Abstract
Human pathogens need to overcome an elaborate network of host defense mechanisms in order to establish their infection, colonization, proliferation and eventual dissemination. The interaction of pathogens with different effector molecules of the immune system results in their neutralization and elimination from the host. The complement system is one such integral component of innate immunity that is critically involved in the early recognition and elimination of the pathogen. Hence, under this immune pressure, all virulent pathogens capable of inducing active infections have evolved immune evasive strategies that primarily target the complement system, which plays an essential and central role for host defense. Recent reports on several bacterial pathogens have elucidated the molecular mechanisms underlying complement evasion, inhibition of opsonic phagocytosis and cell lysis. This review aims to comprehensively summarize the recent findings on the various strategies adopted by pathogenic bacteria to escape complement-mediated clearance.
Electronic supplementary material
The online version of this article (10.1007/s12088-020-00872-9) contains supplementary material, which is available to authorized users.
Keywords: Pathogen, Infection, Virulence, Complement system, Immune evasion
Introduction
Human immunity comprises of the innate and adaptive systems that synergistically target a foreign pathogen leading to its neutralization. Innate immunity non-specifically targets a wide range of pathogens while adaptive immune responses are pathogen specific. Complement system is a critical element of innate immunity that comprises of a coordinated and efficient proteolytic cascade leading to neutralization of the microbial pathogen [1]. The complement system consists of an intricate network of membrane associated and circulating proteins that either function as regulators, functional enzymes or substrates in the extracellular proteolytic cascade [2]. It works in tandem with other immune effector cells and performs various functions such as removal of foreign cells, recycling of cell debris and communication with adaptive responses [3]. Complement activation occurs through three well-described mechanisms namely: classical pathway, alternative pathway and lectin pathway [4]. Each pathway has distinct initiating mechanisms, which enable them to recognize diverse pathogenic structures. The classical pathway activation occurs through the early recognition and binding of the C1q complement protein with the antibodies tethered to the antigen on the foreign cell surface. Lectin pathway is initiated with the binding of carbohydrate moieties on the microbial pathogens with the pathogen recognition molecules (PRMs), such as mannose binding lectins and ficolins. Contrary to both these pathways, the alternative pathway is activated through the spontaneous hydrolysis of C3 on the microbial surface [5].
All these three complement pathways that are activated by distinct initiation stimuli, converge together through the central enzymatic cleavage of C3 (complement protein) and the subsequent formation of C3a and C3b (active fragments of C3) [6]. Covalent attachment of C3b on foreign microbial surfaces fulfills three crucial functions: (1) phagocytosis mediated cell clearance; (2) formation of an amplification loop for the creation of surface associated C3 convertase leading to complement activation; (3) binding of C3 convertase with the increasing concentration of surface bound C3b leads to the creation of C5 convertase [4]. C5 convertase cleaves C5 into its active components; C5a and C5b, which act as a potent anaphylatoxin [7] and an integral component of the membrane attack complex (MAC), respectively. Complement proteins C6, C7 and C8 sequentially interact with C5b to form C5b678, which has the propensity to embed in the cell membrane [8]. The C5b678 complex associates with multiple copies of the complement protein, C9, leading to the formation of MAC and subsequent killing of the cells [8]. Apart from the formation of MAC, surface bound C3b acts as an effector molecule facilitating innate and adaptive immunity [4]. A number of complement receptors present on the immune cells mediate the recognition of opsonized cells and activation of cells involved in adaptive immune response. Interaction of C3b with the members of the RCA family (regulators of complement activation), namely; CD35 (Complement receptor 1-CR1) and CD46, results in the breakdown of C3b to iC3b and/or C3c and C3dg [9]. iC3b and C3dg remains attached to the microbial surface, while C3c (fluid phase protein) gets released in circulation [9]. iC3b binds with CR3 (CD11b) and CR4 (CD11c) that are phagocytic integrin receptors and these interactions facilitates complement-mediated phagocytosis [10]. Furthermore, iC3b also binds with the B cell receptor, CR2 (CD21), that is also present on follicular dendritic cells. The iC3b–CD21 interaction lowers the B cells activation threshold and promotes induction of memory cells [11]. Hence, C3b association and degradation on the microbial surface stimulates multiple immune effector functions; immune adherence (C3b–CR1), phagocytosis (iC3b/C3dg–CR3, iC3b–CR4) and modulating adaptive immune response (iC3b–CD21).
The microbial surface is constituted with pathogen associated molecular patterns (PAMPs), which are recognized by host pattern recognition molecules (PRM) like complement proteins and IgG, resulting in the activation of the innate immune system. Hence, complement proteins are less likely to get activated on host cell surfaces. However, several membrane-bound and circulating regulatory molecules are present in the serum that ensure the prevention of any complement mediated harmful effects on host cells, thus imparting protection and selectivity. Complement regulatory molecules predominantly target C3, its proteolytic fragments and complexes formed [12]. Complement inhibitors typically belong to the RCA family and include both membrane-bound and circulating regulators [4]. CD35 (CR1), CD59, CD46 (membrane co-factor protein, MCP) and CD55 (decay acceleration factor, DAF), are examples of membrane bound regulators [13]. Soluble complement regulators include Factor H (FH), C4b-binding protein (C4BP) and C1 inhibitor (C1-INH). C4BP inhibits both the classical and lectin pathways by cleaving surface associated C4b [14]. FH inhibits the alternative pathway by blocking the association of factor B (FB) with C3b and promoting C3bBb dissociation [15]. C1-INH is a serine protease inhibitor that inactivates MASP-1, MASP-2, C1r and C1s, thus inhibiting both the lectin and classical pathways [16].
The pathogen-neutralizing role of the complement system is crucial for human health, which is substantiated by the observations that humans with complement protein deficiencies are at a higher risk of infections [17]. Infectious diseases pose a great threat to human health and this problem has intensified with the emergence of antibiotic resistance. Thus, it is imperative to understand the complex host–pathogen interactions to develop novel intervention strategies to successfully counter infections. Our current review aims to update the reader about the critical complement evasion strategies adopted by human pathogens, which could be helpful in designing new therapeutic strategies.
Complement Evasion Mechanisms
The successful establishment of infection and pathogenicity by a microbial pathogen is majorly attributed to their ability to evade the sophisticated surveillance mechanisms of the human immune system. Innate immunity is the first line of defense of which the complement system is the primary target of pathogen for immune evasion [18]. C3 is the primary target molecule of most immune evasive strategies of human pathogens. Evolutionary pressure on pathogens has led to the development of complex mechanisms to inhibit C3 mediated effector functions. Identifying pathogenic proteins and their human targets is critical in understanding the pathogenic mechanism that ensures their survival in the human host. In recent years, our knowledge of the molecular mechanisms of complement evasion has expanded remarkably. Although there are a large number of pathogenic complement-binding proteins, their mechanisms of action are common, which could be categorized into a few effective strategies. These strategies include recruitment of the RCA host complement regulatory proteins on the pathogen surface, the expression of cellular components and proteins on the pathogen surface that inhibit complement activation, blocking or destabilization of the C3 convertase and host plasminogen mediated complement degradation. The following sections focus on specific mechanisms underlying complement evasion by human pathogens (Table 1; Fig. 1).
Table 1.
