Abstract
Background
10 days after the first reported case of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in the Netherlands (on Feb 27, 2020), 55 (4%) of 1497 health-care workers in nine hospitals located in the south of the Netherlands had tested positive for SARS-CoV-2 RNA. We aimed to gain insight in possible sources of infection in health-care workers.
Methods
We did a cross-sectional study at three of the nine hospitals located in the south of the Netherlands. We screened health-care workers at the participating hospitals for SARS-CoV-2 infection, based on clinical symptoms (fever or mild respiratory symptoms) in the 10 days before screening. We obtained epidemiological data through structured interviews with health-care workers and combined this information with data from whole-genome sequencing of SARS-CoV-2 in clinical samples taken from health-care workers and patients. We did an in-depth analysis of sources and modes of transmission of SARS-CoV-2 in health-care workers and patients.
Findings
Between March 2 and March 12, 2020, 1796 (15%) of 12 022 health-care workers were screened, of whom 96 (5%) tested positive for SARS-CoV-2. We obtained complete and near-complete genome sequences from 50 health-care workers and ten patients. Most sequences were grouped in three clusters, with two clusters showing local circulation within the region. The noted patterns were consistent with multiple introductions into the hospitals through community-acquired infections and local amplification in the community.
Interpretation
Although direct transmission in the hospitals cannot be ruled out, our data do not support widespread nosocomial transmission as the source of infection in patients or health-care workers.
Funding
EU Horizon 2020 (RECoVer, VEO, and the European Joint Programme One Health METASTAVA), and the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Introduction
In January, 2020, a cluster of patients with pneumonia of unknown cause was reported in Wuhan, China;1 the disease was subsequently named COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The clinical spectrum of COVID-19 varies from asymptomatic or mild symptomatic infections to severe respiratory symptoms and death, with older age groups generally presenting with more severe disease and higher death rates.2, 3 Since its identification, SARS-CoV-2 has rapidly spread across the globe. On June 22, 2020, 177 countries had reported cases of COVID-19, adding up to more than 8·9 million reported cases and 468 000 deaths worldwide.4
Health-care workers are at increased risk of being exposed to SARS-CoV-2 and could potentially have a role in hospital transmission. Nosocomial outbreaks of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus (MERS-CoV) are thought to have played a crucial part in the amplification and spread of these viruses. For MERS-CoV, hospital outbreaks caused approximately 50% of confirmed cases, of which around 40% were in health-care workers.5 Currently, the extent of SARS-CoV-2 transmission and risk factors associated with infection in health-care settings are unclear. During the WHO–China Joint Mission on COVID-19,3 2055 laboratory confirmed cases were reported in health-care workers from 476 hospitals in China, mostly (88%) from Hubei province. Most health-care workers were thought to have been infected within household settings rather than in a health-care setting, although conclusive evidence was scant.3
On Feb 27, 2020, the first patient in the Netherlands tested positive for SARS-CoV-2 RNA after returning from a holiday to Lombardy, Italy.6 In the following week, the number of infections in the country grew to 128, with an increasing proportion of cases without a known source of infection. These cases included nine health-care workers from two hospitals in the province of North Brabant, in the south of the Netherlands.7, 8 The Dutch national outbreak management team advised to extend screening of health-care workers to other hospitals in North Brabant, to assess possible community transmission. From March 6 to March 8, 2020, 1097 employees of nine hospitals were tested, of whom 45 (4%) were positive for SARS-CoV-2.8 A follow-up study was done at three hospitals to assess the clinical presentations of COVID-19 of these health-care workers.7 The impending shortage of personal protective equipment (PPE) and the proposed changes in its use in later phases of the outbreak response also triggered a debate on possible risks to health-care workers.9
Research in context.
Evidence before this study
We searched Google Scholar on April 27, 2020, for articles published since 2020, with the keywords “SARS-CoV-2” AND “healthcare workers” AND “whole genome sequencing”. We did not restrict our search to a publication language. Our search retrieved 13 results. Two reports presented original research of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); no reports were of the role of health-care workers in SARS-CoV-2 transmission or used whole-genome sequencing (WGS). Hospital transmission had an important role in previous outbreaks of Middle East respiratory syndrome and severe acute respiratory syndrome. The scarcity of personal protective equipment led to changes in policy during the initial phases of the SARS-CoV-2 outbreak response, also triggering a debate on possible risks to health-care workers. Up to now, possible SARS-CoV-2 outbreaks in health-care facilities have only been described using traditional molecular diagnostic tools combined with epidemiological data. However, previous studies implementing WGS have shown that hypotheses on virus transmission routes can be incorrect based solely on these data. Moreover, screening of health-care workers can be used to assess the level of local community transmission, but this can only be done if patient-to-health-care worker transmission can be reliably excluded.
