OsATM functions in parallel with homologous recombination to promote accurate meiotic double-strand break repair.
Abstract
ATAXIA TELANGIECTASIA-MUTATED (ATM) protein has been well studied for its roles in the DNA damage response. However, its role in meiosis has not been fully explored. Here, we characterized the functions of the rice (Oryza sativa) ATM homolog during meiosis. Aberrant chromosome associations and DNA fragmentations were observed after the completion of homologous pairing and synapsis in Osatm pollen mother cells (PMCs). Aberrant chromosome associations disappeared in Osspo11-1 Osatm-1 double mutants and more severe defects were observed in Osdmc1 Osatm, suggesting that OsATM functions downstream of OsSPO11-1-catalyzed double-strand break formation and in parallel with OsDMC1-mediated homologous recombination. We further demonstrated that phosphorylation of H2AX in PMCs did not depend on OsATM, in contrast to the situation in somatic cells. Moreover, the removal of OsDMC1 from chromosomes in Osatm PMCs was delayed and the number of HEI10 foci (markers of interference-sensitive crossover intermediates) decreased. Together, these findings suggest that OsATM plays important roles in the accurate repair of meiotic double-strand breaks in rice.
The repair of double-strand breaks (DSBs) by homologous recombination (HR) ensures accurate chromosome segregation in meiosis. This process, profoundly studied in yeast and mammals, starts with the programmed formation of DSBs, which is catalyzed by SPO11, the homolog of subunit A of an archaeal DNA topoisomerase (TopoVIA; Keeney et al., 1997; Robert et al., 2016). Incision sites are further processed by the Mre11-Rad50-Xrs2 complex, to yield 3′ single-stranded DNA tails (Cao et al., 1990; Mimitou and Symington, 2009). The end is initially protected by the replication protein A complex, which is subsequently released when two strand-exchange proteins, RAD51 and DMC1, are loaded onto the end (Fanning et al., 2006; Seeliger et al., 2012). RAD51 and DMC1 promote homology searching, resulting in strand exchange and the formation of joint molecule intermediates (Shinohara et al., 1992; Su et al., 2017). These intermediates are eventually resolved as either crossovers (COs) or noncrossovers (Börner et al., 2004; Osman et al., 2011).
ATM is the gene mutated in the human disease ataxia telangiectasia that causes a number of symptoms (Taylor et al., 1975; Savitsky et al., 1995a). It encodes a large protein with a Pi-3 kinase-like domain at its C terminus and is conserved in mouse (Mus musculus), yeast (Saccharomyces cerevisiae), and Arabidopsis (Arabidopsis thaliana; Savitsky et al., 1995b; Garcia et al., 2000). This protein kinase family was proven to play a central role in the DNA damage response. Hundreds of human (Homo sapiens) proteins are phosphorylated at Ser/Thr-Glu motifs in response to DNA damage in an ATM-dependent manner, and many of these substrates are involved in the DNA damage response process (Matsuoka et al., 2007). Tel1, the ATM homolog in budding yeast, has functions in checkpoint response to DSBs (Mantiero et al., 2007). The Arabidopsis atm mutant displays hypersensitivity to γ-irradiation, and the induction of up-regulated genes in response to γ-irradiation is dependent on AtATM (Garcia et al., 2003; Bray and West, 2005; Culligan et al., 2006). Moreover, AtATM is required for the DNA damage-induced transcription of transposable elements and long noncoding RNAs (Wang et al., 2016b).
ATM homologs play multiple roles during meiosis. ATM-deficient mice are completely infertile due to meiosis arrest and spermatocyte apoptosis during prophase I (Xu et al., 1996). Recent studies provided evidence for the role of ATM in DSB formation. In mouse Atm−/−, levels of SPO11-oligonucleotides, by-products of meiotic DSB formation, are elevated at least 10-fold in spermatocytes (Lange et al., 2011). This may account for the phenotypic rescue of Atm−/− by Spo11+/− (Bellani et al., 2005). In budding yeast, Tel1 prevents clusters of DSBs and further suppresses DSBs within the surrounding chromosomal region (termed DSB interference; Garcia et al., 2015). Amounts of SPO11-oligonucleotide are also increased in the tel1Δ mutant (Mohibullah and Keeney, 2017). ATM family proteins may also influence the formation and distribution of COs. The control of the number and distribution of COs on human autosomes appears to be defective in the absence of ATM, and the lack of Tel1 in budding yeast results in higher recombination and reduced CO interference (Barchi et al., 2008; Anderson et al., 2015). Plants have a unique developmental pattern in life cycles and are ideal systems to dissect the biological functions of proteins that play roles in both mitosis and meiosis. In Arabidopsis atm, pollen mother cells (PMCs) can complete the meiosis process but present aberrant chromosome associations during diakinesis and fragmentation during anaphase I (Garcia et al., 2003). The mutation of other genes, such as AtNBS1, AtATR (ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED), and AtFANCD2, can aggravate the meiotic defects of Atatm (Waterworth et al., 2007; Culligan and Britt, 2008; Kurzbauer et al., 2018). However, the relationship between AtATM and HR remains elusive.
In this study, we present detailed characterization of the function of OsATM in rice (Oryza sativa) meiosis. Our results show that OsATM plays important roles in meiotic DSB repair processes in parallel with OsDMC1-mediated HR.
RESULTS
Characterization of Three Splicing Mutants of OsATM
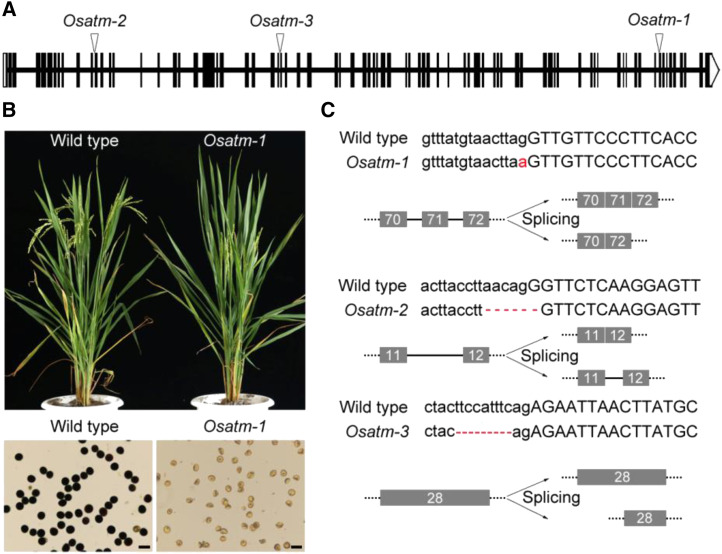
We constructed a library of rice mutants related to meiosis through irradiation mutagenesis. After map-based cloning (Supplemental Fig. S1), three sterile mutants disrupted in OsATM (LOC_Os01g01689) were isolated. These mutants were individually named Osatm-1, Osatm-2, and Osatm-3 (Fig. 1A). Osatm-1 exhibited normal vegetative growth but did not produce any seeds. The pollen grains of Osatm-1 were shrunken and could not be stained by I2-KI, indicating a lack of starch accumulation and sterility (Fig. 1B). When pollinated with wild-type pollen, Osatm-1 produced significantly fewer seeds compared with the wild type, suggesting that female fertility was also affected (Supplemental Table S1).