Complement evasive strategies of human bacterial pathogens
| Evasive strategy | Evasion molecules | Mechanism of action | Bacterial pathogen | References |
|---|---|---|---|---|
| Recruitment of complement regulatory proteins | Vag8 | Interacts with C1-INH | Bordetella pertussis | [19] |
| PspA and PspC | Interacts with C4BP | Streptococcus pneumoniae | [20] | |
| PorA | Interacts with C4BP | Neisseria meningitidis | [21] | |
| Protein H | Interacts with C4BP | Streptococcus pyogenes | [22] | |
| Protein M | Interacts with C4BP | Streptococcus pyogenes | [23] | |
| Lsa 25/30/33 | Interacts with C4BP | Leptospira interrogans | [24] | |
| FHbp | Interacts with Factor H | Neisseria meningitidis | [25, 26] | |
| CbiA | Interacts with Factor H | Borrelia miyamotoi | [27] | |
| CspZ | Interacts with Factor H | Borrelia burgdorferi | [28] | |
| Sht | Interacts with Factor H | Streptococcus agalactiae | [29] | |
| Adr1/Adr2 | Interacts with Factor H | Rickettsia conorii | [30] | |
| SdrE | Interacts with Factor H | Staphylococcus aureus | [31, 32] | |
| Lpd | Interacts with Factor H | Pseudomonas aeruginosa | [33] | |
| BclA | Interacts with Factor H | Bacillus anthracis | [34] | |
| SpeB | Interacts with Factor H | Streptococcus pyogenes | [35] | |
| Blocking host antibodies | Plasmin | Degrades IgG | Staphylococcus aureus | [36] |
| Protein A | Binds to Fc region of IgG |
Group G Streptococci, Staphylococcus aureus |
[37, 38] | |
| Protein G | Binds to Fc region of IgG |
Group G Streptococci, Staphylococcus aureus |
[39] | |
| Sbi | Binds to Fc region of IgG | Staphylococcus aureus | [40] | |
| IdeS | Cleaves IgG from hinge region | Group A Streptococci | [41] | |
| PepO | Binds to C1q and inhibit IgG–C1q | Streptococcus pyogenes | [42] | |
| Vi antigen | Inhibit IgG binding on bacterial surface | Salmonella typhi | [43] | |
| IgA1 hydrolase | Inactivates IgA1 | Streptococcus suis | [44] | |
| IgM Protease | Inactivates IgM | Streptococcus suis | [44] | |
| C3 convertase inhibitors | LPS | Inhibit properdin binding | Escherichia coli, | [45] |
| SpeB | Degrades properdin and C3 | Streptococcus pyogenes | [35, 46] | |
| SCIN | Blocks C3bBb functioning | Staphylococcus aureus | [47, 48] | |
| Efb/Ecb | Stabilize C3bB pro-convertase | Staphylococcus aureus | [49] | |
| Eap | Blocks C4b–C2 interaction | Staphylococcus aureus | [50] | |
| CIP | Blocks C4b–C2 interaction | Streptococcus agalactiae | [51] | |
| BBK32 | Binds to C1r and inhibit C4b2a | Borrelia burgdorferi | [52, 53] | |
| SntA | Binds to C1q and inhibit C4b2a | Streptococcus suis | [54] | |
| MSCRAMM | Inhibits C1q–C1r interaction | Staphylococcus aureus | [55] | |
| Bacterial Proteases | Aureolysin | Cleaves C3 in non-functional form | Staphylococcus aureus | [56] |
| Plasmin | Degrades C3 |
Staphylococcus aureus, Leptospira interrogans, Streptococcus suis |
[36, 57–60] | |
| PaE | Degrades C1 | Pseudomonas aeruginosa | [61, 62] | |
| PaAP | Degrades C1 | Pseudomonas aeruginosa | [61, 62] | |
| EspP | Degrades C3/C3b and C5 | Escherichia coli | [63] | |
| Mirolysin | Degrade ficolin-2, ficolin-3, C4 and C5 | Tannerella forsythia | [64] | |
| AprA | Degrades C1s and C2 | Pseudomonas aeruginosa | [65] | |
| Inhibition of microbial cell Killing | SSL-7 | Inhibits C5–C5 convertase interaction | Staphylococcus aureus | [66] |
| CspA | Inhibit C9 polymerization | Group A Streptococci | [67] | |
| SIC | Prevents C5b-7 insertion in membrane | Borrelia burgdorferi | [68] | |
| CD59 | Inhibits MAC formation |
Borrelia burgdorferi, Helicobacter pylori |
[69] [70] |
|
| CHIPS | Inhibits neutrophil chemotaxis | Staphylococcus aureus | [71] | |
| GAPDH | Inhibits neutrophil chemotaxis | Streptococcus pyogenes | [72] | |
| SepA | Cleaves C5a and prevents chemotaxis | Streptococcus pyogenes | [35] | |
| Ecb | Inhibit C3b interaction with CR1 | Staphylococcus aureus | [73] | |
| Exosporium | Inhibits Oxidative burst | Kingella kingae | [74] | |
| BGA 66/71 | Inhibits MAC formation | Borrelia bavariensis | [75] | |
| LcpA | Blocks C9 polymerization | Leptospira interrogans | [76, 77] |
The various complement evasion strategies employed by human bacterial pathogens have been tabulated. The descriptions of the mechanisms and effector molecules involved for each bacterial pathogen have been listed along with the corresponding references
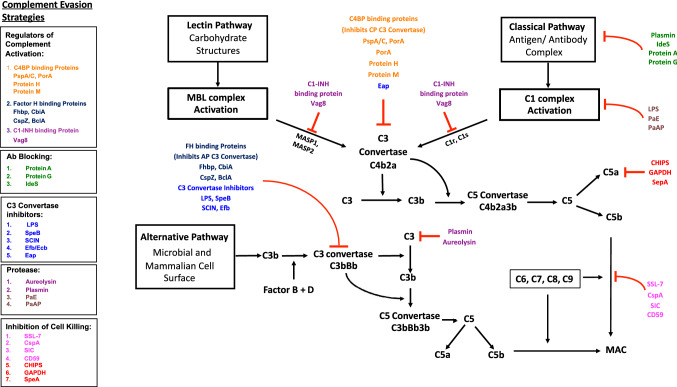
Fig. 1.
Complement evasion strategies adopted by human pathogenic bacteria. Schematic representation of the cascades involved in the classical, alternative and lectin complement pathways have been depicted. The complement evasive strategies exhibited by the different pathogens are listed in the separate boxes that have been classified as per their mechanism of action. The effector molecules under a common complement evasion mechanism have been color-coded. Regulators of complement activation, C4 binding protein (C4BP) and Factor H targets C3 convertase. C1-Inhibitor (C1-INH) targets C1/Mannose binding lectin (MBL) associated proteases. IgG proteases and binding inhibitors (Plasmin, IdeS, Protein A and Protein G) block the activation of the classical pathway. Lipopolysaccharide (LPS), Staphylococcal complement inhibitor (SCIN), Streptococcus pyogenes exotoxin B (SpeB), Extracellular fibrinogen binding protein (Efb) inhibit C3 convertase of the alternative pathway. Pathogenic proteases (Aureolysin, Plasmin) degrade human C3 and host Plasmin degrades IgG. Chemotaxis inhibitory protein (CHIPS) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) degrade the anaphylatoxin molecule C5a. Staphylococcal super antigen like-7 protein (SSL-7), Complement regulator-acquiring surface protein (CspA), Streptococcal inhibitor of complement (SIC) and GPI-Anchored protectin (CD59) inhibit the formation of the membrane attack complex (MAC)
Role of Host Regulatory Proteins in Complement Evasion
RCA proteins play a major role in regulating the activation of complement proteins on self and foreign cells. Human pathogens usually lack self-patterns on their surface. However, they have developed strategies to stably bind with soluble RCA proteins circulating in the human plasma [78]. Among the various complement evasive mechanisms adopted by pathogens, recruiting RCA is the most commonly employed strategy for evading complement activation. RCA recruitment has been widely reported for bacterial pathogens, as well as fungi, viruses and parasites [79–81]. Recruitment of RCA as a complement evasive strategy has certain significant advantages: (1) RCA are host proteins present in relatively higher concentrations in human plasma [4]; (2) RCA have common structural features (short consensus repeats) [3] that engage with various complementary pathogen-derived proteins facilitating their recruitment.
Soluble regulatory proteins such as factor H (FH) and C4-binding protein (C4BP) play a critical role in securing the host cells from undesirable activation of complement proteins. FH binds simultaneously with surface associated C3b and glycosaminoglycans (chondroitin sulphate, heparin sulphate and heparin dermatan) present on the host cell surface through its domains 6–7 and 19–20 [82]. Thus, the complement inhibitory domains of FH (domain 1–4) [82] bind with surface bound C3b and leads to its degradation that is also defined as decay acceleration activity. Due to co-evolution of pathogenic microbes and their hosts, many pathogens have developed the capability to bind with FH. Interestingly, most pathogens interact with FH through the same domains 19–20 and/or 6–7 that binds with C3b and glycosaminoglycans [82]. Like host glycosaminoglycans (GAG), the pathogenic FH binding protein forms a tripartite structure with FH domains and C3b. The Neisseria gonorrhoeae outer membrane porin protein and sialylated lipopolysaccharide (LOS) together interacts with the C-terminus of FH [83]. The FH-binding lipoprotein (fHbp) expressed on Neisseria meningitidis resembles the polyanionic carbohydrates on host surfaces and binds with human FH [25, 26]. The fHbp subvariant is also used as a component of the meningococcal vaccines, Trumenba (Pfizer) and Bexsero (GSK) [84]. Recently, a number of factor H binding proteins from different bacterial pathogens have been reported. The Complement binding and inhibitory protein A (CbiA) of Borrelia miyamotoi has been reported to bind with factor H [27]. Two orthologues of the Borrelia burgdorferi CspA protein have been identified as Factor H binding proteins in Borrelia mayonii [85]. The spirochete Treponema denticola surface lipoprotein FhbB exhibits binding affinity for human FH [86, 87]. The Adr1/Adr2 surface antigens of Rickettsia conorii have been reported to bind with FH [30]. Staphylococcus aureus, the master of immune evasion also expresses a FH binding protein, SdrE that regulates complement activation [31, 32]. The Pseudomonas aeruginosa surface protein dihydrolipoamide dehydrogenase (Lpd) has been reported to bind with serum factor H [33]. Recently, a variety of bacterial proteins such as the CspZ surface protein of Borrelia burgdorferi [28], the Factor H-combining protein of Streptococcus suis [88] and the zinc uptake Sht family of Streptococcus agalactiae [29] have been reported to bind with host FH and impart protection to the bacteria. Binding of FH with Streptococcus pneumoniae PspC protein induces conformational changes in FH that increases its activity than that might be attained through its association with GAG on the host surface [89–91]. The Bacillus anthracis spore surface protein, BclA, has also been reported to bind with human FH in order to counter complement activation [34]. A sequence analysis of the primary amino acid structure of these FH binding proteins shows no conserved homologous binding domains that could mediate the binding function (Figure S1).
