SUMMARY
The flat bones of the skull are densely innervated during development, but little is known regarding their role during repair. We describe a neurotrophic mechanism that directs sensory nerve transit in the mouse calvaria. Patent cranial suture mesenchyme represents an NGF (nerve growth factor)-rich domain, in which sensory nerves transit. Experimental calvarial injury upregulates Ngf in an IL-1β/TNF-α-rich defect niche, with consequent axonal ingrowth. In calvarial osteoblasts, IL-1β and TNF-α stimulate Ngf and downstream NF-κB signaling. Locoregional deletion of Ngf delays defect site re-innervation and blunted repair. Genetic disruption of Ngf among LysM-expressing macrophages phenocopies these observations, whereas conditional knockout of Ngf among Pdgfra-expressing cells does not. Finally, inhibition of TrkA catalytic activity similarly delays re-innervation and repair. These results demonstrate an essential role of NGF-TrkA signaling in bone healing and implicate macrophage-derived NGF-induced ingrowth of skeletal sensory nerves as an important mediator of this repair.
In Brief
Meyers et al. describe the role of skeletal sensory nerves in cranial bone repair. The authors demonstrate several necessary aspects of membranous bone healing, including influx of nerve growth factor (NGF)-expressing macrophages after injury, followed by skeletal sensory nerve ingrowth to positively regulate bone repair.
Graphical Abstract
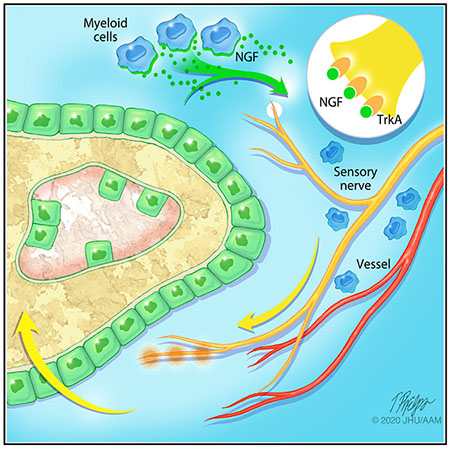
INTRODUCTION
The formation and regeneration of craniofacial bones of the mammalian skeleton requires the action of both intrinsic and extrinsic inductive factors from multiple cell types (Ferguson and Atit, 2019; Lenton et al., 2005; Levi et al., 2012; Pan et al., 2013). Unlike the appendicular skeleton, intramembranous cranial bones form and are healed without a cartilaginous template, by condensations of mesenchymal progenitor cells. During development, these mesenchymal progenitor cells migrate to positions between the brain and surface ectoderm. There, the cells condense and differentiate along an osteogenic lineage that directly generates osteoid that subsequently mineralizes. As membranous bones expand, ossification proceeds in an ordered manner from the skull base toward the skull apex, with bones meeting in the organism’s midline to form the cranial suture complex. Bounded by periosteum (PO) above and the outer meningeal layer below, the cranial suture mesenchyme is a reservoir of skeletal progenitor cells that maintains homeostasis of the uninjured skull or participates in healing the skull after injury (Maruyama et al., 2016; Zhao et al., 2015). Despite the immediate anatomic proximity of the skull and nervous system, little is understood of how neuroskeletal intercommunication informs bone healing after injury.
Many of the cellular events following bone injury resemble those that occur during skeletal development. One notable difference is the sensation of acute pain, which is transmitted by afferent sensory nerves that innervate the appendicular skeleton (Bjurholm et al., 1988; Castañeda-Corral et al., 2011). In this pathway, nerve growth factor (NGF) transmits nociceptive signals either by directly activating tropomyosin receptor kinase A (TrkA)+ sensory nerves or through indirect mechanisms, which intensify alternate nociceptive pathways (Mantyh, 2014; Pezet and McMahon, 2006). In addition to pain sensation, a large body of literature supports an evolutionarily conserved role of TrkA sensory nerves in organogenesis and tissue regeneration (Kumar and Brockes, 2012). For example, sensory or motor neurons are known to drive the subsequent assembly of vascular and tissue components that are responsible for the ability of the starfish and certain amphibians to regenerate entire appendages (Kumar and Brockes, 2012). However, the role of NGF-responsive nociceptive fibers in influencing mammalian bone tissue repair is not yet known.
In comparison with the long bones, in which nerve fiber frequency has been well defined (Bjurholm et al., 1988; Grönblad et al., 1984; Hohmann et al., 1986; Mach et al., 2002), the nerve fibers that innervate the skull are less well understood. Skull-associated nerve fibers are found on both the endocranial and ectocranial surfaces of the calvaria (Alberius and Skagerberg, 1990; Silverman and Kruger, 1989) and have been studied in connection with migraine headaches (Kosaras et al., 2009). These nerve-enriched coverings of calvarial bone, such as dura and PO, are essential for coordinated bone repair (Doro et al., 2017; Levi et al., 2011; Warren et al., 2003). Yet, and despite the immediate anatomic proximity of the skull and nervous system, little is understood of how neuroskeletal intercommunication informs bone healing after injury.
In this study, we characterized the innervation of mouse cranial bone and examined the role of these nerves in a mouse model of bone injury. Our results reveal a neurotrophic mechanism that directs sensory nerve transit in cranial bone repair.
RESULTS
Ngf Expression in Cranial Suture Mesenchyme Coincides with Sensory Nerve Transit
Ngf-expressing cells in the uninjured skull were identified using a previously validated NGF-eGFP reporter animal (Figures 1A–1G) (Kawaja et al., 2011). NGF reporter activity was abundant in mesenchymal cells of all patent cranial sutures (Figures 1A–1C), but not the already fused posterofrontal suture (Figure 1D). Immunocytochemistry performed on parallel sections showed that putative sutural stem cell markers were co-expressed with NGF reporter activity, such as the zinc finger transcription factor Gli1 (Figure 1F) (Zhao et al., 2015) and the Wnt regulatory protein Axin2 (Figure 1G) (Maruyama et al., 2016). Within the bone, NGF-eGFP reporter activity was detected at lower levels in the overlying PO and underlying dura mater (DM) (Figure 1E) but was absent in cortical osteocytes.
Figure 1. Ngf Expression Domains and Innervation of the Uninjured Calvarium.
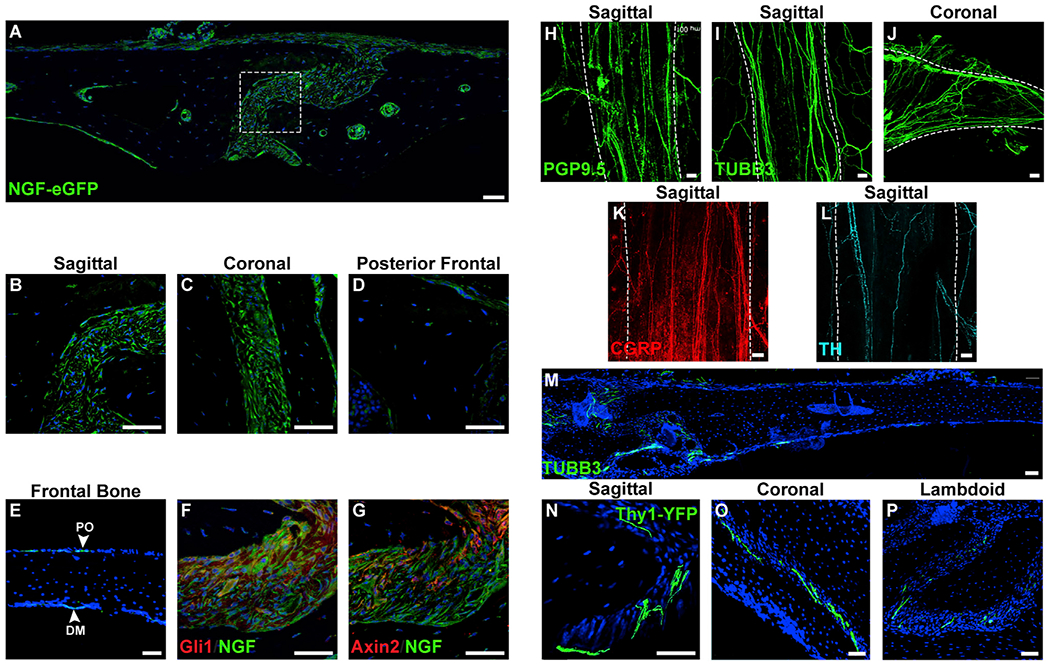
(A) Tile scan of the uninjured parietal bone and sagittal suture (center) in the NGF-eGFP reporter animal.
(B–D) High-magnification images of cranial sutures within NGF-eGFP reporter animals, including (B) patent sagittal suture, (C) patent coronal suture, and (D) fused portion of posterofrontal suture.
(E) NGF reporter activity within the intact frontal bone. PO, periosteum; DM, dura mater.
(F and G) Immunohistochemical staining (appearing red) within the sagittal suture of NGF-eGFP reporter animals, including (F) Gli1 and (G) Axin2.
(H) PGP9.5 whole-mount image of the patent sagittal suture. White dashed lines indicate margins of the patent suture, imaged from the endocranial aspect.
(I and J) TUBB3 (beta III tubulin) whole-mount images of patent cranial sutures, including the (I) sagittal and (J) coronal suture. White dashed lines indicate margins of the patent suture, imaged from the endocranial aspect.
(K and L) Immunohistochemical staining typifying nerve fibers within the sagittal suture, including (K) CGRP+ peptidergic fibers (appearing red) (L) and TH+ sympathetic fibers (appearing cyan). White dashed lines indicate margins of the patent suture, imaged from the endocranial aspect.
(M) Tile scan of TUBB3+ nerve fibers in the uninjured frontal bone. TUBB3+ nerve fibers appear green.
(N–P) Cross-sectional images of nerve fibers within patent cranial sutures using pan-neuronal Thy1-YFP transgenic reporter animals. Thy1-YFP reporter activity within patent cranial sutures, including (N) sagittal, (O) coronal, and (P) lambdoid sutures. Thy1-YFP reporter activity appears green.
DAPI counterstain appears blue in all images. Representative images shown from n = 4 mice. All white scale bar, 50 μm. See also Figure S1.
To identify peripheral nerves within the uninjured skull, we performed immunohistochemical staining using the pan-neuronal markers protein gene product 9.5 (PGP9.5) (Figure 1H) and beta III tubulin (TUBB3) (Figures 1I,1J, and 1M). In whole-mount preparations, PGP9.5+TUBB3+ nerve fibers were observed parallel to patent sagittal and coronal sutures (Figures 1H–1J). In the sagittal suture, the majority of the nerves were positive for CGRP (Figure 1K), but a smaller proportion expressed the sympathetic nerve marker tyrosine hydroxylase (TH) (Figure 1L). In general, uninjured calvarial bones showed comparatively less frequent nerve fibers at the dural and periosteal surfaces in whole-mount preparations (Figure S1). Imaging of coronal sections of the uninjured calvaria showed a clear enrichment of TUBB3+ nerve fibers within the patent cranial sutures (Figure 1M), a pattern confirmed in sections from Thy1-YFP pan-neuronal reporter animals (Figures 1N–1P). Here, abundant YFP-positive nerve fibers are seen traversing or terminating within patent sagittal, coronal, and lambdoid sutures. In contrast, little or no YFP fluorescence was observed at sites of fused calvarial sutures (not shown).
