There is a Blood Commentary on this article in this issue.
Key Points
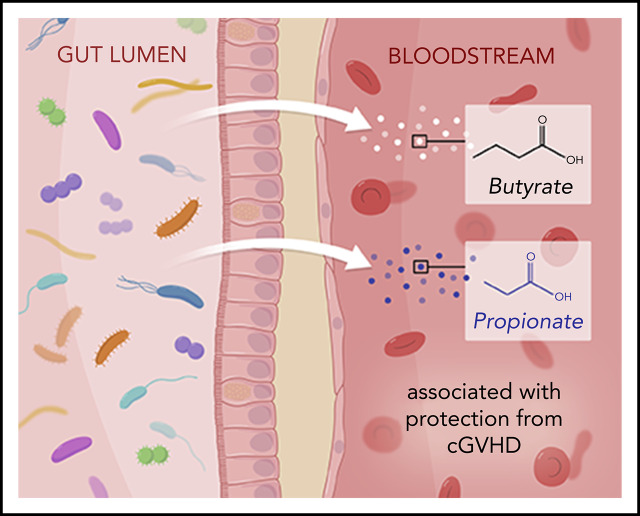
The microbe-derived SCFAs butyrate and propionate in the systemic circulation are associated with protection from cGVHD.
cGVHD is associated with gastrointestinal dysbiosis late after HCT.
Abstract
Studies of the relationship between the gastrointestinal microbiota and outcomes in allogeneic hematopoietic stem cell transplantation (allo-HCT) have thus far largely focused on early complications, predominantly infection and acute graft-versus-host disease (GVHD). We examined the potential relationship of the microbiome with chronic GVHD (cGVHD) by analyzing stool and plasma samples collected late after allo-HCT using a case-control study design. We found lower circulating concentrations of the microbe-derived short-chain fatty acids (SCFAs) propionate and butyrate in day 100 plasma samples from patients who developed cGVHD, compared with those who remained free of this complication, in the initial case-control cohort of transplant patients and in a further cross-sectional cohort from an independent transplant center. An additional cross-sectional patient cohort from a third transplant center was analyzed; however, serum (rather than plasma) was available, and the differences in SCFAs observed in the plasma samples were not recapitulated. In sum, our findings from the primary case-control cohort and 1 of 2 cross-sectional cohorts explored suggest that the gastrointestinal microbiome may exert immunomodulatory effects in allo-HCT patients at least in part due to control of systemic concentrations of microbe-derived SCFAs.
Visual Abstract
Introduction
Chronic graft-versus-host disease (cGVHD) affects 40% to 50% of long-term survivors of allogeneic hematopoietic stem cell transplantation (allo-HCT) and is the leading cause of nonrelapse mortality among patients who survive to 2 years.1 In contrast to acute GVHD (aGVHD), characterized by apoptotic tissue damage, particularly in the intestine, cGVHD is a fibrotic condition predominantly involving the skin and mouth that shares features with the autoimmune condition systemic sclerosis.2 We and others have demonstrated relationships between the microbiota and overall survival, aGVHD mortality, infection, organ toxicity, engraftment, and relapse3-5 in the allo-HCT patient population, but cGVHD remains unexamined.
Preclinical models have demonstrated that T follicular helper and T helper 17 cells are critical for cGVHD development, partly due to production of profibrotic cytokines which have been linked to gastrointestinal (GI) microbiota composition.6-8 Mouse studies also suggest that the GI microbiome modulates alloreactivity in aGVHD, and that these effects are dependent upon microbe-derived metabolites.5,9,10 In preclinical models, the microbe-derived short-chain fatty acid (SCFA) butyrate has been shown to exert local effects on intestinal epithelia and immune populations in the GI tract,8,11,12 and clinical studies have associated colonization with butyrate producers with risk of viral infection.13,14 We have previously demonstrated that allo-HCT is accompanied by a decrease in microbial diversity within the GI tract, particularly loss of commensal anaerobes.3,15,16 We hypothesized that microbiota perturbations may be associated with the development of cGVHD and investigated this using a case-control study design using patient stool and plasma samples collected at our center.
