Abstract
The transcriptomes of the venom glands of 13 closely related species of vermivorous cones endemic to West Africa from genera Africonus and Varioconus were sequenced and venom repertoires compared within a phylogenetic framework using one Kalloconus species as outgroup. The total number of conotoxin precursors per species varied between 108 and 221. Individuals of the same species shared about one-fourth of the total conotoxin precursors. The number of common sequences was drastically reduced in the pairwise comparisons between closely related species, and the phylogenetical signal was totally eroded at the inter-generic level (no sequence was identified as shared derived), due to the intrinsic high variability of these secreted peptides. A common set of four conotoxin precursor superfamilies (T, O1, O2 and M) was expanded in all studied cone species, and thus, they are considered the basic venom toolkit for hunting and defense in the West African vermivorous cone snails. Maximum-likelihood ancestral character reconstructions inferred shared conotoxin precursors preferentially at internal nodes close to the tips of the phylogeny (between individuals and between closely related species) as well as in the common ancestor of Varioconus. Besides the common toolkit, the two genera showed significantly distinct catalogues of conotoxin precursors in terms of type of superfamilies present and the abundance of members per superfamily, but had similar relative expression levels indicating functional convergence. Differential expression comparisons between vermivorous and piscivorous cones highlighted the importance of the A and S superfamilies for fish hunting and defense.
Keywords: conotoxin precursors, transcriptomes, Africonus, Varioconus, vermivorous cones
1. Introduction
Cones (Gastropoda: Conidae) are marine venomous predators that actively hunt on worms, snails and fish [1]. Their venom is a cocktail constituted by hundreds of peptides named conotoxins, as well as by hormones and by other proteins that participate in the synthesis or enhance the activity of the venom [2,3]. Once inside the prey, conotoxins interact with ion channels and neurotransmitter receptors triggering different physiological responses, from sedation to tetanic paralysis [4]. Conotoxin precursors typically present a three domain structure, consisting of signal, pro-peptide and mature (i.e. the functional toxin after processing of the precursor) regions [5]. The signal region is conserved, and it is used to classify the peptides into different ‘superfamilies’ [4]. The composition of the venom is highly variable among species, specimens and even within the same individual depending on its physiological status or ecological interactions [6–13].
Most cone snail venomic studies have been driven preferentially by the pharmacological potential of conotoxins and were limited to the purification of mature peptides and the identification of their function. Thus, they lack the wider evolutionary perspective already applied in the study of other venomous animals [14–16]. Comparing venom cocktails from different cone species within a phylogenetic framework should provide insights on how the rich conotoxin diversity was generated [12,17], to what extent distinct venom repertoires are adapted to different diet specializations [10,18,19], and which are the functional constraints and levels of convergence imposed by this coevolutionary arms race system [8,20], among others.
As in other venomous animals [16,21], dietary breadth has been proposed to be a main factor triggering venom evolution in cones [18,19,22]. Since hunting performance relies on venom specificity, shifts in diet could trigger changes in venom composition [10,23,24] and in general, species with more generalized diets would tend to have more complex venoms [19,22]. Moreover, instances of functional convergence have been shown in the venom cocktails of Atlantic and Indo-Pacific piscivorous cones [8]. Another level of evolutionary complexity comes from the capacity of cones to modulate the composition of their venom depending on its final use, whether to subdue preys or defend themselves against predators [12,25].
The above-mentioned studies explored general venom evolutionary trends at the family (Conidae) level by comparing distantly related lineages. A few studies compared venom cocktails from pairs of species within the same genus but lacked an evolutionary perspective (e.g. [7]). Here, we analyse venom evolution within two radiations of closely related cone species inhabiting West Africa [26,27]: one comprising cones endemic to the Cabo Verde archipelago, ascribed to genus Africonus; the other including cones endemic to Senegal (plus one closely related species inhabiting Canary Islands), recently ascribed to genus Varioconus [28]. Importantly, robust phylogenies based on mitogenomes are available for both clades providing the necessary framework for evolutionary studies [26,27]. The clade of cones endemic to Cabo Verde diversified about 9 Mya into four main lineages and at least 40 endemic species [26]. The clade of cones endemic to Senegal and Canary Islands diversified about 6 Mya into three main lineages and at least 13 endemic species [27]. All species in both clades are vermivorous; Africonus species show little apparent differences in radular tooth morphology whereas the three clades of Varioconus from Senegal and Canary Islands have each distinct radular teeth, suggesting subtle diet specializations [27]. No study has analysed the venom transcriptomes of these endemic cones.
Here, we sequenced the venom gland transcriptomes from 13 species belonging to genera Africonus and Varioconus, as well as one from Kalloconus trochulus, which was used as outgroup. We aimed to (i) describe venom compositions in terms of the presence, member diversity and relative expression levels of the conotoxin precursor superfamilies; (ii) assess the levels of divergence in venom composition at different hierarchical (taxonomic) levels and discern between shared-derived peptides and potential cases of functional convergence; (iii) determine whether there could be instances of differential expression between the two genera as footprint of adaptation; and (iv) compare differential conotoxin expression between these vermivorous species and the piscivorous species Chelyconus ermineus [8] and Pionoconus magus [29] from the Atlantic and the Indo-Pacific oceans, respectively, to further understand the connections between venom evolution to diet specialization and defense.
2. Material and methods
(a). Taxon sampling
Taxon selection was aimed at having at least one representative per main lineage of the two genera plus a close outgroup [26,27]. The complete list of specimens, species, sampling localities and museum vouchers is provided in table 1. Phylogenetic relationships based on mitogenomes are depicted in electronic supplementary material, figure S1. In order to assess intraspecific variability, for the genus Varioconus, we studied two specimens of Varioconus mercator (V_1258 and V_1302). These individuals showed distinct shell phenotypes and in fact, V_1258 could be assigned to the recently described species, Varioconus stimpsonorum [30]. However, its mitogenome sequence divergence to V. mercator is exactly at the threshold used to delimit species in West African cones [27]. Moreover, the results here presented in terms of venom composition (see below) strongly suggest that V. stimpsonorum should be considered a synonym of V. mercator [28], and hence, the specimen V_1258 was treated as V. mercator herein. In the case of Africonus, we included two specimens of Africonus maioensis (A_0055 and A_0039; representing two different shell phenotypes formerly classified as distinct species but now synonymized; [27]). All the specimens were adults and were dissected in a resting stage to remove the venom duct, which was preserved in RNAlater (Invitrogen, Life technologies).
Table 1.
