Abstract
The main neurological manifestation of COVID-19 is loss of smell or taste. The high incidence of smell loss without significant rhinorrhea or nasal congestion suggests that SARS-CoV-2 targets the chemical senses through mechanisms distinct from those used by endemic coronaviruses or other common cold-causing agents. Here we review recently developed hypotheses about how SARS-CoV-2 might alter the cells and circuits involved in chemosensory processing and thereby change perception. Given our limited understanding of SARS-CoV-2 pathogenesis, we propose future experiments to elucidate disease mechanisms and highlight the relevance of this ongoing work to understanding how the virus might alter brain function more broadly.
Impairments of smell and taste perception are two of the main symptoms of COVID–19. In this issue of Neuron, Cooper et al. focus on these chemosensory symptoms and review recent hypotheses of the putative mechanisms mediating the loss of taste and smell through COVID–19.
Main Text
Introduction
Disturbances in smell and taste have emerged as the predominant neurological symptom of the coronavirus disease 2019 (COVID-19), which is caused by SARS-CoV-2. Perhaps as many as 80% or more of patients infected with SARS-CoV-2 report anosmia, hyposmia, ageusia, dysgeusia, or changes in chemesthesis (the ability to sense chemical irritants) (Giacomelli et al., 2020; Kaye et al., 2020; Lechien et al., 2020a; Parma et al., 2020; Spinato et al., 2020; Yan et al., 2020). Self-reported changes in chemical perception can predict whether a subject will test positive for SARS-CoV-2 (Bénézit et al., 2020; Fontanet et al., 2020; Haehner et al., 2020; Moein et al., 2020; Wagner et al., 2020); one recent observational study that included more than two million participants revealed that the loss of smell and taste is more predictive than all other symptoms, including fatigue, fever, or cough (Menni et al., 2020). Most of these studies have lacked objective chemosensory assessment, raising the possibility that chemosensory disturbances are even more prevalent than currently appreciated; indeed, smell testing reveals increased odor detection thresholds in a subset of COVID-19 patients who subjectively report a normal sense of smell (Hornuss et al., 2020; Iravani et al., 2020; Moein et al., 2020; Qiu et al., 2020). These findings have prompted researchers to develop accessible smell tests (in which individuals rate the quality and intensity of scents originating from, e.g., scratch-and-sniff cards or common kitchen items) for potential use as screening tools for COVID-19 (Iravani et al., 2020; Rodriguez et al., 2020).
The close relationship between COVID-19 and changes in chemical sensation raises questions about how SARS-CoV-2 might alter the cells and circuits charged with detecting stimuli and creating perception. Identifying these pathophysiological mechanisms has important implications for the development of possible treatments, as well as for the design of clinical chemosensory assessments to detect SARS-CoV-2 infection. Further, given that the COVID-19 syndrome is associated with neurological symptoms (including dizziness, headache, and altered consciousness) and stroke, characterizing these mechanisms may shed light on how SARS-CoV-2 disrupts neural systems more broadly (Docherty et al., 2020; Helms et al., 2020; Mao et al., 2020). Here we largely focus on interactions between SARS-CoV-2 and the olfactory system, which have been explored in some detail as the pandemic has progressed; as recent data suggest that SARS-CoV-2 may independently target taste and chemesthesis (Parma et al., 2020), we also briefly speculate on possible pathophysiological mechanisms in those systems.
More Than the Common Cold
SARS-CoV-2 belongs to the coronavirus family, which includes the pandemic MERS-CoV and SARS-CoV and the lesser known but more common endemic coronaviruses HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63. The endemic coronaviruses can infect the upper airway and frequently cause the common cold, which in turn is associated with both acute and chronic changes in smell and taste (Dalton, 2004; Mäkelä et al., 1998; Pellegrino et al., 2020; Rowan et al., 2015; Suzuki et al., 2007; Wood et al., 2011). The main proposed mechanisms for acute viral-mediated changes in smell include conductive deficits caused by loss of patency due to swelling of the mucosa and increased mucus production, changes in mucus composition, and secondary changes in olfactory signaling caused by local release of inflammatory intermediates like cytokines (Åkerlund et al., 1995; Chen et al., 2019; Damm et al., 2002; Schlosser et al., 2016; Trotier et al., 2007; Victores et al., 2018; Zhao et al., 2004). While cold-causing viruses likely act through multiple mechanisms to influence smell, recovery from virus-associated olfactory deficits tend to resolve with a time course similar to that of other cold-related symptoms like nasal congestion (Hummel et al., 1998a, 1998b; Zhao et al., 2014).
In a subset of patients, viral infections lead to long-lasting (i.e., months) post-viral anosmia, which is thought to result from direct damage to the olfactory sensory neurons (OSNs) responsible for odor detection in the olfactory epithelium (OE) (Cavazzana et al., 2018; Duncan and Seiden, 1995; Welge-Lüssen, 2005; Welge-Lüssen and Wolfensberger, 2006). Partial or full recovery of olfactory function in these patients is likely due to the recruitment of stem cells in the olfactory epithelium, which can replace damaged OSNs over long timescales. The recovery process is often accompanied by parosmias—distortions of smell perception—associated with wiring errors between newborn OSNs and their post-synaptic targets in the olfactory bulb (OB) (Figure 1 ; Leopold, 2002; Rombaux et al., 2009). Some cases of post-viral anosmia have been hypothesized to be the consequence of viral damage to central nervous system structures; in these cases, coronaviruses and other viruses are thought to gain access to the OB either directly via OSN axons or indirectly by passing through perforations in the cribriform plate (Figure 1; Barnett and Perlman, 1993; Schwob et al., 2001; van Riel et al., 2015).
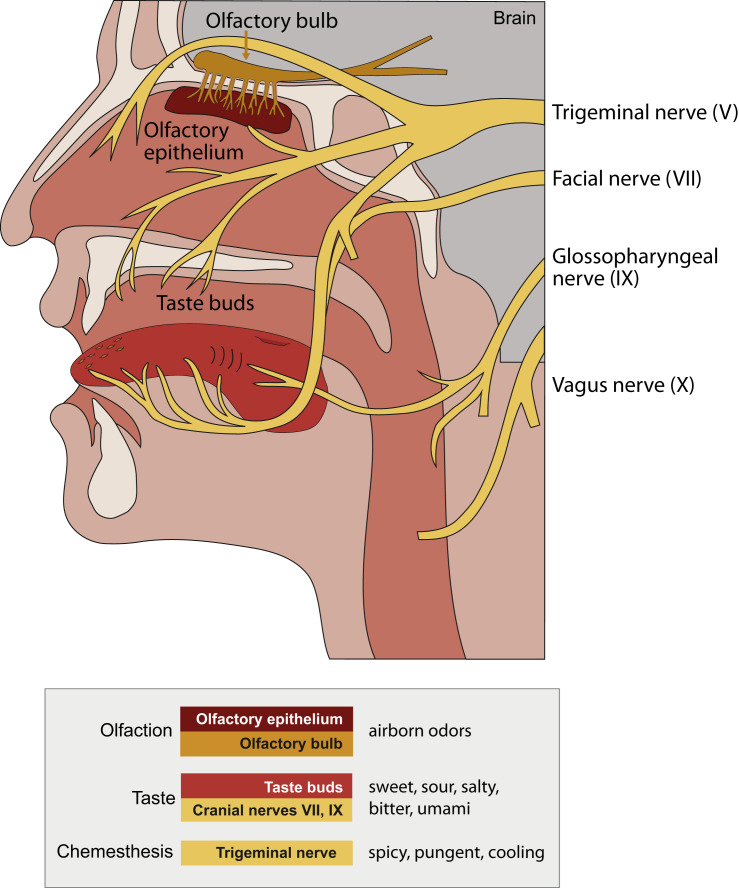
Figure 1.
Chemosensory Anatomy Defines the Potential Attack Surface for SARS-CoV-2
Chemosensation occurs in sensory epithelia in the nose and mouth. Multiple cranial nerves relay the senses of smell, taste, and chemesthesis to the brain. Airborne odors are detected by olfactory sensory neurons that reside in the olfactory epithelium; their axons pierce the bony cribriform plate to enter the olfactory bulb in the brain. Taste buds on the tongue are innervated by sensory afferents from the facial nerve (VII) and glossopharyngeal nerve (IX). The vagus nerve (X) also innervates taste buds residing in the pharynx. The detection of pungent chemicals, also known as chemesthesis, is mediated by both oral and nasal afferents of the trigeminal nerve (V). Although deficits in smell are most commonly reported in COVID-19, all three chemosensory modalities have been reported to be affected.
However, the natural history of COVID-19-associated anosmia argues that SARS-CoV-2 attacks the olfactory system through mechanisms distinct from those used by the more benign endemic coronaviruses. Many patients report anosmia as their first symptom, or in the absence of rhinorrhea or nasal congestion, suggesting that if inflammation is a key component of pathogenesis, it is local rather than generalized (Giacomelli et al., 2020; Kaye et al., 2020; Lechien et al., 2020b; Parma et al., 2020; Spinato et al., 2020; Vaira et al., 2020); indeed, the sensitivity of anosmia as a predictor of COVID-19 increases in patients without other nasal symptoms (Haehner et al., 2020; Shoer et al., 2020). Consistent with this observation, imaging studies of the olfactory system in COVID-19 patients are either normal or reveal focal inflammation (Eliezer et al., 2020; Galougahi et al., 2020). In addition, resolution of anosmia seems more rapid than observed in post-viral olfactory loss, in many cases occurring in weeks (rather than months) after initial symptoms develop (Hopkins et al., 2020; Kaye et al., 2020; Lechien et al., 2020a; Yan et al., 2020). Finally, the limited data currently available suggest that parosmias are infrequent during or after COVID-19 recovery (although this may change as we learn more) (Lechien et al., 2020a; Parma et al., 2020).
Inferring Disease Mechanisms from Patterns of Gene Expression
Coronaviruses are so named because of the halo of spike (S) proteins that decorate their surface (Du et al., 2009; Perlman and Netland, 2009). These S proteins interact with specific cellular receptors to bind host cells; binding is followed by protease-mediated S protein cleavage, which exposes fusion-promoting domains that enable viral entry. SARS-CoV-2 infects cells through interactions between its S protein and the Angiotensin I Converting Enzyme 2 (ACE2) receptor on target cells; ACE2 plays a crucial modulatory role in the renin-angiotensin system, which regulates blood pressure and salt water balance (Bader, 2010; Shang et al., 2020b; Walls et al., 2020; Zhou et al., 2020). Infection requires S protein cleavage, likely by the host cell serine protease TMPRSS2, although other proteases may also be involved (Hoffmann et al., 2020a, 2020b; Zang et al., 2020). With the exception of HCoV-NL63, the endemic coronaviruses do not use ACE2 as their primary cellular receptor (Belouzard et al., 2012; Zumla et al., 2016), a molecular distinction that likely underlies key differences in pathophysiology. SARS-CoV also uses ACE2 as its main receptor, and in one case study SARS-CoV infection was associated with anosmia (Hwang, 2006), although (unlike COVID-19) chemosensory disturbances are not a hallmark of SARS. Differences between SARS-CoV and SARS-CoV-2 in terms of their impacts on chemosensory systems may relate to biophysical differences, as the receptor-binding domain of the SARS-CoV-2 spike protein binds ACE2 with higher affinity and with a different binding mode than that of SARS-CoV (Li et al., 2003; Shang et al., 2020a; Walls et al., 2020). Species variation in the protein sequence of ACE2 significantly affects its affinity for the SARS-CoV-2 S protein, which in turn renders distinct model organisms differentially susceptible to infection (Wan et al., 2020; Zhao et al., 2020).