Added value of this study
Our study aimed to gain insight in possible sources of infection of health-care workers at three hospitals in the Netherlands. All health-care workers with respiratory symptoms or fever in the previous 10 days were screened for SARS-CoV-2 infection. WGS was done of samples obtained from health-care workers and patients at these hospitals and this information was combined with epidemiological data.
Implications of all the available evidence
At the beginning of the SARS-CoV-2 outbreak in the Netherlands, health-care workers were probably infected in the community rather than at the hospitals. Possible nosocomial outbreaks should be carefully investigated using both epidemiological data and WGS to exclude or confirm transmission in health-care facilities.
To understand sources and modes of transmission of SARS-CoV-2 in health-care workers and patients in the same hospitals, we did an in-depth analysis combining epidemiological data with whole-genome sequencing (WGS) of SARS-CoV-2 from clinical samples obtained from health-care workers and patients in three different hospitals
Methods
Study design and participants
We did a cross-sectional study at two teaching hospitals (Amphia Hospital, Breda, Netherlands [700 beds], and Elisabeth-TweeSteden Hospital, Tilburg, Netherlands [800 beds]) and one regional hospital (Bravis Hospital, Roosendaal and Bergen op Zoom, Netherlands [600 beds]), at which 12 022 health-care workers in total were employed. PPE was used according to national guidelines that applied during this period of the outbreak.10, 11 Patients with suspected COVID-19 were nursed under strict isolation precautions and health-care workers applied additional PPE (gowns, gloves, goggles, hair cover, and type IIR surgical masks) on entering the isolation room. When aerosol-generating procedures were done, an FFP2 mask was used.
All health-care workers at these three hospitals who had fever or mild respiratory symptoms in the 10 days before screening for SARS-CoV-2 infection were eligible for testing, which was voluntary. All patients testing positive for SARS-CoV-2 and who had been admitted 2 days or more before the last date of onset of symptoms of health-care workers per hospital were included. All health-care workers with confirmed SARS-CoV-2 infection underwent a structured interview to obtain epidemiological data and to record any history of foreign travel and attendance at public events with more than 50 people, such as the yearly carnival in February, 2020 (appendix 1 p 1).
Ethics approval was obtained from the Medical Ethics Committee Brabant, with a waiver of written informed consent (METC Brabant/20.134/NW2020-26). Verbal informed consent was obtained from all health-care workers for SARS-CoV-2 testing, sequencing, and data collection. Data were deidentified before analysis. For patients, location and sequence data were obtained as part of the routine infection control policy in outbreak situations. All patients are notified of this policy on hospital admission and can actively dissent (opt out).
