Abstract
Purpose:
Progression of prostate cancer is a complex multistep process that involves molecular alterations in cells of the tumor and microenvironment with associated interactions between the stroma and epithelium. We performed genomic expression analyses of stromal infiltration markers to determine the prognostic significance thereof in prostate cancer.
Materials and Methods:
Genome-wide expression profiles of formalin-fixed paraffin-embedded radical prostatectomy samples were evaluated from a prospective registry cohort (n=5,239) and three retrospective institutional cohorts (n=1,135). Two independent stromal gene expression signatures inferred stromal infiltration. Cox proportional hazards regression defined the association between stromal infiltration expression and metastasis-free survival. Cox proportional hazards regression defined the association between stromal infiltration expression and metastasis-free survival.
Results:
Stromal expression scores were correlated with each other and with key stromal markers (CAV1, VIM, TAGLN), basal activity, and CD3 and CD4 immune biomarkers (r>0.5 for all). The top decile of stromal expression was associated with higher genomic-risk score, high CAPRA-S, Gleason 9–10 disease, and a higher risk for metastasis (HR:2.35[1.35–4.08],p=0.002). Higher stromal infiltration score was also associated with decreased expression of DNA repair genes and higher radiation sensitivity genomic scores. Post-operative radiation therapy (RT) was associated with a metastasis-free survival (MFS) benefit for patients with high stromal scores, but not for patients with low stromal scores (Pinteraction=0.02).
Conclustions:
Expression of stromal infiltration markers is correlated with prostate cancer aggressiveness/progression and may be predictive of response to radiation therapy. Stromal infiltration markers should be studied and considered for incorporation into clinical prognostication and decision-making.
Keywords: Prostatic Neoplasms, Genomics, Decipher, Stromal infiltration, Tumor microenvironment
PRECIS FOR TABLE OF CONTENTS:
High genomic expression of stromal infiltration markers was associated with aggressive disease and adverse prostate cancer outcomes. Stromal infiltration markers should be considered for incorporation into clinical prognostication and decision-making.
INTRODUCTION:
Prostate cancer is the most commonly diagnosed non-skin cancer in men.1 Prognostication and treatment decisions have been guided by tumor stage, prostate cancer-specific antigen, and Gleason score for the last several decades.2 Nevertheless, progression of prostate cancer is a complex multistep process that involves molecular alterations in cells of the tumor and microenvironment with associated interactions between the stroma and epithelium that can’t be entirely accounted for by clinical factor risk criteria alone.3
Genomics in prostate cancer has lead to closer investigation and understanding of molecular alterations in cells of the tumor and microenvironment.4–6 As such, genomics are increasingly being incorporated into into prognostication, treatment decisions, and targeted therapy design in prostate cancer, as they enhance our understanding of prostate cancer. Still, the clinical significance and implications of stromal infiltration in primary prostate cancer is not well defined or understood.3
Therefore, we performed genomic expression analyses of stromal infiltration markers and sought to determine the clinical significance thereof in prostate cancer.
MATERIALS AND METHODS:
Study Cohorts
Genome-wide expression profiles of formalin-fixed paraffin-embedded radical prostatectomy (RP) tumor samples were evaluated from a prospective registry cohort (n=5,239) and three retrospective institutional cohorts (n=1,135). The TCGA-prostate cohort was used for validation across platforms (N=498).7 The prospective cohort was comprised of anonymized genome-wide expression profiles from clinical use of the Decipher test between February 2014 to August 2016 retrieved from the Decipher GRID™ ( NCT02609269) and included basic demographic and pathological data. The retrospective cohorts included patients treated with RP at Johns Hopkins University (JHU, n=355) and Mayo Clinic (MC-I, n=545 and MC-II, n=235) and included adequate follow-up for the endpoint of metastasis-free survival (MFS).8–10 Supplemetal Figure 1 summarizes patient cohorts in a flow diagram.
