is expanded in fntb-pggt1b morphants (H, 73% expanded, n=103) and pggt1b morphants (I, 60% expanded, n=89), and reduced in fntb morphants (J, 29% reduced; 71% normal expression, n=17). (K-M) qPCR analysis of scxa, sox9a, col2a1 and myod1 expression at 56 hpf after incubation from 32 to 56 hpf. (K) Cholesterol biosynthesis inhibition (using RO48-8071) reduced col2a1, but had no effect on scxa, sox9a and myod1 compared with controls. (L) ROCK/Rho inhibition (using Y-27632) reduced col2a1, but had no effect on scxa, sox9a and myod1 compared with controls. (M) Cdc42 GTPase inhibition (using ML-141) reduced scxa and col2a1, but did not affect sox9a and myod1. (N) Rac GTPase inhibition (using EHT-1864) expanded scxa expression at 56 hpf after incubation from 32 to 56 hpf. (O) EHT-1864 increased the quantity of scxa+ cells at 56 hpf after incubation from 32 to 56 hpf. (P) qPCR analysis of scxa, sox9a, col2a1 and myod1 expression at 56 hpf after incubation from 32 to 56 hpf. EHT-1864 increased expression of scxa and sox9a, but did not affect col2a1 and myod1. For qPCR (K-M,P), n=3, head region, Welch's two-tailed t-test. Data are mean±s.d. (O) Red bars indicate mean; points represent individual embryos; Mann–Whitney-Wilcoxon test. N.S., not significant; *P<0.05; **P<0.01. Ventral views (B-J,N), anterior towards the left.
Statin promotes the craniofacial tendon program through inhibition of Rac GTPases
The mevalonate pathway metabolizes the biosynthesis of cholesterol and isoprenoids (Goldstein and Brown, 1990). Isoprenoids are covalently attached to proteins by post-translational farnesylation or geranylgeranylation. Farnesylation is the addition of CaaX by farnesyltransferase (FTase); geranylgeranylation is the addition of CAAL by geranylgeranyltransferase I (GGTase I) or GGTase II (McTaggart, 2006). To test the requirement of the cholesterol and/or prenylation branches of the mevalonate pathway in scxa expansion, we incubated embryos with inhibitors of GGTase I (GGTI-286) (Lerner et al., 1995), FTase (FTI-277) (Lerner et al., 1995) and 2,3-oxidosqualene:lanosterol cyclase (Osc; RO48-8071) (Morand et al., 1997). Inhibition of all three branches in combination causes scxa expansion at 56 hpf, which phenocopies that of embryos treated with atorvastatin alone (Fig. S7I,K, arrow). Conversely, microinjection of farnesyl pyrophosphate, a branch-point intermediate for the cholesterol and prenylation branches, rescued scxa expression at 56 hpf following atorvastatin treatment (Fig. 5E).
To dissect the pathways involved in the statin-mediated tendon cell expansion, we targeted specific branches of the mevalonate pathway using genetic and chemical loss- and gain-of-function approaches. To test the requirement of the prenylation branch in the expansion of scxa, we knocked down essential regulators of prenylation, FTase beta (fntb) and protein GGTase I beta (pggt1b) (Mapp et al., 2011). We found that loss of both fntb and pggt1b resulted in an expansion of craniofacial scxa at 60 hpf compared with controls (Fig. 5G,H), suggesting that inhibition of prenylation sufficiently expands craniofacial scxa expression. This expansion of scxa is found in pggt1b morphants alone (Fig. 5I), whereas scxa is reduced in fntb morphants (Fig. 5J), indicating that loss of GGTase I can elicit a tendon cell expansion. Furthermore, microinjection of geranylgeranyl pyrophosphate, an intermediate unique to the geranylgeranylation branch (McTaggart, 2006), rescued scxa expression at 56 hpf following atorvastatin treatment (Fig. 5F). To completely exclude the possibility that the cholesterol branch could be involved in expanding scxa expression, we inhibited cholesterol production. We found RO48-8071 had no significant effect on scxa, sox9a and myod1 transcripts, and significantly decreased col2a1 expression at 56 hpf compared with controls (Fig. 5K), suggesting that inhibition of the cholesterol biosynthesis branch does not expand craniofacial scxa+ tendon progenitors. Together, our data indicate that inhibition of geranylgeranylation alone and GGTase I specifically is required for the statin-mediated expansion of craniofacial scxa+ tendon progenitors in zebrafish.
GGTase I catalyzes the geranylgeranylation of the Rho family of GTPases, regulators of extracellular signaling that control actin cytoskeleton organization, transcriptional activation and mitogenesis (McTaggart, 2006). We examined the role of the Rho family members (Rho, Rac and Cdc42) in patterning the craniofacial musculoskeletal tissues using chemical inhibitors specific to each pathway. ROCK/Rho inhibition (Y-27632) (Uehata et al., 1997) significantly reduced col2a1, while scxa, sox9a and myod1 were not significantly changed at 56 hpf (Fig. 5L). Cdc42 inhibition (ML-141) (Hong et al., 2013) significantly reduced scxa and col2a1, but had no significant effect on sox9a and myod1 expression at 56 hpf (Fig. 5M). In contrast, Rac GTPase inhibition (EHT-1864) (Shutes et al., 2007) caused a qualitative expansion of scxa expression by whole-mount in situ hybridization (Fig. 5N) and significant quantitative increase in scxa+ cells (Fig. 5O) at 56 hpf. Expression of scxa and sox9a was also significantly increased, whereas col2a1 and myod1 were not significantly altered by qPCR upon EHT-1864 treatment at 48 hpf (Fig. 5P). Therefore, the two processes observed upon statin treatment, the expansion of craniofacial scxa+ tendon progenitors and an increase in sox9a expression, can be reproduced through EHT-1864 inhibition of Rac GTPase activity. Although a similar morphological result from pathway inhibition may not represent a true phenocopy, we believe the similarity in gene expression changes caused by Rac inhibition strongly suggests that inhibition of mevalonate through Rac GTPases, results in scxa expansion.
DISCUSSION
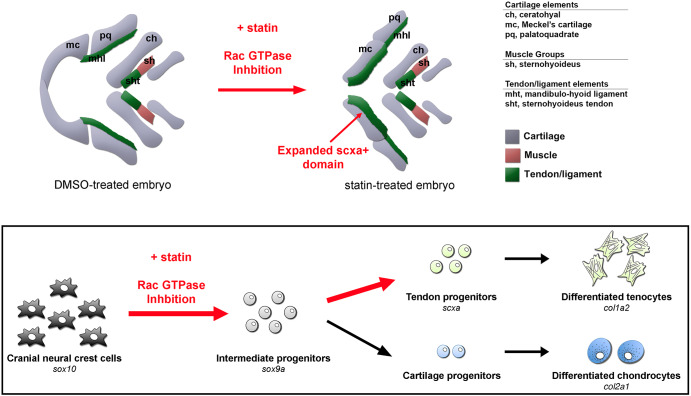
The mechanisms underlying tendon progenitor specification in vertebrates have remained elusive. Although TGFβ and FGF can promote tendon differentiation, no major signaling pathways have been identified to regulate tendon induction (Brent et al., 2003; Pryce et al., 2009; Havis et al., 2016). The small-molecule screen described here identified a new pathway as a crucial regulator of this process. We propose that inhibition of Hmgcr expands the craniofacial tendon progenitor pool through recruitment of neighboring sox9a+ CNC cells, diverting them from a sox10:eGFP+ skeletal fate (Fig. 6). Moreover, our data suggest that the mevalonate pathway acts through Rac GTPases in this context, thereby implicating a unique role for the mevalonate pathway and Rac GTPases in the formation of CNC tendon progenitors.