Factor H (FH) belongs to the family of host proteins comprising of other members such as Factor H like protein (FHL-1) and five factor H related proteins (FHR-1 to FHR-5) [92]. FH and FHRs are encoded by related genes and hence share binding similarities to various ligands. FH inhibits the alternative pathway C3 convertase and acts as negative regulator of complement activation. FH also binds to the monomeric, non-functional C-reactive protein (CRP), whereas FHR-4 binds to the functional pentameric CRP, thus promoting the activation of the classical pathway. FHR-1, FHR-4 and FHR-5 bind with C3b to mediate the formation of the alternative pathway C3 convertase (C3bBb). Binding of FHRs competitively inhibits the C3b binding of FH, thus leading to the activation of the alternative pathway [93].
FH is the negative regulator of the complement pathway, while FHRs positively regulate complement activation. Neisseria meningitidis FH binding protein, Fhbp, recruits host FH and imparts protection to the bacteria from complement mediated clearance. FHR-3 has been reported to compete with FH for its binding with Fhbp, resulting in the reduced FH recruitment on the bacterial surface and subsequent clearance through complement activation [94]. However, in a unique role, FH recruitment on the surface of Mycobacterium bovis mediates its adhesion with phagocytic cells as FH bind with the CR3 receptors on the neutrophils. Similarly, the binding of FHR-1 associated on the M. bovis surface with the CR3 receptors mediates its adhesion with neutrophils. [95].
Similarly, C4BP is another soluble RCA that is abundantly present in human plasma [96], and is responsible for inhibiting complement activation on host cells. Hence, to evade complement mediated clearance, pathogens have adapted to recruit the negative regulator of the complement system, C4BP, on their outer surface. C4BP is a 500 kDa glycoprotein comprising of a 40 kDa β-chain and seven 75 kDa α-chains [14]. C4BP regulates complement activity by binding to activated C4b and C3b and inhibit their functions [96]. C4BP electrostatically interacts with C4b via a group of positively charged amino acids in its α-chain [97]. Similar to FH, C4BP also inhibits the complement activation cascade at a relatively early stage prior to the activation of downstream complement components. Streptococcus pyogenes expresses the M-protein on its surface, which binds to plasma C4BP [23]. M protein is dimeric, α-helical coiled protein having an extracellular N-terminal hypervariable region (HVR) [23]. In spite of its variability, the M protein binds with C4BP, demonstrating the conservation of this essential interaction for the survival of S. pyogenes in the host [98]. M protein and C4b have overlapping binding sites on C4BP. However, the heptameric structure of C4BP allows the binding of both proteins present on the surface of S. pyogenes [99] facilitating its decay accelerating activity that degrades C3b.
Protein H is another S. pyogenes surface expressed protein that binds to C4BP [22]. Protein H interacts with various ligands, including human IgG [100]. However, the binding of protein H with IgG through its Fc region renders it ineffective in activating the complement system and the employment of the FcR [18], leading to inhibition of opsonic phagocytosis. It has been reported that IgG, protein H and C4BP forms a tripartite complex that enhances the binding of C4BP on the bacterial surface [101, 102]. Strains of Neisseria meningitidis express the PorA protein bind with C4BP that inhibits complement activation [21]. Leptospira interrogans expresses the outer membrane proteins Lsa30, Lsa25 and Lsa33 that binds with C4BP and inhibits complement activation [24]. Streptococcus pneumoniae surface proteins, PspA and PspC, bind with C4BP and degrade surface associated C4b to C4dg, which remains associated with the bacterial surface but is unable to form C3 convertase [20]. Certain other non-classical surface associated proteins (moonlighting proteins) have been reported to help in recruiting plasma C4BP on the pathogenic cell surface. For example, Streptococcus pneumoniae utilizes enolase, a glycolytic enzyme, as a C4BP binding protein [103]. The molecular interactions of FH and C4BP with their respective pathogen proteins exhibit a species specificity, which is believed to the basis of host selectivity of pathogens.
Certain bacteria have developed strategies to recruit another serum inhibitor protein C1-INH on their surface. Recently, it has been reported that the Bordetella pertussis Vag8 protein (Virulence associated gene 8) interacts with the host C1-INH, leading to the degradation of the C1r, C1s proteases and the MBL associated serine protease (MASP 2). Recruitment of C1-INH on the bacterial surface results in reduced binding of C4 and C2 on the bacterial surface [19]. Antibodies generated against the Vag8 antigen have been reported to be effective in inhibiting the recruitment of C1-INH on the bacterial surface [104].
Blocking Antibody Binding with Microbial Surfaces
Binding of C1q with antibodies (IgG and IgM) tethered to PAMPs initiates the activation of the classical pathway. The interaction of antibody and complement proteins on the pathogenic surface is crucial for their elimination from the host and several pathogens have developed strategies to disrupt this key interaction [105]. Blocking of the antibody and complement mediated effector mechanism was amongst the first immune evasive strategy described for the bacteria, Staphylococcus aureus and group G Streptococci [37, 39, 106]. Group G Streptococci and Staphylococcus aureus are known to express the cell wall associated Protein A [37, 38] and Protein G [39], respectively, which interacts with various ligands such as IgG. Protein A binds with the Fcγ region of human IgG. It is abundantly expressed on the surface of S. aureus and its interaction with plasma IgG leads to the bacteria being coated with IgG in an inverted or upside-down direction through its Fc region [106]. Protein A prevents IgG from interacting with C1q and FcR of the potent phagocytes due to the inverted configuration of the bound IgG. Another multifunctional protein of Staphylococcus aureus, Sbi also binds to the Fc region of IgG and inhibits complement activation [40]. Group A Streptococci produce immunoglobulin degrading cysteine proteases (IdeS) that cleaves the IgG at the hinge region, thus removing the Fc region and making the bacteria associated IgG inaccessible to the binding of C1q [41]. Recently, it has been reported that Streptococcus suis secretes IgA1 hydrolase and IgM protease, which directly cleave host immunoglobulins [44].
Many bacteria express a capsule on their surface, which helps them in preventing desiccation and resisting host immune responses [107]. The bacterial capsule is primarily composed of repeating units of monosaccharides or amino acids [107, 108]. In an appropriate example of molecular mimicry, the N-acetylneuraminic acid-based capsule of Neisseria meningitidis exhibits poor immunogenicity and a strong structural similarity with the human neural cell adhesion molecule (NCAM) [109]. The N. meningitidis capsule prevents antibody mediated C1q recruitment on the bacterial surface leading to an inhibition of the classical pathway [110]. The capsule also inhibits the alternative pathway by masking the sub-capsular targets for C3b deposition [107]. In addition, the hairy structure of the capsule may create a hindrance in the interaction between the bound C3b with complement receptors present on phagocytes. Some bacteria modify their capsule composition in order to prevent complement activation. Klebsiella pneumoniae serotypes lacking rhamnobiose and mannobiose evade their recognition by the lectin pathway and exhibit greater virulence [111]. Recently, it has been reported that the capsular Vi antigen of Salmonella typhi inhibits bacterial phagocytosis and neutrophil respiratory burst by inhibiting the binding of antibodies on the bacterial surface [43].
In gram-negative bacteria, apart from the capsule, the lipopolysaccharide (LPS) in the outer membrane plays a crucial role in innate immune evasion. LPS is present in both smooth and rough forms that are determined by the presence or absence of the O-antigen, respectively [112]. Bacterial strains with a rough LPS are more sensitive to complement mediated bactericidal activity in comparison to strains displaying a smooth LPS [113]. The O-antigen is an elongated component of LPS that has been reported in Klebsiella pneumoniae to inhibit C1q binding and that of subsequent complement proteins on the outer bacterial membrane. Importantly, the elongated O-antigen has been demonstrated to prevent membrane insertion of the MAC complex [113, 114]. Recently, the Streptococcus pyogenes endopeptidase, PepO has been reported to bind with serum C1q with a higher affinity in comparison to human IgG. PepO inhibits the binding of C1q with pathogen associated IgG and results in the inhibition of classical complement pathway [42].
Inhibition of C3 Convertase Activity and Formation
C3 convertase plays a central role in C3b formation during all the three complement activation pathways. However, the alternative pathway C3 convertase is unstable and has a short half-life. In this regard, the C3 convertase (C3bBb) of the alternative pathway is specifically stabilized by a positive complement regulator, properdin [115]. The association of properdin enhances the half-life of C3 convertase by 5–10-fold [116]. A stable C3 convertase is essential for pathogen clearance from the host and thus, some bacterial species have developed strategies to disturb this stabilization process. Lipopolysaccharide is an integral constituent of the cell membrane of gram-negative bacteria. In many bacterial species, LPS inhibits the binding of properdin with the bacterial surface [18]. O-antigen mutants of LPS in E. coli K12 exhibit higher level of binding of both properdin and C3b on their outer surface in comparison to its wild-type counterparts [45]. However, the mechanism underlying the binding inhibitory properties of LPS remains undefined. Apart from inhibiting the binding of properdin, certain bacteria are able to induce the direct degradation of properdin, for example Streptococcus pyogenes exotoxin B (SpeB) is a cysteine protease that degrades properdin and other complement proteins such as C3 [35, 46].
