Significance
SS-31 is a synthetic peptide that improves mitochondrial function and is currently undergoing clinical trials for treatments of heart failure, primary mitochondrial myopathy, and other mitochondrial diseases. SS-31 interacts with cardiolipin which is abundant in the inner mitochondrial membrane, but mechanistic details of its pharmacological effects are unknown. Here we apply a chemical cross-linking/mass spectrometry method to provide direct evidence for specific interactions between SS-31 and mitochondrial proteins. The identified SS-31 interactors are functional components in ATP production and 2-oxoglutarate metabolism and signaling, consistent with improved mitochondrial function resultant from SS-31 treatment. These results offer a glimpse of the protein interaction landscape of SS-31 and provide mechanistic insight relevant to SS-31 mitochondrial therapy.
Keywords: aging, interactome, mitochondria, cross-linking
Abstract
Mitochondrial dysfunction underlies the etiology of a broad spectrum of diseases including heart disease, cancer, neurodegenerative diseases, and the general aging process. Therapeutics that restore healthy mitochondrial function hold promise for treatment of these conditions. The synthetic tetrapeptide, elamipretide (SS-31), improves mitochondrial function, but mechanistic details of its pharmacological effects are unknown. Reportedly, SS-31 primarily interacts with the phospholipid cardiolipin in the inner mitochondrial membrane. Here we utilize chemical cross-linking with mass spectrometry to identify protein interactors of SS-31 in mitochondria. The SS-31-interacting proteins, all known cardiolipin binders, fall into two groups, those involved in ATP production through the oxidative phosphorylation pathway and those involved in 2-oxoglutarate metabolic processes. Residues cross-linked with SS-31 reveal binding regions that in many cases, are proximal to cardiolipin–protein interacting regions. These results offer a glimpse of the protein interaction landscape of SS-31 and provide mechanistic insight relevant to SS-31 mitochondrial therapy.
Mitochondria are the power stations of the cell and generate cellular energy in the form of adenosine triphosphate (ATP). ATP is produced by oxidative phosphorylation (OXPHOS) through the mitochondrial electron transport system (ETS) comprising five multisubunit protein complexes, complex I (CI) to complex IV, and complex V, ATP synthase. Mitochondrial dysfunction is associated with many human diseases which can manifest in any organ or tissue, including the nervous system, skeletal and cardiac muscles, kidneys, liver, and endocrine system, and present diverse clinical symptoms (1). There are few effective treatments for mitochondrial disease, as dysfunction of the ETS and OXPHOS for ATP production involves multiple proteins and multiple steps (2). A conventional therapeutic strategy of targeting a single protein is unlikely to succeed in restoring function in complicated mitochondrial diseases. Another significant challenge for treating mitochondrial dysfunction is the efficient delivery of therapeutic molecules to the mitochondria. Elamipretide (SS-31) is a synthetic tetrapeptide that is a member of an emerging new class of therapeutics that selectively target mitochondria to restore mitochondrial bioenergetics. SS-31 is currently under clinical trials for multiple mitochondrial disorders, including aging, mitochondrial genetic disease, ischemia, acute kidney injury, and heart failure (3–5). SS-31 has shown promising results for treatment of various mitochondrial diseases; however, its mechanism of action remains unclear.
Mitochondria are composed of two membranes, an outer mitochondrial membrane (OMM) and an inner mitochondrial membrane (IMM) which forms invaginations, commonly named cristae. The cristae membranes constitute the backbone platform where mitochondrial respiratory complexes are located and OXPHOS takes place. Cardiolipin (CL), an unusual anionic lipid with two phosphate head groups and four acyl chains, is almost exclusively localized on the IMM (6). It has long been known that CL has a critical role in bioenergetics, cristae morphology, and assembly of respiratory components into higher order supercomplexes (7, 8). Bound CL molecules are required for the enzymatic activities and stabilities of both individual protein subunits and protein supercomplexes involved in mitochondrial respiration. For example, CL plays an essential role in the oligomerization of the c-rings and lubrication of its rotation in ATP synthase (CV), which can influence the stability of cristae structure through dimerization (9, 10); CL acts as glue holding respiratory supercomplexes (CIII and CIV) together and steering their assembly and organization (11, 12); and the binding sites of CL identified close to the proton transfer pathway in CIII and CIV suggest a role of CL in proton uptake through the IMM (13–15). In the case of the ADP/ATP translocase, ADT, three bound CL molecules securely anchor the carrier protein in the IMM and affect ADP/ATP transport activity (16). Previous studies have shown that SS-31 peptide can penetrate the OMM and concentrate in the IMM by selectively binding to CL (3–5, 17). The leading hypothesis is that the alternating aromatic-cationic motif of this synthetic tetrapeptide binds CL via dual interactions, i.e., hydrophobic interaction with acyl chain and electrostatic interactions with anionic phosphate head groups. This specific binding has been validated by fluorescence spectroscopy, isothermal titration calorimetry, and NMR analysis (18, 19). Though not experimentally demonstrated, it was suggested that binding of SS-31 to CL helps induce tighter curvatures of cristae to stabilize the IMM structure and optimize the organization of respiratory chain supercomplexes for enhanced electron transfer and ATP production (5, 17). Given the numerous lines of evidence showing SS-31–CL and CL–protein interactions, one could speculate direct interactions may exist between SS-31 and mitochondrial proteins. SS-31–protein interactions could play a direct or synergistic role with CL interactions in mitochondrial structure, cristae morphology, and ATP synthesis. To date, no empirical data exist to demonstrate SS-31 interacts with mitochondrial proteins, and the mechanistic details of how SS-31 modulates and rejuvenates mitochondrial function have remained elusive.
Chemical cross-linking mass spectrometry (XL-MS) has evolved to be a valuable and widely used tool for studying protein structures and interactions (20–23). Recent development of protein interaction reporter (PIR)-based XL-MS technology in our group has further enabled large-scale identification of protein interactions in complex mixtures from living cells (24–26) and isolated functional organelles (27, 28). Previous studies have shown that SS-31, when added to permeabilized cells or isolated mitochondria, can decrease generation of H2O2 and increase O2 consumption and ATP production (18, 19, 29–31). To investigate the protein interaction landscape of SS-31, we utilized a N-terminal biotinylated form of SS-31 which allows for affinity enrichment of cross-linked SS-31–protein complexes. PIR-based cross-linkers were applied directly to isolated mitochondria incubated with biotinylated SS-31 (bSS-31) to secure a snapshot of protein interactions in their functional state. Subsequent mass spectrometry analysis uncovered the identities of SS-31 protein interactors with topological information. Here, we present a report of the SS-31 protein interaction network, comprising 12 mitochondrial proteins belonging to nine enzymatic complexes. Interestingly, all 12 proteins were previously reported to bind with CL on the IMM and contribute directly and/or indirectly to mitochondrial respiration. These data provide evidence of direct interactions between SS-31 and CL-associated proteins. Thus, in addition to effects on IMM structure, the interaction between SS-31 and CL may serve to localize SS-31 to the IMM and facilitate its interactions with CL-binding proteins. These SS-31–protein interactions provide insight into the functional role of SS-31 within mitochondria. Furthermore, our research establishes a general approach to investigate the structural details of protein interactions with therapeutic molecules in situ.
Results and Discussion
bSS-31 Interactome.
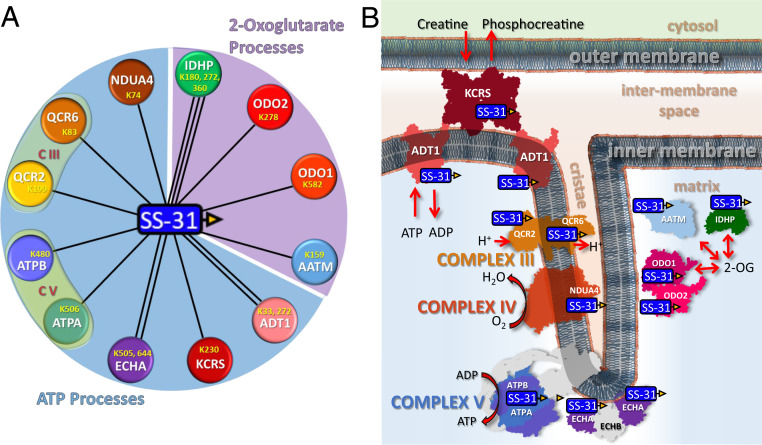
SS-31 has been shown to selectively interact with CL, localizing and concentrating the peptide in the IMM (18). To identify SS-31 protein interaction partners, we utilized an affinity-tagged version containing a biotin group on the peptide N terminus (bSS-31) (SI Appendix, Fig. S1A) and carried out the experimental workflow illustrated in Fig. 1. SS-31 has been demonstrated to have no effect on mitochondrial function in young healthy mitochondria, while increasing mitochondrial ATP production and reducing H2O2 production and redox stress in aged mouse muscle (30, 32). Importantly, bSS-31 also increased mitochondrial respiration and reduced H2O2 production in mitochondrial isolated from aged mice (see Figs. 3 and 4), while not affecting either parameter in mitochondria from young animals. Both SS-31 and bSS-31 reduced superoxide production in aged cardiomyocytes (SI Appendix, Fig. S3B), indicating no discernable difference in the antioxidant activity of SS-31 due to addition to the biotin tag. Isolated mitochondria from aged mouse hearts were incubated with 10 μM bSS-31 for an hour prior to chemical cross-linking with a PIR cross-linker (DP-amide) (Fig. 1 and SI Appendix, Fig. S1B). After cross-linking, mitochondrial proteins were extracted with urea and subjected to a standard tryptic digestion protocol. Peptides cross-linked to bSS-31 were enriched using avidin affinity chromatography and subjected to liquid chromatography-mass spectrometry (LC-MS) analysis employing two different acquisition methods, ReACT (33) and Mango (34) developed in our laboratory for the identification of PIR cross-linked peptide pairs. In total we were able to confidently identify 17 nonredundant cross-linked peptide pairs between bSS-31 and 16 Lys residues on 12 different proteins as shown in Fig. 2 and Dataset S1. The bSS-31 cross-linked peptide identifications were repeatedly observed across biological replicate samples originating from four mice. The cross-linked lysine residues are indicated in Fig. 2. Interestingly the bSS-31 interacting proteins can be grouped into two broad classes, those involved in ATP production and those utilizing 2-oxoglutarate (Fig. 2). Furthermore, each of the 12 proteins is known to directly interact with CL which is important in maintaining the structures and functions for these proteins (Fig. 2).