Fig. 6.
Model for the mechanism of statin-mediated expansion of zebrafish craniofacial tendon program. Schematic illustrates selected craniofacial cartilage, muscle and tendon/ligament groups in ventral view of DMSO- and statin-treated zebrafish at 56 hpf after incubation from 32 to 56 hpf. Exposure of multipotential chondrocyte-tendon progenitors to statins results in re-specification towards a tendon fate – a process mediated through RAC GTPase inhibition. This cell fate expansion is spatially restricted to subsets of CNC-derived elements.
Cranial neural crest lineage specification
CNC cells are a multipotent population (Le Douarin and Kalcheim, 1999), although the time at which these cells become restricted and cues directing the cells towards individual fates are not well understood. Subpopulations of the CNC were postulated to become limited in potential prior to their ventral migration (Le Douarin and Smith, 1988). However, other studies have demonstrated that migrating CNC cells contain both pluripotent and lineage-restricted populations when cultured in vitro (Baroffio et al., 1991). Consistent with these findings, CNC cell fates can be altered in response to different signaling environments, indicating the cells are more plastic than originally believed (Trainor and Krumlauf, 2000). A single cell profiling analysis of mouse neural crest cells showed that cell fate decisions are made through the sequential stages of co-activation of transcriptional programs of competing fates, followed by transcriptional biasing, and finally by resolution towards one of the fates (Soldatov et al., 2019). Within the mesenchymal lineage, the CNC contains osteo-chondrogenic progenitors that become segregated into two distinct fates, cells that give rise to the bones that form either by endochondral ossification or intramembranous ossification (Akiyama et al., 2005; Mori-Akiyama et al., 2003; Nakashima et al., 2002). These studies, along with in vitro differentiation experiments showing osteogenic and chondrogenic differentiation potential of cells derived from embryonic calvarial bone (Grigoriadis et al., 1990; Stringa and Tuan, 1996), support the existence of bi-potential CNC-derived skeletal progenitors. In our studies, we observe changes in scxa+, scxa+/sox9a+ and scxa+/sox10+ cells in embryos treated with statin after the completion of CNC ventral cell migration. Our results are consistent with a model in which the post-migratory CNC cells contain multipotent bone-cartilage-tendon-forming progenitors rather than fate-restricted cells committed to one lineage. Studies have suggested that such multipotent progenitors capable of becoming ligament or bone exist in the zebrafish in a specific craniofacial region in the hyoid arch (Nichols et al., 2016). In our work, expression analysis of tendon markers revealed that the statin-mediated expansion of the craniofacial tendon program is localized to defined domains of the pharyngeal arches, as we never observed the entire skeletal-forming region converting to tendon fates. The restriction in the expansion of the tendon program indicates that either not all populations of CNC-derived mesenchyme are competent to activate a tendon program upon inhibition of the mevalonate pathway, or that skeletal-tendon progenitors are equivalent in their potential throughout the craniofacial region, but a subset of these sit in a microenvironment permissive for statin-mediated activation of tendon fates. These possibilities of cell competence to form tendon or site-specific signaling differences could explain why the axial tendon cells are not expanded upon statin treatment. In considering CNC tendon specification as it relates to other regions of tendon formation, cranial and limb/fin tendon progenitors have been previously suggested to have similar modes of regulation, as these regions do not require muscle for their specification whereas the axial tendon progenitors do (Brent et al., 2005; Edom-Vovard and Duprez, 2004; Grenier et al., 2009). Based on this, it is interesting to note that statin-mediated expansion of scxa expression was observed in fin and craniofacial regions, but not in axial locations. Further experiments are necessary to discern whether this represents a broader similarity in specification programs between the two regions.
Co-expression of sox9a and scxa in tendon development
In addition to increased numbers of scxa+ cells, our study demonstrates that statin promotes a broader scxa+/sox9a+ domain. Previous mouse lineage-tracing studies in the limb have shown that Sox9+ cells contribute to tendon tissues (Huang et al., 2019; Soeda et al., 2010). In the zebrafish head, sox9a and sox10 are expressed throughout the post-migratory CNC cells at 26 hpf prior to the initiation of cranial scxa expression. Previous work and the lineage-tracing studies in this paper show that cranial scxa+ cells descend from a CNC sox10 lineage. Therefore, it is likely that the cranial tendons also form from sox9a+ cells despite there being few scxa+ cells co-expressing either sox9a or sox10 when scxa initiates expression after 40 hpf. This likely reflects the restriction in differentiation of these cells to sox9a+/sox10:eGFP cartilage or scxa+ tendon fates. We observe a few scxa+/sox9a+ cells located at tendon-bone interfaces for Meckel's adductor tendon and palatoquadrate adductor tendon (Fig. 4A-C′, yellow arrow) (McGurk et al., 2017). The localization of scxa+/sox9a+ cells in wild-type embryos could indicate a similarity to the mammalian tendon-bone attachment unit, which also co-expresses these markers (Blitz et al., 2013; Sugimoto et al., 2013). Therefore, the statin-mediated increase in scxa and sox9a transcript levels along with a broadened scxa+/sox9a+ region could be interpreted in two ways: statin results in the persistent expression of sox9a and expansion of scxa, resulting in the conversion of CNC multipotent skeletal progenitors to tendon fates; or there is an increase in tendon-bone attachment unit cell types following statin treatment. We believe our data more strongly support the former interpretation, as our live imaging shows sox10:eGFP+ cells turning on scxa:mcherry expression upon statin treatment, which is not observed in wild-type embryos at this stage. In addition, we observe persistent expanded scxa expression upon statin treatment, whereas in mouse attachment units, Scx expression is not maintained at later stages of development (Blitz et al., 2013). Although our live imaging could suggest a direct transdifferentiation event from sox10:eGFP-expressing chondrocytes to scxa:mcherry-expressing cells, we favor a model where statin mediates the conversion of multipotent sox9a+/sox10:eGFP+ progenitors to tendon fates prior to the initiation of col2a1a:eGFP. Consistent with this interpretation, we do not observe a decrease in col2a1a:eGFP+ cells or cartilage-associated transcripts (col2a1) by qPCR (Fig. 3). This interpretation is also consistent with single cell analysis of murine neural crest cells, showing co-activation of competing fate programs prior to the commitment to a specific cell fate (Soldatov et al., 2019). Notwithstanding, given that statin increases craniofacial sox9a expression (Fig. 3A,B,G) and we did not examine col2a1 expression at later developmental stages, we cannot formally exclude the possibility that statin-mediated modulation of CNC cell fate specification may also direct cells towards a chondrocyte fate. We did observe abnormal cartilage morphology similar to previous reports of statin treatment (Signore et al., 2016). Altered scxa expression pattern was also observed for many of the chemical treatments, which could result from the chemicals targeting pathways associated with cell migration (Eisa-Beygi et al., 2014; Thorpe et al., 2004) and the cytoskeleton (Giger and David, 2017; Kardash et al., 2010; Xu et al., 2012). Interestingly, we found many of the expanded scxa-expressing cells within the periphery of the cartilage elements, corresponding to perichondrium, suggesting the expanded scxa cells arise from perichondrial cells. However, the lack of specific perichondrial markers prior to their morphological appearance makes it difficult to directly test this hypothesis.