Staphylococcus aureus, which is a perfect example of a pathogen exhibiting complement evasion, exhibits another evasion mechanism involving the production of an anti-C3 convertase molecule, Staphylococcal complement inhibitor (SCIN). The SCIN binds with both C3b and the Bb, cleaved fragment of Factor B, forming a link between them and arresting C3bBb in a non-functional conformation [47, 48]. The extracellular fibrinogen binding protein (Efb) and the extracellular complement binding protein (Ecb) stabilize the interaction of C3b and FB increasing the stability of C3bB pro-convertase on the surface of Staphylococcus aureus [49]. Staphylococcus aureus exhibits the ability to not only inhibit the alternative pathway C3 convertase, but also that of the classical and lectin pathways. The extracellular adherence protein (Eap); a soluble complement inhibitor binds with C4b, blocking the C4b–C2 interaction that reduces the levels of the C3 convertase of the classical pathway or the lectin pathway, C4b2a [50]. Streptococcus agalactiae secretes a complement interfering protein (CIP) that has a high binding affinity for C4b and blocks its interaction with C2a, resulting in the reduced formation of C3 convertase, C4b2a [51]. The collagen-binding microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) of Staphylococcus aureus bind with the collagenous domain of C1q and block its interaction with C1r [55]. Recently, the SntA protein of Streptococcus suis [54] and the BBK32 protein of Borrelia burgdorferi have been found to bind with the C1q complex and inhibit the formation of the C3 convertase of the classical pathway [52, 53, 117].
Complement Protein Cleavage by Proteases
The recruitment of active proteases that degrade IgG and complement proteins is another immune evasive strategy adopted by many human bacterial pathogens. These proteases are primarily expressed by the bacteria itself. However, certain bacteria are also able to recruit host plasminogen that gets activated to the protease, plasmin that also degrades the complement molecules. These two distinct sets of proteases involved in complement evasion are described below:
Complement Protein Cleavage by Bacterial Proteases
The bacterial proteases specifically degrade complement proteins into smaller, non-functional fragments that subsequently inhibit complement mediated effector mechanisms for bacterial clearance. Examples of bacterial proteases that degrade complement proteins and effectively inhibit complement activation are described below. Staphylococcus aureus secretes a zinc metalloprotease, aureolysin (Aur), which cleaves C3 at a site located two amino acids away from that of C3 convertase [118]. Pseudomonas aeruginosa alkaline protease (PaAP) and elastase (PaE) degrade C1 and inhibit the activation of the classical pathway [61, 62]. The enterohemorrhagic bacteria, E. coli (EHEC), secrete an extracellular serine protease P (EspP), that cleaves C3/C3b and C5 [63] The C5a peptidase (ScpA) of Streptococci degrades C5a and blocks both pro-inflammatory and chemotactic signaling [56, 119]. The human pathogen Porphyromonas gingivalis secretes an active protease (prtH) that also degrades C3b and IgG [120]. The periodontist pathogen Tannerella forsythia secretes the metalloproteinase mirolysin that inhibits all the three complement pathways by degrading ficolin-2, ficolin-3, C4 and C5 [64]. Pseudomonas aeruginosa expresses an alkaline protease (AprA) that degrades both C1s, C2 and inhibits the classical complement pathway [65].
Complement Protein Cleavage by Activation of Host Plasminogen
A proteolytic strategy adopted by bacterial pathogens is to exploit the host fibrinolytic system that precisely targets activation of plasminogen. Plasminogen is a liver-derived glycoprotein that circulates as a pro-enzyme in human serum and gets activated to plasmin, which further acts in resolving fibrin clots. Plasmin is a serine protease that exhibits a specificity for a broad spectrum of substrates. Besides its primary substrate, fibrinogen, plasmin also cleaves complement proteins, C3b and IgG [121]. Examples of bacteria that recruit plasminogen, which further gets activated to plasmin are described below. Staphylococcus aureus expresses a number of plasminogen binding molecules on their surface. The bound plasminogen gets activated into plasmin by the Staphylococcal enzyme, Staphylokinase (SAK) [36]. Recently, a number of bacteria pathogens have been reported to similarly express the plasminogen binding protein that induces the activation of plasminogen to plasmin and recruits the host machinery to evade complement mediated lysis [57]. The Streptococcus pneumoniae glycolytic enzyme phosphoglycerate kinase (PGK) [58] and Streptococcus suis enolase have been reported to activate host plasminogen [88]. The surface proteins of Leptospira interrogans (LIC11711) [59] and Treponema denticola (FhbB) [86] also bind and activate host plasminogen. Plasmin degrades both IgG and C3b/iC3b on the bacterial surface resulting in reduced opsonization and phagocytosis of the bacteria.
Modulation of Complement Components Involved in Membrane Attack Complex (MAC) Formation, Neutrophil Chemotaxis and Neutrophil Phagocytosis
Terminal molecules involved in the complement cascades are crucial for pathogen killing as they play a crucial role in the construction of MAC, the cytotoxic molecular assembly. MAC formation occurs with the association of C5b6-8 and multiple copies of C9. C5 cleavage by the C5 convertase generates C5b and C5a of which C5b initiates MAC formation and C5a is a potent anaphylatoxin that recruits immune cells at the site of infection. Staphylococcus aureus secretes the staphylococcal super antigen like-7 protein (SSL-7) [60], which binds to C5 and inhibits its interaction with C5 convertase, preventing the initiation of MAC formation. In addition, SSL-7 binds with IgA1 and IgA2 blocking their interaction with FcαR1 and resulting in reduced phagocytosis [66].
Group A streptococci express SIC (streptococcal inhibitor of complement), which binds to soluble C5b-7 complex and prevents its insertion in the cell membrane [68]. An important human pathogen, Helicobacter pylori prevennts MAC formation by binding to the host membrane associated regulator CD59 (protectin) [70]. Leptospiral surface protein LcpA binds with the serum component vitronectin and inhibits the polymerization of C9 that blocks MAC formation [76, 77]. Borrelia burgdorferi expresses two terminal complement inhibitors, CspA and CD59 like proteins. CspA binds simultaneously to C7 and C9 that inhibits ZnCl2 induced C9 polymerization [67, 69]. CspA also binds to FH and is known as the complement regulator acquiring surface protein-1 (CRASP-1) [69, 122, 123]. CspA mutants that lack FH binding activity but retain its C7 and C9 binding activity were unable to protect bacteria from lysis [124]. These findings demonstrate that the inhibition of MAC formation by CspA without FH binding is insufficient in imparting protection to the bacteria. The BGA66 and BGA71 proteins of Borrelia bavariensis interacts with terminal complement components C7, C8, and C9, and inhibit the formation of MAC [75].
Certain pathogens have devised strategies to inhibit neutrophil migration to the site of infection. The S, aureus chemotaxis inhibitory protein (CHIPS) [71] binds with the C5a receptor expressed on neutrophils, reducing neutrophil chemotaxis and helping in the establishment of the infection. The Streptococcus pyogenes glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) also performs moonlighting functions of immune evasion. Bacterial surface GAPDH binds to C5a and inhibits neutrophil recruitment [72]. C5a peptidase (SepA) of S. pyogenes cleaves C5a from C-terminus and prevents chemotaxis of phagocytes [35].
In a recent finding it has been demonstrated that the polysaccharide capsule and exosporium of pediatric pathogen Kingella kingae inhibits the oxidative burst response of neutrophils and also hinders the neutrophil association to the bacteria [74]. The earlier described extracellular complement binding protein (Ecb) of Staphylococcus aureus also inhibits the interaction of C3b with complement receptor 1 (CR1) present on neutrophils. Blocking of C3b–CR1 interaction results in reduced phagocytosis and proteolytic breakdown of C3b [73].
The various complement evasive strategies adopted by the human bacterial pathogens have been summarized in Table 1.
Perspective
The co-existence of humans and pathogens have led to the evolution of a coordinated balance between host immunity and the evasion strategies of the pathogens. Apart from cell adhesion and colonization, evasion of host immunity is another crucial factor for the establishment of infection inside the host. The diverse complement evasive strategies discussed above demonstrates the significance of complement system in elimination of pathogen from the host. Recent findings have elucidated the varied biochemical and structural mechanisms of complement evasion by human pathogens. However, the precise molecular mechanisms that mediate the complement and immune evasion strategies adopted by the many bacterial pathogens still remains a gap in our understanding that warrants further investigations. For example, Bacillus anthracis and Streptococcus pneumoniae have outer capsules composed of poly-γ-d-glutamate and polysaccharides, respectively, that have been reported to evade complement activation. However, the mechanism underlying this evasive strategy that would explain the lack of complement protein binding on the outer capsule still remains unknown.
A number of anti-complement therapeutics are available or in clinical development, some of which are listed below based on their complement target: (1) C1r/s; MASPs—Cinryze, Berinert, Ruconest, Sutimlimab; (2) Factor D—Danicopan, ACH-5228, ACH-5448; (3) Factor B—IONIS-FB-LRX, LNP023, (4) C3—APL-2, APL-9, AMY-101; (5) C5—Eculizumab, Tesidolumab, Pozelimab, Ravulizuma [125, 126].
