Abstract
Cold treatment (vernalization) is required for winter crops such as rapeseed (Brassica napus L.). However, excessive exposure to low temperature (LT) in winter is also a stress for the semi-winter, early-flowering rapeseed varieties widely cultivated in China. Photosynthetic efficiency is one of the key determinants, and thus a good indicator for LT tolerance in plants. So far, the genetic basis underlying photosynthetic efficiency is poorly understood in rapeseed. Here the current study used Associative Transcriptomics to identify genetic loci controlling photosynthetic gas exchange parameters in a diversity panel comprising 123 accessions. A total of 201 significant Single Nucleotide Polymorphisms (SNPs) and 147 Gene Expression Markers (GEMs) were detected, leading to the identification of 22 candidate genes. Of these, Cab026133.1, an ortholog of the Arabidopsis gene AT2G29300.2 encoding a tropinone reductase (BnTR1), was further confirmed to be closely linked to transpiration rate. Ectopic expressing BnTR1 in Arabidopsis plants significantly increased the transpiration rate and enhanced LT tolerance under freezing conditions. Also, a much higher level of alkaloids content was observed in the transgenic Arabidopsis plants, which could help protect against LT stress. Together, the current study showed that AT is an effective approach for dissecting LT tolerance trait in rapeseed and that BnTR1 is a good target gene for the genetic improvement of LT tolerance in plant.
Keywords: rapeseed, associative transcriptomics, photosynthetic gas exchange parameter, tropinone reductase, alkaloid
Introduction
Rapeseed (Brassica napus L.) is one of the major oil crops worldwide, with an average annual cropping area of 35.3 million hectares producing 72.8 million tons of seeds in the past five years (http://www.fao.org/faostat/). Meal cake, the byproduct of rapeseed is also an important source of protein-rich feed for livestock (Wanasundara et al., 2016). Due to the agronomic importance of this oil crop, there is a great interest to boost its yield via genetic improvement of major agronomic traits.
The winter type rapeseed is mainly grown in Europe, which requires strong vernalization and is cold tolerant (O’neill et al., 2019). However, the semi-winter type rapeseed grown in China only needs moderate or weak vernalization, and excessive exposure to low temperature (LT) stress in winter will lead to plant damage at vegetative stage and finally cause yield loss (Liao and Guan, 2001; Zhang et al., 2015; O’neill et al., 2019). Yangtze River basin is the major area for growing semi-winter rapeseed, which accounts for at least 80% of the nation’s total production (Tian et al., 2018). The rapeseed is usually sown in early October shortly after the harvest of rice in this area (Cong et al., 2019). However, in recent years, the delay of rice harvest usually lead to the postpone of rapeseed sowing until late October or early November, which results in poor germination and seedling establishment due to LT (Luo et al., 2019). The biomass of rapeseed seedling is also significantly reduced at overwintering stage, and thus is more susceptible to LT stresses, i.e. chilling (0–15°C) or freezing (<0°C) (Sage and Kubien, 2007; Zhang et al., 2012; Zhang et al., 2016). Moreover, delay of floral initiation and floral bud differentiation processes (Luo et al., 2018) and decrease of effective pod number, pod length, and seed yield (Ozer, 2003) were observed in the late-sowing rapeseed. To cope with LT stresses, plants have evolved several elaborate regulatory mechanisms; among these, balancing or coordinating the photosynthetic processes could be a critical one (Leister, 2019).
It has been established that light-harvesting complex II (LHCII) proteins in higher plants can facilitate their adaption to external biotic or abiotic environmental stresses such as drought stress or blast fungus infection (Andersson et al., 2003; Ganeteg et al., 2004; Xu et al., 2012; Liu et al., 2019b), and the positive function of LHCBs in abscisic acid (ABA) mediated signaling pathway is repressed by WARY40 (Xu et al., 2012; Liu et al., 2013). LT stress induces the accumulation of transcript encoding heat-shock proteins (HSPs), and the persistence of HSPs can enhance the chilling tolerance of tomato fruit (Sabehat et al., 1996; Ding et al., 2001). In Arabidopsis, HSP21 protects the photosynthetic electron transport chain against the deleterious effects imposed by heat stress (Zhong et al., 2013; Bernfur et al., 2017). However, it is unclear whether HSPs function similarly in rapeseed to alleviate injury from LT stress. The other observations have mechanistically described how the photosynthetic organisms maintain their PSII function under stressful conditions with continuing or fluctuating light (Liu et al., 2019a). For instance, the chloroplast protein HHL1 forms a complex with LQY1 to repair and re-assemble PSII, which in turn helps overcome excessive light stress (Jin et al., 2014). In rapeseed, CBF/DREB transcription factors appear to have important roles in maintaining stronger photosynthetic efficiency and higher Calvin circulating enzyme activity under LT conditions (Savitch et al., 2005).
To date, several genetic studies have been reported for quantitative trait loci (QTLs) mapping of photosynthesis (De Miguel et al., 2014; Li et al., 2014; Li et al., 2016; Liu et al., 2017; Oakley et al., 2018; Basu et al., 2019) and LT tolerance in different plant species (Jha et al., 2017). However, very few QTLs have been identified in Brassica species (Ge et al., 2012; Yan et al., 2015). The temperature causing 50% of the maximal damage (LT50) has been regarded as a good index for evaluating LT tolerance (Hincha et al., 1987; Steponkus et al., 1990), which is also significantly correlated with net photosynthesis rate (An) in rapeseed (Urban et al., 2013). Therefore, photosynthetic gas exchange parameters such as An are a suitable index for the evaluation of LT tolerance in rapeseed that can facilitate the follow-up genetic study.
Photosynthesis plays an indispensable role in ensuring adequate energy supply throughout the plant lifecycle. Therefore, enhancing photosynthetic efficiency has been a commonly adopted strategy for crop yield improvement (Long et al., 2006; Lawson et al., 2012; Evans, 2013). In rice, a new photorespiratory bypass was assembled by over-expressing OsGLO3, OsOXO3, and OsCATC genes in the chloroplast, which resulted in obvious increases in photosynthetic efficiency, biomass, and grain yield (Shen et al., 2019). In rapeseed, photosynthetic efficiency has a notable effect on yield, oil content, and fatty acid composition (Ju and Li, 2012; Wang et al., 2015). However, the utilization rate of light energy in rapeseed is only 0.615% to 1.056%, which is much lower than that in rice, wheat or soybean (Zhang et al., 2017a). Therefore, it is possible to further improve the yield by enhancing photosynthetic efficiency in rapeseed.
During the long history of evolution, plants appear to overcome abrupt or mild temperature stresses in winter through a series of changes at molecular, cellular, physiological, and biochemical levels (Zhang et al., 2017b). Alkaloid is one of the major secondary metabolites that is inducible under unfavorable conditions, especially drought stress (Selmar and Kleinwachter, 2013). Heavy metals can also promote the accumulation of alkaloid in Catharanthus roseus L. (Srivastava and Srivastava, 2010). The short-chain dehydrogenase/reductase (SDR) protein, which belongs to the NAD(P)-binding Rossmann-fold superfamily, functions in the biosynthesis of benzylisiquinoline alkaloids (i.e. morphine, codeine) and tropane-derived alkaloids such as scopalamine, atropine, and cocaine. Tropinone reductases (TRs) are a group of SDR proteins which play key roles as a branch point in the biosynthesis pathway of tropane alkaloids (Tonfack et al., 2011). Hence, a study on TRs could help understand how alkaloids function in response to different stresses.
Associative transcriptomics (AT) strategy, which combines association mapping and transcriptome, has greatly facilitated the genetic dissection of complex traits (Bazakos et al., 2017). Considerable progress has been achieved by AT in allopolyploidy crops, such as oilseed rape and wheat, which provided a large number of causative genes or functional markers for molecular marker-assisted breeding. For instance, a transcription factor (HAG1) was identified in rapeseed by AT, which plays an indispensable role in the synthesis of aliphatic glucosinolates (Harper et al., 2012). With AT platform, the genetic studies of many other complex traits in rapeseed were also reported, including homeostasis of nitrate, phosphate, and sulfate anions (Koprivova et al., 2014), calcium and magnesium accumulation (Alcock et al., 2017), lodging resistance (Miller et al., 2018), clubroot resistance (Hejna et al., 2019), erucic acid, and tocopherol (vitamin E) isoform accumulation in seeds (Havlickova et al., 2018), and leaf nutrition concentration (Alcock et al., 2018). Recently, the power of AT was further enhanced by using a much larger panel comprising 383 rapeseed accessions (Havlickova et al., 2018). In bread wheat, two causative genes underlying stem strength variation have also been detected by AT (Miller et al., 2016). Despite the above efforts, the AT approach has not yet been applied to photosynthetic related traits under LT conditions in rapeseed.
The present study aims to identify candidate genes associated with photosynthetic gas exchange parameters including An, stomatal conductance to water vapor (Gsw), internal CO2 concentration (Ci) and transpiration rate/evapotranspiration (E) in rapeseed by AT. Twenty-two candidate genes were obtained, and one was functionally validated. Ectopic expressing tropinone reductase (BnTR1) in Arabidopsis can significantly enhance transpiration rate and LT tolerance, implying its great potential for the genetic improvement of LT tolerance in plant.
Materials and Methods
Plant Materials
A panel comprising 123 rapeseed accessions was used for association study, which is available from the John Innes Centre, Norwich, UK ( Supplementary Table S1 ) (Havlickova et al., 2018). Within this panel, there are 37 winter type, 32 spring type, 47 semi-winter type, and 7 unclassified rapeseed accessions. The panel was sown on 28th Oct in 2016 in Wuhan (114.30°E, 30.57°N), China. All accessions were planted using a completely randomized block design with three replications. The temperature was recorded during the field experiments ( Supplementary Figure S1 ). Compared with those sown on normal occasion (28th Sept in 2016), the late-sown (i.e. 28th Oct) rapeseed seedlings were subjected to LT stress during winter.
