Abstract
Background
Heliconius butterflies are widely distributed across the Neotropics and have evolved a stunning array of wing color patterns that mediate Müllerian mimicry and mating behavior. Their rapid radiation has been strongly influenced by hybridization, which has created new species and allowed sharing of color patterning alleles between mimetic species pairs. While these processes have frequently been observed in widespread species with contiguous distributions, many Heliconius species inhabit patchy or rare habitats that may strongly influence the origin and spread of species and color patterns. Here, we assess the effects of historical population fragmentation and unique biology on the origins, genetic health, and color pattern evolution of two rare and sparsely distributed Brazilian butterflies, Heliconius hermathena and Heliconius nattereri.
Results
We assembled genomes and re-sequenced whole genomes of eight H. nattereri and 71 H. hermathena individuals. These species harbor little genetic diversity, skewed site frequency spectra, and high deleterious mutation loads consistent with recent population bottlenecks. Heliconius hermathena consists of discrete, strongly isolated populations that likely arose from a single population that dispersed after the last glacial maximum. Despite having a unique color pattern combination that suggested a hybrid origin, we found no genome-wide evidence that H. hermathena is a hybrid species. However, H. hermathena mimicry evolved via introgression, from co-mimetic Heliconius erato, of a small genomic region upstream of the color patterning gene cortex.
Conclusions
Heliconius hermathena and H. nattereri population fragmentation, potentially driven by historical climate change and recent deforestation, has significantly reduced the genetic health of these rare species. Our results contribute to a growing body of evidence that introgression of color patterning alleles between co-mimetic species appears to be a general feature of Heliconius evolution.
Keywords: Heliconius, Hybridization, Introgression, Mutation load, Mimicry, Phylogenetics
Background
Butterflies have served as a historically significant model of insect diversity because of the group’s extreme species diversity, containing an estimated 18,000 species, and the even more striking color pattern diversity that defines the subspecies, races, forms, and morphs of many butterfly species. Among butterflies, the Neotropical genus Heliconius has attracted attention for centuries because of their bold and highly variable wing color patterns [1–4], as well as their fascinating behavior and natural history, which includes co-evolved relationships with larval host plants, widespread Müllerian mimicry, adult pollen feeding, and pupal-mating behavior [2, 4–6]. Today, researchers are leveraging the natural diversity of Heliconius butterflies and genomic tools to characterize the molecular genetic basis of mimicry [7–13] and track genome-wide patterns of differentiation and introgression associated with speciation [14–20]. Yet, for all of the attention, a number of fascinating Heliconius species have evaded modern genomic interrogation. Two particularly striking examples are Heliconius nattereri and Heliconius hermathena from Brazil.
Heliconius nattereri is an exceptionally rare, geographically restricted species found only along a section of Brazil’s Atlantic Forest spanning portions of the states of Espírito Santo and Bahia [21–23]. Heliconius nattereri stands out relative to other species in the genus because it is the only Heliconius species that is endangered [24] and it is also the only Heliconius species with pronounced sexually dimorphic color patterns (Fig. 1). Female H. nattereri display a black, yellow, and orange “tiger stripe” pattern that facilitates mimicry with other co-occurring Heliconius and Ithomiini species while males display a non-mimetic yellow and black wing pattern. Prior to Keith Brown’s rediscovery of H. nattereri in the 1960s [21, 22], this species was only known from approximately 20 pinned specimens in museums around the world. Furthermore, males and females had not been collected together, and because of their pronounced sexual dimorphism in both color pattern and behavior, females had been historically described as a different species, H. fruhstorferi Riffarth, 1899. In the 1960s and 1970s, Brown and colleagues extensively surveyed the range of H. nattereri, discovering additional localities and specimens, including one large population that they studied intensively for three years. Based on his work, Brown was not optimistic about the long-term future of H. nattereri because of its limited and still dwindling habitat and his observations that it seemed to be an “inflexible” and “sensitive” species [22].
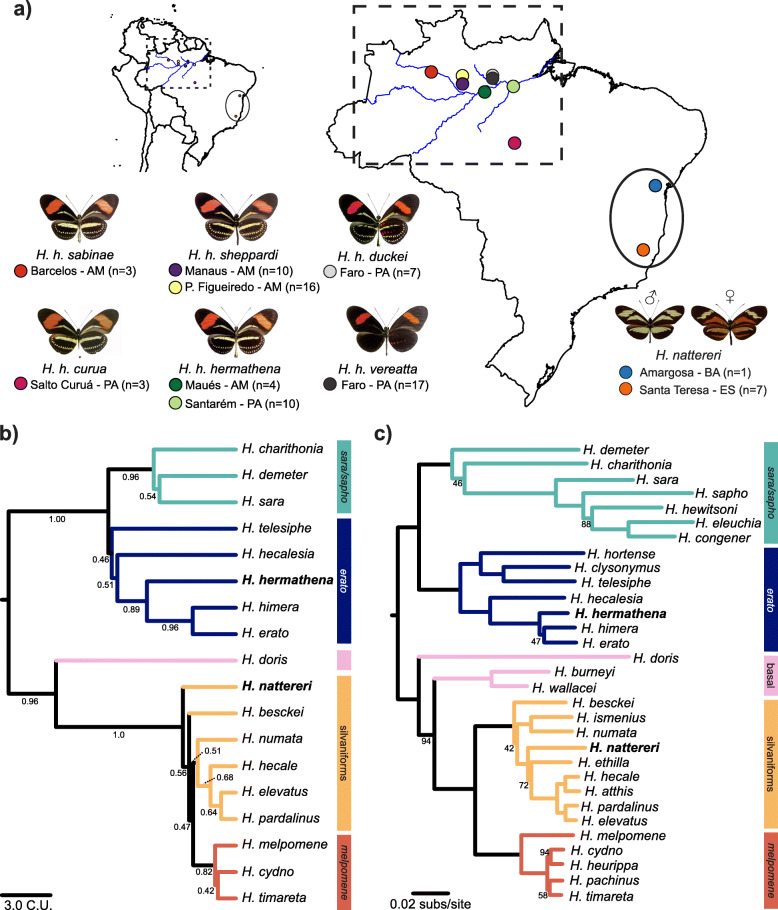
Fig. 1.
Distribution and phylogenetic relationships of Heliconius hermathena and H. nattereri.a Sampling areas and phenotypes of H. hermathena subspecies and H. nattereri. H. hermathena images show dorsal color patterns on the left and ventral on the right. Brazilian states: AM, Amazonas; BA, Bahia; ES, Espírito Santo; PA, Pará. b ASTRAL tree constructed from 8674 trees based on 10-kb autosomal windows, rooted with Eueides tales (not shown). Quartet scores are shown below branches. C.U., coalescent units. c Maximum likelihood tree based on complete mitochondrial DNA sequences. Only branches with less than 100% bootstrap support are labeled. subs/site, substitutions per site. H. nattereri images courtesy of AHB Rosa
Like H. nattereri, H. hermathena has historically been viewed as an unusual and relictual species [25]. Much of what we know about H. hermathena stems from Brown and Benson’s [25] extensive survey, in which they described the systematics, biogeography, natural history, ecology, and behavior of this species. More recently, Seixas et al. [26] described the population ecology of H. hermathena based on a 14-month mark-recapture experiment. Heliconius hermathena is associated with white sand habitats (campina and campinarana) throughout the Brazilian Amazon. These patchy, harsh environments produce a network of seven described H. hermathena subspecies, including H. hermathena curua, which was just described in 2019 [27]. Six H. hermathena subspecies display subtle variations on a shared, putatively non-mimetic color pattern (Fig. 1) while the seventh subspecies, vereatta, lacks much of the yellow patterning on the fore- and hindwings and is a co-mimic of sympatric Heliconius erato and Heliconius melpomene. Heliconius hermathena vereatta has a limited spatial distribution and is known to freely hybridize with adjacent Heliconius hermathena duckei near the town of Faro in the State of Pará [25]. Based on the phenotypic variation among individuals and color pattern frequencies in the field, Brown and Benson [25] inferred that mimicry in H. hermathena was controlled by a single locus with a dominant, melanic allele. The genetics of mimicry in H. hermathena has not been further explored. The evolutionary origin of H. hermathena itself is a topic of interest. The non-mimetic color pattern shared by most H. hermathena subspecies appears to be a combination of features from close relatives H. erato and H. charithonia, and Hewitson [28] reflected this noticeable mixture when naming the species. Today, there are a number of well-characterized instances of introgression and hybrid speciation in Heliconius [15, 19, 20, 29], and while H. hermathena is considered one possible example of hybrid speciation [30–32], the evolutionary origin of this species remains largely unexplored.
As part of an international collaboration, we have been studying the biology, genomics, and population genetics of Brazil’s endemic, and enigmatic, Heliconius fauna. With permission and funding from the Brazilian government, we have performed new surveys of H. nattereri [23] and H. hermathena [27] populations, collected samples, and performed genomic analyses. Here, we present reference genome sequences and whole-genome resequencing datasets for both species. We use these data to infer their modern population structure and historical demography relative to other well-studied and widespread Heliconius species, characterize the genomic consequences of population decline and historical habitat fragmentation, and define the genetic basis of mimicry in H. hermathena and trace its evolutionary origin. These data add to a growing list of genomic resources in Heliconius [15, 20, 33] but crucially yield novel insight into the biology of two unique but critically understudied species.
Results
High-quality reference genome assemblies for H. hermathena and H. nattereri
We first generated high-quality H. hermathena and H. nattereri genome assemblies to facilitate phylogenetic and population genomic analyses using Illumina paired-end and mate-pair sequencing (Table 1; Tables S1-S2, Additional file 1; the “Materials and methods” section). The final H. hermathena assembly comprised 392 Mb in 1913 scaffolds with an N50 of 560 kb, while the H. nattereri assembly comprised 276 Mb in 261 scaffolds with an N50 of 8.8 Mb (Table 1). These values were consistent with the size and heterozygosity estimates of 373 Mb and 0.0057 for H. hermathena and 258 Mb and 0.0075 for H. nattereri, respectively, from analyses of 21-mer frequencies in the raw sequencing data [34]. Furthermore, both genomes were predicted to be among the most complete and least redundant nymphalid assemblies available based on the presence and completeness of universal single-copy orthologs assayed using BUSCO [35] (Table 1).
Table 1.
Heliconius hermathena and H. nattereri genome assembly statistics in comparison with other well-assembled nymphalid genomes
| Speciesa | Length (Mb) | No. of scaffoldsb | N50 (Mb) | BUSCOc | ||
|---|---|---|---|---|---|---|
| Comp. (Dup.) | Frag. (%) | Miss. (%) | ||||
| Bicyclus anynana | 467 | 2423 | 0.66 | 89.6% (0.7%) | 3.9 | 6.5 |
| Danaus plexippus | 245 | 1009 | 0.73 | 96.6% (1.9%) | 2.3 | 1.1 |
| Heliconius charithoniad | 316 | 5162 | 0.11 | 94.3% (0.3%) | 3.8 | 1.6 |
| Heliconius erato demophoon | 383 | 194 | 10.69 | 85.5% (0.7%) | 4.5 | 10.0 |
| Heliconius erato lativitta | 418 | 142 | 5.48 | 73.4% (0.8%) | 5.4 | 21.2 |
| Heliconius hermathena | 392 | 1913 | 0.56 | 96.0% (0.3%) | 2.4 | 1.6 |
| Heliconius melpomene | 275 | 209 | 14.31 | 86.2% (0.4%) | 4.1 | 9.7 |
| Heliconius nattereri | 276 | 261 | 8.84 | 97.3% (0.5%) | 1.2 | 1.5 |
| Hypolimnas misippus | 409 | 1580 | 1.01 | 88.9% (0.3%) | 3.8 | 7.3 |
| Junonia coenia | 586 | 1101 | 1.57 | 96.9% (14.7%) | 1.4 | 1.7 |
| Limenitis arthemis | 306 | 737 | 2.27 | 81.7% (1.4%) | 1.9 | 16.4 |
| Melitaea cinxia | 383 | 5633 | 0.12 | 57.1% (0.2%) | 11.8 | 31.1 |
| Vanessa tameamea | 357 | 1558 | 2.99 | 96.3% (0.4%) | 2.2 | 1.5 |
aAccessions are shown in the “Materials and methods” section
bScaffolds ≥ 5 kb
cCalculated using BUSCO v3 and the Endopterygota database (2440 SCOs) from OrthoDB v9
dAssembled here, see the “Materials and methods” section
Phylogenetic placement of H. hermathena and H. nattereri
Heliconius consists of two major clades with unique characteristics: the erato-sara clade and the melpomene-silvaniform clade (Fig. 1). Morphological studies have placed H. hermathena within the erato-sara clade [25] and H. nattereri within the melpomene-silvaniform clade, but molecular phylogenetics results have been unclear about the fine-scale placement of species. Beltrán et al. [30] used four autosomal and four mitochondrial genes while Kozak et al. [36] used 20 autosomal and three mitochondrial genes to infer Heliconius species relationships. These studies placed H. nattereri and H. ethilla as sister species with low statistical support and H. hermathena as a polytomy with H. himera and H. erato. In a recent pre-print, Kozak et al. [37] used genome-wide SNP calls relative to H. melpomene to reconstruct the Heliconius species tree and found H. hermathena as sister to H. erato and H. himera; they did not include H. nattereri. However, hybridization and gene flow are common within the major Heliconius clades, confounding estimation of species relationships using standard molecular phylogenetic methods based on few loci or a single reference genome [15, 20, 37].
