Summary
Ferroptosis is a type of cell death related to cancer; however, the characteristics of ferroptosis in cancers are still uncertain. Based on the data in The Cancer Genome Atlas, we found that most ferroptosis regulator genes (FRGs) were differentially expressed in tumors, somatic copy number alterations (SCNA) and DNA methylation contributed to their aberrant expression. We established the ferroptosis potential index (FPI) to reveal the functional roles of ferroptosis and noticed that the FPI was higher in tumors than in normal tissues in most cancers and was associated with subtypes and clinical features. The FPI was negatively correlated with several metabolic pathways but positively associated with several important metastasis-related pathways and immune-related pathways. High FPI predicted poor prognosis in several tumors, whereas FPI and FRGs impacted drug sensitivity. Our study presents a systematic analysis of ferroptosis and its regulatory genes and highlights the potential of ferroptosis-based cancer therapy.
Subject Areas: Genomics, Cancer Systems Biology, Cancer, Transcriptomics
Graphical Abstract
Highlights
-
•
The ferroptosis regulator genes were aberrantly expressed in tumor
-
•
The ferroptosis potential index (FPI) was established to model ferroptosis level
-
•
The FPI was correlated with metabolic, metastatic, and immune pathways
-
•
High FPI predicted poor prognosis in many cancer types
Genomics; Cancer Systems Biology; Cancer; Transcriptomics
Introduction
As a newly discovered type of programmed cell death, ferroptosis results from the accumulation of iron-dependent lipid hydroperoxides and leads to cytological changes; the features and mechanisms of ferroptosis are different from those of typical cell death processes, such as apoptosis (Dixon et al., 2012). Previous studies demonstrated that, during the process of ferroptosis, sufficient and available cellular iron is required (Dixon et al., 2012). Inhibition of system XC−, which is a membrane Na+-dependent cysteine-glutamate exchange transporter, and GPX4 can disrupt the oxidation-reduction balance and cause overwhelming lipid peroxidation that ultimately results in cell death (Stockwell et al., 2017; Yang et al., 2014; Yu et al., 2017). The initiation and execution of ferroptosis are affected by multiple factors, including amino acids, lipids, and iron metabolism, and are regulated by various signaling pathways, such as amino acid and glutathione metabolism, lipid metabolism, iron metabolism, and the mevalonate pathway (Stockwell et al., 2017).
As the understanding of ferroptosis has increased, its complex biological function has been revealed (Matsushita et al., 2015; Yang and Stockwell, 2016). Furthermore, ferroptosis has been found to be closely related to various human diseases including periventricular leukomalacia, Huntington's disease, and acute kidney injury (Friedmann Angeli et al., 2014; Inder et al., 2002; Linkermann et al., 2014; Skouta et al., 2014). In addition, the literature has confirmed that ferroptosis suppresses tumor growth and kills tumor cells (Yu et al., 2017) and then plays an important role in cancers such as renal cell carcinomas and liver cancer (Sun et al., 2016b; Yang et al., 2014). For example, erastin-induced ferroptosis decreases the growth of tumors formed from human colorectal cancer cells (Xie et al., 2017). Ductal pancreatic cancer cells with a mutant KRAS gene are more susceptible to ferroptosis than wild-type cells (Eling et al., 2015; Yu et al., 2017). Conversely, inducing tumor cell ferroptosis by small molecules has become an important strategy for the treatment of many tumors, such as hepatocellular carcinoma, kidney cancer, and pancreatic cancer (Eling et al., 2015; Louandre et al., 2013, 2015; Yang et al., 2014). In addition, changes in the gene expression of tumor cells also affect ferroptosis, and a number of genes have been confirmed to regulate ferroptosis. For example, ACSL4 participates in the biosynthesis and remodeling of polyunsaturated fatty acid-Pes, and the downregulation of ACSL4 increases the resistance to ferroptosis (Dixon et al., 2015; Doll et al., 2017). Furthermore, ferroptosis and ferroptosis regulator genes (FRGs) have been identified to be correlated with drug resistance (Lu et al., 2017). For example, it was previously reported that liver cancer cells repress ferroptosis by regulating the expression of NRF2 or MT-1G, which promotes sorafenib resistance in vitro and in tumor xenograft models (Sun et al., 2016a, 2016b). Recently, Wang et al. reported that CD8+ T cells promote tumor ferroptosis during cancer immunotherapy treatment (Wang et al., 2019). Thus, ferroptosis might play important roles during cancer progression and treatment, and a systematic study of ferroptosis and its dysregulation across cancers will be helpful.
In the present study, for the first time, we performed a comprehensive analysis of genomic variations and expression profiles of the FRGs across 20 cancer types. Furthermore, we computationally modeled the ferroptosis level based on FRGs expression and dissected the relations between ferroptosis and cancer clinical features. It was found that ferroptosis was associated with various cancer hallmarks, the immune microenvironment, drug resistance, and patient survival. These results highlight the critical roles of ferroptosis in cancer and should be helpful for further investigations of ferroptosis-related molecular mechanisms and therapy development.