Musculoskeletal development and the cytoskeleton
Despite the consistent statin-mediated increase in scxa+ cells, we observed differing results upon inhibition of branch points downstream in the mevalonate pathway. Inhibition of Rac GTPases alone could produce similar increases in scxa and sox9a transcripts, and had no significant effect on col2a1 expression. In contrast, inhibition of cholesterol synthesis and ROCK/Rho GTPases had no significant effect on scxa and sox9a expression. Inhibition of cholesterol synthesis, ROCK/Rho or Cdc42 had a negative effect on col2a1 expression. Consistent with our findings, conditional knockout of Cdc42 using Prx1-Cre resulted in abnormal chondrogenesis in mice (Aizawa et al., 2012). We also found that ML141-mediated inhibition of Cdc42 reduced scxa expression, whereas EHT1864-mediated inhibition of Rac increased scxa expression and quantity of scxa+ tendon progenitors (Fig. 5N-P), potentially indicating a binary relationship in which Cdc42 is required for and Rac restricts tenogenic fate. Our finding that Cdc42 inhibition reduces scxa expression is consistent with a requirement for Rho/ROCK signals in the expression of tendon markers in mammalian mesenchymal cells in vitro (Maharam et al., 2015).
Previous studies suggest a role for Rac signaling in regulating the cytoskeleton and cell-cell contacts during mesenchymal condensation, perhaps providing a permissive or restrictive environment for cell type specification (Woods et al., 2007). Condensation of mesenchymal progenitors is essential for cartilage formation and precedes their commitment to the chondrogenic lineage (DeLise and Tuan, 2002; Oberlender and Tuan, 1994). Indeed, cell morphology influences the differentiation of mesenchyme towards chondrogenic, osteogenic and adipogenic lineages (McBeath et al., 2004; Solursh et al., 1982). In tenocytes, cell morphology influences gene expression, such as that of collagen type I (Li et al., 2008; von der Mark et al., 1977), and the actin cytoskeleton functions in proper organization of the collagenous extracellular matrix (Canty et al., 2006). This model suggests a cell-autonomous role for the mevalonate pathway and Rac GTPases in tendon fate regulation, but, as these pathways can affect multiple aspects of cell behavior and signaling, it is possible that the mevalonate pathway and Rac inhibition work indirectly on neighboring cells and tissues to alter unknown pathways that result in increased tendon specification events. Nevertheless, it is tempting to speculate that Rac inhibition results in a permissive signaling environment that directly or indirectly leads to mesenchymal progenitors adopting a tendon fate.
Conclusion
Using a zebrafish small-molecule screen, we established that the mevalonate pathway is involved in regulating specification of tenogenic fate based on expansion of a tendon marker domain. Our analyses of the cellular processes governing tendon cell induction provide evidence that, besides cell migration and proliferation, the mevalonate pathway has an important role in specifying cell fate in a developing embryo. Furthermore, we propose that the emergence of the tendon lineage from a common progenitor population occurs through multipotent sox9a+/sox10+ progenitors and is mediated by inhibition of Rac activity. These results could significantly impact the field of regenerative medicine in guiding approaches aimed at generating tendon tissues in vitro. Ultimately, our studies highlight the advantages of the zebrafish system and chemical screening for discovering novel genetic pathways involved in developmental processes.
MATERIALS AND METHODS
Zebrafish husbandry
Zebrafish were staged and maintained as described previously (Kimmel et al., 1995; Westerfield, 2000). All experiments were approved by the HMS and MGH IACUC. sox10:kaede (Dougherty et al., 2012), sox10:mcherry (Kamel et al., 2013), sox10:eGFP (Curtin et al., 2011), col2a1a:eGFP (Dale and Topczewski, 2011), 6xTcf/LefBS-miniP:d2eGFP (Shimizu et al., 2012) and bre:egfp (Laux et al., 2011) were used. hmgcr1bs617 was obtained from Masazumi Tada (University College London, UK) (D'Amico et al., 2007). The scxa:mcherry transgene was generated using a bacterial artificial chromosome (BAC) containing the entire scxa genomic locus (CHORI BACPAC Resources). Using constructs and protocols provided by Stefan Schulte-Merker and Koichi Kawakami (Bussmann and Schulte-Merker, 2011; Suster et al., 2011), mcherry was recombineered into the first exon of the scxa BAC (CH211 251g8) and tol2 sites were introduced into the vector backbone. Properly targeted BACs were injected, embryos were raised to adults, and scxa:mcherry-expressing offspring were identified and raised to establish the line (fb301Tg).
Chemical screen design and validation testing
Wild-type stage-matched embryos were arrayed into 48-well plates (∼10 embryos per well) of individual compounds and exposed from 32 to 56 hpf. The NINDS Custom Collection 2 library (1040 known bioactives) was obtained from the Institute of Chemistry and Chemical Biology, Longwood Screening Facility. The compounds were dissolved in DMSO and subsequently diluted in E3 buffer to a final concentration of ∼13 μM. Each compound was screened in duplicate; DMSO and SU5402 were negative and positive controls, respectively. Whole-mount in situ hybridization for scxa expression was performed to assess craniofacial tendon progenitors. Qualitative scoring (number of embryos with altered scxa expression per number scored) was conducted using the following criteria: normal/unchanged, decreased/absent and increased. Annotations were recorded regarding any changes in the spatial domain of scxa. Most compounds (97.69%; 1016/1040) failed to alter scxa expression, although two (0.19%) were toxic and resulted in death, 10 (0.96%) resulted in developmental delay and unaltered scxa expression, three (0.29%) resulted in general and non-specific morphological abnormalities, two (0.29%) increased scxa expression, six compounds were commercially unavailable and therefore could not be validated, and one (0.10%) decreased scxa expression.
qPCR of embryonic zebrafish
Whole zebrafish embryos (n=15-125) were collected at 56 hpf following atorvastatin treatment (32-56 hpf) and dissected craniofacial regions of zebrafish embryos (n=15-45) were collected at 48 hpf following EHT-1864 treatment (32-48 hpf), at 56 hpf following RO488071, Y27632, ML141 or atorvastatin±SB-431542 treatments (32-56 hpf), and at 72 hpf following atorvastatin (48-72 hpf) or fluvastatin (32-48 hpf) treatments. Owing to high rate of toxicity, embryos collected following fluvastatin were confirmed to have a beating heart and increased scxa:mcherry expression using a dissecting microscope. Embryos were pooled per condition for RNA extraction (Qiagen, 74104). After DNase treatment (Qiagen, 79254), cDNA was synthesized (Roche, 04379012001; Invitrogen, 18091200 and Fisher Scientific, 18-091-050). For TaqMan Fast Universal gene expression assays (Applied Biosystems, 4352042), the FAM-dye probes used were: scxa (Dr03104896) and β-actin (Dr03432610). For Fast SYBR Green PCR assays (Applied Biosystems, 4385618), the primers used were: scxa forward, 5′-GGCGTCCAGTTACATCTCT-3′; scxa reverse, 5′-GTCTTTGCTCATTTTCCTCTGGTT-3′; sox9a forward, 5′-GGCCGACCAGTACCCGCACCTC-3′; sox9a reverse, 5′-GATTCGCTCTGGCCGTTCTTCACC-3′; col2a1 forward, 5′-TGCTGCAGGGCTCCAATGATGT-3′; col2a1 reverse, 5′-CTGCGCCAATGTCCACTCCGAACT-3′; myod1 forward, 5′-GAGGGAGAGGAGGCGACTGAGC-3′; myod1 reverse, 5′-CTGACACGTTGGGCCCATAAAATC-3′; col1a2 forward, 5′-GCCCCGCTGGTAAAGATGGTTCAA-3′; col1a2 reverse, 5′-ACGGCAAGTACGAGCGGGGTTCT-3′); β-actin forward, 5′-GATGCCCCTCGTGCTGTTTTC-3′; β-actin reverse, 5′-TCTCTGTTGGCTTTGGGATTCA-3′).
