Abstract
Current yeast metabolic engineering in isoprenoids production mainly focuses on rewiring of cytosolic metabolic pathway. However, the precursors, cofactors and the enzymes are distributed in various sub-cellular compartments, which may hamper isoprenoid biosynthesis. On the other side, pathway compartmentalization provides several advantages for improving metabolic flux toward target products. We here summarize the recent advances on harnessing sub-organelle for isoprenoids biosynthesis in yeast, and analyze the knowledge about the localization of enzymes, cofactors and metabolites for guiding the rewiring of the sub-organelle metabolism. This review may provide some insights for constructing efficient yeast cell factories for production of isoprenoids and even other natural products.
Keywords: Isoprenoids, Yeast, Compartmentalization, Sub-organelle metabolism
1. Introduction
Isoprenoids, also known as terpenoids, are a large and diverse group of natural compounds used as fragrances and flavors, pharmaceuticals, solvents, cosmetics, food additives and potential advanced biofuels [1]. Yeasts are considered as ideal hosts for production of isoprenoids due to their robustness [2]. In yeast, isoprenoids are synthesized through the mevalonate (MVA) pathway, which have a great demand for acetyl-CoA, NADPH and ATP. Currently, isoprenoids biosynthesis pathways are mainly constructed and optimized in the yeast cytosol by enhancing cofactor supply, and the metabolic fluxes of acetyl-CoA pathway and MVA pathway [[3], [4], [5], [6], [7]]. However, eukaryotic cells are divided into several sub-organelles, which have complex structures with their own membranes, metabolic pathways and in some cases, genomes. In particular, the cellular metabolism is compartmentalized into specialized sub-organelles. For example, oxidative phosphorylation happens in mitochondria and β-oxidation of fatty acids localizes in peroxisomes, which results in the dispersion of cofactors and precursors [8,9]. This subcellular compartmentalization may bring some barriers for substrate channeling in supplying precursors and cofactors.
On the other hand, the membrane boundaries may provide advantages for construction of heterologous pathways. Firstly, concentrating enzymes, cofactors and substrates for improved substrate channeling in compact spaces [10]. Secondly, relieving side-pathway competition for improved biosynthesis selectivity by secluding the biosynthetic pathways from efficient competing enzymes [11]. Thirdly, alleviating the cytoxicity of some hydrophobic products by targeting the biosynthetic pathways into sub-organelles [12]. Though extensive metabolic engineering has been conducted, the production of most isoprenoids in yeast is far behind industrial application, which might be partly attributed to the complex metabolic compartmentalization. Thus, systematic investigation of the cellular sub-compartments and harnessing sub-organelles may provide a feasible approach for further enhancing isoprenoid biosynthesis in yeast and even other eukaryotic cells. In particular, the development of molecular biology makes this more practicable by providing genetic tools such as organelle targeting signals (eg. peroxisomal targeting signals [13,14], the mitochondrial localization signal [15,16]).
In spite of the advantages of organelle engineering, there are still substantial challenges to overcome. The suitable host and organelles should be carefully selected according to the target products to make sure the availability of precursors and cofactors and convenience in construction of the target pathway. However, the knowledge of the subcellular localization of proteins, the membrane transporters for metabolites and cofactors is not totally clear. In this mini-review, we firstly summarize the current exploration in localization and transportation of proteins and metabolites, cofactors across subcellular compartments. Then, we discuss the recent advances and challenges in engineering yeast for isoprenoids biosynthesis in subcellular compartments.
2. Isoprenoids biosynthesis is closely related to subcellular compartments function
Isoprenoids biosynthesis requires acetyl-CoA and reducing power NADPH, which are distributed into various cellular compartments. For example, acetyl-CoA can be cytosolically produced from pyruvate by pyruvate dehydrogenase (Pdh) bypass pathway in the Crabtree-positive Saccharomyces cerevisiae, and generated from citrate by ATP: citrate lyase (Acl) in respiratory oleaginous yeasts. It can also be synthesized in mitochondria by pyruvate dehydrogenase complex (mPdh) pathway as a starting unit in tricarboxylic acid (TCA) cycle and as a degradation product of β-oxidation of fatty acids in peroxisomes [17]. As for NADPH, the cytosolic pentose phosphate pathway (PPP) is the main source of cellular NADPH, which can also be synthesized in peroxisome by isocitrate dehydrogenase (Idp3) through isocitrate/2-oxoglutarate shuttle [18,19]. There is no report on mitochondrial pathways for producing NADPH, which might be imported from cytosol. The sub-organelle distribution of precursors and cofactors suggests that knowing well about localization and regulation of proteins, metabolites and cofactors may help to optimize isoprenoid biosynthesis in yeast by balancing supply of carbon and energy.
2.1. Subcellular localization of isoprenoids related enzymes
Other than acetyl-CoA and NADPH, the isoprenoid biosynthetic enzymes may also localize in different cellular compartments. The knowledge about subcellular localization and abundance of proteins will enhance our understanding of their functions and interactions and also help to optimize the targeted cellular pathways [20,21]. Thus, protein localization information is deeply concerned in recent years [21,22].The model yeast S. cerevisiae has been extensively studied in subcellular protein localization by fluorescence microscopy [21,23]. In addition, computational prediction and mass spectrometry approaches have also been developed for analyzing protein localization [24,25]. Here, we summarize the databases about the collection of green fluorescent protein (GFP) (or otherwise)-tagged proteins for obtaining the localization of proteins. YGFP, the Yeast GFP Fusion Localization Database contained in Saccharomyces Genome Database (SGD), defined 4156 proteins about their subcellular localization, representing 75% of the yeast proteome [21,26]. YPL, the Yeast Protein Localization Database, has two versions (YPL.db, YPL.db2) and harbors 500 sets of image data from high-resolution microscopic analysis of proteins tagged with GFP [27,28]. Moreover, this database can be linked to external databases like SGD. Organelle DB, a web-accessible relational database of organelles and protein complexes, covers over 30 000 proteins from 138 eukaryotic organisms [29]. About S. cerevisiae, SGD database contains compiled protein localization data from large-scale and systematic manual analysis. YRC PIR, the Yeast Resource Center Public Image Repository, is a large database of images depicting the subcellular localization and colocalization of proteins [30]. Other than protein annotation, the YRC PIR aims to providing a plethora of images and their correlating metadata for proteins among multiple organisms. CYCLoPs, the Collection of Yeast Cells Localization Patterns, is a web database that provides retrieval and visualization of yeast cell images and permits the queries of the subcellular localization and abundance profiles of the yeast proteome at single cell level [24]. The data from CYCLoPs also contains computationally derived quantitative profiles of localization and abundance.
According to SGD [26], proteins can be located in 11 different subcellular locations: cell wall, plasma membrane, cytosol, nucleus, mitochondrion, peroxisomes, lipid droplets (LDs), endosome, the endoplasmic reticulum (ER), the Golgi apparatus, and vacuole. We summarize the subcellular localizations of proteins (Erg10, acetoacetyl-CoA thiolase; Erg13, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase; Hmg1,2, HMG-CoA reductase; Erg12, mevalonate kinase; Erg8, phosphomevalonate kinase; Erg19, mevalonate diphosphate decarboxylase; Idi1, isopentenyl diphosphate: dimethylallyl diphosphate isomerase; Erg20, farnesyl diphosphate synthase; Bts1, geranylgeranyl diphosphate synthase; Erg9, squalene synthase; Erg1, squalene epoxidase.) in the MVA pathway (Fig. 1), based on the databases above-mentioned (Table 1). The proteins involved in MVA pathway are distributed in different subcellular compartments: nucleus, cytosol, mitochondrion and ER. It is also noteworthy that the localizations are slightly distinct from different databases, especially the data from Organelle DB in compared to others. The reason may be that there are limitations of fluorescence microscopy in accurately analyzing protein localization, like tagging approaches can result in mis-localizations and the GFP is mostly tagged at the C′ terminus, ignoring the importance of N’ terminus tagging [31].
