Abstract
Synonymous codon usage has been identified as a determinant of translational efficiency and mRNA stability in model organisms and human cell lines. However, whether natural selection shapes human codon content to optimize translation efficiency is unclear. Furthermore, aside from those that affect splicing, synonymous mutations are typically ignored as potential contributors to disease. Using genetic sequencing data from nearly 200,000 individuals, we uncover clear evidence that natural selection optimizes codon content in the human genome. In deriving intolerance metrics to quantify gene-level constraint on synonymous variation, we discover that dosage-sensitive genes, DNA-damage-response genes, and cell-cycle-regulated genes are particularly intolerant to synonymous variation. Notably, we illustrate that reductions in codon optimality in BRCA1 can attenuate its function. Our results reveal that synonymous mutations most likely play an underappreciated role in human variation.
Keywords: synRVIS, synGERP, RVIS, population genetics, codon usage, codon optimality, synonymous mutations, constraint, intolerance, conservation
Introduction
A long-standing assumption in human genetics is that synonymous mutations do not affect fitness because they do not alter the resulting protein sequence. However, recent evidence indicates that synonymous variation is not always neutral and might often have functional consequences.1,2 Synonymous mutations can impact molecular function by disrupting splicing enhancer sites,3,4 mRNA secondary structure,5 and binding sites for regulatory RNA-binding proteins and microRNAs.6,7 Although much less understood, emerging evidence suggests that synonymous mutations can also impact gene expression and translation accuracy. Specifically, biochemical studies indicate that “optimal” codons matching more abundant tRNAs in the cytoplasmic pool can support rapid translation, whereas synonymous but “non-optimal” codons can slow translation.1,8, 9, 10, 11 Importantly, synonymous codon usage also seems to affect human mRNA stability via coupling between mRNA degradation and translation.10,12,13 Indeed, it has long been recognized that the human genome exhibits clear codon usage biases: certain codons are used more frequently than others.14,15
Despite the clear presence of codon usage bias in the human genome, its significance as it relates to human physiology and fitness has been under debate for decades. It is generally accepted that natural selection impacts synonymous codons that impact exon splicing, but it is unclear whether selection shapes codon optimality as it relates to translation efficiency. Although some researchers have argued that selective pressures optimize human codon usage,14,16, 17, 18, 19, 20 others have posited that mutational biases and other neutral factors preclude the role of natural selection in shaping codon optimality.21, 22, 23, 24, 25 These efforts have come to conflicting conclusions because of three main challenges. First, these synonymous mutations are expected to be weakly deleterious because they are more likely to affect protein abundance than function.14,26 Because of the small effective population size of human beings, natural selection is less effective in purging weakly deleterious mutations from the population. Second, codon usage is posited to be functionally linked to tRNA expression.10,27 Because tRNA expression varies widely by tissue,28 each synonymous site is most likely subjected to different evolutionary pressures across tissues. This variation in tRNA expression also makes it difficult to correlate codon usage with tRNA availability. Third, the nucleotide content at synonymous sites strongly correlates with local GC content in nearby non-coding regions. This phenomenon suggests that codon bias is also influenced by evolutionarily neutral processes, such as local variation in mutation rate.14,15,21,29,30 Altogether, these challenges necessitate robust statistical methods that can detect selective constraint on variants of modest effect across a population while controlling for confounding mutational biases.
In this study, we leveraged the unprecedented amount of sequencing data available in two population reference cohorts—TOPMed (62,784 genomes)31 and gnomAD (123,136 exomes)32—to first reaffirm that natural selection optimizes codon content in protein-coding regions in the human genome. This unprecedented amount of sequencing data allowed us to then devise two scores that rank genes by their intolerance to synonymous mutations. The first metric, synRVIS, measures human-specific constraint specifically against changes in codon optimality. The second metric, synGERP, reflects the average phylogenetic conservation at all fourfold degenerate sites across the mammalian lineage in a given gene. These scores, in turn, allow us to identify genes and pathways in which synonymous variants are most likely to affect human fitness.
Methods
Sequence Data
We used summary level allele frequency data from the BRAVO TOPMed database (TOPMed Freeze 5) and gnomAD (release 2.0.2). The TOPMed database contains roughly 463 million variants derived from 62,784 whole genomes and gnomAD contains roughly 15 million variants from 123,136 whole exomes.
We mapped TOPMed variants from hg38 to hg19 by using the LiftoverVcf tool in Picard tools (v2.9.0). We then annotated both the TOPMed and gnomAD VCFs by using Variant Effect Predictor (VEP), version 84.33 To annotate each variant with its most damaging possible effect across all transcripts, we used the VEP “–pick_allele” option with the following order: “rank, canonical, appris, tsl, biotype, ccds, length.”
We then filtered each VCF file so that it contained only variants annotated as “PASS” and removed all variants occurring in repeat regions, as identified by RepeatMasker, version 4.0.5.34 To exclude variants that are expected to disrupt canonical splice sites, we removed all variants occurring within ten intronic nucleotides and three exonic nucleotides of exon-intron boundaries in all Ensembl v75 transcripts. We additionally filtered the gnomAD VCF so that it only retained variants with at least 10-fold coverage in at least 85% of individuals.
Codon Usage Metrics
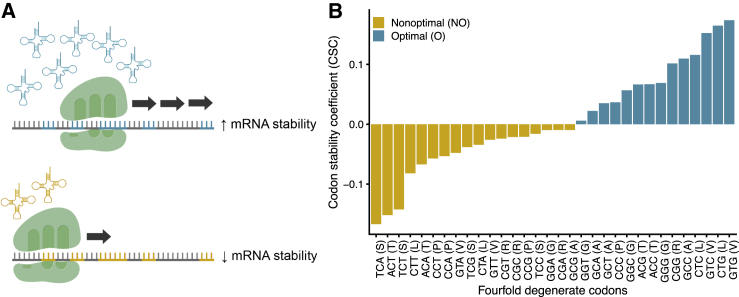
We used two scores for assessing codon usage: the codon stability coefficient (CSC) and the relative synonymous codon usage (RSCU). We obtained CSC scores derived from HEK293T cells.10 Wu et al.[10] also calculated CSC scores for other cell lines, including HeLa and RPE cells, but these scores were very strongly correlated with the HEK293T scores. The CSC represents the Pearson correlation between the frequency of the codon in each transcript and the associated half-life. We classified codons with CSC values greater than 0 as “optimal” and codons with CSC values less than 0 as “non-optimal.”
As an orthogonal measure of codon usage, we calculated RSCU scores for each codon.35 For each codon in each canonical transcript (as defined by Ensembl v75), we calculate the ratio of the observed number of codons to the expected number for a given amino acid. Specifically, for an amino acid i, the RSCU score of its jth amino acid is defined as
where ni denotes the number of synonymous codons for that amino acid. When using RSCU to assess codon optimality, we annotate codons with a value less than 1 as “non-optimal” and greater than 1 as “optimal.” We chose to calculate gene-specific rather than genome-wide RSCU scores, reasoning that gene-specific scores should more adequately reflect tissue-specific sources of constraint.
Site Frequency Spectrum Analyses
We performed all site frequency spectrum (SFS) analyses by using the filtered allele frequency data. We adapted an approach previously employed in Drosophila studies to compare selection on synonymous variation with putatively neutral variants.36,37 Specifically, we matched each observed synonymous variant occurring at fourfold degenerate sites with intronic variants occurring within 10,000 base pairs. We required matched variants to have the same ancestral allele, and in an additional analysis, we required matched variants to also have the same neighboring 5′ and 3′ nucleotides. We matched variants to the direction or strand blindly, such that synonymous mutations were allowed to pair with forward, reverse, and reverse complement intronic sequences. We only considered synonymous variants occurring at fourfold degenerate sites. In a separate analysis, we compared the SFS of synonymous variants that alter codon optimality to loss-of-function variants and missense variants predicted to be “probably” or “possibly” damaging by PolyPhen-238.