Study design
Patient selection
We used the cohort of adult transplant recipients at Memorial Sloan Kettering Cancer Center (MSKCC) who had ≥1 stool sample between day −30 and day +365 (8164 samples, 1081 unique patients). Patients provided written consent to an institutional review board–approved biospecimen-collection protocol. cGVHD cases were identified using National Institutes of Health criteria.17 Diagnoses and onset dates were confirmed by chart review. Controls were randomly selected from the overall cohort on the basis of matched graft type (ie, CD34-selected/T-cell–depleted or unmodified graft) in a 1:3 ratio. Three patients in the cGVHD case group received fecal microbiota transplantation, as did 4 controls as part of a randomized clinical trial.18 Healthy volunteers (plasma samples included in Figure 2A) provided written informed consent prior to the collection of samples for inclusion in this study.
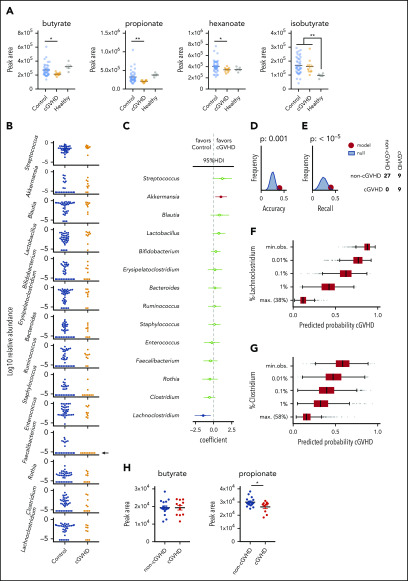
Figure 2.
Circulating microbe-derived metabolites are associated with protection from cGVHD. (A) Metabolomic analysis of SCFAs in peri-day 100 plasma samples from MSKCC (n = 10 cGVHD, 43 control, 5 healthy volunteers). P values were determined on the basis of analysis of variance followed by Mann-Whitney U testing. One sample was included per patient; where multiple plasma samples were available, the closest sample to day 100 was chosen. Three cGVHD patients and 13 control patients had both stool and plasma available in the peri-day 100 window. (B) Log10 relative abundance of 14 candidate microbial genera in cGVHD patients and controls. (C) Logistic regression coefficients of the genus abundances associated with cGVHD or control status (9 and 36 patients, respectively, all recipients of unmodified grafts). Bars represent 95% highest posterior density intervals (HDIs), and intervals highlighted in blue and red are entirely >0 or <0. (D-E) Recall and accuracy of our model compared with 100 000 draws from a binomial null model with P = .2 as per the case-control design. (F-G) Predicted probability of cGVHD at different abundances of Lachnoclostridium or Faecalibacterium in 1000 simulated microbiota communities per abundance level (minimum observed [min. obs.], lowest observed, nonzero family abundance). (H) Duke plasma samples from a cross-sectional cohort of patients who did or did not go on to develop cGVHD (n = 11 cGVHD, 17 no cGVHD).
For the analysis of the Duke University Medical Center (Duke) and University Hospital Regensburg (Regensburg) cohorts, a cross-sectional design was applied, whereby adult allo-HCT recipients were eligible for inclusion on the basis of the availability of paired stool and plasma (Duke) or serum (Regensburg) samples in the time window of interest (days 70 to 130), and without specifying clinical variables or cGVHD outcomes. Patients provided consent in a fashion consistent with each institution’s institutional review board requirements.
Stool sequencing
16S ribosomal-RNA gene sequencing was performed and analyzed as previously described.3,4,16 Shotgun sequencing was analyzed by HUMAnN2,19 and the association between pathway abundances and cGVHD was analyzed using linear discriminant analysis of effect size.20 Stool samples from Duke and Regensburg were aliquoted and frozen locally; DNA extraction and sequencing were performed at MSKCC. Bacterial DNA extraction was performed as previously described for the MSKCC samples15 and with a modified protocol that extracts both bacterial and fungal DNA from stool.21 Duke had 37 unique patients, with 27 evaluable, and Regensburg had 89 unique patients, with 44 stool samples evaluable; missing data were due to sample extraction or sequencing failure.
Plasma metabolomic analysis
Plasma samples were processed as previously described22 and analyzed using quantitative gas chromatography time-of-flight (Agilent 7890B GC System, Agilent 7200A MS Detector). Data analysis was performed using MassHunter Profinder software (version B.08.00, Agilent Technologies).