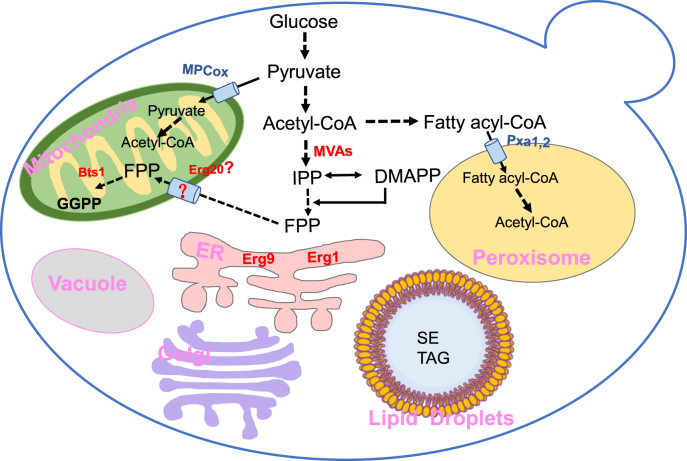
Fig. 1.
Protein localization involved in isoprenoids biosynthesis. The protein localization databases suggest that MVA pathway related proteins are mostly located in the cytosol, while Bts1 locates in mitochondria, Erg9 and Erg1 locate in the ER, and the localization of Erg20 is not verified yet. Bts1, geranylgeranyl diphosphate synthase; Erg20, farnesyl diphosphate synthase; Erg9, squalene synthase; Erg1, squalene epoxidase; MVAs, synthases involved in the MVA pathway; MPCox, mitochondrial pyruvate carrier; Pxa1,2, ATP-binding cassette transporter 1,2; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl diphosphate; ER, endoplasmic reticulum; SEs, sterol esters; TAGs, triaclglycerols.
Table 1.
Subcellular localization of proteins involved in isoprenoids biosynthesis.
| proteins | YGFP | Organelle DB | YPL | CYCLoPs |
|---|---|---|---|---|
| Erg10 | Nulceus | Cytosol | Nulceus | Nulceus |
| Erg13 | Nulceus | ER & Mitochondrion | Nulceus | Nulceus |
| Hmg1 | Nulceus periphery | ER & Mitochondrion & Nucleus | Nulceus periphery | Nulceus periphery & Vac/Vac Membrane |
| Hmg2 | Nulceus periphery | ER & Mitochondrion & Nucleus | Nulceus periphery | Nulceus periphery & Vac/Vac Membrane |
| Erg12 | Cytosol & Nulceus | Cytosol | Cytosol | Cytosol |
| Erg19 | Cytosol | Cytosol | Cytosol | Cytosol |
| Erg8 | Cytosol & Nulceus | Cytosol | Cytosol | Cytosol |
| Idi1 | Cytosol & Nulceus | Cytosol | Cytosol | Cytosol |
| Erg20 | Mitochondrion | Cytosol | ND | ND |
| Bts1 | Mitochondrion | Mitochondrion | Mitochondrion | Mitochondrion |
| Erg9 | ER | ER & Mitochondrion | ER | Cytosol & ER |
| Erg1 | ER | ER | ER & LDs | ER |
ND: not detected.
Recently, a SWAp-Tag (SWAT) method was developed for fast and effortless creation of yeast libraries [31,32]. Besides, the SWAT approach can be used to analyze protein abundance and uncover localization of hundreds of proteins that have never been annotated, and even define a more complete mitochondrial and peroxisomal proteome. This method will facilitate the accurate characterization of enzyme localization of isoprenoid biosynthetic pathways.
2.2. Metabolite transport across subcellular compartments
Though segregated by the membranes, sub-organelles actively communicate and interact with other subcellular compartments to maintain cellular function. Therefore, it is very critical to understand the transferring of metabolites and cofactors across subcellular compartments for rational regulation of the compartmentalized pathways.
The mitochondrion is surrounded by an outer membrane which is permeable to solutes with a molecular mass ≤4–5 kDa and an inner membrane which is rigidly impermeable [33]. The mitochondrial membranes are embedded with a series of mitochondrial carriers (MCs) [34], which transport metabolites, nucleotides, ions and coenzymes. The MCs is defined by the sequence features of its members: a tripartite structure, three tandem homologous sequence repeats of approximately 100 amino acids and six transmembrane α-helices [35]. There are 35 MCs in S. cerevisiae (Table 2), which are classified as nucleotides/dinucleotides transport [[36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]], di-/tri-carboxylates and oxo acids transport [[47], [48], [49], [50], [51], [52], [53], [54], [55], [56]], amino acids transport [[57], [58], [59], [60]], metal ion transport [[61], [62], [63], [64], [65]] and other transporters [66]. Engineering the MCs can regulate the metabolic flux toward targeted products by regulating the metabolites transportation across the mitochondrion. For example, we previously enhanced cytosolic acetyl-CoA for fatty acid biosynthesis by overexpressing MPC1,3 and YHM2(CRC1), which was supposed to increase the transport efficiency of mitochondrial importing of pyruvate and exporting of citrate, respectively [67].
Table 2.
Mitochondrial carriers classified according to substrate specificity.