Central pathology review was performed for all cases. Prior to tissue sampling for the clinical Decipher assay, histologic review of the submitted FFPE block was performed by a pathologist. Details regarding pathology procedures including microarray preprocessing and normalization have been previously described.11–13 Notably, an attempt was made to identify all available FFBE blocks (including lymph node blocks), where the block containing the dominant Gleason tumor was selected for RNA isoloation.11 From there, freshly cut sections from the FFPE blocks (Four 10 μm sections) were deparaffinized before macrodissection of the dominant Gleason tumor for RNA extraction. The acceptance criteria for the Decipher assay include at least 0.5 cm2 of tumor with at least 60% neoplastic cells. Details regarding RNA extraction and laboratory methods have been previously described.4
Statistical analysis
We used the ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data) algorithm of 141 stromal genes to infer stromal infiltration from gene expression data.14 Additionally, we used a 27 gene stromal signature3 with overlap of 9 genes from the 141-gene signature. For the TCGA-PARD cohort, we downloaded the ESTIMATE stromal scores, IHC and Consensus measurement of Purity Estimates (CPE) scores.7 ERG-fusion frequency was examined across deciles of stromal scores as a proxy for tumor purity/tumor signal.15
The distribution of high genomic-risk (Decipher score≥0.6), high CAPRA-S, and Gleason 9–10 disease across deciles of stromal infiltration expression was assessed.
Cox proportional hazards regression defined the association between stromal infiltration expression (high=top decile versus low) and MFS (metastases defined by radiographic evidence) after RP; a multivariable analysis was also performed with adjustment for Gleason score to evaluate the association of stromal infiltration expression and MFS, independent of Gleason score. Lastly, associations between stromal infiltration and radiation response scores were tested using a 24-gene radiation sensitivity signature (PORTOS: Post-Operative Radiation Therapy Outcomes Score)12 and an IFN-related DNA damage resistance signature. Cox proportional hazards examined the association between stromal infiltration (high=top decile versus low) and MFS by receipt of post-RP radiation therapy (RT) with a stromal infiltration*RT interaction term, using a previously-published, matched cohort (n=196; half of the patients received post-RP RT); this cohort was specifically matched (exact 1:1) on preoperative PSA, surgical Gleason score, surgical margin status, extracapsular extension, seminal vesicle invasion, lymph node invasion, and androgen deprivation therapy.12
Spearman’s correlation was used for correlation analysis. Statistical analyses were performed in R v3.3.1, and a 5% significance level was applied for all tests. Local institutional review boards (IRB) approved all data collection.
RESULTS:
Baseline characteristics
In the prospective (n=5,239) and retrospective cohorts (n=1,135), at diagnosis the median age was 65 and 64, median PSA was 6.6 and 9, and there were 18% and 43% of patients with Gleason8–10 scores, respectively.
Distrubtion of stromal genomic expression
In the prospective cohort, there was strong correlation between stromal expression scores (based on ESTIMATE algorithm) and the 141 genes composing that signature (Supplemental Figure 2A). The stromal score was strongly correlated with key well-established stromal markers (genes) not included in the 141 gene stromal signature (CAV1 [r=0.59], VIM [r=0.74], TAGLN [r=0.62], CNN1 [r=0.6]), basal activity (r=0.72), and CD3 (r=0.45) and CD4 (r=0.5) immune biomarkers, and with another independent stromal score based on 27 genes (r=0.84) (Supplemental Figure 2B). Furthermore, ERG-fusion frequency was similar across deciles of stromal scores (Supplemental Figure 2C). Since IHC data for stromal markers were not available in the GRID data, we used IHC data and Consensus Purity Estimates from TCGA-PRAD.7 Both tumor purity measures were negatively associated with stromal score, indicating stromal score reflects stromal infiltration (Supplemental Figure 2D–E). Stromal expression scores were similar across Gleason score and genomic (Decipher) risk score (Supplemental Figure 3).