We folded all allele frequencies: if the alternate allele frequency was greater than 50%, we subtracted it from 100%, meaning the minor allele frequency is always less than or equal to 50%. We then used a two-tailed t test to determine whether SFS distributions were significantly different.38,39 As noted by Keinan et al., this test is conservative because it reflects significant deviation in the mean minor allele frequencies rather than other differences in the shape of the distribution.38
Comparing Phylogenetic Conservation at Synonymous and Intronic Sites
We used a custom script to annotate the TOPMed variants with GERP++ scores, which reflect each genomic site's estimated evolutionary constraint across the mammalian lineage.40 To assess phylogenetic conservation on codon usage, we compared the GERP++ scores of the reference alleles of the synonymous and intronic variants included in the SFS analyses. Because only a fraction of fourfold degenerate sites actually harbor a variant in TOPMed, we also compared the correlation between CSC and GERP++ at all fourfold degenerate sites in the genome. To mitigate confounding due to conservation at splice sites, we excluded codons occurring at exon-intron boundaries in all Ensembl v75 transcripts.
Deriving synRVIS
Synonymous RVIS (synRVIS) is an adaptation of the residual variation intolerance score (RVIS), a previously published score that quantifies genic intolerance for non-synonymous variation.41 Using aggregated allele frequency from gnomAD exomes,32 we defined Y as the total number of common (MAF > 0.5%) synonymous “optimal-to-nonoptimal” (O → NO) SNVs in a gene and X as the total number of synonymous SNVs occurring in a gene. We then regressed Y on X and defined synRVIS as the studentized residual for each gene. The resulting regression line accounts for genic mutation rates, sequence context, and gene size while predicting the expected number of common synonymous variants that result in a non-optimal change. We explored the behavior of the score when we used alternative MAF cutoffs of 1% and 0.1% for defining common variants on the y axis and found that these scores strongly correlated (Pearson’s r = 0.89 and r = 0.74, respectively) (Figures S3A and S3B). We also found strong correlation when we used RSCU instead of CSC to define codon optimality (r = 0.63) (Figure S3C).
synRVIS Permutation Test
We sought to verify that the synRVIS distribution deviates from a null model because the resulting residuals might reflect random noise rather than intergenic patterns of constraint. To perform a permutation test, we randomly assigned synonymous variants to each gene and recalculated the synRVISs. In the presence of intergenic constraint, the real synRVIS distribution should show greater variance than that of the permuted scores. Specifically, we performed 1,000 permutations in which we randomly assigned the gnomAD synonymous variants to genes, controlling for gene size. For each permutation, we fit a regression line and calculated the variance of the studentized residuals. To calculate a p value, we determined the rank of the real synRVIS variance among the variances resulting from these permutations.
Calculating synGERP
We defined the synGERP score as the average GERP++ score40 of all fourfold degenerate sites in a given gene. We excluded all codons immediately adjacent to exon-intron junctions in all Ensembl v75 transcripts to mitigate confounding due to conservation at canonical splice sites.
Gene Set Enrichment Tests
We used logistic regression models to determine the ability of synRVIS and synGERP to predict 360 dosage-sensitive genes contained in the ClinGen Genome Dosage Map and 178 DNA-damage-response genes. We calculated receiver operating characteristic (ROC) curves by using the pROC package in R.42 For the BROCA cancer risk panel genes, we opted to perform a Mann-Whitney U test to compare the intolerance of these genes versus all other genes in the genome rather than evaluate the ROC, given the small sample size of the gene list (n = 66). We additionally performed a permuted Mann-Whitney U test for this particular enrichment test. Specifically, we first computed the actual Mann-Whitney U p value of the observed data. We then randomly permuted the labels of the data and computed additional p values 1,000 times. We defined the permuted p value as the proportion of permuted p values less than or equal to the actual p value derived from the original, unpermuted dataset.
We also compared the distribution of synRVIS and synGERP to LOEUF (loss-of-function observed/expected upper bound fraction), a metric that assesses the observed over expected ratio for loss-of-function variants in gnomAD. Specifically, we computed the median LOEUF score per synRVIS and synGERP decile. We also assessed the median synRVIS and synGERP percentile of genes dynamically expressed during the cell cycle, as identified by CycleBase. All gene lists are available in Table S2.
Gene Ontology Enrichment
We performed gene ontology (GO) enrichment tests of genes falling below the 25th percentile in synRVIS or synGERP to identify classes of genes most intolerant to synonymous variation. We also performed enrichment tests of synRVIS-tolerant but LOEUF-intolerant genes. We defined these genes as genes above the 75th percentile in synRVIS, but below the 25th percentile of LOEUF scores. To perform the enrichment test, we used the PANTHER GO-slim biological process annotation set.43 p values were computed with Fisher’s exact test and corrected via the false discovery rate. We defined corrected p values < 0.05 as significant. The full lists of significant GO enrichment results are available in Table S3.
BRCA1 Function Score Evaluation
We used VEP to annotate the resulting codon changes from synonymous variants assayed in a previous study.44 We then annotated the reference and alternate codons of each variant with their CSC values and removed all variants identified as splice region variants by VEP or occurring within 3 base pairs of exon-intron junctions (Table S4). We annotated variants with function scores less than −0.748 as variants that reduced BRCA1 function. To quantify codon usage changes, we defined ΔCSC as the difference between the CSC value of the alternate codon and the CSC value of the reference codon for each variant.
Data Visualizations
All plots were generated in R using ggplot2.45 Figure 1A was created with BioRender. Color palettes for plots were derived from the wesanderson R package.
Figure 1.
tRNA Availability and Codon Usage Affect mRNA Decay
(A) Codon-optimized transcripts (top panel) exhibit increased mRNA stability compared to less optimized transcripts (bottom panel). Codon optimality modestly correlates with tRNA availability, suggesting optimal codons are decoded by more highly expressed tRNAs and might therefore increase translational efficiency.
(B) Distribution of previously published codon stability coefficient (CSC) scores derived from HEK293T cells10 for fourfold degenerate codons. Colors correspond to optimal and non-optimal codons.
Results
Site Frequency Spectra Reveal Genome-wide Signatures of Purifying Selection on Human Codon Usage
The availability of aggregated human whole-genome allele frequency data from roughly 60,000 individuals contained in the TOPMed database31 provides an unprecedented resource for investigating selective constraint on weakly deleterious variants, such as synonymous mutations. Focusing on synonymous sites where any of the four nucleotides in the third position of the codon encode the same amino acid (i.e., fourfold degenerate), we used this resource to identify potential evidence of natural selection on codon usage. The standard approach for measuring purifying selection is the examination of the allele frequency spectrum. Allele frequency is a powerful proxy of a variant’s phenotypic impact because purifying selection tends to eliminate deleterious variants before they reach a high frequency in the population.46 Hence, the spectrum should skew relative to the neutral mutation rate. The neutral rate is typically defined as the synonymous mutation rate.47 To enable robust comparisons, we generated a neutral reference set of variants by matching each observed synonymous variant to a nearby (<10 kilobases) randomly sampled intronic variant (Figure S1A). This procedure matches the GC content of the neutral reference to the synonymous test set, mitigates regional- and transcription-associated biases in mutation rates, and normalizes the total number of variants included in each set.36,37
Following the classical approach, we first compared the SFS (i.e., the distribution of allele frequencies) of synonymous variants and matched intronic variants (n = 2,896,436) without accounting for changes in codon usage. Consistent with prior studies, the two distributions appeared nearly identical: the synonymous SFS exhibited a very slight skew toward rarer allele frequencies (t test p = 0.02) (Figure S1B). Thus, in aggregate, synonymous variants do not appear to be under significantly more constraint than putatively neutral intronic variants.
