Abstract
Several major factors limit our understanding of hepatocellular carcinoma (HCC). First, human HCCs are infrequently biopsied for diagnosis and thus are not often biologically interrogated. Second, HCC initiation and progression are strongly influenced by the cirrhotic microenvironment, and the exact contributions of intrinsic and extrinsic tumor factors are unclear. A powerful approach to examine the personalized biology of liver cancers and the influence of host tissues is with patient derived xenograft (PDX) models. In Asia, HCCs from hepatitis B virus patients have been efficiently converted into PDXs, but few parallel efforts from the west have been reported. In a large-scale analysis, we implanted 93 HCCs and 8 cholangiocarcinomas (CCAs) to systematically analyze host factors and to define an optimized platform for PDX development from both surgical and biopsy samples. NSG mice that had undergone partial hepatectomy (PHx) represented the best combination of engraftability, growth, and passagability, but overall rates were low and indicative of a unique intrinsic biology for HCCs in the US. PDX models preserved the histology and genetic features of parental tumors, and ultimately, 8 new models were usable for pre-clinical studies. Intriguingly, HCC PDXs were differentially sensitive to regorafenib and sorafenib and CCA PDXs were also highly sensitive to regorafenib. PDX models functionalize early and advanced stage HCCs and revealed unique biological features of liver cancers from the US.
Keywords: Hepatocellular carcinoma, PDX, liver, drug test
INTRODUCTION
Hepatocellular carcinoma (HCC) is the 6th most common cancer and 2nd leading cause of cancer-related death worldwide (1). In the US, its incidence has doubled over the past two decades due to the growing number of patients with advanced hepatitis C virus (HCV) infections and/or non-alcoholic steatohepatitis (NASH) (2,3). Patients with cirrhosis are at high risk for HCC with a 3–8% annual incidence rate (4). Once HCC is diagnosed, the Barcelona-Clinic Liver Cancer (BCLC) staging classification divides patients into five stages (0, A, B, C and D). Stages 0 and A (very early and early) are curable by surgical resection, liver transplantation, or local ablative therapies (4). However, a minority of patients with HCC are diagnosed with early stage tumors and the majority of HCC patients present with intermediate or advanced stage disease, when only palliative therapies are available. While patients diagnosed with early stage tumors can achieve 5-year survival rates exceeding 70% with curative therapies, patients with non-early stage HCC continue to have median survival rates of 1–2 years despite the introduction of novel therapies.
Historically, the diagnosis of HCC is often made radiographically, without the need for histologic confirmation. As a result, most of the characterized HCC tissue is from surgical specimens obtained from early-stage HCC. This lack of available tissue from patients with intermediate or advanced stage HCC has led to a dearth of biological knowledge about non-early stage HCC and likely contributes to slow improvements in palliative therapies over time. It is unknown if intermediate and advanced HCC represent simple progression from early cases or if these HCC populations are comprised of distinct biological entities with unique growth mechanisms, genetic dependencies, and differential sensitivity to systemic therapies.
One way to study the growth dynamics and treatment responses of living tumors is with patient derived xenograft (PDX) models. For HCC, there are only a few published experiences with PDX models, most of which are derived from Asian non-cirrhotic HBV patients who underwent curative resection (5–8). While many HBV-infected patients in Asia develop HCC in the absence of cirrhosis, over 90% of HCV, alcohol, and NASH patients in the U.S. and Europe develop HCC in the presence of cirrhosis (9). Thus, prior PDX models may not be entirely representative of HCCs from the Western world.
In this study, we have established methodologies for PDX development for a large number of American patients with cirrhosis and HCC. Engraftment and passagability rates for HCCs are low, but optimized recipient protocols can increase efficiency. We also found that HCC biopsies can generate PDX models. These HCC PDXs help to characterize the biology of HCC in the Western world and represent an important resource for future studies. This knowledge will help elucidate mechanisms of response to available and experimental therapeutics.
RESULTS
HCCs engraft at low rates in immunodeficient mice without liver damage
First, we implanted fresh surgical liver tumor specimens in the subcutaneous (SC) space of NSG mice (Figure 1A). NSG mice were selected because they are the most immunodeficient mice available. We quantified tumor engraftment defined as growth (at any anatomical location) to greater than 5 mm in diameter within 12 months (Figure 1B). For SC implantation, the engraftment rate was 14.3% (6 of 42 cases; see Table 1 and Figure 1C) and the engraftment time was 27.3 weeks (Figure 1D). Interestingly, in 4 of these 6 cases, engraftment was in the form of metastasis to the liver without detectable growth of tumors in the SC space (Supplementary Table 1 and Figure 1B).
Figure 1. PDX engraftment in immunodeficient recipients with and without liver injuries.
A. Schema of procedure.
B. Representative pictures of tumors engrafted.
C. Engraftment frequency in the subcutaneous space or the liver in different types of immunodeficient murine recipients.
D. Engraftment time of PDXs in different recipients. Fisher’s exact t-tests were used to compare engraftment times.
E. Overall engraftment time of PDXs in all types of recipients.
F. The number of PDX models that engrafted within 5 months in NSG mice plus hepatectomy vs. non-resected NSG mice. Fisher’s exact t-test was used for statistical analysis.
Table 1.
Engraftment of liver cancer PDX models in different locations and recipients.
| Recipients | Route | Implanted | HCC/CCA-PDXs | Engraftment time | Passageable lines | ||
|---|---|---|---|---|---|---|---|
| Amount | Percentage | Weeks | Amount | Percentage | |||
| NSG | SC | 42 | 6 | 14.3% | 27.3 | ||
| Liver | 27 | 4 | 14.8% | 22.9 | |||
| Overall | 44 | 9 | 20.5% | 27.9 | 1 | 2.3% | |
| FRG | SC | 14 | 1 | 7.1% | 13.3 | ||
| Liver | 14 | 4 | 28.6% | 28.7 | |||
| Overall | 14 | 5 | 35.7% | 25.6 | 1 | 7.1% | |
| NSG + PHx | SC | 43 | 7 | 16.3% | 14.4 * | ||
| Liver | 43 | 3 | 9.3% | 26.3 | |||
| Overall | 43 | 9 | 20.9% | 19.5 | 7 | 16.3% | |
| Total | 101 | 23 | 22.8% | 24 | 9 | 8.9% | |
p=0.025 when compared to NSG-SC group.