Outcomes by stromal genomic expression
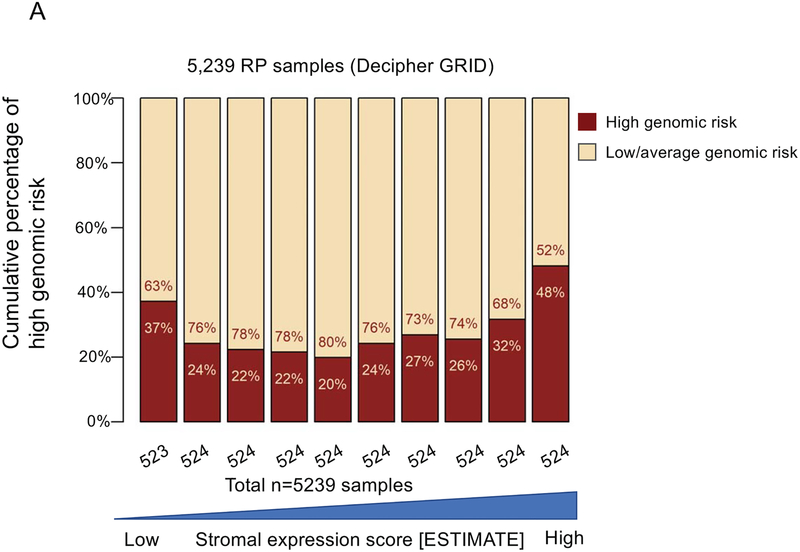
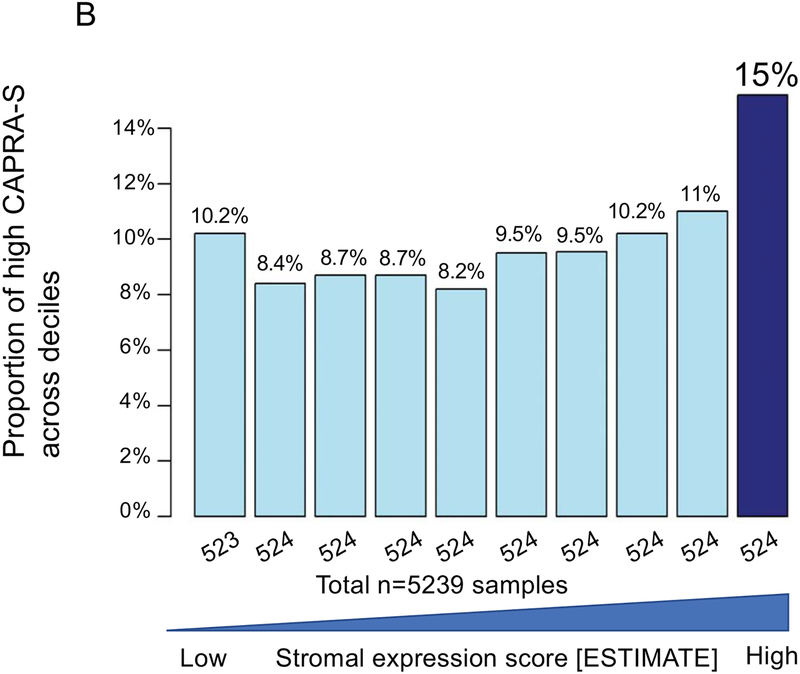
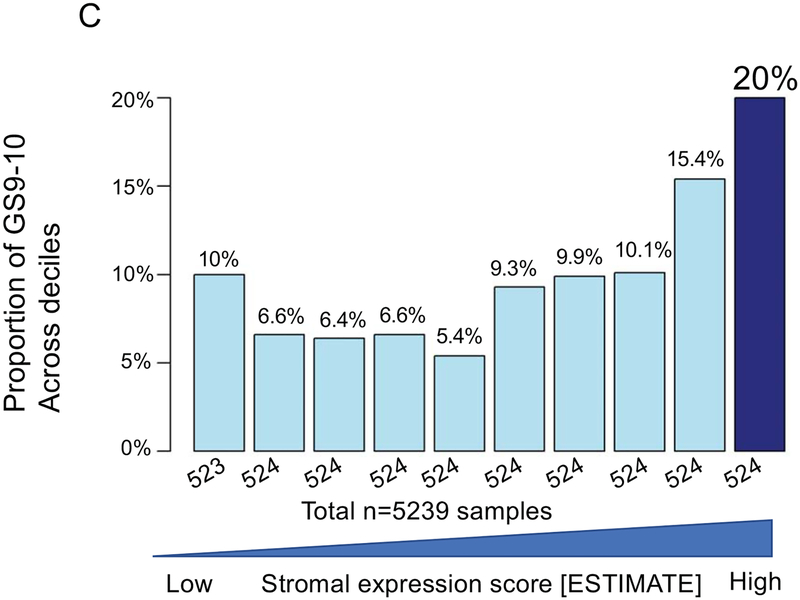
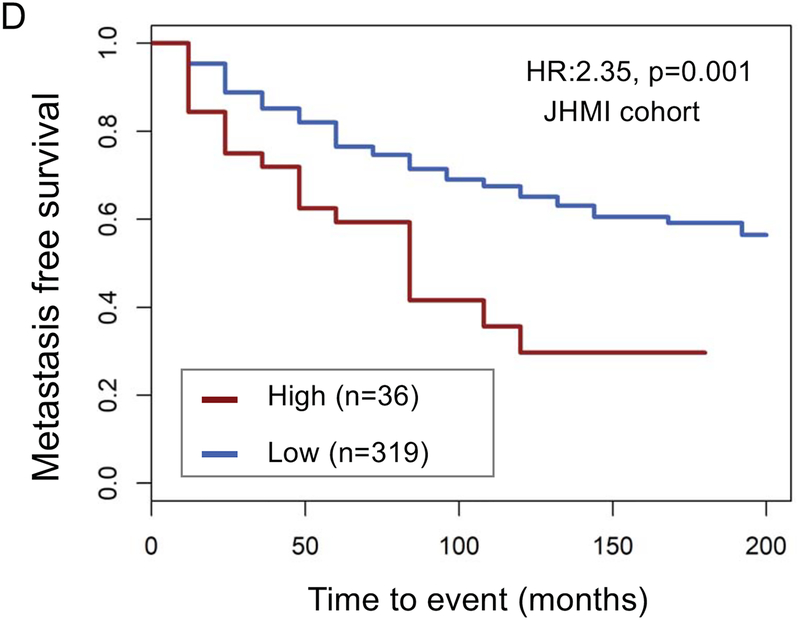
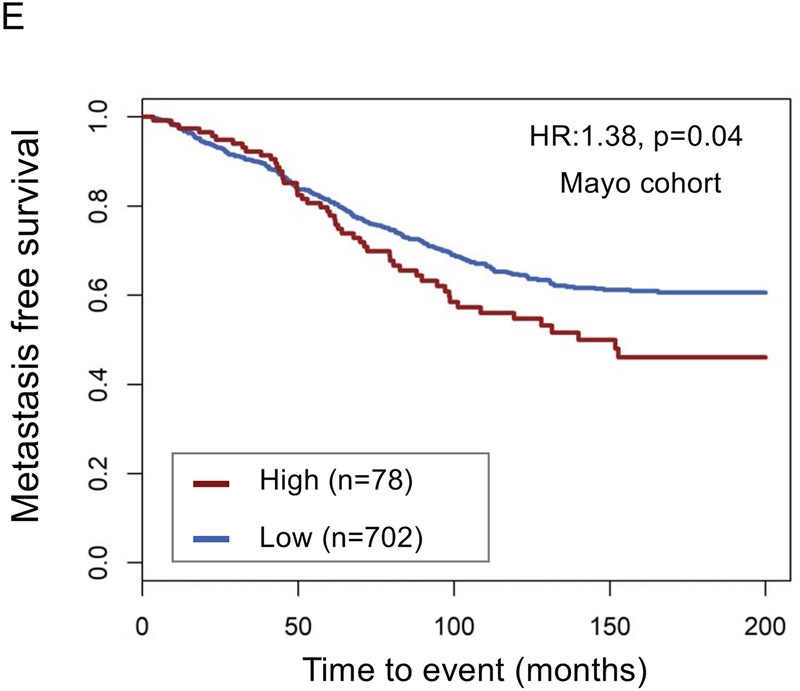
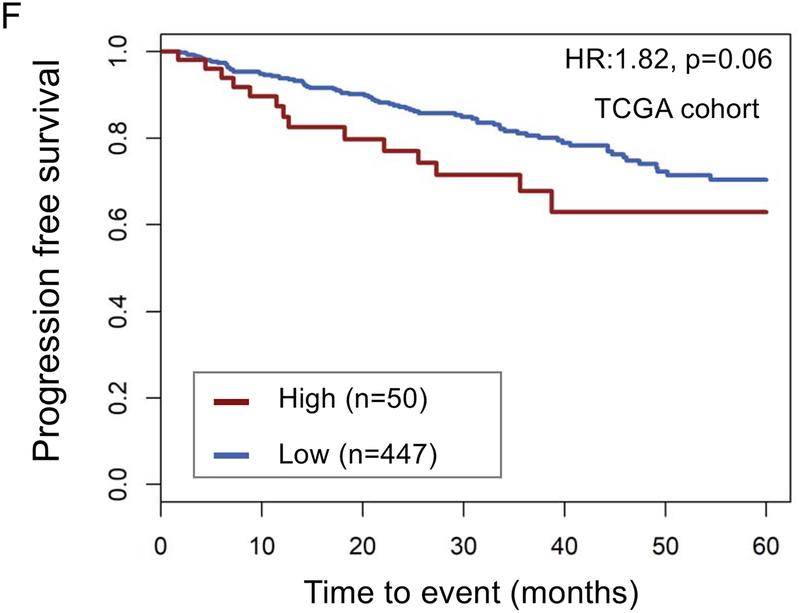
The top decile of stromal expression was associated with high genomic-risk (Decipher≥0.6), high CAPRA-S, and Gleason 9–10 disease (p<0.05 all, Mann-Kendall trend test) (Figure 1A–C). The distribution stromal expression across Gleason and genomic (Decipher) risk scores is displayed in Supplemental Figure 3; notably, 41% of Gleason 