| Mitochondrial carrier | Description | Main substrates | Main metabolic roles | Reference |
|---|---|---|---|---|
| For nucleotides/dinucleotides | ||||
| Ant1 | Peroxisomal adenine nucleotide | ATP, AMP | Peroxisomal fatty acids β-oxidation | [36] |
| Tpc1 | Thiamine pyrophosphate | ThPP, ThMP | Regulation of acetolactate synthase (ALS), pyruvate dehydrogenase (PDH) and oxoglutarate dehydrogenase (OGDH) | [37] |
| Ggc1 | GTP/GDP | GTP, GDP | Intramitochondrial nucleic acid and protein synthesis | [38] |
| Rim2 (Pyt1) | Pyrimidine nucleotides | PyNTPs, PyNMPs | DNA and RNA synthesis | [39] |
| Ndt1-2 | NAD+ | NAD+, (d)AMP, (d)GMP | Redox balance | [40] |
| Apsc1 | Adenosine 5′-phosphosulfate | Adenosine 5′-phosphosulfate |
Thermotolerance, methionine and glutathione synthesis | [41] |
| Aac1-3 | ADP/ATP | ADP/ATP | Oxidative phosphorylation | [[42], [43], [44]] |
| Flx1 | FAD | FAD | Balance of Flavin Nucleotides | [45] |
| Sal1 | ATP-Mg/Pi | ADP, ATP, ATP-Mg, Pi | Oxidative phosphorylation | [46] |
| For di-/tri-carboxylates and oxo acids | ||||
| Dic1 | Dicarboxylate | malate, succinate, malonate, Pi | Krebs cycle, gluconeogenesis, urea synthesis | [47,48] |
| Sfc1 | Succinate-fumarate | Succinate, fumarate | Gluconeogenesis | [49] |
| Oac1 | Oxaloacetate-sulfate | Oxaloacetate, sulfate | Krebs cycle, leucine synthesis | [50,51] |
| Odc1-2 | Oxodicarboxylate | Oxoadipate, oxoglutarate | Lysine and tryptophan metabolism | [52] |
| Yhm2p (Coc1) | Citrate-oxoglutarate | Citrate, oxoglutarate | Citrate-oxoglutarate NADPH shuttle from the mitochondrial matrix to the cytosol | [53] |
| Ctp1 | Citrate | Citrate, isocitrate | Krebs cycle | [54] |
| Mpc1-3 | Pyruvate | Pyruvate | Krebs cycle, lipoic acid synthesis | [55,56] |
| For amino acids | ||||
| Ort1 | Ornithine | Ornithine, arginine, lysine | Amino acid metabolism, urea synthesis | [57] |
| Sam5 | S-adenosylmethionine | S-adenosylmethionine, S-adenosylhomocysteine |
Biotin and lipoic acid synthesis | [58] |
| Crc1 | Carnitine | Carnitine, acylcarnitine | Fatty acids β-oxidation,ethanol oxidation | [59] |
| Agc1 | Aspartate-glutamate | Aspartate, glutamate | Malate/aspartate shuttle | [60] |
| For metal ion | ||||
| Mrs3-4 | Iron | Iron | heme formation and Fe/S protein biosynthesis | [61] |
| Mrs2, Lpe10, Mme1 | Mg2+ | Mg2+ | Energy production, ion metabolism | [[62], [63], [64]] |
| Pic2 | Copper | Copper | Cytochrome c synthesis, protection against oxidative stress | [65] |
| For others | ||||
| Mir1, Pic2 | Phosphate | Pi | Oxidative phosphorylation | [66] |
In contrast to mitochondrion, peroxisomes are surrounded by a single layer of membrane, which is permeable to metabolites up to ~700 Da [68]. This suggests that some essential metabolites and cofactors, such as acetyl-CoA (809 Da) and NADPH (744 Da), are dependent on transporters for crossing the peroxisomal membrane, which has been shown that the peroxisomal membrane of S. cerevisiae is impermeable to NADP(H) and acetyl-CoA in vivo [69]. However, there is only a few of peroxisomal carriers found in S. cerevisiae, including two ATP-binding cassette (ABC) transporters, Pxa1 and Pxa2 for importing long chain fatty acyl-CoA [70] and ATP transporter Ant1 for importing cytosolic ATP into the peroxisomal lumen in exchange with AMP that derived from fatty acid activation [36]. There are no peroxisomal NAD+, NADP+ and acetyl-CoA transporters identified in S. cerevisiae, however, there are a number of shuttle systems involved in the re-generation of peroxisomal cofactors without transportation across the membrane. NAD+ is regenerated in peroxisomes through the malate-oxaloacetate shuttle and malate-aspartate shuttle [71], while NADPH is regenerated via the isocitrate/2-oxoglutarate shuttle [18,19]. Acetyl-CoA generated by peroxisomal β-oxidation is exported by the carnitine shuttle [72] or through the export of citrate. Though the shuttle systems seemly guarantee cofactors regeneration in peroxisomes, it can't be completely excluded the existence of cofactor transporters on peroxisome membrane, since At2g39970 has been characterized as an peroxisomal transporter of NAD+, NADH, AMP and ADP in Arabidopsis thaliana [73,74] and SLC25A17 identified as a peroxisomal transporter of CoA, FAD, FMN and AMP in human [75]. In Fig. 2, we depicted the transmission of acetyl-CoA, energy, cofactors related to isoprenoids synthesis across the cytosol, mitochondria and peroxisomes.
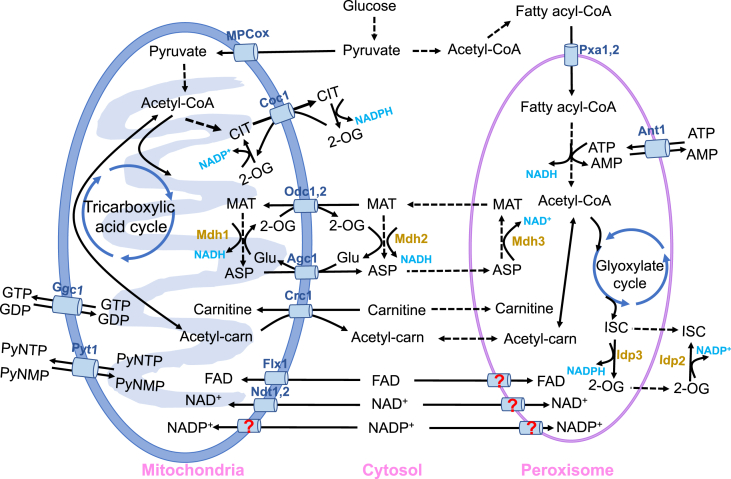
Fig. 2.
Acetyl-CoA, energy, cofactors transmission across the cytosol, mitochondria and peroxisomes.Acetyl-CoA, energy, cofactors are transported across mitochondria and peroxisomes with corresponding transporters and shuttle systems. There are still some transporters not yet identified in yeast for mitochondria NADP+ transportation, and peroxisomal transportation of FAD, NAD+ and NADP+. Mdh1,2,3, malate dehydrogenase1,2,3; Idp2,3, isocitrate dehydrogenase2,3; Pxa1,2, ATP-binding cassette transporter1,2; CIT, citrate; 2-OG, 2-oxoglutarate; MAT, malate; ASP, aspartate; ISC, isocitrate; ATP, adenosine triphosphate; AMP, adenosine monophosphate; GTP, guanosine triphosphate; GDP, guanosine diphosphate; PyNTP, pyrimidine nucleoside triphosphate; PyNMP, pyrimidine nucleoside monophosphate; FAD, flavin adenine dinucleotide; NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate. For mitochondrial carriers, see Table 2.
In addition, organelles could be tethered to each other. These direct connections are termed membrane contact sites (MCSs) that have been measured to be 10–30 nm wide between two organelles [76]. Especially the ER has been found to form contact sites with many other cytoplasmic organelles, including the mitochondria, golgi, peroxisomes, endosomes, LDs and the plasma membrane [77]. One of the MCSs has been well-characterized is the ER-mitochondria encounter structure (ERMES), which is involved in lipid synthesis [78]. In another study, peroxisomal protein Pex11was found to play a role in establishing the contact sites between peroxisomes and mitochondria through the ERMES complex [79]. Engineering the metabolites biosynthesis and transportation among various sub-organelles may provide an alternative strategy for improving the isoprenoids biosynthesis.
3. Metabolic engineering of yeast organelles for isoprenoids biosynthesis
Yeast metabolic engineering for isoprenoids production mostly focused on pathway modification in the cytosol (Fig. 3A), which mainly enhanced the metabolic flux for acetyl-CoA and the MVA pathway. For acetyl-CoA generation, several cytosolic pathways, including PDH-bypass pathway [80,81], pyruvate:formate lyase pathway [82], cytosolic PDH pathway [83], and chimeric phosphoketolase (xPK)-phosphotransacetylase pathway [7], were constructed or optimized. In addition, ATP-citrate lyase derived pathway was reconstructed and optimized for channeling the mitochondrial acetyl-CoA flux toward cytosol via citrate exportation [[84], [85], [86]]. And peroxisomal acetyl-CoA was drove to cytosol by deletions of citrate synthase and malate synthase [87]. With sufficient precursor supply, the MVA pathway was extensively optimized by enhancing the rate-limiting steps [6,88,89], construction of fusion enzymes for substrate channeling [90], introduction of multifunctional enzymes [91,92], and down-regulation of competing pathways [[93], [94], [95]].
