Abstract
The development of B lymphocytes into antibody-secreting plasma cells is central to the adaptive immune system in that it confers protective and specific antibody response against invading pathogen. This developmental process involves extensive morphological and functional alterations that begin early after antigenic stimulation. These include chromatin restructuring that is critical in regulating gene expression, DNA rearrangement and other cellular processes. Here we outline the recent understanding of the three-dimensional architecture of the genome, specifically focused on its contribution to the process of B cell activation and terminal differentiation into antibody-secreting cells.
Keywords: antibody-secreting cells, b lymphocyte, chromatin, gene expression and regulation, genome organisation
Introduction
Upon recognition of microbial-derived antigen, quiescent naïve B lymphocytes undergo rapid clonal expansion. This generates a large population of B cells with identical antigen specificity, engineered to combat the activating pathogen [1,2]. Ultimately proliferation is accompanied by differentiation, where activated B cells terminally differentiate into mature effector cells, such as antibody-secreting plasma cells and memory cells. Concurrent with this clonal expansion, B cells undergo somatic hypermutation (SHM) and class switch recombination (CSR) to diversify their antibody affinity and function [3]. Furthermore, activation induces dramatic transcriptional and morphological changes ensuring the B cells become solely dedicated to their antibody production function. Recent technological advances have shown that the two metres of DNA in every nucleus is packed in an intricate, non-random and critically important three-dimensional network [4,5]. As such, this organisation influences fundamental cellular processes, from transcription, DNA replication and repair to recombination and mitosis. Here we discuss how genome organisation might influence, and adapt to, the molecular and cellular rigors of rapid clonal expansion and differentiation into antibody-secreting plasma cells.
Loops, TADs and compartments—from linear genetics to 3D epigenetics
Enhancers have long been known to regulate gene expression over vast linear distances in a position- and orientation-independent manner [6]. However, for many years the mode of action remained enigmatic until it was shown in erythroid cells that enhancers of the β-globin locus, located 40–60 kb away from the globin promoter, come into close physical proximity with the promoter [7,8]. This DNA loop was erythroid specific. This work was the first evidence that enhancers exert their functions by recruiting transcription factors to stabilise RNA polymerase at the promoter through a three-dimensional contact, with the intervening sequence being looped out.
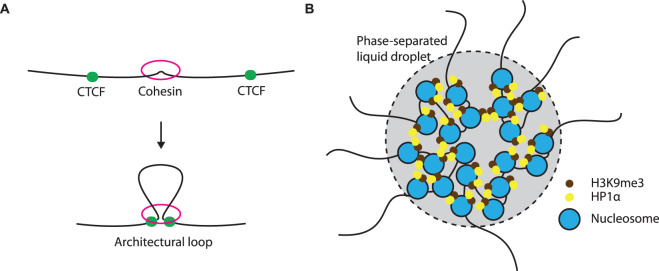
The communication between enhancers and promoters can be disrupted when a cis-element, traditionally known as insulator, is located between the two [9]. Over decades of active searching by scientists, the CCCTC-binding factor (CTCF) appears to be the only example of vertebrate insulator protein [10]. Unlike an enhancer, CTCF works in a position- and orientation-dependent manner and in some cases DNA methylation status is critical in its function [11,12]. Again, the underlying molecular mechanism of insulation remained unclear until the development of the chromosome conformation capture (3C) technique, which utilises a proximity ligation assay to evaluate the three-dimensional proximity of two genomic regions [13]. A high throughput approach using next-generation sequencing (Hi-C) was later developed to assess all DNA–DNA interactions genome-wide [14,15]. Hi-C revealed that complex genomes are spatially segregated into discrete globules of DNA, known as topologically associating domains (TADs). TADs are characterised by preferential interaction within and not between TADs [16,17]. Intriguingly, CTCF binding sites are enriched at the margins, or boundaries, of TADs. Deletion or disruption of these sites dramatically alters TAD structure [17–19], while acute degradation of CTCF completely removes TADs [20,21], indicating that CTCF is essential in the establishment of TADs. Recently it was demonstrated that CTCF, together with cohesin, act as architectural proteins and directly help in the formation of TADs through a loop-extrusion mechanism [18,22–24] (Figure 1A). Therefore, concurring with the traditional view as an insulator protein, CTCF can prevent cross-talk between promoters and enhancers by partitioning them into two different TADs. Thus TAD boundaries play a critical role in gene regulation as they facilitate appropriate interaction between enhancer and promoter, while insulating against inappropriate interactions. For example, recent studies have shown that disruption of TADs can lead to inappropriate interactions between enhancers and promoters, resulting in gene deregulation and disease [25,26].
Figure 1. Two major mechanisms of chromatin organisation.
(A) Loop extrusion model suggests the ring-like cohesin would randomly load onto chromatin with DNA subsequently traverse through the ring and extrudes along [18,22]. Extrusion will progress until it encounters a pair of CTCF in an appropriate orientation. (B) Compartments are speculated to form via a phase separation mechanism [29]. Particular histone modification with its associating chromatin-binding proteins such as H3K9me3 and HP1α, tend to possess unique physical properties, which aggregate and create a phase-separated microenvironment.