No-reverse transcriptase controls were used for each sample. The ratio of the target transcript to β-actin reference was quantified and normalized to the control sample. Statistical analysis was conducted with significance defined as P≤0.05 (Welch's two-sample two-tailed t-test) and error bars represent standard deviation based on all technical replicates and all biological replicates.
Morpholino design, pharmacological treatment and microinjection
Morpholinos (Gene Tools) were injected as described previously: fntβ ATG (Mapp et al., 2011), hmgcr1b ATG (D'Amico et al., 2007) and pggt1β ATG (Mapp et al., 2011). Morpholino concentrations were: fntβ (1.4 ng alone and 2.1 ng in fntβ-pggt1β morphants); pggt1β (5.5 ng alone and 2.8 ng in fntβ-pggt1β morphants); and1.0 ng hmgcr1b. Embryos were incubated in the following compounds: atorvastatin, lovastatin, fluvastatin, FTI-277 and SU5402 (Tocris Bioscience); simvastatin, aphidicolin and GGTI-286 (Sigma-Aldrich); and RO48-8071, EHT-1864, ML-141, Y-27632 and SB-431542 (Cayman Chemical). In the metabolite rescue experiments, the pharyngeal cavity was injected at 28-30 hpf with 1-2 nl of the following compounds (Sigma-Aldrich): 2.6 mM geranylgeranyl pyrophosphate, 10 mM farnesyl pyrophosphate, 1.0 M mevalonolactone and DMSO or ethanol (vehicle control).
Expression and proliferation analysis
Colorimetric whole-mount in situ hybridization was performed as described previously (Brent et al., 2003). Probes include scxa (AL923903 and AL921296), col1a2 (DY559926), tnmd (BC155615 and EV754577), myod1 (BC078421), sox9a (Yan et al., 2005), col2a1 (Yan et al., 1995), runx2a (Burns et al., 2002), hoxa2b (Kong et al., 2014), hoxb2a (Kong et al., 2014), hand2 (Foote et al., 2016), bapx1 (Kamel et al., 2013), tgfbi (CB361081), tgfbr2a (Thisse et al., 2001), pea3 (Münchberg et al., 1999), fzd7a (Kamel et al., 2013), fzd7b (Rochard et al., 2016), shha (Piotrowski and Nüsslein-Volhard, 2000), patched1 (Lewis et al., 1999), bmp4 (Gonzalez et al., 2000), ednraa (Nair et al., 2007) and gsc (Erter et al., 1998). Fluorescent whole-mount in situ hybridization and antibody staining were performed as described previously (Chen and Galloway, 2017). Primary antibodies were 1:100 anti-PSmad3 (Abcam, ab52903); 1:500 anti-kaede (MBL, PM012M); and 1:200 anti-phospho-Histone H3 (Millipore, 06-570). The sensitivity of the anti-kaede antibody marks the descendants of the sox10:kaede+ lineage that have turned off sox10:kaede expression. Secondary antibodies were 1:500 goat anti-mouse IgG2b-HRP (Southern Biotech); 1:200 donkey anti-rabbit Alexa Fluor 488 and 1:500 donkey anti-rabbit IgG-HRP (Jackson ImmunoResearch Laboratories); 1:400 goat anti-mouse Alexa Fluor 488 (Invitrogen); and 1:400 donkey anti-rabbit Alexa Fluor 647 (Abcam). All images captured were of representative embryos for each condition.
Quantification of EdU (5-ethynyl-2′-deoxyuridine)-positive cells
Tg(sox10:mcherry) embryos were chemically treated at 32 hpf, pulsed with EdU for 20 min at 4 h or 5 h intervals, and chased with E3 buffer for 30 min prior to fixation. Embryos were counterstained with Hoechst33342 (Invitrogen). For quantification, we used confocal microscopy and counted the number of proliferative cells in optical sections spanning the depth of a defined craniofacial region of the embryos processed by Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen, C10337). Statistical analysis was conducted with significance defined as P≤0.05 (Welch's two-sample two-tailed t-test) and error bars are standard deviation.
Quantification of scxa-, col1a2- and PH3-expressing cells
For quantification of craniofacial scxa- and col1a2-expressing cells, we counted the number of cells in optical confocal sections spanning the area of the craniofacial region of embryos stained for the transcript of interest by fluorescent whole-mount in situ hybridization and counterstained with Hoechst33342. For quantification of pectoral fin and myoseptal scxa-expressing cells, we counted the number of mcherry+ cells in maximum-intensity projection images spanning the width of the pectoral fin or trunk region, respectively, of Tg(scxa:mcherry) embryos. For quantification of cells co-expressing scxa transcript and PH3 protein (Fig. S3I,L-Q), we counted the number of cells in optical sections spanning the depth of a defined region of the embryos processed by fluorescent whole-mount in situ hybridization and immunohistochemistry. Statistical analysis was conducted using the Mann–Whitney-Wilcoxon test with significance defined as P≤0.05.
Image analysis and live imaging
Embryos were imaged using a Zeiss AxioZoom V16 with ApoTome2 or Nikon Eclipse 80i, and images were acquired using Zeiss Zen or NIS Elements. Confocal time-course images were taken using a Nikon ECLIPSE Ti-E and confocal images were taken using a Zeiss LSM 710 NLO or Nikon ECLIPSE Ti-E. Confocal images were acquired with Zeiss Zen or ImageJ software using the maximum-intensity projection (MIP) feature applied to z-stacks and processed uniformly with Photoshop. Tg(bre:egfp), Tg(scxa:mcherry), Tg(col2a1a:eGFP; scxa:mcherry) and Tg(6XTcf/LefBS-miniP:d2eGFP) embryos were mounted in 0.8-1.0% low melting-point agarose (RPI, 9012-36-6) in E3 buffer. Embryos for live imaging were anesthetized with 0.015-0.2% tricaine in E3 buffer. Embryos, positioned with the ventral side of the head facing the objective, were imaged every 1 h using a z-stack module. Post-imaging, the embryos resumed normal development at 28.5°C in fresh E3 buffer.
Phospho-Smad3 staining
56 hpf embryos were fixed in 4% PFA overnight and then rinsed in PBST (PBS+0.15% Triton X-100) for 3×10 min at room temperature (RT). Embryos were next permeated in 20 μg/ml Proteinase K (Thermo Fisher Scientific, 507203027) for 25 min at room temperature and rinsed in PBST for 3×10 min. Afterwards, the embryos were blocked with 4% BSA (ThermoFisher Scientific, BP1605-100) in PBST for 2 h at room temperature and subsequently incubated with anti-Smad3 antibody overnight at 4°C. Embryos were washed with PBST for at least 2 h, with the PBST changed every 30 min, and then incubated with donkey anti-rabbit Alexa Fluor 647 overnight at 4°C. Hoechst (ThermoFisher Scientific, H3570) was added to the secondary antibody solution for nuclear staining.
Fluorescent whole-mount in situ hybridization
Fluorescent whole-mount in situ hybridization was performed as previously described (Chen and Galloway, 2017) using DIG- (Roche, 11277073910) and fluorescein- (Roche, 11685619910) labeled RNA probes against sox9a and mcherry. Probes were synthesized using Sp6 or T7 RNA polymerase (Roche, 10810274001 or 10881767001). Signals were visualized using a TSA Plus Fluorescein Evaluation Kit (PerkinElmer, NEL741E001KT) and TSA Cyanine 5 Evaluation Kit (PerkinElmer, NEL705A001KT).