Given the long times and low engraftment rates, we aimed to determine what factors influence engraftment, growth rates, and passagability of liver cancer PDX models. The SC approach is convenient to assess for engraftment because you can visualize and palpate for tumor growth, but it is possible that the skin microenvironment was suboptimal for liver cancer engraftment. Since there are likely local pro-growth signals emanating from normal to malignant liver tissues, we also implanted tumors in normal, undamaged NSG livers using an orthograft approach. When tumors were implanted in the liver, the engraftment rate was 14.8% (4 of 27 cases), although this was more challenging to assess because laparotomy was not used to detect early stage PDX tumors prior to terminal liver harvesting. The average engraftment time in liver was 22.9 weeks, but this time reduction (compared to 27.3 weeks for SC) was not deemed to be statistically significant (Figure 1D). Another caveat is that it was more difficult to implant larger or multiple tumor fragments within the liver due to tissue fragility and bleeding risk. Thus, orthografting into the liver is inherently less efficient.
The overall engraftment rate for either SC or liver implantation was 20.5% (9 in 44 cases), and the average engraftment time was 27.9 weeks (Table 1 and Figure 1E). Surprisingly, the growth of tumors often took more than 5 months (27 weeks), making these challenging tumor models to develop (Figure 1E and 1F). This likely reflected the slow growth biology of human HCCs and is consistent with the doubling time of HCCs, which has been reported to be 6–12 months (10).
PDX engraftment is enhanced with genetic and surgically induced liver injuries
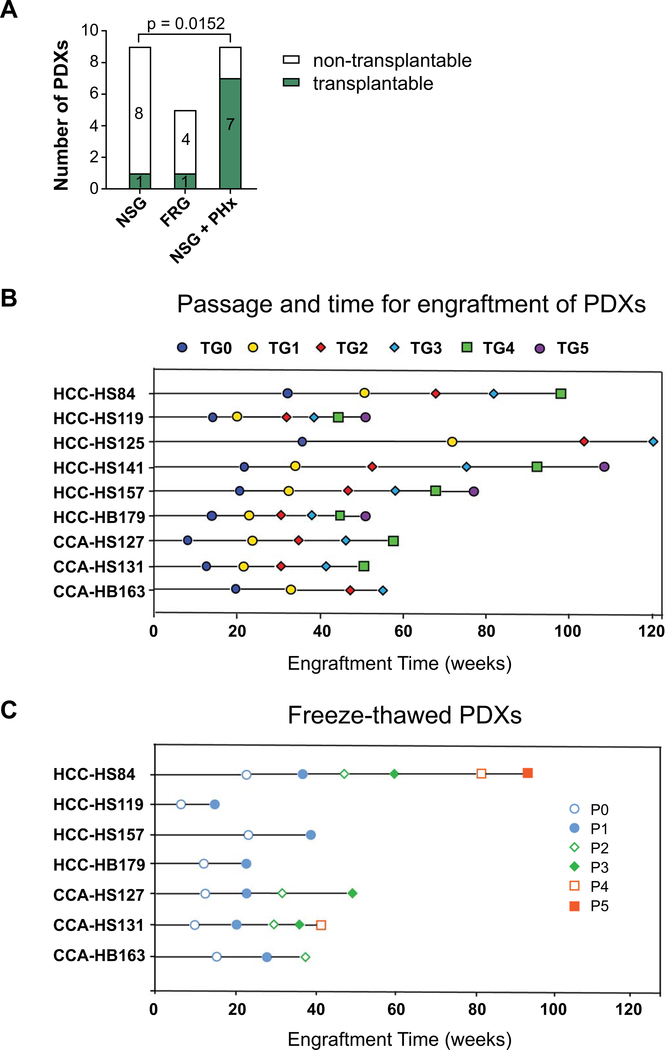
Because liver cancer often arises in the context of diseased liver tissues, we hypothesized that the host environment would be a critical variable. A tumor microenvironment that integrates liver injury, inflammation, and regeneration might accelerate PDX growth. We attempted to transplant tumors into a mouse model with chronic liver damage caused by hereditary tyrosinemia (11). Due to a defect in an enzyme involved in tyrosine metabolism, the livers of Fumarylacetoacetate hydrolase (Fah)−/−; Rag1−/−; IL-2Rγ−/− (FRG) mice accumulate a toxic metabolite called fumarylacetoacetate and die from liver failure within 2–4 months. To effectively treat this liver disease, mice and human patients are normally given nitisilone or NTBC (2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione), a drug that clears fumarylacetoacetate and maintains a healthy liver (11). If kept alive for over 6 months on intermittent treatment, Fah KO mice can develop cirrhosis and HCC (12), indicating that these mice can support the growth of endogenous liver cancers.
14 human liver tumors were transplanted into FRG mice, then NTBC was withdrawn to induce liver damage in the host livers. Tumors were simultaneously implanted in the liver and SC space of the individual FRG mice. Among 14 cases, 5 engrafted (4 in the liver and 1 in the SC space; see Table 1). This increased the engraftment rate in liver from around 14.8% to 28.6%, suggesting that the damaged liver microenvironment could promote HCC engraftment in the liver (Figure 1C). The improvements in overall engraftment frequency between FRG vs. NSG (35.7% vs. 20.5%) was not accompanied by a significantly shortened engraftment time (25.6 vs. 27 weeks) (Table 1 and Figure 1E). The specific engraftment frequency of SC tumors was lower in the FRG vs. NSG mice (7.1% vs. 14.3%) (Figure 1C), suggesting that pro-cancer microenvironmental factors were acting locally and not through the systemic circulation.