At a broader scale, TADs with similar chromatin state tend to aggregate and form compartments [14]. Early Hi-C studies described the genome being partitioned into A- and B-compartments, where cross-talk is common among the same compartment type but minimal between them [14]. Compartment A generally associates with euchromatic regions and harbours active genes, whereas compartment B generally contains inactive heterochromatin. Subsequent Hi-C studies with greater resolution further divided the compartments into six different sub-compartments with distinct histone marks and epigenetic properties [15]. It is therefore speculated that the compartmentalisation is mediated by the similar physical properties of different chromatin states, possibly via phase separation [27–31] (Figure 1B). In support of this, several studies have demonstrated HP1α and other heterochromatic H3K9me3 recognition complexes are able to form phase-separated liquid droplets, containing other macromolecules, such as DNA [29,32,33]. Interestingly, other studies have shown the compartments remain entirely unaffected upon the degradation of CTCF or cohesin, which would completely abrogate TADs [20,21,34,35], demonstrating that compartmentalisation is independent of CTCF and the loop-extrusion mechanism.
While TADs are thought to be largely conserved between cell types, compartments and loops display lineage-specificity [14–16,36]. Genes can switch compartments and this generally associates with the entire TAD flipping from one compartment to another. A compartmental switch from A to B almost always associates with shutting down of transcription. However, switching from B to A does not necessarily link to transcriptional up regulation, but simply endows a permissive environment for looping to occur [37]. Thus promoter-enhancer contact is highly lineage-specific and is believed to be the ultimate factor in contributing cell type and stage specific transcriptional changes.
Early activation—discrete genome reorganisation partitioned from DNA replication and cell division
Shortly after antigen stimulation, naïve quiescent B cells experience a tremendous change in both morphology and function. Early studies documented an exit of G0 phase and entry into cell cycle, accompanied with increases in synthesis of RNA and protein, histone acetylation, and an increase in cell size so dramatic that it is easily observable under a conventional microscope [38–41]. Several recent studies have further shown during this early activation chromatin decondenses, and this requires the transcription factor Myc and energy input in the form of ATP, indicating it is a highly energy-dependent process [42–44]. Intriguingly, the first cell division does not occur until ∼30 h post-activation, relative to the subsequent rapid cell division every 6–10 h thereafter [45,46]. Transcriptomic profiling from our group at different time points post-activation has further stratified the cellular events during this pre-mitotic activation phase [47]. In the hours shortly after activation, gene ontology reveals that B cells dedicate their energies to RNA processing and ribosome biogenesis, and various metabolic processes (Figure 2). Later, just prior to the first division, the focus changes to genes that control DNA replication, packaging, chromatin remodelling and DNA conformational changes.
Figure 2. Cellular events during B cell clonal expansion and differentiation.
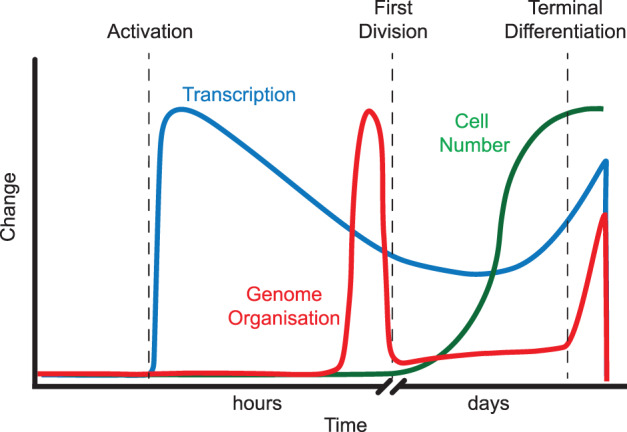
Upon stimulation, massive transcriptional changes prepare B cells for subsequent expansion and differentiation [47]. Just prior to the first cell division, B cells undergo first wave of genome reorganisation, which remains relatively unaltered when cells clonally expand over several days. The second wave of genome restructuring occurs during the final differentiation into plasmablasts. Blue line denotes transcription, red indicates genome organisation whereas green denotes cell number.
In striking contrast with the large number of early transcriptional changes after B cell activation, Hi-C analyses have revealed that genome organisation is relatively unaltered for the first 10 h after activation and undergoes a large genome reorganisation thereafter (Figure 2). Further investigation reveals that the majority of this genome reorganisation event occurs in the G1 phase of the cell cycle, prior to the initiation of DNA replication. Thus, it appears that genome architecture remodelling is independent of DNA synthesis and also mitosis, suggesting a partitioning of genome reorganisation from these physically challenging cellular processes. Like a number of previous studies on B cells [42,47–51], we have found pre-mitotic genome restructuring was associated with remarkable compartment stability [47], suggesting that a majority involve DNA loops. This is in striking contrast with stem cells, where there is extensive A/B compartment switch during lineage specification [36]. Presumably, this is a slower process as genes in the inactive compartment will need to flip to the permissive environment before any transcription could happen.
B cell activation induces the loss of long-range chromosomal contacts and the formation of thousands of DNA loops [42,47]. These short-range contacts mainly consist of CTCF-mediated loops and other enhancer-promoter contacts. It was demonstrated that the promoters gain interactions with multiple non-coding regions at this late stage of pre-mitotic phase and gene expression level is positively correlated with interaction frequency [42,47,49,52]. Thus, it appears that the predominant function of these loops is to drive and augment gene expression. Previously it has be proposed that enhancer-promoter interactions can modulate gene expression in two ways, being either instructive or permissive [53]. The positive correlation between loop formation and gene expression immediately before the first division suggests that these de novo loops are presumably largely instructive [47]. In contrast, in the hours shortly after activation the huge transcriptional burst with that is associated with minimal architectural changes suggests that pre-existing chromatin structures or loops are already poised and permissive, enabling rapid gene activation upon antigen stimulation.
