Significance
Mutations in UBQLN2 cause amyotrophic lateral sclerosis with frontotemporal dementia (ALS/FTD). UBQLN2 regulates proteostasis by clearing misfolded proteins from cells through the proteasome and autophagy degradation pathways. Here, we report on defects in autophagy that results from knockout or expression of WT and ALS/FTD mutant UBQLN2 proteins in cells and mice. We show that loss of UBQLN2 reduces expression of ATP6v1g1, a critical subunit of the ATPase pump that regulates vacuolar acidification and is required for the maturation of autophagosomes. We show that WT but not ALS/FTD mutant UBQLN2 proteins can rescue the acidification defect. Furthermore, WT but not ALS/FTD mutant UBQLN2 proteins bind and stimulate ATP6v1g1 biogenesis, suggesting an important role played by UBQLN2 in V-ATPase regulation.
Keywords: amyotrophic lateral sclerosis, ubiquilin, UBQLN2, autophagy, vacuolar ATPase pump
Abstract
Mutations in UBQLN2 cause amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and other neurodegenerations. However, the mechanism by which the UBQLN2 mutations cause disease remains unclear. Alterations in proteins involved in autophagy are prominent in neuronal tissue of human ALS UBQLN2 patients and in a transgenic P497S UBQLN2 mouse model of ALS/FTD, suggesting a pathogenic link. Here, we show UBQLN2 functions in autophagy and that ALS/FTD mutant proteins compromise this function. Inactivation of UBQLN2 expression in HeLa cells reduced autophagic flux and autophagosome acidification. The defect in acidification was rescued by reexpression of wild type (WT) UBQLN2 but not by any of the five different UBQLN2 ALS/FTD mutants tested. Proteomic analysis and immunoblot studies revealed P497S mutant mice and UBQLN2 knockout HeLa and NSC34 cells have reduced expression of ATP6v1g1, a critical subunit of the vacuolar ATPase (V-ATPase) pump. Knockout of UBQLN2 expression in HeLa cells decreased turnover of ATP6v1g1, while overexpression of WT UBQLN2 increased biogenesis of ATP6v1g1 compared with P497S mutant UBQLN2 protein. In vitro interaction studies showed that ATP6v1g1 binds more strongly to WT UBQLN2 than to ALS/FTD mutant UBQLN2 proteins. Intriguingly, overexpression of ATP6v1g1 in UBQLN2 knockout HeLa cells increased autophagosome acidification, suggesting a therapeutic approach to overcome the acidification defect. Taken together, our findings suggest that UBQLN2 mutations drive pathogenesis through a dominant-negative loss-of-function mechanism in autophagy and that UBQLN2 functions as an important regulator of the expression and stability of ATP6v1g1. These findings may have important implications for devising therapies to treat UBQLN2-linked ALS/FTD.
Mutations in UBQLN2 cause X-linked dominant inheritance of amyotrophic lateral sclerosis (ALS) with frontotemporal dementia (FTD) (1, 2). These mutations affect proteasomal degradation, but it is not clear if they also affect the autophagy–lysosome pathway. UBQLN2 encodes a protein that functions in protein quality control (3). Interestingly, mutations in genes involved in protein quality control are linked to ALS more than any other functional category, strongly suggesting proteostasis disruption may be a key driver of pathogenesis (4, 5). Mutations in UBQLN4 are also linked to ALS (6). However, the UBQLN family of proteins regulate multiple aspects of proteostasis and discovering which of these functions is disrupted is key for therapeutic intervention.
UBQLN2 is one of four homologous UBQLN proteins expressed in humans. Of the four isoforms, UBQLN3 is only expressed in the testis, while the remaining isoforms are differentially expressed throughout the body (7–11). The proteins are all about 600-aa long and contain highly homologous ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains at their N and C termini, respectively. The two domains border a longer, more variable central domain, containing multiple heat-shock protein (HSP)-like STI binding sites (12, 13). The UBA domain functions to bind ubiquitin moieties that are typically conjugated onto misfolded proteins, whereas the UBL domain binds to the S5a subunit in the proteasome cap (14–18). Fittingly, UBQLN proteins function as shuttle factors, facilitating the delivery of misfolded proteins to the proteasome for degradation. Besides acting in delivery, the proteins also function as chaperones, aiding in protein folding, an activity that has been linked to HSP binding to their STI motifs (12, 13, 19).
UBQLNs have also been shown to function in autophagy. The proteins bind and colocalize with LC3 proteins in autophagosomes (20, 21). Moreover, knockdown of human UBQLN proteins, particularly UBQLN1 and 4, leads to a reduction in autophagosome formation (21, 22). Similarly, knockout of the sole ubqln gene in Drosophila leads to severe defects in autophagy (23, 24). However, information on the role of UBQLN2 in autophagy is limited.
Autopsy examination of human UBQLN2-linked ALS/FTD cases show unusual accumulation of UBQLN2 inclusions in the dentate gyrus of the hippocampus, which resemble those seen in C9ORF72-linked cases (25), but which have not been seen in other forms of ALS/FTD or other neurodegenerative disorders (1, 26, 27). The inclusions stained positive for UBQLN2, ubiquitin, and p62 (SQSTM1) but were generally negative for TAR-DNA binding protein 43 (TDP-43) (1). Staining of the spinal cord (SC) revealed large skein-like inclusions that were positive for UBQLN2, ubiquitin, p62, and TDP-43 (1, 28). The accumulation of ubiquitinated protein aggregates is frequently found in most neurodegenerative diseases, suggesting proteostasis dysregulation may be common in the etiology of these diseases (4, 29). Similarly, accumulation of p62 is also indicative of a disruption in proteostasis (30). p62 binds ubiquitinated proteins, packaging them in autophagosomes that then fuse with lysosomes, to activate degradation. p62 binds LC3, an autophagosome-specific protein (31). LC3 protein is synthesized as a precursor called LC3I, which is processed and lipidated to produce LC3II, which is the form tethered to autophagosomes (32, 33). Disturbances in autophagy can therefore be monitored by measuring changes in the levels and/or flux of LC3II proteins (34).
Studies of WT and mutant UBQLN2 proteins in cells and organisms support the idea that the ALS/FTD mutations disrupt both proteasomal degradation and autophagy. The interference in proteasomal degradation appears nondiscriminatory, stalling degradation of both membrane and cytosolic proteins (1, 16, 35, 36). Various mechanisms have been proposed by which UBQLN2 mutations disrupt proteostasis, including an inability to bind factors in the endoplasmic reticulum in order to facilitate endoplasmic reticulum-associated membrane protein degradation (36) and imprecise docking with the proteasome for delivery of their ubiquitinated cargo (1, 16, 35).
Evidence that the UBQLN2 mutants affect autophagy is more circumstantial. The best evidence comes from prominent staining of p62 in UBQLN2 inclusions that are seen in human cases and in different UBQLN2 rodent models of the disease (16, 35, 37). However, it is unknown if the increase reflects an attempt to clear protein aggregates that accumulate because of disruption in proteasomal degradation or because of inefficiencies in autophagy.
Here we show the important requirement of UBQLN2 proteins in autophagy and demonstrate that ALS/FTD mutant UBQLN2 proteins cause a potent defect in autophagosome acidification. We provide evidence that strongly suggests the acidification defect arises from a loss of UBQLN2 function, suggesting the ALS/FTD mutations are antimorphic mutations. Our findings share similarities with a recent report showing that UBQLN proteins regulate vacuolar ATPase (V-ATPase) function (23). However, they differ in the mode in which ALS/FTD mutant UBQLN2 proteins dysregulate V-ATPase function.
Results
Increased Accumulation of p62 and Ubiquitinated Proteins in the P497S UBQLN2 Mouse Model of ALS/FTD.
We previously described transgenic (Tg) mouse lines with neuron-specific expression of human UBQLN2 cDNAs encoding either untagged full-length WT UBQLN2 protein or carrying the ALS/FTD P497S or P506T UBQLN2 mutations, which recapitulated central features of the human disease (38). Mouse lines with equivalent expression of each transgenic protein were identified and called WT356, P497S, and P506T lines. Both lines expressing mutant UBQLN2, but not the WT protein, developed age-dependent motor neuron (MN) disease. Behavioral studies also indicated that the mutant lines developed cognitive deficits, which were milder in the WT356 line. Pathological studies revealed an age-dependent accumulation of ubiquitin-positive UBQLN2 inclusions in the brain and SC only in the mutant lines (38).