While the prior analysis suggests that synonymous sites are not constrained in aggregate, this test treats all synonymous variants as equivalent, ignoring the fact that different variants might experience distinct selective pressures. Specifically, under the codon optimality hypothesis, a synonymous variant that increases codon optimality should be favored, whereas mutations away from optimal codons should be deleterious. While conceptually straightforward, the codon optimality hypothesis is difficult to test because of the challenge of classifying codon optimality. Unlike unicellular organisms, codon optimality cannot be matched to tRNA gene copy number because tRNA expression varies widely by tissue.28,48 Previous studies have relied on the RSCU in the genome as a proxy for codon optimality,35 but these scores do not directly reflect a codon’s effect on translational efficiency. We thus employed the recently published CSC score, defined as the Pearson correlation coefficient between each codon’s frequency in a transcript and the half-life of the transcript in human embryonic kidney 293T (HEK293T) cells.10 CSC scores moderately correlate with tRNA concentrations, suggesting that human codon optimality is partly related to translation elongation speed just like in yeast11 (Figure 1A).
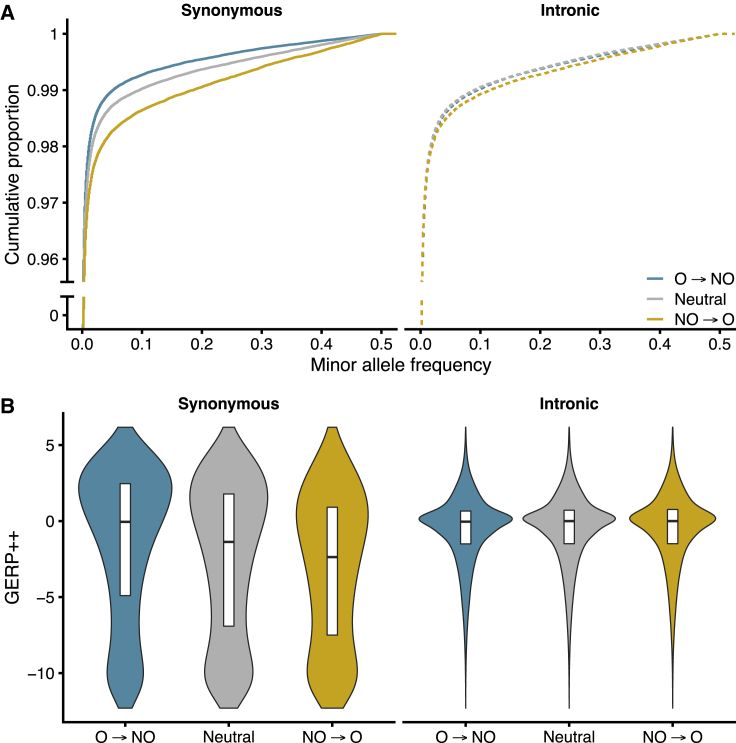
We classified the synonymous variants that resulted in changes from a codon with a positive CSC to a negative CSC as “optimal-to-nonoptimal” (O → NO), the opposite as “nonoptimal-to-optimal” (NO → O), and all others as “neutral.” Strikingly, O → NO synonymous variants segregated at significantly lower frequencies than matched intronic variants (p = 3.3 × 10−33), neutral synonymous variants (p = 1.14 × 10−35), and NO → O synonymous variants (p = 4.2 × 10−88) (Figure 2A). This suggests that synonymous variants that reduce codon optimality are under evolutionary constraint. Furthermore, NO → O synonymous variants segregated at significantly higher allele frequencies than their matched intronic variants (p = 1.9 × 10−14) and neutral synonymous variants (p = 3.0 × 10−32) (Figure 2A), implicating a role of positive selection in optimizing codon content. Similar results were observed when controlling for trinucleotide context (Figure S2A), further supporting that the NO → O and O → NO allele frequency differences cannot be explained by local mutation rate differences.
Figure 2.
SFS and GERP++ Distributions Reflect Selection on Codon Usage
(A) Site frequency spectra of synonymous variants that result in optimal-to-nonoptimal (O → NO), neutral, and nonoptimal-to-optimal codon (NO → O) changes (left panel) and matched intronic variants (right panel).
(B) Distribution of GERP++ scores for the reference alleles of the variants included in (A).
The CSC scores were derived from mRNA stability measurements in a single cell line and therefore might not represent tissue-specific codon optimality patterns. We therefore repeated our SFS analysis by using RSCU, instead of CSC, to define codon optimality instead. Notably, RSCU and CSC are significantly correlated (Pearson’s r = 0.41, p < 10−300), indicating that optimal codons appear more frequently in the human genome. Furthermore, we observe similar evidence of purifying selection against O → NO variants and positive selection on NO → O variants when using RSCU to partition the SFS (Figure S2C). Finally, we compared the allele frequency distributions of codon-optimality-altering synonymous variants to damaging missense and loss-of-function variants (Figure S2E). Unsurprisingly, the missense and loss-of-function variants segregated at lower allele frequencies than O → NO synonymous variants. Thus, although codon-optimality-reducing synonymous variants are subject to negative selection, they are under weaker constraint than other functional variants.
Combined, our results implicate a role of negative selection in purging synonymous variants that reduce codon optimality and a role of positive selection in favoring variants that increase codon optimality. Despite the challenges associated with assessing human codon optimality, these findings strongly suggest that codon usage contributes to human genetic diversity and shapes human evolution.
Optimal Synonymous Sites Are Evolutionarily Conserved across the Mammalian Lineage
The shifts in the site frequency spectrum provide clear evidence of human-specific selective pressures on codon usage. We hypothesized that we should also observe signatures of conservation on codon optimality across related phylogenetic species. We thus assessed conservation at fourfold degenerate sites by using GERP++, a method that assigns each genomic position a score denoting its estimated evolutionary constraint across the mammalian lineage.40
Synonymous sites that are strongly conserved in the human genome have higher GERP++ scores than less conserved sites. Consistent with the hypothesis that optimal codons experience stronger evolutionary conservation, the GERP++ scores of the reference sites of O → NO synonymous variants were significantly higher than the GERP++ scores of matched intronic sites (Mann-Whitney U p = 1.2 × 10−31), neutral synonymous sites (p < 10−300), and NO → O synonymous sites (p < 10−300) (Figure 2B). Moreover, the GERP++ scores of the reference sites of NO → O variants were significantly lower than those of matched intronic (p < 10−300) and neutral synonymous sites (p < 10−300), suggesting weaker phylogenetic constraint at nonoptimal sites. The GERP++ distributions of trinucleotide-matched and RSCU-annotated codon changes corroborated this observation (Figures S2C and S2E). Whereas the prior analysis only considered sites that were variant in the TopMED cohort, we next considered every fourfold degenerate site in the coding genome and found that GERP++ significantly correlated with both CSC (Pearson’s r = 0.26, p < 10−300) and RSCU (r = 0.25, p < 10−300). These results indicate long-term evolutionary pressures on codon usage and are in agreement with prior orthogonal approaches that identified selection on synonymous sites.3,14,18,49, 50, 51, 52
Human Genes Display Differences in Intolerance to Synonymous Variation
Our observations illustrate genome-wide signatures of constraint on codon optimality. However, we suspected that synonymous variation might be under stronger selective constraint in some genes than in others. Therefore, we sought to quantify the strength of selection on synonymous sites per gene. We previously introduced RVIS, a scoring system that quantifies individual genes’ intolerance to missense and loss-of-function mutations by using standing human variation.41 Here, we extended this framework in an approach we term synRVIS. synRVIS quantifies genic constraint against synonymous variants that reduce codon optimality as measured by the codon stability coefficient.