Results
Genetic Alterations of Ferroptosis Regulator Genes in Cancers
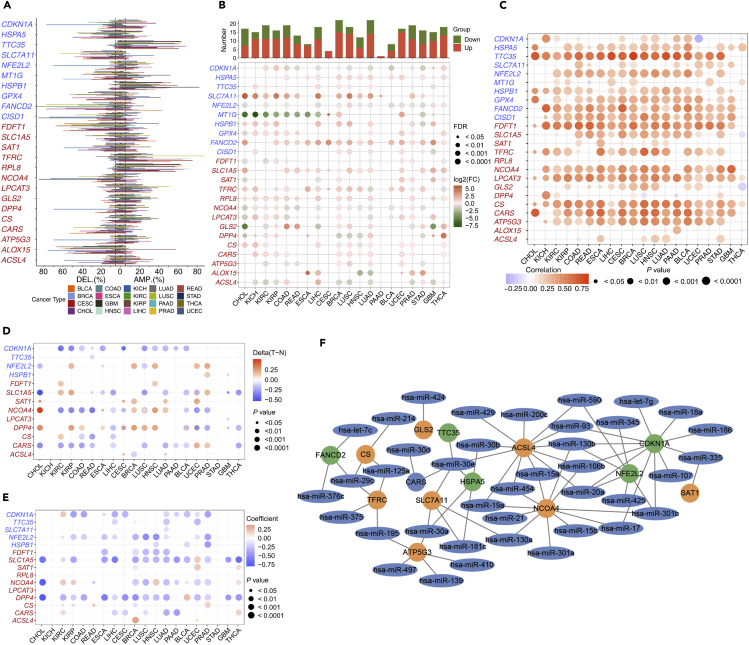
In this study, the twenty four genes that were identified to play critical roles in regulating ferroptosis by previous studies were defined as ferroptosis regulator genes (FRGs), including cyclin-dependent kinase inhibitor 1 (CDKN1A), heat shock protein family A member 5 (HSPA5), ER membrane protein complex subunit 2 (TTC35/EMC2), solute carrier family 7 member 11 (SLC7A11), nuclear factor, erythroid 2 like 2 (NFE2L2), metallothionein-1G (MT1G), heat shock protein beta 1 (HSPB1), glutathione peroxidase 4 (GPX4), Fanconi anemia complementation group D2 (FANCD2), CDGSH iron sulfur domain 1 (CISD1), farnesyl-diphosphate farnesyltransferase 1 (FDFT1), solute carrier family 1 Member 5 (SLC1A5), spermidine/spermine N1-acetyltransferase 1 (SAT1), transferrin receptor (TFRC), ribosomal protein L8 (RPL8), nuclear receptor coactivator 4 (NCOA4), lysophosphatidylcholine acyltransferase 3 (LPCAT3), glutaminase 2 (GLS2), dipeptidyl-dippeptidase-4 (DPP4), citrate synthase (CS), cysteinyl tRNA synthetase (CARS), ATP synthase, H+ transporting, mitochondrial Fo complex subunit C3 (ATP5G3), arachidonate 15-lipoxygenase (ALOX15), and acyl-CoA synthetase long-chain family member 4 (ACSL4) (Stockwell et al., 2017). To determine the patterns of dysregulation of FRGs in cancer, we examined the genomic data, including genetic variation, somatic copy number alternation (SCNA), mRNA expression, and DNA methylation data of tumor and normal tissues from 20 cancer types. The analysis of nonsynonymous mutations in 20 cancer types showed that the mutation frequencies for FRGs were generally low in almost all cancers and relatively high in UCEC (Figure S1A). Furthermore, NFE2L2, which encodes NRF2 and plays a master regulator role in antioxidant responses and has been shown to cause resistance to ferroptosis (Sun et al., 2016b), showed relatively high mutation frequencies in multiple cancers including BLCA, CESC, ESCA, HNSC, LUSC, and UCEC (Figure S1A). However, no mutations of FRGs were significantly related to survival.
To further investigate the genetic aberrations of FRGs in cancer, the percentage of SCNA was examined and the results showed that in general SCNA occurred at high frequencies (with over 5% of all samples) in most cancer types (Figure 1A), but all FRGs in THCA showed a low frequency of SCNA. FRGs presented diverse SCNA profiles. For example, TTC35, HSPB1, TFRC, and RPL8 were more prone to copy number gain than copy number loss in almost all tumors, but SCL7A11 and ALOX15 showed the opposite profile. Furthermore, we analyzed the cooccurrence of mutations and SCNA between FRGs and cancer-specific oncogenes/tumor suppressor genes (TSGs) previously identified by Bailey et al. (2018), in which NFE2L2 was an oncogene in cancer types including BLCA, CESC, HNSC, LIHC, LUSC, and UCEC and CDKN1A was a tumor suppressor gene in BLCA and LIHC. The significance was calculated via the mutual exclusivity test by DISCOVER (Canisius et al., 2016), with a false discovery rate of 1%. A total of 28 oncogenes (Figure S1B) and 43 TSGs (Figure S1C) were found to be altered with FRGs in a mutually exclusive manner in certain cancer types, and FDFT1, RPL8, TFRC, and NFE2L2 were frequently exclusive to oncogenes or TSGs (Figures S1B and S1C).
Figure 1.
The Dysregulation of Ferroptosis Regulator Genes (FRGs)
For which the positive and negative regulators are marked in red and blue, respectively.
(A) Histogram shows the frequency of somatic copy number alterations for each FRG in each cancer type.
(B) Histogram (upper panel) shows the number of significantly differentially expressed genes, and the heatmap shows the fold change and FDR of FRGs in each cancer. Significantly upregulated and downregulated genes are marked in red and green, respectively.
(C) The Spearman's correlation between somatic copy number alterations and the expression of FRGs.
(D) Heatmap shows the differential methylation of FRGs in cancers; hypermethylated and hypomethylated genes are marked in red and blue, respectively (Wilcoxon rank-sum test).
(E) Pearson's correlation of FRGs between transcriptional expression and promoter methylation. Red and blue represent positive and negative correlations, respectively.
(F) The miRNA-mRNA network for FRGs, the orange and green circles are positive and negative FRGs, respectively (Spearman's Rho < −0.1, FDR <0.05).
Aberrant Expression of FRGs among Cancer
Besides genetic alterations, differential expression analysis was performed between tumor and adjacent normal tissues for every cancer type to investigate alterations in the gene expression patterns of FRGs, and the numbers of tumor and normal samples are shown in Table 1. We found that all FRGs were differentially expressed in at least one cancer type. Several FRGs showed consistent expression patterns in cross-cancer analysis. SLC7A11, FANCD2, CARS, SLC1A5, and RPL8 were significantly upregulated in 15, 18, 12, 15, and 14 types of cancers, respectively, whereas NCOA4 was downregulated in 15 cancers (Figure 1B). Additionally, several FRGs showed miscellaneous cancer type-specific patterns that have not been well characterized previously. For example, HSPA5 was upregulated in most cancer types including breast invasive carcinoma (BRCA) (fold change [FC] = 1.87, adjusted p value = 1.42 ×10−42) and LUAD (FC = 1.90, adjusted p value = 2.45 ×10−29) but was significantly downregulated in THCA (FC = 0.68, adjusted p value = 2.67 ×10−7). DPP4, which appears to play a suppressor role in the development of cancer (Masur et al., 2006; Pro and Dang, 2004; Wesley et al., 2005), showed significant upregulation in KIRC, KIRP, ESCA, LUAD, GBM, and THCA, but was downregulated in CHOL, KICH, BRCA, LUSC, HNSC, and STAD. We also noticed that DPP4 had opposite expression profiles in different subtypes of tumors in the lung and kidney. This demonstrated that FRGs might play different roles in different cancers.
Table 1.
