Abstract
Although de novo mutations (DNMs) are known to increase an individual’s risk of congenital defects, DNMs have not been fully explored regarding orofacial clefts (OFCs), one of the most common human birth defects. Therefore, whole-genome sequencing of 756 child-parent trios of European, Colombian, and Taiwanese ancestry was performed to determine the contributions of coding DNMs to an individual’s OFC risk. Overall, we identified a significant excess of loss-of-function DNMs in genes highly expressed in craniofacial tissues, as well as genes associated with known autosomal dominant OFC syndromes. This analysis also revealed roles for zinc-finger homeobox domain and SOX2-interacting genes in OFC etiology.
Keywords: orofacial clefts, whole genome sequencing, de novo mutations, trios
Introduction
Orofacial clefts (OFCs) are the most common craniofacial malformation in humans, affecting 1 in 1,000 live births around the world,1 and cause a significant personal, financial, and societal burden.2 OFCs are phenotypically and etiologically heterogeneous, which presents important opportunities and significant challenges for discovering causal genes. Phenotypically, these malformations can be divided into three general categories: clefting of the lip only (CL), clefting of both the lip and the palate (CLP), and clefting of the palate only (CP). Because CL and CLP (together denoted as CL/P) share a defect in the primary palate, they are often analyzed together, whereas CP is typically analyzed separately, in part, because the secondary palate forms after the lip during development.2, 3, 4, 5 Furthermore, OFCs can also be classified as an isolated (e.g., nonsyndromic) occurrence comprising approximately 70% of CL/P-affected individuals and 50% of CP-affected individuals or as a part of a syndrome comprising the remaining 30% and 50%, respectively.6
These phenotypic classifications inform hypotheses about the type and number of genetic variants controlling an individual’s risk of an OFC. For example, syndromic OFCs are typically attributed to single gene mutations, chromosomal anomalies, or teratogens, whereas nonsyndromic OFCs are considered to have a complex etiology with multiple genetic and environmental risk factors. Similarly, 15% of OFC-affected individuals have some family history of OFCs, suggesting a major contribution of inherited genetic factors, whereas the sporadic nature of most OFCs suggests a possible role for de novo mutations (DNMs). Of course, genetic variants of all types and frequencies plus environmental risk factors combine to influence the penetrance and expression of genes controlling OFCs, making these categories useful but not absolute.
Multiple large-scale genome-wide association studies (GWASs) have been performed in nonsyndromic OFC cohorts to identify ∼45 genetic loci that collectively account for ∼25% of the estimated liability to OFCs.7, 8, 9, 10, 11 Rare coding variants have been examined by exome and limited genome sequencing studies,12, 13, 14, 15, 16, 17, 18, 19 but these approaches have not yet been widely applied, and sample sizes and populations have been limited to about 100 or fewer families with individuals affected by OFCs. In this manuscript, we focus on DNMs as one understudied class of rare variants influencing an individual’s risk to OFCs. Germline DNMs arise spontaneously in either the germ cell of the parents or during the early stages of embryonic development.20 On average, individuals have approximately 100 DNMs throughout their entire genome, and approximately one DNM affects the coding region (exome).21, 22, 23, 24 Genetic studies of DNMs have led to previous successful identifications of genes and pathways underlying multiple congenital disorders, such as congenital heart defects,25, 26, 27 Kabuki syndrome (MIM: 147920),28 and autism.29 However, only a few DNMs have been reported for OFCs to date,15,30, 31, 32, 33, 34 and their overall contribution to OFCs has not been thoroughly assessed in a large sample or on a genome- or exome-wide scale.
Whole-genome sequencing (WGS) of child-parent trios ascertained through several studies of nonsyndromic OFCs generated as part of the Gabriella Miller Kids First (GMKF) Pediatric Research Consortium now makes it feasible to investigate DNMs contributing to OFCs on a genome-wide scale. Therefore, we analyzed the contribution of coding, germline DNMs to various aspects of OFC risk in child-parent trios of European, Colombian, and Taiwanese ancestry.
Methods
Sample of Child-Parent Trios
This study summarizes the initial findings on de novo variants in three samples of child-parent trios. One sample is a set of 415 trios of European ancestry recruited from sites around the United States, Argentina, Turkey, Hungary, and Spain; the second is a set of 275 trios from Medellin, Colombia; and the third is a set of 125 trios from Taiwan. In this study, the three samples are referred to as European, Colombian, and Taiwanese, respectively. Recruitment of participants and phenotypic assessments were done at regional treatment centers after review and approval by each site’s institutional review board (IRB) and the IRB of the affiliated US institutions (HRPO #03-0871, IRB#HSC-MS-03-090, IRB#970405, IRB#200109094, and IRB#200109094). All data collected from human subjects in the Taiwan sample has been monitored and reviewed annually by the IRB at Johns Hopkins School of Public Health since 1991; as of March 12, 2020 the research protocol is no longer active, and the resulting deidentified genomic data has been certified as not “human subjects research.”
Among parents, 88.7% of European, 93.1% of Taiwanese, and 100% of the Colombian parents were unaffected. As most OFC cases are likely to have multifactorial etiology, we consider DNMs to be one of many genetic factors that could influence an individual’s risk, and they need not be limited to affected individuals without any family history of OFCs. Therefore, as in our prior work,35 the cleft status of the parents (Table S1) was not considered in these analyses.
Whole-Genome Sequencing and Variant Calling
The OFC-affected trios were sequenced as part of the GMKF Pediatric Research Consortium, which was established in 2015 with the aim of addressing gaps in the understanding of the genetic etiologies of structural birth defects, such as OFCs, and pediatric cancers. The majority of samples were sequenced from blood samples; however, when blood samples were inaccessible, saliva samples were used for sequencing. The McDonnell Genome Institute (MGI), Washington University School of Medicine in St. Louis carried out the WGS of the European samples, which were subsequently aligned to hg38 and variant called by the GMKF’s Data Resource Center at Children’s Hospital of Philadelphia. Sequencing of the Colombian and Taiwanese samples was conducted at the Broad Institute, and data were aligned to hg38 and called via GATK pipelines22,36,37 at the Broad Institute. Details of the alignment and genotyping workflow used to harmonize these three datasets were recently published;38 in brief, all samples were realigned and recalled via a GATK pipeline at the GMKF Data Resource Center. Overall, the WGS data from these three studies were quite comparable. The average depth per sample for all sequenced individuals was 29.16, ranging from 4.7–50.0.
Quality Control
The WGS data for the 415 European, 275 Latino, 125 Taiwanese child-parent trios were evaluated on the basis of a variety of quality metrics. Individuals with a missingness value, Mendelian error value, or an average read depth outside of three standard deviations from the mean were removed. Additional individuals were removed on the basis of transition/transversion (Ts/Tv), exonic Ts/Tv, silent/replacement, or heterozygotes/homozygotes ratios that were less than or greater than expected while allowing for somewhat lower ratios of heterozygotes/homozygotes ratios in the Colombian sample (which included trios drawn from a number of consanguineous pedigrees). Family relationships were confirmed with identity-by-descent analyses conducted in PLINK (version 1.90b53). X chromosome heterozygosity was used to confirm the sex of all individuals. Finally, after assessing each individual’s quality metrics, only complete child-parent trios were retained, leaving 374 European, 267 Colombian, and 116 Taiwanese child-parent trios for analysis.
Identification of De Novo Variants
Called variants were filtered for minor allele count (MAC)3 ≥ 1, genotype quality (GQ)3 ≥ 20, depth (DP)3 ≥ 10, variant quality scores (QUALs)3 ≥ 200, and quality by depth (QD)3 ≥ 3.0 via VCFtools (version 0.1.13) and GATK (version 3.8); additionally, only bi-allelic variant calls were included in this study. To generate a list of high confidence DNMs, we further filtered variants on the basis of allele balance (AB). An AB filter3 ≥0.30 and ≤0.70 was used for variant calls in the offspring, and an AB filter < 0.05 was used for the corresponding variant calls in the offspring’s parents. Annotation of high confidence DNMs was completed with ANNOVAR (version 201707). One additional European child-parent trio was removed after this annotation because the offspring of European ancestry had 328 whole-genome DNMs, which was greater than three standard deviations from the observed mean. We used a chi-square goodness-of-fit test to confirm that the observed number of coding DNMs for each proband followed the expected Poisson distribution (p = 0.93); the value of lambda was equal to the mean of the number of coding DNMs per proband. The number of DNMs genome-wide for each proband did appear to differ between populations (p < 2.2 × 10−16; Figure S7A); however, this is most likely because of sequencing artifacts or ancestry bias within databases used in the processing and filtering pipeline and not necessarily attributed to some unknown biological event. DNMs were then further filtered on the basis of annotation, and only rare (MAF < 0.1% across all of gnomAD v2.0.1) coding DNMs were kept for further analysis. Although there was a significant difference between populations genome-wide, there was no difference between the number of coding DNMs (p = 0.12; Figure S7B).