Since the architectural changes are independent of DNA synthesis and mitosis, it is possible that the process of transcription or the action of transcription factors mediates genome reorganisation. There is growing evidence that the transcription of long non-coding RNA can mediate the 3D restructuring of chromatin [54,55]. For instance, during early T cell development, the transcription of a non-coding RNA ThymoD at an enhancer of the T cell lineage-specifying factor, Bcl11b, leads to the relocation of locus from nuclear periphery to interior, subsequently allowing proper enhancer-promoter interaction and gene up-regulation [56]. On the other hand, transcription factors can themselves remodel chromatin conformation in a transcription-independent manner. The aforementioned CTCF and cohesin are two examples [20,21,34,35]. In addition, our group has previously demonstrated that the B cell factor Pax5 does not require active transcription in order to wire the genome towards commitment to the B cell lineage [50]. This suggests that some lineage-specific transcription factors are also able to reorganise genome without the need for transcription.
Expansion phase—de novo loop formation during class switch recombination
Once the activated B cells undergo the first cell division, they rapidly divide and expand for many days until terminal differentiation [57]. Despite the time and number of generations that each cell has experienced, Hi-C analyses demonstrated the genome architecture stays remarkably similar [47] (Figure 2). This suggests the loops and structures that were established prior to the first division are preserved over many successive cell divisions. This is in line with cellular studies showing that in B and T lymphocytes fate decisions for terminal differentiation are made within a day after antigen stimulation [46,58–66], probably prior to the first cell division, and the stimuli received afterwards could not intervene. We speculate that the loops and other 3D structures established prior to the first division would pre-program subsequent transcription strength and profile, and the cells thereafter become somewhat refractory to external signals as the genome architecture is already established.
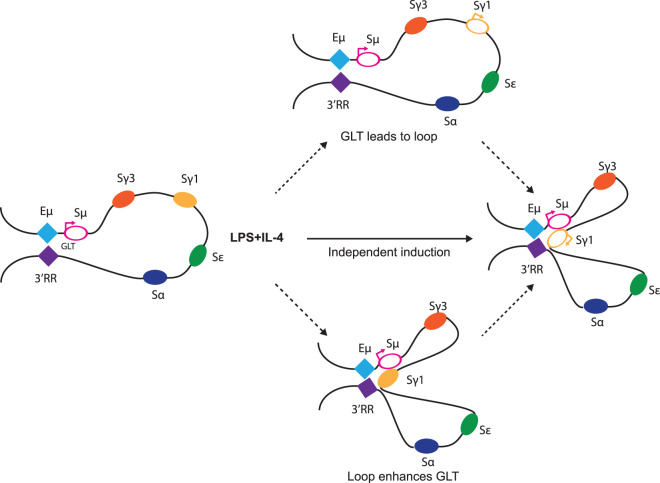
Although the global chromatin conformation remains very similar through successive cell divisions, some local architectural changes are critical. Class switch recombination is the molecular hallmark of B cell activation, where deletional DNA recombination elicits the replacement of the expressed immunoglobulin (Ig) µ and δ heavy chain constant regions (CH) with the downstream γ, ε, or α CH regions [67,68]. This is a tightly regulated process involving the interplay of non-coding transcription, double strand breaks (DSBs), DNA repair and 3D chromatin looping. In a resting B cell, a DNA loop is formed at the IgH locus, between the 3′ regulatory region (3′RR) and the intronic enhancer (Eµ) just upstream of the constant regions (Figure 3 left), which are ∼200 kb apart [69–71]. Downstream of 3′RR are multiple CTCF binding sites and recent studies have suggested that the DNA–DNA interaction is mediated by CTCF via loop-extrusion [72]. Additionally, the interaction co-occurs with a non-coding germline transcription (GLT) around the µ switch (S) region, which plays an important role in unwinding the S region DNA for activation-induced deaminase (AID), a ssDNA mutator, to induce DSBs [3,73]. Once the B cell is activated, depending on the stimulation and cytokines it received, for example LPS with IL-4 will result in a switch to IgG1, the corresponding S region (Sγ1 herein) will come into close proximity to the pre-existing loop, creating a three-point DNA synapsis [71] (Figure 3 right). In the meantime, the stimuli also trigger GLT at the acceptor switch region (Sγ1) followed by recruitment of AID to the interacting foci. DSBs are then induced at the S regions and subsequently resolved by non-homologous end joining (NHEJ) with the intervening DNA deleted. How such activation-induced chromosomal looping is established and its relationship with germline transcription remains unclear, despite several attempts in addressing the question [69,74–78]. As the induced germline transcription and 3D looping is antigen and cytokine dependent, it is conceivable that the downstream factors which elicit the GLT are also contributing to the juxtaposition of the acceptor switching region to the Eµ-3′RR loop [76] (Figure 3, middle path). However, given the emerging evidence about the role of transcription in shaping genome architecture [54,55,78], the germline transcript itself or the process of transcription could plausibly assist the formation of the DNA synapsis (Figure 3, top path). Also, conversely, it is possible that the looping itself allows the 3′RR to gain access and activate the GLT, herein the downstream protein factors induced by cytokine signals play an additional architectural role [76,78] (Figure 3, bottom path).