Procedures
We tested for SARS-CoV-2 infection using oropharyngeal or nasopharyngeal swabs in universal transport medium (Copan, Brescia, Italy) or E-swab medium (Amies; Copan), following local infection control policy during outbreaks. At Amphia Hospital and Bravis Hospital, total nucleic acids were extracted for RT-PCR after an external lysis step (1:1 with lysis binding buffer; Roche Diagnostics, Almere, Netherlands), using MagnaPure96 (Roche) with an input volume of 500 μL and output volume of 100 μL. The extraction was internally controlled by addition of a known concentration of phocine distemper virus (PDV).12 Subsequently, 10 μL extracted nucleic acids was amplified in three singleplex reactions in 25 μL final volume, using TaqMan Fast Virus 1-Step Master Mix (Thermofisher, Nieuwerkerk aan den IJssel, Netherlands), and 1 μL of primers and probe mixture for envelope (E) gene, RNA-dependent RNA-polymerase gene, and PDV.13 Amplification was done in a 7500SDS (Thermofisher) with a cycling profile of 5 min at 50°C, 20 s at 95°C, 45 cycles of 3 s at 95°C, and 30 s at 58°C. At Elisabeth-TweeSteden Hospital, total nucleic acids were extracted, with a known concentration of PDV as internal control, using the QIAsymphony DSP virus pathogen midi kit and pathogen complex 400 protocol of the QIAsymphony Sample Processing system (Qiagen, Hilden, Germany), with an input volume of 400 μL and output volume of 110 μL. The amplification reaction was done in a volume of 25 μL with TaqMan Fast Virus 1-Step Master Mix (Thermofisher) and 10 μL extracted nucleic acids. A duplex PCR for E gene and PDV13, 14 with optimised primer and probe concentrations were done. Amplification with Rotorgene (QIAgen) consisted of 5 min at 50°C and 15 min at 95°C followed by 45 cycles of 15 s at 95°C, 30 s at 60°C, and 15 s at 72°C. Validations of RT-PCR procedures were done according to International Standards Organization guidelines (15189).15
For WGS, samples were selected based on a cycle threshold value less than 32. A SARS-CoV-2-specific multiplex PCR for nanopore sequencing was done, as previously described.16 The resulting raw sequence data were demultiplexed using qcat. Primers were trimmed using cutadapt,17 after which a reference-based alignment to the GISAID (Global Initiative on Sharing All Influenza Data) sequence EPI_ISL_412973 was done using minimap2.18 The consensus genome was extracted and positions with a coverage less than 30 reads were replaced with N using a custom script using biopython software (version 1.74) and the python module pysam (version 0.15.3), as previously described.19 Mutations in the genome were confirmed by manually checking the alignment, and homopolymeric regions were manually checked and resolved, consulting the reference genome. Genomes were included when having greater than 90% genome coverage.
All available full-length SARS-CoV-2 genomes were retrieved from GISAID20 on March 20, 2020 (appendix 1 pp 8–65), and aligned with the newly obtained SARS-CoV-2 sequences in this study using the multiple sequence alignment software MUSCLE (version 3.8.1551).21 Sequences with more than 10% of N position replacements were excluded. The alignment was manually checked for discrepancies, after which the phylogenomic software IQ-TREE (version 1.6.8)22 was used to do a maximum-likelihood phylogenetic analysis, with the generalised time reversible substitution model GTR+F+I+G4 as best predicted model. The ultrafast bootstrap option was used with 1000 replicates. Clusters were ascertained based on visual clustering and lineage designations.23
The code to generate the minimum spanning phylogenetic tree was written in the R programming language. Ape24 and igraph software packages were used to write the code to generate the minimum spanning tree, and the visNetwork software package was used to generate the visualisation. Pairwise sequence distance (used to generate the network) was calculated by adding up the absolute nucleotide distance and indel-block distance. Unambiguous positions were dealt with in a pairwise manner. Sequences that were mistakenly identified as identical, because of transient connections with sequences containing missing data, were resolved.
The multiple sequence alignment was curated and any error-rich sequences or sequences without a date were removed. The alignment was manually inspected and trimmed of the 5′ and 3′ untranslated regions in the bioinformatics software Geneious (version 11.1.3) to include only coding regions. The final length of the alignment was 29 408 nucleotides. Bayesian phylogenetic trees were estimated using BEAST version 1.10.4,25 with a Hasegawa-Kishino-Yano nucleotide substitution model26 and a strict molecular clock. Two independent chains were run for 100 million states, with a Skygrid coalescent prior (appendix 1 p 3).27, 28 and parameters were sampled every 10 000 states. The LogCombiner program was used to combine the independent chains and to remove the burn-in from the tree file, and Tracer29 was used to assess convergence. The maximum clade credibility tree was inferred using the TreeAnnotator program and visualised using baltic code and custom python scripts.
Statistical analysis
Epidemiological data obtained at structured interviews were entered in Castor Electronic Data Capture, version 2019. Continuous variables were expressed as medians and ranges and categorical variables were summarised as numbers and percentages. All analyses were done with SPSS version 25.0 (IBM, Armonk, NY, USA). Because of the descriptive nature of our study, sample size calculations and analyses of significance were not done. Results were reported following STROBE guidelines for observational studies.