Figure 1.
Characterization of the Osatm mutants. A, Gene structure and mutation sites of OsATM in mutants. Exons are shown as black boxes. Untranslated regions are shown in white. B, Comparison of the plants at mature stage and pollen grains of the wild type and Osatm-1. Bars = 50 μm. C, Detailed information of mutation and diagram of defective splicing in Osatm. Uppercase letters present exons and lowercase letters present introns. Mutation sites are shown in red.
The genomic DNA sequence of OsATM spans 35.4 kb. We obtained a 9,614-bp full-length cDNA of OsATM by performing RACE, which consisted of 79 exons (Fig. 1A). There are two featured domains, PI3Kc (phosphoinositide 3-kinase catalytic domain) and FATC (FRAP, ATM, and TRRAP C-terminal), in the C-terminal end of OsATM, which are conserved among ATM homologs in different kingdoms (Supplemental Fig. S2).
Sequencing of the OsATM gene in Osatm-1 revealed a 1-bp substitution in the acceptor site of intron 70 (Fig. 1C). In Osatm-2, a 6-bp deletion occurred at the junction between intron 11 and exon 12. In Osatm-3, a 9-bp deletion occurred at the junction between intron 27 and exon 28. We sequenced the cDNA of OsATM in all three mutants and found that splicing patterns were completely altered. Compared with the wild type, exon 71 was missing in Osatm-1. A part of intron 11 was retained in Osatm-2, and exon 28 was shortened in Osatm-3 (Supplemental Fig. S3). All of these splicing modifications result in frame shifts and premature translation stop.
Aberrant Chromosome Associations and Fragmentations Are Observed in Osatm PMCs
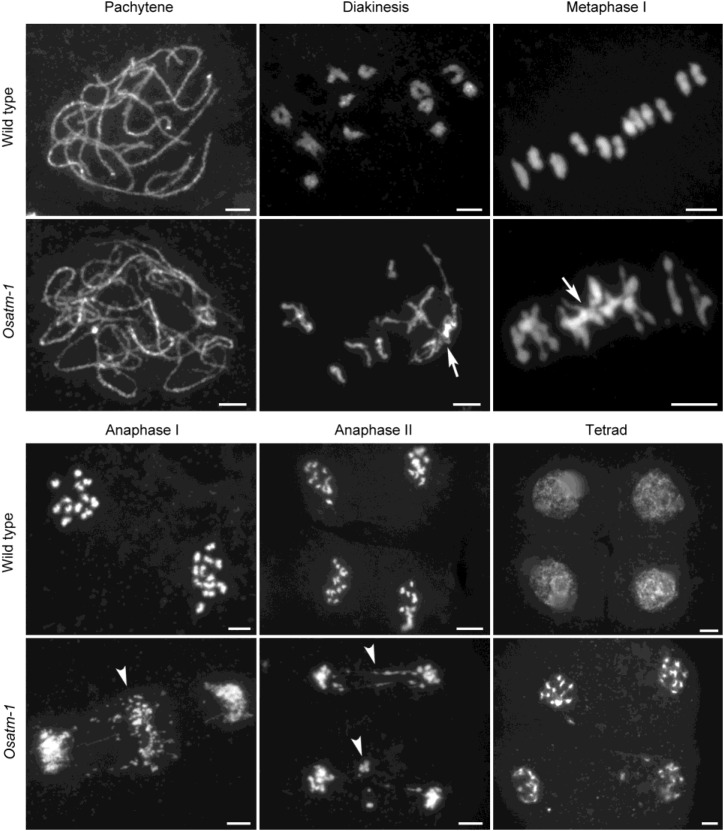
We examined chromosome spreads stained with 4′,6-diamino-phenylindole (DAPI) to show whether Osatm mutants have altered meiotic chromosome behaviors. Chromosome behaviors showed no obvious differences up to pachytene between the wild type and Osatm-1 (n = 30 for both wild-type and Osatm-1 PMCs; Fig. 2). At diakinesis in the wild type (n = 35 for PMCs), 12 highly condensed and well-distinguished bivalents were observed. By contrast, chromosomes entangled to form multivalents in Osatm-1 PMCs at diakinesis (n = 35 for PMCs). These aberrant chromosome associations in Osatm-1 became more evident at metaphase I, although bivalent-like structures existed in most cells. We analyzed the content of chromosome entanglements in Osatm-1 by quantifying the DAPI-stained entities at metaphase I. There are 12 DAPI-stained entities in wild-type PMCs, as they contained 12 isolated bivalents (n = 40 for PMCs). The entangled chromosomes were deemed to be one DAPI-stained entity, and the number of DAPI-stained entities per PMC in Osatm-1 was 4.95 ± 1.97 (n = 40 for PMCs). At anaphase I, when homologous chromosomes separated from each other in the wild type, chromosome bridges and extensive fragmentations were observed in Osatm-1 (n = 32 for both wild-type and Osatm-1 PMCs). Chromosome bridges and fragmentations were also detected at anaphase II, and tetrads with scattered DNA eventually formed in Osatm-1 (n = 35 for Osatm-1 PMCs).
Figure 2.
Comparison of chromosome behaviors between the wild type and Osatm-1 during meiosis. Chromosomes were stained with DAPI. Meiotic stages are indicated above the images. Aberrant chromosome associations are indicated by arrows, and chromosome fragmentations are indicated by arrowheads. Bars = 5 μm.
The chromosome behaviors of Osatm-2 and Osatm-3 resembled that of Osatm-1 (Supplemental Fig. S4). The numbers of DAPI-stained entities were 4.85 ± 1.98 in Osatm-2 (n = 40 for PMCs) and 4.63 ± 1.88 in Osatm-3 (n = 40 for PMCs). These results indicate that mutations of OsATM cause abnormal chromosome interactions during meiosis.