3+3 tumors, 47% of Gleason 3+4 tumors, and 48% of Gleason 4+3 tumors had stromal expression scores above the median and 36% of Gleason 9–10 tumors had stromal expression scores below the median. The top decile of stromal expression (compared with lower stromal expression) was associated with higher risk of metastasis in the JHU (Hazard Ratio [HR] 2.35[1.37–4.02],p=0.001) and MC cohorts (HR1.38[1.02–1.86],p=0.04) [Figure 1 D–E]; there was higher, but non-significant, risk of disease progression in the TCGA cohort (HR1.82[0.94–3.50],p=0.06) (Figure 1F). On multivariable analysis with adjustment for Gleason score, the top decile of stromal score remained independently associated with a higher risk of metastasis (adjusted HR 2.15, 95%CI[1.25–3.7], p=0.005).
Figure 1.
Distribution of (a) high genomic risk score (Decipher scores ≥0.6), (b) high CAPRA-S scores, and (c) Gleason 9–10 disease across deciles of stromal expression scores (p<0.05 Mann-Kendall trend test). Survival analysis stratified by stromal score (high=top decile) of metastasis-free survival over time after radical prostatectomy in the (d) John’s Hopkins University cohort, (e) Mayo Clinic cohorts, and progression-free survival in the (f) TCGA cohort.