To resolve the placement of H. hermathena and H. nattereri within Heliconius, we performed whole-genome alignments and reconstructed species trees based on genome-wide data following Edelman et al. [20]. Edelman et al. [20] inferred the relationships among 13 Heliconius species by aligning de novo-assembled genomes using the progressiveCactus alignment pipeline [38, 39], inferring gene trees for short non-overlapping aligned regions, and summarizing those gene tree topologies using ASTRAL [40, 41]. This approach therefore largely avoids biases introduced by using a single reference genome or a small number of gene trees and therefore more fully captures the different relationships among different genome regions due to gene flow [20]. Support for each branch in the species tree is calculated as the fraction of gene trees that include particular four-taxon topology (ASTRAL’s quartet score) [40]. We aligned our new reference genomes and a new high-quality Heliconius charithonia reference assembly that we generated using publicly available data (Table 1; the “Materials and methods” section) to the multi-species alignment produced by Edelman et al. [20] using progressiveCactus [38, 39]. We then constructed maximum likelihood (ML) trees using autosomal, non-overlapping 10-kb windows and used these trees to infer the Heliconius species tree with ASTRAL-iii (Fig. 1; Figure S1–S10, Additional file 2) [38–42]. The final species tree was inferred using 18 Heliconius genomes and 8674 windows (Fig. 1b). We corroborated these results with smaller coding and non-coding alignment blocks, where the effects of intra-alignment recombination are limited; within the melpomene-silvaniform and erato-sara clades separately; and using only Z-linked windows, as the Z is generally more resistant to gene flow and may better represent the true relationships between the species (Figure S2-S10, Additional file 2; the “Materials and methods” section) [20].
All of these analyses placed H. hermathena as sister to H. erato and H. himera (Fig. 1b; Figure S7-S10, Additional file 2). The branches joining these species together and separating H. hermathena from the other two had quartet scores > 0.89 in analyses of autosomal and Z-linked windows (Fig. 1b; Figure S8-S9, Additional file 2), and somewhat lower scores when using the shorter coding and non-coding blocks (> 0.54; Figure S7, Additional file 2). These results are consistent with the results of Kozak et al. [37] based on genome-wide SNP data. The primary source of discordance (i.e., low quartet scores) in the erato-sara clade is the hybridization event or events between the H. sara/H. demeter ancestor and H. hecalesia described by Kozak et al. [37] and Edelman et al. [20]; this event was reflected by the different relationships between these species in the predominant 10-kb window topologies (Figure S11, Additional file 2) in our analyses.
The placement of H. nattereri remained less certain. First, in contrast to Beltrán et al. [30] and Kozak et al. [36], all of our reconstructions based on autosomal loci place H. nattereri as an outgroup to the remaining members of the melpomene-silvaniform clade (Fig. 1b; Figure S7-S10, Additional file 2). Consistent with previous studies, the melpomene clade nested within the silvaniforms, and there was generally low concordance among trees estimated from different regions (Fig. 1b; Figure S7-S8, Additional file 2). Second, species tree reconstructions based on Z-linked windows recovered a clade comprising H. nattereri, H. numata, and H. besckei as an outgroup to the remaining melpomene-silvaniform clade species (Figure S9, Additional file 2). Kozak et al. [37] did not include H. nattereri but recovered a similar lineage in their analysis of Z-linked markers that included H. numata, H. besckei, and H. ismenius.
Finally, we assembled and analyzed mitochondrial genomes of 33 Heliconius species to more directly compare our results with previous studies. We assembled a typical ~ 15-kb contig for 31 of 33 species by extracting and assembling 26 new mitochondrial genomes from publicly available sequencing data using NOVOPlasty and seven reference mitochondrial genomes (Table S3, Additional file 1) [43]. We then inferred species relationships using these 33 sequences and ML (Fig. 1c) [42]. The relationships we found were similar to those recovered by the smaller mtDNA analyses in Beltrán et al. [30] and Kozak et al. [36], with differences only in clades that historically have been difficult to resolve such as in the melpomene-silvaniform clade (Fig. 1c). This phylogeny was identical to the one recovered by Kozak et al. [37]. Importantly, H. hermathena was inferred to be sister to H. erato and H. himera. However, in sharp contrast with species tree reconstructions based on genome-wide data, the mtDNA analysis recovered the silvaniforms as a monophyletic clade, with H. nattereri nested within.
Small effective population sizes of H. nattereri and H. hermathena result in high deleterious mutation loads
Both H. hermathena and H. nattereri have patchy, limited distributions that make them difficult to find and study in their natural habitats. Heliconius nattereri in particular is restricted to a few pockets of Atlantic Forest in a narrow region of eastern Brazil, usually only above ~ 500 m elevation, and is already listed as endangered by the IUCN [21–24]. To better understand the genetic health of these rare species, we estimated their current genetic diversity, historical population sizes, and deleterious mutation loads using population genomic data.
We sequenced the complete genomes of eight H. nattereri individuals from two locations and 71 H. hermathena individuals spanning six subspecies from seven localities (3–19 individuals per site) and used those data to analyze patterns of variation within and between populations (Fig. 1; Table S1, Additional file 1). We first estimated nucleotide diversity per site (π) in populations of H. hermathena, H. nattereri, and their close relatives H. erato and H. melpomene for comparison (Fig. 2; Tables S4-S5, Additional file 1). Heliconius erato and H. melpomene are widespread, abundant species that have well-characterized genetic history and population structure (e.g., [14, 17]). While average π in H. melpomene and H. erato was 0.0197 and 0.0251, respectively, consistent with previous estimates [14, 17, 44], H. nattereri and H. hermathena carry average π of only 0.0072 and 0.0047 (Fig. 2; Table S5, Additional file 1). H. hermathena populations in particular contain little genetic diversity, with average π ranging from only 0.0011–0.0036, consistent with observations of few individuals in any one locality [25]. The species-wide π values are consistent with analyses of 21-mers performed during reference genome assemblies above. We estimated that the current effective population size (Ne) of H. nattereri is ~ 620,000 (327,000–1,385,000), and the current Ne of H. hermathena is ~ 405,000 (214,000–904,000; Table S5, Additional file 1) using the measured H. melpomene mutation rate of 2.9e−9 per site per generation (μ; 95% CI 1.3e−9 to 5.5e−9) [45] and the classic estimator π = 4Neμ [46]. These estimates should be viewed with caution, however, as the relationship between π and Ne assumes that populations are at equilibrium and that variant sites are evolving strictly neutrally, assumptions that are not likely to be true (see below).
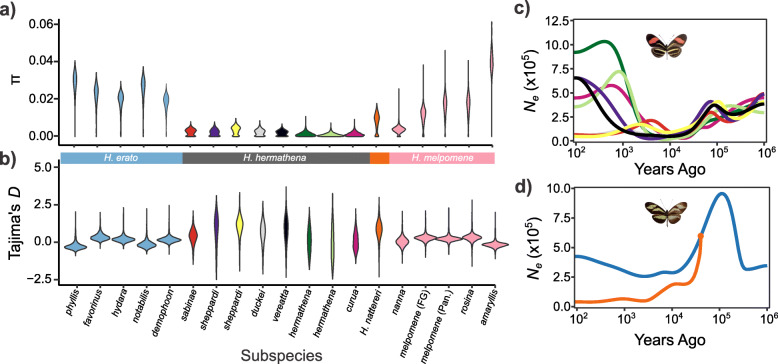
Fig. 2.
Current and historical effective population sizes in Heliconius nattereri and H. hermathena. a Nucleotide diversity per site (π) calculated in non-overlapping 10-kb windows across the autosomes. b Tajima’s D statistic calculated in 10-kb non-overlapping windows across the autosomes. cSMC++ analysis results for H. hermathena. dSMC++ analysis results for H. nattereri. Color schemes follow those in Fig. 1a. Geographical assignments for H. erato and H. melpomene subspecies are shown in Table S4 (Additional file 1)
Current Ne estimates reflect the harmonic mean of population sizes over recent history. To better understand the recent history of H. hermathena and H. nattereri, we estimated historical population sizes using the a multi-sample coalescent approach implemented in SMC++ (Fig. 2) [47]. We found a mixture of H. hermathena population size histories (Fig. 2). However, all H. hermathena populations were predicted to have been small and followed similar trajectories until ~ 10,000 years ago, when the southern populations H. h. vereatta and H. h. sheppardi from Manaus expanded quickly. In contrast to H. hermathena, we found that H. nattereri population sizes reached a peak ~ 100,000 years ago and have declined steadily since. The Santa Teresa population in particular has remained small, Ne ≈ 40,000, for the past 30,000 years. The most recent estimates place the Bahia and Santa Teresa population sizes at 424,000 and 40,000, respectively (Fig. 2; Table S4). The declines in H. nattereri and H. hermathena coincide with the end of the last glacial maximum, about 12,000 years ago.
Previous studies of Heliconius population size histories have only used the single-sample coalescent method implemented in PSMC [48]. However, SMC++ is more powerful and able to accurately infer more recent population size changes than PSMC [47]. We include PSMC results for comparison in Figure S12 (Additional file 2). The PSMC and SMC++ results from 104 to 106 years ago are nearly identical.
We consistently found that H. nattereri and H. hermathena Ne was only 20–25% that of their more widely distributed relatives H. melpomene and H. erato (Fig. 2). Small populations are expected to harbor more slightly deleterious alleles due to the strength of genetic drift relative to natural selection, so we expected H. nattereri and H. hermathena to carry higher deleterious mutation loads than other closely related species with larger effective population sizes. We expected this signal to be especially strong if these species underwent a recent, strong population bottleneck like the one predicted by SMC++ [49, 50]. We therefore calculated the numbers of species-specific substitutions and the site frequency spectrum of derived mutations as qualitative measures of H. nattereri and H. hermathena genetic health.
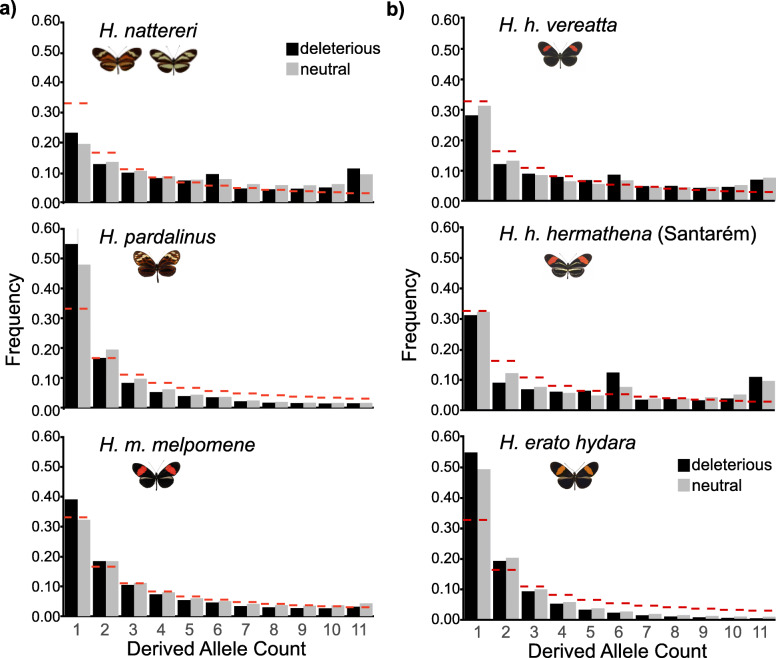
We first compared substitutions and the frequency spectrum of derived polymorphisms in H. nattereri to those in its close relatives H. pardalinus and H. melpomene (Fig. 3a). We called SNPs for each species relative to the H. melpomene genome, inferred the ancestral state for each site, then inferred the impact of each derived mutation on H. melpomene gene models using snpEff [51]. We then calculated the frequency spectra for neutral and deleterious mutations separately (the “Materials and methods” section). H. nattereri has accumulated a significantly higher fraction of deleterious substitutions (7.5% of all substitutions) than either H. pardalinus (5.9%) or H. melpomene (2.0%) since they diverged from their last common ancestor (χ12 > 1272, p < 2e−16 for the three tests; Table S6, Additional file 1). Furthermore, H. nattereri harbors an excess of alleles at intermediate and high frequencies, regardless of their impact (Fig. 3). This excess of intermediate and high-frequency alleles further suggests that H. nattereri recently underwent a strong population bottleneck, consistent with the SMC++ analysis (Fig. 2). This bottleneck hypothesis was supported by a high genome-wide average (+ 0.82) and standard deviation (0.66) of Tajima’s D statistic in H. nattereri (Table S5, Additional file 1) [52]. We did not observe these SFS or Tajima’s D patterns in H. pardalinus or H. melpomene, despite those species carrying similar or higher levels of diversity to H. nattereri (0.0077 and 0.015, respectively; Fig. 3). Our results therefore quantify the threatened status of H. nattereri at the genetic level.