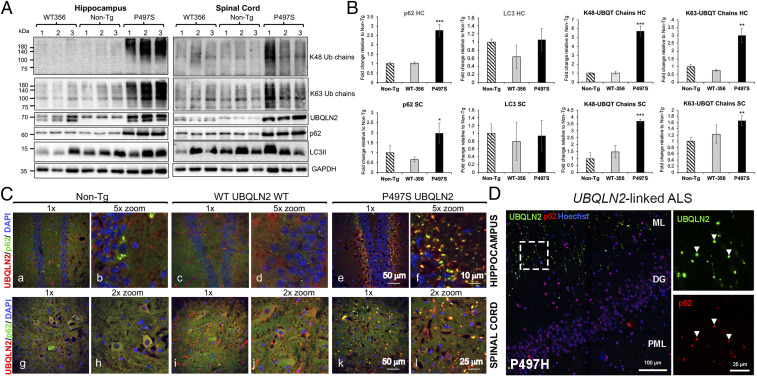
To examine whether ALS/FTD mutations in UBQLN2 affect autophagy, we probed lysates made from the hippocampus and lumbar SC of 8-mo-old P497S, WT356, and nontransgenic (non-Tg) animals for alterations in p62, LC3, and different ubiquitin chains. The P506T line was not used because of difficulty in breeding the line. We used animals between 2 and 12 mo of age in probing and subsequent examinations because P497S Tg animals display robust changes in pathology during this period (38). To ensure reproducibility of our findings, we measured changes in three independent mice (or cell lines for the culture experiments) on the same immunoblot. The immunoblots revealed a massive (approximately threefold) increase in p62 levels, but no significant difference in LC3II, in both the SC and hippocampal samples in P497S Tg animals, compared to WT356 and non-Tg animals (Fig. 1 A and B). Because P497S mice display a build-up of ubiquitinated UBQLN2 inclusions (38), we also probed the lysates for changes in K48- or K63-ubiquitin-chain linkages. The blots revealed variable (1.5- to 5.5-fold) but significant increases in both types of linkages in the hippocampus and SC of P497S animals compared to non-Tg and WT animals, whose chain-linkage levels were similar (Fig. 1 A and B). Taken together, these results strongly suggest that neuronal expression of human UBQLN2 carrying the P497S mutation, but not the WT protein, induces disturbances in proteostasis through possible interference of either the proteasome or autophagy pathways. Our subsequent studies focused on how ALS mutations in UBQLN2 disturb autophagy.
Fig. 1.
UBQLN2 mutations induce pathologic disturbances in autophagy in humans and the P497S mouse model of ALS/FTD. (A) Immunoblots of hippocampus and SC lysates of three independent mice for the different mouse lines probed with the antibodies shown. (B) Quantification of the immunoreactivity for the proteins shown in A. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Confocal microscopy (1× and corresponding 2 or 5× zoomed) images of the merged UBQLN2, p62, and DAPI staining of the dentate gyrus (DG) of the hippocampus (a–f) and ventral horn of the SC (g–l) for the three mouse genotypes at 52 wk of age. (D) Confocal images of the molecular layer (ML), DG, and polymorphic layer (PML) brain region (large image) of a human P497H UBQLN2 patient. Zoomed images demonstrate colocalization between UBQLN2 inclusions (green) and p62 (red) in the molecular layer. Bar sizes shown in this and all subsequent figures.
The Autophagic Marker p62, but Not LC3 or LAMP1, Colocalizes with UBQLN2 Inclusions in P497S Animals.
To determine whether P497S animals display anomalies in localization of autophagy markers, we stained mouse brain and SC sections of all three genotypes for UBQLN2 and different autophagy markers. We focused on p62, LC3, and LAMP1 because of their central function in autophagy. The staining revealed P497S Tg animals contain numerous UBQLN2 inclusions in the brain and SC at 2, 6, 8, and 12 mo of age (Fig. 1C and SI Appendix, Figs. S1 and S2), which were generally larger in size in the older mice. Such inclusions were rare in the non-Tg and WT UBQLN2 Tg mice, although some displayed staining of long structures, which we attributed to artifacts from antibody cross-reaction with blood vessels in nonperfused animals. The staining of UBQLN2 inclusions in P497S animals were seen irrespective of whether animals were perfused or not.
Double-immunofluorescence staining of the sections revealed extensive colocalization of UBQLN2 and p62 in P497S animals. For example, the majority of the UBQLN2 inclusions that decorated the molecular layer of the dentate gyrus also stained positive for p62, although the degree of overlap was quite variable (Fig. 1C and SI Appendix, Fig. S1). The degree of colocalization of the proteins was greater in animals at 12 mo of age than at earlier ages (SI Appendix, Fig. S1B). Similar findings were found upon examination of the CA1 and cortex regions of the brain (SI Appendix, Fig. S1B). In contrast, p62 staining in non-Tg and WT UBQLN2 Tg animals were generally more diffuse, although rare examples of cells with small puncta were found, which we attributed to anomalies in autophagy that typically occur in brain cells.
By comparison to p62, LC3 staining in the brain was more similar across all three genotypes. Its staining was generally diffuse, but closer examination revealed numerous small puncta that were especially noticeable in the granule cell layer of the dentate gyrus (SI Appendix, Fig. S3A). The LC3+ inclusions in the granule cell layer in the 12-mo-old P497S animals were larger in size compared with age-matched non-Tg and WT UBQLN2 Tg animals. However, there was little evidence of colocalization of LC3 staining with UBQLN2 inclusions in P497S animals. Furthermore, double staining of a P497S animal brain section known to contain UBQLN2 inclusions revealed no colocalization of LC3 and p62 staining (using two different antibodies) (SI Appendix, Fig. S4).
Staining of the 12-mo mouse brain sections for LAMP1 indicated strong colocalization of LAMP1 with p62 and UBQLN2 in small puncta in both non-Tg and WT UBQLN2 Tg animals, but not with the large p62 and UBQLN2 puncta found in the P497S UBQLN2 animal (SI Appendix, Fig. S5). Instead, most of the LAMP1+ puncta in the P497S animal were devoid of either p62 or UBQLN2 staining.
Similar findings were made upon examination of stained SC sections for the three genotypes. With rare exceptions, almost all of the UBQLN2 inclusions in the ventral horn of P497S were positive for p62 (Fig. 1C and SI Appendix, Fig. S2 A and B). Close inspection of UBQLN2 inclusions within MNs of P497S animals indicated they were also positive for p62 (SI Appendix, Fig. S2C). As in the brain, we found little sign of colocalization of LC3 with the UBQLN2 inclusions in the SC of P497S animals (SI Appendix, Fig. S3B).
To determine whether the alteration in p62 staining seen in the P497S UBQLN2 mouse line also manifests in humans, we stained three human ALS/FTD UBQLN2 cases carrying different UBQLN2 mutations and found p62 was indeed colocalized with UBQLN2 inclusions in all three cases (Fig. 1D and SI Appendix, Fig. S6A). The number of p62 and UBQLN2 inclusions in the three cases varied considerably, which may reflect different sources and thus processing of the tissues, or intrinsic differences between the UBQLN2 mutations. Further examination of LAMP1 staining in the human P497H case revealed decoration of puncta that were distinct from the UBQLN2+ and p62+ inclusions, which themselves were negative for the protein (SI Appendix, Fig. S6B). The disruption of normal colocalization of these key autophagic proteins in both mutant UBQLN2 mice and human cases strongly suggests UBQLN2 mutations cause defects in autophagy.
Derivation of Cell Lines with Genetic Inactivation of UBQLN2 Expression.
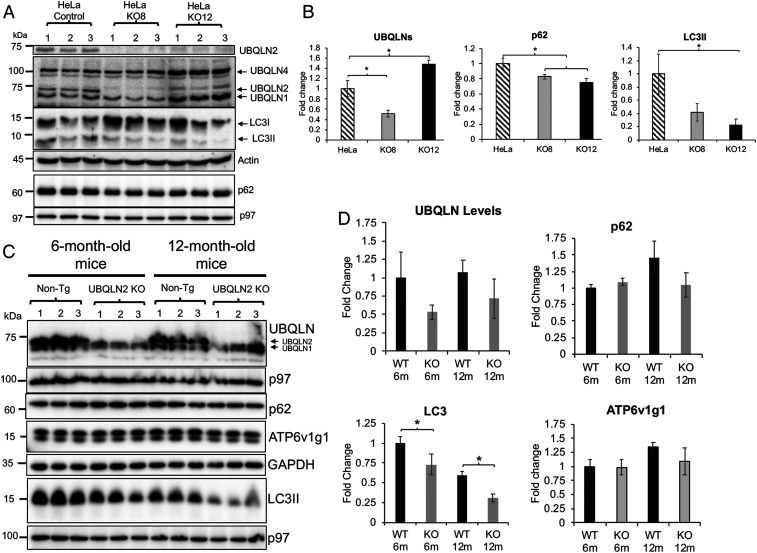
To determine if UBQLN2 is required for autophagy, we utilized CRISPR/Cas9 technology to genetically inactivate UBQLN2 protein expression in both human HeLa and mouse NSC34 MN cells. Two independent clones for each cell type were identified by immunoblot detection to be completely devoid of UBQLN2 expression: HeLa knockout (KO)8 and KO12 lines, and NSC34 cells KO20 and KO69 cell lines (Fig. 2A and SI Appendix, Fig. S7A).
Fig. 2.
Alteration of autophagic proteins in UBQLN2 KO HeLa cells and mouse lines. (A) Immunoblots of three independent lysates from the parental HeLa lines and CRISPR/cas9 UBQLN2 KO8 and KO12 lines for the proteins shown. Detection of the different UBQLN isoforms (second panel) with an antibody that detects all UBQLNs. (B) Quantification of total UBQLN protein expression in the parental and KO HeLa lines. *P < 0.05. (C) Immunoblots of total brain lysates (three independent animals) from non-Tg and UBQLN2 KO animals of 6 and 12 mo of age probed for the proteins shown. (D) Quantification of the changes in the different proteins in the KO relative to the 6-mo-old non-Tg (WT) animals. *P < 0.05.
To understand the consequence of loss of UBQLN2 expression in the KO cells, we first quantified the amount of UBQLN2 protein that was expressed in the parental cells relative to the other UBQLN isoforms. The amount of each UBQLN isoform was calculated by relating the intensity of their respective bands to known amounts of GST-WT UBQLN2 fusion protein that was used as a standard (SI Appendix, Fig. S7 C and D). The measurements revealed that UBQLN2 was the predominant UBQLN isoform in the parental NSC34 cells (representing ∼0.001% of total cell protein). In contrast, normal HeLa cells express approximately one-third of this amount. UBQLN1, 2, and 4 proteins were identified according to their established migration patterns in blots using an antibody that cross-reacts with all three UBQLN isoforms (11). Because UBQLN3 is only expressed in the testis it was not considered further.