Statistical Analysis of De Novo Variants
The statistical analysis used to determine whether there was any excess of coding DNMs by variant class throughout the entire exome and by variant class in individual genes was carried out with the “DenovolyzerByClass” function and the “DenovolyzerByGene” function, respectively, in DenovolyzeR (0.2.0). This R package calculates the enrichment value by dividing the number of observed DNMs by the expected number of DNMs, the latter of which is determined on the basis of the well-established model developed by Samocha et al.39 Under the null model of no association between mutation class and disease status, the number of observed DNMs is expected to fit a Poisson distribution40 with the mean determined by the sequence of the genes in the exome and the fixed sample size. Thus, for each class of mutation, we have a single observation that is Poisson distributed, with “known” mean, M, and therefore known variance, M, because a Poisson distribution must have an identical mean and variance, and standard deviation, sqrt(M). Thus, M here is a fixed “known” constant. Under the alternate model, the number of observed mutations, A, also follows a Poisson distribution, but A does not necessarily equal M. Here, we plot A/M, together with the exact 95% confidence interval of A (the values of A above the 2.5 percentile and below the 97.5 percentile divided by M) determined from the Poisson distribution. The “DenovolyzerByGene” function, which calculates the enrichment and p values for individual genes, was carried out in all OFC-affected trios; this analysis was not divided by cleft subtype. The “DenovolyzerByClass” function paired with the “includeGenes” option in the DenovolyzeR program was used to test whether more DNMs were observed than expected in a particular set of genes. Again, 95% confidence intervals were displayed for the enrichment values, as in the autism study by Satterstrom et al.41 Because prevalence differs between males and females, the rate of observed DNMs is expected to differ between males and females for any mutation exerting the same effect on both sexes on the liability scale. When a rate difference was first observed in children with autism, the observation was dubbed the “female protective effect,” but this general phenomena is best thought of as simply a result of a mutation’s having a constant effect on the liability scale and differing prevalence between the sexes.42 Because our female cases also have a higher rate of DNMs, and a lower overall prevalence, we estimate the effect size of each class of DNMs on a liability scale and observed the difference in DNM rates.
Gene-Set Analyses
A gene set enrichment analysis (GSEA) was carried out with Topp.Fun, available through the ToppGene Suite;43 p values were adjusted for multiple testing via Benjamini and Hochberg,44 a correction method available within ToppFun. The top five most significant terms for each assessed category were then compared. Gene lists of marker genes expressed in ectodermal and mesenchymal cell clusters of the developing lip were identified by Li et al. via single cell RNA sequencing (RNA-seq); these marker genes were used in our analyses and can be found in the supplementary information published in Li et al.45 We generated another set of functionally relevant genes by using processed RNA-seq data generated from human cranial neural crest cell (hCNCC) samples (GEO: GSM1817212, GSM1817213, GSM1817214, GSM1817215, GSM1817216, and GSM1817217), which were downloaded from the Gene Expression Omnibus.46,47 The six hCNCC samples were derived from induced pluripotent stem cells (iPSCs) or embryonic stem cells (ESCs) from three human individuals, and RNA-seq was carried out with NEBNext Multiplex Oligos for sequencing on an Illumna HiSeq 2000. The expression levels from the six samples were averaged and then ranked from highest expression to lowest expression. Genes with probability of being loss-of-function intolerant (pLI) scores3 ≥ 0.95 and in the top 20th percentile for hCNCC expression were prioritized and tested for an excess of DNMs in our trios compared to what would be expected by chance. The clinically-relevant set of genes was constructed after a thorough literature search; known and suggested genes harboring mutations associated with OFCs are summarized in Table S4. The list of suggestive and significant loci used to identify DNMs in genes within ± 500 kb was generated using data generated from Carlson et al.8 and Yu et al.9 Predicted gene interactions were visualized with GeneMANIA.48
In Situ Hybridization
Studies involving mice were conducted in strict accordance with the recommendations in the National Institutes of Health’s “Guide for the Care and Use of Laboratory Animals.” The protocol was approved by the University of Wisconsin-Madison School of Veterinary Medicine Institutional Animal Care and Use Committee (protocol number 13–081.0). C57BL/6J mice (Mus musculus) were purchased from The Jackson Laboratory. Timed pregnancies were established as described previously.49 Embryos at embryonic day 11 (E11) were dissected in PBS and fixed in 4% paraformaldehyde in PBS overnight followed by graded dehydration (1:3, 1:1, 3:1 volume per volume [v/v]) into 100% methanol for storage. After rehydration, embryos were embedded in 4% agarose gel, and 50 μm sections through the lambdoidal junction were made with a vibrating microtome. In situ hybridization (ISH) was performed as previously described.50 Gene-specific ISH riboprobe primers were designed with IDT PrimerQuest and affixed with the T7 polymerase consensus sequence plus a 5-bp leader sequence (CGATGTTAATACGACTCACTATAGGG) to the reverse primer (Table S5). Sections were imaged with a MicroPublisher 5.0 camera connected to an Olympus SZX-10. For each gene, representative images were selected from staining conducted on at least three sections from independent mouse embryos.
Results
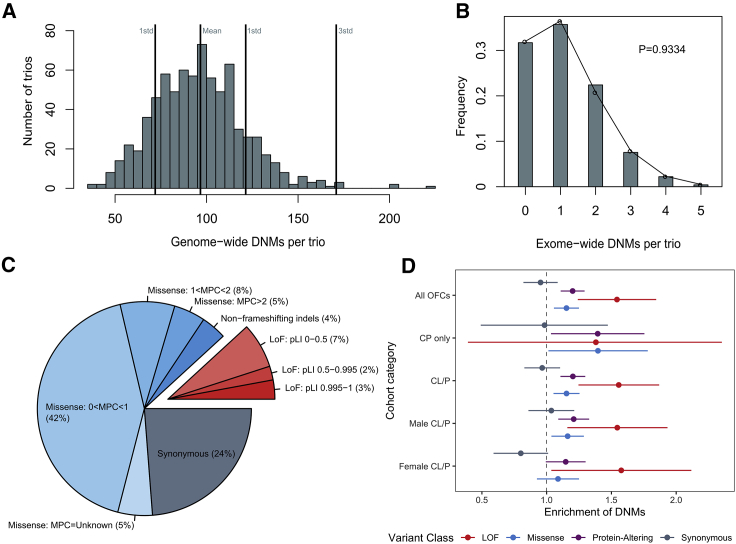
A set of high-confidence DNMs from WGS was generated from 756 complete child-parent trios; counts by cleft subtype, ethnicity, and sex are presented in Table S1. The majority of the offspring had a CL or CLP; 58 European offspring had CP only. While 97% of cases are reported to have an isolated OFC, 3.0% reported other features (e.g., speech delay, hypertelorism, and intellectual disabilities). However, none of these trios have been diagnosed with a recognized genetic syndrome with molecular confirmation. Overall, 73,027 DNMs were identified genome-wide in the 756 child-parent trios with an average of 96.60 DNMs per proband (Figure 1A). Although we identified DNMs genome-wide, the initial analysis reported here focuses on rare, coding DNMs. After filtering for rare (MAF < 0.1% in gnomAD v2.0.1) exonic and splicing variants, 862 coding DNMs in 808 genes were identified, averaging 1.14 DNMs per trio (Tables S2 and S3) and following the expected Poisson distribution (p = 0.93) (Figure 1B).40
Figure 1.
LoF and Missense De Novo Mutations Are Enriched in Individuals with Orofacial Clefts
(A) Distribution of DNMs per trio genome-wide.
(B) Distribution of coding DNMs per trio.
(C) Distribution of rare, coding DNMs by variant class for all OFC-affected trios. DNMs were further divided by MPC score (missense) and gene pLI score (LoF) as a measure of potential deleteriousness.