Role of the funding source
The funder had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
Between March 2 and March 12, 2020, 1796 (15%) of 12 022 health-care workers were voluntarily screened at the three participating hospitals (appendix 1 p 5). At Amphia Hospital, 42 (5%) of 783 health-care workers tested positive for SARS-CoV-2 RNA; at Bravis Hospital, ten (2%) of 443 health-care workers tested positive; and at Elisabeth-TweeSteden Hospital, 44 (8%) of 570 health-care workers tested positive. Characteristics of these 96 health-care workers who tested positive for SARS-CoV-2 RNA are shown in the table . The health-care workers were employed in 58 different departments, including on 42 medical wards. The median age of affected health-care workers was 49 years (range 22–66), and 80 (83%) of 96 were female, reflecting the proportion of female health-care workers among the total population employed in the participating hospitals (ie, 9784 of 12 022 [81%]). 20 staff members who did not have direct contact with patients tested positive for SARS-CoV-2 RNA, of whom six (30%) reported contact with colleagues who had also tested positive. Ten health-care workers reported a history of foreign travel in the 14 days before onset of symptoms, three (30%) of whom had travelled to northern Italy. 60 (63%) health-care workers had celebrated carnival in the 14 days before onset of symptoms, mostly in Breda, Prinsenbeek, and Tilburg. One health-care worker (who reported first symptoms on Feb 21, 2020) attended several carnival events while symptomatic but unaware of having COVID-19. 31 (32%) health-care workers reported close contact with an individual with confirmed COVID-19 in the 14 days before onset of symptoms, either a patient (n=3), colleague (n=18), household member (n=1), or another person outside the hospital (n=9).
Table.
Descriptive characteristics of 96 health-care workers testing positive for severe acute respiratory syndrome coronavirus 2 RNA at three hospitals in the south of the Netherlands in March, 2020
| Health-care workers (n=96) | ||
|---|---|---|
| Sex | ||
| Male | 16 (17%) | |
| Female | 80 (83%) | |
| Age, years | 49 (22–66) | |
| Residence | ||
| Breda | 11 (11%) | |
| Prinsenbeek | 11 (11%) | |
| Tilburg | 24 (25%) | |
| Other city | 50 (52%) | |
| Department | ||
| Medical | 76 (79%) | |
| Staff without direct patient contact | 20 (21%) | |
| Foreign travel, 14 days before onset of symptoms | 10 (10%) | |
| Northern Italy | 3 (3%) | |
| Austria | 3 (3%) | |
| UK | 1 (1%) | |
| Spain | 1 (1%) | |
| Portugal | 1 (1%) | |
| Switzerland | 1 (1%) | |
| Attendance at carnival with 50 people or more, 14 days before onset of symptoms | 60 (63%) | |
| Breda | 7 (7%) | |
| Prinsenbeek | 11 (11%) | |
| Tilburg | 20 (21%) | |
| Other city | 22 (23%) | |
| Attendance at other event with 50 people or more, 14 days before onset of symptoms | 31 (32%) | |
| Close contact with individual with confirmed COVID-19, 14 days before onset of symptoms | 31 (32%) | |
| Patient | 3 (3%) | |
| Colleague | 18 (19%) | |
| Household member | 1 (1%) | |
| Other, outside hospital | 9 (9%) | |
Data are n (%) or median (range).
Between March 2 and March 7, 2020 (Amphia Hospital), March 2 and March 10, 2020 (Bravis Hospital), and Feb 29 and March 9, 2020 (Elisabeth-TweeSteden Hospital), 856 patients were tested for SARS-CoV-2 RNA, of whom 345 were at Amphia Hospital, 228 were at Bravis Hospital, and 283 were at Elisabeth-TweeSteden Hospital (appendix 1 p 5). 23 (3%) patients tested positive for SARS-CoV-2 RNA, nine at Amphia Hospital and 14 at Elisabeth-TweeSteden Hospital.
We obtained complete and near-complete SARS-CoV-2 genomes from 50 of 96 health-care workers (appendix 1 pp 4–5). 30 health-care workers were from Amphia Hospital, six were from Bravis Hospital, and 14 were from Elisabeth-TweeSteden Hospital. We obtained near-complete SARS-CoV-2 sequences from seven patients at Amphia Hospital and three patients at Elisabeth-TweeSteden Hospital.