The Abbreviations and Numbers of Samples for the 20 Types of Tumors Investigated in This Study
| Tumor Type | Abbreviation | Number of Tumor Samples | Number of Normal Samples |
|---|---|---|---|
| Bladder urothelial carcinoma | BLCA | 408 | 19 |
| Breast invasive carcinoma | BRCA | 1,100 | 112 |
| Cervical and endocervical cancers | CESC | 306 | 3 |
| Cholangiocarcinoma | CHOL | 36 | 9 |
| Colon adenocarcinoma | COAD | 459 | 41 |
| Esophageal carcinoma | ESCA | 185 | 11 |
| Glioblastoma multiforme | GBM | 166 | 5 |
| Head and neck squamous cell carcinoma | HNSC | 522 | 44 |
| Kidney chromophobe | KICH | 66 | 25 |
| Kidney renal clear cell carcinoma | KIRC | 534 | 72 |
| Kidney renal papillary cell carcinoma | KIRP | 291 | 32 |
| Liver hepatocellular carcinoma | LIHC | 373 | 50 |
| Lung adenocarcinoma | LUAD | 517 | 59 |
| Lung squamous cell carcinoma | LUSC | 501 | 51 |
| Pancreatic adenocarcinoma | PAAD | 179 | 4 |
| Prostate adenocarcinoma | PRAD | 498 | 52 |
| Rectum adenocarcinoma | READ | 167 | 10 |
| Stomach adenocarcinoma | STAD | 415 | 35 |
| Thyroid carcinoma | THCA | 509 | 59 |
| Uterine corpus Endometrial carcinoma |
UCEC | 544 | 35 |
Since SCNA in tumors plays a critical role in regulating gene expression, we evaluated the effects of SCNA on the gene expression of FRGs. The Pearson correlation between gene expression and copy number from the masked copy number segment of TCGA was examined. The results showed that the expression of most FRGs was obviously correlated with SCNA in most tumors (Figure 1C). For example, the expression of citrate synthase (CS), which participates in oxidative metabolism, was significantly associated with SCNA in all cancers. This result indicates that the aberrance of copy number for FRGs is common in most cancers and can influence gene expression.
In addition to SCNA, the methylation of promoter can regulate gene expression and aberrant DNA methylation of the promoter is associated with tumorigenesis (Shen and Laird, 2013). We observed that FRGs showed complex methylation patterns in the 20 cancer types (Figure 1D), and only CDKN1A consistently showed hypomethylation in 11 tumors. For example, we observed that DPP4 showed hypermethylation in seven cancer types and hypomethylation in four cancer types. Although there were differences in the pattern of methylation for FRGs, a negative relationship was observed between gene expression and DNA methylation overall (Figure 1E). This result demonstrated that promoter DNA methylation may regulate the expression of FRGs in tumors.
Besides SCNA and DNA methylation, microRNAs (miRNAs) can regulate gene expression and become involved in cancer development (Macfarlane and Murphy, 2010). To determine which miRNAs are involved in regulating ferroptosis, we performed an analysis to reveal the network of miRNA-FRGs. The starBase database was used to infer miRNAs that potentially target FRGs, and those miRNAs that were significantly negatively correlated with gene expression were thought to be involved in the regulation of FRGs (Li et al., 2014). As the network showed (Figure 1F), all of the interactions between miRNA and FRGs occurred in more than six cancers. It is obvious that FRGs could be targeted by miRNAs with high frequency, including ACSL4, NCOA4, and CDKN1A (Figure 1F). To further understand the dysregulation of the miRNA-FRG network in tumors, we conducted differential expression analysis of miRNA and quantified the expression aberrances of miRNAs across cancer types (Figure S1D). It was found that several miRNAs had consistent expression trends in tumors, for example, hsa-miR-93 targeted CDKN1A and was downregulated in 11 cancers. On the other hand, miRNAs had different expression trends in different tumors. For example, hsa-miR-375 targeted ACSL4 in five tumors, but it showed upregulation in two cancers and downregulation in three cancers. This suggests that miRNAs play a regulatory role in FRGs expression, and the aberrant expression of miRNAs in tumors could impact ferroptosis.
Since SCNA, DNA methylation, and miRNA expression can all regulate FRGs expression, but their detailed contributions to gene expression are not clear. The linear regression approach was applied to analyze the contribution of each factor and diminish the confounding effects. As the results showed (Figure S1E), SCNA was positively correlated with gene expression, whereas methylation and miRNA expression showed a negative correlation. We further observed multiple regulatory patterns in FRGs, and the expression of several FRGs was only related to a single factor in several tumors, whereas others were related to multiple factors. For example, ACSL4 expression was only associated with miRNA expression in seven cancers, including BLCA, CESC, KIRC, KIRP, LUAD, PRAD, and UCEC. SCNA played the only significant regulatory role for CARS in CHOL, ESCA, and colorectal cancer. However, the expression of NCOA4 and NFE2L2 was significantly regulated by three factors in seven and ten tumors, respectively. Thus, all FRGs show diverse regulation patterns in different cancers. This suggests that the expression regulation patterns of all FRGs were tumor specific.
Computational Modeling of the Ferroptosis Level among Cancers
To further understand the role of ferroptosis in tumorigenesis and investigate the factors or biological processes associated with ferroptosis, the ferroptosis potential index (FPI) was modeled based on the enrichment score (ES) of positive core machine components calculated by ssGSEA minus that of negative core machine components. We evaluated the FPI through three independent GEO gene expression datasets of tumor cell lines that were treated with erastin or withaferin A (WA), which were reported as inducers of ferroptosis (Dixon et al., 2012; Hassannia et al., 2018), or ferrostatin, which was identified as an inhibitor of ferroptosis (Zhang et al., 2019). The FPI was calculated for the gene expression datasets in neuroblastoma cells (GSE112384), clear cell carcinoma cells (GSE121689), and liver cancer cells (GSE104462) (Figure S2). The results showed that erastin and WA increased the FPI markedly in all three cell lines, whereas ferrostatin obviously decreased the FPI compared with the control group (Figures S2A, S2C, and S2E). Because the increases in CHAC1 and PTGS2 mRNA and ACSL4 protein were associated with cells undergoing ferroptosis, but these changes were not consistent in all experiments (Stockwell et al., 2017), we compared the mRNA expression of these three genes in these three cell lines. As the results showed, the mRNA expression of PTGS2 could not significantly distinguish the ferroptosis status in these experiments. For ACSL4, there were no obvious changes in cell lines with ferroptosis induced by erastin or WA, although decrease was observed in the ferroptosis-inhibited cells (Figures S2B and S2D). CHAC1 was upregulated in ferroptosis-induced cell lines treated with erastin or WA but only slightly upregulated in ferroptosis-inhibited cells (Figure S2F). Thus, the FPI could be used to represent the potential level of ferroptosis based on the transcriptome data.