Given data suggesting that ACE2 is necessary for SARS-CoV-2 to infect host cells, researchers have used a variety of approaches to discern the pattern of expression of ACE2 and other viral entry proteins across the tissue landscape, with the goal of inferring possible target cells and disease mechanisms. For example, two studies have reported that cells in the nasal respiratory epithelium (RE) have higher expression of SARS-CoV-2 entry genes than cells in the RE that line the trachea or lungs (Hou et al., 2020; Sungnak et al., 2020). Consistent with this finding, recent work in macaques, ferrets, and cats identifies the nasal epithelium as a major source of viral RNA after SARS-CoV-2 infection (Munster et al., 2020; Shi et al., 2020). These data suggest that the nasal epithelium may act as a major reservoir for the virus (Figures 1 and 2 ).
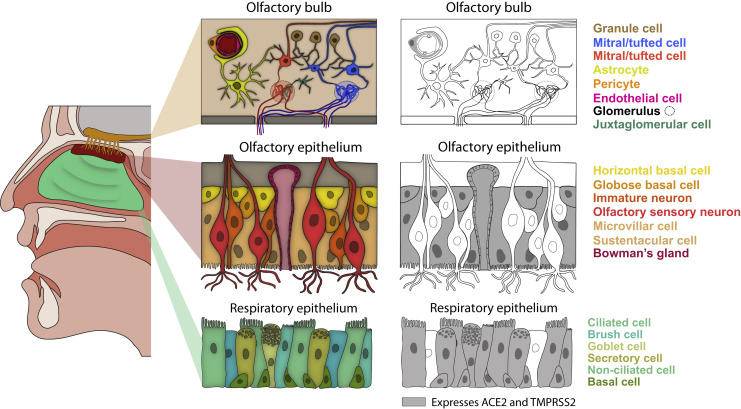
Figure 2.
ACE2-Positive Cells in the Nasal Respiratory Epithelium, Olfactory Epithelium, and the Olfactory Bulb
Schematic of a sagittal view of the human head, in which respiratory and olfactory epithelium as well as the olfactory bulb in the brain are colored (left). For each tissue, a schematic of the anatomy and known major cell types are shown (middle). The nose contains both respiratory and olfactory epithelia. The olfactory epithelium is restricted to medial portion of the superior turbinate and the superior portion of the nasal septum, whereas the respiratory epithelium is continuous with the upper airway (Dahl and Mygind, 1998). Olfactory sensory neurons within the olfactory epithelium are responsible for detecting odors, and in mice and humans they are continuously regenerated from globose progenitors throughout life (Durante et al., 2020; Schwob et al., 2017). The olfactory epithelium also contains other support and non-neuronal cell types, as well as reserve horizontal basal stem cells that respond to injury and can reconstitute olfactory epithelial cell types (Choi and Goldstein, 2018). In the respiratory epithelium, basal progenitor cells generate all epithelial cell types, including ciliated and secretory cells. In the olfactory bulb in the brain (tan), the axons from olfactory sensory neurons coalesce into glomeruli, and mitral/tufted cells innervate these glomeruli and send olfactory projections to downstream olfactory areas. ACE2-positive cell types are indicated in gray (right). Four recent reports have all concluded that ACE2 is not expressed in olfactory sensory neurons (Brann et al., 2020; Chen et al., 2020b; Fodoulian et al., 2020; Ziegler et al., 2020). Pericyte ACE2 expression in the olfactory bulb is inferred from the mouse data; there are currently no available human olfactory bulb sequencing datasets. Figure modified from Brann et al. (2020).
Although much of the human nose is lined with RE, sensory detection occurs in the olfactory epithelium, which houses OSNs and is located in the superior-most regions of the nasal epithelium (Figures 1 and 2). The OE is a complex chemosensory tissue composed of multiple cell types, including immature and mature OSNs, non-neuronal cell types such as the sustentacular, Bowman’s gland, and microvillar cells, and stem cells including globose and horizontal basal cells. Sustentacular cells are particularly intimately associated with OSNs, and they “enwrap” the sensory dendritic cilia that project into the airspace and enable odor detection (Liang, 2020). Four recently published studies—using species ranging from mouse to human—have explored the cell types in the OE that express ACE2 and other viral entry genes (Brann et al., 2020; Chen et al., 2020b; Fodoulian et al., 2020; Ziegler et al., 2020); all four conclude that OSNs do not express ACE2 (Figure 2). Instead, co-expression of ACE2 and TMPRSS2 was observed in key support cells (including sustentacular, Bowman’s gland, and microvillar cells) and stem cells that repopulate the epithelium after damage. Although ACE2 mRNA was identified using single-cell RNA sequencing (scSeq) techniques in only a small subset of these cells, both Brann et al. (2020) and Fodoulian et al. (2020) have demonstrated high level expression of ACE2 protein in a large population of sustentacular cells concentrated in the dorso-medial aspect of the mouse OE, which corresponds to the dorsal “zone” traditionally identified via molecular markers (Gussing and Bohm, 2004; Miyamichi et al., 2005). Consistent with this observation, mouse and human sustentacular cells identified as ACE2 positive by scSeq largely derive from the dorsal zone (Brann et al., 2020).
The co-expression of ACE2 and TMPRSS2 suggests that OE support cells may be the initial targets of SARS-CoV-2 infection. These inference-based conclusions are increasingly being pressure-tested by experiments in which model organisms are directly subject to SARS-CoV-2 infection. One recent paper in the golden hamster reports that SARS-CoV-2 infects sustentacular cells but not OSNs (Bryche et al., 2020). Intriguingly, this phenotype is accompanied by damage to the OSN ciliary layer and an increase in the number of microglia present in the OE, both of which are partially reverted to normal at 14 days after infection. A similar study in the hamster identified a large number of cells in the OE that were infected by SARS-CoV-2, although OE cell types were not definitively identified; inspection of the photomicrographs in that paper reveals that most SARS-CoV-2-positive cells traverse the thickness of the OE, suggesting that sustentacular cells are primary targets for infection (Sia et al., 2020). Human OE samples obtained from COVID-19 patients have similarly been queried for infection by SARS-CoV-2, which has revealed coronaviral antigens present in OE cells; infected cell types were not unambiguously identified, but their shape and position is consistent with the virus targeting sustentacular cells rather than OSNs (Cantuti-Castelvetri et al., 2020; Meinhardt et al., 2020).
What mechanisms might link infection of support cells to the acute changes in smell reported in COVID-19 (Figure 3 )? Localized inflammation in the epithelium might block the olfactory clefts, a pair of narrow passages located in the superior regions of the nasal epithelium through which air must flow to reach the OE, which comprises only 5% of the total nasal epithelium in humans (Besser et al., 2020; Trotier et al., 2007). Consistent with this possibility, a recent CT study of a COVID-19 patient who presented with anosmia revealed blocked olfactory clefts (Eliezer et al., 2020). Alternatively, infection of support cells and the attendant inflammation may cause local increases in inflammatory intermediates such as cytokines, which have been shown to influence OSN function in a non-cell-autonomous manner (Chen et al., 2019). Indeed, a recent study demonstrates elevated levels of inflammatory cytokines in the OE of infected patients (Torabi et al., 2020). Inflammatory intermediates have been suggested to indirectly lower the expression of odorant receptor (OR) genes by OSNs, which could cause significant changes in odor perception; this work also shows that OR expression levels return to normal after cessation of the inflammatory insult (Rodriguez et al., 2020).
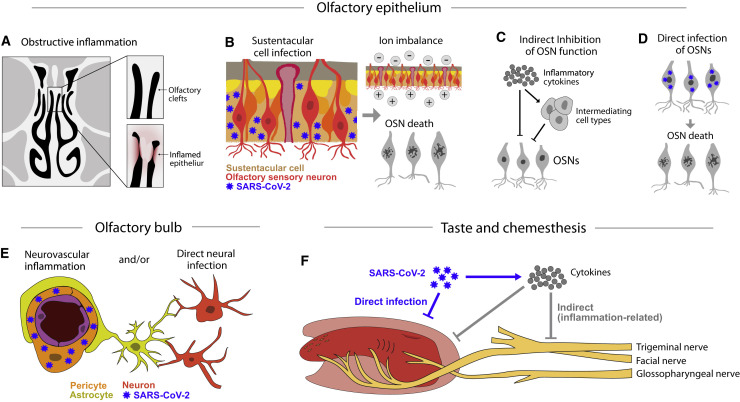
Figure 3.
Possible Mechanisms of Chemosensory Disturbances
(A) COVID-19 patients have reported olfactory loss in the absence of features common to upper respiratory infections like widespread nasal inflammation or obstruction. Such symptoms are consistent with recent CT imaging suggesting that SARS-CoV-2 infection may cause inflammation that is localized to the olfactory clefts (Eliezer et al., 2020).
(B) Sustentacular cells, Bowman’s gland cells, and microvillar cells in the olfactory epithelium express both ACE2 and TMPRSS2 and may be directly infected by SARS-CoV-2. Support cell infection may cause changes in the olfactory mucus or ion imbalances that can inhibit olfactory signaling; such changes may be rapidly reversible. The loss of these support cells in animal models can also result in the death of olfactory sensory neurons.
(C) Inflammatory cytokines may also directly or indirectly inhibit olfactory sensory neuron function.
(D) Although current data suggest that sustentacular and other support cells are the primary targets of SARS-CoV-2, a remaining possibility is that SARS-CoV-2 directly infects OSNs.
(E) Immunostaining for ACE2 protein in the mouse olfactory bulb suggests that ACE2 expression is restricted to vascular pericytes (Brann et al., 2020), which may directly (via perfusion changes) or indirectly (via inflammation) affect chemosensory perception in the brain.
(F) Taste and chemesthetic disturbances may result from the direct infection of cells in the tongue, the secondary consequences of obstruction due to inflammation, or damage following the release of inflammatory cytokines.
Finally, SARS-CoV-2 infection of support cells might alter the OE microenvironment in a manner deleterious to function. Bowman’s glands, for example, secrete mucus, which is essential to normal odor detection (Dear et al., 1991; Kern, 2000; Solbu and Holen, 2012). Sustentacular and microvillar cells remain somewhat mysterious, but seem to structurally support sensory neurons, phagocytose and/or detoxify potentially damaging agents, generate inflammatory cytokines, manage energetics through delivery of glucose to OSNs, and maintain local salt and water balance (Baxter et al., 2020; Lemons et al., 2017; O’Leary et al., 2019; Suzuki et al., 1996; Ualiyeva et al., 2020; Vogalis et al., 2005). Damage to support cells thus has the potential to change ion gradients and fuel availability, which in turn could acutely influence OSN firing rates. Furthermore, the important role of sustentacular cells in anatomically supporting OSN sensory cilia (which are surrounded by sustentacular cell processes) is highlighted by the observation that infection of sustentacular cells by SARS-CoV-2 in the golden hamster is linked to the denuding of OSN cilia, even in the absence of OSN infection (Bryche et al., 2020).