Fig. 3.
Deleterious mutation loads in Heliconius nattereri and H. hermathena. a Derived allele frequency spectra in the Santa Teresa H. nattereri population, its close relative H. pardalinus, and H. melpomene melpomene. b Derived allele frequency spectra from H. erato and H. hermathena populations with the largest sample sizes. Dashed red lines indicate expected fractions based on the coalescent
We found similarly skewed substitution proportions, site frequency spectra, and Tajima’s D distributions in H. hermathena, suggesting a recent bottlenecks and high deleterious mutation load (Figs. 2 and 3). H. hermathena contains an excess of both fixed (χ12 = 5100, p < 2e−16; Table S7, Additional file 1) and intermediate- and high-frequency deleterious alleles relative to H. erato (χ210 > 3,032,371, p < 2e−16 for all tests). The strength of the skew was inversely related to current Ne estimates based on π (Figs. 2 and 3).
Strong population structure in Heliconius hermathena likely causes small Ne
While there is good evidence that H. nattereri is a sensitive and rare species, it was less clear why H. hermathena exhibits such small population sizes and high deleterious mutation loads. However, H. hermathena comprises seven recognized subspecies from white sand habitats (campina and campinarana) scattered around the Amazon River Basin that few other Heliconius species can tolerate [25–27]. This patchy distribution, high habitat fidelity, and observed low dispersal led Brown and Benson [25] to hypothesize that H. hermathena was once widespread but recently fragmented by the expansion of the Amazon rainforest after the last glacial maximum, about 12,000 years ago. We therefore tested whether H. hermathena population fragmentation was contributing to the population genomic patterns we observed.
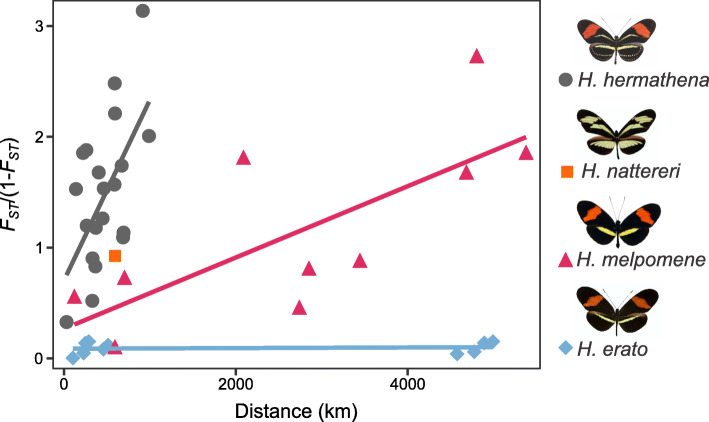
We first assayed genetic differentiation between H. hermathena populations relative to their widespread relatives using FST. Consistent with the hypothesis that their populations are strongly isolated, we found a strong positive correlation between FST and geographical distance between H. hermathena populations (Fig. 4; Table S8, Additional file 1). The rate at which FST increases with geographical distance is nearly four times higher in H. hermathena than in the more widely distributed H. erato or H. melpomene (Fig. 4; Tables S9-S10, Additional file 1).
Fig. 4.
The relationship between genetic distance (FST/(1 − FST)) and geographical distance for Heliconius nattereri, H. hermathena, H. melpomene, and H. erato populations, see Tables S1 and S4 for population and geographical location information (Additional file 1)
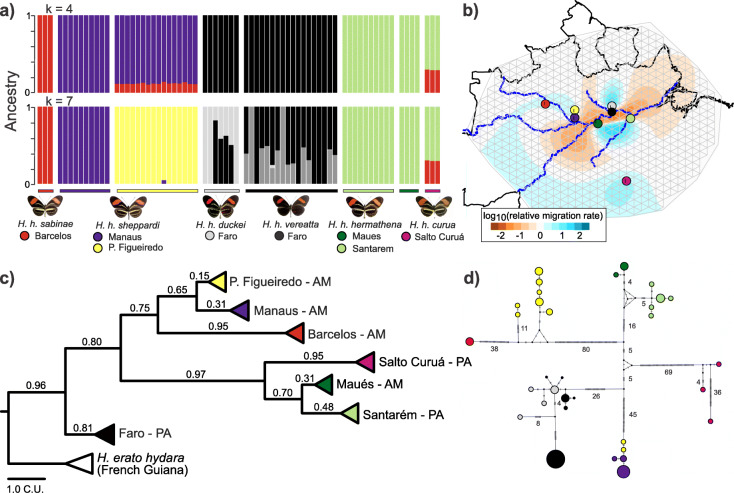
We next inferred H. hermathena population structure using Admixture and a series of expected numbers of populations (k = 2 to k = 10; Fig. 5; Figure S13, Additional file 2) [53]. Figure 5a shows the results for the number of populations with the lowest to cross-validation error (4) and the number of localities/subspecies we sampled (7). Most individuals were well-differentiated by geographical location (Fig. 5), particularly populations separated by the Amazon River. The most admixed individuals are found in the Faro population, consisting of H. h. duckei and the mimetic H. h. vereatta. This strong population structure was also apparent from a haplotype network constructed from whole mtDNA sequences from these 71 individuals, with the exception of two H. h. sheppardi individuals from Presidente Figueiredo (hher39, hher40) that grouped with H. h. sheppardi from Manaus (Fig. 5; Tables S1 and S11). These results are similar to those based on a mtDNA barcode [27].
Fig. 5.
Population structure and gene flow between Heliconius hermathena populations. a Admixture analysis results for the most likely k (4) and the actual number of populations sampled (7). All k values tested are shown in Fig. S13 (Additional file 2). b Relative migration rates across the range of H. hermathena, calculated by EEMS. c Bayesian concordance tree generated from 10-kb autosomal windows. Note that both H. h. vereatta (gray) and H. h. duckei (black) are included in the Faro clade, see Fig. S15 for the full tree (Additional file 2). d Mitochondrial DNA haplotype network constructed using whole mtDNA sequences and popArt. Each hash indicates one mutation, and numbers beside branches indicate total numbers of mutations on that branch. The size of each circle is proportional to the number of individuals sharing that mtDNA haplotype. Circles are colored according to the source population
The fact that there exist few admixed individuals in any of the core populations suggests that migration is rare between H. hermathena populations. Brown and Benson [25] noted that H. hermathena do not disperse more than a few hundred meters from their home ranges, probably due to their habitat fidelity and preference for campina and campinarana over deep forest. This finding further supports the existence of strong barriers to gene flow between geographically close H. hermathena populations (Figs. 4 and 5). We next sought to visualize those barriers to gene flow. We estimated effective migration rates between H. hermathena populations using EEMS [54]. In contrast to analyses of pairwise FST, EEMS estimates migration rates across a geographic region using the geographic locations of and genetic similarity estimates between all populations (Fig. 5b; Figure S14 [54];). Consistent with Admixture results, the Amazon River and its tributaries appear to provide strong isolating barriers between the northern (H. h. sabinae, H. h. sheppardi, H. h. duckei, and H. h. vereatta) and southern (H. h. hermathena and H. h. curua) populations: the narrow strip of extremely low effective migration rates matched the course of the Amazon despite the fact that EEMS is agnostic to topography. The only pairs of populations predicted to frequently share migrants were H. h. duckei and H. h. vereatta from Faro, which are known to frequently hybridize [25] (RRR, personal observation), and the H. h. hermathena populations from Santarém and Maués, which were indistinguishable in Admixture analyses (Fig. 5a; Figure S13, Additional file 2).
Finally, we reconstructed the phylogenetic relationships among H. hermathena populations to begin to understand how H. hermathena originated and became so widespread. We constructed a Bayesian concordance tree using autosomal 10-kb windows (Fig. 5c). Similar to ASTRAL quartet scores, concordance factors (CFs) provide an estimate of the congruence between tree topologies estimated from different genomic windows; low CFs may be caused by a variety of factors, including incomplete lineage sorting and gene flow between populations [55]. The concordance tree shown in Fig. 5c suggests that the subspecies groupings are well-supported by whole-genome data (CFs > 0.65), but that geographically adjacent populations are more weakly differentiated (see Figure S15, Additional file 2, for all individuals). Altogether, we find that H. hermathena is fragmented into small discrete populations that appear to rarely exchange migrants. This reconstruction also placed the Faro population as the most basal H. hermathena clade (Fig. 5c).
No genome-wide evidence that H. hermathena is a hybrid species
We were next interested in determining the origin of H. hermathena. Heliconius hermathena displays a unique combination of red and yellow color patterns that led several authors to posit that it may have been formed by hybridization between H. erato and H. charithonia [25, 31, 32]. In particular, only H. hermathena and the distantly related H. charithonia display characteristic rows of submarginal yellow hindwing spots (Fig. 1a). A single putative H. charithonia x H. erato hybrid has been discovered [56], so it is possible that historical hybridization could have generated a new, distinct species. We tested if H. hermathena was formed by hybridization between these H. erato and H. charithonia by calculating the D statistic in 10-kb windows across the autosomes using the species tree (((H. erato, non-mimetic H. hermathena), H. charithonia), H. melpomene) [57, 58]. Significantly positive D would indicate that H. hermathena shares more derived alleles with H. charithonia than expected due to incomplete lineage sorting since the three focal species diverged, and therefore a hybrid origin of H. hermathena. We found D = − 0.015 ± 0.008, suggesting that this is not the case (Table S11, Additional file 1, comparison 5). The Z chromosome exhibited similar values (D = − 0.027 ± 0.034; Table S11, Additional file 1, comparison 5). Furthermore, no 10-kb window trees from our phylogenetic analyses grouped H. hermathena and H. charithonia, and only 24 of 21,247 autosomal trees (0.11%) and no Z trees grouped H. charithonia with H. erato, H. himera, and H. hermathena. The proportion of autosomal trees grouping these four taxa was similar to the proportion of trees grouping H. sara (0.09%) or H. demeter (0.08%) with H. erato, H. himera, and H. hermathena. We therefore found no genome-wide evidence that H. hermathena was formed by hybridization between H. charithonia and H. erato.
Mimetic H. hermathena vereatta originated via introgression from H. erato
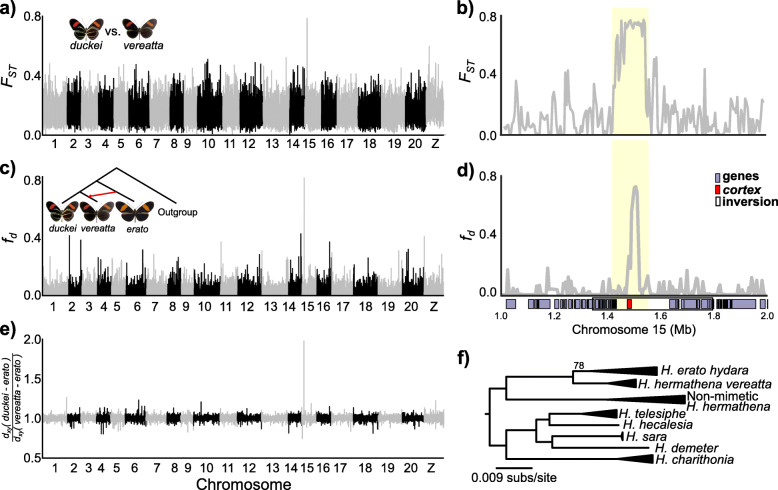
Heliconius hermathena is thought to be one of the rare examples of a non-mimetic Heliconius species because its color pattern does not resemble the color pattern of any other co-occurring species [25, 27, 59]. The exception is H. hermathena vereatta, which lacks yellow patterns and mimics co-occurring H. erato hydara and H. melpomene melpomene near the town of Faro [25]. The mimetic color pattern evolved and is maintained in Faro despite frequent interbreeding between H. h. duckei and H. h. vereatta, suggesting that strong natural selection for mimicry preserves the mimetic form (Fig. 1). We therefore searched for loci associated with yellow presence/absence by scanning for genome regions with high allele frequency differences between vereatta and duckei. We found a single narrow peak of FST on chromosome 15 containing the known color patterning gene cortex. This region showed four-fold higher differentiation than the genome-wide average (Fig. 6). Cortex is part of the H. erato Cr (H. melpomene Yb) locus that controls the presence of the yellow hindwing band across Heliconius, and cortex expression patterns in pupal wing discs prefigure adult melanic patterns [10]. Heliconius h. duckei contained significantly lower π (i.e., higher homozygosity) in the FST peak relative to H. h. vereatta (0.0009 vs. 0.0057), suggesting that the vereatta cortex allele is dominant to the duckei allele and melanizes the yellow patterns typical of the other H. hermathena subspecies.
