Abstract
Objectives
Apoptosis-Inducing Factor (AIF) is a protein involved in mitochondrial electron transport chain assembly/stability and programmed cell death. The relevant role of this protein is underlined because mutations altering mitochondrial AIF properties result in acute pediatric mitochondriopathies and tumor metastasis. By generating an original AIF-deficient mouse strain, this study attempted to analyze, in a single paradigm, the cellular and developmental metabolic consequences of AIF loss and the subsequent oxidative phosphorylation (OXPHOS) dysfunction.
Methods
We developed a novel AIF-deficient mouse strain and assessed, using molecular and cell biology approaches, the cellular, embryonic, and adult mice phenotypic alterations. Additionally, we conducted ex vivo assays with primary and immortalized AIF knockout mouse embryonic fibroblasts (MEFs) to establish the cell death characteristics and the metabolic adaptive responses provoked by the mitochondrial electron transport chain (ETC) breakdown.
Results
AIF deficiency destabilized mitochondrial ETC and provoked supercomplex disorganization, mitochondrial transmembrane potential loss, and high generation of mitochondrial reactive oxygen species (ROS). AIF-/Y MEFs counterbalanced these OXPHOS alterations by mitochondrial network reorganization and a metabolic reprogramming toward anaerobic glycolysis illustrated by the AMPK phosphorylation at Thr172, the overexpression of the glucose transporter GLUT-4, the subsequent enhancement of glucose uptake, and the anaerobic lactate generation. A late phenotype was characterized by the activation of P53/P21-mediated senescence. Notably, approximately 2% of AIF-/Y MEFs diminished both mitochondrial mass and ROS levels and spontaneously proliferated. These cycling AIF-/Y MEFs were resistant to caspase-independent cell death inducers. The AIF-deficient mouse strain was embryonic lethal between E11.5 and E13.5 with energy loss, proliferation arrest, and increased apoptotic levels. Contrary to AIF-/Y MEFs, the AIF KO embryos were unable to reprogram their metabolism toward anaerobic glycolysis. Heterozygous AIF+/- females displayed progressive bone marrow, thymus, and spleen cellular loss. In addition, approximately 10% of AIF+/- females developed perinatal hydrocephaly characterized by brain development impairment, meningeal fibrosis, and medullar hemorrhages; those mice died 5 weeks after birth. AIF+/- with hydrocephaly exhibited loss of ciliated epithelium in the ependymal layer. This phenotype was triggered by the ROS excess. Accordingly, it was possible to diminish the occurrence of hydrocephalus AIF+/- females by supplying dams and newborns with an antioxidant in drinking water.
Conclusions
In a single knockout model and at 3 different levels (cell, embryo, and adult mice) we demonstrated that by controlling the mitochondrial OXPHOS/metabolism, AIF is a key factor regulating cell differentiation and fate. Additionally, by providing new insights into the pathological consequences of mitochondrial OXPHOS dysfunction, our new findings pave the way for novel pharmacological strategies.
Keywords: AIF, Caspase-independent cell death, Hydrocephaly, Metabolism, Mitochondria, OXPHOS
Highlights
-
•
Mitochondrial AIF loss triggers OXPHOS/metabolism impairment.
-
•
AIF-/Y MEFs reprogram their metabolism by enabling an AMPK/GLUT-4 glycolytic pathway.
-
•
OXPHOS dysfunction associated with AIF loss generates a P53/P21 senescent phenotype.
-
•
The lack of metabolic plasticity provokes embryonic lethality in the AIF KO animals.
-
•
AIF+/- females develop immune cell exhaustion and perinatal hydrocephaly.
Abbreviations
- 2-DG
2-Deoxyglucose
- 2-NBDG
2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-d-glucose
- AIF
Apoptosis-inducing factor
- AMPK
5′ adenosine monophosphate-activated protein kinase
- BN-PAGE
Blue native polyacrylamide gel electrophoresis
- BrdU
Bromodeoxyuridine
- CSF
Cerebrospinal fluid
- CDKN1α; P21
Cyclin-dependent kinase inhibitor 1A
- COX
Cyclooxygenase
- DNM1L; DRP1
Dynamin-related protein 1
- ETC
Electron transfer chain
- FCCP
Fluoro-carbonyl cyanide phenylhydrazone
- GLUT-1
Glucose transporter 1
- GLUT-4
Glucose transporter 4
- Hq
Harlequin
- HSC
Hematopoietic stem cells
- HC
Hydrocephalus
- KO
Knockout
- MFI
Mean fluorescence intensity
- OPA1
Mitochondrial dynamin like GTPase
- FIS1
Mitochondrial fission 1 protein
- MFF
Mitochondrial fission factor
- MFN1
Mitofusin 1
- MFN2
Mitofusin 2
- MEFs
Mouse embryonic fibroblasts
- MNNG
N-Methyl-N′-nitro-N-nitrosoguanidine
- TMPD
N,N,N′,N'-tetramethyl-p-phenylenediamine
- OXPHOS
Oxidative phosphorylation
- PFA
Paraformaldehyde
- Xp
Paternal X chromosome
- PS
Phosphatidylserine
- PCD
Programmed cell death
- PI
Propidium iodide
- ROS
Reactive oxygen species
- pRb
Retinoblastoma protein
- STS
Staurosporine
- SDH
Succinate dehydrogenase
- SC
Supercomplex
- SOD1
Superoxide dismutase 1
- SOD2
Superoxide dismutase 2
- WT
Wild-type
1. Introduction
Mitochondria are wide-ranging organelles that through oxidative phosphorylation (OXPHOS) play a key role in the cell by providing energy in the form of ATP and by generating the reactive oxygen species (ROS), which regulate differentiation and fate [[1], [2], [3], [4], [5], [6]]. During OXPHOS, fuel substrates are oxidized and electrons transferred through the mitochondrial electron transfer chain (ETC), a branched chain of multiprotein complexes (complex I to IV). This process is chemiosmotically coupled to the phosphorylation of ADP to ATP by the ATP synthase (complex V). Mitochondrial ROS arise from the 0.1%–2% of electrons that escape from the ETC [7]. The most accepted model of ETC organization is the so-called “plasticity model,” which proposes different stable interactions between complexes (e.g., I and III; or I, III, and IV) in entities named mitochondrial supercomplexes (SCs) [8,9].
AIF is a bifunctional protein, encoded by Aifm1 (Pdcd8) on the X chromosome and implicated in maintaining a functional mitochondrial OXPHOS and, after its translocation to the nucleus, in programmed cell death (PCD) [[10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]]. The role of AIF in mitochondrial complexes assembly seems to relate to its interaction with CHCHD4, a component of the mitochondrial protein import machinery [12,13]. In this manner, AIF or CHCHD4 mutations are concomitant with OXPHOS deficiency-related mitochondriopathies (encephalopathy, infantile motor neuron disease, and early prenatal ventriculomegaly) [[25], [26], [27], [28]]. Moreover, the mitochondrial role of AIF is critical for the inhibition of tumor metastasis [29], and AIF overexpression aggravates hypoxic-ischemic brain injury in neonatal mice [30].
The role of AIF has been mainly assessed in the hypomorphic Harlequin (Hq) mice strain, which is not an AIF KO mouse but presents approximately 70% ubiquitous reduction in AIF expression [10]. Among others, the Hq strain exhibits T-cell development defaults, chronic neurodegeneration, progressive cerebellar ataxia, Purkinje cell degeneration, optic tract dysfunction, early fur abnormalities, and hypertrophic cardiomyopathy [10,16,17,[31], [32], [33]]. Studies on AIF have been complemented by the characterization of tissue-specific knockout models that corroborated T-cell developmental alterations, delayed neurogenesis, skeletal muscle atrophy, and dilated cardiomyopathy [14,15,34]. Finally, in a novel mouse model in which Aifm1 was ablated early during hematopoiesis, we observed hematopoietic stem cell (HSC) loss, thymopoiesis blockade, and delayed development of the T-cell, B-cell, and erythroid lineages [35,36].
Here, by generating a bona fide AIF KO mouse strain, we illustrate in a single model the consequences of the mitochondrial OXPHOS dysfunction associated with the loss of AIF at the cellular, embryonic, and adult mice levels. The generation of veritable AIF KO mice demonstrates new metabolic and phenotypic adaptive responses, reveals a greater role for AIF and mitochondrial OXPHOS in mouse development, and clarifies the AIF function in caspase-independent PCD.
2. Methods
2.1. Mice
Mice were housed at the Cordeliers Center animal facility under strictly controlled, specific-pathogen-free conditions (agreement B75-06-12). Experiments were performed in accordance with ARRIVE ethical guidelines and with the approval of the French Ministry of Agriculture (agreement 1675). Animals were maintained with a rodent diet (R03, Scientific Animal Food & Engineering Diets) and water was available ad libitum in a vivarium with a 12-hour light–dark cycle at 22 °C. In specific experiments, dams and newborns were fed a high-fat ketogenic diet (HFD; Research Diets) supplied or not with riboflavin (5 mg/100 mL) in drinking water.
Aifm1 floxed mice were generated by flanking the exon 11 of Aifm1 with LoxP sequences by using standard gene-targeting techniques (Genoway, France). After 15 backcrosses into the C57BL/6J background, Aifm1 floxed males (Aifm1fl/Y; AIFfl/Y) were crossed with PGK-Cre females (donated by Yvan Lallemand, Pasteur Institute). This crossing induced an excision of exon 11 in Aifm1 that resulted in a frameshift mutation and the creation of a stop codon in exon 12. The resulting Aifm1+/− (AIF+/-) females were identified by genomic PCR assessment (Supplemental Fig. 1C). For hematological analysis, females were euthanized at 6 months old. Spleens, lymph nodes, thymus, and bone marrow were sampled to prepare single cell suspensions by mechanical disruption and passage through a cell strainer. Absolute cell counts were then assessed.
In embryo studies, AIF+/- females were crossed with Aifm1+/Y (AIF+/Y) males. E7.5 to E13.5 embryos were dissected with the help of a stereomicroscope (M80; Leica), and pictures were obtained with an IC80 HD camera (Leica).
2.2. Generation of AIF-/Y mouse embryonic fibroblasts (MEFs)
To generate AIF-/Y MEFs, AIFfl/Y females were crossed with Rosa26-CreERT2 males (provided by Dr. Anton Bernes, NCI, Amsterdam, The Netherlands) [37], and MEFs were generated from a triple E12.5 transgenic male embryo. To obtain AIF-/Y cells, MEFs were treated overnight with tamoxifen (4-OHT; 1 μM).
2.3. Southern blot
Genomic DNA from WT (Co) and AIF-deficient (AIF-/Y) MEFs (4 days post-treatment with tamoxifen; 4-OHT) was extracted by using a Wizard Genomic DNA purification kit (Promega) and digested with Pci1. The genomic DNA fragments obtained, including those encompassing exons 7 to 13 of Aifm1, were separated on a 0.8% agarose gel and transferred to a hybond membrane. The membrane was baked for 2 h at 80 °C to fix DNA and then hybridized with a probe labeled with a Gene Images AlkPhos direct labeling and detection system kit from GE Healthcare. The probe, specific to the 3'end of AIF genomic DNA, was freshly prepared by PCR amplification of genomic DNA by using sense (intron 12 nucleotides 33070–33098) and reverse (intron 13 nucleotides 33433–33459) primers. The membrane was then washed, and the hybridization pattern was revealed by using CDP star (Amersham). Images were acquired on a MF-ChemiBIS 4.2 (DNR Bio-Imaging Systems).
2.4. Mitochondrial and cellular analysis
For mitochondrial ROS, mitochondrial mass, and ΔΨm measurements, MEFs were incubated with MitoSOX (5 μM), Mitotracker Green (100 nM), or Mitotracker Red (100 nM; ThermoFisher Scientific), respectively, before assessment in a FACSCanto II (BD Biosciences) of the total population (10,000 cells). Data were analyzed using FlowJo software and expressed as MFI (mean fluorescence intensity), which referred to the fluorescence intensity of each event on average. Additionally, lysed 15 × 103 WT and AIF-/Y MEFs or 1 × 104 cells from embryos dissociated in trypsin were tested for ATP content with a luciferin-luciferase kit (Abcam) and expressed as an ATP/ADP ratio or RLU (relative light units). In some experiments, MEFs were pretreated with oligomycin (10 μM) before ATP assessment. Measures were performed on an Infinite M100 PRO plate reader (Tecan).
To analyze glucose assimilation, MEFs were incubated (30 min; 37 °C) in glucose-free DMEM with 2-NBDG (100 μM; ThermoFisher Scientific) before the flow cytometry assessment of the total population (10,000 cells).
