Abstract
Giardia lamblia is a causative agent of persistent diarrhoea widespread in regions with low hygienic standards. Laboratory research is based on cloned lines issuing from various patient isolates typed in the late 1980s and 90s using restriction analysis and serology. In the present study, we compared the well-characterized strain WBC6 with another clone of the parent WB isolate termed WBA1 and with a clone from another isolate, GS/M-83-H7, using shotgun mass spectrometry proteomics. We identified 398 proteins differentially expressed between the GS and both WB isolates and 97 proteins differentially expressed between the two WB isolates. We investigated the expression levels of the predominant variant-specific surface proteins (VSPs) in each clone and matched the previously described major VSPs of each strain to the corresponding open reading frame sequences identified by whole-genome sequencing efforts. Furthermore, since the original WB isolate comes from a patient treated with metronidazole, we compared the susceptibilities of the strains to nitro compounds, as well the expression levels of enzymes involved in nitro reduction and on the corresponding enzyme activities and found distinct differences between the three strains.
Key words: Diversity, drug susceptibility, isolates, parasite–host interaction
Introduction
The diplomonadid Giardia lamblia (syn. G. duodenalis, G. intestinalis), an early diverging, anaerobic eukaryote (Adam, 2001; Cernikova et al., 2018), is a causative agent of persistent diarrhoea widespread in regions with low hygienic standards (Hemphill et al., 2019). After stomach passage, cysts ingested via contaminated food or water or via direct personal contact transform into trophozoites which colonize the duodenum and cause the symptoms peaking around 1 week post infection. In general, hosts in good physical condition recover within 2–3 weeks. In rare cases, the infection persists and becomes chronical causing severe damage of the intestinal epithelium (Allain et al., 2017) occasionally resulting in the development of irritable bowel syndrome (Litleskare et al., 2018). Eight different genotypes or assemblages, labelled A to H, have been identified, assemblages A and B having the broadest spectrum identifying animals as well as humans (Heyworth, 2016). Furthermore, assemblage E isolates may cause human giardiasis, as well (Zahedi et al., 2017). Thus, giardiasis occurs in humans as well as in other mammals and can be regarded as a zoonosis (Thompson, 2004). Human giardiasis is treated with the nitroimidazole metronidazole as first-line and the benzimidazole albendazole as second-line drug. Quinacrine, one of the first antigiardial drugs, is not in use, anymore (Gardner and Hill, 2001; Nash, 2001).
The largest part of laboratory research is based on the strain WBC6, an assemblage A1 strain obtained by limited dilution cloning from the WB isolate obtained from duodenal contents from a patient suffering from chronical giardiasis refractory to either quinacrine or metronidazole (MET) treatments (Smith et al., 1982b). The isolate originates from Afghanistan (Smith et al., 1982a) and was axenized at the NIH (Nash et al., 1985). The WBC6 strain, cloned by F. Gillin via limited dilution in 1983 (Campbell and Faubert, 1994), eagerly produces cysts in vitro (Campbell and Faubert, 1994), is amenable to genetic manipulation (Furfine and Wang, 1990; Sun et al., 1998), and its growth in vitro is less affected by the culture medium composition than other strains (own observations, see ‘Materials and methods’ section). Consequently, WBC6 is extensively used to investigate intracellular processes associated with en- and excystation of the parasite (Faso and Hehl, 2011; Zamponi et al., 2017) and has been the first Giardia strain where the complete genome has been sequenced (Morrison et al., 2007). Unfortunately, in many articles before this hallmark, the strain is referred to as WB only. This is insofar confusing, as other clones from the same original isolate with distinct characteristics exist. The most striking of these characteristics is related to a unique feature of G. lamblia, namely antigenic variation (Nash, 2002; Adam et al., 2010). The genome of WBC6 and of other strains that have been sequenced so far (see www.giardiadb.org) contain several hundred open reading frames (ORFs) encoding the so-called variant-specific surface proteins (VSPs). According to a recently published chromosome-scale reference genome, the genes encoding for VSPs, as well as other highly repetitive genes, are distributed over all five chromosomes (Xu et al., 2020). VSPs have the following properties in common: a signal peptide targeting the protein to the surface, a highly variable N-terminus with frequent CXXC-motifs (where X may be any amino acid), a nearly invariant C-terminus of 38 amino acids containing a CRGKA motif, a hydrophobic domain forming the membrane anchor (Adam et al., 2010). The N-terminus of VSPs is highly immunogenic. VSPs therefore constitute the main surface antigens of G. lamblia trophozoites. According to a generally admitted hypothesis, one single trophozoite expresses only one VSP at the same time (Nash et al., 1990b, 2001). During encystation, however, several VSPs may be expressed by the same cell (Carranza et al., 2002). The switching from one VSP to another, thus antigenic variation, at least indirectly depends in all likelihood, on posttranscriptional mechanisms (Kulakova et al., 2006; Prucca et al., 2008, 2011; Prucca and Lujan, 2009).
Studies in the 80s and 90s based on monoclonal antibodies raised against specific epitopes revealed the presence of major VSPs in a given WB trophozoite population. A well-characterized member of these major surface antigens is VSPA6 (in the literature also referred to as CRP170 due to approximate size of 170 kDa) that is expressed by several subclones of the WB isolate (Adam et al., 1988). VSPA6 is recognized by the cytotoxic monoclonal antibody (mAb) 6E7. Trophozoites resistant to mAb 6E7 express a different pattern of VSPs (Nash et al., 1988). Even in the absence of this selective pressure, other VSPs recognized by different monoclonal antibodies appear after many generations (Nash et al., 1990a).
According to further studies, mAb 6E7 recognizes an octapeptide motif (GAAPLYKK) present in 65 amino acid repeats distributed in different numbers over the VSP primary sequence (Mowatt et al., 1994) suggesting an evolution by gene duplication (Yang and Adam, 1995).
Conversely, trophozoites of WBC6 express a predominant VSP, named TSA417, exhibiting a peptide molecular mass of 72.5 kDa as predicted by the corresponding gene sequence (Gillin et al., 1990). Complement-dependent killing of trophozoites during exposure to anti-recombinant TSA417 antiserum provides a selective advantage for minor populations of non-TSA417-expressing trophozoites already present in the original culture (Meng et al., 1993). Furthermore, WBC6 trophozoites bearing non-TSA417-type VSPs appear during the process of excystation (Meng et al., 1993; Svärd et al., 1998).
Another extensively studied G. lamblia strain, namely clone GS/M-83-H7, representing an assemblage B genotype, originates from the human isolate GS obtained in Alaska and axenized by isolation of trophozoites from infected neonatal mice (Nash et al., 1985). This clone expresses a major surface antigen, VSP H7, which is immuno-reactive to a specific, cytotoxic mAb (mAb G10/4). VSP H7 has an apparent molecular mass of ca. 57 kDa (Nash and Mowatt, 1992). For GS/M-83-H7, in vitro antigenic variation replacing VSP H7 by diverse other VSPs on the surface of trophozoites occurs at about one variation event per 6.5 generations in comparison to one variation event per 12–13 generations in the original WB isolate (Nash et al., 1990a). Since GS/M-83-H7 infects humans as well as mice, immune responses to VSP H7 have been investigated not only in vitro (Müller et al., 1996) but also in mouse models (Gottstein et al., 1990; Müller and Gottstein, 1998). These studies show that antigenic variation occurs in vivo as a reaction to humoral immune responses in both hosts (Nash et al., 1990c; Bienz et al., 2001; Müller and von Allmen, 2005).
Recent shotgun mass spectrometry proteome studies have revealed that WBC6 trophozoite populations and trophozoite populations from other assemblage A strains express an impressive number of VSPs at the same time (Emery et al., 2014, 2015). Other studies have revealed that strain-dependent antigenic diversification of trophozoites occurs upon selective drug pressure (Emery et al., 2018; Emery-Corbin et al., 2018; Müller et al., 2019).
Using this sensitive method, we compared the well-characterized strain WBC6 with another clone of the parent WB isolate in the following termed WBA1. This clone grows only in a culture medium with strictly defined components and is refractory to antigenic variation, as previously shown (Nash et al., 1990a), and according to our own observations. Moreover, this strain is refractory to encystation (Campbell and Faubert, 1994). We included the strain GS/M-83-H7 as an outgroup into our comparison expecting that more proteins are differentially expressed between this clone and either WB clone than between both WB clones. We were particularly interested in the expression levels of the predominant VSPs in each clone and matched the previously described major VSPs of each strain to the corresponding ORF sequences identified by whole-genome sequencing efforts. Furthermore, since the original WB isolate comes from a patient treated with the nitroimidazole metronidazole, and since antigenic variation has been shown to be influenced by drug exposure (Müller et al., 2019), we compared the susceptibilities of the strains to nitro compounds and had a closer look on enzymes involved in nitro reduction and on the corresponding enzyme activities.
