Abstract
The phytohormone auxin regulates a wide range of developmental processes in plants. Indole-3-acetic acid (IAA) is the main auxin that moves in a polar manner and forms concentration gradients, whereas phenylacetic acid (PAA), another natural auxin, does not exhibit polar movement. Although these auxins occur widely in plants, the differences between IAA and PAA metabolism remain largely unknown. In this study, we investigated the role of Arabidopsis IAA CARBOXYL METHYLTRANSFERASE 1 (IAMT1) in IAA and PAA metabolism. IAMT1 proteins expressed in Escherichia coli could convert both IAA and PAA to their respective methyl esters. Overexpression of IAMT1 caused severe auxin-deficient phenotypes and reduced the levels of IAA, but not PAA, in the root tips of Arabidopsis, suggesting that IAMT1 exclusively metabolizes IAA in vivo. We generated iamt1 null mutants via CRISPR/Cas9-mediated genome editing and found that the single knockout mutants had normal auxin levels and did not exhibit visibly altered phenotypes. These results suggest that other proteins, namely the IAMT1 homologs in the SABATH family of carboxyl methyltransferases, may also regulate IAA levels in Arabidopsis.
Keywords: auxin, indole-3-acetic acid, metabolism, methyltransferase, phenylacetic acid
1. Introduction
Auxin plays important roles in regulating plant development and environmental responses, including embryogenesis, organ formation, apical dominance, and tropism [1]. These regulations depend on the cellular concentration of auxin in specific tissues, which is determined by biosynthesis, inactivation, and transport, along with signal transduction [2]. Indole-3-acetic acid (IAA) is the most studied naturally occurring auxin, and its regulatory mechanism in plants has been elucidated. IAA is known to be directionally transported from cell to cell through membrane-localized proteins. Among membrane-localized proteins, PIN-FORMED efflux carrier proteins play a crucial role in polar auxin transport and form auxin concentration gradients [3]. In plants, the main biosynthesis pathway converts tryptophan to IAA via indole-3-pyruvate, involving TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) and YUCCA (YUC) families [4–8]. Plants have various IAA inactivation pathways, including oxidation by DIOXYGENASE FOR AUXIN OXIDATION (DAO), amino acid conjugation by the GRETCHEN HAGEN 3 (GH3) family, glucosylation by UDP-GLUCOSYLTRANSFERASE 84B1 (UGT84B1), and methylation by IAA CARBOXYL METHYLTRANSFERASE 1 (IAMT1) [1,9]. Some inactivated forms, e.g., IAA-Ala and -Leu, IAA-glucoside, and IAA-methyl ester (MeIAA) are thought to be storage forms with the potential to be reverted back into IAA for homeostatic regulation [10–12]. Among IAA metabolic enzymes, previous studies demonstrated the physiological importance of IAMT1 in plant growth and physiological responses to environmental cues [13–16]. Qin et al. (2005) demonstrated that the RNAi silencing mutants iamt1 (iamt1-RNAi) showed high-auxin phenotypes. In contrast, the T-DNA insertion mutants of iamt1 (iamt1–1, salk_072125) did not show obvious developmental phenotypes. Therefore, they assumed that products of a truncated transcript of IAMT1 may still have methyltransferase activity [16]. Interestingly, recent studies demonstrated that iamt1–1 mutants exhibited slightly faster opening of the apical hook, impaired gravitropism of hypocotyls, and tolerance to high-temperature male sterility [14,15], although no significant reduction in IAA levels was observed [15]. The reasons for the differences between the phenotype of iamt1-RNAi and iamt1–1 are not clear. The dramatic high-auxin phenotypes of iamt1-RNAi might be caused by simultaneous repression of additional members of the SABATH family, a group of carboxyl methyltransferase genes including IAMT1 [15].
Phenylacetic acid (PAA) is another naturally occurring auxin widely distributed in plants [17,18], but its physiological significance is yet to be demonstrated [18]. PAA is generally less effective than IAA except for the promotion of lateral root formation in peas [19], but endogenous amounts of PAA are higher than those of IAA in various plant species [17,20]. PAA and IAA regulate various genes via the same auxin signaling pathway and are assumed to share the same regulatory roles as auxins [17,21]. A distinctive difference is that PAA does not undergo polar transportation [17]. Based on this evidence, we recently proposed a model wherein IAA acts as a cell to cell communicator and PAA may play a role in the maintenance of steady-state auxin levels required for the preservation of cellular activity in plants [22]. More recently, we demonstrated that the GH3 family alters the ratio of IAA and PAA in Arabidopsis [23]. These observations suggest that various IAA metabolic enzymes may also regulate the endogenous levels of PAA in plants.