Glycolytic and GLUT-4 dependency was verified in MEFs treated or not with indinavir (50 μM; Selleckchem) or 2-Deoxy-d-Glucose (2-DG; 10 mM); AMPK dependency was verified in MEFs pretreated or not with dorsomorphin (Compound C; 25 μM; Selleckchem); the induced cell death rate was assessed by an Annexin-V-APC (0.1 μg/ml; BD Biosciences) and propidium iodide (PI) double labeling on a FACSCanto II in the total population (10,000 cells).
In cell cycle analyses, MEFs were incubated (30 min; 37 °C) with BrdU (10 μM). After fixation and partial DNA denaturation, cells were co-stained with an anti-BrdU-FITC antibody (25 μg/mL; BD Biosciences) and PI before the flow cytometric measurement in the total population (10,000 cells).
Senescence was recorded in MEFs treated with chloroquine diphosphate (300 μM, 2 h) to induce lysosomal alkylinization. Next, C12FDG (Thermofisher Scientific) was added at 33 μM, and the cells were washed twice with PBS before performing the cytofluorometric quantification.
Cell death was assessed by flow cytometry in the total population (10,000 cells) using Annexin-V-APC (assessment of phosphatidylserine exposure; 0.1 μg/ml) and PI (cell viability) co-labeling. In some experiments, WT and AIF-/Y MEFs were pretreated 30 min with MNNG (250 mM, 9 h), staurosporine (STS; 1 μM, 6 h), β-Lapachone (4 μM; 18 h), or etoposide (20 μM, 6 h) before the flow cytometry assessment of cell death. In caspase-dependent control experiments, MEFs were pretreated at 30 min with QVD.OPh (1 μM) before etoposide treatment.
For lactate measurement in MEFs, cells were cultured for 2 h in a serum-free medium and centrifuged. The amounts of lactate in the supernatants were quantified in the Infinite M100 PRO plate reader by using the l-lactate kit from Cayman Chemical. Lactate was also quantified in embryos dissociated in 50 μL of the l-lactate kit assay buffer. After centrifugation, the quantity of lactate in 10 μL of the supernatant was assessed.
2.5. Clark electrode
Oxygen consumption was measured in 5 × 106 WT and AIF-/Y MEFs as previously described [38]. In non-permeabilized cells, respiration was measured under basal conditions in a DMEM medium and in response to the sequential addition of oligomycin (10 μM), fluoro-carbonyl cyanide phenylhydrazone (FCCP; 5 μM), amytal (2 mM), and azide (5 mM). In digitonin-permeabilized cells (1 μg/μL, 5 min), respiration was measured in response to the sequential addition of glutamate/malate (10 mM/4 mM), ADP (2 mM), amytal (2 mM), succinate (10 mM), malonate (10 mM), glycerol-3-phosphate (15 mM), myxothiazol (0.5 μM), tetramethyl-phenylenediamine (TMPD)/ascorbate (0.5 mM/2 mM), and azide (5 mM).
2.6. Mitochondrial supercomplex (SC) assessment
Mitochondria were purified from control (0) or 4-OHT-treated MEFs (4–16 days post-treatment) and permeabilized by digitonin. Solubilized complexes and supercomplexes were separated by 1D BN-PAGE and revealed by the NADH dehydrogenase in-gel activity (0.1 mg/ml NADH and 2.5 mg/ml nitrotetrazolium blue chloride). Alternatively, the mitochondrial complexes I, III, and IV and the SCs were visualized by immunoblot using antibodies against NDUFA9 (20C11B11B11, Life Technologies), UQCRC2 (13G12AF12BB11, Abcam), or COX4I2 (20E8C12, Abcam). Immunoreactive proteins were detected by using HRP-conjugated secondary antibodies and visualized with an ECL chemiluminescence kit (Thermo Scientific). Images were acquired by using the MF-ChemiBIS 4.2 imager.
2.7. Immunofluorescence
MEFs seeded on coverslips were fixed in 2% paraformaldehyde (PFA) and incubated with the mitochondrial marker Mitotracker Red (20 nM; Thermo Fisher) and the nuclear dye Hoechst 33342. Slides were imaged by using an LSM 710 Zeiss confocal microscope and analyzed by ImageJ software.
2.8. Electron microscopy
Control (D0) or tamoxifen-treated MEFs (12–16 days post-treatment) were fixed at 4 °C for 2 h in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3) postfixed for 1 h in 1% buffered osmium tetroxide, dehydrated through a graded ethanol series, and embedded in Epon 812. The ultrathin sections were counterstained with 2% aqueous uranyl acetate for 30 min, and then with lead citrate for 10 min, and finally viewed under a Philips 100X electron microscope. Alternatively, freshly dissected E9.5 WT and AIF-/Y embryos were fixed, processed for ultrathin sections, counterstained, and examined as aforementioned.
2.9. Lentiviral transduction
An in-frame c-terminal tag encoding the V5 epitope was first added to the AIF-wt cDNA (plasmid pcDNA3.1 mAIFV5). The resulting cDNA was then subcloned into the pLVX-IRES-Zs-Green lentiviral vector (Clontech-Takara Bio Europe). Viruses were produced in 293T cells by CaCl2 transient transfection of the lentiviral constructs and the packaging plasmids pCMV-VSV-G and pCMV-dR8.2 dvpr (Addgene plasmids 8454 and 8455, respectively) [39]. Forty-eight hours after transfection, lentiviral supernatants were harvested, clarified by filtration, and used immediately for AIF-/Y MEFs transduction with 6 μg/ml of polybrene. Seventy-two hours after transduction, GFP positive cells were sorted and expanded, and the AIF location and mitochondrial network distribution were visualized by immunofluorescence as aforementioned.
2.10. Immunoblotting
MEFs or freshly dissected embryos were lysed in buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and the protease/phosphatase inhibitor cocktail from Roche. Protein concentration was determined by using the BioRad Protein Assay. Equal amounts of total proteins (50 μg) were loaded on linear SDS-PAGE gels and transferred onto a PVDF or nitrocellulose membrane. Membrane blocking and antibody incubations were performed in TBS 0.1% Tween 20 plus 5% nonfat dry milk. The primary antibodies used were AIF (D-20, Santa Cruz), NDUFA9 (20C11B11B11, Life Technologies), NDUFS3 (3F9DD2, Life Technologies), SDHA (2E3GC12FB2AE2, Life Technologies), UQCRC2 (13G12AF12BB11, Abcam), COX1 (35–8100; Thermo Fisher), COX4I2 (20E8C12, Abcam), ATP5B (3D5AB1, Life Technologies), AMPKalpha (Cell signaling) and phospho-AMPKalpha (Thr172, Cell signaling), p21Cip1 (C-19, Santa Cruz), Phospho-Rb (Cell Signaling), and β-Actin (AC15, Sigma). Immunoreactive proteins were detected by using HRP-conjugated secondary antibodies and visualized with SuperSignal West Dura chemiluminescence kits (Thermo Scientific). Immunoblot images were acquired in the MF-ChemiBIS 4.2 imager.
Brains from the WT, AIF-/X, and AIF-/X HC females were dissected and frozen in liquid nitrogen and used for protein carbonyls detection. After the generation of brain homogenates (2 mg), biotin-hydrazide (60 mM), sodium cyanoborohydride (30 mM), and trichloroacetic acid (10%) were successively added to the samples. After centrifugation, pellets were washed with 1:1 ethanol/ethyl acetate and dissolved in Laemmli buffer. Next, equal amounts of total proteins (50 μg) were loaded on linear SDS-PAGE gels and transferred onto a PVDF membrane. Protein carbonyls were visualized by using streptavidin-HRP revealed by chemiluminescence and quantified by using Multi Gauge 3.0 software (Fujifilm Life Sciences). The optical density was normalized according to an endogenous background and was expressed relative to the data obtained in untreated cells (= 1.0).
2.11. Quantitative RT-PCR
Total RNA from the cells, embryos, or brain was extracted by using a Nucleospin® RNA II kit (Macherey–Nagel). cDNA was prepared by using Superscript® II reverse transcriptase (Life Technologies). Quantitative RT-PCR was performed by using TaqMan® Gene Expression Assays (Life Technologies) for Mfn1 (mitofusin 1), Mfn2 (mitofusin 2), Opa1 (mitochondrial dynamin-like GTPase), Fis1 (mitochondrial fission 1 protein), Dnm1l (dynamin-related protein 1), Mff (mitochondrial fission factor), Sod1 (Super oxide dismutase 1), Sod2 (Super oxide dismutase 2), Glut-1 (Slc2a1, solute carrier family 2 [facilitated glucose transporter], member 1), Glut-4 (Slc2a4, solute carrier family 2 [facilitated glucose transporter], member 4), Tp53 (tumor protein p53), Cdkn1α (cyclin-dependent kinase inhibitor 1A, P21), or Pgc1α (Peroxisome proliferator-activated receptor gamma coactivator 1-alpha). PCR reactions were performed in triplicate by using TaqMan® Fast advanced Master Mix (Life Technologies). The products were amplified in a ViiA7 Real-time PCR System (Life Technologies) at 60 °C for 40 cycles. Data were evaluated by the comparative threshold cycle method. The housekeeping expression of 18S was used to normalize the data.
2.12. Histology
We developed a specific method of embedding without prior macroscopic dissection of the brain to observe both neural tissue with brain envelope and the skull bone on the same histology slide. Wild-type (WT) and AIF-/X hydrocephalus (HC) females were fixed by perfusion in 4% PFA. After decalcification with 0.1 N nitric acid and paraffin embedding, 4 μm thick sections were stained with hematoxylin-eosin. Some tissues were processed without paraffin embedding before vibratome sectioning (for 100 μm thick floating sections). E9.5 WT and AIF-/Y embryos were fixed in 4% PFA and embedded in acrylic resin (historesin acrylic® Leica) before 3 μm sectioning and hematoxylin-eosin staining. Images were obtained by using a Leica photomicroscope.
2.13. Immunohistochemistry
Brains of WT and AIF+/- HC females were fixed in 4% PFA. After decalcification, free-floating brain sections (100 μm) were fixed in PFA before performing the immunostaining with anti-Iba1 (1:100 Dako). We used a long incubation process (24 h) after deterging the lipid with high concentration triton X (1%). Nuclei were counterstained with the nuclear dye Hoechst 33342. Slides were imaged by using an LSM 710 Zeiss confocal microscope and analyzed by using the ImageJ software.
The activities of cyclooxygenase (COX) and succinate dehydrogenase (SDH) were visualized to explore respiratory dysfunction in embryos [40]. Briefly, E9.5 WT and AIF-/Y embryos were dissected, placed in a plastic mold containing cold optimal cutting temperature (OCT) compound, and snap-frozen in liquid nitrogen. Fourteen μm cryosections, obtained by using cryostat CM 3050S (Leica), were incubated for 40 min at 37 °C with 3,3′-diaminobenzidine tetra-hydrochloride, 500 μM cytochrome c, and bovine catalase. The sections were then washed 4 times for 10 min in 0.1 M PBS (pH 7.0). Next, 1.875 mM nitroblue tetrazolium (NBT), 1.30 M sodium succinate, 2.0 mM phenazine methosulfate, and 100 mM sodium azide were applied for 40 min at 37 °C. Sections were finally washed, cover-slipped, and visualized under bright field microscopy.
To evaluate proliferation in embryos, pregnant females were intraperitoneally injected with BrdU (0.1 mg BrdU/g of body weight) 1 h before embryonic dissection. E8.5 and E9.5 WT and AIF-/Y embryos were identified by PCR, dissected, and kept in separated well plates in ice-cold PBS/BSA 0.4%. After fixation in ice-cold EtOH (70%; H2O) overnight, embryos were rehydrated with ice-cold EtOH 50% for 15 min at 4 °C and then washed with ice-cold PBS/BSA 0.4%. Embryos were then submerged in sucrose 30% (diluted in phosphate buffer 0.1 M, pH 7.4) until they reached the bottom of the well. Next, embryos were placed in molds containing cold OCT and snap-frozen in liquid nitrogen. Further, 10–12 μm cryosections were made by using the cryostat CM 3050S and processed for BrdU immunostaining. Briefly, sections were quenched with H2O2 0.6% and MetOH 1% in PBS for 1 h at RT. After washing, slides were unmasked with HCl 3.7% and processed with sodium borate 0.1 M. After a pre-blocking step, an anti-BrdU antibody (BD Biosciences) was added on top of each section and a secondary biotinylated antibody was added. Slides were next incubated with streptavidin-488 and Hoechst. Images were obtained on a fluorescent microscope Axiovert 200M (Zeiss).
For embryonic cell death detection, E8.5 and E9.5 WT and AIF-/Y embryos were dissected, fixed in PFA (4%), embedded in cold OCT and snap-frozen in liquid nitrogen. Next, 10–12 μm cryosections, made with the cryostat CM 3050S, were fixed in methanol/acetic acid solution (2:1) at −20 °C. After permeabilization, TUNEL mix (Roche Diagnostics) was added on top of each section for 1 h at 37 °C. After 2 washing steps, nuclei were colored by Hoechst 33342. Images were obtained on a fluorescent microscope Axiovert 200M (Zeiss) using transmitted light.