Determination of Photosynthetic Gas Exchange Parameters
The fourth true leaf of each of the 123 rapeseed accessions was chosen for the measurement of photosynthetic gas exchange parameters including An, Gsw, Ci, and E in the open field at the 60-d-old seedling stage. Two independent plants of one accession from each block were measured by LI-6400 photosynthesis equipment (Li-Cor 6400; Li-CorInc, Lincoln, NE, USA) as described previously (Yan et al., 2015). The measurements were performed on a sunny day from 27th December to 30th December with a maximum temperature of 9°C in daytime and the lowest temperature of −1°C at night. The phenotypic data were collected from three blocks as biological replications ( Supplementary Table S2 ). Broad-sense heritability was estimated according to a previous study (Kaler and Purcell, 2019).
The photosynthetic gas exchange parameters of Arabidopsis wild type (WT) and transgenic seedlings (ecotype Columbia) were measured on the second functional leaf with the light intensity of 800 μmol m−2 s−1 and CO2 concentration of 400 μl L−1. All of the Arabidopsis plants were grown in the greenhouse (16-h-light/8-h-dark) with the light intensity of 120 μmol m−2 s−1 at 23°C. Each leaf was measured three times as technical replications, and five independent plants from each line were measured as biological replications ( Supplementary Table S2 ).
Genome-Wide Association Study
The association panel and the procedure of AT analysis have been reported in detail previously (Havlickova et al., 2018). In brief, RNA-Seq data were generated from young leaves of the association panel harvested 21 d after sowing under 16-h-light (20°C)/8-h-dark (14°C) glasshouse conditions. The transcriptome data were mapped onto the developed ordered Brassica A and C pan-transcriptomes (He et al., 2015) and resulted in a set of 355,536 SNPs and RPKM values for 116,098 CDS models. Following the removal of SNP markers with minor allele frequencies below 0.01, a total of 256,397 SNPs were retained (http://www.yorknowledgebase.info/) and used as marker input for the Associative Transcriptomics analysis as previously described (Harper et al., 2012; Lu et al., 2014). The current study adopted a compressed mixed linear model including both fixed and random effects according to a previous method (Lipka et al., 2012). The P-values (–log10 converted) for all SNPs were plotted against their physical position in the “pseudo-molecules” to produce a Manhattan plot. The Bonferroni significance threshold was set as P = 3.9 × 10−6 (1/256397) (Duggal et al., 2008). Allelic effects of all candidate SNPs were calculated according to a previous study (Prado et al., 2017); a positive effect indicates that the allele increases the trait value, whereas a negative effect indicates that the alternative allele increases the trait value.
The transcript level was quantified as reads per kb per million aligned reads (RPKM) across the panel. After filtering (RPKM ≤ 0.4), a total of 53,889 gene expression marker (GEM) was obtained. The GEM was regarded as the dependent variable and trait data as the independent variable (Wood et al., 2017). The fixed effect linear model was performed to assess the relationship between gene expression level and the traits (Alcock et al., 2017). The P-value for each GEM was converted (-log10 P) and plotted against its physical position to generate a Manhattan plot. The Bonferroni significance threshold was set at P = 1.85 × 10−5 (1/53889).
Growth Conditions and Stress Treatments
To determine the gene function of BnTR1 on Arabidopsis under freezing conditions, the seeds of WT and transgenic Arabidopsis plants were germinated on 1/2 MS medium and 1/2 MS medium plus 30 μg/ml hygromycin, respectively. After 1 week growth at 23°C (16-h-light/8-h-dark), healthy seedlings with uniform sizes were transplanted into 8×8 cm pots (four plants per pot). Then, the 21-d-old plants were transferred into a growth chamber at −4°C for 4 h after 24 h of cold acclimation and recover at 23°C (16-h-light/8-h-dark). Six pots from each transgenic line or WT plants were used to investigate the survival rate 3 d after recovery. The leaves were sampled for physiological and biochemical measurements.
To determine the effects of alkaloid on Arabidopsis and rapeseed seedlings, WT plants of Arabidopsis were firstly grown under the same conditions as above. Before the freezing treatment, a dosage of 0, 10, and 30 nmol alkaloid (atropine) per seedling was added to the soil for Arabidopsis, while a dosage of 0, 50, and 150 nmol per seedling was added to the soil for rapeseed accession Zhongshuang 11 (ZS11) according to the previous study (Hara and Kurita, 2014). After inoculation overnight, the 21-d-old Arabidopsis plants were transferred into a growth chamber at −4°C for 4 h, the three-leaf-stage rapeseed plants were transferred into the growth chamber at −4°C for 4 h (Yan et al., 2019). The survival rate was investigated in six pots 3 d after recovery ( Supplementary Table S2 ).
To investigate the expression of candidate genes under LT stress conditions, six rapeseed accessions showing extreme photosynthetic efficiency (i.e. Sv706118, Kajsa, Callypso, Libritta, Gefion, and Jupiter; Supplementary Table S1 ) were used for expression analysis of candidate genes. The four-leaf seedlings were transferred into the plant growth chamber with −4°C for 4 h (Yan et al., 2019). The leaves were sampled before, and after freezing treatment, and then used for RNA extraction and molecular analysis. The detailed information was listed in Supplementary Table S2 .
Physiological and Biochemical Measurements
To determine the effects of BnTR1 on Arabidopsis at physiological and biochemical levels under LT stress conditions, the seedlings of transgenic and WT Arabidopsis plants were treated under freezing conditions (−4°C for 4 h) and, the leaves of 21-d-old seedlings were sampled at three time-point (i.e. before freezing treatment, after freezing treatment, and recovery for 3 d at 23°C) for measuring physiological and biochemical characteristics, including Fv/Fm, electrolyte leakage, 3,3′-diaminobenzidine (DAB) staining, proline content, soluble sugar content, reactive oxygen species (ROS) scavenging enzymes activity, H2O2 content, alkaloid content. All measurements were performed with at least three biological replications; detail information for experimental design and plant materials used was listed in Supplementary Table S2 .
The Fv/Fm measurement was performed using the second functional leaves before and after the freezing treatment. The leaves were firstly immersed in 1% agarose overnight avoiding of the dark, then the chlorophyll measurement (Fv/Fm) was measured using the modulated chlorophyll fluorescence instrument (PAM-2500; Walz) as previously reported (Lv et al., 2017).
The electrolyte leakage measurement was performed according to the previous study (Lv et al., 2016). Briefly, six leaves from six were cut and immersed in 8 ml of double-distilled H2O in a 10-ml tube. After shaking overnight, the electrolyte leakage was measured using a model DDS-IIA device (Leici Instrument) as R1; it was measured again and recorded as R2 after boiling at 95°C in a water bath for 15 min and cooling down. The relative electrolyte leakage was calculated as a ratio of R1/R2.
The proline and soluble sugar contents were measured using the kits from Beijing Solarbio Science & Technology as described before (Yan et al., 2019). In brief, 0.1 g fresh tissue was powdered and incubated in 1 ml 3% sulfosalicylic acid (for proline) or ddH2O (for soluble sugar). After centrifuging, 400 μl supernatant was mix with other reaction buffers and incubated at 95°C in a water bath for 15 min, then the absorbance was measured using MULTISCAN FC (Thermo Scientific).
The ROS scavenging enzymes activity was measured by commercial kits according to the manufacturer’s instruction (Beijing Solarbio Science & Technology) with minor modification (Yan et al., 2019). 0.1 g fresh tissue was powdered using 1 ml 0.05 mol/L PBS buffer (pH 7.8). The supernatant was obtained after centrifuging at 8,000g for 10 min at 4°C and used for superoxide dismutase (SOD) activity, peroxidase (POD) activity, catalase (CAT) activity measurement using MULTISCAN FC (Thermo Scientific).
DAB staining was performed as previously described (Ning et al., 2010; Zhang et al., 2011). The fourth functional leaf of each plant was sampled and infiltrated in 0.1 mg/ml 3,3′-diaminobenzidine liquid (50 mM Tris-acetate buffer, pH 5.0). After incubation overnight at 25°C in the dark, the stained leaves were photographed after removing the chlorophyll by absolute ethanol. The H2O2 content was quantified according to the instruction of the kit (Beijing Solarbio Science & Technology) (Yan et al., 2019).
The total alkaloid was extracted as described previously (Zhang et al., 2004; Chen et al., 2013). The freeze-dried leaves were powdered; 0.1 g powder was homogenized overnight with 1.0 ml 70% aqueous methanol at 4°C. Following centrifugation at 10,000 g for 10 min at 4°C, the extracts were absorbed (CNWBOND Carbon-GCB SPE Cartridge, 250 mg, 3 ml; ANPEL) and filtrated (SCAA-104, 0.22 μm pore size; ANPEL). Next, the total alkaloid content was determined using the alkaloid ELISA kit (Hiton) according to the instructions.
RNA Extraction and Gene Expression Analyses
Total RNA was extracted from the Arabidopsis or rapeseed seedlings with TransZol reagent (Trans) and converted to the first-strand cDNA using the EasyScript®One-Step cDNA Synthesis SuperMix (Trans). The Quantitative Real-time PCR (qRT-PCR) was performed using Power SYBR®Green PCR Master Mix according to the manufacturer’s instructions on a StepOnePlusReal-Time PCR System (Applied Biosystems). The primers used for expression analysis of candidate genes either detected by AT approach or involved in the known biosynthetic pathway of alkaloid were listed in Supplementary Table S3 . The relative expression level was determined as previously described (Livak and Schmittgen, 2001).
Vector Construction and Gene Transformation
To generate transgenic lines over-expressing candidate gene (BnTR1), the coding sequence (CDS) of LOC106445422 was amplified from a rapeseed variety ZS11 and ligated into vector pCAMBIA1300 driven by a tobacco cauliflower mosaic virus 35S promoter (CaMV35S). The construct was introduced into Arabidopsis variety Columbia (Col) by Agrobacterium-mediated transformation (Zhang et al., 2006). Three T4 homozygous lines (L1, L3, L5) significantly over-expressing BnTR1 were obtained by screening at 30 μg/ml hygromycin. The primers used for vector construction were listed in Supplementary Table S3 .