In order to develop better intervention strategies against pathogenic infections, it is crucial to understand their immune evasive mechanisms. Certain bacterial complement evasive molecules have also been targeted to facilitate complement-mediated clearance of pathogens [127], which may lead to the development of potential therapeutics or prophylactics. Many pathogens bind with the serum Factor H that inhibits the activation of the alternative pathway. A fusion protein comprising of the cell binding domains of FH and the human IgG Fc region (FH6,7/HuFc) has been reported to induce complement dependent killing in vitro and bacterial clearance in animal models [128–130]. In a pro-vaccine approach based on a bacterial immune evasion protein, it has been demonstrated that the Staphylococcal Sbi protein activated the alternative complement pathway by the recruitment of complement regulators and forming a tripartite complex (Sbi:C3d:FHR-1), Thus, Sbi increases the binding of FHRs with C3b and in the process inhibits FH activity resulting in enhanced complement activation [131]. Further it was reported that a chimera of Sbi, fused with the M. tuberculosis antigen Ag85b, produced effective C3 mediated opsonisation and induced a significantly higher immune response than that observed with the Ag85b alone, thus establishing a proof of principle for a novel vaccine strategy [131]. The Bordetella pertussis autotransporter, Vag8 protein has been reported to inhibit complement activation by recruiting the serum regulator protein C1-INH. Mice immunization with Vag protein has been shown to induce anti-Vag8 antibodies that block the recruitment of C1-INH on the bacterial surface and in turn promoting complement activation that led to a reduction in the B. pertussis load from the mice lungs [104, 132]. Immune responses generated against certain pathogenic molecules involved in immune evasion through vaccine administration have shown promising results in eliciting protection against infections [84, 104].
The complement system has a critical role in innate immunity as the first line of defense against infectious pathogens. However, it also contributes in aggravating disease pathology and thus a fine balance has to be maintained in targeting the complement system. As anti-complement therapy may compromise the ability of opsonization and phagocytosis [133] increase the levels of complement proteins in the human blood stream and even lead to tissue damage [126]. Hence, innovative new generation anti-complement therapeutics are under development to overcome these challenges and improve upon efficacy.
However, the major limitation in developing therapeutics for pathogenic clearance is that certain pathogens have developed multiple complement evasive strategies such as Staphylococcus aureus [38, 134, 135]. Recent findings have also demonstrated that the Staphylococcus aureus can target broad range of hosts by acquiring some genome modification [91]. In a recent study, we have also demonstrated that the negatively charged poly-γ-d-glutamate capsule of Bacillus anthracis inhibits opsonic phagocytosis by impeding complement activation through multiple mechanisms [136]. It would be very challenging to develop a single magic bullet that as a therapeutic can counter all the multiple complement evasion mechanisms of all bacterial pathogens. Nevertheless, the disruption of complement evasion mechanisms is an attractive approach for the development of novel, specific intervention strategies against virulent bacterial pathogens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
The authors would like to acknowledge the funding support of the Department of Biotechnology (BSL-3 facility), Department of Science & Technology (FIST & PURSE) and University Grant Commission (UPE II) to RB and DG.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 2.Killick J, Morisse G, Sieger D, Astier AL. Complement as a regulator of adaptive immunity. Semin Immunopathol. 2018;40:37–48. doi: 10.1007/s00281-017-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricklin D, Reis ES, Dimitrios C, Gros MP, Lambris JD. Complement component C3—the “Swiss Army Knife” of innate immunity and host defence. Immunol Rev. 2016;274:358. doi: 10.1111/imr.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harboe M, Ulvund G, Vien L, Fung M, Mollnes TE. The quantitative role of alternative pathway amplification in classical pathway induced terminal complement activation. Clin Exp Immunol. 2004;138:439–446. doi: 10.1111/j.1365-2249.2004.02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement—a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo RF, Ward PA. Role of C5a in inflammatory responses. Annu Rev Immunol. 2005;23:821–852. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- 8.Serna M, Giles JL, Morgan BP, Bubeck D. Structural basis of complement membrane attack complex formation. Nat Commun. 2016;7:10587. doi: 10.1038/ncomms10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida N, Walz T, Springer TA. Structural transition of complement component C3 and its activation products. Proc Natl Acad Sci USA. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bajic G, Yatime L, Sim RB, Vorup-Jensen T, Anderson GR. Structural insight on the recognition of surface bound opsonin by integrin I domain of complement receptor 3. Proc Natl Acad Sci USA. 2013;110:16426–16431. doi: 10.1073/pnas.1311261110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Giesbrecht BV, Lambis JD. Cutting edge: members of the Staphylococcus aureus extracellular fibrinogen-binding protein family inhibit the interaction of C3d with complement receptor 2. J Immunol. 2008;181:7463–7467. doi: 10.4049/jimmunol.181.11.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nangaku M. Complement regulatory proteins: are they important in disease? J Am Soc Nephrol. 2003;14:2411–2413. doi: 10.1097/01.ASN.0000088010.15313.A1. [DOI] [PubMed] [Google Scholar]
- 13.Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol. 2013;33:479–492. doi: 10.1016/j.semnephrol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ermert D, Blom AM. C4b-binding protein: the good, the bad and the deadly. Novel functions of an old friend. Immunol Lett. 2016;169:82–92. doi: 10.1016/j.imlet.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira VP, Pangburn MK, Cortes C. Complement control protein factor H: the good, the bad, and the inadequate. Mol Immunol. 2010;47:2187–2197. doi: 10.1016/j.molimm.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis AE, Lu F, Mejia P. C1 inhibitor, a multi-functional serine protease inhibitor. Thromb Haemost. 2010;104:886–893. doi: 10.1160/TH10-01-0073. [DOI] [PubMed] [Google Scholar]
- 17.Ghannam A, Fauquert JL, Thomas C, Kemper C, Drouet C. Human complement C3 deficiency: Th1 induction requires T-cell-derived complement C3a and CD46 activation. Mol Immunol. 2014;58:98–107. doi: 10.1016/j.molimm.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Ermerta D, Ram S, Laabei M. The hijackers guide to escaping complement: lessons learned from pathogens. Mol Immunol. 2019;114:49–61. doi: 10.1016/j.molimm.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Hovingh ES, Van den Broek B, Kuipers B, Pinelli E, Rooijakkers SHM, Jongerius I. Acquisition of C1 inhibitor by Bordetella pertussis virulence associated gene 8 results in C2 and C4 consumption away from the bacterial surface. PLoS Pathog. 2017;13:e1006531. doi: 10.1371/journal.ppat.1006531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haleem KS, Ali YM, Yesilkaya H, Kohler T, Hammerschmidt S, Andrew PW, Schwaeble WJ, Lynch NJ. The pneumococcal surface proteins PspA and PspC sequester host C4-binding protein to inactivate complement C4b on the bacterial surface. Infect Immun. 2018;19:87. doi: 10.1128/IAI.00742-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarva H, Ram S, Vogel U, Blom AM, Meri S. Binding of the complement inhibitor C4bp to serogroup B Neisseria meningitidis. J Immunol. 2005;174:6299–6307. doi: 10.4049/jimmunol.174.10.6299. [DOI] [PubMed] [Google Scholar]