Homologous Pairing and Synapsis Occur Normally in Osatm-1
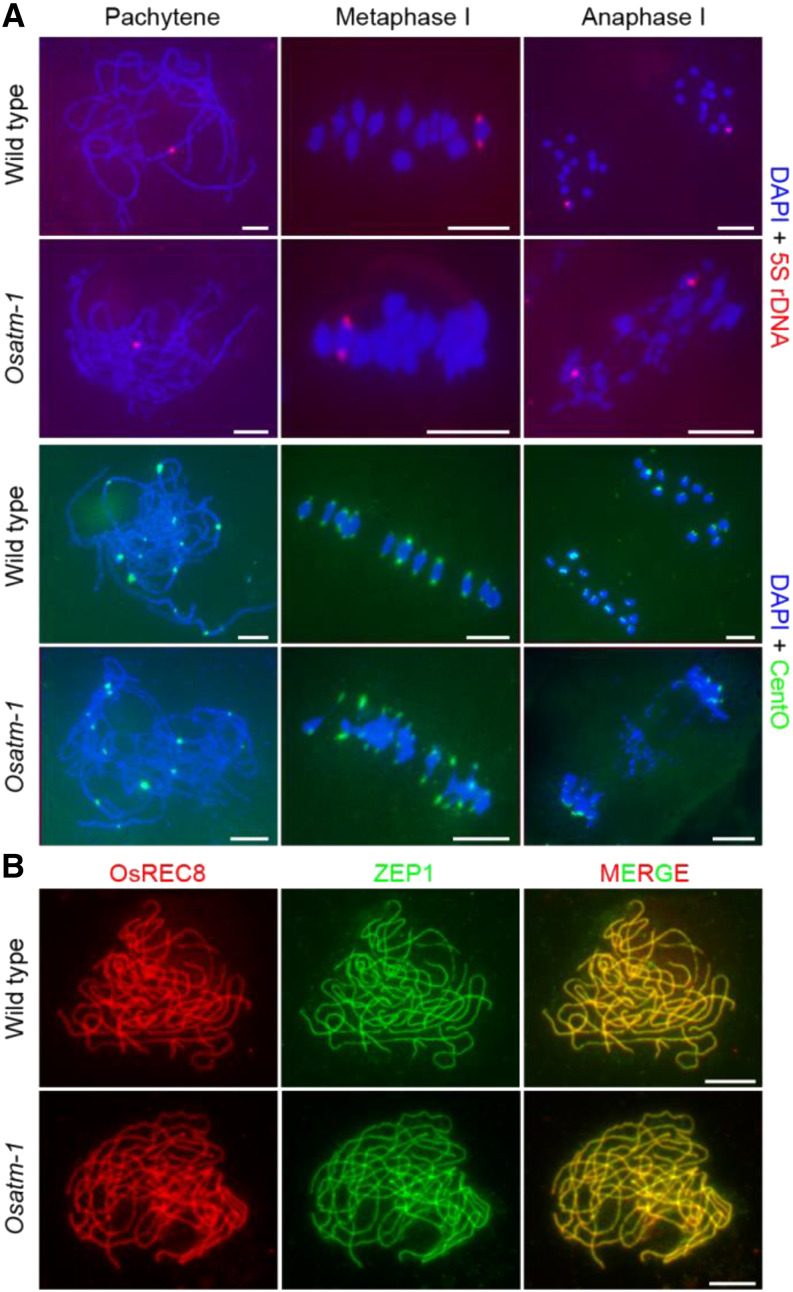
The results of DAPI staining at pachytene in Osatm suggested paired chromosomes (Fig. 2). We performed fluorescence in situ hybridization (FISH) experiments to verify whether the paired chromosomes are homologous (Fig. 3A). 5S rDNA is located on the centromere-proximal region of chromosome 11. Both the wild type and Osatm-1 (n = 30 for both wild-type and Osatm-1 PMCs) exhibited one 5S rDNA signal at pachytene, indicating paired homologous chromosomes. At metaphase I, two signals on one bivalent were observed in Osatm-1 when the bivalent was separated from the entangled chromosomes occasionally (6%; n = 50 for PMCs). The two 5S rDNA foci were pulled toward the opposite poles of PMCs at anaphase I, in accordance with the separation of homologous chromosomes (n = 30 for both wild-type and Osatm-1 PMCs). This indicated that homologous pairing occurs normally in Osatm-1.
Figure 3.
Comparison of chromosome interactions in the wild type and Osatm-1. A, FISH analysis in wild-type and Osatm-1 PMCs using 5S rDNA (red) and CentO (green) probes at different stages. Chromosomes are stained with DAPI (blue). B, Immunostaining of ZEP1 (green) in the wild type and Osatm-1 at pachytene. OsREC8 (red) indicates the chromosomes. Bars = 5 μm.
We also used the centromere probe CentO and the telomere probe pAtT4 to present a detailed morphology of chromosome interactions. At pachytene, in both wild-type and Osatm-1 PMCs (n = 30 for both wild-type and Osatm-1 PMCs), 12 CentO signals were observed (Fig. 3A). At metaphase I, CentO signals were stretched out of the chromosomes due to the pulling force of spindles (n = 30 for both wild-type and Osatm-1 PMCs). We found that all of the CentO signals were pulled to poles of PMCs, and the chromosome fragmentations were completely devoid of CentO at anaphase I in Osatm-1 (n = 20 for Osatm-1 PMCs). In PMCs of Osatm-1 at pachytene, as in the wild type (n = 35 for both wild-type and Osatm-1 PMCs), 24 distinct telomere signals were observed (Supplemental Fig. S5). We also found that pAtT4 signals had a tendency to be situated inside the chromosomes at metaphase I. In contrast to CentO, extensive pAtT4 signals were observed in the chromosome remnants in Osatm-1 at anaphase I (n = 32 for Osatm-1 PMCs).
We used an antibody against ZEP1, a central element of the synaptonemal complex, to check the synapsis status in Osatm-1 (Fig. 3B) by performing immunolocalization. As in the wild type, ZEP1 displayed linear localization patterns that completely merged with cohesion protein OsREC8 in Osatm-1 PMCs at pachytene (n = 30 for both wild-type and Osatm-1 PMCs). These results suggest that mutation of OsATM does not disrupt synapsis.