At least a subset of patients with COVID-19 appear to have longer-lasting anosmia, which may reflect more widespread damage to OSNs and the OE (as is seen in typical post-viral syndromes) (Jafek et al., 1990; Lechien et al., 2020b; Welge-Lüssen and Wolfensberger, 2006). Such damage could be a consequence of high levels of local inflammation, or a secondary consequence of the death of support cells, which in mouse models can lead to cascading changes in the structure and function of the OE (Bergström et al., 2003; Chen et al., 2019; Herrick et al., 2017). Of note, the horizontal basal cells that are normally quiescent but play a particularly important role in regenerating damaged OE also express ACE2 (Brann et al., 2020; Schwob et al., 2017); infection of these cells may therefore slow functional recovery over long time scales.
Is SARS-CoV-2 “Neuroinvasive”? Lessons from the Olfactory Bulb
Recent MRI studies have revealed transient changes in the OB that accompany COVID-related anosmia, consistent with some degree of central involvement in at least a subset of patients (Laurendon et al., 2020; Politi et al., 2020). Certain coronaviruses can pass from the OE through the cribriform plate to infect the OB (Dubé et al., 2018; Durrant et al., 2016). It remains unclear whether SARS-CoV-2 (given that it likely does not directly infect OSNs, and thus cannot pass directly through the olfactory nerve; see Figure 1) has this capacity; nevertheless, this possibility raises the question of whether SARS-CoV-2 can directly target neurons, glia, or other cell types in the OB. To date there are no scSeq datasets for the human olfactory bulb, and thus the distribution of ACE2 and other SARS-CoV-2-specific cell entry genes remains to be determined. However, scSeq and immunostaining of the mouse OB has revealed—as in the nose—that bulb neurons do not express detectable levels of ACE2 (Figure 2; Brann et al., 2020). Consistent with this finding, targeted deep sequencing of dopaminergic juxtaglomerular cells, which receive direct inputs from nose OSNs, failed to reveal Ace2 mRNA (Brann et al., 2020). Furthermore, OB cell targeting was not observed in studies of hamsters infected with SARS-CoV-2 (Bryche et al., 2020).
In contrast, vascular pericytes in the OB expressed high levels of ACE2 protein (Brann et al., 2020), consistent with the reported expression of Ace2 in perivascular cells in the brain and throughout the body (Chen et al., 2020a; He et al., 2020). Pericytes are critical for maintaining the blood-brain barrier, for defining local blood pressure, and for mediating neuroimmune responses (Armulik et al., 2011); infection of these cells thus has the potential to alter perfusion or recruit inflammation, both of which can indirectly influence the function of neural circuits. Thus, while vascular effects and inflammation could influence central brain structures involved in odor perception, current data suggest that it is unlikely that SARS-CoV-2 directly infects OB neurons responsible for processing odor information from the nose and conveying it to the cortex.
Consistent with observations in the OB, there is an increasing amount of data suggesting that ACE2 is not appreciably expressed in neurons in the brain at either the RNA or protein level. Meta-analysis of ten separate mouse deep-sequencing datasets drawn from across the central and peripheral nervous system did not reveal significant expression of either ACE2 or TMPRSS2 in any neural cell type (Brann et al., 2020); scSeq analysis of human prefrontal cortex and hippocampus failed to identify any ACE2-expressing cells (Chen et al., 2020c); and multiple immunostaining studies have demonstrated ACE2 expression in brain vasculature but not in neurons or glia (Brann et al., 2020; Hamming et al., 2004; Kehoe et al., 2016). These expression-based studies are now being complemented by data from a variety of animal models that can be infected with SARS-CoV-2. Infection of macaques, cats, and ferrets with SARS-CoV-2 (which in the monkey and cat resulted in pulmonary infection) did not reveal any virus present in the brain (Munster et al., 2020; Shi et al., 2020). In addition, recent human autopsy studies reveal that the brain contains the least amount of SARS-CoV-2 of any sampled tissue in the body (Puelles et al., 2020).
In contrast to these observations, several reports have suggested that SARS-CoV-2 may be “neuroinvasive,” which we take here to mean the direct infection of neurons or glia in the central nervous system (Baig et al., 2020; Li et al., 2020; Meinhardt et al., 2020; Morris and Zohrabian, 2020; Wu et al., 2020). Arguments supporting this position often refer to data demonstrating that coronaviruses can hop from the nose to the bulb in experimental mouse models (Dubé et al., 2018; Durrant et al., 2016; Perlman et al., 1989; van Riel et al., 2015); however, without exception, the coronaviruses that have this property either do not use ACE2 as a receptor, were explicitly selected during passage for their neurotropic potential, or both (Butler et al., 2006; Cowley and Weiss, 2010). Many lines of evidence for neuroinvasiveness are drawn from prior work on SARS-CoV and its ability to infect ACE2-expressing cells. For example, it has been observed that nasal instillation of SARS-CoV in mouse models expressing human ACE2 (which has much higher affinity for the CoV and SARS-CoV-2 spike protein than does mouse ACE2) can result in widespread infection of brain tissues (Netland et al., 2008; Tseng et al., 2007; Yang et al., 2007); however, these models transgenically express human ACE2 in many cells in which it is normally absent, making it an inappropriate tool to probe viral tropism. Although there is a case report describing detection of SARS-CoV RNA in the CSF of a patient, as this RNA could have originated from many cell types (including vascular cells), it does not clearly demonstrate the neuroinvasive potential of either SARS-CoV or SARS-CoV-2 (Hung et al., 2003). Furthermore, two studies have failed to identify SARS-CoV-2 RNA in the CSF of patients (Farhadian et al., 2020; Schaller et al., 2020). Notably, a recent experiment assessed the consequences of SARS-CoV-2 exposure in mice in which human ACE2 is transgenically expressed under control of the mouse ACE2 promoter (which should largely, although not perfectly, recapitulate endogenous patterns of ACE2 expression); while nasal infection and subsequent spread was sufficient to ultimately kill these mice, post-mortem analysis failed to reveal SARS-CoV-2 in the brain (Bao et al., 2020).
However, other lines of evidence more directly support the possibility that SARS-CoV-2 can directly infect neurons or glia. In one patient autopsy sample, SARS-CoV was identified via antigen staining to be distributed across the brain parenchyma (Xu et al., 2005). Two papers have reported Ace2 staining in astrocytes in the rat cerebellum and brainstem (Gallagher et al., 2006; Gowrisankar and Clark, 2016), a paper from the Human Cell Atlas has reported ACE2 expression in oligodendrocytes (Muus et al., 2020), and a meta-analysis of a single scSeq dataset revealed low-level expression of ACE2 in human middle temporal gyrus and posterior cingulate cortex (although that same dataset included data for only a handful of vascular cells, making it difficult to assess relative expression levels) (Chen et al., 2020c). Two recent papers have observed anti-ACE2 staining in the mouse olfactory bulb, the first in a subset of mitral cells (Bilinska et al., 2020), the second in all mitral cells (Ueha et al., 2020); the latter paper also reports the presence of ACE2 protein in all OSN nuclei (but not OSN cilia, raising the possibility of non-specific staining). Cortical neurons in human brain organoids (which express ACE2) can be infected by SARS-CoV-2 (Gopalakrishnan et al., 2020). Perhaps most convincingly, nasal instillation of SARS-CoV-2 in a mouse with the human ACE2 gene knocked into the mouse Ace2 locus caused infection of brain neurons, although the number, distribution, and identity of these cells was not characterized (Sun et al., 2020).
We are only now beginning to understand SARS-CoV-2 pathogenesis and so it is important to recognize key caveats in interpreting data suggesting SARS-CoV-2 does or does not infect neurons and glia directly. Conclusions drawn based upon the presence or absence of ACE2 message or protein depend upon the specific methods being used: for example, scSeq techniques have low sensitivity but an identifiable noise floor, whereas immunohistochemistry depends upon the vagaries of each anti-ACE2 antibody. Similarly, conclusions based upon SARS-CoV-2 infection of model organisms (transgenic or otherwise) depend upon the expression pattern and S protein affinity of the ACE2 protein in each model, as well as a number of other species-specific factors relating to viral tropism and immune responses. It is clear that there are significant inconsistencies among the datasets that require reconciliation; definitively addressing the question of whether SARS-CoV-2 can infect human neurons (in the bulb or elsewhere) will ultimately require detailed analysis of autopsy tissue. It is also important to note that SARS-CoV-2 in principle has access to the brain through routes independent of the nasal epithelium, including via vasculature and nerves innervating infected tissues; these modes of infection have been observed for other viruses (Dahm et al., 2016; Koyuncu et al., 2013).
In light of this controversy, it is worth noting that many of the observed neurological consequences of COVID-19 (like altered consciousness and stroke) can in principle be explained by a primary vasculopathy and hypercoagulability (Chen et al., 2020d; Helms et al., 2020; Zhang et al., 2020) or by a secondary deficit in brain perfusion caused by elevated cytokines (Poyiadji et al., 2020). In contrast, clinical signs of encephalitis (i.e., infection of neurons), such as increased risk of seizures and inflammatory CSF, have not been commonly observed (Lu et al., 2020). This perspective is consistent with recent MRI findings in COVID-19 patients (Beyrouti et al., 2020; Coolen et al., 2020), and two separate autopsy series that failed to identify any signs of encephalitis (Schaller et al., 2020; Solomon et al., 2020). At this point, therefore, the balance of the clinical evidence points to SARS-CoV-2 influencing the brain indirectly through effects on vasculature, although this view may change as we learn more about viral pathogenesis and we gain access to more detailed clinical information.
SARS-CoV-2 Targets Taste and Chemesthesis
Smell, taste, and chemesthesis are readily distinguishable but synergize to create the perception of the flavor in the mouth. A vast spectrum of volatile food odors is detected retronasally by OSNs, while the taste repertoire is restricted to non-volatile sour, salt, sweet, bitter, and umami stimuli that directly activate taste buds on the tongue. The spiciness of chili peppers and the cool of mint are neither odors nor tastes, but rather activate sensory neurons of chemesthesis that innervate oral epithelia. Most recent reports of smell loss in COVID-19 patients (see above) have generally considered smell and taste loss as unitary and excluded consideration of chemesthesis altogether. Importantly, recent data suggest that taste and chemesthesis may be disturbed independently of smell in COVID-19 patients (Adamczyk et al., 2020; Lechien et al., 2020a; Parma et al., 2020; Vaira et al., 2020). For example, Parma et al. (2020) showed that ∼60% of participants link taste loss (which could be attributed to flavor) to deficits in at least one specific taste quality (e.g., salty taste), suggesting that at least some participants distinguish changes in the taste component of flavor. However, the basic taste modalities were not predictably affected, with some taste qualities being differentially impacted across individuals. What mechanistic hypotheses can explain the specific impact of SARS-CoV-2 on taste function, and which cell populations may be targeted by the virus such that their loss would lead to distorted taste? Taste perception is supported by taste buds, which have a characteristic distribution on the tongue and which house taste receptor cells (TRCs) that are categorized into three morphological types (Figure 4 ): type I for support, type II for sweet, bitter, or umami, and type III for sour) (Chaudhari and Roper, 2010). Like OSNs, individual TRCs are constantly being renewed by stem cells, and thus taste function and perception also depend on rapid and reliable production of the proper proportions of each of the different TRCs (Barlow and Klein, 2015).