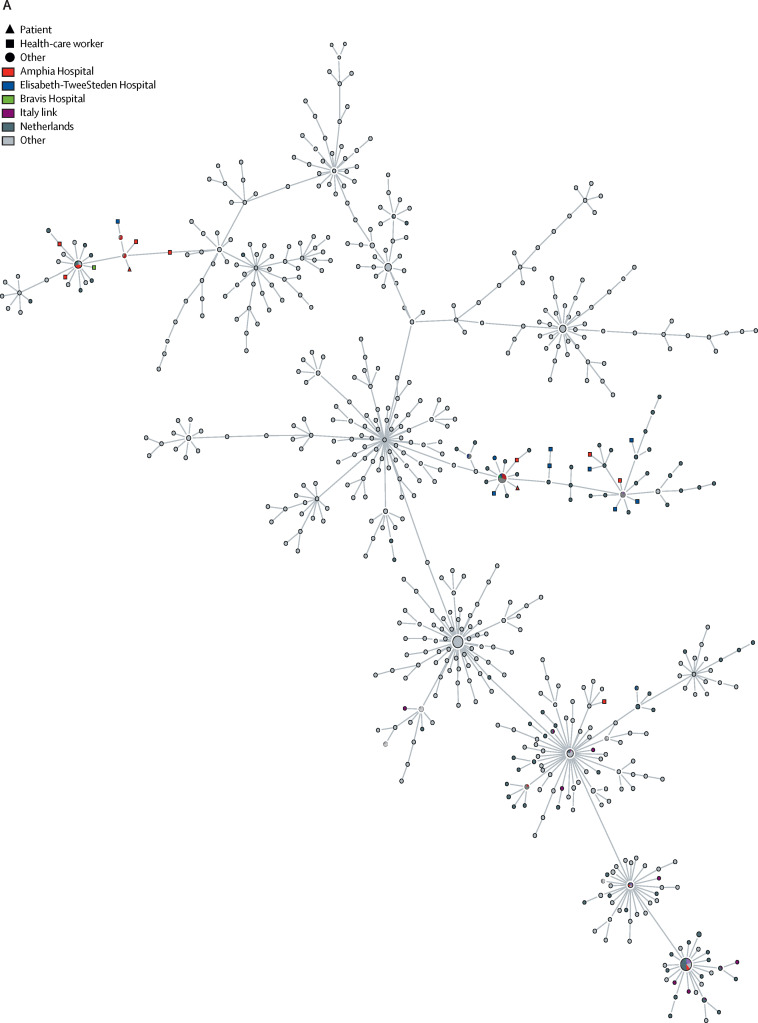
46 (92%) of 50 sequences from health-care workers in this study grouped in three clusters (figure, A ; appendix 1 p 4; appendix 2). Ten (100%) of ten sequences from patients in the study grouped into the same three clusters: seven were in cluster 1, two were in cluster 2, and one was in cluster 3.
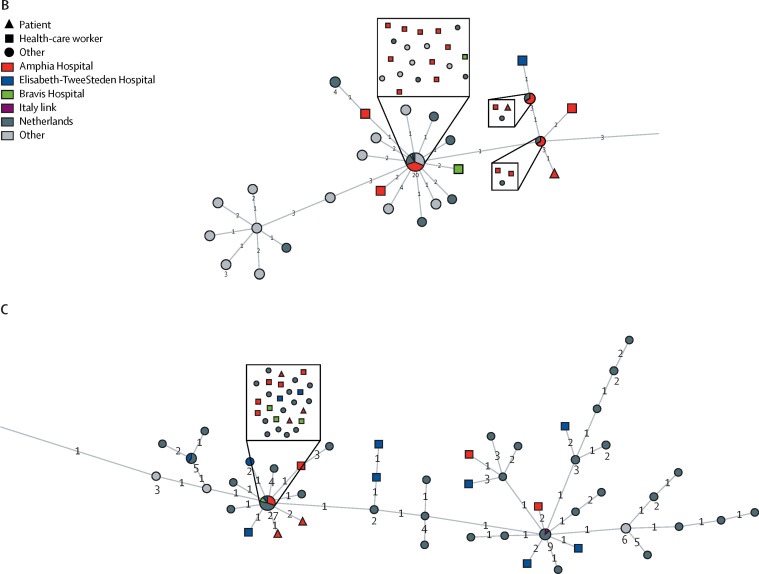
Figure.