2.14. Statistics
The results, displayed as mean ± SEM, were statistically analyzed by the Mann–Whitney or student t tests by using GraphPad Prism software. Significant p values were indicated as follows: ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001. All experiments were independently repeated at least 3 times.
Chemicals and reagents were purchased from Sigma–Aldrich except where otherwise noted.
3. Results
3.1. AIF-/Y MEFs were OXPHOS defective and showed extensive ETC supercomplex disorganization
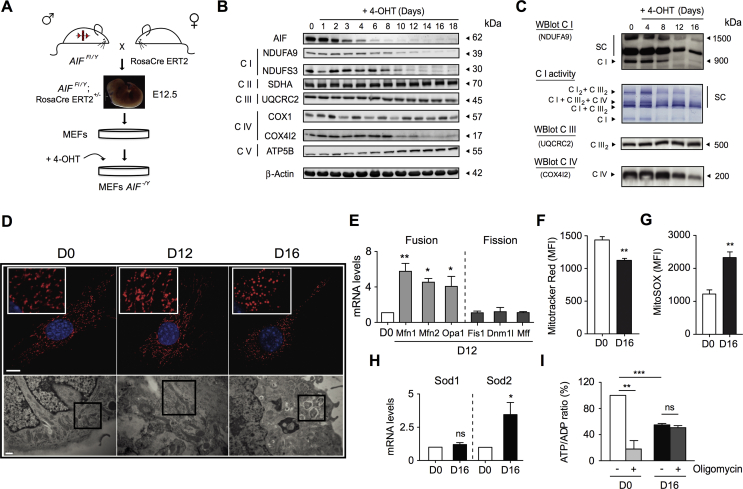
To analyze mitochondrial and cellular alterations associated with AIF loss, we generated a primary mouse embryonic fibroblasts knockout (KO) for AIF (AIF-/Y MEFs) by crossing Rosa26-CreERT2 mice [37] with our novel Aifm1 floxed strain (Figure 1A and Supplemental Fig. 1). Tamoxifen (4-OHT) treatment of AIFFl/Y was conducted; RosaCre ERT2+/− MEFs provoked progressive AIF protein loss (Figure 1B), which destabilized mitochondrial ETC complexes I and IV (e.g., loss of NDUFA9, NDUFS3, and COX4I2 proteins). Complexes II, III, and V (ATP synthase) remained unaltered (Figure 1B). Complementary blue native polyacrylamide gel electrophoresis (BN-PAGE) and immunoblot assessments revealed that AIF deficiency, in addition to a strong reduction in free complex I levels, also provoked ETC SC disorganization with a rapid loss of complex I2 + complex III2 and triple complex I + complex III2 + complex IV structures and a more delayed disruption of complex I + complex III2 organization (Figure 1C). These observations are in accordance with the “plasticity” model of ETC structure in which the alteration of individual complexes directly affects the supercomplex organization [8,9,41].
Figure 1.
AIF loss led to ETC dysfunction and mitochondrial network disorganization. (A) To generate AIF-/Y MEFs, we crossed the Rosa26-CreERT2 mice with the Aifm1 floxed strain [35]. Among the embryos, E12.5 AIFFl/Y; RosaCre ERT2+/− embryos were genetically identified and dissected. After MEFs generation and expansion, cells were treated overnight with tamoxifen (4-OHT; 1 μM) to induce Aifm1 exon 11 excision [35]. (B) Representative time-course immunoblot of untreated (0) or 4-OHT-treated MEFs (1–18 days post-treatment) revealing the progressive loss of AIF and key proteins of the ETC complexes I and IV. Equal loading was confirmed by β-Actin probing. This experiment was repeated three times with similar results. (C) Multiprotein complex assessment by blue native polyacrylamide gel electrophoresis (BN-PAGE) in mitochondria purified from control (0) or 4-OHT-treated MEFs (4–16 days post-addition). Up to down gel pictures: (i) Mitochondrial complex I and complex I-containing supercomplexes visualized by immunoblot (NDUFA9); (ii) Representative result of an NADH dehydrogenase complex I in-gel test revealing ETC supercomplex disorganization after AIF loss; (iii) and (iv) Complex III dimer (UQCRC2) and IV monomer (COX4I2) detected by immunoblot. This experiment was repeated five times with similar results. (D) Analysis of the mitochondrial network changes associated with AIF loss. Upper panels, representative immunofluorescence images of control (D0), and AIF-/Y MEFs (D12 and D16) labeled with Mitotracker Red (mitochondria) and Hoechst (nucleus). Bar: 100 μm. Lower panels, control (D0), and AIF-/Y MEFs (D12 and D16) analyzed by electron microscopy. Representative microphotographs are shown. Black squares underline mitochondrial features. Bar: 0.5 μm. (E) Mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), mitochondrial dynamin-like GTPase (Opa1), mitochondrial fission 1 protein (Fis1), dynamin-related protein 1 (Dnm1l), and mitochondrial fission factor (Mff) mRNA levels determined by quantitative RT-PCR in control (D0) and AIF-/Y MEFs (D12; n = 4). 18S mRNA expression was used to normalize data. Results are expressed as a ratio of mRNA expression relative to control (D0) cells (set at 1.0). (F) Flow cytometry ΔΨm assessment performed by Mitotracker Red labeling in control (D0) and AIF-/Y MEFs (D16) and expressed as a plot (n = 8). Data were obtained in 10,000 cells and expressed as mean fluorescence intensity (MFI). (G) Mitochondrial ROS levels recorded in control (D0) and AIF-/Y MEFs (D16) and graphed (n = 9). Data were obtained in 10,000 cells and expressed as mean fluorescence intensity (MFI). (H) Sod1 and Sod2 mRNA levels determined by quantitative RT-PCR in control (D0) and AIF-/Y MEFs (D16; n = 5). 18S mRNA expression was used to normalize data. Results are expressed as a ratio of mRNA expression relative to control (D0) cells (set at 1.0). (I) ATP/ADP ratio recorded in control (D0) and AIF-/Y MEFs (D16) left untreated or pre-treated with oligomycin (10 μM; n = 7). Results are expressed as a ratio of ATP/ADP relative to control (D0) cells (considered as 100%). Statistical significance was calculated by Mann–Whitney (E, H) or student t (F, G, I) tests. Bars represent mean ± SEM.
Analysis of the oxygen consumption rate confirmed that mitochondrial respiration was significantly altered in AIF-/Y MEFs. As described in Supplemental Fig. 2A, compared to control MEFs (cells before 4-OHT treatment; D0), AIF-/Y MEFs (16 days after tamoxifen treatment; D16) exhibited a significant reduction of oxygen consumption in both phosphorylating (basal) and non-phosphorylating (+oligomycin) conditions. The total oxidative capacity, assessed by the presence of the ionophore FCCP, was also compromised in KO cells. Treatment with the complex I inhibitor amytal abolished the remaining oxygen consumption of AIF-/Y MEFs, indicating that complex I remained slightly functional. Finally, analysis of the individual complex activities revealed that complexes I to IV's activities were affected by AIF deficiency (Supplemental Fig. 2B).
Mitochondrial labeling revealed that the kinetic of AIF disappearance was linked to the progressive disruption of the mitochondrial network (Figure 1D, upper panels). Remarkably, at D12 after tamoxifen, electronic microscopy pictures uncovered elongated mitochondria, suggesting a mitochondrial fusion development (Figure 1D, lower panels). This finding is substantiated by the quantitative RT-PCR analysis of central executioners of mitochondrial dynamics. Indeed, as depicted in Figure 1E, in contrast to the fission factors FIS1, DNM1L (DRP1), and MFF, the mRNA of the key members of the fusion machinery MFN1, MFN2, and OPA1 appeared significantly overexpressed in these AIF-/Y MEFs. Thus, although the inhibition of the mitochondrial fission machinery could also contribute to the elongated mitochondrial phenotype observed in the AIF-/Y MEFs at D12 after tamoxifen treatment, our multiparametric assessment points toward the prevalence of a transient mitochondrial fusion process.
Further, upon complete AIF disappearance (D16 after tamoxifen treatment), we observed a highly fragmented mitochondrial network associated with cellular vacuolization (Figure 1D). These alterations provoked a significant decrease in the cellular mitochondrial transmembrane potential and a high generation of mitochondrial ROS (Figure 1F,G). The excess ROS triggered mRNA overexpression of mitochondrial SOD2 (Superoxide dismutase 2), a protein that partially regulated the excess superoxide OXPHOS products (Figure 1H). Surprisingly, despite mitochondrial dysfunction, ATP levels were relatively preserved in AIF-/Y MEFs (Figure 1I). One possible explanation for this finding is that MEFs normally generate ATP by non-mitochondrial pathways. Alternatively, a possibility is that AIF KO MEFs have reprogrammed their metabolism to counterbalance mitochondrial OXPHOS dysfunction. To evaluate this dichotomy, we treated control and AIF-/Y MEFs with oligomycin, which specifically blocks the mitochondrial ATP synthase and thus inhibits the generation of mitochondrial ATP. In control MEFs, oligomycin treatment dramatically diminished ATP levels, indicating that these cells generate most of the ATP through mitochondria. By contrast, oligomycin marginally modified ATP levels in AIF-/Y MEFs, suggesting that these cells generate ATP through a non-mitochondrial pathway (Figure 1I). Those results suggest that AIF-/Y MEFs were able to modify their metabolism to compensate the mitochondrial OXPHOS dysfunction.
3.2. OXPHOS damage associated with AIF deficiency was counterbalanced by a shift toward anaerobic glycolysis and development of a senescent phenotype
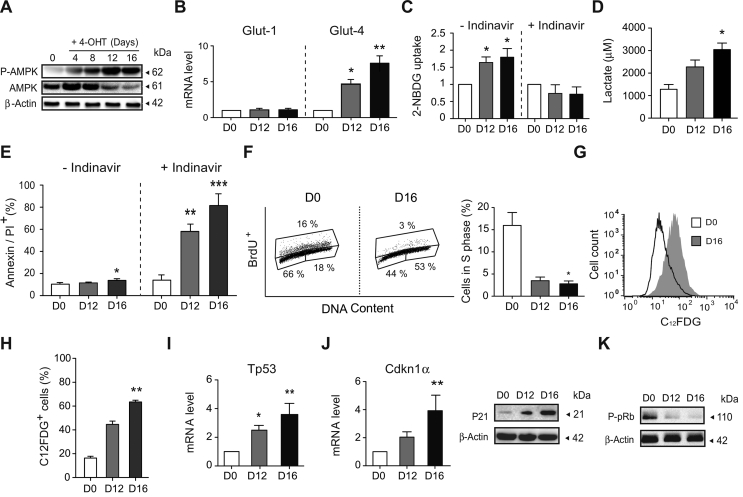
Based on our results thus far, the next step was to analyze the mechanism associated with the adaptive metabolic features of AIF-/Y MEFs. More precisely, by measuring the phosphorylation of AMPK at Thr172 and the mRNA expression of the main glucose transporters expressed in fetal tissues and fibroblasts (GLUT-1 and GLUT-4), we first assessed whether the AIF loss was accompanied by a modification in the glucose assimilation pathway. Phosphorylation of AMPK, a central regulator of energy homeostasis, is a point of convergence of metabolic and genomic stress signals [[42], [43], [44], [45]]. Indeed, AMPK is a major energy-sensing kinase that plays a role in cellular energy homeostasis by activating a variety of catabolic processes in multicellular organisms, such as glucose uptake and metabolism. More precisely, among others, phosphorylated AMPK stimulates glucose uptake by modulating GLUT-4 plasma membrane function [[46], [47], [48], [49]]. As described in Figure 2A,B, the loss of AIF was associated with a significant and specific kinetic phosphorylation/activation of AMPK at Thr172 and the subsequent mRNA overexpression of Glut-4, not Glut-1. This finding suggested an enhancement of glucose uptake mediated by GLUT-4, which was further substantiated by the blockade of 2-NBDG assimilation by the specific GLUT-4 inhibitor indinavir (Figure 2C).
Figure 2.