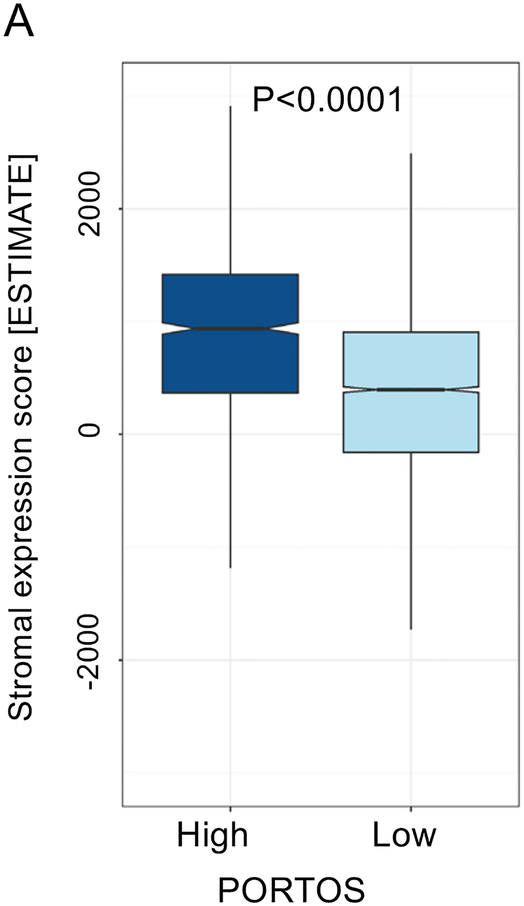
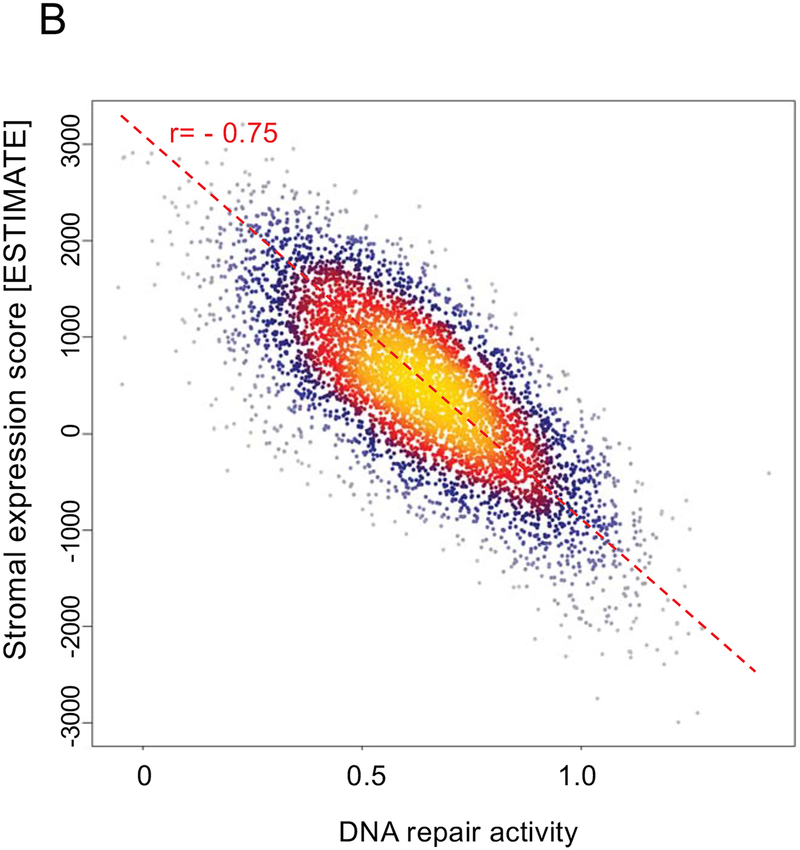
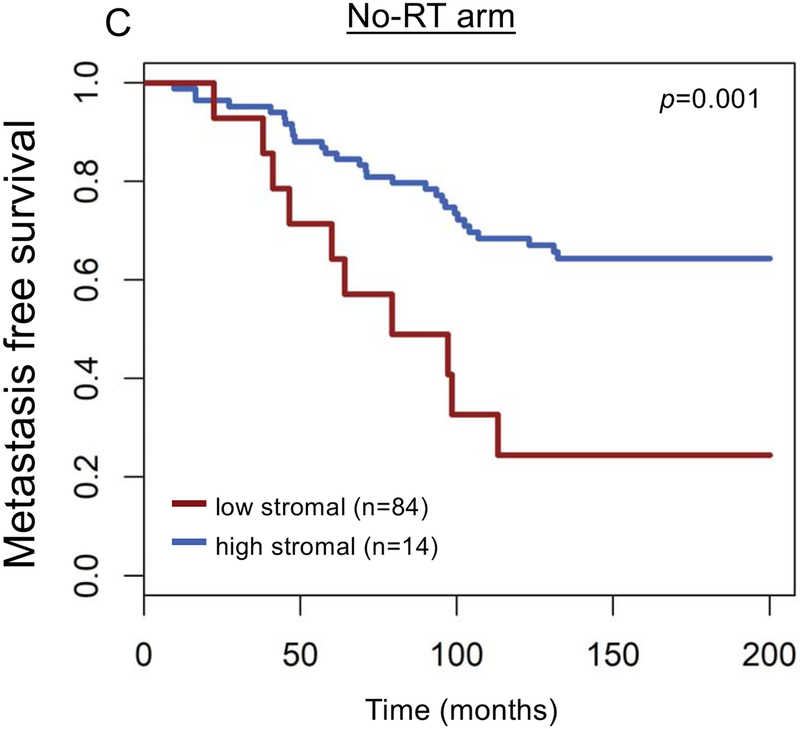
Furthermore, stromal score was correlated with radiation sensitivity PORTOS score (r=0.37), and high PORTOS score (>0) was associated with higher stromal infiltration (p<0.001) (Figure 2A). Stromal score was also negatively correlated with DNA repair activity (r= −0.75) (Figure 2B). On clinical analyses in a matched cohort of patients treated with RT (n=98) and patients with no-RT (n=98), post-operative radiation therapy (RT) was associated with a metastasis-free survival (MFS) benefit for patients with high (top decile) stromal scores, but not for patients with low stromal scores (Pinteraction=0.02; Figure 2C–E); 10-year MFS rates for high versus low stromal scores were 24% versus 68% (P=0.0015) and 50% versus 54% (P=0.45) for patients who did not receive RT versus patients who received RT, respectively.
Figure 2.
Association between stromal expression scores and (a) 24-gene radiation sensitivity signature (PORTOS: Post-Operative Radiation Therapy Outcomes Score), (b) DNA repair activity signature, and (c-e) metastasis-free survival by high (top-decile) versus low stromal expression and receipt of post-radical prostatectomy radiation therapy.
DISCUSSION:
This study highlights the novel findings that expression of stromal infiltration markers is correlated with prostate cancer aggressiveness/progression and may be predictive of response to radiation therapy. Specifically, high expression of stromal infiltration markers was associated with high-risk Decipher genomic-risk score (≥0.6), high CAPRA-S score, Gleason 9–10 disease, and with a higher risk of metastases after RP. Lastly, higher expression of stromal infiltration was associated with high radiation sensitivity genomic scores, low DNA repair activity, and improved MFS with RT. There was an interaction between high stromal expression and receipt of RT such that the significant MFS benefit of RT was limited to patients with high stromal expression. To our knowledge, this study includes the first data to demonstrate such findings.