Minimum spanning tree of available full-length SARS-CoV-2 genomes obtained from GISAID on March 20, 2020
The full tree (A) shows three clusters of SARS-CoV-2 genomes, obtained from sequencing samples from health-care workers and patients in the south of the Netherlands in March, 2020. An interactive version of the full tree can be found in appendix 2; it can be accessed by unzipping and opening the visNetwork.html file. Clusters 2 (B) and 1 (C) are shown in more detail. Numbers next to nodes indicate the number of sequences included. Numbers on branches indicate the difference in number of nucleotides between sequences. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. GISAID=Global Initiative on Sharing All Influenza Data.
Cluster 1 contained 29 sequences (of which 12 were identical) of SARS-CoV-2 in samples taken from health-care workers and patients at all three hospitals (appendix 1 p 5). 13 (45%) sequences were from Amphia Hospital, three (10%) were from Bravis Hospital, and 13 (45%) were from Elisabeth-TweeSteden Hospital (figure, C). 11 (79%) of 14 health-care workers and two (67%) of three patients at Elisabeth-TweeSteden Hospital were in cluster 1.
Cluster 2 contained 20 sequences (of which ten were identical) of SARS-CoV-2 in samples taken from health-care workers and patients at all three hospitals (appendix 1 p 5). 17 (85%) sequences originated from Amphia Hospital, two (10%) were from Bravis Hospital, and one (5%) was from Elisabeth-TweeSteden Hospital (figure, B). Health-care workers in cluster 2 were associated with Prinsenbeek and Breda, either by attendance at the carnival or by residence, more frequently compared with the other clusters (appendix 1 p 4).
Cluster 3 contained seven sequences (of which four were identical) of SARS-CoV-2 in samples taken from health-care workers and patients at all three hospitals. Four sequences were from health-care workers at Amphia hospital and one each was from Bravis Hospital and Elisabeth-TweeSteden Hospital. One sequence from a patient at Elisabeth-TweeSteden Hospital was also included in this cluster. A relatively large proportion of sequences in cluster 3 were from people with a travel history to northern Italy, as described elsewhere.16 However, only two of six health-care workers in this cluster reported recent travel to either Italy or Austria (appendix 1 pp 4, 6–7).
Within each cluster, identical or near-identical sequences in health-care workers at the same hospital, and between patients and health-care workers at the same hospital, were found, but no consistent link was noted among health-care workers on the same ward or between health-care workers and patients on the same ward. Most (81–100%) health-care workers testing positive for SARS-CoV-2 at the three hospitals did not work on a ward with patients with confirmed COVID-19 (appendix 1 p 2). In wards with patients and health-care workers infected with SARS-CoV-2, direct transmission could be excluded in most cases, based on available WGS data (appendix 1 p 2). Notably, in Bravis Hospital, no patients with confirmed SARS-CoV-2 infection were hospitalised within 2 days before health-care workers at that hospital reported onset of symptoms. Additionally, no clusters were reported of more than three health-care workers on the same ward with identical or near-identical (two nucleotide difference or less) sequences. However, we cannot exclude health-care workers being infected in common hospital areas such as staff restaurants.
Discussion
In the present study, we combined epidemiological data with WGS to obtain a deeper understanding of the sources and modes of transmission of SARS-CoV-2 at three hospitals in the south of the Netherlands, which were the first hospitals to identify patients with COVID-19 in the Netherlands. Although possible hospital transmission of SARS-CoV-2 and health-care workers with COVID-19 have been reported,3, 30, 31 to our knowledge, our study is the first to use WGS to analyse possible SARS-CoV-2 nosocomial transmission. Infection of health-care workers could have occurred through foreign travel, community contacts, or nosocomial transmission. The epidemiological data we obtained, combined with the presence of identical viruses in all three hospitals, and with non-hospitalised cases in other locations, indicates widespread community transmission in a very early phase of the outbreak. Mass gatherings, such as carnivals, in which just under two-thirds of health-care workers testing positive for SARS-CoV-2 participated, possibly acted as local super-spreading events.