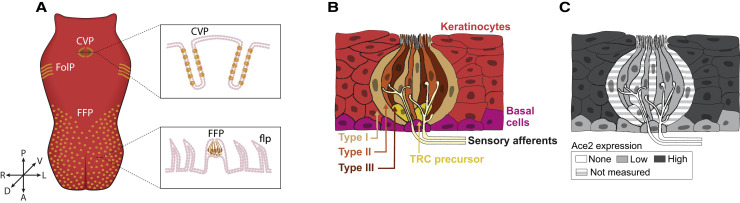
Figure 4.
Organization of the Tongue and Taste Buds
(A) In the front of the tongue, taste buds are situated in fungiform papillae (FFP). FFP are distributed throughout the anterior lingual epithelium, which is covered by mechanosensory filiform papillae (flp). In the posterior tongue, large complex papillae (circumvallate [CVP] and foliate [FolP]) are epithelial invaginations that house hundreds of buds each; rodents have a single CVP, while humans possess 8–12 CVPs in an inverted V arrangement across the posterior tongue (not shown). Modified from Barlow (2015).
(B) Each bud is a collection of ∼100 taste receptor cells (TRCs) categorized into three morphological types (I for support or glial-like, II for sweet, bitter, umami, and III for sour). All TRCs are continuously renewed from basal stem cells adjacent to taste buds (“Basal cells”) (Gaillard et al., 2015; Liu et al., 2013; Okubo et al., 2009). Taste bud-fated daughters generated by basal cells exit the cell cycle and enter buds as postmitotic TRC precursors, which differentiate into each of the TRC types (Miura et al., 2006, 2014). Basal cells also give rise to keratinocytes of the non-taste lingual epithelium including those surrounding taste buds. Individual TRCs have a brief lifespan ranging from 1 to 6 weeks (Beidler and Smallman 1965; Hamamichi et al., 2006; Perea-Martinez et al., 2013). Type III TRCs make conventional presynaptic contacts, while type II TRCs have specialized contacts with sensory afferents to transmit taste information to the CNS (Finger et al., 2005; Romanov et al., 2018; Roper and Chaudhari, 2017; Taruno et al., 2013).
(C) To date, RNA profiling of taste relevant cell populations has been obtained primarily from murine CVP, with two additional datasets for general anterior tongue epithelium in mice and human (see text). A summary of approximate ACE2 expression levels extracted from these studies is depicted here as discussed in the text. Of note, type I TRCs and precursor TRCs have yet to be profiled, and ACE2 is not detected in sensory neuron afferents.
Although formal analyses are only now emerging, a cursory mining of publicly available datasets of type II and III TRCs in mice reveals that ACE2 is expressed by sour-sensing type III TRCs, and to a lesser extent by bitter and sweet/umami-sensing type II TRCs (Han et al., 2020; Lee et al., 2017; Qin et al., 2018; Shigemura et al., 2019; Zhang et al., 2019). While type II and type III TRCs express little to no TMPRSS2, Cathepsins (CTSB, CTSL) are abundant and may function as proteases to cleave SARS-CoV-2 spike protein at type II and III TRCs. Available datasets do not include transcriptional profiling for the type I TRCs, which—much like olfactory sustentacular cells—extend cellular processes that wrap type II and III TRCs (Liang, 2020; Yang et al., 2020). In addition, type I cells degrade ATP, which is used as a neurotransmitter by type II cells to convey taste information to the brain via the gustatory nerves (Bartel et al., 2006; Finger et al., 2005; Vandenbeuch et al., 2013). If type I cells are indeed ACE2 positive and targeted by SARS-CoV-2, the loss of support cells could lead to taste bud collapse via cell death and/or reduced efficacy of taste signals to gustatory nerves (Figure 3). While human taste tissue has yet to be profiled, microarray data comparing macaque taste and non-taste lingual epithelia reveal low levels of ACE2, TMPRSS2, CTSL, and CTSB in both tissue compartments (Hevezi et al., 2009), suggesting that the machinery necessary for SARS-CoV-2 infection of taste-relevant cells may be present in humans.
Adult taste stem cells in the posterior tongue (where they are most abundant) express the marker LGR5 and can give rise to all TRC lineages (Yee et al., 2013). Despite a limited sample size, scSeq profiling revealed that murine LGR5-positive stem cells express ACE2 and TRMPSS2 and thus may be competent for infection by SARS-CoV-2 (Figure 4; Qin et al., 2018). These data raise the possibility that taste dysfunction in COVID-19 patients may be caused or exacerbated by insufficient TRC renewal due to SARS-CoV-2 stem cell damage. In addition, it has been shown that experimentally induced systemic inflammation in mice can reduce taste stem cell output, which in turn leads to depopulated taste buds and perturbed taste function (Cohn et al., 2010; Feng et al., 2014; Kaufman et al., 2018; Kim et al., 2012; Wang et al., 2007, 2009). If SARS-CoV-2 directly infects the tongue, local inflammatory processes could therefore alter stem cell properties and ultimately influence taste perception (Figure 3). Consistent with this possibility, scSeq analysis in mice and humans suggests that ACE2 is highly expressed in subsets of tongue epithelial cells, as are other proteases linked to coronavirus entry, e.g., TMPRSS11D, TMPRSS4, and CTSB (Pisco et al., 2020; Schaum et al., 2018; Venkatakrishnan et al., 2020; Xu et al., 2020).
It is important to note that—unlike OSNs—TRCs are not neurons; thus, all of the cell types identified as ACE2 positive in the tongue to date are either stem or epithelial cells. Transcriptional profiling of sensory neurons in the geniculate ganglion, whose fibers innervate fungiform taste buds, indicate that ACE2 and TMPRSS2 are not expressed (Figure 4; Dvoryanchikov et al., 2017; Zhang et al., 2019). Similarly, the vagal sensory neurons that innervate taste buds in the larynx do not express ACE2 or TMPRSS2 (Prescott et al., 2020). Obtaining more extensive data regarding SARS-CoV-2-relevant gene expression in taste epithelial populations (which have been woefully undersampled compared to similar populations in the olfactory system) will provide a better basis from which to propose mechanistic hypotheses.
In addition to taste and smell perturbations, subsets of COVID-19 patients experience a disruption of chemesthesis. Chemesthesic stimuli are detected by a variety of epithelial sensors and relayed to the brainstem via trigeminal ganglion sensory neurons, distinct branches of which innervate the nasal respiratory RE and the non-taste epithelium of the tongue, among other targets (Frasnelli and Manescu, 2017; Viana, 2011). These trigeminal somatosensory afferents are enriched for TRP channel receptors that detect chemical irritants (Roper, 2014). In the nasal cavity, several irritants are detected by solitary chemosensory cells (SCCs), which express many of the molecules required for bitter taste detection and signal transduction that characterize type II TRCs and are innervated by trigeminal fibers (Finger et al., 2003; Gulbransen et al., 2008; Tizzano et al., 2010). SCCs are capable of driving systemic physiological responses to aversive substances (like reduced respiration) and can recruit a local neurogenic inflammatory response (Saunders et al., 2014; Tizzano et al., 2010). As with the type I TRCs, it is not yet clear from sequencing data whether SCCs express SARS-CoV-2 cell entry genes. Thus how SARS-CoV-2 influences chemesthesis remains uncertain, although these effects are unlikely to be mediated by trigeminal neurons, which do not express ACE2 or TRMPSS2 (Nguyen et al., 2017, 2019).
Conclusions and Outlook
Research into possible mechanisms underlying changes in chemical perception due to COVID-19 has only just begun, and much remains to be learned about the pathophysiology of SARS-CoV-2. That said, current evidence favors a model in which SARS-CoV-2 cell entry genes in the olfactory, gustatory, and chemesthetic systems are not expressed in primary or secondary neurons, but rather are expressed in epithelial, support, and stem cells responsible for maintaining perception. This model suggests that neural function is altered indirectly due to sequelae of SARS-CoV-2 infection of peripheral support cells, including (but not limited to) local inflammation and changes in OSN gene expression and ciliary structure. Observed disease-associated changes in OB MRI intensity suggest that in a subset of patients, there is central involvement as well; given the transient nature of these changes, it is likely that they reflect either local inflammatory processes (that are, for example, a consequence of vascular infection) or more speculatively are a consequence of inflammatory cytokines diffusing from the dorsal epithelium across the perforations in the cribriform plate. Although less likely given current evidence, it also remains possible that SARS-CoV-2 directly infects OSNs or bulb neurons.
The notion that chemical sensory deficits in COVID-19 result from infection of support cells highlights an important lacuna in our understanding: while an enormous amount of effort has been devoted to understanding the molecular mechanisms underlying chemosensation—and the neural pathways that convey information about chemical cues to the brain—we still know little about the non-neuronal cells and structures that support sensory transduction. In this sense, SARS-CoV-2 provides an important opportunity to gain insight into how the complex peripheral tissues that support taste, smell, and chemesthesis enable meaningful interactions with the world.
Much of our current understanding of pathophysiological mechanisms has been inferred from patterns of ACE2 and TMPRSS2 expression. However, these inferences are constrained by our current understanding of the virus and by the predicted relationship between ACE2 mRNA and protein. Although the evidence that ACE2 is obligate for SARS-CoV-2 entry is strong, it has been suggested that other molecules such as BSG, neuropilin-1, or PIKfyve may participate in SARS-CoV-2 entry (Cantuti-Castelvetri et al., 2020; Chen et al., 2005; Kang et al., 2020; Ou et al., 2020; Wang et al., 2020). Furthermore, it has recently been reported that low-level expression of ACE2 may be sufficient to support SARS-CoV-2 infection (Lamers et al., 2020), suggesting that SARS-CoV-2 may infect apparently ACE2-negative cell types. It is also important to note that many of the conclusions drawn about cell entry gene expression (in, for instance, the olfactory bulb) are based largely upon mouse data. Although the mouse and human ACE2 and TMPRSS2 expression data align nearly perfectly in the olfactory epithelium (Brann et al., 2020), there may be clinically relevant divergences between species in the olfactory bulb and brain (Hodge et al., 2019; Maresh et al., 2008). Finally, it is important to note that the ACE2 gene is regulated by inflammation in human cells, and other SARS-CoV-2 entry genes may be similarly modulated by primary infection and inflammation (Ansari et al., 2020; Ziegler et al., 2020). This observation raises the possibility that a broader spectrum of cells expresses ACE2 during SARS-CoV-2 infection than is currently appreciated.