Statistical Analyses
Statistical analyses were conducted with Microsoft Excel (2003) and SPSS (version 22.0) software using one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test or two-tailed Student’s t-test. All data were presented as the means ± standard error (SE) based on three replicates. P<0.05 and P<0.01 were considered statistically significant and highly significant, respectively.
Results
Phenotypic Variation of Photosynthetic Gas Exchange Parameters
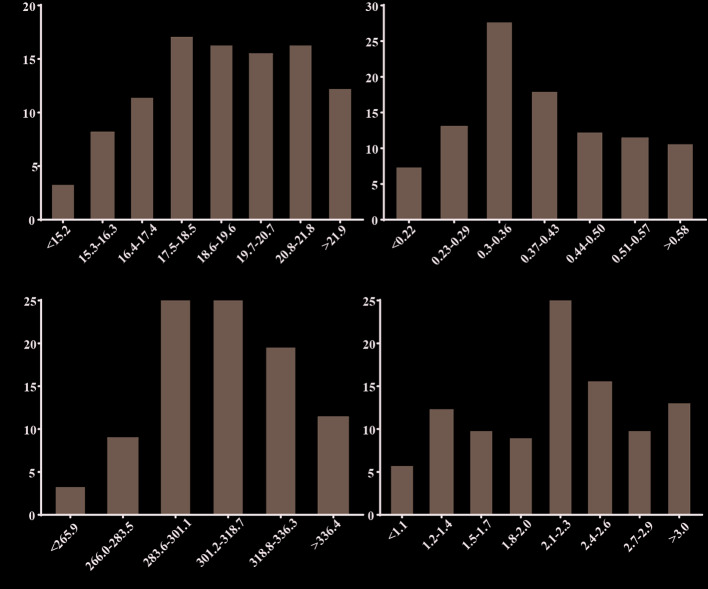
A field-grown rapeseed association panel experienced long-term LT stress in winter ( Supplementary Figure S1 ). The photosynthetic gas exchange parameters such as An, Gsw, Ci, and E were measured since they can reflect photosynthetic efficiency for a plant. Substantial variations for the four traits were observed in 123 rapeseed accessions ( Figure 1 ). An varied from 12.87 to 24.02 with a mean of 19.29 μmol (CO2) m−2 s−1. Similarly, Gsw ranged from 245.21 to 383.28 μmol (CO2) mol−1 and E from 0.96 to 3.75 mmol (H2O) m−2 s−1. Gsw was the most variable trait since it has the largest coefficient of variation (0.69), with a minimum of 0.16 and a maximum of 2.53 mol (H2O) m−2 s−1; the range of broad-sense heritability varied from 49.49% to 68.91% ( Supplementary Table S4 ). Moreover, the four traits in the association panel were positively correlated with each other (P ≤ 0.01) ( Supplementary Table S5 ). For instance, there was a strong correlation between An and E (r = 0.785, P<0.01).
Figure 1.
Phenotypic variation of photosynthetic gas exchange parameters in 123 rapeseed accessions. Trait definition: Net photosynthesis rate (An), Stomatal conductance to water vapor (Gsw), Internal CO2 concentration (Ci), Transpiration rate (E).
Associative Transcriptomics for Photosynthetic Gas Exchange Parameters
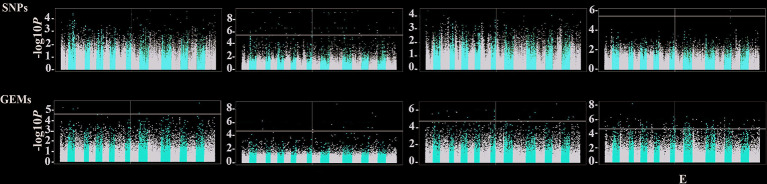
To identify genomic regions controlling photosynthetic related traits, AT was performed in rapeseed. Using the mixed linear model, a total of 201 significant SNPs were detected, which originated from 148 CDSs ( Figure 2 ). Unexpectedly, most of the significant CDSs were associated with Gsw trait, while only one was related to E trait. The detail results including physical positions, P-values, allelic effects were summarized in Supplementary Table S6 . For GEM analysis, a total of 145 CDSs above the corrected Bonferroni thresholds were identified ( Figure 2 ). Of these, a respective of 5, 10, 20, and 110 CDSs were detected for An, Gsw, Ci, and E. The detailed information, including physical positions and P values for all significant GEMs, was listed in Supplementary Table S7 .
Figure 2.
Manhattan plots for AT analysis in 123 rapeseed accessions. Manhattan plots from left to right, represented for An, Gsw, Ci, and E using SNPs (upper section) and GEMs (bottom section), respectively. The -log10 (P values) were plotted against the position of the SNPs or GEMs on 19 chromosomes of Brassica napus. The black line represents the -log10 (P values) converted Bonferroni significance threshold for SNP (5.41) and GEM (4.73), respectively.
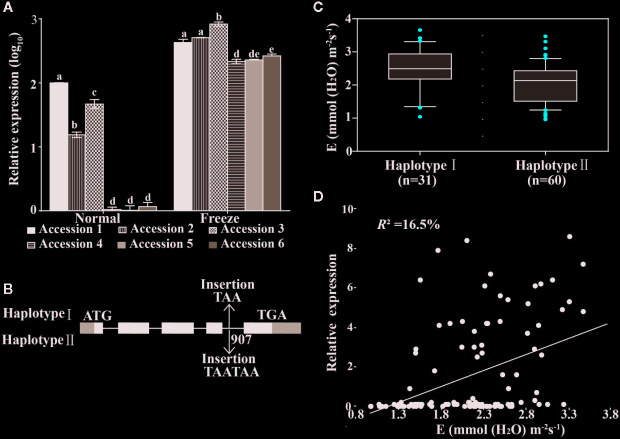
By annotating all the above significant CDS in public database TAIR (https://www.arabidopsis.org/), the present study shortlists the number of candidate genes to only 22 that putatively involved in photosynthesis or LT stress response ( Supplementary Table S8 ). To verify their roles in rapeseed with or without LT stress, six accessions from the association panel were selected; Sv706118, Kajsa, and Callypso (Accessions 1–3) exhibited higher photosynthesis efficiency and were tolerant to freezing stress, while Libritta, Gefion, and Jupiter (Accessions 4–6) presented lower photosynthesis efficiency and were sensitive to freezing stress ( Supplementary Table S1 , Supplementary Figure S2 ). The gene expression profiles were investigated by qRT-PCR in the six accessions under freezing conditions. Results showed that all of these genes exhibited a significantly different expression level in the six accessions, suggesting that these genes indeed involved in freezing stress response. Cab026133.1 seemed to have a much higher expression level in the three tolerant accessions (Accessions 1–3) with or without freezing stress ( Figure 3A ); so did Cab011968.1, Cab022014.2, and Cab007526.2, an ortholog of inorganic carbon transport protein (AT1G70760.1), bZIP transcription factor (AT5G28770.3) and citrate synthase 2 (BnaA10g24440D), respectively. An opposite trend was observed for Bo5g017460.1 and Cab008128.1, an ortholog of F-box family protein (AT2G32560.1) and dehydroascorbate reductase (AT5G16710.1), respectively ( Supplementary Figure S3 ).
Figure 3.
Expression and allelic variation of BnTR1 in rapeseed. (A) Expression analysis of BnTR1 (homolog of Cab026133.1) in six accessions corresponding to Hap 1 and Hap 2 under freezing conditions. The name of accessions 1 to 6 was Sv706118, Kajsa, Callypso, Libritta, Gefion, and Jupiter, respectively. ACTIN gene was used as an internal control. Bars indicate the SE of three biological replicates. Different letters indicate significant differences at P< 0.05 (one-way ANOVA with Tukey’s multiple comparisons test). (B) Correlation analysis between Transpiration rate (E) value and expression level of Cab026133.1 in the association panel (n=123). R2 indicates the coefficient of determination in linear regression. (C, D) Allelic variations at BnTR1 formed two main haplotypes and their effects on E value.
Selection and Characterization of Candidate Gene
Cab026133.1 was selected for further analysis because it not only exhibited the highest P-value (6.33 × 10−9) for E trait in GEM analysis ( Supplementary Table S8 ) but also highly expressed in LT tolerant accessions ( Figure 3A ). Besides, the expression of Cab026133.1 (presented as RPKM) across the rapeseed panel was positively correlated with E level (r=0.406, P < 10−3) and accounted for 16.5% of trait variation ( Figure 3B ). The ortholog of Cab026133.1 in Arabidopsis (AT2g29300) encodes an SDR protein involved in the oxidation-reduction process of secondary metabolites, such as phenols, isoprene, and alkaloid (Selmar and Kleinwachter, 2013). SDR proteins are classified into six subfamilies, and the tropinone reductase subfamily belongs to the major route of alkaloid biosynthesis (Tonfack et al., 2011). The alignment of amino acid sequence clearly illustrated that LOC106445422 shared 87.3% similarity with Cab026133.1 and 57% with CoTR, a known tropinone reductase in Cochlearia officinalis (Brock et al., 2008). LOC106445422 displayed typical SDRs motifs (Gly-X3-Gly-X-Gly) and four conserved residues that form the catalytic tetrad NSYK (N127, S155, Y168, K172) ( Supplementary Figure S4 ). The current study name LOC106445422 as BnTR1 and used as the candidate gene for the follow-up studies.
To assess the effect of allelic variation on BnTR1 in the rapeseed association panel, the genomic region covering the whole gene as well as the 2-kb promoter region was amplified. One TAA/TAATAA insertion was detected in the fourth intron that formed two major haplotypes, i.e. Haplotype I (with TAA insertion) and Haplotype II (with TAATAA insertion) ( Figure 3C ). Haplotype I (n=31) displayed significantly higher E value than Haplotype II (n=60) (P=5.43 × 10−4) ( Figure 3D ). Besides, the LT tolerant Accessions 1 to 3 were determined as Haplotype I while sensitive Accessions 4 to 6 as Haplotype II at BnTR1 locus ( Supplementary Table S1 ). The expression level of BnTR1 in Accessions 1 to 3 was also significantly higher than that in accessions 4 to 6 with or without stress treatment ( Figure 3A ). Therefore, it was evident that E variation may be attributed to expression or allelic variation at BnTR1, which was expressed almost in all tissues of rapeseed at both vegetative and reproductive stages ( Supplementary Figure S5 ).