Materials and methods
Chemicals
If not otherwise stated, all biochemical reagents were from Sigma (St Louis, MO, USA). Nitazoxanide (NTZ) was synthesized at the Department of Chemistry and Biochemistry, University of Bern, Switzerland (Ch. Leumann). NTZ, albendazole (ALB) and MET were kept as 100 mm stock solutions in DMSO at −20°C.
Axenic culture, harvest and storage of G. lamblia trophozoites
Trophozoites from G. lamblia WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) were grown under anaerobic conditions in 10 mL culture tubes (Nunc, Roskilde, Denmark) on modified TYI-S-33 medium as previously described (Clark and Diamond, 2002). In order to ensure the growth of the A1 and H7 clones, the components were as close as possible to the isolation medium, in particular heat-inactivated adult bovine serum (Biofluids, Rockville, MD, USA) and casein peptone (Becton Dickinson, Cockeysville, MD, USA). Prior to shotgun mass spectrometry analysis, the cultures were routinely passaged two times. Subcultures were performed by inoculating 100 μL of cells from a confluent culture detached by cooling (see below) to a new tube containing 10 mL culture medium (Müller et al., 2006). Trophozoites were harvested by incubation on ice for 15 min followed by centrifugation (300 × g, 10 min, 4°C). Pellets were washed three times with ice-cold PBS, counted and stored at −80°C for subsequent proteomic analysis or for enzymatic assays.
Proteomics
Cell pellets were lysed in 100 μL 8 m urea/100 mm Tris/HCl pH 8/cOmpleteTM protease inhibitor cocktail (Roche Diagnostics, Rotkreuz, Switzerland) by incubation for 15 min at room temperature followed by 15 min in an ultrasonic water bath. Proteins were reduced and alkylated with 10 mm DTT for 30 min at 37°C and 50 mm iodoacetamide for 30 min at 37°C. Proteins were precipitated at −20°C by addition of 5 volumes cold acetone and incubation at −20°C overnight. All liquid was carefully removed and the pellet dried in ambient air for 15 min before reconstitution of proteins in 200 μL of 8 m urea, 50 mm Tris-HCl pH 8.0. Protein concentration was determined by Bradford assay and an aliquot corresponding to 10 μg protein was digested by trypsin (1:50 trypsin/protein ratio) for 6 h at 37°C after dilution of urea concentration to 1.6 m with 20 mm Tris-HCl pH 8.0 and 2 mm CaCl2. The digests were acidified with TFA (1%) and analysed by LC-MS/MS. Three repetitive injections of an aliquot corresponding to 500 ng protein digest was separated on an EASY-nLC 1000 coupled to a QExactive mass spectrometer (ThermoFisher Scientific). Peptides were trapped on an Acclaim PepMap100 C18 pre-column (3 μm, 100 A˚, 75 μm × 2 cm, ThermoFisher Scientific, Reinach, Switzerland) and separated by backflush on a C18 column (3 μm, 100 A˚, 75 μm × 15 cm, Nikkyo Technos, Tokyo, Japan) by applying a 60 min gradient of 5% acetonitrile to 40% in water, 0.1% formic acid, at a flow rate of 400 nL min−1. Peptides of m/z 360–1400 were detected with a resolution of 70 000 applying an automatic gain control (AGC) target of 1E06 and a maximum ion injection time of 50 ms. A top 10 data-dependent method for precursor ion fragmentation with a stepped 27% normalized collision energy was applied with the following settings: precursor isolation width of 2 m/z, resolution 17 500, AGC of 1E05 with a minimum target of 1E03, maximum ion time of 110 ms, charge exclusion of unassigned and 1+ ions, peptide match on and dynamic exclusion for 20 s, respectively.
The MS data were obtained from three biological replicates, with three technical replicates for each biological replicate, for each strain. All MS data were processed by MaxQuant (version 1.5.4.1) with matching between runs for the same strain activated, but not between different strains, in order to avoid over-interpretation of the data. The sample sets were interpreted separately by MaxQuant. Fragment spectra were interpreted against a recent Giardia protein sequence database including both WB and GS datasets in fasta format (GiardiaDB-5.0_GintestinalisAssemblageA_AnnotatedProteins; GiardiaDB-5.0_GintestinalisAssemblageB_AnnotatedProteins_v2). The trypsin cleavage rule allowed amide bond cleavage after lysine and arginine but not if a proline follows and up to three missed cleavage sites, fixed carbamidomethylation modification of cysteines, variable oxidation of methionine and acetylation of protein N-termini. Precursor and fragment mass tolerances were set to 10 and 20 ppm, respectively. Peptide spectrum matches, peptide and protein group identifications were filtered to a 1% false discovery rate (FDR) based on reversed database sequence matches, and a minimum of two razor or unique peptides were required to accept a protein group identification. Protein identifications considered as contaminations (e.g. trypsin or BSA) as well as proteins identified only by site (considered by MaxQuant developers as very likely false positives) were removed for statistical validation. The normalized label-free quantification (LFQ) protein group intensities as calculated by MaxQuant were used for relative proteome quantifications. First, we imputed missing protein LFQ values for samples in any condition group when there were at least two LFQ intensities in one group (downshift of 2.5 s.d. with a width of 0.3 s.d.). When comparing VSPs only, peptides unique to a single VSP protein were used for the calculation of protein intensities based on the sum of the three most intense peptides (Top3 approach). Before summing, missing peptide intensities were imputed sample group wise, when there were at least two valid intensities (downshift of 1.8 s.d. with a width of 0.3 s.d.). The resulting protein intensities were named iTop3. This process left proteins without values in one or the other group. For Welch's t-tests, those missing protein intensities were replaced by imputed values from the very low end of intensity distributions in analogy to the LFQ imputation (downshift 2.5 s.d., width of 0.3 s.d.). An FDR-controlled Benjamini–Hochberg procedure was used for the correction of P values. A log2-fold change of at least one and a corrected P value of 0.05 were required to be considered as significant. Statistical testing and imputation were made with a home-made R script run under R-Studio. Our main goal was to identify the VSPs in the H7 proteome. To do this, we had to use the assemblage B database. Since most of the other proteins share high homologies with those from assemblage A strains and are found in both databases, they are called orthologues. We eliminated those proteins that were expressed in both strains sharing the same ORF numbers (e.g. GL50803_10167 in assemblage A and GSB_10167 in assemblage B) via the GenesbyOrthologues tool on the GiardiaDB site (www.giardiabd.org). Small differences in peptide sequences or SNPs were not considered. They can easily be identified by comparing the sequences of orthologues provided by GiardiaDB. For comparative quantification of orthologues, the LFQ values corresponding to GL50803 were used for assemblage A strains and the values corresponding to GSB were used for assemblage B strains.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD017597.
Drug susceptibility assays
Drug susceptibility of G. lamblia trophozoites of the strains C6, A1 and H7 was tested in 96-well plates inoculated with 103 trophozoites per well and MET, NTZ or ALB at various concentrations (dilution factor 2) and incubated in an anaerobic growth chamber (85% N2, 10% H2, 5% CO2). After 72 h (C6, A1) or 96 h (H7), growth of cells was monitored by a vitality assay based on the reduction of resazurin (Alamar Blue) to a pink product that was assayed fluorimetrically (Bénéré et al., 2007). The IC50 values were determined using the logit-log algorithm as described (Müller and Hemphill, 2013).
To determine minimal inhibitory concentrations (MIC), WT and C4 trophozoites were inoculated in the presence of increasing amounts (dilution series by a factor 2) of the nitro compounds MET, NTZ or ALB as a control. The tubes were incubated at 37°C for 96 h (C6, A1) or 120 h (H7). The MIC was determined by observing the wells under the microscope starting from higher to lower concentrations. The concentration at which the first living trophozoites were visible is given as the MIC (Müller et al., 2018).