OsATM Is Required for the Repair of OsSPO11-1-Dependent DSBs
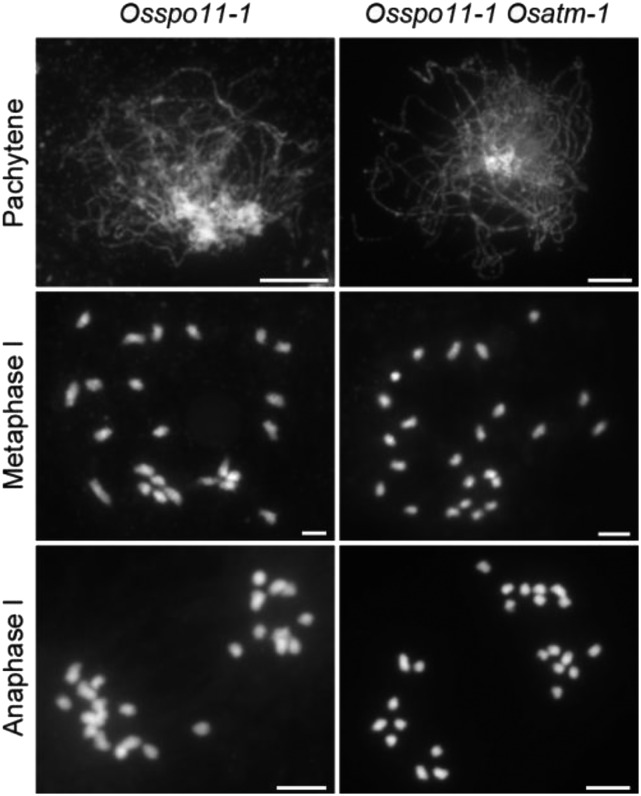
Programmed formation of DSBs initiates HR during meiosis (Keeney et al., 1997). To determine whether the chromosomal abnormalities in Osatm were acquired during meiotic DSB repair, we constructed Osspo11-1 Osatm-1 double mutants to carry out genetic analysis. Due to the absence of meiotic DSBs, homologous chromosomes in Osspo11-1 PMCs failed to pair at pachytene. At metaphase I, PMCs (n = 30 for PMCs) contained 24 scattered univalents. At anaphase I (n = 30 for PMCs), these univalents separated randomly (Fig. 4). The Osspo11-1 Osatm-1 double mutant exhibited the same chromosome phenotype as Osspo11-1, and no aberrant chromosome associations (n = 34 for PMCs at metaphase I) or fragmentations (n = 31 for PMCs at anaphase I) were observed. Thus, the meiotic defects in Osatm-1 are related to the repair of OsSPO11-1-dependent DSBs.
Figure 4.
Genetic analysis of OsATM with OsSPO11-1. DAPI-stained chromosomes of Osspo11-1 and Osspo11-1 Osatm-1 are shown. Meiotic stages are indicated on the left of the images. The chromosome behaviors of Osspo11-1 Osatm-1 are indistinguishable from that of Osspo11-1. Bars = 5 μm.
This result is different from the case of Arabidopsis, in which mutation of AtSPO11-1 only partially eliminates the presence of chromosome fragments in Atatm. However, the residual fragments in Atspo11-1 Atatm may be attributed to residual AtSPO11-1 activity in the Atspo11-1-1 allele (Culligan and Britt, 2008).
OsATM Functions in the Repair of Meiotic DSBs in Parallel with OsDMC1 and OsZIP4
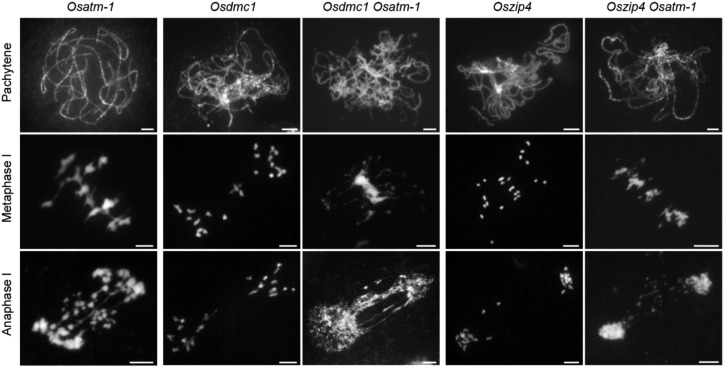
We conducted additional genetic analysis to dissect the relationship between OsATM and HR. DMC1-mediated HR is one of the meiotic DSB repair pathways that leads to the formation of COs. In rice meiosis, the disruption of OsDMC1 results in a typical asynaptic phenotype at pachytene (Wang et al., 2016a). At metaphase I, PMCs (n = 30 for PMCs) contained nearly 24 univalents, and no chromosome fragmentations were detected at anaphase I (n = 30 for PMCs; Fig. 5). When combining the mutation of OsDMC1 and OsATM, interestingly, we observed more severe defects than those of either single mutant. At pachytene, Osdmc1 Osatm-1 displayed destroyed synapsis (n = 40 for PMCs), the same phenotype of Osdmc1. However, chromosomes entangled to mass and no regular chromosome morphology was observed at metaphase I in the double mutants (n = 42 for PMCs). At anaphase I, more chromosome bridges formed due to the stickiness among chromosomes (P < 0.0001, two-tailed Student’s t test; n = 25 for both Osatm-1 and Osdmc1 Osatm-1 PMCs; Supplemental Fig. S6). These data suggest that OsATM and OsDMC1 act in parallel with each other in the meiotic DSB repair process.
Figure 5.
Genetic analyses of OsATM with OsDMC1 and OsZIP4. DAPI-stained chromosomes of Osatm-1, Osdmc1, Osdmc1 Osatm-1, Oszip4, and Oszip4 Osatm-1 are shown. Osdmc1 is the homozygous double mutant of OsDMC1A and OsDMC1B. Meiotic stages are indicated on the left. Aberrant chromosome associations and fragmentations are presented in Osdmc1 Osatm-1 and Oszip4 Osatm-1 double mutants. Bars = 5 μm.
OsZIP4 is one of the ZMM proteins in rice that participates in CO formation, the outcome of HR (Shen et al., 2012). Loss of function of OsZIP4 did not disrupt synapsis but led to abundant univalents (Fig. 5). We constructed Oszip4 Osatm-1 double mutants and found that the synapsis at pachytene was also normal (n = 32 for PMCs), comparable to the two single mutants. Chromosome entanglements at metaphase I and fragmentations at anaphase I were observed in Oszip4 Osatm-1 (n = 35 for PMCs), indicating that aberrant chromosome associations in Osatm-1 arise independently of the ZMM-mediated CO pathway.
OsATM Is Dispensable for Phosphorylation of H2AX during Rice Meiosis
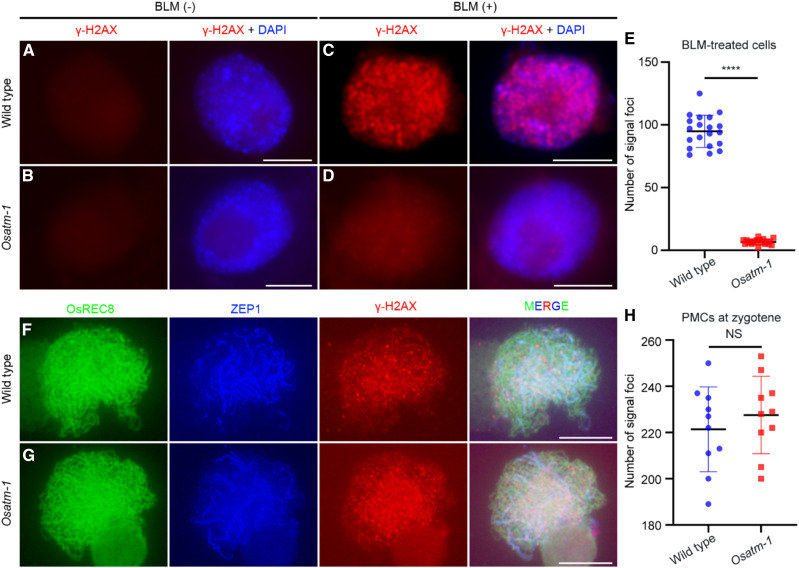
Phosphorylation of histone H2AX, referred to as γ-H2AX, is one of the earliest events in response to DSBs (Rogakou et al., 1998). In Arabidopsis somatic cells, phosphorylation of H2AX is dependent on both ATM and ATR in response to DSBs caused by ionizing radiation (IR), and ATM has a more dominant role (Friesner et al., 2005). Here, we proved that OsATM is also required for the phosphorylation of H2AX in root tip cells when challenged with bleomycin, a DNA-damaging chemical causing single-strand breaks and DSBs. We did not observe any γ-H2AX foci in untreated wild-type cells, but discrete, chromosomally located γ-H2AX foci appeared after bleomycin treatment (94.9 ± 12.8, n = 20 for cells; Fig. 6, A and C). In contrast to the wild type, bleomycin treatment barely induced the formation of γ-H2AX foci in Osatm-1 (6.7 ± 2.11, n = 20 for cells; P < 0.0001, two-tailed Student’s t test; Fig. 6, B, D, and E). As a control, the formation of OsRAD51 foci induced by bleomycin treatment was not affected in Osatm-1 (n = 15 for both wild-type and Osatm-1 cells; P = 0.6470, two-tailed Student’s t test; Supplemental Fig. S7).