Statistical analysis
P values for α-diversity were calculated using a Wilcoxon test using R (version 3.5.0). PERMANOVA testing for β-diversity was performed using the R package vegan (using the adonis function).23 P values for the plasma SCFA analysis were calculated using analysis of variance followed by Mann-Whitney U testing using GraphPad Prism (Figure 2A). Fisher’s exact test, where reported, was also performed using GraphPad Prism. For Figure 2, the log10 abundances of the most abundant microbial genera, as well as genera that have been associated with immune-system dynamics in cancer patients (Faecalibacterium, Clostridium, Rothia, and Staphylococcus),24-26 were analyzed with a Bayesian logistic regression using Python and the pymc3 module,27 further described in supplemental Methods (available on the Blood Web site). P values for accuracy and recall of the trained model were calculated by drawing 100 000 sets of n = 45 binomial samples with P = .2, as per the case-control design and each time calculating accuracy and recall of this null model (for the other centers, the binomial parameter p was set to the fraction of observed cases across all patients per center data set).
Results and discussion
Allo-HCT is accompanied by a loss of fecal microbiota α-diversity.28 Since samples in our previous studies were almost exclusively collected from inpatients, the data from later time points were biased toward patients who either remained admitted for prolonged periods after HCT or were readmitted, perhaps reflecting a more complicated transplant course. Here, we describe a cohort of patients who were sampled both during their inpatient stay and, in some cases, following discharge (n = 1079 patients, 8164 samples; 522 of these samples [6.4%] were collected while the patients were out of the hospital; supplemental Table 1) and demonstrate that loss of α-diversity can be observed for up to 12 months (Figure 1A). We speculate that hospitalization, dietary changes, and exposure to antibiotics and other medications establish lasting changes in microbial communities in the GI tract.29,30
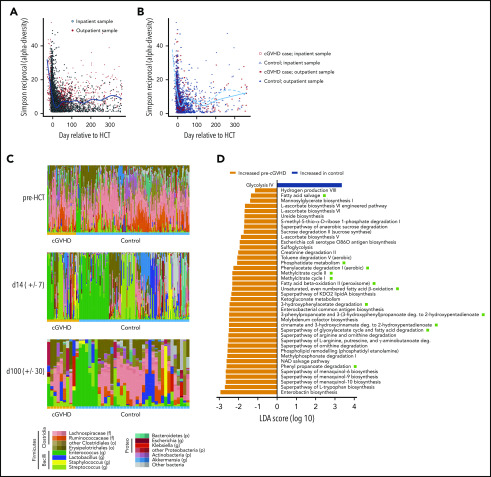
Figure 1.
HCT is characterized by an early dysbiosis that is slow to recover. (A) α-diversity (as measured by the Simpson reciprocal, a parameter that considers the relative abundance of each unique bacterial taxa to summarize overall diversity) in 8073 stool samples from 1079 unique patients who underwent HCT at MSKCC between 2014 and 2018. Demographics are tabulated in supplemental Table 1. (B) α-diversity of 491 stool samples from 53 unique cGVHD patients (red) and 1702 stool samples from 171 unique controls (blue). One patient in the case group had 2 samples available (day 100 and day 7), but both had insufficient reads to include in this analysis. Samples collected from inpatients are shown as filled symbols, and outpatient collections are shown in open symbols. The solid blue line summarizes the cGVHD cases and the dotted line the controls. (C) Fecal microbial taxa pretransplant (day −30 to day −6; n = 43 cases, 168 controls), periengraftment (closest sample to day 14 [d14]; n = 42 cases, 144 controls), and peri-day 100 (closest sample to day 100 [d100], within the day 70 to day 130 window; n = 9 cases, 36 controls). Patient demographics are described in supplemental Table 1. The orange bar below each bar plot denotes cGVHD cases, and the blue denotes controls. Legend describing the color scheme for bacterial taxa of interest is shown below panel D, as we have previously described.15 These color schemes were developed to highlight common taxonomic patterns in microbiota community in allo-HCT patients. Each genus is assigned to a distinct color shade derived from a basal color that is assigned to a higher-rank taxonomic group in the data set, facilitating the visualization of both genus-level and higher-rank taxonomic information. Abbreviations are as follows: (f), family; (g), genus; (o), order; (p), phylum. (D) Shotgun metagenomic sequencing of available day 100 stool samples. Linear discriminant analysis of effect size analysis shown, using a value of P ≤ .05 (n = 8 cGVHD; n = 37 controls). Green squares denote pathways relevant to SCFA metabolism.