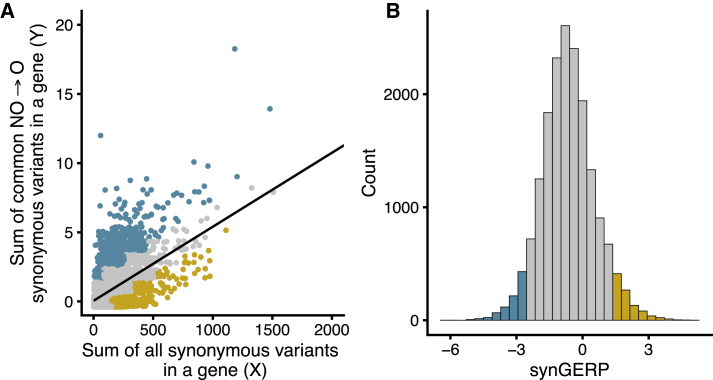
synRVIS only considers variation in the protein-coding genome. Therefore, to increase our sample size for constructing synRVIS, we used sequence data from the 123,136 exomes contained in gnomAD32 rather than the roughly 60,000 genomes contained in TopMED. Specifically, we regressed the number of common (MAF > 0.5%) O → NO synonymous variants (Y) on the total number of observed synonymous variants for each gene (X) (Figure 3A). The resulting regression line predicts the expected number of common O → NO variants accounting for genic mutation rates, sequence context, and gene size. The deviation of each gene from this expectation (more or less variation than expected) is calculated as the studentized residual; a synRVIS below 0 indicates higher intolerance to O → NO synonymous variation. To ensure the resulting residuals reflect intergenic patterns of constraint rather than random noise, we performed a permutation test to verify that these scores deviate from a null model (p = 0.03; see Methods).
Figure 3.
synRVIS Derivation and Distribution of synGERP Scores
(A) synRVIS regression plot in which each point represents a gene. Yellow points represent the bottom fifth percentile (most intolerant) and blue points represent the upper fifth percentile (most tolerant). Two outlier genes with greater than 2,000 synonymous variants are excluded.
(B) The distribution of synGERP scores. As in (A), color coding corresponds to the fifth percentile extremes.
Compared to a weaker constraint, the presence of a strong purifying selection on synonymous sites could reduce overall synonymous polymorphism rates in a gene, which would impact the total number of observed variants in a gene (X). We therefore re-calculated synRVIS by replacing X with each gene’s coding sequence length because the number of observed variants should correlate with gene length. This alternate score strongly correlated with the original synRVIS (Pearson’s r = 0.97). Therefore, overall reductions in polymorphism rates do not seem to limit our power in calculating the score. Furthermore, synRVIS only weakly correlated with the coding length of each gene (Pearson’s r = −0.03), suggesting it is not systematically biased by gene size.
synRVIS provides a direct, gene-specific measure of selection on codon optimality in the human lineage. However, the dynamic range of the synRVIS metric is limited by the comparably small number of mutations at synonymous sites in gnomAD (median of 66 per gene). We therefore created a complementary score, which we termed synGERP, to quantify per-gene conservation at synonymous sites across the mammalian lineage. In order to create a per-gene metric, we took the mean GERP++ score at all fourfold degenerate synonymous sites in a given gene’s canonical transcript, excluding all codons adjacent to exon-intron boundaries. A higher synGERP score signifies overall stronger evolutionary conservation at fourfold degenerate sites for that gene (Figure 3B). Whereas synRVIS specifically considers codon optimality, synGERP reflects evolutionary conservation at fourfold sites regardless of changes in codon usage. Therefore, synGERP reflects additional sources of conservation at synonymous sites beyond codon optimality, such as splicing enhancers, transcription factor binding sites, and RNA secondary structure. To facilitate interpretation of these scores, we calculated genome-wide percentile scores, in which a lower percentile indicates higher intolerance (synRVIS) or higher phylogenetic conservation (synGERP), for synRVIS and synGERP (all scores are available in Table S1).
Interestingly, synRVIS and synGERP were only weakly correlated (Pearson’s r2 = 0.013, p = 2.3 × 10−51). We have similarly observed low correlations between human-specific intolerance scores and GERP-derived scores in prior evaluations of non-coding regulatory regions.53 One possible explanation for this low correlation is that a fraction of codon usage might be under human-specific selection, for example, mirroring human-specific tRNA expression patterns, which would only be captured by synRVIS. Additionally, whereas synRVIS isolates codon optimality effects, synGERP measures the combined constraint on synonymous sites from sources such as splicing enhancers and RNA-binding protein binding sites. Together, these two scores provide a framework for identifying genes that are most intolerant to synonymous variation.
GO enrichment tests revealed that the most synRVIS-intolerant genes (< 25th percentile) were enriched for ontologies related to the cell cycle and transcription; such ontologies included cellular response to DNA damage, microtubule-based processes, and positive regulation of transcription by RNA polymerase II. Furthermore, synGERP intolerant genes were enriched for ontologies such as regulation of proteolysis involved in cellular protein catabolic process, regulation of mRNA stability, and negative regulation of translation (Table S3). These results mirror observations in model organisms that the most codon optimized genes tend to be related to stress responses, translation, and post-transcriptional gene regulation54,55 and therefore underscore the evolutionary significance of codon optimality.
Genes Intolerant to Synonymous Variation Are Enriched for Dosage-Sensitive Genes
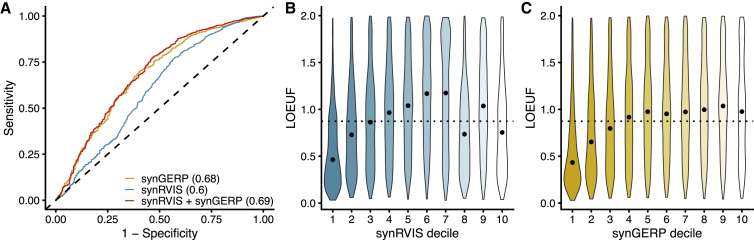
Given the impact of codon usage on mRNA stability and protein expression, we hypothesized that well-established dosage-sensitive genes would be more intolerant to synonymous variation than other genes in the genome. To test this hypothesis, we constructed a logistic regression model to determine whether synRVIS and synGERP could predict the 360 dosage-sensitive genes in ClinGen’s Genome Dosage Map.56 We found that both synRVIS and synGERP significantly predicted this gene set: p = 8.2 × 10−9 (AUC = 0.60) and p = 2.2 × 10−34 (AUC = 0.68), respectively (Figure 4A). A joint model containing both scores achieved an AUC of 0.69, in which both synRVIS and synGERP remain predictive (p = 7.6 × 10−7 and p = 1.4 × 10−31, respectively), indicating that both scores provided significant independent information in predicting dosage-sensitive genes.
