Abstract
The rise of single-cell transcriptomics has created an urgent need for similar approaches that use a minimal number of cells to quantify expression levels of proteins. We integrated and optimized multiple recent developments to establish a proteomics workflow to quantify proteins from as few as 1000 mammalian stem cells. The method uses chemical peptide labeling, does not require specific equipment other than cell lysis tools, and quantifies >2500 proteins with high reproducibility. We validated the method by comparing mouse embryonic stem cells and in vitro differentiated motor neurons. We identify differentially expressed proteins with small fold changes and a dynamic range in abundance similar to that of standard methods. Protein abundance measurements obtained with our protocol compared well to corresponding transcript abundance and to measurements using standard inputs. The protocol is also applicable to other systems, such as fluorescence-activated cell sorting (FACS)-purified cells from the tunicate Ciona. Therefore, we offer a straightforward and accurate method to acquire proteomics data from minimal input samples.
Introduction
Mass spectrometry is a highly sensitive technique capable of detecting peptides at the femtogram scale.1 However, to reach maximal protein identification and reproducibility, the quality and quantity of the sample material hold great importance. Therefore, standard proteomics analysis uses large numbers of cells, typically at 106–108 (ref (2)), to identify a few thousand proteins in a single-shot tandem mass spectrometry (MS/MS) experiment (Table 1). To increase the number of identified proteins, a typical approach involves even more material and extensive fractionation of the sample. However, numerous biological systems, including biopsies, tissue dissections, and other rare and precious samples, do not yield such quantities as they only provide hundreds to thousands of cells. To address this issue, recent efforts have conducted the first proteomics analyses of single cells.3−5 However, these approaches have challenges such as the need for special equipment or a low correlation between replicates.
Table 1. Overview of Requirements and Typical Results for Standard Proteomics Protocols, Single-Cell and Minimal Input Protocolsa.
| number of cells prepared (approx. amount of protein prepared) | approximate number of proteins identified (single-shot) | correlation with standard input sample (ρ) | correlation between replicates (ρ) | correlation between RNA abundance (ρ) | |
|---|---|---|---|---|---|
| standard | ∼106–8 (∼5–500 μg) | 2700 | 0.99 | 0.43 | |
| MiProt6 | n/a (25 μg) | 10 000b | 0.60 | 0.38 | |
| in-StageTip7 | n/a (20 μg) | 1435 | 0.97 | ||
| Myers et al.8 | n/a (2 μg) | 5100b | 0.90 | 0.25–0.50 | |
| Wang et al.9 | 5000 (∼500 ng) | 140 | |||
| Wang et al.10 | 500–5000 (∼50–500 ng) | 105–665 | |||
| SP311 | 1000 (n/a) | 1250 | 0.84–0.91 | ||
| NanoPOTS3 | 1–6 (0.2–1.2 ng) | 669–1153c | 0.95–0.97 | ||
| SCoPe-MS5 | 1 (n/a) | 900 | 0.62 | 0.2–0.4 | 0.25 |
| minimal input protocol | 1000 (∼200 ng) | 2500 | 0.81 | 0.99 | 0.39 |
Information on input amount, number of detected proteins, and correlations are taken from the original publications. Note that some preparations, e.g., the standard input protocol, do not use all of the prepared material for a single injection into the mass spectrometer. ρ—Spearman correlation coefficient.
Fractionated.
Using the MaxQuant match between runs algorithm.
Therefore, other research groups developed protocols to handle samples from more than one cell, but much fewer than the typical number of cells in standard preparations.12 These so-called “nanoproteomics” studies use either single cells or minimal amounts of samples from <5000 cells (providing ∼1 μg of protein).13 The number of cells needed depends on the cell type (size), lysis protocol, and measurement method, to provide enough material for quantitative analysis. For example, using 5000 human breast cancer cells, previous studies identified 105–665 proteins9,10 (Table 1).
Several nanoproteomics methods explored new technologies for minimal sample processing. For example, some custom-designed platforms enable high-resolution proteomics in tens of cells or even single cells.4,14,15 One of the most powerful of these systems is nanoPOTS,3,14,16 which is also used for imaging mass spectrometry.17 Other systems, such as CyTOF,18 CITE-seq.,19 and proximity ligation assays,20 employ antibodies against specific target proteins that create sensitive measurements but are limited to proteins with available reagents. Other methods rely on specialized materials such as collection microreactors8 or paramagnetic beads (SP3)11 to maximize yield from little starting material (Table 1). Microreactors are able to process 2 μg of cellular material and identify >5000 proteins. SP3 can handle samples from as few as 1000 cells; however, the authors indicate that this amount approached the limit of sensitivity and skewed the quantification.11 Further, the SP3 method requires specialized equipment, i.e., paramagnetic beads. Further, an in-StageTip method handles 1 μg of protein, which corresponds to roughly 5000 cells, identifying ∼1435 proteins in a single shot.7,21 Another method, called MiProt, uses much larger samples obtained from biopsies that provide up to 25 μg of protein but conducts both standard proteomic and phosphoproteomic analyses on these samples to identify ∼10 000 proteins in multiple fractions17,6 (Table 1).
Another recent nanoproteomics protocol called SCoPE-MS analyzes proteins from single, hand-picked mammalian cells via tandem mass tagging (TMT) coupled to conventional mass spectrometry5 (Table 1). In a standard 10plex TMT experiment, peptides from 10 different samples are labeled with sample-specific mass tags and then pooled. During the subsequent tandem mass spectrometry experiment, the tags are indistinguishable by mass at the first “precursor” level and therefore isolated together. In the subsequent second level of analysis, the peptides and tags fragment: the peptides can be sequenced and each tag’s channel is quantified through ion intensity measurements. In SCoPE-MS, one channel in the setup is dedicated to a “carrier” with peptides at high abundance that produce enough signal for reliable peptide identification. The remaining channels contain the experimental samples. Their peptides’ abundance is too low for identification, but the intensities of the mass tags are available for quantitation. The fundamental idea of the approach is therefore to separate peptide identification and quantitation. While a breakthrough, results from single cells struggle with proteome coverage, reproducibility, and correlation with corresponding transcript abundances (Table 1).
To provide a simple protocol with reasonable proteome coverage and reproducibility in cases where more than a single cell but not enough for a standard preparation is available, we integrated and optimized multiple steps in the proteomics workflow. Our minimal input method uses 1000 cells, does not require special equipment other than a sonicator for cell lysis, and quantifies >2500 proteins per sample (Table 1). It can detect twofold changes in expression levels with statistical significance. The protocol’s reproducibility and the correlations with transcript abundances are comparable with those of standard proteomics protocols (Table 1). We established and validated the method in a known system of mouse embryonic stem cells and in vitro differentiated motor neurons. We also demonstrated the method’s use in another organism, analyzing the cardiopharyngeal lineage in the tunicate model Ciona robusta.
Results
Simplified Protein Extraction to Maximize Sample Retention
To establish the protocol and assess its performance, we optimized the proteomics workflow at several steps and evaluated proteomic differences in an established in vitro differentiation paradigm comparing mouse embryonic stem cells (ESCs) and differentiated motor neurons (MNs) (Figure 1a).22 First, we tested different sonicators and buffers for cell lysis using 5000 mouse embryonic stem cells purified by fluorescence-activated cell sorting (FACS) and analyzed by label-free mass spectrometry (Figure S1). We compared the results to those from a standard input sample, which contained protein from ∼500 000 mammalian cells. The standard sample preparation included cell lysis with a phosphate-buffered saline without detergent, using the Bioruptor sonicator and cleanup of peptides with reverse-phase filters. For mass spectrometry analysis, we injected all of the peptides derived from the minimal input samples and 600 ng (from ∼60 000 cells) from the standard samples.