In this study, we investigated the role of Arabidopsis IAMT1 in auxin metabolism. We demonstrated that IAMT1 can convert not only IAA to MeIAA but also PAA to PAA-methyl ester (MePAA) in vitro. However, overexpression of IAMT1 decreased the amounts of IAA, but not PAA, in root tips of Arabidopsis. We found that neither alteration of auxin levels nor obvious phenotypes were observed in the CRISPR/Cas9-based null mutants of iamt1 under our growth conditions. Our results suggest that IAMT1 exclusively regulates the levels of IAA in vivo and that members of the SABATH family, a group of carboxyl methyltransferases, may also regulate IAA levels in Arabidopsis.
2. Materials and methods
2.1. Plant materials and growth conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type (WT) plant. Arabidopsis seeds were stratified at 4°C for 2 d in the dark. Seedlings were grown vertically on Murashige and Skoog (MS) agar medium with 1% sucrose under 16 h light condition at 23°C. Transgenic Arabidopsis pMDC7:IAMT1 were used for experiments. For β-estradiol (ER) treatment, seeds were germinated and grown on MS agar medium containing ER (2 μM) for 4 days. MS medium with 0.1% DMSO was used as a mock treatment. For quantitative RT-PCR (qRT-PCR) and auxin metabolite analysis, seedlings were grown vertically on MS agar medium for 10 days, transferred to MS media, underwent shaken cultured at 100 rpm for 2 d, and were then cultured for another 2 days in the presence of ER (2 μM final concentration) or the mock solution. After ER treatment, plants were collected, freeze-dried, and kept at −80°C until use. For the gravitropic response test, the WT and iamt1-c seeds were stratified at 4°C for 2 days in the dark, exposed to light for 6–8 h at 22°C, and were cultivated in the dark for 3 days and then for 12 h after rotating the plate 90°.
2.2. IAMT1 protein preparation in E. coli
DNA fragments of IAMT1 and its homologs were amplified by PCR using genomic DNA from Arabidopsis seedlings as a template using gene-specific primer pairs listed in Table S1. A cDNA of the IAMT1 gene was cloned into the pCold-TF DNA vector (Takara Bio, Kusatsu, Japan) using the In-Fusion system. The E. coli strain BL21 star (DE3) was transformed with the pCold-TF:IAMT1 plasmid. The transformants were incubated at 37°C in TB medium with 50 μg/mL carbenicillin. Protein expression was induced by adding isopropyl β-D-l-thiogalactopyranoside at a final concentration of 1 mM. After incubation at 15°C for 24 h, the cells were collected by centrifugation. The cells were resuspended in sodium phosphate buffer (50 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, pH 7.0) and sonicated to get the cell extract. The TF-fused IAMT1 proteins were purified from the cell extract using TALON metal affinity resin (Takara Bio, Kusatsu, Japan). The TF-fused proteins of IAMT1 homologs were also prepared in a similar manner. The trigger factor region was removed by the Factor-Xa following the manufacturer’s instructions and purified proteins were condensed by ultrafiltration using an Amicon Ultra centrifugal unit (Merck Millipore, Burlington, MA).
2.3. Enzyme assays
Enzymatical assays with IAMT1 and homolog proteins were performed in a 50 μL volume containing 10 μg purified recombinant protein, 50 mM Tris-HCl at pH 7.5, 1 mM IAA or PAA, and 1 mM S-adenosyl-L-methionine (SAM). Enzymes were boiled at 100°C and used as controls. The assays were initiated by the addition of SAM, maintained at 25°C for 4 h, and stopped by the addition of 80 μL of n-hexane. MeIAA and MePAA were extracted by mixing the liquid and the upper layers were collected. These samples were analyzed using the LECO Pegasus HT gas chromatography (GC)-mass spectrum system (LECO, St. Joseph, MI). An SGE BPX35 column (Shimazu, Kyoto, Japan) was used with helium as the carrier gas at a flow rate of 1 mL min−1. The GC program was as follows: 50°C for 2 min, ramp to 320°C at 30°C min−1, followed by a 3 min hold at 320°C. Reaction products were determined by comparison of GC retention time and mass spectrum patterns with those of authentic standards. For the calculation of specific activities, 10 μM of IAMT1 recombinant protein was reacted with 100 μM IAA or PAA, 1 mM SAM, and 50 mM Tris-HCl, at pH 7.5 and 40°C for 30 min at a 50 μL volume. The reactions were initiated by the addition of SAM and stopped by the addition of 50 μL acetonitrile. The reaction products were extracted with 100 μL of n-hexane containing 1 ng/μL ethyl PAA (EtPAA) as an internal standard and were analyzed by GC-MS.