To examine associations between microbiota composition and cGVHD, we identified 54 cGVHD patients from MSKCC with banked stool samples (collected between day −30 and day 365 relative to HCT). The majority of these patients had either classic cGVHD (n = 23; 42.5%) or an overlap syndrome (n = 23; 42.5%), with the minority developing a late-onset aGVHD syndrome (n = 8; 14.8%). The overlap subtype of cGVHD is characterized by the simultaneous presence of the clinical features of aGVHD and cGVHD and has been associated with a poorer overall prognosis than classic cGVHD.31 Most cGVHD cases (29) were mild (53.7%), 9 cases were moderate (16.6%), and 8 were severe (14.8%); the 8 late-acute patients did not have severity recorded. The median time to onset of cGVHD was 184 days (range, 55-451 days), consistent with the reported median onset in the literature of 7.4 months (range, 0.8-45.1 months).32 To draw comparisons between patients who developed cGVHD and those who did not, we employed a case-control study design and randomly selected 171 controls (∼3 controls per case) with stool samples from our institutional database by matching for graft type (CD34-selected or unmodified bone marrow or peripheral blood stem cell grafts). The rate of preceding aGVHD was similar in the 2 groups (45.1% vs 55.1%; P = .36; subtype included in supplemental Table 2). Early exposure to broad-spectrum antibiotics was similar between groups (supplemental Table 3). We observed similar α-diversity patterns over time in the selected cGVHD patients and controls (Figure 1B; supplemental Table 4).
We next analyzed the microbiota composition of samples collected in the pretransplant, periengraftment, and peri-day 100 windows. Prior to allo-HCT, samples from both cases and controls were comparably diverse, and bacteria of the phylum Firmicutes dominated the community compositions (Figure 1C). Consistent with our previous data,18,29 we observed many dysbiotic microbial communities in the periengraftment period, as exemplified by expansion of the Enterococcus genus (shown in green), but these patterns were similar between cases and controls (monodomination incidences: 8/42 cases [19.0%], 38/144 controls [26.4%]; difference was nonsignificant using Fisher’s exact test [P = .68]), and we did not detect significant differences in α-diversity. In the peri-day 100 window, several patient samples remained dominated in a fashion similar to that seen periengraftment, while others reverted to a composition that was more similar to their pretransplant composition, but differences between cases and controls were not significant. A comparison of α-diversity values and β-diversity distances between the groups at the different time points of interest is provided in supplemental Tables 4 and 5.
Since cGVHD is a late complication of allo-HCT, we focused on the peri-day 100 time window to investigate features of the microbiota that may be associated with the development of cGVHD. All day 100 samples for these analyses were collected prior to the onset of cGVHD, but of note, among the 225 patients in this cohort, only 45 had samples available in the day 100 window (36 controls and 9 cases). We performed shotgun metagenomic sequencing on available stool samples, and multiple microbial metabolic pathways were enriched in the cGVHD patients, many of which relate to SCFA metabolism (marked in Figure 1D with green squares). This suggests that there may be functional differences in the microbiome of patients who go on to develop cGVHD compared with those who do not. To examine if this difference in encoded microbial metabolic capacity was correlated with differences in circulating SCFAs, we subjected peri-day 100 plasma samples from cGVHD patients (n = 10) and controls (n = 43) to metabolomic analysis. Of note, these samples were not matched to stool, and in fact only 3 cGVHD plasma samples and 13 control samples are from the same individuals who had stool samples available for analysis in this time window. Plasma concentrations of butyrate and propionate were both significantly lower in patients who went on to develop cGVHD compared with controls (Figure 2A). These molecules have been reported to promote differentiation of regulatory T cells in preclinical models,11,33 and butyrate is thought to protect mice from lethal GVHD due by enhancing the repair of damaged intestinal epithelial cells.9 Hexanoate concentration was also lower in patients who went on to develop cGVHD, and isobutyrate concentration was elevated in both of the transplanted patient groups compared with healthy controls.