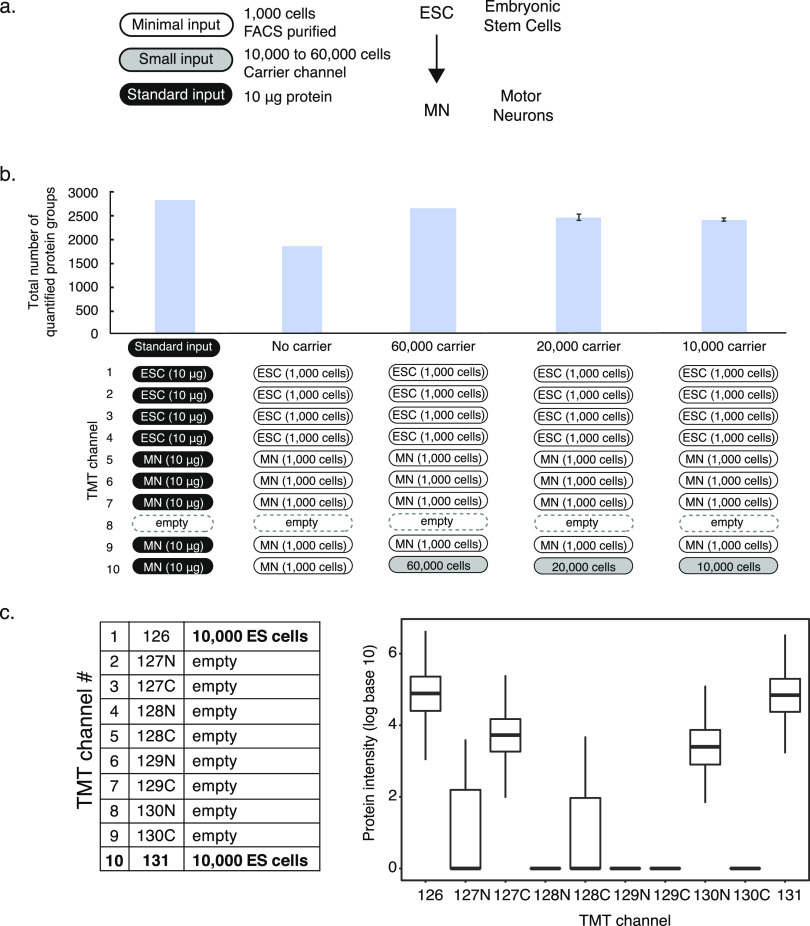
Figure 1.
Optimization of the minimal sample protocol. The panels describe testing of the need for a carrier channel, carrier channel input size, and position. (a) Panel shows an overview of cell numbers used for different purposes in the protocol and comparisons. (b) Comparison of TMT setups for using 1000 cells with and without carrier channel and using 60 000, 20 000, and 10 000 cells for the carrier channel. The graph shows the numbers of protein groups detected using the experimental setup below. The error bars show standard deviation. (c) Effects of carrier channel on neighboring channels at 1 Thompson distance using a 10plex TMT experiment with empty channels 2–8 and a sample from 10 000 cells in channels 1 and 10. ESC—embryonic stem cell, MN—motor neuron, FACS—fluorescence-activated cell sorting, and TMT—tandem mass tag.
As we identified a similar number of peptides and proteins detected in samples from either lysis buffer, we opted for the simpler one, which contained phosphate-buffered saline (PBS) only. In addition, we used the Bioruptor sonicator for the remainder of the experiments as it achieved the best protein identification in our hands. However, different samples or sonicator settings might alter this choice. We speculate that the two sonicators’ different working mechanisms may underlie the differential lysis efficiency: Covaris is a focused ultrasonicator, while Bioruptor uses homogenous energy distributing ultrasounds water bath, which might lead to the difference in results.
The results were reproducible with respect to both identified peptides and proteins (Figure S1). Technical replicates of protein and peptide abundances correlated better than abundances measured across different lysis methods (average R2 = 0.93, Figure S2). We observed higher variation in the number of proteins detected for certain conditions; however, this variance did not translate into protein quantification (Figure S2). For example, even though the number of proteins detected varied in the conditions where Bioruptor was used, the replicates showed a higher correlation in terms of protein quantification compared to conditions where Covaris sonicator was used. These results were similar in the Ciona samples discussed below. We also found that using samples as small as 1000 cells in label-free approaches yielded comparable protein identifications and reproducibility to those using 5000 cells, supporting the use of 5000 cells for optimization of cell lysis (Figure S3).
Experimental Design for Protein Quantitation with Peptide Mass Tags
Next, we optimized the design of the 10plex TMT labeling experiment including a carrier channel. We used a pool of equal proportions of both cell types analyzed in the experimental samples as a carrier. First, we showed that a setup with a carrier channel outperformed a setup without a carrier, i.e., with minimal input in all channels, with respect to the number of detected proteins (“no carrier,” Figure 1b, first three bars). Without a carrier channel, the amount (ng) of protein contributing to the peptide signal is derived from 9 × 1000 = 9000 cells, while for the setup with a carrier channel, the sample is derived from at least 10 000 + (8 × 1000) = 18 000 cells (or more if the carrier is larger). The results suggest that samples from a total of 9000 embryonic stem cells and motor neurons do not provide enough material for substantial peptide identification.
Second, we showed that the carrier should ideally be placed in channel 10 and channel 8 should be left empty. The reason for this setup lies in the carrier channel producing signals “spilling over” into channel 8, as shown in Figure 1c. In Figure 1d setup, all TMT channels are empty except for positions 1 and 10, both contain peptides prepared from 10 000 cells. The carrier in channel 1 produced signals in channels 2, 3, and 5. The carrier in channel 10 did not affect channel 9 immediately adjacent to it. However, we observed a substantial signal at the −1 Thompson distance to the carrier in channel 10, i.e., in channel 8, indicating contamination of the mass tag with the light isotope. Indeed, the observed intensity in channel 8 was about 3–4% of the total intensity in channel 10, consistent with the contamination with the light isotope as reported by the manufacturer. For this reason, we placed the carrier into channel 10 and left channel 8 empty in all subsequent TMT experiments. This setup should be robust even if the reported impurity of the channel 10 label differs.
Finally, we minimized the size of the carrier channel, as a low carrier-to-sample ratio is advantageous with respect to minimizing ratio compression and maximizing reproducibility (Figure S4). We tested carrier channels with peptides derived from 60 000, 20 000, and 10 000 cells. We found that the carrier from 10 000 cells provided similar protein identification and reproducibility to the larger carrier channels. We thus used this carrier size in subsequent experiments, resulting in a final carrier-to-individual sample ratio of 10 000:1000 = 10:1 (Figure 1b, last three bars). Leaving out the carrier channel completely did not improve observed ratios or fold changes further but substantially reduced the number of identified proteins (see above).