Figure 6.
Immunostaining of γ-H2AX in root tip cells and PMCs of the wild type and Osatm-1. A to D, Immunostaining of γ-H2AX in root tip cells of the wild type (A and C) and Osatm-1 (B and D) with and without bleomycin (BLM) treatment. E, Quantifications of foci per cell with bleomycin treatment and statistical analyses from two-tailed Student’s t tests (****P < 0.0001). Means ± sd are shown for the wild type (94.9 ± 12.8; n = 20) and Osatm-1 (6.7 ± 2.11; n = 20). F and G, Immunostaining of γ-H2AX in PMCs of the wild type (F) and Osatm-1 (G) at zygotene. OsREC8 (green) indicates the chromosome and ZEP1 (blue) indicates the meiotic stage. H, Quantifications of foci per PMC and statistical analyses from two-tailed Student’s t tests (P = 0.4408). NS, Not significant. Means ± sd are shown for the wild type (221.4 ± 18.37; n = 10) and Osatm-1 (227.6 ± 16.76; n = 10). Bars = 5 μm.
Programmed DSBs during meiosis also induce the phosphorylation of H2AX in the wild type. We then tested whether γ-H2AX foci formation in Osatm-1 PMCs was abolished. During zygotene in the wild type, accompanied by ZEP1 elongation, extensive γ-H2AX foci were observed (Fig. 6F). In Osatm-1 PMCs at the same stage, we also detected γ-H2AX signals (Fig. 6G). The number of γ-H2AX foci was comparable between the wild type (221.4 ± 18.4, n = 10 for PMCs) and Osatm-1 (227.6 ± 16.8, n = 10 for PMCs; P = 0.4408, two-tailed Student’s t test; Fig. 6H). This result indicates that OsATM is dispensable for the phosphorylation of H2AX during rice meiosis.
Dynamics of OsDMC1 and Number of HEI10 Are Changed in Osatm-1
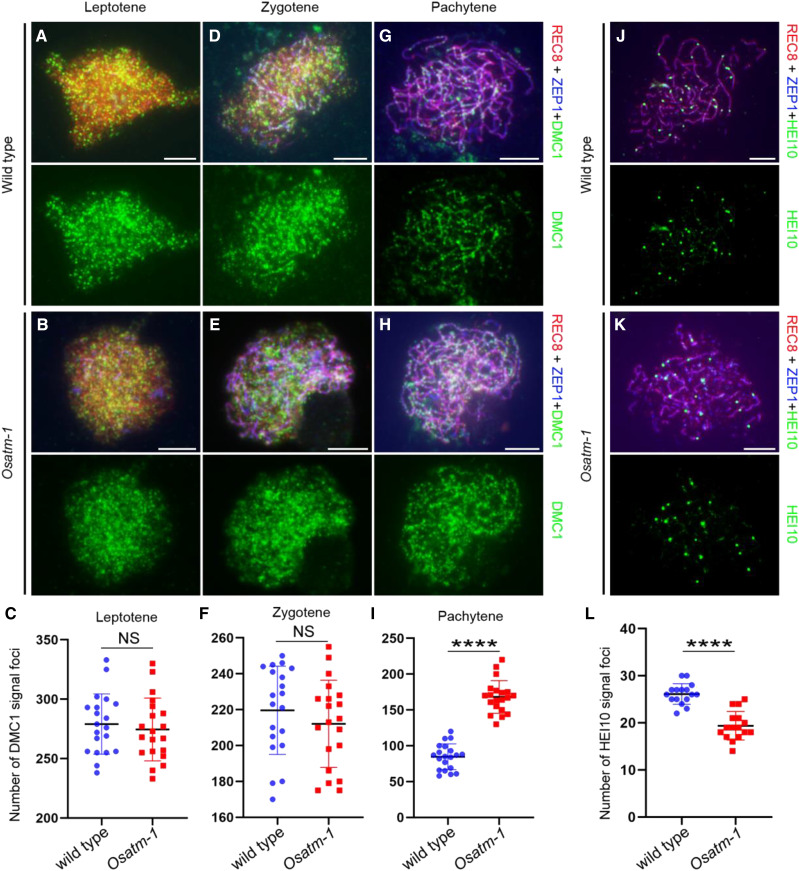
The DSB repair defects in Osatm-1 prompted us to investigate the dynamics of OsDMC1, an important player in HR-mediated DSB repair. At leptotene, as ZEP1 started to be polymerized, abundant OsDMC1 foci were observed in Osatm-1 (Fig. 7, A and B), suggesting that OsATM is not required for the recruitment of OsDMC1 onto chromosomes. The number of OsDMC1 foci in Osatm-1 (279.1 ± 25.3, n = 20 for PMCs) did not deviate significantly from that of the wild type (294.6 ± 26.4, n = 20 for PMCs; P = 0.5859, two-tailed Student’s t test; Fig. 7C). When ZEP1 extended along chromosomes at zygotene, the number of OsDMC1 foci decreased similarly in the wild type (219.6 ± 24.6, n = 20 for PMCs) and Osatm-1 (212.1 ± 24.3, n = 20 for PMCs; P = 0.3378, two-tailed Student’s t test; Fig. 7, D–F). At pachytene, after synapsis completion, OsDMC1 signals depleted rapidly from chromosomes in the wild type (84.7 ± 17.9, n = 20 for PMCs; Fig. 7G). However, the Osatm-1 PMCs maintained significantly more foci of OsDMC1 (168.2 ± 22.8, n = 20 for PMCs) compared with the wild type (P < 0.0001, two-tailed Student’s t test; Fig. 7, H and I). This indicated that the DSB repair process may be delayed in Osatm-1.