Flow cytometry
The Tg(col2a1a:eGFP; scxa:mcherry) embryos were treated with DMSO or atorvastatin from 32 to 56 hpf. At 56 hpf, 15-20 pooled heads from each group were collected for single cell dissociation as previously described (Bresciani et al., 2018). The ratio of each population: scxa:mcherry+ cells, col2a1a:eGFP+ cells and scxa:mcherry+/col2a1a:eGFP+ cells were analyzed using a BD FACS Aria 3 Cell Sorter equipped with ultraviolet, green and red lasers. Gates were set using positive (single transgene for scxa:mcherry and col2a1a:eGFP) and negative (wild-type) control cell populations.
Supplementary Material
Acknowledgements
We thank Caroline Shamu, Jennifer Smith, David Wrobel and Doug Flood, and personnel at the ICCB Longwood, for assistance and guidance with the screen. We are grateful to Jessica Lehoczky, Mor Grinstein, Heather Dingwall, the BIDMC and MGH Simches fish facilities, Trista North, Wolfram Goessling, and North and Goessling lab members.
Footnotes
Competing interests
J.W.C. is currently a full-time employee at Genentech and holds stocks in Roche.
Author contributions
Conceptualization: J.W.C., X.N., C.J.T., J.L.G.; Methodology: J.W.C., X.N., M.J.K., M.-T.N., J.L.G.; Formal analysis: J.W.C., X.N., M.J.K., J.L.G.; Investigation: J.W.C., X.N., M.J.K., M.-T.N., J.L.G.; Resources: C.J.T., J.L.G.; Data curation: J.W.C., J.L.G.; Writing - original draft: J.W.C., X.N., C.J.T., J.L.G.; Writing - review & editing: J.W.C., X.N., M.J.K., M.-T.N., C.J.T., J.L.G.; Visualization: J.W.C., X.N., J.L.G.; Supervision: C.J.T., J.L.G.; Project administration: J.L.G.; Funding acquisition: J.L.G.
Funding
This work was supported by the National Institutes of Health (HD03443 to C.J.T.; AR074541, HD069533 and DE024771), by awards from the Charles H. Hood Foundation and the Harvard Stem Cell Institute to J.L.G., and by a National Science Foundation Graduate Research Fellowship grant (DGE1144152 to J.W.C.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.185389.supplemental
References
- Aizawa R., Yamada A., Suzuki D., Iimura T., Kassai H., Harada T., Tsukasaki M., Yamamoto G., Tachikawa T., Nakao K. et al. (2012). Cdc42 is required for chondrogenesis and interdigital programmed cell death during limb development. Mech. Dev. 129, 38-50. 10.1016/j.mod.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Akiyama H., Kim J.-E., Nakashima K., Balmes G., Iwai N., Deng J. M., Zhang Z., Martin J. F., Behringer R. R., Nakamura T. et al. (2005). Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. USA 102, 14665-14670. 10.1073/pnas.0504750102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroffio A., Dupin E. and Le Douarin N. M. (1991). Common precursors for neural and mesectodermal derivatives in the cephalic neural crest. Development 112, 301-305. [DOI] [PubMed] [Google Scholar]
- Blitz E., Sharir A., Akiyama H. and Zelzer E. (2013). Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development 140, 2680-2690. 10.1242/dev.093906 [DOI] [PubMed] [Google Scholar]
- Brand-Saberi B. and Christ B. (2000). Evolution and development of distinct cell lineages derived from somites. Curr. Top. Dev. Biol. 48, 1-42. 10.1016/S0070-2153(08)60753-X [DOI] [PubMed] [Google Scholar]
- Brent A. E. and Tabin C. J. (2004). FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development 131, 3885-3896. 10.1242/dev.01275 [DOI] [PubMed] [Google Scholar]
- Brent A. E., Schweitzer R. and Tabin C. J. (2003). A somitic compartment of tendon progenitors. Cell 113, 235-248. 10.1016/S0092-8674(03)00268-X [DOI] [PubMed] [Google Scholar]
- Brent A. E., Braun T. and Tabin C. J. (2005). Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development 132, 515-528. 10.1242/dev.01605 [DOI] [PubMed] [Google Scholar]
- Bresciani E., Broadbridge E. and Liu P. P. (2018). An efficient dissociation protocol for generation of single cell suspension from zebrafish embryos and larvae. MethodsX 5, 1287-1290. 10.1016/j.mex.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. E., DeBlasio T., Zhou Y., Zhang J., Zon L. and Nimer S. D. (2002). Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators. Exp. Hematol. 30, 1381-1389. 10.1016/S0301-472X(02)00955-4 [DOI] [PubMed] [Google Scholar]
- Bussmann J. and Schulte-Merker S. (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development 138, 4327-4332. 10.1242/dev.068080 [DOI] [PubMed] [Google Scholar]
- Canty E. G., Starborg T., Lu Y., Humphries S. M., Holmes D. F., Meadows R. S., Huffman A., O'Toole E. T. and Kadler K. E. (2006). Actin filaments are required for fibripositor-mediated collagen fibril alignment in tendon. J. Biol. Chem. 281, 38592-38598. 10.1074/jbc.M607581200 [DOI] [PubMed] [Google Scholar]
- Carney T. J., Dutton K. A., Greenhill E., Delfino-Machin M., Dufourcq P., Blader P. and Kelsh R. N. (2006). A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 133, 4619-4630. 10.1242/dev.02668 [DOI] [PubMed] [Google Scholar]
- Chen J. W. and Galloway J. L. (2014). The development of zebrafish tendon and ligament progenitors. Development 141, 2035-2045. 10.1242/dev.104067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. W. and Galloway J. L. (2017). Using the zebrafish to understand tendon development and repair. Methods Cell Biol. 138, 299-320. 10.1016/bs.mcb.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini A., Mazzotti M., Raiteri M., Soma M. R., Gabbiani G., Fumagalli R. and Paoletti R. (1993). Relationship between mevalonate pathway and arterial myocyte proliferation: in vitro studies with inhibitors of HMG-CoA reductase. Atherosclerosis 101, 117-125. 10.1016/0021-9150(93)90107-6 [DOI] [PubMed] [Google Scholar]
- Couly G. F., Coltey P. M. and Le Douarin N. M. (1993). The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development 117, 409-429. [DOI] [PubMed] [Google Scholar]
- Curtin E., Hickey G., Kamel G., Davidson A. J. and Liao E. C. (2011). Zebrafish wnt9a is expressed in pharyngeal ectoderm and is required for palate and lower jaw development. Mech. Dev. 128, 104-115. 10.1016/j.mod.2010.11.003 [DOI] [PubMed] [Google Scholar]
- Dale R. M. and Topczewski J. (2011). Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev. Biol. 357, 518-531. 10.1016/j.ydbio.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico L., Scott I. C., Jungblut B. and Stainier D. Y. R. (2007). A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr. Biol. 17, 252-259. 10.1016/j.cub.2006.12.023 [DOI] [PubMed] [Google Scholar]
- DeLise A. M. and Tuan R. S. (2002). Alterations in the spatiotemporal expression pattern and function of N-cadherin inhibit cellular condensation and chondrogenesis of limb mesenchymal cells in vitro. J. Cell. Biochem. 87, 342-359. 10.1002/jcb.10308 [DOI] [PubMed] [Google Scholar]
- Dougherty M., Kamel G., Shubinets V., Hickey G., Grimaldi M. and Liao E. C. (2012). Embryonic fate map of first pharyngeal arch structures in the sox10: kaede zebrafish transgenic model. J. Craniofac. Surg. 23, 1333-1337. 10.1097/SCS.0b013e318260f20b [DOI] [PubMed] [Google Scholar]
- Edom-Vovard F. and Duprez D. (2004). Signals regulating tendon formation during chick embryonic development. Dev. Dyn. 229, 449-457. 10.1002/dvdy.10481 [DOI] [PubMed] [Google Scholar]