(D) Enrichment of DNMs ± two standard errors by variant class for all OFCs, probands with CP only, probands with CL/P, male probands with CL/P, and female probands with CL/P. The comparison of trios only affected by CP by sex of the proband was not presented because of small sample sizes.
First, we characterized the distribution of types of coding DNMs among all trios and stratified by OFC subtypes and proband sex. We categorized coding DNMs into four classes on the basis of their predicted function: synonymous, missense (consisting of single amino acid changes and non-frameshifting insertions and deletions), predicted loss-of-function (LoF, made up of stop-gain, frameshifting insertions and deletions, and essential splice site variants), and a combined group of missense and LoF hereafter referred to as protein-altering DNMs. The majority of DNMs (64%) were missense variants (including 4% non-frameshifting indels), 12% were LoF, and the remaining 24% were synonymous (Figure 1C). To assess the severity of specific types of DNMs, missense and LoF DNMs were additionally subcategorized by MPC score and pLI score, respectively; MPC scores are a variant-specific combined measure of predicted deleteriousness that includes missense badness, PolyPhen-2, and constraint. In contrast, the pLI scores are gene specific and represent the tolerance of a gene’s LoF variants; the more intolerant a gene is to a LoF variant, the closer the pLI score is to 1. The overall proportions of DNMs in variant functional classes and subcategories were not significantly different for each OFC subtype (p = 0.20; Figure S1). The same was true for all OFCs when we compared the proportions of DNMs by the sex of the proband (p = 0.48; Figure S2).
We next determined whether trios affected by OFC possessed significantly more coding DNMs than was expected on the basis of mutational models.39 The 756 trios had a significant excess of protein-altering DNMs (enrichment = 1.20; p = 3.22 × 10−6) (Figure 1D and Table S2). The observed excess can be attributed to both LoF DNMs (enrichment = 1.54; p = 2.55 × 10−5) and missense DNMs (enrichment = 1.15; p = 5.92 × 10−4). As anticipated, trios did not possess an excess of synonymous DNMs (enrichment = 0.95; p = 0.76). When stratified by OFC subtypes, a significant excess of protein-altering DNMs was found among both trios affected by CL/P (enrichment = 1.20; p = 6.12 × 10−6) and trios affected by CP (enrichment = 1.39; p = 9.32 x10−3), but the difference in strengths of association is difficult to interpret because of the differing sample sizes (Table S2). Among females with CL/P (n = 254), the excess of protein-altering DNMs (enrichment = 1.15; p = 2.76 × 10−2) was primarily attributed to an excess of LoF DNMs (enrichment = 1.58; p = 7.28 × 10−3). In males with CL/P (n = 444), a similar excess of LoF DNMs (enrichment = 1.55; p = 9.69 × 10−4) and a significant excess of missense DNMs (enrichment = 1.16, p = 4.35 × 10−3) was observed. However, when the effects of DNMs in males and females with CL/P were compared directly on a liability scale, no significant differences were observed between males and females for any variant class (Figure S3).
Next, we performed a GSEA to determine whether genes with LoF or protein-altering DNMs clustered into specific gene sets or pathways relevant to craniofacial development. We also carried out GSEA for genes with synonymous DNMs as a control because these DNMs represent a set of variants most likely to have no effect on an individual’s OFC risk. Protein-altering DNMs were enriched in genes belonging to gene sets broadly related to development and also more specific sets related to OFCs and craniofacial development (Figure S4). For example, a significant enrichment of protein-altering DNMs was identified in genes related to the biological process term “embryo development” (p = 6.11 × 10−6) and human disease terms such as “uranostaphyloschisis” (clefting) and “cleft palate” (p = 3.28 × 10−4 and p = 5.88 × 10−4, respectively). Similarly, an enrichment was identified for multiple terms that described abnormal embryo morphology in mice (Figure S4A, lower panel). Overall, we found that protein-altering DNMs were enriched in genes involved in embryonic development, craniofacial development, and human craniofacial disorders, whereas synonymous DNMs were not enriched in genes relevant to craniofacial development.
Because CL/P and CP have historically been considered distinct disorders, we performed a GSEA for genes with protein-altering DNMs in CL/P and CP separately to determine whether this distinction was reflected in the WGS data. Overall, different gene ontology terms achieved statistical significance for offspring with CL/P and CP (Figure S5). Many biological process and molecular function terms were statistically significant for offspring with CL/P, whereas only one term (“limb bud formation”) was significant for offspring with CP only (p = 6.32 × 10−3) (Figure S5). For disease and human phenotype terms, “cleft palate,” “cleft palate (isolated),” “congenital abnormality,” “hearing problem,” “osteogenesis imperfecta,” and “uranostaphyloschisis” were all significantly enriched for genes with protein-altering DNMs in the CP group but not for those in the CL/P group. In contrast, five mouse phenotypic terms were statistically significant in the CL/P-affected trios, but only one mouse term (“abnormal bone ossification”) was significant for CP-affected trios (p = 3.37 × 10−2). Although both CL/P and CP genes were associated with terms related to embryonic development, the genes with DNMs in CL/P described broad embryonic development, whereas the genes mutated in CP had some features that appeared more specific to the palate. For example, the anterior portion of the secondary palate ossifies during palatogenesis, and hearing problems are commonly reported in individuals with CP. Although not conclusive, the results of these analyses suggest that DNMs play an important role in both CL/P and CP and that potential differences should be further investigated in a larger sample of trios affected by CP only. Because of the small number of CP-affected trios, all analyses hereafter were carried out with all OFC-affected trios and were no longer separated by cleft subtype.
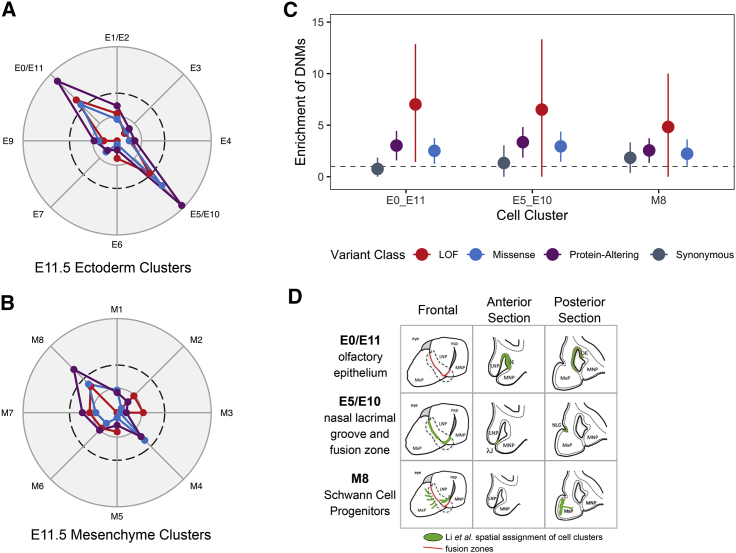
Although the GSEA results are promising, the significant terms represent very general problems with development. To get a more specific view of the impact of genes with DNMs in craniofacial development, we used recently published single-cell RNA-seq data from the lambdoidal junction of the developing murine upper lip.45 The lambdoid junction is the point of fusion of three facial prominences creating the primary palate and upper lip, so we hypothesized that the marker genes for each cell cluster are among the best candidate genes in the genome to be involved in OFCs and could potentially harbor an excess of DNMs. To address this question, we analyzed marker genes belonging to eight clusters of ectodermal cells and nine clusters of mesenchymal cells (Figure 2). The marker genes from two ectodermal cell clusters positioned at the nasal process fusion zone and the olfactory epithelium had a significant excess of protein-altering DNMs (Figures 2A and 2C; p = 1.38 × 10−5 and p = 3.27 × 10−5, respectively). A single mesenchymal cell cluster, classified as Schwann cell progenitors, had a significant excess of protein-altering DNMs (Figures 2B and 2C; p = 5.63 × 10−4); this cluster of cells is derived from neural crest cells and was located adjacent to the fusing lambdoidal junction through mapping the mesenchymal clusters via in situ hybridization (Figure 2D).45,51,52 Overall these analyses point to genes expressed in the cells at the point of fusion as being particularly relevant for an individual’s risk of developing an OFC.
Figure 2.