Together, these results suggest that stromal infiltration marker expression may be both prognostic and predictive in prostate cancer. Notably, though high expression of stromal infiltration markers was associated with high Gleason score, stromal expression was prognostic for risk of metastasis independent of Gleason score on multivariable Cox regression analysis. As such, expression of stromal microenvironment markers may have an important independent role in predicting risk of adverse events in prostate cancer. Furthermore, whether higher expression of stromal infiltration markers is associated with better response to radiation therapy needs further exploration in studies with long clinical follow-up.
Given that infiltrating stromal cells and other immune cells account for a majority of “normal” cells found in solid tumor tissues, these findings have important clinically relevant implications. Mechanisms of prostate cancer development and progression is a complex process that involves alterations of the tumor and microenvironment, where stromal cells likely impact disease progression and treatment response.3 At present, prognostic tools in prostate cancer are principally based on information provided by tumor cells (such as Gleason score, size of tumor, or tumor genomics).16,17 However, increasing evidence suggests that stromal and immune cells are critical for disease progression and drug resistance.18–20
Infiltration of stromal and microenvironment cells may influence genomic or gene expression approaches to prognostic and predictive models given the implications on tumor heterogeneity and purity. The ESTIMATE method uses gene expression data to infer the fractional content of stromal and immune cells in tumor samples, which allows for a straightforward approach to assessing tumor purity and stromal infiltration in tumor samples by using gene expression data.14 Therefore, stromal expression scores can help inform tumor purity/heterogeneity estimates by assessing for the presence of stromal infiltration. Furthermore, the findings in this study suggest that levels of stromal infiltration are likely associated with clinical characteristics and outcomes. With the ongoing shift toward incorporation of genomics into prognostication and trial design in prostate and other cancers, stromal infiltration and other tumor microenvironment markers must be considered.