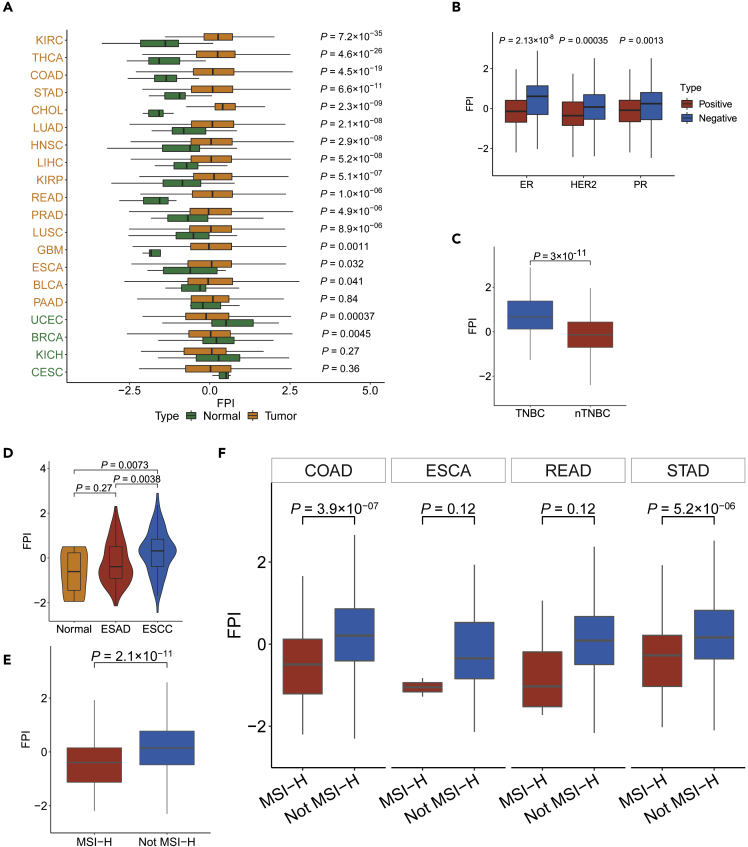
Next, we compared the differences in FPI, the computed marker of ferroptosis, between tumor and normal tissues with the cancer genome atlas (TCGA) data (Figure 2A). Of note, significant differences were found in most cancers such as lung cancer and gastrointestinal cancer and higher FPI was observed in most tumors except for BRCA and UCEC. We also noticed that the FPI of normal tissues in female cancers (BRCA, UCEC, and CESC) was higher than most tumor/normal samples in all cancers. To further dissect the factors involved in these different FPI patterns, we examined the FPI for different subtypes of BRCA. The results presented in Figure 2B showed that all estrogen receptor (ER)-positive, progesterone receptor (PR)-positive, and HER2-positive patients had lower FPIs than the negative patients. Furthermore, triple-negative breast cancer (TNBC) samples showed higher ferroptosis levels than non-TNBC samples (Figure 2C), which was consistent with a previous study (Kettner et al., 2016). Furthermore, among the different subtypes of kidney cancer, the FPI of tumor samples was higher than normal in KIRP and KIRC, whereas the FPI in KICH was lower (Figure 2A). In addition, to further dissect the variances of ferroptosis levels in different histological types, the FPIs in esophageal squamous cell carcinoma (ESCC) and esophagus adenocarcinoma (ESAD) were compared. The results showed that the FPI in ESCC was significantly higher than in ESAD and normal samples, whereas the FPI in ESAD was higher than that in normal but not significant (Figure 2D).
Figure 2.
The Relations between FPI and Histological Types and Molecular Subtypes among Cancers
(A) The different FPIs between tumor and normal tissues among cancers.
(B) Box shows the difference of in FPI between positive and negative receptors in breast cancer.
(C) The difference in FPI between TNBC and nTNBC.
(D) The different FPIs among different histological types of esophageal carcinoma.
(E and F) The FPI for different MSI statuses of overall (E) and detailed (F) digestive system neoplasms. The boxes in (B)–(F) mean the median values ±1 quartile, their whiskers extending from the hinge to the smallest or biggest value, which is 1.5 × interquartile range (IQR) from the box boundaries. All tests were Wilcoxon rank-sum test.
In addition to the cancer types and subtypes, we analyzed the correlation between the FPI and remarkable molecular features such as microsatellite instability (MSI) and driver gene mutations. The FPI decreased obviously in tumors with a high level of MSI (MSI-H) compared with non-MSI-H tumors in gastrointestinal tumors (Figure 2E). The FPI was markedly lower in patients with COAD and STAD with MSI-H, and a slight decrease was also observed in ESCA and READ (Figure 2F). Furthermore, we analyzed the correlation between FPI and 375 driver genes that are frequently mutated in tumors based on regression-based analysis. By controlling for cancer type and tumor mutation burden (TMB), 81 genes were found to be correlated, and most of them were negatively associated with FPI (adj.p < 0.1, Figure S3A). Strikingly, somatic mutations, namely, NFE2L2, BRAF, and TP53, were positively associated with FPI in pancancer associations, but a mutation in TP53 was negatively correlated with ferroptosis in LUSC. This observation was verified by the comparison of FPI between the TP53 mutant and wild-type groups (Figure S3B), consistent with previous findings that showed that TP53 played a dual role in regulating ferroptosis (Kang et al., 2019). The influences of KRAS mutations on FPI were also investigated, and the results showed that the relationship between KRAS mutation and FPI was significantly negative in gastric tumors (Figure S3C) but slightly positive in hepatocellular carcinoma (Figures S3A and S3C). These results indicated that the tumor type and molecular context were critical for the regulation of ferroptosis, which was consistent with a previous study (Tsoi et al., 2018). Furthermore, differential expression of FRGs between wild-type and tumors with mutant TP53 (Figure S3D) and KRAS (Figure S3E) was found to be ubiquitous in most cancers. Taken together, these data indicate that ferroptosis is negatively related to MSI-H in cancer.