Specimens analysed in this study and main statistics of Illumina sequencing and assembly.
| ID | species | country | locality/island | voucher MNCN | SRA accesion | sequencing date (DD/MM/YYYY) | number reads | % clean reads | number contigs | number BLAST hits | number proteins | number conotoxins |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A_0885 | Africonus antoniomonteiroi | Cabo Verde | Pedra Lume, Sal | 15.05/79794 | SRR11807494 | 21-12-2016 | 26 026 957 | 100 | 83 565 | 731 | 214 | 159 |
| A_0520 | Africonus boavistensis | Cabo Verde | Ervatao, Boa Vista | 15.05/80413 | SRR11807497 | 21-12-2016 | 26 715 260 | 100 | 39 935 | 797 | 199 | 168 |
| A_0855 | Africonus cuneolus | Cabo Verde | Fontona, Sal | 15.05/79764 | SRR11807496 | 13-05-2015 | 14 771 346 | 99.48 | 46 983 | 800 | 195 | 154 |
| A_0048 | Africonus galeao | Cabo Verde | Navío Quebrado, Maio | 15.05/78673 | SRR11807500 | 21-12-2016 | 28 109 709 | 100 | 50 811 | 803 | 180 | 151 |
| A_1387 | Africonus grahami | Cabo Verde | Calhau, São Vicente | 15.05/78549 | SRR11807507 | 21-12-2016 | 22 718 525 | 100 | 51 601 | 850 | 183 | 156 |
| A_0025 | Africonus infinitus | Cabo Verde | Ponta do Pau Seco, Maio | 15.05/78650 | SRR11807493 | 13-03-2014 | 35 854 397 | 98.52 | 76 339 | 1091 | 203 | 167 |
| A_0039 | Africonus maioensis | Cabo Verde | Praia Santana, Maio | 15.05/78664 | SRR11807501 | 28-10-2013 | 52 523 501 | 100 | 78 886 | 783 | 237 | 188 |
| A_0055 | Africonus maioensis | Cabo Verde | Navío Quebrado, Maio | 15.05/78680 | SRR11807499 | 28-10-2013 | 44 748 977 | 100 | 105 099 | 850 | 268 | 214 |
| A_0875 | Africonus miruchae | Cabo Verde | Terrinha Fina, Sal | 15.05/79784 | SRR11807495 | 21-12-2016 | 24 097 307 | 100 | 60 347 | 615 | 176 | 136 |
| A_0031 | Africonus raulsilvai | Cabo Verde | Praia da Soca, Maio | 15.05/78656 | SRR11807492 | 28-10-2013 | 56 718 528 | 100 | 99 699 | 1249 | 220 | 183 |
| A_0239 | Africonus verdensis | Cabo Verde | Tarrafal, Santiago | 15.05/78864 | SRR11807498 | 28-10-2013 | 40 237 424 | 100 | 77 906 | 1266 | 239 | 197 |
| V_CG13 | Varioconus guanche | Spain | Playa del Cable, Lanzarote | — | SRR11807502 | 08-03-2016 | 29 973 740 | 100 | 85 276 | 815 | 245 | 195 |
| V_1258 | Varioconus mercator | Senegal | Almadies | 15.05/78419 | SRR11807505 | 08-03-2016 | 28 883 175 | 100 | 66 684 | 631 | 205 | 167 |
| V_1302 | Varioconus mercator | Senegal | Ndayane | 15.05/78463 | SRR11807503 | 08-03-2016 | 28 392 465 | 100 | 75 406 | 783 | 261 | 221 |
| V_1278 | Varioconus reticulatus | Senegal | Ngor | 15.05/78439 | SRR11807504 | 21-12-2016 | 24 263 358 | 100 | 50 358 | 479 | 142 | 108 |
| K_0010 | Kalloconus trochulus | Cabo Verde | Ponta do Pau Seco, Maio | 15.05/78635 | SRR11807506 | 13-05-2015 | 75 347 025 | 96.19 | 69 688 | 1114 | 179 | 141 |
(b). RNA extraction and sequencing
RNA extraction and sequencing were performed as in [8]. Briefly, each venom duct was incubated with 500 µl of TRIzol LS Reagent (Invitrogen, Life Technologies) and grinded with ceramic beads in a Praecellys Evolution homogenizer. Total RNA was purified using the Direct-Zol RNA Miniprep kit (Zymo Research, Irvine) following the manufacturer's instructions. Dual-indexed cDNA libraries were constructed for each sample using the TruSeq RNA library Prep kit v2 (Illumina, San Diego) at Sistemas Genómicos (Valencia, Spain) following the manufacturer's instructions. After the quality of the libraries was checked, they were pooled and split into several runs of paired-end sequencing (2 × 100 bp) in an Illumina HiSeq2500 (each sample divided into two flow cells to avoid sequencing biases) following the standard procedures at Sistemas Genómicos (Valencia, Spain).
(c). Transcriptome assembly and conotoxin identification
The raw reads corresponding to the different individuals were sorted using the library indices, which were removed using Cutadapt v.1.3 [31]. Raw read quality was checked with FastQC v.0.10.1 (www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the assembly was performed using Trinity v.2.6.6 [32] with default settings (minimum contig length = 200 bp, sequence identity threshold = 0.95) and the trimmomatic option active with default parameters. The raw reads of all transcriptomes are available at the SRA database (table 1).
Conotoxin precursors, hormones and associated venom proteins available in GenBank release 222, Uniprot release 2017_09, and ConoServer release 30/10/2017 were downloaded 30 October 2017 and concatenated into a single fasta file. Duplicated sequences were removed, and the resulting file was formatted to create the custom reference database using BLAST+ [33].
Proteins of interest in the assembled transcriptomes were identified using BLASTX over the custom reference database (e-value: 1 × 10−5). The amino acid sequences were manually inspected and those considered as false positives or assembly artefacts (showing internal stop codons and chimeras), those that were duplicated or highly truncated (missing greater than 55% of the estimated length of the reference protein), and those showing low coverage values were discarded. We implemented an extra curation step consisting on TBLASTX searches over the nr database in GenBank to discard wrong open reading frame (ORF) assignations.