De Novo Mutations Are Enriched in Genes Expressed at the Point of Fusion in Lip Development
(A and B) Marker genes for each ectodermal cell sub-cluster (A) and mesenchymal cell sub-cluster (B) were analyzed for an excess of DNMs, and the −log(p value) was calculated. In each radar plot, the dashed circle represents the significance threshold after correcting for multiple tests (p = 2.9 × 10−3). The inner circle represents nominal significance (p = 0.05); the outer circle represents p = 1.0 × 10−5. Each of the clusters are named as reported by Li et al., including three combined clusters where two clusters were merged on the basis of overlapping spatial expression of marker genes: E1/E2 (the combination of clusters E1 and E2), E0/E11 (the combination of clusters E1 and E2), and E5/E10 (the combination of clusters E5 and E10).
(C) Enrichment of DNMs ± two standard errors for significant cell sub-clusters.
(D) Depiction of spatial assignment of cell clusters with a significant excess of DNMs in the frontal view, anterior section, and posterior sections of the lambdoidal junction; adapted from Li et al.45
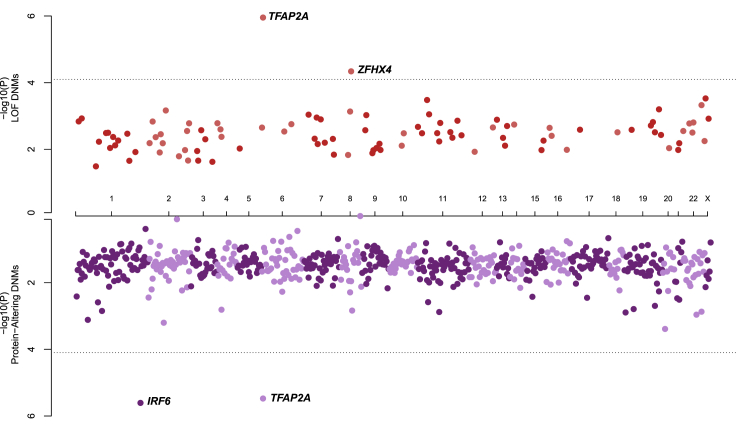
We next identified individual genes with an excess of DNMs by comparing our observed mutation counts with published per-gene mutability models.20,39 Two genes (TFAP2A [MIM: 107580] and ZFHX4 [MIM: 606940]) had significantly more LoF DNMs than expected after correcting for multiple testing (i.e., number of genes with any protein-altering DNM) (Figure 3). Notably, TFAP2A remained significant given a more conservative exome-wide significance threshold suggested by Ware et al.20 (p < 1.3 × 10−6) despite only observing two distinct LoF DNMs (p.Glu104∗ and p.Gly145Glufs∗18; p = 1.11 × 10−6). Although multiple genes had more than one protein-altering DNM that might be critical to the underlying etiology of OFCs, only two genes (TFAP2A and IRF6 [MIM: 607199]) had a significant excess of protein-altering DNMs after correcting for multiple tests (Figure 3). In addition to the two LoF DNMs described above, we identified a third missense variant in TFAP2A (p.Ser247Leu). We also identified three missense variants in IRF6 (p.Gly376Val, p.Asn88Asp, and p.Arg84His).
Figure 3.
De Novo Mutations in IRF6, TFAP2A, and ZFHX4 Are Associated with OFCs
Identification of single genes with an excess of LoF DNMs (top axis) or protein-altering DNMs (missense and/or LoF DNMs; bottom axis). The dashed line indicates the significance threshold after correcting for multiple tests, p < 7.9 × 10−5.
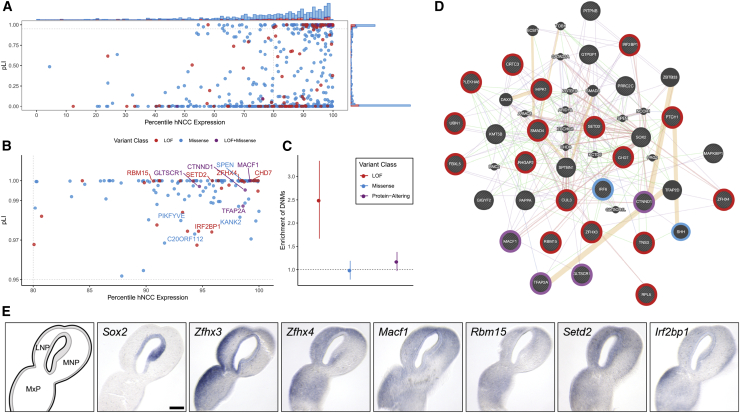
We reasoned that genes highly expressed in craniofacial-relevant tissues that are intolerant to LoF variants would be good candidate genes for OFCs even if they do not individually reach formal statistical significance. To identify such genes, we used RNA-seq data from hCNCC lines (a cell type giving rise to a majority of facial structures). A significant excess of LoF DNMs was observed among genes with pLI > 0.95 in the top 20th percentile of hCNCC expression (enrichment = 2.49; p = 3.46 × 10−6, Figure 4). The 132 genes with protein-altering DNMs in this category were significantly enriched for gene ontology terms related to gene regulation, including RNA Pol II-mediated regulatory elements (i.e., promoters) (p = 9.5 × 10−4) and chromatin binding (p = 9.5x10−4), which is consistent for a multipotent cell type. Among the 31 genes in this category with LoF DNMs, a ToppFun analysis43 using protein interaction data from the NCBI Entrez Gene ftp site53, 54, 55, 56 identified a significant enrichment for genes interacting with SOX2 [MIM: 184429], a gene recognized to play an essential role in controlling progenitor cell behavior during craniofacial development (p = 1.47 × 10−3); the eight genes with LoF DNMs in this category included MACF1 [MIM: 608271], RBM15 [MIM: 606077], SETD2 [MIM: 612778], CHD7 [MIM: 608892], CTNND1 [MIM: 601045], IRF2BP1 [MIM: 615331], ZFHX4, and TFAP2A. Notably, four of these genes (CHD7, TFAP2A, ZFHX3 [MIM: 104155], and ZFHX4) were also ranked in the top 100 expressed genes in the lateral nasal eminence (which contributes to the upper lip) of E10.5 mouse embryos.57 Finally, nine of the genes with pLI > 0.95 in the top 20th percentile of gene expression had multiple protein-altering DNMs, including TFAP2A, CTNND1, ZFHX4, and MACF1. Ultimately, this analysis provided confirmatory evidence for several genes involved in the etiology of OFCs (CHD7 and CTNND1), expanded the evidence for TFAP2A influencing an individual’s risk to OFCs in humans, and implicated additional genes as OFC candidate risk genes (MACF1, SETD2, ZFHX3, and ZFHX4). To confirm a plausible role in orofacial morphogenesis and cleft pathogenesis, we probed expression patterns of these genes in the embryonic tissues forming the upper lip and primary palate in the mouse. As demonstrated previously, Sox2 expression was localized to the nasal pit epithelium at the center of the lambdoidal junction.58 All candidate genes examined were detected in the neural crest mesenchyme, and several exhibited expression in the mesenchyme proximal to the Sox2 domain in the nasal pit, including Zfhx4, Mac1, and Setd2 (Figure 4E).
Figure 4.
DNMs Are Enriched in Genes Expressed in hCNCCs and among SOX-2-Interacting Genes
(A) All genes with at least one protein-altering DNM were ranked by their pLI score for the gene and the expression of the gene in RNA-seq data generated from hCNCCs. The barplot on the top and right axes show the relative counts of genes with missense (blue), LoF (red), and both missense and LoF (purple) DNMs.
(B) Genes with a pLI score > 0.95 and in the top 20th percentile of hCNCC expression. Labeled genes have at least two protein-altering DNMs.
(C) Enrichment of DNMs ± two standard errors in all genes in the top 20th percentile of hCNCC expression with a pLI score > 0.95 for all OFC-affected trios.
(D) Protein-interaction visualization generated in GeneMANIA of the 31 genes with LoF DNMs and a pLI score > 0.95 and that are in the top 20th percentile of hCNCC expression. Pink lines represent physical interactions, orange lines represent predicted interactions, purple lines represent co-expression, green lines represent genetic interactions, and blue lines represent co-localization. Genes are colored on the basis of the types of DNMs present in the OFC dataset.
(E) Sections through the lambdoidal junction of the medial nasal (MNP), lateral nasal (LNP), and maxillary (MxP) processes of gestational day 11 mouse embryos were stained for the indicated gene by in situ hybridization. In the schematic on the lower left, the epithelium is shaded, whereas the neural crest mesenchyme is white.