Association between Ferroptosis and Pathways in Cancer
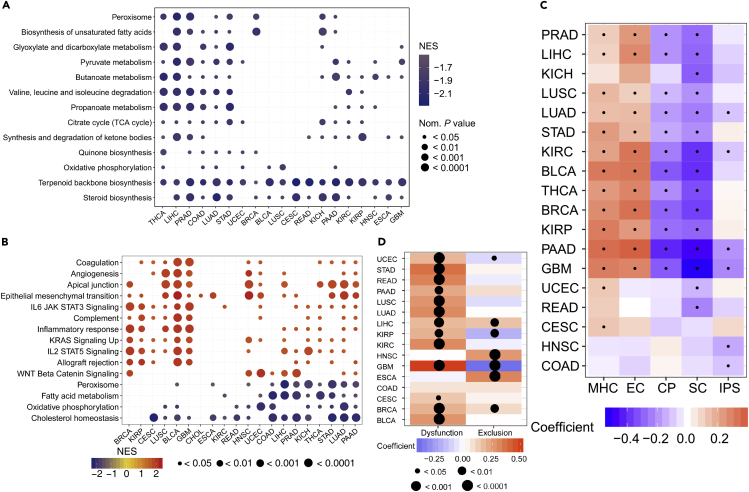
To further elucidate the association between the FPI and other genes and pathways, we calculated Spearman correlation coefficients between FPI and all the genes including FRGs (Table S1). It was found that FPI was generally positively and negatively correlated with the expression of positive and negative FRGs, respectively, but various exceptions were also observed (Table S1). The related cellular signaling of ferroptosis in cancer was investigated by gene set enrichment analysis (GSEA) for each cancer based on the transcriptome of two tumor groups with the top and bottom 30% of FPI. It was observed that metabolism-related pathways in KEGG were usually enriched in tumors with lower FPIs, and pathways frequently enriched (>6 cancers) are presented in Figure 3A. For example, terpenoid backbone biosynthesis and steroid biosynthesis were enriched in the low-FPI group in 19 and 16 cancers, respectively (Figure 3A). Peroxisome, biosynthesis of unsaturated fatty acids, etc. were also significantly correlated with lower FPI in all these cancer types (Figure 3A). Furthermore, the relations between cancer hallmarks and FPI were also analyzed, and the results showed that 15 hallmarks were frequently significantly correlated with FPI (Figure 3B). For example, KRAS signaling, epithelial-mesenchymal transition, IL6 JAK STAT3 signaling, and WNT Beta-catenin signaling were enriched in the high-FPI group, which indicated that ferroptosis was positively related to these oncogenic pathways (Figure 3B). Additionally, metabolism-related hallmarks were observed to be negatively related to ferroptosis, which was consistent with the pathway analysis (Figure 3B).
Figure 3.
Relationships between Ferroptosis and Signaling Pathways and Immunophenotypes
(A and B) Enrichment analysis for metabolism pathway (A) and cancer signaling (B) between high- and low-FPI tumor tissues. NES is the normalized enrichment score in the GSEA algorithm.
(C) The Spearman's correlations between immunophenotypes and the FPI, and dots indicate statistically significant results (p value <0.05).
(D) The Spearman's rank correlation coefficient between the FPI and dysfunction/exclusion scores inferred by TIDE.
Since previous studies showed that ferroptosis was related to the immune response process (Matsushita et al., 2015), we investigated the association between ferroptosis and the immune microenvironment in tumors. The results showed that, in several cancers, FPI was weakly negatively correlated with immunophenoscore (IPS) (Figure 3C), which could predict the response of the immune checkpoint blockade in melanoma tissue (Charoentong et al., 2017). To better understand the relationship between ferroptosis and the response to immunotherapy, we calculated the Spearman coefficients and found that the FPI was positively correlated with the dysfunction score of T cells in 13 cancers. However, no consistent effect of the FPI on the exclusion score of T cells was observed (Figure 3D). Furthermore, FPI was positively correlated with MHC and effector cells (ECs) but negatively correlated with immunomodulators (CP) and suppressor cells (SCs) in most cancers (Figure 3C). The associations between ferroptosis and immune cell types were further evaluated in detail, and the results showed that FPI showed a positive correlation with macrophages in most cancers and a negative association with activated dendritic cells, activated mast cells, plasma cells, and T follicular helper cells (Figure S3F). Thus, there might be close connections between ferroptosis and the immune microenvironment and the function of T cells, and more investigations are needed to reveal the details.
Clinical Relevance of Ferroptosis and Its Regulators
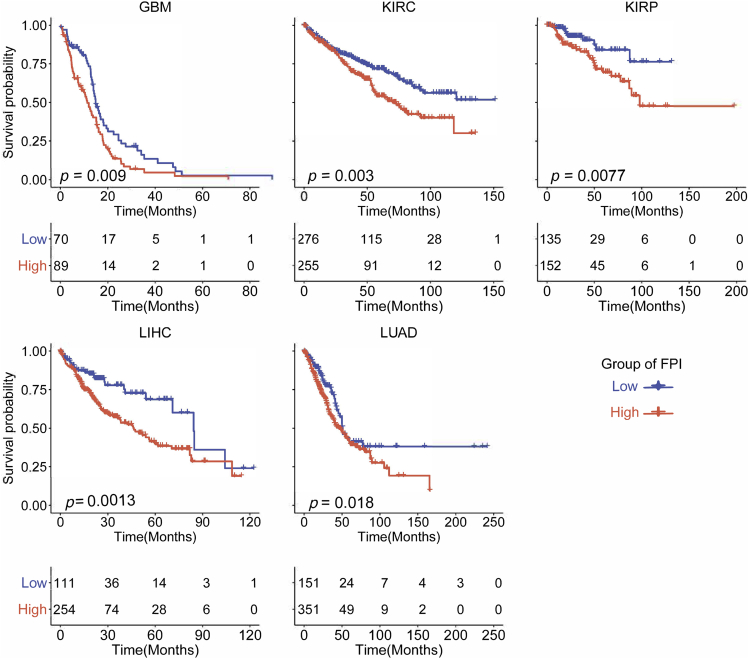
Since ferroptosis is involved in cellular metabolism and the immune microenvironment, it was proposed that ferroptosis and its regulators should be correlated with cancer survival and other clinical characteristics. Based on the clinical data from TCGA, survival analysis was performed and the results were consistent since it was found that the lower FPI predicted a better survival in five cancers including GBM, KIRC, KIRP, LIHC, and LUAD (Figure 4). We further dissected the factors that might impact FPI, including tumor stage, cigarette smoking, age, and sex. As shown in Figure S3G, tumor stages were positively correlated with FPI in LIHC, KIRC, KIRP, and COAD. Only the FPI in LUAD tumors showed a positive correlation with cigarettes smoked per day. Age was positively associated with FPI in GBM and PRAD but showed a negative correlation in seven cancers such as BRCA and LIHC. It was observed that FPI was markedly different between sexes (Figure S3H), FPI for female tumor samples was higher than for male tumor samples in KIRC, LIHC, LUAD, and LUSC but lower in STAD. Significant differences in FPI were also observed among different races in BLCA, BRCA, and ESCA. Furthermore, we found that the FPI was correlated with cancer metastasis, recurrence, and primary and follow-up outcome in several cancers. In addition, we evaluated the association between FPI and other clinical characteristics including alcohol abuse, fatty liver, hemochromatosis, and viral infection (HPV, HBC, HCV, and EBV), but no significant results were observed. These results indicate that ferroptosis might play critical roles in cancer survival and that clinical factors have tumor-specific effects on FPI.
Figure 4.
Kaplan-Meier Analysis of Overall Survival according to the FPI among Cancers
To further dissect the clinical relevance of ferroptosis in cancer, the roles of FRGs in cancer survival were analyzed. The survival analysis showed that all FRGs were associated with overall survival in at least six cancer types (Figure S3I), but the correlations were mixed since most FRGs can serve as a protective or a risk factor in different cancers. For example, patients with higher expression of FANCD2 showed better survival in kidney cancer, LIHC, and LUAD but worse survival in READ, STAD, and BLCA (Figure S3I). However, high expression of CARS and MT1G showed consistently better survival among cancers including KICH, UCEC, PAAD, and HNSC (Figure S3I). Thus, the functional roles of FRGs in cancer survival should be further explored.