The retained sequences constituted our working list of conotoxin precursors, hormones and associated venom proteins (electronic supplementary material, file S1; table S1). The three domain structure and cysteine frameworks of conotoxin precursor alignments were inferred using Conoprec [5]. Proteins were assigned to a given superfamily by comparison with best-hit results using BLASTP searches against GenBank, and in the case of the conotoxin precursors, taking into consideration the percentage of identity in the signal region using a general threshold of 70% [4]. We further checked the correct identification of all conotoxin precursor superfamilies by aligning all the signal regions and building a neighbour-joining dendrogram (electronic supplementary material, figure S2) based on uncorrected p distances using ClustalW [34]. Within each superfamily, sequences were assigned to different groups of paralogy based on the sequence divergence at the pro-peptide region, the presence of different cysteine frameworks in the mature peptide and the recovery of clades in the reconstructed dendrogram. Those sequences that did not match any previously reported conotoxin precursor superfamily were classified into unassigned superfamilies and described here.
(d). Comparative analyses of venom composition
The conotoxin precursors of each species were pairwise compared. All sequences that were common to two or more species were mapped onto the reconstructed phylogeny (electronic supplementary material, figure S1) and analysed using maximum-likelihood ancestral character reconstruction as implemented in BayesTraits v. 2.0.2 (www.evolution.rdg.ac.uk; [35]). The MultiState model was used, and 10 attempts per tree were conducted.
In order to infer venom composition similarities between species and genera, we performed (i) a multiple correspondence analysis (MCA) on the presence or absence of superfamilies (binary data); (ii) a principal component analysis (PCA) on the relative member abundance by estimating the percentage of the different superfamily members; and (iii) a PCA on the relative expression level of the different superfamilies by calculating transcripts per million (TPMs; see expression analyses below). In addition, we ran a PCA comparing relative superfamily member abundances in vermivorous Africonus and Varioconus species against those in piscivorous species C. ermineus and P. magus from the Atlantic and Indo-Pacific oceans, respectively. Both MCA and PCA were performed using PAST 4.01 (https://folk.uio.no/ohammer/past/; [36]). Data were not standardized in the analyses (i.e. the variance-covariance method was used).
(e). Expression analyses
Relative expression levels for each individual were calculated by mapping the raw reads to the nucleotide sequence of each conotoxin precursor using Bowtie 2 [37], and the values were transformed to TPM estimates using RSEM [38] as implemented in Trinity v.2.6.6 [32]. We run the EBSeq software [39] to estimate for each superfamily the posterior probability of being differentially expressed (PPDE) between Varioconus and Africonus, using all the specimens of each genera as biological replicates. We considered as differentially expressed all those conotoxin precursor superfamilies with a PPDE greater than 0.95 and with a fold change above 32 (calculated as log2 RealFC ≥ 5). The same type of analysis was performed to identify those superfamilies differentially expressed in the comparison between vermivory (using the 15 specimens of West Africa as replicates) and piscivory (using the three individuals of C. ermineus [8] and the three individuals of P. magus [29]). The Shapiro–Wilk test rejected the normality of the data, so a Kruskal–Wallis test was run in R [40] over those superfamilies identified as differentially expressed to confirm these results taking variance among replicates into consideration.
3. Results
(a). Venom cataloguing of West African cones
The transcriptomes of the venom glands of 16 individuals corresponding to 14 species of genera Africonus, Varioconus and Kalloconus were assembled. The main statistics associated with the sequencing and assembly procedures are summarized in table 1. Overall, 2254 unique conotoxin precursor transcripts were identified (electronic supplementary material, table S1). The species with the highest and lowest number of conotoxin precursors were V. mercator (V_1302, 221) and V. reticulatus (V_1278, 108), respectively (table 1; figure 1). Conotoxin precursors were classified into 61 known superfamilies and 141 groups of paralogy taking into consideration sequence divergences in the signal and pro-peptide regions and the clades recovered in the reconstructed dendrogram (electronic supplementary material, figure S2). A total of 86 precursors could not be assigned to any known conotoxin superfamily and were grouped into seven new unassigned superfamilies (their signal sequences, cysteine frameworks and best BLAST-P hits are reported in electronic supplementary material, table S2). Several conotoxin precursor superfamilies previously reported as valid such as R, W, Z [41] and Cerm_17 [8], among others, were found to be fragments of other proteins once the right ORFs were identified using TBLASTX (electronic supplementary material, file S2).
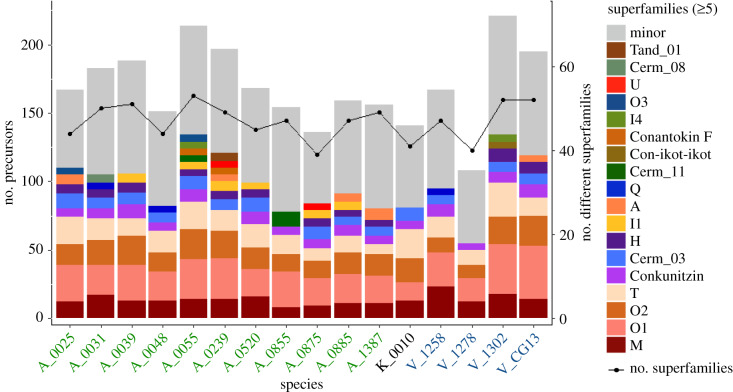
Figure 1.
Venom compositions of the 16 studied specimens. The bars represent the total number of conotoxin precursors. The proportion of superfamilies with five or more members is shown in colours. The dotted line represents the number of different superfamilies identified in the venom. The species codes in green, blue and black belong to Africonus, Varioconus and Kalloconus, respectively.
The diversity of expanded conotoxin precursor superfamilies (i.e. those with five or more members) is represented in figure 1. These expanded conotoxin precursor superfamilies were A, Conantokin F, Con-ikot-ikot, Conkunitzin, H, I1, I4, M, O1, O2, O3, Q, T, U, Cerm_03, Cerm_08, Cerm_11 and Tand_01. The remaining 50 conotoxin precursor superfamilies were considered of minor diversity. The species A. maioensis (A_0055), A. verdensis (A_0239), V. guanche (V_CG13) and V. mercator (V_1302) presented the highest diversity of expanded superfamilies whereas A. galeao (A_0048) and V. reticulatus (V_1278) the lowest (figure 1). The O1, O2, T and M superfamilies had the highest number of members and were present in all individuals (figure 1). The Conkunitzin and Cerm_03 superfamilies also showed high member diversity but were missing in A. verdensis (A_0239) and in A. cuneolus (A_0855) plus V. reticulatus (V_1278), respectively (figure 1). Remarkably, different individuals of the same species could have different expansion patterns. For example, within V. mercator, V_1302 had expanded the H, I4 and Con-ikot-ikot superfamilies, whereas V_1258 showed expansion of the Q superfamily (figure 1).