Figure 3. DNA synapsis during class switch recombination.
Shown on the left, in naïve mature B cells, a long-range interaction is formed between intronic Eµ enhancer and 3′ regulatory region (3′RR) in IgH locus [71]. GLT is constitutively active across Sµ region, indicated as an open pink oval with arrow. Upon LPS and IL-4 stimulation, the switch region for γ1 (Sγ1) starts GLT and comes into close proximity to Eµ (and therefore Sµ) and 3′RR, recombination will then occur between the two S regions with the intervening DNA deleted (depicted on right). In the establishment of CSR the relationship between transcription and 3D looping remains unclear. GLT and looping can be independently induced by the cytokine signal (middle path). Another plausible scenario is the induction of GLT across acceptor switch region (Sγ1) helps the juxtaposition (top path). A third model suggests the 3D looping brought enhancers (3′RR) into close proximity to γ1 and enhance GLT (bottom path).
Other than CTCF, recent studies have unravelled the architectural roles of some other proteins in mediating CSR. The DSB damage response protein p53-binding protein 1 (53BP1) was once thought to be critical in resolving the DSB [79], however, several studies have demonstrated an additional role of 53BP1 in mediating the chromosomal loop in IgH locus [80,81]. In the absence of 53BP1, interaction between 3′RR and Eµ is greatly diminished. Furthermore, Mediator, an evolutionarily conserved protein complex that is essential for PolII transcription, has been shown to facilitate long-range DNA–DNA interactions between the switch regions and the 3′RR, and also the transcriptional activation of GLT at those switch regions [78]. Lastly, a ubiquitous transcription factor YY1 was also revealed to mediate the looping between Eµ and 3′RR [82]. This loop is still present when YY1 lacks its transcriptional activation domain, suggesting YY1 has a direct architectural role in establishing the long-distance loop.
Terminal differentiation—colocalization of genes for maximal antibody production
The prominent endoplasmic reticulum, extensive Golgi apparatus and eccentric position of the nucleus with wheel-like chromatin configuration are all defining features of plasma cells, thought to enhance their massive antibody production [83,84]. In contrast, the plasma cell genome organisation during differentiation and its functional significance to massive antibody synthesis and secretion is poorly understood. Similar to other terminally differentiating cells, heterochromatinization is thought to contribute to locking in the plasma cell transcriptional programme, while shutting down other unnecessary genes, for instance the regular B cell signature genes [85,86]. Deposition of H3K27me3 by EZH2 and the removal of H3K4me1 by LSD1 are implicated in this process [87–89]. Consistent with this, our Hi-C experiments documented a wave of chromatin reconfiguration during this final stage of differentiation, accompanied by substantial transcriptional changes [47] (Figure 2). The restructuring involves enhancer-promoter loops and also a considerable amount of compartment switching [47,90]. For example, the Prdm1 locus, which codes for the master regulator for plasma cell differentiation Blimp-1, exhibits the strongest enhancer-promoter interaction at the plasmablast stage [47]. Whereas the DNA contacts for Bcl6, an important transcription factor during B cell activation, are essentially lost.
Three-dimensional DNA modelling from conformation capture assays revealed that the chromosomes of plasma cells adopt a unique elongated, α-helical structure, resulting in the characteristic cartwheel appearance [90,91] (Figure 4). This is in striking contrast with the spherical-like structure exhibited by the chromosomes in naïve B cells. The cartwheel, elongated configurations are mainly contributed by the larger chromosomes (chr1–9) where the long-range intra-chromosomal interactions are severely diminished. Instead, intriguingly, there is an enrichment of inter-chromosomal contacts between chromosome ends.
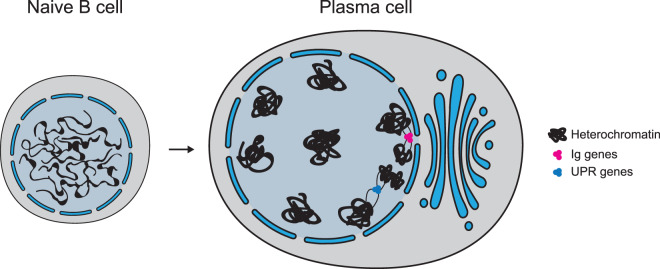
Figure 4. Chromosome reorganisation during terminal plasma cell differentiation.
Extensive heterochromatinization leads to a cartwheel configuration of chromatin in nucleus. Inter-chromosomal transcription hubs for UPR genes and Ig genes are independently formed, with Ig transcription hub located near the nuclear pore for maximal transcript export.
Using 3D imaging along with chromosome conformation capture, another study has shown that in plasma cells the active immunoglobulin genes (Igκ, IgH and IgJ), from three different chromosomes, are colocalised forming an apparent transcription factory [92] (Figure 4). Even more strikingly, this active cluster is located at the nuclear periphery, a region generally associated with gene silencing. More than 30 years ago, Blobel [93] proposed a gene gating hypothesis in which active genes are recruited near the nuclear pores for efficient transcript export. With recent technological advances in sequencing-based methods and electron microscopy, nuclear pores appear to be an exception at the nuclear periphery where gene activation is allowed [94,95]. In humans it has also been demonstrated that the nuclear pore complexes control gene expression by tethering to super-enhancers [96], suggesting an important regulatory role. In plasma cells, the colocalized Ig genes often have their transcripts share the same trafficking channel to the cytoplasm [92]. Therefore, it appears Ig genes colocalize near the nuclear pore for maximal transcript export, which further facilitates efficient and massive antibody production.