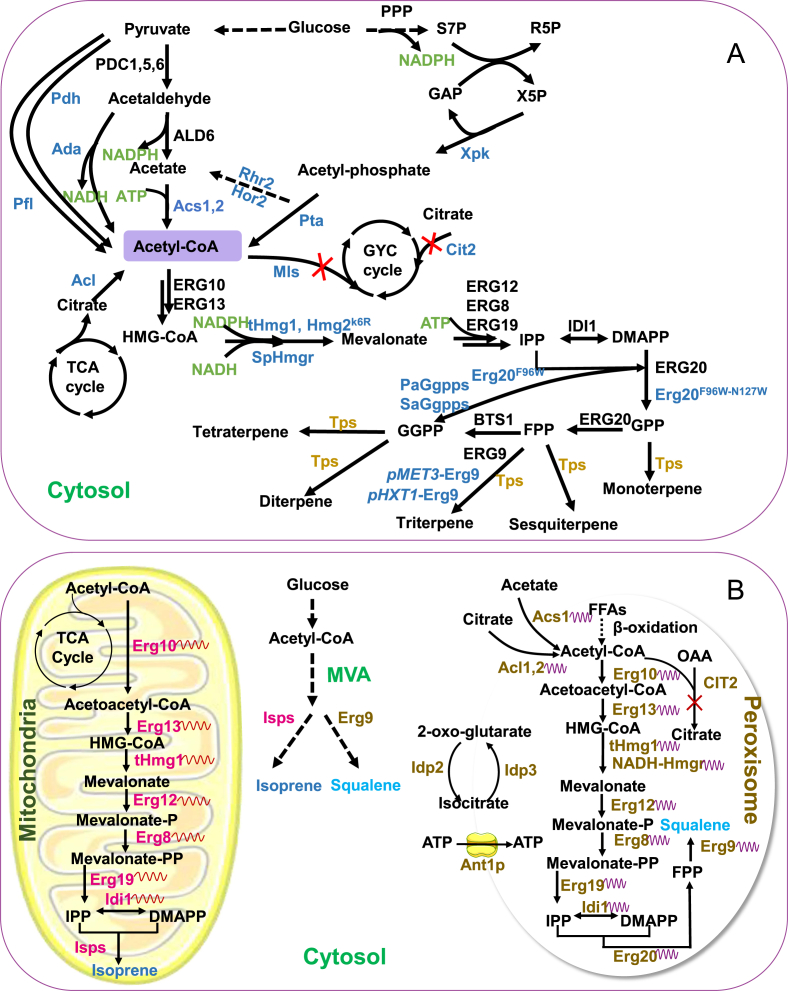
Fig. 3.
Cytosolic and sub-cellular engineering of yeast for isoprenoids biosynthesis. (A) Cytosolic engineering of yeast for isoprenoids biosynthesis. The black font means the native pathway, the blue font means modified pathway, the yellow font means exogenous terpenoids synthase and the green font represents cofactors and ATP. PDH, pyruvate dehydrogenase; Ada, acetaldehyde dehydrogenase; PDC1,5,6, pyruvate carboxy 1,5,6; PFL, pyruvate:formate lyase; Acs1,2, acetyl-CoA synthetase 1,2; Acl, ATP-dependent citrate lyase; TCA, tricarboxylic acid; Rhr2/Hor2, glycerol-3-phosphate phosphatase; Pta, phosphotransacetylase; Mls, malate synthase; Cit2, citrate synthase 2; Xpk, xylulose-5-phosphate phosphoketolase; tHmg1, truncated HMG-CoA reductase 1; SpHmgr, 3-hydroxy-3-methylglutaryl reductase from Silicibacter pomeroyi; NADH-Hmgr, NADH-specific HMG-CoA reductase; Erg10, acetoacetyl-CoA thiolase; Erg13, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase; Erg8, phosphomevalonate kinase; Erg12, mevalonate kinase; Erg19, mevalonate diphosphate decarboxylase; Idi1, dimethylallyl diphosphate isomerase; Erg20, farnesyl diphosphate synthase; Tps, terpene synthase; Bts1, geranylgeranyl diphosphate synthase; PaGgpps, geranylgeranyl diphosphate synthase from Phomopsis amygdali; SaGgpps, geranylgeranyl diphosphate synthase from Sulfolobus acidocaldarius; Erg9, squalene synthase; PPP, pentose phosphate pathway; S7P, sedoheptulose-7-phosphate; R5P, ribose-5-phosphate; GAP, glyceraldehyde-phosphate; X5P, xylulose-5-phosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; NADPH, nicotinamide adenine dinucleotide phosphate; NADH, nicotinamide adenine dinucleotide; ATP, adenosine triphosphate; TCA cycle, tricarboxylic acid cycle; GYC cycle, glyoxylate cycle. (B) Sub-cellular engineering of yeast for isoprenoids biosynthesis. Two examples of isoprenoids synthesized in sub-compartments: dual engineering of cytosol and mitochondria for isoprene biosynthesis, and dual harnessing cytosol and peroxisomes for squalene production. Isps, isoprene synthase; Idp2, isocitrate dehydrogenase 2; Idp3, isocitrate dehydrogenase 3; Ant1, peroxisomal adenine nucleotide transporter; FAs, fatty acids; MVA, mevalonate; OAA, oxaloacetate.
Though cytosolic pathway engineering of terpenoids production in yeast has made a great progress, the current yield is far behind the industrial application except artemisinin [96] and farnesene [7]. Addressing the complicated sub-organelle metabolism may further improve the yeast performance by improving precursor supply and cofactors availability, relieving the side-pathway competition, and balancing the metabolic flux with cell growth. With several advantages as mentioned above, several sub-organelles, including mitochondria, peroxisomes, the ER and LDs, have been harnessed for isoprenoids synthesis.
3.1. Harnessing mitochondria for biosynthesis of isoprenoids
Mitochondria is a cellular sub-organelle for generating cellular energy in the form of adenosine triphosphate (ATP) through the TCA cycle and the oxidative phosphorylation. In addition, mitochondria also participate in some biosynthetic process for a plethora of compounds such as branched-chain amino acids, heme cofactors, lipids, pyruvate and acetyl-CoA. The first report showed that compartmentalization of the P450 enzyme into mitochondria improved hydrocortisone production, since mitochondria have high levels of cofactor heme for maintaining P450 activity [97]. The mitochondrial localization of farnesyl pyrophosphate (FPP) utilizing enzymes such as coenzyme Q-binding protein (Cox10) and geranylgeranyl diphosphate synthase (Bts1), suggests that mitochondria may have sufficient FPP level for biosynthesis of isoprenoid especially sesquiterpenoids (Table 1). Actually, mitochondrial targeting the corresponding sesquiterpene synthases resulted approximately a 3 fold and 7 fold higher production of valencene and amorphadiene, respectively, in compared to that expressed in cytosol [98].
Acetyl-CoA is a critical precursor for isoprenoids biosynthesis and it was estimated that mitochondrial acetyl-CoA level was 20–30 fold higher than that in other compartments [99]. Mitochondrial reconstruction of FPP biosynthesis pathway enabled high level production of amorpha-4,11-diene of 430 mg/L [100]. Dual reconstruction of mitochondrial and cytosolic pathways, for utilizing both mitochondrial and cytosol acetyl-CoA, enhanced isoprene production to 2.5 g/L [15] (Fig. 3B), which was much higher than that solely mitochondrial or cytosolic pathway. Recently, mitochondrial integration of a geranyl diphosphate (GPP) biosynthetic pathway with geraniol synthase (GES), enabled a 6-fold higher geraniol production in compared to that of cytosolic pathways by relieving the cytosolic side-pathway competition for consuming the precursor GPP [101]. These studies suggest that mitochondria had some advantages for production of isoprenoids. However, the heterologous pathways should be carefully balanced in mitochondria, since it is a very crowded sub-organelle with lots of essential components for maintaining cellular function such as respiration [8].