- 22.Ermert D, Weckel A, Agarwal V, Frick IM, Bjorck L, Blom AM. Binding of complement inhibitor C4b-binding protein to a highly virulent Streptococcus pyogenes M1 strain is mediated by protein H and enhances adhesion to and invasion of endothelial cells. J Biol Chem. 2013;288:32172–32183. doi: 10.1074/jbc.M113.502955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh P. The nonideal coiled coil of m protein and its multifarious functions in pathogenesis. In: Linke D, Goldman A, editors. Bacterial adhesion. Advances in experimental medicine and biology. Dordrecht: Springer; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno-Torres A, Malvido-Jiménez IR, de la Peña-Moctezuma A, Castillo Sánchez LO, Fraga TR, Barbosa AS, Isaac L, Sahagún-Ruiz A. Culture-attenuated pathogenic Leptospira lose the ability to survive to complement-mediated-killing due to lower expression of factor H binding proteins. Microbes Infect. 2019;21:377–385. doi: 10.1016/j.micinf.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Schneider MC, Prosser BE, Caesar JJ. Neisseria meningitidis recruits factor H using protein mimicry of the host carbohydrates. Nature. 2009;458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tommassen J, Arenas J. Biological functions of the Secretome of Neisseria meningitidis. Front Cell Infect Microbiol. 2017;16:256. doi: 10.3389/fcimb.2017.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Röttgerding F, Wagemakers A, Koetsveld J, Fingerle V, Kirschfink M, Hovius JW, Zipfel PF, Wallich R, Kraiczy P. Immune evasion of Borrelia miyamotoi: CbiA, a novel outer surface protein exhibiting complement binding and inactivating properties. Sci Rep. 2017;9:7056. doi: 10.1038/s41598-017-00412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcinkiewicz AL, Dupuis AP, Zamba-Campero M, Nowak N, Kraiczy P, Ram S, Kramer LD, Lin YP. Blood treatment of Lyme borrelia demonstrates the mechanism of CspZ-mediated complement evasion to promote systemic infection in vertebrate hosts. Cell Microbiol. 2019;21:e12998. doi: 10.1111/cmi.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moulin P, Rong V, Ribeiro E, Silva A, Pederick VG, Camiade E, Mereghetti L, McDevitt CA, Hiron A. Defining the role of the Streptococcus agalactiae Sht-Family proteins in zinc acquisition and complement evasion. J Bacteriol. 2019;1:1. doi: 10.1128/JB.00757-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garza DA, Riley SP, Martinez JJ. Expression of Rickettsia Adr2 protein in E. coli is sufficient to promote resistance to complement-mediated killing, but not adherence to mammalian cells. PLoS ONE. 2017;12:e0179544. doi: 10.1371/journal.pone.0179544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herr AB, Thorman AW. Hiding in plain sight: immune evasion by the Staphylococcal protein SdrE. Biochem J. 2017;474:1803–1806. doi: 10.1042/BCJ20170132. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Wu M, Hang T, Wang C, Yang Y, Pan W, Zang J, Zhang M, Zhang X. Staphylococcus aureus SdrE captures complement factor H’s c-terminus via a novel ‘close, dock, lock and latch’ mechanism for complement evasion. Biochem J. 2017;474:1619–1631. doi: 10.1042/BCJ20170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallström T, Mörgelin M, Barthel D, Raguse M, Kunert A, Hoffmann R, Skerka C, Zipfel PF. Dihydrolipoamide dehydrogenase of Pseudomonas aeruginosa is a surface-exposed immune evasion protein that binds three members of the factor H family and plasminogen. J Immunol. 2012;189:4939–4950. doi: 10.4049/jimmunol.1200386. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Jenkins SA, Gu C, Shree A. Bacillus anthracis spore surface protein BclA mediates complement factor H binding to spores and promotes spore persistence. PLoS Pathog. 2016;12:e1005678. doi: 10.1371/journal.ppat.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laabei M, Ermert D. Catch me if you can: Streptococcus pyogenes complement evasion strategies. J Innate Immun. 2019;11:3–12. doi: 10.1159/000492944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rooijakkers SH, vanWamel WJB, Ruyken M, van Kessel KPM, van Strijp JA. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005;7:476–484. doi: 10.1016/j.micinf.2004.12.01. [DOI] [PubMed] [Google Scholar]
- 37.Falugi F, Kim HK, Missiakas DM, Schneewind O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio. 2013;4:e00575-13. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Marichannegowda MH, Rakesh KP, Qin HL. Master mechanisms of Staphylococcus aureus: consider its excellent protective mechanisms hindering vaccine development. Microbiol Res. 2018;212–213:59–66. doi: 10.1016/j.micres.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Liang W, Fan H, Yang J, Yang G, Wang X, Chen L, Liang T. Immunological characterization and verification of recombinant streptococcal protein G. Mol Med Rep. 2015;12:6311–6315. doi: 10.3892/mmr.2015.4162. [DOI] [PubMed] [Google Scholar]
- 40.Smith EJ, Visai L, Kerrigan SW, Speziale P, Foster TJ. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun. 2011;79:3801–3809. doi: 10.1128/IAI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pawel-Rammingen UV, Bjorck L. IdeS and SpeB: immunoglobulin-degrading cysteine proteinases of Streptococcus pyogenes. Curr Opin Microbiol. 2003;6:50–55. doi: 10.1016/s1369-5274(03)00003-1. [DOI] [PubMed] [Google Scholar]
- 42.Honda-Ogawa M, Sumitomo T, Mori Y, Hamd DT, Ogawa T, Yamaguchi M, Nakata M, Kawabata S. Streptococcus pyogenes endopeptidase O contributes to evasion from complement-mediated bacteriolysis via binding to human complement Factor C1q. J Biol Chem. 2017;292:4244–4254. doi: 10.1074/jbc.M116.749275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiyoshi H, Wangdi T, Lock G, Saechao C, Raffatellu M, Cobb BA, Bäumler AJ. Mechanisms to evade the phagocyte respiratory burst arose by convergent evolution in typhoidal Salmonella serovars. Cell Rep. 2018;22:1787–1797. doi: 10.1016/j.celrep.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rungelrath V, Weiße C, Schütze N, Müller U, Meurer M, Rohde M, Seele J, Valentin-Weigand P, Kirschfink M, Beineke A, Schrödl W, Bergmann R, Baums CG. IgM cleavage by Streptococcus suis reduces IgM bound to the bacterial surface and is a novel complement evasion mechanism. Virulence. 2018;9:1314–1337. doi: 10.1080/21505594.2018.1496778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179:2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 46.Honda-Ogawa M, Ogawa T, Terao Y, Sumitomo T, Nakata M, Ikebe K, Maeda Y, Kawabata S. Cysteine proteinase from Streptococcus pyogenes enables evasion of innate immunity via degradation of complement factors. J Biol Chem. 2013;288:15854–15864. doi: 10.1074/jbc.M113.469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jongerius I, Kohl J, Pandey MK, Ruyken M, van Kessel KP, van Strijp JA, Rooijakkers SH. Staphylococcal complement evasion by various convertase-blocking molecules. J Exp Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planket KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jongerius I, Garcia BL, Geisbrecht BV, van Strijp JA. Convertase inhibitory properties of Staphylococcal extracellular complement-binding protein. J Biol Chem. 2010;285:14973–14979. doi: 10.1074/jbc.M109.091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. 2015;13:529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pietrocola G, Rindi S, Rosini R, Buccato S, Speziale P, Margarit I. The Group B Streptococcus–secreted protein CIP Interacts with C4, preventing C3b deposition via the lectin and classical complement pathways. J Immunol. 2016;196:385–394. doi: 10.4049/jimmunol.1501954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie J, Zhi H, Garrigues RJ, Keightley A, Garcia BL, Skare JT. Structural determination of the complement inhibitory domain of Borrelia burgdorferi BBK32 provides insight into classical pathway complement evasion by Lyme disease spirochetes. PLoS Pathog. 2019;15:e1007659. doi: 10.1371/journal.ppat.1007659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locke JW. Complement evasion in Borrelia spirochetes: mechanisms and opportunities for intervention. Antibiotics (Basel) 2019 doi: 10.3390/antibiotics8020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng S, Xu T, Fang Q, Yu L, Zhu J, Chen L, Liu J, Zhou R. The surface-exposed protein SntA contributes to complement evasion in zoonotic Streptococcus suis. Front Immunol. 2018;9:1063. doi: 10.3389/fimmu.2018.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang M, Ko YP, Liang X, Ross CL, Liu Q, Murray BE, Höök M. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of gram-positive bacteria inhibit complement activation via the classical pathway. J Biol Chem. 2013;288:20520–20531. doi: 10.1074/jbc.M113.454462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cleary PP, Prahbu U, Dale JB, Wexler DE, Handley J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect Immun. 1992;60:5219–5223. doi: 10.1128/IAI.60.12.5219-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ayón-Núñez DA, Fragoso G, Bobes RJ, Laclette JP. Plasminogen-binding proteins as an evasion mechanism of the host’s innate immunity in infectious diseases. Biosci Rep. 2018 doi: 10.1042/BSR20180705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blom AM, Bergmann S, Fulde M, Riesbeck K, Agarwal V. Streptococcus pneumoniae phosphoglycerate kinase is a novel complement inhibitor affecting the membrane attack complex formation. J Biol Chem. 2014;289:32499–32511. doi: 10.1074/jbc.M114.610212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kochi LT, Fernandes LGV, Souza GO, Vasconcellos SA, Heinemann MB, Romero EC, Kirchgatter K, Nascimento ALTO. The interaction of two novel putative proteins of Leptospira interrogans with E-cadherin, plasminogen and complement components with potential role in bacterial infection. Virulence. 2019;10:734–753. doi: 10.1080/21505594.2019.1650613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bestebroer J, Aerts PC, Rooijakkers SH, Pandey MK, Kohl J, van Strijp JA, de Haas CJ. Functional basis for complement evasion by staphylococcal superantigen-like 7. Cell Microbiol. 2010;12:1506–1516. doi: 10.1111/j.1462-5822.2010.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong YQ, Ghebrehiwet B. Effect of Pseudomonas aeruginosa elastase and alkaline protease on serum complement and isolated components C1q and C3. Clin Immunol Immunopathol. 1992;62:133–138. doi: 10.1016/0090-1229(92)90065-v. [DOI] [PubMed] [Google Scholar]