Validating Accuracy of Measured Protein Abundance
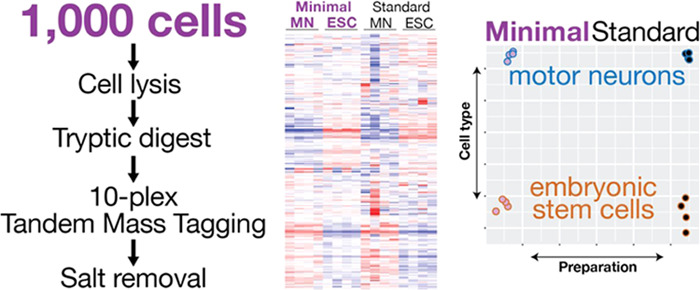
Figure 2a shows an overview of the final protocol that uses 1000 cells in experimental sample channels and 10 000 cells in the carrier channel. We evaluated this workflow with respect to coverage and reproducibility of the measured protein abundance. To do so, we used an established in vitro motor neuron differentiation protocol for mouse embryonic stem cells.22 While requiring only 1/50 of the number of cells, the minimal input protocol’s proteome coverage reached 88% of what we detected in single-shot standard proteome analysis, i.e., 2483 compared to 2828 proteins (Figure 1c). Replicate experiments in the minimal input setup correlated with an average of R2 = 0.99 in log–log abundance plots (Figures S5–S7), indicating high reproducibility. Both minimal and standard input preparations had similar TMT labeling efficiency (>97%). Further, both preparations had similar numbers of proteins identified by unique peptides and quantified by one peptide only (26 and 27% for minimal and standard input, respectively; Figure S8). The reproducibility of protein abundance measurements seemed very similar in proteins from single-peptide quantitation compared to proteins from multipeptide quantitation (Figure S9). Further, ion interference and ratio compression do not appear to impact reproducibility in channels next to the carrier channel.
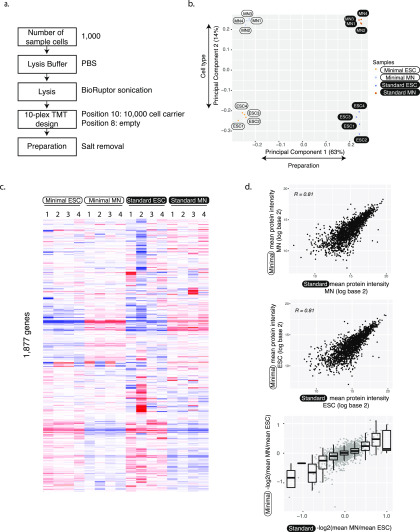
Figure 2.
Accurate and reproducible protein abundance measurements. (a) Flowchart illustrates the minimal input protocol with points of optimization. (b) First principal components of minimal and standard sample input data separate both the protocol but also the two different cell types. The analysis was done using 1763 protein groups that were identified in both minimal and standard preparations. (c) Heat map of protein abundances. Abundances were first scaled to the sum of log base 10 equaling 10 000 in each column. Then, we removed the first principal component and subtracted the row median from each entry to remove the gene-to-gene effect. Note that these normalizations were only used for visualization, not for significance testing for differential expression. We used hierarchical clustering with the Manhattan distance measure. Each experiment has four replicates. (d) Protein abundances (top, middle) and fold changes (bottom) from minimal and standard sample input preparations correlate well. The analysis was done using 1763 protein groups that were identified in both minimal and standard preparations. ESC—embryonic stem cell; MN—motor neuron.
Next, we used principal component analysis to compare results from the standard and minimal input preparations (Figures 2b and S10). As expected, the first component separated the two experimental setups that differed as described above, e.g., through use or omission of FACS purification or different column loading. However, the second component separated the two cell types, indicating that both protocols produce biologically meaningful protein quantitation. Indeed, when removing the first principal component, we observed striking similarities between samples from the minimal and standard input preparations (Figure 2c). Both preparations show similar differences between undifferentiated and differentiated cells. The preparations of minimal and standard were independent and part of the expected sample to sample variation; therefore, preparation “ESC1” in minimal input does not match preparation “ESC1” in the standard input preparation. ESC2 from the standard preparation might be an outlier as it does not cluster as closely with other standard input ESCs.
We further confirmed the consistency between the minimal and the standard input protocol by direct correlation of the measured protein abundances in the two cell types: the Pearson correlation coefficient ranged between 0.81 and 0.77 between the two protocols (Figures 2d, S6, and S7). Both minimal input and standard input preparations also showed a similar correlation with corresponding transcript abundances as taken from bulk RNA sequencing from the same differentiation paradigm,23 with Spearman correlation coefficients ranging from 0.39 to 0.43 (Figure S11).
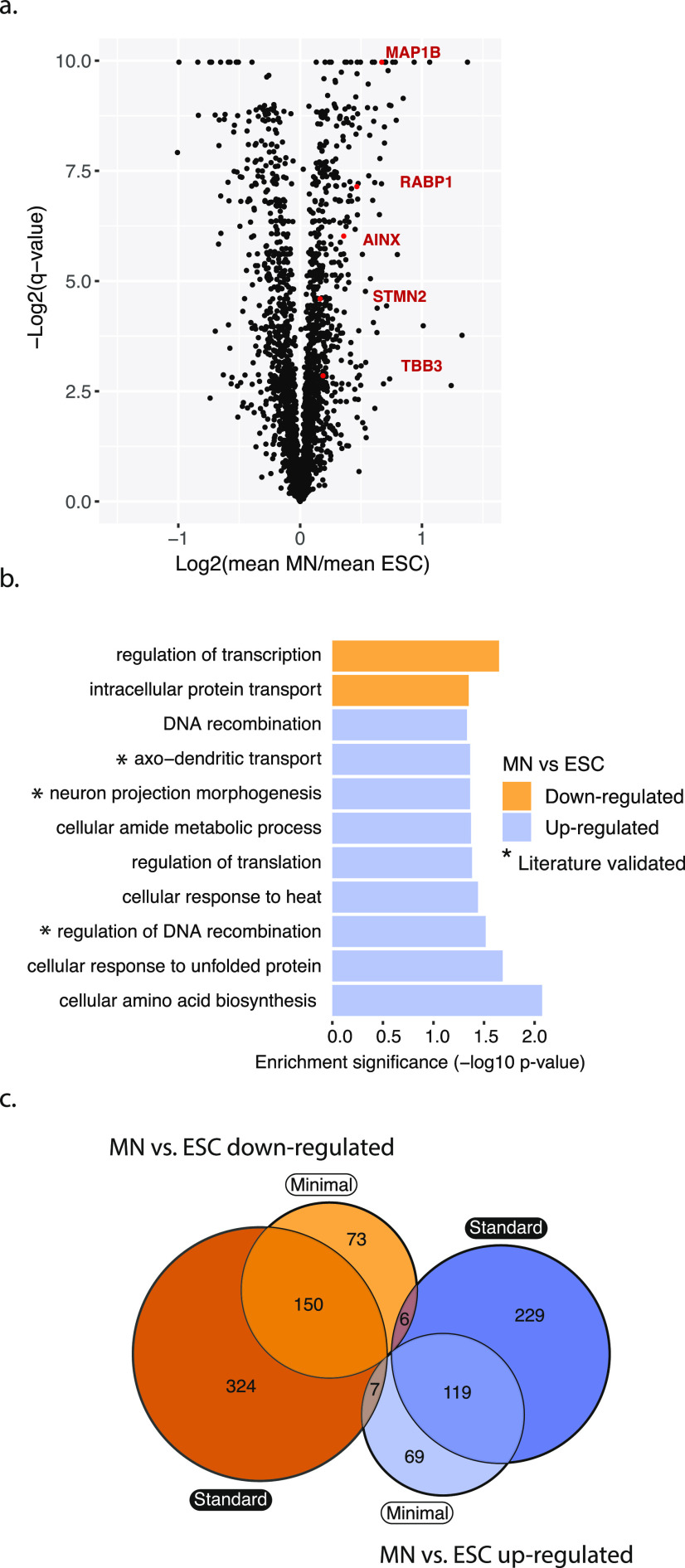
To further validate the protocol’s ability to identify differentially expressed proteins, we turned to known markers of successful differentiation, which we used as positive controls. Several neuron marker genes, e.g., AINX,24 MAP1B,25 RABP1,25,26 STMN2,27 and TBB3,28 were upregulated in MN cells compared to the ESCs in the minimal input preparation (Figure 3a).