Ngf Is Acutely Upregulated following Calvarial Bone Injury
To examine the role of skeletal sensory nerves in cranial bone repair, we first determined the temporo-spatial domains of Ngf expression in the NGF-eGFP reporter animal subjected to experimental injury. In this procedure a small (1.8 mm), circular, full-thickness defect was surgically created in the frontal bone, and re-ossification was followed over a 4 week period using microcomputed tomography (μCT) (Figures 2A–2E). Within the uninjured bone, patchy, weak NGF reporter was present within the PO and DM, but not in cortical osteocytes (Figure 2F). At day 3 (d3) post-injury, the majority of cells within the fibro-inflammatory defect were NGF reporter positive, most notably adjacent to the injured bone (Figure 2G). At later time points corresponding to re-ossification, the majority of eGFP reporter activity was localized to bone-lining cells along the re-growing defect edges (d7 and 14; Figures 2H and 2I). At later times, NGF reporter activity waned but was still present in bone-lining cells (d28; Figure 2J). At all time points, cortical osteocytes remained NGF reporter negative. Semiquantitative analysis of relative NGF-eGFP reporter activity showed a robust (approximately 50-fold) increase in NGF reporter activity at 3 days following injury (Figure 2K). NGF reporter activity remained elevated over baseline throughout the study period but gradually waned to a value ~5.4-fold over uninjured calvaria by the d28 study endpoint (Figure 2K).
Figure 2. NGF-eGFP Reporter Activity following Calvarial Bone Injury.
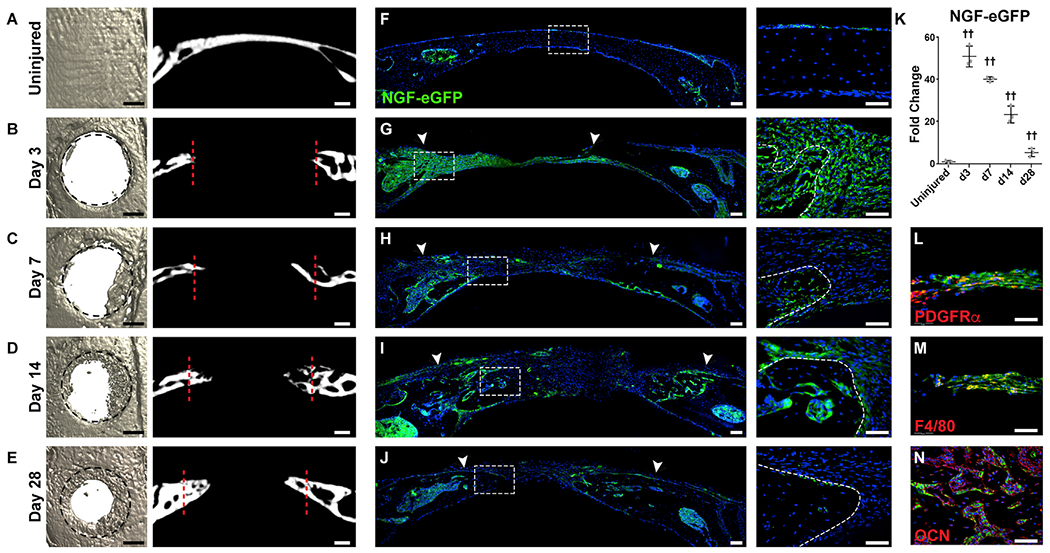
NGF-eGFP reporter animals were subjected to a circular, full-thickness frontal bone defect (1.8 mm in diameter).
(A–E) μCT reconstructions of the defect site in a top-down view (left) and coronal cross-sectional images (right) among (A) uninjured frontal bones and (B–E) d3–d28 post-injury. Margins of original defect are indicated by dashed black or red lines. Black scale bar, 500 μm; white scale bar, 200 μm.
(F–J) Representative tile scans (left) and high-magnification images (right) of NGF-eGFP reporter activity in the calvarial defect site at serial time points, including (F) uninjured frontal bones and (G–J) d3–d28 post-injury. Reporter activity appears green, while nuclear counterstain appears blue. Uninjured control is shown for comparison. White arrowheads indicate margins of defect site. Dashed white lines indicate healing bone edge. White scale bar, 50 μm.
(K) Quantification of relative NGF-eGFP reporter activity from d3–d28 post-injury.
(L–N) Immunohistochemical staining (appearing red) within the defect area of NGF-eGFP reporter animals, including (L) PDGFRα (platelet-derived growth factor receptor α), d3 post-injury; (M) F4/80, d3 post-injury; and (N) osteocalcin (OCN), d14 after injury. In graphs, each dot represents a single animal; n = 3 animals per time point. White scale bar, 50 μm. Data are represented as mean ± SD.
Immunohistochemical staining of NGF-eGFP reporter sections was next performed to delineate Ngf-expressing cell types within the early bone defect niche (Figures 2L and 2M). At early time points post-injury, PDGFRα (platelet-derived growth factor receptor α)+ mesenchymal cells showed high co-expression with eGFP reporter activity (Figure 2L, d3 post-injury). In addition, F4/80+ macrophages also showed substantial NGF reporter activity (Figure 2M, d3 post-injury). At later time points after ossification, we had observed bone-lining cells with the predominant cell type with eGFP reporter activity. To confirm this, osteocalcin (OCN) immunohistochemistry was performed and demonstrated that bone-lining OCN+ osteoblasts were the primary source of Ngf during later stages of defect repair (Figure 2N, d14 shown).
Inflammatory Signals Drive Ngf Expression in Injured Calvaria
A universal response to tissue injury is the induction of pro-inflammatory cytokines, which initiate key signaling cascades critical to tissue repair. To determine the possible role of inflammatory signals on Ngf expression in response to calvarial injury, inflammatory cytokine expression was assessed using immunohistochemistry within NGF-eGFP reporter sections. Robust immunoreactivity for both IL-1β and TNF-α corresponded to domains of NGF reporter activity at d3 post-injury (Figures 3A and 3C, appearing yellow). At d7, less prominent immunoreactivity for IL-1β and TNF-α was observed, primarily in the non-ossified defect site (Figures 3B and 3D, appearing red), while NGF reporter activity had by this point become more restricted to bone-lining osteoblasts (Figures 3B and 3D, appearing green). To determine whether these cytokines directly induced Ngf expression, mouse calvarial cells were exposed to recombinant IL-1β or TNF-α, and Ngf transcripts were assayed using qPCR. Both cytokines induced Ngf mRNA expression in a dose-dependent manner (Figure 3E). Furthermore, both IL-1β and TNF-α induced eGFP reporter in mesenchymal cells isolated from a mouse carrying the NGF-eGFP reporter allele (Figure 3F). These changes were associated with induction of NF-κB signaling activation among mouse calvarial cells (Figure 3G).
Figure 3. Inflammatory Cytokines Induce Ngf Expression within Calvarial Osteoblasts.
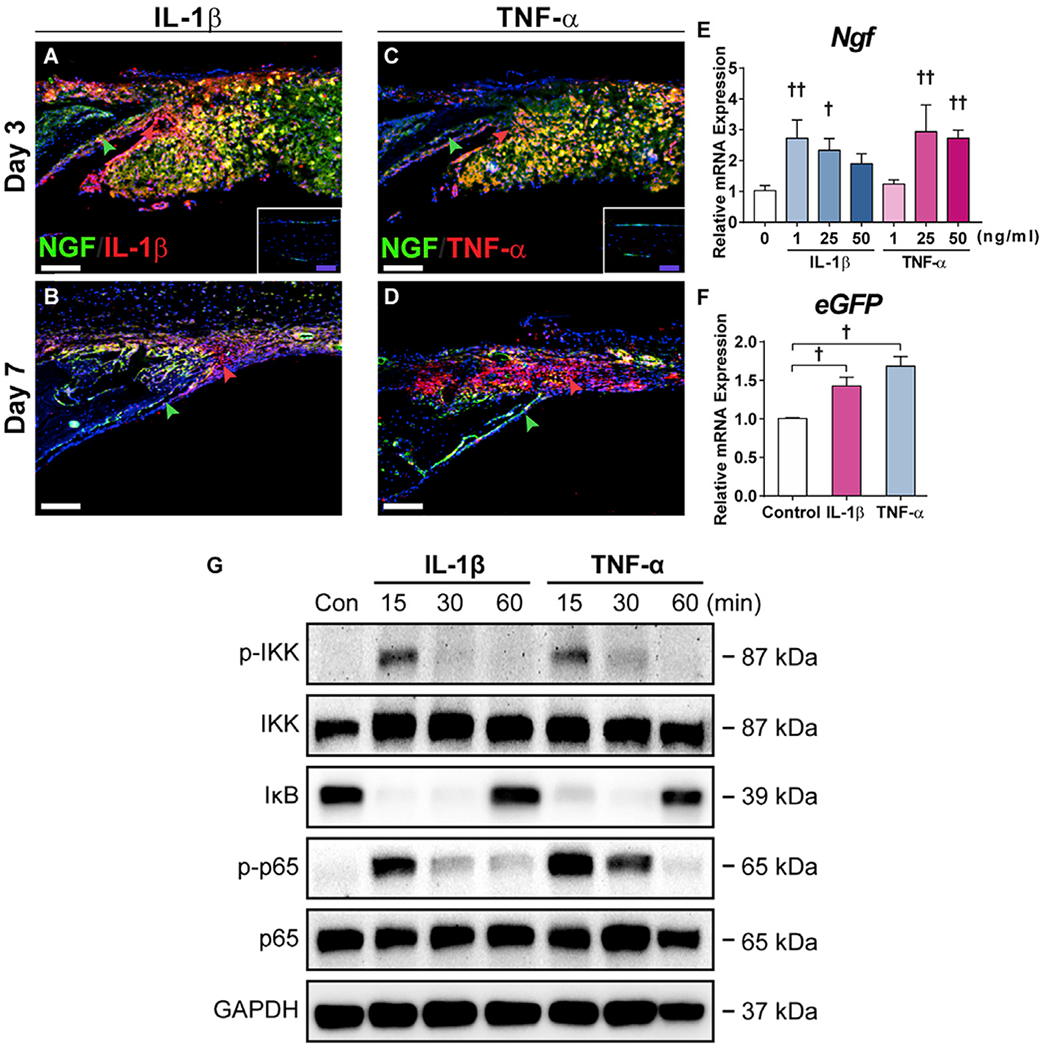
(A and B) Immunohistochemistry for IL-1β, appearing red, on the calvarial injury site within NGF-eGFP reporter sections at (A) d3 and (B) d7 post-injury. Green arrowheads indicate bone-lining eGFP reporter activity, while red arrowheads indicate immunoreactivity within the defect site. White dashed lines indicate bone edges. Inset demonstrates immunoreactivity among uninjured calvarial bone. White scale bar, 50 μm; purple scale bar, 20 μm.
(C and D) Immunohistochemistry for TNF-α, appearing red, on the calvarial injury site within NGF-eGFP reporter sections at (C) d3 and (D) d7 post-injury. Green arrowheads indicate bone-lining eGFP reporter activity, while red arrowheads indicate immunoreactivity within the defect site. White dashed lines indicate bone edges. Inset demonstrates immunoreactivity among uninjured calvarial bone. White scale bar, 50 μm; purple scale bar, 20 μm.
(E) Ngf expression among calvarial osteoblasts 48 h after treatment with recombinant IL-1β or TNF-α, assessed using qRT-PCR.
(F) eGFP expression among NGF-eGFP-derived calvarial osteoblasts 48 h after treatment with IL-1β (1 ng/mL) or TNF-α (25 ng/mL), assessed using qRT-PCR.
(G) NF-κB signaling among calvarial osteoblasts 15–60 min after treatment with IL-1β (1 ng/mL) or TNF-α (25 ng/mL), assessed using western blot.
All experiments were performed in triplicate. Representative histologic images are from n = 3 mice per time point. †p < 0.05 and ††p < 0.01 in comparison with control. Data are represented as mean ± SD.