Immunoblot analysis of the KO cells revealed the absence of the UBQLN2 isoform in all of the KO cell lines, as expected. In the HeLa KO8 cell line, UBQLN2 loss was the only detectable change in UBQLN isoform expression (Fig. 2A). In contrast, in the HeLa KO12 cell line, both UBQLN1 and 4 isoforms were increased, by an unknown mechanism. The net effect of these changes was calculated to reduce the total amount of the different UBQLN protein isoforms in the KO8 cell line by about 50% and to increase their amount in the KO12 line by 150% compared to the parental line (Fig. 2B). The two lines enabled us to distinguish whether effects produced upon elimination of UBQLN2 expression originate from a general reduction in the total UBQLN protein pool or a specific elimination of UBQLN2 expression itself. Meanwhile, quantification of the UBQLN isoforms in the two NSC34 UBQLN2 KO cells revealed complete loss of UBQLN2 expression in the cells with little to no alteration in expression of any other UBQLN isoform (SI Appendix, Fig. S7A).
Germline Deletion of the UBQLN2 Gene in Mouse Can Lead to Up-Regulation of UBQLN Proteins.
The unusual induction of UBQLN1 and 4 proteins in the HeLa KO12 cell line prompted us to examine whether inactivation of the UBQLN2 gene in mice also results in induction of other UBQLN isoforms. Accordingly, we immunoblotted equal amounts of whole-brain lysates made from three normal non-Tg and three UBQLN2 KO mice at 6 and 12 mo of age to monitor UBQLN isoform expression (Fig. 2C). As expected, all of the lysates from mice with germline floxed deletion of UBQLN2 were devoid of the UBQLN2 protein band. Examination of the remaining UBQLN bands revealed little change in any of their levels at 6 mo of age, irrespective of UBQLN2 inactivation. However, at 12 mo of age, two of the UBQLN2 KO mice had increased amounts of a lower migrating UBQLN isoform band. These results suggest that inactivation of UBQLN2 expression can result in up-regulation of other UBQLN isoforms, probably as a result of pressure to compensate for loss of UBQLN2 function.
Inactivation of UBQLN2 in Cells and Mice Leads to Changes in the Expression of Autophagic Proteins.
We next examined whether inactivation of UBQLN2 expression in the cell lines and mice affects autophagy by monitoring changes in p62 and LC3 expression. Immunoblot comparison revealed LC3II levels were decreased in the two HeLa KO lines compared to the parental line, suggesting loss of UBQLN2 reduces autophagy (Fig. 2B). Paradoxically, however, p62 levels were reduced and not increased in the UBQLN2 KO cell lines. We explored the possible reason for the reduction by measuring p62 RNA expression and by examining its cellular staining pattern. Real-time quantitative PCR measurement of p62 mRNA revealed a slight increase, if any, in the UBQLN2 KO lines, but the difference was not significant (SI Appendix, Fig. S8A). The immunofluorescence staining was more instructive, showing p62 had a more punctate distribution in KO8 cells compared to its parental line, where it was more diffuse (SI Appendix, Fig. S8B). Quantification of the staining showed that although the fluorescence intensity was comparable between the two lines, KO8 cells had dramatically higher number of inclusions than normal HeLa cells (SI Appendix, Fig. S8 C and D). We therefore speculate that p62 localization in inclusions may have made the protein more resistant to extraction, leading to gross underestimation of its true levels by immunoblotting. Regardless of the reason, the dramatic intracellular alteration in p62 distribution after UBQLN2 knockdown is also suggestive of perturbation in autophagy.
Similar analysis of the proteins in the NSC34 KO lines revealed contradictory findings (SI Appendix, Fig. S7B). In the KO69 cell line, LC3II and p62 levels were slightly decreased compared to the parental line. In contrast, both proteins were increased in the NSC34 KO20 cell line. We speculate that the increase is due to compensatory changes from pressure to restore autophagy from loss of UBQLN2 expression. Nevertheless, the universal change in LC3 and p62 proteins upon UBQLN2 KO strongly suggests UBQLN2 expression is somehow linked to regulation of autophagy.
Similar analysis of LC3II and p62 proteins in brain lysates from the UBQLN2 KO mice revealed a progressive reduction of LC3II levels in UBQLN2 KO animals at 6 and 12 mo of age compared to control normal non-Tg mice (Fig. 2 C and D). Additional immunoblots revealed only slight differences in p62 levels in animals at the two ages (Fig. 2 C and D). Taken together, the consistent reduction in LC3II accumulation seen upon UBQLN2 knockdown in both mouse and cells strongly suggests UBQLN2 is required for proper regulation of autophagy.
Inactivation of UBQLN2 Expression in HeLa Cells Reduces Autophagic Flux.
To determine if loss of UBQLN2 expression affects autophagy, we examined the parental and UBQLN2 KO cells to see if they have alterations in basal or adaptive autophagy (39, 40). Accordingly, parallel sets of cultures were incubated for different periods of time in normal glucose-containing medium, glucose-lacking medium, or in plain Earle’s balanced salt solution. The former growth condition was used to measure basal autophagy, whereas the latter two conditions were used to measure adaptive autophagy. One set of the cultures was treated with Bafilomycin A1 for different time periods, and a parallel set was left untreated. Bafilomycin A1 inhibits acidification of autolysosomes, stalling maturation and degradation of proteins like p62 and LC3 in autophagosomes (41, 42). Protein lysates from the cultures were probed for changes in LC3 and p62 proteins by immunoblot analysis.
Comparison of the HeLa cell lines revealed both KO8 and KO12 lines induce a smaller increase in LC3II protein accumulation over time after Bafilomycin A1 treatment than the parental cell line (SI Appendix, Fig. S9). This defect was evident under all conditions tested. For example, the extent of autophagic flux, as measured by the increase in LC3II protein accumulation after 4 h of Bafilomycin A1 treatment, was reduced in both the HeLa KO8 and KO12 cell lines compared to their parental HeLa cells (SI Appendix, Fig. S9). Similar comparison of p62 changes revealed more complex regulation of its degradation and induction, as previously reported (42). Nevertheless, these results indicate that knockdown of UBQLN2 in HeLa cells by itself, and irrespective of restoration of total UBQLN levels by other UBQLN isoforms, stalls both basal and adaptive autophagic flux.
Visual Evidence for UBQLN2 in Autophagosomes.
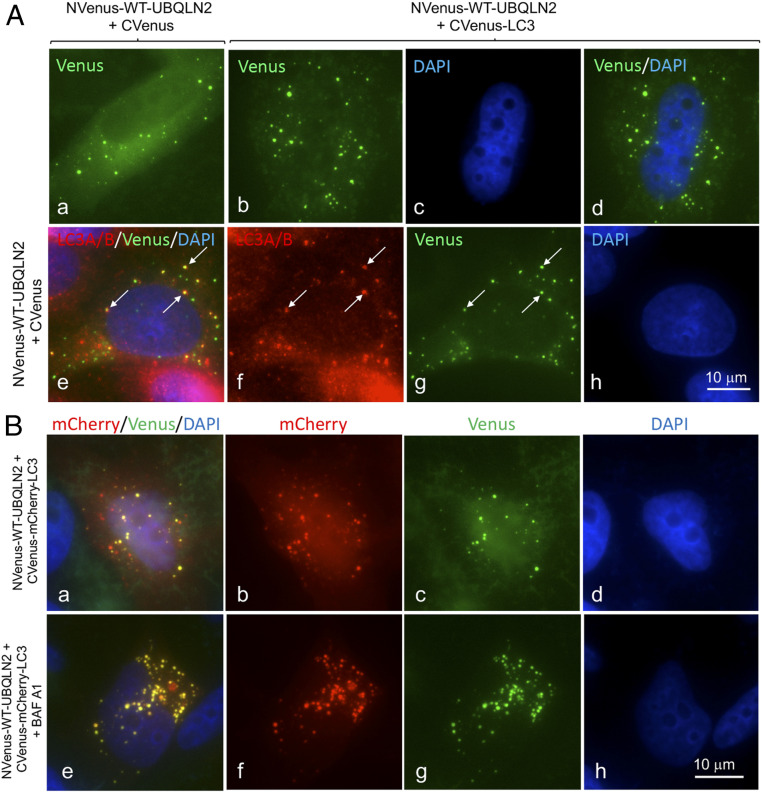
To visualize whether UBQLN2 proteins participate in autophagy we utilized the split-Venus reporter system to determine if UBQLN2 can localize to autophagosomes (43, 44). This reporter system exploits the principle that the Venus fluorescent protein can be split into two nonfluorescent components, which when brought together, such as through binding of their two fusion partners, can reconstitute Venus fluorescence (43). As expected, transfection of HeLa cells with either of the separated N-terminal 173 aa (NVenus) or C-terminal 155 aa Venus (CVenus) expression constructs generated no fluorescence, but cotransfection of the constructs reconstituted widespread fluorescence across the cell, including brighter fluorescence in the nucleus, typical of GFP (SI Appendix, Fig. S10 A2 and E). We next fused the NVenus fragment in-frame with the ATG start codon of WT human UBQLN2 cDNA (NVenus-WT-UBQLN2) and verified its expression along with all subsequent Venus derivatives by immunoblotting (SI Appendix, Fig. S10 A–C). Cells cotransfected with the NVenus-WT-UBQLN2 and CVenus expression constructs contained diffuse fluorescence across the whole cell, interspersed with brighter puncta of different sizes and intensities and together with weaker diffuse fluorescence in the nucleus (Fig. 3 A, a). This pattern is similar to that of endogenous UBQLN2 staining of HeLa cells (SI Appendix, Fig. S10D). To determine if the puncta were autophagosomes, we counterstained the cells to determine if the puncta were positive for LC3 (using antibodies specific for LC3A/B or LC3B), the autophagosome-specific marker. Indeed, double-immunofluorescent microscopy confirmed endogenous UBQLN2 and the analyzed LC3 proteins colocalize in puncta (Fig. 3 A, e–h and SI Appendix, Fig. S10D). Additionally, many of the Venus fluorescent puncta formed in cells transfected with NVenus-UBQLN2 and CVenus also stained positive for LC3 (SI Appendix, Fig. S10F), suggesting they are autophagosomes.