To study the potential clinical implications of ferroptosis, we examined the association between the gene expression of FRGs and 156 clinically actionable genes (CAGs), including 136 genes for targeted therapy and 20 genes for immunotherapy across cancer types (Mak et al., 2016; Van Allen et al., 2014). As an FRG, CDKN1A regulates iron uptake and GPX4 abundance (Stockwell et al., 2017) and is also a CAG targeted by anticancer drugs, such as the inorganic compound arsenic trioxide (Hassani et al., 2018). We found that all 136 CAGs and FRGs had significant co-expression relationships that were significantly different across cancers as shown in Figure S4A. The number of CAGs correlated with FRGs ranged from 45 in LUSC to 132 in THCA (Figure S4B). The FRG-CAG correlation pairs ranged from 69 in LUSC to 581 in THCA (Figure S4C). For example, GPX4 was co-expressed with MAP2K2 in 20 cancer types and showed a significantly positive correlation with TNFRSF4, which are targeted genes for immunotherapy. ACSL4 is significantly correlated with 17 of 20 genes targeted for immunotherapy, suggesting that ACSL4 has a potential effect on cancer immunotherapy (Figure S4D). Furthermore, besides co-expression, the protein-protein interactions among drug-targeted CAGs and FRGs were analyzed, and the results presented in Figure S4E showed that the interactions were obvious. For example, EGFR, the target of lapatinib, gefitinib, and afatinib, was positively associated with FRGs, including DPP4 (R = 0.15) and HSPA5 (R = 0.31), and negatively correlated with FDFT1 (R = −0.24), whereas there were protein-protein interactions between EGFR and DPP4, HSPA5, and FDFT1 (Figure S4E). These results indicated that ferroptosis might be involved in drug effects by interacting with targeted clinically actionable genes. Taken together, our studies suggest that clinically actionable genes are closely related to FRGs, which highlights the significance of ferroptosis in cancer treatment, including both immunotherapy and targeted therapy.
To further understand the correlation between ferroptosis and drug sensitivity, the area under the percent viability curve (AUC) approach was employed to evaluate drug sensitivity (Basu et al., 2013) and calculate the correlation between FPI and drug sensitivity across cancer cell lines. Drugs associated with FPI were also tested for their correlation with FRG expression. We identified that FPI was significantly associated with sensitivity to 64 drugs, including 12 that were negatively correlated and 52 that were positively correlated (Figure S4F). To explore the effects of each FRG on drug sensitivity, we also analyzed the associations between the drug sensitivity of 64 cancer drugs and the expression of FRGs and found 521 significantly correlated pairs (Figure S4F). Among them, the expression of FANCD2 was correlated with the AUC of 57 drugs, whereas no significant relation was observed for CISD1 (Figure S4F). Furthermore, the associations classified the FRGs into two groups: One group included genes such as FANCD2 and ATP5G3, which were positively associated with the AUC of docetaxel, trametinib, etc. (Figure S4F). The other group included SLC7A11, SAT1, and HSPA5, which were negatively associated with the AUC values (Figure S4F). Taken together, these results suggest that ferroptosis is correlated with the sensitivity of multiple drugs.
Discussion
Ferroptosis is driven by damage of cell membranes caused by the loss of activity of GPX4 (Feng and Stockwell, 2018). Researchers identified that ferroptosis is closely related to tumorigenesis and plays an important role in cancer treatment (Shen et al., 2018; Yang et al., 2014). However, there is a lack of systematic studies on ferroptosis and its regulator genes across cancer types. In this study, we employed multi-omics data and clinical data across 20 cancer types from TCGA and revealed the global alterations of ferroptosis regulator genes at genetic, epigenetic, and transcriptional levels. We also used ssGSEA to process expression data to establish FPI to characterize ferroptosis and addressed which genetic and nongenetic factors (including drug and patient phenotype) were related with FPI. Different molecular types affected the ferroptosis in breast cancer and gastrointestinal cancer, which meant that the responses of different molecular subtypes to treatment may be related to ferroptosis. On the other hand, molecular subtypes need to be considered when ferroptosis is applied as a therapeutic strategy.
The mechanism by which ferroptosis regulates tumor cell growth and proliferation is still unclear, but the relationship we observed between FPI and hallmarks of cancer could improve the understanding the role of ferroptosis. The results of GSEA demonstrate that the level of ferroptosis is closely related to tumor-associated hallmarks in most cancers. Ferroptosis genes can play both oncogene and tumor suppressor roles in cancer (Kang et al., 2019), and the FPI acts as a protective or risk factor across cancer types. We also found that several common clinical factors affected ferroptosis, such as cigarette smoking, BMI, and tumor stage. As we know, cigarette smoking is a risk factor for esophageal cancer, lung cancer, and kidney cancer (Chow et al., 2010; Mao et al., 2011; Pesch et al., 2012). In our results, the number of cigarettes per day was positively correlated with the ferroptosis potential index in LUAD, which may be because the cigarette-induced oxidation reaction promoted lipid peroxidation (Barreiro et al., 2010; Guan et al., 2013; Louhelainen et al., 2008). FPI also varied between the sexes in several cancers, including LIHC, LUAD, LUSC, and STAD, and between different races in BRCA, BLCA, and ESCA, which implied that gender and race need to be considered when using ferroptosis as a treatment strategy. We also noticed that better clinical outcomes or status in several cancer types also have lower FPI, which further confirmed the dual role of ferroptosis. Thus, a different strategy of regulating the ferroptosis of tumor cells may benefit patients and improve prognosis.
Furthermore, we showed that FRGs were co-expressed with most clinically actionable genes and interacted with genes modulated by drugs, suggesting that studying ferroptosis may improve the strategy for cancer therapy. We observed that the co-expression of FRGs and CAGs could be roughly divided into two groups and drugs that target clinical genes could have complex regulatory effects, which indicated that clinically actionable gene MAP2K2 may play an important role in ferroptosis (Yang et al., 2014). To further explore drug sensitivity and ferroptosis, we observed that the FPI could characterize the sensitivity of many drugs, the AUC of many drugs was inversely associated with the FPI in cancer cell lines, which implied that regulating the ferroptosis of tumor cells may improve the therapeutic effect of cancer treatments. However, opposite effects were observed regarding the effects of drugs on ferroptosis. Thus, further detailed studies should be carried out to determine the functions and mechanisms of ferroptosis in different cancers. In the present study, we presented a systematical analysis of ferroptosis and its regulator genes across cancers. Most FRGs were aberrantly expressed in tumors among various cancer types, while frequent CNA and differential DNA methylation contributed greatly. We established the FPI to evaluate the ferroptosis level and found that the FPIs were higher in tumors than in the adjacent normal tissues in most cancers and were associated with clinical features, and cancer metastasis, recurrence, outcome, and drug sensitivity. Several well-known anti-cancer drugs were also reported to induce ferroptosis, such as sorafenib, which was used to treat renal cell carcinoma and hepatocellular carcinoma (Louandre et al., 2015; Yang et al., 2014). In addition, small molecule inducers based on the mechanism of system Xc- inhibition or GPX4 inhibition were considered as a therapeutic strategy for cancer (Yang et al., 2014). Thus, our findings highlighted the potential of ferroptosis-based cancer therapy.