2.4. Generation of IAMT1ox plants
The full-length IAMT1 cDNA was provided by RIKEN BioResource Research Center, Japan. The ORF of IAMT1 was amplified by adapter PCR from the RAFL cDNA clone using gene-specific primers (IAMT1-F and IAMT1-R) and adaptor attB primers (adaptor attB1 and adaptor attB2). The IAMT1 fragment was inserted into the pDONR/Zeo vector using the BP clonase II reaction and then this entry clone was transferred into the pMDC7 vector [24] by the LR clonase II reaction (Invitrogen, Waltham, MA). Arabidopsis WT plants were transformed with the resulting construct, pMDC7:IAMT1, using the floral dip method with the Agrobacterium tumefaciens GV3101 pMP90 strain.
2.5. Generation of CRISPR/Cas9-based iamt1 mutants
Knockout mutants for the IAMT1 gene were generated using CRISPR/Cas9-based genome editing methods previously described [25,26]. We designed two guide RNAs to target the IAMT1 gene. The target sequences were GACTTCTCAGCATGAAAGGTGG and CCGAGCCACTAGCCATGCCAAA. The iamt1-c1 and iamt1-c2 mutants were genotyped using the gene-specific primers, IAMT1-GT1 and IAMT1-GT2. The iamt1 mutants would generate a PCR fragment approximately 620 bp when the primer pair IAMT1-GT1/GT2 was used, whereas using the same primers, WT plants produced a 3.7 kb fragment, which often failed to amplify. To determine zygosity, we used IAMT1-GT1 and IAMT1-GT3 primers for PCR reactions. WT plants and heterozygous plants produced a 620 bp fragment, whereas iamt1 mutants could not produce any PCR fragments.
2.6. qRT-PCR analysis
Total RNA was isolated from pER8, IAMT1ox, iamt1-c1, and iamt1-c2 plants using RNeasy (Qiagen, Venlo, Netherlands). The PrimeScriptTM RT reagent kit with a gDNA eraser (Takara Bio, Kusatsu, Japan) was used to generate first-strand cDNA. qRT-PCR was performed on a G8830A AriaMx Real-time PCR system (Agilent Technologies, Santa Clara, CA) using a THUNDERBIRD SYBR qPCR mix (Toyobo, Osaka, Japan) and specific primers are listed in Table S1.
2.7. LC-MS/MS analysis of auxin metabolites
LC-ESI-MS/MS analysis of IAA, oxIAA, PAA, PAA-Asp, and PAA-Glu was performed as previously described [22].
3. Results
3.1. IAMT1 can catalyze the conversion of PAA to MePAA in vitro
Because of the structural similarity of PAA to IAA, we hypothesized that IAMT1 may convert PAA to its methyl ester (Fig. 1). To investigate this hypothesis, we expressed the trigger factor (TF)-fused IAMT1 proteins in E. coli using the pCold-TF vector. After removal of the TF region, recombinant IAMT1 proteins were reacted with IAA or PAA in the presence of the methyl group donor SAM. After the reaction, we analyzed MeIAA and MePAA using gas chromatography-mass spectrometry (GC-MS). MeIAA was identified using a molecular ion at m/z 189 and a base ion at m/z 130 (Fig. 1C, D). Similarly, MePAA was identified using a molecular ion at m/z 150 and a base ion at m/z 91 (Fig. G, H). As shown in Fig. 1E and F, IAMT1 proteins expressed in E. coli formed MeIAA from IAA, as previously reported [27]. We found that IAMT1 proteins also exhibited the carboxyl methyltransferase activity against PAA and formed MePAA (Fig. 1I, J). Furthermore, MePAA was not detected from the reaction mixture with denatured IAMT1 protein. IAMT1 displayed similar specific activity as IAA (0.64 nmol min−1 mg−1 protein) and PAA (0.65 nmol min−1 mg−1 protein), respectively.
Fig. 1.