Enzyme assays
Trophozoite crude extracts were prepared by suspending trophozoite pellets in 50 mm Tris-Cl−, pH 7.0, containing 0.05% triton-X-100 and a protease inhibitor mix (Halt, ThermoFisher Scientific, Waltham, MA) according to the manufacturer's instructions. NAD(P)H oxidase and quinone reductase activities were measured in 96-well microtiter plates containing 100 μL of a reaction mix containing buffer (50 mm Tris-Cl−, pH 7.0), 0.5 mm thiazolyl blue tetrazolium (MTT), 0.5 mm NADH or NADPH and 5 μL of crude extract. To measure quinone reductase activity, 0.1 mm menadione was added to the mix. The plates were incubated at 37°C under aerobic conditions. Substrate and enzyme blanks were included. After different time points, the reaction was stopped by adding 100 μL of pure ethanol thus solubilizing the product formed by the reduction of MTT, formazan (Prochaska and Santamaria, 1988; Müller et al., 2015). Nitroreductase activity was quantified via the reduction of 7-nitrocoumarin (7-NC) to 7-aminocoumarin (7-AC) using the reaction mix as described above, without MTT and 0.1 mm 7-NC, with the same volumes and under the same conditions of incubation. Enzyme and substrate blanks were included. The reaction was stopped by adding 100 μL of 50 mm HCl lowering the pH to allow full protonation of the reaction product (Wagner, 2009). 7-AC was quantified by fluorimetry with excitation at 365 nm and emission at 455 nm (Müller et al., 2015). Absorption at 590 nm (MTT-based assays) and fluorescence intensities were quantified using a 96-well multimode plate reader (Enspire; Perkin-Elmer, Waltham, MA, USA). Protein contents of crude extracts were determined by the Bradford method (Bradford, 1976) using a commercial kit (Biorad, Hercules, CA, USA).
Results and discussion
Mass spectrometry analysis of proteins expressed in G. lamblia trophozoites
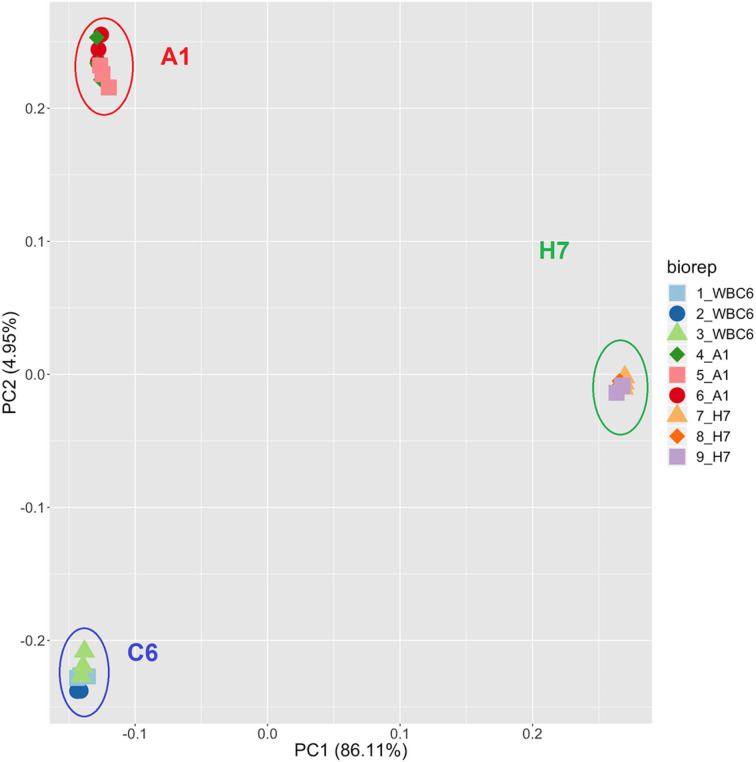
Shotgun mass spectrometry of the proteomes of trophozoites of the strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) allowed the identification of 24 739 unique peptides matching to 2 368 proteins (Table S1). Overall analysis of the data by principal component analysis (PCA) revealed that biological and technical replicates clustered together for each strain. The proteomes of the three strains formed non-overlapping clusters. A1 and C6 were separated by PC2 only, whereas the proteome of H7 was separated by both other strains PC1 and PC2 (Fig. 1). A PCA with strains A1 and C6 only is shown in Fig. S1.
Fig. 1.
Principal component analysis of proteome dataset from G. lamblia trophozoites of different strains. Trophozoites of the strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) were compared by MS shotgun analysis as described in ‘Materials and methods’ section. For each strain, all technical and biological (square, circle, diamond) replicates are shown.
Differentially expressed proteins
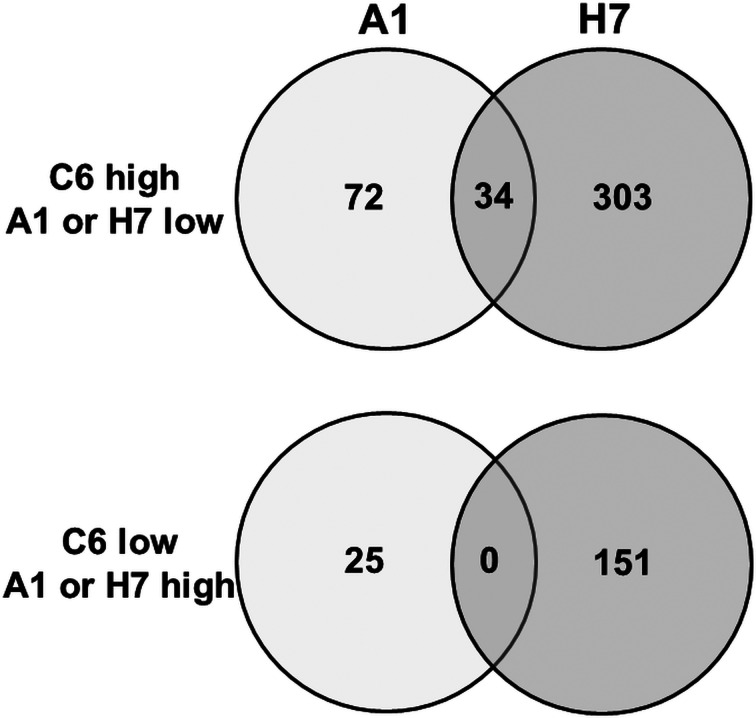
To get a better estimation of ‘true’ differentials between assemblage and B strains, orthologues present in both A and B strains have been subtracted using the GenesbyOrthologues function implemented in the GiardiaDB. Without eliminating these orthologues from the comparison, WBC6 vs H7 would yield 1320 differentials, WBA1 vs H7 would yield 1264 differentials. This analysis revealed that between strains C6 and A1, 97 proteins were differentially expressed, as compared to 454 proteins between strains C6 and H7 (Fig. 2). Comparison of differentially expressed proteins between the strains H7 vs the strains C6 and A1 revealed that 253 proteins had higher levels in both WB strains than in H7, and 146 proteins had higher levels in H7 than in the WB strains. Besides hypothetical proteins, the categories with the highest numbers of differentials comprised proteins involved in cytoskeleton, adhesion and organelle transport, intermediary metabolism, gene expression and development, and signalling (Table 1). Strain H7 expressed a unique set of 13 VSPs different from the VSP sets expressed in both WB strains. H7 and the WB clones belong to different assemblages, it is not surprising that an order of magnitude more differential proteins are identified between H7 and the WB clones than between the two WB clones. Some of the proteins overexpressed in H7, in particular proteins involved in signalling or attachment, may contribute to the broad host spectrum of this strain, which infects humans as well as animals, as reviewed elsewhere (Müller and von Allmen, 2005).
Fig. 2.
Venn diagram detailing the number of differentially expressed proteins in G. lamblia trophozoites of different strains. Trophozoites of the strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) were subjected to MS shotgun analysis as described in ‘Materials and methods’ section. Orthologues present in both A and B strains have been subtracted using the GenesbyOrthologues function implemented in the GiardiaDB.
Table 1.
Number of differentially expressed proteins in strain GS/M-83-H7 as compared to both WBC6 and WBA1 trophozoites
| (Hypothetical) function | Higher levels in WBC6 and WBA1 | Higher levels in GS/M-83-H7 |
|---|---|---|
| Variant-specific surface proteins | 4 | 13 |
| Cytoskeleton, adhesion and organelle transport | 34 | 15 |
| Intermediary metabolism | 54 | 22 |
| Signalling | 16 | 13 |
| Gene expression, cell cycle, development | 48 | 17 |
| Chaperones | 10 | 0 |
| Transport of metabolites | 9 | 5 |
| Hypothetical | 82 | 60 |
| Total | 253 | 145 |
The strains were grown and processed for MS shotgun analysis as described in ‘Materials and methods’ section. The proteins are grouped with respect to their hypothetical functions.