Limitations of the Study
Since current omics data only provide RNA-level quantifications for FRGs, whereas the ferroptosis process relies on proteins, although we tried to infer the ferroptosis status precisely, there could be a variety of inaccuracies. Furthermore, the detailed molecular mechanisms for ferroptosis are still unclear, and currently, the identified FRGs and the genes used for modeling ferroptosis potential have various other functions; thus, the sensitivity and specificity of FPI might be limited. In addition, although the transcriptome datasets of cell lines treated with erastin, withaferin A, or ferrostatin indicated that the FPI was accurate, since the transcriptome datasets with well-characterized ferroptosis status were rare, the bona fide accuracy of the FPI still needs further evaluation. Thus, our study based on current knowledge might need further updates and improvements due to new discoveries.
Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Ze-Xian Liu (liuzx@sysucc.org.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
GSVA (version: 1.30) is available at https://github.com/rcastelo/GSVA.
The TCGA cohort data: The mRNA expression data, copy number alteration thresholded data, masked copy number segmentation data, and DNA methylation 450K data were download from Firehose (http://gdac.broadinstitute.org). Mutation data, miRNA-seq data, and clinical data were downloaded from the Xena Browser (https://xenabrowser.net/datapages/). Tumor suppressor gene lists and oncogene lists were retrieved from Bailey et al. (2018). The information of microsatellite instability of TCGA tumor samples were retrieved from Liu, Y et al. (Liu et al., 2018).
Immune associated data: Immune cell and immunophenotype data were requested from The Cancer Immunome Atlas (https://tcia.at/home) (Charoentong et al., 2017). Dysfunction and exclusion scores were retrieved from TIDE (http://tide.dfci.harvard.edu/) (Fu et al., 2020).
Protein-protein interaction data was download from the Human Protein Reference Database (http://www.hprd.org/) and BioGRID (https://thebiogrid.org).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by grants from National Key R&D Program of China (2018YFC1313300 to RH.X.), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096 to ZX.L.), Tip-top Scientific and Technical Innovative Youth Talents of Guangdong Special Support Program (2019TQ05Y351 to ZX.L.), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-036 to RH.X.), Natural Science Foundation of Guangdong Province (2017A030313485 to RH.X., 2019A1515010634 to ZX.L.), Fundamental Research Funds for the Central Universities (SYSU: 19ykpy184 to Q.Z.), Natural Science Foundation of China (81802438 to Q.Z.), and Science and Technology Program of Guangdong (2019B020227002 to RH.X.).
Author Contributions
ZX.L. and RH.X. designed and supervised the experiments. Z.L., Q.Z., ZX.Z., and SQ.Y. performed the data analysis with contributions from K.Y., Q.Z., X.Z., H.S., HQ.J., H.C., and F.W. ZX.L. and Z.L. wrote the manuscript with contributions of all authors. All authors reviewed the manuscript.
Declaration of Interests
The authors declare that they have no competing interests.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101302.
Contributor Information
Rui-Hua Xu, Email: xurh@sysucc.org.cn.
Ze-Xian Liu, Email: liuzx@sysucc.org.cn.
Supplemental Information
The gene names of the FRGs are marked in red.
References
- Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro E., Peinado V.I., Galdiz J.B., Ferrer E., Marin-Corral J., Sanchez F., Gea J., Barbera J.A., Project E.i.C. Cigarette smoke-induced oxidative stress: a role in chronic obstructive pulmonary disease skeletal muscle dysfunction. Am. J. Respir. Crit. Care Med. 2010;182:477–488. doi: 10.1164/rccm.200908-1220OC. [DOI] [PubMed] [Google Scholar]
- Basu A., Bodycombe N.E., Cheah J.H., Price E.V., Liu K., Schaefer G.I., Ebright R.Y., Stewart M.L., Ito D., Wang S. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canisius S., Martens J.W., Wessels L.F. A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol. 2016;17:261. doi: 10.1186/s13059-016-1114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Chow W.H., Dong L.M., Devesa S.S. Epidemiology and risk factors for kidney cancer. Nat. Rev. Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.J., Winter G.E., Musavi L.S., Lee E.D., Snijder B., Rebsamen M., Superti-Furga G., Stockwell B.R. Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem. Biol. 2015;10:1604–1609. doi: 10.1021/acschembio.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eling N., Reuter L., Hazin J., Hamacher-Brady A., Brady N.R. Identification of artesunate as a specific activator of ferroptosis in pancreatic cancer cells. Oncoscience. 2015;2:517–532. doi: 10.18632/oncoscience.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Stockwell B.R. Unsolved mysteries: how does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16:e2006203. doi: 10.1371/journal.pbio.2006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Li K., Zhang W., Wan C., Zhang J., Jiang P., Liu X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12:21. doi: 10.1186/s13073-020-0721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S.P., Tee W., Ng D.S., Chan T.K., Peh H.Y., Ho W.E., Cheng C., Mak J.C., Wong W.S. Andrographolide protects against cigarette smoke-induced oxidative lung injury via augmentation of Nrf2 activity. Br. J. Pharmacol. 2013;168:1707–1718. doi: 10.1111/bph.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani S., Khaleghian A., Ahmadian S., Alizadeh S., Alimoghaddam K., Ghavamzadeh A., Ghaffari S.H. Redistribution of cell cycle by arsenic trioxide is associated with demethylation and expression changes of cell cycle related genes in acute promyelocytic leukemia cell line (NB4) Ann. Hematol. 2018;97:83–93. doi: 10.1007/s00277-017-3163-y. [DOI] [PubMed] [Google Scholar]
- Hassannia B., Wiernicki B., Ingold I., Qu F., Van Herck S., Tyurina Y.Y., Bayir H., Abhari B.A., Angeli J.P.F., Choi S.M. Nano-targeted induction of dual ferroptotic mechanisms eradicates high-risk neuroblastoma. J. Clin. Invest. 2018;128:3341–3355. doi: 10.1172/JCI99032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inder T., Mocatta T., Darlow B., Spencer C., Volpe J.J., Winterbourn C. Elevated free radical products in the cerebrospinal fluid of VLBW infants with cerebral white matter injury. Pediatr. Res. 2002;52:213–218. doi: 10.1203/00006450-200208000-00013. [DOI] [PubMed] [Google Scholar]