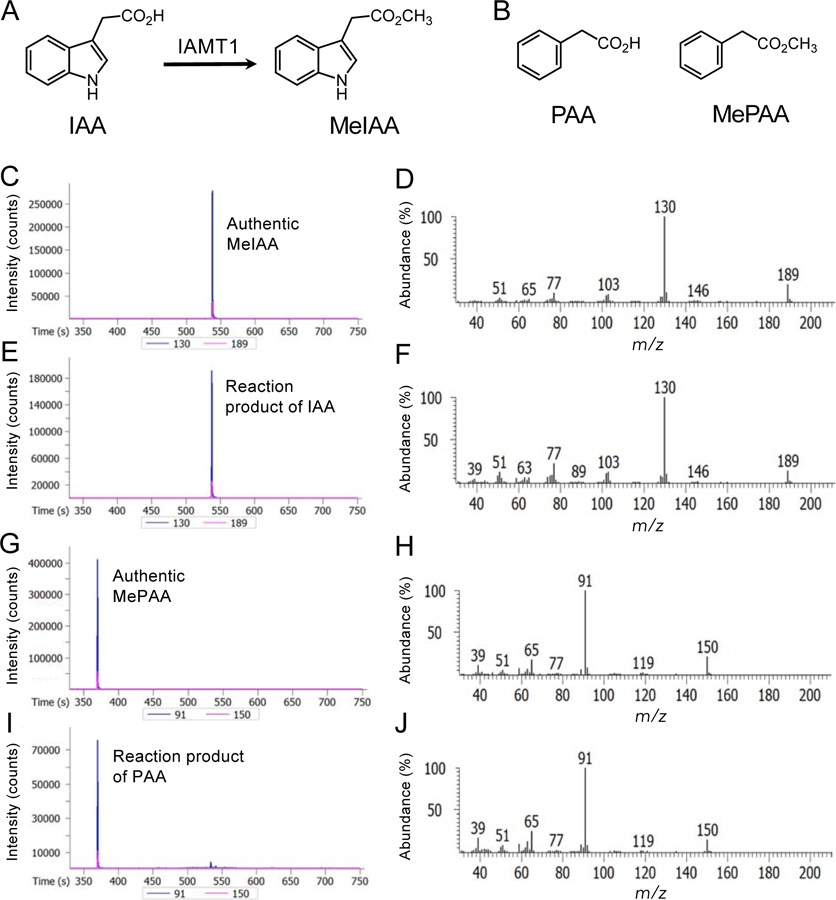
Conversion of IAA and PAA to their methyl esters by IAMT1 in vitro.
A) Conversion of IAA to MeIAA by IAMT1. B) Structures of PAA and MePAA. C) GC-MS chromatogram of authentic MeIAA (m/z 130 and 189). D) Full mass spectra of authentic MeIAA. E) GC-MS chromatogram of the reaction product of IAMT1 with IAA (m/z 130 and 189). F) Full mass spectra of the reaction product of IAMT1 with IAA. G) GC-MS chromatogram of authentic MePAA (m/z 91 and 150). H) Full mass spectra of authentic MePAA. I) GC-MS chromatogram of the reaction product of IAMT1 with PAA (m/z 91 and 150). J) Full mass spectra of the reaction product of IAMT1 with PAA.
3.2. Overexpression of IAMT1 results in the reduction of IAA levels but not PAA levels
A previous study showed that the constitutive overexpression of IAMT1 (35S-IAMT1) results in a series of reduced-auxin phenotypes in Arabidopsis [16]. However, it has not been demonstrated whether the endogenous levels of auxins were altered by the overexpression of IAMT1. The results from our biochemical characterization of IAMT1 suggest that endogenous levels of both IAA and PAA may be reduced by overexpression of IAMT1 in plants. To investigate the role of IAMT1 in auxin metabolism in vivo, we generated transgenic plants harboring the IAMT1 gene in an ER-inducible expression system using a pMDC7 vector (IAMT1ox) (Fig. 2A). Similar to 35S-IAMT1 [16], two independent lines of IAMT1ox1 and IAMT1ox2 exhibited severe auxin-deficient phenotypes, including agravitropic root growth and reduced root length, upon induction of the overexpression of IAMT1 by ER treatment (Fig. 2A, B and Fig. S1). For auxin analysis, we used IAMT1ox1 plants, in which the levels of IAMT1 transcripts were increased by 750-fold in MS medium containing ER (Fig. 2C). We analyzed the endogenous IAA and PAA levels in whole seedlings using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Interestingly, no significant changes in IAA and PAA levels were observed in whole seedlings of IAMT1 overexpressing plants (Fig. 2D). Given that the overexpression of IAMT1 results in an agravitropic root phenotype, we assumed that the auxin levels might be reduced in root apexes, which accumulate relatively higher levels of IAA. To test this hypothesis, we collected root tips of IAMT1ox1 seedings and analyzed the levels of auxins. The IAA levels were reduced by 70% in the root tips of IAMT1 overexpressing plants compared to the levels in mock-treated plants, whereas the PAA levels were not changed (Fig. 2E). Similarly, the levels of 2-oxindole-3-acetic acid (oxIAA), one of the major IAA metabolites in Arabidopsis, were reduced by 65% in the root tips of IAMT1 overexpressing plants (Fig. 2E). These results suggest that IAMT1 selectively metabolizes IAA in Arabidopsis.