The major limitation of this study include the lack of long-term clinical follow-up for the prospective cohort to allow for clinical analyses and the inherent limitations of retrospective analyses in the clinical findings. Second, the study was limited by lack of IHC basd stromal quantification for samples from the Decipher cohort. Nevertheless, the clinical analyses were explored in multiple independent retrospective cohorts and ERG+ distribution and purity analyses support strong tumor signal in the findings. Furthermore, the distribution of stromal scores across well-established prostate cancer risk factors suggests possible non-monotonic behavior where low stromal score may also represent an adverse feature, however this study may be underpowered to detect such differences.
Ultimately, stromal infiltration markers should be further investigated and considered for incorporation into clinical trials and ultimately clinical prognostication and treatment decision-making.
CONCLUSION
Despite any potential limitations, this study demonstrated the novel findings that high genomic expression of stromal infiltration markers was associated with aggressive disease, adverse prostate cancer outcomes, and bette response to radiotherapy. Stromal infiltration markers should be considered for incorporation into clinical prognostication and treatment decision-making.
Supplementary Material
FUNDING:
Dr. Mahal is funded by the Prostate Cancer Foundation-American Society for Radiation Oncology Award to End Prostate Cancer (grant support). Dr Rebbeck reported receipt of grants from the National Institutes of Health. Dr. Nguyen is funded by the Prostate Cancer Foundation (grant support). The work was also supported by the Wood Family Foundation, Baker Family, Freedman Family, Fitz’s Cancer Warriors, David and Cynthia Chapin, Frashure Family, Hugh Simons in honor of Frank and Anne Simons, Campbell Family in honor of Joan Campbell, Scott Forbes and Gina Ventre Fund, and a grant from an anonymous family foundation (grant support).
DISCLOSURES:
Dr Nguyen reported receipt of consulting fees from Ferring, Augmenix, Bayer, Janssen, Astellas, Dendreon, Genome DX, Blue Earth Diagnostics, Cota, and Nanobiotix; grant funding from Janssen and Astellas; and equity in Augmenix. No other disclosures were reported.
REFERENCES:
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. Jama. 1998;280(11):969–974. [DOI] [PubMed] [Google Scholar]
- 3.Tyekucheva S, Bowden M, Bango C, et al. Stromal and epithelial transcriptional map of initiation progression and metastatic potential of human prostate cancer. Nat Commun. 2017;8(1):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spratt DE, Yousefi K, Deheshi S, et al. Individual Patient-Level Meta-Analysis of the Performance of the Decipher Genomic Classifier in High-Risk Men After Prostatectomy to Predict Development of Metastatic Disease. J Clin Oncol. 2017;35(18):1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alshalalfa M, Nguyen PL, Beltran H, et al. Transcriptomic and Clinical Characterization of Neuropeptide Y Expression in Localized and Metastatic Prostate Cancer: Identification of Novel Prostate Cancer Subtype with Clinical Implications. Eur Urol Oncol. 2019;2(4):405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahal BA, Alshalalfa M, Spratt DE, et al. Prostate Cancer Genomic-risk Differences Between African-American and White Men Across Gleason Scores. Eur Urol. 2019. [DOI] [PubMed] [Google Scholar]
- 7.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur Urol. 2016;69(1):157–165. [DOI] [PubMed] [Google Scholar]
- 10.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190(6):2047–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3(5):e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol. 2016;17(11):1612–1620. [DOI] [PubMed] [Google Scholar]
- 13.Zhao SG, Chang SL, Erho N, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol. 2017;3(12):1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muralidhar V, Zhang J, Wang Q, et al. Genomic validation of three-tiered clinical sub-classification of high- risk prostate cancer. Int J Radiat Oncol Biol Phys. 2019. [DOI] [PubMed] [Google Scholar]
- 17.Zhao SG, Chen WS, Das R, et al. Clinical and Genomic Implications of Luminal and Basal Subtypes Across Carcinomas. Clin Cancer Res. 2019;25(8):2450–2457. [DOI] [PubMed] [Google Scholar]
- 18.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–598. [DOI] [PubMed] [Google Scholar]
- 19.Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.