Figure 4.
Dosage-Sensitive Genes Are Intolerant to Synonymous Variation
(A) ROC curve demonstrating the capacity for synRVIS, synGERP, and a joint model to predict ClinGen dosage-sensitive genes. AUCs for the respective models are indicated in parentheses.
(B and C) The distribution of LOEUF scores for each synRVIS (B) and synGERP (C) decile. The black dot indicates the median LOEUF score per synRVIS decile, and the dotted horizontal line indicates the median LOEUF score across all genes.
The ClinGen dosage-sensitive genes included in the prior analysis only include genes implicated in Mendelian disease. Another way to identify dosage-sensitive genes is to identify genes depleted of loss-of-function variants in the human population. To verify that dosage-sensitive genes are intolerant to synonymous variation, we compared synRVIS and synGERP to LOEUF, a metric that represents the ratio of observed/expected loss-of-function variants within gnomAD.32 A lower LOEUF indicates a higher constraint against loss-of-function variation. To compare synonymous and loss-of-function constraint, we plotted the median LOEUF score per synRVIS and synGERP decile (Figures 4B and 4C). We observed that genes more intolerant to synonymous variation tend to be depleted of loss-of-function variation. Furthermore, synRVIS and synGERP both correlated with LOEUF (Pearson’s r = 0.15, p = 3.2 × 10−89 and Pearson’s r = 0.24, p = 3.3 × 10−231, respectively). We were surprised, however, to find that some highly synRVIS-tolerant genes were also enriched for low LOEUF scores (Figure 4B). This discordance implies that certain loss-of-function-intolerant genes are tolerant to changes in codon usage.
A GO enrichment analysis revealed that synRVIS-tolerant (>75th percentile) but LOEUF-intolerant (<25th percentile) genes were significantly enriched for certain neurodevelopmental pathways, such as regulation of dendrite morphogenesis, positive regulation of axonogenesis, and synaptic vesicle endocytosis (Figure S4). Notably, neurons are subject to different translational regulation programs than other cell types are because of mTOR signaling57 and their unique cellular demands, such as local translation at synapses.58 Furthermore, recent evidence suggests that codon optimality may in fact be attenuated in the developing nervous system.54
In summary, both synRVIS and synGERP can broadly predict dosage-sensitive genes. These results emphasize the importance of codon usage in regulating gene expression and demonstrate that natural selection more strongly optimizes codon content in genes where differences in protein abundance strongly impact human physiology.
DNA Damage Genes and Periodically Expressed Cell-Cycle Genes Are Intolerant to Changes in Codon Usage
If codon optimality is important in regulating gene expression, it is most likely to not only be under particularly strong constraint in haploinsufficient genes, but also in genes that are sensitive to tRNA levels. The cytoplasmic tRNA pool changes dynamically in terms of its overall abundance as well as its composition in response to cellular demands.59, 60, 61 We expected that genes that need to be highly expressed when tRNA concentrations are low should be the most intolerant to reductions in codon optimality.
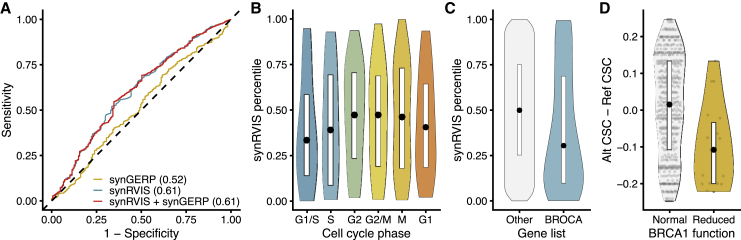
Among classes of genes, we expected DNA-damage-repair genes to be under particularly strong constraint. In yeast, stress due to DNA-damaging compounds results in reduced tRNA export from the nucleus as well as tRNA modifications that enhance translation of key DNA repair proteins.62,63 In mice, knocking out the Elongator complex, which is required for translating codon-biased genes, leads to dysregulation of codon-biased DNA-damage genes.64 Motivated by these findings, we tested whether a previously published list of 178 DNA-damage-response genes were intolerant to synonymous variation.65 In a logistic regression model, synRVIS, but not synGERP, was able to predict genes involved in the response to DNA damage (AUC = 0.61, p = 6.02 × 10−05; AUC = 0.52, p = 0.6, respectively) (Figure 5A). This result implies that codon usage in DNA-damage-repair genes is under human-specific constraint and thus most likely plays a role in regulating this pathway. Although our synGERP analysis suggests that codon optimality is not conserved across eukaryotes, we suspect this discordance between synRVIS and synGERP is due to species-specific variation in the stress-induced tRNA pools.
Figure 5.
Intolerance of DNA-Damage Response and Cell-Cycle-Phase Genes
(A) ROC curve illustrating the capacity of synGERP and synRVIS to predict DNA-damage-response genes. AUCs for the respective models are indicated in parentheses.
(B) synRVIS percentiles of genes periodically expressed in each phase of the cell cycle.
(C) synRVIS distribution for genes contained in the BROCA cancer risk panel versus all other protein-coding genes.
(D) Comparison of changes in CSC scores for synonymous BRCA1 variants that result in normal protein function versus those that reduce protein function.
tRNA levels also oscillate throughout the cell cycle, and genes that are expressed at different phases of the cell cycle have different codon usage.66 In particular, tRNA expression levels are highest in the G2/M phase and lowest at the end of G1 phase. This coupling between tRNA expression and codon usage allows for cell-cycle-dependent oscillations in protein abundance by ensuring that G2 phase genes are less efficiently translated during G1. Accordingly, we hypothesized that genes expressed during the G1 phase should be more intolerant to reductions in codon optimality than G2 genes. Strikingly, the synRVIS distribution for these periodically expressed genes closely matches the oscillatory changes in tRNA abundances; tolerance to reductions in codon optimality is lowest for G1/S-expressed genes and increases stepwise by cell-cycle stage, peaking for G2/M genes (Figure 5B). This finding not only supports previous observations about the codon usage patterns of cell-cycle-related genes, but it provides direct evidence that these patterns are under selective constraint. synGERP scores did not display this pattern (Figure S5A), further suggesting that synRVIS might be more sensitive in detecting human-specific selection on genes that respond to tRNA availability.
The tRNA pool can also be dysregulated in diseases, including certain cancers. Prior studies have found that elevated tRNA concentrations in breast cancer, ovarian cancer, and multiple myeloma promote expression of pro-tumorigenic genes.27,67, 68, 69, 70 We therefore hypothesized that oncogenes that are sensitive to shifts in tRNA abundances should be intolerant to changes in codon usage. To test this hypothesis, we compared the synRVIS and synGERP scores of hereditary breast and ovarian cancer genes included in the BROCA Cancer Risk Panel to all other protein-coding genes in the genome.71,72 This gene list includes 66 genes strongly implicated in hereditary breast and ovarian cancers. Accordingly, synRVISs, but not synGERP scores, were lower for these genes than for the rest of the genes in the genome (synRVIS, Mann-Whitney U p = 0.002, permuted p = 0.002; synGERP, p = 0.80) (Figures 5C and S5B). Taken together, our results demonstrate the importance of codon optimality in mediating gene expression under different physiological states.