Importantly, we learned that an orthograft approach in FRG could increase PDX engraftment frequency but did not shorten engraftment time. We reasoned that the deeper immunodeficiency of NSG as compared to FRG could have provided an important benefit. Because it would be difficult to make FRG more immunodeficient, we asked if introducing liver damage to NSG would allow us to synergize the deeper immunodeficiency of NSG with the liver injury associated with FRG. We stimulated regeneration in NSG mice by performing a 40% PHx of the left lateral lobe at the time of liver tumor implantation. This is a less aggressive surgery than the standard 60–70% PHx, but can be more easily performed and allows for a higher survival rate. For 43 patient cases, we implanted liver tumors into both SC and liver locations in each mouse subjected to PHx. Seven of 43 tumors engrafted in the SC space (16.3%) and 4 of 43 cases engrafted in the liver (9.3%). Two of these cases overlapped, thus resulting in a total of 9 of 43 cases that engrafted (20.9%) (Table 1 and Figure 1C). Although the engraftment frequency did not increase substantially for either SC or liver implantations, the engraftment time for SC cases significantly decreased to 14.4 weeks compared to 27.3 weeks for NSG implantations without hepatectomy (p-value = 0.025; Figure 1D). Seven of the 9 PDX lines generated in NSG mice with hepatectomy engrafted in less than 5 months, while only one of 9 generated in the NSG mice without surgical resection engrafted in less than 5 months (Figure 1F). As mentioned above, FRG did not shorten the time to engraftment. Overall, SC implantation into NSG mice with PHx provided the most advantages in terms of speed and, as will be discussed below, serial passagability of PDX models.
PDX models can also be generated from liver cancer biopsies
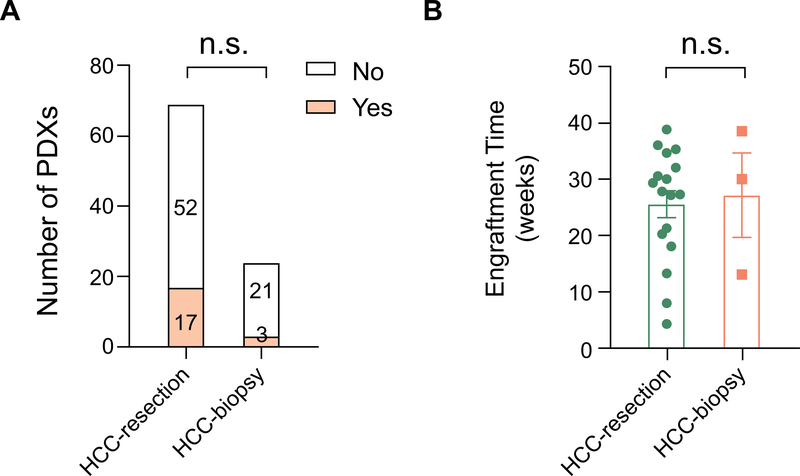
Some patients do not undergo surgery due to metastatic disease or poor functional status but many have undergone biopsy as part of their diagnostic workup. Patients with advance HCCs could benefit from PDX models that reveal therapeutic sensitivities. Because biopsies are derived from advanced BCLC stage HCCs, we hypothesized that these could harbor more aggressive cancer cells that would increase engraftment and PDX growth. To address this, we analyzed the cases where we implanted biopsy derived tissues into the various recipient models described above. Seventeen of 69 HCC surgical (24.6%) and 3 of 24 HCC biopsy samples (12.5%) resulted in engraftment (Supplementary Table 2 and Figure 2A). For biopsies, there was not a significant difference in engraftment frequency in different recipient types, potentially due to low case numbers. The average engraftment time was also similar for surgical and biopsy samples (Figure 2B). Taken together, the data did not support the hypothesis that biopsy cases from advanced HCCs display a more aggressive biology that can be detected in a PDX assay. As a caveat, the reduced mass of biopsy vs. surgically derived tissues could have suppressed engraftment efficiency. We also implanted 6 surgical and 2 biopsies from CCA patients. Two of the surgical and the 1 of the biopsy samples engrafted (Supplementary Table 2), hinting at an overall higher engraftment rate for CCA vs. HCC, regardless if the tissue came from a surgery or biopsy. What is clear is that biopsy samples from liver cancer patients can engraft into useful PDX models. Overall, 23 of 101 (22.8%) primary liver cancers obtained through surgery or biopsy grew macroscopically in the primary PDX transplant setting within 12 months.
Figure 2. Comparison of PDX engraftment in surgical vs. biopsy samples.
A. Engraftment frequency of PDXs in surgical vs. biopsy samples of HCC patients.
B. Engraftment frequency of PDXs in surgical vs. biopsy samples of HCC patients (n = 17, 3).
Fisher’s exact t-test was used for A.
PDXs can be passaged, frozen, thawed, and expanded
For PDX models to be useful for future studies, they not only had to grow in the primary transplant setting, but they needed to be serially passagable. To determine this, we re-implanted fragments of the liver tumor to additional NSG mice without PHx to determine if they could be passaged and how many passages could be sustained. Many PDXs, even if they engrafted initially, did not grow after subsequent passaging. In 9 PDX lines that engrafted in uninjured NSG mice, only one could be serially transplanted (11.1%). In the 5 lines that engrafted in FRG mice, one could be serially transplanted (20%). However, in the 9 lines that engrafted in NSG + PHx mice, 7 could be serially transplanted (Figure 3A) (78%). Thus, host liver resection/regeneration was associated with increased passagability of the PDX lines. The biological mechanisms are unknown, but our data suggested that local or circulating regeneration factors might select for tumor clones that are serially transplantable. Out of the 23 PDXs that engrafted, 15 could be passaged at least once and 9 could be passaged more than 3 to 4 times (Figure 3B). Six of the 15 stopped growing after 2–3 passages. For all passages, we collected and snap froze PDX fragments (~125 mm3) in 10% DMSO/90% FBS to bank tumor stocks. Seven of the lines could be thawed and reimplanted successfully. These lines include HCC-HS84, HCC-HS119, CCA-HS127, CCA-HS131, HCC-HS157, CCA-HB163, and HCC-HB179 (Figure 3C). HS141 could not be successfully thawed but the primary tumor has been passaged for more than 6 times. Altogether, we have generated 8 PDX lines that can be thawed from frozen stocks or maintained as live tumors in mice. In summary, NSG mice undergoing PHx are the optimal hosts for HCC PDX models because of increased engraftment rate, reduced engraftment time, and increased serial passagability after freeze thaw cycles.