- 62.Rooijakkers SH, Van Strijp JA. Bacterial complement evasion. Mol Immunol. 2007;44:23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Abreu AG, Barbosa AS. How Escherichia coli circumvent complement-mediated killing. Front Immunol. 2017;8:452. doi: 10.3389/fimmu.2017.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jusko M, Potempa J, Mizgalska D, Bielecka E, Ksiazek M, Riesbeck K, Garred P, Eick S, Blom AM. A metalloproteinase Mirolysin of Tannerella forsythia inhibits all pathways of the complement system. J Immunol. 2015;195:2231–2240. doi: 10.4049/jimmunol.1402892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, van Strijp JA, Rooijakkers SH. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol. 2012;188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 66.Langley R, Wines B, Willoughby N, Basu I, Proft T, Fraser JD. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J Immunol. 2005;174:2926–2933. doi: 10.4049/jimmunol.174.5.2926. [DOI] [PubMed] [Google Scholar]
- 67.Hallstrom T, Siegel C, Morgelin M, Kraiczy P, Skerka C, Zipfel PF. CspA from Borrelia burgdorferi inhibits the terminal complement pathway. mBio. 2013 doi: 10.1128/mBio.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akesson P, Sjoholm AG, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 69.Marcinkiewicz AL, Kraiczy P, Lin YP. There is a method to the madness: strategies to study host complement evasion by Lyme disease and relapsing fever Spirochetes. Front Microbiol. 2017;8:328. doi: 10.3389/fmicb.2017.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rautemaa R, Rautelin H, Puolakkainen HP, Kokkola A, Karkkainen P, Meri S. Survival of Helicobacter pylori from complement lysis by binding of GPI-anchored Protectin (CD59) Gastroenterology. 2001;120:470–479. doi: 10.1053/gast.2001.21197. [DOI] [PubMed] [Google Scholar]
- 71.de Haas CJ, Veldkamp KE, Peschel A, Weerkamp F, Van Wamel WJ, Heezius EC, Poppelier MJ, Van Kessel KP, van Strijp JA. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial anti-inflammatory agent. J Exp Med. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terao Y, Yamaguchi M, Hamada S, Kawabata S. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J Biol Chem. 2006;281:14215–14223. doi: 10.1074/jbc.M513408200. [DOI] [PubMed] [Google Scholar]
- 73.Amdahl H, Haapasalo K, Tan L, Meri T, Kuusela PI, van Strijp JA, Rooijakkers S, Jokiranta TS. Staphylococcal protein Ecb impairs complement receptor-1 mediated recognition of opsonized bacteria. PLoS ONE. 2017;12:e0172675. doi: 10.1371/journal.pone.0172675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muñoz VL, Porsch EA, St Geme JW. Kingella kingae surface polysaccharides promote resistance to neutrophil phagocytosis and killing. mBio. 2019;25:10. doi: 10.1128/mBio.00631-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brangulis K, Akopjana I, Petrovskis I, Kazaks A, Kraiczy P, Tars K. Crystal structure of the membrane attack complex assembly inhibitor BGA71 from the Lyme disease agent Borrelia bavariensis. Sci Rep. 2018;8:11286. doi: 10.1038/s41598-018-29651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.da Silva LB, Miragaia Ldos S, Breda LC, Abe CM, Schmidt MC, Moro AM, Monaris D, Conde JN, Józsi M, Isaac L, Abreu PA, Barbosa AS. Pathogenic Leptospira species acquire factor H and vitronectin via the surface protein LcpA. Infect Immun. 2015;83:888–897. doi: 10.1128/IAI.02844-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fraga TR, Isaac L, Barbosa AS. Complement evasion by pathogenic Leptospira. Front Immunol. 2016;7:21. doi: 10.3389/fimmu.2016.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kraiczy P, Würzner R. Complement escape of human pathogenic bacteria by acquisition of complement regulators. Mol Immunol. 2006;43:31–44. doi: 10.1016/j.molimm.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 79.Meri T, Blom AM, Hartmann A, Lenk D, Meri S, Zipfel PF. The hyphal and yeast forms of Candida albicans bind the complement regulator C4b binding protein. Infect Immun. 2004;72:6633–6641. doi: 10.1128/IAI.72.11.6633-6641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernet J, Mullick J, Singh AK, Sahu A. Viral mimicry of the complement system. J Biosci. 2003;28:249–264. doi: 10.1007/BF02970145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shao S, Sun X, Chen Y, Zhan B, Zhu X. Complement evasion: an effective strategy that parasites utilize to survive in the host. Front Microbiol. 2019;10:532. doi: 10.3389/fmicb.2019.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meri T, Amdahl H, Lehtinen MJ, Hyvärinen S, McDowell JV, Bhattacharjee A, Meri S, Marconi R, Goldman A, Jokiranta TS. Microbes bind complement inhibitor factor H via a Common site. PLoS Pathog. 2013;9:e1003308. doi: 10.1371/journal.ppat.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madico G, Ngampasutadol J, Gulati S, Vogel U. Factor H binding and function in sialylated pathogenic neisseriae is influenced by gonococcal, but not meningococcal, porin. J Immunol. 2007;178:4489–4497. doi: 10.4049/jimmunol.178.7.4489. [DOI] [PubMed] [Google Scholar]
- 84.Seib KL, Scarselli M, Comanducci M, Toneatto D, Masignani V. Neisseria meningitidis factor H binding protein fHbp: a key virulence factor and vaccine antigen. Expert Rev Vaccines. 2015;14:841–859. doi: 10.1586/14760584.2015.1016915. [DOI] [PubMed] [Google Scholar]
- 85.Walter L, Sürth V, Röttgerding F, Zipfel PF, Fritz-Wolf K, Kraiczy P. Elucidating the immune evasion mechanisms of Borrelia mayonii, the causative agent of Lyme disease. Front Immunol. 2019;26:2722. doi: 10.3389/fimmu.2019.02722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tegels BK, Oliver LD, Jr, Miller DP, Marconi RT. Plasminogen binding and degradation by Treponema denticola: identification of the plasminogen binding interface on the FhbB protein. Mol Oral Microbiol. 2018;33:249–256. doi: 10.1111/omi.12221. [DOI] [PubMed] [Google Scholar]
- 87.Barbosa AS, Isaac L. Complement immune evasion by spirochetes. Curr Top Microbiol Immunol. 2018;415:215–238. doi: 10.1007/82_2017_47. [DOI] [PubMed] [Google Scholar]
- 88.Xia X, Qin W, Zhu H, Wang X, Jiang J, Hu J. How Streptococcus suis serotype 2 attempts to avoid attack by host immune defenses. J Microbiol Immunol Infect. 2019;52:516–525. doi: 10.1016/j.jmii.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 89.Herbert AP, Makou E, Chen ZA, Kerr H. Complement evasion mediated by enhancement of captured factor H: implications for protection of self-surfaces from complement. J Immunol. 2015;195:4986–4998. doi: 10.4049/jimmunol.1501388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pathak A, Bergstrand J, Sender V, Spelmink L, Aschtgen MS, Muschiol S, Widengren J, Henriques-Normark B. Factor H binding proteins protect division septa on encapsulated Streptococcus pneumoniae against complement C3b deposition and amplification. Nat Commun. 2018;9:3398. doi: 10.1038/s41467-018-05494-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dave S, Carmicle S, Hammerschmidt S, Pangburn MK, McDaniel LS. Dual roles of PspC, a surface protein of Streptococcus pneumoniae, in binding human secretory IgA and factor H. J Immunol. 2004;173:471–477. doi: 10.4049/jimmunol.173.1.471. [DOI] [PubMed] [Google Scholar]
- 92.Jozsi M, Tortajada A, Uzonyi B, Goicoechea de Jorge E, Rodriguez de Cardoba S. Factor H-related proteins determine complement-activating surfaces. Trends Immunol. 2015;36:374–384. doi: 10.1016/j.it.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 93.Hebecker M, Jozsi M. Factor H-related protein 4 activates complement by serving as a platform for the assembly of alternative pathway C3 convertase via its interaction with C3b protein. J Biol Chem. 2012;287:19528–19536. doi: 10.1074/jbc.M112.364471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caesar JJ, Lavender H, Ward PN, Exley RM, Eaton J, Chittock E, Malik TH, Goicoechea de Jorge E, Pickering MC, Tang CM, Lea SM. Competition between antagonistic complement factors for a single protein on N. meningitidis rules disease susceptibility. eLife. 2014;3:e04008. doi: 10.7554/eLife.04008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abdul-Aziz M, Tsolaki AG, Kouser L, Carroll MV, Al-Ahdal MN, Sim RB, Kishore U. Complement factor H interferes with Mycobacterium bovis BCG entry into macrophages and modulates the pro-inflammatory cytokine response. Immunobiology. 2016;221:944–952. doi: 10.1016/j.imbio.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 96.Jarva H, Ram S, Vogel U, Blom AM, Meri S. Binding of the complement inhibitor C4bp to serogroup B Neisseria meningitidis. J Immunol. 2018;174:6299–6307. doi: 10.4049/jimmunol.174.10.6299. [DOI] [PubMed] [Google Scholar]
- 97.Blom AM, Zadura AF, Villoutreix BO, Dahlback B. Positively charged amino acids at the interface between α-chain CCP-1 and CCP-2 of C4BP are required for regulation of Classical C3 convertase. Mol Immunol. 1999;37:445–453. doi: 10.1016/S0161-5890(00)00059-6. [DOI] [PubMed] [Google Scholar]
- 98.Persson J, Beall B, Linse S, Lindahl G. Extreme sequence divergence but conserved ligand-binding specificity in Streptococcus pyogenes M protein. PLoS Pathog. 2006;2:e47. doi: 10.1371/journal.ppat.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blom AM, Berggard K, Webb JH, Lindahl G, Villoutreix BO, Dahlback B. Human C4b-binding protein has overlapping, but not identical, binding sites for C4b and streptococcal M proteins. J Immunol. 2000;164:5328–5336. doi: 10.4049/jimmunol.164.10.5328. [DOI] [PubMed] [Google Scholar]