Synonymous Variants that Reduce Codon Optimality in BRCA1 Might Abrogate Protein Abundance
Collectively, our analysis suggests that synonymous mutations that alter codon optimality are under evolutionary constraint, implying that these mutations have functional consequences. In particular, we expect that these variants might affect protein concentration by modulating mRNA translation and stability. To date, synonymous variants have been largely ignored in genetic disease association studies. However, synonymous mutations that reduce codon optimality in genes under strong selection could contribute to Mendelian disease. We have previously demonstrated that non-synonymous intolerance metrics, such as RVIS, facilitate the discovery of disease-associated genes.41,73 synRVIS and synGERP now provide a framework for identifying and prioritizing potential genes in which synonymous variants might also cause disease. Notably, genes with a low synRVIS include genes such as BRCA1 and BRCA2.
Although the functional impact of synonymous variants for most genes is unknown, we took advantage of a unique dataset in which CRISPR was used to perform saturation genome editing to assess the functional consequences of nearly all possible single nucleotide variants in the functionally critical RING and BRCT domains of BRCA1.44 BRCA1 ranked among the most highly intolerant genes (1st percentile synRVIS, 13th percentile synGERP) and loss of this protein predisposes women to breast and ovarian cancer.74,75 Thus, this dataset allows us to systematically answer the question of whether synonymous single nucleotide variants (SNVs) that reduce codon optimality significantly reduce the BRCA1 dosage.
Findlay et al. introduced SNVs in the cell line HAP1, which is critically dependent on BRCA1 for cell survival.44 11 days after introducing the mutations, they sequenced the line to gauge the frequency of each variant within the cell population. Deleterious variants result in cell death by reducing BRCA1 expression or function and were thus less prevalent in the population. These frequencies were converted into a continuous score that reflects protein function. The researchers also measured the expression of BRCA1 to assign RNA scores that directly reflect each variant’s effect on gene expression.
Of roughly 500 introduced synonymous mutations in BRCA1, 19 received scores that signified reduced BRCA1 activity. We hypothesized that synonymous variants that achieved lower functional scores resulted in decreased expression and/or translation. For each synonymous variant, we calculated the difference between the CSC value of the alternate and reference alleles, such that negative changes signify reductions in codon optimality. Accordingly, the 19 synonymous mutations associated with reduced BRCA1 activity were significantly more likely to attenuate codon optimality (Mann-Whitney U p = 0.001) (Figure 5D). We also calculated the correlation between the RNA and function scores and the difference in CSC values for all synonymous variants assayed (Figures S5C and S5D). We found that changes in codon optimality significantly correlated with BRCA1 function scores (Pearson’s r = 0.27, p = 3 × 10−11) and RNA scores (Pearson’s r = 0.15, p = 4.8 × 10−4). Although these correlations are modest, they suggest that at least a fraction of variants that reduce codon optimality may have functional consequences in BRCA1, presumably via the modulation of translation and/or mRNA stability. We note that there are other potential mechanisms by which these variants could functionally impact BRCA1, including via the modulation of splicing enhancers. Therefore, further molecular studies are required to elucidate the precise functional consequences of attenuated codon optimality in BRCA1.
Nonetheless, these results imply that some synonymous variants that affect codon usage can result in large enough effect sizes to cause Mendelian disease. synRVIS thus provides an initial framework for identifying putatively pathogenic synonymous mutations that reduce codon optimality in the interpretation of human genomes, whereby mutations in the most intolerant genes are most likely to be pathogenic. Importantly, three of the 19 synonymous variants that reduce BRCA1 function appear in gnomAD, indicating that some individuals do in fact harbor potentially disease-causing synonymous variants that might be overlooked in standard carrier screens.
Discussion
Through comprehensive analyses, we demonstrate the role of natural selection in optimizing the codon content of the human genome. First, we show that synonymous mutations that reduce codon optimality appear at lower allele frequencies in the human population than neutral variants and variants that increase codon optimality. Supporting this result, we find that optimal codons tend to be more strongly phylogenetically conserved across the mammalian lineage. We introduce two per-gene intolerance scores, synRVIS and synGERP, which assess the strength of selective constraint on synonymous variation in each protein coding gene. synRVIS detects human-specific selection against variants that reduce codon optimality, whereas synGERP reflects the phylogenetic constraint of fourfold degenerate sites in a given gene. We find that these scores predict dosage-sensitive genes, emphasizing the importance of codon usage in mediating protein concentration.
Recent studies have revealed that synonymous codon usage serves as a secondary genetic code that guides translation efficiency and mRNA stability in human cells.10,12,13 In particular, the translation elongation rate, which is partially a function of tRNA abundance, is posited to impact the mRNA degradation rate. Despite these molecular consequences, some population geneticists have argued that the effect size of any single synonymous SNV would be too small to be selected against in the human population. Our results cast doubt on this assumption in two ways.
First, the allele frequency distributions illustrate that there are genome-wide signatures of selection against reductions in codon optimality. This finding shows that some synonymous mutations exert a large enough effect to be selected against even in the context of the small human effective population size. Importantly, we note that the SFS analysis only considers synonymous sites that contain a variant in the reference cohort. Previous analyses have demonstrated that some synonymous sites, such as those in splicing enhancer elements, vary so infrequently that they might not appear in the sample. Therefore, our SFS results might be conservative. In future studies, it would be of value for researchers to complement these analyses with overall polymorphism ratios to estimate the distribution of selection coefficients as they relate to codon optimality.
Second, we demonstrate that some codon-optimality-reducing SNVs in BRCA1 can significantly attenuate protein activity, potentially via reduced mRNA stability and translation. These findings are consistent with a handful of other studies that have implicated synonymous SNVs in human disease.2,76,77 In fact, some synonymous variants that alter codon bias can significantly reduce protein concentration to the same extent as loss-of-function variants76 and most likely represent an underappreciated source of Mendelian disease. However, it is more likely that most synonymous variants only modestly reduce protein output, as the selection on O → NO variants is substantially weaker than loss-of-function mutations. Nonetheless, synonymous SNVs that only modestly reduce protein output could play a significant role in modifying both Mendelian diseases and complex traits, many of which are driven by the cumulative effect of many variants with small effect sizes.78,79
Our results support the functional relevance of the translational regulation of gene expression. Consistent with the effects of translational efficiency on protein output and mRNA stability, we find that dosage-sensitive and loss-of-function-depleted genes tend to be more intolerant to synonymous variation. However, one limitation of our study is that the calculation of synRVIS relies on codon usage metrics derived from a single cell type, whereas tRNA expression varies widely by tissue.28 synGERP, on the other hand, does not rely on codon usage scores but is less sensitive to detecting constraint on potential human-specific tRNA expression dynamics. Indeed, synGERP most likely also detects other sources of conservation, such as constraint on splicing and regulatory motifs. We also note that although synRVIS was built for the assessment of selection on codon optimality, it may detect other confounding sources of constraint that correlate with codon optimality and nucleotide content, such as exonic splicing enhancers.
We found that some loss-of-function-depleted genes involved in neurodevelopment were in fact very tolerant to reductions in codon optimality. Intriguingly, a recent study found that codon optimality is attenuated in genes expressed in the developing Drosophila nervous system.54 This reduced optimality mitigates the effect of codon content on mRNA stability, thereby allowing trans-acting factors, such as RNA-binding proteins and microRNAs, to exert greater influence over mRNA decay in the developing nervous system. If this phenomenon exists in human beings, it could explain our observation that some loss-of-function-depleted genes are tolerant to changes from optimal-to-nonoptimal codons. Additionally, because tRNA expression is most likely markedly different in the brain,9,28,80 synRVIS might be limited in detecting intolerance of neurodevelopmental genes because of its reliance on HEK293T-derived codon stability coefficients. Both of these hypotheses might explain synGERP’s improved ability to predict dosage-sensitive genes because synGERP could detect constraint on binding sites for trans-acting factors and does not rely on CSC in its calculation. Understanding the relationship between tissue-specific codon usage, intolerance, and mRNA decay programs stands as an important goalpost for future studies.