To accommodate massive protein synthesis and secretion, plasma cells elicit the unfolded protein response (UPR) to cope with the ER stress [97]. It is found that in plasma cells the genes involved in the UPR pathway, for instance Xbp1, Atf4 and Bax, also aggregate from different chromosomes and form transcription hubs [90] (Figure 4). Collectively, the inter-chromosomal interactions of Ig and UPR genes in plasma cells enable a co-ordinated and dedicated production of antibody.
Concluding remarks—Long-lived memory formation?
While a large body of research has focussed on understanding the cellular and molecular events governing B cell activation and terminal differentiation into antibody-secreting cells, very little is known about how the genome is wired in long-lived memory cells and how this contributes to their rapid recall upon rechallenge, due to the lack of good surface markers and their low abundance. Current literature suggests that memory B cells still possess a considerable amount of B cell signature and gene programme. For example, transcriptional profiling has shown a relatively similar gene expression landscape between memory and naïve B cells, with noticeable elevations of some anti-apoptotic genes and costimulatory molecules in memory cells [98]. Moreover, during a recall, memory B cells are able to promptly differentiate into plasmablasts and produce antigen-specific antibody, yet surprisingly they are also capable of undergoing a second round of somatic hypermutation and class switch recombination to modify the antibody specificity and isotype [99]. Thus, it is conceivable that the genome organisation of memory B cells would be different from terminally differentiated plasmablasts, while arranging similarly to naïve or activated B cells. Nevertheless, how such chromatin conformation facilitates a rapid memory response is entirely unknown. However, it is likely that it would involve some pre-existing loops poised for rapid activation.
Although the study of transcriptional regulation of memory B cells has long been hampered by their scarcity and heterogeneity, with recent advances in single cell techniques it is now possible to investigate them in greater details. Single-cell RNA-seq [100] and ATAC-seq [101] allow us to characterise the dynamics of transcription and chromatin accessibility, whereas high throughput microscopies and genome architecture mapping (GAM) [102] are able to resolve the chromatin conformation in each individual cell. These single-cell approaches could therefore provide valuable insights into the molecular identity of memory cells, and also other processes during B cell activation.
Perspectives
Three-dimensional genome organisation is critical in regulating a plethora of intricate cellular processes including gene transcription, DNA recombination, repair, replication and mitosis. This is of particular importance to B lymphocytes which, upon antigenic challenge, undergo a massive transcriptional burst, class switch recombination and rapid clonal expansion within a short period of time.
Naïve B cells possess pre-existing chromatin loops poised for rapid transcriptional burst upon stimulation and the subsequent pre-mitotic genome organisation is partitioned from DNA replication and mitosis. The newly established genome organisation will then dictate the transcriptional profiles over the successive clonal expansion until final differentiation into antibody-secreting cells.
Understanding the role of genome architecture in memory B cells is of particular interest to decipher the molecular principles behind their much more rapid secondary response. Also, individual cis-elements such as enhancers, loop anchors and long non-coding RNAs should be studied in depth to elucidate their contributions to genome organisation during B cell activation and differentiation.
Abbreviations
- AID
activation-induced deaminase
- CSR
class switch recombination
- DSBs
double strand breaks
- GLT
germline transcription
- NHEJ
non-homologous end joining
- SHM
somatic hypermutation
- TADs
topologically associating domains
- UPR
unfolded protein response
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access
Open access for this article was enabled by the participation of Walter and Eliza Hall Institute in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
References
- 1.Burnet F.M. (1976) A modification of Jerne's theory of antibody production using the concept of clonal selection. CA Cancer J. Clin. 26, 119–121 10.3322/canjclin.26.2.119 [DOI] [PubMed] [Google Scholar]
- 2.Hozumi N. and Tonegawa S. (1976) Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc. Natl Acad. Sci. U.S.A. 73, 3628–3632 10.1073/pnas.73.10.3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y. and Honjo T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- 4.Bonev B. and Cavalli G. (2016) Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- 5.Bickmore W.A. and van Steensel B. (2013) Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–1284 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 6.Maston G.A., Evans S.K. and Green M.R. (2006) Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7, 29–59 10.1146/annurev.genom.7.080505.115623 [DOI] [PubMed] [Google Scholar]
- 7.Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F. and de Laat W. (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 10.1038/ng1244 [DOI] [PubMed] [Google Scholar]
- 8.Tolhuis B., Palstra R.J., Splinter E., Grosveld F. and de Laat W. (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 10.1016/S1097-2765(02)00781-5 [DOI] [PubMed] [Google Scholar]
- 9.Bell A.C., West A.G. and Felsenfeld G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- 10.Merkenschlager M. and Nora E.P. (2016) CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev. Genomics Hum. Genet. 17, 17–43 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- 11.Bell A.C. and Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- 12.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M. and Tilghman S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 10.1038/35013106 [DOI] [PubMed] [Google Scholar]