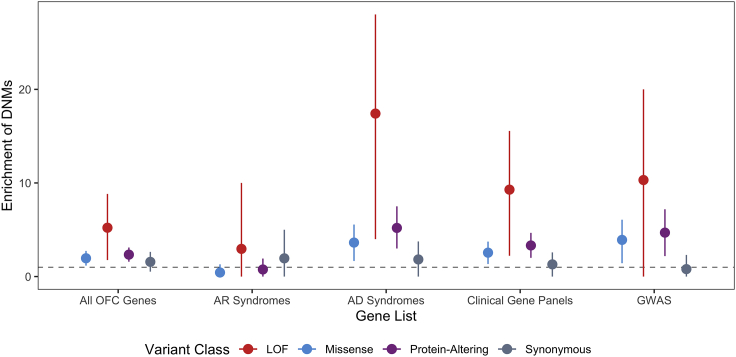
To determine the contribution of DNMs in clinically relevant OFC genes (Table S4), we constructed several gene lists including existing clinical sequencing panels, genes mutated in Mendelian syndromes that include OFCs as a key phenotype, genes previously implicated in candidate gene or exome sequencing studies, and GWAS-nominated genes (see Methods for construction of gene lists). Of the 336 genes comprising this list, we identified 42 individuals with 43 DNMs in 31 genes, representing 6% of all sequenced trios. One individual had two DNMs (a stop-gain in CHD7 and a synonymous variant in SHH [MIM: 600725]), but only the former is predicted to be pathogenic. Overall, 25 missense and 9 LoF DNMs were observed in the list of genes; this was significantly more than expected by chance in this list of genes (p = 8.95 × 10−6; Figure 5). Specifically, the autosomal dominant syndrome gene set had a highly significant excess of protein-altering DNMs (p = 2.29 × 10−9). As expected for DNMs, we did not observe an excess among genes involved in autosomal recessive syndromes (p = 0.74). GWAS-nominated genes also showed a modest excess of protein-altering DNMs (p = 1.45 × 10−6), which in part reflects overlap between these different gene sets (Figure S6).
Figure 5.
GMKF OFC-Affected Trios Have an 18-fold Excess of LoF DNMs in Known Autosomal Dominant OFC Genes
Enrichment of DNMs ± two standard errors for all OFC-affected trios in a clinically relevant gene set related to OFC conditions.
One of the primary challenges in the post-GWAS era is identifying causal genes when the most strongly associated SNPs are non-coding. At some loci, there is an obvious candidate gene where coding mutations can cause Mendelian forms of the same disorder; this is the case for several of the GWAS-nominated genes (e.g., IRF6, GRHL3 [MIM: 608317], ARHGAP29 [MIM: 610496]). Therefore, we hypothesized that identifying a DNM in a gene in the vicinity of a GWAS peak could provide evidence to nominate a gene as truly causal or even to provide additional evidence in support of suggestive loci not yet achieving formal genome-wide significance. To address this question, we evaluated DNMs in genes within 1 Mb (±500 kb) of both suggestive and significant GWAS loci from two recent OFC GWASs.8,9 Overall, 37 protein-altering DNMs were identified within these genes. As expected, several of these DNMs were located within genes already demonstrating strong statistical evidence and implicating them in OFC development. Our results provide confirmatory evidence of a role for such genes (e.g., ARHGAP29, IRF6) in causing OFCs. Protein-altering DNMs were also identified in genes near recently reported GWAS loci (TFAP2A,9 SHROOM3 [MIM: 604570]8), adding evidence in support of their role in OFC etiology. Furthermore, this analysis also identified genes with DNMs not previously suggested as specific candidate genes within the suggestive or significant loci: ZFHX4 at 8q21, RBM15 at 1p13, UBN1 [MIM: 609771] at 16p13.1, and HIRA [MIM: 600237] at 22q11.2, thus providing additional evidence for implicating these genes and loci in an individual’s OFC risk.
Discussion
This analysis represents the largest genetic exploration of coding DNMs to date in 756 child-parent trios affected by nonsyndromic OFCs. This initial study clearly demonstrates that coding DNMs in both biologically relevant and clinically relevant sets of genes might contribute to an individual's risk of nonsyndromic OFC. In addition to providing a unique insight into known OFC risk genes, our results implicate multiple candidate genes and gene interactions. Collectively, these observations and findings provide a better understanding of the genetic architecture of OFCs.
By combining WGS with single-cell sequencing, bulk RNA-seq, and other genomic datasets, we identified multiple genes or sets of interacting genes involved in controlling an individual’s risk to OFCs where mutations had not been reported previously in OFCs. The most promising of these genes include ZFHX3 and ZFHX4, the latter of which was the only gene apart from TFAP2A with multiple LoF DNMs. Both genes had a pLI score of 1, were highly expressed in the hCNCCs (top 20th percentile), and were marker genes for the E0_E11 cell cluster (the olfactory epithelium) analyzed from the single-cell RNA-seq data. ZFHX4 is located on 8q21.11 where microdeletions have been reported in individuals with intellectual disability, facial dysmorphism, and cleft palate.59 In addition, a LoF DNM in ZFHX4 was reported in an individual with disrupted speech development.60 Both ZFHX4 and ZFHX3 were also ranked in the top 100 expressed genes in the lateral nasal eminence (which contributes to the upper lip) of E10.5 mouse embryos, along with other known OFC risk genes (i.e., CHD761 and TFAP2A [see GeneReviews in Web Resources]).57 Moreover, ZFHX4 is located at a suggestive GWAS locus (rs10808812; p = 2.00 × 10−6)8 and is thus implicated in the etiology of OFCs through both LoF DNMs and evidence from common SNPs.
LoF DNMs were also observed in other candidates with pLI > 0.95 in the top 20th percentile of gene expression, including MACF1, RBM15, and SETD2. Along with TFAP2A, CTNND1, and CHD7, all three of these genes are bioinformatically predicted to interact with SOX2, which is known to play an essential role in controlling progenitor cell behavior during craniofacial development.62 While a few of these genes expressed in the mesenchyme were only proximal to the Sox2 domain in the nasal pit, upper lip and palate morphogenesis is dependent upon epithelial-mesenchymal interactions, including signals from the nasal pit that appear to act on the nearby mesenchyme.58 We focused analysis on the tissues that form the midface and upper lip but cannot exclude the possibility that expression of these genes in other tissues (e.g., forebrain neuroectoderm) might also be relevant. Although MACF1 was not individually significant in our analyses (p = 0.028), we notably identified two protein-altering MACF1 DNMs (p.Tyr1357∗ and p.Arg5180Met). MACF1 has recently been reported to play a role in regulating osteogenic differentiation and cranial bone formation in vivo.63 RBM15 is located near a suggestive GWAS locus, providing further evidence for this gene’s involvement in craniofacial pathology. Additionally, mutations in SETD2 have been reported to cause Luscan-Lomish syndrome [MIM: 616831], a craniofacial overgrowth condition resembling Sotos syndrome.64 Identifying a set of possible SOX2-interacting genes anchored by established OFC genes paired with the detection of these genes in the neural crest mesenchyme does provide the strongest evidence to support ZFHX4, ZFHX3, MACF1, RBM15, and SETD2 as OFC candidate risk genes. Therefore, these genes should be further explored in larger samples. Moreover, their role in craniofacial development and any interactions with each other or SOX2 should be evaluated in detail.
A surprising finding of this study was the critical role of TFAP2A DNMs in an individual’s OFC risk. Although there are multiple lines of evidence in the literature to support the role of TFAP2A in craniofacial development and syndromic OFCs, the number of protein-altering DNMs in TFAP2A was unexpected. Complete loss of Tfap2a in mouse models causes severe developmental defects including anencephaly, facial clefts, and thoraco-abdominoschisis.65 On some backgrounds, heterozygous null mutants exhibit anencephaly.66 Although the human phenotype caused by TFAP2A mutations is considerably less severe (see GeneReviews in Web Resources), Branchiooculofacial syndrome (BOFS [MIM: 113620]) is described as a very rare disorder with less than 200 described cases as of 2019,67 corresponding to an estimated prevalence between 1 in 300,000 and 1 in 1,000,000 individuals. BOFS is characterized by problems with branchial arch development leading to skin anomalies on the neck, eye malformations including microphthalmia, anopthalmia, or coloboma, and facial dysmorphism including cleft lip and/or palate, hypertelorism, telecanthus, upslanting palpebral fissures, broad nose, facial muscle weakness, malformed ears, and hearing loss. Given these different phenotypic features, we would expect that the most severely affected individuals with BOFS would have been excluded from this study, so it is unlikely that we would identify three cases of classic BOFS in our 756 trios. We conclude that the phenotype caused by mutations in TFAP2A mutations is broader than previously appreciated; however, on the basis of our observations, TFAP2A mutations could account for ∼0.5% of all OFCs. Additional studies will be required to fully explore the frequency of TFAP2A variants in diverse OFC cohorts.