Mitochondrial OXPHOS dysfunction was counterbalanced in AIF-/YMEFs by a shift towards anaerobic glycolysis and the development of a senescent phenotype. (A) Kinetic phosphorylation of AMPK visualized by immunoblot in left untreated (0) or 4-OHT-treated MEFs (4–16 days post-treatment). Equal loading was confirmed by β-Actin probing. This experiment was repeated three times with similar results. (B) Glut-1 and Glut-4 mRNA levels determined by quantitative RT-PCR in control (D0) and AIF-/Y MEFs (D12 and D16; n = 8). 18S mRNA expression was used to normalize data. Results are expressed as a ratio of mRNA expression relative to control (D0) cells (set at 1.0). (C) Glucose uptake measured by the assimilation of 2-NBDG in control (D0) and AIF-/Y MEFs (D12 and D16) untreated or pre-treated with indinavir (50 μM; n = 6). Results are expressed as a ratio relative to control (D0) cells (set at 1.0). (D) Lactate release, recorded in control (D0) and AIF-/Y MEFs (D12 and D16), was measured as described in the Methods section (n = 4). (E) Cytofluorometric assessment of cell death performed in control (D0) and AIF-/Y MEFs (D12 and D16) untreated or pre-treated with indinavir (50 μM) and labeled with AnnexinV and PI. The frequency of positive staining, which represents dying cells, was recorded and expressed as a plot (n = 6). (F) Flow cytometry cell cycle analysis performed in control (D0) and AIF-/Y MEFs (D12 and D16) by BrdU and PI (DNA content) co-labeling. Left, representative cytometric panels of control (D0) and AIF-/Y MEFs (D16). Right, the percent of cells in phase S was quantified and expressed as a plot (n = 4). (G) Cytometric evaluation of senescence in control (D0) and AIF-/Y MEFs (D12 and D16) using the β-galactosidase substrate C12FDG. Representative flow cytometric profiles of control (D0) and AIF-/Y MEFs (D16). (H) The percentage of C12FDG positive control (D0) and AIF-/Y MEFs (D12 and D16) measured as in (G) was calculated and graphed (n = 4). (I) Tp53 mRNA levels determined by qPCR in control (D0) and AIF-/Y MEFs (D12 and D16; n = 5). 18S mRNA expression was used to normalize data. Results are expressed as a ratio of mRNA expression relative to control (D0) cells (set at 1.0). (J) Left, Cdkn1α mRNA levels determined by qPCR in control (D0) and AIF-/Y MEFs (D12 and D16; n = 6). 18S mRNA expression was used to normalize data. Results are expressed as a ratio of mRNA expression relative to control (D0) cells (set at 1.0). Right, representative immunoblot of control (D0) and AIF-/Y MEFs (D12 and D16) revealing the cell cycle inhibitor P21. Equal loading was confirmed by β-Actin probing (n = 4 experiments with similar results). (K) Representative immunoblot of control (D0) and AIF-/Y MEFs (D12 and D16) revealing the decrease in pRb phosphorylation (P-pRb) associated with AIF loss. Equal loading was confirmed by β-Actin probing (n = 3 experiments with similar results). Statistical significance was calculated by Mann–Whitney (D, F, H, I) or student t (B, C, E, J) tests. Bars represent mean ± SEM.
The measurement of lactate release, a marker of anaerobic glycolysis, indicated that AIF-/Y compensated for the mitochondrial OXPHOS dysfunction through the reinforcement of this glycolytic pathway (Figure 2D). Notably, the anaerobic glycolytic shift appeared to be sufficient to maintain ATP levels (Figure 1I) and preserve AIF-/Y viability (absence of cell death), which surprisingly remained very close to those of control (D0) MEFs (Figure 2E). The central role of AMPK and GLUT-4 in the enhancement of the glucose metabolism and the preservation of ATP levels and cell viability in the AIF KO MEFs was corroborated by the pharmacological inhibition of AMPK or GLUT-4 by dorsomorphin and indinavir, respectively [50,51]. As described in Supplemental Figs. 3 and 2E, AMPK inhibition precluded Glut-4 mRNA overexpression and both Glut-4 mRNA downregulation and inhibition of GLUT-4-mediated glucose uptake/metabolism induced cell death in the AIF KO MEFs. Altogether, these results corroborated that the AIF-/Y MEFs counterbalance the mitochondrial OXPHOS dysfunction by reprogramming their metabolism toward anaerobic glycolysis through an AMPK/GLUT-4 adaptive mechanism.
We next investigated whether the lack of cell death in AIF-/Y MEFs was linked to cell cycle alterations. As demonstrated by BrdU/PI co-staining, contrary to what was reported in embryonic stem cells [52], the loss of AIF provoked cell cycle and cell proliferation arrest in MEFs. More precisely, compared to control MEFs (D0), in which approximately 15%–20% of the cell population was in the proliferative S cell cycle phase, the D12 and D16 AIF KO MEFs showed less than 5% of the population in the S phase (Figure 2F). Surprisingly, the arrest of cell proliferation recorded in AIF-/Y MEFs appeared to be associated with the kinetic activation of a ROS/P53/P21-mediated senescence pathway [53,54], as confirmed by specific C12FDG senescent β-galactosidase staining (with more than 60% positive senescent labeling at D16) and mRNA overexpression of the ROS-induced genetic markers Tp53 and Cdkn1α (Figure 2F,G, H, I, and J). Notably, Cdkn1α expression is tightly controlled by Tp53, and Cdnk1α codes for P21, a potent cyclin-dependent kinase inhibitor [55]. As expected, the kinetic Tp53 and Cdkn1α mRNA overexpression provoked the progressive up-regulation of the P21 protein (Figure 2J). P21 induction by means of cyclin-dependent kinase CDK2 and CDK4/6 inhibition leads to dephosphorylation/activation of retinoblastoma protein (pRB; Figure 2K), a well-established cell cycle regulator that in a non-phosphorylated state inhibits cell cycle progression [[55], [56], [57], [58]]. Indeed, ROS-induced senescence can be identified in vitro by the increased expression of genes/proteins associated with cell cycle arrest (e.g., P53 and P21), pRB dephosphorylation/activation, and presence of senescence-associated β-galactosidase activity, which is attributed to the high lysosomal content of senescent cells [59]. All these features characterized AIF-/Y MEFs.
3.3. AIF-/Y MEFs were resistant to caspase-independent cell death
After remaining senescent for days, approximately 2% of AIF-/Y MEFs spontaneously proliferated. The analysis of these proliferating cells indicated that compared to the senescent AIF-/Y MEFs, cycling AIF KO MEFs diminished the mitochondrial mass and the ROS levels. This finding was correlated to the loss of P21 expression and the subsequent phosphorylation/inactivation of pRb that favors cell cycle progression (Supplemental Fig. 4). These results support the relationship between ROS generation and P21/pRb-mediated cell cycle arrest in the senescent AIF KO cells.
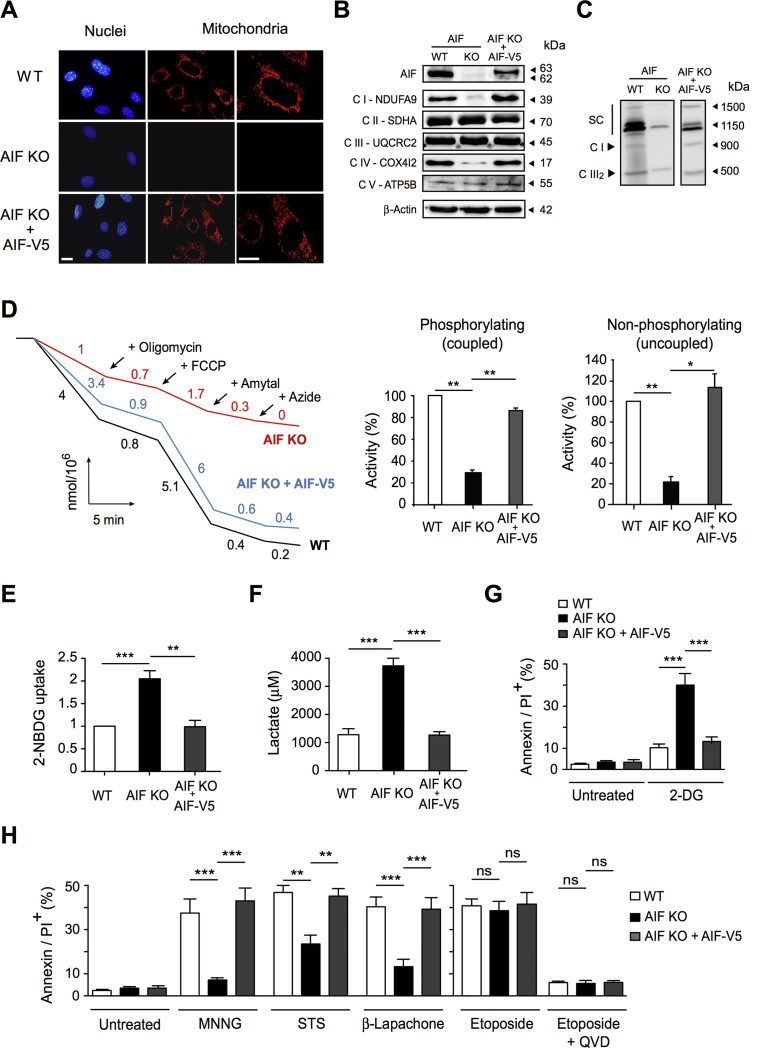
We used proliferating AIF KO MEFs to analyze, among others, the PCD capacities of AIF-/Y cells. To generate a reliable control, we reintroduced the AIF-wt cDNA (V5 tagged) into the AIF-/Y MEFs by lentiviral transduction (AIF KO + AIF-V5 MEFs). AIF KO + AIF-V5 MEFs incorporated AIF into the mitochondria, rescued the normal mitochondrial network (Figure 3A), and reconstituted the ETC proteins (Figure 3B) and SC patterns (Figure 3C). The functionality of mitochondrial OXPHOS in the AIF KO + AIF-V5 MEFs was substantiated by oxygen consumption tests (Figure 3D). Finally, the glycolytic adaptive phenotype of AIF-/Y MEFs was disabled by the lentiviral AIF transduction (Figure 3E,F). Concerning sensitivity to PCD, contrary to primary (WT) and the reconstituted AIF KO + AIF-V5 MEFs, AIF-/Y MEFs were sensitive to the glycolytic inhibitor 2-Deoxyglucose (Figure 3G), underlining its glucose dependency. Additionally, cell lines were exposed to different PCD inducers: MNNG and β-Lapachone (caspase-independent PCD inducers) [10,18,20,21,60], STS (a cytotoxic inducer partially controlled by caspase activation), and etoposide (a typical caspase-dependent agent) [[61], [62], [63]]. As depicted in Figure 3H, AIF-/Y MEFs were highly resistant to MNNG and β-Lapachone, partially resistant to STS, and sensitive to etoposide. We corroborated that the pretreatment of MEFs with the broad caspase inhibitor (QVD.OPh) inhibited etoposide-mediated killing. Finally, as expected, reintroduction of AIF into the AIF-/Y MEFs fully re-sensitized these MEFs to MNNG, β-Lapachone, and STS. Altogether, these results substantiate the specific role of AIF in caspase-independent cell death and corroborate the literature that has described the limited response of AIF-deficient MEFs to STS [14,21]. Finally, those data indicate that AIF-/Y MEFs remain sensitive to caspase-dependent PCD inducers.
Figure 3.
Lentiviral transduction of AIF-/YMEFs with V5 tagged AIF-wt cDNA restored normal mitochondrial OXPHOS/metabolism and corroborated the specific role of AIF in MNNG- and β-Lapachone-mediated cell death. (A) Representative confocal images of AIF+/Y (WT), AIF-/Y (AIF KO), and AIF-/Y expressing AIF-wt MEFs (AIF KO + AIF-V5) labeled with MitoTrackerRed (mitochondria) and Hoechst (nucleus), corroborating that lentiviral transduced AIF-V5 relocalized into mitochondria and reconstituted the mitochondrial network. Bar: 100 μm. (B) Representative immunoblot of the panel of MEFs used in (A) demonstrating the presence of AIF and key proteins of the ETC complexes I to V after the lentiviral transduction of AIF-V5 into the AIF-/Y MEFs. Equal loading was confirmed by β-Actin. This experiment was repeated 5 times with similar results. (C) Mitochondrial supercomplex picturing of WT, AIF KO, and AIF KO + AIF-V5 was performed by 1D BN-PAGE, such as in Figure 1C. Mitochondrial supercomplexes and complex I were detected by NDUFA9 immunoblotting, complex III (dimer) was visualized by UQCRC2 blotting. This experiment was repeated 3 times with similar results. (D) Assessment of basal (coupled) and maximal respiration (uncoupled) in WT, AIF KO, and AIF KO + AIF-V5 MEFs by using a Clark's electrode. Basal respiration corresponds to O2 consumption rate coupled to ATP production, whereas sequential addition of 10 μM oligomycin (ATP synthase inhibitor), 15 μM fluoro-carbonyl cyanide phenylhydrazone (FCCP; uncoupling agent measuring the maximal respiration capacity), 2 mM amytal (complex I inhibitor), or 5 mM sodium azide (mitochondrial OXPHOS inhibitor) enable calculation of the maximal oxygen consumption rate. The percent of activity (relative to that measured in WT cells—considered as a 100%) in coupled and uncoupled conditions was expressed as a histogram (n = 5 independent experiments). Notably, the lentiviral transduction of AIF-V5 into the AIF KO MEFs fully restored mitochondrial respiration. (E) Glucose uptake measured by assimilation of 2-NBDG in WT, AIF KO, and AIF KO + AIF-V5 MEFs (n = 4). Results are expressed as a ratio of mRNA expression relative to control (D0) cells (set at 1.0). (F) Lactate release recorded in WT, AIF KO, and AIF KO + AIF-V5 MEFs as described in the Methods section (n = 4). (G) The panel of MEFs used in (A) to (F) were untreated or treated with 2-Deoxy-d-Glucose (2-DG; 10 mM; 24 h) and labeled with AnnexinV and PI. The frequency of positive staining, which represents dying cells, was recorded and expressed as a plot (n = 4). (H) The panel of MEFs used in (A) to (F) was untreated or treated with N-methyl-N′-nitro-N′-nitrosoguanidine (MNNG; 250 mM; 9 h), staurosporine (STS; 1 μM; 6 h), β-Lapachone (4 μM; 18 h), or etoposide (20 μM; 6 h) in the absence or presence of the broad caspase inhibitor QVD.OPh (1 μM). Next, MEFs were labeled with AnnexinV and PI, and the frequency of positive staining, which represents dying cells, was recorded and graphed (n = 6). Statistical significance was calculated by Mann–Whitney (D, E, F, G) or student t (H) tests. Bars represent mean ± SEM.