- Eisa-Beygi S., Ekker M., Moon T. W., Macdonald R. L. and Wen X.-Y. (2014). Developmental processes regulated by the 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) pathway: highlights from animal studies. Reprod. Toxicol. 46, 115-120. 10.1016/j.reprotox.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Erter C. E., Solnica-Krezel L. and Wright C. V. E. (1998). Zebrafish nodal-related 2 encodes an early mesendodermal inducer signaling from the extraembryonic yolk syncytial layer. Dev. Biol. 204, 361-372. 10.1006/dbio.1998.9097 [DOI] [PubMed] [Google Scholar]
- Foote A. D., Vijay N., Ávila-Arcos M. C., Baird R. W., Durban J. W., Fumagalli M., Gibbs R. A., Hanson M. B., Korneliussen T. S., Martin M. D. et al. (2016). Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat. Commun. 7, 11693 10.1038/ncomms11693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger F. A. and David N. B. (2017). Endodermal germ-layer formation through active actin-driven migration triggered by N-cadherin. Proc. Natl. Acad. Sci. USA 114, 10143-10148. 10.1073/pnas.1708116114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessling W. and North T. E. (2014). Repairing quite swimmingly: advances in regenerative medicine using zebrafish. Dis. Model. Mech. 7, 769-776. 10.1242/dmm.016352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L. and Brown M. S. (1990). Regulation of the mevalonate pathway. Nature 343, 425-430. 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- Gong L., Xiao Y., Xia F., Wu P., Zhao T., Xie S., Wang R., Wen Q., Zhou W., Xu H. et al. (2019). The mevalonate coordinates energy input and cell proliferation. Cell Death Dis. 10, 327 10.1038/s41419-019-1544-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E. M., Fekany-Lee K., Carmany-Rampey A., Erter C., Topczewski J., Wright C. V. and Solnica-Krezel L. (2000). Head and trunk in zebrafish arise via coinhibition of BMP signaling by bozozok and chordino. Genes Dev. 14, 3087-3092. 10.1101/gad.852400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier J., Teillet M.-A., Grifone R., Kelly R. G. and Duprez D. (2009). Relationship between neural crest cells and cranial mesoderm during head muscle development. PLoS ONE 4, e4381 10.1371/journal.pone.0004381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis A. E., Heersche J. N. M. and Aubin J. E. (1990). Continuously growing bipotential and monopotential myogenic, adipogenic, and chondrogenic subclones isolated from the multipotential RCJ 3.1 clonal cell line. Dev. Biol. 142, 313-318. 10.1016/0012-1606(90)90352-J [DOI] [PubMed] [Google Scholar]
- Havis E., Bonnin M.-A., Olivera-Martinez I., Nazaret N., Ruggiu M., Weibel J., Durand C., Guerquin M.-J., Bonod-Bidaud C., Ruggiero F. et al. (2014). Transcriptomic analysis of mouse limb tendon cells during development. Development 141, 3683-3696. 10.1242/dev.108654 [DOI] [PubMed] [Google Scholar]
- Havis E., Bonnin M.-A., Esteves de Lima J., Charvet B., Milet C. and Duprez D. (2016). TGFbeta and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development 143, 3839-3851. 10.1242/dev.136242 [DOI] [PubMed] [Google Scholar]
- Hong L., Kenney S. R., Phillips G. K., Simpson D., Schroeder C. E., Nöth J., Romero E., Swanson S., Waller A., Strouse J. J. et al. (2013). Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem. 288, 8531-8543. 10.1074/jbc.M112.435941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. H., Watson S. S., Wang L., Baker B. M., Akiyama H., Brigande J. V. and Schweitzer R. (2019). Requirement for scleraxis in the recruitment of mesenchymal progenitors during embryonic tendon elongation. Development 146, dev182782 10.1242/dev.182782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. P. and Prince V. E. (2002). Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev. Biol. 247, 367-389. 10.1006/dbio.2002.0701 [DOI] [PubMed] [Google Scholar]
- Hurle J. M., Gañan Y. and Macias D. (1989). Experimental analysis of the in vivo chondrogenic potential of the interdigital mesenchyme of the chick leg bud subjected to local ectodermal removal. Dev. Biol. 132, 368-374. 10.1016/0012-1606(89)90233-9 [DOI] [PubMed] [Google Scholar]
- Hurle J. M., Ros M. A., Gañan Y., Macias D., Critchlow M. and Hinchliffe J. R. (1990). Experimental analysis of the role of ECM in the patterning of the distal tendons of the developing limb bud. Cell Differ. Dev. 30, 97-108. 10.1016/0922-3371(90)90078-B [DOI] [PubMed] [Google Scholar]
- Igel M., Sudhop T. and von Bergmann K. (2001). Metabolism and drug interactions of 3-hydroxy-3-methylglutaryl coenzyme A-reductase inhibitors (statins). Eur. J. Clin. Pharmacol. 57, 357-364. 10.1007/s002280100329 [DOI] [PubMed] [Google Scholar]
- Inman G. J., Nicolás F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J. and Hill C. S. (2002). SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65-74. 10.1124/mol.62.1.65 [DOI] [PubMed] [Google Scholar]
- Istvan E. S. and Deisenhofer J. (2001). Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292, 1160-1164. 10.1126/science.1059344 [DOI] [PubMed] [Google Scholar]
- Jones K. D., Couldwell W. T., Hinton D. R., Su Y. H., He S. K., Anker L. and Law R. E. (1994). Lovastatin induces growth inhibition and apoptosis in human malignant glioma cells. Biochem. Biophys. Res. Commun. 205, 1681-1687. 10.1006/bbrc.1994.2861 [DOI] [PubMed] [Google Scholar]
- Jozsa L. and Kannus P. (1997). Human Tendons: Anatomy, Physiology, and Pathology. Human Kinetics: Champaign, IL. [Google Scholar]
- Kamel G., Hoyos T., Rochard L., Dougherty M., Kong Y., Tse W., Shubinets V., Grimaldi M. and Liao E. C. (2013). Requirement for frzb and fzd7a in cranial neural crest convergence and extension mechanisms during zebrafish palate and jaw morphogenesis. Dev. Biol. 381, 423-433. 10.1016/j.ydbio.2013.06.012 [DOI] [PubMed] [Google Scholar]
- Kardash E., Reichman-Fried M., Maitre J. L., Boldajipour B., Papusheva E., Messerschmidt E. M., Heisenberg C. P. and Raz E. (2010). A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 12, 47-53; sup pp 41-11 10.1038/ncb2003 [DOI] [PubMed] [Google Scholar]
- Kieny M. and Chevallier A. (1979). Autonomy of tendon development in the embryonic chick wing. J. Embryol. Exp. Morphol. 49, 153-165. [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Klotz U. (2003). Pharmacological comparison of the statins. Arzneimittelforschung 53, 605-611. 10.1055/s-0031-1297156 [DOI] [PubMed] [Google Scholar]
- Kong Y., Grimaldi M., Curtin E., Dougherty M., Kaufman C., White R. M., Zon L. I. and Liao E. C. (2014). Neural crest development and craniofacial morphogenesis is coordinated by nitric oxide and histone acetylation. Chem. Biol. 21, 488-501. 10.1016/j.chembiol.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontges G. and Lumsden A. (1996). Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development 122, 3229-3242. [DOI] [PubMed] [Google Scholar]
- Ladstein R. G., Bachmann I. M., Straume O. and Akslen L. A. (2010). Ki-67 expression is superior to mitotic count and novel proliferation markers PHH3, MCM4 and mitosin as a prognostic factor in thick cutaneous melanoma. BMC Cancer 10, 140 10.1186/1471-2407-10-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux D. W., Febbo J. A. and Roman B. L. (2011). Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev. Dyn. 240, 682-694. 10.1002/dvdy.22558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. M. and Kalcheim C. (1999). The Neural Crest, 2nd edn. Cambridge University Press. [Google Scholar]