Health-care workers are at increased risk of being exposed to viruses within hospitals but can also be a source of transmission by introducing a virus into their hospital. SARS-CoV-2 infections in health-care workers can have a substantial effect, because pathogens are introduced into settings with high numbers of individuals with comorbidities, potentially causing high morbidity and mortality among patients. The current study did not find evidence of large-scale nosocomial transmission in the early phase of the Dutch outbreak, and prevailing use of PPE and other infectious disease prevention measures were considered sufficient based on these early analyses and results.32
Outbreaks in health-care settings are traditionally investigated by molecular diagnostic methods combined with epidemiological data. However, previous studies using WGS for hospital outbreak investigations have shown that hypotheses on virus transmission routes can be incorrect based solely on these data. By adding WGS data, particularly if results can be generated in a timely manner, and as long as sufficient reference sequences are available to allow a high resolution of the findings, the sequence analysis can provide essential information and inform subsequent infection control measures.33
The mutation rate of SARS-CoV-2 is estimated to be around 1·16 × 10−3 substitutions per site per year, which corresponds to around one mutation every 2 weeks.34 Therefore, finding identical or near-identical sequences in several locations and hospitals makes it difficult to draw definite conclusions on individual direct health-care worker-to-health-care worker or health-care worker-to-patient transmissions based on sequence data alone in this early stage of the SARS-CoV-2 outbreak, when genetic diversity of the circulating pathogen was negligible. Moreover, we did not obtain WGS of all health-care workers and patients testing positive for SARS-CoV-2 and, because of the small sample size, our analyses should be interpreted with caution. However, the finding of diverse clusters does exclude infection from one source. Moreover, the sequence-based analysis could be biased when sampling and sequencing is not done systematically and when sequence data in some areas are scarce, as is the case for COVID-19 internationally. For the Netherlands, we sequenced a substantial proportion of SARS-CoV-2 genomes as part of the national public health response,16 which was used as a reference set.
In conclusion, the genomic diversity recorded in our study is consistent with multiple introductions through community-acquired infections, and some local amplification related to specific social events in the community, rather than widespread within-hospital transmission. Although direct transmission in hospitals cannot be ruled out, our data do not support widespread nosocomial transmission as the source of infection in patients or health-care workers in our study. Because of the near-real-time sequence generation and analysis, our information was rapidly shared within the Dutch outbreak management team. Partly based on these data, SARS-CoV-2 was concluded to have already spread in the population in the province of North Brabant, which led to a change of policy, in which containment measures were complemented by targeted physical distance measures, starting in the south of the Netherlands initially and later comprising the whole country.16
Acknowledgments
Acknowledgments
We thank David van der Vijver and Miranda de Graaf (Erasmus MC, Rotterdam, Netherlands) for technical support. This study has been partly funded by EU Horizon 2020 projects RECoVer (no 101003589), VEO (no 874735), and the European Joint Programme One Health METASTAVA (no 773830), and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (contract HHSN272201400008C).
Contributors
RSS, SDP, MFQKvdB, and MPGK wrote the report. SDP, MFQKvdB, and JAJWK set up sample and data collection. BBOM, AvdL, IC, MP, PL, SvN, TB, CS, and RJO generated sequence data. WvdB, RGB, MMLvR, AGMB, AJGvO, BMD, AMCB, AvdE, AT, JV, and RM were involved in sample and data collection. RSS, DFN, AO'T, AR, BBOM, MPGK, and MFQKvdB were involved in data analysis and interpretation. RSS, SDP, JAJWK, MFQKvdB, and MPGK designed the study. All authors provided critical feedback.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kluytmans van den Bergh MFQ, Buiting AGM, Pas SD, et al. Prevalence and clinical presentation of health care workers with symptoms of coronavirus disease 2019 in 2 Dutch hospitals during an early phase of the pandemic. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Report of the WHO–China Joint Mission on Coronavirus Disease 2019 (COVID-19) Feb 28, 2020. https://www.who.int/publications/i/item/report-of-the-who-china-joint-mission-on-coronavirus-disease-2019-(covid-19)
- 4.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Regional Office for the Eastern Mediterranean MERS situation update. January, 2020. http://www.emro.who.int/pandemic-epidemic-diseases/mers-cov/mers-situation-update-january-2020.html
- 6.Alderweireld CEABA, Murk JAN, Verweij JJ, Berrevoets MAH, van Kasteren MEE. COVID-19: patiënt nul in Nederland. Ned Tijdschr Geneeskd. 2020;164 [PubMed] [Google Scholar]
- 7.Kluytmans M, Buiting A, Pas S, et al. SARS-CoV-2 infection in 86 healthcare workers in two Dutch hospitals in March 2020. medRxiv. 2020 doi: 10.1101/2020.03.23.20041913. published online March 31. (preprint). [DOI] [Google Scholar]
- 8.Reusken CB, Buiting A, Bleeker-Rovers C, et al. Rapid assessment of regional SARS-CoV-2 community transmission through a convenience sample of healthcare workers, the Netherlands, March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.12.2000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NOS News Care threatens to be squeezed by a shortage of mouth masks. Feb 27, 2020. https://nos.nl/artikel/2324830-zorg-dreigt-in-de-knel-te-komen-door-tekort-aan-mondkapjes.html (in Dutch).