Figure 3. The serial passagability of liver cancer PDX models.
A. This shows the passagability of PDXs generated in three types of recipients. Fisher’s exact t-test was used for statistical analysis.
B. Time required for each PDX model to engraft and passage. Once engrafted, tumors were passaged into additional NSG mice to expand the PDX lines. HCC-HS84 was generated from NSG mice. HCC-HS119 was generated from FRG mice. All other lines were generated from NSG mice that had undergone PHx.
C. Engraftment time for frozen and thawed PDX tumors. CCA-HS127, CCA-HS131, HCC-HS84, HCC-HS119, HCC-HS157, and CCA-HB163 can be expanded.
PDXs maintain parental tumor features at the histologic and transcriptomic levels
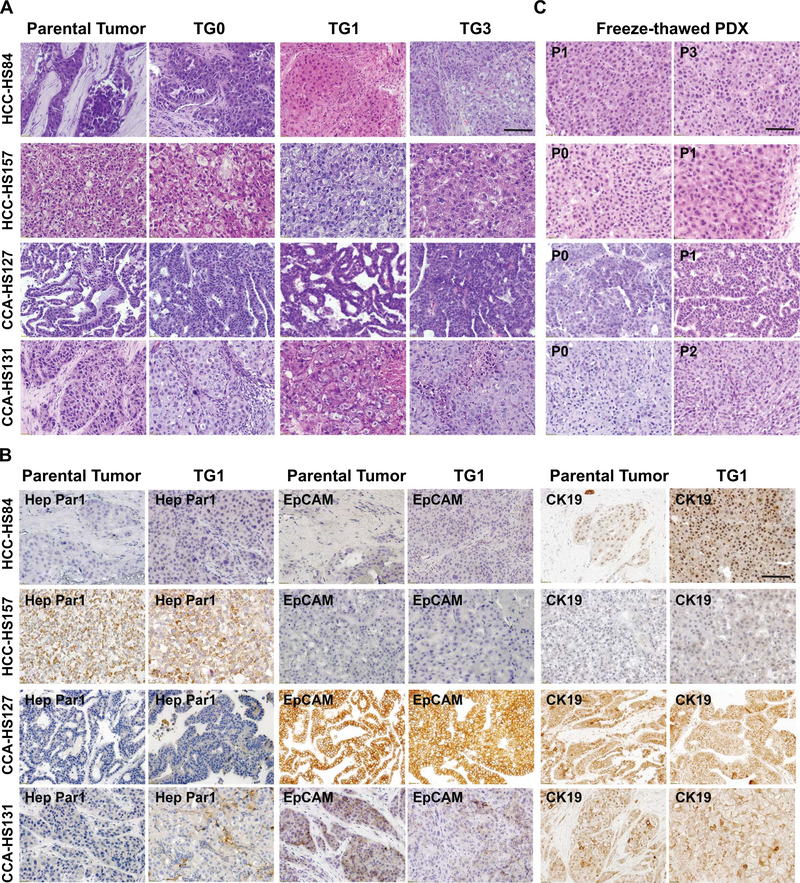
Tumor architecture, histology, growth, and invasiveness of PDX models and their corresponding primary tumors were compared. To confirm the human origin of these PDX tumors, we performed qPCR to show that the PDX lines expressed human rather than mouse genes. We found that 8 originated from human cells (Supplementary Figure 1). We compared the histology of these 8 lines with their parental tumors. H&E staining for PDXs harvested after different passages showed that PDXs and parental tumors were minimally changed over time (Figure 4A, Supplementary Figure 2A, 2C). CD45 staining was used to identify and exclude PDXs that resembled lymphomas (data not shown). Eight pairs of PDXs and parental tumors were stained with Hep Par1, EpCAM, and CK19, markers of hepatic and biliary differentiation. HCC PDXs and primary HCCs expressed the same levels of these markers. As expected, CCA cases were Hep Par1 negative and CK19 positive. Representative images are shown in Figure 4B and Supplementary Figure 2B and 2D. PDX histology was also maintained after thawing and passaging, making it possible to use the PDX models for future studies (Figure 4C, Supplementary Figure 2A, 2C).
Figure 4. PDXs maintain the features of parental tumor histology.
A. H&E staining of parental tumors and passaged PDX lines. Scale bar = 100μm.
B. IHC staining of parental tumor samples and PDXs with anti-Hep Par1, anti-EpCAM, and anti-CK19 antibodies. Scale bar = 100μm.
C. H&E staining showed that thawed PDXs have the similar histology as the primary PDX and patient tumor. Scale bar = 100μm.
H&E = Hematoxylin and eosin; TG = Tumor Graft
Whole-exome and RNA-sequencing (RNA-seq) was used to determine if mutation and expression signatures were preserved. This kind of analysis, along with histologic evaluation, can help to determine if key features of the original cancer are retained. Principal component analysis (PCA) analysis was not able to separate the RNA-seq transcriptomes of seven pairs of tumor and PDX from the same patient (Figure 5A). Clustering based on highly expressed genes in liver cancer resulted in the pairing of PDXs with their corresponding parental tumors (Figure 5B). Whole-exome sequencing analysis also showed that most mutations were retained in the PDXs when compared to parental tumors (Figure 5C). HCC driver mutations were also frequently shared by PDXs and patient tumors (Figure 5D).
Figure 5. HCC PDXs retain the genomic expression and mutational profiles of parental tumors.
A. Principal component analysis (PCA) analysis of RNA-seq data from 7 pairs of parental tumors and PDX samples. T = Tumor, TG = Tumor graft. HS57, HS84, HS119, HS141, HS157 are histologically HCCs. HS127 and HS131 are CCAs.