Figure 3.
Proteins differentially expressed between ESC and MN. (a) Volcano plot showing in red widely recognized neuronal markers overexpressed in MNs compared to ESCs in the minimal input preparation. q-Values were calculated using Student’s t-test, using a permutation-based false discovery rate (FDR) with a cutoff of 0.05. (b) Differentially expressed proteins have similar function enrichments in both the minimal and standard input protocols (p-value < 0.05). (c) Minimal and standard input protocols overlap in their results with respect to identification of differentially expressed proteins: significantly up- or downregulated proteins (MN vs ESC) from either preparation overlap with p = 2.070e–192 or 7.639e–164, respectively (hypergeometric test). There are close to no proteins shared across opposing groups (p = 0.221 and 0.191, hypergeometric test). ESC—embryonic stem cell; MN—motor neuron.
We then tested for differential protein abundances between undifferentiated ESC and differentiated MN cells against variation between replicates, using the Student’s t-test. The approach identified 229 significantly upregulated and 195 significantly downregulated proteins in motor neurons compared to stem cells (q-value < 0.01, Figure 3a). The proteins upregulated in motor neurons were significantly enriched in several categories such as plasma-membrane proteins, axo-dendritic transport, and neuron projection morphogenesis reflecting successful differentiation to motor neuron fate (p-value < 0.05, Fisher’s exact test, Figures 3b and S12).
Next, we compared the significantly differentially expressed proteins between the minimal input and standard sample preparations (Figure 3c). While standard preparation identified more differentially expressed proteins due to lower variability across replicates (Figure 13), the up- and downregulated proteins from either preparation overlapped significantly (p < 0.0001 for MN vs ESC downregulated and upregulated, hypergeometric test). There were virtually no proteins in the opposing groups, further confirming validity of the minimal input preparation (p = 0.22 and 0.19, hypergeometric test).
Due to the semistochastic nature of proteomics experiments, the minimal input protocol identified some differentially expressed proteins that had been missed by the standard input preparation. The proteins specific to the minimal input protocol had abundances ranging over seven orders of magnitude (Figure S14), suggesting that abundance alone does not account for preparation of specific identification. The minimal input-specific proteins included EZRI, PEBP1, and TBB5 that were upregulated in motor neurons and that all have known roles in neuronal cells. Combined, these results support the protocol’s ability to quantify proteins and to identify significantly differentially expressed proteins.29
Applying the Minimal Input Protocol to Sorted C. robusta Cells
To test whether the protocol is generalizable to different systems, we used the model chordate C. robusta. We FACS-purified the cardiopharyngeal lineage using an established protocol30 and applied our method using two different setups. The first experiment employed 1000 cells for each experimental channel with four carrier channels (Figure S15a). It identified 1904 proteins. The carrier channels consisted of whole embryo cells since collecting more than a few thousand cells per condition from the cardiopharyngeal lineage alone was not feasible. In the second experiment, we omitted the carrier channel and used 5000 cells for each experimental condition (Figure S15a), which resulted in 732 identified proteins.
Next, we assessed reproducibility between biological replicates (Figure S15b,c). Both experiments showed a substantial correlation between replicates of log-transformed protein abundances, indicating good reproducibility. As expected, the correlations were slightly lower than those between replicates of log-transformed transcript abundances, as proteomics methods are typically more noisy (Figure S16). When comparing RNA and protein abundances directly, we observed small but statistically significant positive correlations (p-value < 0.05; Figure S17). While the correlation is much higher in studies where transcriptome and proteome data had been collected simultaneously,31 we interpreted the observed positive correlation as an indication that indeed the minimal input protocol provides meaningful protein abundance estimates.
In sum, the reproducibility and coverage of the Ciona experiments were compared with the results that we obtained with the mammalian cells (Figures S5 and S15b,c). However, we detected no statistically significant differentially expressed genes between conditions. While variation between replicates might have contributed to this result, we also speculate that the experimental setup did not allow for detectable proteome changes: the experiment was performed 15 h post fertilization, about an hour after the cells were born, which is enough time to change the transcriptome32,33 but not enough time to translate large numbers of new proteins.
Discussion
We present a straightforward, optimized protocol for proteomics analysis of minimal input samples, i.e., from as few as 1000 cells. The method quantifies ∼2500 proteins in mammalian samples and sensitively identifies differential expression—a result that has not yet been achieved with such few cells and without highly specialized equipment. The minimal input protocol offers an alternative to existing small input methods, such as SP311 or in-StageTip digestion7 (Table 1). Measured protein abundances have high reproducibility across replicates and correlate well with corresponding transcript abundances (Rs = 0.39–0.43, Figure S11). In mouse cell systems where transcriptomic and proteomic data had been collected simultaneously, the correlation can be higher.29,31 Lower correlation in our data is likely due to the fact that we collected transcriptomics and proteomics data collected from different cell batches that may have been at slightly different differentiation stages. Future work might use minimal sample RNA sequencing methods to compare protein and RNA abundances or use protein spike-ins to validate protein abundances directly.
While the number of quantified protein groups in the standard and minimal input protocol are similar (Figure 1b), the standard method identifies twice as many differentially expressed proteins (Figure 3c). The reason for this difference lies in the lower signal-to-noise ratio obtained in measurements from the minimal input sample, resulting likely from increased ratio compression. Ratio compression is a common problem with complex, TMT-based proteomics sample analysis due to coeluting and coisolated peptides.34 The problem is amplified in samples with unequal channel loading,16,35 as is the case for the minimal input setup. For this reason, we minimized the ratio between the carrier and sample channel. We also tested a setup in which we omitted the carrier channel and used equal amounts of sample (from 1000 cells each) per channel (Figure 1b). However, this approach resulted in lower proteome quantitation and high variability.