The 16 studied individuals added up to 80 hormone sequences, which were classified into 10 families: Conopressin (with more than five members in V. guanche, V_CG13), Conorfamide, Insulin-related peptides 1–5, Prohormone-4a and b, Thyrostimulin hormone alpha, and Thyrostimulin hormone beta 5 (electronic supplementary material, file S1; table S1). Insulin-related peptide 5 was only present in K. trochulus (K_0010); Prohormone-4b and Thyrostimulin hormone alpha were only found in Africonus antoniomonteiroi (A_0885). In addition, 326 transcripts were assigned to 15 protein families of various functions, likely associated with venom production. Among these, the Protein Disulfide Isomerase, Conodipine and Ferritin were the most diverse. Interestingly, we identified in the venoms of West African cones several members of the cysteine-rich secretory, antigen 5 and pathogenesis-related 1 (CAP) protein family. These proteins are often secreted and have protease activity with extracellular endocrine or paracrine function in a wide range of animals including venomous ones such as ants, wasps and snakes [42]. These proteins were previously found in the molluscivorous Cylinder textile and Conus marmoreus, and may be important for venom function ([43,44]; electronic supplementary material, figure S3).
(b). Variations in venom composition according to phylogenetic divergence
The venom compositions of the 15 specimens of Africonus and Varioconus were pairwise compared at different taxonomic levels (electronic supplementary material, figure S4). The two specimens of A. maioensis (A_0039 and A_0055) and those of V. mercator (V_1258 and V_1302) shared 51 and 53 conotoxin precursors, respectively, which represented 24–32% of the total sequences (electronic supplementary material, figure S4). The shared sequences between pairs of species from the same lineage within Africonus (see tree in electronic supplementary material, figure S1) were 2–35, with a mean of 11.9 (7% of the mean total sequences); between pairs of species from different lineages within the same genus (either Africonus or Varioconus) were 1–39 with a mean of 8.5 (5% of the mean total sequences); and between genera were 11 (0.7–1.9% of the total sequences (electronic supplementary material, figure S4).
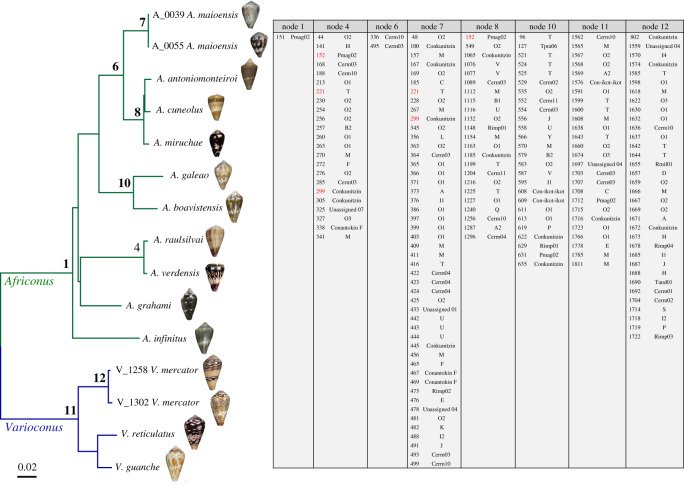
An ancestral character state reconstruction analysis was performed to infer most likely (p > 0.95) conotoxin precursors at the different common ancestors (internal nodes) in the phylogeny using K. trochulus (K_0010) as outgroup (electronic supplementary material, table S3) and to detect potential instances of convergence (see full list of conotoxins found in more than one sample in electronic supplementary material, table S4). Inferred shared conotoxin precursors were preferentially concentrated in the nodes at the tips of the phylogenetic tree (figure 2). Many corresponded to the common ancestors of the pairs of individuals of A. maioensis (A_0039 and A_0055) and V. mercator (V_1258 and V_1302), respectively. In addition, an important number of shared-derived conotoxin precursors was inferred at the ancestors of (i) the closely related species A. verdensis (A_0239) from Santiago and A. raulsilvai (A_0031) from Maio; (ii) A. galeao (A_0048) and A. boavistensis (A_0520) from Boa Vista and (iii) the three species from Sal (A. antoniomonteiroi (A_0885), A. cuneolus (A_0855) and A. miruchae (A_0875); figure 2). Two precursors (Cerm_03 and Cerm_10) were inferred to be present at the ancestor of clade IV of Africonus. One (Pmag_02) and 26 (most prominently M, O1, O2 and T) conotoxin precursors were inferred to be present at the common ancestors of Africonus and Varioconus, respectively (figure 2). According to the inferred phylogeny, there could be several potential cases of convergence, such as transcript 646 (O2), present in A. maioensis (A_0055) and in V. mercator (V_1302); transcript 368 (O1), found in A. maioensis (A_0039) and in V. guanche (V_CG13); or transcript 2124 (O1), shared by A. boavistensis (A_0520) and K. trochulus (K_0010; electronic supplementary material, table S3). Likewise, within Africonus, the common presence of transcript 221 (T) in the common ancestor of A. raulsilvai (A_0031) plus A. verdensis (A_0239) and the distantly related species A. maioensis (A_0039 and A_0055) could be due to convergence.
Figure 2.
Maximum-likelihood ancestral reconstruction of conotoxin precursors along the phylogeny of cones from West Africa (see electronic supplementary material, figure S1). Statistical supports for inferred shared-derived conotoxin precursor are provided in electronic supplementary material, table S3. The table on the right shows the conotoxin precursors that are shared derived at a particular node (see electronic supplementary material, table S4). The red numbers are common conotoxin precursors found in distantly related nodes, which could represent potential instances of convergence. (Online version in colour.)
According to the MCA, species from each genus clustered together in the two-dimensional scatter plot for the presence/ absence of conotoxin superfamilies (figure 3). The species V. reticulatus (V_1278) appeared on the extreme lower end of axis 1, but it was not statistically considered an outlier. Varioconus species had negative Axis 1 scores, whereas Africonus species had positive or very slightly negative scores. The PCA of the relative abundance (percentage of the number of members) of each superfamily revealed no overlapping between genera (figure 3). According to the loadings of PC1 and PC2, Varioconus species had more abundant M, T and O1 superfamilies, whereas in Africonus the pattern was more disperse, with O2, P, Cerm_03 and A superfamilies as major contributors (electronic supplementary material, figure S5). A discriminant function analysis (DFA) using PC1 to PC3 as variables and genus as factor, classified correctly 100% of the cases (also in the jackknifed classification test). Finally, relative expression levels showed no significant differences between both genera, although V. reticulatus (V_1278) and V. mercator (V_1258) were considered outliers, with overexpression of the T superfamily, as indicated by the strong positive loadings along PC1 (figure 3).