B. Clustering of samples based on evaluation of highly expressed genes in liver cancer.
C. Whole-exome sequencing in parental tumors and PDX models.
D. Oncoplot of mutations in parental tumors and PDXs. We focused on 35 genes commonly mutated genes in liver cancer based on the TCGA analysis. COSMIC mutations or exome mutations compared to 1000 genomes are shown.
Clinical or genetic features could not predict engraftment of tumor tissues
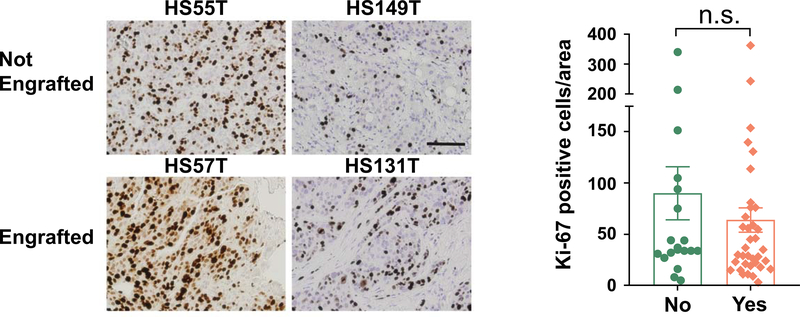
We aimed to determine if clinical variables such as tumor differentiation correlated with engraftment. Among 69 surgical HCC cases, 43 cases were moderately differentiated and 9 of these successfully engrafted (21%, see Supplementary Table 2). Seventeen were poorly differentiated and 5 engrafted (29%). When comparing the engraftment for “moderate”, “moderate to poor”, and “poor” HCCs, there was a non-significant trend of increasing engraftment from 21% to 25% to 29%. However, 3/17 poorly differentiated HCCs were serially transplantable while 0/43 moderately differentiated HCCs were transplantable (Supplementary Table 2, Fisher’s exact test p=0.02). Surprisingly, the well differentiated HCC samples could also engraft, although the number of cases was not high enough to evaluate the engraftment rate. Previous reports of Asian HCC PDX models showed that engraftment correlated with tumor cell proliferation as measured by Ki-67. In our cohort, Ki-67 staining on 20 engrafting and 37 non-engrafting primary tumors showed no significant differences in the frequency of Ki-67 positive cells (Figure 6). To determine if we could identify transcriptomic predictors of engraftment, we also performed deep RNA-seq on cohorts of parental tumors that either did or did not engraft in PDX assays (n = 17 and 19). However, we did not find gene sets that could distinguish between engrafting and non-engrafting cases using gene set enrichment analysis (GSEA). In addition, clinical features such as ALT, AST, and sodium levels could not predict engraftment (data not shown).
Figure 6. Comparison between tumors that did and did not engraft in the PDX assays.
Representative images of Ki-67 staining of patient tumors that did and did not engraft (left). Scale bar = 100μm. Statistical analysis of Ki-67 positive cells (right, n = 20, 37).
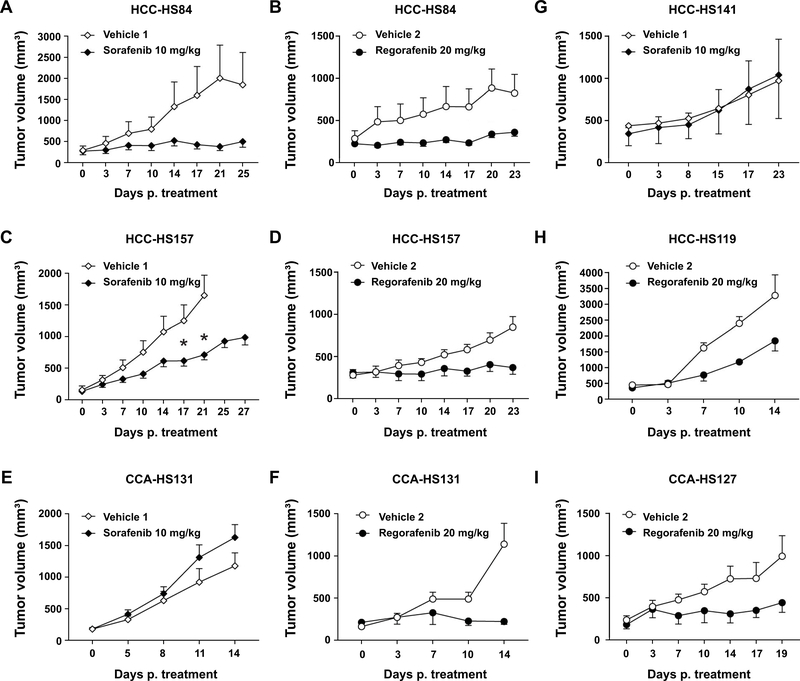
PDX models show differential sensitivity to sorafenib and regorafenib
We sought to ask if liver cancer PDXs could help define therapeutic sensitivities of treatments used in the clinic. Recently, the number of first and second line systemic treatment options for advanced HCC has increased to include four multi-kinase inhibitors and two PD-1 inhibitors, and it has not been clear how to choose which therapy and in which order. Here, we used our PDX models to serve as patient avatars for the choice between the first and second line HCC multikinase inhibitors sorafenib and regorafenib. For sorafenib, two of three HCC PDX models (HCC-HS84, HCC-HS141 and HCC-HS157) showed a substantial response and one did not (Figure 7AC, G), indicating that these models can help to discern sorafenib sensitivity. A CCA PDX model showed no response to sorafenib, as would be expected (Figure 7E). The same PDX models were tested with regorafenib, which is approved for second line HCC treatment in patients who progress on sorafenib (13). More of these models showed sensitivity to regorafenib, but one of the four HCC models was only modestly sensitive (Figure 7B, D, and H). This shows that regorafenib could have a higher response rate in the first line setting and may be appropriate for treatment naive as well as sorafenib resistant patients. The most surprising result was that the CCA-PDX model was exquisitely sensitive to regorafenib, which was previously unknown (Figure 7F and I).
Figure 7. Differential sensitivity of HCC and CCA PDX lines to sorafenib and regorafenib.
A, C, E, G. HCC or CCA PDX lines treated with sorafenib (n = 5 to 10).