Approximately 6% of the trios, which was significantly more than expected by chance, had protein-altering DNMs in clinically-relevant OFC genes. This finding should be interpreted with caution because this was not a population-based genetic screening of individuals with OFCs. This study population was recruited over many years from multiple sites around the world by individuals with a range of clinical skills. Many Mendelian syndromes that include OFCs have variable expressivity, incomplete penetrance, or phenotypic features that might not be apparent at the time of recruitment, especially when cases are often recruited in the first year of life when they present to genetics clinic or begin surgical repairs. Cumulatively, six of the protein-altering DNMs were identified in IRF6 and TFAP2A, accounting for 0.9% of all protein-altering DNMs and 0.8% of sequenced probands. The number of DNMs in IRF6 was expected; dominant mutations in IRF6 cause Van der Woude syndrome (VWS [MIM: 119300]), the most common Mendelian syndrome with an OFC as a key phenotype. 15% of VWS-affected individuals present with only an OFC and are indistinguishable from individuals with nonsyndromic OFCs.68 Despite the number of DNMs in IRF6 and TFAP2A, these DNMs did not alone explain the excess of DNMs observed in the single cell or gene panel analyses (Figures S8A and S8B). Other genes that are associated with dominant Mendelian syndromes and that have LoF DNMs included RPL5 [MIM: 603634], COL2A1 [MIM: 120140], CTNND1, and CHD7. Recognition of the additional features that characterize syndromes caused by mutations in these genes can become clearer with a molecular finding, but it is essential to have a better understanding of how different characteristics of mutations, such as the location within the gene or variant class, contribute to variable expressivity and incomplete penetrance of associated syndromic features. Nonetheless, identifying DNMs in these genes has critical implications for genetic counseling and recurrence risk estimates.
We identified DNMs in CDH1 [MIM: 192090] and CTNND1, two genes recently identified as components of the p120-cadherin complex harboring mutations in families with autosomal dominantly inherited forms of OFC.15,69,70 Mutations in CDH1, CTNND1, PLEKHA5 [MIM: 607770], and PLEKHA7 [MIM: 612686] were reported to account for 14% of multiplex families and 2% of a smaller replication cohort. In this study, we failed to identify DNMs in PLEKHA5 or PLEKHA7; CDH1 and CTNND1 DNMs accounted for less than 1% of the identified variants. Additional studies will be necessary to determine the proportion of OFCs accounted for by these genes and any differences between DNMs and inherited variants described in multiplex families, including detailed phenotyping of individuals and families carrying these variants.71
Historically, CL/P and CP have been considered distinct disorders, so we performed multiple analyses in CL/P and CP separately to determine if this is supported by DNMs.2 Both CL/P and CP had a significant excess of protein-altering DNMs, although the excess of DNMs in the CL/P-affected trios was more apparent. The strength of the significance is most likely due to very different sample sizes; 698 trios were affected by CL/P, and only 58 were affected by CP. Despite having a small number of CP-affected trios, the GSEA still yielded many significant craniofacial disease terms, although none of these terms were significant for the genes with protein-altering DNMs in the CL/P-affected trios. This lends some support to the idea that CL/P and CP could have distinct etiologies, although we were able to identify genes common to both. Because of these results, the contribution of DNMs should be further investigated in larger studies to better understand causal genetic risk factors and how they contribute differently to OFC subtypes.
The CL/P-affected trios were stratified on the basis of proband sex because of the increased prevalence of CL/P among males compared to females (2:1). In developmental disorders with sex biases, it has been suggested that genetic liability to disease is higher in the less-frequently affected sex and would require more severe variants to manifest the disease (i.e., a higher threshold determining affected versus not affected in the sex with lower population prevalence). For OFCs, we would hypothesize that females with CL/P would have an increased burden of DNMs compared to males with CL/P or that they would have more LoF DNMs. However, males and females did not differ significantly by any DNM variant class, and both sexes had a similar significant excess of LoF and protein-altering DNMs. These results suggest that the difference in genetic liability cannot be explained by coding, autosomal DNMs alone.
In conclusion, we have shown the important contribution of rare, coding DNMs in 756 OFC-affected child-parent trios. Through this exploration we have identified multiple candidate risk genes as well as pertinent information regarding well-known and clinically-relevant OFC genes. Although many of the genes identified in this study have only one protein-altering DNM, future studies might identify additional variants in the same genes, providing further evidence for their role in OFC etiology. Like other structural birth defects,26,72,73 both missense and LoF DNMs play a significant role in determining an individual’s risk to OFC, but similar studies in specific OFC subtypes will be necessary to fully understand the genetic architectures specific to each cleft subtype. Our results also suggest that the analysis of rare inherited variants in these same genes will uncover additional risk variants and might identify some of the missing heritability for OFCs. It will be more challenging to explore the implications of noncoding variants and somatic DNMs in relevant tissues that are known to contribute to other congenital defects,74, 75, 76, 77 but this study highlights sets of genes and pathways that should be the focus for identifying possible regulatory elements.
Declaration of Interests
The authors declare no competing interests.
Data and Code Availability
The data analyzed and reported in this manuscript were accessed from the database of Genotypes and Phenotypes (dbGaP; European trios, dbGaP: phs001168.v2.p2; Colombian trios, dbGaP: phs001420.v1.p1; Taiwanese trios, dbGaP: phs000094.v1.p1) and from the Kids First Data Resource Center. Some datasets used in this study are available publicly from the Gene Expression Omnibus (GEO) via the following accession numbers: GEO: GSM1817212, GSM1817213, GSM1817214, GSM1817215, GSM1817216, and GSM1817217.
Acknowledgments
These studies are part of the Gabriella Miller Kids First Pediatric Research Program Consortium, supported by the Common Fund of the Office of the Director of the National Institutes of Health (NIH). Sequencing of the European trios was completed at Washington University’s McDonnell Genome Institute (3U54HG003079-12S1 and X01-HL132363 [M.L.M. and E.F.]), and the Colombian and Taiwanese trios were sequenced at the Broad Institute Sequencing Center (U24-HD090743, X01-HL136465 [M.L.M. and E.F.], and X01-HL140516 [T.H.B. and Azeez Butali]). The sequencing centers plus the Kids First Data Resource Center, supported by the NIH Common Fund through grant U2CHL138346, provided technical and analytical support of this project. The assembling of the sample of child-parent trios, collection of the phenotypic data and samples, and data analysis were supported by the additional grants from the NIH (R01-DE016148 [M.L.M. and S.M.W.], R03-DE026469 [E.F. and M.L.M.], R03-DE027193 [E.J.L.], R00-DE025060 [E.J.L.], R01-DE011931 [J.T.H.], U01-DD000295 [G.W.], and R03-DE027121 [M.A.T., T.H.B., and J.B.]). This study would not be possible without the dedication of many families, study teams, and colleagues worldwide.
Published: June 22, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.018.