Figure 3.
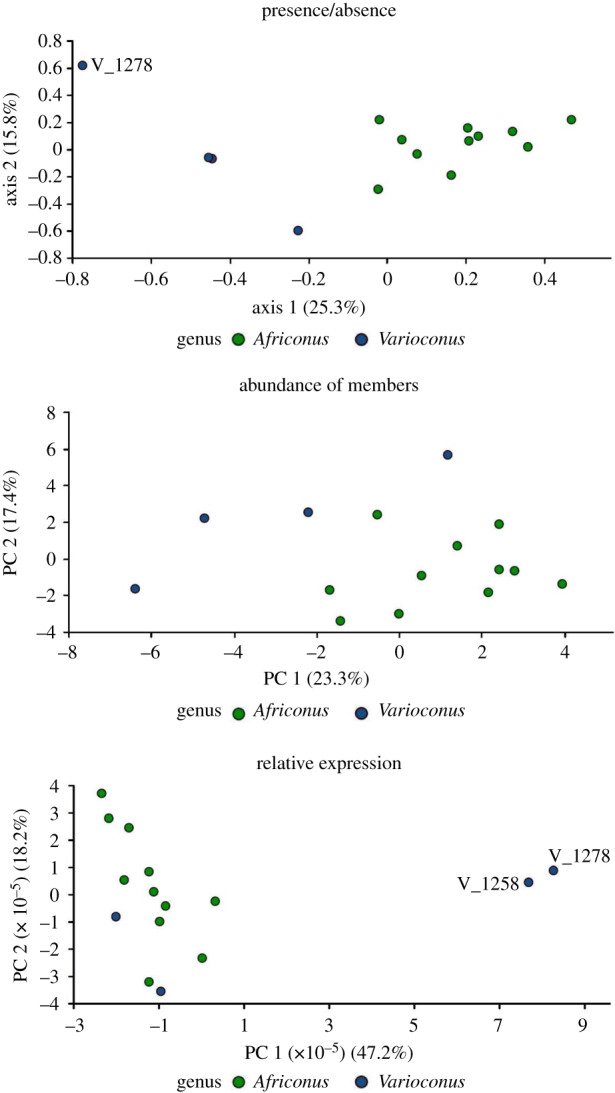
MCA and PCA comparing venom compositions of genera Africonus and Varioconus. The two-dimensional scatter plots are shown. The venom composition was defined as presence/absence of conotoxin precursor superfamilies (a); superfamily conotoxin precursor abundance expressed as percentage over total number of members (b) and relative expression (TPM) levels (c). The percentages of the eigenvalue (MCA) or variance (PCA) for each of the axis are indicated on the corresponding labels. Extremes and outliers are labelled. (Online version in colour.)
(c). Differential conotoxin expression patterns
A total of 11 superfamilies were detected as differentially expressed between Africonus and Varioconus: A2, B1, Cerm_02, Cerm_11, N, Rmil_02, S and V superfamilies were overexpressed in Africonus, whereas Cerm_01, K, and T were in Varioconus. After a Kruskal–Wallis test, only the overexpression of B1, Cerm_01, Cerm_11 and V superfamilies in Africonus remained significant (p < 0.05; electronic supplementary material, table S5).
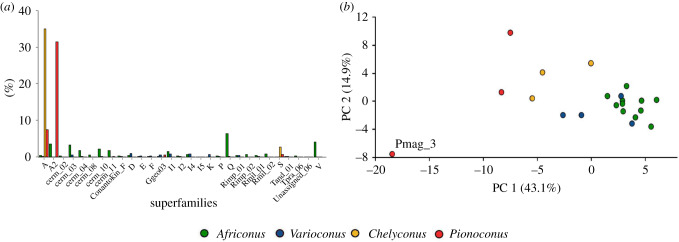
The same tests found 29 superfamilies differentially expressed between vermivorous (Africonus and Varioconus) and piscivorous (Chelyconus and Pionoconus) genera (figure 4a). All but four were confirmed by the Kruskal–Wallis test (figure 4a; electronic supplementary material, table S5). Among those confirmed, the A (p = 0.02) and S (p = 0) superfamilies were overexpressed in both Chelyconus and Pionoconus, and the A2 (p = 0.04) in Pionoconus. The I5 superfamily was overexpressed in P. magus (p = 0.005), but this superfamily has only been reported in this species (figure 4a).
Figure 4.
Differences in venom compositions of vermivorous and piscivorous cones. (a) Average expression (measured in TPMs) of conotoxin precursor superfamilies per genus. Vermivorous genera (Africonus and Varioconus) were compared to piscivorous genera (Chelyconus and Pionoconus). The bar plot depicts those superfamilies differentially expressed between diets. (b) PCA comparing conotoxin precursor abundance per superfamily between vermivorous and piscivorous species. The percentages of variance for each of the axis are indicated on the corresponding labels. In both panels, the genera Africonus, Varioconus, Chelyconus and Pionoconus are depicted in green, blue, yellow and red, respectively.
PCA on the relative abundance of conotoxin superfamily members in the venom clearly separated piscivorous and vermivorous genera (figure 4b). The former showed negative loadings along PC1, with most important contributions from M and Conkunitzin superfamilies, followed by S, A and B1. For the vermivorous genera, Cerm_03 and O2 superfamilies were the most important contributors to the positive loadings (electronic supplementary material, figure S6). DFA using PC1 to PC3 and diet as factor classified correctly 100% of the cases (also in the jackknifed classification test). Additionally, within the piscivorous cones, the PCA separated Pionoconus (Indo-Pacific Ocean) from Chelyconus (Atlantic Ocean). The former had M and Conkunitzin superfamilies as main contributors to the negative loadings whereas O2 and T superfamilies contributed to the separation of Chelyconus (not shown).