Given the emerging central role of support cells in COVID pathophysiology, a key open question is how primary infection of non-neural cells alters chemical perception. Defining the pathophysiological mechanisms underlying anosmia and other SARS-CoV-2-associated disturbances in chemical sensation will require moving past inference based upon gene or protein expression and toward interrogative experiments aimed at falsifying hypotheses about the mechanistic links between, e.g., viral infection, sustentacular cell dysfunction, and altered OSN signaling. For example, the close anatomical apposition of sustentacular cell processes and OSN cilia suggest a direct role in the former in supporting the latter, but the nature of this support and how it might be abrogated after SARS-CoV-2 infection remains enigmatic.
Addressing mechanistic hypotheses will be facilitated by further experiments in mouse models in which human ACE2 is expressed under the control of the mouse ACE2 promoter (Sun et al., 2020) and by more extensive use of non-mouse model organisms, such as hamsters, ferrets, cats, and monkeys, all of which are susceptible to infection by SARS-CoV-2 (Bryche et al., 2020; Munster et al., 2020; Shi et al., 2020; Sia et al., 2020). Similarly, the development and use of benign viruses pseudotyped with the SARS-CoV-2 spike protein can be used to identify targeted cell types without the burden of a BSL-3 containment system. Developing tools to characterize the pattern of infection of SARS-CoV-2 in the olfactory system should be complemented by physiological studies in the OE, OB, and cortex—and parallel behavioral work—exploring how the function of the olfactory system evolves after SARS-CoV-2 infection. Importantly, the olfactory epithelium and tongue can be sampled in live human subjects without dissection, which holds the promise for their use as models for characterizing SARS-CoV-2 interactions with the human nervous system. All of this SARS-CoV-2-specific work will be usefully complemented by additional work exploring the physiological function of support cells in the nose and tongue.
Finally, ongoing basic science efforts will benefit from more and better clinical data (Whitcroft and Hummel, 2020). Given the diversity of symptoms reported by patients, it remains unclear whether COVID-19 attacks chemosensation through one or many pathophysiological mechanisms, or whether specific smell or taste qualities are particularly affected. Similarly, we lack an understanding of how smell, taste, and chemesthesis evolve over the long term in the subset of patients that do not exhibit a quick recovery. Revealing the mechanisms through which SARS-CoV-2 influences chemical sensing will have important implications for our understanding of how viruses can functionally alter sensory systems in specific, and neural circuits more generally.
Acknowledgments
The authors would like to thank the Global Consortium for Chemosensory Research (GCCR) for facilitating the collaborative environment in which this manuscript was conceived. S.R.D. is supported by grants R011DC016222 and U19NS112953 from the National Institutes of Health and by the Simons Collaboration on the Global Brain. L.A.B. is supported by grants R01 DC012383 and R21 CA236480. K.W.C. gratefully acknowledges support from UCI INP. E.D.L. is supported by NIDCD K23 DC014747 to V. Ramakrishnan, R01s DC012555 and DC017679 to S. Kinnamon, and R01 DC014253 to D. Restrepo. M.C.F. is supported by the Yale Medical School Fellowship. P.V.J. is supported by the National Institute of Nursing Research under award number 1ZIANR000035-01. P.V.J. is also supported by the Office of Workforce Diversity, National Institutes of Health, and the Rockefeller University Heilbrunn Nurse Scholar Award. D.H.B. is supported by an NSF Graduate Research Fellowship.
References
- Adamczyk K., Herman M., Fraczek J., Piec R., Szykula-Piec B., Zaczynski A., Wojtowicz R., Bojanowski K., Rusyan E., Krol Z., et al. Sensitivity and specifity of prediction models based on gustatory disorders in diagnosing COVID-19 patients: a case-control study. medRxiv. 2020 doi: 10.1101/2020.05.31.20118380. [DOI] [Google Scholar]
- Åkerlund A., Bende M., Murphy C. Olfactory threshold and nasal mucosal changes in experimentally induced common cold. Acta Otolaryngol. 1995;115:88–92. doi: 10.3109/00016489509133353. [DOI] [PubMed] [Google Scholar]
- Ansari M.A., Marchi E., Ramamurthy N., Aschenbrenner D., Hackstein C.-P., Bowden R., Sharma E., Pedergnana V., Venkateswaran S., Kugathasan S., et al. A gene locus that controls expression of ACE2 in virus infection. medRxiv. 2020 doi: 10.1101/2020.04.26.20080408. [DOI] [Google Scholar]
- Armulik A., Genové G., Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Bader M. Tissue renin-angiotensin-aldosterone systems: Targets for pharmacological therapy. Annu. Rev. Pharmacol. Toxicol. 2010;50:439–465. doi: 10.1146/annurev.pharmtox.010909.105610. [DOI] [PubMed] [Google Scholar]
- Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host-Virus Interaction, and Proposed Neurotropic Mechanisms. ACS Chem. Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online May 7, 2020. [DOI] [PubMed] [Google Scholar]
- Barlow L.A. Progress and renewal in gustation: new insights into taste bud development. Development. 2015;142:3620–3629. doi: 10.1242/dev.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow L.A., Klein O.D. Developing and regenerating a sense of taste. Curr. Top. Dev. Biol. 2015;111:401–419. doi: 10.1016/bs.ctdb.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett E.M., Perlman S. The olfactory nerve and not the trigeminal nerve is the major site of CNS entry for mouse hepatitis virus, strain JHM. Virology. 1993;194:185–191. doi: 10.1006/viro.1993.1248. [DOI] [PubMed] [Google Scholar]
- Bartel D.L., Sullivan S.L., Lavoie E.G., Sévigny J., Finger T.E. Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. J. Comp. Neurol. 2006;497:1–12. doi: 10.1002/cne.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter B.D., Larson E.D., Feinstein P., Polese A.G., Bubak A.N., Niemeyer C.S., Merle L., Shepherd D., Ramakrishnan V.R., Nagel M.A., Restrepo D. Transcriptional profiling reveals TRPM5-expressing cells involved in viral infection in the olfactory epithelium. bioRxiv. 2020 doi: 10.1101/2020.05.14.096016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beidler L.M., Smallman R.L. Renewal of cells within taste buds. J. Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénézit F., Le Turnier P., Declerck C., Paillé C., Revest M., Dubée V., Tattevin P., RAN COVID Study Group Utility of hyposmia and hypogeusia for the diagnosis of COVID-19. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30297-8. Published online April 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström U., Giovanetti A., Piras E., Brittebo E.B. Methimazole-induced damage in the olfactory mucosa: effects on ultrastructure and glutathione levels. Toxicol. Pathol. 2003;31:379–387. doi: 10.1080/01926230390201101. [DOI] [PubMed] [Google Scholar]
- Besser G., Liu D.T., Renner B., Hummel T., Mueller C.A. Reversible obstruction of the olfactory cleft: impact on olfactory perception and nasal patency. Int. Forum Allergy Rhinol. 2020;10:713–718. doi: 10.1002/alr.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyrouti R., Adams M.E., Benjamin L., Cohen H., Farmer S.F., Goh Y.Y., Humphries F., Jäger H.R., Losseff N.A., Perry R.J., et al. Characteristics of ischaemic stroke associated with COVID-19. J. Neurol. Neurosurg. Psychiatry. 2020 doi: 10.1136/jnnp-2020-323586. Published online April 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann D., Tsukahara T., Weinreb C., Lipovsek M., Van den Berge K., Gong B., Chance R., Macaulay I.C., Chou H.-j., Fletcher R., et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. bioRxiv. 2020 doi: 10.1101/2020.03.25.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryche B., St Albin A., Murri S., Lacôte S., Pulido C., Gouilh M.A., Lesellier S., Servat A., Wasniewski M., Picard-Meyer E., et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. bioRxiv. 2020 doi: 10.1101/2020.06.16.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N., Pewe L., Trandem K., Perlman S. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology. 2006;347:410–421. doi: 10.1016/j.virol.2005.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Kallio K., Kaya T., Anastasina M., Smura T., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. bioRxiv. 2020 doi: 10.1101/2020.06.07.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana A., Larsson M., Münch M., Hähner A., Hummel T. Postinfectious olfactory loss: A retrospective study on 791 patients. Laryngoscope. 2018;128:10–15. doi: 10.1002/lary.26606. [DOI] [PubMed] [Google Scholar]
- Chaudhari N., Roper S.D. The cell biology of taste. J. Cell Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Mi L., Xu J., Yu J., Wang X., Jiang J., Xing J., Shang P., Qian A., Li Y., et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Reed R.R., Lane A.P. Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense. Cell Stem Cell. 2019;25:501–513.e5. doi: 10.1016/j.stem.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116:1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Shen W., Rowan N.R., Kulaga H., Hillel A., Ramanathan M., Lane A.P. Elevated ACE2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. bioRxiv. 2020 doi: 10.1101/2020.05.08.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wang K., Yu J., Chen Z., Wen C., Xu Z. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in human and mouse brain. bioRxiv. 2020 doi: 10.1101/2020.04.07.030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi R., Goldstein B.J. Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig. Otolaryngol. 2018;3:35–42. doi: 10.1002/lio2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn Z.J., Kim A., Huang L., Brand J., Wang H. Lipopolysaccharide-induced inflammation attenuates taste progenitor cell proliferation and shortens the life span of taste bud cells. BMC Neurosci. 2010;11:72. doi: 10.1186/1471-2202-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen T., Lolli V., Sadeghi N., Rovai A., Trotta N., Taccone F.S., Creteur J., Henrard S., Goffard J.-C., Dewitte O., et al. Early postmortem brain MRI findings in COVID-19 non-survivors. medRxiv. 2020 doi: 10.1101/2020.05.04.20090316. [DOI] [PubMed] [Google Scholar]
- Cowley T.J., Weiss S.R. Murine coronavirus neuropathogenesis: determinants of virulence. J. Neurovirol. 2010;16:427–434. doi: 10.1007/BF03210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R., Mygind N. Anatomy, physiology and function of the nasal cavities in health and disease. Adv. Drug Deliv. Rev. 1998;29:3–12. doi: 10.1016/s0169-409x(97)00058-6. [DOI] [PubMed] [Google Scholar]