When comparing the set of the 97 differentially expressed proteins between C6 and A1 in more detail, it turned out that – besides 32 hypothetical proteins – 22 VSPs differed between these strains. Eleven differential proteins were involved in signalling, 11 cytoskeleton-related proteins, 10 involved in intermediary metabolism, 10 proteins with functions in gene expression and development, and one with a chaperone-like function (Table 2). Thirty-four proteins were also significantly lower in strain H7 as compared to C6 (Fig. 2), in particular the 15 C6-specific VSPs (see ‘Antigenic variation’ section) and 19 proteins with various functions, as highlighted in Table 2.
Table 2.
Overview of ORFs differentially expressed in WBC6 (C6) as compared to WBA1 (A1) trophozoites
| (Hypothetical) function | Protein | ORF number in Giardia database |
|---|---|---|
| Higher expression in C6 | ||
| Variant-specific surface proteins | VSP-88 | 101 074 |
| VSP-169 | 112 801 | |
| VSP-44 | 113 450 | |
| VSP-5 | 113 797 | |
| VSP-149 | 116 477 | |
| VSP-38 | 13 194 | |
| VSP-160 | 137 612 | |
| VSP-188 | 137 613 | |
| VSP-77 | 137 617 | |
| VSP-8 | 137 618 | |
| VSP-3 | 137 740 | |
| VSP-186 | 14 586 | |
| VSP-100 | 33 279 | |
| VSP-49 | 41 472 | |
| VSP-121 | 5812 | |
| Cytoskeleton, adhesion and organelle transport | Protein 21.1 | 101 168 |
| Ankyrin repeat protein | 14 745 | |
| IFT complex B | 17 223 | |
| Protein 21.1 (Ankyrin repeat) | 17 608 | |
| ENC6 protein | 24 372 | |
| Trichohyalin | 32 375 | |
| Protein 21.1 | 3760 | |
| Cyst wall protein 2 | 5435 | |
| Synaptic glycoprotein SC2 | 88 581 | |
| Dynein heavy chain | 94 440 | |
| Intermediary metabolism | Ubiquitin carboxyl-terminal hydrolase 14 | 102 710 |
| Peroxiredoxin 1 | 14 521 | |
| 2,5-diketo-D-gluconic acid reductase | 151 884 | |
| NAD(P)H:menadione oxidoreductase | 17 150 | |
| Nitroreductase Fd-NR2 | 22 677 | |
| L-serine dehydratase | 24 662 | |
| Glucosamine-6-phosphate deaminase | 8245 | |
| Adenylate kinase | 90 402 | |
| Signalling | Kinase, AGC PKA | 11 214 |
| Kinase, NEK | 13 215 | |
| Kinase, NEK | 137 733 | |
| Kinase, CAMK CAMKL | 14 661 | |
| ARF GAP | 17 561 | |
| AMP-activated protein kinase, gamma-1 subunit | 3414 | |
| Gene expression, cell cycle, development | Chromosome segregation protein SMC | 10 027 |
| Threonyl and alanyl tRNA synthetase second additional domain | 150 018 | |
| CDC19 | 17 413 | |
| Golgi/cell cycle associated protein, putative | 17 472 | |
| Preimplantation protein 3 | 3417 | |
| SBDS C-terminal domain protein, putative | 40 521 | |
| Pelo protein | 5892 | |
| Chaperones | T-complex protein 1, β subunit | 11 397 |
| Hypothetical | (25 proteins) | |
| (12 proteins also in H7) | ||
| Higher expression in A1 | ||
| Variant-specific surface proteins | High cysteine membrane protein VSP-like | 112 135 |
| High cysteine membrane protein Group 1 | 11 309 | |
| VSP-175 | 137 608 | |
| High cysteine membrane protein Group 1 | 16 318 | |
| CXC-rich protein | 17 476 | |
| VSP A6 (annotated as hypothetical protein) | 221 693 | |
| High cysteine membrane protein Group 6 | 41 942 | |
| Cytoskeleton, adhesion and organelle transport | Dynein heavy chain | 17 243 |
| Intermediary metabolism | Glycosaminoglycan polysaccharide lyase | 10 238 |
| Nitroreductase family protein | 15 307 | |
| Signalling | Kinase, NEK | 14 216 |
| Kinase, CAMK CAMKL | 17 566 | |
| Serine/threonine protein phosphatase 5 | 21 498 | |
| Protein kinase NEK family protein | 152 721 | |
| Gene expression, cell cycle, development | MCM2 | 15 344 |
| DNA replication licensing factor MCM6 | 17 390 | |
| RNA binding protein, putative | 92 031 | |
| Hypothetical | (8 proteins) | |
The strains were grown and processed for MS shotgun analysis as described in ‘Materials and methods’ section. For each strain, three biological replicates have been tested (with three technical replicates per biological replicate). The proteins in this list had significantly different expression levels both in iLFQ and iTop3 values. The proteins with different expression levels also in strain GS/M-83-H7 are printed in italics.
Antigenic variation
The strains investigated in this study had been characterized with respect to their (major) surface antigens. Therefore, in a first step, we matched the original antigens by protein–protein blasts to sequences found in the GiardiaDB. It turned out that sequences attributed to TSA417 (strain C6) matched to GL50803_113797, annotated as VSP5 and to GL50803_113450, annotated as VSP44. Concerning strain H7, the sequence published for VSPH7 matched to GSB150963.
Surprisingly, the sequence of CRP170, the predominant surface antigen of strain A1, matched to a sequence annotated as a hypothetical protein, namely GL50803_221693 with 100% identity and a total score of 3491 (Fig. S2). The GL50803_221693 ORF encodes a 228 kDa protein containing 2259 amino acids. The identified peptides of GL50803_221693 covered unique protein sequences near the N terminus (aa 75–151), followed by 26 repeats of the peptide TNPSDPTGTCVSAVDCQGSAGYYTDDS-VSDAKECKKCNAPCTACAGTADKCTKCDANGAAPYLKK (thus containing the GAAPYLKK motif recognized by mAb6E7), and a unique sequence near the C-terminus (aa 1812–2223). Regions where trypsin produces too long/hydrophobic or too short peptides, namely the two transmembrane helices at the C- and N-terminal part of the protein, were missed as can be expected by the used technology (Fig. S3). Proteins encoded by other ORFs sharing high score homologies with CRP170, namely GL50803_137752 and GL50803_37093 (Fig. S2), were not detected in the present dataset.