Re-innervation of the Calvarial Bone Defect Follows Acute Ngf Upregulation
A robust and acute rise in Ngf expression after calvarial injury would be expected to result in a prominent localized ingrowth of skeletal-associated nerve fibers. To test this idea, the temporo-spatial domains of calvarium-associated nerve fibers were next characterized over the same 28 day time interval after bone injury (Figure 4). For this purpose, C57BL/6J wild-type mice were used, and injury sites were subjected to whole-mount immunohistochemical staining with the pan-neuronal marker TUBB3 (Figures 4A–4E). Whole-mount imaging of the endocranial aspect of the uninjured calvaria confirmed our prior findings, including a high density of nerve fibers running along the endocranial surface of the midline suture (denoted by an asterisk) and relatively less prominent nerve fibers forming a meshwork across the calvarial bone (Figure 4A). Upon calvarial bone injury, re-innervation begins on the medial edge of the defect as soon as d3 (Figure 4B), and progressive axonal ingrowth, sprouting, and ingrowth occur over the time course of calvarial repair (Figures 4C–4E). Coronal sections of the calvaria were then visualized by TUBB3 immunohistochemical staining in order to more precisely define the spatial relationships between nerve fibers within the injury site and the regenerating bone (Figures 4F–4I). A representative tile scan of the defect showed that the nerve fibers were most conspicuously the leading bone edges of the healing defect (Figure 4F), which appeared in continuity with nerve fibers within the dura, PO, and adjacent patent suture. High-magnification images confirmed these findings, with prominent small-caliber dendrites wrapping along the healing bone edges (Figures 4G and 4I) and less conspicuous nerve fibers within the defect center (Figure 4H). Further confirmation of small nerve fiber sprouting along the bone defect edge was obtained using immunohistochemical stains for the pan-neuronal marker PGP9.5 (Figure 4J). In orderto further typify the sprouting nerves, additional immunohistochemical stains were performed for the peptidergic fiber marker CGRP and sympathetic marker TH (Figures 4K and 4L). Analysis of serial sections demonstrated that the majority of nerves entering into the healing bone defect were CGRP+ peptidergic nociceptors (Figure 4K), while a smaller subset of nerve fibers represented TH+ sympathetic fibers (Figure 4L). Nerve fibers closely associate with blood vessels during skeletal development and fracture repair (Li et al., 2019; Tomlinson et al., 2016). The spatial relationship between nerve fibers and blood vessels was interrogated by CD31 immunostaining of sections from Thy1-YFP pan-neuronal reporters. Both in uninjured calvaria and during defect healing, YFP+ nerve fibers were observed to course in parallel and close proximity to CD31+ vessels (Figure S2). Thus, injury-associated Ngf elaboration is associated by subsequent neurovascular infiltration of the injured bone, including predominantly CGRP+ fibers at sites of incipient bone formation.
Figure 4. Re-innervation during Calvarial Bone Defect Repair.
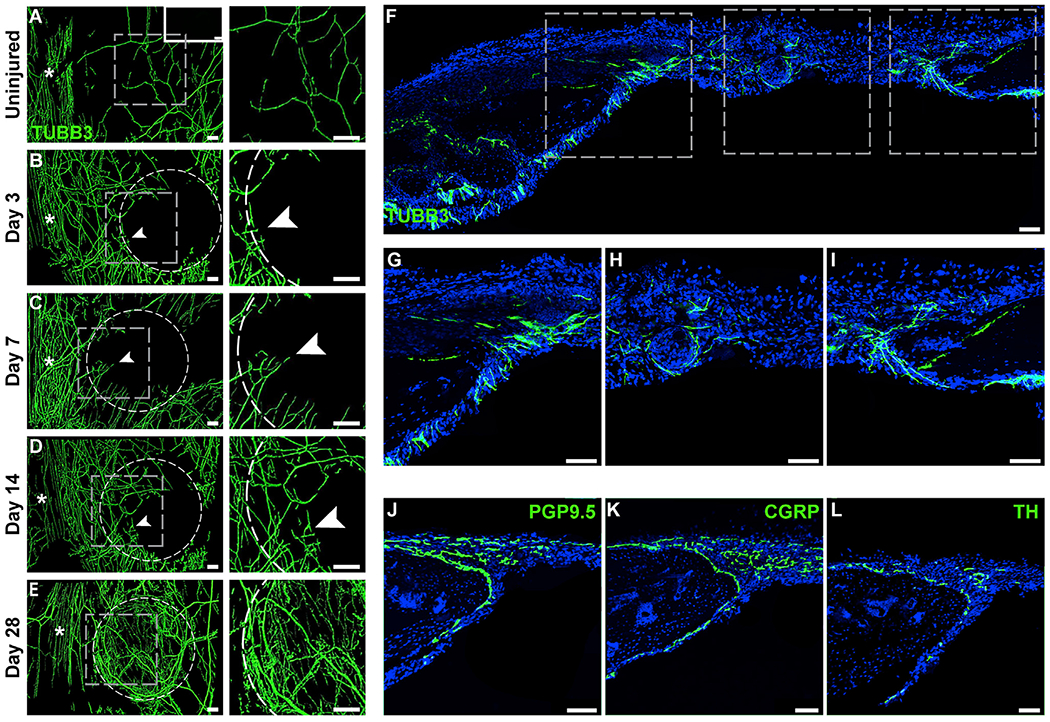
(A–E) Representative whole-mount tile scans (left) and high-magnification images (right) of TUBB3 (beta III tubulin)-stained calvarial defects from (B–E) d3 to d28 post-injury. (A) Uninjured control shown as a comparison. TUBB3+ nerve fibers appear green. Dashed white circles indicates margins of defect. White asterisks indicate midline suture. Upper right-hand inset depicts negative control without primary antibody application. White scale bar, 200 μm.
(F–I) Tile scan image (F) and higher magnification images of TUBB3 immunohistochemical staining of nerve fibers, including (G and I) TUBB3+ nerves at the margins of the defect and (H) center of the defect, d14 post-injury. Asterisk in tile scan indicates the midline suture. White arrowheads indicate sites of prominent re-innervation adjacent to healing bone defect edges.
(J–L) Immunohistochemical staining typifying nerve fibers within the calvarial defect at d14 post-injury, including (J) the pan-neuronal marker protein gene product 9.5 (PGP9.5), (K) the peptidergic fiber marker calcitonin gene-related peptide (CGRP), and (L) the sympathetic marker tyrosine hydroxylase (TH).
Representative images are from n = 3 mice per time point. White scale bar, 50 μm. See also Figure S2.
Disruption of Ngf Attenuates Re-innervation and Impairs Calvarial Regeneration
We next determined the requirement for Ngf expression for normal calvarial bone repair. As multiple cell types showed NGF reporter activity within a defect niche, we first used a locoregional rather than tissue-specific approach for Ngf deletion. For this, Ngffl/fl animals were treated within the defect site with adenovirus encoding Cre recombinase (Ad-Cre), while control littermates received Ad-GFP as a control. Validation of recombination was performed using mT/mG animals, in which Ad-Cre demonstrated significant recombination within the defect site 2 days post-operatively (Figure S3). Healing among Ad-GFP- and Ad-Cre-treated bone defects was examined over a previously validated 4 week period (Zhang et al., 2018) (Figure 5). Results showed a significant reduction in calvarial defect re-ossification among Ad-Cre-treated Ngffl/fl bones, as visualized using a top-down view of μCT reconstruction as well as coronal cross-sectional images (Figures 5A and 5B). μCT quantification was performed to assess bone healing using multiple metrics (Figures 5C–5G). The amount of regenerated bone within the defect site was quantified as bone volume (BV) (Figure 5C; 31.3% reduction), fractional BV/tissue volume (TV) (Figure 5D; 31.0% reduction), and bone fractional area (BFA) (Figure 5E; 19.0% reduction). In agreement with these findings, a semiquantitative bone healing score showed a significant reduction among Ad-Cre-treated bone defect (Figure 5F). Conversely, the mean diameter of the remaining non-ossified bone defect was significantly increased among Ad-Cre-treated samples (Figure 5G; 205.9% increase in comparison with Ad-GFP control). In contrast, no change in bone parameters was observed with Ad-Cre treatment among uninjured calvarial bones within the same animal (Figure S4).
Figure 5. Local Ngf Deletion Impairs Calvarial Bone Defect Repair.
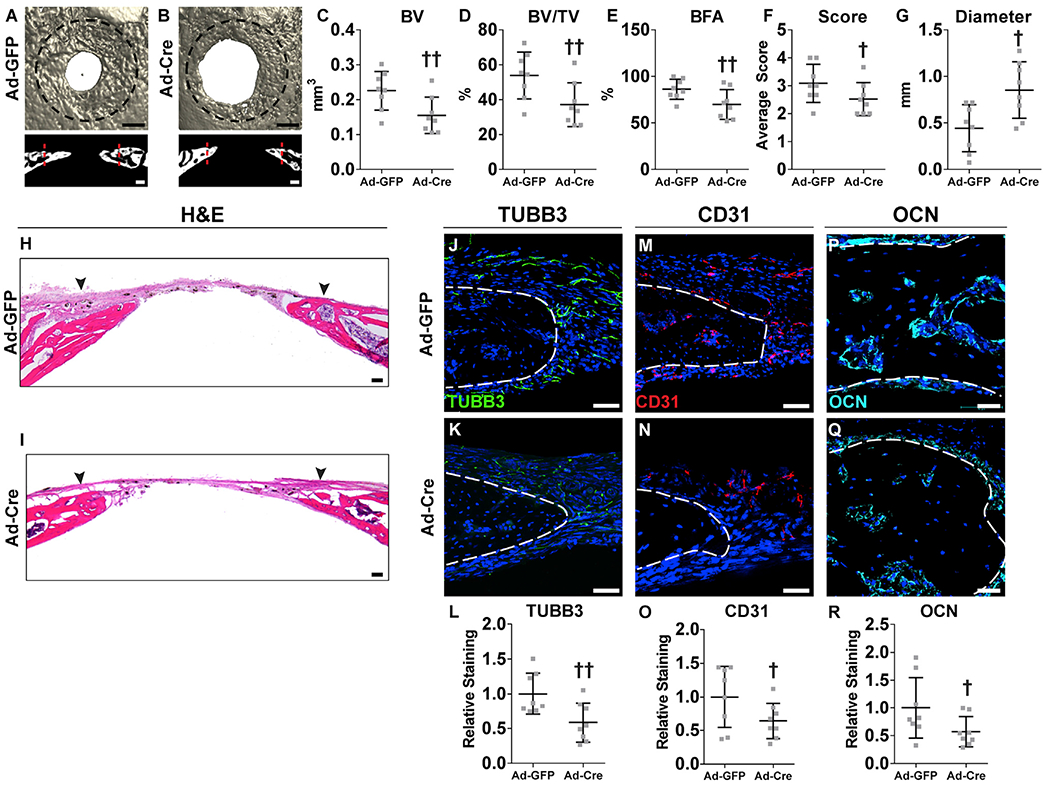
Ngffl/fl animals were locally injected with Ad-GFP or Ad-Cre virus, followed by creation of a 1.8 mm, circular, full-thickness frontal bone defect.
(A and B) μCT reconstructions of the defect site in a top-down view (above) and coronal cross-sectional images (below) among Ngffl/fl animals locally injected with either (A) Ad-GFP or (B) Ad-Cre. Analysis performed at d28 post-injury. Margins of original defect are indicated by dashed black or red lines. Black scale bar, 500 μm; white scale bar, 200 μm.
(C–G) μCT quantification of bone healing among Ad-GFP- and Ad-Cre-injected mice, including (C) bone volume (BV), (D) bone volume/tissue volume (BV/TV), (E) bone formation area (BFA), (F) bone healing score (score), and (G) defect diameter.
(H and I) H&E stain of coronal cross-section of the healing defect site from (H) Ad-GFP- or (I) Ad-Cre-injected mice. Black arrowheads indicate span of initial defect. Black scale bar, 50 μm.
(J–L) Immunohistochemical staining of TUBB3+ (beta III tubulin) nerve fibers at the defect edge from (J) Ad-GFP- or (K) Ad-Cre-injected mice, appearing green and (L) quantification of TUBB3 immunoreactivity within the calvarial defect. Dashed white lines indicate bone edge.