- Dahm T., Rudolph H., Schwerk C., Schroten H., Tenenbaum T. Neuroinvasion and Inflammation in Viral Central Nervous System Infections. Mediators Inflamm. 2016;2016:8562805. doi: 10.1155/2016/8562805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton P. Olfaction and anosmia in rhinosinusitis. Curr. Allergy Asthma Rep. 2004;4:230–236. doi: 10.1007/s11882-004-0031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm M., Vent J., Schmidt M., Theissen P., Eckel H.E., Lötsch J., Hummel T. Intranasal volume and olfactory function. Chem. Senses. 2002;27:831–839. doi: 10.1093/chemse/27.9.831. [DOI] [PubMed] [Google Scholar]
- Dear T.N., Boehm T., Keverne E.B., Rabbitts T.H. Novel genes for potential ligand-binding proteins in subregions of the olfactory mucosa. EMBO J. 1991;10:2813–2819. doi: 10.1002/j.1460-2075.1991.tb07830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., Holden K.A., Read J.M., Dondelinger F., Carson G., et al. Features of 16,749 hospitalised UK patients with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol. medRxiv. 2020 doi: 10.1101/2020.04.23.20076042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M., Le Coupanec A., Wong A.H.M., Rini J.M., Desforges M., Talbot P.J. Axonal Transport Enables Neuron-to-Neuron Propagation of Human Coronavirus OC43. J. Virol. 2018;92 doi: 10.1128/JVI.00404-18. e00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan H.J., Seiden A.M. Long-term follow-up of olfactory loss secondary to head trauma and upper respiratory tract infection. Arch. Otolaryngol. Head Neck Surg. 1995;121:1183–1187. doi: 10.1001/archotol.1995.01890100087015. [DOI] [PubMed] [Google Scholar]
- Durante M.A., Kurtenbach S., Sargi Z.B., Harbour J.W., Choi R., Kurtenbach S., Goss G.M., Matsunami H., Goldstein B.J. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 2020;23:323–326. doi: 10.1038/s41593-020-0587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant D.M., Ghosh S., Klein R.S. The Olfactory Bulb: An Immunosensory Effector Organ during Neurotropic Viral Infections. ACS Chem. Neurosci. 2016;7:464–469. doi: 10.1021/acschemneuro.6b00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvoryanchikov G., Hernandez D., Roebber J.K., Hill D.L., Roper S.D., Chaudhari N. Transcriptomes and neurotransmitter profiles of classes of gustatory and somatosensory neurons in the geniculate ganglion. Nat. Commun. 2017;8:760. doi: 10.1038/s41467-017-01095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer M., Hautefort C., Hamel A.-L., Verillaud B., Herman P., Houdart E., Eloit C. Sudden and Complete Olfactory Loss Function as a Possible Symptom of COVID-19. JAMA Otolaryngol. Head Neck Surg. 2020 doi: 10.1001/jamaoto.2020.0832. Published online April 8, 2020. [DOI] [PubMed] [Google Scholar]
- Farhadian S., Glick L.R., Vogels C.B.F., Thomas J., Chiarella J., Casanovas-Massana A., Zhou J., Odio C., Vijayakumar P., Geng B., et al. Acute encephalopathy with elevated CSF inflammatory markers as the initial presentation of COVID-19. BMC Neurol. 2020;20:248. doi: 10.1186/s12883-020-01812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P., Huang L., Wang H. Taste bud homeostasis in health, disease, and aging. Chem. Senses. 2014;39:3–16. doi: 10.1093/chemse/bjt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger T.E., Böttger B., Hansen A., Anderson K.T., Alimohammadi H., Silver W.L. Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc. Natl. Acad. Sci. USA. 2003;100:8981–8986. doi: 10.1073/pnas.1531172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger T.E., Danilova V., Barrows J., Bartel D.L., Vigers A.J., Stone L., Hellekant G., Kinnamon S.C. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- Fodoulian L., Tuberosa J., Rossier D., Landis B.N., Carleton A., Rodriguez I. SARS-CoV-2 receptor and entry genes are expressed by sustentacular cells in the human olfactory neuroepithelium. bioRxiv. 2020 doi: 10.1101/2020.03.31.013268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanet A., Tondeur L., Madec Y., Grant R., Besombes C., Jolly N., Fernandes Pellerin S., Ungeheuer M.-N., Cailleau I., Kuhmel L., et al. Cluster of COVID-19 in northern France: A retrospective closed cohort study. medRxiv. 2020 doi: 10.1101/2020.04.18.20071134. [DOI] [Google Scholar]
- Frasnelli J., Manescu S. In: Springer Handbook of Odor. Buettner A., editor. Springer International Publishing; Cham: 2017. The Intranasal Trigeminal System; pp. 113–114. [Google Scholar]
- Gaillard D., Xu M., Liu F., Millar S.E., Barlow L.A. β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates. PLoS Genet. 2015;11:e1005208. doi: 10.1371/journal.pgen.1005208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher P.E., Chappell M.C., Ferrario C.M., Tallant E.A. Distinct roles for ANG II and ANG-(1-7) in the regulation of angiotensin-converting enzyme 2 in rat astrocytes. Am. J. Physiol. Cell Physiol. 2006;290:C420–C426. doi: 10.1152/ajpcell.00409.2004. [DOI] [PubMed] [Google Scholar]
- Galougahi M.K., Ghorbani J., Bakhshayeshkaram M., Naeini A.S., Haseli S. Olfactory Bulb Magnetic Resonance Imaging in SARS-CoV-2-Induced Anosmia: The First Report. Acad. Radiol. 2020;27:892–893. doi: 10.1016/j.acra.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli A., Pezzati L., Conti F., Bernacchia D., Siano M., Oreni L., Rusconi S., Gervasoni C., Ridolfo A.L., Rizzardini G., et al. Self-reported olfactory and taste disorders in SARS-CoV-2 patients: a cross-sectional study. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa330. Published online March 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J., Ramani A., Mueller L., Niklas Ostermann P., Gabriel E., Islam Pranty A., Mueller-Shiffmann A., Mariappan A., Goureau O., Gruell H., et al. SARS-CoV-2 targets cortical neurons of 3D human brain organoids and shows neurodegeneration-like effects. bioRxiv. 2020 doi: 10.1101/2020.05.20.106575. [DOI] [Google Scholar]
- Gowrisankar Y.V., Clark M.A. Angiotensin II induces interleukin-6 expression in astrocytes: Role of reactive oxygen species and NF-κB. Mol. Cell. Endocrinol. 2016;437:130–141. doi: 10.1016/j.mce.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Gulbransen B.D., Clapp T.R., Finger T.E., Kinnamon S.C. Nasal solitary chemoreceptor cell responses to bitter and trigeminal stimulants in vitro. J. Neurophysiol. 2008;99:2929–2937. doi: 10.1152/jn.00066.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussing F., Bohm S. NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur. J. Neurosci. 2004;19:2511–2518. doi: 10.1111/j.0953-816X.2004.03331.x. [DOI] [PubMed] [Google Scholar]
- Haehner A., Draf J., Draeger S., With K.d., Hummel T. Predictive value of sudden olfactory loss in the diagnosis of COVID-19. medRxiv. 2020 doi: 10.1101/2020.04.27.20081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamichi R., Asano-Miyoshi M., Emori Y. Taste bud contains both short-lived and long-lived cell populations. Neuroscience. 2006;141:2129–2138. doi: 10.1016/j.neuroscience.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Peng J., Xu H., Chen Q. Taste Cell Is Abundant in the Expression of ACE2 Receptor of 2019-nCoV. Preprints.org. 2020 doi: 10.20944/preprints202004.0424.v1. [DOI] [Google Scholar]
- He L., Mäe M.A., Sun Y., Muhl L., Nahar K., Liébanas E.V., Fagerlund M.J., Oldner A., Liu J., Genové G., et al. Pericyte-specific vascular expression of SARS-CoV-2 receptor ACE2 – implications for microvascular inflammation and hypercoagulopathy in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.05.11.088500. [DOI] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M., et al. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D.B., Lin B., Peterson J., Schnittke N., Schwob J.E. Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc. Natl. Acad. Sci. USA. 2017;114:E5589–E5598. doi: 10.1073/pnas.1701333114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevezi P., Moyer B.D., Lu M., Gao N., White E., Echeverri F., Kalabat D., Soto H., Laita B., Li C., et al. Genome-wide analysis of gene expression in primate taste buds reveals links to diverse processes. PLoS ONE. 2009;4:e6395. doi: 10.1371/journal.pone.0006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C., Surda P., Whitehead E., Kumar B.N. Early recovery following new onset anosmia during the COVID-19 pandemic - an observational cohort study. J. Otolaryngol. Head Neck Surg. 2020;49:26. doi: 10.1186/s40463-020-00423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornuss D., Lange B., Schroeter N., Rieg S., Kern W.V., Wagner D. Anosmia in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.28.20083311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020 doi: 10.1016/j.cell.2020.05.042. Published online May 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T., Rothbauer C., Barz S., Grosser K., Pauli E., Kobal G. Olfactory function in acute rhinitis. Ann. N Y Acad. Sci. 1998;855:616–624. doi: 10.1111/j.1749-6632.1998.tb10632.x. [DOI] [PubMed] [Google Scholar]
- Hummel T., Rothbauer C., Pauli E., Kobal G. Effects of the nasal decongestant oxymetazoline on human olfactory and intranasal trigeminal function in acute rhinitis. Eur. J. Clin. Pharmacol. 1998;54:521–528. doi: 10.1007/s002280050507. [DOI] [PubMed] [Google Scholar]
- Hung E.C.W., Chim S.S.C., Chan P.K.S., Tong Y.K., Ng E.K.O., Chiu R.W.K., Leung C.-B., Sung J.J.Y., Tam J.S., Lo Y.M.D. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin. Chem. 2003;49:2108–2109. doi: 10.1373/clinchem.2003.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C.-S. Olfactory neuropathy in severe acute respiratory syndrome: report of A case. Acta Neurol. Taiwan. 2006;15:26–28. [PubMed] [Google Scholar]
- Iravani B., Arshamian A., Ravia A., Mishor E., Snitz K., Shushan S., Roth Y., Perl O., Honigstein D., Weissgross R., et al. Relationship between odor intensity estimates and COVID-19 population prediction in a Swedish sample. medRxiv. 2020 doi: 10.1101/2020.05.07.20094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafek B.W., Hartman D., Eller P.M., Johnson E.W., Strahan R.C., Moran D.T. Postviral Olfactory Dysfunction. Am. J. Rhinol. 1990;4:91–100. [Google Scholar]
- Kang Y.-L., Chou Y.-Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.21.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman A., Choo E., Koh A., Dando R. Inflammation arising from obesity reduces taste bud abundance and inhibits renewal. PLoS Biol. 2018;16:e2001959. doi: 10.1371/journal.pbio.2001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye R., Chang C.W.D., Kazahaya K., Brereton J., Denneny J.C., 3rd COVID-19 Anosmia Reporting Tool: Initial Findings. Otolaryngol. Head Neck Surg. 2020 doi: 10.1177/0194599820922992. Published online April 28, 2020. [DOI] [PubMed] [Google Scholar]
- Kehoe P.G., Wong S., Al Mulhim N., Palmer L.E., Miners J.S. Angiotensin-converting enzyme 2 is reduced in Alzheimer’s disease in association with increasing amyloid-β and tau pathology. Alzheimers Res. Ther. 2016;8:50. doi: 10.1186/s13195-016-0217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern R.C. Chronic sinusitis and anosmia: pathologic changes in the olfactory mucosa. Laryngoscope. 2000;110:1071–1077. doi: 10.1097/00005537-200007000-00001. [DOI] [PubMed] [Google Scholar]