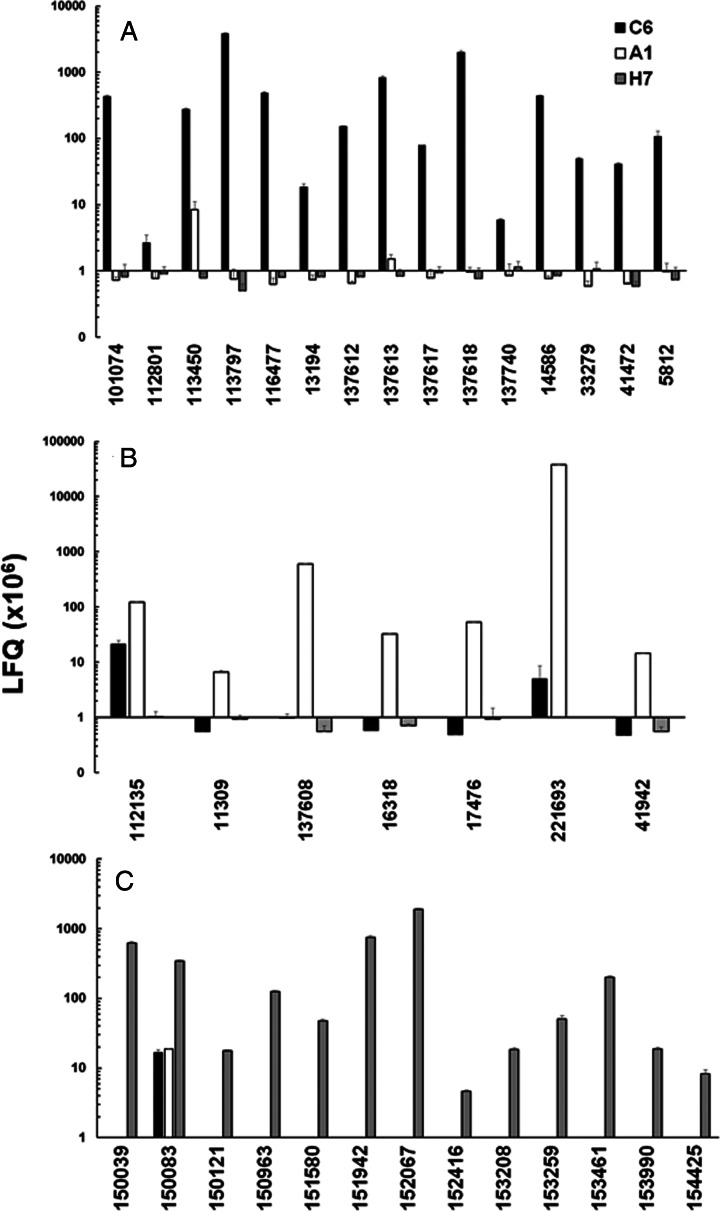
In a second step, we compared the LFQ intensities of the major VSPs in all strains (Fig. 3). In the case of strain C6, TSA417 (VSP5; ORF113797) was still one of the dominating VSPs, followed by VSP-8 (ORF 137618), VSP-188 (ORF 137613) and four other VSPs in the range of 107 LFQ units (Fig. 3A). Amongst these VSPs were the gene products of ORFs 101074 and 137618. ORF 101074 (VSP88) was identical to the sequence of AAG16629, annotated as VSP9B10A (Nash et al., 2001). ORF 137618 (VSP8) covered AF293416, annotated as VSPB10B (Carranza et al., 2002) only to 88% leaving 70 amino acid gaps. In a previously published comparative proteomics study (Emery et al., 2015), a strain WB expressed four of the major VSPs identified in WBC6, namely VSP38 (ORF 13194), VSP49 (ORF 41472), VSP149 (ORF 116477) and the previously mentioned VSP188.
Fig. 3.
Quantitative assessments of the major variant-specific surface proteins (VSPs). Trophozoites of the strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) were subjected to MS shotgun analysis as described in ‘Materials and methods’ section. For all VSPs, mean values ± one standard deviation for LFQ intensities (×106) in three biological replicates are shown. The VSPs are termed by their respective accession numbers in the GiardiaDB (GL50803 for C6 and A1; GSB for H7). A, VSP pattern of strain C6; B, VSP pattern of strain A1; C, VSP pattern of strain H7.
In strain A1, GL50803_221693 (homologous to VSPA6) was – by far – the most predominant VSP with LFQ values above 1011, but not the only one identified. The five next abundant VSPs had, however, expression levels three or more magnitudes lower than VSPA6. Interestingly, VSPA6 was expressed in strain C6 as well, but only at LFQ levels below 107 (Fig. 3B). In strain H7, 13 VSPs with LFQ levels around 107 or higher were identified. VSPH7 (GSB150963) was not predominant, five other VSPs having equal or higher expression levels (Fig. 3C).
Moreover, we have reinvestigated the most predominant VSPs of the three strains using only peptides labelled as being unique to a single VSP by using iTop3 protein intensities (see ‘Materials and methods’ section and Table S2). The corresponding values are shown as Supplementary Fig. S4. Some minor VSPs disappeared, but the overall pattern of the strains did not change as compared to the analysis based on LFQ values.
These results show that A1 had the most ‘homogeneous’ population of trophozoites with respect to their surface proteome. The two other strains yielded mixed trophozoite populations with five or more equally dominant subpopulations.
The fact that VSPA6 was expressed at low levels in strain C6 prompted us to investigate whether this VSP was found in the proteome of the same strain grown on a different medium. The corresponding dataset had been published earlier in a different context (Müller et al., 2019). Since the corresponding ORF (221693) was annotated as a hypothetical protein and not as a VSP, the corresponding expression levels had been overlooked. As shown in Table S3, VSPA6 became the third most abundant VSP in C6 trophozoites grown on a new culture medium differing from the old one merely with respect to the quality of the serum and the peptone. Moreover, VSPA6 had very high expression levels in two Australian assemblage A1 isolates investigated in the same study (Müller et al., 2019). Thus, VSPA6 cannot be regarded as a unique marker for WBA1 and related clones.
Susceptibility to antigiardial drugs and reductase activities
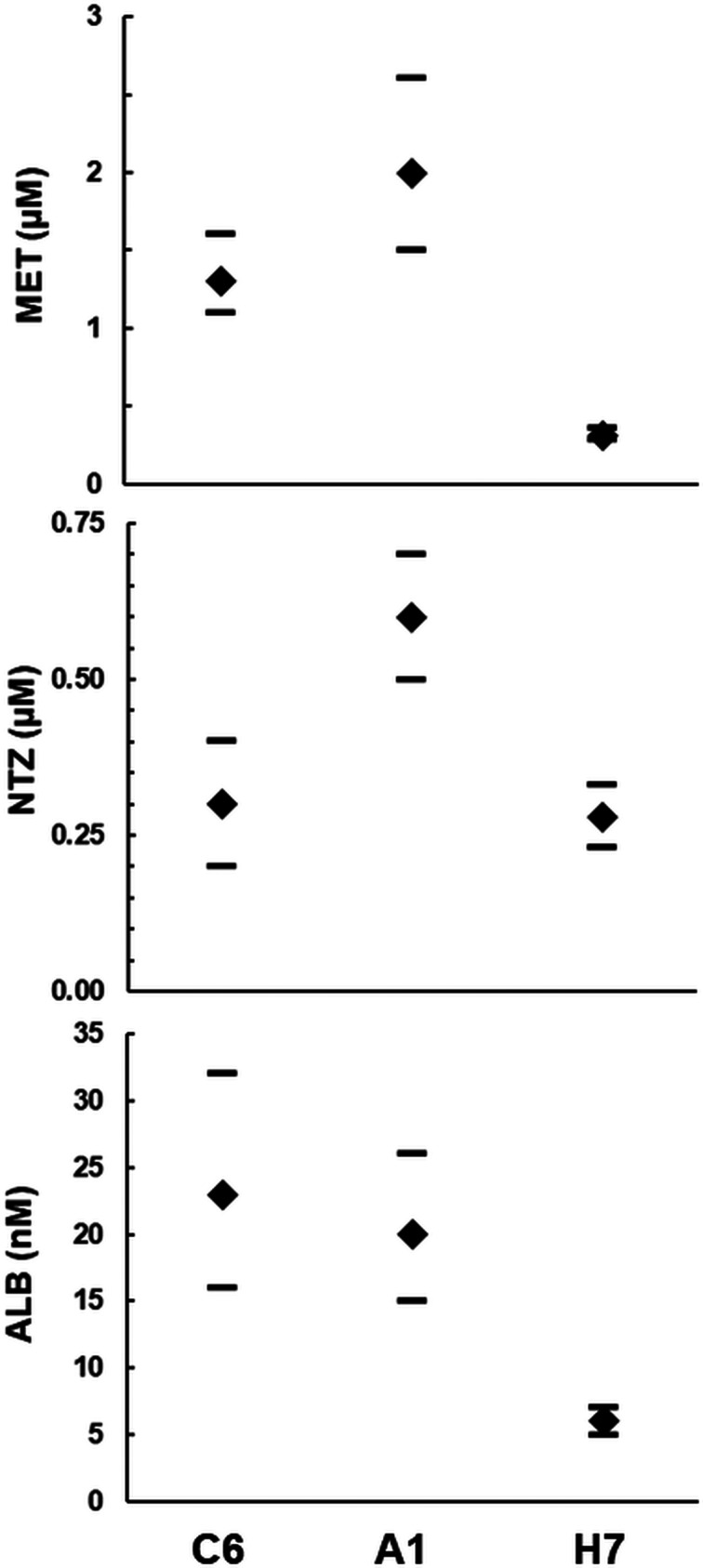
Since the strains C6 and A1 had been isolated from a patient treated with MET, we were curious to investigate whether these strains differed in their susceptibilities to nitro drugs from the strain H7 isolated from a different source. Interestingly, strain A1 had higher IC50s than strain C6 when exposed to MET and to NTZ, but not to ALB. Strain H7 was more susceptible to MET and to ALB than both strains C6 and A1 (Fig. 4). The MIC of both nitro drugs were even nearly one order of magnitude higher on A1 than on both other strains (Table 3). When comparing IC50s and MICs from different strains, one should consider that these constants measure different things, as reviewed elsewhere (Müller and Hemphill, 2013). In the case of bacteria exposed to antibiotics, elevated MICs are regarded as indicative for ‘resistance’, whereas elevated IC50s without elevated MICs indicate ‘tolerance’ (Brauner et al., 2016). In our case, strain A1 would be at the edge of ‘resistance’ with, however, much lower MICs than those of resistant strains generated in the laboratory (Müller et al., 2018).