(M–O) Immunohistochemical staining of CD31+ blood vessels at the defect edge from (M) Ad-GFP- or (N) Ad-Cre-injected mice, appearing red, and (O) quantification of CD31 immunoreactivity within the calvarial defect site. Dashed white lines indicate bone edge.
(P–R) Immunohistochemical staining of OCN+ (osteocalcin) osteoblasts from (P) Ad-GFP- or (Q) Ad-Cre-injected mice, appearing cyan, and (R) quantification of OCN immunoreactivity within the calvarial defect site. Dashed white lines indicate bone edge. White scale bar, 50 μm.
DAPI counterstain appears blue in all images. In graphs, each dot represents a single animal; n = 8 animals per group. †p < 0.05 and ††p < 0.01 in comparison with Ad-GFP control. Data are represented as mean ± SD. See also Figures S3 and S4.
H&E staining confirmed a significant reduction in calvarial re-ossification, including a widened distance between bony fronts (Figures 5H and 5I; black arrowheads indicate original defect). The frequency of TUBB3+ dendrites within the defect site of Ngf floxed animals was next assessed (Figures 5J–5L). TUBB3+ fibers were again noted most prominently at the healing defect edge (Figure 5J). A dramatic reduction in TUBB3+ nerve fibers was observed among Ad-Cre-treated defects (Figure 5K), which when quantified demonstrated a 41.5% reduction in mean TUBB3 immunoreactivity within the defect site (Figure 5L). Vascularity within the bone injury site was next assessed among Ad-GFP- and Ad-Cre-treated animals (Figures 5M–5O). A reduction in CD31+ vascular channels was found within Ad-Cre-treated injury sites (Figures 5M and 5N), demonstrating a 35.8% decrease in comparison with Ad-GFP control (Figure 5O). OCN immunohistochemical staining was next performed and confirmed a significant reduction in OCN+ osteoblastic numbers with Ad-Cre treatment of Ngffl/fl animals (Figures 5P and 5Q), reflecting a 42.9% reduction in OCN immunostaining (Figure 5R). In sum, locoregional deletion of Ngf during bone repair led to significant reduction in injury-associated re-innervation, impaired revascularization, reduced total osteoblastic activity, and overall a significant delay in bone defect healing.
Expression of Ngf in LysM-Expressing Cells Is Necessary for Normal Calvarial Repair
To further assess the requirement for NGF in calvarial defect repair, we sought to identify the cell type responsible for production of this neurotrophin. As reported above (Figures 2L and 2M), NGF reporter activity was observed in both F4/80+ macrophages and PDGFRα+ stromal cells following calvarial injury. We therefore assessed the innervation and bone healing following disruption of NGF in each of these cell types. Ngffl/fl animals were crossed with either LysM-Cre or PDGFRα-CreERT2 lines to yield Ngffl/fl;LysM-Cre (NgfLysM) animals and Ngffl/fl;PDGFRα-CreERT2 (NgfPdgfrα) animals. Ngffl/fl;mT/mG (Ngffl/fl) animals were used as controls (Figure 6). Tamoxifen (TM) was given using previously validated injection schedules (Xu et al., 2019), and the same TM protocol was used across all strains to ensure direct comparability of the results. First, the spatial localization of PDGFRα and LysM reporter activity was confirmed (Figures 6A–6D). In uninjured bone, bone-lining cells (ecto- and endocranial) showed PDGFRα reporter activity (Figure 6A, appearing red), while LysM reporter activity was present within cell more loosely affiliated with the endocranial bone surface (Figure 6C, appearing red). As anticipated and at d3 post-operatively, both PDGFRα-expressing and LysM-expressing cells were abundant in the defect site (Figures 6B and 6D). Validation for Ngf deletion was performed using NGF immunohistochemistry on calvarial injury sites within the Ngffl/fl, NgfPdgfrα, and NgfLysM mice (Figures 6E–6G). NGF immunohistochemical staining (appearing green) was reduced across both NgfPdgfrα and NgfLysM defect sites, in comparison with Ngffl/fl control defects. This was most conspicuous among NgfLysM defects, which showed few NGF immunoreactive cells (Figure 6G). Some residual NGF immunostaining was present within NgfPdgfrα defect sites, which as expected was present only in reporter-negative cells (Figure 6F).
Figure 6. Tissue-Specific Deletion of Ngf in LysM-Expressing Cells but Not Pdgfrα-Expressing Cells Blunts Calvarial Bone Defect Repair.
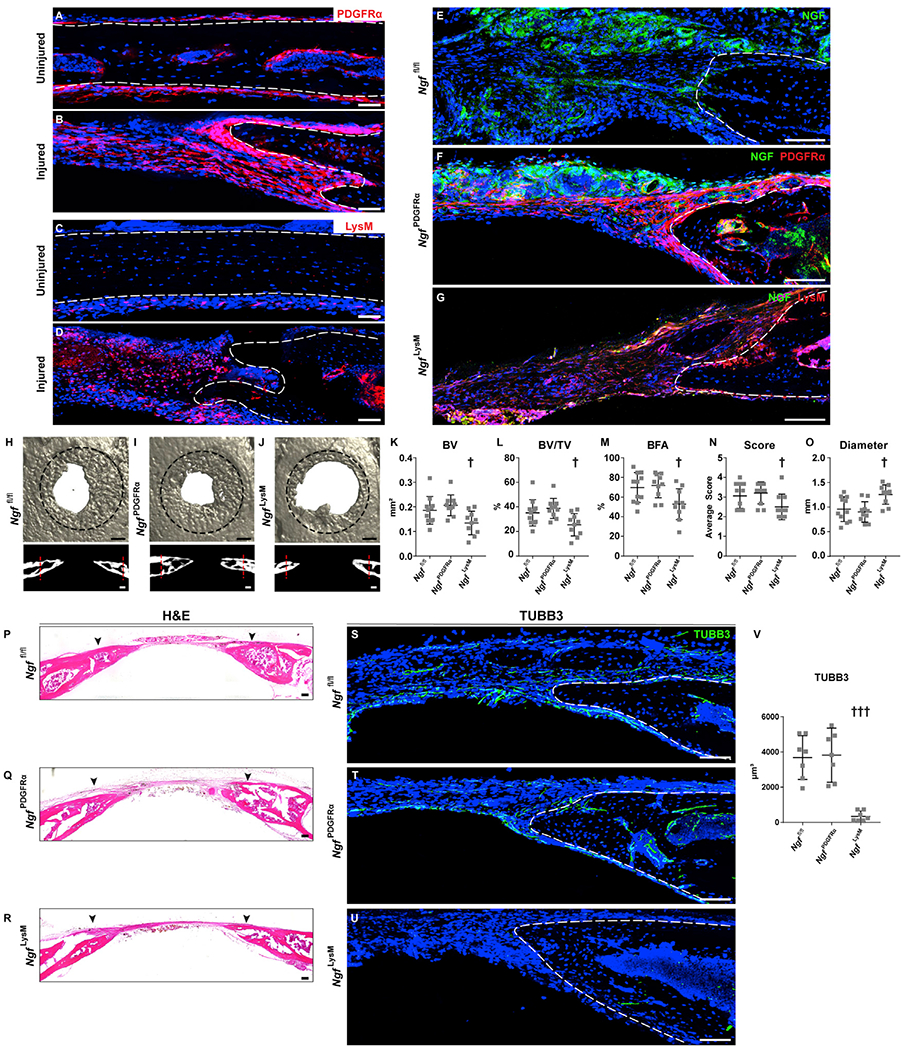
Tissue-specific deletion of Ngf was performed using LysM-Cre (NgfLysM) or Pdgfrα-CreERT2 lines (NgfPdgfrα), and bone healing was compared with Cre− animals (Ngffl/fl). Healing was assessed using a 1.8 mm, circular, full-thickness frontal bone defect.
(A–D) Location of PDGFRα or LysM reporter activity before and after injury. (A and B) PDGFRα reporter activity (PDGFRα-CreERT2-mGFP, recolored red) within the uninjured frontal bone and at d3 post-injury. White scale bar, 50 μm. (C and D) LysM reporter activity (LysM-Cre-tdTomato) within the uninjured frontal bone and at d3 post-injury. White dashed lines indicate bone edges. White scale bar, 50 μm.
(E–G) Validation of Ngf deletion in either PDGFRα-expressing stromal cells (NgfPdgfrα) or LysM-expressing monocytes/macrophages (NgfLysM) in comparison with Ngffl/fl mice, d3 post-injury. NGF immunohistochemistry among (E) Ngffl/fl control bone defects, (F) NgfPdgfrα bone defects, or (G) NgfLysM bone defects. NGF immunostaining appears green, while reporter activity appears red. White scale bar, 50 μm.
(H–J) μCT reconstructions of the defect site in a top-down view (above) and coronal cross-sectional images (below) among (H) Ngffl/fl, (I) NgfPdgfrα, and (J) NgfLysM animals. Margins of original defect are indicated by dashed black or red lines. Black scale bar, 500 μm; white scale bar, 200 μm.
(K–O) μCT quantification of bone healing among Ngffl/fl, NgfPdgfrα, and NgfLysM mice including (K) bone volume (BV), (L) bone volume/tissue volume (BV/TV), (M) bone formation area (BFA), (N) bone healing score (score), and (O) defect diameter.
(P–R) H&E stain of coronal cross-section of the healing defect site from (P) Ngffl/fl, (Q) NgfPdgfrα, and (R) NgfLysM mice. Black arrowheads indicate span of initial defect. Black scale bar, 50 μm.
(S–V) Immunohistochemical staining of TUBB3+ (beta III tubulin) nerve fibers at the defect edge from (S) Ngffl/fl, (T) NgfPdgfrα, and (U) NgfLysM mice, appearing green, and (V) quantification of TUBB3 immunoreactivity within the calvarial defect. Dashed white lines indicate bone edge. White scale bar, 50 μm.
In graphs, each dot represents a single animal. n = 9–11 per group for (K)–(O) and N = 7 per group for (V). †p < 0.05 and †††p < 0.001 in comparison with Ngffl/fl and NgfPdgfrα. Data are represented as mean ± SD. See also Figure S5.
Next, the cell-specific requirements of Ngf in calvaria bone repair were assessed, using Ngffl/fl, NgfPdgfrα, and NgfLysM mice. As in prior experiments, frontal bone healing was assessed over a 4 week period (Figures 6H–6O). Results demonstrated impaired bone healing among NgfLysM but not NgfPdgfrα animals. μCT reconstructions and cross-sectional images demonstrated impaired re-ossification among NgfLysM mice only (Figures 6H–6J). Quantitative μCT metrics of bone healing were reduced among NgfLysM mice only, including BV (Figure 6K; 33% reduction), BV/TV (Figure 6L; 35% reduction), BFA (Figure 6M; 28% reduction), and bone healing score (Figure 6N). Consistent with this observation, the mean diameter of the bone defect area was significantly increased among NgfLysM animals (Figure 6O; 39% increase). In contrast, no statistically significant differences in bone healing were observed between NgfPdgfrα and Ngffl/fl animals.
Histologic examination by H&E staining verified a significant reduction in re-ossification of injury site among NgfLysM mice, as shown by enlarged distances between bony fronts (Figures 6P–6R; black arrowheads indicate original defect). The density of TUBB3+ nerve fibers within the defect site was next evaluated (Figures 6S–6V). Frequent TUBB3+ nerve fibers were again found around the healing bony fronts within Ngffl/fl and NgfPdgfrα defects (Figures 6S and 6T). In contrast, a marked reduction in TUBB3+ nerve fiber frequency was observed within NgfLysM mice (Figure 6U). Quantification of TUBB3+ nerve fibers within the defect span confirmed a 93% reduction within NgfLysM animals (Figure 6V). In sum, Ngf derived from LysM-expressing monocytes/macrophages is essential for bone defect re-innervation and osseous repair. In contrast, Ngf from PDGFRα-expressing stromal cells is dispensable for both defect innervation and bone healing.