Another commonly used remedy for ratio compression is to reduce sample complexity via additional sample fractionation, e.g., with offline high-pH reverse-phase chromatography.36 Exhaustive fractionation can nearly eliminate ratio compression but requires large amounts of sample.37 Fractionation of the minimal input sample, even via specialized spin columns that require less material (e.g., Pierce Thermo Fisher), is impossible without collecting more cells. A possible solution might lie in ion mobility-based separation using the newest front-end technology.36
Other approaches to combat ratio compression include computational algorithms38 and specific data acquisition modes.39−41 While computational methods have had mixed success,36 different data acquisition methods typically require highly advanced technology. One simple approach to reduce interference and ratio compression uses smaller precursor isolation windows;39 a related approach isolates twice, once with standard and once with narrow window size.40 A third approach, requiring specific instrumentation, avoids ion interference by quantifying the mass tags in a second fragmentation step, i.e., by collecting MS3 spectra.38 While all three methods are successful in reducing ratio compression, the collection of additional mass spectra also substantially slows acquisition and therefore reduces proteome coverage.
While affected by ratio compression, the minimal input protocol we developed provides biologically meaningful protein quantitation, as we demonstrated here. The method uses sample amounts relatively easily achievable by, e.g., biopsy, dissection of specific cell types or tissues in vivo or FACS purification of rare cell populations, enabling analysis of highly specific cell populations. The minimal input protocol relies on the use of a carrier channel, as has been pioneered for single-cell analysis.5 In comparison to earlier work,5 we used a much smaller carrier-to-sample ratio, which resulted in less ratio compression, higher reproducibility, and quantitation accuracy. The advantages of the current protocol lie in its independence from specific equipment or reagents and its ability to analyze systems in which it is very difficult to obtain large numbers of cells. Future extensions of the work may involve the use of magnetic beads or hydrostatic pressure techniques to further increase protein yield.
Experimental Procedures
Mammalian Cell Culture
We used the transcription factor cassette Ngn2-Isl1-Lhx3 (NIL) to program motor neurons as previously described.42 We cultured the mouse embryonic stem cell (ESC) line in two-inhibitor-based medium (advanced Dulbecco’s modified Eagle’s medium (DMEM)/F12:neurobasal (1:1) medium (Thermo Fisher, 12634028, 10888022)) supplemented with 2.5% ESC-grade Tet-negative fetal bovine serum (v/v, VWR 35-075-CV), 1× N2 (Thermo Fisher, 17502-048), 1× B27 (Thermo Fisher, 17504-044), 2 mM l-glutamine (Thermo Fisher, 25030081), 0.1 mM β-mercaptoethanol (Thermo Fisher, 21985-023), 1000 U/mL leukemia inhibitory factor (Fisher, ESG1107), 3 μM CHIR99021 (Biovision, 1991), and 1 μM PD0325901 (Sigma, PZ0162-5 MG) and maintained at 37 °C, 8% CO2. We dissociated embryonic stem cells by TrypLE (Gibco, 12605010) and prepared for FACS purification or seeded cells for differentiation into motor neurons.
To differentiate ESCs into motor neurons, we trypsinized ESCs (Thermo Fisher, 25300-120) and seeded single cells at 25 000 cells/mL in the ANDFK medium (advanced DMEM/F12:neurobasal (1:1) medium (Thermo Fisher, 12634028, 10888022)), 10% Knockout SR (v/v) (Thermo Fisher, 10828-028), 2 mM l-glutamine (Thermo Fisher, 25030081), and 0.1 mM 2-mercaptoethanol (Thermo Fisher, 21985-023)) to initiate the formation of embryoid bodies (EBs) (day 4) in the suspension culture using 10 cm untreated dishes (Fisher, 08-772-32) and maintained at 37 °C, 5% CO2. We changed the medium 2 days later (day 2) with the addition of 3 μg/mL doxycycline (Sigma D9891) to induce the NIL transcription factors. We dissociated the EBs using 0.05% trypsin–ethylenediaminetetraacetic acid (EDTA) (Thermo Fisher, 25300-120) and replated, cultured the induced motor neurons for two days, and then prepared samples for FACS purification. We estimated the cell counts used for carrier channels using Countess II FL automated cell counter (Thermo Fisher, AMQAF1000).
C. robusta Handling
Wild C. robusta was obtained from M-REP (Carlsbad, CA) and kept under constant light to avoid spawning. Gametes from several animals were collected separately for in vitro cross-fertilization followed by dechorionation and electroporation as previously described.43 The embryos were cultured in filtered artificial seawater (FASW) in agarose-coated plastic Petri dishes at 18 °C. We electroporated 50 μg of constructs for FAC-sorting (Mesp > tagRFP, MyoD905 > eGFP and Hand-r > tagBFP) and 70 μg of experimental constructs (Mesp > LacZ, Mesp > FgfrDN, Mesp > MekS216D,S220E).
FACS Purification of Mammalian Cells
To purify programmed motor neurons by fluorescence-activated cell sorting (FACS), we prepared single-cell suspensions of each sample to approximately 1–9 × 106 cells/mL. We added 20 μL of 50 μg/mL fluorescein diacetate (FDA) to the cell suspension and isolated motor neurons with a Becton Dickinson ARIA SORP or ARIA II SORP cell sorter. Fluorescein isothiocyanate (FITC) fluorescence was excited with a 488 nm laser and detected with a 530/30 (ARIA) or 525/50 nm (ARIA II) filter. We conducted FACS using a ceramic 100 μm nozzle (Becton Dickinson), sheath pressure of 20 psi, and a low acquisition rate of 1000–4000 events/s. We collected mouse embryonic stem cells and motor neurons in 1.7 mL microcentrifuge tubes containing 20 μL of ice-cold PBS or 0.1% RapiGest (Waters, MA) dissolved in PBS. Total FACS time per experiment was 1–2 h.
FACS Purification of C. robusta Cells
Sample dissociation and FACS were performed as previously described.30,44 Embryos and larvae were harvested at 15 hpf in 5 mL borosilicate glass tubes (Fisher Scientific, Waltham, MA, Cat. no. 14-961-26) and washed with 2 mL of calcium- and magnesium-free artificial seawater (CMF-ASW: 449 mM NaCl, 33 mM Na2SO4, 9 mM KCl, 2.15 mM NaHCO3, 10 mM Tris–Cl pH 8.2, 2.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA)). Embryos and larvae were dissociated in 2 mL 0.2% trypsin (w/v, Sigma, T-4799) in CMF-ASW by pipetting with glass Pasteur pipettes. The dissociation was stopped by adding 2 mL of filtered ice-cold 0.05% bovine serum albumin (BSA) CMF-ASW. Dissociated cells were passed through a 40 μm cell strainer and collected in 5 mL polystyrene round-bottom tube (Corning Life Sciences, Oneonta, New York, REF 352235). Cells were collected by centrifugation at 800g for 3 min at 4 °C, followed by two washes with ice-cold CMF-ASW. Cell suspensions were filtered again through a 40 μm cell strainer and stored on ice. Cell suspensions were used for sorting within 1 h. Cardiopharyngeal lineage cells were labeled by Mesp > tagRFP reporter. The mesenchyme cells were counterselected using MyoD905 > GFP as described.30 Dissociated cells were loaded in a BD FACS Aria cell sorter, 488 nm laser; FITC filter was used for GFP, 407 nm laser; DsRed filter was used for tagRFP, 561 nm laser; and Pacific Blue filter was used for tagBFP.