4. Discussion
(a). Conotoxin precursor assembly and annotation
The use of Illumina short reads to sequence cone venom gland transcriptomes has boosted the identification of conotoxin precursors [13]. However, de novo assembly is not straightforward and the use of various approaches may render strikingly different results. This should be taken into account when reporting the full venom catalogue of a species. In this regard, long read sequencing (paradoxically the now discontinued 454 but not PacBio or ONT technologies, which had not been yet applied to venom gland transcriptomes), which capture full-length peptides, and proteomic approaches have confirmed the exceptional variability of conotoxin precursors and complexity of venom cocktails. Another delicate step is annotation, which entirely relies on the quality of the reference database [45]. On one hand, the actual number of conotoxin precursors could be underestimated. Low-expressed conotoxin precursor transcripts may not be recognized in the absence of counterparts in the reference database, if these were not detected in proteomic analyses, which are less sensitive [13]. On the other hand, we focused here on the possibility that assembly artefacts could overestimate conotoxin diversity [8,19,29]. This is particularly worrisome as, if not detected, these annotation errors could dangerously propagate once incorporated into updated reference databases [7,8,45]. We carefully inspected all the ORFs rendered by the BLASTX searches. At the assembly level, we often found regions of the assembled transcript (particularly at the 5′- and 3′-ends) mapped only by few reads that could lead to frame shifts and generate spurious variability [19]. At the annotation level, we implemented a TBLASTX step, which found two main sources of conotoxin misidentification (electronic supplementary material, file S2): (i) translations into the wrong frame, as exemplified by the R superfamily, originally described in Conus marmoreus [41], which once translated into the correct frame corresponds to the proteasome subunit alpha; and (ii) chimeric transcripts generated during the assembly. This is the case of several conotoxin precursors identified in Darioconus episcopatus [13]. For example, one precursor (BAS24857; named Cerm_18 in [8]) had the typical mature domain associated with T superfamily whereas the putative signal and pro-peptide domains, once translated into the correct frame corresponded to a sodium- and chloride-dependent glycine/GABA transporter. The here identified annotation errors (electronic supplementary material, file S2) should be eliminated from future reference databases.
(b). Venom composition and evolution
The analysed venoms contained 108-221 conotoxin precursors, which is in good agreement with numbers reported for other species of cones [6–8,25,29]. Comparison of venom repertoires revealed a larger set of expanded superfamilies in Africonus than in Varioconus. Although, it has been proposed that larger sets of conotoxins are associated with broader diets [19,22], we could not test this hypothesis, as the breadth of the worm diet of the different Africonus and Varioconus species is largely unstudied. Ecological studies on Miliariconus miliaris showed that the individuals of this species inhabiting the remote Eastern Island presented a considerably broader diet of worms, which could have evolved through ecological release in the absence of congeners [46]. This hypothesis could apply to A. verdensis (A_0239), which had a large conotoxin catalogue with many expanded superfamilies, and lives alone in Santiago Island.
Conotoxins are well known for their accelerated rates of evolution, which in turn generate high-sequence divergences even between individuals of the same species [6,8,25]. This is the basis of the reported general lack of common peptides between cone species, and the extended notion that virtually each species produces a unique venom cocktail [7]. The present study brings, for the first time, the opportunity to test the taxonomic limits of this hypothesis by comparing closely related species sharing relatively recent common ancestors. Individuals of the same species showed around one-fourth common conotoxin precursor sequences. This proportion is similar to those reported for intraspecific comparisons in Dendroconus betulinus [6], Rhombiconus imperialis [25] and C. ermineus [8]. The proportion of shared sequences decreased substantially for the pairwise comparisons between closely related species, within the range of 2–9%, in agreement with that reported for sister species of the genus Turriconus [7]. At the genus level, only 0.7–1.9% of the total sequences were common. Altogether, our results support that a phylogenetic signal remains in venom composition above the species level, but it is quickly eroded as lineages diverge and no identical conotoxin precursors are generally shared between closely related genera [19]. However, it is striking that several identical conotoxin precursor sequences were found between species from distantly related genera within Conidae (electronic supplementary material, file S1), indicating that those sequences are either subjected to strong balancing selection or reflect cases of convergent evolution. The rather erratic distribution of some of these sequences in the phylogeny of Conidae favours the latter hypothesis.
The two genera showed very contrasting results in the ancestral reconstruction analyses: only Pmag_02 could be traced back to the ancestor of Africonus whereas members of 14 conotoxin precursor superfamilies were inferred for the common ancestor of Varioconus. This discrepancy could be due to the larger number of taxa analysed and the greater diversity of members within the expanded superfamilies in Africonus. In any case, M, O1, O2 and T superfamilies were characterized by having five or more members in all studied species. The wider presence of these superfamilies in any cone and always showing similar levels in diversity of members [6–8,19,41] may suggest that the ancestor of living cones already had this core set, and that having members of these superfamilies (not necessarily the same) is essential either for defense (if, as proposed, this was the ancestral role of cone venom; [12]) or for triggering the minimum physiological responses necessary for the capture of a prey, regardless of whether it is a worm, a snail or a fish.
PCA and MCA have been used in several other animal groups to summarize the information related to venom composition [14,15] but not in cones to the best of our knowledge. MCA of presence/absence of superfamilies and PCA of the relative abundance of superfamily members recovered non-overlapping patterns for Africonus and Varioconus, indicating that species that are more closely related tend to have the same conotoxin precursor superfamilies and in similar proportions. By contrast, the PCA for expression levels did not find differences between the two genera, which may indicate functional convergence at this level, in agreement with the common expression patterns of conotoxins found in closely related Indo-Pacific vermivorous cone species that could not be explained by phylogeny but by functional convergence [47].
(c). Differential expression levels of conotoxins between genera and diets
Although the exact worm species eaten by the different species of Varioconus and Africonus are unknown, at least the three clades described within genus Varioconus correlate with different morphologies of the radular teeth suggesting subtle diet specializations [27]. We tested whether the two genera showed differential expression of their venom components, which could be correlated with diet adaptations. The B1, Cerm_01, Cerm_11 and V superfamilies presented significantly different expression between Africonus and Varioconus after the Kruskal–Wallis test was applied. The B1 superfamily (Conantokin) was originally described in the piscivorous G. geographus and reported to provoke a ‘sleeping’ phenotype in vertebrates, but its function in vermivorous species has not been characterized [4]. The V superfamily was first identified in the venom of the vermivorous Virgiconus virgo, but there is no information regarding its function [4]. The Cerm superfamilies were recently described in C. ermineus [8] and their function remains unknown.
Similarly, we tested for differential expression between piscivorous and vermivorous cones. We found four superfamilies differentially overexpressed in the two piscivorous species (A, A2, I5 and S) after the Kruskal–Wallis test. Thus, these superfamilies may be essential for piscivory in cones. The importance of having different members of the A superfamily for hunting fish has been highlighted previously for several cone species, as well as instances of functional convergence between Indo-Pacific and Atlantic piscivorous cones [8]. The S superfamily was first identified in G. geographus and found to inhibit neurotransmitter receptors [4]. Later, it was reported as minor component of different cone species, not all necessarily hunting on fish. The A2 superfamily has been described very recently [7], and its pharmacological function remains unknown. Despite sharing the same cysteine pattern to the I4 superfamily of C. ermineus, the I5 superfamily was defined as new in P. magus because it had a distinct signal region [29]. The functions of both superfamilies are unknown.