- 13.Dekker J., Rippe K., Dekker M. and Kleckner N. (2002) Capturing chromosome conformation. Science 295, 1306–1311 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- 14.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y. et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nora E.P., Lajoie B.R., Schulz E.G.. Giorgetti L., Okamoto I., Servant N. et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I. et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. U.S.A. 112, E6456–E6465 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O. et al. (2015) CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 10.1126/science.1262088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busslinger G.A., Stocsits R.R., van der Lelij P., Axelsson E., Tedeschi A., Galjart N. et al. (2017) Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nora E.P., Goloborodko A., Valton A.L., Gibcus J.H., Uebersohn A., Abdennur N. et al. (2017) Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–44 e22 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N. and Mirny L.A. (2016) Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson I.F., Bauer B., Goetz D., Tang W., Wutz G. and Peters J.M. (2019) DNA loop extrusion by human cohesin. Science 366, 1338–1345 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- 24.Kim Y., Shi Z., Zhang H., Finkelstein I.J. and Yu H. (2019) Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O. et al. (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E. et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X. and Karpen G.H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabari B.R., Dall'Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K. et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falk M., Feodorova Y., Naumova N., Imakaev M., Lajoie B.R., Leonhardt H. et al. (2019) Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399 10.1038/s41586-019-1275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesne A., Baudement M.O., Rebouissou C. and Forne T. (2019) Exploring mammalian genome within phase-separated nuclear bodies: experimental methods and implications for gene expression. Genes (Basel) 10, E1049 10.3390/genes10121049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K. and Sharp P.A. (2017) A phase separation model for transcriptional control. Cell 169, 13–23 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Gao Y., Zheng X., Liu C., Dong S., Li R. et al. (2019) Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76, 646–59 e6 10.1016/j.molcel.2019.08.019 [DOI] [PubMed] [Google Scholar]
- 33.Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L. et al. (2017) Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R. et al. (2017) Cohesin loss eliminates all loop domains. Cell 171, 305–20 e24 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y. et al. (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y. et al. (2015) Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miura H., Takahashi S., Poonperm R., Tanigawa A., Takebayashi S.I. and Hiratani I. (2019) Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization. Nat. Genet. 51, 1356–1368 10.1038/s41588-019-0474-z [DOI] [PubMed] [Google Scholar]
- 38.Jaehning J.A., Stewart C.C. and Roeder R.G. (1975) DNA-dependent RNA polymerase levels during the response of human peripheral lymphocytes to phytohemagglutinin. Cell 4, 51–57 10.1016/0092-8674(75)90133-6 [DOI] [PubMed] [Google Scholar]
- 39.Pogo B.G., Allfrey V.G. and Mirsky A.E. (1966) RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl Acad. Sci. U.S.A. 55, 805–812 10.1073/pnas.55.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogo B.G., Allfrey V.G. and Mirsky A.E. (1967) The effect of phytohemagglutinin on ribonucleic acid synthesis and histone acetylation in equine leukocytes. J. Cell Biol. 35, 477–482 10.1083/jcb.35.2.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlings J.S., Gatzka M., Thomas P.G. and Ihle J.N. (2011) Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 30, 263–276 10.1038/emboj.2010.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieffer-Kwon K.R., Nimura K., Rao S.S.P., Xu J., Jung S., Pekowska A. et al. (2017) Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol. Cell 67, 566–78 e10 10.1016/j.molcel.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouzine F., Wojtowicz D., Yamane A., Resch W., Kieffer-Kwon K.R., Bandle R. et al. (2013) Global regulation of promoter melting in naive lymphocytes. Cell 153, 988–999 10.1016/j.cell.2013.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W. et al. (2012) . c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hawkins E.D., Markham J.F., McGuinness L.P. and Hodgkin P.D. (2009) A single-cell pedigree analysis of alternative stochastic lymphocyte fates. Proc. Natl Acad. Sci. U.S.A. 106, 13457–13462 10.1073/pnas.0905629106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner M.L., Hawkins E.D. and Hodgkin P.D. (2008) Quantitative regulation of B cell division destiny by signal strength. J. Immunol. 181, 374–382 10.4049/jimmunol.181.1.374 [DOI] [PubMed] [Google Scholar]
- 47.Chan W.F., Coughlan H.D., Zhou J.H., Keenan C.R., Bediaga N.G., Hodgkin P.D. et al. (2019) Genome reconfiguration prior to mitosis shapes the generation of adaptive immunity. bioRxiv, 762757 10.1101/762757 [DOI] [Google Scholar]
- 48.Bunting K.L., Soong T.D., Singh R., Jiang Y., Beguelin W., Poloway D.W. et al. (2016) Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity 45, 497–512 10.1016/j.immuni.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S. et al. (2016) Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–84 e19 10.1016/j.cell.2016.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johanson T.M., Lun A.T.L., Coughlan H.D., Tan T., Smyth G.K., Nutt S.L. et al. (2018) Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat. Immunol. 19, 1257–1264 10.1038/s41590-018-0234-8 [DOI] [PubMed] [Google Scholar]