Web Resources
Gabriella Miller Kids First Pediatric Research Program, www.commonfund.nih.gov/KidsFirst
Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/
GeneMANIA, https://genemania.org/
GeneReviews, Lin, A.E., Haldeman-Englert, C.R., and Milunsky, J.M. (1993). Branchiooculofacial Syndrome, https://www.ncbi.nlm.nih.gov/books/NBK55063/
IDT PrimerQuest, http://www.idtdna.com/primerquest
Kids First Data Resource Center, kidsfirstdrc.org
Online Mendelian Inheritance in Man, https://www.omim.org/
ToppFun- Functional Enrichment, https://toppgene.cchmc.org/enrichment.jsp
Supplemental Information
All Coding De Novo Mutations
Summary of Clinically Relevant OFC Genes
References
- 1.Watkins S.E., Meyer R.E., Strauss R.P., Aylsworth A.S. Classification, epidemiology, and genetics of orofacial clefts. Clin. Plast. Surg. 2014;41:149–163. doi: 10.1016/j.cps.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Leslie E.J., Marazita M.L. Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C. Semin. Med. Genet. 2013;163C:246–258. doi: 10.1002/ajmg.c.31381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser F.C. Thoughts on the etiology of clefts of the palate and lip. Acta Genet. Stat. Med. 1955;5:358–369. doi: 10.1159/000150783. [DOI] [PubMed] [Google Scholar]
- 4.Fogh-Andersen P. NytNordisk Forlag; Copenhagen: 1942. Inheritance of Harelip and Cleft Palate. [Google Scholar]
- 5.Marazita M.L., Mooney M.P. Current concepts in the embryology and genetics of cleft lip and cleft palate. Clin. Plast. Surg. 2004;31:125–140. doi: 10.1016/S0094-1298(03)00138-X. [DOI] [PubMed] [Google Scholar]
- 6.Jones M.C. Etiology of facial clefts: prospective evaluation of 428 patients. Cleft Palate J. 1988;25:16–20. [PubMed] [Google Scholar]
- 7.Beaty T.H., Marazita M.L., Leslie E.J. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 2016;5:2800. doi: 10.12688/f1000research.9503.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson J.C., Anand D., Butali A., Buxo C.J., Christensen K., Deleyiannis F., Hecht J.T., Moreno L.M., Orioli I.M., Padilla C. A systematic genetic analysis and visualization of phenotypic heterogeneity among orofacial cleft GWAS signals. Genet. Epidemiol. 2019;43:704–716. doi: 10.1002/gepi.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C., Meng L., Wang W., Song Y., Cheng Y. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig J.M., Zhang D., Xing M., Kim H.S. Meta-analysis: adjusted indirect comparison of drug-eluting bead transarterial chemoembolization versus 90Y-radioembolization for hepatocellular carcinoma. Eur. Radiol. 2017;27:2031–2041. doi: 10.1007/s00330-016-4548-3. [DOI] [PubMed] [Google Scholar]
- 11.Leslie E.J., Carlson J.C., Shaffer J.R., Butali A., Buxó C.J., Castilla E.E., Christensen K., Deleyiannis F.W., Leigh Field L., Hecht J.T. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 2017;136:275–286. doi: 10.1007/s00439-016-1754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bureau A., Parker M.M., Ruczinski I., Taub M.A., Marazita M.L., Murray J.C., Mangold E., Noethen M.M., Ludwig K.U., Hetmanski J.B. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics. 2014;197:1039–1044. doi: 10.1534/genetics.114.165225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu J., Beaty T.H., Scott A.F., Hetmanski J., Parker M.M., Wilson J.E., Marazita M.L., Mangold E., Albacha-Hejazi H., Murray J.C. Whole exome association of rare deletions in multiplex oral cleft families. Genet. Epidemiol. 2017;41:61–69. doi: 10.1002/gepi.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bureau A., Younkin S.G., Parker M.M., Bailey-Wilson J.E., Marazita M.L., Murray J.C., Mangold E., Albacha-Hejazi H., Beaty T.H., Ruczinski I. Inferring rare disease risk variants based on exact probabilities of sharing by multiple affected relatives. Bioinformatics. 2014;30:2189–2196. doi: 10.1093/bioinformatics/btu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox L.L., Cox T.C., Moreno Uribe L.M., Zhu Y., Richter C.T., Nidey N., Standley J.M., Deng M., Blue E., Chong J.X. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 2018;102:1143–1157. doi: 10.1016/j.ajhg.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi M., Hosomichi K., Yamaguchi T., Nagahama R., Yoshida H., Maki K., Marazita M.L., Weinberg S.M., Tajima A. Whole-genome sequencing in a pair of monozygotic twins with discordant cleft lip and palate subtypes. Oral Dis. 2018;24:1303–1309. doi: 10.1111/odi.12910. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y., Patterson K.E., Reinier F., Keesecker S.E., Blue E., Bamshad M., Haddad J., Jr. Copy Number Changes Identified Using Whole Exome Sequencing in Nonsyndromic Cleft Lip and Palate in a Honduran Population. Birth Defects Res. 2017;109:1257–1267. doi: 10.1002/bdr2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoebel A.K., Drichel D., van de Vorst M., Böhmer A.C., Sivalingam S., Ishorst N., Klamt J., Gölz L., Alblas M., Maaser A. Candidate Genes for Nonsyndromic Cleft Palate Detected by Exome Sequencing. J. Dent. Res. 2017;96:1314–1321. doi: 10.1177/0022034517722761. [DOI] [PubMed] [Google Scholar]
- 19.Aylward A., Cai Y., Lee A., Blue E., Rabinowitz D., Haddad J., Jr., University of Washington Center for Mendelian Genomics Using Whole Exome Sequencing to Identify Candidate Genes With Rare Variants In Nonsyndromic Cleft Lip and Palate. Genet. Epidemiol. 2016;40:432–441. doi: 10.1002/gepi.21972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ware J.S., Samocha K.E., Homsy J., Daly M.J. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr. Protoc. Hum. Genet. 2015;87:7.25.1–7.25.15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad D.F., Keebler J.E., DePristo M.A., Lindsay S.J., Zhang Y., Casals F., Idaghdour Y., Hartl C.L., Torroja C., Garimella K.V., 1000 Genomes Project Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besenbacher S., Liu S., Izarzugaza J.M., Grove J., Belling K., Bork-Jensen J., Huang S., Als T.D., Li S., Yadav R. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat. Commun. 2015;6:5969. doi: 10.1038/ncomms6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldmann J.M., Veltman J.A., Gilissen C. De Novo Mutations Reflect Development and Aging of the Human Germline. Trends Genet. 2019;35:828–839. doi: 10.1016/j.tig.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Jin S.C., Homsy J., Zaidi S., Lu Q., Morton S., DePalma S.R., Zeng X., Qi H., Chang W., Sierant M.C. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., Romano-Adesman A., Bjornson R.D., Breitbart R.E., Brown K.K. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner T.N., Coe B.P., Dickel D.E., Hoekzema K., Nelson B.J., Zody M.C., Kronenberg Z.N., Hormozdiari F., Raja A., Pennacchio L.A. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell. 2017;171:710–722e12. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leslie E.J., Taub M.A., Liu H., Steinberg K.M., Koboldt D.C., Zhang Q., Carlson J.C., Hetmanski J.B., Wang H., Larson D.E. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am. J. Hum. Genet. 2015;96:397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riley B.M., Mansilla M.A., Ma J., Daack-Hirsch S., Maher B.S., Raffensperger L.M., Russo E.T., Vieira A.R., Dodé C., Mohammadi M. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu J.M., Leslie E.J., Scott A.F., Murray J.C., Marazita M.L., Beaty T.H., Scharpf R.B., Ruczinski I. Detection of de novo copy number deletions from targeted sequencing of trios. Bioinformatics. 2019;35:571–578. doi: 10.1093/bioinformatics/bty677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Younkin S.G., Scharpf R.B., Schwender H., Parker M.M., Scott A.F., Marazita M.L., Beaty T.H., Ruczinski I. A genome-wide study of de novo deletions identifies a candidate locus for non-syndromic isolated cleft lip/palate risk. BMC Genet. 2014;15:24. doi: 10.1186/1471-2156-15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leoyklang P., Siriwan P., Shotelersuk V. A mutation of the p63 gene in non-syndromic cleft lip. J. Med. Genet. 2006;43:e28. doi: 10.1136/jmg.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peyrard-Janvid M., Leslie E.J., Kousa Y.A., Smith T.L., Dunnwald M., Magnusson M., Lentz B.A., Unneberg P., Fransson I., Koillinen H.K. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet. 2014;94:23–32. doi: 10.1016/j.ajhg.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhyay N., Bishop M., Mortillo M., Chopra P., Hetmanski J.B., Taub M.A., Moreno L.M., Valencia-Ramirez L.C., Restrepo C., Wehby G.L. Whole genome sequencing of orofacial cleft trios from the Gabriella Miller Kids First Pediatric Research Consortium identifies a new locus on chromosome 21. Hum. Genet. 2020;139:215–226. doi: 10.1007/s00439-019-02099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayat M.J., Higgins M. Understanding poisson regression. J. Nurs. Educ. 2014;53:207–215. doi: 10.3928/01484834-20140325-04. [DOI] [PubMed] [Google Scholar]