- Kim A., Feng P., Ohkuri T., Sauers D., Cohn Z.J., Chai J., Nelson T., Bachmanov A.A., Huang L., Wang H. Defects in the peripheral taste structure and function in the MRL/lpr mouse model of autoimmune disease. PLoS ONE. 2012;7:e35588. doi: 10.1371/journal.pone.0035588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyuncu O.O., Hogue I.B., Enquist L.W. Virus infections in the nervous system. Cell Host Microbe. 2013;13:379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. Published online May 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurendon T., Radulesco T., Mugnier J., Gérault M., Chagnaud C., El Ahmadi A.-A., Varoquaux A. Bilateral transient olfactory bulbs edema during COVID-19-related anosmia. Neurology. 2020 doi: 10.1212/WNL.0000000000009850. Published online May 22, 2020. [DOI] [PubMed] [Google Scholar]
- Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., Dequanter D., Blecic S., El Afia F., Distinguin L., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020 doi: 10.1007/s00405-020-05965-1. Published online April 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J.R., Chiesa-Estomba C.M., Place S., Van Laethem Y., Cabaraux P., Mat Q., Huet K., Plzak J., Horoi M., Hans S., et al. COVID-19 Task Force of YO-IFOS Clinical and epidemiological characteristics of 1420 European patients with mild-to-moderate coronavirus disease 2019. J. Intern. Med. 2020 doi: 10.1111/joim.13089. Published online April 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Macpherson L.J., Parada C.A., Zuker C.S., Ryba N.J.P. Rewiring the taste system. Nature. 2017;548:330–333. doi: 10.1038/nature23299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons K., Fu Z., Aoudé I., Ogura T., Sun J., Chang J., Mbonu K., Matsumoto I., Arakawa H., Lin W. Lack of TRPM5-Expressing Microvillous Cells in Mouse Main Olfactory Epithelium Leads to Impaired Odor-Evoked Responses and Olfactory-Guided Behavior in a Challenging Chemical Environment. eneuro. 2017;4 doi: 10.1523/ENEURO.0135-17.2017. ENEURO.0135-0117.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold D. Distortion of olfactory perception: diagnosis and treatment. Chem. Senses. 2002;27:611–615. doi: 10.1093/chemse/27.7.611. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Bai W.Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may be at least partially responsible for the respiratory failure of COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25728. Published online March 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F. Sustentacular Cell Enwrapment of Olfactory Receptor Neuronal Dendrites: An Update. Genes (Basel) 2020;11:11. doi: 10.3390/genes11050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.X., Ermilov A., Grachtchouk M., Li L., Gumucio D.L., Dlugosz A.A., Mistretta C.M. Multiple Shh signaling centers participate in fungiform papilla and taste bud formation and maintenance. Dev. Biol. 2013;382:82–97. doi: 10.1016/j.ydbio.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Xiong W., Liu D., Liu J., Yang D., Li N., Mu J., Guo J., Li W., Wang G., et al. New onset acute symptomatic seizure and risk factors in coronavirus disease 2019: A retrospective multicenter study. Epilepsia. 2020;61:e49–e53. doi: 10.1111/epi.16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä M.J., Puhakka T., Ruuskanen O., Leinonen M., Saikku P., Kimpimäki M., Blomqvist S., Hyypiä T., Arstila P. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:1–9. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresh A., Rodriguez Gil D., Whitman M.C., Greer C.A. Principles of glomerular organization in the human olfactory bulb--implications for odor processing. PLoS ONE. 2008;3:e2640. doi: 10.1371/journal.pone.0002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt J., Radke J., Dittmayer C., Mothes R., Franz J., Laue M., Schneider J., Brünink S., Hassan O., Stenzel W., et al. Olfactory transmucosal SARS-CoV-2 invasion as port of Central Nervous System entry in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.06.04.135012. [DOI] [PubMed] [Google Scholar]
- Menni C., Valdes A.M., Freidin M.B., Sudre C.H., Nguyen L.H., Drew D.A., Ganesh S., Varsavsky T., Cardoso M.J., El-Sayed Moustafa J.S., et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0916-2. Published online May 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H., Kusakabe Y., Harada S. Cell lineage and differentiation in taste buds. Arch. Histol. Cytol. 2006;69:209–225. doi: 10.1679/aohc.69.209. [DOI] [PubMed] [Google Scholar]
- Miura H., Scott J.K., Harada S., Barlow L.A. Sonic hedgehog-expressing basal cells are general post-mitotic precursors of functional taste receptor cells. Dev. Dyn. 2014;243:1286–1297. doi: 10.1002/dvdy.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K., Serizawa S., Kimura H.M., Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J. Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moein S.T., Hashemian S.M.R., Mansourafshar B., Khorram-Tousi A., Tabarsi P., Doty R.L. Smell dysfunction: a biomarker for COVID-19. Int. Forum Allergy Rhinol. 2020 doi: 10.1002/alr.22587. Published online April 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M., Zohrabian V.M. Neuroradiologists, Be Mindful of the Neuroinvasive Potential of COVID-19. AJNR Am. J. Neuroradiol. 2020;41:E37–E39. doi: 10.3174/ajnr.A6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2324-7. Published online May 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C., Luecken M.D., Eraslan G., Waghray A., Heimberg G., Sikkema L., Kobayashi Y., Vaishnav E.D., Subramanian A., Smillie C., et al. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv. 2020 doi: 10.1101/2020.04.19.049254. [DOI] [Google Scholar]
- Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.Q., Wu Y., Bonilla L.S., von Buchholtz L.J., Ryba N.J.P. Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS ONE. 2017;12:e0185543. doi: 10.1371/journal.pone.0185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.Q., Le Pichon C.E., Ryba N. Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. eLife. 2019;8:e49679. doi: 10.7554/eLife.49679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary C.E., Schneider C., Locksley R.M. Tuft Cells-Systemically Dispersed Sensory Epithelia Integrating Immune and Neural Circuitry. Annu. Rev. Immunol. 2019;37:47–72. doi: 10.1146/annurev-immunol-042718-041505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T., Clark C., Hogan B.L.M. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells. 2009;27:442–450. doi: 10.1634/stemcells.2008-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parma V., Ohla K., Veldhuizen M.G., Niv M.Y., Kelly C.E., Bakke A.J., Cooper K.W., Bouysset C., Pirastu N., Dibattista M., et al. More than smell - COVID-19 is associated with severe impairment of smell, taste, and chemesthesis. medRxiv. 2020 doi: 10.1101/2020.05.04.20090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino R., Cooper K.W., Di Pizio A., Joseph P.V., Bhutani S., Parma V. Corona Viruses and the Chemical Senses: Past, Present, and Future. Chem. Senses. 2020 doi: 10.1093/chemse/bjaa031. Published online May 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Martinez I., Nagai T., Chaudhari N. Functional cell types in taste buds have distinct longevities. PLoS ONE. 2013;8:e53399. doi: 10.1371/journal.pone.0053399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Jacobsen G., Afifi A. Spread of a neurotropic murine coronavirus into the CNS via the trigeminal and olfactory nerves. Virology. 1989;170:556–560. doi: 10.1016/0042-6822(89)90446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisco A.O., McGeever A., Schaum N., Karkanias J., Neff N.F., Darmanis S., Wyss-Coray T., Quake S.R. A Single Cell Transcriptomic Atlas Characterizes Aging Tissues in the Mouse. bioRxiv. 2020 doi: 10.1101/661728. [DOI] [Google Scholar]
- Politi L.S., Salsano E., Grimaldi M. Magnetic Resonance Imaging Alteration of the Brain in a Patient With Coronavirus Disease 2019 (COVID-19) and Anosmia. JAMA Neurol. 2020 doi: 10.1001/jamaneurol.2020.2125. Published online May 29, 2020. [DOI] [PubMed] [Google Scholar]
- Poyiadji N., Shahin G., Noujaim D., Stone M., Patel S., Griffith B. COVID-19-associated Acute Hemorrhagic Necrotizing Encephalopathy: CT and MRI Features. Radiology. 2020 doi: 10.1148/radiol.2020201187. Published online March 31, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott S.L., Umans B.D., Williams E.K., Brust R.D., Liberles S.D. An Airway Protection Program Revealed by Sweeping Genetic Control of Vagal Afferents. Cell. 2020;181:574–589.e14. doi: 10.1016/j.cell.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2011400. Published online May 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Sukumaran S.K., Jyotaki M., Redding K., Jiang P., Margolskee R.F. Gli3 is a negative regulator of Tas1r3-expressing taste cells. PLoS Genet. 2018;14:e1007058. doi: 10.1371/journal.pgen.1007058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C., Cui C., Hautefort C., Haehner A., Zhao J., Yao Q., Zeng H., Nisenbaum E.J., Liu L., Zhao Y., et al. Olfactory and Gustatory Dysfunction as An Early Identifier of COVID-19 in Adults and Children: An International Multicenter Study. medRxiv. 2020 doi: 10.1101/2020.05.13.20100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S., Cao L., Rickenbacher G.T., Benz E.G., Magdamo C., Ramirez Gomez L.A., Holbrook E., Dhilla Albers A., Gallagher R., Westover M.B., et al. Innate immune signaling in the olfactory epithelium reduces odorant receptor levels: modeling transient smell loss in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.06.14.20131128. [DOI] [Google Scholar]
- Romanov R.A., Lasher R.S., High B., Savidge L.E., Lawson A., Rogachevskaja O.A., Zhao H., Rogachevsky V.V., Bystrova M.F., Churbanov G.D., et al. Chemical synapses without synaptic vesicles: Purinergic neurotransmission through a CALHM1 channel-mitochondrial signaling complex. Sci Signal. 2018;11:eaao1815. doi: 10.1126/scisignal.aao1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombaux P., Martinage S., Huart C., Collet S. Post-infectious olfactory loss: a cohort study and update. B-ENT. 2009;5(Suppl 13):89–95. [PubMed] [Google Scholar]
- Roper S.D. In: Nilius B., Flockerzi V., editors. Volume II. Springer International Publishing; Cham: 2014. TRPs in Taste and Chemesthesis; pp. 827–871. (Mammalian Transient Receptor Potential (TRP) Cation Channels). [Google Scholar]
- Roper S.D., Chaudhari N. Taste buds: cells, signals and synapses. Nat. Rev. Neurosci. 2017;18:485–497. doi: 10.1038/nrn.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan N.R., Lee S., Sahu N., Kanaan A., Cox S., Phillips C.D., Wang E.W. The role of viruses in the clinical presentation of chronic rhinosinusitis. Am. J. Rhinol. Allergy. 2015;29:e197–e200. doi: 10.2500/ajra.2015.29.4242. [DOI] [PubMed] [Google Scholar]