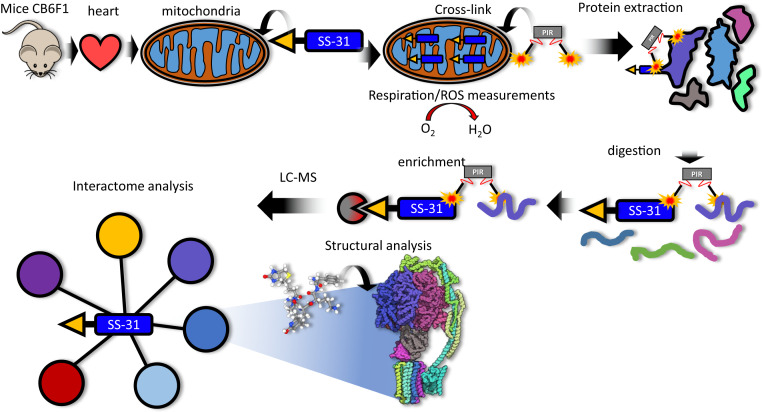
Fig. 1.
Experimental overview. Mice (total of 4), strain CB6F1 (BALB/cBy × C57BL/6), were killed by cervical dislocation. Hearts were excised and processed to isolate mitochondria. Isolated mitochondria were treated with 10 μM biotin SS-31 (bSS-31, blue rectangle with yellow triangle) for 1 h. Oxygen consumption rates and H2O2 production were monitored to evaluate the functional impact of bSS-31 (Fig. 3 B–D). Mitochondria were cross-linked with the PIR cross-linker DP-NHP (gray rectangle with orange stars). Protein was extracted using 8 M urea and digested with trypsin. Enrichment for peptides containing bSS-31 was performed with immobilized monomeric avidin. Peptide samples were analyzed by LC-MS to identify the interactome network for bSS-31 and structural analysis was performed using cross-link distance restraint guided molecular docking.
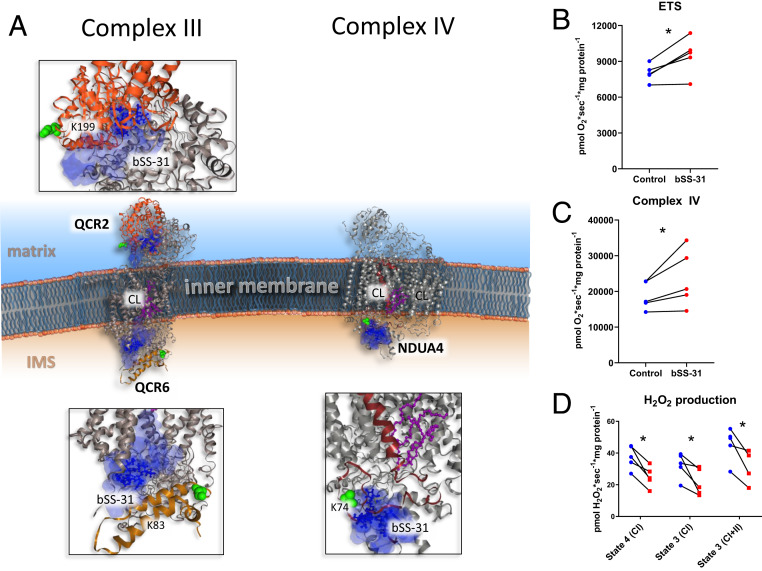
Fig. 3.
Structural and functional impact of bSS-31 interaction with OXPHOS complexes CIII and CIV. (A) Structural view of interaction with subunits QCR2 and QCR6 of CIII and NDUA4 of CIV with Inset showing zoomed view of docked structures. Distance constraints of 0 to 35 Å between the α-carbon of K3 of bSS-31 and the α-carbon of cross-linked lysine were used in molecular docking. Proteins are shown in cartoon view. The top 10 molecular docking results for bSS-31 were included as shown in superimposed semitransparent blue surface view with the top scoring docked position of bSS-31 shown in blue ball-and-stick representation. CL and CL binding residues are shown in magenta ball-and-stick and space-filled representation, respectively. Cross-linked lysines are shown in green spheres. (B) Maximum uncoupled respiration in mitochondria isolated from old mouse hearts in the presence of bSS-31 (red) or vehicle control (blue). See SI Appendix, Fig. S3C for data from young mice. (C) Complex IV activity in the presence of bSS-31 (red) or vehicle control (blue). (D) H2O2 production per O2 consumption in mitochondria isolated in the presence of bSS-31 (red) or vehicle control (blue). *P < 0.05 using a paired t test.
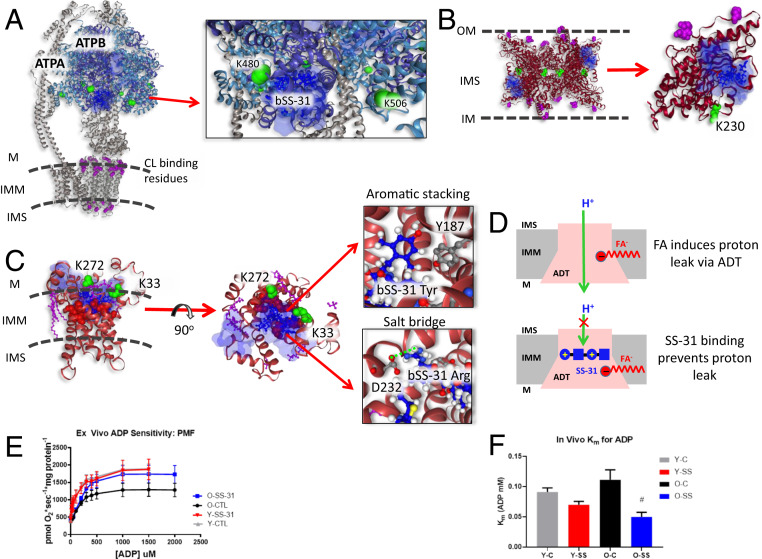
Fig. 4.
Structural and functional impact of bSS-31 interactions with ATP-producing enzymes. Distance constraints of 0 to 35 Å between the α-carbon of K3 of bSS-31 and the α-carbon of cross-linked lysine residues (green spheres) were used in molecular docking. The top 10 docking results are selected to show the ranges of bSS-31 positions in semitransparent blue surface view with the first docking position displayed in blue ball-and-stick view. CL and CL binding residues are shown in magenta ball-and-stick and space-filled representation, respectively. (A) Structure of bSS-31 interaction with ATP synthase with zoomed Inset. (B) A side view of octameric KCRS (PDB: 1crk) bridging outer and inner membranes of mitochondria is shown in cartoon representation at Left. Shown at Right is monomeric chain A in ribbon view with CL binding residues, K408, R418, and K419, in magenta space-filled balls, and cross-linked residue K230 in green spheres. (C) Structural representation of b-SS31 and ADT interaction in the BKA-locked m-state (PDB: 6gci). Cross-linked lysines K33 and K272 are shown in green spheres and CL is shown in magenta ball-and-stick view. The substrate binding sites, K23, R80, R280, Y187, G183, I184, and S228, are shown in red spheres. Interactive versions of the structures can be viewed at xlinkdb.gs.washington.edu/xlinkdb/BiotinylatedSS31_Bruce.php. Insets show detailed view of salt bridge interaction between D232 residue and Arg residue in bSS-31 and aromatic stacking between Y187 and dimethyl tyrosine in bSS-31. (D) Proposed model of FA-dependent proton leak via ADT (Top) (66), SS-31 binding of ADT possibly prevents proton leak with its two positively charged residues (Bottom). (E) SS-31 treatment increases ADP-stimulated respiration in mitochondria isolated from old mice. (F) SS-31 treatment decreases Km for ADP in vivo in old mice. #P < 0.05 using a paired t test compared to old control. Error bars represent the SEM.