Proteome Analysis by Mass Spectrometry
We sonicated cells with Covaris S220 or Diagenode Bioruptor Pico sonicators. The total volume for sonication was about 25 μL for minimal samples (20 μL lysis buffer plus about 5 μL 1× PBS from cell sorting) and 50 μL for standard and carrier samples. Sonication settings used 125 W power for 180 s with 10% peak duty cycle for Covaris and 15 cycles of 30 s on and 30 s off for Bioruptor, in a degassed water bath at 4 °C. After lysis, we heated the samples for 15 min at 90 °C to denature proteins. We then directly added 1 and 0.5 μg of mass spectrometry grade trypsin (Sigma-Aldrich) to standard and minimal samples, respectively, and digested the proteins into peptides at 37 °C overnight. We measured resulting peptide concentrations with the Pierce quantitative fluorometric peptide assay (Thermo Fisher) kit for standard samples.
We dissolved tandem mass tag (TMT) 10plex reagents (Thermo Scientific) in anhydrous acetonitrile (0.8 mg/82 μL). We labeled peptides with 10 μL of the TMT 10plex label reagent per sample. We estimated labeling efficiency as >97% for all samples, based on labeled and unlabeled peptides found in the evidence. txt output file of the mass spectrometry analysis (see below). Following incubation at room temperature for 1 h, we quenched the reactions with 8 μL of 5% hydroxylamine for 15 min. All samples were combined into a new microcentrifuge tube at equal amounts and reduced to remove acetonitrile using an Eppendorf concentrator Vacufuge plus. The salt removal was performed using Pierce C18 Spin Tips (Thermo Scientific, #84850), according to the manufacturer’s instructions.
We used an EASY-nLC 1000 coupled on-line to a Q-Exactive HF spectrometer (both Thermo Fisher Scientific). Solvent A (0.1% formic acid in water) and solvent B (80% acetonitrile, 0.5% acetic acid) were used as mobile phases for gradient separation. The separation was performed using a 50 cm × 75 μm i.d. PepMap C18 column (Thermo Fisher Scientific) packed with 2 μm, 100 Å particles, and heated at 55 °C. We used a 155 min segmented gradient between solvent A and solvent B at a flow rate of 250 nL/min as follows: 2–5% B in 5 min, 5–25% B in 110 min, 25–40% B in 25 min, 49–80% B for 5 min, and 80–95% B for 5 min. Solvent B was held at 95% for another 5 min.
For label-free analysis, the full mass spectrometry (MS) scans were acquired with a resolution of 120 000, an automatic gain control (AGC) target of 3 × 106, with a maximum ion time of 100 ms, and a scan range of 375–1500 m/z. Following each full MS scan, data-dependent high-resolution higher-energy collision dissociation (HCD) MS/MS spectra were acquired with a resolution of 30 000, AGC target of 2 × 105, maximum ion time of 150 ms, 1.5 m/z isolation window, fixed first mass of 100 m/z, and normalized collision energy (NCE) of 27 in centroid mode. For analysis of TMT-labeled samples, the full MS scans were acquired with the same settings. Following each full MS scan, data-dependent high-resolution HCD MS/MS spectra were acquired with a resolution of 60 000, AGC target of 2 × 105, maximum ion time of 100 ms, 1.2 m/z isolation window, fixed first mass of 100 m/z, and NCE of 35 in centroid mode.
Protein Analysis of Raw Data
The RAW data files were processed using MaxQuant45 (version 1.6.1.0) to identify and quantify protein and peptide abundances. The spectra were matched against the Mus musculus Uniprot database (downloaded August 18, 2018) with standard settings for peptide and protein identification that allowed for 10 ppm tolerance, a posterior global false discovery rate (FDR) of 1% based on the reverse sequence of the mouse FASTA file, and up to two missed trypsin cleavages. We found a range from 2.8 to 4.3 and 8% of miscleaved peptides in the minimal and the standard input sample, respectively. We estimated protein abundance using iBAQ31 for label-free experiments and intensity for TMT experiments. 10plex TMT labeling of Lys- and N-terminal amines were considered fixed modifications. TMT quantification was performed at the MS2 level with default mass tolerance and other parameters. We then used the reporter ion intensities as estimates for protein abundance.
Data Analysis
Z-score normalization and Student’s t-test statistics were conducted using Perseus (version 1.5.3.0).46 Student’s t-test analyses were done with permutation-based FDR using a cutoff of 0.05 with 250 randomizations. All further computational analyses were conducted in R. Correlation analysis was conducted with the cor() function. Principal component analysis was conducted with the prcomp() function using log base 2 transformed data. RNA-seq data was used from Velasco et al.23 and analyzed using the EdgeR package.47,48 Function enrichment analyses were done using Panther 14.1.49 Scatterplots and correlation plots were made using ggplot2 package.50
Acknowledgments
B.V. acknowledges funding by American Heart Association Grant #18PRE33990254. D.E.I. acknowledges funding by the NIH/NINDS (1F31NS103447). B.V. and D.E.I. acknowledge Fleur Strand Graduate Fellowship. E.O.M acknowledges funding by Project ALS (A13-0416). L.C. acknowledges funding by NIH/NHLBI R01 HL108643 and by the Leducq Foundation Trans-Atlantic network of excellence award 15CVD01. C.V. acknowledges funding by the NIH/NIGMS (R35GM127089) and the Zegar Family Foundation Fund for Genomics Research at New York University. We thank Judit Villen and Matthias Selbach for productive discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01191.
Label-free mass spectrometry; correlation between different sample preparation techniques; comparison of number of protein groups identified using 1000 and 5000 embryonic stem cells using label-free quantification; optimization of the size of the carrier channel; replicates of minimal input MN and and ESC; correlation coefficients (Spearman) of quadruplicates; distributions of the number of peptides per protein and the number of unique peptides per protein; proteins quantified by a single peptide; graph showing the second and third principal components; minimal and standard input preparations; volcanos plot; proteomics data; transcriptomic data; comparison of transcriptomic and proteomic data (Figures S1–S17) (PDF)
Author Contributions
∥ L.C. and C.V. contributed equally to this work.
The authors declare no competing financial interest.
Notes
The mass spectrometry data including the MaxQuant output files have been deposited to the ProteomeXchange Consortium via the PRIDE51 partner repository with the data set identifier PXD019363.