Fig. 6.
Evolution of the mimetic Heliconius hermathena color pattern. a Genome-wide FST between mimetic H. h. vereatta and non-mimetic H. h. duckei, calculated in non-overlapping 10-kb windows. bFST between H. h. vereatta and H. h. duckei at the chromosome 15 peak, calculated in 5-kb windows (500-bp step). c Genome-wide fd tests for introgression from H. erato hydara into H. h. vereatta, calculated in non-overlapping 10-kb windows. H. melpomene was used as an outgroup. dfd at the chromosome 15 peak. The coordinates of erato-sara clade inversion are also shown (see the “Results” and “Discussion” sections and Fig. S17). e Relative divergence between H. h. duckei and H. h. vereatta to H. erato. a, b Calculated using SNP calls relative to the H. hermathena reference genome while c–e calculated using SNP calls relative to the H. melpomene reference genome to reduce reference bias. For consistency, all results are plotted relative to the H. melpomene reference genome, and all coordinates and gene models are based on the H. melpomene genome (the “Materials and methods” section). f Maximum likelihood tree based on the variation in the cortex region with high FST between mimetic and non-mimetic H. hermathena (highlighted yellow in b and d). Collapsed clades contain multiple individuals of the same species/subspecies. Only branches with less than 100% bootstrap support are labeled. H. erato hydara image: Field Museum of Natural History 124251, CC-BY-NC
Heliconius erato populations have evolved similar color patterns by sharing alleles via hybridization distribution, which mediates mimicry between local H. erato and H. melpomene races [12, 60]. This same process operates in H. melpomene, but mimicry between melpomene-silvaniform clade species has also frequently been mediated by introgression of color patterning alleles between co-mimetic species [15, 18, 61–63]. We next tested whether H. h. vereatta and H. erato converged on their shared color pattern independently or via introgression (Fig. 6d). We specifically tested whether H. hermathena received its melanic cortex allele from sympatric H. erato hydara using the fd statistic (Fig. 6d) [64]. The patterns of fd mirrored the patterns of FST between H. h. duckei and H. h. vereatta: we found a single narrow peak of fd just upstream of cortex, suggesting that the H. h. vereatta color pattern was formed by introgression of a cortex allele from H. erato into a H. h. duckei-like ancestor (Fig. 6c, d). Consistent with this hypothesis, divergence (dxy) between H. h. vereatta and H. erato was significantly lower in this region than genome-wide or between H. h. duckei and H. erato, and ML analysis of variation in this region grouped H. erato and H. h. vereatta (Fig. 6e, f).
While we detected no genome-wide evidence for hybridization between H. hermathena and H. charithonia, it is also possible that these species share color patterns because they shared color patterning alleles via hybridization. Alternatively, hybridization may be so ancient that its signal has been eroded by continued divergence. However, we found no fd signatures suggesting that cortex alleles have been shared between H. hermathena and H. charithonia (Figure S16). Interestingly, the ML tree in Fig. 6f showed that H. charithonia was placed outside a clade containing its sister species H. demeter and H. sara, and H. telesiphe and H. hecalesia. Edelman et al. [20] showed that there was at least one ancient hybridization event between the ancestors of H. sara/H. demeter and H. hecalesia/H. telesiphe that transferred a large (~ 500 kb) inversion centered on cortex. The tree in Fig. 6f suggests that H. charithonia carries the ancestral standard allele in this region, and, indeed, we found (1) two large H. charithonia scaffolds (126 kb and 263 kb) that span the inversion breakpoints in the standard orientation and (2) increased dxy between H. charithonia and species carrying the inversion in this region (Figure S17, Additional file 2). Thus, H. erato, H. himera, H. hermathena, and H. charithonia share ancestral non-inverted haplotypes in the cortex region, perhaps suggesting that H. hermathena and H. charithonia yellow patterning alleles arose independently or are controlled by ancestral variation in this region. The mimetic H. h. vereatta color pattern then appears to be a derived phenotype mediated by introgression of a small upstream region of cortex from co-occurring H. erato.
Discussion
Over 150 years of Heliconius research has provided many key insights into the genetic basis of color patterns, the evolution of Müllerian mimicry, and the fluidity of species boundaries [3, 4, 20, 56, 65]. However, detailed investigations of unique populations continue to provide insight into adaptation, speciation, and the genomic patterns that these processes leave behind. Here, we used whole-genome sequencing data to test hypotheses about the effects of population structure and hybridization on the genetic health and color pattern evolution of two rare Heliconius species.
Influence of population fragmentation and recent bottlenecks on the genetic health of H. nattereri and H. hermathena
Population fragmentation and reduced gene flow can severely reduce genetic diversity within species [66]. The distributions of white sand habitats and Atlantic Forests have significantly shrunk since the end of the last glacial maximum ~ 12,000 years ago, fragmenting populations of species that inhabit them. This habitat loss and fragmentation have only increased over the last century as logging and climate change have rapidly taken their toll [67–70]. It is therefore critical to understand how historical fragmentation and habitat loss have affected the current genetic health of the organisms that inhabit these threatened environments and to use those data to estimate the vulnerability of those species to contemporary habitat loss.
Heliconius nattereri has been found in only four areas of Atlantic Forest that are each surrounded by large areas of unsuitable habitat and deforested areas, leading to the current patchy distribution of this threatened species [21–23, 71]. While local population densities can be high, most collection efforts have produced few specimens from any of these four sites. Interestingly, H. nattereri carries a perhaps surprising amount of genetic diversity (π) considering its small census sizes: 36% of the diversity found in its widespread relative H. melpomene and about the same as the cosmopolitan fly Drosophila melanogaster [72]. However, much of the genetic variation in H. nattereri appears to be deleterious (Fig. 3). Furthermore, mean Tajima’s D, the high variance of D, and historical population size estimates suggest that H. nattereri experienced a recent population bottleneck (Fig. 2). Combined with the continued loss of Atlantic Forest habitats through climate change and deforestation [70], limited host plant ranges and extended larval and pupal development times relative to their competitors, it seems likely that Brown [22] (p. 53) was correct when he wrote that “H. nattereri seems to be a very primitive, inflexible, sensitive, and evidently declining species, with a rather uncertain future.”
In contrast to H. nattereri, H. hermathena is widespread across the Amazon River basin but inhabits unique white sand ecosystems [25–27]. We found that H. hermathena population size and deleterious mutation load estimates are surprisingly similar to those in H. nattereri (Figs. 2 and 3), suggesting that their patchy distribution, strong isolation between patches, and recent population bottleneck have significantly reduced their genetic health. Brown and Benson [25] suggested that H. hermathena was once widespread and then fragmented into isolated populations due to forest growth after the last glacial maximum, ~ 12,000 years ago. Alternatively, H. hermathena may have originated in a single patch and then dispersed to other patches. While our data do not allow us to definitively distinguish between these two scenarios, they suggest the former scenario. Heliconius hermathena population sizes began to diverge ~ 10,000 years ago, suggesting that they were part of a single population until that point (Fig. 2), but those populations are strongly genetically differentiated today (Fig. 5). Genetic diversity is about twice as high in populations north of the Amazon, with the center of diversity near Manaus and Presidente Figueiredo (Figure S14, Additional file 2), yet all populations have similarly low levels of π and Tajima’s D signatures of recent bottlenecks (Fig. 2). If H. hermathena recently spread from a single location, we may expect π to decrease with increasing distance from the source population. Phylogenetic reconstructions suggested that the Faro population is an outgroup to the remainder of the H. hermathena populations (Fig. 5c; Figure S15, Additional file 2).
Low migration rates among populations are likely due to this species’ habitat fidelity and generally low dispersal (a few hundred meters [25];). Dispersal between patches does appear to occur at low rates, though as a single H. h. sheppardi individual from Presidente Figueiredo appears to be slightly admixed with both H. h. sheppardi from Manaus and H. h. vereatta from Faro (according to Fig. 5; Figure S13, Additional file 2). Furthermore, red ventral hindwing spots characteristic of the Faro populations are occasionally found in H. h. hermathena on the southern bank of the Amazon [25]. Overall, the strong population structure we observe in H. hermathena has likely led to the current low effective population sizes and significant deleterious mutation loads (Figs. 4 and 5).
Evolution of non-mimetic H. hermathena
Hybridization between closely related species allows gene flow that may transfer beneficial alleles between species or even produce entirely new species [32, 73]. Heliconius butterflies have provided key insights into the role of hybridization in creating and blurring species boundaries [15, 16, 20, 29]. At least three species (H. heurippa [29], H. hecalesia [20, 37], and H. hortense/H. clysonymus [19]) appear to be hybrid species. Importantly, evidence for the hybrid origin of H. hortense and the ancient hybridizations in the erato-sara clade were only uncovered with whole transcriptome and whole genome data, respectively, highlighting the importance of using genome-wide data to test hypotheses about hybridization. Despite speculation based on their unique color pattern combination and discordant placements in analyses of mitochondrial and nuclear markers [30–32], we found no evidence from whole-genome alignments or polymorphism data that H. hermathena was formed by hybridization between H. charithonia and H. erato. Our results thus reinforce the need to test for introgression with genomic data.
The yellow color patterns of H. hermathena and H. charithonia are therefore controlled by (1) variation evolved independently in the two lineages, (2) shared ancestral variation, or (3) alleles shared via small-scale introgression. We found little evidence for introgression between H. hermathena and H. charithonia, especially near known color patterning genes, including cortex (Figure S11, Additional file 2). The cortex region itself contains multiple color patterning genes, as evidenced by the number of genetically separable color patterning loci in this region (e.g., H. erato Cr, H. melpomene Yb, Sb, and N) and the complex color pattern switches that it controls in H. numata [65, 74]. The fact that H. charithonia and H. hermathena carry the ancestral, non-inverted allele might suggest that their yellow patterns are controlled by ancestral variation (Fig. 6f). In any case, these findings raise the further question of why this color pattern has been maintained over such a long period of time, as these two species diverged ~ 13 million years ago [36]. Non-mimetic H. hermathena primarily inhabit shady undergrowth and fly erratically between shadowy patches, leading Brown and Benson [25] to suggest that the color pattern and flash-disruptive flight behavior that defies “visual following for any distance … would probably be a near optimal means of avoiding predation.” The effectiveness of the aposematic, but non-mimetic color pattern seems to be supported by the narrow distribution of mimetic H. h. vereatta in the southern Faro population, the only location where H. hermathena overlaps the distributions of its co-mimics H. erato hydara and H. melpomene melpomene. On a finer scale, mimetic H. h. vereatta prefers more open habitats on the edges of fields where its co-mimics also fly [25] (RRR, personal observation).