TrkA Signaling Is Required for Normal Calvarial Regeneration
Much like the long bone (Castañeda-Corral et al., 2011), nerve fibers that extend from the trigeminal ganglia to innervate the calvarium are universally TrkA expressing (Han et al., 2019). In order to inhibit TrkA, we used a previously validated chemical-genetic approach, in which TrkA signaling is acutely disrupted over a defined period of time. TrkAF592A mice are homozygous for knockin alleles that encode a phenylalanine-to-alanine substitution in the protein kinase subdomain V, rendering its catalytic activity sensitive to specific inhibition by the membrane-permeable small molecule 1NMPP1 (Chen et al., 2005). Previously we have validated the temporal kinetics of 1NMPP1 inhibition in TrkAF592A and verified that 1NMPP1 has no discernable direct effects on osteoblastic cells (Tomlinson et al., 2016) or on bone repair in wild-type animals (Li et al., 2019).
TrkAF592A mice were now subjected to bone defects with or without 1NMPP1-mediated inhibition of TrkA (Figure 7). Injury sites were again analyzed 4 weeks after defect creation, including analysis of innervation, vascularization, and ossification within the healing bone. μCT demonstrated a reduction in defect re-ossification among 1NMPP1-treated animals at d28 (Figures 7A and 7B). Quantitative μCT metrics of bone formation demonstrated a significant reduction in all parameters among 1NMPP1-treated animals (Figures 7C–7G). This included reductions in BV(Figure 7C; 47.5% reduction), fractional BV (Figure 7D; 47.2% reduction), BFA (Figure 7E; 19.5% reduction), and semiquantitative bone healing score (Figure 7F) and an increase in remaining non-ossified diameter of the defect (Figure 7G; 120.7% increase in comparison with vehicle control). In contrast, μCT analysis of uninjured frontal bones was without detectable change among 1NMPP1-treated TrkAF592A animals (Figure S5).
Figure 7. TrkA Inhibition Impairs Calvarial Bone Defect Repair.
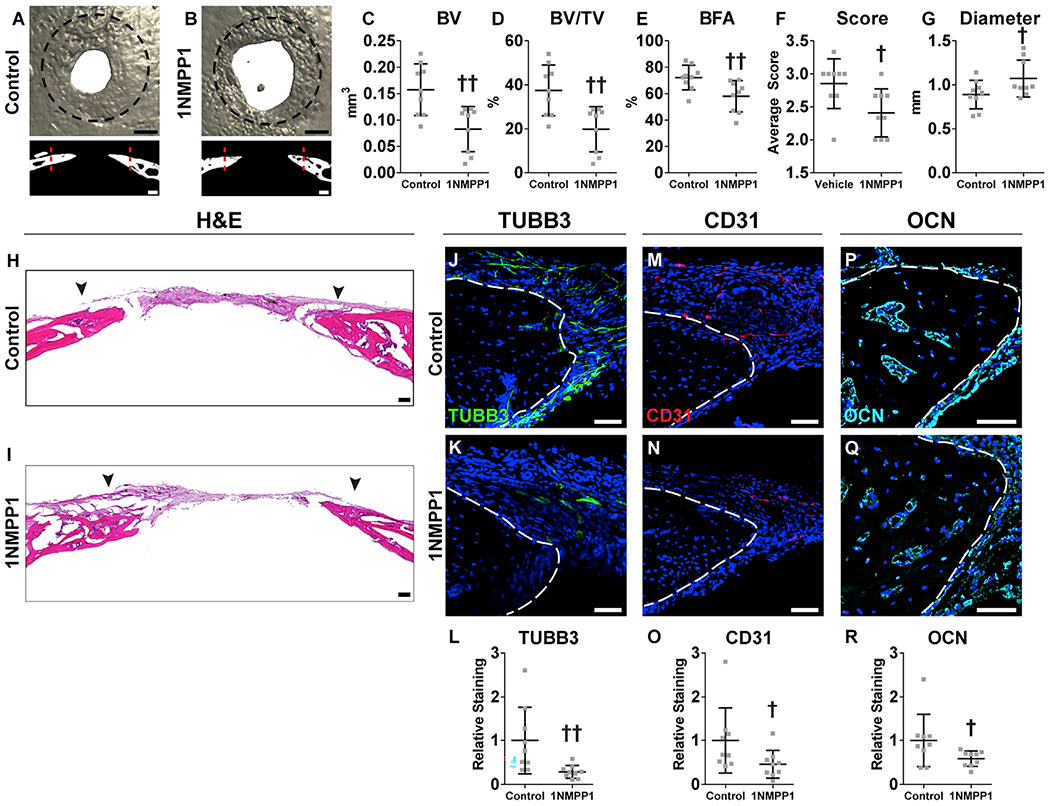
TrkAF592A animals were treated systemically with 1NMPP1 (or vehicle control) to temporally inhibit TrkA catalytic activity, followed by creation of a 1.8 mm, circular, full-thickness frontal bone defect.
(A and B) μCT reconstructions of the defect site in a top-down view (above) and coronal cross-sectional images (below) among TrkAF592A animals treated with (A) vehicle control or (B) 1NMPP1. Black scale bar, 500 μm; white scale bar, 200 μm. Analysis performed at d28 post-injury. Margins of original defect are indicated by dashed black or red lines.
(C–G) μCT quantification of bone healing, among vehicle control or 1NMPP1-injected mice, including (C) bone volume (BV), (D) bone volume/tissue volume (BV/TV), (E) bone formation area (BFA), (F) bone healing score (score), and (G) defect diameter.
(H and I) H&E stain of representative coronal cross-section of the healing defect site from (H) vehicle control and (I) 1NMPP1-injected mice. Black arrowheads indicate span of initial defect. Black scale bar, 50 μm.
(J–L) Immunohistochemical staining of TUBB3+ (beta III tubulin) nerve fibers at the defect edge from (J) vehicle control and (K) 1NMPP1-injected mice, appearing green, and (L) quantification of TUBB3 immunoreactivity within the calvarial defect. Dashed white lines indicate bone edge.
(M–O) Immunohistochemical staining of CD31+ blood vessels at the defect edge from (M) vehicle control and (N) 1NMPP1-injected mice, appearing red, and (O) quantification of CD31 immunoreactivity within the calvarial defect site. Dashed white lines indicate bone edge.
(P–R) Immunohistochemical staining of OCN+ (osteocalcin) osteoblasts from (P) vehicle control and (Q) 1NMPP1-injected mice, appearing cyan, and (R) quantification of OCN immunoreactivity within the calvarial defect site. Dashed white lines indicate bone edge. White scale bar, 50 μm.
DAPI counterstain appears blue in all images. In graphs, each dot represents a single animal; n = 9 per group. †p < 0.05 and ††p < 0.01 in comparison with vehicle control. Data are represented as mean ± SD.
H&E staining confirmed a significant reduction in calvarial re-ossification, including a widened distance between bony fronts (Figures 7H and 7I; arrowheads indicate original defect size). The frequency of TUBB3+ dendrites within the callus of control- and 1NMPP1-treated TrkAF592A;Thy1-YFP animals was next assessed (Figures 7J–7L). TUBB3+ nerve fibers were again noted most prominently at the healing defect edge (Figure 7J). A dramatic reduction in TUBB3+ nerve fibers was observed among 1NMPP1-treated animals (Figure 7K), which when quantified demonstrated a 71.2% reduction in mean TUBB3 immunoreactivity within the defect site (Figure 7L). Vascularity within the bone injury site was next assessed among control- and 1NMPP1-treated TrkAF592A animals (Figures 7M–7O). A reduction in CD31+ vascular channels was found within 1NMPP1-treated animals (Figures 7M and 7N), demonstrating a 54.0% decrease in comparison with vehicle control (Figure 7O). OCN immunohistochemical staining was next performed and confirmed a significant reduction in OCN+ osteoblastic numbers with 1NMPP1-treated TrkAF592A animals (Figures 7P and 7Q), reflecting a 37.2% reduction in OCN immunostaining (Figure 7R). In aggregate, inhibition of TrkA catalytic activity over the time course of bone defect repair led to a significant reduction in injury-associated re-innervation, hampered revascularization, reduced total osteoblastic activity, and led to a significant delay in bone defect healing.
DISCUSSION
In this study, we used both genetic and immunocytochemical approaches to characterize the innervation of the mouse skull and to explore their role in cranial bone healing. Our results in both homeostatic and injury settings are compatible with the classic neurotropic model, in which acute NGF induction in the pre-innervated tissue directs the spatial and temporal ingrowth of nerves (Reichardt, 2006). In this regard, high basal expression of Ngf in patent suture mesenchyme appears to drive nerve ingrowth or promote nerve survival at the suture sites. In the case of calvarial injury, expression of Ngf among defect-associated monocytes/macrophages appears to be responsible for directing tissue re-innervation. Our observations are generally analogous to the sequence of events associated with sensory innervation of mouse long bones, in which induction of Ngf in perichondrial mesenchymal cells directs sensory nerves to regions of incipient mineralization (Tomlinson et al., 2016).
Our finding that sensory nerve regrowth is critical for cranial bone repair is compatible conceptually with a large literature supporting the importance of sensory nerves in regeneration of lower order species. For example, the regeneration of limbs of certain starfish and amphibians, and healing of scales and barbells of fish, are all nerve-dependent processes (Huet, 1975; Kumar et al., 2007; Simões et al., 2014). However, in contrast to the classic neurotropic model, which stipulates that Ngf production is highly restricted, neurotrophin production following injury is more ubiquitous because of the production by invading inflammatory cells. In this setting, defect-associated monocytes/macrophages appear to be the primary cell mediator of Ngf-dependent nerve regrowth. Although not specifically addressed, it is possible that Ngf-expressing macrophages observed in our study represent osteal macrophages or “osteomacs,” which are known regulators of both membranous and endochondral bone formation (Batoon et al., 2019). The distinction between NGF expression in tissue-resident and circulatory monocytes/macrophages would be an interesting avenue for future study. Our observations of macrophage-dependent sensory re-innervation have some support in models of Wallerian degeneration, in which strain-specific differences in macrophage recruitment in mice were linked to differences in sensory axon regeneration (Brown et al., 1991). In the context of the skeleton, our observations are in line with Ngf expression by macrophages in osteoarthritis (Takano et al., 2016, 2017) and suggest common pathophysiologic mechanisms between sclerotic bone changes in osteoarthritis and the anabolic response to bone injury.
An important question that remains to be addressed is the downstream molecular mechanisms that mediate neuroskeletal intercommunication. In particular, the secondary messengers released by nociceptive fibers that regulate skeletal repair. In some regenerative contexts, clear nerve-derived factors have been elucidated. For example and in the salamander blastema, the anterior gradient protein family member nAG is secreted from nerve fibers and activates its receptor Prod 1 on blastemal cells during epimorphic regeneration (Kumar et al., 2007). Various other candidate nerve-derived molecules have been examined in amphibian limb regeneration, including FGFs (fibroblast growth factors), BMPs (bone morphogenetic proteins), GGF (glial growth factor), and neurogelin (Brockes and Kintner, 1986; Farkas et al., 2016; Kumar et al., 2007; Makanae et al., 2016; Mullen et al., 1996). In our prior observations, activation of neurons within the long bone PO is necessary for mechanical load-induced Wnt signaling activity (Tomlinson et al., 2017). Nevertheless, the exact physiologically important secondary signals released from peptidergic nerve fibers within a bone injury site remain unknown and are the subject of continued investigation.