3.2. Harnessing peroxisome for biosynthesis of isoprenoids
Peroxisome could be defined as an organelle containing hydrogen peroxide-generating oxidases and hydrogen peroxide-detoxifying catalase. In addition, fatty acid β-oxidation, the glyoxylic shunt and methanol metabolism also take place in this organelle. Moreover, peroxisome is not essential for cell growth, which makes it a relative orthogonal compartment for construction of heterogenous biosynthetic pathways. Peroxisome was extensively harnessed for biosynthesis of polyhydroxyalkanoate [102,103], fatty acid derivatives [11,104] and prodeoxyviolacein [68], showing several advantages such as relieving side-pathway competition [11,68] and improving precursor supply [11,102,104]. Recently, peroxisome was engineered for production of alkaloid (S)-reticuline by alleviating the cytotoxicity of key enzyme norcoclaurine synthase [105]. The first report on harnessing peroxisomes for isoprenoids production was conducted in yeast Komagataella phaffii (also known as Pichia pastoris) [106]. However, peroxisomal compartmentalization of three carotenogenic enzymes had marginal improvement of lycopene production in compared to that of cytosol-targeted pathway, which might be attributed to the inefficient supply of precursor FPP in the peroxisome. In a recent study, Liu et al. found that peroxisomes could be dynamic depots for squalene storage (Fig. 3B). Actually, completely reconstruction of the full MVA pathway for FPP supply enabled high level production of squalene (1.3 g/L) [107]. Furthermore, dual modulation of cytoplasmic and peroxisomal engineering further improved squalene production of 1.7 g/L in shake flask and 11 g/L in fed-batch fermentation. In another study, peroxisomes were engineered to enhance sesquiterpene α-humulene synthesis in S. cerevisiae. The combination of peroxisomal and cytoplasmic engineering led to 2.5-fold higher production of α-humulene compared to that in cytoplasm-engineered strains [108]. These studies showed that peroxisome is a favorable location for isoprenoids production by improving the supply of the precursors and cofactors.
3.3. Harnessing other organelles for isoprenoid biosynthesis
In addition to mitochondria and peroxisome, some other organelles could also be engineered for isoprenoid production with their own characteristics. For example, endoplasmic reticulum (ER) involves in protein modification and lipid droplets (LDs) contains hydrophobic environment, which may provide some advantages for biosynthesis of hydrophobic isoprenoids by regulating the enzyme abundance.
Expansion of ER, by deleting the phosphatidic acid phosphatas Pah1, boosted the production of triterpenoids β-amyrin, medicagenic acid and medicagenic-28-O-glucoside by 8-, 6- and 16-fold [109]. ER expansion stimulated the expression of biosynthetic enzymes and ultimately boosted triterpenoids accumulation, which was also beneficial for the production of other terpenoids depending on ER-associated enzymes, such as the sesquiterpenoid artemisinic acid. In another study, ER space was expanded by overexpressing a key ER size regulatory factor Ino2, which improved the production of squalene and cytochrome P450-mediated protopanaxadiol by 71- and 8-fold, respectively [110].
In yeast, LDs are about 400 nm in diameter and contain a highly lipophilic core of triacylglycerol (TAG) for energy storage. LDs are bounded by a shell of sterol esters (SE) and a phospholipid monolayer which consists of a distinct set of proteins. It has been shown that the size, number and distribution of LDs are regulated by the expression of diacylglycerol acyltransferase (Dgat) in Yarrowia lipolytica [111]. The hydrophobicity of LDs is beneficial for storing lipotoxic hydrophobic compounds such as isoprenoids, as squalene was reported to be lipotoxic to yeast cells [112]. Indeed, confocal microscopy analysis showed that high level of lycopene aggregated in LDs in engineered S. cerevisiae. And further regulation of LDs size by engineering the TAG metabolism significantly boosted lycopene accumulation [113]. In compared to non-oleaginous yeast, the oleaginous yeast Y. lipolytica was thought to be more suitable for hydrophobic β-carotene production due to the high amounts of cellular LDs. Construction of 12-step biosynthetic pathway in Y. lipolytica enabled the production of 4 g/L β-carotene, which was mainly stored in LDs [12]. Though providing a suitable space for hydrophobic isoprenoid accumulation, the formation of LDs also consumes large amounts of carbons source and thus should be carefully balanced with isoprenoid biosynthesis.
4. Perspective
Though sub-organelles have showed great potential for isoprenoid biosynthesis, there are still some challenges in engineering the subcellular metabolism due to the complication of metabolic communication among sub-organelles. For example, the knowledge on the localization and abundance of proteins is not enough to guide pathway construction and optimization (Table 1), which may lead to flux imbalance. In addition, cargo transportation, cofactors biosynthesis and transportation, organelle permeability are also should be investigated further for rational pathway regulation.
Except for S. cerevisiae, non-conventional yeasts could also be utilized as hosts for isoprenoids production due to their own sub-organelle characteristics. For example, methylotrophic yeasts like K. phaffii and Ogataea (Hansenula) polymorpha possess peroxisomes that can expand up to 80% of the cellular volume [114], which provides enormous protein capacity for heterologous pathway construction. Furthermore, methylotrophic yeasts can use single carbon (C1) feedstocks such as methanol for production of terpene for biofuel application, which should consider the cost and sustainability of substrates and methanol can be synthesized from CO2 [115]. Oleaginous yeasts like Y. lipolytica and Rhodosporidium toruloides are suitable for high-production of hydrophobic isoprenoids like lycopene, astaxanthin since they contain high amounts of LDs for storing isoprenoids with alleviating their lipotoxity.
In summary, the development of synthetic and systems biology will facilitate sub-cellular metabolic engineering for natural products biosynthesis in various yeasts. With clear subcellular metabolism, the dynamic and synergistic engineering of multi-compartment will further improve the natural product biosynthesis with good cellular fitness.
Declaration of competing interest
The authors declare no commercial or financial conflict of interest.