Figure 7.
Immunostaining of OsDMC1 and HEI10 in wild-type and Osatm-1 PMCs. A, B, D, E, G, and H, Dynamic loading patterns of OsDMC1 in the wild type (A, D, and G) and Osatm-1 (B, E, and H) at different meiotic stages. OsREC8 (red) indicates the chromosomes and ZEP1 (blue) indicates the meiotic stages. C, Means ± sd for the wild type (279.1 ± 25.35; n = 20) and Osatm-1 (274.6 ± 26.44; n = 20). F, Means ± sd for the wild type (219.6 ± 24.58; n = 20) and Osatm-1 (212.1 ± 24.28; n = 20). I, Means ± sd for the wild type (84.65 ± 17.89; n = 20) and Osatm-1 (168.15 ± 22.81; n = 20). Quantifications of foci per PMC and statistical analyses from two-tailed Student’s t tests are presented (****P < 0.0001). NS, Not significant. J and K, Number of HEI10 foci in PMCs of the wild type (J) and Osatm-1 (K) at late pachytene. L, Means ± sd for the wild type (26.13 ± 2.187; n = 16) and Osatm-1 (19.38 ± 3.030; n = 16). Quantifications of foci per PMC and statistical analyses from two-tailed Student’s t tests are presented (****P < 0.0001). Bars = 5 μm.
COs are the outcome of HR. We wanted to explore whether the formation of COs was affected by the mutation of OsATM. The bright foci of HEI10, a member of ZMM proteins, are reliable markers of interference-sensitive CO intermediates (Wang et al., 2012). The number of HEI10 foci in Osatm-1 (Fig. 7K; 19.4 ± 3, n = 16 for PMCs) decreased significantly compared with that of the wild type (Fig. 7J; 26.1 ± 2.2, n = 16 for PMCs; P < 0.0001, two-tailed Student’s t test; Fig. 7L). Therefore, the number of interference-sensitive CO intermediates was partially reduced due to the loss of OsATM.
DISCUSSION
The function of ATM in human has received extensive attention since its isolation in 1995 (Savitsky et al., 1995a). In this study, we dissected the meiotic function of OsATM, the ATM homolog in rice, based on the characterization of three allelic mutants. All of these mutants contain nucleotide modifications at the junction between one intron and the following exon. Accordingly, splicing defects (i.e. exon skipping in Osatm-1, retained intron in Osatm-2, and alternate acceptor site in Osatm-3) were detected in these mutants (Fig. 1C). Interestingly, mutations resulting in defective splicing of ATM constitute a significant proportion (∼40%) in patients with ataxia telangiectasia (Teraoka et al., 1999). These data imply that a conserved mechanism may exist that causes the predominance of splicing mutations of ATM in both animals and plants.
OsATM and OsDMC1 Function in Different Pathways in Meiotic DSB Repair
The aberrant chromosome associations in Osatm-1 were reminiscent of the case in meica1. Loss of MEICA1 leads to nonhomologous chromosome association, the formation of massive chromosome bridges, and chromosome fragmentation (Hu et al., 2017). The Osdmc1 meica1 double mutant shows a phenotype similar to Osdmc1, indicating that MEICA1 acts in recombination after strand invasion. Interestingly, Osdmc1 Osatm-1 showed an additive phenotype of these two single mutants (Fig. 5), indicating that OsATM and OsDMC1 function in different pathways during meiotic DSB repair. We proposed a possible mechanism for the more severe phenotype observed in Osdmc1 Osatm. Meiotic DSBs in Osatm were channeled into two pathways for repair. The accurate pathway is OsDMC1-mediated HR that leads to homologous pairing and synapsis. The inaccurate pathway involves interaction between nonhomologous chromosomes. Disruption of OsDMC1 in Osatm blocked HR. Thus, more DSBs were channeled into the inaccurate repair pathway and more ectopic chromosome interactions formed.
Another case of aberrant chromosome associations was observed in the mutant of OsRAD1, which encodes a member of the conserved RAD9-RAD1-HUS1 complex in rice (Hu et al., 2016). OsRAD1 functions in parallel with OsDMC1 during meiotic DSB repair, which resembles the situation of OsATM. Importantly, loss of Ku70 partially reduces the meiotic defects of Osrad1, indicating the function of OsRAD1 in suppressing nonhomologous end joining (NHEJ), another way for DSB repair. The critical determinant between DSB repair pathway choice (i.e. HR and NHEJ) is the initiation of 5′ to 3′ resection of DNA ends (Symington and Gautier, 2011). In budding yeast, Tel1 was reported to promote resection initiation and extension (Mimitou et al., 2017). Thus, OsATM may stimulate DSB resection to inhibit the NHEJ pathway.
An alternative interpretation of the nonhomologous associations in Osatm involves nonallelic HR, which takes place between nonallelic DNA segments that share high sequence identity. Nonallelic HR can markedly alter genome architecture and thus should be repressed (Sasaki et al., 2010). It is tempting to speculate that OsATM participates in the removal of nonallelic HR that may cause nonhomologous associations.
In contrast to mammals (Barlow et al., 1998), the recruitment of OsDMC1 does not depend on OsATM (Fig. 7, A and B). This may account for the normal pairing and synaptonemal complex formation in Osatm-1 (Fig. 3). In the Arabidopsis atm background, mutation of AtATR, another protein kinase involved in responses to DNA damage, prevents complete synapsis of chromosomes. Thus, these two protein kinases may act redundantly in processing meiotic DSBs (Culligan and Britt, 2008). Interestingly, more OsDMC1 foci were present in Osatm-1 than in wild-type PMCs at pachytene (Fig. 7I), which resembles the situation in weak mutant alleles of certain DNA polymerases in Arabidopsis (Wang et al., 2019). This suggests that meiotic DSB repair processes may be delayed due to the loss of OsATM.
OsATM Is Not Essential for the Phosphorylation of H2AX during Rice Meiosis
Studies in mammals clearly established ATM as the major kinase involved in the phosphorylation of H2AX in response to DNA DSBs (Burma et al., 2001). In Arabidopsis, AtATM plays a dominant role in IR-induced γ-H2AX formation (Friesner et al., 2005). Similar results were obtained in this study, as the bleomycin-induced γ-H2AX foci in Osatm-1 root tip cells almost disappeared (Fig. 6, D and E).