Ecotopic Expressing BnTR1 Enhances Freezing Tolerance in Arabidopsis
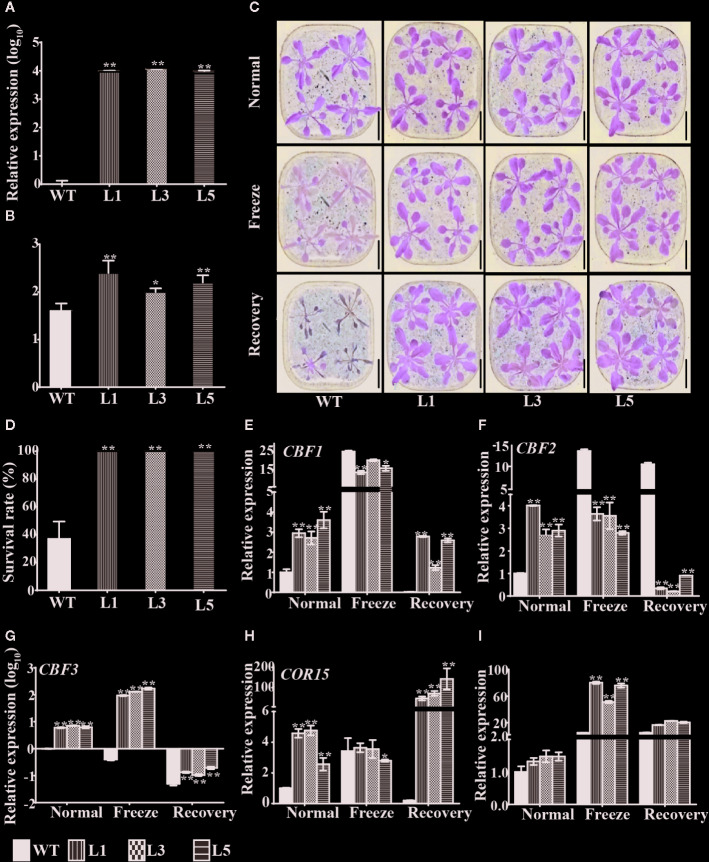
To analyze gene function, three independent Arabidopsis lines (L1, L3, L5) ectopic expressing BnTR1 were generated. All transgenic lines showed an increased expression level of BnTR1 in comparison to the WT plants ( Figure 4A ). At the seedling stage, E value of the transgenic lines was much higher than that of WT plants ( Figure 4B ), thus confirming that BnTR1 controls transpiration rate. Since photosynthetic gas exchange parameters are significantly correlated with cold tolerance (Urban et al., 2013), it was speculated that BnTR1 also involves in LT stress. To test this hypothesis, seedlings were treated at −4°C for 4 h and then recover at 23°C. All of the transgenic lines were shown to be freezing-tolerant because there was no obvious syndrome, while the WT plants were wrinkled and hydrophanous ( Figure 4C ). After recovery for 3 d at 23°C, all transgenic plants survived, but 62% of the WT plants died ( Figure 4D ). These results strongly suggested that BnTR1 enhanced the freezing tolerance of Arabidopsis plants.
Figure 4.
BnTR1 confers freezing tolerance in Arabidopsis. (A) Expression analysis of the BnTR1 transgenic plants (L1, L3, L5) and WT plants under normal condition (i.e. 23°C). (B) Investigation of transpiration rate (E) value in the BnTR1 transgenic lines and WT plants under normal conditions. (C) Performance of the transgenic lines and WT plants before and after freezing treatment (−4°C for 4 h). Scale=2 cm. (D) Survival rates of the transgenic lines and WT plants after freezing treatment. (E–I) Relative expression levels of CBF1 (E), CBF2 (F), CBF3 (G), COR15 (H), RD29A (I) in the transgenic lines and WT plants before and after freezing stress with the Arabidopsis ACTIN gene used as an internal control. Normal represents 23°C, freezing treatment represents 4 h at −4°C, recovery represents 3 d of recovery at 23°C. Bars indicate the SE of three biological replicates. Significant differences are determined by Student’s t-test (*P < 0.05, or **P < 0.01).
To assess whether C-repeat-binding factors (CBFs) contributed to the enhanced freezing tolerance of the transgenic lines, the current study measured the relative expression level of CBFs in transgenic and WT plants after freezing stress ( Figures 4E–G ). In the transgenic lines grown under normal conditions, expressions of CBFs were much higher than that in WT plants, indicating that they were markedly induced by BnTR1. Moreover, the expression of CBF1 and CBF3 was significantly up-regulated and CBF2 down-regulated by BnTR1 during freezing treatment in comparison with those of normal conditions. The current study further examined the expression level of CBFs-targeted cold-responsive genes (COR genes) ( Figures 4H, I ). As expected, the transcript levels of COR genes (namely COR15 and RD29A) in the transgenic lines were highly induced when compared with that in WT plants. These results indicated that BnTR1 influences CBF regulon in the stress-signaling pathway to control freezing tolerance.
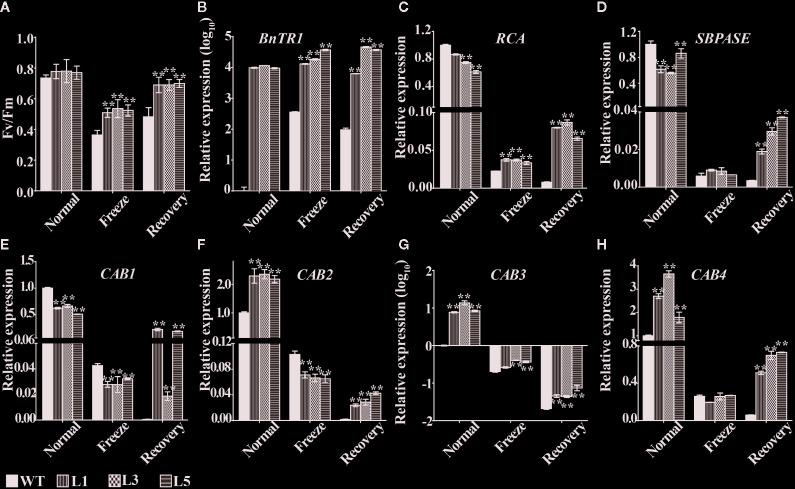
The chlorophyll fluorescence parameter Fv/Fm, an indicator for the potential maximum photosystem II (PSII) capacity of plants, has been widely used to determine the ability of tolerance to environmental stresses under laboratory conditions (Mishra et al., 2014; Thalhammer et al., 2014). Here, markedly higher Fv/Fm ratio was observed in leaves of the transgenic lines under freezing as well as normal conditions ( Figure 5A ). To further clarify whether the difference of photosynthetic capacity was caused by the excessive expression of BnTR1, the current study assessed the expression level of BnTR1 and other genes involved in photosynthetic processes such as RCA, SBPASE (for CO2 fixation or assimilation) and CAB1-4 (for light-harvesting) (Sun et al., 2017; Basu et al., 2019) ( Figures 5B–H ). Under normal conditions, the expression level of RCA, SPASE, and CAB1 were slightly reduced in the transgenic lines compared to WT plants, whereas CAB2, CAB3, and CAB4 were induced. The freezing treatment led to a notable suppression of all genes; however, the expression of genes in the transgenic lines returned to a high level when freezing stress was removed.
Figure 5.
Variation of photosynthetic related traits and genes expression pattern in BnTR1 transgenic plants. (A) Fv/Fm ratio in the transgenic lines and WT plants under freezing stress conditions. (B–H) Relative expression levels of BnTR1 (B), RCA (C), SBPASE (D), CAB1 (E), CAB2 (F), CAB3 (G), CAB4 (H) in the transgenic lines and WT plants before and after freezing stress treatment with Arabidopsis ACTIN gene used as an internal control. L1, L3, L5 represent three independent homozygous lines of BnTR1 transgenic plants. Bars indicate the SE of three biological replicates. Significant differences are determined by Student’s t-test (*P < 0.05, or **P < 0.01).
BnTR1 Contributes to Cell Membrane Protection and Antioxidants
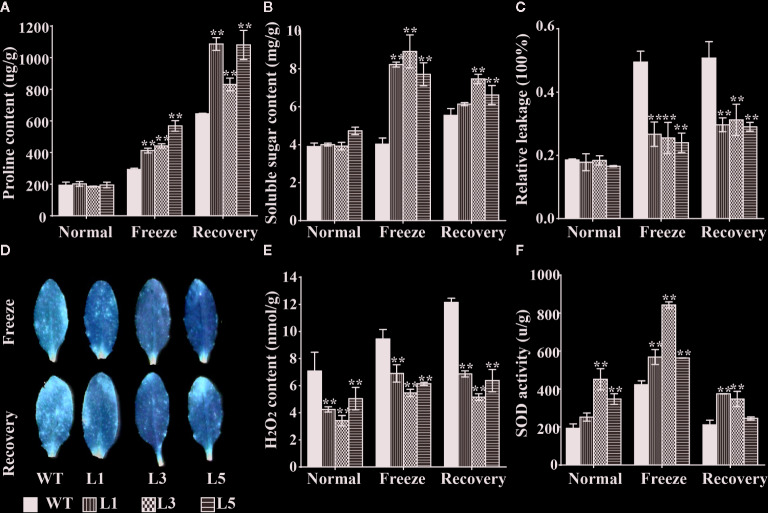
Altering the osmotic balance to maintain the integrity and stability of cell membrane is proposed to be an efficient way for plants adapting to the changing environments (Morsy et al., 2005; Valerio et al., 2011). To test this hypothesis, physiological and biochemical assays were carried out. Results showed that freezing treatment led to only 40% to 200% increase of proline content in WT plants but as high as 100% to 300% in transgenic plants, indicating that the transgenic plants expressing BnTR1 could accumulate more proline ( Figure 6A ). The soluble sugar content showed a similar pattern ( Figure 6B ). However, the electrolyte leakage increased more rapidly in WT plants than in transgenic lines ( Figure 6C ). These results implied that BnTR1 actively responded to freezing stress by maintaining cell membrane stability and osmotic balance.