Fig. 2.
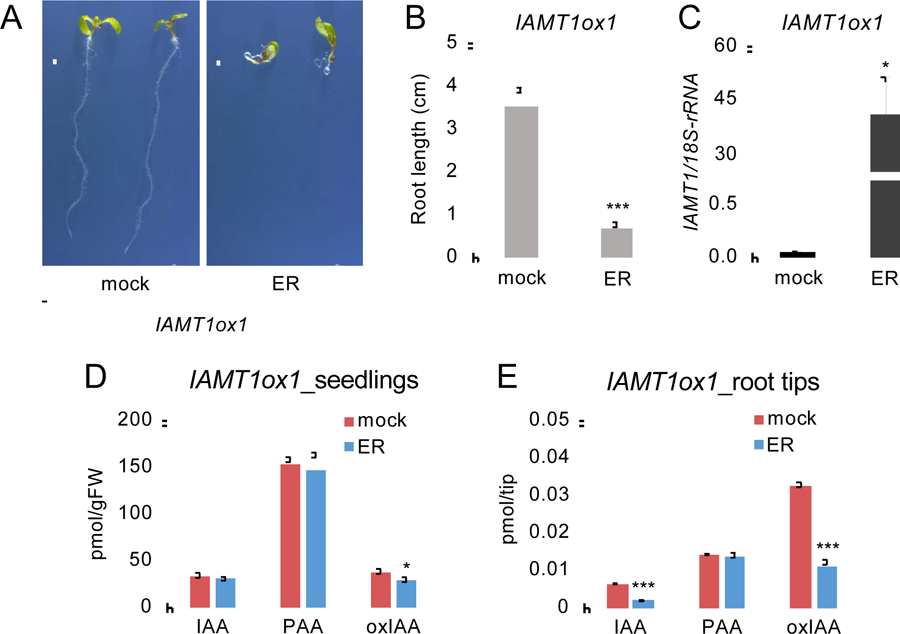
Analysis of IAMT1ox1 plants.
A) Phenotypes of IAMT1ox1 plants. Plants were treated with mock or ER (2 μM) for 4 days. Scale bars = 1 cm. B) Primary root length of IAMT1ox1 plants. Plants were treated with mock or ER for 5 days. Differences between mock- and ER-treated plants are statistically significant at P < 0.05 (***P < 0.001, Student’s t-test). C) Expression levels of IAMT1 in mock- and ER-treated IAMT1ox1 plants. Differences between mock- and ER-treated plants are statistically significant at P < 0.05 (*P < 0.05, Student’s t-test). D) Auxin levels in whole seedlings of mock- and ER-treated IAMT1ox plants. Differences between mock- and ER-treated plants are statistically significant at P < 0.05 (*P < 0.05, Student’s t-test). E) Auxin levels in root tips of mock- and ER-treated IAMT1ox plants. Differences between mock- and ER-treated plants are statistically significant at P < 0.05 (***P < 0.001, Student’s t-test).
We attempted to analyze the levels of MeIAA and MePAA in the WT and IAMT1ox1 plants using GC-MS, but we were not able to quantify these metabolites in this study.
3.3. CRISPR/Cas9-based iamt1 knockout mutants do not exhibit obvious phenotypes
To further investigate a role of IAMT1 in auxin homeostasis, we generated null mutants of IAMT1 (iamt1-c1 and -c2) using a CRISPR/Cas9-based genome editing approach (Fig. 3) [25]. Using genomic DNA sequence analysis, we confirmed that more than 90% of the coding regions of IAMT1 were deleted in the iamt1-c1 and -c2 lines, respectively (Fig. 3 and Fig. S2A). The genotypic analysis further confirmed that iamt1-c1 and -c2 lines are null mutants (Fig. S2B). Although iamt1-RNAi mutants reportedly showed high-auxin phenotypes [16], no obvious phenotypes were observed in our iamt1-c1 and -c2 lines compared to that of WT plants under normal growth conditions (Fig. 3B–D). We analyzed the levels of IAA and PAA in iamt1-c1 seedlings, but these auxin levels were comparable with that of WT plants (Fig. 3E). No increase of oxIAA levels was observed in the iamt1-c1 seedlings either (Fig. 3E). These results suggested that disruption of IAMT1 does not affect auxin levels in Arabidopsis. A previous study demonstrated that the hypocotyls of T-DNA-inserted iamt1–1 mutants showed an abnormal gravitropic response [15]. To test this, we analyzed the gravitropic response of iamt1-c1 and -c2 under dark conditions. However, these mutants showed normal gravitropism of hypocotyls (Fig. S3), suggesting that the iamt1 single knockout mutants did not display obvious phenotypes.