However, γ-H2AX foci were observed in Osatm-1 PMCs at zygotene (Fig. 6, G and H), indicating that OsATM is not essential for the phosphorylation of H2AX in rice meiosis. In mouse meiocytes, ATM is responsible for chromatin-wide phosphorylation of H2AX during leptotene. However, in a particular genotype, Atm–/–Spo11+/–, γ-H2AX signals that coincide with the X and Y chromatin were observed. This demonstrated that ATM is dispensable for the phosphorylation of H2AX in the sex body (Bellani et al., 2005). Moreover, similar levels of γ-H2AX were induced by microbial pathogens in Atatm-2 compared with the wild type in Arabidopsis somatic cells (Song and Bent, 2014). These findings suggest that there are diverse mechanisms of H2AX phosphorylation. In Arabidopsis, ∼10% of IR-induced γ-H2AX requires functional AtATR (Friesner et al., 2005). It is tempting to speculate that OsATR functions in H2AX phosphorylation in rice meiosis when the function of OsATM is compromised. Further studies, such as immunostaining of γ-H2AX in Osatr and Osatm Osatr PMCs, are needed to clarify this issue.
OsATM May Not Be Involved in the Control of Meiotic DSB Number
Studies of Drosophila melanogaster meiosis showed that the number of γ-H2AX foci is dramatically increased in the absence of ATM, suggesting that the number of DSBs is increased (Joyce et al., 2011). More RAD-51 foci were observed in meiotic cells in Caenorhabditis elegans atm-1 mutants compared with the wild type, a result that was consistent with a conserved role of ATM in DSB inhibition (Checchi et al., 2014). Similar results were obtained by immunoblot analysis of SPO11-oligonucleotide complexes in mouse and budding yeast (Lange et al., 2011; Mohibullah and Keeney, 2017). However, we did not observe any significant difference in the foci numbers of γ-H2AX between the PMCs of the wild type and Osatm-1 (Fig. 6H). The foci numbers of OsDMC1, a proxy of DSB sites, were comparable between the wild type and Osatm-1 at leptotene and zygotene (Fig. 7, C and F). Thus, in contrast to animals and yeast, OsATM may not be involved in the control of meiotic DSB number in rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Three mutants of OsATM were isolated from the rice (Oryza sativa) indica variety Guangluai 4 induced by 60Co γ-ray irradiation. The Osspo11-1, Osdmc1, and Oszip4 alleles employed in this study were previously isolated in our lab (Shen et al., 2012; Hu et al., 2016; Wang et al., 2016a). Guangluai 4 was used as the wild type in the related experiments. All plants were grown in paddy fields in Beijing (China), Yangzhou (Jiangsu Province, China), or Sanya (Hainan Province, China) during the natural growing season.
Map-Based Cloning of OsATM
For map-based cloning of OsATM, heterozygous mutant plants of Osatm-1, Osatm-2, and Osatm-3 were crossed with the japonica variety Nipponbare to generate the mapping populations. OsATM was mapped within a 150-kb region on the terminus of chromosome 1 using the mapping population of the F2, F3, and F4 generations (n = 928 in total).
Based on the Michigan State University Rice Genome Annotation Project Database and Resource (http://rice.plantbiology.msu.edu/), a candidate OsATM gene was identified, which was annotated as a phosphatidylinositol 3- and 4-kinase family protein. Sequencing of OsATM in the three mutant lines showed that all mutants were disrupted in this gene.
Full-Length cDNA Cloning of OsATM and Splicing Pattern Analyses in the Mutants
Total RNA extraction from rice young panicles was conducted using the TRIzol reagent (Invitrogen). Reverse transcription was performed with primer Adaptor-T (18) using SuperScript III RNase H reverse transcriptase (Invitrogen). For RACE, 3′-Full RACE Core Set with PrimeScript RTase (TaKaRa) and 5′-Full RACE Kit with TAP (TaKaRa) were used to identify the 3′ and 5′ ends of the cDNA, respectively. PCR was performed to amplify the open reading frame. Then the products were cloned into the PMD18-T vector (TaKaRa) and sequenced. The sequences were then spliced together to obtain the full-length cDNA sequence.
The cDNAs of Osatm-1, Osatm-2, and Osatm-3 (three plants for each mutant) were amplified using primer pairs spanning the mutated site. PCR products were cloned into the PMD18-T vector (TaKaRa) and sequenced (10 clones for each plant).
Meiotic Chromosome Preparation
Fresh young panicles were fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1) at room temperature for 24 h and transferred into a freezer. Anthers of appropriate size were squashed in acetocarmine solution. Cover slips were added and knocked gently by tweezers. After the observation of meiotic stages by light microscopy, acetocarmine was removed by 45% (v/v) acetic acid by wicking with filter paper. Slides with chromosomes were frozen in liquid nitrogen, and the cover slips were removed rapidly. Air-dried samples were counterstained with DAPI in an antifade solution, and new cover slips were added. Images were captured with a Zeiss A2 fluorescence microscope with a micro-CCD camera. The PMCs used for chromosome preparation were obtained from three plants of the wild type and mutants.
FISH
FISH analyses were conducted as described previously (Yang et al., 2016). The PMCs used for FISH analyses were obtained from three plants of the wild type and mutants. Three repetitive DNA elements, pTa794, pAtT4, and CentO, were used as FISH probes. The pTa794 clone contains the coding sequences for the 5S rRNA genes from wheat (Triticum aestivum; Cuadrado and Jouve, 1994). pAtT4 contains telomeric repetitive sequences (Richards and Ausubel, 1988), and CentO contains rice centromeric tandem repeats (Cheng et al., 2002). Chromosome images were captured with the Olympus BX51 fluorescence microscope with a micro-CCD camera using the IPLab 4.0 software.
Bleomycin Treatments and Root Tip Slide Preparation
Husked rice seeds were sown on one-half-strength Murashige and Skoog medium for 5 to 6 d. The seedlings were transferred into bleomycin (20 μg mL−1) for a 2-h treatment. After thorough washing, the root tips were cut and fixed in paraformaldehyde (4% [w/v]) for 30 min. Then, the root tips were digested by cellulase (1% [w/v]) and pectinase (2% [w/v]) for 30 min at 37°C. Root tips were squashed gently onto gelatin-coated slides and cover slips were added.
Immunofluorescence Assay
The primary antibodies against OsREC8, ZEP1, OsDMC1, HEI10, γ-H2AX, and OsRAD51 were generated previously (Wang et al., 2010, 2012, 2016a; Shao et al., 2011; Miao et al., 2013; Xu et al., 2018). The procedure for the immunofluorescence assays was performed as previously described (Shen et al., 2012).
Computational and Database Analyses
Multiple sequence alignment was conducted using MAFFT (https://toolkit.tuebingen.mpg.de/#/tools/mafft) and colored with ESPript (http://espript.ibcp.fr/ESPript/ESPript/). Data analyses were mainly conducted using GraphPad Prism 8 software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsATM (rice), XP_015615410.1; AtATM (Arabidopsis thaliana), NP_001326746.1; ATM (Homo sapiens), XP_011541144.1; Tefu (Drosophila melanogaster), NP_001036712.1; Tel1 (Saccharomyces cerevisiae), AJQ12538.1; ATM-1 (Caenorhabditis elegans), NP_001293214.1; ZEP1 (rice), XP_025880993.1; OsDMC1A (rice), XP_015618986.2; OsDMC1B (rice), XP_025877096.1; and HEI10 (rice), XP_015623604.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Map-based cloning of OsATM.