Fig. 3.
Visual detection of UBQLN2 in autophagosomes. (A) Fluorescent (YFP) images of HeLa cells transfected with NVenus-UBQLN2 WT and CVenus (a). YFP-fluorescence of HeLa cells transfected with NVenus-UBQLN2 WT and CVenus-LC3B (b), DAPI image (c), and the combined YFP and DAPI images (d). (e–h) Cells transfected with NVenus-UBQLN2 WT and CVenus and counterstained for LC3A/B. Arrows show colocalization of the staining in puncta. (B) Images of HeLa cells cotransfected with CVenus-mCherry-LC3B with NVenus-UBQLN2 WT and left untreated (a–d) or treated for 4 h with Bafilomycin A1 (e–h).
To determine if UBQLN2 and LC3 interact with one another in autophagosomes, we fused LC3B with the CVenus reporter construct (SI Appendix, Fig. S10A4). Cotransfection of the resulting CVenus-LC3 with NVenus-WT-UBQLN2 in cells led to reconstitution of fluorescence predominantly in puncta in cells, which by further analysis had properties consistent with autophagosomes (see below) (Fig. 3 A, b–d). The fluorescence outside of the puncta was much weaker, suggesting NVenus-WT-UBQLN2 and CVenus-LC3 accumulate or interact predominantly in autophagosomes.
Generation of a Fluorescent Reporter to Track Maturation of UBQLN2-Containing Autophagosomes.
Because overexpression of UBQLN2 may have resulted in aggregation or phase separation of the protein in cells (45, 46), it was important to demonstrate that the fluorescent puncta seen in our transfections had properties consistent with autophagosomes. An important property of autophagosomes is that they mature into autolysosomes after fusing with lysosomes (42). During this maturation, the pH environment of autophagosomes changes from near neutral to acidic. This change in pH can be exploited to track the fate of fluorescent proteins with different pH sensitivities (42). For example, Pankiv et al. (31) devised a tandem-tagged GFP-mCherry-LC3B fluorescent reporter (SI Appendix, Fig. S10A1) to discriminate autophagosomes from autolysosomes based on quenching of GFP fluorescence, but not mCherry fluorescence of the reporter, during the maturation. Using their reporter, autophagosomes are therefore distinguished by containing both GFP and mCherry fluorescence, the combination of which yields a yellow signal, while autolysosomes are identified by possessing mainly mCherry fluorescence. We devised a similar tandem-tag fluorescent reporter system to detect maturation of UBQLN2-containing autophagosomes to autolysosomes (SI Appendix, Fig. S10A3–5). To do so, we modified our CVenus-LC3 reporter by inserting a cDNA fragment encoding mCherry in-frame between the N-terminal CVenus fragment and LC3B (CVenus-mCherry-LC3B). Cells transfected with this construct alone displayed mCherry, but no Venus fluorescence, as expected (SI Appendix, Fig. S10G). However, when cotransfected with the NVenus-WT-UBQLN2 expression construct, Venus fluorescence was reconstituted and localized mainly in puncta (Fig. 3 B, a–d).
All of the puncta that displayed bright Venus fluorescence also possessed bright mCherry fluorescence, as expected. Additional puncta were found in the cells that had brighter mCherry fluorescence with attenuated Venus fluorescence, consistent with quenching of Venus fluorescence by the low pH environment of autolysosomes or late endosomes. Treatment of the transfected cells with Bafilomycin A1 to inhibit acidification of autophagosomes led to an increase in both the total number of fluorescent puncta and yellow puncta (rose to >90%) in cells (Fig. 3 B, e–h), which is expected if autophagosome-to-autolysosome conversion was blocked. We also counterstained the cells from these transfections for p62, LAMP1, ubiquitin, and UBQLN2 itself and found that the majority of the yellow puncta were positive for all of the markers examined. Interestingly, most of the red puncta, representing autolysosomes were generally negative for the markers, suggesting their disappearance may be intimately linked to autophagosome acidification (SI Appendix, Fig. S11). The loss of ubiquitin staining in the red puncta also suggests that the fluorescent puncta are unlikely to be misfolded UBQLN2 proteins, as one would expect ubiquitin staining of aggregates to persist irrespective of their color. Taken together, these results demonstrate that the Venus reporter constructs are suitable for monitoring maturation of UBQLN2-containing autophagosomes into autolysosomes.
Inactivation of UBQLN2 in HeLa KO8 Cells Impedes Autophagosome Acidification that Can Be Rescued by Reexpression of WT UBQLN2 but Not by ALS-Mutant UBQLN2 Proteins.
We next evaluated whether UBQLN2 proteins carrying ALS/FTD mutations alter maturation of autophagosomes to autolysosomes. We tested this possibility using our Venus reporter assay following expression of the proteins in both normal HeLa and HeLa KO8 cell lines. We first assessed whether autophagosome number and acidification differed in the lines by using the traditional GFP-mCherry-LC3B reporter (31, 47). The quantification revealed a dramatic reduction in both autophagosome number and acidification in the KO8 cell line compared to the normal HeLa line (SI Appendix, Fig. S12 A and B). We assessed if the reduction in autophagosome number in KO8 cells arises from changes in WIPI2 expression or distribution. WIPI2 is required during early steps of autophagosome formation, but is lost once autophagosomes enclose (48). The blots showed KO8 cells have increased WIPI2 levels, but its intracellular distribution in autophagosomes was unremarkable compared to normal HeLa cells (SI Appendix, Fig. S12 C–E). We speculate the up-regulation of WIPI2 expression in KO8 cells may represent a futile attempt made by the cells to restore autophagy. Further characterization of the cell lines showed KO8 cells had reduced lysotracker staining and increased ratio of expression of mature to procathepsin D proteins compared to normal HeLa cells (SI Appendix, Fig. S12 F–I), which is indicative of defects in the endosomal–lysosomal pathway (49). Taken together, the results are consistent with KO8 cells having major defects in autophagy and lysosomal function.
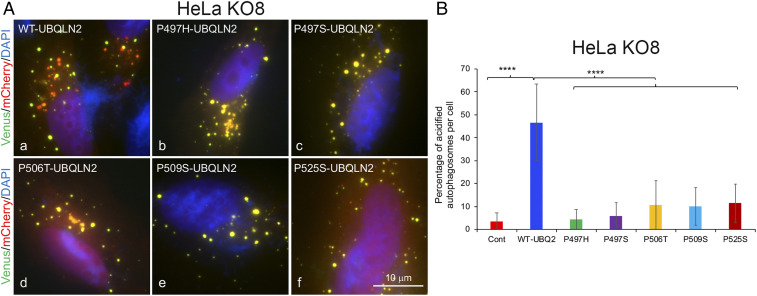
We next determined whether expression of WT and mutant UBQLN2 proteins in the two cell lines alters autophagosome number and acidification. Accordingly, we transfected each of the lines with NVenus-UBQLN2 expression constructs encoding either WT UBQLN2 or carrying one of five different ALS/FTD mutations (16, 50) together with CVenus-mCherry-LC3B and quantified the proportion of autophagosomes that were acidified (mCherry-fluorescent alone due to Venus quenching) in 10 randomly selected cells (SI Appendix, Figs. S13A and S14). Cells with gross overexpression of the reporters were excluded, because high overexpression of UBQLN2 has been found to lead to aggregation of the protein (45, 46, 51). The quantification revealed that normal HeLa cells transfected with WT UBQLN2 had greater autophagosome acidification than for all of the ALS mutants tested (SI Appendix, Fig. S13B). We also stained cells transfected with the P525S-Venus reporter for ubiquitin, UBQLN, and p62 and found frequent colocalization of the markers with yellow puncta, and not red puncta, just like cells transfected with the WT UBQLN2-Venus reporter (SI Appendix, Fig. S11). This staining further suggests that the puncta formed by ALS mutant UBQLN2 proteins are unlikely to be aggregates, otherwise ubiquitin and p62 staining would have persisted irrespective of the color change. By comparison, repetition of the assay in the KO8 line revealed that expression of WT UBQLN2 restores autophagosome acidification to similar levels seen in normal HeLa cells (Fig. 4 and SI Appendix, Fig. S15). In contrast, expression of all five ALS/FTD bearing UBQLN2 mutants failed to restore autophagosome number or acidification, suggesting the ALS/FTD mutations cause loss of function.
Fig. 4.
Defect in rescue of autophagosome acidification by ALS/FTD mutant UBQLN2 proteins. (A) Representative images of the combined YFP, mCherry, and DAPI fluorescent signal in HeLa UBQLN2 KO8 cells cotransfected with CVenus-mCherry-LC3B and either WT or ALS mutant NVenus-UBQLN2 constructs. (B) Quantification of autophagosome acidification. ****P < 0.0001.