Müllerian mimicry mediated by adaptive introgression of a color pattering allele in the erato-sara clade
The evolution of the mimetic H. hermathena color pattern provides a unique example of phenotypic convergence between co-occurring species via sharing of color pattern alleles through small-scale introgression. Numerous examples of this process have been documented in the melpomene-silvaniform clade, including sharing of optix alleles between H. melpomene, H. timareta, and H. elevatus [11, 15, 61]; introgression of H. melpomene optix alleles to the silvaniform H. besckei [18]; introgression of aristaless1 alleles from H. cydno to H. melpomene [13]; introgression of a P supergene allele from H. pardalinus to H. numata [63]; and introgression of cortex alleles between H. melpomene and H. cydno [62]. However, this process has only recently been documented in the erato-sara clade. Van Belleghem et al. [12] and Lewis et al. [60] found evidence that H. erato races converged on similar color patterns via sharing of alleles of these same color patterning genes among populations, while Edelman et al. [20] found evidence for ancient introgression of a large inversion, centered on cortex, between the ancestors of H. sara/H. demeter and H. telesiphe/H. demeter. H. hermathena therefore provides a unique example of recent adaptive introgression of a small color patterning allele between two distinct erato-sara clade species. Our analyses suggest that an ancestral, non-mimetic H. h. duckei-like population hybridized with H. erato and that selection for mimicry maintained the H. erato cortex allele in southern Faro despite continued backcrossing of the novel mimetic H. hermathena with the non-mimetic ancestor (Fig. 6). The narrow interval of high FST, fd, and relative dxy between H. h. duckei and H. h. vereatta suggests that selection for mimicry is very strong in the southern Faro population and/or that hybridization occurred a long time ago. Divergence between the H. erato and H. h. vereatta cortex alleles is 0.0091 relative to the genome-wide average of 0.0153, providing a rough estimate that the introgression occurred ~ 1.5 million years ago (~ 60% of the total time since H. hermathena and H. erato diverged), although divergence in the introgressed region may have been affected by other factors such as natural selection, recombination, or different mutation rates.
Finally, many different loci epistatically interact to produce the final wing color pattern (e.g., [60]). H. h. vereatta and H. h. duckei forewing bands are noticeably redder than other H. hermathena populations, where the bands are more orange (Fig. 1). This could be due to the additional sequence evolution at other color patterning loci, or it is even possible that the H. hermathena/H. erato hybridization event introduced other, small-effect variants and loci other than cortex that produce these more subtle differences between the Faro subspecies and the remainder of H. hermathena populations. However, we do not find strong differentiation at the other major color patterning genes. The broad hump of slightly higher FST on chromosome 10 is centered ~ 3 Mb to the right of WntA and is a collection of widely scattered windows. The 10-kb window containing WntA has FST of 0 (48 SNPs). The window with the highest FST on chromosome 10 (0.28, 104 SNPs) is 370 kb away from WntA. In contrast, the 10-kb windows containing cortex all have FST > 0.70 (> 62 SNPs).
Phylogenetic placement of Heliconius nattereri
Recent species tree reconstructions based on tens to thousands of gene trees have consistently found widespread discordance in the melpomene-silvaniform clade, suggesting that these species, especially the silvaniforms, frequently hybridize with each other [20, 36]. We found the same patterns of discordance among gene trees and between datasets after including our high-quality H. nattereri genome in these analyses, showing that the relationships in this clade remain unresolved [20]. Interestingly, the H. nattereri genome has also been shaped by the rampant gene flow among silvaniforms, which is reflected by the different placements of H. nattereri in species trees based on different datasets: summaries of autosomal gene trees placed H. nattereri as an outgroup to the remaining melpomene-silvaniform clade species (Fig. 1; Figure S7-S8, Additional file 2); Z-linked gene trees placed a clade of H. nattereri, H. numata, and H. besckei as an outgroup to the remaining species (Figure S9), while mtDNA places H. nattereri in the middle of a monophyletic silvaniform clade (Fig. 1). The addition of the H. nattereri, H. hecale, and H. elevatus genomes to the Edelman et al. [20] dataset therefore does little to resolve relationships within this intriguing clade. The Z exhibits generally greater differentiation and more resistance to gene flow than the autosomes, and analyses of the Z can provide better supported gene trees [16, 17, 37]. These observations suggest that, at the least, H. nattereri hybridizes much less frequently with the remaining silvaniforms than the other silvaniforms hybridize with each other. Hybridization within the silvaniforms affects H. nattereri, but perhaps less than the remaining silvaniforms. The mtDNA relationships may be due to mitochondrial capture from one of these hybridization events.
Conclusions
Heliconius butterflies have been studied for over 150 years because of their bold, aposematic color patterns that frequently mediate mimicry among distantly related species. Recent genomics studies of the widespread species H. melpomene and H. erato have provided exciting insights into the important roles that hybridization and introgression play in speciation and color pattern evolution. Our results provide a striking contrast to the patterns that have emerged from years of study focused on abundant and widespread species such as H. erato, H. melpomene, and their close relatives. We showed that both H. nattereri and H. hermathena have substantially reduced genetic diversity and high deleterious mutation loads relative to other Heliconius. These are the result of low effective population sizes, recent population bottlenecks, and pronounced population subdivision. Despite recurrent speculation based on its suggestive color pattern combination, we detect no genomic evidence that H. hermathena is a hybrid species. However, we do find that the one mimetic subspecies of H. hermathena acquired its mimetic phenotype via hybridization with H. erato. We thus provide a unique example of mimicry evolution caused by recent adaptive introgression across species boundaries. Our results further support the conclusion that this process is a general feature of Heliconius color pattern evolution when viewed with other recent analyses of intraspecific and ancient hybridization in the erato-sara clade, and well-documented introgression in the melpomene-silvaniform clade.
Materials and methods
Genome assemblies of H. hermathena and H. nattereri
We isolated DNA from the thorax of four wild-caught H. hermathena adult females, then pooled all four DNA samples before constructing sequencing libraries. DNA was pooled to produce a uniform sample with enough DNA to construct all paired-end and mate-pair libraries, which required 14 μg. We constructed Illumina paired-end (PE) libraries with insert sizes of 250 and 500 bp using the KAPA Hyper Prep Kit (KR0961 – v1.14) and 1 μg genomic DNA. We constructed mate-pair (MP) libraries with insert sizes of 2 kb, 6 kb, and 15 kb using the Nextera Mate Pair Library Prep kit (FC-132-1001) and 4 μg genomic DNA. We then pooled these libraries in a ratio of 50:18:10:17:4 and sequenced them 2 × 100 bp on a single lane of Illumina HiSeq 4000 (Table S2, Additional file 1). We trimmed low-quality regions and remaining adapters from raw PE reads using Trimmomatic v0.36 [75] and from MP libraries using the platanus_internal_trim tool from Platanus v1.2.4 [76]. Trimmed libraries were assembled and scaffolded using Platanus v1.2.4 (default settings) and the assembly polished using Redundans v0.13a (default settings) [77]. We removed scaffolds < 5 kb from the assembly, then generated species-specific repeat libraries and masked repeats using RepeatScout v1.0.5 and RepeatMasker v4.0.8, respectively, to produce the final H. hermathena reference genome assembly [78, 79]. We sequenced and assembled the H. nattereri genome using an identical protocol with genomic DNA extracted from thoraxes of two wild-caught males and two females (Tables S1-S2, Additional file 1). We calculated the frequency spectrum of canonical 21-mers in raw reads from the PE 250 bp libraries using jellyfish 2.2.3 [80], then input the resulting histograms into GenomeScope to estimate genome size, heterozygosity, and sequencing error rates [34].
In addition, we assembled a draft H. charithonia genome using publicly available data for H. charithonia charithonia from Puerto Rico (accessions SRR4032025 and SRR4032026) and Platanus version 2.0.2 [81]. We trimmed adapter sequences using TrimGalore v0.6.1 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and cutadapt v1.18 [82], then identified and removed overrepresented sequences using FastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and RemoveFastqcOverrepSequenceReads.py (last accessed 2019-11-25; available https://github.com/harvardinformatics/TranscriptomeAssemblyTools). GenomeScope analyses showed that SRR4032025 exhibited low heterozygosity (0.67%) and error rates (0.12%) relative to SRR4032026 (3% and 0.41%). The estimated genome size for H. charithonia from SRR4032025 was 303 Mb, similar to its close relatives (Table 1). We therefore assembled contigs using Platanus v2.0.2 and the processed reads from SRR4032025, then scaffolded and phased contigs using reads from both accessions. Finally, we removed scaffolds < 5 kb and identified and masked species-specific repeats using the pipeline outlined above.
We assessed the quality of our assemblies and other well-assembled nymphalid genomes using BUSCO v3 and the endopterygota gene set (2440 single-copy orthologs) from OrthoDB v9 [35, 83]. The accessions of the genome assemblies that we tested are as follows: B. anynana (RefSeq GCF_900239965.1), D. plexippus (GenBank GCA_000235995.2), H. erato demophoon v1 (www.lepbase.org [LepBase] v4), H. e. lativitta v1 (LepBase v4), H. melpomene v2.5 (LepBase v4), Hypolimnas misippus (GenBank GCA_008963455.1), J. coenia v1 (LepBase v4), L. arthemis (BioProject PRJNA556447), M. cinxia (GenBank GCA_000716385.1), and V. tameamea (GenBank GCA_002938995.1).
Finally, we assigned H. hermathena and H. nattereri scaffolds to the H. erato demophoon v1 and H. melpomene v2.5 assemblies, respectively, using RaGOO [84]. We used this chromosome assignment information in plotting chromosome-level plots and to identify sex-linked scaffolds.
Whole-genome alignments and genome-wide consensus phylogeny
We added our new H. nattereri, H. hermathena, and H. charithonia reference genomes to the whole-genome alignment generated by Edelman et al. [20] using the progressiveCactus aligner (commit 348815c from September 19, 2019) [38, 39]. We downloaded the Edelman alignment from Dryad (10.5061/dryad.b7bj832) and extracted all sequences using hal2fasta. We made three modifications to the Edelman alignment based on the mtDNA phylogeny and previous publications (see Figure S1, Additional file 2). First, we added H. hermathena to the branch leading from Anc17 to Anc21 to yield the subclade (((H. erato, H. himera), H. hermathena), H. hecalesia). Next, we added H. charithonia as sister to H. sara to yield the new subclade ((H. charithonia, H. sara), H. demeter). Finally, we re-aligned the silvaniform clade using a polytomy, including H. elevatus and H. hecale (available from LepBase v4) and H. nattereri. These guide trees simply aid the alignment of the sequences and do not greatly affect downstream analyses if the species are closely related [20].
We followed Edelman et al. [20] to infer species trees, using all 18 Heliconius species, as well as just the eight species from the erato clade or 10 species from the melpomene to improve and/or confirm resolution of relationships in those clades. We included Eueides tales in the full Heliconius dataset, H. melpomene in the erato dataset, and H. erato in the melpomene dataset as outgroups. We used the H. melpomene, H. erato, and H. melpomene genomes as reference genomes for these three datasets, respectively, when extracting alignments using the HAL toolkit.
Extensive gene flow among Heliconius species has resulted in a mosaic of gene genealogies across the genome. Edelman et al. [20] inferred gene trees for non-recombining regions, then summarized these gene trees using ASTRAL, which analyzes four-taxon groupings to provide support for each branch in the inferred species phylogeny. ASTRAL and other species tree inference methods assume that each gene tree reflects a single topology, an assumption that is violated if historical recombination has occurred within the gene or window being analyzed. To minimize the number of violations, we inferred phylogenies for small windows with weak evidence for recombination, then summarized them using ASTRAL-iii, similar to Edelman et al. [20]. Linkage disequilibrium is essentially gone within ~ 10 kb in H. melpomene.
For each of the three datasets (all Heliconius, erato clade, and melpomene clade), we extracted alignment blocks from all 10-kb windows from the reference genome, then filtered alignment blocks for single-copy coverage in all species (therefore excluding ambiguously aligned regions and taxon-specific duplications). We stripped all sites with a gap in any species and tested for recombination within each alignment using PhiPack [85], which uses patterns of variation to infer historical recombination events within an alignment. PhiPack reports the probability of NO recombination within the alignment; smaller p values suggest that there is stronger evidence for recombination within the window. We used all autosomal alignments containing sequences for all focal species, at least 1000 aligned bp, at least 100 phylogeny-informative sites, and a probability of recombination p > 1e−10. We chose these cutoffs to include a reasonable number of high-quality windows for each comparison. Figures S2 - S6 (Additional file 2) contain more detailed information on each dataset’s characteristics prior to filtering. These filters produced 8674 valid alignments for the Heliconius dataset, 22,430 windows for the erato clade, and 3601 windows for the melpomene clade. We inferred trees for each valid window using IQtree after selecting the best model using ModelFinder and estimated statistical support using 1000 ultrafast bootstraps. Finally, we inferred the species tree for each dataset using ASTRAL-iii using its exact algorithm [41].
The primary source of the discrepancy in the number of windows is the filter for evidence of recombination within the window, especially in the melpomene clade (Figure S5). We therefore repeated the Heliconius dataset analysis following Edelman et al. [20] using fully aligned blocks from coding and non-coding regions, which tend to be short and therefore have little or no evidence for recombination (Figure S3, S4, and S7, Additional file 2). We constructed trees using all autosomal alignments with at least 150 bp, at least 10 informative sites, and probability of recombination > 1e−4. The final coding and non-coding datasets consisted of 12,369 and 29,071 alignments, respectively. The ASTRAL trees are consistent with the 10-kb window trees and are presented in Figure S7 (Additional file 2).