Fig. 2.
SS-31 interactome. (A) Nodes represented by colored circles indicate the 12 proteins cross-linked to bSS-31. UniProt identifiers are labeled on the nodes along with the lysine residues that were identified as cross-linked to bSS-31. All 12 proteins are known to interact with cardiolipin (CL, red). The proteins are grouped into two major classes: ATP processes (Upper light blue area) and 2-oxogluterate processes (Lower light purple area). (B) Schematic representation of SS-31 peptide cross-linked proteins within the mitochondria. These proteins include creatine kinase (KCRS), ADP/ATP translocase (ADT1), complex III (CIII) subunits QCR2 and QCR6, complex IV (CIV) subunit NDUA4, complex V (CV) subunits ATPA and ATPB, trifunctional enzyme subunit (ECHA), aspartate aminotransferase (AATM), isocitrate dehydrogenase (IDHP), and 2-oxogluterate dehydrogenase complex subunits ODO1 and ODO2. bSS-31 peptide is represented by a blue rectangle with yellow triangle.
We utilized structural data from the Protein Data Bank (PDB) to localize the cross-linked residues and perform cross-link distance restraint guided molecular docking with bSS-31 using PatchDock (35). For isocitrate dehydrogenase (IDHP) and aspartate aminotransferase (AATM), mouse structures were available in the PDB. For the other proteins we employed Clustal Omega (36) to align each mouse protein with homologous structures from other species. Note, all cross-linked Lys residues for the 10 mouse proteins for which no PDB structure exists are conserved in the homologous protein structures from other species. The data have been uploaded into XLinkDB (37) where a table of the cross-linked peptide pairs, interaction network, and docked structures can be viewed (xlinkdb.gs.washington.edu/xlinkdb/BiotinylatedSS31_Bruce.php). For each protein docked with bSS-31, the interaction interface with bSS-31 was defined as those amino acid residues within 5 Å of the surface volume encapsulating the atom positions of bSS-31 from the top 10 scoring docked models. We utilized Composition Profiler (38) to compare the amino acid composition of the bSS-31 interfaces with that of the MitoCarta 2 (MC2) database (39). Statistically significant differences (P value <0.05) included residues Asp and Thr which are enriched in bSS-31 interaction interfaces while Leu was depleted as compared with MC2 (SI Appendix, Fig. S2). This observation suggests that electrostatic and hydrogen bonding interactions through Asp and Thr side chains are likely important for bSS-31–protein binding, consistent with the high number of cationic and H-bonding sites within bSS-31, as shown in crosslink-directed bSS-31–protein structural models discussed below.
ETS Complexes (CIII and CIV).
OXPHOS complexes generate and maintain the mitochondrial membrane potential (ΔΨ) through the ETS (CI to CIV), which powers conversion of ADP to ATP by ATP synthase (CV). The activity of OXPHOS complexes is dependent on cristae morphology and interactions with CL for their proper assembly and function (7). Excitingly, efforts presented here revealed bSS-31 cross-linked with multiple subunits of the OXPHOS complexes CIII, CIV, and CV (Figs. 2–4) from aged mitochondria. Two subunits of CIII were cross-linked to bSS-31. These included K199 of QCR2 and K83 of QCR6 (PDB: 1sqp) (40). Interestingly, the two bSS-31 interacting subunits of CIII lie on opposite sides of the IMM with QCR2 on the electrochemically negative matrix side and QCR6 located on the electrochemically positive intermembrane space side (Fig. 3). Structural details are presented in Fig. 3A. CIII functions to reduce cytochrome c transferring electrons from coenzyme Q through the Q-cycle while shuttling protons across the IMM (41). Structural analysis indicates CIII is normally assembled as a dimer (CIII2), either alone or as part of larger mitochondrial supercomplexes with varying copies of CI and CIV (42). Each monomer of CIII is comprised of 11 subunits in higher eukaryotes (43). QCR2 is a core subunit of CIII and is important for its dimerization. QCR6 is a highly acidic subunit that is important for the interaction of CIII with cytochrome c (44). Reduced cytochrome c carries electrons from CIII to CIV. CIV is the terminal complex of the ETS ultimately transferring electrons from cytochrome c to reduce molecular oxygen to water. bSS-31 was cross-linked to K74 of the CIV subunit NDUA4 which is located in the intermembrane space. Interestingly, the location of NDUA4 within CIV was only recently resolved as the 14th subunit of the complex (45). Its location within CIV, precludes CIV dimer formation, suggesting that in contrast to most crystal structures showing CIV assembled as a dimer of 13-subunit complexes, it is instead a 14-subunit monomer (45). While not essential for assembly of other CIV subunits, NDUA4 plays a critical role in regulating CIV activity (46). Monomeric CIV contains multiple CL binding sites that are important for enzymatic activity (47, 48). Interestingly the PDB structure of human CIV with NDUA4 resolved recently (PDB: 5z62) indicates a CL molecule bound to a deep pocket formed between NDUA4 and cytochrome c oxidative subunits COX2 and COX3, suggesting it may be important for stabilizing the relatively weak interaction of NDUA4 with the rest of CIV (45). The 14-subunit monomer of CIV is also consistent with structures of the respirasome supercomplex (CI1CIII2CIV1) and megacomplex (CI2CIII2CIV2) assemblies which are important for regulation of respiration and display increased electron transport efficiency (45). Remarkably, studies by Chatfield et al., performed with mitochondria from failing and nonfailing human hearts (49), demonstrated that SS-31 improves mitochondrial function in the failing human heart. More specifically, their work showed that SS-31 improved CIV function, specific to CIV that was associated in supercomplex assemblies but not free CIV. Thus our finding that bSS-31 binds NDUA4 in CIV is consistent with data presented by Chatfield et al. and may help explain why they find that SS-31 increases CIV activity, specific in supercomplexes (49).
Functional measurements indicate that maximal flux through the ETS in the presence of 2-[2-[4-(trifluoromethoxy)phenyl]hydrazinylidene]-propanedinitrile (FCCP) was significantly increased in mitochondria from old hearts incubated with bSS-31 compared to those without bSS-31 (Fig. 3B), while there was no effect of bSS-31 in the mitochondria from young hearts (SI Appendix, Fig. S3C). The uncoupling by FCCP in the ETS measurement removes the dependence of respiration on CV. However, the ETS still provides an integrated measure of the electron transport system. In order to focus on one of the specific complexes that interact with bSS-31 we isolated CIV activity. Mitochondrial isolation performed with bSS-31 significantly elevated CIV-specific activity (Fig. 3C), supporting a functional effect of the bSS-31 on this binding partner in mitochondria from aged hearts. While the enhanced ETS flux could be due to improved function of the ETS or increased substrate supply, the reduced H2O2 production reported here in Fig. 3D provided evidence for more efficient electron transfer through the ETS, i.e., fewer electrons escape before the reduction of O2 to water at CIV.
ATP Production and Transport (CV, CK, and ADT).
ATP synthase (CV), creatine kinase (CK), and ATP/ADP translocase (ADT) that we detected as cross-linked with bSS-31 are three key components in participation of ATP production and transport. CV, ADT, together with inorganic phosphate carrier (PiC), form a larger supercomplex structure referred to as the ATP synthasome (Fig. 2) (50). The ETS generated proton gradient and ΔΨ power the phosphorylation of ADP to ATP by CV. ADT and CK, two non-ETS proteins, play critical roles for shuttling metabolites to CV in the form of phosphocreatine (PCr) and ATP. The generation of ATP by OXPHOS is coupled to phosphorylation of creatine (Cr) by CK that utilizes ATP, transported via ADT protein, to synthesize PCr, which provides a temporal and special energy buffer for cells (51).
CV contains three copies each of ATPA and ATPB assembled as a heterohexamer in the F1 region of the complex on the matrix side of the IMM. There are six nucleotide binding sites located at the ATPA/ATPB interface. Cross-links were detected between bSS-31 and K506 of ATP synthase α-subunit (ATPA) and K480 of ATP synthase β-subunit (ATPB). Crosslink-aided molecular docking localized bSS-31 to ATPB facing the IMM near the ATPA/ATPB interface (Fig. 4A). The CV dimer is thought to be essential to the formation of cristae by bending the IMM (52). ATP synthase function depends on CL, which binds specifically, yet intermittently, to the IMM embedded c-ring portion of CV in the F0 region, acting as a lubricant for rotation of the CV rotor (10, 53, 54). CV localizes near the tightly curved ends of cristae, where it assembles as rows of dimers stabilizing the IMM curvature (55–57). Interestingly, isolated CV reconstituted into liposomes self-assemble into rows of dimers inducing membrane curvature (52). Compartments generated by the membrane folds of cristae allow for pools of highly concentrated protons to drive CV rotation. Beyond the relevance of the dimer structure for ATP production by CV, CV dimers have also been implicated in formation of the mitochondrial permeability transition pore (mPTP) (58). SS-31 has been shown to improve mitochondrial ATP production as well as inhibit mPTP formation (19, 31). Prolonged treatment with SS-31 improved function in aged mitochondria, by increasing ATP production, improving flux of the ETS, and reducing oxidative stress (32). The protein interactors in CIII, CIV, and CV identified here are central to all of these processes and may help explain the beneficial effects observed with SS-31 treatment.