- Saunders C.J., Christensen M., Finger T.E., Tizzano M. Cholinergic neurotransmission links solitary chemosensory cells to nasal inflammation. Proc. Natl. Acad. Sci. USA. 2014;111:6075–6080. doi: 10.1073/pnas.1402251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T., Hirschbühl K., Burkhardt K., Braun G., Trepel M., Märkl B., Claus R. Postmortem Examination of Patients With COVID-19. JAMA. 2020;323:2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M.B., et al. Tabula Muris Consortium. Overall coordination. Logistical coordination. Organ collection and processing. Library preparation and sequencing. Computational data analysis. Cell type annotation. Writing group. Supplemental text writing group. Principal investigators Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser R.J., Mulligan J.K., Hyer J.M., Karnezis T.T., Gudis D.A., Soler Z.M. Mucous Cytokine Levels in Chronic Rhinosinusitis-Associated Olfactory Loss. JAMA Otolaryngol. Head Neck Surg. 2016;142:731–737. doi: 10.1001/jamaoto.2016.0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J.E., Saha S., Youngentob S.L., Jubelt B. Intranasal inoculation with the olfactory bulb line variant of mouse hepatitis virus causes extensive destruction of the olfactory bulb and accelerated turnover of neurons in the olfactory epithelium of mice. Chem. Senses. 2001;26:937–952. doi: 10.1093/chemse/26.8.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J.E., Jang W., Holbrook E.H., Lin B., Herrick D.B., Peterson J.N., Hewitt Coleman J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J. Comp. Neurol. 2017;525:1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemura N., Takai S., Hirose F., Yoshida R., Sanematsu K., Ninomiya Y. Expression of Renin-Angiotensin System Components in the Taste Organ of Mice. Nutrients. 2019;11:2251. doi: 10.3390/nu11092251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoer S., Karady T., Keshet A., Shilo S., Rossman H., Gavrieli A., Meir T., Lavon A., Kolobkov D., Kalka I., et al. Who should we test for COVID-19? A triage model built from national symptom surveys. medRxiv. 2020 doi: 10.1101/2020.05.18.20105569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020 doi: 10.1038/s41586-020-2342-5. Published online May 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbu T.T., Holen T. Aquaporin pathways and mucin secretion of Bowman’s glands might protect the olfactory mucosa. Chem. Senses. 2012;37:35–46. doi: 10.1093/chemse/bjr063. [DOI] [PubMed] [Google Scholar]
- Solomon I.H., Normandin E., Bhattacharyya S., Mukerji S.S., Keller K., Ali A.S., Adams G., Hornick J.L., Padera R.F., Jr., Sabeti P. Neuropathological Features of Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2019373. Published online June 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinato G., Fabbris C., Polesel J., Cazzador D., Borsetto D., Hopkins C., Boscolo-Rizzo P. Alterations in Smell or Taste in Mildly Symptomatic Outpatients With SARS-CoV-2 Infection. JAMA. 2020 doi: 10.1001/jama.2020.6771. Published online April 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.05.020. Published online May 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Takeda M., Farbman A.I. Supporting cells as phagocytes in the olfactory epithelium after bulbectomy. J. Comp. Neurol. 1996;376:509–517. doi: 10.1002/(SICI)1096-9861(19961223)376:4<509::AID-CNE1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Saito K., Min W.-P., Vladau C., Toida K., Itoh H., Murakami S. Identification of viruses in patients with postviral olfactory dysfunction. Laryngoscope. 2007;117:272–277. doi: 10.1097/01.mlg.0000249922.37381.1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taruno A., Vingtdeux V., Ohmoto M., Ma Z., Dvoryanchikov G., Li A., Adrien L., Zhao H., Leung S., Abernethy M., et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 2013;495:223–226. doi: 10.1038/nature11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizzano M., Gulbransen B.D., Vandenbeuch A., Clapp T.R., Herman J.P., Sibhatu H.M., Churchill M.E., Silver W.L., Kinnamon S.C., Finger T.E. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc. Natl. Acad. Sci. USA. 2010;107:3210–3215. doi: 10.1073/pnas.0911934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi A., Mohammadbagheri E., Akbari Dilmaghani N., Bayat A.-H., Fathi M., Vakili K., Alizadeh R., Rezaeimirghaed O., Hajiesmaeili M., Ramezani M., et al. Proinflammatory Cytokines in the Olfactory Mucosa Result in COVID-19 Induced Anosmia. ACS Chem. Neurosci. 2020 doi: 10.1021/acschemneuro.0c00249. Published online June 11, 2020. [DOI] [PubMed] [Google Scholar]
- Trotier D., Bensimon J.L., Herman P., Tran Ba Huy P., Døving K.B., Eloit C. Inflammatory obstruction of the olfactory clefts and olfactory loss in humans: a new syndrome? Chem. Senses. 2007;32:285–292. doi: 10.1093/chemse/bjl057. [DOI] [PubMed] [Google Scholar]
- Tseng C.-T.K., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ualiyeva S., Hallen N., Kanaoka Y., Ledderose C., Matsumoto I., Junger W.G., Barrett N.A., Bankova L.G. Airway brush cells generate cysteinyl leukotrienes through the ATP sensor P2Y2. Sci. Immunol. 2020;5:eaax7224. doi: 10.1126/sciimmunol.aax7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueha R., Kondo K., Kagoya R., Shichino S., Ueha S., Yamasoba T. Background mechanisms of olfactory dysfunction in COVID-19: expression of ACE2, TMPRSS2, and Furin in the nose and olfactory bulb in human and mice. bioRxiv. 2020 doi: 10.1101/2020.05.15.097352. [DOI] [Google Scholar]
- Vaira L.A., Deiana G., Fois A.G., Pirina P., Madeddu G., De Vito A., Babudieri S., Petrocelli M., Serra A., Bussu F., et al. Objective evaluation of anosmia and ageusia in COVID-19 patients: Single-center experience on 72 cases. Head Neck. 2020;42:1252–1258. doi: 10.1002/hed.26204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Verdijk R., Kuiken T. The olfactory nerve: a shortcut for influenza and other viral diseases into the central nervous system. J. Pathol. 2015;235:277–287. doi: 10.1002/path.4461. [DOI] [PubMed] [Google Scholar]
- Vandenbeuch A., Anderson C.B., Parnes J., Enjyoji K., Robson S.C., Finger T.E., Kinnamon S.C. Role of the ectonucleotidase NTPDase2 in taste bud function. Proc. Natl. Acad. Sci. USA. 2013;110:14789–14794. doi: 10.1073/pnas.1309468110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A., Puranik A., Anand A., Zemmour D., Yao X., Wu X., Chilaka R., Murakowski D.K., Standish K., Raghunathan B., et al. Knowledge synthesis from 100 million biomedical documents augments the deep expression profiling of coronavirus receptors. bioRxiv. 2020 doi: 10.1101/2020.03.24.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana F. Chemosensory properties of the trigeminal system. ACS Chem. Neurosci. 2011;2:38–50. doi: 10.1021/cn100102c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victores A.J., Chen M., Smith A., Lane A.P. Olfactory loss in chronic rhinosinusitis is associated with neuronal activation of c-Jun N-terminal kinase. Int. Forum Allergy Rhinol. 2018;8:415–420. doi: 10.1002/alr.22053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogalis F., Hegg C.C., Lucero M.T. Ionic conductances in sustentacular cells of the mouse olfactory epithelium. J. Physiol. 2005;562:785–799. doi: 10.1113/jphysiol.2004.079228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Shweta F.N.U., Murugadoss K., Awasthi S., Venkatakrishnan A., Puranik A., Kang M., Pickering B.W., O’Horo J.C., Bauer P.R., Razonable R.R., et al. Augmented Curation of Unstructured Clinical Notes from a Massive EHR System Reveals Specific Phenotypic Signature of Impending COVID-19 Diagnosis. medRxiv. 2020 doi: 10.1101/2020.04.19.20067660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127–e00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhou M., Brand J., Huang L. Inflammation activates the interferon signaling pathways in taste bud cells. J. Neurosci. 2007;27:10703–10713. doi: 10.1523/JNEUROSCI.3102-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhou M., Brand J., Huang L. Inflammation and taste disorders: mechanisms in taste buds. Ann. N Y Acad. Sci. 2009;1170:596–603. doi: 10.1111/j.1749-6632.2009.04480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welge-Lüssen A. Re-establishment of olfactory and taste functions. GMS Curr. Top. Otorhinolaryngol. Head Neck Surg. 2005;4:Doc06. [PMC free article] [PubMed] [Google Scholar]
- Welge-Lüssen A., Wolfensberger M. Olfactory disorders following upper respiratory tract infections. Adv. Otorhinolaryngol. 2006;63:125–132. doi: 10.1159/000093758. [DOI] [PubMed] [Google Scholar]
- Whitcroft K.L., Hummel T. Olfactory Dysfunction in COVID-19: Diagnosis and Management. JAMA. 2020;323:2512–2514. doi: 10.1001/jama.2020.8391. [DOI] [PubMed] [Google Scholar]
- Wood A.J., Antoszewska H., Fraser J., Douglas R.G. Is chronic rhinosinusitis caused by persistent respiratory virus infection? Int. Forum Allergy Rhinol. 2011;1:95–100. doi: 10.1002/alr.20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu X., Chen Z., Duan J., Hashimoto K., Yang L., Liu C., Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhong S., Liu J., Li L., Li Y., Wu X., Li Z., Deng P., Zhang J., Zhong N., et al. Detection of severe acute respiratory syndrome coronavirus in the brain: potential role of the chemokine mig in pathogenesis. Clin. Infect. Dis. 2005;41:1089–1096. doi: 10.1086/444461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.H., Faraji F., Prajapati D.P., Boone C.E., DeConde A.S. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 2020 doi: 10.1002/alr.22579. Published online April 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M., et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Yang R., Dzowo Y.K., Wilson C.E., Russell R.L., Kidd G.J., Salcedo E., Lasher R.S., Kinnamon J.C., Finger T.E. Three-dimensional reconstructions of mouse circumvallate taste buds using serial blockface scanning electron microscopy: I. Cell types and the apical region of the taste bud. J. Comp. Neurol. 2020;528:756–771. doi: 10.1002/cne.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee K.K., Li Y., Redding K.M., Iwatsuki K., Margolskee R.F., Jiang P. Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells. 2013;31:992–1000. doi: 10.1002/stem.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Jin H., Zhang W., Ding C., O’Keeffe S., Ye M., Zuker C.S. Sour Sensing from the Tongue to the Brain. Cell. 2019;179:392–402.e15. doi: 10.1016/j.cell.2019.08.031. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xiao M., Zhang S., Xia P., Cao W., Jiang W., Chen H., Ding X., Zhao H., Zhang H., et al. Coagulopathy and Antiphospholipid Antibodies in Patients with Covid-19. N. Engl. J. Med. 2020;382:e38. doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Scherer P.W., Hajiloo S.A., Dalton P. Effect of anatomy on human nasal air flow and odorant transport patterns: implications for olfaction. Chem. Senses. 2004;29:365–379. doi: 10.1093/chemse/bjh033. [DOI] [PubMed] [Google Scholar]
- Zhao K., Jiang J., Pribitkin E.A., Dalton P., Rosen D., Lyman B., Yee K.K., Rawson N.E., Cowart B.J. Conductive olfactory losses in chronic rhinosinusitis? A computational fluid dynamics study of 29 patients. Int. Forum Allergy Rhinol. 2014;4:298–308. doi: 10.1002/alr.21272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen D., Szabla R., Zheng M., Li G., Du P., Zheng S., Li X., Song C., Li R., et al. Broad and differential animal ACE2 receptor usage by SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.19.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]