Figure 6.
Physiological characterization of BnTR1 transgenic plants under freezing stress conditions. (A–F) Investigation of the proline content (A), soluble sugar content (B), relative leakage (C), DAB staining analysis (D), H2O2 content (E), SOD activity (F) in the transgenic lines and WT plants under freezing stress conditions. L1, L3, L5 represent three independent homozygous lines of BnTR1 transgenic Arabidopsis plants. Bars indicate the SE of three biological replicates. Significant differences are determined by the Student’s t-test (*P< 0.05, or **P < 0.01).
Antioxidants, which function in scavenging the reactive oxygen species (ROS), are generally considered as another effective element in defending abiotic stresses (Choudhury et al., 2017). To determine whether BnTR1 affects the antioxidant system, the accumulation of ROS was determined by DAB staining. The brown precipitate (H2O2) in WT was much larger than that in the transgenic lines ( Figure 6D ), indicating that WT plants had a higher level of H2O2 content than transgenic plants ( Figure 6E ). Oxidoreductases like POD, SOD, and CAT also function in scavenging redundant ROS (Gupta et al., 2016). Here, we found that the transgenic lines exhibited stronger SOD activity than WT ( Figure 6F ), which help plants alleviate oxidation damage from freezing conditions. However, no significant difference was detected for POD and CAT activities. Together, the enhanced freezing adaption for transgenic plants could be attributed to the increased ROS scavenging ability.
BnTR1 Positively Affects Alkaloid Metabolism
BnTR1 (homolog of AT2g29300) is predicted to be a tropinone reductase involved in the biosynthesis of alkaloid (KO00960), which mainly produces atropine. Although all alkaloids (with more than 12,000 different structures) have been well-documented in pharmacology, their roles in abiotic stress remain elusive (Schlager and Drager, 2016). To investigate the specific role of BnTR1 in alkaloid metabolism, the total alkaloids content was quantified. As expected, the transgenic lines led to one- to two-fold increase of alkaloids contents compared with WT plants after freezing stress ( Figure 7A ). To further confirm the effect of alkaloid on stressed plants, exogenous atropine was applied to WT plants, since atropine was considered to be the product of alkaloid metabolism (KO00960) (Hara and Kurita, 2014). Results demonstrated that application of 10 nmol atropine per plant significantly rescued the susceptibility of WT plants, but the protective effect was weakened when dosage increase to 30 nmol ( Figure 7B ). The survival rate increased by three- to four-fold compared with WT plants without atropine treatment ( Figure 7C ).
Figure 7.
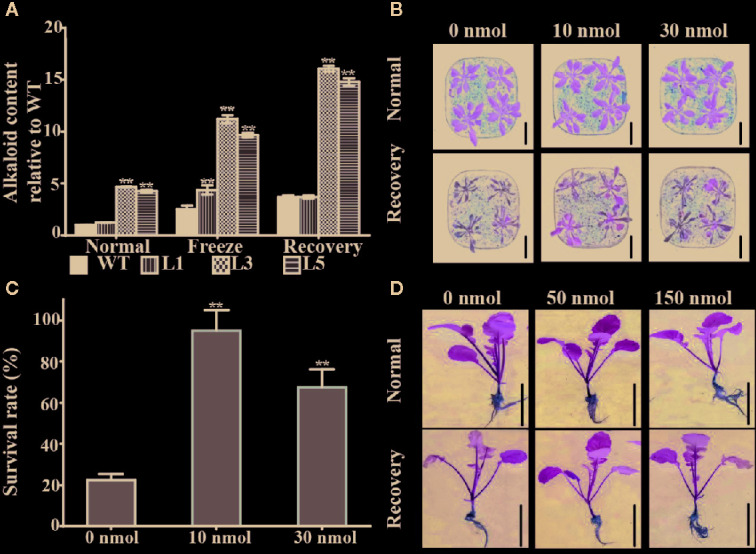
BnTR1 mediates alkaloid accumulation and exogenous atropine application enhances freezing tolerance. (A) Total alkaloids accumulation in BnTR1 transgenic lines and WT plants under freezing stress conditions. L1, L3, L5 represent three independent homozygous lines of BnTR1 transgenic Arabidopsis plants. (B) Phenotypes of Arabidopsis WT plants with exogenous atropine application (0 nmol per plant, 10 nmol per plant, 30 nmol per plant) under freezing stress conditions. Scale = 2 cm. (C) Survival rates of Arabidopsis WT plants with exogenous atropine application after the freezing treatment. (D) Phenotypes of rapeseed WT plants with exogenous atropine application (0 nmol per plant, 50 nmol per plant, 150 nmol per plant) under LT conditions. Scale=5 cm. Bars indicate the SE of three biological replicates. Significant differences are determined by Student’s t-test (*P < 0.05 or **P < 0.01).
To further elucidate the protective role of alkaloid in rapeseed, the current study applied exogenous atropine to a widely cultivated rapeseed variety, ZS11, under freezing conditions. Phenotypic analysis showed that the wilting phenotype of ZS11 plants was partially rescued by exogenous atropine application (50 and 150 nmol per plant) ( Figure 7D ), while no significant difference was observed in the survival rate. It was concluded that alkaloids alleviated the damage on plants from extreme LT stress.
Discussion
Power of AT Approach
Brassica napus originated from the hybridization of Brassica rapa and Brassica oleracea which contribute the A and C genomes, respectively (Cheung et al., 2009). There is only 15% difference in nucleotide structure and 3% difference in transcriptional expression patterns between chromosomes A and C, which limits the development of SNP markers in genome-wide association analysis until the availability of the high throughput next-generation sequencing technology (Adams et al., 2003; Higgins et al., 2012). Over the past years, AT approach based on abundant SNP markers and GEMs, has successfully simplified the complexity of the whole genome (Harper et al., 2012), and has been widely applied in rapeseed, wheat, and other polyploidy crops (Harper et al., 2012; Schuster et al., 2013; Koprivova et al., 2014; Lu et al., 2014; Harper et al., 2016a; Harper et al., 2016b; Miller et al., 2016; Alcock et al., 2017; Havlickova et al., 2018; Miller et al., 2018). However, the genetic basis of photosynthetic-related traits in oil crops remains elusive. Here, the genetic architecture of photosynthetic gas exchange parameters was investigated by AT approach, and a gene termed BnTR1 was confirmed to be responsible for E trait ( Figure 4 ), which might be a promising candidate beneficial to rapeseed in coping with climatic changes. Several other interesting candidates were also identified. For instance, Bo5g155110.1 was found to be significantly associated with E traits (P=1.1×10−5), and down-regulated by freezing stress ( Supplementary Figure S3 ). The homolog in Arabidopsis is Cyclophilin38 (AtCYP38), which functions in the assembly and maintenance of PSII super complex ( Supplementary Table S7 ). The loss-of-function mutant of AtCYP38 shows reduced growth rate and photosynthetic efficiency compared to its wild type. Additionally, the D1 and D2 proteins in PSII reaction center show a short half-life, resulting in susceptibility upon exposure to excessive light (Fu et al., 2007; Sirpio et al., 2008). Bo3g153100.1 was hit by an SNP marker, with -log10 (P-value) value as high as 9.06 ( Supplementary Table S6 ), it was also markedly up-regulated by freezing stress ( Supplementary Figure S3 ). Bo3g153100.1 was homologs to AT4G37930 in Arabidopsis, which has been documented in the photorespiration process (Takahashi et al., 2007). In the knockout mutant of AT4G37930, the photorespiration pathway is destroyed, and the chlorophyll deficiency results in chlorosis (Voll et al., 2006). Therefore, it seems that AT is a powerful tool to identify candidate genes for photosynthesis and LT stress in rapeseed. It is worthy to further study the function of all 22 candidate genes identified here.
The Positive Role of BnTR1 Under LT Conditions
It is generally accepted that photosynthesis is vulnerable to adverse environmental stresses such as extreme temperature, salinity, drought or combined stresses (Sainz et al., 2010; Strzepek et al., 2019). Abiotic stresses lead to photoinhibition as well as excessive generation of ROS, which suppresses the photosynthetic progress and ultimately repress the growth and productivity in plants (Gabriel et al., 2010; Nishiyama et al., 2014). During the long-term evolution, plants have developed a variety of adaptive mechanisms to cope with the stressful conditions (Liu et al., 2019a; Strzepek et al., 2019). The CBF transcription factors in rapeseed are known to be responsible for the photosynthetic performance; CBF5 and CBF17 enhance the energy conversion efficiency under LT conditions (Savitch et al., 2005; Dahal et al., 2012). CBF1-CBF3, also termed dehydration-responsive element-binding factors, have been well-documented in plants. In Arabidopsis, CBF2 represents a negative regulator for LT response, while CBF1 and CBF3 are positive regulators (Novillo et al., 2004; Novillo et al., 2012). Interestingly, increased expression of CBF1 and CBF3 and repressed expression of CBF2 were observed in the BnTR1 transgenic lines ( Figures 4E–G ), indicating that BnTR1 represented a unique influence on CBF members. Both alleviated accumulation of ROS and activated SOD enzyme system was observed in the BnTR1 transgenic lines ( Figure 6 ), suggesting active impacts of BnTR1 on the ROS scavenging system. In addition, ectopic expressing BnTR1 also promote the expression of the genes associated with plant photosynthesis ( Figure 5 ). Specifically, the decrease of RCA transcripts leads to lower An value, which in turn slows down plant growth (Von Caemmerer et al., 2005; Yin et al., 2010). Moreover, RCA enhances growth and photosynthesis under moderate heat stress conditions (Kurek et al., 2007; Kumar et al., 2009). However, overexpression of SBPASE improves sugar accumulation and enhanced photosynthesis efficiency (Miyagawa et al., 2001; Feng et al., 2007; De Porcellinis et al., 2018). In the present study, PSII was severely repressed during freezing treatment, whereas the BnTR1 transgenic Arabidopsis plants still exhibited higher Fv/Fm level compared to WT ( Figure 5A ). These observations suggest that BnTR1 triggered a series of responses including the ROS scavenging system, CBF pathway, and photosynthetic processes. However, further work is required to confirm their roles in LT tolerance.