Supplemental Figure S2. Multiple sequence alignment of the PI3Kc and FATC domains in ATM homologs among species.
Supplemental Figure S3. Detailed sequence modification of cDNA in the mutants.
Supplemental Figure S4. Chromosome behaviors in Osatm-2 and Osatm-3.
Supplemental Figure S5. FISH analysis of telomere in the wild type and Osatm-1.
Supplemental Figure S6. Quantification of chromosome bridges of Osatm-1 and Osdmc1 Osatm-1 at anaphase I.
Supplemental Figure S7. Immunostaining of OsRAD51 in root tip cells of the wild type and Osatm-1 with and without bleomycin treatment.
Supplemental Table S1. Seed setting rate of the wild type and Osatm-1 when pollinated with wild-type pollen grains.
Footnotes
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (grant no. 2016YFD0102001), the National Natural Science Foundation of China (grant no. 31872859), and the Jiangsu Higher Education Institutions of China (grant no. PAPD).
References
- Anderson CM, Oke A, Yam P, Zhuge T, Fung JC(2015) Reduced crossover interference and increased ZMM-independent recombination in the absence of Tel1/ATM. PLoS Genet 11: e1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi M, Roig I, Di Giacomo M, de Rooij DG, Keeney S, Jasin M(2008) ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet 4: e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, Rottinghaus S, Jackson SP, Tagle D, Ried T, et al. (1998) Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 125: 4007–4017 [DOI] [PubMed] [Google Scholar]
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD(2005) SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118: 3233–3245 [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N(2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Bray CM, West CE(2005) DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol 168: 511–528 [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ(2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276: 42462–42467 [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N(1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Checchi PM, Lawrence KS, Van MV, Larson BJ, Engebrecht J(2014) Pseudosynapsis and decreased stringency of meiotic repair pathway choice on the hemizygous sex chromosome of Caenorhabditis elegans males. Genetics 197: 543–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J(2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Jouve N(1994) Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6x-triticale. Chromosome Res 2: 331–338 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Britt AB(2008) Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J 55: 629–638 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB(2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR(2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB(2005) Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A(2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ(2015) Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 520: 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Salanoubat M, Choisne N, Tissier A(2000) An ATM homologue from Arabidopsis thaliana: Complete genomic organisation and expression analysis. Nucleic Acids Res 28: 1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Li Y, Wang H, Shen Y, Zhang C, Du G, Tang D, Cheng Z(2017) Meiotic Chromosome Association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29: 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Tang D, Wang H, Shen Y, Chen X, Ji J, Du G, Li Y, Cheng Z(2016) The exonuclease homolog OsRAD1 promotes accurate meiotic double-strand break repair by suppressing nonhomologous end joining. Plant Physiol 172: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, Pedersen M, Tiong S, White-Brown SK, Paul A, Campbell SD, McKim KS(2011) Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J Cell Biol 195: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N(1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kurzbauer MT, Pradillo M, Kerzendorfer C, Sims J, Ladurner R, Oliver C, Janisiw MP, Mosiolek M, Schweizer D, Copenhaver GP, et al. (2018) Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30: 415–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S(2011) ATM controls meiotic double-strand-break formation. Nature 479: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Clerici M, Lucchini G, Longhese MP(2007) Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep 8: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z(2013) Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS(2009) DNA end resection: Many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Yamada S, Keeney S(2017) A global view of meiotic double-strand break end resection. Science 355: 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N, Keeney S(2017) Numerical and spatial patterning of yeast meiotic DNA breaks by Tel1. Genome Res 27: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC(2011) Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol 190: 523–544 [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM(1988) Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136 [DOI] [PubMed] [Google Scholar]
- Robert T, Nore A, Brun C, Maffre C, Crimi B, Bourbon HM, de Massy B(2016) The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351: 943–949 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM(1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S(2010) Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. (1995a) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268: 1749–1753 [DOI] [PubMed] [Google Scholar]
- Savitsky K, Sfez S, Tagle DA, Ziv Y, Sartiel A, Collins FS, Shiloh Y, Rotman G(1995b) The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet 4: 2025–2032 [DOI] [PubMed] [Google Scholar]
- Seeliger K, Dukowic-Schulze S, Wurz-Wildersinn R, Pacher M, Puchta H(2012) BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol 193: 364–375 [DOI] [PubMed] [Google Scholar]
- Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z(2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tang D, Wang K, Wang M, Huang J, Luo W, Luo Q, Hong L, Li M, Cheng Z(2012) ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T(1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470 [DOI] [PubMed] [Google Scholar]
- Song J, Bent AF(2014) Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLoS Pathog 10: e1004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Cheng Z, Huang J, Lin J, Copenhaver GP, Ma H, Wang Y(2017) Arabidopsis RAD51, RAD51C and XRCC3 proteins form a complex and facilitate RAD51 localization on chromosomes for meiotic recombination. PLoS Genet 13: e1006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J(2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Taylor AM, Harnden DG, Arlett CF, Harcourt SA, Lehmann AR, Stevens S, Bridges BA(1975) Ataxia telangiectasia: A human mutation with abnormal radiation sensitivity. Nature 258: 427–429 [DOI] [PubMed] [Google Scholar]
- Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengüt S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, et al. (1999) Splicing defects in the ataxia-telangiectasia gene, ATM: Underlying mutations and consequences. Am J Hum Genet 64: 1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Huang J, Zhang J, Wang H, Han Y, Copenhaver GP, Ma H, Wang Y(2019) The largest subunit of DNA polymerase delta is required for normal formation of meiotic type I crossovers. Plant Physiol 179: 446–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu Q, Tang D, Liu X, Du G, Shen Y, Li Y, Cheng Z(2016a) OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol 171: 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z(2012) The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z(2010) The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schwacke R, Kunze R(2016b) DNA damage-induced transcription of transposable elements and long non-coding RNAs in Arabidopsis is rare and ATM-dependent. Mol Plant 9: 1142–1155 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Altun C, Armstrong SJ, Roberts N, Dean PJ, Young K, Weil CF, Bray CM, West CE(2007) NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J 52: 41–52 [DOI] [PubMed] [Google Scholar]
- Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D(1996) Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev 10: 2411–2422 [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhang J, Xu M, Ji W, Yu M, Tao Y, Gong Z, Gu M, Yu H(2018) Rice RAD51 paralogs play essential roles in somatic homologous recombination for DNA repair. Plant J 95: 282–295 [DOI] [PubMed] [Google Scholar]
- Yang R, Li Y, Su Y, Shen Y, Tang D, Luo Q, Cheng Z(2016) A functional centromere lacking CentO sequences in a newly formed ring chromosome in rice. J Genet Genomics 43: 694–701 [DOI] [PubMed] [Google Scholar]