- 100.Akesson P, Cooney J, Kishimoto F, Bjorck L. Protein H—a novel IgG binding bacterial protein. Mol Immunol. 1990;27:523–531. doi: 10.1016/0161-5890(90)90071-7. [DOI] [PubMed] [Google Scholar]
- 101.Ermert D, Laabei M, Weckel A, Mörgelin M, Lundqvist M, Björck L, Ram S, Linse S, Blom AM. The molecular basis of human IgG-mediated enhancement of C4b-binding protein recruitment to Group A Streptococcus. Front Immunol. 2019;10:1230. doi: 10.3389/fimmu.2019.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ermert D, Weckel A, Magda M, Mörgelin M, Shaughnessy J, Rice PA, Björck L, Ram S, Blom AM. Human IgG increases virulence of Streptococcus pyogenes through complement evasion. J Immunol. 2018;200:3495–3505. doi: 10.4049/jimmunol.1800090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Agarwal V, Hammerschmidt S, Malm S, Bergmann S, Riesback K, Blom AM. Enolase of Streptococcus pneumoniae binds human complement inhibitor C4b binding protein and contributes to complement evasion. J Immunol. 2012;189:3575–3584. doi: 10.4049/jimmunol.1102934. [DOI] [PubMed] [Google Scholar]
- 104.de Gouw D, de Jonge MI, Hermans PWM, Wessels H, Zomer A, Berends A, Pratt C, Berbers GA, Mooi FA, Diavatopoulos DA. Proteomics-identified Bvg-activated autotransporters protect against Bordetella pertussis in a mouse model. PLoS ONE. 2014;9:e105011. doi: 10.1371/journal.pone.0105011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Diebolder CA, Beurskens FJ, de Jong RN, Koning RI, Strumane K, Lindorfer MA, Voorhorst M. Complement is activated by IgG hexamers assembled at the cell surface. Science. 2014;343:1260–1263. doi: 10.1126/science.1248943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DeDent AC, McAdow M, Schneewind O. Distribution of protein A on the surface of Staphylococcus aureus. J Bacteriol. 2007;189:4473–4484. doi: 10.1128/JB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 108.Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S. Anthrax pathogenesis. Annu Rev Microbiol. 2015;69:185–208. doi: 10.1146/annurev-micro-091014-104523. [DOI] [PubMed] [Google Scholar]
- 109.Hovingh ES, van den Broek B, Jongerius I. Hijacking complement regulatory proteins for bacterial immune evasion. Front Microbiol. 2016;7:2004. doi: 10.3389/fmicb.2016.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Agarwal S, Vasudhev S, DeOliveira RB, Ram S. Inhibition of the classical pathway of complement by meningococcal capsular polysaccharides. J Immunol. 2014;193:1855–1863. doi: 10.4049/jimmunol.1303177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sahly H, Keisari Y, Ofek I. Manno(rhamno)biose-containing capsular polysaccharides of Klebsiella pneumoniae enhance opsono-stimulation of human polymorphonuclear leukocytes. J Innate Immun. 2009;1:136–144. doi: 10.1159/000154812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Steimle A, Autenrieth IB, Frick JS. Structure and function: lipid A modifications in commensals and pathogens. Int J Med Microbiol. 2016;306:290–301. doi: 10.1016/j.ijmm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 113.O’Hara AM, Moran AP, Wurzner R, Orren A. Complement-mediated lipopolysaccharide release and outer membrane damage in Escherichia coli J5: requirement for C9. Immunology. 2001;102:365–372. doi: 10.1046/j.1365-2567.2001.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Doorduijn DJ, Rooijakkers SH, van Schaik W, Bardoel BW. Complement resistance mechanisms of Klebsiella pneumonias. Immunobiology. 2016;221:1102–1109. doi: 10.1016/j.imbio.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 115.Hartmann S, Hofsteenge J. Properdin, the positive regulator of complement, is highly C-Mannosylated. J Biol Chem. 2000;275:28569–28574. doi: 10.1074/jbc.M001732200. [DOI] [PubMed] [Google Scholar]
- 116.Kemper C, Hourcade DE. Properdin: new roles in pattern recognition and target clearance. Mol Immunol. 2008;45:4048–4056. doi: 10.1016/j.molimm.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Garcia BL, Zwarthoff SA, Rooijakkers SH, Geisbrecht BV. Novel evasion mechanisms of the classical complement pathway. J Immunol. 2016;197:2051–2060. doi: 10.4049/jimmunol.1600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laarman AJ, Ruyken M, Malone CL, van Strijp JA, Horswill AR, Rooijakkers SH. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol. 2011;186:6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 119.Lynskey NN, Reglinski M, Calay D, Siggins MK, Mason JC, Botto M, Sriskandan S. Multi-functional mechanisms of immune evasion by streptococcal complement inhibitor C5a peptidase. PLoS Pathog. 2017;13:e1006493. doi: 10.1371/journal.ppat.1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schenkein HA, Fletcher HM, Bodnar M, Macrina FL. Increased opsonization of a prtH-defective mutant of Porphyromonas gingivalis W83 is caused by reduced degradation of complement-derived opsonins. J Immunol. 1995;154:5331–5337. [PubMed] [Google Scholar]
- 121.Bhattacharya S, Ploplis VA, Castellino FJ. Bacterial plasminogen receptors utilize host plasminogen system for effective invasion and dissemination. J Biomed Biotechnol. 2012 doi: 10.1155/2012/482096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kraiczy P, Stevenson B. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: structure, function and regulation of gene expression. Ticks Tick Borne Dis. 2013;4:26–34. doi: 10.1016/j.ttbdis.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tracy KE, Baumgarth N. Borrelia burgdorferi manipulates innate and adaptive immunity to establish persistence in rodent reservoir hosts. Front Immunol. 2017;8:116. doi: 10.3389/fimmu.2017.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hart T, Nguyen NTT, Nowak NA, Zhang F, Linhardt RJ, Diuk-Wasser M, Ram S, Kraiczy P, Lin TP. Polymorphic factor H-binding activity of CspA protects Lyme borrelia from the host complement in feeding ticks to facilitate tick-to-host transmission. PLoS Pathog. 2018;14:e1007106. doi: 10.1371/journal.ppat.1007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mastellos DC, Ricklin D, Lambris JD. Clinical promise of next generation complement therapeutics. Nat Rev Drug Discov. 2019;18:707–729. doi: 10.1038/s41573-019-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zelek WM, Xie L, Morgan BP, Harris CL. Compendium of current complement therapeutics. Mol Immunol. 2019;114:341–352. doi: 10.1016/j.molimm.2019.07.030. [DOI] [PubMed] [Google Scholar]
- 127.Laarman A, Milder F, van Strijp J, Rooijakkers S. Complement inhibition by gram-positive pathogens: molecular mechanisms and therapeutic implications. J Mol Med (Berl) 2010;88:115–120. doi: 10.1007/s00109-009-0572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shaughnessy J, Vu DM, Punjabi R, Serra-Pladevall J, De Oliveira RB, Granoff DM, Ram S. Fusion protein comprising factor H domains 6 and 7 and human IgG1 Fc as an antibacterial immunotherapeutic. Clin Vaccine Immunol. 2014;21:1452–1459. doi: 10.1128/CVI.00444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wong SM, Shaughnessy J, Ram S, Akerley BJ. Defining the binding region in Factor H to develop a therapeutic Factor H-Fc fusion protein against non-typeable Haemophilus influenzae. Front Cell Infect Microbiol. 2016;6:40. doi: 10.3389/fcimb.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ram S, Shaughnessy J, DeOliveira RB, Lewis LA. Utilizing complement evasion strategies to design complement-based antibacterial immunotherapeutics: lessons from the pathogenic Neisseriae. Immunobiology. 2016;221:1110–1123. doi: 10.1016/j.imbio.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang YI, Back CR, Grawert MA, Wahld AA, Denton H, Kildani R, Paulin J, Worner K, Kaiser W, Svergun DI, Sartbaeva Watts AG, Marchbank KJ, van der Elsen JMH. Utilization of Staphylococcal immune evasion protein Sbi as a novel vaccine adjuvant. Front Immunol. 2019;9:3139. doi: 10.3389/fimmu.2018.03139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marr N, Shah NR, Lee R, Kim EJ, Fernandez RC. Bordetella pertussis autotransporter Vag8 binds human C1 esterase inhibitor and confers serum resistance. PLoS ONE. 2011;6:e20585. doi: 10.1371/journal.pone.0020585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Socié G, Caby-Tosi MP, Marantz JL, Cole A, Bedrosian AL, Gasteyger C, Mujeebuddin A, Hillmen P, Vande Walle J, Haller H. Eculizumab in paroxysmal nocturnal haemoglobinuria and atypical haemolytic uraemic syndrome: 10-year pharmacovigilance analysis. Br J Haematol. 2019;185:297–310. doi: 10.1111/bjh.15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pietrocola G, Nobile G, Rindi S, Speziale P. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front Cell Infect Microbiol. 2017;7:166. doi: 10.3389/fcimb.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim HK, Thammavongsa V, Schneewind O, Missiakas D. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr Opin Microbiol. 2012;15:92–99. doi: 10.1016/j.mib.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sharma S, Bhatnagar R, Gaur D. Bacillus anthracis poly-γ-d-glutamate capsule inhibits opsonic phagocytosis by impeding complement activation. Front Immunol. 2020;11:462. doi: 10.3389/fimmu.2020.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.