Reduction in Expression of the ATP6v1g1 Subunit of the V-ATPase Proton Pump in P497S UBQLN2 Mice.
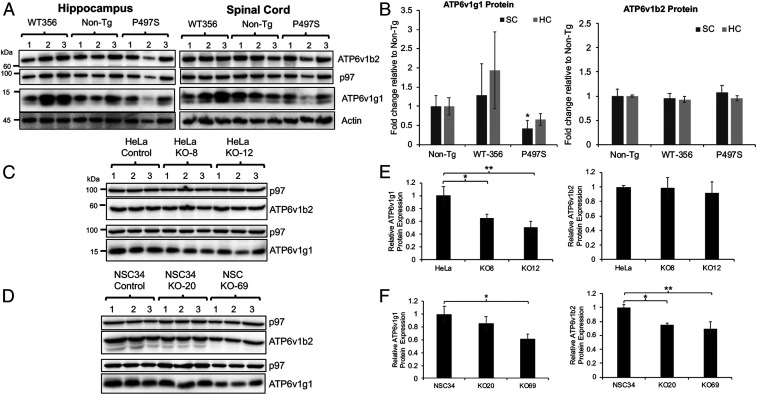
Because ALS/FTD mutant UBQLN2 proteins cause defective autophagosome acidification, we conducted an unbiased proteomics study to determine whether expression of genes that regulate autophagosome acidification were specifically altered in mutant P497S Tg animals. Accordingly, we used isobaric labeling combined with liquid chromatography-mass spectroscopy (LC-MS/MS) to quantify protein expression in the hippocampus and SC (lumbar region) of 2-mo-old P497S, WT356, and non-Tg mice. We then calculated the log2 ratio of the change in expression between P497S vs. WT356, P497S vs. non-Tg, and WT356 vs. non-Tg genotypes (SI Appendix, Fig. S16A). This comparison only includes data for subunits of the V-ATPase proton pump because it regulates autophagosome acidification (42, 52, 53). The proteomics data contained expression information for 15 different subunits of the pump. Of these, expression of only one of them, the ATP6v1g1 subunit, was reduced in P497S animals compared to the control genotypes. The expression of the subunit was slightly decreased in both the hippocampus and SC tissues of P497S mice compared to non-Tg, but was decreased even more when compared to WT UBQLN2 Tg mice. The reason for the greater difference was because expression of the subunit was slightly increased in WT356 animals compared to non-Tg animals. To confirm the changes, we probed hippocampal and SC lysates of 8-mo-old animals of all three genotypes for changes in the subunit. The blots were also probed for ATP6v1b2, another V-ATPase subunit whose expression was calculated to be faintly altered by proteomic analysis (Fig. 5A). The immunoblot results for SC closely matched the proteomics data, showing P497S mutant mice have reduced ATP6v1g1 subunit expression compared to age matched non-Tg and WT UBQLN2 Tg animals. In contrast, expression of the ATP6v1b2 subunit in the same lysates was not altered. However, the blots for the hippocampal lysates for the same set of animals failed to show any significant change in expression of the ATP6v1g1 subunit, although we noticed large fluctuations in expression of the subunit in lysates of different mice (Fig. 5 A and B).
Fig. 5.
Reduction of ATP6v1g1 in P497S mutant mice and UBQLN2 KO cells. (A) Immunoblots of hippocampus and SC lysates of 8-mo-old mice for the three mouse genotypes (three independent mice for each genotype) and probed for the proteins shown. (B) Quantification of protein changes of two different V-ATPase subunits from the blots shown in B. *P < 0.05. (C and D) Immunoblots of HeLa and NSC34 parental and UBQLN2 KO lines for the proteins shown and (E and F) corresponding quantification of the protein levels for the two V-ATPase subunits in the cell lines. *P < 0.05, **P < 0.01.
To confirm that expression of ATP6v1g1 is reduced in P497S animals, we immunoblotted SC lysates made from 14 non-Tg and 14 P497S animals, all at 6 mo of age, for the subunit as well as for ATP6v0a1, another subunit of the V-ATPase pump (SI Appendix, Fig. S16B). Expression of the ATP6v0a1 subunit, particularly fragments of it, were reported to increase following ubqln protein knockdown (23). The blots revealed, after normalizing for protein loading, a 50% reduction in ATP6v1g1 levels in P497S animals compared to the non-Tg animals (SI Appendix, Fig. S16C). By contrast, ATP6v0a1 subunit levels were similar in the two genotypes, including no major difference in fragments of the subunit.
Inactivation of UBQLN2 in Cells Leads to a Reduction in ATP6v1g1 Subunit Accumulation.
We next investigated the reason for the reciprocal change in ATP6v1g1 subunit expression in WT vs. P497S UBQLN2 Tg animals. We reasoned that it could have arisen from functional differences in the properties of WT and mutant UBQLN2 proteins, predicting that WT UBQLN2 may support expression of the subunit, whereas the mutant may have lost this function. To examine this possibility, we immunoblotted equal amounts of proteins from the UBQLN2 KO lines to determine whether loss of UBQLN2 expression affects ATP6v1g1 accumulation (Fig. 5 C and D). Indeed, the immunoblots showed KO of UBQLN2 in both HeLa and NSC34 cells results in reduced accumulation of ATP6v1g1 levels (Fig. 5 E and F). We also examined whether ATP6v1g1 levels were changed in UBQLN2 KO animals and found only a slight reduction in animals at 12 mo of age compared to non-Tg animals, although not significant, matched the trend seen in the KO cells (Fig. 2 C and D and SI Appendix, Fig. S16 D and E).
UBQLN2 and ATP6v1g1 Bind One Another in a Complex: Modulation of ATP6v1g1 Accumulation by Coexpression of WT but Not ALS/FTD Mutant UBQLN2 Proteins.
To determine the mechanism by which UBQLN2 regulates ATP6v1g1 subunit accumulation, we investigated two chief possibilities based on known functions of UBQLN proteins (3): UBQLN2 regulates turnover of the ATP6v1g1 subunit and it functions as a chaperone for the protein. Another possibility was that P497S inclusions may sequester ATP6v1g1, but it was discounted because immunostaining suggested little colocalization of the two proteins in P497S animals (SI Appendix, Fig. S17).
To examine the first possibility, we measured the turnover of ATP6v1g1 in normal HeLa and UBQLN2 KO8 cells by cycloheximide chase experiments (SI Appendix, Fig. S18 A and B). Paradoxically, the results revealed loss of UBQLN2 slows ATP6v1g1 turnover, suggesting UBQLN2 directly or indirectly facilitates ATP6v1g1 degradation. One possibility by which it could regulate ATP6v1g1 stability is through decreased consumption of the subunit from reduced autophagic flux in the line.
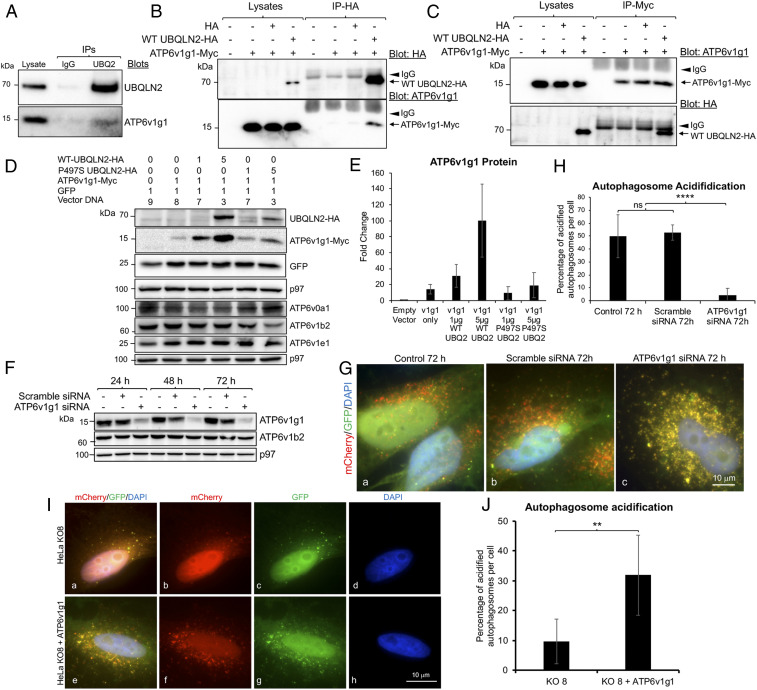
To examine the second possibility, we immunoprecipitated UBQLN2 from NSC34 cells and found endogenous ATP6v1g1 protein was coimmunoprecipitated (Fig. 6A). To confirm their interaction, we coexpressed Myc-tagged ATP6v1g1 with HA-tagged WT UBQLN2 expression constructs in HeLa cells and then immunoprecipitated each of the expressed proteins with antibodies specific for their respective tags (Fig. 6 B and C). The precipitates were then examined for recovery of both proteins by immunoblotting. Immunoprecipitation controls were also performed of cells transfected with neither construct or with cDNAs encoding only the tags. The controls demonstrated the specificity of the antibodies in only precipitating the correct tagged protein. Importantly, analysis of the two precipitates from cells transfected with both constructs revealed coimmunoprecipitation of both proteins, irrespective of the antibody used for the immunoprecipitation (Fig. 6 B and C). These data prove both UBQLN2 and ATP6v1g1 can bind together in cells forming a complex.
Fig. 6.