- 51.Kieffer-Kwon K.R., Tang Z., Mathe E., Qian J., Sung M.H., Li G. et al. (2013) Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155, 1507–1520 10.1016/j.cell.2013.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhri V.K., Dienger-Stambaugh K., Wu Z., Shrestha M. and Singh H. (2020) Charting the cis-regulome of activated B cells by coupling structural and functional genomics. Nat. Immunol. 21, 210–220 10.1038/s41590-019-0565-0 [DOI] [PubMed] [Google Scholar]
- 53.de Laat W. and Duboule D. (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499–506 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- 54.Mele M. and Rinn J.L. (2016) “Cat's cradling” the 3D genome by the act of LncRNA transcription. Mol. Cell 62, 657–664 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 55.van Steensel B. and Furlong E.E.M. (2019) The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 20, 327–337 10.1038/s41580-019-0114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isoda T., Moore A.J., He Z., Chandra V., Aida M., Denholtz M. et al. (2017) Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171, 103–19 e18 10.1016/j.cell.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasbold J., Corcoran L.M., Tarlinton D.M., Tangye S.G. and Hodgkin P.D. (2004) Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat. Immunol. 5, 55–63 10.1038/ni1016 [DOI] [PubMed] [Google Scholar]
- 58.Mercado R., Vijh S., Allen S.E., Kerksiek K., Pilip I.M. and Pamer E.G. (2000) Early programming of T cell populations responding to bacterial infection. J. Immunol. 165, 6833–6839 10.4049/jimmunol.165.12.6833 [DOI] [PubMed] [Google Scholar]
- 59.Wong P. and Pamer E.G. (2001) Cutting edge: antigen-independent CD8T cell proliferation. J. Immunol. 166, 5864–5868 10.4049/jimmunol.166.10.5864 [DOI] [PubMed] [Google Scholar]
- 60.Williams M.A. and Bevan M.J. (2004) Shortening the infectious period does not alter expansion of CD8T cells but diminishes their capacity to differentiate into memory cells. J. Immunol. 173, 6694–6702 10.4049/jimmunol.173.11.6694 [DOI] [PubMed] [Google Scholar]
- 61.van Stipdonk M.J., Lemmens E.E. and Schoenberger S.P. (2001) Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat. Immunol. 2, 423–429 10.1038/87730 [DOI] [PubMed] [Google Scholar]
- 62.van Stipdonk M.J., Hardenberg G., Bijker M.S., Lemmens E.E., Droin N.M., Green D.R. et al. (2003) Dynamic programming of CD8+ T lymphocyte responses. Nat. Immunol. 4, 361–365 10.1038/ni912 [DOI] [PubMed] [Google Scholar]
- 63.Rush J.S. and Hodgkin P.D. (2001) B cells activated via CD40 and IL-4 undergo a division burst but require continued stimulation to maintain division, survival and differentiation. Eur. J. Immunol. 31, 1150–1159 [DOI] [PubMed] [Google Scholar]
- 64.Kaech S.M. and Ahmed R. (2001) Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2, 415–422 10.1038/87720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corbin G.A. and Harty J.T. (2004) Duration of infection and antigen display have minimal influence on the kinetics of the CD4+ T cell response to Listeria monocytogenes infection. J. Immunol. 173, 5679–5687 10.4049/jimmunol.173.9.5679 [DOI] [PubMed] [Google Scholar]
- 66.Iezzi G., Karjalainen K. and Lanzavecchia A. (1998) The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity 8, 89–95 10.1016/S1074-7613(00)80461-6 [DOI] [PubMed] [Google Scholar]
- 67.Xu Z., Zan H., Pone E.J., Mai T. and Casali P. (2012) Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 12, 517–531 10.1038/nri3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stavnezer J. and Schrader C.E. (2014) Igh chain class switch recombination: mechanism and regulation. J. Immunol. 193, 5370–5378 10.4049/jimmunol.1401849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kenter A.L., Feldman S., Wuerffel R., Achour I., Wang L. and Kumar S. (2012) Three-dimensional architecture of the IgH locus facilitates class switch recombination. Ann. N. Y. Acad. Sci. 1267, 86–94 10.1111/j.1749-6632.2012.06604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chatterjee S., Ju Z., Hassan R., Volpi S.A., Emelyanov A.V. and Birshtein B.K. (2011) Dynamic changes in binding of immunoglobulin heavy chain 3′ regulatory region to protein factors during class switching. J. Biol. Chem. 286, 29303–29312 10.1074/jbc.M111.243543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wuerffel R., Wang L., Grigera F., Manis J., Selsing E., Perlot T. et al. (2007) S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27, 711–722 10.1016/j.immuni.2007.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X., Zhang Y., Ba Z., Kyritsis N., Casellas R. and Alt F.W. (2019) Fundamental roles of chromatin loop extrusion in antibody class switching. Nature. 575, 385–389 10.1038/s41586-019-1723-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matthews A.J., Zheng S., DiMenna L.J. and Chaudhuri J. (2014) Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv. Immunol. 122, 1–57 10.1016/B978-0-12-800267-4.00001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yewdell W.T. and Chaudhuri J. (2017) A transcriptional serenAID: the role of noncoding RNAs in class switch recombination. Int. Immunol. 29, 183–196 10.1093/intimm/dxx027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marina-Zarate E., Perez-Garcia A. and Ramiro A.R. (2017) CCCTC-binding factor locks premature IgH germline transcription and restrains class switch recombination. Front. Immunol. 8, 1076 10.3389/fimmu.2017.01076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feldman S., Achour I., Wuerffel R., Kumar S., Gerasimova T., Sen R. et al. (2015) Constraints contributed by chromatin looping limit recombination targeting during Ig class switch recombination. J. Immunol. 