SCFAs are produced in the GI tract as the end products of microbial fermentation by anaerobic taxa. Therefore, to understand what may be causing the changes in plasma SCFA in our patients, we reexamined the peri-day 100 stool samples, focusing on the 10 most-abundant taxa as well as taxa previously associated with immune dynamics in cancer patients, yielding a list of 14 genera (Figure 2B). Of note, the abundance of genus Faecalibacterium, previously associated with success of immune checkpoint blockade therapy25 and increased total white cell count after allo-HCT,26 was below the limit of detection in all peri-day 100 samples from patients who developed cGVHD (Figure 2B, arrow). To explore the association of gut microbiota taxa with cGVHD, we performed a Bayesian logistic regression (supplemental Figures 1 and 2). This revealed that high abundances of the genus Akkermansia (log-odds ratio, 1.0 [0.2, 1.8]; mean and 95% highest posterior density intervals [HDI95], respectively) were associated with cGVHD development, and, conversely, low abundance of the commensal obligate anaerobe Lachnoclostridium was associated with remaining cGVHD-free (−1.5, [−2.6, −0.4] HDI95; Figure 2C). Additionally, we detected associations of the genus Streptococcus with cGVHD (1.2, [−0.1, 2.5] HDI95) and the commensal genus Clostridium (−0.6 [−1.4, 0.2] HDI95) with the absence of cGVHD. The logistic regression model using the stool sample data predicted all cGVHD patients correctly (recall = 1) but also yielded 9 false positives. Accuracy and recall were significantly improved (compared with the null) by including microbiota information in form of 16S relative abundances (Figure 2D-E). The presence of the fermentatively active, butyrate-producing genera Lachnoclostridium, Clostridium, and, to a lesser degree, Faecalibacterium was associated with reduced incidence of cGVHD. To demonstrate the influence of the abundances of these commensal obligate anaerobe genera on the probability of cGVHD, we developed an additional predictive model (described in supplemental Methods). The predictive model demonstrated that as the abundance of Lachnoclostridium, Clostridium, and Faecalibacterium (Figure 2F-G; supplemental Figure 3A) approached 0 in a simulated microbiome, the predicted probability of cGVHD approached 1 (conversely for Akkermansia and Streptococcus, supplemental Figure 3B-C). These metagenomic and microbiota community composition data, combined with the independent plasma data, provide evidence that maintenance of butyrate producers within the GI is correlated with a lower risk of cGVHD; however, given the small study size, we sought to validate these findings in additional patients.
We assembled sets of peri-day 100 samples from 2 additional transplant centers: Duke (United States) and Regensburg (Germany) (demographics in supplemental Table 6). In contrast to the matched case and control design for the MSKCC data set, the evaluable samples from the additional 2 centers are cross-sectional in nature; ie, patients were selected for inclusion on the basis of the availability of paired stool and blood samples within the peri-day 100 window and without regard to their baseline clinical characteristics or their cGVHD outcomes. Initial analysis of stool sample composition was similar to the MSK data, and we did not observe differences in α-diversity or community composition (as assessed by PERMANOVA test of β-diversity distances) between cases and controls in these additional centers (supplemental Figure 4). The Bayesian logistic regression model predicted 8 out of 11 cGVHD cases in the Duke cohort but only 10 out of 23 cGVHD cases in the Regensburg cohort; neither result was statistically significant. The reduced accuracy and recall of our model in the other centers (compared with the MSKCC set) may in part be due to different treatment regimens and patient demographics (not all patients matched the case-control criteria used for MSKCC patients). We also analyzed circulating SCFAs in blood samples from these cross-sectional data sets and observed a similar trend as in MSK patients toward a lower concentration of butyrate in patients who went on to develop cGVHD in the Duke sample set (P = .07) and a significantly lower concentration of propionate (Figure 2H; P = .027). No differences were seen in the Regensburg patient group (supplemental Figure 5); however, these samples were serum, not plasma as in the MSKCC and Duke samples, and the concentrations were lower overall, which may explain the lack of observed difference between groups.