Protective Role of Alkaloids Under LT Conditions
Previous studies have been instrumental in revealing some metabolites underlying stress response mechanisms (Thalmann and Santelia, 2017). So far, definitions of alkaloids are generally focused on strong pharmacological effects, such as antimitotic, antidote, anticancer, and antioxidants (Schlager and Drager, 2016). The concentration of alkaloid compounds is predominantly inducible when plants are subjected to multiple stresses (Srivastava and Srivastava, 2010; Cheng et al., 2018). However, few studies have recognized the positive correlations between alkaloids and stress resistance. Application of sanguinarine for Arabidopsis seedlings under heat stress condition could markedly enhance the tolerance, which presumably by promoting the expression of heat shock proteins like HSP70 and HSP90.1S (Hara and Kurita, 2014; Matsuoka et al., 2016). BnTR1 is predicted to encode a tropinone reductase, which is involved in the metabolic pathway of atropine alkaloids. The current study determined the total alkaloids content under stress conditions, which showed an increased level in BnTR1 transgenic lines under normal and freezing stress conditions compared with WT plants ( Figure 7A ). Moreover, the application of exogenous atropine alleviated the damage caused by extreme temperature in both Arabidopsis and rapeseed seedlings ( Figures 7B, D ), which was in agreement with the observations in sanguinarine under heat stress conditions (Schlager and Drager, 2016). However, more studies are still required to confirm that alkaloids could function as a protectant for plants to confer stronger resistance to LT stresses. The current study has compared the expression level of stress-related genes in Arabidopsis plants treated with exogenous atropine under freezing conditions ( Supplementary Figure S6 ). It was found that atropine could promote the expression of CBF1, CBF3, CAB1, CAB3, CAB4, SPASE before or after freezing treatment, but the extent is much lower than that induced by BnTR1 ( Figures 4 and 5 ). Thus the results confirmed at least in part the protective role of atropine for a plant in adaption to LT stress. It is proposed that BnTR1 works as an effecter via metabolizing alkaloids accumulation, photosynthesis, CBF/DREB pathways, and ROS scavenging system in stressed Arabidopsis, which in turn contributes to the adaptation under LT conditions.
Conclusions
During overwintering for the semi-winter type rapeseed grown in China, the extremely low temperature has a deleterious impact on plant productivity. Therefore, the identification of genes responsible for stress response is the prime interest of researchers. Despite of limited phenotypic data, our associative transcriptomics approach has been successfully used to dissect the genetics of photosynthetic-related traits under low temperature conditions. The first short-chain dehydrogenase/reductase, BnTR1 was identified in rapeseed, which improved the transpiration rate and freezing tolerance of Arabidopsis plants. Taken together, our findings illustrated the molecular mechanism of plant adaption to low temperature stress. Finally, this work sheds light on the way to increase low temperature tolerance in rapeseed by genetic engineering strategies.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding authors.
Author Contributions
Figures, study design: YH, MH. Investigation and data collection: YH, HX, CZ. Data analysis: GL, ZT, LH, IB. Validation: DL. Resources: LH, IB. Writing—original draft: YH, MH. Writing—review and editing: YL, GL. Funding acquisition: XZh, YC, XZ, GL, YL.
Funding
This research was financially supported by funds from the National Key Research and Development Program of China (2018YFD0100905), the Oil Crops Research Institution Basal Research Fund of the Chinese Academy of Agricultural Sciences (CAAS), China (1610172018010), the major science and technology project, Ministry of science and technology, China (2018ZX08020001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00971/full#supplementary-material
References
- Adams K. L., Cronn R., Percifield R., Wendel J. F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. U. S. A. 100, 4649–4654. 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock T. D., Havlickova L., He Z., Bancroft I., White P. J., Broadley M. R., et al. (2017). Identification of candidate genes for calcium and magnesium accumulation in Brassica napus L. by association genetics. Front. Plant Sci. 8, 1968. 10.3389/fpls.2017.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock T. D., Havlickova L., He Z., Wilson L., Bancroft I., White P. J., et al. (2018). Species-wide variation in shoot nitrate concentration, and genetic loci controlling nitrate, phosphorus and potassium accumulation in Brassica napus L. Front. Plant Sci. 9, 1487. 10.3389/fpls.2018.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J., Wentworth M., Walters R. G., Howard C. A., Ruban A. V., Horton P., et al. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II—effects on photosynthesis, grana stacking and fitness. Plant J. 35, 350–361. 10.1046/j.1365-313x.2003.01811.x. [DOI] [PubMed] [Google Scholar]
- Basu U., Bajaj D., Sharma A., Malik N., Daware A., Narnoliya L., et al. (2019). Genetic dissection of photosynthetic efficiency traits for enhancing seed yield in chickpea. Plant Cell Environ. 42, 158–173. 10.1111/pce.13319. [DOI] [PubMed] [Google Scholar]
- Bazakos C., Hanemian M., Trontin C., Jimenez-Gomez J. M., Loudet O. (2017). New strategies and tools in quantitative genetics: how to go from the phenotype to the genotype. Annu. Rev. Plant Biol. 68, 435–455. 10.1146/annurev-arplant-042916-040820. [DOI] [PubMed] [Google Scholar]
- Bernfur K., Rutsdottir G., Emanuelsson C. (2017). The chloroplast-localized small heat shock protein Hsp21 associates with the thylakoid membranes in heat-stressed plants. Protein Sci. 26, 1773–1784. 10.1002/pro.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A., Brandt W., Drager B. (2008). The functional divergence of short-chain dehydrogenases involved in tropinone reduction. Plant J. 54, 388–401. 10.1111/j.1365-313X.2008.03422.x. [DOI] [PubMed] [Google Scholar]
- Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- Cheng T., Hu L., Wang P., Yang X., Peng Y., Lu Y., et al. (2018). Carbon monoxide potentiates high temperature-induced nicotine biosynthesis in tobacco. Int. J. Mol. Sci. 19, 188. 10.3390/ijms19010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F., Trick M., Drou N., Lim Y. P., Park J. Y., Kwon S. J., et al. (2009). Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell. 21, 1912–1928. 10.1105/tpc.108.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury F. K., Rivero R. M., Blumwald E., Mittler R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- Cong R. H., Zhang Z., Lu J. W. (2019). Climate impacts on yield of winter oilseed rape in different growth regions of the Yangtze River Basin. Chin. J. Oil Crop Sci. 41, 894–903. 10.19802/j.issn.1007-9084.2019046 [DOI] [Google Scholar]
- Dahal K., Gadapati W., Savitch L. V., Singh J., Huner N. P. (2012). Cold acclimation and BnCBF17-over-expression enhance photosynthetic performance and energy conversion efficiency during long-term growth of Brassica napus under elevated CO2 conditions. Planta 236, 1639–1652. 10.1007/s00425-012-1710-2. [DOI] [PubMed] [Google Scholar]
- De Miguel M., Cabezas J. A., De Maria N., Sanchez-Gomez D., Guevara M. A., Velez M. D., et al. (2014). Genetic control of functional traits related to photosynthesis and water use efficiency in Pinus pinaster Ait. drought response: integration of genome annotation, allele association and QTL detection for candidate gene identification. BMC Genomics 15, 464. 10.1186/1471-2164-15-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Porcellinis A. J., Norgaard H., Brey L. M. F., Erstad S. M., Jones P. R., Heazlewood J. L., et al. (2018). Overexpression of bifunctional fructose-1,6-bisphosphatase/sedoheptulose-1,7-bisphosphatase leads to enhanced photosynthesis and global reprogramming of carbon metabolism in Synechococcus sp. PCC 7002. Metab. Eng. 47, 170–183. 10.1016/j.ymben.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Ding C.-K., Wang C. Y., Gross K. C., Smith D. L. (2001). Reduction of chilling injury and transcript accumulation of heat shock proteins in tomato fruit by methyl jasmonate and methyl salicylate. Plant Sci. 161, 0–1159. 10.1016/S0168-9452(01)00521-0 [DOI] [Google Scholar]
- Duggal P., Gillanders E. M., Holmes T. N., Bailey-Wilson J. E. (2008). Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 9, 516. 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. R. (2013). Improving photosynthesis. Plant Physiol. 162, 1780–1793. 10.1104/pp.113.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Wang K., Li Y., Tan Y., Kong J., Li H., et al. (2007). Overexpression of SBPase enhances photosynthesis against high temperature stress in transgenic rice plants. Plant Cell Rep. 26, 1635–1646. 10.1007/s00299-006-0299-y. [DOI] [PubMed] [Google Scholar]
- Fu A., He Z., Cho H. S., Lima A., Buchanan B. B., Luan S. (2007). A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 104, 15947–15952. 10.1073/pnas.0707851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel L. T., Michel H., Francois O. (2010). The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J. 60, 691–702. 10.1111/j.1365-313X.2009.03991.x. [DOI] [PubMed] [Google Scholar]
- Ganeteg U., Kulheim C., Andersson J., Jansson S. (2004). Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 134, 502–509. 10.1104/pp.103.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Wang T., Wang N., Wang Z., Liang C., Ramchiary N., et al. (2012). Genetic mapping and localization of quantitative trait loci for chlorophyll content in Chinese cabbage (Brassica rapa ssp. pekinensis). Sci. Horticult. 147, 42–48. 10.1016/j.scienta.2012.09.004. [DOI] [Google Scholar]
- Gupta K., Sengupta A., Chakraborty M., Gupta B. (2016). Hydrogen peroxide and polyamines act as double edged swords in plant abiotic stress responses. Front. Plant Sci. 7, 1343. 10.3389/fpls.2016.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., Kurita I. (2014). The natural alkaloid sanguinarine promotes the expression of heat shock protein genes in Arabidopsis. Acta Physiol. Plant. 36, 3337–3343. 10.1007/s11738-014-1681-y. [DOI] [Google Scholar]