B, D, F, H, I. HCC or CCA PDX lines treated with regorafenib (n = 5 to 10).
Two-tailed T-test were used to determine statistical differences between treatments at the same individual time points (shown by asterisks) and Mann Whitney tests were used to identify statistical differences between growth curves (shown by cross).
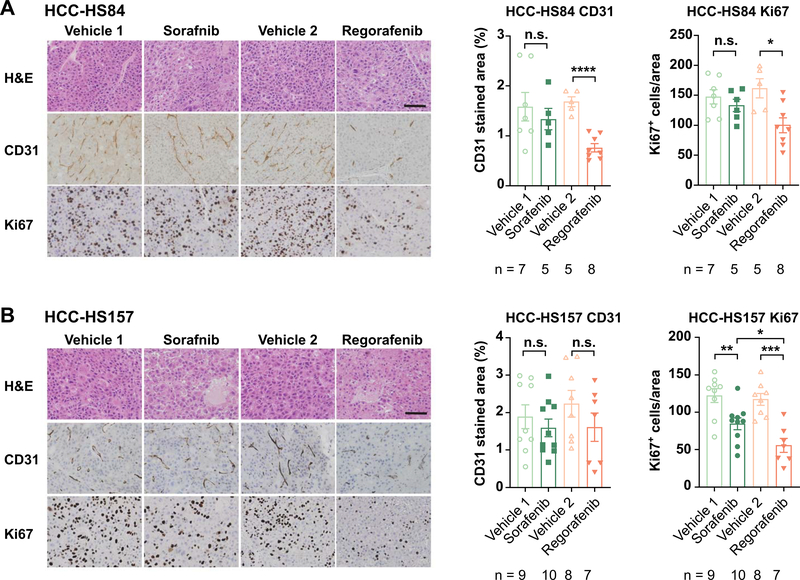
We also performed histological and molecular analyses of treated vs. untreated PDX tumors in an effort to reveal mechanisms by which sorafenib or regorafenib could impede tumorigenesis. We measured the anti-angiogenesis effects by using immunohistochemistry of CD31, which is an endothelial marker that allows us to quantify vessels and we measured proliferation using Ki-67 (Figure 8). Both drugs had an effect on proliferation, but regorafenib had a more pronounced effect in both PDX lines. While sorafenib did not have a significant effect on vascular density as measured by CD31, regorafenib had a significant effect in the HCC-HS84 PDX line. Thus, these drugs have a different magnitude of anti-angiogenesis and anti-proliferative effects in different lines, and this can partially explain the sensitivity of each line.
Figure 8. Effect of regorafenib and sorafenib on angiogenesis and cell proliferation in PDX models.
A. Representative images of H&E, angiogenesis (CD31), and proliferation (Ki-67) of HCC-HS84 PDX treated with Sorafenib or Regorafenib (left). Statistical analysis of CD31 positive area and Ki-67 positive cells (right). Scale bar = 100μm.
B. Representative images of H&E, angiogenesis (CD31) and proliferation (Ki-67) of HCC-HS157 PDX treated with Sorafenib or Regorafenib (left). Statistical analysis of CD31 positive area and Ki-67 positive cells (right). Scale bar = 100μm.
DISCUSSION
PDX models have the potential to uncover new biological information about individual patient derived liver tumors that have not been adulterated by prolonged growth on plastic under artificial nutrient conditions. Our goal was to understand tumor intrinsic and extrinsic features that would impact tumor biology using PDX engraftment, growth, and passagability as in vivo assays. We first attempted to pinpoint the tumor extrinsic host factors that regulate liver cancer growth. In general, it has been difficult to assess the impact of the tumor microenvironment on clinically-relevant liver tumor models. Within genetically engineered mouse models, it is difficult to isolate tumor intrinsic and extrinsic factors. Also, commonly used HCC cell lines have been so intensely selected for rapid growth that they may not respond to more subtle environmental cues that would otherwise have a greater impact on slower growing tumors. In our experience, it was surprising that the location of implantation (SC or liver) did not have a major influence on tumor growth. It is possible that the technical challenges associated with liver orthograft implantation disadvantaged that approach when compared to SC implants, which could accommodate more transplanted tissue.
Not surprisingly, inducing liver injury in immunodeficient mice could promote tumor growth. It is known that tumor initiation is promoted by liver damage, but it is less well understood if the growth of established tumors is enhanced by liver injury. Our data shows that even established HCCs can engraft and grow more efficiently when liver injury is introduced through genetic liver damage (Fah deletion) or surgical resection (PHx). Interestingly, our experiments support a model whereby both signals acting locally in FRG mice and in circulation in the context of hepatectomized NSG mice could promote cancer growth. Deeper immunosuppression was also important, since tumors generally grew better in NSG as compared to FRG mice. This information about non-cell autonomous regulators of HCC growth allowed us to optimize our efforts to functionalize living tumor tissues. We recommend performing 40% PHx in order to promote growth and long-term passagability of HCCs implanted into the SC space of NSG mice.
In regards to comparing host models, there are limitations to our study that need to be highlighted. Because of time constraints that occur after clinical liver resections, we were not able to implant primary patient tumor tissues into the three different host mouse models at the same time. Each host model requires 3–5 mice and thus performing 10–15 surgeries at the end of the clinical workday was not feasible. Instead, our study retrospectively compared the engraftment efficiencies between different hosts, and therefore we cannot say that for each patient, one engraftment approach is definitively better than another. However, we did implant two established PDX models into all three of the hosts and we found that PHx could modestly improve the growth of one of the models within the liver as compared to FRG or no PHx (Supplemental Figure 3). While this indicates that PHx is likely to be an important instigator of engraftment or growth, this is anecdotal data. Another caveat is that we cannot exclude the effects of the surgical procedure itself when we perform PHx in NSG mice. Sham operations without PHx would have provided better controls. Despite these limitations, we believe that there is robust evidence that liver injury exemplified by PHx can promote PDX engraftment in heavily immunosuppressed mice.