Supplementary Material
References
- Sun L.; Zhu G.; Zhao Y.; Yan X.; Mou S.; Dovichi N. J. Ultrasensitive and Fast Bottom-up Analysis of Femtogram Amounts of Complex Proteome Digests. Angew. Chem., Int. Ed. 2013, 52, 13661–13664. 10.1002/anie.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Wang S.; Adhikari S.; Deng Z.; Wang L.; Chen L.; Ke M.; Yang P.; Tian R. Simple and Integrated Spintip-Based Technology Applied for Deep Proteome Profiling. Anal. Chem. 2016, 88, 4864–4871. 10.1021/acs.analchem.6b00631. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Clair G.; Chrisler W. B.; Shen Y.; Zhao R.; Shukla A. K.; Moore R. J.; Misra R. S.; Pryhuber G. S.; Smith R. D.; Ansong C.; Kelly R. T. Proteomic Analysis of Single Mammalian Cells Enabled by Microfluidic Nanodroplet Sample Preparation and Ultrasensitive NanoLC-MS. Angew. Chem., Int. Ed. 2018, 57, 12370–12374. 10.1002/anie.201802843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard-Banek C.; Moody S. A.; Nemes P. Single-Cell Mass Spectrometry for Discovery Proteomics: Quantifying Translational Cell Heterogeneity in the 16-Cell Frog (Xenopus) Embryo. Angew. Chem., Int. Ed. 2016, 55, 2454–2458. 10.1002/anie.201510411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik B.; Levy E.; Harmange G.; Slavov N. SCoPE-MS: Mass Spectrometry of Single Mammalian Cells Quantifies Proteome Heterogeneity during Cell Differentiation.. Genome Biol. 2018, 19, 161 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy S.; Jaehnig E. J.; Krug K.; Kim B.-J.; Saltzman A. B.; Chan D. W.; Holloway K. R.; Anurag M.; Huang C.; Singh P.; Gao A.; Namai N.; Dou Y.; Wen B.; Vasaikar S. V.; Mutch D.; Watson M. A.; Ma C.; Ademuyiwa F. O.; Rimawi M. F.; Schiff R.; Hoog J.; Jacobs S.; Malovannaya A.; Hyslop T.; Clauser K. R.; Mani D. R.; Perou C. M.; Miles G.; Zhang B.; Gillette M. A.; Carr S. A.; Ellis M. J. Microscaled Proteogenomic Methods for Precision Oncology. Nat. Commun. 2020, 11, 532 10.1038/s41467-020-14381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N. A.; Pichler G.; Paron I.; Nagaraj N.; Mann M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Myers S. A.; Rhoads A.; Cocco A. R.; Peckner R.; Haber A. L.; Schweitzer L. D.; Krug K.; Mani D. R.; Clauser K. R.; Rozenblatt-Rosen O.; Hacohen N.; Regev A.; Carr S. A. Streamlined Protocol for Deep Proteomic Profiling of FAC-Sorted Cells and Its Application to Freshly Isolated Murine Immune Cells. Mol. Cell. Proteomics 2019, 18, 995–1009. 10.1074/mcp.RA118.001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Qian W.-J.; Mottaz H. M.; Clauss T. R. W.; Anderson D. J.; Moore R. J.; Camp D. G. 2nd; Khan A. H.; Sforza D. M.; Pallavicini M.; Smith D. J.; Smith R. D. Development and Evaluation of a Micro- and Nanoscale Proteomic Sample Preparation Method. J. Proteome Res. 2005, 4, 2397–2403. 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.; Xu M.; Wang P.; Li L. Development of Mass Spectrometry-Based Shotgun Method for Proteome Analysis of 500 to 5000 Cancer Cells. Anal. Chem. 2010, 82, 2262–2271. 10.1021/ac9023022. [DOI] [PubMed] [Google Scholar]
- Hughes C. S.; Foehr S.; Garfield D. A.; Furlong E. E.; Steinmetz L. M.; Krijgsveld J. Ultrasensitive Proteome Analysis Using Paramagnetic Bead Technology. Mol. Syst. Biol. 2014, 10, 757. 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. A Dream of Single-Cell Proteomics.. Nat. Methods 2019, 809–812. 10.1038/s41592-019-0540-6. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Piehowski P. D.; Kelly R. T.; Qian W.-J. Nanoproteomics Comes of Age.. Expert Rev. Proteomics 2018, 15, 865–871. 10.1080/14789450.2018.1537787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; Piehowski P. D.; Zhao R.; Chen J.; Shen Y.; Moore R. J.; Shukla A. K.; Petyuk V. A.; Campbell-Thompson M.; Mathews C. E.; Smith R. D.; Qian W.-J.; Kelly R. T. Nanodroplet Processing Platform for Deep and Quantitative Proteome Profiling of 10–100 Mammalian Cells. Nat. Commun. 2018, 882 10.1038/s41467-018-03367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Plouffe B. D.; Belov A. M.; Ray S.; Wang X.; Murthy S. K.; Karger B. L.; Ivanov A. R. An Integrated Platform for Isolation, Processing, and Mass Spectrometry-Based Proteomic Profiling of Rare Cells in Whole Blood. Mol. Cell. Proteomics 2015, 14, 1672–1683. 10.1074/mcp.M114.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou M.; Clair G.; Tsai C.-F.; Xu K.; Chrisler W. B.; Sontag R. L.; Zhao R.; Moore R. J.; Liu T.; Pasa-Tolic L.; Smith R. D.; Shi T.; Adkins J. N.; Qian W.-J.; Kelly R. T.; Ansong C.; Zhu Y. High-Throughput Single Cell Proteomics Enabled by Multiplex Isobaric Labeling in a Nanodroplet Sample Preparation Platform. Anal. Chem. 2019, 13119–13127. 10.1021/acs.analchem.9b03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehowski P. D.; Zhu Y.; Bramer L. M.; Stratton K. G.; Zhao R.; Orton D. J.; Moore R. J.; Yuan J.; Mitchell H. D.; Gao Y.; Webb-Robertson B.-J. M.; Dey S. K.; Kelly R. T.; Burnum-Johnson K. E. Automated Mass Spectrometry Imaging of over 2000 Proteins from Tissue Sections at 100-μm Spatial Resolution. Nat. Commun. 2020, 11, 8 10.1038/s41467-019-13858-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orecchioni M.; Bedognetti D.; Newman L.; Fuoco C.; Spada F.; Hendrickx W.; Marincola F. M.; Sgarrella F.; Rodrigues A. F.; Ménard-Moyon C.; Cesareni G.; Kostarelos K.; Bianco A.; Delogu L. G. Single-Cell Mass Cytometry and Transcriptome Profiling Reveal the Impact of Graphene on Human Immune Cells. Nat. Commun. 2017, 8, 1109 10.1038/s41467-017-01015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M.; Hafemeister C.; Stephenson W.; Houck-Loomis B.; Chattopadhyay P. K.; Swerdlow H.; Satija R.; Smibert P. Simultaneous Epitope and Transcriptome Measurement in Single Cells. Nat. Methods 2017, 14, 865–868. 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genshaft A. S.; Li S.; Gallant C. J.; Darmanis S.; Prakadan S. M.; Ziegler C. G. K.; Lundberg M.; Fredriksson S.; Hong J.; Regev A.; Livak K. J.; Landegren U.; Shalek A. K. Multiplexed, Targeted Profiling of Single-Cell Proteomes and Transcriptomes in a Single Reaction. Genome Biol. 2016, 188 10.1186/s13059-016-1045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielaff M.; Kuharev J.; Bohn T.; Hahlbrock J.; Bopp T.; Tenzer S.; Distler U. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017, 16, 4060–4072. 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- Mazzoni E. O.; Mahony S.; Closser M.; Morrison C. A.; Nedelec S.; Williams D. J.; An D.; Gifford D. K.; Wichterle H. Synergistic Binding of Transcription Factors to Cell-Specific Enhancers Programs Motor Neuron Identity. Nat. Neurosci. 2013, 16, 1219–1227. 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S.; Ibrahim M. M.; Kakumanu A.; Garipler G.; Aydin B.; Al-Sayegh M. A.; Hirsekorn A.; Abdul-Rahman F.; Satija R.; Ohler U.; Mahony S.; Mazzoni E. O. A Multi-Step Transcriptional and Chromatin State Cascade Underlies Motor Neuron Programming from Embryonic Stem Cells. Cell Stem Cell 2017, 20, 205.e8–217.e8. 10.1016/j.stem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C. L.; Liem R. K. Characterization of the Mouse Gene Encoding the Neuronal Intermediate Filament Protein Alpha-Internexin. Gene 1994, 149, 289–292. 10.1016/0378-1119(94)90163-5. [DOI] [PubMed] [Google Scholar]