Acknowledgement
This study was financially supported by National Natural Science Foundation of China (Grant no. 21877111 and no. 21922812) and China Postdoctoral Science Foundation (199936).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Vickers C.E., Williams T.C., Peng B., Cherry J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr Opin Chem Biol. 2017;40:47–56. doi: 10.1016/j.cbpa.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 2.Paramasivan K., Mutturi S. Progress in terpene synthesis strategies through engineering of Saccharomyces cerevisiae. Crit Rev Biotechnol. 2017;37:974–989. doi: 10.1080/07388551.2017.1299679. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C., Ju H., Lu C.Z., Zhao F., Liu J., Guo X. High-titer production of 13R-manoyl oxide in metabolically engineered Saccharomyces cerevisiae. Microb Cell Factories. 2019;18:73. doi: 10.1186/s12934-019-1123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broker J.N., Muller B., van Deenen N., Prufer D., Schulze Gronover C. Upregulating the mevalonate pathway and repressing sterol synthesis in Saccharomyces cerevisiae enhances the production of triterpenes. Appl Microbiol Biotechnol. 2018;102:6923–6934. doi: 10.1007/s00253-018-9154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao X., Lv Y.-B., Chen J., Imanaka T., Wei L.-J., Hua Q. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnol Biofuels. 2016;9:214. doi: 10.1186/s13068-016-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y.J., Gao W., Rong Q., Jin G., Chu H., Liu W. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc. 2012;134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 7.Meadows A.L., Hawkins K.M., Tsegaye Y., Antipov E., Kim Y., Raetz L. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature. 2016;537:694–697. doi: 10.1038/nature19769. [DOI] [PubMed] [Google Scholar]
- 8.Malina C., Larsson C., Nielsen J. Yeast mitochondria: an overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Res. 2018:18. doi: 10.1093/femsyr/foy040. [DOI] [PubMed] [Google Scholar]
- 9.Sibirny A.A. Yeast peroxisomes: structure, functions and biotechnological opportunities. FEMS Yeast Res. 2016:16. doi: 10.1093/femsyr/fow038. [DOI] [PubMed] [Google Scholar]
- 10.van Rossum H.M., Kozak B.U., Pronk J.T., van Maris A.J.A. Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab Eng. 2016;36:99–115. doi: 10.1016/j.ymben.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y.J., Buijs N.A., Zhu Z., Gomez D.O., Boonsombuti A., Siewers V. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J Am Chem Soc. 2016;138:15368–15377. doi: 10.1021/jacs.6b07394. [DOI] [PubMed] [Google Scholar]
- 12.Gao S., Tong Y., Zhu L., Ge M., Zhang Y., Chen D. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous beta-carotene production. Metab Eng. 2017;41:192–201. doi: 10.1016/j.ymben.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Lametschwandtner G., Brocard C., Fransen M., Van Veldhoven P., Berger J., Hartig A. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J Biol Chem. 1998;273:33635–33643. doi: 10.1074/jbc.273.50.33635. [DOI] [PubMed] [Google Scholar]
- 14.Rachubinski R.A., Subramani S. How proteins penetrate peroxisomes. Cell. 1995;83:525–528. doi: 10.1016/0092-8674(95)90091-8. [DOI] [PubMed] [Google Scholar]
- 15.Lv X., Wang F., Zhou P., Ye L., Xie W., Xu H. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat Commun. 2016;7:1–12. doi: 10.1038/ncomms12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avalos J.L., Fink G.R., Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol. 2013;31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiltunen J.K., Mursula A.M., Rottensteiner H., Wierenga R.K., Kastaniotis A.J., Gurvitz A. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2003;27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 18.van Roermund C.W., Hettema E.H., Kal A.J., van den Berg M., Tabak H.F., Wanders R.J. Peroxisomal β-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J. 1998;17:677–687. doi: 10.1093/emboj/17.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henke B., Girzalsky W., Berteaux-Lecellier V., Erdmann R. IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the β-oxidation of unsaturated fatty acids. J Biol Chem. 1998;273:3702–3711. doi: 10.1074/jbc.273.6.3702. [DOI] [PubMed] [Google Scholar]
- 20.Ghaemmaghami S., Huh W.-K., Bower K., Howson R.W., Belle A., Dephoure N. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 21.Huh W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 22.Valli M., Grillitsch K., Grünwald-Gruber C., Tatto N.E., Hrobath B., Klug L. A subcellular proteome atlas of the yeast Komagataella phaffii. FEMS Yeast Res. 2020;20 doi: 10.1093/femsyr/foaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh J.L., Chong Y.T., Friesen H., Moses A., Boone C., Andrews B.J. CYCLoPs: a comprehensive database constructed from automated analysis of protein abundance and subcellular localization Patterns in Saccharomyces cerevisiae. G3. 2015;5:1223–1232. doi: 10.1534/g3.115.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen T.N., Brunak S., Von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 26.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2011;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habeler G., Natter K., Thallinger G.G., Crawford M.E., Kohlwein S.D., Trajanoski Z.Y.P.L. Db: the yeast protein localization database. Nucleic Acids Res. 2002;30:80–83. doi: 10.1093/nar/30.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kals M., Natter K., Thallinger G.G., Trajanoski Z., Kohlwein S.D. YPL.db2: the yeast protein localization database. version 2.0Yeast. 2005;22:213–218. doi: 10.1002/yea.1204. [DOI] [PubMed] [Google Scholar]
- 29.Wiwatwattana N., Landau C.M., Cope G.J., Harp G.A., Kumar A., Organelle D.B. An updated resource of eukaryotic protein localization and function. Nucleic Acids Res. 2007;35:D810–D814. doi: 10.1093/nar/gkl1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riffle M., Davis T.N. The yeast resource center public image repository: a large database of fluorescence microscopy images. BMC Bioinf. 2010;11:263. doi: 10.1186/1471-2105-11-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yofe I., Weill U., Meurer M., Chuartzman S., Zalckvar E., Goldman O. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods. 2016;13:371–378. doi: 10.1038/nmeth.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weill U., Yofe I., Sass E., Stynen B., Davidi D., Natarajan J. Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat Methods. 2018;15:617–622. doi: 10.1038/s41592-018-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baccolo G., Stamerra G., Coppola D.P., Orlandi I., Vai M. Mitochondrial metabolism and aging in yeast. Int Rev Cell Mol Biol. 2018;340:1–33. doi: 10.1016/bs.ircmb.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Palmieri F., Pierri C.L. Mitochondrial metabolite transport. Essays Biochem. 2010;47:37–52. doi: 10.1042/bse0470037. [DOI] [PubMed] [Google Scholar]
- 35.Palmieri F., Monne M. Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta. 2016;1863:2362–2378. doi: 10.1016/j.bbamcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Palmieri L., Rottensteiner H., Girzalsky W., Scarcia P., Palmieri F., Erdmann R. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 2001;20:5049–5059. doi: 10.1093/emboj/20.18.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marobbio C., Vozza A., Harding M., Bisaccia F., Palmieri F., Walker J. Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 2002;21:5653–5661. doi: 10.1093/emboj/cdf583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vozza A., Blanco E., Palmieri L., Palmieri F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- 39.Marobbio C.M., Di Noia M.A., Palmieri F. Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution and functional characterization. Biochem J. 2006;393:441–446. doi: 10.1042/BJ20051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todisco S., Agrimi G., Castegna A., Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- 41.Todisco S., Di Noia M.A., Castegna A., Lasorsa F.M., Paradies E., Palmieri F. The Saccharomyces cerevisiae gene YPR011c encodes a mitochondrial transporter of adenosine 5′-phosphosulfate and 3′-phospho-adenosine 5′-phosphosulfate. Biochim Biophys Acta Bioenerg. 2014;1837:326–334. doi: 10.1016/j.bbabio.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Adrian G., McCammon M., Montgomery D., Douglas M. Sequences required for delivery and localization of the ADP/ATP translocator to the mitochondrial inner membrane. Mol Cell Biol. 1986;6:626–634. doi: 10.1128/mcb.6.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawson J., Douglas M.G. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J Biol Chem. 1988;263:14812–14818. [PubMed] [Google Scholar]