3.4. A significant percentage of AIF+/- females developed a hydrocephalus phenotype and exhibited immune cell exhaustion
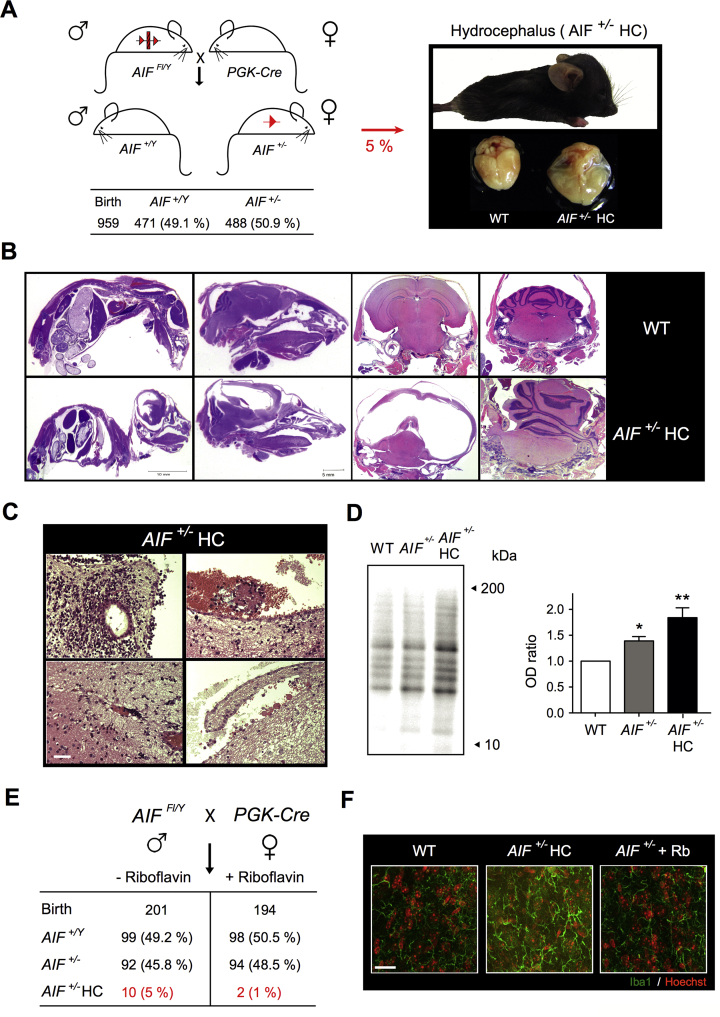
To extend our study toward an in vivo model, we crossed Aifm1 floxed males (AIFFl/Y) with a PGK-Cre strain, which is a driver of Cre recombinase expression very early during embryonic development [64]. This crossbreeding yielded heterozygous female offspring (AIF+/-) in a Mendelian distribution (Figure 4A). Most of the AIF+/- animals did not present external phenotypic changes until they reached 6 months of age, at which stage we detected a significant diminution of bone marrow, spleen, and thymus cellularity (Supplemental Fig. 5). Surprisingly, approximately 10% of AIF+/- females (5% of total newborns) developed a perinatal hydrocephalus phenotype (visible in 2-week-old animals) characterized by an enlarged and dome-shaped head and accompanied by a progressive loss of mobility (Figure 4A). These animals died 4/5 weeks after birth.
Figure 4.
A significant percentage of AIF+/-females developed a hydrocephalus phenotype associated with developmental defaults and an excess of mitochondrial ROS. (A) To generate AIF+/- animals, we crossed Aifm1 floxed males with PGK-Cre females. The table below indicates the offspring distribution. Approximately 10% of AIF+/- females developed a hydrocephalus (HC) phenotype. Photographs depict the phenotype of a 2.5-week-old AIF+/- HC female and a HC brain compared to a WT brain. Note the swelling of the cranial cavity of the mice and the excess of cerebrospinal fluid (CSF) that warps the HC brain. (B) Representative hematoxylin/eosin (HE)-labeled sections of 2.5-week-old WT and AIF+/- HC females showing (left to right) sagittal sections (entire mice and head) and coronal sections (brain and cerebellum). Note the extension of the hydrocephaly, the brain compression exerted by the CSF excess, and the herniated cerebellum in the AIF+/- HC. (C) HE stained brain sections from 2.5-week-old AIF+/- HC animals showing spinal and parenchymatous hemorrhages and loss and defects of ciliation in the ependymal epithelium. Bar: 50 μm. (D) Oxidative modification of proteins assessed by carbonylation immunoblot on brain homogenates from 2.5-week-old WT, AIF+/-, and AIF+/- HC mice. This experiment was repeated 3 times with similar results. The OD ratio depicted in the graph illustrates the levels of protein carbonylation of the different phenotypes. Results are expressed as a ratio of OD relative to WT cells (set at 1.0). (E) PGK-Cre dams were supplied or not with riboflavin in drinking water (5 mg/L) and the number and genotype/phenotype of the progeny were assessed and reported in a table. (F) Characteristic immunofluorescence images of brains from 2.5-week-old WT, AIF+/- HC, and AIF+/- mice supplied with riboflavin (AIF+/- + Rb), showing the microglial (Iba1 labeling) status. Individual cells were visualized by Hoechst (nuclear) co-staining. Bar: 200 μm. Statistical significance in (D) was calculated by a Mann–Whitney test. Bars represent mean ± SEM.
Compared to control animals, the brain of AIF+/-hydrocephalus animals (AIF+/- HC) showed an excess of cerebrospinal fluid (CSF) that warped the brain (right microphotographs in Figure 4A). Hematoxylin/eosin staining of the mice and head sections provided insights into the extent to which the CSF compressed the cerebral mass into the cranial cavity (Figure 4B, left microphotographs). In coronal sections, we observed severe thinning of the brain ventricular wall with an enlargement of the lateral ventricles and meningeal fibrosis (Figure 4B, right panels). Brain development, apart from the compression, seemed normal. Some mice showed a dilated third ventricle and a herniated cerebellum. We also observed strong spinal and parenchymatous hemorrhages. Circulating macrophages engorged with blood could be observed in some sections of the ependymal epithelium (Figure 4C). This epithelium, lining the ventricular cavities, is normally composed of a single cell layer. In the case of AIF+/- HC, the ependymal epithelium lining the ventricles was abnormal with visible defects of ciliation.
Additionally, AIF+/- HC mice presented immature “neuroblast” foci with abnormal structures (Figure 4C). These structures, which normally disappear at the closure of the neural tube, indicated some defects in the development of the AIF+/- HC. The hydrocephalus mice also had a bone structure problem, probably because of the cranial cavity swelling provoked by the CSF pressure. No apoptotic features were observed on the histological sections. AIF+/- HC mice did not exhibit apparent neurodegeneration and had a normal neuronal organization. Notably, a protein carbonylation immunoblot approach performed on brain homogenates from 2.5-week-old animals revealed that compared to AIF+/+ and non-HC AIF+/- females, the brain of the AIF+/- HC females displayed higher ROS levels (Figure 4D). This finding suggested a relationship between ROS excess, generated by OXPHOS dysfunction, and perinatal hydrocephaly. This hypothesis was corroborated by the addition of an antioxidant (riboflavin) in the drinking water of the dams, which significantly reduced the occurrence of the AIF+/- HC phenotype and microglial activation, detected by Iba1 immunolabeling, reflecting the inflammatory process provoked by brain squeezing (Figure 4E,F). The mobility of riboflavin-treated AIF+/- females also appeared normal, indicating that the addition of an antioxidant in the drinking water of AIF+/- mice rescues the WT phenotype.
3.5. AIF loss in mouse embryo induced midgestation lethality associated with energy loss, proliferation arrest, and increased apoptosis
Next, we attempted to generate AIF KO mice by crossing the non-HC AIF+/- females with WT males. This crossing generated AIF+/+ and AIF+/Y animals, suggesting embryonic lethality of the AIF-/Y and AIF+/- progeny (Figure 5A, birth animals). Indeed, AIF-/Y male embryos presented normal development only until E7.5 (Figure 5A,B); then, they displayed growth delay and arrested development between E8.5 and E9.5, leading to embryo death at E11.5 (Figure 5A,B, and C). Female AIF+/- embryos exhibited inconstant developmental phenotypes and died around E13.5 (Figure 5A).
Figure 5.
Mitochondrial AIF deficiency-induced murine embryonic lethality linked to OXPHOS deficiency and energy loss. (A) To generate AIF KO mice, we crossed AIF+/- females with C57BL/6 WT males. After genetic identification, the result of the crossing (progeny and E8.5 to E13.5 embryos) was reported in a table. (B) Light microscope images of E7.5 to E9.5 AIF+/Y and AIF-/Y embryos illustrating the progressive growth delay of the AIF KO embryos. Bar: 500 μm. (C) Hematoxylin/eosin staining performed on the sagittal sections of E8.5 and E9.5 AIF+/Y and AIF-/Y embryos, underlining the morphology and the abnormal nervous development of the AIF KO embryos. Bar: 500 μm. (D) Electron microscopy picturing mitochondria of E9.5 AIF+/Y and AIF-/Y embryos. Representative microphotographs are shown. Arrowheads in the picture mark the mitochondria. White squares show characteristic mitochondria. Bar: 0.5 μm. (E) Immunoblot of AIF and key proteins of the ETC complexes I to V performed in whole protein extracts from E9.5 AIF+/Y and AIF-/Y embryos. Equal loading was confirmed by β-Actin probing. This experiment was repeated 4 times with similar results. (F) Representative results of the histochemical assessment of mitochondrial OXPHOS activity in E9.5 AIF+/Y and AIF-/Y embryos using cytochrome c oxidase/succinate dehydrogenase (COX/SDH) double labeling. Bar: 50 μm. Whereas OXPHOS activity is normal in AIF+/Y embryos, as demonstrated by the brown color of the section, AIF-/Y showed defective OXPHOS activity (absence of brown staining). This experiment was repeated 3 times with similar results. (G) Total ATP levels recorded, as described in the Methods section, in E9.5 AIF+/Y and AIF-/Y embryos (n = 6). Results are expressed as RLU (relative light units). (H) Lactate release measured, as described in the Methods section, in E9.5 AIF+/Y and AIF-/Y embryos (n = 6). Statistical significance in (G) and (H) was calculated by the student t test. Bars represent mean ± SEM.