Strikingly, we not only found a correlation between the strength of selection on codon optimality and disease-relevant genes, but we also found a relationship with the tRNA abundance patterns that prevail when specific genes are expressed. Specifically, changes in tRNA abundance can modulate protein expression in response to different cellular states, including cell-cycle stage, disease, and stress. Previous studies have demonstrated that cellular tRNA concentrations are reduced in response to DNA damage and during the G1 phase of the cell cycle.66 Accordingly, we illustrate that intolerant synRVIS genes are enriched for genes involved in these cellular pathways. synGERP is unable to predict these genes, perhaps implicating a role of human-specific selection on codon optimality in these pathways. We note that tRNA dysregulation also underpins the pathogenesis of other non-cancerous conditions, including some immunodeficiency and neurological disorders.81, 82, 83 Therefore, future work focused on determining potential interspecies variation in dynamic tRNA expression will be crucial in determining whether non-human disease models accurately represent diseases characterized by translational deregulation.
Collectively, our results suggest that codon usage can significantly impact biological traits and might play an underappreciated role in human disease. Just as previously developed intolerance scores have improved our ability to identify disease-associated genes,41,73 synRVIS will aid in prioritizing potential genes in which synonymous variants that reduce codon optimality could cause disease.
We note that synRVIS critically depends on codon usage metrics and the number of individuals sequenced in the reference cohort. Therefore, our resolution to detect intolerance to synonymous variation in the human genome will improve with tissue-specific codon stability coefficients and increased numbers of sequenced individuals.
Data and Code Availability
synRVISs and synGERP scores are available in Table S1. The code for computing these scores is available on GitHub.
Declaration of Interests
D.B.G. is a founder of and holds equity in Praxis, holds equity in Q-State Biosciences, serves as a consultant to AstraZeneca, and has received research support from Janssen, Gilead, Biogen, AstraZeneca, and Union Chimique Belge (UCB). R.S.D. serves as a consultant to AstraZeneca. A.M.M. serves as a consultant to Ribometrix. B.R.C. declares no competing interests.
Acknowledgments
We wish to thank many people for very helpful discussions and comments on the manuscript, including Slavé Petrovski, Chirag Vasavda, Tim Harris, Ayal Gussow, Justin Dhindsa, Daniel Krizay, Sarah Dugger, Evan Baugh, and Gundula Povysil. We also thank Chirag Vasavda, Brian Khoe, Daniel Zhang, and Xinchen Wang for feedback on figure design.
Published: June 8, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.011.
Contributor Information
Ryan S. Dhindsa, Email: rsd2135@cumc.columbia.edu.
David B. Goldstein, Email: dg2875@cumc.columbia.edu.
Web Resources
BioRender, https://biorender.com
BRAVO Database, https://bravo.sph.umich.edu/freeze5/hg38/
BROCA Cancer Risk Panel, https://testguide.labmed.uw.edu/public/view/BROCA
ClinGen Dosage Sensitivity Map, https://dosage.clinicalgenome.org
CycleBase cell cycle scores, https://cyclebase.org/CyclebaseSearch
ExAC Database and constraint scores, https://gnomad.broadinstitute.org/downloads
Human DNA repair genes, https://www.mdanderson.org/documents/Labs/Wood-Laboratory/human-dna-repair-genes.html
PANTHER Gene Ontology, http://geneontology.org
Picard tools, https://broadinstitute.github.io/picard/
Supplemental Information
List of Genes with Their synRVIS and synGERP Scores
Gene Lists Used for Enrichment Tests
GO Enrichment Results
Annotated BRCA1 Variants 5 from Findlay et al. (2018)74
References
- 1.Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Parmley J.L., Chamary J.V., Hurst L.D. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 4.Supek F., Miñana B., Valcárcel J., Gabaldón T., Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 5.Shen L.X., Basilion J.P., Stanton V.P., Jr. Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc. Natl. Acad. Sci. USA. 1999;96:7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brest P., Lapaquette P., Souidi M., Lebrigand K., Cesaro A., Vouret-Craviari V., Mari B., Barbry P., Mosnier J.F., Hébuterne X. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat. Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 7.Capon F., Allen M.H., Ameen M., Burden A.D., Tillman D., Barker J.N., Trembath R.C. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum. Mol. Genet. 2004;13:2361–2368. doi: 10.1093/hmg/ddh273. [DOI] [PubMed] [Google Scholar]
- 8.Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R., Coller J. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini A.A., Del Viso F., Moreno-Mateos M.A., Johnstone T.G., Vejnar C.E., Qin Y., Yao J., Khokha M.K., Giraldez A.J. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016;35:2087–2103. doi: 10.15252/embj.201694699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Q., Medina S.G., Kushawah G., DeVore M.L., Castellano L.A., Hand J.M., Wright M., Bazzini A.A. Translation affects mRNA stability in a codon-dependent manner in human cells. eLife. 2019;8:e45396. doi: 10.7554/eLife.45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radhakrishnan A., Chen Y.H., Martin S., Alhusaini N., Green R., Coller J. The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality. Cell. 2016;167:122–132. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narula A., Ellis J., Taliaferro J.M., Rissland O.S. Coding regions affect mRNA stability in human cells. RNA. 2019;25:1751–1764. doi: 10.1261/rna.073239.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest M.E., Pinkard O., Martin S., Sweet T.J., Hanson G., Coller J. Codon and amino acid content are associated with mRNA stability in mammalian cells. PLoS ONE. 2020;15:e0228730. doi: 10.1371/journal.pone.0228730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamary J.V., Parmley J.L., Hurst L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 15.Eyre-Walker A.C. An analysis of codon usage in mammals: selection or mutation bias? J. Mol. Evol. 1991;33:442–449. doi: 10.1007/BF02103136. [DOI] [PubMed] [Google Scholar]
- 16.Comeron J.M. Weak selection and recent mutational changes influence polymorphic synonymous mutations in humans. Proc. Natl. Acad. Sci. USA. 2006;103:6940–6945. doi: 10.1073/pnas.0510638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavner Y., Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 18.Drummond D.A., Wilke C.O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z., Nielsen R. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 2008;25:568–579. doi: 10.1093/molbev/msm284. [DOI] [PubMed] [Google Scholar]
- 20.Plotkin J.B., Robins H., Levine A.J. Tissue-specific codon usage and the expression of human genes. Proc. Natl. Acad. Sci. USA. 2004;101:12588–12591. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pouyet F., Mouchiroud D., Duret L., Sémon M. Recombination, meiotic expression and human codon usage. eLife. 2017;6:e27344. doi: 10.7554/eLife.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph K.L., Schmitt B.M., Villar D., White R.J., Marioni J.C., Kutter C., Odom D.T. Codon-Driven Translational Efficiency Is Stable across Diverse Mammalian Cell States. PLoS Genet. 2016;12:e1006024. doi: 10.1371/journal.pgen.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sémon M., Lobry J.R., Duret L. No evidence for tissue-specific adaptation of synonymous codon usage in humans. Mol. Biol. Evol. 2006;23:523–529. doi: 10.1093/molbev/msj053. [DOI] [PubMed] [Google Scholar]
- 24.Kanaya S., Yamada Y., Kinouchi M., Kudo Y., Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 2001;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 25.dos Reis M., Savva R., Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keightley P.D., Eyre-Walker A. Deleterious mutations and the evolution of sex. Science. 2000;290:331–333. doi: 10.1126/science.290.5490.331. [DOI] [PubMed] [Google Scholar]