Creatine kinase S-type (KCRS) is a highly symmetric octamer localized in both intermembrane space and cristae space (59). CL plays an important role for the binding of KCRS to the membranes. It has been suggested that this binding is stabilized by electrostatic interactions between the anionic phosphate head group in CL and a conserved basic motif, consisting of three conserved residues, K408, R418, and K419, in the C-terminal tail of KCRS (60). The KCRS crystal structure (PDB: 1crk) (61) revealed two identical, opposite flat surfaces of the oligomers, allowing CK binding to two flat bilayer membranes between OMM and IMM, or the two segments of the cristae junctions (Figs. 2 and 4B) (61, 62). The ability of KCRS to bridge two membranes via CL binding is an important factor for maintaining structural stability and integrity of mitochondria. This bridging is also important for the direct use of ATP, exported via the translocase, to phosphorylate Cr. Additionally, KCRS plays a critical role in clustering and enriching CL in the IMM and facilitating colocalization of ADP/ATP translocase into IMM. Our XL-MS results included the identification of bSS-31 interaction with KCRS at K230 (Fig. 4B). Superimposition of the top 10 molecular docking results localized bSS-31 peptide in the inner membrane space proximal to CL binding regions as displayed in blue cloud in Fig. 4B, consistent with KCRS binding CL and CL interaction with SS-31.
ADT, a transmembrane protein located in the IMM, is a carrier protein for importing ADP from the cytosol and exporting ATP from the mitochondrial matrix through cycling between cytoplasmic-open and matrix-open states (c-state and m-state) (63). While the inhibitor carboxyatractyloside (CATR)-locked ADT c-state crystal structure (PDB: 2c3e) (64) has been available for over a decade, better understanding of the mechanism of ADT for the full transport cycle became possible only recently with resolved crystal structure of bongkrekic acid (BKA) inhibitor-locked ADT in m-state (PDB: 6gci) (65). The six tilted transmembrane α-helices form a bucket-shaped cavity which opens to the intermembrane space (c-state) or the matrix space (m-state). Several CL molecules were found in the crystal structures, attached surrounding the bucket (Fig. 4C). CL molecules tightly bind through several hydrogen bonds and electrostatic interactions to act as an interdomain bridge stabilizing ADT in the membranes (65). The adenine nucleotide binding sites are found in the water-filled central cavity with the phosphate groups bound to K23, R80, and R280; the adenine ring bound with Y187 via aromatic stacking; hydrophobic interactions with G183 and I184; and hydrogen bonding with S228 (63). Our PIR cross-linking experiments identified ADT1, an isoform abundantly expressed in the heart, with two lysine residues (K33 and K272) cross-linked with bSS-31. The top 10 molecular docking models produced using distance constraints from both lysine residues placed bSS-31 (shown in blue cloud) within the water-filled cavity of the translocase in m-state conformation (PDB: 6cgi) (Fig. 4C). bSS-31 is more likely to bind ADT in the m-state for the following reasons. First, lysine residue K272 is only accessible for cross-linking in the m-state as it is exposed to the matrix side of the protein in m-states but buried in the membranes in c-states. Second, both lysine residues K33 and K272 are involved in salt bridge interactions in the c-state with D232 and E265, respectively, and are therefore less likely to be cross-linker reactive in the c-state. Moreover, in the top crosslink-directed docked bSS-31–ADT1 m-state model, D232 forms a salt bridge with the bSS-31 arginine residue (Fig. 4C), and thus, would be unavailable for salt bridge formation with K33 that is important for c-state stabilization (65). And finally, this model also locates the dimethyl tyrosine residue in bSS-31 proximal to Y187 and thus, aromatic stacking analogous to adenine ring interactions may stabilize bSS-31 in this m-state model as well (Fig. 4C). Docking of bSS-31 with c- and m-state conformations is also compared in Movie S1, illustrating c-state stabilizing salt bridges and possible m-state ADT1–bSS-31 salt bridge formation, all of which favor m-state–bSS-31 interactions.
Importantly, ADT was recently identified to support two distinct and competing transport modes involving ADP/ATP exchange and proton leak and thus, ADT connects coupled and uncoupled energy conversion mechanisms in mitochondria, respectively (66). Studies have shown that proton leak through ADT was elevated in aged mitochondria and impairment of ADT was believed to play a central mechanism causing aged-related mitochondria defects (67–69). More significantly, a recent report showed that the mitochondrial trifunctional enzyme-deficient cardiac model system that simulates sudden infant death syndrom (SIDS) disease had a higher level proton leak and SS-31 treatment rescued the increased proton leak (70); another recent report showed that SS-31 can suppress proton leak and rejuvenate mitochondrial function through direct binding with ADT (71). SS-31 shares many common properties with inhibitors BKA and CATR such as larger molecular volume than ADP or ATP, and binding with ADT via multiple polar interactions (65); and additionally, all three molecules were reported to block proton leak in ADT (71). However, unlike SS-31, BKA and CATR decreased the maximal respiratory rate and failed to enhance respiratory control ratio (71). SS-31 binding to the ADT channel yet not functioning as an ADT inhibitor opens the possibility of therapeutic alteration of the balance of conformational states, potentially modulating proton leak and ADP/ATP exchange. As proposed in the recent report, the mechanism of proton leak through ADT is dependent on electrostatic interactions of the positively charged substrate binding site with a fatty acid (FA) cofactor and competes with nucleotide transport through ADT (66). Likewise, one can hypothesize that proton leak is prevented possibly through charge repulsion via two positively charged residues (arginine and lysine) in SS-31 (Fig. 4D). Since favorable SS-31 pharmacological effects on mitochondrial function include increased ATP production and decreased proton leak (3, 71), bSS-31 interaction with ADT in mitochondria identified in this work may be an important mediator of these beneficial effects. Note that addition of the biotin tag does not affect binding specificity of SS-31 with ADT. Zhang et al. demonstrated not only bSS-31 copurification of ADT, but also that bSS-31 is competitive with SS-31 for binding with ADT, indicating both specificity of binding of these peptides with ADT as well as a shared ADT binding site of SS-31 and bSS-31 (71). In addition to direct binding of SS-31 with ADT, the same group also reported that SS-31 associated with the ATP synthasome and stabilized it (71). Our cross-link data provide direct evidence that bSS-31 binds with two members of the ATP synthasome supercomplex, CV and ADT.
To assess the effect of SS-31 binding on the mitochondrial phosphorylation system, we tested the effect of bSS-31 on ADP-stimulated respiration in mitochondria isolated from gastrocnemius muscles. State 3 respiration with complex I substrates was elevated, but the Km for ADP was not affected, in the presence of bSS-31 in mitochondria from the aged gastrocnemius. Neither maximal respiration nor the Km for ADP were affected by bSS-31 in mitochondria from the young (Fig. 4E). This increased state 3 flux could be due to enhanced function anywhere in the oxidative phosphorylation system. In order to more specifically test the effect of SS-31 on ADP sensitivity in vivo we reanalyzed data from Siegel et al. (30) that demonstrated that a single treatment with SS-31 in vivo increased ATPmax and reduced proton leak (higher phosphate/oxygen [P/O] ratio) in aged mouse muscle (SI Appendix, Fig. S3A). In contrast to the data from isolated mitochondria described above (Fig. 4E), the in vivo Km for ADP from the Siegel at al. data was significantly decreased (increased sensitivity for ADP) in the aged mouse muscles following acute treatment with SS-31 with no effect in the young mice (Fig. 4F). This improved ADP sensitivity supports a functional effect of SS-31 on either ADT1, CV, or both. Since favorable SS-31 pharmacological effects on mitochondrial function include increased ATP production and decreased proton leak (3, 71), bSS-31 interaction with CV, ADT, and CK involved in mitochondria respiration identified in this work may be important mediators of these beneficial effects.
Trifunctional Enzyme.