- 41.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584e523. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falconer D.S. The inheritance of liability to diseases with variable age of onset, with particular reference to diabetes mellitus. Ann. Hum. Genet. 1967;31:1–20. doi: 10.1111/j.1469-1809.1967.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Xu H., Aronow B.J., Jegga A.G. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 45.Li H., Jones K.L., Hooper J.E., Williams T. The molecular anatomy of mammalian upper lip and primary palate fusion at single cell resolution. Development. 2019;146:dev174888. doi: 10.1242/dev.174888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prescott S.L., Srinivasan R., Marchetto M.C., Grishina I., Narvaiza I., Selleri L., Gage F.H., Swigut T., Wysocka J. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. 2015;163:68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214-20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heyne G.W., Plisch E.H., Melberg C.G., Sandgren E.P., Peter J.A., Lipinski R.J. A Simple and Reliable Method for Early Pregnancy Detection in Inbred Mice. J. Am. Assoc. Lab. Anim. Sci. 2015;54:368–371. [PMC free article] [PubMed] [Google Scholar]
- 50.Heyne G.W., Everson J.L., Ansen-Wilson L.J., Melberg C.G., Fink D.M., Parins K.F., Doroodchi P., Ulschmid C.M., Lipinski R.J. Gli2 gene-environment interactions contribute to the etiological complexity of holoprosencephaly: evidence from a mouse model. Dis. Model. Mech. 2016;9:1307–1315. doi: 10.1242/dmm.026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furlan A., Adameyko I. Schwann cell precursor: a neural crest cell in disguise? Dev. Biol. 2018;444(Suppl 1):S25–S35. doi: 10.1016/j.ydbio.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 52.Kastriti M.E., Adameyko I. Specification, plasticity and evolutionary origin of peripheral glial cells. Curr. Opin. Neurobiol. 2017;47:196–202. doi: 10.1016/j.conb.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra G.R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T.M. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bader G.D., Betel D., Hogue C.W. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hopper J.J.K., Williams T. FaceBase; 2016. Ectoderm/Mesenchyme mRNA expression by microarray. [Google Scholar]
- 58.Panaliappan T.K., Wittmann W., Jidigam V.K., Mercurio S., Bertolini J.A., Sghari S., Bose R., Patthey C., Nicolis S.K., Gunhaga L. Sox2 is required for olfactory pit formation and olfactory neurogenesis through BMP restriction and Hes5 upregulation. Development. 2018;145:dev153791. doi: 10.1242/dev.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palomares M., Delicado A., Mansilla E., de Torres M.L., Vallespín E., Fernandez L., Martinez-Glez V., García-Miñaur S., Nevado J., Simarro F.S. Characterization of a 8q21.11 microdeletion syndrome associated with intellectual disability and a recognizable phenotype. Am. J. Hum. Genet. 2011;89:295–301. doi: 10.1016/j.ajhg.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eising E., Carrion-Castillo A., Vino A., Strand E.A., Jakielski K.J., Scerri T.S., Hildebrand M.S., Webster R., Ma A., Mazoyer B. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol. Psychiatry. 2019;24:1065–1078. doi: 10.1038/s41380-018-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Félix T.M., Hanshaw B.C., Mueller R., Bitoun P., Murray J.C. CHD7 gene and non-syndromic cleft lip and palate. Am. J. Med. Genet. A. 2006;140:2110–2114. doi: 10.1002/ajmg.a.31308. [DOI] [PubMed] [Google Scholar]
- 62.Mandalos N., Rhinn M., Granchi Z., Karampelas I., Mitsiadis T., Economides A.N., Dollé P., Remboutsika E. Sox2 acts as a rheostat of epithelial to mesenchymal transition during neural crest development. Front. Physiol. 2014;5:345. doi: 10.3389/fphys.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao F., Ma X., Qiu W., Wang P., Zhang R., Chen Z., Su P., Zhang Y., Li D., Ma J. MACF1 Facilitates SMAD7 Nuclear Translocation to Drive Bone Formation in Mice. bioRxiv. 2019 doi: 10.1101/743930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luscan A., Laurendeau I., Malan V., Francannet C., Odent S., Giuliano F., Lacombe D., Touraine R., Vidaud M., Pasmant E., Cormier-Daire V. Mutations in SETD2 cause a novel overgrowth condition. J. Med. Genet. 2014;51:512–517. doi: 10.1136/jmedgenet-2014-102402. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J., Hagopian-Donaldson S., Serbedzija G., Elsemore J., Plehn-Dujowich D., McMahon A.P., Flavell R.A., Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- 66.Kousa Y.A., Zhu H., Fakhouri W.D., Lei Y., Kinoshita A., Roushangar R.R., Patel N.K., Agopian A.J., Yang W., Leslie E.J. The TFAP2A-IRF6-GRHL3 genetic pathway is conserved in neurulation. Hum. Mol. Genet. 2019;28:1726–1737. doi: 10.1093/hmg/ddz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verloes A. Branchio-oculo-facial syndrome. 2009. https://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=en&Expert=1297
- 68.Leslie E.J., Koboldt D.C., Kang C.J., Ma L., Hecht J.T., Wehby G.L., Christensen K., Czeizel A.E., Deleyiannis F.W., Fulton R.S. IRF6 mutation screening in non-syndromic orofacial clefting: analysis of 1521 families. Clin. Genet. 2016;90:28–34. doi: 10.1111/cge.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alharatani R., Ververi A., Beleza-Meireles A., Ji W., Mis E., Patterson Q.T., Griffin J.N., Bhujel N., Chang C.A., Dixit A. Novel truncating mutations in <em>CTNND1</em> cause a dominant craniofacial and cardiac syndrome. bioRxiv. 2019 doi: 10.1101/711184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghoumid J., Stichelbout M., Jourdain A.-S., Frenois F., Lejeune-Dumoulin S., Alex-Cordier M.-P., Lebrun M., Guerreschi P., Duquennoy-Martinot V., Vinchon M. Blepharocheilodontic syndrome is a CDH1 pathway-related disorder due to mutations in CDH1 and CTNND1. Genet. Med. 2017;19:1013–1021. doi: 10.1038/gim.2017.11. [DOI] [PubMed] [Google Scholar]
- 71.Alharatani R., Ververi A., Beleza-Meireles A., Ji W., Mis E., Patterson Q.T., Griffin J.N., Bhujel N., Chang C.A., Dixit A. Novel truncating mutations in CTNND1 cause a dominant craniofacial and cardiac syndrome. Hum. Mol. Genet. 2020:ddaa050. doi: 10.1093/hmg/ddaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Timberlake A.T., Furey C.G., Choi J., Nelson-Williams C., Loring E., Galm A., Kahle K.T., Steinbacher D.M., Larysz D., Persing J.A., Lifton R.P., Yale Center for Genome Analysis De novo mutations in inhibitors of Wnt, BMP, and Ras/ERK signaling pathways in non-syndromic midline craniosynostosis. Proc. Natl. Acad. Sci. USA. 2017;114:E7341–E7347. doi: 10.1073/pnas.1709255114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qi H., Yu L., Zhou X., Wynn J., Zhao H., Guo Y., Zhu N., Kitaygorodsky A., Hernan R., Aspelund G. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14:e1007822. doi: 10.1371/journal.pgen.1007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D’Gama A.M., Walsh C.A. Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci. 2018;21:1504–1514. doi: 10.1038/s41593-018-0257-3. [DOI] [PubMed] [Google Scholar]
- 75.Yuen R.K., Merico D., Cao H., Pellecchia G., Alipanahi B., Thiruvahindrapuram B., Tong X., Sun Y., Cao D., Zhang T. Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med. 2016;1:160271–1602710. doi: 10.1038/npjgenmed.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou J., Park C.Y., Theesfeld C.L., Wong A.K., Yuan Y., Scheckel C., Fak J.J., Funk J., Yao K., Tajima Y. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat. Genet. 2019;51:973–980. doi: 10.1038/s41588-019-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gelb B.D., Chung W.K. Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb. Perspect. Med. 2014;4:a013953. doi: 10.1101/cshperspect.a013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All Coding De Novo Mutations
Summary of Clinically Relevant OFC Genes
Data Availability Statement
The data analyzed and reported in this manuscript were accessed from the database of Genotypes and Phenotypes (dbGaP; European trios, dbGaP: phs001168.v2.p2; Colombian trios, dbGaP: phs001420.v1.p1; Taiwanese trios, dbGaP: phs000094.v1.p1) and from the Kids First Data Resource Center. Some datasets used in this study are available publicly from the Gene Expression Omnibus (GEO) via the following accession numbers: GEO: GSM1817212, GSM1817213, GSM1817214, GSM1817215, GSM1817216, and GSM1817217.