- 10.Rijksinstituut voor Volksgezondheid en Milieu Guidance on infection prevention for hospitals: isolation guidelines. May 13, 2011. https://www.rivm.nl/wip-richtlijn-strikte-isolatie-zkh (in Dutch).
- 11.Rijksinstituut voor Volksgezondheid en Milieu Personal protective equipment directive. September, 2015. https://www.rivm.nl/documenten/wip-richtlijn-persoonlijke-beschermingsmiddelen-zkh (in Dutch).
- 12.Hoek RAS, Paats MS, Pas SD, et al. Incidence of viral respiratory pathogens causing exacerbations in adult cystic fibrosis patients. Scand J Infect Dis. 2013;45:65–69. doi: 10.3109/00365548.2012.708942. [DOI] [PubMed] [Google Scholar]
- 13.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Vries E, Anber J, van der Linden A, et al. Molecular assays for quantitative and qualitative detection of influenza virus and oseltamivir resistance mutations. J Mol Diagn. 2013;15:347–354. doi: 10.1016/j.jmoldx.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 15.ISO Medical laboratories: requirements for quality and competence. November, 2012. https://www.iso.org/standard/56115.html
- 16.Oude Munnink BB, Nieuwenhuijse DF, Stein M, et al. Rapid SARS-CoV-2 whole genome sequencing for informed public health decision making in the Netherlands. bioRxiv. 2020 doi: 10.1101/2020.04.21.050633. published online April 25. (preprint). [DOI] [PubMed] [Google Scholar]
- 17.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 18.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oude Munnink BB, Nieuwenhuijse DF, Sikkema RS, Koopmans M. Validating whole genome nanopore sequencing, using usutu virus as an example. J Vis Exp. 2020;157 doi: 10.3791/60906. [DOI] [PubMed] [Google Scholar]
- 20.Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambaut A, Holmes EC, Hill V, et al. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv. 2020 doi: 10.1101/2020.04.17.046086. published online April 19. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 25.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4 doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 27.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 2013;30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill V, Baele G. Bayesian estimation of past population dynamics in BEAST 1.10 using the Skygrid coalescent model. Mol Biol Evol. 2019;36:2620–2628. doi: 10.1093/molbev/msz172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMichael TM, Currie DW, Clark S, et al. Epidemiology of Covid-19 in a long-term care facility in King County, Washington. N Engl J Med. 2020;382:2005–2011. doi: 10.1056/NEJMoa2005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan M, Qin Y, Xue X, Zhu S. Death from Covid-19 of 23 health care workers in China. N Engl J Med. 2020;382:2267–2268. doi: 10.1056/NEJMc2005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durante-Mangoni E, Andini R, Bertolino L, et al. Low rate of severe acute respiratory syndrome coronavirus 2 spread among health-care personnel using ordinary personal protection equipment in a medium-incidence setting. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.04.042. published online May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houlihan CF, Frampton D, Ferns RB, et al. Use of whole-genome sequencing in the investigation of a nosocomial influenza virus outbreak. J Infect Dis. 2018;218:1485–1489. doi: 10.1093/infdis/jiy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taiaroa G, Rawlinson D, Featherstone L, et al. Direct RNA sequencing and early evolution of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.05.976167. published online March 7. (preprint). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.