Fig. 4.
Determination of drug susceptibilities. The strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) were exposed to serial dilutions of the nitro compounds metronidazole (MET) and nitazoxanide (NTZ), as well as of albendazole (ALB). IC50 values were determined as described in ‘Materials and methods’ section and are given in μm (MET, NTZ) or nm (ALB). Diamonds correspond to the IC50s, bars to the 95% confidence intervals.
Table 3.
Determination of minimal inhibitory concentrations (MICs)
| Compound | C6 | A1 | H7 |
|---|---|---|---|
| MET | 6.25 | 50 | 6.25 |
| NTZ | 1.25 | 10 | 1.25 |
| ALB | 0.125 | 0.125 | 0.125 |
The strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7) were exposed to serial dilutions of the nitro compounds metronidazole (MET) and nitazoxanide (NTZ), as well as of albendazole (ALB). MICs were determined as described in ‘Materials and methods’ section and are given in μm.
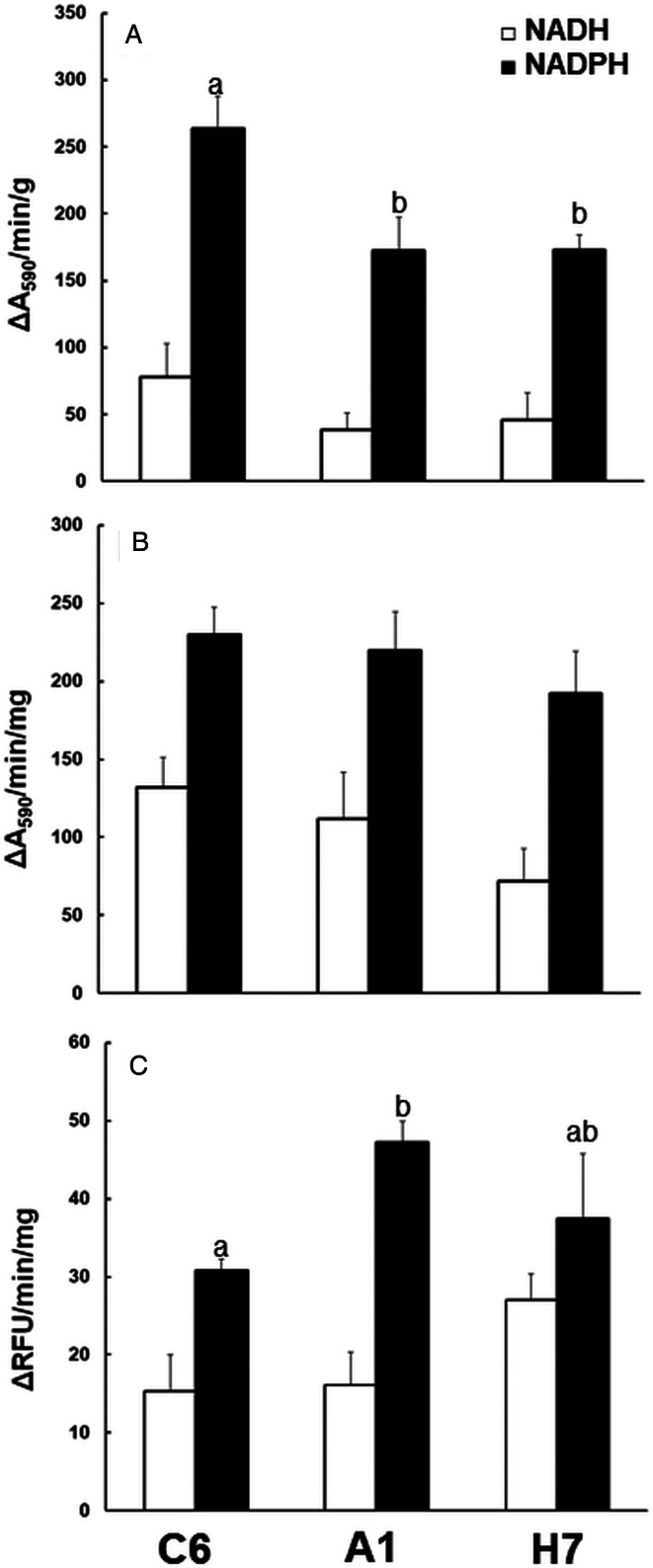
To see whether these differences matched to differences in reductase activities, we assayed NAD(P)H oxidase activity, quinone reductase activity and nitroreductase activity in crude extracts from the three strains (Fig. 5). Crude extracts from strains A1 and H7 had significantly lower NADPH oxidase, but not lower NADH oxidase activities than strain C6 (Fig. 5A). Quinone reductase activity with menadione as a substrate was at equal levels in crude extracts from all three strains (Fig. 5B). Nitroreductase activity, assayed as the full reduction of 7-NC to 7-AC was slightly, but significantly increased in A1 as compared to C6 crude extracts. Crude extracts from strain H7 had an intermediate activity (Fig. 5C).
Fig. 5.
Reductase activities in crude extracts of G. lamblia trophozoites. The assays were performed as described in ‘Materials and methods’ section using either NADH (white bars) or NADPH (black bars) as cofactors. A, NAD(P)H oxidase; B, quinone reductase with menadione as a substrate; C, nitroreductase with 7-nitrocoumarin as a substrate. Mean values ± one standard deviation for three independent assays are given. Values superscribed by different letters are significantly different (two-sided t-tests; P < 0.05).
Enzymes involved in nitro reduction or detoxification processes
These findings prompted us to have a closer look at the expression levels of enzymes involved in nitro reduction and related detoxification processes. The values are listed in Table 4. Strain C6 had significantly higher levels of the nitroreductase NR1 than the two other strains. Conversely, A1 was the only strain expressing the homologous NR family protein without ferredoxin domain at its N-terminus. C6 and A1, but not H7, expressed a 2Fe-2S ferredoxin, the only free ferredoxin detected in the proteome of the strains investigated in this study. Moreover, the ferredoxin-domain protein hydrogenase 1 was identified in all strains with significantly higher levels in strain H7.
Table 4.
Overview of proteins involved in reduction (and thus activation) of nitro compounds and the scavenging of radicals in trophozoites of the strains WBC6 (C6), WBA1 (A1) and GS/M-83-H7 (H7)
| Annotation | DB | C6 | A1 | H7 |
|---|---|---|---|---|
| Fd-NR2 (‘NR1’) | 22 677 | 106 ± 5* | 61 ± 1 | 62 ± 2 |
| Fd 2Fe_2S | 27 266 | 9 ± 1 | 10 ± 0 | nd* |
| Fd-hydrogenase 1 | 6304 | 46 ± 1 | 45 ± 0 | 122 ± 9* |
| NR family (‘NR3’) | 15 307 | nd | 4 ± 0* | nd |
| PFOR 1 | 17 063 | 1713 ± 22 | 1510 ± 29 | 2290 ± 20 |
| PFOR 2 | 114 609 | 4619 ± 48 | 3966 ± 37 | 3938 ± 51 |
| Thioredoxin-reductase | 9827 | 243 ± 26 | 208 ± 20 | 226 ± 7 |
| A-type flavoprotein | 10 358 | 681 ± 11* | 471 ± 10 | 461 ± 13 |
| NAD(P)H oxidase lateral transfer | 33 769 | 2324 ± 56 | 2103 ± 55 | 2032 ± 50 |
| NAD(P)H oxidase | 9719 | 490 ± 16* | 332 ± 11 | 290 ± 3 |
Cells were harvested and subjected to MS shotgun analysis as described in ‘Materials and methods’ section. For each strain, three biological replicates have been tested (with three technical replicates per biological replicate). For all proteins, mean values ± standard errors for LFQ intensities (×106) in three biological replicates are given (nd, below detection limit). DB, number in GiardiaDB; Fd, ferredoxin; LT, lateral transfer; NR, nitroreductase; PFOR, pyruvate-ferredoxin oxidoreductase. *, value is statistically significant from the two others (one-factorial ANOVA, P < 0.01).