- Harper A. L., Trick M., Higgins J., Fraser F., Clissold L., Wells R., et al. (2012). Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat. Biotechnol. 30, 798–802. 10.1038/nbt.2302. [DOI] [PubMed] [Google Scholar]
- Harper A. L., Mckinney L. V., Nielsen L. R., Havlickova L., Li Y., Trick M., et al. (2016. a). Molecular markers for tolerance of European ash (Fraxinus excelsior) to dieback disease identified using Associative Transcriptomics. Sci. Rep. 6, 19335. 10.1038/srep19335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper A. L., Trick M., He Z., Clissold L., Fellgett A., Griffiths S., et al. (2016. b). Genome distribution of differential homoeologue contributions to leaf gene expression in bread wheat. Plant Biotechnol. J. 14, 1207–1214. 10.1111/pbi.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlickova L., He Z., Wang L., Langer S., Harper A. L., Kaur H., et al. (2018). Validation of an updated Associative Transcriptomics platform for the polyploid crop species Brassica napus by dissection of the genetic architecture of erucic acid and tocopherol isoform variation in seeds. Plant J. 93, 181–192. 10.1111/tpj.13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Cheng F., Li Y., Wang X., Parkin I. A., Chalhoub B., et al. (2015). Construction of Brassica A and C genome-based ordered pan-transcriptomes for use in rapeseed genomic research. Data Brief 4, 357–362. 10.1016/j.dib.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejna O., Havlickova L., He Z., Bancroft I., Curn V. (2019). Analysing the genetic architecture of clubroot resistance variation in Brassica napus by associative transcriptomics. Mol. Breed. 39, 112. 10.1007/s11032-019-1021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J., Magusin A., Trick M., Fraser F., Bancroft I. (2012). Use of mRNA-seq to discriminate contributions to the transcriptome from the constituent genomes of the polyploid crop species Brassica napus. BMC Genomics 13, 247. 10.1186/1471-2164-13-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha D. K., Hofner R., Schwab K. B., Heber U., Schmitt J. M. (1987). Membrane rupture is the common cause of damage to chloroplast membranes in leaves injured by freezing or excessive wilting. Plant Physiol. 83, 251–253. 10.1104/pp.83.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha U. C., Bohra A., Jha R. (2017). Breeding approaches and genomics technologies to increase crop yield under low-temperature stress. Plant Cell Rep. 36, 1–35. 10.1007/s00299-016-2073-0. [DOI] [PubMed] [Google Scholar]
- Jin H., Liu B., Luo L., Feng D., Wang P., Liu J., et al. (2014). HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell. 26, 1213–1229. 10.1105/tpc.113.122424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X., Li Z. (2012). Study of different types of rape ( Brassica L.) cultivars (lines) relationship between photosynthetic physiological indices and yield. J. Anhui Agri. Sci. 40, 11213–11215. 10.13989/j.cnki.0517-6611.2012.22.106. [DOI] [Google Scholar]
- Kaler A. S., Purcell L. C. (2019). Estimation of a significance threshold for genome-wide association studies. BMC Genomics 20, 20–33. 10.1186/s12864-019-5992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Harper A. L., Trick M., Bancroft I., Kopriva S. (2014). Dissection of the control of anion homeostasis by associative transcriptomics in Brassica napus. Plant Physiol. 166, 442–450. 10.1104/pp.114.239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Li C., Portis A. R., Jr. (2009). Arabidopsis thaliana expressing a thermostable chimeric Rubisco activase exhibits enhanced growth and higher rates of photosynthesis at moderately high temperatures. Photosynth. Res. 100, 143–153. 10.1007/s11120-009-9438-y. [DOI] [PubMed] [Google Scholar]
- Kurek I., Chang T. K., Bertain S. M., Madrigal A., Liu L., Lassner M. W., et al. (2007). Enhanced Thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell. 19, 3230–3241. 10.1105/tpc.107.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T., Kramer D. M., Raines C. A. (2012). Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr. Opin. Biotechnol. 23, 215–220. 10.1016/j.copbio.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Leister D. (2019). Genetic engineering, synthetic biology and the light reactions of photosynthesis. Plant Physiol. 179, 778–793. 10.1104/pp.18.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang G., Zheng Q., Li B., Jing R., Li Z. (2014). Genetic analysis of biomass and photosynthetic parameters in wheat grown in different light intensities. J. Integr. Plant Biol. 56, 594–604. 10.1111/jipb.12174. [DOI] [PubMed] [Google Scholar]
- Li H., Yang Y., Zhang H., Chu S., Zhang X., Yin D., et al. (2016). A Genetic Relationship between Phosphorus Efficiency and Photosynthetic Traits in Soybean As Revealed by QTL Analysis Using a High-Density Genetic Map. Front. Plant Sci. 7, 924. 10.3389/fpls.2016.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G., Guan C. (2001). Effect of seeding date on yield characteristics of different rapeseed (Brassica napus) genotypes. Chin. J. Appl. Ecol., 12 (6), 853–858. 10.13287/j.1001-9332.2001.0203 [DOI] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M., Bradbury P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- Liu R., Xu Y. H., Jiang S. C., Lu K., Lu Y. F., Feng X. J., et al. (2013). Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 64, 5443–5456. 10.1093/jxb/ert307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fan Y., Mak M., Babla M., Holford P., Wang F., et al. (2017). QTLs for stomatal and photosynthetic traits related to salinity tolerance in barley. BMC Genomics 18, 9. 10.1186/s12864-016-3380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lu Y., Hua W., Last R. L. (2019. a). A new light on Photosystem II maintenance in oxygenic photosynthesis. Front. Plant Sci. 10, 975. 10.3389/fpls.2019.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhang S., Hu J., Sun W., Padilla J., He Y., et al. (2019. b). Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 17572–17577. 10.1073/pnas.1905123116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long S. P., Zhu X. G., Naidu S. L., Ort D. R. (2006). Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29, 315–330. 10.1111/j.1365-3040.2005.01493.x. [DOI] [PubMed] [Google Scholar]
- Lu G., Harper A. L., Trick M., Morgan C., Fraser F., O’neill C., et al. (2014). Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassica napus. DNA Res. 21, 613–625. 10.1093/dnares/dsu024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T., Zhang J., Khan M. N., Liu J., Xu Z., Hu L. (2018). Temperature variation caused by sowing dates significantly affects floral initiation and floral bud differentiation processes in rapeseed (Brassica napus L.). Plant Sci. 271, 40–51. 10.1016/j.plantsci.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Luo T., Xian M., Zhang C., Zhang C., Hu L., Xu Z. (2019). Associating transcriptional regulation for rapid germination of rapeseed (Brassica napus L.) under low temperature stress through weighted gene co-expression network analysis. Sci. Rep. 9, 55–70. 10.1038/s41598-018-37099-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Guo Z., Li X., Ye H., Li X., Xiong L. (2016). New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant Cell Environ. 39, 556–570. 10.1111/pce.12635. [DOI] [PubMed] [Google Scholar]
- Lv Y., Yang M., Hu D., Yang Z., Ma S., Li X., et al. (2017). The OsMYB30 Transcription Factor Suppresses Cold Tolerance by Interacting with a JAZ Protein and Suppressing β-Amylase Expression. Plant Physiol. 173, 1475. 10.1104/pp.16.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka E., Matsubara T., Takahashi I., Murano H., Hara M. (2016). The isoquinoline alkaloid sanguinarine which inhibits chaperone activity enhances the production of heat shock proteins in Arabidopsis. Plant Biotechnol. (Tokyo) 33, 409–413. 10.5511/plantbiotechnology.16.1001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. N., Harper A. L., Trick M., Werner P., Waldron K., Bancroft I. (2016). Elucidation of the genetic basis of variation for stem strength characteristics in bread wheat by Associative Transcriptomics. BMC Genomics 17, 500. 10.1186/s12864-016-2775-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. N., Harper A. L., Trick M., Wellner N., Werner P., Waldron K. W., et al. (2018). Dissecting the complex regulation of lodging resistance in Brassica napus. Mol. Breed. 38, 30. 10.1007/s11032-018-0781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Heyer A. G., Mishra K. B. (2014). Chlorophyll fluorescence emission can screen cold tolerance of cold acclimated Arabidopsis thaliana accessions. Plant Methods 10, 38. 10.1186/1746-4811-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa Y., Tamoi M., Shigeoka S. (2001). Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat. Biotechnol. 19, 965–969. 10.1038/nbt1001-965. [DOI] [PubMed] [Google Scholar]
- Morsy M. R., Almutairi A. M., Gibbons J., Yun S. J., De Los Reyes B. G. (2005). The OsLti6 genes encoding low-molecular-weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 344, 171–180. 10.1016/j.gene.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Ning J., Li X., Hicks L. M., Xiong L. (2010). A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890. 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y., Yamamoto H., Allakhverdiev S., Inaba M., Yokota A., Murata N. (2014). Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 20, 5587–5594. 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Alonso J. M., Ecker J. R., Salinas J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 3985–3990. 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Medina J., Rodriguez-Franco M., Neuhaus G., Salinas J. (2012). Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. J. Exp. Bot. 63, 293–304. 10.1093/jxb/err279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’neill C. M., Lu X., Calderwood A., Tudor E. H., Penfield S. (2019). Vernalization and floral transition in autumn drive winter annual life history in oilseed rape. Curr. Biol. 29, 4300–4306. 10.1016/j.cub.2019.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley C. G., Savage L., Lotz S., Larson G. R., Thomashow M. F., Kramer D. M., et al. (2018). Genetic basis of photosynthetic responses to cold in two locally adapted populations of Arabidopsis thaliana. J. Exp. Bot. 69, 699–709. 10.1093/jxb/erx437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer H. (2003). Sowing date and nitrogen rate effects on growth, yield and yield components of two summer rapeseed cultivars. Eur. J. Agron. 19, 453–463. 10.1016/S1161-0301(02)00136-3. [DOI] [Google Scholar]