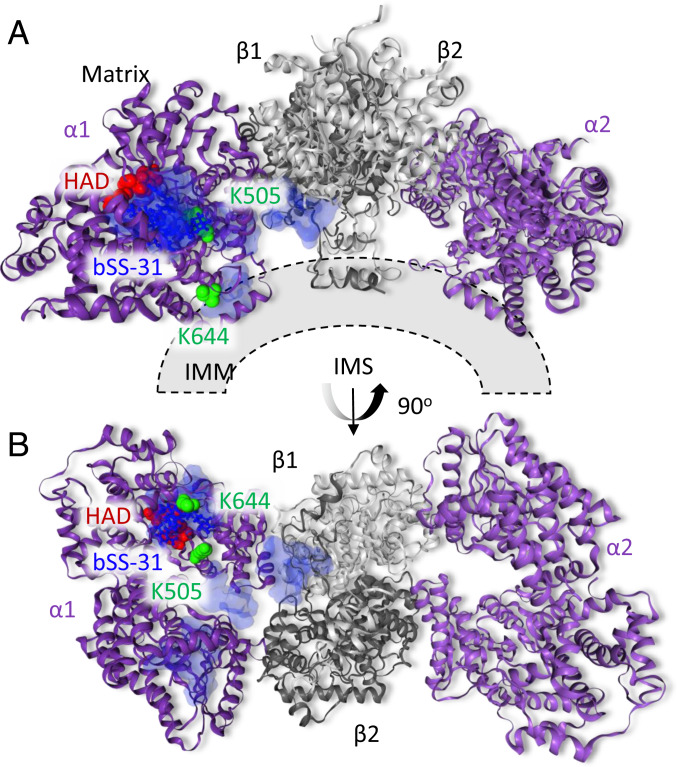
The mitochondrial trifunctional enzyme is a heterooligomeric complex localized on the IMM that catalyzes the latter three reactions of fatty acid β-oxidation for very long chain fatty acids. The trifunctional enzyme consists of the α-subunit (ECHA) which contains 2-enoyl-CoA hydratase (ECH), and 3-hydroxyacyl-CoA dehydrogenase (HAD) activities, and the β-subunit (ECHB) containing 3-ketothiolase (KT) activity. Within mitochondria, the trifunctional enzyme is thought to primarily be assembled as an α2β2 heterotetramer. Recently, structural models for the human trifunctional enzyme have been determined by cryo-EM (72) and X-ray crystallography (73) both highlighting important structural differences between the mammalian mitochondrial trifunctional enzyme and the structures of bacterial orthologs (74, 75). Both structures for the α2β2 mitochondrial trifunctional enzyme reveal a curved structure to the complex where the concave surface is important for interaction with the IMM (72, 73). Beyond functioning in fatty acid β-oxidation, ECHA also functions to remodel CL via monolysocardiolipin acyltransferase activity (76). The α-subunit of the mitochondrial trifunctional enzyme (ECHA) was cross-linked at K505 and K644 to bSS-31. Both of these residues exist on the concave side of the complex near the inner mitochondrial membrane where cardiolipin resides (PDB: 6dv2) (Fig. 5). Cross-linking distance constraint guided docking of bSS-31 with ECHA places bSS-31 into a substrate channel in ECHA near the HAD active side, proposed to be important for cardiolipin remodeling (73) (Fig. 5). Interestingly SS-31 was recently shown to rescue excessive proton leak in cardiomyocytes with mutated ECHA (70).
Fig. 5.
Structural view of bSS-31 interaction with ECHA. Distance constraints of 0 to 35 Å between the α-carbon of K3 of bSS-31 and the α-carbon of cross-linked lysine residues K644 and K505 of PDB structure 6dv2 were used in molecular docking. The top 10 docking results are selected to show the ranges of bSS-31 positions in semitransparent blue surface view with the first docking position displayed in blue ball-and-stick view. The cross-linked Lys residues on ECHA are shown as green space-filled residues. The HAD active site residues proposed to be important for cardiolipin remodeling (S477, H498, E510, and T548) are shown as red space-filled residues. A displays the side view of the complex, while the view from intermembrane space is shown in B. Interactive versions of the structures can be viewed at xlinkdb.gs.washington.edu/xlinkdb/BiotinylatedSS31_Bruce.php.
Two-Oxoglutarate Enzymes.
The metabolite 2-oxoglutarate (also known as α-ketoglutarate, 2-OG) is a key intermediate of the tricarboxylic acid (TCA) cycle, building block for amino acids, and nitrogen transporter. Due to its involvement in both carbon and nitrogen metabolism and multiple signaling pathways, 2-OG has been suggested to act as a master regulator metabolite (77). One example of this is highlighted by the discovery that 2-OG can extend the lifespan of Caenorhabditis elegans by inhibiting ATP synthase and target of rapamycin (TOR) downstream (78). 2-OG also plays a key role in hypoxia signaling serving as a substrate for prolylhydroxylase (PHD) enzymes which regulate the stability of hypoxia-inducible factor (HIF) transcription factors (79, 80). Interestingly, we identified bSS-31 cross-linked with multiple lysine sites across four key proteins, including isocitrate dehydrogenase, 2-OG dehydrogenase (ODO1), dihydrolipoyllysine-residue succinyltransferase component of 2-OG dehydrogenase complex (ODO2), and AATM, all of which produce or utilize 2-OG in their reactions. Excitingly these interactions suggest a potential role for SS-31 in 2-OG signaling.
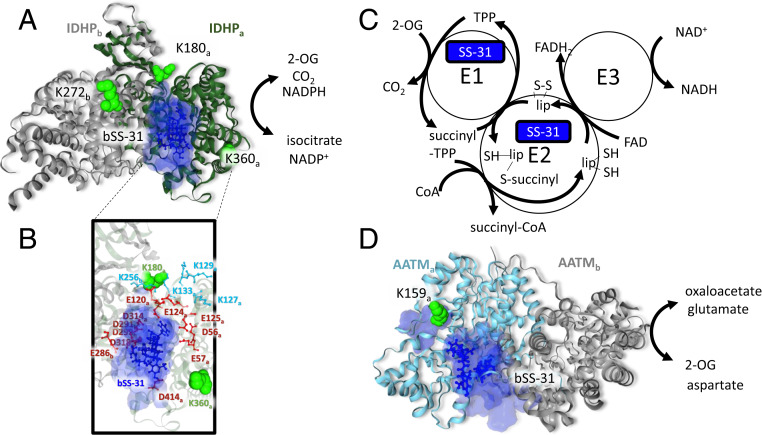
Isocitrate dehydrogenase catalyzes the decarboxylation of isocitrate forming 2-OG and carbon dioxide. There are three types of isocitrate dehydrogenase in mammals, IDH1, IDH2, and IDH3, which differ in structure, function, and subcellular localization (81). IDH1 and IDH2 are NADP+-dependent enzymes that catalyze reversible reactions and are localized in the cytosol and mitochondria, respectively (81). IDH3 is the NAD+-dependent form, localized to the mitochondrial matrix functioning primarily in energy production in the TCA cycle where it catalyzes an irreversible reaction and is allosterically regulated by a number of cofactors (81). The mitochondrial localized NADP+-dependent IDH2 (also known as IDHP) also functions in the TCA cycle but has additional roles in supplying NADPH used in redox homeostasis and fatty acid metabolism (82). This places IDHP at a critical crossroad, regulating energy production through the TCA cycle, maintaining the mitochondrial pool of NADPH needed for the glutathione antioxidant system, and producing 2-OG utilized by PHD proteins in HIF1α signaling. Interestingly, IDHP can also interact with CL-containing membranes that can induce a conformational change in IDHP regulating its enzymatic activity (83). Cross-links were identified between bSS-31 and K180, K272, and K360 of IDHP. Docking of bSS31 with the PDB structure of IDHP (5h3f) results in localization of bSS-31 to a groove between the large and small domains of IDHP near the isocitrate binding pocket (Fig. 6 A and B). Electrostatic repulsion between positively charged Lys residues located in the large domain (K127, K129, K130, and K133) and the small domain (K256) has been suggested to play an important role in IDHP activity by widening the cleft between the domains (84) (Fig. 6B). Neutralization of the charged residues by acetylation or mutation on these Lys residues results in a narrower cleft, smaller isocitrate binding pocket, and lower enzymatic activity (84). A number of acidic residues surround the bSS-31 interface, potentially participating in electrostatic interactions with bSS-31 (Fig. 6B). It is possible that when SS-31 interacts with IDHP, the positive charges in SS-31 would also play a role in widening the cleft between the domains, potentially impacting IDHP activity.
Fig. 6.
SS-31 interaction with 2-oxoglutarate enzymes. (A) bSS-31 interaction with IDHP (PDB: 5h3f) indicated as a homodimer with the IDHPa chain shown as a green ribbon and the IDHPb chain in gray. Lysine residues cross-linked to bSS-31 are displayed as green space-filled residues and labeled with subscript “a” or “b” to indicate which IDHP monomer they are on. bSS-31 (blue ball-and-stick structure) was docked onto the a-subunit using distance restraints for K180 and K360 from the a-subunit and K272 from the b-subunit. The volume encompassing the positions of bSS-31 from the top 10 docking results is displayed as a semitransparent blue surface. (B) View of the bSS-31 interaction interface highlighting surrounding acidic residues (red) and Lys residues (cyan) which are important in maintaining the groove between the large and small domains of IDHP (84). (C) Schematic representation of the 2-oxoglutarate dehydrogenase complex (2-OGDHC). bSS-31 was identified as cross-linked to ODO1 a component of the E1 of 2-OGDHC and ODO2 a component of the E2 portion of 2-OGDHC. (D) bSS-31 interaction with AATM (PDB: 3pdb) shown as a homodimer with the a-chain in teal and the b-chain in gray. Lysine K159 (green space-filled residue) of the a-chain was used to dock bSS-31 into the structure. The volume encompassing the positions of bSS-31 from the top 10 docking results is displayed as a semitransparent blue surface. Interactive versions of the structures can be viewed at xlinkdb.gs.washington.edu/xlinkdb/BiotinylatedSS31_Bruce.php.