The possibility of comparing venom catalogues of closely related species of cones within a phylogenetic framework paves the way to understand how the venom repertoires were assembled and evolve, as well as to discern the relative role of diet and defense as selective forces. Here, we focused on two well-known species radiations of West African cones. For the first time, not only did we established levels of divergence of venom compositions at different taxonomic levels (between individuals, species, main lineages, genera) but also detected shared conotoxin precursors and inferred their potential presence at most recent common ancestors. This allowed (i) disentangling orthologous from paralogous conotoxin precursors; (ii) identifying functionally convergent conotoxin precursors; and (iii) distinguishing shared derived from plesiomorphic conotoxin precursors. Our results demonstrate that different genera (Africonus, Varioconus, Chelyconus and Pionoconus) show distinct venom toolkits in terms of type and member abundance of conotoxin precursor superfamilies but that these differences are less evident when expression levels are analysed. Diet might be the strongest selective factor determining the relative expression of each venom component, as suggested by the differential expression analyses, although the contribution of differential conotoxin expression to defense needs to be further understood.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dr Rui Freitas, Dr Iderlindo Silva dos Santos and Dr Sonia Monteiro de Pina Araujo for their continuous support of our research in Cabo Verde (Autorizações 07/2013, 26/2013, 01/ 2014, 04/2015 and 03/2016); Amadou Gaye and Luigi Tamagnini for their help during sampling in Senegal; and to Francisco Sicilia for assisting material collection in Lanzarote. We thank Jesús Marco and Aida Palacio, who provided access to the supercomputer Altamira (IFCA-CSIC), member of the Spanish Supercomputing Network.
Data accessibility
The transcriptome sequences were deposited at the SRA database of NCBI (https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR11807492-SRR11807507, Bioproject PRJNA631880. The nucleotide sequences of all venom proteins here identified are available in fasta format (electronic supplementary material, file S3). Sequences of all conotoxin precursors and other venom proteins are also available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3n5tb2rdm [48].
Authors' contributions
R.Z. conceived the study; M.J.T., C.M.L.A. and R.Z. obtained the samples; S.A. generated, assembled and annotated the transcriptomes; S.A., M.J.T. and R.Z. analysed the sequence data. All authors participated in the writing of the manuscript.
Competing interests
The authors declare no competing interests.
Funding
This work was funded by the Spanish Ministry of Economy, Industry and Competitiveness (CGL2013-45211-C2-2-P and CGL2016-75255-C2-1-P [AEI/FEDER, UE] to R.Z.; BES-2014-069575 to S.A.). The Doctorate Commission of the University of Salamanca awarded S.A. with funding to partially cover publication expenses.
References
- 1.Tucker JK, Tenorio MJ. 2009. Systematic classification of recent and fossil conoidean gastropods, with keys to the genera of cone shells. Hackenheim, Germany: ConchBooks. [Google Scholar]
- 2.Olivera BM. 2006. Conus peptides: biodiversity-based discovery and exogenomics. J. Biol. Chem. 281, 31 173–31 177. ( 10.1074/jbc.R600020200) [DOI] [PubMed] [Google Scholar]
- 3.Ahorukomeye P, et al. 2019. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. Elife 8, e41574 ( 10.7554/eLife.41574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson SD, Norton RS. 2014. Conotoxin gene superfamilies. Mar. Drugs 12, 6058–6101. ( 10.3390/md12126058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaas Q, Westermann JC, Craik DJ. 2010. Conopeptide characterization and classifications: an analysis using ConoServer. Toxicon 55, 1491–1509. ( 10.1016/j.toxicon.2010.03.002) [DOI] [PubMed] [Google Scholar]
- 6.Peng C, et al. 2016. High-throughput identification of novel conotoxins from the Chinese tubular cone snail (Conus betulinus) by multitranscriptome sequencing. GigaScience 5, 17 ( 10.1186/s13742-016-0122-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, et al. 2017. Divergence of the venom exogene repertoire in two sister species of Turriconus. Genome Biol. Evol. 9, 2211–2225. ( 10.1093/gbe/evx157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abalde S, Tenorio MJ, Afonso CML, Zardoya R. 2018. Conotoxin diversity in Chelyconus ermineus (Born, 1778) and the convergent origin of piscivory in the Atlantic and Indo-Pacific cones. Genome Biol. Evol. 10, 2643–2662. ( 10.1093/gbe/evy150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prator CA, Murayama KM, Schulz JR. 2014. Venom variation during prey capture by the cone snail, Conus textile. PLoS ONE 9, e98991 ( 10.1371/journal.pone.0098991.g001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang D, Duda TF Jr. 2016. Age-related association of venom gene expression and diet of predatory gastropods. BMC Evol. Biol. 16, 27 ( 10.1186/s12862-016-0592-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutertre S, Biass D, Stocklin R, Favreau P. 2010. Dramatic intraspecimen variations within the injected venom of Conus consors: an unsuspected contribution to venom diversity. Toxicon 55, 1453–1462. ( 10.1016/j.toxicon.2010.02.025) [DOI] [PubMed] [Google Scholar]
- 12.Dutertre S, et al. 2014. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Comm. 5, 3521 ( 10.1038/ncomms4521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavergne V, Harliwong I, Jones A, Miller D, Taft R, Alewood PF. 2015. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc. Natl Acad. Sci. USA 112, E3782–E3791. ( 10.1073/pnas.1501334112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs HL, Sanz L, Sovic MG, Calvete JJ. 2013. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp). PLoS ONE 8, e67220 ( 10.1371/journal.pone.0067220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomonte B, et al. 2014. Venomics of New World pit vipers: genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteomics 96, 103–116. ( 10.1016/j.jprot.2013.10.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pekar S, Bocanek O, Michalek O, Petrakova L, Haddad CR, Sedo O, Zdrahal Z. 2018. Venom gland size and venom complexity—essential trophic adaptations of venomous predators: a case study using spiders. Mol. Ecol. 27, 4257–4269. ( 10.1111/mec.14859) [DOI] [PubMed] [Google Scholar]
- 17.Chang D, Duda TF Jr. 2012. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 29, 2019–2029. ( 10.1093/molbev/mss068) [DOI] [PubMed] [Google Scholar]