Fig. 3.
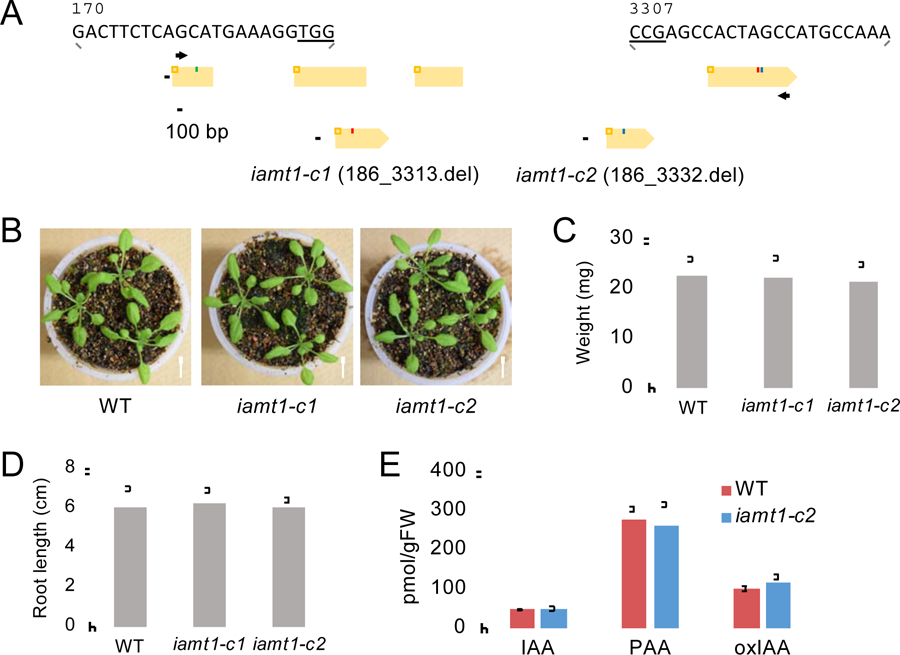
Characterization of the iamt1-c mutants.
A) Mutations in iamt1-c1 and -c2 mutants. Orange boxes represent the exons and black lines represent the introns in the IAMT1 gene. Arrows indicate gene-specific primers used for genotyping. B) Phenotype of 17 d-old iamt1-c seedlings. Scale bars = 1 cm. C) Weight of shoot tissues of 12 d-old iamt1-c seedlings (n = 30). D) Primary root length of 8 d-old iamt1-c seedlings (n = 30). E) Auxin metabolite levels in whole seedlings of 8 d-old WT and iamt1-c2 mutants (n = 4).
3.4. Screening of auxin carboxyl methyltransferases in the SABATH family
There are 24 members in the Arabidopsis SABATH family of carboxyl methyltransferases [15,28]. The results of our iamt1-c and previous RNAi-iamt1 mutant analysis suggest that other auxin carboxyl methyltransferases may play a redundant role in Arabidopsis [16]. Based on the phylogenetic analysis of the Arabidopsis SABATH family [15], we noticed that members of the family are divided into three groups (Table S2). IAMT1 belongs to the same clade as benzoate/salicylate carboxyl methyltransferase (BSMT1), which is different from that of the other two groups with gibberellin methyltransferase (GAMT) or farnesoic acid carboxyl methyltransferase (FAMT) [15]. To explore novel auxin carboxyl methyltransferases, we investigated IAMT1 homologs in the BSMT1 clade (Table S2). Among 15 members in the BSMT1 clade, we successfully expressed six IAMT1 homologs as the TF-fused proteins in E. coli and performed the enzyme activity test using IAA and PAA as substrates (Table S2). However, none of these proteins, including BSMT1, showed methyltransferase activity, whereas IAMT1 catalyzed the methylation of IAA and PAA (Table S2).