- 27.Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., Christophersen N.S., Christensen L.L., Borre M., Sørensen K.D. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duret L., Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hershberg R., Petrov D.A. Selection on codon bias. Annu. Rev. Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 31.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv. 2019 doi: 10.1101/563866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 33.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smit A., Hubley R., Green P. Institute for Systems Biology; 2013. RepeatMasker Open-4.0. [Google Scholar]
- 35.Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 36.Lawrie D.S., Messer P.W., Hershberg R., Petrov D.A. Strong purifying selection at synonymous sites in D. melanogaster. PLoS Genet. 2013;9:e1003527. doi: 10.1371/journal.pgen.1003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Machado H.E., Lawrie D.S., Petrov D.A. Pervasive Strong Selection at the Level of Codon Usage Bias in Drosophila melanogaster. Genetics. 2020;214:511–528. doi: 10.1534/genetics.119.302542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keinan A., Mullikin J.C., Patterson N., Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harpak A., Bhaskar A., Pritchard J.K. Mutation Rate Variation is a Primary Determinant of the Distribution of Allele Frequencies in Humans. PLoS Genet. 2016;12:e1006489. doi: 10.1371/journal.pgen.1006489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickham H. Springer-Verlag New York; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 46.Lappalainen T., Scott A.J., Brandt M., Hall I.M. Genomic Analysis in the Age of Human Genome Sequencing. Cell. 2019;177:70–84. doi: 10.1016/j.cell.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- 48.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savisaar R., Hurst L.D. Exonic splice regulation imposes strong selection at synonymous sites. Genome Res. 2018;28:1442–1454. doi: 10.1101/gr.233999.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y.F., Siepel A. Estimation of allele-specific fitness effects across human protein-coding sequences and implications for disease. Genome Res. 2019;29:1310–1321. doi: 10.1101/gr.245522.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keightley P.D., Halligan D.L. Inference of site frequency spectra from high-throughput sequence data: quantification of selection on nonsynonymous and synonymous sites in humans. Genetics. 2011;188:931–940. doi: 10.1534/genetics.111.128355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bustamante C.D., Nielsen R., Hartl D.L. A maximum likelihood method for analyzing pseudogene evolution: implications for silent site evolution in humans and rodents. Mol. Biol. Evol. 2002;19:110–117. doi: 10.1093/oxfordjournals.molbev.a003975. [DOI] [PubMed] [Google Scholar]
- 53.Petrovski S., Gussow A.B., Wang Q., Halvorsen M., Han Y., Weir W.H., Allen A.S., Goldstein D.B. The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity. PLoS Genet. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burow D.A., Martin S., Quail J.F., Alhusaini N., Coller J., Cleary M.D. Attenuated Codon Optimality Contributes to Neural-Specific mRNA Decay in Drosophila. Cell Rep. 2018;24:1704–1712. doi: 10.1016/j.celrep.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carneiro R.L., Requião R.D., Rossetto S., Domitrovic T., Palhano F.L. Codon stabilization coefficient as a metric to gain insights into mRNA stability and codon bias and their relationships with translation. Nucleic Acids Res. 2019;47:2216–2228. doi: 10.1093/nar/gkz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., ClinGen ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blair J.D., Hockemeyer D., Doudna J.A., Bateup H.S., Floor S.N. Widespread Translational Remodeling during Human Neuronal Differentiation. Cell Rep. 2017;21:2005–2016. doi: 10.1016/j.celrep.2017.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holt C.E., Schuman E.M. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan C.T., Pang Y.L., Deng W., Babu I.R., Dyavaiah M., Begley T.J., Dedon P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saikia M., Wang X., Mao Y., Wan J., Pan T., Qian S.B. Codon optimality controls differential mRNA translation during amino acid starvation. RNA. 2016;22:1719–1727. doi: 10.1261/rna.058180.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torrent M., Chalancon G., de Groot N.S., Wuster A., Madan Babu M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018;11:eaat6409. doi: 10.1126/scisignal.aat6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Begley U., Dyavaiah M., Patil A., Rooney J.P., DiRenzo D., Young C.M., Conklin D.S., Zitomer R.S., Begley T.J. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghavidel A., Kislinger T., Pogoutse O., Sopko R., Jurisica I., Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131:915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 64.Goffena J., Lefcort F., Zhang Y., Lehrmann E., Chaverra M., Felig J., Walters J., Buksch R., Becker K.G., George L. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nat. Commun. 2018;9:889. doi: 10.1038/s41467-018-03221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wood R.D., Mitchell M., Lindahl T. Human DNA repair genes, 2005. Mutat. Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Frenkel-Morgenstern M., Danon T., Christian T., Igarashi T., Cohen L., Hou Y.M., Jensen L.J. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012;8:572. doi: 10.1038/msb.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., Tavazoie S.F. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M.R., Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter A.G., Sourvinos G., Allison S.J., Tosh K., Scott P.H., Spandidos D.A., White R.J. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc. Natl. Acad. Sci. USA. 2000;97:12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y., Goodenbour J.M., Godley L.A., Wickrema A., Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem. Biophys. Res. Commun. 2009;385:160–164. doi: 10.1016/j.bbrc.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh T., Casadei S., Lee M.K., Pennil C.C., Nord A.S., Thornton A.M., Roeb W., Agnew K.J., Stray S.M., Wickramanayake A. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh T., Lee M.K., Casadei S., Thornton A.M., Stray S.M., Pennil C., Nord A.S., Mandell J.B., Swisher E.M., King M.C. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X., Petrovski S., Xie P., Ruzzo E.K., Lu Y.F., McSweeney K.M., Ben-Zeev B., Nissenkorn A., Anikster Y., Oz-Levi D. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet. Med. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall J.M., Lee M.K., Newman B., Morrow J.E., Anderson L.A., Huey B., King M.C. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 75.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., BRCA1 and BRCA2 Cohort Consortium Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 76.Dershem R., Metpally R.P.R., Jeffreys K., Krishnamurthy S., Smelser D.T., Hershfinkel M., Carey D.J., Robishaw J.D., Breitwieser G.E., Breitwieser G.E., Regeneron Genetics Center Rare-variant pathogenicity triage and inclusion of synonymous variants improves analysis of disease associations of orphan G protein-coupled receptors. J. Biol. Chem. 2019;294:18109–18121. doi: 10.1074/jbc.RA119.009253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 78.Boyle E.A., Li Y.I., Pritchard J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bornelöv S., Selmi T., Flad S., Dietmann S., Frye M. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 2019;20:119. doi: 10.1186/s13059-019-1726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morita M., Gravel S.P., Chénard V., Sikström K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 82.Piccirillo C.A., Bjur E., Topisirovic I., Sonenberg N., Larsson O. Translational control of immune responses: from transcripts to translatomes. Nat. Immunol. 2014;15:503–511. doi: 10.1038/ni.2891. [DOI] [PubMed] [Google Scholar]
- 83.Tahmasebi S., Khoutorsky A., Mathews M.B., Sonenberg N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 2018;19:791–807. doi: 10.1038/s41580-018-0034-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Genes with Their synRVIS and synGERP Scores
Gene Lists Used for Enrichment Tests
GO Enrichment Results
Annotated BRCA1 Variants 5 from Findlay et al. (2018)74
Data Availability Statement
synRVISs and synGERP scores are available in Table S1. The code for computing these scores is available on GitHub.