- 44.Kolarov J., Kolarova N., Nelson N. A third ADP/ATP translocator gene in yeast. J Biol Chem. 1990;265:12711–12716. [PubMed] [Google Scholar]
- 45.Tzagoloff A., Jang J., Glerum D.M., Wu M. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J Biol Chem. 1996;271:7392–7397. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 46.Cavero S., Traba J., Del Arco A., Satrústegui J. The calcium-dependent ATP-Mg/Pi mitochondrial carrier is a target of glucose-induced calcium signalling in Saccharomyces cerevisiae. Biochem J. 2005;392:537–544. doi: 10.1042/BJ20050806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lancar-Benba J., Foucher B., Saint-Macary M. Characterization, purification and properties of the yeast mitochondrial dicarboxylate carrier (Saccharomyces cerevisiae) Biochimie. 1996;78:195–200. doi: 10.1016/0300-9084(96)89505-8. [DOI] [PubMed] [Google Scholar]
- 48.Palmieri L., Vozza A., Hönlinger A., Dietmeier K., Palmisano A., Zara V. The mitochondrial dicarboxylate carrier is essential for the growth of Saccharomyces cerevisiae on ethanol or acetate as the sole carbon source. Mol Microbiol. 1999;31:569–577. doi: 10.1046/j.1365-2958.1999.01197.x. [DOI] [PubMed] [Google Scholar]
- 49.Palmieri L., Lasorsa F.M., De Palma A., Palmieri F., Runswick M.J., Walker J.E. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 1997;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- 50.Palmieri L., Vozza A., Agrimi G., De Marco V., Runswick M.J., Palmieri F. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J Biol Chem. 1999;274:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- 51.Marobbio C.M., Giannuzzi G., Paradies E., Pierri C.L., Palmieri F. α-Isopropylmalate, a leucine biosynthesis intermediate in yeast, is transported by the mitochondrial oxalacetate carrier. J Biol Chem. 2008;283:28445–28453. doi: 10.1074/jbc.M804637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmieri L., Agrimi G., Runswick M.J., Fearnley I.M., Palmieri F., Walker J.E. Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J Biol Chem. 2001;276:1916–1922. doi: 10.1074/jbc.M004332200. [DOI] [PubMed] [Google Scholar]
- 53.Castegna A., Scarcia P., Agrimi G., Palmieri L., Rottensteiner H., Spera I. Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J Biol Chem. 2010;285:17359–17370. doi: 10.1074/jbc.M109.097188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaplan R.S., Mayor J.A., Gremse D.A., Wood D.O. High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:4108–4114. doi: 10.1074/jbc.270.8.4108. [DOI] [PubMed] [Google Scholar]
- 55.Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.-C. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herzig S., Raemy E., Montessuit S., Veuthey J.-L., Zamboni N., Westermann B. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 57.Palmieri L., De Marco V., Iacobazzi V., Palmieri F., Runswick M.J., Walker J.E. Identification of the yeast ARG-11 gene as a mitochondrial ornithine carrier involved in arginine biosynthesis. FEBS Lett. 1997;410:447–451. doi: 10.1016/s0014-5793(97)00630-3. [DOI] [PubMed] [Google Scholar]
- 58.Marobbio C., Agrimi G., Lasorsa F., Palmieri F. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 2003;22:5975–5982. doi: 10.1093/emboj/cdg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmieri L., Lasorsa F.M., Iacobazzi V., Runswick M.J., Palmieri F., Walker J.E. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 1999;462:472–476. doi: 10.1016/s0014-5793(99)01555-0. [DOI] [PubMed] [Google Scholar]
- 60.Cavero S., Vozza A., Del Arco A., Palmieri L., Villa A., Blanco E. Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol Microbiol. 2003;50:1257–1269. doi: 10.1046/j.1365-2958.2003.03742.x. [DOI] [PubMed] [Google Scholar]
- 61.Mühlenhoff U., Stadler J.A., Richhardt N., Seubert A., Eickhorst T., Schweyen R.J. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem. 2003;278:40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- 62.Kolisek M., Zsurka G., Samaj J., Weghuber J., Schweyen R.J., Schweigel M. Mrs2p is an essential component of the major electrophoretic Mg2+ influx system in mitochondria. EMBO J. 2003;22:1235–1244. doi: 10.1093/emboj/cdg122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sponder G., Svidova S., Schindl R., Wieser S., Schweyen R.J., Romanin C. Lpe10p modulates the activity of the Mrs2p-based yeast mitochondrial Mg2+ channel. FEBS J. 2010;277:3514–3525. doi: 10.1111/j.1742-4658.2010.07761.x. [DOI] [PubMed] [Google Scholar]
- 64.Cui Y., Zhao S., Wang J., Wang X., Gao B., Fan Q. A novel mitochondrial carrier protein Mme1 acts as a yeast mitochondrial magnesium exporter. Biochim Biophys Acta Mol Cell Res. 2015;1853:724–732. doi: 10.1016/j.bbamcr.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 65.Vest K.E., Leary S.C., Winge D.R., Cobine P.A. Copper import into the mitochondrial matrix in Saccharomyces cerevisiae is mediated by Pic2, a mitochondrial carrier family protein. J Biol Chem. 2013;288:23884–23892. doi: 10.1074/jbc.M113.470674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamel P., Saint-Georges Y., De Pinto B., Lachacinski N., Altamura N., Dujardin G. Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol Microbiol. 2004;51:307–317. doi: 10.1046/j.1365-2958.2003.03810.x. [DOI] [PubMed] [Google Scholar]
- 67.Yu T., Zhou Y.J., Huang M., Liu Q., Pereira R., David F. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174:1549–1558. doi: 10.1016/j.cell.2018.07.013. e1514. [DOI] [PubMed] [Google Scholar]
- 68.DeLoache W.C., Russ Z.N., Dueber J.E. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat Commun. 2016;7:11152. doi: 10.1038/ncomms11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Roermund C., Elgersma Y., Singh N., Wanders R., Tabak H. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD (H) and acetyl-CoA under in vivo conditions. EMBO J. 1995;14:3480–3486. doi: 10.1002/j.1460-2075.1995.tb07354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hettema E.H., Van Roermund C., Distel B., van den Berg M., Vilela C., Rodrigues-Pousada C. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1996;15:3813–3822. [PMC free article] [PubMed] [Google Scholar]
- 71.Mettler I.J., Beevers H. Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol. 1980;66:555–560. doi: 10.1104/pp.66.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Roermund C.W., Hettema E.H., van den Berg M., Tabak H.F., Wanders R.J. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 1999;18:5843–5852. doi: 10.1093/emboj/18.21.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bernhardt K., Wilkinson S., Weber A.P., Linka N. A peroxisomal carrier delivers NAD(+) and contributes to optimal fatty acid degradation during storage oil mobilization. The Plant journal : Cell Mol Biol. 2012;69:1–13. doi: 10.1111/j.1365-313X.2011.04775.x. [DOI] [PubMed] [Google Scholar]
- 74.Agrimi G., Russo A., Pierri C.L., Palmieri F. The peroxisomal NAD+ carrier of Arabidopsis thaliana transports coenzyme A and its derivatives. J Bioenerg Biomembr. 2012;44:333–340. doi: 10.1007/s10863-012-9445-0. [DOI] [PubMed] [Google Scholar]
- 75.Agrimi G., Russo A., Scarcia P., Palmieri F. The human gene SLC25A17 encodes a peroxisomal transporter of coenzyme A, FAD and NAD+ Biochem J. 2012;443:241–247. doi: 10.1042/BJ20111420. [DOI] [PubMed] [Google Scholar]