These data highlight that the lasting damage to the microbiome observed after HCT may be clinically significant. Within a matched case and control group, features of the GI microbiota are associated with subsequent cGVHD onset. The observation that higher circulating concentrations of SCFAs are associated with lower rates of cGVHD in 2 independent patient cohorts supports further exploration of the immunotherapeutic role of SCFAs within the gut lumen and systemic butyrate and propionate.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This research was supported by National Institutes of Health (NIH), National Cancer Institute MSKCC Cancer Center core grants P30 CA008748, R01-CA228358, R01-CA228308, R01-HL147584, and P01-CA023766 (M.R.M.v.d.B.); NIH, National Heart, Lung, and Blood Institute awards R01-HL125571 and R01-HL123340 (M.R.M.v.d.B.) and K08HL143189 (J.U.P.); NIH, National Institute on Aging Project 2 award P01-AG052359 (M.R.M.v.d.B.); NIH, National Institute of Allergy and Infectious Diseases awards U01 AI124275 (M.R.M.v.d.B.) and R01 AI032135, AI095706, and U01 AI124275 (E.G.P.); Tri-Institutional Stem Cell Initiative award 2016-013 (M.R.M.v.d.B.); The Lymphoma Foundation (M.R.M.v.d.B.); The Susan and Peter Solomon Divisional Genomics Program (M.R.M.v.d.B.); the Parker Institute for Cancer Immunotherapy at MSKCC (K.A.M., M.R.M.v.d.B., and J.U.P.); the Sawiris Foundation (J.U.P.); the Society of Memorial Sloan Kettering Cancer Center (J.U.P.); and Seres Therapeutics (M.R.M.v.d.B., J.U.P., J.B.S., A.L.C.G., A.G.C., A.E.S., and A.D.S.). J.S. is supported by NIH, National Institute of Allergy and Infectious Diseases grant U01 AI124275. K.A.M. wishes to acknowledge funding received from the Haematology Society of Australia and New Zealand, The American Australian Association, The Royal Australasian Society of Physicians, Endeavour Australia, and the Parker Institute for Cancer Immunotherapy.
Footnotes
The MSK Fecal Biobank contains >10 000 samples from >1000 allo-HCT recipients and has been used for many retrospective observational studies. There is overlap between the patients in this study and some of these prior papers, but none have examined cGVHD as a clinical outcome. Data sharing requests can be e-mailed to the corresponding author.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.A.M. designed the study, performed research, and wrote the manuscript; J.S. wrote the manuscript and performed Figure 2 analyses; A.L.C.G. and E.R.L. contributed to analyses in Figure 1; A.J.P., B.P.T., P.A.G., D.W., M.D.D., A.D., G.K.A., A.E.S., J.B.S., K.B.N., D.G.B., A.G.C., R.J.R., A.R., A.B., L.B., M.C., M.V.L., D.A.R., and N.C. contributed data and performed research; M.M., C.C., I.P., S.G., Y.T., E.G.P., M.-A.P., E.H., and D.M.P. contributed clinical data and expert review; S.M.D. provided statistical consultation; A.S., J.U.P., J.R.C., and J.B.X. contributed critical data analysis and expertise and edited the manuscript; and M.R.M.v.d.B. contributed to study design and the manuscript.
Conflict-of-interest disclosure: K.A.M. reports consulting with Karius. J.B.S., A.E.S., G.K.A., A.G.C., D.G.B., and P.A.G. report partial salary support from Seres Therapeutics. J.U.P. reports research funding and licensing fees from Seres Therapeutics and consulting with DaVolterra. I.P. has received research funding from Merck and serves on a data and safety monitoring board for ExCellThera. S.G. reports research funding from Amgen, Actinuum, Celgene, Johnson and Johnson, Miltenyi, and Takeda and serves on advisory boards for Amgen, Actinuum, Celgene, Johnson and Johnson, Jazz Pharmaceutical, Takeda, Novartis, Kite, and Spectrum Pharma. M.-A.P. reports honoraria from AbbVie, Bellicum, Bristol-Myers Squibb, Incyte, Merck, Novartis, Nektar Therapeutics, Omeros, and Takeda; serves on data and safety monitoring boards for Servier and Medigene and scientific advisory boards of MolMed and NexImmune; has received research support for clinical trials from Incyte, Kite/Gilead, and Miltenyi Biotec; and serves in a volunteer capacity as a member of the board of directors of the American Society for Transplantation and Cellular Therapy and Be The Match (National Marrow Donor Program), as well as on the Center for International Blood & Marrow Transplant Research Cellular Immunotherapy Data Resource Committee. M.R.M.v.d.B. has received research support from Seres Therapeutics; has consulted for, received honorarium from, or participated in advisory boards for Seres Therapeutics, Flagship Ventures, Novartis, Evelo, Jazz Pharmaceuticals, Therakos, Amgen, Magenta Therapeutics, WindMIL Therapeutics, Merck & Co., Acute Leukemia Forum, and DKMS Medical Council (board); has intellectual property licensing with Seres Therapeutics and Juno Therapeutics; and has stock options with Seres Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Kate A. Markey, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: markeyk@mskcc.org.