- Kang R., Kroemer G., Tang D. The tumor suppressor protein p53 and the ferroptosis network. Free Radic. Biol. Med. 2019;133:162–168. doi: 10.1016/j.freeradbiomed.2018.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner N.M., Voicu H., Finegold M.J., Coarfa C., Sreekumar A., Putluri N., Katchy C.A., Lee C., Moore D.D., Fu L. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell. 2016;30:909–924. doi: 10.1016/j.ccell.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkermann A., Skouta R., Himmerkus N., Mulay S.R., Dewitz C., De Zen F., Prokai A., Zuchtriegel G., Krombach F., Welz P.S. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U S A. 2014;111:16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sethi N.S., Hinoue T., Schneider B.G., Cherniack A.D., Sanchez-Vega F., Seoane J.A., Farshidfar F., Bowlby R., Islam M. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33:721–735.e8. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louandre C., Ezzoukhry Z., Godin C., Barbare J.C., Maziere J.C., Chauffert B., Galmiche A. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int. J. Cancer. 2013;133:1732–1742. doi: 10.1002/ijc.28159. [DOI] [PubMed] [Google Scholar]
- Louandre C., Marcq I., Bouhlal H., Lachaier E., Godin C., Saidak Z., Francois C., Chatelain D., Debuysscher V., Barbare J.C. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015;356:971–977. doi: 10.1016/j.canlet.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Louhelainen N., Myllarniemi M., Rahman I., Kinnula V.L. Airway biomarkers of the oxidant burden in asthma and chronic obstructive pulmonary disease: current and future perspectives. Int. J. Chron. Obstruct. Pulmon. Dis. 2008;3:585–603. doi: 10.2147/copd.s3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Chen X.B., Ying M.D., He Q.J., Cao J., Yang B. The role of ferroptosis in cancer development and treatment response. Front. Pharmacol. 2017;8:992. doi: 10.3389/fphar.2017.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane L.A., Murphy P.R. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak M.P., Tong P., Diao L., Cardnell R.J., Gibbons D.L., William W.N., Skoulidis F., Parra E.R., Rodriguez-Canales J., Wistuba I.I. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin. Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W.M., Zheng W.H., Ling Z.Q. Epidemiologic risk factors for esophageal cancer development. Asian Pac. J. Cancer Prev. 2011;12:2461–2466. [PubMed] [Google Scholar]
- Masur K., Schwartz F., Entschladen F., Niggemann B., Zaenker K.S. DPPIV inhibitors extend GLP-2 mediated tumour promoting effects on intestinal cancer cells. Regul. Pept. 2006;137:147–155. doi: 10.1016/j.regpep.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Matsushita M., Freigang S., Schneider C., Conrad M., Bornkamm G.W., Kopf M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J. Exp. Med. 2015;212:555–568. doi: 10.1084/jem.20140857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch B., Kendzia B., Gustavsson P., Jockel K.H., Johnen G., Pohlabeln H., Olsson A., Ahrens W., Gross I.M., Bruske I. Cigarette smoking and lung cancer--relative risk estimates for the major histological types from a pooled analysis of case-control studies. Int. J. Cancer. 2012;131:1210–1219. doi: 10.1002/ijc.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pro B., Dang N.H. CD26/dipeptidyl peptidase IV and its role in cancer. Histol. Histopathol. 2004;19:1345–1351. doi: 10.14670/HH-19.1345. [DOI] [PubMed] [Google Scholar]
- Shen H., Laird P.W. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Song J., Yung B.C., Zhou Z., Wu A., Chen X. Emerging strategies of cancer therapy based on ferroptosis. Adv. Mater. 2018;30:e1704007. doi: 10.1002/adma.201704007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouta R., Dixon S.J., Wang J., Dunn D.E., Orman M., Shimada K., Rosenberg P.A., Lo D.C., Weinberg J.M., Linkermann A. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J. Am. Chem. Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Niu X., Chen R., He W., Chen D., Kang R., Tang D. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64:488–500. doi: 10.1002/hep.28574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi J., Robert L., Paraiso K., Galvan C., Sheu K.M., Lay J., Wong D.J.L., Atefi M., Shirazi R., Wang X. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell. 2018;33:890–904.e5. doi: 10.1016/j.ccell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen E.M., Wagle N., Stojanov P., Perrin D.L., Cibulskis K., Marlow S., Jane-Valbuena J., Friedrich D.C., Kryukov G., Carter S.L. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley U.V., McGroarty M., Homoyouni A. Dipeptidyl peptidase inhibits malignant phenotype of prostate cancer cells by blocking basic fibroblast growth factor signaling pathway. Cancer Res. 2005;65:1325–1334. doi: 10.1158/0008-5472.CAN-04-1852. [DOI] [PubMed] [Google Scholar]
- Xie Y., Zhu S., Song X., Sun X., Fan Y., Liu J., Zhong M., Yuan H., Zhang L., Billiar T.R. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Guo P., Xie X., Wang Y., Chen G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J. Cell. Mol. Med. 2017;21:648–657. doi: 10.1111/jcmm.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Du L., Qiao Y., Zhang X., Zheng W., Wu Q., Chen Y., Zhu G., Liu Y., Bian Z. Ferroptosis is governed by differential regulation of transcription in liver cancer. Redox Biol. 2019;24:101211. doi: 10.1016/j.redox.2019.101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The gene names of the FRGs are marked in red.
Data Availability Statement
GSVA (version: 1.30) is available at https://github.com/rcastelo/GSVA.
The TCGA cohort data: The mRNA expression data, copy number alteration thresholded data, masked copy number segmentation data, and DNA methylation 450K data were download from Firehose (http://gdac.broadinstitute.org). Mutation data, miRNA-seq data, and clinical data were downloaded from the Xena Browser (https://xenabrowser.net/datapages/). Tumor suppressor gene lists and oncogene lists were retrieved from Bailey et al. (2018). The information of microsatellite instability of TCGA tumor samples were retrieved from Liu, Y et al. (Liu et al., 2018).
Immune associated data: Immune cell and immunophenotype data were requested from The Cancer Immunome Atlas (https://tcia.at/home) (Charoentong et al., 2017). Dysfunction and exclusion scores were retrieved from TIDE (http://tide.dfci.harvard.edu/) (Fu et al., 2020).
Protein-protein interaction data was download from the Human Protein Reference Database (http://www.hprd.org/) and BioGRID (https://thebiogrid.org).