- Prado S. A., Cabrera-Bosquet L., Grau A., Coupel-Ledru A., Millet E. J., Welcker C., et al. (2017). Phenomics allows identification of genomic regions affecting maize stomatal conductance with conditional effects of water deficit and evaporative demand. Plant Cell Environ. 41 (2), 314–326. 10.1111/pce.13083 [DOI] [PubMed] [Google Scholar]
- Sabehat A., Weiss D., Lurie S. (1996). The correlation between heat-shock protein accumulation and persistence and chilling tolerance in tomato fruit. Plant Physiol. 110, 531–537. 10.1104/pp.110.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R., Kubien D. (2007). The temperature response of C 3 and C 4 photosynthesis. Plant Cell Environ. 30, 1086–1106. 10.1111/j.1365-3040.2007.01682.x. [DOI] [PubMed] [Google Scholar]
- Sainz M., Diaz P., Monza J., Borsani O. (2010). Heat stress results in loss of chloroplast Cu/Zn superoxide dismutase and increased damage to Photosystem II in combined drought-heat stressed Lotus japonicus. Physiol. Plant 140, 46–56. 10.1111/j.1399-3054.2010.01383.x. [DOI] [PubMed] [Google Scholar]
- Savitch L. V., Allard G., Seki M., Robert L. S., Tinker N. A., Huner N. P., et al. (2005). The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant Cell Physiol. 46, 1525–1539. 10.1093/pcp/pci165. [DOI] [PubMed] [Google Scholar]
- Schlager S., Drager B. (2016). Exploiting plant alkaloids. Curr. Opin. Biotechnol. 37, 155–164. 10.1016/j.copbio.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Schuster T. M., Setaro S. D., Kron K. A. (2013). Age estimates for the buckwheat family Polygonaceae based on sequence data calibrated by fossils and with a focus on the amphi-Pacific Muehlenbeckia. PloS One 8, e61261. 10.1371/journal.pone.0061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmar D., Kleinwachter M. (2013). Stress enhances the synthesis of secondary plant products: the impact of stress-related over-reduction on the accumulation of natural products. Plant Cell Physiol. 54, 817–826. 10.1093/pcp/pct054. [DOI] [PubMed] [Google Scholar]
- Shen B. R., Wang L. M., Lin X. L., Yao Z., Xu H. W., Zhu C. H., et al. (2019). Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Mol. Plant 12, 199–214. 10.1016/j.molp.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Sirpio S., Khrouchtchova A., Allahverdiyeva Y., Hansson M., Fristedt R., Vener A. V., et al. (2008). AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55, 639–651. 10.1111/j.1365-313X.2008.03532.x. [DOI] [PubMed] [Google Scholar]
- Srivastava N. K., Srivastava A. K. (2010). Influence of some heavy metals on growth, alkaloid content and composition in Catharanthus roseus L. Indian J. Pharm. Sci. 72, 775–778. 10.4103/0250-474X.84592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus P. L., Lynch D. V., Uemura M., Heber U., Pearce R. S. (1990). The Influence of Cold Acclimation on the Lipid Composition and Cryobehaviour of the Plasma Membrane of Isolated Rye Protoplasts [and Discussion]. Phil. Trans. R. Soc Lond. B. 326, 571–583. 10.1098/rstb.1990.0032. [DOI] [Google Scholar]
- Strzepek R. F., Boyd P. W., Sunda W. G. (2019). Photosynthetic adaptation to low iron, light, and temperature in Southern Ocean phytoplankton. Proc. Natl. Acad. Sci. U. S. A. 116, 4388–4393. 10.1073/pnas.1810886116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. N., Wang Q., Sun C. C., Liu F. J., Bi H. G., Ai X. Z. (2017). Response and adaptation of photosynthesis of cucumber seedlings to high temperature stress. Ying Yong Sheng Tai Xue Bao 28, 1603–1610. 10.13287/j.1001-9332.201705.009. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Bauwe H., Badger M. (2007). Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 144, 487–494. 10.1104/pp.107.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A., Hincha D. K., Zuther E. (2014). Measuring freezing tolerance: electrolyte leakage and chlorophyll fluorescence assays. Methods Mol. Biol. 1166, 15–24. 10.1007/978-1-4939-0844-8_3. [DOI] [PubMed] [Google Scholar]
- Thalmann M., Santelia D. (2017). Starch as a determinant of plant fitness under abiotic stress. New Phytol. 214, 943–951. 10.1111/nph.14491. [DOI] [PubMed] [Google Scholar]
- Tian Z., Ji Y. H., Sun L. X., Xu X. L., Fan D. L., Zhong H. L., et al. (2018). Changes in production potentials of rapeseed in the Yangtze River Basin of China under climate change: A multi-model ensemble approach. J. Geogr. Sci. 28, 1700–1714. 10.1007/s11442-018-1538-1 [DOI] [Google Scholar]
- Tonfack L., Moummou H., Latché A., Youmbi E., Benichou M., Pech J.-C., et al. (2011). The plant SDR superfamily: Involvement in primary and secondary metabolism. Curr. Top. Plant Biol. 12, 41–53. 10.1186/1471-2229-12-219. [DOI] [Google Scholar]
- Urban M. O., Klima M., Vitamvas P., Vasek J., Hilgert-Delgado A. A., Kucera V. (2013). Significant relationships among frost tolerance and net photosynthetic rate, water use efficiency and dehydrin accumulation in cold-treated winter oilseed rapes. J. Plant Physiol. 170, 1600–1608. 10.1016/j.jplph.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Valerio C., Costa A., Marri L., Issakidis-Bourguet E., Pupillo P., Trost P., et al. (2011). Thioredoxin-regulated beta-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J. Exp. Bot. 62, 545–555. 10.1093/jxb/erq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll L. M., Jamai A., Renne P., Voll H., Mcclung C. R., Weber A. P. (2006). The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 140, 59–66. 10.1104/pp.105.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Caemmerer S., Hendrickson L., Quinn V., Vella N., Millgate A. G., Furbank R. T. (2005). Reductions of Rubisco activase by antisense RNA in the C4 plant Flaveria bidentis reduces Rubisco carbamylation and leaf photosynthesis. Plant Physiol. 137, 747–755. 10.1104/pp.104.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasundara P., Mcintosh T., Perera S., Withana-Gamage T., Mitra P. (2016). Canola/rapeseed protein-functionality and nutrition. OCL 23, D407. 10.1051/ocl/2016028. [DOI] [Google Scholar]
- Wang J., Jian H., Wei L., Qu C., Xu X., Lu K., et al. (2015). Genome-wide analysis of seed acid detergent lignin (ADL) and hull content in rapeseed (Brassica napus L.). PloS One 10, e0145045. 10.1371/journal.pone.0145045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood I. P., Pearson B. M., Garcia-Gutierrez E., Havlickova L., He Z., Harper A. L. (2017). Carbohydrate microarrays and their use for the identification of molecular markers for plant cell wall composition. Proc. Natl. Acad. Sci. U. S. A. 114, 6860–6865. 10.1073/pnas.1619033114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. H., Liu R., Yan L., Liu Z. Q., Jiang S. C., Shen Y. Y., et al. (2012). Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106. 10.1093/jxb/err315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Qu C., Li J., Chen L., Liu L. (2015). QTL analysis of leaf photosynthesis rate and related physiological traits in in brassica napus. Sci. Agric. Sin. 14, 2095–3119. 10.1016/S2095-3119(14)60958-8. [DOI] [Google Scholar]
- Yan L., Tariq S., Cheng Y., Lü Y., Zhang X. K., Zou X. L. (2019). Physiological and molecular responses to cold stress in rapeseed (Brassica napus L.). J. Integr. Agricult. 18, 2742–2752. 10.1016/S2095-3119(18)62147-1 [DOI] [Google Scholar]
- Yin Z., Meng F., Song H., Wang X., Xu X., Yu D. (2010). Expression quantitative trait loci analysis of two genes encoding rubisco activase in soybean. Plant Physiol. 152, 1625–1637. 10.1104/pp.109.148312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding R., Chai Y., Bonfill M., Moyano E., Oksman-Caldentey K.-M., et al. (2004). Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures. Proc. Natl. Acad. Sci. U. S. A. 101, 6786. 10.1073/pnas.0401391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. R., Henriques R., Lin S. S., Niu Q. W., Chua N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang L., Meng H., Wen H., Fan Y., Zhao J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75, 365–378. 10.1007/s11103-011-9732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. J., Li L., Zhang C. L. (2012). Effects of sowing date and planting density on the seed yield and oil content of winter oilseed rape. Ying Yong Sheng Tai Xue Bao 23, 1326–1332. 10.13287/j.1001-9332.2012.0179. [DOI] [PubMed] [Google Scholar]
- Zhang X., Feng L., Yang T., Xu Z., Hu L. (2015). Effects of Chilling Stress on Physiological Characteristics of Rapeseed Seedlings in Winter. Plant Physiol. J. 51, 737–746. 10.13592/j.cnki.ppj.2014.0411. [DOI] [Google Scholar]
- Zhang Y., Zhang D., Yu H., Lin B., Fu Y., Hua S. (2016). Floral Initiation in Response to Planting Date Reveals the Key Role of Floral Meristem Differentiation Prior to Budding in Canola (Brassica napus L.). Front. Plant Sci. 7, 1369. 10.3389/fpls.2016.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhao X., Guan Z., Wang X., Hou J., Tian J. (2017. a). A review on photosynthetic physiological research and high photosynthetic efficiency breeding of Brassica napus. Chin. Agric. Sci. Bull. 33, 44–54. [Google Scholar]
- Zhang Z., Li J., Pan Y., Li J., Zhou L., Shi H., et al. (2017. b). Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 8, 14788. 10.1038/ncomms14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Zhou W., Wang H., Ding S., Lu Q., Wen X., et al. (2013). Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell. 25, 2925–2943. 10.1105/tpc.113.111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding authors.