There were also tumor intrinsic features of the HCCs that influenced PDX growth. A study by Gu and colleagues showed that more than 60 viable HCC PDX models have been generated from Chinese patients. These PDXs readily engraft, grow, and are used for drug studies (7). Interestingly, our experience with U.S. patients did not mirror that study despite implanting over 100 patient cases. In general, engraftment frequency, growth rates, and passagability were very low in comparison to the Asian studies. We are confident that there were no technical issues impairing PDX engraftment since our group has also generated 30 gastric cancer PDX models from 70 implanted (43% success rate, unpublished), even considering the fact that these PDXs were generated from esophagogastroduodenoscopy derived biopsies that are generally more scant in mass and cellularity. Our data suggests inherent biological differences between liver cancers from different geographic locations, either due to genetic background or etiology.
Overall, we implanted 93 HCC samples and 8 CAA samples in different recipient mice to generate PDX models. Our results showed that immune deficiency and liver injury could improve engraftment and PDX passagability. Using the PDX lines generated with this study, we were able to test the differential sensitivity of sorafenib and regorafenib in both HCC and CCA PDXs. Furthermore, we also found that PDX lines can be generated from surgical samples as well as biopsy samples, making it possible to pre-clinically test drugs for patients with advanced liver cancer. Our study reports an important experience in a large number of liver cancer patients and lays the foundation for future efforts to functionalize HCCs and CCAs from patients.
METHODS
Mice
All mice were handled in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) at UTSW. NSG mice were from UT Southwestern breeding core; FRG mice were from Yecuris Corporation (strain 10–0001). All experiments were done in 6–10 week old male and female mice. All experiments were done in an age and sex controlled fashion unless otherwise noted.
Human samples
HCC and CCA tumor samples were obtained from patients who underwent surgical resection or percutaneous biopsies. All patients provided informed consent under IRB #STU 062013–063 for liver tissues and #STU 092013–010 for blood samples. Pathologic diagnosis was confirmed by a board-certified pathologist specializing in gastrointestinal oncology (P.G.) in a blinded fashion. A total of 101 tumor samples were implanted. Patients have an average age of 62 years old. 77 were male and 24 were female.
Implantation of tumors into recipient mice
For SC experiments, multiple tumor fragments of 2–5 mm3 were implanted with or without PHx. For liver implantation, an incision was made in abdomen and a fragment (1–2 mm3) was implanted and sealed with a small piece of Surgicel to stop bleeding. For FRG mice, NTBC water was removed after the surgery and cycled every 7 to 10 days to induce liver injury. Mice implanted with parental tumors were euthanized before any tumor grew to 2 cm in diameter. Tumor growth in liver was examined at the time of euthanasia. To expand a PDX model, tumors were implanted to multiple NSG mice without partial hepatectomy, snap frozen for future data analysis, fixed with 4% paraformaldehyde for histology, and stocked in 10% DMSO + 90% FBS for future use. Some xenografts were thawed from frozen stocks and re-implanted into NSG mice subcutaneously. After thawing and implanting, mice were maintained the same way as primary xenograft implantation.
Histology
Tissue samples were fixed in 4% paraformaldehyde and paraffin embedded. Primary antibodies used were Hep Par1 (OCH1E5) (Sigma 264M-94), EpCAM (Cell Signaling #14452), CK19 (Abcam ab15463), CD31 (Abcam ab28364), CD45 (Abcam ab10558), and Ki-67 (Abcam ab15580). Detection was performed with the Elite ABC Kit and DAB Substrate (Vector Laboratories), followed by hematoxylin counterstaining (Vector Laboratories).
RNA extraction and RT_qPCR
Total RNA was extracted from tumors and corresponding xenografts using the Invitrogen PureLink RNA mini kit. cDNA was synthesized using iScript reverse transcription reagents (Bio-Rad). Human or mouse gene expression were detected using human or mouse specific primers. A common GAPDH primer set which reacts with both human and mouse GAPDH was used as reference gene. Expression level was normalized to human or mouse sample controls.
Drug studies with PDX models
125 mm3 tumor fragments were implanted in both flanks of 10 NSG mice, with 5 mice for each group. Treatment was initiated when the tumor volume reached 50~200 mm3. Sorafenib and Regorafenib were purchased from LC Laboratories. Sorafenib was dissolved with Cremophor EL: ethanol (1:1) for stock and diluted four-fold with water just before use (14). 10 mg/kg sorafenib was given by oral gavage once a day and control mice were given diluted vehicle. Regorafenib was dissolved in DMSO at 100 mg/ml for stock solution and then diluted using 1:1 mixture of PEG 300 and 30% captisol (15). Mice were gavaged once daily with regorafenib or vehicle at 20 mg/. Long (L) and short diameters (S) were measured for each tumor twice a week. Tumor volume was calculated by V=1/2*L*S2.
Whole exome sequencing, processing, and mutation calling
Genome DNA was extracted from parental tumors and corresponding xenografts using Qiagen all-prep DNA/RNA mini kit. DNA was submitted to Admera for whole-exome sequencing. We used the QBRC pipeline (github.com/Somatic-pipeline/QBRC-Somatic-Pipeline) for somatic mutation calling. Exome-seq reads were aligned to the GRCh38 genome by BWA-MEM (16). Picard was used to add read group information and sambamba was used to mark PCR duplicates. The calculation of read coverage is performed after duplicate removal. GATK toolkit (17–19) was used to perform base quality score recalibration and local realignment around Indels. MuTect (20), VarScan (21), Shimmer (22), SpeedSeq (23), Manta (24), and Strelka2 (25) were used to call SNPs and Indels. A mutation called by >= any 3 of these algorithms was retained. Annovar was used to annotate SNPs and Indels (26). All SNPs and Indels were combined and only kept if there are at least 7 total (wild type and variant) reads in the blood normal sample and at least 3 variant reads in the parental tumor or xenograft sample. Only mutations found in COSMIC (https://cancer.sanger.ac.uk/cosmic) were kept. The visualization of mutations by oncoplot is generated by function oncoplot in R package maftools.