4. Discussion
Despite the severe auxin-deficient phenotypes of IAMT1ox plants, significant changes in auxin levels have not been demonstrated. In this study, we found that IAA levels were reduced in root tip tissues, but not in the entire seedling (Fig. 2D). Although IAMT1 can convert PAA to MePAA in vitro, the levels of PAA were not altered in IAMT1ox plants (Fig. 2D). These results suggest that IAMT1 exclusively metabolizes IAA in Arabidopsis. Therefore, we conclude that auxin-deficient phenotypes of IAMT1ox plants are attributed to a reduction in IAA. Regarding the discrepancy of auxin-deficient phenotypes and normal IAA levels in the entire seedling of IAMT1ox plants, this may be caused by the IAA homeostasis by the Arabidopsis MeIAA esterase (AtMES) family [11]. Yang et al. (2008) demonstrated that members of the AtMES family, including AtMES17, showed esterase activity and converted MeIAA to IAA in vitro. Given that the AtMES family consists of 20 homologs, various AtMES proteins may hydrolyze MeIAA to strictly maintain IAA levels in IAMT1ox plants. We assume that the esterase activity would be relatively low in root tips because a significant reduction of IAA levels was observed in these tissues of IAMT1ox plants. Investigation of the role of AtMES family genes in IAMT1ox plants is important to elucidate metabolic regulation of MeIAA in Arabidopsis.
Here, we demonstrated that CRISPR/Cas9-based iamt1-c1 and -c2 null mutants neither show obvious phenotypes, nor an increase in IAA levels. The reasons for the differences between the phenotype of our CRISPR/Cas9-based iamt1-c mutants and previously described iamt1-RNAi and T-DNA inserted iamt1–1 lines are not clear. A previous study showed that iamt1–1 mutants still produce the same levels of IAA and reduced levels of MeIAA compared to that of WT plants [15]. iamt1-RNAi plants might display high-auxin phenotypes because of the simultaneous repression of an additional member of the SABATH family [16]. Combined with the fact that iamt1-c1 and -c2 do not show obvious phenotypes, we assumed that other auxin carboxyl methyltransferase genes may exert a redundant property in Arabidopsis. We tested the enzyme activity of six IAMT1 homologs in the SABATH family in this study, but no proteins were shown to be auxin carboxyl methyltransferase other than IAMT1 (Table S2). Further biochemical analysis of members in the SABATH family is crucial to understanding IAA homeostasis in Arabidopsis.
Supplementary Material
Supplementary Fig. S1. Phenotypes of IAMT1ox2 plants.
Supplementary Fig. S2. Conformation of iamt1-c mutation in Arabidopsis.
Supplementary Fig. S3. Gravitropic response of iamt1-c mutants.
Supplementary Table S2. Auxin carboxyl methyltransferase activity of IAMT1 homologs.
Supplementary Table S1. Primers used in this study.
Highlights.
Arabidopsis IAMT1 protein can catalyze the methylation of both IAA and PAA in vitro.
Overexpression of IAMT1 reduces IAA levels and results in auxin-deficient phenotypes.
No reduction in PAA levels was observed following overexpression of IAMT1.
Knockout of IAMT1 did not affect auxin levels in Arabidopsis.
IAMT1 homologs may also participate in Arabidopsis IAA homeostasis.
Acknowledgements
We thank Dr. Kazuki Saito for kindly supporting the auxin metabolite measurements.
Funding
This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI [JP18H02457] to H.K. and by KAKENHI [JP19H03253] and Promotion of OUS Research Project [OUS-RP-19-4] to K.H. This work was partially supported by NIH [GM114660] to Y.Z.
Abbreviations:
- gFW
gram fresh weight
- IAA
indole-3-acetic acid
- IAMT1
IAA CARBOXYL METHYLTRANSFERASE 1
- MeIAA
IAA-methyl ester
- MePAA
PAA-methyl ester
- PAA
phenylacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- [1].Ljung K, Auxin metabolism and homeostasis during plant development, Development 140 (2013) 943–950. 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- [2].Kasahara H, Current aspects of auxin biosynthesis in plants, Biosci. Biotechnol. Biochem 80 (2016) 34–42. 10.1080/09168451.2015.1086259. [DOI] [PubMed] [Google Scholar]
- [3].Friml J, Subcellular trafficking of PIN auxin efflux carriers in auxin transport, Eur. J. Cell Biol 89 (2010) 231–235. 10.1016/j.ejcb.2009.11.003. [DOI] [PubMed] [Google Scholar]