Function and interaction of ATP6v1g1 and UBQLN2. (A) Immunoprecipitation of endogenous proteins from NSC34 cells with either an anti-UBQLN2 antibody or IgG control antibody, and subsequent immunoblots for the proteins shown. (B and C) Immunoprecipitation analysis of proteins from HeLa cells transfected with the constructs shown on the top with either anti-HA (B) or anti-Myc (C) antibodies. The arrowheads show the IgG bands and the arrows indicate the respective proteins that were immunoprecipitated (Upper) and coimmunoprecipitated (Lower). (D) Demonstration that UBQLN2 stimulates biogenesis of ATP6v1g1. HeLa cell cultures were cotransfected with identical amounts of total plasmid cDNAs (μg) shown on the top. The next day lysates were prepared from the cultures and immunoblotted for the proteins shown. (E) Quantification of the expressed proteins shows ATPv1g1 expression is increased by UBQLN2 in a dose-dependent manner. (F) Immunoblots of cultures after transfection for different days to knockdown ATP6v1g1. (G) Coverslips showing the different number of autolysosomes (red) collected from the same cultures shown in F. (H) Quantification of autophagososme acidification in the three transfected cultures. ****P < 0.0001. (I) Fluorescent images of HeLa UBQLN2 KO8 cells transfected with GFP-mCherry-LC3B alone (a–d) or cotransfected with ATP6v1g1-myc cDNA (e–h). (J) Quantification showing acidification of HeLa KO8 cells is increased in cells transfected with ATP6v1g1-myc cDNA. **P < 0.01.
We next examined whether ALS/FTD mutant UBQLN2 proteins have altered interaction with ATP6v1g1 using this same assay. Accordingly, HeLa cells were cotransfected with expression constructs encoding ATP6v1g1-Myc and either WT UBQLN2-HA or mutants carrying each of the five different ALS/FTD mutations described by Deng et al. (1). However, immunoblot analysis showed that cells transfected with each of the UBQLN2 mutants had reduced UBQLN2 accumulation compared with cells transfected with WT UBQLN2. Because of this variability, we could not assess changes in binding between the proteins by immunoprecipitation. Instead, we investigated the reason for the variability in expression of ATP6v1g1, predicting that it may stem from differences in chaperone function of WT and mutant UBQLN2 proteins. To investigate this possibility, we transfected HeLa KO8 cells with a constant amount of ATP6v1g1-Myc expression construct (1 μg) and varying amounts (1 and 5 μg) of either WT UBQLN2-HA or a P497S mutant expression construct. A constant amount of a GFP expression construct was included as a control. The immunoblots from these transfections revealed WT-UBQLN2 induced a dose-dependent increase in ATP6v1g1-Myc expression, which was not seen with the P497S mutant (Fig. 6 D and E). Unlike ATP6v1g1-Myc, GFP expression was not changed by transfection of either UBQLN2 construct.
To rule out the possibility that UBQLN2-induced increase in ATP6v1g1-Myc accumulation stems from interference of degradation of the subunit through the proteasome or autophagy pathways, we conducted parallel transfections of the UBQLN2 titrations, but this time treated one set of the cultures with either MG132 or Bafilomycin A1 to inhibit the proteasome or autophagy, respectively. The effectiveness of these treatments was verified by increased accumulation of ubiquitinated proteins and LC3II proteins (SI Appendix, Fig. S18C). Measurement of ATP6v1g1 levels in the treated cultures revealed a 30% increase in the subunit after proteasome inhibition, irrespective of the amount of UBQLN2 transfected (SI Appendix, Fig. S18D). Inhibition of autophagy did not substantially alter ATP6v1g1 levels. These results reveal that the ATP6v1g1 subunit is principally degraded through the proteasome pathway and that the dose-dependent increase of the subunit by UBQLN2 is unlikely to be stem from simple inhibition of the proteasome.
Evidence for Direct Binding between UBQLN2 and ATP6v1g1 Proteins.
To determine if UBQLN2 and ATP6v1g1 bind directly to one another, we conducted GST-pulldown assays assessing binding of His-tagged ATP6v1g1 protein with fixed amounts of WT or mutant GST-UBQLN2 fusion proteins (SI Appendix, Fig. S18E). A negative control examining binding of ATP6v1g1 with GST alone was also conducted. Quantification of the pulldown results (SI Appendix, Fig. S18F), after normalizing for the amount of GST protein that was pulled down, showed ATP6v1g1-His binds stronger with GST-WT UBQLN2 than with any of the five ALS/FTD mutant UBQLN2 proteins. In contrast, binding of ATP6v1g1 with GST alone was negligible.
Knockdown of ATP6v1g1 Expression Reduces Autophagosome Acidification.
To determine whether a reduction of ATP6v1g1 levels could affect autophagosome acidification, we silenced its expression in HeLa cells and quantified autophagosome acidification. Accordingly, a pool of HeLa cells stably expressing the traditional GFP-mCherry-LC3B reporter were transfected with small interfering RNAs (siRNAs) to specifically knockdown ATP6v1g1 expression. Immunoblots revealed that the siRNAs were effective in reducing ATP6v1g1 levels by over 95% over a 72-h period (Fig. 6F). By contrast, ATP6v1g1 levels were relatively unchanged in the cultures transfected with the scrambled siRNAs. Importantly, autophagosome acidification was dramatically reduced only in the cultures in which ATP6v1g1 subunit was knocked down (Fig. 6 G and H and SI Appendix, Fig. S19). These results demonstrate the important requirement of the ATP6v1g1 subunit for autophagosome acidification.
Overexpression of ATP6v1g1 Rescues the Acidification Defect in UBQLN2 KO Cells.
Finally, we tested whether overexpression of ATP6v1g1 is sufficient to rescue the acidification defect in HeLa UBQLN2 KO8 cells. Indeed, we found overexpression of Myc-tagged ATP6v1g1 increased the percentage of acidified autophagosomes compared to cells not transfected with the plasmid (Fig. 6 I and J).
Discussion
Here we show UBQLN2 is required for proper regulation of autophagy and that UBQLN2 proteins carrying ALS/FTD mutations block this function by impeding autophagosome acidification, most likely through a dominant-negative loss-of-function mechanism. These conclusions are based on the major alterations in autophagy that we observed upon KO of UBQLN2 and overexpression of WT and ALS mutant UBQLN2 proteins in cells and animals. Further evidence was found by the abnormal distribution and accumulation of key autophagic proteins in human ALS/FTD UBQLN2 cases and in the P497S UBQLN2 mouse model of ALS/FTD.
KO of UBQLN2 in two independent HeLa cell lines reduced autophagic flux, the number and acidification of autophagosomes, and the levels of ATP6v1g1, a key subunit of the V-ATPase proton pump. In contrast, overexpression of UBQLN2 carrying ALS/FTD mutations in both cells and animals perturbed autophagy, as exemplified by a build-up of p62 and ubiquitin chains, a reduction in autophagosome acidification, and reduction of ATP6v1g1 levels. Mechanistic insight into the underlying reason for the reduction in ATP6v1g1 levels suggests a likely role of UBQLN2 as a chaperone in binding and facilitating ATP6v1g1 biosynthesis. Importantly, this function was abrogated by ALS/FTD mutations in UBQLN2. Indication that disturbances in autophagy may underlie pathogenesis caused by UBQLN2 mutations was evident by the disruption in normal colocalization of LC3 and LAMP1 with p62 and UBQLN2 in a human UBQLN2 case and in the P497S mouse model of ALS/FTD.
Our evidence showing that UBQLN2 is required for proper regulation of autophagy is consistent with prior studies implicating UBQLN proteins function in autophagy, including a recent study showing KO of the sole Drosophila ubqln gene impedes autophagosome acidification (21–24). However, our findings differ from those studies in a number of important ways. First, prior studies showing the importance of UBQLN proteins in autophagy in mammalian cells was demonstrated by KO of multiple UBQLN genes, unlike our study showing KO of UBQLN2 alone is sufficient to induce defects in autophagy. Second, while the defect in autophagosome acidification that we uncovered shares parallels with those reported in Drosophila (23), it differs in two important ways: The identity and nature of the change of the V-ATPase subunit. We found levels of ATP6v1g1 were decreased upon KO of UBQLN2 in two different cell types (HeLa and NSC34 cells) as well as in SC of the P497S mouse model of ALS, whereas KO of ubqln in Drosophila was linked to a toxic increase in expression of ATP6v0a1, a different subunit of the V-ATPase pump (23). Furthermore, we found overexpression of ATP6v1g1 cDNA in UBQLN2 KO8 HeLa cells increased autophagosome acidification, strongly implicating reduction of ATP6v1g1 levels as a key molecular defect. The reason for our divergent findings remains unclear, but may stem from differences in the cells and animals used in the studies. For example, Drosophila has only one ubqln gene, unlike humans and mice, which possess four UBQLN genes. Additionally, the Drosophila ubqln protein is only 47% identical to human UBQLN2 and lacks the PXXP motif where most ALS mutations map. This sequence divergence may explain the functional difference in V-ATPase subunit regulation uncovered in the two studies. It may also explain why expression of human UBQLN2 in Drosophila is toxic (23).
Clear indication that UBQLN2 is required for autophagy was evident by the dramatic reduction in both basal and adaptive autophagy that we observed following KO of UBQLN2 in two different HeLa cell lines. In one of these lines (KO12), UBQLN1 and 4 proteins were up-regulated, resulting in a 1.5-fold increase in the total UBQLN protein pool, probably from pressure to compensate for loss of UBQLN2 expression. Despite this increase, it was insufficient to restore autophagic flux to normal levels. However, a reduction in autophagic flux was not seen following KO of UBQLN2 in NSC34 cell lines, for unclear reasons. One possibility is cell type-specific differences in the mechanisms used by HeLa and NSC34 to compensate for UBQLN2 loss.