RNA-Sequencing and Data Analysis
Total RNA was extracted from tumors using the Invitrogen PureLink RNA mini kit. Libraries were prepared with the Ovation RNA-Seq Systems 1–16 (Nugen) and indexed libraries were multiplexed in a single flow cell and underwent 75 base pair single-end sequencing on an Illumina NextSeq500 using the High Output kit v2 (75 cycles) at the UTSW Children’s Research Institute Sequencing Facility. To compare RNA expression between parental tumor samples that engrafted for did not engraft, RNA was extracted and submitted to Admera for paired-end RNA-seq. The sequence reads were aligned to the GRCm38 with STAR (27,28). Read counts were generated for the annotated genes by featureCounts (29,30). Differential gene analysis was performed use edgeR, using FDR < 0.05 as cutoff (31,32). Heatmaps to visualize the data were generated by using R heat.map2 package. GSEA analysis was performed with a pre-ranked gene list by log fold change (33). PCA analysis was performed by the R prcomp function.
Statistical Analysis
The data in most Figures reflect multiple experiments performed on different days using mice from different litters. Two-tailed Student’s t-tests (two-sample equal variance) were used to test the significance of differences between two groups. Fisher’s exact tests and Mann Whitney tests were used where specifically indicated. In all Figures, statistical significance is represented as mean ± SEM, * (p < 0.05), ** (p < 0.01), *** (p < 0.001). † (p <0.05) and ‡ (p<0.01).
DATA AND SOFTWARE AVAILABILITY
The sequencing data reported in this paper will be deposited into the European Genome-phenome Archive (EGA) database. The EGA access ID will be provided once the uploading is complete.
Supplementary Material
Highlights.
We present the first United States liver cancer PDX biobank from surgical and biopsy cases.
Human HCC engraftment in mice increased with greater immunodeficiency and liver injury.
The frequency and rate of engraftment is low, indicative of unique biological features of HCCs.
These patient avatars show that there are differential sensitivities to sorafenib and regorafenib reflecting heterogeneous responses seen in the clinic.
Acknowledgments
We thank J. Shelton and C. Lewis for histology, the UTSW Bioinformatics and the CRI Sequencing Core for sequencing. P.G. was supported by the UTSW Department of Pathology Intramural Research Program. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Cancer Research Program under Award No. W81XWH-16-1-0158. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. A.G.S. and A.Y. are in part supported by National Cancer Institute (NCI) R01 MD12565. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding agencies had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation of the manuscript.
List of Abbreviations
- HCC
Hepatocellular carcinoma
- PDX
Patient derived xenograft
- CCA
Cholangiocarcinoma
- PHx
Partial hepatectomy
- HCV
Hepatitis C virus
- HBV
Hepatitis B virus
- NASH
Non-alcoholic steatohepatitis
- BCLC
Barcelona-Clinic Liver Cancer
- SC
Subcutaneous
- NSG
NOD Scid IL-2Rγ−/−
- FAH
Fumarylacetoacetate hydrolase
- FRG
Fumarylacetoacetate hydrolase (Fah)−/−; Rag1−/−; IL-2Rγ−/−
- NTBC
2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione
Footnotes
Conflicts of Interest: A.G.S. has served on advisory boards or consulted for Bayer, Eisai, BMS, Exelixis, and TARGET. None of the other authors have any relevant conflicts of interest.
References
- 1.Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol (NY). 2018;43:13–25. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–34. [DOI] [PubMed] [Google Scholar]
- 3.White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin. Gastroenterol. Hepatol. 2012;10:1342–1359.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruix J, Sherman M, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh H, Soo KC, Chow PKH, Panasci L, Tran E. Xenografts of human hepatocellular carcinoma: a useful model for testing drugs. Clin. Cancer Res. 2006;12:4306–4314. [DOI] [PubMed] [Google Scholar]
- 6.Wei W, Wu S, Wang X, Sun CK-W, Yang X, Yan X, et al. Novel celastrol derivatives inhibit the growth of hepatocellular carcinoma patient-derived xenografts. Oncotarget. 2014;5:5819–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Q, Zhang B, Sun H, Xu Q, Tan Y, Wang G, et al. Genomic characterization of a large panel of patient-derived hepatocellular carcinoma xenograft tumor models for preclinical development. Oncotarget. 2015;6:20160–20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan M, Li H, Zhao F, Zhang L, Ge C, Yao M, et al. Establishment of NOD/SCID mouse models of human hepatocellular carcinoma via subcutaneous transplantation of histologically intact tumor tissue. Chin. J. Cancer Res. 2013;25:289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JD, Kim WR, Coelho R, Mettler TA, Benson JT, Sanderson SO, et al. Cirrhosis is present in most patients with hepatitis B and hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 2011;9:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An C, Choi YA, Choi D, Paik YH, Ahn SH, Kim M-J, et al. Growth rate of early-stage hepatocellular carcinoma in patients with chronic liver disease. Clin. Mol. Hepatol. 2015;21:279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, et al. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat. Biotechnol. 2007;25:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grompe M, Lindstedt S, al-Dhalimy M, Kennaway NG, Papaconstantinou J, Torres-Ramos CA, et al. Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 1995;10:453–460. [DOI] [PubMed] [Google Scholar]
- 13.Trojan J, Waidmann O. Role of regorafenib as second-line therapy and landscape of investigational treatment options in advanced hepatocellular carcinoma. J. Hepatocell. Carcinoma. 2016;3:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoshita M, Scopton AP, Ung PMU, Murray MA, Silber L, Maldonado AY, et al. A whole-animal platform to advance a clinical kinase inhibitor into new disease space. Nat. Chem. Biol. 2018;14:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh H, Ong R, Zopf D. Antitumor activity of the multikinase inhibitor regorafenib in patient-derived xenograft models of gastric cancer. J. Exp. Clin. Cancer Res. 2015;34:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;11:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen NF, Gartner JJ, Mei L, Samuels Y, Mullikin JC. Shimmer: detection of genetic alterations in tumors using next-generation sequence data. Bioinformatics. 2013;29:1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods. 2015;12:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 25.Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this paper will be deposited into the European Genome-phenome Archive (EGA) database. The EGA access ID will be provided once the uploading is complete.