- 18.Remigio EA, Duda TF Jr. 2008. Evolution of ecological specialization and venom of a predatory marine gastropod. Mol. Ecol. 17, 1156–1162. ( 10.1111/j.1365-294X.2007.03627.x) [DOI] [PubMed] [Google Scholar]
- 19.Phuong MA, Mahardika GN, Alfaro ME. 2016. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genomics 17, 401 ( 10.1186/s12864-016-2755-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conticello SG, Gilad Y, Avidan N, Ben-Asher E, Levy Z, Fainzilber M. 2001. Mechanisms for evolving hypervariability: the case of conopeptides. Mol. Biol. Evol. 18, 120–131. ( 10.1093/oxfordjournals.molbev.a003786) [DOI] [PubMed] [Google Scholar]
- 21.Pahari S, Bickford D, Fry BG, Kini RM. 2007. Expression pattern of three-finger toxin and phospholipase A2 genes in the venom glands of two sea snakes, Lapemis curtus and Acalyptophis peronii: comparison of evolution of these toxins in land snakes, sea kraits and sea snakes. BMC Evol. Biol. 7, 175 ( 10.1186/1471-2148-7-175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliger CA, Richmond TA, Lebaric ZN, Pierce NT, Sweedler JV, Gilly WF. 2011. Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon 57, 311–322. ( 10.1016/j.toxicon.2010.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duda TF., Jr 2008. Differentiation of venoms of predatory marine gastropods: divergence of orthologous toxin genes of closely related Conus species with different dietary specializations. J. Mol. Evol. 67, 315–321. ( 10.1007/s00239-008-9155-8) [DOI] [PubMed] [Google Scholar]
- 24.Duda TF Jr, Chang D, Lewis BD, Lee T. 2009. Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PLoS ONE 4, e6245 ( 10.1371/journal.pone.0006245). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin AH, Dutertre S, Dutt M, Lavergne V, Jones A, Lewis RJ, Alewood PF. 2019. Transcriptomic–proteomic correlation in the predation-evoked venom of the cone snail, Conus imperialis. Mar. Drugs 17, 3 ( 10.3390/md17030177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abalde S, Tenorio MJ, Afonso CML, Uribe JE, Echeverry AM, Zardoya R. 2017. Phylogenetic relationships of cone snails endemic to Cabo Verde based on mitochondrial genomes. BMC Evol. Biol. 17, 231 ( 10.1186/s12862-017-1069-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abalde S, Tenorio MJ, Afonso CML, Zardoya R. 2017. Mitogenomic phylogeny of cone snails endemic to Senegal. Mol. Phylogenet. Evol. 112, 79–87. ( 10.1016/j.ympev.2017.04.020) [DOI] [PubMed] [Google Scholar]
- 28.Tenorio MJ, Abalde S, Pardos-Blas JR, Zardoya R. 2020. Taxonomic revision of West African cone snails (Gastropoda: Conidae) based upon mitogenomic studies: implications for conservation. Eur. J. Taxon. 663, 1–89. [Google Scholar]
- 29.Pardos-Blas JR, Irisarri I, Abalde S, Tenorio MJ, Zardoya R. 2019. Conotoxin diversity in the venom gland transcriptome of the magician's cone, Pionoconus magus. Mar. Drugs 17, 10 ( 10.3390/md17100553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cossignani T, Allary A. 2019. Lautoconus (Lautoconus) stimpsonorum: nuova specie dal Senegal. Malacol. Mostra Mondiale 104, 21–23. [Google Scholar]
- 31.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. ( 10.14806/ej.17.1.200) [DOI] [Google Scholar]
- 32.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinf. 10, 421 ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. ( 10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagel M, Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 167, 808–825. ( 10.1086/503444) [DOI] [PubMed] [Google Scholar]
- 36.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: Paleontological statistics software package for education and data analyses. Paleontol. Electron. 4, 9. [Google Scholar]
- 37.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323 ( 10.1186/1471-2105-12-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leng N, et al. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. ( 10.1093/bioinformatics/btt087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See https://www.R-project.org/. [Google Scholar]
- 41.Lavergne V, Dutertre S, Jin A-H, Lewis RJ, Taft RJ, Alewood PF. 2013. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genomics 14, 708 ( 10.1186/1471-2164-14-708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fry BG, et al. 2009. The toxicogenomic multiverse: convergent recruitment of proteins Into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511. ( 10.1146/annurev.genom.9.081307.164356) [DOI] [PubMed] [Google Scholar]
- 43.Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. 2003. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J. Biol. Chem. 278, 31 105–31 110. ( 10.1074/jbc.M304843200) [DOI] [PubMed] [Google Scholar]
- 44.Hansson K, Thämlitz AM, Furie B, Furie BC. 2006. A single γ-carboxyglutamic acid residue in a novel cysteine-rich secretory protein without propeptide. Biochemistry 45, 12 828–12 839. ( 10.1021/bi061311a) [DOI] [PubMed] [Google Scholar]
- 45.Salzberg SL. 2019. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 20, 92 ( 10.1186/s13059-019-1715-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duda TF, Lee T. 2009. Isolation and population divergence of a widespread Indo-West Pacific marine gastropod at Easter Island. Mar. Biol. 156, 1193–1202. ( 10.1007/s00227-009-1161-x) [DOI] [Google Scholar]
- 47.Duda TF Jr, Remigio EA. 2008. Variation and evolution of toxin gene expression patterns of six closely related venomous marine snails. Mol. Ecol. 17, 3018–3032. ( 10.1111/j.1365-294X.2008.03804.x) [DOI] [PubMed] [Google Scholar]
- 48.Abalde S, Tenorio MJ, Afonso CML, Zardoya R. 2020. Data from: Comparative transcriptomics of the venoms of continental and insular radiations of West African cones Dryad Digital Repository. ( 10.5061/dryad.3n5tb2rdm) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Abalde S, Tenorio MJ, Afonso CML, Zardoya R. 2020. Data from: Comparative transcriptomics of the venoms of continental and insular radiations of West African cones Dryad Digital Repository. ( 10.5061/dryad.3n5tb2rdm) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The transcriptome sequences were deposited at the SRA database of NCBI (https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR11807492-SRR11807507, Bioproject PRJNA631880. The nucleotide sequences of all venom proteins here identified are available in fasta format (electronic supplementary material, file S3). Sequences of all conotoxin precursors and other venom proteins are also available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3n5tb2rdm [48].