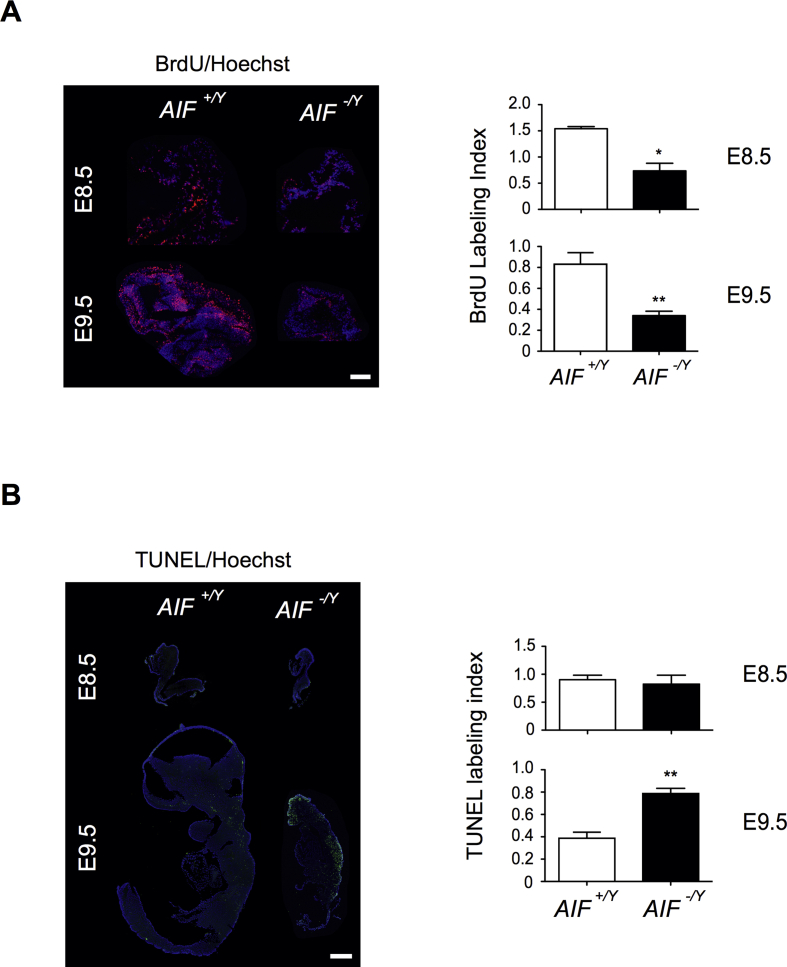
We scrutinized mitochondrial and metabolic alterations associated with the lethality of AIF-/Y embryos. An electronic microscopy approach revealed highly fragmented mitochondria in E9.5 AIF-/Y embryos (Figure 5D). Immunoblot analysis of key mitochondrial ETC proteins showed a dramatic reduction of NDUFA9 (complex I protein) and COX4I2 (complex IV protein; Figure 5E). This was accompanied by an impairment of mitochondrial OXPHOS, as demonstrated by COX/SDH histochemical labeling (Figure 5F): indeed, sections from mutant E9.5 embryos did not show the dark staining characterizing OXPHOS functionality. As a consequence of OXPHOS dysfunction, AIF-/Y embryos exhibited very low ATP levels (Figure 5G). Surprisingly, as shown by the low amount of lactate detected (Figure 5H), AIF-/Y embryos appeared unable to reprogram their metabolism toward anaerobic glycolysis, contrary to AIF-/Y MEFs (Figure 2). Moreover, histological analysis showed that these mitochondrial/metabolic changes were accompanied by a decrease in the proliferation rate starting at E.8.5 (Figure 6A) and by the enhancement of apoptosis at E.9.5 in AIF-/Y embryos (Figure 6B).
Figure 6.
Embryonic AIF loss triggered the arrest of proliferation and enhancement of cellular apoptotic levels. (A) BrdU immunofluorescence analysis of a frozen section of AIF+/Y and AIF-/Y embryos at E8.5 and E9.5. Photographs of the head of a representative embryo. Bar: 500 μm. The BrdU labeling index was calculated as the ratio of BrdU/Hoechst fluorescence measured in a fixed surface. Data in the histogram represent mean ± SEM (n = ratio obtained in at least 5 different surfaces of 3 independent embryonic sections). (B) TUNEL staining of a PFA fixed cryosection of AIF+/Y and AIF-/Y embryos at E8.5 and E9.5. Bar: 500 μm. The TUNEL labeling index was obtained as the ratio of TUNEL/Hoechst fluorescence measured in a fixed surface. Data in the histogram represent mean ± SEM (n = ratio obtained in at least 5 different surfaces of 3 independent embryonic sections). Statistical significance in (A) and (B) was calculated by the student t test. Bars represent mean ± SEM.
Finally, considering the inability of the AIF-/Y embryos to reprogram their metabolism toward anaerobic glycolysis, we attempted to circumvent lethality by eliminating the excess ROS with riboflavin and feeding the dams with an HFD in which the major source of energy changed from carbohydrates to fatty acids, which have proved efficient in other AIF-deficient models [65]. However, even in these alternative conditions, we were unable to generate AIF-/Y and AIF+/- animals (Supplemental Fig. 6). Overall, these data corroborated the mitochondrial OXPHOS/metabolism dysfunction of AIF-/Y embryos, and their incapacity to reprogram their metabolism to grow and differentiate.
4. Discussion
By creating an AIF KO model that enables in vitro and in vivo analysis, we provide insights into new cellular and developmental consequences of the loss of mitochondrial AIF. We show that AIF deficiency triggers destabilization of the mitochondrial ETC complexes I and IV, leading to supercomplex disorganization, mitochondrial transmembrane potential loss, and high levels of ROS generation. AIF-/Y MEFs counterbalance these OXPHOS alterations by a mitochondrial network reorganization implicating a fusion process and the overexpression of the mitochondrial SOD2. A more accurate molecular analysis at the cellular, embryonic, and adult mice levels indicates that the mitochondrial OXPHOS dysfunction associated with AIF loss results in pleiotropic effects: (i) at a cellular level, AIF-/Y MEFs boost their anaerobic glycolytic metabolism and evolve toward a senescence phenotype. The glycolytic phenotype is characterized by AMPK phosphorylation/activation, the overexpression of the glucose transporter GLUT-4, the enhancement of glucose uptake, and anaerobic lactate generation. The energy-sensing kinase AMPK and the glucose assimilation transporter GLUT-4 play central roles in the glycolytic reprogramming of the AIF KO MEFs. Based on these results, further research should analyze the transcriptional drivers activated by AMPK to specifically modulate Glut-4 expression and control the metabolic adaptive responses that run the AIF-/Y MEFs toward senescence.
Concerning the senescent phenotype, the ROS excess associated with OXPHOS dysfunction provokes overexpression of Tp53 and Cdkn1α, genes associated with cell cycle arrest, as well as up-regulation of P21, a protein that by provoking the activation/dephosphorylation of pRB inhibits cell cycle progression. Strikingly, a small percentage of AIF-/Y MEFs spontaneously proliferate. These cycling AIF KO MEFs diminish the mitochondrial mass and regulate ROS levels, inactivating the senescent pathway. Remarkably, the cycling AIF-/Y MEFs are resistant to caspase-independent inducers, underlining the role of AIF in this mode of PCD and avoiding the controversies in the literature on the participation of AIF in cell death; (ii) in whole animal experiments, we have confirmed that the AIF KO animals are embryonic lethal. However, analysis of heterozygous AIF+/- females originally uncovered immune cell exhaustion and the development of perinatal hydrocephalus seemingly associated with an excess of mitochondrial ROS. This phenotype is characterized by an excess of CSF, brain development impairment, meningeal fibrosis, and medullar hemorrhages; (iii) finally, we propose, based on our findings, that AIF-deficient embryonic lethality is linked to the inability of KO embryos to reprogram their metabolism toward anaerobic glycolysis. This provokes dramatic ATP loss, arrest in proliferation, and caspase-dependent embryo death.
The mitochondrial “pro-survival” function of AIF seems linked to its role in the import and assembly of key proteins of the ETC complexes [12,66]. Our results in MEFs and embryos, analyzed within the context of the “plasticity” model of ETC supercomplex organization [8,41], indicate that AIF loss provokes disorganization of the SC architecture, OXPHOS deregulation, ROS overproduction, and very restricted capacity for mitochondrial ATP generation. However, AIF deficiency does not fully yield defective mitochondria in MEFs, as demonstrated by the assessment of individual mitochondrial complex activities, which reveal a limited but still functional ETC (amytal inhibits the residual complex I activity). Indeed, our data on the overgeneration of mitochondrial ROS in the absence of mitochondrial ATP generation strongly suggest that the ETC of the AIF -/Y MEFs is somewhat functional, but the loss of key complex I/IV proteins provokes an impairment. As a consequence, the AIF KO mitochondria appear unable to generate the electrochemical gradient essential to produce ATP, probably because of a premature electron leakage (which generates the excess of ROS detected). Surprisingly, this mitochondrial ETC dysfunction does not lead to cell death. Instead, AIF-/Y MEFs reprogram their metabolism toward AMPK/GLUT-4-mediated anaerobic glycolysis (to generate ATP), overexpress ROS detoxifying enzymes (to regulate the excess of ROS as much as possible), and activate an enzymatically controlled senescent pathway. Therefore, it seems that MEFs have the plasticity to respond to ETC weakening. By contrast, AIF-/Y embryos are unable to perform this metabolic adaptive response and show a drastic developmental defect leading to death. What is the reason for the different MEFs/embryo behavior? We could hypothesize that the in vitro carbohydrate-enriched cell culture condition supports MEFs glycolytic metabolic reprogramming. In vivo, a glycolytic versus OXPHOS metabolic shift around E8.5 has been described to be essential for embryonic survival and fetal development [67,68]. In AIF-/Y OXPHOS dysfunctional embryos, this shift is probably impossible, yielding inevitable embryonic death.
In addition to its mitochondrial OXPHOS function, AIF has a lethal role in its translocation to the nucleus [[18], [19], [20], [21],69]. Strong evidence for an AIF role in PCD has been provided by studies performed with many cell death models [21,22,[70], [71], [72], [73], [74], [75], [76]]. Despite this substantial amount of work, some controversy remains regarding AIF function in PCD. Indeed, studies performed in HeLa cells, SV40-immortalized MEFs, or with caspase-dependent apoptotic inducers, have raised doubts about the apoptogenic AIF potential [14,77,78]. We attempted to elucidate this controversy by verifying the response to typical caspase-dependent and independent inducers in non-transformed WT, AIF-/Y, and AIF KO + AIF reconstituted cells. We observed that AIF-/Y MEFs are significantly resistant to treatment with MNNG and β-Lapachone, classical caspase-independent PCD inducers, but they remain sensitive to caspase-dependent apoptotic drugs. Although a more exhaustive study is required, our results already indicate that the contradictory results on the role AIF plays in PCD could be merely associated with the cellular system employed or the type of PCD inducer tested. In any case, the phenotype of AIF-/Y MEFs confirms the essential role of AIF in the control of caspase-independent programmed cell death.
Genetic inactivation of AIF provokes embryonic death. Consequently, generating an AIF KO progeny is not possible. Other studies in embryoid bodies have suggested that AIF could regulate the initial stages of murine development by controlling the cell death process of cavitation [79]. However, in the study, we show that AIF-/Y embryos develop normally until E7.5 and are morphologically indistinguishable from WT embryos. By E8.5, AIF-/Y embryos are delayed compared to WT littermates. Between E8.5 and E9.5, they stop growing, do not turn, and have abnormal brain development. AIF-/Y embryos die at around E11.5. These results are consistent with another study indicating that AIF and the mitochondrial OXPHOS are not required at the initial stages of mouse development [52]. However, in contrast to this work, we have observed the arrest of proliferation and enhancement of PCD in E8.5 and E9.5 AIF-/Y embryos. In addition, we have observed that OXPHOS dysfunction associated with AIF loss and the incapacity to perform metabolic reprogramming seemed key to the embryonic lethality associated with AIF deficiency.
Further research could help to understand why it is possible to generate AIF+/- offspring from AIFFl/Y; PGK-Cre cross, whereas the resulting AIF+/- females are unable to generate heterozygous AIF progeny. An exciting possibility is the existence of bias in the timing or strength of allele expression depending on the paternal or maternal origin of the X chromosome. Notably, our data suggest that the E8.5 “embryonic-shift” from anaerobic to aerobic metabolism can only occur when the female transmits the WT allele. This has no incidence in the AIFFl/Y and PGK-Cre crossing (having an AIF KO allele generated by PGK-Cre in the floxed X chromosome from the male). In the case of the progeny of AIF+/- females, the AIF KO allele transmitted by the dams would be unable to perform the “embryonic-shift,” resulting in the death of the AIF+/- and AIF-/Y embryos. A second possibility, with similar embryonic death consequences, is the implication of Aifm1 in the particular behavior of the paternal X chromosome (Xp). Indeed, it has been well documented that the Xp chromosome is preferentially inactivated during the first stages of mice development [80,81]. Similarly, there is evidence that some genes are initially expressed from the maternal allelic form [82]. In our case, the Aifm1-mutated maternal allelic form would not be able to generate AIF. If Aifm1 was also inactivated in Xp, the mitochondria of the AIF+/- and AIF-/Y embryos would be dysfunctional and, consequently, the embryo would not be viable. What supports this second possibility is the morphological analysis of the mitochondria from the AIF-mutated embryo (Figure 5D and Supplemental Fig. 7), which reveals the irregular structure associated with mitochondrial dysfunction [35].