A molecule that we believe could be directly linked to dysregulation of autophagy upon KO of UBQLN2 expression or from expression of ALS/FTD mutant UBQLN2 proteins is ATP6v1g1, a critical subunit of the V-ATPase proton pump (54–56). The V-ATPase pump is assembled from a number of subunits which are organized into a membrane-anchored V0 sector and a cytoplasmic V1 sector. V-ATPase activity is regulated by the assembly and disassembly of the two sectors (57–59). The ATP6v1g1 subunit functions to “strap” the two sectors together (57, 58). Thus, loss of the subunit could cause disassembly of the complex and loss of V-ATPase function. Levels of the subunit were reduced not only in UBQLN2 KO cells (both HeLa and NSC34) and mice, but also in P497S UBQLN2 mutant Tg mice. In contrast, slightly elevated levels of ATP6v1g1 were found in Tg mice expressing WT UBQLN2, suggesting ATP6v1g1 levels may be directly regulated by UBQLN2 expression. Indeed, GST-pulldown assays and coexpression studies indicated WT UBQLN2 and ATP6v1g1 bind directly with one another and that biosynthesis of ATP6v1g1 is directly regulated by the amount of UBQLN2 expression. Importantly, we found that UBQLN2 proteins containing ALS/FTD mutations are defective in both binding and stimulating ATP6v1g1 biogenesis. Based on all of these findings we propose UBQLN2 functions as a chaperone, to bind and facilitate ATP6v1g1 biogenesis. Interestingly, degradation of ATP6v1g1 was also dependent on UBQLN2 expression, suggesting multiple levels of regulation.
The idea that UBQLN2 could function as a chaperone coincides with several other findings hinting of such a function. For example, UBQLN proteins bind and are required for insertion of transmembrane proteins, including nuclear encoded mitochondrial proteins (60, 61). An unusual property of this binding involves hydrophobic segments in proteins, which typically need to be shielded from the aqueous cellular environment following synthesis (60, 62). The hydrophobic binding sites in UBQLNs have been mapped to the central domain of the protein, which is also the binding site for HSP chaperone proteins (13, 62–64). It is therefore possible that binding of UBQLN2 with ATP6v1g1 shields the protein prior to assembly of the proton pump. This shielding may not be unique for the ATP6v1g1 subunit, based on the findings that several subunits of the V-ATPase proton pump coimmunoprecipitate with UBQLNs (23). The stimulation of ATP6v1g1 biogenesis by UBQLN2 may be directly or indirectly connected to the shielding and chaperone function. This effect may not be unique to UBQLN2 as a similar effect of binding and stimulating of presenilin biogenesis was shown for UBQLN1 (8).
Strong indication that ALS/FTD mutations lead to failure in autophagosome acidification was clearly evident using the tandem-tag Venus-mCherry-LC3-UBQLN2 reporter system we generated. The reporter system allowed us to directly visualize maturation of UBQLN2-containing autophagosomes to autolysosomes. We confirmed the utility of the reporter system using double immunofluorescent staining methods showing that UBQLN2, LC3, and p62 were all present in autophagosomes decorated by the reporter. Furthermore, because reconstitution of Venus fluorescence in puncta of NVenus-UBQLN2 and CVenus-LC3 (and CVenus-mCherry-LC3) necessitates binding of the two split Venus components, it provided visual demonstration that UBQLN2 and LC3 are capable of interaction within autophagosomes.
A key observation uncovered from use of the Venus UBQLN2-acidification reporter was the defect in acidification with ALS mutant UBQLN2 proteins. The defect was evident upon expression of the mutant proteins in both normal HeLa and UBQLN2 KO8 cells with the effect in the KO8 cells being more potent. The obvious difference between the two cell lines is one has, and the other lacks, endogenous UBQLN2 expression, suggesting that the manifestation of the defect may be influenced by UBQLN2 itself. Importantly, the acidification defect was rescued by expression of WT UBQLN2, but not by any of the five ALS/FTD mutants tested, strengthening the case that the defect is a universal and underlying cause by which UBQLN2 mutations drive pathogenesis. Based on these observations we propose that the ALS/FTD mutations in UBQLN2 are loss-of-function mutations. Furthermore, we propose that this loss of UBQLN2 function results in failure to maintain sufficient ATP6v1g1 subunit accumulation, depletion of which we and others have found is critical for autophagosome acidification (54).
If the loss-of-function hypothesis is correct, the more potent acidification defect seen in the KO8 cells compared to normal HeLa cells may stem from endogenous UBQLN2 protein in normal HeLa cells sustaining sufficient ATP6v1g1 subunit expression to maintain some V-ATPase function, thereby partially obscuring the dominant-negative effects of the mutants. However, we predict that prolonged expression of the mutants in cells would eventually lead to depletion of ATP6v1g1 levels from the normal turnover of the protein without adequate replacement due to increased penetrance of the mutant UBQLN2 protein over time. Although the mechanism by which ALS/FTD mutations in UBQLN2 exert their dominant effects is not known, it is likely to derive from inactivation of the endogenous WT UBQLN2 protein through oligomerization and sequestration by the mutant protein. Such a mechanism of inactivation is possible based on the known properties of UBQLN proteins. For example, UBQLN proteins have been shown to both homo- and hetero-oligomerize, providing a mechanism by which mutant UBQLN2 proteins could bind and deplete other UBQLN protein isoforms like WT UBQLN2 protein, from the functional pool in cells (65, 66). Such a mechanism of coaggregation could also explain the age-dependent concomitant accumulation of both endogenous and transgenic mutant UBQLN2 proteins that are seen in the P497S Tg mouse model of ALS (38). Similar reasoning likely explains how transgenic expression of mutant UBQLN2 proteins in rodents trigger disease. It could also explain the delayed onset of ALS/FTD that is frequently seen in female carriers of UBQLN2 mutations (1, 3), where X-chromosome–linked expression of the mutant UBQLN2 allele could be titrated by that of the normal WT UBQLN2 allele.
If our hypothesis that UBQLN2 is required for ATP6v1g1 biogenesis is correct, it implies that the reduction in ATP6v1g1 subunit levels found in P497S mutant UBQLN2 animals stems from loss of normal UBQLN2 function. Interestingly the UBQLN2 KO mice do not develop any signs of MN disease like the P497S Tg animals. One possibility to explain the discrepancy is if the P497S UBQLN2 mutation (and other ALS mutations in the protein) causes disease through both loss- and gain-of-function mechanisms. Besides depleting WT UBQLN2, the UBQLN2 inclusions could also bind and interfere with the function of other important proteins. One such protein is likely to be p62, a key autophagic protein, mutations in which are also linked to ALS. Immunoblots of brain and SC lysates of 8-mo-old affected P497S mice revealed elevated levels of p62 compared to WT356 and non-Tg mice, while immunostaining revealed p62 localization predominantly in inclusions in the tissues, most of which stained positive for UBQLN2. Similar aberrant mislocalization of p62 with UBQLN2 inclusions was found in all three affected human carriers of different UBQLN2 mutations that we examined. Although the number of UBQLN2 inclusions in the three human cases varied considerably, most of the UBQLN2 inclusions were also positive for p62. The commonality of p62 mislocalization with UBQLN2 inclusions provides further evidence of the validity of the P497S mouse model in faithfully recapitulating important aspects of the human disease.
The lack of LAMP1 staining of the UBQLN2 inclusions seen in the human UBQLN2 case and P497S animals, but its positive staining of puncta in HeLa cells formed by expression of the Venus reporters for the P525S UBQLN2 mutant is especially instructive. We propose that the explanation for the difference in staining is because the puncta in cells are very different to those in the tissues. In HeLa cells, our evidence strongly indicates the UBQLN2 puncta are either autophagosomes or autolysosomes, and therefore would be expected to recruit lysosomes and accordingly be positive for LAMP1. In the case of the human and mouse tissues we propose that the inclusions are most likely to be aggregates that accumulate because of loss of UBQLN2 function in autophagy. We further propose that this disruption of autophagy leads to accumulation of UBQLN2 and p62 aggregates that cannot be packaged and cleared by autophagosomes (consistent with lack of LC3 staining too) and accordingly would not be expected to recruit lysosomes (since they are not autophagosomes).
Our findings that disturbances in autophagy could be a key driver of pathogenesis caused by UBQLN2 mutations add to growing evidence that defects in this degradation pathway could play a central role in triggering ALS (67–69). The impairment in autophagosome acidification found in our studies is also noteworthy, as it adds to growing evidence of similar defects in acidification that have been found within the endosomal–lysosomal system of several neurodegenerative diseases, including Alzheimer’s disease and Down syndrome (53, 70, 71). Our findings may also have important clinical implications for treating humans with UBQLN2 mutations, because they suggest that efforts to stimulate this pathway would be ill-advised as the block in autophagy is downstream of the whole process. Instead, we suggest that efforts focusing on rectifying the acidification defect may prove to be a better strategy to treat ALS and related neurodegenerative disorders.
Materials and Methods
Transgenic mouse lines expressing either WT or ALS P497S mutant UBQLN2 together with human UBQLN2 ALS cases were used to examine the pathologic changes of autophagic proteins. All animal procedures were approved by University of Maryland Baltimore (UBQLN2 Tg mice) and Harvard University (UBQLN2 KO mice) Animal Care and Use Committees and conducted in full accordance with the NIH Guide for the Care and Use of Laboratory Animals (72). We also studied the effects of overexpression of WT and five different ALS mutant UBQLN2 proteins in HeLa cells either containing or lacking UBQLN2 expression. NSC34 cells containing and lacking UBQLN2 expression were also used to evaluate whether loss of UBQLN2 affects autophagy. The detailed experimental procedures and statistical analysis used for all of the studies are provided in SI Appendix, SI Materials and Methods
All of the data, materials, and protocols are described in the main text or SI Appendix.