Finally, we tested consistency between the ASTRAL method and BUCKy using the filtered alignments from the erato and melpomene clade datasets (Figure S9, Additional file 2). We inferred gene trees using MrBayes 3.2 under a GTR model with gamma-distributed rates with two runs of 1000,000 generations and then summarized the results using BUCKy 1.4.4 [86].
Heliconius mitochondrial genome assemblies
We downloaded 31 Heliconius species samples, including seven complete mitochondrial genomes, from NCBI (Table S3, Additional file 1). We used NOVOplasty to extract and assemble mtDNA reads from these accessions using bait sequences indicated in Table S3, a k-mer size of 39, and an expected size range of 12–22 kb [43]. We aligned all mtDNA genomes using the multiple circular sequence alignment program MARS [87]. Finally, we inferred the mtDNA phylogeny using IQtree v1.6.12 using the best-fit model determined using ModelTest and estimated support using 1000 ultrafast bootstraps [42, 88, 89].
Whole-genome resequencing
We collected adult H. hermathena specimens from the states of Amazonas and Pará in northern Brazil between 2010 and 2017 (Fig. 1; Table S1, Additional file 1). We collected adult H. nattereri from the states of Espírito Santo and Bahia in 2015 and 2017. We extracted genomic DNA from thorax tissues from each of these butterflies using a phenol-chloroform DNA extraction protocol, then constructed paired-end libraries using a KAPA Hyper Prep Kit (KAPA Biosystems) and sequenced them to ~ 20X coverage using 2 × 100 bp Illumina HiSeq 2500 (Table S1, Additional file 1). Note that we also re-sequenced H. nattereri individuals that we used for genome assembly because (1) these samples were extremely rare and (2) we had to pool DNA extractions to construct the libraries for genome sequencing and assembly. We did not re-sequence H. hermathena samples used for genome assembly because we had many additional samples (Table S1, Additional file 1).
SNP calling pipeline
We used five different SNP call sets for our analyses: (1) H. nattereri samples relative to the H. nattereri reference genome, (2) H. hermathena samples relative to the H. hermathena reference genome, (3) melpomene-silvaniform clade samples relative to the H. melpomene v2.5 reference genome, (4) erato-sara clade samples relative to the H. erato demophoon v1 reference genome, and (5) erato-sara clade samples relative to the H. melpomene v2.5 reference genome. All SNP call sets were generated using the same pipeline. We trimmed adapters and low-quality regions from raw re-sequencing reads using TrimGalore 0.6.1 and cutadapt v1.18 [82], then removed reads containing overrepresented sequences (identified using FastQC). We then mapped reads to the appropriate reference genome using Bowtie2 v2.3.0-beta7 with the parameter “--very-sensitive-local” [90]. We marked PCR duplicate reads using PicardTools v2.8.1 and realigned around indels using the Genome Analysis ToolKit’s (GATK, v3.8) RealignerTargetCreator and IndelRealigner. Finally, we called SNPs using the GATK UnifiedGenotyper with default settings except the heterozygosity prior was set to 0.02, minimum allowable base quality scores were set to 30, and minimum mapping quality set to 20 [91].
SMC++ analyses
We inferred changes in ancestral population sizes using the sequentially Markovian coalescent implemented in SMC++ v1.15.3 [47]. We used H. hermathena SNP calls relative to the H. hermathena reference genome and H. nattereri SNP calls relative to the H. nattereri genome for these analyses. We filtered out genotypes with quality < 10, then removed SNP sites with any missing genotypes or multiple alternate alleles. We also excluded sex-linked sites from this analysis. We estimated historical population sizes using the SMC++ estimate function, using all samples as distinguished lineages, setting the thinning parameter k = 1000 × ln (no. of samples in analysis × 2), using a mutation rate of 2.9e−9 per bp per generation [45, 47] and assuming four generations per year. We estimated the split time of the northern and southern H. nattereri populations using the SMC++ split function. Previous studies of historical Heliconius population sizes used PSMC (e.g., [17]). We provide PSMC results for H. nattereri and a sample of H. hermathena in Figure S12 (Additional file 2) for comparison.
H. nattereri and H. hermathena mutation load analysis
We used SNP calls from melpomene-silvaniform clade samples aligned to the H. melpomene v2.5 reference genome or erato-sara clade samples aligned to the H. erato demophoon v1 reference genome for this analysis. We calculated nucleotide diversity per site (π), Tajima’s D, and FST in 10-kb non-overlapping windows using VCFtools 0.1.15 [92]. For the H. nattereri mutation load analysis, we determined the ancestral state for each site using H. cydno and H. atthis data, requiring all eight samples called and agreement from a majority of alleles (see Table S4, Additional file 1, for sample information). We annotated the effects and impacts of derived mutations on H. melpomene v2.5 gene models using snpEff 4.3T [51], then summarized fixed and polymorphic derived alleles in H. nattereri, H. melpomene melpomene (French Guiana), H. m. rosina (Panama), and H. pardalinus. We chose six random Santa Teresa H. nattereri samples and six random H. m. rosina samples for this analysis to match the number of H. pardalinus samples. We considered an allele to be fixed in a population if it was present in all individuals from that population. We considered mutations with the following snpEff annotations as “neutral”: synonymous_variant, intron_variant, and NONE. We considered mutations with the following annotations as “deleterious”: missense_variant, start_lost, and stop_gained. We analyzed a subset of H. hermathena populations similarly, including six random individuals from each H. h. sheppardi population, H. h. duckei, H. h. vereatta, and H. h. hermathena (Santarém). We analyzed the six French Guiana H. erato hydara individuals for comparison. We polarized SNP states using H. clysonymus, H. hortense, and H. charithonia (2 samples). We required all four samples to be called and a majority of alleles to agree in order to assign an ancestral state to a particular SNP site.
Heliconius genetic differentiation by distance
We used SNP calls from melpomene-silvaniform clade samples relative to the H. melpomene reference genome and erato-sara clade samples relative to the H. erato reference for this analysis. For each species H. melpomene, H. nattereri, H. hermathena, and H. erato, we estimated the weighted, genome-wide average FST [93] between all pairs of sampled populations using VCFtools v0.1.15. Negative FST estimates were set to 0. We then estimated geodesic distances between populations based on latitude and longitude coordinates using the distGeo() function of the geosphere R package. If a population contained three or more individuals, we used the midpoint calculated using a geographic midpoint calculator (http://geomidpoint.com/) as the population location. In these cases, the midpoint of the segment connecting both populations was used for geodesic distance estimation.
H. hermathena population structure
We estimated the ancestry of H. hermathena individuals using ADMIXTURE v1.3.0 [53]. We used H. hermathena SNP calls relative to the H. hermathena reference genome for this analysis. We first filtered all genotypes with quality < 10, then included only sites with no missing genotypes and minor allele frequency greater than 5%. Finally, we performed linkage disequilibrium-based pruning using PLINK 1.90 [89], keeping only variants with r2 < 0.1 in sliding 50-site windows (10-site step). This resulted in 236,596 sites in the final filtered dataset. The most likely number of clusters was selected based on cross-validation error (CV) and the value of k that minimizes the residuals [94]. We ran ADMIXTURE with cross-validation for values of k from 2 to 10.
We visualized spatial patterns in genetic diversity and regions of gene flow using the program EEMS (estimated effective migration surfaces) [54]. We used the same SNP set generated for ADMIXTURE for this analysis. We calculated the dissimilarity matrix using the bed2diffs program included in the EEMS package. An outer coordinate file was generated using the add path method in the Google Earth Pro (v. 7.3.2.5776) tool. We tested several deme sizes (300, 450, 750) using the runeems_snps version of eems. We set the number of demes to 750 and defined the outer boundary of the region by the polygon (in latitude-longitude coordinates of the focal populations; Table S1, Additional file 1). This number of demes provided fine resolution. We ran the MCMC across 10 independent 10,000,000-step runs logged every 2000 steps and 2,500,000 set as burn-in. The results were combined across the independent analyses using the reemsplots R package (available with EEMS), and convergence of runs was visually assessed (Figure S18, Additional file 2). Using this package, we plotted the geographic distance and genetic dissimilarity across demes and generated surfaces of effective diversity (q; Figure S14, Additional file 2) and effective migration rates (m; Fig. 5).
H. hermathena Bayesian concordance tree construction
We aligned reads and called SNPs for each H. hermathena sample and three H. erato hydara samples (SRR4032061, SRR4032068, and SRR4032074) using the H. hermathena reference genome and the pipeline outlined above. We used the GATK’s FastaAlternateReferenceMaker to generate a consensus fasta for each sample, including IUPAC ambiguities at heterozygous sites. We then split the genome into non-overlapping 10-kb windows, picked 5000 random autosomal windows, and estimated trees for each window using MrBayes 3.2 with a GTRγ substitution model and 6 rate categories [95]. Each MrBayes analysis consisted of two runs of 1000,000 generations and samples every 500 generations. We then used BUCKY 1.4.4 to summarize trees from all 5000 runs, discarding the first 25% of trees from each run, see Figure S15 (Additional file 2).
H. hermathena mtDNA haplotype network
We assembled complete mtDNA sequences for all H. hermathena samples and constructed a haplotype network to infer population structure (Fig. 5d). We performed quality control of raw Illumina reads using FastQC and MultiQC [96]; adapters were removed using Trim Galore. Genomic data was subsequently filtered for mtDNA reads using the package MIRAbait of the MIRA assembler v. 4.0, using the mitogenome of Heliconius melpomene rosina (NCBI Accession Number KP153600) as a reference, and k-mer size 15, since higher k-mer values resulted in poorer mitogenome assembly. Analyses were run in the Galaxy platform [97] with default parameters for each program (except for k-mer value in MIRAbait). We imported all files into Geneious v. 10, where the mitogenome of Heliconius hermathena was assembled (using the Map to Reference command, and H. melpomene rosina mitogenome as reference), based on the paired reads of all individuals. H. hermathena sequencing reads from each individual were then used in a Map to Reference procedure against H. hermathena mitogenome to assemble all individual mitogenomes. We then aligned mitogenomes with the MAFFT [98] plugin in Geneious v. 10 using the FFT-NS-i × 1000 algorithm. Protein coding genes (PCGs) were extracted and concatenated using the Concatenate sequences or alignments command in Geneious resulting in the final alignment comprising all individuals; only the PCGs were used in subsequent analyses. Finally, we visually checked the alignment for possible errors with AliView [99].
The final alignment was imported into DnaSP v. 6 [100] to assess π. Each individual in the alignment was assigned to their specific subpopulation defined by its sampling locality (Data > Define Domain Set). Overall and intrapopulation π values were obtained with the DNA Polymorphism command, first for all subpopulations combined and then for each of the defined subpopulations separately. Population differentiation was measured by overall and pairwise FST using Arlequin v. 3.5 [101] computed for 100 permutations and calculating the distance matrix for pairwise differences. A haplotype network for the mitogenome sequences of all individuals was estimated using the program PopArt v. 1.7 [102], with the median-joining network algorithm (epsilon = 0), see Table S11 (Additional file 1) and Fig. 5d.
Analysis of H. hermathena hybridization and mimicry locus
We searched for evidence of hybridization using the D statistic [57, 58]. We processed and aligned reads and called SNPs using data from H. erato hydara (French Guiana), H. hermathena vereatta, H. hermathena duckei, H. hermathena sheppardi, H. hecalesia, H. telesiphe, H. sara, H. demeter, H. charithonia, and H. melpomene nanna and the pipeline detailed above, but mapped all reads to the H. melpomene v2.5 reference genome assembly to minimize reference bias (Tables S1 and S3, Additional file 1). We randomly sampled six individuals from populations with more than that number. We then calculated D in 10-kb non-overlapping autosomal windows using Simon Martin’s genomics_general toolkit available at https://github.com/simonhmartin/genomics_general (commit 3e9281b from September 24, 2019). Values shown in Table S12 (Additional file 1) are the average and standard deviation (calculated using block jackknifing with 95% of the data).