C6 had significantly higher levels of A-type flavoprotein and of NAD(P)H oxidase than the two other strains. All other proteins investigated within this context, in particular both pyruvate-ferredoxin-oxidoreductases, did not differ between the strains (Table 4).
Conclusions
Our results, in particular those concerning the surface proteome of the investigated strains, show that care must be taken when referring to these strains using the VSP terminology established three decades ago as a result of their reactivity with monoclonal antibodies. A given antibody may cross-react with epitopes on VSPs encoded by a different ORF with high sequence identity and/or with VSPs carrying epitopes with a similar secondary structure, as illustrated by the following example. Using the monoclonal antibody Mab9B10, for instance, two different VSPs, namely VSP9B10A (Nash et al., 2001) and VSP9B10B (Carranza et al., 2002) were identified, which correspond to two major VSPs (VSP88 for sure and VSP8 most likely) of strain WBC6 as shown in our dataset.
Consequently, the hypothesis that one single trophozoite expresses one single VSP could be modified in the sense that one single trophozoite may express VSPs that may be encoded by one or more ORFs. This hypothesis could be verified only by single-cell proteomics (Dou et al., 2019; Liu et al., 2020), a method which has not been established for G. lamblia, so far.
Rich in potential trypsin and chymotrypsin cleavage sites inside a highly repetitive motif (covering three-quarters of its primary sequence in the case of VSPA6), the digest by pancreas proteases (or by proteases released by decaying trophozoites) may yield free epitopes blocking otherwise cytotoxic antibodies. Such a scenario may be relevant for the outcome of G. lamblia infections because cytotoxic anti-VSP antibodies turned out to possess a modulatory function on the proliferative parasite population characteristics in the experimental murine host as reviewed elsewhere (Müller and Gottstein, 1998; Müller and von Allmen, 2005). Moreover, VSPs may have enzymatic functions as evidenced in the case of VSP9B10A (ORF 101074; VSP88), which has cysteine protease activity (Cabrera-Licona et al., 2017). Other functions such as enzyme activities or interactions with receptors on other trophozoites or on host cells remain to be elucidated.
Since the WB isolate issues from a patient treated with MET, the clones A1 and C6 may represent two different strategies to deal with this drug pressure. WBC6 retains a higher flexibility with respect to gene expression and is able to generate a highly nitro drug-resistant offspring in a few selection cycles (Müller et al., 2007) differing from the original clone with respect to appropriate physiological adaptations and an adapted proteome (Müller et al., 2018; Müller et al., 2019). Conversely, WBA1 may represent a ‘freezed’ status with a moderate nitro drug resistance at the price of a loss in gene expression flexibility.
This hypothesis including the identification of elements controlling this flexibility, e.g. in the differential signalome of both strains as evidenced in our dataset, would require further investigations.
Acknowledgements
The Giardia strains used in this study were generous gifts by Bruno Gottstein and Theodore Nash.
Financial support
The work was supported by the Swiss National Science Foundation [grant No. 31003A_163230].
Ethical standards
Not applicable.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020000657.
click here to view supplementary material
Conflict of interest
None of the authors has any competing interests in the manuscript.
References
- Adam RD (2001) Biology of Giardia lamblia. Clinical Microbiology Reviews 14, 447–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam RD, Aggarwal A, Lal AA, de La Cruz VF, McCutchan T and Nash TE (1988) Antigenic variation of a cysteine-rich protein in Giardia lamblia. Journal of Experimental Medicine 167, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam RD, Nigam A, Seshadri V, Martens CA, Farneth GA, Morrison HG, Nash TE, Porcella SF and Patel R (2010) The Giardia Lamblia vsp gene repertoire: characteristics, genomic organization, and evolution. BMC Genomics 11, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain T, Amat CB, Motta JP, Manko A and Buret AG (2017) Interactions of Giardia sp. with the intestinal barrier: epithelium, mucus, and microbiota. Tissue Barriers 5, e1274354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénéré E, da Luz RA, Vermeersch M, Cos P and Maes L (2007) A new quantitative in vitro microculture method for Giardia Duodenalis trophozoites. Journal of Microbiological Methods 71, 101–106. [DOI] [PubMed] [Google Scholar]
- Bienz M, Siles-Lucas M, Wittwer P and Müller N (2001) vsp gene expression by Giardia lamblia clone GS/M-83-H7 during antigenic variation in vivo and in vitro. Infection and Immunity 69, 5278–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brauner A, Fridman O, Gefen O and Balaban NQ (2016) Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nature Reviews Microbiology 14, 320–330. [DOI] [PubMed] [Google Scholar]
- Cabrera-Licona A, Solano-Gonzalez E, Fonseca-Linan R, Bazan-Tejeda ML, Raul A-G, Bermudez-Cruz RM and Ortega-Pierres G (2017) Expression and secretion of the Giardia duodenalis variant surface protein 9B10A by transfected trophozoites causes damage to epithelial cell monolayers mediated by protease activity. Experimental Parasitology 179, 49–64. [DOI] [PubMed] [Google Scholar]
- Campbell JD and Faubert GM (1994) Comparative studies on Giardia lamblia encystation in vitro and in vivo. Journal of Parasitology 80, 36–44. [PubMed] [Google Scholar]
- Carranza PG, Feltes G, Ropolo A, Quintana SM, Touz MC and Luján HD (2002) Simultaneous expression of different variant-specific surface proteins in single Giardia lamblia trophozoites during encystation. Infection and Immunity 70, 5265–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernikova L, Faso C and Hehl AB (2018) Five facts about Giardia lamblia. PLoS Pathogens 14, e1007250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou M, Clair G, Tsai CF, Xu K, Chrisler WB, Sontag RL, Zhao R, Moore RJ, Liu T, Pasa-Tolic L, Smith RD, Shi T, Adkins JN, Qian WJ, Kelly RT, Ansong C and Zhu Y (2019) High-throughput single cell proteomics enabled by multiplex isobaric labeling in a nanodroplet sample preparation platform. Analytical Chemistry 91, 13119–13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery-Corbin SJ, Vuong D, Lacey E, Svard SG, Ansell BRE and Jex AR (2018) Proteomic diversity in a prevalent human-infective Giardia duodenalis sub-species. International Journal for Parasitology 48, 817–823. [DOI] [PubMed] [Google Scholar]
- Emery SJ, van Sluyter S and Haynes PA (2014) Proteomic analysis in Giardia duodenalis yields insights into strain virulence and antigenic variation. Proteomics 14, 2523–2534. [DOI] [PubMed] [Google Scholar]
- Emery SJ, Lacey E and Haynes PA (2015) Quantitative proteomic analysis of Giardia duodenalis assemblage A: a baseline for host, assemblage, and isolate variation. Proteomics 15, 2281–2285. [DOI] [PubMed] [Google Scholar]
- Emery SJ, Baker L, Ansell BRE, Mirzaei M, Haynes PA, McConville MJ, Svard SG and Jex AR (2018) Differential protein expression and post-translational modifications in metronidazole-resistant Giardia duodenalis. Gigascience 7. doi: 10.1093/gigascience/giy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faso C and Hehl AB (2011) Membrane trafficking and organelle biogenesis in Giardia lamblia: use it or lose it. International Journal for Parasitology 41, 471–480. [DOI] [PubMed] [Google Scholar]
- Furfine ES and Wang CC (1990) Transfection of the Giardia lamblia double-stranded RNA virus into giardia lamblia by electroporation of a single-stranded RNA copy of the viral genome. Molecular and Cellular Biology 10, 3659–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TB and Hill DR (2001) Treatment of giardiasis. Clinical Microbiology Reviews 14, 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillin FD, Hagblom P, Harwood J, Aley SB, Reiner DS, McCaffery M, So M and Guiney DG (1990) Isolation and expression of the gene for a major surface protein of Giardia lamblia. Proceedings of the National Academy of Sciences of the USA 87, 4463–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottstein B, Harriman GR, Conrad JT and Nash TE (1990) Antigenic variation in Giardia lamblia: cellular and humoral immune response in a mouse model. Parasite Immunology 12, 659–673. [DOI] [PubMed] [Google Scholar]