Our findings raise the intriguing possibility that sensory nerve signaling might be activated to induce bone repair. The concept that nerve-derived signals could be pro-regenerative is certainly supported by studies in amphibians, in which surgical rerouting of nerve branches can prompt formation of an accessory limb (Maden and Holder, 1984). Delivery of recombinant Ngf itself, however, would represent a pain-evoking stimulus (Hefti, 2020). Moreover, Ngf is susceptible to proteolytic degradation and has a short elimination half-life (Angeletti et al., 1972; Friden et al., 1993; Tria et al., 1994) and is likely not suitable for clinical translation. In a recent report in murine long bone fracture repair, the TrkA agonist gambogic amide (GA) was observed to speed fracture repair (Johnstone et al., 2019). These recent observations provide proof in principle of the pro-regenerative properties of TrkA signaling, which may be harnessed to drive re-innervation, revascularization, and ultimately repair of the mammalian skeleton. The close association of nerves and vessels observed here suggests as well that perivascular precursor cells that contribute to endochondral bone repair (Maes et al., 2010) may likewise be in close proximity to nerve fibers within healing membranous bones. Methods to induce tissue re-innervation without activating pain pathways would be essential for a logical translational approach of the present findings.
On the other hand, it is important to note that skeletal sensory nerves function in both skeletal nociception and anabolism. Consequently, any pharmacological approach to altering sensory nerve function might be expected to affect bone anabolism. This has already been observed in human patients, in whom neutralizing Ngf antibodies predispose to an unusually high rate of subchondral insufficiency fractures that come to clinical attention (Hochberg, 2015). Similarly, missense and nonsense mutations in TRKA (NTRK1) are the cause of CIPA (congenital insensitivity to pain with anhidrosis) syndrome (Indo et al., 1996), characterized in part by delayed fracture healing (Bonkowsky et al., 2003; Toscano et al., 2000). These lines of evidence point to the interrelated nature of pain sensation and skeletal repair in human patients and the central importance of NGF-TrkA signaling in these linked processes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Aaron W. James (awjames@jhmi.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animals were housed and procedures performed under approval of the Animal Care and Use Committee (ACUC) of The Johns Hopkins University (MO16M226 & MO19M366). Mouse lines are summarized in Key Resource Table. NGF-eGFP mice were donated by the Kawaja laboratory, which express eGFP under the control of the mouse NGF promoter (Kawaja et al., 2011). Mice with floxed Ngf alleles were generated in the Minichiello laboratory (Müller et al., 2012). Thy1-YFP mice were used as a pan-neuronal reporter, which harbor a transgene derived from the mouse Thy1 gene that directs expression of YFP in motor and sensory neurons (Feng et al., 2000). TrkAF592A mice were donated from the Ginty laboratory, which are homozygous for a phenylalanine-to-alanine point mutation in exon 12 of the mouse Ntrkl gene (F592A) (Chen et al., 2005). This point mutation in TrkAF592A mice renders the endogenous TrkA kinase sensitive to inhibition by the membrane-permeable small molecule 1NMPP1 (Chen et al., 2005). PDGFRα-CreERT2 mice were donated from the Bergles laboratory and high specificity of PDGFRα reporter activity has been previously validated (Kang et al., 2010; Xu et al., 2019). LysM-Cre mice were purchased from the Jackson Laboratory (Strain #004781, Bar Harbor, ME). PDGFRα-CreERT2 mice were crossed with mT/mG strains to generate PDGFRα-CreERT2;mT/mG reporter animals. LysM-Cre mice were crossed with mT/mG or tdTomato strains to yield LysM-Cre;mT/mG or LysM-Cre;tdTomato mice. Mixed gender, 16 week old animals were used for all experiments unless otherwise stated. Wherever feasible, littermate analysis was performed while blinded to genotype.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal Anti-Axin 2 | Abcam | Cat# ab109307; RRID:AB_10862550 |
| Rabbit polyclonal Anti-CD31 | Abcam | Cat# ab28364; RRID:AB_726362 |
| Rat monoclonal Anti-CD45 | BioLegend | Cat# 103144; RRID:AB_2563458 |
| Rabbit Anti-Calcitonin Gene Related Peptide | Sigma-Aldrich | Cat# C8198; RRID:AB_259091 |
| Rat monoclonal Anti-F4/80 | Abcam | Cat# ab204467; RRID:AB_2810932 |
| Rabbit polyclonal Anti-Human Gli1 | Abcam | Cat# ab49314; RRID:AB_880198 |
| Rabbit polyclonal Anti-IL-1 beta | Abcam | Cat# ab9722; RRID:AB_308765 |
| Rabbit polyclonal Anti-NGF | Abcam | Cat# ab6199; RRID:AB_2152414 |
| Rabbit polyclonal Anti-Osteocalcin | Abcam | Cat# ab93876; RRID:AB_10675660 |
| Rabbit polyclonal Anti-PDGF receptor alpha | Abcam | Cat# ab15501; RRID:AB_301910 |
| Rabbit polyclonal Anti-PGP 9.5 | Agilent Tech | Cat# Z511601-2 |
| Rabbit polyclonal Anti-TNF alpha | Abcam | Cat# ab6671; RRID:AB_305641 |
| Rabbit polyclonal Anti-neuron specific beta III Tubulin | Abcam | Cat# ab18207; RRID:AB_444319 |
| Goat Anti-Mouse IgG H&L (Alexa Fluor® 647) preadsorbed antibody | Abcam | Cat# ab150119; RRID:AB_2811129 |
| DyLight 594 Anti-Rabbit IgG (H+L), made in goat antibody | Vector Laboratories | Cat# DI-1594; RRID:AB_2336413 |
| Bacterial and Virus Strains | ||
| Human Adenovirus Type5 (dE1/E3) | Vector Biosystems | Cat# 1045, Ad-CMV-iCre |
| Human Adenovirus Type5 (dE1/E3) | Vector Biosystems | Cat# 1060, Ad-GFP |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Small molecule 1NMPP1 | Aurora Analytics | Cat# N 0001 |
| DAPI mounting solution | Vector Laboratories | Vectashield H-1500 |
| 14% Ethylenediaminetetraacetic acid | Sigma-Aldrich | Cat# E6511 |
| Critical Commercial Assays | ||
| NF-KappaB Pathway Sampler Kit - 1 Kit | Cell Signaling | 9936T |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat# 1708891 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock #000664 |
| Mouse: mT/mG | The Jackson Laboratory | Stock #007576 |
| Mouse: TdTomato | Donated from Cao laboratory, Jackson Laboratory | Stock #007914 |
| Mouse: NGF-eGFP | Donated from Kawaja laboratory | N/A |
| Mouse: Ngffl/fl | Donated from Minichiello laboratory | N/A |
| Mouse: Thy1-YFP | The Jackson Laboratory | Stock #003709 |
| Mouse: TrkAF592A | Donated from Ginty laboratory, Jackson Laboratory | Stock #022362 |
| Mouse: Pdgfrα-CreERT2 | Donated from Bergles laboratory, Jackson Laboratory | Stock #018280 |
| Mouse: LysM-Cre | Jackson Laboratory | Stock #004781 |
| Oligonucleotides | ||
| Primer: Gapdh Forward: 5′- GAC TTC AAC AGC AAC TCC CAC –3′ | Thermo Scientific | N/A |
| Primer: Gapdh Reverse: 5′- TCC ACC CTG TTG CTG TA –3′ | Thermo Scientific | N/A |
| Primer: Ngf Forward: 5′- ACACTCTGATCACTGCGTTTTTG –3′ | Thermo Scientific | N/A |
| Primer: Ngf Reverse: 5′-CCTTCTGGGACATTGCTATCTGT-3′ | Thermo Scientific | N/A |
| Primer: eGFP Forward: 5′- CAACCACTACCTGAGCACCC –3′ | Thermo Scientific | N/A |
| Primer: eGFP Reverse: 5′- GTCCATGCCGAGAGTGATCC –3′ | Thermo Scientific | N/A |
| Software and Algorithms | ||
| Leica DM6 | Leica Microsystems Inc. | https://www.leica-microsystems.com/ |
| Zeiss LSM780 FCS | Carl Zeiss Microscopy GmbH | https://www.zeiss.com/ |
| SkyScan1172 high-resolution microCT imaging system | Bruker | https://www.bruker.com/ |
| NRecon software 2.0.4.0 | Bruker SkyScan | https://www.bruker.com/ |
| CTVox & CTAn software 1.13 | Bruker SkyScan | https://www.bruker.com/ |
| Photoshop CC, 2017 | Adobe | https://www.adobe.com/ |
| Imaris software v9.3 | Oxford Instruments | https://www.oxinst.com/ |
In order to achieve locoregional NGF deletion within the skull, an adenovirus encoding either Cre Recombinase (Ad-Cre, 1045-HT. Vector Biosystems, Malvern, PA) or GFP (Ad-GFP, 1060-HT, Vector Biosystems) was injected percutaneously overlying the calvaria in mice with NGF floxed alleles (Müller et al., 2012). 1 × 109 PFU particles were diluted in 50 μL normal saline and injected both 48 hr prior to surgery and 1 hr post-surgery. In select experiments, the spatial distribution of recombination was assessed using the same protocol of Ad-Cre injection in mT/mG animals (Muzumdar et al., 2007).
In order to achieve cell specific deletion of Ngf within calvaria bones, Ngf floxed animals were crossed with either LysM-Cre; mT/mG or PDGFRα-CreERT2;mT/mG animals. After backcrossing, this yielded Ngffl/fl;LysM-Cre;mT/mG (NgfLysM) animals and Ngffl/fl;PDGFRα-CreERT2;mT/mG (NgfPdgfrα) animals. Age matched Ngffl/fl;mT/mG animals (Ngffl/fl) were used as controls. In order to control for the potential skeletal effects of tamoxifen (TM), all genotypes were administered TM (150 mg/kg daily for 5 days, i.p.), and surgery performed after a 14 day chase period as we have previously validated in PDGFRα-CreERT2;mT/mG animals (Wang et al., 2020; Xu et al., 2019).
In order to achieve temporal inhibition of TrkA catalytic activity in TrkAF592A animals, the small molecule 1NMPP1 was used (Hanefeld et al., 1996) (Aurora Analytics, LLC, Baltimore, MD). Purity (99.2%) was confirmed by HPLC-UV254, and characterization by 1H NMR (400 MHz, DMSO-d6) was consistent with structure. Stock solution was prepared at 200 mM by dissolving 1NMPP1 in dimethyl sulfoxide (DMSO). For 1NMPP1 administration, intraperitoneal (IP) injections were performed 24 h before, 2 h before and 24 h after calvarial injury using a 5 mM solution at a dosage of 17 ug/g body weight. In all cases, DMSO containing vehicle was used for control treatment. Animals were thereafter maintained on 1NMPP1 containing drinking water (40 uM 1NMPP1 in ddH2O with 1% PBS-Tween 20).
METHOD DETAILS
Calvarial defect creation
Calvarial defect creation was performed based on our prior methods (Chang et al., 2018; Zhang et al., 2018). Before surgery, mice were given an intraperitoneal injection of analgesic (1 mg/kg buprenorphine SR). Briefly, hair overlying the calvaria was clipped, and the skin was aseptically prepared using alternating betadine and alcohol scrubs. Mice were anesthetized by isoflurane gas (1%–3% inh) for the duration of the experiment. After confirming deep anesthesia, a 1 cm sagittal incision was performed over the midline skull to expose the frontal bone. A 1.8 mm diameter, full-thickness, circular frontal bone defect was created using a micro surgical drill and a trephine drill bit. Meticulous care was taken to protect the neighboring sutures and the underlying tissues. Calvarial defect sites were irrigated with normal saline. Finally, the skin was sutured and the animal was monitored per established postoperative protocols. Skulls were harvested up to 28 d after injury.