Catalyzing the rate-limiting fourth step of the TCA cycle is 2-OG dehydrogenase complex (2-OGDHC), which utilizes thiamine pyrophosphate (TPP) as a cofactor and reduces NAD+ to NADH during the conversion of 2-OG to succinyl-CoA and CO2. The 2-OGDHC is a large multiprotein assembly (∼2 MDa) localized on the IMM, comprising multiple copies of three enzymatic components E1 (ODO1), E2 (ODO2), and E3 (dihydrolipoyl dehydrogenase, DLDH) and sharing similar assembly with the pyruvate dehydrogenase complex (PDH) and the branched chain 2-oxoacid dehydrogenase complex (BCKDH). There are no high-resolution structural models available for any components of the mammalian 2-OGDHC. Experimental evidence from multiple sources, including genetic mutational, limited proteolysis and electron microscopy, indicate 2-OGDHC is composed of 24 E2 subunits assembled into a cubic core with varying numbers of E1 and E3 subunits bound to the periphery of the core (85). The activity of 2-OGDHC is tightly regulated, being inhibited by ATP, NADH, and succinyl-CoA, intertwining its function with that of the OXPHOS complexes. Under some conditions it can also be a primary source of mitochondrial reactive oxygen species (ROS) production producing superoxide and hydrogen peroxide at much higher rates than complex I, which is often implicated as the primary source of mitochondrial ROS (86). The E3 component of 2-OGDHC produces ROS in the form of H2O2 and superoxide radicals when electrons from the FADH2 transfer to molecular oxygen. Furthermore, very reactive and damaging lipoate thiyl radicals can form in the E2 component as a result of reaction with a FADH semiquinone (86). We identified cross-links between bSS-31 and K582 of ODO1 (E1) and K278 of ODO2 (E2) Fig. 6C. Interestingly, CL has been shown to be important for the stability of 2-OGDHC (87).
Mitochondrial AATM catalyzes the reversible reaction of aspartate and 2-OG to glutamate and oxaloacetate. AATM is a key enzyme, linking amino acid and carbohydrate metabolism by providing a carbon source (2-OG) to the TCA cycle as well as nitrogen for the synthesis of nonessential amino acids and nucleotides (88). Beyond its canonical function, mitochondrial AATM also functions as kynurenine aminotransferase as well as a transporter for long chain fatty acids (88, 89). AATM also interacts directly with the cardiolipin-containing liposomes, the inner mitochondrial membrane, as well as 2-OGDHC (90, 91). We identified a cross-link between bSS-31 and K159 of AATM. Crosslink-guided docking places bSS-31 near the oxaloacetate binding pocket in the PDB structure 3PDB Fig. 6D. This is on the opposite side of AATM from the long chain fatty acid (LCFA) binding pocket, colored as blue space-filled residues in Fig. 6D.
Overall the enzymes IDHP, ODO1, ODO2, and AATM function in closely connected biological pathways related to regulation of 2-OG levels, mitochondrial redox homeostasis, and HIF1-α signaling in addition to key metabolic pathways. The specific interactions detected between these proteins and bSS-31 suggest a possible role for SS-31 to influence this nexus of mitochondrial function.
Previous efforts to elucidate SS-31 mechanisms have suggested that beneficial therapeutic effects arise from SS-31 interactions with CL. While CL binding may be the primary impact of SS-31 treatment, the present work resulted in identification of the first set of SS-31 interacting proteins from intact, functional mitochondria using PIR XL-MS. Since all identified SS-31 protein interactors also bind CL, untangling the effects of SS-31–CL interactions from SS-31–protein interactions is not possible from the results presented here. However, one possibility is that the association between CL and SS-31 concentrates SS-31 locally at the IMM surface, facilitating its interaction with proteins in this vicinity. Indeed, membrane cardiolipin interactions with SS-31 are responsible for a reported 1,000-fold increase in concentration of SS-31 in this region (31), and yet the nKD for this interaction is 2.9 μM (92). This will result in a relatively high local concentration of free SS-31 proximal to IMM proteins where SS-31–protein interactions could affect function. In this regard, cross-linked site-directed docking of bSS-31 with nearly all relevant protein–cardiolipin cocrystal structures resulted in localization of bSS-31 in solvent accessible regions, away from the membrane spanning regions where known cardiolipin interactions exist. The distance between these cardiolipin–protein interaction sites and identified bSS-31–protein cross-linked sites are beyond the linking distance possible with the cross-linker. Therefore, it is unlikely for most cases that the observed bSS-31–protein cross-linked sites are the secondary result of the association of bSS-31 with specific cardiolipin molecules at cocrystalized sites identified in the figures. An exception is ADT1 where, because of its small size and proximity of cardiolipin, the identified bSS-31 cross-linked sites could result from either direct ADT1–bSS-31 interaction or be a secondary result of bSS-31 localized at the known cardiolipin site. On the other hand, SS-31 interactions reduce ADT1 proton leak (71) and molecular docking based on identified cross-linked sites favor localization of bSS-31 within the pocket of the ADT m-state conformation where binding of the inhibitor BKA also reduces proton leak. Thus, binding of SS-31 with both CL and IMM proteins may be important for observed pharmacological effects and could help rationalize how, despite having only four amino acids, SS-31 can exhibit specificity for beneficial treatment in aged mitochondria.
Moreover, it is important to consider the inherent specificity of the interactions detected by XL-MS. For instance, consider the complex V catalytic subunit ATPA that was linked in all four biological replicates at the singular lysine site, K506. Murine ATPA has 31 lysine sites that could possibly be linked to bSS-31. Therefore, if bSS-31 interaction with ATPA were nonspecific or random, each ATPA K site would have an equivalent random chance probability of being linked to bSS-31 equal to 1 chance in 31. Moreover, if nonspecific or random, the probability that this same lysine site would be observed across all four biological replicates is (1/31)4 or 1 chance in 923,521 (SI Appendix, Fig. S2 C and D). Thus, statistical considerations indicate it is highly unlikely that the observed link between bSS-31 and ATPA K506 observed across all four biological replicates is resultant from nonspecific or random interaction. Similar statistical calculations for most other observed bSS-31 interactor lysine sites yield comparable probabilities counterindicative of random interactions with bSS-31 (SI Appendix, Fig. S2C). These results provide insight and opportunities to better understand the molecular actions of this mitochondrial targeted therapeutic. This set of 12 proteins are all part of a larger energy transfer system for mitochondrial ATP production through the OXPHOS pathway. Four of the proteins are directly involved with 2-OG metabolism and signaling which regulate mitochondrial metabolism, redox homeostasis, and cellular response to hypoxia. Importantly each of the SS-31 interacting proteins also interact with CL, which impacts their structure and function. This observation presents the possibility that SS-31 concentrates in the IMM by interacting with CL, facilitating its interactions with components of the OXPHOS and 2-OG signaling pathways. Numerous studies have demonstrated the beneficial effects of SS-31 to restore healthy mitochondrial function, increase ATP production, and reduce mitochondrial redox stress. Beyond identifying protein interaction partners, the XL-MS data provide structural information, in terms of distance restraints, that were used to localize bSS-31 interacting regions on these proteins. Knowledge of these protein interactors lays the foundation for followup functional studies to evaluate the significance of these interactions. As a general method, employing affinity-tagged drugs with XL-MS should be extensible to investigate the interaction landscapes of therapeutic molecules in complex biological samples such as organelles, cells, and tissues.
Materials and Methods
Animal Husbandry.
This study was reviewed and approved by the University of Washington Institutional Animal Care and Use Committee. Male, 36- to 37-mo-old CB6F1 (BL/6: BalbC) mice were received from the National Institute of Aging’s aged mouse colony. All mice were maintained at 21 °C on a 14/10 light/dark cycle and given standard mouse chow and water ad libitum with no deviation prior to or after experimental procedures. Animals were killed by cervical dislocation with no anesthetic.
Extraction of Tissue, Mitochondrial Isolation, and SS-31 Treatment.
On day of animal killing, the heart (120 to 150 mg wet weight) was resected. The whole heart was homogenized on ice in a glass Dounce homogenizer in 2 mL of ice-cold respiration buffer (RB, 1 mM EGTA, 5 mM MgCl2, 105 mM K-MES, 30 mM KCl, 10 mM KH2PO4, pH 7.1). The homogenates were centrifuged for 10 min at 800 × g at 4 °C. The supernatants were collected and centrifuged for 15 min at 8,000 to 10,000 × g at 4 °C. After the removal of supernatant, the pellet was resuspended in isolation medium. The pellet was centrifuged for 15 min at 8,000 to 10,000 × g at 4 °C and resuspended in isolation medium. Mitochondrial pellets were separated into new centrifuge tubes and diluted with 200 μL RB, 10 mM pyruvate, and 2 mM malate. Either elamipretide or 2 μL saline was added to the mitochondria and incubated for 1 h at room temperature with mild rocking.