- Bouquet C.; Soares S.; von Boxberg Y.; Ravaille-Veron M.; Propst F.; Nothias F. Microtubule-Associated Protein 1B Controls Directionality of Growth Cone Migration and Axonal Branching in Regeneration of Adult Dorsal Root Ganglia Neurons. J. Neurosci. 2004, 24, 7204–7213. 10.1523/JNEUROSCI.2254-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. Retinoic Acid in the Development, Regeneration and Maintenance of the Nervous System. Nat. Rev. Neurosci. 2007, 8, 755–765. 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- Stein R.; Mori N.; Matthews K.; Lo L. C.; Anderson D. J. The NGF-Inducible SCG10 mRNA Encodes a Novel Membrane-Bound Protein Present in Growth Cones and Abundant in Developing Neurons. Neuron 1988, 1, 463–476. 10.1016/0896-6273(88)90177-8. [DOI] [PubMed] [Google Scholar]
- Katsetos C. D.; Legido A.; Perentes E.; Mörk S. J. Class III Beta-Tubulin Isotype: A Key Cytoskeletal Protein at the Crossroads of Developmental Neurobiology and Tumor Neuropathology. J. Child Neurol. 2003, 18, 851–866. discussion 867. 10.1177/088307380301801205. [DOI] [PubMed] [Google Scholar]
- McManus J.; Cheng Z.; Vogel C. Next-Generation Analysis of Gene Expression Regulation – Comparing the Roles of Synthesis and Degradation. Mol. BioSyst. 2015, 2680–2689. 10.1039/C5MB00310E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Racioppi C.; Gravez B.; Christiaen L. Purification of Fluorescent Labeled Cells from Dissociated Ciona Embryos. Adv. Exp. Med. Biol. 2018, 1029, 101–107. 10.1007/978-981-10-7545-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B.; Busse D.; Li N.; Dittmar G.; Schuchhardt J.; Wolf J.; Chen W.; Selbach M. Global Quantification of Mammalian Gene Expression Control. Nature 2011, 473, 337–342. 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Wang W.; Niu X.; Stuart T.; Jullian E.; Mauck W. M. 3rd; Kelly R. G.; Satija R.; Christiaen L. A. Single-Cell Transcriptional Roadmap for Cardiopharyngeal Fate Diversification. Nat. Cell Biol. 2019, 21, 674–686. 10.1038/s41556-019-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razy-Krajka F.; Lam K.; Wang W.; Stolfi A.; Joly M.; Bonneau R.; Christiaen L. Collier/OLF/EBF-Dependent Transcriptional Dynamics Control Pharyngeal Muscle Specification from Primed Cardiopharyngeal Progenitors. Dev. Cell 2014, 29, 263–276. 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauniyar N.; Yates J. R. 3rd. Isobaric Labeling-Based Relative Quantification in Shotgun Proteomics. J. Proteome Res. 2014, 13, 5293–5309. 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L.; Tsai C.-F.; Dirice E.; Swensen A. C.; Chen J.; Shi T.; Gritsenko M. A.; Chu R. K.; Piehowski P. D.; Smith R. D.; Rodland K. D.; Atkinson M. A.; Mathews C. E.; Kulkarni R. N.; Liu T.; Qian W.-J. Boosting to Amplify Signal with Isobaric Labeling (BASIL) Strategy for Comprehensive Quantitative Phosphoproteomic Characterization of Small Populations of Cells. Anal. Chem. 2019, 91, 5794–5801. 10.1021/acs.analchem.9b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swearingen K. E.; Moritz R. L. High-Field Asymmetric Waveform Ion Mobility Spectrometry for Mass Spectrometry-Based Proteomics. Expert Rev. Proteomics 2012, 9, 505–517. 10.1586/epr.12.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M.; Cho J.-H.; Kodali K.; Pagala V.; High A. A.; Wang H.; Wu Z.; Li Y.; Bi W.; Zhang H.; Wang X.; Zou W.; Peng J. Extensive Peptide Fractionation and y1 Ion-Based Interference Detection Method for Enabling Accurate Quantification by Isobaric Labeling and Mass Spectrometry. Anal. Chem. 2017, 2956–2963. 10.1021/acs.analchem.6b04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski M. M.; Mathieson T.; Zinn N.; Sweetman G.; Doce C.; Becher I.; Pachl F.; Kuster B.; Bantscheff M. Measuring and Managing Ratio Compression for Accurate iTRAQ/TMT Quantification. J. Proteome Res. 2013, 12, 3586–3598. 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- McAlister G. C.; Nusinow D. P.; Jedrychowski M. P.; Wühr M.; Huttlin E. L.; Erickson B. K.; Rad R.; Haas W.; Gygi S. P. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal. Chem. 2014, 86, 7150–7158. 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L.; Rad R.; Gygi S. P.; Haas W. MS3 Eliminates Ratio Distortion in Isobaric Multiplexed Quantitative Proteomics. Nat. Methods 2011, 937–940. 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumeliotis T. I.; Weisser H.; Choudhary J. S. Evaluation of a Dual Isolation Width Acquisition Method for Isobaric Labeling Ratio Decompression. J. Proteome Res. 2019, 18, 1433–1440. 10.1021/acs.jproteome.8b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D.; Fujiki R.; Iannitelli D. E.; Smerdon J. W.; Maity S.; Rose M. F.; Gelber A.; Wanaselja E. K.; Yagudayeva I.; Lee J. Y.; Vogel C.; Wichterle H.; Engle E. C.; Mazzoni E. O. Stem Cell-Derived Cranial and Spinal Motor Neurons Reveal Proteostatic Differences between ALS Resistant and Sensitive Motor Neurons. Elife 2019, 8, e44423 10.7554/eLife.44423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L.; Wagner E.; Shi W.; Levine M. Isolation of Sea Squirt (Ciona) Gametes, Fertilization, Dechorionation, and Development. Cold Spring Harbor Protoc. 2009, 2009, db.prot5344 10.1101/pdb.prot5344. [DOI] [PubMed] [Google Scholar]
- Christiaen L.; Wagner E.; Shi W.; Levine M. Isolation of Individual Cells and Tissues from Electroporated Sea Squirt (Ciona) Embryos by Fluorescence-Activated Cell Sorting (FACS). Cold Spring Harbor Protoc. 2009, db.prot5349 10.1101/pdb.prot5349. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Cox J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Sinitcyn P.; Carlson A.; Hein M. Y.; Geiger T.; Mann M.; Cox J. The Perseus Computational Platform for Comprehensive Analysis of (prote)omics Data.. Nat. Methods 2016, 13, 731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Robinson M. D.; McCarthy D. J.; Smyth G. K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. J.; Chen Y.; Smyth G. K. Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288–4297. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H.; Muruganujan A.; Ebert D.; Huang X.; Thomas P. D. PANTHER Version 14: More Genomes, a New PANTHER GO-Slim and Improvements in Enrichment Analysis Tools.. Nucleic Acids Res. 2019, 47, D419–D426. 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H.ggplot2: Elegant Graphics for Data Analysis; Springer Science & Business Media, 2009. [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Pérez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaíno J. A. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.