194, 2380–2389 10.4049/jimmunol.1401170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Braikia F.Z., Oudinet C., Haddad D., Oruc Z., Orlando D., Dauba A. et al. (2017) Inducible CTCF insulator delays the IgH 3’ regulatory region-mediated activation of germline promoters and alters class switching. Proc. Natl Acad. Sci. U.S.A. 114, 6092–6097 10.1073/pnas.1701631114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas-Claudepierre A.S., Robert I., Rocha P.P., Raviram R., Schiavo E., Heyer V. et al. (2016) Mediator facilitates transcriptional activation and dynamic long-range contacts at the IgH locus during class switch recombination. J. Exp. Med. 213, 303–312 10.1084/jem.20141967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manis J.P., Morales J.C., Xia Z., Kutok J.L., Alt F.W. and Carpenter P.B. (2004) 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5, 481–487 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- 80.Rocha P.P., Raviram R., Fu Y., Kim J., Luo V.M., Aljoufi A. et al. (2016) A damage-independent role for 53BP1 that impacts break order and Igh architecture during class switch recombination. Cell Rep. 16, 48–55 10.1016/j.celrep.2016.05.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feldman S., Wuerffel R., Achour I., Wang L., Carpenter P.B. and Kenter A.L. (2017) 53BP1 contributes to Igh locus chromatin topology during class switch recombination. J. Immunol. 198, 2434–2444 10.4049/jimmunol.1601947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mehra P., Gerasimova T., Basu A., Jha V., Banerjee A., Sindhava V. et al. (2016) YY1 controls Emu-3'RR DNA loop formation and immunoglobulin heavy chain class switch recombination. Blood Adv. 1, 15–20 10.1182/bloodadvances.2016000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Albarracin F. and Fonseca R. (2011) Plasma cell leukemia. Blood Rev. 25, 107–112 10.1016/j.blre.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez A., Marin V.G., Junquera S.R., Martinez-Murillo R. and Freire M. (2005) The contributions of Santiago Ramon y Cajal to cancer research - 100 years on. Nat. Rev. Cancer. 5, 904–909 10.1038/nrc1741 [DOI] [PubMed] [Google Scholar]
- 85.Dillon N. and Festenstein R. (2002) Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18, 252–258 10.1016/S0168-9525(02)02648-3 [DOI] [PubMed] [Google Scholar]
- 86.Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A. et al. (2002) Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 17, 51–62 10.1016/S1074-7613(02)00335-7 [DOI] [PubMed] [Google Scholar]
- 87.Scharer C.D., Barwick B.G., Guo M., Bally A.P.R. and Boss J.M. (2018) Plasma cell differentiation is controlled by multiple cell division-coupled epigenetic programs. Nat. Commun. 9, 1698 10.1038/s41467-018-04125-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haines R.R., Barwick B.G., Scharer C.D., Majumder P., Randall T.D. and Boss J.M. (2018) The histone demethylase LSD1 regulates B cell proliferation and plasmablast differentiation. J. Immunol. 201, 2799–2811 10.4049/jimmunol.1800952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo M., Price M.J., Patterson D.G., Barwick B.G., Haines R.R., Kania A.K. et al. (2018) EZH2 represses the B cell transcriptional program and regulates antibody-secreting cell metabolism and antibody production. J. Immunol. 200, 1039–1052 10.4049/jimmunol.1701470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bortnick A., He Z., Aubrey M., Chandra V., Denholtz M., Chen K. et al. (2020) Plasma cell fate is orchestrated by elaborate changes in genome compartmentalization and inter-chromosomal hubs. Cell Rep. 31, 107470 10.1016/j.celrep.2020.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bortnick A., He Z., Aubrey M., Chandra V., Denholtz M., Chen K. et al. (2018) An inter-chromosomal transcription hub activates the unfolded protein response in plasma cells. bioRxiv, 295915 10.1101/295915 [DOI] [Google Scholar]
- 92.Park S.K., Xiang Y., Feng X. and Garrard W.T. (2014) Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev. 28, 1159–1164 10.1101/gad.237479.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blobel G. (1985) Gene gating: a hypothesis. Proc. Natl Acad. Sci. U.S.A. 82, 8527–8529 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blobel G. (2010) Three-dimensional organization of chromatids by nuclear envelope-associated structures. Cold Spring Harb. Symp. Quant. Biol. 75, 545–554 10.1101/sqb.2010.75.004 [DOI] [PubMed] [Google Scholar]
- 95.Buchwalter A., Kaneshiro J.M. and Hetzer M.W. (2019) Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 20, 39–50 10.1038/s41576-018-0063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ibarra A., Benner C., Tyagi S., Cool J. and Hetzer M.W. (2016) Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30, 2253–2258 10.1101/gad.287417.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gass J.N., Gifford N.M. and Brewer J.W. (2002) Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277, 49047–49054 10.1074/jbc.M205011200 [DOI] [PubMed] [Google Scholar]
- 98.Bhattacharya D., Cheah M.T., Franco C.B., Hosen N., Pin C.L., Sha W.C. et al. (2007) Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179, 6808–6819 10.4049/jimmunol.179.10.6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dogan I., Bertocci B., Vilmont V., Delbos F., Megret J., Storck S. et al. (2009) Multiple layers of B cell memory with different effector functions. Nat. Immunol. 10, 1292–1299 10.1038/ni.1814 [DOI] [PubMed] [Google Scholar]
- 100.Usoskin D., Furlan A., Islam S., Abdo H., Lonnerberg P., Lou D. et al. (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]
- 101.Cusanovich D.A., Hill A.J., Aghamirzaie D., Daza R.M., Pliner H.A., Berletch J.B. et al. (2018) A single-cell atlas of In vivo mammalian chromatin accessibility. Cell 174, 1309–24 e18 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beagrie R.A., Scialdone A., Schueler M., Kraemer D.C., Chotalia M., Xie S.Q. et al. (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]