- Le Douarin N. M. and Smith J. (1988). Development of the peripheral nervous system from the neural crest. Annu. Rev. Cell Biol. 4, 375-404. 10.1146/annurev.cb.04.110188.002111 [DOI] [PubMed] [Google Scholar]
- Le Lievre C. S. (1978). Participation of neural crest-derived cells in the genesis of the skull in birds. J. Embryol. Exp. Morphol. 47, 17-37. [PubMed] [Google Scholar]
- Léjard V., Brideau G., Blais F., Salingcarnboriboon R., Wagner G., Roehrl M. H. A., Noda M., Duprez D., Houillier P. and Rossert J. (2007). Scleraxis and NFATc regulate the expression of the pro-α1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 282, 17665-17675. 10.1074/jbc.M610113200 [DOI] [PubMed] [Google Scholar]
- Lerner E. C., Qian Y., Hamilton A. D. and Sebti S. M. (1995). Disruption of oncogenic K-Ras4B processing and signaling by a potent geranylgeranyltransferase I inhibitor. J. Biol. Chem. 270, 26770-26773. 10.1074/jbc.270.45.26770 [DOI] [PubMed] [Google Scholar]
- Lewis K. E., Drossopoulou G., Paton I. R., Morrice D. R., Robertson K. E., Burt D. W., Ingham P. W. and Tickle C. (1999). Expression of ptc and gli genes in talpid3 suggests bifurcation in Shh pathway. Development 126, 2397-2407. [DOI] [PubMed] [Google Scholar]
- Li F., Li B., Wang Q.-M. and Wang J. H.-C. (2008). Cell shape regulates collagen type I expression in human tendon fibroblasts. Cell Motil. Cytoskeleton 65, 332-341. 10.1002/cm.20263 [DOI] [PubMed] [Google Scholar]
- Lin C.-Y., Yung R.-F., Lee H.-C., Chen W.-T., Chen Y.-H. and Tsai H.-J. (2006). Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev. Biol. 299, 594-608. 10.1016/j.ydbio.2006.08.042 [DOI] [PubMed] [Google Scholar]
- Lumsden A., Sprawson N. and Graham A. (1991). Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development 113, 1281-1291. [DOI] [PubMed] [Google Scholar]
- Maharam E., Yaport M., Villanueva N. L., Akinyibi T., Laudier D., He Z., Leong D. J. and Sun H. B. (2015). Rho/Rock signal transduction pathway is required for MSC tenogenic differentiation. Bone Res. 3, 15015 10.1038/boneres.2015.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp O. M., Walsh G. S., Moens C. B., Tada M. and Prince V. E. (2011). Zebrafish Prickle1b mediates facial branchiomotor neuron migration via a farnesylation-dependent nuclear activity. Development 138, 2121-2132. 10.1242/dev.060442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias J. R., Saxena M. T. and Mumm J. S. (2012). Advances in zebrafish chemical screening technologies. Future Med. Chem. 4, 1811-1822. 10.4155/fmc.12.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K. and Chen C. S. (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483-495. 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- McGurk P. D., Swartz M. E., Chen J. W., Galloway J. L. and Eberhart J. K. (2017). In vivo zebrafish morphogenesis shows Cyp26b1 promotes tendon condensation and musculoskeletal patterning in the embryonic jaw. PLoS Genet. 13, e1007112 10.1371/journal.pgen.1007112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart S. J. (2006). Isoprenylated proteins. Cell. Mol. Life Sci. 63, 255-267. 10.1007/s00018-005-5298-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T., Yelon D., Stainier D. Y. R. and Kimmel C. B. (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 130, 1353-1365. 10.1242/dev.00339 [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A.-H. and Le Douarin N. (2000). Duality of molecular signaling involved in vertebral chondrogenesis. Curr. Top. Dev. Biol. 48, 43-75. 10.1016/S0070-2153(08)60754-1 [DOI] [PubMed] [Google Scholar]
- Morand O. H., Aebi J. D., Dehmlow H., Ji Y. H., Gains N., Lengsfeld H. and Himber J. (1997). Ro 48-8.071, a new 2,3-oxidosqualene:lanosterol cyclase inhibitor lowering plasma cholesterol in hamsters, squirrel monkeys, and minipigs: comparison to simvastatin. J. Lipid Res. 38, 373-390. [PubMed] [Google Scholar]
- Mori-Akiyama Y., Akiyama H., Rowitch D. H. and de Crombrugghe B. (2003). Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc. Natl. Acad. Sci. USA 100, 9360-9365. 10.1073/pnas.1631288100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münchberg S. R., Ober E. A. and Steinbeisser H. (1999). Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech. Dev. 88, 233-236. 10.1016/S0925-4773(99)00179-3 [DOI] [PubMed] [Google Scholar]
- Murchison N. D., Price B. A., Conner D. A., Keene D. R., Olson E. N., Tabin C. J. and Schweitzer R. (2007). Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697-2708. 10.1242/dev.001933 [DOI] [PubMed] [Google Scholar]
- Nair S., Li W., Cornell R. and Schilling T. F. (2007). Requirements for Endothelin type-A receptors and Endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development 134, 335-345. 10.1242/dev.02704 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R. and de Crombrugghe B. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17-29. 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- Nichols J. T., Blanco-Sánchez B., Brooks E. P., Parthasarathy R., Dowd J., Subramanian A., Nachtrab G., Poss K. D., Schilling T. F. and Kimmel C. B. (2016). Ligament versus bone cell identity in the zebrafish hyoid skeleton is regulated by mef2ca. Development 143, 4430-4440. 10.1242/dev.141036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden D. M. (1978). The control of avian cephalic neural crest cytodifferentiation: I. Skeletal and connective tissues. Dev. Biol. 67, 296-312. 10.1016/0012-1606(78)90201-4 [DOI] [PubMed] [Google Scholar]
- North T. E., Goessling W., Walkley C. R., Lengerke C., Kopani K. R., Lord A. M., Weber G. J., Bowman T. V., Jang I.-H., Grosser T. et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007-1011. 10.1038/nature05883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlender S. A. and Tuan R. S. (1994). Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development 120, 177-187. [DOI] [PubMed] [Google Scholar]
- Ohashi K., Osuga J.-I., Tozawa R., Kitamine T., Yagyu H., Sekiya M., Tomita S., Okazaki H., Tamura Y., Yahagi N. et al. (2003). Early embryonic lethality caused by targeted disruption of the 3-hydroxy-3-methylglutaryl-CoA reductase gene. J. Biol. Chem. 278, 42936-42941. 10.1074/jbc.M307228200 [DOI] [PubMed] [Google Scholar]
- Oka K., Oka S., Hosokawa R., Bringas P. Jr., Brockhoff H. C. II, Nonaka K. and Chai Y. (2008). TGF-β mediated Dlx5 signaling plays a crucial role in osteo-chondroprogenitor cell lineage determination during mandible development. Dev. Biol. 321, 303-309. 10.1016/j.ydbio.2008.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse R. V. II, Scherz P. J., Campbell J. K. and Tabin C. J. (2007). A cellular lineage analysis of the chick limb bud. Dev. Biol. 310, 388-400. 10.1016/j.ydbio.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T. and Nüsslein-Volhard C. (2000). The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio). Dev. Biol. 225, 339-356. 10.1006/dbio.2000.9842 [DOI] [PubMed] [Google Scholar]