We calculated FST between H. h. vereatta and H. h. duckei in 10-kb non-overlapping (genome-wide) or 5-kb sliding (500-bp step, focal region) windows using VCFtools 0.1.15. We used variant calls from these subspecies relative to the H. hermathena reference assembly. We used the full D statistic call set described in the previous paragraph to test if mimetic H. hermathena alleles originated via introgression from H. erato by calculating the fd in 10-kb non-overlapping windows (genome-wide) and 5-kb sliding windows (500-bp step; focal region) using the tree (((H. h. duckei, H. h. vereatta), H. erato), H. melpomene). Negative fd values were set to 0 before plotting, as these are meaningless [64]. We used the same D statistic callset to calculate dxy in 10-kb non-overlapping windows using the genomics_general toolkit. To be consistent, we plotted all Fig. 6 results relative to the H. melpomene v2.5 reference genome. We assigned H. hermathena scaffolds to H. melpomene scaffolds using RaGOO [84], then used this information during plotting with the gwplotting R package available at https://github.com/nwvankuren/gwplotting. We also used this package to plot H. melpomene gene models.
We reconstructed a maximum likelihood tree for the 100 kb peak of FST between duckei and vereatta using IQtree and the best model chosen by ModelTest, including 1000 ultrafast bootstraps to estimate support. For this analysis, we extracted alternate FASTAs for each sample using the GATK FastaAlternateReferenceMaker, including IUPAC ambiguities at heterozygous sites, then extracted the focal region from those FASTAs as an alignment using samtools faidx.
Supplementary information
Additional file 1: Table S1. Sample and sequencing information for Brazilian Heliconius hermathena and H. nattereri generated in this study. Table S2.Heliconius hermathena and H. nattereri genome sequencing data. Table S3. Samples used for mitochondrial genome assemblies using NOVOplasty (Dierckxens et al., 2017, Nuc Acids Res). Table S4. Sample information for other Heliconius species used in this study. Table S5. Summary statistics of nucleotide diversity per site (pi) and Tajima’s D calculated in 100 kb genome-wide. Table S6. Numbers of substitutions in Heliconius nattereri and relatives. Table S7. Numbers of substitutions in Heliconius hermathena and relatives. Table S8. Whole genome mean FST and 95% empirical confidence interval between H. hermathena populations. Table S9. Whole genome mean FST and 95% empirical confidence interval between H. erato populations. Table S10. Whole genome mean FST and 95% empirical confidence interval between H. melpomene populations. Table S11. Nucleotide diversity, overall and pairwise FST estimated for H. hermathena mitochondrial genomes. Table S12. Patterson’s D statistics calculated in 10 kb windows across all autosomes. H. erato samples are from French Guiana, the geographically closest population to H. hermathena.
Additional file 2: Figure S1. Guide trees used for progressiveCactus alignments in Edelman et al. (2019) and this study. Figure S2. Statistics on unfiltered alignments from the Heliconius dataset of 10 kb windows. Figure S3. Statistics on unfiltered alignments from the Heliconius dataset of coding sequence blocks. Length: length of the raw alignment in base pairs. Figure S4. Statistics on unfiltered alignments from the Heliconius dataset of non-coding sequence blocks. Length: length of the raw alignment in base pairs. Figure S5. Statistics on unfiltered alignments from the melpomene clade dataset of 10 kb windows. Figure S6. Statistics on unfiltered alignments from the erato clade dataset of 10 kb windows. Length: length of the raw alignment in base pairs. Figure S7. ASTRAL trees based on filtered a) coding and b) non-coding blocks from Heliconius dataset. Figure S8. ASTRAL trees based on filtered autosomal 10 kb window alignments from a) the erato clade and b) the melpomene clade. Figure S9. ASTRAL trees based on filtered Z-linked 10 kb window alignments from a) the erato clade and b) the melpomene clade. Figure S10. BUCKy cladograms based on filtered 10 kb window alignments from a) the erato clade and b) the melpomene clade. Figure S11. Predominant topologies for erato clade 10 kb autosomal windows. The top 20 topologies accounted for 80% of all windows. Figure S12. Historical population size estimates for a) H. hermathena and b) H. nattereri using PSMC. Figure S13. ADMIXTURE analysis of H. hermathena polymorphism data for k in 2:10. Figure S14. EEMS diversity estimates for Heliconius hermathena. Figure S15.H. hermathena Bayesian concordance tree based on 5000 autosomal 10 kb windows, showing all individuals. Figure S16. Tests for introgression between Heliconius hermathena and H. charithonia. Figure S17. Divergence (dxy) between Heliconius sara and other erato clade species in the chromosome 15 inversion and 250 kb flanking regions and ocations of scaffolds that mapped to the inversion breakpoints. Figure S18. Convergence of 10 independent EEMS runs.
Acknowledgements
We thank the Brazilian government for the permission to collect samples under SISBIO/ICMBio permits 49941-1, 32853-3, and 10438-1. All H. hermathena and H. nattereri samples are registered in SISGEN under accessions ABE5972 and A951B58. We thank the Amazon people that helped us overcome local challenges, and M Brito, W Silva, V Becker, M Paluch, and F Barreto for helping with the collections in Bahia and Espírito Santo.
Authors’ contributions
DM performed the sequencing, analyzed the data, and wrote the manuscript. NWV analyzed the data, and wrote the manuscript. RRR collected the samples. PG and MMB analyzed the data. MBL collected the samples and performed the sequencing. KLSB performed the sequencing and analyzed the data. AVLF conceived the project and provided the samples. MZC conceived the project and collected the samples. MRK conceived the project and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the following grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), National Science Foundation (NSF), and National Institutes of Health (NIH) grants: CNPq grants 302585/2011-7 and 303834/2015-3, FAPESP grants 2011/50225-3 and 2012/50260-6, and NSF grants DEB-1241056 and DEB-1256742 to AVLF; CNPq grant 400242/2014-1 to MZC; RedeLep-SISBIOTA-Brasil/CNPq grant 563332/2010-7 to MZC and AVLF; CNPq Postdoctoral Fellowship 163700/2017-6 and FAPESP grant 2012/16266-7 to KLSB; FAPESP grant 2011/00417-3 to MMB; and NSF grant IOS-1452648 and NIH grant R35 GM131828 to MRK. This research was also supported by the US Agency for International Development/National Academy of Sciences PEER grant AID-OAA-A-11-00012 to AVLF. The funding agencies had no role in the design, data collection, analysis, or interpretation of the data presented here.
Availability of data and materials
Genome assemblies and sequencing data generated in this study are available at the NCBI associated with BioProjects PRJNA596795 [103] and PRJNA596801 [104]. Genome assemblies are available at the NCBI under BioProjects PRJNA596796 [105] and PRJNA596794 [106].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Darli Massardo and Nicholas W. VanKuren contributed equally to this work.
Contributor Information
Nicholas W. VanKuren, Email: nvankuren@uchicago.edu
Marcus R. Kronforst, Email: mkronforst@uchicago.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12915-020-00797-1.
References
- 1.Bates HW. Contrubutions to an insect fauna of the Amazon valley (Lepidoptera: Heliconidae) Trans Linn Soc London. 1862;23:495–566. [Google Scholar]
- 2.Brown KS. The biology of Heliconius and related genera. Annu Rev Entomol. 1981;26:427–456. [Google Scholar]
- 3.Kronforst MR, Papa R. The functional basis of wing patterning in Heliconius butterflies: the molecules behind mimicry. Genetics. 2015;200:1–19. doi: 10.1534/genetics.114.172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merrill RM, Dasmahapatra KK, Davey JW, Dell’Aglio DD, Hanly JJ, Huber B, et al. The diversification of Heliconius butterflies: what have we learned in 150 years? J Evol Biol. 2015;28:1417–1438. doi: 10.1111/jeb.12672. [DOI] [PubMed] [Google Scholar]
- 5.Hikl A-L, Krenn HW. Pollen processing behavior of Heliconius butterflies: a derived grooming behavior. J Insect Sci. 2011;11:1–13. doi: 10.1673/031.011.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurman TJ, Brodie E, Evans E, McMillan WO. Facultative pupal mating in Heliconius erato: implications for mate choice, female preference, and speciation. Ecol Evol. 2018;8:1882–1889. doi: 10.1002/ece3.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333:1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- 8.Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, et al. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci. 2012;109:12632–12637. doi: 10.1073/pnas.1204800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadeau NJ, Pardo-Diaz C, Whibley A, Supple MA, Saenko SV, Wallbank RWR, et al. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature. 2016;534:106–110. doi: 10.1038/nature17961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallbank RWR, Baxter SW, Pardo-Diaz C, Hanly JJ, Martin SH, Mallet J, et al. Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 2016;14:e1002353. doi: 10.1371/journal.pbio.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Belleghem SM, Rastas P, Papanicolaou A, Martin SH, Arias CF, Supple MA, et al. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 2017;1:1–12. doi: 10.1038/s41559-016-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westerman EL, VanKuren NW, Massardo D, Tenger-Trolander A, Zhang W, Hill RI, et al. Aristaless controls butterfly wing color variation used in mimicry and mate choice. Curr Biol. 2018;28:3469–3474. doi: 10.1016/j.cub.2018.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Counterman BA, Araujo-Perez F, Hines HM, Baxter SW, Morrison CM, Lindstrom DP, et al. Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in Heliconius erato. PLoS Genet. 2010;6:e1000796. doi: 10.1371/journal.pgen.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasmahapatra KK, Walters JR, Briscoe AD, Davey JW, Whibley A, Nadeau NJ, et al. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kronforst MR, Hansen MEB, Crawford NG, Gallant JR, Zhang W, Kulathinal RJ, et al. Hybridization reveals the evolving genomic architecture of speciation. Cell Rep. 2013;5:666–677. doi: 10.1016/j.celrep.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Dasmahapatra KK, Mallet J, Moreira GRP, Kronforst MR. Genome-wide introgression among distantly related Heliconius butterfly species. Genome Biol. 2016;17:25. doi: 10.1186/s13059-016-0889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Leon-Ricardo BX, van Schooten B, Van Belleghem SM, Counterman BA, McMillan WO, et al. Comparative transcriptomics provides insights into reticulate and adaptive evolution of a butterfly radiation. Genome Biol Evol. 2019;11:2963–2975. doi: 10.1093/gbe/evz202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edelman NB, Frandsen PB, Miyagi M, Clavijo B, Davey J, Dikow RB, et al. Genomic architecture and introgression shape a butterfly radiation. Science. 2019;366:594–599. doi: 10.1126/science.aaw2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown KS. Rediscovery of Heliconius nattereri in eastern Brazil. Entomol News. 1970;81:129–140. [Google Scholar]
- 22.Brown KS. The heliconians of Brazil (Lepidoptera: Nymphalidae). Part III. Ecology and biology of Heliconius nattereri, a key primitive species neat extinction, and comments on the evolutionary development of Heliconius and Eueides. Zool Sci Contrib New York Zool Soc. 1972;57:41–69. [Google Scholar]
- 23.Cardoso MZ, Brito MR, Lion MB. Confirmation of the northermost population of the endangered butterfly Heliconius nattereri C. Felder & R. Felder, 1865 (Lepidoptera: Nymphalidae). Biota Neotrop. 2017;17..
- 24.Ministerio do Meio Ambiente. Lista Nacional das Espécies Ameaçadas de Extinção. Diário Oficial da União, Seção I. 2014;245:121–44.