The AIF+/- females show 2 relevant pathological phenotypes: (i) immune exhaustion in the adult animals and (ii) a perinatal hydrocephalus phenotype. Concerning the alterations in the immune system associated with AIF, an initial study was performed with the Hq strain. In this study, low AIF expression led to the high susceptibility of T-cell blasts to activation-induced cell death and reduced sensitivity to neglect-induced cell death. Moreover, thymocytes from Hq mice exhibited higher ROS levels, which impacted thymocyte and T-cell development [31,83]. A more specific work in which Aif was ablated around the DN3 stage of thymocyte development reported that AIF loss had no impact on T-cell development. The use of the CD19-cre mice strain indicated that AIF was not required for B-cell development and function [14]. Our recent work, performed in a bona fide hematopoietic AIF KO strain, where the protein was ablated very early during hematopoiesis, revealed a greater role for AIF in immune cell fate [35,36]. Indeed, the loss of AIF provoked progressive exhaustion of the HSC pool and developmental defaults in the thymocyte, T-cell, B-cell, and erythroid lineages. In agreement with these results, we observed progressive cellular exhaustion in the bone marrow, spleen, and thymus of the AIF+/- females, fully corroborating the relevance of AIF in the fine-tuning of the immune system.
Unreported in the literature is the hydrocephalus phenotype observed in approximately 10% of AIF+/- newborns. The signs of AIF+/- hydrocephaly are readily apparent in young animals that develop the disease before the cranial sutures close (allowing for an enlarged and domed head). This is accompanied by a progressive loss of animal mobility, and the cause is that the brain is completely encased within the bony calvarium. The build-up of CSF causes tissue inflammation (depicted by the microglial activation), hemorrhages of the surrounding brain tissue, enlargement of the ventricles, and herniation of the brain and cerebellum. In a general perspective, hydrocephalus may be caused by blockage of the normal flow of CSF, failure to absorb CSF, or less commonly, overproduction of CSF. CSF circulates throughout the ventricular brain system through the motion of the ciliated ependymal cells that line the ventricles.
In AIF+/- HC, ependymal epithelium was absent from the edge of the ventricles and consequently presented a loss of ciliation. Thus, similar to what has been described in other mice strains [[84], [85], [86], [87], [88]], we propose that AIF+/- hydrocephalus is caused by defects of the normal CSF flow associated with the loss of ciliated ependymal cells. This cellular loss appears to be directly related to the excess of ROS detected in the brain homogenates of the AIF+/- HC animals. Indeed, external control of ROS excess through the supply of riboflavin to the dams and newborns is sufficient to reduce this phenotype to the standards of the C57BL/6J strain [89]. Notably, riboflavin was given to patients with mitochondrial myopathy because of OXPHOS deficiency [[90], [91], [92]]. Notably, riboflavin is a precursor to FMN and FAD, which are cofactors of ETC complexes I and II, respectively. Riboflavin has been proposed to act therapeutically through several potential mechanisms including ROS control and inhibition of the breakdown of complex I by providing more resistance to proteolysis or stabilizing the mitochondrial membrane [92].
5. Conclusion
Our data reveal that the mitochondrial signals regulated by AIF are critical to cellular decision-making. Emerging as a link between mitochondrial metabolism, mouse development, and cell fate, AIF-mediated OXPHOS regulation represents a potential target in the development of new therapeutic approaches. Indeed, further knowledge of how the cells can modify their metabolic pathways may enable their manipulation; ultimately, this might lead to new treatment options for diseases in which the mitochondrial OXPHOS alteration/dysfunction has an instrumental role.
Author contributions
L.D. designed the experiments, performed the in vitro and in vivo studies, interpreted the data, and helped write the manuscript. N.M., L.C., M.-N.B.-N., L.K.L., M.T., A.M., A.P., R.M.-L., M.B., L.V., and D.G. designed the experiments, performed the in vitro and/or in vivo work, and analyzed the results. K.G. conducted electron microscopy. C.L. provided key reagents, organized Clark electrode oxygen consumption assays, and analyzed the results. F.L. provided the PGK-Cre strain and helped in the development of the Aifm1 floxed mice. M.C–S. participated in the development of the Aifm1 floxed mice and helped write the manuscript. P.F.-S. set up the mitochondrial supercomplex analyses and interpreted the data. F.C. provided reagents, designed the assessment of the hydrocephalus animals, and analyzed the results. I.M. provided reagents, performed experiments, designed the embryonic tests, interpreted the data, and helped to write the manuscript. S.A.S. designed the in vitro and in vivo studies, coordinated and supervised the study, interpreted the data, and wrote the manuscript.
Acknowledgments
We thank the animal facility staff (CEF Cordeliers, Paris, France) for mice housing support, Danielle Chateau for help in electron microscopy, Pascal Ezan for help in protein extraction, Patricia Flamant and Patrick Avé for help in brain and embryo immunohistochemistry (Human histopathology and animal models unit, Pasteur Institute), Marie-Annick Thomas for advice on brain assessments, and M. Segade for proofreading. This work was supported by Fondation ARC (PJA20151203407 and PJA20171206551), Fondation pour la Recherche Médicale, French National Research Agency (ANR-09-BLAN-0247), and the SIRIC-CURAMUS (INCA-DGOS-Inserm_12560). L.C. received PhD fellowship support from ENS-Cachan, Société Française d'Hématologie, and Fondation ARC. N.M. received a fellowship from the FRS/FRIA. I.M. is an FNRS research associate and an investigator of WELBIO.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101027.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.MacIver N.J., Michalek R.D., Rathmell J.C. Metabolic regulation of T lymphocytes. Annual Review of Immunology. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearce E.L., Poffenberger M.C., Chang C.H., Jones R.G. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. [Google Scholar]
- 3.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature Reviews Molecular Cell Biology. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 4.Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38(4):633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folmes C.D., Dzeja P.P., Nelson T.J., Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11(5):596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Molecular Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamanaka R.B., Chandel N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends in biochemical sciences. 2010;35(9):505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapuente-Brun E., Moreno-Loshuertos R., Acin-Perez R., Latorre-Pellicer A., Colas C., Balsa E. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 9.Acin-Perez R., Enriquez J.A. The function of the respiratory supercomplexes: the plasticity model. Biochimica et Biophysica Acta. 2014;1837(4):444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Klein J.A., Longo-Guess C.M., Rossmann M.P., Seburn K.L., Hurd R.E., Frankel W.N. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419(6905):367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 11.Pospisilik J.A., Knauf C., Joza N., Benit P., Orthofer M., Cani P.D. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131(3):476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 12.Hangen E., Feraud O., Lachkar S., Mou H., Doti N., Fimia G.M. Interaction between AIF and CHCHD4 regulates respiratory chain biogenesis. Molecular Cell. 2015;58(6):1001–1014. doi: 10.1016/j.molcel.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Meyer K., Buettner S., Ghezzi D., Zeviani M., Bano D., Nicotera P. Loss of apoptosis-inducing factor critically affects MIA40 function. Cell Death & Disease. 2015;6 doi: 10.1038/cddis.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milasta S., Dillon C.P., Sturm O.E., Verbist K.C., Brewer T.L., Quarato G. Apoptosis-inducing-factor-dependent mitochondrial function is required for T cell but not B cell function. Immunity. 2016;44(1):88–102. doi: 10.1016/j.immuni.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishimura R., Martin G.R., Ackerman S.L. Loss of apoptosis-inducing factor results in cell-type-specific neurogenesis defects. Journal of Neuroscience. 2008;28(19):4938–4948. doi: 10.1523/JNEUROSCI.0229-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benit P., Goncalves S., Dassa E.P., Briere J.J., Rustin P. The variability of the harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PloS One. 2008;3(9) doi: 10.1371/journal.pone.0003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung E.C., Joza N., Steenaart N.A., McClellan K.A., Neuspiel M., McNamara S. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. The EMBO Journal. 2006;25(17):4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Artus C., Boujrad H., Bouharrour A., Brunelle M.N., Hoos S., Yuste V.J. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. The EMBO Journal. 2010;29(9):1585–1599. doi: 10.1038/emboj.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baritaud M., Cabon L., Delavallee L., Galan-Malo P., Gilles M.E., Brunelle-Navas M.N. AIF-mediated caspase-independent necroptosis requires ATM and DNA-PK-induced histone H2AX Ser139 phosphorylation. Cell Death & Disease. 2012;3 doi: 10.1038/cddis.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabon L., Galan-Malo P., Bouharrour A., Delavallee L., Brunelle-Navas M.N., Lorenzo H.K. BID regulates AIF-mediated caspase-independent necroptosis by promoting BAX activation. Cell Death & Differentiation. 2012;19(2):245–256. doi: 10.1038/cdd.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moubarak R.S., Yuste V.J., Artus C., Bouharrour A., Greer P.A., Menissier-de Murcia J. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Molecular and Cellular Biology. 2007;27(13):4844–4862. doi: 10.1128/MCB.02141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu S.W., Wang H., Poitras M.F., Coombs C., Bowers W.J., Federoff H.J. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y., Huang S., Liu Z.G., Han J. Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. Journal of Biological Chemistry. 2006;281(13):8788–8795. doi: 10.1074/jbc.M508135200. [DOI] [PubMed] [Google Scholar]
- 24.Benit P., Pelhaitre A., Saunier E., Bortoli S., Coulibaly A., Rak M. Paradoxical inhibition of glycolysis by pioglitazone opposes the mitochondriopathy caused by AIF deficiency. EBioMedicine. 2017;17:75–87. doi: 10.1016/j.ebiom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D'Adamo P. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. The American Journal of Human Genetics. 2010;86(4):639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger I., Ben-Neriah Z., Dor-Wolman T., Shaag A., Saada A., Zenvirt S. Early prenatal ventriculomegaly due to an AIFM1 mutation identified by linkage analysis and whole exome sequencing. Molecular Genetics and Metabolism. 2011;104(4):517–520. doi: 10.1016/j.ymgme.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Diodato D., Tasca G., Verrigni D., D'Amico A., Rizza T., Tozzi G. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. European Journal of Human Genetics. 2016;24(3):463–466. doi: 10.1038/ejhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancho P., Sanchez-Monteagudo A., Collado A., Marco-Marin C., Dominguez-Gonzalez C., Camacho A. A newly distal hereditary motor neuropathy caused by a rare AIFM1 mutation. Neurogenetics. 2017;18(4):245–250. doi: 10.1007/s10048-017-0524-6. [DOI] [PubMed] [Google Scholar]
- 29.Shen S.M., Guo M., Xiong Z., Yu Y., Zhao X.Y., Zhang F.F. AIF inhibits tumor metastasis by protecting PTEN from oxidation. EMBO Reports. 2015;16(11):1563–1580. doi: 10.15252/embr.201540536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Li K., Zhang S., Wang Y., Xu Y., Cronin S.J.F. Overexpression of apoptosis inducing factor aggravates hypoxic-ischemic brain injury in neonatal mice. Cell Death & Disease. 2020;11(1):77. doi: 10.1038/s41419-020-2280-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava S., Banerjee H., Chaudhry A., Khare A., Sarin A., George A. Apoptosis-inducing factor regulates death in peripheral T cells. The Journal of Immunology. 2007;179(2):797–803. doi: 10.4049/jimmunol.179.2.797. [DOI] [PubMed] [Google Scholar]
- 32.van Empel V.P., Bertrand A.T., van Oort R.J., van der Nagel R., Engelen M., van Rijen H.V. EUK-8, a superoxide dismutase and catalase mimetic, reduces cardiac oxidative stress and ameliorates pressure overload-induced heart failure in the harlequin mouse mutant. Journal of the American College of Cardiology. 2006;48(4):824–832. doi: 10.1016/j.jacc.2006.02.075. [DOI] [PubMed] [Google Scholar]