Histology and immunohistochemistry
Calvaria were harvested and placed in 4% paraformaldehyde (PFA) at 4°C for 24 h. After sequential washes in PBS x 3, samples were decalcified in 14% EDTA (1:20 volume, Sigma-Aldrich) for 14 d at 4°C. Coronal sections of the calvaria were obtained using cryosections at 10 or 50 μm thickness. For cryosections, samples were cryoprotected in 30% sucrose overnight at 4°C before embedding in OCT (Tissue-Tek 4583, Torrance, CA). Coronal sections were mounted on adhesive slides (TruBond 380, Matsunami, Bellingham, WA). For immunohistochemistry, sections were washed in PBS x 3 for 10 min. When 50 μm sections were used, sections were next permeabilized with 0.5% Triton-X for 30 min. Next, 5% normal goat serum was applied for 30 min, then incubated in primary antibodies overnight at 4°C in a humidified chamber (see Key Resource Table and Table S1 for a summary of antibodies used). The following day, slides were washed in PBS, incubated in the appropriate secondary antibody, A594-Goat Anti-Rabbit IgG or Goat Anti-Mouse IgG, for 1 h at 25°C, then mounted with DAPI mounting solution (Vectashield H-1500, Vector Laboratories, Burlingame, CA). Digital images of these sections were captured with 10-100 × objectives using upright fluorescent microscopy (Leica DM6, Leica Microsystems Inc., Buffalo Grove, IL) or confocal microscopy (Zeiss LSM780 FCS, Carl Zeiss Microscopy GmbH, Jena, Germany).
Whole mount immunofluorescence
In select experiments, skulls from C57BL/6J mice were used for whole mount imaging. Prior to immunostaining, the skin, muscle, and brain were carefully dissected away from the calvaria under a dissecting microscope, exposing the periosteum and dura mater. The samples were fixed in 4% PFA overnight at 4°C. Samples were then washed with sequential washes in PBS x 3, and permeabilized with 0.5% Triton X-100 at room temperature for 30 min. To block non-specific binding, samples were submerged in 1mL of 10% normal goat serum at room temperature for 30 min. The primary antibody was then applied (1 mL of anti-TUBB3 1:1500; anti-CGRP 1:200; or anti-TH 1:200) overnight at 4°C. Samples were then washed sequential washes in PBS x 3, and submerged in the appropriate secondary antibody (1 mL of A594-Goat Anti-Rabbit IgG, 1:200) for 1 hr at room temperature. Next, calvaria were washed in PBS x 3 and submerged in PBS in a black, glass bottom PELCO dish (Ted Pella, Inc, Redding, CA) for imaging. Imaging was performed using the 10x objective lens on a Zeiss LSM780 FCS, Carl Zeiss Microscopy).
Isolation and culture of mouse calvarial cells
Mouse calvarial cells were collected from C57BL/6J or NGF-eGFP embryos at postnatal day 1. Frontal and parietal bones were subjected to six sequential enzymatic digestions with a mixture containing 1 mg/mL collagenase type I (Worthington Biochemical Corporation, Lakewood, NJ, USA; LS004197) and 1 mg/mL collagenase type II (Worthington Biochemical Corporation; LS004177)(Noda et al., 2016). Cell fractions from sequential digestions 3-5 were collected and cultured in α-MEM supplemented with 15% (vol/vol) FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. To measure cytokine-induced NGF expression, cells were treated with recombinant IL-1β (1-50 ng/mL) or TNF-α (1-50 ng/mL) for 48 h, followed by qRT-PCR analysis.
Ribonucleic acid isolation and quantitative real-time polymerase chain reaction
Total RNA was extracted from the cultured cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. 0.8 κg of total RNA was used for reverse transcription with iScript cDNA synthesis kit (Bio-Rad) following manufacturer’s instructions. Real-time PCR was performed using SYBR Green PCR Master Mix (Thermo Scientific) according to the manufacturer’s protocol. Specific primers for SYBR green qPCR were verified by using Primer-BLAST from National Center for Biotechnology Information (NCBI). Standard cycling condition of amplification (primer Tm ≥ 60°C) was performed; UDG activation, 50°C, 2 min; then Dual-Lock™ DNA Polymerase, 95°C, 2 min; then Denature, 40 cycles of 95°C, 15 s; then Anneal / Extend, 40 cycles of 60°C, 1 min. Further, specificity of amplification for SYBRgreen-based qPCR was validated by the Melt Curve Plot of each reaction that the single peak reaction was observed. Relative gene expression was calculated using a 2-ΔΔCt method by normalization with Gapdh. Primer sequences are presented in Key Resource Table.
Western Blot
Mouse calvarial cells were stimulated with IL-1β (1 ng/mL) or TNF-α (25 ng/mL) and lysed in RIPA buffer (Thermo Fisher Scientific) with protease inhibitor cocktail (Cell Signaling Technologies, Danvers, MA, USA). The separation of proteins was performed using a 10% gradient SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The blotted nitrocellulose membranes were directly blocked with a blocking buffer (5% bovine serum albumin in Tris-buffered saline (TBS) with 0.05% Tween-20 (TBST) for 1 hr and then probed with primary antibodies (see Key Resource Table and Table S1). After washing with TBST three times, membranes were incubated with a horseradish-peroxidase (HPR)-conjugated secondary antibody and visualized with ChemiDoc XRS+ System (Bio-rad, Hercules, CA).
QUANTIFICATION AND STATISTICAL ANALYSIS
MicroCT imaging and analysis
Skulls were dissected free of skin and brain tissue and evaluated using a SkyScan1172 high-resolution microCT imaging system (Bruker, Kontich, Belgium). Scans were obtained at an image resolution of 10 μm, with the following settings: 1 mm of aluminum filter, X-ray voltage of 65 kVP, anode current of 153 μA, exposure time of 65ms, frame averaging of 5, and rotation step of 0.3 degrees. Three-dimensional images were then reconstructed from the 2D X-ray projections by implementing the Feldkamp algorithm using a commercial software package NRecon software (2.0.4.0 SkyScan). For the 3D morphometric analyses of images, CTVox and CTAn software (1.13 SkyScan) were used.
For calvarial defect analysis, a cylindrical volume of interest centered around each defect site was defined as the 1.6 mm in diameter and 1 mm in height with a threshold value of 80. Bone volume (BV) and tissue volume (TV) measurements were calculated from analyses performed for a volume of 100 slices. The amount of bone formation was analyzed and quantified using three methods. First, bone volume (BV) and fractional bone volume (bone volume/tissue volume (BV/TV)) were calculated from binary X-ray images. Second, bone fractional area (BFA) was calculated by using CTVox to create a 3D rendering of the calvarial defect in a top-down view, and then measuring pixels of bone within the defect divided by total defect area using Photoshop (Adobe, San Jose, California). Lastly, a semiquantitative bone healing score from 0 to 4 was assigned by 3 blinded observers according to previous published grading scales for calvarial defect healing (Spicer et al., 2012). Briefly, the grading system was as follows: 0–no bone formation, 1–few bony spicules dispersed through defect, 2–bony bridging only at defect borders, 3–bony bridging over partial length of defect, and 4–bony bridging entire span of defect at longest point.
Histologic image analysis
All images for quantification were obtained either with upright fluorescent microscopy (Leica DM6, Leica Microsystems Inc.) or confocal microscopy (Zeiss LSM780 FCS, Carl Zeiss Microscopy GmbH). NGF-eGFP reporter activity, TUBB3+ nerve fibers, CD31+ blood vessels, and OCN+ osteoblasts were quantified using either the magic wand tool of Photoshop CC, 2017 with a tolerance of 30 (Adobe, San Jose, CA) using five random 40x microscopical fields per sample or three-dimensional volumetric analysis of Imaris software v9.3 (Oxford Instruments, Belfast, UK) using eight serial fields per sample within the defect tissue.
Statistics
Quantitative data are expressed at mean ± SD, with p < 0.05 and p < 0.01 considered significant. The number of samples are indicated in figure legends. A Shapiro-Wilk test for normality was performed on all datasets. Homogeneity was confirmed by a comparison of variances test. Parametric data was analyzed using an appropriate Student’s t test when two groups were being compared, or a one-way ANOVA was used when more than two groups were compared, followed by a post hoc Tukey’s test to compare two groups. Nonparametric data was analyzed with a Mann-Whitney U test when two groups were being compared or a Kruskal-Wallis one-way analysis when more than two groups were compared. Sample size calculations were performed for experiments presented in Figures 5, 6, and 7 based on an anticipated effect size of 1.75, using our previously published data in adult TrkAF592A mice (Tomlinson et al., 2017). For this scenario, with 8 replicates per group a two-sample t test would provide 80% power to detect effect sizes of at least 1.5 assuming a two-sided 0.05 level of significance.
Supplementary Material
Highlights.
Cranial sutures represent an NGF-expressing domain in which sensory nerves transit
Monocyte/macrophage-derived Ngf induces skeletal innervation and bone repair
Blocking NGF-TrkA signaling delays skeletal re-innervation and blunts bone repair
ACKNOWLEDGMENTS
This work was supported by grant R21 DE027922 from the National Institute of Dental and Craniofacial Research (NIDCR) to T.L.C. and A.W.J. Additional support to A.W.J. is provided by NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (grants R01 AR070773 and K08 AR068316), NIH/NIDCR (grant R21 DE027922), the U.S. Department of Defense (U.S. Army Medical Research Acquisition Activity [USAMRAA] grants W81XWH-18-1-0336, W81XWH-18-1-0121, and W81XWH-18-10613), the American Cancer Society (Research Scholar Grant RSG-18-027-01-CSM), the Maryland Stem Cell Research Foundation, and the Musculoskeletal Transplant Foundation. Additional support to T.L.C. is provided by NIH/NIAMS (R01 AR068934), NIH/NIDCR (R21 DE027922), and U.S. Department of Veterans Affairs (VA) Merit Award I01BX001234 and Senior Research Career Scientist Award IK6BX004984. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or Department of Defense. We thank the Johns Hopkins University (JHU) microscopy facility for their technical assistance. The use of the graphical abstract has been authorized by the artist Tim Phelps, whom we thank for his contribution.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107696.
DECLARATION OF INTERESTS
Research unrelated to the work presented herein was supported in the James laboratory by Musculoskeletal Transplant Foundation (MTF) Biologics. A.W.J. is on the scientific advisory board of Novadip, LLC, for unrelated research. The authors have declared that no further conflict of interest exists.