Ex Vivo and In Vivo Functional Assays.
Mitochondrial respiration and H2O2 production were assayed in mitochondria isolated from young (5- to 7-mo-old) and old (36- to 37-mo-old) mouse hearts or gastrocnemius muscles as described above either in the presence or absence of 10 μM bSS-31, using an Oxygraph 2K dual respirometer/fluorometer (Oroboros Instruments). In order to assess in vivo ADP sensitivity, we reanalyzed data from Siegel et al. (30). Mitochondrial superoxide was quantified by the ratio of mitoSOX to mitoTrackerGreen using Leica SP8X confocal microscopy. Further details on these assays are available in SI Appendix, SI Materials and Methods.
Cross-Linking Reaction.
The cross-linker amide-DP-NHP (synthesis described in SI Appendix, SI Materials and Methods) was added at a final concentration of 10 mM (from a 270-mM stock solution) to mitochondria incubated with bSS-31. The reaction mixture was mixed at 800 rpm at room temperature for 30 min. A total of five cross-linked samples were generated including four biological replicates of mouse heart mitochondria incubated with bSS-31 and a negative control sample to which no bSS-31 was added. After cross-linking, the samples were frozen at −80 °C.
Cross-Linked Mitochondrial Sample Preparation.
Cross-linked mitochondrial samples were lysed with 8 M urea in 0.1 M NH4HCO3. Extracted protein was digested with trypsin and bSS-31 cross-linked peptides were enriched with monomeric avidin as described in detail in SI Appendix, SI Methods and Materials.
LC-MS Analysis of Cross-Linked Peptide Pairs.
Samples were analyzed by LC-MS using two methods developed for analysis of cross-linked peptide pairs, namely ReACT (33) and Mango (34). ReACT analysis was carried out on a Velos-FTICR (Fourier-transform ion cyclotron resonance) mass spectrometer coupled with a Waters nanoAcquity UPLC (ultra performance liquid chromatography).
Samples were analyzed by Mango using a Thermo Q-exactive plus mass spectrometer coupled with a Thermo Easy nLC. See SI Appendix, SI Materials and Methods for details on LC-MS.
Data Analysis.
LC-MS data files in.RAW format were converted to .mzXML format using the ReADW tool in the Trans Proteomic Pipeline software suite (93). Comet (94) was used to search the.mzXML files against the Mitocarta 2 database (39) containing both forward and reverse protein sequences (2,084 total sequences) along with the addition of the sequence for bSS-31 (Biotin-D-Arg-dimethyl Tyr-Lys-Phe-NH2). For details see SI Appendix, SI Materials and Methods.
Molecular Docking.
A molecular model for bSS-31 was generated using Avogadro (v. 1.2.0) utilizing a molecular mechanics conformer search of 1,000 conformers followed by geometric optimization. PatchDock (35) was used to dock the model of bSS-31 with structural models in the PDB for bSS-31 interacting proteins (AATM = PDB: 3pdb, ADT c-state = PDB: 2c3e, ADT m-state = PDB: 6gci, ATPA/ATPB = PDB: 5ARA, QCR2/QCR6 = PDB: 1sqp, NDUA4 = PDB: 5z62, ECHA = PDB: 6dv2, IDHP = PDB: 5h3f, and KCRS = PDB: 1crk). Clustal Omega (36) was used to align each mouse protein with homologous structures from other species. For each protein, the top 10 scoring docked models from PatchDock were downloaded and the interaction interface with bSS-31 was defined as those amino acid residues within 5 Å of the surface volume encapsulating the atom positions of bSS-31.
Data Availability.
All biotinylated-SS-31 cross-linking data presented in this paper are available in the database XLinkDB at: xlinkdb.gs.washington.edu/xlinkdb/BiotinylatedSS31_Bruce.php. The LC-MS data have been deposited into the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with accession no. PXD019484.
Supplementary Material
Acknowledgments
We would like to thank Hazel Szeto for her generous gift of biotin-labeled SS-31. We would also like to thank the members of the Bruce Lab and Nathan Alder for helpful input during these experiments. This work was supported by P01 AG001751, an American Federation for Aging Research BIG award, and grants 5R01GM086688, 1R01GM097112, 7S10RR025107, 5U19AI107775, R01HL144778, and R35GM136255 from the US National Institutes of Health.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The LC-MS data have been deposited into the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with accession no. PXD019484.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002250117/-/DCSupplemental.
References
- 1.Craven L., Alston C. L., Taylor R. W., Turnbull D. M., Recent advances in mitochondrial disease. Annu. Rev. Genomics Hum. Genet. 18, 257–275 (2017). [DOI] [PubMed] [Google Scholar]
- 2.El-Hattab A. W., Zarante A. M., Almannai M., Scaglia F., Therapies for mitochondrial diseases and current clinical trials. Mol. Genet. Metab. 122, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szeto H. H., Birk A. V., Serendipity and the discovery of novel compounds that restore mitochondrial plasticity. Clin. Pharmacol. Ther. 96, 672–683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szeto H. H., First-in-class cardiolipin-protective compound as a therapeutic agent to restore mitochondrial bioenergetics. Br. J. Pharmacol. 171, 2029–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szeto H. H., Stealth peptides target cellular powerhouses to fight rare and common age-related diseases. Protein Pept. Lett. 25, 1108–1123 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Pennington E. R., Funai K., Brown D. A., Shaikh S. R., The role of cardiolipin concentration and acyl chain composition on mitochondrial inner membrane molecular organization and function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 1039–1052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paradies G., Paradies V., De Benedictis V., Ruggiero F. M., Petrosillo G., Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta 1837, 408–417 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Ikon N., Ryan R. O., Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 1859, 1156–1163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acehan D. et al., Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 100, 2184–2192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehdipour A. R., Hummer G., Cardiolipin puts the seal on ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 113, 8568–8570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M., Mileykovskaya E., Dowhan W., Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Mileykovskaya E., Dowhan W., Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem. Phys. Lipids 179, 42–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnarez C., Mazat J. P., Elezgaray J., Marrink S. J., Periole X., Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. J. Am. Chem. Soc. 135, 3112–3120 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Arnarez C., Marrink S. J., Periole X., Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci. Rep. 3, 1263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnarez C., Marrink S. J., Periole X., Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci. (Camb.) 7, 4435–4443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann B., Stöckl A., Schlame M., Beyer K., Klingenberg M., The reconstituted ADP/ATP carrier activity has an absolute requirement for cardiolipin as shown in cysteine mutants. J. Biol. Chem. 269, 1940–1944 (1994). [PubMed] [Google Scholar]
- 17.Szeto H. H., Liu S., Cardiolipin-targeted peptides rejuvenate mitochondrial function, remodel mitochondria, and promote tissue regeneration during aging. Arch. Biochem. Biophys. 660, 137–148 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Birk A. V. et al., The mitochondrial-targeted compound SS-31 re-energizes ischemic mitochondria by interacting with cardiolipin. J. Am. Soc. Nephrol. 24, 1250–1261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birk A. V., Chao W. M., Bracken C., Warren J. D., Szeto H. H., Targeting mitochondrial cardiolipin and the cytochrome c/cardiolipin complex to promote electron transport and optimize mitochondrial ATP synthesis. Br. J. Pharmacol. 171, 2017–2028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arora B., Tandon R., Attri P., Bhatia R., Chemical crosslinking: Role in protein and peptide science. Curr. Protein Pept. Sci. 18, 946–955 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Piotrowski C., Sinz A., Structural investigation of proteins and protein complexes by chemical cross-linking/mass spectrometry. Adv. Exp. Med. Biol. 1105, 101–121 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Sinz A., Cross-linking/mass spectrometry for studying protein structures and protein-protein interactions: Where are we now and where should we go from here? Angew. Chem. Int. Ed. Engl. 57, 6390–6396 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Chavez J. D., Bruce J. E., Chemical cross-linking with mass spectrometry: A tool for systems structural biology. Curr. Opin. Chem. Biol. 48, 8–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavez J. D., Hoopmann M. R., Weisbrod C. R., Takara K., Bruce J. E., Quantitative proteomic and interaction network analysis of cisplatin resistance in HeLa cells. PLoS One 6, e19892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavez J. D., Weisbrod C. R., Zheng C., Eng J. K., Bruce J. E., Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteomics 12, 1451–1467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez J. D., Schweppe D. K., Eng J. K., Bruce J. E., In vivo conformational dynamics of Hsp90 and its interactors. Cell Chem. Biol. 23, 716–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweppe D. K. et al., Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 114, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavez J. D. et al., Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 136–141.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson E. J. et al., Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J. Clin. Invest. 