- Hemphill A, Müller N and Müller J (2019) Comparative pathobiology of the intestinal protozoan parasites Giardia lamblia, Entamoeba histolytica, and Cryptosporidium parvum. Pathogens (Basel, Switzerland) 8. doi: 10.3390/pathogens8030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth MF (2016) Giardia duodenalis genetic assemblages and hosts. Parasite 23, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova L, Singer SM, Conrad J and Nash TE (2006) Epigenetic mechanisms are involved in the control of Giardia lamblia antigenic variation. Molecular Microbiology 61, 1533–1542. [DOI] [PubMed] [Google Scholar]
- Litleskare S, Rortveit G, Eide GE, Hanevik K, Langeland N and Wensaas KA (2018) Prevalence of irritable bowel syndrome and chronic fatigue 10 years after Giardia infection. Clinical Gastroenterology and Hepatology 16, 1064–1072 e1064. [DOI] [PubMed] [Google Scholar]
- Liu D, Paczkowski P, Mackay S, Ng C and Zhou J (2020) Single-cell multiplexed proteomics on the IsoLight resolves cellular functional heterogeneity to reveal clinical responses of cancer patients to immunotherapies. Methods in Molecular Biology 2055, 413–431. [DOI] [PubMed] [Google Scholar]
- Meng TC, Hetsko ML and Gillin FD (1993) Antigenic switching of TSA 417, a trophozoite variable surface protein, following completion of the life cycle of Giardia lamblia. Infection and Immunity 61, 5394–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JE, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG and Sogin ML (2007) Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science (New York, N.Y.) 317, 1921–1926. [DOI] [PubMed] [Google Scholar]
- Mowatt MR, Nguyen BY, Conrad JT, Adam RD and Nash TE (1994) Size heterogeneity among antigenically related Giardia lamblia variant-specific surface proteins is due to differences in tandem repeat copy number. Infection and Immunity 62, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N and Gottstein B (1998) Antigenic variation and the murine immune response to Giardia lamblia. International Journal for Parasitology 28, 1829–1839. [DOI] [PubMed] [Google Scholar]
- Müller J and Hemphill A (2013) New approaches for the identification of drug targets in protozoan parasites. International Review of Cell and Molecular Biology 301, 359–401. [DOI] [PubMed] [Google Scholar]
- Müller N and von Allmen N (2005) Recent insights into the mucosal reactions associated with Giardia lamblia infections. International Journal for Parasitology 35, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Müller N, Stäger S and Gottstein B (1996) Serological analysis of antigenic heterogeneity of Giardia lamblia variant surface proteins. Infection and Immunity 64, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Sterk M, Hemphill A and Müller N (2007) Characterization of Giardia lamblia WB C6 clones resistant to nitazoxanide and to metronidazole. Journal of Antimicrobial Chemotherapy 60, 280–287. [DOI] [PubMed] [Google Scholar]
- Müller J, Rout S, Leitsch D, Vaithilingam J, Hehl A and Müller N (2015) Comparative characterisation of two nitroreductases from Giardia lamblia as potential activators of nitro compounds. International Journal for Parasitology. Drugs and Drug Resistance 5, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hemphill A and Müller N (2018) Physiological aspects of nitro drug resistance in Giardia lamblia. International Journal for Parasitology. Drugs and Drug Resistance 8, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Braga S, Heller M and Müller N (2019) Resistance formation to nitro drugs in Giardia lamblia: no common markers identified by comparative proteomics. International Journal for Parasitology: Drugs and Drug Resistance 9, 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash TE (2001) Treatment of Giardia lamblia infections. Pediatric Infectious Disease Journal 20, 193–195. [DOI] [PubMed] [Google Scholar]
- Nash TE (2002) Surface antigenic variation in Giardia lamblia. Molecular Microbiology 45, 585–590. [DOI] [PubMed] [Google Scholar]
- Nash TE and Mowatt MR (1992) Characterization of a Giardia lamblia variant-specific surface protein (VSP) gene from isolate GS/M and estimation of the VSP gene repertoire size. Molecular and Biochemical Parasitology 51, 219–227. [DOI] [PubMed] [Google Scholar]
- Nash TE, McCutchan T, Keister D, Dame JB, Conrad JD and Gillin FD (1985) Restriction-endonuclease analysis of DNA from 15 Giardia isolates obtained from humans and animals. Journal of Infectious Diseases 152, 64–73. [DOI] [PubMed] [Google Scholar]
- Nash TE, Aggarwal A, Adam RD, Conrad JT and Merritt JW (1988) Antigenic variation in Giardia lamblia. Journal of Immunology 141, 636–641. [PubMed] [Google Scholar]
- Nash TE, Banks SM, Alling DW, Merritt JW and Conrad JT (1990a) Frequency of variant antigens in Giardia lamblia. Experimental Parasitology 71, 415–421. [DOI] [PubMed] [Google Scholar]
- Nash TE, Conrad JT and Merritt JW (1990b) Variant specific epitopes of Giardia lamblia. Molecular and Biochemical Parasitology 42, 125–132. [DOI] [PubMed] [Google Scholar]
- Nash TE, Herrington DA, Levine MM, Conrad JT and Merritt JW (1990c) Antigenic variation of Giardia lamblia in experimental human infections. Journal of Immunology 144, 4362–4369. [PubMed] [Google Scholar]
- Nash TE, Luján HT, Mowatt MR and Conrad JT (2001) Variant-specific surface protein switching in Giardia lamblia. Infection and Immunity 69, 1922–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A and Vizcaino JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochaska HJ and Santamaria AB (1988) Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Analytical Biochemistry 169, 328–336. [DOI] [PubMed] [Google Scholar]
- Prucca CG and Lujan HD (2009) Antigenic variation in Giardia lamblia. Cellular Microbiology 11, 1706–1715. [DOI] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elías EV, Rivero FD, Saura A, Carranza PG and Luján HD (2008) Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456, 750–754. [DOI] [PubMed] [Google Scholar]
- Prucca CG, Rivero FD and Luján HD (2011) Regulation of antigenic variation in Giardia lamblia. Annual Review of Microbiology 65, 611–630. [DOI] [PubMed] [Google Scholar]
- Smith PD, Gillin FD, Kaushal NA and Nash TE (1982a) Antigenic analysis of Giardia lamblia from Afghanistan, Puerto Rico, Ecuador, and Oregon. Infection and Immunity 36, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Gillin FD, Spira WM and Nash TE (1982b) Chronic giardiasis: studies on drug sensitivity, toxin production, and host immune response. Gastroenterology 83, 797–803. [PubMed] [Google Scholar]
- Sun CH, Chou CF and Tai JH (1998) Stable DNA transfection of the primitive protozoan pathogen Giardia lamblia. Molecular and Biochemical Parasitology 92, 123–132. [DOI] [PubMed] [Google Scholar]
- Svärd SG, Meng TC, Hetsko ML, McCaffery JM and Gillin FD (1998) Differentiation-associated surface antigen variation in the ancient eukaryote Giardia lamblia. Molecular Microbiology 30, 979–989. [DOI] [PubMed] [Google Scholar]
- Thompson RC (2004) The zoonotic significance and molecular epidemiology of Giardia and giardiasis. Veterinary Parasitology 126, 15–35. [DOI] [PubMed] [Google Scholar]
- Wagner BD (2009) The use of coumarins as environmentally-sensitive fluorescent probes of heterogeneous inclusion systems. Molecules 14, 210–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Jex A and Svard SG (2020) A chromosome-scale reference genome for Giardia intestinalis WB. Scientific Data 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM and Adam RD (1995) Analysis of a repeat-containing family of Giardia lamblia variant-specific surface protein genes: diversity through gene duplication and divergence. Journal of Eukaryotic Microbiology 42, 439–444. [DOI] [PubMed] [Google Scholar]
- Zahedi A, Field D and Ryan U (2017) Molecular typing of Giardia duodenalis in humans in Queensland – first report of assemblage E. Parasitology 144, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Zamponi N, Zamponi E, Mayol GF, Lanfredi-Rangel A, Svard SG and Touz MC (2017) Endoplasmic reticulum is the sorting core facility in the Golgi-lacking protozoan Giardia lamblia. Traffic (Copenhagen, Denmark) 18, 604–621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020000657.
click here to view supplementary material