- 76.Gr Csordás, Renken C., Várnai P., Walter L., Weaver D., Buttle K.F. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rowland A.A., Voeltz G.K. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kopec K.O., Alva V., Lupas A.N. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26:1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mattiazzi Usaj M., Brloznik M., Kaferle P., Zitnik M., Wolinski H., Leitner F. Genome-wide localization study of yeast Pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J Mol Biol. 2015;427:2072–2087. doi: 10.1016/j.jmb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shiba Y., Paradise E.M., Kirby J., Ro D.K., Keasling J.D. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab Eng. 2007;9:160–168. doi: 10.1016/j.ymben.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Chen Y., Daviet L., Schalk M., Siewers V., Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Kozak B.U., van Rossum H.M., Benjamin K.R., Wu L., Daran J.M., Pronk J.T. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab Eng. 2014;21:46–59. doi: 10.1016/j.ymben.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 83.Cardenas J., Da Silva N.A. Engineering cofactor and transport mechanisms in Saccharomyces cerevisiae for enhanced acetyl-CoA and polyketide biosynthesis. Metab Eng. 2016;36:80–89. doi: 10.1016/j.ymben.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 84.Tang X., Feng H., Chen W.N. Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metab Eng. 2013;16:95–102. doi: 10.1016/j.ymben.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 85.Zhou Y.J., Buijs N.A., Zhu Z., Qin J., Siewers V., Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lian J., Si T., Nair N.U., Zhao H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng. 2014;24:139–149. doi: 10.1016/j.ymben.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 87.Chen Y., Siewers V., Nielsen J. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PloS One. 2012;7 doi: 10.1371/journal.pone.0042475. e42475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ignea C., Trikka F.A., Kourtzelis I., Argiriou A., Kanellis A.K., Kampranis S.C. Positive genetic interactors of HMG2 identify a new set of genetic perturbations for improving sesquiterpene production in Saccharomyces cerevisiae. Microb Cell Factories. 2012;11:162. doi: 10.1186/1475-2859-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ignea C., Pontini M., Maffei M.E., Makris A.M., Kampranis S.C. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth Biol. 2014;3:298–306. doi: 10.1021/sb400115e. [DOI] [PubMed] [Google Scholar]
- 90.Ignea C., Trikka F.A., Nikolaidis A.K., Georgantea P., Ioannou E., Loupassaki S. Efficient diterpene production in yeast by engineering Erg20p into a geranylgeranyl diphosphate synthase. Metab Eng. 2015;27:65–75. doi: 10.1016/j.ymben.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 91.Callari R., Meier Y., Ravasio D., Heider H. Dynamic control of ERG20 and ERG9 expression for improved casbene production in Saccharomyces cerevisiae. Front Bioeng Biotechnol. 2018;6:160. doi: 10.3389/fbioe.2018.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dai Z., Liu Y., Huang L., Zhang X. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109:2845–2853. doi: 10.1002/bit.24547. [DOI] [PubMed] [Google Scholar]
- 93.Paradise E.M., Kirby J., Chan R., Keasling J.D. Redirection of flux through the FPP branch-point in Saccharomyces cerevisiae by down-regulating squalene synthase. Biotechnol Bioeng. 2008;100:371–378. doi: 10.1002/bit.21766. [DOI] [PubMed] [Google Scholar]
- 94.Chen H., Zhu C., Zhu M., Xiong J., Ma H., Zhuo M. High production of valencene in Saccharomyces cerevisiae through metabolic engineering. Microb Cell Factories. 2019;18:195. doi: 10.1186/s12934-019-1246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie W., Ye L., Lv X., Xu H., Yu H. Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab Eng. 2015;28:8–18. doi: 10.1016/j.ymben.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 96.Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 97.Szczebara F.M., Chandelier C., Villeret C., Masurel A., Bourot S., Duport C. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- 98.Farhi M., Marhevka E., Masci T., Marcos E., Eyal Y., Ovadis M. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 99.Weinert B.T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S.A., Magnes C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yuan J., Ching C.B. Mitochondrial acetyl-CoA utilization pathway for terpenoid productions. Metab Eng. 2016;38:303–309. doi: 10.1016/j.ymben.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 101.Yee D.A., DeNicola A.B., Billingsley J.M., Creso J.G., Subrahmanyam V., Tang Y. Engineered mitochondrial production of monoterpenes in Saccharomyces cerevisiae. Metab Eng. 2019;55:76–84. doi: 10.1016/j.ymben.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Poirier Y., Erard N., Petetot J.M. Synthesis of polyhydroxyalkanoate in the peroxisome of Saccharomyces cerevisiae by using intermediates of fatty acid beta-oxidation. Appl Environ Microbiol. 2001;67:5254–5260. doi: 10.1128/AEM.67.11.5254-5260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Oliveira V.C., Maeda I., Delessert S., Poirier Y. Increasing the carbon flux toward synthesis of short-chain-length–medium-chain-length polyhydroxyalkanoate in the peroxisome of Saccharomyces cerevisiae through modification of the beta-oxidation cycle. Appl Environ Microbiol. 2004;70:5685–5687. doi: 10.1128/AEM.70.9.5685-5687.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheng J., Stevens J., Feng X. Pathway compartmentalization in peroxisome of Saccharomyces cerevisiae to produce versatile medium chain fatty alcohols. Sci Rep. 2016;6:26884. doi: 10.1038/srep26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grewal P.S., Samson J.A., Baker J.J., Choi B., Dueber J.E. Repurposing the yeast peroxisome to compartmentalize a toxic enzyme enables improved (S)-reticuline production. bioRxiv. 2020 doi: 10.1101/2020.03.23.000851. [DOI] [Google Scholar]
- 106.Bhataya A., Schmidt-Dannert C., Lee P.C. Metabolic engineering of Pichia pastoris X-33 for lycopene production. Process Biochem. 2009;44:1095–1102. [Google Scholar]
- 107.Liu G.-S., Li T., Zhou W., Jiang M., Tao X.-Y., Liu M. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction. Metab Eng. 2020;57:151–161. doi: 10.1016/j.ymben.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 108.Zhang C.B., Li M., Zhao G.R., Lu W.Y. Harnessing yeast peroxisomes and cytosol acetyl-CoA for sesquiterpene α-Humulene production. J Agric Food Chem. 2020;68(5):1382–1389. doi: 10.1021/acs.jafc.9b07290. [DOI] [PubMed] [Google Scholar]
- 109.Arendt P., Miettinen K., Pollier J., De Rycke R., Callewaert N., Goossens A. An endoplasmic reticulum-engineered yeast platform for overproduction of triterpenoids. Metab Eng. 2017;40:165–175. doi: 10.1016/j.ymben.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 110.Kim J.-E., Jang I.-S., Son S.-H., Ko Y.-J., Cho B.-K., Kim S.C. Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway. Metab Eng. 2019;56:50–59. doi: 10.1016/j.ymben.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 111.Gajdos P., Ledesma-Amaro R., Nicaud J.M., Certik M., Rossignol T. Overexpression of diacylglycerol acyltransferase in Yarrowia lipolytica affects lipid body size, number and distribution. FEMS Yeast Res. 2016:16. doi: 10.1093/femsyr/fow062. [DOI] [PubMed] [Google Scholar]
- 112.Valachovic M., Garaiova M., Holic R., Hapala I. Squalene is lipotoxic to yeast cells defective in lipid droplet biogenesis. Biochem Biophys Res Commun. 2016;469:1123–1128. doi: 10.1016/j.bbrc.2015.12.050. [DOI] [PubMed] [Google Scholar]
- 113.Ma T., Shi B., Ye Z., Li X., Liu M., Chen Y. Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab Eng. 2019;52:134–142. doi: 10.1016/j.ymben.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 114.Gleeson M.A., Sudbery P.E. The methylotrophic yeasts. Yeast. 1988;4:1–15. [Google Scholar]
- 115.Duan X.P., Gao J.Q., Zhou Y.J. Advances in engineering methylotrophic yeast for biosynthesis of valuable chemicals from methanol. Chin Chem Lett. 2018;29:681–686. [Google Scholar]