119, 573–581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel M. P. et al., Mitochondrial-targeted peptide rapidly improves mitochondrial energetics and skeletal muscle performance in aged mice. Aging Cell 12, 763–771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao K. et al., Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J. Biol. Chem. 279, 34682–34690 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Campbell M. D. et al., Improving mitochondrial function with SS-31 reverses age-related redox stress and improves exercise tolerance in aged mice. Free Radic. Biol. Med. 134, 268–281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisbrod C. R. et al., In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr J. P., Perumalla P., Chavez J. D., Eng J. K., Bruce J. E., Mango: A general tool for collision induced dissociation-cleavable cross-linked peptide identification. Anal. Chem. 90, 6028–6034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneidman-Duhovny D. et al., PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids. Res. 33, W363–W367 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McWilliam H. et al., Analysis tool web services from the EMBL-EBI. Nucleic Acids. Res. 41, W597–W600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller A., Chavez J. D., Eng J. K., Thornton Z., Bruce J. E., Tools for 3D interactome visualization. J. Proteome Res. 18, 753–758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vacic V., Uversky V. N., Dunker A. K., Lonardi S., Composition profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 8, 211 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvo S. E., Clauser K. R., Mootha V. K., MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esser L. et al., Crystallographic studies of quinol oxidation site inhibitors: A modified classification of inhibitors for the cytochrome bc(1) complex. J. Mol. Biol. 341, 281–302 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Cramer W. A., Hasan S. S., Yamashita E., The Q cycle of cytochrome bc complexes: A structure perspective. Biochim. Biophys. Acta 1807, 788–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letts J. A., Sazanov L. A., Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24, 800–808 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Ndi M., Marin-Buera L., Salvatori R., Singh A. P., Ott M., Biogenesis of the bc1 complex of the mitochondrial respiratory chain. J. Mol. Biol. 430, 3892–3905 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Solmaz S. R., Hunte C., Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J. Biol. Chem. 283, 17542–17549 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Zong S. et al., Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pitceathly R. D. S., Taanman J. W., NDUFA4 (Renamed COXFA4) is a cytochrome-c oxidase subunit. Trends Endocrinol. Metab. 29, 452–454 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Robinson N. C., Functional binding of cardiolipin to cytochrome c oxidase. J. Bioenerg. Biomembr. 25, 153–163 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Malkamäki A., Sharma V., Atomistic insights into cardiolipin binding sites of cytochrome c oxidase. Biochim. Biophys. Acta Bioenerg. 1860, 224–232 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Chatfield K. C. et al., Elamipretide improves mitochondrial function in the failing human heart. JACC Basic Transl. Sci. 4, 147–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C. et al., Mitochondrial ATP synthasome: Three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP. J. Biol. Chem. 279, 31761–31768 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Dzeja P. P., Terzic A., Phosphotransfer networks and cellular energetics. J. Exp. Biol. 206, 2039–2047 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Blum T. B., Hahn A., Meier T., Davies K. M., Kühlbrandt W., Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. U.S.A. 116, 4250–4255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eble K. S., Coleman W. B., Hantgan R. R., Cunningham C. C., Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem. 265, 19434–19440 (1990). [PubMed] [Google Scholar]
- 54.Duncan A. L., Robinson A. J., Walker J. E., Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc. Natl. Acad. Sci. U.S.A. 113, 8687–8692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strauss M., Hofhaus G., Schröder R. R., Kühlbrandt W., Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27, 1154–1160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies K. M., Anselmi C., Wittig I., Faraldo-Gómez J. D., Kühlbrandt W., Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 109, 13602–13607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hahn A. et al., Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell 63, 445–456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giorgio V. et al., Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. U.S.A. 110, 5887–5892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlattner U. et al., Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim. Biophys. Acta 1788, 2032–2047 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Schlattner U. et al., C-terminal lysines determine phospholipid interaction of sarcomeric mitochondrial creatine kinase. J. Biol. Chem. 279, 24334–24342 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Fritz-Wolf K., Schnyder T., Wallimann T., Kabsch W., Structure of mitochondrial creatine kinase. Nature 381, 341–345 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Karo J., Peterson P., Vendelin M., Molecular dynamics simulations of creatine kinase and adenine nucleotide translocase in mitochondrial membrane patch. J. Biol. Chem. 287, 7467–7476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kunji E. R. et al., The transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta 1863, 2379–2393 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Pebay-Peyroula E. et al., Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Ruprecht J. J. et al., The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 176, 435–447.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertholet A. M. et al., H+ transport is an integral function of the mitochondrial ADP/ATP carrier. Nature 571, 515–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tonkonogi M. et al., Reduced oxidative power but unchanged antioxidative capacity in skeletal muscle from aged humans. Pflugers Arch. 446, 261–269 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Kumaran S., Panneerselvam K. S., Shila S., Sivarajan K., Panneerselvam C., Age-associated deficit of mitochondrial oxidative phosphorylation in skeletal muscle: Role of carnitine and lipoic acid. Mol. Cell. Biochem. 280, 83–89 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Serviddio G. et al., Bioenergetics in aging: Mitochondrial proton leak in aging rat liver, kidney and heart. Redox Rep. 12, 91–95 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Miklas J. W. et al., TFPa/HADHA is required for fatty acid beta-oxidation and cardiolipin re-modeling in human cardiomyocytes. Nat. Commun. 10, 4671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang H., et al. , Reduction of elevated proton leak rejuvenates mitochondria in the aged cardiomyocyte. bioAxiv: 10.1101/2020.01.02.893362 (3 January 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang K. et al., Cryo-EM structure of human mitochondrial trifunctional protein. Proc. Natl. Acad. Sci. U.S.A. 115, 7039–7044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia C., Fu Z., Battaile K. P., Kim J. P., Crystal structure of human mitochondrial trifunctional protein, a fatty acid β-oxidation metabolon. Proc. Natl. Acad. Sci. U.S.A. 116, 6069–6074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishikawa M., Tsuchiya D., Oyama T., Tsunaka Y., Morikawa K., Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex. EMBO J. 23, 2745–2754 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venkatesan R., Wierenga R. K., Structure of mycobacterial β-oxidation trifunctional enzyme reveals its altered assembly and putative substrate channeling pathway. ACS Chem. Biol. 8, 1063–1073 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Taylor W. A. et al., Human trifunctional protein alpha links cardiolipin remodeling to beta-oxidation. PLoS One 7, e48628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huergo L. F., Dixon R., The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol. Mol. Biol. Rev. 79, 419–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chin R. M. et al., The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature 510, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuhrmann D. C., Brüne B., Mitochondrial composition and function under the control of hypoxia. Redox Biol. 12, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abboud M. I. et al., 2-Oxoglutarate regulates binding of hydroxylated hypoxia-inducible factor to prolyl hydroxylase domain 2. Chem. Commun. (Camb.) 54, 3130–3133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reitman Z. J., Yan H., Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J. Natl. Cancer Inst. 102, 932–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jo S. H. et al., Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 276, 16168–16176 (2001). [DOI] [PubMed] [Google Scholar]
- 83.Suga K., Hamasaki A., Chinzaka J., Umakoshi H., Liposomes modified with cardiolipin can act as a platform to regulate the potential flux of NADP+-dependent isocitrate dehydrogenase. Metab. Eng. Commun. 3, 8–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Y. et al., Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics. Sci. Rep. 7, 9785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheu K. F., Blass J. P., The α-ketoglutarate dehydrogenase complex. Ann. N. Y. Acad. Sci. 893, 61–78 (1999). [DOI] [PubMed] [Google Scholar]
- 86.Quinlan C. L. et al., The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 289, 8312–8325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Gestel R. A. et al., The influence of the acyl chain composition of cardiolipin on the stability of mitochondrial complexes; an unexpected effect of cardiolipin in alpha-ketoglutarate dehydrogenase and prohibitin complexes. J. Proteomics 73, 806–814 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Jiang X., Wang J., Chang H., Zhou Y., Recombinant expression, purification and crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase. Biosci. Trends 10, 79–84 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Bradbury M. W., Stump D., Guarnieri F., Berk P. D., Molecular modeling and functional confirmation of a predicted fatty acid binding site of mitochondrial aspartate aminotransferase. J. Mol. Biol. 412, 412–422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teller J. K., Fahien L. A., Valdivia E., Interactions among mitochondrial aspartate aminotransferase, malate dehydrogenase, and the inner mitochondrial membrane from heart, hepatoma, and liver. J. Biol. Chem. 265, 19486–19494 (1990). [PubMed] [Google Scholar]
- 91.Donate F., Yañez A. J., Iriarte A., Martinez-Carrion M., Interaction of the precursor to mitochondrial aspartate aminotransferase and its presequence peptide with model membranes. J. Biol. Chem. 275, 34147–34156 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Mitchell W., et al. , Molecular mechanism of action of mitochondrial therapeutic SS-31 (Elamipretide): Membrane interactions and effects on surface electrostatics. bioAxiv: 10.1101/735001 (14 August 2019).
- 93.Keller A. et al., “A uniform proteomics MS/MS analysis platform utilizing open XML file formats” in Mol. Syst. Biol., (2005), Vol. 1, p. 2005 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eng J. K., Jahan T. A., Hoopmann M. R., Comet: An open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All biotinylated-SS-31 cross-linking data presented in this paper are available in the database XLinkDB at: xlinkdb.gs.washington.edu/xlinkdb/BiotinylatedSS31_Bruce.php. The LC-MS data have been deposited into the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with accession no. PXD019484.