Supplementary Material
Acknowledgments
We thank Marika Eszes at the Centre for Brain Research, University of Auckland, New Zealand; and Trong Phung for helpful comments on the manuscript. This work was supported by The Robert Packard Center for ALS Research at Johns Hopkins, the ALS Association, and NIH Grants R01NS098243 and R01NS100008 (to M.J.M.). E.L.S. is supported by Marsden FastStart and Rutherford Discovery Fellowship funding from the Royal Society of New Zealand (15-UOA-157, 15-UOA-003). T.S. is supported by The Les Turner ALS Foundation/Herbert C. Wenske Chair and NIH Grants R01NS078504 and R01NS046535.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917371117/-/DCSupplemental.
Change History
August 30, 2021: Figure 1 and its legend have been updated; please see accompanying Correction for details. The SI Appendix has been updated to coincide with a formal Correction.
References
- 1.Deng H. X., et al., Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen H. P., Van Broeckhoven C., van der Zee J., ALS genes in the genomic era and their implications for FTD. Trends Genet. 34, 404–423 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Higgins N., Lin B., Monteiro M. J., Lou Gehrig’s disease (ALS): UBQLN2 mutations strike out of phase. Structure 27, 879–881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fecto F., Esengul Y. T., Siddique T., Protein recycling pathways in neurodegenerative diseases. Alzheimers Res. Ther. 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahheydari H., et al., Protein quality control and the amyotrophic lateral sclerosis/frontotemporal dementia continuum. Front. Mol. Neurosci. 10, 119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edens B. M., et al., A novel ALS-associated variant in UBQLN4 regulates motor axon morphogenesis. eLife 6, e25453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu A. L., Wang J., Zheleznyak A., Brown E. J., Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol. Cell 4, 619–625 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Mah A. L., Perry G., Smith M. A., Monteiro M. J., Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J. Cell Biol. 151, 847–862 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conklin D., Holderman S., Whitmore T. E., Maurer M., Feldhaus A. L., Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene 249, 91–98 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Kleijnen M. F., et al., The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 6, 409–419 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Safren N., Chang L., Dziki K. M., Monteiro M. J., Signature changes in ubiquilin expression in the R6/2 mouse model of Huntington’s disease. Brain Res. 1597, 37–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaye F. J., et al., A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 467, 348–355 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Hjerpe R., et al., UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 166, 935–949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko H. S., Uehara T., Tsuruma K., Nomura Y., Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 566, 110–114 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Massey L. K., et al., Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J. Alzheimers Dis. 6, 79–92 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Chang L., Monteiro M. J., Defective proteasome delivery of polyubiquitinated proteins by ubiquilin-2 proteins containing ALS mutations. PLoS One 10, e0130162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., et al., Structure of hRpn10 bound to UBQLN2 UBL illustrates basis for complementarity between shuttle factors and substrates at the proteasome. J. Mol. Biol. 431, 939–955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harman C. A., Monteiro M. J., The specificity of ubiquitin binding to ubiquilin-1 is regulated by sequences besides its UBA domain. Biochim. Biophys. Acta, Gen. Subj. 1863, 1568–1574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stieren E. S., et al., Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J. Biol. Chem. 286, 35689–35698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.N’Diaye E. N., et al., The ubiquitin-like protein PLIC-2 is a negative regulator of G protein-coupled receptor endocytosis. Mol. Biol. Cell 19, 1252–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenberg C., et al., Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 19, 3219–3232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee D. Y., Arnott D., Brown E. J., Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep. 14, 373–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Şentürk M., et al., Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat. Cell Biol. 21, 384–396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N’Diaye E. N., et al., PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 10, 173–179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brettschneider J., et al., Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 123, 825–839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scotter E. L., et al., C9ORF72 and UBQLN2 mutations are causes of amyotrophic lateral sclerosis in New Zealand: A genetic and pathologic study using banked human brain tissue. Neurobiol. Aging 49, 214.e1–214.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Gkazi S. A., et al., Striking phenotypic variation in a family with the P497H UBQLN2 mutation including amyotrophic lateral sclerosis, spastic paraplegia, and frontotemporal dementia. Neurobiol. Aging 73, 229.e5–229.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Williams K. L., et al., UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol. Aging 33, 2527.e3–2527.e10 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Labbadia J., Morimoto R. I., The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma S., Attarwala I. Y., Xie X. Q., SQSTM1/p62: A potential target for neurodegenerative disease. ACS Chem. Neurosci. 10, 2094–2114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pankiv S., et al., p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Glick D., Barth S., Macleod K. F., Autophagy: Cellular and molecular mechanisms. J. Pathol. 221, 3–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dikic I., Elazar Z., Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Mizushima N., Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36, 2491–2502 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Gorrie G. H., et al., Dendritic spinopathy in transgenic mice expressing ALS/dementia-linked mutant UBQLN2. Proc. Natl. Acad. Sci. U.S.A. 111, 14524–14529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y., et al., Pathogenic mutation of UBQLN2 impairs its interaction with UBXD8 and disrupts endoplasmic reticulum-associated protein degradation. J. Neurochem. 129, 99–106 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Wu Q., et al., Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol. 129, 417–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le N. T., et al., Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS-FTD-linked UBQLN2 mutations. Proc. Natl. Acad. Sci. U.S.A. 113, E7580–E7589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu M., et al., Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y., Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 (1991). [PubMed] [Google Scholar]
- 42.Yoshii S. R., Mizushima N., Monitoring and measuring autophagy. Int. J. Mol. Sci. 18, e1865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodama Y., Hu C. D., An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques 49, 793–805 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Kodama Y., Hu C. D., Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotechniques 53, 285–298 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Dao T. P., et al., Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander E. J., et al., Ubiquilin 2 modulates ALS/FTD-linked FUS-RNA complex dynamics and stress granule formation. Proc. Natl. Acad. Sci. U.S.A. 115, E11485–E11494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ugolino J., et al., Overexpression of human Atp13a2Isoform-1 protein protects cells against manganese and starvation-induced toxicity. PLoS One 14, e0220849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polson H. E., et al., Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Götzl J. K., et al., Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 127, 845–860 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Gilpin K. M., Chang L., Monteiro M. J., ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum. Mol. Genet. 24, 2565–2577 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Sharkey L. M., et al., Mutant UBQLN2 promotes toxicity by modulating intrinsic self-assembly. Proc. Natl. Acad. Sci. U.S.A. 115, E10495–E10504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maxson M. E., Grinstein S., The vacuolar-type H+-ATPase at a glance—More than a proton pump. J. Cell Sci. 127, 4987–4993 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Lie P. P. Y., Nixon R. A., Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis. 122, 94–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Luca M., et al., RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J. Cell Sci. 127, 2697–2708 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Tomashek J. J., Graham L. A., Hutchins M. U., Stevens T. H., Klionsky D. J., V1-situated stalk subunits of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 272, 26787–26793 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Ohira M., et al., The E and G subunits of the yeast V-ATPase interact tightly and are both present at more than one copy per V1 complex. J. Biol. Chem. 281, 22752–22760 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Breton S., Brown D., New insights into the regulation of V-ATPase-dependent proton secretion. Am. J. Physiol. Renal Physiol. 292, F1–F10 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Marshansky V., Rubinstein J. L., Grüber G., Eukaryotic V-ATPase: Novel structural findings and functional insights. Biochim. Biophys. Acta 1837, 857–879 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Sautin Y. Y., Lu M., Gaugler A., Zhang L., Gluck S. L., Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol. Cell. Biol. 25, 575–589 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itakura E., et al., Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol. Cell 63, 21–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whiteley A. M., et al., Ubiquilin1 promotes antigen-receptor mediated proliferation by eliminating mislocalized mitochondrial proteins. eLife 6, e26435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurlawala Z., Shah P. P., Shah C., Beverly L. J., The STI and UBA domains of UBQLN1 are critical determinants of substrate interaction and proteostasis. J. Cell. Biochem. 118, 2261–2270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuda M., Koide T., Yorihuzi T., Hosokawa N., Nagata K., Molecular cloning of a novel ubiquitin-like protein, UBIN, that binds to ER targeting signal sequences. Biochem. Biophys. Res. Commun. 280, 535–540 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Gavriilidis C., et al., The MTM1-UBQLN2-HSP complex mediates degradation of misfolded intermediate filaments in skeletal muscle. Nat. Cell Biol. 20, 198–210 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Ford D. L., Monteiro M. J., Dimerization of ubiquilin is dependent upon the central region of the protein: Evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem. J. 399, 397–404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dao T. P., et al., ALS-linked mutations affect UBQLN2 oligomerization and phase separation in a position- and amino acid-dependent manner. Structure 27, 937–951.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramesh N., Pandey U. B., Autophagy dysregulation in ALS: When protein aggregates get out of hand. Front. Mol. Neurosci. 10, 263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rudnick N. D., et al., Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc. Natl. Acad. Sci. U.S.A. 114, E8294–E8303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ling S. C., et al., Overriding FUS autoregulation in mice triggers gain-of-toxic dysfunctions in RNA metabolism and autophagy-lysosome axis. eLife 8, e40811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J. H., et al., Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang Y., et al., Lysosomal dysfunction in down syndrome is APP-dependent and mediated by APP-βCTF (C99). J. Neurosci. 39, 5255–5268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.