- [4].Zhao Y, Christensen SK, Fankhauser C, et al. , A role for flavin monooxygenase-like enzymes in auxin biosynthesis, Science 291 (2001) 306–309. 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- [5].Tao Y, Ferrer JL, Ljung K, et al. , Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants, Cell 133 (2008) 164–176. 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stepanova AN, Robertson-Hoyt J, Yun J, et al. , TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development, Cell 133 (2008) 177–191. 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- [7].Mashiguchi K, Tanaka K, Sakai T, et al. , The main auxin biosynthesis pathway in Arabidopsis, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 18512–18517. 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Won C, Shen X, Mashiguchi K, et al. , Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 18518–18523. 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Korasick DA, Enders TA, Strader LC, Auxin biosynthesis and storage forms, J. Exp. Bot 64 (2013) 2541–2555. 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li L, Hou X, Tsuge T, et al. , The possible action mechanisms of indole-3-acetic acid methyl ester in Arabidopsis, Plant Cell Rep 27 (2008) 575–584. 10.1007/s00299-007-0458-9. [DOI] [PubMed] [Google Scholar]
- [11].Yang Y, Xu R, Ma CJ, et al. , Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis, Plant Physiol 147 (2008) 1034–1045. 10.1104/pp.108.118224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ishimaru K, Hirotsu N, Madoka Y, et al. , Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield, Nat. Genet 45 (2013) 707–711. 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- [13].Han W, Han D, He Z, et al. , The SWI/SNF subunit SWI3B regulates IAMT1 expression via chromatin remodeling in Arabidopsis leaf development, Plant Sci 271 (2018) 127–132. 10.1016/j.plantsci.2018.03.021. [DOI] [PubMed] [Google Scholar]
- [14].Abbas M, Hernandez-Garcia J, Blanco-Tourinan N, et al. , Reduction of indole-3-acetic acid methyltransferase activity compensates for high-temperature male sterility in Arabidopsis, Plant Biotechnol. J 16 (2018) 272–279. 10.1111/pbi.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Abbas M, Hernandez-Garcia J, Pollmann S, et al. , Auxin methylation is required for differential growth in Arabidopsis, Proc. Natl. Acad. Sci. U.S.A 115 (2018) 6864–6869. 10.1073/pnas.1806565115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Qin G, Gu H, Zhao Y, et al. , An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development, Plant Cell 17 (2005) 2693–2704. 10.1105/tpc.105.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sugawara S, Mashiguchi K, Tanaka K, et al. , Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants, Plant Cell Physiol 56 (2015) 1641–1654. 10.1093/pcp/pcv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cook SD, An historical review of phenylacetic acid, Plant Cell Physiol 60 (2019) 243–254. 10.1093/pcp/pcz004. [DOI] [PubMed] [Google Scholar]
- [19].Schneider EA, Kazakoff CW, Wightman F, Gas chromatography-mass spectrometry evidence for several endogenous auxins in pea seedling organs, Planta 165 (1985) 232–241. 10.1007/BF00395046. [DOI] [PubMed] [Google Scholar]
- [20].Wightman F, Lighty DL, Identification of phenylacetic acid as a natural auxin in the shoots of higher plants, Physiol. Plant 55 (1982) 17–24. [Google Scholar]
- [21].Shimizu-Mitao Y, Kakimoto T, Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB, Plant Cell Physiol 55 (2014) 1450–1459. 10.1093/pcp/pcu077. [DOI] [PubMed] [Google Scholar]
- [22].Mashiguchi K, Hisano H, Takeda-Kamiya N, et al. , Agrobacterium tumefaciens enhances biosynthesis of two distinct auxins in the formation of crown galls, Plant Cell Physiol 60 (2019) 29–37. 10.1093/pcp/pcy182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Aoi Y, Tanaka K, Cook SD, Hayashi KI, Kasahara H, GH3 Auxin-amido synthetases alter the ratio of indole-3-acetic acid and phenylacetic acid in Arabidopsis, Plant Cell Physiol 61 (2020) 596–605. 10.1093/pcp/pcz223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Curtis MD, Grossniklaus U, A gateway cloning vector set for high-throughput functional analysis of genes in planta, Plant Physiol 133 (2003) 462–469. 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gao X, Chen J, Dai X, Zhang D, Zhao Y, An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing, Plant Physiol 171 (2016) 1794–1800. 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y, Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development, Proc. Natl. Acad. Sci. U.S.A 112 (2015) 2275–2280. 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP, Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family, Plant Cell 15 (2003) 1704–1716. 10.1105/tpc.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang B, Li M, Yuan Y, Liu S, Genome-wide comprehensive analysis of the SABATH gene family in Arabidopsis and rice, Evol. Bioinform Online 15 (2019) 1176934319860864. 10.1177/1176934319860864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Phenotypes of IAMT1ox2 plants.
Supplementary Fig. S2. Conformation of iamt1-c mutation in Arabidopsis.
Supplementary Fig. S3. Gravitropic response of iamt1-c mutants.
Supplementary Table S2. Auxin carboxyl methyltransferase activity of IAMT1 homologs.
Supplementary Table S1. Primers used in this study.