- 33.Chung S.H., Calafiore M., Plane J.M., Pleasure D.E., Deng W. Apoptosis inducing factor deficiency causes reduced mitofusion 1 expression and patterned Purkinje cell degeneration. Neurobiology of Disease. 2011;41(2):445–457. doi: 10.1016/j.nbd.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joza N., Oudit G.Y., Brown D., Benit P., Kassiri Z., Vahsen N. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Molecular and Cellular Biology. 2005;25(23):10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabon L., Bertaux A., Brunelle-Navas M.N., Nemazanyy I., Scourzic L., Delavallee L. AIF loss deregulates hematopoiesis and reveals different adaptive metabolic responses in bone marrow cells and thymocytes. Cell Death & Differentiation. 2018;25(5):983–1001. doi: 10.1038/s41418-017-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertaux A., Cabon L., Brunelle-Navas M.N., Bouchet S., Nemazanyy I., Susin S.A. Mitochondrial OXPHOS influences immune cell fate: lessons from hematopoietic AIF-deficient and NDUFS4-deficient mouse models. Cell Death & Disease. 2018;9(6):581. doi: 10.1038/s41419-018-0583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hameyer D., Loonstra A., Eshkind L., Schmitt S., Antunes C., Groen A. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiological Genomics. 2007;31(1):32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Nuebel E., Wisidagama D.R., Setoguchi K., Hong J.S., Van Horn C.M. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nature Protocols. 2012;7(6):1068–1085. doi: 10.1038/nprot.2012.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross J.M., Oberg J., Brene S., Coppotelli G., Terzioglu M., Pernold K. High brain lactate is a hallmark of aging and caused by a shift in the lactate dehydrogenase A/B ratio. Proceedings of the National Academy of Sciences of the U S A. 2010;107(46):20087–20092. doi: 10.1073/pnas.1008189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudkina N.V., Kouril R., Peters K., Braun H.P., Boekema E.J. Structure and function of mitochondrial supercomplexes. Biochimica et Biophysica Acta. 2010;1797(6–7):664–670. doi: 10.1016/j.bbabio.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nature Reviews Molecular Cell Biology. 2018;19(2):121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabinovitch R.C., Samborska B., Faubert B., Ma E.H., Gravel S.P., Andrzejewski S. AMPK maintains cellular metabolic homeostasis through regulation of mitochondrial reactive oxygen species. Cell Reports. 2017;21(1):1–9. doi: 10.1016/j.celrep.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 44.Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanli T., Steinberg G.R., Singh G., Tsakiridis T. AMP-activated protein kinase (AMPK) beyond metabolism: a novel genomic stress sensor participating in the DNA damage response pathway. Cancer Biology & Therapy. 2014;15(2):156–169. doi: 10.4161/cbt.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmes B.F., Kurth-Kraczek E.J., Winder W.W. Chronic activation of 5'-AMP-activated protein kinase increases GLUT-4, hexokinase, and glycogen in muscle. Journal of Applied Physiology (1985) 1999;87(5):1990–1995. doi: 10.1152/jappl.1999.87.5.1990. [DOI] [PubMed] [Google Scholar]
- 48.McGee S.L., van Denderen B.J., Howlett K.F., Mollica J., Schertzer J.D., Kemp B.E. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57(4):860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 49.Ojuka E.O., Jones T.E., Nolte L.A., Chen M., Wamhoff B.R., Sturek M. Regulation of GLUT4 biogenesis in muscle: evidence for involvement of AMPK and Ca(2+) American Journal of Physiology. Endocrinology and Metabolism. 2002;282(5):E1008–E1013. doi: 10.1152/ajpendo.00512.2001. [DOI] [PubMed] [Google Scholar]
- 50.Bahne E., Sun E.W.L., Young R.L., Hansen M., Sonne D.P., Hansen J.S. Metformin-induced glucagon-like peptide-1 secretion contributes to the actions of metformin in type 2 diabetes. JCI Insight. 2018;3(23) doi: 10.1172/jci.insight.93936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hresko R.C., Hruz P.W. HIV protease inhibitors act as competitive inhibitors of the cytoplasmic glucose binding site of GLUTs with differing affinities for GLUT1 and GLUT4. PloS One. 2011;6(9) doi: 10.1371/journal.pone.0025237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown D., Yu B.D., Joza N., Benit P., Meneses J., Firpo M. Loss of Aif function causes cell death in the mouse embryo, but the temporal progression of patterning is normal. Proceedings of the National Academy of Sciences of the U S A. 2006;103(26):9918–9923. doi: 10.1073/pnas.0603950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munoz-Espin D., Serrano M. Cellular senescence: from physiology to pathology. Nature Reviews Molecular Cell Biology. 2014;15(7):482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Zamudio R.I., Robinson L., Roux P.F., Bischof O. SnapShot: cellular senescence pathways. Cell. 2017;170(4):816–816 e811. doi: 10.1016/j.cell.2017.07.049. [DOI] [PubMed] [Google Scholar]
- 55.Broude E.V., Swift M.E., Vivo C., Chang B.D., Davis B.M., Kalurupalle S. p21(Waf1/Cip1/Sdi1) mediates retinoblastoma protein degradation. Oncogene. 2007;26(48):6954–6958. doi: 10.1038/sj.onc.1210516. [DOI] [PubMed] [Google Scholar]
- 56.Sherr C.J., McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2(2):103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 57.Zheng L., Lee W.H. The retinoblastoma gene: a prototypic and multifunctional tumor suppressor. Experimental Cell Research. 2001;264(1):2–18. doi: 10.1006/excr.2000.5129. [DOI] [PubMed] [Google Scholar]
- 58.Leinicke J.A., Longshore S., Wakeman D., Guo J., Warner B.W. Regulation of retinoblastoma protein (Rb) by p21 is critical for adaptation to massive small bowel resection. Journal of Gastrointestinal Surgery. 2012;16(1):148–155. doi: 10.1007/s11605-011-1747-8. discussion 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorgoulis V., Adams P.D., Alimonti A., Bennett D.C., Bischof O., Bishop C. Cellular senescence: defining a path forward. Cell. 2019;179(4):813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Park E.J., Min K.J., Lee T.J., Yoo Y.H., Kim Y.S., Kwon T.K. beta-Lapachone induces programmed necrosis through the RIP1-PARP-AIF-dependent pathway in human hepatocellular carcinoma SK-Hep1 cells. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hars E.S., Lyu Y.L., Lin C.P., Liu L.F. Role of apoptotic nuclease caspase-activated DNase in etoposide-induced treatment-related acute myelogenous leukemia. Cancer Research. 2006;66(18):8975–8979. doi: 10.1158/0008-5472.CAN-06-1724. [DOI] [PubMed] [Google Scholar]
- 62.Perkins C.L., Fang G., Kim C.N., Bhalla K.N. The role of Apaf-1, caspase-9, and bid proteins in etoposide- or paclitaxel-induced mitochondrial events during apoptosis. Cancer Research. 2000;60(6):1645–1653. [PubMed] [Google Scholar]
- 63.Maas C., de Vries E., Tait S.W., Borst J. Bid can mediate a pro-apoptotic response to etoposide and ionizing radiation without cleavage in its unstructured loop and in the absence of p53. Oncogene. 2011;30(33):3636–3647. doi: 10.1038/onc.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Research. 1998;7(2):105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- 65.Schiff M., Benit P., El-Khoury R., Schlemmer D., Benoist J.F., Rustin P. Mouse studies to shape clinical trials for mitochondrial diseases: high fat diet in Harlequin mice. PloS One. 2011;6(12) doi: 10.1371/journal.pone.0028823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vahsen N., Cande C., Briere J., Benit P., Rustin P., Kroemer G. The absence of apoptosis-inducing factor AIF induces complex I deficiency. Biochimica et Biophysica Acta. 2004;1657:83. [Google Scholar]
- 67.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genetics. 1998;18(3):231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 68.Humble M.M., Young M.J., Foley J.F., Pandiri A.R., Travlos G.S., Copeland W.C. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Human Molecular Genetics. 2013;22(5):1017–1025. doi: 10.1093/hmg/dds506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuste V.J., Moubarak R.S., Delettre C., Bras M., Sancho P., Robert N. Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death & Differentiation. 2005;12(11):1445–1448. doi: 10.1038/sj.cdd.4401687. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y., Dawson V.L., Dawson T.M. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Experimental Neurology. 2009;218(2):193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H., Yu S.W., Koh D.W., Lew J., Coombs C., Bowers W. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. Journal of Neuroscience. 2004;24(48):10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung E.C., Melanson-Drapeau L., Cregan S.P., Vanderluit J.L., Ferguson K.L., McIntosh W.C. Apoptosis-inducing factor is a key factor in neuronal cell death propagated by BAX-dependent and BAX-independent mechanisms. Journal of Neuroscience. 2005;25(6):1324–1334. doi: 10.1523/JNEUROSCI.4261-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ishihara N., Takagi N., Niimura M., Takagi K., Nakano M., Tanonaka K. Inhibition of apoptosis-inducing factor translocation is involved in protective effects of hepatocyte growth factor against excitotoxic cell death in cultured hippocampal neurons. Journal of Neurochemistry. 2005;95(5):1277–1286. doi: 10.1111/j.1471-4159.2005.03446.x. [DOI] [PubMed] [Google Scholar]
- 74.Zong W.X., Ditsworth D., Bauer D.E., Wang Z.Q., Thompson C.B. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes & Development. 2004;18(11):1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hegedus C., Lakatos P., Olah G., Toth B.I., Gergely S., Szabo E. Protein kinase C protects from DNA damage-induced necrotic cell death by inhibiting poly(ADP-ribose) polymerase-1. FEBS Letters. 2008;582(12):1672–1678. doi: 10.1016/j.febslet.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y., Kim N.S., Li X., Greer P.A., Koehler R.C., Dawson V.L. Calpain activation is not required for AIF translocation in PARP-1-dependent cell death (parthanatos) Journal of Neurochemistry. 2009;110(2):687–696. doi: 10.1111/j.1471-4159.2009.06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arnoult D., Parone P., Martinou J.C., Antonsson B., Estaquier J., Ameisen J.C. Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. The Journal of Cell Biology. 2002;159(6):923–929. doi: 10.1083/jcb.200207071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Munoz-Pinedo C., Guio-Carrion A., Goldstein J.C., Fitzgerald P., Newmeyer D.D., Green D.R. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proceedings of the National Academy of Sciences of the U S A. 2006;103(31):11573–11578. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joza N., Susin S.A., Daugas E., Stanford W.L., Cho S.K., Li C.Y. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410(6828):549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 80.Lyon M.F., Rastan S. Parental source of chromosome imprinting and its relevance for X chromosome inactivation. Differentiation. 1984;26(1):63–67. doi: 10.1111/j.1432-0436.1984.tb01375.x. [DOI] [PubMed] [Google Scholar]
- 81.Heard E. Recent advances in X-chromosome inactivation. Current Opinion in Cell Biology. 2004;16(3):247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 82.Krietsch W.K., Fundele R., Kuntz G.W., Fehlau M., Burki K., Illmensee K. The expression of X-linked phosphoglycerate kinase in the early mouse embryo. Differentiation. 1982;23(2):141–144. doi: 10.1111/j.1432-0436.1982.tb01276.x. [DOI] [PubMed] [Google Scholar]
- 83.Banerjee H., Das A., Srivastava S., Mattoo H.R., Thyagarajan K., Khalsa J.K. A role for apoptosis-inducing factor in T cell development. Journal of Experimental Medicine. 2012;209(9):1641–1653. doi: 10.1084/jem.20110306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng X., Lin Q., Liu Y., Jin Y., Druso J.E., Antonyak M.A. Inactivation of Cdc42 in embryonic brain results in hydrocephalus with ependymal cell defects in mice. Protein Cell. 2013;4(3):231–242. doi: 10.1007/s13238-012-2098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Appelbe O.K., Bollman B., Attarwala A., Triebes L.A., Muniz-Talavera H., Curry D.J. Disruption of the mouse Jhy gene causes abnormal ciliary microtubule patterning and juvenile hydrocephalus. Developmental Biology. 2013;382(1):172–185. doi: 10.1016/j.ydbio.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wyss L., Schafer J., Liebner S., Mittelbronn M., Deutsch U., Enzmann G. Junctional adhesion molecule (JAM)-C deficient C57BL/6 mice develop a severe hydrocephalus. PloS One. 2012;7(9) doi: 10.1371/journal.pone.0045619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Park R., Moon U.Y., Park J.Y., Hughes L.J., Johnson R.L., Cho S.H. Yap is required for ependymal integrity and is suppressed in LPA-induced hydrocephalus. Nature Communications. 2016;7:10329. doi: 10.1038/ncomms10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cao M., Wu J.I. Camk2a-Cre-mediated conditional deletion of chromatin remodeler Brg1 causes perinatal hydrocephalus. Neuroscience Letters. 2015;597:71–76. doi: 10.1016/j.neulet.2015.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vogel P., Read R.W., Hansen G.M., Payne B.J., Small D., Sands A.T. Congenital hydrocephalus in genetically engineered mice. Veterinary Pathology Online. 2012;49(1):166–181. doi: 10.1177/0300985811415708. [DOI] [PubMed] [Google Scholar]
- 90.Parikh S., Saneto R., Falk M.J., Anselm I., Cohen B.H., Haas R. A modern approach to the treatment of mitochondrial disease. Current Treatment Options in Neurology. 2009;11(6):414–430. doi: 10.1007/s11940-009-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olsen R.K., Olpin S.E., Andresen B.S., Miedzybrodzka Z.H., Pourfarzam M., Merinero B. ETFDH mutations as a major cause of riboflavin-responsive multiple acyl-CoA dehydrogenation deficiency. Brain. 2007;130(Pt 8):2045–2054. doi: 10.1093/brain/awm135. [DOI] [PubMed] [Google Scholar]
- 92.Avula S., Parikh S., Demarest S., Kurz J., Gropman A. Treatment of mitochondrial disorders. Current Treatment Options in Neurology. 2014;16(6):292. doi: 10.1007/s11940-014-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.