REFERENCES
- Alberius P, and Skagerberg G (1990). Adrenergic innervation of the calvarium of the neonatal rat. Its relationship to the sagittal suture and developing parietal bones. Anat. Embryol. (Berl.) 182, 493–498. [DOI] [PubMed] [Google Scholar]
- Angeletti RH, Aneletti PU, and Levi-Montalcini R (1972). Selective accumulation of (125 I) labelled nerve growth factor in sympathetic ganglia. Brain Res. 46, 421–425. [DOI] [PubMed] [Google Scholar]
- Batoon L, Millard SM, Wullschleger ME, Preda C, Wu AC-K, Kaur S, Tseng H-W, Hume DA, Levesque J-P, Raggatt LJ, and Pettit AR (2019). CD169+ macrophages are critical for osteoblast maintenance and promote intramembranous and endochondral ossification during bone repair. Biomaterials 196, 51–66. [DOI] [PubMed] [Google Scholar]
- Bjurholm A, Kreicbergs A, Brodin E, and Schultzberg M (1988). Substance P- and CGRP-immunoreactive nerves in bone. Peptides 9, 165–171. [DOI] [PubMed] [Google Scholar]
- Bonkowsky JL, Johnson J, Carey JC, Smith AG, and Swoboda KJ (2003). An infant with primary tooth loss and palmar hyperkeratosis: a novel mutation inthe NTRK1 genecausing congenital insensitivityto pain with anhidrosis. Pediatrics 112, e237–e241. [DOI] [PubMed] [Google Scholar]
- Brockes JP, and Kintner CR (1986). Glial growth factor and nerve-dependent proliferation in the regeneration blastema of Urodele amphibians. Cell 45, 301–306. [DOI] [PubMed] [Google Scholar]
- Brown MC, Perry VH, Lunn ER, Gordon S, and Heumann R (1991). Macrophage dependence of peripheral sensory nerve regeneration: possible involvement of nerve growth factor. Neuron 6, 359–370. [DOI] [PubMed] [Google Scholar]
- Castañeda-Corral G, Jimenez-Andrade JM, Bloom AP, Taylor RN, Mantyh WG, Kaczmarska MJ, Ghilardi JR, and Mantyh PW (2011). The majority of myelinated and unmyelinated sensory nerve fibers that innervate bone express the tropomyosin receptor kinase A. Neuroscience 178, 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang L, Xu J, Meyers CA, Li Z, Yan N, Zou E, and James AW (2018). Lineage-specific Wnt reporter elucidates mesenchymal Wnt signaling during bone repair. Am. J. Pathol 188, 2155–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ye H, Kuruvilla R, Ramanan N, Scangos KW, Zhang C, Johnson NM, England PM, Shokat KM, and Ginty DD (2005). A chemical-genetic approach to studying neurotrophin signaling. Neuron 46, 13–21. [DOI] [PubMed] [Google Scholar]
- Doro DH, Grigoriadis AE, and Liu KJ (2017). Calvarial suture-derived stem cells and their contribution to cranial bone repair. Front. Physiol 8, 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas JE, Freitas PD, Bryant DM, Whited JL, and Monaghan JR (2016). Neuregulin-1 signaling is essential for nerve-dependent axolotl limb regeneration. Development 143, 2724–2731. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, and Sanes JR (2000). Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51. [DOI] [PubMed] [Google Scholar]
- Ferguson JW, and Atit RP (2019). A tale of two cities: the genetic mechanisms governing calvarial bone development. Genesis 57, e23248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden PM, Walus LR, Watson P, Doctrow SR, Kozarich JW, Bäckman C, Bergman H, Hoffer B, Bloom F, and Granholm AC (1993). Blood-brain barrier penetration and in vivo activity of an NGF conjugate. Science 259, 373–377. [DOI] [PubMed] [Google Scholar]
- Grönblad M, Liesi P, Korkala O, Karaharju E, and Polak J (1984). Innervation of human bone periosteum by peptidergic nerves. Anat. Rec 209, 297–299. [DOI] [PubMed] [Google Scholar]
- Han HM, Kim TH, Bae JY, and Bae YC (2019). Primarysensory neurons expressing tropomyosin receptor kinase A in the rat trigeminal ganglion. Neurosci. Lett 690, 56–60. [DOI] [PubMed] [Google Scholar]
- Hanefeld U, Rees CW, White AJP, and Williams DJ (1996). One-pot synthesis of tetrasubstituted pyrazoles—proof of regiochemistry. J. Chem. Soc., Perkin Trans 1, 1545–1552. [Google Scholar]
- Hefti F (2020). Pharmacology of nerve growth factorand discovery of tanezumab, an anti-nerve growth factor antibody and pain therapeutic. Pharmacol. Res 154, 104240. [DOI] [PubMed] [Google Scholar]
- Hochberg MC (2015). Serious joint-related adverse events in randomized controlled trials of anti-nerve growth factor monoclonal antibodies. Osteoarthritis Cartilage 23 (Suppl 1), S18–S21. [DOI] [PubMed] [Google Scholar]
- Hohmann EL, Elde RP, Rysavy JA, Einzig S, and Gebhard RL (1986). Innervation of periosteum and bone by sympatheticvasoactive intestinal peptide-containing nerve fibers. Science 232, 868–871. [DOI] [PubMed] [Google Scholar]
- Huet M (1975). [Role of the nervous system during the regeneration of an arm in a starfish: Asterina gibbosa Penn. (Echinodermata, Asteriidae)]. J. Embryol. Exp. Morphol 33, 535–552. [PubMed] [Google Scholar]
- Indo Y, Tsuruta M, Hayashida Y, Karim MA, Ohta K, Kawano T, Mitsubuchi H, Tonoki H, Awaya Y, and Matsuda I (1996). Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat. Genet 13, 485–488. [DOI] [PubMed] [Google Scholar]
- Johnstone MR, Brady RD, Schuijers JA, Church JE, Orr D, Quinn JMW, McDonald SJ, and Grills BL (2019). The selective TrkA agonist, gambogic amide, promotes osteoblastic differentiation and improves fracture healing in mice. J. Musculoskelet. Neuronal Interact 19, 94–103. [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Fukaya M, Yang JK, Rothstein JD, and Bergles DE (2010). NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaja MD, Smithson LJ, Elliott J, Trinh G, Crotty AM, Michalski B, and Fahnestock M (2011). Nerve growth factor promoter activity revealed in mice expressing enhanced green fluorescent protein. J. Comp. Neurol 519, 2522–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaras B, Jakubowski M, Kainz V, and Burstein R (2009). Sensory innervation of the calvarial bones of the mouse. J. Comp. Neurol 515, 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, and Brockes JP (2012). Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 35, 691–699. [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, and Brockes JP (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton KA, Nacamuli RP, Wan DC, Helms JA, and Longaker MT (2005). Cranial suture biology. Curr. Top. Dev. Biol 66, 287–328. [DOI] [PubMed] [Google Scholar]
- Levi B, Nelson ER, Li S, James AW, Hyun JS, Montoro DT, Lee M, Glotzbach JP, Commons GW, and Longaker MT (2011). Dura mater stimulates human adipose-derived stromal cells to undergo bone formation in mouse calvarial defects. Stem Cells 29, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi B, Wan DC, Wong VW, Nelson E, Hyun J, and Longaker MT (2012). Cranial suture biology: from pathways to patient care. J. Craniofac. Surg 23, 13–19. [DOI] [PubMed] [Google Scholar]
- Li Z, Meyers CA, Chang L, Lee S, Li Z, Tomlinson R, Hoke A, Clemens TL, and James AW (2019). Fracture repair requires TrkA signaling by skeletal sensory nerves. J. Clin. Invest 129, 5137–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach DB, Rogers SD, Sabino MC, Luger NM, Schwei MJ, Pomonis JD, Keyser CP, Clohisy DR, Adams DJ, O’Leary P, and Mantyh PW (2002). Origins of skeletal pain: sensory and sympathetic innervation of the mouse femur. Neuroscience 113, 155–166. [DOI] [PubMed] [Google Scholar]
- Maden M, and Holder N (1984). Axial characteristics of nerve induced supernumerary limbs in the axolotl. Wilehm. Roux Arch. Dev. Biol 193, 394–401. [DOI] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, and Kronenberg HM (2010). Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell 19, 329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanae A, Mitogawa K, and Satoh A (2016). Cooperative inputs of Bmp and Fgf signaling induce tail regeneration in urodele amphibians. Dev. Biol 410, 45–55. [DOI] [PubMed] [Google Scholar]
- Mantyh PW (2014). The neurobiology of skeletal pain. Eur. J. Neurosci 39, 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Jeong J, Sheu TJ, and Hsu W (2016). Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat. Commun 7, 10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen LM, Bryant SV, Torok MA, Blumberg B, and Gardiner DM (1996). Nerve dependency of regeneration: the role of Distal-less and FGF signaling in amphibian limb regeneration. Development 122, 3487–3497. [DOI] [PubMed] [Google Scholar]
- Müller M, Triaca V, Besusso D, Costanzi M, Horn JM, Koudelka J, Geibel M, Cestari V, and Minichiello L (2012). Loss of NGF-TrkA signaling from the CNS is not sufficient to induce cognitive impairments in young adult or intermediate-aged mice. J. Neurosci 32, 14885–14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Noda K, Kitami M, Kitami K, Kaku M, and Komatsu Y (2016). Canonical and noncanonical intraflagellar transport regulates craniofacial skeletal development. Proc. Natl. Acad. Sci. U S A 113, E2589–E2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan A, Chang L, Nguyen A, and James AW (2013). A review of hedgehog signaling in cranial bone development. Front. Physiol 4, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet S, and McMahon SB (2006). Neurotrophins: mediators and modulators of pain. Annu. Rev. Neurosci 29, 507–538. [DOI] [PubMed] [Google Scholar]
- Reichardt LF (2006). Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci 361, 1545–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JD, and Kruger L (1989). Calcitonin-gene-related-peptide-immunoreactive innervation of the rat head with emphasis on specialized sensory structures. J. Comp. Neurol 280, 303–330. [DOI] [PubMed] [Google Scholar]
- Simões MG, Bensimon-Brito A, Fonseca M, Farinho A, Valério F, Sousa S, Afonso N, Kumar A, and Jacinto A (2014). Denervation impairs regeneration of amputated zebrafish fins. BMC Dev. Biol 14, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer PP, Kretlow JD, Young S, Jansen JA, Kasper FK, and Mikos AG (2012). Evaluation of bone regeneration using the rat critical size calvarial defect. Nat. Protoc 7, 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Uchida K, Miyagi M, Inoue G, Fujimaki H, Aikawa J, Iwase D, Minatani A, Iwabuchi K, and Takaso M (2016). Nerve growth factor regulation by TNF-α and IL-1β in synovial macrophages and fibroblasts in osteoarthritic mice. J. Immunol. Res 2016, 5706359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Uchida K, Inoue G, Miyagi M, Aikawa J, Iwase D, Iwabuchi K, Matsumoto T, Satoh M, Mukai M, et al. (2017). Nerve growth factor regulation and production by macrophages in osteoarthritic synovium. Clin. Exp. Immunol 190, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RE, Li Z, Zhang Q, Goh BC, Li Z, Thorek DLJ, Rajbhandari L, Brushart TM, Minichiello L, Zhou F, et al. (2016). NGF-TrkA signaling by sensory nerves coordinates the vascularization and ossification of developing endochondral bone. Cell Rep. 16, 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RE, Li Z, Li Z, Minichiello L, Riddle RC, Venkatesan A, and Clemens TL (2017). NGF-TrkA signaling in sensory nerves is required for skeletal adaptation to mechanical loads in mice. Proc. Natl. Acad. Sci. U S A 114, E3632–E3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano E, della Casa R, Mardy S, Gaetaniello L, Sadile F, Indo Y, Pignata C, and Andria G (2000). Multisystem involvement in congenital insensitivity to pain with anhidrosis (CIPA), a nerve growth factor receptor(Trk A)-related disorder. Neuropediatrics 31, 39–41. [DOI] [PubMed] [Google Scholar]
- Tria MA, Fusco M, Vantini G, and Mariot R (1994). Pharmacokinetics of nerve growth factor (NGF) following different routes of administration to adult rats. Exp. Neurol 127, 178–183. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu J, Meyers CA, Gao Y, Tian Y, Broderick K, Peault B, and James AW (2020). PDGFRα marks distinct perivascular populations with different osteogenic potential within adipose tissue. Stem Cells 38, 276–290. [DOI] [PubMed] [Google Scholar]
- Warren SM, Greenwald JA, Nacamuli RP, Fong KD, Song HJ, Fang TD, Mathy JA, and Longaker MT (2003). Regional dura mater differentially regulates osteoblast gene expression. J. Craniofac. Surg 14, 363–370. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang Y, Hsu C-Y, Gao Y, Meyers CA, Chang L, Zhang L, Broderick K, Ding C, Peault B, et al. (2019). Human perivascular stem cell-derived extracellular vesicles mediate bone repair. eLife 8, e48191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chang L, Xu J, Meyers CA, Yan N, Zou E, Ding C, Ting K, Soo C, Pang S, and James AW (2018). Frontal bone healing is sensitive to Wnt signaling inhibition via lentiviral-encoded beta-catenin short hairpin RNA. Tissue Eng. Part A 24, 1742–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Feng J, Ho TV, Grimes W, Urata M, and Chai Y (2015). The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol 17, 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.