REFERENCES
- 1.Wingard J, Majhail N, Brazauskas R, et al. Long-term survival and late deaths after allogeneic hematopoietic cell transplantation. J Clin Oncol. 2011;29(16):2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacDonald K, Hill G, Blazar B. Chronic graft-versus-host disease: biological insights from preclinical and clinical studies. Blood. 2017;129(1):13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenq R, Taur Y, Devlin S, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21(8):1373-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peled J, Devlin S, Staffas A, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol. 2017;35(15):1650-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shono Y, Docampo M, Peled J, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8(339):339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill G, Olver S, Kuns R, et al. Stem cell mobilization with G-CSF induces type 17 differentiation and promotes scleroderma. Blood. 2010;116(5):819-828. [DOI] [PubMed] [Google Scholar]
- 7.Schirmer M, Smeekens SP, Vlamakis H, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167(4):1125-1136.e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163(2):367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathewson N, Jenq R, Mathew A, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease [published correction appears in Nat Immunol. 2016;17(10):1235]. Nat Immunol. 2016;17(5):505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara H, Docampo M, Riwes M, et al. Microbial metabolite sensor GPR43 controls severity of experimental GVHD. Nat Commun. 2018;9(1):3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiko G, Ryu S, Koues O, et al. The colonic crypt protects stem cells from microbiota-derived metabolites [published correction appears in Cell. 2016;167(4):1137]. Cell. 2016;165(7):1708-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haak B, Littmann E, Chaubard J, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood. 2018;131(26):2978-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Huang J, Magruder M, et al. Butyrate-producing gut bacteria and viral infections in kidney transplant recipients: a pilot study. Transpl Infect Dis. 2019;21(6):e13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peled J, Gomes A, Devlin S, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N Engl J Med. 2020;382(9):822-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taur Y, Jenq R, Perales M, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagasia MH, Greinix HT, Arora M, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biol Blood Marrow Transplant. 2015;21(3):389-401.e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taur Y, Coyte K, Schluter J, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med. 2018;10(460):eaap9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franzosa E, McIver L, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai B, Ola M, Rolling T, et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med. 2020;26(1):59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colosimo DA, Kohn JA, Luo PM, et al. Mapping interactions of microbial metabolites with human G-protein-coupled receptors. Cell Host Microbe. 2019;26(2):273-282.e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oksanen J, Blanchet FG, Friendly M, et al. vegan: Community Ecology Package; R package version 2.5-6. https://CRAN.R-project.org/package=vegan. Accessed 24 September 2019.
- 24.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo J, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382-396. [DOI] [PubMed] [Google Scholar]
- 25.Gopalakrishnan D, Koshkin V, Ornstein M, Papatsoris A, Grivas P. Immune checkpoint inhibitors in urothelial cancer: recent updates and future outlook. Ther Clin Risk Manag. 2018;14:1019-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schluter J, Peled JU, Taylor BP, et al. The gut microbiota influences how circulating immune cells in humans change from one day to the next [published online 21 November 2019]. bioRxiv. doi:10.1101/618256. [Google Scholar]
- 27.Salvatier J, Wiecki T, Fonnesbeck C. Probabilistic programming in Python using PyMC3. PeerJ Comput Sci. 2016;2:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taur Y, Xavier J, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55(7):905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morjaria S, Schluter J, Taylor B, et al. Antibiotic-induced shifts in fecal microbiota density and composition during hematopoietic stem cell transplantation. Infect Immun. 2019;87(9):e00206-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pidala J, Vogelsang G, Martin P, et al. Overlap subtype of chronic graft-versus-host disease is associated with an adverse prognosis, functional impairment, and inferior patient-reported outcomes: a Chronic Graft-versus-Host Disease Consortium study. Haematologica. 2012;97(3):451-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora M, Cutler C, Jagasia M, et al. Late acute and chronic graft-versus-host disease after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2016;22(3):449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu J, Kubota T, Takada E, et al. Propionate-producing bacteria in the intestine may associate with skewed responses of IL10-producing regulatory T cells in patients with relapsing polychondritis. PLoS One. 2018;13(9):e0203657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.