- Pryce B. A., Watson S. S., Murchison N. D., Staverosky J. A., Dunker N. and Schweitzer R. (2009). Recruitment and maintenance of tendon progenitors by TGFβ signaling are essential for tendon formation. Development 136, 1351-1361. 10.1242/dev.027342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochard L., Monica S. D., Ling I. T. C., Kong Y., Roberson S., Harland R., Halpern M. and Liao E. C. (2016). Roles of Wnt pathway genes wls, wnt9a, wnt5b, frzb and gpc4 in regulating convergent-extension during zebrafish palate morphogenesis. Development 143, 2541-2547. 10.1242/dev.137000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros M. A., Rivero F. B., Hinchliffe J. R. and Hurle J. M. (1995). Immunohistological and ultrastructural study of the developing tendons of the avian foot. Anat. Embryol. 192, 483-496. 10.1007/BF00187179 [DOI] [PubMed] [Google Scholar]
- Salic A. and Mitchison T. J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105, 2415-2420. 10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. C. and Lehmann R. (2004). Isoprenoids control germ cell migration downstream of HMGCoA reductase. Dev. Cell 6, 283-293. 10.1016/S1534-5807(04)00023-1 [DOI] [PubMed] [Google Scholar]
- Saunders J. W., Jr (1948). The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. J. Exp. Zool. 108, 363-403. 10.1002/jez.1401080304 [DOI] [PubMed] [Google Scholar]
- Schilling T. F. and Kimmel C. B. (1994). Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 120, 483-494. [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Chyung J. H., Murtaugh L. C., Brent A. E., Rosen V., Olson E. N., Lassar A. and Tabin C. J. (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855-3866. [DOI] [PubMed] [Google Scholar]
- Shimizu N., Kawakami K. and Ishitani T. (2012). Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev. Biol. 370, 71-85. 10.1016/j.ydbio.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Shutes A., Onesto C., Picard V., Leblond B., Schweighoffer F. and Der C. J. (2007). Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J. Biol. Chem. 282, 35666-35678. 10.1074/jbc.M703571200 [DOI] [PubMed] [Google Scholar]
- Signore I. A., Jerez C., Figueroa D., Suazo J., Marcelain K., Cerda O. and Colombo Flores A. (2016). Inhibition of the 3-hydroxy-3-methyl-glutaryl-CoA reductase induces orofacial defects in zebrafish. Birth Defects Res. A Clin. Mol. Teratol 106, 814-830. 10.1002/bdra.23546 [DOI] [PubMed] [Google Scholar]
- Soeda T., Deng J. M., de Crombrugghe B., Behringer R. R., Nakamura T. and Akiyama H. (2010). Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635-644. 10.1002/dvg.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov R., Kaucka M., Kastriti M. E., Petersen J., Chontorotzea T., Englmaier L., Akkuratova N., Yang Y., Häring M., Dyachuk V. et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Solursh M., Linsenmayer T. F. and Jensen K. L. (1982). Chondrogenesis from single limb mesenchyme cells. Dev. Biol. 94, 259-264. 10.1016/0012-1606(82)90090-2 [DOI] [PubMed] [Google Scholar]
- Spadari S., Pedrali-Noy G., Ciomei M., Falaschi A. and Ciarrocchi G. (1984). Control of DNA replication and cell proliferation in eukaryotes by aphidicolin. Toxicol. Pathol. 12, 143-148. 10.1177/019262338401200205 [DOI] [PubMed] [Google Scholar]
- Stringa E. and Tuan R. S. (1996). Chondrogenic cell subpopulation of chick embryonic calvarium: isolation by peanut agglutinin affinity chromatography and in vitro characterization. Anat. Embryol. 194, 427-437. 10.1007/BF00185990 [DOI] [PubMed] [Google Scholar]
- Subramanian A. and Schilling T. F. (2014). Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. eLife 3, e02372 10.7554/eLife.02372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y., Takimoto A., Akiyama H., Kist R., Scherer G., Nakamura T., Hiraki Y. and Shukunami C. (2013). Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 140, 2280-2288. 10.1242/dev.096354 [DOI] [PubMed] [Google Scholar]
- Suster M. L., Abe G., Schouw A. and Kawakami K. (2011). Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6, 1998-2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Tan G.-K., Pryce B. A., Stabio A., Brigande J. V., Wang C. J., Xia Z., Tufa S. F., Keene D. R. and Schweitzer R. (2020). Tgfβ signaling is critical for maintenance of the tendon cell fate. eLife 9, e52695 10.7554/eLife.52695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Brugmann S. A., Helms J. A. and Nusse R. (2008). Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135, 3247-3257. 10.1242/dev.023176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X. Q. et al. (2001). Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402). ZFIN Direct Data Submission. https://zfin.org/ZDB-PUB-010810-1
- Thorpe J. L., Doitsidou M., Ho S.-Y., Raz E. and Farber S. A. (2004). Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev. Cell 6, 295-302. 10.1016/S1534-5807(04)00032-2 [DOI] [PubMed] [Google Scholar]
- Trainor P. A. and Krumlauf R. (2000). Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 1, 116-124. 10.1038/35039056 [DOI] [PubMed] [Google Scholar]
- Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., Tamakawa H., Yamagami K., Inui J., Maekawa M. et al. (1997). Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389, 990-994. 10.1038/40187 [DOI] [PubMed] [Google Scholar]
- von der Mark K., Gauss V., von der Mark H. and Müller P. (1977). Relationship between cell shape and type of collagen synthesised as chondrocytes lose their cartilage phenotype in culture. Nature 267, 531-532. 10.1038/267531a0 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide for the Laboratory use of Zebrafish (Danio rerio), 4th edn. Eugene, OR: University of Oregon Press. [Google Scholar]
- Woods A., Wang G., Dupuis H., Shao Z. and Beier F. (2007). Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J. Biol. Chem. 282, 23500-23508. 10.1074/jbc.M700680200 [DOI] [PubMed] [Google Scholar]
- Wortham R. A. (1948). The development of the muscles and tendons in the lower leg and foot of chick embryos. J. Morphol. 83, 105-148. 10.1002/jmor.1050830106 [DOI] [PubMed] [Google Scholar]
- Xu H., Kardash E., Chen S., Raz E. and Lin F. (2012). Gbetagamma signaling controls the polarization of zebrafish primordial germ cells by regulating Rac activity. Development 139, 57-62. 10.1242/dev.073924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Shiraishi Y.-I. and Kuroiwa A. (2013). Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Dev. Biol. 377, 363-374. 10.1016/j.ydbio.2013.02.023 [DOI] [PubMed] [Google Scholar]
- Yan Y.-L., Hatta K., Riggleman B. and Postlethwait J. H. (1995). Expression of a type II collagen gene in the zebrafish embryonic axis. Dev. Dyn. 203, 363-376. 10.1002/aja.1002030308 [DOI] [PubMed] [Google Scholar]
- Yan Y. L., Miller C. T., Nissen R. M., Singer A., Liu D., Kirn A., Draper B., Willoughby J., Morcos P. A., Amsterdam A. et al. (2002). A zebrafish sox9 gene required for cartilage morphogenesis. Development 129, 5065-5079. [DOI] [PubMed] [Google Scholar]
- Yan Y.-L., Willoughby J., Liu D., Crump J. G., Wilson C., Miller C. T., Singer A., Kimmel C., Westerfield M. and Postlethwait J. H. (2005). A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069-1083. 10.1242/dev.01674 [DOI] [PubMed] [Google Scholar]
- Yi P., Han Z., Li X. and Olson E. N. (2006). The mevalonate pathway controls heart formation in Drosophila by isoprenylation of Ggamma1. Science 313, 1301-1303. 10.1126/science.1127704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.