- 25.Brown KS, Benson WW. Evolution in modern Amazonian non-forest islands: Heliconius hermathena. Biotropica. 1977;9:95–117. [Google Scholar]
- 26.Seixas RR, Santos SE, Okada Y, Freitas AVL. Population biology of the sand forest specialist butterfly Heliconius hermathena hermathena (Hewitson) (Nymphalidae: Heliconiinae) in Central Amazonia. J Lepid Soc. 2017;71:133–140. [Google Scholar]
- 27.Freitas AVL, Ramos RR, Silva-Brandão KL, Coutouné N, Magaldi LM, Pablos JL, et al. A new subspecies of Heliconius hermathena (Nymphalidae: Heliconiinae) from southern Amazonia. Neotrop Entomol. 2019;48:467–475. doi: 10.1007/s13744-018-0658-8. [DOI] [PubMed] [Google Scholar]
- 28.Hewitson WC. Exotic butterflies, vol. 1, Heliconia II. London: John van Voorst; 1853. [Google Scholar]
- 29.Mavárez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 30.Beltrán M, Jiggins CD, Brower AVZ, Bermingham E, Mallet J. Do pollen feeding, pupal-mating and larval gregariousness have a single origin in Heliconius butterflies? Inferences from multilocus DNA sequence data. Biol J Linnean Soc. 2007;92:221–239. [Google Scholar]
- 31.Mavárez J, Linares M. Homoploid hybrid speciation in animals. Mol Ecol. 2008;17:4181–4185. doi: 10.1111/j.1365-294x.2008.03898.x. [DOI] [PubMed] [Google Scholar]
- 32.Jiggins CD, Salazar C, Linares M, Mavarez J. Review. Hybrid trait speciation and Heliconius butterflies. Philos Trans R Soc B Biol Sci. 2008;363:3047–3054. doi: 10.1098/rstb.2008.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis JJ, van der Burg KRL, Mazo-Vargas A, Reed RD. ChIP-seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Rep. 2016;16:2855–2863. doi: 10.1016/j.celrep.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 34.Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak KM, Wahlberg N, Neild AFE, Dasmahapatra KK, Mallet J, Jiggins CD. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst Biol. 2015;64:505–524. doi: 10.1093/sysbio/syv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak KM, Mcmillan WO, Joron M, Jiggins CD. Genome-wide admixture is common across the Heliconius radiation doi: 10.1101/414201. [DOI] [PMC free article] [PubMed]
- 38.Armstrong J, Hickey G, Diekhans M, Deran A, Fang Q, Xie D, et al. Progressive alignment with Cactus: a multiple-genome aligner for the thousand-genome era. 10.1101/730531. [DOI] [PMC free article] [PubMed]
- 39.Paten B, Earl D, Nguyen N, Diekhans M, Zerbino D, Haussler D. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 2011;21:1512–1528. doi: 10.1101/gr.123356.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirarab S, Reaz R, Bayzid MS, Zimmermann TS, Swenson M, Warnow T. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30:i541–i548. doi: 10.1093/bioinformatics/btu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Belleghem SM, Baquero M, Papa R, Salazar C, McMillan WO, Counterman BA, et al. Patterns of Z chromosome divergence among Heliconius species highlight the importance of historical demography. Mol Ecol. 2018;27:3852–3872. doi: 10.1111/mec.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keightley PD, Pinharanda A, Ness RW, Simpson F, Dasmahapatra KK, Mallet J, et al. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol Biol Evol. 2015;32:239–243. doi: 10.1093/molbev/msu302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terhorst J, Kamm JA, Song YS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 2017;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46:220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simons YB, Sella G. The impact of recent population history on the deleterious mutation load in humans and close evolutionary relatives. Curr Opin Genet Dev. 2016;41:150–158. doi: 10.1016/j.gde.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cingolani P, Platts A, Wang LLL, Coon M, Nguyen T, Wang LLL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petkova D, Novembre J, Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 2015;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ané C, Larget B, Baum DA, Smith SD, Rokas A. Bayesian estimation of concordance among gene trees. Mol Biol Evol. 2007;24:412–426. doi: 10.1093/molbev/msl170. [DOI] [PubMed] [Google Scholar]
- 56.Mallet J, Beltrán M, Neukirchen W, Linares M. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol Biol. 2007;7:28. doi: 10.1186/1471-2148-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A draft sequence of the neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neukirchen WM. Ein neues Taxon der Gattung Heliconius aus Brasilien (Lepidoptera: Nymphalidae) Entomol Zeitschrift. 1992;102:124–127. [Google Scholar]
- 60.Lewis JJ, Geltman RC, Pollak PC, Rondem KE, Van Belleghem SM, Hubisz MJ, et al. Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc Natl Acad Sci. 2019;116:24174–24183. doi: 10.1073/pnas.1907068116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, et al. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 2012;8:e1002752. doi: 10.1371/journal.pgen.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enciso-Romero J, Pardo-Díaz C, Martin SH, Arias CF, Linares M, McMillan WO, et al. Evolution of novel mimicry rings facilitated by adaptive introgression in tropical butterflies. Mol Ecol. 2017;26:5160–5172. doi: 10.1111/mec.14277. [DOI] [PubMed] [Google Scholar]
- 63.Jay P, Whibley A, Frézal L, Rodríguez de Cara MÁ, Nowell RW, Mallet J, et al. Supergene evolution triggered by the introgression of a chromosomal inversion. Curr Biol. 2018;28:1839–1845. doi: 10.1016/j.cub.2018.04.072. [DOI] [PubMed] [Google Scholar]
- 64.Martin SH, Davey JW, Jiggins CD. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 2015;32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC, Sheppard PM, et al. Genetics and the evolution of muellerian mimicry in Heliconius butterflies. Philos Trans R Soc B Biol Sci. 1985;308:433–610. [Google Scholar]
- 66.Whitlock MC, Barton NH. The effective size of a subdivided population. Genetics. 1997;146:427–441. doi: 10.1093/genetics/146.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laurance WF, Camargo JLC, Luizão RCC, Laurance SG, Pimm SL, Bruna EM, et al. The fate of Amazonian forest fragments: a 32-year investigation. Biol Conserv. 2011;4870:56–67. [Google Scholar]
- 68.Aragão LEOC, Poulter B, Barlow JB, Anderson LO, Malhi Y, Saatchi S, et al. Environmental change and the carbon balance of Amazonian forests. Biol Rev. 2014;89:913–931. doi: 10.1111/brv.12088. [DOI] [PubMed] [Google Scholar]
- 69.Haddad NM, Brudvig LA, Clobert J, Davies KF, Gonzalez A, Holt RD, et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci Adv. 2015;1:e1500052. doi: 10.1126/sciadv.1500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ribeiro MC, Martensen AC, Metzger JP, Tabarelli M, Scarano F, Fortin M-J. The Brazilian Atlantic Forest: a shrinking biodiversity hotspot. Biodivers Hotspots. 2011:405–34.
- 71.Freitas AVL, Marini-Filho OJ. Plano de ação nacional para a conservação dos lepidópteros ameaçados de extinção. Série Espécies Ameaçadas no 13. ICMBio, Bras. Bras. 2011. Available from: http://www.icmbio.gov.br/portal/images/stories/docs-plano-de-acao/pan-lepidoptera/livro_lepidopteras_web2.pdf.
- 72.Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE. A thousand fly genomes: an expanded Drosophila genome nexus. Mol Biol Evol. 2016;33:3308–3313. doi: 10.1093/molbev/msw195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 74.Joron M, Papa R, Beltrán M, Chamberlain N, Mavárez J, Baxter S, et al. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 2006;4:1831–1840. doi: 10.1371/journal.pbio.0040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pryszcz LP, Gabaldón T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 2016;44:e113. doi: 10.1093/nar/gkw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smit, AFA, Hubley, R & Green P. RepeatMasker Open-4.0. Accessed 15 Feb 2016. Available at http://www.repeatmasker.org.
- 79.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 80.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kajitani R, Yoshimura D, Okuno M, Minakuchi Y, Kagoshima H, Fujiyama A, et al. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat Commun. 2019;10:1702. doi: 10.1038/s41467-019-09575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. [Google Scholar]
- 83.Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, et al. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019;47:D807–D811. doi: 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, et al. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 2019;20:224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larget BR, Kotha SK, Dewey CN, Ané C. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics. 2010;26:2910–2911. doi: 10.1093/bioinformatics/btq539. [DOI] [PubMed] [Google Scholar]
- 87.Ayad LAK, Pissis SP. MARS: improving multiple circular sequence alignment using refined sequences. BMC Genomics. 2017;18:86. doi: 10.1186/s12864-016-3477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 94.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ewels P, Ns Magnusson Måand Lundin S, Aller MK. Data and text mining MultiQC: summarize analysis results for multiple tools and samples in a single report. Accessed 15 Mar 2017 Available from: https://academic.oup.com/bioinformatics/article-abstract/32/19/3047/2196507. [DOI] [PMC free article] [PubMed]
- 97.Cock PJA, Grüning BA, Paszkiewicz K, Pritchard L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ. 2013;1:e167. doi: 10.7717/peerj.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 101.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 102.Leigh JW, Bryant D. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- 103.Massardo D, VanKuren N, Nallua S, Rogner Ramos R, Ribeiro PG, Silva-Brandão KL et al. The roles of hybridization and habitat fragmentation in the evolution of Brazil’s enigmatic longwing butterflies, Heliconius nattereri and H. hermathena. Supplementary Datasets. NCBI BioProject PRJNA596795 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA596795]. Accessed 17 May 2020. [DOI] [PMC free article] [PubMed]
- 104.Massardo D, VanKuren N, Nallua S, Rogner Ramos R, Ribeiro PG, Silva-Brandão KL et al. The roles of hybridization and habitat fragmentation in the evolution of Brazil’s enigmatic longwing butterflies, Heliconius nattereri and H. hermathena. Supplementary Datasets. NCBI BioProject PRJNA596801 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA596801]. Accessed 17 May 2020. [DOI] [PMC free article] [PubMed]
- 105.Massardo D, VanKuren N, Nallua S, Rogner Ramos R, Ribeiro PG, Silva-Brandão KL et al. The roles of hybridization and habitat fragmentation in the evolution of Brazil’s enigmatic longwing butterflies, Heliconius nattereri and H. hermathena. Supplementary Datasets. NCBI BioProject PRJNA596801 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA596796]. Accessed 17 May 2020. [DOI] [PMC free article] [PubMed]
- 106.Massardo D, VanKuren N, Nallua S, Rogner Ramos R, Ribeiro PG, Silva-Brandão KL et al. The roles of hybridization and habitat fragmentation in the evolution of Brazil’s enigmatic longwing butterflies, Heliconius nattereri and H. hermathena. Supplementary Datasets. NCBI BioProject PRJNA596801 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA596794]. Accessed 17 May 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sample and sequencing information for Brazilian Heliconius hermathena and H. nattereri generated in this study. Table S2.Heliconius hermathena and H. nattereri genome sequencing data. Table S3. Samples used for mitochondrial genome assemblies using NOVOplasty (Dierckxens et al., 2017, Nuc Acids Res). Table S4. Sample information for other Heliconius species used in this study. Table S5. Summary statistics of nucleotide diversity per site (pi) and Tajima’s D calculated in 100 kb genome-wide. Table S6. Numbers of substitutions in Heliconius nattereri and relatives. Table S7. Numbers of substitutions in Heliconius hermathena and relatives. Table S8. Whole genome mean FST and 95% empirical confidence interval between H. hermathena populations. Table S9. Whole genome mean FST and 95% empirical confidence interval between H. erato populations. Table S10. Whole genome mean FST and 95% empirical confidence interval between H. melpomene populations. Table S11. Nucleotide diversity, overall and pairwise FST estimated for H. hermathena mitochondrial genomes. Table S12. Patterson’s D statistics calculated in 10 kb windows across all autosomes. H. erato samples are from French Guiana, the geographically closest population to H. hermathena.
Additional file 2: Figure S1. Guide trees used for progressiveCactus alignments in Edelman et al. (2019) and this study. Figure S2. Statistics on unfiltered alignments from the Heliconius dataset of 10 kb windows. Figure S3. Statistics on unfiltered alignments from the Heliconius dataset of coding sequence blocks. Length: length of the raw alignment in base pairs. Figure S4. Statistics on unfiltered alignments from the Heliconius dataset of non-coding sequence blocks. Length: length of the raw alignment in base pairs. Figure S5. Statistics on unfiltered alignments from the melpomene clade dataset of 10 kb windows. Figure S6. Statistics on unfiltered alignments from the erato clade dataset of 10 kb windows. Length: length of the raw alignment in base pairs. Figure S7. ASTRAL trees based on filtered a) coding and b) non-coding blocks from Heliconius dataset. Figure S8. ASTRAL trees based on filtered autosomal 10 kb window alignments from a) the erato clade and b) the melpomene clade. Figure S9. ASTRAL trees based on filtered Z-linked 10 kb window alignments from a) the erato clade and b) the melpomene clade. Figure S10. BUCKy cladograms based on filtered 10 kb window alignments from a) the erato clade and b) the melpomene clade. Figure S11. Predominant topologies for erato clade 10 kb autosomal windows. The top 20 topologies accounted for 80% of all windows. Figure S12. Historical population size estimates for a) H. hermathena and b) H. nattereri using PSMC. Figure S13. ADMIXTURE analysis of H. hermathena polymorphism data for k in 2:10. Figure S14. EEMS diversity estimates for Heliconius hermathena. Figure S15.H. hermathena Bayesian concordance tree based on 5000 autosomal 10 kb windows, showing all individuals. Figure S16. Tests for introgression between Heliconius hermathena and H. charithonia. Figure S17. Divergence (dxy) between Heliconius sara and other erato clade species in the chromosome 15 inversion and 250 kb flanking regions and ocations of scaffolds that mapped to the inversion breakpoints. Figure S18. Convergence of 10 independent EEMS runs.
Data Availability Statement
Genome assemblies and sequencing data generated in this study are available at the NCBI associated with BioProjects PRJNA596795 [103] and PRJNA596801 [104]. Genome assemblies are available at the NCBI under BioProjects PRJNA596796 [105] and PRJNA596794 [106].